Abstract
The ability of neural stem cells (NSCs) to switch between quiescent and proliferative states is fundamental for adult neurogenesis and regeneration. Microproteins or short open reading frame (sORF)-encoded peptides (SEPs), are highly abundant yet largely understudied, and their role in brain development remains unclear. Here, we demonstrate that two evolutionarily-conserved microprotein paralogs, Simba1 and Simba2, encoded by sORFs CG15715 and CG18081, respectively, govern the reactivation of Drosophila quiescent NSCs. Both Simba1 and Simba2 function in NSCs and Blood-Brain-Barrier (BBB) glial cells to promote NSC reactivation. Mechanistically, Simba1 and Simba2 act as transcription factors activating the WNT/Wingless signalling pathway during NSC reactivation. We uncover a critical role of Wg signalling molecules in promoting NSC reactivation and the translocation of Wingless from BBB glia to NSCs. Our findings reveal a role for microproteins Simba1/2 in regulating NSC reactivation through Wg signalling. Moreover, our bioinformatic analysis and luciferase assay suggest that ZNF706, the human ortholog of Simba1/2, is required for WNT signaling activation in human cells. The conserved function of Simba1/2/ZNF706 in activating WNT/Wg signalling in Drosophila and humans suggests that this new regulatory paradigm may be applicable to broader cellular processes and disease conditions.
Similar content being viewed by others
Introduction
Neural stem cells (NSCs) are essential for the generation of neurons and glial cells in the central nervous system. In the mammalian adult brain, the majority of adult NSCs reside in a quiescent state, characterized by a lack of proliferation or differentiation1,2. In response to extrinsic stimuli, such as injury, the presence of nutrients, and physical exercise, quiescent NSCs can be reactivated, enabling them to re-enter the cell cycle and resume proliferation3. The ability of NSCs to transit between quiescent and proliferative states is crucial for adult neurogenesis and regenerative capacity. Notably, the population of quiescent NSCs increases in aging brains, while dysregulated reactivation of these cells is linked to defects in adult neurogenesis and neurodevelopmental disorders including microcephaly4,5. Therefore, a comprehensive understanding of the mechanisms governing NSC reactivation is critical for elucidating the complexities of brain development and the underlying causes of neurodevelopmental diseases.
Drosophila larval brain NSCs have emerged as a powerful model for investigating the mechanisms underlying NSC quiescence and reactivation in vivo6,7. In the Drosophila central nervous system, NSCs enter quiescence at the end of embryogenesis8,9,10,11. Following larval hatching, the intake of nutrients triggers quiescent NSCs to resume proliferation within 24 hr after larval hatching (ALH). Dietary amino acids are sensed by the fat body, the invertebrate equivalent of liver and adipose tissue12, which in turn stimulates the Drosophila blood brain barrier (BBB) glial cells13,14. These surface glial cells function as a niche that integrates several signaling pathways, promoting and inhibiting NSC reactivation6,7,13,15. The reactivation of quiescent NSCs depends on the activation of the InR/PI3K/Akt signaling pathway in NSCs by insulin/IGF-like peptides Dilp2 and Dilp6 secreted from the BBB glia13,14,16. In mammalian brains, insulin-like growth factor-1 (IGF-1) and IGF-1 receptor (IGF-1R) have been shown to promote NSC proliferation17. Furthermore, human IGF-1R mutations are associated with microcephaly, a neurodevelopmental disorder18, suggesting that the insulin pathway likely has a conserved function in brain development between flies and humans. On the contrary, the activation of the evolutionarily conserved Hippo pathway keeps NSCs in quiescence19,20,21. Besides the InR/PI3K/Akt and Hippo pathways, the involvement of other signaling pathways in BBB glial cell niche during NSC reactivation remains unclear.
The Wnt/Wingless (Wg) signaling pathway is crucial for the control of cell proliferation, differentiation, and polarization during development22.The activation of the canonical Wnt/Wg pathway is triggered by the binding of a Wnt ligand to the Wnt receptor Frizzled (Fz) resulting in the inactivation of the ‘destruction complex’, composed of the scaffold protein Axin, adenomatous polyposis coli (APC)-glycogen synthase kinase 3 (GSK3/Zw3/Shaggy)/CK1α kinase complex. In the absence of Wnt ligands, the destruction complex kinases phosphorylate cytoplasmic β-catenin and target it for degradation. Inhibition of the destruction complex is mediated by Disheveled (Dsh) that facilitates re-location of Axin to the plasma membrane where GSK3 activity is suppressed, leading to the stabilization and translocation of β-catenin/Arm into the nucleus to regulate downstream target genes23,24. Mutations of Wnt/Wg signaling have been associated with neurodegenerative diseases and cancer formation25,26. Recently, we showed that Drosophila Wg pathway transcriptional co-activator Ebd1 is required for NSC reactivation27. However, the role of the Wg pathway including its core components in NSC reactivation remains to be established.
Microproteins, also known as short open reading frame (sORF)-encoded peptides (SEPs), are comprised of fewer than 100 amino acids and are highly abundant in eukaryotic genomes, including those of humans, mice, and Drosophila, as revealed by the recent advances in proteomics and ribosome profiling28,29. Although more than 900 of SEPS are annotated in the Drosophila genome, only a few have been experimentally investigated30,31,32. Emerging evidence suggests that SEPs can play important roles in various cellular processes, such as cell signaling, metabolism, and cell proliferation and are implicated in diseases like cancer and metabolic disorders28,31,33. Despite many SEPs being expressed in the mammalian brain34,35, they remain largely understudied – particularly in the nervous system.
Here, we demonstrate that two previously uncharacterized Drosophila microproteins/SEPs, Simba1 and Simba2 encoded by the highly conserved sORFs CG15715 and CG18081, respectively, govern the reactivation of quiescent NSCs in Drosophila. Both Simba1 and Simba2 function in NSCs and BBB glial cells to promote NSC reactivation. In vivo protein-DNA interaction profiling using DamID-seq, along with genetic analyses, demonstrated that Simba1 and Simba2 transcriptionally activate Wg during NSC reactivation. We further established the critical role of WNT/Wg pathways in NSC reactivation and the requirement of Wg in both BBB glial cells and NSCs. Importantly, ZNF706, the human ortholog of Simba1/2, is required for the activation of WNT signaling in human cells, suggesting a conserved regulatory mechanism. This new regulatory paradigm may be applicable to a wide variety of cellular processes and disease conditions.
Results
Simba1 and Simba2 are required for quiescent NSC (qNSC) reactivation
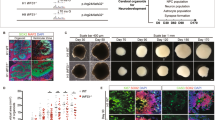
To identify sORFs that are important for quiescent NSC reactivation, we performed a CRISPR KO screen on 25 Drosophila sORFs that are evolutionarily conserved, protein-coding and previously uncharacterized36 (Fig. 1A, B). Dpn, the bHLH repressor Deadpan, which regulates the self-renewal and specification of Drosophila NSCs, is used as a marker for NSCs. Potential reactivation defects were assessed following the EdU (5-ethynyl-2’-deoxyuridine) incorporation assay at 24 h after larval hatching (ALH). From the screening, knockouts of CG15715 and CG18081 and double KO led to reactivation defects, as shown by a markedly increased population of qNSCs that were EdU-negative (30.9%, 32.2% and 33.8% respectively), compared to 9.1 % in control (Fig. 1C, D). Hereafter, CG15715 and CG18081 are referred to as Simba1 and Simba2, respectively, inspired by The Lion King, reflecting their small sizes and roles in development. simba1 KO and simba2 KO are homozygous CRISPR/Cas9 frameshift knockouts induced by an indel with a 4 bp insertion and a 11 bp deletion in simba1 and a 29 bp deletion in simba2, respectively (Fig. 1E). Hemizygotes of simba1 and simba2 over the deficiency line Df(3 L)BSC575 (Df) that removed a few genes including both simba1 and simba2 resulted in similar but slightly more severe phenotypes in NSC reactivation (Figure S1A, B; simba1 KO/Df: 39.7%, simba2 KO/Df: 37.7%, simba1&2 dKO/Df: 46.5%, control: 12.3%), suggesting that simba1 and simba2 KOs are strong loss-of-functions alleles. In addition, the percentage of qNSCs carrying primary protrusions, the hallmark of qNSCs, increased notably in the simba1 KO and simba2 KO brains compared to control (Figure S1C, D; simba1 KO: 21.7%, simba2 KO: 24.0%, simba1&2 dKO: 30.7%, control: 8.5%). Simba1 and Simba2 share 97.1% amino acid identity and are located adjacent to each other in the same genomic region, suggesting that they may be paralogues that evolved by tandem duplication (Fig. 1F). Given that knockout of either gene results in reactivation defects and that the double KO only showed a slight increase in phenotype severity, they do not appear to be functionally redundant (Fig. 1C, D). However, Simba1 and Simba2 do not appear to interact with each other in S2 cells (Figure S1E) or through AlphaFold3 predictions (Figure S1F), suggesting a minimal likelihood of either heterodimer or homodimer formation.
A An illustration of Drosophila NSC reactivation process. Created in BioRender. LIN, J. (https://BioRender.com/s1vy3m3). B Workflow of CRISPR knockout (KO) screen of sORFs in NSC reactivation. C Larval NSCs were labeled with Dpn at 24-h ALH, while active NSCs were labeled by EdU for various genotypes. White arrows point to EdU-negative NSCs. D Quantifications of EdU-negative NSCs in (C). Control (yw): 9.1%, n = 14 Brain Lobes (BL); simba1 KO: 30.9%, n = 16 BL, P = 8.6E-07; simba2 KO: 32.2%, n = 13 BL, P = 7.6E-07; simba1-simba2 double KO: 33.8% n = 16 BL, P = 4.7E-08. E Illustration of genomic lesions in simba1/CG15715 KO, simba2/CG18081 KO and simba1-simba2 double KO. F Amino acid sequence alignment between Drosophila Simba1 and Simba2 and amino acid sequence similarities in various organisms. G, H Larval NSCs were labeled with Dpn and EdU at 24-h ALH for various genotypes. I Quantifications of EdU-negative NSCs in (G) Control (grh > β-galRNAi): 7.9%, n = 20 BL; grh>simba1RNAi: 29.2%, n = 24 BL, P < 1E-15; grh>simba2RNAi: 17.6%, n = 20 BL, P = 4.9E-06. J Quantifications of EdU-negative NSCs in (H) Control (NP6293 > β-galRNAi): 8.0%, n = 11 BL; NP6293>simba1RNAi: 22.6%, n = 11 BL, P = 3.8E-05; NP6293>simba2RNAi: 42.8%, n = 15 BL, P = 3.2E-14. K, L Larval NSCs were labeled with Dpn and EdU at 6-h ALH for various genotypes. M Quantifications of EdU-negative NSCs in (K) Control (grh > β-gal): 15.4%, n = 10 BL; grh>simba1CC: 34.6%, n = 12 BL, P = 2.5E-06; grh>simba1HA: 24.2%, P = 3.2E-02, n = 11 BL; grh>simba2HA: 16.0%, n = 10 BL, P = 0.997. N Quantifications of EdU-negative NSCs in (L) Control (NP6293 > β-gal): 13.5%, n = 14 BL; NP6293>simba1CC: 39.9%, n = 11 BL, P = 7.5E-04; NP6293>simba1HA: 31.0%, n = 10 BL, 4.4E-02; NP6293>simba2HA: 49.8%, n = 13 BL, 2.1E-06. Data are presented as mean ± SD, and statistical significance was determined by one-way ANOVA with multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Scale bars: 10 μm.
