Abstract
Over 75% of failed pregnancies involve implantation defects. Growth factor receptor-bound protein 2 (GRB2) is an adaptor protein involved in signal transduction and cell communication. Here we show that the expression of GRB2 protein is lower in endometrium of infertile women with endometriosis compared to controls. Our mouse endometriosis model revealed that endometriosis development results to GRB2 loss in the eutopic endometrium. To understand the role of GRB2 in the uterus, we generated mice with conditional ablation of Grb2 in the Pgr positive cells (Grb2d/d). Grb2d/d mice were infertile due to implantation failure. Although ovarian functions were normal, Grb2d/d mice had a non-receptive endometrium due to progesterone resistance and dysregulation of steroid hormone and FOXA2 signaling pathways. Furthermore, our results were supported by findings of GRB2 attenuation in primary human endometrial stromal cells from women with endometriosis. Our results demonstrate that GRB2 is critical for endometrial receptivity and decidualization.
Similar content being viewed by others
Introduction
Six percent of women under 44 are infertile, 12% have difficulty getting pregnant or carrying pregnancies to term, and over 75% of failed pregnancies involve implantation defects1. Even the most successful treatments, such as assisted reproductive technologies, still lead to frequent implantation failures. Uterine receptivity, implantation, and decidualization are critical in supporting embryonic growth. These events are regulated by the coordinated actions of estrogen (E2) and progesterone (P4) that are produced and secreted by the ovary. E2 stimulates uterine epithelial cell proliferation, and P4 suppresses E2-induced proliferation. E2 and P4 act on the endometrium and myometrium directly through their cognate nuclear receptors, the progesterone receptor (PGR) and estrogen receptors (ESR1 and ESR2), respectively, and indirectly by stimulation of various growth factors including EGF. During implantation, changes in the expression of adhesion molecules, cytokines, and transcriptional factors are dependent upon E2 and P4. Because of the complexity and dynamic nature of implantation, these molecular changes are still not fully understood.
In rodents, E2 promotes proliferation of epithelial cells in the endometrium to initiate the preparation for pregnancy on gestation day 0.5 (GD 0.5). Subsequently, P4 inhibits E2-induced proliferation of uterine epithelial cells and stimulates stromal cell proliferation on GD 3.52,3. Rapid induction of the cytokine leukemia-inhibitory factor (LIF) by nidatory estrogen with elevated P4 levels is a critical step for uterine receptivity4,5,6. The blastocyst attaches to the uminal epithelium on GD 4.5, and implantation occurs followed by decidualization of the surrounding stroma2,7. During decidualization, uterine stromal cells undergo proliferation and differentiation into large epithelioid decidual cells, which creates a local environment permissive to implantation and supportive of the embryo. Similar to the mouse, the steroid hormones E2 and P4 are critical for the regulation of human endometrium including proliferation, secretion, and menstrual shedding. Defective stromal cell decidualization can result in pregnancy complications including pregnancy loss and miscarriage in mice and human2.
Uterine glands and their secretory products are likely critical regulators of blastocyst implantation, uterine receptivity, and conceptus growth and development in both mice and humans8. During pregnancy, uterine glands present in the endometrium surrounding the embryo implantation site and develop decidua9,10. Defective development of uterine glands can be causing pregnancy loss10. Forkhead box a2 (FOXA2), a member of FoxA family transcription factors, has important roles in the uterine function and implantation11. Foxa2 is only found expressed in the glandular epithelium of the uterus12 and loss of expression defects endometrial gland development resulting in the loss of uterine glands11. Therefore, FOXA2-deficient mice lack of Lif secretion and thus are infertile11,13; however, the upstream regulators of FOXA2 are not completely understood in relation to their uterine functions.
Growth factor receptor-bound protein 2 (GRB2) is an adapter protein critical for the regulation of cell survival, proliferation, differentiation, and metabolism14. GRB2 directly binds EGFR and activates ERK1/2 and AKT in a Ras-dependent manner15. Abnormal expression of GRB2 promotes tumor malignancies by activating both PI3K/AKT and EGFR/ERK pathways16,17. In contrast, downregulation of GRB2 protein expression induced cell growth inhibition18. EGFR and ERK1/2 critically regulate endometrial function during early pregnancy19,20. EGFR plays important roles in epithelial cell proliferation/differentiation and stromal cell decidualization during early pregnancy. However, the role of GRB2 in implantation and decidualization has not been studied.
Here, we show that the amount of GRB2 is decreased in endometrium from infertile women with endometriosis. We used uterine-specific Grb2 knock-out mice and primary human endometrial stromal cells to demonstrate that GRB2 is necessary for implantation and decidualization. Our uterine-specific Grb2 knock-out mice display a non-receptive endometrium due to dysregulation of ovarian steroid hormone signaling as well as FOXA2 signaling.
Results
The levels of GRB2 protein are diminished in endometrium from infertile women with endometriosis
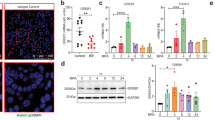
We examined the levels of GRB2 proteins in endometrium from fertile women (n = 19) and infertile women with and without endometriosis (n = 18 and n = 13, respectively) using immunohistochemistry (Fig. 1a). Our immunohistochemistry result revealed strong expression of GRB2 in the endometrial epithelial and stromal cells from control group endometrium at secretory stage. However, the expression of GRB2 proteins was significantly decreased in the epithelial cells (p < 0.001 and p < 0.01, respectively) and stromal cells (p < 0.001 and p < 0.01, respectively) of the eutopic endometrium from infertile women without and with endometriosis compared to controls (Fig. 1a). These results suggest that GRB2 deficiency may be associated with female infertility. To investigate the effect of Grb2 loss in endometriosis and pregnancy, we generated a mouse model with Grb2 conditionally ablated in Pgr-positive cells (Pgrcre/+Grb2f/f; Grb2d/d). Ablation of Grb2 in the uterus was confirmed by Western blot and immunohistochemical analyses (Fig. 1b, c). Our Western blot result showed a remarkable decrease of GRB2 protein in the whole uteri of Grb2d/d mice compared to control (Grb2f/f) mice (Fig. 1b). Immunohistochemical analysis validated that GRB2-postivite cells were undetectable in the uterine epithelial and stromal cells of female Grb2d/d at GD 3.5 (Fig. 1c).
a Representative image of GRB2 immunohistochemistry in the endometrium from infertile women with endometriosis (n = 18), infertile women without endometriosis (n = 13) compared to controls (n = 19) at mild-secretory stage. The quantification of GRB2 immunohistochemistry in the stroma and epithelial cells of endometrium from women with endometriosis, women without endometriosis and control. The results represent the mean ± SEM. ***p = 0.0002, **p = 0.0033, ***p = 0.0007, **p = 0.0086 by Ordinary one-way ANOVA test. b, c The ablation of GRB2 in the uterus were confirmed by Western blot analysis (n = 3 per genotype) and immunohistochemistry in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per genotype). d Representative fluorescence photomicrographs and average total number of endometriotic sites in Grb2f/f and Grb2d/d mice by induced endometriosis based on mT/mG mice (n = 5 per genotype). Endometriotic lesions were visualized by GFP expression in the outside of uterus. White arrow indicates endometriotic lesions. The results represent the mean ± SEM by two-tailed unpaired t-test. e Immunohistochemistry GRB2 analysis from uteri of the endometrium from 1 month and 3 months after endometriosis induction and Sham mice at GD 3.5 (n = 6 per genotype and time point). Semi-quantitative analysis of GRB2 levels in the epithelium and stroma in the stroma and epithelial cells endometrium from 3 months after endometriosis induction and Sham mice at GD 3.5. The results represent the mean ± SEM. *p = 0.0334, *p = 0.0234 and *p = 0.0384, *p = 0.0445, ***p < 0.0001 by Ordinary one-way ANOVA test. Source data are provided in the Source Data file.
To determine whether Grb2 deficient endometrial tissues exhibit different behavior when transplanted into an endometriosis model in mice, we performed endometriosis induction with normal GRB2-positive endometrium (Pgrcre/+Rosa26mTmG/+ control) and endometrium that lacks GRB2 (Pgrcre/+Grb2f/f Rosa26mTmG/+; Grb2d/d Rosa26mTmG/+). While GFP-positive endometriotic lesions were detected in mice with normal GRB2-positive endometrium, we could not detect any endometriotic lesions in mice with Grb2 deficient endometrial tissues (Fig. 1d). We also determine whether GRB2 expression is attenuated after endometriosis development. GRB2 was significantly lower in epithelial (p < 0.05 and p < 0.05, respectively) as well as stromal cells (p < 0.05 and p < 0.001, respectively) of the eutopic endometrium from mice with endometriosis compared to the sham control group at 1 month and 3 months after endometriosis induction (Fig. 1e). Thus, our results suggest that endometriosis development and progression are associated with a loss of GRB2 in the eutopic endometrium that may result in infertility.
