Abstract
Dynamic changes in cell size are associated with development and pathological conditions, including aging. Although cell enlargement is a prominent morphological feature of cellular senescence, its functional implications are unknown; moreover, how senescent cells maintain their enlargement state is less understood. Here we show that an extensive remodeling of actin cytoskeleton is necessary for establishing senescence-associated cell enlargement and pro-inflammatory senescence-associated secretory phenotype (SASP). This remodeling is attributed to a balancing act between the SASP regulator GATA4 and the mechanosensor YAP on the expression of the Rho family of GTPase RHOU. Genetic or pharmacological interventions that reduce cell enlargement attenuate SASP with minimal effect on senescence growth arrest. Mechanistically, actin cytoskeleton remodeling couples cell enlargement to the nuclear localization of GATA4 and NF-κB via the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex. RhoU protein accumulates in mouse adipose tissue under senescence-inducing conditions. Furthermore, RHOU expression correlates with SASP expression in adipose tissue during human aging. Thus, our study highlights an unexpected instructive role of cell enlargement in modulating the SASP and reveals a mechanical branch in the senescence regulatory network.
Similar content being viewed by others
Introduction
Cellular senescence is a multi-functional cell state of stable growth arrest triggered by various intracellular and extracellular stresses1,2,3. Senescence represents a systemic homeostatic response, coordinating the removal of damaged cells and tissue regeneration through the action of intrinsic and extrinsic factors4,5,6. When dysregulated, however, senescence causes numerous aging-related pathologies and eventually contributes to aging7,8,9,10,11,12,13,14,15,16. The key component of these pleiotropic effects of senescence is the senescence-associated secretory phenotype (SASP), which includes many pro-inflammatory cytokines, chemokines, growth factors, and proteases17,18. During the last decade, the molecular mechanisms by which the SASP is initiated are well characterized by identifying its transcriptional activation programs, including NF-κB, GATA4, BRD4, HMGB2, and cGAS-STING19,20,21,22,23,24,25. How senescent cells maintain this massive secretome remains elusive except the sustained mTORC1 activity, which supports the translation of SASP factors and forms a positive feedback loop that enhances SASP expression26,27,28,29,30,31.
Cell enlargement is the most prominent characteristic of senescence; senescing cells increase in size, significantly deviating from the optimal cell size of their proliferating counterparts32,33. Recent studies demonstrated that such enlargement decreases the DNA-to-cytoplasm ratio, globally affecting the cell proteome composition and contributing to proliferation defects34,35,36. Although this elegant series of studies suggested that cell enlargement could be a causal, rather than just consequential, factor for senescence, they did not fully address another biologically and physiologically important question of senescence: why do senescent cells need to increase in size? Specifically, we still do not sufficiently know the functional role of cell enlargement, nor do we understand how senescent cells maintain their enlargement state.
Yes-associated protein (YAP, also known as YAP1) is a core effector of cellular mechanosignaling, which senses various mechanical cues, such as cell stretching, membrane tension, and extracellular/intracellular stiffness via actin stress fibers37,38,39,40,41,42,43. Such cytoskeletal reorganization increases YAP nuclear import by stretching the nucleus and remodeling nuclear pores via the Linker of Nucleoskeleton and Cytoskeleton (LINC) complex, activating its downstream transcriptional program44,45,46. Interestingly, a recent study showed that stromal YAP activity declines during physiological aging, and sustaining YAP function rejuvenates old cells by limiting senescence and its associated inflammation47. YAP has also been shown to overcome oncogene-induced senescence by promoting dNTP biosynthesis48. Despite this significant advance, it is still unclear what the reciprocal relationship between YAP activity and senescence-associated cell enlargement is. Specifically, whether YAP modulates cell enlargement and, if so, what the functional consequences of such modulation remain elusive.
In the current work, we showed that senescent cells extensively remodel the actin cytoskeleton network to support and maintain cell enlargement. The SASP regulator GATA4 and the mechanosensor YAP antagonistically regulate the expression of the Rho family GTPase RHOU to set such remodeling off during senescence. Interestingly, genetic or pharmacological interventions that reduce cell enlargement ablate the SASP with minimal effect on senescence growth arrest, suggesting an instructive role for actin remodeling and cell enlargement in the secretory axis of the senescence regulatory network. Mechanistically, cell enlargement-associated actin remodeling signals through the LINC complex to promote the nuclear localization of GATA4 and NF-κB, forming a positive feedback loop to maintain the SASP transcriptional program. Physiologically, RhoU protein abundance increases in the adipose tissue of male mice during natural aging and high-fat diet (HFD)-induced senescence; RHOU expression correlates with SASP expression in human adipose tissue during aging. Therefore, our study proposes that the mechanistic branch of the senescence regulatory network is essential for the pro-inflammatory secretory program of senescence.
Results
Actin stress fibers support cell enlargement and SASP
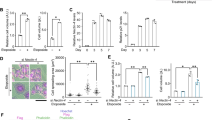
To investigate the temporal regulation of cell enlargement during senescence, we leveraged DNA damage-induced senescence, in which senescence induction can be tightly regulated17,49,50 (Fig. 1a). Cell cycle arrest, as assessed by the expression of two main cyclin-dependent kinase inhibitors (CDKN1A and CDKN2A), was established at the earlier stage; the expression of several pro-inflammatory SASP genes (e.g., IL1A, IL8, and CXCL1) displayed the delayed pattern of activation, as previously reported (Fig. 1a)17. By contrast, the kinetics of cell enlargement fall between those of cell cycle arrest and the SASP, as assessed by forward scatter intensity (FSC) using flow cytometry, which is proportional to cell diameter (Fig. 1a).
a Abundance of the indicated mRNAs for the indicated day after senescence induction by 12 Gy IR in IMR90 cells expressing hTERT (IMR90-hTERT cells) analyzed by RT-qPCR (left, n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). Cell size for the indicated day analyzed by Fluorescence-Activated Cell Sorting (right, n = 4 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). b Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, and phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 57, 36, 46, and 47 randomly chosen cells from 3 biologically independent experiments for PRO, SEN, SEN+LatA, and SEN + Y27632, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test, AFU denotes arbitrary fluorescence units). c Quantification of the percentage of SA-β-gal positive cells (n = 9 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). d Abundance of the indicated proteins analyzed by immunoblotting (left, n = 3 biologically independent experiments) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). e Abundance of the indicated proteins analyzed by immunoblotting (left, n = 3 biologically independent experiments) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. LatA denotes Latrunculin A. Source data are provided as a Source Data file.
While verifying this observation with microscopic analysis, interestingly, we found that the formation of actin stress fibers resembles such kinetics: it becomes prominent after the establishment of cell cycle arrest, but before the development of full SASP activation (Fig. 1a; Supplementary Fig. 1a). This change was accompanied by an increased ratio of filamentous actin (F-actin) to free globular actin (G-actin), with increased levels of F-actin and total actin (Supplementary Fig. 1b). This finding, together with previous studies showing that actin cytoskeleton is an integral part of cellular mechanical dynamics and plays a role in supporting diverse changes in cell morphology and shape51,52,53,54,55, suggests that actin stress fibers may contribute to senescence-associated cell enlargement and SASP regulation.
To test this hypothesis, we employed Latrunculin A, an inhibitor of actin polymerization, to disrupt actin stress fibers56. Treatment with Latrunculin A suppressed the formation of actin stress fibers during senescence, as expected; it also reduced the size of senescent cells as assessed by FACS analysis and confirmed by microscopic analysis, suggesting a role for actin stress fibers in cell enlargement (Fig. 1b; Supplementary Fig. 1c). Consistently, phalloidin intensity normalized to cell size did not differ significantly between proliferating and senescent cells, with or without Latrunculin A treatment. Disruption of actin stress fibers did not affect either senescence-associated β-galactosidase (SA-β-Gal) activity (Fig. 1c), a well-established marker of senescence, or cell cycle arrest, as assessed by expression of cyclin-dependent kinase (CDK) inhibitors (i.e., CDKN1A and CDKN2A) and Bromodeoxyuridine (BrdU) incorporation (Fig. 1d; Supplementary Fig. 1d). By contrast, expression of several SASP genes, including IL1A, IL1B, IL8, and CXCL1, was strongly suppressed upon treatment with Latrunculin A (Fig. 1d), suggesting that actin stress fibers are somehow involved in the SASP induction or maintenance. The same was true for another human fibroblast, BJ cells (Supplementary Fig. 2a–c).
To corroborate these results, we continuously inhibited the Rho-associated protein kinase (ROCK) with its specific inhibitor Y27632 during senescence. ROCK activity has been shown to be required for the optimal assembly of actin stress fibers51. We found that ROCK inhibition suppressed cell enlargement and SASP expression in two different human fibroblast cell lines (i.e., IMR90 and BJ cells), as did Latrunculin A treatment (Fig. 1b, e; Supplementary Fig. 1c, 2d, e). ROCK inhibition partially reduced p21 and p16 expression in senescent cells, but they were still significantly higher than those in proliferating cells. Consistently, Y27632 treatment had negligible effects on BrdU incorporation (Fig. 1e; Supplementary Fig. 1d, 2e). Of note, treatment with Y27632 also decreased SA-β-Gal activity (Fig. 1c; Supplementary Fig. 2f), consistent with its previously reported anti-senescence effects57,58. These data suggest that treatment with Y27632 may have additional downstream targets besides actin stress fibers to modulate senescence. By contrast, pre-treatment with jasplakinolide51,52, an actin polymerization stabilizer, has negligible effects on actin stress fibers, cell enlargement, and SASP expression during senescence (Supplementary Fig. 2g). These data suggest that jasplakinolide treatment acts on the same pathway of actin stress fiber formation during senescence. Collectively, these data suggest that actin stress fibers support cell enlargement and that such a morphological change is distinctively associated with the SASP.
