Abstract
Drosophila larval hematopoiesis takes place at the lymph gland, where blood cell progenitors differentiate into two possible cell types: Plasmatocytes, analogous to mammalian macrophages, or crystal cells that share features with mammalian megakaryocytes; a third cell type, the lamellocytes, develop only upon specific immune challenges. Here we show that autophagy inhibition in blood cell progenitors results in augmented crystal cell differentiation due to Notch accumulation. Notch activation during hematopoiesis depends on the endocytic pathway, which crosstalks with autophagy: While Notch activation depends on endocytosis and endosomal maturation, Notch lysosomal degradation requires autophagy. TOR signaling inhibits autophagosome biogenesis that in turn prevents the formation of Notch-containing amphisomes, which are necessary for Notch lysosomal destruction. Reduction of Notch lysosomal degradation shifts the balance towards Notch activation at endosomal membranes, thereby enhancing differentiation of crystal cells. Our work therefore defines a mechanism of regulation of immune cell differentiation in response to the nutritional status of the organism.
Similar content being viewed by others
Introduction
Drosophila larval hematopoiesis takes place at the lymph gland, where blood cell progenitors localized centrally at the so-called medullary zone undergo differentiation to give rise to myeloid-like lineage cells, which include plasmatocytes and crystal cells (CCs), while a third cell-type, the lamellocytes, differentiates only upon certain specific immune challenges, such as parasitic wasp egg invasion1,2. Plasmatocytes are macrophage-like cells that represent around 95% of total circulating blood cells, while CCs account for the remaining 5%3. CCs display large cytoplasmic prophenoloxidase crystalline inclusions, which breakdown after the immune insult, releasing phenoloxidases to the blood stream, thus contributing to melanization and destruction of the pathogen4,5.
The balance between maintenance and differentiation of blood cell progenitors of the lymph gland is tightly controlled by a wide array of factors ranging from signaling proteins6,7,8,9,10 and metabolites9,11, to chemical and mechanical cues12,13, which ultimately determine the progenitor fate. Particularly, differentiation of progenitors into CCs depends largely on the Notch pathway, activated, at least in part, by its ligand Serrate, which is expressed in specialized scattered cells localized at the outer-most region of the medullary zone13,14,15,16,17. Two populations of blood cell progenitors occur at the medullary zone of the lymph gland: Internally localized core progenitors, and externally localized distal progenitors18,19. Distal progenitors, but not core progenitors, are competent to undergo terminal differentiation, acquiring either a plasmatocyte fate in response to MAP kinase signaling mediated by the transcription factor Pointed2, or a crystal cell fate in response to Notch activation18. Notch pathway inhibition in blood cell progenitors provokes reduction or complete loss of CCs, while genetic overactivation of the pathway in progenitors leads to excess of CCs18,20.
Macroautophagy (referred hereafter as autophagy) is an evolutionary conserved self-degradative process, essential for housekeeping functions, such as recycling of macromolecules and organelles, as well as for removing protein aggregates. During autophagy, the cytoplasmic components to be degraded are sequestered in a double membrane organelle called autophagosome, which then fuses with lysosomes, where hydrolytic enzymes execute degradation of the cargo21,22,23,24. Alternatively, an intermediate step may take place, as autophagosomes can fuse with late endosomes/multivesicular bodies to generate amphisomes, which in turn fuse with lysosomes, where degradation of the cargo takes place25,26,27. Although autophagy is essentially a bulk degradation process, selectivity is attained by autophagy adapter proteins that bind to certain cargoes, such as organelles and aggregation-prone misfolded proteins or protein aggregates28,29.
In spite of autophagy being an essentially housekeeping recycling process, it is of fundamental importance in various cell differentiation events across eukaryotic organisms30,31,32,33,34,35,36,37. Particularly, in mammalian hematopoiesis, loss-of-function experiments have revealed that autophagy participates in differentiation or maintenance of most immune cell types from both the myeloid and lymphoid lineages38,39,40,41,42. However, the mechanisms by which autophagy regulates hematopoiesis are poorly defined at the molecular and cellular level.
In the current study, we show that basal autophagy in the Drosophila lymph gland is particularly active, and restrains differentiation of CCs. Inhibition of autophagy in blood cell progenitors results in increased levels of Notch, whose regulation depends largely on the endocytic pathway: While endocytosis and endosome maturation are necessary for Notch activation, formation of multivesicular body vesicles and fusion of the multivesicular body with lysosomes are required for Notch lysosomal degradation. Notch lysosomal degradation is regulated by autophagy through amphisome formation, and autophagy is in turn regulated by nutrient availability through the TOR pathway. Our study therefore establishes a mechanistic link between the nutritional status of the organism and blood cell differentiation through the regulation of autophagy and Notch activation.
Results
High levels of basal autophagy in the lymph gland are required for normal hematopoiesis
To investigate the role that autophagy plays in blood cell differentiation in the lymph gland of third instar larvae, we first analyzed the level of autophagy in this organ. Three different criteria, namely nucleation of the autophagosome reporter 3xmCherry-Atg8a, Lysotracker staining43 and Ref(2)P accumulation, revealed that autophagy levels in both fed and starved larvae are high at the lymph gland, in comparison with other Drosophila larval tissues (Supplementary Figs. 1 and 2). Interestingly, autophagy activation was higher at the medullary zone in comparison with the cortical zone, as revealed by Atg8a nucleation (Supplementary Fig. 1A). These observations suggest that autophagy may play a specific role in blood cell progenitors (see below). Canonical autophagy pathway regulators were required for basal autophagy in the lymph gland, as indicated by RNAi-mediated knock-down of different components of the autophagy pathway44 (Supplementary Fig. 3).
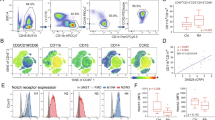
To investigate a possible role of autophagy in the differentiation of blood cells, we performed loss-of-function experiments of autophagy pathway genes and analyzed blood cell specific markers. In Atg1Δ3D homozygous mutant larvae, autophagy was suppressed (Fig. 1A) and crystal cells increased dramatically (Fig. 1D, E), while the proportion of blood cell progenitors and plasmatocytes were not significantly modified (Fig. 1B, C).
Confocal images of individual lobes of larval lymph glands; primary lobes indicated in white dashed lines. In Atg1Δ3D homozygous mutants, autophagy activation was reduced, as indicated by 3xmCherry-Atg8a nucleation (A n = 10 primary lobes for wild type, n = 8 for Atg1Δ3D; p < 0.0001). Progenitor (domeMESO > GFP; B n = 13 for wild type, n = 14 for Atg1Δ3D) or plasmatocyte (P1; C n = 24 for wild type, n = 28 for Atg1Δ3D) populations were unaffected in Atg1 mutants (p = 0.2210; p = 0.8221, respectively). Crystal cells increased in Atg1Δ3D mutant larvae as assessed by either anti-Lozenge (Lz; D n = 7 for each genotype; p = 0.0009) or anti-prophenoloxidase (PPO; E n = 10 for each genotype; p = 0.0016). Box plots were used to visualize data distribution, where the box represents the interquartile range (25th and 75th percentiles), the central line indicates the median, and whiskers indicate the minimum and maximum values in the dataset. Individual data points are shown. Statistical analysis was performed using an unpaired two-tailed Student’s t-test. **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significant (p > 0.05). Source data for plots are provided as a Source Data file.
Given the increase of crystal cell differentiation in the Atg1Δ3D mutant lymph gland, we next wondered if autophagy is required tissue-autonomously in blood cell progenitors for normal crystal cell differentiation, so we expressed double stranded RNAs against various autophagy regulators specifically in this cell type by using a domeMESO-Gal4 driver. Knock-down (KD) of Atg1 or Atg17 (components of the autophagy initiation complex) (Fig. 2A–C), of Vps15 or Vps34 (components of the nucleation complex) (Fig. 2A, D, E), or of Atg18 (involved in phagophore expansion)24 (Fig. 2A, F) provoked clear increase of CC differentiation (Fig. 2G), as visualized with anti-Lozenge (Lz) or anti-prophenoloxidase (PPO) antibodies17,18. In contrast, silencing of autophagy genes with an Hml-Gal4 driver in partially or totally differentiated cells did not provoke significant alterations in the number of crystal cells (Supplementary Fig. 4). These results indicate that autophagy is required in blood cell progenitors to restrain CC differentiation.
