Abstract
Neuronal TDP-43 aggregates are a hallmark ALS pathology. The integrated stress response (ISR) occurs downstream of TDP-43 pathology and may promote neurodegeneration. Here we demonstrate that a CNS penetrant small molecule eIF2B activator inhibits the ISR in cellular models of ALS and the brain of an inducible mouse model of TDP-43 pathology, where it transiently slowed progression of locomotor deficits and neurodegeneration. ISR activation was observed in ALS patient spinal cord and CSF. The investigational drug DNL343 was advanced into Phase 1 and Phase 1b randomized, double-blind, placebo-controlled trials in healthy and ALS participants, respectively (NCT04268784/NCT05006352); the primary objective in both studies was to investigate the safety and tolerability DNL343. DNL343 demonstrated a half-life supporting once-daily dosing and showed extensive CSF distribution. DNL343 was generally well tolerated and reduced ISR biomarkers in peripheral blood mononuclear cells and CSF of ALS participants. Therefore, DNL343 is a useful investigational drug to explore the effects of ISR inhibition in ALS models and individuals with neurological diseases.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease, marked by progressive loss of motor neurons, muscle weakness, paralysis, and ultimately death. Roughly 10% of ALS have a defined genetic basis, while 90% of ALS is sporadic with no known genetic contribution. While the exact molecular mechanisms underlying ALS pathogenesis remain unclear, several genes modifying ALS risk mediate RNA metabolism, including TARDBP, TIA1, FUS, and C9orf721,2,3,4,5. TARDBP’s gene product, TDP-43, is an essential, predominantly nuclear RNA binding protein (RBP) involved in RNA processing6,7. Although TARDBP mutations account for 1−5% of ALS cases, predominantly cytoplasmic TDP-43 inclusions are found in brains and spinal cords in ∼97% of people with ALS, including most individuals with sporadic ALS and C9orf72 repeat expansions8,9. Nuclear loss of TDP-43, which is also observed in disease, likely contributes to pathogenesis by de-repressing insertion of cryptic exons into hundreds of mRNAs, leading in most cases to nonsense-mediated RNA decay and downregulation of mRNA expression10,11,12,13,14,15,16,17. Therefore, altered RNA metabolism or RBP function may be a common driver of ALS pathogenesis and these cellular dysregulations may be superimposed onto toxic gain of function mechanisms triggered by TDP-43 aggregates1,7,18,19.
Growing evidence indicates that the integrated stress response (ISR) pathway contributes to ALS pathogenesis and is intimately linked to the biology of disease-associated RBPs, including TDP-43. Aberrant ISR pathway activation was observed in several ALS models, involving expression of mutant TDP-4320,21,22,23 and the presence of a C9orf72 hexanucleotide (G4C2) repeat expansion (HRE) or its dipeptide repeat protein (DPR) products23,24,25,26,27,28,29. ISR is mediated by four kinases (PERK, GCN2, PKR and HRI) that phosphorylate eukaryotic Initiation Factor 2 alpha (eIF2α) in response to various cellular insults, including defects in proteostasis, oxidative stress and nutrient deprivation30,31,32,33,34,35,36,37. Phosphorylated eIF2α in turn inhibits the guanine nucleotide exchange factor (GEF) activity of eukaryotic Initiation Factor 2B complex (eIF2B), rendering it inactive. Global protein translation stalling ensues, and untranslated mRNAs induce formation of membrane-less RNA-rich organelles termed stress granules36,38,39. ISR activation also involves the upregulation of stress response genes, such as activating transcription factor 4 (ATF4), which eludes this translational inhibition. ATF4 subsequently increases transcription of other stress response genes, including regulators of metabolism (e.g., CHAC1), nutrient uptake (e.g., amino acid transporters SLC1A5 and SLC7A5) and apoptosis (e.g., TRIB3 and DDIT3)31,32,40. Acutely induced ISR is commonly viewed as a protective adaptive response, whereas chronic activation of this pathway can be maladaptive and deleterious21,41,42,43. Neurons appear to be particularly sensitive to ISR activation, which can lead to neurodegeneration, for instance upon nerve injury36 or with LoF EIF2B5 mutations causing vanishing white matter disease (VWMD), a rare degenerative leukodystrophy44,45,46,47,48. ISR pathway activation has also been reported in spinal cord from people with ALS, based on higher levels of phosphorylated eIF2α, CHOP/DDIT3 and other markers49.
Generally, various stress factors have been shown to trigger ISR in ALS models, including oxidative stress, unfolded protein response and mitochondria dysfunction, and TDP-43 proteinopathy may account in part for ALS-linked ISR activation49,50,51. While cytoplasmic accumulation of TDP-43 can promote ISR, growing evidence indicates ISR itself may exacerbate TDP-43 aggregation, pointing to potential feedforward pathogenic mechanisms. Indeed, TDP-43 is recruited to stress granules via its COOH-terminal low complexity domain upon ISR activation and this association can facilitate cytoplasmic TDP-43 aggregation30,31,32,33,34,35,36,37,52. Additionally, stress granule formation is sufficient to drive TDP-43 aggregation31,32,40. Conversely, deletion of stress granule-associated proteins, such as ataxin-2 (ATXN-2), or pharmacological inhibition of stress granule formation can attenuate TDP-43 aggregation and neurodegeneration in various models of ALS21,41,42,43. Collectively, these studies suggest inhibition of the ISR may represent an attractive therapeutic approach for ALS36,39,53.
Pharmacological modulation of the ISR pathway via eIF2B activation using small molecules has been demonstrated in the literature44,46,54,55,56. The original eIF2B activator, ISRIB, was shown to stabilize the eIF2B complex through a direct interaction and stimulate its GEF activity despite the presence of phosphorylated eIF2α57,58. The ISRIB analog 2BAct has also exhibited neuroprotective activity in an eIF2B mutant mouse model of VWMD44,46. However, ISRIB exhibits poor solubility and oral bioavailability44,59 and 2BAct is associated with safety findings46, complicating their clinical development. We are now developing DNL343, a CNS-penetrant, potent small molecule eIF2B activator, which demonstrates a desirable drug profile, including a long half-life and high oral bioavailability across preclinical species60. DNL343 reduced CNS ISR activation and neurodegeneration in both the optic nerve crush injury and eIF2B mutant mice48. In the latter model, ISR pathway biomarker Growth Differentiation Factor-15 (GDF-15), a member of the transforming growth factor-β (TGF-β) superfamily61, was corrected in a dose-dependent fashion in the mouse brain and CSF after chronic dosing of DNL343, which also reduced plasma neurofilament light (NfL) and rescued neurological deficits48. While the eIF2B mutant mouse model enabled proof-of-concept efficacy and biomarker studies, whether eIF2B activators can attenuate ISR in the CNS of ALS preclinical models and ultimately people with ALS remains a critical unanswered question.
In this study, we show that ISR activation by pathological TDP-43 and stress granule-induced TDP-43 phase transitions are rescued by DNL343 in cellular models. We then demonstrate that reduction of ISR activation via eIF2B activation in the brain of rNLS8 transgenic mice, a preclinical model of TDP-43 pathology22, results in a mild and transient delay in developing locomotor deficits and a transient reduction in plasma NfL levels. We advanced DNL343 into clinical trials and present pharmacokinetics (PK), pharmacodynamics (PD), and safety results of both a Phase 1 trial in healthy participants and a Phase 1b trial in participants with ALS. We demonstrate ISR pathway engagement using a PD biomarker assay in PBMCs and reduced levels of the ISR marker GDF-15 protein in the CSF of ALS participants at exposures that were generally safe and well tolerated, supporting the potential for DNL343 to be used to explore the effects of ISR inhibition in participants with ALS.
Results
DNL343 prevents ISR induced by expression of cytoplasmic TDP-43 and C9orf72 HRE
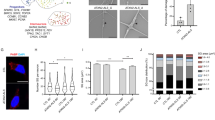
Expression of mutant ALS-linked genes including TARDBP and C9orf72 HRE have been shown to induce ISR activation22,23,24,25,26,27,28,29. The first step in the ISR is the phosphorylation of eIF2α by the ISR kinases (PERK, PRK, HRI, GCN2) (Fig. 1a)62. To assess the effect of our eIF2B activator, DNL343, we first engineered H4 neuroglioma cells inducibly expressing a truncated GFP-tagged TDP-43 fusion protein lacking the NH2-terminus that harbors the nuclear localization signal (NLS), GFP-TDP-43(86-414), and thus modeling the disease-associated cytoplasmic mislocalization of TDP-43. A full-length, wildtype TDP-43 GFP fusion (GFP-TDP-43FL) was expressed in parallel as a control to distinguish the effect of TDP-43 cytoplasmic mislocalization on ISR from its ectopic expression. Induction of GFP-TDP-43(86-414) with doxycycline (Dox) treatment for 24 h increased phosphorylation of eIF2α (Supplementary Fig. 1a,b) and nuclear levels of ATF4 (Fig. 1b, c) compared to expression of GFP control and GFP-TDP-43FL. Pretreatment with DNL343 did not alter the levels of phospho-eIF2α (p-eIF2α) in the GFP-TDP-43 expressing cells, consistent with the fact eIF2B activation is downstream of eIF2α phosphorylation (Fig. 1a and Supplementary Fig. 1b). In contrast, DNL343 prevented the upregulation of both nuclear and total ATF4 protein (Fig. 1b-d). Interestingly, expression of GFP-TDP-43FL increased total ATF4 to levels observed with GFP-TDP-43(86-414) whereas nuclear ATF4 remained unchanged (Fig. 1c-d). To further evaluate expression changes downstream of ATF4 in both conditions, we performed bulk RNA-seq on H4 cells expressing inducible GFP-TDP-43FL, GFP-TDP-43(86-414) or GFP alone. Compared to the limited number of ISR genes upregulated with full length TDP-43 induction, cytoplasmic TDP-43 expression caused upregulation of most ISR gene transcripts, including MTHFD2, GDF15, SLC7A11, and ATF3, relative to the GFP expressing samples (Fig. 1e-f). Thus, robust ISR activation was observed with both cytoplasmically mislocalized TDP-43 and overexpression of full-length wildtype TDP-43 itself. To assess differential expression at the pathway level, we curated 30 previously reported ISR genes from the literature (Supplementary Data 1). This gene set is significantly upregulated by GFP-TDP-43(86-414) expression (Fig. 1g, GSEA p-value: 3.8×10-8). Moreover, transcript level changes induced by GFP-TDP-43(86-414) were prevented by DNL343 treatment, with significant downregulation of ISR genes, including CHAC1, ATF4, and DDIT3 (Fig. 1h).
a Schematic depicting the ISR pathway, including kinases mediating the phosphorylation of eIF2α, and activation of eIF2B by DNL343. b Confocal microscopy images showing induced expression of GFP-TDP-43FL and GFP-TDP-43(86-414) variant in H4 cells increased immunoreactivity of ATF4 (anti-ATF4, red) compared to GFP control. This ATF4 signal was reduced with 1 µM DNL343 treatment. Scale bar: 20 µm. c Quantification of ATF4 immunoreactivity in (b) from n = 3 (GFP Control) or 4 biological replicates (GFP-TDP-43 variants). a.u.: arbitrary units. d ECLIA-based assay result demonstrating elevated ATF4 protein levels with induced expression of GFP-TDP-43FL and GFP-TDP-43(86-414), compared to GFP control cells. Pre-treatment with DNL343 reduced ATF4 protein levels in each cell line (n = 6 biological replicates). e Volcano plot of differentially regulated genes in H4 cells induced with GFP-TDP-43FL compared to GFP control. Representative ISR genes with significant expression changes are labeled as red (upregulation) or blue (downregulation) and insignificant genes as gray. Significance cutoff, adjusted p-value < 0.05. f Volcano plot of differentially regulated genes in H4 cells induced with GFP-TDP-43(86-414) compared to GFP control. Representative ISR genes are labeled as in (e). Significance cutoff, adjusted p-value < 0.05. g Gene set enrichment analysis (GSEA) plot for ISR gene comparing truncated GFP-TDP-43(86-414) from GFP control expressing cells. h Volcano plot of differentially regulated genes 24 hours after DNL343 treatment in H4 cells induced with GFP-TDP-43(86-414). The same set of representative ISR genes from (e) and (f) are labeled. All data are shown as mean ± SEM (c, d). Statistical significance was determined with one-way ANOVA, with Tukey’s multiple comparison, ns P > 0.05. Source data are provided in Source Data file.
We next investigated whether DNL343 can also block C9orf72 HRE-induced ISR. HEK293 cells were engineered to stably express either GFP or a C9orf72 repeat expansion linked to GFP ((G4C2)71-GFP). Expression of (G4C2)71-GFP significantly increased ATF4 protein levels by ∼2.3 fold compared to GFP expression alone and DNL343 treatment prevented this increase (Supplementary Fig. 1c). Expression of (G4C2)71-GFP upregulated most of the pre-defined ISR genes compared to GFP expression alone (Supplementary Fig. 1d, e). These transcriptional changes were largely corrected by DNL343 treatment (Supplementary Fig. 1f).
DNL343 prevents de novo stress granule formation and dissolves preformed stress granules
To understand whether ISR activation also contributes to cytoplasmic TDP-43 inclusions as a result of stress granule formation, H4 cells were engineered to express inducible GFP-tagged versions of wildtype (WT) or mutated forms of TDP-43, as well as other RBPs previously shown to associate with stress granules, such as TIA1 and FUS. Mutated forms of RBPs involved NLS-truncated TDP-43 (TDP-43(86-414)) with the additional ALS-linked mutation M337V (TDP-43(86-414, M337V)), ALS-linked mutation of TIA1 (TIA1(A381T)), and NLS-truncated version of FUS(G515X). Following treatment with Dox for 24 h, GFP signal for all proteins was diffuse throughout the cytoplasm. Treatment of cells with the ISR-activating agent sodium arsenite (NaAsO2) induced GFP-positive punctate subcellular structures for all RBPs that colocalized with the stress granule marker G3BP1 tagged with mCherry (merged as yellow signal, Fig. 2a, Supplementary Fig. 2a and Supplementary Movie 1). Quantification of colocalization between cytoplasmic TDP-43 and G3BP1 showed that >60% of TDP-43(86-414) or mutant TDP-43(86-414, M337V) granules are found within stress granules at 30 min, and this increased to >90% colocalization at 1 h and 2 h (Fig. 2b).
a Fluorescence microscopy of fixed H4 cells expressing mCherry-tagged G3BP1 (red) and GFP-tagged RBPs (TDP-43, TIA1, and FUS variants) (green). In the presence of 250 µM NaAsO2 for 1 h, these RBPs colocalized to G3BP1 positive stress granules (shown in yellow). Nuclei were labeled with DAPI (blue). Scale bar: 20 µm. See Supplementary Fig. 2a for single channel images. b Live-cell imaging quantification shown in Supplementary Movie 1 of the colocalization between GFP-tagged TDP-43 variants and G3BP1-mCherry in the presence of 200 µM NaAsO2 over time. Colocalization was quantified as a percentage of the TDP-43 puncta found within stress granules out of the total number of TDP-43 puncta identified per cell. n = 3 independent experiments. Data shown as mean ± SEM. Statistical significance was determined by one-way ANOVA, with Tukey’s multiple comparison, ns P > 0.05. c Microscopy images of H4 inducible cells pre-treated with 1 µM DNL343 or vehicle, followed by 200 µM NaAsO2 for 2 h and immunostained with an antibody recognizing the epsilon subunit of eIF2B. DNL343 prevented the puncta formation of truncated TDP-43 (green), G3BP1 (magenta), and eIF2B (red). Arrowheads indicate colocalization of each marker. Nuclei were stained with DAPI (blue). Scale bar: 20 µm. Three independent experiments were performed with similar results. d−f Inset of single cell from merged image in (c). Fluorescent line intensity histograms of GFP (d) or GFP-TDP-43(86-414) (e, f), G3BP1, and eIF2B across the yellow lines in the images. Source data are provided in Source Data file.
