Abstract
Bone metastsis in advanced breast cancer patients are usually osteolytic. A better understanding of the mechanisms in osteolytic metastasis is critical for the development of new therapies. YTH domain-containing family protein 3 (YTHDF3) has been reported to function as an N6-methyladenosine (m6A)-modified mRNA regulator. In this study, we found YTDHF3 expression was associated with clinical characteristics of breast cancer patients. YTHDF3 expression influenced the migration and invasion capacity of breast cancer cells in vitro and in vivo, and low expression of YTHDF3 suppressed cancer cell-induced osteoclast differentiation and osteolytic bone destruction. Moreover, we found YTHDF3 enhanced the translation of zinc finger E-box-binding protein 1 (ZEB1) and SMAD family member 5 (SMAD5) by reading the m6A modification sites in their mRNAs and further promoted the epithelial-mesenchymal transition (EMT) of breast cancer cells. Enhanced expression of ZEB1 promoted the transcription of bone morphogenetic protein inhibitors such as NOG, FST and CCN2, which boosts osteolytic metastasis. Furthermore, we newly found Wnt family member 5B (WNT5B) expression was regulated by ZEB1, also involved in osteolytic process. In conclusion, YTHDF3 plays an important role in osteolytic metastasis and it may serve as a potential prognostic biomarker and therapeutic target for breast cancer bone metastasis.
Similar content being viewed by others
Introduction
Among females, breast cancer is one of the leading causes of cancer-related death worldwide [1]. A high proportion of breast cancer deaths are caused by metastatic disease. It was reported that the 5-year survival rate of patients with metastatic breast cancer is approximately 26% based on Surveillance, Epidemiology, and End Results (SEER) data [2]. Multiple organs, such as bone, brain, lungs and liver, may be involved in breast cancer metastasis. Bone is one of the most predominant sites of breast cancer metastasis, accounting for nearly 50% of advanced breast cancer [3]. Bone lesions in advanced breast cancer patients are usually osteolytic, leading to severe pain, neurological dysfunction, and even pathological fracture [4]. Due to the influence of bone metastasis on survival and quality of life, an improved understanding of the mechanism is essential for survival and the development of new treatments.
As one of the most abundant modifications in eukaryotic mRNA, N6-methyladenosine (m6A) plays a critical role in biological processes through involvement with RNA splicing, translation, and stability [5,6,7]. In addition, m6A modification plays regulatory roles in numerous cellular functions, such as cell self-renewal, differentiation, invasion, and apoptosis [8]. The impact of m6A modification on cancer cells depends on cancer-associated genes. A growing amount of evidence suggests that m6A modification plays a dual role in cancer development [9, 10]. Cancer progression may be influenced by m6A modification through regulating the expression of oncogenes or anti-oncogenes. In addition, the regulation of m6A levels may also impact the role of m6A in cancer development. Current studies tend to assume that the way m6A affects cancer progression depends on the function of target genes (oncogenes or anti-oncogenes), the m6A level in cancer and the regulation of m6A-methylated RNA [8]. Several m6A binding proteins called “readers” are able to regulate m6A-methylated RNAs, for example, YTH domain-containing proteins (such as YTHDF1-3, YTHDC1 and YTHDC2). As an important reader of m6A regulators, YTHDF3 promotes the translation efficiency of target genes by recognizing m6A sites in target RNA [11]. Recent studies have revealed that YTHDF3 regulates progression, proliferation, and invasion in various cancers. In ocular melanoma, YTHDF3 improves the translation of catenin beta 1 (CTNNB1) by reading the m6A sites in CTNNB1 mRNA, thereby enhancing the migration and invasion of ocular melanoma cells [12]. A similar promotion phenomenon was found in hepatocellular carcinoma and bladder cancer [13, 14]. YTHDF3 improves the translation of genes that are critical to brain metastasis (ST6GALNAC5 and GJA1) and facilitates breast cancer brain metastasis [15]. However, the clinical correlation and expression of YTDHF3 in breast cancer bone metastasis and its function in the interaction between breast cancer cells and the bone microenvironment still require further study.
In the present study, YTDHF3 expression gradually increased in normal tissue, primary breast cancer and metastatic lesions and was also associated with overall survival (OS) and metastasis-free survival. Our results demonstrated that YTHDF3 expression was associated with the migration and invasion ability of breast cancer cells and that knockdown of YTHDF3 resulted in a decrease in metastasis to bone and lungs in mouse models. We also found that low expression of YTHDF3 suppressed cancer cell-induced osteoclast differentiation and osteolytic bone destruction. Moreover, YTHDF3 enhanced the translation of zinc finger E-box-binding protein 1 (ZEB1) and SMAD family member 5 (SMAD5) mRNAs and further promoted the epithelial mesenchymal transformation (EMT) of breast cancer cells. Enhanced expression of ZEB1 increased the transcription of osteoclastogenesis-associated secretory proteins such as bone morphogenetic protein (NOG, FST, CCN2) and Wnt family member 5B (WNT5B), thus promoting osteolytic bone metastasis.
Results
YTHDF3 expression in primary and metastatic breast cancer lesions and its relationship with prognosis
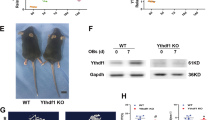
Differences in YTHDF3 expression between breast and paracancerous tissues in the TCGA BRCA dataset were analyzed using the GEPIA online database. Gene expression data from three datasets, GSE2034, were downloaded from the GEO database to analyze the differences in YTHDF3 expression in breast cancer tissue and bone metastases [16,17,18]. Although a previous article merged multiple datasets for expression analysis, in this study, we applied a single GEO dataset with 286 samples to exclude batch effects. The results from TCGA and GEO datasets revealed that the YTHDF3 mRNA expression level was significantly higher in breast cancer tissues than in paracancerous tissues. The expression of YTHDF3 was also significantly higher in bone metastases than in primary breast cancer foci (Fig. 1A, B). To further validate our findings, a breast cancer tissue microarray (No. HBreD055CD01) was purchased from Shanghai Xinchao Biotechnology Co., Ltd. The expression levels of YTHDF3 in breast cancer paracancerous tissues, and breast cancer primary tissues and metastatic tissues were analyzed by immunohistochemical techniques. The results showed that YTHDF3 was predominantly expressed in the cytoplasm of the tumor tissue. Notably, the protein levels of YTHDF3 exhibited a gradual increase from paraneoplastic to primary and metastatic tissues, aligning with the database analysis results (Fig. 1C, Supplementary Fig. S1A). Additionally, There was a significant elevation in YTHDF3 protein expression in matched bone metastasis lesions compared to primary tumor tissues (Fig. 1D). Furthermore, we investigated the promoter methylation level of YTDHF3 between normal tissue and tumors in the UALCAN database (https://ualcan.path.uab.edu/), and we found that the methylation level of YTHDF3 was higher in tumor tissue (Supplementary Fig. S1B). Through GISTIC online analysis, we explored YTHDF3 putative copy number variations (Supplementary Fig. S1C). GEPIA database analysis and GEO microarray analysis demonstrated that the expression level of YTHDF3 was significantly and negatively correlated with the overall survival and metastasis-free survival of breast cancer patients. Further analysis using the GEO microarray data suggested that YTHDF3 expression also significantly influenced metastasis-free survival in specific organs, such as bone metastasis-free survival, brain metastasis-free survival, and lung metastasis-free survival (Fig. 1E, F). We conducted WB assays to determine the differences in YTHDF3 expression between breast cancer wild-type cells and metastatic cells using 231 wild-type cells, 231 highly metastatic cells (231-HM) and two strains of 231 bone metastatic cells (231-BO-1, 231-BO-2). YTHDF3 expression in the normal breast epithelial cell line MCF10A and breast cancer cell lines 4T1, MCF7, 453, ZR-75-1, BT549, 468, BT20, and HS578T was further investigated through WB assays. Compared with wild-type 231 cells, metastatic cell lines were found to have significantly high expression of YTHDF3, which was more pronounced in bone metastatic cell lines (Fig. 1G). Moreover, YTHDF3 expression was higher in most of the breast cancer cell lines than in MCF10A cells (Fig. 1H). The qPCR assay revealed a similar expression trend of YTHDF3 in breast cancer cell lines (Supplementary Fig. S1D).
