Abstract
Studying how single nucleotide polymorphisms (SNPs) crosstalk with non-autologous factors to cause complex autoimmune diseases is challenging. An amino acid replacement in the neutrophil cytosolic factor 1 (NCF1-339/NCF1R90H) leading to lower reactive oxygen species induction has been reported as the major SNP for systemic lupus erythematosus (SLE). Here we show that infection with the murine norovirus (MNV) contributes to the induction of lupus in Ncf190H mice. Mutant NCF190H upregulates the IFN-α/JAK1/STAT1 pathway in macrophages and anti-MNV-antibody production. In parallel, the MNV infection of NCF190H mice upregulates Toll-like receptor 7 in macrophages, plasmacytoid dendritic cells and B220+ splenocytes, thereby promoting germinal center formation and lupus-associated autoantibodies production. These compounded effects lead to protection against MNV infection but also glomeruloneph ritis with proteinuria and lupus arthritis in the absence of chemical inducers such as pristane. Our data thus suggest that this SLE-associated SNP, NCF190H, synergizes with MNV infection to induce the development of mouse lupus.
Similar content being viewed by others
Introduction
Systemic lupus erythematosus (SLE) is a common and complex autoimmune disease, caused by an interplay of many unknown genes and environmental factors. Yet, it has been difficult to precisely pinpoint and functionally confirm the causative factors. Thanks to genome-wide association studies (GWAS), hundreds of lupus disease-associated loci in the genome have been identified, which has strengthened the view that SLE is indeed a polygenic disease most likely regulated by variants in loci determining tolerance and immune responsiveness of the immune system1. However, the identification of the causative SNPs and the inducing environmental factors, remains largely unknown2,3. An alternative approach to identify the underlying genes has been to use genetic crosses of inbred animals susceptible to SLE. We have approached this problem by positioning the causative genes in mice and rats, and with these studies, we could identify some of the most important SNPs for complex autoimmune diseases.
The first autoimmune disease-associated gene cloned was the neutrophil cytosolic factor (Ncf1), encoding a protein (NCF1, alias p47phox) critically involved in the formation of the NADPH oxidase 2 (NOX2) complex, responsible for inducing reactive oxygen species (ROS) responses4,5. In rats, an amino acid replacing SNP (Ncf1T153M) associated with autoimmune diseases was positioned4,6, and an effect was reproduced in mice with a spontaneous Ncf1m1J mutation7. In humans, the NCF1 locus has been difficult to study due to copy number variations (CNV), but after exon sequencing a functionally important SNP could be identified8. This SNP (rs201802880 or NCF1-339, here denoted NCF1R90H) replaces arginine with histidine at position 90 (R90H), affecting NCF1 interaction with the plasma cell membrane5. Analysis of SLE cohorts showed a high odds ratio (OR) and allelic frequency for NCF1-339 (NCF190H)9,10. Importantly, the NCF190H allele caused a decreased ROS response in phorbol myristate acetate (PMA)-stimulated cells from patients with SLE in vitro, and it was also associated with an activated interferon signaling response, similar to previous findings in the Ncf1m1J mutated mice10,11,12. BALB/c mice with the Ncf1m1J mutation spontaneously showed signs of lupus with autoantibodies and glomerulonephritis and developed severe pristane-induced lupus, a commonly used mouse model of SLE12,13. The Ncf1m1J mutation was associated with the activation of interferon signaling genes, including the STAT1 pathway12. An important remaining question is why and how the interferon response, and the development of lupus, are triggered in the presence of a low ROS-producing NCF1 variant. The detected antibodies to murine norovirus (MNV) in ROS-deficient Ncf1m1J mutated mice with spontaneous lupus preliminarily suggested norovirus as a possible environmental factor12. In humans, there is no consensus on specific causative environmental challenges or virus infections, although DNA viruses, such as Epstein Barr virus (EBV), BK-virus (BKV), and retroviruses (RV) have been suggested to have disease association14,15,16. No substantial evidence for triggering lupus as a result of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) epidemic has been observed, although scattered cases have been reported. However, Covid-19 has been shown to be able to trigger the presentation or exacerbation of autoimmune diseases as well as a longstanding inflammatory syndrome in genetically predisposed patients17,18,19.
Genetic predisposition accounts for approximately thirty percent of the risk of developing autoimmune diseases and interacting environmental factors could include toxic agents and diet but also infections20. The NCF1-regulated differences in the interferon signaling responses were seen in germ-free mice only after immunization, and they were also obvious after pristane injection, which triggered a more severe lupus-associated response12,13,21. Like several other mouse lupus models, pristane-induced lupus has been extensively studied for pathogenic mechanisms, and different downstream causative mechanisms have been suggested22. To understand the pathogenic role of the NCF1 polymorphism relevant to humans, we have addressed the role of the Ncf1R90H alleles in mice.
Here we study the Ncf1R90H alleles in mice, to understand the pathogenic role of the NCF1 polymorphism relevant to humans. To our surprise, the Ncf190H allele does not enhance pristane-induced lupus in the mice under specific pathogen-free conditions. However, infection with MNV causes the development of severe lupus in Ncf190H mice irrespective of whether these were naïve or injected with pristane. Thus, a synergy between a defined gene mutation and a specific non-autologous factor for the induction of an autoimmune disease is described.
Results
Environmental MNV induces lupus in BALB/c.Ncf1 90H mice
To determine lupus susceptibility, we followed C57BL/6NQ.Ncf190H and BALB/c.Ncf190H mice for 12 months, however, we did not find any signs of lupus. Thereafter, we injected them with pristane to establish the pristane-induced lupus (PIL) model but we did not see a significant effect by the Ncf190H allele. However, years later after establishing the colony in our isolated SPF facility, we noticed a sudden outbreak of lupus in a cage of eight-week-old Ncf190H BALB/c mice, with arthritis, enlarged spleens, and proteinuria (Supplementary Figs. 1a–d). The only changed condition was a new virus infection and we found both antibodies and detection of live viruses, indicating a recent infection with murine norovirus (MNV) (Supplementary Table 1). We isolated virus RNA from mouse feces and confirmed the presence of a specific MNV strain 59591 by sequencing (Supplementary Table 2, Supplementary Fig. 2).
To confirm the causative role of MNV, we transferred the BALB/c.Ncf1R90H littermates from an SPF facility to an isolated unit, and later added MNV-positive feces in the cages. We followed the mice for signs of virus infection, immune responses, and disease symptoms (Fig. 1a). Five weeks after MNV infection, Ncf190H mice started to develop lupus arthritis (Fig. 1b). Eight weeks later, proteinuria, and sera autoantibodies against dsDNA, Sm/RNP, phospholipid (PLs), and antigens of β2-GP1 were increased in MNV-infected Ncf190H mice, compared to MNV-infected Ncf1R90 wild-type mice or non-MNV infected Ncf190H mice (Fig. 1c, d). As antibodies are important for clearing MNV infection23 we monitored virus RNA, and anti-MNV antibodies. We found that the levels of virus RNA, 7 days post infection, were lower in the Ncf190H mice compared with Ncf1R90 mice. The levels of anti-MNV antibodies were positively correlated with the maximum arthritis score, levels of proteinuria, and anti-dsDNA antibodies in Ncf190H mice, with a trend of correlation with other investigated lupus-associated phenotypes (Fig. 1e, Supplementary Fig. 3a–c). Besides, we also noted a pronounced splenomegaly in MNV-infected Ncf190H mice, with an increased spleen index (Fig. 1f). Inflammatory cell infiltration was observed in the joint tissues of MNV-Ncf190H mice (Fig. 1g). Glomerulonephritis and interstitial mononuclear cell infiltrations were observed in the kidneys, with increased IgG and C3 deposits in the glomeruli (Fig. 1g, h). We found gene expression of Ifnα and anti-viral ISGs including Irf7, Stat1, Mx1, Ip-10, and Isg15 in both spleens and kidneys, and also Irf1 in the spleens to be increased in MNV infected Ncf190H mice, compared with wild-type Ncf1R90 (Fig. 1i, j). A pronounced phosphorylation of JAK1 and STAT1 was detected in the kidneys of MNV-Ncf190H mice compared to Ncf1R90 littermates (Fig. 1k, Supplementary Fig. 4a, b). We found decreased MNV load in the gastrointestinal tract, and sera antibodies clearing MNV displayed a positive correlation with lupus associated signs in the Ncf190H mice. These results suggest that the NCF190H variant protects against MNV infection but allows the immune response against MNV to cause lupus.
a Timeline of the MNV infection experiment with male BALB/c. Ncf190H mice after transferring from the SPF facility to the MNV-infected facility (day 1). b The representative appearance of arthritis in the ankle of hind paws on day 49 and mean arthritis scores from days 1 to 56 (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. c The levels of proteinuria (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. d The level of anti-dsDNA, anti-Sm/RNP, anti-phospholipid (anti-PLs) specific antibodies, and β2-GP1 on day 56 (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. e Correlation with anti-MNV antibodies (OD): maximum arthritis score (p = 0.0138), proteinuria (p = 0.0049), anti-dsDNA antibodies (p = 0.0055) in Ncf190H mice on day 56 post of MNV infection (n = 8 per group). Data were analyzed using the Pearson correlation test. f Enlarged spleen and spleen index in mice (n = 7/group). The spleen index is defined by the spleen weight (mg) divided by the body weight (g) and then multiplied by 10 (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. g Representative H&E stained joint and kidney sections (magnification x 10). Histological scoring of joint and kidney inflammation using a scale of 0-3 (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. h Immunofluorescence images (magnification x 20) and histology scores of deposits of IgG and C3 in the glomerulus from wild-type (WT) R90 and 90H mice on day 56 (n = 4 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. i Relative expression of Ifnα and anti-viral ISGs (Irf1, Irf7, Stat1, Mx1, Ip10 and Isg15) within spleens (n = 4 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. j Relative expression of Ifnα and anti-viral ISGs (Irf1, Irf7, Stat1, Mx1, Ip10 and Isg15) within kidneys. The expression of mRNAs was normalized to the housekeeping gene β-actin (n = 4 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. k Immunoblot analysis of p-JAK1/JAK1 and p-STAT1/STAT1 proteins in the kidneys on day 56 after MNV infection (n = 6 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM.
