Abstract
Genome-wide association studies have identified many loci for brain disorders, but most non-coding variants fail to colocalize with bulk expression quantitative trait loci. Single-cell expression quantitative trait loci studies capture cell-type-specific regulation but are often underpowered. We developed Bulk And Single cell expression quantitative trait loci Integration across Cell states (BASIC) to combine bulk and single-cell expression quantitative trait loci through “axis-quantitative trait loci,” which decompose bulk-tissue effects along orthogonal axes of cell-type expression. BASIC better distinguishes shared versus cell-type-specific effects and increases power. Analyzing single-cell expression quantitative trait loci with cortex bulk data from MetaBrain using BASIC identified 5644 additional gene with quantitative trait loci (74.5%), equivalent to a 76.8% increase in sample size. Integrating axis-quantitative trait loci with 12 brain-related traits improved colocalization by 53.5% versus single-cell studies and 111% versus bulk studies, revealing risk genes such as DEDD for Alzheimer’s disease and drug candidates including cabergoline.
Similar content being viewed by others
Introduction
Genome-wide association studies (GWAS) have successfully unveiled numerous genetic associations with brain-related traits, including tobacco and alcohol use1, psychiatric disorders2,3,4,5, and neurodegenerative disease3,6,7,8. These associations reveal the genetic landscape of complex diseases and provide a new foundation for therapeutic development. However, a substantial proportion of identified genetic variants reside within non-coding regions9,10. Integrating functional genomics data to decode the molecular mechanism of GWAS loci would be critical for advancing our understanding of disease pathology.
Expression quantitative trait loci (eQTL) link regulatory variants to inter-individual differences in gene expression levels11. This approach has been effective in deciphering the functional roles of genetic variants within brain tissues, where the complexity of gene regulation is most profound12. For instance, de Klein et al. and Sieberts et al. respectively profiled genetic regulatory variants for gene expression across seven brain regions with a total of more than 8500 brain RNASeq samples13,14. These studies provide novel insights into gene regulation in the brain and pinpoint disease susceptibility genes for different disorders.
Despite the progress12,15, eQTLs from bulk tissues may miss regulatory variants that only affect certain cell types, especially for less common ones. Indeed, even with large sample sizes, many GWAS loci for brain-related traits failed to be colocalized with bulk(bk)-eQTLs. Moreover, studies have shown that the same cell types from different brain tissues tend to share effects, making the study of eQTLs from cell types (sc-eQTLs) more compelling16. Due to the advancement of single-cell or single-nuclei RNASeq technologies, it is now cost-effective to sequence hundreds of samples and profile cell type-specific eQTLs across the brain17. This is particularly important for neurodegenerative disorders like Alzheimer’s disease (AD)18 and addiction-related traits1,19, which affect a subset of cell types. Indeed, even though sc-eQTLs datasets are much smaller than bk-eQTLs, they can already colocalize a comparable number of GWAS loci. Further improving the power of sc-eQTL studies and refining eQTL effect sizes can potentially resolve more GWAS loci and identify risk genes across brain cell types.
To enhance the efficacy of sc-eQTL studies, we propose a novel method that integrates both bulk13 and single-cell RNA sequencing data20,21 to improve the detection of cell type-specific eQTLs. This method leverages two key insights: First, there is growing recognition that the cell states vary continuously22,23. Clustering cells into discrete cell types may lose information. Here, we calculate the principal components of sc-eQTLs effects and use them as a proxy for continuous cell states. Biologically similar cells tend to have similar PC coordinates. Importantly, we can project sc-eQTLs onto the PCs and calculate the “axis-QTLs”. Axis-QTLs decompose sc-eQTLs and bk-eQTLs in orthogonal directions. They allow us to look beyond cell types and identify shared and distinct regulatory effects across cell types. A second idea is to model bulk eQTLs (bk-eQTLs) as weighted averages of axis-eQTLs to utilize the large sample sizes of bk-eQTLs studies and improve power. These two ideas employed in BASIC allow us to greatly improve the power for identifying regulatory variants for brain gene expression levels. Integrating BASIC-refined sc-eQTLs and axis-QTLs with GWAS can further link regulatory variants to risk genes for various diseases, improving the power of colocalization analysis.
Results
Method overview
We introduce the BASIC framework to jointly analyze bk-eQTL and sc-eQTLs to improve the detection of sc-eQTLs and identify target genes for GWAS loci. BASIC relies on two key ideas. First, principal components of sc-eQTLs effects can help cluster cell types with similar biological functions. We employ meta-regression to model sc-eQTLs effects using PCs of sc-eQTLs effects as covariates (Fig. 1 and Supplementary Data 1). It decomposes sc-eQTLs effects in orthogonal directions. In this case, the model’s intercept represents shared effects across cell types. At the same time, the coefficients for other PCs measure distinct effects between major brain cell clusters, e.g., barrier cells, glial cells, and neurons. A second idea is to integrate sc-eQTLs and bk-eQTLs to improve the inference of axis-QTLs. Extending our previous work, we theoretically prove that bk-eQTLs can be decomposed into orthogonal axis-eQTLs. Leveraging this compositional relationship and jointly modeling bk-eQTLs and axis-eQTLs can improve the power (Fig. 2).
We calculate the principal components (PCs) of significant sc-eQTLs in the meta-analysis of datasets from Fujita et al. 21 and Bryois et al. 20. Each data point in the plot represents a distinct cell type. Biologically similar cell types tend to be clustered together in the plot, e.g., neurons (e.g., inhibitory and excitatory neurons), and glial cells (e.g., oligodendrocytes, astrocytes, and microglia). Using PCs as covariates, BASIC will borrow strength from biologically similar cell types and improve the power of identifying regulatory variants. As we show in Supplementary Notes Section 2, the PCs of the sc-eQTLs effects are concordant with the PCs of the covariance matrix of cell type gene expression levels.
BASIC consists of three key steps, including 1) estimating cell-type weights from bulk eQTLs and sc-eQTLs; 2) using principal components of sc-eQTLs effects to cluster biologically similar cell types and cell states; 3) joint modeling axis-eQTLs and bk-eQTLs using a meta-regression model with principal components as covariates. The axis-QTLs can improve the discovery of risk genes for brain-related traits in downstream colocalization and TWAS analyses. This figure is created in BioRender. W, L. (2025) https://BioRender.com/r75v850.
BASIC substantially improves the power for identifying regulatory variants for gene expression levels. The improvement is substantial when the eQTLs effects are shared in biologically similar cell types, but remains sizable in scenarios where eQTL effects are present in only one or a few random cell types. The refined sc-eQTLs and axis-QTLs effects from BASIC can substantially improve all downstream analyses, including transcriptome-wide association studies (TWAS), co-localization, and drug repurposing analyses13.
In the following sections, we will first describe the real data application and compare different methods in real data. We will leave the details of simulation studies in Supplementary Notes.
Principal component analysis reveals distinct cell type clusters
We first perform fixed effect meta-analysis of sc-eQTLs from Bryois et al. 20 and Fujita et al. 21 datasets for 7 overlapped cell types, to maximize the sample sizes. We name the resulting sc-eQTLs “meta-sc-eQTLs”. Using the meta-sc-eQTLs dataset, we performed principal component analysis (PCA) of sc-eQTL effects across cell types (see Methods for details), and show the plots of top PCs in Fig. 1.
We can view the PCs as weighted combinations of sc-eQTLs effects from different cell types. The plots of top PCs of sc-eQTLs effects reveal clusters of cell types with shared biology. For instance, the first two PCs separate barrier cells (i.e., pericytes and endothelial cells) from glial and neuronal cells. Endothelial cells carry specialized functions in blood-brain barrier formation, blood flow regulation, and brain homeostasis24, while pericytes, cells that wrap around the walls of small blood vessels, play key roles in the blood-brain barrier and blood vessel formation25. On the other hand, there are close cell-cell interactions between glial and neuronal cells. Forming their own clusters will help borrow strength from related cell types. PCs 3 to 5 further separate glial cells from neurons and excitatory neurons from inhibitory neurons. Instead of analyzing associations between cell type gene expression levels, we propose to project sc-eQTLs effects across cell types onto the PCs, and name them axis-eQTLs. Axis-QTLs measure how the regulatory effects vary along the axis of variation of gene expression levels (i.e., the PCs of sc-eQTL effects, which are also the PCs of gene expression levels) and can better retrieve shared effects across cell types. As shown below, BASIC and axis-QTLs can lead to more eSNPs and eGenes (genes with at least one eSNP) and colocalize more GWAS loci.
BASIC substantially improves the number of eQTLs across brain cell types
Using the dataset from Bryois et al. as input, we first compare the power of identifying eSNPs associated with gene expression levels across cell types. A SNP is considered associated if the two-sided p-values < 1 × 10⁻⁶, which is roughly the lower bound of Bonferroni threshold, adjusting for the number of SNPs tested for each gene in a cell type. We compare BASIC, JOBS26 (which shares a similar idea to IBSEP27, and jointly analyzes sc- and bk-eQTLs), mashr28, and the method that analyzes sc-eQTLs data from Bryois et al. 20 alone for detecting regulatory effects across brain cell types. For mashr, we apply it to analyze sc-eQTLs (mashr-sc) or jointly analyze both single-cell and bulk eQTLs (mashr-sc+bk). By analyzing axis-QTLs, BASIC can borrow strength from shared effects across cell types and bk-eQTLs. It has the highest power. By analyzing axis-QTLs and using the Cauchy combination test to combine results across different PCs, BASIC identifies 80,8976 SNPs and 8597 genes associated with at least one PC, which is 38.19% and 22.22% higher than JOBS-identified eSNPs and eGenes (Fig. 3A, B). Among the axis-QTLs, 20775/13554 SNPs and 589/364 genes were associated with the first and second PC, representing distinct effects between barrier cells and glial/neuronal cell types. 18714/17295/9600 SNPs and 349/338/206 genes were associated with the 3rd to 5th PCs, representing eQTLs with distinct effects between glial or neuronal cell types (Fig. 3C, D, Supplementary Data 2).
