Abstract
Abnormalities in ether lipid metabolism as well as the formation of neutrophil extracellular traps have recently been recognized as detrimental factors affecting tumorigenesis and progression. However, the role of abnormal ether lipid metabolism in colorectal cancer (CRC) evolution has not been reported. Here we show that the lipid metabolism-related gene enoyl-CoA δ-isomerase 2 (ECI2) plays a tumor-suppressor role in CRC and is negatively associated with poor prognosis in CRC patients. We mechanistically demonstrate that ECI2 reduces ether lipid-mediated Interleukin 8 (IL-8) expression leading to decreased neutrophil recruitment and neutrophil extracellular traps formation for colorectal cancer suppression. In particular, ECI2 inhibits ether lipid production in CRC cells by inhibiting the peroxisomal localization of alkylglycerone phosphate synthase (AGPS), the rate-limiting enzyme for ether lipid synthesis. These findings not only deepen our understanding of the role of metabolic reprogramming and neutrophil interactions in the progression of CRC, but also provide ideas for identifying potential diagnostic markers and therapeutic targets for CRC.
Similar content being viewed by others
Introduction
Tumor metabolic reprogramming plays an important role in CRC, and altered lipid metabolism is considered a hallmark feature of malignancy1. Lipid metabolic reprogramming is similar to the Warburg effect, with typical features affecting the biology of tumor proliferation, metastasis, and apoptosis1. It was found that abnormalities in ether lipid metabolism may be closely related to tumor evolution and metastasis2. However, there is still a considerable gap in our understanding of the molecular pathways involved in ether lipid metabolism related gene expression and action in CRC.
ECI2 proteins promote the re-entry of enoyl coenzyme A into the β-oxidation cycle3 and also participate in peroxisomal lipid metabolism4,5. There are still relatively few studies on the relevant functions of ECI family proteins in tumors6. Ether lipids (including alkyl acyl phospholipids and enoyl phospholipids, also known as acetylated phospholipids) are a unique and important class of glycerophospholipids. Ether lipids contain a glycerol backbone, an alkyl bond, or a vinyl bond, and their alkyl chains are attached to the sn-1 position by an ether bond7. Ether lipids are initially synthesized in the peroxisome and processed in the endoplasmic reticulum7. Key rate-limiting enzymes involved include FAR1/2, GNPAT, and AGPS7. Ether lipids may regulate cell differentiation, influence cell signaling, and reduce oxidative stress through their ability to act as potential endogenous antioxidants7,8,9. Ether lipids have been found to be increased in a variety of cancers and associated with higher invasiveness10. Targeted inhibition of peroxisomal lipid metabolic pathways that lead to specific ether lipid production in cancer cells could treat malignancies10,11. Recent studies have identified peroxisome-driven ether lipids as being involved in ferroptosis in cancer12. ECI2, a protein present in both mitochondria and peroxisomes, is thought to be involved in the β-oxidation of polyunsaturated fatty acids4,5. We wondered whether ECI2 is aberrantly expressed in CRC and whether it could affect fatty acid oxidation through the CRC. we also wanted to know whether it could regulate peroxisomal ether lipid metabolism.
Neutrophils are increasingly recognized as playing an important role in tumors. One defense response of abnormally triggered neutrophils is known as NETosis, in which activated neutrophils expel their DNA and intracellular contents in a meshwork called neutrophil extracellular traps (NETs)13. In cancer, NETosis is associated with increased disease progression, metastasis, and complications14,15,16. Studies have shown that reactive oxygen species (ROS) production is critical for all forms of NETosis17. NETs formation in solid tumors is dependent on IL-818. CXCL8 belongs to the CXC family of classic chemokines and is responsible for neutrophil recruitment and activation to sites of inflammation19,20. The CXCL8-CXCR1/2 axis plays an important role in tumor growth, angiogenesis, metastasis, stemness, and recruitment of immune cells to the tumor microenvironment21,22. Overexpression of CXCL8 has been observed in a variety of cancers20,23, including colorectal cancer23. Lipids were found to regulate IL-8 transcription in endothelial cells24. Ether lipids regulate inflammation by maintaining neutrophil membrane phospholipid composition and viability25. Ether glycerol lipids promote Interleukin 1β (IL-1β) release and the progression of experimental autoimmune encephalomyelitis26. Recent studies have suggested that hyperoxidation of ether-linked phospholipids accelerates NETs formation27. We wondered whether ether-bonded phospholipids could regulate NETosis and IL-8 in CRC, and if so, what is the mechanism of regulation?
The relationship between ether lipid metabolism and NETosis in CRC is still unknown, despite relevant studies suggesting that ether lipids regulate NETs formation. In the present study, we found that ECI2 levels were negatively associated with poor prognosis in clinical CRC patients. Mechanistically, ECI2 inhibits neutrophil extracellular traps-mediated CRC progression in the tumor microenvironment by regulating ether lipid-mediated IL-8 in CRC cells. Furthermore, we found that ECI2 inhibits ether lipid production by inhibiting the peroxisomal localization of AGPS, the rate-limiting enzyme for ether lipid synthesis in CRC. Taken together, these findings emphasize an important role between ECI2, ether lipid metabolism, IL-8 transcription, and NETosis. These findings not only expand our understanding of the roles of metabolic reprogramming and neutrophil-neutrophil interactions in CRC pathogenesis but also provide ideas for finding potential diagnostic markers and therapeutic targets for CRC.
Results
ECI2 downregulation predicts poor prognosis in CRC
We first downloaded the colorectal cancer dataset (Supplementary Data 1) from the TCGA database and analyzed it by GO analysis and KEGG analysis using the Metascape database. The fatty acid metabolic pathway genes enriched in the Reactome gene set in the GO form were analyzed (Supplementary Fig. 1a). Genes differentially expressed in colorectal cancer were screened by GEPIA database. Our aim was to screen for differential genes that are closely related to lipid metabolism in colorectal cancer. Three genes, FASN, PTGS1, and SCD, which were more studied among the first 15 most significantly differentiated genes, were excluded, and the remaining 12 differentially differentiated genes were subjected to RT-qPCR to detect their mRNA levels in 12 pairs of paired colorectal cancer tissues (Supplementary Data 1). The gene ECI2 was screened to be significantly differentially expressed in this pathway. ECI2 was found to be differentially expressed in most tumors and normal tissues by the GEPIA database and the Kaplan–Meier database (Supplementary Fig. 1b), The expression of ECI2 in CRC was significantly lower than that in normal tissues, and the expression of ECI2 was negatively correlated with the prognosis of patients with CRC (Supplementary Fig. 1c–e). Application of RT-qPCR and Western blots revealed that ECI2 expression in CRC cells was lower than that in normal colorectal epithelial cells FHC (Fig. 1a, b). Immunofluorescence experiments revealed that ECI2 was expressed in mitochondria and peroxisomes in SW620 cells (Fig. 1c). Application of RT-qPCR and Western blots revealed that the expression of ECI2 in fresh CRC tissues was significantly lower than that in normal tissues (Fig. 1d, e and Supplementary Data 2). Immunohistochemistry showed that ECI2 expression in CRC tissues was significantly lower than that in normal tissues (Fig. 1f), and was negatively correlated with CRC metastasis (Fig. 1g) and staging (Fig. 1h). Analysis of the relationship between ECI2 expression and clinicopathological parameters revealed that the expression level of ECI2 was negatively correlated with CRC tumor size, TNM stage, lymph node metastasis, and distant metastasis (Supplementary Tables 1 and 2). Survival analysis showed that disease-free survival (P = 0.011) and overall survival (P = 0.044) were higher in colorectal cancer patients with high expression of ECI2 than in those with low expression of ECI2 (Fig. 1i, j). The above results suggest that ECI2 has an inhibitory effect on colorectal cancer growth and metastasis.
RT-qPCR (a) (Representative data from n = 3 independent experiments, P < 0.0001) and Western blots (b) experiments to detect ECI2 in seven CRC cell lines and one normal colorectal epithelial cell (FHC) line. b The samples derive from the same experiment but different gels for ECI2, and another for Tubulin were processed in parallel. Tubulin served as loading control (The quantification provided under the blots is for the representative blot from n = 3 independent experiments). c ECI2 distribution in SW620 cells. Mitotracker is a mitochondrial probe and PMP70 is a peroxisomal marker (Representative images from n = 3 independent experiments, scale bar 10 μm). RT-qPCR (d) (n = 16 human samples per group, P < 0.0001) and Western blots (e) (n = 18 human samples per group, P < 0.0001) experiments to detect ECI2 in fresh CRC (T) and normal tissues (N). Right panel shows the statistics. The samples derive from the same experiment but different gels for ECI2, and another for Tubulin were processed in parallel (e). Tubulin served as loading control. Representative data from n = 3 independent experiments. f Immunohistochemical detection of ECI2 expression in paired CRC tissues (n = 210 pairs human samples, 3 fields assessed per sample). Right panel shows the statistics (P < 0.0001). g ECI2 levels in 24 distant metastatic and 186 non-metastatic patients (n = 186 non metastasis group, n = 24 metastasis group, P < 0.0001). h TNM staging for 107 stage III + IV and 103 stage I + II ECI2 levels (n = 103 stage I + II group, n = 107 stage III + IV group, P < 0.0001). i, j Kaplan–Meier method for statistical analysis of disease-free survival (i) and overall survival (j) (n = 152 low ECI2 group, n = 58 high ECI2 group). Data are expressed as mean ± s.d. (a, d, e, f, g, h). Statistical significance was determined by two-tailed Paired t-test (d, e), two-tailed unpaired Student’s t-test (f, g, h), one-way ANOVA with Tukey multiple comparison test (a) or log-rank test (i, j). Source data are provided as source data files.
ECI2 inhibits CRC invasion and metastasis through tumor cancer microenvironment
Based on the results of ECI2 expression in CRC cells (Fig. 1a, b), stable overexpression cell lines of ECI2 were constructed by screening HCT116 and RKO, which expressed ECI2 at a low level, and stable silencing cell lines of ECI2 were constructed by screening DLD1 and SW620, which expressed ECI2 at a high level (Supplementary Fig. 1f, g and Fig. 2a, b). In vitro functional experiments showed that ECI2 had insignificant effects on the proliferation and invasion of CRC cells. (Fig. 2c–g and Supplementary Fig. 1h–l). Subcutaneous tumor models showed that stable upregulation of ECI2 significantly inhibited mouse colorectal cancer cell line MC38 growth in mice in vivo (Supplementary Fig. 1m–q), while silencing ECI2 did the opposite (Fig. 2h–k). A significant increase in liver metastatic nodules was observed after further intrasplenic injection of MC38 cells with silenced ECI2 (Fig. 2l, m). The marked difference between the results of the in vivo and in vitro experiments suggests that ECI2 may have an effect on the proliferation and metastasis of CRC cells through the tumor microenvironment.
