Abstract
Early-onset lung cancer in young adults represents a less studied clinical entity with increasing incidence which still affects a large number of cancer patients. Here we perform a comprehensive analysis of early-onset lung cancer for the clinicopathological features, genomic alterations, gene expression, and immune landscape by establishing a cohort enrolling 421 non-small cell lung cancer (NSCLC) patients from ten medical centers in China. Comparative analysis reveals a distinct genomic alteration between younger and elder patients with NSCLC, with ERBB2 mutations and ALK-rearrangement strikingly more frequent in younger group. Transcriptome profiling indicates altered cellular metabolism and immune-related genes in tumors from younger patients. Immunological analysis reveals a decreased infiltration of immune cells (notably T cells) in tumors from younger patients. Cellular and mechanistic studies show that the prevalent ERBB2 mutants in cancer from younger patients can indeed drive tumorigenesis by elevating AKT signaling. Importantly, meta-analysis of clinical trials and our clinical practice further validate the promise of HER2-targeted therapy to treat early-onset NSCLC in East Asian patients. Our comprehensive and integrative analysis not only reveal multiple unrecognized characteristics of early-onset lung cancer, but also inform actionable therapeutics to manage this type of cancer.
Similar content being viewed by others
Introduction
Lung cancer is the most commonly diagnosed human cancer with a poor prognosis and the leading cause of cancer-related mortality1. Non-small cell lung cancer (NSCLC) overwhelmingly remains a disease of the elderly population2. However, the incidence of early-age onset lung cancer (usually <40-50 years) is considerably increasing3,4,5, possibly due to changes in environment and lifestyle factors as well as the popularity of CT scans in regular check-up program6. Compared to late-onset counterparts, lung cancers in younger patients were quite less studied. The survival and prognosis for this group of patients are still controversial. Some studies report that the survival is similar between younger and elders, whereas others show that younger patients have worse prognosis7,8. Epidemiological investigation indicates that non-smokers, females, and adenocarcinoma are predominant in early-onset lung cancers9,10. In a few studies with limited genetic profiling data, a varied mutational spectrum across different cohorts for NSCLC at young age was reported. For instance, ALK and ROS1-rearrangement lung cancers are significantly associated with young age in multiple studies across different ethnic groups11,12,13,14,15,16,17. In contrast, EGFR mutations were not consistently associated with cancer from younger patients17,18. A more comprehensive scanning using bigger cohorts and defined ethnic population is needed to reveal these discrepancies in terms of genomic alterations in younger patients with NSCLC. Furthermore, other molecular features such as the transcriptome, signaling pathways, and immune features have not been systematically described for NSCLC in younger patients before. Little is known about the underlying biology of early-onset NSCLC. Accordingly, there remains some uncertainty and lack of consensus on the management strategy for this type of cancer in the clinic.
In this study, we performed a comprehensive multicenter cohort study to systematically analyze the mutational, transcriptional, and immunological profiles of early-onset lung adenocarcinoma in younger patients and assessed the impact of these characteristics on the prognosis, tumor progression, and treatment strategies for this type of cancer. Our integrative analysis presents a comprehensive view on the molecular features of early-age onset lung cancer in younger East Asian patients and provides important insights on therapeutical interventions to manage these tumors clinically.
Results
Clinicopathologic characteristics of the patients in the cohort
In this study, a total of 421 patients were enrolled (Younger Group, YG: n = 215; Elder Group, EG: n = 206) in the cohort from ten medical centers in China, who were diagnosed with primary lung adenocarcinoma (pLUAD) between 2016 and 2023. To make a better comparison between YG and EG, we employed a stringent age cutoff at diagnosis in defining YG (age between 20 and 40) and EG (age between 60-80). Their clinical characteristics are presented in Table 1. Similar to elder ones, younger patients were more likely to be female (145/215, 67%), less likely to have a family history (9/215, 4.2%). However, compared to elder group, young patients had a lower smoking rate (P = 0.01), were more likely to be asymptomatic at diagnosis (P < 0.0001), and usually presented with earlier tumor stages (P < 0.0001) (Tumor size, P < 0.0001; Lymphatic metastasis, P < 0.001; Distant metastasis, P = 0.02, respectively). To explore whether the difference in the probability of symptoms between YG and EG was influenced by staging, we conducted separate chi-square tests for early-stage (Stage I and II) and late-stage (Stage III and IV) (Supplementary Table 1). The results indicate that younger patients were less likely to experience symptoms in the early stages (P = 0.002). Furthermore, logistic regression analysis revealed that the probability of symptom occurrence increased with the stage progression in both groups, but the range of increase for younger patients was higher than elder patients (YG: P < 0.001, OR = 10.68, 95% CI: 3.34 to 34.12; EG: P < 0.001, OR = 5.8, 95% CI: 2.99 to 11.25) (Supplementary Fig. 1). These results demonstrate that younger patients were less likely to experience symptoms only in the early stages, while were comparable to elder patients in the advanced stages. Following the International Association for the Study of Lung Cancer (IASLC) grading system, we categorized the patients with pathological results. However, some late-stage patients who were ineligible for surgery lacked available histopathological morphology assessments, and their results were categorized as “undefinable”. YG had a significantly less percentage of “undefinable” late-stage patients than EG (P < 0.0001). Nevertheless, upon excluding the undefinable subgroup, no significant differences in pathological staging were observed between YG and EG (P = 0.55, Supplementary Table 2).
Distinct gene mutational profiles between NSCLC of younger and elder patients
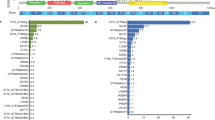
To investigate genomic alterations, we conducted targeted DNA sequencing analysis on both YG and EG by detecting 808 cancer-related genes (see “Methods”). The top 10 mutated genes and their frequency varied significantly between these two groups (Fig. 1a). Both EGFR and TP53 were highly mutated in both YG and EG. In contrast, ERBB2 (also known as HER2) and ALK stood in the 3rd and 4th place in YG but not among the top 10 hits in EG. Additionally, we observed higher mutation frequencies in genes related to cancer susceptibility (NQO1, MTHFR, and KMT2B), genomic stability maintenance (BRCA1 and BRCA2), and base excision repair (XRCC1) in YG (Fig. 1a).
a The mutation frequency of the top 10 mutated genes in younger group (YG, n = 215) and elder group (EG, n = 206). b The detailed mutational profile and clinical characteristics for each patient in both cohorts. The upper part displays the mutation frequency and types of 11 driver genes recommended by the NCCN guidelines (Point, point mutation; Del, deletion; Ins, insertion; CNV, copy number variation). The lower part displays the corresponding clinical information for each patient. c Comparative analysis of mutation frequencies in 11 driver genes across two cohorts (EG, n = 206; YG, n = 215). The P values were calculated by two-sided chi-square test or two-sided Fisher’s exact test as appropriate. d The heatmap illustrates the co-mutation and exclusion of 11 driver genes in YG (left, n = 215). The P values were obtained from two-sided Fisher’s exact test, and the color intensity reflects the magnitude of the P value with * indicating the statistical significance of the P value. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Based on the odds ratio (OR) calculated by two-sided Fisher’s exact test, the relationship between two genes can be determined: OR < 1 indicates an exclusionary relationship between two genes, while an OR > 1 indicates a co-mutation relationship (right). The forest plot displays the OR values (dots) and 95% confidence intervals (whiskers). e The mutation subtypes and locations of ERBB2 mutations in YG. f Differences in ERBB2 mutation types between the EG and YG groups, and the ERBB2 exon 20 mutation subtypes in YG (n = mutational events). g Differences in ALK mutation types between the EG and YG groups, and the types of fusions and the subtypes of EML4::ALK fusion in YG (n = mutational events). Source data are provided as a Source Data file.
