Abstract
Lethal encephalitis caused by rabies virus (RABV) in mammals is known to be associated with the production of several pro-inflammatory cytokines, but the mechanism of such induction remains unclear. In this study, we establish that the laboratory strain CVS-11 infects astrocytes which are the most abundant glial cell population and the dominant source of inflammatory factors in the central nervous system (CNS). A screen identifies the E3 ubiquitin ligase FBXL18 as a critical factor responsible for RABV-induced inflammation. Mechanistically, infection by RABV upregulates FBXL18, which induces K11-type ubiquitination of BST2 on Lys109 and Lys110, two residues that are also ubiquitinated for degradation via K33-type ubiquitination by a yet unknown E3 ligase. FBXL18-mediated ubiquitination stabilizes BST2, leading to hyperphosphorylation of IκBα and excessive NF-κB activation. Knockdown of FBXL18 effectively inhibits IL-6 production and RABV replication in astrocytes and neurons, thereby mitigating the virulence of RABV in mice. Our findings suggest that targeting FBXL18 is a potentially effective strategy for rabies treatment.
Similar content being viewed by others
Introduction
Rabies is a highly fatal zoonosis caused by rabies virus (RABV) and characterized with encephalomyelitis associated with damage of the central nervous system (CNS) in mammals. As a neurotropic virus, plentiful studies indicated that RABV spreads trans-synaptically from infected to connected neurons, and the viral replication occurs primarily in neurons1. Recent studies suggest that virions and viral antigen can be detected in astrocytes in RABV-infected mouse brains2,3,4.
Astrocytes, as the most abundant glial cell population, are closely aligned with neuronal synapses, blood vessels, and ventricle cavities. In addition to their functional interactions with neurons and endothelial cells, astrocytes are endowed with the ability to secrete important inflammatory mediators, such as interleukin-6 (IL-6), CXC chemokine ligand 10 (CXCL-10), and tumor necrosis factor (TNF-α), which can either facilitate the recruitment of immune cells to clear invading viruses or contribute to neurons injury5,6. EV71 (human Enterovirus 71) has been reported to preferentially infect astrocytes and induce IL-6 release, leading to neural pathogenesis in mice7. The lab-attenuated RABV variant CVS-B2c had been reported to produce more double-stranded RNA (dsRNA) than its WT counterpart DRV-AH08, resulting in the activation of the RIG-I signaling pathway and the suppression of viral replication in astrocytes4.
Ubiquitination regulates virtually all cellular processes by affecting the stability, localization, or activity of target proteins8,9,10,11, among which, E3 ubiquitin ligases (E3s) play a critical role in substrate selection. Numerous studies have shown that E3s are important for the outcome of virus-host interactions. For example, ubiquitination of RIG-I by TRIM25 is essential for the initiation of antiviral responses, which activates the MAVS-TRAF6 antiviral axis and modulates NF-κB activation12. Similarly, RNF26 is known to temporally regulate virus-triggered type-I interferon induction by promoting K11-linked polyubiquitination of MITA13. Considering the observation that inflammation activated by IL-6 is crucial in the pathophysiology of encephalomyelitis4,14, we set out to identify E3 ligases involved in RABV replication and IL-6-associated neural pathogenesis.
In this study, we identify FBXL18 as a critical factor that positively regulates RABV replication by screening 182 predicted E3 genes by RNAi. This E3 ligase stabilizes BST2 by competitive ubiquitination, promoting the production of IL-6 and CXCL-10 in astrocytes. Moreover, knocking down FBXL18 in mice attenuates RABV virulence, suggesting that it is a potential therapeutic target in treating RABV infection.
Results
Robust induction of IL-6 and CXCL-10 in astrocytes by RABV
To investigate the response of astrocyte to RABV, we examined its infection of cultured astrocytes. Our results revealed that both mouse astrocytes MA-C and human astrocytes SVGP12 can be infected by RABV, and the infection efficiency of CVS-11 in N2a cells is detectably higher than that in MA-C or SVGP12 cells (Fig. 1A). We also determined viral replication using TCID50 and found that CVS-11 replicated at significantly lower rates in astrocytes than in N2a cells (Fig. 1B). Results obtained by RT-qPCR and immunoblotting were consistent with viral titers data, each showing that the copy number and protein levels of viral N/P in N2a cells were higher than in MA-C cells from 24 to 72 h (Fig. 1C, D).
A MA-C, SVGP12, and N2a cells were infected with CVS-11 at different MOIs (N2a, MOI = 0.1; MA-C and SVGP12, MOI = 1) for 24 h or 48 h. Viral particles were detected by direct fluorescence antibody assay (DFA); red fluorescence represents cells and apple-green fluorescence represents fluorescein isothiocyanate (FITC)-labeled RABV. Scale bar, 25 µm. B–D N2a and MA-C cells were infected with CVS-11 at different MOIs for indicated time, the titers of extracellular viral particles, copy numbers and protein level of N P were analyzed using DFA (B), RT-qPCR (C), and IB (D) individually. E, F Six-weeks old C57BL/6 mice were challenged intramuscularly (IM) with 10 LD50 of lethal CVS-11, the mouse brains were isolated at 7 d.p.i. and the infection in primary neurons and astrocytes were detected by indirect immunofluorescence. RABV (green), glial fibrillar acidic protein (GFAP) (red), and neuronal nuclei (NeuN) (red) were shown. Nuclei were counterstained with DAPI (blue). G MA-C cells were infected with CVS-11 for indicated time, the concentration of cytokines was analyzed using ELISA. Results in (E and F) are one representative from three biologically independent experiments with similar results. Scale bar, 100 µm or 10 µm. The quantitative results (mean ± s.e.) shown in (B, C, and G) were from three independent experiments each done in triplicate. Unpaired two-tailed Student’s t-test were performed. Source data are provided as a Source Data file.
Furthermore, examination of brains from infected mice showed that the signals of GFAP colocalized extensively with those of viruses, especially in ectocinerea and hippocampus (Figs. S1A and 1E), and the colocalization (white) between GFAP (red) and RABV (green) in representative images were showed (Fig. S2A), indicating that RABV infection could occur in astrocytes at 7 d.p.i. In all experiments, NeuN and RABV-infected cells were used as a positive control (Figs. S1B, 1F, and S2B).
We next determined whether RABV infection induces inflammatory response in astrocytes by examining the levels of secreted IL-6 and other pro-inflammatory cytokines. Results from ELISA experiments demonstrated that IL-6 and CXCL-10 were significantly induced by CVS-11, while the level of IFN-β only exhibited a slight increase (Fig. 1G). Such increase in cytokines also occurred in MA-C cell lines and primary astrocytes in response to infection, as demonstrated by RT-qPCR (Fig. S3A, B). These results indicate that the slow replication of RABV in astrocytes also triggers robust production of proinflammatory cytokines.