Grb2 loss results in female infertility
We examined the expression of GRB2 to determine the cell-specific and temporal expression profiles of GRB2 in uteri of pregnant wild-type female mice from GD 0.5 to 7.5 using immunohistochemistry analysis. Our immunohistochemistry showed dynamic expressions of GRB2 proteins during early pregnancy (Fig. 2a). GRB2 proteins were weakly detected in uterine stromal cells of wild-type mice at GD 0.5. The levels of GRB2 proteins gradually increased from GD 2.5 to GD 3.5. At implantation stage GD 4.5, we detected strong GRB2 expression in decidual cells surrounding implantation sites. At GD 5.5, stromal cells underlying the implantation chamber differentiate into epithelioid cells to form the primary decidual zone (PDZ)21. GRB2-positive cells were detected in the PDZ at GD 5.5, but the intensity was decreased in the PDZ at GD 7.5. In contrast to the dynamic changes in the stroma, GRB2 expression was consistently strong in luminal and glandular epithelial cells from GD 0.5 to GD 4.5. These results suggest that GRB2 may play an important role during early pregnancy.
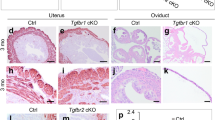
a Representative image of GRB2 immunohistochemistry in the uterine tissue from GD 0.5 to GD 7.5 (n = 5 per time point). b Visualizations of implantation sites by Chicago Sky Blue 6B dye in Grb2f/f and Grb2d/d uteri at GD 4.5 (n = 6 per genotype) after intravenous injection of Chicago Sky Blue 6B dye. c Hematoxylin and eosin analysis of implantation sites from uteri of Grb2f/f (i–iii) and Grb2d/d (iv–ix) mice at GD 4.5. d Immunohistochemistry PTGS2 analysis from uteri of Grb2f/f (i and ii) and Grb2d/d (iii–vi) mice at GD 4.5 (n = 3).
To explore the functional role of uterine Grb2 in pregnancy, we performed a fertility test where female Grb2f/f and Grb2d/d mice were mated with wild-type male mice, and their litters and pups were examined for 6 months (n = 5). Grb2f/f control mice gave birth to an average of 4.37 ± 0.24 litter per mouse and an average of 6.19 ± 0.28 pups per litter, whereas all Grb2d/d mice were sterile (Table 1). The results of the fertility test suggest that Grb2 plays an important role in pregnancy.
Because of reported Cre recombinase expression within the pituitary, ovary, uterus, and mammary gland of the Pgrcre/+ mouse22, we reasoned that Grb2d/d infertility might result from ovarian defects. To determine whether Grb2d/d mice had any ovarian phenotypes, we examined levels of serum E2 and P4 as well as ovarian histology (n = 5). The serum level of E2 and P4 showed no significant difference between Grb2f/f (E2 = 7.87 ± 0.94 pg/ml and P4 = 8.01 ± 1.20 ng/ml) and Grb2d/d (E2 = 6.85 ± 0.95 pg/ml and P4 = 6.48 ± 1.01 ng/ml) mice at GD 3.5 (Supplementary Fig. 1a). Grb2d/d mice showed normal ovarian morphology comparable to Grb2f/f mice (Supplementary Fig. 1b). These results suggest that Grb2d/d female mice have normal ovarian functions and that the infertility of Grb2d/d female mice results from uterine dysfunction.
Grb2 d/d female mice fail to support pregnancy past implantation
To determine the cause of infertility in Grb2d/d mice, ultrasound imaging was used to monitor pregnancy in Grb2f/f and Grb2d/d mice during early pregnancy. The embryos were detected in Grb2f/f but not Grb2d/d mice at GD 5.5, 7.5, and 9.5 by ultrasonography (Supplementary Fig. 2a), and the 3D reconstructions were generated to reveal the shape and size of implantation sites (Supplementary Fig. 2b). The sizes of embryos in Grb2f/f mice gradually increased during early pregnancy, the average sizes being 2.19 ± 0.34 mm3 at GD 5.5, 6.86 ± 0.83 mm3 at GD 7.5 and 24.87 ± 1.3 mm3 at GD 9.5 (Supplementary Fig. 2c). However, we could not detect any embryos in Grb2d/d mice at GD 5.5, GD 7.5, or GD 9.5 (Supplementary Fig. 2d). Our results suggest that Grb2d/d mice were infertile due to early pregnancy loss.
Grb2 d/d mice exhibit implantation defects
In order to determine the cause of early pregnancy loss in Grb2d/d mice, female Grb2f/f and Grb2d/d mice were examined at implantation stage GD 4.5. Implantation sites were visualized by intravenous injection of Chicago blue (Fig. 2b; n = 6). Although Grb2f/f mice revealed normal 8.29 ± 0.49 implantation sites, Grb2d/d mice did not show any Chicago blue-stained implantation sites (Fig. 2b). Next, our histological analysis revealed that embryos in Grb2f/f mice were attached to the luminal epithelium and surrounded by decidualized cells in (Fig. 2c). However, H&E staining for Grb2d/d mice revealed two groups of implantation sites in the uteri of (Fig. 2c). In group #1 (n = 3), the uterine lumen was not closed, and the embryos were free-floating within the uterine lumen. (Fig. 2c). In group #2 (n = 3), the uterine lumen was closed; however, the blastocysts were loosely affixed to the luminal epithelium and decidual cells were not observed (Fig. 2c). We performed IHC for prostaglandin-endoperoxide synthase 2 (PTGS2; COX2), a decidualization marker23, to examine decidualization at implantation sites. Grb2f/f mice showed strong PTGS2 staining in the decidual cells surrounding the embryos (Fig. 2d). However, we could not observe any PTGS2 expression in luminal epithelium and implantation sites in Group #1 or #2 of Grb2d/d mice (Fig. 2d). These results show Grb2d/d mice were infertile due to defective embryo implantation and decidualization.
Grb2 d/d mice exhibit dysregulation of PGR/FOXO1 and EGFR/ERK signaling in implantation
PGR and FOXO1 are important transcription factors regulating epithelial integrity and decidualization during implantation, and they are expressed reciprocally to each other24,25. We assessed the expression of FOXO1 and PGR in the implantation sites of Grb2f/f and Grb2d/d mice at GD 4.5 (Fig. 3; n = 5). FOXO1 proteins were strongly expressed in the nuclei of luminal epithelial cells in Grb2f/f mice (Fig. 3a, b). However, the expression of FOXO1 was significantly decreased (p < 0.001 and p < 0.001, respectively) in both Group #1 and Group #2 of Grb2d/d mice compared to control (Fig. 3a, b). Importantly, the localizations of FOXO1 were mainly in cytoplasm instead of nuclear. Moreover, its expression in Group #1 was significantly 2.6 times lower (p < 0.01) than in Group#2 (Fig. 3b). The nuclear expression of FOXO1 proteins at implantation sites of controls coincides with PGR loss in luminal epithelium and strong expression of PGR in stroma (Fig. 3c, d). However, the levels of nuclear PGR in luminal epithelium were highly increased in both Group #1 and Group #2 of Grb2d/d mice compared to controls (Fig. 3d). In contrast to luminal epithelium, stromal PGR protein was significantly decreased in both Group #1 and Group #2 of Grb2d/d mice compared to control mice (Fig. 3d; p < 0.001 and p < 0.05, respectively). Moreover, its expression in Group #2 was significantly higher (p < 0.05) than in Group#1 (Fig. 3d). Our IHC results suggest that the dysregulation of PGR and FOXO1 at implantation sites causes implantation failure of Grb2d/d mice.
a Immunohistochemistry analysis of FOXO1 from the uteri of Grb2f/f (i and ii) and Grb2d/d (iii–vi) mice at GD 4.5. b Semi-quantitative analysis of FOXO1 levels in the epithelium of Grb2f/f and Grb2d/d mice at GD 4.5 (n = 5 per group). The results represent the mean ± SEM. ***p < 0.0001, ***p < 0.0001, and **p = 0.0033 by Ordinary one-way ANOVA test. c Immunohistochemistry analysis of PGR from the uteri of Grb2f/f (i and ii) and Grb2d/d (iii–vi) mice at GD 4.5. d Semi-quantitative analysis of PGR levels in the epithelium and stroma of Grb2f/f and Grb2d/d mice at GD 4.5 (n = 5 per group). The results represent the mean ± SEM. ***p < 0.0001, *p = 0.0125, and *p = 0.0163 by Ordinary one-way ANOVA test. Source data are provided in the Source Data file.
