Abstract
D-type cyclins (hereafter, cyclin D) are central regulators orchestrating G1/S cell cycle transition. Accordingly, aberrant expression of cyclin D is strongly correlated with proliferation-related diseases such as cancer. However, the mechanisms regulating cyclin D turnover are incompletely elucidated. Here we identify FBXO32, namely atrogin-1, as the E3 ubiquitin ligase that targets all three cyclin D for ubiquitination and stabilization. Specifically, FBXO32 catalyzes the lysine (Lys/K)27-linked polyubiquitination of cyclin D1 at the K58 site and subsequent stabilization. Moreover, GSK-3β inactivation-mediated dephosphorylation of cyclin D1 facilitates its interaction with FBXO32 and subsequent ubiquitination. Furthermore, FBXO32 exhibits tumor-promoting effect in mouse models and increased FBXO32 is associated with poor prognosis of cancer patients. Additionally, disrupting the FBXO32-cyclin D axis enhances the tumor-killing effect of cyclin-dependent kinase (CDK)4/6 inhibitor palbociclib. Collectively, these findings reveal that FBXO32 enhances the protein stability of cyclin D via K27-linked ubiquitination, and contributes to cancer progression and the limited response of cancer cells to CDK4/6 inhibitors.
Similar content being viewed by others
Introduction
D-type cyclins, containing three isoforms cyclin D1, D2, and D3, which share high structural and functional similarities, play a pivotal role in defining cell cycle exit and progression by regulating G1/S cell cycle transition1. The dysregulation of D-type cyclins, especially cyclin D1, has been proved to be correlated with various proliferation-related diseases such as cancer2,3,4. Apart from regulating cell cycle, cyclin D have a much broader tumor-promoting effects, including invasion and metastasis5, angiogenesis6, cancer metabolism7,8, anti-senescence properties9 and immune evasion10,11.
D-type cyclins play a key role in transmitting signals from extracellular stimulations to the downstream effectors to exert different kinds of cell functions1. Cyclin D act as a direct target and vital growth factor sensor of mitogenic signaling cascades, including Ras/Raf/MAPK12,13, phosphatidylinositol 3-kinase (PI3K)-Akt14,15,16, and β-catenin/TCF-LEF pathways17,18. After receiving mitogenic signals, cyclin D heterodimerize with CDK4/6 to form kinase complexes, phosphorylating and inactivating retinoblastoma (Rb) protein, which leads to the release and nuclear translocation of the E2F transcription factor, thereafter, the expression of downstream genes to drive cell cycle progression19. In addition to the Rb family members, a plethora of substrates such as the transforming growth factor-β (TGFβ)-responsive transcriptional modulator SMAD3, FOXM1 and the MEF2 family have been reported to be phosphorylated by cyclin D-CDK4/6 heterodimer to drive cell growth or migration20,21,22. Besides that, cyclin D itself can bind with a large number of functional molecules including p27 and STAT3 in a CDK4 or CDK6 independent manner to regulate cell migration and cell cycle progression23,24. Notably, it has been demonstrated that overexpression of cyclin D1 is a driving feature in various cancer types, including leukemia, head and neck, breast, non–small cell lung cancer, and prostate25,26. Given the importance of cyclin D in cell cycle and cancer progression regulation, there is no doubt that a better understanding of the regulatory mechanisms for cyclin D will provide critical information on how to block cell cycle progression and related processes in cancer development.
Ubiquitin-proteasome system (UPS) pathway plays a vital role in regulating the protein level of cyclin D based on the fact the D-type cyclins are highly labile proteins with a half-life of less than 30 min. Over the past decades, a variety of ubiquitin ligases including FBXO4, FBXO31, FBXW8, β-TRCP, SKP2, FBXL2, FBXL8 and the anaphase-promoting complex/cyclosome (APC/C) have been reported to target cyclin D for degradation19. Importantly, AMBRA1 was identified as the master regulator that ubiquitinates and degrades cyclin D27,28,29. More recently, MG53 (also named TRIM72) was found to facilitate the ubiquitination and degradation of cyclin D130. These studies mainly focus on ubiquitination-mediated degradation of cyclin D; nevertheless, it is still unclear for the role of nondegradative form of ubiquitination in regulating cyclin D function.
Here, we define an important role of lysine (Lys/K)27-linked polyubiquitination in enhancing the protein stability of cyclin D. Especially, we identify FBXO32 as a bona fide ubiquitin ligase facilitating the K27-linked ubiquitination of cyclin D at K58 site, and investigate the role of this nondegradative form of ubiquitination in cyclin D stabilization and the tumorigenesis. Moreover, GSK-3β-mediated threonine (Thr/T)-286 and Thr-288 phosphorylation of cyclin D1 has been proved to participate in the process of FBXO32-mediated cyclin D1 ubiquitination and stabilization. These findings reveal complicated layers of cyclin D turnover regulation coordinated by FBXO32-mediated cyclin D ubiquitination to drive tumorigenesis. Therefore, targeting the FBXO32-cyclin D axis may be a potential strategy for suppressing tumorigenesis and sensitizing cancer cells to CDK4/6 inhibitors.
Results
FBXO32 physically interacts with cyclin D1
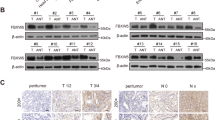
To determine the role of nondegradative form of ubiquitination in regulating cyclin D function, we constructed two mutants of cyclin D1 (namely cyclin D1T286A and cyclin D1T288A) insensitive to ubiquitin-mediated recognition and degradation and tested their ubiquitination levels14,19,29,31,32. Strikingly, the dephosphorylation state of cyclin D1, cyclin D1T286A and cyclin D1T288A, remains to sustain a high level of ubiquitination (Fig. 1a), suggesting that there exist(s) additional ubiquitin ligase(s) regulating cyclin D1 function rather than triggering degradation. We then explored the potential E3 ligases that bind with both cyclin D1T286A and cyclin D1T288A. Proteomics approach and co-immunoprecipitation (Co-IP) assay revealed that FBXO32 interacted with both cyclin D1T286A and cyclin D1T288A (Fig. 1b and Supplementary Fig. 1a, b). Moreover, Co-IP assay showed that endogenous cyclin D1 could be immunoprecipitated by FBXO32 and vice versa (Fig. 1c). To verify the above findings, Huh7 cells were transfected with Flag-FBXO32, HA-cyclin D1, or both, followed by IP assays using anti-Flag or anti-HA antibodies. In this manner, we confirmed the interaction between FBXO32 and cyclin D1 (Fig. 1d). Furthermore, immunofluorescent staining assay revealed that cyclin D1 and FBXO32 mainly colocalized in the nucleus of the cells, and this colocalization was significantly enhanced after FBXO32 overexpression (Fig. 1e). These assays revealed that FBXO32 interacts with cyclin D1. To explore whether FBXO32 binds to cyclin D1 directly, glutathione-S-transferase (GST) pull-down assay was utilized and the result showed that GST-tagged FBXO32 efficiently coprecipitated HA-cyclin D1 from cell lysates (Fig. 1f). In addition, Surface Plasmon Resonance (SPR) assay also indicated a direct interaction between FBXO32 and cyclin D1 with a high binding activity (KD = 3.25 × 10−6 M) (Fig. 1g). Collectively, these results indicated that FBXO32 physically interacts with cyclin D1. Lastly, we mapped the interaction domains between FBXO32 and cyclin D1 using a series of truncations. As shown in Fig. 1h, FBXO32 mutant (mutant 1–115), but not mutant 1–60, could interact with HA-tagged cyclin D1, indicating that cyclin D1 interacts with domain 60–115 of FBXO32. Meanwhile, we found that domain 95–162 of cyclin D1 was responsible for binding with FBXO32 (Fig. 1i). Taken together, our results indicate that FBXO32 directly interacts with cyclin D1.
a 293T cells were co-transfected with His-Ub and wild-type (WT) HA-cyclin D1, or various HA-cyclin D1 mutants (T286A and T288A), or a Vector control for 48 h. Then the cells were lysed with ubiquitination assay lysis buffer, followed by pull-down using Ni-NTA beads, and the precipitates were analyzed by western blot. b Schematic of the screening workflow to identify FBXO32 as a protein candidate interacting with dephosphorylated cyclin D1. c Co-immunoprecipitation (Co-IP) assay was utilized to determine the interaction between endogenous FBXO32 and cyclin D1 in Huh7 cells. IgG indicates the control antibody group. d Co-IP analysis of the interaction between Flag-tagged FBXO32 and HA-tagged cyclin D1 in Huh7 cells transfected with the indicated expression constructs. e Huh7 or CFPAC-1 cells were transfected with Flag-FBXO32 plasmid or control Vector for 48 h. Immunofluorescence assay was utilized to measure the colocalization of FBXO32 and cyclin D1. Hoechst was used to stain DNA. Scale bar, 10 μm. f Huh7 cells transfected with HA-cyclin D1 construct were lysed and incubated with GST or GST-FBXO32 conjugated to beads. Pull-down samples and 5% of the input were analyzed by western blot and Ponceau S staining. g SPR assay revealed the kinetic interaction between His-tagged recombinant human FBXO32 and cyclin D1. h 293T cells were co-transfected with WT HA-cyclin D1 and various domain-deleted Flag-FBXO32 plasmids for 48 h. MG132 (10 μM) was added to incubate with the cells for 4 h before being harvested. Then co-IP assay was performed using an anti-HA antibody to pull down HA-tagged cyclin D1, followed by western blot analysis. i WT HA-cyclin D1 and cyclin D1 truncation mutants were co-transfected with Flag-FBXO32 into 293T cells for 48 h. MG132 was added to incubate with the cells for 4 h before being harvested. Then, cells were collected for the co-IP assay using anti-Flag antibody, followed by immunoblotting. Representative immunoblot was shown from 3 biologically independent experiments (a, c, d, f, h, i). Source data are provided as a Source Data file. WCE whole cell extraction, OE overexpression.
