Abstract
Emerging evidence indicates important interconnections between autophagy and the secretion of small extracellular vesicles. However, our understanding of these secretory vesicles remains incomplete. Here, we identify a subtype of small extracellular vesicles, termed autophagic extracellular vesicles (AEVs), which are distinct from exosomes. Extracellular AEVs characterized by a size less than 100 nm exhibit increased secretion when the autophagy response is induced. Amphisomes, the hybrid organelles, are essential for the secretion of these types of vesicles. Further exploration reveals that autophagic cargos, certain ESCRT Ⅲ components and the Rab13 serve as distinctive markers for distinguishing AEVs from exosomes. Moreover, we find that the biogenesis of AEVs functionally requires components of the ESCRT Ⅲ complex and the GTPase Rab27a. Finally, we confirm that the enteroviral particles or genomes can be encapsulated into AEVs and subsequently infect receptor-negative cells with high efficiency. This model represents a prominent pattern of EV-mediated virus transmission.
Similar content being viewed by others
Introduction
Mammalian cells secrete extracellular vesicles (EVs), which can be distinguished on the basis of their origin, size, and other factors. Small extracellular vesicles (sEVs) are EVs smaller than 200 nm in diameter, and are the most widely studied types of EVs because of their many roles in different physiological and pathological conditions1,2. As an important subpopulation of sEVs, exosomes begin with inward budding of the plasma membrane to form early endosomes followed by maturation into multivesicular bodies (MVBs), which involves the formation of intraluminal vesicles (ILVs) via inward budding of the endosomal membrane. MVBs fuse with the plasma membrane and then release ILVs into the extracellular space as exosomes3. The mechanisms of exosome biogenesis are fairly well understood and importantly involve subunits of the endosomal sorting complex required for transport (ESCRT). The ESCRT machinery as a driver of membrane shape and scission, and is involved in the formation of MVBs and ILVs. As a result, some ESCRT components can act selectively on MVB and ILV subpopulations fated for secretion as exosomes, meanwhile, other components, such as ESCRT Ⅲ, remain on the outside of ILVs until they are recycled4.
The canonical pathway of autophagy involves the coordinated degradation and recycling of intracellular components and pathogenic material. This occurs through degradation upon the fusion of autophagosomes with lysosomes. Unlike degradative autophagy, the autophagic machinery can, through a shared yet partially divergent pathway, result in the secretion of cytoplasmic components rather than their degradation. This process involves the fusion of autophagosomes and MVBs to create a secretory amphisome, which subsequently releases its cargo upon binding to plasma membrane in an event termed secretory autophagy5,6,7. The consequences of this secretory amphisome release may be the presence of sEVs positive for both exosomal markers and autophagy-associated proteins, which may contain poorly characterized cargos and functionalities that are distinctly different from those of conventional exosomes. The common step in exosome biogenesis and secretory amphisome formation is the formation of MVBs. Nevertheless, extracellular secretion of autophagy components and autophagy cargo receptors including p62/SQSTM1 and LC3B can be markedly enhanced in response to lysosomal inhibition or the accumulation of autophagosomes, but these autophagic dysfunction status has a negligible effect on the secretion of exosomal markers8,9. In addition, autophagy-related LC3 conjugation machinery regulates selective packaging and secretion of the secretome extracellularly10. These observations suggest that the secretion of autophagy-related proteins may occur in distinct extracellular vesicle populations. Importantly, many infectious pathogens such as viruses, specifically disrupt the lysosomal system, preventing clearance by autophagy-dependent degradation and impacting sEV or exosome secretion11,12,13,14. Therefore, exploring the relationship between secretory autophagy and exosomes not only offers a framework for a clearer understanding of extracellular vesicle heterogeneity, but also provides insights into disturbances in homeostasis such as during viral infection.
In this study, we identified a subtype of small extracellular vesicles, termed AEVs (autophagic extracellular vesicles) that were distinct from conventional exosomes. Extracellular AEVs characterized by a size less than 100 nm exhibited increased secretion when autophagy response was triggered in vitro and in vivo. Amphisomes, formed by the fusion of MVBs and autophagosomes, are essential for the secretion of these types of vesicles. Further analysis revealed that autophagic cargos, ESCRT Ⅲ components and Rab13 serve as distinctive markers for distinguishing AEVs from exosomes. Moreover, we determined that the biogenesis of AEVs functionally requires certain ESCRT Ⅲ components and the small GTPase Rab27a. Finally, using two enterovirus strains, we confirmed that the AEVs could carry and deliver viral particles or whole genomes into receptor-negative host cells with high infection efficiency, thereby broadening viral tropism and facilitating viral transmission.
Results
Starvation-induced autophagy affects the size distribution of small extracellular vesicles
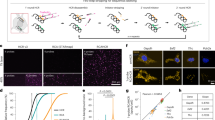
Previous studies revealed that interconnections between the autophagy process and small extracellular vesicle (sEV)/exosome production is closely connected15. To investigate how the autophagic response affects the secretion yield and particle size of small extracellular vesicles, HEK-293T cells were cultured under serum-starved conditions to induce an autophagic response as determined by autophagic flux of cargos such as p62 and LC3B-Ⅱ (Fig. 1A). The vacuolar-type ATPase (v-ATPase) inhibitor bafilomycin A1 (BafA1), which disrupts lysosome acidification, was employed to facilitate the accumulation of autophagosomes (Fig. 1A). The sEV fractions were isolated from the culture supernatant using the traditional method that is the main one used in published studies (Fig. 1B). Nanoparticle tracking analysis (NTA) revealed a singular bell-shaped size distribution with peaks at 143 nm in the sEVs derived from 293 T cells under normal conditions (Fig. 1C). Unexpectedly, a double-bell-shaped size distribution was detected in sEVs isolated from serum-starved 293 T cells, with peaks at 92 nm and 138 nm. Similar double-peak contours were also observed in sEVs derived from serum-starved 293 T cells after BafA1 treatment, which correlated with the accumulation of autophagosomes, with peaks at 99 nm and 136 nm (Fig. 1C). Further analysis revealed that the percentage or number of sEVs with a size smaller than 100 nm derived from serum-starved 293 T cells, regardless of BafA1 treatment, was significantly greater than that from normally cultured cells (Fig. 1D–F). However, the secretion of sEVs with a size range of 100-200 nm varied minimally among normal cells, nutrient-deprived cells and cells treated with BafA1 (Fig. 1G). The double-bell-shaped size distributions of sEVs were also observed in other cell lines (Neuro-2a, THP−1 and SH-SY5Y cells) when autophagy was induced by serum deprivation, and the percentage of sEVs with a size less than 100 nm from serum-starved cells increased (Fig. S1). These results suggested that the induction of autophagy due to nutrient deprivation affects the size distribution of sEVs and that this phenomenon is a generalized response. Additionally, the increase in sEVs from starved 293 T cells with a size smaller than 100 nm was predominantly within the range of 70-90 nm (Fig. 1H). To further characterize the increase in sEVs smaller than 100 nm induced by autophagy in vivo, we detected sEVs from mouse serum after the mice were maintained under normal feeding or starvation conditions. An induction of the autophagic response in the livers of starved mice was observed, as evidenced by a decrease in the expression of the autophagic cargo protein p62 (Fig. 1I). As shown in Fig. 1J, the percentage of sEVs less than 100 nm in size was notably greater in serum from starved mice than in serum from normal feeding mice. However, the quantity of sEVs in serum with a size range of 100-200 nm differed little between normal feeding mice and starved mice (Fig. 1K). Collectively, these findings suggest that the starvation-induced autophagic response influences the size distribution of small EVs, which might be attributed to autophagosome formation.
A Whole cell lysates from HEK293T cells under normal conditions or serum - starved, with or without BafA1 treatment, were collected and subjected to blotting for autophagic marker proteins. B Schematic illustration of the differential centrifugation protocol for isolating small EVs (sEVs). Created in BioRender. Ruan, H. (2025) https://BioRender.com/zrc5x19. C, D The particle size distribution for sEVs from various treatment of 293T cells as determined by nanoparticle tracking analysis (NTA). The red frame represented the proportion of particles with size less than 100 nm (C). Data presented as mean ± SD from three independent experiments (D). E–G The quantity of sEVs derived from 293 T cells subjected to various treatments was quantified using NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ***p < 0.0001 (E), ***p = <0.0001 (F). ns: no significance. H The detailed particle size distribution of sEVs with a size below 100 nm, derived from 293 T cells cultured under normal conditions or serum - starved, as determined by NTA detection. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. *p = 0.0210 (60-70 nm), **p = 0.0016 (70-80 nm),***p = 0.0069 (80-90 nm). I Tissue lysates were prepared from the liver tissues of fasted and normally fed mice and then blotted for the indicated proteins. Each group consisted of six mice (n = 6). J, K Serum samples were collected from fasted and normally fed mice to assess the particle size distribution of sEVs using NTA. Each group consisted of six mice (n = 6). Data are presented as the mean ± SD. Error bars denote the maximum and minimum values. For the box plots, the horizontal line inside represents the median, while the bottom and top of the box correspond to the 25th and 75th percentiles, respectively. Statistical analysis was performed using a two-tailed unpaired Student’s t-test. ***p < 0.0001 (J). ns: no significance (K). Source data are provided as a Source Data file.
The stress-induced autophagy response led to increased secretion of sEVs less than 100 nm in size
To further investigate whether other inducers of the autophagy response affect the size distribution of sEVs, cells were treated with the autophagy inducer rapamycin to trigger an autophagy response, and the corresponding secreted sEVs exhibited a double-bell-shaped size distribution (Fig. 2A, B). Compared with the sEVs secreted from mock cells, the percentage of sEVs smaller than 100 nm increased (Fig. 2C, D). The quantity of sEVs with a size range of 100–200 nm differed little between mock cells and rapamycin-treated cells (Fig. 2D).
A Whole cell lysate from 293 T cells in the presence or absence of rapamycin were collected and blotted for the indicated proteins. B, C The particle size distribution for sEVs from 293 T cells treated with or without rapamycin as determined by NTA. The red frame represented the proportion of particles with a size less than 100 nm, and data presented as mean ± SD from three independent experiments. D, E The quantity of sEVs derived from 293 T cells either with or without rapamycin treatment was quantified by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. ***p = 0.0008 (D). F Varicella zoster virus (VZV)-infected ARPE−19 cells with or without acyclovir treatment were collected and blotted for the indicated proteins. G, H The quantity of sEVs in the supernatant of variously treated cells was determined by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ** p = 0.0027(VZV) and 0.0020(VZV + ACV) (G), ***p = 0.0001(VZV) and 0.0001(VZV + ACV) (H). ns: no significance. I NTA detection for particle size distribution of sEVs with a size below 100 nm, derived ARPE−19 cells with or without VZV infection. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. *p = 0.0230 (60-70 nm),***p = 0.0006 (70-80 nm), **p = 0.0012 (80-90 nm), **p = 0.0019 (90−100 nm). J The quantity of sEVs derived from ARPE−19 cells either with or without VZV infection was quantified by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ns: no significance. K The percentage of sEVs in the serum of healthy young men aged 18-35 (n = 39) and healthy older men aged over 65 years (n = 53) was determined by NTA. Data are presented as the mean ± SD. Error bars denote the maximum and minimum values. For the box plots, the horizontal line inside represents the median, while the bottom and top of the box correspond to the 25th and 75th percentiles, respectively. Statistical analysis was performed using a two-tailed unpaired Student’s t-test. ***p < 0.0001 Source data are provided as a Source Data file.