Environmental MNV aggravates pristane-induced lupus, with arthritis, in C57 black mice
We have previously shown that C57 black mice are susceptible to pristane-induced lupus by an NCF1-associated effect, mediated by hyperactive plasmacytoid dendritic cells (pDCs) by activating type I interferon and STAT1 pathways24. We found that MNV induces lupus in BALB/c.Ncf190H mice, and we investigated whether environmental factor could affect pristane-induced lupus (PIL) in B6N.Q.Ncf190H (BQ.Ncf190H) mice. Before that, we found an intraperitoneal injection of pristane in SPF-housed BQ.Ncf190H mice led to hyperactivated macrophages and neutrophils (Fig. 2a, b, Supplementary Fig. 5). MNV-infected homozygous female and male BQ.Ncf190H mice, and also heterozygous mice developed more severe lupus arthritis as compared with MNV-infected BQ.Ncf1R90 littermates in PIL (Fig. 2c, Supplementary Fig. 6a, b). The levels of anti-dsDNA antibodies were also increased in both female and male MNV-BQ.Ncf190H mice, compared with MNV-Ncf1R90 mice, seven months post-injection of pristane (Fig. 2d, Supplementary Fig. 6a–c). It demonstrates that MNV indiscriminately aggravates the diseases in both the female and male Ncf190H mice in the PIL model. We even detected slightly increased anti-dsDNA antibodies after MNV-infection, but before pristane injection, in Ncf190H compared with Ncf1R90 mice (Supplementary Fig. 6d). Anti-MNV antibodies were detected at seven-month post-injection of pristane, with and without MNV infection (Fig. 2e, Supplementary Fig. 6e). Ncf190H mice exhibited higher histology scores of joints, with more infiltrating cells, and higher histology scores of kidneys with increased mesangial stromal and cellularity, compared with wild-type mice seven months after pristane injection (Fig. 2f, Supplementary Fig. 6f, g). Given the above, we found that environmental MNV infection enhanced the development of pristane-induced lupus in Ncf190H C57 black mice.
Gating and population of immune cells in the peritoneal cavity of the male 90H mice 3 days after pristane injection. a Macrophages (CD11b+ F4/80+ Ly6C−) (R90: n = 5; 90H; n = 10). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. b Neutrophils (CD11b+ Ly6G+ Ly6C−) (R90: n = 5; 90H; n = 10). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. c Mean arthritis scores before and after pristane injection (0–7 months) in female (R90: n = 9; 90H: n = 7; MNV-R90: n = 12; MNV-90H: n = 8) and male (R90: n = 7; 90H: n = 5; MNV-R90: n = 7; MNV-90H: n = 6) mice, with and without MNV infection. MNV-R90 vs MNV-90H. Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. d Levels of anti-dsDNA antibodies in non-MNV infected female mice (Pristane-R90: n = 7 and Pristane-90H: n = 6) and male mice (Pristane-R90: n = 7 and Pristane-90H: n = 5) and MNV infected female mice (MNV-Pristane-R90: n = 9 and MNV-Pristane-90H: n = 10) and male mice (MNV-Pristane-R90: n = 5 and MNV-Pristane-90H: n = 8) seven months post-injection of pristane. Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. e Levels of anti-MNV antibodies in non-MNV infected female mice (Pristane-R90: n = 10 and Pristane-90H: n = 10) and male mice (Pristane-R90: n = 10 and Pristane-90H: n = 10) and MNV-infected female mice (MNV-Pristane-R90: n = 11 and MNV-Pristane-90H: n = 17) and male mice (MNV-Pristane-R90: n = 5 and MNV-Pristane-90H: n = 12). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. f Statistics of histology scores of joints and kidneys seven months post-injection of pristane (n = 8 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM.
The NCF190H allele upregulates JAK1/STAT1 in mouse macrophages
Naïve SPF mice with the Ncf190H allele were healthy and did not differ in behavior or appearance compared to mice with the wild-type allele. They also had normal Ncf1 mRNA expressions and normal numbers of mature macrophages (CD11b+ F4/80+ % Live CD45+) (Supplementary Fig. 7a, b). As expected, bone marrow-derived macrophages (BMDMs) from BQ.Ncf190H mice had lower intracellular and extracellular ROS production, as measured in naïve BMDMs or PMA-stimulated BMDMs. The NOX2 inhibitor GSK2795039 blocked both intracellular and extracellular ROS in Ncf190H and Ncf1R90 macrophages (Fig. 3a, b). To investigate functional downstream effects, we measured expression and phosphorylation of STAT1 (p-STAT1) in mouse macrophages, one of the key transcriptional factors activating ISG expression. Naïve macrophages from mice expressing NCF190H or with deficient NCF1 expression due to the Ncf1m1J mutation had a higher level of JAK1, STAT1, and STAT3 expression than wild-type BQ.Ncf1R90 mice. IFN-α treatment led to higher level phosphorylation of these molecules, which was more robust in Ncf190H or Ncf1m1J macrophages (Fig. 3c–e) and confirmed by immunofluorescent staining (Supplementary Fig. 7c, d). NOX2 blocker treatment of macrophages from wild-type Ncf1R90 mice enhanced IFN-α-induced phosphorylation of JAK1, STAT1, and STAT3, while H2O2 exposed macrophages from Ncf190H and Ncf1m1J mice, had diminished phosphorylation (Fig. 3f–i). Investigation of the expression of STAT1 and p-STAT1 in peritoneal exudates macrophages (CD45+ CD11b+ F4/80+ Ly6C−) one day post-MNV intraperitoneal injection showed that p-STAT1 was upregulated in Ncf190H mice compared to wild-type Ncf1R90 (Fig. 3j, k). Taken together, the JAK/STAT signaling pathway in macrophages is profoundly activated by MNV infection in Ncf190H mice due to low ROS production.
a Intracellular ROS in BMDMs without (Blank) and with (Rosup) stimulation (n = 8 per group). Intracellular ROS in BMDMs with NOX2 inhibitor GSK2795039 (n = 5 per group). R90 vs 90H, or m1J. Data were analyzed using one-way ANOVA and presented as mean ± SEM. R90 vs GSK-R90, 90H vs GSK-90H, or m1J vs GSK-m1J, Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. b Extracellular ROS production in BMDMs without (Blank) and with (PMA) stimulation (n = 5 per group). Extracellular ROS in BMDMs after treatment with NOX2 inhibitor GSK2795039 (n = 3 per group). R90 vs GSK-R90, 90H vs GSK-90H, m1J vs GSK-m1J. R90 vs 90H or m1J, 90H vs Hank’s Balanced Salt Solution (HBSS). Data were analyzed using one-way ANOVA and presented as mean ± SEM. R90 vs GSK-R90, 90H vs GSK-90H, or m1J vs GSK-m1J, Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. c Western blots of JAK1, STAT1, and STAT3 proteins in naïve macrophages, and after IFN-α stimulation from 90H and m1J male mice, compared to wild-type (R90) male mice (n = 5 per group). d, e Quantification of relative protein expression, normalized to β-actin (n = 4 per group). Quantitative comparisons between samples on the same blots or the samples derive from the same experiment and that blots were processed in parallel. R90 vs 90H, R90 vs m1J. IFN-α R90 vs IFN-α 90H, IFN-α R90 vs IFN-α m1J. Data were analyzed using one-way ANOVA and presented as mean ± SEM. f Western blots of p-JAK1, p-STAT1, and p-STAT3 in IFN-α stimulated macrophages, together with adding Ncf1/NOX2 blocker to cells from wild-type mice (n = 5 per group), or adding H2O2 to cells from 90H or m1J mice (n = 5 per group). g–i Quantification of relative protein expression, normalized to β-actin (n = 5 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. j Representative histograms of STAT1 and p-STAT1 in peritoneal exudates macrophages (CD45+ CD11b+ F4/80+ Ly6C−) one- day post-MNV injection (1dpi) by IP. k Geometric mean of STAT1 and p-STAT1, and relative expression of p-STAT1/STAT1 within peritoneal exudates macrophages (MNV-R90: female: n = 4, male: n = 4; MNV-90H: female: n = 5, male: n = 4). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. BMDMs were obtained from the differentiation of monocytes recovered from the femur and tibia of 6 to 8- week- old male B6N.Q.Ncf1R90, B6N.Q. Ncf190H, B6N.Q and B6N.Q.Ncf1m1J mice.
