Abstract
Modification of guanosine to N7-methylguanosine (m7G) in the variable loop region of tRNA is catalyzed by the METTL1/WDR4 heterodimer and stabilizes target tRNA. Here, we reveal essential functions of Mettl1 in Drosophila fertility. Knockout of Mettl1 (Mettl1-KO) causes no major effect on the development of non-gonadal tissues, but abolishes the production of elongated spermatids and mature sperm, which is fully rescued by expression of a Mettl1-transgene, but not a catalytic-dead Mettl1 transgene. This demonstrates that Mettl1-dependent m7G is required for spermatogenesis. Mettl1-KO results in a loss of m7G modification on a subset of tRNAs and decreased tRNA abundance. Ribosome profiling shows that Mettl1-KO led to ribosomes stalling at codons decoded by tRNAs that were reduced in abundance. Mettl1-KO also significantly reduces the translation efficiency of genes involved in elongated spermatid formation and sperm stability. Germ cell-specific expression of Mettl1 rescues disrupted m7G tRNA modification and tRNA abundance in Mettl1-KO testes but not in non-gonadal tissues. Ribosome stalling is much less detectable in non-gonadal tissues than in Mettl1-KO testes. These findings reveal a developmental role for m7G tRNA modification and indicate that m7G modification-dependent tRNA abundance differs among tissues.
Similar content being viewed by others
Introduction
Post-transcriptional epigenetic modifications of RNA are together known as the “epitranscriptome” and are being revealed to have diverse and widespread functions in gene regulation. To date, over 170 RNA modifications have been identified, approximately 80% of which are found in tRNAs1,2. Chemical modifications regulate tRNA structure, dynamics, and interactions with other molecules, and result in altered translation3,4. While some tRNA modifications play a fundamental role in cell proliferation, an increasing number of studies show that aberrant tRNA modification can result in severe developmental defects and human diseases, indicating that tRNA modifications play roles in specific tissues during animal development4,5,6,7.
N7-methylguanosine (m7G) is an RNA modification observed in mRNA caps8 and at internal positions within tRNAs, rRNAs, mRNAs and miRNAs9,10,11,12,13,14,15. m7G in tRNAs is widely conserved in prokaryotes, eukaryotes and some archaea16. In eukaryotes, Methyltransferase-like 1 (METTL1) catalyzes the addition of m7G at internal sites of tRNAs, mRNAs, and miRNAs10,12,15,17. METTL1 interacts with the non-catalytic subunit WD repeat domain 4 (WDR4)17,18,19,20,21, and both are widely conserved from yeast to mammals. m7G modification occurs at the 46th guanosine in the variable loop region of tRNAs, within the conserved motif, “RAGGU” (R = A or G, G represents m7G)10. In Saccharomyces cerevisiae, transfer RNA methyltransferase 8 (Trm8), a homolog of METTL1, introduces the m7G modification onto tRNAs, such as tRNA ValAAC, and promotes tRNA stability and cell growth at higher temperatures17,18,22. Hypomodified tRNAs are degraded by the 5′–3′ exonucleases, Rat1 and Xrn1, which form the rapid tRNA decay (RTD) pathway in yeast22,23,24. Rat1 and Xrn1 are highly conserved among animals, indicating that a similar degradation system is present in animals25. Indeed, METTL1-depletion in mammalian cells leads to decreased steady-state levels of several tRNAs that are targets of METTL1, retardation of cell growth26,27 and impaired translation of genes associated with neural differentiation10. Conversely, increased levels of METTL1 result in increased m7G modification of specific tRNAs, such as tRNA ArgTCT, tRNA stabilization, cell cycle acceleration through enhanced translation of cell cycle related genes, such as CDK6 and CDK8, and promotion of oncogenicity27. The primary let-7e miRNA is also methylated by METTL112. Internal m7G modifications in mRNAs are recognized by Quaking proteins, which enable their transport into stress granules. This modulates mRNA stability and the cellular stress response in mammals28. These studies have revealed the role of m7G at the single cell level; however, its roles at tissue and organ levels during animal development are less well characterized.
Recent clinical studies have revealed that variations in METTL1 and WDR4 cause microcephalic primordial dwarfism, Galloway–Mowat syndrome29,30,31, multiple sclerosis32,33, and male infertility34, indicating a potential link between internal m7G regulation and pathological characteristics35. Furthermore, knockout of Wdr4 (a mouse homolog of Drosophila melanogaster Wuho (Wh) in mice results in embryonic lethality36. These observations indicate that the m7G modification machinery is critical for mammalian development; however, it is unclear whether m7G modification alone is responsible for the above developmental defects. Indeed, WDR4 physically interacts with proteins other than METTL1. WDR4 is proposed to be involved in genome stability through interaction with Flap endonuclease 136 and in cerebellar development and locomotion through interaction with ATRGAP1737. Therefore, it is not known whether the developmental disorders caused by METTL1 or WDR4 variants are directly linked to defective m7G modification.
In D. melanogaster, Wh is required for gonad development38,39. Wh mutants show sterility with arrested spermatogenesis in males and semi-sterile phenotypes with abnormal egg chamber and decreased germline stem cell number in females38,39. Wh is considered to control ovary function through interaction with partner proteins, including Mei-P26, Nanos, and Bgcn38. However, it remains to be addressed whether Wh associates with the Drosophila METTL1 ortholog and if the fertility defects in Wh mutants are caused by a loss of m7G modification or via defective association with interactors of Wh other than Mettl1.
Here, we demonstrated that candidate gene 4045 (CG4045) (hereafter referred to as Mettl1) is required for Drosophila male and female fertility. Mettl1 associates with Wh in vivo and in vitro and catalyzes m7G-modification on a subset of tRNAs in a Wh-dependent mechanism. Depletion of Mettl1 results in a loss of m7G modification and decreased abundance of nearly all of the tRNAs targeted by Mettl1. Consistent with the decrease in tRNA abundance, we found increased occurrence of ribosome pausing and subsequent collisions of trailing ribosomes in Mettl1-KO testes at the codons decoded by the reduced tRNAs, leading to the reduction of the translation efficiency of genes involved in mature sperm formation. Taken together, our work reveals the developmental role of Mettl1-dependent m7G in multicellular organisms.
Results
Mettl1, the Drosophila ortholog of the m7G-modifier, METTL1, is essential for Drosophila fertility
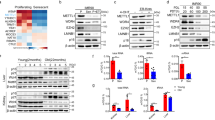
We studied the physiological functions of Mettl1 in D. melanogaster. The Drosophila gene, CG4045 (herein referred to as Mettl1), is an ortholog of mammalian METTL1 and contains an RNA methyltransferase-like domain that is highly conserved across animals (Fig. 1a, Supplementary Fig. 1a)40. To generate a Mettl1-knock out (KO) strain using the CRISPR/Cas9 system, a guide RNA (gRNA) was selected to target the N-terminal part of Mettl1 (Fig. 1a). Two different transgenic strains harboring either a 1 nucleotide deletion (Mettl1KO1) or a 23-nucleotide deletion with an 8-nucleotide insertion (Mettl1KO2) were generated, both of which lose a large part of the methyltransferase-like domain because of frame-shift (Fig. 1a, Supplementary Fig. 1b). To detect Mettl1 and the Mettl1-interacting partner, Wh, mouse-monoclonal antibodies were generated. Western blot analyses showed a strong ~32 kDa Mettl1 signal in heterozygous ovaries and control (yw) testes (Fig. 1b, c). In contrast, this signal was undetectable in Mettl1KO1 and Mettl1KO2 ovaries and Mettl1KO1 testes (Fig. 1b, c, Supplementary Fig 1c), indicating that Mettl1 was absent in Mettl1-KO animals. Notably, a Wh signal was still observed in Mettl1-KO flies (Fig. 1b), indicating that Wh stability is unaffected by the absence of its heterodimer partner, Mettl1.
a Schematic diagram of the genomic structure of Mettl1 and a gRNA targeting Mettl1 exon 2 for CRISPR-mediated generation of Mettl1-KO lines. Mettl1 sequences of the WT (yw) and the Mettl1-KO mutants (Mettl1KO1 and Mettl1KO2). b Western blot showing Mettl1 and Wh in Drosophila ovaries. c Western blot showing Mettl1 in Drosophila testes. d Male fertility assay at 25 °C using WT (yw), WT with Mettl1 transgene (yw; [Mettl1]), Mettl1-KO (Mettl1KO1) and Mettl1-rescue (Mettl1KO1; [Mettl1]) males. e Female fertility assay at 25 °C using control (Mettl1KO1/FM7, Mettl1KO2/FM7), Mettl1-KO (Mettl1KO1, Mettl1KO2) and Mettl1-Rescue group (Mettl1KO1; [Mettl1], Mettl1KO2; [Mettl1]) females. FM7 is a balancer chromosome. f Male fertility assay at 25 °C using (yw; m + /z + ), and zygotic Mettl1-KO (Mettl1KO1; m + /z–), maternal and zygotic Mettl1-KO (Mettl1KO1; m–/z–) males. Each western blots were reproduced three times with similar results (b, c). In box plots of fertility assays, central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. The number (n = ) below the box plot indicates sample size. Upper and lower edges of boxes represent third and first quartiles, respectively (d, e). Two-sided P-values were calculated using Tukey’s HSD test (d, e). Source data of Fig. 1b–e are provided as a Source Data file.
Basic analyses of adult morphology (Supplementary Fig. 1d), body weight (Supplementary Fig. 1e), rate of emerging adult flies (Supplementary Fig. 1f) and locomotive activity (Supplementary Fig. 1g), indicated that Mettl1KO1 and Mettl1KO2 flies developed normally to the adult stage. However, we observed that mutant males and females had significantly reduced fertility (Fig. 1d, e, Supplementary Fig. 2a–c). Mutant males produced one tenth and mutant females one third of the progeny produced by control flies (Fig. 1d, e, Supplementary Fig. 2a–c). Consistent with the reduced fertility of Mettl1-KO females, the size of Mettl1-KO ovaries was significantly reduced (Supplementary Fig. 2d). To confirm whether this was caused by loss of Mettl1 function, we introduced the Mettl1 genomic region into the second chromosome. The [Mettl1] transgene rescued fertility and ovary size to the levels of the control groups (Fig. 1d, e, Supplementary Fig. 2b, d). Moreover, to evaluate the maternal contribution to fertility, we generated maternal and zygotic mutant males. Small numbers of offspring (less than 10) were detected in three of seven crosses from zygotic mutant males, whereas no offspring were produced by maternal and zygotic mutant males, indicating a maternal contribution of Mettl1 (Fig. 1f). Taken together, these data reveal a critical role of Mettl1 in male fertility.