A Comparison of YTDHF3 expression in breast cancer tissues and paracancer tissues using TCGA database data. B Comparison of YTDHF3 expression in primary breast cancer lesions and metastatic tissues using GEO microarray data. C Representative images of IHC showing the YTHDF3 expression in breast cancer paracancer tissues, primary breast cancer foci tissue, and the results of H-score statistical analysis. D IHC image of YTHDF3 expression in primary breast cancer tissue and matched bone metastasis lesion. E YTHDF3 expression levels were negatively correlated with overall survival of breast cancer patients. F YTHDF3 expression was negatively correlated with metastasis-free survival and bone metastasis free survival of breast cancer patients. G YTHDF3 expression in different breast cancer cell lines. H YTHDF3 expression in 231 cells and its metastatic subtypes (231-HM: highly metastatic cell line; 231-BO: bone metastasis subtype).
YTHDF3 expression is related to the migration and invasion of breast cancer cells and the associated osteoblast differentiation-inducing ability
Using the YTHDF3 expression differences in various cell lines, 231 cells and 231-BO-1 cells were selected to construct YTHDF3-overexpressing and YTHDF3-knockdown stable cells, respectively. Among other breast cancer cells, 4T1 cells with low YTHDF3 expression and BT549 cells with high YTHDF3 expression were also chosen to construct stable overexpressing and knockdown cells, respectively. The stably transfected cells were constructed by a lentiviral infection technique and verified by WB assays (Fig. 2A, B, Supplementary Fig. S2A, B). Then, the effect of YTHDF3 overexpression on the migration and invasion ability of breast cancer cells was investigated through Transwell assays. The results showed that the overexpression of YTHDF3 in 231 and 4T1 cells significantly enhanced the migration and invasion ability of breast cancer cells (Fig. 2C, D, Supplementary Fig. S2C, D). In contrast, the migration and invasion ability of breast cancer cells significantly decreased after the knockdown of YTHDF3 in 231BO and BT549 cells (Fig. 2C, D, Supplementary Fig. S1C, D). From the wound healing assays, it was found that the migration of breast cancer cells increased after the overexpression of YTHDF3 but significantly decreased after the knockdown of YTHDF3 (Fig. 2E, G, Supplementary Fig. S2E, F). In addition, to investigate the effect of YTHDF3 on proliferative ability, we performed a plate colony formation assay and found no significant difference between the shNC and shYTHDF3 groups (Supplementary Fig. S4C). Angiogenesis assays found that the downregulation of YTDHF3 reduced angiogenesis in 231BO cells (Supplementary Fig. S4E). We hypothesize that YTDHF3 may promote bone metastasis by adhering to cells in the bone microenvironment. However, the cell adhesion assay revealed no significant change after YTHDF3 knockdown (Supplementary Fig. S4F).
A WB and qPCR verified overexpression of YTHDF3 in 231 cells. B WB and qPCR verified knockdown of YTHDF3 in 231BO cells. C, D Transwell assay revealed that YTHDF3 overexpression promoted migration and invasion ability of 231 cells, while knockdown of YTHDF3 in 231BO cells reversed these effects. E, G Wound heal assay revealed that YTHDF3 overexpression increased migration rate of 231 cells, while YTHDF3 knockdown decreased migration rate of 231BO cells. F, H Osteoclast-induced differentiation assay examining the effect of YTHDF3 knockdown or overexpression on the ability of breast cancer cell lines to induce osteoclast differentiation.
To test whether YTHDF3 expression in breast cancer cells could influence osteoclast differentiation, we performed qPCR to investigate changes in the expression of osteoclast differentiation-associated genes after YTDHF3 knockdown but found no positive results (Supplementary Fig. S4D). Furthermore, we treated primary mouse bone marrow cells with RANKL and M-CSF together with conditioned medium from various stably transfected breast cancer cells. YTHDF3 overexpression enhanced the ability of conditioned medium from breast cancer cell culture to induce osteoblast differentiation; the inducing effects were not observed in the conditioned medium from YTHDF3-knockdown breast cancer cells (Fig. 2F, H). Finally, we performed WB to explore changes in the expression of EMT markers in the abovementioned stable cell lines and found that Slug expression was positively correlated with YTDHF3 expression (Supplementary Fig. S2G).
Knocking down YTHDF3 expression inhibits bone metastasis, lung metastasis of breast cancer cells and associated osteolytic bone destruction in vivo
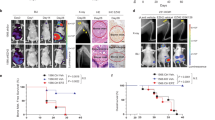
To construct the in vivo breast cancer bone metastasis model, the plasmids encoding dual luciferase were first introduced into the 4 cell lines (231BO-shNC, 231BO-YTHDH3sh1, 231HM-shNC, and 231HM-YTHDH3sh1) via lentivirus transfection. Then, the 231BO-shNC and 231BO-YTHDH3sh1 cell lines were injected into the arterial circulatory system of NCG mice by ultrasound-guided left ventricular injection to construct a breast cancer bone metastasis mouse model. The successful injection was verified by IVIS imaging, and the fluorescence intensity between the two groups was almost identical. Notably, a significant reduction in bone metastases was found in the mice injected with 231BO-YTHDH3sh1 cells 4 weeks after injection (Fig. 3A). In additon, we constructed breast cancer lung metastasis mouse models in NCG mice by injecting 231HM-shNC and 231HM-YTHDH3sh1 cell lines via tail vein injgection. Four weeks post-injection, luciferase imaging of the lungs revealed revealed markedly fewer lung metastases in mice injected with 231HM-YTHDH3sh1 cells compared with controls (Fig. 3B). H&E staining and Cytokeratin 18 (CK18) immunohistochemistry of lung sections confirmed the reduced tumor burden in 231HM-YTHDF3-sh1 mice relative to 231HM-shNC mice (Fig. 3C). To explore whether the expression of YTHDF3 in breast cancer cells was correlated with the osteolytic destruction of bone metastases in vivo, an in situ bone marrow cavity tumor model was established by tibial plateau injection with breast cancer cells. It was found that there were some distant metastases in the mice injected with NC cells; moreover, both the fluorescence intensity of bone metastases and the extent of bone destruction were also higher than those in the YTHDF3 knockdown groups (Fig. 3D).