The Ncf1 90H allele protects against infection but induces lupus with an isolated MNV strain
To confirm the lupus-inducing effect of MNV and to understand its pathogenic function, we administered the MNV sub-strain Berlin/06/06DE S99 through oral gavage25 (Fig. 4a). We could detect virus RNA in the small intestines and colons in all mice but not in the spleen, confirming that the MNV infection was localized to the gastrointestinal tract and did not spread systemically. Importantly, the levels in the small intestine and colon were dramatically lower in the BQ.Ncf190H mice compared with BQ.Ncf1R90 mice, measured seven days after MNV infection, showing that the lower capacity to produce ROS by NCF190H limits the infection (Fig. 4b). The BQ.Ncf190H mice, developed higher levels of IgG, compared to wild-type littermates, measured in sera 56 days after MNV infection, and induced an antibody response to MNV, in both males and females (Fig. 4c, d). The levels of anti-MNV antibodies in Ncf190H mice correlated with the increase in total IgG levels (Fig. 4e. Supplementary Fig. 8a, b). The Ncf190H mice had increased levels of autoantibodies against ssRNA, dsDNA and Sm/RNP, and also proteinuria (Fig. 4f–i). Based on the dramatically increased levels of IgG and higher titers of autoantibodies in MNV-infected Ncf190H mice, we investigated the activation of T and B cells at the end of the experiment on day 84. The NCF190H allele did not affect the numbers of T helper cells (Th) (CD4+) or regulatory T cells (Tregs) (CD4+ FOXP3+) cells among live CD45+ cells, but we found an increased expression of CD62L and CD69, indicating a higher activation (Supplementary Fig. 8c, d). The relative frequency of central or virtual memory T cells (Tcm/Tvm) (CD62L+ CD44+) in CD4+ Foxp3+, T follicular cells (Tfh) (CXCR5+ PD-1+) among CD4+ FOXP3− cells and T follicular regulatory (Tfr) (CXCR5+ PD-1+) among CD4+ Foxp3+ cells, were increased whereas tissue-resident memory T cells (Trm) (CD62L− CD69hi) among CD4+ CD44hi cells, were decreased in Peyer’s patches (PPs) of Ncf190H mice (Fig. 4j–m, Supplementary Fig. 8e). Instead, the number of Tfh and Tfr cells were not changed in the small intestine outside PPs (Supplementary Fig. 8f–h). Both Tfh and Tfr cells regulate the generation of antigen-specific antibody-secreting cells (ASCs) in the germinal center (GC). The population of ASCs (IgDlo CD138+ Sca-1+) from PPs or the small intestine was not changed (Supplementary Fig. 8f, i, j). Analysis of the B cell compartment showed that GC-B cells were increased in the PPs of Ncf190H mice, indicating a strong B cell activation in response to the MNV infection (Fig. 4n, Supplementary Fig. 8f). We found an increase of long-lived plasma cells (LLPCs) in the PPs of Ncf190H mice (Fig. 4o, Supplementary Fig. 8f, k). We conclude that the MNV gastrointestinal infection in Ncf190H mice trigger a strong activation of T and B cells, both in the intestine and systemically in germinal centers, leading to the expansion of LLPCs.
a Timeline of MNV infection by gavage. b The quantification of MNV in the small intestine, colon, and spleen on day 7 by RT-qPCR (n = 9 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. c Titer of anti-MNV antibodies on days 0, 14, 28, 56, 70, and 84 in females (MNV-R90: n = 6; MNV-90H: n = 13) and males (MNV-R90: n = 5; MNV-90H: n = 6). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. d IgG production in femals (R90: n = 6; 90H: n = 13; MNV-R90: n = 6; MNV-90H: n = 13) and males (R90: n = 5; 90H: n = 6; MNV-R90: n = 5; MNV-90H: n = 6). Data were analyzed using one-way ANOVA and presented as mean ± SEM. e The correlation between IgG and anti-MNV antibodies in wild-type Ncf1R90 mice (n = 11) and Ncf190H mice (n = 12). Data were analyzed using the Pearson correlation test. f Anti-ssRNA antibodies on day 56 in females (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 13) and males (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 13). Data were analyzed using one-way ANOVA and presented as mean ± SEM. g Anti-dsDNA antibodies on day 56 in females (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 13) and males (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 6). Data were analyzed using one-way ANOVA and presented as mean ± SEM. h Anti-Sm/RNP antibodies on day 56 in females (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 13) and males (R90: n = 5; 90H: n = 5; MNV-R90: n = 5; MNV-90H: n = 6). Data were analyzed using one-way ANOVA and presented as mean ± SEM. i Proteinuria on day 56 in females (R90: n = 6; 90H: n = 13; MNV-R90: n = 6; MNV-90H: n = 13) and males (R90: n = 5; 90H: n = 6; MNV-R90: n = 5; MNV-90H: n = 6). Data were analyzed using one-way ANOVA and presented as mean ± SEM. j The ratio of Tcm or Tvm cells in CD4+ Foxp3+ in the PPs (MNV-R90: n = 11; MNV-90H: n = 19). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. k, l The ratio of Tfh cells and Tfr cells in CD4+ FOXP3+ cells (MNV-R90: n = 11; MNV-90H: n = 19). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. m The ratio of Trm cells in CD4+ CD44hi cells (MNV-R90: n = 11; MNV-90H: n = 18). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. n The ratio of GC-B cells in IgDlo B cells (MNV-R90: n = 11; MNV-90H: n = 19). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. o The ratio of LLPCs in IgDlo CD138+ Sca-1+ cells (MNV-R90: n = 11; MNV-90H: n = 19). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM.
Non-mucosal MNV infection induces lupus arthritis in Ncf1 90H mice
The virus oral infection was localized to the gastrointestinal tract but also triggered the immune responses in draining immune organs like the PPs and mesenteric lymph nodes (mLNs). Importantly, it also activated the immune system systemically, as indicated by a strong response in the spleen. We were curious to see if the virus could trigger a similar immune response by extra-intestinal exposure and thereby induce lupus. We therefore infected the mice with the MNV strain intravenously and intraperitoneally (Fig. 5a). An immune response to the MNV developed in both groups of mice, as seen by an induced antibody response to MNV (Fig. 5b). Similar to the per orally infected mice, we found dramatically increased titers of anti-ssRNA autoantibodies and production of IgG in mice with the Ncf190H allele (Fig. 5c, d). All BQ.Ncf190H mice developed arthritis, but none of the wild-type BQ.Ncf1R90 mice, with onset as early as day 14, reaching maximum severity on day 35 (Fig. 5e–h). We also measured the levels of antibodies to type II collagen (COL2) since these are prominent natural autoantibodies in mouse lupus but may also occur as a response to joint inflammation26,27. Mild but significant increased IgG, IgG2b, and IgM antibodies to COL2 were detected in MNV-Ncf190H mice on day 21 compared with naive Ncf190H mice (Fig. 5i–k). B cell-ELISpot data showed consistent results of increased numbers of anti-COL2 IgG and IgM antibody secreting cells (ASCs) in the spleen of Ncf190H mice after injection of MNV when compared with naive mice or MNV-Ncf1R90 mice (Fig. 5l, m). In the late stage, on day 56, we also found increased anti-dsDNA and anti-Sm/RNP antibodies, as well as proteinuria in MNV-injected mice compared to naïve mice (Supplementary Fig. 9). Taken together, we showed that extraintestinal injection of MNV trigger a more rapid activation of the immune system, leading to lupus arthritis and induction of typical lupus autoantibodies in Ncf190H mice.
a Timeline of MNV infection in male mice by i.v. and i.p. injection. b Anti-MNV antibodies on days 0, 14, 28, and 56. c Anti-ssRNA antibodies on day 21 (R90: n = 6; 90H: n = 6; MNV-R90: n = 11; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. d IgG production on day 21 (R90: n = 8; 90H: n = 12; MNV-R90: n = 11; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. e Representative paws on day 21. f Mean arthritis score (MNV-R90: n = 8; MNV-90H: n = 11). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. g Arthritis incidence (n = 9 per group). h Representative HE staining of the joints (n = 4, magnification x 2.5 and x 10). i Anti-collagen II (COL2) IgG antibodies (R90: n = 7; 90H: n = 7; MNV-R90: n = 11; MNV-90H: n = 20). Data were analyzed using one-way ANOVA and presented as mean ± SEM. j Anti-COL2 IgG2b antibodies (R90: n = 7; 90H: n = 7; MNV-R90: n = 9; MNV-90H: n = 20). Data were analyzed using one-way ANOVA and presented as mean ± SEM. k Anti-COL2 IgM antibodies (R90: n = 8; 90H: n = 11; MNV-R90: n = 11; MNV-90H: n = 18). Data were analyzed using one-way ANOVA and presented as mean ± SEM. The level of anti-COL2 antibodies was measured in mouse sera on day 21 by ELISA. l, m Representative B cell-ELISpot and statistics of total anti-COL2 IgG and IgM ASCs in the spleen were shown (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM.
