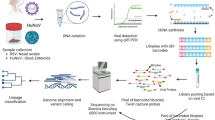
Abstract
Juruaça virus (JUAV), previously unclassified, was isolated from bats and administered to neonatal and adult BALB/c mice to investigate acute and chronic disease progression. In this study, we conducted genomic sequencing to achieve taxonomic classification and utilized these models to explore the inflammatory response and sickness behavior in both neonatal and adult mice. Neonates received a single intranasal instillation of infected brain homogenate (20 µL), whereas 31-day-old mice were given the same volume intranasally for three consecutive days. Control groups were administered equal volumes of uninfected brain homogenate. Our findings reveal that intranasal JUAV infection-induced acute meningoencephalitis and death in neonates, while adult mice exhibited chronic infection with variable clinical signs, inflammatory mediator production, histopathological changes, and neuropathological features. Interestingly, only some adult mice showed sickness behavior post-infection, and among these, a subset continued to decline and die. The differential tissue damage observed in mice with and without overt disease symptoms suggests mechanisms of resistance or tolerance, where exceeding tolerance capacity resulted in pathological outcomes, including chronic dysfunction or death. This study provides the first evidence of JUAV’s capability to infect mammals, demonstrating its distinct impact on bats and variable effects in neonatal and adult mice. We provisionally classified JUAV as closely related to the clade containing tombus-like virus 6 found in mute swan feces. Our research highlights the importance of understanding viral–host interactions and the inflammatory responses that contribute to disease variability, offering insights into tolerance and resistance mechanisms based on inflammatory response modulation.
Similar content being viewed by others
Introduction
Viruses can trigger various host defense mechanisms such as avoidance, resistance, and tolerance to maintain homeostasis1,2,3. Avoidance involves detecting and steering clear of pathogens, often through social behaviors4,5. Resistance relies on the immune system to recognize and eliminate pathogens3. In contrast, tolerance involves controlling the damage caused by pathogens without necessarily eliminating them6. Recent reviews associate these concepts with the modulation of the inflammatory response (e.g. ref. 7).
The immune system’s response to viral infections involves early detection and activation of antiviral mechanisms, such as the production of type I interferons (IFNs) and the activation of various immune cells, including natural killer (NK) cells, B cells, and T cells8,9,10. Neonates are particularly vulnerable to infections due to their developing immune systems, which favor anti-inflammatory responses and allow the establishment of microbiota without disease11,12. In contrast, adults exhibit diverse immune responses and sickness behaviors due to the complex mechanisms viruses use to infect host cells13.
Juruaça virus (JUAV) is a newly discovered virus, and this study represents the first investigation into its pathogenicity in mammals. We previously found that intracerebral inoculation of JUAV in neonatal mice caused acute fatal infections, while adult mice developed chronic infections with varying clinical signs, including ruffled fur, a hunched posture, body weight loss, hindlimb clasping, and paralysis. This study aims to taxonomically classify JUAV, explore immune responses in neonatal and adult BALB/c mice, and determine the correlation between sickness behavior and inflammatory responses. We hypothesize that modulation of the inflammatory response is linked to tolerance, resistance, and sickness behavior, influencing the infection outcome.
Methods
All procedures were conducted in strict adherence to international biosafety guidelines and the established protocols for the care and utilization of laboratory animals, as outlined by the Ethics Committee for the Use of Animals in Research (CEUA) of the Instituto Evandro Chagas. The experimental protocols were granted prior approval (Protocol No. 0022/2012/CEUA/IEC/CENP/SVS/MS for neonate experiment and Protocol No. 27/2017 CEUA/IEC/CENP/SVS/MS for adult experiment), and biosafety level 3 standards were meticulously followed throughout. The unclassified JUAV strain BE AN 401933 was obtained from bats at the Igarapé do Batata, situated 15 km from Porto Trombetas in the Oriximiná municipality, located in the middle Amazon region of Brazil.
This strain was specifically selected to induce prototypical acute and chronic viral disease progression in neonatal and adult BALB/c mice, respectively. BALB/c mice are widely recognized for their well-characterized immune system, making them a standard model for immunological studies (e.g. refs. 14,15,16). They exhibit a Th2-biased immune response, which is crucial for understanding the mechanisms of viral tolerance and resistance17,18. This predisposition allows for the detailed examination of cytokine profiles and immune cell activation relevant to Juruaça virus (JUAV) infections. Furthermore, BALB/c mice are known for their susceptibility to various viral infections with contrasting immune responses in the neonate and adult mouse models, (e.g. refs. 19,20), making them an ideal model for studying viral pathogenesis and host immune responses. This susceptibility facilitates the clear observation of disease progression, immune responses, and potential mechanisms of tolerance and resistance.
Furthermore, female BALB/c mice were deliberately selected to mitigate the deleterious effects of home-cage aggression, thereby safeguarding both animal well-being and the integrity of scientific findings derived from group-housed specimens21. These mice were accommodated within a meticulously regulated animal facility, where environmental parameters such as a 12-h light–dark cycle and ad libitum access to food and water were rigorously upheld.
Viral stock production and titration
The process of viral stock production and titration adhered to a stringent protocol. Brain homogenates obtained from infected mice euthanized at 8 dpi underwent centrifugation at 10,000 rpm for 15 min at 4 °C to isolate viral suspensions. These suspensions, diluted at a ratio of 1:10 in 0.8% phosphate-buffered saline (PBS) supplemented with 0.75% bovine serum albumin, streptomycin (100 μg/mL), and penicillin (100 U/mL) (pH 7.2), were subsequently intracerebrally inoculated into two-day-old neonatal mice. The infected neonates were meticulously monitored until 7–8 dpi, upon which the onset of severe clinical signs prompted euthanasia, and the specimens were preserved at −80 °C to generate a viral stock. Control uninfected mice underwent identical euthanasia and storage conditions for comparative purposes.
The determination of viral titer utilized the median lethal dose (LD50), representing the concentration of virus causing mortality in 50% of genetically identical hosts under uniform environmental conditions2. Serial dilutions of the viral suspensions (ranging from 10−2 to 10−10) were prepared and administered to separate groups of neonatal mice (six mice per group). The LD50/0.02 mL, indicating the lethal dose for 50% of infected mice, was computed following the method outlined by Reed and Muench (1938)3.
Subsequently, equivalent volumes (0.02 mL) of brain suspensions from infected or uninfected mice were intranasally delivered to 2-day-old BALB/c mice allocated into experimental and control groups. Daily observations were conducted to monitor sickness behavior in each cohort.
In a distinct experiment involving adult mice infected with the same JUAV strain, chronic disease progression was instigated by administering three consecutive intranasal instillations of 0.02 mL (once daily for three consecutive days) of the infectious suspension (5.6 LD50/0.02 mL).
This experimental design in adult mice was informed by preliminary findings suggesting the absence of apparent sickness behavior following the administration of lower doses. In both neonatal and adult mouse experiments, subsequent sickness behavior was meticulously recorded and correlated with cellular and humoral innate immune responses22.
Genomic sequencing
Genomic sequencing commenced with the homogenization of 5 mg of brain tissue from neonatal mice infected with JUAV, employing a TissueLyser II system (QIAGEN, Hilden, Germany) operated at 25 Hz for 2 min, using 600 µL of 1-thioglycerol/homogenization solution and one 5 mm tungsten bead. RNA extraction was then carried out using the Maxwell® 16 LEV simply RNA Tissue Kit (Promega, Madison, WI, USA) in the Maxwell® 16 System (Promega) following the manufacturer’s protocol. Subsequently, sequencing was conducted utilizing the Ion Torrent platform (Life Technologies, Applied Biosystems).
Construction of genomic libraries from the obtained cDNA was automated, facilitated by the AB Library Builder™ System and the Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific), adhering to the manufacturer’s recommendations. During library construction, cDNA molecules were cleaved into selectable size fragments of 200 base pairs by an included enzyme. Adapters and barcode sequences were then automatically ligated to the 200-base pair cDNA fragments. Following this, library size selection and purification were performed using 2% agarose gel electrophoresis (E-Gel® SizeSelect™).
Real-time polymerase chain reaction (PCR) was conducted using an Ion Library TaqMan Quantitation Kit (Thermo Fisher Scientific) on a LightCycler 480 instrument (Roche) to assess the concentration of each library for subsequent pooling, ensuring an equal quantity of molecules per sample. Samples with low concentrations were subjected to conventional PCR amplification using Platinum PCR Supermix High Fidelity and Ion Plus Fragment Library Adapters (Thermo Fisher Scientific).