A, B compare the number of significant eSNPs and eGenes identified by five approaches: (i) using Bryois et al. only; (ii) mashr-sc (mashr applied to sc-eQTLs data), (iii) mashr-sc+bk (mashr applied to analyze sc-eQTLs and bk-eQTLs data), (iv) JOBS, and (v) BASIC. To facilitate the comparison of different methods, we use the Cauchy combination test to combine the p-values of different cell types or PCs. We also replicate significant eSNPs and eGenes in Fujita et al. C, D show the numbers of discovered and replicated eGenes and eSNPs based on axis-QTLs.
JOBS integrates sc-eQTLs with bk-eQTLs datasets but does not model shared effects between cell types. JOBS is not as powerful as BASIC, but outperforms other methods. Specifically, JOBS increases the number of eSNPs by 304% to 1085% compared to analyzing sc-eQTLs alone, 79% to 764% compared to mashr-sc, and 73% to 978% compared to mashr-sc+bk. The improvement is higher for more common cell types. For instance, in the most abundant cell type, excitatory neurons, JOBS identifies 460,329 eSNPs, compared to 69,064 using sc-eQTLs alone (567% increase), to 76,329 using mashr-sc (503% increase), and 101,159 using mashr-sc+bk (359% increase). The improvement remains substantial in less abundant cell types, e.g., endothelial cells. In this case, JOBS identifies 49,599 eSNPs compared to 5900 using sc-eQTL alone (741% increase), 7276 using mashr-sc (582% increase), and 6035 using mashr-sc+bk (722% increase) (Supplementary Fig. 1, Supplementary Data 3).
We then validated the eSNPs identified by each method in two independent datasets: Fujita et al. 21 which included seven overlapping cell types as Bryois et al. 20, and Lopes et al. 29, which includes only microglia. We first compare the number of eSNPs identified across cell types and PCs based on Cauchy combined p-values. BASIC replicated 447,952 eSNPs, which led to 196.12%, 406.12%, 336.02% and 18.83% improvement over using sc-eQTLs data alone, mashr-sc, mashr-sc+bk, and JOBS, respectively. For axis-QTLs, 323,613, 8547, 5803, 8350, 6986, 3629 and 1285 eSNPs are replicated for PC0 to PC6 (Fig. 3A, C, Supplementary Data 2). When it comes to cell type level eQTLs, the replication results confirmed a significant increase of power of JOBS. For instance, in excitatory neurons, JOBS replicated 218,077 eSNPs in Fujita et al. 21 compared to 56,643 using Bryois et al. 20 alone (285% increase), 61,703 eSNPs using mashr-sc (253% increase) and 83,412 eSNPs using mashr-sc+bk (161% increase). Moreover, many of these eSNPs were novel, and not found in Bryois et al. 20 or MetaBrain13, showing that JOBS does not simply copy results from bk-eQTLs (Supplementary Fig. 1, Supplementary Data 3). BASIC does not identify eQTLs specific to a given cell type and was not included in this comparison. The patterns remain similar when comparing the power of eGene discovery (Fig. 3B, D, Supplementary Data 2) and replications (Supplementary Fig. 2, Supplementary Data 4).
Finally, using meta-sc-eQTLs dataset as input, we compared the number of eGenes identified by BASIC, JOBS, mashr. Meta-sc-eQTLs data alone identified 7575 eGenes, while mashr-sc+bk and JOBS identified 9227 and 10,006 eGenes. By analyzing axis-QTLs, BASIC further increases the number of eGenes to 13,219, representing a 75% improvement in eGene discovery power compared to meta-sc-eQTLs alone. We use BASIC and JOBS results based on meta-sc-eQTL for all downstream analyses (Supplementary Figs. 3–5, Supplementary Data 5–7).
Simulation studies highlight the improved power of BASIC
Baseline read counts for sc-RNA-Seq data were simulated using a negative binomial distribution with parameters estimated from Velmeshev et al. 30. The baseline read counts for each individual were further adjusted based on causal eQTLs effect sizes and genotypes. Bulk RNA-Seq data were simulated by aggregating the reads from constituent cell types, weighted by cell type proportions sampled from real data estimates. Notably, the cell type proportions may differ between individuals. Based on simulated read counts, we calculated transcript per million (TPM), performed inverse normal transformation of TPMs, and used them to conduct bk- and sc-eQTL analyses, following conventional sc-eQTL analysis pipelines31. A genetic variant is deemed to be associated if the Cauchy-combined p-values from different cell types or different PCs (for BASIC) are smaller than \(1\times {10}^{-6}\), which is roughly the Bonferroni threshold for testing multiple SNPs in the cis-region. We consider a bk-eQTLs sample size of 2000 or 3000, mimicking the sample sizes of MetaBrain, and sc-eQTLs sample sizes of 300 and 400, mimicking the sample sizes of sc-eQTLs datasets from brain cell types. We compare the performance of the following methods: BASIC, JOBS, bulk eQTLs alone, sc-eQTLs alone, mashr using sc + bulk eQTLs, and mashr using sc-eQTLs alone. To evaluate type I errors and power, we calculate the fraction of significant eQTLs or eGenes with two-sided p-values < 1×10⁻⁶ for each method. Detailed information on the simulation settings is provided in the Supplementary Notes.
We first verified that the type I error for eQTLs detection was well controlled (Supplementary Data 8). When no causal SNPs were present, all methods had controlled type I error rates. We next compare the power of different methods. We consider scenarios with different combinations of cell types with eQTLs effects, i.e., eQTLs effects are present in a) two randomly chosen cell types, b) in two biologically similar cell types (astrocytes and microglia), c) in four randomly chosen cell types, and d) in all cell types. In all simulation scenarios, BASIC consistently outperforms alternative approaches, especially when compared to the use of sc-eQTLs data alone. When eQTLs effects are present in only a few cell types, bk-eQTLs may fail to identify the association and the effects in uncommon cell types may be masked. By analyzing axis-QTLs, BASIC borrows strength between similar cell types and from bk-eQTLs datasets. For example, in the scenario where eQTLs effects are present in microglia and astrocytes, two glial cell types, when analyzing sc-eQTL datasets with N = 400 and bk-eQTLs dataset with N = 3000, the power for BASIC is 46.2% which is 25% higher than the power for the second most powerful method JOBS (36.9%). JOBS and BASIC are both much more powerful than analyzing sc-eQTLs alone (31.4%), using mashr to analyze multiple cell types (23.9%), and using mashr to analyze sc-eQTLs and bk-eQTLs (27.6%). While mashr-sc+bk increases the power over analyzing sc-eQTLs alone in many scenarios, it has lower power than BASIC. This is because mashr was developed to analyze eQTLs from multiple tissues and it only considers a few scenarios where eQTLs are shared or independent across tissues. Yet, it does not properly model the compositional relationship between axis-eQTLs and bk-eQTLs, which is key to the improved power. This pattern is consistent across all scenarios where BASIC remains the top-performing method (Supplementary Figs. 6–9, Supplementary Data 9).
Colocalization using axis-QTLs substantially increases the fraction of colocalized GWAS loci
We perform colocalization to dissect GWAS loci and identify target genes for regulatory variants. For input of eQTLs effects, we use bk-eQTLs from MetaBrain, meta-sc-eQTL dataset, and also consider refined sc-eQTLs effects from JOBS, mashr-sc, mashr-sc+bk. Moreover, we also use axis-QTLs from BASIC as input. Unlike sc-eQTLs, axis-QTLs are combinations of sc-eQTLs effects across cell types, which can better capture how regulatory effects change along the axis of expression variation and allow borrowing strength across cell types. We employed the COLOC32 method to colocalize GWAS loci with sc-eQTLs or axis-QTLs effects. A locus is deemed colocalized if the posterior probability of the GWAS locus and eQTLs sharing a causal variant exceeds 0.9 (i.e., PP4 > 0.9).
We first verified that the type I error was well controlled in our colocalization analysis through extensive simulations (Supplementary Data 10). Next, we demonstrated that using axis-QTLs as input colocalizes the most loci across all simulation settings, when compared to using meta-sc-eQTLs alone, bk-eQTLs alone, JOBS, mashr-sc, and mashr-sc+bk. (Supplementary Data 11).
We next perform colocalization analysis using for 12 brain-related GWAS phenotypes. Our study focuses on tobacco and substance use1,19, neuropsychiatric disorders (e.g., attention deficit hyperactivity disorder (ADHD)33, bipolar disorder (BiPo)3, major depressive disorder (MDD)7, Schizophrenia (SZ)4), and neurodegenerative disease, i.e., amyotrophic lateral sclerosis (ALS)5, Alzheimer’s disease (AD)34, and Parkinson’s Disease (PD)35 (Supplementary Data 12, 13).
