Abstract
Epigenetic dysregulation, including accumulation of Histone H3 lysine 27 acetylation (H3K27ac), is a hallmark of pVHL-deficient clear cell Renal Cell Carcinomas (ccRCCs). Using an in vivo positive selection ORF screen in poorly tumorigenic pVHL-proficient cells and mechanistic studies in pVHL-deficient cells, we discovered that the aspartate (Asp) and glutamate (Glu) transporter, SLC1A1/EAAT3, is a metabolic dependency in ccRCC. pVHL loss promotes Hypoxia Inducible Factor (HIF)-independent SLC1A1 expression via H3K27ac dysregulation. SLC1A1 inactivation, genetically or pharmacologically, depletes Asp/Glu-derived metabolites (e.g., Tricarboxylic acid cycle and nucleotide intermediates), impedes ccRCC growth, and sensitizes ccRCCs to anti-metabolite drugs (e.g., glutaminase blockers). In human tumors, higher SLC1A1 expression is associated with reduced immune infiltration, oncogenic metabolic programs, and advanced stage/metastatic disease. Finally, in ccRCC animal models, SLC1A1 inactivation diminishes lung metastasis and the outgrowth of established renal tumors. Altogether, our studies credential SLC1A1 as an actionable, HIF-independent, metabolic dependency in pVHL-deficient ccRCCs.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is among the most common forms of cancer in men and women, accounting for nearly 80,000 new diagnoses annually1. RCC occurs in three major subtypes: clear cell RCCs (ccRCCs), which have the hallmark loss of the von Hippel–Lindau tumor suppressor protein (pVHL); and the papillary (pRCC) and chromophobe (chRCC) cancers, which are typically pVHL-proficient2.
pVHL canonically functions in a ubiquitin ligase (E3) complex3, which is perhaps best-known for the oxygen-dependent proteolysis of the α-subunit of the hypoxia inducible factor (HIFα) transcription factors4,5. Therefore, pVHL-deficient ccRCCs exhibit chronic HIFα activation, and these hyperactivated HIF transcription programs, particularly those controlled by HIF2α, drive ccRCC oncogenesis6,7. These observations have justified the development of pharmacological HIF2α inhibitors8,9, one of which is now FDA approved for the treatment of both familial (VHL syndrome-associated) and sporadic ccRCC10. However, because of genetic heterogeneity and/or acquired resistance, HIF2α inhibition alone is often not curative, and HIF-independent therapeutic strategies that complement HIF2α blockade are clinically relevant.
Chromatin dysregulation is another hallmark of ccRCC and occurs via both HIF-dependent and independent mechanisms11. HIF-dependent mechanisms include the transcriptional activation of several JumonjiC-family histone demethylases (KDMs)12, whereas HIF-independent mechanisms include the genetic inactivation of SWI/SNF-family chromatin modifiers, such as PBRM113; or inactivation of histone modifiers, such as SETD2, MLL3, KDM5C, and KDM6A14. These changes select for oncogenic epigenetic programs in ccRCC, but also render the tumors dependent on the compensatory function of other chromatin/histone modifiers for survival15. Epigenetic dysregulation, therefore, represents an exploitable vulnerability in kidney cancer.
To address the oncogenic importance of epigenetic dysfunction, we previously profiled changes in histone modification patterns in pVHL-deficient ccRCCs (vs the pVHL-proficient pRCCs and chRCCs) and observed that ccRCCs have elevated levels of acetylated lysine 27 on Histone H3 (H3K27ac)15. H3K27ac is associated with transcriptional activation and its accumulation at cis-regulatory elements (e.g., promoters and enhancers/super-enhancers) marks master regulators of cellular identity and oncogenesis in many cancers16,17. In ccRCC, specific alterations in H3K27ac have been previously linked to tumorigenesis18 and metastatic progression19. Importantly, these earlier studies largely relied on the HIF2α-dependent 786-O cells (or their metastatic derivatives), perhaps inadvertently missing the importance of HIF-independent epigenetic programs. Altogether, we hypothesized that H3K27ac marks critical genes in pVHL-deficient ccRCCs that sustain tumorigenic and metastatic programs via both HIF-dependent and independent mechanisms. Here, using genomics analysis and functional in vivo and cell-based studies, we linked pVHL-dependent epigenetic dysregulation to HIF-independent control of the SLC1A1/EAAT3 transporter, establishing an actionable target in ccRCC.
Results
Dysregulated H3K27ac patterns identify candidate ccRCC oncogenes
Reintroduction of pVHL into pVHL-deficient ccRCC cell lines does not prominently impact their in vitro growth under standard culture conditions. Therefore, lentivirally transducing pVHL (or empty vector, as a control) into pVHL-deficient ccRCCs has allowed us to previously generate isogenic pairs of a number of ccRCC cell lines15. Using one representative isogenic pair—the HIF2α-independent UMRC-2 cells, with or without pVHL expression – we probed pVHL-dependent changes in H3K27ac using ChIP-Seq analysis. We annotated 72,243 peaks, including 8359 peaks that were decreased [Fold Change (FC) ≤ −1.5] in pVHL-proficient (VHL) vs pVHL-deficient (VEC) cells (Fig. 1a).
a Heatmap representing the H3K27ac peaks that were decreased (≥1.5 fold) in UMRC-2 cells upon expression of pVHL (VHL) vs empty vector (VEC) control. b–d H3K27ac signal at the CA9 (b), SHMT2 (c), or TLR3 (d) proximal regions in pVHL-proficient (VHL) and pVHL-deficient (VEC) versions of two ccRCC cell lines (UMRC-2 and 786-O). e–g H3K27ac signal at the CA9 (e), SHMT2 (f), or TLR3 (g) proximal regions, as described in (b–d), in human ccRCC tumors vs their paired adjacent normal tissue. h, i Log2[fold-change (FC)] in H3K27ac signal between pVHL-proficient vs pVHL-deficient UMRC-2 cells at promoter regions (h) or enhancers/super-enhancers (SE) (i). j Volcano plot of RNA-Seq data showing differentially expressed genes in cells described in (h, i). Adjustments for multiple comparisons were made using the quasi-likelihood pipeline implemented by EdgeR. k Venn diagram indicating the overlap between pVHL-dependent changes in H3K27ac in the promoter and enhancer/super-enhancers (SE) and changes in mRNA abundance to generate a candidate gene list. For Promoter and SE datasets, FC ≥ 1.5 was used as a cut-off; whereas for the mRNA dataset, FC ≥ 2.0 was used as a cut-off.
The genes harboring elevated H3K27ac in the pVHL-deficient cells not only included the expected canonical HIFα target genes (e.g., CA9, NDRG1, and EGLN3), but also previously characterized ccRCC oncogenes (e.g., SHMT220,21 and ZHX222) and genes that had not been associated with ccRCC (e.g., TLR3 and SLC28A1) (Fig. 1b–d; Supplementary Fig. 1a–d; and Supplementary Data 1). We then compared the UMRC-2-derived H3K27ac maps with the 786-O human ccRCC cell lines. Despite the lower signal in 786-O cells, we found virtually identical pVHL-dependent H3K27ac marking at all HIF target genes (Fig. 1b–d and Supplementary Fig. 1a–d), ~60% overlap of the H3K27ac peaks at superenhancers (Supplementary Fig. 1e), and ~36% overlap at all other H3K27ac sites (Supplementary Fig. 1f). Importantly, qPCR analysis of a select subset of H3K27ac-marked genes confirmed elevated mRNA expression levels in pVHL-deficient cells (VEC, Supplementary Fig. 1g), validating the functional relevance of our H3K27ac dataset.
Next, using a previously published dataset18, we compared H3K27ac deposition in human renal tumors (vs adjacent normal tissue). This comparison showed nearly 70% of the genomic regions marked by elevated H3K27ac in pVHL-deficient UMRC-2 cells were, likewise, decorated with higher H3K27ac in human ccRCC tumors (Fig. 1e–g, Supplementary Fig. 1h–m, and Supplementary Data 1). Together, these observations validated our cell-based findings in human tumors and justified further investigation into the functional importance of the relatively understudied H3K27ac-marked loci.
We then overlapped the transcriptional output of the ccRCC cells (with or without pVHL) with the H3K27ac data. By comparing H3K27ac at annotated cis-regulatory elements, including promoters (Fig. 1h) and enhancers/super-enhancers (Fig. 1i), with pVHL-dependent transcriptional changes determined by RNA-Seq (Fig. 1j), we established a candidate list of ~100 genes that had higher H3K27ac and elevated mRNA levels in pVHL-deficient cells, and also encoded druggable gene products, such as cell-surface proteins, enzymes, etc. (Fig. 1k).
Functional in vivo screens score SLC1A1 as an oncogenic driver in ccRCC
pVHL-proficient cells, unlike their pVHL-deficient counterparts, do not efficiently form tumors in vivo. To functionally interrogate the importance of our candidate genes, we performed an in vivo positive selection (up) screen and sought to identify H3K27ac-marked genes whose expression was sufficient to confer tumorigenic potential onto otherwise poorly tumorigenic pVHL-proficient cells. We manually curated an ORF library corresponding to our (~100) candidate genes and included additional controls, such as BFP, GFP, and HcRed. The individual clones were procured from the Broad Institute in the pLX317 backbone, which has a unique 20-nucleotide barcode associated with each gene. We generated an equimolar pool of the individual clones, packaged this library into lentiviral particles, and transduced pVHL-proficient versions of UMRC-2 cells at low MOI. After expansion, a small aliquot (~10%) of transduced cells was set aside to measure representation at the assay start (T0), and 5e6 of the remaining cells were inoculated subcutaneously (flank xenografts) into immunodeficient NCRnu/nu mice (Fig. 2a).
a Schema of the in vivo positive selection screen to identify genes whose expression is sufficient to promote tumorigenesis in pVHL-proficient UMRC-2 ccRCCs. b–d Photographs showing tumor growth (b) and H&E-stained sections of two representative tumors (c, d) of the indicated versions of the UMRC-2 cells that were inoculated subcutaneously to form flank xenografts in NCRnu/nu mice. The primary endpoint for pVHL-deficient (VEC) UMRC-2 cells was reached at 8 weeks post-injection, and for the pVHL-proficient cells expressing the ORF library (VHL; ORF lib) at 12 weeks post-injection. e Heatmap depicting the Log2(fold change) in the relative abundance of barcodes representing the indicated genes, as measured by next-gen sequencing from genomic DNA harvested from the pVHL-proficient versions of the UMRC-2 cells that were lentivirally transduced with the ORF library (VHL; ORF lib). Each column represents an independent tumor. f–h Immunoblot analysis (f) and subcutaneous tumor growth (g, h) measured in pVHL-proficient versions of UMRC-2 cells lentivirally transduced to express the indicated genes and then inoculated subcutaneously into flanks of NCRnu/nu mice. The numbers in (g) indicate the number of independent biological replicates analyzed for each gene.
Eight tumors representing four independent biological replicates were analyzed for tumor growth. As expected, the pVHL-deficient UMRC-2 cells (VEC, our positive control) formed tumors at 100% penetrance (8/8 injections), whereas pVHL-proficient UMRC-2 cells (VHL, our negative control) failed to form any tumors (0/8 injections) (Fig. 2b). Interestingly, introduction of the ORF-library into pVHL-proficient cells led to tumor growth in 5/8 injections (~60% penetrance) (Fig. 2b–d). Tumors derived from these ORF-library transduced cells were harvested to measure ORF representation at the assay endpoint (Tend). The barcode was PCR amplified from the genomic DNA of the T0 and Tend samples and compared using next-gen sequencing.
We anticipated that barcodes representing oncogenes would be enriched in tumors, whereas barcodes representing either neutral bystanders or tumor suppressors would be depleted. To account for biological differences, we chose to pursue candidates that were enriched in at least two of the five tumors. While our negative controls (e.g., BFP, GFP, and HcRed) were depleted in all five tumors, we identified thirteen enriched candidate genes, many of which were known targets of HIF, such as BNIP3, NDRG1, and STC2 (Fig. 2e and Supplementary Data 2).
