Abstract
The immune checkpoint protein, Lymphocyte activation gene-3 (LAG3), binds Major Histocompatibility Complex Class II (MHC-II) and suppresses T cell activation. Despite the recent FDA approval of a LAG3 inhibitor for the treatment of melanoma, how LAG3 engages MHC-II on the cell surface remains poorly understood. Here, we determine the 3.84 Å-resolution structure of mouse LAG3 bound to the MHC-II molecule I-Ab, revealing that domain 1 (D1) of LAG3 binds a conserved, membrane-proximal region of MHC-II spanning both the α2 and β2 subdomains. LAG3 dimerization restricts the intermolecular spacing of MHC-II molecules, which may attenuate T cell activation by enforcing suboptimal signaling geometry. The LAG3-MHC-II interface overlaps with the MHC-II-binding site of the T cell coreceptor CD4, implicating disruption of CD4-MHC-II interactions as a mechanism for LAG3 immunosuppressive function. Lastly, antibody epitope analysis indicates that multiple LAG3 inhibitors do not recognize the MHC-II-binding interface of LAG3, suggesting a role for functionally distinct mechanisms of LAG3 antagonism in therapeutic development.
Similar content being viewed by others
Introduction
Over the last decade, immune checkpoint blockade emerged as a transformative strategy for the treatment of cancer. The first FDA-approved checkpoint inhibitors, antibodies targeting the immune inhibitory receptors PD1 and CTLA4, block ligand-mediated activation to boost the function of effector T cells in the tumor environment1. The clinical success of PD1 and CTLA4 antagonism has prompted the assessment of additional immune checkpoint proteins as therapeutic targets, especially given that existing immunotherapies are only effective for a subset of patients and cancer types2. To this end, a monoclonal antibody targeting the immune checkpoint protein Lymphocyte Activation Gene-3 (LAG3) was recently approved by the FDA as a combination therapy with anti-PD1 for the treatment of advanced melanoma3. Unlike the PD1 and CTLA4 systems, where a large body of research provided detailed insight into their inhibitory mechanisms, we have limited molecular-level understanding of LAG3-mediated immunosuppression4.
LAG3 is expressed on a wide range of immune cell subsets and is best characterized for its role as a negative regulator of activated CD4+ and CD8+ T cells5,6,7,8. Interactions between LAG3 and its primary ligands MHC class II9,10 and Fibrinogen-like protein 1 (FGL1)11 inhibit antigen-stimulated T cell activation, although the relative importance of each ligand is unclear. LAG3 mutations that disrupt FGL1 binding had no effect in mouse models of autoimmunity and cancer, whereas mutations that disrupted MHC-II binding nullified the suppressive activity of LAG3 in both systems12. These data suggest that MHC-II interactions more broadly affect LAG3 function, and that FGL1-mediated effects may be context-dependent. LAG3 may also exert ligand-independent functions by disrupting interactions between the TCR signaling kinase Lck and the intracellular domains of CD4 and CD813.
Structural studies of LAG3 ectodomains, either alone or in complex with inhibitory antibodies, have provided insight into the molecular architecture of the LAG3 protein14,15,16. These structures revealed that the four immunoglobulin-like domains of LAG3 (D1-4) adopt an extended conformation and homodimerize through D2-D2 interactions14,16. LAG3 homodimerization has been shown to be functionally important, as mutations and antibodies that disrupt the D2-D2 interface inhibit LAG3-mediated immunosuppression16,17. Inhibitory antibodies have been found to recognize epitopes in all four LAG3 domains, and the clinically approved antibody relatlimab has been shown to bind a linear epitope in a non-conserved “Loop 1” insertion (residues S69 to G93) within D1 of human LAG3 (hLAG3)14,15,16,18,19. An agonist antibody, IMP-761, has also been shown to bind to D1 of human LAG320,21, implying that there are functional or therapeutic differences associated with targeting different domains or epitopes of the LAG3 ECD.
Here, we report the structure of the ligand-binding D1-D2 domains of mouse LAG3 (mLAG3) in complex with the MHC-II molecule I-Ab. Structural analysis reveals that the MHC-II-binding sites of LAG3 and CD4 overlap such that LAG3 competitively inhibits MHC-II-CD4 interactions, LAG3 dimerization is predicted to enforce suboptimal spacing of MHC-II-TCR complexes, and LAG3 antagonist antibodies may function through MHC-II-blocking and non-blocking mechanisms. These findings address several longstanding questions about LAG3 molecular function and provide a blueprint for the structure-guided development of targeted immunotherapies.
Results
Crystal structure of LAG3 bound to MHC-II
We used a single-chain fusion approach to stabilize low affinity (∼5–10 µM KD) LAG3-MHC-II interactions14,22 for structure determination. In this construct, the ligand binding D1-D2 region of mouse LAG3 (mLAG3, Fig. 1a) was fused to the N-terminus of the I-Ab beta chain using a flexible (GGGGS)4 linker (Fig. 1b). Class II-associated invariant chain peptide (CLIP) was also fused to the N-terminus of the MHC-II alpha chain as previously described23, and the two constructs were co-expressed to reconstitute the LAG3-peptide-MHC-II complex. The single-chain protein, henceforth referred to as scLAG3-MHC-II, eluted as a single monodisperse peak following purification on a size exclusion column (Fig. 1c).
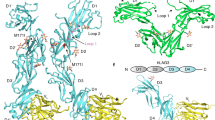
a Cartoon depicting LAG3 (cyan) in the context of an MHC-II (violet/pink) bound to the TCR-CD3 complex (grey). D1 of LAG3 binds to MHC-II, and D2 mediates LAG3 dimerization. b Schematic of the scLAG3-MHC-II construct used for structure determination. c A size-exclusion chromatogram from purification of scLAG3-MHC-II indicates that the construct elutes as a monodisperse peak. The inset SDS-PAGE gel shows the purified LAG3D12-MHC-IIα and CLIP-MHC-II β single-chain fusion proteins. This data is representative from one of at least three independent scLAG3-MHC-II purifications. Source data are provided as a Source Data file. d Crystal structure of the scLAG3-MHC-II construct consisting of the LAG3 D1 and D2 domains (teal) bound to I-Ab (violet/salmon) loaded with CLIP (yellow).
The 3.84 Å-resolution crystal structure of scLAG3-MHC-II reveals that D1 of LAG3 engages a surface spanning the membrane-proximal α2 and β2 subdomains of I-Ab (Fig. 1d, Supplementary Fig. 1, Supplementary Table 1). The structure was solved by molecular replacement with 2.1 Å-resolution models for both mLAG3 and I-Ab, which guided our analysis of putative contact residues in the medium-resolution scLAG3-MHC-II structure14,23. LAG3 interactions with α2 are largely mediated by inter-strand loops at the tip of mLAG3 D1, whereas its interactions with β2 are mediated by residues along the edge of the D1 β-sheet (Fig. 2a). A cluster of polar residues in two D1 loops, N54, R57, R121, Q124 and R125, are located opposite a charge-complementary surface of MHC-II α2 containing S152, D189, E198 and E199 (Fig. 2b, Supplementary Fig. 1a). The mLAG3-β2 interface contains a mixture of polar and hydrophobic amino acids and is centered on the flexible Loop 2 region (G103 to P111) of D1 (Fig. 2a). Loop 2 lacks secondary structure in unliganded structures of LAG3, and MHC-II engagement rearranges this region into a more compact β-turn that packs against a ridge of hydrophobic residues in α2 (F119) and β2 (I176, L186, M188) (Fig. 2a)14,16.
