Abstract
Background
Radioresistance is an objective biological factor that affects the efficacy of clinical radiotherapy in hepatocellular carcinoma (HCC). However, the mechanism involved has not yet been fully understood.
Methods
Integrative analysis of HCC patient data with RNA-seq and IHC. Radiosensitivity assessed by colony formation assays using HCC cell lines and xenograft models established in B-NDG mice. USP11 promoter activity measured by dual luciferase reporter. Protein interactions analyzed by Co-IP/GST pulldown; post-translational modifications by ubiquitylation assays. Rac1 activity was quantified by measuring GTP-bound levels. Rac1 structural dynamics simulated through molecular dynamics. Functional validation performed using pharmacological inhibitors, genetic depletion and site-directed mutagenesis.
Results
Elevated activated Rac1(Rac1-GTP) predicted poor radiotherapeutic response. Ionizing radiation (IR) activated Rac1 and induced its nuclear translocation. Rac1-GTP is required for the transcriptional upregulation of USP11. USP11 stabilised Rac1-GTP via deubiquitination at residues K123/K147/K183, forming a self-reinforcing Rac1-USP11 amplification loop driving radioresistance. USP11 knockdown or mutation of Rac1 deubiquitination sites (K123/K147/K183) destabilized Rac1-GTP and reversed radioresistance. Elevated Rac1-GTP and USP11 correlated with adverse clinical outcomes. Critically, combined NSC23766/Mitoxantrone treatment showed enhanced radiosensitization.
Conclusion
This study identifies the Rac1-USP11 reciprocal feedback loop as a novel, self-reinforcing mechanism driving radioresistance in HCC. Targeting this loop via combined Rac1-GTP/USP11 inhibition represents a promising therapeutic strategy for radiosensitizing HCC.
Similar content being viewed by others
Introduction
HCC is the fourth leading cause of cancer-related death worldwide [1]. Potentially curative hepatic resection is the optimal therapy for HCC, but most patients are not candidates for resection [2]. With the development of radiotherapy technology, radiotherapy has shown promising outcomes in lowering the clinical stage and reducing the size of tumours in HCC [3,4,5]. However, the current therapeutic outcomes for HCC patients are still unsatisfactory because of the high frequency of resistance to treatment, and the molecular mechanisms underlying radioresistance in HCC remain poorly understood.
Recent studies have identified dysregulated signalling pathways, including DNA damage repair, metabolism, and redox signalling (ROS), as key contributors to HCC radioresistance [6,7,8]. Notably, members of the Rho GTPase family, which regulate cellular processes such as migration and DNA repair, have emerged as potential mediators of radioresistance [9,10,11]. Among these, Rac1, a critical regulator of actin dynamics and ROS, has been implicated in cancer progression and therapy resistance [12,13,14]. Our next-generation sequencing analysis of HCC patients revealed Rac1 as one of top differentially expressed genes between radiotherapy non/poor responders and good responders, suggesting its pivotal role in modulating radiosensitivity. To elucidate how Rac1 governs radiosensitivity, we next dissected its dynamic regulatory landscape.
Rac1 functions as a molecular switch in cells, exhibiting an active state when it is bound to guanosine triphosphate (GTP) and an inactive state when it is associated with guanosine diphosphate (GDP) [15]. As a signalling nexus, Rac1-GTP coordinates oncogenic transcription through multilayered control of effector cascades like PI3K-AKT, PAK-MAPK, and NF-κB pathway, establishing it as a central rheostat of malignant gene expression [16,17,18]. The activity of Rac1 is subjected to regulation through multiple mechanisms. These include the regulation by guanine nucleotide exchange factors (GEFs), GTPase activating proteins (GAPs) and guanine nucleotide dissociation inhibitors (GDIs), as well as posttranslational modifications (PTMs) of Rac1, which include phosphorylation, SUMOylation and ubiquitination [19,20,21]. Ubiquitination of Rac1, a critical regulatory mechanism controlling its stability and activity, has emerged as active research areas. While E3 ligases mediating Rac1 ubiquitination have been partially characterised [21], the deubiquitinating enzymes (DUBs) responsible for stabilising Rac1-GTP remain poorly explored. Identifying such DUBs is crucial to understanding how Rac1-GTP accumulates in tumours and drives therapy resistance [22].
This study revealed that IR-activated Rac1 drives its nuclear translocation to transcriptionally upregulate USP11. Newly synthesised USP11 then stabilises Rac1-GTP through site-specific deubiquitination, establishing a self-reinforcing Rac1-USP11 signalling circuit that orchestrates acquired radioresistance in HCC. Clinically, high USP11 and Rac1-GTP levels correlate with poor radiotherapy response and prognosis in HCC patients. Functionally, genetic or pharmacological inhibition of USP11 and Rac1-GTP significantly impaired HCC progression and radioresistance both in vitro and in vivo. These findings establish the USP11-Rac1 axis as a therapeutic target to overcome radioresistance in HCC.
Results
Rac1 activation status is associated with radioresistance of HCC
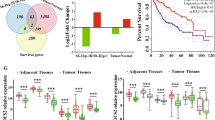
Elucidating the molecular mechanisms driving radioresistance in HCC is essential to overcoming therapeutic inefficacy. HCC patients after radiotherapy were stratified as good responders or non/poor responders based on pathological scores and imaging data. Next-generation sequencing of HCC patient samples revealed marked gene expression differences between non-/poor responders and good responders. Rac1 ranked among the top ten differentially expressed genes (Fig. 1a). Global transcriptomic analysis demonstrated that Rac1 transcripts were significantly elevated in hepatic tumours compared to normal liver tissue (Fig. 1b). High Rac1 expression also correlated with a poor HCC prognosis, as shown by Kaplan–Meier overall survival analysis using GEPIA (Fig. 1c). And Rac1-GTP was highly expressed in HCC tissues of patients with poor prognosis (Fig. 1d).
a Gene expression analysis from patients with HCC (n = 3). Three-dimensional conformal RT (3DCRT) was delivered to patients. The planned total dose was 18 Gy, with a fractional size of 3.0 Gy, using 6-MV x-rays with a linear accelerator (Elekta Synergy; Elekta, Stockholm, Sweden) at five fractions per week. b Analysis of the GEPIA database revealed that Rac1 is upregulated in liver tumor tissues compared with normal tissues. c Kaplan–Meier curves of HCC survivals based on the expression status of Rac1 gene according to TCGA and GEPIA dataset. d Representative images and IHC scores of Rac1-GTP expression in tumor tissues of radioresistant HCC patients (Non/poor responders) (n = 8) and radiosensitive HCC patients (Good responders) (n = 11). e, f Dose responses of survival factions of Huh7 and HepG2 cells treated with DMSO or NSC23766. g–k B-NDG male mice were subcutaneously injected with Huh7 cells, then NSC23766 injected by intraperitoneal injection (1.5 mg/kg body weight), a 160 KeV X-ray linear accelerator (Rad Source Technologies Inc., TX, USA) with dose rate of 1.214 Gy/min (total 8 Gy) was used for animal irradiation (n = 5 per group). Tumor volumes were calculated (g). Tumor weights were determined (h). Representative images were shown (i). Data represent the mean ± s.d. Statistical significance was determined by two-tailed unpaired Student’s t test (h) and two-way ANOVA (g). **P < 0.01; ***P < 0.001; ****P < 0.0001. H&E and IHC analyses of mouse tumor tissues were performed with the indicated antibodies (j, k). Representative images were shown (j). The expression levels of the indicated proteins were quantified in eight microscopic fields of the tumor samples (k). Scale bars, 50 μm.