We prioritized genes that were not canonical HIF-targets and, in secondary validation experiments, subcutaneously inoculated pVHL-proficient UMRC-2 and 786-O cells individually expressing these candidate genes into NCRnu/nu mice (Fig. 2f and Supplementary Fig. 2a). PKLR expression promoted tumor growth at 50% penetrance (2/4 injections) in pVHL-proficient UMRC-2 cells, but not in 786-O cells. However, in both 786-O and UMRC-2 cells, the reintroduction of SLC1A1 conferred tumorigenic potential, albeit at 50% penetrance in 786-O cells (2/4 injections) and 100% penetrance in UMRC-2 cells (4/4 injections) (Fig. 2g, h and Supplementary Fig. 2b, c). SLC1A1 over-expression conferred a 2-fold growth advantage even in the pVHL-deficient UMRC-2 ccRCCs, despite their already elevated levels of endogenous SLC1A1, when using a weaker promoter (e.g., EF1a); however, we noted a fitness cost upon supra-physiological expression of SLC1A1 when using a strong promoter (e.g., CMV) (Supplementary Fig. 2d–g). Finally, this oncogenic effect was specific because overexpression of human SLC1A1, which shares 94% similarity to the mouse ortholog, did not promote tumorigenesis in the non-ccRCC mouse Renca cells (Supplementary Fig. 2h–k).
pVHL inactivation induces SLC1A1 expression in a HIF-independent manner
SLC1A1 [also called Excitatory Amino Acid Transporter 3 (EAAT3)] encodes a dicarboxylic amino acid transporter, which is a member of a family of five asp/glu transporters (EAAT1-5) (Supplementary Fig. 3a). Our studies showed that H3K27ac marking at the SLC1A1 locus was diminished upon pVHL expression in the ccRCC cell lines (Fig. 3a, top). Interestingly, we also confirmed increased H3K27ac marking at the SLC1A1 locus in a majority of human ccRCC tumors (e.g., #1 to #5 and #9) vs the normal adjacent tissue (Fig. 3a, bottom). However, we also noted H3K27ac marking in some normal tissues (e.g., #7–#10). This possibly represents the basal SLC1A1 expression in kidney tubules to perform its canonical role in dicarboxylic amino acid uptake. Importantly, these observations were specific to SLC1A1 because neither H3K27ac marking nor pVHL dependence was robustly detected for other members of the SLC1/EAAT family in human cell lines or patient tumors (Supplementary Fig. 3b–k).
a H3K27ac signal at the SLC1A1 promoter proximal region in pVHL-proficient (VHL) and pVHL-deficient (VEC) versions of two ccRCC cell lines (UMRC-2 and 786-O) (top); and in human ccRCC tumors (T) or their paired adjacent normal tissue (N) (bottom). b, c Immunoblotting and real-time qPCR of the indicated genes in pVHL-deficient (VEC) and pVHL-proficient (VHL) versions of UMRC-2 (b) and 786-O (c) cells. For (b, c), data indicate mean ± S.D. of mRNA levels (n > 4 biological replicates) compared using one-way ANOVA with Sidak’s multiple comparison test. d Immunoblotting of the indicated proteins in the indicated kidney lineage cells. e Heatmap of the relative expression of the indicated proteins as determined using ImageJ quantification of (d) and correlation of the relative expression of SLC1A1 and either HIF1α (top) or HIF2α (bottom). The highest protein expression value was normalized to 1. f, g Immunoblot analysis of the indicated proteins (f) and real-time qPCR of the indicated genes (g) in pVHL-deficient or pVHL-proficient versions of UMRC-2 cells treated for 16 h with the indicated ‘hypoxia mimetics’ (e.g., CoCl2, DMOG, or DFO) or environmental hypoxia (1% O2). h, i Immunoblot analysis of the indicated proteins (h) and real-time qPCR of the indicated genes (i) in pVHL-proficient UMRC-2 cells lentivirally transduced to express non-degradable versions of HIF (HIF1αdPA and HIF2αdPA), or pVHL-deficient UMRC-2 cells. For (g, i), data indicate the mean ± S.D. of mRNA levels (n ≥ 3 biological replicates) were compared using one-way ANOVA with Dunnett’s multiple comparison test. *p < 0.05; **p < 0.01; #p < 0.0001; ns non-significant.
We next addressed the role of the HIFα isoforms in driving SLC1A1 expression, given that pVHL inactivation leads to the chronic accumulation of both HIF1α and HIF2α. Using western blotting and real-time qPCR analysis, we confirmed in two human ccRCC cell lines (UMRC-2 and 786-O) that reintroduction of pVHL decreased both mRNA and protein expression of SLC1A1 (Fig. 3b, c). Interestingly, using a large panel of kidney lineage cells, we noted a strong correlation in the expression levels of SLC1A1 and HIF1α, but not HIF2α (Fig. 3d, e), leading to the hypothesis that chronic HIF1α activation in pVHL-deficient cells drove SLC1A1 expression, as has been suggested previously in Fibrosarcoma cells23.
To address the functional links between HIFα and SLC1A1, we performed a series of genetic and pharmacological studies. HIFα proteolysis is primed by EGL-9 homolog (EglN)-dependent prolyl hydroxylation at two key proline residues in the presence of oxygen. Hydroxylated HIFα is then recognized by pVHL and targeted for ubiquitin-dependent proteolysis24. Inactivation of the EglNs in pVHL-proficient cells attenuates HIFα hydroxylation and thus stabilizes HIFα, thereby offering a biochemical tool to annotate HIFα targets. Consistent with this, pVHL-proficient cells subjected to pharmacological (e.g., CoCl2, DMOG, and DFO) or physiological (e.g., 1% oxygen) conditions that attenuate EglN activity, stabilized HIFα and restored expression of the HIF target genes (e.g., DDIT4 and NDRG1). However, these treatments failed to restore SLC1A1 levels (Fig. 3f, g). Next, we lentivirally infected pVHL-proficient cells to express dual proline to alanine mutant (dPA) versions of either HIF1α or HIF2α. These mutants are not recognized by pVHL and are thus stable and constitutively active. Expressing HIF1αdPA and HIF2αdPA in pVHL-proficient cells restored the expression of their corresponding target genes (e.g., DDIT4, SLC2A1/GLUT1, and NDRG1), but failed to restore SLC1A1 expression (Fig. 3h, i). Finally, using CRISPR/Cas9, we inactivated HIF1β (or ARNT), the obligate binding partner for HIF1α and HIF2α. Unlike the canonical HIF target, NDRG1, ARNT loss failed to diminish SLC1A1 expression in two pVHL-deficient cells, CAKI-2 and UMRC-2 (Supplementary Fig. 3l). Together, these observations demonstrated that SLC1A1 expression in ccRCCs was pVHL-dependent, but HIF-independent.
SLC1A1 function is necessary for the survival of pVHL-deficient ccRCCs
SLC1A1 drives the cellular uptake of Asp and Glu (and to a lesser extent Cys)25,26. Despite being ‘non-essential’ under normal physiological conditions, Asp and Glu are known to provide an essential carbon skeleton to support the dysregulated biosynthetic needs of transformed cells27,28. CRISPR/Cas9-mediated inactivation of SLC1A1 using multiple sgRNAs in cell lines with evident SLC1A1 expression, showed that SLC1A1 was necessary for survival in many pVHL-deficient ccRCC cells (e.g., A498, CAKI-2, and UMRC-2). Surprisingly, the pVHL-proficient TK-10 renal adenocarcinoma cell line was also SLC1A1-dependent (Fig. 4a–c). Notably, unlike the other cell lines tested, the dependency in UMRC-2 cells was evident only upon chronic SLC1A1 loss (>10 days). Importantly, this dependence was not impacted by culture conditions because SLC1A1 deficiency conferred fitness defects in nutrient-rich cell culture media (e.g., DMEM and RPMI 1640), physiologically balanced media (e.g., HPLM29) (Supplementary Fig. 4a, b), and under nutrient-limited anchorage-independent growth (Fig. 4d).
a, b Immunoblot analysis (a) and normalized cell numbers [relative to control sgRNA #1 (sgCon 1)] (b), in the indicated cell lines that were lentivirally transduced to express sgRNAs targeting SLC1A1 (1A1sg) or non-targeting controls (sgCon). Cell counts for A-498 and UMRC-2 were performed 7- and 14-days post selection, respectively. Counts plotted as mean ± S.D. were compared to sgCon 1 using one-way ANOVA with Dunnett’s multiple comparison test. For A498, n = 2 biological replicates. For UMRC-2, n = 3 biological replicates. c Crystal violet staining of indicated cell lines transduced to express sgRNAs targeting SLC1A1 (1A1sg4, 1A1sg5) or non-targeting control (sgCon). d Full well scans and representative photomicrographs of INT-stained colonies formed after 6 weeks of growth in soft agar using UMRC-2 cells lentivirally transduced to express sgRNAs targeting SLC1A1 (1A1sg4, 1A1sg5) or non-targeting controls (sgCon). e Structural modeling of Asp and Glu within the SLC1A1 ligand-binding pocket, indicating certain loss-of-function mutations found in dicarboxylic aminoaciduria. f Immunoblot analysis of wild-type (WT) SLC1A1, Ile395 (I395Δ) and Arg445 ->Trp [R445W (RW)] SLC1A1 mutants, or BFP control, in pVHL-deficient or proficient versions of UMRC-2 cells, as indicated. g Normalized abundance, relative to the total ion count, of the listed metabolites, measured using LC-MS/MS from pVHL-proficient versions of UMRC-2 cells lentivirally transduced with the indicated forms of SLC1A1 or miscellaneous BFP control. Ion counts plotted as mean ± S.D. were compared to BFP control using two-way ANOVA with Dunnett’s multiple comparison test; n = 3 biological replicates. h, i Immunoblot analysis (h) and cell counts plotted as mean ± S.D. i of UMRC-2 cells first lentivirally transduced to express the indicated forms of SLC1A1 (or GFP control) and then transduced to express either a sgRNA targeting SLC1A1 (1A1sg5) or a non-targeting control (sgCon). Immunoblots were done 3 days post-selection in Puromycin (2 µg/ml), and cell numbers were measured 14 days post-selection. Counts in the sg5-expressing arms in (i) were compared using two-way ANOVA with Sidak’s correction for multiple testing; n = 4 biological replicates. Immunoblots in (h) were run on the same gel; however, because of the significantly greater expression of GFP, lower exposures of the first two lanes are cut and presented alongside exposures with similar intensities for WT and RW SLC1A1. All cell counts were measured using a Vi-Cell (Beckman). In (b, g, i), *p < 0.05; **p < 0.01; ^p < 0.001; #p < 0.0001; ns non-significant.
Modeling the ligand-binding pocket of SLC1A1 (PDB: 6X2Z), we identified the key residues that directly interacted with Asp and Glu (e.g., Ser331-333, Asp444, and Arg447) (Fig. 4e), consistent with the recently published Cryo-EM structure of SLC1A130. We compared our model with SLC1A1 loss-of-function mutations, such as the Ile395 deletion (I395Δ) and the Arg445 to Trp mutants [R445W (RW)], which have been described in a human pathology called Dicarboxylic Aminoaciduria31. Interestingly, although not directly involved in substrate binding, we found that Arg445 likely stabilizes the electrostatic interactions in the substrate-binding pocket (Fig. 4e).
Unlike our controls, BFP and the functionally dead RW mutant, reintroduction of wild-type (WT) SLC1A1 into pVHL-proficient cells increased intracellular levels of Asp, Glu, and their metabolic derivatives [e.g., Glutamine (Gln), α-ketoglutarate (α-KG), and Fumarate (Fum)], but did not impact pools of unrelated amino acids, such as Alanine (Ala) or Cysteine (Cys) (Fig. 4f, g). We then engineered a sgRNA-resistant version of WT SLC1A1 [5 R (WT)], which had silent mutations in the sgRNA recognition site. Expressing 5 R (WT), but not the inactive RW mutant [5 R (RW)], rescued the fitness defects associated with SLC1A1 loss in pVHL-deficient cells (Fig. 4h, i and Supplementary Fig. 4c). Altogether, these studies confirmed that the fitness defects triggered by the SLC1A1 sgRNAs were ‘on-target’ and suggested that SLC1A1’s Asp/Glu transport activity was necessary for its oncogenic function in ccRCC.