a LAG3-MHC-II binding interfaces on the structure of scLAG3-MHC-II are indicated with black boxes. The Loop 1 insertion in LAG3 D1 was disordered in the structure and is indicated by a dotted line, and the residues of Loop 2 are shown as blue spheres. The MHC-II α and β chains are colored salmon and violet, respectively, and mLAG3 is colored cyan. The left zoom panel depicts Loop 2 interactions with the α2 and β2 subdomains of I-Ab. LAG3 residues associated with loss-of-binding mutations are labeled in red, with asterisks designating mutations that were tested in this study. The Cα positions of G103R and P111A mutations are marked with red spheres. The right zoom panel depicts the charge-complementary surface between strand-connecting loops of LAG3 and the α2 domain of I-Ab. b SPR isotherms comparing the binding of mLAG3-Fc and mLAG3P111A-Fc to immobilized I-Ab. The mLAG3 and mLAG3P111A proteins were each produced as N-terminal fusions to human IgG1 Fc. c SPR isotherms comparing the binding of mLAG3 and mLAG3G103R or mLAG3R125A to immobilized I-Ab, and the binding of mLAG3 to I-Ab containing E198A and E199A mutations. d Size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) was used to measure the solution molecular weight of scLAG3-MHC-II, which corresponds to a 2:2 complex. The molecular weight indicated is an average of three independent experiments. e A 2:2 complex of mLAG3D12 (cyan and teal) bound to I-Ab (violet/salmon), generated through crystallographic symmetry, is depicted as it may occur between two cells. Engagement of MHC-II by LAG3 dimers separates the MHC-II molecules by ∼105 Å. APC: antigen-presenting cell. f A 2:2 model of LAG3-MHC-II was generated by superimposing two copies of scLAG3-MHC-II onto the structure of dimeric mouse LAG3 (PDB 8DGG)16.LAG3 is in green and MHC-II is in violet/salmon. Source data are provided as a Source Data file.
The established Loop 2 mutations, G103R12, R106A24, and P111A10 disrupt LAG3 binding to MHC-II-expressing cells and can be explained by the scLAG3-MHC-II structure. The G103 residue is buried at the interface such that steric clashes would occur upon the introduction of a bulky arginine side chain, and replacement of the charged side-chain of R106 with alanine likely perturbs polar interactions with opposing MHC-IIβ2 residues (Fig. 2a). The effect of the P111A mutation is less obvious given that P111 is located outside the LAG3-MHC-II interface (Fig. 2a). However, Alphafold modeling25,26 suggests that substitution of this rigid proline with alanine creates a helix in Loop 2 that distorts the positions of MHC-II-binding residues (Supplementary Fig. 2b). SPR measurements support the above predictions and show a loss of binding between mLAG3P111A or mLAG3G103R and I-Ab (Fig. 2b, c). We also performed an SPR experiment to investigate the contributions of residues in the charge-complementary mLAG3-MHC-IIα2 interface. We determined that an R125A mutation in mLAG3, or an E198A/E199A double mutation in I-Ab, each diminish LAG3-MHC-II binding (Fig. 2c). These findings agree with published data showing that an R121A mutation (analogous to R103 in the mature hLAG3 ECD) attenuates binding between LAG3-expressing and MHC-II-expressing cells24.
Loop1 of LAG3 has been implicated in MHC-II binding18,24, but this region is disordered in the complex structure and faces away from MHC-II (Fig. 2a). Through our efforts to study LAG3-MHC-II interactions, we also determined a 4.66 Å-resolution structure of a single-chain Loop 1 deletion mLAG3 construct bound to MHC-II (scLAG3ΔL1-MHC-II). We found that truncation of Loop 1 did not affect the ability of mLAG3 to co-crystallize with MHC-II, and the positions of the mLAG3 ΔL1 and I-Ab domains were unchanged compared to scLAG3-MHC-II (Supplementary Fig. 2c, Supplementary Table 1), suggesting that Loop1 is not essential for LAG3-MHC-II complex formation. To test this hypothesis, we compared the binding I-Ab to mLAG3 and a Loop 1 deletion construct (mLAG3ΔL1) using surface plasmon resonance (SPR), and we evaluated the ability of mLAG3ΔL1 to inhibit mLAG3 binding to MHC-II-expressing cells. In the SPR study, we found that mLAG3 binds to MHC-II with 2-fold higher affinity (7.4 µM KD) than mLAG3ΔL1 (14.8 µM KD) (Supplementary Fig. 2d). However, in the cellular assay, unlabeled mLAG3ΔL1 strongly inhibited the binding of fluorescently-labeled mLAG3 to MHC-II+ Raji cells (Supplementary Fig. 2e). Taken together, these data suggest that Loop 1 plays a minor role in LAG3-MHC-II engagement. Loop 1 is positioned outside of the LAG3-MHC-II interface (Fig. 2a), and we posit that it indirectly promotes MHC-II binding by stabilizing the “kinked” MHC-II-bound conformation of D1-D2 by sterically restricting the motion of the opposing D1 in LAG3 homodimers (Supplementary Fig. 3a)14,16.
Modeling dimeric LAG3 interactions with MHC-II
The scLAG3-MHC-II construct provides a unique tool for determining LAG3-MHC-II stoichiometry, as the single-chain format prevents the dissociation of low affinity LAG3-MHC-II complexes in solution. Analysis of scLAG3-MHC-II by size exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) indicates that its molecular weight corresponds to a 2:2 complex, which is consistent with the binding of two monomeric MHC-II molecules to one LAG3 homodimer (Fig. 2d). Although scLAG3-MHC-II crystallized as a 1:1 complex, a dimer formed between mLAG3 protomers from adjacent unit cells, enabling us to model the 2:2 interaction. The dimer interface is located within the same hydrophobic surface as those of published LAG3 structures14,15,16, except that the orientation of D2-D2 is skewed by ∼ 110° relative to D2-D2 in unliganded mLAG3 (Supplementary Fig. 3b). By contrast, the D1-D2 interdomain angles are nearly identical between the MHC-II-bound and unliganded structures (Supplementary Fig. 3c). In the crystal lattice, the mLAG3 dimer forms a V-shaped wedge between MHC-II proteins that separates their peptide-binding domains by >100 Å (Fig. 2d). Previous studies of T cell signaling have shown that increased separation between MHC-II molecules, starting at distances of 60 Å, is associated with diminished TCR activation27. Thus, LAG3 could potentially inhibit antigen-mediated T cell signaling by inducing suboptimal TCR-MHC-II geometry.
The mLAG3 ECD has crystallized in multiple dimer conformations14,16, and we cannot rule out the possibility that the presence of a linker perturbs scLAG3-MHC-II dimerization. To consider alternative possibilities, we modeled a 2:2 interaction based on the conserved D2-D2 orientation observed in three structures of the full-length LAG3 ECD (Fig. 2e, Supplementary Fig. 3a), including a cryoEM structure in which LAG3 is not constrained by crystal packing14,15,16. In this 2:2 model, the MHC-II proteins are oriented differently with respect to one another, but inter-MHC-II distance is still approximately 100 Å (Fig. 2f). This analysis indicates that LAG3-enforced separation of MHC-II proteins is maintained regardless of which dimer conformation is adopted on the cell surface.