To confirm the localization of cytoplasmic TDP-43 to stress granules, we performed super-resolution microscopy in our H4 lines. Treatment with NaAsO2 caused both TDP-43(86-414) and mutant TDP-43(86-414, M337V) to localize within granules marked by either G3BP1-mCherry, anti-ATXN2, or anti-UBAP2L (Supplementary Fig. 2b-g). Line profile intensity histograms intersecting the granules showed that cytoplasmic TDP-43 associates with the periphery of stress granules and is also found throughout the core structure of the granules, as defined by each of the three stress granule markers (Supplementary Fig. 2c, e, and g). NaAsO2 treatment also caused association of eIF2B epsilon subunit with stress granules marker G3BP1-mCherry and GFP-TDP-43(86-414) (Fig. 2c, e). Intensity profiles showed that G3BP1-mCherry and eIF2B epsilon colocalize regardless of the expression of GFP-TDP-43(86-414) (Fig. 2d–f). DNL343 pretreatment caused G3BP1-mCherry, eIF2B epsilon and GFP-TDP-43(86-414) to remain diffusely localized despite NaAsO2-mediated ISR activation (Fig. 2c, f). Further analysis of G3BP1-mCherry- positive stress granules in NaAsO2-treated H4 cells showed that GFP-TDP-43(86-414) expression significantly increases the average stress granule size (mean area) relative to GFP expression, with a trend for an increase in colocalization of GFP-TDP-43 with G3BP1-mCherry (Supplementary Fig. 3a, p-value = 0.0538). DNL343 pretreatment abolished the effect of GFP-TDP-43(86−414) expression on stress granule size and significantly reduced its colocalization with G3BP1-mCherry (Supplementary Fig. 3a). DNL343 also induced rapid dissolution of pre-existing stress granules upon addition 1 h after treatment with NaAsO2 (Supplementary Movie 2 and Supplementary Fig. 3b, c). Taken together, these results indicate that DNL343 can modulate the localization of cytoplasmic TDP-43 by either preventing formation of new stress granules or dissolving preformed granules.
DNL343 prevents stress granule formation in ALS-linked human iPSC-derived neurons
To examine stress granule formation in an ALS relevant cell type, human iPSC-derived motor neurons from a healthy individual and a TARDBP mutant carrier (TDP-43(G298S)) were treated with NaAsO2 or thapsigargin as previously described43. Robust formation of G3BP1-positive stress granules was observed in both the healthy control and TDP-43(G298S) neurons (Supplementary Fig. 3d). Pretreatment with the DNL343 analog DN2736 (Supplementary Fig. 3e, IC50 = 3 nM from an ATF4 reporter assay in H4 cells) dose-dependently prevented formation of stress granules in both genotypes in the presence of either NaAsO2 or thapsigargin, while the structurally related but much less potent analog DN9052 (Supplementary Fig. 3e, IC50 = 910 nM) had no effect on stress granule formation (Supplementary Fig. 3d, f). In an independent paradigm, forebrain neurons from a C9orf72 HRE carrier (6–8 kb expansion) with ALS formed more stress granules than isogenic control neurons with CRISPR-mediated excision of the repeat expansion63 following a 2 h treatment with thapsigargin (Supplementary Fig. 3g, h), suggesting ISR activation is altered in this ALS model. Pretreatment with DNL343 prevented stress granule formation regardless of genotype (Supplementary Fig. 3g, h).
Acute dosing of a CNS penetrant eIF2B activator reduces ISR activation in an ALS mouse model
To assess the activity of a CNS penetrant eIF2B activator in a preclinical model of ALS, we utilized the previously described rNLS8 mouse model that expresses human TDP-43 with an ablated NLS under an inducible neuronal promoter (hTDP-43-ΔNLS, rNLS8)64, thus replicating in vivo the cytoplasmic targeting of TDP-43 achieved in our cellular models. Upon removal of Dox from the diet, mice express cytoplasmic TDP-43 in the CNS within 1 week and ultimately develop ALS/FTD-like phenotypes over the course of 4−10 weeks off Dox (WOD). Recently, this model was shown to exhibit ISR activation from 1 WOD, prior to disease onset22. To test whether an eIF2B activator can modulate ISR in the brain of this mouse model, we employed DN9058, a CNS penetrant analog of DNL343. Of note, tool compounds are typically used for exploratory in vivo studies when a compound is actively being evaluated in clinical trials under an approved Investigational New Drug (IND). As with all analogs of DNL343 used in this study, DN9058 is derived from the same scaffold as DNL343 and shares a highly similar structure and mechanism of action, exhibiting a low nM cellular potency in the ATF4 reporter assay (IC50 = 3.2 nM) (Supplementary Fig. 4a, b). After 2 WOD, rNLS8 and single transgenic (sTg) control animals (which lack the ability to induce human TDP-43 in response to Dox removal) received acute administration of DN9058 (50 mg/kg) or its vehicle for two consecutive days by oral gavage (Fig. 3a). PK analysis showed that in both transgenic lines, DN9058 mean exposure in the brain was higher than in plasma, indicating DN9058 is a brain penetrant molecule (Supplementary Fig. 4c). Unbound DN9058 exposure was well above the IC90 observed in the ATF4 reporter in vitro assay (i.e., 85 nM). Human TARDBP transcript levels in the brain were unaltered by DN9058 in both sTg and rNLS8 mice (Supplementary Fig. 4d). Similarly, total TDP-43 protein levels in the soluble fraction and phospho-S409/410 TDP-43 (p-TDP-43) levels in the insoluble fraction of caudal cortex lysates were not affected by acute dosing of DN9058 (Supplementary Fig. 4e-h), confirming the validity of the transgenic model to test the drug activity.
a Experimental design of acute DN9058 dosing of rNLS8 transgenic mice. All animals were fed Dox-containing diet until 8 weeks of age, including non-transgenic (nTg) controls, single transgenic (sTg) controls and double transgenic (rNLS8) mice, then Dox was removed from their diet except for group 6 (double transgenic (rNLS8) on Dox for two additional weeks). On 13th and 14th day off Dox, indicated groups were dosed with DN9058 by oral gavage at 50 mg/kg per animal weight. b-c Quantification of p-eIF2α normalized to loading control eIF2α (b) and ATF4 level normalized to GAPDH (c) from Supplementary Fig. 4i and j (n = 8 sTg Control or 12 rNLS8 mice). Data are shown as fold-changes relative to vehicle-treated control mice. d Transcriptional fold change of pre-selected ISR genes in rostral cortex of indicated mouse line (n = 7 (nTg Ctrl), 8 (rNLS8 Dox), 9 (sTg Ctrl) and 12 (rNLS8) mice). Data are shown as mean ± SEM (b−d). Statistical significance was determined with Kruskal-Wallis test with Dunn’s multiple comparisons (b), ordinary One-way ANOVA with Tukey’s multiple comparison (c, d). Source data are provided in Source Data file.
As previously observed22, rNLS8 brains showed ISR activation based on increased levels of phosphorylated eIF2α and ATF4 proteins from immunoblot analyses (Fig. 3b, c, Supplementary Fig. 4i, j). Acute dosing of DN9058 significantly reduced levels of these two ISR markers in rNLS8 mouse brains (Fig. 3b, c). Of note, as eIF2B acts downstream of eIF2α, the downregulation of p-eIF2α by the eIF2B activator suggests the ISR may amplify via feed forward mechanisms. Upregulation of transcripts indicative of ISR activation was confirmed in rNLS8 mouse brains based on quantitative PCR and was generally reduced by acute DN9058 dosing (Fig. 3d). Specifically, expression of Chac1, Gdf15 and Mthfd2 returned to control levels, whereas that of Ddit3 and Atf4 was partially corrected after acute DN9058 dosing (Fig. 3d). Interestingly, previously defined later-disease stage ISR genes, such as Ccl12 and Il6, were unaffected (Supplementary Fig. 4k)22, suggesting that longer eIF2B activation may be required to correct neuroinflammatory pathways.
Chronic dosing of a CNS penetrant eIF2B activator reduces ISR and transiently delays progression of locomotor deficits in an ALS mouse model
rNLS8 mice were chronically dosed with DN9058 via in-diet chow (50 mg of DN9058 per kg of chow, estimated dosing to be ~7.5 mg per kg of body weight per day) to investigate impact on ALS-relevant phenotypes over 6 WOD (Fig. 4a). Plasma samples were collected at 0, 2, 4, and 6 WOD to monitor the DN9058 intake and its exposure to the animals. PK analysis confirmed the constant exposure of DN9058 to the dosed animals, with a slight drop at 6 WOD (Supplementary Fig. 5a) and unbound brain exposure was generally well above the IC50 (3.2 nM) observed in the ATF4 reporter in vitro assay, with levels maintained near the IC90 (85 nM). Higher levels of DN9058 in the brains than in plasma from both sTg control and rNLS8 further validated the high brain penetrant characteristics of DN9058 (Supplementary Fig. 5b). As expected, human TARDBP transcript levels were selectively upregulated in rNLS8 mouse brain at 6 WOD, though at lower levels than those observed at 2 WOD (Supplementary Fig. 5c). Total TDP-43 protein and insoluble phospho-TDP-43 (p-TDP-43) levels were increased in rNLS8 mouse brains (Supplementary Fig. 5d-h). While chronic DN9058 dosing did not affect total TDP-43 protein levels, it caused a mild, but significant increase in p-TDP-43 levels specifically in the brains from rNLS8 female mice (Supplementary Fig. 5e, f).
a Experimental design of chronic DN9058 dosing for 6 weeks off dox (WOD). DN9058 was formulated at 50 mg/kg per chow for the chronic administration to the indicated conditions. b-c Quantification of p-eIF2α normalized to loading control eIF2α (b) and ATF4 level normalized to GAPDH (c) from Supplementary Fig. 5i and j, respectively (n = 8 (sTg Control), 10 (rNLS8 Veh), and 13 (rNLS8 DN9058) mice). Data are shown as fold-changes relative to vehicle-treated control mice. d-e Transcript level changes of pre-selected ISR genes (d) and ISR genes indicated in later stage of the disease (e) in rostral cortex (n = 9 (sTg Control), 10 (rNLS8 Dox), 11 (rNLS8 Veh), and 16 (rNLS8 DN9058) mice). f Time (weeks) to show clasping for individual rNLS8 mice that demonstrated the clasping phenotype (n = 13 (Veh) and 18 (DN9058) mice). g Plasma NfL concentration at baseline (0 WOD), 2, 4, and 6 WOD in rNLS8 mice (n = 11 (Veh) and 16 (DN9058) mice). The same figure legend for rNLS8 + Veh (yellow) and rNLS8 + DN9058 (orange) is shared with panel (f). Source data are provided in Source Data file. Data are shown as mean ± SEM (b−e) and individual values overlayed on violin plot (f, g). Statistical significance was determined with Kruskal-Wallis test with Dunn’s multiple comparisons (b, c), ordinary One-way ANOVA with Tukey’s multiple comparison (d, e), Two-tailed Mann-Whitney test (f), and Two-way ANOVA with multiple comparisons (g).
Next, we assessed the ISR pathway with chronic dosing of DN9058. Both phospho-eIF2α and ATF4 protein levels remained increased at later stage of the disease progression in rNLS8 mouse brains. DN9058 dosing caused a significant reduction in p-eIF2α levels but only a trend for a decrease in ATF4 protein in the brain (Fig. 4b, c, Supplementary Fig. 5i, j). ISR gene transcripts, such as Atf4, Chac1, and Gdf15, were also upregulated in rNLS8 brains at 6 WOD and chronic dosing of DN9058 significantly reduced their levels (Fig. 4d). Importantly, in contrast to acute dosing studies, later-disease state, inflammatory genes, Ccl12 and Il6, were effectively reduced with chronic DN9058 treatment (Fig. 4e). These data demonstrate DN9058 successfully inhibits ISR activation throughout disease progression in rNLS8 mice.
To determine the functional impact of chronic ISR inhibition, we evaluated behavioral performances of our mouse cohort with chronic DN9058 dosing. Both vehicle- and DN9058-treated rNLS8 mice began to show weight loss around 2 WOD and exhibited progressive weight loss until 6 WOD with no significant drug effect (Supplementary Fig. 6a). While rNLS8 mice demonstrated rapid progression of motor deficits, characterized by collapsing splay or clasping of hindlimbs, DN9058 dosing slightly delayed progression to high neurological score before 3 WOD and development of the clasping phenotype (Supplementary Fig. 6b). The time that rNLS8 mice with DN9058 dosing took to show clasping (median = 2.3 WOD) was significantly longer than the rNLS8-vehicle group (median = 2 WOD) (Fig. 4f). However, the mild beneficial effect of DN9058 was transient, as all rNLS8 mice displayed clasping phenotype before reaching 4 WOD (Supplementary Fig. 6c). Furthermore, DN9058 dosing temporarily improved the rotarod performance of rNLS8 mice, increasing the latency to fall compared to vehicle-treated rNLS8 mice at 2 WOD (Supplementary Fig. 6d, e). Similarly, the impaired performance of rNLS8 mice in the inverted grid suspension test was significantly but transiently improved by DN9058 dosing in the grip strength and coordination at 2 WOD (Supplementary Fig. 6f, g). Nearly half of the rNLS8-DN9058 mice were able to remain on the grid for 60 s, whereas the majority of rNLS8-Veh mice showed decreases in the latency to fall at 2 WOD. Like in the clasping paradigm, however, all the rNLS8 mice ultimately failed the locomotor function tests, suggesting that blocking ISR with DN9058 slows but does not prevent the progression of the motor deficits in this mouse model. Finally, plasma NfL was assessed as a marker of neurodegeneration. Levels progressively increased from baseline to 4 WOD and remained high at 6 WOD, albeit slightly lower than at 4 WOD in rNLS8 mice (Fig. 4g). DN9058 dosing led to significantly lower plasma NfL levels in rNLS8 mice at 4 WOD relative to rNLS8-Veh mice (Fig. 4g), suggesting eIF2B activation in the brain confers partial neuroprotection.
ISR activation is detectable in the spinal cord and CSF of individuals with ALS
To better understand ISR activation in people with ALS, we examined the expression of the ISR gene set (Supplementary Data 1)44,46,65 in RNA isolated from ALS spinal cord tissue samples (Target ALS, March 2022 release, demographics and characteristics of the samples can be found in Supplementary Table 1). The ISR gene set was upregulated in the cervical region of the spinal cord from people with ALS compared to controls (Fig. 5a, GSEA p-value: 0.0006), with BCL2L11, CEBPA, MTHFD2 and SLC1A5 showing the most significant gene-level increases (Fig. 5b). Increased ISR gene set expression was also observed in the lumbar region (Supplementary Fig. 7a, b, GSEA p-value: 0.0161). There was no significant upregulation of the ISR gene set in the thoracic region of the spinal cord (GSEA p = 0.1358), although individual ISR genes, such as CEBPA, were significantly upregulated (Supplementary Fig. 7c, d).
a Density plot of ISR gene set (orange line) in cervical spinal cord tissue from Target ALS dataset. b Normalized ISR gene expression plots comparing Control and ALS from cervical spinal cord. Samples are from the Target ALS dataset and include cervical spinal cord tissue with n = 116 for ALS samples and n = 16 for control samples. c Non-age adjusted CSF GDF-15 concentration (pg/mL) in healthy participants (n = 47) and ALS participants (n = 27) at baseline. d Age adjusted CSF GDF-15 concentration (pg/mL) in MAD healthy participants (n = 47) and ALS participants (n = 27) at baseline. Data shown as Min-to-Max Box plots overlayed with individual values (b−d). Statistical significance was determined with Two-tailed Mann-Whitney test. Source data are provided in Source Data file.
Next, we assessed levels of secreted protein GDF-15 in the CSF of people with ALS, based on preclinical studies in the VWMD mouse model and optic nerve crush paradigm showing it is upregulated in the CNS as part of ISR activation and corrected with eIF2B activators48. GDF-15 protein concentration was higher in the CSF of participants with ALS than in healthy volunteers without age adjustment (Fig. 5c). Since GDF-15 is known to increase with age in plasma66,67 and a correlation between age and CSF GDF-15 concentration was seen in both ALS and healthy participants (Supplementary Fig. 7e), CSF GDF-15 levels were examined after age adjustment, yielding comparable results (Fig. 5d). These data corroborate the activation of ISR pathway in the CNS of individuals with ALS.
