Abstract
Intellectual disability affects 1–3% of the global population, with many unidentified genetic causes. This study investigates the role of CAPZA2, an actin cytoskeleton regulator, in neurodevelopmental disorders using CAPZA2 heterozygous knockout (CAPZA2+/−) and heterozygous point mutant (CAPZA2c.G776T/+) mice. CAPZA2+/− and CAPZA2c.G776T/+ mice demonstrate notable decreases in CAPZA2 expression within the hippocampus and prefrontal cortex (PFC), crucial for learning and memory. Interestingly, the reduction of CAPZA2 in CAPZA2c.G776T/+ mice is less than 50%. Behavioral assays revealed that CAPZA2+/− mice exhibited motor dysfunction and anxiety-like behaviors, along with impairments in both spatial and non-spatial memory, accompanied by deficits in social interactions. These phenotypic manifestations are also mirrored in the CAPZA2c.G776T/+ mice, thus validating the genotype-phenotype correlation. Morphological analyses of these gene-edited mice indicate structural synaptic impairments, with increased dendritic spine density, altered spine morphology in the hippocampus, and decreased dendritic complexity in the PFC. Single-cell RNA-seq analysis of hippocampal tissue in CAPZA2+/− mice shows widespread transcriptional dysregulation, affecting neurodevelopment and synaptic function genes. The altered PSD95 and glutamate receptor levels support these findings. These insights highlight the gene’s essential role in brain development and function, with potential implications for understanding and treating neurodevelopmental disorders.

Similar content being viewed by others
Introduction
Intellectual disability (ID) represents a diverse clinical and genetic spectrum, impacting 1–3% of the global population, characterized by significant challenges in intellectual functioning, adaptive behaviors, and practical skills1,2,3,4. This condition profoundly affects interpersonal relationships and environmental interactions5. Despite identifying mutations in ~25% of individuals with ID, the genetic underpinnings remain elusive in up to 60% of cases6. The quest for novel causative genes and the investigation into the roles of their encoded proteins are vital for advancing our understanding of ID pathogenesis, which, in turn, will inform clinical diagnosis and therapeutic approaches.
Recent discoveries have pinpointed mutations in the CAPZA2 gene as potential contributors to ID7,8,9. CAPZA2, situated on chromosome 6, encodes the α2 subunit of the Capping Protein (CapZ), a crucial element of the actin cytoskeleton across eukaryotes10. CapZ regulates actin filament dynamics, essential for cellular structure and movement11,12,13. It binds to actin filaments, preventing further monomer addition and facilitating the formation of branching actin networks14,15. This significantly impacts processes such as cellular morphological changes, neuron growth, and migration. Especially during neurodevelopment, CapZ plays a crucial role in establishing neural networks and refining neural functions by regulating dendritic structure and synaptic plasticity16. Therefore, CAPZA2, as an important gene regulating the actin cytoskeleton, may cause neurodevelopmental abnormalities and lead to ID when mutated. Recent clinical studies have shown a significant association between CAPZA2 gene mutations and the occurrence of various neurodevelopmental disorders. In a previous study, two pediatric patients were identified as carrying disruptive heterozygous de novo mutations in CAPZA2 (c.776 G > T, p.Arg259Leu and c.766 A > G, p.Lys256Glu), which may lead to neurological symptoms including motor and speech developmental delays, intellectual disability, hypotonia, and a history of epilepsy7. These mutations may affect the function of CapZ protein, leading to dendritic development disorders and synaptic plasticity abnormalities, thereby impacting neuron growth and connectivity. Additionally, a novel in-frame deletion variant within the CAPZA2 domain may lead to secondary microcephaly and global developmental delay, further supporting the critical role of CAPZA2 in neurodevelopment8. Recently a study identified a fourth case of non-syndromic neurodevelopmental disorder associated with a mutation in CAPZA2. Whole exome sequencing detected a novel de novo splicing variant c.219+1 G > A in the CAPZA2 gene, which results in an exon 4 skipping mutation in mRNA splicing, and manifests itself in the form of epileptic seizures, mild intellectual disability, and impaired movement in addition to respiratory tract infections immune system disorders9. The evidence extends to the DECIPHER database, which shows that chromosomal deletions affecting a specific region have been identified in 20 children. These deletions encompass several genes, including the CAPZA2 gene. They are associated with symptoms such as language delays, cognitive deficits, aphasia, and autism, reflecting the manifestations seen in the previously mentioned cases17. These above correlations suggest a significant role for CAPZA2 in neurodevelopment, yet the direct causality and clinical implications of CAPZA2 disruption in developmental disorders warrant further exploration. Therefore, there is an urgent need to validate further the causal relationship between CAPZA2 gene mutations and ID using animal models and to explore the underlying molecular mechanisms.
Our study aims to thoroughly investigate the relationship between CAPZA2 deficiency and ID by employing two distinct mouse models: CAPZA2 heterozygous knockout (CAPZA2+/−, Het) mice and heterozygous mutant (CAPZA2c.G776T/+, MUT) mice. The CAPZA2+/− mice, in which one allele of the CAPZA2 gene is deleted, are used to evaluate whether the loss of CAPZA2 directly contributes to neurodevelopmental abnormalities, particularly the behavioral and pathological phenotypes associated with ID. The CAPZA2c.G776T/+ mice are developed to mimic the mutation identified in clinical patients, enabling us to examine whether this specific mutation directly drives the onset of ID, thereby validating its pivotal role in the pathogenesis of ID.
This study is the first to report an ID mouse model attributed to CAPZA2 mutations. Preliminary evidence demonstrates that CAPZA2 deficiency impairs neuronal synaptic structures in the prefrontal cortex (PFC) and hippocampus, leading to neuronal loss, glial cell dysregulation, and altered expression of synaptic-related proteins. These findings highlight the pathological basis of behavioral abnormalities in CAPZA2-deficient mice, offering new insights into the mechanisms of ID and providing a new direction for targeted therapeutic strategies.
Results
Expression pattern of CAPZA2 and establishment of gene-edited mice
Expression patterns of CAPZA2 in mouse brains
In the continuous developmental stages, we meticulously mapped the ontogenetic trajectory of the CAPZA2 protein in the brains of normal mice. We then extended our study to multiple brain regions in adult mice, systematically assessing the expression patterns of this protein (Fig. 1A–D). Immunofluorescence results further revealed the spatial expression patterns of CAPZA2 in coronal brain sections (Fig. 1E). Experimental results indicate that CAPZA2 protein exhibits a distinct prenatal peak on embryonic day 16, with expression levels gradually decreasing thereafter and remaining stable at a lower level postnatally. Further investigation revealed that CAPZA2 is widely expressed in various brain regions of adult mice, including the olfactory bulb, cortex, hippocampus, thalamus, cerebellum, and brainstem. Strong expression was observed in the hippocampus, which is crucial for spatial learning and memory. CAPZA2 expression was also detected in the basolateral amygdala (BLA), a brain region vital for emotion and fear learning18.
A, B Western blot representative bands and quantitative analysis describe the CAPZA2 levels at different developmental stages, from mouse embryos to postnatal stages (Male mice were selectively utilized upon confirmation of reliable sex determination, n = 3). C, D Western blot representative bands and statistical analysis of CAPZA2 in various brain regions of adult mice (5 months old, n = 3 male). E Representative coronal sections of WT mice. CAPZA2 expression in the brain was examined by immunofluorescence in WT mice (5 months old, n = 3 male). Scale bar = 500 μm, blue: DAPI; red: CAPZA2. F Schematic describing strategy to generate CAPZA2+/− and CAPZA2c.G776T/+ mice. G, H Western blot representative bands and quantification of CAPZA2 expression in the hippocampus and PFC of CAPZA2+/− and WT mice. n = 6 (WT/Het, 3 males and 3 females). Lane arrangement note: Blank lanes in Western blot indicate molecular weight marker positions. Uncropped original images are provided in the Supplementary Information. I The mRNA expression level of CAPZA2 in the hippocampus and PFC of CAPZA2+/− and WT mice. n = 6 (WT/Het, 3 males and 3 females). J, K Western blot representative bands and the quantification of CAPZA2 expression in the hippocampus and PFC of CAPZA2c.G776T/+ and WT mice. n = 5 (WT/MUT, 3 males and 2 females). Lane arrangement note: Blank lanes in Western blot indicate molecular weight marker positions. Uncropped original images are provided in the Supplementary Information. L The mRNA expression level of CAPZA2 in the hippocampus and PFC of CAPZA2c.G776T/+ and WT mice. n = 6 (WT/MUT, 3 males and 3 females). M Representative images of immunofluorescence staining for CAPZA2 in the hippocampus and PFC of CAPZA2+/- and WT mice (blue: DAPI; red: CAPZA2). N Representative images of immunofluorescence staining for CAPZA2 in the hippocampus and PFC of CAPZA2c.G776T/+ and WT mice (blue: DAPI; red: CAPZA2). O Significant differences in CAPZA2 fluorescence intensity in the hippocampus and PFC of mice in the CAPZA2+/− and WT groups. n = 6 (WT/Het, 3 males and 3 females). P Significant differences in CAPZA2 fluorescence intensity in the hippocampus and PFC of mice in the CAPZA2c.G776T/+ and WT groups. n = 6 (WT/MUT, 3 males and 3 females). Unpaired t test was performed for statistical analysis. The data represented as mean ± S.E.M; *P < 0.05, **P < 0.01, ***P < 0.001.
Generation and characterization of Gene-edited mice
To understand CAPZA2’s role in vivo, we generated a gene-edited mouse through precise CRISPR/Cas9 gene editing (Fig. 1F, Supplementary Fig. 1A–D)19. Given the lethal outcome observed in homozygous CAPZA2 knockout mice and the heterozygous nature of mutations in affected patients, our study focused on heterozygous variants, ensuring equal representation of males and females within each experimental group. Subsequent validation techniques, including Western blotting, qPCR, and immunofluorescence, confirmed a notable reduction in CAPZA2 expression within the hippocampus and PFC of these mouse models. Compared with the WT littermates, the CAPZA2 protein levels in the hippocampus of CAPZA2+/− mice decreased to approximately 80%, and those in the PFC decreased to ~60% (Fig. 1G–I). Compared with the control group, the levels of CAPZA2 protein in the hippocampus and PFC of CAPZA2c.G776T/+ mice decreased to ~50% and 30%, respectively (Fig. 1J-L). These findings are complemented by diminished mRNA levels and attenuated fluorescence intensity in immunostaining (Fig. 1M–P). The significant reduction in CAPZA2 expression in both CAPZA2+/− and CAPZA2c.G776T/+ model mice fully validates the successful establishment of the two heterozygous models. Despite these molecular variations, both transgenic lines maintained typical phenotypical attributes, such as standard fur pigmentation, normal appetite, and consistent growth patterns. Notably, the CAPZA2c.G776T/+ variant exhibited a significant increase in brain mass (Supplementary Fig. 2A–C).
CAPZA2 gene defects lead to developmental abnormality and social behavioral deficits in mice
Given the clinical symptoms of patients with CAPZA2 mutations, we hypothesize that CAPZA2 deficiency may be associated with anxiety behavior, motor defects, social impairments, and cognitive abnormalities. Therefore, we conducted the OFT, EPM, RRT, MWM, NORT, Y-maze, and three-chamber social interaction tests.
