Abstract
Alveolar Type 1 and Type 2 cells are vital for lung gas exchange and become compromised in several diseases. While key differentiation signals are known, their emergence and fate plasticity are unclear. Here we show in the embryonic lung that single AT2s emerge at intermediate zones, extrude, and connect with nearby epithelium via interlumenal junctioning. We observe AT2s retain fate plasticity until the bZIP transcription factor C/EBPα suppresses Notch signaling at a novel Dlk1 enhancer. Both Dlk1 and Cebpa are regulated by the polycomb repressive complex (PRC2), which together form a “pulse generator” circuit that times Dlk1 expression and thus Notch activation, resulting in a “salt and pepper” pattern of AT1 and AT2 fate. In injured adult lungs, C/EBPα downregulation is required to re-access AT2 fate plasticity and is mediated by the dominant negative C/EBP family member CHOP. Finally, Cebpa loss also activates a “defender” AT2 state, distinct from its reparative state, and we propose AT2s toggle between either state following infection to protect and repair alveoli.
Similar content being viewed by others
Introduction
Alveoli are the sites of gas exchange in the lung that are lined by two distinct epithelial cell types. The flat surface of alveolar type 1 (AT1) cells facilitates gas exchange, while cuboidal alveolar type 2 (AT2) cells secrete surfactant to prevent alveolar collapse. Defective alveoli underlie several maladies—bronchopulmonary dysplasia (BPD) in infants, chronic obstructive pulmonary disease (COPD)/emphysema1, pulmonary fibrosis2, lung adenocarcinoma3,4, and respiratory infections like Severe Acute Respiratory Syndrome (SARS)5,6 and COVID-197. Deciphering how these crucial alveolar cell types emerge as well as the mechanisms that regulate their fate is imperative for developing therapies that will not only slow or stop disease progression but may even promote lung repair.
As alveologenesis begins, AT1 and AT2 cells arise from a common pool of distal progenitors (DP)4,8,9,10,11, with differentiation detected as early as embryonic day 16.5 (E16.5) in mice, save for one study reporting an earlier timepoint12. Upon differentiation, AT1 and AT2 cells are arranged in a salt and pepper pattern13 along a lumenal surface, which, as alveoli mature, interconnect with lumens of anatomically distinct branches to facilitate gas conduction between alveoli14. After development, mature AT2 cells can act as antigen presenting cells in an immune response15,16 (partly directed interferon signaling17,18), as well as facultative stem cells following injury4,11,19,20,21 (directed by Wnt22 and Notch23 signaling). While both AT2 states are critical after infection for alveolar recovery, a recent study found interferon stimulation suppresses repair of the lung epithelium following infection24,25, suggesting that signals driving these states may oppose one another at the cellular level.
Here we show nascent AT2 cells are first detected molecularly at E15.5 as singletons at stereotyped intermediate regions of distal branches. Following basal extrusion, we observe nascent AT2s can form de novo attachments to nearby but anatomically separate epithelia, a step that is likely critical in stitching together alveolar lumen and that we term “interlumenal junctioning”, followed by sporadically patterned AT2 maturation, distinct from the “proximal-to-distal wave” pattern reported to initiate differentiation of AT1s and AT2s earlier13.
We determine that nascent AT2s retain fate plasticity into the first perinatal week, revising the lineage model to include a “window of fate plasticity” spanning from distal progenitors to the AT2 lineage. From screening transcriptional regulators around this window, we identify a critical negative regulator of AT2 fate plasticity, the bZIP transcription factor C/EBPα, that when lost results in AT2-to-AT1 conversion. Previously, C/EBPα has been reported to play a role in epithelial homeostasis26 and is implicated in fate regulation in multiple tissues, including the blood and liver27. Further, we found that the observed AT2-to-AT1 differentiation was mediated in a non-cell autonomous manner by the Notch regulator DLK1, a cell-surface transmembrane protein28 that has been associated with brain and muscle development, stem cell maintenance, lung cancer, and abnormal tissue repair23,29. Observing a pulsed expression of Dlk1 during alveologenesis, we find Dlk1 and Cebpa are both regulated by the polycomb repressive complex (PRC2), and determine these components together constitute an incoherent feed-forward loop. Upon abrogation of this “pulsed generator” circuit, we observe a disruption of AT1/AT2 differentiation. PRC2 is a well-established transcriptional repressor that has been shown to regulate DLK1 expression in neuronal fate selection30,31,32 and recent evidence suggests it also modulates C/EBPα expression in the context of adipocyte differentiation33.
Following injury in the adult lung C/EBPα must be downregulated in AT2s for conversion to AT1. We identify that the dominant negative member of the C/EBP family, CHOP, is induced in AT2s following injury and its upregulation promotes AT2-to-AT1 differentiation. Finally, we observe Cebpa loss also activates a “defensive” AT2 state that is enriched in antipathogen response genes, likely regulated by interferon signaling, and is mutually exclusive from the AT1-associated DATP state34.
Results
Timing and pattern of AT2 cell emergence
To delineate newly forming AT2 cells from distal progenitors and their mature counterparts, we analyzed scRNAseq data of the distal epithelial lineage from a prior study sampling the embryonic, perinatal, and adult timepoints of mouse lung development35 (Fig. 1a). Clustering analysis of the distal epithelium identified both DP and AT1 populations, as well as stratified AT2s into two groups (Fig. 1b) that are largely separate in time (Fig. 1c). While these AT2 groups positively correlate in gene expression (Pearson coefficient = 0.6, Supplementary Fig. 1a), their distinction as a separate cluster was confirmed by coassignment probability (Supplementary Fig. 1b). Taken together with the temporal pattern, we refer to the earlier cluster as the nascent AT2 (nAT2) state, and the later as the mature (mAT2) AT2 state. From this distinction, we were able to identify hundreds of genes unique to either DP, nAT2, or mAT2 cluster (Fig. 1d, Supplementary Fig. 1c and Supplementary Tables 1, 2), and initially focused on two for protein-level validation: Retnla expressed by nAT2s and Cd74 by mAT2s (Fig. 1e and Supplementary Fig. 1d). Retnla encodes a regulator of inflammation, resistin-like molecule (RELM) α, while CD74 is critical in MHC-II antigen processing. In contrast to commonly used markers that label both DP and AT2 lineage starting around E17.5 (such as SFTPC)4,10, RELMα uniquely labels nAT2s at this stage and can be used to distinguish them from both DPs and AT1s, as can be seen at an E17.5 transition zone where the mAT2 marker CD74 is not highly expressed (Fig. 1f and Supplementary Fig. 1e). Further, flow cytometry confirmed at E18.5 that a substantial proportion of marker-expressing AT2s are RELMα⁺ CD74− (Supplementary Fig. 1g), which shifts to RELMα− CD74+ in adult AT2s (Supplementary Fig. 1h).
a scRNAseq UMAP generated from a published timecourse study (GSE119228)35. b Leiden clustering identifies mature AT2s (mAT2), nascent AT2s (nAT2), distal progenitors (DP), and AT1s. c Cluster proportion of AT2 lineage across development shows their separation over time. d Venn diagram indicates the number of genes shared or restrictively expressed. e Dot plot of stage-restricted marker gene Retnla (nAT2) and Cd74 (mAT2). f E17.5 immunostain of RELMα (green), CD74 (red), and DP/AT1-restrictive luminal marker PDPN (white). Bar, 25 µm. g E15.5 (left) and E16.5 (right) immunostain for smooth muscle actin (SMA, red), RELMα (green), E-cadherin (ECAD, blue), and PDPN (white). Close-up images for E15.5 lungs of rare single nAT2s (asterisk) emerging at intermediate regions, not terminal end buds (arrowheads). Bars, 100 µm (left and right panels), 20 µm (close-ups). Mesothelium is marked with white dash. h Quantification of RELMα+ nAT2 immunostains from E15.5 to E17.5 (n represents branches scored per timepoint; data are represented as median with whiskers extending to the upper and lower quartiles. p values determined using Kruskal–Wallis test). i Quantification of RELMα+ nAT2 morphology (columnar versus basally extruded). Extruded values presented as mean ± SD (n represents cells scored at each timepoint). j Schematic of the timing and sequence of nAT2 emergence. At E15.5 the first nAT2 emergent zone (bright green) occurs between the terminal end bud and stalks of the undifferentiated distal epithelium (blue), which ~E16.5 expands during branching morphogenesis. Around E17.5, nAT2s next emerge more proximally (dark green) as airway smooth muscle remodels into myofibroblasts—leaving the final emergence of nAT2s to occur at the distal tips after birth. Source data are provided as a Source Data file.
Given the distinct labeling of nAT2s by RELMα in the embryonic lung, we used it to identify when and where AT2 cells first emerge during alveologenesis. Careful immunostaining revealed nAT2s emerge as early as E15.5 (Fig. 1g), which steadily increases over time (Fig. 1h). At first emergence, SOX9low/RELMα+ and nAT2s are singletons (Supplementary Fig. 1e, f) with retained columnar morphology (Fig. 1i) positioned at anatomical intermediate zones, located between airway smooth muscle (ASM)-covered distal branch stalks and terminal end buds (Fig. 1g, asterisks). After their initial emergence, nAT2s next arise (~E17.5) within distal stalks following ASM to myofibroblast remodeling as previously reported36, then finally arise at terminal end buds soon after birth4. These observations support a sequential, three-zone model of AT2 differentiation (Fig. 1j, left and Supplementary Fig. 1d, e) that integrates previously conflicting findings that reported AT2 differentiation first occurring either at terminal end buds12 or solely in a proximal-to-distal pattern4,9.
Nascent AT2s junction with nearby but anatomically distinct epithelium
While imaging RELMα at these timepoints, we also observed nAT2s forming connections to lumens across anatomically separate branches (Fig. 2a), a behavior which increased in frequency over time (Fig. 2b). While it is known that during alveolar maturation AT2 cells are capable of simultaneously integrating into lumens of multiple alveoli14,36, the process by which it occurs has yet to be described. Careful immunostaining of junctioning nAT2s revealed their negativity for AT1 markers (PDPN, RAGE, and HOPX) and positivity for AT2 markers (SFTPC, LAMP1, and MUC1) that is often higher than non-junctioned AT2s, suggesting they may be further along in differentiation (Supplementary Fig. 2a, b). Using high-resolution imaging and 3D reconstruction, we observed that junctions occurred either directly with the second lumen, or indirectly by attachment to another extruded nAT2 (Fig. 2c, Supplementary Movies 1–3 and Supplementary Fig. 2c).
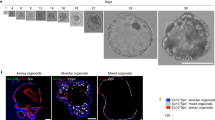
a E17.5 immunostain of RELMα (green), PDPN (white), and DAPI (blue) depicting where nAT2s are observed traversing the interstitium to form interlumenal junctions (ILJs). Magnified confocal sections (yellow box, i) of a representative nAT2 cell embedded in lumen A, at z-position +18 µm (top right), as well as in lumen B, at z-position +24 µm (bottom right). Bar, 50 µm (left), 10 µm (close-ups; right). b Quantification of ILJs during development (n represents nAT2 cells scored at each timepoint). ****p = 4.8 × 10−11 (one-way ANOVA, data as mean ± SD). c 3D rendering immunostaining of a direct (upper, AT2-to-AT1) and indirect (lower, AT2-to-AT2) ILJ from an immunostain for E-cadherin (green), RELMα (red), PDPN (white), and DAPI. Lumen boundaries are marked in yellow dashes. d Schematics of approach for timelapse confocal microscopy of Nkx2.1Cre; Rosa26mTmG lineage labeled precision cut lung slices (Adapted from BioRender) depicted in (e, f). e Representative direct nAT2 junctions between two lumens in 12-h timeperiod. f Two nAT2s established contact at 14.5 h and junction between two lumens in 15-h snapshots, contact marked with white arrows. g Representative MUC1 domain length in non-junctioning (left) and junctioning nAT2s. h Quantification of (g) shows a reduction in MUC1 domain length following ILJ (n represents number of nAT2 cells scored at each timepoint). ****p = 4.8 × 10−11 (Student’s two-sided t test, data as mean ± SD). i Observed sequence of nAT2 basal extrusion and apical constriction. j Schematic of junctioning outcomes (none, indirect, or direct) as interstitial thickness decreases. Source data are provided as a Source Data file. Created in BioRender. Cai, J. (2025) https://BioRender.com/9wf54yt.
To further investigate the dynamics of nAT2 junctioning, we performed timelapse confocal microscopy on Precision Cut Lung Slices (PCLS) isolated from E17.5 murine lungs wherein the distal lung epithelium was labeled with a membrane-tethered GFP (TgNkx2.1-Cre Rosa26mTmG) so that nAT2s could be readily visualized (Fig. 2d). We observed both direct and indirect nAT2 junctioning—direct when the interstitial distance was short (Fig. 2e) and indirect when the nAT2 was greater than a cell length away (Fig. 2f, Supplementary Fig. 2d and Supplementary Movie 4). During time-lapse microscopy, we also observed cases wherein nAT2s junctioned only temporarily (Supplementary Fig. 2diii). Finally, we observed that for most, if not all junctioning nAT2s, a significant reduction in length of MUC1 domains (Fig. 2g, h). Taken together, we believe our observations are of an initial “interlumenal junctioning” event critical in the process of integrating alveolar lumenal surfaces together (Fig. 2i, j) initiated upon basal extrusion of nAT2s. Further, we hypothesize the manner of junctioning (direct or indirect) is dependent on interstitial distance, and that defective nAT2 junctioning could result in alveolar simplification similar to what is observed in BPD and COPD.