Human PBMCs can be used to model ISR pathway activation
To determine if PBMCs could be used as a proxy to measure ISR activity, ISR gene regulation in human PBMCs was evaluated using qPCR and in human forebrain-like iNeurons for comparison. PBMCs treated ex vivo and iNeurons treated in vitro with NaAsO2 showed a concordance in upregulated genes, along with some unique changes in each cell type (Supplementary Fig. 8a). The top five upregulated ISR genes were ATF3, CHAC1, DDIT3, JUN, and TRIB3. Upregulated ISR genes were not completely overlapping between both cell types with BTG2 exclusively upregulated in iNeurons and the amino acid transporters SLC7A5 and SLC7A11 only upregulated in PBMCs. The effect of DNL343 on ISR gene expression was then examined in PBMCs after co-stimulation with NaAsO2 ex vivo. Of the 26 ISR genes assessed, 19 were significantly downregulated by DNL343 (Supplementary Fig. 8b). CHAC1 transcript and ATF4 protein levels were reduced by DNL343 in a dose-dependent manner with geometric mean unbound EC50 values of 3.53 nM and 3.95 nM, respectively (Supplementary Fig. 8c, d). PBMCs from healthy and ALS participants generally showed comparable profiles (Supplementary Fig. 8e–g). Therefore, PBMCs are responsive to ex vivo ISR inhibition mediated by DNL343 and can be used to monitor DNL343 effects in clinical biomarker assays.
DNL343 clinical trials in healthy participants and participants with ALS
Safety and tolerability, PK and PD of DNL343 were assessed in a randomized, double-blind Phase 1 study (NCT04268784; see Methods and Supplementary Information for site locations and PIs) consisting of a single ascending dose (SAD, Part A) and multiple ascending dose (MAD, Part B) cohorts in healthy participants (Fig. 6a, Supplementary Fig. 9a). DNL343 or matching placebo was administered orally. In the SAD cohorts (Part A), participants received a single dose of DNL343 ranging from 15−800 mg (n = 36 total) or placebo (n = 12 total). In the MAD cohorts (Part B), participants (n = 47) were randomized to groups receiving DNL343 for 14 days at doses ranging from 45 mg once-daily (QD) to 260 mg QD, or placebo (n = 13). The baseline characteristics of the healthy participants are presented in Supplementary Table 2.
a CONSORT diagram describing Phase 1 screening and randomization. b CONSORT diagram describing Phase 1b screening and randomization. c Healthy participants plasma concentration-time profiles upon single doses of DNL343 in the fasted state (n = 6 in each group). Presented as the geometric mean (95% CI) and colored by dose level. d, e Healthy participants (n = 6, 7, 7, 7, and 6 participants, for the 45 mg, 100 mg, 145 mg, 200 mg, and 260 mg group respectively) (d) and participants with ALS (n = 9, 7 participants in the 100 mg and 200 mg groups, respectively) (e) plasma pharmacokinetics upon QD dosing of DNL343 at steady state. Presented as the geometric mean (95% CI) and colored by dose level. f, g Healthy participants (n = 6, 7, 7, 7, and 6 participants, for the 45 mg, 100 mg, 145 mg, 200 mg, and 260 mg group respectively) (f) and participants with ALS (n = 8, 7 participants in the 100 mg and 200 mg groups respectively) (g) DNL343 CSF:unbound plasma concentration ratios after multiple doses. Presented as individual datapoints colored by dose level overlayed over a boxplot summarizing all the data in the panel. The middle line of the boxplot displays the median, the lower and upper limits of the box display the first and third quartiles, and the whiskers extend from the box to the largest and smallest values no further than 1.5-fold the interquartile range from the box. The lower and upper limits of quantification (LLOQ and ULOQ) for CSF concentrations was 0.005 and 5 µM, respectively. The LLOQ and ULOQ for plasma concentrations was 0.002 and 2 µM, respectively. Note: In the 100 mg group, data were not collected and/or analyzed for two participants due to dose reduction (1 participant), and no collection of CSF samples (1 participant). In the 200 mg group, data were not collected or analyzed for two participants due to dose reduction (1 participant) and early discontinuation of treatment (1 participant). Source data are provided in Source Data file.
In the randomized, double-blind Phase 1b study (NCT05006352; see Methods and Supplementary Information for site locations and PIs), the safety, tolerability, PK and PD of DNL343 was investigated in a total of 29 people with ALS receiving standard-of-care therapy (Fig. 6b, Supplementary Fig. 9b). The participants were randomized to a 28 day regimen of DNL343 100 mg QD (n = 10), or DNL343 200 mg QD (n = 9) or matching placebo (n = 10), followed by an optional 18 month Open Label Extension (OLE). The baseline characteristics of the participants with ALS are presented in Supplementary Table 3.
DNL343 plasma pharmacokinetics and CSF distribution in healthy participants and participants with ALS
Plasma DNL343 PK data was collected after single oral doses in healthy participants and after multiple oral doses in both healthy participants and participants with ALS. During the SAD part of the study, over the 15−800 mg dose range, in the fasted state, plasma concentrations increased in a dose-dependent manner with low to moderate PK variability and mean t1/2 values ranging from ~31−45 h across the dose range with monoexponential elimination (Fig. 6c, Supplementary Table 4).
During the MAD part of the study in healthy participants, after administration of a first oral dose of DNL343 on Day 1, in the fasted state, the median tmax was ~14−24 h; mean area under the concentration-time curve from time zero to 24 h (AUC24) and maximum concentration (Cmax) values increased across the 45- to 260 mg QD dose range. The variability in DNL343 AUC24 and Cmax on Day 1 and Day 14 was low to moderate across all doses (Supplementary Table 5). At steady state on Day 14, plasma concentrations increased in a dose-dependent manner across the dose range (Fig. 6d). Mean t1/2 after the last dose ranged from ~38−46 h across the 45−260 mg QD dose range. Additionally, peak-to-trough fluctuation of plasma concentrations was low across the 24 h dosing interval at steady state in healthy participants (Supplementary Table 5).
In participants with ALS after administration of a first oral dose of DNL343 on Day 1 in the fasted state, the median tmax was ~8 h (range 2−24 h); mean AUC24 and Cmax values increased across the 100−200 mg doses. The variability in DNL343 AUC24 and Cmax on Day 1 and on Day 28 was moderate across all doses (Supplementary Table 6). Steady state plasma concentrations increased in a dose-dependent manner (Fig. 6e). Trough concentration (Ctrough) appeared to reach steady state by Day 14, consistent with a slow absorption and slow elimination compound (as identified in healthy participant SAD and MAD studies). Peak-to-trough fluctuation of plasma concentrations was low across the 24 h dosing interval at steady state in participants with ALS (Supplementary Table 6).
CSF DNL343 PK data was collected after multiple oral doses in both healthy participants and participants with ALS. In healthy participants (MAD) median DNL343 CSF-to–unbound plasma concentration ratios, measured on day 12 following multiple doses, ranged from 0.689 to 0.795, with no dose-related trends across the 45−260 mg QD dose range (Fig. 6f). In participants with ALS median DNL343 CSF-to–unbound plasma concentration ratios, measured on Day 28 following multiple doses, were 0.927 and 1.09 for the 100 and 200 mg QD doses, respectively (Fig. 6g). These data in healthy and ALS participants indicate that DNL343 readily crosses the blood-brain barrier where it can reach its target in the CNS.
DNL343 inhibits ISR activity in healthy participants and participants with ALS
In the Phase 1 SAD cohort, ISR pathway inhibition was measured at baseline and after oral dosing of healthy participants by evaluating ATF4 protein and CHAC1 gene expression, alongside a broader ISR gene panel, in freshly isolated PBMCs stimulated with NaAsO2 ex vivo. Reduction in ATF4 protein levels and CHAC1 gene expression was observed at all dose levels ≥ 45 mg through 48 h post-dose, with levels trending back to baseline at 168 h post-dose (Supplementary Fig. 10a, b, Supplementary Table 7). Reductions in gene expression were also observed across a panel of ISR-related genes (Supplementary Fig. 10c). Similarly, in the MAD cohorts DNL343 achieved >50% inhibition of ATF4 protein levels and CHAC1 gene expression in freshly isolated PBMCs at all dose levels studied 24 h after the final dose (Fig. 7a, b, Supplementary Table 8). A broader set of ISR-related genes, including TRIB3 and PSAT1, was also notably inhibited (Supplementary Fig. 10d).
a−d ATF4 protein (a) and CHAC1 gene (b) expression in ex vivo stimulated PBMCs from healthy participants. ATF4 protein (c) and CHAC1 gene (d) expression in ex vivo stimulated PBMCs from ALS participants. PBMCs were freshly isolated from participants in each dose group shown at the times indicated and analyzed by either ECLIA (a, c) or multiplex qPCR (b, d). Values shown as median (IQR = interval from the first to the third quartile, shown as error bars) percent change from baseline. For MAD healthy participants ATF4 protein, n = 12, 6, 7, 7, 7, 6 participants for placebo, 45 mg, 100 mg, 145 mg, 200 mg and 260 mg groups respectively; CHAC1 gene, n = 11, 6, 7, 5, 7, 6 participants for placebo, 45 mg, 100 mg, 145 mg, 200 mg and 260 mg groups respectively. For ALS participants ATF4 protein, n = 6, 7, 6 participants for placebo, 100 mg, and 200 mg groups respectively; CHAC1 gene, n = 5, 7, 6 participants for placebo, 100 mg, 200 mg groups respectively through Day 28 for each dose group. e Heat map depicting relative change from baseline for a panel of ISR genes. Gene expression measured by multiplex qPCR in freshly isolated ex vivo stimulated PBMCs from ALS patients in each dose group. Values grouped based on median percent change from baseline and genes rank ordered based on percent change from baseline at Day 28 in the highest dose cohort (200 mg). f Percent change from baseline in CSF GDF-15 protein concentration at Day 28 in ALS participants, n = 9, 8, 8 participants for placebo, 100 mg, 200 mg groups respectively per group. Data are shown as box plot with individual percent change values overlayed. The middle line of the boxplot displays the median, the lower and upper limits of the box display the first and third quartiles, and the whiskers extend to the largest and smallest values. Source data are provided in Source Data file.
We next explored the PD effect of DNL343 in a Phase 1b study in participants with ALS. ATF4 protein levels and CHAC1 gene expression were reduced in ex vivo stimulated PBMCs throughout the dosing period in both DNL343 dose groups, with the 200 mg dose group demonstrating ∼80% median reduction of both biomarkers at trough on the last day of dosing, Day 28 (Fig. 7c, d, Supplementary Table 9). As in the Phase 1 SAD and MAD studies, gene expression of additional ISR genes was inhibited by DNL343 treatment (Fig. 7e). Finally, 28 days of DNL343 administration to ALS participants appeared to reduce CSF GDF-15 by ~10% from baseline in both dose groups (median 11.2% and 9.8% in the 100 mg and 200 mg arms, respectively), whereas a similar reduction was not observed in the placebo group in the phase 1b study (Fig. 7f).
DNL343 is generally well tolerated in healthy participants and participants with ALS
DNL343 was generally well tolerated in the Phase 1 healthy participant SAD and MAD cohorts at all dose levels. The majority of the treatment-emergent adverse events (TEAEs) were mild in severity. There were no deaths, serious or severe AEs. In the SAD cohorts, TEAEs occurred at a similar frequency overall in DNL343-treated (64%) vs. placebo-treated (67%) participants (Supplementary Table 10). All TEAEs in the DNL343 treatment groups were mild. The most frequent TEAE in DNL343-treated vs. placebo in healthy volunteers across all single-doses was postural dizziness (19 vs 17%). In the MAD cohorts, TEAEs occurred at a similar frequency overall in DNL343-treated (91%) vs placebo-treated (92%) participants (Supplementary Data 2). Most multiple-dose TEAEs were mild in severity. The most common multiple-dose TEAE was headache appearing in similar frequency between DNL343-treated (56%) vs. placebo-treated (54%) participants. There were no dose-dependent or clinically meaningful trends in safety laboratory tests, ECGs, or vital signs. No findings of clinical concern were seen on physical, neurological examination or psychiatric surveillance screening.
In the Phase 1b study, DNL343 was generally well tolerated in participants with ALS at doses of 100 or 200 mg administered daily for 28 days (Supplementary Data 2). There were no deaths, serious or severe AEs. All TEAEs were National Cancer Institute - Common Terminology Criteria for Adverse Events (NCI-CTCAE) Grade 1 or 2 (i.e. mild or moderate) in severity and occurred at a similar frequency overall in DNL343-treated (74%) vs. placebo-treated (89%) participants. The most frequent TEAEs that were non-procedure related, occurred in two or more DNL343 participants and were more common in DNL343-treated than in placebo were: headache (37% vs. 22%), fatigue (32% vs. 22%), and hypogeusia (11% vs. 0%); the highest severity in DNL343-treated participants was Grade 1. There was one adverse event of rash (200 mg group) that was Grade 2 in severity and considered related to study drug by the investigator, leading to discontinuation of study drug. Two blinded dose reductions occurred. In one participant, a TEAE of cognitive disorder that was Grade 2 experienced on Day 19 led to dose reduction from 200 to 100 mg after which the TEAE resolved. One participant described a TEAE of chills (Grade 1) after which the dose was reduced in a blinded manner (from 100 mg to placebo) and remained ongoing during the remainder of the 28-day double-blind treatment period. In ALS participants there were no dose-dependent or clinically meaningful trends on safety parameters, including clinical laboratory tests, ECGs, vital signs, routine physical and neurological examinations.
Discussion
Neuronal TDP-43 aggregates are a cardinal ALS pathology and, when taken along with causative mutations in TARDBP and other genes involved in RNA metabolism, it suggests that disruption of this process is a key driver of disease8,9,68,69,70,71. Intimately connected to this notion is the hypothesis that chronic ISR activation is central to ALS pathogenesis through various mechanisms, including formation of RNA-rich stress granules that facilitate TDP-43 aggregation36 and exacerbation of ISR downstream of pathological TDP-4322. The emergence of CNS penetrant small molecule modulators of the ISR pathway has set the stage for testing this therapeutic hypothesis in the clinic. In this study, we provide proof-of-concept data demonstrating that DNL343 and structurally-related compounds sharing an identical mode of action are potent eIF2B activators that attenuate the ISR pathway in various cellular models of ALS and in the brain of transgenic mice overexpressing pathogenic TDP-43, resulting in a subtle and temporary delay in progression of locomotor symptoms and neurodegeneration. DNL343 was also examined at a range of doses in both healthy participants as well as participants with ALS and demonstrated ISR pathway inhibition and, more generally, a pharmacological profile suitable for progression into larger clinical studies.
Several possible approaches to inhibit the ISR have been explored, including targeting the eIF2α kinases or direct activation of eIF2B49,72,73. Selecting one of the four eIF2α kinases to inhibit the ISR may limit the therapeutic utility of the intervention as other kinases may compensate for the loss of function, and the most advanced example of this approach, targeting PERK, revealed on-target toxicity74,75. Direct activation of eIF2B was first demonstrated with ISRIB, a molecule that reduces ISR activation independently of eIF2α phosphorylation54 and stabilizes the eIF2B complex76. Later studies showed ISRIB or analogs thereof are well tolerated and have potent effects on ISR biology both in vitro and in vivo53. Pharmacological activation of eIF2B also resulted in rescue of neurological defects, including electrophysiologic and behavioral deficits in a traumatic brain injury model55 and in an Alzheimer’s disease mouse model77. Compelling protective activity of eIF2B activators was observed in the VWMD mouse model, where phenotypes induced by disease-causing LoF homozygous Eif2b5 mutations, such as ISR activation, white matter pathology and locomotor deficits, were ameliorated by ISRIB44 or an ISRIB-like compound, 2BAct46.
Though an exciting molecule with potent effects, ISRIB suffers from poor solubility and undesirable physicochemical properties for drug development44,59 and 2BAct is associated with preclinical safety findings46. We have now developed DNL343 as a potent, CNS penetrant, eIF2B activator with excellent PK properties to suppress the ISR in preclinical and clinical settings60. DNL343 was previously shown to decrease ISR activation and neurodegeneration in the optic nerve crush injury paradigm and in the eIF2B LoF mouse mutant48, similar to studies with other eIF2B activators44,46. Our more recent study explored in greater depth the impact of eIF2B activation on VWMD model-associated peripheral and CNS biomarkers, which were also dysregulated in VWMD patients’ samples, including various cytokines, GDF-15 and NfL, and demonstrated DNL343 can prolong eIF2B mutant mouse survival48.
