Abstract
Long-read sequencing boosts alternative splicing analysis but faces technical and computational barriers in single-cell and spatial settings. High Nanopore error rates compromise cell barcode and UMI recovery, while read truncation and misalignment undermine isoform quantification. Downstream, a statistical framework to assess splicing variation within and between cells or spatial spots is lacking. We introduce Longcell, a statistical and computational pipeline for isoform quantification from single-cell and spatially barcoded Nanopore long reads. Longcell efficiently recovers cell barcodes and UMIs, corrects sequencing errors, and models splicing diversity within and between cells or spots. Applied across multiple datasets, Longcell allows accurate identification of spatial isoform switching. Longcell also reveals widespread high intra-cell isoform heterogeneity for highly expressed genes. Finally, on a perturbation experiment for 9 splicing factors, Longcell identifies regulatory targets that are validated by targeted sequencing.
Similar content being viewed by others
Introduction
Alternative splicing is a pervasive gene regulatory mechanism which affects more than 90% of multi-exon human genes. It is a key regulatory step in gene expression that allows a limited genome to express an impressive diversity of coding and non-coding RNAs1. Alternative splicing plays an important role in key biological processes such as cell differentiation, lineage determination2 and tumorigenesis3, and its dysregulation has been connected to many diseases such as cancer4, intellectual disability and autism spectrum disorders (ASDs)5. While alternative splicing has been widely studied at tissue level2,6,7, it remains poorly understood at the single cell level8,9.
Technical and computational challenges have limited our ability to explore alternative splicing in the single-cell and spatial setting. Commonly used droplet-based scRNA-seq protocols10,11,12,13 are based on short-read sequencing that only captures the 3’ or 5’ ends of RNA transcripts. The Smart-seq protocol14,15,16 can achieve full-length transcript coverage but requires further assembly and has limited throughput. Advances in full-length sequencing methods provide new opportunities for single-cell isoform analysis, and recent studies have demonstrated their feasibility17,18,19,20. The two most widely used full-length sequencing protocols are provided by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (Nanopore). While full-length sequencing overcomes the read length limitations of short-read sequencing and enables the full-length RNA isoform detection, these methods also have their own challenges that impact their scalability and accuracy. Current versions of Pacbio’s protocol have higher sequencing accuracy but limited sequencing capacity21. In contrast, current Nanopore protocols can achieve higher sequencing capacity to profile a large number of transcripts22 across a large number of cells/spots, but generally exhibit lower sequencing accuracy. In recent years, various experimental strategies have been developed to address the above problems. For PacBio, a novel technique called Multiplexed Arrays Isoform Sequencing (MAS-ISO-seq) is designed to boost sequencing throughput by over 15-fold23. For Nanopore, the Rolling Circle Amplification to Concatemeric Consensus (R2C2) method is developed to improve sequencing accuracy through consensus error correction24. Our method complements these advances by reducing the noise introduced by Nanopore’s lower sequencing accuracy through computational approaches. In this paper, we develop a computational pipeline for pre-processing and analysis of single-cell and spatially barcoded Nanopore sequencing data. The data we analyze were generated by the 10× single-cell and Visium platforms, but the model should be generalizable to other platforms.
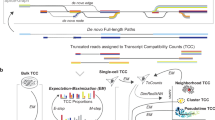
In broad strokes, a pipeline for isoform quantification on long-read sequenced single-cell libraries requires four pivotal steps (Supplementary Fig. 1): (1) Recovery of the cell barcode and unique molecular identifier (UMI) from each read. (2) Within each cell, group reads with similar alignment based on their recovered UMIs, as the UMI count, not the read count, better reflects the true quantity of a transcript. (3) Within each UMI-based read cluster, collapse reads into a consensus alignment that best represents the isoform of origin. (4) Derive isoform-level quantification based on the consensus alignments of each UMI cluster. It is important to note that the UMI, a unique feature in single-cell and spatial experiments to mitigate PCR amplification bias, is pivotal to accurate quantification in these experiments. Tools developed for bulk sequencing data, when applied to the single-cell and spatial settings, ignore the UMI. Recent studies that analyze single-cell long-read RNA-seq data also ignore the noise in UMI assignment25,26,27.
There are two key challenges in developing this pipeline for Nanopore long-read sequencing data due to the low per-base sequencing accuracy (85–99%)22,28,29,30. First, the lower sequencing accuracy makes it difficult to recover the cell barcode and UMI from the read, even as we show in the latest improved version of the technology. These barcodes are short sequence tags that are not tolerant of sequencing errors. While many methods now successfully address the cell barcode recovery problem19,20,31 (Supplementary Fig. 2), the importance and difficulty of UMI recovery have not been addressed. Different from cell barcodes, which are designed following a whitelist of known sequences, UMIs are shorter and randomly generated, which makes their recovery more vulnerable to sequencing errors. Existing methods like Sicelore219 and FLAMES20 collapse UMIs within a fixed edit distance to quantify isoform expression, but we will show that such a strategy is not foolproof: If sequencing error is high, existing methods overestimate the number of UMIs, causing inflated quantification. Such inflation due to sequencing error can be more severe with increased PCR amplification.
In addition to inaccurate UMI recovery, low sequencing accuracy, and read truncation can also lead to misidentification of isoforms. With the accumulation of sequencing errors in long reads, small internal exons are sometimes wrongly mapped, leading to the false-positive discovery of novel isoforms. Read truncation, due to pore-block and early stop in reverse transcription32, is also pervasive in Nanopore sequencing (Supplementary Fig. 3). These issues need to be addressed to allow reliable downstream alternative splicing analysis with single-cell and spatially barcoded long-read sequencing.
To address these significant challenges in single-cell and spatially barcoded Nanopore sequencing, we developed Longcell, a computational pipeline tailored for isoform quantification. Key advances in Longcell include (1) precise recovery of UMIs and their use in correcting truncation and mapping errors, and (2) a robust statistical framework for analyzing inter- and intra-cell splicing heterogeneity. By incorporating these features, Longcell overcomes the technical barriers posed by sequencing errors and variable sparse per-cell coverage, enabling reliable downstream analyses of alternative splicing.
We validated Longcell using simulated and real datasets, demonstrating its accuracy in isoform quantification across single-cell and spatial platforms. We then applied Longcell to single-cell and Visium Nanopore sequencing datasets to quantify splicing variation at the single-cell/spot level. For single-cell data, we decompose heterogeneity in the usage of an exon (or splice-site) into intra-cell and inter-cell splicing heterogeneity. These analyses revealed widespread intra-cell splicing heterogeneity in highly expressed genes and spatially resolved isoform switching events. Longcell also enabled the identification of splicing factor regulatory targets in a CRISPR perturbation experiment. Together, these results demonstrate Longcell’s potential to uncover splicing dynamics and regulatory mechanisms with single-cell and spatial resolution.
Results
Single-cell isoform quantification through UMI retrieval and UMI-based denoising
UMIs are a distinctive feature of single-cell and spatial RNA sequencing. A UMI barcode is attached to each transcript prior to amplification and should be identifiable from the sequenced read. Accurate recovery of UMIs from sequenced reads enables the removal of amplification noise and can facilitate the correction of truncation and mapping errors. In this section, we begin by presenting empirical evidence of the challenges in UMI recovery and then detail how Longcell addresses these issues to improve isoform quantification.
To illustrate how sequencing errors can impact UMI identification, we extracted the 10 base-pair sequence located directly adjacent to confidently assigned cell barcodes, which should be part of the Illumina R1 adapter (Fig. 1A). Since the adapter sequence is fixed and known, we calculate the edit distance between the original adapter 10-mer with the extracted sequences. While most extracted sequences exhibit a low edit distance from the original, ~5–10% of the sequences have edit distance greater than 2, even for data generated by the latest R10 protocol (Fig. 1A bottom left, Supplementary Fig. 4C). These high edit distances exceed typical thresholds used in barcode clustering algorithms, resulting in inflated UMI counts per gene or isoform.
A Top: The structure of a read in 10 × 5’ toolkit. Bottom left: Edit distance distribution between 10mers aside the confidently identified cell barcode and the original sequences. Bottom right: UMI graph constructed using this distribution. UMIs highlighted in green originate from the same molecule but fail to be connected due to sequencing errors. B Application of DBSCAN (blue) and Longcell (coral) to sampled 10-mer adapters across simulated amplification levels (n = 20 replicates per level). C UMI cluster size distribution for gene ENSG00000269590, which has only one isoform. Compared to 10× short reads, DBSCAN applied to Nanopore reads produces more singleton and small clusters. D Correction of wrongly mapped exons for VPS28-201 after UMI deduplication. The misaligned region is indicated by a black arrow. E Bulk-level percent-spliced-in (\(\psi\)) estimation for constitutive exons before (blue) and after (coral) Longcell correction. Each point represents an exon (n = 1146 from 325 genes); lines connect values for the same exon pre- and post-processing. F Comparison for isoform quantification of VPS28 across methods. The y-axis shows the proportion of misaligned reads mapped to VPS28-201. G Overview of Longcell: ① simulated isoforms for a gene, including two true isoforms (a and b) and a misaligned (n). ② UMIs are first clustered within each cell. In the example cell, three UMI clusters are formed. ③ Misalignment correction is performed using an isoform attribution table built per cell based on UMI clusters. Using cluster 1 as an example, isoform (a) is the majority, so the misaligned read (n) is attributed to isoform a, and (n, a) is recorded as 1 in the table. The table is then aggregated across cells and isoforms which are frequently assigned to other isoforms are classified as misalignment. ④ Truncation errors are corrected by comparing to complete read within each cluster. In the example, the truncated 5’ end for the third read in cluster 1 and the second read in cluster 3 is corrected. ⑤ Small UMI clusters are pruned based on cluster size distributions for each isoform. All boxplots show the median (center line), the 25th and 75th percentiles (bounds of the box), and whiskers extend to 1.5× the interquartile range from the box limits. Outliers beyond 1.5× IQR are shown as individual points. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
To understand this inflation, based on empirically observed sequencing error distributions, we simulated UMIs with comparable edit distances from a random set of “true” UMIs (Supplementary Fig. 4C). Frequently, the observed UMIs corresponding to a single true UMI split into many small or singleton clusters due to sequencing errors. As a visualization, Fig. 1A (bottom right) presents a toy example of a UMI graph simulated under the observed edit distance distribution. Each node represents a UMI generated through amplification and subject to simulated Nanopore-like sequencing errors, with colors indicating their true molecular origin. Edges are drawn between UMIs with a pairwise edit distance less than 2. Due to sequencing errors, many UMIs fail to connect with others from the same original molecule and instead appear as isolated singletons. In Fig. 1B we use a simulation to show the performance of the regular clustering method for the UMI clustering under the Nanopore sequencing noise. As the real UMI sequences are not known, we use the 10mer adapter adjacent to the cell barcode to mimic the PCR replicates of a single UMI. We extracted 10mer adapters from all reads with a confidently identified cell barcode. To simulate different levels of PCR amplification, we repeatedly sampled varying numbers of adapters (ranging from 2 to 50) from this pool and applied UMI clustering algorithms to each sample. In an ideal scenario, clustering would return a single group, as all sampled adapters originate from the same true sequence. However, commonly used clustering methods such as DBSCAN (eps = 2, merge UMIs with edit distance lower than 2, which has the best performance in clustering accuracy in our test) tend to overestimate the number of UMIs at higher amplification levels, resulting in inflated UMI counts. This phenomenon, which we call UMI scattering, leads to an inflated estimation of gene expression. Since the degree of scattering depends on PCR amplification fold and differs across UMI sequences, the severity of inflation is random and differs across transcripts. The consequences of UMI scattering can be assessed in real data by considering genes with only a single isoform and comparing the UMI cluster size distributions between short-read sequencing and Nanopore long-read sequencing of the same cDNA library. For single-isoform genes, the distribution of true UMI cluster sizes should be comparable between the short and long read data, but that is not what is observed: The observed UMI cluster sizes for long reads show inflation at and near a size of 1, compared to that for short read data (Fig. 1C). This provides direct evidence for the UMI scattering phenomenon.