The E3 ubiquitin ligase FBXL18 promotes IL-6 production in RABV-infected astrocytes
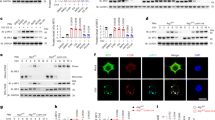
As one of the most widely used post-translational modifications, ubiquitination regulates critical processes such as DNA repair, protein quality control, and immunity. Among the enzymes involved in ubiquitination, E3 ubiquitin ligases (E3s) are critical in the final step of the reaction and in substrate recognition. To identify potential E3 ligases involved in RABV-related encephalitis, we knocked down 182 genes predicted to code for E3 ubiquitin ligases by transfecting MA-C cells with a pair of short interfering RNAs (siRNAs) (Dharmacon) that targets candidate genes (Fig. S4A). Transfected cells were infected with CVS-11 for 24 h and the effects of the knockdown were measured by determining secreted IL-6. The initial round of screening identified seven genes, including FBXL18, FBXL22, ASB2, Hrd1, WWP1, UBE2D1, and NEDD4 that detectably affected RABV-induced IL-6 production (Fig. 2A). The importance of these genes was further evaluated by measuring the mRNA levels of IL-6, N and P genes of the virus and virus replication after gene knockdown (Figs. S4B and S5). These efforts revealed that silencing FBXL18 caused consistent suppression IL-6 production accompanied by a reduction in viral replication (Fig. 2B). Conversely, ectopic expression of FBXL18 in infected MA-C cells led to an increase in the mRNA levels of both IL-6 and CXCL-10 (Fig. 2C).
A MA-C cells transfected with specific siRNAs were infected with CVS-11 for 24 h, the level of IL-6 was analyzed by ELISA. B, C MA-C cells were transfected with the siRNA against FBXL18, plasmid for HA-FBXL18 expression, or negative control and then infected with CVS-11 for 24 h. The mRNA levels of FBXL18, IL-6, CXCL-10 were analyzed by RT-qPCR; FBXL18, viral N/P protein were analyzed using IB with the corresponding antibodies. D–F The MA-C cells and N2a cells were infected with CVS-11 for indicated time, the FBXL18 protein level was analyzed using IB (D), and the mRNA level was analyzed using RT-qPCR (F). The relative expression level of FBXL18 normalizing to GAPDH or β-actin using Image J (E). Data show in results (A–C and E, F) were mean ± s.e. from three biologically independent experiment each done in triplicate. Unpaired two-tailed Student’s t-test were performed. Source data are provided as a Source Data file.
FBXL18 is a component of the Skp-Cullin-F-box (SCF) complex that functions as an E3 ubiquitin ligase15. We next examined whether CVS-11 affects the expression of FBXL18, FBXL22, WWP1, or UBE2D1 by measuring their mRNA and protein levels by RT-qPCR and IB in both MA-C and N2a cells, which revealed that infection by CVS-11 markedly induced the protein level of FBXL18 and UBE2D1, without affecting their mRNA levels (Figs. 2D–F and S6). CVS-11 infection only modestly increased the mRNA levels of FBXL22 but caused a reduction in its protein level. In contrast, WWP1 exhibited negligible changes at both transcriptional and translational levels in CVS-11-infected cells (Fig. S6). Taken together, these results reveal that the E3 ligase FBXL18 is induced by RABV infection, leading to induction of IL-6 production in astrocytes.
FBXL18 induces IL-6 production by activating the NF-κB pathway
NF-κB activation has been shown to induce IL-6 expression16, we thus examined whether the above-obtained candidate proteins (including FBXL18, FBXL22, UBE2D1, and WWP1) affect the activation of NF-κB using the luciferase reporter assay. Knockdown of FBXL18 led to inhibition of NF-κB activation in response to RABV infection. Additionally, the results indicated that depletion of FBXL18 led to more significant reduction in NF-κB activation than depletion of the other candidates (Figs. 3A and S7A). In contrast, ectopic expression of FBXL18 led to increased NF-κB activation (Fig. 3B). In line with these results, infection by CVS-11 led to an increase in phospho-p65, which was accompanied by a decrease in IκBα. Importantly, silencing of FBXL18 did not affect the level of p-IKKα/β, suggesting that FBXL18 activates NF-κB pathway by modulating the cellular level of IκBα via a pathway that does not involve IKKα/β activation (Fig. 3C).
A, B MA-C cells were transfected with combinations of plasmids for HA-FBXL18 expression, NF-κB-responsive luciferase and the Renilla luciferase for 24 h. Samples containing siRNA against FBXL18 were similarly established. Following this, luciferase reporter activities were induced by CVS-11 for 24 h. NF-κB luciferase activity was normalized to those of Renilla luciferase and presented as a fold induction. Fractions of cell lysates were analyzed by IB and probed with antibodies of IκBα and FBXL18, β-actin was used as a loading control. C MA-C cells transfected with siRNA against FBXL18 for 24 h were infected with CVS-11 for 2 h. The levels of p-IKKα/β, IκBα and P-p65 were analyzed using IB with corresponding antibodies. GAPDH was probed as loading controls. The levels of IκBα and P-p65 were quantified by using Image J. D–F MA-C cells were transfected with siRNA against FBXL18 or negative control for 24 h, then infected with CVS-11. D Fractions of cell lysates were analyzed by IB and probed with antibodies of IκBα and FBXL18, β-actin was used as a loading control. E Fixed cells were sequentially stained with antibodies specifically for p65 followed by DAPI to identify the nuclei. Images were captured using confocal laser scanning microscopy (Scale bar, 20 µm). F The ratio of nuclear localization was determined by counting at least 300 cells each sample. G MA-C cells transfected with siRNA against FBXL18 or negative control, after CVS-11 stimulated for 2 h, subjected to fractionation and the presence of the relevant proteins in the cytosolic (Cyt) and nuclear (Nuc) fraction was detected using IB with corresponding antibodies. GAPDH and PCNA were used for controls. Data shown in (G) was a representative of three biologically independent experiments with similar results, and the results (mean ± s.e.) shown in (A–C, and F) was from three independent experiments. Unpaired two-tailed Student’s t-test (A, B, and F) and One-way ANOVA (C) were performed for data analysis. Source data are provided as a Source Data file.
A critical event associated with NF-κB activation is degradation of IκBα and the translocation of p65 into the nuclei. We thus determined the role of FBXL18 in nuclear translocation of p65 in RABV-infected cells. As expected, after infection with RABV for 24 h, nuclear translocation of p65 occurred in almost all infected cells (Fig. 3D, E). Importantly, in FBXL18 knockdown samples p65 nuclear translocation occurred in less than 20% of the cells upon RABV challenge (Fig. 3F). However, the depletion of other candidates exhibited negligible effects on the nuclear translocation of p65 in RABV-infected cells (Fig. S7B). Furthermore, the time-lapse imaging at 20, 40, 60, 120, 180, and 240 min and movies covering 12 time points clearly demonstrated nuclear translocation of p65 was suppressed in FBXL18-silenced N2a cells in response to CVS-11 infection (Fig. S7C and Supplementary Movies 1–4). Conversely, knockdown of FBXL18 caused the retention of p65 in the cytoplasm in RABV-infected cells.
We further analyzed the localization of p65 by cell fractionation using proliferating cell nuclear antigen (PCNA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the marker for nuclear and cytosolic protein, respectively17,18. In FBXL18-silenced cells, only weak signals of p65 were detected in the nuclear fraction, which is consistent with the results from immunostaining assays (Fig. 3G). Thus, FBXL18 promotes IL-6 production by inducing NF-κB activation in RABV-infected astrocytes.
