Abstract
Oral squamous cell carcinoma (OSCC) progresses from epithelial cell proliferation to malignancy. Given the higher proportion of male patients compared to female patients, the androgen signaling pathway is believed to play a significant role in promoting epithelial cell proliferation. However, the underlying molecular mechanisms remain unclear. Here, we identified RBAP48 as a novel androgen receptor (AR) co-activator in OSCC cells. Our results show that RBAP48 was highly expressed in OSCC tumor tissues from patients with a poor prognosis. Further, RBAP48 knockdown decreased genome-wide oncogene transcription. RBAP48 and AR interacted to activate CCND1 and RAB31 transcription, and upregulated RELA and CCNE1 mRNA expression through an AR-independent pathway. Additionally, RBAP48 promoted OSCC cell proliferation and was involved in the cellular response to drugs and external compounds in vitro, ultimately driving cancer progression. Our results indicate that RBAP48 is a novel oncogene and a promising target for predicting and treating OSCC progression.
Similar content being viewed by others
Introduction
Oral squamous cell carcinoma (OSCC) is a significant health concern with a poor 5-year prognosis1,2,3. Global Cancer Statistics from 2020 have revealed notable discrepancies in OSCC outcomes between male and female4. Owing to an elevated overall risk, male patients exhibit markedly higher rates of morbidity and mortality. This disparity is largely attributed to external factors such as smoking and alcohol consumption5,6. In South-Central Asia, the high incidence of OSCC is linked to the widespread practice of betel nut chewing7. Clinical research has also suggested that long-term use of alcohol or tobacco triggers abnormal proliferation of oral epithelial cells, increasing susceptibility to OSCC8,9. These external stimuli directly harm oral keratinocytes and fibroblasts, causing DNA damage and the release of reactive oxygen species (ROS)10,11. Furthermore, the complex interactions between epithelial and mesenchymal cells amplify the pathogenic cascade. Oral epithelial cells undergo a series of pathologic changes involving external stimulation, fibrosis, proliferation, and carcinogenesis12,13. Through a multistep process that accumulates genetic and epigenetic alterations in oncogenes, several signaling pathways are disrupted, affecting the cell cycle, cell proliferation, and apoptosis regulation14,15. Thus, novel biomarkers are needed to enhance our understanding of OSCC and identify new therapeutic targets.
The androgen receptor (AR) is a critical ligand-dependent transcription factor that specifically binds to androgens in vivo and activates downstream gene transcription. AR contains several functional domains, including a DNA-binding domain (DBD), a ligand-binding domain (LBD), a ligand-independent activation function domain (AF-1), and a ligand-dependent activation function domain (AF-2). Upon binding to androgen, AR translocates to the nucleus, where it is recruited to the gene promoter and enhancer regions containing specific androgen response elements (AREs)16,17. AR facilitates the recruitment of co-regulators, such as chromatin remodeling and histone-modifying proteins, to regulate the transcription of dihydrotestosterone (DHT)-related genes18,19. Although the precise role of AR in OSCC remains unclear, it is believed that AR signaling pathways contribute to OSCC proliferation and metastasis. Moreover, AR coregulators and signaling pathways may function differently in OSCC compared to those in prostate and liver cancers20,21,22,23. Therefore, further exploration of the function of the AR and its coregulators in OSCC is crucial.
Retinoblastoma-associated protein 48 (RBAP48), also known as retinoblastoma-binding protein 4 (RBBP4), is a highly conserved histone chaperone found in several species24,25. RBAP48 features a WD40-repeat structure that is speculated to act as a scaffold for protein-protein interactions in multiprotein complexes26,27. As a histone chaperone, RBAP48 participates in chromatin assembly and remodeling, histone post-translational modifications, and gene expression regulation28. Notably, RBAP48 is a component of several transcription-related complexes, including the nucleosome remodeling and deacetylase (NuRD), switch-independent 3 A (Sin3A), and polycomb repressive 2 (PRC2) complex29,30,31,32. RBAP48 is also part of the nucleosome-remodeling factor (NURF) complex, where it interacts with WRAD subunits33. Dysregulation of RBAP48 (RBBP4) has been linked to the etiology of several cancers, including liver, breast, and gastric cancers34,35,36,37. In two OSCC studies, RBAP48 has been implicated in DNA repair and human papillomavirus (HPV) infection38,39. Despite such progress, the molecular mechanisms underlying the effects of RBAP48 on OSCC remain poorly understood.
In this study, we found that RBAP48 was highly expressed in patients with OSCC, based on both tumor tissue samples and data from The Cancer Genome Atlas (TCGA). The prognosis of OSCC patients was positively correlated with high RBAP48 expression. We identified RBAP48 as a novel AR coactivator that upregulates AR-mediated gene transcription in OSCC cells in the presence of DHT. In OSCC cells, RBAP48 interacts with the WRAD subunits to enhance the transcription of AR target genes. Interestingly, our results demonstrated that RBAP48 was also associated with SP-1 to upregulate the transcription of its downstream target genes in the absence of DHT. Furthermore, RBAP48 significantly enhanced the proliferation and sensitivity of OSCC cells to external stimuli. In conclusion, our data demonstrate that RBAP48 promotes OSCC progression through a regulatory network involving both AR/DHT-dependent and independent pathways. The expression of RBAP48 could serve as a robust biomarker for the early diagnosis and classification of OSCC.
Results
RBAP48 is highly expressed in OSCC patients
We obtained 14 pairs of fresh OSCC tissue (T) and non-cancerous oral epithelial tissue (NC) to investigate the role of RBAP48 in OSCC. Western blot analysis revealed that RBAP48 protein expression was elevated in most OSCC tumor tissues (11/14, Fig. 1A). The mRNA levels of RBAP48 were markedly higher in OSCC tissues than those in NC tissues (Fig. 1B). Furthermore, there was a positive correlation between RBAP48 mRNA and protein levels in both NC and tumor tissues, indicating an oncogenic role for RBAP48 in OSCC (Supplementary Fig. 1A). The protein expression of RBAP48 in patients was positively correlated with their pathological grades, especially in the middle and late stages (Grade III/IV), indicating its potential involvement in promoting and sustaining malignancies (Fig. 1C-D). Additionally, univariate analysis suggested that increased RBAP48 expression was not associated with age, lymphatic metastasis, distant metastasis, tumor location, tobacco or alcohol. However, it exhibited strong correlations with sex, original tumor size, and clinical stage (Table 1). A cohort of 499 patients with HNSC was obtained from TCGA database. We further analyzed the correlation between RBAP48 and AR transcript levels with clinical variables in the TCGA-HNSC cohort, the chi-square test indicated that RBAP48 is associated with male gender and clinical stage in the TCGA-HNSC cohort (Table 2), while AR is not associated with the three clinical variables analyzed (Table 3). Notably, patients with unclassified HNSCs and higher RBAP48 expression showed reduced survival rates (Fig. 1E).