The contribution of Rac1 activity to HCC radiosensitivity was further examined in vitro using NSC23766, a Rac1 inhibitor. Pretreatment with 10 µM NSC23766 effectively reduced Rac1 activity in human HCC cells without inducing cytotoxicity (Fig. S1a, b). Inhibition of Rac1 markedly decreased the survival of Huh7 and HepG2 cells following IR, yielding sensitizer enhancement ratio (SER) of 1.34 and 1.28, respectively (Figs. 1e, f and S1c, d). A significant increase in γ-H2AX-positive nuclei occurred post-IR, followed by a gradual decline. HCC cells pretreated with NSC23766 exhibited more γ-H2AX foci and a delayed decline compared with those treated with DMSO. These findings suggest that Rac1 inactivation accelerates IR-induced DNA damage and impairs repair in HCC in vitro (Fig. S2a–d). Flow cytometry further revealed a higher proportion of Annexin V-positive apoptotic cells in the IR group compared with controls. NSC23766 pretreatment further enhanced apoptosis (Fig. S2e–h).
To assess the role of activated Rac1 in vivo, an HCC xenograft model was used to test whether NSC23766 improves therapeutic outcomes after IR. IR reduced xenograft volume, with a more pronounced effect observed in the NSC23766-treated group compared to saline controls (Fig. 1g, i). Rac1 inhibition also diminished xenograft weight following IR (Fig. 1h). IHC analysis confirmed that NSC23766 suppressed Rac1-GTP and its downstream effector p-PAK1, demonstrating a potent inhibitory effect on Rac1 activity in the murine model. IR elevated cleaved caspase-3 expression while reducing Ki67 levels. Furthermore, NSC23766 amplified the IR-induced decrease in Ki67 and increase in cleaved caspase-3 (Fig. 1j, k). In conclusion, these results indicate that Rac1 inactivation enhances radiosensitivity both in vitro and in vivo.
IR Ignites Rac1 activation and nuclear translocation to drive USP11 transcription
Previous research demonstrated that Rac1-GTP increases after radiation [23]. A similar pattern was observed in HCC cells, where Rac1-GTP rose within 24 h of irradiation, despite no significant changes in total Rac1 protein levels (Fig. 2a). Immunofluorescence staining of Rac1-GTP in Huh7 cells revealed its accumulation in the nucleus within 24 h post irradiation (Fig. 2b). A cytoplasm-nucleus separation assay showed consistent results (Fig. 2c). Given that Rac1 has been reported to indirectly modulate transcriptional pathways, our proteomic analysis of Rac1-knockdown Huh7 cells demonstrates that USP11 transcription was profoundly suppressed (Fig. S3a–c). Data from the GEPIA database also revealed a positive correlation between Rac1 and USP11 mRNA (Fig. S3d).
a The levels of Rac1-GTP and Rac1 in Huh7 cells treated with 8 Gy X-ray radiation for different durations. b Confocal microscopy was used to analyze the localization of Rac1-GTP in Huh7 cells treated with 8 Gy X-ray radiation for 24 h. c The levels of Rac1-GTP in nucleus and cytoplasm of Huh7 cells treated with 8 Gy X-ray radiation for different durations. d Dual-luciferase reporter assays were performed by transfecting pGL3-USP11 into Rac1 knockdown and its control of Huh7 cells. e ChIP assay was performed in Huh7 cells using an anti-Rac1 antibody. The enrichment of the USP11 promoter was assessed by PCR with specific primers. Input, total genomic DNA. f RT‒qPCR analysis of USP11 in Rac1 knockdown and its control of Huh7 cells. g Western blot of USP11 in Rac1 knockdown and its control of Huh7 cells. h Dual-luciferase reporter assays were performed via cotransfection of pGL3-USP11, Vector/Flag-Rac1-WT/Flag-Rac1-Q61L/Flag-Rac1-T17N into HEK293T cells. i RT‒qPCR analysis of USP11 after the transfection of Vector/Flag-Rac1-WT/Flag-Rac1-Q61L/Flag-Rac1-T17N into HEK293T cells. j Western blot analysis of USP11 expression in HEK293T cells transfected Vector/Flag-Rac1-WT/Flag-Rac1-Q61L/Flag-Rac1-T17N. k Dual-luciferase reporter assays were performed by transfecting pGL3-USP11 into Huh7 cells, followed by 8 Gy X-ray radiation for 24 h. l Western blot analysis of USP11 expression in Huh7 cells treated with 8 Gy X-ray radiation for different durations.
Afterwards, the dual-luciferase assays were conducted to confirm Rac1 governs USP11 expression through transcriptional regulation, showing that Rac1 knockdown significantly reduced USP11 transcription (Fig. 2d). ChIP assay was conducted to investigate the recruitment of Rac1 to the chromatin of the USP11 gene. As demonstrated in Fig. 2e, Rac1 was specifically associated with chromatin at the USP11 promoter, suggesting its potential role in the transcriptional regulation of USP11, either directly or indirectly. Consequently, both the mRNA and protein levels of USP11 were significantly lower (Fig. 2f, g). Functional characterisation of Rac1 mutants reveals that the Q61L variant sustains constitutive activation of Rac1-GTP, whereas the T17N mutation severely impairs GTPase activity [24]. mutants Rac1-Q61L and Rac1-T17N were used, and results showed increased USP11 transcription in Rac1-WT and Rac1-Q61L groups, while the Rac1-T17N group exhibited no significant difference from controls (Fig. 2h). Similarly, Rac1-WT and Rac1-Q61L promoted both USP11 transcription and translation, whereas Rac1-T17N did not (Fig. 2i, j). Further luciferase assays indicated increased transcription of USP11 following IR (Fig. 2k). Notably, USP11 expression also increased progressively within 24 h of irradiation (Fig. 2l). Collectively, these results demonstrate that IR induces Rac1 activation and subsequent nuclear translocation, where Rac1-GTP is required for the transcriptional upregulation of USP11.
