Abstract
High-grade serous ovarian carcinoma (HGSOC) is one of the most lethal gynecological cancer. Genetic studies have revealed gene copy number alterations (CNAs) frequently occurred in HGSOC pathogenesis, however the function and mechanism of CNAs for microRNAs are still not fully understood. Here, we show the dependence on gene copy number amplification of MIR937 that enhances cell autophagy and dictates HGSOC proliferative activity. Data mining of TCGA database revealed MIR937 amplification is correlated with increased MIR937 expression and cell proliferation of HGSOC. Deletion of MIR937 in HGSOC cells led to impaired autophagy and retarded cell proliferation, and the extent for its inhibitory effects scaled with the degree of MIR937 copy loss. Rescue assay confirmed miR-937-5p, a mature product of MIR937, was sufficient to restore its oncogenic function. Mechanistically, MIR937 amplification raised the expression of miR-937-5p, enhanced its binding to 3′ UTR of FBXO16 transcript, and thereby restricting FBXO16 degradative effects on ULK1. Our results demonstrate that MIR937 amplification augments cell autophagy and proliferation, and suggest an alternative strategy of MIR937/FBXO16/ULK1 targeting for HGSOC treatment.

Similar content being viewed by others
Introduction
Ovarian cancer (OV) is a malignant tumor that poses serious threats to women’s health [1]. Among various histotypes of OV, high-grade serous ovarian cancer (HGSOC) is the most prevalent, accounting for 60–80% of new cases each year [2, 3]. It is estimated in clinic that more than 75% of HGSOC patients are diagnosed at advanced stages [4], which makes it most deadly and tough to be interfered with. So, the 5-year survival rate for HGSOC is less than 30% and has not been significantly improved over the last three decades for lack of early diagnostic markers and efficient therapeutic targets [5]. The need for more rigorous study has been strengthened to enhance our understanding of HGSOC tumor initiation and progression to provide with novel intervening strategies.
HGSOC patients are characterized by high frequency of somatic copy number alterations (CNAs), including deletion of tumor suppressors genes and amplification of oncogenes [6]. CNAs in most cases are typically correlated with changes of gene expression and protein abundances [7,8,9]. Increasing evidences have showed that gene amplification always causes proliferative phenotypes, attributing to enhanced expression of specific genes or the cumulative effects of gene collections [10]. In HGSOC, gain of function for gene amplification in tumorigenesis has been extensively studied. For instance, OV cells with genomic amplified URI relied on enhanced URI protein to maintain its survival [11]. USP13 amplification and over-expression reprogramed cancer cell metabolism, and aggravated tumor progression of HGSOC [12]. In addition, amplified RAD21 strengthened its interaction with YAP/TEAD4 to suppress interferon signaling and facilitated immune evasion of HGSOC cells [13]. So, researchers have been focusing on revealing the precision medical targets for related genomic alterations in clinic. HER2 antibody (trastuzumab), has got its way on the therapy of HER2 amplified breast cancer [14]. Although not for HGSOC, such success brings an inspiring possibility that targeting of amplified tumor drivers holds great potential for cancer treatment. Not only for protein-coding genes, CNAs of small non-coding microRNAs (miRNAs) also represent a type of genomic aberrations with cancer driving capabilities [15]. However, lack interpretation of the underlying mechanism for miRNA amplification in carcinogenesis has seriously limited the progress for HGSOC treatment.
miRNAs are composed of 19–24 nucleotides in length, and have profound biological functions in the aspects of cell survival, proliferation, and differentiation primarily by complementary binding of mRNA targets [16]. Functional studies have confirmed that miRNA can also act as tumor suppressors or oncogenes, and genomic aberrations of miRNAs can lead to miRNAs dys-regulation and constantly associate with cancer progression [17, 18]. The notion has been validated in HGSOC that copy number gain of miR-569 increased its expression and boosted tumor survival and proliferation by attenuating TP53INP1 transcription [19]. miRNA intervention by mimics or inhibitors (with anti-miR function) has shown great potential in clinic treatment for cancer and related diseases. For example, a mimic of tumor suppressor miR-16 (MesomiR-1), and an anti-miR targeting miR-155 (MRG-106) both reached phase I trial for treating malignant pleural mesothelioma and cutaneous T cell lymphoma, respectively [20, 21]. Given the prevalence of miRNA CNAs in cancers [22, 23] and the feasibility of manipulating miRNA expression in recipient, it encourages us to unravel the functional mechanism for amplified miRs in HGSOC to provide with prospective therapeutic targets.
In this study, we screened the amplified miRs in the frequently amplified locus of 8q24.3 in HGSOC patients by TCGA database. We found MIR937 amplification can promote proliferation of HGSOC cells. Mechanistically, MIR937 amplification increased miR-937-5p expression, reduced FBXO16 and its degradative effects on ULK1 protein, and thus enhanced autophagy. Importantly, our study revealed the regulatory axis for HGSOC progression and shed light on the novel combinational therapeutic targets for the patients.
Materials and methods
Cell culture and tissue samples
HEK293T, OVCAR3, A2780, TOV21G, and SKOV3 cells were obtained from Procell Life Science & Technology Co., Ltd (Wuhan, China). HEK293T cells were maintained in low-glucose Dulbecco’s Modified Eagle Medium (DMEM) with 10% fetal bovine serum at 37 °C in 5% CO2. OVCAR3, A2780, TOV21G, and SKOV3 cell lines were grown in RPMI medium with 10% fetal bovine serum at 37 °C in 5% CO2. All the cell lines were confirmed to have no contamination of mycoplasma. Frozen tissue specimens of normal ovary and ovary with epithelial malignant tumor were received from Cancer Hospital Affiliated to Shandong First Medical University. And the informed consent was obtained from all subjects.
Plasmids, antibodies, and reagents
Human ULK1, FBXO16, CUL1, SKP1, and RBX1 genes were amplified and subcloned into the pcDNA3.1 vector with Flag or Myc tag. HA-tagged WT, K48-, and K63- Ub encoding plasmids were subcloned into the pcDNA3.1 vector. Antibodies against ULK1 (catalog 8054S), p62 (catalog 23214S), LC3A/B (catalog 4108S), CUL1 (catalog 4995S), ATG7 (catalog 8558S), ATG13 (catalog 13273S), Beclin-1 (catalog 3495S), Flag (catalog 14793S), Myc (catalog 2278S), and HA (catalog 3724S) were purchased from Cell Signaling Technology. Antibody against GAPDH (catalog ab181602) was obtained from Abcam. Anti-FBXO16 antibody (catalog LS-C82721) was purchased from LSBio. Cell counting kit-8 (CCK-8) (catalog C0038), d-luciferin potassium salt (catalog ST196), and crystal violet staining solution (catalog C0121) were purchased from Beyotime Biotechnology. CHX was purchased from MedChemExpress (catalog HY-12320). Cell-Light EdU Apollo567 In Vitro Kit (catalog C10310-1) was purchased from Ribobio.
Generation of MIR937 KO cell lines
For MIR937 knockout, pLentiCRISPR v2 (Addgene, catalog 52961) was digested with KpnI (NEB, catalog R3142V) and EcoRI (NEB, catalog R0101V). Two sgRNAs flanking MIR937 genomic locus were designed to delete MIR937 encoding sequence. The sequence for the 1st sgRNA is TGCCCCCGGTGAGTCAGGGT, and the sequence for the 2nd sgRNA is GTTCCCGAGCTCCTGCAGGT, which were driven by U6 or H1 promoter, respectively. The encoding cassette for U6 promoter-sgRNA1-H1-promoter-sgRNA2 was synthesized, amplified, and ligated into pLentiCRISPR v2. The constructed plasmid, pMD2.G (addgene, catalog 12259) and psPAX2 (addgene, catalog 12260) were co-transfected into HEK293T cells for virus package. OVCAR3 and A2780 cells were infected, selected with puromycin (MCE, catalog HY-B1743A), and single cell was screened for MIR937 knockout validation.