In the emulsion step, both aqueous and organic phases were prepared, with each microreactor producing identical copies of a single fragment attached to a microsphere. After ligation of the P1 and A adapters, monoclonal microspheres underwent evaluation using the Ion Sphere™ Quality Control Assay. Microsphere release was facilitated in the enrichment step, retaining only those microspheres with library fragments attached. Primers and sequencing polymerase were subsequently added to suspensions of microspheres loaded with the samples, followed by deposition onto a sequencing chip. Sequencing was conducted on the Ion Torrent PGM platform using the Ion PGM Hi-Q Sequencing Kit (Thermo Fisher Scientific) with the Ion 318™ Chip Kit v2 BC as per the manufacturer’s guidelines.
Assembly and phylogenetic analysis
The reads obtained from the sequencing step were assembled utilizing MIRA3 software (https://www.chevreux.org/projects_mira.html) with default parameters. Subsequently, the contigs were compared to the nonredundant (NR) database using the BlastX program, and contigs containing virus sequences were selected for phylogenetic analysis.
Phylogenetic inference utilized amino acid sequences from various virus strains sourced from the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). Specifically, the functional domains of the viral RNA-dependent RNA polymerase (RdRp; PF00998) from multiple genera within the Tombusviridae family, as well as unclassified species related to this family, were included in the analysis.
The dataset derived from the study samples underwent multiple sequence alignment (MSA) employing the MAFFT v.7 program23. Alignment data underwent manual inspection for necessary adjustments using the Geneious v.9.1.8 program (https://www.geneious.com/).
Initially, the aligned dataset underwent analysis to determine the optimal nucleotide substitution method. Subsequently, phylogenetic trees were constructed utilizing the maximum likelihood (ML) method24. Both methodologies were implemented using the IQ-TREE v.1.6.12 program25. Additionally, bootstrapping with 1000 replicates was conducted to enhance the reliability of the cluster values26.
The viruses utilized for ML phylogenetic analysis are detailed in Table S1. Phylogenetic tree visualization was executed using the FigTree v.1.4.4 program (https://github.com/rambaut/figtree/releases/tag/v1.4.4). The midpoint rooting method, available in the phylogenetic visualization software, was applied to the dataset. Following phylogenetic evaluation and refinement, a Scalable Vector Graphics (.svg) format file was generated for image editing and manipulation using the Inkscape v.1.1 program (https://inkscape.org/release/inkscape-1.1/).
Experimental timeline for neonates and adults
Two-day-old mice were intranasally instilled with 0.02 mL of a 1:10 dilution of virus-infected brain homogenate. Control mice received an equivalent volume of uninfected brain homogenate. The neonatal mice were weighed daily until 12 days post-infection (dpi). At 4, 8, and 12 dpi, both infected and control mice were euthanized to evaluate the acute innate immune response.
Subsequently, 31-day-old mice were intranasally instilled with equal volumes and titers of brain homogenates from either JUAV-infected or uninfected mice. This procedure was carried out once daily for three consecutive days. Following instillations, all mice were observed twice a day throughout the experiment.
Detection of JUAV viral antigens and microglial immunolabeling in neonatal mice
In neonatal mice, the presence of JUAV antigens was identified using a commercial immunoperoxidase kit (mouse-on-mouse, MOM; Vector Laboratories) in following the manufacturer’s instructions, with slight modifications. Initially, tissue sections were washed in 0.1 M phosphate-buffered saline (PBS; pH 7.2) and subjected to antigen retrieval in 0.2 M boric acid (pH 9.0) for 60 min at 70 °C. Subsequently, the sections were permeabilized in PBS containing Triton X-100 (0.5%), followed by blocking of nonspecific binding sites with mouse IgG blocking reagent in conjunction with a concentrated protein solution from the MOM kit for 24 h.
The sections were then incubated in a solution containing the anti-Juruaça polyclonal primary antibody (diluted 1:100), provided by the Arbovirology and Hemorrhagic Fevers Section of the Evandro Chagas Institute. The specificity of this primary antibody was validated using hemagglutination inhibition and complement fixation assays. To ensure accurate immunostaining targeting both microglia and macrophages, sections were subjected to treatment with a solution containing an anti-Iba1 antibody (Wako©, #019-19741) diluted 1:500 in PBS (pH 7.2) for 24 h. Following PBS washes, secondary antibodies (MOM biotinylated anti-mouse IgG and biotinylated goat anti-rabbit IgG) were applied to detect viral and microglial antigens, respectively.
To inhibit endogenous peroxidase activity, the sections were immersed in a 0.3% hydrogen peroxide solution for 15 min. Subsequently, incubation with avidin-biotin complex (ABC) solution (Vector Laboratories) for 3 h was performed. The diaminobenzidine (DAB)/nickel/glucose oxidase method was utilized to reveal positively labeled sites, employing a specific protocol aimed at reducing the signal-to-noise ratio and enhancing contrast27. Finally, the sections were mounted on slides, coverslipped with Entellan, and examined using an Axiophot microscope (Carl Zeiss, Germany). Images were captured using an AxioCam HRC digital color camera (Carl Zeiss, Germany).
Viral antigen immunolabeling in adult mice
Antigen retrieval was performed using bacterial Type VIII proteinase (Sigma-Aldrich, USA) in sodium phosphate buffer (PB) (pH 7.8) in a water bath at 37 °C for 3 min. Nonspecific sites were blocked with skim milk (Nestlé®/Switzerland) and horse serum from the Vectastain® ABC HRP Kit (Peroxidase, Mouse IgG, Vector Laboratories, USA) in PBS for 20 min in the dark at room temperature. The samples were then incubated with the primary anti-JUAV antibody diluted 1:100 in PBS for 1 h in the dark at room temperature. Sections were washed 3× with PBS and incubated first with a biotinylated anti-mouse IgG secondary antibody (Vector Laboratories, USA) in PBS (1:5) for 1 h at room temperature and then with streptavidin conjugate and alkaline phosphatase (Invitrogen™, USA) in PBS (1:200) for 1 h at room temperature. Subsequently, the samples were washed three times with Tris(hydroxymethyl)-aminomethane (Tris) buffer (Sigma, USA) (pH 7.4) and incubated for 30–40 min with a HistoMark® Red phosphatase system kit (KPL, USA)28. The tissues were then washed three times with distilled water, counterstained with Harris hematoxylin for 1 min, dehydrated in increasing concentrations of 95% alcohol and absolute ethanol I and II, and cleared in xylene I and II (1 min per step). The sections were then coverslipped with Entellan (Merck/Germany) and analyzed under an optical microscope (Axiophot, Zeiss, Germany) equipped with a digital camera (AxioCam HRC, Zeiss, Germany).
To validate the specificity of our polyclonal antibody for virus antigen detection, we initially conducted specificity assays and cross-reactivity assessments across a spectrum of viral species. No instances of cross-reactivity were discerned, thereby affirming the antibody’s selectivity towards JUAV. Subsequently, a battery of serological evaluations was undertaken, encompassing complement fixation, immunofluorescence, and viral particle immunoprecipitation assays, supplemented by immunoelectron microscopy utilizing the polyclonal antibody. These supplementary analyses serve to reinforce the antibody’s specificity and its suitability for the objectives of our investigation.
Microglia immunolabeling in adult mice
Horizontal 80-μm-thick sections (1:4) containing all areas of interest were sliced with a vibratome. The sections in each group were labeled with selective immunomarkers. The anti-IBA1 antibody was used as a selective marker for microglia and macrophages.
Immunoperoxidase staining was used to reveal antigenic sites. Antigen retrieval was performed with 0.2 M boric acid (pH 9.0) for 1 h in a water bath at 60 °C. The sections were then permeabilized with 0.5% Triton X-100 in PBS. Nonspecific sites were blocked with 10% goat serum and 10% horse serum in PBS containing 2% azide for 24 h. The sections were incubated in the abovementioned blocking solution containing a primary antibody (Monoclonal Anti-IBA1, Wako, Japan; diluted 1:500) for 72 h. Next, the sections were incubated in the abovementioned blocking solution containing biotinylated goat anti-rabbit IgG as the secondary antibody (Vector Laboratories, USA; diluted 1:250) for 12 h. Endogenous peroxidase was inactivated by incubation with 3% hydrogen peroxide and distilled water for 10 min. The sections were washed twice with 0.1 M PB (pH 7.4) and incubated in 1% acetate buffer (pH 6.0, 3 min). Finally, the sections were transferred into the avidin–biotin ABC complex solution from the Vectastain® Kit (Vector Laboratories, USA) and incubated for 3 h. The sections were washed, and horseradish peroxidase (HRP) was visualized via the DAB/nickel/glucose oxidase metho27. This procedure increased the background and foreground contrast. For the DAB/nickel/glucose oxidase method, all sections were washed again in PBS before incubation in 0.2 M acetate buffer (pH 6.0) for 5 min and incubation in GND solution (0.6 mg/mL DAB, 2.5 mg/mL ammonium nickel chloride, and 1 mg glucose oxidase) for visualization. All steps were carried out under gentle and constant agitation. The brain sections were mounted on gelatinized slides and dried at room temperature. The dried sections were dehydrated by sequential incubation in gradient solutions of 95% alcohol and absolute ethanol I and II and cleared in xylene (100%) I and II (1 min per step). The sections were then coverslipped with Entellan (Merck/Germany) and analyzed under an optical microscope (Axiophot, Zeiss, Germany) equipped with a digital camera (AxioCam HRC, Zeiss, Germany).
