Abstract
Here we describe ProtacID, a flexible BioID (proximity-dependent biotinylation)-based approach to identify PROTAC-proximal proteins in living cells. ProtacID analysis of VHL- and CRBN-recruiting PROTACs targeting a number of different proteins (localized to chromatin or cellular membranes, and tested across six different human cell lines) demonstrates how this technique can be used to validate PROTAC degradation targets and identify non-productive (i.e. non-degraded) PROTAC-interacting proteins, addressing a critical need in the field of PROTAC development. We also demonstrate that ProtacID can be used to characterize native, endogenous multiprotein complexes without the use of antibodies, or modification of the protein of interest with epitope tags or biotin ligase tagging.
Similar content being viewed by others
Introduction
One of the most exciting and fastest-moving areas of modern drug discovery, Targeted Protein Degradation (TPD) harnesses the power of the endogenous ubiquitin proteasome system to specifically degrade proteins linked to disease1,2. PROteolysis TArgeting Chimeras (PROTACs) are bivalent, cell-permeable TPD compounds consisting of a warhead moiety that interacts with one or more target proteins, and a ubiquitin E3 ligase recruiter component, connected via a chemical linker (Fig. 1a, inset). PROTACs thus direct an endogenous E3 ligase to neo-substrates to effect ubiquitylation, resulting in 26S proteasome-mediated degradation. The majority of the >5000 PROTACs3 developed to date utilize just two ubiquitin E3 ligases: (i) VHL (Von Hippel-Lindau), the substrate targeting receptor subunit of a Cullin 2 (CUL2)-RING E3 Ligase complex (CRL2VHL) or (ii) CRBN (cereblon), a substrate receptor subunit of a CUL4-based complex (CRL4CRBN). PROTACs serve as important research tools, and multiple PROTAC-based drugs have recently entered clinical trials1.
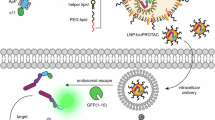
a A PROTAC molecule consists of a warhead and E3 recruiter moieties connected by a chemical linker (inset). ProtacID is conducted by (i) stably expressing a relevant E3 ligase subunit (e.g., VHL or CRBN) fused with a BioID tool such as miniTurbo12. miniTurbo is an abortive biotin ligase that generates chemically reactive biotin (biotinoyl-AMP, red circles), which diffuses away from the enzyme and reacts with amine groups on lysine residues in nearby proteins. (ii) In the presence of a PROTAC, neo-substrates are recruited to the E3-miniTurbo protein, but are not ubiquitylated and degraded in the presence of the neddylation inhibitor MLN4924. Lysine residues in proteins within ~10 nm (an average protein has a Stokes radius of 4–5 nm) are susceptible to biotinylation. (iii) Following cell lysis, biotinylated proteins are isolated using streptavidin-sepharose and identified using mass spectrometry. Created in BioRender. Duan, S. (2025) https://BioRender.com/5kqqsu9. b Schematic diagram of the FLAG-miniTurbo (FmT)-tagged VHL fusion protein variants tested here. All vector variants were created with and without an N-terminal nuclear localization signal (NLS; see “Methods” for details). Created in BioRender. Duan, S. (2025) https://BioRender.com/x7rd333. c Schematic of the CRBNmidi-mTF fusion protein used for ProtacID. Created in BioRender. Maitland, M. (2025) https://BioRender.com/ykrihkh.
Importantly, however, while PROTAC development has surged in popularity, our ability to characterize PROTAC specificity in living cells has lagged behind. A common method for assessing TPD tool specificity is global proteome analysis, which identifies proteins that decrease in abundance (via so-called productive interactions) following PROTAC treatment4. While powerful, this approach has some important limitations. The loss of a particular protein in response to PROTAC treatment does not necessarily indicate that it is a direct PROTAC target. For example, if a PROTAC-targeted protein plays an important regulatory role upstream of one or more additional short-lived polypeptide(s), or happens to be a key structural component of a protein complex, its loss can trigger the secondary loss of multiple additional proteins5,6,7. Conversely, PROTAC binding does not always result in the degradation of a protein interactor, yet could still affect its activity, localization, and/or interactions with binding partners. These so-called non-productive PROTAC-target interactions are not detected in global proteome analyses.
Proximity-dependent labeling techniques such as BioID are now widely used to identify protein-protein interactions in living cells8,9. As previously demonstrated with CRL4CRBN, tagging an E3 ligase with a BioID tool can also be used to effect PROTAC (or molecular glue)-mediated recruitment of a biotin ligase to an endogenous neo-substrate10. We reasoned that applying a similar approach to the VHL protein and the CRBNmidi tool (a recently developed stable CRBN protein variant11) could be used to better characterize PROTAC binding partners.
Here we demonstrate that our ProtacID approach can be used to: (i) validate PROTAC neo-substrates identified in standard global proteome analyses, (ii) identify non-productive PROTAC interactors, and (iii) characterize PROTAC target interacting protein partners.
Results
ProtacID tool development
To conduct ProtacID, a FLAG-tagged miniTurbo12 (FmT) enzyme bearing a nuclear localization signal (NLS) was fused to the N- or C-terminus of full-length VHL or a VHL variant lacking unstructured residues 1–53 (ΔVHL, as in ref. 13) (Fig. 1b). The resulting fusion proteins were stably expressed (Supplementary Fig. 1a) in human embryonic kidney 293 Flp-In cells. Cells were treated with: (a) the neddylation inhibitor MLN4924 to block cullin activity (and thus rescue CRL2VHL targets), (b) DMSO (negative control) or the PROTAC ACBI114, and (c) biotin (Fig. 2a). Following cell lysis, biotinylated proteins were affinity-purified using streptavidin-sepharose beads.