Behavioral impacts of reducing CAPZA2 expression in mice
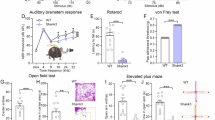
In the OFT, CAPZA2+/− mice exhibited a notable increase in both total and outer zone distance traveled, suggesting abnormally heightened locomotor activity, and potential anxiety-like behavior (Fig. 2A, Supplementary Fig. 3A). Further supporting this observation, in the EPM, CAPZA2+/− tended to spend less time in the open arms than WT mice. Additionally, we recorded the movement distance of the mice in both the open and closed arms. We found that CAPZA2+/− mice covered a relatively shorter distance in the open arms, while traveling a longer distance in the closed arms. These findings provide further evidence of the anxiety-like behavior manifested by CAPZA2+/− mice (Fig. 2B, Supplementary Fig. 3B, C).
A Representative locomotor trajectories and anxiety-like behaviors in WT mice and CAPZA2+/− mice, expressed as distance of movements in the outer region. B Shortened residence time in the open arm of the EPM in CAPZA2+/− mice exhibits behavioral anxiety. C The RRT showed that CAPZA2+/− mice had a reduced residence time on the rotarod indicating a malfunctioning balance. D–G CAPZA2+/− mice showed reduced spatial memory capacity in the MWM, as evidenced by a prolonged latency to reach the platform during training (D), a prolonged latency to find the target location during testing (K), a reduced number of traversals through the target location (F), and a shortened search time in the target quadrant (G). H Representative trajectories and summary plots showing time spent sniffing new and old objects, with significant differences in cognitive indices between CAPZA2+/− and WT mice. I CAPZA2+/− mice had significantly lower rates of spontaneous alternation in the Y-maze, but there was no difference in the total number of arms they traversed. Representative trajectories and summary plots (J) showing time spent sniffing unfamiliar mice and empty cages during the social interaction phase (K) and time spent sniffing different unfamiliar mice during the social preference phase (L). n = 12 (WT/Het, 6 males and 6 females). M Locomotor trajectories and movement distance in the outer zone of WT mice and CAPZA2c.G776T/+ mice. N CAPZA2c.G776T/+ mice have a shortened dwell time in the open arm indicating behavioral anxiety. O The RRT showed that CAPZA2c.G776T/+ mice had a reduced residence time on the rotarod indicating a malfunctioning balance. P Latency to find the hidden platform during the training session of the MWM. CAPZA2c.G776T/+ mice differed significantly from WT mice. CAPZA2c.G776T/+ mice showed reduced spatial memory capacity during the test phase, as evidenced by elevated latency time to find the target location during the test period (Q), a lower number of traversals through the target location (R), and a lower search time in the target quadrant (S). T Representative trajectories and summary plots showing time spent sniffing new and old objects, with significant differences in cognitive indices between CAPZA2c.G776T/+ and WT mice. U CAPZA2c.G776T/+ mice had significantly lower rates of spontaneous alternation in the Y-maze, but there was no difference in the total number of arms they traversed. Representative trajectories (V) and summary plots showing time spent sniffing unfamiliar mice and empty cages during the social interaction phase (W) and time spent sniffing different unfamiliar mice during the social preference phase (X). n = 10 (WT/MUT, 5 males and 5 females). Two-way ANOVA and unpaired t test were performed for statistical analysis. The data represented as mean ± S.E.M; n.s. not significant. *P < 0.05, **P < 0.01, ***P < 0.001.
We conducted the RRT to assess motor balance and coordination. The CAPZA2+/−mice demonstrated significantly shorter latencies to fall from the rotarod, pointing to compromised motor balance compared to their WT counterparts (Fig. 2C).
Cognitive abilities were further evaluated through the MWM, Y-maze, and NORT, focusing on spatial learning, memory, and recognition. In the MWM test, despite following a similar learning curve pattern as WT mice, CAPZA2+/− mice took significantly longer to locate the platform, especially on the fifth day of training, indicating spatial learning difficulties (Fig. 2D). This was corroborated by their performance in a subsequent probe test, where they displayed both longer latencies to reach the target location for the first time and fewer passes over the target, alongside spending less time in the target quadrant (Fig. 2E–G). In the NORT, these mice showed an impaired ability to distinguish between novel and familiar objects, as evidenced by a lower recognition index, highlighting deficits in recognition memory (Fig. 2H). Furthermore, the Y-maze evaluated spatial working memory and revealed decreased spontaneous alternations in CAPZA2+/− mice without affecting arm entry counts, suggesting a specific deficit in this cognitive domain (Fig. 2I). Social behaviors were explored using a three-chambered social interaction test. Unlike WT mice, which showed a preference for the chamber containing another mouse, CAPZA2+/− mice did not display this sociability tendency, nor did they differentiate between familiar and novel mice during the social novelty phase, indicating impairments in social interaction and novelty recognition (Fig. 2J–L).
In summary, our findings paint a comprehensive picture of the behavioral disruptions in CAPZA2+/− mice, spanning from increased anxiety and motor balance issues to marked deficits in spatial learning, memory, and social interaction. These results highlight the critical role of CAPZA2 in maintaining normal cognitive function and behavior.
Neurological phenotypes in CAPZA2c.G776T/+ mice mirror patient clinical symptoms
To elucidate the link between CAPZA2c.G776T/+ mutations and associated neurological phenotypes, we employed CRISPR-Cas9 technology to generate heterozygous mutant mice. These mice underwent behavioral tests, revealing key insights into their neurological profile. In the OFT, CAPZA2c.G776T/+ mice demonstrated hyperactivity compared to WT mice (Supplementary Fig. 4A). A closer examination of their movement patterns showed an increase in the outer zone, a behavior typically associated with heightened anxiety (Fig. 2M). These anxiety-like tendencies were further confirmed by the EPM, where mutant mice entered the open arm for a reduced time (Fig. 2N), with a corresponding significant reduction in the distance traveled in the open arm and a significantly higher distance traveled in the closed arm. (Supplementary Fig. 4B, C). The RRT exposed clear motor balance issues in CAPZA2c.G776T/+ mice, as indicated by their reduced latency to fall (Fig. 2O). This was paralleled by their performance in the MWM, where mutant mice exhibited significantly longer escape latencies from the fourth day of training onwards, underscoring spatial learning challenges (Fig. 2P). Probe tests further revealed deficits in hippocampus-dependent spatial memory, as evidenced by a lack of quadrant preference and reduced platform crossings (Fig. 2Q–S). NORT and Y-maze outcomes indicated recognition and impaired spatial working memory in CAPZA2c.G776T/+ mice (Fig. 2T, U). Additionally, social interaction tests unveiled significant deficits in the CAPZA2c.G776T/+ mice (Fig. 2V–X).
In summary, CAPZA2c.G776T/+ mice recapitulate key neurological phenotypes observed in patients with pronounced deficits in motor control, spatial learning, memory, and social interaction.
Key characteristics of synapses in the brains of CAPZA2+/- mice are altered
Structural impairments in synaptic plasticity in CAPZA2+/- mice
Synaptic plasticity, encompassing both structural and functional facets, is fundamental for the proper execution of cognitive processes20. Aiming to connect pathological defects in CAPZA2+/− mice with potential spine formation or synaptic maturation disruptions, we employed Golgi staining to assess synaptic branching complexity, spine density, and morphology within the hippocampal DG region and PFC. Despite Sholl analysis revealing similar dendritic complexity in the hippocampal DG neurons compared to control mice, we observed a pronounced increase in dendritic spine density of CAPZA2+/− mice (Fig. 3A–E). Further detailed analyses pointed to significant morphological changes in the spine of CAPZA2+/− mice, showing more elongated, thin, and curvilinear structures suggesting immature spine development, which in combination with an increased number of overall dendritic spines, suggests that CAPZA2 deficiency may affect the pruning of dendritic spines in relation to the maturation of their morphology (Fig. 3F). This shift suggests a potential hindrance in synaptic maturation and synapse pruning within the hippocampus. Examination of the PFC revealed a significant reduction in the number of neuronal dendritic branches at varying distances from the soma in CAPZA2+/− mice, hinting at disrupted dendritic architecture (Fig. 3G, H). This was further compounded by a marked decrease in dendritic spine density and the presence of fewer mature spines, underscoring a structural injury (Fig. 3I–K). Notably, we also identified breaks in dendrites and their branches across both the hippocampus and PFC, indicating neuronal damage that could directly impede synaptic transmission efficiency.
A Representative confocal images (Golgi staining) of the pyramidal neuronal dendrites in the hippocampus and images of dendritic skeletonization obtained by imageJ analysis. (Scale bar, 50 μm.) B Sholl profile reporting almost no significant difference in dendritic branching complexity of hippocampal neurons in groups of CAPZA2+/− and WT mice. n = 15 neurons from 5 mice (WT/Het, 3 males and 2 females). C Schematic diagram of different morphology of dendritic spines. They can be categorized as filopodia spine, long-thin spine, thin spine, stubby spine, mushroom spine and branched spine, with two or more heads. Confocal micrographs of hippocampus dendrites from CAPZA2+/- and WT mice (D) and bar graphs with quantification of spine density (E) and spine morphology (F). n = 18 neurons from 6 mice (WT/Het, 3 males and 3 females). (Scale bar, 10 μm.) (G) Representative confocal images of the pyramidal neuronal dendrites in the PFC and images of dendritic skeletonization obtained by imageJ analysis. (Scale bar, 50 μm). H Sholl profile reporting significantly reduced dendritic branching complexity in the PFC of CAPZA2+/- mice. n = 15 neurons from 5 mice. (WT/Het, 3 males and 2 females). Confocal micrographs of PFC dendrites from CAPZA2+/- and WT mice (I) and bar graphs with quantification of spine density (J) and spine morphology (K). n = 18 neurons from 6 mice. (WT/Het, 3 males and 3 females, Scale bar, 10 μm.) L–N PSD95 representative immunoblots in CAPZA2+/− and WT mice, along with quantifying histograms and mRNA expression levels. n = 6 (WT/Het, 3 males and 3 females). Lane arrangement note: Blank lanes in Western blot indicate molecular weight marker positions. Uncropped original images are provided in the Supplementary Information. O–Q GRIN2A representative immunoblots in CAPZA2+/− and WT mice, along with quantifying histograms and mRNA expression levels. n = 6 (WT/Het, 3 males and 3 females). R–T GRIN2B representative immunoblots in CAPZA2+/− and WT mice, along with quantifying histograms and mRNA expression levels. n = 6 (WT/Het, 3 males and 3 females). U–Y GLUR2 and P-GLUR2 representative immunoblots in CAPZA2+/− and WT mice, along with quantifying histograms and mRNA expression levels. n = 6 (WT/Het, 3 males and 3 females). Two-way ANOVA and unpaired t test were performed for statistical analysis. The data represented as mean ± S.E.M; n.s. not significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Dysregulation of synaptic proteins in CAPZA2+/- mice
The integrity of postsynaptic proteins is crucial for synaptic maturation and functionality, with disturbances often linked to neurodevelopmental disorders21,22. To delve into the molecular aspects of the observed synaptic alterations, we assessed the expression of key synaptic markers, including PSD95, Synapsin-1 (Syn-1), Synaptophysin (SYP), GLUR1, GLUR2, GRIN2A, and GRIN2B, across the hippocampal and PFC regions. In the hippocampus of CAPZA2+/− mice, PSD95 protein and mRNA expression notably increased, contrasting with a reduction observed in the PFC (Fig. 3L–N). Levels of Syn-1 and SYP remained consistent with controls in both regions, indicating that the basic function of synaptic vesicles was unaffected. (Supplementary Fig. 5A–E). Notably, PFC expression of NMDA receptor subunits GRIN2A and GRIN2B was significantly impacted, with a reduction in GRIN2A and an elevation in GRIN2B, highlighting a potential shift in receptor composition (Fig. 3O–T). For AMPA receptor subunits GLUR1 and GLUR2, our analysis did not reveal significant differences in GLUR1 levels in both the hippocampus and PFC. However, hippocampal analysis indicated a significant change in GLUR2 and its phosphorylation level, with an increase in GLUR2 and a decrease in its phosphorylated form, pointing to changes in receptor phosphorylation states. (Fig. 3U–Y, Supplementary Fig. 5F–I).