NCF190H allows development of lupus through T cell-dependent germinal center response
We could confirm that the natural peroral MNV infection was restricted locally to the gastrointestinal tract, but in BQ.Ncf190H mice the virus infection was more restricted. However, it distorted the systemic immune response driving it to convert to pathogenic autoimmunity. Therefore, we focused on the maturation and differentiation of B and T cells in the spleen of the Ncf190H mice. Already in naïve Ncf190H mice, we found an increased population of mature B cells (CD93−, gated on CD19+ B220+), but not B2 cells, along with an increased number of follicular B (FOB) cells (CD23+ CD21+, gated on CD19+ B220+ CD93−), and naive FOB cells (IgDhi IgMlo, gated on CD23+ CD21+ CD19+ B220+ CD93−). Only the ratio of marginal zone B cells (MZB) in live cells from Ncf190H mice showed an increase, and not the IgD− IgMhi MZB cells (Fig. 6a–c, Supplementary Fig. 10a–d).
a The frequency of B and T cells in the spleen of naïve mice. The ratio of mature B cells (CD93−, gated on CD19+ B220+) in B2 cells (CD19+ B220+) (R90: n = 5; R90H: n = 9; 90H: n = 13). Data were analyzed using one-way ANOVA and presented as mean ± SEM. b The ratio of follicular B cells (CD23+ CD21+, gated on CD19+ B220+ CD93−) in B2 cells (R90: n = 5; R90H: n = 9; 90H: n = 13). Data were analyzed using one-way ANOVA and presented as mean ± SEM. c The ratio of IgDhi IgMlo follicular B cells (IgDhi IgMlo, gated on CD23+ CD21+ CD19+ B220+ CD93−) in B2 cells (R90: n = 5; R90H: n = 9; 90H: n = 13). Data were analyzed using one-way ANOVA and presented as mean ± SEM. d Mice were infected with MNV for 21 days by i.v. and i.p. injection. The frequency of IgDhi IgMlo from B2 cells (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. e The histogram and MFI value of MHCII, and CXCR4 on B2 cells (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. f The expression of PD-1, CXCR5, CD44, CD69 and CD62L on Th cells (CD4+) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. g Naïve Tregs (CD4+ FOXP3+), Tem cells (CD62L− CD44+, gated on CD4+ FOXP3+), Tcm/Tvm cells (CD62L+ CD44+, gated on CD4+ FOXP3+) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. h Tfh (CXCR5+ PD-1+, gated on CD4+ FOXP3−) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. i Tfr cells (CXCR5+ PD-1+, gated on CD4+ FOXP3+) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. j Trm cells (CD62L− CD69hi, gated on CD4+ CD44hi) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. k GC-B cells (CD38lo GL7+ cells, gated on IgD− B220+ CD19+ population) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM. l ASCs (CD138+ Sca-1+ IgD−) subpopulation: PBs (CD19+ B220+), NEW PCs (CD19+ B220−), and LLPCs (CD19− B220−) (R90: n = 5; 90H: n = 5; MNV-R90: n = 8; MNV-90H: n = 12). Data were analyzed using one-way ANOVA and presented as mean ± SEM.
Next, we investigated the effect of MNV infection on the B cells and T cells in the spleen. We found that the population of naïve B2 cells (IgDhi IgMlo B2 cells) was increased by the MNV infection but the increase was not influenced by the genotype (Fig. 6d, Supplementary Fig. 10e). However, the expression of MHCII and CXCR4 on B2 cells were increased in Ncf190H mice (Fig. 6e). MNV infection led to a decrease of regulatory T cells (% CD4+ FOXP3+ in live cells), but an increase of CD44 expression on Tregs in Ncf190H mice (Supplementary Fig. 10f, g). In the Ncf190H mice, MNV infection reduced the number of Th cells, but with an increased expression of PD-1, CXCR5, CD44, and CD69 (Fig. 6f, Supplementary Fig. 10h, i). Tem cells (CD44+ CD62L−, gated on CD4+ FOXP3+) were increased by MNV infection in Ncf190H mice, whereas naive Tregs (CD44− CD62L+, gated on CD4+ FOXP3+) and Tcm/ Tvm cells (CD62L+ CD44+, gated on CD4+ FOXP3+) were decreased (Fig. 6g). MNV infection also led to an expansion of follicular T cells, in particular in Ncf190H mice, in both Tfh (CXCR5+ PD-1+, gated on CD4+ FOXP3−) and Tfr (CXCR5+ PD-1+, gated on CD4+ FOXP3+) compartments (Fig. 6h, i). Similarly, the frequency of Trm (CD62L− CD69hi, gated on CD4+ CD44hi) were increased by MNV infection in Ncf190H mice (Fig. 6j). Considering the importance of the GC response in lupus, we explored the impact of NCF1-mediated ROS burst on GC-B cells. We found a dramatically increased population of GC-B cells (CD38lo GL7+ cells, gated on B220+ CD19+ IgD− population) in Ncf190H mice, compared to wild-type Ncf1R90 or non-MNV injected Ncf190H mice (Fig. 6k, Supplementary Fig. 10j). We also measured ASC (IgDlo CD138+ Sca-1+) sub-populations including PBs (CD19+ B220+), newly formed PCs (NEW PCs) (CD19+ B220−), and LLPCs (CD19− B220−). Importantly, the number of ASCs was increased in Ncf190H mice after MNV infection (Supplementary Fig. 10k). We also observed an increased population of PBs, NEW PCs, and LLPCs in MNV-infected Ncf190H mice, compared with MNV-infected wildtypes and also increased numbers of LLPCs in MNV-Ncf190H mice, compared to naive Ncf190H mice (Fig. 6l). Taken together, the Ncf190H allele promoted maturation and differentiation of B and T cells, and the NCF190H allele, in response to MNV infection, mediated a T cell-dependent GC response.
The Ncf1 90H allele promotes the upregulation of TLR7
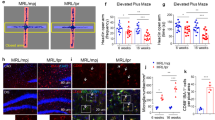
TLR7 is thought to localize in endolysosomes and are also present on the cell surface of immune cells, with responses in dendritic cells, macrophages and B cells28. A translocation from the X chromosome onto the Y chromosome causes overexpression of the translocated genes, which includes the ssRNA-recognizing TLR7 member of the TLR family of receptors in male mice bearing the y-linked autoimmune accelerating (Yaa) locus, which is sufficient to enhance TLR7-mediated activation of innate immune responses and lupus development29,30,31. To investigate the role of TLR7 in ssRNA virus- MNV-induced lupus. We took advantage of the Yaa locus and confirmed that intact TLR7 is upregulated in the spleen of naïve Ncf190H mice with the Yaa locus (Fig. 7a). We also found expression of intact TLR7 to be increased in environmental MNV-infected homozygous Ncf190H mice with Yaa locus (Fig. 7b). In littermate mice, without the Yaa locus, non-mucosal MNV infection had an increased ratio of cleaved TLRs to intact TLRs and increased levels of intact TLR7 (Fig. 7c). Similarly, mucosal-MNV infection upregulated intact TLR7 expression in Ncf190H mice (Fig. 7d). We did not observe the effect by Yaa locus on the ratio of immune cells but we found dramatically increased populations of macrophages (CD45+ CD11b+ F4/80+) and pDCs (CD11clo/int Ly6Chi PDCA-1+ B220+) in MNV infected spleens, compared to spleen without infections, in particular after the mucosal infection (Supplementary Fig. 11 and 12a, b). TLR7 is expressed in multiple immune cell subsets, including B cells, cDCs, pDCs, macrophages, and monocytes. Based on this, we investigated the TLR7 and TLR9 expressions on macrophages, pDCs, and B220+ splenocytes. It displayed an increased level of TLR7 in macrophages, pDCs, and B220+ cells from naïve Ncf190H mice with the Yaa locus, compared to mice without the Yaa locus (Fig. 7e–g). TLR9 was not changed in macrophages, pDCs, or B220+ cells by the Yaa locus or the NCF190H variant (Supplementary Fig. 11 and 12c–e). TLR7 expression was increased in macrophages, pDCs, and B220+ splenocytes after mucosal MNV infection in Ncf190H mice. An increase only in macrophages by non-mucosal infection was observed in Ncf190H mice. In addition, it showed a dramatic increase of TLR7 in macrophages and B220+ cells by mucosal MNV infection compared with non-mucosal infection (Fig. 7h–j). In contrast, the Ncf190H allele had no significant effect on TLR9 expression with or without MNV infection (Supplementary Fig. 12f–h). Taken together, the NCF190H variant upregulates the MNV-induced TLR7 signaling pathway, to mediate activation of innate immune responses and lupus development.
a Immunoblot analysis of intact TLR7 protein and MyD88 protein in the spleen of naïve male 90H mice with or without Yaa locus (n = 6 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. b Immunoblot analysis of TLR7 and MyD88 protein in environmentally MNV-infected 90H mice with Yaa locus on day 70 (n = 4 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. c Immunoblot analysis of intact TLR7 protein, cleaved TLR7 fragment (TLR7-F), and MyD88 in non-mucosal MNV-infected 90H mice (n = 4 per group). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. d Immunoblot analysis of intact TLR7 and MyD88 proteins in non-MNV and mucosal MNV infected mice (n = 6 per group). Data were analyzed using one-way ANOVA and presented as mean ± SEM. e–g Flow cytometric analysis of TLR7 in the spleen of naïve mice with and without Yaa locus in macrophages (CD45+ CD11b+ F4/80+) (R90: n = 6; 90H: n = 6; R90.Yaa: n = 5; 90H.Yaa: n = 8), pDCs (CD45+ CD11clo/int Ly6Chi PDCA-1+ B220+) (R90: n = 4; 90H: n = 6; R90.Yaa: n = 5; 90H.Yaa: n = 8), and CD45+ B220+ splenocytes (R90: n = 4; 90H: n = 6; R90.Yaa: n = 5; 90H.Yaa: n = 8). Data were analyzed by the Mann-Whitney test (two-tailed) and presented as mean ± SEM. h–j Flow cytometric analysis of TLR7 in macrophages (Non MNV-R90: n = 2; Non MNV-90H: n = 2; Non-muco MNV-R90: n = 4; Non-muco MNV-90H: n = 8; Muco MNV-R90: n = 5; Muco MNV-90H: n = 6), pDCs (Non MNV-R90: n = 2; Non MNV-90H: n = 2; Non-muco MNV-R90: n = 4; Non-muco MNV-90H: n = 8; Muco MNV-R90: n = 5; Muco MNV-90H: n = 6), and B220+ cells (Non MNV-R90: n = 2; Non MNV-90H: n = 2; Non-muco MNV-R90: n = 4; Non-muco MNV-90H: n = 8; Muco MNV-R90: n = 5; Muco MNV-90H: n = 6) from the spleen of mice without MNV infection, with non-mucosal MNV infection, and mucosal MNV infection. Data were analyzed using one-way ANOVA and presented as mean ± SEM.
