Abstract
Emerging imaging spatial transcriptomics (iST) platforms and coupled analytical methods can recover cell-to-cell interactions, groups of spatially covarying genes, and gene signatures associated with pathological features, and are thus particularly well-suited for applications in formalin fixed paraffin embedded (FFPE) tissues. Here, we benchmark the performance of three commercial iST platforms—10X Xenium, Vizgen MERSCOPE, and Nanostring CosMx—on serial sections from tissue microarrays (TMAs) containing 17 tumor and 16 normal tissue types for both relative technical and biological performance. On matched genes, we find that Xenium consistently generates higher transcript counts per gene without sacrificing specificity. Xenium and CosMx measure RNA transcripts in concordance with orthogonal single-cell transcriptomics. All three platforms can perform spatially resolved cell typing with varying degrees of sub-clustering capabilities, with Xenium and CosMx finding slightly more clusters than MERSCOPE, albeit with different false discovery rates and cell segmentation error frequencies. Taken together, our analyses provide a comprehensive benchmark to guide the choice of iST method as researchers design studies with precious samples in this rapidly evolving field.
Similar content being viewed by others
Introduction
Spatial transcriptomics (ST) tools measure the gene expression profiles of tissues or cells in situ. These approaches overcome the limitations of single-cell RNA-sequencing (scRNA-seq) methods by negating the need for cell dissociation, thus maintaining both local and global spatial relationships between cells within a tissue. ST can recover cell-cell interactions, groups of spatially covarying genes, groups of cells predictive of cancer survival, and gene signatures associated with pathological features1,2. These advantages, coupled with rapidly emerging computational and analytical methods, have led to substantial excitement about deploying these platforms in fundamental biology studies and in the clinic for research and diagnostic purposes3,4,5.
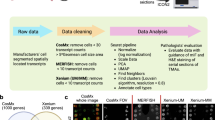
ST tools can be split into two broad categories: sequencing (sST) and imaging (iST) based modalities. sST methods tag transcripts with an oligonucleotide address indicating spatial location, most commonly by placing tissue slices on a barcoded substrate; isolating tagged mRNA for next-generation sequencing; and computationally mapping transcript identities to locations6. In contrast, iST methods most commonly use variations of fluorescence in situ hybridization (FISH) where mRNA molecules are tagged with hybridization probes, which are detected in a combinatorial manner over multiple rounds of staining with fluorescent reporters, imaging, and de-staining (Fig. 1a)7. Computational reconstruction then yields maps of transcript identity with single-molecule resolution. Compared to sST methods, iST methods are targeted to subsets of the transcriptome due to their reliance on pre-defined gene panels and they adopt the higher spatial resolution and sensitivity of FISH, yielding single-cell resolution data8.
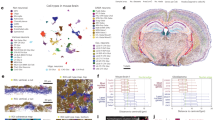
a Overall approach for generating iST data. b Different amplification approaches for Xenium, MERSCOPE, and CosMx. c Overview of the tissue types and numbers of cores used in this study. BlC bladder cancer, BrC breast cancer, CRC colorectal cancer, HNSCC head and neck squamous cell carcinoma, Mel Melanoma, NSCLC non-small cell lung cancer, OvC ovarian cancer, SCC squamous cell carcinoma. d DAPI images from the Xenium run of each TMA, including Tumor TMA 1(tTMA1), Tumor TMA 2(tTMA2), and Normal TMA(nTMA).
While the iST methods share some similarities, significant differences arise in primary signal detection and amplification, sample processing, and the subsequent fluorescent cycling chemistry (Fig. 1b)9,10,11. The need for amplification of signal is coupled to the sample processing, namely whether the sample is cleared, gel-embedded, or photobleached to quench autofluorescence. There are tradeoffs due to differences in sample processing for each iST method. For example, clearing of the sample increases signal quality but can prevent follow-up H&E staining and complicate immunostaining, which, in turn, can make cell segmentation more challenging. Finally, there are tradeoffs between imaging time, molecular plex, and imaging area covered, which result from the particular combination of the molecular protocol and the imaging hardware implementation12.
A key historic limitation in the widespread use of iST methods with human clinical samples was the incompatibility of most methods with formalin-fixed, paraffin-embedded (FFPE) tissue samples13,14. FFPE is the standard format for clinical sample preservation for pathology due to its ability to maintain tissue morphology and sample stability at room temperature for decades, accounting for over 90% of clinical pathology specimens15. The ability to process FFPE samples with iST will enable the use of archival tissue banks for studies and obviate the need for specialized sample harvesting workflows. However, FFPE samples tend to suffer from decreased RNA integrity, particularly after having been stored in archives for extended periods of time16.
Three companies recently released the first FFPE compatible commercial iST platforms: 10x’s Xenium, NanoString’s CosMx, and Vizgen’s MERSCOPE9,10,11,17. These three platforms each use different protocols, probe designs, signal amplification strategies, and computational processing methods, and therefore may potentially yield different sensitivities and downstream results. The main chemistry difference lies in transcript amplification: 10x Xenium uses a small number of padlock probes with rolling circle amplification; CosMx uses a low number of probes amplified with branch chain hybridization; and MERSCOPE uses direct probe hybridization but amplifies by tiling the transcript with many probes (Fig. 1b). However, no head-to-head performance comparisons on matched samples have been published. Understanding the key differences across platforms will allow users to make better-informed decisions regarding panel design, method choice, and sample selection as they design costly experiments, often on precious samples that have been bio-banked for years18.
In this study, we compared currently available FFPE-compatible iST platforms on matched tissue samples. We prepared a set of samples representative of typical archival FFPE tissues, comprised of 33 different tumor and normal tissue types, and acquired matched data from sequential sections according to the manufacturer’s best practices at the time of writing, generating a dataset of >5.0 M cells. We analyzed the relative sensitivity and specificity of each method on shared transcripts and further quantified the concordance of the iST data across each platform with paired scRNA-seq data collected by 10x Chromium Single Cell Gene Expression FLEX. Then we focused on cell-level comparisons, evaluating the out-of-the-box segmentation for each platform based on detected genes and transcripts and coexpression patterns of known disjoint markers. Finally, we cross-compared the ability of each platform to identify cell type clusters with breast and breast cancer tissues as an example use case. Taken together, our work provides the first head-to-head comparison of these platforms across multiple archival healthy and cancerous FFPE tissue types.
Results
Collection of matched iST data across 33 FFPE tissue types reveals high transcript counts obtained by Xenium and CosMx
To test the performance of the latest generation of FFPE-compatible iST tools, we sought to match gene expression and sample as much as possible given available panel configurations and manufacturer guidelines. To accomplish this, we used three previously generated multi-tissue tissue microarrays (TMAs) from multiple types of clinical discarded tissue (see Methods). We focused on FFPE tissues as the standard method for sample processing and archival in pathology. Tumor TMA 1 (tTMA1) consisted of one hundred and seventy 0.6 mm diameter cores (i.e., sampled regions) from seven different cancer types, with 3-6 patients per cancer type, and 3–6 cores per patient (Fig. 1c, d, Supplementary Table 1). Tumor TMA 2 (tTMA2) consisted of forty-eight 1.2 mm diameter cores from nineteen different cancer types, with each tissue type coming from one or two patients and represented in 2–3 cores (Fig. 1c, d, Supplementary Table 2). A normal tissue TMA (nTMA) contained forty-five 1.2 mm diameter cores spanning sixteen normal tissue types isolated with each tissue type coming from one patient and represented in 2–3 cores (Fig. 1c, d, Supplementary Table 3). CosMx and Xenium suggest pre-screening samples based on H&E, while MERSCOPE recommends a DV200 > 60%. Since our goal was to determine the compatibility of iST platforms under typical workflows for standard biobanked FFPE tissues from clinical pathology labs, and since TMAs are challenging to assay by DV200, samples were not prescreened based on RNA integrity. Samples were screened by H&E in the process of TMA assembly. TMAs were sliced into serial sections for processing by 10x Xenium, Vizgen MERSCOPE, and NanoString CosMx, following manufacturer instructions (see Methods).
The three different iST platforms offer different degrees of customizability and panel compositions. In terms of panel design, MERSCOPE and Xenium offer either fully customizable panels or standard panels with optional add-on genes, while CosMx offers a standard 1 K (substantially larger plex) panel with optional add-on genes (while this paper was in review Xenium and CosMx began providing options for 5000 and 6000 gene, respectively). We opted to run the CosMx 1 K panel as available commercially, as well as the Xenium human breast, lung, and multi-tissue off-the-shelf panels. We then designed two MERSCOPE panels to match the pre-made Xenium breast and lung panels, by filtering out any genes which could potentially lead to high expression flags in any tissue in the Vizgen online portal. This resulted in a total of six panels, with each panel overlapping the others on > 65 genes (Supplementary Table 4).
We carried out multiple runs with each panel following manufacturer instructions as provided in 2023 and 2024 (Supplementary Table 5), with efforts made to ensure that head-to-head comparisons were available at similar time points for each pair of platforms. 2023 data on tTMA1 and the nTMA were acquired with lung, breast, and multi-tissue panels for Xenium, MERSCOPE lung and breast panels, and the CosMx 1k panel. 2024 data was acquired on tTMA1 and tTMA2 with only the breast and 1k panels, along with reference single-cell transcriptomic data from sequential slices. In an intentional deviation from manufacturer instructions, the 2024 round of tTMA1 was intentionally carried out with matched baking times after slicing for a head-to-head comparison on equally prepared tissue slices. We note that between 2023 and 2024, CosMx updated its detection algorithms and Xenium improved its segmentation capabilities by adding additional membrane staining. The main part of this manuscript is thus based mostly on the 2024 data as more representative of the current capabilities of the technologies, while the results from 2023 are included in the Supplementary Figs. and tables as useful comparisons for previous datasets acquired on these platforms.