Genomic DNA isolation, PCR, and copy number validation
Total genomic DNA was extracted by a DNA isolation Kit (Vazyme, DC112-01) for both in vitro cultured OV cell lines and human ovary tissue. For MIR937 knockout analysis, genomic DNA of cultured OV cell lines was used as template for PCR amplification and sequencing with the following pair of primers: F: GTGGGGGCGTATAGTCTCTTG, R: GCATCGGTTAGTGTGCCTG. For clinical samples, FAM-labeled probe and the amplifying primers for genomic MIR937 sequence were designed, VIC-labelled Rnase-P probe and primers were used to quantify the copy number of the reference gene, and the genomic DNA from TOV21G cells was used as diploid control. TaqMan copy number assay was performed with TaqMan Fast Advanced Master Mix (ThermoTM Fisher Scientific, catalog 4444554). The results were collected on Quantstudio 1 Plus, and analyzed by CopyCaller Software (ThermoTM Fisher Scientific).
Generation of stable cell lines
FBXO16 coding sequence was sub-cloned into pCDH-GFP vector with Flag tag. ShRNA oligonucleotides targeting FBXO16 were annealed into pLKO.1-EGFP vector. The sequences for shRNA are listed as follows: FBXO16-shRNA1: 5′-GCTATTGAATGACCGGGTA-3′; FBXO16-shRNA2: 5′-CAAGCTTCCAAGGGTGTTA-3′. The related plasmids were packaged into lentivirus with psPAX and pMD2.G plasmids for OVCAR3 infection, and the infected OVCAR3 cells were selected with puromycin at 1 μg/ml.
MiRNA, siRNA transfection
miR-937-5p/3p mimics, mimics negative control (NC), inhibitors, or inhibitor NC oligonucleotides were transfected into OVCAR3, A2780, TOV21G, and SKOV3 cells, by HiperFect Transfection Reagent (Qiagen, catalog 301704) according to manufacturer’s instructions. For transient silencing of FBXO16, duplexs of siRNA were transfected into OV cells with LipofectamineTM RNAiMAX reagent (Thermo Fisher Scientific, catalog 13778075) according to the protocol. Target sequences used for FBXO16 knockdown were purchased from GenePharma. All the sequences used are available in Supplementary Table 1.
RNA isolation and quantitative RT-PCR
Total RNA from tissues or cultured cells were isolated by FastPure Cell/Tissue Total RNA Isolation Kit (Vazyme, catalog RC101-01). RNA was reversed transcribed with the PrimeScript RT reagent Kit for mRNA (Toyobo, catalog RR037A) or with the miRNA 1st Strand cDNA Synthesis Kit (Vazyme catalog MR101) for microRNA following the manufacturers’ instructions. qRT-PCR assay was performed by using SYBR green (CWBIO, catalog CW2601H) on a Quantstudio 1 Plus instrument. Relative RNA expression was quantification via the comparative 2−ΔΔCt method. The RT-qPCR primer sequences for relevant genes were listed in Supplementary Table 2. Primers for miRNAs detection were purchased from GenePharma, and U6 was used as control for microRNA expressional normalization.
Cell proliferation and colony formation assay
OVCAR3, A2780, and TOV21G cells were seeded in 96-well dishes at 2000 cells per well, and SKOV3 cells were seeded with 1500 cells/well. Cell Counting Kit-8 (CCK-8) kit (Beyotime, catalog C0038) was used to determine cell count and viability, and the absorbance was measured at 450 nm by a spectro-photometric reader (MD, Spectramax 190). For colony formation assay, MIR937 knockout OVCAR3 and A2780 cells were seeded into 6-well plates (600 cells/well), and cultured for 10 days. The colonies were fixed with 4% paraformaldehyde (PFA), and stained with 0.5% crystal violet. The number of colonies was counted by ImageJ software, and the data were represented as mean ± SD. Each experiment was repeated three times.
5-ethynyl-2′-deoxyuridine (EdU) incorporation assay
Transient transfected or MIR937 knockout OVCAR3 and A2780 cells were seeded in 96-well plates with 1 × 104 cells per well. Cell-Light EdU Apollo 567 In Vitro Imaging Kit (RiboBio, catalog C10310-1) were used to perform EdU assay according to the manufacturer’s instructions. In brief, after being incubated with 10 mM EdU for 2 h, the cells were fixed with 4% PFA, permeabilized in 0.5% Triton X-100, and labelled with fluorescent dye of Apollo 567. The nuclei were stained with DAPI (Solarbio, catalog C0065) for 15 min. Images were captured with a fluorescence microscope (Nikon, TS2R). EdU-positive cells were quantified with ImageJ software, and summarized as mean ± SD with at least three random fields.
Dual-luciferase reporter assay
pGL3-3M-Luc vector was used to insert the 3′ un-translated region (UTR) of FBXO16 transcript or its mutant 3′ UTR without miR-937-5p binding site. HEK293T cells were seeded in 24-well dishes one day before transfection. The WT or the mutant plasmids (with FBXO16 3′ UTR) were co-transfected with NC or miR-937-5p mimics. Forty-eight hours later, the cells were harvested and lysed, and Dual-Luciferase® Reporter Assay System (Promega, catalog E1910) was used to measure the luciferase activity by a GloMax 20/20 Luminometer. The relative luciferase activity was normalized with Renilla activity. Three independent experiments were conducted.
Immunoblotting and immunoprecipitation
Immunoblotting was performed as previously described [24]. In short, the cells were lysed with RIPA lysis buffer supplemented with PMSF (Solarbio, catalog P0100) and a protease inhibitor cocktail (Solarbio, catalog P1260). After centrifugation, the cell lysate was collected, boiled, and subjected to SDS-PAGE, and transferred to nitrocellulose (NC) filter membrane for immunoblotting analysis. As for immunoprecipitation analysis, IP buffer (50 mM Tris-HCl, 150 mM NaCl, 1.0% NP-40) with protease inhibitor cocktail was used to split the cells on ice for 15 min. The supernatant was collected and immunoprecipitated with agarose conjugated anti-Flag-Tag antibody (abmart, catalog M20018) overnight at 4 °C. The immunoprecipitates were washed with IP buffer three times, boiled in SDS sample buffer and analyzed by immunoblotting with indicated antibodies.
In vitro binding and ubiquitination assays
ULK1 and FBXO16 recombinant proteins were prepared using a TNT Quick Coupled Transcription/Translation System (Promega). For the binding assay, ULK1 and FBXO16 were mixed together, followed by immunoprecipitation with the FBXO16 antibody and western blotting with the ULK1 antibody. The FBXO16-associated E3 complex was purified from the 293T cells transfected with Flag-CUL1/Myc-SKP1/Myc-RBX1 with the anti-Flag M2 affinity gel from Abmart. For in vitro ubiquitination assay, the purified E3 complex and FBXO16 were incubated in 30 μl reaction buffer containing 300 mM HEPES, pH 7.2, 50 mM MgCl2, and 2 mM dithiothreitol. Additional components included recombinant E1 activating enzyme (UBA1) and recombinant E2 (UBCH5). Purified recombinant monoubiquitins (WT, K48, K63) and 20 mM ATP were added to the reaction system, which was incubated for 1 h at 30 °C with shaking at 450 r.p.m. The reactions were terminated by adding Western blotting loading buffer and the products were resolved on SDS-PAGE.