FBXO32 upregulates D-type cyclins protein level by ubiquitin-mediated stabilization
The interaction between the E3 ubiquitin ligase FBXO32 and cyclin D1 prompts us to explore the resulting influence on cyclin D1 protein level. Interestingly, ectopic expression of FBXO32 upregulated the protein level of cyclin D1 in a dose-dependent manner (Fig. 2a). Conversely, knockdown of FBXO32 decreased the protein level of cyclin D1 (Fig. 2b). Moreover, neither FBXO32 overexpression (Supplementary Fig. 2a, b) nor FBXO32 knockdown (Supplementary Fig. 2c) affected the mRNA level of cyclin D1 in Huh7 and CFPAC-1 cells, suggesting that FBXO32 upregulates cyclin D1 through post-translational or translational regulation. Furthermore, immunoblotting analysis showed that proteasome inhibitor (MG132) weakened silencing FBXO32-mediated downregulation of cyclin D1 (Supplementary Fig. 2d), indicating that proteasome was involved in the regulation of cyclin D1 by FBXO32. We further determined whether FBXO32 upregulates cyclin D1 via its reported E3 ligases. As shown in Supplementary Fig. 3a, the protein levels of those reported ubiquitin ligases for cyclin D (FBXO4, FBXO31, FBXW8, β-TRCP, SKP2, FBXL2, FBXL8, APC/C, MG53 and AMBRA1) displayed no obvious changes after FBXO32 overexpression, implying that these reported ubiquitin ligases are not involved in FBXO32-mediated cyclin D1 stabilization. Meanwhile, we found that the binding of FBXO32-cyclin D1 is stronger than that of AMBRA1-cyclin D1 in Huh7 cells (Supplementary Fig. 3b), suggesting that FBXO32 may play an important role in sustaining the balance of cyclin D1 protein level between degradation and stabilization.
a Huh7 and CFPAC-1 cells were transfected with the indicated concentrations of plasmids expressing Flag-FBXO32 and negative control for 48 h, followed by western blot analysis to detect the protein levels of cyclin D1. b After transfection with FBXO32-specific shRNA or negative control (sh-NC) for 48 h, Huh7 and CFPAC-1 cells were lysed for western blot analysis to determine cyclin D1 protein levels. c 293T cells were co-transfected with His-Ub and HA-cyclin D1 expression plasmid together with either a Vector control, Flag-FBXO32 or FBXO32ΔF plasmid for 48 h, followed by ubiquitination assay. d, e Huh7 (d) and CFPAC-1 (e) cells were transfected with Flag-tagged FBXO32 expression plasmid (Flag-FBXO32) or negative control (Vector) for 48 h, followed by treatment with 100 μg/mL cycloheximide (CHX) for the indicated times. The protein level of cyclin D1 was detected by western blot and quantified using Image J software. f, g After co-transfection with plasmids expressing HA-cyclin D1 and Flag-FBXO32 or negative control (Vector) for 48 h, Huh7 (f) and CFPAC-1 (g) cells were treated as in (d). The protein level of HA-cyclin D1 was measured by western blot and quantified using Image J software. h, i Plasmids expressing HA-cyclin D2 or HA-cyclin D3 were co-transfected with Flag-FBXO32 or negative control (Vector) into Huh7 cells for 48 h, followed by treatment as in (d). The protein levels of HA-cyclin D2 (h) and HA-cyclin D3 (i) were measured by western blot and quantified using Image J software. j 293T cells were co-transfected with Flag-FBXO32 and HA-cyclin D1 together with WT His-Ub or various His-Ub mutants (M1, K6, K11, K27, K29, K33, K48, K63) for 48 h, followed by ubiquitination assay. k 293T cells were co-transfected with Flag-FBXO32 and HA-cyclin D1 together with WT His-Ub or His-Ub mutant (K27R), followed by ubiquitination assay. Representative immunoblot was shown from 3 biologically independent experiments (a–k). Data are presented as mean ± SEM, and P-values were calculated using two-tailed unpaired Student’s t-test (d–i). Source data are provided as a Source Data file. LV lentivirus.
Subsequently, we explored whether FBXO32 upregulates cyclin D1 via direct ubiquitination. As expected, ectopic expression of FBXO32 increased the ubiquitination level of cyclin D1, whereas an F-box domain, essential for the formation of Skp1–Cullin1–F-box (SCF) E3 ligase complexes, deletion mutant of FBXO32 (hereafter referred as FBXO32ΔF) was unable to elevate cyclin D1 ubiquitination as much as the full-length FBXO32 (Fig. 2c). To exclude the possibility that FBXO32 promotes cyclin D1 ubiquitination through an E3 ligase-independent way, siRNA specifically targeting cullin1 necessary for sustaining the E3 ligase activity of FBXO32 was transfected into Huh7 cells in the presence of Flag-FBXO32 or not. The results showed that cullin1 knockdown completely abrogated FBXO32-mediated ubiquitination of cyclin D1 (Supplementary Fig. 4a). Meanwhile, Co-IP assay showed a slight interaction between FBXO32ΔF and cullin1 in Huh7 cells (Supplementary Fig. 4b), which may be the reason why FBXO32ΔF cannot abolish the ubiquitination of cyclin D1 completely. Together, these results indicated that FBXO32 upregulated cyclin D1 through an E3 ligase-dependent manner. Considering that there are three D cyclins in mammals (namely cyclins D1, D2, and D3), we further detected the effect of FBXO32 to cyclin D2 and D3. As shown in Supplementary Fig. 5a–c, ectopic expression of FBXO32 enhanced the protein and ubiquitination levels of cyclins D2 and D3. These results implied that FBXO32 may upregulate cyclin D via ubiquitination.
Next, we explored whether FBXO32 influences the protein stability of cyclin D. The cycloheximide (CHX) pulse-chase assay showed that ectopic expression of FBXO32 prolonged the half-life of endogenous cyclin D1 in both Huh7 and CFPAC-1 cells (Fig. 2d, e). Consistently, FBXO32 had a similar effect on exogenously expressed cyclin D1 with HA tag (Fig. 2f, g). Moreover, we also observed that FBXO32 extended the half-lives of cyclin D2 and cyclin D3 (Fig. 2h, i). Collectively, these results indicated that FBXO32 delayed the turnover of all three D-type cyclins.
Based on the fact that ubiquitin can be conjugated by another ubiquitin through any of its seven Lys residues (K6, K11, K27, K29, K33, K48, and K63) or, alternatively, the methionine (Met/M)1, we then characterized which of these eight linkage types is responsible for FBXO32-mediated cyclin D1 ubiquitination by generating a panel of ubiquitin mutants which substitutes arginine(Arg/R) for lysine residues at all sites except the indicated one. The ubiquitination assay revealed that FBXO32-mediated K27-linked ubiquitination of cyclin D1 (Fig. 2j). Complementarily, K27R ubiquitin, in which only K27 was replaced with arginine, was unable to facilitate FBXO32-mediated cyclin D1 ubiquitination modification (Fig. 2k). Collectively, these results indicated that FBXO32 stabilizes cyclin D1 protein via K27-linked polyubiquitination.
FBXO32 mediates cyclin D1 protein stabilization by ubiquitination at lysine 58
To identify the ubiquitination sites of cyclin D1, Flag-FBXO32 and HA-cyclin D1 plasmids were transfected into 293T cells for 48 h and then HA-cyclin D1 was immunoprecipitated and subjected to LC-MS/MS analysis (Fig. 3a). An analysis of purified HA-cyclin D1 protein using mass spectrometry revealed that 5 lysine residues (K58, K72, K114, K123 and K180) of cyclin D1 might be conjugated to ubiquitin (Fig. 3b and Supplementary Fig. 6). We mutated all five Lys residues to Arg respectively, and observed that only K58R mutant displayed an obvious reduction of cyclin D1 ubiquitination induced by FBXO32, whereas other mutants showed no obvious changes (Fig. 3c). Moreover, overexpression of Flag-FBXO32 led to an apparent increase in the protein level of wild-type cyclin D1 but had no obvious effect on the K58R mutant (Fig. 3d). Additionally, cyclin D1K58R showed a markedly decreased half-life than WT cyclin D1 in the presence of FBXO32 (Fig. 3e, f), which further strengthens that FBXO32 ubiquitinates cyclin D1 at K58 site and leads to the stabilization of cyclin D1.
a Schematic of the workflow for identifying the potential ubiquitination sites of cyclin D1 influenced by FBXO32 with mass spectrometry analysis. Schematic illustration was created in BioRender (https://BioRender.com/m10t423). b Lysine 58 (K58) ubiquitination sites in cyclin D1 identified by mass spectrometry analysis. c 293T cells were co-transfected with plasmids expressing Flag-FBXO32 and His-Ub together with WT HA-cyclin D1 or various mutants HA-cyclin D1 (K58R, K72R, K114R, K123R, K180R) for 48 h. Then the cells were lysed with ubiquitination lysis buffer, followed by pull-down using Ni-NTA beads, and the precipitates were analyzed by western blot. d Flag-FBXO32 or control plasmid was co-transfected with HA-cyclin D1 WT or K58R in Huh7 and CFPAC-1 cells, followed by immunoblotting with indicated antibodies. e, f Huh7 cells (e) and CFPAC-1 cells (f) were co-transfected with Flag-FBXO32 and WT HA-cyclin D1 or mutant HA-cyclin D1K58R for 48 h, followed by treatment with 100 μg/mL CHX for the indicated times. The protein levels of HA-cyclin D1 and HA-cyclin D1K58R were detected by western blot and quantified using Image J software. The immunoblotting experiments were repeated three times with similar results (c–f). Data are presented as mean ± SEM, and P-values were calculated using two-tailed unpaired Student’s t-test (e, f). Source data are provided as a Source Data file.
Interestingly, we observed that cyclin D1K58R seems to be more susceptible to degradation (Fig. 3e, f). Therefore, we investigated whether the degradation of cyclin D1K58R is dependent on the phosphorylation of T286 and T288, which is important for cyclin D1WT degradation. First, we assessed the phosphorylation of T286 and T288 of cyclin D1K58R and found that there is no obvious elevation of pT286 (or pT288) of cyclin D1K58R along with its degradation (Supplementary Fig. 7a). Second, the double points mutant plasmids of cyclin D1 (carrying K58R and T286A or K58R and T288A) were constructed and co-transfected with Flag-FBXO32 into Huh7 cells. The results revealed that neither T286A nor T288A mutation rescued the protein level of cyclin D1K58R (Supplementary Fig. 7b). These results indicated that the degradation of cyclin D1K58R is not dependent on the phosphorylation of T286 or T288. Furthermore, we observed that the proteasome inhibitor MG132, not CQ (chloroquine, autophagy inhibitor), rescued the degradation of cyclin D1 K58R mutant (Supplementary Fig. 7c), indicating that proteasome participates in the degradation of cyclin D1K58R. Additionally, we have determined the half-life of cyclin D1K58R in various liver cancer cells with different levels of FBXO32. The results showed that HepG2 cell has the lowest FBXO32 level (even cannot be detected), while SNU475 has the highest FBXO32 level (MeWo cell line was utilized as positive control33) (Supplementary Fig. 8a–c). Moreover, we found that K58R mutation shortened the half-life of cyclin D1 in Huh7 and SNU475 cells, but had no obvious changes in HepG2 cells (Supplementary Fig. 8d–f). These results indicated that FBXO32 stabilized cyclin D1 via ubiquitination at K58 site only in FBXO32-expressing cells; in other words, the K58 ubiquitination-mediated stabilization of cyclin D1 protein is dependent on FBXO32 levels.