A Western blotting staining showed the expression of RBAP48 in 14 pairs of fresh OSCC tissues. The relative expression (/β-actin) is shown numerically below each band. B qPCR results showed the mRNA expression of RBAP48 in 14 pairs of patients. Student t-test is used, and p < 1e-2**. C Immunohistochemical staining showed the expression of RBAP48 in non-cancerous oral tissues (n = 4) and OSCC tissues of different grades (n = 90). D Immunohistochemical statistical results were obtained using the student t-test between groups. Grade I, (n = 6); Grade II, (n = 25); Grade III, (n = 34); Grade IV, (n = 25). p < 1e-4****, ns indicates no statistical significance. E–I HNSC Patient survival analysis showed the effect of RBAP48 or AR expression on survival in different types of patients from the TCGA database. (E) RBAP48 in all patients; (F) AR in all patients; (G) RBAP48 in AR-positive patients; (H) RBAP48 in AR-negative patients; and (I) RBAP48 in AR-negative patients beyond 900 days.
Given the disproportionally larger number of male patients with HNSC and the significant activation of the AR pathway in males, we investigated the effect of AR expression on survival of patients with HNSC. Statistical analysis, excluding sex variables, showed that patients with AR-positive expression (ARpositive) had a poorer prognosis, suggesting a contributory role of AR in HNSC (Fig. 1F). Among ARpositive patients, those with higher RBAP48 expression had worse survival outcomes, indicating that ARpositive/RBAP48high HNSC patients may exhibit increased malignancy (Fig. 1G). Notably, RBAP48 mRNA expression positively correlated with that of classical AR target genes, such as CCND1 and RAB31 (Supplementary Fig. 1B-1C). RBAP48 did not affect the short-term survival of patients with low AR expression (ARnegative) (Fig. 1H). However, high RBAP48 expression significantly reduced the expected survival of ARnegative patients in advanced stages (beyond 900 days) (Fig. 1I). The multivariate analysis results for overall survival (OS) using TCGA data were performed. Significant risk factors were lymph node status (HR = 1.797, p = 0.001) and distant metastasis (HR = 16.399, p = 0.013). Tumor size showed a trend (HR = 1.446, p = 0.089), while gender, AR, RBAP48, and stage were not significant (Supplementary Fig. 1F). These data demonstrate that RBAP48 is highly expressed in HNSC and OSCC, correlates with poor survival outcomes, and underscores its multifaceted role in OSCC.
RBAP48 promotes AR-mediated transactivation
RBAP48 is a component of the PRC2 repressed-transcription complex40,41,42, suggesting its potential involvement in inhibiting genes downstream of AR. However, we found that RBAP48 actually increased AR-mediated transactivation. Specifically, RBAP48 increased AR-mediated transactivation following DHT treatment of HEK-293 cells (Fig. 2A). Furthermore, DHT treatment significantly enhanced RBAP48-induced transactivation via the full-length AR (AR-FL) and AR-AF2 domains (Fig. 2B), indicating that RBAP48 upregulates AR-mediated transcription in the presence of DHT. In both the CAL-27 and SCC-9 cell lines, RBAP48 upregulated AR-mediated transcription in the presence of DHT, but did not enhance AR-mediated transcription in the absence of DHT (Fig. 2C, D). These data demonstrate that RBAP48 promotes AR-induced transactivation.
A Luciferase assays indicated that RBAP48 up-regulated AR-mediated gene transcription in HEK-293 cells with DHT. Student t-tests were used, and p < 1e-2**. B Multiple luciferase assays showed RBAP48 increased the AR full-length and AR-AF2 domain transactivation. C-D Relative luciferase assays indicated the function of RBAP48 on AR-mediated transactivation in CAL-27 cells and SCC-9 cells. E RNA-seq heatmap showed that RBAP48 significantly affected genes in the absence and presence of DHT (p < 5e-2*, FC > 2), and gene clusters were divided into four groups according to expression levels. F The Venn diagram showed the number of genes up-regulated and down-regulated by RBAP48 in DHT-untreated and DHT-treated conditions.
RBAP48 regulates genome-wide mRNA expression in OSCC cells
To investigate the impact of RBAP48 on global mRNA expression in OSCC, we used siRNA to decrease RBAP48 expression in CAL-27 and SCC-9 cells (Supplementary Fig. 1G, H), followed by RNA-seq analysis of CAL-27 cells (Fig. 2E). Under DHT treatment, RBAP48 depletion resulted in the downregulation of 654 genes (489 + 165) and the upregulation of 704 genes (546 + 158). In the absence of DHT, RBAP48 knockdown significantly increased the mRNA expression of 1518 genes (1360 + 158) and decreased the expression of 1748 genes (1583 + 165) (Fig. 2F). Analysis of transcriptome regulation in the absence of DHT highlighted the substantial impact of RBAP48 on AR/DHT-independent genes.
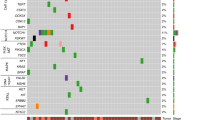
We further examined these significantly altered genes for their potential involvement in signaling pathways. Genes regulated by RBAP48 were associated with processes such as cell cycle, cellular response to stimulation, chemical responses, and DNA damage in the absence of DHT. This indicated a strong association between RBAP48 and OSCC cell proliferation, as well as their responses to external chemical stimuli (Fig. 3A, B). In the presence of DHT, genes reduced by RBAP48 were mainly involved in the cell cycle, AR signaling pathway, ATP metabolism, and cellular sensitivity to chemicals (Fig. 3B–C). After DHT treatment, RBAP48 appeared to exert a stronger influence on OSCC proliferation. Based on volcano plot statistics, we filtered the genes implicated in these pathways with substantial changes (Fig. 3D and Supplementary Fig. 2A). Crucial genes promoting cancer progression, including CAL, CCNE1, and RELA, were regulated by RBAP48, even in the absence of DHT treatment (Fig. 3E and Supplementary Fig. 2B). We then performed a correlation analysis with a series of target genes in the TCGA-HNSC data. The results showed that RBAP48 mRNA positively correlated with the mRNAs of CCND1, RAB31, RELA and CCNE1 in TCGA cohort (Supplementary Fig. 1B–E). Moreover, we examined the protein expression of a subset of target genes in clinical OSCC tissues and matched adjacent NC tissues using western blotting. The results demonstrated that RBAP48 protein levels was positively correlated with the protein levels of CCND1, RAB31, and BAG1 in OSCC samples (Supplementary Fig. 2D–G). Overall, RBAP48 significantly upregulated the mRNA expression of key oncogenes that promotes OSCC cell proliferation.