Transmission electron microscopy (TEM)
Two paraformaldehyde-fixed brains (harvested from neonatal mice at 8 dpi and adult GI3 mice at 59 dpi) from each experimental group were serially sectioned (300 µm thickness) using a vibratome and were then postfixed in a solution containing 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer (TCS) (pH 7.2) supplemented with 3 mM calcium chloride (CaCl2) for 2 h at room temperature29.
The sections were washed 4 times in buffered saline solution with 0.1 M sodium cacodylate (CBS) and CaCl2 (3 mM), and the areas of interest were postfixed with 1% osmium tetroxide (OsO4) supplemented with 0.8% potassium ferrocyanide and 5 mM CaCl2 in cacodylate buffer (0.1 M) (pH 7.2) for 1 h at 4 °C. Contrast blocking was performed with 2.5% uranyl acetate in 25% acetone for 1 h at 4 °C. Dehydration was performed by incubation with increasing concentrations of acetone (50%, 70%, 90%, and 100% I, II and III) for 10 min each, and the samples were then incubated in increasing concentrations of Epon diluted in acetone (1:2 for 12 h; 1:1 for 8 h; 2:1 for 24 h; Epon plus DMP-30 for 24 h) at 4 °C. The samples were finally incubated in Epon plus DMP-30 in appropriate molds in an oven at 60 °C for 48 h for resin polymerization (LUFT, 1961).
Electron microscopy negative contrasting
Supernatants from cultured cells infected with JUAV were used for negative contrasting. Primary glial cell cultures were prepared as described elsewhere30. A drop of the material was placed on a 400-mesh copper grid covered with a formvar plastic film reinforced by a thin layer of carbon31. After one minute, the excess material was removed with a strip of filter paper, after which a drop of 2% phosphotungstic acid (PTA) (pH 6.6) was added to the grid for one minute32. The excess PTA was then removed with a strip of filter paper, and the remaining material was observed using TEM (Zeiss, EM 900).
Immunoelectron microscopy
The culture supernatants of infected and uninfected (control) cells were centrifuged at 12,000 revolutions per minute (rpm) for 15 min at 4 °C. The supernatant was used to dilute the antibody suspension to match the concentration of the sample of interest (1:250, 1:500, and 1:750). The mixtures were incubated in the homogenizer overnight at 4 °C. Then, each mixture was centrifuged at 12,000 rpm for 30 min at 4 °C, the supernatant was discarded, and the pellet was resuspended in distilled water and processed for negative contrasting32.
Flow cytometry
The production of cytokines in serum and brain homogenates was analyzed according to the manufacturer’s recommendations (BD™ Cytometric Bead Array (CBA) Mouse Th1/Th2/Th17 Kit, BD Biosciences, USA) and BD™ Cytometric Bead Array (CBA) Mouse Inflammation Kit, BD Biosciences, USA).
The standard curve was generated with serial dilutions of the cytokine standard solutions. Twenty-five-microliter serum and brain samples and each serial dilution of the standards were transferred to labeled flow tubes. The samples were incubated for two hours, after which the cytokines of interest were quantified via flow cytometry (BD FACSCanto II). The data were processed using FACS DIVA software and analyzed using FCAP Array 3.0 software.
Indirect nitric oxide detection
To quantify nitrite, which indirectly detects NO, in brain homogenates from infected and control mice, Griess reagent was used according to the manufacturer’s instructions (Molecular Probes, USA).
A standard curve was generated using the nitrite concentrations predicted in the kit in PHEM 2X buffer. First, 50 μL of each serial dilution of the standard and the supernatants of mouse brain suspensions were added to a 96-well plate in triplicate. Then, 50 μL of Griess reagent, prepared as a 1:1 ratio of solution A [N-(1-naphthyl) ethylenediamine dihydrochloride] to solution B (sulfanilic acid), was added to each well. The plate was incubated for 20 min at room temperature.
After incubation with the reagents, 5 μL of 2% vanadium chloride (VCl3) in 6 N hydrochloric acid solution was added, and the plate was incubated for 25 min at 60 °C in a water bath. The plate was then gradually cooled to room temperature, and the absorbance of each sample was measured with a spectrophotometer (EL 800, Bio-Tek) using a 550 nm wavelength filter33. For this method, the detection limit of nitrite is 1.0 μm.
Histopathological analysis
After macroscopic analysis, the left cerebral hemispheres harvested from two animals from each experimental group at 59 dpi were subjected to histopathological processing. The tissues were dehydrated in increasing alcohol concentrations (70%, 80%, 95%, and absolute ethanol I–IV) and cleared in xylene (I–III). The sections were immersed in a paraffin bath at 60 °C three times for 1 h and 20 min each. The tissue sections used for histopathological analysis were sliced from paraffin-embedded tissue blocks28.
The blocks were sectioned using a rotary microtome (RM2235, Leica, Switzerland). The sections (5 µm thick) were mounted on glass slides and incubated in an oven at 60 °C overnight. The following day, the sections were dewaxed in xylene I and II (100%, for 3 min each), rehydrated in decreasing concentrations of alcohol (absolute ethanol I and II, 95%, 80% and 70%) and in distilled water (1 min each), and then stained with Harris hematoxylin for 2 min. The sections were then washed with distilled water, immersed in 70% alcohol, and stained with eosin for 3 min. This process was followed by dehydration in increasing concentrations of alcohol (70%, 80%, 95%, and absolute ethanol I and II) and xylene (I and II) (1 min of immersion in each)34.
The sections were mounted on slides and coverslipped using Entellan (Merck/Germany) prior to evaluation by optical microscopy (Axiophot-Zeiss, Germany). Images were acquired with a digital camera (AxioCam HRC, Zeiss, Germany).
Results
Taxonomic classification of JUAV
For taxonomic classification, we investigated morphological and genomic characteristics. We evaluated the ultrastructural features of the viral particles in the mouse brain at 8 dpi and in JUAV-infected cultured primary glial cells. The viral particles were spherical in shape and nonenveloped, with a diameter of approximately 30 nm. The particles were clustered in the cytoplasm, and their presence was associated with histopathological alterations and changes in inflammatory cytokine production in infected mouse brain tissue and behavioral changes in the infected mice (Fig. 1A–D).
Electron micrographs illustrating JUAV particles. A Negative staining of primary culture supernatant from mouse central nervous system (CNS) cells, highlighting viral particles. B Ultrathin sections of the mouse brain at 8 days post-infection (dpi), showing nonenveloped viral particles clustered in the cytoplasm. C and D Immunoelectron micrographs demonstrating immunocomplexes formed between the anti-JUAV antibody and viral particles (white arrows). These images are representative of three independent experiments.
Via molecular analysis, a total of 406,963 reads were obtained, and in the second run, 1,671,628 reads were obtained. After assembly, 89,562 reads were generated, resulting in a total of 13,615 contigs, with the largest contig containing 6753 nucleotides (nt). The sequence coverage exceeded 3000 fold (coverage = genome size/number of reads). An RNA genome containing 5020 nt was obtained and was shown to harbor three open reading frames (ORFs) and two intergenic regions (IGRs) flanked by 5’ and 3’ untranslated regions. Protein sizes and isoelectric points were predicted using Geneious software. The genome of JUAV is depicted in Fig. 2A and B. This organization of this genome is similar to that of other tombusviruses.
A Illustration of protein functional domains in various viruses from the Tombusviridae family. The yellow arrow indicates the protein-coding region, with functional domains highlighted in purple. The “ps_ssRNAv_Tolivirales_RdRp” sequence represents the catalytic core domain of the RNA-dependent RNA polymerase (RdRp) enzyme in positive-sense single-stranded RNA (+ssRNA) viruses of the order Tolivirales. B Features of the JUAV genome, detailing the lengths of individual genes, and the position and isoelectric point (pI) of each putative protein. Data shown are representative of three independent experiments.
BLASTX results revealed sequence identities of 54.6%, 47.6%, and 31.7% corresponding to ORF1, ORF2, and ORF3, respectively, with mute swan feces-associated tombus-like virus 635,36.