Immunohistochemistry (IHC)
The ovary tissues collected from control donors or the OV patients were formalin-fixed and paraffin-embedded (FFPE). The FFPE ovarian samples were sectioned and mounted on poly-lysine-coated slides. The sections were de-paraffinized and re-hydrated by xylene and gradient concentrations of ethanol, respectively. Then, antigen retrieval was fulfilled by 100 °C boiling of the section in 10 mM citrate buffer (Solarbio, catalog C1010) for 20 min. Then, a universal two-step detection kit (ZSGB-bio, catalog PV-9000) was used according to the manufacturer’s instructions for Immunohistochemistry (IHC) analysis. In detail, the endogenous peroxidase activity was blocked, and ULK1 antibody (Boster, catalog A00584-1) (1:250) diluted was incubated with the section overnight at 4 °C. The reaction enhancer reagent was used for 20 min before diaminobenzidine detection (ZSGB-bio, ZLI-9018).
Confocal imaging
Autophagic flux was monitored by confocal microscopy. In brief, OVCAR3 cells with stable knockdown of FBXO16 were infected with GFP-RFP-LC3 adenovirus. Zeiss laser scanning confocal (LSM880) was used to measure autophagosomes (yellow puncta in merged images) and autolysosomes (red puncta in merged images) in normoxia and hypoxia conditions.
Ovarian cancer xenograft mice model
Six-week-old female BALB/c athymic nude mice were obtained from the Huafukang BioScience company and bred under pathogen-free conditions. To establish subcutaneous xenograft models, OVCAR3 and A2780 cells (5 × 106/mice) in 100 μl phosphate buffered saline were subcutaneously injected into the right flank of the nude mice. The size for xenograft tumor generated by MIR937 wild type or genomic deleted OV cells was monitored every three days after its first appearance. For rescue assay, 4 × 106 OVCAR3 or A2780 cells in PBS were subcutaneously injected, and agomiR-NC, agomiR-937-5p, and agomiR-937-3p (RiboBio) were directly injected into the implanted tumor at the dose of 1 nmol in 20 μl PBS every 4 days for five times. Tumor volume (V) was monitored by measuring the length (L) and width (W) with a Vernier caliper and calculated with formula V (cm3) = W2 × L/2. Finally, the mice were euthanized, and all tumors were excised, weighed. To establish intraperitoneal xenograft model, MIR937+/+, MIR937+/−, MIR937−/− OV cells were transfected with pCDNA3.1-Luc plasmid, and screened with G418 for three days, and single cell was propagated with luciferase activity. 5 × 106 luciferase labelled OVCAR3 or A2780 cells were injected intraperitoneally, and in vivo bioluminescence signals were collected by an animal imager (PerkinElmer, IVIS Lumina XRMS III) at day 7, 14, and 21 after inoculation by intraperitoneal injection of D-luciferin (150 mg/kg) (Beyotime, catalog ST196). All animal experiments were carried out in accordance with the Guide for the Care and Utilization of Laboratory Animals (Shandong University of Traditional Chinese Medicine) and were approved by the Institutional Animal Care and Use Committee of Shandong University of Traditional Chinese Medicine.
Quantitative proteomics analysis
Proteomics analysis was executed by Jingjie PTM BioLabs (Hangzhou, China). In brief, 4D Label free proteomics analysis was conducted following the standard procedures: protein preparation, trypsin digestion, HPLC fractionation, LC–MS/MS analysis, and bioinformatics analysis.
Statistics
All experiments were repeated at least three times, unless otherwise stated. Values are presented as the mean ± SD. Statistical differences between two groups was assessed by two-tailed Student’s t-test. While for multiple groups comparisons, statistical significance was evaluated by one-way analysis of variance (ANOVA) using GraphPad Prism 9.0 software. For correlation analysis, Pearson’s correlation was used for calculating r and p. P ≤ 0.05 was considered statistically significant.
Results
MIR937 is amplified in HGSOC patients and correlates with increased expression of MIR937
Somatic gene CNAs frequently occurred in the whole genome of HGSOC patients, which indicated that CNA genes might contribute to the pathogenesis of HGSOC. Genome-wide association study has been conducted and revealed that the genomic fragment of 8q24 is a susceptible locus for HGSOC patients [25]. Further, in this locus, 8q24.3 was one of the most significantly amplified fragments across the genome, which comprised 75 typical genes [26]. However, the large proportion of the genes located has not been functionally clarified in HGSOC. microRNA (miR) is small and short in genome sequence, and what interests us most is how the genomic alterations of such little fragments affects the progression of HGSOC. Impressively, there are three miRs located in 8q24.3 loci, MIR937, MIR939, and MIR661 (Fig. 1a). Firstly, we checked the absolute expressional level for the three miRs in the HGSOC patients from The Cancer Genome Atlas (TCGA) database, and found the expression of MIR937 is relatively high. At the same time, the expression of MIR939 and MIR661 is quite low (Fig.1b). We also examined the expression for these miRs in all OV cell lines from Cancer Cell Line Encyclopedia (CCLE) database. Consistent results were obtained (Supplementary Fig. 1a). So, we chose MIR937 for further study. We systematically screened the amplification frequency of MIR937 across different cancer types by TCGA database, and discovered MIR937 amplified with the highest frequency in patients with HGSOC (Fig. 1c).
a Schematic diagram of three miRs in the genome locus of 8q24.3. b The expressional data of three miRs were retrieved from cBioportal tools for TCGA database analysis. c Genomic amplification of MIR937 across human cancers retrieved from cBioportal tools for TCGA databases analysis. The correlations between MIR937 copy number and its expression were performed by the data from TCGA database (d) or CCLE OV cancer cell lines (e). f MIR937 expression was quantified in HGSOC patients and the comparison between MIR937 diploid and amplified patients was performed. g Pearson correlation between the MIR937 copy number and the proliferation score calculated by ssGSEA. h Pearson correlation between the MIR937 mRNA expression and the proliferation score calculated by ssGSEA. The gene set named “positive regulation of cell population proliferation” was obtained from the Gene Ontology (GO) knowledgebase (https://www.geneontology.org/). ssGSEA: single sample gene set enrichment analysis. Statistical analysis was performed using one-way ANOVA in (b), and two-tailed Student’s t-test in (f); Pearson correlation coefficients (R) were employed to evaluate the correlations between two continuous variables in (d, e, g, h). ****P < 0.001.
To further demonstrate the clinical significance of MIR937, we firstly conducted a correlation analysis between DNA copy number and expression of MIR937 in HGSOC patients and cancer cell lines, and it was revealed that RNA level of MIR937 positively correlated with its DNA copy number (Fig. 1d, e). In addition, the positive correlation was also observed in all cancer cells in CCLE (Supplementary Fig. 1b). Meanwhile, If the patients or OV cells lines were grouped according to CNA status of MIR937, it can also be found that the abundance of MIR937 transcript elevated significantly upon its amplification (Supplementary Fig. 1c, d). To verify the correlation, we collected HGSOC patient samples, and by Taqman qPCR analysis, 7 out of 45 samples were validated with MIR937 amplification, which showed significant higher expression of MIR937 (Fig. 1f). The data indicated MIR937 upregulation is a major consequence of MIR937 amplification. Moreover, we performed gene ontology (GO) enrichment analysis by TCGA database, and discovered that MIR937 copy number and expression are positively correlated with GO terms of “positive regulation of cell population proliferation” (Fig. 1g, h) and “negative regulation of programmed cell death” (Supplementary Fig. 1e, f). Collectively, our results implied that miR937 genomic amplification accounts much for its upregulation, and thus may aggravates its pathological effects in HGSOC occurrence.