GSK-3β inactivation-mediated dephosphorylation of cyclin D1 facilitates FBXO32-induced cyclin D1 protein ubiquitination and stabilization
While previous studies have demonstrated that phosphorylation or dephosphorylation plays an important role in E3 ligase-mediated substrate recognition, we further explored whether phosphorylation participates in FBXO32-mediated cyclin D1 ubiquitination and stabilization. As expected, either T286A or T288A mutation of cyclin D1, mimicked the dephosphorylation state of cyclin D1, enhanced its interaction with FBXO32 (Fig. 4a). Moreover, we observed a marked enhancement of ubiquitination after T286A or T288A mutation of cyclin D1 in the presence of FBXO32 (Fig. 4b). Conversely, T286D or T288D mutation of cyclin D1, mimicked the phosphorylation of cyclin D1, attenuated FBXO32-induced ubiquitination of cyclin D1 (Supplementary Fig. 9). These results indicated that FBXO32 stabilizes and ubiquitinates cyclin D1 by recognizing dephosphorylated cyclin D1. Furthermore, CHX pulse-chase experiment revealed that dephosphorylation of cyclin D1 reinforced FBXO32-mediated cyclin D1 protein stabilization (Fig. 4c, d and Supplementary Fig. 10a, b). In addition, we found that both mutants of cyclin D1 (HA-cyclin D1T286A and HA-cyclin D1T288A) displayed a marked increase of ubiquitination and a prolonged half-life after ectopic expression of FBXO32 (Fig. 4e–h), indicating that not only the WT, but also dephosphorylated mutants of cyclin D1 were ubiquitinated and stabilized by FBXO32.
a 293T cells were co-transfected with Flag-FBXO32 and WT HA-cyclin D1, or various HA-cyclin D1 mutants (T286A, T288A) or a Vector control for 48 h, followed by co-IP assay. b 293T cells were co-transfected with Flag-FBXO32 and His-Ub together with WT HA-cyclin D1, various HA-cyclin D1 mutants (T286A, T288A) or a Vector control for 48 h, then the cells were lysed to detect ubiquitinated cyclin D1. c, d Huh7 and CFPAC-1 cells were co-transfected with Flag-FBXO32 and WT HA-cyclin D1 or various HA-cyclin D1 mutants for 48 h, followed by treatment with 100 μg/mL CHX for the indicated times, then cyclin D1 was determined by western blotting and quantified by Image J software. e, f After co-transfection with His-Ub and HA-cyclin D1T286A (or HA-cyclin D1T288A) together with Flag-FBXO32 or a Vector control for 48 h, 293T cells were lysed for ubiquitination assay. g, h Huh7 cells were co-transfected with HA-cyclin D1T286A (g) or HA-cyclin D1T288A (h) and Flag-FBXO32 or a Vector control for 48 h, followed by a CHX pulse-chase assay. i 293T cells were co-transfected with HA-cyclin D1 and His-Ub together with Flag-FBXO32 or a Vector control for 36 h, followed by treatment with GSK-3β inhibitor TWS119 (5 μM), DYRK1B inhibitor AZ191 (1 μM) or MEK1/2 inhibitor trametinib (100 nM) for another 12 h, followed by ubiquitination assay. j After co-transfected with HA-cyclin D1, His-Ub and Flag-FBXO32 together with GSK-3β or negative control siRNA for 48 h, 293T cells were lysed for ubiquitination assay. k 293T cells were co-transfected with HA-cyclin D1, His-Ub and Flag-FBXO32 together with WT GSK-3β or various GSK-3β mutants, followed by ubiquitination assay. l 293T cells were transfected with WT GSK-3β, or various GSK-3β mutants, or negative control vector for 48 h, followed by GSK-3β activity measurement. (n = 3 independent experiments). Representative immunoblot was shown from 3 biologically independent experiments (a–k). Data are presented as mean ± SEM, and P-values were calculated using one-way ANOVA with Dunnett’s (c, d) or Tukey’s (l) multiple comparisons or two-tailed unpaired Student’s t-test (g, h). Source data are provided as a Source Data file.
Next, we determined which phosphokinase participated in the regulation of FBXO32-mediated cyclin D1 ubiquitination. Considering that three kinases (GSK-3β, DYRK1B, and MEK1/2) have been reported to participate in the phosphorylation of cyclin D1 at T286 or T288, three inhibitors (TWS119, AZ191 and trametinib) separately targeting GSK-3β, DYRK1B and MEK1/2 were utilized. The ubiquitination assay showed a GSK-3β inhibition enhanced FBXO32-induced ubiquitination of cyclin D1 (Fig. 4i). Consistently, knockdown of GSK-3β with small interference RNA (siRNA) markedly promoted the ubiquitination of cyclin D1 induced by FBXO32 (Fig. 4j). By contrast, GSK-3β overexpression attenuated FBXO32-mediated ubiquitination of cyclin D1 (Fig. 4k). Furthermore, previous researches have reported that the serine(Ser/S)9 phosphorylation of GSK-3β plays a vital role in regulating cyclin D1 phosphorylation34. Thus, we constructed constitutively active mutant GSK-3βS9A (where serine 9 to alanine substitution mimics the dephosphorylation) and constitutively inactive mutant GSK-3βS9E which substitutes glutamate for serine 9 residues to mimic the phosphorylation of GSK-3β successfully (Fig. 4l). As shown in Fig. 4k, l, ectopic expression of GSK-3βS9A markedly alleviated FBXO32-induced cyclin D1 ubiquitination, whereas GSK-3βS9E overexpression had an opposite effect. Taken together, these results indicated that GSK-3β inactivation-dependent dephosphorylation of cyclin D1 facilitates its recognition and ubiquitination by FBXO32.
FBXO32 promotes tumor growth and metastasis in vitro and in vivo
We further explored the biological effect of FBXO32 in vivo and in vitro. First, we constructed orthotopic xenograft tumor models using hepatocellular carcinoma (HCC) and pancreatic ductal adenocarcinoma (PDAC) cell lines with stable knockdown or overexpression of FBXO32. As shown in Fig. 5a, b, knockdown of FBXO32 dramatically inhibited the tumor growth of liver cancer, as indicated by smaller tumor volume and lower proliferation marker Ki-67. Interestingly, we observed a lower incidence of lung metastasis in FBXO32-knockdown mice compared with the negative control (Fig. 5c). Consistently, FBXO32 knockdown also suppressed the tumor growth of PDAC orthotopic xenograft (Fig. 5d, e), as well as liver metastasis of pancreatic cancer (Fig. 5f). By contrast, overexpression of FBXO32 had an opposite effect on tumor growth and metastasis. As shown in Supplementary Fig. 11a–h, overexpression of FBXO32 leaded to a larger tumor volume (Supplementary Fig. 11a, c), higher positive rate of Ki-67 (Supplementary Fig. 11b, d) and metastasis rate (lung metastasis for liver cancer and liver metastasis for pancreatic cancer) (Supplementary Fig. 11e–h). These results indicated that FBXO32 promoted tumor growth and metastasis in orthotopic xenograft tumor models.
a–f Huh7 or CFPAC-1 cells (5 × 106) stably expressing sh-FBXO32 or sh-NC were injected subcutaneously to axilla of BALB/c nude mice. After 4 weeks, the masses of tumor formed from Huh7 or CFPAC-1 cells were divided equally into 1 mm3 pieces and implanted orthotopically into BALB/c nude mice (n = 6). The mice were sacrificed 4 weeks later, then the tumor volumes were monitored (a, d). H&E staining was performed to determine the tumor tissue, and immunohistochemistry (IHC) staining was utilized to measure the protein levels of FBXO32 and Ki-67 (b, e). Scale bar, 50 μm. Representative lung (or liver) metastasis nodule images were presented, and the incidence of lung (or liver) metastasis from liver (or pancreatic) cancer was calculated. H&E and IHC staining were used to determine the metastatic tumor from liver or pancreas (c, f). Top scale bar, 200 μm, middle and bottom scale bar, 50 μm. g, h Hepa1-6 cells with constitutively expressing luciferase were injected orthotopically into C57BL/6N mice (n = 4). Then AAV-shFbxo32 and AAV-shNC were injected intraperitoneally into the mice after one week. Bioluminescence device was utilized to monitor the tumor size at the first and fifth week, and representative bioluminescence images were shown (g). QPCR was performed to detect Fbxo32 mRNA level of tumor tissues from three groups of mice (h). i–k CCK-8 and colony formation assays were used to measure the proliferation of Huh7 and CFPAC-1 cells after infected with LV-shFBXO32 and LV-shNC, or transfected with OE-FBXO32 and control plasmids. (n = 3 independent experiments). l, m Migration (l) and invasion (m) assays were performed to determine the metastasis of Huh7 and CFPAC-1 cells with stable expression of sh-FBXO32 or sh-NC by lentivirus infection. (n = 3 independent experiments). Scale bar, 100 μm. Data are represented as mean ± SEM and statistical analysis was performed using two-tailed Student’s t-test (a, b, d, e, i–m) or one-way ANOVA with Dunnett’s multiple comparisons (g, h). Source data are provided as a Source Data file. AAV adeno-associated virus.
Moreover, we utilized a DEN/CCl4-induced primary liver cancer mouse model to test the tumor-suppressive effect of Fbxo32 knockdown using adeno-associated virus (AAV8)-mediated shRNA delivery. As shown in Supplementary Fig. 12a–f, AAV8-mediated Fbxo32 knockdown leads to the inhibition of liver cancer growth and lung metastasis, as well as proliferation marker Ki-67 and cyclin D1. Meanwhile, the orthotopic tumor mouse model was constructed using mouse HCC cell line Hepa1-6, constitutively expressing luciferase, to test the therapeutic effect of targeting Fbxo32 using AAV8-mediated shRNA delivery. As shown in Fig. 5g, h, suppression of Fbxo32 expression resulted in smaller primary liver tumors, suggesting that Fbxo32 could be regarded as a novel potential target for cancer treatment. Regarding pancreatic cancer, we also observed a similar result in the KPC (genotype: LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre) mouse model, a widely used mouse model developing pancreatic cancer spontaneously with KRAS activation and p53 inactivation. We found that the KPC mice injected with AAV8-shFbxo32 showed decreased tumor growth and liver metastasis, as well as proliferation marker Ki-67 and cyclin D1 (Supplementary Fig. 12g–k). These results indicated that Fbxo32 promotes tumor growth and metastasis in vivo.