Crystal cells were visualized by anti-Lozenge (Lz) or anti-prophenoloxidase (PPO) immunofluorescence in Z-stack projections of lymph gland lobes (white dashed lines) where the indicated double-stranded RNAs, affecting different autophagy genes, have been expressed under control of a domeMESO-Gal4 driver (A–F). Crystal cell differentiation increased in comparison to wild type controls (UAS-LacZ). Quantification of the normalized number of crystal cells per lobe is depicted on panel (G) where box plots show the data distribution, with the box representing the interquartile range, the central line indicating the median, and whiskers extending to the minimum and maximum values. For Lz+ cells, statistical analysis was performed using two-sided likelihood ratio test (Chi-squared) followed by Dunnett’s test for treatment versus control comparisons (p < 0.0001; p < 0.0001; p = 0.0862; p < 0.0001; p = 0.0010, respectively). For PPO+ cells, ANOVA followed by Dunnett’s test for treatment versus control comparisons was used (p < 0.0001; p < 0.0001; p = 0.0002; p < 0.0001; p < 0.0001, respectively). For wild type, n = 65 primary lobes; for Atg1RNAi, n = 24; for Atg17RNAi, n = 25; for Vps15RNAi, n = 19; for Vps34RNAi, n = 13; for Atg18RNAi, n = 21 for Lz, n = 20 for PPO. Source data for plots are provided as a Source Data file. ***p < 0.001; ****p < 0.0001; ns, not significant (p > 0.05).
Autophagy sets a limit to Notch pathway activation and crystal cell differentiation
It has been shown that differentiation of CCs largely depends on activation of the Notch pathway, stimulated at least in part by its ligand Serrate15,16,17,18,20. We analyzed whether ligand-independent activation of Notch45 also contributes to CC differentiation in the lymph gland. This seems to be the case, as progenitor-specific overexpression of the E3 ubiquitin ligase deltex (dx), which promotes ligand-independent Notch endocytosis and Notch pathway activation46,47,48, provoked significant increase of CC differentiation (Supplementary Fig. 5A, B, D). Consistent with this, CC differentiation was also enhanced by the KD of Suppressor of deltex (Su(dx)), another E3 ligase that in this case inhibits non-canonical Notch pathway activation by promoting Notch lysosomal degradation49,50 (Supplementary Fig. 5A, C, D). These observations support the notion that ligand-independent Notch pathway activation contributes to CC differentiation in the lymph gland.
Next, we conducted genetic interaction experiments to explore if autophagy-dependent regulation of Notch may account for the effect of autophagy on CC differentiation. As expected, inhibition of autophagy in progenitors failed to promote CC differentiation when Notch expression was simultaneously silenced (Fig. 3A–D, N). Interestingly, the increase of CCs observed upon KD of Atg1 was suppressed in Notch (Fig. 3A, B, E, F, N) or Suppressor of Hairless (Fig. 3A, B, G, H, N) heterozygous mutant larvae, suggesting that autophagy antagonizes Notch activation during CC differentiation. Similarly, genes that participate of the non-canonical (ligand-independent) Notch pathway also showed antagonistic genetic interactions with autophagy: KD of dx, which on itself did not modify CC differentiation, suppressed the enhancement of CC differentiation induced by KD of Atg1 (Supplementary Fig. 6A–D, G). Likewise, overexpression of Su(dx), which on itself had no effect on differentiation, suppressed the enhancement of CCs provoked by Atg1 silencing (Supplementary Fig. 6A, B, E–G). Taken together, these results suggest that autophagy sets a limit to Notch pathway activation in the lymph gland, thereby restraining CC differentiation.
Crystal cells were visualized in Z-stack projections of lymph gland single lobes (dashed lines) by anti-PPO immunofluorescence (A–H); RNAi expression was driven with domeMESO-Gal4. I–M upper row: Anti-Notch immunofluorescence in wild type larvae and lymph gland lobes with reduced autophagy. N Quantification of the normalized number of crystal cells per lymph gland lobe. Kruskal-Wallis test followed by Dunn’s test for multiple comparisons. Mean values of genotypes marked with different letters are significantly different (p < 0.05). For wild type, n = 33 primary lobes; for Atg1RNAi, n = 43; for NRNAi, n = 18; for NRNAi/Atg1RNAi, n = 23; for N55e11/+, n = 14; for N55e11/+; Atg1RNAi, n = 28, for Su(H)1/+, n = 18; for Su(H)1/+; Atg1RNAi, n = 18. I–M lower row: Notch transcriptional activity was evaluated with an E(spl)mβ-HLH-GFP transcriptional reporter in each genotype. O Mean fluorescence intensity of anti-Notch staining was quantified in single slices, and relative change of immunofluorescence respect to the control was calculated and plotted. Statistical analysis in was performed using one-way ANOVA followed by Dunnett’s test for comparisons of treatments versus control (p < 0.0001 for each genotype). For wild type, n = 37 primary lobes; for Atg1RNAi, n = 25; for Atg17RNAi, n = 16; for Vps15RNAi, n = 16; for Atg18RNAi, n = 25. P Quantification of the normalized number of GFP-positive cells per lobe. For wild type, n = 42 primary lobes; for Atg1RNAi, n = 21; for Atg17RNAi, n = 20; for Vps15RNAi, n = 55; for Atg18RNAi, n = 13. Statistical analysis in was performed using Kruskal-Wallis test followed by Dunn’s test for comparisons of treatments versus control (p = 0.0036; p = 0.0004; p < 0.0001; p < 0.0001, respectively). N–P Box plots show the data distribution, with the box representing the interquartile range (25th and 75th percentiles), the central line indicating the median, and whiskers extending to the minimum and maximum values. Source data for plots are provided as a Source Data file. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Then, we investigated if the effect of autophagy on Notch pathway activation may rely on direct control of Notch receptor abundance. This was indeed the case, as KD of autophagy genes in blood cell progenitors provoked significant increase of Notch protein levels, as revealed by anti-Notch immunofluorescence (Fig. 3I–M, O), without affecting Notch transcription (Supplementary Fig. 7). Downregulation of autophagy also provoked increased Notch-dependent transcription, as indicated by the activity of an Enhancer of Split transcriptional reporter51 (Fig. 3I–M, P). These results indicate that upon autophagy inhibition, Notch protein accumulates, and the Notch pathway is overactivated. Interestingly, autophagy seems to impinge specifically on Notch signaling, as the JAK-STAT and Wnt pathways were not significantly affected after autophagy inhibition in blood cell progenitors, while Hedgehog signaling exhibited a modest reduction (Supplementary Fig. 8).
Crosstalk between the endocytic and autophagy pathways accounts for autophagy-dependent regulation of Notch and crystal cell differentiation
We next wondered about the mechanism by which autophagy controls Notch abundance. Autophagy is essentially a bulk degradation process and, with few exceptions52,53,54, does not usually target specific signaling molecules. We therefore reasoned that a crosstalk between the autophagy machinery and the endocytic pathway–essential for Notch signaling in some biological contexts45,55–might account for autophagy-dependent Notch degradation. In the Notch canonical pathway, after binding its ligand, the Notch receptor is cleaved at the plasma membrane, first by the metalloprotease Kuzbanian (Kuz), and then by the γ-secretase complex, releasing the Notch Intracellular Domain (NICD) that enters the nucleus and controls gene expression56 (Fig. 4A). However, in some biological contexts, Notch is activated by the γ-secretase not only at the plasma membrane, but also at the membrane of endosomes (Fig. 4B): After being cleaved by Kuz, Notch undergoes endocytosis, ending up at the membrane of early endosomes (EEs), which then acidify and mature to late endosomes (LEs), and finally evolve into multivesicular bodies (MVBs)57. In this context, the γ-secretase can cleave Notch either at the membrane of LEs, or at the limiting membrane of MVBs, releasing the NICD that then enters the nucleus and regulates transcription58 (Fig. 4B). This mechanism of Notch activation can also take place in a ligand-independent manner (Fig. 4C), in which case full-length Notch undergoes endocytosis without being cleaved by Kuz, and then the same process as in the canonical (ligand-dependent) Notch pathway takes place59 (Fig. 4C).