Using axis-QTLs from BASIC as input colocalizes the most loci (287), followed by JOBS (246). In general, using single-cell eQTLs helps colocalize more loci than using the bk-eQTLs dataset, highlighting the importance of resolving cellular heterogeneity of regulatory effects. By borrowing strength from bk-eQTLs datasets, methods such as JOBS and mashr-sc+bk can refine eQTLs effect estimates and lead to a bigger number of colocalized loci compared to using meta-sc-eQTLs dataset alone (Supplementary Figs. 10, 11, Supplementary Data 14, 15).
Colocalization using axis-QTLs from BASIC leads to the discovery of many interesting risk genes that are missed by other approaches. One example is CACNA2D2, a gene that encodes the alpha-2/delta subunit of the voltage-dependent calcium channel complex. The worm ortholog of CACNA2D2, known as tag-180, is involved in nicotine-motivated behaviors36. Loss-of-function mutations in tag-180 result in impaired development of nicotine-conditioned cue preference, indicating that the alpha-2/delta subunit of calcium channels is involved in nicotine-seeking behavior. Together, evidence suggests that CACNA2D2 may influence smoking behavior by modulating neuronal circuits involved in reward and addiction through its role in calcium channel function.
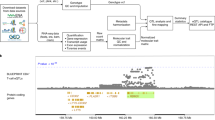
BASIC axis-eQTLs also uniquely colocalize with (Death Effector Domain-Containing DNA-Binding Protein) for AD (Fig. 4). DEDD is a protein involved in apoptotic processes, which may influence AD through its role in cell death and inflammation37,38. DEDD functions by influencing the nuclear factor-kappa B (NF-κB) pathway, which is crucial in the activation of glial cells like microglia39. Dysregulated DEDD activity might amplify neuroinflammatory responses, contributing to disease progression. DEDD is also implicated in cellular responses to oxidative stress40. Dysfunctional regulation of oxidative stress responses can lead to increased neuronal damage41. DEDD may be involved in AD etiology through its role in apoptosis and cellular signaling.
We present locus zoom plots (negative log10-based two-sided p-value) of a 1-Mb region surrounding the DEDD gene (chromosome 1:160,722,754–161,717,430 in hg38 genomic positions) across multiple datasets: AD GWAS, meta-sc-eQTLs (from seven cell types), MetaBrain bk-eQTLs, and BASIC axis-QTLs from PC0 to PC6. The darkness of the point reflects its linkage disequilibrium (LD) coefficient (R2) with the sentinel variant in the locus. The posterior probability of colocalization (PP4 in coloc) is indicated in each panel. Interestingly, this locus cannot be colocalized with any sc-eQTLs, shows moderate evidence of colocalization with bk-eQTLs, but can be colocalized with axis-QTLs from PC0 and PC1. As shown in Supplementary Fig. 16, the sc-eQTLs effects for many variants tend to have opposite effects across cell types and can cancel each other in bk-eQTLs analysis. This example demonstrates the importance of separating shared and distinct regulatory effects across cell types using axis-QTLs.
Enhanced detection of disease-associated loci through refined sc-eQTLs and axis-QTLs in TWAS
Utilizing refined sc-eQTLs and axis-QTLs as input, we can also improve TWAS34,42,43. Compared to colocalization, TWAS provides a complementary approach to link GWAS hits to regulated target genes. TWAS can also identify novel gene-level associations. We extended the framework of summary statistics-based TWAS method EXPRESSO44 to take axis-QTLs as input and perform axis-TWAS (Method). We also proposed a novel summary statistics-based procedure to validate prediction accuracy when external individual-level genotype, gene expression data, or eQTLs summary statistics are not available.
We define each locus as a 1 MB genomic window centered on the sentinel gene, extending 500 KB on each side. Genes are ranked by their TWAS p-values in ascending order, with the initial locus encompassing the 1 MB region around the gene with the lowest p-value. Subsequent loci are similarly defined, using significant genes falling outside already established loci. We will also merge overlapping loci to eliminate redundancies.
First, using JOBS-refined sc-eQTLs as inputs, we identified 7.50% and 94.55% more significant prediction models compared to results using meta-sc-eQTLs and MetaBrain2 inputs, respectively (Supplementary Data 16). Based on generated prediction models, the TWAS analyses have controlled genomic control values and identified 289 loci, representing 17.95% and 52.91% increases compared to results using meta-sc-eQTLs and MetaBrain2 (Supplementary Data 17, 18). These findings highlight the enhanced power of using JOBS-refined sc-eQTLs in TWAS, particularly in identifying loci beyond those reported in the GWAS catalog.
Next, using axis-QTLs as input, we can perform axis-TWAS, which allows us to identify a similar number of risk loci (260) as JOBS that are either shared between cell types or have distinct effects between cell types. We also identified 282 novel risk genes that JOBS misses. The largest improvement occurs for the SmkInit trait with 78 novel genes (Supplementary Data 17, 18). We also apply lassosum45 to build gene expression prediction models using sc-eQTLs and axis-QTLs as input. Our results show that the improvement of BASIC remains when alternative methods are used to generate prediction models and perform TWAS (Supplementary Data 16–18).
To differentiate causal genes and the neighboring associated genes due to linkage disequilibrium, we focused on genes confirmed as targets in TWAS fine mapping (with posterior inclusion probability > 95%). We are most interested in the genes identified uniquely in cell type-specific TWAS or axis-TWAS, but missed in MetaBrain TWAS (Fig. 5, Supplementary Fig. 12, Supplementary Data 19).
Each column represents a distinct gene, and each row corresponds to a cell type or principal component. Bi-clustering uncovers disease biology by jointly identifying genes and cell type modules that exhibit similar effects. The color scale indicates effect direction and magnitude: darker colors represent stronger associations. Genes shown in red font are missed by TWAS using bk-eQTLs, and an asterisk (*) marks genes located at least 1 million basepair away from any reported GWAS hits (i.e., novel genes). Only genes identified as target genes in both TWAS (two sided p-value < 0.05/[number of significant expression prediction models]) and colocalization analyses (PP4 > 0.9) are shown.
Some interesting hits are worth highlighting. First, we identify USP35 as a risk gene for ALS in inhibitory neurons (p-value = \(1.2\times {10}^{-6}\), and PIP = 1). Inhibitory neurons are known to be affected in early-stage ALS46. USP35 encodes a de-ubiquinating enzyme, important for beta-2 adrenergic receptor (β₂-AR) signaling47. In addition to its roles in skeletal and heart muscle, β₂-adrenergic receptor signaling activates the cAMP, PKA, CREB pathway48, which regulates genes essential for neuronal survival49, oxidative stress resistance50, and mitochondrial function51. These functions are critical for ALS etiology. By stabilizing these pathways, β₂-AR activity may enhance motor neuron resilience. β2-AR agonists have been shown as safe and potentially effective therapies for ALS52.
Interesting risk genes for ALS also include ATF6B (with axis-QTLs for the third PC being p = \(3.7\times {10}^{-8}\)). While direct associations between ATF6B and ALS have not yet been established, ATF6B is a well-recognized key regulator of the cellular response to endoplasmic reticulum (ER) stress through its activation of the unfolded protein response (UPR) pathway53. Extensive research suggests that ER stress and UPR activation play a significant role in the pathogenesis of ALS54,55, highlighting the potential relevance of ATF6B in the disease mechanism.
Another interesting target gene is CAAP1, the caspase activity and apoptosis inhibitor 1 gene, with the axis-QTLs of the 6th PC being the most significant association (p = \(3.3\times {10}^{-10}\)). CAAP1 is involved in inhibiting caspase activity and apoptosis. Apoptotic pathways are relevant in ALS, which can induce motor neuron death. In transgenic mSOD1 mice, caspases were shown to play an instrumental role in neurodegeneration, which suggests that caspase inhibition may have a protective role in ALS56. Intracerebroventricular administration of zVAD-fmk, a broad caspase inhibitor, was shown to delay disease onset and mortality for ALS. It also inhibits caspase-1 activity as well as caspase-1 and caspase-3 mRNA up-regulation, providing evidence for a non-cell-autonomous pathway regulating caspase expression. Together, the evidence provides support for the functional roles of CAAP1 in ALS.
JOBS and BASIC refined eQTLs also lead to the discovery of Leucine Zipper and EF-Hand Containing Transmembrane Protein 1 (LETM1) for Parkinson’s disease. LETM1 is a mitochondrial protein involved in calcium homeostasis and mitochondrial ion transport. Mitochondrial dysfunction is a central pathological feature of PD57. In PD, mitochondrial Ca2+ signaling contributes to the death of nigral dopaminergic neurons, mainly by regulating the production of adenosine triphosphate (ATP) and mitochondrial oxidant stress58. Targeting calcium homeostasis has emerged as a potential therapeutic approach in Parkinson’s disease59. Therefore, LETM1’s role in mitochondrial health makes it an interesting gene for functional follow-up.