Discussion
We have observed that infection with MNV, a normally non-pathogenic virus, triggers a disease mimicking lupus in mice with SNP associated with SLE in humans. On the other hand, the lupus causative SNP leading to a lower ROS response, protected the mice from the virus infection by restricting the spread of the virus. MNV is a single-stranded RNA virus infecting intestinal epithelial cells but which is not spreading systemically. Importantly, it activates not only the local immune response but also the immune system systemically leading to activation of both T and B cells with enlargement of spleens. Upon infection in low ROS-producing Ncf190H mice, the virus releases ssRNA, triggering the interferon signaling pathway, possibly through activation of TLR7 and the STING pathway32,33. NCF1 is expressed mainly in phagocytic and antigen-presenting cells, including macrophages, dendritic cells, and B cells. Activation of these cells triggers the NOX2 complex to produce ROS. And the downstream oxidative metabolites such as hydrogen peroxide will oxidize regulatory cysteines in many proteins intracellularly and protect the cell from an exhaustive activation of JAK/ STAT mediated activation of interferon signaling34. However, a NOX2 complex with a lower capacity to produce ROS, due to NCF190H, will allow phosphorylation of STAT1 and activate the interferon signaling cascade. We have earlier shown that the critical target cell for the development of mouse lupus is the plasmacytoid dendritic cell, which downstream lead to B cell activation and autoantibody production24. From the perspective of immune defense against viruses, it makes sense as it will lead to more efficient virus neutralization. A genetically determined lower capacity to produce ROS from the NOX2 complex could therefore be beneficial for the protection of virus infection, particularly ssRNA viruses, known to efficiently activate interferon signaling through TLR7 or STING. However, if the infection is prolonged and turns into chronic inflammation, the lack of ROS allows an exaggerated immune activation, which may lead to systemic autoimmunity.
The SNP coding for the NCF190H variant was detected by exon sequences of NCF1 copies in humans. Targeted mutations leading to Ncf1R90H replacement in mice have independently been made by several groups in parallel24,35,36,37. The disease-causative effect in humans has been partly reproduced based on the commonly used protocol of pristane or imiquimod-induced lupus but with the suggestion of different downstream mechanisms. In one report, mild spontaneous lupus developed in aged (>5 months) B6 mice but could be exacerbated after pristane injection, and the disease development was dependent on decreased efferocytosis leading to an expansion of Tfh2 cells35. In another study, the Ncf190H mice did not develop spontaneous lupus but had severe disease after imiquimod (IMQ) treatment36. Clearly, the induced disease manifestations associated with the NCF190H variant are dependent on the environmental trigger. For example, intraperitoneal injection of mannan induces severe psoriasis and psoriatic arthritis37. Acidification of phagosomes, due to low ROS in phagosomes of macrophages and pDCs, has been suggested as the initial effect35,36. On the other hand, we have shown that NCF1 deficiency due to the Ncf1m1J mutation modifies regulation of cysteine oxidation and could thereby affect different pathways such as interferon signaling, activation of antigen receptors, or affecting antigen processing12,34,38,39,40.
Mutations in different NOX2 components have been shown to enhance various autoimmune diseases, but with different downstream mechanisms13,41,42,43. Symptoms mimicking lupus, with autoantibodies and glomerulonephritis, appeared spontaneously in naïve ROS-deficient BALB/c. Ncf1m1J mutated mice, which could be explained as mice in the SPF animal house for these experiments were exposed to recurrent MNV infections as they had antibodies to MNV12. Pristane injection has been shown to induce mild lupus in C57 black mice even in an MNV-free SPF facility, and the disease was enhanced by the Ncf1m1J mutation24. In comparison with the Ncf1m1J mutation, the Ncf190H mutation has more remaining ROS-inducing capacity. In our study, pristane-injection did not induce significant signs of lupus, or lupus-associated antibodies, in Ncf190H mice neither having C57 black nor BALB/c genes predominantly. Instead, significantly increased lupus diseases only developed after MNV infection, regardless of whether they had been injected with pristane or not, and the development of the disease with MNV infection was completely dependent on the NCF190H allele. Thus, as compared with pristane injections, it is possible that MNV triggers a different mechanism signaling pathway leading to lupus symptoms.
Since MNV is normally not a pathogenic virus in common mouse strains, it has previously not been included in many SPF protocols and its importance was identified only recently. There are different substrains of MNV and here is shown that two different substrains can induce lupus, however, it might not apply for all substrains. MNV is known to primarily affect the intestinal epithelial cells, similar to the human norovirus, but it is unclear whether the infection spreads systemically, and infecting macrophages, dendritic cells, and B cells in vivo44. It was earlier reported that noroviruses can infect B cells and impair B cell development within the bone marrow in a STAT1-dependent but IFN signaling-independent manner45. Type I IFN (IFN-α/β) controls systemic replication, and type III IFN (IFN-λ) controls MNV persistence in the intestinal epithelium31,46,47. In our work, we clarified that MNV infection triggers TLR7 and activates IFN signaling in the background of ROS deficiency. The NCF190H variant could activate IFN signaling by several mechanisms, including lowering endosomal pH and direct inhibition of oxidation in the regulatory cysteines present in JAK1 and STAT1. This work is compatible with that the virus infects the intestinal epithelia, including macrophages and dendritic cells surveilling incoming infections, which in turn activates the immune system in PPs, draining lymph nodes, and subsequently the systemic immune system.
The key event is the exposure of viral ssRNA, which under reducing conditions activates interferon signaling pathways leading to a strong activation of antigen presenting cells including B cells, and subsequently autoreactive T cells. This pathogenic event is possible due to an interaction between a commonly occurring cytosolic protein, NCF190H, through the formation of the NOX2 complex, and a commonly occurring environmental factor, an ssRNA virus. The corresponding human norovirus-induced infections are widely spread, but transient and asymptomatic in most individuals and an association with SLE is therefore difficult to determine48. Both the structure and function of NCF1 and the NOX2 complex are, however, highly conserved.
Most importantly, even the functional role of the NCF1 polymorphism is similar between rodents and humans. It is therefore likely that a functional ROS response by the NOX2 complex gives relative protection against norovirus infections, but also an increased risk of developing SLE and other autoimmune disorders. Our discovery of MNV as a trigger of lupus in Ncf190H mice does not exclude other viruses, environmental factors or genes, to be causative for SLE. To the best of our knowledge, this is a disease-causative interaction has been shown between a defined genetic and a specific environmental factor to control a complex autoimmune disease.
Methods
Construction of mice
Balb/cByJ (stock 001026) and C57BL/6NJ (stock 005304) were originally from The Jackson Laboratory. B6N.Q (C57/B6N.Q/rhd) and B10.Q (C57/B10N.Q/rhd) have been fully backcrossed into B6N and B10 genomes but with an MHCII Aq containing haplotype from DBA/1 to establish the BQ.Ncf190H and BQ.Ncf1m1J strains37,49. The backcrossing onto the BALB/c to establish the BALB/c.Ncf190H strain. Age- and sex-matched littermates from heterozygous intercrosses were used in all experiments. For MNV-infected experiments in vivo, Ncf190H (abbreviated as 90H) refers to BALB/c.Ncf190H/90H, Ncf1R90H (abbreviated as R90H) refers to BALB/c.Ncf1R90/90H and Ncf1R90 (abbreviated as R90) refers to BALB/c.Ncf1R90/R90 mice. Unless otherwise stated, Ncf190H (abbreviated as 90H) refers to B10.Q.Ncf190H/90H and Ncf1m1J (abbreviated as m1J) refers to B10.Q.Ncf1m1J/m1J mice. B6.SB-Yaa/J (stock 000483) from the Jackson Laboratory has been fully backcrossed onto B10.Q, denoted as B10.Q.Yaa. The Yaa-carrying strain with fully functional NCF1 (R90.Yaa) or with human NCF190H allele (90H.Yaa) was obtained by crossing BQ.Ncf190H/90H with B10.Q.Yaa. The mice were kept and bred in the specific pathogen-free facility of Laboratory Animal Center of Southern Medical University and Comparative Medicine Annex (KM-A) of Karolinska Institutet except the infection experiments which were operated in the barrier of Comparative Medicine’s Annex (KM-F) of Karolinska Institutet. The facilities have a climate-controlled environment with a 14 h light/10 hrs dark cycle. The animals were housed in individually ventilated polystyrene cages containing enrichments with standard chow and water given ad libitum. For all the experiments, 4- to 8-week-old age- and sex-matched mutated mice and wild-type littermate controls were used. Experimental mice were killed by cervical dislocation. All the experimental procedures were approved by the Southern Medical University and Karolinska Institutet Animal Ethics Board (Guangzhou, China, permit number: L2020013 or Stockholm, Sweden, permit number: Dnr 23517-2022, 10523-2022 and 2660-2019). All animal experiments were performed according to the ARRIVE guidelines50.