Virus infection induced host autophagy has been widely reviewed16. Our results confirmed that the Varicella-Zoster virus (VZV), a human herpes virus responsible for chicken pox, could induce an autophagy response in human retinal pigment epithelium (RPE) cells, as determined by observing autophagic cargos such as NBR1, p62 and LC3B-Ⅱ (Fig. 2F). This autophagy process could be inhibited by treatment with acyclovir (ACV), an acceptable anti-herpesvirus drug (Fig. 2F). NTA revealed that the percentage of sEVs less than 100 nm in size from VZV-infected cells clearly increased compared with those from mock cells (Fig. 2G, H). The antiviral drug acyclovir, which inhibited VZV-induced autophagy, significantly decreased the secretion of sEVs smaller than 100 nm. Like in the starvation treatment group, the number of sEVs with a size less than 100 nm, which increased in the VZV-infected cells, was predominantly in the 70–100 nm range (Fig. 2I). In addition, VZV infection did not alter the number of secreted sEVs within the size range of 100–200 nm (Fig. 2J). Similarly, other human viral strains, such as coxsackievirus B4 (CVB4) and Herpes simplex virus type 1 (HSV−1), which have been confirmed to induce a cellular autophagy response (Fig. S2A, D), showed increased secretion of sEVs smaller than 100 nm (Fig. S2B, C, E, F).
Dysfunction of the autophagy-lysosome pathway is frequently associated with aging and age-related diseases17. Therefore, we detected the concentration of sEVs smaller than 100 nm in serum samples isolated from healthy older men aged over 65 years and observed a significantly greater percentage of sEVs less than 100 nm in size in the older population than in younger individuals aged 18–40 (Fig. 2K) (Supplementary Tables 1 and 2).
Although nanoparticle tracking analysis (NTA) is widely used to detect the size and concentration of extracellular vesicles, resistive pulse sensing (RPS) techniques that are calibrated with size-standards, and limit of detection can be reported in terms of diameter, and are emerging as new methods for particle sizing and counting18. Therefore, the size distribution of sEVs from mock, starved, CVB4-infected and rapamycin-treated 293 T cells was detected by RPS techniques through a nanopore chip with a measurement range of 50–240 nm. Compared with that from mock cells, the secreted sEVs in the size range of 50–100 nm was increased from starved, CVB4-infected and rapamycin-treated 293 T cells compared to that from mock cells (Fig. S2G, K), which is consistent with the above NTA results. Moreover, when RPS was used, both Neuro-2a cells and SH-SY5Y neuronal cells presented higher percentages of sEVs ranging from 50–100 nm in size under starvation conditions (Fig. S2H, I, K). Compared with that from mock cells, the quantity of secreted sEVs smaller than 100 nm was significantly greater from VZV-infected RPE cells (Fig. S2J). Overall, our results indicated that induction of the autophagy response modulated the size distribution of small EVs, resulting in increased secretion of sEVs with sizes smaller than 100 nm.
Autophagic extracellular vesicles (AEVs) represent a subtype of small extracellular vesicles and are distinct from exosomes
Exosomes are typically described as having a diameter of 30−150 nm. The ESCRT pathway is involved in exosome biogenesis2. To investigate whether secreted exosomes contribute to the increase in the quantity of sEVs with size less than 100 nm induced by autophagy, we generated TSG101 and ATG7 gene knockout (TSG101-KO and ATG7-KO) HEK-293T cells, which were either subjected to starvation treatment or not. TSG101 plays a role in the process of exosome biogenesis and ATG7 is essential for autophagosome formation. Western blot analysis revealed that the vesicles secreted from starved wild-type cells (WT cells) or TSG101-KO cells contained high levels of autophagic cargos, including NBR1, p62 and lipidated LC3B (LC3B-II). In contrast, these autophagic cargos were barely detected in the vesicles secreted from either normal or starved ATG7-KO cells (Fig. 3A, B). CD63 and CD9, which are often considered markers associated with exosomes, were largely decreased in secreted vesicles from TSG101-KO cells compared with those from WT cells, whereas the expression of the marker CD81 decreased slightly (Fig. 3A, B). Importantly, compared with those in WT cells, the levels of both CD63 and CD9 in vesicles secreted from ATG7-KO cells were unaltered, whereas the level of CD81 was reduced. These results suggested that inhibition of exosome biogenesis did not affect the secretion of autophagic cargos via small extracellular vesicles. Moreover, the secretion of sEVs less than 100 nm in size from TSG101-KO cells was greater than that from WT cells under starvation conditions, but this secretion was markedly suppressed in ATG7-KO cells (Fig. 3C). These findings indicated that the inhibition of autophagosome formation, rather than exosome biogenesis, contributed to the release of sEVs less than 100 nm in size.
A The vesicle fractions from an equal number of wild-type (WT), TSG101-, or ATG7-knockout (KO) 293 T cells, following culture under normal or serum-starved conditions, were collected and subjected to blotting. B Quantification of proteins in vesicle fractions from TSG101-KO or ATG7-KO cells under serum-starved conditions relative to those from WT cells. The relative expression was calculated by normalizing the expression levels of proteins from WT cells in each experimental group. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. ***p < 0.0001 (NBR1), ***p < 0.0001 (p62), ***p < 0.0001 (LC3B-II), ***p < 0.0001 (CD63), ***p < 0.0001 (CD9), ***p < 0.0001 and ***p = 0.0005 (CD81), ***p < 0.0001 and ***p = 0.0002 (TSG101). ns: no significance. C The percentage of sEVs with size below 100 nm from WT and KO cells under normal or serum-starved conditions, was determined by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. ns: no significance. D Cell lysates from Rab11 knockout (KO) 293 T cells under normal or serum-starved conditions were collected and subjected to blotting. E The percentage of sEVs with indicated size derived from an equal number of WT or Rab11-KO 293 T cells that were subjected to various treatments, was determined by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ns: no significance. F The vesicle fractions from WT or Rab11-KO 293 T cells under serum-starved culture condition were collected and blotted for the indicated proteins. G The vesicle fractions from WT or CD63-KO 293 T cells under serum-starved conditions were collected and subjected to blotting. H The vesicle fractions from WT or ATG7-KO 293 T cells, following culture under normal or serum-starved conditions, were collected and subjected to blotting. I, J The sEV subpopulations from normally cultured and serum-starved 293 T cells were determined via FlowSOM algorithm. Data were shown in t-distributed stochastic neighbor embedding (t-SNE) plot (I). The frequency of top10 proteins in the cluster was listed (J). Source data are provided as a Source Data file.
The Rab11 protein is required for the fusion of MVBs and autophagosomes to generate hybrid amphisomes, which are subsequently released into the extracellular space as vesicles19. To explore whether the fusion of MVBs and autophagosomes affects the sEVs smaller than 100 nm, we generated Rab11 gene knockout (Rab11-KO) cells. Deletion of Rab11 did not affect the fusion of autophagosomes with lysosomes under starvation conditions, as evidenced by decreased p62 levels and increased LC3B-II accumulation (Fig. 3D). We found that the secretion of vesicles less than 100 nm in size from Rab11-KO cells failed to increase despite the induction of autophagy by means of starvation, rapamycin treatment and CVB4 infection (Fig. 3E). Moreover, the autophagic cargos NBR1, p62 and LC3B-II were barely detected in the secreted vesicles of starved Rab11-KO cells (Fig. 3E). In contrast, the levels of the exosome markers CD63 and CD9 remained unchanged, with the exception of those of CD81 (Fig. 3F). Similarly, compared with those in starved WT cells, the levels of p62 and LC3B-II in the secreted vesicles of starved CD63 gene knockout (CD63-KO) cells were not affected (Fig. 3G). A previous study revealed that glutamine-depleted conditions could induce the secretion of Rab11a-exosomes that were unlike late endosomal exosomes20. To determine whether Rab11a-exosomes are involved in autophagy-induced vesicle formation, we detected the levels of Rab11a and caveolin−1 in vesicles secreted from WT and ATG7-KO cells, both of which were enriched in Rab11a-exosomes. These proteins were elevated in the secreted vesicles from starved ATG7-KO cells compared with those from normal culture conditions (Fig. 3H), suggesting that the subtypes of the Rab11a-exosomes were not the same as those of the autophagy-induced vesicles. Therefore, these results demonstrated that amphisomes, formed by the fusion of MVBs and autophagosomes, were essential for the secretion of vesicles less than 100 nm in size that carried autophagic cargos. Importantly, these amphisome-associated extracellular vesicles were distinct from exosomes derived from ILVs because of the inward budding of MVBs. Therefore, we termed these vesicles autophagic extracellular vesicles (AEVs), a subtype of small extracellular vesicles.
To further confirm that AEVs represent a subpopulation of small extracellular vesicles, we utilized a proximity barcoding (PBA) assay, a high-throughput method for single-sEV analysis, to simultaneously profile approximately 260 surface proteins on a single sEV. The proteomic characteristics of sEVs isolated from normal or starved 293 T cells were determined via the FlowSOM algorithm, an unsupervised machine learning process, to generate a sEV cluster map. The clustering of sEVs from samples is shown in the t-distributed stochastic neighbor embedding (t-SNE) plot via FlowSOM in Fig. 3I. Next, we found a distinct subpopulation of vesicles (blue dots) present in sEVs derived from starved cells, accounting for 99.24% of the total, compared with only 0.76% of the sEVs from normally cultured cells. CD81 protein expression was relatively high in this subpopulation (Fig. 3J). These results may explain why the level of CD81, rather than that of CD63 and CD9, decreased in the sEV fraction from starved Rab11-KO cells, as shown in Fig. 3E. Taken together, our results suggest that AEVs, characterized by their transport of autophagy cargo proteins and sizes less than 100 nm, represent a subtype of small extracellular vesicles. They were distinguishable from both exosomes and the traditionally recognized small EVs.