According to the National Comprehensive Cancer Network (NCCN) guidelines, we further narrowed our focus on 11 key driver genes in lung cancer (Fig. 1b, c). We found significant differences in the mutation frequency in EGFR, ERBB2, ALK, and KRAS between YG and EG. In YG, the mutation frequencies of EGFR and KRAS were significantly lower than those in EG (P < 0.01 for both), while the frequencies of ALK and ERBB2 alterations were notably higher (ALK: P < 0.01; ERBB2: P < 0.0001) (Fig. 1c). Moreover, insertion mutations in ERBB2 and fusions in ALK were notably more prevalent in YG compared to EG, while other driver genes did not exhibit difference between the two groups (Fig. 1b, c). We further examined whether these genetic alterations tend to occur together or mutually exclusive. The heatmap indicates that there were six pairs of genes showing either co-mutation or mutually exclusive mutation relationships in YG (Fig. 1d). For example, EGFR was mutually exclusive with ERBB2, ALK, and KRAS, whereas ALK with NTRK1, KRAS with NTRK3, and ROS1 with RET exhibited co-mutation relationships (Fig. 1d). To eliminate the impact of clinical risk factors on the results, we performed propensity score matching (PSM) on two cohorts (Supplementary Table 3), and similar conclusions were obtained (Supplementary Fig. 2).
Considering the dramatic enrichment of mutational ERBB2 and ALK, we conducted a detailed analysis of the mutation patterns for ERBB2 and ALK between the two groups. Statistical analysis revealed that, among the 44 younger patients with ERBB2 mutations, there were eight different mutation patterns (Supplementary Fig. 3a). The mutations predominantly occurred as exon 20 insertions (38/44, 86%), with the most common mutation type being Y772_A775dup (30/38, 79%), followed by G776delinsVC (7/38, 18%) (Fig. 1e, f and Supplementary Fig. 3a). In contrast, for elder patients, there were 13 ERBB2 gene mutations among 12 patients, with copy number variations (CNVs) being the prevalent type (5/13, 39%). The exon 20 mutations were only identified in two patients (2/13, 15%) (Fig. 1f and Supplementary Fig. 3a). For ALK alterations, there were also highly significant differences between the two groups (Fig. 1g and Supplementary Fig. 3b). In elder patients, point mutations were predominant (15/18, 83%) and only two patients having fusions (2/18, 11%). In contrast, ALK-altered younger patients predominantly had fusions (34/56, 61%), and the majority of them were EML4::ALK fusions (26/34, 77%) (Fig. 1g and Supplementary Fig. 3b). In addition, among the EML4::ALK fusions, the predominant mutation types were V3 (E6::A20, 46%) and V1 (E13::A20, 39%) (Fig. 1g).
Comparative analysis with public database
To further validate our findings and explore the potential differences among various races, ethnicities, and ages, we extracted public data from cBioPortal database which includes 11,652 lung cancer patients19. After filtering, a total of 3322 patients were identified as pLUAD with 117 of Asian ethnicity (Fig. 2a, b). Among these 3322 patients, 1748 had age data available, including 30 patients between 20 and 40 years old (30/1748, 1.7%) and 1137 patients between 60 and 80 years old (1137/1748, 65%) (Fig. 2b). Except for those with missing race and ethnicity information, the majority of the patients were of White ethnicity (Fig. 2c). The lack of data from Asian ethnicity from major public cancer consortium further demonstrates the value of our dataset in this study. Using this public dataset, we compiled the top 10 mutation genes for all patients or for Asian patients (Fig. 2d). Apart from the tumor suppressor gene TP53, the driver gene KRAS occupied a dominant position (993/3322, 30%) for all patients, followed by EGFR (774/3322, 22%). In contrast, EGFR was the most frequently mutated gene in Asian patients, far higher than that in White-dominant public cohorts (68/117, 58%). The mutation frequency of KRAS ranked in the 3rd place among Asian group (16/117, 14%) and was lower than that in all patients. In addition, ERBB2 and NTRK3 also ranked in the top ten mutated genes among Asian patients (Fig. 2d). These differences highlight the importance of ethnicity and genetic backgrounds in cancer studies, and may impact the clinical management of lung cancer (e.g., the choices of targeted therapy) on a large nation-wide or region-wide scale.
a–c Overview of data and case selection process from database (a), and age distribution of patients with age data (b). Proportion of race and ethnicity among with primary lung adenocarcinoma (pLUAD) patients and those with age data in the database, Other: American Indian or Alaska Native, Native Hawaiian or Other Pacific Islander; NA: race and ethnicity details unavailable (c). d Top 10 mutated genes in all patients (n = 3322) or Asian patients (n = 1748) in the public database. e Comparison of EGFR, ERBB2, ALK, and KRAS mutations between EG (n = 1137) and YG (n = 30) in public database. f Comparison of mutations in four genes between EG (n = 1137) and YG (n = 30) in public database and our cohort (EG, n = 206; YG, n = 215). g Comparison of mutations in four genes between Asian patients in public database (n = 117) and elder patients in our data (n = 206). For (e–g) P values were calculated by two-sided chi-square test or two-sided Fisher’s exact test as appropriate. Source data are provided as a Source Data file.
Next, we compared the mutation frequencies of EGFR, ERBB2, ALK, and KRAS among elder patients, younger patients, and Asian patients in the public database with our data. We firstly conducted an internal age group comparison within the public database. A significantly higher mutation frequency of EGFR was observed in YG than in EG (30% vs. 14%, P < 0.05), while the KRAS mutation frequency was lower in YG compared to EG (13% vs. 28%) (Fig. 2e). These data suggest that EGFR, but not KRAS, ERBB2 nor ALK, may be primary driver gene for early-onset lung cancer in White ethnicity. Then, we incorporated our data into comparison. No matter in YG or EG, the mutation frequencies of EGFR, ERBB2 and ALK were all consistently higher in our Asian data than those in White-dominant public data, while the KRAS mutation frequency was lower (Fig. 2f). By directly comparing our data with 117 Asian patients from the public dataset, we found a consistency of the mutation frequencies for most of the interrogated driver genes, further validating the accuracy of our data (Fig. 2g). These results consolidate the above findings using an independent dataset, and underscore the differences between ethnic populations.
Transcriptomic and immunological analysis of NSCLC of younger and elder patients
To compare the gene expression patterns between NSCLC from younger and elder patients, we performed bulk RNA-seq analysis for 14 tumors (YG: n = 7; EG: n = 7) and made sure that these were specimens with well-matched factors other than age (Supplementary Table 4). A total of 109 differentially expressed genes (DEGs) were identified between YG and EG, with 69 genes down-regulated and 40 genes up-regulated (Fig. 3a and Supplementary Data 1). Some of the cancer-related genes (e.g., ARNT2), immune-related genes (e.g., CXCL9, CXCL10, and CXCR5) and metabolism-related genes (e.g., PLA2G2D and APOA1) were highlighted (Fig. 3a). Functional gene enrichment analysis of these DEGs indicated a significant enrichment for terms related to immune or metabolism functions (Fig. 3b). These data suggest that younger tumors may possess different immune and metabolism properties compared to elder cancers.