Additionally, we tested the tumor-promoting role of FBXO32 in vitro. First, CCK-8 assay revealed that knockdown of FBXO32 inhibited the growth of Huh7 and CFPAC-1 cells, whereas overexpression of FBXO32 had the opposite effects (Fig. 5i). Second, colony formation assay showed that inhibition of FBXO32 expression resulted in a decreased number of colonies while ectopic expression of FBXO32 upregulated cell colonies (Fig. 5j, k). Third, trans-well assays represented that cell migration and invasion were significantly reduced upon FBXO32 knockdown and enhanced upon FBXO32 overexpression (Fig. 5l, m and Supplementary Fig. 13a, b). Lastly, unbiased RNA-seq has been utilized to show the effect of FBXO32 loss on pathway deregulation using FBXO32 gene knockout Huh7 cell line (KO-FBXO32-Huh7) (Supplementary Fig. 14a–c). The downregulated top 100 genes expression results were shown as in Supplementary Fig. 14d. Subsequently, a deeper investigation of the downregulated genes uncovered a major enrichment of cell cycle-related genes (Supplementary Fig. 14e), which was further consolidated with qPCR (Supplementary Fig. 14f). Collectively, these results indicated that FBXO32 exerts a tumor-promoting role in HCC and PDAC.
FBXO32-mediated cyclin D elevation activates CDK4/6 and drives tumor development
D-type cyclins have been shown to initiate cell division via forming a complex with CDK4/6 and activating CDK4/6, which subsequently phosphorylates and inactivates the retinoblastoma tumor suppressor. Accordingly, increased level and activity of cyclin D-CDK4/CDK6 complex are critical for unchecked cell proliferation of tumor cells. Hence, we tested whether FBXO32-mediated cyclin D1 upregulation could activate CDK4/6 kinase. As shown in Fig. 6a, b, in the presence of FBXO32, more cyclin D bound to CDK4 and CDK6. Furthermore, western blot (WB) revealed that FBXO32 overexpression enhanced the protein levels of phosphorylated Rb (p-RbS780, p-RbS795, p-RbS807/811) (Fig. 6c). Moreover, in vitro kinase assay revealed that CDK4 or CDK6 kinase activity (reflected by phosphorylated Rb) showed no obvious changes after FBXO32 recombinant protein replenishment (Supplementary Fig. 15a, b), suggesting that FBXO32 itself has little effect on CDK4/6 kinase without cyclin D1 elevation. Meanwhile, we performed another kinase assay for Rb by immune-purified endogenous CDK4/6 with or without FBXO32 overexpression. The results showed that more cyclin D1 bound to CDK4 and CDK6 after FBXO32 overexpression in 293T cells, which contributes to the enhancement of CDK4/6 kinase activity (reflected by elevated phosphorylated Rb) (Supplementary Fig. 15c, d). Taken together, these results indicated that FBXO32 activated the CDK4/6-Rb signaling pathway via upregulating D-type cyclins.
a, b 293T cells were transfected with Flag-tagged FBXO32 expression plasmid (Flag-FBXO32) or negative control (Vector) for 48 h. The co-IP assay was performed using anti-CDK4 (a) or anti-CDK6 (b) antibody to pull down the target protein complex, followed by western blot analysis. The immunoblotting experiments were repeated three times with similar results. c Huh7 and CFPAC-1 cells were transfected with the indicated concentrations of plasmids expressing Flag-FBXO32 and negative control (Vector) for 48 h, followed by western blot analysis to detect phosphorylated Rb. The immunoblotting experiments were repeated three times with similar results. d CCK-8 assays were performed to measure the proliferation of Huh7 and CFPAC-1 cells transfected with Flag-FBXO32 or Vector plasmids in the presence of palbociclib (10 μM) or not. (n = 3 independent experiments). e Cell cycle analysis of Huh7 and CFPAC-1 cells with stable expression of sh-FBXO32 or sh-NC using lentivirus was performed using flow cytometry in the absence or presence of nocodazole (0.33 μM) for 16 h. (n = 3 independent experiments). f, g Trans-well assays were performed to measure migration (f) and invasion (g) of Huh7 and CFPAC-1 cells treated as described in (d). Scale bar, 100 μm. (n = 3 independent experiments). Data are represented as mean ± SEM, and statistical analysis was performed using one-way ANOVA with Tukey’s multiple comparisons (d, f, g). Source data are provided as a Source Data file.
Next, we determined whether FBXO32 promoted cell proliferation and metastasis via activating CDK4/6. As shown in Fig. 6d and Supplementary Fig. 16, inhibition of CDK4/6 by palbociclib abolished FBXO32-induced cell proliferation in Huh7 and CFPAC-1 cells. Meanwhile, cell cycle analysis with flow cytometry showed that inhibition of FBXO32 expression prevented the nocodazole-induced G2/M block and instead substantially increased the fraction of cells in G1 (Fig. 6e). Moreover, trans-well assay showed that palbociclib treatment dramatically inhibited FBXO32-induced migration and invasion of cancer cells (Fig. 6f, g). Taken together, our results indicated that FBXO32-mediated cyclin D elevation activates CDK4/6 and drives tumor development.
FBXO32-cyclin D axis limits the sensitivity of palbociclib for cancer treatment
To investigate whether FBXO32 could influence the cancer therapeutic effect of palbociclib, a series of experiments was utilized. CCK-8 and colony formation assay showed that palbociclib suppressed the proliferation of cancer cells, which was strengthened by FBXO32 knockdown (Fig. 7a–c). Meanwhile, we found that FBXO32 knockdown synergizes with palbociclib to suppress the growth of Huh7 and CFPAC-1 cells (Supplementary Fig. 17a–d). Moreover, trans-well assay showed that FBXO32 knockdown dramatically exaggerated palbociclib-induced inhibition of migration and invasion in Huh7 and CFPAC-1 cell lines (Fig. 7d, e). These results indicated that silencing FBXO32 enhanced the therapeutic effect of palbociclib for cancer treatment in vitro. Next, we consolidated these findings in orthotopic tumor mouse model. The results showed that FBXO32 knockdown enhanced the growth inhibition effect of palbociclib for HCC(Fig. 7f), accompanied with the down-expression of cyclin D1 (Fig. 7g). Interestingly, we found a decreased ratio of lung metastasis of liver cancer in the group of combined usage sh-FBXO32 and palbociclib compared to use them alone (Supplementary Fig. 18a, b). Consistently, we observed a similar result in the orthotopic xenograft tumor model of PDAC, which showed that silencing FBXO32 strengthened the inhibition effect of palbociclib on tumor growth and liver metastasis (Fig. 7h, i and Supplementary Fig. 18c, d). In aggregate, our results demonstrated that inhibition of FBXO32 expression enhanced the therapeutic effect of palbociclib for cancer treatment.
a–e CCK-8, colony formation and trans-well assays were performed to measure the proliferation (a–c), migration (d) and invasion (e) of Huh7 and CFPAC-1 cells with stably expressing FBXO32 shRNA (sh-FBXO32) or negative control (sh-NC) by lentivirus infection in the presence of palbociclib (10 μM) or not. Scale bar, 100 μm. Statistical analysis was performed using one-way ANOVA with Tukey’s multiple comparisons, and data are represented as mean ± SEM (n = 3 independent experiments). f–i Huh7 and CFPAC-1 cells with stably expressing FBXO32 shRNA (sh-FBXO32) or negative control (sh-NC) were constructed through lentivirus infection, and then were injected subcutaneously to axilla of BALB/c nude mice. After 4 weeks, the masses of tumor formed from Huh7 or CFPAC-1 cells were divided equally into 1 mm3 pieces and implanted orthotopically into BALB/c nude mice. Both sh-NC and sh-FBXO32 group mice were treated with palbociclib (100 mg/kg) or Vehicle (sodium lactate) daily by oral gavage administration for 14 days. The mice were sacrificed after the last administration. Representative images of primary liver (f) and pancreatic (h) tumors were presented, and the tumor volumes were monitored. Statistical analysis was performed using one-way ANOVA with Dunnett’s or Tukey’s multiple comparisons, and data are represented as mean ± SEM (For HCC, n = 7 biologically independent animals per group; For PDAC, n = 5 biologically independent animals per group). H&E staining was performed to determine the tumor tissue, and IHC staining was utilized to measure the protein levels of FBXO32 and cyclin D1 in HCC (g) and PDAC (i) tumor tissues. Scale bar, 50 μm. Source data are provided as a Source Data file.
Positive correlation of FBXO32 and p-GSK-3βSer9 with cyclin D1 in clinical liver and pancreatic cancer samples
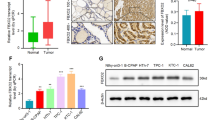
To investigate the relationship between FBXO32 and cancer, TCGA database was utilized and the results showed that FBXO32 was upregulated in tumor tissues of HCC, PDAC, and cholangiocarcinoma (CCA) (Supplementary Fig. 19a). We also found that ectopic expression of FBXO32 increased the protein levels of cyclin D1, D2 and D3 in CCA cell line HuCCT1 (Supplementary Fig. 19b), suggesting the positive correlation between FBXO32 and cyclin D in CCA. Considering that CCA and PDAC are rich in stroma, we explored how much of the FBXO32 expression comes from cancer cells. Double-immunofluorescent staining in PDAC tissues showed that approximately 90% of FBXO32-positive cells co-stained with CK19 (Supplementary Fig. 19c), a general human PDAC cell marker. We also found a similar result that FBXO32 colocalized with CK7 (a general marker of human CCA cells) in human CCA tissues (Supplementary Fig. 19d). These results indicated that FBXO32 was predominantly expressed in CCA and PDAC cells. Moreover, we observed that the mRNA and protein levels of FBXO32 were upregulated in 30-paired samples of HCC and PDAC (Supplementary Fig. 20), which was further consolidated by the immunostaining results in 131-paired HCC and 90-paired PDAC samples (Fig. 8a, b; Supplementary Fig. 21a–d). The analysis of clinical influence of FBXO32 revealed that high FBXO32 expression was related to aggressive tumor phenotypes (e.g., tumor recurrence and metastasis) in HCC, while FBXO32 was associated with larger tumor size and poorer histologic grade in PDAC (Supplementary Tables 1, 2). Multivariate analyses identified that FBXO32 expression was an independent prognostic factor for HCC and PDAC (Supplementary Tables 3, 4). The median overall survival (OS) of both HCC patients and PDAC patients with FBXO32 high expression was shorter than that with FBXO32 low expression (Supplementary Fig. 21e, f). Meanwhile, we found a similar effect of FBXO32 in 89-paired CCA tissues, which showed that FBXO32 is upregulated in CCA tissues and negatively correlated with poor prognosis of CCA (Supplementary Fig. 22a–d). Taken together, these data indicated that FBXO32 is elevated in tumor tissues and could be a valuable indicator for poor prognosis in hepatobiliary and pancreatic cancer.