Grb2 d/d mice exhibit decidualization defects
Decidualization is a process of stromal differentiation during embryo implantation26. However, it is difficult to examine the effect of GRB2 loss on decidualization during early pregnancy because Grb2d/d mice have implantation failure. To investigate the role of Grb2 in decidualization, we applied a mouse artificial decidualization model in Grb2d/d mice. Ovariectomized female Grb2f/f and Grb2d/d mice were hormonally and physically stimulated to artificially induce decidualization, mimicking an invading embryo in the left horn of the uterus. The other uterine horn was left unstimulated as a control. Although Grb2f/f mice displayed a decidual uterine horn that responded well to this artificial induction, Grb2d/d mice displayed a defect of decidual response (Fig. 4a). The weight ratio of stimulated to control horn was significantly decreased (p < 0.001) in Grb2d/d mice (1.06 ± 0.07, n = 4) compared to Grb2f/f mice (16.42 ± 1.74, n = 5) (Fig. 4a). Histological analysis confirmed that the stimulated uterine horn from control mice showed enlarged, cuboidal decidual cells, but the Grb2d/d mice did not detect decidual cells in the stimulated uterine horns (Fig. 4b). Further, the expression of the known decidualization markers, bone morphogenetic protein 2 (Bmp2) (p < 0.001), follistatin (Fst) (p < 0.001), FK506-binding protein 5 (Fkbp5) (p < 0.01), and wingless-related mouse mammary tumor virus integration site 4 (Wnt4) (p < 0.01), were significantly decreased in the decidual uterine horn of Grb2d/d mice compared to the decidual uterine horn of Grb2f/f (n = 4) (Fig. 4c).
a A decrease of the stimulated/control uterine weight ratio in Grb2f/f and Grb2d/d mice at decidualization day 5 (n = 5 for Grb2f/f and n = 4 for Grb2d/d mice). The results represent the mean ± SEM. ***p = 0.0001 by two-tailed unpaired t-test. b Hematoxylin and eosin (H&E) staining in control and stimulated horn of Grb2f/f and Grb2d/d mice at decidualization day 5. c RT-qPCR analysis for the expression of decidualization marker genes, Bmp2, Fst, Fkbp5, and Wnt4, in the uteri of Grb2f/f and Grb2d/d mice (n = 4 per genotype). The results represent the mean ± SEM. ***p < 0.0001, ***p < 0.0001, ***p < 0.0001, ***p < 0.0001, *p = 0.0435, **p = 0.0045, **p = 0.0034, **p = 0.0042 by Ordinary one-way ANOVA test. d The expression of GRB2 in hESCs from control women without (Ctrl) and with (Eosis) endometriosis (n = 22 per group). The expression of GRB2, IGFBP1, and PRL levels in hESCs and after silencing with siRNA in decidualized HESCs. Each treatment was performed with at three biological replicates (n = 3 per group and time point). The results represent the mean ± SEM. ***p = 0.0001, **p = 0.0085, *p = 0.0200, **p = 0.0035, ***p = 0.0004, and ***p < 0.0001, by Ordinary one-way ANOVA test. Source data are provided in the Source Data file.
Infertile women with endometriosis display markedly reduced decidualization and impaired uterine receptivity27. Therefore, we examined GRB2 levels in primary human endometrial stromal cells (hESCs) from women with or without endometriosis using RT-qPCR. GRB2 levels were significantly lower (p < 0.001) in hESCs from women with endometriosis compared to controls (Fig. 4d). hESCs can be cultured in vitro and treated with a hormone cocktail of E2, P4, and cAMP (EPC) to induce decidualization28. To determine the effect of GRB2 attenuation on human decidualization, we combined this system with a siRNA loss of function approach in control hESCs. The GRB2 knock-down efficiency was assessed by RT-qPCR before inducing decidualization, confirming that GRB2 mRNA levels were significantly decreased in hESCs treated with GRB2 siRNA compared with non-targeting pool siRNA (Fig. 4d). hESC transfected with a non-targeting pool of control siRNA and treated with EPC showed robust induction of classic decidual markers IGFBP1 and PRL (Fig. 4d). Conversely, in cells transfected with GRB2 siRNA, the induction of these markers was significantly reduced compared to the control (Fig. 4d). These results indicated that GRB2 plays an important role for decidualization in mice as well as hESCs.
Mice with Grb2 conditional deletion in the uterine epithelium showed normal fertility
Uterine epithelial-stromal crosstalk is crucial for implantation and decidualization7. GRB2 proteins are strongly expressed in uterine epithelial and stromal cells at pre-implantation stage. To examine the role of Grb2 in uterine epithelium, we conditionally ablate Grb2 in the adult mouse endometrial endometrium by crossing Ltficre/+29 and Grb2f/f mice. Ltficre/+Grb2f/f mice displayed Grb2 deletion in the luminal and glandular epithelium, while retaining Grb2 expression in the stroma in contrast to Grb2d/d mice (Supplementary Fig. 3a). To assess overall fecundity, control Grb2f/f and Ltficre/+Grb2f/f females were mated with wild-type male mice (n = 5). Grb2f/f and Ltficre+Grb2f/f mice had normal number of pups with an average of 5.89 ± 0.078 and 5.71 ± 0.079 pups per litter, respectively (Supplementary Fig. 3b). Ltficre+Grb2f/f mice revealed normal ovarian functions including ovulation, fertilization and histology (Supplementary Fig. 3c, d). We also monitored pregnancy and embryo growth in Ltficre+Grb2f/f mice using high frequency ultrasound. Normal sizes and numbers of embryos were detected at GD 5.5, GD 7.5, and GD 9.5 in both Grb2f/f and Ltficre+Grb2f/f mice (Supplementary Fig. 4a). Furthermore, the number of implantation sites were no different between Ltficre+Grb2f/f and Grb2f/f mice using Chicago blue at GD 4.5, GD 5.5 and GD 9.5 mice (Supplementary Fig. 4b). In addition, both Grb2f/f and Ltficre+Grb2f/f mice revealed normal decidual responses by artificial decidualization experiment (Supplementary Fig. 4c). These results demonstrated that epithelial Grb2 ablation does not impair implantation and decidualization.
Grb2 loss results in a non-receptive endometrium due to dysregulation of ovarian steroid hormone signaling
Loss of epithelial E2 action is essential for implantation at the secretory phase in all eutherian mammal species studied30,31,32. Proliferation is markedly reduced in epithelial cells at the peri-implantation stage for embryo attachment20,33. We examined expression of Ki67, a proliferation marker, at GD 3.5, which marks the pre-implantation stage2 (Fig. 5a). While cell proliferation in the luminal epithelium was significantly increased (p < 0.001) in Grb2d/d mice (95.21 ± 1.91 %) compared to control Grb2f/f mice (3.99 ± 1.58 %), cell proliferation in the stroma was significantly decreased (p < 0.001) in Grb2d/d mice (7.49 ± 3.04 %) compared with Grb2f/f mice (63.77 ± 4.59 %). Next, we investigated whether Grb2 ablation caused excess E2 signaling. The expression of Clca3, Ltf, and Muc-1, E2 target genes, were significantly increased (p < 0.01, p < 0.05, and p < 0.001, respectively) in Grb2d/d mice compared to control mice (Fig. 5b). We also detected a significant decrease (p < 0.01) of ESR1 levels in stromal cells of Grb2d/d mice compared to control Grb2f/f mice but not in epithelial cells (Fig. 5c). These results demonstrate that E2 activity is enhanced in uterine epithelial cells of the Grb2d/d mice.
a Immunohistochemistry of Ki67 analysis in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per genotype). The results represent the mean ± SEM. ***p < 0.0001 and ***p < 0.0001 by two-tailed unpaired t-test. b RT-qPCR analysis for E2 target genes, Clca3, Ltf, and Muc-1, in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 4 per genotype). The results represent the mean ± SEM. **p = 0.0063, *p = 0.0221, and ***p = 0.0006 by two-tailed unpaired t-test. c Immunohistochemistry of ESR1 analysis in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per genotype). The results represent the mean ± SEM. ** p = 0.0014 by two-tailed unpaired t-test. d RT-qPCR analysis for P4 target genes, Il13ra2, Fst, Areg, and Lrp2, in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 4 per genotype). The results represent the mean ± SEM. **p = 0.0015, ***p = 0.0009, **p = 0.0019 and ***p < 0.0001 by two-tailed unpaired t-test. e Immunohistochemistry of PGR analysis in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per genotype). The results represent the mean ± SEM. ***p < 0.0001 by two-tailed unpaired t-test. f Immunohistochemistry of stromal maker protein COUP-TFII and g Vimentin in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per genotype). The results represent the mean ± SEM. *** p < 0.0001, and *** p < 0.0001 by two-tailed unpaired t-test. Source data are provided in the Source Data file.