Cell type aware drug repurposing and validation pipeline identifies promising drugs for treating diseases
We developed a cell type-aware computational drug repurposing pipeline (Method) leveraging BASIC results to identify drugs that can consistently reverse disease-associated gene expression in disease-relevant cell types. Using TWAS results as input, CMap web portal calculates a τ score for each drug x disease pair. A more negative τ score indicates that the candidate drug can more consistently reverse gene expression, implicating its potential to be repurposed for treating the disease (Fig. 6 and Supplementary Fig. 13). To assess the robustness of the results against the choice of gene sets, we conducted a drug repurposing sensitivity analysis using alternative gene sets, including the top 10, 15, or 20 genes from each TWAS. The resulting τ scores remained highly correlated across these analyses. Notably, over 92% of the drugs identified using our original approach (based on significant genes under the Bonferroni threshold) were consistently re-identified with the alternative gene sets, demonstrating the robustness of our results (Supplementary Fig. 14).
AD risk genes are enriched in excitatory neurons. We use the significant AD TWAS associations in excitatory neurons to identify drugs that could potentially reverse the disease gene expressions. CMap Drug clusters are obtained from the L1000FWD study. Each point in the scatter plot represents a drug-induced signature in HT29 cells—chosen for their proximity to excitatory neurons based on Euclidean distances of transcriptome-wide gene expression levels. CMap computes a τ score to quantify the correlation between each query signature and the reference profile, measuring whether the molecule can consistently reverse the gene expression levels of AD risk genes. An empirical one-sided p-value is calculated for each compound, measuring the significance of τ scores (Supplementary Data 20). Points shown as diamonds indicate significant CMap τ scores. Colors represent drug classes based on their mechanism of action. Common drug classes and representative drugs are labeled. The new drug cabergoline, identified for AD treatment, is highlighted as a dopamine receptor agonist.
To validate the drugs identified by BASIC, we further employed several orthogonal strategies: (1) using enrichment analysis to assess whether drug target pathways are enriched with TWAS hits following Chen et al. 60, and (2) utilizing Mendelian Randomization (MR) methods to determine whether the expression levels or protein abundance of drug target genes causally impact disease risk61 (Supplementary Data 20).
First, several drugs show a strong negative τ score for SmkInit, including zonisamide (τ= −92.89, p = 0.0229) and loxapine (τ= −86.64, p = 0.0424) in oligodendrocyte progenitor cells (OPCs). Both drugs have been validated by six orthogonal methods, which offer multiple lines of evidence to support the discovery. Zonisamide is an anti-seizure medication with a multifaceted mechanism of action, including inhibiting voltage-gated sodium channels and glutamate-mediated neurotransmission, and enhancing inhibitory GABAergic and serotonergic pathways62. Additionally, it elevates dopamine levels in the striatum, which may help mimic the rewarding effects of addictive substances like nicotine63. Studies have shown that zonisamide could decrease nicotine withdrawal and craving64. When combined with bupropion, zonisamide may be an effective way to help smokers quit smoking63. Conversely, we identified suggestive causal relations between protein levels of the drug target genes CA2 (p = 0.0302) and CA7 (p = 0.0115), as well as gene expression levels of CA3 (p = 0.0436) in OPCs, and SmkInit.
Loxapine is another interesting target, which is a typical antipsychotic medication primarily used to manage conditions such as schizophrenia and, occasionally, bipolar disorder, particularly in cases involving acute agitation65,66. Although it may not be the first-line treatment for smoking disorders, its anxiolytic properties could be beneficial since many people smoke as a self-medication to cope with stress or anxiety67. To our knowledge, at least two other antipsychotics, clozapine68,69 and bupropion70 have been shown to reduce nicotine use. Additionally, we identified a significant causal association (p = 0.0468) between the protein levels of ADRA1A, a gene targeted by loxapine, and SmkInit.
We identified memantine as a potential candidate for drinking addiction. Memantine exhibited a significantly low τ in excitatory neurons (τ = −98.96, p = 0.0051) and inhibitory neurons (τ = −92.31, p = 0.0178), which is validated by five independent methods. Memantine is a non-competitive antagonist of ionotropic NMDA receptors and is FDA-approved for the treatment of moderate to severe Alzheimer’s disease71. It has been shown to inhibit ethanol-induced upregulation of NMDA receptors72. Studies in rats indicate that memantine may reduce alcohol cravings73,74. Recent clinical studies suggest that memantine can suppress alcohol cravings in moderate drinkers experiencing deprivation75. In our orthogonal validation, we found a causal association (p = 0.0064) between the expression levels of the target gene, GRIN1, in inhibitory neurons and the drinks per week phenotype.
Moreover, we identified cabergoline as a potential candidate for Alzheimer’s disease (AD). Cabergoline exhibited a significantly low τ in excitatory neurons (τ = −93.32, p = 0.0127, Fig. 6), which is further validated by five independent methods. Cabergoline, a dopamine D2 receptor agonist, is known for its high affinity for the D2 receptor, which plays key roles in dopamine replacement therapy for conditions like Parkinson’s disease76, restless legs syndrome77, and more recently, Alzheimer’s disease78. Moreover, dopamine D2 receptor agonists have been reported to exert neuroprotective effects against oxidative stress79, which play a key role in neurodegenerative diseases, including AD80. Cabergoline has been shown to enhance cognitive functions, such as processing speed, working memory, visual learning, and problem-solving, in patients with hyperprolactinemia81. As validation, we showed that the expression level in excitatory neurons of the ADRA1A gene, a drug target gene for cabergoline that encodes the alpha-1A adrenoceptor, is causative for AD (p = 0.0057).
Finally, alfacalcidol markedly reverses disease-associated gene expression for schizophrenia (SZ) in excitatory neurons (τ = −93.6, p = 0.0220). Notably, the SZ risk genes identified using the axis-TWAS method, including VDR and CYP27B1, show significant enrichment in the alfacalcidol-targeted pathway (p = 0.0002). Furthermore, a previous study indicates that VDR is overexpressed in SZ patients compared to healthy controls82. Alfacalcidol may be a potential therapeutic for SZ by reversing disease-associated gene expression levels.
Discussion
In this article, we present BASIC, a novel method that substantially improves the statistical power to detect eQTLs by integrating sc-eQTLs and bk-eQTLs datasets from brain. As a conceptual innovation, BASIC looks beyond cell types and seeks to identify axis-QTLs associated with major axes of gene expression variations, which are linear combinations of gene expressions across cell types. Axis-QTLs can capture shared and distinct regulatory effects across cell types and substantially improve the power of identifying regulatory variants.
The improved power of BASIC motivates us to consider a new paradigm for analyzing eQTLs using sc-RNASeq data. The conventional approach is to group cells into cell types and calculate sc-eQTLs measuring regulatory effects in each cell type. Just as all clustering methods, grouping cells into distinct cell groups can sometimes be an arbitrary procedure, especially for cells that are “on the boundary” of two annotated cell type groups. There is a growing consensus in the field that the cell states vary in a continuum and should be modeled that way23. Our axis-QTLs approach shares similarity with those ideas, yet our approach works directly with sc-eQTLs summary statistics and jointly analyzes bk-eQTLs to improve power.
Moreover, while different cell types have different gene expression profiles, it does not necessarily suggest the regulatory effects are different across these cell types. The axis-QTLs project sc-eQTLs effects onto the PCs of gene expression levels, so it can better tease apart shared and distinct effects across cell types. It is particularly useful for improving the power of detecting eQTLs when the cell type containing causal variants is rare or lowly expressed. The advantage of axis-QTLs can be further revealed by the intersection between axis-QTLs and sc-eQTLs (Supplementary Fig. 15). We observe that a substantial proportion of sc-eQTLs from different cell types were identified by axis-QTLs. Yet, sc-eQTL analyses in rare cell types (e.g., microglia and endothelial cells) may fail to detect any axis-QTLs, including those with shared effects across cell types, e.g., the axis-QTLs from PC0, which shows sc-eQTL analysis can be very underpowered for rare cell types. The identified sc-eQTLs and axis-QTLs also have similar patterns of enrichment for functional scores, suggesting axis-QTLs might similarly regulate gene expression levels as regular sc-eQTLs.
The performance of BASIC is robust across different genetic architectures of regulatory effects. BASIC offers the largest improvement when biologically similar cell types share regulatory variants. This is evident from our simulation studies. This advantage is recapitulated in real data analysis, where analyzing axis-QTLs identifies and replicates a larger number of regulatory variants. When biologically similar cell types do not share regulatory variants, BASIC still performs comparably to JOBS, which do not model the shared effects. We also applied BASIC to sc-eQTLs from immune cell types in the OneK1K study and demonstrated that it continues to outperform alternative methods (e.g., identifying a larger number of eQTLs and eGenes), highlighting its broad applicability beyond brain cell types (Supplementary Data 21).
