Abstract
Effective sex separation remains a critical challenge for mosquito genetic control. Genetic sexing strains (GSS) address this by linking maleness with selectable traits, enabling efficient female removal. Here, we present a versatile platform for GSS development in the invasive Aedes albopictus mosquito that integrates CRISPR-engineered selectable phenotypes with sex conversion via nix, the male-determining factor. As a proof-of-concept, we disrupt the yellow pigmentation gene and restore its function in males using nix-containing transgenes, producing a stable strain with yellow females and dark males. Beyond serving as a vivid marker, yellow confers added advantages: GSS females pupate later than wild females, enhancing protandry-based sorting, and lay desiccation-sensitive eggs, lowering accidental female release risk. The strain is compatible with size-based separation, improving sexing accuracy through the integration of natural and engineered dimorphisms. To our knowledge, this represents the first engineered sex-linked selectable trait in mosquitoes based on endogenous genes, establishing a foundation for scalable GSS development.
Similar content being viewed by others
Introduction
Aedes albopictus, the Asian tiger mosquito, has emerged as one of the world’s most invasive disease vectors, capable of transmitting arboviruses such as Dengue, Zika, and Chikungunya1. Traditional control strategies, including insecticide application and habitat management, have struggled to contain mosquito populations, driving increased interest in genetic control approaches2,3. Genetic control strategies have now been developed based on a number of different underlying technologies, including irradiation-based sterilization of the Sterile Insect Technique (SIT), and more recently the use of symbionts, genetic modification, and gene drives4. Most mosquito control strategies rely on the mass release of males, necessitating the complete removal of females. Released females can spread diseases, increase nuisance biting and interfere with the dispersal of released males. In Wolbachia-based suppression programs, female releases can compromise the sustainability of control programs. However, existing sex-separation methods, such as pupal size sorting, remain inefficient, costly, and difficult to scale for large-scale mosquito control programs5,6.
Genetic sexing strains (GSS) provide a scalable and cost-effective solution by linking maleness to a selectable trait, allowing for the efficient or automated removal of females. A GSS typically consists of at least two components: (1) an autosomal recessive mutation that disrupts a visible or conditional selectable trait, and (2) linkage of a wild-type (WT) rescue allele of this gene to the male-specific chromosome, ensuring trait restoration exclusively in males7,8. Historically, the development of a GSS in mosquitoes has been hampered by the laborious and unpredictable nature of classical mutagenesis and translocation approaches. Recent advances in CRISPR/Cas9 gene editing offer a predictable engineering approach, enabling precise, targeted modifications.
Aedes mosquitoes lack morphologically distinct sex chromosomes and sex is determined by a small, non-recombining autosomal region known as the M-locus9. Within this region, the male-determining factor nix functions as a master regulator of sex determination and defines male development10. Engineered insertions of nix can function as synthetic neo-sex chromosomes, converting genetic females into fertile pseudomales and resulting in the loss of the ancestral M-locus in these populations11,12. We reasoned that nix-mediated sex conversion could be leveraged to develop a robust GSS, based on linking selectable rescues to masculinized pseudomales in a mutant background.
In this work, we demonstrate this approach by engineering a GSS in Ae. albopictus that integrates CRISPR-disruption of an endogenous gene resulting in a visible phenotype (yellow) with nix-mediated male rescue. The yellow (y) gene, a well-documented but biochemically unresolved melanization factor, produces a viable and vivid mutant phenotype in Ae. aegypti and Ae. albopictus13,14. We disrupted yellow to generate a stable, homozygous mutant strain in which all mosquitoes display the yellow phenotype. We then restored yellow function with engineered rescues containing both mini-yellow and nix, yielding a pure-breeding GSS where all males are dark and all females remain yellow. We evaluate this GSS in terms of its stability of sex conversion, impact on sex-biased gene expression, and practical utility for sex separation. We find that pseudomales exhibit gene expression profiles closely resembling WT males, confirming successful sex conversion. We also show that GSS females display delayed larval development, enhancing protandry-based sex sorting, and produce desiccation-sensitive eggs, which could contribute to genetic containment of unintentionally released females in field applications. This work provides a versatile platform for engineering GSSs in Aedes mosquitoes and highlights a path forward for the development of mosquito population suppression strategies.
Results
Generating a yellow mutant as a selectable trait
To develop a genetically encoded sex-separation marker, we used CRISPR/Cas9 to generate a yellow mutant strain in Ae. albopictus. The yellow gene is located on chromosome 1, approximately 81.7 Mb from the M-locus. It has a relatively simple two-exon structure, producing a single mRNA isoform (Fig. 1A). We targeted the first exon with sgRNA134 and injected pre-assembled Cas9-RNP complexes into syncytial embryos. High rates of injected mosquitoes displayed mosaic pigmentation (Supplementary Fig. 1) and those surviving to the adult stage were backcrossed to WTs to establish isofamilies. As low recombination rates were expected between the yellow locus and the nearby non-recombining region of chromosome 1, we intercrossed F1 adults from different isofamilies to obtain F2 homozygous yellow mutants, which were crossed to establish a stable mutant strain. As expected from the intercrossing of F1s, amplicon sequencing revealed four different circulating null alleles, all containing frameshift mutations that resulted in premature stop codons before the yellow Major Royal Jelly Protein (MRJP) domain and in the loss of AlphaFold-predicted tertiary structure of Yellow (Fig. 1B).
A Structure of the yellow gene showing the location of the sgRNA134 target in the first exon, the Major Royal Jelly Protein (MRJP) domain and the stop codon (TAG). The target site of sgRNA134 (red) and PAM site (blue) is shown followed by AlphaFold2-predicted 3D structure of Yellow. B CRISPR-induced yellow mutant alleles isolated. Alignment view of mutant alleles containing deletions or insertions causing frameshifts and premature termination. All alleles encode proteins lacking the MRJP motif and correct tertiary structure. C–G Comparison of WT and yellow phenotypes across development indicating pigmentation differences in key structures highlighted with arrows: C–D Fourth instar larvae - trachea (Tr), siphon (Si), saddle (Sa), lateral hairs (LH), mental plate (MP), and antennae (An). E One-day-old male pupae - terga (Te) and mesothorax (M). F Adult females. G One-day-old eggs laid by WT and yellow females.
Consistent with the role of yellow in melanin biosynthesis15, mutants failed to properly deposit melanin, resulting in a striking pigmentation phenotype throughout development (Fig. 1C–G). Larvae displayed reduced cuticular melanin, particularly in antennae, mental plates, lateral hairs, trachea, siphons and saddles (Fig. 1C, D). In pupae, pigmentation differences were most pronounced in the dorsal mesothorax and terga, irrespective of pupal age (Fig. 1E). In adults the characteristic black-and-white “tiger stripes” of Ae. albopictus appeared yellow (Fig. 1F). Eggs laid by yellow females failed to fully melanize, remaining golden-brown instead of black as WT eggs (Fig. 1G), consistent with previous studies16. Notably, this egg melanization phenotype was maternally inherited and was not restored by crossing yellow females to WT males.
Transgenic rescue of the yellow phenotype
To restore pigmentation, we designed a minimal yellow (mini-yellow) rescue construct that removed the large 25-kbp intron between the two exons. To do so, a 1.1 kbp sequence of upstream regulatory region followed by exon 1 were amplified and cloned in-frame with exon 2 flanked by 660 bp of presumed terminator (Fig. 2A). This mini-yellow rescue, called EnYR (Endogenous yellow rescue), was cloned into a piggyBac transformation construct also containing an eGFP transformation marker, driven by OpiE2 (Fig. 2A). Given the untested nature of these regulatory regions, we also constructed a second mini-yellow rescue, called HspYR (Heatshock yellow rescue), where the mini-yellow was driven by the ubiquitous Ae. albopictus hsp83 promoter, previously validated using a DsRed reporter (Supplementary Fig. 2).