AEVs exhibited a unique composition consisting of Rab GTPases and ESCRT components
To identify the characteristics of AEVs, we designed two separate strategies for AEV collection using differential centrifugation coupled with serial filtration (Method A and Method B in Fig. 4A). Using sequential filtration in conjunction with ultracentrifugation at 100,000 g (Method A), approximately 60% of the total vesicles were less than 100 nm in size, suggesting that this method was insufficient for isolating high-purity AEVs (Fig. 4B). After many rounds of testing and improvement, we performed an additional ultracentrifugation at 40,000 g to rule out large-sized vesicles from the supernatant after passage through a 0.1 μm filter, followed by a ultracentrifugation step at 100,000 g, referred to as Method B. The proportion of AEVs with sizes less than 100 nm exceeded 80%, as determined by NTA (Fig. 4B). To further support these findings, we analyzed the autophagic markers associated with AEVs in collected vesicle fractions using either Method A or Method B and revealed that the levels of NBR1, p62 and LC3B-II were similarly enriched in these vesicle fractions (Fig. 4C). Importantly, the levels of CD63 and CD9 were markedly lower in the vesicle fractions isolated by Method B than in those obtained through Method A, whereas this effect was not observed for CD81 (Fig. 4C). In addition, the p62-positive AEVs with fluorochrome labeling mainly displayed sizes less than 100 nm, as determined by NTA (Fig. 4D). These results suggested that Method B for collecting purified AEVs was sufficiently valid.
A Schematic presentation of the differential centrifugation protocol used to isolate autophagic extracellular vesicles (AEVs) from the cell culture supernatant. The majuscule A and B represented two methods for AEV isolation. Created in BioRender. Ruan, H. (2025) https://BioRender.com/pouij2q. B The percentage of AEVs with a size below 100 nm from supernatant using method A or method B were determined by NTA. 293 T cells were cultured under serum-starved condition for 24 h and the supernatant was collected. Data were presented as the mean ± SD of four independent experiments. p-values were determined by unpaired two tailed Student’s t tests. ***p = <0.0001. C The AEV fraction from serum-starved 293 T cells using method A or method B were collected and subjected to blotting for the indicated proteins. D The particle size distribution for p62-labelled AEVs were determined by NTA. The AEVs were isolated from serum-starved 293 T cells and incubated with p62 antibody conjugating Alexa Fluor 488. Data presented as mean ± SD from three independent experiments. GO enrichment analysis of proteins enriched in AEV fraction from serum-starved wild-type 293 T cells (starv-WT) relative to AEV fraction from normally cultured wild-type 293 T cells (nm-WT) with the top 20 terms for cellular component (E) and biological processes (F). Each group contained three independent biological replicates. G, H GO enrichment analysis of proteins decreased in AEV fraction from serum-starved Rab11-KO 293 T cells (starv-KO) relative to AEV fraction from serum-starved WT 293 T cells (starv-WT) with the top 20 terms for cellular component (G) and biological processes (H). I Table of significantly changed proteins in different originated AEVs from quantitative proteomic analysis between starv-WT relative to nm-WT and starv-KO relative to strav-WT. Numbers in the table represented the multiples of change, indicating either an increase or a decrease. Source data are provided as a Source Data file.
To define the protein composition of the AEVs, we performed a quantitative proteomic analysis on the AEVs from starved HEK-293T cells and the vesicles from normally cultured cells, and Method B was used for isolation. Gene Ontology (GO) terms related to biological processes highlighted a profound enrichment in AEV proteins that localize to autophagy/macroautophagy, autophagosome maturation, the MVB assembly/sorting pathway and the ESCRT complex in control cells versus vesicles from normally cultured cells (Fig. 4E). This finding was consistent with our prior findings that revealed that the production of AEVs depended on the fusion of autophagosomes with MVBs. In addition, GO terms related to molecular function revealed a significant enrichment of AEV proteins associated with protein binding and RNA binding terms (Fig. 4F), suggesting that the loading and contents of proteins in AEVs were associated with specific proteins, such as protein and RNA binding proteins.
To further corroborate these results, we performed proteomics and compared vesicles purified from starved Rab11-KO cells with those from starved WT cells (Supplementary Data 1). We found that many proteins were dramatically decreased in vesicles from starved Rab11-KO cells and that the proteins whose expression was reduced were associated mainly with autophagy, the MVB functional pathway and ESCRT complex activity, as annotated by GO terms for biological processes (Fig. 4G), which is consistent with the role of Rab11 in controlling the fusion of autophagosomes with MVB to release their contents outside the cells as AEVs. The proteins whose expression was significantly reduced in vesicles from Rab11-KO cells exhibited a highly interconnected network enriched in protein binding, as annotated by GO molecular function terms (Fig. 4H). Notably, proteomic analyses of the AEV fraction from starved WT cells revealed that the expression of Rab13, a small GTPase, was greater in the AEV fraction than in the vesicle fraction from normally cultured cells. In addition, Rab11 knockout resulted in a significant reduction in Rab13 levels in AEVs (Fig. 4I), indicating that Rab13 acts as a cargo marker involved in the formation of AEVs. Moreover, we observed that several components of ESCRT, including VPS4A/B, CHMP2A/1 A/2B, VAT1, TSG101, MVB12A and PDCD6IP/Alix, were also enriched in the AEV fraction derived from starved WT cells. In contrast, these ESCRT components were largely decreased in vesicle fractions from Rab11-KO cells (Fig. 4I). These observations highlight that specific ESCRT components, which are not typically present in defined exosomes, may be involved in AEV formation or secretion. Notably, the majority of these components were the ESCRT III complex and its associated accessory proteins. Taken together, our findings confirmed that the isolation of purified AEVs can be effectively achieved using a specific separation method. Moreover, this type of extracellular vesicle is enriched in Rab GTPases and ESCRT III components.
Autophagic cargos, ESCRT-III components, and Rab13 serve as distinct markers that differentiate AEVs from conventional exosomes
We next assessed whether the Rab13 and ESCRT components identified via proteomics analysis were released in association with AEVs in response to autophagy. AEV or vesicle fractions were isolated from starved WT, TSG101-KO, ATG7-KO or Rab11-KO cells using Method B, as described above. As shown in Fig. 5A, TSG101 knockout did not affect the secretion of Rab13, CHMP2A or VPS4A/B, or the release of AEVs under starvation conditions. In contrast, the knockout of ATG7 and Rab11 led to a decrease in the expression of these proteins in the vesicle fraction, suggesting that the expression of the Rab13 and ESCRT-III components was indeed coupled with AEV secretion.
A The vesicle fraction from supernatant of wild-type (WT) or gene knockout (KO) 293 T cells with or without starvation induction was collected and blotted for the indicated proteins. Similar results were obtained in three independent experiments. B–D The iodixanol density gradient procedure to separate sEV fraction secreted from serum-starved 293 T cells (C) and subcellular fractionations of these cells (D). A 5-40% Optiprep gradient was collected in 12 fractions (F1-F12) and analyzed by western blot. Similar results were obtained in three independent experiments. B Created in BioRender. Ruan, H. (2025) https://BioRender.com/al3dqi8. E Western blot of indicated proteins from untreated AEVs or AEVs incubated with 100 μg/ml trypsin and/or 1% Triton X-100 (TX-100) for 30 minutes at 4 °C. Similar results were obtained in three independent experiments. F Immunogold-stained electron microscopy images of AEVs from starved 293 T cells. The gold particles (black dots) indicated the presence of LC3B in the vesicles, while p62 and Rab13 were present on the vesicle membranes. Bar =100 nm. Similar results were obtained in three independent experiments. The AEV fractions from THP-1 (G) and Neuro−2a (H) cell lines under normally cultured or serum-starved condition were collected and blotted for the indicated proteins. Cell lines were cultured in serum-free medium to induce autophagy. Similar results were obtained in three independent experiments. I Schematic of distinct biomarkers for classic exosomes and AEVs, both of which were two subtypes of small EVs. Source data are provided as a Source Data file.
We further attempted to separate the AEVs from total small EVs on the basis of density differences, and total small EVs prepared from starved 293 T cells using differential centrifugation, as described in Fig. 1B, were sedimented in a density gradient (5%–40% iodixanol) for 18 h at 100,000 g, leading to equilibration according to vesicle density. Western blot analysis revealed that the exosome/sEV markers CD63, CD9 and CD81 were present in fractions 7 and 8 (Fig. 5C). These fractions were also enriched in autophagic cargos, including the LC3B-II, Rab13, p62, and ESCRT III components VPS4A/B and CHMP2A, suggesting that the density of the AEVs is similar to that of the exosomes in iodixanol gradients, making it challenging to effectively separate the AEVs from the exosomes (Fig. 5C). Moreover, we identified lipidated LC3B-II in fraction 9, which was cofractionated with both Rab13 and Rab11 following continuous density gradient ultracentrifugation of the cell lysate. The early endosome marker EEA1 (fractions 4–5), endoplasmic reticulum marker calreticulin (fractions 2–5) and the cytoskeleton marker α-tubulin did not colocalize with the lipidation of LC3B-II (Fig. 5D). In addition, the late endosome/MVB marker CD63 was present mainly in fractions 6–8, as illustrated in Fig. 5D.
Protease protection assays were conducted in which starved 293T-derived AEVs were treated with either trypsin or trypsin plus detergent (Triton X−100). The autophagic cargos (p62 and NBR1), ESCRT-III components (VPS4A/B and CHMP2A) and Rab13 were dramatically digested by trypsin in the absence of detergent. However, LC3B-II was resistant to trypsin digestion in the absence of detergent but was degraded upon exposure to detergent. The protein TSG101, known for its presence in the luminal compartment of exosomes, maintained a stable abundance in AEVs regardless of trypsin digestion (Fig. 5E). Consistently, the immunoelectron microscopy showed that the AEVs isolated from starved 293 T cells exhibited immunoreactivity with antibodies against p62 and Rab13 on the vesicle membranes, while antibodies against LC3B were present inside the vesicles (Fig. 5F).
Next, we determined whether these specific contents are present in AEVs from other cell types. The levels of autophagic cargos (NBR1, p62 and LC3-II), ESCRT-III components (VPS4A/B and CHMP2A) and Rab13 were markedly greater in the AEVs derived from leukemia monocytic THP−1 and neuroblastoma Neuro2a cells under starved conditions than in those derived from cells maintained under normal culture conditions (Fig. 5G, H). Overall, on the basis of these results, we proposed that the AEVs originating from intracellular hybrid amphisomes were rich in autophagic cargos, ESCRT III components and small GTPases. These elements serve as distinctive markers for distinguishing AEVs from conventional exosomes (Fig. 5I).