USP11 directly interacts with Rac1 and stabilizes Rac1-GTP
Notably, co-immunoprecipitation (co-IP) coupled with Liquid Chromatography–Tandem Mass Spectrometry (LC‒MS/MS) in Huh7 cells revealed USP11 as a critical Rac1-interacting protein. And the interaction between Rac1 and USP11 was confirmed with co-IP assays from whole Huh7 cell lysates (Fig. 3a). Exogenous Flag-USP11 and Myc-Rac1 co-expression in HEK293T cells further validated their association by co-IP and western blot analysis (Fig. S4a). GST pull-down assays with purified Flag-USP11 and GST-Rac1 recombinant proteins demonstrated a direct interaction in vitro (Fig. 3b). These findings verify that Rac1 physically associates with USP11 in cells.
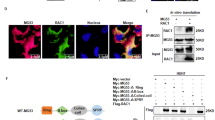
a Co-immunoprecipitation analysis of the interaction between endogenous USP11 and Rac1 in Huh7 cells. b GST pulldown analysis using purified bacteria-expressed GST-Rac1 protein and Flag-USP11 were performed in the presence of GTP. c Co-immunoprecipitation analysis in HEK293T cells expressing the indicated Flag-Rac1proteins. d Confocal microscopy was used to analyze the expression of USP11 (red) and Rac1-GTP (green) in Huh7 cells, and nuclei were stained with DAPI (blue). Scale bar, 20 µm. e The levels of Rac1-GTP and Rac1 in Huh7 and HepG2 cells with or without USP11 overexpression. f The levels of Rac1-GTP and Rac1 in Huh7 and HepG2 cells with or without USP11 knockdown. g The levels of Rac1-GTP and Rac1 in indicated cells. MG-132 (10 μM) were added for 4 h. h Rac1 and Rac1-GTP levels in HepG2 cells with or without USP11 knockdown followed by treatment with CHX (10 µM) for the indicated times. i Rac1 and Rac1-GTP levels in Huh7 cells with or without USP11 knockdown followed by treatment with CHX (10 µM) for the indicated times.
USP11 showed stronger binding to constitutively active Rac1-Q61L than to dominant-negative Rac1-T17N, suggesting a preference for GTP-bound Rac1 (Fig. 3c). Immunofluorescence staining in Huh7 cells showed that USP11 and Rac1-GTP share similar localisation patterns (Fig. 3d). These results suggest that USP11 functions through its interaction with Rac1-GTP. Rac1-GTP levels were measured in HCC cells following USP11 modulation to investigate the functional consequences of the Rac1-GTP-USP11 interaction. Overexpression of USP11 significantly increased Rac1-GTP levels without affecting total Rac1 expression (Fig. 3e). In contrast, USP11 knockdown reduced Rac1-GTP levels, while total Rac1 expression remained unchanged (Fig. 3f). The decrease in Rac1-GTP following USP11 knockdown was reversible by proteasome inhibition, suggesting that USP11 stabilises Rac1-GTP by preventing proteasomal degradation (Fig. 3g). To further explore the role of USP11 in Rac1-GTP stability, cells were treated with cycloheximide (CHX) to block protein synthesis. Rapid degradation of Rac1-GTP was observed in USP11-knockdown HCC cells, highlighting USP11’s role in protecting Rac1-GTP from proteasomal degradation (Fig. 3h, i).
USP11 deubiquitinates Rac1 at the K123, K147 and K183 residues
Given the deubiquitinase activity of USP11, the ubiquitination levels of Rac1 were assessed. USP11 overexpression reduced Rac1 ubiquitination in HEK293T cells (Fig. 4a), while USP11 knockdown increased ubiquitinated Rac1 in Huh7 cells (Fig. 4b). These results confirm that USP11 functions as a deubiquitinase, targeting Rac1 for deubiquitination in HCC cells. Additionally, HEK293T cells were transfected with lysine-specific ubiquitin mutants (K6R, K11R, K27R, K29R, K48R, K63R), revealing that USP11 specifically cleaves K11- and K63-linked polyubiquitin chains on Rac1 (Fig. 4c).
a Anti-Ub immunoblotting assay of Myc-Rac1 ubiquitination in HEK293T cells with or without USP11 overexpression treated with MG-132 (10 μM) for 4 h. b Anti-Ub immunoblotting assay of Flag-Rac1 ubiquitination in Huh7 cells with or without USP11 knockdown treated with MG-132 (10 μM) for 4 h. c Immunoprecipitation and immunoblotting assay were selected to detect the type of ubiquitination of Myc-Rac1 in HEK293T cells transfected with wild-type or K6/K11/K27/K29/K33K48/K63 mutant Ub. Cells were treated with MG-132 (10 μM) for 4 h before harvesting. d Mass spectrometry analysis was employed to identify changes in the ubiquitination sites of Rac1 in overexpressed USP11 cells compared to controls. e Immunoprecipitation and immunoblotting to detect the ubiquitination of Rac1 mutants in HEK293T cells co-transfected with Myc-Rac1 mutants, Flag-USP11, and HA-Ub. Cells were treated with MG132 (10 μM) for 4 h before harvesting. f Immunoprecipitation and immunoblotting to detect the ubiquitination of Rac1 mutants in Huh7 cells with USP11 knockdown or not. Cells were co-transfected with Myc-Rac1 mutants and HA-Ub and treated with MG132 (10 μM) for 4 h before harvesting.
LC‒MS/MS data were reanalysed to identify the potential deubiquitination sites on Rac1 [23]. Lysines K123, K133, K147, and K183 showed reduced di-glycine modification, suggesting these residues as potential deubiquitination sites (Fig. 4d). These residues are highly conserved across species (Fig. S5a). Mutants K123R, K133R, K147R, and K183R were constructed, and K123R, K147R, and K183R mutants significantly reversed the decrease in Rac1 polyubiquitination in the presence of USP11, resembling the results observed in USP11-knockdown Huh7 cells (Fig. 4e, f). These results elucidate the mechanism by which USP11 deubiquitinates Rac1, identifying the specific ubiquitin chains and sites involved in this process.