The mechanosensor YAP restrains cell enlargement and SASP
Enlarged cells experience mechanical stress, which they need to overcome for homeostasis59,60,61,62. One critical regulator of mechanical stress responses is YAP37,39,40,41. During cell spreading, actin stress fibers signal through the LINC complex to stretch the nucleus and remodel nuclear pores. YAP then enters the nucleus to initiate a transcriptional program that determines cell fate, such as proliferation, survival, and differentiation44,46. Interestingly, however, gene set enrichment analysis (GSEA) of differentially expressed genes during DNA damage-induced senescence indicated suppression, not activation, of the YAP pathway in senescent cells (Fig. 2a, Supplementary Data 1-3). The same held true for replicative senescence and oncogene-induced senescence (Supplementary Fig. 3a)63. Western blot analysis verified this notion, showing that phosphorylation of YAP at Ser127, a key regulatory event of YAP inhibition37,40, was enhanced and its protein abundance decreased during senescence (Fig. 2b). This observation is in line with recent studies showing that YAP activity declines in stromal cells during physiological mouse aging, contributing to the activation of the cGAS-STING pathway, chronic inflammation, and senescence47. Thus, these data demonstrate that YAP activity decreases in senescent cells despite their enlarged status, suggesting a senescence-specific regulation of YAP that is decoupled from mechanotransduction.
a Gene Set Enrichment Analysis (GSEA) of differentially expressed genes during senescence (nominal p-value and normalized enrichment score (NES) calculated by GSEA, one-sided permutation test). b Abundance of the indicated proteins analyzed by immunoblotting (left, n = 5 biologically independent experiments). Densitometric analysis performed to determine either the YAP/VINCULIN ratio or the p-YAP/YAP ratio (right, mean ± SEM, two-sided one sample t-test). c Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). The arrowhead indicates endogenous YAP. d Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, and phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 67, 52, 40, and 38 randomly chosen cells from 3 biologically independent experiments for PRO, PRO + YAP5SA, SEN, and SEN + YAP5SA, respectively). e Quantification of the percentage of SA-β-gal positive cells (left, n = 6 biologically independent samples) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples). f Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). g Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 99, 79, 71, and 70 randomly chosen cells from 3 biologically independent experiments for PRO, PRO+shYAP#1, SEN, and SEN+shYAP#1, respectively). h Quantification of the percentage of SA-β-gal positive cells (left, n = 6 biologically independent samples) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples). Box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values; one-way ANOVA test with Tukey’s multiple comparisons test; AFU denotes arbitrary fluorescence units for d, g. Mean ± SEM; one-way ANOVA test with Tukey’s multiple comparisons test for e, h. PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. Source data are provided as a Source Data file.
To examine the potential role of YAP reduction in senescence, we ectopically expressed the YAP5SA mutant, a constitutively active form of YAP37,40, to maintain its activity during senescence (Fig. 2c). Interestingly, expression of YAP5SA prevented cell enlargement as well as the formation of actin stress fibers during senescence (Fig. 2d); however, neither SA-β-Gal activity nor CDKN1A/2A expression changed upon YAP5SA expression in senescent cells (Fig. 2e). Consistent with the close association between cell enlargement and the SASP, expression of YAP5SA halted the expression and secretion of several SASP factors during senescence (Fig. 2c, e; Supplementary Fig. 3b). Of note, expression of YAP5SA tended to increase p16 expression and SA-β-Gal activity in proliferating cells (Fig. 2c, e), indicating either its potentially dichotomous effect on senescence or an oncogene-induced senescence-like state. Ectopic expression of wild-type YAP (YAPWT) had similar effects on cell enlargement and SASP expression, although to a lesser extent than YAP5SA (Supplementary Fig. 3c). YAPWT expression did not significantly increase p16 or SA-β-Gal activity in proliferating cells, in contrast to the effect of YAP5SA expression. These data suggest that YAP mainly counteracts a subset of the senescence regulatory network.
Effects of YAP5SA expression on cell enlargement and SASP depended largely on the transcriptional regulation, as evidenced by the observation showing that expression of a transcriptionally inactive YAP5SA/S94A exerted negligible effects64 (Supplementary Fig. 3d–f). By contrast, depletion of YAP1 further enhanced actin stress fiber formation and cell enlargement (Fig. 2f, g). These were accompanied by enhanced SASP expression and secretion, but without altering SA-β-Gal activity or CDKN2A expression (Fig. 2f, h; Supplementary Fig. 3g). These phenotypes were verified with additional YAP1 shRNA (Supplementary Fig. 3h). Furthermore, the effects of YAP5SA expression on cell enlargement and SASP were conserved in senescent BJ cells (Supplementary Fig. 4a–c).
Lastly, ectopic expression of TAZ, a paralogue of YAP, negligibly affected cell enlargement and actin remodeling (Supplementary Fig. 5a); interestingly, ectopic expression of TAZ enhanced rather than suppressed the SASP (Supplementary Fig. 5a, b), suggesting distinct roles for YAP and TAZ in senescence that have been also described in other biological conditions65. Collectively, these data show that gain of function of YAP inhibits the morphological changes associated with senescence and the SASP, while its loss of function enhances them, suggesting that YAP is a focal node linking cell enlargement and the secretory phenotype.
RHOU mediates senescent actin remodeling and enlargement
To explore the mechanism by which YAP modulates actin remodeling and cell enlargement in senescence, we investigated how sustaining YAP activity influences gene expression during senescence (Fig. 3a; Supplementary Data 1–3). Genes that are upregulated during senescence but reversed by YAP5SA expression (cluster 3) showed significant enrichment for the terms “Defense response to virus”, “Innate immune response”, and “Inflammatory response”, as expected for a role of YAP in the suppression of the SASP (Fig. 3a; Supplementary Data 4). We also found the term “Aging” to be enriched, verifying recent studies showing that sustaining YAP function opposes the emergence of aging-associated traits47,48,66. Interestingly, we also found significant enrichment for cellular processes related to actin cytoskeleton remodeling and cell morphology (e.g., cell adhesion, cell migration) (Fig. 3a; Supplementary Data 4). This observation supports the possibility that YAP may be able to suppress morphological changes associated with senescence.
a Clustering analysis of differentially expressed genes upon ectopic expression of YAP5SA during senescence (left, normalized log2(fold change) in mRNA expression across samples). GO term analysis of genes belonging to cluster 3 (right top, hypergeometric test with Benjamini-Hochberg correction). Expression plot of genes belonging to cluster 3 and the GO term “actin cytoskeleton organization” (right bottom, normalized log2(fold change) in mRNA expression across the indicated samples). b Abundance of the indicated mRNAs analyzed by RT-qPCR (left, n = 6 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test) and abundance of the indicated proteins analyzed by immunoblotting (right, n = 2 biologically independent experiments). The arrowhead indicates endogenous YAP. c Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). d Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 57, 32, 28, and 23 randomly chosen cells from 3 biologically independent experiments for PRO, PRO+shRHOU#1, SEN, and SEN+shRHOU#1, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test, AFU denotes arbitrary fluorescence units). e Quantification of the percentage of SA-β-gal positive cells (left, n = 6 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. Source data are provided as a Source Data file.
To identify a key downstream effector of YAP in mediating such effects, we searched for genes that met the following criteria: (1) upregulated during senescence; (2) downregulated by ectopic expression of YAP5SA; and (3) belonging to the GO term “Actin cytoskeleton organization”. From a total of 9 satisfied genes, we focused on the Ras Homolog Family Member U (RHOU) based on its strongest response to YAP5SA expression (Fig. 3a). We verified that RHOU is induced during DNA damage-induced, oncogene-induced, and replicative senescence, and expression of YAP5SA suppressed its induction (Fig. 3b; Supplementary Fig. 6a–d). By contrast, expression of YAP5SA/S94A failed to suppress RHOU expression in senescence (Supplementary Fig. 6d), further strengthening our conclusion that YAP exerts its effects on cell enlargement during senescence through transcriptional regulation.
RHOU belongs to a family of Rho GTPases that modulate actin organization, cell migration, and cell shape67,68,69,70. Thus, we reasoned that RHOU is involved in establishing cell enlargement during senescence. To this end, we depleted RHOU during senescence and examined its biological consequences. Depletion of RHOU partially suppressed the formation of actin stress fibers and cell enlargement during senescence (Fig. 3c, d). Consistent with a prominent role for cell enlargement in SASP regulation, depletion of RHOU significantly suppressed expression and secretion of several SASP factors but marginally increased expression of cell cycle regulators and SA-β-Gal activity (Fig. 3c, e; Supplementary Fig. 6e). These effects were verified with multiple different shRNAs against RHOU (Supplementary Fig. 6f) and in another cell line (i.e., senescent BJ cells) (Supplementary Fig. 7a–c).
By contrast, ectopic expression of RHOU exhibited opposite phenotypes: enhancing actin stress fibers, cell enlargement, and SASP expression during senescence (Fig. 4a–c). Of note, ectopically expressed RHOU did not show noticeable changes in cellular localization during senescence (Supplementary Fig. 7d). Furthermore, RHOU modulation has only a negligible effect on actin stress fiber formation in proliferating cells, suggesting a senescence-specific function distinct from its previously reported role71. We leveraged these gain-of-function phenotypes to investigate the epistatic relationship between RHOU and YAP in senescence. Ectopic expression of YAP5SA failed to suppress cell enlargement, actin stress fibers, and the SASP in senescent cells sustaining RHOU expression (Fig. 4d–f); this is in sharp contrast to its effects on cells that do not ectopically express RHOU (Fig. 2c–e). Collectively, these data suggest that RHOU mediates senescence-associated actin remodeling and cell enlargement, as well as SASP regulation, which is negated by YAP.
a Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). The arrowhead indicates GFP-RHOU. b Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, and phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 96, 80, 70, and 62 randomly chosen cells from 3 biologically independent experiments for PRO, PRO + RHOU, SEN, and SEN + RHOU, respectively). c Quantification of the percentage of SA-β-gal positive cells (left, n = 6 biologically independent samples, except SEN + RHOU where n = 5) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples). d Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). The arrowhead indicates endogenous YAP. e Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Scale bar, 50 μm. Phalloidin intensity, cell size, phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 56, 53, 51, and 66 randomly chosen cells from 2 biologically independent experiments for PRO, PRO + YAP5SA/RHOU, SEN, and SEN + YAP5SA/RHOU, respectively). Double denotes Flag-YAP5SA + GFP-RHOU. f Quantification of the percentage of SA-β-gal positive cells (left, n = 6 biologically independent samples) and abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples). Box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values; one-way ANOVA test with Tukey’s multiple comparisons test; AFU denotes arbitrary fluorescence units for b, e. Mean ± SEM; one-way ANOVA test with Tukey’s multiple comparisons test for c,f. PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. Source data are provided as a Source Data file.