A The dotted graph showed the gene pathways enriched in different groups of gene clusters. Red indicates positive regulation of this pathway, and blue indicates negative regulation of this pathway. The size of the dot plot represented the enrichment degree. B-C GESA analysis represented an increased enrichment of these pathways. (B) the significantly enriched pathway state in the non-treated DHT state, and (C) the situation in the DHT treatment. D Volcanic dots represented significantly altered genes with siRBAP48 up-regulated (red) or down-regulated (green) under non-treated DHT conditions (p < 1e-2, FC > 2). E Genes that typically change in enrichment pathways were shown using heatmaps.
Following the RNA-seq analysis, we assessed the depletion efficiency of different siRNAs against RBAP48 at the mRNA level in CAL-27 and SCC-9 cells, and RBAP48 mRNA expression was significantly reduced (Supplementary Fig. 2C). In CAL-27 RBAP48 knockdown notably decreased the mRNA expression of CCND1, RAB31, BAG1, CAT, and several other oncogenes following DHT treatment (Fig. 4A and Supplementary Fig. 3A–3B). Similar results were obtained in SCC-9 cells, where RBAP48 positively upregulated the expression of these oncogenes (Fig. 4B and Supplementary Fig. 3C, D). Additionally, silencing of RBAP48 led to a significant reduction in the mRNA expression levels of ARF4, ATF5, BIN1, and BSG without DHT treatment in both OSCC cell lines (Fig. 4C, D and Supplementary Fig. 4A–D). Together, these findings demonstrate that RBAP48 plays a crucial role in the regulating a series of downstream genes to promote OSCC progression.
A-B siRNA was used to reduce the expression of RBAP48, and the significant changed genes were detected with qPCR assays in (A) CAL27 cells and (B) SCC9 cells with DHT treatment. C, D qPCR assays detected the expression of genes that changed significantly only in the absence of DHT in (C) CAL-27 cells and (D) SCC-9 cells. All qPCR statistics used the student t-test, and all experiments were independently repeated at least three times. p < 5e-2*, p < 1e-2**, p < 1e-3***, p < 1e-4****, and ns represented non-significant.
RBAP48 interacts with AR, SP1, and other transcription factors in OSCC cells
Next, we investigated the transcription factors that interact with RBAP48. In the absence of DHT, RBAP48 primarily coregulated its downstream genes with SP1, RELA, MYCN, and other transcription factors (Fig. 5A). Notably, significant enrichment of regulatory activity was observed in the downstream pathways of SP1 and RELA. These regulatory genes also showed marked enrichment in KMT2D target genes, independent of DHT (Fig. 5B). Given that KMT2D is an essential histone modifier of the WRAD subcomplex, we hypothesized that RBAP48 participates in transactivation mediated by the aforementioned transcription factors through its interaction with WRAD and KMT2D. Surprisingly, the expected interactions between the subcomplex members and SP1 or RELA were not detected by protein interaction website in the presence of AR (Fig. 5C). Furthermore, the genes regulated by RBAP48 were enriched in AR, VEGF, and AP1 following DHT treatment (Supplementary Fig. 5A, B). Notably, ASH2L was identified as a key protein capable of interacting with AR and SP-1 (Fig. 5C, D). These findings suggest that ASH2L and RBAP48 play crucial roles in the protein complex.
A-B (A) Go-TRRUST and (B) Go-Transcription Factor Targets detection revealed the Transcription factors enriched by the above significantly changed genes and the downstream of the potential Transcription factors enriched without DHT treated, respectively. C-D STRING analysis revealed the potential protein interaction network of RBAP48 when interacting with (C) AR or (D) SP1, respectively. E co-IP assays indicated the endogenous interaction between RBAP48 and AR with DHT treated in CAL-27 cells. F In CAL-27 cells, co-IP assays with the RBAP48 knockdown represented the AR and WRAD subcomplex interaction changes. G The impact of ASH2L depletion on the interplay between RBAP38 and AR protein in CAL-27 cells. H RBAP48 reduction on the interaction among the transcription factors SP1/RELA with RBAP48 in CAL-27 cells.
To elucidate the endogenous protein interactions in OSCC cells, we performed endogenous co-immunoprecipitation assays. In both OSCC cell lines, RBAP48 interacted with the AR in the presence of DHT (Fig. 5E and Supplementary Fig. 5C). Remarkably, RBAP48 also interacted with the AR in the absence of DHT, suggesting the potential influence of other proteins on this phenomenon. Silencing RBAP48 in CAL-27 cells for co-immunoprecipitation assays revealed a partial weakening of the interaction between AR, WDR5, RBBP5, ASH2L, and DPY30, indicating that RBAP48 may affect the interaction between AR and WRAD complex subunits (Fig. 5F). Conversely, ASH2L knockdown did not significantly affect the interaction between RBAP48 and the AR. These results suggest that RBAP48 independently influences AR and WRAD interactions, underscoring its crucial role in gene transactivation (Fig. 5G). To mimic AR signal inactivation, siRNA was used to silence AR without DHT stimulation. RBAP48 maintained its interaction with SP1 or RELA regardless of the presence of the AR protein (Fig. 5H). These results suggest that AR was not the dominant protein in the participation of RBAP48 in SP1/RELA-mediated gene transcription, and RBAP48 could still interact with other transcription factors to promote the transcription of downstream target genes in the absence of AR signaling.
RBAP48 modulates the recruitment of WRAD subcomplexes to gene promoter regions
Next, we investigated the molecular mechanisms underlying the effects of RBAP48 on gene transcription with or without AR activation. In the context of classical AR-mediated gene transcription, we identified two AREs in the promoter regions of CCND1 and RAB31 (Fig. 6A). A ChIP assay was performed to detect protein recruitment and histone modification levels at the CCND1-ARE1 and ARE2 regions (Fig. 6C, D). Silencing of RBAP48 led to reduced recruitment of AR and ASH2L in the ARE1 region, while no such effect was observed in the ARE2 region, concomitant with a decrease in H3K4me3 levels. Similar ChIP assays were performed on the promoter region of RAB31. RBAP48 significantly influenced AR and ASH2L recruitment and H3K4me3 modification in both ARE1 and ARE2 (Fig. 6E, F). These data underscore the vital role of RBAP48 in modulating the WRAD/AR complex on AR cis-regulatory elements of the target genes.