Western blots to verify the efficiency of overexpressing (a) and silencing (b) ECI2. The samples derive from the same experiment but different gels for ECI2, and another for Tubulin were processed in parallel (a, b). Tubulin served as loading control. The quantification provided under the blots is for the representative blot from n = 3 independent experiments. CCK8 assay to detect changes in proliferation of ECI2-silenced CRC cells DLD1 (c) and SW620 (d) (Representative data from n = 3 independent experiments). e Colony formation assay to detect the colonies formed by ECI2-silenced CRC cells. Right panel shows the statistics (Representative images from n = 3 independent experiments). f Transwell invasion assay to detect the invasion by ECI2-silenced CRC cells (scale bar 50 μm). Right panel shows the statistics (Representative images from n = 3 independent experiments). g The wound healing assay to detect migration of ECI2-silenced CRC cells (scale bar 100 μm). Right panel shows the statistics (Representative images from n = 3 independent experiments). h–k Subcutaneous tumorigenic model of C57BL/6J mice constructed by silencing ECI2-silenced MC38 cells (18 days post injection, n = 6 mice per group, shNC was the control group). h Photographs of the tumors at the end of the experiment. i Representative pictures of HE staining and ki-67 histochemistry of the tumors (3 fields assessed per sample, scale bar 20 μm). Weight (j) (P < 0.0001) and volume (k) of the tumors at the end of the experiment (n = 6 mice per group). l, m A mouse CRC liver metastasis model was constructed by intrasplenic injection of the mouse CRC cell line MC38, which silences ECI2 (n = 7 mice per group). l Photographs of liver tissues at the end of the experiments (scale bar 1 cm) and HE staining of liver metastatic tumors (3 fields assessed per sample, scale bar 100 μm). m Quantification of liver metastatic nodules (n = 7 mice per group). Data are expressed as mean ± s.d. (c, d, e, f, g, j, k, m). Statistical significance was determined by two-way ANOVA with Tukey or Sidak’s multiple comparison test (c, d, e, f, g, k) or two-tailed unpaired Student’s t-test (j, m). Source data are provided as source data files.
ECI2 inhibits IL-8 expression, neutrophil infiltration and NETosis in CRC
To explore the molecular mechanisms by which ECI2 inhibits CRC progression, we performed RNA-seq, GO analysis, and KEGG enrichment analysis. The pathway with the highest enrichment score was found to be the downregulated cytokine-cytokine receptor interaction pathway (Fig. 3a and Supplementary Fig. 2a). The genes of this pathway were mapped as heatmaps (Fig. 3b). RT-qPCR experiments on the 12 most significantly changed differential genes showed that ECI2 overexpression resulted in the most pronounced downregulation of CXCL8 (IL-8), followed by CCL5 and IL15 (Supplementary Fig. 2b). ELISA assay for the highly changed cytokines CXCL8, CCL5, CSF1, and IL15 in CRC cells revealed that ECI2 downregulated the CRC cell levels of IL-8 (Fig. 3c, d), but did not affect the levels of CCL5, CSF1 and IL15 (Supplementary Fig. 2c–e). To explore whether this cytokine pathway affects CRC immunomodulation, immunohistochemistry was performed to detect immune cells in 80 CRC tissues. The results revealed that the infiltration of neutrophils was most pronounced in CRC tissues with low expression of ECI2 (Supplementary Fig. 2f), and the number of neutrophils infiltrated was negatively correlated with the expression of ECI2 (Supplementary Fig. 2g). In vivo experiments revealed an increase in neutrophil infiltration after intrasplenic injection of MC38 cells silencing ECI2 in mice (Fig. 3e, f). Since IL-8 is not expressed in mice, but CXCL1/KC, CXCL2/MIP-2, and CXCL5-6/LIX are thought to be functional IL-8 homologs28. RT-qPCR examination of the mRNA content of CXCL1, 2, and 5 in MC38 silenced with ECI2 revealed that CXCL1 was upregulated the most significantly (Supplementary Fig. 2h). We subsequently performed in vivo mouse studies using CXCL1 as a homologue of mouse IL-8. Immunohistochemical examination of mouse liver tissues after intrasplenic injection of MC38 cells that silenced ECI2 revealed that silencing ECI2 promoted CXCL1 production in mouse liver metastatic tissues (Fig. 3e). The above in vivo and in vitro functional experiments and RNA-seq results further suggested that ECI2 may exert an oncogenic role in CRC by affecting neutrophil infiltration and cytokine IL-8.
a, b ECI2-overexpressing CRC cells were analyzed via RNA-seq and GO functional analysis; cytokine pathway-enriched genes heatmap shown (n = 3 samples (per group) generated after independent generation of cells and processed on different days). c, d ELISA was used to measure IL-8 (Representative data from n = 3 independent experiments, P < 0.0001). e, f Immunohistochemistry of Ly6G+ neutrophils and KC (CLCX1) in liver metastatic tumors (20 μm scale); neutrophils quantified (f). n = 7 mice per group, 3 fields assessed per sample. g, h ECI2-overexpressing CRC cells co-cultured with dHL-60; CCK8 and Transwell assays conducted (50 μm scale, P < 0.0001). Representative data from n = 3 independent experiments. i Conditioned medium (CM) of ECI2-overexpressing CRC cells induced dHL-60 chemotaxis. n = 3 independent experiments. j–l Immunofluorescence (Representative images from n = 3 independent experiments, scale bar 200 μm, P < 0.0001) and Western blots assays were used to detect NETs after co-culture. l The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel. H3 served as loading control. The quantification provided under the blots is for the representative blot from n = 3 independent experiments. NETs in liver metastases (m, n) (200 μm scale, n = 7 mice per group, 3 fields assessed per sample)and mice serum (o) (n = 7 mice per group); n Quantification of Fig. 3m. p–t Mice were injected intrasplenically with MC38 cells that silenced ECI2 and treated with anti-Ly-6G (IgG was the control, n = 7 mice per group). p Photos of liver tissue (scale bar 1 cm), HE staining (50 μm scale), and immunohistochemistry of neutrophil (50 μm scale; magnification scale bar 150 nm) and KC (CLCX1) (50 μm scale) (3 fields assessed per sample). q Quantification of liver metastatic nodules (n = 7 mice per group). r Quantification of neutrophils (n = 7 mice per group, 3 fields assessed per sample, P < 0.0001). s ELISA assay for the NETs in mice serum (P < 0.0001, n = 7 mice per group). t Immunofluorescence and quantification (right panel) of NETs in liver metastatic tumors (n = 7 mice per group, 3 fields assessed per sample, 200 μm scale, P < 0.0001). Data are expressed as mean ± s.d. (c, d, f, g, h, i, j, k, n, o, q, r, s, t). Statistical significance was determined by two-way ANOVA with Tukey or Sidak’s multiple comparison test (c, d, g, i), one-way ANOVA with Tukey multiple comparison test (h), two-tailed unpaired Student’s t test (f, j, k, n, o, q, r, s, t) or Hypergeometric Test (a). Source data are provided as source data files.
Because human neutrophils begin to undergo apoptosis within 6 to 12 h of isolation, making their use in experimental studies limited by their very short lifespan29. The human myeloid neutrophil-like cell line, HL-60 cell line, is commonly used as a replacement for human neutrophils, and the gene expression of its differentiated cells bears a resemblance to that of the primary neutrophil30. HL-60 was differentiated into neutrophils with 1.25% DMSO in 5% CO2 at 37 °C (differentiated neutrophils were noted as dHL-60). Giemsa staining (Supplementary Fig. 2i) and flow cytometry (Supplementary Fig. 2j) assays showed successful differentiation. ECI2 significantly inhibited CRC cell proliferation (Fig. 3g and Supplementary Fig. 2k) and invasion (Fig. 3h and Supplementary Fig. 2l) after co-culturing CRC cells with neutrophil dHL-60 cells. Neutrophil dHL-60 chemotaxis assays showed that conditioned medium (CM) of CRC cells overexpressing ECI2 significantly inhibited dHL-60 migration (Fig. 3i), while the opposite was true for conditioned medium silencing ECI2 (Supplementary Fig. 2m). Immunofluorescence staining (Fig. 3j, k and Supplementary Fig. 2n, o) and Western blots (Fig. 3l) revealed that the NETosis was inhibited when CRC cells overexpressing ECI2 were co-cultured with dHL-60, and the contrary was true for silencing ECI2. Immunofluorescence detection of liver tissues of mice after intrasplenic injection of MC38 cells with silenced ECI2 revealed that silencing ECI2 promoted the NETosis in metastatic tumor tissues (Fig. 3m, n). ELISA assay revealed that mice in the silenced ECI2 group had increased levels of cit-H3 in the serums (Fig. 3o). In mice injected intrasplenically with MC38 cells that silenced ECI2 and treated with anti-Ly-6G, decreases in the number of liver metastasis nodules (Fig. 3p, q), neutrophil infiltration (Fig. 3p, r), KC (CXCL1) expression (Fig. 3p), and the content of cit-H3 in the serums and liver metastasis tissues were observed (Fig. 3s, t). The above experimental results indicated that ECI2 significantly inhibited IL-8 production, neutrophil infiltration, and NETosis in CRC.