In general, our study provides highly valuable preliminary evidence for the generation of pathological phenotypes: CAPZA2 deficiency may lead to a series of abnormal changes in synaptic plasticity, which are clearly reflected in the alterations of synaptic structures and the abnormalities of synaptic protein compositions.
Impaired neuronal morphology and glial reactivity in CAPZA2 deficiency
After discovering the impairment of synaptic plasticity, we explored the potential link between CAPZA2 deficiency and nerve cell health. We used immunostaining to label Microtubule-associated protein 2 (MAP2), Doublecortin (DCX), Ionized calcium-binding adapter molecule 1 (IBA1), and Glial fibrillary acidic protein (GFAP). MAP2 immunostaining highlighted significant disparities in MAP2 intensity within the hippocampal DG. Notably, CAPZA2+/− mice exhibited marked deviations in neuronal morphology compared to WT. WT neurons exhibited intact neuronal shapes and extensive apical dendrites, while the neurons of CAPZA2+/− mice were characterized by disrupted, fragmented, and disorganized dendritic structures. This morphological disruption indicates severe neuronal disorders due to CAPZA2 insufficiency (Fig. 4A, B). DCX immunostaining, which is indicative of neurogenesis levels, revealed reduced neurogenesis within the hippocampal DG of CAPZA2+/- mice (Fig. 4C, D). The observed decrease in both MAP2 and DCX expression, coupled with disturbed neuronal morphology, underscores the detrimental impact of CAPZA2 deficiency on neuronal structure and regeneration. Furthermore, we noted significant alterations in glial markers: GFAP expression decreased, whereas IBA1 expression increased in the hippocampus of CAPZA2+/− mice. A reversed pattern was observed in the PFC, indicating intricate glial reactivity changes and suggesting a nuanced neuropathological condition accompanying CAPZA2 haploinsufficiency (Fig. 4E–L).
A, B Representative fluorescence micrographs and statistical analysis of MAP2 showing the morphology and quantities of mature neuron in the hippocampus. (Enlarged image: Scale bar, 100 μm; Detailed image: Scale bar, 50 μm). n = 5 (WT/Het, 3 males and 2 females). C, D Representative fluorescence micrographs and statistical plot of DCX showing the morphology and quantities of neurons in the hippocampus. (Scale bar, 100 μm). n = 5 (WT/Het, 3 males and 2 females). E, F Representative fluorescence micrographs and statistical analysis showing the quantities of astrocytes the hippocampus. (Scale bar, 100 μm). n = 5 (WT/Het, 3 males and 2 females). G, H Western blot of GFAP in the hippocampus and PFC of CAPZA2+/− and WT mice with bands and statistical analysis. n = 6 (WT/Het, 3 males and 3 females). I The mRNA expression level of GFAP in the hippocampus and PFC of CAPZA2+/− and WT mice. n = 6 (WT/Het, 3 males and 3 females). J–L Representative fluorescence micrographs and statistical analysis showing the quantities of microglia in the hippocampus and PFC. (Scale bar, 100 μm). n = 5 (WT/Het, 3 males and 2 females). Unpaired t test were performed for statistical analysis. The data represented as mean ± S.E.M; n.s. not significant. *P < 0.05, **P < 0.01, ***P < 0.001.
Pathological similarities in CAPZA2c.G776T/+ and CAPZA2+/− mice highlight synaptic and neuronal disruptions
We next explored the molecular and cellular phenotypes in the CAPZA2 point mutation mice. Golgi staining revealed maintained dendritic complexity yet increased spine density with a tendency towards immature spine morphologies in the hippocampus of CAPZA2c.G776T/+ mice (Fig. 5A–E). This contrasts with the diminished dendritic branching and spine density observed in PFC regions, suggesting impaired synaptic connections and signaling (Fig. 5F–J). Neuronal counts and morphological assessments further confirmed disruptions in the hippocampus of CAPZA2c.G776T/+ mice, coupled with decreased neurogenesis (Fig. 5K–N). The expression patterns of GFAP and IBA1 indicated altered glial activity in the CAPZA2c.G776T/+ mice, reflecting a complex state of neuropathology (Fig. 5O–T). Protein and mRNA levels of synaptic markers such as PSD95, Syn-1, SYP, and GLUR2 highlighted synaptic protein dysregulation in the CAPZA2c.G776T/+ mice, particularly noted by changes in PSD95 and GLUR2 expressions in the hippocampus and altered NMDA receptor subunit levels in the PFC (Fig. 5U–X, Supplementary Fig. 6).
A Representative confocal images of pyramidal neuronal dendrites in the hippocampus and images of dendritic skeletonization obtained by imageJ analysis. (Scale bar, 50 μm). B Sholl profile reporting almost no significant difference in dendritic branching complexity of hippocampal neurons in groups of CAPZA2c.G776T/+ and WT mice. n = 15 neurons from 5 mice (WT/MUT, 3 males and 2 females). Confocal micrographs of hippocampal dendrites from CAPZA2c.G776T/+ and WT mice (C) and bar graphs with quantification of spine density (D) and spine morphology (E). n = 18 neurons from 6 mice (WT/MUT, 3 males and 3 females, Scale bar, 10 μm). F Representative confocal images of pyramidal neuronal dendrites in the PFC and images of dendritic skeletonization obtained by imageJ analysis. (Scale bar, 50 μm). G Sholl profile reporting significantly reduced dendritic branching complexity in the PFC of CAPZA2c.G776T/+ mice. n = 15 neurons from 5 mice. (WT/MUT, 3 males and 2 females). Confocal micrographs of PFC dendrites from CAPZA2c.G776T/+ and WT mice (H) and bar graphs with quantification of spine density (I) and spine morphology (J). n = 18 neurons from 6 mice (WT/MUT, 3 males and 3 females, Scale bar, 10 μm). K, L Representative fluorescence micrographs and statistical analysis of MAP2 showing the morphology and quantities of mature neuron in the hippocampus. (Enlarged image: Scale bar, 100 μm; Detailed image: Scale bar, 50 μm). n = 5 (WT/MUT, 3 males and 2 females). M, N Representative fluorescence micrographs and statistical analysis of DCX showing the morphology and quantities of neurons in the hippocampus. (Scale bar, 100 μm). n = 5 (WT/MUT, 3 males and 2 females. O, P Representative fluorescence micrographs and statistical analysis showing the quantities of astrocytes in the hippocampus. (Scale bar, 100 μm). n = 5 (WT/MUT, 3 males and 2 females). Q Western blot with bands and statistical analysis and the mRNA expression level of GFAP in the hippocampus and PFC of CAPZA2c.G776T/+ and WT mice. n = 6 (WT/MUT, 3 males and 3 females). R–T Representative fluorescence micrographs and statistical plot showing the quantities of microglia in the hippocampus and PFC. (Scale bar, 100 μm). n = 5, (WT/MUT, 3 males and 2 females). U PSD95 representative immunoblots in CAPZA2c.G776T/+ and WT mice, quantifying histograms as well as mRNA expression levels. n = 6 (WT/MUT, 3 males and 3 females). V GRIN2A representative immunoblots in CAPZA2c.G776T/+ and WT mice, quantifying histograms as well as mRNA expression levels. n = 6 (WT/MUT, 3 males and 3 females). W GRIN2B representative immunoblots in CAPZA2c.G776T/+ and WT mice, quantifying histograms as well as mRNA expression levels. n = 6 (WT/MUT, 3 males and 3 females). X GLUR2 and P-GLUR2 representative immunoblots in CAPZA2c.G776T/+ and WT mice, quantifying histograms as well as mRNA expression levels. n = 6 (WT/MUT, 3 males and 3 females). Two-way ANOVA and unpaired t test were performed for statistical analysis. The data represented as mean ± S.E.M; n.s. not significant. *P < 0.05, **P < 0.01, ***P < 0.001.
These findings suggest that the CAPZA2c.G776T/+ mouse model exhibits synaptic and neuronal disruptions akin to those observed in CAPZA2+/- mice, underscoring the critical role of CAPZA2 in neuronal integrity and synaptic networking, with implications for neurodevelopmental and brain disorders.
Transcriptomic insights unveil broad molecular disruptions in CAPZA2 deficiency
In order to establish a basic profile of the cell population, we performed a single-cell transcriptome on the hippocampus of CAPZA2+/− mice (Fig. 6A), which showed that the cell population of CAPZA2+/− mice was altered, with the proportion of excitatory neurons significantly lower than that in WT mice, and the proportion of inhibitory neurons significantly higher than that in WT mice. This suggests that the cellular composition type is skewed, with implications for hippocampal function and structure (Fig. 6B–D). Differential gene expression analysis across brain cell types identified thousands of genes whose altered expression patterns contribute to the observed pathological phenotype (Fig. 6E, F). Pathway enrichment analysis and functional annotation of differentially expressed genes (DEGs) revealed that upregulated pathways included mRNA splicing, mRNA processing, and RNA splicing, etc., suggesting the potential involvement of compensatory molecular mechanisms. (Fig. 6G). Specifically, they are part of the post-transcriptional gene expression regulation, which involves processing and modifying precursor mRNA to generate mature mRNA, further influencing its stability and translation efficiency. This process regulates neuronal development and maturation, including axon and dendrite development, synapse formation, and functional maturation23,24. In contrast, these genes were also involved in key neurodevelopmental and synaptic processes, with significant downregulation in pathways related to axonogenesis, synapse organization, and cognition (Fig. 6H). Subsequently, we validated the differential genes in CAPZA2+/− and CAPZA2c.G776T/+ mice and control mice using qRT-PCR. The results showed a significant downregulation of genes involved in neurotransmitter release, synapse formation, and stability in the hippocampus of the model mice, such as Dgki25, Dlg126, Tiam127, Ctnna228,29, and Nrcam30. Genes associated with synaptic plasticity regulation, mRNA processing, and mRNA stability, including Nrgn31, Rpl832, Ndufa233, and Ubb34, were significantly upregulated (Fig. 6I–L). This further confirms the profound impact of CAPZA2 deficiency on neuronal circuits and functions.