In addition to hindering UMI recovery, sequencing errors can also mislead read alignment and introduce noise into downstream isoform identification. This issue is particularly pronounced for small exons, where sequencing errors can lead to misalignment. For example, the exon 8 in the isoform VPS28-201, of length just 29 bases, is often misaligned to in our Nanopore single cell sequencing data, which leads to two false positive splicing sites (Fig. 1D). Another example is given in the exon 2 of RPL41-204, which has length 23 bases (Supplementary Fig. 3B). Quantitatively assessing the impact of these errors on genes is challenging, as numerous factors contribute, including data quality, exon length and sequence, and the characteristics of the flanking regions. To circumvent this limitation, we examined the splicing of short internal constitutive exons. To avoid the influence of end truncations, we calculated the percent-spliced-in (PSI) (ψ) values by considering only reads fully spanning the target exon. As these exons are constitutive in the annotation, we expected their ψ values at the bulk level to approach 1. However, in a human colorectal metastasis sample sequenced by Nanopore, we observed an average ψ of only 0.67 across 1,146 constitutive exons from 325 genes with read counts exceeding 30 (Fig. 1E). This finding suggests that misalignment of short internal exons is a common issue.
To address these challenges, we developed a robust UMI recovery procedure within Longcell (Long-read single-cell transcriptomics), designed to handle UMI scattering noise and simultaneously leverage the UMI to correct for truncation and mapping inaccuracies. The Longcell pipeline includes four main steps (Fig. 1G): (1) extraction of cell barcodes and putative UMI sequences, (2) clustering of UMI sequences, (3) correction of misalignments and truncations, and (4) filtering of UMI clusters. Steps (2) and (4) improve UMI recovery, while step (3) corrects misaligned reads and generates improved consensus read alignments for downstream isoform identification (Fig. 1D).
In step (1), after cell barcode assignment, as the UMI is located adjacent to the cell barcode, we extract the adjacent sequence that putatively contains the UMI, together with short flanking sequences on both sides so as to account for possible insertions and deletions. Using the cell barcode, we then group all putative UMI sequences assigned to the same cell.
In step (2), ideally, we should only need to disambiguate between UMIs mapping to the same isoform within the same cell, however, since isoform assignment is often misled by read truncation and misalignment (Supplementary Fig. 3), the putative UMIs extracted for each cell are first grouped at a meta-isoform-group level. A meta-isoform group comprises all isoforms that can be transformed to each other through end truncations or misalignment of short internal exons. Within each meta-isoform group, we apply an iterative clustering procedure to cluster the putative UMI sequences, allowing for sequencing errors (Supplementary Fig. 5A). We then take each resulting UMI cluster as representing one unique original UMI, which corresponds to one RNA molecule of origin.
In step (3), after UMI clustering, we order the isoforms in each UMI cluster by their count, length, and mapping quality. In most cases, the highest-ranked isoform should be taken as the representative isoform for that cluster. As misalignments are mainly due to errors introduced into the read after UMI barcoding, wrongly mapped reads often coexist with correctly mapped reads in the same UMI cluster. Although we expect more correctly mapped reads as compared to wrongly mapped reads at the bulk level, within one UMI cluster, wrongly mapped read(s) can dominate due to random sampling. To prevent wrongly mapped reads from being chosen as the representative isoform of a UMI cluster, Longcell identifies isoforms which frequently coexist in the same UMI cluster and compares their bulk level counts. The isoform with the smaller bulk proportion is then designated as misalignments and corrected within each UMI cluster. More details for the complete algorithm can be found in the “Methods” section.
To demonstrate how this mapping correction step improves isoform quantification, we revisit the issue noted earlier in Fig. 1E: constitutive internal exons, whose PSI should be 1, exhibited an observed average value of only 0.67 in the colorectal metastasis data set. After applying the mapping correction, the average ψ for constitutive exons increased to 0.92 (Fig. 1E), a value that aligns better with expectations. The importance of this step is also exemplified by the quantification of VPS28-201, where a small middle exon was wrongly mapped in 20% of the reads (Fig. 1D, F). Longcell recovered the existing isoform annotation for this gene without prior knowledge. In contrast, existing methods either erroneously preserve 5–20% of the misaligned reads and assign them to an incorrect isoform or discard these reads entirely. Both approaches can bias the relative expression estimates between isoforms. Among the existing methods, IsoQuant, which does not utilize UMI information to refine read alignment, was most affected by this issue. However, as we will show later, the UMI-corrected reads from Longcell can be used as input to IsoQuant to derive accurate isoform quantification. Overall, step (3) of Longcell helps correct the mapping errors and assign reads to the true isoform.
In step (4), to address UMI scattering, we hypothesize that all expressed transcripts of the same isoform should have a similar amplification fold, as they share the same RNA sequence (Supplementary Fig. 5B). Consequently, their UMI cluster sizes should also be similar. Thus, within each isoform, we rank UMI clusters by size and adaptively prune those in the left tail of the empirical distribution (e.g., singletons), as the small clusters are likely due to errors (Supplementary Fig. 5C). More details for the adaptive pruning method can be found in the “Methods” section. To validate this step, we revisit Fig. 1B, C. In Fig. 1B, Longcell demonstrates stable quantification across all PCR amplification folds, addressing the inflated quantification caused by UMI scattering in dbscan. Figure 1C shows the comparison of UMI cluster size distribution of a single-isoform gene to the counterpart in matched Illumina sequencing of the same cDNA library. For such genes, the UMI cluster size distributions should be the same between Nanopore and Illumina data sets. As Fig. 1C shows, Longcell effectively correct the left tail inflation due to erroneous singleton clusters. After Longcell correction, the two distributions are comparable.
The UMI-corrected reads from each UMI cluster can be used for isoform identification, quantification, and differential splicing analysis. For annotation-based isoform identification and quantification, Longcell includes a module that assigns the representative read alignment of each UMI cluster to a specific isoform in the annotation (Supplementary Fig. 6B). Additionally, the UMI-collapsed and corrected reads from Longcell can be used as input to other isoform identification and quantification methods that accept single-cell data format. The benchmarking presented in the next section explores the strengths of each approach.
However, the incompleteness of current annotations introduces potential biases in annotation-based isoform-level analyses, as unknown isoforms not included in the annotation can be incorrectly assigned. A detailed example of this issue is provided in the next section. To reduce the impact of such biases and to reduce the number of hypothesis tests, we recommend conducting differential splicing analyses at the meta splice site level, as described in “Methods”.
Longcell improves isoform quantification for single-cell and spatial Nanopore sequencing
In Fig. 1B–F, we have already shown empirically how each step of Longcell’s pipeline effectively reduces the specific technical noise/biases that were identified. We evaluated the isoform quantification accuracy of the entire Longcell pipeline. For comparison, we benchmarked Longcell with other existing approaches for single-cell long-read-based isoform quantification: Sicelore219, FLAMES20, IsoQuant33, and Nanopore’s official method wf-single-cell34. Additionally, we integrated Longcell’s UMI recovery and UMI-based read preprocessing with IsoQuant, which focuses more on the downstream transcript discovery and quantification steps. The comparison of IsoQuant versus this hybrid pipeline, which we call Longcell+IsoQuant, specifically allows us to evaluate the effectiveness of Longcell’s UMI pre-processing step.
We benchmarked the methods in three ways: first, we generated simulation data across a range of low, medium, and high data quality, and across different sequencing depths, where the true isoform expression is known and used as a gold standard for all methods. Second, we generated library-matched Pacbio and Nanopore sequencing data on a 5’ cDNA library from a cancer cell line and compared the isoform quantifications derived from the two orthogonal technologies. Third, we considered library-matched Nanopore and saturated Illumina sequencing of a 3’ Visium library of mouse brain and compared each method’s isoform quantification to that derived using traditional methods aided by the Illumina-based UMI whitelist. We only considered the quantification of known isoforms and did not consider de novo isoform identification, which is usually done at the pseudobulk level where existing bulk-level tools can be applied33,35,36,37,38,39,40,41.
First, in simulation-based benchmarks, we simulated from two real data sets as a foundation. In the first simulation, we generated 1000 isoforms from 187 genes across 220 cells extracted from a real cancer sequencing dataset to be analyzed in detail later. We assumed that the observed expression of these isoforms is the true expression and introduced PCR duplicates, sequencing errors, and end truncations to mimic the data quality in different Nanopore sequencing platforms and Guppy versions (Fig. 2A and Supplementary Fig. 4A–C), see “Methods” for details of the simulation procedure. Since we know the true expression level of each isoform, we used that as the gold standard, against which we compared all methods. For each method, we computed both the Spearman and Pearson correlation between the method’s output and the gold standard, for each cell across isoforms (Supplementary Fig. 6C). The results show that, across different data quality, Longcell achieves the highest correlation with gold standard compared to existing methods (mean Spearman correlation = 0.76, 0.80, 0.82 for low-, middle- and high-quality data; Fig. 2C and Supplementary Fig. 8B). The performance of IsoQuant in isoform identification and quantification is also significantly improved (13.73% increase in Spearman correlation) when combined with Longcell across different data qualities and sequencing depth (mean Spearman correlation = 0.78, 0.82, 0.83 for low-, middle- and high-quality data; Fig. 2C and Supplementary Fig. 8A, B).
A The procedure to simulate single-cell long reads sequencing as a benchmark dataset. Different number of transcripts are first simulated under the guidance of a cell isoform expression dataset. Each transcript is then amplified according to their GC ratio. Then, the sequencing errors and truncations are introduced to mimic the Nanopore sequencing data quality. B The procedure to generate the real benchmark dataset. The left plot shows the sequencing for Jurkat cells: The 10× full-length cDNA library was randomly split into two parts, one for Pacbio and one for Nanopore. Sequences from Pacbio was processed by their official tool isoseq, while Nanopore sequencing was processed by other methods for single-cell isoform quantification. The output from each method is then compared with the isoform quantification from Isoseq by correlation. The right plot shows the sequencing for the mouse olfactory bulb: The Visium full-length cDNA library was randomly split into two parts, one for Illumina and one for Nanopore. The Ilumina sequencing is used to guide the cell barcode and UMI recovery to generate a confident single-cell isoform quantification. Methods to be benchmarked are applied only to the Nanopore data. The output from each method is then compared with the confident isoform quantification by correlation. C The per cell Spearman correlation on simulated data across different data quality for 220 cells. D The per-cell Spearman correlation on the full transcriptome simulated data across different down-sampling rates for 918 cells. E The per cell Spearman correlation on the real data: the upper one is the result on the Jurkat cells (5881 cells) and the bottom one is the result on the MOB (918 cells). Boxplots are defined as in Fig. 1. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
In the second simulation, we focused on testing the methods on a full transcriptome data set sequenced at varying depths. Starting from a mouse olfactory bulb (MOB) Visium data set that we will introduce in detail later, we applied the same procedure as simulation 1 to generate full transcriptome datasets containing 23,560 isoforms from 13,242 genes across 918 cells under the Nanopore R10 data quality setting (Supplementary Fig. 9A, B). We downsampled the data at rates decreasing from 1 to 0.2 to obtain data sets of decreasing coverage, then applied each of the methods to each of the downsampled data sets. In this simulation, Longcell+IsoQuant achieves the overall best performance (mean Spearman correlation = 0.87), while Longcell (mean Spearman correlation = 0.85) and wf-single-cell (mean Spearman correlation = 0.85) were ranked second and third, respectively (Fig. 2D, Supplementary Fig. 9C, D).