USP11 promotes radioresistance in HCC by stabilizing Rac1-GTP at the K123, K147 and K183 residues
Mutations at K123, K147, and K183, but not K133, reduced Rac1-GTP expression. Additionally, K123, K147, and K183 mutations reversed USP11-induced Rac1-GTP upregulation (Fig. 5a). These results demonstrate that K123, K147, and K183, particularly K123, are key to Rac1-GTP stability mediated by USP11.
a The levels of Rac1-GTP in HEK293T cells co-transfected with Myc-Rac1 mutants (K123R, K133R, K147R, and K183R) and Flag-USP11 or the corresponding vector. b Initial structure of the Rac1 wide type and double-mutant (K123R-K147R). The Rac1 protein is depicted in an azure ribbon, with key structural regions highlighted: Switch I in red and Switch II in blue. The K123R and K147R mutations are represented as spheres, while the binding ligand GDP and the cofactor Mg²⁺ are shown in stick and sphere models, respectively. c Root mean square deviation (RMSD) over the simulation time for the WT and double-mutant systems, illustrating differences in global structural stability. d Root mean square fluctuation (RMSF) by residue for the WT and double-mutant systems, highlighting increased flexibility in specific regions of the double-mutant structure. e, f Colony formation assay in the indicated cells, and the quantification data represent the mean ± SD.
500-ns molecular dynamics simulations revealed that the K123R-K147R double mutation induced conformational instability at Rac1’s GTP-binding interface (Fig. 5b). This mutation resulted in an increased Root Mean Square Deviation (RMSD) (0.17 nm vs. 0.12 nm for wild-type Rac1) and enhanced flexibility in the Switch I region (Fig. 5c, d). The K123R-K147R double mutation destabilized Rac1 globally, increasing flexibility in critical Switch I and II regions. This disruption likely affects the stable binding of GDP and Mg²⁺, impairing GTP binding to Rac1 (Fig. S5b, c).
Furthermore, in USP11-overexpressing HepG2 cells, mutation of K123, K147, and K183 to arginine significantly decreased survival after IR. The SERs were 1.34, 1.25, and 1.22, respectively (Figs. 5e, f, and S5d, e). These findings suggest that USP11 enhances radioresistance in HCC by stabilising Rac1-GTP, primarily through deubiquitination at the K123, K147, and K183 residues.
Synergistic inhibition of USP11 and Rac1 restores HCC radiosensitivity in vivo
Additionally, the role of USP11 in HCC radioresistance was examined. Depletion of USP11 increased radiosensitivity in both in vitro and in vivo models (Figs. S6a–i and S7a–i). Notably, USP11 expression was significantly higher in non/poor responders compared to good responders (Fig. 6a). Considering the possible feedback regulation of Rac1 and USP11, we wondered whether simultaneous inhibition of USP11 and Rac1 activity may further increase efficacy. We next explored the effects of combination treatment with small-molecule inhibitors on the radiosensitivity of HCC. The small-molecule Rac1 inhibitor NSC23766 and Mitoxantrone (MIX) lipid nanoparticles were used for subsequent experiments. We examined the effects of combination treatment with MIX and NSC23766 on tumour growth and radiosensitivity in xenograft tumour models. Compared with single inhibitor treatment, combination treatment had a more pronounced inhibitory effect on the growth and radioresistance of tumours (Fig. 6c–g). Collectively, these results demonstrate that combination treatment can overcome radioresistance in HCC in vivo, indicating that the combination of NSC23766 and MIX may be a potential radiosensitization strategy for HCC.
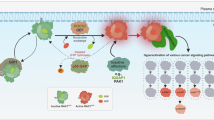
a Representative images and IHC scores of Rac1-GTP expression in tumor tissues of radioresistant HCC patients (Non/poor responders) (n = 8) and radiosensitive HCC patients (Good responders) (n = 11). b A schematic model for the mechanisms of Rac1-USP11 feedback loop in HCC radioresistance. IR induces Rac1 activation and nuclear translocation, where Rac1-GTP transcriptionally upregulates USP11. USP11 then stabilizes Rac1-GTP through deubiquitination at residues K123/K147/K183, establishing a self-reinforcing Rac1-USP11 amplification loop that drives radioresistance. c–g Huh7 cells were subcutaneously injected into B-NDG male mice, and then NSC23766 (1.5 mg/kg/day for 1 week) and MIX (3 mg/kg) lipid nanoparticles were injected by intraperitoneal injection 10 days later; then, a 160 KeV X-ray linear accelerator (Rad Source Technologies Inc., TX, USA) with a dose rate of 1.214 Gy/min (total 8 Gy) was used for animal irradiation (n = 5 per group). Representative images were showed (c). Tumor volumes were calculated (d). Tumor weights were determined (e). Data represent the mean ± s.d. Statistical significance was determined by two-way ANOVA (d). Statistical significance was determined by two-tailed unpaired Student’s t test (e, g). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.H&E and IHC analyses of mouse tumor tissues were performed with the indicated antibodies (f, g). Representative images were shown (f). The expression levels of the indicated proteins were quantified for eight microscopic fields of the tumor samples (g). Scale bar, 50 μm.
Discussion
Rac1 activity plays a crucial role in the development of several cancers through various mechanisms [25,26,27]. In the present study, we discovered that IR instigates Rac1 activation and nuclear translocation, enabling Rac1-GTP to transcriptionally upregulate USP11 in HCC. Newly synthesised USP11 then sustains Rac1-GTP stability through deubiquitination, forging a self-reinforcing Rac1-USP11 signalling circuit and driving HCC radioresistance (Fig. 6b). The Rac1-USP11 loop may synergise with known radioresistance mechanisms such as ROS scavenging and DNA repair, as Rac1-GTP is known to regulate NOX-dependent ROS production and promote nuclear DNA damage response pathways [28].
A growing body of evidence supports that activated Rac1 regulates cell motility and cytoskeleton polymerization and affects the transcription of downstream genes. The overexpression and hyperactivation of Rac1 are correlated with the aggressive growth of tumour cells and therapeutic resistance [13, 29]. Our data indicated that activated Rac1 is potentially an attractive radiosensitization target in HCC.
Rac1 undergoes a conformational change upon binding to GTP, exposing its hidden nuclear localisation signal (NLS) and allowing it to be recognized by the nuclear import receptor karyopherin α for active translocation into the nucleus [30]. And Rac1 in nucleus has been reported to recruit multiple transcription factors to exert its effects [31, 32]. Among these, several essential transcription factors, such as E2F, NF-κB, and STAT3, have been identified as downstream effectors of Rac1 [33,34,35]. Here, we found that the abundance of Rac1 exhibited a positive correlation with USP11 in HCC, implying a potential regulatory relationship between Rac1 and USP11. The results demonstrated that Rac1-GTP accumulates in cells, especially in the nucleus, after stimulation with IR, and the expression of USP11 is correspondingly increased.
