Abstract
Endometriosis involves ectopic growth of endometrial-like tissue, yet the spatial transcriptomic and metabolic landscape of ovarian endometriomas remains poorly understood. This investigation presents a comprehensive multi-omics analysis of ovarian endometriomas incorporating single-cell RNA sequencing in conjunction with Digital Spatial Profiler-Whole Transcriptome Atlas for spatial transcriptomics, and non-targeted Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging for spatially resolved metabolomics. Our integrated analysis confirms the importance of cell adhesion, ECM-receptor interaction, and focal adhesion pathways in disease context. We identify XBP1, VCAN, and CLDN7 as key markers in epithelial cells, and THBS1 in perivascular cells. Spatially resolved metabolomics further reveals altered activity of cytochrome P450 enzymes, lipoprotein particles, and cholesterol metabolism in mesenchymal regions, along with several undefined metabolites enriched in epithelial areas of endometriomas compared to ovarian cortex controls. These findings reveal potential markers and metabolic pathways linked to ovarian endometriomas, offering a foundation for future diagnostic and therapeutic strategies.
Similar content being viewed by others
Introduction
Endometriosis, an estrogen-responsive benign gynecological disorder, is characterized by the ectopic growth of endometrial-like tissue beyond the confines of the uterus, leading to persistent pain, dysmenorrhea, and reproductive challenges1,2. This condition is categorized into three subtypes based on histopathological and anatomical criteria: ovarian endometriotic cysts (referred to as endometriomas or colloquially as chocolate cysts), superficial peritoneal endometriosis, and deep infiltrating endometriosis3,4. It predominantly affects women in their reproductive years, with an estimated incidence of approximately 10%5. Symptoms such as dysmenorrhea are present in 40–60% of cases, while infertility impacts 20–30% of individuals diagnosed with endometriosis6. Ovarian endometriotic cysts necessitate dedicated molecular investigation owing to its distinctive microenvironment, heightened hormonal sensitivity, and consequential clinical relevance7. In contrast to superficial or deep infiltrating endometriotic lesions, ovarian endometriotic cysts demonstrate dysregulated expression of immune-inflammatory mediators and estrogen-dependent signaling pathways7. Ovarian endometriotic cysts are further recognized as the well-established direct precursors of clear-cell and endometrioid ovarian carcinomas, supported by histopathological and molecular evidence8. Such molecular heterogeneity underscores the imperative for subtype-specific mechanistic exploration for endometriosis to advance precision therapeutics and optimize prognostic trajectories.
In a previous study, Fonseca et al. utilized single-cell RNA sequencing (scRNA-seq) to develop a cellular atlas of endometriosis, uncovering the molecular profiles of endometrial-type epithelial and stromal cells characterized by disrupted pro-inflammatory pathways and increased expression of complement proteins9. More recently, Marečková et al. launched the Human Endometrial Cell Atlas, a comprehensive single-cell reference repository assembling data from 63 women with and without endometriosis, integrating published and novel endometrial single-cell transcriptomics10. These investigations offer detailed and thorough transcriptional expression patterns via high-throughput scRNA-seq, laying a strong groundwork for subsequent in-depth studies on endometriosis. Nonetheless, our current understanding of the development and causes of ovarian endometriomas remains limited, despite extensive research efforts spanning several decades. Several crucial questions remain unanswered. There exists a notable research gap concerning the transcriptional features of specific regions or cell types within ovarian endometriomas, and the domain of spatial single-cell omics beyond transcriptomics remains only partially elucidated.
In our study, we focused primarily on ovarian endometriotic cysts, a prominent subtype of endometriosis. We constructed a multi-omics landscape of ovarian endometriomas by combining scRNA-seq, spatial transcriptomics utilizing Digital Spatial Profiler-Whole Transcriptome Atlas (DSP-WTA), and spatial-resolved metabolomics based on non-targeted Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging (MALDI-MSI) at a precision of 50 μm. Through this comprehensive methodology, we unveiled numerous distinctive markers featuring innovative core targets or metabolites that may potentially influence human ovarian endometriomas. These findings from the analysis of human ovarian endometriomas suggest potential avenues for identifying transcriptomic and metabolic signatures that may serve as candidate targets for clinical diagnosis, aligning with the need to address the challenges of this subtype, where early detection remains elusive despite severe clinical consequences.
Results
Single-cell transcriptomics of ovarian endometriomas
Initially, we generated a single-cell atlas of ovarian endometriomas, encompassing three endometriomas and three unaffected normal ovarian cortices (Supplementary Fig. 1). Patients with laparoscopically confirmed endometriosis (endometriomas group) and control group were recruited between 20 and 40 years old. All patients in the endometriomas group were classified as Stage 4 for endometriosis. Postmenopausal individuals were excluded from both groups. After applying stringent filtering criteria, 41,434 high-quality cells were retained for subsequent analysis, demonstrating an average of approximately 4722 unique molecular identifiers (UMIs) per cell and identifying roughly 2225 genes per cell.
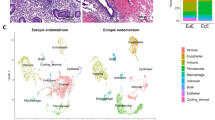
Cellular heterogeneity was explored through uniform manifold approximation and projection (UMAP), and cells were categorized based on the similarity of their distinctive gene expression profiles (Fig. 1A, B). Utilizing the expression patterns of marker genes, violin plot visualizations revealed the unsupervised categorization of individual cell transcriptomes into six distinct primary cellular clusters, encompassing mesenchymal cells, perivascular cells, epithelial cells, endothelial cells, myeloid cells, and T/NK cells (Fig. 1C). It was worth mentioning that perivascular markers include RGS5, NOTCH3, and PDGFRB, while epithelial cells are characterized by KRT8, KRT18, and KRT19. Upon examination of the UMAP plot, it is evident that epithelial cells and mesenchymal cells do not overlap, leading us to infer that epithelial cell populations consist purely of epithelial cells. In the previous study for endometriosis9, epithelial cells were also found to express mesenchymal markers, aligning with our findings. Meanwhile, the scarcity of oocytes and their low abundance, likely below the detection limit of scRNA-seq, make their capture inherently challenging. The top 5 most abundantly expressed genes in each cellular population were depicted using a heatmap display to identify potential markers for ovarian cortex (Supplementary Fig. 2A and Supplementary Data 1). Furthermore, UMAP visualizations were generated for each cell cluster in ovarian cortex with and without endometriosis (Fig. 1D and Supplementary Fig. 2B).
A UMAP visualization illustrating human ovarian cell populations. B UMAP visualization of ovarian cell populations, color-coded for the control (control; n = 3) and endometriosis (EM; n = 3) groups. C Visualization showcasing marker genes within each subcluster. D UMAP visualizations specifically focusing on perivascular cells and epithelial cells. E Fraction of cells within each subcluster. F Comparative cellular ratios of each subcluster between the control and EM groups. Control (Ctrl); Endometriosis (EM); Uniform manifold approximation and projection (UMAP).
Among these prominent cell populations, the cellular composition analysis concerning both the absolute count and proportion of cells captured in our scRNA-seq revealed a notable decrease in mesenchymal cells, endothelial cells, and myeloid cells, whereas perivascular cells, epithelial cells, and T/NK cells demonstrated escalating tendencies in the ovarian endometriomas group compared to the control ovarian group (Fig. 1E and Supplementary Fig. 2C, D). Particularly noteworthy was the substantial rise in the proportion of perivascular cell populations from 3.45% (control group) to 22.59% (ovarian endometriomas group). Although the capture of epithelial cells was relatively low, their proportions across samples remained roughly even (Supplementary Fig. 2D). Furthermore, a noticeable increase in epithelial cell proportion was observed, rising from 0.14% in controls to 0.63% in ovarian endometrioma cases.
We also obtained previously published datasets on endometriosis9, comprising high-quality one normal ovarian sample and three endometrioma samples. Notably, cell populations with higher cellular abundance in single-cell sequencing samples, such as mesenchymal and endothelial cells, demonstrated strong consistency across studies. Conversely, low-abundance populations may exhibit divergent proportional trends due to inter-individual heterogeneity and technical biases in cell capture efficiency. Our integrated analysis may overcome these limitations by enabling the identification of candidate biomarkers even in sparsely represented cell populations.