MIR937 deficiency attenuates proliferation of ovarian cancer cells
To investigate the potential effect of MIR937 on OV progression, we designed CRISPR-Cas9-based knockout strategy for MIR937 in OVCAR3 and A2780 cells (Fig. 2a). In detail, two sgRNAs were used flanking the MIR937 genome locus without affecting the adjacent exons for SCRIB encoding. Single cells with MIR937 knockout were screened, validated by genomic DNA PCR amplification and Sanger sequencing. Homozygous and heterozygous loss of MIR937 cells (MIR937−/−, MIR937+/−) were selected (Fig. 2b). We firstly monitored the genomic loss of MIR937 on OV proliferation by CCK8 assay in vitro, and realized that MIR937+/− grew slower than MIR937+/+ cells, while MIR937−/− extended the inhibitory effect for both OVCAR3 and A2780 cells (Fig. 2c, d). In addition, colony formation assay revealed the capacity of the cells to form colonies declined gradually with MIR937 gene loss (Supplementary Fig. 2a, c). The statistical results also demonstrated the significant loss for the colony formation ability with MIR937 knockout (Supplementary Fig. 2b, d). EdU incorporation assay was also performed, which further proved that MIR937 deficiency inhibited the proliferative features for both OVCAR3 and A2780 cells (Supplementary Fig. 2e). The statistical data showed EdU positive cells were reduced by heterozygous loss of MIR937 to 78% or 70%, but reduced by homozygous loss of MIR937 to 42% or 44% respectively for OVCAR3 and A2780 cells (Supplementary Fig. 2f, g). The in vitro data highly indicated that deficiency degree of MIR937 correlated with inhibitory extent for proliferation of OV cells.
a CRISPR-Cas9 based knockout strategy for MIR937 in OVCAR3 and A2780 cells. b Sanger sequencing for determined the genotype of MIR937+/− and MIR937−/− OVCAR3 and A2780 cells. Growth curve of MIR937+/+, MIR937+/−, and MIR937−/− cells for OVCAR3 (c) and A2780 (d). The cells were seeded in 96-well plates with the initial number of 2000, and the growth status was continuously monitor for 5 days. Xenograft tumors derived from MIR937+/+, MIR937+/−, and MIR937−/− OVCAR3 cells (e), and A2780 cells (f) were displayed. 5 × 106 for each cell were s.c. injected into the right flank of nude mice (n = 5). The mice were sacrificed three weeks later after the inoculation. Growth curve for the xenograft tumors for OVCAR3 (g) and A2780 (h) cells with the indicated MIR937 gene loss. The volume of tumors was measured every three days after their appearances (day 6). i, j Tumor weights were measured after mice were sacrificed. k Representative bioluminescence images of mice intraperitoneal injected with luciferase-expressing OVCAR3 and A2780 cells (MIR937+/+, MIR937+/−, and MIR937−/−) were collected at day 12 after inoculation. Data are shown as the mean ± SD. Similar results were obtained in three independent experiments. Statistical analysis was performed using one-way ANOVA in (c, d, g–j); *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
To further validate the function of MIR937, we firstly subcutaneously injected MIR937+/+, MIR937+/−, MIR937−/− cells to the right flank of the nude mice to establish xenograft models. The mice were sacrificed when the tumor grew to the appropriate size to visualize the differences of tumor growth for different groups (Fig. 2e, f). By monitoring the growth rate, and the weight for the xenograft tumor, we consistently found MIR937−/− cells exhibited more restricted cell growth than MIR937+/− cells (Fig. 2g–j). Next, we generated luciferase labelled OVCAR3 and A2780 cells with MIR937 deficiency, and injected these cells intraperitoneally to mimic native environment for OV tumor growth. The in vivo luciferase signal captured consistently denoted that MIR937 deficiency could severely suppress proliferation of OV cells (Fig. 2k). In general, these data indicated that MIR937 might play an oncogenic role in OV development.
miR-937-5p accounts for MIR937 loss of function in ovarian cancer cells
MIR937 transcribed as a precursor, and finally resulted in the formation of two mature products, miR-937-5p and miR-937-3p. Whether miR-937-5p or miR-937-3p is responsible for MIR937 oncogenic role remains to be illustrated. So, firstly, mimics or inhibitors of the two miRs were used to over-express or repress miR-937-5p and miR-937-3p expression in OVCAR3 and A2780 cells (Supplementary Fig. 3a–d). By CCK8 analysis, we discovered overexpression of miR-937-5p significantly enhanced the proliferation rate for both cell lines, while miR-937-3p has no effect (Fig. 3a, b). Consistently, the inhibitors of miR-937-5p but not miR-937-3p could inhibit the proliferation of OVCAR3 and A2780 cells (Fig. 3c, d). In addition, miR-937-5p could also improve the proliferation ability of TOV21G and SKOV3 cells (Supplementary Fig. 3e, f). Meanwhile, EdU assay was also performed, and the results showed that miR-937-5p over-expression could significantly increase the proportion of EdU-positive cells (Supplementary Fig. 3g, h). It highly suggested that miR-937-5p was the major product responsible for MIR937 oncogenic effects.
Growth curves for OVCAR3 (a) and A2780 (b) cells were monitored by CCK8 analysis upon miR-937-5p or 3p mimics transfection. c, d Growth curves for OVCAR3 and A2780 cells were detected after transfection of miR-937-5p or 3p inhibitors. Growth curves for OVCAR3 (e) and A2780 (f) comparison of MIR937+/+ cells with MIR937−/− cells, into which miR-937-5p or 3p mimics were transfected. MIR937+/+ cells mock transfected, while MIR937−/− cells were transfected with miR-937-5p or 3p mimics (g) The graphical summary for agomiR injection into the xenograft tumors. Xenograft tumors derived from OVCAR3 (h) and A2780 (i) cells were displayed. MIR937+/+ tumors were mock treated with agomiR-NC, and for MIR937−/− tumors, agomiR-937-5p and -937-3p were used to interfere with tumor growth. Growth curve for the xenograft tumors affected by agomiR-937-5p and -937-3p for OVCAR3 (j) and A2780 cells (k). Tumor weights for OVCAR3 (l) and A2780 (m) cells were measured at day 25 when the mice were sacrificed after five rounds of agomiR injection. Data are shown as the mean ± SD. Similar results were obtained in three independent experiments. Statistical analysis was performed using one-way ANOVA in (a–f, j–m); **P < 0.01; ***P < 0.001; ****P < 0.0001.
Then, we over-expressed miR-937-5p or miR-937-3p in MIR937−/− cells to further investigate which product could rescue MIR937 deficiency caused arrest of OV cell growth. We found that only miR-937-5p mimics transfection could rescue the growth ability of MIR937−/− cells close to that of MIR937+/+ cells (Fig. 3e, f). Finally, we established subcutaneous xenograft tumors by MIR937+/+, and MIR937−/− cells of OVCAR3 and A2780, into which agomiR-937-5p and agomiR-937-3p were injected every 3 days for five times (Fig. 3g). At day 25, the mice were sacrificed, and the xenograft tumors were measured in size and weight to confirm the effects for both miRs. When being injected with agomiR-937-5p, but not agomiR-937-3p, the size for MIR937−/− xenografts increased significantly (Fig. 3h, i). Furthermore, agomiR-937-5p rescued the proliferation rate and the weight for the xenograft tumors of MIR937−/− OVCAR3 and A2780 cells (Fig. 3j–m). Taken together, our data consolidated that miR-937-5p is the product of MIR937 gene for its oncogenic roles.