a The volcano plot displays differentially expressed genes (DEGs) between NSCLC tumors of younger and elder patients by RNA-seq analysis. b The top 10 enriched KEGG pathways and GO (BP) terms of DEGs. Two-sided hypergeometric test or two-sided Fisher’s exact test for P value. Benjamini and Hochberg's method was used for false discovery rate (FDR) correction. c The heatmap illustrates the results of immune infiltration estimation using TIMER2.0 on the expression profiles of 14 tumor samples. The estimation adopts the XCELL algorithm, displaying the infiltration status of important immune cell subtypes in the TME. Color-coded scores represent the infiltration level. d The Tukey’s box plot displays the median (thick line), interquartile range (box limits), 1.5× the interquartile range span (whiskers), and outliers (dots) of positive cells by each IHC staining in the tumor or interface in samples from EG (red, n = 110) and YG (blue, n = 108). T: tumor zone; I: interface zone. Unpaired two-sided t-test for P value. PCD4-T = 0.0054; PCD4-I = 0.0032; PCD8-T = 0.0239; PCD8-I < 0.0001; PCD3-T < 0.0001; PCD3-I = 0.0003. e Heatmap showing immune cell infiltration in the tumor or interface zones of each case in EG and YG samples. Red indicates higher density of corresponding marker staining than the mean level, while blue denotes lower density. f Representative cases showing the IHC staining patterns of biomarkers (CD4, CD8, and CD3) with statistically significant differences in positive cell counts in the tumor or interface zones. The cases were derived from patients who underwent IHC (n = 218). Scale bar, 200 μm. g Representative cases showing the multiplex immunofluorescence staining patterns of biomarkers (CD3, CD4, CD8, FOXP3, and PANCK) with statistically significant differences of intensity and density in composite and single-stained images. The cases were derived from patients who underwent multiplex immunofluorescence (n = 20). Scale bar, 200 μm. h The comparison of CD3+(EG: n = 10; YG: n = 8), CD4+(EG: n = 10; YG: n = 9), CD8+(EG: n = 8; YG: n = 10), FOXP3+(EG: n = 10; YG: n = 10) and PANCK+(EG: n = 10; YG: n = 9) intensity (H-Scores) between EG and YG patients. i The comparison of CD3+(EG: n = 9; YG: n = 10), CD4+(EG: n = 10; YG: n = 9), CD8+(EG: n = 8; YG: n = 10), FOXP3+(EG: n = 10; YG: n = 10) and PANCK+(EG: n = 10; YG: n = 9) density (Cells/mm²) between EG and YG patients by cell counts. j Cell density of CD4+CD3+ T cells (EG: n = 10; YG: n = 9), CD8+CD3+ T cells (EG: n = 8; YG: n = 7), and CD4+CD3+FOXP3− T cells (EG: n = 10; YG: n = 9) between EG and YG patients. For (h–j) the dot-line plots indicate Mean ± SD. The outliers were excluded by applying a normality test. Unpaired two-sided t-test for P value. k The comparison of CD4+, CD8+, and CD3+ T cell infiltration levels between samples with ERBB2 (n = 14) and ALK (n = 11) mutations and the overall levels of EG (n = 110) and YG (n = 108). Tukey’s box plot displays the median (thick line), interquartile range (box limits), 1.5× the interquartile range span (whiskers), and outliers (dots). Unpaired two-sided t-test for P value. Source data are provided as a Source Data file.
To further explore the immune differences between the two groups, we utilized TIMER2.0 immune estimation function20 and our RNA-seq data to assess the infiltration levels of representative immune cells in tumor microenvironment (TME). The results showed that the immune infiltration in elder patients was generally higher than that in younger patients, with the differences mostly notable in the T cell and B cell subtypes (Fig. 3c). To validate this, we assessed the infiltration levels of lymphocytes by immunohistochemistry (IHC) analysis on 218 stage I patient samples (EG: n = 110; YG: n = 108) (see Methods and Supplementary Table 5). We found that the cell counts of positively stained helper T cells (CD4+) and cytotoxic T cells (CD8+) in both the tumor zone (T) and the tumor/non-tumor interface zone (I) were significantly lower in YG compared to EG (Fig. 3d and Supplementary Table 6). Furthermore, the count of mature T cells (CD3+) also exhibited a highly significant difference in both zones with YG lower than EG (Tumor: P < 0.0001; Interface: P < 0.001). On the other hand, there were no significant differences observed in B cells (CD20+), plasma cells (CD138+), and T regulatory cells (Treg, FOXP3+) (Fig. 3d). The differences in CD4, CD8, and CD3 results were further visualized in the heatmap as well as representative staining images (Fig. 3e, f). To eliminate the impact of clinical risk factors on the IHC analysis, we performed PSM adjustment by 1:1 ratio on these 218 patients and obtained a total of 116 matched patients (YG: n = 58 vs. EG: n = 58) (Supplementary Table 7). Again, consistent results were also obtained after PSM adjustment as the count of CD3, CD4, and CD8 T cells was significantly lower in YG compared to EG (Supplementary Fig. 4). Furthermore, we conducted multiplex immunofluorescence analysis on 20 representative PSM-matched samples (EG: n = 10; YG: n = 10) (Fig. 3g). Consistent with original immunohistochemistry analysis, multiplex immunofluorescence results showed that the infiltration of CD4+, CD8+, and CD3+ T cells were significantly lower in YG compared to EG patients (Fig. 3g–j).
Next, we compared the immune cell infiltration levels in patients with ERBB2 (n = 14) and ALK (n = 11) alterations to the overall levels in EG and YG. The results showed that the levels of immune cell infiltration in both ERBB2- and ALK-mutant patients were significantly lower than those in EG (Fig. 3k). Despite no statistical significance being reached, ERBB2- and ALK-mutant tumors also generally had lower levels of immune cell infiltration than the whole YG (Fig. 3k). Particularly, patients with ALK alterations exhibited a quite strong downward trend in CD4, CD8, and CD3 levels. These data suggest that younger tumors tended to have an immunosuppressive TME and low levels of immune cell infiltration, and ERBB2 and ALK mutations may play a driving role in shaping such TME.
Functional and mechanistic characterization of ERBB2 mutations in tumors from younger patients
Considering the high prevalence of ERBB2 mutations in NSCLC of younger patients, we then tried to determine whether the highly mutated ERBB2 underlies the tumorigenesis of early-onset NSCLC. We exogenously expressed the two major ERBB2 mutation subtypes (YVMA, encoded by Y772_A775dup; insVC, encoded by G776delinsVC) as well as the wild type (WT) form in two human NSCLC cell lines NCI-H322 and PC-9. Compared to WT, the two mutants apparently increased the phosphorylation level of ATK but not ERK in NCI-H322 cells. In contrast, only YVMA mutant could do so in PC-9 cells (Fig. 4a). As PC-9 cells already contain oncogenic mutation of EGFR (exon 19, delE746-A750) while NCI-H322 did not have this known cancerous driver, such genetic background difference may cause varied responses to ERBB2 mutant expression. Indeed, we found that the YVMA mutant could promote NCI-H322 cell growth in 2D monolayer cell culture conditions while no such effect in PC-9 cells (Fig. 4b). It is possible that other oncogenic events in PC-9 cells play a pivotal role in driving their 2D cell growth. Interestingly, in a soft agar-based 3D colony formation assay, the promotive effect of ERBB2 mutants was evident in both cell lines compared to GFP control (Fig. 4c). In addition, both YVMA and insVC ERBB2 mutants could significantly accelerate cell migration (Fig. 4d, e). To further examine the mutant function in vivo, we employed a mouse xenograft model by injecting ERBB2-expressing or control (GFP) NCI-H322 cells into nude mice. Compared to WT-ERBB2 or control group, YVMA mutant significantly gave rise to more incidence and bigger size of tumors (Fig. 4f–h). These data demonstrate that the dominant ERBB2 mutants found in early-onset NSCLC can indeed serve as oncogenic drivers for tumorigenesis.
a Immunoblotting analysis of indicated targets in control (GFP) or ectopic ERBB2-expressing NCI-H322 and PC-9 cells. YVMA stands for Y772_A775dup mutant, and insVC represents G776delinsVC mutant. p, phosphorylated; t, total. b Cell growth effect of indicated cell lines in 2D monolayer cell culture condition. Mean ± SD with n = 3 biological replicates. Unpaired two-sided t-test for P value. c 3D colony formation assay for indicated cells. Mean ± SD with n = 3 biological replicates. Unpaired two-sided t-test for P value. d, e Wound healing assay for indicated NCI-H322 cells to examine cell migration ability (d) and histogram shows the statistical analysis of the results (e). Scale bar, 250 μm. Mean ± SD with n = 3 biological replicates. Unpaired two-sided t-test for P value. f Tumor images of NCI-H322 cell-based mice xenografts for each group. g Tumor volume measured at indicated time points after xenograft implantation. n = 12 for each group. (n, number of tumors). h Tumor weight of mice xenografts for each group. (n = 6 for GFP, n = 10 for ERBB2-WT, n = 10 for YVMA) Mean ± SEM. Unpaired two-sided t-test for P value. i The number of DEGs caused by ectopic expression of ERBB2-WT or YVMA mutant in NCI-H322 cells via RNA-seq analysis. j Venn diagrams showing the overlaps of DEGs for the indicated groups, the P values were obtained from two-sided Fisher’s exact test. k Heatmap of DEGs in ERBB2-WT- or YVMA-expressing NCI-H322 cells. I Scatter plot highlighting individual DEGs. m, n Top 10 enriched KEGG categories of ERBB2-WT (m) or YVMA (n) groups. One-sided Fisher’s exact test for P value. Source data are provided as a Source Data file.