We also performed simulations to examine the performance of methods on the challenging scenarios of micro-exons and NAGNAGs. The misalignment of microexons, e.g., the example in 1D, was part of the motivation behind the UMI-based misalignment correction procedure in Longcell. The full transcriptome simulation data set above includes 464 genes containing alternatively spliced exons shorter than 50 base pairs, which in total contribute to 2014 alternatively spliced isoforms. In Supplementary Fig. 10, we show the summary statistics of these microexons, whose length varies from <10 to 50 bases, and our sensitivity in their detection at varying downsampling rates. Similar to other alternative splicing events, Longcell+IsoQuant was the most accurate, followed by Longcell and wf-single-cell. For the most extreme case of alternative splicing events at tandem acceptor splice sites (within the sequence motif NAGNAG), we note that Longcell does allow mismatches of three nucleotides for internal exons in reference-based isoform alignment. To verify the ability of Longcell to quantify such events, we did another simulation over the MYL6 gene. This gene contains 4 isoforms, MYL6-201, MYL6-202, MYL6-207, and MYL6-218, where there is a 3nt difference in the second exon between MYL6-201, MYL6-202, and between MYL6-207 and MYL6-218. We simulated 240 cells in total and divided them into four groups, each group would mainly express two types of isoforms. We then apply Longcell, Longcell+IsoQuant, and wf-single-cell to this data to show if those isoforms can be correctly identified and quantified (Supplementary Fig. 10). All 3 three methods correctly detected the four isoforms without false positive identification. We further compare their single cell isoform quantification by mean square error (MSE) between the estimation and the ground truth. Overall, Longcell (mean MSE = 1.02) and Longcell+IsoQuant (mean MSE = 1.22) have better performance over wf-single-cell (mean MSE = 1.48).
In the Pacbio-Nanopore benchmark, we sequenced 18 genes, including 110 isoforms in 5881 Jurkat cells on both Pacbio and Nanopore R10 platforms (Fig. 2B). Given the low sequencing error rates in Pacbio (Supplementary Fig. 7), we processed the Pacbio data with their official tool Isoseq42 and used the resulting isoform quantification as a cross-technology validation. Then we applied all tools on the Nanopore sequencing data and, for each cell, computed the per-cell correlation across isoforms between the Isoseq quantification and the isoform expression output from each method (Fig. 2E and Supplementary Fig. 8C). Longcell significantly improves upon the correlations of existing methods for data generated by the Nanopore R10 platform, producing the most accurate single cell isoform quantification across the board (mean Spearman correlation = 0.43, 8.72% improvement compared to the second accurate method Sicelore2).
In the final benchmark, we analyzed a full transcriptome sequencing for MOB slice (Fig. 2B). Lebrigand et al. sequenced the sample with Nanopore sequencing and library-matched 10× short read sequencing near saturation27. They recovered the cell barcodes and UMIs in Nanopore in a supervised fashion by referring to the cell barcodes and UMIs in the short reads sequencing data set, and ended up with 40142 isoforms in 18724 genes for 918 cells. Such supervised recovery for cell barcodes and UMIs should be more accurate, and thus, we applied all tools on the Nanopore sequencing data and compared their output with Legibrand et al.’s isoform quantification. Among all methods, Longcell-IsoQuant (mean Spearman correlation = 0.70) and wf-single-cell (mean Spearman correlation = 0.66) have the best performance (Fig. 2E and Supplementary Fig. 8C). Compared to IsoQuant (mean Spearman correlation = 0.58), Longcell-IsoQuant shows 20% improvement.
Note that the cancer cell line data for benchmark 2 comes from a 5’ targeted library and the MOB data set for benchmark 3 comes from a 3’ full transcriptome library. Overall, these benchmarks show that Longcell’s UMI recovery and UMI-based misalignment and truncation correction improve isoform quantification across different sequencing designs.
Biases from incomplete annotations and meta-splice site-based quantification
The results from simulation data, cell-matched Oxford Nanopore–Pacific Biosciences sequencing data, and spatial validation data demonstrate that Longcell’s innovations improve isoform quantification, both independently and in combination with IsoQuant. However, it is important to note that the metrics presented here are reference-based and may not fully capture the complexity of transcript splicing, as unknown isoforms that are not in the reference may confuse all algorithms. To illustrate such complexity, we use an example from the Jurkat dataset (Supplementary Fig. 11). For the gene VIM, which has three isoforms (201, 206, and 209), different methods yield conflicting conclusions about their expression: IsoQuant attributes most reads to VIM-209, Isoseq assigns the majority to VIM-201, and wf-single-cell divides the reads roughly equally between VIM-201 and VIM-206.
Examining the reads mapped to VIM reveals significant 3′ truncations, with most reads covering exons 2–5, which are shared by VIM-209, VIM-201, and VIM-206. This overlap limits isoform assignment. However, a subset of reads covering both the 5′ and 3′ ends provides additional clues. For instance, three reads span four exons at the 3′ end, shared only by VIM-201 and VIM-209, potentially explaining why IsoQuant and Isoseq exclude VIM-206. Additionally, tens of reads span exon 1 at the 5′ end. Among these, 24 reads exhibit a splice site corresponding to exon 1 of VIM-209, while 175 align with a splice site corresponding to exon 1 of VIM-204. This combination of splice sites, absent in the isoform annotation, suggests the existence of a novel isoform, here referred to as VIM-new. Consequently, reads spanning exons 2–5 could correspond to VIM-201, VIM-209, or 5′-truncated VIM-new. Current quantification methods overlook this potential isoform: IsoQuant assigns all such reads to 5′-truncated VIM-209, whereas Isoseq attributes them to VIM-201. Given the observed alignments, it remains challenging to determine which method is accurate.
All annotations are incomplete, and it is important to remember that unannotated isoforms, in addition to truncations and mapping errors, can confuse all current algorithms. To mitigate the undesired effects of unannotated isoforms, the remaining analyses in this paper quantify alternative splicing events at the splice site, rather than the isoform level. For each gene, Longcell identifies differences between reads that cannot be attributed to truncation and treats these differences as putative alternative splice sites (Supplementary Fig. 12A). Due to the prevalence of read truncation in Nanopore sequencing, differences at the 5’ are excluded, and for all reads lacking polyA tail, differences at the 3’ are also discarded (see Supplementary Fig. 6 for coverage biases for data from 3’ and 5’ protocols). To reduce dimensionality and collinearity, putative alternative splice sites are grouped into “meta splice sites” based on co-variation across reads. This grouping effectively reduces the number of features for downstream analysis (Supplementary Fig. 12B–E). Importantly, meta splice sites are defined adaptively based on the observed covariance between the individual splicing sites in the data, thus distilling the data into its sufficient statistics.
Quantification of intra versus inter-cell splicing variation in spatial and single-cell data
When an alternative splicing event is detected at the bulk-tissue level, a basic question is whether the two isoforms are co-expressed within the same cells or in separate cells. This question has been examined by single-cell short-read sequencing43,44, where the conclusion was that alternative isoforms predominantly originate from different cells. However, existing studies did not fully account for the variation in total gene expression and sequencing coverage across cells45, and current single-cell data sets all suffer from low per-cell sequencing coverage that limits the sensitivity to detect multiple isoforms within the same cell. Here, we propose a statistical model to explicitly quantify inter- versus intra-cell splicing variation at a given exon/splice site, accounting for the variation in total read coverage across sites and across cells. For an exon/site, intra-cell heterogeneity refers to the case where both types of isoforms—those including the exon and those excluding it—co-exist within the same cell. In contrast, inter-cell heterogeneity refers to the degree to which the percent of transcripts that include the exon (PSI) varies across cells (Fig. 3C). For spatial transcriptomic data, similar definitions could be made for inter- versus intra-spot heterogeneity.
A The UMAP of the colorectal cancer metastasis to the liver (CRCLM) single cell data, each point is a cell colored by its cell type definition. B The paired VISIUM sequencing for the same CRCLM sample. The left plot shows the histology, indicating the tumor region. The right plot shows the dominant cell type in each spot. C The relationship between \(\bar{\psi }\) and \(\phi\), \(\bar{\psi }\) means the mean of percent-spliced-in for an exon across the cell population, \(\phi\) means the inter-cell heterogeneity of this exon. The histograms of \(\psi\) are colored by their \(\bar{\psi }\). The dots above each histogram show the alternative splicing across the cell population. The dot filled in red means the target exon is preserved in the isoforms in this cell, while the dot filled in white means the target exon is spliced out. The circle filled in red gradient means the cell expresses both isoforms. D ϕ vs. \(\bar{\psi }\) distribution for alternative spliced exons in CRCLM single cell data, the color indicates the confidence interval of ϕ estimation. E The \(\psi\) distribution for exon 6 of MYL6, which has a very high ϕ, indicating high intercell heterogeneity. In this histogram, x-axis shows the exon \(\psi\) and y-axis shows the cell frequency whose \(\psi\) value falls in this bin. As \(\psi\) estimation is influenced by the gene expression, the average gene expression for is bin is shown in color gradient. F The left plot shows the ψ distribution for exon 6 of MYL6 across cells, epithelial show the highest inclusion-level of this exon. The expression of two dominant isoforms across different cells are shown in the right heatmap, epithelial has higher expression of MYL6-218 compared to other cell types. G The sashimi plot shows the comparison of bulk expression of MYL6-207 and MYL6-218 between epithelial cells and other immune cells. H The spatial view of the expression for MYL6-218 (top) and MYL6-207 (bottom). The regions for myeloid cells are marked by black circles. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
Formally, for cell \(c\), let \({X}_{c}^{e}\) be the count of reads with exon \(e\) retained. Here, we will describe the model in terms of exons, since that is simpler and more intuitive, but in our analysis, we apply this model on the level of meta-splice sites, as described in the previous section. As noted there, the grouping of splice sites into meta-splice sites retains the sufficient statistics in the data while reducing the number of redundant features, increasing the power of any downstream transcriptome-wide screens. We assume that \({X}_{c}^{e}\sim {Binomial}({N}_{c}^{e},{\psi }_{c}^{e})\), where \({N}_{c}^{e}\) is the total number of reads mapping to the gene containing the exon in cell \(c\), and \({\psi }_{c}^{e}\) is the unknown cell-specific PSI value for exon \(e\), i.e., the proportion of transcripts of cell \(c\) with exon \(e\) retained.
First, it is important to note that if we fix the bulk PSI (i.e., the mean of \({\psi }_{c}^{e}\) across cells), there is a clear trade-off between inter-cell heterogeneity and mean intra-cell heterogeneity: for example, consider the case where bulk PSI is 0.5. Then, in one extreme, all cells could be at 0.5 (maximizing intra-cell heterogeneity and minimizing cross-cell heterogeneity), or, in the other extreme, half of the cells could be at 0 with the other half of the cells at 1 (no intra-cell heterogeneity, and maximum cross-cell heterogeneity). Imagine this as trying to distribute the two isoforms amongst all cells. If we put the two isoforms in the same cell, then across cells, there will necessarily be fewer differences between cells. If we put the two isoforms in different cells, then there will be more inter-cell heterogeneity but less intra-cell heterogeneity.
With the cell-specific PSI \({\psi }_{c}^{e}\) unknown, our goal is to estimate its distribution across cells. Since \({\psi }_{c}^{e}\in \left[{{\mathrm{0,1}}}\right],\) we use the Beta distribution to flexibly model its variation across cells. The mean of the Beta distribution, \({\mu }^{e}=E[{\psi }_{c}^{e}]\), is the bulk-level PSI for this exon, while the variance of the Beta distribution measures the degree of inter-cell heterogeneity in exon usage. Since the variance of the Beta distribution scales with the mean in the form \({\mu }^{e}\left(1-{\mu }^{e}\right)\), we compute the dispersion \({\phi }^{e}={var}\left({\psi }_{c}^{e}\right){\left[{\mu }^{e}\left(1-{\mu }^{e}\right)\right]}^{-1}\in \left[{{\mathrm{0,1}}}\right]\, (1)\) as a mean-invariant measure of inter-cell splicing heterogeneity. A low value of \({\phi }^{e}\) indicates that the distribution of \({\psi }_{c}^{e}\) across cells is unimodal. Thus, for example, if the bulk PSI of exon \(e\) is \({\mu }^{e}=1/2\), and \({\phi }^{e}\) is low, then we would expect the single cell PSI’s are also around \(1/2\) and so there would be high intra-cell heterogeneity and low inter-cell heterogeneity. In contrast, a high \({\phi }^{e}\) indicates that \({\psi }_{c}^{e}\) is bimodal, meaning that individual cells either splice this exon in or out, with low intra-cell heterogeneity (Fig. 3C). Let \({\hat{\mu }}^{e},{\hat{\phi }}^{e}\) be our maximum likelihood estimates of \({\mu }^{e},{\phi }^{e}\), respectively, based on Longcell’s isoform quantification. Standard errors and confidence intervals of \({\hat{\mu }}^{e},{\hat{\phi }}^{e}\), which we compute by Bootstrap, allow the assessment of estimation uncertainty due to low sequencing coverage. For a given dataset, a plot of \({\hat{\mu }}^{e}\) versus \({\hat{\phi }}^{e}\) across genes and exons gives a birds-eye-view of the bulk-level mean exon inclusion rates versus the degree of inter-cell heterogeneity across the transcriptome. We recommend using either error bars or color shading to reflect the estimation uncertainty. See Methods for details on estimation and inference.