A Structure of EnYR (Endogenous yellow rescue) and HspYR (Heatshock yellow rescue) rescue constructs containing the mini-yellow coding sequence (CDS) driven either by endogenous yellow regulatory elements or the hsp83 promoter and P10 terminator. Both constructs also contain the OpIE2:eGFP transformation marker and piggyBac arms. B Crossing scheme to test complementation. Transgenic males homozygous for the native yellow (y+) allele were crossed to homozygous yellow (y-) mutant females, producing F1 progeny heterozygous at the yellow (y+/y-) locus and carrying the mini-yellow rescue construct. F1 transgenic males were then test-crossed with homozygous yellow (y-) mutant females to evaluate pigmentation restoration in F2 progeny. As a control non-transgenic F1 heterozygous males were crossed to homozygous y- females. C Distribution of pigmentation phenotypes for dark and yellow among transgenic and non-transgenic individuals. Significant reduction from a default 50% yellow (z range: –4.82 to –8.38; p < 0.00000146, two-tailed z-test for proportions) is indicated. Source data are provided as a Source Data file.
Microinjections of eggs of the yellow strain with mini-yellow rescue constructs resulted in extremely low post-injection survival. Among the few hatching larvae, none displayed transient expression of the fluorescent marker and were likely individuals not successfully injected. We therefore injected WT eggs and introgressed transgenes into the yellow background. Three independent transgenic strains were established for each mini-yellow construct and transgenic males from each of the six isofamilies were sequentially crossed to yellow females for two generations (Fig. 2B).
In F2, we evaluated the overall frequency of the yellow phenotype and its co-occurrence with the eGFP marker, which would indicate a lack of genetic rescue from the transgene. In crosses involving the EnYR construct, both yellow and dark progeny containing the transgene (eGFP-positive) were observed and the overall frequency of the two phenotypes was similar to control crosses, involving non-transgenic F1 males (Fig. 2C). These results suggested that EnYR failed to rescue pigmentation (β = 0.207, SE = 0.154, z = 1.341, p = 0.180). In contrast, HspYR fully restored pigmentation, with all transgenic individuals displaying WT-like dark coloration (Fig. 2C). The proportion of dark F2 individuals increased to 81.78 ± 1.24%, indicating successful genetic rescue in all lines (β = –1.384, SE = 0.169, z = –8.208, p < 0.001). These results confirmed that the yellow regulatory regions we selected failed to correctly express mini-yellow, whereas the hsp83 promoter drove sufficient expression for effective yellow complementation.
Combining sex conversion and yellow rescue
To genetically associate mini-yellow rescue to maleness, we next inserted nix within HspYR, generating construct YRN (yellow rescue with nix) that was injected into WT embryos (Fig. 3A). The nix isoform we used was previously shown to transform genetic females into fertile males, which are referred to as pseudomales from here on11,12. Seven independent transgenic isofamilies (YRN1-7) were established from independent founders. Males or pseudomales from each isofamily were individually backcrossed to yellow females for multiple generations until a pure-breeding sexing strain was isolated producing exclusively dark, eGFP-positive males and yellow, eGFP-negative females (Fig. 3B–D).
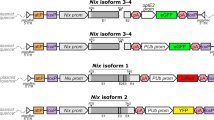
A Structure of the YRN (yellow rescue with nix) construct created by adding nix 3&4 isoform, flanked by its native regulatory region and an SV40 poly(A) terminator motif (red), to the HspYR (Heatshock yellow rescue) plasmid, which contains the hsp83-driven mini-yellow and the eGFP transformation marker. B–D Crossing scheme for isolating functional integrations. B Transgenic male isofounders are crossed with yellow females. C F1 progeny were analyzed for sex and transgenic status. D F2 transgenic males from isofamilies with significant sex bias (χ² ≥ 15.1, p < 0.0001; indicated by *) were individually crossed to yellow females to exclude subfamilies producing transgenic females or yellow individuals and to purify M-locus free male lineages by molecular genotyping for the endogenous nix and yellow. E Sex conversion and yellow rescue in crosses between YRN and yellow females, using HspYR as a control. The frequency of pigmentation phenotypes (dark and yellow), separated by sex, is presented as the mean proportion ± SEM across three gonotrophic cycles from two cages. Numbers indicate the total individuals recorded per group. Source data are provided in the Source Data file.
YRN-6 produced exclusively dark transgenic males and yellow non-transgenic females (Fig. 3C). In contrast, YRN-4 and YRN-5 exhibited high rates of transgenic females, suggesting failure of the transgene to masculinize genetic females. In families YRN-1, 2, 3 and 7 both transgenic and non-transgenic males were recovered, implying that their male founders were true males, transmitting both the M-linked nix and the YRN-linked nix. Despite multiple backcrossing rounds, these families did not yield pure-breeding pseudomales deprived of the endogenous M-locus.
We therefore focused on transgenic males from family YRN-6. Molecular genotyping confirmed that YRN-6 males lacked the endogenous M-locus (Supplementary Fig. 3). To validate the function of YRN as a sexing system, we crossed yellow females either to YRN-6 pseudomales or HspYR-1 males that harbor only the hsp83-driven mini-yellow. In crosses involving HspYR-1, dark and yellow phenotypes segregated equally in male and female pupae (χ²(1) ≈ 2.21, p = 0.137). In YRN-6, all female pupae recovered were yellow, while all phenotypically male pupae were dark (χ²(1) ≈ 1747, p < 0.001) (Fig. 3E). A small proportion (4.2%) of pseudomales displayed intersex phenotypes, including intermediate genitalia, antennae and mouthparts. These intersexes were unable to blood-feed or mate. Based on these results, we established YRN-6 as our first pure-breeding GSS strain that has now been maintained stably for more than 20 generations.
Masculinization of gene expression in GSS pseudomales
To explore the effect of nix-based sex conversion on sex-biased gene expression, we performed RNA-seq on adult whole bodies from WT males, WT females, GSS pseudomales, and GSS females. First, we assessed the correlation of genome-wide gene expression profiles, finding that GSS pseudomales closely resembled WT males (Spearman rs = 0.955 ± 0.001) despite being genetically identical to GSS females besides the insertion of the YRN construct (Fig. 4A). Next, we compared the expression of well-known sex-biased genes - nix, doublesex, fruitless, myo-fem, myo-sex (Fig. 4B). For nix we found no reads mapping from either WT or GSS females, as expected. Both males and pseudomales on the other hand had good coverage over exons, but pseudomale coverage was limited to the two exons included in the nix transgene (isoform 3-4). Consistent with expression of nix, sex-specific splicing of the downstream sex-determination genes doublesex and fruitless was faithfully executed in pseudomales, mirroring the splicing in WT males and resulting in the skipping of female-specific exons. The female-specific flight muscle protein myo-fem was not expressed in pseudomales, but its male-specific paralog myo-sex was17. Mapping to the yellow gene indicated that pseudomales express the yellow rescue at much higher levels than endogenous yellow, which is consistent with our use of the ubiquitous hsp83 promoter. As expected, we observed a dip in mapped reads from GSS females in the position corresponding to the sgRNA target site, which is likely an outcome of indels contained in the yellow mutant alleles (Fig. 1B). These results supported the conclusion of successful phenotypic masculinization and pigmentation encoded from the two rescue transgenes.
A Spearman correlation matrix of genome-wide expression in sequenced samples. Higher correlation values indicate greater similarity in transcriptomic profiles. B RNA-seq coverage plot for sex-linked markers (myo-fem, myo-sex, yellow) and key sex-determination genes (nix, doublesex, fruitless). Exons are indicated in gray, except for female-specific exons that are indicated in pink. Introns not to scale. Coverage for male samples is shown on the positive Y-axis, while female coverage is reflected on the negative Y-axis for clarity. C Differential expression analysis comparing wild-type (WT) females versus WT males (top) or genetic sexing strain (GSS) pseudomales (bottom), indicating changes or retention of sex-biased direction between analyses. Source data are provided as a Source Data file.