LAG3 and CD4 bind to overlapping interfaces on MHC-II
There has been conflicting evidence regarding the ability of LAG3 and CD4 to compete for MHC-II interactions5,8,12,28. LAG3 was initially classified as a CD4 homolog based on structural and genomic similarity29, and LAG3-Fc fusion proteins have been shown to inhibit CD4/MHC-II-mediated cellular aggregation9. On the other hand, the D1 domains of LAG3 and CD4 are poorly conserved (∼20% amino acid identity), and recent cell staining data indicated that LAG3 and CD4 can simultaneously bind to MHC-II molecules on the cell surface10. Comparison of scLAG3-MHC-II and CD4-MHC-II30 structures demonstrates that LAG3 and CD4 bind to overlapping interfaces on the MHC-II protein (Fig. 3a). Superimposing the MHC-II components of each structure shows that LAG3 and CD4 engage MHC-II via similar binding modes, with D1-D2 of mLAG3 approaching MHC-II at a shallower angle than D1-D2 of CD4 (Fig. 3a). LAG3 covers nearly double the amount of surface area buried at the CD4 interface (1012 Å2 vs. 512 Å2) and occludes the CD4 binding sites on both MHC-II α2 and β2 (Fig. 3b). This larger surface area is consistent with the higher MHC-II-binding affinity of LAG3 (KD, 2 to 14 µM)14,22 compared to CD4 (KD > 400 µM)31,32.
a Alignment of scLAG3-MHC-II and CD4-MHC-II (PDB 1JL4)30 structures indicates that LAG3 (cyan) and CD4 (yellow) share a binding site on MHC-II. The MHC-II α and β chains are shown in light and dark grey, respectively. The approach angle of CD4 D1-D2 is tilted by ∼25° compared to LAG3 D1-D2. b The regions surfaces bound by LAG3 (left, cyan) and CD4 (right, yellow) are painted onto the surface of MHC-II molecules. The LAG3-MHC-II interface buries nearly double the amount of surface area (1012 Å2) of that buried by the CD4-MHC-II interface (512 Å2). c Raji APC cells were stained with fluorescently labeled LAG3 tetramers (20 nM) in the presence or absence of 1 µM CD4HA competitor. The control represents Raji APC cells incubated with streptavidin 647 only. Data are shown as normalized median MFI ± SEM based on triplicate wells from two independent experiments (n = 2). Open and closed circles represent data points from each independent experiment. Statistics were obtained using a one-way ANOVA multiple comparison in Prism 9 (Version 9.5.1). d BLI sensograms depict the binding of immobilized CD4HA to HLA-DR4 molecules loaded with an HIV peptide (DR4HIV)42 in the presence of hLAG3-Fc protein (5 μM) or Fc control protein (5 μM). The plot depicts a representative sensogram from two independent experiments. Source data are provided as a Source Data file.
Analysis of the scLAG3-MHC-II structure reveals that both sides of the interface are conserved. On the LAG3 side, the MHC-II-binding interface is conserved in mouse and human LAG3 despite relatively low overall sequence identity (67%) between their D1 domains (Supplementary Fig. 4a). By contrast, comparison of CD4-MHC-II (PDB ID: 1JL4)30 and scLAG3-MHC-II structures revealed that there is minimal conservation among the putative MHC-II-binding residues in CD4 and LAG3 (Supplementary Fig. 4b). On the MHC-II side, the LAG3-binding interface is conserved in the mouse I-Ab and I-Ek MHC-II molecules and in the human DR, DP, and DQ MHC-II molecules (Supplementary Fig. 5), indicating that the LAG3-MHC-II binding mode is similar between mouse and human MHC-II subtypes.
We directly tested the ability of LAG3 to compete for CD4-MHC-II interactions using biophysical and cellular approaches. Because low affinity CD4-MHC-II interactions are difficult to detect in most formats31, we used a high-affinity variant of human CD4 containing the mutations Q65Y, T70W, S85R and D88R (CD4HA) for our competition assays32. We first evaluated the ability of CD4HA to compete with a hLAG3 (hLAG3) for binding the human MHC-II-expressing Raji cell line. We found that pre-treatment of the cells with 1 µM of unlabeled CD4HA led to a ∼ 50% reduction in the binding of fluorescently labeled hLAG3 tetramers (Fig. 3c). In a second assay, we tested whether a human LAG3-Fc fusion protein (hLAG3-Fc) inhibits the binding of CD4HA and MHC-II using biolayer interferometry (BLI). We found that CD4HA detectably bound to the human MHC-II molecule HLA-DR4HIV, and that addition of hLAG3-Fc inhibited CD4HA interactions with HLA-DR4 (Fig. 3d). These structural and functional data suggest that competition for CD4-MHC-II binding is a potential mechanism for LAG3-mediated T cell suppression.
Analysis of epitopes bound by MHC-II- and LAG3-targeting antibodies
We analyzed the epitopes of several MHC-II- and LAG3-targeting antibodies to investigate their ability to modulate LAG3-MHC-II interactions. Inspection of the structure of the antibody 44H10 bound to HLA-DR1 (PDB ID: 8EUQ)33,34 suggests that 44H10 blocks MHC-II interactions with both LAG3 and CD4 (Fig. 4a). To test this hypothesis, we evaluated the ability of 44H10 to disrupt hLAG3 tetramer binding to APC cells. We found that the addition of 44H10 almost completely inhibited the binding hLAG3 to APC cells (Fig. 4b), and that LAG3 did not stain HEK293T cells that do not express MHC-II (Supplementary Fig. 6a), which is consistent with our structural prediction. In parallel, we used BLI to determine that 44H10 inhibits interactions of both hLAG3-Fc and CD4HA with HLA-DR4 (Figs. 4c, d). To our knowledge, this is the first demonstration of a dual inhibitor of MHC-II interactions with CD4 and LAG3 and identifies 44H10 an intriguing tool for immunological studies.
a Structural alignments showing that the antibody 44H10 (blue)34 occupies the MHC-II interface bound by LAG3 (cyan, left) and CD4 (yellow, right). b Raji APC cells were stained with a fluorescently labeled LAG3 tetramers (20 nM) in the presence or absence of a 44H10 single-chain variable fragment (scFv)-Fc fusion protein. The control represents Raji APC cells incubated with streptavidin-647 alone. Data shown as normalized median MFI ± SEM based on triplicate wells from one representative experiment. The experiment was independently repeated two times. Statistics were obtained using a one-way ANOVA multiple comparison in Prism 9 (Version 9.5.1). c BLI sensograms depict the binding of immobilized human LAG3 protein to HLA-DR4HIV in the presence of increasing concentrations of 44H10 scFv-Fc fusion protein. d BLI sensograms depict the binding of immobilized CD4HA protein to HLA-DR4HIV in the presence of increasing concentrations of 44H10 scFv-Fc fusion protein. The plots in c and d each depict a representative sensogram from two independent experiments. Source data are provided as a Source Data file.