Each data set was processed according to the standard base-calling and segmentation pipeline provided by each manufacturer. The resulting count matrices and detected transcripts were then subsampled and aggregated to individual cores of the TMA (Methods). Across all datasets we generated 394,635,679 transcripts and 5,017,397 cells. Overall, we found that the cores from each TMA were generally well adhered to the tissue and detected transcripts. The total number of transcripts recovered for each run in 2024 was highest for CosMx, followed by Xenium, and then MERSCOPE (Supplementary Table 6). Based on the initially reported number of transcripts, tTMA1 appeared to provide more counts than tTMA2 and nTMA, which we ascribed to differences in tissue quality (Supplementary Table 6). We note that the total number of transcripts from the MERSCOPE nTMA run was below what would be typically thought of as a successful run. Such a sample would normally be excluded from analysis, but we continued with the data through all analyses to illustrate how low transcript capture affects downstream results (Supplementary Table 6, Supplementary Figs. 1–4).
Xenium shows higher transcript counts per gene without sacrificing specificity
We next sought to directly compare the performance of each iST platform on matched genes. We began with a pseudo-bulk-based approach at the core level since this would not depend on differences in cell segmentation performance (see Methods)19.
First, we examined the reproducibility of the data across experiments using tTMA1 (23) and nTMA (23). When the same genes were evaluated by Xenium and MERSCOPE in sequential sections across different panels, we found that the total transcript count of all shared genes was highly correlated across data sets acquired with different panels, regardless of the tissue of origin (Supplementary Fig. 1a) Deviation in total transcript count was primarily driven by changes in morphology of a core from section to section (Supplementary Fig. 1b,c). However, explorations on a gene-by-gene basis, even in morphologically well-matched cores showed that some genes consistently had higher or lower expression in the breast or lung panel, up to 3-fold (Supplementary Fig. 1d,e). We ascribe these differences primarily to slight competition between probes but note that the magnitude of this effect is comparable to findings in bulk and single cell RNA-seq methods in FFPE tissue20. To explore technical replicability further we examined pseudo-bulk gene expression correlation for cores from the same patient and same tissue type in the same dataset and found that correlation was high (Spearman’s r > 0.74) in almost all cases (Supplementary Fig. 2a,b), indicating good sample-to-sample reproducibility within a given platform. These correlation was similar to that observed when comparing the same core profiled across slides with different panels, suggesting that each platform was highly self-consistent and matching previous reports for MERSCOPE finding high section to section correlation21,22.
We also examined the correlation of the data from each platform between 2023 and 2024 using tTMA1 (23) and tTMA1 (24). Despite several differences in acquisition (age of sample, changes to the Xenium and CosMx analysis algorithms, and using matched baking conditions in 2024 vs manufacturer recommended conditions in 2023) we nonetheless found that the total transcript count of every shared gene from all matched cores was highly correlated (Spearman r = 0.99, 0.94, 0.95 for Xenium, MERSCOPE, and CosMx, respectively, Supplementary Fig. 2c–e). The correlation plots showed an average of only 18% median fold decrease in expression across all cores for Xenium in 2024 vs 2023; a four-fold increase for CosMx, and an eight-fold decrease for MERSCOPE—differences we ascribe primarily to the improved CosMx algorithm and changes in the baking time affecting MERSCOPE. Breaking down the correlations by core showed similar results regardless of the tissue of origin, though with more variance between the datasets for MERSCOPE and CosMx than for Xenium (Supplementary Fig. 2f–j). Thus, the correlation of gene expression across matched samples is high in all three technologies across a variety of conditions, but sensitivities could show significant differences.
To evaluate the relative sensitivity of each platform, we plotted the total transcript counts of every shared gene between all combinations of platform and panel, summed across all matched cores. We found generally linear relationships between all pairs of platforms (Fig. 2a–c, Supplementary Fig. 3). When we initially examined CosMx compared to Xenium data using tTMA1 (23) we noted an upward curve in the lower expression regime indicative of higher-than-expected calls associated with the low expression regime by CosMx, though this is no longer present in tTMA1(24) likely because of the updated base calling algorithm (Supplementary Fig. 3a,d,e). Since the three methods use variable number of probes per transcript, we broke these analyses down by gene length, but found no noticeable differences between the performance on short, medium, or long transcripts (Supplementary Fig. 4a–i). Xenium consistently showed higher expression levels on the same genes than CosMx in tTMA1 (24) when all slides were treated the same way, with the Xenium breast dataset having 2-fold more counts than the CosMx multi-tissue data sets (Fig. 2a). CosMx showed higher expression levels than MERSCOPE (median of 11-fold). (Fig. 2b). Finally, Xenium showed 20-fold higher median expression with the breast panels than MERSCOPE. In tTMA2 (24), where slide histology was carried out according to manufacturer protocols, we nonetheless found generally consistent results, though the MERSCOPE data set showed lower expression levels (Supplementary Fig. 3a–c). Intriguingly, in the earlier round of experimentation, tTMA1 (23), while Xenium still showed higher counts than the other two platforms (Supplementary Fig. 3d–j), the MERSCOPE breast panel showed higher expression levels than CosMx levels, which were only 2.6-fold lower than Xenium’s (Supplementary Fig. 3i). Considering the overall higher transcripts per cell across platforms for tTMA1 over tTMA2 and nTMA, and the differences in sample handling across rounds of experimentation (Supplementary Table 5), this suggests that MERSCOPE’s ability to detect transcripts is highly sensitive to both sample quality and processing.
a Scatter plots of summed gene expression levels (on a logarithmic scale) of every shared gene between Xenium (breast) and CosMx (1k) data, captured from matched cores from tTMA1(24). Each data point corresponds to a gene. The red line represents the fitted regression (center), obtained from a first-order polynomial fit in log–log space and back-transformed to the original scale. The shaded band shows ±1.96× the standard deviation of residuals (linear scale) around this line. b Same as (a) but between MERSCOPE (breast) and CosMx(1k). c Same as (a) but between Xenium(breast) and MERSCOPE(breast). d Violin plot of percentage of all transcripts corresponding to genes relative to the total number of calls (including negative control probes and unused barcodes) averaged across cores of the same tissue type. Results are from Xenium’s breast panel; MERSCOPE’s breast panel; and CosMx’s multi-tissue 1k panel. Each data point represents a TMA-tissue type combination, such as tTMA1(24)-BrC or tTMA2(24)-BlC. Violins show kernel density; interior lines denote quartiles (median = 2nd quartile). The full data is shown in Supplementary Fig. 5a. e Violin plot of false discovery rate (FDR) where FDR(%) = (blank barcode calls / total transcript calls) x (Number of panel genes /Number of blank barcode) x 100. f Same as (e) but using negative control probes to replace blank barcodes. MERSCOPE is missing in this bar plot as it does not have negative control probes by design. g Violin plot of number of genes detected above noise, estimated as two standard deviations above average expression of the negative control probes. h Same as (g) but normalized to the number of genes in a panel or in percentage. Pairwise differences between platforms were assessed with two-sided Mann–Whitney U tests for (d–h); brackets show unadjusted p-values for each comparison.
We next wanted to assess the specificity of each platform. Each of the three platforms includes negative controls which are used to evaluate sample quality23,24. Xenium and CosMx include both negative probes (e.g., real probes targeting nucleic acids that are not present in human tissue) and negative barcodes (e.g., algorithmically allowable barcodes that are not associated with any probe in the experimental panel). MERSCOPE includes only negative barcodes by default. To determine specificity, first we calculated the fraction of all transcripts corresponding to targeted genes relative to the total number of calls (Fig. 2d). We found that Xenium consistently showed the highest on-target fraction, while CosMx was lowest across most tissue types except for bladder cancer and tonsil, where CosMx showed a higher on-target fraction than MERSCOPE (Supplementary Fig. 5a). However, this measurement is biased because of the relative numbers of controls and target barcodes. We therefore also adopted a false discovery rate (FDR) calculation which normalizes for these differences and is calculated against both the negative probes and negative barcodes (see Methods, Fig. 2e, f). We found that Xenium consistently showed the lowest FDR while CosMx showed the highest FDR in most cancer types (15 out of 22 TMA-cancer type combinations, see Supplementary Fig. 5b,c) regardless of whether we standardized to negative control barcodes or probes.
Finally, we used the negative control barcodes to evaluate the number of genes reliably detected by each platform in each tissue type. For each core, we calculated the number of genes that were detected two standard deviations above the average expression of the negative control probes. These numbers were then averaged for cores of the same tissue type. Because the CosMx panel was almost three times larger, it yielded a larger absolute number of detected genes in all 22 TMA-cancer type combinations (Fig. 2g, Supplementary Fig. 5d). CosMx also detected the highest fraction of genes in 15 out of 22 TMA-cancer type combinations, followed by Xenium (5 cancer types across two TMAs: breast cancer, melanoma, pancreatic cancer, kidney cancer, and SCC, see Supplementary Fig. 5e). The conclusions above are broadly the same when using 2023 protocols (Supplementary Figs. 6–7), except that the 2024 version of CosMx chemistry shows an almost 10-fold lower FDR in some tissues, with a corresponding increase in number of genes above noise, regardless of whether sample histology was carried out the same way or following manufacturer instructions—again in line with increased sensitivity from CosMx’s updated data processing.