The interpretation of axis-QTLs warrants discussion. Unlike standard sc-eQTLs analysis, which identifies regulatory variants for each cell type, axis-QTLs seek to identify genetic variants associated with PCs of cell type gene expression levels. While sc-eQTLs are intuitively appealing, they have several limitations when we aim to maximize the power to identify regulatory variants. Sc-eQTL studies are known to be underpowered for uncommon cell types, since the number of reads covering rare cell types is lower and the gene expression measurements can be noisier. On the other hand, many variants have shared regulatory effects across cell types. Axis-QTLs analyze genetic associations with PCs of gene expression levels and can naturally borrow information from shared effects across cell types. There are strong biological interpretations of axis-QTLs as well. For example, the intercept from the BASIC model captures shared effects across all cell types, while PCs 1-3 can capture regulatory effects that are different between barrier, glial, and neuronal cell types. Importantly, the loadings for different cell types can be of opposite signs, which will allow us to aggregate signals across cell types where the eQTLs effects have opposite directions (e.g., DEDD gene, Supplementary Fig. 16). This conceptually novel way to model regulatory effects allows us to identify regulatory variants and risk genes that are otherwise missed by standard sc-eQTLs analysis. Furthermore, we calculated the variance explained by each PC (Supplementary Fig. 17). Interestingly, the variance explained by each PC remains relatively similar, and no single PC dominates, e.g., the intercept term explains ~25% of the variance, and the first to fifth PC each explain 10%-15% of the variance. It shows that a larger fraction of variants has shared effects across cell types, and variants with distinct effects across cell types tend to be captured evenly by axis-QTLs from PCs 1-5.
BASIC and axis-QTLs can also be easily modified to work with sc-eQTLs datasets only without matched bk-eQTLs datasets. In this case, the meta-regression model can be used to analyze sc-eQTLs only, and the likelihood term involving the bk-eQTL data can be dropped. Similar to the improvement of BASIC over JOBS, we anticipate that axis-QTLs can identify more regulatory variants and eGenes than sc-eQTLs data alone, even without matched bk-eQTLs.
Despite these strengths, several limitations warrant further attention and improvement in future research. First, when integrating sc-eQTLs and bk-eQTLs datasets, we assume regulatory variants of each cell type have similar effects between datasets. While this assumption seems restrictive intuitively, it works well in our application, leading to replicable eQTL and eGenes. Yet, sources of heterogeneity may exist due to different disease conditions, age distribution, or ancestries between datasets, which may become more apparent as diverse datasets are generated in the near future. One possible extension in the future is to incorporate a random effect in the model of sc- and bk-eQTLs effects, to accommodate the heterogeneity. We hypothesize that the extension may further improve the power and the precision of the eQTLs effect estimates.
Second, due to the lack of non-European functional genomic datasets from the brain, BASIC and axis-QTLs focus on samples of European ancestry. We have developed methods such as TESLA60 that can optimally integrate eQTLs datasets from European ancestry with a multi-ancestry GWAS. This will ensure our results can yield optimal power to identify causal variants and genes in samples of European ancestry. Yet, this does not take away the need to generate functional genomic datasets from non-European populations, which is necessary to identify causal genes in those populations. When samples from diverse populations become available, we can extend our meta-regression framework to include principal components of genome-wide allele frequencies to account for the effect size differences between populations. Similar ideas have been exploited in our TESLA and MEMO1 methods.
In summary, we present a new method to calculate axis-QTLs and use it to identify regulatory variants and risk genes for various brain-related traits. By integrating with bk-eQTLs data from the brain, we substantially augment the sample sizes and improve the power. The research community has been actively generating single cell or single nuclei RNASeq datasets. The method will continue to be useful to combine single cell and bulk eQTLs datasets from different tissues, and contribute to our understanding of the genetic basis of complex traits.
Methods
Below we introduce key ideas of BASIC, which uses meta-regression models to borrow strength from shared effects across cell types and then integrates with bk-eQTLs datasets to enlarge sample sizes and further improve power. We also propose the new concept of axis-QTLs which captures how the effects of regulatory variants vary along the PCs for cell type specific gene expression or sc-eQTLs. Axis-QTLs allow us to look beyond cell types and identify disease risk genes that may be missed by conventional methods.
Calculation of principal components of sc-eQTLs effects across cell types
To calculate the PCs of sc-eQTLs, we construct a matrix \({{\boldsymbol{F}}}\) of sc-eQTLs effects across variant sites and cell types, where each row represents a cell type and each column represents a gene x SNP pair. This calculation will only use eQTLs commonly measured across all cell types. To calculate the PCs, we perform singular value decomposition for \({{\boldsymbol{F}}}\), i.e.,
Where \({{{\boldsymbol{C}}}}_{{{\boldsymbol{F}}}}\) and \({{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\) are orthogonal matrices and \({{{\boldsymbol{D}}}}_{{{\boldsymbol{F}}}}\) is a diagonal matrix. The columns of the matrix \({{{\boldsymbol{FE}}}}_{{{\boldsymbol{F}}}}\) are the PCs, whereas the diagonal entries of \({{{\boldsymbol{D}}}}_{{{\boldsymbol{F}}}}\) are the eigenvalues. We arrange the eigenvalues in decreasing order and arrange the corresponding eigenvectors accordingly, so that the first PC captures the largest variation. We can also similarly perform eigenvalue decomposition of the covariance matrix of \(F\), i.e.,
It is clear from Fig. 1 that PCs can cluster biologically similar cell types together. We denote the \({l}^{{th}}\) PC for the \({k}^{{th}}\) cell type as the \({Z}_{{kl}}\). For notational simplicity, we can set \({Z}_{k0}=1\), so its corresponding coefficient represents the intercept.
Given a set of PCs pre-calculated from one dataset, we can project sc-eQTLs effects from another dataset onto the pre-calculated PCs. Specifically, given the matrix of sc-eQTLs effects in another dataset \({{{\boldsymbol{F}}}}^{{{\boldsymbol{\#}}}}\), we can project the effects on each PC by \({{{\boldsymbol{F}}}}^{{{\boldsymbol{\#}}}}{{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\), where \({{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\) is the orthogonal matrix in the singular value decomposition of \({{\boldsymbol{F}}}\) and also the same orthogonal matrix in the eigenvalue decomposition of the covariance matrix of \({{\boldsymbol{F}}}\). Similar to the PC of the matrix \({{\boldsymbol{F}}}\), \({{{\boldsymbol{F}}}}^{{{\boldsymbol{\#}}}}{{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\) is the weighted sum of sc-eQTLs effects across cell types and captures the major axes of variation of the regulatory effects.
Meta regression model of sc-eQTLs across cell types
Using principal components as covariates in meta-regression models, we can characterize the heterogeneity between cell types and borrow strength from shared effects to improve the detection of sc-eQTLs.
Given the sc-eQTLs effects and the PCs, the model takes the following form:
\({\epsilon }_{{jk}}\) is the residual and follows a normal distribution. \({{{\boldsymbol{\gamma }}}}_{{{\boldsymbol{j}}}{{\boldsymbol{.}}}}=({\gamma }_{j0},\,{\gamma }_{j1},\ldots,{\gamma }_{{jL}})\) are the regression coefficients. In our analyses, we can vary the number of PCs included in the model between 1 and \(K-1\), where \(K\) is the number of cell types. We further denote the vector of sc-eQTL effects for variant \(j\) as \({{{\boldsymbol{b}}}}_{{{\boldsymbol{j}}}}=({b}_{j1},\ldots,{b}_{{jK}})\). Under the model, the sc-eQTL effect follows:
where \({{\boldsymbol{Z}}}\) is the matrix of PCs and \({{{\boldsymbol{V}}}}_{{{\boldsymbol{j}}}}\) is the covariance matrix between sc-eQTLs effects of variant \(j\) across cell types, which may be induced by sample overlaps of different cell types.
As we showed in Supplementary Note section 2, the PCs of the sc-eQTLs effects approximate the PCs of the gene expression levels and can separate biologically distinct cell types. The axis-QTLs can be viewed as the genetic effects on the PCs of cell type gene expression levels.
Similar to linear mixed models83, we consider the “leave one chromosome out (LOCO)” strategy: For each chromosome we analyze, we use the PCs of sc-eQTLs from the rest of the chromosomes to avoid “contaminations” of the signals from nearby variants. The results using PCs calculated with LOCO closely resemble those calculated from all chromosomes, though.
Integrate bulk-eQTLs to improve the power to detect axis-QTLs
We showed theoretically, under very general conditions, bk-eQTLs can be approximated by a weighted linear combination of sc-eQTLs effects from constituent cell types26. So, the mean values of bk-eQTLs are also weighted linear combinations of the mean values of axis-QTLs. This approximation would allow us to borrow strength from the large sample sizes of bk-eQTLs datasets, such as MetaBrain2 data. Specifically, we have
where \({\hat{w}}_{k}\) denotes the cell weights estimated from non-negative least square method, i.e.,
Under the constraint of \({w}_{k} > 0\) and \({\sum }_{k=1}^{K}{w}_{k}=1\). We focus on SNPs that are measured in all cell types and bulk tissue to estimate cell type weights. As we show in Supplementary Fig. 18 and Supplementary Data 22, the cell type weights closely resemble the average cell type proportions. The uncertainty of estimated cell type weights can be quantified using parametric bootstrap (Supplementary Notes 3).
The distribution of bk-eQTL and sc-eQTLs satisfies:
The log-likelihood takes the form:
where C denotes the collection of likelihood terms that do not contain parameters of interest. As we show in the Supplementary Notes, the model can be fitted using weighted linear regression in closed form. We call each \({\gamma }_{{jl}}\) the \({l}^{{th}}\) axis-QTLs for variant \(j\). To test for association, we use the score statistic to test whether \({\gamma }_{{jl}}\) is equal to 0. The test statistic follows a chi-square distribution with 1 degree of freedom.
Cauchy combination of p-values from different axis-QTLs
To facilitate the comparison with other methods that analyze each cell type separately, we also consider an omnibus test of whether the association is significant for any axis-QTLs. Here, to properly test for the omnibus hypothesis and control for multiple comparisons, we propose using Cauchy combinations84 of p-values from different cell types or PCs.