A, B, C Schematic representation of the Notch pathway. A After Ser binding, Kuzbanian (Kuz) and the γ-secretase complex (γ-sec) cleave Notch, and the Notch Intracellular Domain (NICD) enters the nucleus to regulate gene expression together with Suppressor of Hairless (Su(H)). B After cleavage by Kuz, Notch can undergo endocytosis that depends on the dynamin shibire (shi), and remains inserted in the membrane of early endosomes that mature by the action of Hrs and Rab5 to late endosomes. Formation of intraluminal vesicles of the multivesicular body is mediated by ESCRT complexes 0-III, which include the subunits TSG101, Vps25 and Shrub; Vps4 is the effector ATPase. Notch can be cleaved at the limiting membrane of late endosomes or multivesicular bodies, and enters the nucleus to regulate transcription. Notch present in intraluminal vesicles of the multivesicular body is degraded at the lysosome (not shown in the scheme). C Ligand-independent activation of Notch: Notch can undergo endocytosis without previous ligand binding; in this case, the entire (uncleaved) receptor ends up in the membrane of endosomes. The subsequent steps of the pathway are identical to those described in B. The E3 ubiquitin ligase deltex (dx) stimulates Notch endocytosis; Suppressor of deltex (Su(dx)) stimulates the formation of Notch-containing intraluminal vesicles. D–G Crystal cells visualized by anti-Lozenge (Lz) or anti-prophenoloxidase (PPO) immunofluorescence after knock-down of shi (E), Hrs (F) or Rab5 (G), with a domeMESO-Gal4 driver. White dashed lines mark lymph gland lobes. H Quantification of the results shown in panels (D–G) boxes represent the interquartile range, the central line indicates the median, and whiskers extend to the minimum and maximum values. For wild type, n = 62 primary lobes; for shibireRNAi, n = 24; for HrsRNAi, n = 20; for Rab5DN, n = 22. The statistical analysis was performed in both cases using Kruskal-Wallis test, followed by Dunn’s test for treatment versus control comparisons (p = 0.0017 for Rab5DN Lz+ cells; p < 0.0001 for every other comparison). Source data for plots are provided as a Source Data file. **p < 0.01; ****p < 0.0001.
We therefore explored whether normal progression of the endocytic pathway is required for Notch activation and CC differentiation in the lymph gland, and then, if the endocytic pathway may crosstalk with autophagy, thereby accounting for autophagy-dependent regulation of Notch (Fig. 4). Indeed, blood cell progenitor-specific KD of Shibire, a dynamin essential for completing endocytosis60, decreased CC differentiation (Fig. 4D, E, H), suggesting that endocytosis is required for Notch activation in this context. Likewise, KD of Hrs, required for EE to evolve into LE, or expression of a dominant-negative form of the small GTPase Rab5, required for EE formation and maturation58,61, both significantly reduced CC differentiation (Fig. 4D, F–H).
Invagination of the limiting membrane of the MVB, leading to intraluminal vesicle formation (Fig. 4B, C), is promoted by four ESCRT complexes (Endosomal Sorting Complex Required for Transport 0 to III)62,63, and incorporation of Notch to intra luminal vesicles results in Notch lysosomal degradation64,65. To gather further evidence that the endocytic pathway is necessary for Notch regulation, and particularly to assess if ESCRT complexes promote Notch degradation in blood cell progenitors, we silenced the expression of TSG101, a component of ESCRT-I complex, of Vps25, component of ESCRT-II, of Shrub, member of ESCRT complex III, or the expression of Vps4, the effector ATPase of all ESCRT complexes66,67 (Fig. 4B, C). Silencing of any of the above genes, expected to inhibit intraluminal vesicle formation and Notch lysosomal degradation, provoked enhancement of CC differentiation (Fig. 5A–G), and increased Notch protein levels (Fig. 5A–E, H). These results further support the notion that the endocytic pathway regulates Notch protein levels in blood cell progenitors, thereby controlling CC differentiation.
Anti-Lozenge (Lz) and anti-prophenoloxidase (PPO) stainings to visualize crystal cells in Z-stack projections of lymph gland primary lobes (white dashed lines), and assess the effect of domeMESO-Gal4-driven knock-down of genes of different ESCRT complexes (B–D) or the gene Vps4 (E) that encodes the effector ATPase of ESCRT complexes, in comparison with wild type individuals (A). F, G Quantification of normalized crystal cells per lobe as assessed by anti-Lz or anti-PPO staining respectively. F One-way ANOVA followed by Dunnett’s test for treatment versus control comparisons (p < 0.0001; p = 0.0125; p < 0.0001; p < 0.0001, respectively). G Two-sided likelihood ratio test (Chi-squared) followed by Dunnett’s test for treatment versus control comparisons (p < 0.0001 for each genotype). For wild type, n = 86 primary lobes; for TSG101RNAi, n = 24 for Lz and n = 22 for PPO; for Vps25RNAi, n = 24; for shrubRNAi, n = 19; for Vps4RNAi, n = 33. Notch immunofluorescence performed with an antibody targeting the Notch extracellular domain shows that Notch protein levels in each genotype (A–E) parallels the increase in differentiation of crystal cells. H Quantification of Notch immunofluorescence expressed as relative change in fluorescence intensity respect of the wild type genotype. For wild type, n = 20; for TSG101RNAi, n = 16; for Vps25RNAi, n = 19; for shrubRNAi, n = 15; for Vps4RNAi, n = 14. One-way ANOVA followed by Dunnett’s test for treatment versus control comparisons (p < 0.0001; p = 0.0038; p < 0.0001; p = 0.0732, respectively). F–H Box plots show the data distribution, with the box representing the interquartile range, the central line indicating the median, and whiskers extending to the minimum and maximum values. Source data for plots are provided as a Source Data file. *p < 0.05; **p < 0.01; ****p < 0.0001; ns, not significant (p > 0.05).
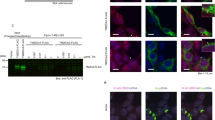
Having established that the endocytic pathway plays a central role in Notch regulation in blood cell progenitors during CC differentiation, we next wondered whether autophagy impinges on Notch activation through the endocytic pathway. It is well known that autophagosomes can fuse with MVBs to generate amphisomes, and that amphisomes in turn fuse with lysosomes to generate autolysosomes25,68 (Fig. 6A). Moreover, it has been proposed that autolysosomes, where degradation of the autophagosome cargo takes place68, are formed exclusively through this intermediate fusion event69,70. We therefore investigated if inhibition of autophagosome formation may lead to accumulation of MVBs, stalling Notch lysosomal degradation, and shifting the balance towards Notch activation. In agreement with this hypothesis, high-resolution confocal imaging (AiryScan) revealed that in wild type larvae, a proportion of Notch protein appeared within or associated with Rab5-positive vesicles (Fig. 6B; Supplementary Fig. 9A), or Rab7-positive vesicles (Fig. 6C, D; Supplementary Fig. 9B), which correspond respectively to EE, and LE/MVBs71,72. Upon autophagy inhibition in blood cell progenitors, Rab7-positive vesicles (LE/MVBs) significantly enlarged (Fig. 6C, F, G), and contained increased amounts of Notch protein (Fig. 6C, F, H), supporting the notion that autophagy mediates lysosomal degradation of Notch, which is present in endosomes. Furthermore, in wild type individuals Notch colocalized and associated by proximity ligation assays with the autophagosomal marker Atg8a (Fig. 6D, E; Supplementary Fig. 9B, C and Supplementary Fig. 10) and/or with the endolysosomal marker Lamp73 (Fig. 6E; Supplementary Fig. 9C), suggesting that Notch traffics through amphisomes and autolysosomes prior to lysosomal degradation. Taken together, our results support a model in which autophagy regulation, through the fusion of autophagosomes with MVBs, sets a balance between Notch activation at the limiting membrane of LE/MVBs, and Notch lysosomal degradation once it has been incorporated in intraluminal vesicles of the MVB. Consistent with such model, silencing of the Vha44 subunit of the V-ATPase, required for acidification of endolysosomal compartments (Fig. 7A, B, F), of the phosphatidylinositol(3)P-5-kinase Fab1 (Fig. 7A, C, F), required for endolysosomal maturation and function74 (Fig. 6A), enhanced CC differentiation. Likewise, silencing of the SNAREs Vamp7 or Syntaxin-17, required for fusion of autophagosomes with MVBs, as well as for fusion of autophagosomes or amphisomes with lysosomes75,76 (Fig. 6A), enhanced CC differentiation (Fig. 7A, D–F). These results suggest that prevention of autolysosome formation inhibits Notch lysosomal degradation, thus favoring instead Notch activation at the limiting membrane of LEs/MVBs.
A Schematic representation of endosomal maturation and fusion with autophagosomes. Endosomes mature into multivesicular bodies, in which limiting membrane the Notch Intracellular Domain (NICD) can be cleaved and enter the nucleus (nucleus not shown). Alternatively, the NICD can be sorted to intraluminal vesicles of the multivesicular body that fuses with autophagosomes to give rise to amphisomes, which fuse with lysosomes, resulting in autolysosomes where the NICD is degraded. B–F High-resolution (AiryScan) confocal images of Notch associated to endosomal or autophagic vesicles in the lymph gland. Part of Notch protein (cyan, white arrowheads) localized in Rab5-positive early endosomes (red, yellow arrowheads) (B) Rab7-positive late endosomes/multivesicular bodies (red, yellow arrowheads) (C), amphisomes positive for Rab-7 (green, yellow arrowheads) and Atg8a (red, pink arrowheads) (D) or autolysosomes decorated with Lamp (green, yellow arrowheads) and Atg8a (red, pink arrowheads) (E). F After autophagy inhibition through the expression of Atg1RNAi, Rab7-positive vesicles significantly enlarged (red, yellow arrowheads) and contained increased amounts of Notch protein (cyan, white arrowheads). G Quantification of the size of Rab7-positive vesicles in the experiments of panels (C, F) and after silencing the indicated autophagy genes. Wild type, n = 1431 vesicles; Atg1RNAi, n = 753; Atg17RNAi, n = 628; Vps15RNAi, n = 713; Atg18RNAi, n = 1085. H Quantification of Notch in Rab7 vesicles in the experiments of panels (C, F) and after silencing of the indicated autophagy pathway genes. Wild type, n = 966 vesicles; Atg1RNAi, n = 1989; Atg17RNAi, n = 1564; Vps15RNAi, n = 1857; Atg18RNAi, n = 1048. Boxes represent the interquartile range, the central line indicates the median, and whiskers extend to 10th and 90th percentiles. Mean values are shown in G as a dash. The statistical analysis performed in both cases was Kruskal-Wallis test followed by Dunn’s test for treatment versus control comparisons (p = 0.0067 for Atg18 RNAi in G; p < 0.0001 for every other comparison). Source data for plots are provided as a Source Data file. **p < 0.01; ****p < 0.0001.