Concordance with reference RNA-seq data
We next sought to determine whether a higher number of expressed genes is representative of increased sensitivity to real biology or increased false positive rates. First, we evaluated the correlation of iST data to reference RNA-seq data. We aggregated pseudo-bulk both tTMA1 (23 and 24) data from all panels of the three platforms and compared them to data from the TCGA25 program (see Methods)11. In Breast Cancer from tTMA1 (24), the Spearman correlation coefficients were 0.64 for Xenium, 0.55 for MERSCOPE, and 0.80 for CosMx (Fig. 3a). The performances across all other cancer types followed a similar trend, with CosMx showing the highest correlation coefficient for a particular tissue type followed by Xenium and then MERSCOPE (Supplementary Table 8). These results stand in contrast to results on tTMA1 (23) where each method was generally comparable in correlation coefficient (Supplementary Fig. 8a Supplementary Table 8). We also observed similar correlation coefficients across lung and multi-tissue gene panels relative to bulk RNA-seq expression data (Supplementary Fig 8b).
a Scatter plots of common genes, showing the averaged expression of a gene across breast cancer cores profiled by the indicated panel, normalized to 100,000 vs the average FPKM from TCGA for all samples of a matched tissue type from tTMA1 (24). 1st-order polynomial fitting was performed and is shown as a black line. Insets are Spearman correlation coefficients. b Same as (a) but showing breast cores from nTMA (23) dataset versus averaged nTPM values from GTEx breast samples. c Scatter plots of overlapping genes, showing the aggregated expression of a gene in tTMA1 (23) across smooth muscle cells profiled by the indicated panel vs the gene expression from scRNA-seq. d Same as (c) but for tTMA1 (24). e Same as (c) but for tTMA2 (24). f Comparisons of residuals between single-cell and tTMA1 (23)/tTMA1 (24) for all platforms.
We also compared the pseudo-bulk results from nTMA(23) with bulk RNA-seq data obtained from GTEx12,26 The Xenium breast and CosMx data sets showed similar correlations to breast data obtained from GTEx, while the MERSCOPE had significantly lower correlation, consistent with a run which doesn’t pass QC (Spearman’s correlation coefficients of 0.33 vs 0.46, 0.57, respectively, Fig. 3b. Supplementary Table 9, Supplementary Fig 8c). These relative trends remained true across most normal tissue types, though we found that thyroid, pancreas, and lymph nodes showed the lowest correlations across all panels while prostate, tonsil, and liver showed the highest correlations (Supplementary Table 9). We also evaluated the expression of tissue-specific transcript markers across each platform by selecting marker genes from the GTEx database (see Methods). In nTMA (23), we found tissue-specific expression patterns of several of these markers across all selected panels when visualized across each healthy tissue type with Xenium showing the most distinct pattern followed by CosMx and finally MERSCOPE (Supplementary Fig. 8d). Overall, our comparison to TCGA and GTEx 2023 data suggests that while platforms may be more highly correlated to reference datasets in some cases, all are within a similar correlation regime regardless of tissue type. We next sought to validate the consistently higher correlation of CosMx vs Xenium vs MERSCOPE. Since TCGA and GTEx data are not generated from matched samples, we also performed a more stringent comparison by generating single-cell transcriptomic data for both tTMA1(24) and tTMA2(24), acquired on sequential sections (Fig. 3c–e, Supplementary Table. 5). This resulted in a matched single-cell reference dataset comprised of 14,945 and 17,749 high-quality single cells respectively for tTMA1 and tTMA2. Since TMAs include many cell types which are difficult to separate in single cell workflows, we decided to subset out vascular smooth muscle cells based on canonical marker genes identified (Methods). This cell type is readily recognizable in each platform and thus serves as a good basis of comparison of relative gene expression across modalities. The observed population’s showed high expression of genes known to be associated with smooth muscle cells (but not used for selection) such as MYH11, DST, and LUM; while showing low expression of genes not associated with smooth muscle cells such as CSTG, CD86, and PDCD1LG2.
We then aggregated pseudo-bulk expression from all three spatial platforms and single-cell data and performed correlation analyses across all spatially measured genes in this cell type27. When we compared Spearman correlation coefficients, we recapitulated the trends observed in 2024 data compared to reference bulk RNA-seq: namely, CosMx had the highest correlation coefficients, followed by Xenium, and finally MERSCOPE (Fig. 3c–e). Importantly, this is true for both tTMA1 (23) and tTMA1 (24). We found that on tTMA1 (24), CosMx has significantly higher correlation coefficients relative to Xenium (0.77 vs 0.53, p = 1.2 × 10−10, Fisher’s z transformation throughout) and MERSCOPE (0.6, p = 1.7 × 10−6) on tTMA1. This was consistent with results on tTMA2 (24): (CosMx: 0.78, Xenium: 0.72, p = 0.02, MERSCOPE: 0.23, p = 0) (Fig. 3d, e). However, when we restricted the same correlation analysis to only the expression of the common genes measured by each spatial platform relative to single-cell, the differences in correlation to single-cell data among the platforms were reduced and were no longer statistically significant on tTMA1 (24) (CosMx: 0.60, Xenium: 0.53, p = 0.23, MERSCOPE: 0.54, p = 0.20) while on tTMA2 (24) only the MERSCOPE difference remained significant (CosMx: 0.61, Xenium: 0.69, p = 0.84, MERSCOPE: 0.3, p = 1.9 × 10−3) (Supplementary Fig. 8e–g). We therefore conclude that the primary factor contributing to the higher correlation of CosMx was the larger panel size, which encompassed genes with a broader range of expression values.
The availability of a matched reference data set across several replicates allowed us to probe whether there were platform specific systematic biases in measuring certain genes. By comparing the residuals of the fits to single cell data we found that all three platforms consistently over- or undercounted- certain genes—largely non-overlapping sets—relative to single-cell measurements, presumably due to differences in sensitivities among probe designs and decoding chemistries (Fig. 3f). While the random noise between replicates could be of equal magnitude to this effect as evidence by genes with large residuals in one data set or another, this systematic effect could explain the spread in gene expression observed between platforms in Fig. 2a–c.
Out of the box segmentation and filtration from each platform yields cells with comparable numbers of detected transcripts and genes
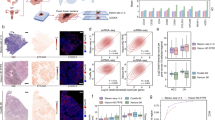
Next, we compared the performance of each iST method on a single-cell level. As of 2024, all three platforms generate cell boundaries based on a DAPI image combined with a membrane marker (Fig. 4a). Raw images are provided for these stains to facilitate custom cell segmentation approaches, but we opted to use out-of-the-box cell segmentation method for each individual platform to replicate a real-world use-case of these platforms. We evaluated the cell segmentation accuracy by comparing the segmentation outputs against manual annotations of a subset of tTMA1 (24). This subset included three TMA cores selected for their morphological similarity across platforms and their representation of different cellular structures (see Methods) and yielded > 31,400 annotated cells (see Methods) in total. We then computed precision, recall, and F1 score, across three distinct cellular scenarios: dense cells, sparse cells, and elongated cells (Fig. 4b). In dense cell distributions, CosMx and Xenium achieved significantly higher precision (0.90) than MERSCOPE (0.83, p < 0.01). Recall and F1 score results are consistent with precision across each platform. The segmentation accuracy of all three platforms decreased in elongated cells and followed by sparse cells but the relative performance of the platforms remained the same. The reduced performance is likely due to the complex and irregular shapes of elongated cells and the reduced contextual information surrounding sparse cells, increasing the likelihood of false positive and negatives. No significant performance difference was detected between CosMx and Xenium across the three scenarios.
a Top row: Subset of data showing DAPI (blue fill) and membrane staining (green fill) overlaid with cell segmentation boundaries (white outline) and manually annotated cell centroid (red point). Middle row: all the transcripts in green dots and EPCAM in blue dots. Bottom row: segmented cell boundaries (white outline) before and after filtration (Cyan outline: Cells kept after quality control; Orange outline: Cells excluded after quality control). We acquired imaging data from 263 TMA cores (170 from tTMA1, 48 from tTMA2, and 45 from nTMA) using Xenium, MERSCOPE, and CosMx, respectively. Segmentation was performed on three representative TMA cores for each IST platform, yielding a total of 9 cores and 31,384 annotated cells. b Segmentation accuracy evaluated by three metrics: Precision, Recall, and F1 Score, in various scenarios including dense cells, sparse cells, and elongated cells. Pairwise platform differences were tested using two-sided Tukey’s HSD following one-way grouping by platform (per core and scenario). Reported p-values are Tukey-adjusted for multiple comparisons. Asterisks indicate significance thresholds (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001). Exact adjusted p-values are shown above brackets. c Heatmap of transcripts per cell after filtration. All available genes are considered here for each panel. We filtered out cells with fewer than 10 transcripts for Xenium and MERSCOPE, and fewer than 20 transcripts for CosMx, in accordance with each platform’s recommended threshold. d Same as (c) but showing unique genes per cell. e Same as (c) but reanalyzed using only shared genes. f Same as (d) but reanalyzed using only shared genes. g Co-expression density map for three pairs of disjoint genes (rows) from all three platforms (columns) from tTMA1 (24). MERSCOPE breast dataset does not have enough cells to generate the 2D histogram for PPARG vs. CD68. All cells across all tissues which include at least one detected transcript of either of the indicated genes are plotted together, with color indicating the number of cells at the indicated expression levels of each gene. Data throughout is from tTMA1 (24) and tTMA2 (24).
We next filtered out empty regions of space and cells without any transcripts for downstream examination and quantified the fraction of cells containing differing numbers of transcripts per cell (Supplementary Fig. 10a). We chose a permissive threshold of removing cells with fewer than 10 transcripts for Xenium and MERSCOPE, and 20 transcripts for CosMx from downstream analysis as recommended by each technology11,28,29. tTMA1 (24) consistently had a greater fraction of cells passing filtration, with CosMx retaining the most cells (95.97%) followed by Xenium (94.28%) and MERSCOPE (27.97%) (Supplementary Table 6) while tTMA2 (24) had lower cell retention in Xenium (92.66%) and MERSCOPE (2.98%) but slightly higher cell retention in CosMx (96.30%). The relative order of the platforms based on the fractions of cells remained the same. Unsurprisingly, filtration decreased the number of retained cells per unit area for all platforms, with the smallest decrease coming for CosMx and Xenium (Supplementary Fig. 10b). The cells retained from CosMx and Xenium had similar areas, while filtration of the MERSCOPE data sets resulted in a higher average cell area (Supplementary Fig. 10c). This effect is largely driven by the removal of low-quality cells, specifically those with fewer than 10 detected transcripts.