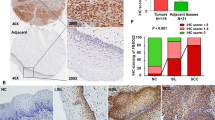
a, b Representative IHC staining images of FBXO32 in tumor and adjacent non-tumor tissues of HCC (a) and PDAC (b) (n = 30 samples). Left scale bar, 200 μm, and right scale bar, 50 μm. c–g Representative IHC images of consecutive sections of HCC tissues stained with antibodies against p-GSK-3βS9, FBXO32 and cyclin D1, respectively (c), followed by scoring according to the staining intensity and positive proportion of target protein (n = 131 samples). Then the correlations between cyclin D1 and FBXO32 or p-GSK-3βS9 were evaluated (d, e). Statistical analysis was performed using two-tailed Spearmanʹs rank correlation test. Kaplan–Meier survival curves were performed to analyze the outcome of 4 subgroups HCC patients with different FBXO32/cyclin D1 (or FBXO32/p-GSK-3βS9) protein levels (f, g). Statistical analysis was performed using Log-rank statistic test. h–l PDAC samples were utilized to detect the protein levels of p-GSK-3βS9, FBXO32 and cyclin D1 by IHC staining (n = 90 samples). Representative images were shown in (h). Correlation analysis (i, j) between cyclin D1 and FBXO32 or p-GSK-3βS9 and Kaplan–Meier survival analysis (k, l) was performed as described in (c–g). Scale bar, 50 μm. Statistical analysis was performed using two-tailed Spearman’s rank correlation test and log-rank statistic test, respectively. Source data are provided as a Source Data file.
To determine the relationship between FBXO32 (or p-GSK-3βSer9) and cyclin D1 in clinical specimens, Immunohistochemistry (IHC) staining was employed and the results showed that the protein level of cyclin D1 was positively correlated with FBXO32 and p-GSK-3βSer9 in 131-paired HCC samples (Fig. 8c–e). Furthermore, we observed a poor survival in HCC patients with high expression of either FBXO32 and cyclin D1 or FBXO32 and p-GSK-3βSer9 (Fig. 8f, g). Consistently, IHC analysis of PDAC and CCA samples also displayed a similar effect, which showed that FBXO32 and p-GSK-3βSer9 positively correlated with cyclin D1 protein level (Fig. 8h–j and Supplementary Fig. 22e, f). PDAC and CCA cancer patients with FBXO32 and cyclin D1 (or p-GSK-3βSer9) high expression showed a markedly decreased OS (Fig. 8k, l and Supplementary Fig. 22g). These results indicated that FBXO32 and p-GSK-3βSer9 positively correlated with cyclin D1 protein level in hepatobiliary and pancreatic cancer, and high FBXO32 and p-GSK-3βSer9 protein level may contribute to cancer initiation and progression.
Discussion
In this study, we revealed a key role of the nondegradative form of ubiquitination in cyclin D stabilization. Ubiquitination, the most common post-translational modification of protein, is essential for signal transduction as well as cell proliferation and differentiation in all eukaryotes. Although the primary function of ubiquitination is to generate degrons (degradation signals) on short-lived proteins and misfolded/damaged proteins, more and more studies revealed that the nondegradative form of ubiquitination also plays a vital role in regulating protein function, such as activity, subcellular location and protein-protein interaction. Cyclin D1, as a central regulator of cell cycle, has been reported to be ubiquitinated and degraded by several E3 ligases containing FBXO4, FBXO31, FBXW8, β-TrCP, SKP2, FBXL2, FBXL8, APC/C, MG53 and AMBRA1. Nevertheless, it is still unclear for the role of nondegradative form of ubiquitination in regulating cyclin D function. In the study, we first found that FBXO32 is the ubiquitin ligase stabilizing cyclin D1 rather than triggering degradation without influencing the protein levels of the reported E3 ligases. Interestingly, we observed that cyclin D1 could interact with AMBRA1 (a well-established tumor suppressor acting as an E3 ligase promoting the ubiquitination-mediated degradation of cyclin D1), which is weaker than the binding with FBXO32. These results suggested that cyclin D1 protein sustains a dynamic balance in response to various degrading and stabilizing factors. Once this balance is broken by different kinds of internal (e. g. gene mutation) or external (e. g. growth factors) factors, it will lead to the dysregulation of cyclin D1 and consequent diseases. Therefore, it would be very interesting to decipher the mechanism of the dynamic regulation of cyclin D1 between its degraders (such as AMBRA1) and stabilizers (such as FBXO32).
We demonstrated FBXO32 as a bona fide E3 ligase targeting cyclin D1 for ubiquitination and stabilization based on the following evidence. First, FBXO32 promotes K27-linked ubiquitination of cyclin D1, while the ubiquitin ligase activity-dead mutant of FBXO32 (FBXO32ΔF) fails to potentiate this modification. Second, K58 of cyclin D1 is ubiquitinated by FBXO32, and mutation of this site (K58R) almost blocks its ubiquitination by FBXO32. Third, FBXO32 dramatically prolongs the half-life of cyclin D1, which is weakened after K58R mutation. Although different ubiquitin linkages including K63 and K48 have been reported to be added to cyclin D1, we first revealed the role of K27-linked polyubiquitination in stabilizing cyclin D1. Consistently, a previous study also uncovered WWP1-mediated K27-linked polyubiquitination in stabilizing substrate protein DVL235, and another study revealed that NLRP14 maintained the stability of NCLX by K27-linked polyubiquitination36. Notably, we identified the ubiquitination site, K58, of cyclin D1 and consolidated this site plays an essential role in FBXO32-mediated cyclin D1 stabilization. By contrast, K269 of cyclin D1 has been demonstrated to be ubiquitinated by Fbx437 and AMBRA129, leading to cyclin D1 degradation and cell growth inhibition. Furthermore, we found that FBXO32 prolongs the half-lives of cyclin D1, cyclin D2 and cyclin D3 via ubiquitination. However, sequence alignment of three cyclin D revealed that cyclin D1 and cyclin D3 shared a common K58 site, whereas cyclin D2 had not, suggesting that FBXO32 ubiquitinates cyclin D2 through other lysine site. Of course, the mechanism(s) by which FBXO32 enhances the protein stability and ubiquitination of cyclin D2 and cyclin D3 need to be further investigated.
FBXO32 belongs to the F-box protein super family that functions as substrate recognition subunit in SCF E3 ligase complex and was initially identified as a key regulator in muscle homeostasis and heart development38,39. Recently, emerging evidence shows that FBXO32 is also involved in the process of tumorigenesis. However, the role of FBXO32 in tumorigenesis is still controversial in different human cancers. FBXO32 has been reported to inhibit ovarian cancer40 and lung cancer41, whereas it promotes tumor development in breast cancer42, glioma43 and melanoma33. In this study, we first uncovered the tumor-promoting role of FBXO32 in hepatobiliary and pancreatic cancer. Through in vitro and in vivo assays, we found that FBXO32 promotes the proliferation, invasion and metastasis of tumor cells, while knockdown of FBXO32 has opposite effects. In line with its role as a positive regulator of the well-established pro-oncoprotein cyclin D1, higher FBXO32 expression is associated with poorer survival in hepatobiliary and pancreatic cancer patients. From the above results and previous studies, we can see that FBXO32 exhibits a different role in regulating tumorigenesis, possibly through the regulation of different sets of substrates in various tissues. Of note, what contributes to the discrepancies of FBXO32 in regulating tumorigenesis among different tissues would be very interesting and needs to be further studied.
Phosphorylation is the most common post-translational modification involved in F-box proteins-mediated substrate recognition and ubiquitination, although other post-translational modifications, such as glycosylation, have been reported44,45,46,47. In our research, we strikingly found that dephosphorylation of cyclin D1 enhanced its interaction with FBXO32 and subsequent ubiquitination, while phosphorylation of cyclin D1 by GSK-3β decreased this interaction and ubiquitination, which is consistent with recent research that phosphorylation is not always promoting the substrate recognition by E3 ligases. Mounting evidence showed that dephosphorylation facilitates the substrate recognition by E3 ligases. For example, two recent studies separately reported that phosphorylation of IRE1α prevents its binding with the SEL1L/HRD1 E3 ligase complex48 and the ubiquitination of PD-L1 is also abrogated after its phosphorylation at T194/T210 residues by NEK2 (never in mitosis gene A-related kinase 2)49, which is in line with our findings that dephosphorylation of cyclin D1 promoted its recognition and ubiquitination by FBXO32. However, we cannot rule out the possibility that other PTMs, such as glycosylation, may be involved in FBXO32-mediated cyclin D1 recognition.
In conclusion, we revealed that FBXO32 promoted the K27-linked polyubiquitination of cyclin D and stabilizes it, and then enhanced cyclin D-CDK4/6 complex formation, leading to cancer progression and the limited response of cancer cells to palbociclib. Specifically, we identify that K58 is the ubiquitination site of cyclin D1, and mutation of this site remarkably attenuates FBXO32-mediated ubiquitination and stabilization of cyclin D1. Moreover, phosphorylation of GSK-3β has been proved to participate in FBXO32-mediated ubiquitination of cyclin D1 (Fig. 9). Our study not only reveals a critical ubiquitination-mediated mechanism enhancing cyclin D1 stability, but also uncovers the tumor-promoting role of FBXO32 in liver and pancreatic cancer. Therefore, targeting FBXO32-cyclin D axis may be a promising avenue for the treatment of human cancers with FBXO32 overexpression, such as HCC, CCA and PDAC.