A The pattern showed potential ARE sites in the promoter regions of CCND1 and RAB31 genes. B The pattern showed the potential SP1 binding sites (SPSs) in the promoter region of the RELA gene and the potential RELA binding sites (RSs) in the CCNE1 promoter. C–F The ChIP qPCR results revealed the recruitment of RBAP48, AR and ASH2L and the modification level of H3K4me3 at the ARE site after RBAP48 depletion treated with DHT in CAL-27 cells. The total amount of DNA measured by qPCR represented relative enrichment (%input), and the student t-tests were used. IgG represented the positive control of antibodies. G, H ChIP assays demonstrated the recruitment of RBAP48, AR and ASH2L, as well as changes in H3K4me3 modification levels in the SPS regions of (G) RELA promoter and RS regions of (H) CCNE1 promoter.
Interestingly, there are two SP1 binding sites (SPS) in the promoter region of RELA, which functions without DHT. In addition, there were two RELA protein-binding sites (RS) on the CCNE1 promoter, which were significantly affected by RBAP48 in the absence of DHT in OSCC (Fig. 6B). ChIP assays showed that silencing of RBAP48 decreased the recruitment of SP1 and ASH2L and the modification level of H3K4me3 at the promoter region (Fig. 6G). Similarly, diminished recruitment of RBAP48 near RS2 on the CCNE1 promoter was associated with decreased recruitment of RELA and ASH2L proteins as well as decreased H3K4me3 modification levels (Fig. 6H). These results indicate that RBAP48 plays an essential role in the transcriptional activity of gene promoter regions. Notably, our analysis revealed that RBAP48-regulated genes were enriched in both SP1 and RELA, with over half of these genes regulated by both transcription factors. Additionally, DNA motif analysis unveiled striking similarity at the 5’ end of the DNA binding of SP1 and RELA, suggesting potential co-regulation of their target genes by the two transcription factors (Supplementary Fig. 5D).
Further exploration identified shared recruitment sites for SP1 and RELA in the promoter regions of RHOC and CSRP1. ChIP assay results showed that SP1, RELA, and ASH2L recruitment was significantly reduced upon RBAP48 silencing, accompanied by decreased histone H3K4me3 modification (Supplementary Fig. 5E, F). Notably, the facilitating role of RBAP48 in SP1-mediated transcription prompted a re-evaluation of the impact of SP1 and RBAP48 expression on the survival of AR-negative patients. RBAP48high/SP1high patients exhibited the poorer survival, indicating a role for RBAP48 in promoting SP1-mediated transcription and cancer progression in the absence of AR (Supplementary Fig. 6A). Additionally, a positive correlation was observed between RBAP48 and RELA mRNA expression in AR-negative patients (Supplementary Fig. 6A). While the above correlation in AR-positive patients has not been observed (Supplementary Fig. 6B). All these results suggested that the relationship between RBAP48 and SP1 expression levels could serve as a vital prognostic indicator for OSCC.
RBAP48 promotes OSCC cell growth and resistance to external stimulation
Finally, we explored the effects of RBAP48 on the biological functions of OSCC cells. We established stable RBAP48 knockdown in CAL27 cells using lentiviral vectors. In the xenograft tumorigenesis assay, RBAP48 knockdown significantly reduced the size of xenograft tumors (Fig. 7A, B). Similarly, RBAP48 knockdown reduced the ectopic tumor growth rate and final tumor weight (Fig. 7C, D). Subsequent qPCR assays showed that RBAP48 knockdown notably decreased the mRNA expression of key oncogenes including CCND1, RAB31, CCNE1, and RELA (Fig. 7E). Notably, RBAP48 knockdown did not significantly affect P21 expression but reduced CCND1 expression in mouse tissues and OSCC cells, indicating a dominant role of RBAP48 in cell proliferation rather than apoptosis or senescence (Supplementary Fig. 6C, D). Similarly, we analyzed the expression of RBAP48 and that of the classical cellular senescence genes in the RNA-seq data. The results showed that TP53 and CDKN1A were affected by RBAP48 knockdown, while the majority of senescence-related genes were not regulated by RBAP48 (Supplementary Fig. 6E). These results suggested that cellular senescence might be not mainly influence by RBAP48.
A-B Typical illustration of ectopic tumorigenesis in mice and tumor size. Tumor cells were completed by stably silencing RBAP48 CAL-27 cells with lentivirus. C-D Comparison of tumor-bearing volume and final tumor-bearing weight in mice. The student t-tests were used between two groups. E The mRNA expression of RBAP48 and its regulated genes were detected by qPCR assay using RNA extracted from tumor tissue as template. F Cell clone formation assay showed the effect of RBAP48 silencing on cell colony size and number. G-H Cell growth curve experiments showed the effects of RBAP48 silencing on cell growth under DHT treatment and DHT treatment, respectively. I Cell growth curve assay showed that overexpression of RBAP48 reversed the growth inhibition effect of CDK4/6 inhibitor. J Schematic representation of the key role of RBAP48 different transcription patterns jointly promote the biological function of OSCC cells biological function such as stimulation response and cell growth in the absence of DHT treatment and DHT treatment.
We examined the effects of RBAP48 on biological functions at the cellular level in vitro. In CAL-27 cells, the silencing RBAP48 significantly reduced the size and number of clones in a DHT-independent manner (Fig. 7F). Similarly, cell growth curve experiments showed that RBAP48 depletion significantly reduced cell growth in both DHT-free and DHT-treated cells (Fig. 7G and H). Overexpression of RBAP48 increases the proliferation rate in two OSCC cell lines (Supplementary Fig. 6F, G). We then assessed the effects of RBAP48 on cellular tolerance to external stimuli. Tumor cells often develop adaptive resistance to therapeutic drugs. In OSCC, the transition from fibrosis to keratinization can be induced by excessive external stimulation13,43,44, with palbociclib, a common CDK4/6 inhibitor, significantly inhibited the growth rate of OSCC cells. Our data showed that the ectopic expression of RBAP48 significantly increase the proliferation of OSCC-derived cells in palbociclib treatment (Fig. 7I). Treatment of CAL-27 cells with doxorubicin and cisplatin was used to simulate the DNA damage repair state. Western blot results showed an enhancement of γ-H2AX expression induced by DNA damage was markedly reduced by the ectopic expression of RBAP48 in CAL-27 cells. These results suggest that RBAP48 may be involved in DNA damage repair process, subsequently decreasing cellular sensitivity to drug treatments (Supplementary Fig. 6H). In addition, exposure of OSCC cells to guvacoline as simulation external stimulation resulted in fibrotic morphological changes in CAL-27 cells (Supplementary Fig. 6I). In cells of RBAP48 overexpression, the genes RRAGA and WDR45, which positively regulate the cellular response to external stimuli, were significantly downregulated. These results suggest that RBAP48 may participate in regulation of cell response to perceive external stimuli (Supplementary Fig. 6I, J). The analysis of the GSE173855 dataset for HNSC patients who received radiotherapy revealed a significant positive correlation between RBAP48 expression in primary tumors and the time to relapse, indicating that higher RBAP48 expression in primary tumors is associated with a longer time to relapse (Supplementary Fig. 7A). In contrast, RBAP48 expression in relapsed tumors does not predict the time to relapse (Supplementary Fig. 7B). High RBAP48 expression in primary tumors is associated with higher expression levels of numerous DNA damage repair genes but not in relapsed tumors (Supplementary Fig. 7C, D). These findings suggest that RBAP48 may play a role in DNA damage repair mechanisms in HNSC, but its predictive value for relapse time is specific to primary tumors.