Finally, we asked if SLC1A1’s oncogenic function occurred via alterations in HIFα’s transcriptional output. Neither inactivation of SLC1A1 in pVHL-deficient cells (Supplementary Fig. 4d, e) nor reintroduction of SLC1A1 into pVHL-proficient cells (Supplementary Fig. 4f) had any discernible effect on the expression of the canonical HIFα target genes, including CA9, DDIT4, and NDRG1. Moreover, we found that the SLC1A1 loss had additive cytotoxicity when combined with the selective HIF2ɑ inhibitor, PT-2385 (Supplementary Fig. 4g, h). Therefore, we concluded that HIFα and SLC1A1 drove two non-overlapping oncogenic programs in kidney cancer.
SLC1A1 drives oncogenic metabolic reprogramming in ccRCC
SLC1A1’s canonical substrates, Asp/Glu, feed into many metabolic pathways. Therefore, we next interrogated the metabolic impact of CRISPR/Cas9-mediated SLC1A1 inactivation in ccRCCs, using a LC-MS/MS-based targeted metabolomics assay that measured the steady-state abundance of ~300 polar metabolites representing central carbon, amino acid, and nucleotide metabolism32. To this end, we optimized the use of the UMRC-6 human ccRCC cells. These cells also showed reduced SLC1A1 levels upon pVHL expression (Supplementary Fig. 5a); however, unlike the A-498 cells, which began showing fitness loss within 1–3 days of SLC1A1 inactivation, UMRC-6 tolerated SLC1A1 loss for ~4–6 days (Supplementary Fig. 5b), enabling metabolomics studies.
Analyzing the metabolic alterations caused by SLC1A1 loss, using MetaboAnalyst33, we noted the expected decrease in intracellular Asp/Glu, their modified derivatives [e.g., N-acetyl aspartyl glutamate (NAAG), Carbamoyl aspartate (Car-Asp), and Pyroglutamate (PyroGlu)], and their downstream products in the Tricarboxylic acid (TCA) cycle [e.g., α- KG, Fum, Malate (Mal), and Succinate (Suc)]. Interestingly, SLC1A1 depletion also reduced the abundance of several metabolites in the nucleotide/1-carbon metabolism pathways (Fig. 5a–e and Supplementary Data 3).
a–e Heatmap of fold change in normalized ion counts (a) and in the abundance of representative metabolites in the indicated pathways (b–e), as measured using LC-MS/MS analysis from UMRC-6 cells that were lentivirally transduced with sgRNAs targeting SLC1A1 (1A1sg4 or 1A1sg5) or non-targeting control (sgCon). Metabolites were harvested one day post-selection with puromycin (2 μg/ml) before the emergence of overt cytotoxicity. Ion counts in (b–e) were plotted as mean ± S.D. and compared using two-way ANOVA (sgCon vs 1A1sg4 and sgCon vs 1A1sg5) with Dunnett’s multiple comparison test; n = 3 biological replicates, *p < 0.05; **p < 0.01; ^p < 0.001; #p < 0.0001. f–k Crystal violet staining and normalized cell counts (relative to sgCon), of UMRC-6 (f, g), UMRC-2 (h, i), and A-498 (j, k) cells that were supplemented with 100 μM of the indicated esterified amino acids beginning three days prior to infection, and then lentivirally transduced with the indicated sgRNAs, and cultured for 7 days post-selection with puromycin (2 μg/ml). In g, i, k, cell counts were measured using a Vi-Cell (Beckman), plotted as mean ± S.D., and compared using two-way ANOVA (relative to sgCON within each group) with Dunnett’s multiple comparison test. For UMRC-6, n = 2 biological replicates. For UMRC-2, n = 2 biological replicates. For A-498, n = 3 biological replicates. *p < 0.05; **p < 0.01; ^p < 0.001; #p < 0.0001; ns = non-significant.
To address the importance of Asp and/or Glu in SLC1A1 function, we performed metabolic rescue experiments. We cultured pVHL-deficient UMRC-6 cells in media supplemented with cell-permeable, esterified, versions of SLC1A1’s substrates (e.g., Asp-Ome or Glu-OMe), which can be transported into cells independent of SLC1A1. We then lentivirally transduced these cells with CRISPR/Cas9 constructs to inactivate SLC1A1. Significant fitness defects were observed upon SLC1A1 loss in cells that were left untreated or supplemented with cell-permeable versions of (control) Ala; however, supplementation of cell-permeable Asp and Glu was sufficient to rescue SLC1A1-dependent fitness defects (Fig. 5f, g). Similar results were observed using three additional cell lines: UMRC-2 (Fig. 5h, i), A-498 (Fig. 5j, k), and 786-O cells (Supplementary Fig. 5c, d). These observations orthogonally established the importance of SLC1A1’s canonical metabolic function as the underlying driver of its oncogenic role.
SLC1A1 reprograms the tumor metabolome and the immune microenvironment
What might be the functional relevance of high SLC1A1 expression in human tumors? To begin addressing this question, we annotated TCGA renal tumors based on SLC1A1 gene expression and, as a control, expression of the canonical HIF target gene, CA9. Whereas CA9 expression was predominantly limited to ccRCCs, SLC1A1 mRNA expression was less specific and occurred in ccRCCs, but also in pRCC and normal tissues (Fig. 6a), consistent with our previous observation (Fig. 3a). Using the ccRCC subset, we then performed gene set enrichment analysis (GSEA)34 on tumors annotated into the top vs bottom quartile of CA9 or SLC1A1 expression. As expected, high CA9 expression was associated with signatures of HIF/hypoxia (Fig. 6b and Supplementary Data 4); however, consistent with the HIF-independent expression of SLC1A1, we found SLC1A1 expression was strongly correlated with widespread metabolic reprogramming but not with hypoxic programs (Fig. 6b and Supplementary Data 5).
a Principal components analysis and T-stochastic neighbor embedding of the indicated TCGA kidney cancer cohorts (KIRC = clear cell; KIRP = papillary; KICH = chromophobe) compared to normal samples. Color overlay indicates level of expression of CA9 (top) and SLC1A1 (bottom). Tumors were stratified by mRNA abundance between high (n = 135) and low (n = 135) groups. CPM counts per million. b GSEA of the high vs low CA9 and SLC1A1 tumors, as indicated, from the annotated dataset described in (a). NES normalized enrichment score. c Pathway analysis, using metaboanalyst, of LC-MS/MS derived metabolomics data from human ccRCC tumors from combined analysis of four MSKCC patient cohorts (RC12, n = 32; RC18Flow, n = 22; RC18MR, n = 67; and RC20, n = 51). Samples were stratified using SLC1A1 mRNA expression into top (high) and bottom (low) quartiles. p-value corrections for multiple testing were performed using Metaboanalyst’s pipeline. d–f Pearson correlation coefficient (ρ) of SLC1A1 expression level to the ImmuneScore signature (141 genes) (d), Volcano plot (e), and heatmap (f) showing the average correlation of the top 13 unique immune cell type-specific gene sets to SLC1A1 abundance. All analysis was done using RNA-Seq data from the indicated ccRCC cohort (RC18 MR, RC18 Flow, or RC20). Multiple comparison corrected p-values were averaged using Fisher’s method, and correlation coefficients were averaged using Fisher’s Z-transformation.
We then performed a treatment-agnostic meta-analysis of four Memorial Sloan Kettering Cancer Center (MSKCC) human tumor datasets (RC12, n = 32; RC18Flow, n = 22; RC18MR, n = 67; and RC20, n = 51), generated upon consent (MSKCC IRBs 06-107 and 12-237), using steady-state metabolomics and gene expression datasets. We first sorted tumors based on SLC1A1 expression and then compared metabolic and immunological signatures using the top (SLC1A1-high) and bottom (SLC1A1-low) quartiles. In the metabolomics dataset, we found that SLC1A1-high tumors showed significant enrichment in many of the same pathways that were depleted upon SLC1A1 loss in our cell-based studies, including aspartate, glutamate, and nucleotide metabolism (Fig. 6c). Moreover, deconvoluting immune cell signatures from the RNA-Seq datasets showed that SLC1A1 expression was negatively correlated with immune cell signatures and that tumors with higher SLC1A1 were associated with lower infiltration of T helper (Th2), Dendritic (DC), and T effector memory (TEM) cells (Fig. 6d–f). Together, these correlative findings in human tumors lent physiological credibility to our cell-based observations. We concluded that SLC1A1 expression did not correlate with HIF activity but showed a strong association with Asp/Glu/nucleotide metabolism and was negatively associated with key immunological signatures.
SLC1A1 activity confers an actionable metabolic vulnerability in ccRCC
SLC1A1 expression was observed most prominently in the normal mouse brain and kidney tissue (Supplementary Fig. 6a). However, homozygous Slc1a1 knockout in mice lacks any significant growth defects under normal (unstressed) conditions35. Moreover, humans with loss-of-function mutations in SLC1A1 have normal lifespans and show limited clinical symptoms except for the urinary excretion of Asp/Glu and minor cognitive defects31. This tissue-restricted expression and lack of phenotype in normal tissue support the existence of a therapeutic index when using SLC1A1 inhibitors against cancer cells.
SLC1A1 modulates glutamatergic neuronal transmission by clearing post-synaptic extracellular glutamate36,37, and pharmacologically targeting this function has been of interest in neuro-pathologies. Presently, SLC1A1 inhibitors are based on two scaffolds: an aspartate-like backbone38,39,40,41,42, which lacks specificity and pharmaceutical properties for systemic dosing; and a bicyclic imidazopyridine aniline43, as represented by compound 3e (Cmpd 3e) (Supplementary Fig. 6b), which has poor solubility and presents challenges for in vivo use because of the presence of a labile furan ring.
Despite these limitations, we began experimentally characterizing the utility of Cmpd 3e in ccRCC. We synthesized Cmpd 3e using the reported Groebke−Blackburn−Bienaymé condensation. After quality control steps to confirm desired purity and structure (Supplementary Fig. 6c), we tested the direct binding of Cmpd 3e with SLC1A1 using a Cellular Thermal Shift Assay (CETSA). Membrane-enriched fractions harvested from renal cells expressing V5-tagged SLC1A1 were treated with 3e (100 µM) and then exposed to increasing temperatures. We saw increased SLC1A1 thermal stability upon 3e treatment (relative to control DMSO), in line with direct interaction (Fig. 7a). We then tested the cytotoxic effects of 3e. Consistent with our genetic experiments, we noted significant cytotoxicity in the SLC1A1 hyper-dependent A-498 cells (IC50: ~0.9 µM). In contrast, UMRC-2 cells, which showed a delayed response to genetic inactivation of SLC1A1, were largely insensitive to Cmpd 3e at early time points (IC50: ~30 µM). Based on prior reports44, the Glutaminase inhibitor CB-839 was studied in parallel as a positive control and showed comparable cytotoxic effects in both cell lines (IC50: <1 µM) (Fig. 7b, c). Finally, 3e treatment also led to a significant decrease in anchorage-independent growth in both A-498 and UMRC-2 cells (Fig. 7d, e), albeit at higher doses in UMRC-2 cells, consistent with our genetic studies.
a SLC1A1 immunoblots in whole cell extracts treated ex vivo with 100 µM Cmpd 3e or vehicle (DMSO) and then heated to the indicated temperature for 5 min. Quantification per condition of the relative signal intensity at each temperature compared to 25 °C, determined using ImageJ. b, c Cell viability curves, plotted using the dose-response regression models in Graphpad (b) and heatmaps, relative to viability in DMSO (c), in the indicated ccRCC cell lines treated with a dose range of Cmpd 3e (0–50 μM) for 7 days or CB-839 (0–4 μM) for 3 days. For (b, c), viability was measured using Cell-titer Glo and IC50 values were determined using GraphPad. d, e Full well scans and representative photomicrographs of INT-stained colonies formed after 6 weeks of growth in soft agar using A-498 (d) or UMRC-2 (e) cells treated with the indicated concentrations of Cmpd 3e. Colony counts were compared using one-way ANOVA with Dunnett’s multiple comparison test, n = 3 biological replicates. f, g Heatmap (f) and bar graph (mean ± S.D.) (g) of normalized abundance, relative to total ion count (TIC), of the indicated metabolites, measured using LC-MS/MS in A-498 cells treated with 5 μM Cmpd 3e, or DMSO, for 3-5 days in HPLM and then harvested for metabolomics. Comparisons were made for each metabolite relative to DMSO control using multiple t tests with Benjamini, Krieger, and Yekutieli FDR correction; n = 3 biological replicates. h Crystal violet staining of A-498 cells supplemented with the indicated esterified-amino acid for 72 h and then treated with 12 μM Cmpd 3e or vehicle for 7 days. i Gating strategy used to determine the fraction of CD3/CD28 responsive T-cells in human peripheral blood mononuclear cell (PBMC) samples. j Histogram comparing carboxyfluorescein diacetate succinimidyl ester (CFSE) signal in T-cells from PBMC samples treated with or without CD3/CD28 dynabeads and low-dose (5 µM) or high-dose (25 µM) Cmpd 3e. *p < 0.05; **p < 0.01; ^p < 0.001.