Anti-Lozenge (Lz) and anti-prophenoloxidase (PPO) staining were used to visualize crystal cells in Z-stack projections of the lymph gland (white dashed line). After knock-down of the V-ATPase subunit Vha44 or the lipid kinase Fab1 with a domeMESO-Gal4 driver, an increase of crystal cell differentiation occurred in comparison with wild type control larvae (A–C). Knock-down of the SNAREs Vamp7 or Syntaxin-17 also led to augmented crystal cell differentiation (D, E). F Quantification of normalized crystal cells per lobe, as assessed with anti-Lz and anti-PPO antibodies respectively. Box plots show the data distribution, with the box representing the interquartile range, the central line indicating the median, and whiskers extending to the minimum and maximum values. Wild type, n = 22; Vha44RNAi, n = 24; fab1RNAi, n = 21; Vamp7RNAi, n = 20; Syx17RNAi, n = 19. Brown-Forsythe and Welch ANOVA followed by Dunnett’s T3 test for treatments against control comparisons (for Lz, p = 0.0037, p < 0.0001, p < 0.0001, p < 0.0001, respectively; for PPO, p < 0.0001, p = 0.0209, p = 0.0002, p = 0.0013, respectively). Source data for plots are provided as a Source Data file. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001.
Nutrient availability regulates autophagy through the TOR pathway, thereby controlling crystal cell differentiation
In eukaryotic cells, nutrient availability mediates regulation of autophagy through the TOR pathway77,78,79, so we sought to assess if alterations in the composition of the larval culture medium impinge on CC differentiation by modulating the autophagy-Notch axis in blood cell progenitors. After increasing the amount of yeast in the medium, which augments amino acid availability80, autophagy was significantly reduced (Supplementary Fig. 11A, B), while Notch abundance increased significantly (Fig. 8A, B), and CC differentiation was enhanced (Fig. 8C, G). The increase of CC differentiation in the yeast-rich medium was suppressed upon Atg1 overexpression (Fig. 8D, G), a treatment that increases autophagy81 Additionally, the increase of CCs in yeast-rich medium (Fig. 8C) was mitigated after knocking-down the amino acid transporter Slimfast (Slif)82 (Fig. 8E, G). Conversely, upon overexpression of Slif in normal larval medium, strong increase of CC differentiation was observed (Fig. 8C, F, G). Overall, these results indicate that an amino acid-rich diet inhibits autophagy in the lymph gland, leading to Notch accumulation, and increased crystal cell differentiation.
Anti-Notch immunofluorescence (A) indicated that Notch protein levels were higher in lymph glands of larvae grown in culture medium containing high yeast (4% w/v) in comparison to larvae grown in normal medium (2% yeast w/v). B Notch immunofluorescence intensity (arb. units) relative to the one of larvae reared in normal food. Unpaired two-tailed Student’s t-test (****, p < 0.0001). Normal food, n = 13 primary lobes; high yeast, n = 14. Crystal cell differentiation increased in larvae grown in the yeast-rich medium, in comparison to normal fly food (C), as assessed by anti-Lozenge (Lz) immunofluorescence in Z-stack projections of lymph gland lobes (dashed line). The increase of crystal cell differentiation observed in (C) was suppressed after overexpression of Atg1 (D) with domeMESO-Gal4 driver. RNAi-mediated knock-down of the amino acid transporter Slimfast (Slif) prevented the increase of crystal cell differentiation provoked by the addition of extra-yeast in the fly food (E). Overexpression of Slif led to increased crystal cell differentiation in larvae reared in normal food, compared with wild type animals (C, F). G Quantification of the results of panels (C–F). One-way ANOVA followed by Tukey’s test for multiple comparisons. Different letters represent statistically significant differences (p < 0.05). For the wild type genotype, n = 144 primary lobes in normal food and n = 51 in high yeast; for UAS-Atg1, n = 24 in normal food and n = 24 in high yeast; for slifRNAi, n = 75 in normal food and n = 44 in high yeast; for UAS-slif, n = 77. B, G Box plots show the data distribution, with the box representing the interquartile range, the central line indicating the median, and whiskers extending to the minimum and maximum values. Source data for plots are provided as a Source Data file.
The increase of CC differentiation observed in response to augmented nutrient availability was conveyed by the TOR pathway, since in Rheb PΔ1 or TOR2L19 heterozygous loss-of-function mutants, CC differentiation was mitigated (Fig. 9A–D). Consistent with this, overexpression of Akt or Rheb, as well as KD of Tsc1, treatments that inhibited autophagy (Supplementary Fig. 11C–H), significantly enhanced CC differentiation (Fig. 9E–I). These results suggest that amino acid availability regulates activation of the TOR pathway in blood cell progenitors, thereby controlling autophagy and crystal cell differentiation.
The number of crystal cells in the lymph gland (white dashed line) was assessed using anti-Lozenge (Lz) and anti-prophenoloxidase (PPO) staining in Z-stack projections. The increase of crystal cell differentiation induced by a high-yeast diet (A) was mitigated in RhebPΔ1 (B) or TOR2L19 (C) heterozygous larvae. D Quantification of the above results. Two-way ANOVA followed by Tukey’s test for multiple comparisons. When boxes are marked with the same letter, the difference between the mean values is not statistically significant (p > 0.05). If two boxes are marked with different letters, the mean values are significantly different (p < 0.05). For the wild type genotype, n = 48 primary lobes in normal food and n = 46 in high yeast; for RhebPΔ1/+, n = 13 in normal food and n = 14 in high yeast; for TOR2L19/+, n = 23 in normal food and n = 23 in high yeast. E–H Overexpression of Akt, silencing of Tsc1 or overexpression of Rheb driven by domeMESO-Gal4 led to increased crystal cell differentiation. I Quantification of these results, represented as the ratio between crystal cells and total cortical zone (CZ) hemocytes, which were calculated as the sum of P1+ cells (mature plasmatocytes) and Lz + /PPO+ cells (crystal cells). One-way ANOVA followed by Dunnett’s test for treatments versus control comparisons (p < 0.0001 for each comparison). For wild type, n = 27 for Lz and n = 71 for PPO; for UAS-Akt, n = 18 for Lz and n = 16 for PPO; for Tsc1RNAi, n = 35 for Lz and n = 36 for PPO; for UAS-Rheb, n = 22 for Lz and n = 26 for PPO. D, I Box plots show the data distribution, with the box representing the interquartile range, the central line indicating the median, and whiskers extending to the minimum and maximum values. Source data for plots are provided as a Source Data file. ****p < 0.0001.
In summary, our results indicate that amino acid abundance can control the extent of crystal cell differentiation in the larval lymph gland by modulating the TOR pathway, which in turn inhibits autophagy. Autophagy inhibition provokes an increase of Notch protein levels, which in turn promotes crystal cell differentiation (Fig. 10). Autophagy-dependent regulation of Notch therefore emerges from a crosstalk between the endocytic and autophagy pathways: In conditions of reduced autophagy (high amino acids), Notch lysosomal degradation is hindered, thus favoring Notch accumulation at the limiting membrane of late endosomes/multivesicular bodies, leading to increased Notch signaling and crystal cell differentiation (Fig. 10).
Amino acids enter the cell through the transporter Slimfast (Slif), thereby stimulating the kinase TOR, which results in inhibition of autophagosome formation (yellow lines). Notch is endocytosed after ligand (Ser) binding, and Kuzbanian (Kuz)-dependent cleavage. The Notch Intracellular Domain (NICD) remains inserted in the membrane of early endosomes, which then mature into late endosomes and further into multivesicular bodies (black arrows). The NICD can be processed by the γ-secretase (γ-sec) at the limiting membrane of late endosomes or multivesicular bodes, and released to enter the nucleus, where it regulates transcription (green arrows). Alternatively, the NICD present at the limiting membrane of multivesicular bodies, can be internalized to end up in intraluminal vesicles. Fusion of multivesicular bodies with autophagosomes results in Notch-containing amphisomes, which fuse with lysosomes to form autolysosomes (red arrows), where Notch is finally degraded. Thus, nutrient availability regulates the abundance of autophagosomes (yellow arrows), which ultimately determines the extent of Notch lysosomal degradation (red arrows). If Notch is not degraded, it can be activated to control gene expression (green arrows).