A Living imaging results 0 week and 4weeks after injection of 231-BO-shNC-luc (n = 6) and 231-BO-shYTHDF3-1-luc (n = 6) cells via left ventricle. Tumor fluorescence signal was also quantified. B Living imaging photos 4 weeks after infection of 231-HM-shNC-luc (n = 5) and 231-HM-shYTHDF3-luc (n = 5) cells injection via tail vein and tumor fluorescence was quantified. C Representative H&E and CK18 IHC images of mice lung metastasis lesion showed different tumor burden between two groups. D Living imaging photos 4 weeks after infection of 231-BO-shNC-luc (n = 6) and 231-BO-shYTHDF3-1-luc (n = 6) cells into the tibial plateau and the bone lesion flux was indicated by fluorescence signals.
YTHDF3 binds to the mRNAs of ZEB1 and SMAD5 and enhances their translation
Through functional analysis by in vivo and in vitro experiments, we initially confirmed that YTHDF3 enhanced the migratory invasive ability of breast cancer cells and their osteolytic bone destructive effects in the bone microenvironment. To further investigate the specific mechanism by which YTHDF3 affects breast cancer functioning, we sequenced the stably transduced cell lines 231BO-shNC and 231BO-shYTHDF3-1 using RNA-seq and m6A-seq and downloaded and analyzed the RIP-seq data of GSE130171 from the GEO database [15]. Using p value < 0.01 and fold_enrichment >3.5, the top 500 peaks were selected from the m6A-seq results. Next, motif analysis was performed for the top 500 peaks, and the DRACH domain (human-derived highly conserved sequence most likely to be m6A methylation modified) was mapped. It was demonstrated that m6A-seq successfully detected the m6A modification site (Fig. 4A). Differential analysis of RNA-seq found a total of 334 differential genes (FDR < 0.05, fold change > 1.50 & <0.67), with 197 upregulated genes and 137 downregulated genes (Supplementary Fig. S3A, B). GO analysis of the differential genes revealed that expression changes in YTHDF3 were associated with GO terms such as response to zinc ion, extracellular space and cytokine activity (Supplementary Fig. S3C). KEGG pathway analysis revealed that the differentially expressed genes were correlated with pathways such as the IL17 signaling pathway, hematopoietic cell lineage and osteoclast differentiation (Supplementary Fig. S3D). In addition, GO analysis of differential m6A peaks found that these peaks were related to GO terms such as intracellular protein transport, cytoplasmic vesicle and transcription coactivator activity (Supplementary Fig. S3E). KEGG pathway analysis of differential peaks revealed that they were associated with the Wnt signaling pathway, RNA transport and parathyroid hormone synthesis secretion and action (Supplementary Fig. S3F). Further annotation of the differential m6A peaks showed that most of them were located in the exon region (Supplementary Fig. S3G).
A Motif analysis of DRACH domain for meRIP-seq results of NC and shYTHDF3 groups; B Venn plots of selected genes in RNA-seq, meRIP-seq, and RIP-seq. C Visualization of representative m6A peaks in the ZEB1 and SMAD5 gene in NC and shYTHDF3 groups. D mRNA levels of ZEB1 and SMAD5 after YTHDF3 knockdown detected by qPCR. E RIP-qPCR showing the mRNA binding levels of ZEB1 and SMAD5 to YTHDF3 protein. F WB detection of protein changes of ZEB1 and SMAD5 after YTHDF3 knockdown.
YTHDF3, as a m6A reader protein, can recognize mRNAs with m6A methylation and promote their translation, suggesting that YTHDF3 affects downstream molecules by means of posttranscriptional regulation. Then, if the expression of YTHDF3 protein is altered, the RNA levels of the downstream molecules it directly targets will be not changed, but the m6A methylation levels and protein levels of these molecules will be changed. Therefore, in this study, we intersected genes without significant differential expression in RNA-seq with genes differentially expressed in m6A-seq and genes predicted to show direct protein‒RNA interactions in RIP-seq analysis. We expected to identify downstream targets in the intersection.
After comprehensively analyzing these three datasets and consulting relevant studies, we identified two EMT-related transcription factors, ZEB1 and SMAD5, as possible downstream targets of YTHDF3. The mRNA levels of these two transcription factors were not regulated by YTHDF3, but their m6A methylation levels were associated with YTHDF3, and these transcription factors could directly bind to the YTHDF3 protein (Fig. 4B). Differential m6A peaks of ZEB1 and SMAD5 mRNA were also detected between the shNC and shYTHDF3 groups (Fig. 4C). Survival analysis using the GEO microarray data (GSE2034) suggested that ZEB1 and SMAD5 expression was also associated with overall survival (P < 0.05) and that there was no statistically significant association with bone metastasis-free survival (Supplementary Fig. S4A). In addition, the expression level of ZEB1 was higher in bone metastasis lesions than in primary lesions, with no significant expression change in SMAD5 (Supplementary Fig. S4B). Taken together, the bioinformatics analysis revealed that YTHDF3 might enhance the translation of ZEB1 by reading the m6A modification information on its mRNA.
We then conducted qPCR assays to detect the mRNA levels of ZEB1 and SMAD5 in 231BO cell lines and verified that they were not affected by the level of YTHDF3 protein (Fig. 4D), which is consistent with the analysis results. We also performed RIP-qPCR for RNA immunoprecipitation using the YTHDF3 antibody in 231BO cells and found that the mRNA of ZEB1 was more significantly enriched in YTHDF3 protein than in IgG (Fig. 4E). In addition, WB experiments showed that the protein levels of ZEB1 and SMAD5 were positively correlated with YTHDF3 (Fig. 4F).
YTHDF3 enhances the metastatic and osteoclast differentiation inducing ability of breast cancer cells by upregulating ZEB1 and SMAD5 expression
To verify whether YTHDF3 exerts its specific functions through ZEB1 and SMAD5, we performed cell migration and osteoclast differentiation induction assays using a series of YTHDF3-overexpressing 231 cell lines after ZEB1 and SMAD5 knockdown (Fig. 5A, B). We found that the knockdown of ZEB1 or SMAD5 reduced the increased migration and invasion ability caused by YTHDF3 overexpression (Fig. 5C–H). In addition, the knockdown of ZEB1 also reversed the osteoclast differentiation-inducing ability elevated by YTHDF3 overexpression (Fig. 5I, J). ZEB1, as a transcription factor, can directly regulate downstream genes by binding to promoter regions. Encouraged by the results obtained, we next investigated which genes that affect the metastasis and osteolytic bone destruction abilities of breast cancer cells are directly regulated by ZEB1.