For example, to combine different axis-QTLs, we use the following formula:
The statistic \({T}_{{BASIC}}\) follows a Cauchy distribution, and the p-values can be evaluated accordingly. Similarly, we can also use the Cauchy combination test to combine the p-values of sc-eQTLs from different cell types for methods that analyze each cell type separately.
Colocalization analysis
We utilized the COLOC32 R package, a Bayesian statistical method for colocalization analysis, to evaluate whether GWAS loci and sc-eQTLs signals share a common causal variant. To ensure consistency, all GWAS and eQTLs datasets were lifted over to the GRCh38 reference genome. For each GWAS locus, we examined the cis-eQTLs within the same genomic region, testing for colocalization. Loci with the posterior probability PP4 exceeding 0.9 are considered colocalized in our analyses, indicating that both GWAS and eQTLs signals may share the same causal SNP.
Generalized TWAS using axis-QTLs
TWAS was developed as a framework to identify the association between genetically regulated gene expression levels and the trait of interest. It first creates gene expression prediction models using a dataset that measures both genotypes and gene expression levels. It then tests for the association between predicted gene expression levels and the phenotype of interest (possibly in another dataset). Recent development of TWAS can also be applied to datasets with only eQTLs summary statistics or GWAS summary statistics44.
As we show in earlier sections, the PCs of sc-eQTLs across cell types are concordant with the PCs of gene expression levels, which captures the major direction of expression variation. Using axis-QTLs as input, we will predict the projected gene expression levels to different PCs for each individual, and test their association with disease outcomes. In our analysis, we use EXPRESSO44 to generate gene expression prediction models. Our method takes QTL summary statistics as input and also uses 3D genome and epigenetic information to prioritize causal variants and improve the prediction accuracy. The prediction weights for gene \(g\) are denoted by \({{{\boldsymbol{w}}}}_{{{\boldsymbol{g}}}}={({w}_{g1},\ldots,{w}_{{gJ}})}^{{\prime} }.\) To further ensure the comparison of BASIC is not affected by the choice of gene expression prediction method, we also use lassosum45 to generate gene expression prediction models and perform TWAS.
We will next assess the accuracy of predicted expression levels along the PCs. If an independent test dataset is available, we first need to project its cell type gene expression levels for each gene \(g\), i.e., \({{{\boldsymbol{y}}}}_{{{\boldsymbol{g}}}}\), onto the PCs, by \({\widetilde{{{\boldsymbol{y}}}}}_{{{\boldsymbol{g}}}}={{{\boldsymbol{y}}}}_{{{\boldsymbol{g}}}}{{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\), where \({{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\) is orthogonal matrix used in the singular value decomposition of the sc-eQTLs effect size matrix. The projected expression level of gene \(g\) across different PCs is denoted by \({\widetilde{{{\boldsymbol{y}}}}}_{{{\boldsymbol{g}}}}\). We could estimate the correlation between predicted gene expression and \({\widetilde{{{\boldsymbol{y}}}}}_{{{\boldsymbol{g}}}}\) by
Notably, \({\widetilde{{{\boldsymbol{y}}}}}_{{{\boldsymbol{g}}}}^{{{\boldsymbol{T}}}}{{{\boldsymbol{X}}}}_{{{\boldsymbol{g}}}}\) can be approximated by the external sc-eQTLs effect size \({{{\boldsymbol{\beta }}}}_{{{\boldsymbol{g}}}}{{{\boldsymbol{E}}}}_{{{\boldsymbol{F}}}}\) and \({\mathrm{var}}({{{\boldsymbol{X}}}}_{{{\boldsymbol{g}}}}{{{\boldsymbol{w}}}}_{{{\boldsymbol{g}}}})\) could also be derived from the reference LD panel. So \(r\) can still be calculated if only sc-eQTLs summary statistics are available from the test data.
To calculate the p-value of the derived correlation, we use Fisher’s transformation to obtain the z-score from correlation coefficients:
where n is the sample size. The converted Z-score follows a normal distribution. The prediction model is deemed significant if \(r > 0.1\) and p-value < 0.05, following the standard procedure in TWAS43.
Finally, when an independent sc-eQTLs dataset is unavailable, we propose to simulate another set of eQTLs from the original data as an external validation set. Assume we have the effect size \({{{\boldsymbol{b}}}}_{{{\boldsymbol{g}}}}={({b}_{g1},\ldots,{b}_{{gJ}})}^{{\prime} }\) and standard error \({{{\boldsymbol{s}}}}_{{{\boldsymbol{g}}}}={({s}_{g1},\ldots,{s}_{{gJ}})}^{{\prime} }\) from the original eQTLs summary statistics. We use parametric bootstrap and simulate a new set of effect sizes from a multivariate normal distribution.
where\(\,{{\boldsymbol{R}}}\) is the LD correlation matrix of cis-SNPs for gene g. For each simulated summary statistic \({{{\boldsymbol{\alpha }}}}_{{{\boldsymbol{g}}}}\) and its standard deviation \({{{\boldsymbol{s}}}}_{{{\boldsymbol{g}}}}\), we will calculate \(r\) using the formula above and evaluate its statistical significance. We repeat the simulation 100 times and report the average \(r\) and p-values.
Bi-clustering identifies relevant cell types/axis of gene expression variation and risk genes
We utilize bi-clustering algorithms to identify risk genes and disease-relevant cell types or axes of gene expression variations.
We construct a matrix of TWAS effect sizes, with each row representing a cell type or PC of gene expressions, and each column representing a gene. Bi-clustering is a method that clusters a matrix’s rows and columns simultaneously. We use the two-dimensional Euclidean distance as the dissimilarity metric for bi-clustering. To be more specific, for a given gene x cell type cluster, we label the TWAS effect sizes across different genes and cell types/gene expression PCs in a cluster as \({T}_{1},\ldots,{T}_{m}\). The dissimilarity metric for the set of genes and cell types is given by:
where \({T}_{{j}_{1}}\) and \({T}_{{j}_{2}}\) are the TWAS effect sizes.
Bi-clustering can simultaneously identify cell types/PCs with shared risk genes as well as risk gene clusters with similar effects. To further assess the statistical significance of identified gene x cell type/PC clusters, we use permutation test by shuffling the rows and columns of the TWAS effect matrix to generate an empirical distribution for the dissimilarity measures for a given set of genes and cell types/PCs. To evaluate p-values, we compare the dissimilarity metric obtained from the original data with the generated empirical distribution. The p-values are calculated based on the fraction of generated dissimilarity metrics that are smaller than the value obtained from the original data.
Cell-type-aware computational drug repurposing pipeline and validation
We modified the cell-type-aware drug repurposing pipeline (CADRE) from our previous work44. We first cluster the RNASeq data of cell lines from the CMap database85 with brain sn-RNASeq data. For each brain-related cell type, we identify the cell line whose transcriptome profiles most closely resemble the cell type (based on Euclidean distance of the expression levels of all measured genes). We focus on the Touchstone subset, which includes gene expression profiles from nine cell lines treated with ~3000 well-annotated small-molecule drugs. Our drug repurposing analysis was restricted to four traits with sufficient numbers of associated loci (i.e., smoking initiation, drinks per week, Alzheimer’s disease, and schizophrenia), since CMap requires at least 10 positively, and 10 negatively associated genes as input85,86. CMap computes a τ score to link disease states with drug-induced gene expression profiles. A more negative τ score indicates that the drug can normalize the gene expression profile associated with the trait, suggesting its potential for disease treatment.
We propose a permutation procedure to evaluate the statistical significance of the τ scores. Specifically, for each permutation, we shuffle the gene names for significant TWAS genes (under the Bonferroni threshold for testing multiple genes in each cell type), pair the Z-scores with shuffled gene names, and use them as input. Since permutation breaks the link between gene names and their TWAS Z-scores, τ scores calculated from permuted datasets will form a null distribution. The p-values can be calculated by the fraction of τ-scores from the permuted dataset that are more negative than those from the original dataset.
We also employ two different classes of methods (7 methods in total) to validate the putative drugs identified by our pipeline. These methods include:
-
(1)
Enrichment analysis examining whether drug-targeted pathways are enriched with TWAS hits, based on the hypothesis that an effective drug may exert its therapeutic effect by targeting disease-associated genes. For this purpose, we utilized eTESLA60, a published method that leverages meta-regression to assess whether a drug target pathway is enriched with TWAS hits. We curated gene sets for drug-targeted pathways using DrugBank87, a database of the mechanisms of action for approved drugs. We constructed a design matrix where each row represents a gene and each column represents a pathway. Each entry of the matrix is assigned a value of 1 if the gene is part of a specific pathway and 0 otherwise. eTESLA then regresses Z-scores over the design matrix, and computes p-values of regression coefficients to determine whether the corresponding drug-targeted pathways show significant enrichment with the TWAS hits.