While eIF2B activators can correct effects of the genetic lesion in the VWMD mouse model, it is unclear if they can attenuate CNS ISR activation and slow disease progression in ALS preclinical models and critically, in people with ALS. We selected the rNLS8 ALS mouse model because of evidence of CNS ISR activation22, which is facilitated by cytoplasmic accumulation of TDP-43 due to removal of its NLS, as also supported by our cellular studies. While artificial, this inducible transgenic mouse model has the merit to replicate the ISR pathway activation we and others have observed in the CNS of patients with ALS. Our acute and chronic studies employing a CNS-penetrant tool compound (DN9058) amenable to PK/PD and efficacy studies in mice and chemically related to the investigational drug DNL343 indeed showed this compound can reduce ISR protein and mRNA markers in the brain of rNLS8 mice after induction of the TDP-43 transgene. In a chronic dosing study, DN9058 was able to transiently slow progression of motor phenotypes in rNLS8 mice in two independent behavioral paradigms, the rotarod and the inverted grid suspension test. Mild treatment benefits were observed at 2 WOD and were sex-independent, but they were lost at later timepoints, likely due to the aggressive disease phenotypes of this model. Similarly, DN9058 dosing resulted in lower plasma NfL levels specifically at 4 WOD, suggesting eIF2B activation confers some level of neuroprotection in the CNS of rNLS8 mice, at least temporarily. Chronic dosing of DN9058 caused a mild increase in insoluble p-TDP-43 levels specifically in female brains, suggesting inhibiting ISR does not prevent or may even slightly increase aggregation of cytoplasmically-restricted TDP-43 in the rNLS8 model. However, given the reduction in NfL levels, this subtle TDP-43 phenotype is unlikely to promote neurodegeneration and increases in p-TDP-43 could also potentially be a result of reduced degeneration in neurons with TDP-43 pathology. We note that silencing specific components of the ISR pathway, such as Chop/Ddit3, via antisense oligonucleotides did not slow symptom progression in the same model22, suggesting that eIF2B activation is more protective, likely via modulation of additional branches of the ISR pathway. Notably, DN9058 treatment caused an unexpected decrease in phosphorylation of eIF2α, given that eIF2B is downstream of eIF2α. This potentially suggests that prolonged ISR inhibition can trigger crosstalk between branches of ISR, perhaps via downregulation of eIF2α kinases or upregulation of phosphatases, which may eventually block the feed-forward regulatory loops of ISR in vivo. In summary, our studies, together with other studies in the field (including evidence of ISR activation in ALS patients’ spinal cord and CSF), provided incentive to test the therapeutic hypothesis that eIF2B activators may be beneficial for individuals with ALS in clinical settings.
In this study, we report clinical data on DNL343, an investigational small molecule eIF2B activator evaluated in a Phase 1 trial in healthy participants (NCT04268784) and in two trials in individuals living with ALS (NCT05006352, NCT05842941). In our Phase 1 and Phase 1b studies, DNL343 PK in healthy participants and participants with ALS similarly demonstrated low variability, long plasma half-life, extensive CSF distribution evidenced by similar CSF and unbound plasma concentrations of DNL343, and predictable dose-related increases in exposure (AUC and Cmax). DNL343 PD showed reduced ISR biomarkers in ex vivo stimulated PBMCs. In the Phase 1 MAD and Phase 1b ALS studies, DNL343 distributed to a similar extent in plasma and CSF. Taken together, DNL343 exposures that maximize peripheral ISR biomarker responses in PBMCs are hypothesized to induce similar responses in the CNS. DNL343 was also generally safe and well-tolerated in both healthy and ALS participants across a broad range of doses that robustly inhibited the ISR in PBMCs evaluated in both our Phase 1 study and the Phase 1b study (Supplementary Data 2) and showed evidence of CNS ISR inhibition in subjects with ALS based on the trend of reduction of GDF-15 protein levels. Given the limited number of participants and short duration of the Phase 1 and Phase 1b studies reported here, later stage studies are needed to further characterize safety and to assess the impact of DNL343 on clinical outcomes in people with ALS. DNL343 has been investigated in the HEALEY ALS Platform Trial (NCT05842941). While the favorable PK, PD, and safety profile supported continued development of DNL343 as a potential treatment for ALS, recent topline clinical data indicated that primary clinical endpoints were not met after 6 months, suggesting that ISR pathway inhibition may not modify disease outcome, at least in this time period. Further studies will be needed to better understand the impact of ISR inhibition in ALS and other neurological indications. These preclinical data suggest that pharmacological modulation of this pathway can modestly impact disease progression in rNLS8 mice, and identification of ALS patient subsets or other disease contexts with a strong ISR signature may lead to more optimal treatment paradigms.
Methods
Study Design
This translational study set out to understand the involvement of stress granule biology and the ISR pathway in ALS to support the development of DNL343, an investigational small molecule that targets eIF2B and inhibits the ISR. We employed a variety of cell-based model systems spanning from ALS-related gene expressing immortalized cells (HEK293 and H4), human iPSC-derived neurons from people with ALS (motor and forebrain neurons), and human PMBCs. ISR activation was observed with expression of both cytoplasmic TDP-43 and C9orf72 HRE, and RBPs associated with ALS were present in stress granules, both of which were ameliorated with DNL343 treatment. Inducible animal model expressing cytoplasmic TDP-43, rNLS8 transgenic mouse, was utilized to present the capacity of eIF2B agonist in correcting the elevated ISR markers in the CNS. Human PMBCs were used to develop a peripheral pathway engagement assay for DNL343 using ISR genes that are also upregulated by the ISR in human iNeurons. Clinical studies of DNL343 in healthy participants (Phase 1) and participants with ALS (Phase 1b) were conducted to evaluate the pharmacokinetics, peripheral ISR modulation, and safety in humans. IRB reviewed informed consent was obtained from every participant after the nature and possible consequences of the studies were explained.
Small molecules eIF2B activators
The discovery and preclinical profiles of DNL343 and DN2736 were previously described48,60. DN9058 and DN9052 are described in this study. ATF4 NanoLuc reporter cellular assay was used to determine the cellular potency of newly reported small molecules as previously described60.
Cell line development and maintenance
Lentiviruses encoding G3BP1-mCherry in a pLVX-CMV-IRES-Hygro vector (Clontech) and GFP-tagged RBPs [GFP-TDP-43(WT), GFP-TDP-43(86-414), GFP-TDP-43(86-414, M337V), TIA1-GFP, TIA1(A381T)-GFP, FUS(G515X)-GFP] in a pLVX-TetOne_IRES_Puro vector (Clontech) were first expressed in HEK293T-17 cells to produce virus using Lenti-x Single Shot (ClonTech, #631275). H4 cells, a human neuroglioma cell line, (ATCC HTB-148) were transduced with these viruses under the selection of hygromycin B (Invitrogen, #10687010) and puromycin (Gibco, #A1113803) to generate stable cell clones. All inducible cell lines were grown in DMEM (Gibco, #11965-092), supplemented with 10% FBS (Clontech, #631106) and 1X sodium pyruvate (Gibco, #11360-070), 1 µg/mL puromycin and 150 µg/mL hygromycin B at 37 °C in 5% CO2.
HEK293 cells stably expressing EGFP or (G4C2)71-EGFP were generated as previously described78. Briefly, HEK293 cells were transfected with pcDNA3.1 + EGFP or (G4C2)71-EGFP with Lipofectamine 3000 (Invitrogen) and selected with G418 (Corning, #30234CI) to produce stable clones. Stable HEK293 cells were maintained in DMEM (Gibco, #11965-092), supplemented with 10% FBS (VWR, #97068-085), and 1X sodium pyruvate (Gibco, #11360-070) at 37 °C in 5% CO2.
Chemical stressors and DNL343 treatments in H4 inducible cells and HEK293 cells
For H4 inducible cells, each line was expressed for 24 h by inducing with 1 μg/mL doxycycline (Clontech, #NC0424034). After doxycycline induction, cells were treated as follows: for qPCR, ATF4 ECLIA, anti-p-eIF2α and anti-ATF4 immunostaining, cells were treated with 1 μM DNL343 or DMSO for 2.5 h at 37 °C in 5% CO2. For fixed stress granule assays, cells were pretreated with 1 μM DNL343 or DMSO for 30 min and stressed with NaAsO2 (Sigma, #S7400-100G) (see figure legends for concentration and duration). For live-cell stress granule formation assays, cells were induced for 24 h with doxycycline. Nuclei were then labeled with NucBlue Live ReadyProbes reagent (Hoechst 33342) (Invitrogen, #R37605) for 20 min 37 °C, 5% CO2. Then 200 μM NaAsO2 was added to the cells and imaged immediately as described below. For stress granule dissolution assays, cells were induced for 24 h with doxycycline. After induction, cells were treated with 250 μM NaAsO2 for 1 h, followed by 1 μM DNL343 or DMSO and immediately imaged for 130 min as described below. For HEK293 cells stably expressing (G4C2)71-GFP or GFP control, samples were treated with 1 μM DNL343 or DMSO for 2.5 h at 37 °C in 5% CO2. Following incubation, samples were collected for qPCR or ATF4 ECLIA as described below.
Maintenance and generation of iPSC-derived motor neurons
Human induced pluripotent stem cells (iPSCs) previously reported43, were utilized in this study. iPSCs were cultured on Matrigel-coated plates using mTeSR Plus (Stem Cell Technologies) at 37 °C with 5% CO2. Cells were dissociated using Gentle Cell Dissociation Reagent (Stem Cell Technologies) and passaged as clumps.
Motor neurons (MNs) were differentiated as previously described42,43. Briefly, iPSCs were dissociated as single cells using Accutase (Stem Cell Technologies) and plated on a Matrigel-coated 6-well plate in mTeSR Plus supplemented with 10 μM ROCK inhibitor Y-27632 (RI). The following day, RI was withdrawn from media and iPSCs were cultured to 70−90% cell confluency. On day 1 of MN differentiation, the medium was replaced with N2/B27 medium (DMEM/F12 + GlutaMAX, 1X N2 supplement, 1X B27 supplement, 150 μM ascorbic acid, and 1% Penicillin-Streptomycin) supplemented with 1 μM Dorsomorphin, 10 μM SB431542, 3 μM CHIR99021, and 5 μM RI. Cells were maintained in this medium with daily changes for 6 days. On day 7, medium was changed to N2/B27 medium supplemented with 1 μM Dorsomorphin, 10 μM SB431542, 1.5 μM retinoic acid (RA), 200 nM Smoothened Agonist (SAG) and medium changes were performed daily. At day 18, the cells were dissociated with Accutase and expanded in motor neuron progenitor (MNP) medium (N2/B27 medium supplemented with 3 μM CHIR99021, 2 μM DMH1, 2 μM SB431542, 0.1 μM RA, 0.5 μM Purmorphamine and 0.5 mM valproic acid) on Matrigel-coated plates, as previously described79. MNPs were maintained in this medium and passaged with Accutase for up to 8 passages. For continued differentiation, MNPs were harvested with Accutase and plated at ~2 × 106 cells per well on a 6-well plate serially coated overnight with 0.001% (0.01 mg/mL) poly-D-lysine (PDL, Sigma) and poly-L-ornithine (PLO, Sigma) followed by an overnight incubation with 20 μg/mL laminin (Life Technologies). The cells were plated into N2/B27 maturation medium (N2/B27 medium containing 2 ng/mL GDNF, 2 ng/mL BDNF, and 2 ng/mL CNTF) supplemented with 1.5 μM RA, 200 nM SAG, and 10 μM RI. On day 20 of motor neuron differentiation (2 days post MNP dissociation), RI was reduced to 2 μM. On day 22, medium was changed into N2 B27 maturation medium supplemented with 2 μM DAPT and 2 μM RI. Cells were maintained with full medium changes every 2 days hereafter. On day 26, DAPT was withdrawn from the media.
For stress granule assays, cells were replated onto 384-well plates on day 27 as previously described43. In brief, 384-well plates were coated in advance with 0.001% PDL/PLO overnight at 37 °C, washed twice with 1X PBS, and incubated with 20 μg/mL laminin overnight at 37 °C. To harvest iPS-MNs, Accumax was added to day 27 motor neurons and incubated for 60 min at 37 °C with 5% CO2. Cells were then gently triturated 15 times with a P1000 pipette and incubated at 37 °C, 5% CO2 for an additional 15 min to complete dissociation. DMEM/F12 + GlutaMAX medium was added to dilute the dissociation reagent and the cell containing solution was transferred to a 15 mL conical tube. Cells were pelleted by centrifuging for 5 min at 200 × g and supernatant was aspirated. Cells were resuspended in N2/B27 maturation medium supplemented with 10 μM RI and passed through a 40 μm mesh filter to remove clumps. 104 iPS-MN cells were plated into each well of a PDL/PLO/laminin-coated 384 well in N2/B27 maturation media supplemented with 10 μM RI. After 24 h, cells were fed by adding N2/B27 maturation medium to each well. After allowing the cells to adhere for 48 h, the cells were treated as described.
Generation of iPSC-derived NGN2 neurons
The human iPS cell line, BIONi010-C-13, was purchased from Millipore Sigma (#66540561) and expresses Neurogenin-2 (NGN2) upon doxycycline treatment. The neuron differentiation protocol was adapted from a previous study80, with modifications described. Briefly, three days prior to differentiation, iPS cells were released into single cells by Accutase and were resuspended in N2 pre-differentiation medium (1X Knockout DMEM/F12, 1X NEAA, 1X N2 supplement, 10 ng/mL NT-3, 10 ng/mL BDNF, 2 μg/mL doxycycline). Cells were counted and seeded at ~8 × 103 cells/cm2 (7.5 × 105 cells/well of 6-well plate) in N2 pre-differentiation medium containing 10 nM RI (Tocris, #125410). A full pre-differentiation medium change was performed over the next 2 days (with doxycycline and no RI). For neuronal differentiation, the pre-differentiated cells were resuspended in the classic N2/B27 differentiation medium (1X DMEM/F12, 1X Neurobasal-A or Neurobasal medium, 1X NEAA, 0.5X GlutaMAX, 0.5X N2 supplement, 0.5X B27-VA supplement, 10 ng/mL NT-3, 10 ng/mL BDNF, 1 μg/mL mouse laminin, 2 μg/mL doxycycline). The cells were plated onto pre-PDL coated 96 well plates (Corning, #354640) coated with additional laminin (15 μg/ml) for 2 h at 37 °C in 5% CO2. Starting from DIV7, weekly 75% medium changes were done with the classic N2/B27 medium containing laminin (1 μg/mL). NGN2 neurons were ready to use by DIV14.
Generation of iPSC-derived forebrain neurons
C9orf72-expanded, CS52iALS-C9n6 (RRID:CVCL_JC27), and its isogenic control, CS52iALS-C9n6.ISO363,81,82, were purchased from Cedars-Sinai iPSC Core facility to differentiate into forebrain neurons. Forebrain iPSC-derived Neurons (iNeurons) were derived using a small molecule induced differentiation protocol from Stemcell Technologies. Briefly, embryoid body (EB) formation was initiated from iPSC lines in Aggrewell plates (Stemcell Technologies, #34811). EBs were plated onto Matrigel coated plates on day 5. On day 11/12 after EB formation, neural rosette selection was performed according to the protocol. Some lines required far less incubation –as few as 30 min at 37 °C—in rosette selection medium to remove neural rosettes. Neural rosettes were replated on Matrigel coated plates and expanded as neural progenitor cells for 1–2 additional weeks to allow for high enough yield to advance to forebrain neuron differentiation or for cryopreservation at this stage. Neural progenitor cells were seeded for forebrain neuron differentiation (Stemcell Technologies, #08600) and medium changes performed according to the protocol. Five to 7 days later forebrain progenitors were lifted and seeded according to the vendor protocol for the final iNeuron maturation (Stemcell Technologies, #08605). Every 2–3 days, 75% medium changes were completed. By ~DIV14, mature cells expressed predominantly TUJ1 with low levels of GFAP-positive astrocytes (~10%) in the culture and were treated with compounds as described.
Stress and eIF2B compound treatments in iNeurons
For C9orf72 ALS iPSC-derived and isogenic control forebrain iNeurons, cells were pretreated with 1 μM DNL343 or vehicle for 30 min, followed by 0.01 μM or 0.1 μM thapsigargin for 2 h at 37 °C, 5% CO2. Following incubation, cells were fixed and stained as described below.
For TDP-43(G298S) and healthy control iMNs, cells were pretreated with 1, 0.1, 0.01, or 0.001 μM eIF2B compounds or DMSO for 1.5 h, followed by 100 μM NaAsO2 or 0.25 μM thapsigargin for 2 h at 37 °C, 5% CO2. Cells were fixed and stained for G3BP1 as described below in immunocytochemistry.