FBXL18 activates NF-κB by a mechanism involved in BST2 and IκBα
To identify the cellular targets of FBXL18, we used quantitative proteomic analysis to compare protein abundance between MA-C cells and those that have received FBXL18-specific siRNAs. These efforts revealed that 17 proteins were significantly downregulated in FBXL18-silenced cells. Among these, the level of bone marrow stromal cell antigen 2 (BST2) decreased approximately 0.617 folds in FBXL18-silenced cells. BST2, also known as tetherin or CD317, is an IFN-induced type II transmembrane protein19,20. BST2 had been previously reported to play a role in NF-κB signaling21,22,23, we thus examined whether it is involved in NF-κB activation mediated by FBXL18 in RABV-infected cells. Knocking down of BST2 effectively suppressed NF-κB activation, which was accompanied by a reduction in nuclear translocation of p65 in infected cells (Fig. 4A). Furthermore, knockdown of BST2 suppressed NF-κB activation in response to RABV infection. Conversely, ectopic expression of this protein promoted NF-κB activation (Fig. 4B, C). To address the roles of BST2 in NF-κB signaling, the mRNA levels of cytokines IL-6 and CXCL10, two genes known to be regulated by NF-κB were measured, which revealed that BST2 induced the expression of IL-6 and CXCL-10 (Fig. 4D).
A MA-C cells transfected with siRNA against BST2, after CVS-11 stimulated for 2 h. Samples were processed as described in Fig. 3G for IB analysis. B, C MA-C cells were transfected with combinations of plasmids for Flag-BST2 expression, NF-κB-responsive luciferase and the Renilla luciferase for 24 h and then induced by CVS-11 for 24 h. Samples containing siRNA against BST2 were similarly established. Samples were processed as described in Fig. 3A, B for luciferase activity measure and IB analysis. D MA-C cells transfected with siRNA against BST2 or the plasmid expressing Flag-BST2 for 24 h, and then infected with CVS-11 for 24 h. The level of relevant genes was analyzed by RT-qPCR. E, F MA-C cells were transfected with siRNA against BST2 or plasmids for HA-FBXL18 expression, NF-κB-responsive luciferase, and the Renilla luciferase individually or in combination for 24 h and then induced by CVS-11 for 24 h. Samples were processed as described in this figure (A, B) for luciferase activity measure and IB analysis. G, H MA-C cells transfected with BST2-expressing plasmids or specific siRNA or in combination with FBXL18-expressing plasmid were infected with CVS-11 for 2 h. Confocal microscopy analysis of p65 nuclear translocation in MA-C cells. Samples were processed as described in Fig. 3D, E. Scale bar, 25 µm. I Quantification of normalized mCherry-p65 nuclear localization from experiment (G) by using Olympus Cell Sens Dimension with Measurement module. Statistical analysis in each panel was performed by Student’s t test. Data shown in (A) was a representative from three biologically independent experiments with similar results. And the quantitative results (mean ± s.e.) shown in (B–D, F, and I) were from three independent experiments each done in triplicate. Unpaired two-tailed Student’s t-test were performed for data analysis. Source data are provided as a Source Data file.
Importantly, we also observed that silencing BST2 suppressed NF-κB activation caused by overexpression of FBXL18 (Fig. 4E, F), suggesting that BST2 was downstream of FBXL18 in this signaling cascade. The role of BST2 in FBXL18-induced NF-κB signaling was further examined by determining nuclear translocation of p65. The rates of nuclear-localized p65 in RABV-infected cells became diminished upon FBXL18 silencing and such decrease was reversed by overexpressing BST2. In agreement with these results, silencing of BST2 significantly inhibited nuclear translocation of p65 in cells overexpressing FBXL18 and the intensity of nuclear p65 signal was quantified (Fig. 4G–I). These results establish that in RABV-infected astrocytes, FBXL18 promotes inflammation via the NF-κB pathway by targeting BST2.
Stabilization of BST2 by FBXL18 requires its F-box domain
The observed upregulation of BST2 by FBXL18 may occur at the transcriptional or translational level. To distinguish between these two possibilities, we measured its impact on mRNA and protein levels of BST2, respectively. Knocking down FBXL18 caused a slight reduction in BST2 mRNA. Conversely, overexpression of this protein increased the expression of BST2, and such effects occurred independent of RABV infection (Fig. 5A, B). Yet, when the abundancy of BST2 was measured, silencing FBX18 caused a severe reduction in its protein level, which can be reversed by ectopic expression of the E3 ligase. When measured by immunostaining, the increase of BST2 caused by RABV infection was suppressed by FBXL18 silencing (Fig. 5C), and this phenotype occurred in multiple cell lines, including N2a and HEK293 cells (Fig. 5D).
A, B MA-C transfected with siRNA against FBXL18 or plasmid expressing HA-FBXL18 were infected with CVS-11 for 24 h. The mRNA level of BST2 was analyzed by RT-qPCR (A), the protein level of BST2 was detected using IB (B). C Confocal microscopy analysis of BST2 location in MA-C cells, which transfected with siRNA against FBXL18 or negative control, after CVS-11 or mock stimulated for 24 h. D N2a or HEK293 cells were transfected with plasmids expressing FBXL18, BST2 or negative control for 24 h, the level of BST2 was analyzed using IB. E Immunoprecipitation (with anti-Flag) and IB analysis the direct interaction of FBXL18 and BST2 in HEK293 cells, which transfected with Flag-BST2, HA-FBXL18 or negative control. F FBXL18 or the FBXL18ΔF-box mutant was co-expressed with Flag-BST2 in HEK293 cells for 24 h, the level of relevant proteins was analyzed using IB. G Immunoprecipitation (with anti-Flag) and IB analysis the direct interaction of HA-FBXL18ΔF-box and BST2 in HEK293 cells, which transfected with Flag-BST2, HA-FBXL18ΔF-box, or negative control. The quantitative results (mean ± s.e.) shown in (A) were from three independent experiments each done in triplicate. Unpaired two-tailed Student’s t-test were performed. Results in (C–G) were one representative from three biologically independent experiments with similar results. Source data are provided as a Source Data file.
Next, we explored the possibility that BST2 is a substrate of FBXL18. First, we determined their interactions by immunoprecipitation using Flag antibody, which showed that complexes formed by these two proteins in lysates of cells transfected to express HA-FBXL18 and Flag-BST2 can be captured by Flag beads, indicating binding between these two proteins (Fig. 5E).
FBXL18 harbors an F-box domain important for the formation of the SCF complex required for protein ubiquitination. To examine the roles of the F-box domain of FBXL18 in the regulation of BST2, we generated an F-box domain mutant (ΔF-box). Whereas expression of HA-FBXL18 in HEK293 cells with Flag-BST2 increased BST2 abundancy, similarly expressed FBXL18ΔF-box did not have such effect (Fig. 5F). Moreover, we found that the mutant FBXL18ΔF-box had lost the ability to interact with BST2 (Fig. 5G). Taken together, these results indicate that FBXL18 likely directly interacts with BST2 to increase its cellular abundancy by a mechanism that requires its F-box domain.
FBXL18 catalyzes K11-linked ubiquitination of BST2 at Lys109 and Lys110
The requirement of the F-box domain for the impact of FBXL18 on BST2 prompted us to examine the latter as a ubiquitination substrate for the E3 ligase. We first expressed Flag-BST2 and HA-Ub with His-FBXL18 or FBXL18ΔF-box in HEK293 cells and isolated Flag-BST2 by IP. Detection of the IP products by immunoblotting with the HA antibody revealed patterns characteristic of ubiquitination in Flag-BST2 co-expressed with FBXL18 but not its mutant FBXL18ΔF-box (Fig. 6A).
A Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination of BST2 induced by FBXL18, in which HEK293 cells transfected with combination of Flag-BST2, HA-Ub, His-FBXL18, or His-FBXL18ΔF-box. B Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination of BST2, in which HEK293 cells transfected with Flag-BST2, WT HA-Ub, or Ub-mutants along with His-FBXL18. C Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination style of BST2 induced by FBXL18, in which HEK293 cells transfected with His-FBXL18, Flag-BST2, and HA-ubiquitin (K11) or its mutants K11R. D–F HEK293 or N2a cells were transfected with Flag-BST2, BST2 harboring lysine mutants, or HA-FBXL18 for 24 h, then the level of relevant proteins was analyzed using IB. G Co-immunoprecipitation (with anti-Flag) and IB analysis of HEK293 cells were transfected with His-FBXL18, HA-ubiquitin, Flag-BST2, and its mutants, in which the lysine residues at position 122, 165, 109/110 were replaced with arginine. A–C and G After transfected for 24 h, the cells were treated with 10 µM MG132 for 10 h before harvest. Results shown in (A–G) are one representative from three biologically independent experiments with similar results. Source data are provided as a Source Data file.
To determine the type of polyubiquitin chains conjugated to BST2 by FBXL18, we co-expressed Flag-BST2 and His-FBXL18 with each of a series of ubiquitin mutants each containing only one of the seven lysine residues24 (−K6, −K11, −K27, −K29, −K33, −K48, −K63), followed by detection of ubiquitination on isolated Flag-BST2. Ubiquitination by K11, K29, and K33 mutants was abundant with K11 displaying the highest level of modification (Fig. 6B). Consistent with this observation, inclusion of the K11R ubiquitin mutant in this assay did not lead to detectable BST2 ubiquitination (Fig. 6C).
To determine the modification sites of BST2 by FBXL18, we performed mass spectrometric analysis of BST2 isolated from cells co-expressing FBXL18. A modified peptide -K110VSQALEQQAR120- was detected and MS/MS spectra showed that the ubiquitination mapped to residue Lys110 (Fig. S8). We next confirmed the role of Lys110 in FBXL18-mediated stabilization of BST2 by testing the response of the BST2K110R mutant to the E3 ligase, and found that the mutant can still be stabilized (Figs. 6D, E and S9), suggesting the existence of additional ubiquitination sites on BST2.
To determine such sites, we introduced substitution mutations in each of the seven lysine residues and the mutants were individually co-expressed with FBXL18 in HEK293 cells, and found that the protein levels of mutants K109–K110, K122R and K165R were lower than those of the wild-type (Fig. 6D). In N2a cells, the stability of K109–K110 and K165R mutants was not enhanced by FBXL18 (Fig. 6E). Moreover, these mutations did not detectably impact the stability of BST2 in the absence of FBXL18 overexpression (Fig. 6F). Importantly, when co-expressed with FBXL18, the BST2K109R-K110R mutant can no longer be ubiquitinated. In contrast, mutations in K122 or K165 did not detectably affect FBXL18-induced ubiquitination of BST2 (Fig. 6G). These results establish that K109 and K110 on BST2 are the recipient sites for FBXL18-induced ubiquitination.
FBXL18 protects BST2 from K33-linked ubiquitination and degradation
Our results suggested that K11-linked ubiquitination of BST2 by FBXL18 enhanced the stability of BST2 (Fig. 6B). Intriguingly, ubiquitination of the BST2 was most robustly detected in reactions receiving the K33-and K27-only ubiquitin variants under conditions of no FBXL18 overexpression (Fig. 7A). We thus hypothesized that FBXL18 may induce an alternative type of the ubiquitination on BST2. Indeed, expression of the UbK33R mutant with FBXL18 in HEK293 cells led to robust BST2 ubiquitination and no modification occurred when the ubiquitin mutant Ub-K33 that only harbored K33 was used. Of note, both ubiquitin variants can be used to modify BST2 in cells when FBXL18 was not overexpressed (Fig. 7B). Concurrently, RABV infection induced a shift from K33-linked to K11-linked polyubiquitination on BST2, which was accompanied by an increase in the protein level of FBXL18 (Fig. 7C).
A Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination of BST2, in which HEK293 cells were transfected with Flag-BST2, WT HA-Ub, or Ub-mutants. B Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination of BST2, in which HEK293 cells were transfected with His-FBXL18, Flag-BST2, and HA-ubiquitin (K33) or its mutants K33R. C Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination of BST2, in which N2a cells were transfected with Flag-BST2 and HA-ubiquitin (K11 or K33) for 24 h and infected with CVS-11 for further 24 h. D, E Co-immunoprecipitation (with anti-Flag) and IB analysis of the ubiquitination and lysine residues of BST2, HEK293 cells were transfected with His-FBXL18, HA-ubiquitin (K11), HA-ubiquitin (K33), and Flag-BST2 or its mutants. A–E After transfected for 24 h, the cells were treated with 10 µM MG132 for 10 h before harvest. Results shown in (A–E) were one representative from three biologically independent experiments with similar results. Source data are provided as a Source Data file.
We next determined whether K11-linked ubiquitination outcompetes K33-linked ubiquitination on K109 and K110 of BST2 in cells in which FBXL18 was overexpressed. High levels of FBXL18 promoted K11-linked ubiquitination of BST2, which was accompanied by a decrease in K33-linked ubiquitination. Importantly, K33-linked ubiquitination of BST2 also required both K109 and K110 (Fig. 7D, E).
Collectively, these results suggest that the K11-type ubiquitin chains on BST2 became dominant in RABV-infected cells, which leads to its stabilization and subsequent activation of NF-κB.
FBXL18 promotes viral replication in RABV-infected N2a cells
To determine the role of FBXL18 in RABV virulence in neuron cells, we first examined the expression level of cytokines IL-6 and CXCL-10 in RABV-infected N2a cells following FBXL18 silencing. Consistent with results from experiments using astrocyte cells, silencing of FBXL18 significantly inhibited the expression of viral protein and a reduction of viral titers (Fig. 8A, B). NF-κB activation, and increased production of IL-6 and CXCL-10 were also detected in neuron cells (Fig. 8C, D). Thus, in both astrocytes and neuron cells, RABV infection induces the expression of FBXL18 to promote inflammation and viral replication.
A, B N2a cells transfected with siRNA against FBXL18 or plasmid expressing HA-FBXL18 were infected with CVS-11 (MOI = 0.1) for 24 h, the levels of cytokines, FBXL18, and viral N/P were analyzed by RT-qPCR, the relevant protein levels were analyzed using IB with corresponding antibodies and the viral titers of extracellular were analyzed by DFA. C N2a cells were transfected with combinations of plasmids for HA-FBXL18 expression or siRNA against FBXL18, the NF-κB luciferase reporter, and the Renilla luciferase for 24 h. Samples were processed as described in Fig. 3A for luciferase activity measure. D The concentration of cytokines was tested by ELISA. Data shown in (A–D) were mean ± s.e. from three biologically independent each done in triplicate and unpaired two-tailed Student’s t-test were performed. Source data are provided as a Source Data file.