a ProtacID treatment timeline. See “Methods” for details. Created in BioRender. Duan, S. (2025) https://BioRender.com/8lrbx5r. b Schematic diagrams of the canonical (cBAF), polybromo-associated (PBAF), and non-canonical (ncBAF) BAF complex variants. Components specific to each BAF variant are color-coded as in panel d. Created in BioRender. Duan, S. (2025) https://BioRender.com/7p89ex4. c FmT-VHL proteins form a ternary complex with ACBI1 and SMARCA4. MLN4924 and increasing doses of ACBI1 (0, 10, 100, 1000 nM) were applied to 293 Flp-In cells expressing the indicated FmT-tagged VHL proteins. After biotin treatment, cells were lysed, and biotinylated proteins were isolated using streptavidin-sepharose. Isolated proteins (PD, pulldown) or whole cell lysates (input) were subjected to western blotting with the indicated antibodies. IB immunoblot. n = 3 biological replicates. d FmT-VHL tool testing. ProtacID was conducted on 293 Flp-In cells expressing the indicated FmT-tagged VHL protein variants, and treated with either DMSO (−) or 200 nM ACBI1 (+). The Dot Plot depicts proteins identified by ProtacID, where dot size indicates relative peptide abundance across experiments, dot shade indicates average peptide counts, and dot border indicates protein identification confidence level. Standard BioID (right) was also conducted in 293 Flp-In cells expressing FmT-SMARCA2 or FmT-SMARCA4. PROTAC neo-substrates are indicated by arrowheads (right). Proteins grouped according to the BAF variant. See “Methods” for a description of SAINTexpress statistical analysis, which provides a Bayesian false discovery rate (BFDR) for all mass spectrometry data. Source data are provided as a Source data file.
BAF (BRG1/BRM-associated factor) protein complexes are ~2 MDa multi-subunit SWI/SNF chromatin remodelers15. Three distinct BAF complex variants have been described: canonical (cBAF), non-canonical (ncBAF), and polybromo-associated (PBAF)15 (Fig. 2b). A number of VHL-recruiting PROTACs targeting BAF complex components have been developed, including ACBI114, which targets the bromodomains of the SMARCA2 and SMARCA4 ATPases (interchangeable components of all three BAF complex variants) and PBRM1 (a PBAF-specific protein).
An aliquot (~10%) of the protein-bound beads was subjected to SDS-PAGE and western blotting analysis. SMARCA4 was isolated (and therefore biotinylated) following ACBI1 treatment (but not in the absence of PROTAC), and SMARCA4 biotinylation increased in a PROTAC dose-dependent manner (Fig. 2c, Supplementary Fig. 2). FmT-tagged VHL/ΔVHL can thus form a ternary complex with SMARCA4 and ACBI1 to effect SMARCA4 biotinylation.
The remainder of the beads was treated with trypsin, and the eluted peptides were identified using mass spectrometry. To first identify VHL-specific interactors, the DMSO-treated (i.e., no PROTAC) samples were compared to data from 293 Flp-In cells expressing the FmT protein alone, using the SAINTexpress algorithm16. High confidence proximity interactors of FmT-tagged VHL/ΔVHL were defined as those with a Bayesian false discovery rate (BFDR) ≤ 0.01. As expected, FmT-VHL/ΔVHL interacted specifically with other components of the CRL2VHL complex (CUL2, ELOB/C), the well-characterized VHL substrates HIF1A and ARNT/HIF1B, and other previously reported VHL interactors17,18 (Supplementary Data 1). Consistent with these observations, immunofluorescence (IF) and Cell Map analyses (based on similarity to BioID data generated from ~200 standard intracellular localization markers19) indicated that the FmT-VHL/ΔVHL proteins harboring NLSs are localized primarily to the nucleus (Supplementary Fig. 3, Supplementary Data 1).
PROTAC-mediated interactions were defined as proteins detected in PROTAC-treated samples that displayed a ≥2-fold (log2) increase in peptide counts over those observed in DMSO-treated samples. As expected, SMARCA2, SMARCA4, and PBRM1 were identified as high-confidence interactors following ACBI1 treatment in all four ProtacID experiments (i.e., full-length or ΔVHL, FmT-tagged at the N- or C-terminus; all hits are presented in Fig. 2d, all peptide count data in Supplementary Data 1). For comparison, standard BioID20 was also conducted using 293 Flp-In cells stably expressing FmT-SMARCA2 or FmT-SMARCA4 (Fig. 2d, Supplementary Data 2). ACBI1 ProtacID conducted with the FmT-ΔVHL tool yielded the highest number of high-confidence proximity interactor identifications, comprising 21 of the 22 BAF subunits identified in the standard SMARCA2/4 BioID analyses (Fig. 2d, Supplementary Data 1 and 2). FmT-ΔVHL was thus used in all subsequent experiments.
ProtacID can distinguish between BAF complex variants
Like ACBI1, ACBI221 and AU-1533022 are VHL-recruiting PROTACs that target the bromodomains of SMARCA2, SMARCA4, and PBRM1. VZ18523 is a VHL-based PROTAC directed against the bromodomains of the BRD7 and BRD9 proteins, subunits specific to the PBAF and ncBAF complexes, respectively15. ProtacID conducted with ACBI2 and AU-15330 displayed extensive (100% and 85%, respectively) overlap with the ACBI1 dataset (Fig. 3, Supplementary Data 3). As expected, ProtacID conducted with VZ185 identified both BRD7 and BRD9, along with additional components of the ncBAF and PBAF complexes (Fig. 3, Supplementary Data 3). Importantly, however, cBAF-exclusive subunits (ARID1A/B and DPF1/2) were not identified in the VZ185 ProtacID analysis (Fig. 3, Supplementary Data 3). Together, these data demonstrate that ProtacID can be used to validate PROTAC neo-substrates and identify proximal interacting protein partners of PROTAC target proteins in living cells, and indicate that ProtacID can distinguish between closely related multiprotein complexes.
Five different BAF PROTACs were subjected to ProtacID analysis across three different human cell lines (293, 697, and HAP1), as indicated. AU-24118 (blue font) is a CRBN-recruiting PROTAC; all others are VHL-recruiting PROTACs. green—high confidence (BFDR ≤ 0.01) protein identification. white—not identified. Proteins grouped according to BAF variant, as in Fig. 2.
ProtacID can be used across cell lines
ProtacID conducted in FmT-ΔVHL-expressing 697 cells (a human pre-B leukemia line) using ACBI1 and ACBI2 yielded a dataset with >95% overlap with that observed in 293 cells (Fig. 3, Supplementary Fig. 1b, Supplementary Data 4). ACBI1 ProtacID was also conducted in FmT-ΔVHL-expressing wild-type (WT) HAP1 cells (a near-haploid human leukemia cell line), and HAP1 SMARCA2 and ARID1A knockout lines. The WT HAP1 BAF ProtacID dataset was identical to that identified in 293 cells, and HAP1 KO cell datasets specifically lacked the proteins encoded by each KO gene (Fig. 3, Supplementary Fig. 1c, Supplementary Data 5). ProtacID can thus yield reproducible and highly accurate PROTAC proximity data across cell lines, and in the same cell line with discrete genetic changes.