Transcriptional regulation in ovarian endometriomas
In our investigation of transcriptional regulation in ovarian endometriomas at the single-cell level, we conducted an analysis of differentially expressed genes (DEGs) across various cell populations in both the control and ovarian endometriomas groups (Supplementary Data 2). This analysis unveiled a number of significantly enriched DEGs (Fold change > 2, P < 0.05), which were visualized through volcano plots and heatmaps (Fig. 2A, B and Supplementary Fig. 3A). Notably, the epithelial cell population exhibited a notably higher number of DEGs compared to other cell types.
A Comparison of the quantity of upregulated and downregulated DEGs within each subcluster between the control and EM groups. B Representation of the fold changes of upregulated and downregulated DEGs within each subcluster between the control and EM groups. DEGs with expression level of zero in either the control or EM group were excluded. C KEGG pathway enrichment analysis of DEGs in epithelial cells. D Violin plot illustrating the expression of C3 in epithelial cells from the control and EM groups (The number of Cells: n = 30 in control group; n = 123 for EM group). E Co-immunofluorescence staining of C3 and PanCK in human ovarian cortex from the control and EM groups. White arrows label lesional epithelial regions. F Relative fluorescence intensity of C3 (n = 3 samples per group). G KEGG pathway enrichment analysis of DEGs in perivascular cells. H Violin plot displaying the expression of CD44 in perivascular cells from the control and EM groups (The number of Cells: n = 760 in control group; n = 4383 for EM group). I Co-immunofluorescence staining of CD44 and α-SMA in human ovarian cortex from the control and EM groups. J Relative fluorescence intensity of CD44 (n = 3 samples per group). DNA was stained with DAPI. Data are presented as mean ± SD. Student’s t-test was applied to two-group analyses (two-sided). The statistical significance in KEGG analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Scale bar: 2000 μm. For (D and H), the plot displays a combination of a violin plot and a box plot, with gene expression levels on the y-axis. Violin Plot: The outline represents the kernel density distribution of the data, with its upper and lower extremes indicating the data’s maximum and minimum values. Internal Box Plot: The box depicts the interquartile range (IQR); The upper hinge represents the 75th percentile (third quartile); The central line indicates the median (50th percentile); The lower hinge represents the 25th percentile (first quartile). Control (Ctrl); Endometriosis (EM); Differentially expressed genes (DEGs); Kyoto Encyclopedia of Genes and Genomes (KEGG); Standard deviation (SD); Single-cell RNA sequencing (scRNA-seq). Source data are provided as a Source Data file.
Upon applying a more stringent screening criterion (Fold change > 2, FDR < 0.05), a substantial reduction in the number of DEGs within the epithelial cell population was observed, while DEGs in other cell subsets did not exhibit significant changes (Supplementary Fig. 3B, C). This discrepancy could be attributed to the relatively smaller size of the epithelial cell population. Given our research’s primary objective of identifying potential targets within minor cell populations through comprehensive single-cell multi-omics analysis, we chose a relatively lenient threshold (Fold change > 2, P < 0.05) in our scRNA-seq analysis to facilitate seamless integration with other omics datasets. Subsequent analysis primarily focused on examining the impact of perivascular and epithelial cell populations on the pathogenesis of ovarian endometriomas.
The epithelial cells, despite being the least abundant in the ovarian cortex regardless of ovarian endometriomas status, displayed the highest number of DEGs between the two groups. Upon closer examination of the epithelial cells, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis revealed a series of intricate events related to protein synthesis and degradation. These events encompassed functions such as ribosome, spliceosome function, protein processing within the endoplasmic reticulum (ER), and ubiquitin-mediated proteolysis (Fig. 2C). Additionally, Gene Ontology (GO) analysis highlighted a significant enrichment of processes like cellular component organization or biogenesis, protein targeting to ER, protein binding, and RNA binding (Supplementary Fig. 3D). A previous study suggested that endometrial-type epithelium exhibited increased activation of complement signaling within the context of endometriomas, with the expression of complement proteins, such as C3, indicating dysregulated innate immunity9. We utilized C3 as a validated marker for the epithelial cells of ovarian endometriomas. Within the DEGs in epithelial cells, our investigation pinpointed a notable upregulation of C3 [Log2(FC): 2.39 and P-value: 5.68 × 10−06] in ovarian endometriomas in comparison to control group (Fig. 2D). This finding was further supported by immunofluorescence staining of C3 in corresponding ovarian cortex, validating the consistent expression patterns within epithelial cells (Fig. 2E, F).
Furthermore, the analyses of KEGG and GO illuminated a pronounced enrichment of pathways and biological processes associated with the extracellular matrix, focal adhesion, and ECM-receptor interaction within the DEGs of perivascular cells (Fig. 2G and Supplementary Fig. 3E). Of particular interest was the identification and verification of CD44, a pivotal cell-surface glycoprotein crucial for cell-cell interactions and adhesion11. CD44 exhibited a significant upsurge in the perivascular cell population of ovarian endometriomas group when contrasted with control group (Fig. 2H–J). Taken together, our scRNA-seq analysis underscores the critical involvement of transcriptional regulation in both epithelial cells and perivascular cells within the context of ovarian endometriomas.
Spatial transcriptomic analysis of epithelial cell cluster
In order to delve deeper into the transcriptional regulatory network of specific cellular populations from a spatial vantage point, we performed a spatial transcriptomic analysis based on DSP-WTA in ovarian cortex with and without endometriosis (Fig. 3A). Initially, haematoxylin and eosin (H&E) staining revealed the architecture of ovarian cortex in control and ovarian endometriomas groups (Fig. 3B). Epithelial cell populations were distinguished with PanCK labeling, and regions of interest (ROI) containing epithelial cells were selected within the ovarian cortex with and without endometriosis (Fig. 3C). Quality control of the selected ROIs for epithelial cells was assessed through Correlation plot view, leading to the identification of 496 DEGs (Supplementary Data 3), comprising 467 upregulated and 29 downregulated DEGs in the selected epithelial cells (Fig. 3D and Supplementary Fig. 4A). The Volcano plot illustrated key DEGs exhibiting significant differences in fold change and P-value, encompassing PAEP, MMP7, and MUC5B (Fig. 3E). Consistent with scRNA-seq analysis, GO enrichment also highlighted processes such as cellular component organization or biogenesis, protein targeting to ER, protein binding, and RNA binding (Supplementary Fig. 4B–D). Additionally, KEGG analysis unveiled enrichment in processes related to protein synthesis and degradation, including ribosome and spliceosome pathways, ubiquitin-mediated proteolysis, oxidative phosphorylation, and reactive oxygen species (Fig. 3F). Immunofluorescence staining was employed to validate representative DEGs, revealing a significant increase in Claudin 3 (CLDN3), Claudin 4 (CLDN4), and Heat shock protein family A (Hsp70) member 8 (HSPA8) expression levels in ovarian endometriomas compared to control group (Fig. 3G–J and Supplementary Fig. 4E, F). In summary, our study successfully established a DSP-based spatial transcriptomics approach, demonstrating a high degree of enrichment similarity between DSP-WTA-based spatial transcriptomic and scRNA-seq analyses within epithelial cell populations.
A Concise illustration of the DSP-WTA methodology (control group, n = 3; EM group, n = 3). B H&E staining of human ovarian cortex in the control and EM groups. C PanCK staining in human ovarian cortex of control and EM groups, with ROIs selected for the epithelial cell region. D Quantification of DEGs for the selected ROIs of epithelial cells between control and EM groups. E Volcano plot of DEGs for the selected ROIs of epithelial cells between control and EM groups, highlighting representative DEGs. F KEGG pathway enrichment analysis of DEGs for the selected ROIs of epithelial cells between control and EM groups. Yellow line represents for the percent of gene raito and column represents for −log10 (P-value). G Co-immunofluorescence staining of CLDN3 and PanCK in human ovarian cortex of control and EM groups. H Co-immunofluorescence staining of CLDN4 and PanCK in human ovarian cortex of control and EM groups. I Relative fluorescence intensity of CLDN3 (n = 3 samples per group). J Relative fluorescence intensity of CLDN4 (n = 3 samples per group). DNA was stained with DAPI. White arrows label lesional epithelial regions. Data are presented as mean ± SD. Student’s t-test was applied to two-group analyses (two-sided). Statistical significance for DEGs was determined by exact test. The statistical significance in KEGG analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Scale bar: 100 μm for (B and C); 2000 μm for (G and H). Control (Ctrl); Endometriosis (EM); Differentially expressed genes (DEGs); Kyoto Encyclopedia of Genes and Genomes (KEGG); Standard deviation (SD); Regions of interest (ROI); Digital Spatial Profiler-Whole Transcriptome Atlas (DSP-WTA); Haematoxylin and eosin (H&E). Source data are provided as a Source Data file.