To illustrate the procedure, we first consider a sample of colorectal cancer metastasis to the liver (CRCLM). The sample is processed by both 10× Chromium (single cell) and 10× Visium (spatial), with the cDNA libraries from both single cell and spatially barcoded samples split and sequenced by both Illumina and Oxford Nanopore46 (Fig. 3A, B). In the single-cell sample, we identified four main cell types based on gene expression. The UMAP derived from Illumina short read gene expression and Nanopore long read isoform expression reveal similar geometries, indicating that after Longcell’s cell barcode extraction and UMI recovery, cell types visible by short read sequencing are also detectable from the long read data (Supplementary Fig. 13A). For the CRCLM spatial data, the clusters identified based on isoform expression map to histologically distinct regions on the slide (Supplementary Fig. 13B). Through spot decomposition based on the gene expression we get 3 main cell types for the CRCLM spatial data (Fig. 3B and Supplementary Fig. 13C). For downstream alternative splicing analysis, given that existing isoform annotations are incomplete, we applied Longcell in annotation-free mode. After preprocessing, we applied the Beta-Binomial model above to the meta-splice sites identified by Longcell. Figure 3D shows the \({\hat{\mu }}^{e}\) versus \({\hat{\phi }}^{e}\) plot for exons \(e\) in highly expressed genes in the single cell CRCLM dataset. The standard error of \({\hat{\phi }}^{e}\) are shown as continuous color scale. We filtered out \({\hat{\phi }}^{e}\) with high standard error and preserve confident \({\hat{\phi }}^{e}\) estimation for 88 meta splicing sites in 57 genes. We found that most alternatively spliced exons for highly expressed genes have low estimated \(\phi\), indicating a low inter-cell heterogeneity but high intra-cell heterogeneity (Fig. 3D). Thus, for highly expressed genes, the dominant pattern is for different isoforms to be co-expressed at a similar ratio within each cell across the cell population. For example, we observed an isoform switching event involving exon 4 of RBIS, which shows a low \(\phi\) and thus a unimodal distribution for the cell-specific PSI (Supplementary Fig. 14A). Indeed, the two isoforms, RBIS-208 and RBIS-213, are co-expressed in each tumor epithelial cell with a relative 1:1 ratio. (Supplementary Fig. 14B–D). Another example gene ZFAS1 also shows such low intra-cell heterogeneity (Supplementary Fig. 14E). We detected a unimodal distribution of cell-specific PSI for its exon 4. Two isoforms, ZFAS1-201 and ZFAS1-205 are involved in this alternative splicing event and they are also co-expressed in each cell with a relative 1:1 ratio and without any obvious cell-type-specific distinction (Supplementary Fig. 14F–H).
We identified a few cases of alternatively spliced exons with high \(\phi\), indicating high inter-cell variation and a bimodal distribution for ψ. An example is exon 6 in MYL6. The two isoforms of MYL6 are enriched in different cell groups, with co-expression of MYL6-218 and MYL6-207 in tumor epithelial cells and dominance of MYL6-207 in macrophages and CD8 cells (Fig. 3E–G). Just as highly variable genes analysis identifies genes with high variation in total expression across cells, the Beta-Binomial above identifies exons with high variation in percent-splice-in across cells, which could be informative for revealing cell sub-populations distinguished by alternative splicing. The alternative splicing of MYL6 is also observed in the paired Visium dataset, in which the myeloid cells also predominantly express MYL6-207 while tumor epithelial cells have high expression in both MYL6-207 and MYL6-218 (Fig. 3H, Supplementary Fig. 15). For a comprehensive list of the estimations for inter-cell splicing heterogeneity, see Supplementary Data 1.
We further applied Longcell to Nanopore sequencing data of mouse embryonic brain cells19. This data describes the differentiation process from neuroblasts to glutamatergic and GABAergic neurons (Fig. 4A). In this dataset we preserved confident \({\hat{\phi }}^{e}\) for 268 meta splicing sites in 156 genes. As in the CRCLM data, high intra-cell and low inter-cell splicing heterogeneity was observed in this dataset for most of the alternatively spliced events (Fig. 4B). For example, the Serbp1 gene is alternatively spliced at exons 4 and 5, leading to 4 main isoforms. These four isoforms are co-expressed in single cells across the neuroblast to glutamatergic neuron differentiation trajectory, with no obvious cell type-specific distinction (Fig. 4D–F). To identify genes with the most significant differential isoform usage across cells, we ranked exons by \({\hat{\phi }}^{e}\) value and found two meta splice sites in the Pkm gene that display very high inter-cell splicing heterogeneity (Fig. 4B). This is consistent with the published result that single cells tend to predominantly express one isoform for the alternatively spliced Pkm gene in this tissue19. Our analysis found that exon 9 for Pkm has a continuously increasing trend towards retention as cells progress from neuroblasts to glutamatergic neurons (Fig. 4G, H). This indicates a continuous switch in splicing from the Pkm-201 isoform to the Pkm-202 isoform (Fig. 4I, J) as neurons mature towards the differentiated state.
A Umap of cells in the mouse embryo brain colored by cell types. B ϕ vs. \(\bar{\psi }\) distribution for alternative spliced exons, color indicates the confidence interval of ϕ. C The ψ distribution for exon 4 of Serbp1, which has a relatively low ϕ and shows a unimodal distribution, indicating a low inter-cell heterogeneity. D Umap of cells in mouse embryo brain. Cells are colored by ψ for exon 4 of Serbp1. Cells that have low expression (<3) of this gene and could not give a confident ψ estimation are the smallest points colored in gray. The gene expression for each cell is shown by the point size while the ψ estimation is shown by color gradient. Three differentiation stages from neuroblast to glutamatergic cells are highlighted by three circles (red: early, blue: middle, and green: late). E, F The alternative splicing for Serbp1 in above three circled groups. The alternative splicing of exon 4 and 5 in Serbp1 mainly leads to 4 isoforms: Serbp1-201, Serbp1-203, Serbp1-207, and Serbp1-211. Each single cell co-expressed part of those 4 isoforms and there is no obvious cell-type-specific pattern of the alternative splicing from both single cell view (E heatmap) and bulk level (F sashimi plot). G The ψ distribution for exon 9 of Pkm, which has a relatively high ϕ and shows a bimodal distribution, indicating a high inter-cell heterogeneity. H Umap of cells in mouse embryo brain. Cells are colored by ψ for exon 9 of Pkm. I, J The alternative splicing for Pkm in above three circled groups. The alternative splicing of exon 9 in Pkm mainly leads to 2 isoforms: Pkm-201 ad Pkm-202. An obvious transition of the expression of two isoforms can be identified both in both bulk (J sashimi plot) and single cell level (I heatmap). Source data are provided in https://doi.org/10.5281/zenodo.15320816.
Detection of differential splicing events between cell populations
To identify differentially spliced genes between two cell populations, we developed a test that accounts for both the mean splicing difference between populations as well as the inter-cell heterogeneity within each subpopulation. Consider the exon colored yellow in Fig. 5A. As in the last section, we will describe the model for each exon, but our analysis applies the model at the meta-splice-site level. We let \({{X}_{c}}^{e}\) be the count of the number of reads retaining the exon for each cell \(c\). Given the total count \({N}_{c}\) of reads for the gene in cell \(c\), we model \({{X}_{c}}^{e}\) with a Beta-Binomial distribution, \({{X}_{c}}^{e}\sim {Binomial}({N}_{c},{{\psi }_{c}}^{e})\), with \({{\psi }_{c}}^{e}\sim {Beta}({{\alpha }_{1}}^{e},{{\beta }_{1}}^{e})\) for cells in population 1, and \({{\psi }_{c}}^{e}\sim {Beta}\left({{\alpha }_{2}}^{e},{{\beta }_{2}}^{e}\right)\) for cells in population 2. Based on this model, we perform a generalized likelihood ratio test of the null hypothesis \({H}_{0}:{{\alpha }_{1}}^{e}={{\alpha }_{2}}^{e},{{\beta }_{1}}^{e}={{\beta }_{2}}^{e}\), which corresponds to the scenario where the two cell populations have the same Beta distribution parameters for the cell-specific PSI values. This model can account for uneven total gene expression across cells and, by testing for changes in both shape parameters of the underlying Beta distribution, can sensitively detect both a shift in mean and a change in dispersion in the population distribution of \({{\psi }_{c}}^{e}\). After correction for multiple hypothesis testing, exons that display differences in the distribution of \({{\psi }_{c}}^{e}\) across the two cell populations are reported. The detected signals will then be decomposed into mean-level and variance-level changes for better interpretability (Fig. 5A).
A The principle of generalized likelihood ratio test to identify differentially expressed isoforms. The mean change vs. variance change of ψ for all significant meta splicing sites identified in Jurkat (B) and stimulated Jurkat cells (C) after knock-out of splicing factors. Each point is a significant meta-splicing site labeled by its gene identity. Meta splicing sites with too low mean or variance change are not labeled. D Correspondence of significant meta-splicing sites between original and stimulated Jurkat cells. The line represents the fitted linear regression; shaded areas indicate the 95% confidence interval around the fit. After stimulation, there is a significant change of gene expression and alternative splicing in Jurkat cells, but the regulation of splicing factors keeps the same direction. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
Longcell reveals targets of splicing regulators in single cell CRISPR experiment
We applied Longcell, with the differential expression module, to a perturbation experiment in naïve and stimulated Jurkat human cell lines. The Jurkat cell line was derived from a T cell leukemia and stably expressed Cas9. On this cell line, we transduced a multiplexed gRNA lentiviral library targeting 9 splicing factor genes (two gRNAs per gene), including well-known factors from the HNRNPLL and SRSF families and less-studied factors such as PCBP2 and CWC27 (Supplementary Fig. 16A). After 14 days, we harvested the cells, generated single-cell libraries, and conducted full transcriptome Nanopore sequencing47. We identified 7 meta-splicing sites in 5 genes in Jurkat cell lines and 11 sites in 6 genes in stimulated Jurkat cell lines that were affected by the knock-out of splicing factors after FDR control at threshold 0.05 (Fig. 5B, C and Supplementary Data 2). For example, we observed that knock-out of HNRPLL significantly promoted the inclusion of exons 4 and 6 in PTPRC, leading to the transition from the expression of PTPRC-214 to PTPRC-209, which corroborates existing knowledge about these factors47,48 (Supplementary Fig. 17A–D).
We also compared the regulation patterns of splicing factors between stimulated and unstimulated T cells. Although the stimulation significantly changed the expression profile of T cells, we were still able to identify overlapping regulation signals between the naïve and perturbed settings. For these overlapping targets, the effect sizes of splicing factor knockout correlate highly between the stimulated and unstimulated group (\(p=0.001\)), validating each other and indicating that these specific regulatory relationships of the splicing factors are not changed by the stimulation (Fig. 5D).
Besides the well-known regulation of PTPRC mentioned above, we also identified novel regulation patterns such as PCBP2 promoting the inclusion of exons 3 and 4 in DGUOK (Fig. 6A). Knock-out of this splicing factor can lead to a transition from expression of isoform DGUOK-208 to expression of isoform DGUOK-202 and DGUOK-203 (Fig. 6C, D). We also verified this regulation pattern between PCBP2 and DGUOK in a targeted sequencing experiment where these genes were enriched and thus received much higher coverage (Supplementary Fig. 18). Another example is the alternative splicing of the exon 4 in the gene PTS was found to be regulated by CELF2. CELF2 was shown to promote the removal of exon 4 and knock-out of CELF2 leads to the transition of expression from PTS-201 to PTS-207 (Supplementary Fig. 17E–H). We also found that the alternative splicing of ARHGEF1 is regulated by CELF2 (Fig. 6E, F). But converse to PTS, our result shows that CELF2 can suppress the removal of exons 14 and 15, thus promoting the transition from ARHGEF1-203 to ARHGEF1-227 (Fig. 6G, H).