CRBNmidi ProtacID
The WT CRBN protein is unstable in the absence of its CRL binding partner DDB124, limiting its use as an analytical tool in living cells. CRBNmidi, a stable CRBN protein variant that displays an extended half-life, was recently developed by the Ciulli lab11. N- and C-terminally tagged FmT-CRBNmidi proteins were stably expressed in 293 Flp-In cells (Supplementary Fig. 1g) and tested in ProtacID with the CRBN-recruiting BAF PROTAC AU-2411825, which targets SMARCA2/4 and PBRM1 via the same warhead present in ACBI1 and AU-15330. The CRBNmidi-mTF (C-term tagged) variant (Fig. 1c, Supplementary Fig. 1b) yielded the most robust dataset, identifying SMARCA2, SMARCA4, PBRM1, and the same set of BAF complex components identified with VHL-directed BAF PROTACs (Fig. 3, Supplementary Data 6). Consistent with these data, IF and Cell Map analyses indicated that the CRBNmidi-mTF protein is localized to the nucleus (Supplementary Fig. 3, Supplementary Data 6). ProtacID can thus be used for the analysis of both VHL- and CRBN-based PROTACs, together comprising the vast majority of current TPD tool compounds.
ProtacID identifies a non-productive PROTAC-protein interaction
Notably, KIF20B (a kinesin motor protein linked to cell division26) was detected as a high-confidence interactor in ACBI1 ProtacID conducted with all four FmT-VHL fusion variants, and in all three cell lines tested here, but was not detected in ACBI2, AU-15330, VZ185, or AU-24118 ProtacIDs. KIF20B was also not detected in standard FmT-SMARCA2 or FmT-SMARCA4 BioID analyses (Fig. 4a, Supplementary Data 1–6). Consistent with these observations, KIF20B was biotinylated in an ACBI1 dose-dependent manner, but did not appear to undergo ternary complex formation in the presence of ACBI2 or AU-15330 (Fig. 4b). Consistent with the original report14, our own 293 Flp-In cell global proteome analysis (Fig. 4c, Supplementary Data 7) and western blotting (Fig. 4b) indicated that KIF20B is not degraded following ACBI1 treatment. Finally, while SMARCA2, SMARCA4, and PBRM1 were not detected, KIF20B was also detected as an ACBI1 interactor in ProtacID conducted in the absence of MLN4924, via both western blotting and mass spectrometry (Fig. 4d, Supplementary Data 8). Together, these data indicate that KIF20B is a non-productive ACBI1 interactor (Fig. 4e). ProtacID can thus also be used to identify non-productive PROTAC-protein interactions.
a Dot Plot (dot attributes as in Fig. 2d) highlighting KIF20B, SMARCA2, and SMARCA4 identification in ProtacID analyses (293 cells) with the indicated PROTAC, or standard BioID analysis. n = 3 biological replicates. b 293 cells expressing FmT-ΔVHL were treated with increasing concentrations (0, 10, 100, 1000 nM) of the indicated PROTACs, biotin was added, and streptavidin pulldowns were conducted (as in Fig. 2c). Isolated proteins (pulldown, PD) or whole cell lysates (input) were subjected to western blotting with the indicated antibodies. n = 3 biological replicates. c Global proteomics analysis of ACBI1-treated 293 Flp-In cells. Cells were treated for 3 h with DMSO or ACBI1 (1 μM, n = 5 biological replicates/condition). Five hundred nanograms of whole cell lysate were analyzed on a Bruker TIMS-TOF mass spectrometer in DIA mode. Differential abundance analysis was performed using linear modeling with empirical Bayes moderation (limma), applying a two-sided moderated t-test with Benjamini-Hochberg multiple testing correction. d (left) Dot Plot highlighting KIF20B, SMARCA2 and SMARCA4 identifications in an ACBI1 ProtacID experiment conducted in the absence of the neddylation inhibitor MLN4924. (Dot attributes as indicated in Fig. 2d.) (right) Western blotting results of the same experiment (ACBI1 gradient 0, 10, 100, 1000 nM). n = 3 biological replicates. e ProtacID workflow for the identification of productive and non-productive PROTAC-protein interactions. ProtacID conducted in the presence of a PROTAC and MLN4924 can identify both productive and non-productive PROTAC interactors (along with their interacting protein partners). ProtacID conducted in the presence of a PROTAC, but in the absence of MLN4924, will identify only non-productive PROTAC interactors. Source data are provided as a Source data file.
ProtacID characterization of BET-targeting PROTACs
Bromodomain and extraterminal domain (BET) family proteins such as BRD2, BRD3, and BRD4 are epigenetic readers that interact with acetylated lysine residues in histones to regulate transcription. Based principally on the JQ1 compound27, a number of different BET PROTACs have been developed, including the VHL-recruiting ARV-77128 and MZ129 tools, and the CRBN-recruiting dBET630 compound.
As expected, ProtacID conducted in 293 cells with ARV-771 and MZ1 PROTACs identified BRD2, BRD3, and BRD4 (Fig. 5, Supplementary Data 9), along with a number of additional chromatin-associated proximity interactors that were also identified in a standard BioID analysis of FmT-BRD2, FmT-BRD3, and FmT-BRD4 (Fig. 5, Supplementary Data 10 and see ref. 31). ProtacID conducted with the same two VHL-recruiting BET PROTACs in 697 cells displayed 100% overlap within this interactor set (Fig. 5, Supplementary Data 11). Consistent with the presence of the same warhead, the CRBN-recruiting PROTAC dBET6 generated an analogous interactome in 293 cells (Fig. 5, Supplementary Data 12).
ProtacID characterization of EED-targeting PROTACs
The EED (embryonic ectoderm development) protein is a component of polycomb repressive complex 2 (PRC2), responsible for epigenetic silencing via methylation of histone H3K2732. The PRC2 complex comprises the core subunits EED, EZH2, and SUZ12, along with additional variable accessory factors33. Two VHL-directed EED PROTACs, UNC770034 and PROTAC235, effect the degradation of EED, EZH2, and SUZ12 in the patient-derived non-Hodgkin lymphoma cell line Karpas-422. As expected, ProtacIDs conducted with EED PROTACs identified EED, EZH2, and SUZ12 as high confidence PROTAC proximity partners in Karpas-422 cells (Fig. 6a, Supplementary Fig. 1d, Supplementary Data 13). Consistent with previous AP-MS analyses36,37, the PRC2-associated AEBP2, MTF2, and PHF19 proteins were also identified in both ProtacID analyses (Fig. 6a).