We subsequently utilized an intersection approach that integrates DEGs obtained from both DSP-WTA and scRNA-seq methodologies, as identified within epithelial cells from ovarian endometriomas and control cohorts, unveiling a total of 206 shared DEGs, comprising 181 commonly upregulated DEGs and 7 commonly downregulated DEGs (Fig. 4A–C). Subsequently, detailed enrichment analyses were conducted for all common DEGs, upregulated common DEGs, and downregulated common DEGs. Notably, KEGG analysis highlighted ribosome pathways as the most enriched among the all common DEGs (Fig. 4D). Additionally, KEGG and GO analyses underscored the enrichment of cell adhesion molecules within the all common DEGs identified in epithelial cells through integrated analysis of DSP-WTA and scRNA-seq methodologies (Fig. 4D and Supplementary Fig. 5A). Further enrichment analyses of upregulated and downregulated common DEGs in epithelial cells unveiled a significant enrichment of cell adhesion molecules and associated processes, indicating fundamental alterations present in both upregulated and downregulated common DEGs (Supplementary Fig. 5B–E). Noteworthy core DEGs, including X-box binding protein 1 (XBP1), Versican (VCAN), and Claudin 7 (CLDN7), exhibited substantial increases in expression within epithelial cells of ovarian endometriomas (Fig. 4E–G). Immunofluorescence staining corroborated these findings, demonstrating consistent expression patterns in epithelial cells that aligned with the data identified through DSP-WTA and scRNA-seq analyses (Fig. 4H–M). These findings indicate that the combined analysis of both methods can effectively filter out DEGs with inconsistent expression patterns. Meanwhile, it is observed that upregulated common DEGs play a significant role for ovarian endometriomas.
A Venn diagram illustrating all common DEGs identified in both DSP-WTA and scRNA-seq analyses. B Venn diagram showing upregulated common DEGs identified in both DSP-WTA and scRNA-seq analyses. C Venn diagram displaying downregulated common DEGs identified in both DSP-WTA and scRNA-seq analyses. D KEGG pathway enrichment analysis of all common DEGs. E Violin plot visualization of XBP1 expression in epithelial cells of control and EM groups (The number of Cells: n = 30 in control group; n = 123 for EM group). F Violin plot visualization of VCAN expression in epithelial cells of control and EM groups (The number of Cells: n = 30 in control group; n = 123 for EM group). G Violin plot visualization of CLDN7 expression in epithelial cells of control and EM groups (The number of Cells: n = 30 in control group; n = 123 for EM group). H Co-immunofluorescence staining of XBP1 and PanCK in human ovarian cortex of control and EM groups. I Co-immunofluorescence staining of VCAN and PanCK in human ovarian cortex of control and EM groups. J Co-immunofluorescence staining of CLDN7 and PanCK in human ovarian cortex of control and EM groups. K Relative fluorescence intensity of XBP1 (n = 3 samples per group). L Relative fluorescence intensity of VCAN (n = 3 samples per group). M Relative fluorescence intensity of CLDN7 (n = 3 samples per group). DNA was stained with DAPI. White arrows label lesional epithelial regions. Data are presented as mean ± SD. Student’s t-test was applied to two-group analyses (two-sided). The statistical significance in KEGG analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Scale bar: 2000 μm. Control (Ctrl); Endometriosis (EM); Differentially expressed genes (DEGs); Kyoto Encyclopedia of Genes and Genomes (KEGG); Standard deviation (SD); Digital Spatial Profiler-Whole Transcriptome Atlas (DSP-WTA); Single-cell RNA sequencing (scRNA-seq). Source data are provided as a Source Data file.
Spatial transcriptomic analysis of perivascular cell population
A previous study has indicated that perivascular cells can influence disease progression through various mechanisms12,13. Subsequently, we conducted a detailed analysis to explore the role of perivascular cell populations in ovarian endometriomas by DSP-WTA. Perivascular cell populations were identified using α-SMA labeling, followed by the ROI selection within ovarian cortex with and without endometriosis (Fig. 5A). Correlation plot view indicated the quality control of ROI selection, and identified 59 upregulated and 3 downregulated DEGs in perivascular cell populations (Fig. 5B–D and Supplementary Data 4). GO enrichment of cellular component, biological process, and molecular function revealed the involvement of focal adhesion-related processes such as extracellular matrix, extracellular structure organization, and extracellular matrix structural constituent, in the transcriptional regulatory network mediated by perivascular cells (Fig. 5E–G). The integrated analysis of DSP-WTA and scRNA-seq data in perivascular cells unveiled 12 common DEGs between ovarian endometriomas and control ovaries (Fig. 5H). Additionally, KEGG enrichment analysis highlighted the participation of these DEGs in pathways including ECM-receptor interaction and focal adhesion (Fig. 5I). Notably, our investigation further revealed an upregulation of Thrombospondin 1 (THBS1) in both the quantity and expression levels of perivascular cells in ovarian endometriomas compared to control ovaries (Fig. 5J–M). In conclusion, the integrated analysis of DSP-WTA and scRNA-seq data in ovarian endometriomas has identified THBS1 as a key factor within perivascular cells.
A Immunostaining of α-SMA in human ovarian cortex from control and EM groups, with ROIs selected for the perivascular cell region. B Correlation plot depicting selected ROIs of perivascular cells in human ovarian samples. C Number of DEGs identified in selected ROIs of perivascular cells between control and EM groups. D Volcano plot illustrating DEGs in selected ROIs of perivascular cells between control and EM groups, highlighting representative DEGs. E GO enrichment analysis of cellular components for DEGs in selected ROIs of perivascular cells between control and EM groups. F GO enrichment analysis of biological processes for DEGs in selected ROIs of perivascular cells between control and EM groups. G GO enrichment analysis of molecular functions for DEGs in selected ROIs of perivascular cells between control and EM groups. H Venn diagram showing all common DEGs identified in DSP-WTA and scRNA-seq analyses. I KEGG pathway enrichment analysis of all common DEGs identified in DSP-WTA and scRNA-seq. J UMAP visualization of THBS1 expression patterns in control and EM groups. K Violin plot visualization of THBS1 expression in perivascular cells of control and EM groups (The number of Cells: n = 760 in control group; n = 4383 for EM group). L Co-immunofluorescence staining of THBS1 and α-SMA in human ovarian cortex from control and EM groups. M Relative fluorescence intensity of THBS1 (n = 3 samples per group). DNA was stained with DAPI. Data are presented as mean ± SD. Student’s t-test was applied to two-group analyses (two-sided). Statistical significance for DEGs was determined by exact test. The statistical significance in the enrichment analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Scale bar: 100 μm for (A); 2000 μm for (L). Control (Ctrl); Endometriosis (EM); Differentially expressed genes (DEGs); Kyoto Encyclopedia of Genes and Genomes (KEGG); Gene Ontology (GO); Standard deviation (SD); Regions of interest (ROI); Digital Spatial Profiler-Whole Transcriptome Atlas (DSP-WTA); Single-cell RNA sequencing (scRNA-seq). Source data are provided as a Source Data file.