Collectively, these findings suggest that RBAP48 is involved in regulation of cell cycle processes and cell response to external stimuli to influence OSCC progression.
Discussion
Recent studies have focused on WD-40 repeat-containing proteins, highlighting their importance in cancer processes and their potential as therapeutic targets45,46. RBAP48, a member of this protein family, plays a unique role in tumors. In this study, we reveal that RBAP48 acts as a key player in OSCC development by acting as a transcriptional co-activator. We found that RBAP48 was highly expressed in OSCC tissues, and it promoted cell growth and the response of cells to external signals through various transcription factors and signaling pathways. Mechanically, RBAP48 facilitated gene transcription via both AR/DHT-dependent and independent pathways. This complex role suggests that RBAP48 is an important target for diagnosing and treating OSCC (Fig. 7J).
RBAP48 and another protein, RBAP46, share a high level (92%) of sequence similarity and often coexist in different chromatin-modifying complexes47. Although these two proteins are part of separate complexes, these proteins likely have similar role as they are mutually exclusive in these complexes48. Both RBAP48 and RBAP46 have been linked to several types of cancer, making them potential targets for new cancer treatments. For example, RBAP46 is upregulated in cancers such as lung, kidney and, and breast cancers49,50,51. Similarly, RBAP48 is overexpressed in liver cancer (hepatocellular carcinoma) and acute myeloid leukemia52,53. In a brain cancer study, RBAP48 knockdown in human glioblastoma cells sensitized them to temozolomide (TMZ) chemotherapy by downregulating the activity of key DNA repair genes (e.g., RAD51), which is crucial for homologous recombination repair54. These findings suggest that RBAP48 promotes cancer progression by affecting DNA repair and the chromatin landscape. In agreement, our data showed that RBAP48 significantly affected the cellular response to DNA damage and external stimuli in OSCC cells. Our RNA-seq data showed that RBAP48 knockdown significantly downregulated the expression of key genes (e.g., CDKN1A, BAX, and CRIP1) involved in DNA repair (Fig. 3). Thus, our study supports the idea that prolonged exposure to harmful substances like alcohol, tobacco, betel nut affects DNA repair in oral epithelial cells, which likely contribute to OSCC development.
We also found that RBAP48 regulates the cellular response of oral epithelial cells to external stimuli in both DHT-treated and untreated conditions, with the regulated genes showing both commonalities and specificity in these situations. Based on our RNA-seq results, RBAP48 was identified as a novel marker of DNA repair and the response of cells to external stimuli in OSCC (Figs. 2–4). The processes of DNA repair and response to external signals represent a long-term change in the cell phenotype, progressing from fibrosis to hyperplasia and eventually to cancer. RBAP48 appears to exert AR signaling in a way that promotes cancer, particularly when AR activation is low. This observation may offer a plausible explanation for the poor prognosis observed in patients with OSCC with low AR levels but high RBAP48 expression (Fig. 1). RBAP48 and AR did not emerge as significant predictors of overall survival in the TCGA dataset (Supplementary Fig. 1F). This finding may be attributed to several limitations. Firstly, the sample size and heterogeneity of the TCGA dataset may not be sufficient to detect subtle associations. Secondly, measurement variability in RBAP48 and AR levels could introduce noise, reducing the ability to identify significant effects. Additionally, the complex biological roles of these proteins, involving interactions with other pathways, may not be fully captured in the TCGA data. Furthermore, unaccounted clinical factors could confound the relationship between RBAP48, AR, and survival. Lastly, the analysis may lack the statistical power to detect significant associations due to small effect sizes. Future studies with larger sample sizes and comprehensive clinical data are needed to further investigate the predictive value of RBAP48 and AR (Supplementary Fig. 1F).
Additionally, RBAP48 interacts with certain transcription factors in OSCC revealing intriguing insights. Our RNA-seq analysis showed that transcription factors enriched in RBAP48 regulated genes involved in the KMT2D pathway, which is important for regulating the cell cycle and changes in cell phenotypes (Fig. 5)55,56,57. KMT2D is a critical modification enzyme in the WRAD subcomplex of the COMPASS complex. The way RBAP48 regulates KMT2D target genes depends on whether cells are treated with DHT. Under normal (non-DHT) conditions, RBAP48 works with WRAD/KMT2D complex and interacts primarily with SP1 to regulate target gene transcription. However, in DHT-treated cells, RBAP48 helps control the activity of AR target genes (Fig. 5). These observation led us to speculate that RBAP48 has a significant impact on the regulation of gene activity.
Furthermore, RBAP48 acted as a scaffolding protein to facilitate assembly among complex proteins, as well as the accessibility of histones to RBAP48-chromatin binding sites. In agreement with previous studies58,59, we found that RBAP48 interacted directly with histone residues (Fig. 5). This high-level regulation of the chromatin landscape is specific to OSCC, and our findings suggest that RBAP48 could be targeted for new treatments. Inhibiting the ability of RBAP48 recruit proteins to chromatin may offer as a novel approach to treat OSCC.
In conclusion, our study shows that RBAP48 plays an important role in the progression of OSCC by regulating gene activity through both AR-dependent and independent pathways. Its role in DNA repair, response to external stimuli, and chromatin regulation makes it a promising target for diagnosing and treating OSCC. Our study has several limitations. First, due to the lack of specific classification for OSCC in public datasets, further multicenter validation is required to determine whether RBAP48 serves as a risk factor for cancer progression in HNSC patient cohorts such as TCGA. Second, although TP53 and CDKN1A were significantly regulated by RBAP48, their corresponding biological phenotypes were not prominently observed. These findings will be further clarified in future studies.