Unexpectedly, USP11 was identified as a direct Rac1-interacting protein, suggesting functional crosstalk beyond transcriptional regulation within the Rac1-USP11 axis. Previous studies indicate that activated Rac1 is labile and is subject to ubiquitination and degradation. For example, it has been reported that E3 ubiquitin protein ligase 1 (HACE1) directly interacts with GTP-bound Rac1 to catalyse the polyubiquitylation and degradation of Rac1-GTP [21]. Another E3 ubiquitin ligase, MG53, has also been shown to act as a direct inhibitor of Rac1 to catalyse the ubiquitination of Rac1 and further inhibit Rac1 activity in HCC cells [20]. Notably, the substrate protein is not only ubiquitinated by E3 ubiquitin ligases but also removes bound ubiquitin molecules through deubiquitinating enzymes. Therefore, we investigated whether Rac1 serves as a direct substrate for USP11 and if USP11 regulates Rac1 activity through deubiquitination-mediated stabilisation. In the present study, we demonstrated that USP11 could directly interact with and stabilised Rac1-GTP through deubiquitination.
For USP11, ubiquitin chain cleavage assays with all eight linkages revealed its preference for Lys63, Lys6, Lys33, and Lys11-linked chains as well as linear chains over Lys27, Lys29, and Lys48-linked chains [36, 37]. Additionally, we demonstrated that USP11 preferentially cleaves the K11-linked and K63-linked polyubiquitin chains on the Rac1 protein. Furthermore, USP11 was shown to deubiquitinate Rac1 at lysine residues 123, 147, and 183, thereby stabilising Rac1-GTP by antagonising its ubiquitin-proteasome system (UPS)-mediated degradation.
USP11 has been reported to play key roles in regulating the cell cycle, DNA repair, signal transduction, tumour development, and other critical biological processes [38,39,40]. For example, USP11 has been found to potentiate HGF/AKT signalling to drive metastasis in HCC by deubiquitinating and stabilizing the eEF1A1 protein [38]. Notably, the downregulation of USP11 induced by Notch/Hey1 not only confers multiple malignant characteristics of aggressive gliomas but also enhances the self-renewal and tumour-forming abilities of patient-derived glioma-initiating cells [41]. These studies suggest that USP11 may play distinct roles in tumour progression, depending on the cancer type or targeted substrate. Our data demonstrated that USP11 silencing inhibited HCC growth and proliferation in vitro and in vivo, suggesting that USP11 functions as an oncoprotein that promotes HCC progression. Furthermore, our rescue experiments revealed that the USP11 overexpression-induced radioresistance of HCC cells is reversed by Rac1 knockdown. Mutations in Rac1 at K123, K147, and K183 also abolished the radioresistance phenotype, indicating that these residues may be attractive radiosensitization targets in HCC.
Inevitably, there are some limitations to our study. Firstly, while elevated Rac1-GTP and USP11 levels correlate with inferior radiotherapy response in our HCC patient cohort, further validation with broader patient cohorts is still needed. Secondly, although activated Rac1 has been demonstrated to promote USP11 transcription, the precise molecular mediators of this transcriptional cascade require experimental elucidation. Notably, Rac1 has been reported to modulate E2F1 and STAT3 functions through distinct signalling cascades [34, 42, 43], and E2F1 itself is a documented transcriptional activator of USP11, suggesting a plausible mechanism linking Rac1 activation to USP11 upregulation [44, 45]. Additionally, our study is limited to HCC, so further research is needed to understand the role of the Rac1-USP11 feedback loop in other cancers. Investigating the expression and function of these genes in different types of cancer would improve the relevance and potential applications of our findings.
In summary, this work clarified the pivotal role of the Rac1-USP11 feedback loop in HCC radioresistance. We found that Rac1-GTP accumulates in cells, especially in the nucleus, after stimulation with IR, which subsequently contributes to the transcription of USP11. Rac1 is deubiquitinated to maintain its activity by USP11, thereby, thus a reciprocal feedback loop is formed between USP11 and Rac1-GTP. And the synergistic effect of NSC23766 and MIX highlights the therapeutic potential of concurrently targeting Rac1-GTP and USP11 to overcome compensatory feedback loops. Additionally, this study also provides valuable clues for the future development of specific small-molecule inhibitors targeting USP11 and Rac1-GTP. To realize these therapeutic possibilities and fully understand their clinical implications, comprehensive validation through preclinical and clinical studies is essential.
Methods
Patient samples
A total of nineteen HCC specimens were obtained from patients who underwent either hepatectomy or ultrasonically guided liver biopsy before receiving radiotherapy at the Eastern Hepatobiliary Surgery Hospital, Navy Military Medical University of China. The progression and prognosis of these patients were evaluated using CT scans conducted before and after radiotherapy, in accordance with the Response Evaluation Criteria in Solid Tumours version 1.1 (RECIST 1.1). Patients who showed no reduction or experienced progression in HCC volume following IR were classified as non/poor responders (8 specimens). In contrast, those who exhibited substantial tumour reduction after irradiation were categorised as good responders (11 specimens). All clinical experiments involving tissue samples adhered to international ethical standards and the guidelines outlined by the World Medical Association (Declaration of Helsinki). Written informed consent was obtained from all participants prior to their involvement. The study was approved by the Institutional Research Ethics Committee of the Eastern Hepatobiliary Surgery Hospital, Navy Military Medical University (approval number: EHBHKY2020-K-012).
RNA-seq analysis
Total EV RNA isolated from plasma was treated with DNase I. Strand-specific RNA-seq libraries were prepared using the SMARTer Stranded Total RNA-Seq Kit—Pico Input Mammalian, followed by library quality assessment. Sequencing was performed on an Illumina platform (150 bp PE). Raw reads were filtered, aligned to GRCh38 using HISAT2, and gene expression quantified in TPM. mRNA and lncRNA annotations were sourced from GENCODE. Differential exLRs were identified, annotated, and subjected to KEGG pathway enrichment analysis using DAVID, as described previously [46].
Cell culture
Human cell lines Huh7, HepG2, and HEK293T were procured from the American Type Culture Collection (ATCC). The cells were cultured in DMEM (Hyclone, Utah, USA) supplemented with 10% fetal bovine serum (Gibco, California USA), 100 µ/mL penicillin, and 100 µg/mL streptomycin (Beyotime Biotechnology, Shanghai, China), in a humidified incubator set to 5% CO2 and 95% air. The cultures were maintained until passage 20. Authentication of cell identity was performed using short tandem repeat (STR) profiling. Routine mycoplasma contamination testing was conducted using MycoBlue Mycoplasma Detector (Vazyme, Nanjing, China), and only cells testing negative for contamination were used.