Nascent and mature AT2 state differ in fate plasticity and BH3 regulation
Following their initial specification, nAT2s undergo maturation in the first week or so after birth, which can be observed by the switch from RELMα to CD74 expression (Fig. 3a, b). As RELMα expression is lost in mAT2s, we observe its concomitant upregulation in the airway epithelium (Supplementary Fig. 3a). In contrast to the initial proximal-to-distal wave pattern that AT1/AT2 fate selection is reported to occur4,13, we observe AT2 maturation does not follow this pattern but rather occurs in a sporadic salt-and-pepper pattern (Fig. 3c, d and Supplementary Fig. 3b), suggesting maturation may be governed by a distinct mechanism.
a PN1 and adult (≥PN60) lungs immunostained for AT2 marker MUC1 (white), RELMα (green), CD74 (red), and DAPI. Bar, 10 µm. b Timecourse quantification of the nAT2 transition to mAT2. p values determined using Brown–Forsythe and Welch ANOVA test (n represents AT2s scored per timepoint in experimental triplicate, values are mean ± SD). c PN1 immunostains for RELMα (green), CD74 (red), and DAPI, showing the pattern of mAT2s and nAT2s. Bar, 50 µm. d Schematic contrasting the reported proximal-distal pattern of AT1/AT2 differentiation with the sporadic pattern of AT2 maturation. e scRNAseq PAGA velocity analysis of E15.5 and E17.5 distal lung epithelial cells. Arrow thickness represents the relative cell fraction along a depicted cluster trajectory. A trajectory is observed from nAT2s to nAT1s (red arrow). f Velocity magnitude and AT1/AT2 gene score for the nAT2 fraction from (e). g Velocity analysis of the nAT2 fraction from e depicting AT1/AT2 score (cutoff 0.5). h UMAP of scRNAseq timecourse dataset (GSE149563)39 of the distal epithelium. i Leiden clustering identifies the AT2-to-AT1 transitional cluster (tAT2 > 1; brown) whose AT1 trajectory is confirmed by velocity analysis (red box). j Timepoint distribution within the tAT2 > 1 cluster. k Representative organoids cultured on Matrigel for 4 days in the presence of FGF7 (50 ng/mL), derived from either E15.5 DPs, E18.5 nAT2s, PN3 mAT2s, or adult mAT2s. AT2 and AT1 fate is detected by immunostaining for PDPN (white), RAGE (red), DAPI (blue), and AT2 marker SFTPC (green). DPs express both the AT1 (RAGE, PDPN) and AT2 (SFTPC) markers. Bars, 20 µm. l Quantification of (k) cell percentage per spheroid that differentiated into AT1. Data are represented as box plots showing median, upper, and lower quartiles, and the whiskers extend to the minima and maxima. p values determined using Kruskal–Wallis test. (n = spheroids per condition pooled from 3 independent experiments. m A revised model alveolar epithelial differentiation wherein a window of fate plasticity exists during which AT2s retain the ability to rapidly differentiate into AT1s. All experiments were repeated at least three times. Source data are provided as a Source Data file.
Given the molecular distinction between nAT2 and mAT2 states, we next sought to determine whether functional differences also exist. One manner in which time-dependent cell states can vary is in fate plasticity, as has been observed in neutrophils37 in vivo and lung epithelial iPSCs38 in culture. To determine whether and how nAT2s and mAT2s differ in fate plasticity, we analyzed two previously generated scRNAseq datasets that sample timepoints either during12 (Fig. 3e–g and Supplementary Fig. 3c–g) or far beyond39 (Fig. 3h–j) alveolar epithelial differentiation. Analyzing first the distal epithelium prior to (E15.5) and during (E17.5) alveolar epithelial differentiation, we performed standard clustering as well as UMAP visualization and found that distal epithelial cells isolated from these timepoints only rarely co-clustered (Supplementary Fig. 3e), indicating no E15.5 progenitor population had significantly adopted either AT1 or AT2 transcriptional program, a central tenet of the early lineage specification model proposed in a prior study12. A weak correlation between one of the E15.5 progenitor clusters (DE3) and the E17.5 progenitor cluster (0.42) was observed (Supplementary Fig. 3e, yellow box), suggesting a shared transcriptional program.
Single-cell transcriptional dynamics analysis was next analyzed to determine when and how the AT1 and nAT2s clusters segregate. Performing pseudotime (Supplementary Fig. 3f), velocity (Supplementary Fig. 3g), as well as integrated PAGA-Velocity analysis (Fig. 3e) all identified E15.5 clusters as the starting timepoint, as well as identified the AT1 cluster as the predicted endpoint (Supplementary Fig. 3f, g). Further, velocity analysis found a substantial proportion of nAT2s to be on a transcriptional trajectory towards the AT1 cluster, suggesting fate plasticity was retained (Fig. 3e, red arrow). To determine whether a subset of nAT2s displayed a biased trajectory towards AT1 differentiation, we established an AT1 and AT2 score based on well-established marker genes9 and observed a clear stratification within the nAT2 cluster (Fig. 3f). Separating the nAT2 cluster by this ratio into two subgroups (High AT1 or Low AT1 gene expression, >0.5 = High AT1/AT2 score), we compared velocity trajectories and observed a clear separation (Fig. 3g), indicating that a nAT2 subset with a high AT1/AT2 ratio is leaving the nAT2 state and transitioning to AT1.
After observing that embryonic nAT2s possibly retain fate plasticity, we sought to determine how long AT2s retain fate plasticity after birth. Using a previously reported dataset that samples across the lifetime of the distal epithelium of the mouse lung39, we performed similar processing as before to remove duplicates and non-distal epithelial cells before performing UMAP visualization and clustering analysis (Fig. 3h, i). Analyzing more widely over time, we again observed earlier progenitor populations, mature AT1 and AT2 clusters, as well as a cluster transitioning between them as previously reported39. Velocity analysis of this intermediate cluster found the cellular trajectories are oriented away from AT2 and towards AT1 (Fig. 3i, red box) similar to our prior observation. Finally, analysis of the timepoints of cells within the AT2→AT1 transitioning clusters revealed most were from the early perinatal period, suggesting fate plasticity was lost as AT2s mature within the first week after birth (Fig. 3j). To confirm this, we performed alveolosphere differentiation assays following our previously described protocol10 and found that both nAT2s as well as early mAT2s (PN3) could give rise to AT1 cells (Fig. 3k, l), confirming fate plasticity was lost primarily in a time-dependent manner, independent of nAT2 or mAT2 state. Taken together, our analysis indicates that fate plasticity, a property of DPs, is retained in newly formed AT2 cells for a period of 1–2 weeks after birth, revising the model of the alveolar epithelial fate selection model to include a window of plasticity (Fig. 3m).
Another property known to vary from early life to maturity is mitochondrial sensitivity to apoptotic stimuli (apoptotic priming)40, which is reduced during maturation in several organs but can be retained in certain stem cell populations41,42. To determine whether apoptotic sensitivity is retained in nAT2s, BH3 profiling was performed on isolated DPs, nAT2s, as well as adult mAT2s. As expected, DPs and nAT2s exhibited heightened sensitivity to pro-apoptotic BH3 peptides as indicated by higher rates of cytochrome c release, while mAT2s were largely unresponsive (Supplementary Fig. 3h). Taken together, our findings indicate the nAT2 state retains both fate plasticity as well as sensitivity to BH3-regulated apoptosis despite being functionally differentiated.
C/EBPα is required to maintain but not select AT2 fate
Given the loss of fate plasticity as nAT2s mature into mAT2s, we next sought to determine how this shift is regulated. Referencing the scRNAseq data we previously used to distinguish nAT2 and mAT2 clusters (Fig. 1a, b) as well as AT2s sequenced more deeply by the Tabula Muris Consortium43, we screened putative transcriptional and epigenetic regulators from a curated list (1678 genes) and identified 67 genes with ≥2-fold enrichment along the AT2 (Supplementary Table 3) lineage (DP→nAT2→mAT2) compared to AT1 (Fig. 4a). Of these, we then considered only the 32 genes with known embryonic lethal or lung defects when knocked out in mice (Fig. 4a and Supplementary Fig. 4a). These genes can be stratified into four categories based on expression pattern over time: constitutive, DP-restrictive, DP/nAT2-restrictive, and nAT2/mAT2-restrictive (Fig. 4a, lower and Supplementary Fig. 4a). Of these, we focused on genes expressed in the observed window of fate plasticity (Supplementary Fig. 4b), which narrowed the list to four genes, Etv5 and Elf5 (constitutive) as well as, Cebpa and Nupr1 (nAT2/mAT2-Restrictive) (Fig. 4b). While both constitutively expressed genes (Etv5 and Elf5) have been investigated in the lung, findings suggest regulatory roles other than fate plasticity, as ETV5 is required primarily to maintain AT2 survival44 while ELF5 appears to broadly regulate aspects of lung epithelial differentiation45. Further, we excluded study of Nupr1 given its primary role is in death regulation, not differentiation46,47. This left Cebpa as the putative regulator, for which we validated the timing of expression via immunostaining in the embryonic lung (Fig. 4c).
a Bioinformatic screen of 1678 transcriptional or epigenetic regulators, of which 67 are enriched along AT2 lineage. 32 of these have known embryonic lethal or lung defects upon knockout in mice and can be stratified into four temporal categories: DP, DP-nAT2, nAT2-mAT2, and constitutive. The observed plasticity window is depicted in red. b Dot plot of Sox9 (DP restrictive; yellow), Cebpa and Nupr1 (nAT2-mAT2 restrictive; pink), Etv5 and Elf5 (constitutive; gray) temporal expression (plasticity window in red). c E16.5 and E17.5 lungs immunostained for PDPN (white), C/EBPα (green), ECAD (red), and DAPI (blue). C/EBPα+ cells are marked (arrowheads) within the proximal (P) and distal (D) regions. Bars, 100 µm. d Alveolar regions of PN0 control Nkx2.1Cre; Rosa26mTmG; Cebpawt/fl (left) and Nkx2.1Cre; Rosa26mTmG; Cebpafl/fl (right) lungs immunostained for DP lineage reporter (GFP) and MUC1 (white). Note GFP+ AT2s and AT1s (flat and MUC1−, labeled “1”) form even after Cebpa deletion. Bars, 50 µm. e Alveolar regions of adult control LyzMCre; Rosa26mTmG; Cebpawt/fl (left) and LyzMCre; Rosa26mTmG; Cebpafl/fl (right) lungs immunostained for MUC1 (white) and CD74 (blue). AT2-lineage derived GFP+ cells differentiate into AT1s (asterisk) upon Cebpa deletion. Bars, 100 µm. f Quantification of (e) showing percent GFP+ cells that differentiate into AT1s upon Cebpa deletion (n represents number of GFP+ cells sampled for the Cebpawt/fl and Cebpafl/fl conditions in experimental triplicate). ****p = 1.4 × 10−14 (Student’s two-sided t test, data as mean ± SD). All experiments were repeated at least three times. Source data are provided as a Source Data file.
Previous Cebpa research in the developing mouse lung performed knockouts via a Tet-On approach and concluded it was required for alveolar epithelial differentiation48,49. These findings are conflicting with the expression pattern of Cebpa, which is first expressed after AT2 differentiation, not before (Fig. 4b, c and Supplementary Fig. 6b). Given this discrepancy, as well as a subsequent study that questioned the viability of the prior Tet-On approach in the lung50, we first sought to confirm whether Cebpa was required for alveolar epithelial differentiation via a different method of gene deletion. Using a sparse labeling with efficient recombination approach (Nkx2.1-Cre; Rosa26mTmG/mTmG; Cebpafl/fl) wherein a subset of distal bipotent progenitors is both fluorescently labeled and floxed gene recombined10, we tested whether Cebpa loss indeed resulted in suppressed differentiation. Surprisingly, we observed that both AT2 and AT1 fate selection occurred despite the absence of Cebpa (Fig. 4d and Supplementary Fig. 4c), both at the molecular and morphological level.
As we expected Cebpa to play a role later in development consistent to when it is expressed, we next performed a sparse but efficient deletion experiment wherein Cebpa was deleted in a subset of AT2 cells starting in the early perinatal period (~PN5, LyzMCre; Rosa26mTmG/mTmG; Cebpafl/fl). While lineage-traced control AT2 cells maintain their fate into adulthood, a significant portion of AT2 cells lacking Cebpa converted into AT1 cells (Fig. 4e, f), suggesting an increase in AT2 fate plasticity. Finally, we confirmed the result using an AT2-restrictive tamoxifen-inducible Cre recombinase line (SftpcCreER) which again found an increase in conversion to AT1 fate both morphologically and molecularly (Supplementary Fig. 4d). Taken together, we determined that C/EBPα, a bZIP transcription factor expressed following AT2 cell specification is required during the perinatal period to maintain, not select, AT2 fate, suggesting it plays a role in suppressing fate plasticity during the transition from nascent to mature AT2 state.