Cell type-specific functions of Simba1 and Simba 2 during NSC reactivation
To elucidate cell type-specific roles of Simba1 and Simba2 in NSC reactivation, we knocked down Simba1 and Simba2 specifically in NSCs or blood-brain-barrier (BBB) glial cells (surface glia). simba1 and simba2 share high similarity in coding sequences, while their noncoding regions, particularly the 3′ UTR, are distinct. Thus, we used RNAi lines that target their 3’UTR region, respectively (Figure S1G). BLAST analysis confirmed that both RNAi targets are transcript-specific, with no detectable off-target matches, indicating precise knockdown of each gene. At 24 h ALH, Simba1 knockdown in NSCs using grainyhead-Gal4 (grh-Gal4) driver caused an increase in the number of EdU-negative qNSCs (29.2%), compared with 7.9% in control, while its knockdown in BBB glia by NP6293-Gal4 driver caused an increase in the number of qNSCs (22.6%), compared with 8.0% in the control (Fig. 1G–J). This observation suggested that simba1 knockdown in either NSCs or BBB glia cells led to defects in NSC reactivation, with a slightly stronger phenotype using the NSC-specific driver. Consistent with this observation, at 24 h ALH, Simba1 (Simba1CC, CC: C-terminal cerulean tag) overexpression in NSCs and BBB glia cells partially rescued simba1 KO-induced reactivation defects, with a significantly higher suppression when Simba1CC was overexpressed using the NSC-specific driver (Figure S1I, J).
On the other hand, Simba2 knockdown in NSCs caused a mild increase in the number of qNSCs (17.6%), compared with 7.9% in control, while its knockdown in BBB glia caused a prominent increase in the number of qNSCs from 8.0% in control to 42.8% (Fig. 1G–J). This result suggested that knocking down simba2 in either NSCs or BBB glia also leads to defects in NSC reactivation, with a stronger phenotype in BBB glia. Consistent with this observation, overexpression of HA-tagged Simba2 (Simba2HA) in NSCs and BBB glia partially rescued simba2 KO-induced reactivation defects, with greater rescue extent when overexpressing in BBB glia (Figure S1K, L).
To examine the transcriptional level of simba1 and simba2 in NSCs and BBB glia, we used a public scRNA-seq dataset of Drosophila larval brains at 16 h ALH37. Cell types were identified using established marker genes: dpn for NSCs, repo for pan-glial cells, CG6126 for surface glia, wrapper for cortex/chiasm glia, and wun2 for astrocyte/neuropil glia. Both simba1 and simba2 are expressed in NSCs and glial cells. Consistent with our in vivo knockdown experiments, simba1 has higher expression levels in NSCs, while simba2 shows higher expression levels in BBB glia (surface glia) (Figure S1H). These differential expression patterns further support the notion that Simba1 appears to be more important for NSCs, while Simba2 may function primarily in BBB glia, to promote NSC reactivation.
Taken together, Simba1 and Simba2 appear to act both cell-autonomously and non-autonomously in NSCs and BBB glial cells for NSC reactivation.
Overexpression of Simba1 and Simba2 triggers premature NSC reactivation
To explore the potential overexpression phenotype, we overexpressed Simba1 and Simba2 using either NSC- or BBB glial cell-specific drivers. At 6 h ALH, overexpression of Simba1 (both Simba1CC and Simba1HA) in NSCs by grh-Gal4 driver was sufficient to trigger NSC premature reactivation, evident with an increase in the number of EdU-positive NSCs (34.6% and 24.2%), compared with 15.4% in the control (Fig. 1K, M). Likewise, its overexpression in BBB glial cells by NP6293-Gal4 driver resulted in an increase in the number of EdU-positive NSCs (39.9% and 31.0%), compared with 13.5% in control (Fig. 1L, N). Thus, Simba1 overexpression in either NSCs or BBB glial cells can trigger premature NSC reactivation.
In contrast, Simba2 (Simba2HA) overexpression in BBB glia, but not in NSCs, promoted premature NSC reactivation with an increase in the number of EdU-positive NSCs from 13.5% in control to 49.8% (Fig. 1L, N). We observed that Simba2-HA localization was excluded from the nucleus in NSCs upon overexpression in NSCs (refer to the following section; Figure S2B). Given the prediction that Simba1 and Simba2 are nuclear proteins with a Zinc-finger motif, overexpression of Simba2 in NSCs might be non-functional and is unable to trigger premature reactivation. In addition, overexpression of either Simba1 or Simba2 in nutrition-restriction conditions failed to trigger NSC premature reactivation (Figure S1M, N). Taken together, overexpression of Simba1 and Simba2 can trigger premature NSC reactivation in the presence of nutrition.
Simba1 and Simba2 exhibit nuclear and cytoplasmic localization in NSCs and glial cells
To analyze the domains and structures of Simba1 and Simba2, we performed InterPro domain analysis and Alphafold structure prediction38,39. Simba1 and Simba2 consist of a small EDRK-rich factor (SERF) domain in the N-terminal, a zinc-finger domain, and a short disordered amino acid sequence in the C-terminal (Figure S2A). The SERF domain can bind to nucleic acids, and zinc-finger domains can mediate DNA-binding and protein-protein interactions40,41. To determine the subcellular localization of Simba1 and Simba2 in the Drosophila larval brain, we analyzed the localization of transgenic HA-tagged Simba1 and Simba2 in NSCs (by grh-gal4) and glial cells (by pan-glial driver repo-gal4), respectively. At 24 h ALH, Simba1HA was localized to the nucleus of both NSCs and glial cells (Figure S2B, C). Similarly, Simba2HA localized to the nucleus of glial cells when overexpressed by the pan-glial driver repo-gal4 (Figure S2C). However, Simba2HA overexpressed in NSCs, was only detected in the cytoplasm but not the nucleus, consistant with its minimal activity in NSCs (Figure S2B).
To validate these findings on endogenous protein, we generated polyclonal antibodies against full length Simba2 (AbMart), which recognized both Simba1 and Simba2. Endogenous Simba1 and Simba2 detected by the antibodies exhibit both nuclear and cytoplasmic localization in the larval brain cells (Figure S2D, F). Simba1/2’s fluorescent signal intensity was significantly reduced to 0.4-fold and 0.41-fold in simba1 and simba2 KOs, respectively, and largely diminished (0.17-fold) in the double KO (Figure S2D, E). Similarly, western blot confirmed reduced Simba levels in simba1 KO (0.39-fold), simba2 KO (0.22-fold) with a nearly complete loss (0.01-fold) in the double KO (Figure S2G, H), supporting the specificity of the antibodies.
To further examine the expression patterns of Simba1/2, we performed immunostaining of Simba1/2 using anti-Simba antibodies in larval brains at the timepoints of 0h-, 6h-, 12h-, 24h- and 72h- ALH (Figure S2I). Quantifications of Simba1/2 protein levels showed that Simba1 and Simba2 overall protein levels in larval brains increased from 0 h to 6 h ALH and remained high levels from 6 h to 24 h ALH. At 72 h ALH, Simba1/2 protein levels decreased (Figure S2J). We have also quantified Simba1 and Simba2 levels in NSCs at these timepoints and found that the level increases from 0 h ALH to 12 h ALH, maintains at 24 h ALH, and dropped at 72 h ALH (Figure S2J).
Taken together, Simba1 and Simba2 can localize to both the nucleus and cytoplasm in NSCs and glial cells and there expressions are upregulated during NSC reactivation.
In vivo protein-DNA profiling by TaDa revealed WNT/Wingless signaling as a potential primary target of Simba1 and Simba2 in Drosophila NSCs
Given the presence of SERF domain and zinc-finger domain, as well as the nuclear localization of Simba1 and Simba2, we speculated that Simba1 and Simba2 act as transcriptional regulators. To profile the in vivo protein-DNA interactions of Simba1 and Simba2 in NSCs, we generated transgenic flies expressing Simba1 and Simba2 fusion proteins with Dam and performed Targeted DNA adenine methyltransferase (Dam) identification (TaDa) in 24 h ALH larval brains. Simba1 and Simba2 fused with Dam and a nuclear localization sequence were expressed by grh-Gal4; Gal80ts to achieve NSC-specific and temporal (0h-24h ALH) resolution. Transgenic flies expressing Dam alone under the same driver was used as a negative control (Fig. 2A). As a result, we identified 7391 peaks in Simba1 group and 9154 peaks in Simba2 group (FC > 2, p value < 0.001, between 2 replicates). The peak correlations between 2 replicates are 0.82 and 0.94 for Simba1 and Simba2, respectively, indicating a good reproducibility of this genomic profiling (Figure S2K). Proximity-based nearest gene annotation of the peaks showed that Simba1 targets 4551 genes while Simba2 targets 4429 genes, with 2623 genes overlapping (Fig. 2B). KEGG pathway enrichment analysis was performed to identify pathways that were over-represented in Simba 1 and Simba2 target genes. Strikingly, WNT/Wingless signaling is the top enriched pathway in all Simba1, Simba2, and their commonly targeted genes, followed by MAPK signaling, Lysosome, mTOR signaling, etc. (Fig. 2C, D). Our data suggests that Simba1 and Simba2 potentially target the WNT/Wingless signaling pathway in NSCs.
A Workflow of Simba1 and Simba2 Tada (DamID)-seq. B Venn diagram of Simba1 and Simba2 target genes (2 replicates each) from DamID-seq. C Dot plot of KEGG pathway enrichment results on Simba1 target genes. D Dot plot of KEGG pathway enrichment results on Simba2 target genes. For KEGG enrichment analysis in (C, D), a one-sided hypergeometric test was used to assess enrichment, with Benjamini–Hochberg correction applied for multiple comparisons.