Murine norovirus preparation
The MNV strain Berlin/06/06DE S99 was cultured in the permissive macrophage mouse cell line RAW 264.7 grown in Dulbecco’s minimum essential medium (DMEM, no pyruvate; catalog no. FG 0435, Biochrom), supplemented with 10% low endotoxin fetal bovine serum (Hyclone FBS, SH30088), 1% non-essential amino acids (Life Technologies, 11140-050) and 5% penicillin/streptomycin (Life Technologies, 15140122). Overnight cell cultures in T75 flasks were infected with MNV and incubated for 24 h, after which the supernatant was harvested, and cells were removed by centrifugation. Virus stock was stored at −80 °C. Viral preparations were derived from cell cultures. Viral infectivity was evaluated by a cell infectivity assay, determining the 50% tissue culture infective dose (TCID50) using 24-well plates and negative sense RNA detection for confirmation of infection51.
Establishment of lupus and arthritis models
To develop the pristane-induced lupus model, mice were injected intraperitoneally with a single dose of 500 μL pristane (MilliporeSigma, P2870) and followed for 6 months. For environmental MNV infections, bedding from the MNV-positive cage was added weekly in equal amounts as clean autoclaved bedding to the cages that housed 8-week-old MNV-free Ncf190H and Ncf1R90 mice obtained from the SPF animal facility52. MNV RNA was isolated from the feces and sequenced (Table S2, 3). For the mucosal MNV-induced model, all mice were infected by the MNV-S99 strain via oral gavage inoculation at a dose of 2 × 105 TCID50 per mouse. For the non-mucosal MNV-induced model, the mice were injected with 3 × 105 TCID50 by both intravenous and intraperitoneal injections, respectively.
Evaluation of arthritis and lupus
Arthritis was monitored using a macroscopic scoring system in which 1–5 points were given7. Briefly, 5 points were given to each visibly inflamed (erythema and swelling) ankle or wrist and 1 point to each inflamed toe. Histopathological evaluation on the ankle joints and kidneys collected at the endpoint by H&E staining was also performed. For assessing the glomerular deposits, PBS-flushed kidneys were embedded in OTC Tissue-Tek (Sakura, 4583) compound, snap frozen, and stored at −80°C. Five-micrometer cryo-sections were cut, fixed, permeabilized with acetone, and stained with Alexa Fluor 488-conjugated anti-mouse complement component C3 (1:200 dilutions; Cedarlane, CL7503AF4) or Alexa Fluor 488-conjugated goat anti–mouse IgG specific for Fcγ fragment (1:500 dilutions, Jackson ImmunoResearch, 115-545-071). Glomerular deposits were recorded by LSM880 with Airyscan, and evaluated by two blinded individual scorers using immunofluorescence microscopy Zen Blue v3.1 and a semiquantitative scoring system. Proteinuria was determined with semiquantitative urine testing strips (Uristix, 2857) using midstream urine.
ELISA and ELISpot
For DNA, RNA and Sm/RNP ELISA, plates were coated with 1 µg mL−1 Sm/RNP (Avivasysbin, OPMA04153-1000 IU) or 20 µg mL−1 poly-L-lysine (MilliporeSigma, P2658) before the addition of 20 µg mL−1 of calf thymus DNA (Sigma-Aldrich, D7290), 25 µg mL−1 RNA (Thermo Scientific, AM7120G). Plates were then blocked in PBS/2% FBS (Thermo Fisher Scientific, 26140079) for 2 h at room temperature. Sera were 1:50 diluted in PBS/2% FBS. Bound IgG was detected with HRP-conjugated goat anti-mouse IgG (H + L) (1:4,000 dilutions; Southern Biotech, 1031-05) followed by addition of substrate solution (Seramun Diagnostica, S-100-TMB). Samples were acquired with Biotek Gen5 v1. 05.
For anti-COL2 antibody or IgG detection, plates were coated with 10 µg mL−1 of purified COL2, goat anti-mouse IgG, or human ads-UNLB (SouthernBiotech, 1030-01) in PBS at 4 °C overnight. After blocking with 5% BSA in PBS at room temperature (RT) for 2 h, diluted polyclonal antibodies purified from QD mouse antibodies or 1:1000 diluted serum samples were incubated at RT for 2 h. The antibody titers were evaluated using HRP-conjugated goat anti-mouse-IgG, -IgG2b, and -IgM (1:4,000 dilutions; Southern Biotech, 1030-05, 1090-05, 1020-05). Samples were acquired with Biotek Gen5 v1. 05.
Detection of anti-MNV antibodies in the sera was performed using an ELISA Kit (Express Biotech, MD, 595-631C) or plates coated with 104 copies/100 μL of MNV-S99 solution, after blocking with 3% BSA at 37 °C for 2 h. Sera was diluted 1:1000 in PBS/2% FBS. Bound IgG was detected with goat anti-mouse IgG, human ads-BIOT (1:4000 dilutions; Southern Biotech, 1030-08), followed by Streptavidin-HRP (1:800 dilutions, Southern Biotech, 7105-05) and substrate solution (Seramun Diagnostica, S-100-TMB)53. Samples were acquired with Biotek Gen5 v1. 05.
For ELISpot, plates were activated with 15 µL of 35% ethanol, washed two times with PBS, and coated with 10 μg mL−1 COL2 in PBS at 4 °C overnight. After washing, 1 × 106 splenocytes were plated per well in complete RPMI media containing 10% FCS and penicillin/streptomycin and incubated for 6 h at 37°C. After washing, biotinylated goat anti-mouse IgG (1:1000 dilutions, Southern Biotech, 1030-08) or IgG2b (1:1000 dilutions, Southern Biotech, 1090-08) was added to the wells and incubated at RT for 2 h. ExtrAvidin conjugated alkaline phosphatase (Sigma-Aldrich, E2636) diluted in PBS was added and incubated for 30–45 min. 100 μL of substrate BCIP/Nitroblue Tetrazolium (Sigma-Aldrich, B3804) per well (1 tablet in 10 mL ddH2O and pre-filtered with 0.45 µm) was added and incubated in the dark for 10–15 min. When visible spots appeared, the wells were rinsed thoroughly with tap water. The plastic bottom from the plate was removed, rinsed further, and dried in darkness. Scanned wells (ImmunoScan) were recorded by BioSpot Software, and analyzed with ImmunoSpot software (Cellular Technology).
Flow cytometry
Flow cytometry analyzes were performed54. Briefly, organs were collected, mashed, and filtered through 45 µM filter to obtain single-cell suspensions in PBS. To prepare single-cell suspensions, the perfused kidneys were digested with 1 mg/mL collagenase (Roche, 11088866001) and 0.1 mg/mL DNase I (Roche, 10104159001) in a 37 °C water bath for 45 min. Red blood cells were lysed using ammonium-chloride-potassium (ACK) buffer (homemade), cells were counted on a Sysmex, and purified rat anti-mouse Fc-block (CD16/CD32, 24G2, homemade, BD Biosciences; ≤ 1 μg/million cells in 100 μl) was added for 10 min at RT. Surface antigens were stained with fluorescently labeled antibodies. All the antibodies were used with 0.2 µg per million cells in 100 µl volume. To stain immune cells in the peritoneal cavity, spleens, and kidneys, CD45-HV500, Ly6C-BV605, Ly6G-APC, F4/80-PE, B220-Fluor50, CD19-PE, CD3-APC, CD4-PerCP/Cyanine5.5 and CD8a-FITC antibodies (Biolegend) were used. For the staining B and T cells, CD19-PE-Cy7, B220-PB, CD93-PE, CD21-APC, CD23-PerCP-Cy5.5, IgM-BV605, IgD-BV650, B220-APC, MHCII-FITC, CXCR4-PerCP-Cy5.5, CXCR5-BV421, CD4-BV605, CD44-AF700, CD62L-FITC, CXCR5-BV421, PD-1-PE-Cy7, FOXP3-APC, CD93-PE, CD19-AF700, CD138-BV605, GL7-APC, CD38-PE, Sca-1-PE-Cy7, CD69-APC, CXCR5-PB, PD-1-PE, CCR6-PE-Cy7, CXCR3 (CD183)-PerCP-Cy5.5, CD69-PE antibodies (Biolegend) were used. For detection of STAT1/p-STAT, peritoneal exudates cells were collected one day after MNV intraperitoneal injection at the dose of 3 × 105 TCID50, stained with CD45-HV500, CD11b-PE-Cy7, Ly6C-BV605, Ly6G-APC, F4/80-FITC (Biolegend), washed and fixed with Cytofix buffer (BD Biosciences) for 10 min at 37 °C, permeabilized with Phosflow Perm Buffer III on ice for 30 min, washed twice and then stained with PE Mouse anti-Total Stat1 (1:50 dilutions; BD Phosflow, N-Terminus, 558537) and BV421 Mouse anti-p-STAT1Tyr701 antibody (1:50 dilutions; BD Phosflow, 566238). For the detection of TLR7 and 9, splenocytes were collected, CD45-PerCP-Cy5.5, CD11b-PB, F4/80-APC, Ly6C-BV605, CD11c-PE-Cy7, CD19-AF700, B220-PB, PDCA1 (CD317)-APC antibodies (Biolegend) were used. Cells were fixed, permeabilized, and stained with TLR7-PE (BioLegend, clone: A94B10, 160003), and TLR9-FITC (BioLegend, clone: S18025A, 159107) antibodies. Dead cells were excluded using FVS780 (BD Biosciences, 565388) or a fixable near-IR dead cell stain kit (Thermo Fisher Scientific, L10119). Samples were either acquired with an Attune v5.2.0 flow cytometer (Thermo Fisher) or using LSR Fortessa (BD Biosciences). Samples were analyzed with FlowJo version 10.6.