Rab13 contributes to the sorting of cargo proteins into AEVs without affecting their secretion
Given the significant enrichment of the small GTPase Rab13 in AEVs, we aimed to investigate its functional role in AEV biogenesis. Super-resolution (SR) microscopy revealed that endogenous Rab13 was localized around MVBs with CD63 labeling under normal conditions in 293 T cells (Fig. 6A). Notably, upon autophagic activity induced by starvation, the structures of Rab13 closely bind to CD63-labeled MVBs (Fig. 6A), indicating that Rab13 may be involved in the amphisome generation process. Unexpectedly, Rab13 knockout did not affect the secretion of total sEVs regardless of whether the cells were subjected to starvation stimulation (Fig. 6B). Moreover, the secretion of AEVs differed little between WT and Rab13-KO cells under starvation conditions (Fig. 6C). These results suggest that Rab13 does not contribute to the formation or release of AEVs. Nevertheless, western blot analysis revealed that the levels of NBR1, p62 and LC3B-II in AEVs from starved Rab13-KO cells were largely lower than those in AEVs from starved WT cells, suggesting that the depletion of Rab13 disturbed the secretion of autophagic cargos via AEVs (Fig. 6D). The ESCRT components VPS4A/B, CHMP2A and TSG101 were also reduced in AEVs from starved Rab13-KO cells. In support of this observation, the colocalization of Rab13 with LC3B or p62 was observed in starved WT cells. In contrast, this colocalization was rarely observed in Rab11-KO cells (Fig. 6E). These results demonstrated that the interaction between Rab13 and autophagic cargos depended on amphisome formation, subsequently facilitating the sorting of these cargos into AEVs. Moreover, in starved WT cells, LC3B exhibited aggregate structures in close proximity to CD63-labeled MVBs without complete colocalization (Fig. 6F). However, those LC3B-positive aggregates were localized around CD63-labeled MVBs in starved Rab13-KO cells (Fig. 6F). Our results confirmed that LC3B was present inside AEVs.
A Representative confocal immunofluorescence images from normally cultured and serum-starved 293 T cells, which were immunostained for endogenous Rab13 (green) and CD63 (red). Bar = 10 μm. Enlarged insets were presented at the bottom with a bar of 1 μm. B, C The secretion of sEVs and AEVs from wild-type (WT) or Rab13 knockout (KO) 293 T cells, with or without starvation treatment, was determined by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test (B) or unpaired two tailed Student’s t tests (C). ns: no significance. D Western blot for the AEV fraction from serum-starved WT and Rab13-KO 293 T cells. E Representative images of WT and Rab13-KO 293 T cells immunostained for endogenous Rab13 (green), LC3B (red) and p62(red). Bar = 10 μm. Enlarged insets were displayed with the bar = 1 μm. Percentage colocalization of Rab13 with LC3B or /p62 was quantified by determining Pearson’s correlation coefficients providing a measure for the relative degree of co-localization of proteins. For each sample, five cells were quantified using ImageJ software. Three replicate samples from independent experiments were analyzed for each group. p-values were determined by unpaired two tailed Student’s t tests. ***p = <0.0001 (above), ***p = <0.0001 (below). F Confocal images and enlarged insets of LC3B (green) and CD63(red) in serum-starved WT and Rab13-KO 293 T cells. Bar = 10 μm. Scale bars for enlarged insets: 1 μm. G Representative images of TEM from 293 T cells after starving for 24 h. The green arrows represent the compact multivesicular structure and the yellow arrows represent intraluminal vesicles (ILVs). Bar = 500 nm. Similar results were obtained in three independent experiments. H, I Schematic illustration for flow cytometric sorting of CD63 and LC3B labelled amphisomes. The mCherry-GFP-positive amphisomes (I) were coated onto cover slip intended for confocal imaging. Bar = 2 μm. H Created in BioRender. Ruan, H. (2025) https://BioRender.com/98rha9d. J The secreted sEVs were isolated from 293 T cells co-transfected with CD63-mCherry and LC3B-GFP, and then imaged under super-resolution microscopy. Bar = 200 nm. Similar results were obtained in three independent experiments. K The number of amphisomes in 293 T cells under normal or starved culture conditions, with or without BafA1 treatment, was calculated using flow cytometric analysis. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ***p = <0.0001, *p = 0.024. L A volcano plot of the changed proteins identified in WT and Rab13-KO 293 T cells. The proteins were plotted according to their -log10 P values as determined by two-tailed t test and log2 fold change. Each group contained three independent biological replicates. M GO enrichment analysis of decreased proteins in AEV fraction from serum-starved Rab13-KO 293 T cells relative to AEV fraction from serum-starved WT 293 T cells with the terms for biological processes. Source data are provided as a Source Data file.
On the basis of these results, we speculated that Rab13 may be involved in controlling the transfer of lipidated LC3B from the amphisome membrane to the interior of AEVs during the fusion of autophagosomes with MVBs. Depletion of Rab13 did not affect this fusion but redirected the localization of lipidated LC3B residing on the amphisome membrane.
Transmission electron microscopy (TEM) analysis of starved 293 T cells revealed two distinct lumens in the amphisomes. One lumen consisted of a compact multivesicular structure (green arrow), possibly formed from autophagosomes after fusing with MVBs, while the other was filled with typical intraluminal vesicle (ILV) structures (yellow arrow), known as exosomes (Fig. 6G). These findings may explain why disruption of ILV formation through the deletion of ESCRT genes did not affect AEV secretion. To further corroborate these results, 293 T cells were cotransfected with the LC3B-GFP fusion vector to label autophagosomes and the CD63-mCherry fusion vector to label MVBs. The presence of amphisomes could then be scored on the basis of both GFP and mCherry. After starvation, the transfected cells were disrupted using mechanical milling, and the GFP-mCherry-positive amphisomes were sorted by flow cytometry (Fig. 6H). As shown in Fig. 6I, the amphisomes, coexpressing CD63 and LC3B, exhibited two distinct groups, each labeled with either mCherry or GFP fluorescence. In support of this observation, the sEVs isolated from the cotransfected cells clearly presented two subtypes of vesicles, one labeled with GFP and the other with mCherry, as detected under super-resolution (SR) microscopy (60 nm SR) (Fig. 6J). These findings suggested that the amphisomes resulting from autophagosome‒MVB fusion consisted of two distinct types: one type comprised the ILV–exosomes formed by the inward budding of MVBs, while the other constituted AEVs derived from autophagosomes. Both types could be released extracellularly as sEV-sized exosomes or sEV-sized AEVs when the amphisomes fused with the plasma membrane. In addition, amphisomes can also fuse with lysosomes to form autolysosomes, followed by degradation of the cargo. Therefore, we quantified intracellular amphisome numbers upon starvation or BafA1-mediated lysosome inhibition and observed an increase in the number of amphisomes when lysosomal function was compromised (Fig. 6K). Our data presented above in Fig. 1 show no significant alteration in AEV secretion in BafA1-treated cells, suggesting that the intracellular accumulation of amphisomes did not increase their fusion with the plasma membrane as an alternative route for extracellular release. These results suggest that the different fates of amphisomes, including lysosomal degradation and extracellular release, are independent.
Given that Rab13 affects the secretion of autophagic cargos and ESCRT components via AEV release, we subsequently investigated whether Rab13 plays a role in determining the composition of AEVs. Quantitative proteomics analysis of AEVs from starved Rab13-KO 293 T cells compared with those from WT cells revealed that the levels of LC3B (MAP1LC3B) and ESCRT components were significantly decreased (Fig. 6K) (Supplementary Data 2), which is consistent with the results of the western blot analysis presented in Fig. 6D. GO terms of biological processes highlighted a significant decrease in AEV proteins from starved Rab13-KO cells that localize to autophagy-associated processes such as macroautophagy and autophagosome maturation, indicating that Rab13 depletion could disturb the packaging of autophagy-related proteins into AEVs (Fig. 6L). Importantly, this category also included a significant term related to protein localization to the plasma membrane, suggesting that Rab13 may be involved in regulating the localization of target proteins, such as autophagic cargo and ESCRT-III components, to the membrane. Taken together, our results suggest that Rab13 contributes to the sorting of autophagic cargos and ESCRT components into AEVs without significantly affecting the formation or secretion of these vesicles.
The biogenesis of AEVs depends on ESCRT-III/VPS4 and Rab27a
The ESCRT III complex is a conserved membrane remodeling machinery essential for the budding of endosomal vesicles and exosome formation21,22. Since the AEVs contained ESCRT III components, we aimed to investigate whether these components were involved in the membrane fission process that resulted in the formation of AEVs during amphisome formation. As shown in Fig. 7A, when the ESCRT-III components CHMP4A and CHMP4B were knocked down via transfection of small interfering RNAs(siRNAs), there was a marked decrease in AEV secretion relative to control or negative control siRNA (si-NC). The double knockdown of CHMP4A and CHMP4B resulted in an enhanced effect. Furthermore, when the ESCRT accessory proteins VPS4A and VPS4B were knocked down, the secretion of AEVs also decreased. Knockdown of other components, such as CHMP1B, CHMP2A and CHMP2B, did not influence AEV secretion (Fig. 7A). TEM analysis of CHMP4A and CHMP4B double-knockdown 293 T cells under starvation conditions revealed the absence of a compacted multivesicular structure in amphisomes (green arrow, Fig. 7B). In addition, we also observed abnormal morphology for MVBs lacking ILVs (yellow arrow, Fig. 7B). In support of this observation, the compact multivesicular structure within the amphisomes reappeared when the interference effect of the siRNA was exhausted (Fig. 7C, red arrow). Furthermore, by using SR microscopy, the lipidated LC3B was found to not form aggregate structures, as shown in Fig. 7D, but instead colocalized with CD63-labeled MVBs in a punctate distribution, indicating that the lack of these ESCRT-III components led to the abnormal location of AEV cargos. These observations suggested that AEV biogenesis in amphisomes was required for CHMP4A-, CHMP4B- and VPS4A/B-mediated membrane fission.
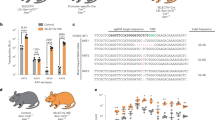
A The secretion of AEVs from mock control or siRNA-transfected 293 T cells was detected by NTA. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. **p = 0.0027(si-CHMP4A vs si-NC), ***p = 0.0008(si-CHMP4B vs si-NC), ***p = 0.0005(VPS4A vs si-NC), **p = 0.0069(VPS4B vs si-NC). Representative images of TEM from 293 T cells after co-transfecting with CHMP4A and CHMP4B siRNAs for 24 h (B) or 10 days (C). Both green arrows and red arrows indicated the compact multivesicular structure in amphisomes. The yellow arrow represented MVB structure. Bar = 500 nm. Similar results were obtained in three independent experiments. D Confocal images of LC3B (green) and CD63 (red) in 293 T cells following co-transfecting with siRNAs targeting CHMP4A and CHMP4B. Bar = 10 μm. Similar results were obtained in three independent experiments. The secretion of AEVs from wild-type (WT) or Rab27a knockout (KO) 293 T cells were detected by NTA (E) or western blot (F). Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. **p = 0.0017. G The percentage of AEVs in serum samples from normally fed and fasted Rab27a knockout (KO) mice was determined by NTA. Each group contained six mice (n = 6). Data are presented as the mean ± SD. Error bars denote the maximum and minimum values. For the box plots, the horizontal line inside represents the median, while the bottom and top of the box correspond to the 25th and 75th percentiles, respectively. Statistical analysis was performed using a two-tailed unpaired Student’s t-test. ns: no significance. Source data are provided as a Source Data file.