The antibodies relatlimab18, favezelimab35, 1501136, M8-4-616, and ieramilimab37 antagonize LAG3 function and have each been reported to disrupt LAG3-MHC-II binding. We mapped the epitopes of each antibody in the context of the LAG3-MHC-II structure to gain insight into their inhibition mechanisms (Fig. 5). Surprisingly, the LAG3 peptide recognized by the therapeutic antibody relatlimab18, G83-G93, is not predicted to contact MHC-II. Docking the peptide-bound structure of relatlimab onto its predicted position in the LAG3-MHC-II complex places it adjacent to both MHC-II and the plasma membrane (Fig. 5a). Despite its non-interface epitope, relatlimab has been shown to inhibit LAG3 binding to MHC-II+ cells. This observed inhibition may occur because flexible motion in Loop 1 introduces clashes between relatlimab and MHC-II or the membrane plasma membrane, either of which could indirectly hinder LAG3 access to MHC-II.
a Structural modeling predicts that relatlimab (green) binds an epitope (red spheres) distal to the LAG3-MHC-II interface. The disordered Loop 1 region was derived from an Alphafold25,53 model of hLAG3 D1, and the epitope-bound structure of relatlimab (PDB 7UM3)18 was aligned to the corresponding Loop 1 residues to place relatlimab in the context of LAG3 and MHC-II. b Aligning favezelimab-LAG3 and scLAG3-MHC-II structures predicts that favezelimab binding is incompatible with LAG3-MHC-II interactions due to steric clashes with MHC-II β2 (red). c Mapping the epitopes of 15011 (yellow spheres) and M8-4-6 (orange spheres) onto the structure of scLAG3-MHC-II reveals that the antibodies engage LAG3 residues in or adjacent to the MHC-II binding interface. d Comparison of the MHC-II-binding interface or mLAG3 (top) and the ieramilimab-binding interface of hLAG3 (bottom). The D1-D2 domains of mLAG3 from the scLAG3-MHC-II structure and hLAG3 (PDB: 7TZG) are depicted in surface representation, and the residues bound by MHC-II or ieramilimab. e Residues bound by the mouse LAG3-specific antibody C9B7W (red spheres) are not predicted to disrupt LAG3-MHC-II interactions. To emphasize the position of these residues at the dimer interface, they are shown on scLAG3-MHC-II structures that were modeled as a 2:2 complex. f Model of the D4-specific scFv F7 bound to LAG3-MHC-II. To generate the model, D1 from the human LAG3-F7 complex structure (PDB ID:7TZG) was docked onto D1 from the scLAG3-MHC-II structure. F7 binds a membrane-proximal site in LAG3 D4 and is not predicted to disrupt LAG3-MHC-II interactions.
Aligning the LAG3-favezelimab15 and scLAG3-MHC-II structures reveals that favezelimab is incompatible with LAG3-MHC-II binding due to clashes between the MHC-IIβ2 domain and the favezelimab light chain (Fig. 5b). Disruption of LAG3-MHC-II by the human/mouse cross-reactive antibody, 15011, is explained by 15011 engagement of Loop 2 residues present at the LAG3-MHC-II interface (Fig. 5c). Similarly, M8-4-6 binds to several mouse LAG3 residues T146, R148, N151, and R152 in the D1 β-sheet that engages MHC-II β2 (Fig. 5c)16. Comparison of the MHC-II-binding interface of mLAG3 and the ieramilimab-binding interface of hLAG3 indicates that the ieramilimab epitope overlaps with predicted MHC-II-binding residues R57, Q128, and R129 in hLAG3 (Fig. 5d). On the other hand, the mouse-specific LAG3 antagonist antibody, C9B7W, binds the D2-D2 dimer interface (Fig. 5e)16, and the human-specific antagonist, F7, binds to a membrane-proximal epitope in D4 (Fig. 5f)14 such that neither antibody inhibits LAG3-MHC-II interactions14,38.
Comparison of LAG3-MHC-II and LAG3-FGL1 binding interfaces
In addition to MHC-II, the secreted ligand FGL1 stimulates LAG3-mediated immunosuppression, and it is currently unclear whether these proteins bind distinct or overlapping interfaces11. Although no LAG3-FGL1 complex structure is available, LAG3 residues that influence FGL1 binding have been identified through mutagenesis studies12,14. We mapped the putative FGL1-binding residues K27, R109, H113, V116, and M16712,14 onto the structure of scLAG3-MHC-II to compare the LAG3-MHC-II and LAG3-FGL1 binding interfaces. The FGL1-binding residues are located along a large surface of LAG3 D1 that is adjacent to the MHC-II binding region (Fig. 6a). To test whether FGL1 inhibits LAG3-MHC-II binding, we stained Raji cells with fluorescently labeled hLAG3 in the presence or absence of recombinant human FGL1. We found that high concentrations of an FGL1-Fc fusion protein strongly inhibited the binding of fluorescently labeled hLAG3 tetramers to the cells (Fig. 6b). These data indicate that MHC-II and FGL1 have partially overlapping binding sites on LAG3, which is consistent with preclinical data showing that the antibodies relatlimab and 15011 can block LAG3 interactions with both FGL1 and MHC-II14,18.
a Mutations known to affect LAG3 binding to FGL1 (blue) were mapped onto the structure of scLAG3-MHC-II. LAG3 is shown in gray surface representation, and a boundary surrounding the putative FGL1-binding residues is shown as a dotted line. b Raji cells were stained with fluorescently labeled LAG3 tetramers (50 nM) in the presence of the FGL1 fibrinogen-like domain fused to the C-terminus of human IgG1 Fc (FGL1FD-Fc) or IgG1 Fc. Background-subtracted median MFI was normalized to Fc control. Data shown as normalized median MFI ± SEM based on triplicate wells from one representative experiment. The negative control shows Raji cells stained with Streptavidin 647 (SA-647) alone. Statistics were obtained using a one-way ANOVA multiple comparison in Prism 9 (Version 9.5.1). A representative data plot from one of two independent experiments is shown. Source data are provided as a Source Data file.
Discussion
Our structure-function study provides important insight into several aspects of LAG3 molecular biology and raises new questions about the role of ligand engagement in LAG3-mediated immunosuppression. The finding that LAG3 occupies the MHC-II-CD4 binding site resolves discrepancies about the ability of LAG3 to compete with CD4 for MHC-II interactions. As the LAG3 ICD was previously shown to disrupt intracellular interactions between Lck and CD413, our data suggest that LAG3 also utilizes a complementary, steric competition mechanism to disrupt extracellular CD4 signaling. Characterization of the MHC-II-specific antibody 44H1034 revealed that it blocks MHC-II interactions with both CD4 and LAG3. Given that the LAG3 binding site on MHC-II extends outside of the CD4-MHC-II interface, it is conceivable that antibodies targeting precise regions of MHC-II could selectively inhibit LAG3-MHC-II interactions for biomedical applications. It is notable that the LAG3-binding interface is distant from the peptide-binding groove, as it has been claimed that LAG3 binding is dependent on peptide-MHC-II stability10. We speculate that stabilizing peptides prolong the half-life of MHC-II on the cell surface to allow for a larger number of LAG3 interactions.
LAG3 dimerization is important for multiple aspects of LAG3 function16,17, and our SEC-MALS analysis suggested that LAG3-MHC-II form a 2:2 complex. One probable role of LAG3 dimerization is to strengthen LAG3-MHC-II binding through avidity enhancement. However, modeling the 2:2 LAG3:MHC-II complex predicts that LAG3 dimers may also act as wedge that perturbs TCR signaling by altering the surface geometry of MHC-II molecules27,39. Considering that TCRs, MHC-II, CD4 and other coreceptors form a spatially sensitive network of interactions39,40, it will be interesting to observe how LAG3 functions in the more complex environment of the immune synapse.