Mettl1 depletion impairs the formation of elongated spermatids and results in the loss of mature sperm
Drosophila spermatogenesis involves germ cells communicating with somatic cells, proceeding through mitotic and meiotic cell division, differentiating into elongating spermatids, and forming bundle structures in the testis. Each step in this process is tightly regulated (Fig. 2a)41. To study the expression patterns of Mettl1 in the testis, we generated a transgenic strain carrying C-terminally Flag-tagged Mettl1, [Mettl1-Flag] (Fig. 2b) and confirmed that [Mettl1-Flag] rescued Mettl1KO1 fertility to a similar extent as untagged [Mettl1] (Fig. 1d, Supplementary Fig. 3a). We observed Flag-tag signals in almost all testicular cells, indicating that Mettl1 is ubiquitously expressed in the various gonadal cell types (Fig. 2b). Wh-GFP shows a similar ubiquitous expression pattern through spermatogenesis, indicating the presence of the Mettl1/Wh heterodimer in all testicular cells39. To study whether spermatogenesis was affected by Mettl1-KO, we assessed mature sperm generation. We found no mature sperm in the seminal vesicles of Mettl1-KO testes (Fig. 2c), which is consistent with the decreased fertility of Mettl1-KO males (Fig. 1d). To examine the spermatogenesis defects in more detail, we studied the germ cell marker proteins, Aubergine, a marker of germline cells, and Boule, a Drosophila orthologue of vertebrate DAZL, a marker of 32- and 64-cell cysts and elongating spermatids within Drosophila testes42. Aubergine was observed in a wild-type pattern within Mettl1-KO testes (Supplementary Fig. 3b). Moreover, Boule was also detected in a wild-type pattern in 32- and 64-cell cysts within Mettl1-KO testes (Fig. 2d, Supplementary Fig. 3c). Boule-positive cysts defective in elongation were accumulated near the seminal vesicle region (Fig. 2d), indicating that the loss of Mettl1 did not affect meiosis but may have affected the elongation step of spermatogenesis. Furthermore, we observed no elongated spermatids (Fig. 2e). We then examined the distribution of acetylated microtubules, a marker of elongated spermatids43. Most Mettl1-KO testes (23/31) showed a decreased acetylated microtubule signal (indicating an elongation defect, Fig. 2f). The remainder had no elongated spermatids (8/31, Fig. 2f). Together, these results indicate that elongated spermatid formation and the later stages of spermatogenesis are severely impaired in Mettl1-KO testes.
a Schematic diagram of Drosophila spermatogenesis. The genes indicated below are markers used to identify each stage of spermatogenesis. (GSC; germline stem cell, CySC; cyst stem cell) (b) Immunostaining of Mettl1-Flag in the testis. yw is a wild-type control. (Left) Overall view of testis. (Right) Zoom-in view of the white box in panel. c Transparent image of seminal vesicle (arrowheads) from control (yw) and Mettl1-KO (Mettl1KO1) males. d Representative images of Boule localization in testes from WT (yw) and Mettl1-KO (Mettl1KO1) adult males. Testes were stained for Boule (green). Dotted line surrounds the region of spermatid elongation defects. e Differential interference microscopy image of spermatid bundles (arrowheads) in control (yw) and Mettl1KO1 testes. f Immunostaining of acetylated-Tub in WT (yw) and Mettl1-rescue (Mettl1KO1; [Mettl1]) testes. Arrowheads show elongated spermatids. g Male fertility assay at 25 °C using WT (yw), WT with Mettl1 transgene driven by Nanos promoter (yw; [nosP-Mettl1]), Mettl1-KO (Mettl1KO1) and Mettl1-KO with Nanos-driven Mettl1 (Mettl1KO1; [nosP-Mettl1]). In box plots, central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. Two-sided P-values were calculated using Tukey’s HSD test. The number (n = ) below the box plot indicates the sample size. Each immunostaining were reproduced three times with similar results. Source data of Fig. 2g is provided as a Source Data file. Scale bars of b (left), c, d, e, and f = 100 μm, and b (right) = 50 μm.
We observed no change in overall morphology or in the distribution of somatic niche cells (hub cells) or somatic cyst cells in Mettl1-KO testes, indicating the requirement for Mettl1 in testicular germ cells (Supplementary Fig. 3d). Mettl1 is still detected in the carcasses of males after gonad removal (Supplementary Fig. 3e), indicating that Mettl1 may play a critical role in elongated spermatid formation in tissues other than the testis. We assessed the copulation behavior of Mettl1-KO males and tumorous testis (tut)-KO males, which are also deficient in spermatogenesis44. Interestingly, both Mettl1-KO and tut-KO males showed significantly decreased copulation rate (Supplementary Fig. 3f), indicating that disrupted spermatogenesis impairs sexual behaviors via unknown signaling from the testes to the brain. To directly examine whether Mettl1 acts in a cell-autonomous or non-cell autonomous manner, we prepared a Mettl1-transgene driven by a germ-specific Nanos promoter and conducted a fertility assay. The Nanos-Mettl1-transgene [nosP-Mettl1] fully rescued the sterile phenotype and acetylated microtubule signal of Mettl1-KO males (Fig. 2g, Supplementary Fig. 3g). Moreover, somatic cell-specific knockdown (KD) of Mettl1 did not affect fertility (data not shown). Taking these findings together, we concluded that Mettl1 plays a critical role in elongating spermatid formation via a cell-autonomous mechanism.
Catalytic activity of Mettl1 is required for Drosophila fertility
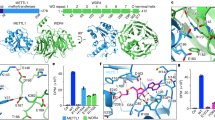
Although mammalian METTL1 and its yeast ortholog, Trm8, catalyze the m7G modification on tRNAs, it is not known whether Drosophila Mettl1 possesses this activity10,17. Given that METTL1 acts as the catalytic partner in a heterodimer with WDR410,17,20,21, we investigated the formation of a homologous complex in flies. We conducted immunoprecipitation from [Mettl1-Flag] ovaries or Oregon R wild-type ovaries using two different buffer conditions and performed liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis of the samples (Fig. 3a, Supplementary Fig. 4a, b). Mettl1-Flag co-immunoprecipitated with Wh irrespective of buffer conditions, indicating that the Mettl1/Wh heterodimer is formed in vivo, as in other organisms (Fig. 3a, Supplementary Fig. 4a, b). Notably, we did not detect Mei-P26, Nanos or Bgcn proteins, which can associate with Wh38, indicating that Wh makes a distinct protein complex with either Mettl1 or these other proteins. To confirm direct interaction, we purified MBP-Mettl1 and GST-Wh from Escherichia coli and performed in vitro GST-pull down assays (Supplementary Fig. 5a, b). MBP-Mettl1 but not MBP alone associated with GST-Wh but not with the GST moiety (Supplementary Fig. 5a, b).
a Volcano plot showing enrichment rates and significance levels of each protein as log2 fold change (anti-Flag M2/negative control) versus negative log 10 of the Fisher’s exact test two-sided P-value. The four red dots represent Mettl1 interactors, and the blue dot represents Mettl1 in HEPES-NP40 buffer. b (Top) Schematic diagram showing the positions of the amino acid residues important for methylation activity of Mettl1. Mettl1 harboring L157A and D160A mutations is termed Mettl1-Cd. Blue box indicates catalytic domain of Mettl1. (Bottom) In vitro m7G modification assay for testing Mettl1-Cd activity. The m7G containing tRNAs were visualized by 14C. Toluidine blue O staining was performed to visualize RNAs. MBP (negative control) or MBP-fused protein was incubated with the substrate tRNA TrpCCA and GST-Wh. c Male fertility assay for WT (yw), Mettl1-KO (Mettl1KO1), and transgenes, [Mettl1] and [Mettl1-Cd], in Mettl1KO1 background. In box plots, central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. The number (n = ) below the box plot indicate the sample size. Two-sided P-values were calculated using Tukey’s HSD test. Source data of Fig. 3b, c are provided as a Source Data file.
We then conducted in vitro m7G methylation assays using the above recombinant proteins with a synthesized tRNA and a shortened tRNA fragment designed according to a previous study45, both of which include the RAGGU motif, a conserved target sequence of eukaryotic METTL1 (Fig. 3b, Supplementary Fig. 5c, d). Drosophila Mettl1 required Wh for tRNA methylation, consistent with a recently proposed structural model of m7G methylation (Supplementary Fig. 5d)20,21. Mettl1 did not introduce m7G to a target RNA in which the 4th guanosine of the RAGGU motif was converted to cytosine, indicating that Drosophila Mettl1/Wh methylates target tRNAs in a similar manner to that of METTL1/WDR4 in other eukaryotes (Supplementary Fig. 5d)10. To create a catalytic dead mutant of Mettl1, L157A and D160A mutations (Mettl1-Cd: Cd), both of which are located in the methylation consensus motif, were engineered (Supplementary Fig. 1a). We confirmed that these mutations abolished methylation activity of Mettl1 in vitro (Fig. 3b).
This finding led us to test the requirement of Mettl1 catalytic activity for fertility. We prepared a catalytic dead Mettl1-transgene [Mettl1-Cd], harboring L157A and D160A mutations in Mettl1 (Fig. 3c) and conducted fertility rescue experiments (Fig. 3c). The sterile phenotype of Mettl1KO1 was rescued by the wild-type Mettl1-transgene [Mettl1], but not by [Mettl1-Cd], indicating that male fertility is dependent on the catalytic activity of Mettl1 (Fig. 3c). [Mettl1-Cd] transgene shows slightly lower levels of the mutant protein than wild-type (Supplementary Fig. 5e). Thus, we can not rule out the possibility that the failure of the rescue by [Mettl1-Cd] transgene is due to a slight level of difference of the mutant proteins. [Mettl1-Cd] did not show any dominant-negative effects on fertility in wild-type backgrounds (Supplementary Fig. 5f).