C/EBPα represses a non-cell autonomous AT1 differentiation program driven by DLK1
To determine more precisely how C/EBPα regulates AT2 fate plasticity, we conducted scRNAseq of the alveolar epithelium from both control (Cebpawt/fl) and AT2-floxed (Cebpafl/fl) lungs, wherein Cre-mediated deletion was induced in the perinatal period using SftpcCreER in combination with the iSure-Cre-tdT (TgiSureCre-tdT)51 allele. Retrospective identification of cell types via Louvain clustering using Seurat was able to distinguish control (AT2 – Cebpawt/fl) and Cebpa floxed (AT2 – Cebpafl/fl) AT2s as well as AT1s (Fig. 5a-i, ii and Supplementary Fig. 5a), a subset of which we confirmed were derived from AT2 cells following Cre induction by expression of the Cre-inducible reporter TgiSureCre-tdT (Supplementary Fig. 5b, c), confirming our prior lineage tracing experiments. We observed a substantial down- and upregulation of 117 and 116 genes, respectively, upon loss of Cebpa (Supplementary Fig. 5d). To determine how AT2 and AT1 gene expression overall (Supplementary Fig. 5e) was impacted following Cebpa loss, we first used gene scores to broadly assess changes in either AT2 or AT1 related genes (Supplementary Fig. 5g). While no significant upregulation of AT1 genes was observed (suggesting C/EBPα likely does not repress fate plasticity by directly repressing the AT1 gene program), a reduction in overall AT2 genes was observed in AT2s lacking Cebpa (Fig. 5a-ii). To further determine how AT2-associated genes were altered, we separately assessed genes restrictively enriched in either nAT2 or mAT2 state (Supplementary Fig. 5f). Upon Cebpa loss, AT2 cells reduce expression of mAT2 genes while concomitantly upregulating nAT2 gene expression (Fig. 5a-ii), suggesting re-accessing of a plastic, nascent-like AT2 state.
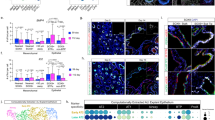
a Design of scRNAseq experiment (upper). UMAP (i, lower left) with Louvain clustering depicts AT1s and AT2s (both control and Cebpa floxed) groups. Dot plot (ii, lower right) of gene score corresponding to cell type (AT1, AT2) or state (nAT2, mAT2) from all clusters. Note the change in scores following Cebpa deletion, suggesting some reacquisition of nAT2 state (red dashed box). b Design of iSuRe-Cre experiment (above). Representative immunostains for tdT-lineage (red) and EdU (green) from Cebpa deleted lungs with either high (~99% AT2s) or sparse dose (~20% AT2s) TMX (lower). Note lineage labeled cells that have proliferated (asterisk) or are AT1s (arrowhead). Bars, 50 µm. c Quantification of (b) showing percent tdT+ AT1s upon Cebpa deletion in either High Dose (fl/fl—H.D.) or Sparse (fl/fl—S) deletions compared to control (wt/fl). p values determined using One-way ANOVA with Tukey’s multiple comparisons testing (n represents tdT+ cells sampled for each condition in experimental triplicate; data as mean ± SD). d Screen for upregulated genes encoding a non-cell autonomous signal (left). Of the 107 genes upregulated following Cebpa deletion (log2 FC ≥ 0.7), 22 encode extracellular proteins, 6 of which are expressed in nAT2s. The 4 genes that encode either a secreted (green) or membrane bound (red) protein are plotted by fold change (right). e Representative regions of control or Cebpa deleted mouse lungs immunostained for DLK1 (green) and tdT (red). Bars, 20 µm. f Quantification of (e) DLK1 expression by tdT+ AT2s in control, sparse, and high dose conditions. Note similar amounts in high dose and sparse deletion conditions, 47.2% and 43.9% respectively. p values determined using One-way ANOVA with Tukey’s multiple comparisons testing (n represents tdT+ cells sampled for each condition in experimental triplicate; data as mean ± SD). All experiments were repeated at least three times. g Bulk ATACseq tracks showing chromatin accessibility of Cebpawt/fl (blue) and Cebpafl/fl (red) AT2 cells (n = 3 replicates for each condition), showing the Dlk1 transcriptional start site and intergenic differentially methylated region (IG-DMR). The closest C/EBPα binding site to Dlk1 is observed at a distal trans-regulatory element (TRE, yellow) downstream of the IG-DMR, which displays reduced chromatin accessibility upon Cebpa loss. ChIP-seq of AT2s for C/EBPα shows increased binding at the putative TRE (red box). Source data are provided as a Source Data file.
To better understand the behavior of these plastic AT2s as well as focus our investigation on their molecular regulation, we next sought to determine whether the AT2-to-AT1 differentiation observed upon Cebpa loss was mediated by a cell-autonomous or non-autonomous mechanism. While a common experimental approach in Drosophila research52,53 to delineate between mechanisms targeting internal signaling or proliferation (cell autonomous) versus expression of an extracellular signal (non-cell autonomous), such experiments are less common in the mouse and have been difficult using inducible Cre lines until the development of transgenic alleles such as iSure-Cre51. Usage of a CreER line alone under sparse labeling conditions can result in incomplete or mosaic recombination of alleles and label “escaper cells” 54, confounding interpretations of the gene’s necessity. To overcome this, we next performed sparse labeling with efficient Cebpa deletion using the iSure-Cre allele (SftpcCreER; TgiSuRe-Cre; Cebpafl/fl) at a tamoxifen dose we determine labels ~20% of AT2s validated by immunostaining (Fig. 5b, c and Supplementary Fig. 5h, i). This method of sparse labeling with efficient recombination can be used to delineate whether a phenotype is regulated in a cell autonomous (i.e. via a cell intrinsic mechanism, in which case the phenotype would occur in both sparse and non-sparse labeling experiments) or non-cell autonomous manner (i.e. via a cell extrinsic mechanism, in which case the phenotype would occur significantly less if at all in the sparse labeling experiment). In contrast to when Cebpa is removed from most AT2s (~99%), we observed that removal of Cebpa from sparse numbers of AT2s (~20%) resulted in a substantial reduction of AT2-to-AT1 differentiation as assessed by percentage of lineage labeled AT1s and proliferation (Fig. 5b and Supplementary Fig. 5j). We also confirmed that all lineage labeled AT1 cells in the sparse TMX condition were found to have either direct contact or present within 10 µm of a lineage positive AT2 cell (Supplementary Fig. 5k, l). This finding strongly indicates that AT2 fate plasticity acts via a cell non-autonomous mechanism, likely an extracellular signal, to promote AT2-to-AT1 differentiation.
Given this, we focused our screen on genes upregulated in Cebpa deleted AT2s that encode proteins with an extracellular localization (GO:0005615), which returned 22 genes, 6 of which were also expressed in the nAT2s from the prior scRNAseq timecourse, and of these 4 are localized on the cell membrane or secreted (Dlk1, Col23a1, Cd14, and Apoc1) (Fig. 5d and Supplementary Fig. 5m, n). The most highly upregulated of these genes is Dlk1, which encodes a single-pass type I membrane protein localized to the outer cell surface that is known to regulate Notch signaling55 and AT2-to-AT1 differentiation23 (Fig. 5d, red).
Finally, we confirmed DLK1 is also upregulated at the protein level following Cebpa loss in culture (Supplementary Fig. 5o), and further observe its expression is maintained under sparse labeling with efficient Cebpa deletion in vivo, suggesting C/EBPα directly regulates DLK1 in a cell autonomous manner (Fig. 5e, f). Taken together, we hypothesize that C/EBPα begins to block fate plasticity of specified AT2s by direct suppression of Dlk1 and its subsequent activation of Notch signaling.
After establishing C/EBPα likely directly represses Dlk1 expression, we sought to determine where within the genome it is mediated. We performed bulk ATAC-seq on both control and Cebpa floxed AT2s in the same experiment timeline as for scRNAseq (Supplementary Fig. 5p), but AT2-lineage cells were further purified by FACS using the genetically encoded fluorophore tdTomato. After confirming correlation between ATACseq replicates (Supplementary Fig. 5q), we identified 23,764 sites within the genome that significantly changed in accessibility following Cebpa loss (Supplementary Fig. 5r), with decreased accessibility associated with 3,986 genes, consistent with C/EBPα’s well-established role as a positive regulator of gene expression56. We observed 369 DEGs shared between our ATACseq and scRNAseq datasets (Supplementary Fig. 5s) and mapped the proportions of genomic features that are up- and downregulated (Supplementary Fig. 5t). We also observed a smaller but substantial number of genes (2384) with associated upregulation in chromatin accessibility following Cebpa loss, indicating repressive regulation, including the transcriptional start site (TSS) of Dlk1 (Fig. 5g). Further, the change in enrichment of transcription factor binding sites following Cebpa loss is consistent with prior work on Nkx2.1 and AP-1 acting as a bivalent co-factor to reinforce fate of both AT1 (via YAP/TAZ interactions) and AT2 (via C/EBPα) cells57 (Supplementary Fig. 5u–w).
In investigating putative regulatory sites near Dlk1, we initially considered the nearby intergenic differentially methylated region (IG-DMR) previously demonstrated to strongly regulate Dlk1 expression58. While no change in accessibility was detected in the IG-DMR locus, a region nearby was found to decrease following Cebpa loss (Fig. 5g). Further, this region included a predicted regulatory site containing a C/EBPα binding motif EM10E0548430 previously identified by ENCODE (Fig. 5g, dashed red), where it could possibly act to repress Dlk1 and that we validated C/EBPα binds to in AT2 cells following analysis of a previously published ChIPseq dataset54 (Fig. 5g), lower.
PRC2 and C/EBPα act as a Dlk1 “pulse generator” critical for patterning alveolar epithelial fate
Given the perinatal repression of Dlk1 by C/EBPα, we were curious of its expression earlier in lung development. As previously reported59,60, we observed that Dlk1 is lowly expressed at stages prior to alveolar epithelial differentiation (e12.5-E15.5), both at the protein (Fig. 6a) and RNA level (Fig. 6b, dashed gray box). Further, Dlk1 expression appeared to peak in the period from E17.5 to PN1 (Fig. 6a, b and Supplementary Fig. 6a). As Cebpa is not detected at earlier stages (≤E15.5) (Supplementary Fig. 6b), we were curious how Dlk1 downregulation is mediated. Suppression of Dlk1 has also been found to be mediated by the polycomb repressive complex (PRC2), including the H3K27 methyltransferase EZH261. We observed that genes encoding PRC2 members were enriched at earlier stages (Fig. 6b, red text), and further that EZH2 protein is substantially downregulated after E16.5 (Fig. 6c).
a E15.5 (top) and E17.5 (bottom) lungs immunostained for DLK1 (green) and ECAD (red), close-ups show DLK1 intensity (fire LUT). Bars, 10 µm. b Expression of Cebpa, Dlk1, and PRC2 member genes (red) over time in distal lung epithelium. Note nominal Cebpa and Dlk1 expression before E16.5 (gray box). c E16.5 and E17.5 distal branches immunostained for PDPN (white), EZH2 (green), and ECAD (red) with distal tips indicated (D). Bars, 50 µm. d Immunostained E15.5 DPs differentiated with FGF7 alone without (Control) or with GSK126 (10 µM). Bars, 20 µm. e Quantification of (d) of DLK1+ cells per spheroid at day 4 (n represents spheroid number per condition in experimental triplicate). Data as mean ± SD (Student’s two-sided t test). f Cistrome analysis of the Cebpa locus identifies EZH2 (red) and the other PRC2 members (gray) with enriched binding. g E15.5 DP spheroids cultured as in (d) immunostained for C/EBPα. Bars, 20 µm. h Quantification of (g) percent C/EBPα+ cells per spheroid (n represents number of spheroids for each condition in experimental triplicate). Data as mean ± SD (Student’s two-sided t test). i Distal epithelial branch of an E15.5 Nkx2.1Cre; Rosa26mTmG; Ezh2fl/fl mouse lung with sparsely labeled DPs (GFP+) wherein Ezh2 is deleted that is immunostained for C/EBPα (white). Bar, 10 µm. j Quantification of (i) percent lineage negative (RFP+) or lineage positive (GFP+) DPs expressing C/EBPα in the distal alveolar buds. Data are represented as box plots showing median, upper, and lower quartiles, and the whiskers extend to the minima and maxima. p values determined using two-tailed Mann–Whitney U-test (n represents number of distal alveolar buds sampled for both groups). k Proposed incoherent, type 2 Feed Forward Loop (I2-FFL) model. l Fitting of observed Dlk1 expression pattern from scRNAseq timecourse study to a mathematical I2-FFL Model (red line, RMSE ≤ 0.75 represents high goodness of fit). m E15.5 DP spheroids cultured as in (d), immunostained for PDPN (white), SFTPC (green), RAGE (red), and DAPI. Bars, 20 µm. n Quantification of (m) showing proportion of DPs (white), AT1s (gray) and AT2s (black). Data as mean ± SD (n represents number of spheroids per condition in experimental triplicate; one-way ANOVA). o Schematic of working model. A DLK1 pulse occurs upon loss of PRC2 in nAT2s, stimulating Notch signaling in DP or plastic neighboring cells. This pulse followed by lateral inhibition results in the observed pattern of AT1/AT2 fate. All experiments were repeated at least three times. Source data are provided as a Source Data file.