Defective P4 action is the primary cause of the defects in ESR1 downregulation, and has been called P4 resistance34. Next, we used RT-qPCR and immunohistochemistry to assess expression of P4 target genes and PGR in Grb2d/d mice. mRNA levels of P4 target genes Il13ra2, Fst, Areg, and Lrp2 were significantly down-regulated (p < 0.01, p < 0.001, p < 0.01, and p < 0.001, respectively) in Grb2d/d mice compared to control mice (Fig. 5d). Interestingly, stromal PGR expression was significantly reduced (p < 0.001) in Grb2d/d mice compared to control mice (Fig. 5e). However, the expression of epithelial PGR did not change in Grb2d/d mice (Fig. 5e). Moreover, expression of the stromal markers, vimentin and COUP-TFII, was significantly decreased (p < 0.001 and p < 0.001, respectively) in the stromal cells of Grb2d/d mice compared to control mice (Fig. 5f, g). These results suggest that GRB2 loss results in a non-receptive endometrium due to dysregulation of ovarian steroid hormone signaling.
Grb2 regulates the E2/P4-related as well as EGF-related genes
To identify the signaling pathways that Grb2 regulates at pre-implantation, we performed RNA-seq analysis in the uterine tissue of Grb2f/f and Grb2d/d mice at GD 3.5. Our transcriptomic analysis identified 2243 differentially expressed genes (DEGs) (fold change > 2, p < 0.05 and FDR < 0.05) including 917 upregulated genes and 1326 down-regulated genes in the uterus of Grb2d/d mice compared with Grb2f/f mice (Supplementary Data 1). Upstream regulator analysis by Ingenuity Pathway Analysis (IPA) showed that major altered pathways in the Grb2d/d uterus included 17β-estradiol/ESR1, progesterone/PGR, STAT3, FOXO1, and EGF/EGFR signaling (Fig. 6a). Our pathway analysis by IPA identified 508 differentially expressed E2-related genes (Supplementary Data 2) and 191 differentially expressed P4-related (Supplementary Data 3) genes in Grb2d/d uteri compared with Grb2f/f uteri (Fig. 6b). Importantly, 246 EGF/EGFR-related genes were identified in Grb2d/d mice compared with Grb2f/f mice (Fig. 6c and Supplementary Data 4). Because EGFR critically regulates endometrial function and decidualization during early pregnancy19, and GRB2 interacts with EGFR to activate Ras and its downstream kinases, ERK1/214, we validated our transcriptomic results using RT-qPCR. Our RT-qPCR results revealed that the mRNA expression of Egfr was significantly (p < 0.05) decreased (−1.78-fold) in the Grb2d/d mice compared to the Grb2f/f mice (Fig. 6d). The levels of Erbb2 and Erbb3 mRNA were significantly (p < 0.05 and p < 0.001, respectively) increased (2.38-fold and 2.98-fold) in the Grb2d/d mice compared to the Grb2f/f mice, respectively. In addition, the expression of Egf mRNA was no different between Grb2d/d (1.82 ± 0.26) and Grb2f/f mice (1.51 ± 0.28) (Supplementary Fig. 5). The expression of ERBB2 protein was not different in the uterus of both Grb2f/f and Grb2d/d mice (Supplementary Fig. 5) but EGFR protein level was remarkably decreased in the epithelial and stromal cells of Grb2d/d mice (Fig. 6d).
a GO terms of the upregulated genes in the uteri of Grb2d/d mice compared with that of Grb2f/f mice from RNA-seq data. b Heatmap of E2-related genes and P4-related genes in the uterus of Grb2f/f and Grb2d/d mice at GD 3.5. c Heatmap of uterine EGFR-related genes and d RT-qPCR and immunohistochemistry analysis of EGFR in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 4 per genotype). The results represent the mean ± SEM. *p = 0.0221 by two-tailed unpaired t-test. e Immunohistochemistry analysis of EGFR and pERK1/2 in the uteri of Grb2f/f and Grb2d/d mice at GD 4.5. f Semi-quantitative analysis of EGFR and pERK1/2 levels in the uteri of Grb2f/f and Grb2d/d mice at GD 4.5 (n = 5 per genotype). The results represent the mean ± SEM. ***p < 0.0001, ***p < 0.0001, *p = 0.0300, *p = 0.0455, ***p < 0.0001, and ***p < 0.0001 by Ordinary one-way ANOVA test. Source data are provided in the Source Data file.
Mice lacking Egfr in the uterus failed to maintain and process decidualization during implantation19. Therefore, we next examined the expression of EGFR proteins at implantation sites of Grb2f/f and Grb2d/d mice at GD 4.5 (Fig. 6e, f). While EGFR proteins were strongly expressed at stromal cells near implantation sites of Grb2f/f mice, it was not detected at luminal epithelium. The levels of stromal EGFR proteins were significantly (p < 0.001 and p < 0.001, respectively) reduced in both Group #1 and Group #2 of Grb2d/d mice compared to control (Fig. 6f). However, the levels of epithelial EGFR proteins were significantly (p < 0.05 and p < 0.05, respectively) increased in both Group #1 and Group #2 of Grb2d/d mice compared to control. Furthermore, pERK1/2 proteins were strongly expressed in stromal cells near implantation sites of Grb2f/f mice, while they were very weakly detected in luminal epithelium. The levels of stromal pERK1/2 proteins were significantly (p < 0.001 and p < 0.001, respectively; 3.6-fold and 12.9-fold, respectively) decreased in both Group #1 and Group #2 of Grb2d/d mice compared to control (Fig. 6e, f). These results revealed that GRB2 loss causes dysregulation of ovarian steroid E2/P4 signaling as well as EGFR/ERK signaling in the uterus.
Next, we examined whether tert-Butylhydroquinone (TBHQ), an ERK1/2 activator, rescues the phenotypes of Grb2 deficient mice. Grb2f/f and Grb2d/d mice were treated daily with TBHQ for 1 week (50 mg/kg via intraperitoneal injection). Unfortunately, TBHQ resulted in implantation failure of control and Grb2d/d mice (Supplementary Fig. 6). Our previous study showed that the expression of ERK1/2 signaling molecules are dynamically regulated during early pregnancy and play a critical role in decidualization20. In addition, we examined the effect of U0126, an ERK1/2 inhibitor, during early pregnancy. Control mice were treated with vehicle or U0126 (150 μmol/kg one time/week via intraperitoneal injection) for 1 month and then ultrasound imaging was performed to monitor pregnancy. U0126-treated mice showed a significant decrease (p = 0.0088) in the number of implantation sites compared to vehicle group at GD 9.5 (Fig. 7a–c). Therefore, our results suggest that ERK1/2 signaling molecules should be tightly regulated for implantation and decidualization during early pregnancy.
a The number of implantation sites were analyzed by 3D reconstruction of implantations from mice treated with U0126 at GD 5.5, GD 7.5, and GD 9.5 (n = 5 per treatment). b Representative image of implantation site of mice treated with vehicle and U0126 at GD 9.5. c Mice treated with U0126 shows a decreased number of implantation sites compared with mice treated with vehicle at GD 9.5 (n = 5 per treatment). The results represent the mean ± SEM. **p = 0.0088 by two-tailed unpaired t-test. Source data are provided in the Source Data file.
Grb2 is critical for endometrial gland development and function in mouse uterus
Endometrial glands expressing FOXA2 play an important role in the implantation process6,11,35. Therefore, we examined gland formation and function in Grb2d/d mice. Grb2d/d mice (8.67 ± 1.67) had significantly fewer endometrial glands compared to controls at GD 3.5 (26.67 ± 2.11, 3.0-fold, p < 0.001; Fig. 8a). IHC analysis revealed a significant reduction (p < 0.001) of FOXA2, a glandular specific marker, in glands of Grb2d/d mice compared to control mice (Fig. 8b). In order to understand the molecular dysregulation in glands of Grb2d/d mice, we utilized our transcriptomic data from GD 3.5 Grb2d/d mouse uteri to compare the dysregulated genes to those dysregulated in Pgrcre/+Foxa2f/f (Foxa2d/d) uteri at GD 3.536. Out of a total of 2,243 genes differentially expressed due to Grb2 loss, 593 (26.44%) were also differentially expressed in Foxa2d/d mice (Fig. 8c, Supplementary Data 5). Further, a clustering heatmap of the 593 genes identified 343 down-regulated genes in both Grb2d/d and Foxa2d/d mice (Fig. 8d). Importantly, the levels of Lif and several glandular epithelium-specific genes (Prss29, Spink3, Ttr, Wfdc3) were significantly reduced in the uteri of both Foxa2d/d and Grb2d/d mice compared to controls (Fig. 8e). However, the uterine weight and the expression of FOXA2 were not different (p = 0.4213) in the uteri of 3–4-week-old Grb2d/d mice (1.52 ± 0.19) compared to the Grb2f/f mice uteri (1.74 ± 0.16) (n = 4) (Supplementary Fig. 7).
a The number of grands in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 6). The results represent the mean ± SEM. ***p < 0.0001 by two-tailed unpaired t-test. b Immunohistochemistry analysis of FOXA2 in the uteri of Grb2f/f and Grb2d/d mice at GD 3.5 (n = 5 per treatment). The results represent the mean ± SEM. ***p < 0.0001 by two-tailed unpaired t-test. c Venn diagram of identifying genes that overlapped in RNA-Seq data between Foxa2d/d and Grb2d/d mice at GD 3.5. d A heatmap of differentially expressed genes in uterus of control, Foxa2d/d and Grb2d/d mice at GD 3.5 from RNA-Seq data (n = 4 per genotype). e Heatmap of Grb2 and Foxa2-related genes in the uteri of Foxa2d/d and Grb2d/d mice at GD 3.5 (n = 4 per genotype). Source data are provided in the Source Data file.