Drosophila Mettl1 mediates m7G modification of tRNA
To understand the molecular mechanism underlying the Mettl1-KO fertility defects, we surveyed m7G modification sites on tRNAs because tRNAs are efficiently methylated by METTL1 in mammalian cells10. We performed an m7G site-specific cleavage assay using sodium borohydride (NaBH4) and aniline46,47. The resulting 5′ and 3′ fragments were detected by northern blotting using specific tRNA probes (Fig. 4a, b, Supplementary Fig. 6a, b). We detected 3′ fragments of tRNAs isolated from testes (Fig. 4b) and ovaries (Supplementary Fig. 6a) of wild-type Mettl1 but not Mettl1-KO flies, supporting Mettl1 deposition of m7G onto tRNAs in both tissues (Fig. 4b, Supplementary Fig. 6a). Moreover, the 3′ fragments from the same tRNAs were lost in Wh mutant testes (Fig. 4c), consistent with the collaborative role of Wh for m7G modification of tRNA in vitro (Fig. 4c, Supplementary Fig. 5c).
a Schematic diagram of m7G site-specific reduction and cleavage. Cleaved 3′ fragments were detected by northern blotting. b Northern blot of chemically-treated total RNAs from wild-type control (yw) and Mettl1-KO (Mettl1KO1) testes. The probe was designed at the 3′ end of TrpCCA. Arrowhead denotes the position of the cleaved 3′ fragment. c Northern blot of chemically-treated total RNAs from WT (FM7c) and Wh-KO (Wh56) testes. The probe was designed around the 3′ end of tRNA TrpCCA. Arrowhead denotes the position of the cleaved 3′ fragment. d TRAC-seq scheme using Drosophila gonads. e Mettl1-dependent m7G modified tRNAs identified in testes by TRAC-seq. f Sequence motif of m7G modification sites identified in testes by TRAC-seq. The arrowhead corresponds to the m7G site. g Representative plot of the cleavage score (difference between cleavage score of WT and Mettl1-KO) of tRNA CysGCA (Cys-GCA-2) between WT and Mettl1-KO, which showed the highest score difference by TRAC-seq. Pink shading represents the variable loop region of tRNA CysGCA. h Quantitative comparison of cleavage scores (CleavageScore) of identified m7G-modified tRNAs (n = 36) between WT and Mettl1-KO testes. yw is treated as a wild-type control. Two-sided P-value was calculated by the Mann–Whitney U-test are indicated. Central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. In our TRAC-seq, 3 biological replicates were set for each sample. Each northern blot was reproduced for three times (Fig. 4b, c). Source data of Fig. 4b, c, g and h are provided as a Source Data file.
We next comprehensively investigated the cleaved tRNA fragments by deep sequencing [i.e., tRNA reduction and cleavage sequencing (TRAC-seq)10,48] (Fig. 4d, Supplementary Fig. 6c). TRAC-seq identified 23 tRNAs in testes and 22 tRNAs in ovaries. In our analysis, tRNA ArgTCT was categorized as m7G-modified in testis but not in ovary. This difference can be attributed to the limited number of tRNA reads from the Drosophila ovary. The m7G-modified tRNAs have the RAGGU motif (Fig. 4f) located in the variable loop (Fig. 4g), which is similar to the findings of a previous study10. Consistent with the northern blot data (Fig. 4b, c), the TRAC-seq cleavage score was markedly decreased in Mettl1-KO samples, further supporting the function of Mettl1 in m7G modification of tRNA (Fig. 4h). Together, these data indicate that Mettl1/Wh is responsible for introducing the m7G modification onto a subset of tRNAs in the testis.
Drosophila Mettl1 regulates the abundance of a subset of tRNAs
In S. cerevisiae, loss of m7G modification on tRNAs results in decreased tRNA abundance17,22. This mechanism for this is the rapid tRNA decay (RTD) pathway mediated by exonucleases22,23,24. We therefore reasoned that a similar RTD pathway acts in Drosophila upon the loss of Mettl1. In testes, Mettl1 depletion resulted in changes in the levels of some tRNAs (Fig. 5a, b). In Drosophila testis, the mean steady-state levels of 15 m7G-modified tRNAs (ProAGG, ProCGG, ProTGG, ValAAC, ValCAC, ValTAC, LysCTT, LysTTT, ArgTCT, AlaAGC, AlaCGC, AlaTGC, CysGCA, TyrGTA, and iMetCAT) were significantly decreased, whereas the abundances of non-m7G modified tRNAs (22 of 45 tRNAs) showed limited changes (Fig. 5a). We divided m7G-modified tRNAs into two groups according to steady-state levels of tRNA abundance in Mettl1-KO testes (Fig. 5b, c, Supplementary Fig. 7a): destabilized in Mettl1-KO (group 1) and unaltered in Mettl1-KO (group 2). We also defined m7G-hypomodified tRNAs as group 3 (Fig. 5a, c, Supplementary Fig. 7a). Consistent with the TRAC-seq data (Fig. 5a–c), northern blot analysis validated the reduced levels of group 1 tRNAs (ProTGG and ValCAC) and the absence of change in group 2 tRNA (TrpCCA) and group 3 tRNA (LeuAAG) in Mettl1-KO testes (Fig. 5d) (see Fig. 6 for further characterization of these tRNA groups for translation).
a Volcano plot representing changes in tRNA abundance between Mettl1-KO (Mettl1KO1) and wild-type (yw) testes. Each dot shows the change in abundance of one tRNA. The two-sided P-values were calculated by applying the Wald test with the Benjamini-Hochberg correction implemented in DEseq2 module of the tRAX pipeline. b Heatmap showing changes in m7G-modified tRNA abundance between Mettl1-KO (Mettl1KO1) and WT testis. Drosophila tRNAs were classified into three groups according to the difference in abundance between Mettl1-KO (Mettl1KO1) and wild-type (WT), and P-values indicate significant differences; group 1 (m7G-modified tRNAs and significantly decreased abundance, log2FC < 0, P < 0.05); group 2 (other m7G-modified tRNAs, P ≥ 0.05); group 3 (non-m7G tRNAs). The P-values indicated correspond to those of (a). c Quantitative comparison of changes in tRNA abundance between the three groups of tRNAs (Group1: n = 15, Group2: n = 8, Group3: n = 22) defined in this study. Two-sided P-values were calculated by the Mann–Whitney U-test are indicated. Box plots represent maximum, median and minimum values with outliers. Central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. In our tRNA expression analysis, 3 biological replicates were set for each sample. d Northern blot analysis of tRNA abundance in Mettl1-KO (Mettl1KO1) and WT (yw) testes. 2S rRNA was used as a loading control. Each northern blot was reproduced for three times. Source data of Fig. 5 are provided as a Source Data file.
a Metagene plot for ribosome footprints around start codons. The 5′ ends of 29 nt reads are depicted. Pearson’s correlation coefficients (r), ribosome footprints (b), and RNA-Seq reads (c) on each ORF. The color scales for r are represented. d Ribosomal occupancy analysis across A-site codons in the Mettl1-KO testis. Source data is provided as a Source Data file. e Ribosome occupancy analysis for the codons decoded by the indicated tRNA groups (Group1: n = 15, Group2: n = 8, Group3: n = 22). Central bands, upper and lower edges of box plots represent median, first and third quartiles, respectively. Upper and lower whiskers represent maximum, and minimum values. f Metagene plot for ribosome occupancy around CCA codons. The positions of paused and collided ribosomes are highlighted by arrows. g Differences in ribosome occupancy changes on A-site codons in Mettl1-KO testes between all analyzed genes and genes that are essential for fertility (Supplementary Table S1).
To gain insight into how the levels of hypomodified tRNAs are decreased in testes, we examined aminoacylation levels of tRNAs by acid urea polyacrylamide gel electrophoresis and northern blot analysis. We found that the majority of tRNA ValCAC, an m7G-tRNA, was aminoacylated in Mettl1-KO testes, indicating that the decrease of tRNA in Mettl1-KO testes is not caused by loss of aminoacylation (Supplementary Fig. 7b). The reduced levels of a subset of tRNAs in Mettl1-KO flies also led us to investigate whether the RTD pathway explains the sterile phenotype. A genetic approach revealed that Trm8, a METTL1 ortholog in S. cerevisiae, is important for m7G tRNA modification and stability and for temperature sensitive growth18,22. It is noteworthy that the RTD pathway can be suppressed by overexpression of eukaryotic translational elongation factor 1 A (eEF1A), encoded by the TEF1 gene in yeast, because eEF1A binds directly to tRNAs and protects them from the RNA decay machinery49,50. Therefore, we asked whether overexpression of eukaryotic translational elongation factor 1 alpha 1 (eEF1α1, CG8280), a fly homolog of eEF1A, rescues the male sterility in Mettl1-KO flies (Supplementary Fig. 7c). Actin mRNAs are highly expressed in various cell types including in testes; therefore, we expressed eEF1A under the control of Act-Gal4. Overexpression of eEF1α1 partially rescued the fertility of Mettl1-KO males, indicating that a pathway similar to the S. cerevisiae RTD pathway may play a role and regulate tRNA abundance in Drosophila.
In mammalian A549 cells, let-7e-5p miRNA biogenesis is affected by m7G methylation12 because m7G modification in the primary transcript of let-7e disrupts G-quadruplex formation and facilitates precursor miRNA processing12. In D. melanogaster, depletion of let-7 causes a spermatogenesis defect51; therefore, the phenotype that we observed in Mettl1-KO flies can be theoretically explained by a defect in let-7 biogenesis. However, the steady-state level of let-7 miRNA was not significantly changed in Mettl1-KO testes, indicating that the sterile phenotype of Mettl1-KO males is not caused by m7G modification of the let-7 primary miRNA (Supplementary Fig. 7d). Notably, sequence comparison between mouse and D. melanogaster indicated that the RAGGU motif in the mouse let-7e primary miRNA is not identical to that in D. melanogaster (UAGGU) and that the G bases that make up the G-quadruplex are A bases in D. melanogaster. This might explain why let-7 was not m7G-modified in Drosophila (Supplementary Fig. 7e). Taking these observations together, we conclude that the Drosophila let-7 primary miRNA is not modified by Mettl1 and is unrelated to the sterile phenotype of Mettl1-KO males (Fig. 2, Supplementary Fig. 7d).