InterProScan analysis revealed a high sequence identity between the protein encoded by ORF2 (480 aa) and the domain of the RdRp protein between positions 16 and 475.
The phylogenetic tree contained several monophyletic clades corresponding to each genus within the Tombusviridae family (Fig. 3). However, JUAV was grouped into the most external monophyletic clade with other unclassified viruses within this family.
Phylogenetic tree of different strains of viruses belonging to the Tombusviridae family. The maximum likelihood (ML) method was used based on the amino acid sequences of the functional domains of the viral RNA-dependent RNA polymerase (RdRp) (PF00998), with the LG + F + R5 matrix identified as the best model for amino acid substitution. Known genera are shown in different colors. The sample identified in this study is indicated by a yellow arrow. The numbers at each main node represent bootstrap values as percentages (1000 replicates). The scale bar indicates the amino acid divergence per site between sequences. JUAV was clustered into a clade of ungrouped viruses outside this family, supported by a bootstrap value of 94%. The genome organization of the identified genus and ungrouped viruses, such as Mute swan virus and Serdyukov virus, shows a similar genome organization to JUAV, clustering in the same clade in the ML phylogenetic tree. Phylogenetic tree: TCV/NC_003821: Turnip crinkle virus; CCFV/NC_001600/Australia: Cardamine chlorotic fleck virus; HCRSV/NC_003608/ ElSalvado: Hibiscus chlorotic ringspot virus; JINRV/NC_002187: Japanese iris necrotic ring virus; MNSP/NC_001504: Melon necrotic spot virus; PSNV/NC_004995/Japao/2002: Pea stem necrosis virus; SoYMMV/NC_011643/Coreiado: Soybean yellow mottle mosaic virus; CPMoV/NC_003535/Nigeria: Cowpea mottle virus; NLVCV/NC_009017/Alaska/20: Nootka lupine vein-clearing virus; SCV/NC_001780: Saguaro cactus virus; CbMV/NC_021926/USA/2002: Calibrachoa mottle virus; AnFBV/NC_007733/USA/2006: Angelonia flower break virus; CarMV/NC_001265: Carnation mottle virus; PFBV/NC_005286 /Espanha/20: Pelargonium flower break virus; HnRSV/NC_014967/USA/2011: Honeysuckle ringspot virus; PelRSV/NC_026240: Pelargonium ringspot virus; ELV/NC_026239: Elderberry latent virus; PCRPV/NC_005985: Pelargonium chlorotic ring pattern virus; RrLDV/NC_020415/USA/2009: Rosa rugosa leaf distortion virus; MCMV/NC_003627: Maize chlorotic mottle virus; PLPV/NC_007017/Alemanha/2: Pelargonium line pattern virus; CMMV/NC_0111108/ReinoUnid: Cocksfoot mild mosaic virus; TPAV/NC_021705/USA/2005: Thin paspalum asymptomatic virus; PMV/NC_002598/USA/1957: Panicum mosaic virus; GaMV/NC_001818: Galinsoga mosaic virus; FNSV/NC_020469/Colombia/2: Furcraea necrotic streak virus; TNVA/NC_001777: Tobacco necrosis virus A; OMMV/NC_006939/Portugal/2: Olive mild mosaic virus; OLV1/NC_001721/Italia: Olive latent virus 1; TLeV1/NC_015227/USA/2006: Trailing lespedeza virus 1; CLSV/NC_007816/Canada/200: Cucumber leaf spot virus; YSV/NC_022895/Nigeria/199: Yam spherical virus; PoLV/NC_000939: Pothos latent virus genes; MWLMV/NC_009533/USA/2007: Maize white line mosaic virus; JCSMV/NC_005287/Ira/2006: Johnsongrass chlorotic stripe mosaic virus: BBSV/NC_004452/China/1980: Beet black scorch virus; TNVD/NC_003487/Hungria/19: Tobacco necrosis virus D; LWSV/NC_001822/Franca/199: Leek white stripe virus; MNeSV/NC_007729: Maize necrotic streak virus; CBLV/NC_004725/Bulgaria/2: Cucumber Bulgarian latent virus; EMCV/NC_023339/Israel/200: Eggplant mottled crinkle virus; PLV/AY100482: Pear latent virus; LNV/DQ011234/Taiwan: Lisianthus necrosis virus; LNSV/JN700748/USA/1998: Lettuce necrotic stunt virus; MPV/NC_020073/Marrocos/19: Moroccan pepper virus; CNV/NC_001469: Cucumber necrosis virus; CyRSV/NC_003532: Cymbidium ringspot virus; PeNSV/NC_005285: Pelargonium necrotic spot virus; PNSV/AJ607402: Pelargonium necrotic spot virus; AMCV/NC_001339: Artichoke mottled crinkle virus; CIRV/GQ259480/Italia/2009: Carnation Italian ringspot virus; CIRV/NC_003500: Carnation Italian ringspot virus; GALV/NC_011535/Japao: Grapevine Algerian latent virus; TBSV/NC_001554: Tomato bushy stunt virus; PEMV2/NC_003853: Pea enation mosaic virus-2; CMoV/KF533713/ReinoUnido: Carrot mottle virus; CMoV/NC_011515/Alemanha/2: Carrot mottle virus; CMoMV/NC_001726 /Australia: Carrot mottle mimic virus; GRV/NC_003603/AfricadoSul: Groundnut rosette virus; ETBTP/NC_024808/Ethiopian: Ethiopian tobacco bushy top virus; TBTV/NC_004366/China/2001: Tobacco bushy top virus; OPMV/NC_027710/NovaZeland: Opium poppy mosaic virus; OCST/NC_003633: Oat chlorotic stunt virus; BYDVGAV/NC_004666: Barley yellow dwarf virus-GAV; DRVX1/AB033715: Dianthovirus RVX1; CRV/NC_003530: Carnation ringspot virus; RCNMV/NC_003756: Red clover necrotic mosaic virus; SCNMV/NC_003806: Sweet clover necrotic mosaic virus; Juruaca_BEAN401933: Virus Juruaçá; OBDV/NC_001793: Oat blue dwarf virus; GFkV/NC_003347/Italia/199: Grapevine fleck virus; APLV/NC_020470/Colombia/1: Andean potato latent virus; NCBV/NC_001990/AfricadoSu: Nudaurelia capensis beta virus; DpTV/NC_005898/China/2005: Dendrolimus punctatus tetravirus; HEV/KR872417: Hepatitis E virus; CuTV/NC_015521/USA/1988: Cutthroat trout virus; PLRV/NC_001747: Potato leafroll virus; PEMV1/NC_003629: Pea enation mosaic virus-1; SCPMV/NC_001625: Southern cowpea mosaic virus; HcRNAV/NC_007518: Heterocapsa circularisquama RNA virus; NV/NC_002690: Nodamura virus; SJNNV/NC_003448: Striped Jack nervous necrosis virus; EPMS2/NC_001417: Enterobacterio phage MS2; EPQB/NC_001890: Enterobacteria phage Qbeta; TAstV1/NC_005790/USA/2000: Turkey astrovirus 2; RA/NC_025346/USA/2010: Rabbit astrovirus; TaV/AF282930: Thosea asigna virus; WAC/NC_004541: Walrus calicivirus; SaV/NC_010624/Tailandia/2: Sapovirus Mc10; NAV1/NC_007916/ReinoUnido: Newbury agent 1 virus; RHDV/NC_001543/China/1991: Rabbit hemorrhagic disease virus; PRRSV/NC_001961/Americado: Porcine respiratory and reproductive syndrome virus; ToTV/NC_009013/Espanha/20: Rice tungro spherical virus; CRLVpot/NC_006271/USA/200: Broad bean wilt virus 2; RTSV/NC_001632: Bean pod mottle virus; PYFV/NC_003628: Parsnip yellow fleck virus; SDV/NC_003785/Japao/1979: Satsuma dwarf virus; TomRSV/NC_003840: Tomato ringspot virus; BPMV/NC_003496: Cherry rasp leaf virus; BBWV2/NC_002690: Tomato torrado virus; IAPV/NC_009025/Israel/200: Cricket paralysis virus; CrPV/NC_003924/Australia: Israel acute paralysis virus of bees; ssRNAV/NC_007522/Japao/20: Schizochytrium single-stranded RNA virus; RsRNAV1/NC_018613/Japao: Rhizosolenia setigera RNA virus 01; PnPV/NC_003113/Taiwan/199: Perina nuda virus; ChPV2/NC_024766/HongKong: Chicken picornavirus 2; DHV1/NC_008250/USA/1949: Duck hepatitis A virus 1; HPeV/NC_001897: Human parechovirus; SPaV1/NC_018226/Franca/20: Swine pasivirus 1; SePV1/NC_009891/Canada/20: Seal picornavirus type 1; KuV/KC935379/Hungria/2011: Kunsagivirus 1; HAV/NC_001489: Hepatitis A virus; AEV/NC_003990: Avian encephalomyelitis virus; PSV1/NC/003987: Porcine sapelovirus 1; EVC96/KF495604/China/2011: Human enterovirus C96; SVV001/NC_011349/USA/2002: Seneca valley virus; EMCV/NC_001479: Encephalomyocarditis virus; MSP1/JQ814851/China/2010: Miniopterus schreibersii picornavirus 1; ERAV/NC_003982: Equine rhinitis A virus; HCoV/NC_023984/Holanda/19: Human cosavirus; ERBV1/NC_003983: Equine rhinitis B virus 1; PTV1/NC_003985: Porcine teschovirus 1; BHuV1/NC_018668/Hungria/2: Bovine hungarovirus 1; >MoVA2/NC_023987/Hungria/2: Mosavirus A2; RoV2/NC_024070/Gambia/200: Rosavirus 2; CPDV/NC_021178/HongKong/2: Canine picodicistrovirus; TVH/NC_021201/USA/2008-20: Turkey hepatitis virus 2993D; SaliV-FHB/NC_025114/China: Salivirus FHB; TV3/NC_014413/China/2006: Turdivirus 3; SakoVA/NC_022802/Portugal: Feline sakobuvirus A; AV/NC_001918/Paquistao/19: Aichi virus; TuGV/NC_018400/Hungra/201: Turkey gallivirus; TV1/NC_014411/China/2007: Turdivirus 1; JSY/NC_028380/China/2015: Chicken sicinivirus JSY. Phylogenetic analysis was repeated independently three times with consistent results.