Discussion
Our study reveals that the expression of GRB2 is decreased in eutopic endometrium from infertile women with endometriosis compared to controls. Uterine epithelial-stromal crosstalk is crucial for implantation and decidualization7. GRB2 proteins are strongly expressed in uterine epithelial and stromal cells at the pre-implantation stage. However, Ltficre+Grb2f/f mice revealed normal uterine functions. Interestingly, GRB2 knock-down in hESC resulted in a significant reduction of decidual markers IGFBP1 and PRL compared to the control (Fig. 4d) and stromal PGR expression was only significantly reduced in Grb2d/d mice compared to control mice (Fig. 5e). Moreover, expression of the stromal markers, vimentin and COUP-TFII, was significantly decreased in the stromal cells of Grb2d/d mice compared to control mice (Fig. 5f, g). Our results suggest that stromal GRB2 plays a critical function for regulation of receptive endometrium and ovarian steroid hormone signaling.
GRB2 binds to EGFR for various biological processes by activating PI3K/AKT and ERK/MAPK pathways16,17. Our RNA-seq analysis identified 246 dysregulated EGFR-related genes in the uterus of Grb2d/d mice compared to control. EGFR is critical for endometrial function during early pregnancy19. EGFR is expressed in the stroma surrounding the implanting blastocyst37,38. However, the expressions of EGFR and pERK1/2 were remarkably decreased in the uterus of Grb2d/d mice at implantation sites of GD 4.5. The defective decidualization responses of eutopic and ectopic endometrial stromal cells in endometriosis patients39,40 are similar to those observed in Grb2d/d mice as well as hESCs from women without disease with the knock-down of GRB2. Importantly, endometrial epithelium-specific Grb2 knockout mice (Ltficre/+Grb2f/f) exhibited normal implantation and decidualization. Our results suggest that the attenuation of GRB2 in infertile women with endometriosis causes infertility due to implantation failure based on evidence from Grb2d/d mice. However, the molecular mechanism of GRB2 attenuation in the etiology and pathophysiology of endometriosis-related infertility will require further study.
Our previous results showed that mice lacking Egfr expression (Pgrcre/+Egfrf/f mice) are severely subfertile19 as we observed in Grb2d/d mice. Early pregnancy failure and implantation site demise is a key factor in subfertility of both female Grb2d/d and Pgrcre/+Egfrf/f mice. They have the same phenotype that pregnancy demise occurred shortly after blastocyst implantation due to defects in decidualization. Modest reductions in litter size observed in the absence of uterine Erbb2 or Erbb3 indicates that they may facilitate EGFR signaling as dimerizing partners but do not play a critical role in pregnancy individually19. While Egf expression was not changed in Grb2d/d mice, Erbb2 and Erbb3 were significantly increased in Grb2d/d mice. These results suggest that Grb2 targets EGF signaling to regulate uterine function.
Grb2d/d mice exhibited aberrant activation of epithelial proliferation and a decrease in stromal proliferation. Activation of epithelial proliferation at the pre-implantation stage leads to implantation failure in several mutant mouse studies41,42. Moreover, the aberrant activation of epithelial proliferation is found in the endometrium of infertile women with endometriosis43,44. In contrast, increased stromal cell proliferation is a crucial step for normal uterine receptivity45 and is considered to be an initiator of decidualization46. In the Grb2d/d uterus on GD 4.5, the expression of PTGS2 was significantly reduced in the stromal cells surrounding the embryo at failed implantation sites. This finding suggests that endogenous decidualization was disrupted at the molecular level by loss of GRB2.
The regulation of P4 and E2 is critical for the regulation of the molecular and cellular events leading to the uterine receptivity for implantation47. P4 suppresses E2 action on epithelial proliferation and promotes stromal cell proliferation to prepare uterine receptivity for implantation and decidualization48. Loss of E2 action is essential for the implantation at the secretory phase in all eutherian mammal species43. The imbalance of P4 and E2 action is found in the endometrium of women with endometriosis49. Grb2d/d mice revealed dysregulation of PGR and ESR1 in uterine stroma at GD 3.5. As a result, the expression of P4 target genes were significantly reduced, while E2 target genes were significantly upregulated in Grb2d/d mice. Failure to downregulate ESR1 signaling is a sign of a homeostasis imbalance in both endometriosis and infertility50, and is likely the cause of excessive proliferation in the endometrium of women with endometriosis51.
Interestingly, we found that the number of uterine glands was significantly decreased in the uterus of Grb2d/d mice. Uterine glands are critical for implantation and decidualization as reported in uterine Foxa2-deleted mice (Foxa2d/d)11,35. FOXA2 is explicitly expressed in the glandular epithelium of uterus in both mice and human52,53. FOXA2 was decreased in eutopic endometrium from women with endometriosis54. In Foxa2d/d mice, the absence of uterine glands led to severe defects of implantation and decidualization35,53. We observed that the expression of FOXA2 was significantly decreased in the glandular epithelium in the uterus of Grb2d/d mice. Grb2d/d and both FOXA2-deficient mouse models (Foxa2d/d and Ltficre/+Foxa2f/f) are infertile due to defects in embryo attachment on GD 4.511,35. Furthermore, our comparative analysis of transcriptomic data from Grb2d/d and Foxa2d/d mice confirmed the molecular impact of uterine Grb2 loss on FOXA2-regulated genes.
In conclusion, our findings demonstrate that GRB2 plays a critical role for the successful progression of early pregnancy. As we observed attenuation of GRB2 in human eutopic endometrium from infertile women with endometriosis, loss of Grb2 in the mouse uterus resulted in infertility due to implantation and decidualization defects. Interestingly, the ablation of Grb2 in the uterus impaired uterine gland function normally supported by FOXA2. Therefore, our studies suggest a conceptual framework for understanding abnormal endometrial homeostasis, with implications for the diagnosis and treatment of a non-receptive endometrium in early pregnancy loss.
Methods
Study design
The main objective of this study was to evaluate the role of GRB2 in infertility. First, the expression of GRB2 was assessed in eutopic endometrium of infertile women with or without endometriosis compared to fertile women. To determine whether endometriosis affects GRB2 expression, GRB2 protein expression were examined in a mouse model of endometriosis. Subsequently, we determine that impact of Grb2 ablation on the infertility implantation, and decidualization in Grb2d/d and littermate control mice. Finally, transcriptomic analysis was applied to dissect the molecular mechanisms of Grb2 in the uterus. The control and treatment groups and the number of biological replicates (sample sizes) for each experiment are specified in the figure legends. The number of animals for each study type were determined by the investigators on the basis of previous experience with the standard disease models that were used or from pilot studies. Animals were randomly allocated to the control and treatment groups and housed together to minimize environmental differences and experimental bias. Analysis of endpoint readouts was carried out in a blinded fashion.
Ethics statement and declarations
The institutional review boards of University of Missouri, Wake Forest University, and University of North Carolina approved this study. The Institutional Animal Care and Use Committee at University of Missouri approved all experiments relating to mice.
All animal experiments were approved by the University of Missouri Animal Care and Use Committee. Mice were housed and bred in a designated animal care facility at University of Missouri with controlled humidity and temperature conditions and a 12 h light/dark cycle.
Human endometrium samples
The human endometrial samples used to examine GRB2 expression patterns were obtained from Wake Forest University and University of North Carolina. Written informed consent was obtained from all participants. The study design and conduct complied with all relevant regulations regarding the use of human study participants and was conducted in accordance with the criteria set by the Declaration of Helsinki. Control endometrial tissues were laparoscopically negative for endometriosis and had not been on any hormonal therapies for at least 3 months prior to surgery. Histologic dating of endometrial samples was done on the basis of the criteria of Noyes et al.55 and confirmed by subsequent histopathological examination by an experienced fertility specialist (B.A.L.). Normally cycling women without infertility who were free of hormones for at least 60 days served as fertile controls. Tubal ligation was a source for normal subjects as it allows us to rule out endometriosis laparoscopically. All endometriosis-related infertility cases were undergoing endometrial sampling prior to surgical removal of endometriosis. All patients did not have uterine leiomyoma and adenomyosis. To investigate GRB2 amounts in the endometrium from women, 18 infertile women with endometriosis, 13 infertile women without endometriosis and 19 control eutopic endometrium were used. Samples used for immunohistochemistry were fixed in 10% buffered formalin prior to embedding in paraffin wax.