Translatome analysis in Mettl1-KO testis
To examine whether the reduced abundance of tRNAs affects mRNA translation, we performed ribosome profiling of dissected testes52,53. Given that dissected testes provide limited material, we employed a method tailored for low input with RNA-dependent RNA amplification54. Remarkable 3-nt periodicity (Fig. 6a) and high reproducibility (Fig. 6b, c) confirmed high quality data were obtained from the small amounts of tissue. We observed remarkably high ribosome occupancy on A-site codons with the loss of Mettl1 (Fig. 6d). Many of these codons corresponded to codons decoded by the m7G-modified tRNAs (Fig. 6d). Notably, among the m7G-modified tRNAs, the sub-group with reduced abundance (group 1, Fig. 5c) showed prominent effects on ribosome traversal (Fig. 6e). In support of these findings, ribosome profiling of testes from another Mettl1-KO (Mettl1KO2) showed similar results (Supplementary Fig. 8a–d). Strikingly, ribosome pausing on codon CCA, recognized by group 1 tRNA ProTGG, induced collisions with trailing ribosomes to form disomes and trisomes (Fig. 6f). We observed that more pronounced ribosome stalling on CCA, GUG, CCU, and CCG codons occurred on a subset of transcripts, such as ones encoding genes essential for fertility (Fig. 6g, Supplementary Table 1).
In addition to the codon-wise assessment, we also investigated the impact of m7G tRNA modification on translation efficiency across open reading frames (ORFs). We measured translation efficiency (TE) after normalizing the ribosome footprints against RNA abundance measured by RNA-Seq and found that TE for a notable number of transcripts (1078) was down-regulated by the loss of METTL1 (Fig. 7a). Although these TE down-regulated transcripts were more prone to possess codons decoded by group 1 tRNAs (Fig. 7b), we did not detect a correlation between genes that were downregulated in TE and their Pro codon usage (Supplementary Fig. 9a). Gene ontology (GO) terms associated with the TE down-regulated genes included the key terms for gonadal functions, such as sperm individualization, sperm axoneme assembly, and spermatid development (Fig. 7c), consistent with the observed phenotype of Mettl1-KO. We noticed translational impairment of Don Juan (Dj), which is expressed in elongated spermatids and individualized sperms55, in Mettl1-KO testis (Fig. 7d). We validated this observation using a GFP-tagged Dj transgene; Dj-GFP expression was significantly decreased in Mettl1-KO testis (Fig. 7d) compared with a control GFP-tagged transgene (Vasa-GFP, Supplementary Fig. 9b).
a MA (M, log ratio; A, mean average) plot showing the fold change of translation efficiency by Mettl1-KO and the averaged read number. Each dot represents one transcript. Significantly changed transcripts (up-regulated, log2 fold change ≥ 1 and FDR ≤ 0.05; down-regulated, log2 fold change ≤ −1 and FDR ≤ 0.05) are highlighted. b Fraction of codons decoded by group 1 tRNAs in the indicated transcript group. Two-sided P-value was calculated using Pearson’s χ2 test. c Gene ontology analysis of mRNAs with TE down-regulated genes in Mettl1-KO testes (P < 0.01). The color scales for significance and the size scales for the number of genes in each category are shown. One-sided P-values were calculated by Fisher’s exact test. d Comparison of Dj-GFP expression between WT (w; Dj-GFP) and Mettl1-KO (Mettl1KO1; Dj-GFP) testes. Dotted line surrounds the outline of testis. Observation for each sample was reproduced for three times. Scale bars is 100 μm. e Northern blots of the chemically-treated total RNAs from testis and male body (testis removed) samples. The probe was designed at the 3′ end of TrpCCA. Arrowheads denote the position of the cleaved 3′ fragment. f Northern blot analysis of tRNA ProTGG abundance in WT (yw), Mettl1-KO (Mettl1KO1) and Mettl1-KO with Nanos-driven Mettl1 (Mettl1KO1; [nosP-Mettl1]) testes. 2S rRNA was used as a loading control. Each northern blot was reproduced for three times. Source data of Fig. 7a, e and f are provided as a Source Data file.
In addition to transcripts with down-regulated TE, we found a small fraction of transcripts with higher TE in Mettl1-KO testes (Fig. 7a). These mRNAs included Crc, a Drosophila homolog of ATF4, which has an upstream ORF (uORF) (Supplementary Fig. 9c)56. The uORF typically traps scanning ribosomes and inhibits the complex reaching the main downstream ORF; however, reduced abundance of the initiator tRNA (Fig. 5b) may explain the induction of leaky scanning of the uORF, as represented by eIF2 inactivation in the integrated stress response57. Taken together, these data indicate that translational changes in the late stage of spermatogenesis lead to infertility of Mettl1-KO males (Fig. 1d).
Phenotypic analyses showed that Mettl1-KO had no major effect on development except for in gonads (Supplementary Fig. 1d–g). We, therefore, investigated whether this phenotype correlates with tRNA modification status and TE in testes and non-testes tissue. We prepared body samples without testes (body) and analyzed the status of m7G-tRNA modification (Fig. 7e). As expected from the Mettl1 protein levels in body samples (Supplementary Fig. 3e), the 3′ shorter fragment was observed at similar extents in body and testis samples in a Mettl1-dependent manner (Fig. 7e). However, loss of Mettl1 had little effect on ribosome stalling in body samples. Ribosome occupancy change at CCA codons in body samples was remarkably lower than that found in testis (for example, as log2 ratio, 2.1 vs. 0.6 in Mettl1KO1 and 2.4 vs. 0.6, in Mettl1KO2) (Supplementary Fig. 9d–f), indicating that ribosome stalling is more prominent in testis compared with non-gonadal tissues. Notably, Nanos-driven Mettl1 failed to rescue the status of m7G modification in body samples (Fig. 7e) but recovered the 3′ shorter fragment and tRNA Pro abundance in testis (Fig. 7e, f), further supporting the critical role of Mettl1 in male fertility via a cell-autonomous mechanism (Fig. 2g).
Discussion
Here, we addressed the essential role of Mettl1 in the formation of elongated spermatids and male fertility through m7G tRNA modification. We found that ribosome pausing occurs at Pro and Val codons decoded by m7G-modified tRNAs in Mettl1-KO testes. Consistent with defective elongated spermatid formation in Mettl1-KO testes, Mettl1 might play an important role in mRNA translation involved in spermatid development, sperm axoneme assembly and sperm individualization (Fig. 7c). During late-stage spermatogenesis, chromatin modifiers cause chromosome compaction to shut down transcription. This stage therefore relies on translation of stored mRNAs, which is regulated by RNA binding proteins58,59. Our data show that epitranscriptomic regulation of tRNA, m7G modification, also plays a critical role during this process. Furthermore, apart from the fertility phenotype, no other developmental defect was observed despite the presence of Mettl1 and m7G modified tRNAs in non-gonadal tissues. This indicates that different tissues vary in their susceptibility to abnormal development in response to changes in m7G modification.
The finding that Mettl1-KO resulted in decreased expression of the majority, but not all, m7G-modified tRNAs (Fig. 5a, b) was consistent with previous studies in mammals and yeast10,22,27. Critical components of the yeast RTD pathway, Rat1/Dhp1, Met22, and Xrn1, are all present in higher eukaryotes60. Therefore, the molecular machinery reported in the yeast RTD pathway may also function in other organisms, although this has not been confirmed in animals. eEF1α1 overexpression partially rescued Mettl1-KO fertility, which implicates a similar system working in animals. Interestingly, our data showed the m7G-modified tRNA species to be largely similar between Drosophila and mammals, although there were some differences in Mettl1 dependency on tRNA stability. Although m7G-modified tRNAs have a RAGGU motif (Fig. 4f) that is similar to a motif described in a previous study10, our TRAC-seq analysis showed that tRNA ArgTCT is categorized as m7G-modified in testis but not in ovary. This difference might be caused by different read numbers used to calculate cleavage score. Thus, except for ArgTCT, our analysis showed that 22 m7G-modified tRNAs were observed in both testis and ovary (Fig. 4e, Supplementary Fig. 6d), indicating that the repertoire of m7G-modified tRNAs is largely consistent among tissues. Moreover, we found that m7G-modified tRNA PheGAA was stable in Drosophila Mettl1-KO (Fig. 5a, b). This is clearly different from that in observed in METTL1-depleted mammalian cells10,27. Therefore, it would be interesting to conduct a similar analysis in various animals to determine common and different features among m7G-modified tRNAs. The RTD in yeast acts on tRNAs lacking one or more of several different modifications49; therefore, we speculate that these differences might be caused by varying combinations of other tRNA modifications. Indeed, a subset of hypomodified tRNAs is reduced in Drosophila adult flies lacking cytosine-5 tRNA methyltransferases61; therefore, it is likely that tRNA decay can occur in diverse cell types, depending upon the combinations of RNA modifications.
It was surprising to find that the only obvious abnormality in Mettl1-KO flies was restricted to fertility because Mettl1 is ubiquitously expressed and m7G modification of tRNAs also occurs in nongonadal tissues (Fig. 7e). Notably, there is no post-meiotic transcription in Drosophila testes, which is conserved in mammals62. Therefore, proteins involved in spermatid differentiation must be translated from previously produced mRNAs that are stored in a translationally repressed state. During this process, numerous mRNAs are translated and spermatids undergo dynamic morphological changes, which include nuclear condensation and axoneme formation, without supplementation of fundamental translation factors, such as translation elongation factors. Based on our analysis of eEF1α1, we speculate that low eEF1α1 expression may sensitize testes more than other tissues to Mettl1 deficiency. However, we have not reached a definitive conclusion for this important point. It is therefore necessary to understand how hypomodified tRNAs are destabilized in testes. We also showed that Mettl1-dependent m7G methylation requires Wh in vitro and that m7G modification is lost in wh mutants (Figs. 3a, 4c, Supplementary Figs. 4, 5a–c), indicating that a collaborative role between Mettl1 and Wh is critical for spermatogenesis. However, we do not rule out the possibility that a m7G-independent function of Mettl1 or Wh causes part of the observed phenotypes.