To catalog sex-biased gene expression, we ran differential expression analysis comparing WT males and females to compile a list of Ae. albopictus sex-associated markers, containing genes expressed in a sex-specific or significantly sex-biased manner. This catalog included 12.1% of the 17189 expressed genes, comprising 859 male-markers and 1228 female-markers (Supplementary Data 1). We then ran a second differential expression analysis comparing gene expression between GSS pseudomales and WT females, to track changes in sex-biased expression with WT females acting as a reference (Fig. 4C). Overall, most markers retained their sex-bias—623 (72.53%) and 917 (74.67%) for male- and female-markers, respectively. Levels of sex-biased expression (fold change) of markers that retained their sex-specificity were significantly higher than markers losing sex-biased expression (Tukey-Kramer HSD α = 0.05), indicating that retention of sex-bias was more likely for strongly sex-biased genes (Supplementary Fig. 4). The majority of markers that changed sex-bias classification in the pseudomale-female analysis became unbiased (222 male-markers and 309 female-markers; Fig. 4C). Only two female-markers were more highly expressed in pseudomales, one of which was yellow. Eleven male-biased genes were completely absent in pseudomales, suggesting their M-locus linkage or in trans regulation by M-locus factors. To evaluate M-locus linkage, we used the chromosome quotient method18 and male-specific k-mers19,20 using male and female whole-genome sequencing data21. Five of the eleven genes were confirmed to be M-locus linked by PCR using male and female genomic DNA (Supplementary Fig. 5).
Benchmarking the yellow-GSS for sex sorting
To begin exploring the potential use of our color-based sex separation marker for Ae. albopictus, we quantified the pigmentation intensity of male and female pupae from GSS and WT strains to assess variation between individuals. To control for life-history factors like age, food availability, density, etc. and to enable precise comparisons between WT and GSS individuals, larvae from each strain were co-reared in shared containers under high-density protocols, approximating conditions of mass-rearing22. Image analysis of pupal pigmentation intensity confirmed that GSS female pupae were significantly paler than GSS pseudomales and all WT pupae (ANOVA F(3, 1450) = 843.6, p < 0.0001; Tukey-Kramer HSD α = 0.05) (Fig. 5A). This pigmentation difference was so pronounced that optical sex separation of GSS pupae based on color and size, as observed by the naked eye, was faster and less error-prone than sex separation of the WT strain using a dissecting microscope (Supplementary Figs. 6 and 7).
Measurements of (A) Pupal pigmentation intensity (mean gray value) and (B) pupal width (mm). Data for both metrics are shown as boxplots indicating the median (line), mean (cross), interquartile range (box), and whiskers extending to ± 1.5×IQR. Statistical significance was assessed using ANOVA followed by Tukey’s HSD test, with significant differences indicated by letter groups (p < 0.05). C Combined analysis of width and pigmentation levels. Each point represents an individual pupa, with histograms showing the distribution of phenotypes. D Development time, measured as the average number of days until pupation. Data are presented as the mean value ± SEM. Statistical significance was determined by a Welch’s two-tailed t test (* denotes t(2369) = –3.07, p = 0.0021; *** denotes t(2048) = 11.28, p < 2.2 × 10⁻¹⁶). E Pupation dynamics over time. Kernel density plots show daily pupation probability. Semi-transparent areas represent each of the four replicates in the experiment and solid lines indicate the cumulative probability. Source data are provided as a Source Data file.
We also collected data on pupal size and larval development dynamics for both strains, as proxies for mosquito fitness. We found no significant difference in the width, or any other size-related metric, of male pupae between the GSS and WT strain. Interestingly, GSS female pupae were slightly larger than female WT pupae (ANOVA F(3, 1450) = 320.1, p < 0.0001; Tukey-Kramer HSD α = 0.05) (Fig. 5B), suggesting that this GSS should be compatible with sex separation methods leveraging size differences, including approaches like glass plate separation23 or automated robotic sorting24. The intersection of size and color metrics enhanced the separation of males and females in the GSS strain compared to WTs (Fig. 5C). Insofar as insect size can reliably estimate mosquito fitness25, these results suggest no detectable costs for GSS individuals compared to WTs.
In terms of developmental dynamics, GSS pseudomales became pupae slightly faster than WT males and GSS females took significantly longer to pupate than WT females (Welch’s two-tailed t test: t(2369) = 3.074, p = 0.0021; t(2048) = −11.28, p < 0.0001, respectively) (Fig. 5D, E). Combined, faster development of GSS pseudomales and slower development of GSS females increased the protandry index (accelerated male developmental time) to 27.8 ± 0.82 in the GSS strain, compared to 17.4 ± 0.29 in WTs. To test whether this enhanced protandry could be practically useful for operational use, we seeded triplicate WT and GSS high-density rearing trays (3 larvae per ml), this time rearing each strain separately. Pupae were collected multiple times per day to improve the resolution of pupation dynamics (Fig. 6A). Consistent with previous observations, significant differences in pupation timing were observed between males and females in the two strains (Welch’s two tailed t test for strain:sex, t(3896) = 11.282, p < 0.001). There was an approximately 40% delay in the time needed for pupation of GSS females (Fig. 6B), resulting in an improved protandry index of 26.2 ± 0.08 for the GSS strain, compared to 19.4 ± 0.59 for WT strain. This translated to a mean delay of nine hours in female pupation, resulting in higher male recovery rates (proportion of recovered males from total males) for any rate of female contamination compared to the WT strain (Fig. 6C).
A Pupation dynamics tracking cumulative male recovery and female contamination for the wild-type (WT) and genetic sexing strain (GSS). B Delay in the pupation time of females relative to males expressed as a percentage increase in development time. C Male recovery rates at three levels of female contamination comparing WT and GSS males. All panels (A–C) originate from the same experiment performed with three independent biological replicates per strain (Replicate sample numbers are indicated). Bar plots represent mean ± SEM. Source data are provided as a Source Data file.
The accelerated pupation of GSS pseudomales could imply differences in reproductive fitness, given trade-offs between development time and fecundity. To test whether GSS pseudomale mating performance was compromised, we evaluated their ability to induce post-mating refractoriness in females using both group and single-female assays. In group mating experiments, females mated to either pseudomales (eGFP/+) or true males (dsRED/+) were subsequently exposed to males of the opposite type, and the paternal origin of offspring was scored based on larval fluorescence. Across >5268 screened larvae, no evidence of re-insemination was detected. In complementary single-female assays, in which individual copulations were directly observed and mated females were then transferred to cages with males of the opposite type, rare cases of sequential insemination were detected (3/40 females). However, the frequency of re-insemination did not significantly differ between females mated first with pseudomales versus females mated first to true males (β = –0.747, SE = 1.268, z = –0.589, p = 0.556). Together, these results indicate that GSS pseudomales are equally capable as true males in inducing post-mating refractoriness in females, suggesting high reproductive fitness despite their lack of the native male-specific M-locus (Supplementary Data 2).
Desiccation-sensitivity of GSS eggs
During the establishment of stable yellow and GSS populations, we observed considerable variability in the hatching rates of eggs laid by yellow females. Previous studies have shown that following oviposition the chorion of Ae. albopictus eggs undergo melanization via a process mediated by several yellow-family genes, which hardens the eggshell and confers desiccation resistance16,26. Considering that yellow eggs did not darken like WT eggs (Fig. 1G) and were too fragile for microinjection, we hypothesized that the absence of maternal yellow could compromise desiccation resistance and/or embryonic dormancy, traits that have enabled the long-distance dispersal of this mosquito as desiccated, dormant eggs27,28.