Spatially resolved metabolomics of ovarian endometriomas
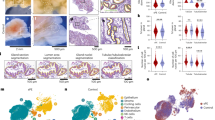
Previous investigation has presented evidence suggesting that numerous metabolites may play roles in the pathogenesis of ovarian endometriomas, potentially offering diagnostic, prognostic, and staging capabilities14,15. To delve deeper into this, our initial step involved conducting non-targeted metabolomics coupled with MALDI-MSI, which distinctly illustrated the spatial distribution of discriminative metabolites within ovarian cortex affected by ovarian endometriomas compared to unaffected ovarian cortex (Fig. 6A). MALDI-MSI emerges as a powerful tool for investigating metabolic heterogeneity in situ by directly visualizing the spatial arrangement of metabolites16,17 To visually represent the ovarian tissue samples from both control and ovarian endometriomas groups utilized in spatially resolved metabolomic analysis, we employed H&E staining, as depicted in Fig. 6B. Each pixel within the MSI of ovarian cortex carries its unique metabolic signature specific to its region, reflecting the structural and functional distinctions between tissues affected by ovarian endometriomas and control ovarian tissue18. Our investigation proceeded to assess the metabolic similarities across different pixels within ovarian cortex affected by ovarian endometriomas and control ovarian tissue using bisecting K-means clustering. Figure 6C showcased the resulting label maps generated from sections of ovarian cortex, revealing the segregation of image pixels into seven distinct clusters based on their metabolic profiles. Notably, the label maps vividly delineate epithelial cell regions highlighted in red (2676; Cluster 5) and mesenchymal cell regions in blue (5585; Cluster 4). Furthermore, we delved into the pathways enriched by differential metabolites within the overall area analyzed through spatially resolved metabolomics in ovarian cortex affected by ovarian endometriomas and control ovarian tissue. Our findings unveiled enriched pathways such as nicotinate and nicotinamide metabolism, along with metabolism of xenobiotics by cytochrome P450 (Fig. 6D). Additionally, we identified several key differential metabolites, notably nicotinic acid and thioacetic acid, exhibiting significant upregulation in ovarian endometriomas compared to control ovarian tissue (Fig. 6E, F and Supplementary Data 5).
A Schematic representation of MALDI-MSI procedure for human ovarian cortex with and without EM (control group, n = 2 spatial samples; EM group, n = 2 spatial samples). B H&E staining of human ovarian cortex for MALDI-MSI analysis. Blue arrows label lesional epithelial regions. The experiment was repeated independently a minimum of three times. C Application of K-means clustering to ovarian sections, delineating seven distinct clusters. D KEGG pathway enrichment analysis of differential metabolites across all regions of ovarian sections in control and EM groups. E Identification of differential metabolites across all regions of ovarian sections in control and EM groups. Normalize the metabolite expression levels using the Z-score method. F Visualization of metabolite ion distributions depicting nicotinuric acid (m/z 179.0428) and thiodiacetic acid (m/z 184.9694) in ovarian sections of control and EM cohorts. Scale bar: 500 μm. The statistical significance in the enrichment analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Control (Ctrl); Endometriosis (EM); Kyoto Encyclopedia of Genes and Genomes (KEGG); Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging (MALDI-MSI); Haematoxylin and eosin (H&E).
Utilizing MALDI-MSI, we observed alterations in the spatial distribution of cytochrome P450, the singularly enriched pathway within the mesenchymal cell region (Fig. 7A and Supplementary Data 6). Figure 7B illustrated substantial increases in Felbamate, 4-Hydroxycyclo-phosphamide, and Ifosfamide within the mesenchymal cell region of ovarian endometriomas compared to the control ovarian tissue. Subsequently, we examined the expression patterns of DEGs of associated pathways using scRNA-seq data, pinpointing an enrichment of cytochrome P450 in the mesenchymal cell population. Heatmap and violin plot visualizations unveiled DEG encoding specific enzymes known to participate in cytochrome P450. Notably, we observed a significant upregulation of GSTM5, which encoded a glutathione S-transferase, in mesenchymal cells of ovarian endometriomas compared to control ovaries (Supplementary Fig. 6A). Importantly, GO and KEGG analyses of DEGs in scRNA-seq of mesenchymal cell populations revealed highly enriched events related to lipoprotein particles and cholesterol metabolism (Fig. 7C, D). Additionally, DEGs associated with lipid metabolism (e.g., GGT5 and PTGDS) exhibited increased expression in mesenchymal cells of ovarian endometriomas (Supplementary Fig. 6B). UMAP visualizations demonstrated that GSTM5 and PTGDS were highly enriched in mesenchymal cells, reinforcing the idea that these genes might play particularly roles within this cellular compartment. Moreover, GGT5 was also highly expressed among the top cell types, including mesenchymal cells (Supplementary Fig. 6C). Subsequently, we identified significant elevations of lipids and lipid-like molecules, including PE (42:8), PC (32:4), and polyporusterone C, in the mesenchymal cell region of ovarian endometriomas tissue relative to the control ovarian tissue (Fig. 7E). Sphingomyelin, a key player in sphingolipid metabolism, exhibited a marked increase in the mesenchymal cell region of the ovarian endometriomas group compared to the control group (Fig. 7F). Additionally, several metabolites in glycerophospholipid metabolism, such as phosphatidate, PE (36:4), and phosphatidylserine, displayed increases in the mesenchymal cell region of ovarian endometriomas tissue in contrast to the control ovarian tissue (Fig. 7G).
A KEGG pathway enrichment analysis and representation of differential metabolites in the mesenchymal cell region of ovarian sections within control and EM groups. Normalize the metabolite expression levels using the Z-score method. B Visualization of metabolite ion distributions for Felbamate (m/z 237.0877), 4-Hydroxycyclo-phosphamide (m/z 275.0135), and Ifosfamide (m/z 259.0185) in ovarian sections of control and EM groups. C GO enrichment analysis of DEGs in mesenchymal cell populations identified through scRNA-seq. Yellow line represents for the percent of gene raito and column represents for −log10 (P-value). D KEGG pathway enrichment analysis of DEGs in mesenchymal cell populations identified through scRNA-seq. E Visualization of metabolite ion distributions for PE 42:8 (m/z 796.5388), PC 32:4 (m/z 724.4959), and polyporusterone C (m/z 475.3099) in ovarian sections of control and EM groups. F Visualization of metabolite ion distributions for sphingomyelin (m/z 821.6228) in ovarian sections of control and EM groups. G Visualization of metabolite ion distributions for phosphatidate (m/z 671.4597), PE 36:4 (m/z 738.5015), and phosphatidylserine (m/z 846.4998) in ovarian sections of control and EM groups. The statistical significance in the enrichment analysis is calculated using the hypergeometric test. P < 0.05 is considered statistically significant. Control (Ctrl); Endometriosis (EM); Gene Ontology (GO); Kyoto Encyclopedia of Genes and Genomes (KEGG); Matrix-Assisted Laser Desorption/Ionization-Mass Spectrometry Imaging (MALDI-MSI); Phosphatidylethanolamine (PE); Phosphatidylcholines (PC).
Due to the limited number of epithelial cells located at the periphery of the ovarian cortex, the differential metabolites identified using a spatially resolved metabolomic approach did not show a statistically enriched pathway. This lack of enrichment could be attributed to the unclear specific functions of certain newly identified differential metabolites. However, during our analysis, we fortuitously discovered several undefined differential metabolites that displayed significant fold changes between the control and ovarian endometriomas groups within the epithelial cell region (Supplementary Data 7). Noteworthy among these differential metabolites were compounds with m/z values of 150.8838 and 429.9879, demonstrating significant downregulation, while others, including those with m/z values of 214.8328, 342.0397, 835.525, and 862.5466, exhibited elevations in the epithelial cell region of ovarian endometriomas tissue compared to the control ovarian tissue (Supplementary Fig. 7).
To exclude the potential confounding effects of technical variability and confirm that the observed metabolic alterations were disease-mediated, we expanded our sample size by adding one additional case of endometrioma and one unaffected ovarian tissue sample. The subsequent comprehensive analysis of the combined datasets (2 samples for the ovarian endometriomas group vs 2 samples for the control group) in our study consistently revealed identical directional shifts in these metabolites (Supplementary Fig. 8), further bolstering the ovarian endometriomas-related essence of these alterations. Our findings collectively unveil the potential regulatory roles of pivotal metabolites in ovarian endometriomas through the application of a MALDI-MSI-based spatially resolved metabolomic methodology.
Discussion
The overarching aim of this study is to elucidate the spatiotemporal dynamics of human ovarian endometriomas through a multi-dimensional analytical framework, with a specific focus on spatial transcriptomics and metabolomics. Our selection of ovarian endometrioma sample was strategically based on two key considerations: (1) As the most prevalent and surgically accessible form of infiltrating endometriosis, these lesions represent a critical clinical endpoint of disease progression; (2) Their well-demarcated cystic structure provides an optimal spatial reference that facilitates precise correlation between cellular transcriptomic profiles and regional metabolic microenvironments.