We further tried to identify downstream target genes affected by ectopic ERBB2 expression by RNA-seq in NCI-H322 cells (Supplementary Data 2). Compared to GFP control, either WT or YVMA mutant induced a significantly more portion of up-regulated genes than down-regulated genes, and YVMA affected more genes than WT (Fig. 4i). Despite that the overlap of targets was significant between the two ERBB2 isoforms (Fig. 4j), there were still many genes only uniquely regulated by the YVMA mutant (Fig. 4k). These genes may mediate the oncogenic functions of ERBB2 mutations. For instance, FGF21, a gene only up-regulated by YVMA mutant (Fig. 4l), was reported to promote the progression of NSCLC via SIRT1/PI3K/AKT signaling axis21. Functional gene enrichment analysis showed that “ErbB signaling pathway” was the most enriched term in WT sample, consistent with classic ERBB2 function (Fig. 4m). In contrast, for YVMA group, the top term became “PI3K-Akt signaling pathway” and the enrichment significance was higher than that in WT group (Fig. 4n). These results were in line with enhanced AKT phosphorylation by the YVMA mutant (Fig. 4a), indicating that ERBB2 mutations may promote tumorigenesis primarily through AKT signaling.
Clinical responses of lung cancer to anti-HER2 targeted therapy
The high prevalence of ERBB2 mutations in early-onset lung cancer implies that anti-HER2 targeted therapy may serve as a primary treatment option for a significant portion of younger NSCLC patients. However, given the low mutation frequency of ERBB2 in overall lung cancer (2–3% in NSCLC patients)22,23,24, the clinical application of HER2-targeted drugs was rare. To fairly evaluate their potential clinical response in lung cancer, we performed a systematic meta-analysis by collecting data from 18 published clinical trials containing anti-HER2 therapies (pyrotinib and poziotinib) (Fig. 5a). All these clinical trials were conducted in NSCLC. Among these trials, five provided ERBB2 mutation subtypes data, four of them were on Chinese patients and one was on US patients25,26,27,28,29. As we failed to obtain age information, we just compared the objective response rate (ORR) of the two drugs for patients with ERBB2 mutations. Different studies achieved varied ORR ranging from 16 to 54.4% for either drug (Supplementary Table 8). When breaking down the ERBB2 mutation subtypes, we observed superior efficacy of HER2-targeted drugs for G778_P780dup mutation, especially for poziotinib (Fig. 5b, c).
a The workflow of meta-analysis for evaluating the clinical response of HER2-targeted therapy in NSCLC patients. b, c Treatment efficacy analysis of pyrotinib and poziotinib on the three most common ERBB2 mutation subtypes in NSCLC (b) and a detailed data breakdown from five clinical trials (c). d Diagram of the treatment scheme for Patient A. IM imaging, SD stable disease, PD progressive disease, PR, partial response. e Representative CT images of tumor lesion of Patient A to therapy. f Primary tumor size of Patient A during the entire treatment process. g Serum NSCLC associated-tumor markers (CEA carcinoembryonic antigen, NSE neuron-specific enolase, CYFRA cytokeratin fragment) of Patient A during the multiple-line therapy. h Representative CT images of tumor lesions of Patient B. i Diagram of the treatment scheme for Patient B. j Metastatic tumor size of Patient B during the pyrotinib treatment process. IM imaging, PR partial response. Source data are provided as a Source Data file.
Encouraged by the above results, we identified two early-onset NSCLC patients (Patient A: a 52-year-old male patient with stage IV cancer; Patient B: a 33-year-old female patient with stage IIIA cancer) during our clinical practice whose tumors from pathological biopsies harbor ERBB2 Y772_A775dup mutation. Notably, other typical lung cancer driver mutations were not identified. Patient A received front-line platinum-based chemotherapy (docetaxel for 2 months, then pemetrexed for 6 months) and second-line combined immune checkpoint inhibitor (ICI) immunotherapy (camrelizumab for 2 months) (Fig. 5d). Although the tumor was controlled upon early-line treatment, it showed disease progression after 10 months, with growth in primary tumor and increase in metastatic nodules (Fig. 5e). We speculate that this tumor may be driven by the ERBB2 mutation and this patient may be benefited from HER2-targted therapy. Since the patient took pyrotinib, this HER2-targeted therapy resulted in a rapid decrease in tumor size and serum tumor markers (Fig. 5e–g), indicating a partial response. Notably, the patient remained progression-free under the pyrotinib therapy for >12 months during the follow-up. Patient B was diagnosed with lung adenocarcinoma and underwent radical lobectomy for cancer. Postoperative routine chemotherapy (pemetrexed plus platinum) was given (Fig. 5i). After seven years, the patient received biopsy due to multiple new nodules in the left visceral pleura which was confirmed as recurrence (pleural metastasis) (Fig. 5h, i). After receiving oral pyrotinib targeted treatment for 6 months, the patient achieved sustained disease remission (Fig. 5h, j). These results highlight the potential of HER2-targeted therapy to treat ERBB2-mutant NSCLC, especially, for younger patients whose tumors are largely driven by mutant ERBB2.
Discussion
In this study, we presented a comprehensive resource and analysis on a less characterized clinical entity, early-onset lung cancer, in Chinese younger patients. By comparing to late-onset cancers in elder populations, we systematically revealed unique features of younger tumors in terms of their clinical characteristics, genetic mutations, transcriptome, immune microenvironment, oncogenic drivers, and underlying mechanisms. Based on these findings, we further interrogated potential therapeutic interventions to manage this type of cancer in clinic.
In our multicenter cohort, younger NSCLC patients were mostly non-smoking females and were unlikely to have a family history. Notably, compared to their elder counterparts, younger patients were less likely to present symptoms at early tumor stage. This, combined with certain social and psychological factors, may contribute to misdiagnosis30,31. When these patients seek medical attention after experiencing symptoms, they might have been already in the advanced stages of cancer. This may explain why some previous studies reported that younger patients are primarily diagnosed at an advanced stage32,33,34. Nevertheless, it is worth noting that in our study, the majority of younger patients were in Stage I, which contradicts previous findings32,33,34. We surmised that the widespread adoption of low-dose CT screening for high-risk populations or general population during regular check-ups has enabled these patients to identify suspicious lung nodules and subsequently receive post-operative pathological confirmation. Therefore, these findings underscore the importance of popularizing CT screening in regular check-up programs among general populations in addition to applying CT screening for high-risk populations.
Genetic profiling revealed a significant prevalence of ERBB2 and ALK alterations in younger NSCLC of our cohort, which was further confirmed by independent interrogation of public datasets. In addition, the mutational pattern of these genes was also different between younger and elder tumors, suggesting a divergent etiology for early- and late-onset lung cancer. ERBB2, also known as HER2 (Human Epidermal Growth Factor Receptor 2), belongs to the epidermal growth factor receptor family, which is structurally and functionally similar to EGFR35. ERBB2 amplification/overexpression can be found in 15–20% invasive breast cancer and has important prognostic and therapeutic values36,37. However, in NSCLC, only less than 3% of patients bearing ERBB2 mutations and about 2.5% of patients exhibit HER2 overexpression38,39,40. Therefore, ERBB2 is not regarded as an impactful gene in lung cancer. Here we speculate that ERBB2 mutations may substitute EGFR as an oncogenic driver in a significant portion of early-onset NSCLC in East Asian populations, because that: (1) previous and our data here proved an oncogenic role for those predominant ERBB2 mutations in lung cancer41,42; (2) we further showed oncogenic ERBB2 mutation primarily functions through activation of AKT pathway, rather than ERK signaling shown in previous studies42,43; (3) EGFR and ERBB2 exon 20 mutations were mutually exclusive in YG, and (4) the combined mutation frequencies of EGFR and ERBB2 in YG were approximately equal to the mutation frequency of EGFR in EG. In this regard, ERBB2 might be a primary and impactful gene for the onset and progression of younger NSCLC affecting many patients.