To investigate this, we collected twelve egg bowls from cages containing yellow males crossed to WT and GSS females and recorded for each egg bowl, the total number of eggs laid by WT (dark eggs) and GSS (yellow eggs) females. During storage, we randomly selected three egg batches every seven days for hatching and screened hatched larvae for cuticle color to calculate relative hatching rates. Overall, GSS eggs exhibited significantly lower hatching than WT eggs at all time points (paired two-tailed t test, t(11) = 10.6, p < 0.0001; ANCOVA phenotype, F(1,18) = 10.97, p = 0.0039; storage duration, F(1,18) = 9.71, p = 0.0060; Tukey-Kramer HSD α = 0.05) (Fig. 7A). While the viability of WT eggs began to decline only after the third week of storage, remaining above 50% until the end of the experiment, GSS eggs were completely unviable by the fourth week (Fig. 7A). To test whether the increased mortality of GSS eggs was due to increased desiccation sensitivity, we transferred fully-matured, 4-day-old GSS and WT eggs from moist filter papers to glass slides and monitored their morphology as the surrounding environment dried via evaporation under the microscope. Within a few minutes, most yellow eggs displayed visible signs of desiccation, ultimately resulting in the irreversible collapse of their chorionic structure rendering them nonviable. Conversely, WT eggs retained their structural integrity and remained unaffected (Fig. 7B). These findings show that the yellow mutant background of GSS females imparts significant desiccation sensitivity that is maternally defined, a trait that could serve as a useful barrier to prevent gene flow or establishment of incompatible symbionts in field populations from the unintentional, rare release of contaminating females. Importantly, since identifying the desiccation sensitivity of the GSS eggs, we modified our protocols for egg storage to maintain optimal moisture and to hatch stored eggs within two weeks from oviposition - practices that ensure a healthy and stable maintenance of the GSS population in our insectary for the last one and a half years.
A Hatching rates of stored wild-type (WT) and genetic sexing strain (GSS) eggs tracked over one month. Hatching rates were calculated as the proportion of eggs that successfully developed into L2 larvae, based on three independent biological replicates per time point. Data are presented as the mean value ± SEM. Significant effects of strain and storage duration were detected (ANCOVA: F₁,₁₈ = 10.97, p = 0.0039; 0.0060, respectively). Letter groupings were determined using post-hoc Tukey’s HSD tests (α = 0.05). Source data are provided as a Source Data file. B Time-lapse images of mature, 4-day-old eggs from WT and GSS cages mounted on a dry glass slide and photographed one- and six-minutes following removal from moist egg papers.
Discussion
The VIENNA-8 GSS of the Mediterranean fruit fly (Ceratitis capitata) has for decades now served as the gold standard for insect sex separation, providing a reliable and scalable solution for medfly SIT programs7,8. Despite extensive efforts and coordinated research to develop similar GSSs in other insect species of medical and agricultural importance, progress has been limited, primarily due to the unpredictability of classical genetic approaches. As a result, genetic control programs for Aedes mosquitoes currently rely on naturally occurring sexual dimorphisms for female elimination6,24,29. The most widely used method is size-based sorting at the pupal stage, where female pupae are slightly larger than males. However, this approach presents several challenges: (1) size dimorphism is a continuous trait with significant overlap between the sexes leading to female contamination30,31; (2) sexing at the pupal stage requires rearing both sexes until pupation resulting in wasted resources; and (3) manual size-based sorting is labor-intensive and difficult to scale for mass-production. While recent advances in automated sorting technologies, such as robotic systems leveraging size or other morphological differences, may help address some of these limitations, they remain costly and complex to implement6,24,32. In contrast, a well-designed GSS could offer a more practical, cost-effective and scalable solution for mosquito sex separation33.
In this study, we introduce a versatile platform for the development of GSSs in the important arboviral vector Ae. albopictus. Our platform is based on the CRISPR-mediated disruption of native mosquito genes encoding selectable phenotypes, followed by their genetic rescue using sex-converting transgenes. As a proof-of-concept, we selected the highly-conserved yellow gene - one of the first phenotypic mutants discovered in the fly room of Thomas Hunt Morgan34. We selected this gene for its relatively simple structure and lack of multiple isoforms. Based on previous work in Ae. albopictus, homozygous null mutants were expected to display a clearly visible phenotype throughout development, and to be both viable and fertile14, which we confirmed by generating a mutant strain using sgRNA134 targeting the first exon and establishing a stable homozygous strain.
To genetically rescue cuticle pigmentation, we generated constructs containing the spliced yellow coding sequence that we called mini-yellow. The EnYR construct containing mini-yellow flanked by its native regulatory regions failed to complement pigmentation in all transgenic strains generated, suggesting that the selected regions lack critical promoter and/or enhancer regions. A second construct, HspYR, expressing mini-yellow from validated regulatory regions of the Ae. albopictus hsp83 gene successfully restored pigmentation in the yellow mutant background. RNA-seq expression data revealed that the hsp83 regulatory regions drove high levels of expression of the mini-yellow in adults, suggesting that this promoter could be useful for the development of other GSSs with selectable traits requiring ubiquitous expression.
Building on recent work11,12, which demonstrated that nix alone is sufficient to masculinize genetic females into fertile males, we incorporated nix into our mini-yellow construct, effectively coupling dominant pigmentation with masculinizing sex conversion. Among the seven independent strains generated, YRN-6 was the only one in which all phenotypically dark males carried the transgene, confirming exclusive linkage between male sex and the nix transgene. In contrast, three other strains produced both transgenic and non-transgenic males, indicating that their founders were likely genetic males rather than sex-converted pseudomales. Attempts to isolate males lacking the native M-locus from these strains, by individually crossing fluorescent males to yellow females, failed to recover any family containing exclusively sex-converted males, suggesting incorrect expression of the nix transgene from these integrations and lack of pseudomale fertility in these strains.
Overall, our results confirm that nix-based sex conversion offers a practical alternative to other methods for tightly linking rescue constructs to maleness, overcoming key technical challenges associated with reciprocal translocations and Y-chromosome knock-ins. Reciprocal translocations impose significant male fertility costs due to gamete aneuploidy35. Y-chromosome knock-ins, on the other hand, remain technically challenging due to the highly repetitive nature of male-specific chromosomes, making precise insertions difficult. To the best of our knowledge, successful Y-chromosome knock-ins have only been achieved in D. melanogaster, despite significant efforts—both by our group and others—to adapt this approach for other insect species36,37. Additionally, even when Y-linked insertions are obtained, integrations frequently suffer from reduced or variegated transgene expression, likely due to the heterochromatic environment of the Y chromosome36,38,39. In Ae. albopictus, nix-based sex conversion bypasses these limitations by enabling a stable, autosomal approach to masculinization, providing a more reliable and scalable solution for genetic sexing.
Most insect species are not as malleable as Ae. albopictus in manipulations of their sex determination pathway. Even when master sex determination genes are known, their expression in genetic females can be insufficient to fully convert them into fertile males, for example by requiring functions of other male-specific genes present in Y-chromosomes or M-loci40. Alternatively, transgenic expression of primary male determiners can be lethal, for example by interfering with the dosage compensation pathway in genetic females and resulting in misexpression of X-linked genes41,42,43. Our transcriptomic analysis confirmed that despite their genetic female background, pseudomales exhibited a gene expression profile highly similar to that of WT males. This included the correct splicing of well-known sex-determination genes like dsx and fru44,45, but also most other sex-biased genes, which are cataloged here. Among the male-markers we found an interesting subset of eleven genes, whose expression was not detected in GSS pseudomales, suggesting M-locus linkage, or their transcriptional regulation by M-locus linked factors other than nix. We confirmed the localization of five of these within the M-locus, of which one (LOC134287234) is predicted as non-coding and four as protein-coding genes. LOC109432934 contains a leucine-rich repeat domain, LOC134285312 a sperm tail domain, LOC134289344 a YqaJ-like viral recombinase domain and LOC134285028 lacks any known motifs.