Mapping epitopes onto the scLAG3-MHC-II structure indicates that antibodies antagonize LAG3 through MHC-II-dependent and independent mechanisms, and that the LAG3 inhibitors favezelimab35, 1501136, M8-4-616, and ieramilimab37 engage the MHC-II-binding interface of LAG3. Additionally, we found that the therapeutic antibody relatlimab3 is likely to disrupt LAG3-MHC-II binding indirectly, rather than through direct steric inhibition. This antibody mapping data is consistent with mutational studies showing that disruption of the LAG3-MHC-II binding site12, LAG3 ICD41, and the LAG3 dimer interface16 each inhibit LAG3 activity in various contexts. This suggests that LAG3 either employs multiple immunosuppression mechanisms, or that coordinated action between various LAG3 domains is required for optimal function. Future studies comparing the relative importance of functional interactions such as dimerization, MHC-II-engagement, and association with TCR-CD3 complexes will be important for the optimal design of LAG3-based immunotherapies.
Methods
Protein expression and purification
For the single-chain mLAG3-MHC-II construct (scLAG3-MHC-II), the LAG3D12-MHC-IIα component was generated by fusing domains 1-2 of mouse LAG3 (mLAG3, amino acids 23–254) to the N-terminus of the ECD of the I-Ab alpha chain using a (GGGGS)4 linker. The CLIP-MHC-IIβ component was generated by fusing CLIP (aa 86–100: PVSKMRMATPLLMQA) to the N-terminus of the ECD of the I-Ab β chain using a thrombin-cleavable linker23. To enforce heterodimerization, the α and β chain components of scLAG3-MHC-II were fused to 3C-cleavable acid/base zippers from C-jun or C-fos transcription factors, respectively42. LAG3D12-MHC-IIα includes a C-terminal biotin-acceptor peptide tag (BAP-tag: GLNDIFEAQKIEW) followed by a 6xHis tag. To purify scLAG3-MHC-II, the LAG3D12-MHC-IIα and CLIP-MHC-IIβ constructs were individually cloned into the Baculovirus expression vector pAcGp67A and insect cells (T. Ni cells, Expression Systems, 94-002 F) were co-infected with recombinant Baculovirus from the two constructs (see below for purification information). The Loop 1 deletion construct, scLAG3ΔL1-MHC-II was designed the same way as scLAG3-MHC-II except that it was truncated to remove residues from Q71 to P92 of mLAG3 D1(Supplementary Table 2).
The HLA-DR4HIV construct was generated by co-expressing a fusion of a thrombin-cleavable p24 HIV Gag peptide (aa 164–183: AFSPEVIPMFSALSEGATPQ) to in the N terminus the ECD of the HLA-DR4 β chain with the ECD of HLA-DR4α14. The proteins contain the same acid/base zippers and BAP/6xHis tags as described for scLAG3-MHC-II, except the β-chain portion of HLA-DR4HIV does not contain a 6xHis-tag. The two constructs were cloned into pAcGp67A and insect cells were coinfected with recombinant Baculovirus incorporating the α and β chains described above to generate HLA-DR4HIV proteins.
The 44H10 scFv34 was generated by connecting the VH and VL domains with a (GGGGS)3 linker. The scFv was then fused at its C-terminus to the Fc domain of human IgG1 followed by a 6xHis tag. The CD4HA construct contains the D1 and D2 domains of CD4 modified with four affinity-enhancing mutations32 (Q65Y, T70W, S85R and D88R). CD4HA was cloned into pAcGp67A and contains a C-terminal BAP/6xHis tag.
The mLAG3P111A mutant was also generated with a C-terminal Fc/6xHis tag and was purified as described above for the 44H10 scFv-Fc construct. The mLAG3 mutants mLAG3G103R and mLAG3R125A, as well as mLAG3ΔL1 (residues 71-92 deleted) were cloned into the pAcGp67A vector. Human LAG3 constructs (amino acids 63-312) were cloned into the pAcGp67A vector containing either a C-terminal BAP/6xHis tag or a Fc/6xHis tag. Human FGL1 (amino acids: 63-312) was fused with an N-terminal Fc tag and a C-terminal BAP/6xHis tag. The fragment was subcloned into the pAcGp67A vector.
The ECDs of human RNF43 (amino acids 24–197) and Nectin-2 ECD (amino acids 32-350) were cloned into the pAcGp67A vector with a C-terminal BAP/6xHis tag.
All recombinant proteins above were expressed using Baculovirus by infecting Trichoplusia ni cells (Expression Systems, Cat# 94-002 F) at a density of 2 × 106 cells/mL followed by 48–72 h of incubation at 27 °C. For proteins subjected to N-glycan removal, Kifunensine (Toronto Research Chemicals, Cat# TRC-K450000-50 MG), was added to a final concentration of 5 µM at the time of infection. The proteins were retrieved from culture supernatant by nickel affinity purification using nickel Nickel-NTA resin (Qiagen, Cat# 30250). Size-exclusion chromatography (SEC) was performed on a Superdex 200 Increase 10/300 GL column by FPLC (Äkta Pure, GE Healthcare).
All LAG3 proteins and the CD4 protein were purified in PBS buffer, pH 7.2. The scLAG3-MHC-II protein complex was stored in HBS200 buffer (20 mM HEPES and 200 mM NaCl, pH 7.2). The HLA-DR4HIV and the 44H10 scFv proteins were stored in HBS buffer (20 mM HEPES and 150 mM NaCl, pH 7.2). The FGL1FD-Fc protein was stored in HBS buffer supplemented with 1 mM CaCl2. Biotin labeled proteins were site-specifically biotinylated at the BAP-tag by using BirA ligase kit (Avidity, LLC, Cat# BIRA-500) followed by SEC purification to remove excess biotin reagent. RNF43, Nectin-2, and Endoglycosidase F1 were stored in HBS buffer.
Streptomyces avidinii Streptavidin (SA, aa37-157) was cloned into pET21a (Novagen, Cat# 69740) with the following modification: a C-terminal GGSG linker followed by a Cysteine for Maleimide labeling, a Proline, and a stop codon. The construct was transformed into BL21-CodonPlus (DE3)-RIL cells (Agilent, Cat# 230245) and induced with 1 mM IPTG (RPI, Cat# I56000-25.0) once cultures reached an optical density at 600 nm (OD600) of 0.6 to 0.8. Cells were pelleted following 4 h of induction at 37 °C, and inclusion bodies were purified from cell lysates43. Cells were resuspended in lysis buffer (50 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0, 0.01 % sodium azide, 1 mM DTT, 200 mM sodium chloride, 1.0% sodium deoxycholate, and 1.0% Triton X-100) and then the insoluble fraction was pelleted by centrifugation. The pellet was washed twice with 50 mM Tris–HCl pH 8.0, 1 mM EDTA pH 8.0, 0.01 % sodium azide, 1 mM DTT, 100 mM sodium chloride, and 0.5 % Triton X-100, and then once with the same buffer without Triton X-100 to isolate purified inclusion bodies. Inclusion bodies were dissolved in 8 M Guanidine Hydrochloride (pH 1.5) with 500 uM TCEP and refolded by rapid dilution into PBS containing 0.2 mM PMSF and 0.500 mM TCEP. The refolded SA protein was then labeled with Alexa Fluor 647 through maleimide coupling with the Sulfo-C2-Maleimide kit (Invitrogen, Cat# A20347) using manufacturer’s instructions to generate SA-647, and excess dye was removed by SEC purification.