Vectors and reagents
Lentiviral vectors (pGWLV13-Flag-USP11, pLVX-Flag-Rac1 and pGWLV13-Myc-Rac1) were purchased from Genewiz (Suzhou, China). The empty vector was used as the negative control. The pLVX-Flag-Rac1-WT/Q61L/T17N plasmid, HA-ubiquitin plasmid and its mutant (K6R, K11R, K27R, K29R, K33R, K48R, K63R) plasmid were purchased from MiaoLing Biotechnology (Wuhan, China). The RNAi-resistant Rac1 WT, K123R, K133R, K147R and K183R constructs contain non-sense T371C, T377C, and A383G mutations. The target sequences for shRNA were as follows: USP11, CCGTGATGATATCTTCGTCTA; Rac1, GGATACAGCTGGACAAGAATTCAAGAGATTCTTGTCCAGCTGTATCCTTTTTT. NSC23766 was obtained from APE×BIO (TX, USA). Mitoxantrone hydrochloride liposome injection was sourced from CSPC (Beijing, China).
Colony formation assay
The radiosensitivity of HCC cells was evaluated through a colony formation assay. A range of 500–4000 cells per well were plated in 6-well plates and exposed to different doses of IR (0, 2, 4, 6, 8 Gy) in triplicate. Following irradiation, the cells were incubated for 14 to 20 days to allow colony formation, then fixed and stained with crystal violet. Colonies containing more than 50 cells were counted, and the survival fraction was determined. Survival curves were constructed using formula SF = 1−(1−exp(−k*D)) ^ N, based on the single-hit multi-target model.
Xenograft tumor mouse model
Male B-NDG mice (NOD.CB17-PrkdcscidIl2rgtm1/Bcgen), aged 5 to 6 weeks, were purchased from Biocytogen (Beijing, China) and used to establish the xenograft tumour model. All animal procedures were carried out following institutional and national ethical guidelines. The study was approved by the Institutional Research Ethics Committee of the State Key Laboratory of Radiology and Radiation Protection, School of Radiation Medicine and Protection, Medical College of Soochow University, Jiangsu Province, China. In brief, 2 × 106 Huh7 cells in 50 µL of serum-free DMEM mixed with 50 µL of matrix gel were injected subcutaneously. Tumour growth was monitored every other day, and tumour volume was calculated using the formula V (mm³) = 0.5 × length × width².
Cell and tumor irradiation
HCC cells were irradiated with a 160 kV X-ray irradiator at different dose levels, using a dose rate of 1.214 Gy/min (Rad Source Technologies Inc., TX, USA). For in vivo tumour irradiation, mice with HCC xenografts were anaesthetised with a ketamine/xylazine mixture (100 mg/kg + 10 mg/kg) and positioned in a custom lead mold for localized X-ray irradiation (8 Gy).
Luciferase reporter assay
Cells were transfected with luciferase reporter and internal control pRL-TK (Promega, Madison, WI, USA) using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, Massachusetts, USA). Luciferase activity was detected using a Dual-Luciferase Reporter Assay System. Promoter activities were expressed as the ratio of Firefly luciferase activity to Renilla luciferase activity as previously described [47].
Chromatin immunoprecipitation (ChIP) assays
ChIP assay was carried out using a commercial ChIP kit (ACE, BK0045-01) according to the manufacturer’s instructions as described previously [48]. Immunoprecipitation was performed using either an anti-Rac1 antibody or a control IgG antibody. Quantitative PCR (qPCR) was subsequently conducted using primers specifically designed to amplify target sequences within the human USP11 promoter as follow: F: 5′- GGTCTAGAACTCACCCAACTGC -3′, R: 5′- ATCTGGGCAGCCAGGTAAATC -3′.
Co-immunoprecipitation
Co-immunoprecipitation (co-IP) was performed to investigate the interaction between Rac1 and USP11. Cell lysates (300–500 μg) were prepared using NP40 buffer (Thermo Fisher Scientific, USA). The lysates were used for immunoprecipitation with anti-Myc, anti-Flag, or the specified antibody, and then incubated by Protein A/G Magnetic Beads (MedChemExpress, New Jersey, USA). The eluted proteins were then analysed by western blotting.
Liquid chromatography - tandem mass spectrometry assays
For the identification of interacting proteins, a protein band visualised by Coomassie blue staining was excised from an SDS-PAGE gel and subjected to in-gel digestion. The digestion was performed overnight at 37 °C in 50 mM ammonium bicarbonate buffer containing RapiGest (Waters Corporation) and 200 ng of sequencing-grade modified trypsin (Promega, USA). The digested peptides were then analysed using high-sensitivity LC-MS/MS with an Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, USA). Protein identification was carried out by matching the fragment spectra to the UniProt protein database (EMBL-EBI) using the Mascot search engine (v.2.3; Matrix Science) and Proteome Discoverer software (v.1.4; Thermo Fisher).
Glutathione-S-transferase pulldown assay
GST pulldown assays were performed as previously described. Briefly, equal amounts of Flag-tagged USP11 (300 ng) were incubated with GST-Rac1 (100 ng) and glutathione magnetic beads in a modified binding buffer containing 50 mM Tris-HCl (pH 7.5), 1% Triton X-100, 150 mM NaCl, 1 mM DTT, 0.5 mM EDTA, 100 μM PMSF, 100 μM leupeptin, 1 μM aprotinin, 100 μM sodium orthovanadate, 100 μM sodium pyrophosphate, and 1 mM sodium fluoride. The mixture was incubated overnight at 4 °C. The beads were washed three times with binding buffer and then analysed by immunoblotting.
Analysis of Rac1 activity
Rac1 activation was assessed by quantifying the ratio of GTP-bound Rac1 to total Rac1 protein levels. Cells were lysed in ice-cold lysis buffer. Lysates were incubated with PAK-PBD-conjugated GST beads (Millipore Sigma, USA) for 1 h at 4 °C with rotation to pull down active GTP-bound Rac1. Beads were washed once with lysis buffer containing 1% NP-40, followed by two washes with NP-40-free buffer, and resuspended in 1× Laemmli loading buffer (YEASEN, China). Precipitated GTP-bound Rac1 and total Rac1 in whole-cell lysates were resolved by SDS-PAGE and immunoblotted using an anti-Rac1 monoclonal antibody. Protein bands were quantified by densitometry (ImageJ), and Rac1 activity was normalized to total Rac1 and α-tubulin (loading control) levels.