Bone marrow-derived macrophages (BMDMs)
Mouse BMDMs were obtained from the differentiation of monocytes recovered from the femur and tibia of 6 to 8- week- old male B6N.Q.Ncf1R90, B6N.Q. Ncf190H, B6N.Q and B6N.Q.Ncf1m1J mice. Bone marrow was flushed from the cavity, and single-cell suspensions were generated and cultured in RPMI-1640 medium (Gibco, 21875091) with M-CSF (20 ng/mL, Peprotech, 315-02-10) and 10% fetal bovine serum for obtaining macrophages. The mature macrophages (M0) were confirmed by identifying the makers (F4/80+ CD11b+) using flow cytometry. For ROS measurement, BMDMs were cultured, collected, washed with PBS and used. To measure the intracellular ROS production, a DCFH-DA fluorescent probe kit (Byotime, S0033S) was used with or without Rosup stimulation. For extracellular ROS measurement, BMDMs were treated with 100 μL of isoluminol reagent buffer, as previously described10. Data was collected using a Biotek plate reader.
Immunofluorescence
BMDMs from the naïve and IFN-α stimulated mice were cultured in confocal dishes, fixed with 4% paraformaldehyde, blocked with 10% FCS/PBS buffer, and then incubated with the primary antibodies: mouse anti-p47phox (D-10) antibodies (Santa Cruz Biotechnology, sc-17845), or STAT1 (CST, clone: D1K9Y, 65748), p-STAT1Tyr701 (CST, clone: 58D6, 88845), STAT3 (CST, clone: 79D7, 4904), and p-STAT3Tyr705 (CST, clone: Tyr705, 9131) specific rabbit antibodies (1:1000 dilutions). Subsequently, cells were incubated with either goat anti-mouse IgG (H + L) (1:1000 dilutions; CST, 4408) or goat anti-rabbit IgG (H + L)-Alexa Fluor 488 (1:1000 dilutions; CST, 4412), followed by counterstaining with 4’, 6-diamidino-2-phenylindole DAPI (Vector, CA) and dried for 30 min before scanning under a confocal microscope (LSM880 with Airyscan, CarlZeiss, Germany).
Western blot
For experiments in vitro, 1000 U/mL of IFN-α (BioLegend, 752804) was used to stimulate BMDMs (M0 cells) for 30 min, 1000 U mL−1 of IFN-α and 25 µM of GSK2795039 (MCE, HY-18950) or 1000U mL−1 of IFN-α and 10 µM of H2O2 was used. Total protein lysates from cells or tissues were extracted using the radioimmunoprecipitationassay (RIPA) solution with a cocktail of protease and phosphatase inhibitors (Roche, 04693116001, 04906837001). The final protein concentration of each sample was determined with a BCA kit (ThermoFisher Scientific, 23225). The supernatants from protein lysates were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) (Invitrogen, NP0321BOX). The following primary antibodies were used: mouse ani-NCF1 (p47phox) (D-10) antibodies (Santa Cruz Biotechnology, sc-17845) or JAK1 (CST, 3332), p-JAK1 (CST, 3331), JAK2 (CST, 3230), p-JAK2 (CST, 3771), STAT1 (CST, clone: D1K9Y, 65748), p-STAT1Tyr701 (CST, clone: 58D6, 88845), STAT3 (CST, clone: 79D7, 4904), and p-STAT3Tyr705 (CST, clone: Tyr705, 9131) specific rabbit antibodies (1:1000 dilutions), TLR-7 (CST, D7, 5632) and MyD88 (CST, D80F5, 4283) specific rabbit antibodies, and β-Actin rabbit antibody (CST, 4967) at 4°C overnight. The signal was detected using the goat anti-mouse IgG conjugated with HRP (1:4000 dilutions; Southern Biotech, 1031-05) or anti-rabbit IgG conjugated with HRP (1:4000 dilutions; Southern Biotech, 4030-05) secondary antibody. The data were collected by Bio-Rad CFX Manager v3.1 and analyzed by Image J.
Quantitative real-time PCR
Spleen, kidney, and bone marrow cells were obtained from age- and sex-matched mutated and wild-type mice after carbon dioxide asphyxiation. Bone marrow macrophages were harvested and cultured as described above. Total RNA was isolated by using TRIzol Reagent (Invitrogen, 15596026CN), and complementary DNA (cDNA) was synthesized using the First Strand cDNA Synthesis Kit (ThermoFisher Scientific, K1622). RT-qPCR was performed by Agilent Strata gene Mx3005P with FastStart Universal SYBR Green Master (Roche, 4913914001) to assess gene expression. The data were analyzed with Microsoft Office 16. The relative gene expression normalized by β-actin was calculated using the 2−△△CT method. The primers used are described in Supplementary Table 3 and 4.
The small intestine, colon, and spleen were prepared for RNA extraction52. Detection of MNV was done with an MNV-specific primer pair targeting a 396 bp region at the 5′end of MNV ORF2 (VP1) using an RT-PCR kit (Thermo Fisher Scientific)55. E.Z.N.A.R Viral RNA Kit (Omega, R6874-01) was used to extract the viral RNAs from the feces of MNV-infected mice. The SuperScript® First-Strand Synthesis System for RT-PCR (Invitrogen, 11904-018) was used to synthesize first-strand cDNA. Different regions of the MNV genomes were amplified by applying 12 pairs of oligonucleotide primers designed based on the sequence from the MNV BJ 10-2062 strain (Table S3). After purification and cloning into the pMD18-T vector (TaKaRa, Japan), the PCR products were sequenced by Beijing Sunbiotech Co., Ltd. (Beijing, China). Lasergene software (DNAstar Inc., USA) was used to assemble and analyze the sequence data. Clustal X 2.1 and MEGA 6 programs were used to perform multiple sequence alignments and phylogenetic analyzes, respectively. The genome sequence of the isolated strain was registered in GenBank (Accession Number: MT358379). Strand-specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA as described previously56.
Statistical analysis
Quantitative data were expressed as mean ± SEM. The statistical analysis of differences between the experimental groups was performed using a one-way analysis of variance (ANOVA) or Student’s t-test. ROS assays were calculated using Mann-Whitney or Kruskal-Wallis tests. Clinical arthritis and histology scores were analyzed using the Mann-Whitney U-test, whereas cellular analyzes were analyzed with the One-Way ANOVA combined with the Tukey post-test. Pearson’s correlation test was used for analyzing the correlation of antibody responses using GraphPad Prism Software Version 9.5.0 (GraphPad Software, Inc.). All data shown are mean ± SEM. The p values less than 0.05 were considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are included in the Supplementary Information or available from the authors, as are unique reagents used in this Article. The raw numbers for charts and graphs are available in the Source Data file whenever possible. Source data are provided with this paper.
References
Fava, A. & Petri, M. Systemic lupus erythematosus: diagnosis and clinical management. J. Autoimmun. 96, 1–13 (2019).
Visscher, P. M., Yengo, L., Cox, N. J. & Wray, N. R. Discovery and implications of polygenicity of common diseases. Science 373, 1468–1473 (2021).
Lappalainen, T. & MacArthur, D. G. From variant to function in human disease genetics. Science 373, 1464–1468 (2021).
Olofsson, P. et al. Positional identification of Ncf1 as a gene that regulates arthritis severity in rats. Nat. Genet 33, 25–32 (2003).
Li, X. J., Marchal, C. C., Stull, N. D., Stahelin, R. V. & Dinauer, M. C. p47phox Phox homology domain regulates plasma membrane but not phagosome neutrophil NADPH oxidase activation. J. Biol. Chem. 285, 35169–35179 (2010).
Hultqvist, M. et al. Positioning of a polymorphic quantitative trait nucleotide in the Ncf1 gene controlling oxidative burst response and arthritis severity in rats. Antioxid. Redox Signal 14, 2373–2383 (2011).
Hultqvist, M. et al. Enhanced autoimmunity, arthritis, and encephalomyelitis in mice with a reduced oxidative burst due to a mutation in the Ncf1 gene. Proc. Natl Acad. Sci. USA 101, 12646–12651 (2004).
Olsson, L. M. et al. Copy number variation of the gene NCF1 is associated with rheumatoid arthritis. Antioxid. Redox Signal 16, 71–78 (2012).
Zhao, J. et al. A missense variant in NCF1 is associated with susceptibility to multiple autoimmune diseases. Nat. Genet 49, 433–437 (2017).
Olsson, L. M. et al. A single nucleotide polymorphism in the NCF1 gene leading to reduced oxidative burst is associated with systemic lupus erythematosus. Ann. Rheum. Dis. 76, 1607–1613 (2017).
Linge, P. et al. NCF1−339 polymorphism is associated with altered formation of neutrophil extracellular traps, high serum interferon activity and antiphospholipid syndrome in systemic lupus erythematosus. Ann. Rheum. Dis. 79, 254–261 (2020).
Kelkka, T. et al. Reactive oxygen species deficiency induces autoimmunity with type 1 interferon signature. Antioxid. Redox Signal 21, 2231–2245 (2014).
Kienhöfer, D. et al. Experimental lupus is aggravated in mouse strains with impaired induction of neutrophil extracellular traps. JCI Insight 2, e92920 (2017).