miR-937-5p directly targets the tumor suppressor gene of FBXO16
Since miR-937-5p is functional in promoting tumor growth, we hypothesized that miR-937-5p might target to some tumor suppressor genes (TSGs) in OV. To ensure accurate screening of miR-937-5p target genes, we conducted a detailed literature search using keywords of “HGSOC” and “tumor suppressor genes”. We manually curated 45 tumor suppressor genes, which had been verified by different research teams, for target screening. We also used the TargetScan database to get the list of potential miR-937-5p targets. When overlapping the genes from the two lists, eleven TSGs were revealed to be likely targeted by miR-937-5p (Fig. 4a). To verify whether the genes were truly targeted by miR-937-5p, we transfected miR-937-5p mimics in OVCAR3 cell, quantified the expression of the indicated genes, and found miR-937-5p could only reduce the expression of FBXO16 (Fig. 4b). Conversely, deficiency of MIR937 in the OVCAR3 cells greatly elevated mRNA level of FBXO16 (Supplementary Fig. 4a). In addition, one binding site in the 3′ UTR of FBXO16 was discovered, and luciferase reporter analysis displayed that miR-937-5p inhibited luciferase activity with WT 3′ UTR but did not affect that with mutant 3′ UTR (Fig. 4c, d). To directly test whether miR-937-5p binding of FBXO16 3′ UTR inhibited its protein translation, we transfected mimics and inhibitors and detected the influence of miR-937-5p on protein levels of FBXO16 in OVCAR3 and A2780 cells. It showed that miR-937-5p mimics remarkably reduced, while the inhibitors enhanced the expression of FBXO16 (Fig. 4e). Consistently, MIR937 loss coupled with FBXO16 protein increase (Supplementary Fig. 4b). What’s more, we also submitted the samples of xenograft tumors for western blot analysis, and found that agomiR-937-5p could reduce FBXO16 protein levels that had been enhanced by MIR937 loss (Supplementary Fig. 4c). Collectively, these data confirmed that FBXO16 is the bona fide target of miR-937-5p in OV cancer cells.
a Venn diagram showing the overlapping 11 potential targets by lists of TSGs and targets of miR-937-5p acquired from TargetScan database. b The expression of 11 predicted genes were detected by qPCR analysis upon miR-937-5p mimics transfection in OVCAR3 cells. c Potential miR-937-5p binding sequence in the 3′ UTR of FBXO16 mRNA predicted by TargetScan. The sequence of the mutant construct for luciferase reporter assay was also indicated. d Relative luciferase activity was monitored after the former described reporter plasmids co-transfected with miR-937-5p mimics in HEK293T cells. The firefly luciferase activity was normalized to renilla activity. e FBXO16 protein level was detected following transfection of miR-937-5p mimics or inhibitors in OVCAR3 and A2780 cells. GAPDH was used as a loading control. f Xenograft tumors derived from OVCAR3 cells were displayed. Two cell lines of OVCAR3 with FBXO16 stable knockdown and one control cell line (vector) were s.c. injected into nude mice. g Growth curve for the xenograft tumors generated by OVCAR3 cells with stable knockdown of FBXO16. h Tumor weights were measured on day 21 after the mice being sacrificed. Data are shown as the mean ± SD. Similar results were obtained in three independent experiments. Statistical analysis was performed using two-tailed Student’s t-test in (b, d) and one-way ANOVA in (g, h); *P < 0.05; ***P < 0.001; ****P < 0.0001.
In addition, the tumor suppressive effects of FBXO16 for OV have been further verified in our model of study. We silenced FBXO16 expression by three small interfering RNAs (siRNAs) (Supplementary Fig. 4d) and detected the effect of FBXO16 on cell proliferation. By CCK8 analysis, we found that all the siRNAs targeting FBXO16 could significantly enhanced the proliferative activity for OVCAR3 cells (Supplementary Fig. 4e). Besides, two short hairpin RNAs (shRNAs) targeting FBXO16 were also constructed to generate stable cell lines with FBXO16 silencing (Supplementary Fig. 4f). It was also revealed that stable knockdown of FBXO16 could consistently improve the proliferation rate of OVCAR3 cells (Supplementary Fig. 4g). For in vivo assay, both shRNAs could remarkably increase the size, growth rate, and weight of tumors by the subcutaneous xenograft model (Fig. 4f–h). In summary, our data demonstrated that miR-937-5p targeted FBXO16, thus reduced its tumor suppressive effects and played oncogenic roles.
FBXO16 negatively regulates autophagy in ovarian cancer
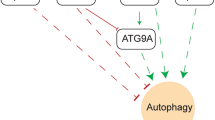
To demonstrate the molecular mechanism by which MIR937/FBXO16 regulate OV proliferation, we firstly tested the influence of MIR937 loss on hnRNPL, a formerly reported target of FBXO16. The results showed no dramatic changes of hnRNPL in MIR937−/− compared with that of MIR937+/+ cells (Supplementary Fig. 5a). It indicated that MIR937 might exert its oncogenic effects through a distinctive role of FBXO16. So, we sought to uncover novel targets of FBXO16 in OV. 4D label-free quantitative proteomic analysis was performed to screen the biological processes potentially affected in OVCAR3 cells with stable knockdown of FBXO16. GO enrichment analysis for differentially expressed proteins was performed and the results showed that cell cycle, and autophagy-related pathways were significantly upregulated upon FBXO16 silencing, whereas cell apoptosis and amino acid metabolic-related processes were downregulated (Fig. 5a). Since autophagy is an essential process that regulates cell proliferation, cell death, and cell metabolism, we asked whether FBXO16 affected the expression of key regulatory proteins for autophagy. By western blot analysis, we found FBXO16 knockdown increased the protein level of ULK1, but not ATG7, ATG13, or Beclin-1, and could simultaneously enhance the ratio of LC3II/I, but reduce p62 under both normal and hypoxic conditions (Fig. 5b). We further monitored autophagic flux by confocal microscopy in OVCAR3 cells infected with GFP-RFP-LC3 adenovirus. The results showed that both shRNA targeting FBXO16 could enhance basic and hypoxia-induced autophagy flux (Fig. 5c–e). Stable over-expression of FBXO16 displayed remarkable reduction of ULK1 protein, and suppression of cellular autophagy reflected by LC3II/I ratio and p62 (Fig. 5f). In addition, the result that FBXO16 only targeted ULK1 was further confirmed with transient knockdown of FBXO16 by three siRNA in both A2780 and TOV21G cells (Supplementary Fig. 5b, c). Furthermore, we assessed the impact of FBXO16 on the phosphorylation level of ULK1. The results indicated that the knockdown of FBXO16 significantly elevated ULK1 phosphorylation, whereas the over-expression of FBXO16 resulted in a reduction of ULK1 phosphorylation under both normal and hypoxic conditions (Supplementary Fig. 5d, e). Collectively, our findings suggest that FBXO16 modulates ULK1 activation by influencing its protein abundance, thereby regulating the initiation of autophagy.
a Gene ontology enrichment analysis of upregulated (Up) and downregulated (Down) gene sets in OVCAR3 cells with stable knockdown of FBXO16. b IB analysis for autophagy-related proteins affected by stable knockdown of FBXO16 in OVCAR3 cells under normal and hypoxia conditions. The cells were placed in the anaerobic incubator with 1% oxygen at 37 °C for 12 h. Similar results were obtained in three independent experiments. c–e OVCAR3 cells with stable knockdown of FBXO16 were infected with GFP-RFP-LC3 adenovirus for 48 h, then cells were placed in the anaerobic incubator with 1% oxygen at 37 °C for 12 h. Autophagic flux was monitored by confocal microscopy (c). The mean number of autophagosomes per cell (yellow puncta in merged images) (d) and autolysosomes (red puncta in merged images) (e) were counted. f IB analysis for autophagy-related proteins affected by over-expression of FBXO16 in OVCAR3 cells under normal and hypoxia conditions.