Materials and methods
Cell lines and cell culture
The oral squamous cell carcinoma cell lines were obtained from the ATCC cell repository. Squamous-cell carcinoma epithelial cells (CAL-27 and SCC-9 cells) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific). At 37°C and 5% CO2, all cells were grown with 10% fetal bovine serum (FBS; Gibco), 50 units/mL penicillin, and 50 units/mL streptomycin. 5α-Dihydrotestosterone (DHT; Sigma-Aldrich; D-073; final concentration: 10-7M), Doxorubicin (Dox; MCE, HY-15142), Cisplatin (Cis; MCE, HY-17394), Palbociclib (MCE; HY-50767; final concentration: 10-6M) and Guvacine (MCE; HY-N2482; final concentration: 10-8M) were dissolved in DMSO or ethanol (EtOH; Aladdin). When it was necessary to add a drug stimulus, all the cells were grown in DMEM without phenol red. Culture equipment was purchased from Jet Bio-Filtration Co., Ltd., Guangzhou, China.
Plasmids and antibodies
Full-length RBAP48 was cloned into pcDNA3.1, containing FLAG tags, which were digested with the dual enzymes EcoRI and NotI. The primers used for PCR amplification were: F, TCAGAATTCGCCACCATGGATGGCCGACAAGGAAGC and R, GACTGACAGCGGCCGCCTAGGACCCTTGTCCTTCTGG. Our previous work characterized the plasmids used in the luciferase experiments60. The antibodies used in this study were as follows: anti-RBAP48 (Abcam; #ab79416), anti-Androgen receptor (Proteintech; #22089-1-AP), anti-ASH2L (Abcam, #ab181117), anti-WDR5 (Abcam, #ab56919), anti-RBBP5 (Abcam, #ab154755), anti-DPY30 (Abcam, #ab126352), anti-SP1 (Abcam, #ab227383), anti-RELA (Proteintech, #10745-1-AP), anti-H3K4me3 (Millipore; #04-745), anti-β-actin (Proteintech, #20536-1-AP), anti-Rabbit/Mouse (ABclonal), anti-IgG (Proteintech, #10238-1-AP), anti-FLAG (Proteintech, #20543-1-AP).
siRNA and lentivirus
Supplementary Table 1 lists the siRNA sequences against RBAP48, ASH2L, and AR designed in this study. All siRNAs were transfected using the transfection agent jet-PRIME (Polyplus). RBAP48 lentivirus was synthesized using the same target sequence as siRBAP48. Lentiviruses were purchased from the GeneChem Company, Shanghai, China and the manufacturer’s recommendations were followed while transfecting the system into the cells.
Luciferase reporter assays
HEK-293, CAL-27, and SCC-9 cells were co-transfected with AR or AR-truncated mutants (20 ng), ARE-tk-Luc plasmids (200 ng), control Renilla luciferase (pRL) plasmids (5 ng) and FLAG-tagged RBAP48 in the indicated amounts. After co-transfection for 4 h, the cells were grown in a medium containing 5% FBS stripped of hormones. After an additional day, the cells were harvested for the luciferase reporter assay (Promega) after 12 h of androgen stimulation. The final relative activity was determined as the luciferase fluorescence intensity normalized to the pRL fluorescence intensity.
Western blotting and co-immunoprecipitation (co-IP) analysis
Western blotting was performed according to the standard procedures. Whole-cell lysis was performed using an anti-IgG antibody for 2 h. Sepharose-condensed Protein G beads (GE Healthcare) were used to rotate antibody-protein interactions. After rinsing thrice and boiling the lysate, western blotting was performed.
RNA and quantitative real-time PCR (qPCR)
cDNAs were reverse transcribed using the PrimeScript RT-PCR kit after total RNA was extracted using TRIzol (Takara). Real-time qPCR experiments on a LightCycler 96 instrument (Roche) were performed using the SYBR Premerase Taq kit (Roche). Supplementary Table 2 describes each qPCR primer. The results of at least three different trials were analyzed using Student’s t-tests.
Chromatin immunoprecipitation (ChIP) assays
Using established techniques from Nature Protocols, we performed ChIP assays. RBAP48 expression was silenced by transfecting siRNA to block its expression in CAL-27 cells. The cells were grown in DMEM without phenol. After reaching 80% confluence, the cells were exposed to 100 nM DHT or comparable amounts of EtOH for 1 h. DNA was used as a qPCR template after the experiments, and the primers used are listed in Supplementary Table 3.
Cell growth and colony formation assays
Before being stained with trypan blue and counted using a hematocytometer, all OSCC cells were grown for a predetermined period. Approximately 500 cells per 35 mm plate were used for cell drug-treatment experiments, while 1000 cells per column were used for cell growth-line experiments. Cells were subjected to a 14 day DHT or EtOH treatment period to complete the colony formation experiment.
Xenograft tumor
Eight BALB/c nude male mice (6 weeks old) were obtained from Charles River Laboratories, Beijing, China. The mice were subcutaneously inoculated with OSCC cells. Before injections, 50 μL of medium and 50 μL of Matrigel (BD Biosciences) mixture was added to about 5.0 million OSCC cells in which RBAP48 was silenced (shRBAP48), and its negative control (shCtrl). The tumor size was monitored for 49 days. The following formula was used to calculate the tumor volume: V = (π/6) × (L × W)3/2, where V stands for volume (mm3), L for the greatest diameter (mm), and W for the most minor diameter (mm). The Animal Ethics Committee of the China Medical University approved and supervised the animal experiments (KT20240575). We have complied with all relevant ethical regulations for animal use. The maximal tumor did not exceed 15 mm in any one dimension.
Tissue samples and immunohistochemistry (IHC)
The Hospital of Stomatology, affiliated with China Medical University, provided all OSCC samples and noncancerous tissues, and all patients received curative care. Patients with OSCC underwent surgical operations and tumor sample collection before receiving any main treatments, such as radiation, chemotherapy, or adjuvant therapies. General outpatient procedures were performed to obtain noncancerous oral epithelial tissues (noncancerous tissues). Gingival and localized epithelial hyperplastic tissues were among the non-cancerous tissues evaluated in this study. IHC studies were conducted as previously explained61. The Human Research Ethics Committee of China Medical University authorized the collection of tissue samples and all studies. For each sample, informed permission was obtained from the Hospital of Stomatology at China Medical University provided clinical OSCC sections. The Approval Number for Scientific Research Ethics is No.12. All ethical regulations relevant to human research participants were followed. Informed consent was obtained from the patients.
RNA-seq
CAL-27 cells were treated with 10 nM DHT and EtOH for 24 h. Each experiment was repeated thrice. Total RNA was extracted using an RNeasy kit (Qiagen), and the RNA concentration was quantified. The procedures for analyzing RNA-seq data were introduced in our previous study60, and graphs were generated using GraphPad 8 and OriginLab. The RNA-seq data were uploaded to the GEO database (accession number GSE210765).