After filtration, we compared the number of transcripts and the number of unique genes per retained cell across all tissues and all panels, focusing on tumor cores that were sampled by all three platforms (Fig. 4c–f). As expected, given its larger panel size, CosMx detected the highest number of transcripts per cell and the highest number of unique genes per cell in all tissue types, followed by Xenium breast panel and then MERSCOPE breast panel (Fig. 4c, d). If these analyses were restricted to only the shared genes across all panels, numbers were much lower (Fig. 4e, f), with the Xenium breast panel giving the highest numbers of transcripts per cell in 21 out of 22 TMA-cancer type combinations. The CosMx data showed the highest number of transcripts per cell in liver cancer and comparable transcript counts in testicular cancer to the Xenium breast panel. The MERSCOPE data generally had the lowest number of transcripts per cell, though pancreatic cancer approached the results from Xenium, and had higher transcripts per cell than CosMx (Fig. 4e). Xenium had the highest number of unique genes per cell across all tissue types and followed by CosMx and MERSCOPE (Fig. 4f). When we performed similar analyses for the 2023 datasets, even though these did not include membrane segmentation for Xenium (Supplementary Fig. 10d–g) and note that MERSCOPE showed significantly higher numbers of transcripts. This is consistent with our previous results (Supplementary Fig. 2i) showing that in certain conditions MERSCOPE could approach Xenium transcript counts.
We then determined how different iST platforms’ segmentation algorithms perform by assessing the expression of canonical markers. We examined the co-expression of CD3E, a canonical T-cell marker, and EPCAM, a marker for epithelial cells30,31 across all filtered cells; the co-expression of CD4 and CD8A, markers of T-cell subsets32,33,34; and the co-expression of PPARG, a marker for adipocytes and CD68, a marker for macrophages35,36,37. We reasoned that all these marker gene pairs are disjointly expressed, and a well-performing segmentation algorithm should yield few cells expressing both markers. We pooled all the filtered cells from matched tTMA1 (24) cores of each platform and plotted the expression of one gene against the other and converted the scatter plot to a 2D histogram showing cell numbers in each co-expression bin. We found that Xenium showed clear patterns of disjoint expression, separating cells from different lineages, while MERSCOPE showed such a pattern for CD3E vs. EPCAM but not for the other two pairs (Fig. 4g). Higher quality MERSCOPE data from tTMA1 (23) showed such a pattern for CD3E vs. CD19, CD4 vs. CD8A, and CD3E vs. EPCAM (Supplementary Fig. 10h–i). Similarly, CosMx only showed such a pattern for PPARG vs. CD68 but not for the other two pairs. Nevertheless, since the CosMx panel is much higher plex, and retained similar numbers of transcripts and genes to Xenium, we next wondered how these two methods performed in terms of cell type recovery.
Clustering analyses reveal differences in cell type recovery across platforms
In a typical iST workflow, a key step is reducing the dimensionality of the data by identifying cell types, their unique states, and their expression patterns for further analysis leveraging spatial information38. To compare across platforms, we clustered the data from the filtered cells from all the cores for each TMA with a focus on breast tissues. The initial clustering of whole TMA datasets (except MERSCOPE normal tissue) showed expected batch effects caused by patients and tissue types with broadly similar cluster arrangements around morphological tissue features (Supplementary Fig. 11). We removed batch effects (see Methods) and then performed targeted clustering and cell type annotation for breast samples from tTMA1 (23), tTMA1 (24) and tTMA2 (24) for Xenium breast panel, MERSCOPE breast panel and CosMx multi-tissue panel (Supplementary Fig. 12).
When looking at tTMA1 (23), in breast cancer, after batch effect removal (Supplementary Fig. 12d–f), Xenium resulted in nine cell types (alveolar cells, B cells, basal cells, fibroblast, hormone-sensing cells, immune cells, myeloid, T cells, and vascular & lymphatic cells) (Fig. 5a). On the other hand, CosMx resulted in eight cell types (alveolar cells, basal cells, epithelial cells, fibroblast cells, hormone-sensing cells, immune cells, myeloid, and vascular & lymphatic cells). MERSCOPE resulted in six cell types, including alveolar cells, fibroblast cells, hormone-sensing cells, myeloid cells, T cells, and vascular & lymphatic cells. The cell type annotation of Xenium and CosMx was comparable in terms of both transcriptomic profile and subtype depth, with CosMx only unable to annotate immune cell subtypes (B cell and T cell). Gene expression of the same cell type from both platforms correlated well (Fig. 5a). MERSCOPE, on the other hand, identified most, but not all, the cell types recognized by Xenium and CosMx, including alveolar cells, fibroblast cells, hormone-sensing cells, myeloid, T cells, and vascular & lymphatic cells. MERSCOPE and Xenium showed a high correlation for almost all matching clusters. The correlation map shows a clearer one-to-one mapping between MERSCOPE and Xenium clusters than Xenium and CosMx clusters (Fig. 5a).
a Clustering results of breast cancer samples in tTMA1 (23) from Xenium breast panel, MERSCOPE breast panel, and CosMx multi-tissue panel. Correlation plot showing the correlation between cell types identified in CosMx and Xenium as well as MERSCOPE and Xenium. b Clustering results of breast cancer samples in tTMA1 (24) from Xenium breast panel, MERSCOPE breast panel, and CosMx multi-tissue panel. Correlation plot showing the correlation between cell types identified in CosMx and Xenium as well as MERSCOPE and Xenium. c Heatmaps show high cell type annotation correlation between experiments conducted for Tumor TMA 1 of Xenium (top), MERSCOPE (middle), CosMx (bottom). d Clustering results of breast cancer samples in tTMA2 (24) from Xenium breast panel and CosMx multi-tissue panel. Correlation plot showing the correlation between cell types identified in CosMx and Xenium.
Results remained similar when the same sample was rerun with all spatial methods receiving the same slicing protocol a year later (Fig. 5b). Xenium identified all nine cell types as well as one additional cell type, epithelial cells, while CosMx and MERFISH identified the exact same set of cell types. Further, the cell types identified from tTMA1 (23) and tTMA1 (24), the two experiments, correlated well (Fig. 5c), where all cell types from tTMA1 (23) show high correlation with the same identifications in tTMA1 (24). However, CosMx, Xenium, and MERSCOPE exhibit differences in cell type annotations, particularly in the identification of cell subtypes. For example, in Core 99, Xenium appears blue in the center because many cells are identified as alveolar, while CosMx appears yellow due to the identification of epithelial cells. Alveolar cells are a subtype of epithelial cells, and closer examination of the heatmaps reveals that the transcriptomic profiles of alveolar cells identified in Xenium often exhibit high correlation with epithelial clusters in CosMx and MERSCOPE (Fig. 5b). While performing the cell type annotations independently for each platform, the markers provided by CosMx and MERFISH cannot support a confident classification of the cells as alveolar, unlike Xenium, which shows clear alveolar markers (Supplementary Fig. 13). This lack of specificity is also observed with immune cells: Xenium identifies more subtypes with distinct markers, whereas CosMx and MERSCOPE often only produce clusters broad enough to indicate immune cells without further subtyping. These limitations result in correlation heatmaps where cell type clusters do not always align one-to-one between platforms, such as seen in tTMA1 (24), where the Xenium alveolar cluster correlated with both alveolar and epithelial clusters in CosMx and MERSCOPE.
The results in tTMA1(24) stand in contrast to those in tTMA2 (24). In the Invasive Breast Cancer samples in tTMA2 (24) CosMx found seven cell types, including alveolar cells, basal cells, fibroblasts, hormone-sensing cells, immune cells, myeloid, and vascular & lymphatic cells; whereas Xenium only annotated five cell types, including basal cells, fibroblast cells, hormone-sensing cells, immune cells and vascular & lymphatic cells. MERSCOPE is not included due to its low number of transcripts. High correlation is again found between the cell types of CosMx and Xenium. In conclusion, for all TMAs in 23 and 24, all platforms are capable of generating results that support reasonable, consistent cell type annotations that would allow further biological analyses. We find that CosMx or Xenium can resolve higher numbers of cell types, with the relative performance likely dictated by the presence of key marker genes in the panel (more likely in the larger CosMx panel) vs the higher sensitivity and lower false positives (more likely in Xenium measurements).
Discussion
In this study, we compared data obtained with three commercially available iST platforms with archival FFPE tissues to assess overall technical performance and help guide experimental design with human patient-derived samples that represent an important use case of these platforms. We focused our analyses on technical performance as a function of tissue source, including 17 different tumor types and 16 normal tissue types. Overall, we found that each iST platform presented various tradeoffs in terms of implementation, panel design and panel options, and resulting total transcript quantification and downstream analyses, including cell segmentation, cell quality, and biological interpretation. All these factors must be considered when designing iST experiments.
There are significant workflow differences between the different platforms which factor into the choice of method. Cutting samples onto MERSCOPE coverslips is more technically challenging than on standard microscope slides. The total hands-on time for running a slide on Xenium is 2–3 days compared to 5–7 days for MERSCOPE and 2 days on CosMx. We found that MERSCOPE and CosMx are well set up for batch processing in the wet lab, either due to built-in pause points or the instrument’s ability to run multiple samples. Xenium is limited for batch processing by a need for a separate thermocycler for each slide pair processed in parallel. After staining, selecting regions of interest (ROIs) presented a surprising challenge for some systems: the Xenium platform could readily image the entire slide as a single ROI which easily covered entire TMAs, but the MERSCOPE ran into a 1 cm2 imaging area limit which meant cores in the addressable region were left unimaged, while the CosMx workflow required a demanding manual selection of ROIs for each core. These factors are likely to change as each company updates its protocol, but currently, Xenium offers the shortest, least hands-on workflow.