Notably, it remains controversial whether METTL1 depletion affects global translation efficiency. METTL1 depletion in human glioblastoma LNZ308 cells27, HuCCT1, and RBE cells26, and in mouse ES cells10, reduces global translation efficiency. In contrast, METTL1-depletion did not affect overall translation levels in A549, HepG2, and HeLa cells12,15,28. Considering that initiator tRNA was classified into group 1 tRNA in our analysis, the reduction of initiator tRNA might be presumed to affect global ribosome initiation. However, the abundance of iMet tRNA was only modestly affected in Mettl1-KO flies (Fig. 5b). Therefore, it remains unclear to what extent this change can explain the reduction of TE (Fig. 7a) and the observed phenotypes (Fig. 2) in Mettl1-KO flies. Ribosome profiling data indicate that ribosome collisions occur at codons decoded by group 1 tRNAs (Fig. 6d, e). There appeared to be a weak correlation between codon use of group 1 tRNAs and changes in TE (Fig. 7b); however, that correlation was not supported in codon frequency analysis limited to the Pro codons (Supplementary Fig. 9a). Currently, we do not yet have a clear answer to why we could not detect a correlation between the Pro codon frequency and changes in TE, however, it would be interesting to analyze whether a small number of Pro codons are sufficient to induce ribosome collisions and/or whether the position of Pro codons affects TE.
The GO analysis of cellular component terms of genes that were down-regulated in Mettl1-KO testes revealed that Mettl1 is critical for translation of mRNAs involved in elongated spermatid and mature sperm formation, which is supported by our experimental observations (Fig. 2). Analysis of GO biological process terms of genes that were down-regulated in Mettl1-KO testes revealed that Mettl1 is critical for the translation of mRNAs involved in the catabolism of fructose, which acts as an energy source for spermatozoa63. These data indicate that Mettl1-KO severely impairs protein synthesis from a subset of mRNAs during late rather than early stages of spermatogenesis, resulting in male sterility. We did not find any enrichment or depletion of the conserved m7G motif in genes with Mettl1-dependent TE (data not shown), indicating TE changes caused by decreased tRNA abundance rather than direct m7G modification of mRNAs are likely to account for the spermatogenesis defects in Mettl1-KO testes.
Interestingly, our ribosome profiling analyses showed increased translation efficiency for 50 transcripts (Fig. 7a). A recent report revealed that internal m7G-modified mRNAs with GA-rich motifs are directed to stress granules via Quaking proteins in human cells28. Stress granules contain non-translating mRNAs64; therefore, the high TE in Mettl1-KO testes might result from depression of stress granule-stored, m7G-modified mRNAs.
Activation of Mettl1 can occur in cancer cells, resulting in stabilization of tRNA (ArgTCT4-1) and increased translation of cell-cycle gene mRNAs, such as CDK6 and others, which promotes tumor progression26,27. Notably, not only METTL1/WDR4 but also eEF1α1 expression is elevated in hepatocellular carcinoma cell lines and clinical samples65, indicating that tRNA stability (ArgTCT4-1) might be synergistically increased in hepatocellular carcinoma cells. Tumor progression depends on the promotion of tRNA (ArgTCT4-1) methylation by METTL1; therefore, modulating tRNA levels in some way might attenuate tumor progression. A combination of drugs targeting the tRNA decay machinery may enhance Mettl1 inhibitor effects.
Methods
Fly stocks
A complete list of fly stocks used in this study is presented in Supplementary Table 2. All stocks were maintained at 25 °C on standard medium. Mettl1-knockout (Mettl1-KO) lines were generated using the transgenic CRISPR-Cas9 method66. To generate mutants, we used the following strains: y1 v1 nos-phiC31; attP40 host (NIG-FLY stock TBX-0002), y2 cho2 v1; Sp hs-hid/CyO (NIG-FLY stock TBX-0009), y1 w1118; +; attP2{nos-cas9}67, and Df(1)JA27/FM7c, P{w[+mC]=GAL4-Kr.C}DC1, P{w[+mC]=UAS-GFP S65T}DC5, sn[+] (Bloomington Drosophila Stock Center #5193). Disruption of the Mettl1 region in mutants was validated by PCR and Sanger sequencing. All crosses were performed at 25 °C. Oligonucleotide sequences for Mettl1-KO line generation are shown in Supplementary Table 3. To confirm the requirement of Wh in Mettl1-dependent m7G methylation, we used the following strain: w* wuho56/FM7c, sn+ (Bloomington Drosophila Stock Center #41112). w* / Y; P{Dj-GFP.S}AS1 / CyO (Bloomington Drosophila Stock Center #5417) was used to validate expression of Dj (don juan) in testes. To generate yw; Vasa-GFP and Mettl1KO1; [Vasa-GFP] males (Supplementary Fig. 9b), we used the following strain: w*; TI{TI}vasEGFP.KI (KYOTO Stock Center 118616).
Fertility assay
To test male or female fertility, a single fly was mated with three control (y1 w1118) opposite sex flies at 25 °C for three or 10 days. After mating, the parental flies were removed, and incubation continued for 11 (male) or 4 days (female). Emerged adult flies (progeny) were counted and the average number per vial calculated and evaluated by Tukey’s HSD test. Ten independent vials for each strain were prepared. Each fertility assay was reproduced for at least 2–3 times. The complete list of flies used in these assays is shown in Supplementary Table 2.
Climbing assay
A climbing assay was performed as described previously68. Groups of 20 adult male flies (3-days after eclosion) were collected using CO2 anesthesia and then transferred into a 250 ml graduated glass cylinder. The flies were allowed 120 s to climb past the 200 ml line (17.5 cm from the cylinder bottom). The number of flies above the 200 ml line at 120 sec was counted and the ratio of flies able to climb the cylinder calculated. Five separate assays were performed and subjected to Tukey’s HSD test.
Weight analysis
Groups of 10 adult male flies were weighed on an electronic balance. Three replicates were performed for each strain (y1w1118 and Mettl1KO1). Two-tailed Student’s t-test was performed for statistical analysis.
Courtship assay
All flies were collected within 8 h after eclosion and kept in vials filled with Drosophila standard yeast-based food at 25 °C in 40–60% relative humidity under a 12-h light/dark cycle. Male flies were maintained individually. Virgin females were maintained in groups of 10–30 flies. y1w1118 females were used as mating partners for male flies. Males and females 5–8 days after eclosion were used for video recordings. All video recordings were performed during the light phase of the light/dark cycle. Video annotation software BORIS was used for analysis69.
To quantify the copulation rate, a pair of male and female flies were introduced into each artificial chamber (diameter 10 mm, height 4 mm, bottom coated with Drosophila standard food), and video recording was performed using a CMOS camera (DFK 33UP1300, The Imaging Source Asia Co., Ltd) equipped with a zoom lens (M0814-MP2, CBC Optics Co., Ltd.) for 24 h at one fps. y1w1118, Mettl1KO1, or tut-KO males were used to serve as the test flies. We calculated the copulation rate by determining whether each pair had copulated or not in 24 hours.
Measurement of adult progeny emerging rate
Test crosses were performed using heterozygous female (Mettl1KO1/FM7 or y1w1118/FM7) and wild-type male flies. One virgin female and three males were crossed and removed 5 days after mating. Emerged offspring were counted 14 days after mating and then evaluated by two-tailed Student’s t-test.
Western blotting
Western blotting was performed as described previously70. Ovaries were collected from 3–4-day-old adult females. An anti-Mettl1 monoclonal antibody, an anti-Wh monoclonal antibody (both generated in this study), and an anti-Tubulin antibody [E7 culture supernatant, Developmental Studies Hybridoma Bank] were used as primary antibodies. A goat anti-mouse IgG (H + L) Poly-HRP secondary antibody (Thermo Fisher, USA, #32230, 1:5000–10000) was used as secondary antibody.
Measurement of ovary size
Ovaries were dissected from 7-day-old adult females. Ovaries were fixed in 4% paraformaldehyde phosphate buffer solution (Nacalai Tesque, Japan) for 15 min at room temperature or overnight at 4 °C. After washing with PBS-T three times, ovary size was measured using a stereomicroscope and cellSens (Olympus, Japan) imaging software. Significant differences in ovary size were evaluated by Tukey’s HSD test.
Immunofluorescence
Immunostaining analysis was conducted as described previously71. Testes from 3-day-old adult males and ovaries from 7-day-old adult females were dissected in ice-cold PBS. Tissues were fixed in 4% paraformaldehyde phosphate buffer solution (Nacalai Tesque #09154-85) for 15 min at room temperature. The fixed tissues were washed five times with PBS containing 0.1% Tween 20 (PBS-T), and then incubated twice in PBS-T containing 0.2% Triton X-100 and 0.1% BSA (PBS-BT) for 10 min. The washed tissues were then incubated with primary antibodies in PBS-BT at 4 °C for 1 day. Primary antibodies used were: anti-Aub (4D10)72, anti-acetylated tubulin (Sigma, 1:100 dilution), anti-boule (1:250 dilution, a kind gift from S. Wasserman, University of California San Diego), anti-FLAG (M2, Sigma, 1:1000 dilution; to detect Mettl1-3×Flag), anti-Fas3 (7G10, Developmental Studies Hybridoma Bank), and anti-Tj (10009Ab-1, NIG-FLY, Japan). Incubated tissues were then washed five times and further washed twice with PBS-BT. Next, the tissues were incubated at 4 °C for 1 day with the following secondary antibodies as appropriate: Alexa 546-conjugated anti-mouse IgG (Thermo Fisher, A11030, 1:250 dilution), Alexa 488-conjugated anti-mouse IgG (Thermo Fisher, A11029, 1:250 dilution), Alexa 488-conjugated anti-rabbit IgG (Thermo Fisher, A11008, 1:250–1000 dilution), and Alexa 546-conjugated anti-rabbit IgG (Thermo Fisher, A11071, 1:250–1000 dilution). After washing three times for 10 min in PBS-T, tissues were mounted in Vectashield Mounting Medium containing DAPI (Vector Laboratories). Images of testes and ovaries were taken using a confocal microscope (Zeiss LSM900). 405 nm or 488 nm laser were used for excitation. 400–505 nm and 505–700 nm filters were used for fluorescence collection. Seminal vesicles and elongated spermatids in testes were fixed by the same method and observed by light field confocal microscopy (Zeiss LSM900). Using independent fluorescence channels, we validated that the green signals in Fig. 2b, d, f did not come from autofluorescence of the Drosophila tissue (data not shown).
Production of monoclonal antibodies
Recombinant full-length Drosophila Mettl1 protein tagged with maltose binding protein and Wh (N-terminal 200 amino acids) tagged with glutathione S-transferase (GST) were prepared as antigens and purified from E. coli. Mice were immunized with each antigen. The production and selection of hybridomas were performed as previously described73.