YAP and GATA4 compete to balance RHOU and SASP expression
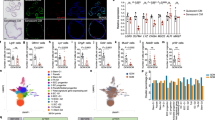
Despite the fact that knockdown of YAP1 enhanced RHOU expression in senescent cells, it failed to do so under proliferating conditions (Supplementary Fig. 8a). Thus, the induction of RHOU requires an additional positive input that is initiated at the onset of senescence. Furthermore, given the genetic association between RHOU and SASP expression (Figs. 3, 4), it is plausible to assume that such an input contributes to SASP regulation. We have previously identified GATA4 as a key regulator of the SASP, which accumulates upon senescence induction by evading autophagic degradation and acts in part through the NF-κB pathway20,72. Consistent with the observation that RHOU is necessary but not sufficient to regulate the SASP (Figs. 3e, 4c), ectopic expression of GATA4 increased RHOU expression along with several SASP genes (Fig. 5a–c), indicating that GATA4 is an upstream regulator of RHOU. We verified this by performing ChIP-qPCR analysis (Fig. 5b). Ectopic expression of YAP5SA suppressed these positive effects of GATA4 on RHOU and SASP expression (Fig. 5c). Consistent with a role for RHOU in cell enlargement, GATA4 expression enhanced the formation of actin stress fibers and caused cell enlargement, which was negated by ectopic expression of YAP5SA (Fig. 5d). Such antagonism was further supported by the significant overlap between genes upregulated by GATA4 expression20 and genes downregulated by YAP5SA expression (Supplementary Fig. 8b), including RHOU.
a Abundance of RHOU mRNA analyzed by RT-qPCR (n = 3 biologically independent samples, mean ± SEM, two-sided unpaired t-test). b Schematic representation of the RHOU promoter region with the indicated qPCR primers (left). Potential GATA4 motifs are shown. Signals obtained from chromatin immunoprecipitation (ChIP) were normalized to signals obtained from an input sample (right, n = 3 biologically independent samples, % Input, mean ± SEM, two-sided unpaired t-test). c Abundance of the indicated proteins analyzed by immunoblotting (left, n = 3 biologically independent experiments). The arrowhead indicates endogenous YAP. Abundance of the indicated mRNAs analyzed by RT-qPCR (right, n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). d Representative images of actin stress fiber staining (left, n = 2 biologically independent experiments). Phalloidin intensity, cell size, and phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 61, 47, and 43 randomly chosen cells from 2 biologically independent experiments for PRO, PRO + GATA4-HA, and PRO + GATA4-HA+Flag-YAP5SA, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test). AFU denotes arbitrary fluorescence units. e, f Immunocytochemistry of GATA4 protein (left). Quantitative analysis of immunofluorescence of the ratio of nuclear to cytosolic GATA4 protein, the nuclear GATA4 protein, and the cytosolic GATA4 protein (right, n = 38, 38, 42, and 43 randomly chosen cells from 3 biologically independent experiments for PRO, PRO + YAP5SA, SEN, and SEN + YAP5SA, respectively, n = 31, 31, 26, and 26 randomly chosen cells from 2 biologically independent experiments for PRO, PRO+shYAP#1, SEN, and SEN+shYAP#1, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test). The inset in the images is magnified. PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. Scale bar, 50 μm. Source data are provided as a Source Data file.
YAP is a transcriptional cofactor that does not itself have any DNA binding activity37,38,40,42,43; thus, we hypothesized that YAP may bind to GATA4 to alter its transcriptional activity. However, while YAP binds to its well-characterized binding partner TEAD, it failed to do so for GATA4 or NF-κB family member RELA, ruling out such a possibility (Supplementary Fig. 8c).
Interestingly, we found that ectopic expression of YAP5SA decreased nuclear GATA4 and RELA but increased their cytosolic levels, which results in a reduced ratio of nuclear to cytosolic GATA4 and RELA (Fig. 5e; Supplementary Fig. 9a). By contrast, knockdown of YAP1 showed opposite trends (Fig. 5f; Supplementary Fig. 9b), consistent with its effect on SASP expression (Fig. 2e, h). Thus, these data suggest that YAP may suppress the expression of RHOU and SASP genes, at least in part, by preventing the nuclear localization of GATA4 and NF-κB.
While examining genes upregulated by GATA4 expression, we found that ectopic expression of GATA4 increased the expression of BTRC and FBXW7, E3 ligases previously shown to target YAP for degradation73,74, which was also observed during senescence (Supplementary Fig. 10a, b). Consistently, ectopic expression of GATA4 reduced YAP protein abundance (Supplementary Fig. 10c). Thus, we hypothesized that these E3 ligases might be responsible for the reduced YAP activity during senescence. Ectopic expression of BTRC, but not FBXW7, further decreased YAP protein abundance in senescent cells while enhancing SASP expression (Supplementary Fig. 10d, e), suggesting that BTRC is involved in modulating the YAP-RHOU-SASP axis. Consistent with this notion, knockdown of BTRC partially rescued the reduction of YAP expression in senescent cells, while partially suppressing the expression of RHOU and SASP genes (Supplementary Fig. 10f, g). Thus, these data suggest that BTRC contributes, at least in part, to the senescence-dependent decline in YAP activity.
Collectively, these data suggest a genetic antagonism between GATA4 and YAP that converges on the regulation of RHOU and SASP expression during senescence.
Cell enlargement enhances GATA4 and NF-κB nuclear transport
Next, we sought to elucidate the potential mechanism by which YAP prevents the nuclear localization of GATA4 and NF-κB. We hypothesized that cell enlargement may be involved in this process for the following reasons: (1) YAP expression inversely dictated cell enlargement during senescence (Fig. 2d, g); (2) cell enlargement alters the mechanical coupling of the cytoskeleton to the nucleus via the LINC complex54,61,62; and (3) such an alteration is proposed to stretch the nucleus, remodel nuclear pores, and modify the nucleocytoplasmic transport44,45,61.
Indeed, we found that disruption of actin stress fibers by treatment with Latrunculin A or Y27632 or RHOU knockdown decreased nuclear GATA4 and RELA but increased their cytosolic levels, resulting in a reduced ratio of nuclear to cytosolic GATA4 and RELA, similar to the case of ectopic expression of YAP5SA. Furthermore, such treatments did not affect their overall levels in senescent cells (Supplementary Figs. 11, 12). Thus, cell enlargement may promote the nuclear localization of GATA4 and NF-κB through this mechanical axis, which is negated by ectopic expression of YAP5SA. Of note, this mechanical regulation applied to YAP itself, as previously reported44 and as assessed by the increased ratio of nuclear to cytosolic YAP in senescent cells (Supplementary Fig. 11d). However, the overall levels of YAP (both nuclear and cytosolic YAP) decreased during senescence, negating an increased nuclear localization of YAP and contributing to its reduced functional outcome as shown above.
To test this hypothesis, we examined whether knockdown of SYNE1, a central component of the LINC complex61,75,76,77,78, affects the nuclear localization of GATA4 and NF-κB during senescence and, if so, the SASP. We reasoned that the disruption of the LINC complex decouples the mechanical link between the cytoskeleton and the nucleus during senescence, thereby suppressing senescence-associated changes in the nucleus. Knockdown of SYNE1 did not affect cell enlargement, but it partially reversed senescence-associated changes in nuclear size (Fig. 6a, b; Supplementary Fig. 13a). These data suggest that actin stress fibers fail to stretch the nucleus in senescent cells when the LINC complex is disrupted. Of note, knockdown of SYNE1 increased the overall intensity of phalloidin staining, partially due to increased formation of actin stress fibers in the nuclear and perinuclear regions; this could be due to a role of SYNE1 in inside-out mechanical coupling79.
a Abundance of the indicated proteins analyzed by immunoblotting (n = 3 biologically independent experiments). b Representative images of actin stress fiber staining (left, n = 3 biologically independent experiments). Phalloidin intensity, cell size, and phalloidin intensity normalized to cell size analyzed by ImageJ software (right, n = 61, 50, 44, and 43 randomly chosen cells from 3 biologically independent experiments for PRO, PRO+shSYNE1#1, SEN, and SEN+shSYNE1#1, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test, AFU denotes arbitrary fluorescence units). The inset in the images is magnified. c Immunocytochemistry of GATA4 protein (left, n = 2 biologically independent experiments). The inset in the images is magnified. Quantitative analysis of immunofluorescence of the ratio of nuclear to cytosolic GATA4 protein, the nuclear GATA4 protein, the cytosolic GATA4 protein, and the total GATA4 protein (right, n = 15 for PRO; n = 15 for PRO+shSYNE1#1; n = 21 for SEN; n = 20 for SEN+shSYNE1#1 randomly chosen cells from 2 biologically independent samples, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test). d Abundance of the indicated mRNAs analyzed by RT-qPCR (n = 3 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). e Quantification of the percentage of SA-β-gal staining (n = 6 biologically independent samples, mean ± SEM, one-way ANOVA test with Tukey’s multiple comparisons test). PRO and SEN denote proliferating and senescent IMR90-hTERT cells, respectively. Scale bar, 50 μm. Source data are provided as a Source Data file.
Furthermore, we found that knockdown of SYNE1 decreased nuclear GATA4 and RELA but increased their cytosolic levels, which results in a reduced ratio of nuclear to cytosolic GATA4 and RELA. Furthermore, such treatments did not affect their overall levels in senescent cells (Fig. 6c; Supplementary Fig. 13b). Consistent with these observations, knockdown of SYNE1 significantly suppressed expression and secretion of several SASP factors during senescence (Fig. 6a, d; Supplementary Fig. 13c). Of note, neither SA-β-Gal activity nor expression of CDK inhibitors (e.g., CDKN1A, CDKN2A) was reduced (Fig. 6d, e), indicating that not all transcriptional programs are perturbed by SYNE1 knockdown during senescence, including the p53-p21 pathway. We also found that knockdown of SYNE1 did not affect the expression of SYNE261,76,77, another component of the LINC complex that acts redundantly with SYNE1 in some contexts (Supplementary Fig. 13d). Interestingly, SYNE2 expression was significantly reduced during senescence. Consistently, knockdown of SYNE2 has negligible effects on either SASP or p16 expression (Supplementary Fig. 13e). Thus, these data suggest that SYNE1 plays a more critical role in senescence than SYNE2. We verified these results with an additional SYNE1 shRNA (Supplementary Fig. 13f).
To corroborate our mechanical model, we examined whether cells plated on soft substrates respond differently to senescence stimulation. We found that cells plated on a low attachment plate (2 kPa) failed to increase actin stress fibers and cell size during senescence, suggesting that senescent cell enlargement depends on the mechanical environment (Supplementary Fig. 14a). Induction of several SASP factors, but not p21, was also attenuated on a low attachment plate, consistent with a role for cell enlargement in SASP regulation (Supplementary Fig. 14b). Interestingly, RHOU induction was intact under the same conditions, highlighting its unique position, distinct from most SASP genes, in this regulatory circuit. These data suggest that SASP expression is mechanically sensitive and may be differentially regulated in a manner that depends on the tissue microenvironment80.
Lastly, we examined whether this mechanistic RHOU-LINC axis is specific to SASP regulation or plays a more general role in NF-κB activation. To this end, we employed two different models of acute NF-kB activation81: (1) TNFα treatment in proliferating cells and (2) LPS stimulation in proliferating cells. We found that TNFα treatment induced the nuclear translocation of NF-κB in proliferating cells as expected, but neither RHOU nor SYNE1 knockdown affected it (Supplementary Fig. 15a, b). Consistently, neither RHOU nor SYNE1 knockdown suppressed the expression of NF-κB downstream targets (Supplementary Fig. 15a, b). The same was true for the stimulation of proliferating cells by LPS: neither RHOU nor SYNE1 knockdown affected acute NF-κB activation (Supplementary Fig. 15c, d). Thus, these data further solidify a senescence-specific role of the RHOU-LINC axis in modulating the SASP.