We analyzed each resulting dataset with a combination of manufacturer recommended processes for each platform and computational tools that can be implemented by the user downstream. These pipelines each result in count matrices and detected transcripts that can be analyzed using a suite of emerging tools. To facilitate the comparisons of iST platforms at a high level and enhance readability, we synthesized our findings into a qualitative summary table (Table 1).
When analyzed at a core level to abrogate the effects of individual cell-segmentation performance, we found that the total number of transcripts varied substantially across iST platform. With 2023 chemistry, Xenium yielded the highest number of transcripts, followed by CosMx and MERSCOPE, but with updated chemistry in 2024, CosMx showed higher total numbers of transcripts, consistent with a larger overall panel size. However, when this analysis was restricted to shared genes, Xenium generally had higher sensitivity for the same genes over CosMx and MERSCOPE across each tissue type, panel, sample, histological preparation, and acquisition round. The Xenium platform also showed the highest specificity by several false discovery rate metrics relative to CosMx and MERSCOPE.
The most prominent observation across these comparisons for CosMx was the improvement from 2024 to 2023 chemistry. With 2024 protocols, CosMx showed the smallest difference relative to Xenium, a roughly two-fold decrease in transcript counts in samples prepared the same way (tTMA1) and in samples prepared according to manufacturer instructions (tTMA2). This is a dramatic improvement over the ~12-15 fold difference in sensitivity observed with 2023 protocols. The improvement in sensitivity also comes with an improvement in specificity, which, for some tissues, reached almost a 10-fold difference. This suggests that the new version of the CosMx algorithm presents a substantial improvement and presents a choice in slightly lower specificity and sensitivity relative to Xenium vs higher molecular plex.
The most prominent observation for MERSCOPE was the large degree of variability between samples, TMAs, and protocols. Relative to Xenium, MERSCOPE sensitivities for shared genes varied between 2.6- and 100-fold lower. However, the highest MERSCOPE sensitivity obtained from the highest quality samples (tTMA1) measured at the earlier time point (2023) and following manufacturer recommended protocols. While the test case here was for typical archival tissues, these results underscore the importance of pre-screening samples for RNA-integrity for MERSCOPE to ensure high quality input tissue. This would require more tissue and limit the number of compatible samples, which can be weighed against the benefit of highly customizable panel design.
Across all methods, we note that because of the small number of replicates from each tissue we stop short of making blanket statements about relative performance across a particular tissue type. Instead, we conclude that Xenium shows the highest per-gene sensitivity on matched samples, though with updated protocols CosMx’s sensitivity approaches Xenium and provides more overall transcripts when considering the larger panel size. MERSCOPE can achieve similar sensitivities relative to Xenium, but is highly sensitive to sample input quality and protocol deviations. While this paper was in review, two other comparison studies came to similar conclusions. A similar head-to-head comparison in FFPE tissue between CosMx and Xenium by Cook et al. found a similar difference in sensitivity to our results39. Interestingly, a comparison of public (unmatched) data from high quality mouse brain by Hartman and Satija found that MERSCOPE provided the highest sensitivities40, in line with the idea that this technique is highly sensitive to input tissue quality.
Regardless of these differences in specificity and sensitivity we found that on matched samples, all platforms were highly correlated to each other. Genes profiled across different panels within the same technology, at different time points, with different histology protocols, or even with different slicing rounds were all generally highly correlated. Similarly cores from the same patient were also highly correlated. This suggests that all three platforms can be trusted for normalized measurements of gene expression within a sample. Furthermore, all three platforms also show similar performance relative to orthogonal (non-spatial) technologies, finding comparable correlation of pseudo-bulk data to RNA-seq data from GTEx or the TCGA, as well as scRNA-seq data generated for tTMA1 and tTMA2, across each panel and platform. The correlation to orthogonal data was high but not perfect, and we found that each platform appeared to repeatedly over- or under-estimate the expression of a small number of genes (Fig. 3F)—suggesting, in some cases, middlingly-expressed genes could be seen as highly or lowly expressing.
A significant advantage of spatial transcriptomics data is the ability to map expression in single cells. We compared each platform on a cell-level basis by assessing cell identification and cell clustering. Overall, it appears that the out-of-the-box segmentation from CosMx and Xenium achieved better accuracy than MERSCOPE in terms of precision, recall, and F1 score across dense, sparse, and elongated cells. All platforms now use a membrane marker for segmentation, but the specific markers and segmentation algorithms are different, which could affect cell typing41,42,43,44, by misattributing marker genes to adjacent cells or by missing cells whose nuclei are out of the imaging plane. It is likely that segmentation performance could be improved on a sample-by-sample or tissue-type-by tissue-type basis, and future work should seek to assess cell segmentation tools and their performance across data from each platform to help inform the choice of analytical method where needed. After applying an expression level filter, CosMx overall retained the highest number of cells across various filtering stringencies. Xenium was able to effectively separate cells from different lineage markers, as judged by finding minimal coexpression of disjoint markers, while CosMx and MERSCOPE either showed more double positive cells or had fewer cells expressing the target genes overall).
To determine whether clearer identification of lineage markers resulted in improved ability to identify cell types, we performed clustering analyses specifically in breast cancer samples. We found that all three platforms had similar cell type annotation results and similar transcriptomic profile for tTMA1 (23) and tTMA1 (24). This consistency across platforms and over 2 years of sequencing highlights the reproducibility and robustness of our cell type annotations and transcriptomic profiling for TMAs. On the highest quality cores in tTMA1, MERSCOPE still successfully identified cell groups, capturing the patterns seen in other platforms, consistent with the trends in the overall transcript counts which show that high quality samples could achieve similar performance to the other platforms. We note that while similar numbers of cell types are recovered, the spatial component of the data shows that occasionally low expression of key marker genes can result in misannotation of a cell subtype which is identified in another technique.
Examining the latest protocols (2024 data), in some cases, Xenium showed larger number of annotated cell types while in others CosMx did. Since we used the full panel, not only the shared genes, when performing these clustering analyses, these differences are likely due to a balance between the benefit of more sensitive detection of tissue relevant genes and the possibility of missing informative genes in the larger panel. The number of genes included in a panel or plex is an important factor in ST experiments, and we recommend subsampling existing atlas data to determine whether the gene set which can be studied will be sufficient to cluster the cell types of interest and identify the necessary biological programs. We note that each of the manufacturers has released new products and now offer increased panel sizes since the completion of these experiments with panels now reaching 5000 genes for Xenium, 6000 genes for CosMx, and 1000 genes for MERSCOPE.
There are several limitations of our study. Our panel design for MERSCOPE required the removal of genes so the panel was compatible with all tissues, lowering the plexity slightly. This could have compromised MERSCOPE’s ability to identify cell types relative to Xenium. Due to insufficient tissue, we were unable to run the same TMA across multiple years and multiple slicing and fixation protocols, requiring us to split our efforts between tTMA1 and tTMA2. Nonetheless, we were able to show both the effects on matched sample preparation and sample preparation following manufacturer recommendations. A similar limitation affected our ability to compare to single cell data. Because of insufficient tissue and the TMA design we could not achieve an equal level of sampling with single cell approaches as with spatial methods. Thus, while we were able to identify similar cell subpopulations for the purposes of exploring relative gene expression, we leave the question of the ability of spatial methods to perform generalized cell type clustering relative to single cell to other investigations. This limit did not apply to comparing clustering between spatial platforms, where we had similar numbers of cells and could identify tissues of origin based on TMA location.
Most importantly, we only attempted to compare the performance of iST platforms under typical use cases for clinical samples obtained from archival biobanks. Our results don’t necessarily extend to non-human samples, frozen samples, and even FFPE samples which have been extensively validated for high RNA integrity. Indeed, there have been reports that MERSCOPE, in previous studies of the mouse brain, shows comparable or even superior results to those reported by 10x Xenium45. Given the large change in data quality between TMAs, and even the same sample run multiple times, we cannot exclude the possibility that in the highest quality samples MERSCOPE would provide higher transcript numbers, with the associated downstream benefits relative to Xenium and CosMx. However, the current guidance of DV200 > 0.6 restricts studies to the upper regime sample quality and limits archival investigations. Finally, we note that this study only compares the ability of spatial transcriptomic platforms to reveal transcriptional information. Specifically, we do not answer whether ST can predict protein expression of clinical biomarkers. Our preliminary exploration based on PD-L1 status annotation in tTMA1 did not show significant expression differences for any platform, but rigorous testing of this would require larger powered cohorts in each individual tissue type and clinical annotation.
Despite these limitations, our overall interpretation of these results is that amplification of RNA signal is especially important for recovery of transcript counts by iST in low-quality samples where RNA may be highly degraded and fewer landing sites are available for probes. Platforms (such as Xenium and CosMx) which rely on small numbers of landing sites and are subsequently heavily amplified are robust to RNA degradation and are thus more broadly compatible with a broad range of samples. On the other hand, when sample quality is high (as in some of our tumor samples) the gap between amplified and unamplified platforms’ performance closes and most platforms can yield useful data for subsequent downstream spatial analysis.
Methods
Sample choice and TMA construction
Three TMAs were constructed using FFPE clinical discards at Brigham and Women’s Hospital Pathology Core and were acquired with a waiver of consent for non-sequencing based readouts under IRB 2014P001026. The project adheres to all applicable ethical regulations, and the study protocol has been approved by the Human Research Committee of the Institutional Review Board (IRB) at Mass General Brigham. The samples included:
-
1.
Tumor TMA 1(tTMA1): A tumor TMA of 170 cores, 0.6 mm in diameter, including a variety of cancer samples and healthy lymphoid tissue as a positive staining control. The TMA samples were selected from samples previously characterized by ImmunoProfile and were selected to encompass both high and low levels of the biomarkers in the ImmunoProfile panel [CD8, PD-1, PD-L1, Foxp3, tumor marker (Cytokeratin, Sox10, or PAX8)]. Annotations were performed by KF and SR based on H&E and immunofluorescence staining. Cores included both tumor and healthy control annotation, though for the purpose of this study, all were combined under their tumor label. Tumors were also chosen to be a mixture of PD-L1 high and PD-L1 low. This TMA had previously been studied by both H&E, and several highly multiplexed immunostaining approaches, and was known to be of high morphological integrity.