Drosophila Wingless/WNT signaling pathway is required for Drosophila NSC reactivation
Given that Wingless/WNT signaling is the top candidate pathway targeted by Simba1 and Simba2, we sought to determine if Wingless/WNT pathway is required for NSC reactivation. We first analyzed the function of Wingless (Wg), a well-studied ligand of WNT/Wg signaling and examined wgts, a temperature-sensitive allele at the restrictive temperature, 29 °C42, in NSC reactivation. At 24 h ALH, the vast majority of NSCs in wgts (Fig. 3A, B; 84.2%) failed to incorporate EdU, compared with that in control (Fig. 3A, B; 13.1%), In addition, the number of NSCs retaining protrusions was significantly increased in wgts larval brains (Fig. 3C; 28.0%), in contrast to that in control (Fig. 3C; 14.0%). Moreover, at 24 h ALH, 32.4% of the NSCs failed to incorporate EdU in wgI-16, a hypomorphic wg allele carrying an ~17kb P-element insertion at the 3’ regulatory region (Fig. 3A, B), compared with control (13.1%). Thus, Wg is required for NSC reactivation.
A, D Larval brains were labeled with EdU, Dpn, and Mira at 24-h ALH. B Quantifications of EdU-negative NSCs in (A). Control (yw): 13.1%, n = 27 BL; wgts: 84.2%, n = 9 BL, P = 6E-15; wgI-16: 32.4%, n = 14 BL, P = 1.2E-11. C Quantification of NSCs with primary protrusion at 24-h ALH. Control (yw): 14.0 %, n = 12 BL; wgts: 28.0%, n = 11 BL, P = 9.9E-11. E Quantifications of EdU-negative NSCs in (D). Control (yw): 11.2 %, n = 17 BL; fz21: 50.9 %, n = 10 BL, P = 1.5E-07; dsh75: 88.7%, n = 11 BL, P < 1E-15; dsh3: 39.6%, n = 10 BL, P = 7.5E-05. F Larval brains were labeled with EdU and Dpn at 24-h ALH. G Quantifications of EdU-negative NSCs for (F) Control (grh > β-gal): 11.6%, n = 8 BL; grh>sggS9A: 31.8%, n = 16 BL, P = 1.3E-07. H, I Larval brains were labeled with EdU, Dpn and Mira for various genotypes at 24-h ALH. J Quantifications of EdU-negative NSCs in (H). Control (grh > β-gal): 7.3%, n = 12; grh>wg RNAi: 27.3%, n = 12, P = 7.0E-10. K Quantifications of NSCs with protrusions for various phenotypes. Control (grh > β-gal): 6.4%, n = 9 BL; grh>wg RNAi: 16.3%, n = 6 BL, P = 1.9E-07. L Quantifications of EdU-negative NSCs in (I). Control (NP6293 > β-gal): 9.7%, n = 10 BL; NP6293>wg RNAi#1: 17.6%, n = 10 BL, P = 1.7E-10; NP6293>wg RNAi#2: 15.7%, n = 10 BL, P = 4.9E-08. Data are presented as mean ± SD, and statistical significance was determined by one-way ANOVA with multiple comparisons in (B, E, L), and two-sided t-test for binary comparisons in (C, G, J–K). ∗p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Scale bars: 10 μm.
Next, we tested the function of several core components of WNT/Wg pathway, including Frizzled (Fz) and Dishevelled (Dsh) in NSC reactivation. Loss-of-function mutants fz21, dsh75 and dsh3 all exhibited a marked increase in the number of qNSCs that failed to incorporate EdU, compared to control (Fig. 3D, E; Control, 11.2%; fz21, 50.9%; dsh75, 88.7%; dsh3, 39.6%). In addition, knockdown of fz1, fz2, dsh, and dally under the control of NSC driver grh-Gal4 at 24 h ALH resulted in NSC reactivation defects (Figure S3A, B). These data indicate that WNT/Wg signalling is required for qNSC reactivation.
Next, we investigated whether Arm/β-catenin, the key effector of canonical Wg signaling, is involved in NSC reactivation. Since arm alleles are embryonically lethal, which precluded us from analyzing NSC reactivation in homozygous mutant larval brains, we knocked down arm in NSCs by RNAi. Upon knocking down arm by RNAi #1(VDRC#107344) under the control of grh-Gal4 at 24 h ALH, 18.8% of NSCs failed to incorporate EdU, significantly higher than 5.8% in control brains (Figure S3C, D). Two other arm RNAi lines also had a significant increase in the number of EdU-negative NSCs in larval brains (Figure S3C, D). Moreover, all three arm RNAi lines displayed a significant increase in the number of NSCs retaining primary protrusion compared with the control (Figure S3E). In contrast, knocking down arm RNAi lines with repo-Gal4, NP6293-Gal4 (perineural glial/PG driver), or NP2276-Gal4 (subperineural glial/SPG driver) did not cause any delay of NSC reactivation (Figure S3F). These observations suggested that Arm/β-catenin is required in NSCs, but not glial cells, for their reactivation. Consistent with these observations, overexpression of sggS9A, a constitutively active form of GSK3β/sgg43, in NSCs led to a defect in NSC reactivation, as 31.8% of NSCs remain quiescence as compared to 11.6% in control at 24 h ALH (Fig. 3F, G).
Taken together, canonical WNT signaling, Wg/Fz/β-catenin axis plays an important role in NSC reactivation.
Wg is required in both NSCs and glial cells for Drosophila NSC reactivation
Next, we investigated which cell type Wg function is required for NSC reactivation. Knocking down wg in NSCs under the control of grh-Gal4 resulted in NSC reactivation defects at 24 h ALH, as shown by a significant increase in the number of NSCs failed to incorporate EdU (Fig. 3H, J; Control, 7.3%; wg RNAi, 27.3%). Consistent with this phenotype, wg knockdown in NSCs results in a significant increase in the number of NSCs retaining primary protrusions compared to control (Fig. 3K). Upon wg knockdown in BBB glial cells by PG driver NP6293-Gal4 at 24 h ALH, a significant higher percentage of qNSCs was observed in wg RNAi (Fig. 3I, L; Control, 9.7%; wg RNAi#1, 17.6%, wg RNAi#2, 15.7%). Consistent with this observation, wg knockdown in glial cells driven by a pan-glial driver repo-Gal4, but not neuronal drivers by elav-Gal4, pros-Gal4, or nSyb-Gal4, resulted in NSC reactivation defects shown by EdU incorporation assay (Figure S3G, H). Depletion of Wntless, a protein necessary for Wg secretion44, in BBB glia cells is sufficient to induce a significant increase in the number of quiescent NSCs compared to control, further supporting that Wg from the BBB glia is necessary for NSC reactivation (Figure S3I, J).
Taken together, these results indicate that Wg can function both non-cell autonomously in BBB glial cells and intrinsically in NSCs to promote NSC reactivation.
Wg is present in BBB glial cells and translocated to NSCs during larval brain development
To understand the subcellular localization of Wg in the larval brains, we analyzed a membrane marker CD8-GFP driven by wgKO-Gal4 containing the endogenous regulatory sequence of wg45. Remarkably, Wg was predominantly detected in the superficial layer of larval brains at 0 h ALH where BBB glia is presumably located (Fig. 4A, B), implying that Wg might be present in BBB glial cells. The CD8-GFP signal was reduced dramatically at 24 h ALH and almost undetectable at 48 h ALH (Fig. 4B), suggesting that abundant Wg was only observed before 24 h ALH and its levels reduced over larval development. Next, we examined Wg localization in larval brains expressing CD8-GFP under wgKO-Gal4 at 0 h ALH. Similar to Wg pattern shown by CD8-GFP driven by wgKO-Gal4, Wg protein was primarily located at the superficial layer of both ventral nerve cord (VNC) and central brain (CB), as well as presumptive NSCs located right beneath the surface (white arrows) (Fig. 4C, D). The 3-dimensional (3D) view of the larval brain further supported this notion (Fig. 4E). To confirm the cell identity in these Wg-expressing cells, we co-labelled Wg with BBB glia or NSCs (Fig. 4F). At 16 h ALH, Wg localized in BBB glial cells and NSCs (Fig. 4F, boxes and arrows).
A Schematic diagram of Drosophila early larval brain, including qNSC, aNSC, Blood-brain-barrier (BBB) and Neuropil. Created in BioRender. LIN, J. (2025) https://BioRender.com/s1vy3m3. B Laval brains at 0 h ALH (n = 8), 24 h ALH (n = 4), and 48 h ALH (n = 3) from wgKO-Gal4 driven UAS-CD8-GFP were detected with Wg and GFP. n = number of biological replicates. Scale bars: 20 μm. C, D Ventral nerve cord (VNC) and central brain (CB) from 0 h ALH larval brains of wgKO-Gal4 driven UAS-CD8-GFP (n = 8) were examined with Wg and GFP. Representative regions showing the localization of GFP and Wg at the outer layer of VNC and CB were boxed and zoomed in on the right. n number of biological replicates. Scale bars: 10 μm. E Three-dimensional view (3D) of 0 h ALH brains expressing CD8-GFP under the control of wgKO-Gal4. Scale bars: 20 μm. F Laval brains at 16 h ALH from NP6293-Gal4 (n = 5)/grh-Gal4 (n = 4) driven UAS-CD8-GFP were labeled with Wg, GFP, and Dpn. n number of biological replicates. Scale bars: 10 μm. G At 24 h ALH, larval brains were labeled with Wg and GFP for various genotypes. Genotypes: NP6293-gal4 > CD8GFP (n = 5), NP6293-gal4>WgGFP (n = 12), NP2276-gal4>WgGFP (n = 4), nvr2-gal4>WgGFP (n = 4) and arlm-gal4>WgGFP (n = 3). Surface glia driver: NP6293-gal4, sub-perineurial glia driver: NP2276-gal4, cortex glia driver: nvr2-gal4, astrocyte-like glia driver: alrm-gal4. Scale bars: 10 μm. H Larval brains at 0- (n = 3), 2- (n = 3), 6- (n = 3), 12- (n = 4), 16- (n = 4), and 24-h ALH (n = 12) were labelled with GFP and Wg for NP6293-gal4> WgGFP. Scale bars: 10 μm.
To further examine Wg localization in the larval brain, transgenic wingless fused with GFP was overexpressed by different glial subtype specific drivers, including BBB glia, cortex glia, astrocyte-like/Neuropil glia, followed by the detection of Wg by anti-Wg antibodies at 24 h ALH. Cortex glia- and astrocyte like glia-sourced WgGFP was detected to be restricted to the cell body of the WgGFP-expressing cells. Surprisingly, however, BBB glia (NP6293-Gal4 driver) -sourced WgGFP was mainly detected in NSCs, which colocalized with Wg detected by anti-Wg antibodies (Fig. 4G). In control brains in which NP6293-Gal4 driver was crossed to CD8GFP, Wg remained at low levels and CD8GFP was largely restricted to the BBB glia (Fig. 4G). These observations suggest that Wg can be translocated from BBB glial cells to NSCs. However, WgGFP could still translocate from BBB glia to NSCs under nutrient-restriction conditions (Figure S3K), suggesting that this translocation is independent of nutritional status.