ProtacID characterization of an epidermal growth factor receptor (EGFR) PROTAC
Finally, it was important to determine whether the ProtacID approach can be applied to PROTACs that target proteins localized to other (non-nuclear) intracellular locations. The VHL-recruiting PROTAC MS3938 (employing the tyrosine kinase inhibitor gefitinib39 as the warhead) can target EGFR mutant proteins that drive non-small cell lung cancer and other diseases40. MS39 was subjected to ProtacID, in this case using an FmT-ΔVHL tool lacking an NLS, and which thus localizes to both the cytoplasmic and nuclear compartments (Supplementary Fig. 3). As expected, MS39 ProtacID identified EGFR proteins expressed in both the HCC827 (EGFRExdel19) and NCI-H3225 (EGFRL858R) cell lines. Two additional previously reported EGFR-interacting partners41,42 were also identified in both analyses (Fig. 6b, Supplementary Fig. 1e, f, Supplementary Data 14 and 15).
Together, these data indicate that ProtacID is a versatile proximity-dependent biotinylation approach that can be used to characterize VHL or CRBN-recruiting PROTACs directed against protein targets in different intracellular locations and across different human cell types.
Discussion
The development of TPD compounds has emerged as an important strategy to target proteins previously considered to be “undruggable” by small molecule inhibitors1. PROTACs thus hold great promise for the treatment of currently intractable diseases. However, as more PROTAC (and molecular glue)-based compounds progress to clinical use, it will become increasingly important to more fully characterize their interactomes in vivo.
The specificity of warhead compounds is generally first characterized in vitro using recombinant target proteins or protein domains, to demonstrate that the compound of interest does not interact with (or displays significantly lower affinity for) related recombinant proteins in the same protein family. If the compound is used to create a PROTAC, cell lysates treated with the resulting tool may then be subjected to a standard global proteome analysis, which identifies proteins that change in abundance in response to the compound. Importantly, however, this approach alone does not provide direct evidence for target engagement in vivo, nor can it identify non-productive PROTAC-protein interactions. Here we describe a simple strategy (Fig. 4e) that can be used as a complementary approach to address this critical need.
PROTAC development can be slow and challenging. Even once highly specific warheads have been identified and optimized, combining them with standard linkers and E3 recruiter moieties does not always result in efficient target protein degradation (much less result in a compound with drug-like properties). More rapid and straightforward methods for in vivo compound testing—such as ProtacID—could help to streamline this process, identifying compounds with promising specificity at earlier stages of development.
Future ProtacID development will focus on developing tools for the analysis of PROTACs and molecular glue compounds that recruit other ubiquitin E3 ligases, such as BIRC2, DCAF16, MDM2, XIAP, and FBXO221,2,43,44,45. And finally, while the quality of a ProtacID analysis will be dependent on the properties (e.g., solubility, cell permeability and specificity) of the associated PROTAC, we also report here that ProtacID can be used to generate highly accurate proximity interactome data similar to standard BioID approaches, identifying native, endogenous protein complexes without the use of antibodies, epitope tags or large biotin ligases fused to the protein of interest. This approach could be used to, e.g., characterize how endogenous protein complexes are “re-wired” by specific coding mutations, or by other drugs or compounds, without any modification to the structure or expression level of the protein of interest.
Methods
Cloning
VHL and ΔVHL cDNAs were amplified by PCR from the VHL-pGex2TK plasmid (Addgene 20790; all cloning primers listed in Supplementary Data 16). The amino acid sequence PKKKRKVEDPKKKKKV was also added to all ProtacID constructs to generate FmT-VHL/ΔVHL/CRBNmidi proteins with or without an NLS. To generate inducible 293 Flp-In cell lines, VHL/ΔVHL sequences were cloned into the pcDNA5 FRT/TO vector containing N-term and C-term 3xFLAG-miniTurbo using in-fusion HD EcoDry Mix (Takaro 639689). For C-term miniTurbo variants, HindIII and NotI restriction sites were used. NsiI and EcoRV restriction sites were used to generate N-term miniTurbo constructs. FLAG-miniTurbo-ΔVHL was also cloned into the pSTV646 lentiviral vector using Xho1 and EcoR1 restriction sites. The CRBNmidi (Addgene 215330) coding sequence was cloned into pcDNA5 FRT/TO as above to generate a CRBNmidi-miniTurbo-FLAG construct. A 12-amino acid linker (GGGGGSGGGGGS) was added between mini-turbo and CRBNmidi by PCR.
Reagents
ACBI2 and VZ185 were obtained through the Boehringer Ingelheim opnMe program. UNC7700 was a kind gift from the L James laboratory (University of North Carolina, Chapel Hill, NC, USA). ACBI1 (catalog no. HY-128359, lot 66200), AU-15330 (HY-145388, lot 145137), AU-24118 (HY-163410, lot 535238), dBET6 (HY-112588, lot 113183), ARV-771 (HY-100972, lot 380611), and PROTAC2, EED degrader (HY-130615, lot 113670) were purchased from MedChemExpress. MZ1 (catalog no. 21622, batch 0664999-3) and MLN4924 (15217, batch 0712600-6) were purchased from Cayman Chemical Company. MS39 (7397, batch 1A/293284) was purchased from Tocris Bioscience. All PROTACs were solubilized in DMSO and used as indicated.
Cell culture
HEK 293 Flp-In T-REx cells (Thermo Fisher R78007, female) were cultured in DMEM (Wisent) supplemented with 10% FBS (Wisent). 697 cells (DSMZ ACC42, male) were cultured in RPMI medium (Wisent) supplemented with 10% heat-inactivated FBS (Gibco). The HAP1 (male) parental cell line was a kind gift from the A Schimmer lab (Princess Margaret Cancer Centre). HAP1 KO lines (SMARCA2KO and ARID1AKO) were kind gifts from the S Kubicek lab (CeMM, The Research Center for Molecular Medicine, Vienna, Austria). HAP1 cells were cultured in IMDM (Iscove’s Modified Dulbecco Medium) with 10% FBS. The Karpas-422 (female) cell line was a kind gift from the R Kridel lab (Princess Margaret Cancer Centre), and cultured in RPMI medium supplemented with 10% FBS. HCC827 (female) and NCI-H3255 (female) cell lines were kind gifts from the M Tsao lab (Princess Margaret Cancer Centre) and cultured in RPMI medium supplemented with 10% FBS. All media was supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin (Wisent). Cells were cultured at 37 °C in a humidified chamber with 5% CO2, following standard protocols, and tested for mycoplasma twice/year (MycoAlert Mycoplasma Detection kit, Lonza).