Another important feature of early-onset NSCLC is the severe deficiency in the cell-mediated immunity of younger patients revealed by transcriptomic and immunological analysis. A significant reduction of CD4+, CD8+, and CD3+ T lymphocytes at both the tumor and tumor/non-tumor interface zones in younger patients indicated an immunosuppressive microenvironment. Current immunotherapy for NSCLC focuses on immune checkpoint inhibitors (ICIs), which act by releasing the functional suppression on immune cells, especially T cells, to enhance the anti-tumor activity of the immune system44,45. However, based on our findings, the effectiveness of immunotherapy in younger patients might be limited. From the therapeutic perspective, in addition to EGFR, targeting oncogenic ERBB2 or ALK might be attractive strategy to clinically manage these early-onset NSCLCs. We corroborated the possibility of targeting HER2 in younger NSCLC patients using both meta-analysis of multiple clinical trial data and our own patient data. Notably, in addition to pyrotinib and poziotinib, drugs that can target ERBB2 exon 20 insertion mutations have made new progress, including Sunvozertinib (DZD9008)46, Zongertinib (BI 1810631)47,48 and BAY 292708849 as well as others50. Future work is needed to fully evaluate these HER2-targeted therapies in the treatment of ERBB2 mutant-driven NSCLC.
There are several limitations to our current study. First, our work mainly focused on the patients of East Asian ethnicity and some of the conclusions may not apply to other ethnic groups. Second, we did not perform whole genome sequencing for this cohort to establish the complete landscape of genomic features. Third, the immune analysis was limited only to several major immune cell types. Single-cell RNA-seq profiling of these tumors may be helpful in resolving this issue in the future. Finally, we did not discriminate germline mutations as they were unlikely to affect the major conclusions. In summary, our work comprehensively characterizes multiple features of early-onset lung cancer in East Asian younger patients and provides deep insights on the tumorigenesis and clinical management of this type of cancer.
Methods
Ethical approval
This study has received approval from the Institutional Review Board of The Fourth Affiliated Hospital of China Medical University, Shenyang, China (EC-2022-KS-027 and EC-2023-HS-054) and has obtained written informed consent from patients and/or their families. All studies using mice were performed in accordance with our animal protocols (#NEU-EC-2024A030S) from Care and Use of Laboratory Animals and approved by the Biological and Medical Ethics Committee of Northeastern University (China).
Patient cohort
In this study, the patients were recruited from ten medical centers in China between January 2016 to December 2023 (Supplementary Fig. 5). We defined younger patients as individuals aged between 20 and 40 years old. Those in this age group have already passed adolescence with their bodily functions generally stabilized, and they have not yet experienced the effects of metabolic decline, immune system deterioration, or chronic diseases51. Elder patients were defined as individuals aged between 60 and 80 years old. According to statistical data from the Surveillance, Epidemiology, and End Results (SEER, 2017–2021), the median age at diagnosis for lung and bronchus cancer is 71 years, and patients between the ages of 60 and 80 make up over 70% of the total patient population2. Therefore, patients in this age range are representative of the typical lung cancer patient population. In addition to the age restriction, enrolled patients were pathologically and radiologically confirmed with primary lung adenocarcinoma (pLUAD) and not having received neoadjuvant treatment before surgery. Moreover, patients with other serious illnesses that could potentially affect the study results were excluded. Collected clinical information included patient age, sex, smoking history (yes or no), family history (any cancer), symptoms at diagnosis, tumor location, size, staging, and main histological subtypes (Supplementary Data 3). Information on the sex and/or gender of participants was collected by investigators based on patient’s self-report. The race and ethnicity of all patients in this study were identified through patient’s self-report. For analysis using the public cBioPortal database, the race and ethnicity information was directly extracted from patient’s records deposited in the database. The baseline data and Formalin-Fixed Paraffin-Embedded (FFPE) tissue samples of all patients in the study were collected from ten medical centers in China. All fresh tissue samples for transcriptome sequencing were obtained from the Fourth Affiliated Hospital of China Medical University. The study was approved by a multicenter research ethics committee and each hospital’s local research ethics, and has obtained explicit consent from patients and/or their families. Each patient had signed consent to publish clinical information potentially identifying individuals.
Genetic profiling
The DNA panel of this study comprised two versions: one contains 168 tumor-related hotspot genes, and the other includes 808 genes (605 genes with full exon coding regions and 203 genes with hotspot mutation regions). The 168 panels were used for 159 younger patients and 78 elder patients, while the 808 panels were used for 56 younger patients and 128 elder patients. The gene panels used for all tests are listed in Supplementary Data 4. The samples used for genetic testing were either FFPE tissue (10 sections of 10 µm paraffin slides) or tissue obtained through biopsy. For FFPE tissue, DNA was extracted using Ion AmpliSeq Direct FFPE DNA kit. The level of contamination and degradation was assessed using a QIAxcel fragment analyzer, and DNA concentration was measured using a Qubit DNA Assay Kit and Qubit 4.0 Fluorometer. To prepare DNA libraries, a minimum of 0.5 μg of genomic DNA with high molecular weight (greater than 20 Kb single bands) was required for each sample.
The Truseq Nano DNA HT Sample Prep Kit was used to generate sequencing libraries and unique index codes were added to each sample. Genomic DNA samples were fragmented to approximately 350 bp using an ultrasonic disruptor, followed by end repair and PCR amplification. After purification using the AMPure XP system, library size distribution was analyzed using a bioanalyzer, and then real-time PCR quantification (3 nmol/L) was performed. Subsequently, the indexed samples were clustered using the HiSeq X PE Cluster Kit V2.5 system and sequenced on the Illumina Novaseq 6000 platform which generates 150 bp paired-end reads. The raw sequencing data, including gene sequences and sequencing quality information, were recorded in FASTQ format files. After removing contaminated, low-quality, and unidentifiable reads, the data were then aligned to the human genome reference to obtain results in BAM format. The MuTect and Strelka tools were employed to detect somatic single nucleotide variants (SNVs) and insertions-deletions (InDels). Copy number variations (CNVs) were identified using the Control-FREEC system, and the GISTIC algorithm was applied to infer regions of recurrent amplification or deletion in the genome. Significant CNV regions were defined based on G-scores exceeding 0.1. As the samples were collected from different hospitals across a long time duration, the raw sequencing data for some of the samples were not available. In such cases, the processed data were used for downstream analysis. The sequencing results and parameters for each patient are presented in Supplementary Data 5.
Gene expression profiling by RNA-seq
For tumor samples, we conducted gene expression analysis using fresh tumor tissue samples. A total of 14 samples were available for the study (YG: n = 7 vs EG: n = 7). These samples were sent to BerryGenomics (Beijing, China) Co., Ltd for transcriptome profiling by RNA-seq method. The samples were quantified using a NanoDrop2000 spectrophotometer and determined sample RINe values and concentrations using the Agilent4200 TapeStation system. Library construction and sequencing were performed using the PE150 sequencing mode on the Illumina NGS platform. The reads were aligned to the reference genome using HISAT2 software, and the alignment rate was calculated. For gene-level quantification, FeatureCount was applied to each sample, resulting in a matrix of gene expression values represented as FPKM values. DESeq2 package in R was utilized for conducting differential gene expression analysis. The DEGs were defined as |log2(fold change)|>1 and adjusted P value (Q value) <0.05. KOBAS and topGO software were used for KEGG and GO enrichment analysis, respectively. TIMER2.0 Database was utilized to validate the correlation between the expression levels of DEGs and the infiltration levels of immune cells.
For cellular samples, a minimum of 3 × 106 cells were collected, and RNA extraction was performed using TRIzol reagent. Each condition was performed in triplicates. Strand-specific libraries were prepared by BIOWEFIND (Wuhan, China) Co., Ltd and sequenced on Illumina PE150 platform. To quantify gene expression levels, pre-trimmed reads were aligned to the hg38 human reference genome with the UCSC known gene transcript annotation using HISAT2 with the default parameters. Gene counts were assessed using HTSeq, and DEGs were defined using DESeq2 with the criteria as |log2(fold change)| >1 and P value < 0.05. Heatmaps were created using pheatmap from the “ggplot2” package. Scatterplots were created using the ggplot2 package in R. KEGG analysis of DEGs was performed in R using the package clusterprofiler with default settings (P < 0.05).