ECI2 inhibits CRC invasion and metastasis by reducing IL-8-mediated NETosis
To explore whether ECI2 regulates the NETosis in CRC by modulating IL-8, we performed the following experiments. Western blots and immunofluorescence experiments showed when dHL-60 was cultured with a conditioned culture of CRC cells silenced with ECI2, the NETosis enhanced by silencing of ECI2 was reversed by the addition of DNase I, a nuclease capable of degrading NET-DNA (Fig. 4a, b). Whereas, when dHL-60 was cultured with a conditioned culture of CRC cells overexpressing ECI2, the NETosis suppressed by overexpressing of ECI2 was reversed by the addition of PMA (phorbol-12-myristate-13-acetate, a potent inducer of NETosis) (Supplementary Fig. 2p, q). After addition of DNase I to dHL-60 co-cultured with ECI2-silenced CRC cells, the promotion of CRC cell proliferation (Fig. 4c) and invasion (Fig. 4d) after silencing of ECI2 was reversed by addition of DNase I to dHL-60. After addition of PMA to dHL-60 co-cultured with CRC cells overexpressing ECI2, the proliferation (Supplementary Fig. 2r), and invasion (Supplementary Fig. 2s) of CRC cells inhibited by overexpression of ECI2 were reversed by addition of PMA to dHL-60. After intrasplenic injection of MC38 cells silencing ECI2 in mice treated with DNase I, a decrease in the number of liver metastasis nodules (Fig. 4e, f), and in the content of cit-H3 in serums and liver metastasis tissues of mice were observed (Fig. 4g, h). These results suggest that ECI2 inhibits CRC progression by suppressing NETosis of neutrophils.
a, b ECI2-silenced CRC cell medium cultured dHL-60 with DNase I (0.25 μ/ml). NETs detected by Western blots (The quantification provided under the blots is for the representative blot from n = 3 independent experiments) and immunofluorescence (200 μm scale, representative images from n = 3 independent experiments). The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel (a). H3 served as loading control. c, d Co-cultured ECI2-silenced CRC cells with dHL-60 and DNase I; CCK8 (P < 0.0001) and Transwell assays conducted (50 μm scale). Representative data from n = 3 independent experiments. e–h Mice injected intrasplenically with ECI2-silenced MC38 cells and DNase I (n = 7 mice per group); liver tissue and HE staining (1 cm and 50 μm scales, 3 fields assessed per sample); f metastatic nodule quantification (n = 7 mice per group). g, h Serum and tumor cit-H3 levels measured by ELISA (n = 7 mice per group) and immunofluorescence (200 μm scale, n = 7 mice per group, 3 fields assessed per sample). i Anti-IL-8 (500 ng/ml) treated ECI2-silenced CRC cell supernatants induced dHL-60 chemotaxis (n = 3 independent experiments). j, k Co-cultured ECI2-silenced CRC cells with anti-IL-8 and dHL-60 were analyzed for NETs using Western blots (The quantification provided under the blots is for the representative blot from n = 3 independent experiments) and immunofluorescence (200 μm scale, representative images from n = 3 independent experiments, P < 0.0001). The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel (j). H3 served as loading control. l, m Silenced ECI2 CRC cells with anti-IL-8 were added and co-cultured with dHL-60, then CCK8 proliferation and Transwell invasion assays were conducted (50 μm scale, P < 0.0001). Representative data from n = 3 independent experiments. n–q Mice were intrasplenically injected with MC38 cells that silenced ECI2 and silenced CXCL1 (n = 6 mice per group). n Photos of liver tissue and HE staining (1 cm and 50 μm scales, 3 fields assessed per sample). o Quantification of liver metastatic nodules (n = 6 mice per group). p, q Cit-H3 levels in tumors and serum were measured by immunofluorescence (200 μm scale, n = 6 mice per group, 3 fields assessed per sample) and ELISA (n = 6 mice per group) (P < 0.0001). Data are expressed as mean ± s.d. (b, c, d, f, g, h, i, k, l, m, o, p, q). Statistical significance was determined by two-way ANOVA with Tukey or Sidak’s multiple comparison test (c, i, l), one-way ANOVA with Tukey multiple comparison test (d, m) or two-tailed unpaired Student’s t-test (b, f, g, h, k, o, p, q). Source data are provided as source data files.
The ability of different doses of IL-8 to induce NETosis in primary human neutrophils was detected by SYTOX Green immunofluorescence staining and MPO-DNA complex. It was found that 100 ng/ml IL-8 induced the strongest NETosis of neutrophils (Supplementary Fig. 3a, b). Next, we will apply 100 ng/ml IL-8 for in vitro experiments. ECI2 overexpressing CRC cells with addition of recombinant IL-8 cytokines were co-cultured with neutrophil dHL-60 to examine the proliferation and invasion of CRC cells, the migration of dHL-60, and the NETosis of neutrophils. The results showed that the dHL-60 cells migration (Supplementary Fig. 3c), the NETosis (Supplementary Fig. 3d, e), CRC cell proliferation (Supplementary Fig. 3f), and invasion (Supplementary Fig. 3g), which were inhibited by ECI2 overexpression, were all reversed by the cytokine IL-8. While the addition of anti-IL-8 in ECI2-silenced CRC cells were co-cultured with neutrophil dHL-60, the proliferation and invasion of CRC cells, migration of dHL-60, and the NETosis of neutrophils were detected. The results showed that the dHL-60 cell migration (Fig. 4i), the NETosis (Fig. 4j, k), CRC cell proliferation (Fig. 4l) and invasion (Fig. 4m), which were promoted by ECI2 silencing, were all reversed by anti-IL-8. After intrasplenic injection of ECI2-silenced MC38 cells in mice, a reduction in the number of liver metastatic nodules (Supplementary Fig. 3h, i), neutrophil infiltration (Supplementary Fig. 3h, j), KC (CXCL1) expression (Supplementary Fig. 3h), and cit-H3 content in the serums of the mice and in the liver metastatic tissues (Supplementary Fig. 3k, l) were observed by anti-KC (CXCL1) treatment. Similarly, after intrasplenic injection of MC38 cells with simultaneous silencing of ECI2 and CXCL1 in mice, a reduction in the number of liver metastatic nodules (Fig. 4n, o), cit-H3 content in the serums of the mice and in the liver metastatic tissues (Fig. 4p, q) were observed. To exclude whether ECI2 regulates CRC cell proliferation and invasion via IL-8 in the absence of neutrophils, we added recombinant IL-8 cytokines to CRC cells overexpressing ECI2 or anti-IL-8 to cells silencing ECI2 but in the absence of neutrophil dHL-60 co-culture to detect CRC cell proliferation and invasion. It was found that in the absence of neutrophils present, ECI2 exerted insignificant effects on CRC cell proliferation (Supplementary Fig. 3m, o) and invasion (Supplementary Fig. 3n, p) via IL-8. The above results suggest that ECI2 inhibits CRC invasion and metastasis by reducing the IL-8-mediated NETosis.
ECI2 inhibits ether lipid metabolism-mediated CRC progression by inhibiting AGPS peroxisome localization
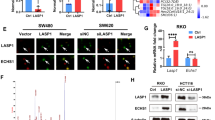
To explore whether the lipid metabolism-related gene ECI2 regulates lipid metabolism, lipidomic analysis revealed that the 20 lipids with significant changes induced by overexpression of ECI2 were mainly concentrated in ether phospholipids, with ether-lipid-linked phosphatidylethanolamine (PE-O) and ether-lipid-linked phosphatidylcholine (PC-O) the most obvious ones (Fig. 5a). The ether lipids with the most pronounced differences were further confirmed by plotting the thermograms of ether lipid-linked lipids as PE-O and PC-O (Fig. 5b). ECI2 has been reported to be involved in the β-oxidation of fatty acids. ECI2 was found not to affect fatty acid oxidation in CRC cells by fatty acid oxidation kit assay (Supplementary Fig. 4a, b). This result is similar to the metabolomics results (Supplementary Data 3). Addition of ether lipids (18:0-18:1 PE-O) in the absence of neutrophils had no significant effect on CRC cell proliferation and invasion (Supplementary Fig. 3m, n). In contrast, the significant inhibitory effect of ECI2 overexpression on CRC cells was significantly reversed by the addition of 18:0-18:1 PE-O to ECI2 overexpressing CRC cells co-cultured with neutrophils (Supplementary Fig. 3f, g). After co-culturing dHL-60 with ECI2 overexpressing CRC cells treated with 18:0-18:1 PE-O, both the inhibition of NETosis (Fig. 5c, d) and the attenuation of dHL-60 cell migration (Fig. 5e) were reversed. The above results suggest that ECI2 inhibits ether lipid production. The peroxisome plays a central role in the metabolism of reactive oxygen species, bile acids, and ether lipids. Studies of individual peroxisomal proteins and metabolites support their pro-tumorigenic function2. At least three peroxisomal proteins (Pex3, Pex16, and Pex19) are essential for peroxisomal membrane assembly and peroxisomal membrane protein (PMP) import11. PMP70 is a marker for peroxisomal membranes2. To explore the mechanism of ECI2 inhibition of ether lipid production, Western blots detection of peroxisome-related markers revealed that ECI2 did not affect the expression of PEX19, PEX16, and PMP70 (Supplementary Fig. 4c), suggesting that ECI2 did not affect peroxisome biogenesis and abundance. The expression of ether lipid metabolism rate-limiting enzymes FAR1, GNPAT, and AGPS was detected by RT-qPCR (Supplementary Fig. 4d) and Western blots (Supplementary Fig. 4e), and it was found that ECI2 did not affect the changes in their mRNA and protein expression levels. By peroxisome isolation assay and Western blots, it was found that ECI2 did not affect the distribution of FAR1 and GNPAT in peroxisomes and cytoplasm, but ECI2 affected the distribution of AGPS (Fig. 5f, g), which resulted in a significant decrease in peroxisomes and a significant increase in cytoplasm (Fig. 5f, g). Immunofluorescence staining also revealed the inhibitory effect of ECI2 on AGPS peroxisomal localization (Supplementary Fig. 4f). The above results indicated that ECI2 inhibited the peroxisome localization of AGPS, the ether lipid-mediated NETosis and the proliferation and invasion of CRC cells.
a, b Lipidomic analysis of the overexpressing ECI2 group (ECI2) and the control group (NC) (n = 3 samples (per group) generated after independent generation of cells and processed on different days). a The top 20 lipids with the most pronounced differences among the downregulated lipids were detected. b Heatmap of ether-associated lipids. c, d After addition of 18:0-18:1 PE-O (10 μg /ml) of CRC cells overexpressing ECI2 co-cultured with dHL-60, NETs formation was detected by applying immunofluorescence (c) (scale bar 200 μm, representative images from n = 3 independent experiments) and Western blots (d) (The quantification provided under the blots is for the representative blot from n = 3 independent experiments). The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel (d). H3 served as loading control. e Addition of supernatants from ECI2 overexpressing CRC cells cultured in 18:0-18:1 PE-O (10 μg /ml) induced chemotaxis assay of dHL-60 cells. n = 3 independent experiments. f, g Peroxisome isolation assay and Western blots to detect changes in FAR1, GNPAT, and AGPS in peroxisome (P) and cytoplasm (C) of CRC cells overexpressing (f) and silencing ECI2 (g). The samples derive from the same experiment but different gels for AGPS, GAPDH, another for FAR1, another for GNPAT and another for PMP70 were processed in parallel (f, g). GAPDH and PMP70 served as loading control. The quantification provided under the blots is for the representative blot from n = 3 independent experiments. h–l Mice were intrasplenically injected with MC38 cells transfected with shECI2 and AGPS-WT or AGPS-Mu lentivirus (n = 7 mice per group). h Representative images of liver tissue (scale bar 1 cm, 3 fields assessed per sample), HE staining of liver metastatic tumors (scale bar 50 μm), Ly6G-labeled neutrophils of liver metastatic tumors (scale bar 50 μm. Enlarged image scale bar 150 nm), and KC (CLCX1) of liver metastatic tumors (scale bar 50 μm), respectively, from left to right. i Quantification of liver metastatic nodules (n = 7 mice per group). j Quantification of neutrophils in liver metastatic tumors (n = 7 mice per group, 3 fields assessed per sample). k ELISA assay for the level of cit-H3 in the serum of mice (P < 0.0001, n = 7 mice per group). l Representative pictures of NETs in liver metastatic tumors by applying immunofluorescence assay (n = 7 mice per group, 3 fields assessed per sample, scale bar 200 μm, P < 0.0001). Data are expressed as mean ± s.d. (c, e, i, j, k, l). Statistical significance was determined by two-way ANOVA with Tukey or Sidak’s multiple comparison test (e) or two-tailed unpaired Student’s t-test (c, i, j, k, l). Source data are provided as source data files.