Prior studies have laid the groundwork with foundational single-cell transcriptomic atlases9,10, while our research specifically addresses knowledge gaps through methodological innovation. Investigating the utilization of multi-omics methodologies to dissect the pathogenic mechanisms of ovarian endometriomas presents a significant challenge. Within this research endeavor, we undertook a multi-omics appraisal of ovarian endometriomas employing scRNA-seq, augmented by DSP-WTA and MALDI-MSI methodologies (Supplementary Fig. 9). By integrating MALDI-MSI (50 μm precision) with DSP-WTA spatial transcriptomics and scRNA-seq, we establish the metabolite-transcript correlation map across distinct histological zones of ovarian endometriomas. Moreover, this research tackles the spatial confinement of distinct cellular populations (e.g., epithelial cells and perivascular cells) by utilizing DSP-WTA technology to conduct targeted investigations on spatial domains for transcriptomic data collection. By integrating a comprehensive analysis with specific spatial areas and single-cell transcriptomic data within related clusters, the study discerns more reliable potential biomarkers, presenting fresh opportunities for single-cell spatial co-transcriptomic inquiries encompassing minute cell populations. These strategies methodically revealed pathogenic markers associated with ovarian endometriomas through the lenses of spatial transcriptomics and spatial metabolomics.
Utilizing DSP-based spatial transcriptomic analysis within specific cellular locales unveiled intricate spatial gene expression patterns19,20. Through the implementation of DSP-WTA technology, we successfully captured and scrutinized the spatial heterogeneity and functional diversity among cell populations residing in distinct regions. While preliminary studies have established a significant association between cell adhesion and endometriosis21,22, the precise targets of these adhesion molecules within particular regions contributing to endometriosis in certain individuals remain enigmatic. Our investigation surprisingly identified the upregulation of cell adhesion molecules, including CLDN3, CLDN4, CLDN7, and VCAN, in the epithelial cell domains of ovarian endometriomas. Integrated analysis with scRNA-seq in epithelial cell populations further validated CLDN7 and VCAN as pivotal epithelial cell adhesion-related factors in ovarian endometriomas. Notably, we observed a consistent upregulation trend in these Claudin subunits within epithelial cell lineages of ovarian endometriomas tissues, implying that the Claudin protein family may represent a core target in ovarian endometriomas pathogenesis. Our methodologies also suggest that epithelial cell adhesion plays a critical role in the pathogenesis of ovarian endometriomas, offering valuable insights into key pathogenic factors through single-cell transcriptome and spatial transcriptomic data within specific epithelial cell regions. Notably, CLDN3, CLDN4, and CLDN7, encoding members of the Claudin family, represent integral membrane proteins forming tight junction strands crucial for maintaining cell polarity and signal transductions23,24,25. Prior research offered clues into aberrantly expressed hub Claudins in both infertile individuals and patients with endometriosis26. Additionally, VCAN, encoding a member of the aggrecan/versican proteoglycan family, serves as a key constituent of the extracellular matrix, participating in cell adhesion, migration, and proliferation27,28. Studies have indicated significantly elevated VCAN levels in endometriotic peritoneum, with its expression potentially modulated by TGF-β1 in HMrSV5 cells29. Our investigation further underscores the central roles of VCAN, showcasing substantial increases in ovarian endometriotic cysts. Moreover, the upregulations of PAEP, MMP7, and MUC5B within the epithelial cells of ovarian endometriomas, as unveiled by ROI-specific transcriptomic profiling utilizing DSP-WTA, corresponds with the implantation theory of ovarian endometriomas. Initial research findings indicated that these genes might be expressed and functioned in endometrial or cervical tissues30,31,32. This molecular signature implied either the ectopic implantation of endometrial/cervical-derived epithelial cells or the adoption of endometrial/cervical-like characteristics by the indigenous ovarian epithelia, thereby furnishing transcriptomic evidence that bolstered the implantation hypothesis at a single-cell resolution.
Within perivascular cells, our analysis revealed 12 common DEGs intricately linked to ECM-receptor interaction and focal adhesion pathways. Notably, THBS1 emerged as a pivotal factor exhibiting a significant increase in ovarian endometriomas, with consistent expression profiles observed in both DSP-WTA and scRNA-seq analyses. THBS1, a secretory adhesive glycoprotein, plays a crucial role in mediating cell-to-cell and cell-to-matrix interactions33,34. While prior research has elucidated THBS1’s involvement in vascular inflammation by modulating the migration and adhesion of mononuclear cells in mouse models of vascular-related disorders35, its specific roles in ovarian endometriomas remain uncharted. We hypothesize, based on our integrated profiling, that THBS1 may also play a contributory role in the pathogenesis of ovarian endometriomas. By amalgamating these cutting-edge technological methodologies and groundbreaking findings, our study not only enriches our comprehension of the molecular intricacies within epithelial and perivascular cells but also provides a perspective on the spatial orchestration of transcriptional regulation within distinct cellular microenvironments.
Additionally, studies revealed that ovarian endometriomas exerted adverse effects on the contiguous normal ovarian cortex36,37. The impact of endometriomas on the ovarian cortex was a progressive, multifactorial process that typically did not result in complete destruction of the ovarian cortex, involving mechanisms such as local invasion, chronic inflammation, oxidative stress, and fibrosis, leading to a gradual decline in ovarian function38. Spatially resolved metabolomics offers a precise method for mapping metabolic changes within specific regions of ovarian cortex affected by endometriosis. This sophisticated approach facilitates a detailed exploration of metabolic profiles in distinct spatial areas39,40, offering valuable insights into the metabolic alterations characteristic of ovarian endometriomas. While our spatially resolved metabolomic analysis has successfully identified numerous previously unrecognized metabolic alterations between ovarian cortex with and without endometriosis, we acknowledge that many of these metabolites currently lack functional annotations in existing databases. Conventional KEGG pathway analysis demonstrated limited capacity to enrich metabolism-associated pathways, likely due to the spatial complexity of ovarian endometriomas. To overcome this limitation, our study employed the integrated analysis of scRNA-seq and MALDI-MSI, which has shed light on the significance of cytochrome P450, lipoprotein particles, and cholesterol metabolism in mesenchymal cells when comparing ovarian cortex affected by endometriosis with those from control subjects. GSTM5 has been demonstrated to be involved in the regulation of glutathione S-transferase, playing crucial roles in detoxifying electrophilic compounds via glutathione conjugation41. Integrated analysis incorporating MALDI-MSI finding collectively suggests that cytochrome P450 plays pivotal pathogenic roles in ovarian endometriomas. Moreover, mesenchymal cellular populations exhibited pronounced enrichment in cholesterol metabolism and lipoprotein particle-related pathways, providing a biologically grounded framework for spatial metabolomic validation. Our scRNA-seq data further elucidated that lipid metabolism-associated genes GGT5 and PTGDS exhibited significantly heightened expression in mesenchymal cells of ovarian endometriomas. GGT5, responsible for encoding an enzyme that facilitated the conversion of leukotriene C4 to leukotriene D4, was identified as a pivotal regulator in the dysregulation of lipid metabolism within mesenchymal cell populations42,43. PTGDS acted as a glutathione-independent prostaglandin D synthase, catalyzing the conversion of prostaglandin H2 (PGH2) to prostaglandin D2 (PGD2), playing dual roles in the regulation of lipid transport and prostaglandin metabolic pathways, while also maintaining crucial pathophysiological functions in lipid metabolism-related human disorders44,45. Guided by these transcriptomic signatures, we systematically mapped metabolic perturbations within mesenchymal cell-enriched regions through spatial metabolomics, such as PE, PC, polyporusterone C, sphingomyelin, phosphatidate, phosphatidylethanolamine, and phosphatidylserine. These spatially resolved metabolic alterations, particularly the coordinated dysregulation of Lipids and lipid-like molecules, Sphingolipid and glycerophospholipid metabolism, suggest potential mechanistic links to ovarian endometriomas pathogenesis. By merging spatial information with metabolomic data and scRNA-seq results, a comprehensive understanding of the metabolic and molecular dysregulations underlying ovarian endometriomas can be achieved.