Lentivirus production
One day before transfection, 0.5 million 293T cells were seeded in a 6-well plate. The next day, cells were transfected with the FLAG-miniTurbo-ΔVHL pSTV646 plasmid, along with the lentiviral transfer plasmid and the packaging plasmids pMDG VSVG and PAX2 (X-tremeGENE Millipore Sigma 6366244001) according to the manufacturer’s instructions. Viral supernatants were collected 48 h after transfection, combined, and filtered through a 0.45 μm Millex-HA syringe filter (Millipore Sigma SLHA033SS). Filtered supernatants were added to target cells in the presence of 8 μg/ml polybrene (Sigma-Aldrich TR-1003-G). After 1 day of incubation, viral supernatants were removed and replaced with normal media. Puromycin (2 μg/ml) was added to the cells 48 h post-infection to initiate selection for stably transduced cells.
Generation of stable lines
HEK 293 Flp-In T-REx cells were co-transfected with pOG44 (Flp-recombinase expression vector) and pcDNA5 FRT/TO plasmid containing the coding sequences for FLAG-miniTurbo (FmT) fused in-frame with VHL, ΔVHL, or CRBNmidi. Control cells were also generated, expressing FmT alone. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Following transfection, cells were selected with 200 μg/ml hygromycin. All other cell lines were transduced with the pSTV6 lentiviral vector encoding FLAG-miniTurbo-ΔVHL, CRBNmidi-FmT, or FmT alone. 50–60% confluent cells in 6-well plates were infected with 350 μL of unconcentrated virus and polybrene to a final concentration of 8 μg/ml. Following transduction, cells were selected with 2 μg/ml puromycin.
ProtacID
Cell lines stably expressing FmT-tagged VHL, ΔVHL, or CRBNmidi proteins were divided into two treatment conditions, each comprising 5 × 15 cm2 plates at ~80% confluency. Expression of the fusion protein was induced by treating the cells overnight with 2 μg/ml doxycycline. The following day, the cells were treated with 0.5 μM MLN4924 (Cayman Chemical 15217). After 1 h, cells were treated with either PROTAC (based on DC50 values; ACBI1 0.2 μM, ACBI2 0.1 μM, AU-15330 0.4 μM, VZ185 0.1 μM, AU-24118 1 μM, MZ1 0.5 μM, ARV-771 0.2 μM, dBET6 1 μM, PROTAC2 1 μM, UNC7700 1 μM, MS39 1 μM) or DMSO for 3 h, followed by the addition of biotin to a final concentration of 50 μM for 60 min. At the endpoint, culture medium was removed, cells were washed with chilled PBS, and harvested by scraping. Cells were pelleted by centrifugation at 300 × g for 5 min. The pellet was washed once with 10 ml PBS and centrifuged again at 300 × g for 5 min. PBS was removed, the cell pellet was flash-frozen in liquid nitrogen, and stored at −80 °C.
BioID
BioID was conducted as previously described47. In brief, the cell pellet was resuspended in 10 ml lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA), 1% Triton X-100, 0.1% SDS, 0.5% sodium deoxycholate, 1:500 protease inhibitor cocktail (Sigma-Aldrich), 1:1000 benzonase nuclease (Novagen 71205-M) and incubated on an end-over-end rotator at 4 °C for 1 h. The lysate was briefly sonicated to disrupt any visible aggregates, then centrifuged at 45,000 × g for 30 min at 4 °C. Supernatant was transferred to a fresh 15 ml conical tube. Thirty microliters of packed, pre-equilibrated streptavidin-sepharose beads (Cytiva 17-5113-01) were added, and the mixture was incubated for 3 h at 4 °C with end-over-end rotation. Beads were pelleted by centrifugation at 376 × g for 2 min and transferred with 1 ml of lysis buffer to a fresh Eppendorf tube.
An aliquot (~10%) of the beads was saved for immunoblotting. The remainder of the beads was washed four times with 50 mM ammonium bicarbonate (ammbic, pH 8.3) and transferred to a fresh centrifuge tube for two additional washes with 1 ml of ammbic. Tryptic digestion was performed by incubating the beads with 1 μg MS-grade TPCK trypsin (Promega V5280) in 200 μl 50 mM ammbic (pH 8.3) overnight at 37 °C. The following morning, an additional 0.5 μg trypsin was added, and the beads were incubated for another 2 h at 37 °C. Beads were then pelleted by centrifugation at 2000 × g for 2 min, and the supernatant was transferred to a fresh Eppendorf tube. Beads were washed twice with 200 μL 50 mM ammbic, and the washes were pooled with the eluate. The sample was lyophilized and resuspended in buffer A (0.1% formic acid). One-fifth of each sample was analyzed per MS run.
Two biological replicates (i.e., two separate sets of cultured cells) were used for each experiment. Each biological replicate was analyzed by mass spectrometry twice, to generate two technical replicates each, for a total of four MS analyses/BioID dataset. The datasets reported here comprise the combined (total) peptide (spectral) counts for these four analyses. Peptide counts for all four individual runs, and peptide totals (SpecSum) are reported in Supplemental Data 1–16.
Mass spectrometry (ProtacID)
High-performance liquid chromatography (HPLC) was conducted using a 2 cm pre-column (Acclaim PepMap 50 mm × 100 μm inner diameter) and 50 cm analytical column (Acclaim PepMap, 500 mm × 75 μm diameter; C18; 2 μm; 100 Å, Thermo Fisher Scientific, Waltham, MA) running a 120 min reversed-phase buffer gradient at 225 nl/min on a Proxeon EASY-nLC 1000 pump in-line with a Thermo Q-Exactive HF quadrupole-Orbitrap mass spectrometer. A parent ion scan was performed using a resolving power of 60,000, then up to the twenty most intense peaks were selected for MS/MS (minimum ion count of 1000 for activation) using higher energy collision-induced dissociation (HCD) fragmentation. Dynamic exclusion was activated such that MS/MS of the same m/z (within a range of 10 ppm; exclusion list size = 500) detected twice within 5 s were excluded from analysis for 15 s.