We wanted to understand whether ECI2 affects peroxisome localization of other PTS2-containing proteins. Based on the current literature, we detected other PTS2-containing proteins (PHYH, DEPP, Kichip4, P7BP2) other than AGPS by peroxisome isolation assays31. We found that ECI2 did not affect other identified PTS2-containing proteins other than AGPS (Supplementary Fig. 4g). To investigate whether there is a physical interaction between ECI2 and AGPS. And, whether ECI2 disrupts the interaction between AGPS and the PTS2 receptor PEX7. The CO-IP experiments revealed that ECI2 did not physically interact with AGPS (Supplementary Fig. 4h); instead, AGPS interacted with PEX7 (Supplementary Fig. 4i). Furthermore, we found that AGPS binding to PEX7 was reduced in ECI2 overexpressing CRC cells. The opposite was true for silencing ECI2 (Supplementary Fig. 4j). It is suggested that ECI2 selectively inhibits the peroxisomal localization of AGPS, possibly by inhibiting the interaction of AGPS with PEX7. AGPS lentiviral vector mutated (AGPS-Mu) in the PTS2 sequence (responsible for introducing the relevant proteins into the peroxisome) and control wild-type lentiviral vector (AGPS-WT) were constructed (Supplementary Fig. 4k). Simultaneous transfection of silencing ECI2 (shECI2) and AGPS-WT or AGPS-Mu revealed that AGPS-Mu in the absence of neutrophils had no obvious effect on CRC cell proliferation and invasion after silencing ECI2 (Supplementary Fig. 4l, m). However, CRC cells co-transfected with both shECI2 and AGPS-Wu viruses were found to significantly reverse the enhancement of CRC cell proliferation and invasion after ECI2 knockdown (Supplementary Fig. 4n, o), the enhancement of NETosis (Supplementary Fig. 4p, q), and the migration of dHL-60 (Supplementary Fig. 4r) induced by ECI2 knockdown, when co-cultured with neutrophil dHL-60. The continued addition of 18:0-18:1 PE-O counteracted the effect of AGPS-Mu (Supplementary Fig. 4n–r). After intrasplenic injection of MC38 cells in mice transfected with both ECI2 silencing and AGPS-WT or AGPS-Mu, a reduction in the number of liver metastatic foci (Fig. 5h, i), neutrophil infiltration (Fig. 5h, j), KC (CXCL1) expression (Fig. 5h), and cit-H3 content in serums and liver metastatic tissues of the mice were observed in the AGPS-Mu group (Fig. 5k, l). The above results tentatively suggest that ECI2 reduces the production of ether lipids by inhibiting the localization of peroxisomes in AGPS, which in turn inhibits the NETosis of neutrophil and the proliferation and invasion of CRC.
ECI2 inhibits NETosis and CRC progression by suppressing ether lipid metabolism-mediated IL-8 transcription
To investigate whether ECI2 regulates IL-8 through ether lipids, addition of 18:0-18:1 PE-O revealed significant upregulation of IL-8 in CRC cells overexpressing ECI2 (Fig. 6a). Transfection of AGPS-WT or AGPS-Mu in CRC cells silencing ECI2 revealed that AGPS-Mu significantly downregulated IL-8 (Fig. 6b). To exclude whether ether lipids regulate neutrophil IL-8 production, addition of ether lipids to dHL-60 cells revealed that ether lipids did not affect the amount of IL-8 (Fig. 6c). To determine whether IL-8 accumulation occurs at the transcriptional level, CRC cells were pretreated with actinomycin D followed by 18:0-18:1 PE-O. RT-qPCR quantitative analysis showed that actinomycin D almost completely inhibited IL-8 transcription (85% and 95% inhibition in the control and 18:0-18:1 PE-O groups, respectively) (Fig. 6d). To determine whether the differences in IL-8 post-transcriptional regulation explained the differences in IL-8 kinetics induced by the two groups, CRC cells were treated with 18:0-18:1 PE-O for 3 h prior to actinomycin D treatment and RT-qPCR was performed to detect IL-8 mRNA at different time points. The results showed that IL-8 transcripts of the two groups were almost completely degraded at around 1 h (Fig. 6e), indicating that the two groups of IL-8 mRNA half-life was similar. Application of the luciferase reporter plasmid system with human IL-8 promoter −1481 to +44 bp showed that 18:0-18:1 PE-O induced transcriptional activation of the IL-8 promoter (Fig. 6f). These data suggest that 18:0-18:1 PE-O-induced IL-8 expression in CRC cells is regulated at the transcriptional level.
a ECI2-overexpressing CRC cells ± dHL-60 co-culture treated with 18:0-18:1 PE-O, and ELISA detected IL-8 (P < 0.0001). b shECI2 and AGPS-WT or AGPS-Mu transfected CRC cells ± dHL-60 co-culture treated with 18:0-18:1 PE-O had IL-8 levels measured by ELISA (P < 0.0001). c dHL-60 cells with 18:0-18:1 PE-O were assessed for IL-8 by ELISA. d CRC cells pretreated with actinomycin D then 18:0-18:1 PE-O for 3 h were analyzed for IL-8 by RT-qPCR. e CRC cells treated with 18:0-18:1 PE-O for 3 h before actinomycin D were sampled at intervals for IL-8 by RT-qPCR (P < 0.0001). f Transfected CRC cells with the human IL-8 promoter and 18:0-18:1 PE-O had luciferase activity measured (P < 0.0001). g, i ECI2-overexpressing CRC cells successively treated with 18:0-18:1 PE-O, anti-IL-8 and PMA were co-cultured with dHL-60. NETs and invasion detected by Western blots (The quantification provided under the blots is for the representative blot from n = 3 independent experiments) and Transwell (Representative data from n = 3 independent experiments, 50 μm scale, P < 0.0001). The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel (g). H3 served as loading control. h, j shECI2 and AGPS-Mu transfected CRC cells treated with recombinant IL-8 and DNase I were co-cultured with dHL-60. NETs and invasion detected by Western blots (The quantification provided under the blots is for the representative blot from n = 3 independent experiments) and Transwell (Representative data from n = 3 independent experiments, 50 μm scale, P < 0.0001). The samples derive from the same experiment but different gels for cit-H3, and another for H3 were processed in parallel (h). H3 served as loading control. k–n Mice with ECI2-silenced MC38 cells treated with Reparixin (n = 6 mice per group) were analyzed for liver tissue (1 cm and 50 μm scale, 3 fields assessed per sample), quantified metastatic nodules (P < 0.0001, n = 6 mice per group), tumors NETs by immunofluorescence (200 μm scale, n = 6 mice per group, 3 fields assessed per sample) and serum NETs by ELISA (n = 6 mice per group). Data are expressed as mean ± s.d. (a, b, c, d, e, f, i, j, l, m, n). Statistical significance was determined by two-way ANOVA with Tukey or Sidak’s multiple comparison test (a, b, d, e, f), one-way ANOVA with Tukey multiple comparison test (i, j) or two-tailed unpaired Student’s t-test (c, l, m, n). Representative data from n = 3 independent experiments (a–f). Source data are provided as source data files.
After different treatments of ECI2 overexpressing CRC cells in advance, they were co-cultured with dHL-60 cells (with or without PMA) to detect CRC cell invasion and NETosis in dHL-60 cells. The results showed that the inhibitory effect of ECI2 overexpression on CRC cell invasion and NETosis in dHL-60 was reversed by 18:0-18:1 PE-O, and then CRC cell invasion and NETosis in dHL-60 were reversed and reduced again by anti-IL-8, and finally PMA acting on dHL-60 cells reversed the effect of anti-IL-8 (Fig. 6g, i). CRC cells invasion and NETosis of dHL-60 cells were examined after transfection of ECI2 gene-silenced CRC cells (with or without IL-8) with AGPS-WT or AGPS-Mu and co-culturing with neutrophil dHL-60 (with or without DNase I). The results showed that AGPS-Mu reversed the promotional effect of silencing ECI2 on CRC cell invasion and NETosis in dHL-60, the addition of cytokine IL-8 in turn reversed and increased CRC cell invasion and NETosis in dHL-60, and finally DNase I reversed the effect of cytokine IL-8 (Fig. 6h, j). The above results suggest that ECI2 inhibits NETosis by suppressing ether lipid metabolism-mediated IL-8 transcription, which in turn inhibits CRC progression.