Collectively, these data suggest that cell enlargement selectively modulates the SASP transcriptional program by remodeling the nuclear import of GATA4 and NF-κB through the LINC complex, which could be counteracted by sustained expression of YAP.
RHOU expression associates with SASP and senescence in vivo
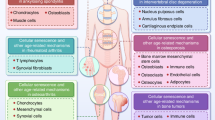
To corroborate our in vitro findings, we employed two different models of senescence in male mice: natural aging and HFD-induced senescence. Previous studies have shown that senescent cells accumulate in the adipose tissue of mice during natural aging and HFD82,83,84,85. We verified this as assessed by increased levels of well-established senescence markers, including p16, p21, and Gata4, as well as SASP factors, such as Il1a and Il6. RhoU was also highly induced under these conditions (Fig. 7a), which correlated with the ratio of Gata4 to Yap proteins across samples. Thus, these data suggest that RhoU expression may be associated with senescence and the SASP in mice.
a Adipose tissues were taken from young (4.5-month-old) and old (26-month-old) male C57BL/6 mice and abundance of the indicated proteins was analyzed by immunoblotting (left, n = 3 biologically independent samples). Adipose tissues were taken from normal chow-fed and high-fat diet (HFD)-fed male C57BL/6 mice and abundance of the indicated proteins was analyzed by immunoblotting (right, n = 4 biologically independent samples). b Box plot analysis comparing the abundance of the indicated genes between RHOUlow and RHOUhigh adipose tissue samples in human aging. Samples from adipose tissues (subcutaneous) in the Genotype-Tissue Expression (GTEx) database were divided into two age groups [‘Young’ (20–39 years old) and ‘Old’ (60–79 years old)], and subsequently categorized into 2 groups [RHOUlow (bottom 10%) and RHOUhigh (top 10%)] according to RHOU expression (n = 10, 11, 23, and 24 biologically independent individuals for Young RHOUlow, Young RHOUhigh, Old RHOUlow, and Old RHOUhigh, respectively, box plots show the 1st and 3rd quartiles; whiskers show the minimum and maximum values, one-way ANOVA test with Tukey’s multiple comparisons test, TPM denotes transcripts per million). c Model of how cell enlargement antagonistically regulated by GATA4 and YAP supports the SASP program142. Source data are provided as a Source Data file.
Next, we investigated whether RHOU expression is associated with the SASP in human adipose tissue aging. To this end, we employed the Genotype-Tissue Expression (GTEx) database, which contains gene expression profiles of non-diseased human tissues across different age groups86. First, we divided the adipose tissue samples into two age groups as previously30: “Young (20 ~ 39 years old)” and “Old (60 ~ 79 years old)”. Then each group was categorized into 2 subgroups according to RHOU expression and the association in expression between RHOU and SASP was analyzed. We found that the level of RHOU expression correlated with the expression of several SASP genes in adipose tissue from the old age group, including CXCL1, MMP2, CLL20, EREG, VEGFA, and IGFBP4. These trends were lost in the young group (Fig. 7b). Thus, this analysis suggests that the RHOU-SASP axis may function in adipose tissue during human aging. Future studies will need to verify our correlative in vivo findings using genetically engineered mouse models.
Discussion
Cell plasticity allows mammalian cells to change their phenotype, enabling their functional adaptation in response to intrinsic and extrinsic cues87,88. One such plasticity is an increase in cell size; several different cell types during development, especially those involved in secretion, exhibit cell size plasticity to meet their functional needs59,60,62,89,90. Recent studies also showed that increasing cell size beyond the physiological threshold can lead to senescence34,35; cell enlargement is also closely associated with the decline in function of mammalian cells during aging36, and contributes to senescence through osmotic stress, replication stress, and an impaired DNA damage response91,92,93. Our work adds to the growing list of cell size-dependent functional plasticity by providing a mechanistic logic for how cell enlargement modulates SASP expression through altered nuclear trafficking of SASP regulators, independent of the previously characterized autophagic regulation of GATA4 (Fig. 7c). This is partially supported by the fact that knockdown of p62, an autophagy adaptor that targets GATA4 for degradation20,72, and YAP depletion have a synergistic effect on IL1A and IL1B expression in proliferating cells (Supplementary Fig. 16a). However, these results should be interpreted with caution because p62 has additional substrates besides GATA450,94,95,96,97,98. Interestingly, this appears to be a selective process because the p53-p21 axis–a main regulator of cell cycle arrest in senescence3,32,99–still operates properly in senescent cells whose cell enlargement is genetically or pharmacologically suppressed. How such a selective remodeling of the nuclear import is achieved remains unclear and is the subject of future investigation.
The YAP transcriptional program coordinates the complex interplay between cell proliferation, survival, differentiation, metabolism, and biomechanics37,40,41,42,43,100,101,102,103. Previous studies have shown that YAP responds to various mechanical cues, including cell spreading, stretching, and cytoskeletal tension, by changing its localization to the nucleus to activate the downstream transcriptional program39. Although senescent cells also experience these YAP-stimulating mechanical stimuli, YAP activity appears to be reduced during senescence. The same holds true during aging in mice47. What drives such a reduction in YAP activity during senescence or aging, or both, is less understood. Here, we showed that GATA4-dependent upregulation of BTRC partially contributes to the senescence-dependent decline of YAP activity and its decoupling from mechanotransduction during senescence. This regulation may cooperate with an uncharacterized phosphorylation event of YAP in senescence for its inhibition (Fig. 2b) and potentiate the GATA4-SASP axis through the genetic feedback circuit (Fig. 7c).
Previous studies suggest that YAP expression suppresses senescent growth arrest and the SASP by modulating CDK666,104. We verified that ectopic expression of YAP5SA increased expression of CDK6 in both proliferating and senescent cells (Supplementary Fig. 16b). However, CDK6 expression did not show a significant reduction during senescence. In addition, YAP5SA expression only marginally rescued the reduced levels of lamin B1, a well-characterized marker of senescence32. We are currently unsure of the causes of this discrepancy; however, previous studies maintain their cells under atmospheric O2 concentration (~20%), which is not a physiological O2 level and is known to cause oxidative stress105. By contrast, we used a physiological O2 ( ~ 3%) to avoid such a complication. Future studies are warranted to examine whether oxidative stress is at least partially responsible for this discrepancy.
Cell hypertrophy is a complex process that requires changes in multiple cellular processes such as growth signaling, metabolism, and mechanical networks59,60,62. It is well known that senescent cells upregulate and sustain the mTORC1 pathway, which in turn supports cellular overgrowth and massive secretion during senescence26,27,28,29,106. Senescent cells also alter their metabolism, including cholesterol metabolism, which help meet metabolic and structural needs for the production of cell membranes and organelles during their hypertrophy30,107,108,109,110,111,112,113,114. Previous studies have shown that senescent cells exhibit extensive formation of actin stress fibers57,115,116. Our current study establishes such an alteration as a functional module of senescence that not only supports cell enlargement but also promotes the secretory phenotype. The GATA4-RHOU-LINC axis governs this module, which is opposed by YAP. It is also possible that RHOU indirectly modulates actin stress fibers during senescence through NF-κB regulation117. YAP appears to exert such effects in part by transcriptionally repressing RHOU in a TEAD-binding dependent manner, thereby suppressing senescence-associated actin remodeling, in contrast to its well-characterized role as a transcriptional co-activator118.
Recently, an elegant study demonstrated that YAP activity in stromal cells, which declines during aging, preserves nuclear envelope integrity and suppresses senescence47. Disruption of nuclear envelope integrity has been shown to generate cytoplasmic chromatin fragments and contribute to senescence119,120,121. Yet, what causes detrimental effects in the absence of YAP activity is not fully understood. We propose that a mechanical branch of the senescence regulatory network, mainly regulated by the GATA4-RHOU pathway, plays an antagonistic role to YAP during senescence and potentially also during aging. These double negative feedback loops are further supported by the observations that (1) either disruption of actin stress fibers by LatA treatment or SYNE1 depletion potently or partially suppressed RHOU expression, and (2) RHOU depletion rescued the reduced levels of YAP during senescence (Supplementary Fig. 16c–e). These effects are likely due in part to reduced nuclear localization of GATA4 under these conditions, thereby lowering the transcription of RHOU and BTRC. Thus, maintaining the fine balance between the GATA4-RHOU and YAP pathways may be the key to healthy aging.
Recent advances in senescence-based therapies make it as a promising strategy for treating aging4,9,11,13,14,122,123,124,125,126,127. In particular, several different classes of senolytics have been developed that selectively eliminate senescent cells and have shown efficacy in preclinical mouse models of various age-related diseases, including neurodegenerative diseases, osteoarthritis, and metabolic syndrome12,14,18,123. Despite this success, most senolytics have significant limitations such as lack of specificity/universality and side effects12,13,123,128, hindering their clinical translation. In addition, senescent cells play a context-dependent role in health and disease, suggesting that not all senescent cells are deleterious and that their indiscriminate removal may therefore cause unanticipated problems4,6,13,125,129,130,131,132,133,134. An alternative approach to senolytics is senomorphics or senostatics, which aims to suppress the deleterious effects of senescent cells, such as chronic pro-inflammatory SASP, without killing them10,12,18,127,135. However, currently available senostatic drugs are limited and most of them directly target the NF-κB pathway, which is also essential for acute inflammation; thus, it may not be ideal. Our results suggest that modulating either cell enlargement or YAP activity may be exploited to reduce pro-inflammatory SASP. How to achieve this in a senescence-specific manner is an exciting avenue for further research, as its tailored modulation may expand the inventory of senostatic options. It is noteworthy that GATA4 forms a feed-forward activation loop with RHOU for cell enlargement and the SASP, providing an attractive treatment modality for senescence-associated diseases.
In conclusion, we show that cell enlargement plays an instructive role in the establishment of pro-inflammatory SASP and reveal a genetic circuit, the GATA4-RHOU-LINC pathway, that contributes to a mechanical branch in the senescence regulatory network. YAP activity counteracts this circuit under physiological conditions and its decline may fully unleash the potential of senescence and its associated inflammation to drive aging. Therefore, disabling the GATA4-RHOU-LINC pathway or sustaining YAP activity may represent a therapeutic strategy for healthy aging.