As EZH2 and the PRC2 complex are enriched earlier when Dlk1 is downregulated, we next tested whether EZH2 activity is required to suppress DLK1 in culture. Isolated E15.5 DPs were cultured as organoids and treated either with FGF7 alone or with the EZH2 inhibitor GSK126 (10 µM) for 4 days (Fig. 6d). While control organoids had detectable DLK1 protein (~12% DLK1+ cells), levels significantly increased when treated with the EZH2 inhibitor (~22%, Fig. 6e), indicating EZH2 methyl transferase activity is required for DLK1 suppression.
When characterizing the expression of Ezh2 alongside Cebpa, we noticed a consistent mutually exclusive pattern (Fig. 6b and Supplementary Fig. 6c), suggesting EZH2/PRC2 might also suppress Cebpa. We analyzed a published ENCODE epigenetic dataset that mapped the PRC2 modification H3K27me3 across development62 and observed its time-dependent reduction at the Cebpa locus concomitant to when Cebpa expression is upregulated (Supplementary Fig. 6d). Further, analysis of ChIPseq database of mouse tissues and cells63 confirmed significant enrichment in binding of EZH2 as well as other PRC2 components near the Cebpa locus (Fig. 6f). To determine whether EZH2/PRC2 is also necessary for repressing C/EBPα, we again conducted organotypic culture with E15.5 DPs during which differentiation was promoted by addition of FGF7 as previously described10. Blocking of EZH2 activity by GSK126 significantly increased the percentage of C/EBPα+ cells compared to control (55% versus 34% respectively, Fig. 6g, h). Finally, we confirmed whether Ezh2 is required in vivo to suppress C/EBPα by using a sparse deletion approach wherein a Cre line we previously found to sparsely label DPs10 (TgNkx2.1-Cre) was bred to a transgenic line to delete Ezh2 and lineage label by GFP (TgNkx2.1-Cre Rosa26mTmG/mTmG; Ezh2fl/fl). Immunofluorescence conducted at E15.5, a timepoint where no C/EBPα is observed (Supplementary Fig. 6b), found significant upregulation of C/EBPα in Ezh2 deleted (GFP+) versus control (RFP+) DPs residing within the same end buds (Fig. 6i, j).
The interactions between EZH2/PRC2, C/EBPα, and DLK1 we observed appeared to fit the definition of a type 2 incoherent feed-forward loop64 (I2-FFL, Fig. 6k). An incoherent feedforward loop (IFFL) is a type of network motif found in biological systems, such as gene regulatory networks. It contains three components, a regulator, an intermediate, and a target—we propose these components in our model to be PRC2, C/EBPα, and Dlk1, respectively (Fig. 6k). The term incoherent means that the regulator (PRC2) acts in a conflicting manner to the target—by both inhibiting (directly on Dlk1) and activating it (indirectly through repression of the repressive intermediate C/EBPα. One emergent property of an I-FFL is the capacity to generate a pulse of the target gene’s expression (Dlk1) following downregulation of the upstream regulator (EZH2/PRC2). To more precisely assess whether the observed temporal pattern of Dlk1 expression matched the pulse predicted by an I2-FFLbiocircuits package (https://biocircuits.github.io/). After establishing reasonable values for parameters describing the relationship of factors in the I2-FFL (see “Methods”), the model was found to fit well to the observed Dlk1 expression pattern as assessed by root mean squared error (RMSE = 0.12, Fig. 6l and Supplementary Fig. 6e). Next, we wanted to assess the importance of this molecular circuit in patterning alveolar epithelial differentiation. Immunostaining of E15.5 DPs in organotypic culture for markers of AT2 and AT1 fate (SFTPC and RAGE, respectively) revealed that blocking EZH2 activity, with or without FGF7 treatment, significantly increased the proportion of molecularly specified AT2s while concomitantly reducing AT1s compared to FGF7 alone (Fig. 6m, n). Finally, we wanted to assess whether lateral inhibition of Notch signaling occurs in AT1 cells, as it has been well demonstrated to promote a spatial “salt and pepper” pattern of cell fates65. Analysis of scRNAseq timecourse datasets found that nascent AT1 cells upregulate the Notch target gene Hes1 suggesting signaling upregulation as expected from prior studies66 as well as downregulation of Notch ligands Jag1 and Dll4 that is consistent with lateral inhibition of Notch signaling (Supplementary Fig. 6f, g). Further, we also observe in nAT1s changes in Notch regulators that is also consistent with lateral inhibition, specifically downregulation of Dlk1 and upregulation of Dlk2 (Supplementary Fig. 6f, g)—the latter has been found to act as a cis-inhibitor of DLK167. Therefore, our findings suggest the following potential model of fate regulation (Fig. 6o): Following PRC2 downregulation a DLK1 pulse promotes a subset of AT2s to send Notch signals which, through lateral inhibition, resolves into a salt and pepper pattern of alveolar epithelial fate. Taken together, our findings support the model that EZH2/PRC and C/EBPα act as a molecular circuit (specifically an I2-FFL) to generate a DLK1 pulse during late-stage embryogenesis that is critical to patterning the proportion of AT2 and AT1 cells.
C/EBPα suppression is required for accessing both reparative and defensive AT2 states
Given the role of C/EBPα in suppressing plasticity in AT2 cells, we sought to determine if and how it is overridden following injury in the adult lung, where AT2s are well established to act as facultative progenitors to regenerate lost AT1 cells4,22. To first determine if C/EBPα downregulation is required following injury in adulthood for AT2-to-AT1 conversion, we infected adult mouse lungs harboring a Cre-dependent fluorescent reporter (Rosa26mTmG/mTmG) with AAVs targeting AT2 cell expression via a minimal Sftpc promoter as previously described10 to express either Cre alone or Cre and C/EBPα. After 5 days, we injured lungs via intratracheal instillation of the irreversible FGFR inhibition Fiin1 (14.75 mg/kg, as previously characterized by our group10) or intraperitoneal injection of BHT (225 mg/kg) and performed immunostaining 14 days afterwards (Fig. 7a upper and Supplementary Fig. 7a, b). Intratracheal instillation of AAVs efficiently labeled AT2s with high specificity (Supplementary Fig. 7c). While injured lungs showed AT2-to-AT1 conversion when treated with the control AAV, it was significantly reduced in those given C/EBPα overexpressing AAVs (Fig. 7a, b).
a Experimental design for AAV Cebpa deletion study (top). Immunostained lungs 14 days post injury (bottom) showing GFP+ AT1s (asterisk). Bars, 100 µm. b Quantification of (a) percent GFP+ AT1s (n represents GFP+ cells sampled per condition in experimental triplicate). Data as mean ± SD (Student’s two-sided t test). c Adult mouse lung 16 days post Bleomycin injury immunostained for ECAD (green), C/EBPα (red), and DAPI (blue). Lower panel depicts C/EBPα expression (fire LUT) in injured versus uninjured sites. Bars, 100 µm. d Quantification of (c) mean C/EBPα intensity of AT2s within uninjured or injured regions. Data are represented as box plots showing median, upper, and lower quartiles, and the whiskers extend to the minima and maxima. p values determined using two-tailed Mann–Whitney U-test (n represents number of distal alveolar buds sampled per condition). e Dot plot of AT2s from published scRNAseq 1X Bleomycin timecourse study (GSE141259) depicting genes of known C/EBPα suppressors (Ddit3, Atf3, Atf4, and Ezh2—upregulation indicated in red) as well as DATP gene score. f Experimental design for stimulating CHOP in vivo. Control or CT020312 treated PN25 lungs immunostained for AT2 lineage (GFP) and MUC1 (white). Bars, 50 µm. g Quantification of (f) showing percent GFP+ AT1s in control and treated condition (n represents GFP+ cells sampled per condition). Data as mean ± SD (Student’s two-sided t test). h Leiden clustering identifies two Cebpafl/fl AT2 clusters: AT2DLK1+ (green) and AT2Def (red) which distinct velocity trajectories. i Gene Ontology (GO) analysis of the AT2Def cluster (Fisher’s Exact Test). j UMAP with overlaid DATP and Defender gene scores. k SPRING plot of clusters in (h), showing AT2Def state (red) in between the control AT2 (orange) and AT2DLK1+ (green) clusters. l SPRING plot of the DATP-gene score. m Immunostained PN3 mAT2 spheroids cultured with FGF7 alone (Control, top) or with interferon-γ (20 ng/mL; bottom). Bars, 20 µm. n Quantification of (m) showing percent of AT1s per spheroid (n represents spheroids sampled per condition). Data as mean ± SD (Student’s two-sided t test). o Schematic working model of AT2s oscillating between AT2Def (purple) and AT2DATP (yellow) states, both suppressed by C/EBPα. Upon AT2 loss (gray AT2s) and C/EBPα downregulation following infection, remaining AT2s could oscillate (left) between either state allowing for both a proper response to infection and regeneration. In a static model (right), inability to access both AT2Def and AT2DATP states would result in either impaired pathogen response or repair, respectively. All experiments were repeated at least three times (adapted from BioRender). Source data are provided as a Source Data file.
Next, we examined whether C/EBPα expression is downregulated following injury using the 1X Bleomycin model, which results in patchy regions of injury and that resolve within 4–6 weeks. Bleomycin-treated lungs were imaged 16 days after administration and injured areas detected by alveolar simplification and reduction in E-cadherin as previously observed68. Immunostaining for C/EBPα revealed a significant downregulation in injured versus uninjured regions (Fig. 7c, d). This finding is in alignment with analysis of a recent scRNAseq timecourse study20 following single-dose bleomycin, during which Cebpa expression is downregulated in AT2 cells as early as 2 days post bleomycin (Fig. 7e).
After establishing the presence and necessity of C/EBPα downregulation following injury, bioinformatic screening of the bleomycin injury timecourse for transcriptional regulators known to suppress C/EBPα that are also upregulated in the previously identified AT2-to-AT1 transitionary20 state (Krt8+ ADI, also known as DATP34 or PATS69) identified three genes, Atf3, Atf4, and Ddit3 (Fig. 7e and Supplementary Fig. 7e). Both activating transcription factors 3 and 4 are known to bind the Ddit3 promoter to drive expression70,71, and DNA damage-inducible transcript 3 protein, also known as C/EBP homologous protein (or CHOP), has been well established to directly bind and act as a dominant negative inhibitor of C/EBP transcription factors in a prior study72. Given its direct interaction and inhibition of C/EBPα, we next wanted to determine whether induction of CHOP is alone sufficient to drive AT2-AT1 conversion. A day after lineage labeling AT2 cells in lungs of perinatal mice (PN11, SftpcCreER; Rosa26mTmG), intraperitoneal injections of either vehicle alone or a small molecule inducer of CHOP via PERK activation CCT020312 (2 mg/kg) were administered every 4 days and lungs isolated at PN25 (Fig. 7f). While little to no lineage labeled AT1s were observed in vehicle treated lungs, there was a significant increase in those treated with CCT020312 (Fig. 7f, g), suggesting PERK/CHOP induction can promote AT2-to-AT1 conversion.
To precisely map AT2 transcriptional trajectories and programs following Cebpa loss, we conducted Velocity analysis as well as Leiden clustering on our scRNAseq dataset and confirmed a cellular trajectory from AT2 to AT1 cluster as expected (Fig. 7h). Unexpectedly, we observed another distinct trajectory terminating on a distinct AT2 cell cluster that was enriched in genes associated with pathogen defense response and interferon signaling by GO analysis (Fig. 7i and Supplementary Table 4), leading us to name it the “defender” state AT2def (Fig. 7h, red). Visualizing the expression of AT2def enriched genes versus those associated with the damage-associated transitionary progenitor (DATP) state34, we observed that these programs were largely mutually exclusive, apparent by both UMAP (Fig. 7j) and SPRING plot (Fig. 7k, l). Given the distinct trajectories, as well as prior research indicating interferon signaling negatively impacts lung epithelial repair24, we were interested whether stimulation of the AT2def state by interferon signaling would block AT1 conversion in culture. Given the expression pattern of interferon receptors as well as transcriptional targets in the AT2def cluster (Supplementary Fig. 7d, e), we treated cultures with interferon-γ (Fig. 7m). While perinatal AT2s could give rise to AT1s following FGF7 treatment in organotypic culture, addition of exogenous interferon-γ resulted in a significant reduction AT1 conversion, consistent with the model that AT2def and AT2-to-AT1 molecular program are antagonistic to each other at the cellular level (Fig. 7m, n). To assess the degree to which these programs are present in the same or separate AT2 cells following injury, we analyzed scRNAseq datasets wherein lungs were injured in distinct ways, including diphtheria toxin ablation of AT1 cells73, influenza74, LPS75, and Bleomycin20. In support of our findings following Cebpa deletion, DATP and Defender states were found to primarily exist in separate AT2 cell clusters during the repair process across all forms of injury (Supplementary Fig. 7f).