Next, we measured the metabolic impact of Cmpd 3e using steady-state LC-MS/MS-based metabolomics. Treatment with sub-lethal regimens of Cmpd 3e led to a measurable decrease in Asp, Glu, and their derivatives (e.g., Glutathione and nucleotide intermediates) (Fig. 7f, g). Importantly, supplementing esterified versions of Asp or Glu, or providing high-dose Glutathione, was sufficient to partially mitigate the cytotoxic effects of 3e (Fig. 7h). These metabolic alterations and the rescue experiments, together, support the ‘on-target’ effects of Cmpd 3e. Finally, we tested if Cmpd 3e has any undesirable effects on T cell fitness, given their relevance in ccRCC immunotherapy. We noted that even high doses of Cmpd 3e were not deleterious to T cells, as evidenced by the proliferative index of ex vivo CD3/CD28-stimulated human T cells (Fig. 7i, j).
Unfortunately, 3e had poor pharmacological features. Acute intraperitoneal (IP) administration of 3e in NSG mice at 10 mg/kg (Supplementary Fig. 6d) led to a plasma Cmax of only ~90 nM and a relatively short half-life (<3 h) (Supplementary Fig. 6e). Cmpd 3e accumulated in the kidney to some degree (Cmax: 1.25 µM) but preferentially accumulated in the CNS (Cmax: 5.84 µM) (Supplementary Fig. 6e). Finally, in vitro microsomal metabolic stability profiles in human and rat indicated high turnover (human and rat intrinsic clearance > 80% hepatic, human CLint > 142 ml/min/kg, rat CLint 495 ml/min/kg) (Supplementary Fig. 6f). Altogether, we found that SLC1A1 is an actionable metabolic dependency in ccRCC; however, Cmpd 3e was not an ideal pharmacological agent to efficiently engage SLC1A1 in in vivo tumor efficacy studies.
HIF activation/hypoxia rewires the TCA cycle and drives Glutaminase-dependent Gln conversion into Glu, and subsequent reductive carboxylation to generate Citrate45,46. Glutaminase blockade (e.g., using CB-839) diminishes Asp/Glu/Citrate pools and represents a metabolic vulnerability in kidney cancer44,47. We hypothesized that SLC1A1 overexpression (and consequent elevated Asp/Glu pools) would reduce dependence on Glutaminase, whereas inactivation of SLC1A1 (and consequent depleted Asp/Glu pools) would sensitize cells to Glutaminase blockade. Indeed, we observed that overexpression of WT SLC1A1, but not the functionally dead RW mutant, conferred resistance to CB-839 (Supplementary Fig. 6g, h). Conversely, CRISPR/Cas9-mediated inactivation of SLC1A1 sensitized ccRCC cells to CB-839 treatment (Supplementary Fig. 6i, j). These studies highlighted the metabolic importance of SLC1A1 and the functional interaction between SLC1A1 and other targetable metabolic pathways in kidney cancer.
Elevated SLC1A1 expression correlates with advanced/metastatic ccRCC
To validate SLC1A1’s oncogenic function in an independent clinical cohort, we developed IHC assays to detect SLC1A1 in tumor cores. Using cell lines that express either low SLC1A1 (UMRC-2; VHL) or high SLC1A1 (UMRC-2; VEC), we first optimized a SLC1A1 IHC staining protocol (Supplementary Fig. 7a). In normal human kidney tissue, stained using this optimized protocol, we found notable SLC1A1 expression on the apical, lumen-facing, side of the renal cortical epithelium, consistent with SLC1A1’s role in amino acid reabsorption; however, no SLC1A1 staining was seen in the hypoxic renal medulla. In tumor tissue, SLC1A1 expression was redistributed throughout the entire cell surface (Supplementary Fig. 7b), perhaps driven by a loss of apico-basal polarity in renal tumors.
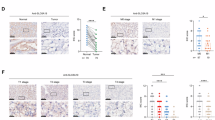
We then analyzed a tumor/normal tissue microarray (TMA) generated from renal cancer patients seen at DFCI and BWH (IRB 01-130). We qualitatively measured the ‘H-score’, a multiplication product of membranous SLC1A1 staining intensity (0–3, with 3 being highest) and the percentage of tumor section stained. This score, ranging between 0–300, is a standard measure of staining intensity employed by pathologists. First, we found higher SLC1A1 expression in (advanced) stage 4 disease (H-score: ~200; Fig. 8a, b). Consistent with this, we found higher SLC1A1 expression in metastatic (M1) compared to non-metastatic (M0) ccRCCs (Fig. 8c, d). Unlike both ccRCC and papillary RCC tumors (H-score: ~120–150), we noted that benign renal oncocytomas and renal chromophobe tumors had no detectable membranous staining of SLC1A1 (Fig. 8e and Supplementary Fig. 7c). These expression patterns were also consistent with TCGA data2,48, which indicated significantly higher SLC1A1 mRNA expression in both ccRCC and pRCC, compared to the chromophobe tumors (Supplementary Fig. 7d and Supplementary Data 6). Lastly, using the M0 vs M1 cohort, we tested the prognostic importance of SLC1A1 expression. These results were confusing. We found that higher SLC1A1 expression trended with worse overall survival (OS) in the M0 cohort, without reaching statistical significance. However, in the M1 cohort, which already had higher SLC1A1 expression and poorer survival compared to M0, a further stratification based on SLC1A1 expression indicated an association of higher SLC1A1 with better OS (Supplementary Fig. 7e–g). In summary, these studies established an unexpected redistribution of SLC1A1 away from the apical surface on the membranes of tumor cells, indicating a qualitative difference in normal vs tumor SLC1A1 expression. Furthermore, we noted an association of SLC1A1 expression with advanced/metastatic disease. Finally, we concluded that SLC1A1 expression was not an ideal prognostic marker for OS.
a, b Representative photomicrographs (a) and H-scores (b) showing SLC1A1 levels in tumors across the indicated ccRCC stages (I = 19; II = 8; III = 7; and IV = 48). *p < 0.05, determined by comparing all data to Stage I using two-way ANOVA with Dunn’s multiple comparison test. c, d Representative photomicrographs (c) and H-scores (d) determined using sections of ccRCC tumors from patients with either metastatic (M1, n = 48) or non-metastatic (M0, n = 34) disease. **p < 0.01, determined using the Mann–Whitney test. e H-scores determined from the indicated human renal tumors were compared to the ccRCC arm using two-way ANOVA with Dunn’s multiple comparison test; ccRCC (n = 82); pRCC (n = 13); chRCC (n = 5); Benign tumors (n = 7). f Immunoblotting (top) and crystal violet (bottom) of 786-M1A cells that were lentivirally transduced to express a sgRNA targeting SLC1A1 (1A1sg5) or a non-targeting control (sgCon). g–i Day 2 flux values (g), representative bioluminescence images taken at indicated timepoints (h), and fold change (relative to day 2) in flux intensity of tumor burden in the lungs of mice (i), measured using the 786-M1A cells, described in (f), that were inoculated into the tail veins of NSG mice. Experimental groups were compared using a two-tailed, unpaired t-test in (g) and using 2-way Anova with Bonferroni correction in (i). For sgCon, n = 4 biological replicates. For 1A1sg5, n = 5 biological replicates. One mouse in the control arm (g) was prematurely euthanized, unrelated to tumor burden, and images for this mouse were used in the first 3 data points in (i). j Histograms indicating the relative expression of SLC1A1 in A-498 cells expressing SLC1A1-targeting shRNAs (1A1sh1 and 1A1sh2) compared to control (LacZ). Cells were treated with 1 µg/ml Dox for 4 days or vehicle (day 0). k Relative levels of SLC1A1 mRNA (mean ± S.D.) in A-498 cells as described in (j), compared using 2-way ANOVA with Sidak correction; n = 3 biological replicates. l, m Crystal violet staining (l) and quantification (m) of the A-498 cells described in (j), grown with or without Dox (1 µg/ml) for 14 days. For (m), all conditions were normalized to vehicle-treated LacZ cells and plotted as mean ± S.D. Statistical comparisons were made using two-way ANOVA with Dunn’s multiple comparison test; n = 3 biological replicates. n Crystal violet staining of A-498 cells expressing a 1A1sh2-resistant form of SLC1A1, 5 R (WT), or vector control and subsequently infected with either SLC1A1-targeting shRNAs (1A1sh1 and 1A1sh2) or non-targeting control (LacZ). All conditions were treated with Dox (1 µg/ml) for 14 days. o–q Bioluminescence flux values at the indicated timepoints prior to Dox treatment (o), bioluminescence images taken at indicated timepoints (days) (p), and log2(fold change - relative to day 16) in flux intensity in the kidneys of mice (i), inoculated with the A-498 cells transduced with dox-inducible shRNAs targeting SLC1A1 (1: 1A1sh1 and 2: 1A1sh2) or a non-targeting control (L: shLacZ) that were injected orthotopically into the kidneys of NCRnu/nu mice. Following 16 days, all mice were placed on the Dox diet (625 mg/kg). Experimental groups were compared using 1-way ANOVA with Holm-Sidak correction; n = 10 biological replicates. In (q), statistical comparisons were made following outlier removal using the ROUT method (one datapoint in the 1A1sh1 cohort, marked by § and indicated in red in both (p, q), was excluded). *p < 0.05; **p < 0.01; ^p < 0.001; #p < 0.0001; ns non-significant. In (b, d, e) black line is plotted at the population median. In (g, o, q), the black line is plotted at the population mean.
SLC1A1 is required for ccRCC metastasis and tumor maintenance
To test the functional relevance of SLC1A1 in tumor metastasis, we utilized the metastatic derivative of the 786-O ccRCC cell lines—786-M1A, which also express firefly luciferase19,49. SLC1A1 inactivation caused notable fitness defects within 4–7 days in these cells (Fig. 8f). We then repeated infections, expanded, and inoculated these cells via tail vein injections into NSG mice prior to the emergence of overt fitness defects. Early timepoints post-injection confirmed similar tumor cell burdens in mouse lungs in cells expressing sgRNAs targeting SLC1A1 (1A1sg5) and the non-targeting (sgCon) controls (Fig. 8g, h). However, SLC1A1 loss had a profound deleterious effect on the growth of metastatic lung nodules at the experimental endpoint (day 61; Fig. 8g–i).