Analysis of the tumor microenvironment by immunohistochemistry (IHC)
Due to the high proportion of stage I patients in YG, we chose stage I patient samples for IHC to ensure well representation of the results and a sufficiently large sample size for both YG and EG. Notably, some IA1 stage patients had insufficient tumor tissues, which were excluded from collecting specimens from this subset of patients. In total, samples from 218 patients (EG: n = 110; YG: n = 108) were undergone IHC analysis. Six antibodies were employed for IHC analysis: anti-CD4 (1:200 dilutions, #ab183685, Abcam), anti-CD8 (1:500 dilutions, #ab217344, Abcam), anti-CD3 (1:200 dilutions, #ab16669, Abcam), anti-CD20 (1:100 dilutions, #ab64088, Abcam), anti-CD138 (1:1000 dilutions, #ab128936, Abcam) and anti-FOXP3 (1:200 dilutions, #ab215206, Abcam). Continuous sections (3.5 µm) were prepared from the FFPE tissues of these patients. Two pathologists, who were blinded to the grouping information, independently reviewed each slide to ensure that the tumor tissue content was suitable for statistical analysis. Subsequently, we randomly selected 4–6 regions of 1 mm² each in tumor area and tumor/non-tumor interface area, respectively. Estimated counts of positively stained cells in the selected boxes were determined by the ImageJ software, and the final results were expressed as cell density per mm². To visually present the differences in immune cell infiltration between the two groups in two distinct regions, we generated an immune heatmap to visualize the IHC data. The calculation formula is as follows:
“Nt” represents the number of positive cells in the tumor area, “Ni” denotes the number of positive cells in the interface area, “T” stands for the total number of cases, and “Ntix” signifies the count of positive cells in the tumor or interface area for a specific case. Finally, these fold-change values were log2-transformed, with “0” as the reference point, and the range was defined between −4 and 2. In the heatmap, values greater than 0 were defined as “hot” (red), while values less than 0 were defined as “cold” (blue).
Multiplex immunofluorescence
We utilized PSM to select ten pairs of patients with well-matched major risk factors for multiplex immunofluorescence (EG: n = 10; YG: n = 10). The slides were incubated with blocking antibody diluent at room temperature for 10 min and then incubated overnight at 4 °C with primary antibodies. The slides were then incubated with the secondary antibody (HRP polymer, anti-mouse/rabbit IgG) at room temperature for 10 min. Subsequently, fluorophore (tyramide signal amplification or TSA plus working solution) was applied to the sections, followed by heat treatment with microwave. The primary antibodies were applied sequentially, followed by incubation with the secondary antibody and TSA treatment. The following antibodies were used: anti-CD4 (1:50 dilutions, #ZM0418, ZSGB-BIO), anti-CD8 (1:400 dilutions, #CST70306, Cell Signaling technology), anti-CD3 (1:400 dilutions, #ab135372, Abcam), anti-FOXP3 (1:200 dilutions, #BX50188, Biolynx) and anti-PANCK (1:1000 dilutions, #C2562, Sigma-Aldrich). Nuclei were stained with DAPI.
To obtain multispectral images, stained slides were scanned using the Polaris System (PerkinElmer, Massachusetts, USA). The images were processed using OLYVIA software (OLYMPUS). The analysis included calculating tissue area, cell count, positive rate, and the density of positive cells. For each indicator, an H-score was calculated based on its intensity: H-Score = [(Intensity 1 + percentage of positive cells) × 1 + (Intensity 2 + percentage of positive cells) × 2 + (Intensity 3 + percentage of positive cells) × 3] × 100. Outliers were identified using the normal distribution method and excluded when analyzing the differences between the two groups of data.
Cell culture
Human non-small cell lung cancer NCI-H322 and PC-9 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. HEK293FT cells were maintained in DMEM medium containing 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were cultured at 37 °C under a humidified atmosphere containing 5% CO2. All cells were regularly tested negative for mycoplasma contamination.
Plasmid construction
The coding region of human ERBB2 (NM_004448) was PCR amplified from pLV3-CMV-ERBB2-Puro plasmid (#P48113, MiaoLingBio) and then inserted into pHAGE-EF1α-BSD lentiviral expression vector between XbaI and BamHI restriction enzyme sites. The FLAG tag was added via PCR primer in the C-tail of the ERBB2 transcript. The ERBB2 mutants (Y772_A775dup, YVMA; G776delinsVC, insVC) were created by site-directed mutagenesis under the same pHAGE-EF1α-BSD vector as ERBB2-wild type (WT). A schematic diagram of the construction of the expression plasmid used in this study is presented in Supplementary Fig. 6. The primers used for molecular cloning are listed in Supplementary Table 9.
Ectopic expression of ERBB2 in NSCLC cells
WT and mutant ERBB2 were introduced into NCI-H322 or PC-9 cells by lentiviral infection. To package lentivirus, DNA transfection was performed in HEK293FT cells cultured in 15 cm plates using Lipofectamine 2000 transfection reagent (#11668027, Thermo Fisher Scientific) with a mix of 22.5 μg ERBB2-expressing lentiviral vector, 11.25 μg pCMVR8.74 and 6 μg pMD2.G plasmids. Two days after transfection, the virus-containing supernatant was collected and centrifuged at 3000 r.p.m. for 5 min to remove the cell debris. The virus supernatant was aliquoted and stored at −80 °C before use. To make fair comparison between different ERBB2 mutants, the infected NSCLC cells were examined by immunoblotting before functional assays to keep relatively even levels of introduced WT or mutant ERBB2.
Cell growth assay
For 2D monolayer cell growth, different ERBB2-expressing NCI-H322 or PC-9 cells were seeded in 12-well plates in triplicates. The initial plating cell number and density were the same between different comparison groups (8 × 103 cells per well for NCI-H322; 6 × 103 cells per well for PC-9). After 9 days, cell counting was performed using a hemocytometer. For 3D cell growth, soft agar colony formation assay was performed. 1% and 0.6% melting agarose were prepared and mixed with 20% fetal bovine serum supplemented RPMI-1640 media to make 0.5% and 0.3% agarose at 40 °C. Two milliliters of 0.5% agarose was added to the bottom of the 6-well plate and was incubated at 37 °C for more than 30 min. Cells were trypsinized to produce a single-cell suspension (5 × 103 cells per well) and mixed with 2 mL of 0.3% agarose before seeding on top of the 0.5% agarose base. Add 2 mL 20% fetal bovine serum containing RPMI-1640 media on top of the solidified agarose layers. The cells were then cultured routinely for 30 days, with the medium changed three times a week. The colonies were stained with 0.1% crystal violet before taking pictures and ImageJ was employed for colony counting. All the experiments were performed in triplicate.
Wound healing assay
NCI-H322 cells were seeded at a density of 3 × 106 per well on 6-well plates and incubated overnight. Gently drag a p200 pipet tip across the cell monolayer in a straight line to generate an incision, resulting in the formation of a “scratch”. Three horizontal scratches and three vertical scratches are evenly distributed across the 6-well plate. Remove the debris and smooth the edge of the scratch by washing the cells twice with 1× phosphate-buffered saline (PBS) and replacing with fresh medium. Cells were allowed to migrate across the wounds and pictures were taken at 0 h and 48 h. Each experiment was performed in triplicates.
Mouse xenograft
All studies using mice were performed in accordance with our animal protocols (#NEU-EC-2024A030S) from Care and Use of Laboratory Animals and approved by the Biological and Medical Ethics Committee of Northeastern University (China). Six-week-old BALB/c nude female mice were purchased from Beijing HFK Bioscience Co., Ltd (Beijing, China). NCI-H322 cells (6 × 106) infected with GFP or ERRB2-expressing lentivirus were re-suspended in a 7:3 ratio of PBS and Matrigel (#356234, Corning) and injected into female athymic mice subcutaneously at bilateral flanks. When the tumor volume reached 100 mm3 (4 days after cell inoculation), start to monitor the tumor size twice a week. Tumor volume was calculated by length × width2/2. At the endpoint of the experiment, mice were sacrificed, and tumors were removed for photos and weighting. All the tumors were within the maximal size (<1.5 cm) and volume (<2000 mm3) limit allowed by the related guidelines of the Biological and Medical Ethics Committee of Northeastern University.