-
(2)
Mendelian randomization (MR) methods to investigate whether the expression levels or secreted protein levels of drug target genes causally influence disease risk. To test our hypotheses that gene expression or protein levels regulated by the drug target genes have a causal effect on disease, we applied three complementary MR approaches: inverse variance weighted MR88 (MR-IVW), MR-Egger89, and MR-RAPS90. These methods are known for their distinct strengths; when used together, they may enhance the robustness of the findings. We name our analyses based on the type of molecular QTLs used and the MR approaches employed, i.e., eQTL-MR-IVW, eQTL-MR-Egger, eQTL-MR-RAPS for gene expression QTLs, and pQTL-MR-IVW, pQTL-MR-Egger, pQTL-MR-RAPS for protein QTLs.
eQTLs datasets.
We utilized several eQTLs datasets in our analysis. For single-cell data, we used two published sc-eQTLs datasets: Fujita et al21. and Bryois et al.20. The Fujita et al21. dataset consists of single-nucleus RNA sequencing (snRNA-seq) data derived from the dorsolateral prefrontal cortex (DLPFC) of 424 elderly individuals. It includes gene expression profiles from seven major neocortical cell types and 64 subtypes, enabling cell-type-specific cis-eQTLs mapping. For our analyses, we focused on the seven major cell types. The Bryois et al. dataset includes single nucleus RNA sequencing (sn-RNASeq) data from 196 donors of European ancestry, covering eight major central nervous system (CNS) cell types, including oligodendrocytes, excitatory neurons, inhibitory neurons, astrocytes, microglia, oligodendrocyte precursor cells (OPCs), endothelial cells, and pericytes within the cortical region of the brain. For bulk eQTLs data, we leveraged the MetaBrain2 dataset, which consists of 2683 cortical samples of European ancestry aggregated from 14 independent studies. We perform fixed effect meta-analysis of sc-eQTLs for the seven overlapping cell types between the Fujita et al.21 and Bryois et al.3 to maximize sample sizes, which we call meta-sc-eQTLs in the manuscript. This meta-analyzed sc-eQTLs dataset, along with the bulk eQTL data from MetaBrain2, are jointly analyzed using the BASIC framework and alternative methods (e.g., mashr) and integrated with GWAS for all downstream analyses.
To validate identified sc-eQTLs, we used the dataset from Lopes et al.,29 which contains 255 primary human CD11b+ microglia samples isolated from several brain regions in 100 individuals of European ancestry. While there are other sc-eQTLs datasets, they do not contain full eQTLs summary statistics, making it impossible to validate identified eQTLs summary statistics. We therefore advocate for the release of full summary statistics from published studies to maximize their utility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The MetaBrain eQTLs summary statistics13 can be obtained from https://www.metabrain.nl/cis-eqtls.html. Fujita et al. cell type-level eQTLs21 are available from https://vmenon.shinyapps.io/rosmap_snrnaseq_eqtl/. Bryois et al. cell type-level eQTLs20 are available from https://doi.org/10.5281/zenodo.5543734. Lopes et al. microglial eQTLs16 can be downloaded from https://doi.org/10.5281/zenodo.4118605. Velmeshev et al. single cell RNAseq data91 are available from https://autism.cells.ucsc.edu. The BASIC axis-QTLs and sc-eQTLs summary statistics are available from https://liugroupstatgen.shinyapps.io/BASIC-axis-QTL/ and the linked Zenodo repository92 (https://doi.org/10.5281/zenodo.17221192).
Code availability
The software implementing the BASIC method is available at https://github.com/LidaWangPSU/BASIC under the MIT License and the linked Zenodo repository92 (https://doi.org/10.5281/zenodo.17221192).
References
Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022).
Meng, X. et al. Multi-ancestry genome-wide association study of major depression aids locus discovery, fine mapping, gene prioritization and causal inference. Nat. Genet. 56, 222–233 (2024).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
van Rheenen, W. et al. Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat. Genet. 53, 1636–1648 (2021).
New insights into the genetic etiology of Alzheimer’s disease and related dementias | Nature Genetics.
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
Identification of common genetic risk variants for autism spectrum disorder | Nature Genetics.
Consortium, G. T. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585 (2013).
Zhang, F. & Lupski, J. R. Non-coding genetic variants in human disease. Hum. Mol. Genet. 24, R102–R110 (2015).
Nica, A. C. & Dermitzakis, E. T. Expression quantitative trait loci: present and future. Philos. Trans. R. Soc. B: Biol. Sci. 368, 20120362 (2013).
Aguet, F. et al. Genetic effects on gene expression across human tissues. Nature 550, 204–213 (2017).
de Klein, N. et al. Brain expression quantitative trait locus and network analyses reveal downstream effects and putative drivers for brain-related diseases. Nat. Genet. 55, 377–388 (2023).
Sieberts, S. K. et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci. Data 7, 340 (2020).
Yazar, S. et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041 (2022).
Lopes, K. P. et al. Genetic analysis of the human microglial transcriptome across brain regions, aging and disease pathologies. Nat. Genet 54, 4–17 (2022).
Westra, H.-J. & Franke, L. From genome to function by studying eQTLs. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 1842, 1896–1902 (2014).
Patel, D. et al. Cell-type-specific expression quantitative trait loci associated with Alzheimer disease in blood and brain tissue. Transl. Psychiatry 11, 1–17 (2021).
Liu, M. et al. Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat. Genet. 51, 237–244 (2019).
Bryois, J. et al. Cell-type-specific cis-eQTLs in eight human brain cell types identify novel risk genes for psychiatric and neurological disorders. Nat. Neurosci. 25, 1104–1112 (2022).
Fujita, M. et al. Cell subtype-specific effects of genetic variation in the Alzheimer’s disease brain. Nat. Genet. 56, 605–614 (2024).
Marshall, L., Raychaudhuri, S. & Viatte, S. Understanding rheumatic disease through continuous cell state analysis. Nat. Rev. Rheumatol. 21, 323–335 (2025).
Nathan, A. et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature 606, 120–128 (2022).
Jeong, H.-W. et al. Single-cell transcriptomics reveals functionally specialized vascular endothelium in brain. eLife 11, e57520 (2022).
Attwell, D., Mishra, A., Hall, C. N., O’Farrell, F. M. & Dalkara, T. What is a pericyte? J. Cereb. Blood Flow. Metab. 36, 451–455 (2016).
Wang, L. et al. An atlas of single cell eQTLs dissects autoimmune disease genes and identifies novel drug classes for treatment. Cell Genomics 5, 100820 (2025).
Yu, X. et al. A unified framework for cell-type-specific eQTL prioritization by integrating bulk and scRNA-seq data. Am. J. Hum. Genet. 112, 332–352 (2025).
Urbut, S. M., Wang, G., Carbonetto, P. & Stephens, M. Flexible statistical methods for estimating and testing effects in genomic studies with multiple conditions. Nat. Genet. 51, 187–195 (2019).
Lopes, K. P., Snijders, G. J. L., Humphrey, J., de Witte, L. D. & Raj, T. Atlas of genetic effects in human microglia transcriptome across brain regions, aging and disease pathologies. Alzheimer’s. Dement. 17, e050942 (2021).
Velmeshev, D. et al. Single-cell genomics identifies cell type-specific molecular changes in autism. Science 364, 685–689 (2019).
Ding, R. et al. xQTLbiolinks: a comprehensive and scalable tool for integrative analysis of molecular QTLs. Brief Bioinform. 25, bbad440 (2023).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. 55, 198–208 (2023).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Salim, C., Batsaikhan, E., Kan, A. K., Chen, H. & Jee, C. Nicotine motivated behavior in C. elegans. Int. J. Mol. Sci. 25, 1634 (2024).
Alcivar, A., Hu, S., Tang, J. & Yang, X. DEDD and DEDD2 associate with caspase-8/10 and signal cell death. Oncogene 22, 291–297 (2003).
Lee, J. C. et al. DEDD regulates degradation of intermediate filaments during apoptosis. J. Cell Biol. 158, 1051–1066 (2002).
Colonna, M. & Butovsky, O. Microglia Function in the Central Nervous System During Health and Neurodegeneration. Annu Rev. Immunol. 35, 441–468 (2017).
Ni, Y. et al. Death effector domain-containing protein induces vulnerability to cell cycle inhibition in triple-negative breast cancer. Nat. Commun. 10, 2860 (2019).
Flury, A. et al. A neurodegenerative cellular stress response linked to dark microglia and toxic lipid secretion. Neuron 113, 554-571.e14 (2024).
Gamazon, E. R. et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 47, 1091–1098 (2015).
Khunsriraksakul, C. et al. Integrating 3D genomic and epigenomic data to enhance target gene discovery and drug repurposing in transcriptome-wide association studies. Nat. Commun. 13, 3258 (2022).
Wang, L. et al. Integrating single cell expression quantitative trait loci summary statistics to understand complex trait risk genes. Nat. Commun. 15, 4260 (2024).
Mak, T. S. H., Porsch, R. M., Choi, S. W., Zhou, X. & Sham, P. C. Polygenic scores via penalized regression on summary statistics. Genet Epidemiol. 41, 469–480 (2017).
Montanana-Rosell, R. et al. Spinal inhibitory neurons degenerate before motor neurons and excitatory neurons in a mouse model of ALS. Sci. Adv. 10, eadk3229 (2024).
Berthouze, M., Venkataramanan, V., Li, Y. & Shenoy, S. K. The deubiquitinases USP33 and USP20 coordinate beta2 adrenergic receptor recycling and resensitization. EMBO J. 28, 1684–1696 (2009).