Methods
Cell lines and senescence induction
Human diploid fibroblasts [IMR90 (female, CCL-186) and BJ (male, CRL-2522)] and human embryonic kidney cells (HEK-293T, CRL-3216) were obtained from the American Type Culture Collection. IMR90 and BJ cells were maintained in 3% O2 and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 15% fetal bovine serum (FBS), penicillin/streptomycin and 0.1 mM non-essential amino acids. HEK-293T cells were maintained in 5% CO2 and cultured in DMEM supplemented with 10% FBS and penicillin/streptomycin. Senescence was induced by ionizing radiation (12 Gy) or treatment with bleomycin (10 μg/ml for 24 h; Cayman Chemical) for IMR90 and BJ cells expressing human telomerase reverse transcriptase (hTERT) (DNA damage-induced senescence, SEN), or H-RasV12 expression for IMR90 and BJ cells expressing hTERT [oncogene-induced senescence, SEN (OIS)], or replicative exhaustion for IMR90 cells [replicative senescence, SEN (Rep)]136. For culturing cells on soft substrates, cytosoft (Advanced BioMatrix, 5141) and cytosoft imaging plates were used (Advanced BioMatrix, 5185). For ectopic expression of either open reading frames (ORFs) or short hairpin RNAs (shRNAs), cells carrying a Dox-inducible (Tet-On) vector expressing ORFs or shRNAs were grown with doxycycline (Dox) at 1 μg/ml unless otherwise indicated. For Latrunculin A (Sigma, L5163) treatment, cells were pretreated with Latrunculin A at 100-200 nM for 24 h prior to senescence induction. For Y27632 (Cayman Chemical, 10005583) treatment, cells were pretreated with Y27632 at 10-20 μM for 2 h prior to senescence induction, and then media were refreshed with Y27632 every 2 d until sampling. For TNFα (Cell to Bio, TNF0501) and LPS (Sigma-Aldrich, L3129) treatment, cells were treated with TNFα at 10 ng/ml for 30 m and LPS at 5 μg/ml for 3 h, respectively. For jasplakinolide (Cayman Chemical, 11705) treatment, cells were pretreated with jasplakinolide at 100 μM for 6 h prior to senescence induction.
Lentiviral vectors and siRNAs
Lentiviral vectors used in this study were shRNAs targeting YAP1, RHOU and SYNE1 subcloned from the GIPZ Lentiviral shRNA Library (Dharmacon) into pINDUCER10 vectors137. The shRNA targeting sequences are as follows: YAP1 shRNA#1, AGCTCAGATCCTTTCCTTA; YAP1 shRNA#2, CAATCACTGTGTTGTATAT; RHOU shRNA#1, AGGACTCCAGATAAAATGA; RHOU shRNA#2, AGATGAAATCGATATTAGA; RHOU shRNA#3, GGAGTATAGTGCAAATTCA; RHOU shRNA#4, CCCAGTTACTTATGATTTA; SYNE1 shRNA#1, AGCAAAGTTCTAGAAGCAA; SYNE1 shRNA#2, CCGGTGACAGCAAAGTTCT; SYNE2 shRNA#1, AGCTTGGAGATCTTCACTA; firefly luciferase (used as a control) shRNA, CCCGCCTGAAGTCTCTGATTAA. YAP5SA and YAP5SA/S94A cDNAs were provided by Prof. Dae-Sik Lim (KAIST) and subcloned into pINDUCER20-N-term-Flag vector50. YAP 1 (Open Biosystems), GATA4 (Open Biosystems), H-RasV12 (Addgene, plasmid 18749) cDNAs were subcloned into the pINDUCER20 vector137. WWTR1, BTRC, FBXW7 (Open Biosystems) cDNAs were subcloned into the pINDUCER20-N-term-Flag vector. RHOU cDNA was cloned from the cDNA library derived from IMR90 cells using the following primers: forward, 5′-CACCATGCCCCCGCAG-3′; and reverse, 5′-TACGAAACAGCAGTACTTCTTCC-3′. RHOU cDNA was then subcloned into the pINDUCER20-N-term-GFP vector (which was generated in this study). siRNAs were transfected into cells at 5 nM for the individual siRNA using Lipofectamine RNAiMAX transfection reagents (Invitrogen) according to the manufacturer’s instructions. The following siRNAs were used: firefly luciferase siRNA, CGUACGCGGAAUACUUCGAUU; BTRC siRNA, AAGUGGAAUUUGUGGAACAUC; p62 siGENOME siRNA, Dharmacon, D-010230-04.
Lentiviral production
Lentiviral gene delivery was performed as described50. Lentiviruses were produced by co-transfection of HEK-293T cells with lentiviral backbone constructs and packaging vectors using PolyJet (SignaGen Laboratories, SL100688) transfection protocol. The lentiviral supernatant was collected 48 h after transfection. The cells were transduced with lentiviral supernatant with 10 μg/ml polybrene (Sigma, H9268) for 1 day. Infected cells were selected using geneticin (200 μg/ml; Gibco) for 7 days or puromycin (2 μg/ml; Clontech) for 3 days. For co-infections, cells were sequentially selected.
Animals
7-week-old C57BL/6 mice were obtained from Central Lab Animal Incorporation (Seoul, Korea). Mice were housed in ventilated cages in a specific pathogen-free animal facility at a constant temperature (22 °C) and humidity (56%) with a 12 h light/dark cycle. For the natural aging model, young and old adipose tissue was obtained from 4.5- and 26-month-old male mice. For the HFD-induced senescence model, adipose tissue was obtained from 8-week-old C57BL/6 male mice fed either normal chow or high-fat diet (HFD, 60% fat, Research diet D12492) for 16 weeks. We considered only male mice as research subjects because estrogen in female mice can affect natural aging and HFD-induced senescence and act as a confounding factor. All mouse experiments were approved by the Seoul National University Institutional Animal Care and Use Committee (SNUIACUC, SNU-210315-3-4). The study complied with all relevant ethical regulations for animal testing and research.
Gene expression profiling
Total RNAs were isolated using the acidic phenol extraction method. One microgram of high-quality RNA samples (RNA integrity number, RIN > 7.0) was used to construct RNA sequencing (RNA-seq) libraries using the TruSeq Stranded mRNA LT Sample Prep Kit (Illumina). Total RNA integrity was checked using an Agilent 2200 TapeStation. RNA-seq was performed on an Illumina NovaSeq 6000 sequencer at Macrogen. The sequence reads were trimmed with Trimmomatic and mapped against the human reference genome (hg38) using Hisat2. Read counts per gene were calculated by using featureCounts. Differential expression analysis was conducted by using the DESeq2 R package (v4.1.3). Genes with adjusted p-value < 0.05 and |fold change| > 1 were selected as differentially expressed genes (DEGs). Hierarchical clustering analysis was conducted using SeqMonk (v1.48.1, Babraham Bioinformatics). The Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 was used for GO term analysis138.
Gene set enrichment analysis
Gene Set Enrichment Analysis (GSEA) was performed by using the default settings of the GSEA software (v4.1.0): weighted enrichment statics and Signal2Noise metric for ranking genes. Additional transcriptome data used in this study for GSEA were GEO: GSE130727 (WI-38 replicative and oncogene-induced senescence). The YAP target gene set (a total of 54 genes) was derived from previous studies139. The YAP signature gene set (a total of 57 genes, Cordenonsi YAP conserved signature) has been obtained from the Molecular Signature Database. The complete lists of genes are presented in Supplementary Data 5.
RT-qPCR
Total RNAs were extracted using RNAiso plus lysis reagent (Takara), and cDNA was synthesized using ReverTra Ace qPCR RT Master Mix (Toyobo) according to the manufacturer’s instructions. RT-qPCR was performed in triplicate using SYBR TOPreal qPCR 2X PreMIX (Enzynomics) on a QuantStudio 5 Real-Time PCR System (Applied Biosystems)140. The data were analyzed using QuantStudio Design and Analysis software (v.1.4.3). GAPDH served as an endogenous normalization control. RT-qPCR primer sequences used in this study are listed in Supplementary Data 6.
Western blotting
Cells were harvested and lysed in 1x Laemmli sample buffer. For tissue sampling, adipose tissues were homogenized using a TissueLyser II (Qiagen) in lysis buffer (25 mM Tris, pH 7.6, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) with protease/phosphatase inhibitors (Roche). The homogenate was centrifuged at 16,000 × g for 10 min at 4 °C and the supernatant was collected. Protein content was determined using a BCA protein assay kit (Pierce). Protein lysates were loaded onto the SDS-polyacrylamide gel electrophoresis. After electrophoresis, the proteins were transferred to a nitrocellulose membrane and subjected to immunoblotting with the indicated antibodies. Immunoblotting images were acquired using an iBright CL1000 imaging system (Thermo Fisher). Immunoblotting signals were normalized to control (either VINCULIN, GAPDH or ACTB) unless otherwise indicated. The following antibodies were used for immunoblotting in this study: anti-VINCULIN (Sigma, v9131, dilution 1:20,000), anti-GAPDH (Santa Cruz Biotechnology (SCBT), sc25778, dilution 1:2,000), anti-ACTB (SCBT, sc47778, dilution 1:1,000), anti-total YAP (SCBT, sc-101199, dilution 1:1,000), anti-p-YAP (S127) (Cell Signaling Technology (CST), 13008, dilution 1:1,000), anti-WRCH1 (RHOU) (Abcam, ab80315, dilution 1:200), anti-WRCH1 (RHOU) (Atlas Antibodies, HPA049592, dilution 1:75), anti-pan-TEAD (CST, 13295, dilution 1:1,000), anti-TAZ (CST, 4883, dilution 1:1,000), anti-βTrCP (CST, 4394, dilution 1:500), anti-p21 (Calbiochem, OP64, dilution 1:200), anti-p16 (BD Biosciences, 554079, dilution 1:1,000), anti-p16 (Proteintech, 10883-1-AP, dilution 1:1,000), anti-p16 (SCBT, sc-56330, dilution 1:200), anti-IL-1B (Proteintech, 16806-1-AP, dilution 1:1,000), anti-IL-6 (R&D Systems, MAB2061, dilution 1:400), anti-IL-8 (R&D Systems, MAB208, dilution 1:500), anti-IL-1A (R&D Systems, MAB200, dilution 1:500), anti-p-RELA (CST, 3033, dilution 1:1,000), anti-total RELA (CST, 8242, dilution 1:1,000), anti-total RELA (SCBT, sc-372, dilution 1:1,000), anti-FLAG (Sigma, F1804, dilution 1:1,000), anti-GATA4 (Proteintech, 19530-1-AP, dilution 1:1000), anti-Nesprin1 (SYNE1) (Invitrogen, MA5-18077, dilution 1:500), anti-CDK6 (CST, 3136, dilution 1:1,000), anti-Lamin B1 (CST, 12586, dilution 1:750), anti-GFP (SCBT, sc-9996, dilution 1:5,000), HRP-conjugated anti-mouse IgG secondary antibodies (Jackson ImmunoResearch, 115-035-003, dilution 1:5,000) and HRP-conjugated anti-rabbit IgG secondary antibodies (Jackson ImmunoResearch, 111-035-003, dilution 1:5,000).
Immunoprecipitation
Cells were harvested and lysed in lysis buffer (25 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol and protease inhibitors). Anti-Flag-conjugated magnetic beads (Sigma, M8823) were used to enrich bait proteins. Equal amounts of protein lysates were subjected to affinity-purification for 2 h at 4 °C. The beads were washed six times with lysis buffer, and the proteins bound to the beads were eluted with 2× Laemmli sample buffer and subjected to SDS-PAGE.