Schematic illustration of the protein stability of cyclin D coordinated by FBXO32 and GSK-3β. In normal cells, cyclin D protein is phosphorylated by GSK-3β kinase at the Thr-288 and Thr-286 site, then recognized and added the K48-linked ubiquitin chains by other E3 ligases, which leads to the degradation of cyclin D in a proteasome-dependent manner. While in cancer cells, GSK-3β is phosphorylated and then inactivated, which contributes to the dephosphorylation of cyclin D. Highly expressed FBXO32 in tumor tissues recognizes dephosphorylated cyclin D and catalyzes the K27-linked polyubiquitination of cyclin D1 at the K58 site, suppressing the degradation of cyclin D1, which leads to uncontrolled cell cycle and cancer progression. Graphical abstract was created in BioRender (https://BioRender.com/h63r237).
Methods
All animal experiments were approved by the Laboratory Animal Welfare and Ethics Committee of the Army Medical University. The human study referring to HCC patients was approved by the Ethics Committee of Southwest Hospital. The human studies for PDAC and cholangiocarcinoma CCA were approved by the Ethics Committee of Shanghai Outdo Biotech Company. All the animal and human studies strictly adhered to the relevant ethical regulations of the corresponding ethics committee. Sex/gender was not considered in the study design and analysis because this study was not designed to detect sex difference.
Patients and specimens
A total of 131 patients (104 men and 27 women of age 18–75 years) with pathological diagnosis of hepatocellular carcinoma (HCC) who had received surgical resection from Southwest Hospital (Chongqing, China) were incorporated into the study. Tissue microarray of 90-paired PDAC (49 men and 41 women of age 39–83 years) and 89-paired CCA (50 men and 39 women with age of 36–79 years) were obtained from Shanghai Outdo Biotech Company (Shanghai, China). Data of clinicopathological and post-surgical follow-up were collected with harmonized standard. OS was defined as the time from the date of surgery to death or the last follow-up date. This study was approved by the Ethics Committee of Southwest Hospital and the Ethics Committee of Shanghai Outdo Biotech Company. Informed consent was obtained from every patient. Sex/gender was not considered in the study design and analysis.
Reagents and antibodies
MG132 (S2619) and CHX (S7418) were obtained from Selleck Corporation. Lipofectamine 2000 (L3000015) was purchased from Thermo Fisher Scientific. TWS119 (HY-10590), AZ191 (HY-12277), trametinib (HY-10999) were bought from MedChemExpress (MCE) corporation. GSK3-β activity detection kit (GMS50161.3.3) was obtained from GENMED. The multiplex immunofluorescence (IF) staining kit (AFIHC024) was purchased from AiFang Biotechnology company. GST-tag Protein Purification Kit (P2262) was purchased from Beyotime Biotechnology. Ni-NTA agarose beads (30210) were bought from Qiagen company. Primers used for qPCR and site-directed mutagenesis were synthesized from Sangon Biotech, and the sequences are listed in Supplementary Tables 5 and 6, respectively. The siRNA and shRNA oligos were purchased from GenePharma and Cyagen Biotechnology Corporation, respectively. The sequences of these oligos are listed in Supplementary Table 7.
Antibody against FBXO32 (ab168372, 1:1000 for WB), CK19 (ab52625, 1:200 for IF), CK7 (ab181598, 1:200 for IF) were purchased from Abcam. Antibody against HA (11867423001, 1:1000 for WB) was obtained from Roche. Antibodies against β-actin (20536-1-AP, 1:5000 for WB), α-Tubulin (11224-1-AP, 1:5000 for WB), Cyclin D1 (60186-1-Ig, 1:250 for IP), FBXO32 (67172-1-Ig, 1:200 for IHC), CDK4 (11026-1-AP, 1:1000 for WB, 1:250 for IP), GSK3β (22104-1-AP, 1:1000 for WB), AMBRA1 (13762-1-AP, 1:1000 for WB), FBXO31 (27294-1-AP, 1:1000 for WB), APC/C (10918-1-AP, 1:1000 for WB) were bought from Proteintech. Antibodies against Cyclin D1 (2978, 1:1000 for WB, 1:200 for IHC), Ki-67 (9449, 1:200 for IHC), CDK6 (13331, 1:1000 for WB, 1:250 for IP), p-RbS780 (8180, 1:1000 for WB), p-RbS795 (9301, 1:1000 for WB), p-RbS807/811 (8516, 1:1000 for WB), p-GSK3βS9 (9323, 1:1000 for WB, 1:200 for IHC), Myc tag (2278, 1:1000 for WB), β-TRCP (4394, 1:1000 for WB), SKP2 (4358, 1:1000 for WB), mouse IgG isotype (5145, 1:250 for IP) were purchased from Cell signaling Technology. Secondary antibodies containing rabbit IgG (7074, 1:5000 for WB), mouse IgG (7076, 1:5000 for WB) and rat IgG (7077, 1:5000 for WB) were purchased from Cell Signaling Technology. Antibody against Flag (F1804-1MG, 1:1000 for WB, 1:250 for IP) was obtained from Sigma. Antibodies against Cyclin D1 (sc-450, 1:500 for IF), Cyclin D2 (sc-452, 1:1000 for WB), Cyclin D3 (sc-6283, 1:1000 for WB), Rb (sc-102, 1:1000 for WB), FBXL8 (sc-390682, 1:1000 for WB) were purchased from Santa Cruz. Antibody against FBXO32 (A6825, 1:200 for IF), FBXL2 (A10296, 1:1000 for WB), MG53 (A16751, 1:1000 for WB), FBXO4 (A9968, 1:1000 for WB), FBXW8 (A18122, 1:1000 for WB) were purchased from ABclonal Biotechnology.
Plasmid construction and site-directed mutagenesis
The eukaryotic expression plasmids containing Flag-FBXO32, HA-cyclin D1, HA-cyclin D2, HA-cyclin D3, Myc-GSK-3β, ubiquitin (Ub), Flag-FBXO32ΔF, various truncations of Flag-FBXO32 and HA-cyclin D1 were constructed by the Genechem corporation using pcDNA3.1 as vector. HA-cyclin D1K58R, HA-cyclin D1 K58R, HA-cyclin D1 K72R, HA-cyclin D1 K114R, HA-cyclin D1 K123R, HA-cyclin D1 K180R, HA-cyclin D1 T286A, HA-cyclin D1 T288A, His-UbK27R, GSK-3βS9A and GSK-3βS9E were generated by PCR amplification using the QuikChange Lightning Site-Directed Mutagenesis Kit (Agilent, USA). The primers for site-directed mutagenesis were designed with an online tool supported by Agilent Corporation. A series of Ub mutants (including M1, K6, K11, K27, K29, K33, K48, K63), HA-cyclin D1K58R/T286A and HA-cyclin D1K58R/T288A were purchased from Genechem corporation.
Cell culture
Cell lines containing Huh7, HepG2, CFPAC-1, 293T were purchased from the Cell Bank of Type Culture Collection of Fudan University. Mewo cell line was purchased from Pricella Biotechnology Company. SNU449 and SNU475 cell lines were obtained from BeNa Culture Collection. HuCCT1 cell line was purchased from Bohu Biological Technology. Mouse HCC cell line Hepa1-6 was purchased from the American Type Culture Collection (ATCC). These cells were all cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C and 5% CO2.
Cell cycle analysis
Huh7 and CFPAC-1 cells with stable expression of sh-FBXO32 or sh-NC were treated with 0.33 μM Nocodazole for 16 h before harvest, then the cells were taken out for cell cycle analysis using the Cell Cycle Detection Kit (Beyotime) according to manufacturer’s instruction. Briefly, the cells were trypsinized and collected into centrifuge tubes, then washed with phosphate-buffered saline (PBS) for 2 times, and then resuspended in 75% ethanol and fixed overnight at 4 °C. Next, the cells were incubated with RNaseA (50 μg/mL) (Sigma) in PBS at 37 °C for 30 min and incubated with propidium iodide (PI, 50 μg/mL) at darkness for 10 min, and then the cell cycle was assayed by Flow cytometer (Becton, Dickinson and Company). The gating strategy of flow cytometry data was presented in Supplementary Fig. 23.
Co-IP assay
After treatment by corresponding methods, the cells were lysed with co-IP lysate buffer (Beyotime) containing protease inhibitor cocktail (Selleck) for 20 min on ice. The cell lysates were centrifuged at 12,000×g for 10 min at 4 °C, then 20% of supernatant protein solution was used as the whole cell extraction (WCE) for western blot analysis and the rest of solution was used for Co-IP assay. To reduce non-specific binding, the remaining solution was incubated with 20 μL Protein A/G Plus-agarose (Santa Cruz Biotechnology) for 1 h with constant rotation at 4 °C, followed by centrifugation at 1000×g for 5 min at 4 °C. The supernatant was incubated with 2 μg primary antibody for 4–6 h at 4 °C, and then 50 μL Protein A/G Plus-agarose was added and incubated on a rotating device overnight at 4 °C. After that, the precipitated proteins were washed with lysis buffer and boiled with 2× SDS-PAGE loading buffer, and then the supernatant was used for western blotting.
Mass spectrometry (MS) analysis
After transfection with HA-cyclin D1T286A and HA-cyclin D1T286A or negative control vector for 48 h, Huh7 cells were lysed for Co-IP assay to pull down HA-cyclin D1T286A and HA-cyclin D1T286A interacting proteins using anti-HA antibody. After boiling with 2× SDS-PAGE loading buffer, the immune-precipitates were separated on SDS-PAGE gel, followed by Coomassie blue staining (Proteintech). The region on the gel showing different bands between control and experimental group was resected and then obtained samples including experimental group (HA-cyclin D1T286A or HA-cyclin D1T286A) and control group (vector) were subjected to LC-MS/MS analysis in Applied Protein Technology company. In the liquid chromatography (LC), peptides from each example were separated by using Easy nLC system (Thermo Scientific). Eluted peptides were analyzed using the Q-Exactive mass spectrometer (Thermo Scientific). The mass spectrometer was operated in positive ion mode. Full scan MS spectra (m/z 300–1800) were acquired with a resolution of 70,000 at 100 m/z. Raw data of the mass spectrum signal was analyzed by Proteome Discoverer Daemon (Thermo Scientific, version 2.5). The search parameters included searching against UniProt human protein database (Ver. 2023-01) with a maximum of two missed cleavages, fixed modifications of carbamidomethylation on cysteine, variable modification of oxidization on methionine. Regarding the identification of cyclin D1 ubiquitination sites induced by FBOX32, 293T cells were co-transfected with Flag-FBXO32 and HA-cyclin D1 expression plasmids for 48 h, then lysed for Co-IP assay to pull down HA-cyclin D1 (containing the ubiquitinated HA-cyclin D1) using anti-HA antibody. Thereafter, the immunoprecipitates were denatured and separated on SDS-PAGE gel. After staining with Coomassie blue, the targeted protein band was sent to Applied Protein Technology company for LC-MS/MS analysis. Uniprot_human database was searched by Mascot 2.2 soft for modification of identification.