For NGN2 iNeurons, cells were pretreated with 2 μM DNL343 for 30 min, followed by 50 μM for 4 h 37 °C, 5% CO2. Total RNA was then isolated using the RNeasy Plus Micro Kit (Qiagen) following the manufacturer’s protocol. Gene expression was then measured as described below in Fluidigm assays.
Immunocytochemistry
Following treatments in H4 inducible cells and C9orf72 iNeurons, samples were washed with 1X PBS and fixed in 4% paraformaldehyde for 15 min at room temperature (RT). Cells were then permeabilized with 0.1% Triton X-100 in PBS for 20 min at RT, and then blocked in 0.1% Triton X-100 and 3% BSA in PBS for 1 h at RT. The following primary antibodies were diluted in blocking buffer and incubated overnight at 4 °C: eIF2B epsilon (Novus, #NBP1-30449, 1:500), ATXN2 (BD Biosciences, #611378, 1:200), UBAP2L (Bethyl, #A300-533A-M, 1:500), G3BP1 (Bethyl, #A302-033A, 1:500), phospho-EIF2α (Cell Signaling, #3398, 1:200), and ATF4 (Cell Signaling, #11815, 1:200). Following washes with 1X PBS 3 times for 5 min each, cells were then diluted in block solution containing Alexa Fluor 647 secondary antibodies (Invitrogen, #A-31573 and #A32787) diluted 1:500 each and DAPI (ThermoFisher, #62248, 1:5000) at RT for 1 h. Cells were then washed 4 times for 5 min each in 1X PBS before image acquisition.
For iMNs, after stress treatment, cells were fixed by adding 4% paraformaldehyde (PFA) for 60 min at RT. Cells were then washed three times with 1X PBS. Next, cells were simultaneously blocked and permeabilized with 0.1% Triton-X and 5% goat serum diluted in 1X PBS for 45 min at RT. A wash with wash buffer (0.01% Triton-X in 1X PBS) was performed and cells were incubated overnight at 4 °C with primary antibody anti-G3BP1 (MBL #RN048PW, 1:500) diluted in wash buffer containing 5% goat serum. The plates were then washed five times with wash buffer. Secondary antibody Alexa Fluor 488 (Invitrogen, #A11034, 1:1000) diluted in wash buffer containing 5% goat serum was added to the cells and incubated overnight at 4 °C. Plates were washed 10 times with wash buffer and nuclei were stained with DAPI (1:5000 in PBS) overnight at 4 °C. Finally, cells were washed twice with wash buffer and preserved by adding 50% v/v glycerol in 1X PBS. Plates were imaged in this solution as described below in imaging of 384-well plate.
Electrochemiluminescence-based detection (ECLIA) for the quantification of ATF4 protein
A non-competitive sandwich immunoassay employing a biotinylated CREB-2 (ATF4) mAb (Santa Cruz Biotechnology, #sc-390063 LS) as capture and a SULFO TAG labeled (ruthenylated) anti-ATF4 pAb (Proteintech, #10835-1-AP) for detection was used to quantify levels of ATF4 protein. Biotinylation and ruthenylation were performed by B2S Life Sciences. MSD GOLD 96-well small spot streptavidin SECTOR plates (Meso Scale Diagnostics (MSD), #L45SA) were washed 3 times with 300 μL/well with wash buffer (1X TBST) using a plate washer. To each well 25 μL of 2 μg/mL Biotinylated Mouse anti-CREB-2 coating antibody diluted in Diluent 100 (MSD, #R50AA-2) was added and the plate was incubated at RT for 1 h on an orbital shaker at 700 rpm. ATF4 protein (Origene, TP760367) calibration standards were prepared in 1/50 Diluted Jurkat Cell Lysates (1×107 cells/mL) using CST lysis buffer at 3X dilutions between 360 pg/mL and 10 pg/mL and then 2x dilutions down to 2.5 pg/mL. Plates were washed as above and 25 μL of each calibration standard or sample, undiluted, were added to replicate wells. Plates were incubated overnight (16−24 h) at 4 °C on an orbital shaker at 700 rpm. Plates were washed as above and 25 μL of 0.5 μg/mL Rabbit anti-ATF4 Sulfo Tag polyclonal detection antibody in 1X MSD Blocker A (MSD, #R93AA-2) + 0.1 mg/mL Rabbit Gamma Globulin (Rockland Immunochemicals, #D610-1000) + 1 mg/mL Mouse Gamma Globulin (Rockland Immunochemicals, #D609-0100) in 1x TBST was added to each well. Plates were incubated for 1 h at RT on an orbital shaker at 700 rpm. Plates were washed again and 150 μL per well of MSD GOLD Read Buffer (MSD, #R92TG) was added to each well. Plates were read on a 1201 MSD SECTOR 600MM plate reader (MSD, #IC1AA-0) within 30 min of adding buffer and data was captured in Methodical Mind (MSD, version Fall 2020 (1.0.38)). Phase 1 and Phase 1b ATF4 assays were performed at B2S Life Sciences (Franklin, IN).
RNA-sequencing in H4 and HEK293 cells
Total RNA from H4 and HEK293 cells was extracted using the RNeasy Plus Micro Kit (Qiagen) and resuspended in nuclease-free water. Following the protocol defined by the manufacturer, RNA-seq libraries were generated using the QuantSeq 3’ mRNA-seq Library Prep Kit FWD for Illumina (Lexogen A01173) with high quality total RNA (RINe > 8) as input template. Briefly, total RNA was primed with oligo(dT) for reverse transcription, followed by RNA removal. Lexogen’s UMI Second Strand Synthesis Module was added to introduce unique molecular identifiers to identify and remove PCR duplicates. Amplified cDNA was purified using magnetic beads followed by 12 cycles of PCR with dual indexes and PCR purification. Library quantity and quality were assessed with High Sensitivity D1000 ScreenTape (Agilent 5067-5584). Libraries were pooled in equimolar ratios and the sequencing pool was purified with magnetic beads to remove residual adapter dimers. Sequencing reads were generated on an Illumina NovaSeq 6000 instrument (75 bp single end) by SeqMatic (Fremont, CA, USA).
Microscopy and image analysis
High-content confocal microscopy for fixed and live cells
For imaging H4 cells, cells were plated at 20 K cells/well in a PDL-coated 96-well cell carrier ultra plate (Perkin Elmer, #6055302) and induced with 1 μg/mL doxycycline for 24 h. For imaging C9orf72 iNeurons, cells were plated at 20 K cells/well on a 96-well plate (Corning, #354640). Stress and DNL343 treatments were performed as described above. Cells were then imaged on an Opera Phenix (Perkin Elmer) high-content spinning disk confocal microscope using a 40X/1.1 NA water immersion objective. To independently quantify stress granules (i.e. G3BP1-mCherry) and TDP-43-GFP puncta, a spot-based segmentation algorithm was implemented by using the Harmony HCS software (Perkin Elmer, version 4.9). Then, a masking step determined the TDP-43-positive spots found within G3BP1. Spot colocalization was quantified by calculating the percent overlap between double positive spots and the total number of TDP-43 spots. To demonstrate colocalization between stress granules, TDP-43 puncta, and eIF2B puncta, line intensity profiles were manually drawn intersecting each channel using FIJI/ImageJ. To measure cellular expression of p-EIF2α and ATF4, cells were segmented and identified by the Harmony software, and the mean fluorescence intensity was measured automatically.
For imaging iMNs, 384-well plates were imaged using a Nikon Eclipse Ti2 microscope system operated with NIS Elements High Content Software (Nikon). A 20X/0.75 NA objective was used to collect 7 μm focal range with 8 z slices for each of 6 field of views acquired per well. The excitation lasers used were 395 nm and 470 nm for DAPI and GFP, respectively. The slice images of each field of view were merged in FIJI/ImageJ using the maximum intensity projection function. Stress granules and nuclei were segmented and quantified using a custom CellProfiler pipeline83. Nuclei were identified and segmented using an object diameter of 23−80 pixels. To identify cell boundaries, the G3BP1 granule channel was overlayed, and a trace was propagated from the nuclei mask outward to the edge limit of the cytoplasmic fluorescent signal. To identify G3BP1-positive stress granules within cell bodies, punctate structures were processed to enhance speckle-like features that were 1−10 pixels in diameter. The total area enclosed by each identified feature (stress granule or nuclei) were calculated to quantify relative stress granule abundance.
Super resolution microscopy
H4 cells were plated at 20 K cells/well on a PLL-coated 96 w glass bottom plate (CellVis, #P96-1.5H-N) and induced with doxycycline for 24 h. Cells were treated, fixed, and stained as described above. Images were acquired with a scanning confocal microscope (Leica SP8; Leica Microsystems) using the Lightning super resolution mode and acquired with a 40 × /1.3 NA oil objective at a voxel size of 50 µm XY and 300 µm Z. The excitation lasers used were 405 nm (DAPI) and a supercontinuum white light laser, tuned to 488 nm (GFP), 561 nm (mCherry), and 633 nm (Alexa Fluor 647). Images were processed using the Adaptive Lightning processing algorithm. Two independent confocal z-stacks of 10 μm were acquired for each channel using sequential scan settings from each of 2 duplicate wells per treatment group. To visually confirm colocalization of TDP-43 within stress granules, as marked by G3BP1-mCherry, anti-ATXN2, or anti-UBAP2L, the signal from each channel was masked using an intensity-based segmentation of positive pixels and visualized using Bitplane Imaris using 3D surface rendering. Raw image stacks were opened in FIJI/ImageJ and line intensity histograms intersecting each channel were generated to demonstrate colocalization.
Animals
rNLS8 transgenic mice and littermate controls were produced by intercrossing of homozygous tetO-hTDP-43-ΔNLS line mice with hemizygous NEFH-tTA line mice both on a C57BL/6JAusb background following >10 generations of backcrossing. The mice were fed with Dox-containing chow (200 mg/kg chow, Specialty Feeds, Australia). Male and female mice at ~8 weeks of age were housed in temperature- and humidity- controlled conditions (21 ± 1 °C, 55 ± 5 %) with a 12 h light / dark cycle (lights on at 6:00 h). Experiments were conducted with approval of the Animal Ethics Committee of The University of Queensland (#2023/AE000484). Upon the initiation of the experiment (Off-Dox), mice were switched to normal chow to induce expression of hTDP-43-ΔNLS. Mice were randomly allocated to treatment groups. Both sexes were included and balanced between groups. Monogenic littermate and non-transgenic C57BL/6JAusb animals were used as controls.
Nonclinical drug (DN9058) formulation and dosing experimental procedures
We conducted drug formulation for DN9058 following the preparation procedure. Briefly, DN9058 was dissolved in 50% volume 0.2% Polysorbate (Tween-80, Sigma-Aldrich #P4780) solution of the final volume. Solution was sonicated at 60% power for 5 min with 50 s / 50 s intervals twice. An equal volume of 1% methyl cellulose (Sigma #09963) solution was added to reach the final targeted volume. The solution was mixed for additional 30 min and kept at 2−5 °C on a rotator until administration. DN9058 solution was prepared freshly and administered on the same day.
The acute dosing experiment procedures were conducted starting from 13th day off Dox for two consecutive days. On 13th and 14th day off Dox, all mice received vehicle (0.1% Polysorbate 80, 0.5% Methylcellulose in water) and 50 mg/kg (body weight) DN9058 by oral gavage (p.o.) with 24 h intervals. On 14th day, 3–4 h after the vehicle and DN9058 administration, mice were euthanized for tissue collection.
For the chronic dosing of DN9058, DN9058 was formulated in the rodent food chow by Specialty feed (Australia) at a dose of 50 mg per kg of diet chow, with a goal of 7.5 mg DN9058 consumption per kg of body weight in a day. The normal rodent food chow (Specialty feed, Australia) was used for the vehicle treatment.
Mouse plasma and tissue collection procedure
The whole blood of all mice was collected through submandibular bleeding by 0.5 M EDTA pre-coated syringes and immediately transferred to 1.5 mL Eppendorf tubes containing 4 μL of 0.5 M EDTA. The blood samples were immediately spun in a refrigerated centrifuge at 15,000 × g for 5 min following Beckman’s method. The supernatants were collected as plasma samples immediately, snap frozen on dry ice, and transferred to –80 °C freezer for storage. For tissue collection, the mice were euthanized by injection of 50 μL Lethabarb (Virbac #LETHA450) and underwent transcardiac perfusion using 50 mL PBS. The brains were immediately micro-dissected, frozen on dry ice, and transferred to –80 °C freezer for storage.
DN9058 pharmacokinetics
Concentrations of DN9058 in plasma and brain were measured using LC-MS/MS by Quintara Discovery. Brain samples were homogenized in 2–3 volumes of 20% acetonitrile. Plasma samples and brain homogenate samples were diluted 2-fold in blank mouse plasma. A 20 µL aliquot of diluted plasma or plasma diluted brain homogenate was extracted with 100 µL of acetonitrile containing the internal standard (DN1206). The mixture was vortexed on a shaker for 15 min and subsequently centrifuged at 3220 × g for 15 min. A 15 µL aliquot of the supernatant was mixed with 150 µL of 35% acetonitrile with 0.1% formic acid for injection to the LC/MS/MS. Calibration standards and quality control samples were prepared by spiking the test compound into blank mouse plasma and then processed with the unknown samples in the same batch. Total DN9058 concentration determined for each sample was adjusted to unbound fraction protein by multiplying with plasma and brain fraction unbound value (fu) of 0.102 and 0.245, respectively.
qPCR gene expression analysis from mouse brain tissue
Mouse rostral cortices were utilized for the validation of mouse model and the analysis of gene expression changes after DN9058 dosing. RNA from the brains were isolated using RNeasy Plus Mini Kit (Qiagen, #74134). The isolated RNAs were reverse transcribed to cDNA with SuperScript IV VILO Master Mix (Invitrogen, #11756050). Taqman probes (Thermo Fisher, #4331182; human TARDBP, Hs00606522_m1; Mouse Atf4, Mm00515325_g1; Ddit3, Mm01135937_g1; Chac1, Mm00509926_m1; Ccl12, Mm01617100_m1; Mthfd2, Mm00485276_m1; Ppp1r15a, Mm01205601_g1; Gdf15, Mm00442228_m1; Il6, Mm00442228_m1) were utilized for specific transcript target amplifications, including human TARDBP and key ISR genes, via QuantStudio 6 Flex System. Quantitative PCR (qPCR) reaction was carried out for each sample containing 1 μL sample cDNA, 5 μL Taqman Fastman Advanced Master Mix (Thermo Fisher, #4444557), 0.5 μL of 20 × Taqman probe for target gene with FAM, 0.5 μL of 20× HPRT probe (Thermo Fisher, #4331182; Mm03024075_m1) with VIC-PL as a loading control, and 3 μL of nuclease free water. The samples went through pre-amplification hold stage at 95 °C for 20 s, followed by 40 cycles of amplification stage with (1) 95 °C for 1 s and (2) 60 °C for 20 s. The relative expression of genes was determined using ΔΔCt method, having the relative fold change of transcript expression from non-transgenic control sample calculated as 2-ΔΔCt.
Protein extraction and immunoblotting analysis from mouse brain tissue
Mouse caudal cortices were used for immunoblotting. Briefly, the caudal cortical tissues were thawed on ice and homogenized in 5 × v/w RIPA lysis buffer (50 mM Tris, 150 mM NaCl, 1% NP-40, 5 mM EDTA, 0.5% sodium deoxycholate, and 0.1% SDS, pH 8.0) containing 1 mM PMSF, phosphatase inhibitor cocktail (Sigma #4906845001), and protease inhibitor cocktail (Sigma #11836170001) with three 1.4 mm Zirconium oxide beads (Bertin Instruments, #P000927-LYSK0-A) using Precellys tissue homogenizer (Bertin Instruments, #P000669-PR240-A). Samples were centrifuged at 4 °C, 100,000 xg for 30 min, and the supernatant was taken as the RIPA-soluble fraction. The remaining pellet was washed with RIPA buffer once, and the resulting pellet was dissolved in 5 × v/w urea buffer (7 M urea, 2 M thiourea, 4% CHAPS, and 30 mM Tris, pH 8.5) using the Precellys homogenizer and centrifuged at 22 °C, 100,000 × g for 30 min. This supernatant was taken as the RIPA-insoluble fraction. Protein concentrations of the RIPA-soluble fractions were determined using the PierceTM BCA Protein Assay Kit (Thermo Fisher Scientific #23225).