A Single-nucleus transcriptome profiling workflow. B Number of seven cell types detected by transcriptome sequencing. UMAP plot (C) and bar plot (D) showing the proportions of the seven major cell types found in the CAPZA2+/− and WT mice of hippocampus. E Venn diagram showing the 19 coregulated DEGs among all seven cell types. The numbers of DEGs specific to each cell type are also presented. F Volcano diagram showing differentially expressed genes in seven cell types. G, H Gene ontology analysis of the top 10 functional clusters of up-regulated. G and down-regulated (H) differentially expressed genes in the hippocampus of CAPZA2+/− compared to WT mice. I, J Decreased mRNA expression levels of differently expressed genes in the hippocampus in CAPZA2+/− and CAPZA2c.G776T/+ mice compared with WT mice. K, L Increased mRNA expression levels of differently expressed genes in the hippocampus in CAPZA2+/− and CAPZA2c.G776T/+ mice compared with WT mice. n = 6 (WT/Het/MUT, 3 males and 3 females). Unpaired t test was performed for statistical analysis. The data represented as mean ± S.E.M; *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
Developmental disorders, which are characterized by impairments in cognitive, behavioral, and social functions, are highly prevalent worldwide, underscoring the urgent need to investigate their underlying pathological mechanisms. Through clinical reports, we have identified neurodevelopmental disorders associated with mutations in the CAPZA2 gene as a rare condition, with only four clinical cases documented to date. These cases offer preliminary insights into the complex relationship between CAPZA2 mutations and neurological disorders. However, the small sample size and the lack of comprehensive biological studies necessitate further research to better understand the pathogenesis, clinical manifestations, and potential therapeutic strategies for these conditions. To address these gaps, we first established CAPZA2 heterozygous knockout and CAPZA2 heterozygous missense mutant mouse models. The development of these gene-edited mouse models not only allows for the investigation of the direct effects of CAPZA2 gene loss on disease but also replicates the gene defects observed in human patients. This provides a controlled experimental platform to study disease symptoms, progression, pathological features, and gene functions. Importantly, these models are pivotal for translating basic research into clinical applications and advancing therapeutic strategies.
Previous clinical cases have highlighted neurodevelopmental disruptions in patients with CAPZA2 missense mutations. Based on this, we hypothesize that CAPZA2 deficiency may induce intellectual disability-related behavioral and phenotypic abnormalities by interfering with synaptic plasticity. To test this hypothesis, we comprehensively investigated the effects of CAPZA2 abnormalities on neurodevelopment, utilizing behavioral assessments, pathological examinations, and molecular biological analyses. The results show that CAPZA2 heterozygous knockout or missense mutation mice exhibit significant impairments in learning and memory, along with reduced social interactions, increased motor impairments, and anxiety-like behaviors. These behavioral abnormalities are highly consistent with the ID phenotypes observed clinically, further supporting the critical role of CAPZA2 deficiency in disease development. Additionally, pathological changes and structural damage to synapses observed in the model mice provide further evidence of a direct association between CAPZA2 dysfunction and neurodevelopmental disorders. Therefore, our study provides strong evidence for the causal relationship between CAPZA2 mutations and neurodevelopmental disorders.
A key finding emerged when breeding offspring from the model mice: the absence of viable CAPZA2−/− offspring. This result underscores the indispensable role of CAPZA2 in normal developmental processes, indicating that complete knockout of this gene leads to non-viability in living organisms. Our findings corroborate previous studies. Myriam Hemberger et al. screened 103 embryonic lethal and sub-viable mouse knockout lines from the Deciphering the Mechanisms of Developmental Disorders (DMDD) program project for placental phenotype studies. They found that 21 of these lines were sub-viable, with mutant pups accounting for 13% or fewer of all offspring obtained, a proportion significantly lower than the 25% expected from heterozygous crosses. CAPZA2 was among these lines (Supplementary Data 1)35. We further traced the Rare Disease Data Center (RDDC) established by members of the Guangzhou Rare Disease Gene Therapy Alliance, which explicitly states that CAPZA2 homozygous knockout mice exhibit preweaning lethality and display defects in multiple organ systems, including the nervous and immune systems (https://www.informatics.jax.org/allele/MGI:5638561). We next assessed the expression of CAPZA2 in the CAPZA2+/− and CAPZA2c.G776T/+ mice. Compared to WT mice, CAPZA2 expression in the hippocampus and PFC of CAPZA2+/− mice exceeded 50%. This observation led us to hypothesize that, since each cell carries two alleles, the CRISPR/Cas9 system used for heterozygous knockdown has a probabilistic 50% chance of targeting one allele while leaving the other unaffected. This could result in compensatory upregulation of the unaffected allele, potentially mitigating the genetic disruption and explaining the observed knockdown efficiency of over 50%. Further analysis shows that, compared to WT mice, CAPZA2 expression in the hippocampus of CAPZA2c.G776T/+ mice was reduced to approximately 50%, while expression in the PFC was reduced to about 30%. This finding corroborated a study on fruit flies when combined with the manifestation of multiple abnormal behaviors in CAPZA2c.G776T/+ mice. This study reported two children with heterozygous de novo mutations in CAPZA2 (c.776 G > T, p.Arg259Leu and c.766 A > G, p.Lys256Glu) who exhibited neurological symptoms. Using a fruit fly model, it was demonstrated that human CAPZA2 could partially replace the function of the fruit fly cpa gene, but the variants were less efficient in rescuing the lethality of cpa null mutants and displayed a dominant-negative effect in bristle morphogenesis. They indicated that under normal conditions or partial loss of cpa function, the human CAPZA2 did not cause phenotypic abnormalities, whereas the variants led to defects. This suggests that the variants not only impair their own function but also interfere with normal physiological processes, consistent with the characteristics of a dominant-negative effect7. Building on this, we propose that the missense mutation in CAPZA2c.G776T/+ mice may exert a dominant-negative effect. We hypothesize that, in the case of a heterozygous mutation, the mutant subunit may aberrantly occupy critical binding sites on actin filaments that are typically reserved for the wild-type CapZ complex, thereby hindering the proper function of the wild-type complex. This interference may prevent the mutant α-subunit from effectively regulating actin filament growth and, through competition with the wild-type subunit for binding sites, exert a dominant-negative effect on the normal function of the wild-type CapZ complex. This hypothesis is supported by prior studies, reinforcing the potential for a dominant-negative mechanism36,37. Both CAPZA2+/− and CAPZA2c.G776T/+ mice exhibited abnormalities in behavioral activity, anxiety-like behavior, motor ability, social interaction, and cognitive function. Interestingly, the MWM analysis of CAPZA2c.G776T/+ mice revealed a gradual decline in learning efficiency throughout the training, with significant differences from WT mice on the second, fourth, and fifth days. This suggests that the CAPZA2c.G776T/+ mice exhibit substantial learning deficits. Additionally, spatial memory impairments in these mice were pronounced, as evidenced by their performance in both the target and initial quadrants, indicating a widespread impact on spatial memory. Together, these findings underscore the critical role of CAPZA2 in cognitive function and overall organism vitality. Furthermore, the differential expression and impact of CAPZA2 mutations highlight the complexity of gene function regulation, emphasizing the importance of this research in elucidating the mechanisms underlying neurodevelopmental disorders.
Currently, there is limited research on CAPZA2 in the central nervous system (CNS). However, existing studies have demonstrated that CapZ plays a critical role in synaptogenesis, including neurogenesis, synaptic stability, and structural remodeling38,39,40,41. Brain development is accompanied by a cycle of synapse formation and elimination, which is fundamental to learning and memory consolidation42,43. CapZ’s involvement in memory encoding highlights its central role in synaptic plasticity. Research has revealed a close relationship between memory and synaptic adaptability, with dendritic spines playing a key role in regions such as the hippocampus and PFC44,45. Changes in the morphology and density of dendritic spines, that is, dendritic spine remodeling, are indispensable for synaptic - level functional modifications that support cognitive abilities46,47,48,49,50,51. To validate the hypothesized role of CAPZA2 in synaptic plasticity, we conducted studies on two mouse models and reached definitive conclusions. Employing Golgi staining to examine neuronal structures within these mice revealed distinct synaptic alterations in both the PFC and hippocampus. Notably, in the hippocampus, we observed an increased density of dendritic spines and a marked shift in spine morphology, including a decrease in mushroom-type spines and an increase in filamentous protrusions and elongated immature neurons. This suggests that CAPZA2 deficiency may lead to an increase in dendritic spine density in the hippocampus of mice, a change that may be due to enhanced spine formation or reduced synaptic elimination, a biological process known as synaptic pruning52. Conversely, in the PFC, in addition to obvious morphological changes in the spines themselves, both mouse models exhibited reduced dendritic branching and spine density. This pattern indicates a diminished synaptic complexity within the PFC architecture, leading to fewer neural connections between synapses and compromised efficiency in neurotransmission. Thus, our findings not only align with but also extend previous studies, underscoring the role of CapZ and, by extension, CAPZA2 dysfunction—in the pathogenesis of neurological disorders53,54.
The early weeks after birth represent a critical period for brain development, during which the formation, development, maturation, pruning, and regression of neurons and dendritic spines are highly active55,56,57,58,59. We speculate that CAPZA2 deficiency may affect synaptic plasticity, leading to abnormal development of brain neurons and dendritic spines, which in turn causes abnormal accumulation of synaptic structures. Therefore, we examined several proteins related to synaptic composition. The results revealed dysregulated synaptic proteins in CAPZA2-deficient mice, further supporting the notion that synaptic plasticity plays a central role in the behavioral phenotypes resulting from CAPZA2 deficiency. Notably, we observed a significant increase in PSD95 levels in the hippocampus, alongside an elevation in the AMPA receptor subunit GLUR2 in these mouse models. Conversely, there was a notable reduction in PSD95 and the NMDA receptor subunit GRIN2A in the PFC, while GRIN2B levels were significantly increased. These findings imply that CAPZA2 deficiency disrupts synaptic density across different brain regions, affecting the postsynaptic membrane’s composition and its associated receptors. Correlating these molecular findings with histomorphometric observations, we noted an increase in dendritic spine density in the hippocampus, corresponding with the elevated expression of PSD95 and higher levels of GLUR2 receptors. Combining the abnormal changes in glial cells observed through molecular detection and single-cell sequencing, we speculate that this process may be linked to insufficient microglia-mediated synaptic pruning. A study on Fmr1 knockout mice suggests that, under normal conditions, microglia mediate synaptic pruning, maintaining the homeostatic balance of the nervous system by identifying and phagocytosing excess dendritic spines. However, in Fmr1 knockout mice, the pruning function is impaired, accompanied by abnormalities such as abnormal ubiquitination of PSD95 and an increase in dendritic spine density. As a result, the normal development and functional maintenance of neuronal synapses are disrupted60. In contrast, the PFC exhibited a decrease in neuronal dendritic complexity, and synaptic spine density, aligning with the observed reduction in PSD95 and GRIN2A. This process may affect the signaling of the postsynaptic membrane and the normal network connectivity of neurons61. These results indicate that CAPZA2 deficiency can affect the synaptic density and receptor composition in different brain regions, thereby influencing synaptic structural plasticity. This finding holds profound significance for understanding the underlying mechanisms of CAPZA2-related neurodevelopmental and behavioral phenotypes.