Flow cytometry with mammalian cells
MHC-II positive human APC Raji cells (Promega, Cat# JA1111) and HEK293T cells (a gift from Dr. Eric Lau, original commercial source ATCC, Cat# CRL-3216) were cultured at 37 °C, with a humidified atmosphere of 5% CO2. Cells were cultured in growth media consisting of DMEM with high glucose, sodium pyruvate and L-glutamine (Gibco, Cat# 11995065), supplemented with 10% FBS (Peak serum, Cat# PS-FB1), 0.1% Penicillin-Streptomycin (Gibco, Cat# 15-140-122). For MHC-II-LAG3 blocking assays, 5 × 104 Raji APC cells were incubated with 50 µl growth media with 1 µM of 44H10 scFv targeting MHC-II in 96-well (V-well, Cat# Corning 3894) plates. Separately, biotinylated LAG3 protein was incubated with streptavidin 647 (SA-647) (produced in-house and described in protein purification) at a molar ratio of 4.5:1 of protein to streptavidin to form protein-streptavidin tetramers. After a 15 min cell incubation, 20 nM of LAG3-tetramers (80 nM effective protein concentration) were incubated with cells that contained 44H10 scFv, or growth media only, on a plate shaker at 500 rpm for 1 h at 4 °C. Cells were then washed three times with DPBS (Corning, Cat# 21-031-CV). After washes, the cells were resuspended in DPBS and analyzed with an Accuri C6 (BD, Bioscience) flow cytometer. The FGL1-LAG3 competition assay to Raji MHC-II cells were performed in the same way as the scFv blocking but with a 30 min cell incubation of FGL1FD-Fc of varying concentrations (1-10 µM) before incubation of 50 nM LAG3-tetramers to 1 × 105 cells per replicate well. Comparison of unspecific binding of biotinylated LAG3-tetramers to MHC-II-negative HEK293T cells, including 44H10, was performed as described above, but utilizing a 50 nM concentration of LAG3-tetramers. For MHC-II-scFv (44H10) binding to Raji MHC-II cells, an anti-Fc 647 antibody (Southern Biotech, Cat# 2048-31) (Supplementary Table 4) was utilized by pre-incubating 44H10-Fc with anti-Fc 647 (1:50 final concentration) on rotation for 30 min in +4 °C. The pre-mixed scFv-Fc-647 was then mixed at 100 nM with 5 × 104 Raji APC cells per well in 50 µl growth media in a 96-well (V-well, Cat# birCorning 3894) plate shaking at 500 rpm for 1 h in +4 °C. Cells resuspended in DPBS were analyzed after 3 washes in DPBS (Corning, Cat# 21-031-CV) using an Accuri C6 (BD Bioscience) flow cytometer. For CD4-LAG3 blocking assays, 5 × 104 Raji APC cells were incubated with or without 1 µM CD4HA protein. Separately, biotinylated LAG3 protein was incubated with streptavidin 647 (SA-647) at a ratio of 4.5:1 of protein to streptavidin to form protein-streptavidin tetramers. After a 15 min incubation, 20 nM of LAG3-tetramer was incubated with cells that contained CD4HA protein or growth media for 1 h at 4 °C at 500 rpm. Cells resuspended in DPBS were analyzed after 3 washes in DPBS (Corning, Cat# 21-031-CV). Cells were analyzed with an Accuri C6 (BD Bioscience) flow cytometer. All flow data was analyzed using FlowJo (version 10.6.1).
SPR binding studies
Surface plasmon resonance experiments were performed on a BIAcore T100 instrument (GE Healthcare). For analyzing the binding of mLAG3-Fc and mLAG3P111A-Fc to I-Ab, approximately 400 resonance units (RU) of biotinylated I-Ab protein was immobilized on a streptavidin-coated sensor chip (Cytiva, Cat# BR100531) and increasing concentrations of mLAG3-Fc protein in HBS-BSA-P buffer (25 mM HEPES, 200 mM NaCl, 0.1% BSA and 0.005% surfactant P20) was injected over the chip. The chip was regenerated after each injection with 30-second washes of regeneration buffer (10% ethylene glycol, 500 nM NaCl). The binding responses were reference-subtracted from a flow cell containing a similar immobilization level of a non-related negative control protein (mouse RNF43, described in protein purification section above). For analyzing the binding of mLAG3 or mLAG3R125A or mLAG3G103R or mLAG3R125A or mLAG3ΔL1to I-Ab and mLAG3 binding to I-Ab(E198/199A), approximately 400 RU of biotinylated I-Ab protein or I-Ab(E198/199A) mutant was immobilized on a SA chip. mLAG3 or mLAG3 mutants were injected at gradient concentrations. The signal for each flow cell was reference-subtracted from a flow cell containing a similar immobilization level of control protein (human Nectin-2, in-house prepared, described in the protein purification). The maximum response (RU max) for each experiment was plotted as a function of concentration using Prism 9 (GraphPad, Version 9.5.1). Steady-state binding parameters were determined based on a 1:1 Langmuir model using the BIAcore T100 evaluation software.
Biolayer interferometry (BLI) based competition assays
BLI assay was performed using the Gator™ Label-Free Bioanalysis instrument (Gator Bio, Palo Alto, CA, USA) and Gator One 2.7 (software version 2.15.5.1221). HBS buffer consisting of 0.1% BSA, 0.005% P20, was used in the BLI experiments shown in Fig. 4c, d. PBS buffer with similar supplements was used in the BLI experiments shown in Fig. 3d. Streptavidin (SA) sensor probes (Gator Bio, USA, Cat# #160002) were used for the sample measurements. The BLI-based competition of hLAG3-Fc protein and CD4HA for binding to DR4HIV, the CD4HA-coated probes were incubated with 2.5 µM HLA-DR4HIV in the absence and presence of either 5 µM hLAG3-Fc or 5 µM Fc control, with hLAG3-Fc, Fc and buffer as reference wells. The BLI-based competition assay using 44H10 scFv and LAG3 or CD4HA for binding to MHC-II was performed by loading SA probes with biotinylated hLAG3 ECD or CD4HA. The binding shift (nm) was then detected by incubating the probes with a fixed concentration of HLA-DR4HIV (5 µM) in the presence or absence of 44H10 scFv (2.5 µM), with 44H10 scFv and buffer alone as references.
Multi-angle light scattering (MALS)
For SEC-MALS analysis, 10 µL of scLAG3-MHC-II (2 mg/mL) was subjected to size-exclusion chromatography using a Superdex 200 Increase 5/150 GL column followed by UV, multi-angle light scattering, and refractive index detection using a Wyatt Technologies MALS configuration (Wyatt Technologies, Santa Barbara, CA). Data analysis was performed using Astra software package (version 7.3) (Wyatt Technology Corporation, Santa Barbara, CA). Prior to the MALS experiment, the construct was pre-treated with Endoglycosidase F1 (in-house prepared, described in the protein purification), 3 C protease (1:200 (w/w), GenScript, Cat# Z03092), and carboxypeptidase A (1:100 (w/w), Sigma, Cat# C9268-500UN) to remove N-linked glycans, acid/base zippers, and C-terminal His tags, respectively.