For protein identification, Thermo .RAW files were converted to the .mzXML format using Proteowizard48, then searched using X!Tandem49 and COMET50 against Human RefSeq Version 45 database (containing 36,113 entries). Data were analyzed using the trans-proteomic pipeline51 via the ProHits software suite (v3.3)52. Search parameters specified a parent ion mass tolerance of 10 ppm, and an MS/MS fragment ion tolerance of 0.4 Da, with up to 2 missed cleavages allowed for trypsin. Variable modifications of +16@M and W, +32@M and W, +42@N-terminus, and +1@N and Q were allowed. Proteins identified with an iProphet cut-off of 0.9 (corresponding to ≤1% FDR) and at least two unique peptides were analyzed with SAINT Express v.3.6.1316,53. Twenty control runs (from 10 or more biological replicates of cells stably expressing the FmT epitope tag only) were collapsed to the four highest spectral counts for each prey and compared to two biological replicates (each with two technical replicates) for each experiment. High confidence interactors were defined as those with a BFDR ≤ 0.01. High confidence PROTAC-mediated interactions were defined as those with >5 average peptide counts/MS run and >2-fold (log2) spectral counts in the presence versus absence of PROTAC.
Global proteome analysis
Cell pellets (derived from a single well of a 6-well plate, n = 5 individual wells analyzed per condition) were resuspended in 100 μL lysis buffer (5% SDS, 50 mM triethylammonium bicarbonate, pH 8.5) and sonicated. Following centrifugation, cleared lysates were reduced (5 mM DTT 60 °C 30 min), alkylated (20 mM iodoacetamide RT 15 min), then acidified to a final concentration of 1.2% H3PO4. Lysates were diluted (1/7) with 100 mM TEAB/90% methanol, and 192.5 μL were loaded onto S-trap™ micro columns (ProtiFi, Fairport, NY) followed by washing according to the manufacturer’s instructions. On-column digest was conducted with 2 μg MS-grade TPCK trypsin (Promega V5280) overnight at 37 °C. Peptides were eluted from the S-Trap (50 mM TEAB, 0.2% HCOOH, and CH3CN, successively) and desalted, and 500 ng of sample was loaded onto an Evotip Pure™ column (Evosep EV2011). Liquid chromatography was performed with the Evosep One (Evosep) pump with an SPD30 method using a PepSep™ C18 HPLC column (15 cm × 150 μm ID, 1.5 μm; Bruker, Bremen). The TIMS-TOF HT (Bruker, Bremen) mass spectrometer was operated in PASEF-DIA54,55 positive ion mode (MS scan range 100–1700 m/z). Ion mobility range was 1/K0 = 1.6–0.6 Vs cm−2 using equal ion accumulation and ramp time in the dual TIMS analyzer of 100 ms each. DIA (scan range 400–1201 m/z) mass window width was 26 Da with 1 Da overlap and 32 steps per cycle. Collision energy was lowered stepwise as a function of increasing ion mobility, starting from 20 eV for 1/K0 = 0.6 Vs cm−2 and 59 eV for 1/K0 = 1.6 Vs cm−2. The ion mobility dimension was calibrated linearly using three ions from the Agilent ESI LC/MS tuning mix (m/z, 1/K0: 622.0289, 0.9848 Vs cm−2; 922.0097, 1.1895 Vs cm−2; and 1221.9906, 1.3820 Vs cm−2). Data files were analyzed on the Fragpipe platform(v20.0) using DIA-NN56,57 (v1.8.2_beta_8) with a Bruker spectral library generated from K562 and Molt4 trypsin-digested cell lysates fractionated using high pH reversed-phase chromatography and analyzed on a TIMS-TOF HT.
Global proteome data analysis
Protein groups were loaded into R (4.3.1, R Core Team (2020), https://www.R-project.org/). Proteins labeled as known contaminants in the spectral library and proteins not quantified in all replicates of at least one condition were removed from analysis. Imputation of missing data (random draws from a manually defined left-shifted Gaussian distribution (shift = 1.8, scale = 0.3)) and testing for protein differential abundance were done using the R package differential enrichment analysis of proteomics data (DEP) (1.24.0). Differential abundance analysis was performed using linear modeling with empirical Bayes moderation (limma), applying a two-sided moderated t-test with Benjamini-Hochberg multiple testing correction.
Western blotting
ProtacID blots. Aliquots (~10%) of protein-bound beads from ProtacID samples were washed 4× with cell lysis buffer, heated in 50 μL of 4×NuPAGE buffer at 95 °C for 5 min, then centrifuged in a tabletop centrifuge at max speed for 10 min. The supernatant was transferred to a fresh Eppendorf tube for immunoblotting analysis.
For blotting of whole cell lysates, cells were collected as above, and pellets were lysed for 3 min RT in 20 mM Tris-HCl (pH 8), 150 mM NaCl, 10 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, protease inhibitor (Roche 11873580001), and benzonase (Millipore Sigma E1014). SDS was added to a final concentration of 1%. Lysates were then spun in a tabletop centrifuge at max speed for 10 min, and supernatants transferred to fresh tubes. Protein concentrations were measured (Thermo Scientific Pierce™ BCA Protein Assay Kit 23225) according to the manufacturer’s protocol. 10-30 μg total protein was loaded into Invitrogen NuPAGE Bis-Tris Gels and run in 1X NuPAGE MOPS buffer (Invitrogen NP0001) for 90 min at 120 V. Proteins were transferred to 0.2 μm polyvinylidene difluoride (PVDF, Cytiva) membrane using a Bio-Rad Mini Trans-Blot Cell at 70 V/500 mA for 1.5 h in Tris-Glycine transfer buffer (3 g/L Tris, 14.4 g/L glycine, and 10% methanol) on ice. Membranes were blocked in 5% BSA in PBS-T for 1 h at room temperature, followed by overnight incubation with primary antibodies: FLAG (1:5000 Sigma-Aldrich F1804, Mouse), SMARCA4 (BRG1 1:1000; Cell Signaling Technology 49360T, Rabbit), MPHOSPH1 (KIF20B 1:1000; Invitrogen PA5-120351, Rabbit), CRBN (1:1000, Cell Signaling Technology D8H3S, Rabbit Monoclonal), VHL (1:1000 Santa Cruz Biotechnology sc-135657, mouse), Vinculin (1;10,000 Sigma-Aldrich V9131, mouse) diluted in 5% BSA. After primary antibody incubation, membranes were washed 3 × 10 min with PBS-T, then incubated for 1 h at RT with LI-COR secondary antibodies IRDye 800CW Goat anti-Rabbit (LI-COR 926-32211) or 680RD Goat anti-Mouse (926-68070), all diluted 1:5000 in 5% BSA. Membranes were washed 3 × 10 min with PBS-T, then imaged using a LI-COR Odyssey CLx scanner.