Knockdown of FBXL18 reduces RABV virulence and IL-6 production in vivo
To investigate the role of FBXL18 in RABV-associated neural pathogenesis in vivo, the FBXL18-knockdown mice model was established25,26,27,28 by stereotaxically infusing AAV-shFBXL18 or AAV-shNC into the both lateral ventricles of 5 weeks-old C57BL/6 (Fig. 9A, B)29. The effectiveness of FBXL18 silencing in mice brains were evaluated by using RT-qPCR, IB, and IF (Fig. 9C–E).
A Schematic diagram of AAV-shNC and AAV-shFBXL18 structures. B 5-weeks old female mouse were administrated with AAV-shNC and AAV-shFBXL18 for 21 d, and then infected with CVS-11 at a lethal dosage of 10 LD50 via intramuscular (i.m). The models of (A and B) were created with BioGDP.com and Adobe Illustrator. C, D The silencing efficiency of FBXL18 in brains were confirmed by using RT-qPCR (C), IB (D). E Representative photomicrographs of injection sites in both lateral ventricles. F–H Body weight change (F), clinical score (G), and survival ratio (H) were monitored daily for 21 d. I–K The brains of infected mice were collected at 7/12 d and analyzed the levels of IL-6, CXCL10, and CVS-11 N/P genes by RT-qPCR (I-J) or ELISA (K). Data shown in (E) was a representative of three independent experiments with similar results. Scale bar, 100 µm. Results in C, F, G, and I–K (mean ± s.e.) shown were from five or three biologically independent experiments (C, n = 5; F, G and I–K, n = 3) and unpaired two-tail Student’s t test (C), Ordinary two-way ANOVA with multiple comparisons (F, G), Ordinary one-way ANOVA with multiple comparisons (I–K) were performed for data analysis. The log-rank (Mantel–Cox) test was used to analysis the statistical difference in survival rates between the shNC-CVS-11 and shFBXL18-CVS-11 in H. Source data are provided as a Source Data file.
To better evaluate the effect of FBXL18 silencing on viral virulence, we measured the body weight, clinical score, and survival of infected mice daily for a total of 21 d. Our results showed that the body weight decreased drastically from 5 to 11 or 6 to 12 d.p.i. in groups receiving shNC or untreated. In contrast, the body weight in mice receiving shFBXL18 only showed a slight decrease from 6 d to 13 d, then gradually recovered to normal body weight (Fig. 9F). Importantly, the infected mice receiving shNC and control groups showed incoordination at 5 d.p.i. or 6 d.p.i. then rapidly developed overt clinical symptoms and body weight loss. Typical neurological symptoms were observed at 9–12 d.p.i., presenting hypothermia, abundant white discharge from the eyes, respiratory failure, and death. However, for mice receiving shFBXL18, only 2 out 10 displayed clinical symptoms and death (Fig. 9G). Finally, 80% of the mice receiving shFBXL18 survived post CVS-11 infection, whereas only 10% of the mice receiving shNC survived and no mice in the untreated control group survived (Fig. 9H).
When relevant cytokines and viral gene abundancy were measured, the levels of IL-6, CXCL-10, and CVS-11 N and P genes in mice brains displayed a drastic reduction in shFBXL18-AAV treated groups when compared to shNC-AAV or untreated control groups at both 7 and 12 d.p.i. (Fig. 9I–K). Taken together, these results indicate that knockdown of FBXL18 can effectively protect mice from lethal infection by RABV.
Discussion
As the most abundant immune cells in the CNS, astrocytes are essential to mount proper or sometime excessive anti-viral and inflammatory responses by regulating various host signaling pathways. Earlier studies revealed that RABV infection induces robust expression of IL-6 in the CNS2,4,14. The NF-κB pathway is also activated by RABV infection in humans and canines30,31. Yet, the mechanism of such immune induction remains poorly characterized. We performed a screen of 182 predicted E3 ubiquitin ligase genes and identified several positive regulators of NF-κB activation and IL-6 production in RABV-infected astrocytes, including FBXL18, FBXL22, WWP1, and UBE2D1. In particular, we found that FBXL18 markedly induced by RABV infection in astrocytes and neuronal cells, and knockdown of this E3 ligase more pronouncedly impaired NF-κB activation and virus replication than depletion of UBE2D1, WWP1 or FBXL22. Further, we identified BST2 as an important cellular target of FBXL18 and demonstrated that FBXL18 likely directly interacts with and ubiquitinates BST2 to enhance its stability, thereby enhancing NF-κB-mediated IL-6 production. These several lines of evidence prompted us to select FBXL18 as the focus of this study. Nevertheless, it is very likely that other host factors also govern the life cycle of the rabies virus. More work is needed to firmly establish the role of these factors in viral virulence. It will be of great interest to determine whether simultaneous knockdown of these factors and FBXL18 will allow more effective treatment of rabies virus infection.
As a member of the SCF superfamily E3 ligases, FBXL18 is known to regulate the abundance of several proteins via the ubiquitin proteasome pathway, including LRRK2, AKT, PRS15A, and Morf4l1, thereby contributing to the progression of familial Parkinson’s disease and malignant tumors32,33,34. To our knowledge, this study is the first report to identify the dual role of FBXL18 as an E3 ubiquitin ligase in modulating RABV replication and associated encephalomyelitis.
Our results establish that FBXL18 targets BST2, a protein known to be involved in inhibition of viral replication via various mechanisms, including tethering enveloped virions to cell surface and restricting the release of HIV particles35,36, promoting PEDV-N protein degradation through the BST2-MARCHF8-CALCOCO2-autophagosome pathway37, and activating NF-κB through the recruitment of TRAF2/6, TAB, and the mitogen-activated kinase TAK122. Viruses have also been found to use various strategies to counter the inhibitory effect of BST2. For example, ORF7a of SARS-CoV inhibits BST2 glycosylation by direct interactions, thus reducing its tethering function38.
In sharp contrast to these studies, we found that silencing of BST2 inhibited RABV replication both in MA-C and N2a cells (Fig. S10A). Similar knockdown of BST2 in A549 cells allowed more robust replication of the H1N1 virus (Fig. S10B), further confirming its opposite roles in the virulence of RABV and such viruses as H1N1 and SARS-CoV. Overexpression of FBXL18 led to more extensive ubiquitination and higher protein level of BST2 (Fig. 6A), which is counterintuitive as ubiquitination often causes protein degradation. Our results support a model whereby RABV infection induces the expression of FBXL18, which catalyzes K11-type ubiquitination on K109 and K110 in BST2, thus preventing it from being modified by K33-type ubiquitin chains and subsequent degradation (Fig. 10). Although the E3 ligases responsible for adding K33-type polyubiquitin chains to BST2 is unknown, our results suggest that interference of such E3s will phenocopy the effect of FBXL18 overexpression.
A model was created using Adobe Illustrator and described the role of FBXL18 in RABV-related lethal encephalitis by triggering induction of NF-κB. The lethal RABV infection induces the expression of FBXL18 in astrocytes, subsequently, FBXL18 interacts with and stabilizes substrate BST2 by catalyzes K11-linked ubiquitination on its K109 and K110, the two residues that are also the recipient sites of K33-type ubiquitination (by a yet unknown E3). Obviously, the FBXL18-induced ubiquitination outcompetes the degradation prone K33-type ubiquitination, thus stabilizing BST2, which in turn activates NF-κB and promotes IL-6 production.