Discussion
These results show that AT2 fate plasticity exists in a time-window from late embryonic to early postnatal life. The timing and length of this window is dictated by a molecular circuit driven by PRC2 and C/EBPα that together generate a pulse of Dlk1, a known regulator of adult AT2 fate plasticity. Downregulation of C/EBPα occurs following injury in the adult lung, is required to regenerate AT1 cells, and is mediated in part by the dominant negative C/EBP family member CHOP. C/EBPα also suppresses an AT2 defensive state that is mutually exclusive from fate plasticity that appears to be driven by interferon signaling.
From characterizing its nascent state, we determined AT2 cells first emerge around E15.5 at intermediate regions between the tips and stalks of the distal branches. Our findings consolidate differing models on AT2 emergence, with our observed timing supporting the bipotent model4,9,10,35,39 and our observed pattern suggesting an intermediate starting point, distinct from that proposed in both the bipotent (more proximal) and earlier differentiation12 (distal tip) models. After initial emergence (E15.5-16.5), subsequent AT2s next arise at proximal regions coinciding with ASM-to-myofibroblast differentiation as previously reported36. Taken together, these findings suggest more than one cue may be present to suppress alveolar epithelial differentiation, one distal and another proximal in position. While it has been established that distal tips are enriched in Wnt activity and thus retain stemness76, it is unclear how more proximally positioned DPs are suppressed. One compelling possibility is that ASM directly suppresses differentiation of these proximally localized distal progenitors through direct interaction on the basal epithelial surface, although it is unclear how said suppression is molecularly mediated.
Our findings on AT2 emergence have also provided insight into the sequence of their morphological changes. Given that basal extrusion occurs after AT2 molecular specification, it is likely downstream of fate selection rather than an upstream regulator as previously proposed77. This observation is supported by prior work wherein AT2 fate selection was found to occur under culture conditions that block basal extrusion10. If a byproduct of fate selection, what purpose does basal extrusion serve? Following extrusion, we observe that some extruded AT2 cells engage with nearby epithelium from anatomically distinct branches, forming what we name interlumenal junctions, or ILJs. We hypothesize that basal extrusions are required to form these junctions, that they are mediated in part by contraction of the AT2 lumenal surface, and likely play a critical early role in postnatal alveologenesis, specifically the stitching together of alveolar lumens78. While this process has been observed at the tissue level14, as has the multilumenal nature of mature AT2s36, many questions remain as to the underlying cellular and molecular mechanisms governing it. Further work on interlumenal junctions will be critical to understanding this stage of alveolar maturation and assessing whether its disruption could underlie diseases such as BPD and COPD.
Following their emergence, nascent AT2 cells undergo transition towards their mature AT2 transcriptional state over the course of ~7 days. This process occurs in a sporadic, salt and pepper pattern distinct from the reported proximal-to-distal wave of alveolar epithelial fate selection13,79, suggesting the cues driving maturation might arise from a different source, which is likely to be immune, given the genes enriched in mAT2s as well as prior studies demonstrating interactions underlying one another’s maturation35 and function15. While AT2s have been shown to impact macrophage maturation35, an immune cell type has not been implicated in driving AT2 maturation during postnatal development. More work is needed to better understand which immune cell types interact with AT2 cells during this process and determine how they impact differentiation. While AT2 cells from both nascent and mature states retained plasticity, mAT2s quickly lose this capability within the first week or so after birth, suggesting fate plasticity is somewhat negatively correlated to AT2 maturation. Given these findings, we revise our model of alveolar epithelial differentiation to include a fate plasticity window extending from E15.5 distal progenitors into AT2 nascency, and that tapers down over 1–2 weeks after birth. It is likely this window is critical for postnatal alveologenesis and might be involved in the observed age-dependent severity of COVID1980.
While our bioinformatic screening for transcriptional regulators of the plasticity window identified C/EBPα as a possible repressor, previous genetic deletions studies concluded that Cebpa is required earlier for both AT1 and AT2 differentiation48,49. As these findings conflict with the observed expression pattern of C/EBPα, which is upregulated following AT2 fate selection, we conducted sparse Cebpa deletion experiments in both DP and AT2 populations. From these studies we found that Cebpa is dispensable for the selection but required for the maintenance of AT2 fate, with the later finding recently confirmed in a manuscript by other groups54,81. The discrepancy is likely the result of the field’s prior use of genetic tools now found to cause toxicity in the developing mouse lung50. While our subsequent scRNAseq and ATACseq analysis reveals repressive targets of C/EBPα, its positive transcriptional regulation of gene expression has been demonstrated including recent publications54,81. While Cebpa loss does result in a reduced expression of several AT2 mature genes, identified in both studies, it is important to note that these AT2 cells do not concomitantly increase the expression of AT1 genes nor directly convert into AT1 cells, making it less likely that AT2s are undergoing rapid, direct transdifferentiation to AT1 fate. Rather the AT2 program reverts to the nascent AT2 state, again suggesting a repressive mechanism is at play that targets fate plasticity. While Sox9 was reported in the other study to be upregulated following Cebpa loss, we only detected a modest increase at the RNA level and screening of scRNAseq data referencing a DP gene list revealed no strong upregulation, supporting our model that loss resulted in reversion to a more nAT2 state rather than full dedifferentiation back to DP. Recent work has found that AT1s in the perinatal period also retain fate plasticity and when stretch and YAP/TAZ signaling is lost they are capable to AT1-to-AT2 differentiation82. While more work is needed to integrate those findings with ours and others54, one compelling model is that both perinatal AT2 and AT1 cells retain fate plasticity for a period of time but are each suppressed by distinct repressors – C/EBPα for AT2s and YAP/TAZ for AT1s.
Our investigation into the key gene(s) repressed by C/EBPα that could regulate fate plasticity identified Dlk1, which encodes a cis-regulator of Notch signaling previously implicated in AT2-to-AT1 conversion23. Notch is distinct from other signaling pathways in its use of cis-regulators (like DLK1) for activation and has been shown to be critical in generating its well-characterized “salt and pepper” pattern83. While regulation of the Dlk1 locus is well documented to be mediated by an intergenic differentially methylation region (IG-DMR) downstream of the gene58,84, ATAC-seq revealed no change in its accessibility nor any changes immediately upstream of the gene’s transcriptional start site upon loss of Cebpa. Rather, a dramatic change in accessibility was observed in an annotated but not well-characterized region roughly 2 kilobases downstream of the IG-DMR that the UCSC genome browser predicts is an enhancer. Since we did not measure changes in enhancer activity upon Cebpa deletion, more work is needed to determine how this element is regulated during development as well as repair following injury in the adult lung.
Our investigation into developmental regulation of Dlk1 expression determined that PRC2 and C/EBPα act as a type 2 incoherent feedback loop to generate of pulse of DLK1 following downregulation of the PRC2 complex and likely its repressive epigenetic mark, H3K27me3. While several feedback loops have been postulated and constructed in the field of systems biology64, most of the loops observed in eukaryotic cells are either type 1 coherent or incoherent in configuration64. While the identified loop type (I2-FFL) is observed less frequently, we believe the PRC2-C/EBPα regulation plays a critical role in timing and pattern of alveolar epithelial differentiation, and future studies are needed to test whether reactivation of said loop could be used to re-access AT2 fate plasticity in diseased lungs. We believe this feedback loop constitutes a novel molecular circuit—a “pulse generator” of Notch activation, resulting in spatial patterning of AT1 and AT2 differentiation through lateral inhibition (Supplementary Fig. 5x). Further in vivo investigation of PRC2’s regulation of DLK1 in the lung would help better characterize the I2-FFL loop and could provide insight into how this balance changes in a diseased state. It is also conceivable that disruption of this molecular circuit could result in an aberrant AT2 state that is disease-associated, such as those observed in models of IPF85,86. It also remains to be determined whether AT2s are heterogeneous in their capacity to generate AT1s.
Finally, our investigation into the adult lung demonstrates that C/EBPα downregulation following injury is required for repair and is partly mediated by a C/EBP family member with dominant negative activity, CHOP72. As CHOP is upregulated from various types of injury that cause ER stress87, it is still unclear how C/EBPα downregulation is stimulated following injury of alveolar epithelial cell death, especially as we observe little to no C/EBPα downregulation upon sparse AT2 ablation (Supplementary Fig. 7e). One compelling model requiring further inquiry is that AT1 death sends a specific cue that downregulates C/EBPα to promote fate plasticity in nearby AT2 cells and thus alveolar repair. Interestingly, we found that Cebpa loss also allowed AT2 cells to access what appears to be a pathogen defensive state driven by interferon signaling. While the presence of such a state is expected, as AT2 cells have been implicated in playing a role in immune response in the lung15, said state was not expected to be suppressed by C/EBPα nor to be a mutually exclusive transcriptional state from the well-established regenerative DATP state69,88.
Taken together, these observations suggest that C/EBPα might suppress a larger program that alternates between two states—one regenerative and the other defensive. If true, said program would represent a more robust response to pathogen-mediated injury then previously appreciated, as alternating between regenerative and defense states could ensure enough AT2s are available at a given time to both promote clearance of the pathogen as well as repair damaged alveoli (Fig. 7o).
We will briefly cover the limitations of this study to motivate and direct future work. First, we dissociate murine lungs to form organoids, which, while a common approach in the field10,22,36, might alter cellular phenotypes and promote fate plasticity. While the observed behaviors of cultured DPs and AT2s largely match in vivo experiments10,22,36 and velocity analysis (Fig. 3e–j), future work investigating fate plasticity more precisely in vivo would be informative. Differences observed by normalized gene expression alone would benefit from validation via ISH or immunostaining. As our study does not investigate whether AT1s derived from DPs are distinct from those from AT2s, more work is needed to make said delineation. As conclusions of scRNAseq cluster similarity can be impacted by cut-off points, more work delineating differences in E15.5 DPs in an unbiased manner would be informative. While we focused on the role of C/EBPα, future work on other genes from our screen would likely identify other critical regulators of AT1/2 fate plasticity (see Supplementary Table 3). As BH3 death regulation is studied only to determine its similarity between DPs, nAT2s, and mAT2s, more research is needed to determine its role during development and repair. As CHOP expression was induced using a smaller activator of PERK (CCT020312) that may stimulate off-target molecules, conflating our experiments, further work is needed to precisely investigate the function of CHOP in vivo. Finally, as our experiment revealing C/EBPα’s regulation of a defensive AT2 state did not directly introduce a pathogen, future work is needed to confirm that said state is upregulated following infection.
Methods
Ethics
All mouse experiments followed applicable regulations and guidelines and were approved by the Institutional Animal Care and Use Committee at the Mayo Clinic (Protocol# A00006319-21). Mice were housed under a 12h light/12h dark cycle, and lights were not used during the dark cycle. Mouse housing temperature was kept at 65–75 °F ( ~ 18–23 °C) and humidity at 40–60%. Mice were housed in filtered cages and all experiments were performed in accordance with approved Institutional Animal Care and Use Committee protocols and ethical considerations at the Mayo Clinic.
Mouse strains
Timed-pregnant C57BL/6J females (abbreviated B6; Jackson Laboratories) were used for all embryonic time points, with gestational age verified by crown–rump length. For studies of adult wild-type lungs, B6 males and females were used. Sparse labeling and recombination studies were conducted by Cre recombinase expression using gene-targeted alleles BAC-Nkx2.1Cre, LyzM (also called Lyz2)-Cre, and SftpcCre-ERT2-rtTA58. Cre-dependent target genes were the conditional “floxed” Cebpa89 and Ezh290 for removal of Cebpa and Ezh2, respectively. Two Cre reporters were used: Rosa26mTmG 91 (JAX stock #037456) and TgiSuRe-Cre 51. Upon recombination, the iSure-Cre allele labels the cell with cytoplasmic tdTomato (tdT) and constitutively expresses Cre recombinase in the targeted cell, thus ensuring that all Cre-conditional alleles are recombined, even under sparse or clonal conditions. For initiating recombination with SftpcCre-ERT2 (SftpcCreER), intraperitoneal injection of tamoxifen was administered either at 20 mg/kg for complete (high dose) recombination in adult mice (>PN60), or at 30 ng/kg for sparse recombination in perinatal mice (P0-P21), except where noted. EdU (Cayman Chemicals 20518) was administered via drinking water at 0.1 mg/mL for the indicated period of time. Genomic DNA was extracted from ear punches, followed by Proteinase K (KAPA Biosystems) digestion, and genotyping was performed by polymerase chain reaction (PCR) using published primer sets.