Finally, we addressed the necessity of SLC1A1 in established tumors. To this end, we transduced luciferase-expressing A-498 cells with doxycycline (Dox) inducible short hairpin RNA (shRNA) constructs to target SLC1A1 (e.g., 1A1sh1 and 1A1sh2) or LacZ (non-specific control). Dox treatment decreased cell surface SLC1A1 expression, as observed by flow cytometry (Fig. 8j and Supplemental Fig. 7h); and SLC1A1 mRNA levels, as measured by qPCR analysis (Fig. 8k). However, both 1A1sh1 and 1A1sh2 modestly reduced SLC1A1 even in the absence of Dox, suggesting some leakiness. Dox treatment of cells expressing 1A1sh1 and 1A1sh2, but not shLacZ, led to fitness defects in vitro (Fig. 8l, m). The 1A1sh2 targeting sequence overlaps the 1A1sg5 sequence used in our earlier studies. Consistent with this, the expression of the 5 R (WT) SLC1A1, which harbors silent mutations in this recognition sequence, rescued the fitness defects associated with 1A1sh2 (Fig. 8n). These observations confirmed the performance and on-target effects of our conditional inactivation system.
Inoculating these cells orthotopically into the kidneys of immunodeficient nude mice (NCRnu/nu) on regular chow allowed tumors to engraft efficiently in all three cohorts (Fig. 8o, no difference in luc signal between 1A1sh and shLacZ, day 2 to day 16). Following this, all mice were switched to Dox chow. Notably, by the end of the assay (day 70), we found a significantly lower tumor burden in cells expressing the SLC1A1-targeting shRNAs (vs the shLacZ control), with the more efficient 1A1sh1 having a stronger biological effect than the less efficient 1A1sh2 (Fig. 8p, q). Altogether, these studies established the importance of SLC1A1 in both efficient metastasis and established tumor maintenance.
Discussion
Epigenetic dysregulation drives widespread transcriptional changes, which often makes the identification of downstream oncogenic targets technically challenging. A typical strategy to fish for such oncogenic targets is by deploying a negative-selection or ‘dropout’ genetic screen, which scores for the loss of sg/shRNA representation in unhealthy/dying cells in vitro. This approach is noisy and scores many false positive hits due to ‘off-target’ cytotoxicity. Performing this analysis under nutrient-rich in vitro conditions also limits the discovery of physiologically relevant metabolic targets, whose dependencies are often influenced by nutrient availability in the tumor environment.
We modified our target discovery pipeline to mitigate these concerns. First, instead of a ‘dropout’, we designed a positive-selection approach, which scored for a gene’s ability to confer tumorigenic potential. Second, we performed our screen in vivo, to enable the identification of physiologically relevant metabolic targets. Third, we rigorously validated the hits from our initial screen using secondary in vivo assays and cell-based studies. All primary observations relied on multiple ccRCC cell lines, using multiple sgRNAs or shRNAs targeting SLC1A1, which were confirmed to be ‘on-target’ using rescue experiments, and supported by validations in human tumor samples. This discovery pipeline represented a rigorous experimental strategy that led to the discovery of an epigenetically dysregulated metabolic target.
The role of SLC1A1 is poorly understood in cancer biology. Much of the biological characterization of SLC1A1 has been done in neurological models, where this transporter limits neurotransmission by removing Glu from the post-synaptic space. However, SLC1A1’s oncogenic role has been recently suggested in human tumors, including lung50, T-cell lymphomas51, and gliomas52. The importance of Asp/Glu uptake in tumor biology is also supported by the oncogenic importance of SLC1A1 paralogs (e.g., SLC1A3) in hypoxic tumors27. Recent clinical studies in ccRCC have also established correlations between SLC1A1 expression and advanced53/metastatic disease54. Unfortunately, these studies have provided opposite associations, with one reporting better prognosis with high SLC1A1 expressing tumors53, and the other showing bad prognosis54. Both studies relied on mRNA levels to establish associations, and neither provided any functional studies to establish causality. Our own work suggests that SLC1A1 protein expression itself is a poor prognostic marker. We saw opposite trends in OS in M0 vs M1 patient cohorts and could not exclude the potential confounding effects of SLC1A1 expression in normal kidney tubules. Importantly, as has been established recently55,56, the association with good or bad prognosis itself has limited utility in defining actionable targets in cancer, with many important oncogenic targets having an association with good prognosis (e.g., ESR1, which encodes ER, in breast cancer and EPAS1, which encodes HIF2α, in ccRCC). In summary, we conclude, based on this growing body of work and our rigorous functional studies, that SLC1A1 does indeed present a viable metabolic target in ccRCC, but its expression patterns alone do not offer any prognostic value.
Metabolic studies have shown the significant elevation of one-carbon intermediates in human renal tumors (vs normal adjacent tissue), which likely supports nucleotide biosynthesis57. Our studies suggest that SLC1A1 imports Asp to fuel the biosynthesis of these key nucleotide intermediates. Recent studies, including one in kidney cancer, have also demonstrated that relative to their primary counterparts, metastatic tumors have higher TCA metabolite pools58,59. SLC1A1 inactivation in ccRCC leads to a prominent depletion of TCA cycle intermediates and leads to a suppression of metastatic outgrowth. We speculate that these two features are biologically linked. Our findings justify future studies addressing the importance of SLC1A1-dependent TCA cycling as an underlying vulnerability in metastatic tumors, especially focusing on the tropism of tumor metastasis (e.g., brain vs lung).
Chronic HIF2α activation has directly influenced our drug discovery efforts in kidney cancer, offering HIF-dependent angiogenesis, HIF-dependent nutrient-sensing pathways (e.g., mTOR), and, most recently, direct HIF2α inhibitors as therapies. Unfortunately, these approaches do not cure ccRCC, suggesting the existence of HIF-independent oncogenic escape routes. Our discovery suggests that SLC1A1 represents one such HIF-independent metabolic pathway. It will be important in the future to understand the HIF-independent mechanism by which pVHL-deficient tumors elevate SLC1A1 expression and also address the relative importance of SLC1A1 as a target in both HIF2α-dependent and independent ccRCC tumors. Importantly, we find that SLC1A1 function was relevant in both HIF2α-dependent cells (e.g., A-498 and 786-O) and HIF2α-independent cells (e.g., UMRC-2 and UMRC-6); and that SLC1A1 loss had additive cytotoxic effects when combined with the HIF2α inhibitor.
Our clinical correlations of SLC1A1 expression with immune infiltration signatures have certain caveats. First, these signatures were derived by deconvoluting bulk RNA-Seq signatures. Although current computational algorithms enable such deconvolution, a thorough validation of these signatures using single-cell sequencing or spatial studies is necessary. Additionally, these correlative differences will be functionally tested using ex vivo immunological assays (e.g., T-cell killing, ELISpot, etc.) and in immune-proficient mouse models in future studies.
One caveat of our study was that the existing SLC1A1 inhibitor (Cmpd 3e) had poor solubility and low potency (IC50 in the μM range); and lacked the pharmacological properties to effectively target the transporter in vivo. However, we noted cytotoxic responses in cell-based studies where pharmacological SLC1A1 blockade phenocopied SLC1A1’s genetic inactivation and established, using metabolic studies and rescue experiments, that the effects of the inhibitor were likely ‘on-target’. Future studies could focus on optimizing assay conditions (e.g., use glutamine starvation, etc.) to exacerbate the dependency on extracellular Asp/Glu, as is relevant in the nutrient-starved/hypoxic tumor microenvironment; and/or develop 3e analogs with improved potency and metabolic stability that offer oral delivery and are not brain-penetrant to avoid targeting SLC1A1 in the CNS. Altogether, our studies establish a strong rationale to launch future medicinal chemistry campaigns that develop improved SLC1A1 blockers.
Methods
All experiments and analyses performed in this study were done upon institutional approval and maintaining compliance with the regulatory requirements. All relevant protocol numbers (IBC, IRB, and IACUC) are provided in the relevant sub-sections.
Cell lines
Cell-based experiments were done under an institutionally approved IBC protocol (Cleveland Clinic, 1907; PI: Chakraborty). A-498 (RRID: CVCL_1056) and 786-O (RRID:CVCL_1051) renal carcinoma cells were obtained from the American Type Culture Collection. UMRC-2 (RRID: CVCL_2739) and UMRC-6 (RRID: CVCL_2741) cells were obtained from Dr. Bert Zbar and Dr. Marston Linehan (National Cancer Institute). HEK293T (RRID:CVCL_0063) was obtained from the Lerner Research Institute’s Cell Media Core. The 786-M1A cells were obtained from Dr. Joan Massagué (Memorial Sloan Kettering Cancer Center). 293FT cells were procured from Thermofisher (ThermoFisher, R70007, RRID:CVCL_6911). All cells were maintained in Dulbecco’s Modified Eagle's Medium (DMEM) (Life Technologies 11995073) supplemented with 10% Fetal Bovine Serum (FBS) (Life Technologies 10437-028), and 1X Penicillin-Streptomycin (Life Technologies 15140163). Cells stably expressing cDNAs or sgRNA were generated by lentiviral infection followed by selection with Puromycin (2 µg/ml) or Blasticidin (10 µg/ml), as appropriate. All cells were grown at 37 °C in 5% CO2.
Plasmids
The individual clones represented in the ORF library encoding the candidate oncogenes and miscellaneous controls were provided by Dr. David Root from the Broad Institute’s Gene Perturbation Platform in the pLX317 backbone, which has a unique 20 nt barcode associated with each gene. The barcode descriptions are provided in Supplementary Data 2. The clones were transformed and amplified in HB101 E. coli (Promega L2015), quantified using the nanodrop, and combined into an equimolar pool. The CRISPR/Cas9 constructs to target SLC1A1 were based on sequences provided in the Gecko library and generated by annealing the appropriate sgRNAs (described in the Oligo list) and cloning them into BsmBI-digested pLentiCRISPRv2 plasmids (RRID: Addgene_52961). The shRNA constructs to conditionally knockdown SLC1A1 were generated using either the sequence derived from the Broad TRC collection (TRCN0000255383; named 1A1sh1) or designed in-house using a target sequence that overlapped with 1A1sg5 (described in the Oligo list, named 1A1sh2). These sequences were annealed and cloned into AgeI- and EcoRI-digested pLKO-puro-TetOn plasmid.
Lentiviral infections
Lentiviruses were generated by packaging desired constructs into either 293FT or 293 T packaging cells. Typically, the packaging cells were seeded into 6 cm plates (~1.8E06—2.6E06 cells), attached overnight, and then transfected. The standard transfection mixture included 3 μg of total DNA [1.5 µg lentiviral plasmid + 1.5 µg helper plasmids - psPAX2 (RRID: Addgene_12260) and pMD2.G (RRID: Addgene_12259) mixed in a 3:1 ratio], combined with 9 µl Lipofectamine 2000 (Invitrogen 11668019). Transfection mixtures were incubated at room temperature for 15 min, added dropwise onto cells, and then incubated for sixteen hours. Following this, the media was changed, and viral supernatant was collected at 48-h and 72-h post-transfection and combined. Lentiviral supernatants were filtered (0.45 μm), aliquoted, and then frozen at −80 °C. Cells were seeded for viral infections into 6-well plates (150,000–200,000 per well). The following day, cells were transduced with 0.5 ml of the lentiviral supernatant in the presence of either 8 μg/ml polybrene (A-498 and 786-O cells) or 2 μg/ml polybrene (UMRC-2 and UMRC-6 cells). The plates were centrifuged at 300\(\times g\) for 40 min and returned to the incubator for 8 h, after which the media was replaced, and cells were grown for an additional 24 h prior to selection with the appropriate antibiotic.
Generation of SLC1A1 mutant constructs
The SLC1A1 gene was sub-cloned from the pLX317-SLC1A1 construct (obtained from the Broad Institute) into the pDONR223 backbone to generate the pEntry-SLC1A1 construct using Gateway cloning (Invitrogen). Mutations encoding I395Δ, R445W, and mismatches that conferred resistance to sg5 were generated by site-directed mutagenesis (Quickchange II XL; Agilent 200521) using the oligos described in the primer list and verified by Sanger sequencing. Expression constructs were generated by cloning the SLC1A1 mutant constructs into the pLX304 destination vector (RRID: Addgene_25890) using Gateway cloning (Invitrogen). The plasmids were packaged into lentiviral particles, as described above. For rescue experiments, cells were first engineered to express sgRNA-resistant versions of either WT or mutant SLC1A1 (or GFP, as a control), selected with blasticidin (10 μg/ml, 7–10 days), and then lentivirally transduced with sgRNAs targeting either SLC1A1 or a non-targeting control and selected with puromycin (2 µg/ml, 3 days).