Lorton, D. & Bellinger, D. L. Molecular mechanisms underlying beta-adrenergic receptor-mediated cross-talk between sympathetic neurons and immune cells. Int. J. Mol. Sci. 16, 5635–5665 (2015).
Ryu, H., Lee, J., Impey, S., Ratan, R. R. & Ferrante, R. J. Antioxidants modulate mitochondrial PKA and increase CREB binding to D-loop DNA of the mitochondrial genome in neurons. Proc. Natl. Acad. Sci. USA 102, 13915–13920 (2005).
Zhou, Q. et al. Upregulation of postsynaptic cAMP/PKA/CREB signaling alleviates copper(II)-induced oxidative stress and pyroptosis in MN9D cells. Toxicology 494, 153582 (2023).
Dagda, R. K. & Das Banerjee, T. Role of protein kinase A in regulating mitochondrial function and neuronal development: implications to neurodegenerative diseases. Rev. Neurosci. 26, 359–370 (2015).
Bartus, R. T. et al. beta2-Adrenoceptor agonists as novel, safe and potentially effective therapies for Amyotrophic lateral sclerosis (ALS). Neurobiol. Dis. 85, 11–24 (2016).
Lei, Y. et al. Molecular mechanism of ATF6 in unfolded protein response and its role in disease. Heliyon 10, e25937 (2024).
Jeon, Y. M., Kwon, Y., Lee, S. & Kim, H. J. Potential roles of the endoplasmic reticulum stress pathway in amyotrophic lateral sclerosis. Front. Aging Neurosci. 15, 1047897 (2023).
Walker, A. K. & Atkin, J. D. Stress signaling from the endoplasmic reticulum: a central player in the pathogenesis of amyotrophic lateral sclerosis. IUBMB Life 63, 754–763 (2011).
Li, M. et al. Functional role of caspase-1 and caspase-3 in an ALS transgenic mouse model. Science 288, 335–339 (2000).
Henrich, M. T., Oertel, W. H., Surmeier, D. J. & Geibl, F. F. Mitochondrial dysfunction in Parkinson’s disease - a key disease hallmark with therapeutic potential. Mol. Neurodegener. 18, 83 (2023).
Zhang, J. et al. Calcium homeostasis in parkinson’s disease: from pathology to treatment. Neurosci. Bull. 38, 1267–1270 (2022).
Kaur, S. et al. Targeting calcium homeostasis and impaired inter-organelle crosstalk as a potential therapeutic approach in Parkinson’s disease. Life Sci. 330, 121995 (2023).
Chen, F. et al. Multi-ancestry transcriptome-wide association analyses yield insights into tobacco use biology and drug repurposing. Nat. Genet. 55, 291–300 (2023).
Namba, S. et al. A practical guideline of genomics-driven drug discovery in the era of global biobank meta-analysis. Cell Genom. 2, 100190 (2022).
Biton, V. Clinical pharmacology and mechanism of action of zonisamide. Clin. Neuropharmacol. 30, 230–240 (2007).
Rose, J. E., Willette, P. N., Botts, T. L., Botts, D. R. & Behm, F. M. Bupropion/zonisamide combination to assist smokers to switch from combustible cigarettes to electronic nicotine delivery systems (ENDS). Drug Alcohol Depend. 234, 109346 (2022).
Dunn, K. E. et al. Zonisamide reduces withdrawal symptoms but does not enhance varenicline-induced smoking cessation. Nicotine Tob. Res. 18, 1171–1179 (2016).
Lehman, A. F. et al. Practice guideline for the treatment of patients with schizophrenia, second edition. Am. J. Psychiatry 161, 1–56 (2004).
Pacciardi, B., Calcedo, A. & Messer, T. Inhaled loxapine for the management of acute agitation in bipolar disorder and schizophrenia: expert review and commentary in an era of change. Drugs R. D. 19, 15–25 (2019).
Hughes, J. R., Stead, L. F. & Lancaster, T. Anxiolytics for smoking cessation. Cochrane Database Syst. Rev. 2000, CD002849 (2000).
George, T. P., Sernyak, M. J., Ziedonis, D. M. & Woods, S. W. Effects of clozapine on smoking in chronic schizophrenic outpatients. J. Clin. Psychiatry 56, 344–346 (1995).
McEvoy, J. et al. Clozapine decreases smoking in patients with chronic schizophrenia. Biol. Psychiatry 37, 550–552 (1995).
Hays, J. T., Ebbert, J. O. & Sood, A. Treating tobacco dependence in light of the 2008 US Department of Health and Human Services clinical practice guideline. Mayo Clin. Proc. 84, 730–735 (2009).
Gortelmeyer, R. & Erbler, H. Memantine in the treatment of mild to moderate dementia syndrome. A double-blind placebo-controlled study. Arzneimittelforschung 42, 904–913 (1992).
Maler, J. M. et al. Memantine inhibits ethanol-induced NMDA receptor up-regulation in rat hippocampal neurons. Brain Res. 1052, 156–162 (2005).
Escher, T., Call, S. B., Blaha, C. D. & Mittleman, G. Behavioral effects of aminoadamantane class NMDA receptor antagonists on schedule-induced alcohol and self-administration of water in mice. Psychopharmacology 187, 424–434 (2006).
Holter, S. M., Danysz, W. & Spanagel, R. Evidence for alcohol anti-craving properties of memantine. Eur. J. Pharm. 314, R1–R2 (1996).
Bisaga, A. & Evans, S. M. Acute effects of memantine in combination with alcohol in moderate drinkers. Psychopharmacology 172, 16–24 (2004).
Baas, H. K. & Schueler, P. Efficacy of cabergoline in long-term use: results of three observational studies in 1,500 patients with Parkinson’s disease. Eur. Neurol. 46, 18–23 (2001).
Scholz, H. et al. Dopamine agonists for restless legs syndrome. Cochrane Database Syst. Rev. 2011, CD006009 (2011).
Koch, G. et al. Effect of rotigotine vs placebo on cognitive functions among patients with mild to moderate Alzheimer disease: a randomized clinical trial. JAMA Netw. Open 3, e2010372 (2020).
Kitamura, Y., Taniguchi, T., Shimohama, S., Akaike, A. & Nomura, Y. Neuroprotective mechanisms of antiparkinsonian dopamine D2-receptor subfamily agonists. Neurochem Res. 28, 1035–1040 (2003).
Feng, Y. & Wang, X. Antioxidant therapies for Alzheimer’s disease. Oxid. Med. Cell Longev. 2012, 472932 (2012).
Montalvo, I. et al. Improvement in cognitive abilities following cabergoline treatment in patients with a prolactin-secreting pituitary adenoma. Int. Clin. Psychopharmacol. 33, 98–102 (2018).
de Jong, S. et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS One 7, e39498 (2012).
Loh, P.-R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed model association for biobank-scale data sets. bioRxiv (2018).
Liu, Y. & Xie, J. Cauchy combination test: a powerful test with analytic p-value calculation under arbitrary dependency structures. J. Am. Stat. Assoc. 115, 1–18 (2019).
Subramanian, A. et al. A next generation connectivity map: l1000 platform and the first 1,000,000. Profiles Cell 171, 1437–1452.e17 (2017).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Wishart, D. S. et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Burgess, S., Butterworth, A. & Thompson, S. G. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665 (2013).
Burgess, S. & Thompson, S. G. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur. J. Epidemiol. 32, 377–389 (2017).
Zhao, Q., Wang, J., Hemani, G., Bowden, J. & Small, D. S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 48, 1742–1769 (2020).
Velmeshev, D. et al. Single-cell analysis of prenatal and postnatal human cortical development. Science 382, eadf0834 (2023).
Wang, L. G., Shuang; Chen, Siyuan; Markus, Havell; Wang, Gao; Carrel, Laura; Zhan, Xiang; Liu, Dajiang; Jiang, Bibo. Integrating axis-QTL and GWAS looks beyond cell types and unveils novel insights into brain-related traits. (Zenodo, 2025).
Acknowledgements
The research in this project is supported by NIH grants R01ES036042 (D.J.L. and B.J.), R01HG011035 (D.J.L and B.J.), R01AI174108 (D.J.L. and L.C.), R01HL173869 (D.J.L), U01AI185638 (D.J.L.), R21AI160138 (L.C. and D.J.L.), F30GM151848(H.M.), and Finkelstein Memorial Trainee Research Award(L.W.). Figure 2 is created with BioRender.com.
Author information
Authors and Affiliations
Contributions
X.Z., D.J.L., and B.J. designed, led, and oversaw the study. L.W. and S.G. led the data analysis and carried out software development. L.W. and S.G. contributed to process sc-eQTLs and generated GWAS summary statistics. L.W. and S.G. contributed to simulation and integrative genomic analysis. S.C. developed the RShiny App for result compilation. H.M. and L.W. conducted drug repurposing analysis. L.C. and G.W. contributed to data interpretation and evaluation. L.W., S.G., X.Z., D.J.L., and B.J. prepared the manuscript. All authors contributed to manuscript editing and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Inge Holtman and the other anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, L., Gao, S., Chen, S. et al. Integrating axis quantitative trait loci looks beyond cell types and offers insights into brain-related traits. Nat Commun 16, 10606 (2025). https://doi.org/10.1038/s41467-025-65643-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-65643-w
This article is cited by
-
Integrating axis quantitative trait loci looks beyond cell types and offers insights into brain-related traits
Nature Communications (2025)