Discussion
In this work, we have shown that autophagy is critically required in blood cell progenitors of the Drosophila lymph gland to set a limit to crystal cell differentiation. Autophagy exerts this control by promoting Notch degradation through a crosstalk with the endocytic pathway (Fig. 10). As previously reported in other biological settings, we have found that both Notch activation and Notch degradation take place in compartments of the endocytic pathway of blood cell progenitors48,58,83,84,85. Upon genetic blockage of endocytosis, or hindering maturation of early endosomes in blood cell progenitors, Notch-dependent differentiation of CCs was impaired. Conversely, by blocking components of ESCRT complexes or of its effector ATPase Vps4, which mediate formation of intraluminal vesicles of the MVB, Notch degradation was impaired, leading to increased CC differentiation. Noteworthy, silencing of the subunit Vha44 of the V-ATPase led to modest although significant increase of crystal cell differentiation. This suggests that, unlike in other organs86,87, reduction of endolysosomal acidification in blood cell progenitors leads to reduction of Notch degradation in these cells. We found that autophagosomes are necessary for Notch degradation. This is consistent with a model in which fusion of MVBs with autophagosomes giving rise to amphisomes is necessary for Notch lysosomal degradation. According to this model, autophagy inhibition is expected to reduce Notch lysosomal degradation, leading to increased Notch signaling, and augmented CC differentiation. In agreement with such mechanism, genetic or physiologic graded overactivation of the TOR pathway, which mediates inhibition of autophagy77,78, increases CC differentiation. Further studies are required to assess whether the transcription factor Mitf/TFEB, an important regulator of autophagy acting in opposition to the mTOR pathway, contributes to modulate Notch activation in blood cell progenitors in response to nutrient availability88. Our results therefore establish a mechanistic link between the nutritional status of the organism and differentiation of cells of the immune system, in which the Notch pathway is regulated by TOR signaling through the modulation of autophagy (Fig. 10).
It is increasingly evident that autophagy is required for proper differentiation or maintenance of most mammalian hematopoietic lineages35,89, although the mechanisms involved are incompletely defined. Paralleling our observations in Drosophila blood cell progenitors, autophagy is also highly active in mammalian hematopoietic stem cells90, where autophagy is presumably required for lowering reactive oxygen species through mitophagy91. In T-lymphocytes, as well as in some myeloid lineages, autophagy is believed to control the OXPHOS/glycolytic ratio, thereby impinging on differentiation92,93. During B-cell maturation to antibody-producing plasma cells, autophagy is required to limit the unfolded protein response at the ER, and accumulation of protein aggregates, which in turn has an impact on differentiation94.
Whereas in Drosophila hematopoiesis the primary role of Notch is to promote crystal cell differentiation and maturation14,15,17,18,19,95, in mammalian hematopoiesis Notch has a more complex and incompletely understood function. Notch has been reported to act either as a tumor suppressor or as a tumor promoter in different types of leukemia96,97, and consistent with this, Notch fulfills different roles depending on the hematopoietic lineage98,99,100,101,102,103.
TOR dependent regulation of Notch was previously reported in other Drosophila organs, even though the mechanisms involved were not completely defined. In Drosophila enteroblasts, regulation of Notch by the TOR pathway was shown to depend on miRNAs104, which does not exclude the possibility that TOR might regulate Notch through an autophagy-dependent mechanism in the Drosophila gut as well. Autophagy-dependent degradation of Notch apparently takes place in Drosophila follicle cells by mid-oogenesis105, opening the possibility that the complete mechanism described here may be more general, perhaps operating in other Drosophila cell types besides hematopoietic progenitors.
Evidence for autophagy-dependent destruction of Notch has also been reported in mammalian cells106, although the mechanism in that context involves direct uptake of Notch into Atg16L1-positive vesicles, rather than into regular Rab5/Rab7-positive endosomes. Consistent with this, human Notch1 NICD, but not Notch2-4 NICD, can bind directly to p62, which promotes its autophagic degradation107. We have shown in the current work that Notch degradation strongly depends on ESCRT complexes, and therefore, on the endocytic pathway. Thus, if direct uptake of Notch into autophagosomes does occur in cells of the Drosophila lymph gland, this mechanism is not expected to play a central role in limiting Notch protein accumulation. In any case, it remains to be assessed if the whole mechanism controlling Notch activity, described here in Drosophila, operates in mammalian systems as well.
What is the physiological significance of crystal cell differentiation being dependent on amino acid availability? This is still unclear, and may relate to enhancement of immune responses according to nutrient availability. Interestingly, totally unexpected new functions of crystal cells in Drosophila physiology are starting to emerge5. The adaptive meaning of nutrient-dependent regulation of their differentiation might therefore relate to novel functions that these intriguing cells may play in fly development or physiology.
Methods
Flies were reared at 25 °C on a standard cornmeal/agar medium. For Gal4-mediated genetic manipulations, all experimental crosses were conducted at 25 °C, with F1 first instar larvae subsequently incubated at 29 °C to maximize Gal4 activity. To assess the nutritional impact on CC differentiation, standard cornmeal medium supplemented with 4% or 2% yeast was utilized. For starvation assays, larvae were incubated on agar plates for a period of 6 h prior to dissection. RNAi lines against autophagy genes have been previously validated: Atg1RNAi (BL 26731)108, Atg17RNAi (BL 36918)109, Vps15RNAi (BL 34092)110, Vps34RNAi (BL 33384)109. The following fly stocks were obtained from the Bloomington Drosophila Stock Center (https://bdsc.indiana.edu): w1118 (BL 3605), Canton-S (BL 9515), UAS-GFP (BL 1521), YFP-Rab7 (BL 62545), YFP-Rab5 (BL 62543), Notch55e11 (BL 28813), Su(H)1 (BL 417), Atg1RNAi (BL 26731), Atg17RNAi (BL 36918), Atg18RNAi (BL 34714), Vps15RNAi (BL 34092), Vps34RNAi (BL 33384), NotchRNAi (BL 33611), Su(dx)RNAi (BL 67012), E(spl)mß-HLH-GFP (BL 65294), shibireRNAi (BL 28513), HrsRNAi (BL 33900), UAS-Rab5DN (BL 9772), Tsg101RNAi (BL 38306), Vps25RNAi (BL 54831), Fab1RNAi (BL 35793), Vamp7RNAi (BL 38300), Syx17RNAi (BL 25896), UAS-Atg1 (BL 60734), slimfastRNAi (BL 64922), UAS-slimfast (BL 52661), UAS-Akt (BL 8191), Tsc1RNAi (BL 52931), UAS-Rheb (BL 9688), Hml-Gal4 (BL 30140), Stat92ERNAi (BL 33637), 10xStat92E-GFP (BL 26197), AxnRNAi (BL 31705) and UAS-CiHA (BL 32571). shrubRNAi (v108557) and Vps4RNAi (v105917) were obtained from the Vienna Drosophila Resource Center (https://stockcenter.vdrc.at). The following stocks were kindly provided by colleagues: domeMESO-Gal4 (Utpal Banerjee), 3xmCherry-Atg8a and Atg1Δ3D (Gábor Juhász), UAS-LacZ (Ben Shilo), UAS-GFP-Ref2P (Thomas Neufeld), UAS-GFP-Lamp (Helmut Kramer), UAS-deltex (Spyros Artavanis-Tsakonas), RhebPΔ1 and TOR2L19 (Sean Oldham).