The boxplot (Fig. 4) demonstrates the evolutionary distances between species within this genus. It shows several outliers within the unclassified clade, probably due to the high diversity among these viruses. The supplementary figures and tables provide additional information related to the JUAV’s molecular analysis.
Juruaça virus NCBI code is PP911331. The BLAST results indicated a relationship between JURV and tombo-like viruses, suggesting an affiliation with the Tombusviridae family.
Changes in cytokine production in the brains of infected neonatal mice
Graphical representations of the progressive increase in inflammatory cytokine production in the brain samples collected from infected neonatal mice at 4, 8, and 12 dpi are shown in Fig. 5.
Cytokine levels in brain homogenates from control (n = 27) and JUAV-infected neonatal mice collected at 4 days post-infection (dpi) (n = 9), 8 dpi (n = 9), and 12 dpi (n = 9). The Y-axis represents cytokine concentrations expressed in pg/mL. Data are presented as means ± standard error of the mean (SEM). Cytokine measurements include: A IFN-γ, B TNF-α, C IL-1β, D IL-6, E IL-12p40, F IL-10, and G IL-4, and H NO. Notably, IFN-γ and TNF-α levels were highest at 12 dpi, whereas IL-1β, IL-6, and IL-12p40 peaked at 8 dpi and then decreased. IL-6 and IL-12p40 remained significantly elevated compared to control values at 12 dpi. Conversely, IL-10 and IL-4 concentrations decreased as the disease progressed. A significant reduction in NO production was observed only at 12 dpi. Statistical significance was determined using two-way ANOVA with Bonferroni correction for multiple comparisons (*p < 0.05; **p < 0.01; ***p <0.001). Data are representative of three independent experiments.
Sickness behavior in neonates with acute infection
Table 1 shows the relationships between clinical signs and inflammatory events in the neonatal mouse brain. Sickness behavior in the BALB/c mice included ruffled fur, tremors, a hunched posture, weight loss, and hindlimb paralysis. The clinical signs were more intense at 12 dpi, which coincided with increased TNF-α and IFN-γ production and decreased IL-10 production. At this stage of infection, the production of the proinflammatory cytokines IL-6 and IL-1β had already declined.
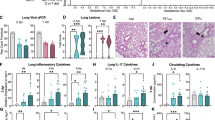
As the disease progressed, contrasting body weight patterns were observed between neonatal and adult JUAV-infected BALB/c mice (Fig. 8). Whereas the body weight changes in the neonates occurred shortly after infection (days post-infection), those in adult mice occurred late (weeks post-infection).
Neuropathological features of infected neonatal mice
Mice intracerebrally inoculated with Juruaça virus (JUAV) exhibited acute fatal infections, characterized by detectable symptoms such as tremors and subsequent flaccid paralysis, evident by 4 days post-infection (dpi). The clinical signs exhibited by neonate mice are the result of a complex interplay between neuronal virus infection and the subsequent inflammatory response elicited by the host’s immune system. Furthermore, our primary cell culture studies have revealed that both glial cells and neurons are susceptible to JUAV infection. The presence of nonenveloped viral particles in the brain was concomitant with the onset of these clinical manifestations. At 8 dpi, the infected neonatal BALB/c mice exhibited significant JUAV antigen immunolabeling in the olfactory bulb, cortex, hippocampus, and cerebellum (Fig. 6). Histopathological analysis at 12 dpi revealed vascular congestion in the meninges, choroid plexus, and olfactory bulb and severe tissue damage in cortical areas. The observed cortical tissue damage included vacuolar edema and coagulative necrosis across the parenchyma, pyknosis, karyorrhexis and apoptotic neuronal and glial cell bodies. Activated macrophages and swollen reactive astrocytes exhibited eccentric nuclei, an abundant hyaline cytoplasm, and prototypical processes (gemistocytic astrocytes) in response to tissue damage, and leukocyte infiltration and perivascular hemorrhagic foci with red blood cells outside the vascular bed were visible (Fig. 7).
Photomicrographs of brain sections from control (A and B, n = 9) and JUAV-infected neonatal mice (C–F, n = 9) at 8 days post-infection (dpi), immunostained with an anti-JUAV polyclonal antibody. A and B Control brain sections showing the absence of immunolabeling. C–F Infected brain sections showing increased viral antigen immunolabeling in cell bodies within the olfactory bulb (C), cortex (D), hippocampus (E), and cerebellum (F). The same individuals were used for both histopathology and immunohistochemistry. Data shown are representative of two independent experiments with similar results.
Photomicrographs of hematoxylin and eosin (HE)-stained histological sections of the brains of control (A, n = 3) and JUAV-infected neonatal mice (B–H, n = 9) at 12 days post-infection (dpi). A Control brain section showing normal histology. B Infected brain sections showing: C vascular congestion in the olfactory bulb (white arrows). D Diencephalic hemorrhagic focus (circle) and karyorrhexis (black arrows). E Vascular congestion in the 4th ventricle choroid plexus (white arrows). F Red blood cells outside the vascular lumen in the cortical region (circle). G Typical apoptotic cell bodies in the olfactory bulb (yellow arrowheads). H Severe necrotic damage, apoptotic cell bodies, and leukocytes in the cortical areas. Histopathological findings were consistent across three independent experiments.
The observed histopathological changes at 12 dpi were consistent with a reactive microglial response, with microglia exhibiting enlarged cell bodies, shortened and thickened processes, and a macrophage-like rounded morphology, in contrast to a surveillant nonreactive microglial response characterized by smaller cell bodies and thinner and ramified processes. These morphological responses of microglia were found in all cortical areas, the olfactory bulb, the anterior olfactory nucleus, and near the lateral ventricles of the diencephalic region (Fig. 8).
Photomicrographs of IBA-immunostained brain sections from JUAV-infected neonatal mice (n = 9) at 12 days post-infection (dpi). A Low-magnification image showing microglia in the olfactory bulb. C Increased microglial density near the lateral ventricle. E Increased microglial density in the cerebellum. G Increased microglial density at the borders of adjacent cortical and mesencephalic regions. B, D, F, and H High-magnification views of the rectangular regions indicated in the low-power images, demonstrating morphological changes in microglia at various stages of reactivity. White arrows indicate microglia with thinner and thicker processes, while black arrows and arrowheads indicate round macrophage-like microglia and meningeal microglia, respectively. Control mouse sections (not shown) exhibited non-activated microglia. Scale bars: A, C, E, and G = 550 µm; B, D, F, and H = 100 µm; I = 1 mm. Microglial activation patterns were confirmed in two independent experiments.
Sickness behavior and neuropathological features associated with chronic JUAV infection in adult mice
Table 2 demonstrates the marked variability in sickness behavior among infected adults as the infection progressed. Some mice were not sickened after inoculation, and among those who did become ill, some continued to decline and die, while others recovered and returned to a healthy state. Table 2 shows the data of 10 individual mice observed daily for 100 consecutive days.