CHX-pulse chase analysis
After being transfected for 48 h, cells in 6-well plates were treated with CHX (100 μg/mL) for the indicated times. Then the cells were lysed for western blotting.
In vivo ubiquitination assays
293T cells were planted in 10 cm dishes and co-transfected with His-Ub, Flag-FBXO32 and HA-cyclin D1 plasmids. After 48 h of transfection, the cells were lysed in buffer A (6 M guanidinium-HCl, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris-HCl (pH 8.0), 5 mM imidazole and 10 mM β-mercaptoethanol) on ice for 1 h, followed by exposure to 50 μL Ni-NTA beads (Qiagen, Valencia, CA) overnight. Then beads were washed in sequence with buffer A, buffer B (8 mM urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris/HCl (pH 8.0), 10 mM β-mercaptoethanol), buffer C (8 mM urea, 0.1 M Na2HPO4/NaH2PO4, 10 mM Tris/HCl (pH 6.3), 10 mM β-mercaptoethanol, 0.2% Triton X-100), and buffer C containing 0.1% Triton X-100. His-tagged ubiquitinated proteins were then eluted by buffer D (200 mM imidazole, 0.15 M Tris-HCl (pH 6.7), 30% glycerol, 0.72 M β-mercaptoethanol, 5% SDS). The eluate was analyzed using western blotting.
GST pull-down
The lysates of E. coli BL21 transformed with GST or GST-FBXO32 plasmid were incubated with Glutathione-Agarose beads and purified according to the manufacturer’s instruction of GST-tag Protein Purification Kit (Beyotime). Thereafter, the purified GST or GST-FBXO32 were mixed with cell lysates of 293T cells transfected with HA-cyclin D1 expression plasmid, then incubated with Glutathione-Agarose beads in lysis buffer at 4 °C overnight. After that, the beads were washed three times with lysis buffer, and the precipitates were analyzed by western blotting.
Surface plasmon resonance assay
Recombinant human-derived cyclin D1 and FBXO32 protein labeled with 6×His tag in their N terminal were expressed in E. coli and purified. Subsequently, 20 μg cyclin D1 protein (50 μg/mL, diluted with sodium acetate, pH 4.0) was immobilized on a CM5 sensor chip (#BR-1005-30) after its activation by a mixture of 200 μM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and 50 μM N-hydroxysuccinimide (NHS) at 10 μL/min for 10 min. Then the surface of the chip was blocked with 1 M ethanolamine (10 μL/min for 10 min). Recombinant human FBXO32 protein was diluted to a series of concentrations (10, 5, 1.25, 0.625, 0.3125 and 0.078125 μM (all in PBS)), and was flowed at 30 μL/min for 150 s in each run. At the end of each flow, cells were regenerated for 5 min with 10 mM glycine-HCl (pH 2.0) solution at 10 μL/min. The association/dissociation phases were monitored using a BIAcore T200 system (GE Healthcare), and the affinity constants were calculated with T200 evaluation software (version 2.0, GE Healthcare). Data were exported to Origin 7 software (v.7.0552, OriginLab) for generating the final figures50,51.
GSK3β activity assay
The GSK3β activity assay kit (GENMED) was used to determine the activity of GSK3β according to the manufacturer’s instructions. The principle of the colorimetric method is based on the ability of GSK3β to phosphorylate the target sequence GPHRSTPESRAAV, and then the phosphorylated polypeptide participates in reactions that pyruvate kinase and lactate dehydrogenase oxidize NADH (reduced nicotinamide adenine dinucleotide) into NAD (nicotinamide adenine dinucleotide) in the presence of ATP and GSK3α inhibitor Aloisine A. The absorbance was read at 340 nm.
In vitro kinase assay
Purified kinases cyclin D1-CDK4 (HY-P702682, MCE) (0.5 μg) or cyclin D1-CDK6 (HY-P702687, MCE) (0.5 μg) and kinase substrate Rb C-terminal (769–921) recombinant protein (HY-P7S0127, MCE) (0.2 μg) were incubated in the presence or absence of purified FBXO32 in 30 μL kinase buffer (50 mM Tris-HCl pH 7.5, 100 mM KCl, 50 mM MgCl2, 1 mM Na3VO4, 1 mM dithiothreitol (DTT), 5% glycerol, 0.5 mM ATP) at 30 °C for 1 h. The reaction was terminated by the addition of SDS-PAGE loading buffer and heating to 100 °C for 5 min, and immunoblotting using antibodies against Rb phosphorylated at Ser 780, Ser 795 and Ser 807/811. Kinase and Rb (769–921) were detected by Ponceau S staining. Regarding the kinase assay for Rb by immune-purified endogenous CDK4/6 from 293T cells with or without FBXO32 overexpression. Briefly, Cyclin D1-CDK4 or Cyclin D1-CDK6 complex in 293T cells were immunoprecipitated by CDK4 or CDK6 antibody, followed by twice washing with lysis buffer and kinase buffer, respectively. After preincubation with kinase buffer, the precipitates were added with Rb C-terminal recombinant protein and incubated for 1 h at 30 °C. Then the kinase assay was performed as described above.
Animal studies
All mice were maintained in the animal room of Southwest Hospital in specific pathogen-free (SPF) conditions under a 12/12 h dark/light cycle and appropriate temperature (22–24 °C) and humidity (30–70%). Animals were randomly allocated into experiment groups. All BALB/C nude mice and wild-type C57BL/6N mice used in this study were obtained from GemPharmatech.
Cell lines with FBXO32 stable knockdown and overexpression (or the negative control) were utilized to construct orthotopic tumor models. Briefly, 5 × 106 tumor cells were subcutaneously implanted in the right axilla of male and female BALB/C nude mice (4–6 weeks old). Next, the tumor tissues were harvested at day 21 after implantation and cut into 1 mm3 pieces immediately. Then these pieces were transplanted into the livers or pancreases of anesthetized nude mice by laparotomy. The livers, pancreases and lung tissues were obtained after 28 days. The weights and volumes of liver, pancreas and excised tumors were measured, and the lung metastasis rate of HCC or the liver metastasis rate of PDAC was acquired by counting the number of metastatic mice. The tumor volume was calculated with the following formula: Volume = 1/2 L1 × (L2)2. L1 represents the long axis, while L2 indicates the short axis of the tumor. Tumor tissues were collected for H&E and IHC staining. For animal study to investigate the effect of FBXO32 knockdown combined with palbociclib, cell lines with FBXO32 stable knockdown (or the negative control) were utilized and the orthotopic tumor model was constructed as described above. When tumors reached a size of 200 mm3, the mice of FBXO32 knockdown and control groups were both treated daily by oral gavage administration with vehicle or palbociclib (100 mg/kg) for 14 days. Then the tumors were harvested for further analysis as described above.
To determine the therapeutic effect of targeting FBXO32, the orthotopic tumor mouse model was constructed using mouse HCC cell line Hepa1-6 constitutively expressing luciferase. Hepa1-6 cells (5 × 106) were orthotopically implanted into the livers of male and female C57BL/6N mice (4- to 6-week-old). Then the mice were randomized into three groups and intraperitoneally injected with AAV-shNC or AAV-shFBXO32 (1 × 1011 virus particles per mouse) after one week. Then the tumor loads were evaluated by bioluminescence (BLI) at day 7 (the day of randomization for baseline) and day 35 (the observation end point). Then the tumor tissues were collected for qPCR analysis.
To test the tumor-suppressive effect of FBXO32 knockdown in vivo, the DEN/CCl4-induced primary liver cancer mouse model and AAV8 containing sh-FBXO32 or sh-NC were constructed and utilized. Briefly, male and female C57BL/6N mice (4- to 6-week-old) were intraperitoneally injected with AAV-null or AAV-shFBXO32 (6 × 1010 virus particles per mouse) at day 7 postpartum, and then HCC was induced by the combination of DEN (25 mg/kg i.p.) (Sigma-Aldrich, N0756) given at day 14 postpartum and 14 weekly injections of CCl4 (0.5 mL/kg i.p., dissolved in corn oil). At the age of 24 weeks, mice were euthanized. The tumor number was counted and the largest tumor diameter was measured. Tumor tissues were collected for H&E and IHC staining.
Primary pancreatic cancer mouse model used for determining the tumor-suppressive effect of FBXO32 knockdown in vivo. Male and female KPC (LSL-KrasG12D/+; LSL-Trp53R172H/+; Pdx-1-Cre) transgenic mice (2- to 3-week-old) were purchased from Cyagen Biosciences (Guangzhou). At the age of 4 weeks, KPC mice were randomized into 2 groups: AAV-shNC group and AAV-shFBXO32 group (1 × 1011 virus particles per mouse, i.p.). After another 16 weeks, mice were euthanized and the tumor number and the largest tumor diameter was evaluated. Tumor tissues were collected for H&E and IHC staining. For mouse tumor experiments, the maximal tumor burden (15 mm) permitted by the Laboratory Animal Welfare and Ethics Committee of the Army Medical University was not exceeded in this study.
Statistical analysis
All results were presented as the means ± standard errors of the mean (SEMs). The statistical analyses of clinical data were analyzed by SPSS 24.0 (IBM, Armonk, NY) and all graphs were made on GraphPad Prism 8.0 software (GraphPad software, San Diego, CA). Pearson Chi-squared test and Fisher’s exact test were used to assess the correlation between FBXO32 expression and clinicopathological characteristics, and Spearman correlation test was used to investigate the association between expression of FBXO32 and Cyclin D1. Kaplan–Meier curves were used to compare the survival probabilities between groups by the log-rank test. The Cox proportional hazard regression model was used to analyze the hazard ratios (HRs) and 95% confidence intervals (CIs) of variables. Multivariate Cox’s regression models were applied to identify independent prognostic factors. Two-tailed unpaired Student’s t-test was performed for comparison between two groups. ANOVA with Tukey’s test was performed for comparison among multiple groups. Statistical significance was determined at P < 0.05.