Dissection, fixation and immunostaining of lymph glands
Wandering third instar larvae were utilized for dissection of lymph glands in PBS. Using precision forceps (Dumont #5), lymph glands were extracted along with the larval brain and mouth hooks, and placed in microfuge tubes containing ice cold PBS, as previously described111. Dissected tissue was fixed in 4% formaldehyde for 30 min and washed three times in PBS with 0.4% Triton-X (0.4% PBST) for 15 min. Blocking was carried out in 10% normal goat serum (Sigma-Aldrich) in PBST for 30 min, followed by overnight incubation with primary antibodies at 4 °C in 10% normal goat serum. Thereafter, lymph glands were washed three times in 0.4% PBST for 15 min, and incubated with secondary antibodies for 1.5 h at room temperature in 10% normal goat serum. Finally, samples were washed three times in 0.4% PBST before mounting on glass slides in mounting medium (Mowiol 4-88 anti-fade agent, EMD Millipore Corp., Billerica, MA). The following primary antibodies were used: Mouse anti-Lozenge 1:10 (anti-lozenge, Developmental Studies Hybridoma Bank), mouse anti-Notch Extracellular Domain 1:10 (C458.2H, Developmental Studies Hybridoma Bank), mouse anti-P1 1:100 (gift from I. Ando), rabbit anti-PPO 1:2000 (generous gift from G. Christophides), mouse anti-Ci 1:10 (2A1, Developmental Studies Hybridoma Bank), mouse anti-Armadillo 1:50 (N2 7A1, Developmental Studies Hybridoma Bank). Alexa Fluor 488, Alexa Fluor 647 and Cy3-conjugated secondary antibodies 115-545-062, 115-605-166, and 111-165-144, respectively, Jackson ImmunoResearch) were utilized in 1:250 dilutions.
Image acquisition and processing
Lymph glands were imaged using a Zeiss LSM 880 confocal microscope, acquiring either Z-stacks or single confocal planes. Image processing was conducted using Fiji/ImageJ software (NIH). Lobe area quantification was performed by manually delimiting lobe perimeter. Automatic thresholding was applied using the Default method in Fiji software to measure domeMESO and P1 areas. For crystal cell quantification, the total number of CCs was quantified using Fiji’s “Find Maxima” function based on nuclear markers such as Lz and E(spl)mβ-HLH-GFP, and manually when using the cytoplasmic marker PPO. These values were then corrected for lobe area in comparison with the control treatment whenever a significant difference in lobe size was found between the treatments (whole Z-stack projections). In the experiments where the TOR pathway was genetically activated (Fig. 9E–I), crystal cell differentiation was assessed by calculating the ratio of crystal cells to total cortical zone hemocytes (P1-positive for plasmatocytes + Lz-positive or PPO-positive for crystal cells) on a single confocal plane. In Figs. 3 and 5, Notch levels were calculated as mean immunofluorescence intensity of the whole primary lobe. In Fig. 6, Notch intensity was measured inside Rab7-positive vesicles, which were delimited by automatically thresholding using the Moments method and measured using the “Analyze Particles” function. 3xmCherry-Atg8a and GFP-Ref(2)P foci were quantified using the “Find Maxima” function of the Fiji software. For comparing multiple tissues (Supplementary Fig. 1B–G), 3xmCherry-Atg8a foci were normalized to cell density relative to salivary gland, which showed the lowest density. The Lysotracker area fraction was calculated by automatically thresholding the Lysotracker positive area and dividing it by the total area of the tissue. For measuring Notch localization in endolysosomal/autophagosomal compartments, Notch foci were identified using the “Find Maxima” function, then counted inside vesicles using masks generated by automatic thresholding on Fiji. Proximity Ligation Assays puncta were counted using the “Find Maxima” function in Fiji, normalized by the total area of the lobe.
Western blots and proximity ligation assays
Protein samples were obtained from Drosophila larval tissues (lymph gland, n = 100 larvae; imaginal disks, n = 100; salivary gland, n = 30; fat body, n = 20; gut, n = 20) and manually homogenized in RIPA buffer supplemented with protease inhibitors; protein concentrations were determined using the BCA Protein Assay Kit (Sigma-Aldrich, B9643) according to the manufacturer’s protocol. For Atg8a detection, proteins were separated on 15% SDS-PAGE gels, while for Ref(2)P-GFP detection, 10% SDS-PAGE gels were utilized. Proteins were transferred to nitrocellulose membranes (Millipore) using a wet transfer system at 100 V for 1 h. Membranes were then blocked in 5% skim milk in TBS-T (Tris-buffered saline with 0.1% Tween-20) for 1 h at room temperature, after which they were incubated overnight at 4 °C with the following primary antibodies diluted in TBS-T with 3% BSA: For Atg8a, rabbit anti-GABARAP 1:1000 (ab109364, Abcam); for GFP-Ref(2)P, rabbit anti-GFP 1:1000 (Life Sciences); tubulin was detected with a mouse anti-alpha-tubulin antibody 1:5000 (12G10, Developmental Studies Hybridoma Bank, 12G10). Following three washes with TBS-T, membranes were incubated with IRDye® 800CW secondary antibodies (LI-COR, anti-mouse catalog # 926-32212; anti-rabbit # 926-32213) diluted 1:5000 in TBS-T with 5% skim milk for 1 h at room temperature. Fluorescent signal was detected at 800 nm using an Odyssey CLx Imager (LI-COR). Band intensities were quantified using Empiria Studio software (LI-COR).
Proximity Ligation Assays were carried out with a Duolink in Situ Orange Starter Kit Mouse/Rabbit, (DUO92102, Sigma-Aldrich), utilizing the above rabbit anti-GABARAP antibody 1:100, mouse anti-Notch antibody 1:10, and mouse anti-GFP 1:100 (A-1112C, Invitrogen).
Quantitative real-time PCR (qPCR)
Total RNA was extracted from 150 lymph glands per genotype using TRIzol reagent (Thermo Fisher Scientific) and quantified with a NanoDrop spectrophotometer. One microgram of RNA was reverse transcribed into cDNA using random primers and M-MLV Reverse Transcriptase (Promega, # M17080). qPCR was conducted on a LightCycler® 480 Instrument II (Roche) with LightCycler® 480 SYBR Green I Master in a 10 µL reaction volume, including cDNA template, 0.2 µM of each primer, and SYBR Green I master mix. Thermal cycling consisted of 40 cycles of denaturation, annealing, and extension, followed by melting curve analysis. Relative expression was calculated using the ∆∆Ct method, normalized to the housekeeping gene tubulin. Reactions were performed in technical triplicates. The primer sequences used were: Tubulin: Fw: ATCCCCAACAACGTGAAGAC Rv: GCCTGAACATAGCGGTGAAC; Notch: Fw: CCGCCGTACTCGAATCAGTC; Rv: CGATGGCGAGCCCAAGTAG
Statistical analysis
Statistical analysis were performed on GraphPad Prism version 8 and RStudio (R version 4.2.3). The threshold for statistical significance was established as p < 0.05. Each experiment was independently repeated at least twice.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data generated in this study are provided in the Source Data file. Source data are provided with this paper.
References
Cho, B. et al. Single-cell transcriptome maps of myeloid blood cell lineages in Drosophila. Nat. Commun. 11, 4483 (2020).
Girard, J. R. et al. Paths and pathways that generate cell-type heterogeneity and developmental progression in hematopoiesis. eLife 10, e67516 (2021).
Banerjee, U., Girard, J. R., Goins, L. M. & Spratford, C. M. Drosophila as a genetic model for hematopoiesis. Genetics 211, 367–417 (2019).
Binggeli, O., Neyen, C., Poidevin, M. & Lemaitre, B. Prophenoloxidase activation is required for survival to microbial infections in drosophila. PLOS Pathog. 10, e1004067 (2014).
Shin, M. et al. Drosophila immune cells transport oxygen through PPO2 protein phase transition. Nature https://doi.org/10.1038/s41586-024-07583-x (2024).
Benmimoun, B., Polesello, C., Waltzer, L. & Haenlin, M. Dual role for insulin/TOR signaling in the control of hematopoietic progenitor maintenance in Drosophila. Development 139, 1713–1717 (2012).
Morin-Poulard, I. et al. Vascular control of the Drosophila haematopoietic microenvironment by Slit/Robo signalling. Nat. Commun. 7, 11634 (2016).
Terriente-Félix, A., Pérez, L., Bray, S. J., Nebreda, A. R. & Milán, M. A. Drosophila model of myeloproliferative neoplasm reveals a feed-forward loop in the JAK pathway mediated by p38 MAPK signalling. Dis. Model Mech. 10, 399–407 (2017).
Goyal, M., Tomar, A., Madhwal, S. & Mukherjee, T. Blood progenitor redox homeostasis through olfaction-derived systemic GABA in hematopoietic growth control in Drosophila. Development 149, dev199550 (2021).
Kapoor, A., Padmavathi, A., Madhwal, S. & Mukherjee, T. Dual control of dopamine in Drosophila myeloid-;like progenitor cell proliferation and regulation of lymph gland growth. EMBO Rep. 23, e52951 (2022).
Dragojlovic-Munther, M. & Martinez-Agosto, J. A. Multifaceted roles of PTEN and TSC orchestrate growth and differentiation of Drosophila blood progenitors. Development 139, 3752–3763 (2012).
Tian, Y., Morin-Poulard, I., Liu, X., Vanzo, N. & Crozatier, M. A mechanosensitive vascular niche for Drosophila hematopoiesis. Proc. Natl Acad. Sci. 120, e2217862120 (2023).