ChIP-Seq analysis
Isogenic human ccRCC lines, lentivirally transduced to express HA-pVHL (VHL) or Empty Vector (VEC) control, were expanded. ~20 million cells (typically 3 × 15 cm dishes) were washed twice with ice-cold 1X PBS and then fixed using 1% methanol-free formaldehyde (Thermofisher 5701) in 1× PBS. Excess formaldehyde was quenched by the addition of 125 mM Glycine. Fixed cells were harvested and lysed in SDS lysis buffer [1% SDS, 10 mM EDTA, 50 mM Tris-HCl (pH 8) + protease inhibitor cocktail (PIC, cOmplete Mini, Roche)] and sonicated under optimized conditions that yielded an average DNA length of ~300 bp, typically 6 cycles of 30 s each using a microprobe sonicator at 30% output. Chromatin was quantified, and 10 μg total chromatin was used for each H3K27ac ChIP. ChIP-Seq libraries were prepared using the Illumina TruSeq library kit and analyzed using either a published ChIP-Seq analytical pipeline60, or by calling significant peaks using MACS2 broadpeaks (Version 2.2.7.1), where super-enhancers were identified using Homer findpeaks (Version 3.12) (-style super-minDist 12500). Bam files were normalized for sequencing depth (readCount), and log2 fold changes between VHL and VEC samples were determined using Deeptools bamCompare (Version 3.5.1). Finally, log2fold changes at significant peaks and superenhancers were determined using Encode (Version 130.2) bigWigAverageOverBed. Data from both approaches largely overlapped and is provided in GSE241864.
For patient samples, raw read files of H3K27ac ChIP-seq, published previously18, were downloaded from NCBI SRA. Reads with low quality were trimmed using Cutadapt (Version 4.0) and a quality cutoff > 20. Reads smaller than 20 bp were removed. Trimmed reads were aligned to hg19 using RNA STAR (Version 2.7.8a), and only uniquely mapped reads with mapQ ≥ 60 were used for downstream analysis. Multiple BAM files representing the same sample were merged using Samtools merge (Version 1.16.1). Merged bam files were normalized for sequencing depth (SES) and background subtracted using Deeptools bamCompare (Version 3.5.1) against input samples. Log2 fold changes between tumor and normal samples were then calculated using Deeptools bigwigCompare. Karyotype plots showing the abundance of H3K27ac at specific genomic regions of interest were generated using KaryoplotR.
RNA-Seq analysis
Total RNA, extracted from the isogenic (pVHL-proficient vs deficient) ccRCC cell lines using Trizol reagent (Life Technologies), was used for library preparation with or without polyA enrichment. Following standard QC (e.g., quantification on Qubit and RIN determination on a Bioanalyzer), cDNA libraries were prepared using Illumina TruSeq RNA Library Prep Kit with 50–100 ng of total RNA and sequenced on the Illumina HiSeq 4000. Raw sequences (.fastq files) were quality control tested using FastQC (FastQC, RRID:SCR_014583). The 40 bp single-end reads had, on average, high-quality metrics (>30 Phred score) and nucleotide distributions. Reads with Phred (Phred, RRID:SCR_001017) scores <28 were trimmed using Cutadapt (cutadapt, RRID:SCR_011841) v1.16. While FPKM values derived using Illumina’s BaseSpace RNA-Seq Alignment workflow were initially used to determine log2 fold-changes, the RNA-Seq datasets were also reanalyzed using more contemporary pipelines. To this end, reads were aligned to the UCSC hg19 build of the human transcriptome using HISAT2 (HISAT2, RRID:SCR_015530) v2.1.0 and total read counts for genomic features (i.e., genes) were measured using featureCounts (featureCounts, RRID:SCR_012919) v1.6.4. Counts were filtered, normalized (Trimmed Mean of M-values), and differential expression was determined using EdgeR (v3.12).
ORF library screen and validation
pVHL-proficient UMRC-2 cells were transduced in four independent infections (R1 to R4) with the ORF library (described in Supplemental Table S2) and selected using Puro (2 µg/ml). Cells were harvested, and approximately 10% were set aside to measure initial barcode representation (T0), whereas the remaining 90% of the cells were expanded and injected subcutaneously into mice on both flanks at 5E6 cells per injection into immunodeficient NCRnu/nu mice, giving two tumor injections per biological replicate (totaling eight injections). Following 8–12 weeks of growth, tumors were harvested, cut into two pieces to either be fixed in 10% formalin or processed for downstream sequencing (Tend). The barcode region of the integrated lentiviral cassettes was amplified from the genomic DNA of T0 and Tend samples, indexed, and sequenced on an Illumina MiSeq sequencer. Read counts from each sample were normalized to the total counts from the corresponding sample, and then representation in Tend samples was compared to the corresponding T0 to identify candidate genes that showed enrichment (log2fold change ≥ 1.0) in at least two of the five tumors. Seven of the top hits (BCKDHA, EFNA1, NEK6, PKLR, RNASET2, SLC1A1, or TLR3) were validated in a secondary screen.
Immunoblotting
Cells were washed in ice-cold 1× PBS and then lysed on ice in lysis buffer [50 mM Tris.Cl (pH 7.5), 400 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, and 10% glycerol] freshly supplemented with a PIC tablet for 30 min at 4 °C. Extracted proteins were quantified using the Bradford Assay (Biorad 5000006), and ~30–50 μg protein was analyzed by SDS-PAGE. The following primary antibodies were used for immunoblot analysis: ARNT (Cell Signaling Technology, #5537; RRID:AB_10694232), GLUT1 (Cell Signaling Technology, #12939, RRID:AB_2687899), HIF1α (Cell Signaling Technology, #14179, RRID:AB_2622225), HIF2α (Cell Signaling Technology, #71565), NDRG1 (Cell Signaling Technology, #5196, RRID:AB_10626626), SLC1A1/EAAT3 (Cell Signaling Technology, #14501, RRID:AB_2798499), TBP (Cell Signaling Technology, #44059, RRID:AB_2799258), Tubulin (Cell Signaling Technology, #2128, RRID:AB_823664), Vinculin (Cell Signaling Technology, #13901, RRID:AB_2728768), V5 Tag (Cell Signaling Technology, #80076, RRID:AB_2920661). All antibodies were used at 1:1000 dilutions. Primary antibodies were detected with HRP-conjugated secondary antibodies (Pierce, 1:5000) and chemiluminescent HRP substrates (Supersignal West Pico PLUS, Thermo Fisher Scientific; Pierce ECL Plus, Thermo Fisher Scientific; or Immobilon ECL Ultra, Millipore) (Thermo Fisher Scientific, RRID:SCR_008452).
Esterified-amino acid rescue of SLC1A1-KO cells
UMRC-6, UMRC-2, A-498, or 786-O cells (3E05 per 10 cm dish) were treated with a mixture of methyl esterified amino acids (100 μM aspartate, 100 μM glutamate, 100 μM alanine, and 1 mM glutathione) for 72 h, with medium replenished every 72 h through the experiments. Post-treatment, cells were seeded in 6-well plates at 1.5E05 cells/well in triplicate. Lentiviral transduction was performed using 500 μl of lentivirus containing either a control guide sgRNA or one of two SLC1A1 sgRNAs, along with 500 μl of polybrene-supplemented media [final concentration, either 8 μg/ml (A-498 and 786-O cells) or 2 μg/ml (UMRC-2, and UMRC-6 cells)]. Medium was replaced with fresh DMEM 24 h post-transduction. Puromycin selection (2 μg/ml) was initiated 48 h post-transduction. At 72 h, one third of the cell suspension from each well was transferred to a new 6-well plate for continued culture, while the remaining cells were harvested for Western blot analysis to confirm knockout. Cells were passaged at low density (~1:20) in supplemented DMEM for 7–10 days (UMRC-6 and A-498) or 2–3 weeks (UMRC2 and 786-O), depending on the cell line. Cell counts were performed using a Vi-Cell to assess the effects of treatment.
Soft agar assay
2.0E04 UMRC-2 or 5.0E04 A-498 cells were plated in 0.4% agarose on top of a 0.8% agarose base layer in a 6-well plate. Once agarose solidified, 1 ml of media with or without Cmpd 3e at the indicated concentrations was added on top. Cells were left to grow for 6 weeks with continuous media replacements every 3–4 days. At the assay endpoint, surviving colonies were stained using iodonitrotetrazolium chloride (INT) at 1 mg/ml overnight and counted manually.
T-cell proliferation
Fresh human PBMCs from three healthy donors (3.0E05 cells) were labeled with CFSE (CellTrace™ CFSE Cell Proliferation Kit) (Invitrogen, C34554) according to the manufacturer’s protocol. Briefly, Cells were washed with PBS and resuspended in 1 µM staining solution. Cells were incubated for 20 min at 37 °C protected from light, followed by 5 min incubation. Next, cells were stimulated with 4ul CD3/CD28 dynabeads (Gibco, 11161D) in the presence of Cmpd 3e at 0 µM, 5 µM, and 25 µM for 5 days. The proportion of CD3/CD28 stimulated T-cells was measured by flow cytometry using CD3 expression to subset the T-cell population and CFSE intensity to mark proliferating cells.
Animal studies: subcutaneous and orthotopic tumor model
All animal experiments were done in compliance with the IACUC policies at the Cleveland Clinic (Protocol 2168; PI: Chakraborty). The studies utilized two immunodeficient strains of mice: athymic nude mice derived from the Balb/c background [either Taconic (NCRNu) or Jackson laboratory (JAX 2019), as per availability] or the NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (JAX 005557)]. All mice were housed inside a barrier facility with the following environmental conditions: 325 lux max light intensity, 14 h light: 10 h dark cycle, temperature within ±2 °F of the 70–74 °F set point, and humidity between 30% and 70%. Kidney cancer occurs preferentially in males (~75% of the cases); however, based on our experience, we have noted no difference in tumor growth in immunodeficient mice. Therefore, all experiments were conducted agnostically to sex. The institutional recommendations are to euthanize mice when tumor burden ≥10% of body weight or when animals are moribund and/or show poor body conditioning score (<2). All experiments were done in compliance with these human endpoints.
A-498 cells expressing luciferase and a doxycycline-inducible shRNA (pLKO-TetON: shLacZ, 1A1sh1, or 1A1sh2) were expanded in media containing Tet-free serum, and 1E6 cells were injected orthotopically into the kidneys of 6–8-week-old immune-deficient male NCRnu/nu mice in a 30 µl volume of PBS. After 2 days, initial engraftment in the kidney was measured using an IVIS Lumina following luciferin injection at 150 mg/kg. At day 16, tumor growth was evaluated again, and animals were removed from a normal diet (Inotiv, 2918; Teklad Irradiated Global 18% Protein Rodent Diet) and placed on a diet containing doxycycline (Inotiv, TD.03106; 625 mg/kg). Tumor growth was routinely monitored until experimental endpoint (before humane endpoint; tumor weight <10% body weight, which typically shows a maximal tumor signal of >10E9). Total flux was determined using Living Image software.
Metastasis assay
3E05 luciferase-expressing 786-M1A cells were injected into the lateral caudal vein of 6–8-week-old male NSG mice in a 100 µl volume of PBS. After 2 days, initial engraftment in the lungs was evaluated using an IVIS Lumina following luciferin injection at 150 mg/kg. Tumor burden in the lung was routinely monitored until experimental endpoint (before humane endpoint; tumor weight < 10% body weight, which typically results in a maximal tumor signal > 10E9). Total flux was analyzed using Living Image software and normalized to the values recorded on day 2.