It is known that the receptor for IL-8 and CXCL1 on neutrophils is CXCR1/2, and in order to show the efficacy of CXCR1/2 receptor antagonists, we applied the CXCR1/2 receptor antagonist, Reparixin, for in vivo experiments to improve the clinical significance of the results of this study. In mice injected intrasplenically with MC38 cells that silenced ECI2 and treated with Reparixin, decreases in the number of liver metastasis nodules (Fig. 6k, l), the content of cit-H3 in the liver metastasis tissues and the serums were observed (Fig. 6m, n). For the rigor of the study, we used primary human peripheral blood neutrophils and primary mouse bone marrow neutrophils instead of dHL-60 for in vitro experimental studies. CM of CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; supplemented with anti-IL-8 or 18:0-18:1 PE-O) were used to culture primary neutrophils (1 × 105, with or without DNase I), then the purified NETs-DNA were used to culture CRC cells to detect alterations in the invasive and proliferative capacity of CRC cells. Similar to the results of the dHL-60 study, ECI2 modulated primary neutrophil NETosis-mediated CRC invasion and proliferation by modulating peroxisome localization of AGPS, ether lipids, and IL-8 in CRC cells (Supplementary Figs. 5a–c, 6a–c). ROS production is known to be critical for all forms of NETosis, and we performed ROS assays after CM of CRC cells cultured under different conditions were used to culture primary human neutrophils. It was found that silencing ECI2 and AGPS mutations suppressed ROS levels in neutrophils, whereas supplementation with 18:0-18:1 PE-O reversed the effect of AGPS-Mu and finally anti IL-8 counteracted the effect of 18:0-18:1 PE-O (Supplementary Fig. 5d). Unexpectedly, we found that treatment of neutrophils with 18:0-18:1 PE-O alone did not alter neutrophils ROS levels, whereas IL-8 alone increased neutrophil ROS levels (Supplementary Fig. 5d). The results suggest that ether lipids need to affect ROS by affecting IL-8. After culturing primary neutrophils (1 × 105, with or without DNase I) with CM of CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; supplemented with anti-IL-8 or 18:0-18:1 PE-O), Sytox Green dye staining and MPO-DNA complex assay revealed that, similar to dHL-60 cells, NETosis of primary neutrophils was affected by ECI2, AGPS, ether lipids and IL-8 in CRC cells (Supplementary Figs. 5e, f, 6f, g). In agreement with the in vitro and in vivo results, we examined AGPS distribution in ECI2-silenced mouse MC38 cells and found that ECI2 silencing promoted peroxide media of AGPS in MC38 cells (Supplementary Fig. 6d). Further by ELISA assay, we found that AGPS mutation in MC38 cells silenced with ECI2 suppressed the level of CXCL1, while the addition of 18:0-18:1 PE-O counteracted the effect of AGPS-Mu (Supplementary Fig. 6e).
Discussion
Altered lipid metabolism is considered a hallmark feature of malignant tumors. Ether lipids2, as a specific type of lipid, have received increasing attention for their role in tumors2,12. Recently, a relationship between abnormal ether lipid metabolism in peroxisomes and neutrophil function has been reported27,32. However, whether ether lipid metabolism abnormality plays a role in CRC and its aberrant molecular mechanisms are still largely overlooked. In this study, we screened a lipid metabolism-associated protein, ECI2, and characterized its function in CRC progression. Mechanistically, we found that ECI2 inhibits NETosis-mediated CRC progression in the microenvironment by suppressing ether lipid production-mediated transcription of IL-8 in colorectal cancer cells. In which ECI2 is inhibiting ether lipid production by inhibiting the peroxisomal localization of the ether lipid metabolism rate-limiting enzyme AGPS (Supplementary Fig. 7c).
In order to discover the changes of genes related to abnormal lipid metabolism in CRC, we screened the differential gene ECI2 using public databases. Our study found that ECI2 expression was downregulated in CRC tissues and negatively correlated with poor prognosis in CRC patients. Further in vivo functional experiments revealed that ECI2 had insignificant effects on the proliferation and invasion of CRC cells in vitro. In contrast, in vivo experiments in mice revealed that ECI2 significantly inhibited the proliferation and metastasis of CRC. This suggests that we ECI2 may act by affecting the tumor microenvironment of CRC. Further, we found that ECI2 may play a role through neutrophils in the microenvironment. By co-culturing with neutrophils, the proliferation and invasive of CRC cells were enhanced. Studies of neutrophil infiltration into the tumor microenvironment (TME) have revealed their diversity and plasticity in cancer development33. Tumor-associated neutrophils (TAN) exert both pro- and anti-cancer roles16,34,35, but the mechanisms are not fully understood. There are a large number of published reports supporting the tumor-promoting role of neutrophils16,35. It has been reported that neutrophils can undergo a unique form of cell death known as NETosis (the process of NETs formation), which involves the extrusion of their nuclear DNA content and cleaved proteins into the extracellular space in the form of so-called NETs36. NETs are involved in the onset and progression of a wide variety of diseases17,35, but the presence of NETs in patients with cancer has not been extensive and in-depth studies until recently14,37, including colorectal cancer38, breast cancer39, and pancreatic cancer40. Recent reports have shown that NETs increase tumor growth by driving mitochondrial homeostasis in tumor cells41. NETs may also impair tumor cell contact with cytotoxic immune cells, leading to evasion of immunological surveillance42. Our study found that ECI2, through inhibition of the CRC NETs to inhibit the proliferation and metastasis of CRC. The duality of neutrophils in cancer extends to cancer treatments such as radiotherapy, chemotherapy, and immunotherapy43. A recent study elaborated that chemotherapy via IL-1β triggers NETosis to cause epithelial mesenchymal transition and chemoresistance44. However, a recent study by Li et al. demonstrated that a combination of drugs induced NETs via ROS in order to enhance chemotherapeutic efficacy in PI3KCA-mutant CRC45. These two processes differ in terms of triggering mechanism and downstream effects46. In addition, the chemotherapeutic agents used in these studies were different. These findings call into question the conventional wisdom that NETs act primarily as impediments to apoptosis47 and provoke thought about what specific conditions and factors influence the actions of NETs. The role that NETs play in cancer may depend on a variety of factors, including the substances used to induce NETs, the type of cells targeted, and the specific kind of NETs46. Mutational landscapes of cancer may also modulate different roles of neutrophils in chemotherapeutic response or malignant behavior of tumors45,46. Li et al. used a combination of drugs that specifically induced NETs in PI3KCA-mutated CRC, suggesting the presence of a mutational effect45.
So, what is the mechanism by which ECI2 affects NETosis. A variety of cytokines have been found to play a role in NETs formation39,42,48,49. For example, NETs formation in solid tumors is partially dependent on IL-818,48. CXCL8 is produced by various cancer cells and regulates cancer progression in an autocrine manner, and overexpression of CXCL8 has been observed in a variety of cancers20,23. Recently, it has been found that the tumor-secreted protease tissue protease C (CTSC) promotes breast-lung metastasis through pro-activation of neutrophil membrane-bound protease 3 (PR3), which facilitates IL-1β processing and NF-κB activation, thereby upregulating IL-6 and CCL3 for neutrophil recruitment and NETs formation39. The secreted protease PRSS35 inhibits hepatocellular carcinoma by disabling CXCL2-mediated NETs49. Through transcriptomics, we found that ECI2 could enrich in cytokine and cytokine receptor action pathways, we speculated whether ECI2 acted through the relevant cytokine action on neutrophils yet. Interestingly, we observed in this study that decreased levels of ECI2 led to an increase in the chemokine CXCL8 (IL-8), resulting in greater accumulation of neutrophils and increased NETs formation in TME.
As a lipid metabolism gene, especially a fatty acid oxidation-related gene4,5, we found that ECI2 did not affect fatty acid oxidation in CRC cells by fatty acid oxidation kit assay. Further, we found that ECI2 overexpression significantly decreased multiple ether phospholipids without affecting fatty acid changes by lipid quantitative histology assay, which was consistent with our results. Ether lipids represent an important group of lipids7, and abnormalities in their metabolic processes are associated with immune disorders, inflammation, and cancer32. Targeted inhibition of peroxisomal lipid metabolic pathways leading to specific ether lipid production in cancer cells can treat malignant tumors10,11. Recent studies have reported the involvement of peroxisome-driven ether lipids in the ferroptosis process in cancer12. Ether lipids also play a role in the regulation of cytokines. Ether lipids were found to affect IL-1β release and experimental autoimmune encephalomyelitis26. Increased IL-8 transcription in endothelial cells was also found to be regulated by oxidized phospholipids24. Peroxisome beta-oxidation and ether lipid synthesis control the development and activation of innate and adaptive immune cells32. Ether lipids are autoantigens that stimulate invariant natural killer T cells in the thymus8. Acute ether lipid deficiency affects neutrophil function in mice50. High oxidation of ether-linked phospholipids accelerates NETs formation27. Ether-linked phospholipids have also been found to be required for mitochondrial reactive oxygen species homeostasis51. Combined with the literature, we speculated whether ECI2 would regulate IL-8 and thus affect NETosis in the microenvironment through ether lipids in the CRC. Consistent with our conjecture, our study found that ECI2 inhibits ether lipid production, which in turn regulates IL-8 transcription in CRC cells. Further, ECI2 was found to inhibit NETosis-mediated CRC progression in the tumor microenvironment by suppressing IL-8 in CRC cells. Additionally, upon antagonizing IL-8, ECI2-influenced ether lipid production did not cause NETosis in neutrophils, which further suggests that ECI2-regulated ether lipids are required to influence NETosis via IL-8 in CRC cells.
Since ECI2 regulates ether lipid metabolism in CRC cells to regulate NETosis, we wondered by what mechanism ECI2 regulates ether lipid metabolism. According to reports, peroxisomes are the main organelles producing ether lipids7,12. So far, FAR1/2, GNPAT, and AGPS have been found to be the key rate-limiting enzymes for major ether lipid metabolism7,12. Among them, the sequence near the N-terminus (PTS2) of alkylglycerol phosphate synthase (AGPS) is essential for AGPS to enter into the peroxisome and function52. AGPS has been reported to alter the structural balance between signaling lipids to enhance cancer pathogenicity10. Our findings indicate that ECI2 abnormalities in CRC cells do not affect peroxisome biogenesis and abundance; nor do they affect the expression of ether lipid metabolism rate-limiting enzymes FAR1, GNPAT, and AGPS; but they inhibit the distribution of AGPS. That is, ECI2 overexpression promotes an increase in AGPS in peroxisomes and a decrease in its distribution in the cytoplasm. We wanted to explore why ECI2 selectively inhibits peroxisomal localization of AGPS but no other proteins involved in ether lipid synthesis. It was found that ECI2 disrupts the interaction between AGPS and the PTS2 receptor PEX7.
To summarize our findings, ECI2 inhibits ether lipid production by inhibiting the peroxisome localization of AGPS in CRC cells. It further inhibits neutrophil extracellular trap-mediated colorectal cancer progression by suppressing ether lipid-mediated transcription of IL-8, which in turn inhibits neutrophil extracellular trap-mediated CRC progression. Thus, our study provides strong evidence that ECI2 is associated with NETosis in the tumor microenvironment, expanding our understanding of the mechanisms controlling neutrophil recruitment and activity in the TME. In addition, ECI2 was also associated with ether lipid metabolism and IL-8 transcription, which provides ideas about the relationship between tumor metabolic reprogramming and immune regulation.