TCGA data analysis and survival analysis
Data of 499 patients with head and neck squamous cell carcinoma (HNSC) were obtained from TCGA database for correlation analysis of mRNA or protein expression. The data are presented in Supplementary Table 4. The data included patient ID, age, grade (partial), survival status, elapsed time, and gene expression. Gene expression data were provided in fragments per kilobase of transcript per million mapped reads (FPKM) for all patients include RBAP48, AR, SP1, and other genes. Survival analysis was performed using GraphPad Prism 10 software. Overall survival was used as the clinical endpoint in this study.
Statistical analysis and reproducibility
In this study, dichotomization was performed as follows: values lower than the mean of the overall data was considered low expression of each gene, and values higher than the mean were considered high expression. Statistical analyses were performed using SPSS version 19.0 software. Continuous variables are reported as mean ± standard deviation (SD). Student’s two-tailed t-test was used to determine the statistical significance between groups. The Mann–Whitney U test was used to analyze clinical specimens. All experiments were independently repeated at least three times. For qPCR results from triplicate wells, PRISM GraphPad 8 was used for statistical analysis. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data were uploaded to the GEO database (accession number GSE210765) and are available at the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE210765. The uncropped and unedited blot images are shown in Supplementary Fig. 8. The TCGA data of Supplementary Table 4 is contained within the Supplementary Data 1, and the numerical source data can be found in the Supplementary Data 2. Supporting data related to this work are available upon request.
References
Fang, Y., Yang, Y. & Liu, C. Evolutionary relationships between dysregulated genes in oral squamous cell carcinoma and oral microbiota. Front Cell Infect. Microbiol. 12, 931011 (2022).
Li, R. et al. Induction chemotherapy of modified docetaxel, cisplatin, 5-fluorouracil for laryngeal preservation in locally advanced hypopharyngeal squamous cell carcinoma. Head. Neck 44, 2018–2029 (2022).
Thawani, R. et al. The contemporary management of cancers of the sinonasal tract in adults. CA. Cancer J. Clin. 73, 72–112 (2022).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 71, 209–249 (2021).
Gupta, S. et al. Relationship between type of smokeless tobacco & risk of cancer: a systematic review. Indian J. Med. Res. 148, 56–76 (2018).
Bagnardi, V. et al. Alcohol consumption and site-specific cancer risk: a comprehensive dose-response meta-analysis. Br. J. Cancer 112, 580–593 (2015).
Han, A. Y. et al. Epidemiology of squamous cell carcinoma of the lip in the united states: a population-based cohort analysis. JAMA Otolaryngol. Head. Neck Surg. 142, 1216–1223 (2016).
Turati, F. et al. A meta-analysis of alcohol drinking and oral and pharyngeal cancers. Part 2: results by subsites. Oral. Oncol. 46, 720–726 (2010).
Justice, A. C. Alcohol and the global burden of cancer: what are we missing?. Lancet Oncol. 22, 1048–1049 (2021).
Rehman, A. et al. Areca nut alkaloids induce irreparable DNA damage and senescence in fibroblasts and may create a favourable environment for tumour progression. J. Oral. Pathol. Med. 45, 365–372 (2016).
Prime, S. S. et al. Fibroblast activation and senescence in oral cancer. J. Oral. Pathol. Med. 46, 82–88 (2017).
Bijai, L. K. & Muthukrishnan, A. Potential role of fibroblast senescence in malignant transformation of oral submucous fibrosis. Oral. Oncol. 127, 105810 (2022).
Tilakaratne, W. M. et al. Oral submucous fibrosis: review on aetiology and pathogenesis. Oral. Oncol. 42, 561–568 (2006).
Cheng, R. H. et al. Genetic susceptibility and protein expression of extracellular matrix turnover-related genes in oral submucous fibrosis. Int. J. Mol. Sci. 21, 8104 (2020).
Kondaiah, P., Pant, I. & Khan, I. Molecular pathways regulated by areca nut in the etiopathogenesis of oral submucous fibrosis. Periodontol. 2000 80, 213–224 (2019).
Ishii, S. The role of histone deacetylase 3 complex in nuclear hormone receptor action. Int. J. Mol. Sci. 22, 9138 (2021).
Scholtes, C. & Giguere, V. Transcriptional control of energy metabolism by nuclear receptors. Nat. Rev. Mol. Cell Biol. 23, 750–770 (2022).
Perissi, V. et al. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116, 511–526 (2004).
O’Malley, B. W. & Kumar, R. Nuclear receptor coregulators in cancer biology. Cancer Res. 69, 8217–8222 (2009).
Tomasovic-Loncaric, C. et al. Androgen receptor as a biomarker of oral squamous cell carcinoma progression risk. Anticancer Res. 39, 4285–4289 (2019).
Liu, X. et al. Androgen receptor promotes oral squamous cell carcinoma cell migration by increasing EGFR phosphorylation. Onco. Targets Ther. 12, 4245–4252 (2019).
Wu, T. F. et al. The oncogenic role of androgen receptors in promoting the growth of oral squamous cell carcinoma cells. Oral. Dis. 21, 320–327 (2015).
Colella, G. et al. Expression of sexual hormones receptors in oral squamous cell carcinoma. Int J. Immunopathol. Pharm. 24, 129–132 (2011).
Qian, Y. W. et al. A retinoblastoma-binding protein related to a negative regulator of Ras in yeast. Nature 364, 648–652 (1993).
Huang, S., Lee, W. H. & Lee, E. Y. A cellular protein that competes with SV40 T antigen for binding to the retinoblastoma gene product. Nature 350, 160–162 (1991).
Jain, B. P. & Pandey, S. WD40 repeat proteins: signalling scaffold with diverse functions. Protein J. 37, 391–406 (2018).
Schapira, M. et al. WD40 repeat domain proteins: a novel target class?. Nat. Rev. Drug Discov. 16, 773–786 (2017).
Moody, R. R. et al. Probing the interaction between the histone methyltransferase/deacetylase subunit RBBP4/7 and the transcription factor BCL11A in epigenetic complexes. J. Biol. Chem. 293, 2125–2136 (2018).
Millard, C. J. et al. The structure of the core NuRD repression complex provides insights into its interaction with chromatin. Elife 5, e13941 (2016).
Zhang, Y. et al. Histone deacetylases and SAP18, a novel polypeptide, are components of a human Sin3 complex. Cell 89, 357–364 (1997).
Ciferri, C. et al. Molecular architecture of human polycomb repressive complex 2. Elife 1, e00005 (2012).
Liu, X. A structural perspective on gene repression by polycomb repressive complex 2. Subcell. Biochem 96, 519–562 (2021).