K11-type polyubiquitination appears to differently impact protein stability. On the one hand, it promotes caspase-1 degradation, an event that is counteracted by the NS1 protein of ZIKV, which protects caspase-1 from being degraded by recruiting the DUB USP8 to cleave the K11-linked ubiquitin chains39. On the other hand, K11-ubiquitin chains can promote protein stability. For example, RNF26 catalyzes K11-linked polyubiquitination on MITA and protects it from K48-type polyubiquitination and subsequent degradation induced by RNF-5 on the same lysine residue (K150)13. The mechanism of the opposite effect of the same ubiquitin chain on protein stability awaits further investigation.
The findings that knockdown of FBXL18 not only suppresses IL-6 production but also inhibits viral replication, thereby attenuates RABV virulence in mice. Thus, targeting this E3 ligase can be an effective strategy for rabies treatment. Interestingly, inhibitors against the SCFCdc4 ubiquitin ligase and Met30, which is also an SCF complex, have been identified40,41. It will be of great importance to determine whether such platforms allow the screening of small molecule inhibitors against FBXL18.
Methods
Mice
Female C57BL/6 mice (5 and 8 weeks-old) were purchased from Shanghai Lingchang Biotechnology Co., Ltd. and Spearhead (Beijing) Biotechnology Co., Ltd., respectively. All mice were housed in insulator cages under specific pathogen-free conditions with free access to food and water and the housing environment was maintained at a temperature of 22 ± 2°C with 55 ± 5% relative humidity, accompanied with a 12 h light-dark cycle (07:00–19:00 light phase). All animal experiments were approved by the Animal Care and Use Committees of the Changchun Veterinary Research Institute (Approval number, IACC of AMMS-11-2022-022).
Cells culture and viruses
Mouse astrocyte cells (MA-C) were purchased from Shanghai Qingqi biological company and was validated with GFAP (a cell-specific marker of astrocytes). Mouse Neuro-2aA (N2a), human astrocyte (SVGP12), and A549 were preserved by our laboratory and HEK293 cell lines was a gift from Prof. Jinliang, Wang (Harbin Veterinary Research Institute)42. The primary astrocytes were isolated, routinely maintained, and characterized using GFAP in our laboratory. These cells were cultured in DMEM (Corning, cat#10-013-CVRC), all mediums were supplemented with 10% fetal bovine serum (FBS; Cell-Box, AUS-01S-02), 100 U/mL of penicillin, and 100 mg/mL of streptomycin (HyClone, cat#SV30010) at 37 °C in 5% CO2. Rabies virus challenge virus standard (CVS)-11 strain (GenBank No:LT839616) was obtained from the UK OIE Reference Laboratory14. The viral strain was propagated in N2a cells and stored at −80 °C. Virus titers were determined as the 50% tissue culture infective doses (TCID50) per milliliter by direct immunofluorescence assays (DFA) in N2a cells43,44.
Reagents and antibodies
The mouse antibodies against viral N and P were prepared and stored in our laboratory14. The antibodies against Phospho-NF-κB (CST, cat#3033), Phospho-IKKα/β (CST, cat#2697), PCNA (CST, cat#13110), GAPDH (CST, cat#2118), NF-κB (CST, cat#8242), IκBα (Beyotime Biotechnology, cat#AF1282), HA (Beyotime Biotechnology, cat#AF2858), β-actin (Beyotime Biotechnology, cat#AF2815), BST2 (mouse, Abcam, cat#ab272169), BST2 (human, Abcam, cat#ab243229), FBXL18 (Bioss, cat#bs-16039R; and was validated using IB/IP in Fig. S11), GFAP (Abcam, cat#ab279291), NeuN (Abcam, cat#ab177487), UBE2D1 (proteintech, cat#15475-1-SP), ASB2 (ABclonal, cat#A17923), WWP1 (proteintech, cat#6704-1-Ig), Hrd1 (ABclonal, cat#A2605), FBXL22(abcam, cat#223059), NEDD4 (Proteintech, cat#21698-1-SP), Flag (Proteintech, cat#66008-4-Ig), NP Antibody (Sino Biological, cat#11675-T62), Alexa Fluor 594 goat anti-chicken antibody (Thermo, cat#A32759), horseradish peroxidase (HRP)-labeled IgG mouse secondary antibodies (Beyotime Biotechnology, cat#A0216) and rabbit secondary antibodies (Beyotime Biotechnology, cat#A0208) were obtained. FITC Anti-Rabies Monoclonal Globulin (Fujirebio, cat#800-092) was obtained. MG132 (cat#HY-13259) were purchased from Med Chem Express.
Immunofluorescence assays
Immunofluorescence samples were fixed with 4% paraformaldehyde (PFA) (Solarbio, cat#P1110) for 15 min at room temperature, blocked with 0.25% Triton X-100 (Amresco, cat#0694-1L) and 5% bovine serum albumin (Solarbio, cat#SW3015) for 20 min and 1 h, respectively. After incubation with appropriate primary antibodies overnight at 4 °C, samples were washed with PBS and incubated with secondary antibodies for 1 h. Images were captured by confocal laser scanning microscopy.
Small interfering RNAs, plasmids, and transfection
Short interfering RNAs targeting 182 specific E3 ligase obtained from Dharmacon (USA) were delivered into MA-C cells using the Lipofectamine RNAi-MAX reagent (Thermo, cat#13778075) according to the manufacturer’s instructions. All the siRNA sequences were purchased from Gene Pharm (Shanghai, China) and listed in Table S1.
The pHA-FBXL18 and pHA-BST2 plasmids were generated by amplifying the open reading frame (ORF) of FBXL18 (NCBI Gene ID: 231863) and BST2 (NCBI Gene ID: 69550) genes and cloning them into the pcDNA3.1(+) vector. And a series of ubiquitin mutants were a gift from Prof. Zhao-Qing Luo (Purdue University), originally described in Qiu et al., Nature, 2016. For transfection, cells cultured to 70%–80% confluence were transfected with siRNA or plasmids using Lipofectamine 2000 (Thermo, cat#11668019) according to manufacturer’s instructions.
Quantitative real-time reverse transcription-PCR (qRT-PCR)
RNA was isolated using TRIzol regents (Thermo, cat#15596018) and the cDNA was synthesized using M-MLV reverse transcriptase (TaKaRa, cat#2641B) according to the manufacturer’s protocols. The genome copies were quantified by RT-qPCR using TB Green Premix Ex Taq II (TaKaRa, cat#RR820A200), and they were conducted by using an Agilent Stratagene Mx3000P Q-PCR system and following the previously described amplification condition14. The mRNA levels were normalized using GAPDH. Fold variations between RNA samples were calculated by the 2−ΔΔCT threshold cycle (CT) method after normalization14. All oligonucleotide primers used for qPCR in this study were listed in Table S2.
Immunoblot (IB) analysis
Cells and mice brains were lysed with RIPA buffer (Merck, cat#20-188) containing Halt Protease & Phosphatase Inhibitor Cocktail (Thermo, cat#78442). Nuclear and cytoplasmic proteins were separated using a nuclear and cytoplasmic protein extraction kit (Beyotime Biotechnology, cat#P0028). The obtained protein was quantified using BCA protein assay kit (Beyotime Biotechnology, cat#P0010) and 30 μg protein samples separated by SDS-PAGE were transferred onto polyvinylidene fluoride (PVDF) membranes (Merck, cat#ISEQ00010) and incubated overnight with primary antibody. After a further incubation with HRP-conjugated secondary antibody, the blots were visualized using the Omni-ECL Femto Light Chemiluminescence Kit (EpiZyme, cat#SQ201) and Tanon-5200 system (Tanon, Shanghai, China), the densitometry was quantified using ImageJ (version 1.36b; NIH, MD, USA) and normalized to signals of bands of GAPDH or β-actin.