Immunocytochemistry
Cells were plated on the Lab-Tek II chamber slide (Thermo Fisher) and treated as indicated. Cells were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature, washed three times with PBS, permeabilized with ice-cold PBS containing 0.5% Triton X-100 for 15 min, washed with ice-cold PBS containing 0.1% Triton X-100 (PBS-T), blocked with 3% bovine serum albumin (BSA) in PBS-T for 1 h at room temperature, and subjected to immunostaining with the indicated antibodies overnight at 4 °C. The following antibodies were used in this study: anti-GATA4 (Proteintech, 19530-1-AP, dilution 1:200), anti-total RELA (CST, 8242, dilution 1:200), anti-GFP (SCBT, sc-9996, dilution 1:1000), and anti-YAP (SCBT, sc-101199, dilution 1:200). Cells were washed three times with PBS-T, and incubated with Alexa Fluor-488 conjugated secondary antibody (Invitrogen, A11008, dilution 1:1000) for 1 h at room temperature in the dark. When relevant, Rhodamine phalloidin (Invitrogen, R415, dilution 1:200) were used for filamentous-actin visualization. 4,6-diamidino-2-phenylindole (DAPI) was used to stain the nuclei. Finally, the coverslips were washed four times with PBS-T and mounted using anti-fade mounting reagent (Vector Laboratories, H-1200). Images were obtained on a single focal plane using an LSM 700 laser scanning confocal microscope (ZEN v2.3, Zeiss) with the same laser intensity for comparative intensity analysis within the same set of experiments. The intensity was either reported as is or normalized as indicated. Analyses of cell area, phalloidin intensity, and GATA4/RELA/YAP intensity were performed using ImageJ software (1.51k, NIH). The cell boundary was delineated based on phalloidin staining and a differential interference contrast (DIC) image used to define the region of interest (ROI). Both cell area and phalloidin staining intensity within the ROI were quantified. To quantify the normalized nuclear and cytosolic intensity of GATA4, RELA, and YAP, four and ten square regions of interest (5 × 5 pixels) were randomly selected for the nucleus and cytosol of each cell, respectively, and the intensity value measured within the regions was averaged to represent the nuclear and cytosolic intensity of GATA4 and RELA. Total GATA4 and RELA intensities were measured within ROIs delineated based on the GATA4 and RELA immunostaining signals.
BrdU incorporation assay
Cells were plated in the 48-well plates and labeled with 10 μM BrdU (BD Biosciences) for 3 h. BrdU-incorporated nuclei were visualized by immunofluorescence using Alexa Fluor 488 Mouse anti-BrdU (BD Biosciences, 558599, dilution 1:10) according to the manufacturer’s instructions. The percentage of positive stained cells was calculated as (number of positive cells) / (total number of cells) × 100 in 3 randomly selected area per sample at ×100 magnification.
SA-β-galactosidase assay
SA-β-gal activity was determined using the C12FDG staining kit according to the manufacturer’s instructions. Cells were pretreated with 100 nM bafilomycin A1 (Cayman Chemical, 11038) for 1 h, followed by incubation with 20 μM C12FDG (Invitrogen, D2893) for 1 h. SA-β-gal activity was analyzed by fluorescence-activated cell sorting (FACS) (Attune NxT Flow Cytometer, Thermo Fisher Scientific). Data were acquired using Attune NxT software (v3.1.2) and analyzed using FlowJo software (v10.6.2). The gating strategy is provided in Supplementary Data 7.
FACS analysis for cell volume
Cell volume was determined by fluorescence-activated cell sorting (FACS) (Attune NxT Flow Cytometer, Thermo Fisher Scientific). Forward scatter area (FSC-A) was used to estimate the relative cell volume of each population141. Data were acquired using Attune NxT software (v3.1.2) and analyzed using FlowJo software (v10.6.2). Cell volume was expressed as a fold change of the proliferating cell volume.
Actin segmentation by ultracentrifugation
Actin stabilization buffer (50 mM PIPES (pH 6.9), 50 mM NaCl, 5 mM MgCl2, 5 mM EGTA, 2 mM ATP, 5% glycerol, 0.1% Nonidet P-40, 0.1% Triton X-100, 0.1% Tween 20, 0.1% β-mercaptoethanol, protease inhibitors, and phosphatase inhibitors) was added to culture dishes for cell lysis. Cells were incubated in the buffer for 10 min at 37 °C and then collected by scraping into 1.5 ml microcentrifuge tubes. After collection, the samples were centrifuged at 300 x g at room temperature to remove insoluble particles. The protein concentration of each lysate was determined by BCA assay (Pierce). Equal amounts of cell lysates were diluted with actin stabilization buffer and subjected to subsequent fractionation. 10% of the volume of the diluted cell lysates was set aside as total protein input. The F-actin and G-actin pools in the diluted cell lysates were separated by ultracentrifugation at 100,000 x g for 1 h at 37 °C. After centrifugation, F-actin sedimented while G-actin remained in the supernatant due to their distinct molecular weights. The supernatant containing the G-actin pool was collected in a new tube, while the F-actin pellet was resuspended in cold distilled water with 1 μM cytochalasin D (Sigma-Aldrich) and incubated on ice for 45 min to facilitate the dissolution of F-actin. Laemmli buffer was then added to both fractions and boiled at 95 °C for 10 min. The amounts of F/G actin were analyzed by Western blotting with antibody against β-actin.
Trichloroacetic acid (TCA) protein precipitation
IMR90-hTERT cells carrying a Dox-inducible (Tet-On) vector expressing the indicated constructs were grown with or without Dox and treated with bleomycin to induce senescence in six-well plates. Seven days after bleomycin treatment, cells were washed with PBS and incubated with DMEM containing 0.5% FBS, penicillin/streptomycin and 0.1 mM non-essential amino acids with or without Dox. After 3 days of incubation, the conditioned medium was collected and cellular material was removed by centrifugation at 200 × g for 5 min. To precipitate proteins from the conditioned medium, 25% (v/v) trichloroacetic acid (TCA) was added to the conditioned medium, vortexed, and incubated at 4 °C for 1 h. Protein precipitates were pelleted by centrifugation at 17,000 × g for 10 min at 4 °C and washed twice with ice-cold acetone. The pellets were dried at 95 °C for 10-15 min, dissolved in Laemmli buffer, boiled at 95 °C for 20 min, and then subjected to Western blotting.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed using the X-ChIP protocol (Abcam) with modifications. In brief, 6 × 106 cells were crosslinked with 1% formaldehyde for 15 min at room temperature, quenched with 125 mM glycine, and washed twice with cold PBS. The cells were then scraped, collected into 1 ml of cellular lysis buffer (20 mM Tris pH 8.0, 85 mM KCl, 0.5% NP-40, and protease inhibitors), and centrifuged at 800 × g for 5 min. Cell pellets were resuspended with 0.3 ml of cold nuclear lysis buffer (1% Triton X-100, 0.1% sodium deoxycholate, 0.25% SDS, 1 mM EDTA, 50 mM Tris, pH 7.5, 140 mM NaCl, and protease inhibitors). Nuclei were then sonicated using a Q800R instrument (Qsonica) set to 50% amplitude for 30 s (followed by 30 s rest) for a total time of 10 min, and centrifuged at 13,000 × g for 10 min. Thirty micrograms of sheared chromatin was diluted in ice-cold dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris, pH 8.0, and protease inhibitors) to a total of 1 ml, and 1% of chromatin was set aside as an input control. Chromatin was incubated with anti-HA-conjugated magnetic beads (Thermo Fisher, 88837) for 2 h at 4 °C with mild agitation. The beads were washed sequentially with 1 ml of low-salt buffer, high-salt buffer, LiCl buffer, and TE buffer, eluted with elution buffer (1% SDS, 100 mM NaHCO3). Eluted DNA was incubated with 5 mM NaCl overnight at 65 °C, treated with RNase A at 37 °C for 30 min and proteinase K at 55 °C for 1 h, and purified using a PCR purification kit (Bioneer). ChIP samples were then subjected to qPCR. Primer sequences used in this study are listed in Supplementary Data 6.
Gene expression analysis in human adipose tissue aging
The transcripts per million normalized RNA-seq datasets from the publicly available Genotype-Tissue Expression (GTEx) database (v8) were used. Samples from the subcutaneous adipose tissues in the GTEx database were divided into two age groups [Young (20–39 years old) and Old (60–79 years old)]. Then, in each group, samples were categorized into two subgroups [RHOUlow (bottom 10%) and RHOUhigh (top 10%)] according to RHOU expression. In both age groups, we compared the expression levels of the indicated SASP genes between the RHOUlow and RHOUhigh subgroups.
Statistical analysis
No statistical methods were used to predetermine sample sizes. Data met the assumptions of the statistical tests used. Statistical parameters including the exact value of n, precision measures and statistical significance are reported in the figures and figure legends. Data analyses were performed using GraphPad Prism (v10.2.3, Graphpad Software Inc) and Microsoft Excel software (2013).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data supporting the results of this study are available within the paper and its Supplementary Data. The RNA-seq data generated by this study are deposited in the Gene Expression Omnibus database under project accession number GSE278460. Publicly available datasets used in this study are as follows: GSE130727 (Supplementary Fig. 3a) [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE130727] and GTEx v8 (Fig. 7b) [https://www.gtexportal.org/home/]. Uncropped scans of all blots and gels in Figures and Supplementary Figs. are provided in the Source Data file. Source data are provided with this paper.
References
Salama, R., Sadaie, M., Hoare, M. & Narita, M. Cellular senescence and its effector programs. Genes Dev. 28, 99–114 (2014).
Herranz, N. & Gil, J. Mechanisms and functions of cellular senescence. J. Clin. Invest 128, 1238–1246 (2018).
Gorgoulis, V. et al. Cellular senescence: defining a path forward. Cell 179, 813–827 (2019).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169, 1000–1011 (2017).
Calcinotto, A. et al. Cellular senescence: aging, cancer, and injury. Physiol. Rev. 99, 1047–1078 (2019).
Ring, N. A. R., Valdivieso, K., Grillari, J., Redl, H. & Ogrodnik, M. The role of senescence in cellular plasticity: Lessons from regeneration and development and implications for age-related diseases. Dev. Cell 57, 1083–1101 (2022).
Campisi, J. Aging, cellular senescence, and cancer. Annu Rev. Physiol. 75, 685–705 (2013).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Childs, B. G. et al. Senescent cells: an emerging target for diseases of ageing. Nat. Rev. Drug Discov. 16, 718–735 (2017).
Kirkland, J. L. & Tchkonia, T. Cellular senescence: a translational perspective. EBioMedicine 21, 21–28 (2017).
Campisi, J. et al. From discoveries in ageing research to therapeutics for healthy ageing. Nature 571, 183–192 (2019).