A WB detection of protein changes of ZEB1 after knockdown ZEB1 in YTHDF3-overexpression 231 cell lines. B WB detection of protein changes of SAMD5 after knockdown SMAD5 in YTHDF3-overexpression 231 cell lines. C, D Transwell assay revealed migration and invasion ability change in YTHDF3-overexpression 231 cell lines after ZEB1 knockdown. E, F Transwell assay revealed migration and invasion ability change in YTHDF3-overexpression 231 cell lines after SMAD5 knockdown. F Wound healing assay revealed migration rate change in YTHDF3-overexpression 231 cell lines after SMAD5 knockdown. G, H Wound healing assay revealed migration rate change in YTHDF3-overexpression 231 cell lines after ZEB1 or SMAD5 knockdown. I, J Effect of knockdown of ZEB1 expression on the ability to induce osteoclast differentiation in breast cancer cells that overexpress YTHDF3.
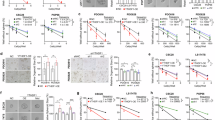
From previously reported studies, we learned that ZEB1 can promote EMT in cells by binding to the promoter region of E-cad as a transcriptional repressor and downregulating its expression [19]. However, this is not sufficient to explain the ability of ZEB1 to promote osteolytic bone destruction. ELISA was performed to detect changes in the secretion of IL6, which is associated with osteoclast differentiation, after YTHDF3 knockdown in 231BO cells, but no significant change was found (Supplementary Fig. S4G). To explain this phenomenon, we analyzed the RNA-seq data after the knockdown of ZEB1 in breast cancer [20] and CHIP-seq data of ZEB1 [21] from previous studies. WNT5B, a secreted protein that can promote osteoclast differentiation [22, 23], was identified as a downstream target of ZEB1 (Fig. 6A). Furthermore, we found that several bone morphogenetic protein (BMP) inhibitors, which have been reported to be correlated with osteoclast differentiation, were positively correlated with YTDHF3 expression (Supplementary Fig. S3H–J). By using the GEPIA database, we found a correlation between the expression of ZEB1 and WNT5B (Fig. 6B). However, the WB assay results indicated no correlation between WNT5B and YTHDF3 expression in the cell lysate (Fig. 6C). Therefore, we analyzed WNT5B secretion content in culture medium supernatant from collected breast cancer 231 cells and stably transfected cells by WB. It was found that the change in WNT5B secretion was positive with the expression of YTHDF3 (Fig. 6D). Furthermore, we performed a luciferase reporter assay to investigate the possible correlation between WNT5B and ZEB1. We found that the knockdown of ZEB1 reversed the elevated luciferase activity induced by YTDHF3 overexpression (Fig. 6E). qPCR showed that the RNA level of WNT5B was positively correlated with YTHDF3 expression (Fig. 6F). WB assays revealed that WNT5B secretion decreased after the knockdown of ZEB1 in YTHDF3-overexpressing cells (Fig. 6G). Therefore, we concluded that ZEB1 might promote the secretion of WNT5B and BMP inhibitors, such as NOG, FST, and CCN2, by binding to their promoter regions, further influencing the osteolytic differentiation-inducing ability of breast cancer cells.
A Venn plot of selected genes in RNA-seq and Chip-seq. B Correlation of ZEB1 and WNT5B expression. C Western Blot detection of changes of WNT5B after YTHDF3 knockdown and overexpression in cell lysate. D Western Blot detection of changes of WNT5B after YTHDF3 knockdown and overexpression in cellular supernatant. E Relative luciferase change after ZEB1 knockdown in YTHDF3-overexression 231 cells by luciferase reporter assay. F q-PCR shows the RNA level of WNT5B after YTHDF3 knockdown and overexpression. G WB assays showing WNT5B secretion contents in the culture medium supernatant from breast cancer cells 231 and its stable transfection cells.
Discussion
Breast cancer is one of the most common malignant cancers in the female population around the world, has a high mortality rate, and seriously affects the health of patients [24]. Distant metastasis is one of the main reasons for its poor prognosis [25]. Distant metastasis of breast cancer, particularly to the bone, poses one of the most formidable challenges in cancer therapy and is a leading cause of mortality among cancer patients [25]. Therefore, delving deeper into the mechanisms underlying breast cancer bone metastasis and identifying novel therapeutic targets is of paramount importance for improving clinical outcomes. In this study, we conducted a comprehensive analysis of public datasets and clinical tissue samples] and found that YTHDF3 is markedly upregulated in both primary and metastatic breast cancer lesions, and its overexpression correlates strongly with metastatic progression and poor clinical outcomes in patients with breast cancer. The above results suggested that YTHDF3 may promote breast cancer progression and potentially play an enhancing role in the metastasis of breast cancer.
YTHDF3, one of the key m6A recognition proteins, is aberrantly expressed and plays different roles in various cancers [12,13,14]. Here, we found that the increase in YTHDF3 expression enhanced the metastatic ability of breast cancer cells using various breast cancer cell lines. Furthermore, we also found overexpression of YTHDF3 was associated with metastasis to various distant organs, such as the brain, lung, and bone in GEO microarray data. We further constructed bone metastasis and lung metastasis mouse models through left ventricular injection and tail vein injection of breast cancer cells and examined their growth and metastasis in vivo using an IVIS system. The results showed that the knockdown of YTHDF3 reduced the metastatic ability of breast cancer cells to bone and lungs. Collectively, our findings demonstrated that YTHDF3 promotes the multiorgan metastatic ability of breast cancer cells and enhances the osteolytic bone destruction of bone metastasis through in vivo and in vitro experiments. Indeed, a recent study has uncovered an essential role of YTHDF3 in controlling the interaction between cancer cells and brain microenvironment, thereby inducing brain metastatic competence [15]. Therefore, our study focused on the role of YTHDF3 in the bone metastasis of breast cancer and found the increase in YTHDF3 expression enhanced the ability of breast cancer cells to induce osteoclast differentiation, and vice versa. Additionally, we also established an osteolytic destruction model by the bone marrow cavity injection of breast cancer cells, the findings of which further validated the results of the in vitro results.