Animals and tissue collection
Mice were housed and bred in a designated animal care facility at the University of Missouri with controlled humidity (30-70%) and temperature conditions (68–79 °F) and a 12 h light/dark cycle. Grb2 conditional knockout mice were generated crossing Pgrcre/+22 or Ltficre/+56 males with female mice carrying the Grb2f/f allele (Pgrcre/+Grb2f/f; Grb2d/d). All control (Grb2f/f), Pgrcre/+Grb2f/f, and Ltficre/+ Grb2f/f mice were mixed background C57BL/6J and 129S1/SvImJ strains and 8-12-week-old female mice from. were used for all experiments. For the early pregnancy study, female Grb2f/f and Grb2d/d were mated with C57BL/6 male mice, and uterine samples from pregnant mice were obtained at different days of pregnancy. The morning of vaginal plug is designated as GD 0.5. Uteri were collected at GD 3.5 and 4.5, and implantation sites were visualized on GD 4.5 by intravenous injection of 1% Chicago Sky Blue 6B dye (Sigma-Aldrich, St. Louis, MO). Fertility was assessed by mating female Grb2f/f and Grb2d/d mice with wild-type C57BL/6 male mice for 6 months. The number of litter and pups born during that period was recorded. U0126 was purchased from Selleck (Pennsylvania, USA) and tert-Butylhydroquinone (TBHQ) was purchased from MedChemExpress (New Jersey, USA). For ERK inhibitor and activator study, U0126 and TBHQ were dissolved in solution 5% DMSO + 40% PEG400 + 45% Saline + 5% Tween80. For U0126 treatment study, female control mice at 8 weeks old were received with either vehicle (PBS) and U0126 (150 μmol/kg) every week by intraperitoneal (i.p) injection for 2 weeks. For TBHQ treatment study, Grb2f/f and Grb2d/d mice were daily received with TBHQ (50 mg/kg) by intraperitoneal (i.p) injection for 1 week. After the last injection, mice were mated with wild-type male mice. Vevo F2 LAZR-X ultrasound system was used at GD 5.5, 7.5, and 9.5. Uterine tissues were collected at the time of dissection and immediately either fixed with 4% (vol/vol) paraformaldehyde for histology or immunohistochemistry or snap frozen and stored at −80 °C for RNA/protein extraction. The levels of progesterone and estrogen in serum were measured by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core.
Induction of endometriosis in mouse
The endometriosis was induced in 8-week-old female control (Pgrcre/+Rosa26mTmG) and Grb2d/d Rosa26mTmG mice which have conditional double-fluorescent Cre reporter gene had daily 1 μg/ml of E2 injection for 3 days and then autologous endometriosis induction were performed57. After 1 month post endometriosis induction, the mice were euthanized, and endometriotic lesions were counted under fluorescence microscope.
Reverse transcription-quantitative PCR
1 µg of RNA was used for reverse transcription with MMLV Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. mRNA expression levels of genes of interest were measured by real-time PCR TaqMan or SYBR green analysis using an Applied Biosystems StepOnePlus system (Applied Biosystems, Foster City, CA, USA). The mRNA quantities were normalized against the housekeeping gene, 60S ribosomal protein L7. Analysis of mRNA expression was first undertaken by the standard curve method, and results were corroborated by cycle threshold values assessing gene expression. Primer sequences used in these studies are shown in Supplementary Table 1.
Histology and immunostaining
Histology and immunohistochemistry analyses were performed as previously described54,58.
For immunohistochemistry, dewaxed hydrated paraffin-embedded tissue sections were blocked with 10% normal goat serum in PBS (pH 7.5) and then incubated with primary antibody diluted in 10% normal goat serum in PBS (pH 7.5) overnight at 4 °C anti-GRB2 (1:2000 dilution; ab32037; Abcam), anti-PTGS2 (1:500 dilution; 160106; Cayman), anti-FOXO1 (1:500 dilution; CS-2880; Cell Signaling), anti-PGR (1:2000 dilution; CS-8757S; Cell Signaling), anti-EGFR (1:500 dilution; CS-2646; Cell Signaling), anti-ERBB2 (1:1000 dilution; CS-4290S; Cell Signaling), anti-pERK (1:500 dilution; CS-4370; Cell Signaling), anti-COUP-TFII (1:1000 dilution; PP-H7147-10; Perseus Proteomics), anti-Vimentin (1:1000 dilution; ab92547; Abcam), anti-FOXA2 (1:1000 dilution; WRAB-FOXA2; Seven Hills BioReagent), anti-Ki67 (1:1000 dilution; BD550609; BD Pharmingen), anti-ERα (1:1000 dilution; sc-8002; Santa Cruz). On the next day, the sections were incubated with the appropriate species specific HRP-conjugated secondary antibody (2 μg/ml; Vector Laboratories) for 1 h at room temperature. Immunoreactivity was detected using the Vectastain Elite DAB kit (Vector Laboratories). A semi-quantitative grading system (H-score) was used to compare the immunohistochemical staining intensities as previously described59. The overall score ranged from 0 to 300.
Western blot analysis
Proteins were extracted using lysis buffer (10 mM Tris-HCl (pH 7.4), 150 mM NaCl, 2.5 mM EDTA, and 0.125% Nonidet P-40 (vol/vol)) supplemented with both a protease inhibitor cocktail (Roche, Indianapolis, IN) and a phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO). Ten micrograms of total protein was electrophoresed via 8% SDS–PAGE gels and transferred into a polyvinylidene difluoride membrane (Millipore Corp., Bedford, MA, USA). Membrane was blocked with Casein (0.5% w/v) in PBS with 0.1% Tween 20 (v/v; Sigma-Aldrich) prior to exposure to anti-GRB2 (1:20,000 dilution; CS-3972; Cell Signaling) antibody. Total β-Actin (1:20,000 dilution; SAB5600204; Sigma) levels were examined for loading controls. Immunoreactivity was visualized by incubation with a horseradish peroxidase-linked secondary antibody (anti-Rabit IgG (1:5000 dilution; PI-9500; Vector Laboratories)) followed by exposure to Electrochemiluminescence reagents (ECL) according to the manufacturer’s instructions (GE Healthcare Biosciences).
Ultrasound imaging
To examine the size of embryo during the early pregnancy, the Vevo F2 LAZR-X small animal ultrasound system (FUJIFILM VisualSonics Inc., Toronto, Canada), a non-invasion high resolution imaging tool, was used at GD 5.5, 7.5 and 9.5. Mice were anesthetized using isoflurane and positioned supine on a heated imaging stage with the face toward the scientist. ECG, body temperature, and respiratory physiology were always controlled. Eye creme was put on mice’s eyes to protect and maintain the physiological ocular welfare. The hair on the abdomen was removed with commercial hair removal cream. Then pre-warmed ultrasound gel was applied directly onto the depilated skin. First, the transducer 47 MHz (MS550D-0421) was used to identify the bladder as reference point. B-Mode or brightness mode was used to render a two-dimensional gray scale image of anatomical structures. A motor was attached to the transducer to translate the transducer from the left to the right side of the abdomen with step sizes of scanning (10 μm). The acquired 2D images were processed with Vevo Lab software (Fujifilm, VisualSonics, version 5.5.1) to reconstruct and analysis the 3D volume of implantations to obtain transverse, sagittal, and coronal representations of implantations.