As a malignant tumor, CRC still faces challenges in precise treatment and prevention of recurrence and metastasis, which leads to high mortality and recurrence rates in patients. Therefore, the identification of reliable biomarkers is an important step in reducing the mortality and recurrence rates of CRC patients. However, the progress of our work remains limited by the complexity and variability of tumor patients and precision therapy medicine. In this study, we analyzed differential genes related to tumor metabolism and found a strong association between ECI2, ether chain phospholipid metabolism, NETosis, and CRC prognosis. By applying CXCR1/2 receptor antagonists in mice also provided clinical implications for this study. The combination of detecting low expression of ECI2 in colorectal cancer and high levels of the NETosis markers cit-H3 and ether phospholipids in serum could be of potential value as a potential candidate target for CRC therapeutic strategies.
Methods
Ethics statement
Our study complied with all relevant codes of ethics of Southern Medical University. All animal protocols were approved by the Animal Research Ethics Committee of Southern Medical University and were conducted in accordance with the guidelines for the use of laboratory animals. The collection and use of clinical materials and human samples were approved by the Institutional Research Ethics Committee of Nanfang Hospital of Southern Medical University.
Cell culture
The MC38 cell line (C57BL/6J mouse colon adenocarcinoma cells) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS. All CRC cell lines were cultured in RPMI 1640 medium with 10% FBS. Neutrophils (primary human neutrophils, primary mouse bone marrow neutrophils, HL-60) were cultured in Roswell Park Memorial Institute (RPMI)1640 medium (Thermo Scientific, Waltham, MA, USA) with 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA).
CCK8, colony formation assay, cell wound healing assay, Transwell invasion assay
For Cell proliferation assay53, 1 × 103 cells seeded in 96-well plates, cultured 24 h, treated with CCK-8, 2 h incubation, read at 450 nm, repeated 3 times. For Colony formation assay53, Cells inoculated in 6-well plates, cultured 2 weeks, fixed with 4% paraformaldehyde, stained with 1% Giemsa, colonies >50 counted, 3 experiments done. For Cell wound healing assay53, 1.2 × 106 cells cultured 24 h, wounded, washed, RPMI 1640, closure at 0, 72 h, images for migration, 3 fields measured. For Transwell invasion assay53, pre-coated Transwell chamber with Matrigel, 2 × 105 cells in serum-free media, 8-μm pores, 10% FBS attractant, 37 °C, 2 days, stained with crystal violet, 5 fields counted. Under different culture conditions, CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; supplemented with 500 ng/ml anti-IL-8, 100 ng/ml recombinant IL-8 cytokine or 10 μg/ml ether lipid (18:0-18:1 PE-O)) were co-cultured with dH-60 cells (1 × 105, with or without 25 ng/ml PMA, 0.25 μ/ml DNase I), and CRC cells were assayed by CCK8 assay and Transwell invasion assay.
Clinical human tissue specimens
The 18 paired colorectal cancer lesions and adjacent non-cancerous clinical tissue samples were obtained from colorectal cancer patients (Supplementary Tables 4). All patients were from the Department of General Surgery, Nanfang Hospital, Southern Medical University. Supplementary Table 1 details the clinicopathologic characteristics, of the 210 CRC patients utilized for IHC testing (Fig. 1f). In order to use these clinical materials for research purposes, we obtained prior written informed consent from the patients and approval from the Institutional Research Ethics Committee of Nanfang Hospital of Southern Medical University.
Immunohistochemistry
Processed with heat, dewaxing, rehydration, antigen repair in citrate buffer, peroxide block, sealed with FBS. Overnight incubation with primary antibody at 4 °C, secondary antibody at 37 °C, DAB, and hematoxylin staining, then observed53.
Co-immunoprecipitation (co-IP) assay, RNA extraction, RT-qPCR, and Western blots analysis
For co-IP assay, 1 × 106 cells cultured, washed with cold PBS, lysed with cold RIPA, scraped, oscillated, centrifuged. Protein G beads removed, target antibody incubated, protein A added, washed, boiled, SDS-PAGE, Western blots analyzed53. For RNA extraction and RT-qPCR53, Trizol extracted RNA; PrimeScript RT synthesized cDNA; RT-qPCR on ABI7500 with SYBR PreMix; GAPDH as control. For Western blots53, proteins separated by SDS-PAGE, transferred to PVDF, blocked, incubated with primary/secondary antibodies, detected by chemiluminescence. RT-qPCR primer sequences are shown in the Supplementary Table 3.
Lentiviral and plasmid transduction and transfection
Silencing ECI2 (shECI2) lentivirus was constructed from Obio Technology Co. (Y18163, GL413NC, Shanghai, China), and overexpression of ECI2, AGPS wild-type (AGPS-WT), and AGPS mutant (AGPS-Mu) lentivirus was constructed from WZ Biosciences Inc (Shandong, China). Lentiviral and blank vectors transfected, puromycin selected, protein and mRNA analyzed by RT-qPCR and Western blots53.
Mice
Mice were housed at the animal facility of Southern Medical University. Four-week-old female C57BL/6J and 8-week-old female C57BL/6J were purchased from Guangdong Animal Center, China.
Animal studies
All animals were housed at a suitable temperature (18–23 °C) and humidity (22–50%) under a 12/12-h light/dark cycle. All animals were allowed to eat and drink freely during the experiment. All animal experiments were approved by the Animal Research Ethics Committee of Southern Medical University. For the mouse subcutaneous tumorigenic model, each group (NC/ECI2, shNC/shECI2) of MC38 cells (4 × 106) was injected subcutaneously into four-week-old female mice (C57BL/6J; Guangdong Animal Center, China). Tumor volume (length × width2 × 0.5) was measured every 4 days in mice. The tumor load was less than the maximum diameter (15 mm) approved by the University Animal Research Ethics Committee of Southern Medical University. The experiment was terminated when the tumors appeared about 1.5 mm, and all mice were euthanized by inhalation of carbon dioxide.
For the mouse colorectal cancer liver metastasis model, 1 × 106 MC38 shNC or MC38 shECI2 cells were added to 100 μL PBS using a 28G insulin syringe and injected into the spleen of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China) via a 3 cm midline open abdomen. For the role of AGPS-Mu, MC38 cells transfected with both shECI2 and AGPS-WT or AGPS-Mu were injected into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China). For the role of mouse CXCL1, MC38 cells transfected with both shECI2 and shCXCL1 were injected into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China). For the effect of DNase I, after injection of MC38 shECI2 cells into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China), mice were injected with DNase I (2.5 mg/kg, intraperitoneally, Roche) once a day, and the control group was given PBS. For the effect of anti-KC (CXCL1), after injection of MC38 shECI2 cells into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China), mice were injected twice a week intraperitoneally with anti-KC (CXCL1) (1.2 μg/kg, PeproTech), and isotype control IgG was given to the control group. For the effect of anti-Ly6G, after injection of MC38 shECI2 cells into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China), anti-mouse Ly6G (5 mg/kg, EBioscience) was injected intraperitoneally twice a week, and isotype control IgG was given to the control group. For the effect of mouse CXCR 1/2, after injection of MC38 shECI2 cells into the spleens of 8-week-old female mice (C57BL/6J; Guangdong Animal Center, China), mice were injected every two days until the completion of the study intraperitoneally with CXCR1/2 antagonists Reparixin (15 mg/kg, Selleck). Mice were euthanized 25 days after injection of the cells, when the mice lost 20–25% of their body weight or when the mice developed cachexia. Liver metastases (including the size and number of metastatic cancer nodules on the surface of the liver) were observed by dissection. Whole liver tissues were cut and fixed with l0% neutral formalin, and the number of metastatic foci were counted after paraffin embedding, dehydration, serial tissue sections, and HE staining for light microscopic observation. The infiltration of each immune cell and the expression of KC (CXCL1) in the liver tissue were detected by immunohistochemistry; NETs formation (MPO, cit-H3) was detected by immunofluorescence in the liver tissue; and the level of citrullinated histone 3 (cit-H3), which labels NETs, in the serums of mice was detected by ELISA.
Lipidomic analysis
Lipid metabolite analysis of cell samples was performed by the targeted quantitative lipidomic method by Wuhan Metware Biotechnology Co., Ltd (Wuhan, China). Previously frozen colorectal cancer cells (1 × 107) were homogenized in 1 ml of lipid extract (methyl tert-butyl ether: methanol = 3:1). The samples were stirred for 5 min at 4 °C. Then, 200 µl of deionized water was added to the mixture, followed by centrifugation at 12,000 × g for 10 min at 4 °C. The extract supernatant was dried and redissolved. Metabolites were quantified using an ultra-performance liquid chromatography-electrospray tandem mass spectrometry (UPLC-MS/MS) system (UPLC, ExionLC™AD, https://sciex.com.cn/; MS/MS, QTRAP® 6500 +, https://sciex.com.cn/). Analysis was accomplished using triple quadrupole mass spectrometry in Multiple Reaction Monitoring (MRM) mode. The software Analyst 1.6.3 was utilized to process the mass spectrometry data. The samples were quality controlled and a total of 834 lipid metabolites were detected.
RNA sequencing and data processing
Total RNA was extracted using Trizol (Life Technologies) according to the manufacturer’s instructions, and RNA quality was assayed by both the NanoDrop 2000 spectrophotometer and the Agilent 2100/4200 assay. A total of 3 μg of RNA per sample was used for analysis. Sequencing was sampled from a single replicate. Libraries were constructed using the common library method. After library construction, Qubit 3.0 was used for preliminary quantification, and RT-qPCR was used to accurately quantify the effective concentration of the libraries. RNA-seq was performed by BerryGenomics (Beijing, China) on the Illumina NovaSeq 6000 sequencing platform in PE150 mode. After obtaining the sequencing data, basic data quality control was performed first, and then these high-quality sequences were aligned to the reference genome and analyzed for gene expression level and gene structure. The reference genome for this project can be downloaded from: Genome Download Link (please use IE browser to open this link).
Enzyme-linked immunosorbent assay (ELISA)
Blood samples obtained from mice by venipuncture and cellular serum samples were aliquoted and stored immediately at −80 °C according to our institutional specimen storage protocol. Quantification was performed according to the kit manufacturer’s instructions, respectively. The kits for human IL-8, human CCL5, human CSF1, human IL15, and mouse CXCL1 were purchased from Proteintech (China). The kit for mouse Cit-H3 was purchased from Ruixin Bio (China).