A, B ψ distribution of exons 3 and 4 of DGUOK in nontarget and PCBP2 knock-out cells estimated from the full transcriptome sequencing data. A significant decrease of \(\psi\) (\({fdr}=2.16\times {10}^{-7}\)) can be observed after the knock-out. C Comparison of expression for 3 main DGUOK isoforms between nontarget and PCBP2 knock-out cell populations by two-sided Wilcoxon rank sum test. The exact p-values are shown without adjustment for multiple tests. D Sashimi plot for 3 main DGUOK isoforms in Nontarget and PCBP2 knock-out cell populations. The detected exons are marked by a black box. E, F ψ distribution of exons 14 and 15 of ARHGEF1 changes after knock-out of CELF2. A significant decrease of \(\psi\) (\({fdr}=3.77\times {10}^{-9}\)) can be observed. G Comparison of expression for 2 main ARHGEF1 isoforms between nontarget and CELF2 knock-out cell populations using the same test setting as (C). H Sashimi plot for 2 main ARHGEF1 isoforms in Nontarget and CELF2 knock-out cell populations. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
Spatial isoform switching in mouse olfactory bulb identified via Longcell
To further illustrate spatial splicing analysis with Longcell, we revisit the VISIUM Nanopore sequencing of the MOB slice from the experiment conducted by Lebrigand et al. 27. The spatial domains of this data are shown in Fig. 7A. In our benchmarks in Fig. 2, we showed that Longcell gives accurate isoform-level quantification for this data as compared to using the UMI whitelist of library-matched saturated Illumina sequencing. We further applied Longcell to quantify intra vs. inter-spot splicing variation, where we obtained confident estimates of the inter-cell dispersion \({\hat{\phi }}^{e}\) for 185 meta splicing sites in 131 genes, all of which are highly expressed. Like the CRCLM and mouse embryonic brain samples, we found that, for these highly expressed genes, most meta splicing sites to have substantial intra-spot splicing heterogeneity and, relatively, low inter-spot splicing heterogeneity (Fig. 7B). For example, \({\hat{\phi }}^{e}\) for a meta site in the gene Apod is 0.06, which indicates low inter-spot heterogeneity (Supplementary Fig. 19A, B). This meta-site corresponds to the exon 5 in Apod, and the alternative splicing of this exon leads to 2 isoforms: Apod-202 and Apod-203. This gene is highly expressed in the Olfactory Nerve Layer (ONL), and each cell expresses around ¾ Apod-203 and ¼ Apod-202 (Supplementary Fig. 19C, D). Such low inter-cell heterogeneity indicates that the two isoforms may have different but important functions in this tissue.
A The spatial plot for the VISIUM slice, each spot is colored by the layer identification. B ϕ vs. \(\bar{\psi }\) distribution for alternative spliced exons, color indicates the confidence interval of ϕ. C The mean change vs. variance change of ψ for all significant meta-sites which are alternatively spliced across different layers. The point size indicates the significance after FDR control, while the color indicates in which layer this meta-site is alternatively spliced. D The ψ distribution for exon 3 of Plp1, which has the highest ϕ and shows a bimodal distribution, indicating a low inter-cell heterogeneity. E Spatial plot of spots in the slice of mouse olfactory bulb. The spots are colored by ψ for exon 3 of Plp1. Cells which have low expression (<3) of this gene and could not give a confident ψ estimation are the smallest points colored in gray. The gene expression for each spot is shown by the point size, while the ψ estimation is shown by the color gradient. F, G The alternative splicing for Plp1 in different layers. H The ψ distribution for exon 4 of Mapre3, which has a relatively high ϕ and show a bimodal distribution, indicating a high inter-cell heterogeneity. I Spatial plot of spots in the slice of mouse olfactory bulb. The spots are colored by ψ for exon 4 of Mapre3. J, K The alternative splicing for Mapre3 in different layers. Source data are provided in https://doi.org/10.5281/zenodo.15320816.
Of the meta-sites that show high inter-spot heterogeneity, the one with the highest inter-cell dispersion is a meta-site in the Plp1 gene (Fig. 7B, D). This agrees with the experiment from Lebrigand et al. 27 which identified and validated this isoform switch of Plp1 between different layers of the olfactory bulb.
To detect if there are any other isoform switching events between different layers, we applied Longcell’s differential splicing test on this sample. In total, 312 meta splicing sites in 224 genes are significantly alternatively spliced in different layers at FDR threshold 0.05 (Fig. 7C). The Plp1 gene is also one of the most significant signals in this differential test. There exists an alternative 3’ splice site in the exon 3 of Plp1 gene, which leads to 2 isoforms Plp1-201 and Plp1-202. The outer layers like ONL have high expression of both Plp1-201 and Plp1-202, while inner layers, such as the Granule Cell Layer (GCL + RMS), exclusively express Plp1-201 (Fig. 7E–G). For a comprehensive list of the identified isoform switching events, see Supplementary Data 2.
To what extent is inter-spot splicing heterogeneity driven by tissue domain (layer)? Interestingly, through a global analysis, we find that across genes, the inter-spot heterogeneity of a meta-site is not strongly correlated to whether it is differentially spliced between layers (Supplementary Fig. 19E). One example is exon 4 of the Mapre3 gene. There is an alternative 3’ splice site in this exon, leading to the two isoforms Mapre3-201 and Mapre3-202. The splicing of this exon shows high inter-cell heterogeneity that is not aligned with the tissue layer (Fig. 7H, I). Instead, we found that within each layer, each single cell exclusively expresses Mapre3-201 or Mapre3-202, but the choice to express which isoform is not correlated to the layer identity (Fig. 7J, K). Thus, something else is driving the splicing of this gene. This highlights the importance of unsupervised inter-spot splicing heterogeneity quantification that is not dependent on known tissue domains.
Discussion
Long-read sequencing is a powerful tool for detecting and characterizing alternative splicing in single-cell and spatial transcriptomics. However, the influence of sequencing errors, read truncation, and mapping errors on single-cell long-read-based isoform quantification has not been characterized. In this study, we started with a detailed examination of these sources of noise and described how UMI scattering affects single-cell isoform quantification. We also observed that misalignment and technical truncation of exons can lead to spurious alternative splicing detections, especially in the absence of a reference isoform annotation. To overcome these issues, we developed Longcell, a novel preprocessing method that efficiently provides accurate isoform quantification for long-read sequencing with single-cell and spatial barcodes. Through simulated and real data sets, Longcell is found to improve isoform quantification and allow precise identification of isoform switching between cell populations and spatial domains.
Our results inform experimental design for future single-cell long-read studies: It is important to note that we correct for read truncation and mapping errors by taking a “consensus” among reads assigned to the same UMI, which correspond to PCR duplicates. Thus, PCR replicates are both a blessing and a curse, and our simulations show that a mean PCR amplification fold of 5 already allows for the correction of many truncation and mapping errors. Thus, a shallow PCR amplification before sequencing is still recommended.
For downstream analyses, we proposed a novel metric to evaluate the exon- and splice-site usage heterogeneity within versus across single cells. Armed with this metric, we re-examined the question of intra-cell splicing heterogeneity that was the focus of previous studies43,44,45: do cells predominantly express only one isoform, or do they co-express multiple isoforms of the same gene? Across multiple datasets, we found that same-cell co-expression of multiple isoforms is common across highly expressed genes. Further research is required to reveal the biological mechanisms of intra-cell isoform switching.
We also developed a new test for differential splicing analysis that detects, for an exon or splice-site, a change in the distribution of its cell-specific PSI between two groups of cells. The test adjusts for differences in read coverage between cells and goes beyond shifts in mean to detect changes in the shape of the PSI distribution. To illustrate this method, we performed a splicing factor knock-out experiment and applied Longcell to identify targets for multiple splicing factors.
Methods
This study used only cell lines and public data and did not involve human participants, animal subjects, or other procedures requiring ethical approval.
Jurkat cell culture
Jurkat (ATCC TIB-152) and Cas9-stable Jurkat (SL555, GeneCopoeia, Inc., Rockville, MD, USA) cells were maintained in Roswell Park Memorial Institute (RPMI) 1640 medium (A3160702, Gibco) supplemented with 10% FBS (A3160702, Gibco).
CRISPR knock-out
Pooled splicing factor knock-out was performed. Briefly, the oligonucleotide pool for gRNA library targeting splicing factors was cloned to the lentiGuide-Puro plasmid (Addgene plasmid #52963), and the produced lentiviruses were transduced to Cas9 stable Jurkat cells using spinoculation at 800 g for 30 min at 32 °C. After that, cell pellets were resuspended to fresh media and plated in a six-well plate. After 72 h, transduced cells were selected by puromycin (Life Technologies, CA, USA)47.
Single-cell library preparation
Single-cell cDNA and gene expression libraries of cell lines and patient samples are generated by Chromium Next GEM single cell 5’ library & Gel Bead Kit v2 (PN-1000263, 10× Genomics, Pleasanton, CA, USA) as per the manufacturer’s protocol. Then, 6 pmol of gRNA scaffold binding primer (oJR160; AAGCAGTGGTATCAACGCAGAGTACCAAGTTGATAACGGACTAGCC) was added to RT master mix directly before droplet generation. After cDNA amplification, we performed 0.6× left-sided SPRI cleanup reaction for cDNAs and 0.6×–1.8× double-sided SPRI selection for gRNA using SPRIselect (Beckman Coulter Life Sciences, CA, USA). The sgRNA fractions were amplified by primers oJR163 and oJR165 (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT, CAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGAGTACCAAGTTGATAACGGACTAGCC) and sequenced with the gene expression library. We amplified cDNA and gene expression libraries with 16 and 14 cycles of PCR, respectively.
The quality of libraries was confirmed using 2% E-Gel (A42135, ThermoFisher Scientific, Waltham, MA, USA). They were then quantified by Qubit (Invitrogen).
Then, the single-cell cDNA libraries were converted to Oxford Nanopore libraries using a ligation sequencing kit (SQK-LSK110, Oxford Nanopore Technologies).
Illumina sequencing and preprocessing
Gene expression libraries for single-cell samples were sequenced using NovaSeq 6000 platforms using S4 flow cell (20028313, Illumina, San Diego, CA, USA) with recommended read cycles (Single-cell: 26, 8, 0, 91).
Cell Ranger (10× Genomics) version 5.0.0 “mkfastq” command was used to generate Fastq files, and “count” command was used with default parameters to do alignment to GRCh38 and to generate a matrix of UMI counts per gene and associated cell barcode.
DNA amplification for targeted sequencing
We used 3’-end phosphorothioate-modified primers for multiplexed PCR targeting transcript sequences (Supplementary Data 3). In the PCR procedure, single-cell cDNA (from Jurkat Cells in Fig. 2B, 10 μL) was added with 100 μM (total) phosphorothioate-modified primers (0.5 μL, resulting in 1 μM in the final PCR mix) with A1 adapter sequence (CTGGCTCCTCTGTATGTTGGAGAAT). Additionally, 10 μM partial R1 (CTACACGACGCTCTTCCGATCT) with 5’ phosphate (5 μL), Kapa master mix (25.0 μL) (KK2602, Roche), and water were added to make up the total volume to 50 μL. The thermal cycling conditions included denaturation at 62 °C for 40 s, extension for 60 s, and 50 cycles. The resulting PCR product underwent 1.2× Mag-Bind® TotalPure NGS bead purification (M1378-02, Omega Bio-tek), and the elution was done in 35 μL of water. For the Lambda nuclease step, purified DNA in distilled water (42.5 μL) was combined with Lambda exo buffer (5 μL) and Lambda exo nuclease (2.5 μL) (M0262S, New England Biolabs). The mixture was incubated at 37 °C for 1 h, followed by the addition of 1 μL of 0.5 M EDTA. Subsequently, the solution underwent a heat treatment at 75 °C for 10 min. The final step involved 1× Mag-Bind® TotalPure NGS bead purification, and the elution was performed in 22 μL of water. In the Final PCR, DNA (22 μL), primers which was used for previous PCR amplification, including 10 μM partial R1 (5 μL) with A1 sequence (0.5 μL), and Kapa master mix (25 μL) were combined. The thermal cycling conditions included a denaturation step at 62 °C, an extension step for 60 s, and 35 cycles. The resulting PCR product underwent 0.8× bead purification, and the elution was done in 30 μL of water.