Kang, C. Senolytics and senostatics: a two-pronged approach to target cellular senescence for delaying aging and age-related diseases. Mol. Cells 42, 821–827 (2019).
Chaib, S., Tchkonia, T. & Kirkland, J. L. Cellular senescence and senolytics: the path to the clinic. Nat. Med. 28, 1556–1568 (2022).
Zhang, L. et al. Cellular senescence: a key therapeutic target in aging and diseases. J. Clin. Invest. 132. https://doi.org/10.1172/JCI158450 (2022).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: An expanding universe. Cell 186, 243–278 (2023).
Funk, M. C. et al. Aged intestinal stem cells propagate cell-intrinsic sources of inflammaging in mice. Dev. Cell 58, 2914–2929.e2917 (2023).
Coppe, J. P. et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 6, 2853–2868 (2008).
Birch, J. & Gil, J. Senescence and the SASP: many therapeutic avenues. Genes Dev. 34, 1565–1576 (2020).
Chien, Y. et al. Control of the senescence-associated secretory phenotype by NF-kappaB promotes senescence and enhances chemosensitivity. Genes Dev. 25, 2125–2136 (2011).
Kang, C. et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science 349, aaa5612 (2015).
Tasdemir, N. et al. BRD4 Connects Enhancer Remodeling to Senescence Immune Surveillance. Cancer Discov. 6, 612–629 (2016).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Aird, K. M. et al. HMGB2 orchestrates the chromatin landscape of senescence-associated secretory phenotype gene loci. J. Cell Biol. 215, 325–334 (2016).
Narita, M. et al. Spatial coupling of mTOR and autophagy augments secretory phenotypes. Science 332, 966–970 (2011).
Herranz, N. et al. mTOR regulates MAPKAPK2 translation to control the senescence-associated secretory phenotype. Nat. Cell Biol. 17, 1205–1217 (2015).
Laberge, R. M. et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat. Cell Biol. 17, 1049–1061 (2015).
Rabanal-Ruiz, Y. et al. mTORC1 activity is supported by spatial association with focal adhesions. J. Cell Biol. 220. https://doi.org/10.1083/jcb.202004010 (2021).
Roh, K. et al. Lysosomal control of senescence and inflammation through cholesterol partitioning. Nat. Metab. 5, 398–413 (2023).
Kim, J. et al. Autophagy-dependent splicing control directs translation toward inflammation during senescence. Dev Cell. https://doi.org/10.1016/j.devcel.2024.10.008 (2024)
Hernandez-Segura, A., Nehme, J. & Demaria, M. Hallmarks of cellular senescence. Trends Cell Biol. 28, 436–453 (2018).
Davies, D. M., van den Handel, K., Bharadwaj, S. & Lengefeld, J. Cellular enlargement - A new hallmark of aging? Front Cell Dev. Biol. 10, 1036602 (2022).
Neurohr, G. E. et al. Excessive cell growth causes cytoplasm dilution and contributes to senescence. Cell 176, 1083–1097.e1018 (2019).
Lanz, M. C. et al. Increasing cell size remodels the proteome and promotes senescence. Mol. Cell 82, 3255–3269.e3258 (2022).
Lengefeld, J. et al. Cell size is a determinant of stem cell potential during aging. Sci. Adv. 7, eabk0271 (2021).
Hansen, C. G., Moroishi, T. & Guan, K. L. YAP and TAZ: a nexus for Hippo signaling and beyond. Trends Cell Biol. 25, 499–513 (2015).
Lin, K. C., Park, H. W. & Guan, K. L. Regulation of the hippo pathway transcription factor TEAD. Trends Biochem Sci. 42, 862–872 (2017).
Panciera, T., Azzolin, L., Cordenonsi, M. & Piccolo, S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 18, 758–770 (2017).
Totaro, A., Panciera, T. & Piccolo, S. YAP/TAZ upstream signals and downstream responses. Nat. Cell Biol. 20, 888–899 (2018).
Zheng, Y. & Pan, D. The hippo signaling pathway in development and disease. Dev. Cell 50, 264–282 (2019).
Piccolo, S., Panciera, T., Contessotto, P. & Cordenonsi, M. YAP/TAZ as master regulators in cancer: modulation, function and therapeutic approaches. Nat. Cancer 4, 9–26 (2023).
Franklin, J. M., Wu, Z. & Guan, K. L. Insights into recent findings and clinical application of YAP and TAZ in cancer. Nat. Rev. Cancer 23, 512–525 (2023).
Elosegui-Artola, A. et al. Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores. Cell 171, 1397–1410.e1314 (2017).
Andreu, I. et al. Mechanical force application to the nucleus regulates nucleocytoplasmic transport. Nat. Cell Biol. 24, 896–905 (2022).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Sladitschek-Martens, H. L. et al. YAP/TAZ activity in stromal cells prevents ageing by controlling cGAS-STING. Nature 607, 790–798 (2022).
Santinon, G. et al. dNTP metabolism links mechanical cues and YAP/TAZ to cell growth and oncogene-induced senescence. EMBO J. 37. https://doi.org/10.15252/embj.201797780 (2018)
Lee, Y. et al. A flow-cytometry-based assessment of global protein synthesis in human senescent cells. STAR Protoc. 2, 100809 (2021).
Lee, Y. et al. Coordinate regulation of the senescent state by selective autophagy. Dev. Cell 56, 1512–1525.e1517 (2021).
Tojkander, S., Gateva, G. & Lappalainen, P. Actin stress fibers-assembly, dynamics and biological roles. J. Cell Sci. 125, 1855–1864 (2012).
Stricker, J., Falzone, T. & Gardel, M. L. Mechanics of the F-actin cytoskeleton. J. Biomech. 43, 9–14 (2010).
Hansson, B. et al. Adipose cell size changes are associated with a drastic actin remodeling. Sci. Rep. 9, 12941 (2019).
Basso, M. et al. Actin-based deformations of the nucleus control mouse multiciliated ependymal cell differentiation. Dev Cell. https://doi.org/10.1016/j.devcel.2024.11.008 (2024).
Kim, Y. et al. Actin depolymerizing factor destrin governs cell migration in neural development during Xenopus embryogenesis. Mol. Cells 47, 100076 (2024).
Fujiwara, I., Zweifel, M. E., Courtemanche, N. & Pollard, T. D. Latrunculin A Accelerates Actin Filament Depolymerization in Addition to Sequestering Actin Monomers. Curr. Biol. 28, 3183–3192.e3182 (2018).
Mu, X. et al. Cytoskeleton stiffness regulates cellular senescence and innate immune response in Hutchinson-Gilford Progeria Syndrome. Aging Cell 19, e13152 (2020).
Park, J. T. et al. A crucial role of ROCK for alleviation of senescence-associated phenotype. Exp. Gerontol. 106, 8–15 (2018).
Lloyd, A. C. The regulation of cell size. Cell 154, 1194–1205 (2013).
Miettinen, T. P., Caldez, M. J., Kaldis, P. & Bjorklund, M. Cell size control - a mechanism for maintaining fitness and function. Bioessays 39. https://doi.org/10.1002/bies.201700058 (2017)
Carley, E., King, M. C. & Guo, S. Integrating mechanical signals into cellular identity. Trends Cell Biol. 32, 669–680 (2022).
Xie, S., Swaffer, M. & Skotheim, J. M. Eukaryotic Cell Size Control and Its Relation to Biosynthesis and Senescence. Annu Rev. Cell Dev. Biol. 38, 291–319 (2022).
Casella, G. et al. Transcriptome signature of cellular senescence. Nucleic Acids Res 47, 7294–7305 (2019).
Zhao, B. et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 22, 1962–1971 (2008).
Reggiani, F., Gobbi, G., Ciarrocchi, A. & Sancisi, V. YAP and TAZ Are Not Identical Twins. Trends Biochem Sci. 46, 154–168 (2021).
Xie, Q. et al. YAP/TEAD-mediated transcription controls cellular senescence. Cancer Res 73, 3615–3624 (2013).
Raftopoulou, M. & Hall, A. Cell migration: Rho GTPases lead the way. Dev. Biol. 265, 23–32 (2004).
Loebel, D. A. et al. Rhou maintains the epithelial architecture and facilitates differentiation of the foregut endoderm. Development 138, 4511–4522 (2011).
Hodge, R. G. & Ridley, A. J. Regulation and functions of RhoU and RhoV. Small GTPases 11, 8–15 (2020).
Shutes, A., Berzat, A. C., Cox, A. D. & Der, C. J. Atypical mechanism of regulation of the Wrch-1 Rho family small GTPase. Curr. Biol. 14, 2052–2056 (2004).
Tao, W., Pennica, D., Xu, L., Kalejta, R. F. & Levine, A. J. Wrch-1, a novel member of the Rho gene family that is regulated by Wnt-1. Genes Dev. 15, 1796–1807 (2001).
Kang, C. & Elledge, S. J. How autophagy both activates and inhibits cellular senescence. Autophagy 12, 898–899 (2016).
Zhao, B., Li, L., Tumaneng, K., Wang, C. Y. & Guan, K. L. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(beta-TRCP). Genes Dev. 24, 72–85 (2010).
Tu, K. et al. Fbxw7 is an independent prognostic marker and induces apoptosis and growth arrest by regulating YAP abundance in hepatocellular carcinoma. Mol. Cancer 13, 110 (2014).
Mejat, A. & Misteli, T. LINC complexes in health and disease. Nucleus 1, 40–52 (2010).
Martins, R. P., Finan, J. D., Guilak, F. & Lee, D. A. Mechanical regulation of nuclear structure and function. Annu Rev. Biomed. Eng. 14, 431–455 (2012).
Isermann, P. & Lammerding, J. Nuclear mechanics and mechanotransduction in health and disease. Curr. Biol. 23, R1113–R1121 (2013).
Martino, F., Perestrelo, A. R., Vinarsky, V., Pagliari, S. & Forte, G. Cellular mechanotransduction: from tension to function. Front Physiol. 9, 824 (2018).
Schwartz, C. et al. Lamins and nesprin-1 mediate inside-out mechanical coupling in muscle cell precursors through FHOD1. Sci. Rep. 7, 1253 (2017).
Fuchs, E. et al. Understanding the microenvironment and how this controls cell fate. Dev. Cell 58, 2819–2821 (2023).
Liu, T., Zhang, L., Joo, D. & Sun, S. C. NF-kappaB signaling in inflammation. Signal Transduct. Target Ther. 2, 17023 (2017).
Ogrodnik, M. et al. Obesity-induced cellular senescence drives anxiety and impairs neurogenesis. Cell Metab. 29, 1061–1077.e1068 (2019).
Schafer, M. J. et al. Exercise prevents diet-induced cellular senescence in adipose tissue. Diabetes 65, 1606–1615 (2016).