Previous studies have demonstrated that Rab27a is involved in the release of exosomes originating from MVBs23. Thus, Thus, we tested whether Rab27a was required for AEV secretion. Compared with that from WT cells, the release of AEVs from stable Rab27a-knockout 293 T cells (Rab27a-KO) was significantly decreased (Fig. 7E). Similarly, depletion of Rab27a suppressed the secretion of p62, LC3B-Ⅱ, p62, CHMP2A, VPS4A/B and TSG101 via AEVs (Fig. 7F). In further support of this finding, we detected no increase in AEV secretion from the serum of Rab27a-KO mice under starvation conditions compared with that under normal feeding conditions (Fig. 7G). These results revealed that Rab27a was functionally required for the extracellular secretion of autophagic cargos and ESCRT-III components via AEV populations.
AEVs that encapsulate enterovirus infectious elements exhibit infectivity in non-permissive cells, thereby expanding viral tropism
Viral infection not only induces a host autophagy response but also impacts the secretion of exosomes/sEVs24,25. Coxsackievirus B4 (CVB4), with one of the highest proportions of fatal outcomes among enterovirus serotypes, was confirmed in this study to induce an autophagy response and increase AEV secretion (Fig. S2A, C). Therefore, exploring the role of AEVs during CVB4 infection events is logical. Purified AEVs from CVB4-infected WT cells contained virus-like particles enclosed within membrane structures, as observed by TEM. Similar results were also obtained for AEVs from CVB4-infected TSG101-KO cells, with the presence of virus-like virions encapsulated within the vesicles (Fig. S4). In contrast, the AEVs from the CVB4-infected Rab11-KO cells were identified by TEM to not contain virus-like particles inside these vesicles (Fig. 8A). In further support of this observation, the CVB4 structural protein VP1, along with LC3B-II and p62, was detected in the AEV fraction derived from infected WT cells, suggesting that the virus-like particles enclosed within these AEVs were indeed authentic CVB4 virions (Fig. 8B). To determine whether the virions enclosed within the AEVs retain their infectivity, we utilized purified AEVs derived from CVB4-infected WT, TSG101-KO or Rab11-KO cells to treat susceptible HeLa cells. As shown in Fig. 8C, the level of viral RNA in HeLa cells did not significantly differ following treatment with various sources of AEVs at different time points. However, interestingly, we observed that the AEVs from infected WT or TSG101-KO cells, but not from Rab11-KO cells, efficiently entered and replicated over time in NIH 3T3 cells, a non-permissive mouse cell lacking viral receptors (Fig. 8D), suggesting that AEVs containing the CVB4 virion exhibit high infectivity in nonpermissive cells. Moreover, CVB4 RNA was significantly more enriched in both the brains and spleens of mice injected with AEVs from WT cells than in those of mice injected with AEVs from Rab11-KO cells (Fig. 8E). Compared with those from mice injected with WT cells, the survival rate from mice injected with AEVs from Rab11-KO cells was greater (~70%) (Fig. 8F). These findings indicated that CVB4 virions enclosed within AEVs have greater infection efficiency, thereby overcoming restrictions to viral tropism.
A Representative TEM images of AEVs from CVB4-infected WT or Rab11-KO 293 T cells. Green arrows represented the encapsulated CVB4 virion. Bar = 200 nm. Enlarged inset was presented with a bar of 100 nm. Similar results were obtained in three independent experiments. B Western blot analysis for AEV fractions from CVB4-infected WT or Rab11-KO 293 T cells. The copy number of CVB4 RNA in HeLa (C) or NIH 3T3 (D) cells after treating with different sources of AEVs were determined by real-time PCR. The AEV fractions from CVB4-infected WT, TSG101-KO or Rab11-KO 293 T cells were incubated HeLa or NIH 3T3 for indicated times. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. ***p = 0.0003 (24 h), 0.0001 (48 h), <0.0001 (72 h). ns: no significance. E The copy number of CVB4 RNA for various mouse tissues after injecting with corresponding AEVs. Each group contained six mice (n = 6). Data are presented as the mean ± SD and p-values were determined by unpaired two tailed Student’s t tests. ***p < 0.0001 (brain), ***p = <0.0001 (spleen). F The survival rate of mouse was monitored daily until 10 days post-infection. The asterisks indicate statistical significance by log rank (Mantel-Cox) test. **p = 0.0014. G Western blot analysis for HEK293T cells in the presence or absence of EV71 infection. Similar results were obtained in three independent experiments. H Schematic illustration for isolating sEVs and AEVs from EV71-infecetd cells. Created in BioRender. Ruan, H. (2025) https://BioRender.com/b94llbk. I Western blot analysis for sEVs(EV71-sEVs) and AEVs (EV71-AEVs) from EV71-infected cells. J Agarose gel electrophoresis for full length EV71 genome in different sources of AEVs. Mock uninfected cells as the negative control. K Real-time PCR analysis for EV71 RNA copy number in sEVs and AEVs from EV71-infecetd cells. Data are presented as the mean ± SD of three independent experiments. p-values were determined by unpaired two tailed Student’s t tests. L The copy number of EV71 RNA in treated L929 cells were determined by real-time PCR. Data are presented as the mean ± SD of three independent experiments. p-values were determined by one-way ANOVA with Dunnett’s multiple comparisons test. Source data are provided as a Source Data file.
Our previous study demonstrated that the accumulation of enterovirus 71 (EV71)-induced autophagosomes facilitated their fusion with MVBs, leading to the packaging of the viral RNA genome into amphisomes for extracellular release26. Therefore, investigating whether AEVs from EV71-infected cells contain a viral genome is meaningful. EV71 infection triggered an autophagy response in 293 T cells characterized by no alteration in p62 but an increase in LC3B-II, suggesting that incomplete autophagy was induced by EV71, whose accumulated autophagosomes did not fuse with lysosomes for degradation. In the following step, we isolated sEVs (EV71-sEVs, containing the AEVs) or AEVs (EV71-AEVs) from the culture medium derived from EV71-infected cells (Fig. 8H). Neither EV71-sEVs nor EV71-AEVs showed detectable levels of the viral structural protein VP1, indicating that no complete virus particles were present within these vesicles (Fig. 8I). However, we observed that both EV71-sEVs and EV71-AEVs contained the viral full-length genome (~7000 bp). Moreover, the levels of the EV71 RNA genome did not significantly differ between EV71-sEVs and EV71-AEVs from an equal number of infected cells (Fig. 8K). Our published study demonstrated that preventing autophagosome–MVB fusion via Rab11 knockdown could abolish the sorting of EV71 RNA into amphisomes, leading to a marked decrease in viral RNA for extracellular release26. These results collectively indicated that extracellular vesicle-associated EV71 genomes were primarily encapsulated within the AEVs. Importantly, the viral genomes encapsulated within AEVs exhibited high infectivity in a non-permissive L929 cells (a mouse fibroblast line), in contrast to EV71 free virions (Fig. 8L). Taken together, our data supported that AEVs could carry and deliver viral particles or whole genomes into receptor-negative host cells with high infection efficiency, overcoming restrictions to viral tropism.
Discussion
The common step in exosome biogenesis and the formation of secretory amphisomes is the formation of MVBs, often leading to confusion among secretory amphisomes, exosomes, and small extracellular vesicles (sEVs)6,8,27. Here, we observed that a subtype of small EVs with a size less than 100 nm exhibited increased secretion during autophagy induction in vitro and in vivo. This phenomenon depended on the formation of amphisomes and their subsequent fusion with the plasma membrane. Genetic loss-of-function experiments demonstrated that blocking exosome biogenesis had little effect on the release of these vesicles, which were size less than 100 nm in size. In support of this observation, the amphisomes in starved cells exhibited distinct compartment filled with multivesicular structures, distinct from those of typical ILVs. These key observational data suggested that these autophagy-associated vesicles within amphisomes represented a subpopulation of sEVs, with independent pathways for their origins that differ from those of exosomes (Fig. 9). Notably, a recent study demonstrated that large multivesicular body-like structures that originate from intracellular amphisomes, can be released en bloc by a plasma membrane-enclosed compartment termed “amphiectosome”28. These amphiectosomes release their intraluminal vesicles (ILVs) through a “torn-bag mechanism”. Unfortunately, we were unable to collect amphiectosomes from the cell medium in our study, and the authors of the recent study did not provide their isolation procedure. We speculate that amphiectosome structures may be highly fragile and susceptible to rapid rupture upon extracellular release. In addition, unlike under our experimental conditions, in the previous study, the cells were cultured in the presence of fetal bovine serum (FBS) in the culture medium without triggering autophagy. Given that FBS contains a substantial number of EVs, distinguishing the origin of EVs may be challenging. Notably, the authors of that study observed that the ILVs of amphiectosomes were either exclusively LC3B positive or CD63 positive, which is consistent with our findings indicating that the LC3B positive AEVs formed from autophagosomes, whereas CD63 positive exosomes originated from ILVs. Furthermore, we determined that AEVs originating from intracellular hybrid amphisomes were rich in autophagic cargos, ESCRT III components, and Rab13, distinguishing them from conventional exosomes. Nevertheless, the characteristics of AEVs indicate that they still belong to the category of sEVs not only because of their similar sizes and densities, but also because both exhibit identical biomarkers such as CD81 and TSG101. Notably, a previous study reported that the release of Rab11a-enriched exosomes, induced by nutrient deprivation, resulted in the release of different cargos such as accessory ESCRT-III proteins and the ILV-associated mechanism was not required for their biogenesis20. Our findings revealed that the production of these Rab11a-exosomes was not affected by the suppression of autophagy, indicating that AEVs and Rab11a-exosomes are distinct types of small EVs. All of those findings demonstrate that sEVs exhibit high heterogeneity because of their different mechanisms of biogenesis. Understanding this heterogeneity provides valuable insights into the clinical application of sEVs. Additionally, our results confirmed that Rab27a was functionally required for the secretion of AEVs. This small GTPase plays a confirmed role in the fusion of MVBs with the plasma membrane, resulting in the release of exosomes23. Therefore, the overlapping biogenesis and secretion processes of AEVs and exosomes often lead to them being conflated. Ultracentrifugation at 100,000 g is currently the most widely used method for isolating small extracellular vesicles29,30,31. Our study revealed that traditional ultracentrifugation, as well as iodixanol gradient method, were insufficient for separating AEVs from sEVs, including exosomes. This explains the frequent detection of autophagic proteins such as LC3B and p62 in these isolated vesicles, which are considered exosomes or sEVs across many studies.
Upon autophagy induction, mature autophagosomes fuse with multivesicular bodies (MVBs) via Rab11 to form hybrid amphisome organelles. Autophagy cargos such as lipidated LC3B (LC3B-II) and p62, along with several ESCRT components, intermix with intraluminal vesicles through a process mediated by Rab13. This process involves intraluminal budding mechanisms that depend on the interaction between ESCRT-III components (CHMP4A and CHMP4B) and VPS4A/B. Subsequently, hybrid amphisomes release two subpopulations of small extracellular vesicles (sEVs) through Rab27a-dependent exocytosis: one consisting of autophagic extracellular vesicles (AEVs) containing autophagy cargos, ESCRT components and Rab13, the other being traditional exosomes. Created in BioRender. Ruan, H. (2025) https://BioRender.com/rhh7l0k.