Nanopore reads sequencing and preprocessing
cDNA libraries from the Jurkat CRISPR experiment and CRCLM sample were sequenced by Oxford Nanopore platform using Promethion R9.4.1 chemistry flow cell (FLO-PRO002, Oxford Nanopore Technologies) as per the manufacturer’s protocol.
The amplified cDNA library GSP based Jurkat targeted experiment was sequenced by ONT long-read sequencing platform (Oxford Nanopore Technology) using Promethion R10.4.1 chemistry flow cell (FLO-PRO114M, Oxford Nanopore Technologies) as per the manufacturer’s protocol.
We performed basecalling on the raw fast5 data using models of super accuracy (SUP) of Guppy5 for the Jurkat CRISPR experiment, and using Guppy6 with high accuracy models for the CRCLM sample and GSP-based Jurkat targeted sequencing.
Pacbio reads sequencing and preprocessing
cDNA or amplified DNA samples are sent to prepare the library for PacBio sequencing on SMRT cell to Azenta Life Sciences. The PacBio sequencing library was prepared according to the manufactured instruction (The BluePippin size selection system) using the PacBio prep kit 1.0 (100-222-300, Pacific Bioscience) followed by DNA damage repair and end repair are performed to ensure the integrity of fragmented DNA, then adapters with barcodes are ligated to both ends of the DNA fragments, facilitating the identification of individual sequences during the sequencing process. Following this, the prepared library is bound to a polymerase, forming a complex that is then loaded into the SMRT Cell for sequencing. The SMRT Cell, equipped with millions of zero-mode waveguides (ZMWs), serves as the platform for single-molecule sequencing. During the sequencing process, DNA polymerase synthesizes complementary strands in real-time, and the system detects the fluorescence signals emitted. The libraries were sequenced using PacBio P6C4 chemistry in one SMRT cell V3 (101-820-200, Pacific Bioscience) in PacBio Sequel II with 10 h movie time.
Data simulation
For the first simulation on 1000 genes, the cell-by-isoform expression matrix was simulated from the gene expression matrix from the 10X CRCLM sample. The expression for 1000 highly variable genes are referred to simulate the expression for 1000 isoforms, which come from 187 genes (Supplementary Fig. 4B).
Full length transcripts are first generated according to the simulated cell isoform expression matrix. Adapters, cell barcodes, and UMIs are attached to the 5’ end of each transcript to simulate the 10× single-cell 5’ library. Then, the first round of truncation at both ends is introduced to mimic the RNA degradation and early stop of reverse transcription. The length of truncated reads is guided by a Bernoulli-geometric distribution (By default the for each read, the probability to be truncated is 0.6, the length of truncation follows a geometric distribution G(0.05)). Then each read is amplified, and the amplification fold is according to the GC ratio of the read. We simulate the function between amplification fold (\(A\)) and GC ratio (\(G\)) with \(A=k\left({sigmoid}\left(0.5\left(G-50\right)\right)\right)+2\), in which \(k\) is a constant scaling factor (by default \(k=20\)). Another round of truncation will be introduced later to mimic the pore block during Nanopore long-read sequencing. Finally, each read is introduced to sequencing errors. The introduction of sequencing errors is guided by a Markov chain with four states: correct, indel, del, and wrong base. Different transition matrices are used to simulate different data quality.
For the second full transcriptome simulation, the cell-by-isoform expression matrix is simulated from the MOB Nanopore Visium sample. The simulation step is the same as the above process, but we focused on read downsampling in the high data quality setting. After the reads are simulated, the seqtk v1.3 is used to randomly sample the fastq files with sampling rates from 0.2 to 1 to simulate different sequencing depths.
Cell barcode and UMI assignment
-
1.
Search for the adapter sequence and polyA: For each read in the raw fastq, Longcell first searches for the adapter sequence aside the transcript according to the adapter design. For time efficiency, Longcell will first try searching the whole sequence of the adapter along the reads. If the full adapter could not be found, Longcell will next try searching the substring of the adapter in a sliding window way (with window size as 10 bp and step as 2 bp by default). Reads without or with over 1 valid adapters would be discarded. Reads with one valid adapter in the reverse strand will be reverse complemented. Overall, 50 ~ 80% reads can be found with a valid adapter in this step (Supplementary Fig. 20A).
For the 5’ library, the part of read upstream the position of the adapter would be trimmed. We preserve 55 bp at the 3’ end of the trimmed part as the tag region for further identification of cell barcode and UMI. Longcell will further search for polyA at the 3’ end of the trimmed reads (reads with over 15 A found in a 20 bp window are claimed to have a valid polyA). For the 3’ library, the part after polyA would be trimmed and 55 bp nearby the polyA would be preserved. This step would output a fastq with polished reads and table with three columns: read name, tag regions and the existence of the polyA.
-
2.
Cell barcode match (Supplementary Fig. 2A): The reference barcodes can be obtained from a matched Illumina sequencing of the cDNA library, or, in the absence of this Illumina run, one can use the 10X barcode pool that contains all possible barcodes. All barcodes will be vectorized into k-mers to build a k-mer dictionary (in our settings, we use k = 6). At the first iteration of barcode alignment, a prior distribution for barcode start position will be set as a normal distribution which could cover the whole tag region. The 95% confidence interval for this distribution should indicate the search region. Barcodes with the highest number of k-mer overlaps (top 5 by default) with the search region will be preserved as candidate barcodes. Each candidate barcode will slide over the search region to find the best match. The barcode with the minimum edit distance will be aligned and its start position will be used to update the start position distribution. After all reads get aligned with a cell barcode, low-quality alignments would be filtered out. There are two types of low-quality alignments. The first one is too many mismatches (with high edit distance (over 3 by default)). The second one is and the alignment has a deviant start position. More specifically, we would fit a normal distribution for confident cell barcode alignments (edit distance = 0). For other barcode alignments with mismatches, if their start positions are outside the 95% confidence interval of the above distribution, they would be filtered out as deviant cell barcodes. Overall, 50–70% reads out of all can be found with a valid cell barcode in this step (Supplementary Fig. 20A). As UMI is known to be located beside the cell barcode, after cell barcode alignment, we extract this region that putatively contains the UMI, with 1 bp flanking bases to be tolerant of insertions and deletions. A region with the same length as UMI from the adapter nearby the confidently identified cell barcode (edit distance = 0) will be extracted. We further calculate the Needleman-Wunsch score between the extracted region and its originally known sequence as an evaluation of data quality.
-
3.
Reads mapping: reads were aligned to the human Genome (GRCh38) with minimap2 v2.24 in spliced alignment mode (command: “minimap2 -ax splice -t $thread –junc-bed $bed –secondary=no –sam-hit-only $refer $fastq > $output”). The splice junction bed file was generated from the Gencode v39 GTF using paftools.js, a companion script of minimap2.
-
4.
Read filtering: Given the mapping result, each read is recorded as a set of splicing sites and its start and end position, for example \(s,{s}_{1}|{s}_{2},{s}_{3}|\ldots |{s}_{n},e\), in which \(s\) means the start position (starting from 5’ in the positive strand) and \(e\) means end position, and \({s}_{i}\) is a middle splice site. The count for each splicing site is summed across all reads for a gene. Reads with the splicing sites whose count is lower than a threshold (10 by default) will be filtered out.
-
5.
UMI clustering (Fig. 1G, step 2): We define a meta-isoform group as the set of reads representing isoforms which could transform to each other by end truncations and misalignment of small internal exons (shorter than 100 bp). For example, for two reads \({R}_{1},{R}_{2}\) with overlapping regions, here we define \(s\) as the start point of the read, and \(e\) as the end point of the read, \({base}\left({R}_{i},a,b\right)\) as the bases for read \({R}_{i}\) within point \(a\) and \(b\), then the overlapping region for the two reads should be \({s}_{o}=max\left({s}_{1},{s}_{2}\right),{e}_{o}=min\left({e}_{1},{e}_{2}\right)\). \({R}_{1},{R}_{2}\) belong to the same meta-isoform group if:
$$ {base}\left({R}_{1},{s}_{o},{e}_{o}\right)+{base}\left({R}_{2},{s}_{o},{e}_{o}\right)-2\times {intersect}\left[{base}\left({R}_{1},{s}_{o},{e}_{o}\right)\right.\\ \left.+{base}\left({R}_{2},{s}_{o},{e}_{o}\right)\right] < {thresh}$$For each meta-isoform group within each cell, we calculate Needleman-Wunsch scores between UMIs to serve as similarity. We then build a graph based on the Needleman-Wunsch similarity. Each node is a UMI and the edge weight is the Needleman-Wunsch score. For UMIs with length as \(l\), edges with weight lower than \(\frac{l}{2}+2\) will be filtered out. We then apply an iterative Louvain algorithm on this graph using the R package igraph v 1.5.1. Each time UMIs within the same cluster will be separated as a sub-graph. The iteration continues until all subgraphs are highly connected (min_cut > 3) or the minimum edge weight for all sub-graphs is larger than \(\frac{l}{2}+4\).
-
6.
Misalignment correction (Fig. 1G, step 3): During the mapping step, some reads can be wrongly mapped due to sequencing errors, especially for some small internal exons. As misalignment is mainly due to sequencing errors that occur after PCR amplification, wrongly mapped reads could coexist with correctly mapped reads in the same UMI cluster. Thus, we could correct misalignment cases during the UMI clustering step. We expect in the bulk level, there would be more correctly mapped reads compared to wrongly mapped reads, but within one UMI cluster, there can be more wrongly mapped reads as it is a random sampling process to split the bulk pool into each UMI cluster. Thus, simply collapsing reads in a UMI cluster into a consensus read could not completely correct for misalignment.
In detail, to correct for such misalignment cases, for a gene, within each UMI cluster, we order isoforms in the UMI cluster by their read counts, length and mapping quality. The isoform ranked first has the highest probability to represent the original transcript of the UMI cluster so it would be chosen as the representative for this cluster. An attribution table will be built to count how many reads for each isoform are assigned to this representative in this cluster. Such an attribution table will be summed across all UMI clusters across all cells for this gene as an integration. Thus, the attribution table would be a square matrix with the number of rows and columns equal to the number of kind of isoforms (\(n\)) for this gene. Here we call it as matrix \(A\) with \(n\) rows and columns. \({A}_{{ii}}\) means the number of reads for isoform \(i\) attributed to itself, while \({A}_{{ij}}{|i}\ne j\) means the number of isoform \(i\) attributed to isoform \(j\). As many wrong mapped reads coexist with the correct ones in the same UMI cluster and correct mapped reads should have quantity advantage, wrong mapped reads should be mostly assigned to the correctly mapped reads. Thus isoform \(i\) would be treated as a wrong mapped case when \(\frac{{A}_{{ii}}}{{\sum }_{i\ne j}^{n}{A}_{{ij}}} < {thresh}\) (2 by default). For each isoform \(i\) classified as a mis-mapping case, the associated reads are reassigned to isoform \(j={argma}{x}_{j}({A}_{{ij}})\). Following this correction, most UMI clusters are expected to contain a single isoform, allowing these clusters to be collapsed into a consensus read. For UMI clusters that still contain multiple isoforms, only the most prevalent isoform, determined by majority voting, is retained, and the reads for this isoform are collapsed into a consensus read.
As an illustration of the above, we have shown a toy example in Fig. 1D. There, we have three UMI clusters: cluster 1 contains two reads mapping to isoform (a) and a misaligned read (n), so the representative isoform for the cluster 1 should be isoform (a) and the misaligned read (n) should be attributed to isoform (a). Then, in the attribution table for this cell in step 3, the (n, a) is 1. Similarly, in cluster 1 the two reads of isoform (a) are attributed to isoform (a). In cluster 2, 1 read of isoform (a) is attributed to isoform (a), and thus in the single cell attribution table, the value for (a, a) is 3. After we build attribution tables for all single cells, we sum them up to get a cell population level statistic. In our example, in the attribution table across all cells, 20 reads of the isoform (n) are attributed to isoform (a) while only 10 reads are attributed to itself. That means isoform (n) always coexist and is dominated by isoform (a) within the same UMI cluster, across cells. Since each UMI cluster corresponds to the same RNA molecule, we thus classify isoform (n) as a misalignment and correct it into isoform (a). This use of UMI information for the detection and harmonization of alternative mappings within the same UMI cluster allows us to distinguish read misalignment versus true alternative splicing.