To elucidate the time window of the translocation of Wg from BBB glial cells to NSCs, we performed a time-course experiment from 0 h ALH to 24 h ALH. At 0 h ALH, WgGFP signal, colocalized with Wg antibody, was detected mainly in the BBB glia. At 2 h ALH and onwards, strikingly, the majority of WgGFP started to be detected in NSCs, colocalized with Wg antibody signal (Fig. 4H).
Taken together, our results suggest that Wg is abundant only in early larval brains and can be rapidly translocated from BBB glial cells to NSCs.
Wg is a direct transcriptional target of both Simba1 and Simba2
Our TaDa analysis suggested that Simba1 and Simba2 bind to wg regulatory region, as multiple peaks were found near wg 5’ UTR region in both Simba1 and Simba2 binding profile (Fig. 5A), Peaks 1085, 1548 and 1549 contain DNA sequences in the wg promoter region near the transcription start site (TSS) that are known to be bound by transcription factors that regulate wg gene expression, such as da (daughterless) and twi (twist)46, suggesting that the binding of Simba1/2 at this regions may regulate Wg expression. This protein-DNA interaction at peaks1548 and peak1549 in wg promoter region was further validated by chromatin immunoprecipitation (ChIP)-quantitative polymerase chain reaction (qPCR) analysis in S2 cells, with 5.52-fold and 6.94-fold increase of the binding affinity respectively, compared to control (Figure S4A-B). Our result suggests that wg is a direct transcriptional target of both Simba1 and Simba2.
A Normalized fold changes of Simba1 and Simba2 binding peaks near wg transcription start site (TSS). Peak1085, peak1548 and peak1549 were annotated. B At 24-h ALH, quantitative PCR quantification of wg RNA: Control:1-fold, n = 6; simba1 KO: 0.32-fold, n = 6, P = 7.8E-13; simba2 KO: 0.30-fold, n = 5, P = 1.0E-12; simba1/2 double KO: 0.30-fold, n = 6, P = 3.5E-13. senseless RNA: Control:1-fold, n = 5; simba1 KO: 0.20-fold, n = 5, P = 9.8E-14; simba2 KO: 0.26-fold, n = 5, P = 9.5E-13; simba1/2 double KO: 0.15-fold, n = 3, P = 8.2E-13. C At 24 h ALH, larval brain lobes were labeled with Wg, Dpn and Msps. While NSCs were marked by Dpn. D Whole brain lobe quantifications of Wg intensity normalized to Dpn in (C), Control:1-fold, n = 9 BL; simba1 KO: 0.38-fold, n = 11, P = 4.4E-05; simba2 KO: 0.28-fold n = 10, P = 5.9E-06; simba1&2 double KO: 0.30-fold n = 15, P = 1.8E-06. E Single NSC quantification of Wg intensity normalized to Dpn in (C), Control:1-fold, n = 127 NSCs; simba1 KO: 0.31-fold, n = 178, P < 1E-15; simba2 KO: 0.17-fold, n = 118, P < 1E-15; simba1/2 double KO: 0.13-fold, n = 192, P < 1E-15. F At 24 h ALH, larval brain lobes were labeled with Sens and Dpn. White arrows point to nucleus of NSCs. G Single NSC quantification of Sens intensity normalized to Dpn in (F), Control:1-fold, n = 130 NSCs; simba1 KO: 0.63-fold, n = 246, P < 1E-15; simba2 KO: 0.42-fold, n = 226, P < 1E-15; simba1/2 double KO: 0.53-fold, n = 224, P < 1E-15. H At 24 h ALH, quantification of wg RNA: Control (repo > β-gal):1-fold, n = 3; repo>simba1: 1.76-fold, n = 3, P = 3.10E-03; repo>simba2: 1.48-fold, n = 3, P = 3.02E-02. senseless RNA: Control:1-fold, n = 3; repo>simba1: 3.12-fold, n = 3, P = 5E-04; repo>simba2: 2.11-fold, n = 3, P = 2.9E-02. I, J Larval NSCs were labeled with Dpn and EdU at 24-h ALH for various genotypes. K Quantifications of EdU-negative NSCs per brain lobe for genotypes in (I) Control: 9.6%, n = 12 BL; simba1 KO: 44.9%, n = 10, P = 1.6E-14; simba1 KO, grh>wgGFP: 24.1%, n = 11, P = 1.1E-07; simba1 KO, NP6293>wgGFP: 26.6%, n = 15, P = 4.3E-07. L Quantifications of EdU-negative NSCs in (J) Control: 7.7%, n = 11 BL; simba2 KO: 46.3%, n = 14, P < 1E-15; simba2 KO, grh>wgGFP: 32.7%, n = 14, P = 1.6E-07; simba2 KO, NP6293>wgGFP: 20.4%, n = 16, P < 1E-15. Data are presented as mean ± SD, and statistical significance was determined by one-way ANOVA with multiple comparisons. Scale bars: 10 μm.
We further performed quantitative PCR (qPCR) to probe wg mRNA level in simba1 KO and simba2 KO brains. Strikingly, wg mRNA levels decreased to 0.32-fold in simba1 KO and 0.30-fold in simba2 KO and to 0.30-fold in the double KO larval brains at 24 h ALH, compared to control (Fig. 5B). Consistent with this observation, Wg protein levels were also drastically reduced to 0.31-, 0.17-, and 0.13-fold in simba1 KO, simba2 KO, and the double KO larval brains, respectively (Fig. 5C–E). In addition, knocking down simba1 in NSCs or simba2 in glial cells is sufficient to reduce Wg protein levels in larval brains (Figure S5A–C). Next, we examined mRNA level of senseless, a well-known Wg target gene in Drosophila imaginal discs47,48, in simba1 KO, simba2 KO, and double KO larval brains at 24 h ALH by qPCR. Depletion of simba1 and simba2 resulted in a dramatic reduction of senseless mRNA level to 0.20-, 0.26-, and 0.15-fold, respectively during NSC reactivation (Fig. 5B). Likewise, Senseless protein level was significantly reduced specifically in NSCs in simba1 KO and simba2 KO brains, suggesting impaired Wg signaling in NSCs (Fig. 5F, G).
Conversely, overexpressing Simba1 and Simba2 by pan glia-specific driver, repo-Gal4, increased wg expression to 1.76- and 1.48- fold, respectively (Fig. 5H). Similarly, at 24 h ALH overexpression of Simba1 and Simba2 in glial cells under repo-Gal4 driver resulted in a dramatic increase in senseless mRNA levels (3.12-fold and 2.11-fold), compared to control (Fig. 5H). Consistent with this result, overexpression of Simba1 or Simba2 in glial cells enhances fluorescent intensity of Wg protein in larval brains at 24 h ALH (Figure S5D, F). Overexpression of Simba1 or Simba2 in NSCs, however, shows a slight but non-significant increase of Wg protein (Figure S5D-E). This observation is consistent with our finding that Wg is translocated from BBB glial cells to NSCs (Fig. 4G). Thus, both Simba1 and Simba2 are critical for the upregulation of wg expression in larval brains.
Simba1 and Simba2 promote qNSC reactivation through Wg/WNT signaling activation
In wgts mutant brains at 24 h ALH, Simba1 and Simba2 protein level exhibit non-significant decrease (0.8-fold) compared to control (Figure S5 G, H), suggesting that Wg has no obvious effect on Simba1 and Simba2 abundance. To determine the genetic epistasis between wg and simba1/2, we overexpressed Wg in simba1 KO brains. At 24-h ALH, the number of EdU-negative NSCs in simba1-depleted brains overexpressing Wg from NSCs was dramatically reduced to 24.1% compared with 44.9% in simba1 KO control brains (Fig. 5I, K). Likewise, Wg overexpression in BBB glia exhibited a similar rescue effect from 44.9% to 26.6% in simba1 KO control brains (Fig. 5I, K). Next, we tested whether Wg overexpression could also suppress simba2 KO phenotype. In simba2 KO brains, overexpressing Wg in BBB glial cells largely rescued reactivation defect caused by simba2 KO from 46.3% to 20.3% in simba2 KO control brains (Fig. 5J, L). Overexpressing Wg in NSCs showed partial, albeit weaker rescue effect on simba2 KO from 46.3% to 32.7% in simba2 KO control brains (Fig. 5J, L). The greater suppression in simba2 KO by Wg overexpression in glial cells than in NSCs supports the notion that Simba2 primarily functions in glial cells. Since Wg overexpression in BBB glial cells and NSCs could rescue simba1 KO and simba2 KO induced NSC reactivation defects, we concluded that Wg signaling is a physiological -relevant target of Simba1 and Simba2 in both cell types for NSC reactivation.
WNT signaling pathway is also a potential target of ZNF706, the human ortholog of Simba1/2
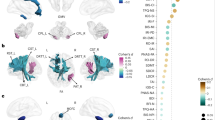
Simba1/2 is highly conserved, as both show 79% amino acid similarity and 64% identity with their mouse and human homologs, Zfp706 and ZNF706, respectively (Fig. 1F), with the latter two sharing identical amino acids. We investigated whether the DNA-binding profile of Simba1 and Simba2 is conserved between Drosophila and humans. We re-analyzed human ChIP-seq data of ZNF706 in a human colon cancer cell line LoVo from a public dataset49. We assessed the DNA-binding profile conservation by comparing KEGG enrichment results between Simba common target genes and ZNF706 target genes. Remarkably, major signaling pathways revealed by Drosophila TaDa, namely WNT signaling, MAPK signaling, Hippo pathway, Lysosome, and Hedgehog pathway were also identified as top enriched pathways in human ZNF706 target genes (Fig. 6A, B). These data suggest that the WNT signaling pathway may be a potential target of ZNF706, the human ortholog of Simba1/2.