Artificial decidualization response
To test the uterine response to an artificial decidual stimulus, female Grb2f/f and Grb2d/d mice were ovariectomized at 6 weeks of age, allowed to recover for 2 weeks as described previously study60. Then mice were subjected to the following hormonal regimen: 100 ng of E2 per day for 3 days; 2 days rest; then three daily injections of 1 mg of P4 plus 6.7 ng of E2. To induce artificial decidualization, the left uterine horn was mechanically stimulated by scratching the full length of the anti-mesometrial side with a burred needle 6 h following the third P4 and E2 injection. The other horn was left unstimulated as a control. Daily injections of P4 (1 mg/mouse) + E2 (6.7 ng/mouse) were continued for 5 days to maximize the decidual response. Then, mice were sacrificed on day 5. The uteri were then excised, weighed, and fixed in 4% paraformaldehyde for histological analysis. For RNA or protein extractions, uterine tissues were collected and immediately frozen in dry ice.
hESC culture and in vitro decidualization
Human primary endometrial stromal cells (hESCs) were isolated from pure endometrial tissue by collagenase digestion20,61,62. Briefly, endometrial tissue was minced and digested twice at 37 °C in buffered saline supplemented with 0.5% collagenase (Sigma-Aldrich, St. Louis, MO) and 0.002% DNase (Sigma-Aldrich, St. Louis, MO). Then endometrial tissues were filtered through 70 µm sterile sieves to exclude epithelium. The remaining hESCs were cultured at 37 °C in 5% CO2, and incubated with RPMI-1640 phenol red–free medium (Gibco, Grand Island, NY) contained 0.1 mM sodium pyruvate (Gibco, Grand Island, NY), 10% fetal bovine serum (FBS; Gibco, Grand Island, NY), and 1% penicillin streptomycin (P/S; Gibco, Grand Island, NY). Small interfering RNA (siRNA) of GRB2 (L-019220-00-0020, Dharmacon, Lafayette, CO) was used for the knock-down of GRB2 gene expression and non-targeting scrambled siRNA (NC1486135, Dharmacon) was used as a control. Transfection of hESC were conducted using lipofectamine 2000 (Invitrogen, Carlsbad, CA) based on the manufacturer’s instructions. In brief, after 4 h of serum starvation, hESCs were incubated in serum-free media with transfection complex for 6 h before replacing with growth media and incubating for 2 days before in vitro decidualization induction. To induce in vitro decidualization, hESC were incubated with differentiation media contained OPTI-MEM medium (Gibco) with 2% FBS depleted of steroids by pretreatment with dextran-coated charcoal (CSFBS; Gibco), 10 nM 17β-estradiol (E2; Sigma-Aldrich, St. Louis, MO), 1 μM medroxyprogesterone acetate (Sigma-Aldrich), 50 μM Cyclic adenosine monophosphate (cAMP; Sigma-Aldrich), and 1% Penicillin and streptomycin (P/S; Gibco). Differentiation media were replaced every 2 days until the 6 days.
RNA-sequencing analysis
Total RNA was extracted from the uterine tissues using the RNeasy total RNA isolation kit (Qiagen, Valencia, CA). NanoDrop was used to determine RNA purity and for an initial estimate of RNA concentration. For RNA-Seq, 500 ng of total RNA were used. RNA-Seq libraries were prepared by the Van Andel Genomics Core using the KAPA mRNA Hyperprep kit (v4.17) (Kapa Biosystems, Wilmington, MA, USA). RNA was sheared to 300–400 bp. Prior to PCR amplification, cDNA fragments were ligated to IDT for Illumina unique dual adapters (IDT DNA Inc). Quality and quantity of libraries were assessed using a combination of Agilent DNA High Sensitivity chip (Agilent Technologies, Inc.) and QuantiFluor® dsDNA System (Promega Corp., Madison, WI, USA). Individually indexed libraries were pooled and 50 bp, paired end sequencing was performed using an Illumina NovaSeq6000 sequencer and S2, 100 cycle sequencing kit (Illumina Inc., San Diego, CA, USA). Each library was sequenced to an average raw depth of 50 M reads. Base calling was done by Illumina RTA3 and output of NCS was demultiplexed and converted to FastQ format with Illumina Bcl2fastq v1.9.0. The raw reads were initially filtered by removing low quality reads (average quality scores <20) and aligned to the mm39 genome using Star 2.7.9a63. The differentiated expressions of genes (DEGs) were determined by the exact test of EdgeR64. The DEGs were determined as maximum counts ≥1; fold change of >2 (upregulated) or <−2 (down-regulated), FDR < 0.05. The Principle Component Analysis (PCA) map were generated by plotMDS in EdgeR64. The expressions of DEGs were normalized, scaled, clustered to generate the heatmap using the ComplexHeatmap package65. The significantly enriched pathways and upstream regulators based on the DEGs were identified using Ingenuity pathway analysis (IPA, Qiagen). The dotplot was generated by Tidyverse package66.
Comparative transcriptomic analysis
Comparisons of GD 3.5 Grb2d/d to GD 3.5 Foxa2d/d uterine dysregulated genes were performed by comparing differentially expressed genes determined between differentially expressed genes of Grb2d/d mice with differentially expressed genes determined by previously reported transcriptomics analysis of Foxa2d/d mice publicly available from the GEO database (GSE48339)36. The differentially expressed gene lists of Foxa2d/d mice used for comparative transcriptomics were prepared by original authors of the studies according to their published methods of analysis36. Duplicate genes present in the gene lists (due to the presence of multiple array probes for some genes) were removed based on GeneBank ID or gene symbol. Briefly, the transcriptome of Grb2d/d at GD 3.5 overlapped with Foxa2d/d transcriptome at GD 3.5.
Statistical analysis
To assess statistical significance of parametric data, the Student’s t-test was used for data with only two groups. For data containing more than two groups, one-way ANOVA was used, followed by Tukey’s post hoc test for multiple comparisons. All data are presented as means ± SEM. p < 0.05 was considered statistically significant. All statistical analyses were performed using the Instat package from GraphPad (San Diego, CA, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-seq data generated in this study have been deposited in the NCBI Gene Expression Omnibus database under accession code GSE260505 and previously reported transcriptomics analysis of Foxa2d/d mice is publicly available under accession code GSE4833936. The all data generated in this study are provided in the Supplementary Information/Source Data file. Source data are provided with this paper.
References
Messaoudi, S. et al. 15 years of transcriptomic analysis on endometrial receptivity: what have we learnt? Fertil. Res. Pr. 5, 9 (2019).
Cha, J., Sun, X. & Dey, S. K. Mechanisms of implantation: strategies for successful pregnancy. Nat. Med. 18, 1754–1767 (2012).
Patel, B. et al. Role of nuclear progesterone receptor isoforms in uterine pathophysiology. Hum. Reprod. Update 21, 155–173 (2015).
Stewart, C. L. et al. Blastocyst implantation depends on maternal expression of leukaemia inhibitory factor. Nature 359, 76–79 (1992).
Kelleher, A. M., DeMayo, F. J. & Spencer, T. E. Uterine glands: developmental biology and functional roles in pregnancy. Endocr. Rev. 40, 1424–1445 (2019).
Kelleher, A. M., Milano-Foster, J., Behura, S. K. & Spencer, T. E. Uterine glands coordinate on-time embryo implantation and impact endometrial decidualization for pregnancy success. Nat. Commun. 9, 2435 (2018).
Hantak, A. M., Bagchi, I. C. & Bagchi, M. K. Role of uterine stromal-epithelial crosstalk in embryo implantation. Int. J. Dev. Biol. 58, 139 (2014).
Spencer T. E. Biological roles of uterine glands in pregnancy. Semin. Reprod. Med. 32, 346–357 (2014).
Yuan, J. et al. Tridimensional visualization reveals direct communication between the embryo and glands critical for implantation. Nat. Commun. 9, 603 (2018).
Burton, G., Jauniaux, E. & Charnock-Jones, D. Human early placental development: potential roles of the endometrial glands. Placenta 28, S64–S69 (2007).
Jeong, J.-W. et al. Foxa2 is essential for mouse endometrial gland development and fertility. Biol. Reprod. 83, 396–403 (2010).
Besnard, V., Wert, S. E., Hull, W. M. & Whitsett, J. A. Immunohistochemical localization of Foxa1 and Foxa2 in mouse embryos and adult tissues. Gene Expr. Patterns 5, 193–208 (2004).
Dhakal, P., Fitzgerald, H. C., Kelleher, A. M., Liu, H. & Spencer, T. E. Uterine glands impact embryo survival and stromal cell decidualization in mice. FASEB J. 35, e21938 (2021).
Wee, P. & Wang, Z. Epidermal growth factor receptor cell proliferation signaling pathways. Cancers 9, 52 (2017).
Lim, S.-J., Lopez-Berestein, G., Hung, M.-C., Lupu, R. & Tari, A. M. Grb2 downregulation leads to Akt inactivation in heregulin-stimulated and ErbB2-overexpressing breast cancer cells. Oncogene 19, 6271–6276 (2000).
Ijaz, M. et al. The role of Grb2 in cancer and peptides as Grb2 antagonists. Protein Pept. Lett. 24, 1084–1095 (2017).
Sattler, M. et al. Critical role for Gab2 in transformation by BCR/ABL. Cancer Cell 1, 479–492 (2002).
Modi, H. et al. Inhibition of Grb2 expression demonstrates an important role in BCR–ABL-mediated MAPK activation and transformation of primary human hematopoietic cells. Leukemia 25, 305–312 (2011).
Large, M. J. et al. The epidermal growth factor receptor critically regulates endometrial function during early pregnancy. PLoS Genet. 10, e1004451 (2014).
Lee, C. H. et al. Extracellular signal-regulated kinase 1/2 signaling pathway is required for endometrial decidualization in mice and human. PLoS ONE 8, e75282 (2013).
Krehbiel, R. H. Cytological studies of the decidual reaction in the rat during early pregnancy and in the production of deciduomata. Physiological Zool. 10, 212–234 (1937).