FBXO16 interacts with ULK1 and facilitates its K48-linked poly-ubiquitination
FBXO16 always acts as an adapter that recruits the substrate to SKP1-cullin 1 (CUL1)-RBX1 complex for its ubiquitination and subsequent degradation (Fig. 6a). So, we checked whether FBXO16 could interact with ULK1 to possibly affect its protein stability. By co-immunoprecipitation assay, we found ectopically expressed Flag-tagged FBXO16 strongly interacted with Myc-tagged ULK1 (Fig. 6b). Meanwhile, Flag-tagged ULK1 could also bind to Myc-tagged FBXO16 (Fig. 6c). We also detected the interaction between endogenous FBXO16 and ULK1 in OVCAR3 cells (Fig. 6d). To verify that FBXO16 associated with ULK1 directly, FBXO16 and ULK1 recombinant proteins were prepared and in vitro immunoprecipitation was performed. These results revealed that FBXO16 could form a complex with ULK1 directly (Fig. 6e). To determine the binding requirements for FBXO16-ULK1 complex, we generated three and four deletion constructs of FBXO16 and ULK1, respectively, and performed the co-IP assay. The results showed that the C-terminal domains (CTD) of FBXO16 and ULK1 are responsible for their interaction (Fig. 6f, g). Then, we transfected A2780 and OVCAR3 cells with plasmids encoding FBXO16 (WT) and FBXO16 (∆CTD) proteins, and by CCK8 analysis, we confirmed the necessity of CTD domain for FBXO16 inhibitory function on OV cell proliferation (Fig. 6h, i). We further detected ULK1 interaction with the key components of SCF complex by co-IP analysis, and unveiled that Flag-tagged ULK1 could bind to Myc-tagged CUL1, SKP1, and RBX1 (Supplementary Fig. 6a–c). Besides, knockdown of FBXO16 could significantly suppress the interaction of ULK1 and CUL1 or RBX1 (Supplementary Fig. 6d, e). This highly implied that ULK1 was recruited to SCF complex by FBXO16.
a Schematic presentation of predicted interaction between ULK1 and CUL1-SKP1-FBXO16 complex. Co-IP analysis with anti-Flag antibody to detect the interaction between Flag-FBXO16 and Myc-ULK1 (b), and Myc-FBXO16 and Flag-ULK1 (c) in HEK293T cells co-transfected with the indicated plasmids. d Lysates from OVCAR3 cells were subjected to immunoprecipitation analysis with anti-FBXO16 antibody, followed by immunoblotting analysis with anti-FBXO16 and anti-ULK1 antibodies, respectively. e Recombinant FBXO16 and ULK1 proteins were prepared in an in vitro transcription and translation system, immunoprecipitation analysis was performed using anti-FBXO16 antibody, immunoblot analysis was followed with anti-ULK1 antibody. Wild type (WT) and truncated mutant schematic structures of ULK1 (f) and FBXO16 (g) were presented (upper), and co-IP with Flag-antibody was conducted to visualize the interaction between FBXO16 with ULK1 truncations (lower), and ULK1 with FBXO16 truncated mutants (lower). OVCAR3 (h) and A2780 (i) cells were infected with lenti-virus to over-express FBXO16 (WT) and FBXO16 (∆CTD) proteins, and CCK8 assay was performed to detect the proliferation of both cells. Co-IP with anti-Flag antibody followed by immunoblot analysis (IB) to detect ULK1 ubiquitination affected by FBXO16 in HEK293T cells. WT, K48, and K63 mutant forms of HA-Ubs were used in (j), while WT and K48R mutant of HA-Ubs were used in (k). Similar results were obtained in three independent experiments. l Recombinant ULK1 and FBXO16 proteins were prepared in an in vitro transcription and translation system. In vitro ubiquitination assay was performed in the presence of E1, E2, FBXO16, ULK1, Ubs (WT, K48, K63), and immunoprecipitated CUL1-SKP1-RBX1 complex. The ubiquitination of ULK1 was examined by immunoblot analysis with anti-ULK1 antibody.
Further, we co-transfected HEK293T cells with Flag-ULK1, Myc-FBXO16, and HA-Ubs (WT, K48, and K63). Cell lysates were immunoprecipitated with Flag antibody, and poly-ubiquitinated ULK1 was detected by western blot. It was revealed FBXO16 could significantly increase K48, but not K63, linked poly-ubiquitination of ULK1 (Fig. 6j). Consistently, FBXO16 knockdown in OVCAR3 cells showed reduction of both WT and K48 linked poly-ubiquitination of ULK1 (Supplementary Fig. 6f). On the other hand, we transfected Flag-ULK1, Myc-FBXO16, and HA-Ubs (WT, K48R) into HEK293T cells, and the cell lysate were subjected to co-IP analysis. The results illustrated that lysine-48 (K-48) mutation to arginine (R) of Ub blocked FBXO16-mediated ubiquitination of ULK1, which further validated that FBXO16 facilitated K48-linked poly-ubiquitination of ULK1 (Fig. 6k). We also performed in vitro ubiquitination assay in the presence of recombinant FBXO16 and ULK1 proteins, and the results showed that FBXO16 is responsible for K48 poly-ubiquitination of ULK1 (Fig. 6l). To further demonstrate that FBXO16 facilitated ULK1 degradation via ubiquitination, we transiently knockdown FBXO16, and treated the cells with cycloheximide (CHX) to detect the stability of ULK1 protein affected by FBXO16. The data showed that FBXO16 knockdown increased ULK1 protein level and slowed down its degradation in both OVCAR3 (Supplementary Fig. 6g, h) and A2780 cells (Supplementary Fig. 6i, j). Altogether, our data proved that FBXO16 accelerated ULK1 degradation by enhancing its K48-linked poly-ubiquitination.
Clinical MIR937 amplification correlates with FBXO16 and ULK1 expression
We then asked whether MIR937 amplification in OV functions through FBXO16/ULK1 axis. Firstly, level of ULK1 and autophagy (manifested by p62 and LC3II/I) were detected by western blot in MIR937 loss OVCAR3 and A2780 cells, and the results showed both ULK1 and autophagy levels were slightly downregulated in MIR937+/− cells, while notably decreased in MIR937−/− cells in comparison with that of MIR937+/+ cells (Fig. 7a). To further clarify the dependence of miR-937-5p by MIR937 loss in inhibiting autophagy, miR-937-5p expression was enhanced and inhibited by mimics and inhibitors, and consistently we found miR-937-5p overexpression promoted cell autophagy, while its knockdown inhibited cell autophagy (Supplementary Fig. 7a). In addition, we checked the ULK1 expression in xenograft tumors derived from MIR937-/- OVCAR3 and A2780 cells, which were interfered with agomiR-937-5p or 3p. The results showed agomiR-937-5p restored ULK1 expression by MIR937 loss (Supplementary Fig. 7b). Furthermore, we also tested whether miR-937-5p effects on ULK1 and autophagy was through its downregulation of FBXO16. So OVCAR3 and A2780 cells were firstly transfected with miR-937-5p inhibitors to transiently increase FBXO16, and then with siRNA targeting FBXO16, to detect whether FBXO16 knockdown could rescue ULK1 and autophagy, which had been suppressed by miR-937-5p inhibition mediated FBXO16 elevation. The results revealed that the restrain on ULK1 and autophagy by miR-937-5p inhibitors could be both ameliorated by FBXO16 knockdown (Supplementary Fig. 7c). So, we concluded that MIR937 functioned through miR-937-5p/FBXO16/ULK1 to regulate OV cancer progression.
a IB analysis of protein levels for ULK1, P62, LC3II/I in MIR937+/+, MIR937+/−, and MIR937−/− OVCAR3 (upper) and A2780 (lower) cells. GAPDH was used as loading control. b IB analysis for comparing FBXO16, ULK1 protein abundances in clinical OV with control samples. Ctrl: normal ovary, MIR937 WT: OV patient samples without amplification of MIR937. MIR937 AMP: OV patient samples with MIR937 amplification. c Representative images for IHC analysis of ULK1 protein in OV patient samples or the normal control, Scale bar: 100 μm. Kaplan–Meier survival analysis of OV patients from GSE26193 dataset grouped by FBXO16 (d) and ULK1 (e) expression. f Kaplan–Meier survival analysis of OV patients grouped by both FBXO16 and ULK1 expression in GSE26193. Similar results were obtained in three independent experiments.