In terms of sex separation, we benchmarked the fitness and performance of the yellow-GSS, measuring differences in pigmentation intensity between male and female pupae. Our results confirmed that GSS females were significantly paler than GSS pseudomales, making sex sorting based on color highly efficient. The pronounced contrast in pigmentation allowed for rapid and more reliable visual sorting compared to the traditional method of pupal sexing under a dissecting microscope or based on size. This suggests that yellow-GSS provides a robust improvement to current sorting methods, providing a clear, visible indicator of female contamination. Color-based sorting could be useful as a standalone trait for imaging- and algorithm-assisted automated sorting, or could be used as a secondary marker in future designs involving high-throughput traits like temperature-sensitive lethals.
Beyond pigmentation, we also examined pupal size and larval development dynamics to evaluate potential fitness costs associated with the GSS genotype. Pseudomale pupal size and development were comparable to WT males. GSS female pupae were slightly larger than WT females, a feature that enhanced sex separation accuracy, when combined with pigmentation differences. This size difference may reflect reduced larval competition at later larval stages given the early pupation of pseudomales, or that the delay in female pupation results in overall larger female larvae and thus larger pupae. Interestingly, homozygosity for yellow in GSS females led to slower larval development, significantly increasing protandry—the earlier emergence of males relative to females. In WT Ae. albopictus, protandry alone is usually insufficient for reliable sex separation due to overlapping male and female development times46,47. However, the enhanced protandry observed in the GSS expanded the temporal gap between male and female pupation, improving sorting efficiency and reducing female contamination. This aligns with findings by Bellini et al.30, who noted that while natural protandry exists, it is not pronounced enough for effective sex separation. By amplifying this developmental difference, yellow-GSS offers an additional trait that could have practical applications for mosquito control programs.
To evaluate the suitability of our GSS pseudomales for genetic control programs, we assessed their ability to induce post-mating female monogamy, also known as mating refractoriness48. We found that pseudomales induced levels of female post-copulatory refractoriness comparable to those of true males, supporting their potential utility in genetic control strategies. Male mating competitiveness can generally be divided into two categories: pre-mating and post-mating competitiveness. Pre-mating competitiveness refers to the ability of released males to successfully mate with females in competition with wild males, reflecting factors such as courtship vigor and attractiveness. Post-mating competitiveness, in contrast, refers to the ability of released males to successfully fertilize eggs and induce refractoriness in females, preventing or significantly reducing subsequent matings with wild males. Post-mating competitiveness is particularly critical for genetic control programs targeting insect species whose females naturally exhibit strong mating refractoriness under field conditions, as is the case with many mosquitoes49,50,51,52. Even if released males exhibit normal pre-mating competitiveness, population suppression efforts will fail if these males do not effectively induce refractoriness. While suboptimal pre-mating competitiveness can typically be compensated for by increasing male release ratios, low post-mating competitiveness cannot be easily mitigated. Previous studies evaluating nix-masculinized pseudomales reported reduced mating competitiveness in laboratory cages using 1:1 ratios with true males11,12. In these experiments, competitiveness was measured as the fraction of offspring sired by pseudomales compared to true males, a variant of Fried’s index typically used for measuring sterile male competitiveness53,54. However, it is now generally well established that such measurements of mating competitiveness can result in high levels of variation in the calculated competitiveness from each cage55, with competitiveness changing according to the male release ratio56,57, even though it theoretically should not, and that small-cage experiments promote the otherwise rare occurrence of multiple insemination events58. Therefore, reliable evaluations of GSS pseudomale fitness will require additional testing including multiple release ratios and ideally larger, semi-field conditions, which were outside the scope of this study. Our results demonstrate that GSS pseudomales possess post-mating competitiveness comparable to true males, justifying further investigations into their overall competitiveness and fitness. Future studies should explicitly assess pre- and post-mating competitiveness separately across a range of ecologically relevant release ratios and evaluate their interactions with genetic control technologies, such as sterilization, in conditions more representative of natural populations.
A notable characteristic of the yellow mutant is the desiccation sensitivity of GSS eggs. The failure of eggs laid by GSS females to melanize properly resulted in a marked reduction in their viability over time. While this trait posed an initial challenge that required adjustments to rearing protocols, simple measures such as maintaining optimal humidity of egg papers and hatching stored eggs within two weeks have ensured stable colony maintenance in the laboratory. We anticipate that these protocols could be easily adapted for future mass-rearing operations, ensuring the strain’s scalability59. In the context of field deployment, this trait could serve as an inherent containment mechanism, minimizing the risk of accidental female releases and reducing the potential for unintended gene flow or Wolbachia establishment in target populations.
Looking ahead, our findings suggest that color-based sex sorting can be both faster and more accurate than pupal size sorting alone. The yellow mutant, besides being an excellent color marker, also offers useful secondary phenotypes like slow larval development and egg desiccation sensitivity. Future studies could focus on developing high-throughput sorting technologies to fully exploit the advantages of this strain. While the yellow phenotype offers a strong standalone visual marker, future iterations of this GSS platform could integrate next-generation selectable traits, such as temperature-sensitive lethals, to enhance scalability further. A key strength of our GSS development framework is its adaptability selectable marker mutants and their corresponding rescues can be iteratively introduced and expanded, enabling the system to evolve alongside future advancements in genetic control technologies.
Methods
Mosquito strains and rearing
The FPA (Foshan) Ae. albopictus strain was used as the laboratory WT strain in all experiments. All strains were reared at 27 ± 1 °C, 75 ± 5% humidity with 14:10 (LD) photoperiod regime. Larvae were reared in 160 × 135 mm plastic containers in 300 mL demineralized water at a density of up to 1.33 larvae per milliliter. The rearing tank was provided with a daily supplement of fish food (‘Essence’, Alltech-Coppens, Leende, NL). Pupae were manually collected every 48 h, and transferred to the designated cages. Adult mosquitoes were maintained in cages and given access to sugar solution (10% sucrose, 0.1% methylparaben). Female blood meals consisted of heparinized bovine blood augmented with 5 mg/mL ATP. Blood meals were provided using Hemotek feeding system (Discovery Workshops, Accrington, UK). Four days after blood feeding, an egg-bowl was introduced into each blood-fed cage. After 24 hours, egg-bowls were removed and egg papers were transferred to Petri dishes. Petri dishes were left to dehydrate for 24 h before being Parafilm-sealed to maintain some moisture for long-term storage. To synchronize hatching, mature stored eggs (≥96 h old) were immersed in deoxygenated hatching solution consisting of 0.25 g nutrient broth (Merck-Sigma Aldrich, USA) and 0.05 g baking yeast dissolved in 750 mL of distilled water and incubated overnight at room temperature. Hatched larvae were collected after 6 hours as L1, or the following day as L2 and moved to rearing containers.
Generating a yellow mutant strain
We selected sgRNA-134 (CACGTAGTCCCCACTGGCCA-GGG) to target the first exon of the yellow gene (LOC109402083 in AalbF5) using CRISPOR60. Ribonucleoprotein injection mixes containing SpCas9 (300 ng/μL) and sgRNA (80 ng/μL) (IDT, Coralville, IA) were pre-incubated at 37 °C for 30 min and microinjected into syncytial blastoderm embryos following standard protocols61. Mosaic individuals were backcrossed to WT mosquitoes to establish isofamilies. To minimize fitness costs and account for the proximity of yellow to the non-recombining region of chromosome 1 (which contains the M and m loci), heterozygotes from different families were crossed to establish a homozygous yellow line. DNA from a mixed-sex pool of F5 yellow individuals was extracted, and the target site was PCR amplified and sequenced using next-generation sequencing (NGS; Hy-labs, Israel). Mutant alleles were characterized and quantified, and predicted protein structures were generated using AlphaFold62.