Immunofluorescence (IF)
Cells were washed with PBS and fixed in 4% PFA for 10 min at RT, followed by permeabilization in 0.1% Triton X-100 for 10 min at RT, and blocked with 3% BSA diluted in PBS for 1 h at RT. Fixed cells were incubated with primary antibody (anti-Flag M2 antibody, Sigma F1804, 1:1000 dilution) at 4 °C overnight. After three washes in PBS, cells were incubated with secondary antibody (anti-mouse IgG (Alexa Fluor® 647, Cell Signaling #4410, 1:1000 dilution)) and Streptavidin Alexa Fluor™ 488 Conjugate (Invitrogen S11223) with 1:1000 dilution for 1 h at RT. After three PBS washes in the dark, cells were stained with DAPI and mounted with VECTASHIELD® Vibrance™ Antifade Mounting Medium (Vector Laboratories, VECTH17002). For NCI-H3255, HCC827, HAP1, Karpas-422, and 697 cells, images were acquired on a Leica SP8 confocal microscope with a 63× objective oil lens. For HEK293 cells, images were acquired on a Leica MICA Microhub using the confocal module with a 63X objective water lens.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. The raw mass spectrometry data generated in this study have been deposited in the MassIVE (massive.ucsd.edu) database under accession code MSV000095096 [https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=27365aa7aa87450e93b171f6c7e5de9d]. This dataset is also cross-referenced on ProteomeXchange (proteomexchange.org) as accession code PXD053263. Source data are provided with this paper.
Materials availability
ProtacID plasmids will be made available at Addgene.
References
Bekes, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Schapira, M., Calabrese, M. F., Bullock, A. N. & Crews, C. M. Targeted protein degradation: expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 (2019).
Ge, J. et al. PROTAC-DB 3.0: an updated database of PROTACs with extended pharmacokinetic parameters. Nucleic Acids Res. 53, D1510–D1515 (2025).
Cowan, A. D. & Ciulli, A. Driving E3 ligase substrate specificity for targeted protein degradation: lessons from nature and the laboratory. Annu. Rev. Biochem. 91, 295–319 (2022).
Mermet-Meillon, F. et al. Protein destabilization underlies pathogenic missense mutations in ARID1B. Nat. Struct. Mol. Biol. 31, 1018–1022 (2024).
Redler, R. L., Das, J., Diaz, J. R. & Dokholyan, N. V. Protein destabilization as a common factor in diverse inherited disorders. J. Mol. Evol. 82, 11–16 (2016).
Swamy, K. B. S., Schuyler, S. C. & Leu, J. Y. Protein complexes form a basis for complex hybrid incompatibility. Front. Genet. 12, 609766 (2021).
Gingras, A. C., Abe, K. T. & Raught, B. Getting to know the neighborhood: using proximity-dependent biotinylation to characterize protein complexes and map organelles. Curr. Opin. Chem. Biol. 48, 44–54 (2019).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Yamanaka, S. et al. A proximity biotinylation-based approach to identify protein-E3 ligase interactions induced by PROTACs and molecular glues. Nat. Commun. 13, 183 (2022).
Kroupova, A. et al. Design of a Cereblon construct for crystallographic and biophysical studies of protein degraders. Nat. Commun. 15, 8885 (2024).
Branon, T. C. et al. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 36, 880–887 (2018).
Gadd, M. S. et al. Structural basis of PROTAC cooperative recognition for selective protein degradation. Nat. Chem. Biol. 13, 514–521 (2017).
Farnaby, W. et al. BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design. Nat. Chem. Biol. 15, 672–680 (2019).
Varga, J., Kube, M., Luck, K. & Schick, S. The BAF chromatin remodeling complexes: structure, function, and synthetic lethalities. Biochem. Soc. Trans. 49, 1489–1503 (2021).
Teo, G. et al. SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteom. 100, 37–43 (2014).
Tarade, D. & Ohh, M. The HIF and other quandaries in VHL disease. Oncogene 37, 139–147 (2018).
Tabaro, F. et al. VHLdb: a database of von Hippel-Lindau protein interactors and mutations. Sci. Rep. 6, 31128 (2016).
Go, C. D. et al. A proximity-dependent biotinylation map of a human cell. Nature 595, 120–124 (2021).
Gupta, G. D. et al. A dynamic protein interaction landscape of the human centrosome-cilium interface. Cell 163, 1484–1499 (2015).
Kofink, C. et al. A selective and orally bioavailable VHL-recruiting PROTAC achieves SMARCA2 degradation in vivo. Nat. Commun. 13, 5969 (2022).
Xiao, L. et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 601, 434–439 (2022).
Zoppi, V. et al. Iterative design and optimization of initially inactive proteolysis targeting chimeras (PROTACs) identify VZ185 as a potent, fast, and selective von Hippel-Lindau (VHL) based dual degrader probe of BRD9 and BRD7. J. Med. Chem. 62, 699–726 (2019).
Fischer, E. S. et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature 512, 49–53 (2014).
He, T. et al. Development of an orally bioavailable mSWI/SNF ATPase degrader and acquired mechanisms of resistance in prostate cancer. Proc. Natl. Acad. Sci. USA 121, e2322563121 (2024).
Janisch, K. M., McNeely, K. C., Dardick, J. M., Lim, S. H. & Dwyer, N. D. Kinesin-6 KIF20B is required for efficient cytokinetic furrowing and timely abscission in human cells. Mol. Biol. Cell 29, 166–179 (2018).
Filippakopoulos, P. et al. Selective inhibition of BET bromodomains. Nature 468, 1067–1073 (2010).
Raina, K. et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 113, 7124–7129 (2016).
Zengerle, M., Chan, K. H. & Ciulli, A. Selective small molecule induced degradation of the BET bromodomain protein BRD4. ACS Chem. Biol. 10, 1770–1777 (2015).