We next examined in clinical whether MIR937 amplification was correlated with FBXO16 and ULK1 in OV patients. In our study, seven out of all the enrolled HGSOC patients (n = 45) were validated with MIR937 amplification. And the representative data of FBXO16 and ULK1 protein expression in MIR937 amplified patients was presented in comparison with that of normal or MIR937 diploid patients. The results indicated that FBXO16 expression was relatively high in normal ovary tissues, slightly reduced in MIR937 diploid OV samples, while decreased dramatically in MIR937 amplified patients (Fig. 7b). And inversely, ULK1 expression increased somewhat in MIR937 diploid OV samples, but was further augmented in MIR937 amplified patients (Fig. 7b). The results were also validated by IHC assay that ULK1 abundance was reinforced more by MIR937 amplification than MIR937 diploid in HGSOC patients (Fig. 7c). Finally, we reviewed the clinical significance for FBXO16 downregulation or ULK1 upregulation in HGSOC patients. For lacking of clinical samples in hand, we take the advantage of Kaplan plotter database to investigate FBXO16 and ULK1 correlation with prognosis of HGSOC patients. Individually, ULK1 high expression predicted worse survival in GSE3149 dataset, while FBXO16 high expression favored HGSOC patient survival in GSE63885 dataset (Supplementary Fig. 7d, e). Moreover, survival analysis by GSE26193 datasets showed the same results (Fig. 7d, e). And when the patients from GSE26193 dataset were grouped according to both FBXO16 and ULK1 expression, we can see that the survival rate for ULK1low/FBXO16high patients were greatly improved, while ULK1high/FBXO16low patients exhibited the worst survival capabilities (Fig. 7f). Collectively, our study implied that MIR937 amplification induced ULK1 high expression and FBXO16 low expression simultaneously contribute to malignant progression of HGSOC patients. Finally, to consolidate the clinical relevance and shed light on the precise therapeutic prospect in accordance with MIR937 CNAs and FBXO16 expression, we divided HGSOC patients into chemotherapy sensitive and insensitive groups from TCGA database. By correlation analysis, we found in the complete/partial response group, MIR937 CNAs showed no correlation with FBXO16, while in the stable/progressive OV group, MIR937 CNAs negatively correlated with FBXO16, which implied that the combination of MIR937 CNAs and FBXO16 expression might be a good indicator of sensitivity to chemotherapy for OV patients (Supplementary Fig. 7f, g).
Discussion
The data presented here demonstrate that MIR937 copy number changes functioned through miR-937-5p to dictated autophagy and cell proliferation of HGSOC cells. MIR937 deficiency abolished miR-937-5p inhibitory effect on FBXO16, resulted in accumulation of ULK1 protein, and enhanced autophagy, thereby facilitating OV cell proliferation and accelerating tumor progression. Mechanistically, MIR937 copy number amplification upregulated miR-937-5p, which strengthened its binding with 3′ UTR of FBXO16, and attenuating FBXO16 mediated ULK1 degradation via SCFFBXO16 complex. Importantly, FBXO16 is downregulated and ULK1 is upregulated in MIR937 amplified HGSOC samples, and FBXO16highULK1low favors survival for HGSOC patients. Collectively, these results unveiled a regulatory axis for promoting OV carcinogenesis elicited by MIR937 amplification.
Somatic CNAs are hallmarks of many cancers [27,28,29]. Somatic CNAs generally refers to gain or loss of large fragments of genomic DNA. From this sight, CNAs of a single genes did not always occur exclusively with that of its adjacent genes. From this point, the oncogenic or the tumor suppressive effects of amplification or lost CNAs might be a combinational result for a cluster of genes. The matter is the same for miRNAs despite their minimal nucleotides in length. Although tumorigenesis function of CNAs for protein-coding genes has been extensively studied, the studies for miRNAs CNAs were still limited which hindered the processes for precise targeting of patients with specific alteration. Using TCGA database for systematic analysis, we observed that MIR937 amplification correlated with enhanced proliferation with HGSOC tumors. Correspondingly, deletion of MIR937 in HGSOC cells greatly restricted cell proliferation and tumor formation in mouse xenograft models. Importantly, homo-loss of MIR937 exacerbated the inhibitory effects exerted by heterozygous loss of MIR937 for tumor growth. These results identify a dose effect of MIR937 copy number in tumor promotion.
By rescue and bioinformatics analysis, we confirmed miR-937-5p is responsible for MIR937 oncogenic roles and interfered with FBXO16 function. FBXO16 belongs to F-box protein that participates in the assembly of cullin-RING ubiquitin ligase (CRL) [30]. A typical member of CRL is SKP1/CUL1/F-box (SCF) E3 ubiquitin ligase complex, which controlled protein turnover in a proteasome-dependent manner. Generally, SKP1 and F-box protein act as adapter, and recruit the substrate to CUL1, where RBX1 promotes poly-ubiquitination of the targeted protein [31]. Due to alternative cullin members and diversity of F-box proteins, nearly 200 unique CRLs existed in the eukaryotic cells [32]. Although FBXO16 has been revealed to suppress OV progression [33]; how MIR937 amplification directs FBXO16 function in HGSOC and whether MIR937 attenuated FBXO16 containing SCF activity remains elusive. Our experiments showed that MIR937 deficiency highly increased FBXO16 expression, and upon mutation of its putative binding site in 3′ UTR of FBXO16, the inhibition on FBXO16 expression was diminished. Notably, 4D label-free quantitative proteomics study revealed that FBXO16 might negatively regulate autophagy related pathways, and knockdown of FBXO16 restored autophagy in OV cells suppressed by miR-937-5p inhibitors. Based on these findings, we hypothesized MIR937 amplification improve autophagy though FBXO16. Indeed, we found FBXO16 interacted with ULK1 and accelerated its degradation by promote its K48 linked poly-ubiquitination. Further, our data also defined the interaction between ULK1 and SKP1, CUL1, and RBX1. So, we propose that MIR937 amplification function through SCFFBXO16 to modulate ULK1 mediated autophagy.
Autophagy is a complicated process that is capable of recycling intracellular components to maintain metabolism and survival. Recent studies assumed that autophagy might play opposite roles in cancer to suppress initiation, while promote growth and progression for established tumors [34, 35]. So it is delicate in targeting autophagy for cancer treatment. In HGSOC, autophagy has been reported to promote metastasis and chemo-resistance [36], and inhibition of autophagy might induce apoptosis [37]. Besides, ULK1 inhibitor can reduce autophagy and then viability of HGSOC cells [38]. In our study, we found MIR937 positively regulated ULK1-mediated autophagy and ULK1 protein accumulation is accompanied with reduced FBXO16 protein and MIR937 amplification in HGSOC patient samples. In particular, clinical data revealed patients expressing with high FBXO16 and low ULK1 favors better prognosis. Collectively, our data provided an alternative therapeutic strategy targeting ULK1-autophagy in MIR937 amplified HGSOC patients.
In summary, our findings identify MIR937 amplification as an oncogenic feature of HGSOC patients. It not only defines the fundamental effects of MIR937 in tumor promotion in mouse models, but also highlights the correlation of MIR937 amplification and ULK1 mediated autophagy in HGSOC. The results suggest that targeting of miR-937-5p holds great prosperities for treatment of HGSOC with MIR937 amplification.
Data availability
The supporting data for the findings of this study can be obtained from the corresponding author upon reasonable request.
References
Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68:284–96.
Vaughan S, Coward JI, Bast RC Jr., Berchuck A, Berek JS, Brenton JD, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer. 2011;11:719–25.