Soyal, S. M. et al. Cre‐mediated recombination in cell lineages that express the progesterone receptor. Genesis 41, 58–66 (2005).
Sun, X. et al. Kruppel-like factor 5 (KLF5) is critical for conferring uterine receptivity to implantation. Proc. Natl. Acad. Sci. USA 109, 1145–1150 (2012).
Vasquez, Y. M. et al. FOXO1 regulates uterine epithelial integrity and progesterone receptor expression critical for embryo implantation. PLoS Genet. 14, e1007787 (2018).
Takano, M. et al. Transcriptional cross talk between the forkhead transcription factor forkhead box O1A and the progesterone receptor coordinates cell cycle regulation and differentiation in human endometrial stromal cells. Mol. Endocrinol. 21, 2334–2349 (2007).
Wetendorf, M. & DeMayo, F. J. The progesterone receptor regulates implantation, decidualization, and glandular development via a complex paracrine signaling network. Mol. Cell. Endocrinol. 357, 108–118 (2012).
Klemmt, P. A., Carver, J. G., Kennedy, S. H., Koninckx, P. R. & Mardon, H. J. Stromal cells from endometriotic lesions and endometrium from women with endometriosis have reduced decidualization capacity. Fertil. Steril. 85, 564–572 (2006).
Kim, J. J., Jaffe, R. C. & Fazleabas, A. T. Comparative studies on the in vitro decidualization process in the baboon (Papio anubis) and human. Biol. Reprod. 59, 160–168 (1998).
Liang, X. et al. The uterine epithelial loss of Pten is inefficient to induce endometrial cancer with intact stromal Pten. PLoS Genet. 14, e1007630 (2018).
Tan, J., Paria, B. C., Dey, S. K. & Das, S. K. Differential uterine expression of estrogen and progesterone receptors correlates with uterine preparation for implantation and decidualization in the mouse. Endocrinology 140, 5310–5321 (1999).
Brenner, R. M., West, N. B. & McClellan, M. C. Estrogen and progestin receptors in the reproductive tract of male and female primates. Biol. Reprod. 42, 11–19 (1990).
Lessey, B. A. et al. Immunohistochemical analysis of human uterine estrogen and progesterone receptors throughout the menstrual cycle. J. Clin. Endocrinol. Metab. 67, 334–340 (1988).
Yoshinaga, K. Uterine receptivity for blastocyst implantation. Ann. N. Y. Acad. Sci. 541, 424–431 (1988).
Young, S. L. & Lessey, B. A. Progesterone function in human endometrium: clinical perspectives. Semin. Reprod. Med. 28, 5–16 (2010).
Kelleher, A. M. et al. Forkhead box a2 (FOXA2) is essential for uterine function and fertility. Proc. Natl. Acad. Sci. USA 114, E1018–E1026 (2017).
Filant, J., Lydon, J. P. & Spencer, T. E. Integrated chromatin immunoprecipitation sequencing and microarray analysis identifies FOXA2 target genes in the glands of the mouse uterus. FASEB J. 28, 230–243 (2014).
Das, S., Tsukamura, H., Paria, B., Andrews, G. & Dey, S. Differential expression of epidermal growth factor receptor (EGF-R) gene and regulation of EGF-R bioactivity by progesterone and estrogen in the adult mouse uterus. Endocrinology 134, 971–981 (1994).
Tong, B. J., Das, S., Threadgill, D., Magnuson, T. & Dey, S. Differential expression of the full-length and truncated forms of the epidermal growth factor receptor in the preimplantation mouse uterus and blastocyst. Endocrinology 137, 1492–1496 (1996).
Bui, A. H., Timmons, D. B. & Young, S. L. Evaluation of endometrial receptivity and implantation failure. Curr. Opin. Obstet. Gynecol. 34, 107–113 (2022).
Lessey, B. A. & Young, S. L. What exactly is endometrial receptivity? Fertil. Steril. 111, 611–617 (2019).
Kim, T. H. et al. ARID1A is essential for endometrial function during early pregnancy. PLoS Genet. 11, e1005537 (2015).
Kim, T. H. et al. Role of SIRT1 and progesterone resistance in normal and abnormal endometrium. J. Clin. Endocrinol. Metab. 107, 788–800 (2022).
Fox, C., Morin, S., Jeong, J.-W., Scott, R. T. Jr & Lessey, B. A. Local and systemic factors and implantation: what is the evidence? Fertil. Steril. 105, 873–884 (2016).
Lessey, B. A. & Kim, J. J. Endometrial receptivity in the eutopic endometrium of women with endometriosis: it is affected, and let me show you why. Fertil. Steril. 108, 19–27 (2017).
Daikoku, T. et al. Conditional deletion of Msx homeobox genes in the uterus inhibits blastocyst implantation by altering uterine receptivity. Dev. Cell 21, 1014–1025 (2011).
Das, S. K. Regional development of uterine decidualization: molecular signaling by Hoxa‐10. Mol. Reprod. Dev. Incorp. Gamete Res. 77, 387–396 (2010).
Dey, S. et al. Molecular cues to implantation. Endocr. Rev. 25, 341–373 (2004).
Li, Q. et al. The antiproliferative action of progesterone in uterine epithelium is mediated by Hand2. Science 331, 912–916 (2011).
Lessey B. A. & Young S. L. Homeostasis imbalance in the endometrium of women with implantation defects: the role of estrogen and progesterone. Semin. Reprod. Med. 32, 365–375 (2014).
Lessey, B. A., Palomino, W. A., Apparao, K. B., Young, S. L. & Lininger, R. A. Estrogen receptor-alpha (ER-alpha) and defects in uterine receptivity in women. Reprod. Biol. Endocrinol. 4(Suppl 1), S9 (2006).
Park, J. S. et al. Endometrium from women with endometriosis shows increased proliferation activity. Fertil. Steril. 92, 1246–1249 (2009).
Villacorte, M. et al. β-Catenin signaling regulates Foxa2 expression during endometrial hyperplasia formation. Oncogene 32, 3477–3482 (2013).
Filant, J. & Spencer, T. E. Endometrial glands are essential for blastocyst implantation and decidualization in the mouse uterus. Biol. Reprod. 88, 93 (2013).
Marquardt, R. M. et al. Endometrial epithelial ARID1A is critical for uterine gland function in early pregnancy establishment. FASEB J. 35, e21209 (2021).
Noyes, R. W., Hertig, A. T. & Rock, J. Dating the endometrial biopsy. Am. J. Obstet. Gynecol. 122, 262–263 (1975).
Daikoku, T. et al. Lactoferrin-iCre: a new mouse line to study uterine epithelial gene function. Endocrinology 155, 2718–2724 (2014).
Yoo, J.-Y. et al. Loss of MIG-6 results in endometrial progesterone resistance via ERBB2. Nat. Commun. 13, 1101 (2022).
Kim, B. G. et al. Aberrant activation of signal transducer and activator of transcription-3 (STAT3) signaling in endometriosis. Hum. Reprod. 30, 1069–1078 (2015).
Ishibashi, H. et al. Sex steroid hormone receptors in human thymoma. J. Clin. Endocrinol. Metab. 88, 2309–2317 (2003).
Finn, C. & Martin, L. Endocrine control of the timing of endometrial sensitivity to a decidual stimulus. Biol. Reprod. 7, 82–86 (1972).
Yoo, J.-Y. et al. G-protein coupled receptor 64 is required for decidualization of endometrial stromal cells. Sci. Rep. 7, 5021 (2017).
Kim, T. H. et al. Loss of HDAC3 results in nonreceptive endometrium and female infertility. Sci. Transl. Med. 11, eaaf7533 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Gu, Z. G., Eils, R. & Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32, 2847–2849 (2016).
Wickham, H. et al. Welcome to the tidyverse. J. Open Source Softw. 4, 1686 (2019).
Acknowledgements
This work was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Numbers R01HD102170 and R01HD101243 (to J.W.J.) and the National Reserch Foundation of Korea founded by Korea Government under Award Number RS-2023-NR077285 (to Y.P.C). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Y.P.C. and J.W.J. were responsible for the concept of the study; S.L.Y. and B.A.L. collected human samples; D.N.T., Y.J.H., K.C.K., and T.H.K. carried out experiments; R.M.M., R.L., T.H.K. and J.W.J. analyzed data; C.C. provided transgenic mice; D.N.T., R.M.M., T.H.K., Y.P.C., and J.W.J. contributed to writing the manuscript. All authors contributed to the final manuscript version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tran, D.N., Hwang, Y.J., Kim, K.C. et al. GRB2 regulation of essential signaling pathways in the endometrium is critical for implantation and decidualization. Nat Commun 16, 2192 (2025). https://doi.org/10.1038/s41467-025-57173-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57173-2