Ubiquitylation assay
Huh7 and HEK293T cells were treated with 10 µM MG132 (MedChemExpress, USA) for 4 h. After treatment, the cells were washed with PBS and lysed for immunoprecipitation using 4 µg of anti-Rac1, anti-Myc, or anti-Flag antibodies. Ubiquitination of Rac1 was then detected by Western blotting with anti-HA or anti-Ub antibodies.
Quantification and statistical analysis
Data are presented as the mean ± SD from three independent experiments. Statistical analysis was performed using GraphPad Prism 6 software. The t-test and ANOVA was used to compare means. Overall survival curves were plotted via the Kaplan–Meier method and compared by the log-rank test. Bars and errors represent the mean ± standard deviation (SD) of repeated measurements; Significance levels were indicated as follows: *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Data availability
The data that support the findings of this study are available in the main text, supplemental figures and supplemental information file. Data are also available from the corresponding author upon reasonable request.
References
European Association for the Study of the Liver. EASL Clinical Practice Guidelines on the management of hepatocellular carcinoma. J Hepatol. 2025;82:315–74.
Vogel A, Meyer T, Sapisochin G, Salem R, Saborowski A. Hepatocellular carcinoma. Lancet. 2022;400:1345–62.
Lee S, UKim TH. Current evidence and the potential role of proton beam therapy for hepatocellular carcinoma. Clin Mol Hepatol. 2023;29:958–68.
Choi S, HSeong J. Strategic application of radiotherapy for hepatocellular carcinoma. Clin Mol Hepatol. 2018;24:114–34.
Apisarnthanarax S, Barry A, Cao M, Czito B, DeMatteo R, Drinane M, et al. External beam radiation therapy for primary liver cancers: an ASTRO Clinical Practice Guideline. Pract Radiat Oncol. 2022;12:28–51.
Geng L, Zhu M, Luo D, Chen H, Li B, Lao Y, et al. TKT-PARP1 axis induces radioresistance by promoting DNA double-strand break repair in hepatocellular carcinoma. Oncogene. 2024;43:682–92.
Liao J, Yi Y, Yue X, Wu X, Zhu M, Chen Y, et al. Methyltransferase 1 is required for nonhomologous end-joining repair and renders hepatocellular carcinoma resistant to radiotherapy. Hepatology. 2023;77:1896–910.
Wen J, Xiong K, Aili A, Wang H, Zhu Y, Yu Z, et al. PEX5, a novel target of microRNA-31-5p, increases radioresistance in hepatocellular carcinoma by activating Wnt/β-catenin signaling and homologous recombination. Theranostics. 2020;10:5322–40.
Seshacharyulu P, Halder S, Nimmakayala R, Rachagani S, Chaudhary S, Atri P, et al. Disruption of FDPS/Rac1 axis radiosensitizes pancreatic ductal adenocarcinoma by attenuating DNA damage response and immunosuppressive signalling. eBioMedicine. 2022;75:103772.
Liu N, Cui W, Jiang X, Zhang Z, Gnosa S, Ali Z, et al. The critical role of dysregulated RhoB signaling pathway in radioresistance of colorectal cancer. Int J Radiat Oncol Biol Phys. 2019;104:1153–64.
Pranatharthi A, Thomas P, Udayashankar AH, Bhavani C, Suresh SB, Krishna S, et al. RhoC regulates radioresistance via crosstalk of ROCK2 with the DNA repair machinery in cervical cancer. J Exp Clin Cancer Res. 2019;38:392.
Zhou W, Zhao Z, Lin A, Yang JZ, Xu J, Wilder-Romans K, et al. GTP signaling links metabolism, DNA repair, and responses to genotoxic stress. Cancer Discov. 2024;14:158–75.
Wu J, Wu Y, Zhao T, Wang X, Guo Q, Wang S, et al. Targeting RAC1 reactivates pyroptosis to reverse paclitaxel resistance in ovarian cancer by suppressing P21-activated kinase 4. MedComm. 2024;5. e719.
Huang H, Tsui Y-M, Ho DW-H, Chung CY-S, Sze KM-F, Lee E, et al. LANCL1, a cell surface protein, promotes liver tumor initiation through FAM49B-Rac1 axis to suppress oxidative stress. Hepatology. 2024;79:323–40.
Nguyen LK, Kholodenko BN, von Kriegsheim A. Rac1 and RhoA: networks, loops and bistability. Small GTPases. 2018;9:316–21.
Liu H, Pan D, Li P, Wang D, Xia B, Zhang R, et al. Loss of ZBED6 protects against sepsis-induced muscle atrophy by upregulating DOCK3-mediated RAC1/PI3K/AKT signaling pathway in pigs. Adv Sci. 2023;10:e2302298.
Feddersen CR, Schillo JL, Varzavand A, Vaughn HR, Wadsworth LS, Voigt AP, et al. Src-dependent DBL family members drive resistance to vemurafenib in human melanoma. Cancer Res. 2019;79:5074–87.
Cheng X, Barakat R, Pavani G, Usha MK, Calderon R, Snella E, et al. Nod1-dependent NF-kB activation initiates hematopoietic stem cell specification in response to small Rho GTPases. Nat Commun. 2023;14:7668.
Oberoi-Khanuja T, KRajalingam K. Ubiquitination of Rac1 by inhibitors of apoptosis (IAPs). Methods Mol Biol. 2014;1120:43–54.
Ma X, Ma X, Zhu L, Zhao Y, Chen M, Li T, et al. The E3 ubiquitin ligase MG53 inhibits hepatocellular carcinoma by targeting RAC1 signaling. Oncogenesis. 2022;11:40.
Torrino S, Visvikis O, Doye A, Boyer L, Stefani C, Munro P, et al. The E3 ubiquitin-ligase HACE1 catalyzes the ubiquitylation of active Rac1. Dev Cell. 2011;21:959–65.
Majolée J, Podieh F, Hordijk P, LKovačević I. The interplay of Rac1 activity, ubiquitination and GDI binding and its consequences for endothelial cell spreading. PLoS ONE. 2021;16:e0254386.
Tang Y, Wang T, Gu L, Xu Y, Yang Z, Zhu W, et al. USP11 exacerbates radiation-induced pneumonitis by activating endothelial cell inflammatory response via OTUD5-STING signaling. Int J Radiat Oncol Biol Phys. 2024;119:1261–74.
Alan J, KLundquist EA. Mutationally activated Rho GTPases in cancer. Small GTPases. 2013;4:159–63.
Ma N, Xu E, Luo Q, Song G. Rac1: a regulator of cell migration and a potential target for cancer therapy. Molecules. 2023;28:2976.