Traditional exosomes are known primarily for their role in cell communication, and transport of various cargos including proteins, lipids and nucleic acids. They are derived from MVBs and released upon MVB-plasma membrane fusion3. In contrast, AEVs are associated with autophagy processes. Our findings suggest that AEVs may play a unique role in maintaining cellular homeostasis during stress conditions, such as starvation and virus infection. For example, while exosomes might transfer signaling molecules to neighboring cells under normal conditions, AEVs could be more involved in removing damaged cellular components or misfolded proteins during stress. This difference in function could be related to their distinct biogenesis pathways. Future studies, such as those focusing on the contents of exosomes and AEVs, could further elucidate these functional differences.
A rapidly growing area of interest in EV research is the potential application of EVs as diagnostic biomarkers, and the key issue to be addressed in their application for disease diagnosis is their heterogeneity1. We proposed an improved procedure for isolating purified AEVs with sizes less than 100 nm. Although the Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018) guidelines recommend that the term “small EVs” (sEVs) to describe vesicles with a diameter less than 200 nm32, our data revealed that under normal conditions, the percentage of sEVs with a size less than 100 nm was low in various cell lines and mouse peripheral blood. Considering that the cellular basal autophagic response under normal conditions induces a slight AEV release, we hypothesized that small EVs could be classified as AEVs with sizes less than 100 nm and exosomes or other vesicles with sizes ranging from 100–200 nm. Nevertheless, more precise and comprehensive detection methods, such as single-vesicle capture, are needed. The secretion of AEVs dramatically increased after triggering autophagy. Autophagy is triggered by numerous conditions, such as cancer, neurodegeneration, aging and viral infection24,33,34,35. Indeed, our results revealed that several human viruses, such as Varicella-Zoster virus (VZV), coxsackievirus B4 (CVB4) and Herpes simplex virus type 1 (HSV−1), induced a host autophagy response, resulting in increased AEV secretion. In addition, a higher level of AEVs was observed in the aging population. Therefore, the detection of AEV secretion has the potential to serve as a diagnostic indicator or therapeutic response for many disease pathologies. Notably, a previous study revealed that the number of small EVs with sizes <100 nm, which they considered exosomes or exosome-like vesicles (ELVs), was significantly greater in stage IIA breast cancer patients than in healthy donors36. Nonetheless, the diagnostic potential of AEVs across various diseases likely stems from their specific cargos, including but not limited to proteins and microRNAs.
Mechanisms of exosome biogenesis are fairly well understood and involve subunits of the ESCRT. The ESCRT machinery is composed of ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III components and the ATPase VPS4, which cooperate in a stepwise manner. ESCRT-III components drive MVB membrane budding and fission to ultimately form ILVs. ESCRT-III remains on the outside of the MVB membrane without being involved in the ILV lumen until it is recycled by VPS421,37. Therefore, ESCRT-III components generally cannot be detected in exosomes. However, our study revealed that several ESCRT-III components, such as CHMP2A and CHMP2B, were enriched in AEVs. Yoshinori et al. reported that CHMP2A is translocated to the phagophore and controls the formation of double-membrane autophagosomes. In addition, CHMP2B is associated with autophagosome maturation, as supported by several studies38,39. As a result, these components are involved in AEV formation, which originates from the fusion of autophagosomes with MVBs. Our data confirmed that other ESCRT-III components CHMP4A and CHMP4B, along with VPS4A/B, contribute to membrane fission in amphisomes, leading to intracellular AEV biogenesis. However, CHMP4A and CHMP4B were absent from the AEV fractions. ESCRT-III/VPS4 complexes can promote both inside-out (positively curved membranes) and outside-in (negatively curved membranes) membrane fission reactions, and emerging mechanistic models for ESCRT-mediated membrane fission have been reported22,40,41. A previous study demonstrated that CHMP4B preferentially binds negatively curved membranes, resulting in 100 nm deep membrane concavities42. Moreover, other studies have confirmed that both CHMP4A and CHMP4B assemble into circular membrane-associated polymers that can be engaged to deform the membrane to which they are attached40. Although lack of the direct experiment evidences, we hypothesize the ESCRT-III components CHMP4A and CHMP4B, in conjunction with VPS4, could target to the side of autophagosome membranes during amphisome formation and promote negatively curved membranes, resulting in generation of the luminal vesicles known as AEVs.
The family of Rab proteins plays a significant role in the secretion of EVs such as Rab27a/b and Rab3123,43. Our work revealed that the Rab13 is involved in the sorting of AEV-specific cargos into AEVs but does not affect their formation or secretion. Several studies have shown that Rab13 controls the delivery of cargo trafficking through recycling endosomes to the plasma membrane, resulting in high protein–protein affinity44,45,46,47. Importantly, cellular Rab13 traffics to two distinct populations of vesicles, those carrying transferrin receptors from recycling endosomes and Rab7-positive vesicles originating from late endosomes/MVBs45. Notably, a study Su et al. demonstrated that Rab13 acts as a positive regulator of autophagy and is closely related to core autophagy regulators including LC348. Significant alterations have been observed in the localization and morphology of Rab13 during the autophagy process were observed in our study. We therefore hypothesized that MVB-localized Rab13 is solubilized from the membrane and regulates protein cargo sorting into AEVs through its high protein-protein affinity when amphisome formation. In particular, previous work has shown that Rab13 additionally regulates the secretion of a subclass of β1-integrin+/Rab13+ sEVs that are distinct from classical CD63+ exosomes in mutant KRAS colorectal cancer cells49. Given the upregulation of autophagy in KRAS-mutant tumors50,51, it is tempting to speculate that these β1-integrin+/Rab13+ sEVs could be categorized as AEVs.
Enteroviruses have historically been considered non-enveloped and rely on cell lysis to exit52,53. The presence of EV-enclosed virus particles has been confirmed for several types of naked viruses, including picornaviruses, hepatitis E virus, rotavirus, and norovirus, and raises important questions regarding their role in infection54,55,56. Our data suggested that coxsackievirus B4 (CVB4) particles were enclosed by the AEVs and could be transferred to receptor-negative host cells with high infection efficiency. Studies in recent years have reported that several picornavirus species are encapsulated in EVs containing the autophagosomal marker LC357,58,59,60. Many picornaviruses actively induce autophagosome accumulation to support virus replication or exocytosis5. On the basis of these findings and our findings, we propose that autophagosomes engulf nascent virions and then fuse with MVBs to form amphisomes, resulting in the secretion of AEV-enclosed virions into the extracellular environment. According to this model, the biological properties of AEV-enclosed viruses may differ from those of their naked counterparts. In addition, we observed that the extracellular vesicle-associated enterovirus 71 genomes were primarily encapsulated within AEVs and exhibited a high infectivity in non-permissive cells. Unfortunately, our previous study demonstrated that the EV71-induced formation of amphisomes facilitated viral genome packaging into exosomes, rather than relying on the process of exosome biogenesis26. We must recognize that owing to the limitations of experimental operation and understanding at that time, we were unable to differentiate between AEVs and exosomes, and we lacked data to verify whether the viral genome was present in other types of sEVs, excluding AEVs. Our findings raise important questions about the model of AEV-enclosed virions for non-enveloped viruses. This model may represent a primary pattern of EV-mediated virus invasion, immune escape and transmission, at least for enteroviruses.
Methods
Cell culture, virus strains and plasmids
HEK-293T, ARPE-19, HeLa, SH-SY5y, THP-1, Neuro 2a, NIH 3T3 and L929 cell lines were obtained from the American Type Culture Collection (ATCC, USA). The HEK-293T, ARPE-19, NIH 3T3, HeLa and L929 cells were maintained in DMEM medium (HyClone, USA), the THP-1 cells were cultured in RPMI 1640 medium (HyClone, USA), the SH-SY5y and Neuro-2a cells were grown in DMEM/F12 medium (HyClone, USA). The cell culture supplemented with 10% fetal bovine serum (FBS, ExCell Bio, China) and incubated at 37 °C in a 5% CO2 humidified incubator. FBS used in these cultures were depleted of EVs by ultracentrifugation at 120,000 g for 3 h. All the cell lines used in this study tested negative for mycoplasma contamination using the reagent PlasmoTest (InvivoGen).
For serum starvation, cells were cultured in serum-free medium for 24 hours. If needed, bafilomycin A1 (20 nM, TargetMol, China) or rapamycin (100 nM, TargetMol, China) was added into the serum-free DMEM and incubated for 24 hours. To assess the extent of cell death induced by starvation, LDH release into the culture medium was measured using a commercially available LDH assay kit (DOJINDO, #CK12), to ensure that the cell mortality rate remained below 5%.
Human herpesvirus 1 strain (HSV-1, GenBank: GU734771.1), Coxsackievirus B4 strain (CVB4, GenBank: X05690.1) and Enterovirus 71 strain (EV71, GenBank: HQ882182.1) were kindly provided by Prof. Zhiwei Wu (Nanjing University, China) and passaged in Vero E6 cells. Varicella-zoster virus strain (VZV, GenBank: AB097933.1) was a kind gift from Prof. Tong Cheng (Xiamen University) and propagated in ARPE-19 cells. Both titers of viruses were determined by the 50% tissue culture infection dose (TCID50) assay and stored at -80 °C until used.
The expression plasmid pCMV-GFP-LC3B (#D2815) and pCMV-CD63-mCherry (#D2841) were purchased from Beyotime Biotechnology (Shanghai, China). All the sequence of inserted genes in plasmids were verified by DNA Sequencing.
Generation of CRISPR-Cas9 KO cell lines
The guide RNAs (sgRNAs) targeting the human genes were designed using the CRISPick online software (Broad institute), and subcloned into the pSpCas9(BB)-2A-Puro plasmid (Addgene no. 48139). Forward and reverse oligos for sgRNAs were custom synthesized (Tsingke Biotech, Beijing, China) and annealed to generate sticky ends. pSpCas9 plasmid was cleaved with BbsI enzyme (#R3539; New England Biolabs) and ligated with annealed sgRNA using T4 ligase (Takara). HEK-293T cells were transfected with the corresponding sgRNA-ligated plasmids using Lipofectamine 3000 (Thermo Fisher Scientific) for 48 hours, followed by treating with 2 μg/mL puromycin for 3 days to select gene-knockout cells. Single-cell clones were selected for three weeks via limiting dilution assay. Knockout of corresponding genes were validated by western blot using specific antibodies. The sequences of sgRNAs used for CRISPR knockout are as follows: human TSG101: 5’-CTGTTCTGTTTTCAGGCCG-3’; human SDCBP :5’-GACAGAATGTCATTGGATTGA-3’; human CD63: 5’-AACGAGAAGGCGATCCATAAGG-3’; human Rab11: 5’-GAGAGTGATCTACGTCATCTC-3’; human Rab13: 5’-GTATACTAGGATAATGCCCT-3’;
human Rab27a:5’-CCAAAGCTAAAAACTTGATG-3’.