Protein samples of RIPA-soluble or -insoluble fractions were separated by electrophoresis (120 V for 90 min) on a 12% polyacrylamide gel in the presence of reducing agent (2-mercaptoethanol, Sigma #63689). After SDS-PAGE, proteins were transferred to nitrocellulose membranes (LI-COR Biosciences, #P/N926-31092) and incubated in blocking solution (5% (w/v) BSA, 0.05% (w/v) Tween-20 in TBS (TBST)), then incubated overnight with primary antibodies diluted in the blocking solution. Primary antibodies used for immunoblotting were rabbit anti-TDP-43 recognizing both human and mouse protein (Proteintech #10782-2-AP), rat anti-phospho-S409/410 TDP-43 (Biolegend # 829901), mouse anti-GAPDH (Proteintech #60004-1-Ig), rabbit anti-phosphorylated eIF2α (Cell Signaling, #3398), rabbit anti-eIF2α (Cell Signaling, #9722), and rabbit anti-ATF4 (Cell Signaling, #11815). The nitrocellulose membranes were washed with TBST and incubated with IRDye secondary antibodies for 1 h at RT. Protein bands were visualized using the Odyssey CLx Imaging System (LI-COR Biosciences, Lincoln, NE, USA) and quantified using Image Studio Lite software (LI-COR Biosciences). The relative changes of protein levels were calculated by quantifying the bands in RIPA soluble fractions relative to the band of the internal reference GAPDH protein, or by quantifying the bands in RIPA-insoluble fractions relative to total protein and subsequently normalized to the control groups.
Mouse monitoring for body weight and neurological score evaluation
Mice were monitored and weighed three times per week after Dox feed. For the observation of collapsing splay or clasping of hindlimbs, mice were suspended by the tail for >5 s. The failure to extend both hindlimbs was recorded as a positive response of collapsing splay and holding hindlimbs together was recorded as a presence of clasping splay as described previously.
Rotarod test
Mice were placed on a rotarod apparatus (Ugo Basile SRL, Gemonio, Italy) at a speed of 5 rpm with acceleration up to 40 rpm within 300 s. The time each mice took to fall from the apparatus was recorded. If mice were still running at the end of the testing session, their times were recorded as 300 s. Three training sessions were performed one week prior to time off Dox, and two test sessions were conducted biweekly, with the final score being the highest time of the two test sessions.
Inverted grid suspension test
To evaluate the grip strength and coordination of mice, the inverted grid suspension test was utilized. Mice were placed on a grid net, which slowly turned upside down so they were suspended above a padded surface. The latency to fall for each mice (in seconds) was recorded for three sessions at indicated time points. If mice were running at the end of the testing session, the latency was recorded at 60 s. The maximal performance was recorded for further analysis.
Detection of mouse plasma NfL
Plasma samples for chronic DN9058 dosing study were collected at the beginning of dosing (Baseline, 0 Weeks Off Dox (WOD)), 2 weeks after (2 WOD), 4 weeks after (4 WOD), and 6 weeks after (6 WOD) as terminal point. Neurofilament light (NfL) protein concentration in the plasma samples was measured with Simoa Nf-Light Advantage Kit (Quanterix, #104073) using Quanterix HD-X instrument. The assay was carried out as described by the manufacturer. Briefly, mouse plasma was diluted with Sample Diluent (Quanterix, #102252) at dilution factor of 20 and loaded to Simoa 96-well microplate along with kit calibrators and controls. Based on the calibration curve generated, NfL concentration from each sample was interpolated.
Gene expression in human ALS spinal cord samples
RNA was isolated from sections of thoracic spinal cord from ALS individuals and controls obtained from Target ALS (see Supplementary Table 1 for demographics and characteristics of human spinal cord samples). In brief, frozen tissue was homogenized in Buffer RLT Plus (Qiagen, #1053393) with 1% β-mercaptoethanol (Sigma, #M3148-25ML) at a ratio of 10 μL of buffer for every 1 mg of tissue using a Qiagen Tissue LyserII for 3 min at 30 kHz for 2 cycles. Lysate was centrifuged at 21,000 × g for 5 min and 300 μL of supernatant was combined with 300 μL of buffer before transferring to a QIA shredder (Qiagen, #79656) and centrifuging at 16,000 × g for 30 s. Flow-through was then processed using the protocol for purification of total RNA from animal cells included with the RNeasy plus mini kit (Qiagen, #74136). Gene expression was then measured using the Fluidigm protocol described in this section.
ISR GSEA analysis of bulk RNA-seq from spinal cord tissue of ALS vs healthy controls (Target ALS)
FASTQ files included in the Target ALS March 2022 RNA-seq release were retrieved from the NY Genome Center’s Collaborator Portal. Reads were trimmed with skewer (version 0.2.2)84 and aligned with the STAR aligner (version 2.7.1a)85. A STAR index was built for the human reference genome (GRCh38_p23) with –sjdbOverhan 101 and Gencode transcript models (release 28) were provided via the –sjdbGTFfile argument. Reads were aligned the following STAR arguments: --readFilesCommand zcat --outFilterType BySJout --outFilterIntronMotifs RemoveNoncanonicalUnannotated --outSAMunmapped Within --outSAMattributes NH HI AS nM MD XS --outSAMstrandField intronMotif --outSAMtype BAM Unsorted --outBAMcompression 6. Gene expression was quantified with Salmon (version 0.13.1)86 using Gencode transcript models (release 28). Downstream analysis of bulk RNA-seq data was carried out in R version 4.3.0 (2023-04-21) (Team, 2021). All software versions for the RNA-seq analysis correspond to Bioconductor release 3.17. Lowly expressed and non-protein-coding genes were removed, and differential expression analysis was performed using the limma/voom pipeline87,88. Linear models were constructed to identify genes differentially expressed between the ALS and healthy controls using the following fixed effects: subject_group, sex and age_at_death. We used gene set enrichment analysis (GSEA) to assess differential gene expression at the pathway level. GSEA statistics were generated using the fgsea function from the eponymous R package using the t-statistic as the gene ranking statistic89. The constituent genes of the ISR gene set are provided in Supplementary Data 1.
Preclinical ISR gene expression in PBMCs from freshly collected whole blood
Whole blood was collected in CPT sodium heparin tubes (BD, #BD362753) at Denali Therapeutics following manufacturer’s protocol. The studies were approved by local IRBs, and all participants signed informed consent. Following collection, CPT tubes were inverted 10 times to mix, and, for each subject, half of the tubes were treated with DNL343 (2 μM), and half were treated with an equivalent volume of DMSO and incubated at RT for 30 min. Tubes were inverted 10 times then centrifuged at 1800 × g for 30 min at RT. The plasma/PBMC layer was then poured through a 100 μm filter (Corning, #431752) into a 50 mL conical tube (one tube for DNL343-treated samples and one tube for DMSO-treated samples for each subject) and an aliquot of cells was counted using ViaStain AOPI Staining Solution (Nexcelom, #CS2-0106) on a Nexcelom Cellometer Auto 2000 (Nexcelom). Tubes were centrifuged at 500 × g for 15 min at RT and plasma and pellet were separated. Autologous treated plasma was used to resuspend the PBMC cell pellet and 1–1.5 M cells per well were transferred to 96-well round bottom plates (Costar, #3879) with 2 to 3 replicate wells for each sample. For each subject and each treatment condition plates were divided into non-incubated and incubated. For non-incubated plates, samples were immediately processed for RNA and protein as described below. Incubated plates (DMSO and DNL343) were incubated in 5% CO2 at 37 °C for 30 min. After incubation, samples were either stimulated with 50 μM NaAsO2 (Sigma Aldrich, #S7400) or not stimulated and treated with an equal volume of water. Samples were incubated for 4−6 h in 5% CO2 at 37 °C. Plates were then centrifuged for 5 min at 500 × g at RT and supernatant was removed.
For RNA collection, cell pellets were resuspended in 150 μL Buffer RLT Plus (Qiagen, #1053393) with 1% β-mercaptoethanol and frozen at -80 °C. After thawing samples, 150 μL additional Buffer RLT Plus+ 1% β-mercaptoethanol was added to each sample and RNA was then isolated using RNeasy Plus Mini Kit (Qiagen, #74136) or RNeasy Plus 96 Kit (Qiagen, #74192).
CHAC1 gene expression and ATF4 protein level EC50 assays in PBMCs
Cryopreserved human PBMCs were thawed and resuspended in RPMI complete media with glutamax (Gibco, #61870-036) supplemented with 10% fetal bovine serum (Sigma, #F8317) and penicillin-streptomycin (Gibco, #15140-122). In technical triplicate for each condition for each subject, 1.5 million cells were plated in each well of a 96 well clear polypropylene round bottom plate (Costar, #3879) and maintained at 37 °C under 5% CO2, pre-treated with a range of DNL343 concentrations between 15.3 pM and 4 μM for 30 min, and then stimulated with 50 μM NaAsO2 (Sigma Aldrich, #S7400) for 4 h. Plates were then centrifuged for 5 min at 500 × g at RT and supernatant was removed.
For CHAC1 gene expression, cell pellets were lysed in Buffer RLT Plus (Qiagen, #1053393) with 1% β-mercaptoethanol (Sigma, #M3148-25ML) and frozen at -80 °C. RNA was then isolated using the RNeasy Plus 96 Kit (Qiagen, #74192). Gene expression was measured using Fluidigm multiplex qPCR as described. For ATF4 protein levels, cell pellets were resuspended in 150 mL of CST lysis buffer (CST, #9803S) containing benzonase (Millipore, #70746-10KUN), complete mini protease inhibitor (Roche, #04 693 159 001), and PhosSTOP phosphatase inhibitor (Roche, #04 906 837 001), referred to as “CST lysis buffer” throughout, and incubated on ice for 30 min. Plates were then centrifuged at 2500 × g for 20 min at 4 °C. Supernatant was then collected and frozen. ATF4 ECLIA analysis was performed as described in technical replicates for each sample. Concentrations of ATF4 for each sample were determined by MSD analysis software using a 4-parameter logistic model. The dose-response curve for both assays was fitted using log(inhibitor) vs response-variable slope (four parameters) with GraphPad Prism. Any donors with a Hill slope outside of -0.5 to -2.0 were excluded from analysis. Unbound fraction in 10% fetal bovine serum calculated using ultracentrifugation method is 0.426. Unbound EC50 was calculated as the geometric mean of the EC50 values for each individual donor. EC50 ATF4 assays were performed at Denali Therapeutics.
Preclinical ISR gene expression in cryopreserved PBMCs from ALS patients
Cryopreserved human PBMCs were thawed and resuspended in RPMI media as described above. For each subject and each treatment condition plates were divided into non-incubated and incubated. For each subject 0.25 million cells were plated in each well of a 96 well clear polypropylene round bottom plate (Costar, #3879) in technical triplicate when possible, for each condition described. For non-incubated plates, samples were immediately processed for RNA described above. Incubated plates were maintained at 37 °C under 5% CO2, pretreated with 2 μM DNL343 or equivalent volume of DMSO for 30 min, and then stimulated with 50 μM sodium arsenite (Sigma Aldrich, #S7400) or not stimulated and treated with an equal volume of water for 4 h. Plates were then centrifuged for 5 min at 500 × g at RT and supernatant was removed. Cell pellets were resuspended in 150 μL Buffer RLT Plus (Qiagen, #1053393) with 1% β-mercaptoethanol and frozen at -80 °C. After thawing samples, 200 μL additional Buffer RLT Plus+ 1% β-mercaptoethanol was added to each sample and RNA was then isolated using RNeasy plus micro kit protocol (Qiagen, #74034).
Fluidigm assays
The concentration of RNA samples was measured using a Nanodrop 8000 Spectrophotometer (Thermo Scientific, #ND-8000-GL). For each sample, 20−50 ng of RNA (normalized to the same concentration within a given experiment) was reverse transcribed into cDNA using Reverse Transcriptase Master Mix (Fluidigm, now called Standard BioTools, #100-6298) in a 5 μL reaction (Fluidigm, protocol 100-6472 B1). Next, gene expression preamplification with Fluidigm Preamp Master Mix (Fluidigm, #100-5744 (now 100−5580)) using TaqMan Assays (Thermo Scientific) was performed per the manufacturer’s protocol (Fluidigm, protocol 100-5876 C2) for 14−17 cycles depending on starting RNA concentration. For each preamplified cDNA sample, 5 μL of sample was added to 20 μL of DNA Suspension Buffer (Teknova, #T0221) and then combined with 20X GE Sample Loading Reagent (Fluidigm, catalog #85000735 (now 100-7610)) and Taqman Fast Advanced Master Mix (Applied Biosciences, #4444557)). Taqman assays are combined with assay loading reagent (Fluidigm, #85000736 (now 100-7611)). Gene expression analysis was carried out in triplicate for each combination of sample and TaqMan assay using the 96.96 dynamic array IFC plates (Fluidigm, #BMK-M-96.96) on the Biomark HD system (Fluidigm, #BMKHD-BMKHD) according to manufacturer’s protocol (Fluidigm, PN 100-2638 E1). Data was collected with BioMark Data Collection software (Fluidigm) and processed with Real-Time PCR Analysis Software (Fluidigm).
Preclinical ISR gene expression analysis in PBMCs and ALS spinal cord samples
Each sample was run in technical triplicate with an ISR gene panel according to Fluidigm protocol described in this section. For each gene and sample pair, data was analyzed as follows. Technical triplicate Ct values for a given sample and gene pair were averaged and, for PBMCs and spinal cord samples, if the standard deviation was >1, or, for iNeurons, if the range is greater than 1.5 Ct, the value furthest from the median was excluded. The delta Ct was calculated by subtracting the geometric mean of the housekeeping genes ACTB, RPLP0, and RPS17L expression from that sample. The delta delta Ct was calculated compared to specified controls using the 2−ΔΔCt method.
Phase 1 study design
The Phase 1, randomized, double-blind, placebo-controlled single ascending dose (SAD) and multiple ascending dose (MAD) oral dose trial (NCT04268784, first participant signed Informed Consent Form: 24 January 2020, last participant completed: 03 August 2021) was conducted in healthy participants (Fig. 6a and Supplementary Fig. 9a) at a single center. The study was reviewed and approved by the local health authority and ethics committee prior to study conduct. Primary objectives were to investigate safety, tolerability, and plasma pharmacokinetics (PK) in single and multiple oral doses of DNL343. Secondary objectives included characterization of the concentration of DNL343 in CSF following multiple oral doses, in plasma PK after a high-fat meal after up to two oral doses, in urine PK after a single-dose, and pharmacodynamics (PD) of ATF4 protein expression and CHAC1 gene expression in peripheral blood mononuclear cells (PBMCs) after single and multiple doses. Exploratory marker measures were to assess changes in other blood and CSF biomarkers. The study consisted of two parts: Part A was a randomized, placebo-controlled, double-blind, SAD escalation study incorporating a food-effect evaluation of DNL343. Part B was a randomized, placebo-controlled, double-blind, 14 day MAD escalation stage with a single oral formulation. A Dose Escalation Committee composed of the investigator, other investigative site representatives, and Sponsor representatives reviewed safety and tolerability prior to each dose escalation.
The Phase 1 study was conducted at a single clinical research unit (CRU) in the Netherlands and included six SAD dosing cohorts (DNL343 15, 45, 100, 200, 400, 800 mg daily), five MAD dosing cohorts (45, 100, 145, 200, 260 mg daily). Eligible participants were randomized to receive DNL343 or placebo in a 6:2 ratio in SAD cohorts and in a between 8:2 or 7:3 ratio in MAD cohorts for a total of 14 days. Eligible participants were male and female and female of non-childbearing potential healthy volunteers aged 18−55 years with a body mass index between 18.5 and 32.0 kg/m2 weight of at least 50.0 kg.