Cho, B. et al. Systemic control of immune cell development by integrated carbon dioxide and hypoxia chemosensation in Drosophila. Nat. Commun. 9, 2679 (2018).
Ferguson, G. B. & Martinez-Agosto, J. A. Yorkie and scalloped signaling regulates Notch-dependent lineage specification during Drosophila hematopoiesis. Curr. Biol. 24, 2665–2672 (2014).
Duvic, B., Hoffmann, J. A., Meister, M. & Royet, J. Notch signaling controls lineage specification during Drosophila larval hematopoiesis. Curr. Biol. 12, 1923–1927 (2002).
Lebestky, T., Jung, S. H. & Banerjee, U. A Serrate-expressing signaling center controls Drosophila hematopoiesis. Genes Dev. 17, 348–353 (2003).
Mukherjee, T., Kim, W. S., Mandal, L. & Banerjee, U. Interaction between Notch and Hif-alpha in development and survival of Drosophila blood cells. Science 332, 1210–1213 (2011).
Blanco-Obregon, D., Katz, M. J., Durrieu, L., Gándara, L. & Wappner, P. Context-specific functions of Notch in Drosophila blood cell progenitors. Dev. Biol. 462, 101–115 (2020).
Ho, K. Y. L., Carr, R. L., Dvoskin, A. D. & Tanentzapf, G. Kinetics of blood cell differentiation during hematopoiesis revealed by quantitative long-term live imaging. eLife 12, e84085 (2023).
Lebestky, T., Chang, T., Hartenstein, V. & Banerjee, U. Specification of Drosophila hematopoietic lineage by conserved transcription factors. Science 288, 146–149 (2000).
Cuervo, A. M. Autophagy: Many paths to the same end. Mol. Cell Biochem 263, 55–72 (2004).
Yorimitsu, T. & Klionsky, D. J. Autophagy: molecular machinery for self-eating. Cell Death Differ. 12, 1542–1552 (2005).
Feng, Y., He, D., Yao, Z. & Klionsky, D. J. The machinery of macroautophagy. Cell Res 24, 24–41 (2014).
Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 21, 439–458 (2020).
Berg, T. O., Fengsrud, M., Strømhaug, P. E., Berg, T. & Seglen, P. O. Isolation and characterization of rat liver amphisomes. evidence for fusion of autophagosomes with both early and late endosomes. J. Biol. Chem. 273, 21883–21892 (1998).
Nakamura, S. & Yoshimori, T. New insights into autophagosome-lysosome fusion. J. Cell Sci. 130, 1209–1216 (2017).
Fader, C. M., Sánchez, D., Furlán, M. & Colombo, M. I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250 (2008).
Bjørkøy, G. et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 171, 603–614 (2005).
Anding, A. L. & Baehrecke, E. H. Cleaning house: selective autophagy of organelles. Developmental Cell 41, 10–22 (2017).
Vázquez, P. et al. Atg5 and Ambra1 differentially modulate neurogenesis in neural stem cells. Autophagy 8, 187–199 (2012).
Zhang, J. et al. FRS2α-mediated FGF signals suppress premature differentiation of cardiac stem cells through regulating autophagy activity. Circ. Res 110, e29–e39 (2012).
Wang, S. et al. Autophagy-related gene Atg5 is essential for astrocyte differentiation in the developing mouse cortex. EMBO Rep. 15, 1053–1061 (2014).
Chen, X., He, Y. & Lu, F. Autophagy in stem cell biology: a perspective on stem cell self-renewal and differentiation. Stem Cells Int. 2018, 9131397 (2018).
Bankston, A. N. et al. Autophagy is essential for oligodendrocyte differentiation, survival, and proper myelination. Glia 67, 1745–1759 (2019).
Clarke, A. J. & Simon, A. K. Autophagy in the renewal, differentiation and homeostasis of immune cells. Nat. Rev. Immunol. 19, 170–183 (2019).
Varga, V. B. et al. Autophagy is required for spermatogonial differentiation in the Drosophila testis. Biol. Futur 73, 187–204 (2022).
Tan, J. Y. K., Chew, L. Y., Juhász, G. & Yu, F. Interplay between autophagy and CncC regulates dendrite pruning in Drosophila. Proc. Natl Acad. Sci. 121, e2310740121 (2024).
Riffelmacher, T. et al. Autophagy-dependent generation of free fatty acids is critical for normal neutrophil differentiation. Immunity 47, 466–480.e465 (2017).
Jacquel, A. et al. Autophagy is required for CSF-1-induced macrophagic differentiation and acquisition of phagocytic functions. Blood 119, 4527–4531 (2012).
Riffelmacher, T., Richter, F. C. & Simon, A. K. Autophagy dictates metabolism and differentiation of inflammatory immune cells. Autophagy 14, 199–206 (2018).
Metur, S. P. & Klionsky, D. J. Adaptive immunity at the crossroads of autophagy and metabolism. Cell Mol. Immunol. 18, 1096–1105 (2021).
Menshikov, M., Zubkova, E., Stafeev, I. & Parfyonova, Y. Autophagy, Mesenchymal stem cell differentiation, and secretion. Biomedicines 9, https://doi.org/10.3390/biomedicines9091178 (2021).
Hegedűs, K. et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol. Biol. Cell 27, 3132–3142 (2016).
Chang, Y. Y. & Neufeld, T. P. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol. Biol. Cell 20, 2004–2014 (2009).
Hounjet, J. & Vooijs, M. The Role of Intracellular Trafficking of Notch Receptors in Ligand-Independent Notch Activation. Biomolecules 11, https://doi.org/10.3390/biom11091369 (2021).
Matsuno, K., Diederich, R. J., Go, M. J., Blaumueller, C. M. & Artavanis-Tsakonas, S. Deltex acts as a positive regulator of Notch signaling through interactions with the Notch ankyrin repeats. Development 121, 2633–2644 (1995).
Yamada, K. et al. Roles of Drosophila deltex in Notch receptor endocytic trafficking and activation. Genes Cells 16, 261–272 (2011).
Shimizu, H., Hosseini-Alghaderi, S., Woodcock, S. A. & Baron, M. Alternative mechanisms of Notch activation by partitioning into distinct endosomal domains. J. Cell Biol. 223, e202211041 (2024).
Fostier, M., Evans, D. A., Artavanis-Tsakonas, S. & Baron, M. Genetic characterization of the Drosophila melanogaster Suppressor of deltex gene: A regulator of notch signaling. Genetics 150, 1477–1485 (1998).
Cornell, M. et al. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152, 567–576 (1999).
Rodrigues, D., Renaud, Y., VijayRaghavan, K., Waltzer, L. & Inamdar, M. S. Differential activation of JAK-STAT signaling reveals functional compartmentalization in Drosophila blood progenitors. Elife 10, https://doi.org/10.7554/eLife.61409 (2021).
Gao, C. et al. Autophagy negatively regulates Wnt signalling by promoting Dishevelled degradation. Nat. Cell Biol. 12, 781–790 (2010).
Mancias, J. D., Wang, X., Gygi, S. P., Harper, J. W. & Kimmelman, A. C. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 509, 105–109 (2014).
Simpson, J. E. et al. Autophagy supports PDGFRA-dependent brain tumor development by enhancing oncogenic signaling. Developmental Cell 59, 228–243.e227 (2024).
Le Borgne, R., Bardin, A. & Schweisguth, F. o. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development 132, 1751–1762 (2005).
Bray, S. J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 17, 722–735 (2016).
Huotari, J. & Helenius, A. Endosome maturation. EMBO J. 30, 3481–3500 (2011).
Vaccari, T., Lu, H., Kanwar, R., Fortini, M. E. & Bilder, D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol. 180, 755–762 (2008).
Schnute, B., Troost, T. & Klein, T. Endocytic trafficking of the notch receptor. Adv. Exp. Med Biol. 1066, 99–122 (2018).
van der Bliek, A. M. & Meyerowrtz, E. M. Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature 351, 411–414 (1991).
Bucci, C. et al. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70, 715–728 (1992).
Katzmann, D. J., Babst, M. & Emr, S. D. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106, 145–155 (2001).
Hurley, J. H. ESCRTs are everywhere. EMBO J. 34, 2398–2407 (2015).
Jia, L., Yu, G., Zhang, Y. & Wang, M. M. Lysosome-dependent degradation of Notch3. Int. J. Biochem. Cell Biol. 41, 2594–2598 (2009).
Yamamoto, S., Charng, W.-L. & Bellen, H. J. in Current Topics in Developmental Biology. 92, (ed. Raphael Kopan) 165–200 (Academic Press, 2010).
Williams, R. L. & Urbé, S. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8, 355–368 (2007).
Vietri, M., Radulovic, M. & Stenmark, H. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21, 25–42 (2020).
Zhao, Y. G., Codogno, P. & Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 22, 733–750 (2021).