Plasmid construction and transgenesis
For the EnYR mini-yellow rescue, the yellow coding sequence was amplified in two fragments: exon 1 (with 1100 bp upstream regulatory region) and exon 2 (660 bp), omitting the large intron. These fragments were assembled using Gibson assembly into an OpiE2:eGFP piggyBac transformation plasmid. For the HspYR plasmid, a 2076 bp fragment of the hsp83 promoter (LOC109407918) was cloned upstream of the mini-yellow coding sequence followed by the P10 terminator13, from Addgene plasmid #100580. To generate the YRN plasmid, the nix3&4 isoform11, from Addgene plasmid #173667, was inserted along with its native regulatory sequence into the HspYR plasmid. Cloning was performed using HiFi DNA Assembly (New England Biolabs) and Q5 Hot Start High-Fidelity PCR. Plasmids were validated by sequencing and microinjected at 200 ng/μL along with 40 ng/μL of a mosquito codon-optimized piggyBac transposase helper plasmid11. Individuals transiently expressing eGFP were backcrossed to WT to establish transgenic isofamilies.
Genotyping for endogenous and transgenic components
Primer sets were designed using Geneious Prime (v2023.1) to distinguish between endogenous and exogenous components (Supplementary Fig. 3A, Supplementary Data 3). For the yellow gene, forward primers targeting the 5′ UTR and hsp83 promoter were paired with reverse primers matching the unaltered sgRNA134 cut site and yellow exon 2. To differentiate between endogenous and transgenic nix, a triplex PCR was employed using a forward primer for exon 1 and two reverse primers, one targeting an endogenous intronic gap and another specific to the transgenic SV40 terminator. These assays were used to genotype DNA from transgenic dark male isofounders and to identify lineages deprived of endogenous elements (Supplementary Fig. 3B).
Testing yellow rescue from transgenic constructs
Ten transgenic males carrying either the YRN construct or the non-masculinizing HspYR construct in the WT genetic background were crossed to ten yellow females. Two replicate cages were used for each cross, with each cage undergoing three rounds of blood feeding and egg laying (technical replicates). The sex, phenotype, and transgenic profile of pupae from each gonotrophic cycle were recorded to calculate sex-specific phenotype distributions.
Sex-biased gene expression analysis
Four replicate samples, each comprising three 4-day-old flying males or females from the WT and GSS strains, were collected. Anesthetized mosquitoes were transferred to tubes containing 2.3 mm zirconium-silicate beads (BioSpec Products, USA) and ice-cold TRI-reagent (Zymo Research Corp., USA). Whole-body RNA was extracted using a Minilys bead-beater (Bertin Corp., USA), followed by two chloroform washes, ethanol suspension, and purification on a Zymo-spin IICR column (Zymo Research Corp., USA). mRNA libraries were prepared using poly-A selection and sequenced on an Illumina NovaSeq 6000 with 100 bp paired-end reads, yielding approximately 45 million reads per sample. Sequencing data are available in NCBI under bioproject accession PRJNA1100762. Adapter sequences and poly-A/T stretches were removed using cutadapt63, and reads shorter than 30 bp were discarded. Remaining reads were aligned to the Ae. albopictus reference genome (AalbF5) using the STAR aligner64 in EndToEnd mode with an outFilterMismatchNoverLmax parameter of 0.04. Duplicate reads were removed using PICARD MarkDuplicates65, and gene expression was quantified with htseq-count66 using the AalbF5 GTF annotation. Principal component analysis (PCA) was used to select the three most similar samples per group for differential expression analysis with DESeq267, filtering for a log₂ fold change >2 and adjusted p-value < 0.05. Genes were classified as specifically expressed if they were detected in all samples of one group (with a cumulative count ≥5) and absent in the other.
Analysis of M-locus linkage of male-associated genes
To confirm the linkage to the M-locus of genes expressed in WT males but not in pseudomales, two computational approaches were used: the chromosome quotient (CQ) method18 and male-specific k-mers19,20. CQ values were calculated by mapping male and female whole-genome sequencing reads21 to transcripts and coding sequences in the AalbF5 assembly (GCF_035046485.1) using bowtie with parameters -v0, -a, and --suppress 1,2,4,5,6,7,8,968. The CQ for each sequence was determined as the ratio of female to male normalized reads. Additionally, 25 bp k-mers were generated from each sex-specific library using jellyfish69, k-mers present at least ten times in male libraries but absent in female libraries were identified as male-specific. These k-mers were mapped to putative M-linked sequences using bowtie, and the total length covered by male-specific k-mers was measured. Finally, PCR on genomic DNA from pools of 20 males or females was used to validate M-locus linkage predictions (Supplementary Fig. 5).
Quantifying pupal color, size, and protandry
Images of pupae in water-filled Petri dishes were captured using a Nikon D7100 camera with a Laowa 60 mm f/2.8 2X Ultra-Macro lens mounted on a tripod. Image analysis was performed with ImageJ. Size measurements were calibrated using the Petri dish diameter as a reference. The script used for region-of-interest (ROI) identification, the full set of analyzed images, and the complete measurement datasets are publicly available through the figshare repository associated with this project. Additionally, representative sample images from each experimental group are provided as supplementary materials (Supplementary Fig. 6).
Synchronized larvae were obtained by simultaneously immersing WT and GSS eggs in hatching solution. L1 larvae from each strain were counted, and three high-density rearing trays of 1,500 larvae per strain were seeded into 500 mL of water (3 larvae/mL) in trays with a 325×175 mm surface area (2.6 larvae/cm²). Larvae were fed daily with TetraBits Complete fish food (Tetra, Melle, Germany), and trays were monitored for the onset of pupation. Once formed, and over a five-day period, pupae were collected at ten time points, manually sexed, and counted. Recovery rates for males and females were calculated based on the total number collected from each tank. Protandry was calculated for each replicate using an adaptation of the SBM (sexual bimaturism) index calculation following70 using the mean pupation time of females (F) and males (M) with the following formula: SBM = 100 x [2(F-M)]/(F + M).
Mating refractoriness assays
To evaluate whether GSS pseudomales successfully induce post-mating refractoriness in females, we conducted two reciprocal mating assays. In the first experiment a group of 50 unmated, 5-day-old females were introduced to a cage containing either 25 GSS pseudomales (fluorescently labeled with eGFP) or 25 true males carrying a fluorescent marker (OpiE2:dsRED). Two hours after introduction, females were blood-fed and transferred to a cage containing 25 males of the opposite type for the remainder of the experiment. Four days later, three egg bowls were collected from each cage and the paternal origin of L2 larvae was determined using fluorescent screening. In the second experiment, in which the single female insemination status was assessed, 25 females were individually introduced into a cage containing either 25 GSS pseudomales or transgenic true males. Each female was monitored from the moment of introduction for interception and mating. Following at least one successful copulation (lasting at least 20 seconds) the mated female was transferred to a cage containing 25 males of the opposite type for the remainder of the experiment. The cages were blood-fed on the same day, and females were individually collected four days later for egg laying. Single-female clutch analysis based on the L2 transgenic profile was subsequently performed for 20 females from each group.
Evaluating desiccation-sensitivity of GSS and WT eggs
Six egg bowls were placed in a blood-fed cage containing an equal number (50 each) of WT and GSS females, both mated to yellow males. Prior to storage, the egg papers were divided in half, and the total number of eggs was documented on the 12 egg papers, distinguishing between yellow and WT eggs. Over the course of one month, three stored egg papers were randomly hatched each week. Larval phenotypes were evaluated three days post-hatching at the L2–3 stage, and hatching rates were calculated based on the total number of recovered larvae per strain.
Statistics and reproducibility
No statistical method was used to predetermine sample size; sample numbers were maximized within the practical limits of each experimental setup. Data normality was assessed using the Shapiro–Wilk test (for datasets with n < 30), and homogeneity of variance was evaluated using Levene’s test. Image analysis outliers were identified using the interquartile range (IQR) method and excluded only when values fell more than 1.5 × the IQR below the first quartile or above the third quartile. Their removal did not alter the overall statistical trends. The statistical tests applied in each experiment are detailed in the corresponding Results sections. Statistical significance was defined as α = 0.05 for all tests. All analyses were performed in R version 4.4.1 (2024-06-14 ucrt) using the car, agricolae, and emmeans packages, with figures generated in ggplot2 and D3.js.