Protein modeling
Glutamate (green) was docked into the co-crystal structure of SLC1A1/EAAT3 + aspartate (PDB: 6X2Z; gray) and subsequently energy minimized and refined using MOE. 2D ligand interaction diagrams were generated in MOE, while the 3D renderings were generated in ChimeraX (h-bonds from aspartate in pink). Frequently mutated Arg445, which does not directly interact with the amino acid substrates, is highlighted in cyan.
Metabolomics (LC-MS/MS)
Sample preparation. Fresh media was added to the plates 2 h before metabolite extraction. After 2 h, the media was aspirated, cells were rinsed with normal saline (0.9% NaCl), and the plates were placed on dry ice before adding 4 ml of 80% (vol/vol) methanol (cooled to −80 °C). The plates were incubated at −80 °C for 20 min and then scraped on dry ice with a cell scraper. The cell lysate/methanol mixture was transferred to a 15-ml conical tube on dry ice and centrifuged at 14,000×g for 5 min at 4–8 °C to pellet the cell debris. The metabolite-containing supernatant was transferred to a new 15-ml conical tube on dry ice, and 500 μl 80% (v/v) methanol (−80 °C) was added to the pellet in a 15-ml tube and vortexed for 1 min at 4–8 °C. The tubes were spun the tubes at 14,000×g for 5 min at 4–8 °C, and the supernatant was transferred to a 50-ml conical tube on dry ice. The sample was divided into several 1.5-ml microcentrifuge tubes (1.5 ml in each tube) and a refrigerated Centri-vap (Labconco) was used to pellet metabolites.
Targeted Mass Spectrometry. Samples were re-suspended using 20 μl HPLC-grade water for mass spectrometry. The 5–7 μl were injected and analyzed using a hybrid 6500 QTRAP triple quadrupole mass spectrometer (AB/SCIEX) coupled to a Prominence UFLC HPLC system (Shimadzu) via selected reaction monitoring (SRM) of a total of 298 endogenous water-soluble metabolites for steady-state analyses of samples. Some metabolites were targeted in both positive and negative ion mode for a total of 309 SRM transitions using positive/negative ion polarity switching. ESI voltage was +4950 V in positive ion mode and –4500 V in negative ion mode. The dwell time was 3 ms per SRM transition, and the total cycle time was 1.55 s. Approximately 9–12 data points were acquired per detected metabolite. Samples were delivered to the mass spectrometer via hydrophilic interaction chromatography (HILIC) using a 4.6 mm i.d × 10 cm Amide XBridge column (Waters) at 400 μl/min. Gradients were run starting from 85% buffer B (HPLC grade acetonitrile) to 42% B from 0 min to 5 min; 42% B to 0% B from 5 min to 16 min; 0% B was held from 16 min to 24 min; 0% B to 85% B from 24 min to 25 min; 85% B was held for 7 min to re-equilibrate the column. Buffer A was comprised of 20 mM ammonium hydroxide/20 mM ammonium acetate (pH = 9.0) in 95:5 water:acetonitrile. Peak areas from the total ion current for each metabolite SRM transition were integrated using MultiQuant v3.0 software (AB/SCIEX).
TCGA analysis
HTSeq counts files for clear cell (TCGA-KIRC), papillary (TCGA-KIRP), and chromophobe (TCGA-KICH), RCCs were downloaded from the Genomic Data Commons Data Portal. Counts were filtered and normalized using EdgeR. Principal component analysis was performed on RCC tumors using all genes, and clustering by K-means was based on RCC subtype. ccRCC samples were stratified using either CA9 or SLC1A1 expression into the top 75%tile (representing the high group) and bottom 25%tile (representing the low group). The high and low groups were compared using GSEA to identify functional enrichment and EdgeR to determine differentially expressed genes.
Functional enrichment analysis
GSEA software was downloaded from the GSEA website [http://www.broad.mit.edu/gsea/downloads.jsp]. GSEA was performed on the EdgeR normalized counts per million using the ‘Hallmark’ gene sets for identification of enriched/depleted signatures. Gene Sets with an FDR < 0.10 and a nominal p-value < 0.05 were considered significant.
Human tumor analysis
Human tumor data were compiled from either public datasets (e.g., TCGA and NCBI SRA) or from human renal cancer biospecimens that were collected upon consent in accordance with approved institutional IRBs (MSKCC IRB: 06-107 and 12-237; DFCI/BWH IRB: 01-130).
In the MSKCC cohorts, data from four ccRCC cohorts from MSKCC (32 samples in RC12, 22 in RC18Flow, 67 in RC18MR, and 51 in RC20) with metabolomics and RNA-sequencing data were used to compare the association between SLC1A1 and metabolic and Immune Score signature, as described previously61. For the metabolic studies, a meta-analysis was done by combining the data from all four cohorts. First, RNA-seq-derived gene expression data were used to sort the tumor samples into quartiles based on SLC1A1 expression. LC-MS/MS-based targeted metabolomics datasets of the SLC1A1-top vs SLC1A1-bottom quartiles were compared using Wilcoxon signed-rank tests, and p-values were adjusted using Benjamini–Hochberg correction. Metabolites were mapped to pathways using KEGG annotations using the metaboanalyst package33.
To measure immune correlations, three cohorts were analyzed individually. After stratifying tumors into high- vs low-SLC1A1 expression, using the RNA-Seq datasets, correlation coefficients and p-values were calculated within each cohort separately, and coefficients were averaged using Fisher’s Z, and p-values were averaged using Fisher’s method. The analysis was then repeated among 23 unique immune signatures, described previously62.
Differential abundance (DA) scores evaluated whether a metabolic pathway was differentially abundant between the two groups. DA scores were calculated by applying a Wilcoxon rank-sum test to all metabolites in a pathway, and p-values were then corrected using the Benjamini–Hochberg correction (p < 0.05). The DA score for each pathway is calculated as:
Only pathways with three or more significantly altered metabolites were scored.
In the DFCI/BWH cohorts, the RCC tissue microarray (TMA) analysis used two previously constructed RCC TMAs. The representation in the first TMA was: 82 ccRCCs (19 stage I, 8 stage II, 7 stage 3, and 48 stage 4 tumors), 13 pRCCs, 5 chRCCs, and 6 benign oncocytoma tumors. The representation in the second TMA was: 33 M0 tumors and 49 M1 tumors. In both TMAs, each RCC case was represented by four cores taken from its tumor tissue. The average H-score from all 4 cores was calculated and statistically compared by the two-way ANOVA or Mann–Whitney test, as described in the figure legend. In addition, multiple cores representing normal kidney parenchyma and benign tumor entities were included in both TMAs.
Immunohistochemical staining of TMA sections with SLC1A1/EAAT3 antibody (Cell Signaling Technology, #14501 clone, RRID:AB_2798499, 1:100) was performed using an automated staining system (Bond III, Leica Biosystems). Semi-quantitative scoring was performed for the intensity of staining of tumor cell membranes (0 = negative; 1 = weak; 2 = moderate; 3 = strong). The H-score was then calculated as the sum of intensity scores, each multiplied by the percentage of positive tumor cells with that intensity level. The cores in the TMA were scored independently by two pathologists who subsequently reviewed the scores together to reach final consensus scores.
Pharmacological studies
The pharmacological properties of Cmpd 3e were analyzed by Q2 Solutions as a fee-for-service by using turbo ion spray liquid chromatography/tandem mass spectrometry (LC/MS/MS). All sample tissues were extracted using protein precipitation in the presence of Diclofenac (100.00 ng/ml in 1:1 MeOH/ACN), used as an internal standard. Supplementary Table 5 describes the detailed parameters used in the analytical pipeline.
Statistics and reproducibility
All statistical analysis was performed in GraphPad Prism 10 (v. 10.1.0) or R (v.4.2.1). For all experiments, a biological replicate represented the average values of the technical replicates. Multiple biological replicates were then compared by statistical testing. All experiments were typically performed three (or more) independent times. In instances where statistical conclusions were drawn from two biological replicates, the experiment was simultaneously performed using multiple cell lines and confirmed to show the same findings. Parametric data was analyzed using a two-sided t-test or one-way/two-way ANOVA based on the number of groups being compared. Non-parametric data was analyzed using Mann–Whitney U-test or the Kruskal–Wallis test based on the number of groups being compared. In all cases of multiple comparisons, additional tests were performed as appropriate to correct for false discovery rates. There is one instance of outlier removal. Here, the robust regression and outlier removal (ROUT) method was used in Graphpad. Sample size was determined based on power analysis and/or previous experience to reach the desired p-value of 0.05 at a power of 0.8. Animal studies were randomized by inoculating the different tumor cell types at random into the left vs right kidney (e.g., in the dox-induced shRNA experiment) or at random into mouse tails (e.g., in the metastasis experiment), following which the investigators were blinded to the sample identity during the course of luciferase imaging, until the assay endpoint. Stratification by sex was not necessary because in immune-deficient mice, we have not noted any sex-based differences in kidney cancer growth.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All the data necessary to evaluate the conclusions of the manuscript are provided in the paper and/or the Supplementary Materials. All ChIP-Seq and gene expression data are deposited into the Gene Expression Omnibus [Gene Expression Omnibus (GEO), RRID:SCR_005012] under accession number GSE241864. The ccRCC patient H3K27ac data were obtained from the previously deposited dataset GSE8609518. All metabolomics data are available at the NIH Common Fund’s National Metabolomics Data Repository (NMDR), the Metabolomics Workbench63 (RRID:SCR_013794), where it has been assigned Study ID ST004135. The data can be accessed directly via its Project DOI. All processed and raw data, including Graphpad Prism files with statistical analysis, are provided within the Supplementary Information/Source Data file. All reagents are available commercially, and published constructs will be deposited in Addgene to ensure public distribution. Source data are provided with this paper.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 72, 7–33 (2022).
Ricketts, C. J. et al. The cancer genome atlas comprehensive molecular characterization of renal cell carcinoma. Cell Rep. 23, 313–326.e5 (2018).
Kaelin, W. G. Jr. The von Hippel–Lindau tumour suppressor protein: O2 sensing and cancer. Nat. Rev. Cancer 8, 865–873 (2008).
Ivan, M. et al. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 (2001).
Jaakkola, P. et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Kondo, K., Kim, W. Y., Lechpammer, M. & Kaelin, W. G. Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 1, E83 (2003).
Gordan, J. D., Bertout, J. A., Hu, C. J., Diehl, J. A. & Simon, M. C. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 11, 335–347 (2007).
Cho, H. et al. On-target efficacy of a HIF-2alpha antagonist in preclinical kidney cancer models. Nature 539, 107–111 (2016).
Chen, W. et al. Targeting renal cell carcinoma with a HIF-2 antagonist. Nature 539, 112–117 (2016).
Jonasch, E. et al. Belzutifan for renal cell carcinoma in von Hippel–Lindau disease. N. Engl. J. Med 385, 2036–2046 (2021).
Liao, L., Testa, J. R. & Yang, H. The roles of chromatin-remodelers and epigenetic modifiers in kidney cancer. Cancer Genet. 208, 206–214 (2015).
Xia, X. et al. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc. Natl. Acad. Sci. USA 106, 4260–4265 (2009).
Varela, I. et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 (2011).
Dalgliesh, G. L. et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 463, 360–363 (2010).
Chakraborty, A. A. et al. HIF activation causes synthetic lethality between the VHL tumor suppressor and the EZH1 histone methyltransferase. Sci. Transl. Med. 9, eaal5272 (2017).
Loven, J. et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Yao, X. et al. VHL deficiency drives enhancer activation of oncogenes in clear cell renal cell carcinoma. Cancer Discov. 7, 1284–1305 (2017).
Rodrigues, P. et al. NF-kappaB-dependent lymphoid enhancer co-option promotes renal carcinoma metastasis. Cancer Discov. 8, 850–865 (2018).
Huo, F. C., Xie, M., Zhu, Z. M., Zheng, J. N. & Pei, D. S. SHMT2 promotes the tumorigenesis of renal cell carcinoma by regulating the m6A modification of PPAT. Genomics 114, 110424 (2022).