Liu J, Zhang C, Zhang T, Chang C-Y, Wang J, Bazile L, et al. Metabolic enzyme LDHA activates Rac1 GTPase as a noncanonical mechanism to promote cancer. Nat Metab. 2022;4:1830–46.
Zhou K, Rao J, Zhou Z-H, Yao X-H, Wu F, Yang J, et al. RAC1-GTP promotes epithelial-mesenchymal transition and invasion of colorectal cancer by activation of STAT3. Lab Invest. 2018;98:989–98.
Goldschmidt-Clermont PJ, Sevilla BA. Redox and actin, a fascinating story. Redox Biol. 2025;83:103630.
Li Q, Qin T, Bi Z, Hong H, Ding L, Chen J, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11:1456.
Sandrock K, Bielek H, Schradi K, Schmidt GKlugbauer N. The nuclear import of the small GTPase Rac1 is mediated by the direct interaction with karyopherin alpha2. Traffic. 2010;11:198–209.
Michaelson D, Abidi W, Guardavaccaro D, Zhou M, Ahearn I, Pagano M, et al. Rac1 accumulates in the nucleus during the G2 phase of the cell cycle and promotes cell division. J Cell Biol. 2008;181:485–96.
Kawashima T, Bao YC, Nomura Y, Moon Y, Tonozuka Y, Minoshima Y, et al. Rac1 and a GTPase-activating protein, MgcRacGAP, are required for nuclear translocation of STAT transcription factors. J Cell Biol. 2006;175:937–46.
Xu L, Zhang L, Zhang S, Yang J, Zhu A, Sun J, et al. Taxifolin inhibits melanoma proliferation/migration impeding USP18/Rac1/JNK/β-catenin oncogenic signaling. Phytomedicine. 2024;123:155199.
Simon AR, Vikis HG, Stewart S, Fanburg BL, Cochran B, HGuan KL. Regulation of STAT3 by direct binding to the Rac1 GTPase. Science. 2000;290:144–47.
Shi X, Chen W, Yin Y, Cao H, Wang X, Jiang W, et al. RAC1high NK cell-based immunotherapy in hepatocellular carcinoma via STAT3-NKG2D axis. Cancer Lett. 2024;592:216909.
Ideguchi H, Ueda A, Tanaka M, Yang J, Tsuji T, Ohno S, et al. Structural and functional characterization of the USP11 deubiquitinating enzyme, which interacts with the RanGTP-associated protein RanBPM. Biochem J. 2002;367:87–95.
Harper S, Gratton HE, Cornaciu I, Oberer M, Scott DJ, Emsley J, et al. Structure and catalytic regulatory function of ubiquitin specific protease 11 N-terminal and ubiquitin-like domains. Biochemistry. 2014;53:2966–78.
Chen J, Ning D, Du P, Liu Q, Mo J, Liang H, et al. USP11 potentiates HGF/AKT signaling and drives metastasis in hepatocellular carcinoma. Oncogene. 2024;43:123–35.
Qiao L, Zhang Q, Sun Z, Liu Q, Wu Z, Hu W, et al. The E2F1/USP11 positive feedback loop promotes hepatocellular carcinoma metastasis and inhibits autophagy by activating ERK/mTOR pathway. Cancer Lett. 2021;514:63–78.
Xu Y, Zeng J, Liu K, Li D, Huang S, Fu S, et al. USP11 promotes lipogenesis and tumorigenesis by regulating SREBF1 stability in hepatocellular carcinoma. Cell Commun Signal. 2024;22:550.
Wu H-C, Lin Y-C, Liu C-H, Chung H-C, Wang Y-T, Lin Y-W, et al. USP11 regulates PML stability to control Notch-induced malignancy in brain tumours. Nat Commun. 2014;5:3214.
Shi Y, Bollam SR, White SM, Laughlin SZ, Graham GT, Wadhwa M, et al. Rac1-mediated DNA damage and inflammation promote Nf2 tumorigenesis but also limit cell-cycle progression. Dev Cell. 2016;39:452–65.
Karnoub AE, Der C, JCampbell SL. The insert region of Rac1 is essential for membrane ruffling but not cellular transformation. Mol Cell Biol. 2001;21:2847–57.
Liu L, Zhang H, Shi L, Zhang W, Yuan J, Chen X, et al. Inhibition of Rac1 activity induces G1/S phase arrest through the GSK3/cyclin D1 pathway in human cancer cells. Oncol Rep. 2014;32:1395–400.
Zhou F, Deng Z, Shen D, Lu M, Li M, Yu J, et al. DLGAP5 triggers proliferation and metastasis of bladder cancer by stabilizing E2F1 via USP11. Oncogene. 2024;43:594–607.
Yu S, Li Y, Liao Z, Wang Z, Wang Z, Li Y, et al. Plasma extracellular vesicle long RNA profiling identifies a diagnostic signature for the detection of pancreatic ductal adenocarcinoma. Gut. 2020;69:540–50.
Feng Y, Yuan P, Guo H, Gu L, Yang Z, Wang J, et al. METTL3 mediates epithelial-mesenchymal transition by modulating FOXO1 mRNA N6 -methyladenosine-dependent YTHDF2 binding: a novel mechanism of radiation-induced lung injury. Adv Sci. 2023;10:e2204784.
Adams DR, Ron DKiely PA. RACK1, A multifaceted scaffolding protein: structure and function. Cell Commun Signal. 2011;9:22.
Funding
This work was supported by the National key R&D Program of China (2022YFC2503700, 2022YFC2503703), the Natural Science Foundation of China (82103483, 82102941), the Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX23_3262), the Key Scientific Research Projects of Jiangsu Provincial Health Commission (ZD2021053), the Collaborative Projects of Collaborative Innovation Center of Radiological Medicine of Jiangsu Higher Education Institutions (FY202401), the Suzhou Fundamental Research Project (SJC2023001), and A Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Contributions
Kaixiao Zhou, Yang Jiao, Yabo Jiang designed research; Kaixiao Zhou, Yabo Jiang, Jiahao Guo, Haobo Zhang, Yuhao Hu, Xuanyu Meng, Yecheng Li, Shaohua Wei, and Jian Wang performed research; Kaixiao Zhou and Yang Jiao analyzed data; Xubiao Wei contributed experimental facilities and analytic tools; Kaixiao Zhou wrote the paper, and Yabo Jiang, Shuqun Cheng, Jianping Cao and Yang Jiao revised the content. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, K., Jiang, Y., Guo, J. et al. The Rac1-USP11 feedback amplification loop: a radiation-activated engine driving radioresistance in hepatocellular carcinoma. Br J Cancer 134, 391–403 (2026). https://doi.org/10.1038/s41416-025-03265-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41416-025-03265-1