Palmer, A. K. et al. Targeting senescent cells alleviates obesity-induced metabolic dysfunction. Aging Cell 18, e12950 (2019).
Nguyen, T. T. & Corvera, S. Adipose tissue as a linchpin of organismal ageing. Nat. Metab. 6, 793–807 (2024).
Consortium, G. T. The genotype-tissue expression (GTEx) project. Nat. Genet 45, 580–585 (2013).
Ge, Y. & Fuchs, E. Stretching the limits: from homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet 19, 311–325 (2018).
Jessen, K. R., Mirsky, R. & Arthur-Farraj, P. The Role of Cell Plasticity in Tissue Repair: Adaptive Cellular Reprogramming. Dev. Cell 34, 613–620 (2015).
Yang, X. & Xu, T. Molecular mechanism of size control in development and human diseases. Cell Res 21, 715–729 (2011).
Ovrebo, J. I. & Edgar, B. A. Polyploidy in tissue homeostasis and regeneration. Development 145. https://doi.org/10.1242/dev.156034 (2018).
Crozier, L. et al. CDK4/6 inhibitor-mediated cell overgrowth triggers osmotic and replication stress to promote senescence. Mol. Cell 83, 4062–4077.e4065 (2023).
Foy, R. et al. Oncogenic signals prime cancer cells for toxic cell overgrowth during a G1 cell cycle arrest. Mol. Cell 83, 4047–4061.e4046 (2023).
Manohar, S. et al. Genome homeostasis defects drive enlarged cells into senescence. Mol. Cell 83, 4032–4046.e4036 (2023).
Johansen, T. & Lamark, T. Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J. Mol. Biol. 432, 80–103 (2020).
Kurusu, R. et al. Integrated proteomics identifies p62-dependent selective autophagy of the supramolecular vault complex. Dev. Cell 58, 1189–1205.e1111 (2023).
Kelly, G. et al. Suppressed basal mitophagy drives cellular aging phenotypes that can be reversed by a p62-targeting small molecule. Dev. Cell 59, 1924–1939.e1927 (2024).
Kim, J., Lee, Y., Jeon, T., Kim, M. S. & Kang, C. All cells are created equal in the sight of autophagy: selective autophagy maintains homeostasis in senescent cells. Autophagy 17, 3260–3261 (2021).
Kwon, Y., Kim, J. W., Jeoung, J. A., Kim, M. S. & Kang, C. Autophagy Is Pro-Senescence When Seen in Close-Up, but Anti-Senescence in Long-Shot. Mol. Cells 40, 607–612 (2017).
Yan, J. et al. The role of p21 in cellular senescence and aging-related diseases. Mol. Cells 47, 100113 (2024).
Moroishi, T., Hansen, C. G. & Guan, K. L. The emerging roles of YAP and TAZ in cancer. Nat. Rev. Cancer 15, 73–79 (2015).
Kim, M. K. et al. The TGFbeta->TAK1->LATS->YAP1 Pathway Regulates the Spatiotemporal Dynamics of YAP1. Mol. Cells 46, 592–610 (2023).
Choi, S. et al. Hippo-YAP/TAZ signalling coordinates adipose plasticity and energy balance by uncoupling leptin expression from fat mass. Nat. Metab. 6, 847–860 (2024).
Collins, J. M. et al. YAP and TAZ couple osteoblast precursor mobilization to angiogenesis and mechanoregulation in murine bone development. Dev. Cell 59, 211–227.e215 (2024).
Xu, X. et al. YAP prevents premature senescence of astrocytes and cognitive decline of Alzheimer’s disease through regulating CDK6 signaling. Aging Cell 20, e13465 (2021).
Coppe, J. P. et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One 5, e9188 (2010).
Carroll, B. et al. Persistent mTORC1 signaling in cell senescence results from defects in amino acid and growth factor sensing. J. Cell Biol. 216, 1949–1957 (2017).
Wiley, C. D. & Campisi, J. The metabolic roots of senescence: mechanisms and opportunities for intervention. Nat. Metab. 3, 1290–1301 (2021).
Kim, Y., Jang, Y., Kim, M. S. & Kang, C. Metabolic remodeling in cancer and senescence and its therapeutic implications. Trends Endocrinol. Metab. https://doi.org/10.1016/j.tem.2024.02.008 (2024).
Accumulation of G3P and pEtN drives cellular senescence via altered lipid metabolism. Nat. Metab. 6, 205-206 https://doi.org/10.1038/s42255-024-00975-3 (2024).
Tighanimine, K. et al. A homoeostatic switch causing glycerol-3-phosphate and phosphoethanolamine accumulation triggers senescence by rewiring lipid metabolism. Nat. Metab. 6, 323–342 (2024).
Dou, X. et al. PDK4-dependent hypercatabolism and lactate production of senescent cells promotes cancer malignancy. Nat. Metab. 5, 1887–1910 (2023).
Maus, M. et al. Iron accumulation drives fibrosis, senescence and the senescence-associated secretory phenotype. Nat. Metab. 5, 2111–2130 (2023).
Thompson, C. B. et al. A century of the Warburg effect. Nat. Metab. 5, 1840–1843 (2023).
Kuehnemann, C. & Wiley, C. D. Cholesterol drives inflammatory senescence. Nat. Metab. 5, 355–356 (2023).
Chen, Q. M. et al. Involvement of Rb family proteins, focal adhesion proteins and protein synthesis in senescent morphogenesis induced by hydrogen peroxide. J. Cell Sci. 113, 4087–4097 (2000).
Tsai, C. H. et al. Up-regulation of cofilin-1 in cell senescence associates with morphological change and p27(kip1) -mediated growth delay. Aging Cell 20, e13288 (2021).
Oh, A. et al. NF-kappaB signaling in neoplastic transition from epithelial to mesenchymal phenotype. Cell Commun. Signal 21, 291 (2023).
Kim, M., Kim, T., Johnson, R. L. & Lim, D. S. Transcriptional co-repressor function of the hippo pathway transducers YAP and TAZ. Cell Rep. 11, 270–282 (2015).
Ivanov, A. et al. Lysosome-mediated processing of chromatin in senescence. J. Cell Biol. 202, 129–143 (2013).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Miller, K. N. et al. Cytoplasmic DNA: sources, sensing, and role in aging and disease. Cell 184, 5506–5526 (2021).
McHugh, D. & Gil, J. Senescence and aging: Causes, consequences, and therapeutic avenues. J. Cell Biol. 217, 65–77 (2018).
van Deursen, J. M. Senolytic therapies for healthy longevity. Science 364, 636–637 (2019).
Kim, J., Lee, Y., Roh, K., Kim, M. S. & Kang, C. Targeting the stress support network regulated by autophagy and senescence for cancer treatment. Adv. Cancer Res 150, 75–112 (2021).
Schmitt, C. A., Wang, B. & Demaria, M. Senescence and cancer - role and therapeutic opportunities. Nat. Rev. Clin. Oncol. 19, 619–636 (2022).
Wang, T. W. & Nakanishi, M. Immune surveillance of senescence: potential application to age-related diseases. Trends Cell Biol https://doi.org/10.1016/j.tcb.2024.06.007. (2024).
Yoon, J. H. & Han, H. J. Senotherapeutics: Different approaches of discovery and development. Mol. Cells 47, 100002 (2024).
Kim, E. J. et al. A focused natural compound screen reveals senolytic and senostatic effects of Isatis tinctoria. Anim. Cells Syst. (Seoul.) 26, 310–317 (2022).
Yu, Q. et al. Cellular senescence promotes progenitor cell expansion during axolotl limb regeneration. Dev. Cell 58, 2416–2427.e2417 (2023).
Demaria, M. et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733 (2014).
Helman, A. et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat. Med 22, 412–420 (2016).
Mosteiro, L. et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science 354. https://doi.org/10.1126/science.aaf4445 (2016).
Grosse, L. et al. Defined p16(High) senescent cell types are indispensable for mouse healthspan. Cell Metab. 32, 87–99.e86 (2020).
Reyes, N. S. et al. Sentinel p16(INK4a+) cells in the basement membrane form a reparative niche in the lung. Science 378, 192–201 (2022).
D’Ambrosio, M. & Gil, J. Reshaping of the tumor microenvironment by cellular senescence: An opportunity for senotherapies. Dev. Cell 58, 1007–1021 (2023).
Kang, E., Kang, C., Lee, Y. S. & Lee, S. V. Brief guide to senescence assays using cultured mammalian cells. Mol. Cells 47, 100102 (2024).
Meerbrey, K. L. et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl Acad. Sci. USA 108, 3665–3670 (2011).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37, 1–13 (2009).
Kurppa, K. J. et al. Treatment-induced tumor dormancy through yap-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell 37, 104–122.e112 (2020).
Bong, D., Sohn, J. & Lee, S. V. Brief guide to RT-qPCR. Mol. Cells 47, 100141 (2024).
Song, Y. & Lee, Y. Brief guide to flow cytometry. Mol. Cells 47, 100129 (2024).
Joung, J. Created in BioRender. https://BioRender.com/w61y314 (2025).
Acknowledgements
We thank Dae-Sik Lim (KAIST), Hyun Woo Park (Yonsei University), and Seung Joon Baek (Seoul National University) for sharing reagents and providing valuable advice. We are also grateful to all of the members of the Kang lab for their support. This work was supported by the Suh Kyungbae Foundation (SUHF-17020068), the National Research Foundation of Korea (NRF-2020R1A5A1018081), and the Korea Evaluation Institute of Industrial Technology (1415181231) to Chanhee Kang. M.-S.K. was supported by the National Research Foundation of Korea (RS-2023-00246366). J.M.S. was supported by the National Research Foundation of Korea (RS-2024-00440824, RS-2024-00346395). Y.J. was supported by the National Research Foundation of Korea (RS-2023-00275353).
Author information
Authors and Affiliations
Contributions
J.J., M.-S.K., and C.K. conceived and designed the project. J.J., Y.H., Y.K., J.K., H.C., T.J., Y.J., and E.-J.K. performed most of the experiments and analyzed the data. S.H.L. and J.M.S. provided mouse samples for analysis. S.J.E. provided intellectual contributions to the project. J.J., J.K., M.-S.K., and C.K. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks David Kast and Masashi Narita for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Joung, J., Heo, Y., Kim, Y. et al. Cell enlargement modulated by GATA4 and YAP instructs the senescence-associated secretory phenotype. Nat Commun 16, 1696 (2025). https://doi.org/10.1038/s41467-025-56929-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56929-0
This article is cited by
-
VIRMA/IGF2BP3-mediated ANLN upregulation promotes intrahepatic cholangiocarcinoma growth by forming a positive feedback loop with RhoA/YAP1/TEAD1 signaling pathway
Cell Death & Disease (2026)
-
Regulation of cellular senescence in tumor progression and therapeutic targeting: mechanisms and pathways
Molecular Cancer (2025)