Filimonenko, M. et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J. Cell Biol. 179, 485–500 (2007).
Razi, M., Chan, E. Y. W. & Tooze, S. A. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185, 305–321 (2009).
Behnia, R. & Munro, S. Organelle identity and the signposts for membrane traffic. Nature 438, 597–604 (2005).
Dunst, S. et al. Endogenously tagged rab proteins: a resource to study membrane trafficking in Drosophila. Developmental Cell 33, 351–365 (2015).
Akbar, M. A., Ray, S. & Krämer, H. The SM Protein Car/Vps33A Regulates SNARE-mediated Trafficking to Lysosomes and Lysosome-related Organelles. Mol. Biol. Cell 20, 1705–1714 (2009).
Rusten, T. E. et al. Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol. Biol. Cell 17, 3989–4001 (2006).
Itakura, E., Kishi-Itakura, C. & Mizushima, N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151, 1256–1269 (2012).
Takáts, S. et al. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J. Cell Biol. 201, 531–539 (2013).
Scott, R. C., Schuldiner, O. & Neufeld, T. P. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Developmental cell 7, 167–178 (2004).
Neufeld, T. P. TOR-dependent control of autophagy: biting the hand that feeds. Curr. Opin. Cell Biol. 22, 157–168 (2010).
Kim, J., Kundu, M., Viollet, B. & Guan, K.-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Lee, K. P. et al. Lifespan and reproduction in Drosophila: new insights from nutritional geometry. Proc. Natl Acad. Sci. 105, 2498–2503 (2008).
Scott, R. C., Juhász, G. & Neufeld, T. P. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr. Biol. 17, 1–11 (2007).
Colombani, J. et al. A nutrient sensor mechanism controls Drosophila growth. Cell 114, 739–749 (2003).
Hori, K. et al. Drosophila deltex mediates suppressor of Hairless-independent and late-endosomal activation of Notch signaling. Development 131, 5527–5537 (2004).
Schneider, M., Troost, T., Grawe, F., Martinez-Arias, A. & Klein, T. Activation of Notch in lgd mutant cells requires the fusion of late endosomes with the lysosome. J. Cell Sci. 126, 645–656 (2013).
Johnson, S. A., Zitserman, D. & Roegiers, F. Numb regulates the balance between Notch recycling and late-endosome targeting in Drosophila neural progenitor cells. Mol. Biol. Cell 27, 2857–2866 (2016).
Yan, Y., Denef N. Fau - Schüpbach, T. & Schüpbach, T. The vacuolar proton pump, V-ATPase, is required for notch signaling and endosomal trafficking in Drosophila. 3, 387–402 (2009).
Vaccari, T. et al. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor.137, 1825–32 (2020).
Tognon, E. et al. Control of lysosomal biogenesis and Notch-dependent tissue patterning by components of the TFEB-V-ATPase axis in Drosophila melanogaster.13, 499–514 (2016).
Riffelmacher, T. & Simon, A. K. Mechanistic roles of autophagy in hematopoietic differentiation. Febs j. 284, 1008–1020 (2017).
Warr, M. R. et al. FOXO3A directs a protective autophagy program in haematopoietic stem cells. Nature 494, 323–327 (2013).
Jensen, K. S. et al. FoxO3A promotes metabolic adaptation to hypoxia by antagonizing Myc function. Embo j. 30, 4554–4570 (2011).
Suda, T., Takubo, K. & Semenza, G. L. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell 9, 298–310 (2011).
Rožman, S. et al. The generation of neutrophils in the bone marrow is controlled by autophagy. Cell Death Differ. 22, 445–456 (2015).
Pengo, N. et al. Plasma cells require autophagy for sustainable immunoglobulin production. Nat. Immunol. 14, 298–305 (2013).
Terriente-Felix, A. et al. Notch cooperates with Lozenge/Runx to lock haemocytes into a differentiation programme. Development 140, 926–937 (2013).
Kushwah, R., Guezguez, B., Lee, J. B., Hopkins, C. I. & Bhatia, M. Pleiotropic roles of Notch signaling in normal, malignant, and developmental hematopoiesis in the human. EMBO Rep. 15, 1128–1138 (2014).
Láinez-González, D., Serrano-López, J. & Alonso-Dominguez, J. M. Understanding the Notch signaling pathway in acute myeloid leukemia stem cells: from hematopoiesis to neoplasia. Cancers (Basel) 14, https://doi.org/10.3390/cancers14061459 (2022).
Stier, S., Cheng, T., Dombkowski, D., Carlesso, N. & Scadden, D. T. Notch1 activation increases hematopoietic stem cell self-renewal in vivo and favors lymphoid over myeloid lineage outcome. Blood 99, 2369–2378 (2002).
Burns, C. E., Traver, D., Mayhall, E., Shepard, J. L. & Zon, L. I. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19, 2331–2342 (2005).
Mercher, T. et al. Notch signaling specifies megakaryocyte development from hematopoietic stem cells. Cell Stem Cell 3, 314–326 (2008).
Lee, D. et al. ER71 Acts Downstream of BMP, Notch, and Wnt Signaling in Blood and Vessel Progenitor Specification. Cell Stem Cell 2, 497–507 (2008).
Wang, L. et al. Notch-dependent repression of miR-155 in the bone marrow niche regulates hematopoiesis in an NF-κB-dependent manner. Cell Stem Cell 15, 51–65 (2014).
Suresh, S. & Irvine, A. E. The NOTCH signaling pathway in normal and malignant blood cell production. J. Cell Commun. Signal 9, 5–13 (2015).
Foronda, D., Weng, R., Verma, P., Chen, Y. W. & Cohen, S. M. Coordination of insulin and Notch pathway activities by microRNA miR-305 mediates adaptive homeostasis in the intestinal stem cells of the Drosophila gut. Genes Dev. 28, 2421–2431 (2014).
Barth, J. M. I., Hafen, E. & Köhler, K. The lack of autophagy triggers precocious activation of Notch signaling during Drosophila oogenesis. BMC Developmental Biol. 12, 35 (2012).
Wu, X. et al. Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat. Commun. 7, 10533 (2016).
Zhang, T., Guo, L., Wang, Y. & Yang, Y. Macroautophagy regulates nuclear NOTCH1 activity through multiple p62 binding sites. IUBMB Life 70, 985–994 (2018).
Zirin, J., Nieuwenhuis, J. & Perrimon, N. Role of autophagy in glycogen breakdown and its relevance to chloroquine myopathy. PLOS Biol. 11, e1001708 (2013).
Xu, T., Nicolson, S., Denton, D. & Kumar, S. Distinct requirements of autophagy-related genes in programmed cell death. Cell Death Differ. 22, 1792–1802 (2015).
Sopko, R. et al. Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in drosophila embryos. Developmental Cell 31, 114–127 (2014).
Evans, C. J., Liu, T. & Banerjee, U. Drosophila hematopoiesis: markers and methods for molecular genetic analysis. Methods 68, 242–251 (2014).
Acknowledgements
We are grateful to many colleagues of the Drosophila community, the Bloomington Stock Center and the Vienna Drosophila Resource Center for fly strains. A special thanks to Berta Pozzi and Anabella Srebrow from IFIByNE for their assistance with Proximity Ligation Assays, Andrés Rossi and Esteban Miglietta, from FIL microscopy facility, for technical support; Andrés Liceri for fly food preparation; the FIL personnel for assistance and members of the Wappner lab for fruitful discussions. This work was supported by grants from Agencia Nacional de Promoción de Científica y Tecnológica: PICT-2018-1501 and PICT-2021-I-A-00240 to PW and PICT-2019-0621 and PICT-2015-0225 to MJK.
Author information
Authors and Affiliations
Contributions
Conceptualization, M.J.K., F.R., F.E., N.S., M.M. and P.W.; Methodology, M.J.K., F.R., and F.E.; Formal Analysis, M.J.K., F.R., F.E., A.G.B., S.P.P., T.P., and N.S.; Investigation, M.J.K., F.R., F.E., A.G.B., S.P.P., T.P., and N.S.; Resources, M.J.K., G.L.B., M.M., and P.W.; Writing–Original Draft, P.W.; Writing–Review & Editing, M.J.K., F.R., F.E., A.G.B., S.P.P. and M.M.; Visualization, M.J.K., F.R., F.E., A.G.B., and T.P.; Supervision, M.J.K., G.L.B., M.M., and P.W.; Project Administration, M.J.K. and P.W.; Funding Acquisition, M.J.K. and P.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Thomas Vaccari, Tina Mukherjee, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Katz, M.J., Rodríguez, F., Evangelisti, F. et al. Autophagy controls differentiation of Drosophila blood cells by regulating Notch levels in response to nutrient availability. Nat Commun 16, 5858 (2025). https://doi.org/10.1038/s41467-025-58389-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58389-y