A Dot plot of KEGG pathway enrichment results on ZNF706 target genes from ChIP-seq dataset in LoVo cells. B Dot plot of KEGG pathway enrichment results on Simba1 and Simba2 common target genes from Tada (DamID)-seq dataset in Drosophila NSCs at 24-h ALH. C Visualization of ZNF706 target genes enriched in WNT signaling pathway. Created in BioRender. LIN, J. (https://BioRender.com/s1vy3m3). D, E p value and fold enrichment quantifications of ZNF706 targeting WNT signaling key components. F A list of Simba1/Simba2/ZNF706 commonly targeted WNT pathway related genes from Simba Tada (DamID)-seq data and ZNF706 ChIP-seq data. G Quantitative PCR quantification of ZNF706 mRNA levels in STF3A cells: siNT: 1.00-fold; siZNF706#1: 0.20-fold; siZNF706#2: 0.27-fold; siNT+BIO: 1.39-fold; siZNF706#1 + BIO: 0.23-fold; siZNF706#2 + BIO: 0.19-fold. BIO: 6-Bromoindirubin-3’-oxime, GSK3 inhibitor. Experiments were performed in 3 batches, each with 3 technical replicates. H Luciferase activity of WNT reporter normalized to cell viability in STF3A cells: siNT: 1.00-fold; siZNF706#1: 0.39-fold, P = 1.4E-05; siZNF706#2: 0.40-fold, P = 2.0E-05; siNT+BIO: 4.15-fold, P < 1E-15; siZNF706#1 + BIO: 2.17-fold, P = 7.2E-13; siZNF706#2 + BIO: 1.42-fold, P = 2.9E-03. Experiments were performed in 3 batches, each with 3 technical replicates. Data are presented as mean ± SD, and statistical significance was determined by one-way ANOVA with multiple comparisons. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
ZNF706, the human ortholog of Simba, is required for WNT signaling activation in human STF3A cells
Simba1 and Simba2 share 79.1% amino acid sequence similarity with their human homolog ZNF706. Mouse homolog Zfp706 and human homolog ZNF706 have identical protein sequences. Alpha-fold structure prediction of Simba and ZNF706 yielded nearly identical structural models, suggesting that their structures might be conserved (Figure S6A). Given this conservation, we investigated whether ZNF706, the human ortholog of Simba, also regulates WNT signaling in human cells. Firstly, we further analyzed individual target genes under WNT signaling KEGG ontology that were targeted by ZNF706 (Fig. 6C). We found that multiple core components of WNT signaling were predicted to be targeted by ZNF706 including WNT ligands, FZD receptors, LRP5, GSK3B, CK1alpha and TCF7/LEF1 (Fig. 6C-E). Moreover, a considerable portion of ZNF706 target genes in WNT signaling appear to be Simba target genes in Drosophila as well, indicating a strong conservation of Simba in regulating WNT signaling (Fig. 6F).
To test whether human ZNF706 regulates WNT signaling, we performed a superTOPFlash assay that measures β-catenin transcriptional activity using STF3A cells that contain an integrated superTOPFlash luciferase reporter and stable expression of murine Wnt3A in HEK293 cells, as previously described50. Knockdown efficiency of ZNF706 siRNAs were evaluated and validated by qPCR (Fig. 6G). In STF3A cells, knocking down ZNF706 strongly reduced WNT signaling activity to 0.39- and 0.40-fold in two independent siRNAs targeting ZNF706 (Fig. 6H). This result suggests that human ZNF706 is required for WNT signaling in STF3A cells.
To determine whether the reporter activity reduction upon ZNF706 is due to impaired WNT signaling activity, STF3A cells were treated with BIO, a GSK3 inhibitor in ZNF706 knockdown condition51. As expected, this treatment exhibited elevated WNT signaling reporter activity compared to control (Fig. 6H). Importantly, BIO treatment reversed the effect of ZNF706 knockdown on the reduction of WNT signaling reporter activity (Fig. 6H). Therefore, ZNF706 has a conserved function in activation of WNT signaling activity in human STF3A cells (Fig. 7).
In the Drosophila larval brain, Simba1 and Simba2 are required for NSC reactivation by transcriptionally activating Wingless in BBB glia and NSCs. Human homolog ZNF706 might be required for activating WNT signaling in colon cancer cells and HEK293T cells. Created in BioRender. LIN, J. (https://BioRender.com/s1vy3m3).
Discussion
This study provides insights into the function of two conserved microprotein/peptide paralogs, Simba1 and Simba2, in Drosophila. Specifically, we demonstrated their function in NSCs and BBB glial cells in promoting the reactivation of quiescent NSCs. We have also uncovered the conserved WNT/Wg pathway as a developmental signaling pathway that is required for Drosophila NSC reactivation. Simba1/2 directly bind to the promoter region of the wg gene, promoting its expression in Drosophila NSCs. Our genetic analysis indicated that wg is a physiological relevant target of Simba1/2 in Drosophila NSC reactivation (Fig. 7). Bioinformatic anlysis in human colon cancer cells suggest that this regulation between ZNF706, the human homolog of Simba/12, and Wg is likely conserved (Fig. 7). The Simba1/2-Wg axis may be applied to other cellular processes in development and diseases.
The function of microproteins in brain development
Among more than 900 microproteins in the Drosophila genome, only a handful of those have been investigated experimentally36,52,53. In this study, we identified Simba1 and Simba2 as the first two micropeptides that function in NSC reactivation during Drosophila brain development. Notably, similar to the tandemly duplicated sloth1/sloth2 sORFs54, Simba1/2 paralogs exhibit non-redundant roles despite their sequence similarities. However, Sloth1 and Sloth2 physically interact54, whereas Simba1 and Simba2 do not. We speculate that Simba1 and Simba2 might interact with distinct co-factors to regulate gene expression in BBB glial cells and NSCs, contributing to their non-redundant functions. Simba1/2 and ZNF706 display sequence, structure and functional similarities, highlighting a great conservation of these proteins during evolution. Increasing evidence suggests that mammalian Zfp706 /ZNF706 play a role in stem cell development. Mouse Zfp706 is required for the cell fate transitions in embryonic stem cells55, while zebrafish Znf706 regulates development of primordial germ cells, precursors of germline stem cells56. Simba1/2 contain an N-terminal SERF domain (1-40 aa) that was recently shown to participate in protein aggregation57,58,59,60,61, C-terminal zinc-finger domain, and an IDR domain at the C-terminal end. Based on the binding of Simba1/2 to the promoter region of wg locus, this zinc-finger domain likely mediates the binding to DNA sequence. The function of the IDR domain in Simba1/2 remains to be investigated. The involvement of the Simba1/2 homolog ZNF706 in amyloid and a-synuclein aggregation suggests ZNF706 may be a target of neurodegenerative diseases62. Since the SERF domain is either mostly present in Simba1 KO (removing 5 aa) or intact in Simba2 KO, the SERF domain may not be critical for NSC reactivation. Our study in Simba1/2 demonstrated that they have an earlier function during brain development, which is likely independent of the potential function in protein aggregation. Interestingly, many microproteins have enriched expression in the human brain35,63,64. The function of mammalian homolog of Simba1/2 and other microproteins in brain development and function warrants further investigation.
WNT/Wg signaling in NSC reactivation
It is well-established that InR/mTor/Akt and Hippo pathways regulate Drosophila NSC reactivation at the BBB glial cells, the major niche for NSCs13,14,19,20,21,65. It is of great interest to identify other signaling pathways that control NSC reactivation. Here, we have shown that WNT/Wg signaling pathway functions in both BBB glial cells and NSCs and plays a critical role in NSC reactivation. Consistent with this finding, Drosophila Wg pathway transcriptional co-activator Ebd1 is required for NSC reactivation27. Since canonical WNT/Wg signaling is required for NSC reactivation, future study on the target genes of the transcription factor β-catenin will be important to elucidate how Wg pathway regulates NSC reactivation.
Wg protein is readily detected in the Drosophila larval brain upon larval hatching and quickly diminished by 24 h ALH, suggesting a transient Wg signaling for NSC reactivation. Interestingly, the ligand Wg is at first present in BBB glial cells and can be translocated to NSCs within 2 h, suggesting that BBB glial cells may serve as a potential source of Wg. Wg can be secreted by Golgi, diffuse and form a gradient for long-range signaling in the Drosophila wing disc or secreted via extracellular vesicles for short-range signaling66,67,68. Given that BBB glial cells and NSCs are juxtaposed, Wg likely translocates from BBB glial cells to NSCs via extracellular vesicles.
The WNT signaling pathway is known to regulate mouse brain development and patterning69,70,71, as well as NSC stemness, self-renewal, and differentiation72,73,74. Moreover, WNT signaling has been reported to regulate the reactivation of mouse adult quiescent NSCs in the hippocampus75,76 and axonal regeneration, guidance and differentiation77,78,79. Interestingly, although Wg is not normally expressed in adult neurons, injury can induce its expression to activate quiescent NSCs in the Drosophila adult optic lobe80, supporting its role in neuronal regeneration. Therefore, the role of the WNT/Wg signaling pathway in brain development, NSC reactivation, and regeneration is likely to be conserved between Drosophila and mouse.
Microproteins Simba1/2/ZNF706 are conserved regulators of WNT/Wg signaling
Simba1/2 contain a single zinc-finger domain, which may have diverse functions in DNA and RNA binding, protein-protein interactions, and etc. Here, we demonstrated that Simba1/2 are capable of binding to DNA by NSC-specific DamID analysis in vivo and ChIP experiments in vitro. Their nuclear localization in NSCs and glial cells also supports this role. Human ZNF706 also has DNA binding activity as shown in assays including ChIP and Cut & Tag56,62,81, which is consistent with our luciferase assay. Therefore, Simba1, Simba2, and their human homolog ZNF706, likely function as transcription factors. Consistent with this finding, ZNF706 has been shown recently to directly target SLC7A11 expression in human hepatocellular carcinoma81.
We have identified Wg signaling as a direct target of Simba1 and Simba2 in Drosophila NSCs and provided evidence that ZNF706 has a conserved function in regulating WNT signaling in human cells. The in vivo function of mammalian ZNF706/Zfp706 remains to be investigated. Besides WNT/Wg pathway, other key developmental signaling pathways such as MAPK signaling and Hippo signaling are also potential targets of Simba1/2/ZNF706. Given that WNT/Wg signaling is one of the key pathways that plays crucial roles in numerous cellular processes, including development and homeostasis, and has a strong association with diseases such as cancer and neurological disorders, the link between Simba1/2/ZNF706 and Wnt/Wg pathway will have extensive impact on understanding these critical processes from flies to humans.
Limitations of study
Besides Wg, other components of WNT/Wg signaling, such as Wnt10 and Fz, have also been identified as potential targets of Simba1/2 based on in vivo protein-DNA profiling. However, their functional validation in vivo remains to be conducted. Given that there are more than ten such candidates, they will be validated in Drosophila in future studies. WNTs are typically expressed at low levels across various tissues, often falling below detection thresholds82. This has posed a challenge in analyzing WNTs expression and function in different cell types. Human STF3A cells -HEK293 cells that stably express TCF/LEF1 luciferase reporter and murine Wnt3A - have been used to assess whether ZNF706 is required for activation of WNT signaling. While our results support such a role for ZNF706, the specific WNT ligands regulated by ZNF706 in human cells remain unclear. Given that humans have 19 WNTs, each likely exhibits cell-type-specific expression, future work is warranted to determine how ZNF706 regulates these WNTs, particularly in NSCs. Since mammalian WNT1, WNT3A, WNT7A, WNT8, WNT11 have known functions during various stages of brain development83,84, some of these WNTs may represent conserved targets of ZNF706. In our superTOPFlash assay, BIO treatment is not able to fully reverse WNT signaling activity upon ZNF706 knockdown, suggesting that ZNF706 might also regulate WNT pathway components downstream of GSK3.