ELISA assays
The mice were anesthetized and brain tissues were obtained at 7 d.p.i. and 12 d.p.i. The brain was mechanically homogenized in PBS with Halt Protease & Phosphatase Inhibitor Cocktail (Thermo, cat#78442) and centrifuged at 12,000 × g for 10 min at 4 °C, the supernatants were then collected45. MA-C and N2a cells were cultured in 12-well plates and infected with RABV for 24 h, then the supernatants were collected at indicated times. The concentrations of IL-6, CXCL-10, and IFN-β were measured using ELISA using kits (Thermo, cat#BMS603-2TWO; BMS6018; 424001) following manufacturer’s instructions. The final concentration of cytokines was determined using an enzyme-linked immunosorbent assay (Infinite M200PRO, TECAN) with OD value at 450 nm.
Dual-luciferase reporter assays
MA-C and N2a cells were seeded in 24-well plates and transfected with NF-κB luciferase reporter (300 ng), TK Renilla reporter (100 ng), and the indicated plasmids by using Lipofectamine 2000 for 24 h. Cells were infected with RABV for 24 h and luciferase activities were measured with a dual-specific luciferase assay kit (Promega, cat#E1910) and luminometer (GloMax® 20/20 Single Tube Luminometer, Promega) according to manufacturer’s instructions. Firefly luciferase activities were normalized on the basis of Renilla luciferase activities. All reporter assays were performed at least three times.
Co-immunoprecipitation (Co-IP)
The indicated plasmids were transfected into HEK293 cells for 48 h, the cells were washed by cold PBS and harvested by lysis buffer (ddH2O containing 50 mM Tris-HCl pH 7.5, 150 mM NaCl and 1% Triton X-100) on ice for 5 min. Cell lysates were cleared by centrifugation at 12,000 × g for 10 min at 4 °C in three times, and then the cell supernatants were incubated with anti-HA agarose beads or anti-Flag agarose beads (Sigma, cat#A2220; Thermo, cat#26181) at 4 °C for 12 h. The beads were washed four times with lysis buffer and the proteins were eluted by using HA or Flag peptide (Thermo, cat#26184; Sigma, cat#F4799-4MG).
Live cell imaging
N2a cells were seeded in 12-well plates (Corning, cat#3513) for 24 h, and transfected with siRAN against FBXL18 or control plasmid in combination with pCMV-mCherry-P65 (Beyotime Biotechnology, cat#D2821). After transfection for 24 h, cells were infected with strain CVS-11, and were stained with Hoechst (Beyotime Biotechnology, cat#33342) for 10 min. After washing with PBS, cells were maintained in fresh culture medium. Live-cell imaging was performed using an Operetta high-content imaging system equipped with a live-cell incubation chamber maintained at 37 °C and 5% CO2. Images were captured using an LWD 20× objective in wide field fluorescence mode. After adding CVS-11 viruses for 1 h, cells were imaged every 20 min over a period of 4 h to monitor the translocation of p65 into the nucleus46.
Adeno-associated virus (AAV) injection
Mouse FBXL18 was silenced by using an adeno associated viral (AAV) vector delivery system27,28. Briefly, the adeno-associated virus vector expressing short hairpin RNA (shRNA) targeting the sequence of FBXL18 and negative control were synthesized and cloned into GV480 (pAAV-U6-shRNA-CAG-mCherry) vector with BsmBI sites (purchased from Shanghai Genechem), recombinant vector was detected by DNA sequencing. The sequences were listed in Table S3. AAV-shFBXL18 and negative control viral particles (1.3 × 1011 v.g for each mouse) were infused into the both lateral ventricles of 5 weeks-old C57BL/6 (coordinates: 2.30 mm posterior to the bregma, ± 3.00 mm bilateral to the midline, and 4.00 mm deep to the skull surface) by using a microsyringe pump at a constant rate of 1 μL/min, followed by a 3 min of rest to ensure proper diffusion.
CVS-11 challenge experiment in mice
The total sixteen C57BL/6 mice (5 weeks-old, female) that had been administered with AAV-shNC or AAV-shFBXL18 for three weeks were intramuscularly (IM) injected with a lethal dose of CVS-11 (10 LD50, 107 TCID50) into the hind legs, the C57BL/6 mice (8 weeks-old, female) were synchronously injected with equivalent CVS-11 or DMEM as control. The mouse brains (n = 3) were collected at 7 d.p.i. and 12 d.p.i. for RT-qPCR, IB, Immunofluorescence (IF), and ELISA analysis. Survival percent, body weight changes, and clinical score of mice (n = 10) were recorded daily for 21 d.
Statistical analysis
Data are presented as the mean ± standard error of the mean (SEM), and the difference between groups was evaluated by Student’s t-test, One-way ANOVA, or Two-way ANOVA test. The log-rank (Mantel–Cox) method was used for survival analysis. The statistical tests used for each figure are list in the corresponding figure legends. Each assay was performed by at least three independent experiments, and all data were performed using GraphPad Prism software (version 8.0, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentralproteomexchange.org) via the iProX partner repository with the dataset identifier PXD058432 for identification of BST2 ubiquitination sites and PXD058558 for quantitative proteomic of FBXL18. Source data are provided with this paper.
References
Dietzschold, B., Li, J., Faber, M. & Schnell, M. Concepts in the pathogenesis of rabies. Fut. Virol. 3, 481–490 (2008).
Feige, L. et al. Susceptibilities of CNS cells towards rabies virus infection is linked to cellular innate immune responses. Viruses 15, 88 (2022).
Potratz, M. et al. Astrocyte infection during rabies encephalitis depends on the virus strain and infection route as demonstrated by novel quantitative 3D analysis of cell tropism. Cells 9, 412 (2020).
Tian, B. et al. Lab-attenuated rabies virus causes abortive infection and induces cytokine expression in astrocytes by activating mitochondrial antiviral-signaling protein signaling pathway. Front. Immunol. 8, 2011 (2017).
Farina, C., Aloisi, F. & Meinl, E. Astrocytes are active players in cerebral innate immunity. Trends Immunol. 28, 138–145 (2007).
Soung, A. & Klein, R. S. Viral encephalitis and neurologic diseases: focus on astrocytes. Trends Mol. Med. 24, 950–962 (2018).
Luo, Z. et al. EV71 infection induces neurodegeneration via activating TLR7 signaling and IL-6 production. PLoS Pathog. 15, e1008142 (2019).
Popovic, D., Vucic, D. & Dikic, I. Ubiquitination in disease pathogenesis and treatment. Nat. Med. 20, 1242–1253 (2014).
Guo, Y., Cui, S., Chen, Y., Guo, S. & Chen, D. Ubiquitin specific peptidases and prostate cancer. PeerJ 11, e14799 (2023).
Bednash, J. S. & Mallampalli, R. K. Regulation of inflammasomes by ubiquitination. Cell Mol. Immunol. 13, 722–728 (2016).
Kravtsova-Ivantsiv, Y. & Ciechanover, A. Ubiquitination and degradation of proteins. Methods Mol. Biol. 753, 335–357 (2011).