The field of biomarker research for endometriosis is rapidly advancing, yet there remains a need to further emphasize prediction targets. Given the diverse subtypes of endometriosis, which are likely to necessitate tailored approaches to both treatment and diagnosis, ongoing large-scale profiling endeavors are clearly justified. The multitude of newly discovered metabolites through spatial metabolomics demands additional annotation and validation to ascertain their roles in modulating the pathogenicity of endometriosis. We also recognize that ribosome, which fundamentally regulates protein synthesis, is likely to exert pivotal roles in the molecular mechanisms underlying this disorder in epithelial cells. However, it is still necessary to systematically study which downstream proteins are specifically affected by ribosomal changes to elucidate their precise regulatory dynamics in epithelial cell populations of ovarian endometriomas. Meanwhile, it must be acknowledged that the current analysis did not model patient-level variability, and a pseudobulk approach would be more suitable for future studies.
In summary, our study collectively presents a comprehensive multi-omics approach to understanding ovarian endometriomas, incorporating scRNA-seq in conjunction with DSP-WTA and MALDI-MSI. This integrated methodology serves as a valuable resource for pinpointing early clinical targets of ovarian endometriomas derived from spatial transcriptomics and spatially resolved metabolomics.
Methods
Subjects for scRNA-seq, DSP-WTA, and MALDI-MSI
Fresh ovarian specimens obtained from individuals with and without ovarian endometriomas were collected from Xuzhou Central Hospital. Our study complies with all relevant ethical regulations. This study was approved by Reproductive Medicine Ethics Committee of Xuzhou Central Hospital (XZZXYY-SZZX-20230106-002). Written informed consent was obtained from each participant for the use of tissue samples for multi-omic analysis, and reporting identifying clinical information, in this case, hospital, diagnosis, age, sex, ethnicity, and number of pregnancies. No monetary compensation was provided to the participants, whose voluntary involvement contributed to the advancement of knowledge with the potential to inform future diagnostic and therapeutic strategies for ovarian endometriomas. Patients with endometriosis were required to have histopathologically confirmed ovarian endometriomas through laparoscopic surgery. Controls were chosen from women undergoing laparoscopic procedures for non-endometriosis conditions, such as benign ovarian cysts, devoid of visual or histological signs of endometriosis. Exclusions comprised individuals with concurrent pelvic pathologies (e.g., adenomyosis, uterine fibroids), recent hormonal therapy, malignancies, autoimmune disorders, postmenopausal status, or pregnancy at recruitment. Menstrual cycle stage was not used as a specific inclusion or exclusion criterion for participant selection in this study. In line with standard clinical practice for non-urgent gynecological surgery, procedures were scheduled to avoid the menstrual period. Comprehensive participant characteristics for the multi-omics investigation were delineated in Supplementary Data 8. Samples were taken from tissues of approximately 100 mg, encompassing the cystic wall and cavity of endometrioma and a modest portion of neighboring ovarian tissue, which originated from the ovarian cortical compartment. Each ovarian tissue sample from the same patient was divided into three sections, earmarked for scRNA-seq, DSP-WTA, and MALDI-MSI. Following rigorous quality assessments, 6 samples were designated for scRNA-seq analysis, 6 samples were allocated for DSP-WTA analysis, and 4 samples were employed for MALDI-MSI analysis.
H&E staining
The ovarian samples were fixed in formalin, dehydrated using a gradient of ethanol concentrations (70%, 80%, 90%, and 100%), and subsequently embedded in paraffin. Subsequently, the tissues were sectioned into slices measuring 5 μm in thickness, deparaffinized in xylene for 20 min, rehydrated in a series of ethanol solutions (100, 90, 80, and 70%), and rinsed with distilled water for 15 min. Following these procedures, the sections were stained with hematoxylin for 5 min and eosin for 1 min. Full scan images of ovarian cortex were captured by using Nikon Eclipse Ti2. H&E staining image for MALDI-MSI assay was conducted using Leica Aperio CS2.
Immunofluorescence staining
Immunofluorescence staining was carried out on paraffin-embedded sections. The sections underwent deparaffinization, rehydration, and antigen retrieval procedures before being blocked with 2% bovine serum albumin (w/v) and exposed to the specified primary antibodies or IgG (Supplementary Data 9 and Supplementary Fig. 10) overnight at 4 °C. After three washes with phosphate-buffered saline (PBS), each lasting 15 min, the slides were then treated with corresponding Alexa Fluor-conjugated secondary antibodies (Thermo Scientific, Waltham, MA, USA) for 2 h at room temperature. Following this, the samples were counterstained with DAPI and mounted using VECTASHIELD Mounting Medium. Imaging was performed using the Axioscan7 Multispectral Imaging System (Carl Zeiss).
Single-cell suspensions, library preparation, and sequencing
After the surgical procedure, freshly resected tissues were washed with Hanks’ Balanced Salt Solution (HBSS), then minced into smaller fragments on ice using collagenase I/IV in HBSS. The fragments were incubated for 30 min at 37 °C with manual agitation every 10 min. Subsequently, the digested tissues were filtered through a 40-μm nylon mesh to obtain a cell suspension, which was centrifuged at 500 × g for 5 min at 4 °C. Following removal of the supernatant, the pelleted cells were treated with red blood cell lysis buffer, washed with HBSS, and then resuspended in a buffer (0.04% BSA + PBS). The cell count and viability of the single-cell suspensions were assessed using Trypan Blue staining. scRNA-seq was performed using a Chromium instrument (10× Genomics, Pleasanton). The samples were processed with a Chromium Single Cell 3′ Library & Gel Bead Kit v3 (10× Genomics). Subsequently, the gene expression libraries were sequenced by Guangzhou OmicsMaster Biotechnology Co., Ltd, using a NovaSeq 6000 sequencer (Illumina).
scRNA-seq data processing
The raw BCL files were converted to FASTQ files, aligned, and the counts quantified utilizing Cell Ranger software (version 6.1.0; 10× Genomics). Reads with low-quality barcodes were excluded, and UMIs were filtered and aligned to the reference genome (Ensembl_release93). Reads uniquely mapping to the transcriptome and overlapping an exon by at least 50% were selected for UMI counting. Before quantification, UMI sequences underwent correction for sequencing errors, and the EmptyDrops method was applied for valid barcode identification46. Cell barcode assignment and UMI counting generated the cell-by-gene matrices. For subsequent analysis of each sample, the cell-by-gene matrices were individually imported into Seurat version 4.1.047. Cells with abnormally high UMI counts (≥ 40,000) or mitochondrial gene percentages (≥ 20%) were removed. Cells with fewer than 200 or more than 7200 detected genes were also excluded. Furthermore, doublet gel beads in emulsion (GEMs) were filtered out using DoubletFinder (v2.0.3)48. UMAP was employed for visualizing the cell clusters. Upregulated enriched genes were identified as significant using the default threshold of ln(fold change) > 0.25 and P-value < 0.01. DEGs were considered significant via Seurat 4.1.0 with a fold change greater than 2 and a P value below 0.05.
DSP-WTA assay
Methods for GeoMx RNA assays have been delineated in a prior investigation49. Formalin-fixed, paraffin-embedded tissue slides were affixed to positively charged histology slides. Sections were heated at 65 °C for 45 min in a Hyb EZ II hybridization oven (Advanced Cell Diagnostics). The slides were deparaffinized using Citrisolv (Decon Labs, 1601), rehydrated in an ethanol gradient, and rinsed in 1× PBS (pH 7.4). Subsequently, the slides were immersed for 15 min in 1× Tris-EDTA pH 9.0 buffer (Sigma Aldrich, SRE0063) at 100 °C under low pressure in a TintoRetriever Pressure Cooker (bioSB, 7008). After washing, the slides were treated with 1 µg/mL proteinase K (Thermo Fisher Scientific, AM2546) in PBS for 15 min at 37 °C, followed by another PBS wash. The tissues were then fixed in 10% neutral-buffered formalin (Thermo Fisher Scientific, 15740) for 5 min, treated with NBF stop buffer (0.1 M Tris Base, 0.1 M Glycine, Sigma) for 5 min twice, and washed for 5 min in PBS. Subsequently, the tissues were incubated overnight at 37 °C with GeoMx™ RNA detection probes in Buffer R (NanoString Technologies) using a Hyb EZ II hybridization oven (Advanced Cell Diagnostics). During the incubation, the slides were shielded with HybriSlip Hybridization Covers (Grace BioLabs, 714022). After the incubation, the HybriSlip covers were gently removed, and stringent 25-min washes were carried out twice in 50% formamide and 2× SSC at 37 °C. The tissues were then washed for 5 min in 2× SSC and blocked in Buffer W (NanoString Technologies) for 30 min at room temperature within a humidity chamber. PanCK + Syto13 (NanoString Technologies, GMX-RNA-MORP-HST-12) and α-SMA (Novus Biologicals, NBP2-34522AF647) in Buffer W were applied to each section for 1 h at room temperature. Following this, the slides were rinsed twice in fresh 2× SSC and loaded onto the GeoMx™ DSP.