In conclusion, our study indicates that CAPZA2 deficiency directly leads to neurodevelopmental disorders, manifesting as pathological phenotypes such as behavioral anxiety, motor imbalance, social impairments, and cognitive abnormalities. The mechanisms underlying the defects induced by the two mutations seem to differ. CapZ is a heterodimeric complex composed of α and β subunits encoded by CAPZA and CAPZB, respectively. These subunits tightly bind to form a stable complex that effectively interacts with the barbed ends of actin filaments to regulate their function. We propose that the CAPZA2c.G776T/+ mutation may exhibit a dominant-negative effect, leading to structural changes in the CAPZA2 subunit and disrupting the interaction between the α and β subunits. This would prevent the normal assembly of the CapZ complex, ultimately affecting the intracellular transport of normal proteins and interfering with the function and expression levels of wild-type proteins. Additionally, this mutation may demonstrate a dominant-negative effect by occupying critical binding sites on actin filaments or by inducing the formation of abnormal actin filament structures. On the other hand, the CAPZA2+/- variant may affect gene transcription or mRNA stability due to the direct reduction in gene dosage, leading to insufficient formation of functional CapZ complexes. However, the commonality is that the pathogenic processes of both mutants may involve mechanisms such as neuronal damage, glial cell expression dysregulation, and synaptic plasticity dysfunction. Specifically, CAPZA2 insufficiency causes abnormalities in brain neurons and is sufficient to induce corresponding consequences at the synaptic level. The distinct abnormal manifestations observed in the hippocampus and PFC further confirm that synaptic plasticity regulation is a complex process, which can differentially affect presynaptic structures, postsynaptic structures, and subsets of synapses.
In summary, our study has successfully established CAPZA2 heterozygous knockout and point mutation mouse models; it will help us to further understand the pathogenesis of neurodevelopmental disorders caused by CAPZA2. Therefore, we have not only replicated the developmental disorder phenotypes of clinical patients in the mouse model also identified multiple abnormal indicators through meticulous pathological exams, linking CAPZA2 deficiency to behavioral and cognitive phenotypes. This highlights the gene’s impact on synaptic plasticity and reveals a direct association between CAPZA2 gene mutations and neurodevelopmental disorders, providing a molecular-level exploration of the series of clinical symptoms triggered by these gene mutations.
In addition, based on the current development trends in the field of gene therapy and our understanding of the pathogenic mechanism of CAPZA2, we are optimistic about the potential of using adeno-associated virus (AAV) vectors for gene therapy. AAV vectors, known for their high gene transfer efficiency, low immunogenicity, and good safety profile, have become the preferred tool in the field of gene therapy. In clinical trials, AAV has been extensively used to treat various genetic disorders including but not limited to hemophilia62, retinal diseases63,64, and neuromuscular diseases65,66. For instance, in the study of retinal degenerative diseases, the P23H mutation in the RHO gene is associated with a dominant-negative effect67, researchers used AAV to deliver the wild-type rhodopsin gene to treat the P23H mutation in the rhodopsin gene. They found that in the P23H transgenic mouse model, the expression of rhodopsin increased and retinal function improved to some extent after the successful delivery of the RHO301 gene via AAV5. This suggests that exogenous normal gene introduction can alleviate retinal functional defects caused by the mutation in this disease model. Although overexpression of the gene in WT mice has negative effects, it showed positive therapeutic effects in the mutant mouse model, providing strong evidence for the application of gene therapy in genetic diseases. The research by Hugo J. Bellen and colleagues also demonstrated that overexpression of the reference CAPZA2 in fruit fly cpa null mutants can rescue lethality7. We hypothesize that after the normal CAPZA2 gene is delivered into cells via AAV, the expressed protein may preferentially participate in the assembly of normal protein complexes or signaling pathways due to its correct structure and function. Thus, delivering the normal CAPZA2 gene to patients via AAV vectors may effectively compensate for or restore functional deficits caused by gene mutations. This approach could potentially improve patients’ clinical symptoms and, in some cases, even lead to a cure for the disease.
Our current study has several limitations. In this study, we utilized mouse models with different genetic backgrounds: CAPZA2+/- was constructed on the C57BL/6 N (B6N) strain, while CAPZA2c.G776T/+ was constructed on the C57BL/6 J (B6J) strain. Although both strains are commonly used inbred lines, research has shown that genetic background may significantly influence disease phenotypes. Jennifer A. Kearney et al. found that, compared to B6J, the Dravet syndrome model mice on the B6N background exhibited milder seizure severity and higher survival rates, suggesting that genetic background differences may potentially impact experimental outcomes68. In the future, we will unify the genetic background to reduce experimental bias.
As a capping protein, CAPZA2 primarily regulates the dynamic stability of microtubules and microfilaments. It binds to actin to control dendritic spine formation and synaptic structural stability, which are crucial for neuronal synaptic connections and synaptic plasticity. Our research mainly explored that CAPZA2 deficiency can lead to abnormal behaviors and molecular pathological phenotypes. These abnormalities are closely related to neuronal damage, dysregulation of glial cell expression, disruption of synaptic structural plasticity, and imbalance in the composition of synaptic proteins. However, we did not find direct evidence for a deeper understanding of the functional role of the CAPZA2 protein and its mechanism of action on synaptic plasticity. Future studies should further explore the specific role of CAPZA2 in regulating synaptic plasticity, such as increasing functional assessments of synapses to clarify the impact of CAPZA2 deficiency on synaptic transmission efficiency and neuronal communication. It is essential to analyze the regulatory networks underlying the observed phenotypic changes. Additionally, the mediating role of microglia in this process should be further investigated to better understand the impact of synaptic pruning imbalance on neurodevelopment. Continuing to explore how to restore CAPZA2 function through pharmacological interventions, gene therapy, or by modulating other molecular mechanisms related to synaptic plasticity to alleviate symptoms remains a crucial direction for future research.
Materials and methods
Animal preparation
All animal experiments in this study were conducted in accordance with the National Institutes of Health Laboratory Animal Care and Use Guidelines (NIH, 2011) and approved by the Animal Care and Use Committee of Minzu University of China. We have complied with all relevant ethical regulations for animal use. The animals were raised at 23 ± 1 °C and 50 ± 1% relative humidity under a 12 h light/dark cycle (lights on from 8 a.m. to 8 p.m.) and provided ad libitum access to a standard diet and drinking water. For Western blot, qPCR and snRNA-seq experiments, mice were anesthetized with 3% isoflurane, followed immediately by transcardial perfusion (left ventricular perfusion with ice-cold PBS at 4 °C, ~30 mL). The entire perfusion process was completed within ~1 min. Brain tissue was then rapidly dissected, flash-frozen in liquid nitrogen, and stored at −80 °C, ensuring that the entire procedure from perfusion to freezing was completed within 3 min. For Golgi staining, mice were deeply anesthetized with 3% isoflurane and euthanized via 5% isoflurane overdose in accordance with AVMA guidelines. Death was confirmed by dual verification including ≥1 min of respiratory arrest and cessation of cardiac activity by palpation. Brains were harvested within 2 min post-euthanasia and immediately immersion-fixed in Golgi staining solution. No signs of distress or pain responses were observed throughout the procedure.
All CAPZA2+/− mouse lines (MGI:7792458, Cyagen Biosciences allele designation: Capza2em1Cya) were generated in C57BL/6NCya background (strain code: C001072). CAPZA2 is located on the 6th chromosome of mice (Chr6: 17637009-17666971 bp, GRCm39 genome assembly). Using CRISPR/Cas9 technology, sgRNAs were designed to generate CAPZA2+/− mice through high-throughput electroporation of fertilized eggs. Sperm samples were collected and cryopreserved from these mice after sexual maturity. The CAPZA2+/− mice, where the CAPZA2 gene has been deleted from exon 3 to exon 6 (exon 3 - exon 6, based on transcript NM_007604.3/ENSMUST00000015877.13), were created by introducing two sgRNAs (gRNA-1: ATGTGCCTTAAACGGTGCAATGG, gRNA-2: ACCAGTTTAGGCTCTAACCAGGG) with Cas9 to induce the repair of the resulting two double-strand breaks (DSBs) by non-homologous end joining (NHEJ), resulting in a deletion of the intervening DNA sequence. Briefly, two sgRNAs were designed to target the region upstream of exon 3 or downstream of exon 6 of CAPZA2. All CAPZA2+/− mice used in the experiments were from heterozygous breeder pairs. Cyagen Biotech produced the F1 generation of CAPZA2+/− mice. For all behavioral and biological experiments in the C57BL/6NCya background, F1 CAPZA2+/− mice were crossed with wild-type (WT) mice to generate offspring of CAPZA2+/− heterozygotes and littermate control WT mice. The genetic lineage was confirmed by genotyping PCR of mouse tail DNA using the primers F1 (TGTGGTGTGGCTTGTTCTTAGTT) and R1 (CTCACAAGGGAAGTCAGTCTGTAAA) for the knockout allele (795 base pairs).
All CAPZA2c.G776T/+ mouse lines (MGI:106222, Shanghai Model Organisms Center, Inc. allele designation: Capza2em1(R259L)Smoc) were generated in C57BL/6Smoc background (strain code: SM-001). The CAPZA2c.G776T/+ mice were generated using CRISPR/Cas9 technology and homology-directed repair to introduce the desired point mutation. Cas9 mRNA and guide RNA (gRNA) were obtained through in vitro transcription, and oligo donor DNA was synthesized. Cas9 mRNA, gRNA, and donor DNA were microinjected into C57BL/6Smoc mice fertilized eggs to generate F0 mice. The F0 mice identified as positive by PCR amplification and sequencing were mated with C57BL/6Smoc mice to obtain positive, stable, and heritable F1 mice. All CAPZA2c.G776T/+ mice used in the experiments were from heterozygous breeder pairs. For all behavioral and biological experiments in the C57BL/6Smoc background, F1 CAPZA2c.G776T/+ heterozygous mice were bred with WT mice to generate offspring of CAPZA2c.G776T/+ heterozygotes and littermate control WT mice. Genotyping PCR and sequencing were performed on mouse tail DNA samples to confirm the presence of the mutation. The genomic DNA was extracted from the mouse tail, and PCR amplification was carried out using primers (CAPZA2 - MUT - F1: CCATCTGTCTTTGCCTCTGC; CAPZA2 - MUT - R1: GGGATCACCTCTTCATTGTA). The PCR products were separated and detected by 1% agarose gel electrophoresis. Subsequently, the PCR products were sequenced (CAPZA2 - MUT: CCATCTGTCTTTGCCTCTGC), and the genotype of the mouse was determined based on the peak pattern at the 776th base site in the sequencing result (the peak pattern at the mutation site of the heterozygous mouse is a double peak, and that of the WT mouse is a single peak).