Immunoblotting
Cells were washed with PBS and lysed with RIPA buffer supplemented with complete protease inhibitor and phosphatase inhibitor on ice. Protein lysates were separated by 10 to 12% bis-tris polyacrylamide gel and then transferred to a polyvinylidene fluoride (PVDF) membrane. Then, membranes were blocked for 1 h at room temperature (RT) in 5% skim milk in 1 × Tris-buffered saline Tween-20 (TBST). Primary antibodies diluted in 1 × TBST were incubated overnight at 4 °C: anti-HER2/ErbB2 (1:2000 dilutions, #18299-1-AP, Proteintech Group), anti-phosphorylated Akt (Ser473) (pAkt473, 1:2000 dilutions, #4060, Cell Signaling Technology), anti-phosphorylated Akt (Thr308) (pAkt308, 1:1000 dilutions, # 2965, Cell Signaling Technology), anti-Akt (1:1000 dilutions, # 10176-2-AP, Proteintech Group), anti-phosphorylated Erk 1/2 (Thr202, Tyr204) (pErk; 1:2000 dilutions, # 110441-R0072, Sino Biological), anti-p44/42 MAPK (Erk1/2) (Erk; 1:2000 dilutions, #4695, Cell Signaling Technology), and anti-GAPDH (1:1000 dilutions; #SC-32233, Santa Cruz). After incubation with primary antibody, the membranes were washed with 1 × TBST three times for 30 min before incubation with Rabbit anti-Goat lgG Fc Secondary Antibody, HRP (1:20000 dilutions, #31433, Thermo Fisher Scientific) or Rabbit anti-Mouse IgG (H + L) Secondary Antibody, HRP (1:20000 dilutions, #31450, Thermo Fisher Scientific) for 1 h at RT. The membranes were then washed with 1 × TBST three times for 30 min. Finally, a chemiluminescence substrate was added to the membranes and the result was resolved using a Tanon 5200 Chemiluminescent Imaging System.
Meta-analysis
The PubMed, Web of Science, Embase databases, and the Cochrane Central Register of Controlled Trials were searched until 13 April 2024. The records were systematically evaluated using inclusion and exclusion criteria. Two researchers independently performed the initial search, and controversies were resolved by discussion. Keywords and search strategies for two drugs are present in Supplementary Data 6. Studies were included if they met the following criteria: (1) the literature focused on the pathological type of non-small cell lung cancer; (2) the intervention was pyrotinib or poziotinib; (3) the study type was clinical trials, including observational studies or randomized controlled trials; (4) The outcome is overall survival duration; and (5) the full texts were available in English. Studies were excluded based on the following criteria: (1) the research subject is small cell lung cancer; (2) The outcome does not match; and (3) The literature type includes case reports, reviews, Meta-analysis, letters, conference papers, and consensus.
Patient treatment with pyrotinib
The two eligible patients had lung adenocarcinoma with diagnoses confirmed histologically. Prior to treatment, written informed consent with pyrotinib was obtained from the patient. The patient meet the following criteria: (1) signed informed consent document; (2) ECOG performance status 0–2; (3) pathologically-confirmed, locally advanced or metastatic lung adenocarcinoma; (4) ERBB2 Y772_A775dup mutation meanwhile without other mutations in those 11 key driver genes in lung cancer, as detected by local testing of tumor and circulating tumor nucleic acid in blood; and (5) Have adequate organ function (the following laboratory values at the visit: absolute neutrophil count (ANC) ≥ 1.0 × 109/L; platelets ≥75 × 109/L; hemoglobin (Hgb) ≥ 9.0 g/dL; calculated creatinine clearance ≥40 mL/min; total bilirubin <1.5 × ULN; Aspartate transaminase (AST) ≤ 3 × ULN and alanine transaminase (ALT) ≤ 3 × ULN). The measurable lesions of the patients were evaluated by modified Response Evaluation Criteria in Solid Tumors (mRECIST) during the treatment process. The chemo- or targeted- therapy was given at the normal clinical dose. The treatment protocol was approved by Institutional Review Board of The Fourth Affiliated Hospital of China Medical University (EC-2023-HS-054) and the patient gave written informed consent, according to CARE guidelines and in compliance with the Declaration of Helsinki principles. Each patient had signed consent to publish clinical information potentially identifying individuals.
Software and algorithms
The software and algorithms used in this study are as follows: SPSS 29.0.1.0 (171), KOBAS 3.0, topGO 2.56.0, pheatmap 1.0.12, clusterProfiler 4.8.2, dplyr 1.1.0, DESeq2 1.40.2, ggplot2 3.4.2, ggrepel 0.9.2, bioMart 2.56.1, org.Hs.eg.db 3.15.0, ImageJ 1.54 g.
Statistical and reproducibility
Categorical variables were presented as absolute values (N) or percentages (%) and were analyzed using the Pearson chi-square test and Fisher’s exact test (n < 5). Univariate and multivariate correlation analyzes were conducted using logistic regression analysis. Statistical significance was expressed as Mean ± SD or Mean ± SEM. Two-sided t-tests were used to analyze the mean differences between the two groups of data. At least two or three independent replicates were performed in general to ensure reproducibility, and all attempts at reproducibility were successful. For statistically significant results, we use * to denote P < 0.05, ** for P < 0.01, *** for P < 0.001, and **** for P < 0.0001. All statistical analyzes were performed using SPSS software (IBM Corp., USA), GraphPad Prism 9 software, and R package. Propensity score matching (PSM) was applied to perform a 1:1 matched case-control analysis, with a caliper width equal to 0.02 of the standard deviation of the logit of the propensity score. The PSM analysis was conducted in R using the MatchIt 4.5.5 package.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the results in this study are available within the paper and its Supplementary information. The raw sequencing data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences under the following accessions: GSA-human PRJCA026454 for DNA sequencing of clinical samples and GSA-human HRA007083 for RNA-seq in cell lines. The DNA data are available under controlled access and may be requested by completing the application form via GSA-Human System. Data acquisition is granted by the corresponding Data Access Committee (DAC). The approximate response time for accession requests is about 2 weeks. The raw sequencing data for RNA-seq in clinical samples were deposited in NCBI GEO database under the accession number GSE226481. Source data are provided with this paper.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 74, 229–263 (2024).
Sagerup, C. M., Smastuen, M., Johannesen, T. B., Helland, A. & Brustugun, O. T. Increasing age and carcinoma not otherwise specified: a 20-year population study of 40,118 lung cancer patients. J. Thorac. Oncol. 7, 57–63 (2012).
Teng, Y. et al. Lung cancer burden and trends from 2000 to 2018 in China: comparison between China and the United States. Chin. J. Cancer Res 35, 618–626 (2023).
Zhou, Y. et al. Long-term trends of lung cancer incidence and survival in southeastern China, 2011-2020: a population-based study. BMC Pulm. Med. 24, 25 (2024).
Ugai, T. et al. Is early-onset cancer an emerging global epidemic? Current evidence and future implications. Nat. Rev. Clin. Oncol. 19, 656–673 (2022).
Chinese Expert Group on Early, D., Treatment of Lung, C. & China Lung Oncology, G. [China National Lung Cancer Screening Guideline with low-dose computed tomography (2023 Version)]. Zhongguo Fei Ai Za Zhi 26, 1–9 (2023).
Arnold, B. N. et al. Lung cancer in the very young: treatment and survival in the national cancer database. J. Thorac. Oncol. 11, 1121–1131 (2016).
Jiang, W. et al. Comparisons of multiple characteristics between young and old lung cancer patients. Chin. Med J. 125, 72–80 (2012).
Subramanian, J. et al. Distinctive characteristics of non-small cell lung cancer (NSCLC) in the young: a surveillance, epidemiology, and end results (SEER) analysis. J. Thorac. Oncol. 5, 23–28 (2010).
Hsu, C. L. et al. Advanced non-small cell lung cancer in patients aged 45 years or younger: outcomes and prognostic factors. BMC Cancer 12, 241 (2012).
Luo, W. et al. Characteristics of genomic alterations of lung adenocarcinoma in young never-smokers. Int J. Cancer 143, 1696–1705 (2018).
Cai, L. et al. The genomic landscape of young and old lung cancer patients highlights age-dependent mutation frequencies and clinical actionability in young patients. Int. J. Cancer 149, 883–892 (2021).
Hughes, D. J. et al. Non-small cell lung cancer (NSCLC) in young adults, age < 50, is associated with late stage at presentation and a very poor prognosis in patients that do not have a targeted therapy option: a real-world study. Cancers 14, 6056 (2022).