During the procedure, complete slides were captured at a 20× magnification, and ROIs with diameters ranging from 300 to 600 μm were designated. The GeoMx software was utilized to delineate areas of illumination (AOIs or segments) within each ROI. Subsequently, the DSP exposed the AOIs to 385 nm light (UV), liberating the indexing oligos, which were then harvested using a microcapillary. These indexing oligos were then placed in a 96-well plate for subsequent processing. Following an overnight desiccation, the indexing oligos were reconstituted in 10 μL of DEPC-treated water. Sequencing libraries were produced through PCR using the photo-released indexing oligos, AOI-specific Illumina adapter sequences, and distinct i5 and i7 sample indices. The PCR reactions were combined and purified twice using AMPure XP beads (Beckman Coulter, A63881) in accordance with the manufacturer’s instructions. The consolidated libraries were sequenced at 2 × 75 base pairs employing the single-index workflow on an Illumina NovaSeq6000 to yield raw reads. The raw data were imported into the GeoMx NGS Pipeline & Convert software to transform FASTQ files into DCC files compatible with DSP analysis, followed by sequence alignment and sequencing statistics. Each ROI (AOI) was assessed for sequencing quality based on three key metrics: The number of raw reads must exceed 1000; The proportion of reads aligned to the reference sequence should be greater than 75%; Sequencing saturation must surpass 50%. Only ROIs that met these criteria were preserved for further analysis. Given variations in ROI (AOI) area size, sequencing data volume, and sequencing saturation, the Q3 method was employed to standardize gene expression levels within each ROI (AOI) to facilitate subsequent comparative analysis. DEGs between the two groups were deemed significant with a fold change exceeding 2 and a false discovery rate (FDR) value below 0.05.
Sample preparation for MALDI-MSI assay
The fresh samples were first washed with pre-cooled PBS to eliminate any excess blood stains on the surface. Subsequently, they were placed in an embedding box and embedded with 10% carboxymethyl cellulose (CMC) embedding agent (Sigma Aldrich, C5678). The embedding box was then placed on dry ice, allowing the CMC to solidify and attain a white appearance, indicating completion of the embedding process. Once solidified, the samples were transferred to a −80 °C freezer for storage. Tissue sections, each 20 μm thick, were prepared using the Leica CM1950 cryostat (Leica Microsystems GmbH). H&E staining was performed on standard glass slides, while the adjacent tissue section was then thaw-mounted onto ITO-coated microscopic slides (Bruker Daltonics) for MALDI-MSI. Following mounting, the tissue sections were desiccated for 30 min in a desiccator before the application of matrix. Subsequently, they were transferred to a −80 °C refrigerator for sealed storage after vacuum packaging.
MALDI matrix preparation and application
A solution of 5 mg/mL CHCA was prepared by dissolving it in 70% HPLC grade CAN containing 0.1% TFA. The 20 µm thick tissue sections were then sprayed using a TM-sprayer (HTX Technologies). The parameters for the matrix application set in the TM-sprayer were as follows: spray nozzle velocity (1200 mm/min), track spacing (2 mm), flow rate (0.12 mL/min), spray nozzle temperature (72 °C), and nitrogen gas pressure (10 psi).
Mass spectrometry imaging
The metabolites present in the samples were visualized using a timsTOF fleX MALDI 2 (Bruker Daltonics), which was outfitted with a 10 kHz smartbeam 3D laser. MALDI2 MSI was conducted in negative ion mode utilizing a full scan mode covering the m/z range of 50–1500. Imaging was carried out at a spatial resolution of 50 µm. During the MALDI2 MSI process, the laser operated at a frequency of 10,000 Hz, delivering 62% of the laser energy with 100 laser shots per 50 µm pixel.
MALDI-MSI data analysis
MSI data was processed using SCiLS Lab software (version 2021c premium, Bruker Daltonics) with Root Mean Square normalization. The K-means analysis function within the software was utilized to cluster the imaging pixels in the target area. This analysis assigned the same color label to areas exhibiting similar metabolite expression patterns. Metabolites were identified by comparing their accurate m/z values (within <10 ppm accuracy) with the Human Metabolome Database. Metabolites meeting the criteria of fold change ≥1.5 and P-value < 0.05 from the rank sum test were deemed to be indicative of differential metabolites between two groups. KEGG enrichment was used to compare and analyze the core biochemical, metabolic and signaling transduction pathways for differential metabolites between two groups. Heatmap was used to observe metabolic changes between two groups.
Statistics and reproducibility
Data are presented as mean ± standard deviation (SD). Each experiment was repeated independently a minimum of three times. Statistical analyses were conducted using GraphPad Prism 6.01 (GraphPad Inc., La Jolla, CA, USA). For comparisons, student’s t-test was applied to two-group analyses. P < 0.05 is considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The raw scRNA-seq and DSP-WTA data generated in this study have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences under accession codes HRA009006 and HRA010960. The raw spatial MALDI-MSI metabolome data generated in this study have been deposited in the METASPACE platform (https://metaspace2020.org/project/yu-2025). Source data are provided with this paper.
Code availability
Codes used to perform the analysis and reproduce the figures are available on GitHub (https://github.com/ycli1995/scRNAseq20250408) and Zenodo (https://doi.org/10.5281/zenodo.17421060).
References
Shim, J. Y. et al. Evaluation and management of endometriosis in the adolescent. Obstet. Gynecol. 143, 44–51 (2024).
Agarwal, S. K. et al. Clinical diagnosis of endometriosis: a call to action. Am. J. Obstet. Gynecol. 220, 354.e351–354.e312 (2019).
Wang, Y., Nicholes, K. & Shih, I. M. The origin and pathogenesis of endometriosis. Annu. Rev. Pathol. 15, 71–95 (2020).
Uzelli Şimşek, H., Şimşek, T., Duruksu, G., Furat Rençber, S. & Yazır, Y. Different effect of dienogest on endometrium mesenchymal stem cells derived from healthy and endometriosis tissues. Balk. Med. J. 41, 484–490 (2024).
Symons, L. K. et al. The immunopathophysiology of endometriosis. Trends Mol. Med. 24, 748–762 (2018).
Wang, Z. et al. Integrated bioinformatics analysis uncovers characteristic genes and molecular subtyping system for endometriosis. Front. Pharmacol. 13, 932526 (2022).
Zhou, J. et al. Peritoneal fluid cytokines reveal new insights of endometriosis subphenotypes. Int. J. Mol. Sci. 21, 3515 (2020).
Anglesio, M. S. et al. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848 (2017).
Fonseca, M. A. S. et al. Single-cell transcriptomic analysis of endometriosis. Nat. Genet. 55, 255–267 (2023).
Marečková, M. et al. An integrated single-cell reference atlas of the human endometrium. Nat. Genet. 56, 1925–1937 (2024).
Yang, X. et al. Bioinformatics analysis of aggressive behavior of breast cancer via an integrated gene regulatory network. J. Cancer Res. Ther. 10, 1013–1018 (2014).
Xu, G. et al. Mapping single-cell transcriptomes of endometrium reveals potential biomarkers in endometrial cancer. Immunotargets Ther. 13, 349–366 (2024).
Nathan, A. et al. Single-cell eQTL models reveal dynamic T cell state dependence of disease loci. Nature 606, 120–128 (2022).
Samare-Najaf, M., Razavinasab, S. A., Samareh, A. & Jamali, N. Omics-based novel strategies in the diagnosis of endometriosis. Crit. Rev. Clin. Lab. Sci. 61, 205–225 (2024).