-
2.
Tumor TMA 2 (tTMA2): A tumor TMA of 48 cores, 1.2 mm in diameter, including a variety of cancer. This TMA was chosen for the breadth of tissue lineages included and the relatively large core size.
-
3.
Normal TMA(nTMA): A normal TMA of 45 cores 1.2 mm in diameter representing a broad range of normal tissues. Samples were sourced from the same patient in either duplicate or triplicate. This TMA was chosen for the breadth of tissue lineages included and the relatively large core size.
All samples were fully de-identified before assembly into TMAs. The breakdown of the number of samples per tissue and the number of cores per tissue is included in Supplementary Tables 1–3.
Preparation of sequential sections
Sequential sections were prepared according to manufacturer instructions (“Tissue Preparation Guide Demonstrated Protocol CG000578” for Xenium, “91600112 MERSCOPE User Guide Formalin-Fixed Paraffin-Embedded Tissue Sample Preparation RevB” for Vizgen, and “MAN-10159-01CosMx SMI Manual Slide Preparation Manual” for CosMx) at the Brigham and Women’s Hospital Pathology Core. Prior to collecting samples, ~50 µm of each TMA were faced off to reach deeper into the sample where RNA integrity was likely higher. 5 µm sequential sections were then collected, floated in a 37 °C water bath, and adhered to Xenium slides (10x, PN 1000460), Vizgen FFPE coverslips (Vizgen, PN 10500102), or standard Superfrost+ slides for CosMx (Leica BOND PLUS slides, Leica Biosystems S21.2113.A). TMAs were sliced as close to the center of the active area as possible for each platform. Samples were baked at 42 °C for 3 h for Xenium, 55 °C for 15 minutes for MERSCOPE, and 60 °C for 16 h for CosMx. Sections were stored according to manufacturer instructions prior to processing, with 10x Xenium stored in a desiccator at room temperature, Vizgen MERSCOPE coverslips stored at −20 °C, and NanoString CosMx slides stored at 4 °C. To benchmark technologies under same preparation protocol, samples of tTMA1 (24) data were baked overnight and stored at room temperature. Samples for 10x Xenium and Vizgen MERSCOPE were brought to the Spatial Technology Platform at the Broad Institute for processing, while samples for NanoString CosMx were processed at the Wei lab at Brigham and Women’s Hospital.
Vizgen MERSCOPE probe selection
Pre-designed probe panels from Vizgen were not available at the time of the experiment. Therefore, we ordered custom gene panels to match the pre-released gene panels from 10x for the human breast and human lung panels. Gene lists were uploaded to the Vizgen panel design portal and were checked against all profiled tissues, removing genes that were overexpressed in any individual tissue based on Vizgen’s design guidelines (FPKM > 900), and ensuring that the total panel FPKM did not exceed the allowed limit in any individual sample type. Panels were manufactured at the 300 gene scale as custom panels BP0892 and BP0893. The final gene lists, for all three iST modalities are available in Supplementary Table 4.
Vizgen MERSCOPE data acquisition
MERSCOPE samples were imaged according to manufacturer protocol “9160001 MERSCOPE Instrument User Guide RevF”. Samples were processed in two batches, the first of four samples, two of each TMA and with each library prepped in parallel; and a follow up sample of each TMA re-run with the breast panel. Samples were first hybridized with anchoring probes overnight before being embedded in a polyacrylamide gel. Samples were incubated for two hours with a digestion solution at 37 °C and then overnight at 47 °C overnight in a detergent clearing solution and proteinase K to remove native proteins while the anchoring probes kept nucleic acids bound to the gel. After clearing, samples were additionally photobleached using Vizgen’s MERSCOPE Photobleacher for three hours at room temperature in the clearing solution. Samples were hybridized with encoding probes and a cell boundary stain (PN 10400118) and then imaged with imaging kits (PN 10400005). Samples were stored at 37 °C in clearing solution after hybridization and before final imaging. After an initial examination of the data, a second batch of both TMAs was run a second time with the human breast panel, increasing the set imaging capture thickness from 5 µm to 10 µm to capture more tissue from cores that had lifted during the gel embedding process. MERSCOPE data acquired with a 10 µm imaging depth (Supplementary Table 5), resulted in a median 3.0-fold increase in expression across all transcripts. We excluded the 5 µm MERSCOPE breast panel data from most comparisons have left references to the lung panel data in as an illustrative example of an unsuccessful run (indicated as such throughout the figures). Data was processed on premises through the standard Vizgen workflow to generate cell by gene and transcript by location matrices. We segmented the data with a built-in Cellpose method on the most accurate looking cell boundary stain.
10x Xenium data acquisition
10x Xenium samples were processed in three batches according to manufacturer protocols “Probe Hybridization, Ligation & Amplification, User Guide CG0000582” and “Decoding & Imaging, User Guide CG000584”. Samples were stained utilizing 10x’s predesigned Human Breast (10x, PN 1000463), Human Multi-Tissue and Cancer (10x, PN 1000626), and Human Lung panels (10x, PN 1000601), as they became available from the manufacturer. Slides for both TMAs were processed in pairs according to which probe library they were receiving. Slides were stained with a Xenium imaging kit according to manufacturer instructions (10x, PN 1000460). Briefly, padlock probes were incubated overnight before rolling circle amplification and native protein autofluorescence was reduced with a chemical autofluorescence quencher. Slides were processed on a 10 Xenium Analyzer, with ROIs selected to cover the entire TMA region. Data was processed on premises through the standard 10x workflow to generate cell by gene and transcript by location matrices. The 10x multimodal segmentation kit and protocol was used for 2024 but not for 2023 data acquisition.
NanoString CosMx
NanoString CosMx samples were prepared with one 1000 plex panel. Samples were hybridized with probes and stained with cell markers. Samples were loaded onto the CosMx SMI at the same time for imaging, during which branched fluorescent probes were hybridized onto the samples to amplify the signal above the background.
NanoString CosMx samples were prepared with Human Universal Cell Characterization 1000 Plex Panel (part number 122000157) according to manufacturer protocol “MAN-10159-01 CosMx SMI Manual Slide Preparation Manual”. Firstly, slides were baked at 60°C overnight for better tissue adherence. After baking, slides were treated sequentially with deparaffinization, target retrieval (15 min at 100°C), permeabilization (3 µg/mL proteinase K, 15 min at 40 °C), fiducials application, post-fixation, NHS-acetate application and then hybridized with denatured probes from universal panel and default add-on panel. After in situ hybridization (18 h at 37 °C), slides were washed and incubated with DAPI (15 min at RT) and marker stain mix (with PanCK, CD45, CD68 and cell segmentation marker CD298/B2M). Slides were washed and loaded onto the CosMx SMI for UV bleaching, imaging, cycling and scanning. Raw images were decoded by default pipeline on Atomx SIP (cloud-based service). Machine: CosMx_0020. Serial Number: INS2301H0020.
iST Data preprocessing
After data acquisition, the resulting outputs were uploaded to a Google bucket associated with a terra.bio Workspace for distribution and follow on analysis.
To facilitate standardized data formatting and subsequent analytical processes, we built a data ingestion pipeline with the following objectives: (a) to grab cell-level and transcript-level data from diverse platforms and normalize the data structure; (b) to tag each cell and transcript with essential metadata including tissue type, tumor status, PD-L1 status, among others (Supplementary Fig. 14); and (c) to transform the data into various formats tailored to the requirements of particularized analyses. Specifically, to tag the data, core centers in the TMA were manually identified using DAPI images (Xenium) or cell metadata that contains global coordinates (MERSCOPE and CosMx) using QGIS(version:3.16.10-Hannover). Cells or transcripts within a specified radius were then labeled with core metadata via spatial joining (implemented by GeoPandas, version:0.13.0). In instances where cores are in close proximity or when a uniform radius cannot be applied effectively, we manually generated the core boundary masks.
Single-cell RNA sequencing
For each sample, two formalin-fixed paraffin-embedded (FFPE) curls (25 μm each) were dissociated using the Miltenyi Biotech FFPE Tissue Dissociation Kit (CG000632 RevA, 10X Genomics). The resulting cell suspension was divided equally into four centrifuge tubes, each containing approximately 300,000 cells. Cells in each tube were hybridized with a unique Probe Barcode, as per the instructions in the “Chromium Fixed RNA Profiling Reagent Kits for Multiplexed Samples” user guide (CG000527, 10X Genomics). Post-hybridization, cells from the four tubes were washed, counted, and pooled in equal proportions. Approximately 40,000 cells from the pooled suspension were loaded onto a Chromium Q chip (PN-1000422, 10X Genomics). Sequencing libraries were prepared and sequenced on an Illumina NovaSeq platform using paired-end dual-indexing (49 cycles for Read 1, 10 cycles for i7, 10 cycles for i5, and 90 cycles for Read 2). The sequencing data was demultiplexed using bcl2fastq (Illumina). The resulting FASTQ files were processed with Cell Ranger v7.0.1 (10X Genomics) using the multi pipeline and the GRCh38-2020-A reference genome.
Statistics and reproducibility
Reproducibility comparison
To evaluate panel to panel reproducibility using tTMA1 (23) and nTMA (23) we summed the expression level of shared genes between indicated panels (breast vs. multi-tissue and breast vs. lung panels from Xenium and breast vs. lung panels from MERCOPE) over an individual core and plotted all cores present in each panel, before calculating a Spearman’s correlation. The format of the data used is shown in Supplementary Table 10. To evaluate core to core reproducibility, the individual gene counts of replica 1 were plotted against those of replica 2 and a Spearman’s correlation was calculated.