Phase 1b study design
The multicenter, randomized, double-blind, and placebo-controlled clinical trial (NCT05006352 first participant signed Informed Consent Form: 11 August 2021, last participant completed last visit: 05 June 2024) evaluated placebo, DNL343 100 mg and 200 mg daily in participants with ALS (Fig. 6b and Supplementary Fig. 9b). The primary objective was to evaluate the safety and tolerability of DNL343 administered daily for 28 days. Secondary objectives were to characterize plasma DNL343 PK and CSF concentrations, PD of ATF4 protein and CHAC1 gene expression in peripheral blood mononuclear cells (PBMCs). Exploratory PD biomarker measurements in urine, plasma and CSF are planned. The study allowed enrollment of up to 45 participants with ALS with a minimum of ~30 participants required to complete the double-blind period. An interim analysis of safety, PK, and available PD data was to be performed after ~15 or more participants had completed the double-blind period, to determine if enrollment of additional participants, up to a maximum of 45 participants, was needed for analysis of the primary and secondary endpoints. Slow enrollment during the COVID-19 epidemic and the early available data led to conclusion of enrollment with the minimum protocol planned sample size of ~30.
The study was conducted at 10 clinical sites of which 6 sites recruited participants for the study (University Medical Center Utrecht, Netherlands; Center for Human Drug Research, Netherlands; California Pacific Medical Center, CPMC; University of California at San Diego, CA; Emory University, GA; Hospital for Special Care, CT). The study was reviewed and approved by local health authorities and ethics committees prior to study conduct. Eligible participants were men or women aged 18−80 years with a diagnosis of laboratory-supported probable, probable, or definite (sporadic or familial) ALS according to the El Escorial World Federation of Neurology revised research diagnostic criteria90; years since symptom onset are <4 years; SVC or FVC > 50% predicted during screening. The study allowed for locally approved ALS treatments (e.g. riluzole). Participants with unstable psychiatric, endocrine, pulmonary, cardiovascular, gastrointestinal, hepatic, pancreatic, renal disorders, metabolic, hematologic, immunologic, or allergic disease (hypersensitivity to two or more medication allergies) or neurological disorders (including stroke, cognitive impairment, or seizure within 5 years) other than ALS were excluded.
Study outcomes
In both studies, safety and tolerability were assessed by AE monitoring, clinical laboratory tests (blood chemistries, hematology, and urinalysis), vital signs, electrocardiograms, physical, pulmonary function tests, neurological examinations and psychiatric assessments including the Columbia Suicide Severity Rating Scale and pharmacokinetic parameters in plasma and CSF were assessed. Pharmacodynamic assessments included percent change from baseline of ATF4 and CHAC1 in PBMCs.
Pharmacokinetic assessments from clinical trials
For PK sampling in Part A (SAD) of the healthy participant study, participants were required to fast for at least 8 h before and 4 h after receiving the dose of study intervention. Whole blood for plasma PK was collected on Day 1 predose, and 0.5, 1, 1.5, 2, 4, 8, and 12 h postdose, and on Days 2, 3, 4, 8, and 15 at ~24, 48, 72, 168, and 336 h postdose for full profiles of single-dose PK.
For PK sampling in Part B (MAD) of the healthy participant study, participants were assessed after multiple QD doses in the fasted condition for 14 days. For PK sampling, participants were required to fast for at least 8 h before and 4 h after receiving the dose of study intervention. Whole blood for plasma PK was collected for full PK profiles on Days 1 and 14 predose, at 0.5, 1, 1.5, 2, 4, 8, 12, and 24 h post-dose; additional timepoints at 24, 48, 72, 168 and 336 h after the last dose. A CSF sample was collected for PK 4–8 h after the morning dose on Day 12.
For the ALS participant study multiple doses of DNL343 100-, 200- mg or placebo QD were assessed on an empty stomach (i.e., ≥ 1 h before or ≥ 2 h after a meal) for 28 days. Whole blood for plasma PK was collected for full PK profiles on Days 1 predose, at 1, 2, 4, 8, and 12 h post-dose; predose Days 2, 7, 14 and 21 at ~24 h after the previous dose Day 28 predose, at 1, 2, 4, and 8 h post-dose (a 24 h post-dose sample was imputed from the predose sample on Day 28). A CSF sample was collected for PK around 4 h after the morning dose on Day 28.
Plasma and CSF concentrations of DNL343 were measured using LC-MS/MS. The bioanalysis method was fully validated and met the acceptance criteria for intra- and inter-run precision and accuracy defined in the 2018 FDA Bioanalytical Method Validation Guidance (https://www.fda.gov/media/70858/download).
Noncompartmental analysis (NCA) was performed on individual plasma concentration data using Phoenix 64 WinNonlin (versions 8.1 and 8.3). CSF PK data and CSF-to-plasma ratios were summarized with R (version 4.2.1). Plasma and CSF PK graphics were generated in R.
Clinical PD PBMC assays
Whole blood was collected in CPT sodium heparin tubes (BD, #BD362753) following manufacturer’s protocol. Following collection, CPT tubes were inverted 10 times to mix and then centrifuged at 1800 × g for 30 min at RT.
Freshly isolated PBMC assay was performed at Denali Therapeutics or CHDR. The CPT tubes were inverted to mix and the plasma/PBMC layer was then poured through a 100 μm filter (Corning, #431752) into a 50 mL conical tube and an aliquot of cells was counted using ViaStain AOPI Staining Solution (Nexcelom, #CS2-0106) on a Nexcelom Cellometer Auto 2000 at Denali or counted using propidium iodine (Miltenyi Biotec, #130-093-233) and the MACSQuant analyzer (Miltenyi Biotec) at CHDR. 50 mL tubes were centrifuged at 500 × g for 15 min at RT and plasma and pellet were separated. Autologous plasma was used to resuspend the PBMC cell pellet and 1–1.5 M cells per well were transferred to 96-well round bottom plates (Costar, #3879) with 2 to 3 replicate wells for each participant/condition. Plates were incubated in 5% CO2 at 37 °C for 30 min. After incubation, samples were stimulated with 50 µM NaAsO2 (Sigma Aldrich, #S7400). Samples were incubated for 4 h in 5% CO2 at 37 °C. Plates were then centrifuged for 5 min at 500 × g at RT and supernatant was removed.
For RNA collection, cell pellets were resuspended in 150 μL Buffer RLT Plus (Qiagen, #1053393) with 1% β-mercaptoethanol, sealed (Corning Axygen PCR-AS-600), and frozen at -80 °C. After thawing samples, 150 μL additional Buffer RLT Plus+ 1% β-mercaptoethanol was added to each sample and RNA was then isolated using RNeasy Plus 96 Kit (Qiagen, #74192).
For protein, cell pellets were resuspended in 150 mL of CST lysis buffer (CST, #9803S) containing benzonase (Millipore, #70746-10KUN), complete mini protease inhibitor (Roche, #04 693 159 001), and PhosSTOP phosphatase inhibitor (Roche, #04 906 837 001), referred to as “CST lysis buffer” throughout, and incubated on ice for 30 min. Plates were then centrifuged at 2500 × g for 20 min at 4 °C. Supernatant was then collected and frozen.
Clinical ISR gene expression analysis in human PBMCs
Each sample was run in technical triplicate with an ISR gene panel according to Fluidigm protocol described in this section. For each gene and sample pair, data was analyzed as follows. All Ct values greater than 28, labeled as NA, or flagged were excluded. For each gene and sample combination the median and range across the technical triplicates was determined. If 3 values were present and if the Ct range was greater than 1.5, the value furthest from the median was dropped. The arithmetic mean of Cts across the remaining technical replicates was calculated. The geometric mean across the housekeeping genes (ACTB, RPLP0, RPS17L) was calculated for each sample. Samples where the geometric mean of the housekeeping genes was greater than 12 (Phase 1) or 15 (Phase 1b) were excluded. For each sample the delta Ct was calculated by subtracting the geometric mean of the housekeeping genes from the averaged Ct value from the step above. The arithmetic mean of dCts across the biological replicates for each participant/condition was calculated. Percent change from baseline per participant and sample was calculated using the equation (2^(-1 *(dct value of timepoint-dct of baseline) – 1) *100. Data was analyzed in R version 4.2.2. Heatmaps were generated in GraphPad Prism 9.
Clinical ATF4 protein level analysis in human PBMCs
Samples are plated in technical duplicate on the assay plate. If technical duplicate RLU values for a sample had a CV value > 30% then the sample was excluded from analysis. Pg/ml concentrations of ATF4 for each sample were determined by Meso Scale Discovery (MSD) analysis software Discovery Workbench (MSD version LSR_4_0_13) using a 4-parameter logistic model. BLQ and values less than LLOD (10) were assigned ½ LLOD which is 5 pg/mL. A correction factor to adjust for cell count was applied to values greater than or equal to LLOQ and not BLQ.
The mean concentration (pg/mL) of the remaining samples was taken across technical replicates. If there were at least 3 biological replicate values, the range and the median across each group of biological replicates was calculated. If (range/median) > 0.5, then the biological replicate furthest from the median was dropped. The mean concentrations (pg/mL) of the remaining samples across the biological replicates was averaged. The percent change from baseline per participant and sample was calculated.
GDF-15 protein level measurements and analysis in human CSF
CSF samples were diluted 50x in Diluent 100 (MSD, #R50AA-4) and assayed according to manufacturer’s protocol using the R-PLEX Human GDF-15 Antibody Set (MSD, # F21YD-3) and MSD GOLD 96-well small spot streptavidin SECTOR plates (MSD, #L45SA). Samples were plated in technical duplicate on the assay plate. Plates were read on a 1201 MSD SECTOR 600MM plate reader (MSD, catalog # IC1AA-0) within 30 min of adding buffer and data was captured in Methodical Mind (MSD, version Fall 2020 (1.0.38)). Pg/ml concentrations for each sample were determined by MSD analysis software Discovery Workbench (MSD, version LSR_4_0_13) using a 4-parameter logistic model. The mean concentrations (pg/mL) of the technical replicates were averaged. The percent change from baseline per participant and sample was calculated. Data was analyzed in R version 4.2.2.
Statistics and reproducibility
Preclinical studies
Cellular preclinical studies were independently conducted with 2–3 technical replicates per independent experiment. A total of 7−12 animals per group were used for biochemical analyses and 8−16 animals per group were used for behavioral studies. No statistical method was used to predetermine sample size. Animals used in the preclinical studies were randomly assigned to experimental groups. Researchers conducting the in vivo preclinical studies were blinded to the treatment assignment.
Data have been shown as ± SEM or SD, as indicated in the figure legends. Statistical analyses were performed in either GraphPad Prism 9 or R. For studies completed in H4 cells, HEK293 cells or C9orf72 iNeurons, analysis was done using one-way analysis of variance (ANOVA) with multiple comparison adjustments, as indicated in the figure legends. Nominal P values were presented. For ALS and control spinal cord gene expression analysis (Fluidigm), fold change relative to average of control samples was calculated and comparison between ALS and control samples assessed using multiple t-tests on log2 fold change data, with the false discovery rate controlled at 5%. For iNeuron Fluidigm gene expression analysis, fold change data relative to average of non-stimulated, DMSO-treated samples was calculated and comparison between stimulated samples treated with DNL343 or DMSO is performed using multiple paired t-tests on log2 fold change data, with the false discovery rate controlled at 5%. For gene expression analysis in PBMCs from healthy participants, fold change relative to non-incubated control within each donor was calculated and comparison between stimulated samples treated with DNL343 or DMSO was assessed using multiple paired t-tests on log2 fold change data, with the false discovery rate controlled at 5%. For ALS PBMC gene expression analysis of CHAC1 and TRIB3, fold change relative to the average of the non-incubation condition in healthy controls was calculated and comparison between DMSO and DNL343 treated samples was assessed using multiple ratio-paired t-tests with Holm-Sidak correction, 0.05 alpha. For these experiments comparison of ISR gene expression between non-incubated samples from ALS and healthy controls was performed using multiple t-tests on log2 fold change data, with the false discovery rate controlled at 5%.
Clinical studies
No statistical method was used to predetermine sample size for the Phase 1 and Phase 1b studies. The selected sample sizes were considered adequate to meet the primary and secondary objectives of the studies, without exposing an undue number of participants to the experimental drug. Both studies randomized participants to receive placebo or study intervention (at multiple dose levels), using a block-randomization algorithm (implemented by unblinded CRO personnel), with block size depending on cohort specific randomization ratio for the Phase 1, and block size of 3 for the Phase 1b study. Both participants and the investigators were blinded to the treatment assignment. No data was excluded from the analysis described in this manuscript, except as specifically noted.
In the Phase 1 study, data were summarized by treatment group (pooled placebo group and each DNL343 dose group) for each study part. In the Phase 1b study, data were summarized by treatment group for the double-blind period. Demographic and other baseline characteristics were summarized descriptively for the Randomized participants population. Treatment-emergent adverse events (TEAEs) were defined as AEs that occurred or worsened after initiation of study drug. The incidence and severity of TEAEs was summarized descriptively for the Safety population (all treated participants). PK data were summarized using descriptive statistics for all DNL343 treated participants, with evaluable PK data. Pharmacodynamic data were summarized using descriptive statistics for all treated participants with evaluable PBMC data. No imputation for missing data was performed.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data associated with this research is present in the paper or in the Supplementary Information. Raw RNA-seq data (FASTQ files) and metadata for tissue samples were originally downloaded from the NY Genome Center’s Collaborator Portal on 2021/09/09. The TargetALS consortium has since made this data publicly available via the NCBI GEO repository as part of Superseries GSE137810. Bulk RNA-seq data from in vitro models has been deposited at the NCBI GEO repository as Superseries GSE281960. DNL343 analogs can be made available through a material transfer agreement by submitting a request at https://denalitherapeutics.com/contact. All individual participant data collected during the clinical trials, that underlie the results of this manuscript that do not include identifying information, will be available to investigators whose proposed use of the data has been approved by the study sponsor. The data is available beginning 9 months and ending 36 months following the publication of this manuscript. Proposals may be submitted directly to the study sponsor at data-requests@dnli.com. The study protocol will be available upon request to the study sponsor. Source data are provided with this paper.
Code availability
The code used for bulk RNA-seq analysis in this manuscript is available via Zenodo under DOI: 10.5281/zenodo.14041444.
References
Kim, G., Gautier, O., Tassoni-Tsuchida, E., Ma, X. R. & Gitler, A. D. ALS Genetics: gains, losses, and implications for future therapies. Neuron 108, 822–842 (2020).
Mejzini, R. et al. ALS Genetics, mechanisms, and therapeutics: where are we now?. Front Neurosci. 13, 1310 (2019).
Goutman, S. A. et al. Emerging insights into the complex genetics and pathophysiology of amyotrophic lateral sclerosis. Lancet Neurol. 21, 465–479 (2022).
Fernandes, N., Eshleman, N. & Buchan, J. R. Stress granules and ALS: a case of causation or correlation?. Adv. Neurobiol. 20, 173–212 (2018).
Li, Z., Liu, X. & Liu, M. Stress granule homeostasis, aberrant phase transition, and amyotrophic lateral sclerosis. ACS Chem. Neurosci. 13, 2356–2370 (2022).
Ratti, A. & Buratti, E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J. Neurochem. 138, 95–111 (2016).
Keating, S. S., San Gil, R., Swanson, M. E. V., Scotter, E. L. & Walker, A. K. TDP-43 pathology: from noxious assembly to therapeutic removal. Prog. Neurobiol. 211, 102229 (2022).
Neumann, M. et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006).
Arai, T. et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 351, 602–611 (2006).
Ma, X. R. et al. TDP-43 represses cryptic exon inclusion in the FTD-ALS gene UNC13A. Nature 603, 124–130 (2022).
Ling, J. P., Pletnikova, O., Troncoso, J. C. & Wong, P. C. TDP-43 repression of nonconserved cryptic exons is compromised in ALS-FTD. Science 349, 650–655 (2015).
Brown, A. L. et al. TDP-43 loss and ALS-risk SNPs drive mis-splicing and depletion of UNC13A. Nature 603, 131–137 (2022).
Melamed, Z. et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019).
Mehta, P. R., Brown, A. L., Ward, M. E. & Fratta, P. The era of cryptic exons: implications for ALS-FTD. Mol. Neurodegener. 18, 16 (2023).
Akiyama, T., Koike, Y., Petrucelli, L. & Gitler, A. D. Cracking the cryptic code in amyotrophic lateral sclerosis and frontotemporal dementia: towards therapeutic targets and biomarkers. Clin. Transl. Med. 12, e818 (2022).
Baughn, M. W. et al. Mechanism of STMN2 cryptic splice-polyadenylation and its correction for TDP-43 proteinopathies. Science 379, 1140–1149 (2023).