Winter, G. E. et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol. Cell 67, 5–18.e19 (2017).
Lambert, J. P. et al. Interactome rewiring following pharmacological targeting of BET bromodomains. Mol. Cell 73, 621–638.e617 (2019).
Margueron, R. & Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 469, 343–349 (2011).
Bharti, H., Han, S., Chang, H. W. & Reinberg, D. Polycomb repressive complex 2 accessory factors: rheostats for cell fate decision? Curr. Opin. Genet. Dev. 84, 102137 (2024).
Bashore, F. M. et al. PROTAC linkerology leads to an optimized bivalent chemical degrader of Polycomb Repressive Complex 2 (PRC2) components. ACS Chem. Biol. 18, 494–507 (2023).
Hsu, J. H. et al. EED-Targeted PROTACs degrade EED, EZH2, and SUZ12 in the PRC2 complex. Cell Chem. Biol. 27, 41–46.e17 (2020).
Hauri, S. et al. A High-density map for navigating the human Polycomb complexome. Cell Rep. 17, 583–595 (2016).
Huttlin, E. L. et al. Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509 (2017).
Cheng, M. et al. Discovery of potent and selective epidermal growth factor receptor (EGFR) bifunctional small-molecule degraders. J. Med. Chem. 63, 1216–1232 (2020).
Herbst, R. S., Fukuoka, M. & Baselga, J. Gefitinib—a novel targeted approach to treating cancer. Nat. Rev. Cancer 4, 956–965 (2004).
Sharma, S. V., Bell, D. W., Settleman, J. & Haber, D. A. Epidermal growth factor receptor mutations in lung cancer. Nat. Rev. Cancer 7, 169–181 (2007).
Li, J. et al. Perturbation of the mutated EGFR interactome identifies vulnerabilities and resistance mechanisms. Mol. Syst. Biol. 9, 705 (2013).
Tong, J., Taylor, P. & Moran, M. F. Proteomic analysis of the epidermal growth factor receptor (EGFR) interactome and post-translational modifications associated with receptor endocytosis in response to EGF and stress. Mol. Cell. Proteom. 13, 1644–1658 (2014).
Liu, Z. et al. An overview of PROTACs: a promising drug discovery paradigm. Mol. Biomed. 3, 46 (2022).
Nie, D. Y. et al. Recruitment of FBXO22 for targeted degradation of NSD2. Nat. Chem. Biol. 20, 1597–1607 (2024).
Basu, A. A. et al. A CRISPR activation screen identifies FBXO22 supporting targeted protein degradation. Nat. Chem. Biol. 20, 1608–1616 (2024).
Samavarchi-Tehrani, P., Abdouni, H., Samson, R. & Gingras, A. C. A versatile lentiviral delivery toolkit for proximity-dependent biotinylation in diverse cell types. Mol. Cell. Proteom. 17, 2256–2269 (2018).
Coyaud, E. et al. BioID-based identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteom. 14, 1781–1795 (2015).
Kessner, D., Chambers, M., Burke, R., Agus, D. & Mallick, P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534–2536 (2008).
Craig, R. & Beavis, R. C. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 (2004).
Eng, J. K., Jahan, T. A. & Hoopmann, M. R. Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 (2013).
Deutsch, E. W. et al. Trans-proteomic pipeline: robust mass spectrometry-based proteomics data analysis suite. J. Proteome Res. 22, 615–624 (2023).
Liu, G. et al. ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat. Biotechnol. 28, 1015–1017 (2010).
Mellacheruvu, D. et al. The CRAPome: a contaminant repository for affinity purification-mass spectrometry data. Nat. Methods 10, 730–736 (2013).
Meier, F. et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Mol. Cell. Proteom. 17, 2534 (2018).
Meier, F. et al. Parallel Accumulation-Serial Fragmentation (PASEF): Multiplying Sequencing Speed and Sensitivity by Synchronized Scans in a Trapped Ion Mobility Device. J. Proteome Res. 14, 5378 (2015).
Demichev, V. et al. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat. Methods. 17, 41 (2020).
Demichev, V. et al. dia-PASEF data analysis using FragPipe and DIA-NN for deep proteomics of low sample amounts. Nat. Commun. 13, 3944 (2022).
Acknowledgements
We thank the Gingras lab for the pDEST-pcDNA5 vector. S.S. was supported by a Princess Margaret Cancer Centre post-doctoral fellowship. M.E.R.M. is supported by a Canadian Institutes of Health Research (CIHR) post-doctoral fellowship. L.J. is supported by the MITACS Accelerate Award IT21126. D.Y.N. was supported by a Canada Graduate Scholarship—Doctoral Research Award from CIHR (494204) and a Doctoral Training Scholarship from Fonds de recherche du Québec—Santé (320128). J.S.-G. is supported by generous funding from The Princess Margaret Cancer Centre Foundation. C.H.A. was supported by funding from CIHR grants FDN154328 and OGB190363. The Structural Genomics Consortium is a registered charity (no: 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, Bristol Myers Squibb, Genentech, Genome Canada through The Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and US), Pfizer and Takeda. B.R. holds the Philip S. Orsino Chair in Leukemia Research, administered through the University of Toronto and The Princess Margaret Cancer Centre. Work in the B.R. lab was supported by the CIHR project grant program and additional generous funding from the Princess Margaret Cancer Centre Foundation.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.S., M.E.R.M., D.Y.N., D.B.-L., C.H.A., B.R. Investigation: S.S. performed all VHL ProtacID experiments. M.E.R.M. performed CRBNmidi ProtacID experiments, CRBN Western blotting, and data analysis for global proteomics. M.E.R.M. and M.K. performed IF analysis on 293 cells. L.J. performed all standard BioID experiments, VHL protein expression level Western blotting, and IF on all non-293 cells. S.D. conducted extensive cloning and cell culture work. D.Y.N. generated multiple cell lines and conducted western blotting. J.S.-G. performed all mass spectrometry and BioID/ProtacID MS data analysis. S.S., C.H.A., and B.R. wrote the paper. All authors contributed to the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tatsuya Sawasaki and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shrestha, S., Maitland, M.E.R., Jing, L. et al. Characterization of PROTAC specificity and endogenous protein interactomes using ProtacID. Nat Commun 16, 8089 (2025). https://doi.org/10.1038/s41467-025-63357-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-63357-7