Plasmid construction
To produce plasmids expressing Drosophila Mettl1, Mettl1-Catalytic dead (Cd), Mettl1-3×Flag, nosp-Mettl1, UASt-Mettl1, and UASt-eEF1α1, Mettl1 and eEF1α1 coding regions were amplified using KOD plus DNA polymerase (Toyobo #KOD-201). The Nanos promoter region was amplified from pBFv-nosp-Cas9. The amplified fragments and vector pBFv-UAS3 were fused using the In-Fusion® HD Cloning Kit (TaKaRa). The oligonucleotide sequences used are shown in Supplementary Table 2. To produce the plasmids expressing Mettl1 and Wh in E. coli, Mettl1 and Wh coding sequences were amplified using KOD plus DNA polymerase (Toyobo) and were then integrated into pGEX 5x-1 (Amersham), or pMAL (NEB). The oligonucleotide sequences used are shown in Supplementary Table 3.
GST pull-down assays
GST pull-down assays were performed as previously described74. Purified GST-tagged proteins were incubated in methylation buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.2 mM DTT, 10 mM EDTA, 0.2 mM MgCl2) with Glutathione Sepharose™ 4B (GE healthcare 17-0756-01) for 1 h at 4 °C to immobilize GST-tagged proteins. Immobilized proteins were washed three times with methylation buffer. After washing, MBP or MBP-tagged protein was added and incubated for 4 h at 4 °C. After incubation, proteins were washed five times with methylation buffer, and then eluted with 2×Sample buffer [100 mM Tris-HCl (pH 6.8), 4% (w/v) SDS, 0.2% (w/v) bromophenol blue, 20% (v/v) glycerol, and 200 mM DTT]. Samples were incubated for 5 min at 95 °C, and were then resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The resolved proteins were detected by Coomassie brilliant blue staining.
Preparation of protein samples for LC-MS/MS analysis
To prepare immunoprecipitated samples for LC-MS/MS, the anti-3×FLAG antibody used for immunoprecipitation was cross-linked to beads using dimethyl pimelimidate (Thermo Fisher). Immunoprecipitation was performed using mouse anti-FLAG M2 or mouse non-immune IgG (negative control) and ovaries from negative control (Oregon R) or 3×Flag-tagged Mettl1 (attp40[Mettl1-Flag]) flies. We used two different buffer conditions: Tris-Triton buffer [20 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 0.1% TritonX-100] and Hepes-NP40 buffer [30 mM Hepes-KOH (pH 7.3), 150 mM KOAc, 5 mM MgOAc, 5 mM DTT, 0.1% NP40]. LC-MS/MS analysis was performed as described previously75. The immunoprecipitated proteins were separated on 4–12% SDS-PAGE gels and stained with SimplyBlue (Thermo Fisher). Gel containing proteins was excised and cut into pieces of approximately 1 mm2. Proteins in the gel pieces were reduced with DTT (Thermo Fisher), alkylated with iodoacetamide (Thermo Fisher), and digested with trypsin and lysyl endopeptidase (Promega) in a buffer containing 40 mM ammonium bicarbonate, pH 8.0, overnight at 37 °C. The resultant peptides were analyzed on an Advance UHPLC system (AMR/Michrom Bioscience) connected to a Q Exactive mass spectrometer (Thermo Fisher). The raw mass spectrum was processed using Xcalibur (Thermo Fisher). The raw LC-MS/MS data were analyzed against the UniprotKB database restricted to Drosophila melanogaster using Proteome Discoverer version 1.4 (Thermo Fisher) with the Mascot search engine version 2.5 (Matrix Science). A decoy database comprising either randomized or reversed sequences in the target database was used for false discovery rate (FDR) estimation, and Percolator was used to evaluate false positives. Search results were filtered against 1% global FDR for high confidence level. Quantitative analyses were performed using scaffold 5 (Proteome Software Inc, Portland, OR), and volcano plots of identified proteins were generated using Student’s t-test or Fisher’s exact test under a normalization scheme. Single replicate was analyzed following to standards in the field.
Northern blotting
Northern blotting was performed as described previously76. Sample RNAs were denatured with Gel loading buffer II (Invitrogen #AM8546G) for 3 min at 65 °C. Sample RNAs were resolved by electrophoresis on 12% urea-PAGE gels, which contain 7 M urea, 10× TBE (1 M Tris base, 1 M boric acid, 0.02 M EDTA), and 12% acrylamide. To assess m7G site-specific cleavage of tRNAs and to confirm tRNA levels, 1.25 μg (from ovaries) and 0.5 μg (from testes) sample RNAs were resolved by electrophoresis and then transferred to Hybond N+ membranes (Cytiva). Transferred RNAs were UV-crosslinked with 1200 × 100 mJ/cm2 irradiance at 254 nm. After crosslinking, hybridization was conducted at 42 °C overnight in 7% SDS, 0.2 M sodium phosphate (pH 7.2), and 1 mM EDTA with 32P-labeled DNA probes. Membranes were then washed with 2× saline sodium citrate containing 0.1% SDS at 42 °C. To detect in vitro m7G methylation, the hybridization step was skipped. The oligonucleotide sequences used are shown in Supplementary Table 3.
In vitro methylation assay
GST, GST-Wh, MBP, MBP-Mettl1, and MBP-Mettl1-Cd were produced and purified from E. coli. An in vitro tRNA methylation assay was performed essentially as described previously with some modifications77. Purified GST and MBP protein (1 μg each) were incubated with chemically synthesized RNA (0.3 nmol) at 26 °C in a buffer containing 50 mM Tris-HCl (pH 8.0), 100 mM KCl, 5 mM MgCl2, 2 mM DTT and 2 μCi/mL S-adenosyl-L-[methyl-14C] methionine (PerkinElmer) for 3 h. RNAs were then isolated with phenol:chloroform and ethanol precipitated. The resultant RNAs were separated on 7 M urea-containing denaturing polyacrylamide gels. The gels were stained with Toluidine Blue O in 0.5 × TBE, destained with 0.5 × TBE, and dried. The 14C-labeled RNA bands were visualized on a Typhoon FLA900 (GE Healthcare).
m7G site-specific reduction and cleavage
Total RNA from ovaries and testes were isolated using Isogen (Nippon Gene, 311-02501) following the manufacturer’s instructions. Ten micrograms of total RNA were incubated with 0.1 M NaBH4 and 1 mM free m7GTPfor 30 min on ice in the dark. Reduced RNAs were precipitated with 3 M sodium acetate pH 5.2 (Thermo Fisher), glycogen (Nacalai Tesque) and ethanol at −20 °C for at least 1 h. RNA samples reduced by NaBH4 were then reacted with aniline-acetate solution (H2O:glacial acetate acid:aniline, 7:3:1) at room temperature in the dark for 2 h to cause m7G site-specific cleavage46,47. The cleaved RNAs were detected by northern blotting. Total RNA from Wh-KO males was isolated from 3-day-old adult male testes of w* wh56/FM7c sn+ flies (Bloomington Drosophila Stock Center #5417).
TRAC-seq and profiling of tRNA expression
Sequencing and data analysis were performed as previously described48 with some modifications. Small RNAs (<200 nt) were isolated from whole ovary (one sample) and testis (three replicates) RNAs using a mirVana miRNA Isolation Kit (Thermo Fisher, AM1561) following the manufacturer’s instructions. To exclude methylation inhibiting the reverse transcription of tRNAs, 2.5 μg of extracted small RNAs were treated with recombinant ALKB and ALKB D135S proteins. Demethylated small RNAs were reduced and cleaved by the method described in the “m7G site-specific reduction and cleavage” section of the manufacturer’s protocol. After cleavage and ethanol precipitation, small RNA sequencing libraries of m7G tRNAs were constructed using a NEBNext Multiplex Small RNA Library Prep Set for Illumina (NEB, E7300S). In preparing the library, TGI-RT (Thermostable Group II intron reverse transcriptase, TGIRT-III-10, InGex) was used instead of the reverse transcriptase in the library kit. The prepared libraries were sequenced on a Novaseq 6000 system. The 290 reference sequences of mature Drosophila tRNAs (i.e., dm6-mature-tRNAs) were downloaded from GtRNAdb78. Identical sequences were removed to yield 84 unique sequences and a “CCA” sequence was added to the 3′-end of each tRNA (i.e., “dm6-mature-tRNAs_unique_CCA”). The 5′- and 3′-adapter sequences of the sequenced reads were trimmed and low-quality sequences were filtered using FASTP ver. 0.23.279 to obtain “clean reads”. The clean reads were mapped to the reference “dm6-mature-tRNAs_unique_CCA” using BOWTIE ver. 1.3.180. The number of reads mapped on each tRNA (i.e., “TotalReads”) was obtained from the mapping data (bam) using PICARD ver. 3.0.0 (https://broadinstitute.github.io/picard/command-line-overview.html). The number of reads starting at each site of each tRNA (i.e., “StartReads”) was obtained using the custom shell script. The ratio between StartReads and TotalReads as an indicator of the m7G-dependent read cleavage (i.e., “StartRatio”) was calculated for each site i of each tRNA of chemically-treated and non-treated samples as:
The cleavage score as an overall indicator of the m7G modification (i.e., “CleavageScore”) was calculated for each site i of each tRNA of WT and Mettl1-KO samples as:
The difference in cleavage score between WT and Mettl1-KO samples (i.e., “Cleavage score”) was defined as another indicator of Mettl1-dependent m7G modification as:
For testes, these feature values were averaged from the data of three replicates. The sites with StartRatioWT,treat ≥ 0.02 were regarded as candidates. Those with CleavageScorei,WT ≥ 2 and Cleavage scorei ≥ 2 were extracted as the final candidate m7G-dependent cleavage sites. The sequences of ± 10 bases around the candidate m7G site were analyzed to detect the conserved motif for the m7G modification using MEME ver. 5.5.281.
The chemically non-treated testis samples for TRAC-seq (WT: three replicates; Mettl1-KO: three replicates) were processed to profile tRNA expression using the tRAX pipeline82.
Acid urea PAGE
Acid urea PAGE was performed as previously described83. Briefly, total RNAs were treated with 0.1 M Tris pH 9.0 at 42 °C for 1 hr. Treated and control RNA samples were then separated by acid urea PAGE.