Real-time quantitative PCR
Total RNA from the mouse PFC and hippocampus was extracted using trizol reagent (Thermo Fisher, MA, USA, 15596018CN). The concentration and purity of the total RNA were assessed using SpectraMax QuickDrop (Molecular Devices, USA). A StarScript RT Mix kit (GenStar, Beijing, China, A230) synthesized the first-strand cDNA. The expression levels of the target genes and the endogenous control gene (GAPDH) were analyzed by real-time quantitative PCR using 2×RealStar Fast SYBR qPCR Mix (GenStar, Beijing, China, A301) on a Light Cycler 96. Finally, the data were transformed using the ∆∆CT method with GAPDH as the reference gene and normalized to the wild-type samples for comparison. The primer sequences are available in the Supplementary Table 1.
Western blotting
Mouse PFC (20 mg) and hippocampal (15 mg) were collected and homogenized with RIPA lysis buffer (Solarbio, Beijing, China, R0020) and protease/phosphatase inhibitors (Bimake, Texas, USA, B14001, B15001). Total protein concentration was measured using the BCA Protein Assay Kit (Beyotime, Shanghai, China, P0009). 30 µg of total proteins were separated by SDS-PAGE and transferred to a 0.45 μm polyvinylidene fluoride membrane (ThermoFisher Scientific). Following membrane blocking, samples were incubated overnight with primary antibodies at 4 °C. The membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG (H + L) (1:5000, Zhongshan Jin Qiao, Beijing, China, catalog no. ZB-2301) or HRP-conjugated goat anti-mouse IgG (H + L) (1:5000, Zhongshan Jin Qiao, Beijing, China, catalog no. ZB-2305) for 1 h at room temperature. Protein signals were visualized and quantified using the BeyoECL Star Kit (Beyotime, Shanghai, China, P0018AS) for chemiluminescence detection (Tanon 4200, Shanghai, China). The following are the detailed information of the antibodies we used: anti-CAPZA2 antibody (1:1000, Abcam, Cambridge, UK, Ab175378), anti-β-Tubulin antibody (1:1000, Jinpulai, Beijing, China, P01L07), anti-GAPDH antibody (1:1000, Cell Signaling, Massachusetts, USA, #5174), anti-Synapsin-1 antibody (1:1000, Cell Signaling, Massachusetts, USA, 5297S), anti-Synaptophysin antibody (1:1000, Cell Signaling, Massachusetts, USA, 36406S), anti-PSD95 antibody (1:1000, Cell Signaling, Massachusetts, USA, 3409S), anti- GFAP antibody (1:1000, Beyotime, shanghai, China, AF1177), anti-GLUR1 antibody (1:1000, BOSTER, Wuhai, China, PB9204), anti-Phospho GLUR1(S845) antibody (1:1000, BOSTER, Wuhai, China, BM4637), anti-GLUR2 antibody (1:1000, Abclonal, Wuhai, China, A0111), anti-Phospho GLUR2(S880) antibody (1:1000, Abclonal, Wuhai, China, AP0257), anti-GRIN2B antibody (1:1,000, Beyotime, shanghai, China, AF7029), anti-GRIN2A antibody (1:1,000, Beyotime, shanghai, China, AF7026).
Immunohistochemistry
Mice were deeply anesthetized with 3% isoflurane and immediately subjected to cardiac perfusion via left ventricular puncture using pre-cooled PBS (~30 mL). The entire perfusion procedure was rigorously limited to 1 min. Within 2 min post-perfusion, brain tissues were rapidly dissected and fixed overnight in 4% paraformaldehyde (PFA) at 4 °C. Subsequent dehydration was performed sequentially: once in 20% sucrose solution and twice in 30% sucrose solution. Whole-brain tissue was embedded in OCT and sectioned (30 μm thickness) using a freezing microtome (Leica CM3050S). Sections were then incubated overnight at 4 °C in primary antibody solution. The next day, the sections were incubated with secondary antibodies for 1 h at room temperature: Alexa Fluor 488-conjugated rabbit anti-goat IgG (H + L) (1:500, Zhongshan Jin Qiao, Beijing, China, catalog no. ZF-0514). Sections were mounted onto glass slides (Citotest, Jiangsu, Beijing) using fluorescent mounting medium (containing DAPI, Zhongshan Jin Qiao, Beijing, China, ZLI-9557). Images were captured with a Leica TCS SP8 confocal microscope (Leica, Solms, Germany). The following are the detailed information of the antibodies we used: anti-CAPZA2 antibody (1:200, Abcam, Cambridge, UK, Ab175378), anti-IBA1 antibody (1:100, Beyotime, shanghai, China, catalog no. AF7143), anti-GFAP antibody (1:100, Beyotime, shanghai, China, catalog no. AF1177), anti-MAP2 antibody (1:50, Cell Signaling, Massachusetts, USA, #4542), and anti-DCX antibody (1:800, Cell Signaling, Massachusetts, USA, #4604).
Behavioral tests
Mice used in the behavioral tests were all three months old, with an equal number of males and females in each group. The experimenters should familiarize themselves with the mice in advance to prevent stress reactions caused by handling before the experiment. Before the start of the experiment, the mice should be placed in the behavioral testing room for at least one hour for acclimatization. The tests are conducted in a randomized order. After each test, 75% ethanol is used to clean away any odors.
Open field test
The open field test (OFT) assessed the experimental animals’ basic motor abilities and mental state by recording their free exploration behavior in an open environment. The device for the OFT (Taimeng, Chengdu, China) is a 50 × 50 × 40 cm large iron box with a square bottom and an upward opening. Rodents naturally tend to explore a new environment, and the total distance traveled by mice in the arena reflects their spontaneous activity level. At the beginning of the experiment, the mice are placed in the arena for 1 min for adaptation, followed by a 5-min period of free exploration. The software records the movement trajectory and automatically generates information such as time and distance.
Elevated plus maze
The elevated plus maze (EPM) is one of the most used rodent tests for assessing anxiety-like behavior. The apparatus comprises two opposing open arms (50 × 10 cm) and two opposing wall-enclosed arms (50 × 10 × 40 cm). The maze is placed on a 50 cm high pedestal (Taimeng, Chengdu, China). Each mouse was placed on the central platform facing one of the open arms. An observer blinded to the animal groups recorded the time spent in each arm in 8 min.
Morris water maze
The morris water maze (MWM) is a classical behavioral test used to evaluate spatial learning and memory abilities of mice. Approach-avoidance behavior is innate in animals, and this experiment stimulates the survival instinct of mice in adverse environments to help them find a safe environment in the shortest possible time. The water maze consists of a circular pool (diameter: 120 cm) and a circular platform (diameter: 10 cm) (Taimeng, Chengdu, China). During the experiment, the water temperature is kept constant (18–22 °C), and the mouse must face the pool wall during testing while the position of the safe platform remains unchanged. The experiment comprises two stages: place navigation and spatial exploration. From the first to the fifth day of place navigation, white pigment is added to the water to make the water surface opaque, and the safe platform is located 1 cm below the water surface. The experimenter places the mouse in each quadrant in turn and records the time for 60 s. If the mouse finds the safe platform within 60 s, the software stops tracking and allows the mouse to stay on the platform for 10 s. If the mouse still hasn’t found the platform in 60 s, the experimenter guides it to the platform and lets it stay there for 30 s. Ensure that each mouse receives training in all four quadrants per day, record the time required for each mouse to reach the platform during each training, and take the average of the four training times as the data for that day. On the sixth day, the spatial exploration test period begins. Before the experiment, the target safe platform is removed, and the mouse is placed in the opposite quadrant of the target platform. Indicators are recorded reflecting their learning and memory abilities, such as the time spent in each quadrant and the number of platform crossings within 60 s.
Rotarod
The rotarod test (RRT) is used to evaluate mice’s motor coordination and balance ability. This test requires a 3-day training period, during which the rotarod rotates at a uniform speed of 10 rpm/min. Each mouse was trained thrice daily, and the mice were allowed to move on the rotating rod for more than 90 s. After the three-day training period, testing begins. During the test, the rotarod accelerates from 4 rpm/min to 40 rpm/min within 5 min, and the time that the mouse stays on the rod is recorded. Each mouse is tested three times, with a 1-h interval between each test. The average of the three test results is taken as the final time.
Novel object recognition task
Novel object recognition task (NORT) is an experiment used to detect the visual-spatial memory ability of animals. The detection principle uses the natural tendency of mice to explore new things. Two different objects are placed in the environment during the experiment (Taimeng, Chengdu, China), and whether there is a clear preference for the novel object is observed. The experiment process is divided into two stages: training and testing. During the training stage, two cone-shaped objects, A and B, are placed at the left and right ends of one side of the wall. The mouse is placed in the box with its back facing the two objects, and the distance between the tip of the mouse’s nose and the two objects must be consistent. The mouse then undergoes 10 min of free exploration during the training period. After the 10-min training period, the mouse is immediately returned to the breeding cage and given a 1-h rest before the testing phase begins. During the testing phase, object B in the box is replaced with a differently shaped and colored ball, C. The mouse is once again placed in the box and allowed to explore for 10 min freely, and the time that the mouse spends exploring or touching the two objects is recorded.
Y-maze test
The Y-maze test is used to test the spatial logical memory ability of experimental animals. The device defines its three arms as A, B, and C arms (arm length: 30 cm, arm width: 6 cm, height of the wall: 15 cm). The detection principle uses the natural tendency of mice to explore new environments. It tests the mouse’s working memory capacity in the hippocampus through the limitation of narrow space and three arms (Taimeng, Chengdu, China). At the beginning of the experiment, the mouse is placed at the end of one arm, facing away from the central zone, and allowed to explore for 8 min freely. The total number of times the mouse enters each arm and the order in which it enters the three arms are recorded. If the mouse enters three consecutive arms without repeating, it is counted as one correct spontaneous alternation behavior (SAPs). The proportion of the total number of spontaneous alternation behaviors to the total number of entries reflects whether the mouse’s spatial exploration has logical memory.
Three-chamber social interaction test
The three-chamber social test box is a rectangular device (60 × 40 × 22 cm) made of transparent polycarbonate divided into three chambers, with each side chamber connected to the center chamber through a movable door (RWD Life Science, Shenzhen, China). The experiment consists of three stages: adaptation, phase 1 (social interaction), and phase 2 (social preference).
In the adaptation stage, the mouse is placed in the center chamber and allowed to explore the whole apparatus for 5 min freely. In phase 1, a stranger mouse S1 that is age, sex, and strain-matched is enclosed in a resin-made small cage as a social stimulus and placed in one of the side chambers, while the other side chamber is kept empty but has a similar cage. The experimental subject is placed in the center chamber and allowed to explore for 10 min freely. In phase 2, another stranger mouse S2 that is age, sex, and strain-matched is placed in an empty cage as a new social stimulus and the mouse is again placed in the center chamber for 10 min of free activity. Each mouse undergoes the entire experiment for 25 min, with software automatically recording the mouse’s movement trajectory and the time spent sniffing the experimental mice.