Liu, Z. et al. Serine hydroxymethyltransferase 2 knockdown induces apoptosis in ccRCC by causing lysosomal membrane permeabilization via metabolic reprogramming. Cell Death Dis. 14, 144 (2023).
Zhang, J. et al. VHL substrate transcription factor ZHX2 as an oncogenic driver in clear cell renal cell carcinoma. Science 361, 290–295 (2018).
Yang, Z. et al. HIF-1alpha drives resistance to ferroptosis in solid tumors by promoting lactate production and activating SLC1A1. Cell Rep. 42, 112945 (2023).
Kaelin, W. G. Jr. & Ratcliffe, P. J. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 (2008).
Chen, Y. & Swanson, R. A. The glutamate transporters EAAT2 and EAAT3 mediate cysteine uptake in cortical neuron cultures. J. Neurochem. 84, 1332–1339 (2003).
O’Kane, R. L., Martinez-Lopez, I., DeJoseph, M. R., Vina, J. R. & Hawkins, R. A. Na(+)-dependent glutamate transporters (EAAT1, EAAT2, and EAAT3) of the blood-brain barrier. A mechanism for glutamate removal. J. Biol. Chem. 274, 31891–31895 (1999).
Garcia-Bermudez, J. et al. Aspartate is a limiting metabolite for cancer cell proliferation under hypoxia and in tumours. Nat. Cell Biol. 20, 775–781 (2018).
Raho, S. et al. KRAS-regulated glutamine metabolism requires UCP2-mediated aspartate transport to support pancreatic cancer growth. Nat. Metab. 2, 1373–1381 (2020).
Cantor, J. R. et al. Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258–272.e17 (2017).
Qiu, B. & Boudker, O. Symport and antiport mechanisms of human glutamate transporters. Nat. Commun. 14, 2579 (2023).
Bailey, C. G. et al. Loss-of-function mutations in the glutamate transporter SLC1A1 cause human dicarboxylic aminoaciduria. J. Clin. Invest. 121, 446–453 (2011).
Yuan, M. et al. Ex vivo and in vivo stable isotope labelling of central carbon metabolism and related pathways with analysis by LC-MS/MS. Nat. Protoc. 14, 313–330 (2019).
Xia, J., Mandal, R., Sinelnikov, I. V., Broadhurst, D. & Wishart, D. S. MetaboAnalyst 2.0–a comprehensive server for metabolomic data analysis. Nucleic Acids Res. 40, W127–W133 (2012).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Peghini, P., Janzen, J. & Stoffel, W. Glutamate transporter EAAC-1-deficient mice develop dicarboxylic aminoaciduria and behavioral abnormalities but no neurodegeneration. EMBO J. 16, 3822–3832 (1997).
Bunch, L., Erichsen, M. N. & Jensen, A. A. Excitatory amino acid transporters as potential drug targets. Expert Opin. Ther. Targets 13, 719–731 (2009).
Nieoullon, A. et al. The neuronal excitatory amino acid transporter EAAC1/EAAT3: Does it represent a major actor at the brain excitatory synapse? J. Neurochem 98, 1007–1018 (2006).
Bridges, R. J. & Esslinger, C. S. The excitatory amino acid transporters: pharmacological insights on substrate and inhibitor specificity of the EAAT subtypes. Pharm. Ther. 107, 271–285 (2005).
Esslinger, C. S. et al. The substituted aspartate analogue L-beta-threo-benzyl-aspartate preferentially inhibits the neuronal excitatory amino acid transporter EAAT3. Neuropharmacology 49, 850–861 (2005).
Liu, N., Jensen, A. A. & Bunch, L. Beta-indolyloxy functionalized aspartate analogs as inhibitors of the excitatory amino acid transporters (EAATs). ACS Med Chem. Lett. 11, 2212–2220 (2020).
Denton, T., Seib, T., Bridges, R. & Thompson, C. Synthesis and preliminary evaluation of trans-3,4-conformationally-restricted glutamate and pyroglutamate analogues as novel EAAT2 inhibitors. Bioorg. Med. Chem. Lett. 12, 3209–3213 (2002).
Shimamoto, K. et al. Syntheses of optically pure beta-hydroxyaspartate derivatives as glutamate transporter blockers. Bioorg. Med. Chem. Lett. 10, 2407–2410 (2000).
Wu, P., Bjorn-Yoshimoto, W. E., Staudt, M., Jensen, A. A. & Bunch, L. Identification and structure-activity relationship study of imidazo[1,2-a]pyridine-3-amines as first selective inhibitors of excitatory amino acid transporter subtype 3 (EAAT3). ACS Chem. Neurosci. 10, 4414–4429 (2019).
Gameiro, P. A. et al. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 17, 372–385 (2013).
Jiang, L. et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258 (2016).
Metallo, C. M. et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature 481, 380–384 (2011).
Okazaki, A. et al. Glutaminase and poly(ADP-ribose) polymerase inhibitors suppress pyrimidine synthesis and VHL-deficient renal cancers. J. Clin. Invest. 127, 1631–1645 (2017).
Linehan, W. M. & Ricketts, C. J. The Cancer Genome Atlas of renal cell carcinoma: findings and clinical implications. Nat. Rev. Urol. 16, 539–552 (2019).
Vanharanta, S. et al. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat. Med. 19, 50–56 (2013).
Guo, W. et al. Dysregulated glutamate transporter SLC1A1 propels cystine uptake via Xc(-) for glutathione synthesis in lung cancer. Cancer Res. 81, 552–566 (2021).
Xiong, J. et al. SLC1A1 mediated glutamine addiction and contributed to natural killer T-cell lymphoma progression with immunotherapeutic potential. EBioMedicine 72, 103614 (2021).
Wang, X. et al. SLC1A1-mediated cellular and mitochondrial influx of R-2-hydroxyglutarate in vascular endothelial cells promotes tumor angiogenesis in IDH1-mutant solid tumors. Cell Res. 32, 638–658 (2022).
Chen, Y. et al. Identification of prognostic metabolism-related genes in clear cell renal cell carcinoma. J. Oncol. 2021, 2042114 (2021).
Ergun, S., Gunes, S., Buyukalpelli, R. & Aydin, O. Glutamate transporter SLC1A1 is associated with clear cell renal cell carcinoma. Turk. J. Med. Sci. 49, 531–537 (2019).
Smith, J. C. & Sheltzer, J. M. Genome-wide identification and analysis of prognostic features in human cancers. Cell Rep. 38, 110569 (2022).
Kaelin, W. G. Jr. Common pitfalls in preclinical cancer target validation. Nat. Rev. Cancer 17, 425–440 (2017).
Hakimi, A. A. et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell 29, 104–116 (2016).
Bezwada, D. et al. Mitochondrial complex I promotes kidney cancer metastasis. Nature 633, 923–931 (2024).
Bartman, C. R. et al. Slow TCA flux and ATP production in primary solid tumours but not metastases. Nature 614, 349–357 (2023).
Orlando, D. A. et al. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep. 9, 1163–1170 (2014).
Yoshihara, K. et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4, 2612 (2013).
Bindea, G. et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity 39, 782–795 (2013).
Sud, M. et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 44, D463–470 (2016).
Acknowledgements
A.A.C. is supported by seed money from the Cleveland Clinic Foundation, the DoD’s KCRP-ECI award (W81XWH-20-1-0804), two Velosano pilot awards, the V Foundation Scholar Award (V2020-011), the ACS-Research Scholar Grant (RSG-22-067-01-TBE) and the NCCN-YIA. W.G.K. is supported by an NIH R35CA210068 and NIH P50CA101942 and is an HHMI Investigator. S.S. was supported by NIH P50CA101942. S.V. is supported by the Sigrid Jusélius Foundation. R.R.K. is supported, in part, by a Department of Defense Early Career Investigator Grant (KCRP-AKCI, W81XWH-21-0942). The NIH Common Fund’s National Metabolomics Data Repository (NMDR) website, the Metabolomics Workbench, is supported by NIH grant U2C-DK119886 and OT2-OD030544 grants.
Author information
Authors and Affiliations
Contributions
Conceptualized, designed, and analyzed experiments: T.G., P.K., S.M., F.G., C.T., E.R., J.K., C.S., N.D., D.A.O., M.G.G., G.P., R.M.S., S.R.M., J.A.C., J.M.A., T.A.C., S.V., S.R.S., S.S., W.G.K. and A.A.C. Performed experiments: T.G., P.K., S.M., F.G., J.K., C.S., N.D., D.A.O., M.G.G., G.P., R.S., S.R.M., J.A.C., C.T. and A.A.C. Generated patient datasets: E.R., R.R.K., A.A.K., M.M., E.S., T.K.C. and S.S. Wrote manuscript: T.G. and A.A.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflicts of interest with the data presented in this study. W.G.K. is a paid advisor to Casdin Capital, Circle Pharma, Fibrogen, Nextech and Tango Therapeutics and serves on the boards of IcoOVir Bio, Lifemine Therapeutics and Eli Lilly. T.K.C. reports institutional and/or personal, paid and/or unpaid support for research, advisory boards, consultancy and/or honoraria past 5 years and ongoing, from Alkermes, AstraZeneca, Aravive, Aveo, Bayer, Bristol Myers-Squibb, Calithera, Circle Pharma, Deciphera Pharmaceuticals, Eisai, EMD Serono, Exelixis, GlaxoSmithKline, Gilead, IQVA, Infinity, Ipsen, Jansen, Kanaph, Lilly, Merck, Nikang, Nuscan, Novartis, Oncohost, Pfizer, Roche, Sanofi/Aventis, Scholar Rock, Surface Oncology, Takeda, Tempest, Up-To-Date, CME events (Peerview, OncLive, MJH, CCO and others), outside the submitted work; institutional patents filed on molecular alterations and immunotherapy response/toxicity and ctDNA; equity in Tempest, Pionyr, Osel, Precede Bio, CureResponse, InnDura; committee service at NCCN, GU Steering Committee, ASCO/ESMO, ACCRU, KidneyCan; medical writing and editorial assistance support may have been funded by Communications companies in part; no speaker’s bureau; mentored several non-US citizens on research projects with potential funding (in part) from non-US sources/Foreign Components; The institution (Dana-Farber Cancer Institute) may have received additional independent funding of drug companies or/and royalties potentially involved in research around the subject matter of T.K.C’s findings (not related to this study). T.K.C. is supported in part by the Dana-Farber/Harvard Cancer Center Kidney SPORE (2P50CA101942-16) and Program 5P30CA006516-56, the Kohlberg Chair at Harvard Medical School and the Trust Family, Michael Brigham, Pan Mass Challenge, Hinda and Arthur Marcus Fund and Loker Pinard Funds for Kidney Cancer Research at DFCI. S.S. reports receiving commercial research grants from Bristol-Myers Squibb, AstraZeneca, Exelixis and Novartis; is a consultant/advisory board member for Merck, AstraZeneca, Bristol-Myers Squibb, CRISPR Therapeutics AG, AACR and NCI; receives royalties from Biogenex; and mentored several non-US citizens on research projects with potential funding (in part) from non-US sources/Foreign Components. R.R.K reports consulting for Eisai, and institutional research funding from Pfizer, Takeda, Novartis, Exelixis, Xencor, Arsenal Bio and Allogene Therapeutics.
Peer review
Peer review information
Nature Communications thanks Yilong Zou, Qing Zhang, Chengheng Liao, and the other anonymous reviewer for their contribution to the peer review of this work. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Affiliation where work was originally done: Sayed Matar, Fatme Ghandour, erise Tang, Jesminara Khatun, David A. Orlando, Matthew G. Guenther, Steven R. Martinez.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Grubb, T., Koochaki, P., Matar, S. et al. The SLC1A1/EAAT3 dicarboxylic amino acid transporter is an epigenetically dysregulated nutrient carrier that sustains oncogenic metabolic programs. Nat Commun 16, 10012 (2025). https://doi.org/10.1038/s41467-025-64983-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64983-x