Methods
Fly stocks and culture
Drosophila stocks were cultured at 25 °C on standard medium unless otherwise stated. Yellow white (yw) and UAS–β-gal RNAi were used as controls for most experiments. Most experiments were carried out at 25 °C, except for RNAi knockdown and overexpression that were performed at 29 °C, unless otherwise indicated.
The following fly strains were used in this study: grh-GAL4 (Andrea Brand), UAS-LT3-Ndam (Andrea Brand), UAS-Dam-Simba1 (generated in this work), UAS-Dam-Simba2 (generated in this work), wgKO-Gal4 (J. P Vincent), UAS-WgGFP (J. P Vincent), simba1 KO (Norbert Perrimon), simba2 KO (Norbert Perrimon) simba1-simba2 dKO (Norbert Perrimon), Df(3 L)BSC575 (simba1/2 deficiency; BDSC, 27587), repo-GAL4 (BDSC, #7415), wgts (BDSC, #7000), wgI-16 (BDSC, #2977), fz21 (BDSC#41787), dsh75 (BDSC, #68165), dsh3 (BDSC, #5299), UAS-arm RNAi #3 (BDSC, #35004), fz1 RNAi (BDSC, #31311), fz2 RNAi (BDSC, #31390), dsh RNAi (BDSC, #31307), UAS-SggS9A (BDSC, #5255) UAS-CD8GFP (BDSC, #5137), nSyb-Gal4 (BDSC, #51635), nrv2-GAL4 (BDSC, #6797), alrm-GAL4 (BDSC, #67032), UAS-simba1RNAi (VDRC, #108387), UAS-simba2RNAi (VDRC, #106387), UAS-arm RNAi #1 (VDRC, #107344), UAS-arm RNAi #2 (VDRC, #7767), wls RNAi (VDRC, #5215), wls RNAi II (VDRC, #5214), wg RNAi #1 (VDRC, #13352), wg RNAi #2 (VDRC, #104579), NP2276-Gal4 (DGRC, #112853) and NP6293-Gal4 (DGRC, #105188), UAS-Simba1CC (FlyORF, #F003456), UAS-Simba1HA (FlyORF, #F000909), UAS-Simba2HA (FlyORF, #F003792).
RNAi Knockdown
Transgenic flies carrying a UAS–RNAi construct targeting the gene of interest were crossed with driver lines expressing GAL4 under a cell type–specific promoter, together with Dcr2 to enhance RNAi efficiency. Crosses were maintained at 25 °C for 4 h to allow synchronized egg laying, after which the adults were removed. 24 h later, newly hatched larvae were shifted to 29 °C. At 6 h or 24 h ALH, brains were dissected for downstream analysis. For simba1/2 knockdown experiments, we used UAS–simba1 RNAi (VDRC #108387) and UAS–simba2 RNAi (VDRC #106387), respectively. The simba1 RNAi targets the 3′ UTR of the simba1 transcript (nucleotides 1352–1570), while simba2 RNAi targets the 3′ UTR of the simba2 transcript (nucleotides 1064–1433).
Immunohistochemistry
Drosophila larvae were dissected in PBS, and the larval brains were fixed in 4% EM-grade formaldehyde in PBT (PBS + 0.3% Triton-100) for 22 min. The samples were processed for immunostaining as previously described by Li et al. DNA was labeled with DAPI (1:1,500, Sigma, 28718-90-3). The images were collected on a Zeiss LSM 710 confocal microscope (Axio Observer Z1, ZEISS) using a Plan-Apochromat 40×/1.3NA (numerical aperture) oil differential interference contrast objective. They were then processed with Zen software (2010 version), Fuji ImageJ.
The primary antibodies used in this paper, were guinea pig anti-Dpn (1:1000), rabbit anti-Dpn (1:200), mouse anti-Mira (1:50; F. Matsuzaki), rabbit anti-GFP (1:3000; F. Yu), guinea pig anti-Sens (1:1000; H. Bellen), mouse anti-GFP (1:3000; F. Yu), rabbit anti-Msps (1:500, J. Raff), rabbit anti-HA (1:1000, Sigma-Aldrich, H6908), rabbit anti-Simba (1:200, generated in this work by AbMart), mouse anti-Repo (1:20; DSHB, 8D12), mouse anti-Wg (1:10, DSHB, 4D4). The secondary antibodies used were conjugated with Alexa Fluor 488, 555 or 647 (Jackson laboratory).
EdU incorporation assay
Larvae of various genotypes were fed with food supplemented with 0.2 mM EdU from ClickiT® EdU Imaging Kits (Invitrogen) for 4 h. The larval brains were dissected in PBS and fixed with 4% EM-grade formaldehyde in PBS for 22 min, followed by washing thrice (each wash for 10 min) with 0.3% PBST, and blocked with 3% BSA in PBST for 30 min. The incorporated EdU was detected by Alexa Fluor azide, according to the Click-iT EdU protocol (Invitrogen). The brains were rinsed twice and subjected to standard immunohistochemistry.
Antibody generation
Simba antibody was generated by AbMart (Shanghai, China). Full length of Simba2 coding sequence was used to synthesize polypeptides as immunogen and then injected to Rabbit. The antibody was then subjected to affinity purification to obtain purified polyclonal Simba antibodies.
Quantitative PCR
25-30 first instar larvae per group were dissected in cell culture medium, and RNA from brains was extracted using TRI reagent (Sigma-Aldrich) following the manufacturer protocol. ProtoScript First Strand cDNA Synthesis kit (New England Biolabs, Inc.) was used for reverse transcription. qPCR was carried out with different primer pairs. The primers used are listed in the key resource table. Reference genes used as an internal control were as follows: rp49/Rpl32(ribosomal protein L32), Sdh (succinate dehydrogenase), and Tbp1/Rpt5 (regulatory particle triple-A adenosine triphosphatase 5).
For human cells, total mRNA was extracted by RNA isolation kit V2 (Vazyme). HiScript II Reverse Transcriptase (Vazyme) was used for reverse transcription. PCR was carried out with different primer pairs. The primers used are listed below. Reference gene used as an internal control was ACTIN.
The primers pairs used for RT-qPCR for Drosophila genes were the following:
wg forward: 5’- TCAGGGACGCAAGCATAATAG-3’,
wg reverse: 5’- CGAAGGCTCCAGATAGACAAG-3’.
senseless forward: 5’- CCGAAAAGGAGCATGAACTC-3’,
senseless reverse: 5’- CGCTGTTGCTGTGGTGTACT-3’.
rp49 forward: 5’-TGTCCTTCCAGCTTCAAGATGACCATC-3’,
rp49 reverse: 5’-CTTGGGCTTGCGCCATTTGTG-3’.
sdh forward: 5’-GTCTGAAGATGCAGAAGACC-3’,
sdh reverse: 5’-ACAATAGTCATCTGGGCATT-3’.
Tbp-1 forward: 5’-AAGCCCGTGCCCGTATTATG-3’,
Tbp-1 reverse: 5’-AAGTCATCCGTGGATCGGGAC-3’.
actin5C forward 5’- GAGCGCGGTTACTCTTTCAC −3’
actin5C reverse 5’- GCCATCTCCTGCTCAAAGTC −3’.
The primers pairs used for RT-qPCR for human genes were the following:
ZNF706 forward: 5’- CACAAATGCCAGACCCTAAGA −3’,
ZNF706 reverse: 5’- GCTAATTCTGGAGGAAGTGGAG −3’.
ACTIN forward: 5’- AAGGATTCCTATGTGGGCGACG-3’,
ACTIN reverse: 5’- GCCTGGATAGCAACGTACATGG-3’.
Generation of DamID plasmids and transgenic flies
In-fusion cloning (Takara) was used for the generation of UAS-Dam-Simba1 and UAS-Dam-Simba2 plasmids. CDS sequences of simba1 and simba2 were inserted into pUAST attB-LT3-NDam vectors (a gift from Andrea Brand). Purified plasmids were then sent for P-element-mediated transformation and injected into w1118 embryos (BestGene lnc) to obtain UAS-NDam-Simba1 and UAS-NDam-Simba2 transgenic flies.
Primers used in cloning were the following:
Simba1 Dam cloning forward: 5’- CGCAGATCTGCGGCCGATGGCACGTGGACAC −3’,
Simba1 Dam cloning reverse: 5’- CCTCGAGCCGCGGCCGCTCAGACCTCCTTCAG −3’.
Simba2 Dam cloning forward: 5’- CGCAGATCTGCGGCCGATGGCACGTGGACAC-3’,
Simba2 Dam cloning reverse: 5’- CCTCGAGCCGCGGCCGCTCAGACCTCCTTCAG-3’.
TaDa in vivo profiling
To identify in vivo Simba1/2-bound DNA fragments, UASNDam-Simba1/2) or UAS-Dam control (pUAST attB-LT3-NDam) was expressed under the control of grh-Gal4, tub-Gal80ts driver. Embryos were collected in a 4 h period and transferred into 18 °C (restrictive temperature) for 44 h till hatching and then shifted into 29 °C for 24 h before dissection. Genomic DNA of Dam-Simba1/2 or Dam control was extracted and amplified as previously described85. Libraries were constructed from the amplified DNA samples using the illumina TruSeq DNA PCR-free library preparation kit according to the manufacturer’s protocol and sequenced on the illumina MiSeq equipment. Paired reads (2 × 250 bp) were sequenced from Dam-Simba1/2 or Dam-Control library. Raw sequencing reads were quality-checked by FastQC86 and were aligned on Drosophila melanogaster (dm6) reference genome using Bowtie 287. The peak calling was done by MACS388, and the nearest genes were annotated using ChIPseeker89. Two biological replicates were performed for the profiling and analysis. The genome-wide correlation analysis between replicates were performed by Galaxy software90. The genome is divided into bins (1 kb), and the Tada peak signal in each bin from each dataset is counted. These numbers are then compared using Pearson correlation coefficients that reflect global similarity.
TaDa downstream analysis
Nearest genes from Simba1 and Simba2 all replicates were filtered by the following criteria: p value < 0.001 and fold change>2. Venn diagram analysis were performed between four replicates by R91 in Rstudio92. Simba1 and Simba2 nearest genes in both replicates were then subjected to KEGG enrichment analysis by clusterProfiler93. Results were visualized by Dot plot. Binding peaks profiles of Simba1 and Simba2 near wg promoter region were visualized by Integrative Genomics Viewer (IGV)94. Fold change of Simba1/2 binding peaks signals were normalized to control (Dam).