To further leveraged data from tTMA1 (24) to evaluate run-to-run reproducibility or check how consistent each platform’s results were across different experiments, we compared the total gene expression levels of the same genes measured in 2023 and 2024 from the same panel using scatter plots. We did this for all matching tissue samples and calculated Spearman’s correlation to quantify the reproducibility. The format of the data used is shown in Supplementary Table 10. To assess consistency between different but similar samples or sample-to-sample reproducibility, we performed the same comparison on a core basis for all the cores that have matching shapes and cellular structures when imaged in 2023 and 2024. We then summarize the Spearman’s correlation values in boxplot to show the relative performance between different iST platforms. While a high correlation value shows that the gene expression patterns are similar, it doesn’t account for the overall levels of expression. To get a more complete picture, we also calculated the average log2 fold change values of gene expression between 2024 and 2023 of a core and presented the results in a boxplot (Supplementary Fig. 2i,j).
On target rates and false-discovery measurements
To compare across panels and platforms, we subset all datasets to include only cores assayed in all runs. The fraction of on-target barcodes was calculated as a percentage of all transcripts corresponding to genes relative to the total number of calls (including negative control probes and unused barcodes or blank barcodes). These measurements were performed on individual cores and averaged across all cores of the same tissue type.
Because the difference in relative numbers of controls and target barcodes across different platforms, we adopted the false discovery rate (FDR) calculation to evaluate the specificity in a more normalized way (Fig. 2e, f, Supplementary Fig. 6b,c). We calculated the FDR of platform p panel m data in tissue t using the following Eq. (1) and cell level data (see example in Supplementary Table 11):
Where N is the total number of cores that belong to tissue type t, I is the total number of unique genes, J is the total number of negative control probes, \({g}_{{in}}\) is the gene expression of gene i in core n, \({{neg}}_{{jn}}\) is the total calls negative control probe j in core.
Since MERSCOPE does not include negative control probes, FDR was recalculated by substituting negative control with blank barcodes (Fig. 2f, Supplementary Fig. 6c) using the following Eq. (2):
Where N is the total number of cores that belong to tissue type t, I is the total number of unique genes, L is the total number of unused barcodes or blank barcodes, \({g}_{{in}}\) is the gene expression of gene i in core n, \({{blank}}_{{{\mathrm{ln}}}}\) is the total calls of unused barcode or blank barcode l in core n, specifically, we used “BLANK” for Xenium, “Blank” for MERSCOPE, and “SystemControl” for CosMx. We only used the data from matched cores, so N is same for different platform p.
Sensitivity comparison
Sensitivity was measured by the percentage of the total number of unique genes detected above noise level, where the noise was estimated as two standard deviations above average expression of the negative control probes.
Single-cell RNA sequencing analysis
Cell Ranger from 10x Genomics was used to demultiplex the raw sequencing data into FASTQ and to align the FASTQ files. R version 4.2.2 and Seurat version 4 were used for all downstream analyses. RNA sequencing results for tTMA1 and tTMA2 went through quality filtering where cells with greater than 5% mitochondrial gene expression or less than 200 expression counts were removed, resulting in 14,945 and 17,749 cells respectively. The number of features and percent mitochondrial can be seen in Supplementary Fig. 9. The data was scaled and normalized, and PCA with dim 1:20 as well as FindCluster with resolution 0.5 were used to find the cell clusters. No batch effect is observed. Markers for each cluster were found and used for cell type annotation. Cells that are used for RNA correspondence analysis are ACTA2-active cells (ACTA > 0), that are 526 and 1117 cells for tTMA1 and tTMA2 respectively (Supplementary Fig. 5c).
Orthogonal RNA-Seq concordance analysis
RNA TCGA cancer sample gene data summarizes 7932 samples from 17 different cancer types, and it provides FPKM for each gene documented. We used all samples which were annotated as BRCA (Breast cancer), BLCA (Bladder cancer), COAD and READ (colorectal cancer), HNSC (head and neck squamous cell carcinoma), LUAD and LUSC (non-small cell lung cancer), SKCM (melanoma), and OV (ovarian cancer). For GTEx, we selected the tissue types matching the annotation in our normal tissue TMA. For each panel, the genes probed by iST were averaged across all patients with the matching tissue label from the RNA-seq database.
To get pseudo-bulked iST values, the expression level of each gene in each core was normalized to the sum of all genes in that core and scaled by 100,000. We then averaged these scaled pseudo-bulk expression values across cores and plotted them against the averaged FPKMs from reference RNA-seq data sets.
ACTA2-active cells (ACTA > 0, 526 and 1117 cells for tTMA1 and tTMA2 respectively) from single-cell RNA sequencing experiment are also used to compare to the iST data. The single-cell gene expression is aggregated by summing the counts. We used the Cell x Gene portal46. to identify the top gene identifying smooth muscle cells which was contained in each panel: ACTA2-active cells (ACTA > 0) for Xenium and CosMx or MYLK-active cells (MYLK > 0) for MERSCOPE are extracted from the iST datasets. (ACTA2 was too highly expressed to include in the MERSCOPE panel.) Then, gene expression is also aggregated by summing the transcript counts. We use Spearman’s correlation to compute the correlation coefficient, and lm (linear model) is used to fit the line.
Tissue marker enrichment analysis
To determine the assay’s ability to specifically identify known lineage markers, we focused on the normal tissue TMA profiled with multi-tissue panel of Xenium, breast panel of MERSCOPE, and 1 K panel of CosMx. We selected genes with known canonical expression patterns using based on transcriptomics data from GTEx. If a gene had 20-fold higher expression in a specific tissue than every other tissue combined, this gene was considered to be a tissue marker and was used for assessing specificity for each platform. Counts for each gene were normalized to the total counts within the core, and then the Z-score of this gene across tissue types was plotted in a heatmap (Supplementary Fig. 8d). We calculated average expression of a gene across cores of the same tissue type and normalized to the total averaged expression of all genes. Z-scores were calculated with the mean and standard deviation across all averaged genes.
Evaluation of cell segmentation accuracy
In this study, the cell segmentation accuracy of three iST platforms was systematically evaluated using manually annotated ground truth data (31384 cells in total) from 3 TMA cores. These cores were selected to represent 3 distinct cellular scenarios—dense, sparse, and elongated cell distributions—and were morphologically matched across the platforms. We employed both DAPI and membrane staining to identify cells, then marked their centroids using the open-source software QGIS (version 3.16.10-Hannover). The resulting shapefiles were subsequently processed in Python (version 3.10) with GeoPandas (version 0.13.1) for data analysis.
For each platform across three distinct cellular scenarios: dense cells, sparse cells, and elongated cells. Multiple segmentation instances were generated to account for variability and ensure statistical robustness. We used precision, recall, and F1 score as performance metrics. Precision was calculated as the ratio of correctly identified positive segments to the total predicted positive segments, reflecting the accuracy of positive predictions. Recall measured the ratio of correctly identified positive segments to the actual positive segments, indicating the platform’s ability to capture all relevant segments. The F1 score, representing the harmonic mean of precision and recall, provided a balanced assessment of the segmentation performance by combining both metrics. To compare the performance across different platforms and core types, statistical analyses were performed using one-way Analysis of Variance (ANOVA), followed by Tukey’s Honestly Significant Difference (HSD) post-hoc tests for pairwise comparisons.
Evaluation of cell segmentation biological performance
To evaluate the biological performance of the segmentation, we plotted coexpression plots of gene pairs that are mutually exclusive including CD3E vs. EPCAM, CD4 vs. CD8, and PPARG vs. CD68. We pooled all the filtered cells from matched cores of each platform from tTMA1, dropped cells which do not express either gene, plotted the expression of one gene against the other, and converted the scatter plot to a 2D histogram showing cell numbers in each co-expression bin (Fig. 4g, Supplementary Fig. 10h,i).
Cells per area quantification
Segmented cells were aggregated by TMA cores. For Xenium and MERSCOPE data, the estimation of tissue area was performed by calculating the area of a discernible circle, utilizing respective radius of 0.3 µm, 0.6 µm, and 0.6 µm for tTMA1, tTMA2, and nTMA, respectively. Conversely, for the CosMx dataset, the tissue area estimation was approached differently due to its square-like data presentation, a result of the FOV selection process. Here, the tissue area was deduced by multiplying the number of FOVs covered by each core with the area of a single FOV.
Clustering
For cell filtering, cells with less than 10 transcript counts in MERFISH and Xenium datasets were removed, and cells with less than 20 transcript counts in CosMx datasets were removed. We followed standard processes to then cluster and annotate cell types across each dataset using Scanpy47. Briefly, data was normalized and scaled, dimensionality reduction was performed and cell clusters were identified48,49. To identify the cell type for each cluster, we used a t-test to find the markers for each Leiden cluster and annotated them according to previous literature50,51,52,53,54,55,56,57. These are some of the example markers used for cell type annotation: in breast samples, PIGR and KIT for alveolar cells, for B cells, KRT5, DST, and MYLK for basal cells, LUM, MMP2 and CXCL12 for fibroblast, etc. Heatmaps of the top 3 markers for each cluster are drawn for each dataset from all three panels (refer to Supplementary Fig. 13a–c). For datasets that showed batch effect with patients, Harmony was used to remove this variance58. Correlation heatmaps were generated over overlapping genes that exist in both datasets, and the Spearman correlation coefficient was calculated.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw data used in this study have been deposited in NCBI’s Gene Expression Omnibus59, and are accessible through GEO Series accession numbers GSE308145, GSE308146, GSE308147, GSE308148 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSExxx). The processed data generated in this study have been deposited in the Zenodo database (DOI: 10.5281/zenodo.16848917), including cell-level data, gene-level data, reconstructed.h5ad files, and the Source_Data.zip archive containing the datasets used for figure generation. In addition, we developed a web portal (https://broadinstitute.github.io/ist_benchmarking_showcase_portal/) to visualize the IST data used in this project with Celldega (https://broadinstitute.github.io/celldega/). Source data are provided with this paper.