Jog, N. R. & James, J. A. Epstein barr virus and autoimmune responses in systemic lupus erythematosus. Front Immunol. 11, 623944 (2020).
Zhao, J., You, X. & Zeng, X. Research progress of BK virus and systemic lupus erythematosus. Lupus 31, 522–531 (2022).
Ukadike, K. C. & Mustelin, T. Implications of endogenous retroelements in the etiopathogenesis of systemic lupus erythematosus. J. Clin. Med 10, 856 (2021).
Larionova, R. et al. SARS-Cov2 acute and post-active infection in the context of autoimmune and chronic inflammatory diseases. J. Transl. Autoimmun. 5, 100154 (2022).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880 e821 (2019).
Bodansky, A. et al. Molecular mimicry in multisystem inflammatory syndrome in children. Nature 632, 622–629 (2024).
Vojdani, A., Pollard, K. M. & Campbell, A. W. Environmental triggers and autoimmunity. Autoimmune Dis. 2014, 798029 (2014).
Wing, K., Klocke, K., Samuelsson, A. & Holmdahl, R. Germ-free mice deficient of reactive oxygen species have increased arthritis susceptibility. Eur. J. Immunol. 45, 1348–1353 (2015).
Satoh, M. & Reeves, W. H. Induction of lupus-associated autoantibodies in BALB/c mice by intraperitoneal injection of pristane. J. Exp. Med 180, 2341–2346 (1994).
Chachu, K. A. et al. Antibody is critical for the clearance of murine norovirus infection. J. Virol. 82, 8 (2008).
Luo, H. et al. NCF1-dependent production of ROS protects against lupus by regulating plasmacytoid dendritic cell development and functions. JCI Insight, 8, e164875 (2023).
Roth, A. N. et al. Norovirus infection causes acute self-resolving diarrhea in wild-type neonatal mice. Nat. Commun. 11, 2968 (2020).
Tarkowski, A. et al. Patterns of autoreactivity to collagen type II in autoimmune MRL/l mice. Clin. Exp. Immunol. 63, 441–449 (1986).
Aoun, M. et al. Antigen-presenting autoreactive B cells activate regulatory T cells and suppress autoimmune arthritis in mice. J. Exp. Med. 220, e20230101 (2023).
Kanno, A. et al. Targeting cell surface TLR7 for therapeutic intervention in autoimmune diseases. Nat. Commun. 6, 6119 (2015).
Subramanian, S. et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc. Natl Acad. Sci. USA 103, 9970–9975 (2006).
Ramanujam, M. et al. Interferon-alpha treatment of female (NZW x BXSB)F(1) mice mimics some but not all features associated with the Yaa mutation. Arthritis Rheum. 60, 1096–1101 (2009).
Nice, T. J. et al. Type I interferon receptor deficiency in dendritic cells facilitates systemic murine norovirus persistence despite enhanced adaptive immunity. PLoS Pathog. 12, e1005684 (2016).
Enosi Tuipulotu, D., Netzler, N. E., Lun, J. H., Mackenzie, J. M. & White, P. A. TLR7 agonists display potent antiviral effects against norovirus infection via innate stimulation. Antimicrob. Agents Chemother. 62, e02417–e02417 (2018).
Yu, P. et al. cGAS-STING effectively restricts murine norovirus infection but antagonizes the antiviral action of N-terminus of RIG-I in mouse macrophages. Gut Microbes 13, 1959839 (2021).
James, J. et al. Redox regulation of PTPN22 affects the severity of T-cell-dependent autoimmune inflammation. Elife 11, e74549 (2022).
Geng, L. et al. Human SLE variant NCF1-R90H promotes kidney damage and murine lupus through enhanced Tfh2 responses induced by defective efferocytosis of macrophages. Ann. Rheum. Dis. 81, 255–267 (2022).
Meng, Y. et al. The NCF1 variant aggravates autoimmunity by facilitating the activation of plasmacytoid dendritic cells. J. Clin. Invest 132, e153619 (2022).
Li, Y., Li, Z., Nandakumar, K. S. & Holmdahl, R. Human NCF1(90H) variant promotes IL-23/IL-17-dependent mannan-induced psoriasis and psoriatic arthritis. Antioxid. (Basel) 12, 1348 (2023).
James, J. et al. Redox regulation of LAT enhances T cell-mediated inflammation. Antioxid. (Basel) 13, 499 (2024).
Savina, A. et al. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell 126, 205–218 (2006).
Yang, M. et al. Cutting edge: processing of oxidized peptides in macrophages regulates T cell activation and development of autoimmune arthritis. J. Immunol. 199, 3937–3942 (2017).
Jacob, C. O. et al. Haploinsufficiency of NADPH oxidase subunit neutrophil cytosolic factor 2 is sufficient to accelerate full-blown lupus in NZM 2328 mice. Arthritis Rheumatol. 69, 1647–1660 (2017).
Winter, S., Hultqvist Hopkins, M., Laulund, F. & Holmdahl, R. A reduction in intracellular reactive oxygen species due to a mutation in NCF4 promotes autoimmune arthritis in mice. Antioxid. Redox Signal 25, 983–996 (2016).
He, C. et al. NCF4 dependent intracellular reactive oxygen species regulate plasma cell formation. Redox Biol. 56, 102422 (2022).
Wobus, C. E. et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2, e432 (2004).
Hsu, C. C. et al. Murine norovirus inhibits B cell development in the bone marrow of STAT1-deficient mice. Virology 515, 123–133 (2018).
Donaldson, E. F., Lindesmith, L. C., Lobue, A. D. & Baric, R. S. Norovirus pathogenesis: mechanisms of persistence and immune evasion in human populations. Immunol. Rev. 225, 190–211 (2008).
Rocha-Pereira, J. et al. Interferon lambda (IFN-lambda) efficiently blocks norovirus transmission in a mouse model. Antivir. Res 149, 7–15 (2018).
Nordgren, J. & Svensson, L. Genetic susceptibility to human norovirus infection: an update. Viruses 11, 226 (2019).
Bäcklund, J. et al. C57BL/6 mice need MHC class II Aq to develop collagen-induced arthritis dependent on autoreactive T cells. Ann. Rheum. Dis. 72, 1225–1232 (2013).
Kilkenny, C., Browne, W. J., Cuthill, I. C., Emerson, M. & Altman, D. G. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 8, e1000412 (2011).
Alsved, M. et al. Aerosolization and recovery of viable murine norovirus in an experimental setup. Sci. Rep. 10, 15941 (2020).
Pearson, J. A. et al. Norovirus changes susceptibility to type 1 diabetes by altering intestinal microbiota and immune cell functions. Front Immunol. 10, 2654 (2019).
Hwang, S. et al. Murine norovirus: propagation, quantification, and genetic manipulation. Curr. Protoc. Microbiol. 33, 11–61 (2014).
Sareila, O., Hagert, C., Rantakari, P., Poutanen, M. & Holmdahl, R. Direct comparison of a natural loss-of-function single nucleotide polymorphism with a targeted deletion in the Ncf1 gene reveals different phenotypes. PLoS One 10, e0141974 (2015).
Kim, J. R. et al. Prevalence of murine norovirus infection in Korean laboratory animal facilities. J. Vet. Med Sci. 73, 687–691 (2011).
Vashist, S., Urena, L. & Goodfellow, I. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J. Virol. Methods 184, 69–76 (2012).
Acknowledgements
This study was supported by the KI Foundation for Virus Research (2023-00122, YL), KI Foundation funds for rheumatology research (2023-02710, YL), Science and Technology Major Project District-School Cooperation Outstanding Youth Fund (Shenzhen Nanshan District Health System (NSZD) (NSZD2023062, ZL), the EU COSMIC Marie Curie grant (765158, RH), the Swedish Research Council (2023-06482, RH), Southern Medical University (SMU) grant (C1034211, RH), the Natural Science Foundation of China (No.32070913, 82471830, W2431021, RH), Vetenskapsrådet (VR) (2024-02575, RH), NovoNordisk (NNF24OC0090035, RH), Leo Foundation (LF-OC-22-001023, RH), Cancer foundation (222350Pj01H, RH), and KAW (2019.0059, RH). We thank prof. Patrik Medstrand for collaboration of MNV strain Berlin/06/06DE S99 production.
Funding
Open access funding provided by Karolinska Institute.
Author information
Authors and Affiliations
Contributions
Y.L. and R.H. designed the experiments. Y.L. established models and did most of the experiments. A.C. did the flow cytometry and ELISPOT. Z.L. did histology immunofluorescence staining and western blot. J.L. supervised virus production. M.A. produced and quantified MNV. Q.L., R.X., and H.L. participated in the collection of samples, and the scoring of mice. H.L. helped with the collection of samples and the detection of proteinuria. D.L. and J.X. participated in breeding mice. KSN contributed to mice breeding and mice injection. L.M. was involved in the genotyping of the mice. Y.L. wrote the first draft of the paper with the help of R.H. R.H. supervised the project. All authors approved the study and revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
All authors agree to publish the data in this paper. The authors declare no competing interests. The authors declare an absence of any commercial or financial relationships.
Peer review
Peer review information
Nature Communications thanks Clifford Lowell and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, Y., Coelho, A., Li, Z. et al. The systemic lupus erythematosus-associated NCF190H allele synergizes with viral infection to cause mouse lupus but also limits virus spread. Nat Commun 16, 1593 (2025). https://doi.org/10.1038/s41467-025-56857-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56857-z
This article is cited by
-
Macrophage retrotransposon expression is associated with lupus
Genes & Immunity (2025)