Cannistra SA. Cancer of the ovary. N Engl J Med. 2004;351:2519–29.
Matsuo K, Machida H, Matsuzaki S, Grubbs BH, Klar M, Roman LD, et al. Evolving population-based statistics for rare epithelial ovarian cancers. Gynecol Oncol. 2020;157:3–11.
Jacobs IJ, Menon U, Ryan A, Gentry-Maharaj A, Burnell M, Kalsi JK, et al. Ovarian cancer screening and mortality in the UK collaborative trial of ovarian cancer screening (UKCTOCS): a randomised controlled trial. Lancet. 2016;387:945–56.
Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905.
Henrichsen CN, Vinckenbosch N, Zöllner S, Chaignat E, Pradervand S, Schütz F, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–9.
Stranger BE, Forrest MS, Dunning M, Ingle CE, Beazley C, Thorne N, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53.
Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z. Global analysis of genome, transcriptome, and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol. 2012;8:608.
Tang YC, Amon A. Gene copy-number alterations: a cost-benefit analysis. Cell. 2013;152:394–405.
Theurillat JP, Metzler SC, Henzi N, Djouder N, Helbling M, Zimmermann AK, et al. URI is an oncogene amplified in ovarian cancer cells and is required for their survival. Cancer Cell. 2011;19:317–32.
Han C, Yang L, Choi HH, Baddour J, Achreja A, Liu Y, et al. Amplification of USP13 drives ovarian cancer metabolism. Nat Commun. 2016;7:13525.
Deng P, Wang Z, Chen J, Liu S, Yao X, Liu S, et al. RAD21 amplification epigenetically suppresses interferon signaling to promote immune evasion in ovarian cancer. J Clin Invest. 2022;132:e159628.
Baselga J, Norton L, Albanell J, Kim YM, Mendelsohn J. Recombinant humanized anti-HER2 antibody (Herceptin) enhances the antitumor activity of paclitaxel and doxorubicin against HER2/neu overexpressing human breast cancer xenografts. Cancer Res. 1998;58:2825–31.
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–69.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90.
Okada N, Lin CP, Ribeiro MC, Biton A, Lai G, He X, et al. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–50.
Chaluvally-Raghavan P, Zhang F, Pradeep S, Hamilton MP, Zhao X, Rupaimoole R, et al. Copy number gain of hsa-miR-569 at 3q26.2 leads to loss of TP53INP1 and aggressiveness of epithelial cancers. Cancer Cell. 2014;26:863–79.
van Zandwijk N, Pavlakis N, Kao SC, Linton A, Boyer MJ, Clarke S, et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017;18:1386–96.
Smith ES, Whitty E, Yoo B, Moore A, Sempere LF, Medarova Z. Clinical applications of short non-coding RNA-based therapies in the era of precision medicine. Cancers. 2022;14:1588.
Soh J, Cho H, Choi CH, Lee H. Identification and characterization of microRNAs associated with somatic copy number alterations in cancer. Cancers. 2018;10:475.
Marcinkowska M, Szymanski M, Krzyzosiak WJ, Kozlowski P. Copy number variation of microRNA genes in the human genome. BMC Genom. 2011;12:183.
Zhang Z, Zhang L, Wang B, Wei R, Wang Y, Wan J, et al. MiR-337-3p suppresses proliferation of epithelial ovarian cancer by targeting PIK3CA and PIK3CB. Cancer Lett. 2020;469:54–67.
Goode EL, Chenevix-Trench G, Song H, Ramus SJ, Notaridou M, Lawrenson K, et al. A genome-wide association study identifies susceptibility loci for ovarian cancer at 2q31 and 8q24. Nat Genet. 2010;42:874–9.
Bell D, Berchuck A, Birrer M, Chien J, Cramer DW, Dao F, et al. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15.
Steele CD, Abbasi A, Islam SMA, Bowes AL, Khandekar A, Haase K, et al. Signatures of copy number alterations in human cancer. Nature. 2022;606:984–91.
Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–40.
Jakubek YA, Chang K, Sivakumar S, Yu Y, Giordano MR, Fowler J, et al. Large-scale analysis of acquired chromosomal alterations in non-tumor samples from patients with cancer. Nat Biotechnol. 2020;38:90–96.
Paul D, Islam S, Manne RK, Dinesh US, Malonia SK, Maity B, et al. F-box protein FBXO16 functions as a tumor suppressor by attenuating nuclear β-catenin function. J Pathol. 2019;248:266–79.
Bennett EJ, Rush J, Gygi SP, Harper JW. Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell. 2010;143:951–65.
Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20.
Ji M, Zhao Z, Li Y, Xu P, Shi J, Li Z, et al. FBXO16-mediated hnRNPL ubiquitination and degradation plays a tumor suppressor role in ovarian cancer. Cell Death Dis. 2021;12:758.
Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–30.
White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12:401–10.
Ma H, Li Y, Wang X, Wu H, Qi G, Li R, et al. PBK, targeted by EVI1, promotes metastasis and confers cisplatin resistance through inducing autophagy in high-grade serous ovarian carcinoma. Cell Death Dis. 2019;10:166.
Salvi A, Young AN, Huntsman AC, Pergande MR, Korkmaz MA, Rathnayake RA, et al. PHY34 inhibits autophagy through V-ATPase V0A2 subunit inhibition and CAS/CSE1L nuclear cargo trafficking in high grade serous ovarian cancer. Cell Death Dis. 2022;13:45.
Singha B, Laski J, Ramos Valdés Y, Liu E, DiMattia GE, Shepherd TG. Inhibiting ULK1 kinase decreases autophagy and cell viability in high-grade serous ovarian cancer spheroids. Am J Cancer Res. 2020;10:1384–99.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (82274575), Major Basic Research Project of Shandong Natural Science Foundation (ZR2023ZD56), the Natural Science Foundation of Shandong Province (ZR2021MH175, ZR2022LZY011), Co-construction project of State Administration of TCM (GZY-KJS-SD-2023-034, GZY-KJS-SD-2023-046), Taishan Scholars Program (tstp20240513), Central Government Guides Local Science and Technology Development Fund Projects of Shandong Province (YDZX20203700001407). Key Laboratory of Traditional Chinese Medicine Classical Theory, Ministry of Education, Open Research Project. National Youth Qihuang Scholar Training Program and Shandong Province Traditional Chinese Medicine High Level Talent Cultivation Project.
Author information
Authors and Affiliations
Contributions
ZZ and XL conceptualized the study. ZZ, CC and WL performed the experiments, and analyzed the data. XL and XY performed bioinformatics analysis. YZ, QH and HS helped with the xenograft mouse experiments. YZ, XZ, RW, NF, LC and MZ performed in vitro proliferation analysis. ZZ and XL wrote the manuscript. XL supervised the research project. The order of co-first authors was based on the order with which the individuals joined the project.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interest.
Ethics approval and consent to participate
All experimental procedures involving the mice were approved by the Institutional Animal Care and Use Committee of the Laboratory Animal Center of Shandong University of Traditional Chinese Medicine. The approval number is SDUTCM20221009382. Human OV tissues were obtained at Cancer Hospital Affiliated to Shandong First Medical University, and this study was approved by the local ethics committee with approval number of SDTHEC2023004016. The informed consent was obtained from all participants.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Angelo Peschiaroli
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, Z., Liu, X., Chu, C. et al. MIR937 amplification potentiates ovarian cancer progression by attenuating FBXO16 inhibition on ULK1-mediated autophagy. Cell Death Dis 15, 735 (2024). https://doi.org/10.1038/s41419-024-07120-8
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41419-024-07120-8
This article is cited by
-
F-box proteins in cancer: from cancer cells to the tumor microenvironment
Cell Communication and Signaling (2025)