Single-cell RNA-seq bioinformatic analysis
Cell isolation for library preparation
Cells from PN25 SftpcCreER;iSuReCre;Cebpawt/fl and SftpcCreER;iSuReCre;Cebpafl/fl mice (2 weeks after TMX) were isolated and enriched (EpCAM+) as per the protocol described below.
scRNAseq library preparation and processing
Single-cell RNA sequencing was performed using the Chromium Single Cell 3’ system (10x Genomics) at the single-cell sequencing core at Mayo Clinic according to manufacturer’s instruction (10x Genomics). Sequencing depth is an average of 75k reads per cell to detect lowly expressed genes. Fastq files were generated using Cellranger v 7.2. The sequenced files were mapped to the GRCm38/mm10, supplemented with tdTomato, PhiMut, and WPRE transcripts as per the iSuRe-Cre construct51.
scRNAseq data analysis
We used Scanpy92 and Seurat v.4.2.393 for processing and downstream analysis. In the Scanpy pipeline, we identified highly variable genes using the scanpy function scanpy.pp.highly_variable_genes with arguments min_mean = 0.0125, max_mean = 3, and min_disp = 0.5. We restricted to this set of genes for downstream analysis. We further used scanpy.pp.regress_out to regress out unwanted variation in the count of Unique Molecular Identifiers (UMIs) and the percentage of mitochondrial reads. We ran scanpy’s built in Principle Component Analysis (PCA) algorithm with sc.tl.pca and default parameters. We then created a nearest neighbor graph using sc.pp.neighbors with 200 neighbors and 50 Principle Components. We then produced a UMAP dimensionality reduction with scanpy.tl.umap using min_dist = 0.001. We identified cell clusters through unsupervised clustering using the Leiden clustering algorithm through with resolution = 0.8. We assigned cell labels to the clusters based on marker gene expression, and removed several contaminating mesenchymal clusters. These clusters were then visualized on the UMAP.
In the Seurat pipeline, doublets were estimated and removed using the DoubletFinder Package94. Dead cells or outliers were excluded by removing cells with less than 500 features and with more than 10% mitochondria gene content. Post alignment, the Seurat objects were merged, normalized with LogNormalize method, and scaled. Top 3000 variable genes were used to anchor. After PCA dimensional reduction with 30 principal components, we mapped our data on UMAP projection with Louvain clustering algorithm. Subsequently, canonical marker genes for AT1 and AT2 cells95 and sample conditions were used to assign cellular groups.
For RNA velocity, velocyto was used with default run command and run10x sub-command to obtain velocity read counts96. For velocity analysis, we used Scanpy and scVelo packages. The datasets were filtered by expression of <200 genes or >3200 genes, and genes were expressed in <3 cells. The dataset was subsequently normalized and the top 3000 highly variable genes with a minimum 20 expressed count threshold were used in preprocessing. A dynamical model was used to recover velocity estimate following Scvelo default parameters (pp.moments(n_neighbors = 30), tl.recover_dynamics(fit_scaling = False), tl.velocity(mode = ‘dynamical’)).
We then produced velocity stream overlay on UMAP projection and latent time. We used kmeans clustering from the SKlearn package to identify the two predominant directions of velocity vectors within the nAT2 cell cluster. We then calculated an AT1/AT2 score in these clusters based on expression of cell markers. We further performed partition-based graph abstraction (PAGA)97 based on the velocity vectors of each cell type to determine broader cell type linkages98.
For STITCH analysis (to generate SPRING plots), we ported in formatted datasets and followed steps of log normalization, and determined highly variable genes as instructed in a previous example99 in MATLAB 2023a. The visualization was done using ForcedAtlas layout in Gephi v7.2.1.
For analysis of lung development through embryonic, perinatal, and adult timepoints in published scRNAseq datasets, the processed mRNA counts for each cell were used from developing mouse lung datasets, (GSE119228)35, (GSE113320)12, and (GSE149563)39. Analysis of bleomycin-induced lung injury on AT2 cells was performed on a published scRNAseq dataset (GSE141259)20.
Lung isolation and processing
For prenatal time points, individual embryos were staged by fetal crown–rump length before sacrifice and lungs removed en bloc. For postnatal time points, mice were euthanized by either carbon dioxide inhalation or intraperitoneal injection of Pentobarbital (Fatal-Plus Solution) and dissected to both expose the lungs and allow for exsanguination via the abdominal aorta. 5-10 mL of phosphate-buffered saline (PBS; Ca2+ and Mg2+ free, pH 7.4) was gently perfused into the right ventricle of the heart by syringe with a 25-gauge needle until the lungs appeared white. For postnatal time points (≥8 days), the trachea was then cannulated with either a blunt 22- or 25-guage catheter, and the lungs were gently inflated to full capacity with molten low-melting-point agarose (Sigma, 2% in PBS). For all time points, lungs were then separated by lobe and fixed in 4% PFA at 4 °C for either 1 h (E15.5–E16.5), 1.5 h (E17.5), 2 h (E18.5–PN10), or 4 h (>PN10). For sectioning, a vibrating microtome (Leica) was used to generate embryonic (150 µm) or adult (250 µm) tissue sections of uniform thickness.
Immunostaining
Immunohistochemistry was performed as previously described in Treutlein et al.9 using primary antibodies against the following epitopes (used at 1:500 dilution unless otherwise noted): proSPC (rabbit, Sigma AB3786), RAGE (rat, R&D MAB1179), E-cadherin (rat, Life Technologies ECCD-2), PDPN (hamster, DSHB 8.1.1), MUC1 ((hamster, Thermo Scientific HM1630 and rabbit (GeneTex GTX100459)), Alpha-SMA (1:100, mouse IgG2a, Invitrogen 50-9760-82), EZH2 (ENX-1; mouse IgG2b, Santa Cruz Biotechnology sc-166609), KI67 (rat, Thermo Scientific 14-5698-82), RELMα (rabbit, PeproTech 500-P214), CD74 (rat, BD Biosciences 555317), DLK1 (rabbit, R&D Systems MAB8634), C/EBPα (rabbit, Cell Signaling 8178), LAMP1 (rat, DSHB 1D4B-s), and HOPX (rabbit, Abcam ab307671). Briefly, fixed lung slices were permeabilized and blocked overnight in blocking buffer (5% goat serum/PBS/0.5% Triton X-100) at 4 °C. Primary antibody incubation steps were conducted in blocking buffer over 3 nights and washing steps were performed in five rounds (30, 30, 60, 90, and 90 min, respectively) using the blocking buffer. Primary antibodies were subsequently detected using Alexa Fluor-conjugated secondary antibodies used at 1:1000 dilution unless otherwise noted, including Goat anti-rabbit IgG Alexa 555 conjugated (Invitrogen, A21428), Goat anti-rabbit IgG Alexa 647 conjugated (Invitrogen, A32733), Goat anti-rat IgG Alexa 488 conjugated (Invitrogen, A11006), Goat anti-rat IgG Alexa 647 conjugated (Invitrogen, A21247), Goat anti-hamster Alexa 647 (Invitrogen, A21451), and Goat anti-hamster Alexa 594 (Invitrogen, A21113) unless noted otherwise. Secondary antibody incubation steps were conducted in blocking buffer over 2 nights and washing steps were performed in five rounds (30, 30, 60, 90, and 90 min respectively) using PBT (0.1% Tween-20/PBS). For co-staining using antibodies from the same rabbit origins, we performed sequential staining with 1-day anti-rabbit blocking between processes. In brief, we stain with primary and secondary antibodies of the 1st marker as described, and then we block the stained tissue with 40 μg/mL Fab Fragment Goat Anti-Rabbit IgG (Jackson ImmunoResearch, AffiniPure™, 111-007-008) prior to the 2nd marker staining. EdU was detected using the standard Click-iT reaction (Thermo Fisher). Lung slices were then mounted in Vectashield Mounting Medium (Vector labs, H1000) and sealed with a coverslip. Images were acquired using a laser-scanning confocal microscope (Zeiss LSM 780 and Leica Stellaris 8) and subsequently processed using ImageJ.
Timelapse imaging
Fluorescently lineage labeled mouse lung were sectioned at 200 μm thick using compresstome. When imaging, we image sectioned slices in serum-free DMEM media on top of Matrigel for stability, using Timelapse setup in LAS-X software for 24 hr consecutively.
Confocal images 3D reconstruction
We reconstructed 3D representation of cell imaging using IMARIS 10.2.1. The z-stack images are imported as.tif images and reconstructed into 3D representation of tissue images. Marker intensities are individually normalized thresholds for intensity and contrast, smoothing cellular structures for interpretation accuracy, and removal background noise.
Cell isolation and culture
For time points E15.5, E18.5, and PN3, lungs were isolated en bloc without perfusion and pooled by litter (6–8 lungs) for further processing. The lungs were then dissociated in 50 U/mL Dispase (Corning 354235) and triturated with flame-sterilized glass Pasteur pipettes of decreasing diameters (1000 µm, 800 µm, and 600 µm) to achieve a single-cell suspension. For PN25 and adult time points, the lung was perfused with DPBS (Ca2+ and Mg2+ free, pH 7.4) as described above. The trachea was punctured and then cannulated with a blunt 22-guage catheter, and the lungs were inflated to full capacity with Dispase (~1 mL), and the trachea was sealed using a non-absorbable silk suture. The inflated lobes were incubated in Dispase for 45 min at 25 °C on a shaker.
For all time points, digested lungs were minced with a razor blade, suspended in 7 mL of digestion buffer (RPMI-1640 containing FBS (10%, Corning), penicillin-streptomycin (1 U/mL, Thermo Scientific), and DNase I (0.33 U/mL, Roche)), incubated at 4 °C for 10 min on a flat shaker and triturated briefly with a 5-mL pipette.
To deplete red blood cells, an equal volume of AT2 isolation media (RPMI-1640 supplemented with penicillin-streptomycin (1 U/mL, Thermo Scientific) and DNase I (0.33 U/mL, Roche)) was added to the lung single-cell suspension. The suspension was passed through a 100 µm mesh filter to remove residual tissue fragments and centrifuged at 400 × g, 4 °C for 10 min. Pelleted cells were resuspended in 5 mL of red blood lysis buffer (BD Biosciences), incubated for 2 min at room temperature, and neutralized with an equal volume of AT2 isolation media. The suspension was passed through a 40 µm mesh filter (Fisher), centrifuged at 400 × g, 4 °C for 10 min, and then resuspended in MACS buffer (2 mM EDTA, 0.5% BSA in PBS, filtered and degassed) for purification.
The resultant single-cell suspension was incubated with Mouse Fc Blocking Reagent (Miltenyi Biotec, 130-092-575) for 5 min at 4 °C. Other cell types were first depleted with antibodies against CD45 (eBioscience, 13-0451-85), CD31 (BD Biosciences, 557355), Ter119 (eBioscience, 13-5921-85), CD140a (Miltenyi Biotec, 130-101-905), and CD104 (Miltenyi Biotec, 130-106-922), followed by anti-biotin (Miltenyi Biotec, 130-090-485) magnetic MicroBeads. Cell suspensions were passed through a 35 µm cell strainer (BD Biosciences) prior to being loaded on LD columns (Miltenyi Biotec) as per the vendor’s protocol. The flow-through was centrifuged at 300 × g, 4 °C for 5 min and the pelleted cells were resuspended in MACS buffer.
For positive selection of cell types, the following antibodies were used: anti-EpCAM (clone G8.8, eBioscience, 13-5791-82) for E15.5 distal bipotent progenitors, anti-RELMα (PeproTech, 500-P214BT) for E18.5 nascent AT2 cells, and either anti-CD74 or anti-MHC-II (clone M5/114.15.2, eBioscience, 13-5321-82) for mature AT2 cells. Following antibody incubation, the cells were then incubated with either anti-biotin or anti-rat IgG MicroBeads and loaded on MS columns (Miltenyi Biotec) as per the vendor’s protocol. The flow-through was discarded and the column was taken off the magnet. The cells remaining in the column were eluted with MACS buffer, resulting in a highly enriched preparation of either bipotent progenitors (EpCAM+), nascent AT2s (RELMɑ+), or mature AT2s (MHC-II+), unless noted otherwise.
For culturing bipotent progenitors, nAT2s, or mAT2s, cell density and viability were calculated using a Vi-CELL XR Cell Viability Analyzer (Beckman Coulter). Cells were cultured in eight-well #1.5 coverglass chambers (Cellvis, C8-1.5H-N), pre-coated with growth factor-reduced Matrigel (85 µL, Corning 354230) for 30 min at 37 °C. The cells were plated at varying densities: 30,000 cells/well (for E15.5 and E18.5), 60,000 cells/well for PN3), and 80,000 cells/well (for PN25 and adult). Cells were supplemented with FGF7 (50 ng/mL, R&D Systems 5028-KG-025), GSK126 (10 µM, ApexBio A3446), KL001 (12 µM, MedChemExpress HY-108468), and interferon-γ (20 ng/mL, PeproTech 315-05) to the indicated concentrations in 400 µL of DMEM/F12 supplemented with penicillin-streptomycin (1 U/mL, Thermo Scientific). Excluding FGF7, all other small molecules listed above were added after a 12 h incubation period to allow for cells to seed and adhere to the layer of Matrigel. Cells were maintained in an incubator at 37 °C with 5% CO2/air, with media changes every other day, typically for 4 days, except where indicated otherwise.