Chen, Y. et al. Immunomodulation in endometriosis: investigating the interrelationship between VISTA expression and Escherichia.Shigella-associated metabolites. Int. Immunopharmacol. 137, 112366 (2024).
Sun, C., Wang, F., Zhang, Y., Yu, J. & Wang, X. Mass spectrometry imaging-based metabolomics to visualize the spatially resolved reprogramming of carnitine metabolism in breast cancer. Theranostics 10, 7070–7082 (2020).
Zhao, Y. M. et al. Matrix-assisted laser desorption ionization mass spectrometry imaging reveals the spatial distribution of compounds that may exacerbate inflammation in garden ginseng and ginseng under forest. Talanta 279, 126594 (2024).
Sun, C., Liu, W., Mu, Y. & Wang, X. 1,1’-binaphthyl-2,2’-diamine as a novel MALDI matrix to enhance the in situ imaging of metabolic heterogeneity in lung cancer. Talanta 209, 120557 (2020).
Pelka, K. et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 184, 4734–4752.e4720 (2021).
Brady, L. et al. Inter- and intra-tumor heterogeneity of metastatic prostate cancer determined by digital spatial gene expression profiling. Nat. Commun. 12, 1426 (2021).
Belmonte, B. et al. PARP-1, EpCAM, and FRα as potential targets for intraoperative detection and delineation of endometriosis: a quantitative tissue expression analysis. Reprod. Biol. Endocrinol. 22, 92 (2024).
Wang, Z. et al. CADM2 participates in endometriosis development by influencing the epithelial-mesenchymal transition. Reprod. Sci. 31, 3049–3057 (2024).
Hu, P. et al. CLDN4 as a novel diagnostic and prognostic biomarker and its association with immune infiltrates in ovarian cancer. Med. Inflamm. 2023, 1075265 (2023).
Sugimoto, K. et al. Cell adhesion signals regulate the nuclear receptor activity. Proc. Natl. Acad. Sci. USA 116, 24600–24609 (2019).
Belardi, B. et al. Claudin-4 reconstituted in unilamellar vesicles is sufficient to form tight interfaces that partition membrane proteins. J. Cell Sci. 132, jcs221556 (2018).
Li, Q. et al. Identification of candidate gene signatures and regulatory networks in endometriosis and its related infertility by integrated analysis. Reprod. Sci. 29, 411–426 (2022).
Long, X., Deng, Z., Li, G. & Wang, Z. Identification of critical genes to predict recurrence and death in colon cancer: integrating gene expression and bioinformatics analysis. Cancer Cell Int. 18, 139 (2018).
Rahmani, M. et al. Versican: signaling to transcriptional control pathways. Can. J. Physiol. Pharmacol. 84, 77–92 (2006).
Tani, H. et al. Role of versican in the pathogenesis of peritoneal endometriosis. J. Clin. Endocrinol. Metab. 101, 4349–4356 (2016).
Richter, C. et al. Expression of the glycodelin A gene and the detection of its protein in tissues and serum of ovarian carcinoma patients. Anticancer. Res. 27, 2023–2025 (2007).
Hamutoğlu, R. et al. The regulation of trophoblast invasion and decidual reaction by matrix metalloproteinase-2, metalloproteinase-7, and metalloproteinase-9 expressions in the rat endometrium. Reprod. Med. Biol. 19, 385–397 (2020).
Hebbar, V., Damera, G. & Sachdev, G. P. Differential expression of MUC genes in endometrial and cervical tissues and tumors. Bmc. Cancer 5, 124 (2005).
Wilson, Z. S. et al. Critical role of thrombospondin-1 in promoting intestinal mucosal wound repair. JCI Insight 9, e180608 (2024).
Li, Y., Qin, J., Chen, G., Wu, W. & Sun, X. Plasma THBS1 as a predictive biomarker for poor prognosis and brain metastasis in patients with HER2-enriched breast cancer. Int. J. Clin. Oncol. 29, 427–441 (2024).
Liu, Z. et al. Thrombospondin-1 (TSP1) contributes to the development of vascular inflammation by regulating monocytic cell motility in mouse models of abdominal aortic aneurysm. Circ. Res. 117, 129–141 (2015).
Emidio, G. et al. Increased levels of oxidative and carbonyl stress markers in normal ovarian cortex surrounding endometriotic cysts. Gynecol. Endocrinol. 30, 808–812 (2014).
Gao, X., Jin, Y. & Zhang, G. Systematic review and meta-analysis: impact of various hemostasis methods on ovarian reserve function in laparoscopic cystectomy for ovarian endometriomas. Altern. Ther. Health Med. 30, 312–319 (2024).
Kitajima, M. et al. Association between ovarian endometrioma and ovarian reserve. Front. Biosci. (Elite Ed.) 10, 92–102 (2018).
Wang, Z. et al. Spatial-resolved metabolomics reveals tissue-specific metabolic reprogramming in diabetic nephropathy by using mass spectrometry imaging. Acta Pharm. Sin. B 11, 3665–3677 (2021).
Li, W. et al. 3-O-Acetyl-11-keto-β-boswellic acid ameliorated aberrant metabolic landscape and inhibited autophagy in glioblastoma. Acta Pharm. Sin. B 10, 301–312 (2020).
Liu, H. et al. Investigation on intestinal proteins and drug metabolizing enzymes in simulated microgravity rats by a proteomics method. Molecules 25, 4391 (2020).
Miyata, J. et al. Dysregulated fatty acid metabolism in nasal polyp-derived eosinophils from patients with chronic rhinosinusitis. Allergy 74, 1113–1124 (2019).
Wang, Y., Yao, J., Zhang, Z., Wei, L. & Wang, S. Generation of novel lipid metabolism-based signatures to predict prognosis and immunotherapy response for colorectal adenocarcinoma. Sci. Rep. 14, 17158 (2024).
Chen, C., Zhao, Z., Han, J., Zhang, Y. & Nie, G. Ptgds downregulation protect vestibular hair cells from aminoglycoside-induced vestibulotoxicity. PLoS ONE 20, e0320634 (2025).
Liao, Y. et al. Lipid metabolism patterns and relevant clinical and molecular features of coronary artery disease patients: an integrated bioinformatic analysis. Lipids Health Dis. 21, 87 (2022).
Lun, A. T. L. et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
McGinnis, C. S., Murrow, L. M. & Gartner, Z. J. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 8, 329–337.e324 (2019).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Acknowledgements
The authors wish to thank all study participants, research staff, and students who assisted with this work. We are grateful to Guangzhou OmicsMaster Biotechnology Co., Ltd. for scRNA-seq, DSP-WTA, and MALDI-MSI in this study. The authors also wish to thank Yu Tang (Guangzhou OmicsMaster Biotechnology Co., Ltd.) and Di Shen (Guangzhou OmicsMaster Biotechnology Co., Ltd.) for their valuable contributions to this work. This work was supported by the National Natural Science Foundation of China (82271633), Natural Science Foundation of Jiangsu Province (BK20241858), Qinglan Project of Jiangsu Province of China, Fund of Xuzhou Science and Technology (KC23189, KC20092 and KC22038), Xuzhou Municipal Health Commission General Program Project (XWKYHT20240081), Open Fund of Jiangsu Key Laboratory for New Drug Research and Clinical Pharmacy, and Xuzhou Pengcheng Talents - Medical young reserve talents (XWRCHT20220011).
Author information
Authors and Affiliations
Contributions
J.Y., B.Z., Y.C., and M.X. initiated the project, designed the study, coordinated the experiment, and wrote the manuscript. Y.Q., X.C., S.Z., Z.L., and T.W. performed the experiments and provided conceptual inputs for the paper. J.C., X.Y., W.T., J.G., Q.S., N.C., Xiangnan-C., J.L., Q.H., L.H., C.W., X.W., Q.H., Q.X., Yi-Z., J.R., W.Q., L.K., Y.H., Y.W., C.Q., X.Z., and Y.Z. analyzed the data. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qi, Y., Chen, X., Zheng, S. et al. Single-cell and spatially resolved omics reveal transcriptional and metabolic signatures of ovarian endometriomas. Nat Commun 16, 11539 (2025). https://doi.org/10.1038/s41467-025-66706-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66706-8