As the infection progressed, the sickness behavior and body weight loss of the adult mice varied. Indeed, while three distinct clinical groups (G1, G2, and G3) were established based on the manifestation of clinical signs, only the individuals with severe signs (G3) exhibited significant weight loss as the infection progressed (Fig. 9).
A Body weight changes in neonatal mice following JUAV infection. Neonatal mice (control, n = 10; infected, n = 10) received a single intranasal instillation of 20 µL of brain homogenate from infected or uninfected (control) mice. Significant weight loss was observed in the infected neonatal mice starting at 7 days post-instillation. B Body weight changes in adult mice following JUAV infection. Adult mice (control, n = 5; infected, n = 15) received daily intranasal administrations over three consecutive days. Significant weight loss was observed only in the infected group, coinciding with severe clinical signs at 35 days post-infection. Statistical analysis for neonatal mice (A) was performed using two-way ANOVA with Bonferroni correction for multiple comparisons (***p < 0.001; p < 0.01; p < 0.05). Statistical analysis for adult mice (B) was performed using two-way ANOVA with Tukey’s post hoc test (***p < 0.001; **p < 0.01; *p < 0.05). Weight measurements were recorded from three independent cohorts of mice.
Sickness behavior was highly variable in JUAV-infected adult mice. Some infected mice exhibited no clinical signs (G1), and other infected mice exhibited many clinical signs, such as ruffled fur, a hunched posture, body weight loss, hindlimb clasping, and paralysis. In adult mice, the initial sign—ruffled fur in the cervical region—appeared at 30 dpi in two of the 10 infected adult mice. In the following days, fur ruffling spread to other regions of the body in four of the individual mice, whose tails also showed hindlimb clasping behavior upon tail suspension (Fig. 9B and F). At 50 dpi, 5 mice exhibited reduced mobility, paralysis of one or both hindlimbs, a hunched posture, and weight loss. Three mice died between 53 and 65 dpi. All the dead animals exhibited ruffled fur, hindlimb clasping, severely hunched posture, hindlimb paralysis, and severe weight loss. The survivors (n = 7) exhibited ruffled fur, hindlimb clasping, weight loss, and slightly hunched posture. Hindlimb paralysis occurred in only three of the survivors. Two of the seven survivors exhibited no clinical signs.
Counterintuitively, the mice in group 2 showed significant decreases in brain mass, specifically in the telencephalon, cerebellum, and brainstem. These animals did not experience weight loss (G2) by the time of sacrifice (100 dpi) (Fig. 10G, right) but did exhibit hindlimb clasping behavior. All symptomatic mice in group 3 exhibited CNS atrophy, which appeared to be associated with reductions in both gray and white matter.
Panels showing variability in sickness behavior in JUAV-infected adult mice: A and E Mice from Group 1 (G1) exhibiting no clinical signs. B and F An infected mouse from Group 2 (G2) showing moderate clinical signs, including ruffled fur, hunched posture, and hindlimb clasping. C and D Mice from Group 3 (G3) displaying severe clinical signs such as hindlimb paralysis, significant weight loss (C), and pronounced hunched posture (D). G Comparison of brain mass between a control uninfected brain (left, n = 13) and an infected brain from a G2 mouse (right), showing severe brain mass loss. Both G2 and G3 animals exhibited similar brain shrinkage (n = 36). Clinical observations were replicated in two independent experiments.
JUAV antigen expression in the brains of infected adult mice
Only infected mice in groups G2 and G3 showed immunolabeling of viral antigens in the brain. Immunolabeling was more intense in the spinal cord and the temporal and frontal neocortical regions, but many other CNS regions, including the thalamus, hypothalamus, cerebellum, hippocampus, and striatum, also exhibited immunolabeling (Fig. 11).
Photomicrographs of brain tissue sections from control (n = 5, A–C) and JUAV-infected (n = 15, D–L) mice, immunolabeled for viral antigens and counterstained with hematoxylin. A–C Control mice showing no viral antigen immunolabeling in the cerebral cortex (A), cerebellum (B), or spinal cord (C). D–F Infected Group 1 (G1) mice showing no viral antigen immunolabeling in the cerebral cortex (D), cerebellum (E), or spinal cord (F). G–I Infected Group 2 (G2) mice showing conspicuous viral antigen immunolabeling (arrows) in the cerebral cortex (G), cerebellum (H), and spinal cord (I). J–L Infected Group 3 (G3) mice showing conspicuous viral antigen immunolabeling (arrows) in the cerebral cortex (J), cerebellum (K), and spinal cord (L). Scale bar = 50 µm. Immunolabeling data were consistent across three independent experiments.
Surveillant and reactive microglia
Compared with the brains of control uninfected and infected mice in G1, the brains of infected adult animals in G2 and G3 showed intensely IBA-1-positive reactive microglia (Fig. 12).
Photomicrographs of IBA-1-immunolabeled brain sections from control (n = 5) and JUAV-infected (n = 10) adult BALB/c mice. A–C Surveillant microglia in control mice: A cerebral córtex, B hippocampus, C spinal cord, D–F microglia from Group 1 (G1)-infected mice displaying minimal reactivity: D cerebral córtex, E hippocampus, F spinal cord, G–I reactive microglia in Group 2 (G2) infected mice: G cerebral cortex, H hippocampus, I spinal cord, J–L reactive microglia in Group 3 (G3) infected mice: J cerebral cortex, K hippocampus, and L spinal cord. Scale bar = 50 µm. Observations were confirmed in three independent experiments.
Indeed, smaller cell bodies with thinner and longer processes were observed in microglia from the control and G1 mice, while microglia with the typical reactive morphology (larger somata, thicker processes, and multiple shorter processes) were found at greater densities in the olfactory bulb, olfactory nucleus, putamen, cerebral cortex, mesencephalon, thalamus, cerebellum, spinal cord, and meninges in the G2 and G3 mice.
Cytokine production and ultrastructural changes in the brain in chronically infected adult mice
Counterintuitively, the levels of inflammatory cytokines (TNF-α, IL-6, IL-1β, and IFN-γ) in the brains of uninfected control mice and in the brains of chronically infected mice were indistinguishable. Indeed, in chronically infected animals with severe clinical signs, which exhibited reactive microglia and viral particles in the brain parenchyma, the levels of IL-12p70, TNF-α, IFN-γ, IL-6, and IL-10 in the blood and brain tissue did not differ significantly from those in control mice. However, the brain tissue level of the chemokine monocyte chemoattractant protein-1 (MCP-1) was closely associated with the severity of clinical signs (Fig. 13G).
Top: Electron micrographs of ultrathin brain sections from uninfected (n = 3, A) and severely infected BALB/c G3 (n = 3) mice, potentially exhibiting JUAV-induced encephalomyelitis. A Frontal cortex of an uninfected animal showing normal cell preservation. B The frontal cortex of a severely infected G3 mouse shows a necrotic cell with cytoplasmic destruction (arrow). C and D Thalamic sections from an infected G3 mouse showing myelin sheath damage (arrows). E Frontal cortex of an infected G3 mouse demonstrating an area of edema due to cell death. F Spherical electron-dense particles in the thalamus of an infected G3 mouse, suggesting the presence of JUAV particles (ellipse). Bottom: G Average MCP-1 levels in the brain in relation to disease severity. Control group (CG), Group 1 (G1) with no clinical signs, Group 2 (G2) with moderate clinical signs, and Group 3 (G3) with severe clinical signs. Fifty percent of infected mice exhibited aggravated sickness behavior after 50 days post-infection (dpi), including hunched posture, hindlimb paralysis, weight loss, and a mortality rate of 30%. Statistical analysis was performed using the Mann–Whitney U test. **p < 0.01. Electron microscopy and MCP-1 measurements were performed in two independent experiments.
While the levels of NO (measured with Griess reagent) and proinflammatory cytokines (measured by flow cytometry) were significantly increased in the brains of acutely infected neonatal mice, those in the brains of chronically infected animals did not differ significantly from those in uninfected control mice.
However, the level of MCP-1 in the brain was directly proportional to the intensity of clinical signs, with higher levels of this chemokine in G3 mice. We suggest that the increased production of MCP-1 in the brains of JUAV-infected adult mice may play a major role in the associated chronic neuroinflammation and variability in sickness behavior.
From 70 to 100 dpi, when all the remaining mice were sacrificed, no worsening or improvement of clinical signs was observed in the animals that survived the viral infection.
Ultrastructural analysis revealed necrosis and myelin sheath damage in infected mice in G2 and G3. Indeed, the presence of spherical electron-dense particles in the thalamus in G3 mice suggested the presence of JUAV-like viral particles in the parenchyma (Fig. 13A–F).
The supplementary figures and tables contain additional information regarding cytokine production and body weight changes during JUAV disease progression.