Ding, L. et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer 18, 45 (2019).
Li, L. et al. Epigenetic modification of MiR-429 promotes liver tumour-initiating cell properties by targeting Rb binding protein 4. Gut 64, 156–167 (2015).
Guo, Q. et al. Expression of HDAC1 and RBBP4 correlate with clinicopathologic characteristics and prognosis in breast cancer. Int. J. Clin. Exp. Pathol. 13, 563–572 (2020).
Cui, F. et al. Grifola frondosa glycoprotein GFG-3a arrests s phase, alters proteome, and induces apoptosis in human gastric cancer cells. Nutr. Cancer 68, 267–279 (2016).
Zhu, P. et al. ZIC2-dependent OCT4 activation drives self-renewal of human liver cancer stem cells. J. Clin. Invest. 125, 3795–3808 (2015).
Ishimaru, N. et al. Expression of the retinoblastoma protein RbAp48 in exocrine glands leads to Sjogren’s syndrome-like autoimmune exocrinopathy. J. Exp. Med. 205, 2915–2927 (2008).
Lohavanichbutr, P. et al. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch. Otolaryngol. Head. Neck Surg. 135, 180–188 (2009).
Liu, Z. et al. Design and Synthesis of EZH2-Based PROTACs to Degrade the PRC2 Complex for Targeting the Noncatalytic Activity of EZH2. J. Med. Chem. 64, 2829–2848 (2021).
Ferreira, M. et al. The deletion of the protein phosphatase 1 regulator NIPP1 in testis causes hyperphosphorylation and degradation of the histone methyltransferase EZH2. J. Biol. Chem. 293, 18031–18039 (2018).
Han, Z. et al. Dual-acting peptides target EZH2 and AR: a new paradigm for effective treatment of castration-resistant prostate cancer. Endocrinology 164, bqac180 (2022).
Shieh, D. H., Chiang, L. C. & Shieh, T. Y. Augmented mRNA expression of tissue inhibitor of metalloproteinase-1 in buccal mucosal fibroblasts by arecoline and safrole as a possible pathogenesis for oral submucous fibrosis. Oral. Oncol. 39, 728–735 (2003).
Moutasim, K. A. et al. Betel-derived alkaloid up-regulates keratinocyte alphavbeta6 integrin expression and promotes oral submucous fibrosis. J. Pathol. 223, 366–377 (2011).
Xu, C. & Min, J. Structure and function of WD40 domain proteins. Protein Cell 2, 202–214 (2011).
Tian, J. et al. Discovery and structure-based optimization of potent and selective WD repeat domain 5 (WDR5) inhibitors containing a dihydroisoquinolinone bicyclic core. J. Med. Chem. 63, 656–675 (2020).
Cao, R. & Zhang, Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet Dev. 14, 155–164 (2004).
Verreault, A. et al. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr. Biol. 8, 96–108 (1998).
Kim, D. S. et al. Panel of candidate biomarkers for renal cell carcinoma. J. Proteome Res. 9, 3710–3719 (2010).
Thakur, A. et al. Aberrant expression of X-linked genes RbAp46, Rsk4, and Cldn2 in breast cancer. Mol. Cancer Res. 5, 171–181 (2007).
Wang, C. L. et al. Discovery of retinoblastoma-associated binding protein 46 as a novel prognostic marker for distant metastasis in nonsmall cell lung cancer by combined analysis of cancer cell secretome and pleural effusion proteome. J. Proteome Res. 8, 4428–4440 (2009).
Song, H. et al. Genes encoding Pir51, Beclin 1, RbAp48 and aldolase b are up or down-regulated in human primary hepatocellular carcinoma. World J. Gastroenterol. 10, 509–513 (2004).
Sakhinia, E. et al. Routine expression profiling of microarray gene signatures in acute leukaemia by real-time PCR of human bone marrow. Br. J. Haematol. 130, 233–248 (2005).
Kitange, G. J. et al. Retinoblastoma binding protein 4 modulates temozolomide sensitivity in glioblastoma by regulating DNA repair proteins. Cell Rep. 14, 2587–2598 (2016).
Fagan, R. J. & Dingwall, A. K. COMPASS ascending: emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Lett. 458, 56–65 (2019).
Alam, H. et al. KMT2D deficiency impairs super-enhancers to confer a glycolytic vulnerability in lung cancer. Cancer Cell 37, 599–617.e7 (2020).
Zhang, Y. et al. Genome-wide CRISPR screen identifies PRC2 and KMT2D-COMPASS as regulators of distinct EMT trajectories that contribute differentially to metastasis. Nat. Cell Biol. 24, 554–564 (2022).
Murzina, N. V. et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure 16, 1077–1085 (2008).
English, C. M. et al. Structural basis for the histone chaperone activity of Asf1. Cell 127, 495–508 (2006).
Sun, S. et al. BAP18 coactivates androgen receptor action and promotes prostate cancer progression. Nucleic Acids Res. 44, 8112–8128 (2016).
Wang, X. et al. BAP18 is involved in upregulation of CCND1/2 transcription to promote cell growth in oral squamous cell carcinoma. EBioMedicine 53, 102685 (2020).
Acknowledgements
We appreciate Professor Shigeaki Kato for valuable comments. We thanks Dr. Yunlong Huo for kind technique support, and SeqHealth Tech Co., Ltd Wuhan, China, for RNA-seq analysis. This study was supported by the National Natural Science Foundation of China (32370634, 32170603 for Yue Zhao, 82273123 for Chunyu Wang, 32100440 for Ge Sun); China Postdoctoral Science Foundation (2021M693529 for Ge Sun); Foundation of Liaoning Province of China (LJKZ0756 for Shengli Wang, LJKMZ20221152 for Chunyu Wang), Science and Technology Project of Liaoning Province (2022JH2/20200027 to Shengli Wang).
Author information
Authors and Affiliations
Contributions
Y.Z. and G.S. conceived the project, designed the experiments and wrote the paper. X.W. and G.Y. performed the main experiments and collected the data. H.L., C.W., Y.K. and S.W. participated in the experiments. W.L., L.L. and R.Z. drafted the figures. K.Z., M.W. and R.L. analyzed the data. B.Z., Y.B., D.Y. and B.N. reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Jochen Hess, Matias Eliseo Melendez, and Hiromu Suzuki for their contribution to the peer review of this work. Primary Handling Editor: Johannes Stortz. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, X., Yan, G., Li, H. et al. RBAP48 facilitates the oral squamous cell carcinoma process in an androgen receptor-dependent and independent manners. Commun Biol 8, 829 (2025). https://doi.org/10.1038/s42003-025-08215-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08215-4