The development of bone metastasis is a complex process that broadly involves 1. colonization, i.e., the entry of circulating tumor cells into the bone marrow microenvironment; 2. dormancy, i.e., tumor cells adapt to the bone marrow microenvironment and enter dormancy; 3. activation, i.e., tumor cells switch from a dormant to a proliferative state; and 4. bone destruction, i.e., tumor cells destroy the original skeletal structure and function [26]. As a key initial step that increases the metastatic ability of cancer cells and their detachment from the primary site, EMT allows breast cancer cells to be more invasive and more metastatic [27]. EMT is a cell phenotype alteration that occurs during embryonic development and tissue remodeling. It transforms epithelial-like phenotype cells into mesenchymal-like phenotype cells and enhances the migratory invasive ability of cancer cells by weakening their intercellular adhesion [28]. To elucidate the potential mechanism of YTHDF3-mediated metastasis, we performed an integrated multi-omics analysis incorporating RNA-seq, m6A-seq, and RIP-seq data. Through this comprehensive approach, we systematically identified downstream targets of YTHDF3, with a particular emphasis on the EMT-associated effectors ZEB1 and SMAD5. ZEB1 is closely related to EMT, and it can promote the EMT process of cancer cells by repressing the expression of the epithelial phenotype marker E-cadherin and correspondingly increasing the expression of the mesenchymal phenotype markers vimentin and N-cadherin [19]. SMAD5, as an intracellular protein, is involved in the transforming growth factor β (TGF-β) signaling pathway in cancer progression and metastasis [29]. Phosphorylated SMAD5 binds to SMAD4, translocates to the nucleus, and binds to other transcription factors to activate or repress the expression of target genes [30]. SMAD5 is associated with EMT and promotes cancer cell proliferation and metastasis in nasopharyngeal and gastric cancers [29, 31, 32]. In this study, our findings confirmed that, as a classic m6A reader, YTHDF3 could read m6A sites of ZEB1 and SMAD5 mRNAs to promote their protein translation. These prompted us to performe a functional reversion assay by knocking down ZEB1 and SMAD5 in YTHDF3-overexpressing cells. It was found that knocking down ZEB1 and SMAD5 reversed the phenotype of increased migratory invasive ability enhanced by YTDHF3 overexpression. These findings suggested that YTHDF3 might promote the translation of ZEB1 and SMAD5 by binding to the methylation sites in the mRNA, thus promoting cancer metastasis by EMT in breast cancer.
Moreover, we found that genes associated with osteoclast differentiation were enriched in down-regulated genes upon YTHDF3 knockdown in 231BO cells. We further confirmed via in vitro and in vivo experiments that the increased osteoclast differentiation-inducing ability caused by YTHDF3 overexpression could also be reversed by ZEB1 knockdown. The formation of metastasis depends on multiple interactions between cancer cells and the tumor microenvironment. During osteolytic bone metastasis, osteoclast differentiation plays a crucial role; therefore, understanding the mechanism by which tumor cells influence osteoclast differentiation is essential for improving the treatment of osteolytic metastasis. ZEB1, as a transcription factor, can directly regulate downstream genes by binding to promoter regions. Here, we found that enhanced expression of ZEB1 promoted the transcription of bone morphogenetic protein inhibitors, such as NOG, FST, and CCN2, which enhanced osteolytic metastasis. To further investigate the specific mechanism by which YTHDF3 affects bone metastasis, we analyzed the RNA-seq data after the knockdown of ZEB1 in breast cancer [20] and CHIP-seq data of ZEB1 [21] from previous studies. WNT5B, a secreted protein that can promote osteoclast differentiation [22, 23, 33], was identified as a downstream target of ZEB1. WNT5B is involved in many aspects of bone physiology, including bone formation and degradation [34]. WNT5B enhances bone degradation by increasing triiodothyronine receptor auxiliary protein (TRAP) activity through the receptor-like tyrosine kinase (RYK) receptor and inhibiting β-catenin signaling in osteoclasts [23]. CHIP-seq data suggest that ZEB1 may bind to the promoter regions in WNT5B genes [21, 35]. In this study, we found that WNT5B secretion in the culture medium supernatants from 231 breast cancer cells and stably transfected cell lines were enhanced by YTHDF3 overexpression but decreased with the knockdown of ZEB1 even in the presence of YTHDF3 overexpression. It suggested that YTHDF3 in breast cancer cells promoted promotes osteoclast differentiation by enhancing the secretion of WNT5B in a ZEB1-dependent manner.
In summary, we summarized the following mechanism by which YTHDF3 promotes the osteolytic bone metastasis of breast cancer cells (Fig. 7). In breast cancer cells, YTHDF3 enhances the translation of ZEB1 and SMAD5 by reading and recognizing m6A-modified sequences in mRNAs. As a result, the enhanced expression of these two proteins increases the invasiveness of breast cancer cells by promoting the EMT process. More critically, we uncovered that YTHDF3 drives breast cancer bone metastasis by upregulating the key effector ZEB1. As a transcription factor, enhanced expression of ZEB1 promoted the transcription of bone morphogenetic protein inhibitors such as NOG, FST, and CCN2, which boosts osteolytic metastasis. Furthermore, we newly found that ZEB1 boosts WNT5B secretion, thereby effectively enhancing the osteoclast differentiation-inducing ability of breast cancer cells.
Overexpression of YTHDF3 in breast cancer cells upregulates the expression of ZEB1 and SMAD5 by reading and recognizing m6A modified sequences in mRNAs. The increasing level of ZEB1 and SMAD5 promotes the EMT process so that increasing the invasiveness of breast cancer cells. Furthermore, in bone microenvironment, ZEB1 boosts the expression of WNT5B, thus increasing the osteoclast differentiation and finally causing osteolytic lesions.
Materials and methods
Gene expression and clinical analysis
Microarray datasets of normal tissue and breast cancer tissue were extracted from The Genotype-Tissue Expression (GTEx) database (https://gtexportal.org/home/) and The Cancer Genome Atlas (TCGA) database ((https://portal.gdc.cancer.gov/). The differential expression and OS of YTHDF3 were analyzed online using Gene Expression Profiling Interactive Analysis [36] (GEPIA).
The remaining microarray data were obtained from the Gene Expression Omnibus (GEO) database under accession number GSE2034. A total of 286 breast cancer samples were included to analysis the correlation of YTHDF3 expression with the metastasis-free survival rate. The above analyses were performed using R software (4.1.1).
Tissue samples
A tissue microarray (Product number: HBreD055CD01) was purchased from Shanghai Xinchao Co., Ltd. The tissue microarray contains a variety of specimens, including fibrocystic breast disease, primary lesions, metastatic lymph nodes, and metastatic lesions of breast cancer, as well as clinical characteristics such as pathological grade, age, tissue site and TNM staging. In addition, matched human breast samples (primary lesions and bone metastatic lesions) were obtained from Shanghai Cancer Center. The use of human tissue was approved by the Ethics Committee of Fudan University Shanghai Cancer Center, and written informed consent was obtained from each patient. Clinical characteristics of samples included in this study were listed at Supplementary Tables S1 and S2.