Golgi staining and morphological analysis of dendrites and dendritic spines
To assess the impact of CAPZA2 on dendrite and dendritic spine formation in the mouse brain, we performed Golgi staining using the FD Rapid GolgiStain Kit (FD Neurotechnologies, Ellicott City, MD, USA, PK401) as described previously69. Briefly, mice were deeply anesthetized with 3% isoflurane and euthanized via 5% isoflurane overdose. Euthanasia confirmation was established by ≥1 min respiratory arrest and cessation of cardiac activity via thoracic palpation. Without perfusion, brain tissues were dissected within 2 min post-euthanasia and immediately immersed in pre-mixed (24 h) Golgi staining solution A/B. After 24 h, the solution was replaced with fresh A/B mixture, followed by incubation at room temperature in the dark for 14 days. Subsequently, the solution was changed to reagent C, refreshed the next day, and the tissues were further incubated at room temperature in the dark for 72 h. Subsequently, the brain tissue was sectioned into 100 µm thick slices. These sections were mounted on gelatin-coated glass slides and left to dry naturally for 1 day. The Golgi staining reaction was performed according to the manufacturer’s instructions. Dendritic images were captured using a Leica fluorescence microscope (Leica TCS SP8 STED 3X, Germany) with a 20X objective under brightfield microscopy. Dendritic spine images were captured using a 63X oil immersion lens under brightfield microscopy. Dendrite and dendritic spine analysis followed the FD Rapid Golgi Stain™ Kit protocol. Briefly, dendrites were quantified using ImageJ and the NeuronJ plugin to calculate branch point numbers and dendrite lengths. Sholl analysis was used to assess dendritic morphology, particularly the branching distribution of dendrites at different distances, to analyze dendritic complexity further. Spine density was calculated by counting the number of spines along every 1 μm segment of the dendrites. This procedure was repeated for each neuron within the selected images.
snRNA-Seq analysis
Single-nucleus RNA sequencing (snRNA-seq) analysis was performed using an improved nuclear isolation method, successfully extracting nuclei from mouse hippocampal tissue70. Using optimized reagents and techniques ensured the isolated nuclei’s high purity, and the single-nucleus suspension’s purity was assessed using flow cytometry. The libraries for single-nucleus RNA sequencing were constructed using the Chromium Single Cell 3′ Library Kit v3, ensuring adequate raw data. The data processing involved cell-level quality control and filtering, generating high-quality data for further analysis. To explore cellular differentiation and development processes, we applied pseudo-time analysis, utilizing Monocle2 for trajectory analysis, followed by Gene Ontology (GO) and pathway analyses to reveal the biological processes and potential regulatory mechanisms underlying gene expression in the mouse hippocampus. Further details of the experimental procedures and data analysis methods are as follows.
Nucleus isolation
A modified protocol was employed to isolate nuclei from mouse hippocampal tissues. Frozen hippocampal tissues were placed directly into a prechilled Dounce homogenizer containing an ice-cold homogenization buffer. The buffer consisted of specific components such as 0.25 M sucrose, 25 mM KCI, 5 mM MgCl2, 20 mM tricine-KOH (pH 7.8), 1 mM dithiothreitol, 0.15 mM spermine, 0.5 mM spermidine, protease inhibitors, 5 μg/mL actinomycin, 0.32% Nonidet P-40, and 0.04% bovine serum albumin. After 25 strokes with a loose pestle, the homogenate was mixed 1:1 in OptiPrep and centrifuged at 10,000 × g for 20 min at 4 °C. The separated nuclei, which pelleted at the bottom of the centrifuge tube, were collected, washed once to remove OptiPrep, and resuspended in Dulbecco’s modified Eagle medium/F12 supplemented with 10% fetal bovine serum. The nuclei were diluted to an appropriate concentration, ensuring the dilution was accurate by counting with a hemocytometer. The purity of the single - nucleus suspensions was assessed by flow cytometry to obtain high - purity samples. All buffers and gradient solutions for nuclei extraction contained 60 U/mL RNAsin (Promega) to protect RNA integrity.
Single-Nuclei RNA sequencing
The single-nuclei RNA sequencing (snRNA-seq) libraries were generated using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1 (1000121; 10× Genomics), following the manufacturer’s instructions. For library construction, 40 μL diluted nucleus suspension (with a specific concentration of nuclei per μL, e.g., 400 nuclei per μL) was mixed with reverse-transcription reagent mix and loaded into a chip for single-cell encapsulation. Subsequently, the encapsulated nuclei were immediately incubated on a thermocycler to enable reverse transcription of the RNA and generate barcoded cDNA. The cDNA was then used for library construction as per the kit instructions. The concentrations of the final libraries were determined by Qubit (Thermo Fisher Scientific), and the fragment lengths were measured by a Fragment Analyzer (Advanced Analytical Technologies). The libraries were subjected to paired-end sequencing on a NovaSeq 6000 system (Novogene), aiming to obtain a sufficient amount of raw data per library, for example, at least 100 GB of raw data.
Statistical analysis of snRNA-Seq
Preprocessing and quality control were crucial steps. Since snRNA-seq libraries capture reads from both unspliced pre - messenger RNAs (mRNAs) and mature mRNAs, a pre-mRNA reference genome was first generated according to the instructions provided by 10× Genomics. The demultiplexed FASTQ files were then aligned to the GRCm38/mm10 pre-mRNA reference genome using Cell Ranger (version 7.1.0) with default settings. After alignment, the default quality control settings in Cell Ranger were used as an initial step to retain only barcodes with unique molecular identifier counts within the top 10% of the 99th percentile of unique molecular identifier values among all barcodes and sort them into cell-associated matrixes. The remaining barcodes were excluded as background. For the second round of quality control in Seurat (version 5.0.3), the distributions of the numbers of genes, numbers of unique molecular identifier counts, and percentages of mitochondrial genes for each sample were controlled. Nuclei with ≤200 genes, ≥20,000 unique molecular identifiers, or ≥5% mitochondrial genes were filtered out to exclude potential dead cells and cell debris. The final filtered matrix was obtained for further analysis.
Pseudo-time analysis
We utilized Monocle2 (http://cole-trapnell-lab.github.io/monocle-release) with DDR-Tree and default parameters for trajectory analysis. Before conducting the Monocle analysis, we carefully selected the marker genes from the Seurat clustering results and the raw expression counts of the filtered cells. Based on this pseudo-time analysis, we further applied branch expression analysis modeling (BEAM Analysis) to analyze the genes that determine branch fate. This approach allowed us to explore the potential developmental trajectories and differentiation processes of the cells in the mouse hippocampus, providing valuable insights into the dynamic changes of gene expression during cell development or in response to the experimental treatment.
Gene ontology analysis
The genes identified from the snRNA - seq data were functionally annotated using Gene Ontology (GO) analysis. The analysis was conducted through online platforms or specific software tools related to GO analysis (geneontology.org). The genes were classified into different GO categories such as biological process, molecular function, and cellular component. This allowed for a comprehensive understanding of the potential functions of the genes expressed in the mouse hippocampal tissues and provided insights into the biological processes and molecular mechanisms involved in the normal physiology or potential pathological conditions of the hippocampus.
Pathway analysis
Pathway analysis was also performed to explore the biological pathways associated with the genes in the mouse hippocampal tissues. Ingenuity pathway analysis (QIAGEN) and STRING analysis (https://string.db.org) were utilized. By mapping the genes to known pathways, the interactions and relationships among the genes were investigated. This analysis helped in identifying the key pathways that were active in the hippocampal cells and understanding how these pathways might be involved in maintaining the normal functions of the hippocampus or potentially contributing to any observed changes or dysregulations in the tissue.
Ethics and inclusion statement
This study did not involve clinical research or patient data. All animal experiments were performed in strict compliance with the Guide for the Care and Use of Laboratory Animals (NIH, 2011) and reported following the ARRIVE 2.0 guidelines (PLoS Biol 18:e3000411). The experimental protocols were approved by the Institutional Animal Care and Use Committee of Minzu University of China (Approval No. ECMUC2022010AA). Euthanasia was conducted in accordance with the AVMA Guidelines for the Euthanasia of Animals (2020). We have complied with all relevant ethical regulations for animal use.
Statistics and reproducibility
All data analyses were performed with GraphPad Prism 8.0 software. Biochemical experiments were measured using optical density using ImageJ 1.54 g. Results were expressed as the mean ± SEM (standard error of the mean), with the biological replicate number (n) indicated in each figure legend. Each experiment was performed in triplicate. The normal distribution test (D ‘Agostino-Pearson omnibus normality test) and the homogeneous variance test (F test) were conducted to ensure that the subsequent tests could be completed using the parameter test method. According to the number of factors and whether the pairing was reasonable, the t-test or analysis of variance (ANOVA) method was selected for statistical analysis. Specifically, the statistical significance between the two groups was assessed using an unpaired two-tailed independent samples t-test. Two-way ANOVA with Tukey’s multiple comparisons test was applied for multiple factor comparisons. P-values less than 0.05 (P ≤ 0.05) were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the article and its Supplementary Information files. The source data for figures are provided in Supplementary Data 2-3. The raw single-cell RNA sequencing data have been deposited in the GEO database under accession number GSE295653. The data supporting this study’s findings are available from the corresponding author upon reasonable request.
References
Maia, N., Nabais Sá, M. J., Melo-Pires, M., de Brouwer, A. P. M. & Jorge, P. Intellectual disability genomics: current state, pitfalls and future challenges. BMC Genom. 22, 909 (2021).
Jia, X. et al. Importance of glutamine in synaptic vesicles revealed by functional studies of SLC6A17 and its mutations pathogenic for intellectual disability. eLife 12, https://doi.org/10.7554/eLife.86972 (2023).
Maulik, P. K., Mascarenhas, M. N., Mathers, C. D., Dua, T. & Saxena, S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res. Dev. Disabil. 32, 419–436 (2011).
Strømme, P. & Valvatne, K. Mental retardation in Norway: prevalence and sub-classification in a cohort of 30037 children born between 1980 and 1985. Acta Paediatr. 87, 291–296 (1998).
Oliveira, L. F. et al. Etiology of intellectual disability in individuals from special education schools in the south of Brazil. BMC Pediatr. 20, 506 (2020).
Rauch, A. et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am. J. Med. Genet. 140, 2063–2074 (2006).
Huang, Y. et al. Variants in CAPZA2, a member of an F-actin capping complex, cause intellectual disability and developmental delay. Hum. Mol. Genet. 29, 1537–1546 (2020).
Pi, S., Mao, X., Long, H. & Wang, H. A de novo inframe deletion variant in CAPZA2 tentacle domain with global developmental delay and secondary microcephaly. Clin. Genet. 102, 355–356 (2022).
Zhang, X. M. et al. Heterozygous CAPZA2 mutations cause global developmental delay, hypotonia with epilepsy: a case report and the literature review. J. Hum. Genet. 69, 197–203 (2024).
Hart, M. C., Korshunova, Y. O. & Cooper, J. A. Mapping of the mouse actin capping protein alpha subunit genes and pseudogenes. Genomics 39, 264–270 (1997).
Mejillano, M. R. et al. Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end. Cell 118, 363–373 (2004).
Jo, Y. J., Jang, W. I., Namgoong, S. & Kim, N. H. Actin-capping proteins play essential roles in the asymmetric division of maturing mouse oocytes. J. Cell. Sci. 128, 160–170 (2015).
Amatruda, J. F. & Cooper, J. A. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J. Cell. Biol. 117, 1067–1076 (1992).
Schafer, D. A. & Cooper, J. A. Control of actin assembly at filament ends. Annu. Rev. Cell. Dev. Biol. 11, 497–518 (1995).