Ribosome footprinting and RNA-Seq
Cell lysates were prepared as previously reported with modifications84. Testes or male body (with testis removed) were dissected from 2–3-day-old adult males in ice-cold PBS containing 100 µg/ml cycloheximide. Dissected testes were immediately frozen in liquid nitrogen and stored at −80 °C. We prepared 600 pairs of testes per sample and two replicates from wild-type (yw) and Mettl1-KO (Mettl1KO1) males, respectively. Male bodies were prepared from 100 male adults for each strain. Frozen testes or body samples were mixed with 300 µl of frozen lysis buffer droplets (20 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 1% Triton X-100, 100 µg/ml chloramphenicol, and 100 µg/ml cycloheximide), and then ground at 3000 rpm for 15 s in a Multi-beads Shocker (YASUI KIKAI). The lysate was slowly thawed at 4 °C and treated with 10 U Turbo DNase (Thermo Fisher) on ice for 10 min to remove genome DNA. The supernatant was further centrifuged at 20,000 g for 30 min at 4 °C.
Library construction of ribosome profiling (or Thor-Ribo-Seq) was conducted as described previously54,84. Cell lysate containing 8.5 µg total RNA was treated with 20 U RNase I (Epicenter) in a 300 µl reaction at 25 °C for 45 min. Then, the reaction was stopped with 200 U of SUPERase•In RNase Inhibitor (Thermo Fisher). Ribosomes were isolated by sucrose cushion ultracentrifugation at 100,000 rpm at 4 °C for 1 h using a TLA110 rotor (Beckman Coulter). RNAs in the ribosome pellet were isolated with TRIzol-LS (Thermo Fisher) and a Direct-zol RNA MicroPrep kit (Zymo Research). Purified RNA fragments ranging from 17 to 34 nt were selected by gel electrophoresis, dephosphorylated by T4 polynucleotide kinase (New England Biolabs), and ligated to preadenylated linkers containing the T7 promoter region using T4 RNA ligase 2, truncated KQ (New England Biolabs). Before ligation, the linkers were preadenylated by Mth RNA Ligase (NEW England Biolabs). Linker-ligated RNAs were treated with riboPOOL for D. melanogaster (siTOOLs Biotech) to remove fragments of ribosomal RNAs. In vitro transcription was then performed using a T7-Scribe Standard RNA IVT Kit (CELLSCRIPT). RNAs were dephosphorylated by T4 polynucleotide kinase (New England Biolabs) and ligated with linker for reverse transcription. The cDNAs were synthesized by ProtoScript II (New England Biolabs) and amplified using Phusion High-Fidelity DNA Polymerase. The DNA libraries were sequenced on the HiSeq X platform (Illumina) with the paired-end 150-bp option.
For RNA-Seq, we extracted total RNA from the lysate originally used for ribosome profiling using TRIzol LS (Thermo Fisher) and a Direct-zol RNA MicroPrep Kit (Zymo Research). Sequencing libraries were prepared using 1 µg of the total RNA and the SEQuoia Express Standard RNA Library Prep Kit (Bio-Rad). rRNA depletion was performed with riboPOOL for D. melanogaster (siTOOLs Biotech). Sequence data were processed using previously established methods85,86. Translation efficiency change was determined using the DESeq2 package87. Gene ontology (GO) analysis was performed with DAVID88,89.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon request. The original raw data in this study are publicly available. The mass analyses data generated in this study have been deposited in ProteomeXchange (PXD044501) / jPOST repository (JPST002287). The TRAC-Seq data have been deposited in the DNA Data Bank of Japan (DDBJ) (PRJDB16512). The ribosome profiling, and RNA-Seq data obtained in this study are deposited in the National Center for Biotechnology Information (NCBI) database (GSE241519). Source data are provided with this paper.
Code availability
The codes for data analysis of TRAC-seq and tRNA expression are available in Zenodo (https://doi.org/10.5281/zenodo.13340795)90. Source codes of softwares for ribosome profiling analysis were shared at GitHub (https://github.com/ingolia-lab/RiboSeq). The codes deposited in Zenodo (https://doi.org/10.5281/zenodo.7477706)91 were used for data analyses of ribosome footprinting.
References
Boccaletto, P. et al. MODOMICS: a database of RNA modification pathways. 2021 update. Nucleic Acids Res. 50, D231–D235 (2022).
Roundtree, I. A., Evans, M. E., Pan, T. & He, C. Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017).
Pan, T. Modifications and functional genomics of human transfer RNA. Cell Res. 28, 395–404 (2018).
Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 22, 375–392 (2021).
Blaze, J. & Akbarian, S. The tRNA regulome in neurodevelopmental and neuropsychiatric disease. Mol. Psychiatry 27, 3204–3213 (2022).
Chujo, T. & Tomizawa, K. Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J. 288, 7096–7122 (2021).
Orellana, E. A., Siegal, E. & Gregory, R. I. tRNA dysregulation and disease. Nat. Rev. Genet 23, 651–664 (2022).
Furuichi, Y. Discovery of m(7)G-cap in eukaryotic mRNAs. Proc. Jpn Acad. B Phys. Biol. Sci. 91, 394–409 (2015).
Chu, J. M. et al. Existence of internal N7-methylguanosine modification in mRNA determined by differential enzyme treatment coupled with mass spectrometry analysis. ACS Chem. Biol. 13, 3243–3250 (2018).
Lin, S. et al. Mettl1/Wdr4-mediated m(7)G tRNA methylome is required for normal mRNA translation and embryonic stem cell self-renewal and differentiation. Mol. Cell 71, 244–255.e5 (2018).
Malbec, L. et al. Dynamic methylome of internal mRNA N(7)-methylguanosine and its regulatory role in translation. Cell Res. 29, 927–941 (2019).
Pandolfini, L. et al. METTL1 promotes let-7 MicroRNA processing via m7G methylation. Mol. Cell 74, 1278–1290.e9 (2019).
Sprinzl, M., Horn, C., Brown, M., Ioudovitch, A. & Steinberg, S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998).
White, J. et al. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol. Cell Biol. 28, 3151–3161 (2008).
Zhang, L. S. et al. Transcriptome-wide Mapping of Internal N(7)-Methylguanosine Methylome in Mammalian mRNA. Mol. Cell 74, 1304–1316.e8 (2019).
Tomikawa, C. 7-Methylguanosine Modifications in Transfer RNA (tRNA). Int. J. Mol. Sci. 19, 4080 (2018).
Alexandrov, A., Martzen, M. R. & Phizicky, E. M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 8, 1253–1266 (2002).
Alexandrov, A., Grayhack, E. J. & Phizicky, E. M. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 11, 821–830 (2005).
Leulliot, N. et al. Structure of the yeast tRNA m7G methylation complex. Structure 16, 52–61 (2008).
Li, J. et al. Structural basis of regulated m(7)G tRNA modification by METTL1-WDR4. Nature 613, 391–397 (2023).
Ruiz-Arroyo, V. M. et al. Structures and mechanisms of tRNA methylation by METTL1-WDR4. Nature 613, 383–390 (2023).
Alexandrov, A. et al. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 21, 87–96 (2006).
Chernyakov, I., Whipple, J. M., Kotelawala, L., Grayhack, E. J. & Phizicky, E. M. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5’−3’ exonucleases Rat1 and Xrn1. Genes Dev. 22, 1369–1380 (2008).
Whipple, J. M., Lane, E. A., Chernyakov, I., D’Silva, S. & Phizicky, E. M. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev. 25, 1173–1184 (2011).
Watanabe, K. et al. Degradation of initiator tRNAMet by Xrn1/2 via its accumulation in the nucleus of heat-treated HeLa cells. Nucleic Acids Res. 41, 4671–4685 (2013).
Dai, Z. et al. N(7)-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 81, 3339–3355 e8 (2021).
Orellana, E. A. et al. METTL1-mediated m(7)G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 81, 3323–3338.e14 (2021).
Zhao, Z. et al. QKI shuttles internal m(7)G-modified transcripts into stress granules and modulates mRNA metabolism. Cell 186, 3208–3226.e27 (2023).
Braun, D. A. et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med Genet A 176, 2460–2465 (2018).
Shaheen, R. et al. Mutation in WDR4 impairs tRNA m(7)G46 methylation and causes a distinct form of microcephalic primordial dwarfism. Genome Biol. 16, 210 (2015).
Trimouille, A. et al. Further delineation of the phenotype caused by biallelic variants in the WDR4 gene. Clin. Genet 93, 374–377 (2018).
Hadjigeorgiou, G. M. et al. Replication study of GWAS risk loci in Greek multiple sclerosis patients. Neurol. Sci. 40, 253–260 (2019).
Zhao, Y. et al. m7G methyltransferase METTL1 promotes post-ischemic angiogenesis via promoting VEGFA mRNA translation. Front Cell Dev. Biol. 9, 642080 (2021).
Wang, Y. J. et al. Genetic association of the functional WDR4 gene in male fertility. J. Pers. Med. 11, 760 (2021).
Xia, X., Wang, Y. & Zheng, J. C. Internal m7G methylation: a novel epitranscriptomic contributor in brain development and diseases. Mol. Ther. Nucleic Acids 31, 295–308 (2023).
Cheng, I. C. et al. Wuho is a new member in maintaining genome stability through its interaction with flap endonuclease 1. PLoS Biol. 14, e1002349 (2016).
Wu, P. R. et al. Wdr4 promotes cerebellar development and locomotion through Arhgap17-mediated Rac1 activation. Cell Death Dis. 14, 52 (2023).
Rastegari, E. et al. WD40 protein Wuho controls germline homeostasis via TRIM-NHL tumor suppressor Mei-p26 in Drosophila. Development 147, dev182063 (2020).
Wu, J., Hou, J. H. & Hsieh, T. S. A new Drosophila gene wh (wuho) with WD40 repeats is essential for spermatogenesis and has maximal expression in hub cells. Dev. Biol. 296, 219–230 (2006).
Campeanu, I. J. et al. Multi-omics integration of methyltransferase-like protein family reveals clinical outcomes and functional signatures in human cancer. Sci. Rep. 11, 14784 (2021).
Demarco, R. S., Eikenes, A. H., Haglund, K. & Jones, D. L. Investigating spermatogenesis in Drosophila melanogaster. Methods 68, 218–227 (2014).
Maines, J. Z. & Wasserman, S. A. Post-transcriptional regulation of the meiotic Cdc25 protein Twine by the Dazl orthologue Boule. Nat. Cell Biol. 1, 171–174 (1999).