Flow cytometry
We perform flow cytometry of AT2 cells using Miltenyi’s Tyto flow sorter. Filtered and RBC depleted lung cell isolates are to be incubated with 20 µL of each of the following 1:50 diluted primary antibodies: anti-RELMɑ, anti-CD74, and anti-MHC-II for 30 min at 4 °C. Cells are washed with sorting buffer (Miltenyi, 130-107-207) and subsequently incubated with 20 µL of each of the following secondary antibodies: (Anti-rabbit Alexa Fluor 647, anti-rat Alexa Fluor 405) for 30 min at 4 °C. Subsequently, the samples are washed, filtered, and resuspended in 1 mL sorting buffer and transferred to the sorting compartment of the cartridge (Miltenyi, 130-104-791). Gates for markers positivity are drawn using unstained cells as negative controls.
BH3 analysis
Lung samples from mice of different ages (E15.5, E18.5, and ≥PN60) were dissociated into a single-cell suspension and enriched using relevant markers as described above. Cells were stained on ice for 25 min away from light, then centrifuged at 200 × g for 5 min, and subjected to flow cytometry–based BH3 profiling as previously described by the Sarosiek lab80. Briefly, cells were treated with activator or sensitizer BH3 peptides (New England Peptide) for 60 min at 28 °C in mannitol experimental buffer (MEB) [10 mM Hepes (pH 7.5), 150 mM mannitol, 50 mM KCl, 0.02 mM EGTA, 0.02 mM EDTA, 0.1% BSA, and 5 mM succinate] with 0.001% digitonin. Peptide sequences are as follows: BIM (Ac-MRPEIWIAQELRRIGDEFNA-NH2), BID (Ac-EDIIRNIARHLAQVGDSMDRY-NH2), PUMA (p53 upregulated modulator of apoptosis) (Ac-EQWAREIGAQLRRMADDLNA-NH2), BAD (Ac-LWAAQRYGRELRRMSDEFEGSFKGL-NH2), Hrk (Harakiri) (Ac-WSSAAQLTAARLKALGDELHQ-NH2), and MS1 (MCL-1 specific) (Ac-RPEIWMTQGLRRLGDEINAYYAR-NH2). After peptide exposure, cells were fixed in 2% PFA for 15 min which was then neutralized by addition of N2 buffer [1.7 M Tris base and 1.25 M glycine (pH 9.1)]. Cells were stained overnight with DAPI (1:1000, Abcam) and anti–cytochrome c–Alexa Fluor 647 (1:2000, clone 6H2.B4, BioLegend) in a saponin-based buffer (final concentration, 0.1% saponin; 1% BSA) and then analyzed by flow cytometry. Cytochrome c release in response to BIM treatment was measured on an Attune NxT flow cytometer (Thermo Fisher Scientific).
Conditional deletion of Cebpa and Ezh2 in vivo
For conditional and efficient recombination of floxed alleles (Cebpaflox or Ezh2flox) at different time points and cell types, we bred the floxed allele into one of four mouse lines, each with a distinct combination of Cre and reporter alleles. For in vivo sparse cell labeling with efficient recombination of either Cebpaflox or Ezh2flo alleles in distal progenitors, the TgNkx2.1-Cre; Rosa26mTmG/mTmG mouse line was used as we have previously found it sparsely labels DPs with efficient flox allele recombination10. For sparse labeling of nAT2s with efficient recombination of Cebpaflox, the LyzMCre; Rosa26mTmG/mTmG mouse line was used as previously described10. For broad labeling of AT2 cells with efficient Cebpaflox recombination following a high dose of tamoxifen (20 mg/kg), the SftpcCre-ERT2; Rosa26mTmG/mTmG mouse line was used. Finally, to sparsely label AT2s and retain efficient Cebpaflox recombination following a low dose of tamoxifen (30 ng/kg), the SftpcCre-ERT; TgiSuRe-Cre mouse line was used. Upon its recombination, the TgiSuRe-Cre allele constitutively expresses both the fluorescent lineage label tdTomato as well as Cre recombinase, an approach that has been well established to efficiently recombine multiple floxed under lower tamoxifen doses for targeting sparse cells51. To determine the proliferation of AT2 cells, EdU (1 mg/mL) was administered in the drinking water for the indicated period of time.
ATAC-seq and CHIP-seq analysis
Cells from PN25 SftpcCreER; TgiSuRe-Cre; Cebpawt/fl and SftpcCreER; TgiSuRe-Cre; Cebpafl/fl mice (2 weeks after TMX) were isolated and enriched (EpCAM+) as per the protocol described above. Raw sequencing reads were trimmed of sequencing adapters and bad quality bases with cutadapt version 2.8 in Trim Galore version 0.6.6 using default parameters. Bowtie2 version 2.5.0 (parameters: --end-to-end --very-sensitive --no-mixed --no-discordant --phred33 -I 10 -X 700) was used to map trimmed reads to the reference genome Mus_musculus.GRCm38. Picard MarkDuplicates (v.2.26.10; http://broadinstitute.github.io/picard) was used for duplicate removal. Reads were shifted by +4 bp for those mapping to the positive strand and −5 bp for those mapping to the negative strand. ATAC-seq analysis was performed with HOMER(v 4.11). Tag directories were made for each ATAC-seq run using HOMER100 “makeTagDirectory”. Peaks for individual replicates were identified using the “findPeaks” command. To generate high-confidence peak lists across both replicate samples for downstream analysis, ATAC-seq peaks were identified using the HOMER “getDifferentialPeaksReplicates.pl” command. This command uses DESeq2101,102 and identifies peaks that pass 2-fold enrichment and false discovery rate (FDR) < 0.05 cutoffs. For these analyses, peak lists combining all peaks from the comparison conditions were assembled. One condition was used as the target, and the other was used as background. To characterize enriched motifs near differentially enriched peaks, we used HOMER’s “findMotifsGenome.pl” command to scan the entire peak by using “-size given” with standard background. ATAC-seq peak annotation was performed using the homer annotatePeaks.pl function to assign the nearest gene name to the peaks. To estimate the enrichment of transcription factors and epigenetic modifications on the murine genome, we used the Cistrome DB Toolkit analysis browser63,103 using default settings.
Incoherent feed-forward loop type 2 model fitting
To assess whether the temporal expression pattern of Dlk1 fit that predicted by a feed-forward loop we used the publicly available biocircuits package (https://biocircuits.github.io/) to numerically solve the differential equations for a type 2, incoherent feed-forward loop following a step reduction of PRC2. After solving for a function over time for Dlk1, an R script was generated to align its peak value to that observed in the scRNAseq timecourse and calculate a goodness of fit by root mean squared error (RMSE). Parameters of the I2-FFL model were independently varied until a good fit was reached (beta = 1.9, gamma = 5.5, and kappa = 3).
Mouse lung injury experiments and viral transfections
Lung injury was induced in adult mice by single-dose administration of bleomycin sulfate (0.25 mg/kg; VWR J60727-MA), which was dissolved in sterile saline and administered via intratracheal (i.t.) administration. Lungs were isolated 16 days post-injury and processed for immunostaining.
To test whether C/EBPα is required for proper injury repair, adult Rosa26mTmG/mTmG mice were administered either an AAV-Sftpc-Cre (control), or an AAV-Sftpc-Cebpa-T2A-Cre (prepared by the Boston Children’s Hospital Viral Vector Core (1.5 × 1014 GC (genome copies)/mL, serotype 2/9) and intratracheally instilled (5 µL of viral solution diluted in 45 µL PBS)) to overdrive Cebpa expression and induce GFP labeling of AT2 lineage. Details on plasmid construction and composition is provided in Supplementary Data 1. A minimal 320 bp Sftpc promoter element was used for both AAV preps. Five days later, all mice were administered Fiin1 hydrochloride (14.75 mg/kg; Tocris 4002) via intratracheal instillation as described in our prior work10 or BHT (225 mg/kg; Sigma B1378) via intraperitoneal injection73 to induce lung injury. Lungs were isolated 14 days post-injury and processed for immunostaining. For all intratracheal instillation experiments, mice were anesthetized with isoflurane and kept in recovery under a heat lamp.
Small molecule induction of CHOP
To induce CHOP expression in AT2 cells, SftpcCreERT2-rtTA; Rosa26mTmG/mTmG mice were first administered tamoxifen (20 mg/kg) at PN11 for labeling of AT2 cells with GFP. Twenty-four hours later, mice were either given corn oil (vehicle) or CCT020312 (MedChem Express HY-119240), a PERK activator found to upregulate CHOP expression104, at 2 mg/kg dissolved in corn oil via intraperitoneal injection every 4 days (4 doses total). Lungs were isolated 2 weeks later and processed for immunostaining.
Statistics and reproducibility
Data analysis and all statistical tests were performed with GraphPad Prism software or R. Biological replica of 3 were conducted unless specified otherwise. Replicate experiments were biological replicated with a mix of male and female mice randomly selected, and the quantitative values are presented as mean ± SD unless indicated otherwise. In graphs displayed as box plots, the horizontal lines represent the median and error bars depict the 75th–25th interquartile range (IQR). Two-sided Student’s t tests (for normally distributed data), Mann–Whitney U-tests (for data not normally distributed), Brown–Forsythe, Welch ANOVA or Kruskal–Wallis (for comparisons between ≥3 groups) with Dunnett’s multiple comparisons test were used to determine p values. No statistical method was used to predetermine sample size, and data distribution was tested for normality prior to statistical analysis and plotting.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors confirm that all relevant data are available in the paper or its Supplementary Information files. The scRNAseq datasets are available in the GEO repository under accession numbers “GSE119228”, “GSE113320”, “GSE109774”, and “GSE149563” for developing and adult mouse lung cells, “GSE295269” for CEBP/α knockout mouse lung epithelial cells, “GSE141259” for adult bleomycin injury, “GSE218666” for AT1 injury, “GSE184384” for influenza injury, and “GSE113049” for LPS injury models. The bulk ATACseq datasets are available in the GEO repository under accession number “GSE296786”. Source data are provided with this paper.
Code availability
Code to reproduce the analyses described in this manuscript can be accessed at https://github.com/JimmyMayoGit/Sawhney_2025.
References
Barnes, P. J. et al. Chronic obstructive pulmonary disease. Nat. Rev. Dis. Prim. 1, 15076 (2015).
Martinez, F. J. et al. Idiopathic pulmonary fibrosis. Nat. Rev. Dis. Prim. 3, 17074 (2017).
Siegel, R., Naishadham, D. & Jemal, A. Cancer statistics, 2013. CA Cancer J. Clin. 63, 11–30 (2013).
Desai, T. J., Brownfield, D. G. & Krasnow, M. A. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–194 (2014).
Franks, T. J. et al. Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34, 743–748 (2003).
Hwang, D. M. et al. Pulmonary pathology of severe acute respiratory syndrome in Toronto. Mod. Pathol. 18, 1–10 (2005).
Tian, S. et al. Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 15, 700–704 (2020).
Rawlins, E. L., Clark, C. P., Xue, Y. & Hogan, B. L. The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–3745 (2009).
Treutlein, B. et al. Reconstructing lineage hierarchies of the distal lung epithelium using single-cell RNA-seq. Nature 509, 371–375 (2014).
Brownfield, D. G. et al. Alveolar cell fate selection and lifelong maintenance of AT2 cells by FGF signaling. Nat. Commun. 13, 7137 (2022).
Zacharias, W. J. et al. Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251–255 (2018).
Frank, D. B. et al. Early lineage specification defines alveolar epithelial ontogeny in the murine lung. Proc. Natl. Acad. Sci. USA 116, 4362–4371 (2019).
Vila Ellis, L. & Chen, J. A cell-centric view of lung alveologenesis. Dev. Dyn. 250, 482–496 (2021).
Weibel, E. R. On the tricks alveolar epithelial cells play to make a good lung. Am. J. Respir. Crit. Care Med. 191, 504–513 (2015).
Shenoy, A. T. et al. Antigen presentation by lung epithelial cells directs CD4+ TRM cell function and regulates barrier immunity. Nat. Commun. 12, 5834 (2021).
Choi, J. et al. Inflammatory signals induce AT2 cell-derived damage-associated transient progenitors that mediate alveolar regeneration. Cell Stem Cell 27, 366–382.e367 (2020).
Malik, A. et al. Epithelial IFNγ signalling and compartmentalized antigen presentation orchestrate gut immunity. Nature 623, 1044–1052 (2023).
Gessani, S., Conti, L., Del Cornò, M. & Belardelli, F. Type I interferons as regulators of human antigen presenting cell functions. Toxins 6, 1696–1723 (2014).
Rock, J. R. et al. Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. Proc. Natl. Acad. Sci. USA 108, E1475–E1483 (2011).
Strunz, M. et al. Alveolar regeneration through a Krt8+ transitional stem cell state that persists in human lung fibrosis. Nat. Commun. 11, 3559 (2020).