Hu, M. et al. Comprehensive comparative molecular characterization of young and old lung cancer patients. Front. Oncol. 11, 806845 (2021).
Pan, X. et al. Frequent genomic alterations and better prognosis among young patients with non-small-cell lung cancer aged 40 years or younger. Clin. Transl. Oncol. 20, 1168–1174 (2018).
Tanaka, K. et al. Unique prevalence of oncogenic genetic alterations in young patients with lung adenocarcinoma. Cancer 123, 1731–1740 (2017).
Sacher, A. G. et al. Association between younger age and targetable genomic alterations and prognosis in non-small-cell lung cancer. JAMA Oncol. 2, 313–320 (2016).
Hou, H. et al. Comprehensive molecular characterization of young chinese patients with lung adenocarcinoma identified a distinctive genetic profile. Oncologist 23, 1008–1015 (2018).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020).
Yu, X. et al. FGF21 promotes non-small cell lung cancer progression by SIRT1/PI3K/AKT signaling. Life Sci. 269, 118875 (2021).
Fan, Y. et al. Clinical and molecular characteristics of Chinese non-small cell lung cancer patients with ERBB2 transmembrane domain mutations. Mol. Oncol. 14, 1731–1739 (2020).
Shigematsu, H. et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 65, 1642–1646 (2005).
Tomizawa, K. et al. Prognostic and predictive implications of HER2/ERBB2/neu gene mutations in lung cancers. Lung Cancer 74, 139–144 (2011).
Zhou, C. et al. Pyrotinib in HER2-mutant advanced lung adenocarcinoma after platinum-based chemotherapy: a multicenter, open-label, single-arm, phase II study. J. Clin. Oncol. 38, 2753–2761 (2020).
Song, Z. et al. Pyrotinib in patients with HER2-amplified advanced non-small cell lung cancer: a prospective, multicenter, single-arm trial. Clin. Cancer Res. 28, 461–467 (2022).
Liu, S. M. et al. First-line pyrotinib in advanced HER2-mutant non-small-cell lung cancer: a patient-centric phase 2 trial. Nat. Med. 29, 2079–2086 (2023).
Elamin, Y. Y. et al. Poziotinib for patients With HER2 exon 20 mutant non-small-cell lung cancer: results from a phase II trial. J. Clin. Oncol. 40, 702–709 (2022).
Le, X. et al. Poziotinib in non-small-cell lung cancer harboring HER2 exon 20 insertion mutations after prior therapies: ZENITH20-2 trial. J. Clin. Oncol. 40, 710–718 (2022).
Bryant, A. S. & Cerfolio, R. J. Differences in outcomes between younger and older patients with non-small cell lung cancer. Ann. Thorac. Surg. 85, 1735–1739 (2008).
Kang, J. et al. Risk factors for early-onset lung cancer in Korea: analysis of a nationally representative population-based cohort. Epidemiol. Health 45, e2023101 (2023).
Rich, A. L. et al. Non-small cell lung cancer in young adults: presentation and survival in the English National Lung Cancer Audit. QJM 108, 891–897 (2015).
Garrana, S. H. et al. Clinical and imaging features of non-small-cell lung cancer in young patients. Clin. Lung Cancer 22, 23–31 (2021).
Etzel, C. J. et al. An epidemiologic study of early onset lung cancer. Lung Cancer 52, 129–134 (2006).
Yarden, Y. & Sliwkowski, M. X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2, 127–137 (2001).
Sapino, A., Goia, M., Recupero, D. & Marchio, C. Current challenges for HER2 testing in diagnostic pathology: state of the art and controversial issues. Front. Oncol. 3, 129 (2013).
King, C. R., Kraus, M. H. & Aaronson, S. A. Amplification of a novel v-erbB-related gene in a human mammary carcinoma. Science 229, 974–976 (1985).
Mazieres, J. et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J. Clin. Oncol. 31, 1997–2003 (2013).
Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Li, B. T. et al. HER2 amplification and HER2 mutation are distinct molecular targets in lung cancers. J. Thorac. Oncol. 11, 414–419 (2016).
Perera, S. A. et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc. Natl. Acad. Sci. USA 106, 474–479 (2009).
Wang, S. E. et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell 10, 25–38 (2006).
Fan, Y. X. et al. Acquired substrate preference for GAB1 protein bestows transforming activity to ERBB2 kinase lung cancer mutants. J. Biol. Chem. 288, 16895–16904 (2013).
Dal Bello, M. G., Alama, A., Coco, S., Vanni, I. & Grossi, F. Understanding the checkpoint blockade in lung cancer immunotherapy. Drug Discov. Today 22, 1266–1273 (2017).
Forde, P. M., Kelly, R. J. & Brahmer, J. R. New strategies in lung cancer: translating immunotherapy into clinical practice. Clin. Cancer Res. 20, 1067–1073 (2014).
Wang, M. et al. Sunvozertinib, a selective EGFR inhibitor for previously treated non-small cell lung cancer with EGFR exon 20 insertion mutations. Cancer Discov. 12, 1676–1689 (2022).
Heymach, J. et al. A phase I, open-label, dose confirmation, escalation, and expansion trial of BI 1810631 as monotherapy in patients with advanced or metastatic solid tumors with HER2 aberrations. Clin. Lung Cancer 24, e65–e68 (2023).
Wilding, B. et al. Zongertinib (BI 1810631), an irreversible HER2 TKI, spares EGFR signaling and improves therapeutic response in preclinical models and patients with HER2-driven cancers. Cancer Discov. 15, 119–138 (2024).
Kim, T. et al. Safety and anti-tumor activity of BAY 2927088 in patients with HER2-mutant NSCLC: Results from an expansion cohort of the SOHO-01 phase I/II study. J. Clin. Oncol. 42, LBA8598 (2024).
Piotrowska, Z. et al. Safety, tolerability, and antitumor activity of zipalertinib among patients with non-small-cell lung cancer harboring epidermal growth factor receptor exon 20 insertions. J. Clin. Oncol. 41, 4218–4225 (2023).
Fidler, M. M. et al. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: a population-based study. Lancet Oncol. 18, 1579–1589 (2017).
Acknowledgements
The authors appreciate Dr. Wenjun Shang (Department of Thoracic Surgery, Beilun Branch Hospital of the First Affiliated Hospital of Medical School of Zhejiang University), Dr. Yongzhi Wang (Department of Thoracic Surgery, Dandong First Hospital), Dr. Zhaokun Shen (Department of Thoracic Surgery, Panjin Liao-Oil Field Gem Flower Hospital), Dr. Zhilong Zhao (Department of Thoracic Surgery, Dalian Zhongshan Hospital), Dr. Haiyang Chen (Department of Thoracic Surgery, Liaoyang Central Hospital) for their sincere support of this project. This study was supported by the Scientific Research Foundation of Education Department of Liaoning Province (#JCZR2020016) to Y.T., Key Program of Liaoning Provincial Department of Science and Technology Foundation (#2020JH2/10300060) to X.Y., the National Natural Science Foundation of China (32470673), the 111 Project (B16009) and the Construction Project of Liaoning Provincial Key Laboratory, China (2022JH13/10200026) to T.F.
Author information
Authors and Affiliations
Contributions
Y.T., X.Y., and T.F. conceived the study and designed the research. R.M. and W.Z. performed most of the detailed analytic and experimental work. S.W., C.Z., W.W., B.Y., H.X., H.W., P.L., R.L., and C.L. assisted with the acquisition of multi-center data. Y.L., Q.Y., C.S., H.Z., R.L., L.Z., D.Z., X.Z., and Z.W. analyzed the data. Y.Q and R.W. provided technical support in the field of pathology. X.Y., T.F., and Y.T. wrote the manuscript with the input from R.M. and W.Z., X.Y., and T.F. supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yoon Soo Chang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tian, Y., Ma, R., Zhao, W. et al. Comprehensive characterization of early-onset lung cancer, in Chinese young adults. Nat Commun 16, 1976 (2025). https://doi.org/10.1038/s41467-025-57309-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-57309-4
This article is cited by
-
Development of PET/CT-clinical nomograms for predicting lymph node metastasis in primary lung cancer
European Radiology (2025)