Discussion
Disease tolerance and host resistance represent crucial mechanisms within the innate immune response that mitigate the negative impact of pathogenic infections1,37. This complex interplay is characterized by variability in immune responses among individuals38, influenced by genetic, epigenetic, and environmental factors39,40,41,42,43.
We utilized neonatal and young adult BALB/c mice to explore sickness behavior and neuropathological features following acute and chronic Juruaça virus (JUAV) infection. Sequencing of the JUAV genome revealed molecular similarities with members of the unclassified Tombusviridae family and mute swan feces-associated tombus-like virus 635,36. However, the provisional classification of JUAV requires further validation.
Our in vivo investigations into JUAV infection unveiled significant intra- and intergroup variability, encompassing clinical signs, inflammatory cytokine production, histopathology, and neuropathology in both neonatal and adult mice.
The Tombusviridae family, encompassing diverse positive-sense RNA plant viruses, shares common genomic characteristics across its genera44. Mute swan feces-associated tombus-like virus 6, identified in mute swan feces in China, belongs to this family36. While interspecies virus transmission is rare, adaptive mutations can facilitate cross-species infections45. If JUAV indeed shares ancestry with mute swan feces-associated tombus-like virus 6, it suggests a potential origin from a plant virus, with subsequent adaptation for mammalian infection, impacting bats and neonatal and adult mice differently46,47,48,49.
During acute JUAV infection in neonates, injury to infected cells initiates local inflammation, marked by increased proinflammatory cytokines (IL-1β, IL-6, IFN-γ, TNF-α) and the chemokine MCP-1. Some neonatal mice exhibited resolution and recovery, while others displayed dysfunctional responses, leading to widespread inflammation and multiorgan failure50.
The presence of red blood cells in Fig. 7 suggests significant tissue damage within the central nervous system. This damage is likely due to severe necrosis, which compromises the integrity of blood vessels, leading to hemorrhage and the infiltration of red blood cells into the surrounding tissue. Our analysis of proinflammatory cytokines in the brain supports this conclusion, indicating a strong inflammatory response that exacerbates tissue damage and contributes to the observed hemorrhage51.
In contrast, adult mice exhibited a subdued cytokine response but sustained MCP-1 levels, correlating with inflammatory infiltration in various organs. Chronic infection persisted in adults, potentially exceeding tolerance thresholds52,53.
Transcriptomic analyses in rodent models highlight differential responses to inflammation, with P. leucopus exhibiting greater tolerance, underscoring the importance of inflammatory modulation in resistance and tolerance mechanisms54. Similarly, diverse host defense gene programs in response to influenza infections illustrate varying tolerance and resistance mechanisms55.
Microglia are pivotal cells governing the innate immune response within the brain. When triggered by external stimuli, they undergo activation, releasing proinflammatory mediators associated with neurotoxicity and neuroinflammation. Alternatively, microglia can adopt a neuroprotective phenotype, wherein they release anti-inflammatory mediators56,57,58. Neonatal mice displayed heightened proinflammatory cytokines, while adults exhibited attenuated responses, associated with brain and spinal cord atrophy and motor dysfunction59. Chronic infections can induce T-cell exhaustion and hinder memory T-cell generation, exacerbating disease progression60,61.
Though consistent with findings in other chronic viral infections, our study did not explore T-cell responses, necessitating further investigations into underlying mechanisms62,63,64.
While our study provides significant insights into the taxonomic classification of Juruaça virus (JUAV) and its effects on neonatal and adult BALB/c mice, several limitations warrant consideration. Firstly, the lack of a reference genome and potential gaps in sequencing coverage contribute to taxonomic uncertainty. Enhancing genomic sequencing methodologies to obtain a complete reference genome is crucial for a more robust classification of JUAV.
Secondly, the intranasal approach used for viral infection, though commonly employed, may need to be more consistent in viral delivery and distribution. This method only partially replicates natural infection routes, potentially affecting the generalizability of our findings. Future studies should explore alternative infection routes to improve the accuracy and reliability of infection models.
Furthermore, the unusually high variability in clinical signs among infected adult mice complicates data interpretation. This variability underscores the inherent biological differences in virus-host interactions and emphasizes the need for larger sample sizes and rigorous statistical analyses to account for such variability.
Our study also lacked comprehensive immune cell analyses and the expansion of the cytokine panel to include additional interleukins and interferons. Future research should include these analyses to provide a more detailed understanding of the immune response to JUAV.
Additionally, the absence of studies on the potential transmission of JUAV between mice and the presence of the virus in fecal samples limits our understanding of its transmission dynamics. Investigating these aspects in future studies will be crucial for a comprehensive understanding of JUAV’s epidemiology.
Lastly, while our validation of the anti-JUAV polyclonal antibody was thorough, more extensive validation and characterization of immune responses, including T cell infiltration and peripheral macrophage involvement, are necessary to elucidate JUAV infection’s pathogenesis fully.
Addressing these limitations in future research will enhance the robustness and reliability of our findings and contribute to a deeper understanding of JUAV’s biology and its impact on mammalian hosts. By refining our methodologies and expanding our analyses, we aim to provide more comprehensive insights into viral pathogenesis, host immune responses, and potential therapeutic interventions1,65,66.
Data availability
All relevant data are provided within the manuscript and its supporting information files. Additional data are publicly available on NCBI under the accession code PP11331.
References
Medzhitov, R., Schneider, D. S. & Soares, M. P. Disease tolerance as a defense strategy. Science 335, 936–941 (2012).
Rivas, F. V., Chervonsky, A. V. & Medzhitov, R. ART and immunology. Trends Immunol. 35, 451 (2014).
Reed, H. & Muench, H. A simple method of estimating fifty per cent endpoints. Am. J.f Epidemiol. 27, 493–497 (1938).
Kavaliers, M., Choleris, E., Agmo, A. & Pfaff, D. W. Olfactory-mediated parasite recognition and avoidance: linking genes to behavior. Horm. Behav. 46, 272–283 (2004).
Curtis, V. A. Infection-avoidance behaviour in humans and other animals. Trends Immunol. 35, 457–464 (2014).
Råberg, L., Graham, A. L. & Read, A. F. Decomposing health: tolerance and resistance to parasites in animals. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 37–49 (2009).
Pereira, P. D. C. et al. Genes, inflammatory response, tolerance, and resistance to virus infections in migratory birds, bats, and rodents. Front. Immunol. 14, 1239572 (2023).
Sadler, A. J. & Williams, B. R. Interferon-inducible antiviral effectors. Nat. Rev. Immunol. 8, 559–568 (2008).
Lang, R. et al. Expression and mechanisms of interferon-stimulated genes in viral infection of the central nervous system (CNS) and neurological diseases. Front. Immunol. 13, 1008072 (2022).
Shah, V. K., Firmal, P., Alam, A., Ganguly, D. & Chattopadhyay, S. Overview of immune response during SARS-CoV-2 infection: lessons from the past. Front. Immunol. 11, 1949 (2020).
Cuenca, A. G., Wynn, J. L., Moldawer, L. L. & Levy, O. Role of innate immunity in neonatal infection. Am. J. Perinatol. 30, 105–112 (2013).
Clemens, E. A. & Alexander-Miller, M. A. Understanding antibody responses in early life: baby steps towards developing an effective influenza vaccine. Viruses 13, (2021). https://doi.org/10.3390/v13071392
Diggins, N. L. & Hancock, M. H. Viral miRNA regulation of host gene expression. Semin. Cell Dev. Biol. 146, 2–19 (2023).
Fairweather, D., Kaya, Z., Shellam, G. R., Lawson, C. M. & Rose, N. R. From infection to autoimmunity. J. Autoimmun. 16, 175–186 (2001).
Phelps, A. L. et al. Aerosol infection of Balb/c mice with eastern equine encephalitis virus; susceptibility and lethality. Virol. J. 16, 2 (2019).
Ortiz-Perez, A. et al. Innate immunity and pathogenesis of biliary atresia. Front. Immunol. 11, 329 (2020).
Vahlenkamp, T. W., Tompkins, M. B. & Tompkins, W. A. The role of CD4+CD25+ regulatory T cells in viral infections. Vet. Immunol. Immunopathol. 108, 219–225 (2005).
Jarnagin, K., Alvarez, O., Shresta, S. & Webb, D. R. Animal models for SARS-Cov2/Covid19 research-A commentary. Biochem. Pharmacol. 188, 114543 (2021).
Campetella, O. E., Barrios, H. A. & Galassi, N. V. Suppressor T-cell population induced by Junin virus in adult mice. Immunology 64, 407–412 (1988).
Eichinger, K. M. et al. Alveolar macrophages support interferon gamma-mediated viral clearance in RSV-infected neonatal mice. Respir. Res. 16, 122 (2015).