ChIP-qPCR assay
The ChIP assay was performed using Drosophila S2 cells according to the manufacturer’s protocol (Cell Signaling, #9005). Cells were transfected by Flag-tagged Simba1 and Simba2, with empty Flag as a control. Rabbit-anti Flag antibodies (Sigma-Aldrich, F1804-1MG) were used to pull down the chromatin-protein complex, and 1% of the precipitated DNA was used for real-time qPCR per reaction. To validate Dam-Simba1/2-binding fragments, primers for ChIP-qPCR are designed to target the peaks that located in proximity to the wg transcription start site.
The primers pairs used for RT-qPCR for human genes were the following:
Intergenic control forward: 5’- CTGCAGTGAAGGAACGATAGAG −3’,
Intergenic control reverse: 5’- CTTGAACCCGAATCCGATGT −3’.
wg promoter 1 forward: 5’- CAAAGCTATGACGAAATTCATGAGG-3’,
wg promoter 1 reverse: 5’- CCCAATAAGAAATGGCGGAAATG-3’.
wg promoter 2 forward: 5’- CAATGCAGGAGTCAGGGTATAG −3’,
wg promoter 2 reverse: 5’- CATCGTCGGTCTTCGTCTTC-3’.
wg promoter 3 forward: 5’- GCGAGCCGAGTGTATTCTAT-3’,
wg promoter 3 reverse: 5’- GCCGATATTGCTGCTCATATTT-3’.
Cell lines and transfection
STF3A cell (Cellosaurus, CVCL_C6NQ, stably express WNT signaling reporter and murine Wnt3a)95 was used for super top-flash assay. Cells at 70–80% confluency were transfected using a calcium phosphate-DNA precipitate prepared by mixing DNA with 2× HEPES-buffered saline and 0.25 M CaCl₂. The mixture was incubated for 20 minutes to form a precipitate and added dropwise to cells. After 16 h, the media was replaced, and cells were harvested for analysis 48 h later.
Super TOP flash assay
The Super TOP Flash assay was performed to assess Wnt/β-catenin signaling activity. STF3A cells were seeded in 6-well plates and after 24 h were transfected with ZNF706 siRNAs (50 nM) or non-targeting siRNA (50 nM) as control for 16 h. Luciferase activity was measured using the ONE-Glo™ Luciferase Assay System (Promega) and normalized to cell viability by TC20 Automated Cell Counter (Bio-Rad). Data were reported as fold changes relative to non-targeting control. For GSK3 inhibitor treatment, cells were treated with BIO (6-Bromoindirubin-3’-oxime, 500 nM) 16 h after siRNA transfection. siRNAs used in this work were the following: siNT (Sigma-Aldrich, SIC001), siZNF706#1 (Sigma-Aldrich, #0239), siZNF706#2 (Sigma-Aldrich, #0249).
Western blot
Drosophila larval brains were dissected at 24-h ALH and were collected and lysed using PierceTM RIPA buffer (Thermo Fisher Scientific, cat no: 89901) with protease inhibitors. The proteins samples were separated using SDS-PAGE and were transferred onto the nitrocellulose membrane. The membranes were blocked with low-fat dry milk in PBS with 0.1% Tween20 (PBST) for 1 h at RT. Subsequently, the membranes were incubated with respective primary antibodies in 5% BSA with PBST overnight at 4 °C. The membranes were washed thrice with PBST and incubated with HRP-conjugated secondary antibodies to probe the target proteins for 1 h at RT. The membranes were washed and the proteins were detected using SuperSignal West Pico Chemiluminescence Substrate (Protein Biology, cat no: 34580). Antibody used in western blot were: mouse anti-Actin (1:2000; DSHB, JLA20), rabbit anti-Simba (1:500, generated in this work by AbMart).
Quantification and statistical analysis
Statistical analysis was performed using GraphPad Prism 10. P values were calculated from two-tailed unpaired Student’s t test for comparison of two samples. One-way analysis of variance (ANOVA), followed by Sidak’s multiple comparisons test, was used for comparison of more than two sample groups. In all graphs, ∗indicates 0.05 < p < 0.01, ∗∗indicates p < 0.01, ∗∗∗indicates p < 0.001, ∗∗∗∗indicates p < 0.0001, and ns indicates p > 0.05. Cell numbers were quantified and represented as mean ± SD and/or percentage of phenotype. Each experiment has at least three independent replications unless otherwise stated.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and the source data for figures are provided with the paper. TaDa Dam-ID sequencing dataset generated in this work has been deposited in Gene Expression Omnibus (GEO) with accession number: GSE290649. Source data are provided with this paper.
References
Morshead, C. M. et al. Neural stem cells in the adult mammalian forebrain: a relatively quiescent subpopulation of subependymal cells. Neuron 13, 1071–1082 (1994).
Doetsch, F., Caillé, I., Lim, D. A., García-Verdugo, J. M. & Alvarez-Buylla, A. Subventricular zone astrocytes are neural stem cells in the adult mammalian brain. Cell 97, 703–716 (1999).
Fabel, K. & Kempermann, G. Physical activity and the regulation of neurogenesis in the adult and aging brain. Neuromolecular Med. 10, 59–66 (2008).
Cloëtta, D. et al. Inactivation of mTORC1 in the developing brain causes microcephaly and affects gliogenesis. J. Neurosci. 33, 7799–7810 (2013).
Baser, A., Skabkin, M. & Martin-Villalba, A. Neural stem cell activation and the role of protein synthesis. Brain Plast. 3, 27–41 (2017).
Ding, W. Y., Huang, J. & Wang, H. Waking up quiescent neural stem cells: molecular mechanisms and implications in neurodevelopmental disorders. PLoS Genet 16, e1008653 (2020).
Otsuki, L. & Brand, A. H. Quiescent neural stem cells for brain repair and regeneration: lessons from model systems. Trends Neurosci. 43, 213–226 (2020).
Isshiki, T., Pearson, B., Holbrook, S. & Doe, C. Q. Drosophila neuroblasts sequentially express transcription factors which specify the temporal identity of their neuronal progeny. Cell 106, 511–521 (2001).
Tsuji, T., Hasegawa, E. & Isshiki, T. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Dev. Camb. Engl. 135, 3859–3869 (2008).
Lai, S.-L. & Doe, C. Q. Transient nuclear Prospero induces neural progenitor quiescence. eLife 3, e03363 (2014).
Otsuki, L. & Brand, A. H. Cell cycle heterogeneity directs the timing of neural stem cell activation from quiescence. Science 360, 99–102 (2018).
Britton, J. S. & Edgar, B. A. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Dev. Camb. Engl. 125, 2149–2158 (1998).
Chell, J. M. & Brand, A. H. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143, 1161–1173 (2010).
Sousa-Nunes, R., Yee, L. L. & Gould, A. P. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471, 508–512 (2011).
Gujar, M. R. & Wang, H. A fly’s eye view of quiescent neural stem cells. Oxf. Open Neurosci. 1, kvac001 (2022).
Spéder, P. & Brand, A. H. Gap junction proteins in the blood-brain barrier control nutrient-dependent reactivation of Drosophila neural stem cells. Dev. Cell 30, 309–321 (2014).
Arsenijevic, Y., Weiss, S., Schneider, B. & Aebischer, P. Insulin-like growth factor-I is necessary for neural stem cell proliferation and demonstrates distinct actions of epidermal growth factor and fibroblast growth factor-2. J. Neurosci. J. Soc. Neurosci. 21, 7194–7202 (2001).
Juanes, M. et al. Three novel IGF1R mutations in microcephalic patients with prenatal and postnatal growth impairment. Clin. Endocrinol. (Oxf.) 82, 704–711 (2015).
Huang, J., Wu, S., Barrera, J., Matthews, K. & Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 122, 421–434 (2005).
Ding, R., Weynans, K., Bossing, T., Barros, C. S. & Berger, C. The Hippo signalling pathway maintains quiescence in Drosophila neural stem cells. Nat. Commun. 7, 10510 (2016).
Poon, C. L. C., Mitchell, K. A., Kondo, S., Cheng, L. Y. & Harvey, K. F. The hippo pathway regulates neuroblasts and brain size in Drosophila melanogaster. Curr. Biol. CB 26, 1034–1042 (2016).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205 (2012).
MacDonald, B. T., Tamai, K. & He, X. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17, 9–26 (2009).
Bejsovec, A. Wingless/Wnt signaling in Drosophila: the pattern and the pathway. Mol. Reprod. Dev. 80, 882–894 (2013).
Nusse, R. Wnt signaling in disease and in development. Cell Res 15, 28–32 (2005).
Bejsovec, A. Wingless signaling: a genetic journey from morphogenesis to metastasis. Genetics 208, 1311–1336 (2018).
Huang, J. et al. Histone lysine methyltransferase Pr-set7/SETD8 promotes neural stem cell reactivation. EMBO Rep. 22, e50994 (2021).
Saghatelian, A. & Couso, J. P. Discovery and characterization of smORF-encoded bioactive polypeptides. Nat. Chem. Biol. 11, 909–916 (2015).
Martinez, T. F. et al. Accurate annotation of human protein-coding small open reading frames. Nat. Chem. Biol. 16, 458–468 (2020).
Aspden, J. L. et al. Extensive translation of small open reading frames revealed by Poly-Ribo-Seq. eLife 3, e03528 (2014).
Andrews, S. J. & Rothnagel, J. A. Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15, 193–204 (2014).
Couso, J.-P. & Patraquim, P. Classification and function of small open reading frames. Nat. Rev. Mol. Cell Biol. 18, 575–589 (2017).
Merry, T. L. et al. Mitochondrial-derived peptides in energy metabolism. Am. J. Physiol. Endocrinol. Metab. 319, E659–E666 (2020).
Budamgunta, H. et al. Comprehensive peptide analysis of mouse brain striatum identifies novel sORF-encoded polypeptides. Proteomics 18, e1700218 (2018).
Duffy, E. E. et al. Developmental dynamics of RNA translation in the human brain. Nat. Neurosci. 25, 1353–1365 (2022).
Bosch, J. A. et al. Molecular and functional characterization of the Drosophila melanogaster conserved smORFome. Cell Rep. 42, 113311 (2023).
Brunet Avalos, C., Maier, G. L., Bruggmann, R. & Sprecher, S. G. Single cell transcriptome atlas of the Drosophila larval brain. eLife 8, e50354 (2019).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res 51, D418–D427 (2023).
Tunyasuvunakool, K. et al. Highly accurate protein structure prediction for the human proteome. Nature 596, 590–596 (2021).
Leon, O. & Roth, M. Zinc fingers: DNA binding and protein-protein interactions. Biol. Res. 33, 21–30 (2000).
Laity, J. H., Lee, B. M. & Wright, P. E. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 11, 39–46 (2001).
Nüsslein-Volhard, C., Kluding, H. & Jürgens, G. Genes affecting the segmental subdivision of the Drosophila embryo. Cold Spring Harb. Symp. Quant. Biol. 50, 145–154 (1985).