-
7.
Scatter reduction (Fig. 1G, step 4, Supplementary Fig. 5C): Molecules of the same isoform should have similar PCR amplification fold, as they share the same oligonucleotide sequence (Supplementary Fig. 5B). Thus, small clusters (e.g., singletons) that deviate from the “median” cluster size are likely to be due to the UMI scattering effect caused by sequencing errors. To estimate how many UMI singletons may be generated, we first evaluate the sequence data quality via the distribution of Needleman score for 12mer adapters upstream of the confidently identified cell barcode. Here we suppose the minimum threshold of Needleman score for a UMI to be clustered is \(k\) (by default \(k=\frac{l}{2}+2,l\) is the length of UMI), the ratio of adapters \(r\) with Needleman score lower than \(k\) is an estimation of the ratio of singletons. Suppose for an isoform in a cell, if all UMIs are correct, the size of each UMI cluster \(i\) is \({s}_{i}\). Now with the sequencing error, we estimate that within each UMI cluster, \(r{s}_{i}\) UMIs will get enriched with sequencing errors and couldn’t be clustered correctly, which lead to a smaller UMI cluster with size \(\left(1-r\right){s}_{i}\). The left \(r{s}_{i}\) UMIs will become singletons or small UMI clusters. Given such an assumption, suppose we have \(m\) UMI clusters for an isoform in a cell, we can rank them by their sizes in an increasing order as \({s}_{1},{s}_{2},\ldots,{s}_{m}\). Then the smallest cluster will be iteratively removed until \({\sum }_{i=1}^{p}{s}_{i}\ge \frac{r{\sum }_{j=p+1}^{m}{s}_{j}}{1-r}\). Then the UMI count would be \(m-p\). This strategy only uses the information from the sequencing data itself. Thus, it can be generalized.
Isoform identification and quantification
Longcell includes a module to assign the representative read alignment of each UMI cluster to a unique isoform in the annotation. For this section, we use “read” to denote the representative read of a UMI cluster. Due to the end truncations in Nanopore sequencing (Supplement Fig. 6A), many reads cannot be unambiguously assigned to a unique isoform, Longcell applies an iterative isoform assignment scheme (Supplement Fig. 6B), where the counts of unambiguously assigned reads serve as prior to guide the probabilistic assignment of the ambiguous reads. Besides using our isoform identification and quantification module, we also combine Longcell with IsoQuant to show the improvement of isoform quantification by denoising.
The isoform identification and quantification would be processed in 2 different ways given if the isoform annotation is provided.
-
1.
With reference isoform annotation: The Gencode v39 GTF was used here to guide the isoform assignment of reads. In the preprocessing step, each read has already been aligned to the reference genome template, giving a set of aligned segments, which we call putative exon regions, separated by gaps. Similarly, each isoform in the reference annotation also corresponds to a set of exons buttressed by the 5’ and 3’ untranslated regions. For a read to be assigned to an annotated isoform, the putative exon regions contained by the read must be a subset of the exon regions in the annotated isoform. A maximum total number of small offsets are allowed at the starts and ends of the middle exons (default = 3 bp). Reads that have no compatible isoform assignments, at this step, are discarded. The remaining reads may be assigned to multiple compatible isoforms in the reference annotation. Let \({U}_{{ri}}\) be the indicator of whether read \(r\) is compatible with isoform \(i\), which is known at this point. Let \({Z}_{{ri}}\) be the indicator of whether read \(r\) originated from isoform \(i\), which is unknown and is the variable that we would like to impute. Let \({p}_{i}\) be the true proportion of transcripts from the gene that are from isoform \(i\). Let \({F}_{i}\left(r\right)\) be the probability of a transcript of isoform \(i\) being observed in our data in the form of read \(r\), e.g., through truncation and misalignment. Then, treating \({Z}_{{ri}}\) as missing data, the complete data log-likelihood is of the form
$$L=\log P\left(U,Z|p\right)={\sum }_{r,i}{Z}_{{ri}}\left[\log \left({F}_{i}\left(r\right)+\log \left({p}_{i}\right)\right)\right]$$(1)Treating \(Z\) as missing data, we can estimate the isoform proportions \(p\) by Expectation-Maximization (EM). In the E-step of EM, with the current iteration’s estimate \({p}^{(t)}\) of \(p\), we compute
$$E\left[{Z}_{{ri}}|U,{p}^{\left(t\right)}\right]=\frac{{F}_{i}\left(r\right){p}_{i}}{{\sum }_{{i}^{{\prime} }}{U}_{{i}^{{\prime} }r}{F}_{{i}^{{\prime} }}\left(r\right){p}_{{i}^{{\prime} }}}$$(2)In the M-step of EM, the estimates of \(p\) are updated by a weighted tabulation based on \(E[{Z}_{{ri}}{|U},{p}^{(t)}]\). In actual computation, we perform smart initialization by first pre-computing an ordering \(R\) of the reads based on the number of compatible isoform assignments, with the uniquely assigned reads coming first, the reads with two compatible isoforms coming next, etc. The count of uniquely assigned reads (reads which are only compatible to one annotated isoform) are tabulated to serve as the initial values of \(p\). The values of \({Z}_{{ri}}\) are then updated based on the pre-computed ordering \(R\), with the values of \(p\) updated after the update \([{Z}_{{ri}}:i=1,\ldots,I]\) of of each read.
For the emission distribution \({F}_{i}(r)\), we simply assumed that truncation can happen anywhere within a transcript with equal probability, and thus assumed a uniform assignment distribution of each read to its compatible isoforms. This is because we could not observe a reliable pattern for the length and position of truncations in our data, and thus feel that a uniform prior is more appropriate.
-
2.
Without isoform annotation: Each transcript would be recorded as a vector of splicing sites in a truncation-aware model to be tolerant of end truncations. The truncation-aware model means if an exon is not covered by the range for a read, we treat it as missing instead of a spliced-out case. As an example, for a gene with splicing sites \({s}_{1},{s}_{2},\ldots,{s}_{n}\), a transcript can be represented as \(s,{s}_{k1}|{s}_{k2},{s}_{k3}|\ldots |{s}_{{km}},e\), in which \(s\) is the start sites at 5’ while \(e\) is the end site at 3’ in the positive strand. This transcript is then transformed to a vector \(v\) with length \(n\). Suppose \({s}_{i} < s\) and \({s}_{i+1} > s\), then \(v\left[1:i\right]={NA}\), as it’s hard to know if it’s an alternative 5’ event or just a technical truncation. For the 3’ end, suppose \({s}_{j} < e\) and \({s}_{j+1} > e\), if the polyA exists, \(v\left[j+1:n\right]=0\) while if not, \(v\left[j+1:n\right]={NA}\). For the middle part, \(v\left[{k}_{i}\right]=1,i={{\mathrm{1,2}}},\ldots,m\) while other sites are filled with \(0\). Based on this way we could get a read by splice site matrix with number of rows as number of reads, and number of columns as number of all splicing sites for this gene, and this matrix is filled with \({{\mathrm{0,1}}},{NA}\).
Splice sites which always coexist or are mutually exclusive would further be merged as meta-splice sites to avoid repetitive calculations in the downstream analysis. To be more specific, for two columns in the splice site matrix, one represents splicing sites \({s}_{i}\) and the other represents site \({s}_{j}\), we use \({agree}({s}_{i},{s}_{j})\) to represent the count of reads in which \({s}_{i}\) and \({s}_{j}\) are concomitant, and \({disagree}({s}_{i},{s}_{j})\) to represent the count of reads in which \({s}_{i}\) and \({s}_{j}\) are mutually exclusive. we then define the similarity between them as
As \({NA}\) won’t be counted in the \({agree}({s}_{i},{s}_{j})\) and \({disagree}({s}_{i},{s}_{j})\), to avoid the splicing sites are merged into sites with many \({NA}\), here we use \({nonNA}\left({s}_{i}\right)\) to indicate the number of reads in which \({s}_{i}\) is not \({NA}\). And we require \({agree}({s}_{i},{s}_{j})+{disagree}({s}_{i},{s}_{j})\ge \frac{{nonNA}({s}_{i})+{nonNA}({s}_{j})}{2}\), otherwise we set \({sim}({s}_{i},{s}_{j})=0\).
Then we do clustering based on the similarity between each pair of splicing with the cutoff \({sim}({s}_{i},{s}_{j})\ge 0.95\). Different splicing site similarity above 0.95 doesn’t affect too much for the differential alternative splicing test (Supplementary Fig. 12F). Splicing sites within one cluster would be merged as a meta site.
To model the alternative splicing based on such a quantification, for each meta-splicing site \(i\) in a cell \(c\), the count of reads with \({non}-{NA}\) value for this site is its gene count \({{G}_{c}}^{i}\), while the count of reads with \(1\) for this site is the spliced-in count \({{X}_{c}}^{i}\).
Comparison with other methods
All methods (FLAMES (git commit hash: 774e16ae53a1430e03081970827e93f1fbaecead), sicelore2 v1, wf-single-cell v0.2.9, IsoQuant v3.3) are applied to the benchmark datasets with their default parameters. To be fair, all methods used the same mapping software (minimap2 v2.24 and samtools v1.18), reference genome (GRCh38), and isoform annotations (Gencode v39 GTF).
For the benchmark based on simulation, simulated fastqs are used as input for all methods except for isoquant. Barcode whitelist is also input for FLAMES, sicelore2, and Longcell. As wf-single-cell searches for cell barcodes without the barcode whitelist, the cell barcode recovery is not compared. Isoquant is not specifically designed for single cells and does not have a cell barcode identification module. Here, we used the barcode-identified fastq from sicelore2 as the input for isoquant. Each method outputs a cell-by-isoform count matrix, and then all matrices are fully joined with the ground truth matrix by isoform. The Pearson correlation for each cell between the ground truth and output was calculated to evaluate the performance of each method.
For the benchmark based on cell-matched PacBio and Nanopore sequencing, we processed the Pacbio HIFI reads by their official tool isoseq v3.4.0 with default parameters except the coverage when collapsing reads with the same UMI into an isoform (--min-aln-coverage = 0.5, --min-aln-identity = 0.9). The Nanopore sequencing for this sample was input to the methods we compared in the same way as the simulation. And the evaluation is also the same as simulation.
For the time and RAM usage comparison (Supplementary Fig. 21), we used the down-sampled CRCLM single-cell long reads data as the low cell number benchmark (490 genes in 282 cells) and GSP-based Jurkat targeted sequencing as the high cell number benchmark (18 genes in 5881 cells). The benchmark datasets were further down-sampled into 5 datasets with a gradient number of reads (600k, 1200k, 1800k, 2400k, 3000k). We applied FLAMES, sicelore2, Longcell, and IsoQuant on the benchmark datasets on a high-performance computing grid (Rocky Linux 8 × 86_64 on a variety of AWS EC2 instance types). (Unfortunately, we could not install wf-single-cell on our high-performance computing grid. We ran the analysis based on wf-single-cell on a Windows platform, and thus, the running time and RAM usage are not comparable. Sicelore2 did not finish for some of the data within the benchmark).