Preparation of small extracellular vesicles (sEVs) and autophagic extracellular vesicles (AEVs)
The wild cell lines and knockout cell lines were seeded in 10 cm tissue culture dishes. Upon reaching approximately ~90% confluence, the medium containing 10% FBS was carefully removed. Cells were then washed with phosphate-buffered saline (PBS) slightly and subsequently cultured in 10% EV-depleted FBS medium or serum-free medium for 24 h. As for virus infection experiment, cells were infected with CVB4 at 0.05 TCID50 or EV71 at 0.03 TCID50 for 24 h under 10% EV-depleted FBS medium to avoid cell necrosis and apoptosis. The culture medium was centrifuged at 3000 × g for 15 min at 4 °C and 10,000 × g for another 30 min at 4 °C to remove cells, debris and large vesicles.
Small extracellular vesicles (sEVs) isolation according to standard protocols with some adaptations: the processed supernatants were filtered through 0.22 μm pore membrane filters (Millipore) and subjected to ultracentrifugation by 100,000 × g at 4 °C for 90 minutes using Optima L-100 XP ultracentrifuge (Beckman Coulter) to pellet sEVs. Crude sEVs pellets were then gently diluted in sterile PBS and ultracentrifuged for an additional 90 min at 100,000 × g. The final sEV pellets was resuspended in sterile PBS for further analysis.
For autophagic extracellular vesicles (AEVs) isolation, the processed supernatants were sequentially filtered through 0.22 μm and 0.1 μm pore filters. The filtered supernatants were centrifuged at 40,000 × g for 60 minutes at 4 °C to remove large vesicles with size >100 nm, followed by ultracentrifuged by 100,000 × g for 90 minutes at 4 °C. The resulting AEV pellets were resuspended in sterile PBS and centrifuged again at 100,000 × g for 90 minutes at 4 °C to remove soluble factors for a series of subsequent experiments.
For serum isolation, the blood samples from mouse or donor were kept at 4 °C overnight without anticoagulant to ensure serum separation. Equal volume of serum was centrifuged at 5000 × g for 10 min and then diluted with appropriate PBS. The diluted serum samples were filtered through 0.22 μm pore membrane filters and subjected to ultracentrifugation by 100,000 × g at 4 °C for 90 minutes. The crude sEV pellets were cleaned by an additional 90 min at 100,000 × g to obtain purified sEVs for subsequent analysis.
Of note, for NTA assays, infectious activity, digestion protection and iodixanol gradient separation, the suspension of sEVs or AEVs were used within 12 h. For western blot, MS and PBA, they were stored at -80 °C and used within three months. In addition, for all comparisons of sEVs or AEVs between experimental conditions, results from individual cohorts were corrected as indicated on the basis of total cell number to ensure that vesicle quantification was not confounded by seeding differences.
Nanoparticle tracking analysis (NTA) for particle size and concentration of EVs
The Nano NS300 instrument (Malvern Instruments, UK) equipped with laser-based light scattering system was used to track the size and number of EVs. Samples resuspended in PBS were diluted to achieve concentrations between 1×107/ml and 1×109/ml, and then manually pushed into the sample chamber, making sure there were no bubbles in the sample chamber and tubing. Laser light scattering was performed to visualize EVs in collected supernatants using a conventional optical microscope perpendicular to the beam axis. During this process, EVs were recorded and analyzed based on Brownian motion and the diffusion coefficient. Each sample was subjected to three 60-second video recordings under the following conditions: cell temperature: 25 ~ 26 °C; Syringe speed: 40 µl/s. The data were subsequently analyzed in triplicate to confirm reproducibility using NTA 2.3 software to calculate the number and size of EVs in each sample.
For detection of p62-positive fluorescent AEVs, the AEVs isolated from serum-free, 24-hour cultured HEK 293 T cells and incubated with SQSTM1/p62 antibody conjugating Alexa Fluor 488(#ab185015, Abcam,1:500) at room temperature for 2 h. Then the AEVs were subjected to ultracentrifugation by 100,000 × g at 4 °C for 90 minutes to remove excess antibody and were cleaned by PBS with an additional ultracentrifugation at 100,000 × g for 90 min. The particle size of p62-positive fluorescent AEVs was analyzed using NTA instrument in fluorescence mode (laser wavelength 488 nm). Sample was manually loaded into the sample chamber, and a fluorescence filter was inserted into the optical path. Camera levels and focus settings were adjusted to optimize particle tracking within the optimal threshold range. Subsequent analysis steps followed the standard NTA procedure.
Resistive pulse sensing (RPS) for particle size analysis
Particle size distribution was measured by Nanocoulter Counter (Resen Technology, Shenzhen, China) through a single-particle analysis approach, and the nanopore chip with the measuring range of 50-240 nm was employed. Samples underwent differential centrifugation and filtration as previously, followed by dilution in PBS to achieve an optimal concentration within the instrument’s detection range (1 × 10⁷/mL to 1 × 10⁹/mL), and then introduce into the sample port. The resistive pulse sensing technique relied on an electric induction zone generated by electrodes on either side of the nanopore in electrolyte solution. As individual particles traversed the nanopore, the particle displaced an equal volume of conductive solution, instantly increasing the resistance of the induction zone and generating a pulse signal. The signal was then analyzed by Nanocoulter software to obtain multi-dimensional particle information. The RPS assays were performed by Resen Technology (Shenzhen, China).
Iodixanol gradient ultracentrifugation
For subcellular fractionation, HEK-293T cells were serum starved for 24 h, harvested in tris-sucrose buffer (0.25 M sucrose, 10 mM Tris, pH 7.5) with Protease Inhibitor Cocktail (APExBIO, USA). Cells were mechanically lysed using the grinding tube and gently ground for 5 minutes to release cellular contents. The lysate was cleared of cellular debris and nuclei by centrifugation at 10,000 g and the post-processing extract was loaded on a continuous iodixanol density gradient. To generate the 12 fractions continuous iodixanol density gradient, 5%, 10%, 20%, and 40% w/v iodixanol solutions (Sigma‑Aldrich) were diluted in tris-sucrose buffer and incubated at 4 °C overnight to generate a continuous gradient. The post-processing extract was loaded on the top of the gradient and centrifuged at 100,000 × g for 16 hours at 4 °C in SW40 Ti rotor (Beckman Coulter). After centrifugation, fractions were collected and ultracentrifuged at 110,000 × g for 60 min at 4 °C, and pellets were resuspended in 5×lysis buffer (Fudebiotech, China) for western blot analysis.
For small EV fractionation, sEV pellets were generated by differential centrifugation as described above, then resuspended in sterile PBS and centrifuged at 100,000 g for 16 hours at 4 °C. Fractions were carefully extracted from each gradient layer, diluted in PBS and further centrifuged at 100,000 × g for 90 min to obtain purified sEVs for subsequent analysis.
Trypsin protection assay
Autophagic extracellular vesicles (AEVs) were isolated from HEK-293T cell supernatants treated after 24 h of serum-free DMEM treatment. AEVs were purified as described and divided equally into three tubes (20 µL per tube). To each, 20 μL of PBS, 20 μL of 100 μg/ml trypsin (Gibco, USA) in PBS, or 20 μL of 100 μg/ml trypsin with 2% Triton X-100 (Sigma‑Aldrich, USA) was added. After incubating at 4 °C for 30 min with occasional flicking, the reactions were terminated by adding 5 μL 5× loading buffer (Fudebiotech, China) and heating at 95 °C for 5 min. Each sample (25 µL) was then loaded for western blot analysis.
Surface proteins of single EV by proximity-dependent barcoding assay (PBA)
To profile single EV surface proteins, PBA was carried out using antibody-DNA conjugates and next-generation sequencing. PBA probes were prepared through chemical conjugation of antibodies with DNA oligonucleotides containing unique protein and molecular tags and a universal primer-binding sequence. For EVs capture, biotinylated cholera toxin subunit B (CTB) was added to streptavidin-coated 96-well plates, incubated at room temperature for 20 minutes, and washed with PBST three times. Next, the sample-antibody mixture was diluted to 20 μL in PBA buffer and transferred to each CTB-coated well, allowing EVs to bind via ganglioside GM1 (which is enriched in membrane lipid rafts). Captured EVs were fixed with 4% paraformaldehyde.
For EV proteomic analysis, DNA polymerase extended the antibody-conjugated DNA, generating specific EV tags. The extended DNA fragments were compiled into a library and sequenced. Tag matching then produced EV counts and EV-protein data matrices, identifying individual EV surface protein profiles. For subpopulation analysis, FlowSOM was applied for dimensionality reduction and clustering of PBA data, with clusters visualized using tSNE and UMAP methods, enabling detailed analysis of EV subtypes at the single-vesicle level. The PBA assays were designed according to the protocols published by Vesicode (Solna, Sweden) and performed by Echobiotech (Beijing, China).
Mass spectrometry (MS) of extracellular vesicles
Small EVs or AEVs from wild-type, Rab11 knockout or Rab13 knockout HEK-293T cells were lysed using SDT buffer (4% SDS, 100 mM Tris-HCl, pH 7.6). Proteins were further purified by SDS-PAGE gels, and gel bands were excised and eluted. The proteins were then digested into peptide fragments by trypsin incubation at 37 °C for 15-18 h, and the resulting peptides were collected as a filtrate. The peptides of each sample were desalted on C18 Cartridges (Thermo scientific). For mass spectrometry analysis, 15 μL of the prepared protein sample was loaded into the injection tube and subjected to liquid chromatography-mass spectrometry (LC-MS). Peptides were separated via liquid chromatography, ionized, and analyzed by Orbitrap Astral mass spectrometer (Thermo Scientific) connected to an Vanquish Neo system liquid chromatography (Thermo Scientific) in the data-independent acquisition (DIA) mode. DIA data was analyzed with DIA-NN 1.8.1. All reported data were based on 99% confidence for protein identification as determined by false discovery rate (FDR) ≤ 1%. For protein identification and quantification, the recorded spectra from three independent biological replicates were searched by UniProt human proteome database. GO enrichment analyses were performed using DAVID (Database for Annotation, Visualization and Integrated Discovery; https://david.ncifcrf.gov/).