Lee, N. R., Kim, H. I., Choi, M. S., Yi, C. M. & Inn, K. S. Regulation of MDA5-MAVS antiviral signaling axis by TRIM25 through TRAF6-mediated NF-kappaB activation. Mol. Cells 38, 759–764 (2015).
Qin, Y. et al. RNF26 temporally regulates virus-triggered type I interferon induction by two distinct mechanisms. PLoS Pathog. 10, e1004358 (2014).
Tu, Z. et al. Pentagalloylglucose inhibits the replication of rabies virus via mediation of the miR-455/SOCS3/STAT3/IL-6 pathway. J. Virol. 93, e00539-19 (2019).
Cardozo, T. & Pagano, M. The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 (2004).
Zheng, Y. et al. Mettl14 mediates the inflammatory response of macrophages in atherosclerosis through the NF-κB/IL-6 signaling pathway. Cell Mol. Life Sci. 79, 311 (2022).
Chai, X. et al. Hypoxia induces pulmonary arterial fibroblast proliferation, migration, differentiation and vascular remodeling via the PI3K/Akt/p70S6K signaling pathway. Int. J. Mol. Med. 41, 2461–2472 (2018).
Timmins, J. M. et al. Calcium/calmodulin-dependent protein kinase II links ER stress with Fas and mitochondrial apoptosis pathways. J. Clin. Invest. 119, 2925–2941 (2009).
Kupzig, S. et al. Bst-2/HM1.24 is a raft-associated apical membrane protein with an unusual topology. Traffic 4, 694–709 (2003).
Mahauad-Fernandez, W. D. & Okeoma, C. M. The role of BST-2/Tetherin in host protection and disease manifestation. Immun. Inflamm. Dis. 4, 4–23 (2016).
Sauter, D. et al. A rare missense variant abrogates the signaling activity of tetherin/BST-2 without affecting its effect on virus release. Retrovirology 10, 85 (2013).
Tokarev, A. et al. Stimulation of NF-κB activity by the HIV restriction factor BST2. J. Virol. 87, 2046–2057 (2013).
Liu, W., Cao, Y., Guan, Y. & Zheng, C. BST2 promotes cell proliferation, migration and induces NF-κB activation in gastric cancer. Biotechnol. Lett. 40, 1015–1027 (2018).
Qiu, J. et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature 533, 120–124 (2016).
Li, M. X. et al. Gene deficiency and pharmacological inhibition of caspase-1 confers resilience to chronic social defeat stress via regulating the stability of surface AMPARs. Mol. Psychiatry 23, 556–568 (2018).
Li, Y. et al. Inhibition of Cdk5 rejuvenates inhibitory circuits and restores experience-dependent plasticity in adult visual cortex. Neuropharmacology 128, 207–220 (2018).
Li, B. et al. FTO-dependent m(6)A modification of Plpp3 in circSCMH1-regulated vascular repair and functional recovery following stroke. Nat. Commun. 14, 489 (2023).
Deng, S. et al. Optogenetic stimulation of mPFC alleviates white matter injury-related cognitive decline after chronic ischemia through adaptive myelination. Adv. Sci. (Weinh.) 10, e2202976 (2023).
Jiang, S. et al. Generic diagramming platform (GDP): a comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 53, D1670–d1676 (2025).
Liu, S. Q., Xie, Y., Gao, X., Wang, Q. & Zhu, W. Y. Inflammatory response and MAPK and NF-kappaB pathway activation induced by natural street rabies virus infection in the brain tissues of dogs and humans. Virol. J. 17, 157 (2020).
Liu, S. Q., Gao, X., Xie, Y., Wang, Q. & Zhu, W. Y. Rabies viruses of different virulence regulates inflammatory responses both in vivo and in vitro via MAPK and NF-kappaB pathway. Mol. Immunol. 125, 70–82 (2020).
Yu, H. et al. Elevated FBXL18 promotes RPS15A ubiquitination and SMAD3 activation to drive HCC. Hepatol. Commun. 7, e00198 (2023).
Zou, C. et al. Mortality factor 4 like 1 protein mediates epithelial cell death in a mouse model of pneumonia. Sci. Transl. Med. 7, 311ra171 (2015).
Zhang, J. et al. The F-box protein FBXL18 promotes glioma progression by promoting K63-linked ubiquitination of Akt. FEBS Lett. 591, 145–154 (2017).
Perez-Caballero, D. et al. Tetherin inhibits HIV-1 release by directly tethering virions to cells. Cell 139, 499–511 (2009).
Olety, B., Peters, P., Wu, Y., Usami, Y. & Göttlinger, H. HIV-1 propagation is highly dependent on basal levels of the restriction factor BST2. Sci. Adv. 7, eabj7398 (2021).
Kong, N. et al. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy 16, 1737–1752 (2020).
Taylor, J. K. et al. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J. Virol. 89, 11820–11833 (2015).
Zheng, Y. et al. Zika virus elicits inflammation to evade antiviral response by cleaving cGAS via NS1-caspase-1 axis. EMBO J. 37, e99347 (2018).
Orlicky, S. et al. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat. Biotechnol. 28, 733–737 (2010).
Aghajan, M. et al. Chemical genetics screen for enhancers of rapamycin identifies a specific inhibitor of an SCF family E3 ubiquitin ligase. Nat. Biotechnol. 28, 738–742 (2010).
Ni, Z. et al. Influenza virus uses mGluR2 as an endocytic receptor to enter cells. Nat. Microbiol. 9, 1764–1777 (2024).
Roy, S., Mukherjee, S., Pawar, S. & Chowdhary, A. Evaluation of in vitro antiviral activity of datura metel linn. Against rabies virus. Pharmacogn. Res. 8, 265–269 (2016).
Ramakrishnan, M. A. Determination of 50% endpoint titer using a simple formula. World J. Virol. 5, 85–86 (2016).
Lai, N. et al. Systemic exosomal miR-193b-3p delivery attenuates neuroinflammation in early brain injury after subarachnoid hemorrhage in mice. J. Neuroinflammation 17, 74 (2020).
Duran, C. L. et al. Cooperative NF-κB and Notch1 signaling promotes macrophage-mediated MenaINV expression in breast cancer. Breast Cancer Res. 25, 37 (2023).
Acknowledgements
This work was supported by grants from National Key Research and Development Program of China (No. 2022YFD1800100), and the National Natural Science Foundation of China (Nos. 32273093, 31972720, 31902307). We thank Dr. Zhao-Qing Luo for helpful discussion and for critical review of the manuscript. Mass spectrometric analysis was performed in the core facility of the First Hospital of Jilin University and Jingjie PTM BioLab.
Author information
Authors and Affiliations
Contributions
J.W., J.G., N.L., Z.T., S.-Y.L., J.M., X.Z., Z.H., Y.F., S.-N.L. and G.T.conducted the experiments and analyzed the data; J.W., J.G., J.F., C.T. and Y.L. designed the experiments. J.W., Z.T., H.C., Y.Z., J.F. and Y.L. wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
All authors contributed to the article and approved the submitted version. The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, J., Guo, J., Li, N. et al. The E3 ubiquitin ligase FBXL18 stabilizes BST2 to promote inflammation in RABV-infected astrocytes. Nat Commun 16, 11324 (2025). https://doi.org/10.1038/s41467-025-66427-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66427-y