Cell culture
The MDA-MB-231, MCF7, MDA-MB-453, MDA-MB-468, BT549, BT20, HS578T, 4T1, and MCF10A cell lines were obtained from the American Type Culture Collection (ATCC). MDA-MB-231 HM cells (highly metastatic derivatives) and MDA-MB-231 BO cells (bone metastatic derivatives) were gifts from the laboratory of Pro. Yang at Shanghai Cancer Center. MCF10A cells were cultured in MEGM (Lonza/Clonetics Corporation, Switzerland) with the MEGM growth kit (Lonza/Clonetics Corporation, Switzerland) and 100 ng/ml cholera toxin. BT549 cells were grown in RPMI-1640 medium (Sigma-Aldrich, America) containing 10% FBS and 1% penicillin‒streptomycin. MDA-MB-468 cells were cultured in L-15 medium (Sigma-Aldrich, America) containing 10% FBS and 1% penicillin‒streptomycin and incubated without CO2. The rest of the cell lines were cultured in DMEM (Gibco, America) containing 10% FBS and 1% penicillin‒streptomycin. All cell lines except MDA-MB-468 were incubated at 37 °C in 5% CO2. The cell lines mentioned were identified using STR profiling and confirmed to be negative for mycoplasma.
Immunohistochemistry (IHC)
Paraffin-embedded tissue slides were deparaffinized using xylene and rehydrated through an alcohol series (100% alcohol, 90% alcohol, 80% alcohol, 70% alcohol). Slides were subjected to antigen retrieval using sodium citrate solution at 100 °C for 10 min followed by incubation with 3% H2O2 for 15 min. Tissue sections were blocked with 5% normal goat serum for 30 min at room temperature and incubated with 1/100 dilution of anti-YTHDF3 antibody (Abcam, Britain, Rabbit monoclonal, ab220161) at 4 °C overnight, followed by incubation with 1/10000 dilution of secondary antibodies (Abcam, Britain, Goat Anti-Rabbit IgG, ab205718) and DAB (Solarbio, China) for IHC staining. The staining scores of microarrays were calculated using the H-score. The H-score is one of the widely used digital image analysis tools to quantify biomarker expression, enabling fast, objective, and highly reproducible quantification of biomarkers from whole-slide IHC images [37, 38]. For the tissue microarray, the staining was quantified by 2 independent investigators according to the H-score. In the H-score evaluations, the staining intensity of positive cells was calculated from 0 to 3, corresponding to negative (no color), weak positive (light yellow), moderate positive (brown‒yellow), and strong positive (brown), respectively. H-score = (% of weak positive area × 100) + (% of moderate positive area × 200) + (% of strong positive area × 300). The H-score is a value between 0 and 300, with a higher value indicating a stronger overall positive intensity [39, 40].
Western blotting (WB)
Cells were collected and lysed with RIPA lysis buffer (Beyotime, China). Protein samples were quantified using a BCA protein assay kit (Epizyme, America) and boiled with SDS‒PAGE Protein Loading Buffer (Thermo Fisher) at 100 °C for 10 min. Protein samples were separated by SDS‒PAGE and then transferred to PVDF membranes (Thermo Fisher, America). The PVDF membranes were blocked at room temperature for 60 min and incubated with primary antibodies at 4 °C overnight, followed by incubation with secondary horseradish peroxidase (HRP)-conjugated antibodies. An ECL western blotting detection kit (Thermo Fisher, America) was applied to visualize the immunoblot. The antibody information is listed in Supplementary Table S3.
Plasmid and transfection
Human YTHDF3 was cloned and inserted into pcDNA3.1 (Lingke, China) to generate a YTDHF3-overexpressing plasmid. For YTDHF3, ZEB1, and SMAD5 knockdown, shRNA oligonucleotides were cloned and inserted into pLKO.1 (Lingke, China). All target sequences are listed in additional file: Supplementary Table S4. Vectors in lentiviral packaging plasmids psPAX2 and pMd2.G were packaged into 293FT cells. Breast cancer cells were transduced with virus and polybrene for 24–48 h, followed by culture with puromycin or bleomycin. Transfection efficiency was examined by WB. The shRNA sequences are provided in Supplementary Table S4.
RNA isolation and quantitative real-time PCR (qRT‒PCR)
Total RNA was extracted from cells using NucleoZOL (Takara, Japan) reagent, followed by the synthesis of cDNA using a Color Reverse Transcription Kit (EZBioscience, America) following the manufacturer’s protocol. qRT‒PCR was performed on an ABI 7500 Real-time PCR system (Thermo Fisher, America) using AceQ SYBR qPCR Master Mix (Vazyme, China). Primer sequences are listed in additional file: Supplementary Table S4. Target gene expression values were normalized to 18S rRNA or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels. Primers sequences are listed in Supplementary Table S4.
Wound healing migration assays
Breast cancer cells were seeded onto 6-well plates and grown until the plate was covered, and straight wound lines were made vertically in the wells. The suspended cells were washed with PBS buffer, and serum-free fresh medium was added to culture the remaining cells for 24 h. Cells were imaged at 0 h and 24 h.
Clone formation assay
Breast cancer cells in logarithmic phase were re-suspended and inoculated with 1000 cells/holes in a 6-well plate. Cells were cultured for 14 days and culture medium was changed every 3 days. After cloning, cells were fixed with 1 ml 4% paraformaldehyde per well for around 30 min and washed twice with PBS. Cells were stained with crystal violet solution for 10–20 min and washed several times with PBS.
Angiogenesis assay
For angiogenesis assay, the 48-well plates were previously coated with Matrigel (Sigma, America) for 30 min in 37 °C. HUVEC cells (ATCC, America) in logarithmic phase were harvested and plated on 48-well plates (50,000 cells/well) and cultured with conditioned medium from cancer cells (mixed with DMEM at a 1:10 ratio). The 48-well plates were incubated in an incubator at 37 °C, 5% CO2 condition, and photographed at 2, 4, 6, 8 h for observation.
Cell adhesion assay
C3H10 (Mouse mesenchymal stem cells, ATCC, America), MG63 (Osteoblastic osteosarcoma cells, ATCC, America) and RAW264.7 (Mouse mononuclear macrophages, ATCC, America) were cultured to 100% density in 24-well plates. Then MDA-231-B cells with stable expression of GFP were inoculated on a 24-well plate with cells mentioned above and shake cultured for 4 h (120 rpm). Removing culture supernatant and washing twice carefully with PBS, cells were fixed with formaldehyde for 15 min. The density and number of cells with green fluorescence were calculated using fluorescence microscope.
Transwell assays
Breast cancer cells (2 × 105) were plated in the upper chamber of Transwell migration chambers (Corning, America), and DMEM supplemented with 30% FBS was placed in the lower chamber. For Transwell invasion assays, the Transwell chambers were previously coated with Matrigel (Sigma, America). After 24 h of incubation at 37 °C, cells on the inner side were removed, and the membrane was fixed with 4% paraformaldehyde for at least 15 min followed by staining with crystal violet. Images of the membrane stained with crystal violet were captured for cell counting.