Barkauskas, C. E. et al. Type 2 alveolar cells are stem cells in adult lung. J. Clin. Investig. 123, 3025–3036 (2013).
Nabhan, A. N., Brownfield, D. G., Harbury, P. B., Krasnow, M. A. & Desai, T. J. Single-cell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123 (2018).
Finn, J. et al. Dlk1-mediated temporal regulation of notch signaling is required for differentiation of alveolar type II to type I cells during repair. Cell Rep. 26, 2942–2954.e2945 (2019).
Major, J. et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science 369, 712–717 (2020).
Jiang, P. et al. Ineffectual type 2-to-type 1 alveolar epithelial cell differentiation in idiopathic pulmonary fibrosis: persistence of the KRT8(hi) transitional state. Am. J. Respir. Crit. Care Med. 201, 1443–1447 (2020).
Lourenco, A. R. et al. C/EBPa is crucial determinant of epithelial maintenance by preventing epithelial-to-mesenchymal transition. Nat. Commun. 11, 785 (2020).
Suh, H. C. et al. C/EBPalpha determines hematopoietic cell fate in multipotential progenitor cells by inhibiting erythroid differentiation and inducing myeloid differentiation. Blood 107, 4308–4316 (2006).
Baladron, V. et al. dlk acts as a negative regulator of Notch1 activation through interactions with specific EGF-like repeats. Exp. Cell Res. 303, 343–359 (2005).
Grassi, E. S. & Pietras, A. Emerging roles of DLK1 in the stem cell niche and cancer stemness. J. Histochem. Cytochem. 70, 17–28 (2022).
Yen, Y. P. et al. Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity. Elife 7, e38080 (2018).
Shivram, H., Le, S. V. & Iyer, V. R. PRC2 activates interferon-stimulated genes indirectly by repressing miRNAs in glioblastoma. PLoS ONE 14, e0222435 (2019).
Kojima, S. et al. Epigenome editing reveals core DNA methylation for imprinting control in the Dlk1-Dio3 imprinted domain. Nucleic Acids Res. 50, 5080–5094 (2022).
Zhao, J. et al. PPARgamma and C/EBPalpha enable adipocyte differentiation upon inhibition of histone methyltransferase PRC2 in malignant tumors. J. Biol. Chem. 300, 107765 (2024).
Auyeung, V. C. et al. IRE1alpha drives lung epithelial progenitor dysfunction to establish a niche for pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 322, L564–L580 (2022).
Cohen, M. et al. Lung single-cell signaling interaction map reveals basophil role in macrophage imprinting. Cell 175, 1031–1044.e1018 (2018).
Gillich, A. St. et al. Alveoli form directly by budding led by a single epithelial cell. bioRxiv https://doi.org/10.1101/2021.12.25.474174 (2021).
Blanco, M. A. et al. Chromatin-state barriers enforce an irreversible mammalian cell fate decision. Cell Rep. 37, 109967 (2021).
Hurley, K. et al. Reconstructed single-cell fate trajectories define lineage plasticity windows during differentiation of human PSC-derived distal lung progenitors. Cell Stem Cell 26, 593–608.e598 (2020).
Zepp, J. A. et al. Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371, eabc3172 (2021).
Sarosiek, K. A. et al. Developmental regulation of mitochondrial apoptosis by c-Myc governs age- and tissue-specific sensitivity to cancer therapeutics. Cancer Cell 31, 142–156 (2017).
Singh, R. et al. Radiotherapy-induced neurocognitive impairment is driven by heightened apoptotic priming in early life and prevented by blocking BAX. Cancer Res. 83, 3442–3461 (2023).
Basundra, R. et al. Constitutive high expression of NOXA sensitizes human embryonic stem cells for rapid cell death. Stem Cells 40, 49–58 (2022).
The Tabula Muris Consortium. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Zhang, Z. et al. Transcription factor Etv5 is essential for the maintenance of alveolar type II cells. Proc. Natl. Acad. Sci. USA 114, 3903–3908 (2017).
Metzger, D. E., Stahlman, M. T. & Shannon, J. M. Misexpression of ELF5 disrupts lung branching and inhibits epithelial differentiation. Dev. Biol. 320, 149–160 (2008).
Santofimia-Castaño, P. et al. Inactivation of NUPR1 promotes cell death by coupling ER-stress responses with necrosis. Sci. Rep. 8, 16999 (2018).
Liu, J. et al. NUPR1 is a critical repressor of ferroptosis. Nat. Commun. 12, 647 (2021).
Basseres, D. S. et al. Respiratory failure due to differentiation arrest and expansion of alveolar cells following lung-specific loss of the transcription factor C/EBPalpha in mice. Mol. Cell Biol. 26, 1109–1123 (2006).
Martis, P. C. et al. C/EBPalpha is required for lung maturation at birth. Development 133, 1155–1164 (2006).
Morimoto, M. & Kopan, R. rtTA toxicity limits the usefulness of the SP-C-rtTA transgenic mouse. Dev. Biol. 325, 171–178 (2009).
Fernández-Chacón, M. et al. iSuRe-Cre is a genetic tool to reliably induce and report Cre-dependent genetic modifications. Nat. Commun. 10, 2262 (2019).
Germani, F., Bergantinos, C. & Johnston, L. A. Mosaic analysis in Drosophila. Genetics 208, 473–490 (2018).
Burke, R. & Basler, K. Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development 122, 2261–2269 (1996).
Hassan, D. & Chen, J. CEBPA restricts alveolar type 2 cell plasticity during development and injury-repair. Nat. Commun. 15, 4148 (2024).
Nueda, M. L., Baladrón, V., Sánchez-Solana, B., Ballesteros, M. A. & Laborda, J. The EGF-like protein dlk1 inhibits notch signaling and potentiates adipogenesis of mesenchymal cells. J. Mol. Biol. 367, 1281–1293 (2007).
Ramji, D. P. & Foka, P. CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 (2002).
Little, D. R. et al. Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo. Nat. Commun. 12, 2509 (2021).
Aronson, B. E. et al. A bipartite element with allele-specific functions safeguards DNA methylation imprints at the Dlk1-Dio3 locus. Dev. Cell 56, 3052–3065.e3055 (2021).
Hagan, J. P., O’Neill, B. L., Stewart, C. L., Kozlov, S. V. & Croce, C. M. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS ONE 4, e4352 (2009).
Visel, A., Thaller, C. & Eichele, G. GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–D556 (2004).
Sanli, I. et al. Meg3 non-coding RNA expression controls imprinting by preventing transcriptional upregulation in cis. Cell Rep. 23, 337–348 (2018).
He, Y. et al. Spatiotemporal DNA methylome dynamics of the developing mouse fetus. Nature 583, 752–759 (2020).
Zheng, R. et al. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 47, D729–D735 (2018).
Alon, U. An Introduction to Systems Biology: Design Principles of Biological Circuits 2nd edn (CRC Press, Taylor & Francis Group, 2020).
Chitnis, A. B. The role of Notch in lateral inhibition and cell fate specification. Mol. Cell Neurosci. 6, 311–321 (1995).
Ochi, S., Imaizumi, Y., Shimojo, H., Miyachi, H. & Kageyama, R. Oscillatory expression of Hes1 regulates cell proliferation and neuronal differentiation in the embryonic brain. Development 147, dev182204 (2020).
Nueda, M. L., Naranjo, A. I., Baladron, V. & Laborda, J. The proteins DLK1 and DLK2 modulate NOTCH1-dependent proliferation and oncogenic potential of human SK-MEL-2 melanoma cells. Biochim. Biophys. Acta 1843, 2674–2684 (2014).
Milara, J. et al. Sphingosine-1-phosphate is increased in patients with idiopathic pulmonary fibrosis and mediates epithelial to mesenchymal transition. Thorax 67, 147–156 (2012).
Kobayashi, Y. et al. Persistence of a regeneration-associated, transitional alveolar epithelial cell state in pulmonary fibrosis. Nat. Cell Biol. 22, 934–946 (2020).
Jiang, H. Y. et al. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell Biol. 24, 1365–1377 (2004).
Ma, Y., Brewer, J. W., Diehl, J. A. & Hendershot, L. M. Two distinct stress signaling pathways converge upon the CHOP promoter during the mammalian unfolded protein response. J. Mol. Biol. 318, 1351–1365 (2002).
Ron, D. & Habener, J. F. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 6, 439–453 (1992).
Konkimalla, A. et al. Transitional cell states sculpt tissue topology during lung regeneration. Cell Stem Cell 30, 1486–1502.e1489 (2023).
Beppu, A. K. et al. Epithelial plasticity and innate immune activation promote lung tissue remodeling following respiratory viral infection. Nat. Commun. 14, 5814 (2023).
Riemondy, K. A. et al. Single cell RNA sequencing identifies TGFbeta as a key regenerative cue following LPS-induced lung injury. JCI Insight 5, e123637 (2019).
Goss, A. M. et al. Wnt2/2b and beta-catenin signaling are necessary and sufficient to specify lung progenitors in the foregut. Dev. Cell 17, 290–298 (2009).
Li, J. et al. The strength of mechanical forces determines the differentiation of alveolar epithelial cells. Dev. Cell 44, 297–312.e295 (2018).
Konkimalla, A. et al. Multi-apical polarity of alveolar stem cells and their dynamics during lung development and regeneration. iScience 25, 105114 (2022).
Alanis, D. M., Chang, D. R., Akiyama, H., Krasnow, M. A. & Chen, J. Two nested developmental waves demarcate a compartment boundary in the mouse lung. Nat. Commun. 5, 3923 (2014).
Inde, Z. et al. Age-dependent regulation of SARS-CoV-2 cell entry genes and cell death programs correlates with COVID-19 severity. Sci. Adv. 7, eabf8609 (2021).
Hassan, D. & Chen, J. CEBPA restricts alveolar type 2 cell plasticity during development and injury-repair. bioRxiv https://doi.org/10.1101/2023.10.10.561625 (2023).
Penkala, I. J. et al. Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell 28, 1775–1789.E1775 (2021).
Sprinzak, D. et al. Cis-interactions between Notch and Delta generate mutually exclusive signalling states. Nature 465, 86–90 (2010).
Kagami, M. et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet. 6, e1000992 (2010).
Yao, C. et al. Senescence of alveolar type 2 cells drives progressive pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 203, 707–717 (2021).
Wang, F. et al. Regulation of epithelial transitional states in murine and human pulmonary fibrosis. J. Clin. Invest. 133, e165612 (2023).
Kaufman, R. J. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev. 13, 1211–1233 (1999).
Toth, A. et al. Alveolar epithelial progenitor cells require Nkx2-1 to maintain progenitor-specific epigenomic state during lung homeostasis and regeneration. Nat. Commun. 14, 8452 (2023).
Zhang, P. et al. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity 21, 853–863 (2004).
Shen, X. et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell 32, 491–502 (2008).
Muzumdar, M. D., Tasic, B., Miyamichi, K., Li, L. & Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605 (2007).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
Haber, A. L. et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498 (2018).
Wolf, F. A. et al. PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59 (2019).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414 (2020).
Wagner, D. E. et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018).
Heinz, S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010).
Gontarz, P. et al. Comparison of differential accessibility analysis strategies for ATAC-seq data. Sci. Rep. 10, 10150 (2020).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Mei, S. et al. Cistrome Data Browser: a data portal for ChIP-Seq and chromatin accessibility data in human and mouse. Nucleic Acids Res. 45, D658–D662 (2016).
Lei, Y., He, L., Yan, C., Wang, Y. & Lv, G. PERK activation by CCT020312 chemosensitizes colorectal cancer through inducing apoptosis regulated by ER stress. Biochem. Biophys. Res. Commun. 557, 316–322 (2021).
Acknowledgements
The authors thank members of the Brownfield laboratory for helpful discussions and critical reading of the manuscript. We thank Daniel Tenen (Cebpafl/fl) for the transgenic mouse strain; Hannah Golding for performing organotypic culture experiments; and Gamaliel Taengwa for data quantification. This work was supported by R00HL127267, R01HL171056, and a Pilot Award from Mayo Clinic’s Center for Biomedical Discovery (D.G.B.).
Author information
Authors and Affiliations
Contributions
A.S.S., B.J.D. and D.G.B. conceived the study and designed the experiments. A.S.S., B.J.D., J.C., D.G., G.A., A.O., H.H., L.S., X.Q., S.H., R.S. and A.U. performed the experiments. A.S.S., B.J.D. and A.O. analyzed and interpreted the data. J.C., A.H., E.L. and D.G.B. performed the scRNAseq analysis. D.G., L.W. and Z.W. performed the ATACseq analysis. R.B. provided the iSuRe-Cre mouse line. R.W. and S.D.L. performed bleomycin injury experiments, and K.A.S. and Z.I. performed BH3 profiling experiments. D.G.B., A.S.S. and B.J.D. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sawhney, A.S., Deskin, B.J., Cai, J. et al. A molecular circuit regulates fate plasticity in emerging and adult AT2 cells. Nat Commun 16, 8924 (2025). https://doi.org/10.1038/s41467-025-64224-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64224-1