Theil, J. H. et al. The epidemiology of fighting in group-housed laboratory mice. Sci. Rep. 10, 16649 (2020).
Dantzer, R. Cytokine-induced sickness behaviour: a neuroimmune response to activation of innate immunity. Eur. J. Pharmacol. 500, 399–411 (2004).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Myung, I. Tutorial on maximum likelihood estimation. J. Math. Psychol. 47, 90–100 (2003).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Felsenstein, J. Phylogenies and the comparative method. Am. Nat. 125, 1–15 (1985).
Shu, S., Ju, G. & Fan, L. The glucose oxidase-DAB-nickel method in peroxidase histochemistry of the nervous system. Neurosci. Lett. 85, 169–171 (1988).
Matos, G. C. et al. Experimental infection of golden hamsters with Guama virus (Peribunyaviridae, Orthobunyavirus). Microb. Pathog. 135, 103627 (2019).
Karnovsky, M. A formaldehyde–glutaraldehyde fixative of high osmolality for use in electron microscopy. J. Cell Biol. 27, 1A–149A (1965).
Moura Neto, V., Mallat, M., Jeantet, C. & Prochiantz, A. Microheterogeneity of tubulin proteins in neuronal and glial cells from the mouse brain in culture. EMBO J. 2, 1243–1248 (1983).
Almeida, J. D. Practical aspects of diagnostic electron microscopy. Yale J. Biol. Med. 53, 5–18 (1980).
Brenner, S. & Horne, R. A negative staining method for high resolution electron microscopy of viruses. Biochim. Biophys. Acta 34, 103–110 (1959).
García-Robledo, E., Sokratis & Papaspyrou, A. A fast and direct spectrophotometric method for the sequential determination of nitrate and nitrite at low concentrations in small volumes. Mar. Chem. 162, 30–36 (2014).
Cardiff, R. D., Miller, C. H. & Munn, R. J. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb. Protoc. 2014, 655–658 (2014).
Park, J., Kim, J., Kim, J., Park, J. & Kim, J. Mute swan feces associated tombus-like virus 6, a novel virus in the family Tombusviridae. bioRxiv (2022).
Schoch, C. L. et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford) 2020, (2020). https://doi.org/10.1093/database/baaa062.
Råberg, L., Sim, D. & Read, A. F. Disentangling genetic variation for resistance and tolerance to infectious diseases in animals. Science 318, 812–814 (2007).
Stegelmeier, A. A. et al. Type I Interferon-mediated regulation of antiviral capabilities of neutrophils. Int. J. Mol. Sci. 22, (2021). https://doi.org/10.3390/ijms22094726
Ivashkiv, L. B. & Donlin, L. T. Regulation of type I interferon responses. Nat. Rev. Immunol. 14, 36–49 (2014).
Liu, J., Qian, C. & Cao, X. Post-translational modification control of innate immunity. Immunity 45, 15–30 (2016).
Chen, K., Liu, J. & Cao, X. Regulation of type I interferon signaling in immunity and inflammation: a comprehensive review. J. Autoimmun. 83, 1–11 (2017).
Zhou, Y., He, C., Wang, L. & Ge, B. Post-translational regulation of antiviral innate signaling. Eur. J. Immunol. 47, 1414–1426 (2017).
Barrat, F. J., Crow, M. K. & Ivashkiv, L. B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat. Immunol. 20, 1574–1583 (2019).
Hyodo, K. & Okuno, T. Hijacking of host cellular components as proviral factors by plant-infecting viruses. Adv. Virus Res. 107, 37–86 (2020).
Li, Q. et al. Cross-species transmission, evolution and zoonotic potential of coronaviruses. Front. Cell. Infect. Microbiol. 12, 1081370 (2022).
Colson, P. et al. Pepper mild mottle virus, a plant virus associated with specific immune responses, Fever, abdominal pains, and pruritus in humans. PLoS ONE 5, e10041 (2010).
Balique, F., Lecoq, H., Raoult, D. & Colson, P. Can plant viruses cross the kingdom border and be pathogenic to humans? Viruses 7, 2074–2098 (2015).
Jiwaji, M. et al. Providence virus: an animal virus that replicates in plants or a plant virus that infects and replicates in animal cells? PLoS ONE 14, e0217494 (2019).
Kim, J. S., Yoon, S. J., Park, Y. J., Kim, S. Y. & Ryu, C. M. Crossing the kingdom border: Human diseases caused by plant pathogens. Environ. Microbiol. 22, 2485–2495 (2020).
Frank, M. G., Fleshner, M. & Maier, S. F. Exploring the immunogenic properties of SARS-CoV-2 structural proteins: PAMP:TLR signaling in the mediation of the neuroinflammatory and neurologic sequelae of COVID-19. Brain Behav. Immun. 111, 259–269 (2023).
Carod-Artal, F. J. Stroke in central nervous system infections. Ann. Indian Acad. Neurol. 11, S64–S78 (2008).
Shi, C. & Pamer, E. G. Monocyte recruitment during infection and inflammation. Nat. Rev. Immunol. 11, 762–774 (2011).
Fantuzzi, L., Tagliamonte, M., Gauzzi, M. C. & Lopalco, L. Dual CCR5/CCR2 targeting: opportunities for the cure of complex disorders. Cell. Mol. Life Sci. 76, 4869–4886 (2019).
Balderrama-Gutierrez, G. et al. An infection-tolerant mammalian reservoir for several zoonotic agents broadly counters the inflammatory effects of endotoxin. mBio 12, https://doi.org/10.1128/mBio.00588-21 (2021).
Cohn, O. et al. Distinct gene programs underpinning disease tolerance and resistance in influenza virus infection. Cell Syst. 13, 1002–1015.e1009 (2022).
Garaschuk, O. & Verkhratsky, A. Physiology of microglia. Methods Mol. Biol. 2034, 27–40 (2019).
Jurga, A. M., Paleczna, M. & Kuter, K. Z. Overview of general and discriminating markers of differential microglia phenotypes. Front. Cell. Neurosci. 14, 198 (2020).
Paolicelli, R. C. et al. Microglia states and nomenclature: a field at its crossroads. Neuron 110, 3458–3483 (2022).
Pirko, I. et al. Brain atrophy correlates with functional outcome in a murine model of multiple sclerosis. Neuroimage 54, 802–806 (2011).
Utzschneider, D. T. et al. T cell factor 1-expressing memory-like CD8(+) T cells sustain the immune response to chronic viral infections. Immunity 45, 415–427 (2016).
Snell, L. M. et al. CD8. Immunity 49, 678–694.e675 (2018).
Ye, B. et al. T-cell exhaustion in chronic hepatitis B infection: current knowledge and clinical significance. Cell Death Dis. 6, e1694 (2015).
Jofra, T. et al. Extrinsic protein tyrosine phosphatase non-receptor 22 signals contribute to CD8 T cell exhaustion and promote persistence of chronic lymphocytic choriomeningitis virus infection. Front. Immunol. 8, 811 (2017).
Apol, Á., Winckelmann, A. A., Duus, R. B., Bukh, J. & Weis, N. The role of CTLA-4 in T cell exhaustion in chronic hepatitis B virus infection. Viruses 15, (2023). https://doi.org/10.3390/v15051141
Martins, R. et al. Disease tolerance as an inherent component of immunity. Annu. Rev. Immunol. 37, 405–437 (2019).
Ayres, J. S. Surviving COVID-19: a disease tolerance perspective. Sci. Adv. 6, eabc1518 (2020).
Acknowledgements
Brazilian National Research Council—CNPq INCT-VER, 406360/2022-7; Brazilian National Research Council—CNPq, 101716/2024-9; Brazilian National Research Council—CNPq, 303270/2021-7; Brazilian National Research Council—CNPq, 310295/2021-1; Brazilian National Research Council—CNPq, 407075/2021-6; Brazilian National Research Council—CNPq, 301268/2019-3.
Author information
Authors and Affiliations
Contributions
T.S.C., N.C.C., T.P.A., I.T.E.P., and S.P.S. equally contributed to the experimental data collection and analysis. J.A.P.D., J.A.S.Q., P.F.C.V., D.B.A.M., A.P.D.R., and S.P.S. provided data analysis and interpretation, K.G.B.C. and T.S.P. helped to collect experimental data, D.G.D. format figures according to the NPJ rules; D.G.D., J.A.P.D., D.B.A.M., and C.W.P.D. wrote the draft and the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
da Silva Cabral, T., Cayuela, N.C., Carvalho, K.G.B. et al. Juruaça virus taxonomy, tolerance and resistance to infection, and inflammatory response modulation in murine model. npj Viruses 2, 46 (2024). https://doi.org/10.1038/s44298-024-00056-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s44298-024-00056-y