Quantification of single-cell isoform heterogeneity
\(\phi\) estimation: for each meta-splicing site \(i\) in a gene in a cell \(c\), we computed its gene count \({{G}_{c}}^{i}\) and spliced-in count \({{X}_{c}}^{i}\) during the isoform quantification without annotation in Longcell. We modeled the spliced-in count with a binomial distribution \({X}_{c}^{i}\sim {Binomial}({G}_{c}^{i},{\psi }_{c}^{i})\). Then, we modeled the \({\psi }_{c}^{i}\) with a beta distribution \({Beta}({\alpha }^{i},{\beta }^{i})\). Thus, we modeled the spliced-in count with a beta-binomial distribution as \({X}_{c}^{i}\sim {BetaBinomial}({G}_{c}^{i},{\alpha }^{i},{\beta }^{i})\). \({\alpha }^{i},{\beta }^{i}\) are estimated by maximum likelihood estimation and then we estimate \({\phi }^{i}=\frac{1}{{\alpha }^{i}+{\beta }^{i}+1}\). To estimate the confidence interval of \({\phi }^{i}\) estimation, we applied Bootstrap by estimating \({\alpha }^{i},{\beta }^{i}\) multiple times. For time efficiency, as the estimation of \({\phi }^{i}\) for many lowly expressed genes has high variance and is not informative, here we only estimate the \({\phi }^{i}\) for highly expressed genes. Only meta-sites with \({{G}_{c}}^{i}\ge 10\) in over 30 cells would be used to calculate \({\phi }_{i}\). Meta splice sites with \(0.1 < \bar{{\psi }^{i}} < 0.9\) and \({CI}({\phi }^{i}) < 0.2\) are preserved as valid results.
Differential alternative splicing analysis
We also modeled the spliced-in count with a binomial distribution \({X}_{c}^{i}\sim {BetaBinomial}({G}_{c}^{i},{\alpha }^{i},{\beta }^{i})\). Based on this distribution, we could do a generalized likelihood ratio test to identify if two groups \(({g}_{1}\,{and}\,{g}_{2})\) share the same parameters of \(\alpha\) and \(\beta\). The distance between the \({\psi }_{i}\) distributions from two groups is quantified as the Wasserstein distance. The confidence interval for the distribution distance is calculated by bootstrap. FDR control is applied to significant meta splice sites and signals with \({fdr} > 0.05\) were filtered out.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The single-cell RNA-seq for Jurkat targeted genes (used for benchmark, including ONT and PacBio), and CRISPR experiment on Jurkat and stimulated Jurkat cells (used for differential alternative splicing test) in this study are available from the Gene Expression Omnibus with the accession GSE295932. The single-cell RNA-seq and Visium for the CRCLM sample are available at https://www.ncbi.nlm.nih.gov/gap/sstr/report/phs001818.v4.p1 with the entry id as 8640. The single-cell RNA-seq for the mouse embryonic brain sample is available in Gene Expression Omnibus under accession number GSE130708 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi]. The Visium for the MOB sample is available in Gene Expression Omnibus under accession number GSE153859 [https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi]. Source data are provided with this paper in https://doi.org/10.5281/zenodo.15320816.
Code availability
The scripts for the preprocessing of single-cell long reads are wrapped and collected in two R packages: Longcellsrc49 and LongcellPre50. Longcellsrc is available at https://github.com/yuntianf/Longcellsrc (https://doi.org/10.5281/zenodo.15318856) under the MIT License. This project incorporates functions adapted from kseqpp (licensed under the MIT License: https://github.com/cartoonist/kseqpp.git). LongcellPre is available at https://github.com/yuntianf/LongcellPre (https://doi.org/10.5281/zenodo.15318854) under the MIT License. The scripts for downstream analysis are wrapped into the R package Longcell51: https://github.com/yuntianf/Longcell (https://doi.org/10.5281/zenodo.15318807).
References
Scotti, M. M. & Swanson, M. S. RNA mis-splicing in disease. Nat. Rev. Genet 17, 19–32 (2016).
Baralle, F. E. & Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017).
Zhang, Y., Qian, J., Gu, C. & Yang, Y. Alternative splicing and cancer: a systematic review. Signal Transduct. Target Ther. 6, 78 (2021).
Stanley, R. F. & Abdel-Wahab, O. Dysregulation and therapeutic targeting of RNA splicing in cancer. Nat. Cancer 3, 536–546 (2022).
Zhang, X. et al. Cell-type-specific alternative splicing governs cell fate in the developing cerebral cortex. Cell 166, 1147–1162 e1115 (2016).
Yeo, G., Holste, D., Kreiman, G. & Burge, C. B. Variation in alternative splicing across human tissues. Genome Biol. 5, R74 (2004).
Marasco, L. E. & Kornblihtt, A. R. The physiology of alternative splicing. Nat. Rev. Mol. Cell Biol. 24, 242–254 (2023).
Westoby, J., Artemov, P., Hemberg, M. & Ferguson-Smith, A. Obstacles to detecting isoforms using full-length scRNA-seq data. Genome Biol. 21, 74 (2020).
Arzalluz-Luque, A. & Conesa, A. Single-cell RNAseq for the study of isoforms-how is that possible? Genome Biol. 19, 110 (2018).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Keren-Shaul, H. et al. MARS-seq2.0: an experimental and analytical pipeline for indexed sorting combined with single-cell RNA sequencing. Nat. Protoc. 14, 1841–1862 (2019).
Hashimshony, T. et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-seq. Genome Biol. 17, 77 (2016).
Hagemann-Jensen, M. et al. Single-cell RNA counting at allele and isoform resolution using Smart-seq3. Nat. Biotechnol. 38, 708–714 (2020).
Picelli, S. et al. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014).
Picelli, S. et al. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods 10, 1096–1098 (2013).
Byrne, A. et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 8, 16027 (2017).
Gupta, I. et al. Single-cell isoform RNA sequencing characterizes isoforms in thousands of cerebellar cells. Nat. Biotechnol. 36, 1197–1202 (2018).
Lebrigand, K., Magnone, V., Barbry, P. & Waldmann, R. High throughput error corrected Nanopore single cell transcriptome sequencing. Nat. Commun. 11, 4025 (2020).
Tian, L. et al. Comprehensive characterization of single-cell full-length isoforms in human and mouse with long-read sequencing. Genome Biol. 22, 310 (2021).
Rhoads, A. & Au, K. F. PacBio sequencing and its applications. Genom. Proteom. Bioinform. 13, 278–289 (2015).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Al’Khafaji, A. M. et al. High-throughput RNA isoform sequencing using programmed cDNA concatenation. Nat. Biotechnol. 42, 582–586 (2024).
Volden, R. et al. Improving nanopore read accuracy with the R2C2 method enables the sequencing of highly multiplexed full-length single-cell cDNA. Proc. Natl. Acad. Sci. USA 115, 9726–9731 (2018).
Joglekar, A. et al. Single-cell long-read sequencing-based mapping reveals specialized splicing patterns in developing and adult mouse and human brain. Nat. Neurosci. 27, 1051–1063 (2024).
Kabza, M. et al. Accurate long-read transcript discovery and quantification at single-cell, pseudo-bulk and bulk resolution with Isosceles. Nat. Commun. 15, 7316 (2024).
Lebrigand, K. et al. The spatial landscape of gene expression isoforms in tissue sections. Nucleic Acids Res. 51, e47 (2023).
Philpott, M. et al. Nanopore sequencing of single-cell transcriptomes with scCOLOR-seq. Nat. Biotechnol. 39, 1517–1520 (2021).
Volden, R. & Vollmers, C. Single-cell isoform analysis in human immune cells. Genome Biol. 23, 47 (2022).
Ni, Y., Liu, X., Simeneh, Z. M., Yang, M. & Li, R. Benchmarking of Nanopore R10.4 and R9.4.1 flow cells in single-cell whole-genome amplification and whole-genome shotgun sequencing. Comput. Struct. Biotechnol. J. 21, 2352–2364 (2023).
You, Y. et al. Identification of cell barcodes from long-read single-cell RNA-seq with BLAZE. Genome Biol. 24, 66 (2023).
Amarasinghe, S. L. et al. Opportunities and challenges in long-read sequencing data analysis. Genome Biol. 21, 30 (2020).
Prjibelski, A. D. et al. Accurate isoform discovery with IsoQuant using long reads. Nat. Biotechnol. 41, 915–918 (2023).
Technologies, O.N. wf-single-cell (v0.2.9). https://github.com/epi2me-labs/wf-single-cell.git (2023).
Kovaka, S. et al. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 20, 278 (2019).
Tang, A. D. et al. Full-length transcript characterization of SF3B1 mutation in chronic lymphocytic leukemia reveals downregulation of retained introns. Nat. Commun. 11, 1438 (2020).
Pardo-Palacios, F. J. et al. SQANTI3: curation of long-read transcriptomes for accurate identification of known and novel isoforms. Nat. Methods 21, 793–797 (2024).
Wyman, D. et al. A technology-agnostic long-read analysis pipeline for transcriptome discovery and quantification. Preprint at biorxiv, 672931 (2019).
Chen, Y. et al. Context-aware transcript quantification from long-read RNA-seq data with Bambu. Nat. Methods 20, 1187–1195 (2023).
Volden, R. et al. Identifying and quantifying isoforms from accurate full-length transcriptome sequencing reads with Mandalorion. Genome Biol. 24, 167 (2023).
Gao, Y. et al. ESPRESSO: Robust discovery and quantification of transcript isoforms from error-prone long-read RNA-seq data. Sci. Adv. 9, eabq5072 (2023).
Biosciences, P. Iso-Seq (v3.4.0) Analysis documentation. https://isoseq.how/ (2024).
Shalek, A. K. et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 498, 236–240 (2013).
Song, Y. et al. Single-cell alternative splicing analysis with expedition reveals splicing dynamics during neuron differentiation. Mol. Cell 67, 148–161.e145 (2017).
Buen Abad Najar, C. F., Yosef, N. & Lareau, L. F. Coverage-dependent bias creates the appearance of binary splicing in single cells. Elife 9, e54603 (2020).
Sathe, A. et al. Colorectal cancer metastases in the liver establish immunosuppressive spatial networking between tumor-associated SPP1+ macrophages and fibroblasts. Clin. Cancer Res. 29, 244–260 (2023).
Kim, H. S., Grimes, S. M., Hooker, A. C., Lau, B. T. & Ji, H. P. Single-cell characterization of CRISPR-modified transcript isoforms with nanopore sequencing. Genome Biol. 22, 331 (2021).
Topp, J. D., Jackson, J., Melton, A. A. & Lynch, K. W. A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14, 2038–2049 (2008).
Fu, Y. Single-cell and spatial alternative splicing analysis with Nanopore long read sequencing. Longcellsrc https://doi.org/10.5281/zenodo.15318856 (2025).
Fu, Y. Single-cell and spatial alternative splicing analysis with Nanopore long read sequencing. LongcellPre https://doi.org/10.5281/zenodo.15318854 (2025).
Fu, Y. Single-cell and spatial alternative splicing analysis with Nanopore long read sequencing. Longcell https://doi.org/10.5281/zenodo.15318807 (2025).
Acknowledgements
This work was supported by US National Institutes of Health grants U01CA217875 (HPJ and AS), R33CA247700 (HPJ), R35HG011292-01 (BTL, HSK), and 5R01-HG006137 (YF, NRZ). HPJ also received support from the American Cancer Society Mission Boost award (MBGI-21-109-01—MBG). Additional support was received from the Clayville Foundation. AS received additional support from the Stanford University Translational Research and Applied Medicine (TRAM) pilot grant program. This work with the Stanford Cancer Institute biobank was supported by a National Cancer Institute Cancer Center Support Grant (P30CA124435).
Author information
Authors and Affiliations
Contributions
Conceptualization: Y.F., H.P.J., N.R.Z. Initial data analysis and preprocessing: Y.F., S.H. Algorithm for cell barcode recovery: Y.F., S.M.G. Algorithm for UMI recovery: Y.F. Model formulation for downstream analyses: Y.F., N.R.Z. Algorithm development and implementation: Y.F. Simulation design and data analysis: Y.F. Data generation for liver metastasis sample: H.K., A.S., B.T.L. Single cell perturbation experiment: H.K. Data analysis: Y.F. Benchmarking: Y.F., J.I.A., S.H. Targeted sequencing validation: H.K. Pacbio sequencing validation: S.R. Manuscript writing: Y.F., N.R.Z. with feedback from J.I.A., H.K., and H.P.J. Supervision: H.P.J. and N.R.Z.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fu, Y., Kim, H., Roy, S. et al. Single cell and spatial alternative splicing analysis with Nanopore long read sequencing. Nat Commun 16, 6654 (2025). https://doi.org/10.1038/s41467-025-60902-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-60902-2
This article is cited by
-
Mapping isoforms and regulatory mechanisms from spatial transcriptomics data with SPLISOSM
Nature Biotechnology (2026)