Osteoclastogenesis assays
Primary bone marrow cells were harvested from the femora and tibiae of 4–6-week-old C57/BL6 mice and plated in MEM-α supplemented with 10% FBS and 30 ng/ml M-CSF for 48 h. The adherent cells were harvested and plated on 48-well plates (50,000 cells/well) and then cultured with conditioned medium from cancer cells (mixed with MEM-α at a 1:10 ratio) supplemented with 30 ng/ml RANKL and 30 ng/ml M-CSF for osteoclast differentiation. Tartrate-resistant acid phosphatase (TRAP) staining was performed on days 7–10 (conditioned medium mixture was changed every 3 days). TRAP-stained images were captured to count the number of mature osteoclast cells (TRAP+ multinucleated cells).
Mouse model and in vivo imaging system (IVIS)
Luciferase-expressing plasmids were purchased from Lingke Biotechnology Co. Lentivirus packaging was performed as described above and was used to infect successfully constructed 231-BO-shNC, 231-HM-shNC, 231-BO-shYTHDF3-1, and 231-HM-shYTHDF3 cells. The stably transduced cell lines were screened by the addition of blasticidin S for ~1 week. To a 96-well plate containing 50 μL of cell suspension in each well, luciferase substrate (15 mg/mL, 50 μL each well) was added. After reaction for 5 min, the chemiluminescence signal (luminescence, RLU) of each well was measured using a microplate reader (Thermo Scientific, America)) to detect the expression level of luciferase in the stably transfected cell lines.
231-HM-shNC-luc and 231-HM-shYTHDF3-luc cells at the logarithmic growth stage were trypsinized, counted, and diluted to a concentration of 1 × 107 cells/mL. NCG-immunodeficient mice (4 weeks old, female, purchased from GemPharmatech Co., Ltd. China) were randomly divided into two groups of 5 mice each and injected with 231-HM-shNC-luc and 231-HM-shYTHDF3-1-luc cells (1 × 106 cells/mouse) via the tail vein. The status of the mice was observed regularly after cell injection. Four weeks post-injection, the distribution of metastasis in the lungs of the mice was detected by in vivo imaging. The whole lung was fixed in formalin for paraffin embedding and sectioning. Then, they were stained H&E and IHC (CK18, 1:800, Rabbit monoclonal, ab181597, Abcam, Britain) to observe the size and shape of the metastases.
For the cell injection experiments involving 231-BO-shNC-luc and 231-BO-shYTHDF3-1-luc cells, NCG-immunodeficient mice were randomly divided into two groups of 6 mice. Prior to the injection procedure, the mice were anesthetized using isoflurane, and the location of the left ventricle of the mice was probed using an ultrasound probe. After the location of the left ventricle was determined, 100 μL of 231-BO-shNC-luc and 231-BO-shYTHDF3-1-luc cell suspensions (1 × 107 cells/mL) were injected into the left ventricle from the thorax of the mouse under the guidance of an ultrasound probe. Successful injection was indicated by a large number of white noise spots in the left ventricle on ultrasound. After injection, live images of the mice were taken when the mice were in a stable state. The luciferase substrate was injected into each mouse at a dose of 10 μL of luciferase substrate/g, and the mice were scanned and photographed using IVIS Spectrum (Lumina III, PerkinElmer, America) after 5 min. After 4 weeks, the mice were photographed again.
Four-week-old female BALB/c nude mice were divided into two groups of 6 mice each and injected with 20 μL of 231-BO-shNC-luc or 231-BO-shYTHDF3-1-luc cells (5 × 107 cells/mL) into the tibial bone marrow cavity. After injection, the mice were kept in standard housing conditions for 6–8 weeks and photographed using IVIS Spectrum (Lumina III, PerkinElmer, America) as described above. The experimental protocols were approved by the rules of the Institutional Animal Care and Use Committee of Fudan University Shanghai Cancer Center.
RNA immunoprecipitation (RIP)
The RIP assay was conducted with Magna RIP kits (Merck Millipore, America) following the manufacturer’s instructions. In the RIP experiments, RNA-binding protein‒RNA complexes were immunoprecipitated using YTHDF3 antibody (Abcam, America, ab220161), in which the murine IgG antibody and the input group were set as controls. RNA expression levels were measured using RT‒qPCR as described above.
Detection of secreted protein
Cells grown to a density greater than 90% were cultured with serum-free DMEM for 48 h. The medium was transferred to a centrifuge tube, centrifuged at 1000 rpm for 3 min, filtered through a 0.45 μm filter, added to an ultrafiltration tube and centrifuged at 7000 × g for 40 min at 4 °C. The remaining medium was mixed with loading buffer and heated to 100 °C for 10 min. The proteins in the medium were separated by SDS‒PAGE, transferred to PVDF membranes, and stained with Ponceau S. The stained protein bands were photographed, and the stained darker proteins served as an internal reference control. WB was conducted to detect the contents of secreted protein in the medium.
RNA-seq and m6A-seq
YTHDF3 knockdown 231BO stable cells and control cells were cultured to logarithmic growth phase and collected. Total RNA was extracted and subjected to mRNA and m6A-seq sequencing (Ouyi Biomedical Technology Co., Ltd, Shanghai, China).
Statistical analysis
All quantitative experiments were performed using at least three independent biological repeats, and the results are presented as means ± standard deviation (S.D.) or means ± standard error of the mean (S.E.M.). Survival curves were plotted using the Kaplan‒Meier method, and differences between subgroups were compared using log-rank tests. Statistical analysis was performed using GraphPad Prism and R software (version 4.1.1). The fluorescence values in IVIS were quantified by Aura software. Comparisons between two groups for continuous variables with a normal distribution and chi-squared variance were performed using t tests, and data between multiple groups were compared using ANOVA. Data for continuous variables that did not qualify for the t test were analyzed using the Mann‒Whitney U test. For levels of significance, the following designations were used within this manuscript: *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns not significant. Differences were considered statistically significant when p < 0.05.
Data availability
The data that support the findings of this study are available on request from the corresponding author, Wangjun Yan, upon reasonable request.
References
Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: burden and trends. Cancer Epidemiol Biomark Prev. 2017;26:444–57.
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30.
Awolaran O, Brooks SA, Lavender V. Breast cancer osteomimicry and its role in bone specific metastasis; an integrative, systematic review of preclinical evidence. Breast. 2016;30:156–71.
Yang M, Liu C, Yu X. Skeletal-related adverse events during bone metastasis of breast cancer: current status. Discov Med. 2019;27:211–20.
Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187.
Paramasivam A, Vijayashree PJ, Raghunandhakumar S. N6-adenosine methylation (m6A): a promising new molecular target in hypertension and cardiovascular diseases. Hypertens Res. 2020;43:153–4.
Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. m6A-dependent regulation of messenger RNA stability. Nature. 2014;505:117.
He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176.