Lopez-Erauskin, J. et al. Stathmin-2 loss leads to neurofilament-dependent axonal collapse driving motor and sensory denervation. Nat. Neurosci. 27, 34–47 (2024).
Ling, S. C., Polymenidou, M. & Cleveland, D. W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron 79, 416–438 (2013).
Ito, D., Hatano, M. & Suzuki, N. RNA binding proteins and the pathological cascade in ALS/FTD neurodegeneration. Sci. Transl. Med. 9, eaah5436 (2017).
de Mena, L., Lopez-Scarim, J. & Rincon-Limas, D. E. TDP-43 and ER Stress in Neurodegeneration: Friends or Foes?. Front Mol. Neurosci. 14, 772226 (2021).
Kim, H. J. et al. Therapeutic modulation of eIF2alpha phosphorylation rescues TDP-43 toxicity in amyotrophic lateral sclerosis disease models. Nat. Genet 46, 152–160 (2014).
Luan, W. et al. Early activation of cellular stress and death pathways caused by cytoplasmic TDP-43 in the rNLS8 mouse model of ALS and FTD. Mol. Psychiatry 28, 2445–2461 (2023).
Cheng, W. et al. C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2alpha phosphorylation. Nat. Commun. 9, 51 (2018).
Green, K. M. et al. RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun. 8, 2005 (2017).
Kramer, N. J. et al. CRISPR-Cas9 screens in human cells and primary neurons identify modifiers of C9ORF72 dipeptide-repeat-protein toxicity. Nat. Genet 50, 603–612 (2018).
Zhang, Y. J. et al. Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med. 24, 1136–1142 (2018).
Westergard, T. et al. Repeat-associated non-AUG translation in C9orf72-ALS/FTD is driven by neuronal excitation and stress. EMBO Mol. Med. 11, e9423 (2019).
Parameswaran, J. et al. Antisense, but not sense, repeat expanded RNAs activate PKR/eIF2alpha-dependent ISR in C9ORF72 FTD/ALS. Elife 12, e85902 (2023).
Zu, T. et al. Metformin inhibits RAN translation through PKR pathway and mitigates disease in C9orf72 ALS/FTD mice. Proc. Natl. Acad. Sci. USA 117, 18591–18599 (2020).
Liu-Yesucevitz, L. et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS ONE 5, e13250 (2010).
Parker, S. J. et al. Endogenous TDP-43 localized to stress granules can subsequently form protein aggregates. Neurochem. Int. 60, 415–424 (2012).
Meyerowitz, J. et al. C-Jun N-terminal kinase controls TDP-43 accumulation in stress granules induced by oxidative stress. Mol. Neurodegener. 6, 57 (2011).
Colombrita, C. et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 111, 1051–1061 (2009).
Dewey, C. M. et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol. Cell Biol. 31, 1098–1108 (2011).
McDonald, K. K. et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum. Mol. Genet 20, 1400–1410 (2011).
Cui, Q., Liu, Z. & Bai, G. Friend or foe: the role of stress granule in neurodegenerative disease. Neuron 112, 2464–2485 (2024).
Shorter, J. Phase separation of RNA-binding proteins in physiology and disease: an introduction to the JBC Reviews thematic series. J. Biol. Chem. 294, 7113–7114 (2019).
Johnson, B. S. et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 284, 20329–20339 (2009).
Li, Y. R., King, O. D., Shorter, J. & Gitler, A. D. Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013).
Zhang, P. et al. Chronic optogenetic induction of stress granules is cytotoxic and reveals the evolution of ALS-FTD pathology. Elife 8, e39578 (2019).
Becker, L. A. et al. Therapeutic reduction of ataxin-2 extends lifespan and reduces pathology in TDP-43 mice. Nature 544, 367–371 (2017).
Markmiller, S. et al. Context-Dependent and disease-specific diversity in protein interactions within stress granules. Cell 172, 590–604 e513 (2018).
Fang, M. Y. et al. Small-molecule modulation of TDP-43 recruitment to stress granules prevents persistent TDP-43 accumulation in ALS/FTD. Neuron 103, 802–819 e811 (2019).
Abbink, T. E. M. et al. Vanishing white matter: deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 6, 1407–1422 (2019).
Hanson, F. M., Hodgson, R. E., de Oliveira, M. I. R., Allen, K. E. & Campbell, S. G. Regulation and function of elF2B in neurological and metabolic disorders. Biosci. Rep. 42, BSR20211699 (2022).
Wong, Y. L. et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. Elife 8, e42940 (2019).
Wong, Y. L. et al. The small molecule ISRIB rescues the stability and activity of vanishing white matter disease eIF2B mutant complexes. Elife 7, e32733 (2018).
Yulyaningsih, E. et al. DNL343 is an investigational CNS penetrant eukaryotic initiation factor 2B activator that prevents and reverses the effects of neurodegeneration caused by the integrated stress response. Elife 12, RP92173 (2024).
Marlin, E., Viu-Idocin, C., Arrasate, M. & Aragon, T. The role and therapeutic potential of the integrated stress response in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 23, 7823 (2022).
Ilieva, E. V. et al. Oxidative and endoplasmic reticulum stress interplay in sporadic amyotrophic lateral sclerosis. Brain 130, 3111–3123 (2007).
Mead, R. J., Shan, N., Reiser, H. J., Marshall, F. & Shaw, P. J. Amyotrophic lateral sclerosis: a neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 22, 185–212 (2023).
Walker, A. K. et al. ALS-associated TDP-43 induces endoplasmic reticulum stress, which drives cytoplasmic TDP-43 accumulation and stress granule formation. PLoS One 8, e81170 (2013).
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Sidrauski, C. et al. Pharmacological brake-release of mRNA translation enhances cognitive memory. Elife 2, e00498 (2013).
Chou, A. et al. Inhibition of the integrated stress response reverses cognitive deficits after traumatic brain injury. Proc. Natl. Acad. Sci. USA 114, E6420–E6426 (2017).
Krukowski, K. et al. Small molecule cognitive enhancer reverses age-related memory decline in mice. Elife 9, e62048 (2020).
Tsai, J. C. et al. Structure of the nucleotide exchange factor eIF2B reveals mechanism of memory-enhancing molecule. Science 359, eaaq0939 (2018).
Zyryanova, A. F. et al. Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B. Science 359, 1533–1536 (2018).
Wang, C. et al. Inhibiting the integrated stress response pathway prevents aberrant chondrocyte differentiation thereby alleviating chondrodysplasia. Elife 7, e37673 (2018).
Craig, R. A. et al. A potent, selective, and brain-penetrant eIF2B activator designed for the treatment of neurodegenerative diseases. J. Med Chem. 67, 5758–5782 (2024).
Lockhart, S. M., Saudek, V. & O’Rahilly, S. GDF15: A hormone conveying somatic distress to the brain. Endocr. Rev. 41, bnaa007 (2020).
Donnelly, N., Gorman, A. M., Gupta, S. & Samali, A. The eIF2alpha kinases: their structures and functions. Cell Mol. Life Sci. 70, 3493–3511 (2013).
Sareen, D. et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci. Transl. Med. 5, 208ra149 (2013).
Walker, A. K. et al. Functional recovery in new mouse models of ALS/FTLD after clearance of pathological cytoplasmic TDP-43. Acta Neuropathol. 130, 643–660 (2015).
Pakos-Zebrucka, K. et al. The integrated stress response. EMBO Rep. 17, 1374–1395 (2016).
Tanaka, T. et al. Plasma proteomic signature of age in healthy humans. Aging Cell 17, e12799 (2018).
Lehallier, B., Shokhirev, M. N., Wyss-Coray, T. & Johnson, A. A. Data mining of human plasma proteins generates a multitude of highly predictive aging clocks that reflect different aspects of aging. Aging Cell 19, e13256 (2020).
Van Deerlin, V. M. et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 7, 409–416 (2008).
Kabashi, E. et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet 40, 572–574 (2008).
Sreedharan, J. et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science 319, 1668–1672 (2008).
Hofmann, J. W., Seeley, W. W. & Huang, E. J. RNA binding proteins and the pathogenesis of frontotemporal lobar degeneration. Annu Rev. Pathol. 14, 469–495 (2019).
Hetz, C., Axten, J. M. & Patterson, J. B. Pharmacological targeting of the unfolded protein response for disease intervention. Nat. Chem. Biol. 15, 764–775 (2019).
Mallucci, G. R., Klenerman, D. & Rubinsztein, D. C. Developing therapies for neurodegenerative disorders: insights from protein aggregation and cellular stress responses. Annu Rev. Cell Dev. Biol. 36, 165–189 (2020).
Atkins, C. et al. Characterization of a novel PERK kinase inhibitor with antitumor and antiangiogenic activity. Cancer Res 73, 1993–2002 (2013).
Yu, Q. et al. Type I interferons mediate pancreatic toxicities of PERK inhibition. Proc. Natl. Acad. Sci. USA 112, 15420–15425 (2015).
Sidrauski, C. et al. Pharmacological dimerization and activation of the exchange factor eIF2B antagonizes the integrated stress response. Elife 4, e07314 (2015).
Oliveira, M. M. et al. Correction of eIF2-dependent defects in brain protein synthesis, synaptic plasticity, and memory in mouse models of Alzheimer’s disease. Sci. Signal 14, eabc5429 (2021).
Naguib, A. et al. SUPT4H1 depletion leads to a global reduction in RNA. Cell Rep. 26, 45–53 e44 (2019).
Du, Z. W. et al. Generation and expansion of highly pure motor neuron progenitors from human pluripotent stem cells. Nat. Commun. 6, 6626 (2015).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron 104, 239–255 e212 (2019).
Andrade, N. S. et al. Dipeptide repeat proteins inhibit homology-directed DNA double strand break repair in C9ORF72 ALS/FTD. Mol. Neurodegener. 15, 13 (2020).
Lee, J. et al. LSM12-EPAC1 defines a neuroprotective pathway that sustains the nucleocytoplasmic RAN gradient. PLoS Biol. 18, e3001002 (2020).
Carpenter, A. E. et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006).
Jiang, H., Lei, R., Ding, S. W. & Zhu, S. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. BMC Bioinforma. 15, 182 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015).
Korotkevich, G. et al. Fast gene set enrichment analysis. bioRxiv https://doi.org/10.1101/060012 (2021).
Ludolph, A. et al. A revision of the El Escorial criteria - 2015. Amyotroph. Lateral Scler. Front. Degener. 16, 291–292 (2015).
Acknowledgements
We thank the volunteers, patients, and their families who participated in the clinical studies. We dedicate this publication to Patrick Carl Gustav Haddick, who spearheaded the clinical biomarker work in this study and who reminded us that time is precious, and how we spend it matters. We thank Gabby Lunkes de Melo for help with the RNA-seq experiments on cellular models, Annette Garza-Meilandt for managing the DNL343 program, Mark Kafka for critical reading of the manuscript as well as Amber Kampen and Mai Thayer for help with pharmacology data management in Vortex. We acknowledge the Target ALS Human Postmortem Tissue Core, New York Genome Center for Genomics of Neurodegenerative Disease, Amyotrophic Lateral Sclerosis Association and TOW Foundation. We thank the Cedars-Sinai Medical Center’s David and Janet Polak Foundation Stem Cell Core Laboratory for their iPSC cell lines. We would like to thank the members of the Nikon Imaging Center at UC San Diego for help with imaging experiments. We thank Johann Chow for technical assistance for cell-based assays, Nick Propson for iPSC technical support, Alisa Arata for reagent preparation for pharmacodynamic assays, Roni Chau for advice in handling mouse samples and running in vivo experiments, Anil Rana for drawing schematic figures, and Benjamin Rogers for drawing the chemical structure of DN9058. We also thank the Denali clinical operations team for execution of the Phase 1 and Phase 1b studies. Untreated cryopreserved ALS PBMCs were obtained from DNLI-D-0003 study (NCT03757351). The research in this study was funded by Denali Therapeutics Inc. G.W.Y. is supported by NIH R01 HG004659, U24 HG009889 and an Allen Distinguished Investigator Award, a Paul G. Allen Frontiers Group advised grant of the Paul G. Allen Foundation.
Author information
Authors and Affiliations
Contributions
Discovery and development of the compound: MO, AAE. Preclinical study design: BNF, SBY, IVC, MHF, WL, MEKC, WED, TM, MO, AQV, GWY, AAE, KS-L, EY, AKW, GDP, LAK, JWL. Preclinical study execution: BNF, SBY, MHF, WL, SB, MEKC, MYC, SKD, CLH, AQV, LMSM, CX. Preclinical interpretation of the reported results: BNF, SBY, IVC, MHF, WL, MEKC, MYC, TM, AQV, GWY, AAE, KS-L, EY, AKW, GDP, LAK, JWL. Preclinical statistical analysis and data visualization: BNF, SBY, MHF, WL, MEKC, KS, DT, AQV, GWY, TS, KS-L, LAK, JWL. Supervision of preclinical studies: WED, GWY, KS-L, AKW, GDP, LAK, JWL. Development of the clinical study designs: IVC, MHF, LDS, RMT, AB, SD, DJ, CH, KS-L, MDT. Execution of the phase 1 (including generation of results): IVC, MHF, RDM, RMT, MV, LS, TMB, LHvdB, AB, MF, JAH, GJG, SD, CH, MDT. Execution of the phase 1b (including generation of results): IVC, MHF, RDM, LDS, RMT, MV, LS, TMB, LHvdB, MF, JAH, GJG, SD, CH, MDT. Interpretation and analysis of the reported results of the phase 1: IVC, MHF, RDM, LDS, RMT, AB, MF, SD, CH, KS-L, MDT. Interpretation and analysis of the reported results of the phase 1b: IVC, MHF, RDM, LDS, MF, FH, SD, CH, KS-L, MDT. Design of clinical statistical analyses: IVC, MHF, RDM, LDS, RMT, SD, MDT. Execution of clinical statistical analyses: IVC, MHF, RDM, SD. Analysis and interpretation of the clinical pharmacokinetics data: IVC, MF, SD. Oversight for execution of the phase 1 and phase 1b: CH, MDT. Writing – original draft: BNF, SBY, IVC, MHF, LDS, SD, GDP, LAK, JWL. Writing – review & editing: All authors. Additional authors with equal contribution: WL, RDM.
Corresponding authors
Ethics declarations
Competing interests
SBY, IVC, RDM, RMT, AB, SB, MEKC, MYC, MF, FH, MO, KS, CLH, TM, SD, AAE, DJ, CH, GDP, MDT and JWL are current employees and shareholders of Denali Therapeutics. BNF, MHF, LDS, SKD, WD, CX, and KS-L, EY, and LAK are former employees of Denali Therapeutics. MV, LS, GJG. have appointments at CHDR. LS, GJG have appointments at LUMC. TB and LvB. have appointments at UMCU. LHvdB served in advisory boards for Biogen, Amylyx, Ferrer, Corcept, QurAlis, Cytokinetics, Argenx, VectorY, Zambon paid to institution. LHvdB has participated as principal investigator to clinical trials on ALS sponsored by Biogen, Cytokinetics, Ferrer, Amylyx, Wave Life Sciences, Corcept therapeutics, Sanofi, AB Science, IONIS Pharmaceuticals, Apellis Pharmaceuticals, Alexion Pharmaceuticals, Orphazyme, Orion Pharma and Denali. GWY is a co-founder, member of the Board of Directors, on the SAB, equity holder, and paid consultant for Eclipse BioInnovations. GWY is a visiting professor at the National University of Singapore. GWY’s interests have been reviewed and approved by the University of California, San Diego in accordance with its conflict-of-interest policies. The aforementioned authors declare no other competing financial and non-financial interests. The remaining authors declare no competing interests. DNL343 is an investigational drug that has not been approved by any Health Authority.
Peer review
Peer review information
Nature Communications thanks Hongqiu Gu and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Flores, B.N., Yu, S.B., Cohen, I.V. et al. Investigational eIF2B activator DNL343 modulates the integrated stress response in preclinical models of TDP-43 pathology and individuals with ALS in a randomized clinical trial. Nat Commun 16, 7690 (2025). https://doi.org/10.1038/s41467-025-63031-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63031-y
This article is cited by
-
Twin pairs discordant for incident coronary artery disease reveal epigenetic and transcriptomic differences by gene region
Molecular Genetics and Genomics (2026)
-
Biomolecular condensates: molecular structure, biological functions, diseases, and therapeutic targets
Molecular Biomedicine (2025)