Code availability
All code used in this manuscript for data processing and analysis are available on GitHub (https://github.com/broadinstitute/ist_benchmarking/).
References
Chen, W. T. et al. Spatial transcriptomics and in situ sequencing to study Alzheimer’s disease. Cell 182, 976–991 (2020).
Hunter, M. V. et al. Spatially resolved transcriptomics reveals the architecture of the tumor-microenvironment interface. Nat. Commun. 12, 6278 (2021).
Lein, E., Borm, L. E. & Linnarsson, S. The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 0358, 0064–0069 (2017).
Lewis, S. M. et al. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012 (2021).
Rao, A. et al. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Haque, A. et al. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 9, 75 (2017).
Williams, C. G. et al. An introduction to spatial transcriptomics for biomedical research. Genome Med 14, 68 (2022).
Tian, L., Chen, F. & Macosko, E. Z. The expanding vistas of spatial transcriptomics. Nat. Biotechnol. 41, 773–782 (2023).
Janesick, A. et al. High resolution mapping of the breast cancer tumor microenvironment using integrated single cell, spatial and insitu analysis of FFPE tissue. bioRxiv 10, 510405 (2022).
He, S. et al. High-plex multiomic analysis in FFPE tissue at single-cellular and subcellular resolution by spatial molecular imaging. bioRxiv 11, 467020 (2021).
Moffitt, J. R. et al. Molecular, spatial, and functional single-cell profiling of the hypothalamic preoptic region. Science 362, eaau5324 (2018).
Du, J. et al. Advances in spatial trasnciptomics and related data analysis strategies. J. Transl. Med. 21, 330 (2023).
Phan, H. V. et al. High-throughput RNA sequencing of paraformaldehyde-fixed single cells. Nat. Commun. 12, 5636 (2021).
Xu, Z. et al. High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat. Commun. 14, 2734 (2023).
Gnanapragasam, V. J. Unlocking the molecular archive: the emerging use of formalin-fixed paraffin-embedded tissue for biomarker research in urological cancer. BJU Int. 105, 274–278 (2010).
Matsunaga, H. et al. Reproducible and sensitive micro-tissue RNA sequencing from formalin-fixed paraffin-embedded tissues for spatial gene expression analysis. Sci. Rep. 12, 19511 (2022).
Chen, K. H. et al. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa 6090 (2015).
Walsh, L. A. & Quail, D. F. Decoding the tumor microenvironment with spatial technologies. Nat. Immunol. 24, 1982–1993 (2023).
Junttila, S. et al. Benchmarking methods for detecting differential states between conditions from multi-subject single-cell RNA-seq data. Brief. Bioinform. 23, bbac286 (2022).
Newton, Y. et al. Large scale, robust, and accurate whole transcriptome profiling from clinical formalin-fixed paraffin-embedded samples. Sci. Rep. 10, 17597 (2020).
Liu, J. et al. Concordance of MERFISH spatial transcriptomics with bulk and single-cell RNA sequencing. Life Sci. Alliance 6,e202201701(2023).
Zhang, M. et al. Molecularly defined and spatially resolved cell atlas of the whole mouse brain. Nature 624, 343–354 (2023).
Zimmerman, S. M. et al. Spatially resolved whole transcriptome profiling in human and mouse tissue using digital spatial profiling. Genome Res. 32, 1892–1905 (2022).
Trevethan, R. Sensitivity, specificity and predictive values: foundations, liabilities, and pitfalls in research and practice. Front Public Health 5, 307 (2017).
Tomczak, K., Czerwinska, P. & Wiznerowicz, M. Review The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemporary Oncology/Współczesna Onkologia 2015, 68–77 (2015).
GTEx Consortium. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
Squair, J. W. et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12, 5692 (2021).
He, S. et al. High-plex imaging of RNA and proteins at subcellular resolution in fixed tissue by spatial molecular imaging. Nat. Biotechnol. 40, 1794–1806 (2022).
Garrido-Trigo, A. et al. Macrophage and neutrophil heterogeneity at single-cell spatial resolution in human inflammatory bowel disease. Nat. Commun. 14, 4506 https://doi.org/10.1038/s41467-023-40156-6 (2023).
Nieto, P. et al. A single-cell tumor immune atlas for precision oncology. Genome Res. 31, 1913–1926 (2021).
Elmentaite, R. et al. Single-cell atlases: shared and tissue-specific cell types across human organs. Nat. Rev. Genet. 23, 395–410 (2022).
Van Pul, L. J. C. et al. Double positive T cells: more than just a developmental stage? Immunol. Lett. 218, 72–80 (2020).
Zaunders, J. J. & Munier, M. L. CD4+ C. T. L. immune warriors in the battle against viruses and cancer. FEBS J. 286, 1291–1303 https://doi.org/10.1111/febs.14793 (2019).
Zaunders, J. J. et al. CD4+CD8+ T cells: biological confusion and serendipity. Part I: Synthesis of the data on dual T-cell phenotype. Immunol. Cell Biol. 96, 733–749 (2018).
Miao, J. et al. PPARγ in adipocyte biology and systemic metabolism. Trends Endocrinol. Metab. 30, 883–897 (2019).
Tremblay, A. M. & Huot, J. R. Adipose tissue infiltration by macrophages in obesity: crosstalk with adipocytes. Cell. Mol. Immunol. 17, 116–118 (2020).
Zhang, H. et al. Tissue-resident macrophages in adipose tissue. Immunol. Cell Biol. 99, 229–244 (2021).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet 24, 550–572 (2023).
Cook, D. P. et al. A comparative analysis of imaging-based spatial transcriptomics platforms. bioRxiv (2023).
Hartman, A. & Satija, R. Comparative analysis of multiplexed in situ gene expression profiling technologies. eLife 13 https://doi.org/10.1101/2024.01.11.575135 (2024).
Greenwald, N. F. et al. Whole-cell segmentation of tissue images with human-level performance using large-scale data annotation and deep learning. Nat. Biotechnol. 40, 555–565 (2022).
Stringer, C. et al. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Petukhov, V. et al. Cell segmentation in imaging-based spatial transcriptomics. Nat. Biotechnol. 40, 345–354 (2022).
Chen, H., Li, D. & Bar-Joseph, Z. SCS: cell segmentation for high-resolution spatial transcriptomics. Nat. Methods 20, 1237–1243 (2023).
Salas, S. M. et al. Optimizing Xenium In Situ data utility by quality assessment and best-practice analysis workflows. Nat. Methods. 22, 813–823 (2025).
CZI Cell Science Program et al. CZ CELLxGENE Discover: a single-cell data platform for scalable exploration, analysis and modeling of aggregated data. Nucleic Acids Res. 53.D1, D886–D900 (2025).
Wolf, F., Angerer, P. & Theis, F. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Dries, R. et al. Advances in spatial transcriptomic data analysis. Genome Res. 31, 1706–1718 (2021).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019).
Kumar, T. et al. A spatially resolved single-cell genomic atlas of the adult human breast. Nature 620, 181–191 (2023).
Gray G. K. et al. A human breast atlas integrating single-cell proteomics and transcriptomics. Dev. Cell 57, 1400–1420.e7 (2022).
Reed, A. D. et al. A single-cell atlas enables mapping of homeostatic cellular shifts in the adult human breast. Nat. Genet. 56, 652–662 (2024).
Travaglini, K. J. et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 587, 619–625 (2020).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Wu, S. Z. et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet 53, 1334–1347 (2021).
Dave, A. et al. The breast cancer single-cell atlas: defining cellular heterogeneity within model cell lines and primary tumors to inform disease subtype, stemness, and treatment options. Cell Oncol. (Dordr.) 46, 603–628 (2023).
Yeo, S. K. et al. Single-cell RNA sequencing reveals distinct patterns of cell state heterogeneity in mouse models of breast cancer. eLife 9, e58810 (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41, D991–D995 (2012).
Acknowledgements
We thank Nir Hacohen, Ilya Korsunsky, Roopa Madhu, and Kseniia Anufrieva for helpful discussions, as well as Patricia Rogers and Natan Pierete for assistance with the 10x Xenium data acquisition. We appreciate 10x Genomics, Vizgen, and NanoString Technologies for reviewing data and analyses for quality. This work is supported by a Broad Institute SPARC grant, by a HTAN grant (3U2CCA233195-05S1), and by a Brigham and Women’s Hospital Department of Medicine - Broad Institution collaborative research Award. K.W. is supported by a NIH-NIAMS K08AR077037, a Burroughs Wellcome Fund Career Awards for Medical Scientists, and a Doris Duke Charitable Foundation Clinical Scientist Development Award. B.A.G. is supported in part through the Geisel School of Medicine at Dartmouth’s Center for Quantitative Biology through a grant from the National Institute of General Medical Sciences (NIGMS, P20GM130454) of the NIH.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.Y., S.F., tissue-microarray construction: K.F., K.P., T.B., S.R., pathological annotation: K.P., S.R., gene selection: A.Y., S.F., Xenium and MERSCOPE data acquisition: J.N., CosMx data acquisition: C.G., M.T., K.W., analysis: H.W., R.H., B.G., S.F., S.K., figure generation: H.W., R.H., B.G., S.F., writing original draft: H.W., R.H., J.N., B.G., S.F., draft reviewing and editing: H.W., R.H., K.W., B.G., S.F., supervision: K.W., B.G., S.F., funding acquisition: S.F., K.W.
Corresponding authors
Ethics declarations
Competing interests
K.W. receives research support from Merck Sharp & Dohme, 10X Genomics, and research collaboration agreement with NanoString. Consumables used in this study from both companies were purchased at full price. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Muhammad Dawood and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, H., Huang, R., Nelson, J. et al. Systematic benchmarking of imaging spatial transcriptomics platforms in FFPE tissues. Nat Commun 16, 10215 (2025). https://doi.org/10.1038/s41467-025-64990-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-64990-y