Ethics declarations
All animals were handled in accordance with and under the supervision of the ARO Institutional Animal Care and Use Committee approval number 2307-118-2-VOL-IL. All insect work was performed in facilities maintaining Arthropod Containment Level 2. This work received Institutional Approval and relevant authorizations from the Israel Ministry of Environmental Protection and Ministry of Agriculture (#31/2019).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Source data are provided with this paper. The experimental data generated in this study have been deposited in the figshare database under accession code 8054761. The sequences of the constructed plasmids are included in the Source Data file of this manuscript in genebank format. The transcriptomic data generated in this study have been deposited in the NCBI Sequence Read Archive under bioproject PRJNA1100762. Source data are provided with this paper.
References
Nie, P. & Feng, J. Niche and range shifts of Aedes aegypti and Ae. albopictus suggest that the latecomer shows a greater invasiveness. Insects 14, 810 (2023).
Alphey, L. Genetic control of mosquitoes. Annu. Rev. Entomol. 59, 205–224 (2014).
Raban, R., Marshall, J. M., Hay, B. A. & Akbari, O. S. Manipulating the destiny of wild populations using CRISPR. Annu. Rev. Genet. 57, 361–390 (2023).
Wang, G.-H. et al. Combating mosquito-borne diseases using genetic control technologies. Nat. Commun. 12, 4388 (2021).
Balatsos, G. et al. Sterile insect technique (SIT) field trial targeting the suppression of Aedes albopictus in Greece. Parasite 31, 17 (2024).
Crawford, J. E. et al. Efficient production of male Wolbachia-infected Aedes aegypti mosquitoes enables large-scale suppression of wild populations. Nat. Biotechnol. 38, 482–492 (2020).
Franz, G. in Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management (eds Dyck, V. A., Hendrichs, J. & Robinson, A. S.) 427–451 (Springer, 2005).
Augustinos, A. A. et al. Ceratitis capitata genetic sexing strains: laboratory evaluation of strains from mass-rearing facilities worldwide. Entomol. Exp. Appl. 164, 305–317 (2017).
Newton, M. E., Southern, D. I. & Wood, R. J. X and Y chromosomes of Aedes aegypti (L.) distinguished by Giemsa C-banding. Chromosoma 49, 41–49 (1974).
Hall, A. B. et al. A male-determining factor in the mosquito Aedes aegypti. Science 348, 1268–1270 (2015).
Lutrat, C., Olmo, R. P., Baldet, T., Bouyer, J. & Marois, E. Transgenic expression of Nix converts genetic females into males and allows automated sex sorting in Aedes albopictus. Commun. Biol. 5, 210 (2022).
Zhao, Y. et al. The AalNix3&4 isoform is required and sufficient to convert Aedes albopictus females into males. PLoS Genet. 18, e1010280 (2022).
Li, M. et al. Germline Cas9 expression yields highly efficient genome engineering in a major worldwide disease vector, Aedes aegypti. Proc. Natl. Acad. Sci. USA 114, E10540–E10549 (2017).
Liu, T. et al. Construction of an efficient genomic editing system with CRISPR/Cas9 in the vector mosquito Aedes albopictus. Insect Sci. 26, 1045–1054 (2019).
True, J. R. Insect melanism: the molecules matter. Trends Ecol. Evol. 18, 640–647 (2003).
Noh, M. Y., Mun, S., Kramer, K. J., Muthukrishnan, S. & Arakane, Y. Yellow-y functions in egg melanization and chorion morphology of the Asian tiger mosquito, Aedes albopictus. Front. Cell Dev. Biol. 9, 769788 (2021).
O’Leary, S. & Adelman, Z. N. CRISPR/Cas9 knockout of female-biased genes AeAct-4 or myo-fem in Ae. aegypti results in a flightless phenotype in female, but not male mosquitoes. PLOS Negl. Trop. Dis. 14, e0008971 (2020).
Hall, A. B. et al. Six novel Y chromosome genes in Anopheles mosquitoes discovered by independently sequencing males and females. BMC Genomics 14, 273 (2013).
Carvalho, A. B. & Clark, A. G. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 23, 1894–1903 (2013).
Papathanos, P. A. & Windbichler, N. Redkmer: an assembly-free pipeline for the identification of abundant and specific X-chromosome target sequences for X-shredding by CRISPR endonucleases. CRISPR J. 1, 88–98 (2018).
Palatini, U. et al. Comparative genomics shows that viral integrations are abundant and express piRNAs in the arboviral vectors Aedes aegypti and Aedes albopictus. BMC Genomics 18, 512 (2017).
Balestrino, F., Puggioli, A., Gilles, J. R. L. & Bellini, R. Validation of a new larval rearing unit for Aedes albopictus (Diptera: Culicidae) mass rearing. PLoS ONE 9, e91914 (2014).
Malfacini, M. et al. Aedes albopictus sterile male production: influence of strains, larval diet and mechanical sexing tools. Insects 13, 899 (2022).
Mamai, W. et al. Efficiency assessment of a novel automatic mosquito pupae sex separation system in support of area-wide male-based release strategies. Sci. Rep. 14, 9170 (2024).
Beukeboom, L. W. Size matters in insects - an introduction. Entomol. Exp. Appl. 166, 2–3 (2018).
Noh, M. Y. et al. Yellow-g and Yellow-g2 proteins are required for egg desiccation resistance and temporal pigmentation in the Asian tiger mosquito, Aedes albopictus. Insect Biochem. Mol. Biol. 122, 103386 (2020).
Swan, T. et al. A literature review of dispersal pathways of Aedes albopictus across different spatial scales: implications for vector surveillance. Parasit. Vectors 15, 303 (2022).
Girard, M. et al. Human-aided dispersal and population bottlenecks facilitate parasitism escape in the most invasive mosquito species. PNAS Nexus 3, gae175 (2024).
Lutrat, C. et al. Sex sorting for pest control: it’s raining men!. Trends Parasitol. 35, 649–662 (2019).
Bellini, R., Puggioli, A., Balestrino, F., Carrieri, M. & Urbanelli, S. Exploring protandry and pupal size selection for Aedes albopictus sex separation. Parasit. Vectors 11, 650 (2018).
Zacarés, M. et al. Exploring the potential of computer vision analysis of pupae size dimorphism for adaptive sex sorting systems of various vector mosquito species. Parasit. Vectors 11, 656 (2018).
Lutrat, C. et al. Combining two genetic sexing strains allows sorting of non-transgenic males for Aedes genetic control. Commun. Biol. 6, 646 (2023).
Papathanos, P. A. et al. A perspective on the need and current status of efficient sex separation methods for mosquito genetic control. Parasit. Vectors 11, 654 (2018).
Morgan, T. H. The origin of nine wing mutations in Drosophila. Science 33, 496–499 (1911).
Benet, J., Oliver-Bonet, M., Cifuentes, P., Templado, C. & Navarro, J. Segregation of chromosomes in sperm of reciprocal translocation carriers: a review. Cytogenet. Genome Res. 111, 281–290 (2005).
Gamez, S., Antoshechkin, I., Mendez-Sanchez, S. C., Campo, C. H. & Akbari, O. S. Exploiting a Y chromosome-linked Cas9 for sex selection and gene drive. Nat. Commun. 12, 5434 (2021).
Buchman, A. & Akbari, O. S. Site-specific transgenesis of the Drosophila melanogaster Y-chromosome using CRISPR/Cas9. Insect Mol. Biol. 28, 65–73 (2019).
Dimitri, P. & Pisano, C. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics 122, 793–800 (1989).
Berloco, M., Palumbo, G., Piacentini, L., Pimpinelli, S. & Fanti, L. Position effect variegation and viability are both sensitive to dosage of constitutive heterochromatin in Drosophila. G3 (Bethesda) 4, 1709–1716 (2014).