Riparbelli, M. G., Persico, V. & Callaini, G. The microtubule cytoskeleton during the early drosophila spermiogenesis. Cells 9, 2684 (2020).
Chen, D. et al. Three RNA binding proteins form a complex to promote differentiation of germline stem cell lineage in Drosophila. PLoS Genet 10, e1004797 (2014).
Matsumoto, K. et al. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett. 581, 1599–1604 (2007).
Wintermeyer, W. & Zachau, H. G. Tertiary structure interactions of 7-methylguanosine in yeast tRNA Phe as studied by borohydride reduction. FEBS Lett. 58, 306–309 (1975).
Zueva, V. S., Mankin, A. S., Bogdanov, A. A. & Baratova, L. A. Specific fragmentation of tRNA and rRNA at a 7-methylguanine residue in the presence of methylated carrier RNA. Eur. J. Biochem. 146, 679–687 (1985).
Lin, S., Liu, Q., Jiang, Y. Z. & Gregory, R. I. Nucleotide resolution profiling of m(7)G tRNA modification by TRAC-Seq. Nat. Protoc. 14, 3220–3242 (2019).
Dewe, J. M., Whipple, J. M., Chernyakov, I., Jaramillo, L. N. & Phizicky, E. M. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 18, 1886–1896 (2012).
Turowski, T. W., Karkusiewicz, I., Kowal, J. & Boguta, M. Maf1-mediated repression of RNA polymerase III transcription inhibits tRNA degradation via RTD pathway. RNA 18, 1823–1832 (2012).
Toledano, H., D’Alterio, C., Czech, B., Levine, E. & Jones, D. L. The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 485, 605–610 (2012).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Iwasaki, S. & Ingolia, N. T. The growing toolbox for protein synthesis studies. Trends Biochem Sci. 42, 612–624 (2017).
Mito, M., Shichino, Y. & Iwasaki, S. Thor-Ribo-Seq: ribosome profiling tailored for low input with RNA-dependent RNA amplification. bioRxiv, 2023.01.15.524129 (2023).
Hempel, L. U., Rathke, C., Raja, S. J. & Renkawitz-Pohl, R. In Drosophila, Don Juan and Don Juan like encode proteins of the spermatid nucleus and the flagellum and both are regulated at the transcriptional level by the TAF II80 cannonball while translational repression is achieved by distinct elements. Dev. Dyn. 235, 1053–1064 (2006).
Vasudevan, D. et al. Translational induction of ATF4 during integrated stress response requires noncanonical initiation factors eIF2D and DENR. Nat. Commun. 11, 4677 (2020).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Idler, R. K. & Yan, W. Control of messenger RNA fate by RNA-binding proteins: an emphasis on mammalian spermatogenesis. J. Androl. 33, 309–337 (2012).
Linn, E., Ghanem, L., Bhakta, H., Greer, C. & Avella, M. Genes regulating spermatogenesis and sperm function associated with rare disorders. Front Cell Dev. Biol. 9, 634536 (2021).
Nagarajan, V. K., Jones, C. I., Newbury, S. F. & Green, P. J. XRN 5’–>3’ exoribonucleases: structure, mechanisms and functions. Biochim Biophys. Acta 1829, 590–603 (2013).
Genenncher, B. et al. Mutations in cytosine-5 tRNA methyltransferases impact mobile element expression and genome stability at specific DNA repeats. Cell Rep. 22, 1861–1874 (2018).
Barreau, C., Benson, E., Gudmannsdottir, E., Newton, F. & White-Cooper, H. Post-meiotic transcription in Drosophila testes. Development 135, 1897–1902 (2008).
Mukai, C. & Okuno, M. Glycolysis plays a major role for adenosine triphosphate supplementation in mouse sperm flagellar movement. Biol. Reprod. 71, 540–547 (2004).
Buchan, J. R. & Parker, R. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 (2009).
Chen, S. L. et al. eEF1A1 overexpression enhances tumor progression and indicates poor prognosis in hepatocellular carcinoma. Transl. Oncol. 11, 125–131 (2018).
Kondo, S. & Ueda, R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195, 715–721 (2013).
Kondo, S. et al. Neurochemical organization of the drosophila brain visualized by endogenously tagged neurotransmitter receptors. Cell Rep. 30, 284–297.e5 (2020).
Madabattula, S. T. et al. Quantitative analysis of climbing defects in a Drosophila model of neurodegenerative disorders. J. Vis. Exp. 100, e52741 (2015).
Friard, O., Gamba, M. & Fitzjohn, R. BORIS: a free, versatile open‐source event‐logging software for video/audio coding and live observations. Methods Ecol. Evol. 7, 1325–1330 (2016).
Miyoshi, K., Tsukumo, H., Nagami, T., Siomi, H. & Siomi, M. C. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 19, 2837–2848 (2005).
Saito, K. et al. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 24, 2493–2498 (2010).
Nishida, K. M. et al. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13, 1911–1922 (2007).
Nishida, K. M. et al. Respective functions of two distinct Siwi complexes assembled during PIWI-interacting RNA biogenesis in Bombyx germ cells. Cell Rep. 10, 193–203 (2015).
Saito, K. et al. Pimet, the Drosophila homolog of HEN1, mediates 2’-O-methylation of Piwi- interacting RNAs at their 3’ ends. Genes Dev. 21, 1603–1608 (2007).
Tani, N., Tanno, N. & Ishiguro, K. I. Tandem immuno-purification of affinity-tagged proteins from mouse testis extracts for MS analysis. STAR Protoc. 3, 101452 (2022).
Saito, K. et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461, 1296–1299 (2009).
Okamoto, H. et al. Substrate tRNA recognition mechanism of tRNA (m7G46) methyltransferase from Aquifex aeolicus. J. Biol. Chem. 279, 49151–49159 (2004).
Chan, P. P. & Lowe, T. M. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Langmead, B. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics Chapter 11, Unit 11 7 (2010).
Bailey, T. L. et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208 (2009).
Holmes, A. D., Howard, J. M., Chan, P. P. & Lowe, T. M. tRNA Analysis of eXpression (tRAX): a tool for integrating analysis of tRNAs, tRNA-derived small RNAs, and tRNA modifications. bioRxiv, 2022.07.02.498565 (2022).
Kohrer, C. & Rajbhandary, U. L. The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods 44, 129–138 (2008).
Mito, M., Mishima, Y. & Iwasaki, S. Protocol for disome profiling to survey ribosome collision in humans and zebrafish. STAR Protoc. 1, 100168 (2020).
Han, P. et al. Genome-wide survey of ribosome collision. Cell Rep. 31, 107610 (2020).
Kashiwagi, K. et al. eIF2B-capturing viral protein NSs suppresses the integrated stress response. Nat. Commun. 12, 7102 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2008).
Huang, D. W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009).
Center for Genome Informatics, Joint Support-Center for Data Science Research Organization of Information and Systems (ROIS). Scripts used in Kaneko et al. Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster (1.0). Zenodo (2024).
Shichino, Y., & Iwasaki, S. Custom scripts for A parasitic fungus employs mutated eIF4A to survive on rocaglate-synthesizing Aglaia plants. Zenodo https://doi.org/10.5281/zenodo.6787991 (2022).
Acknowledgements
We thank T. Suzuki for critical suggestions on this project, A. Terui, A. Masuda, Y. Kitahara, Y. Nagashima, and T. Miyoshi for technical assistance, Y. Shichino and M. Mito for technical advice in ribosome footprinting, and M. Masukawa and S. Kobayashi of the Drosophila Genetic Resource Center in Kyoto Institute of Technology and the Bloomington stock center, respectively, for providing the fly strains. We also Jeremy Allen, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript. This work was partially supported by JSPS KAKENHI under grant numbers JP18H02379, JP16H06279 (both PAGS), JP23H02415 (SI) and JP22H02669 (KS), the Takeda Science Foundation, the Yamada Science Foundation, the Mochida Memorial Foundation (KS), The Naito Science & Engineering Foundation and the Uehara Memorial Foundation (KS), AMED under grant number JP20gm1410001 (SI), and the Program of the Joint Usage/Research Center for Developmental Medicine and High Depth Omics, IMEG, Kumamoto University (KS). K.T. was a recipient of the RIKEN Student Researcher program and a World-leading Innovative Graduate Study Program in Proactive Environmental Studies (WINGS-PES) from The University of Tokyo. This study used the HOKUSAI SailingShip supercomputer facility at RIKEN.
Author information
Authors and Affiliations
Contributions
Shunya Kaneko: conceptualization, formal analysis, investigation, methodology, visualization, validation, writing-original draft, writing-review and editing. Keita Miyoshi: investigation; methodology; visualization; validation, writing-review and editing. Kotaro Tomuro: data curation, investigation; methodology; writing-review and editing. Makoto Terauchi: data curation, software; investigation; methodology; writing-original draft; writing-review and editing. Ryoya Tanaka: investigation, methodology and editing. Shu Kondo: resource; methodology and editing. Naoki Tani: LC/MS/MS; methodology and editing. Kei-Ichiro Ishiguro: Investigation; methodology and editing. Atsushi Toyoda: methodology. Azusa Kamikouchi: investigation, methodology and writing-review and editing. Hideki Noguchi: data curation, investigation; methodology; writing-original draft; writing-review and editing. Shintaro Iwasaki: data curation, investigation; methodology; writing-original draft; writing-review and editing. Kuniaki Saito: conceptualization; funding acquisition; investigation; methodology; project administration; supervision; writing-original draft; writing-review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Richard Gregory and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kaneko, S., Miyoshi, K., Tomuro, K. et al. Mettl1-dependent m7G tRNA modification is essential for maintaining spermatogenesis and fertility in Drosophila melanogaster. Nat Commun 15, 8147 (2024). https://doi.org/10.1038/s41467-024-52389-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52389-0
This article is cited by
-
Emerging roles of tRNA modification-mediated codon-specific translational reprogramming in cancer biology
Cell Death & Disease (2026)
-
Lysyl-tRNA synthetase orchestrates translation and hormone signaling during reproduction in Aedes aegypti mosquitoes
Communications Biology (2025)
-
Hallmarks and mechanisms of cellular senescence in aging and disease
Cell Death Discovery (2025)
-
Human DUS1L catalyzes dihydrouridine modification at tRNA positions 16/17, and DUS1L overexpression perturbs translation
Communications Biology (2024)