He L, Li J, Wang X, Ying Y, Xie H, Yan H, et al. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J Cell Mol Med. 2018;22:4630–9.
Wang S, Chai P, Jia R, Jia R. Novel insights on m6A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17:101.
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27:315–28.
Xu Y, He X, Wang S, Sun B, Jia R, Chai P, et al. The m6A reading protein YTHDF3 potentiates tumorigenicity of cancer stem-like cells in ocular melanoma through facilitating CTNNB1 translation. Oncogene. 2022;41:1281–97.
Guo JC, Liu Z, Yang YJ, Guo M, Zhang JQ, Zheng JF. KDM5B promotes self-renewal of hepatocellular carcinoma cells through the microRNA-448-mediated YTHDF3/ITGA6 axis. J Cell Mol Med. 2021;25:5949–62.
Jin H, Ying X, Que B, Wang X, Chao Y, Zhang H, et al. N6-methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195–207.
Chang G, Shi L, Ye Y, Shi H, Zeng L, Tiwary S, et al. YTHDF3 induces the translation of m6A-enriched gene transcripts to promote breast cancer brain metastasis. Cancer Cell. 2020;38:857.
Azzam DJ, Zhao D, Sun J, Minn AJ, Ranganathan P, Drews-Elger K, et al. Triple negative breast cancer initiating cell subsets differ in functional and molecular characteristics and in γ-secretase inhibitor drug responses. EMBO Mol Med. 2013;5:1502–22.
Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–9.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9.
Wu HT, Zhong HT, Li GW, Shen JX, Ye QQ, Zhang ML, et al. Oncogenic functions of the EMT-related transcription factor ZEB1 in breast cancer. J Transl Med. 2020;18:51.
Mock K, Preca BT, Brummer T, Brabletz S, Stemmler MP, Brabletz T. The EMT-activator ZEB1 induces bone metastasis associated genes including BMP-inhibitors. Oncotarget. 2015;6:14399–412.
Feldker N, Ferrazzi F, Schuhwerk H, Widholz SA, Guenther K, Frisch I, et al. Genome-wide cooperation of EMT transcription factor ZEB1 with YAP and AP-1 in breast cancer. EMBO J. 2020;39:e103209.
Kim B, Kim H, Jung S, Moon A, Noh DY, Lee ZH, et al. A CTGF-RUNX2-RANKL axis in breast and prostate cancer cells promotes tumor progression in bone. J Bone Min Res. 2020;35:155–66.
Santiago F, Oguma J, Brown AMC, Laurence J. Noncanonical Wnt signaling promotes osteoclast differentiation and is facilitated by the human immunodeficiency virus protease inhibitor ritonavir. Biochem Biophys Res Commun. 2012;417:223–30.
Liu L, Zhang Y, Lu J. The roles of long noncoding RNAs in breast cancer metastasis. Cell Death Dis. 2020;11:749.
Bray F, McCarron P, Parkin DM. The changing global patterns of female breast cancer incidence and mortality. Breast Cancer Res. 2004;6:229–39.
Coleman RE, Croucher PI, Padhani AR, Clezardin P, Chow E, Fallon M, et al. Bone metastases. Nat Rev Dis Prim. 2020;6:83.
Ottewell PD, O’Donnell L, Holen I. Molecular alterations that drive breast cancer metastasis to bone. Bonekey Rep. 2015;4:643.
Sims NA, Martin TJ. Coupling the activities of bone formation and resorption: a multitude of signals within the basic multicellular unit. Bonekey Rep. 2014;3:481.
Drabsch Y, Dijke P, ten. TGF-β signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31:553–68.
Orlowski J. SMAD5 signaling: more than meets the nuclei. Cell Res. 2017;27:1075–6.
Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration, and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered. 2021;12:225–39.
Zheng YJ, Zhao JY, Liang TS, Wang P, Wang J, Yang DK, et al. Long noncoding RNA SMAD5-AS1 acts as a microRNA-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma. FASEB J. 2019;33:12915–28.
Choi Y, Yoo JH, Lee JH, Lee Y, Bae MK, Kim YD, et al. Connective tissue growth factor (CTGF) regulates the fusion of osteoclast precursors by inhibiting Bcl6 in periodontitis. Int J Med Sci. 2020;17:647–56.
Suthon S, Perkins RS, Bryja V, Miranda-Carboni GA, Krum SA. WNT5B in Physiology and Disease. Front Cell Dev Biol. 2021;9:667581.
Katsura A, Tamura Y, Hokari S, Harada M, Morikawa M, Sakurai T, et al. ZEB1-regulated inflammatory phenotype in breast cancer cells. Mol Oncol. 2017;11:1241–62.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102.
Aeffner F, Wilson K, Martin NT, Black JC, Hendriks CLL, Bolon B, et al. The gold standard paradox in digital image analysis: manual versus automated scoring as ground truth. Arch Pathol Lab Med. 2017;141:1267–75.
Meyerholz DK, Beck AP. Principles and approaches for reproducible scoring of tissue stains in research. Lab Invest. 2018;98:844–55.
Guo R, Berry LD, Aisner DL, Sheren J, Boyle T, Bunn PA Jr., et al. MET IHC is a poor screen for MET amplification or MET Exon 14 mutations in lung adenocarcinomas: data from a tri-institutional cohort of the lung cancer mutation consortium. J Thorac Oncol. 2019;14:1666–71.
Paschalis A, Sheehan B, Riisnaes R, Rodrigues DN, Gurel B, Bertan C, et al. Prostate-specific membrane antigen heterogeneity and DNA repair defects in prostate cancer. Eur Urol. 2019;76:469–78.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (No. 82072972).
Author information
Authors and Affiliations
Contributions
LX, SEW, and PHX were responsible for the design of the study and the writing the manuscript. QY conducted the additional experiments and performed data analysis in response to the reviewers’ comments, and completed the revision of the manuscript. QLY and XF were responsible for validating the immunohistochemical findings and providing expert pathological insights. WJY, SEW, YS, and KL were responsible for experiment design. BNW, MC, and WDH conducted data analysis work. SLS, SEW, and WJY were responsible for the edit of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All methods employed in this study were conducted in strict accordance with the relevant guidelines and regulations. The analysis of human samples complied with the principles outlined in the Declaration of Helsinki and was approved by the institutional research Ethics Committee of Fudan University Shanghai Cancer Center (IRB Number: 050432-4-2108*). Written informed consent was obtained from each participant. All animal experiments were perfomed in accordance with the NC3Rs ARRIVE guidelines and were approved by the Institutional Animal Care and Use Committee of Fudan University Shanghai Cancer Center (IRB Number: FUSCC-IACUC-S20210366).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, L., Yu, Q., Peihang, X. et al. YTHDF3 promotes breast cancer osteolytic bone metastasis by enhancing the translation of ZEB1 and SMAD5. Oncogenesis 14, 41 (2025). https://doi.org/10.1038/s41389-025-00583-0
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41389-025-00583-0