Isolation of peroxisomes by gradient centrifugation
2 × 108 cells were collected. Peroxisome extraction buffer (5 mM MOPS, pH 7.65, containing 0.25 M sucrose, 1 mM EDTA, and 0.1% ethanol, protease inhibitor mixture) was added (sigma). The cell suspension was then transferred to a Dounce homogenizer tube and carefully homogenized 25 times on ice using a mortar and pestle B. The tube was centrifuged at 1000 × g for 10 min at 4 °C, and the supernatant was transferred and centrifuged at 2000 × g for 10 min at 4 °C to obtain a pellet (i.e., mitochondrial layer.). The supernatant was transferred to a new centrifuge tube and centrifuged at 25,000 × g for 20 min. The supernatant (peroxisome-free fraction) was transferred to a new tube and the suspension (precipitate) was the crude peroxisome fraction. Peroxisome extraction buffer was added to the precipitate and centrifuged at 100,000 × g for 1.5 h. The substrate was collected for further analysis in Western blots experiments.
Immunofluorescence detection of NETs
CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; with or without 500 ng/ml anti-IL-8, 100 ng/ml recombinant IL-8 cytokine or 10 μg/ml 18:0-18:1 PE-O) conditioned medium was added to 24-well plates with polylysine-coated Cicle Microsop Cover Glass and incubated with dHL-60 (1 × 105, with or without 25 ng/ml PMA, 0.25 μ/ml DNase I). NETs of dHL-60 cells were detected after 4 h of incubation.
For cell samples, cells were fixed using 4% paraformaldehyde for 15 min. permeabilized in 0.1% Triton X-100 for 5 min. The cover glasses were then closed in PBS containing 2% bovine serum albumin for 1 h at room temperature. Then the cover glasses were incubated overnight at 4 °C in a mixture of the two primary antibodies. The following primary antibodies cit-H3 (anti-rabbit antibody, Abcam), MPO (anti-mouse antibody, Immunoway) were used. The next day cover glasses were washed using cold PBS and incubated for 1 h in the dark at room temperature with a mixture of two secondary antibodies produced in different species. The following secondary antibodies were used: anti-rabbit labeled with Alexa Fluor 488 and anti-mouse labeled with Alexa Fluor 594. Slides and coverslips were restained using 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich) to visualize nuclei. The cover glasses were sealed with a sealer containing an anti-fluorescence quencher. Each sample was examined under a fluorescence microscope and photographed. NETosis was determined to be the percentage of field-of-view positivity for citrullinated histone 3 (cit-H3). For paraffin-embedded tissue samples, sections were made at a thickness of 4 mm. After deparaffinization, antigen repair was performed in a citrate solution in a microwave oven (95 °C, 30 min). Then closed in PBS containing 2% bovine serum albumin for 1 h at room temperature. The next steps are similar to the staining of cell samples.
Chemotaxis assay
The dHL-60 cells (1 × 105 cells) were added to the upper chamber added to the Transwell device (Corning, 3402). Conditioned medium of differently treated CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; with or without 500 ng/ml anti-IL-8, 100 ng/ml recombinant IL-8 cytokine or 10 μg/ml 18:0-18:1 PE-O) was added to the lower chamber as a chemoattractant. And after 3.5 h of incubation optical microscopy was observed to count the number of cells located in the lower chamber of the number of dHL-60.
Induction of dHL-60 cell differentiation
HL-60 (ATCC: CCL-240), a human myeloid neutrophil-like cell, was cultured in RPMI-1641 (Sigma, USA) with 20% fetal bovine serum (FBS) and 1% penicillin/streptomycin and maintained in a 5% CO2 incubator at 37 °C. The cells were incubated with 1.25% DMSO in 5% CO2 at 37 °C for 5 days to allow them to differentiate into neutrophils, which were recorded as dHL-60. Differentiation was verified by flow cytometry and Giemsa staining.
Flow cytometry
Pre-differentiated and post-differentiated HL-60 cells (5 × 106 cells) were collected. Goat serum was blocked on ice for 30 min, and the cells were incubated with goat anti-rat CD11b antibody coupled with Alexa fluor647 dye (BD Biosciences) and goat anti-rabbit CD16 antibody coupled with FITC (BD Biosciences) for 30 min at 4 °C protected from light. Finally, the cells were resuspended in 100 μl PBS and the cell suspension was transferred to flow cytometry tubes for flow cytometry analysis (CytoFLEX S, Beckman). Analysis was performed using Flow-jo (V10.8.1) software (BD, USA).
Actinomycin D assay
To determine whether IL-8 accumulation occurs at the transcriptional level, CRC cells were pretreated with actinomycin D (2 μg/ml; FDbio, China) for 30 min, followed by treatment with 18:0-18:1 PE-O (10 μg/ml) for 3 h, and RNA was extracted for RT-qPCR. To determine whether IL-8 post-transcriptional regulation differed in the experimental and control groups, CRC cells were treated with 18:0-18:1 PE-O (10 μg/ml) for 3 h prior to actinomycin D (2 μg/ml) treatment. RNA was extracted at different time points for RT-qPCR detection of IL-8 mRNA. IL-8 mRNA abundance was analyzed with GAPDH as an endogenous control.
Luciferase assay
IL-8 activation was measured using the IL-8-dependent luciferase reporter gene (pGL3-IL8-Luc (−1481 + 44)). All experiments were performed according to the kit instructions (Promega). Cells were inoculated in 96-well plates and cell lysis buffer was transferred to black microtiter plates. Firefly luciferase reaction solution was then added and firefly luciferase activity was measured. The activity was measured after incubation with Renilla luciferase reaction solution.
Neutrophil isolation and NETosis detection
Neutrophils were isolated from adult human peripheral blood by density gradient separation using dextran-Ficoll-Paque Premium (GE Healthcare) and from bone marrow of 7–8-wk-old mice using Percoll (GE Healthcare)54. Isolated neutrophils were confirmed to be of >95% purity by flow cytometric analysis of staining of neutrophil markers. Unless noted otherwise, neutrophils were cultured in RPMI 1640 medium containing 10% FBS.
For visualization, neutrophils cultured with CM from different CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; with or without 500 ng/ml anti-IL-8, 100 ng/ml recombinant IL-8 cytokine or 10 μg/ml 18:0-18:1 PE-O) were inoculated in 96-well plates for the corresponding incubation, and the cell-impermeable DNA dye SytoxGreen (Thermo Fisher Scientific, 1:10,000) and the cell-permeable DNA dye Hoechst33342 (Thermo Fisher Scientific, 1:1000) were added to the incubation system. At the end of the incubation, the plates were moved directly to a fluorescence microscope for NETs formation visualization. For NETs purification, CM-cultured neutrophil supernatants from different CRC cells were centrifuged at 18,000 × g for 10 min at 4 °C. The precipitate was then resuspended in 100 μl cold PBS. For each assay, three independent experiments were performed where each experiment used neutrophils from a different donor.
Measuring MPO-DNA complexes
To quantify NETosis, MPO-DNA complexes were identified using a capture ELISA. 96-wellmicrotiter plates were coated with 5 μg/ml anti-MPO monoclonal antibody (Proteintech, 22225-1-AP) as the capturing antibody (75 µl per well) overnight at 4 °C. After blocking in 1% BSA (125 µl per well), 40 µl of samples together with peroxidase-labeled anti-DNA monoclonal antibody was added (component No.2 of the Cell Death Detection ELISA kit, Roche, 11774425001), incubated at room temperature for 2 h and then washed with PBS three times. The peroxidase substrate (ABTS) (Roche, 11774425001) was added. After incubation at 37 °C in the dark for 40 min, the optical density was measured at 405 nm using a microplate reader.
ROS analysis
Neutrophils were treated with different conditioned medium of CRC cells (transfected with shECI2 or AGPS-Mu/AGPS-WT; with or without 500 ng/ml anti-IL-8, 100 ng/ml recombinant IL-8 cytokine or 10 μg/ml 18:0-18:1 PE-O) for 2 h. Subsequently, the cells were incubated with 10 mM CM-H2DCFDA (Yeasen), for 15 min and then washed with pre-chilled PBS, and the fluorescence of each well was monitored at 480 nm excitation and 540 nm emission.
Edu assay
CRC cells were inoculated in 24-well plates and cultured for 24 h prior to administration of the Edu assay kit (Beyotime Biotechnology, China). Subsequently, cells were fixed, permeabilized and stained according to the manufacturer’s instructions. Image acquisition and analysis using fluorescence microscopy.
Fatty acid oxidation assay
CRC cells were inoculated in 96-well plates and cultured for 24 h. Subsequently, the cells were subjected to FAO assay according to the manufacturer’s instructions (AssayGenie, China). O.D. 492 readings were measured using a microplate reader.
Statistics and reproducibility
Error lines indicate mean ± SD. Statistical significance was assigned at P values of <0.05 and detected by Prism9 (GraphPad software), ImageJ software, or SPSS software Statistics 22 (IBM Corp.). The Shapiro-Wilk test was used to determine the sample distribution type. A two-tailed Student’s t-test was used to evaluate statistical significance between two groups for normal distribution. For the nonparametric tests, the two-tailed Mann–Whitney test was used to evaluate statistical significance between two groups. For more than two groups, the significance was calculated by the Kruskal-Wallis test or ordinary one-way ANOVA depending on the sample distribution type; a post hoc Tukey test was used to conduct multiple comparisons. For more than two factors, the significance was calculated by the two-way ANOVA; a post hoc Tukey or Sidak’s test was used to conduct multiple comparisons. Correlations between ECI2 expression and immune cells were tested with Pearson correlation. A two-sided χ2 test was used. Survival curves were plotted using the Kaplan–Meier method. Correlation analysis was performed using Correlation of Ozone correlations. The relationship between ECI2 expression and clinicopathological characteristics was analyzed using the chi-square test and Pearson correlation. The experiments were not randomized, except that mice were randomly grouped before different treatments. Data collection and analysis were not performed blind to the conditions of the experiments, except for IHC score analysis. Each experiment was repeated at least three times independently. Some studies present the results of a representative experiment chosen from independent experiments, where independent experiments refer to experiments conducted on different days. All data in the article, supplementary Information are available.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the Sequence Read Archive (SRA) under the accession code PRJNA999604. The metabolomics data generated in this study have been deposited in the MetaboLights database under the accession code MTBLS10323. All data in the article, supplementary Information are available. Source data are provided with this paper.
References
Chen, D. et al. Lipid metabolism reprogramming in colorectal cancer. J. Cell. Biochem. 124, 3–16 (2022).