Hazelett, D. J., Bourouis, M., Walldorf, U. & Treisman, J. E. decapentaplegic and wingless are regulated by eyes absent and eyegone and interact to direct the pattern of retinal differentiation in the eye disc. Dev. Camb. Engl. 125, 3741–3751 (1998).
Bänziger, C. et al. Wntless, a conserved membrane protein dedicated to the secretion of Wnt proteins from signaling cells. Cell 125, 509–522 (2006).
Alexandre, C., Baena-Lopez, A. & Vincent, J.-P. Patterning and growth control by membrane-tethered Wingless. Nature 505, 180–185 (2014).
modENCODE Consortium et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 (2010).
Franch-Marro, X. et al. Wingless secretion requires endosome-to-Golgi retrieval of Wntless/Evi/Sprinter by the retromer complex. Nat. Cell Biol. 10, 170–177 (2008).
Nolo, R., Abbott, L. A. & Bellen, H. J. Senseless, a Zn finger transcription factor, is necessary and sufficient for sensory organ development in Drosophila. Cell 102, 349–362 (2000).
Yan, J. et al. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154, 801–813 (2013).
Coombs, G. S. et al. WLS-dependent secretion of WNT3A requires Ser209 acylation and vacuolar acidification. J. Cell Sci. 123, 3357–3367 (2010).
Meijer, L. et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem. Biol. 10, 1255–1266 (2003).
Hoskins, R. A. et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25, 445–458 (2015).
Chanut-Delalande, H. & Zanet, J. Small ORFs, big insights: Drosophila as a model to unraveling microprotein functions. Cells 13, 1645 (2024).
Bosch, J. A. et al. Two neuronal peptides encoded from a single transcript regulate mitochondrial complex III in Drosophila. eLife 11, e82709 (2022).
Leeb, M., Dietmann, S., Paramor, M., Niwa, H. & Smith, A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem Cell 14, 385–393 (2014).
Zhang, W. et al. Znf706 regulates germ plasm assembly and primordial germ cell development in zebrafish. J. Genet. Genomics 52, 666–679 (2025).
Sahoo, B. R. & Bardwell, J. C. A. SERF, a family of tiny highly conserved, highly charged proteins with enigmatic functions. FEBS J. 290, 4150–4162 (2023).
Falsone, S. F. et al. SERF protein is a direct modifier of amyloid fiber assembly. Cell Rep. 2, 358–371 (2012).
Pras, A. et al. The cellular modifier MOAG-4/SERF drives amyloid formation through charge complementation. EMBO J. 40, e107568 (2021).
Meinen, B. A., Gadkari, V. V., Stull, F., Ruotolo, B. T. & Bardwell, J. C. A. SERF engages in a fuzzy complex that accelerates primary nucleation of amyloid proteins. Proc. Natl Acad. Sci. USA. 116, 23040–23049 (2019).
Balasubramaniam, M., Ayyadevara, S. & Shmookler Reis, R. J. Structural insights into pro-aggregation effects of C. elegans CRAM-1 and its human ortholog SERF2. Sci. Rep. 8, 14891 (2018).
Sahoo, B. R. et al. Protein G-quadruplex interactions and their effects on phase transitions and protein aggregation. Nucleic Acids Res. 52, 4702–4722 (2024).
Sandmann, C.-L. et al. Evolutionary origins and interactomes of human, young microproteins and small peptides translated from short open reading frames. Mol. Cell 83, 994–1011 (2023).
Duffy, E. E., Assad, E. G., Kalish, B. T. & Greenberg, M. E. Small but mighty: the rise of microprotein biology in neuroscience. Front. Mol. Neurosci. 17, 1386219 (2024).
Liu, J., Spéder, P. & Brand, A. H. Control of brain development and homeostasis by local and systemic insulin signalling. Diabetes Obes. Metab. 16, 16–20 (2014).
Beckett, K. et al. Drosophila S2 cells secrete wingless on exosome-like vesicles but the wingless gradient forms independently of exosomes. Traffic Cph. Den. 14, 82–96 (2013).
Mulligan, K. A. et al. Secreted Wingless-interacting molecule (Swim) promotes long-range signaling by maintaining Wingless solubility. Proc. Natl Acad. Sci. Usa. 109, 370–377 (2012).
Swarup, S. & Verheyen, E. M. Wnt/wingless signaling in Drosophila. Cold Spring Harb. Perspect. Biol. 4, a007930 (2012).
McMahon, A. P., Joyner, A. L., Bradley, A. & McMahon, J. A. The midbrain-hindbrain phenotype of Wnt-1-/Wnt-1- mice results from stepwise deletion of engrailed-expressing cells by 9.5 days postcoitum. Cell 69, 581–595 (1992).
Lee, S. M., Tole, S., Grove, E. & McMahon, A. P. A local Wnt-3a signal is required for development of the mammalian hippocampus. Dev. Camb. Engl. 127, 457–467 (2000).
Stuebner, S., Faus-Kessler, T., Fischer, T., Wurst, W. & Prakash, N. Fzd3 and Fzd6 deficiency results in a severe midbrain morphogenesis defect. Dev. Dyn. Publ. Am. Assoc. Anat. 239, 246–260 (2010).
Kalani, M. Y. S. et al. Wnt-mediated self-renewal of neural stem/progenitor cells. Proc. Natl Acad. Sci. 105, 16970–16975 (2008).
Munji, R. N., Choe, Y., Li, G., Siegenthaler, J. A. & Pleasure, S. J. Wnt signaling regulates neuronal differentiation of cortical intermediate progenitors. J. Neurosci. J. Soc. Neurosci. 31, 1676–1687 (2011).
Bengoa-Vergniory, N. & Kypta, R. M. Canonical and noncanonical Wnt signaling in neural stem/progenitor cells. Cell. Mol. Life Sci. CMLS 72, 4157–4172 (2015).
Varela-Nallar, L. & Inestrosa, N. C. Wnt signaling in the regulation of adult hippocampal neurogenesis. Front. Cell. Neurosci. 7, 100 (2013).
Piccin, D. & Morshead, C. M. Wnt signaling regulates symmetry of division of neural stem cells in the adult brain and in response to injury. Stem Cells Dayt. Ohio 29, 528–538 (2011).
Patel, A. K., Park, K. K. & Hackam, A. S. Wnt signaling promotes axonal regeneration following optic nerve injury in the mouse. Neuroscience 343, 372–383 (2017).
Keeble, T. R. et al. The Wnt receptor Ryk is required for Wnt5a-mediated axon guidance on the contralateral side of the corpus callosum. J. Neurosci. J. Soc. Neurosci. 26, 5840–5848 (2006).
Zhang, X. et al. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat. Cell Biol. 9, 743–754 (2007).
Simões, A. R. et al. Damage-responsive neuro-glial clusters coordinate the recruitment of dormant neural stem cells in Drosophila. Dev. Cell 57, 1661–1675.e7 (2022).
Chu, J. et al. A novel MYC-ZNF706-SLC7A11 regulatory circuit contributes to cancer progression and redox balance in human hepatocellular carcinoma. Cell Death Differ. 31, 1333–1348 (2024).
Wexler, E. M., Paucer, A., Kornblum, H. I., Palmer, T. D. & Geschwind, D. H. Endogenous Wnt Signaling Maintains Neural Progenitor Cell Potency. Stem Cells 27, 1130–1141 (2009).
Harrison-Uy, S. J. & Pleasure, S. J. Wnt Signaling and Forebrain Development. Cold Spring Harb. Perspect. Biol. 4, a008094 (2012).
Mulligan, K. A. & Cheyette, B. N. R. Wnt Signaling in Vertebrate Neural Development and Function. J. Neuroimmune Pharmacol. 7, 774–787 (2012).
Southall, T. D. et al. Cell-type-specific profiling of gene expression and chromatin binding without cell isolation: assaying RNA Pol II occupancy in neural stem cells. Dev. Cell 26, 101–112 (2013).
Babraham Bioinformatics - FastQC A Quality Control tool for High Throughput Sequence Data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Yu, G., Wang, L.-G. & He, Q.-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinforma. Oxf. Engl. 31, 2382–2383 (2015).
Galaxy Community The Galaxy platform for accessible, reproducible, and collaborative data analyses: 2024 update. Nucleic Acids Res. 52, W83–W94 (2024).
R Core Team. R: A Language and Environment for Statistical Computing. (2024).
Allaire, J. RStudio: integrated development environment for R. Boston MA 770, 165–171 (2012).
Yu, G., Wang, L.-G., Han, Y. & He, Q.-Y. clusterProfiler: an R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 16, 284–287 (2012).
Thorvaldsdóttir, H., Robinson, J. T. & Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192 (2013).
McCulloch, M. W. B. et al. Psammaplin A as a general activator of cell-based signaling assays via HDAC inhibition and studies on some bromotyrosine derivatives. Bioorg. Med. Chem. 17, 2189–2198 (2009).
Acknowledgements
We thank A. Brand, J. P Vincent, F. Matsuzaki, H. Bellen, J. Raff and F. Yu; the Bloomington Drosophila Stock Center, Vienna Drosophila Resource Center, and Kyoto Stock Centre DGGR; the Developmental Studies Hybridoma Bank for fly stocks and antibodies; Yanhui Hu, James B. Brown, and Susan E. Celniker for the annotation of Drosophila smORFs; the Transgenic RNAi Project at Harvard Medical School for sgRNA-expressing fly lines; Felipe Escobedo, James Thai LaGraff, and Jorden Rabasco for help generating smORF KO flies; Brinda Palliyana for technical assistance; and M Gujar and W.H. Chooi for critically reading the manuscript. This work is supported by Singapore Ministry of Education (MOE) Academic Research Fund (AcRF) Tier 2 (MOE-T2EP30123-0018) to H.W. N.P. is an HHMI investigator. J.A.B. was supported by the Damon Runyon Foundation (DRG-2258-16) and a ‘Training Grant in Genetics’ T32 Ruth Kirschstein-National Research Service Award institutional research training grant funded through the NIH/National Institute of General Medical Sciences (T32GM007748).
Author information
Authors and Affiliations
Contributions
Conceptualization, J.L. and H.W.; methodology, data curation, and formal analysis, J.L., Q.D., Y.G., Y.S.T., Z.Z., L.K.A.T., B.O.P.T., D.V., J.B., and N.P.; writing–original draft, writing–review and editing, J.L., D.V., and H.W.; funding acquisition, H.W.; supervision, H.W.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Itaru Imayoshi, Tzumin Lee, and Fumio Matsuzaki for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, J., Deng, Q., Gao, Y. et al. Microproteins Simba1 and Simba2 activate Wingless signaling during the reactivation of neural stem cells in Drosophila. Nat Commun 16, 11651 (2025). https://doi.org/10.1038/s41467-025-66727-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66727-3