Akin, O. & Mullins, R. D. Capping protein increases the rate of actin-based motility by promoting filament nucleation by the Arp2/3 complex. Cell 133, 841–851 (2008).
Menna, E. et al. Eps8 controls dendritic spine density and synaptic plasticity through its actin-capping activity. EMBO J. 32, 1730–1744 (2013).
Firth, H. V. et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using ensembl resources. Am. J. Hum. Genet. 84, 524–533 (2009).
Atsak, P. et al. Endocannabinoid signaling within the basolateral amygdala integrates multiple stress hormone effects on memory consolidation. Neuropsychopharmacol 40, 1485–1494 (2015).
Song, T. J. et al. Altered behaviors and impaired synaptic function in a novel rat model with a complete Shank3 deletion. Front. Cell. Neurosci. 13, 111 (2019).
Fan, M. et al. JADE2 is essential for hippocampal synaptic plasticity and cognitive functions in mice. Biol. Psychiat. 92, 800–814 (2022).
Amador, A. et al. Modelling and treating GRIN2A developmental and epileptic encephalopathy in mice. Brain 143, 2039–2057 (2020).
Brechet, A. et al. AMPA-receptor specific biogenesis complexes control synaptic transmission and intellectual ability. Nat. Commun. 8, 15910 (2017).
Apicco, D. J. et al. Dysregulation of RNA Splicing in Tauopathies. Cell. Rep. 29, 4377–4388.e4374 (2019).
Chapman, R. M. et al. Convergent evidence That ZNF804A Is a regulator of pre-messenger rna processing and gene expression. Schizophr. Bull. 45, 1267–1278 (2019).
Bartsch, V. B., Lord, J. S., Diering, G. H. & Zylka, M. J. Mania- and anxiety-like behavior and impaired maternal care in female diacylglycerol kinase eta and iota double knockout mice. Genes Brain Behav. 19, e12570 (2020).
Yoshida, S. et al. Knockdown of synapse-associated protein Dlg1 reduces syncytium formation induced by human T-cell leukemia virus type 1. Virus Genes 37, 9–15 (2008).
Zou, W. et al. A dendritic guidance receptor complex brings together distinct actin regulators to drive efficient F-Actin assembly and branching. Dev. Cell. 45, 362–375.e363 (2018).
Terracciano, A. et al. Meta-analysis of genome-wide association studies identifies common variants in CTNNA2 associated with excitement-seeking. Transl. Psychiatry 1, e49 (2011).
Smith, A., Bourdeau, I., Wang, J. & Bondy, C. A. Expression of Catenin family members CTNNA1, CTNNA2, CTNNB1 and JUP in the primate prefrontal cortex and hippocampus. Brain Res. Mol. Brain Res. 135, 225–231 (2005).
Bianchi Porro, G., Ardizzone, S. & Petrillo, M. Cetraxate is not superior to placebo in the treatment of duodenal ulcer. J. Clin. Gastroenterol. 12, 234–236 (1990).
Zhang, Y. et al. Association between NRGN gene polymorphism and resting-state hippocampal functional connectivity in schizophrenia. BMC Psychiatry 19, 108 (2019).
Zhang, J. et al. Exploration of RNA-binding proteins identified RPS27 as a potential regulator associated with Kaposi’s sarcoma development. BMC Cancer 25, 362 (2025).
Perrier, S. et al. Recessive mutations in NDUFA2 cause mitochondrial leukoencephalopathy. Clin. Genet. 93, 396–400 (2018).
van Tijn, P. et al. Alzheimer-associated mutant ubiquitin impairs spatial reference memory. Physiol. Behav. 102, 193–200 (2011).
Perez-Garcia, V. et al. Placentation defects are highly prevalent in embryonic lethal mouse mutants. Nature 555, 463–468 (2018).
Gerasimavicius, L., Livesey, B. J. & Marsh, J. A. Loss-of-function, gain-of-function and dominant-negative mutations have profoundly different effects on protein structure. Nat. Commun. 13, 3895 (2022).
Bergendahl, L. T. et al. The role of protein complexes in human genetic disease. Protein Sci. 28, 1400–1411 (2019).
Korobova, F. & Svitkina, T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol. Biol. Cell. 21, 165–176 (2010).
Ziv, N. E. & Smith, S. J. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron 17, 91–102 (1996).
Yoshihara, Y., De Roo, M. & Muller, D. Dendritic spine formation and stabilization. Curr. Opin. Neurobiol. 19, 146–153 (2009).
Fan, Y., Tang, X., Vitriol, E., Chen, G. & Zheng, J. Q. Actin capping protein is required for dendritic spine development and synapse formation. J. Neurosci. 31, 10228–10233 (2011).
Lee, J. H. et al. Astrocytes phagocytose adult hippocampal synapses for circuit homeostasis. Nature 590, 612–617 (2021).
Rongo, C. & Kaplan, J. M. CaMKII regulates the density of central glutamatergic synapses in vivo. Nature 402, 195–199 (1999).
Howland, J. G. & Wang, Y. T. Synaptic plasticity in learning and memory: stress effects in the hippocampus. Prog. Brain. Res. 169, 145–158 (2008).
Eriksson, J., Vogel, E. K., Lansner, A., Bergström, F. & Nyberg, L. Neurocognitive architecture of working memory. Neuron 88, 33–46 (2015).
Cingolani, L. A. & Goda, Y. Actin in action: the interplay between the actin cytoskeleton and synaptic efficacy. Nat. Rev. Neurosci. 9, 344–356 (2008).
Pelucchi, S. et al. Cyclase-associated protein 2 dimerization regulates cofilin in synaptic plasticity and Alzheimer’s disease. Brain Commun. 2, fcaa086 (2020).
Sekino, Y., Kojima, N. & Shirao, T. Role of actin cytoskeleton in dendritic spine morphogenesis. Neurochem. Int. 51, 92–104 (2007).
Hotulainen, P. & Hoogenraad, C. C. Actin in dendritic spines: connecting dynamics to function. J. Cell. Biol. 189, 619–629 (2010).
Ethell, I. M. & Pasquale, E. B. Molecular mechanisms of dendritic spine development and remodeling. Prog. Neurobiol. 75, 161–205 (2005).
Nishiyama, J. Plasticity of dendritic spines: molecular function and dysfunction in neurodevelopmental disorders. Psychiatr. Clin. Neurosci. 73, 541–550 (2019).
Faust, T. E., Gunner, G. & Schafer, D. P. Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 22, 657–673 (2021).
Nelson, T. J., Backlund, P. S. Jr. & Alkon, D. L. Hippocampal protein-protein interactions in spatial memory. Hippocampus 14, 46–57 (2004).
Gulesserian, T., Kim, S. H., Fountoulakis, M. & Lubec, G. Aberrant expression of centractin and capping proteins, integral constituents of the dynactin complex, in fetal down syndrome brain. Biochem. Biophys. Res. Co. 291, 62–67 (2002).
Petanjek, Z. et al. Extraordinary neoteny of synaptic spines in the human prefrontal cortex. Proc. Natl. Acad. Sci. USA 108, 13281–13286 (2011).
Penzes, P., Cahill, M. E., Jones, K. A., VanLeeuwen, J. E. & Woolfrey, K. M. Dendritic spine pathology in neuropsychiatric disorders. Nat. Neurosci. 14, 285–293 (2011).
Aceti, M. et al. Syngap1 haploinsufficiency damages a postnatal critical period of pyramidal cell structural maturation linked to cortical circuit assembly. Biol. Psychiatr. 77, 805–815 (2015).
Bastrikova, N., Gardner, G. A., Reece, J. M., Jeromin, A. & Dudek, S. M. Synapse elimination accompanies functional plasticity in hippocampal neurons. Proc. Natl. Acad. Sci. USA 105, 3123–3127 (2008).
Andersen, S. L. & Teicher, M. H. Delayed effects of early stress on hippocampal development. Neuropsychopharmacol 29, 1988–1993 (2004).
Su, P. et al. EF1α-associated protein complexes affect dendritic spine plasticity by regulating microglial phagocytosis in Fmr1 knock-out mice. Mol. Psychiatr. 29, 1099–1113 (2024).
Chen, W. et al. AMPA glutamate receptor subunits 1 and 2 regulate dendrite complexity and spine motility in neurons of the developing neocortex. Neuroscience 159, 172–182 (2009).
George, L. A. et al. Multiyear Factor VIII Expression after AAV Gene Transfer for Hemophilia A. N. Engl. J. Med. 385, 1961–1973 (2021).
Pavlou, M. et al. Novel AAV capsids for intravitreal gene therapy of photoreceptor disorders. Embo. Mol. Med. 13, e13392 (2021).
He, X. et al. AAV for gene therapy in ocular diseases: progress and prospects. Research 6, 0291 (2023).
Arbab, M. et al. Base editing rescue of spinal muscular atrophy in cells and in mice. Science 380, eadg6518 (2023).
Cheng, W. et al. Intrathecal delivery of AAV-NDNF ameliorates disease progression of ALS mice. Mol. Ther. 31, 3277–3289 (2023).
Mao, H. et al. AAV delivery of wild-type rhodopsin preserves retinal function in a mouse model of autosomal dominant retinitis pigmentosa. Hum. Gene. Ther. 22, 567–575 (2011).
Kang, S. K., Hawkins, N. A. & Kearney, J. A. C57BL/6J and C57BL/6N substrains differentially influence phenotype severity in the Scn1a (+/-) mouse model of Dravet syndrome. Epilepsia Open 4, 164–169 (2019).
Chae, S. et al. The dopamine analogue CA140 alleviates AD pathology, neuroinflammation, and rescues synaptic/cognitive functions by modulating DRD1 signaling or directly binding to Abeta. J. Neuroinflamm. 21, 200 (2024).
Lau, S. F., Cao, H., Fu, A. K. Y. & Ip, N. Y. Single-nucleus transcriptome analysis reveals dysregulation of angiogenic endothelial cells and neuroprotective glia in Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 117, 25800–25809 (2020).
Acknowledgements
This paper was supported by the National Key R&D Program of China (No. 2021YFC1005300), the Major Scientific and Technological Projects for Collaborative Prevention and Control of Birth Defects in Hunan Province of China (No. 2019SK1010, 2019SK1014), and the Clinical Medical Research Center for Hereditary Birth Defects and Rare Diseases in Hunan Province (No. 2023SK4053), the Science and Technology Projects of the Xizang Autonomous Region, China (No. XZ202501YD0018); the National Natural Science Foundation of China (82471560).
Author information
Authors and Affiliations
Contributions
Mei Guo, Xiao Mao, Xianglan Piao, Hua Wang, Yang Du, and Yong Cheng designed research; Mei Guo, Liming Liu, and Manyu Xiao. performed research; Qibin Li, Xiaobin He, and Xing Pan contributed new reagents/analytic tools; Yuewen Chen and Wanying Yi analyzed single-cell sequencing data; Mei Guo and Yong Cheng wrote the paper, and all authors discussed the experiments and final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, M., Liu, L., Mao, X. et al. Deciphering the role of CAPZA2 in neurodevelopmental disorders: insights from mouse models. Commun Biol 8, 1048 (2025). https://doi.org/10.1038/s42003-025-08385-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-025-08385-1