Proteolytic processing, deglycosylation, and reductive methylation of proteins
Kifunensine-sensitized scLAG3-MHC-II or scLAG3ΔL1-MHC-II complexes were individually purified as described above and then treated overnight at 4 °C with 3 C protease (1:200 (w/w), GenScript, Cat# Z03092), Endoglycosidase F1 (1:200, w/w), bovine carboxypeptidase A (1:100 (w/w)) to remove C-terminal tags, N-linked glycans and disordered residues at the C-terminus of the proteins. Both proteins were further subjected to reductive methylation using borane-dimethylamine (Thermo Scientific Chemicals, Cat# 177310250) and paraformaldehyde (Thermo Scientific Chemicals, Cat# 043368.9 M) at 4 °C degree for overnight. The reactions were quenched at 200 nM Tris, pH 8.0 followed by buffer exchange to HBS200 buffer prior to sparse matrix crystallization screening.
Crystallization scLAG3-MHC-II or scLAG3ΔL1-MHC-II complexes
Purified scLAG3-MHC-II or scLAG3ΔL1-MHC-II complexes were concentrated to 18 mg/mL and 16 mg/ml, respectively. All protein crystals were grown by sitting drop vapor diffusion. Drops containing 0.1 μL of protein were combined with 0.1 μL of mother liquor. Crystals were observed from the PEG/ION screen (Hampton research, Cat# HR2-139) and ProPlex screen (Molecular Dimensions, Cat# MD1-42. Crystals of scLAG3-MHC-II were obtained from mother liquor containing 0.15 M Ammonium sulfate, 0.1 M Tris 8.0 and 15 % w/v PEG 4000. Crystals of scLAG3ΔL1-MHC-II were obtained from mother liquor containing 1% w/v Tryptone, 0.001 M Sodium azide, 0.05 M HEPES sodium pH 7.0, 12% w/v Polyethylene glycol 3350 and 3% w/v Dextran sulfate sodium salt (Hampto research, Cat# HR2-138).
The crystals were cryoprotected in mother liquor containing 25% ethylene glycol prior to plunge-freezing in liquid nitrogen. The diffraction data for scLAG3-MHC-II was collected at Stanford Synchrotron Radiation Lightsource (SSRL), beam line 12-1. The diffraction data for scLAG3ΔL1-MHC-II was collected at National Synchrotron Light Source-II, beamline 19-ID (NYX). Datasets were indexed, integrated, scaled and merged with the XDS package44,45,46.
Structure determination and analysis
The structures were solved using molecular replacement (MR) in Phaser47 using the structures of mouse LAG3 D1-D2 (PDB ID: 7TZE)14 and I-Ab23 (PDB ID: 1MUJ) as models. The models were first subjected to rigid body refinement using the Phenix.refine program48,49, followed by iterative rounds of building in and several rounds of positional and B-factor refinement. Model building and structure refinement were performed in Coot50 and Phenix48,49, respectively. Buried surface areas calculations, and interface residues identification, were performed using PDBe-PISA web server51 followed by manual inspection. Figures were generated in Pymol software (Schrödinger)52. The model of LAG3 with full loop structure were obtained from ColabFold53.
Structural modeling
The model of LAG3 D1 containing the P111A mutation was generated using the Alphafold 3 web server26. To model the 2:2 LAG3:MHC-II complex, two copies scLAG3-MHC-II were aligned to the D2-D2 dimer in the structure of the full-length LAG3 mouse ECD (PDB 8DGG)16. The D3-D4 domains were modeled based on their position in the full-length structure. The relatlimab-LAG3-MHC-II model was generated iteratively. First, Alphafold modeling was used to generate a model of LAG3 D1 containing Loop 1, as this region is disordered in published structures of LAG325,53. The Loop 1-containing D1 model was then superimposed onto the D1 domain from the structure of human LAG3 (PDB 7TZG)14, and the modeled Loop 1 residues were engrafted onto the structure. The epitope-bound structure of relatlimab18 was then docked onto the corresponding residues (G83-G93) in the modeled Loop 1 region. Lastly, the relatlimab-LAG3 complex was aligned to scLAG3-MHC-II to predict the relative positions of relatlimab, LAG3, and MHC-II.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. X-ray crystallography data for the structures of scLAG3-MHC-II and scLAG3ΔL1-MHC-II have been deposited to the Protein Data Bank under the accession codes 9CYM and 9CYL, respectively. All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 11, 3801 (2020).
Seidel, J. A., Otsuka, A. & Kabashima, K. Anti-PD-1 and Anti-CTLA-4 Therapies in Cancer: Mechanisms of Action, Efficacy, and Limitations. Front Oncol. 8, 86 (2018).
Tawbi, H. A. et al. Relatlimab and Nivolumab versus Nivolumab in Untreated Advanced Melanoma. N. Engl. J. Med. 386, 24–34 (2022).
Graydon, C. G., Mohideen, S. & Fowke, K. R. LAG3’s Enigmatic Mechanism of Action. Front Immunol. 11, 615317 (2021).
Aggarwal, V., Workman, C. J. & Vignali, D. A. A. LAG-3 as the third checkpoint inhibitor. Nat. Immunol. 24, 1415–1422 (2023).
Blackburn, S. D. et al. Coregulation of CD8+ T cell exhaustion during chronic viral infection by multiple inhibitory receptors. Nat. Immunol. 10, 29–37 (2009).
Woo, S.-R. et al. Immune inhibitory molecules LAG-3 and PD-1 synergistically regulate T-cell function to promote tumoral immune escape. Cancer Res. 72, 917–927 (2012).
Mariuzza, R. A., Shahid, S. & Karade, S. S. The immune checkpoint receptor LAG3: Structure, function, and target for cancer immunotherapy. J. Biol. Chem. 300, 107241 (2024).
Huard, B., Prigent, P., Tournier, M., Bruniquel, D. & Triebel, F. CD4/major histocompatibility complex class II interaction analyzed with CD4- and lymphocyte activation gene-3 (LAG-3)-Ig fusion proteins. Eur. J. Immunol. 25, 2718–2721 (1995).
Maruhashi, T. et al. LAG-3 inhibits the activation of CD4 + T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat. Immunol. 19, 1415–1426 (2018).
Wang, J. et al. Fibrinogen-like Protein 1 Is a Major Immune Inhibitory Ligand of LAG-3. Cell 176, 334–347.e12 (2019).
Maruhashi, T. et al. Binding of LAG-3 to stable peptide-MHC class II limits T cell function and suppresses autoimmunity and anti-cancer immunity. Immunity 55, 912–924.e8 (2022).
Guy, C. et al. LAG3 associates with TCR-CD3 complexes and suppresses signaling by driving co-receptor-Lck dissociation. Nat. Immunol. 23, 757–767 (2022).
Ming, Q. et al. LAG3 ectodomain structure reveals functional interfaces for ligand and antibody recognition. Nat. Immunol. 23, 1031–1041 (2022).
Mishra, A. K. et al. CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker. Structure 31, 1149–1157.e3 (2023).
Silberstein, J. L. et al. Structural insights reveal interplay between LAG-3 homodimerization, ligand binding, and function. Proc. Natl. Acad. Sci. 121, e2310866121 (2024).
Adam, K. et al. Cutting Edge: LAG3 Dimerization Is Required for TCR/CD3 Interaction and Inhibition of Antitumor Immunity. J. Immunol. 2300673 https://doi.org/10.4049/jimmunol.2300673. (2024).
Thudium, K. et al. Preclinical Characterization of Relatlimab, a Human LAG-3–Blocking Antibody, Alone or in Combination with Nivolumab. Cancer Immunol. Res. 10, 1175–1189 (2022).