Dahabieh, M. S. et al. Peroxisomes and cancer: the role of a metabolic specialist in a disease of aberrant metabolism. Biochim. Biophys. Acta Rev. Cancer 1870, 103–121 (2018).
Houten, S. M., Violante, S., Ventura, F. V. & Wanders, R. J. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu. Rev. Physiol. 78, 23–44 (2016).
Fan, J., Li, X., Issop, L., Culty, M. & Papadopoulos, V. ACBD2/ECI2-mediated peroxisome-mitochondria interactions in Leydig cell steroid biosynthesis. Mol. Endocrinol. 30, 763–782 (2016).
Fan, J., Liu, J., Culty, M. & Papadopoulos, V. Acyl-coenzyme A binding domain containing 3 (ACBD3; PAP7; GCP60): an emerging signaling molecule. Prog. Lipid Res. 49, 218–234 (2010).
Dundr, P. et al. HNF1B, EZH2 and ECI2 in prostate carcinoma. Molecular, immunohistochemical and clinico-pathological study. Sci. Rep. 10, 14365 (2020).
Dean, J. M. & Lodhi, I. J. Structural and functional roles of ether lipids. Protein Cell 9, 196–206 (2018).
Facciotti, F. et al. Peroxisome-derived lipids are self antigens that stimulate invariant natural killer T cells in the thymus. Nat. Immunol. 13, 474–480 (2012).
Hossain, M. S. et al. Plasmalogens rescue neuronal cell death through an activation of AKT and ERK survival signaling. PLoS ONE 8, e83508 (2013).
Benjamin, D. I. et al. Ether lipid generating enzyme AGPS alters the balance of structural and signaling lipids to fuel cancer pathogenicity. Proc. Natl Acad. Sci. USA 110, 14912–14917 (2013).
Lodhi, I. J. & Semenkovich, C. F. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 19, 380–392 (2014).
Zou, Y. et al. Plasticity of ether lipids promotes ferroptosis susceptibility and evasion. Nature 585, 603–608 (2020).
Ronchetti, L. et al. Neutrophil extracellular traps in cancer: not only catching microbes. J. Exp. Clin. Cancer Res. 40, 231 (2021).
Cristinziano, L. et al. Neutrophil extracellular traps in cancer. Semin. Cancer Biol. 79, 91–104 (2022).
Masucci, M. T., Minopoli, M., Del Vecchio, S. & Carriero, M. V. The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Front. Immunol. 11, 1749 (2020).
Hedrick, C. C. & Malanchi, I. Neutrophils in cancer: heterogeneous and multifaceted. Nat. Rev. Immunol. 22, 173–187 (2022).
Papayannopoulos, V. Neutrophil extracellular traps in immunity and disease. Nat. Rev. Immunol. 18, 134–147 (2018).
de Andrea, C. E. et al. Heterogenous presence of neutrophil extracellular traps in human solid tumours is partially dependent on IL-8. J. Pathol. 255, 190–201 (2021).
Bakouny, Z. & Choueiri, T. K. IL-8 and cancer prognosis on immunotherapy. Nat. Med. 26, 650–651 (2020).
Raza, S. et al. Multifaceted role of chemokines in solid tumors: from biology to therapy. Semin. Cancer Biol. 86, 1105–1121 (2022).
Ha, H., Debnath, B. & Neamati, N. Role of the CXCL8-CXCR1/2 axis in cancer and inflammatory diseases. Theranostics 7, 1543–1588 (2017).
Liu, Q. et al. The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Factor Rev. 31, 61–71 (2016).
Casasanta, M. A. et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 13, eaba9157 (2020).
Yeh, M. et al. Increased transcription of IL-8 in endothelial cells is differentially regulated by TNF-alpha and oxidized phospholipids. Arterioscler. Thromb. Vasc. Biol. 21, 1585–1591 (2001).
Lodhi, I. J. et al. Peroxisomal lipid synthesis regulates inflammation by sustaining neutrophil membrane phospholipid composition and viability. Cell Metab. 21, 51–64 (2015).
Boomkamp, S. D. et al. Effect of ether glycerol lipids on interleukin-1β release and experimental autoimmune encephalomyelitis. Chem. Phys. Lipids 194, 2–11 (2016).
Yotsumoto, S. et al. Hyperoxidation of ether-linked phospholipids accelerates neutrophil extracellular trap formation. Sci. Rep. 7, 16026 (2017).
Hol, J., Wilhelmsen, L. & Haraldsen, G. The murine IL-8 homologues KC, MIP-2, and LIX are found in endothelial cytoplasmic granules but not in Weibel-Palade bodies. J. Leukoc. Biol. 87, 501–508 (2010).
Kolaczkowska, E. & Kubes, P. Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159–175 (2013).
Collins, S. J. The HL-60 promyelocytic leukemia cell line: proliferation, differentiation, and cellular oncogene expression. Blood 70, 1233–1244 (1987).
Kunze, M. The type-2 peroxisomal targeting signal. Biochim. Biophys. Acta Mol. Cell Res. 1867, 118609 (2020).
Di Cara, F., Savary, S., Kovacs, W. J., Kim, P. & Rachubinski, R. A. The peroxisome: an up-and-coming organelle in immunometabolism. Trends Cell Biol. 33, 70–86 (2023).
Jaillon, S. et al. Neutrophil diversity and plasticity in tumour progression and therapy. Nat. Rev. Cancer 20, 485–503 (2020).
Coffelt, S. B., Wellenstein, M. D. & de Visser, K. E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 (2016).
Xiong, S., Dong, L. & Cheng, L. Neutrophils in cancer carcinogenesis and metastasis. J. Hematol. Oncol. 14, 173 (2021).
Brinkmann, V. et al. Neutrophil extracellular traps kill bacteria. Science 303, 1532–1535 (2004).
Zhang, Y. et al. A signature for pan-cancer prognosis based on neutrophil extracellular traps. J. Immunother. Cancer 10, e004210 (2022).
Yang, L. et al. DNA of neutrophil extracellular traps promotes cancer metastasis via CCDC25. Nature 583, 133–138 (2020).
Xiao, Y. et al. Cathepsin C promotes breast cancer lung metastasis by modulating neutrophil infiltration and neutrophil extracellular trap formation. Cancer Cell 39, 423–437.e427 (2021).
Deng, J. et al. DDR1-induced neutrophil extracellular traps drive pancreatic cancer metastasis. JCI Insight 6, e146133 (2021).
Yazdani, H. O. et al. Neutrophil extracellular traps drive mitochondrial homeostasis in tumors to augment growth. Cancer Res. 79, 5626–5639 (2019).
Teijeira, Á. et al. CXCR1 and CXCR2 chemokine receptor agonists produced by tumors induce neutrophil extracellular traps that interfere with immune cytotoxicity. Immunity 52, 856–871.e858 (2020).
Shahzad, M. H. et al. Neutrophil extracellular traps in cancer therapy resistance. Cancers 14, 1359 (2022).
Mousset, A. et al. Neutrophil extracellular traps formed during chemotherapy confer treatment resistance via TGF-β activation. Cancer Cell 41, 757–775.e710 (2023).
Li, Y. et al. Neutrophil extracellular traps induced by chemotherapy inhibit tumor growth in murine models of colorectal cancer. J. Clin. Invest. 134, e175031 (2024).
Mousset, A. & Albrengues, J. NETs unleashed: neutrophil extracellular traps boost chemotherapy against colorectal cancer. J. Clin. Invest. 134, e178344 (2024).
Cools-Lartigue, J., Spicer, J., Najmeh, S. & Ferri, L. Neutrophil extracellular traps in cancer progression. Cell. Mol. Life Sci. 71, 4179–4194 (2014).
Ogawa, R. et al. Loss of SMAD4 promotes colorectal cancer progression by recruiting tumor-associated neutrophils via the CXCL1/8-CXCR2 axis. Clin. Cancer Res. 25, 2887–2899 (2019).
Wang, T. et al. Secreted protease PRSS35 suppresses hepatocellular carcinoma by disabling CXCL2-mediated neutrophil extracellular traps. Nat. Commun. 14, 1513 (2023).
Lodhi, I. J., Link, D. C. & Semenkovich, C. F. Acute ether lipid deficiency affects neutrophil biology in mice. Cell Metab. 21, 652–653 (2015).
Chen, Z. et al. Ether phospholipids are required for mitochondrial reactive oxygen species homeostasis. Nat. Commun. 14, 2194 (2023).
Kunze, M. et al. Mechanistic insights into PTS2-mediated peroxisomal protein import: the co-receptor PEX5L drastically increases the interaction strength between the cargo protein and the receptor PEX7. J. Biol. Chem. 290, 4928–4940 (2015).
Chen, L. et al. CSRP2 suppresses colorectal cancer progression via p130Cas/Rac1 axis-meditated ERK, PAK, and HIPPO signaling pathways. Theranostics 10, 11063–11079 (2020).
Kuang, Y., Parthasarathy, U. & Martinelli, R. Protocol for density gradient neutrophil isolation and flow cytometry-based characterization from human peripheral blood. STAR Protoc. 4, 102497 (2023).
Acknowledgements
This work is supported by the National Natural Science Foundation of China (grant number 82072705、81874074). Please address all the correspondence and requests for materials to X.L. (nfydlxn@126.com).
Author information
Authors and Affiliations
Contributions
X.L. conceived the study and supervised experiments. L.C. and X.L. designed experiments. L.C., P.D., Y.C., and L.L. performed experiments. L.C. and H.W. analyzed RNA-seq data and lipid metabolomics data. Y.L. and X.L. provided clinical specimens. Y.L., Q.Y., and L.Z. provided constructive advice. X.L. and L.C. wrote the paper. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, L., Dai, P., Liu, L. et al. The lipid-metabolism enzyme ECI2 reduces neutrophil extracellular traps formation for colorectal cancer suppression. Nat Commun 15, 7184 (2024). https://doi.org/10.1038/s41467-024-51489-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51489-1
This article is cited by
-
Remodeling the tumor dormancy ecosystem to prevent recurrence and metastasis
Signal Transduction and Targeted Therapy (2026)
-
From biosynthesis to function: the roles of ether lipids in cancer development and treatment
Lipids in Health and Disease (2025)
-
Single-cell RNA sequencing reveals neutrophil differentiation-related genes for immunotherapy response prediction in colorectal cancer
BMC Cancer (2025)
-
CRIP1 inhibits cutaneous melanoma progression through TFAM-mediated mitochondrial biogenesis
Scientific Reports (2025)
-
CRLS1 influences liver metastasis in colon cancer by regulating lipid metabolism pathways
Functional & Integrative Genomics (2025)