Aryan, A. et al. Nix alone is sufficient to convert female Aedes aegypti into fertile males and myo-sex is needed for male flight. Proc. Natl. Acad. Sci. USA 117, 17702–17709 (2020).
Krzywinska, E. & Krzywinski, J. Effects of stable ectopic expression of the primary sex determination gene Yob in the mosquito Anopheles gambiae. Parasit. Vectors 11, 646 (2018).
Criscione, F., Qi, Y. & Tu, Z. GUY1 confers complete female lethality and is a strong candidate for a male-determining factor in Anopheles stephensi. eLife 5, e19281 (2016).
Qi, Y., Criscione, F., Biedler, J. K., Sharakhov, I. V. & Tu, Z. Guy1, a Y-linked embryonic signal, regulates dosage compensation in Anopheles stephensi by increasing X gene expression. eLife 8, e43570 (2019).
Salvemini, M. et al. Genomic organization and splicing evolution of the doublesex gene, a Drosophila regulator of sexual differentiation, in the dengue and yellow fever mosquito Aedes aegypti. BMC Evol. Biol. 11, 41 (2011).
Salvemini, M. et al. The orthologue of the fruitfly sex behaviour gene fruitless in the mosquito Aedes aegypti: evolution of genomic organisation and alternative splicing. PLoS ONE 8, e48554 (2013).
Puggioli, A. et al. Efficiency of three diets for larval development in mass rearing Aedes albopictus (Diptera: Culicidae). J. Med. Entomol. 50, 819–825 (2013).
Mamai, W. et al. Optimizing larval mass-rearing techniques for Aedes mosquitoes: enhancing production and quality for genetic control strategies. Parasite 32, 29 (2025).
Sutter, A., Price, T. A. & Wedell, N. The impact of female mating strategies on the success of insect control technologies. Curr. Opin. Insect Sci. 45, 75–83 (2021).
Alphey, L. et al. Sterile-insect methods for control of mosquito-borne diseases: an analysis. Vector Borne Zoonotic Dis. 10, 295–311 (2010).
Knipling, E. F. Possibilities of insect control or eradication through the use of sexually sterile males. J. Econ. Entomol. 48, 459–462 (1955).
Lance, D. R. & McInnis, D. O. in Sterile Insect Technique (eds Dyck, V. A., Hendrichs, J. & Robinson, A. S.) 113–142 (CRC Press, 2021).
Whitten, M. & Mahon, R. in Sterile Insect Technique (eds Dyck, V. A., Hendrichs, J. & Robinson, A. S.) 601–626 (Springer, Berlin/Heidelberg, 2006).
Fried, M. Determination of sterile-insect competitiveness. J. Econ. Entomol. 64, 869–872 (1971).
Hooper, G. H. S. & Horton, I. F. Competitiveness of sterilized male insects: a method of calculating the variance of the value derived from competitive mating tests. J. Econ. Entomol. 74, 119–121 (1981).
Bond, J. G. et al. Sexual competitiveness and induced egg sterility by Aedes aegypti and Aedes albopictus gamma-irradiated males: a laboratory and field study in Mexico. Insects 12, 145 (2021).
Chen, C., Qualls, W. A., Xue, R., Gibson, S. & Hahn, D. A. X-rays and gamma rays do not differ in their effectiveness for sterilizing pupae and adults of the mosquito Aedes aegypti (Diptera: Culicidae). J. Econ. Entomol. 118, toaf030 (2025).
Kittayapong, P., Ninphanomchai, S., Thayanukul, P., Yongyai, J. & Limohpasmanee, W. Comparison on the quality of sterile Aedes aegypti mosquitoes produced by either radiation-based sterile insect technique or Wolbachia-induced incompatible insect technique. PLoS ONE 20, e0314683 (2025).
Yamada, H. et al. Sperm storage and use following multiple insemination in Aedes albopictus: encouraging insights for the sterile insect technique. Insects 15, 721 (2024).
Zheng, M.-L., Zhang, D.-J., Damiens, D. D., Lees, R. S. & Gilles, J. R. L. Standard operating procedures for standardized mass rearing of the dengue and chikungunya vectors Aedes aegypti and Aedes albopictus (Diptera: Culicidae) - II - egg storage and hatching. Parasit. Vectors 8, 348 (2015).
Concordet, J.-P. & Haeussler, M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245 (2018).
Fuchs, S., Nolan, T. & Crisanti, A. Mosquito transgenic technologies to reduce Plasmodium transmission. Methods Mol. Biol. 923, 601–622 (2013).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Dobin, A. & Gingeras, T. R. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinform. 51, 11.14.1–11.14.19 (2015).
Broad Institute. Picard toolkit. http://broadinstitute.github.io/picard/ (accessed 3 March 2024).
Putri, G. H., Anders, S., Pyl, P. T., Pimanda, J. E. & Zanini, F. Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics 38, 2943–2945 (2022).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Marçais, G. & Kingsford, C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011).
Blanckenhorn, W. U., Stillwell, R. C., Young, K. A., Fox, C. W. & Ashton, K. G. Proximate causes of Rensch’s rule: does sexual size dimorphism in arthropods result from sex differences in development time? Am. Nat. 169, 245–257 (2007).
Acknowledgements
The authors would like to thank Dor Perets, Guy Ostrovsky, Shira Kehat, Gleb Ens, and Ezra Bohbot for technical assistance. Special thanks to Kostas Bourtzis for his invaluable insights, longstanding support and guidance, which have greatly contributed to this work. We thank Mariangela Bonizzoni and Francesca Scolari for the Foshan strain. We thank Amir Szitenberg of the Mantoux Bioinformatics Institute of the Nancy and Stephen Grand Israel National Center for Personalized Medicine, Weizmann Institute of Science for technical support in the RNA-seq analysis. This study benefited from discussions at meetings for the Coordinated Research Project D44003 on the “Generic approach for the development of genetic sexing strains for SIT applications”, funded by the International Atomic Energy Agency (IAEA).
This research was supported by grants from the Ministry of Science & Technology, Israel to PAP (grant agreement numbers 3-16795 and 3-17985). Funding was also provided by the German-Israeli Middle East Project Cooperation of the German Research Foundation (SCHE 1833/7-1 to PAP and MFS) and the European Union’s Horizon Europe Research and Innovation Program (REACT - grant agreement number 101059523 to PAP and MFS) and the Agence Nationale de la Recherche (GC-TiMO - grant agreement number ANR-23-CE35-0003 to EM and PAP). Initial support was generously provided in the form of an International Fellowship to FK from the Research Fund for International Cooperation, Robert H. Smith Faculty of Agriculture, Food and Environment, HUJI and startup funds to PAP.
Author information
Authors and Affiliations
Contributions
Conceptualization—D.S.Y.Z., P.A.P.; Methodology—D.S.Y.Z., P.A.P.; Resources—D.S.Y.Z., F.K., E.M., P.A.P.; Validation—D.S.Y.Z., D.A.H., Y.A., P.A.P.; Formal Analysis—D.S.Y.Z., D.A.H., P.A.P.; Investigation—D.S.Y.Z., O.T., F.K., D.A.H., D.G., N.G., I.H., M.F.S., Y.A., P.A.P.; Writing—Original Draft Preparation—D.S.Y.Z., P.A.P.; Writing—review and editing—D.S.Y.Z., Y.A., E.M., M.F.S., P.A.P.; Visualization—D.S.Y.Z., P.A.P.; Supervision—P.A.P.; Funding Acquisition—E.M., M.F.S., P.A.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Nigel Beebe, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zaada, D.S.Y., Toren, O., Krsticevic, F. et al. Mosquito sex separation using complementation of selectable traits and engineered neo-sex chromosomes. Nat Commun 16, 11175 (2025). https://doi.org/10.1038/s41467-025-66940-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-66940-0