Other materials and methods
Details on generation of stable cell pools, construction of FBXO32-knockout cell lines, cell proliferation and colony formation assay, cell migration and invasion assay, cell cycle analysis, immunofluorescence, IHC, western blotting, RNA extraction and quantitative PCR were described in the Supplementary materials and methods.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The proteomics data generated in this study have been deposited in the ProteomeXchange Consortium via the iProX partner repository with the dataset identifier PXD052332. The transcriptomic data sets generated in this study have been deposited in the Gene Expression Omnibus (GEO) database with the accession code GSE284506. Source data are provided as a Source Data file. The remaining data are available within the Article, Supplementary Information or Source Data file. Source data are provided with this paper.
References
Musgrove, E. A., Caldon, C. E., Barraclough, J., Stone, A. & Sutherland, R. L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 11, 558–572 (2011).
Koch, L. M. et al. Cytosolic pH regulates proliferation and tumour growth by promoting expression of cyclin D1. Nat. Metab. 2, 1212–1222 (2020).
Jeong, K. et al. Nuclear focal adhesion kinase controls vascular smooth muscle cell proliferation and neointimal hyperplasia through GATA4-mediated cyclin D1 transcription. Circ. Res. 125, 152–166 (2019).
Schwabe, R. F. et al. c-Jun-N-terminal kinase drives cyclin D1 expression and proliferation during liver regeneration. Hepatology 37, 824–832 (2003).
Fuste, N. P. et al. Cytoplasmic cyclin D1 regulates cell invasion and metastasis through the phosphorylation of paxillin. Nat. Commun. 7, 11581 (2016).
Marsaud, V. et al. Cyclin K and cyclin D1b are oncogenic in myeloma cells. Mol. Cancer 9, 103 (2010).
Wang, H. et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 546, 426–430 (2017).
Luo, C. et al. Obesity/type 2 diabetes-associated liver tumors are sensitive to cyclin D1 deficiency. Cancer Res. 80, 3215–3221 (2020).
Anders, L. et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 20, 620–634 (2011).
Goel, S. et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 548, 471–475 (2017).
Deng, J. et al. CDK4/6 inhibition augments antitumor immunity by enhancing T-cell activation. Cancer Discov. 8, 216–233 (2018).
Perry, J. E., Grossmann, M. E. & Tindall, D. J. Epidermal growth factor induces cyclin D1 in a human prostate cancer cell line. Prostate 35, 117–124 (1998).
Won, K. A., Xiong, Y., Beach, D. & Gilman, M. Z. Growth-regulated expression of D-type cyclin genes in human diploid fibroblasts. Proc. Natl. Acad. Sci. USA 89, 9910–9914 (1992).
Diehl, J. A., Cheng, M., Roussel, M. F. & Sherr, C. J. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev. 12, 3499–3511 (1998).
Takuwa, N., Fukui, Y. & Takuwa, Y. Cyclin D1 expression mediated by phosphatidylinositol 3-kinase through mTOR-p70(S6K)-independent signaling in growth factor-stimulated NIH 3T3 fibroblasts. Mol. Cell Biol. 19, 1346–1358 (1999).
Daniel, P. et al. Selective CREB-dependent cyclin expression mediated by the PI3K and MAPK pathways supports glioma cell proliferation. Oncogenesis 3, e108 (2014).
Shtutman, M. et al. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 96, 5522–5527 (1999).
Tetsu, O. & McCormick, F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398, 422–426 (1999).
Qie, S. & Diehl, J. A. Cyclin D degradation by E3 ligases in cancer progression and treatment. Semin. Cancer Biol. 67, 159–170 (2020).
Zelivianski, S., Cooley, A., Kall, R. & Jeruss, J. S. Cyclin-dependent kinase 4-mediated phosphorylation inhibits Smad3 activity in cyclin D-overexpressing breast cancer cells. Mol. Cancer Res. 8, 1375–1387 (2010).
Rader, J. et al. Dual CDK4/CDK6 inhibition induces cell-cycle arrest and senescence in neuroblastoma. Clin. Cancer Res. 19, 6173–6182 (2013).
Lazaro, J. B., Bailey, P. J. & Lassar, A. B. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 16, 1792–1805 (2002).
Leslie, K. et al. Cyclin D1 is transcriptionally regulated by and required for transformation by activated signal transducer and activator of transcription 3. Cancer Res. 66, 2544–2552 (2006).
Sherr, C. J. & Roberts, J. M. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 13, 1501–1512 (1999).
Kim, J. K. & Diehl, J. A. Nuclear cyclin D1: an oncogenic driver in human cancer. J. Cell Physiol. 220, 292–296 (2009).
Sauter, E. R. et al. Cyclin D1 is a candidate oncogene in cutaneous melanoma. Cancer Res. 62, 3200–3206 (2002).
Maiani, E. et al. AMBRA1 regulates cyclin D to guard S-phase entry and genomic integrity. Nature 592, 799–803 (2021).
Chaikovsky, A. C. et al. The AMBRA1 E3 ligase adaptor regulates the stability of cyclin D. Nature 592, 794–798 (2021).
Simoneschi, D. et al. CRL4(AMBRA1) is a master regulator of D-type cyclins. Nature 592, 789–793 (2021).
Fang, M. et al. E3 ligase MG53 suppresses tumor growth by degrading cyclin D1. Signal Transduct. Target. Ther. 8, 263 (2023).
Zou, Y., Ewton, D. Z., Deng, X., Mercer, S. E. & Friedman, E. Mirk/dyrk1B kinase destabilizes cyclin D1 by phosphorylation at threonine 288. J. Biol. Chem. 279, 27790–27798 (2004).
Takahashi-Yanaga, F. et al. Involvement of GSK-3beta and DYRK1B in differentiation-inducing factor-3-induced phosphorylation of cyclin D1 in HeLa cells. J. Biol. Chem. 281, 38489–38497 (2006).
Habel, N. et al. FBXO32 links ubiquitination to epigenetic reprograming of melanoma cells. Cell Death Differ. 28, 1837–1848 (2021).
Takahashi-Yanaga, F. & Sasaguri, T. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cell Signal 20, 581–589 (2008).
Zhao, D. et al. Targeting E3 ubiquitin ligase WWP1 prevents cardiac hypertrophy through destabilizing DVL2 via inhibition of K27-linked ubiquitination. Circulation 144, 694–711 (2021).
Meng, T. G. et al. NLRP14 safeguards calcium homeostasis via regulating the K27 ubiquitination of Nclx in oocyte-to-embryo transition. Adv. Sci. 10, e2301940 (2023).
Barbash, O., Egan, E., Pontano, L. L., Kosak, J. & Diehl, J. A. Lysine 269 is essential for cyclin D1 ubiquitylation by the SCF(Fbx4/alphaB-crystallin) ligase and subsequent proteasome-dependent degradation. Oncogene 28, 4317–4325 (2009).
Bodine, S. C. & Baehr, L. M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 307, E469–E484 (2014).
Sukari, A., Muqbil, I., Mohammad, R. M., Philip, P. A. & Azmi, A. S. F-BOX proteins in cancer cachexia and muscle wasting: emerging regulators and therapeutic opportunities. Semin. Cancer Biol. 36, 95–104 (2016).
Chou, J. L. et al. Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab. Invest. 90, 414–425 (2010).
Zhang, N. et al. FBXO32 targets PHPT1 for ubiquitination to regulate the growth of EGFR mutant lung cancer. Cell Oncol. 45, 293–307 (2022).
Sahu, S. K. et al. FBXO32 promotes microenvironment underlying epithelial-mesenchymal transition via CtBP1 during tumour metastasis and brain development. Nat. Commun. 8, 1523 (2017).
Liao, X. et al. Downregulation of BASP1 promotes temozolomide resistance in gliomas via epigenetic activation of the FBXO32/NF-kappaB/MGMT axis. Mol. Cancer Res. 21, 648–663 (2023).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004).
Skaar, J. R., Pagan, J. K. & Pagano, M. SnapShot: F box proteins I. Cell 137, 1160–1160.e1161 (2009).
Yoshida, Y. et al. E3 ubiquitin ligase that recognizes sugar chains. Nature 418, 438–442 (2002).
Yoshida, Y. et al. Fbs2 is a new member of the E3 ubiquitin ligase family that recognizes sugar chains. J. Biol. Chem. 278, 43877–43884 (2003).
Lv, X. et al. Modulation of the proteostasis network promotes tumor resistance to oncogenic KRAS inhibitors. Science 381, eabn4180 (2023).
Zhang, X. et al. NEK2 inhibition triggers anti-pancreatic cancer immunity by targeting PD-L1. Nat. Commun. 12, 4536 (2021).
Ma, T. et al. Low-dose metformin targets the lysosomal AMPK pathway through PEN2. Nature 603, 159–165 (2022).
Wang, P. et al. Erianin suppresses constitutive activation of MAPK signaling pathway by inhibition of CRAF and MEK1/2. Signal Transduct. Target. Ther. 8, 96 (2023).
Acknowledgements
This research was funded by the National Natural Science Foundation of China (31900449 to H.X. and 82273112 to F.H.), the Chongqing Medical Youth Outstanding Talent Project (YXQN202497 to H.X.), and the Chongqing Natural Science Foundation (CSTB2023NSCQ-MSX0590 to H.X.).
Author information
Authors and Affiliations
Contributions
H.X. conceived the project and designed the study. F.L., H.Y., Y.Z., Y.M., X.C., J.Z. and L.S. performed experiments, analyzed and interpreted data. H.X., C.X., F.H., and L.Z. supervised experiments and interpreted data. R.G., Y.W., P.Z., X.W., P.B. and L.Z. collected clinical samples and analyzed data. H.X. and F.L. wrote the original manuscript. H.X. and F.L. revised and edited the manuscript. All authors discussed the results and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Thomas Bonacci and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, F., Yu, H., Zhang, Y. et al. F-box protein FBXO32 ubiquitinates and stabilizes D-type cyclins to drive cancer progression. Nat Commun 16, 4060 (2025). https://doi.org/10.1038/s41467-025-59407-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59407-9
This article is cited by
-
Antitumor efficacy and molecular mechanism of lenvatinib combined with vitamin K2 against hepatocellular carcinoma
World Journal of Surgical Oncology (2026)
-
Evaluation of reference genes for accurate qRT-PCR normalization in bitter gourd across different tissues and stress conditions
BMC Plant Biology (2025)
-
MicroRNAs as the critical regulators of ubiquitin-proteasome system through F-box protein targeting during tumor progression
Cancer Cell International (2025)