Ascione, A. et al. Development of a novel human phage display-derived anti-LAG3 scFv antibody targeting CD8+ T lymphocyte exhaustion. BMC Biotechnol. 19, 67 (2019).
Angin, M., Brignone, C. & Triebel, F. A LAG-3–Specific Agonist Antibody for the Treatment of T Cell–Induced Autoimmune Diseases. J. Immunol. https://doi.org/10.4049/jimmunol.1900823. (2020)
Agnihotri, P. et al. Epitope Mapping of Therapeutic Antibodies Targeting Human Inhibitory Receptor Lymphocyte Activation Gene 3 Protein (LAG3). J. Immunol. 209, 1586–1594 (2022).
MacLachlan, B. J. et al. Molecular characterization of HLA class II binding to the LAG-3 T cell co-inhibitory receptor. Eur. J. Immunol. 51, 331–341 (2021).
Zhu, Y., Rudensky, A. Y., Corper, A. L., Teyton, L. & Wilson, I. A. Crystal Structure Of MHC Class II I-Ab in Complex with a Human CLIP Peptide: Prediction of an I-Ab Peptide-binding Motif. J. Mol. Biol. 326, 1157–1174 (2003).
Huard, B. et al. Characterization of the major histocompatibility complex class II binding site on LAG-3 protein. Proc. Natl. Acad. Sci. USA 94, 5744–5749 (1997).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Cebecauer, M. et al. CD8+ cytotoxic T lymphocyte activation by soluble major histocompatibility complex-peptide dimers. J. Biol. Chem. 280, 23820–23828 (2005).
Workman, C. J., Dugger, K. J. & Vignali, D. A. A. Cutting edge: molecular analysis of the negative regulatory function of lymphocyte activation gene-3. J. Immunol. 169, 5392–5395 (2002).
Triebel, F. et al. LAG-3, a novel lymphocyte activation gene closely related to CD4. J. Exp. Med. 171, 1393–1405 (1990).
Wang, J. et al. Crystal structure of the human CD4 N-terminal two-domain fragment complexed to a class II MHC molecule. PNAS 98, 10799–10804 (2001).
Davis, S. J. et al. The nature of molecular recognition by T cells. Nat. Immunol. 4, 217–224 (2003).
Wang, X. X. et al. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4–HLA-DR1 complex. Proc. Natl. Acad. Sci. USA 108, 15960–15965 (2011).
Dubiski, S., Cinader, B., Chou, C. T., Charpentier, L. & Letarte, M. Cross-reaction of a monoclonal antibody to human MHC class II molecules with rabbit B cells. Mol. Immunol. 25, 713–718 (1988).
Kassardjian, A. et al. Modular adjuvant-free pan-HLA-DR-immunotargeting subunit vaccine against SARS-CoV-2 elicits broad sarbecovirus-neutralizing antibody responses. Cell Rep. 42, 112391 (2023).
Garralda, E. et al. A first-in-human study of the anti-LAG-3 antibody favezelimab plus pembrolizumab in previously treated, advanced microsatellite stable colorectal cancer. ESMO Open 7, 100639 (2022).
Grandal, M. M. et al. Anti-lag-3 antibodies and compositions. WIPO(PCT) Patent No. US20220056126A3 (2019).
Schöffski, P. et al. Phase I/II study of the LAG-3 inhibitor ieramilimab (LAG525) ± anti-PD-1 spartalizumab (PDR001) in patients with advanced malignancies. J. Immunother. Cancer 10, e003776 (2022).
Workman, C. J., Rice, D. S., Dugger, K. J., Kurschner, C. & Vignali, D. A. A. Phenotypic analysis of the murine CD4-related glycoprotein, CD223 (LAG-3). Eur. J. Immunol. 32, 2255–2263 (2002).
Mossman, K. D., Campi, G., Groves, J. T. & Dustin, M. L. Altered TCR signaling from geometrically repatterned immunological synapses. Science 310, 1191–1193 (2005).
Dustin, M. L. & Depoil, D. New insights into the T cell synapse from single molecule techniques. Nat. Rev. Immunol. 11, 672–684 (2011).
Maeda, T. K., Sugiura, D., Okazaki, I.-M., Maruhashi, T. & Okazaki, T. Atypical motifs in the cytoplasmic region of the inhibitory immune co-receptor LAG-3 inhibit T cell activation. J. Biol. Chem. 294, 6017–6026 (2019).
Su, L. F., Del Alcazar, D., Stelekati, E., Wherry, E. J. & Davis, M. M. Antigen exposure shapes the ratio between antigen-specific Tregs and conventional T cells in human peripheral blood. Proc. Natl Acad. Sci. USA 113, E6192–E6198 (2016).
Nelson, C. A., Lee, C. A. & Fremont, D. H. Oxidative refolding from inclusion bodies. Methods Mol. Biol. 1140, 145–157 (2014).
Kabsch, W. Integration, scaling, space-group assignment and post-refinement. Acta Cryst. D. 66, 133–144 (2010).
Otwinowski, Z. & Minor, W. [20] Processing of X-ray diffraction data collected in oscillation mode. in Methods in Enzymology 276 307–326 (Elsevier, 1997).
Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. H. K. L. 3000: the integration of data reduction and structure solution – from diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 (2006).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl Crystallogr. 40, 658–674 (2007).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst. D. 68, 352–367 (2012).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
The PyMOL Molecular Graphics System. Schrödinger, LLC.
Mirdita, M. et al. ColabFold: Making Protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Acknowledgements
We thank the staff at the 19-ID beamline of the Advanced Light Source and the 12-1 beamline of the Stanford Synchrotron Light Source for assistance with remote X-ray data collection. We thank Aashish Manglik and Christian Billesbølle for preliminary structural screening, and we thank Chris Garcia and Gaya Amarasinghe for helpful advice. V.C.L. and Q.M. are supported by Bankhead Coley Grant 22B07 from the Florida Department of Health, and V.C.L. is a Rita Allen Scholar. D.W.L. is supported by NIH R01AI140758, R01AI159678, and P01AI120943. K.S. is supported by Bankhead Coley Grant 24B04 from the Florida Department of Health. Shared resources were provided by the Moffitt Cancer Center Support Grant NIH P30CA076292.
Author information
Authors and Affiliations
Contributions
V.C.L., D.A. and Q.M. wrote the manuscript. K.S. assisted with data analysis and experimental design. V.C.L., Q.M., D.A., D.A.P. and D.W.L. designed the experiments. Q.M., A.M., E.M., S.S. and C.M. purified recombinant proteins. Q.M. crystallized the proteins and collected the x-ray diffraction data. Q.M. and T.H.T. performed the structural studies, including data processing, structure solution, and refinement. Q.M. performed SPR experiments. Q.M. and D.A. performed cellular binding studies and BLI experiments. V.C.L. and Q.M. performed the structural analysis. D.A.P. and D.W.L. performed MALS experiments and assisted with data analysis.
Corresponding author
Ethics declarations
Competing interests
V.C.L. is a consultant on unrelated projects for Cellestia Biotech, Remunix, and Curie.Bio. The remaining authors have no competing interests.
Peer review
Peer review information
Nature Communications thanks Roy Mariuzza, Lawrence Stern and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ming, Q., Antfolk, D., Price, D.A. et al. Structural basis for mouse LAG3 interactions with the MHC class II molecule I-Ab. Nat Commun 15, 7513 (2024). https://doi.org/10.1038/s41467-024-51930-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51930-5