Western blot
Samples of treated cell and sEV fractions were lysed in RIPA lysis buffer (phenylmethyl sulfonyl fluoride, NaF, NaVO4 and protease inhibitor, Santa Cruz) for 15 minutes on ice and then subjected to a centrifugation at 12,000 × g for 10 minutes at 4 °C. Lysates were quantified by BCA assay (#23225,Thermo Fisher Scientific), boiled in 5×sample loading buffer(Fudebiotech, China), separated by 12% SDS-PAGE gels(Bio-Rad, USA), and transferred onto polyvinylidene fluoride (PVDF) membrane (Millipore, USA). Membranes were blocked with 5% nonfat dry milk in TBST and then incubated with appropriate primary antibodies overnight at 4 °C, followed by incubating with HRP-conjugated secondary antibody (Southern Biotech, 1:5000) for 1 hour at room temperature and exposed using an enhanced chemiluminescence system (NCM Biotech, Suzhou, China). Primary antibodies included the following: SQSTM1/p62(#ab109012, Abcam,1:2000), CD9(#ab236630, Abcam,1:1000), CD63(#ab193349,Abcam,1:1000), CHMP2A(#10477-1-AP, Proteintech,1:500), VPS4A/B(#17673-1-AP, Proteintech,1:1000), LC3B(#A19665, Abclonal,1:1000), NBR1(#16004-1-AP, Proteintech, 1:500), TSG101(#14497-1-AP, Proteintech, 1:500) SDCBP/Syntenin-1(#2239-1-AP, Proteintech, 1:500), CD81(#sc-166029, Santa Cruz Biotechnology,1:200). Rab13(#A10571, Abclonal, 1:500), GAPDH (#AC033, Abclonal, 1:2000). CD81(#MAB4615, R&D,1:500), Calreticulin (#12238, Cell Signaling technology,1:1000), Rab7(#9367, Cell Signaling technology,1:1000), α-tubulin (#5335, Cell Signaling technology, 1:2000), Rab11(#5589, Cell Signaling technology,1:1000), Rab5 (#3547, Cell Signaling technology,1:1000), EEA1(#sc-137130, Santa Cruz Biotechnology,1:500). Enterovirus 71 VP1(#GTX132339, GeneTex, 1:1000), Anti-Coxsackievirus B4 (This antibody developed in our laboratory through the immunization of mice with inactivated CVB4 particles, the serum obtained was purified and verified using Enzyme-linked immunosorbent assay (ELISA)).
Fluorescence and immunofluorescence microscopy
The wild-type, Rab13 knockout or CHMP4A/B knockdown HEK-293T cells were seeded to cover glass slides. After the designed treatment, the cells were washed for 3 times with PBS and fixed with 4% paraformaldehyde for 15 minutes at room temperature. Subsequently, cells were permeabilized with 0.5% Triton X-100 for 15 minutes, followed by blocking with 5% BSA in PBS for 60 min. For endogenous protein detection, cells were incubated with diverse groups of primary antibodies as following: mouse anti-CD63(#ab193349, Abcam,1:500) and Rabbit anti-Rab13(#A10572,1:200); mouse anti-LC3B(#A17424, Abclonal, 1:400) and Rabbit anti-Rab13(#A10572, Abclonal,1:200); mouse anti-p62(#56416, Abcam,1:500) and Rabbit anti-Rab13(#A10572, Abclonal, 1:200); Rabbit anti-CD63(#ab252919, Abcam,1:500) and mouse anti-LC3B(#A17424, Abclonal, 1:400). All primary antibodies were diluted in antibody dilution buffer (# 003218, Life Technologies) and incubated overnight at 4 °C. Coverslips were washed in PBS before being incubated with species-specific Alexa Fluor-conjugated (488 and 647) secondary antibodies (diluted 1:1000 in PBS) for 1 h at room temperature and washed again in PBS. DAPI dyes (Beyotime Biotechnology, Shanghai, China) was used for cell nucleic acid stains. Fluorescence microscopy was performed using a Nikon Eclipse Ti confocal microscope (Tokyo, Japan) equipped with s a Nikon A1R-HD25 confocal microscope (ran by NIS-Elements) equipped with an Apo TIRF 60x/1.49 NA oil immersion lens.
Transmission electron microscopy (TEM)
Wild-type cells and knockdown cells were fixed using 2% w/v PFA and 1.5% glutaraldehyde (#G5882, Sigma-Aldrich) in 0.1 M sodium cacodylate buffer (pH 7.4) at room temperature. Cells were washed in ice-cold distilled water and placed in 2% buffered osmium tetroxide at 4 °C overnight. Then cells were incubated in graded alcohol concentrations (20%, 50%, 70%, 90%, 100%, 100%; 2 min in each), and embedded in epoxy resin (#45345, Sigma-Aldrich) according to standard protocols. Ultrathin sections of 70 nm thickness were prepared using a Leica Ultramicrotome UC7.
The morphology of purified AEVs was measured by TEM as our previous description26. Images were collected on a Hitachi HT7700 transmission electron microscope (Tokyo, Japan).
Immunoelectron microscopy
A 10 μL drop of the sample solution containing AEVs was adsorbed to a glow-discharged carbon-coated copper grid (ZhongJingKeYi Ltd., Beijing, China), blocked with 0.5% BSA for 20 min at room temperature and incubated with an antibody against LC3B(#M152-3, MBL,1:100), p62((#PM045, MBL, 1:100) and Rab13 (#A10571, Abclonal, 1:50) in 1% BSA for 2 hour at room temperature. The samples were washed 3 times with PBS for 5 min and incubated with a 10 nm anti-mouse (#G7777, Sigma‑Aldrich) or anti-rabbit (#7402, Sigma‑Aldrich) IgG-Gold antibody diluted 1:100 in 1% BSA for 1 hour at room temperature. After washing 3 times with PBS and deionized water for 5 min, samples were stained with freshly prepared 1% uranyl acetate. Samples were imaged on a Hitachi H-7700 transmission electron microscope and operated at an acceleration voltage of 80 kV.
Flow cytometric sorting of CD63 and LC3 positive amphisomes
HEK 293 T cells were co-transfected with expression vectors CD63-mCherry and LC3-GFP using Lipofectamine 3000 (Life Technologies, USA) at 70% confluence. After 24 hours, the cells were cultured in serum-free DMEM for an additional 24 hours. The cells were collected and disrupted mechanically using a glass homogenizer in cold PBS on ice. The lysates were centrifuged at 10000 × g for 15 minutes, suspended and filtered through a 0.8 μm membrane filter (Millipore). The mCherry and GFP double-positive amphisomes were sorted using a flow cytometry (FACSAria Ⅲ Cell Sorter, BD). The sorting sample were further suspended in PBS and centrifugation at 40,000 × g for 30 minutes to precipitate the amphisomes. The mCherry-GFP-positive amphisome pellets were resuspended in PBS and were coated onto a cover slip intended for confocal imaging. Fluorescence microscopy was performed using a Nikon Eclipse Ti confocal microscope (Tokyo, Japan) equipped with s a Nikon A1R-HD25 confocal microscope equipped with an Apo TIRF 60x/1.49 NA oil immersion lens.
Analysis of vesicles-enclosed enterovirus infection
For detecting the vesicle-enclosed virion infection, equal number of wild-type or Rab11-knockout 293 T cells infected with CVB4 at 0.05 TCID50 or EV71 at 0.03 TCID50 for 24 h under 10% EV-depleted FBS medium to avoid cell necrosis and apoptosis, and corresponding AEV or sEV pellets were isolated as described above. An immune-selection method was used to remove the free enteroviral virion that mixed in AEV or sEV pellets, the AEVs or sEV pellets were resuspended in 1 mL sterile PBS and incubated with excess CVB4- or EV71-spectific antibodies at 4 °C overnight. Then the resuspension solutions mixed with Protein A/G magnetic beads (Millipore) for 30 minutes at room temperature. The tubes placed into the magnetic stand to capture the beads resulted in collecting free virions, then transferred supernatants to new centrifuge tubes (purified AEVs or purified vesicles) for TEM analysis or treating with various cell lines (NIH 3T3 or L929 cells).
For determination of viral RNA copy number, the total RNA was isolated from purified AEV, sEV pellets or treated cells using TRIzol reagent (TaKaRa) according to the manufacturer’s specification and then reverse transcribed using random hexamers with the reverse-transcription kit (TaKaRa). The cDNA was subjected to quantitative PCR using SYBR-Green Master Mix (Life Technologies) on an ABI QuantStudio 6 Flex for 40 cycles. The primers were CVB4-Forward (5’-CGGGAGCGTCCTCTTACATC-3’) and CVB4-Reverse (5’-GACGTCAAAAACTGCGTGCT-3’); EV71-Forward (5’-AGATAGGGTGGCAGATGTAATTGAAAG-3’) and EV71- Reverse (5’- TAGCATTTGATGATGCTCCAAT-3’). The fragment corresponding to nucleotides 1648-1780 of CVB4 and 2462-2635 of EV71were adjusted to a concentration gradient (1 × 101 copies/μL to 1 × 108 copies/μL) and were used as the standard to calculate the copy number of CVB4 and EV71 RNA, respectively.
For determination of EV71 complete genome in sEVs and AEVs, the primers used were EV71-Forward (5’-AGAGCTTCGTTCAGCACTCC-3’) and EV71- Reverse (5’- GGAAGGTCGCATTACCCCAA-3’).
Clinical specimens
This study adhered to the ethical principles of the Declaration of Helsinki. Human specimens were obtained from voluntarily consenting patients at Affiliated Hospital of Medical School Nanjing University (Suzhou Hospital, Suzhou, China). To qualify as a healthy donor, individuals must fulfill health criteria, including the absence of chronic or communicable diseases and demonstration of normal findings in standard blood tests, urinalysis, liver function tests, and renal function evaluations. The serum samples from healthy individuals were selected and divided into two groups. The age distribution of the young group was 18-40 years old, and the age of the elderly group was over 65 years old. This study was approved by the Ethics Committee of the Suzhou Hospital of Nanjing University.
Animal experiment
Wild-type C57BL/6 J mice were purchased form Jihui Laboratory Animal Care Co.,Ltd (Shanghai, China). Rab27a-konckout (Rab27a-KO) mice (C57BL/6 J background) were constructed by Cyagen Biosciences (Suzhou, China). Mice were maintained in a temperature-controlled environment at 25 °C with a 12 h light and 12 h dark cycle. For starvation experiment, ten weeks-old of wild-type and Rab27a-KO mice were fasted for 48 hours but supplied with drinking water. At the end of the fasting period, the mice were euthanized, livers were collected for WB analysis and blood samples were collected for the assessment of the particle size distribution of sEV or AEVs. For vesicles-enclosed CVB4 infection experiment, each wild-type mouse (n = 6) was injected with AEVs or vesicles isolated from supernatant of 5 × 107 CVB4-infected wild-type or Rab11-KO 293 T cells through intra-peritoneally. The injected mice were continuously observed to record survival status. Mice were dissected at 24 h post-injection to collect brain, heart, spleen, lung, kidney, pancreas and small intestine tissues to screen virus replication. Total RNA was extracted from individual tissues using TRIzol (Life Technologies, USA) and the viral RNA copy number was determined as described above.
All animal experiments were performed in accordance with the recommendations of the Guide for the Care and Use of Medical Laboratory Animals (Ministry of Health, PR China, 1998) and the guidelines of the Laboratory Animal Ethical Commission of Soochow University (#202309A0476).
Statistical analyses
All data were depicted using scatter plots where the mean ± SD were shown superimposed over the individual data points using GraphPad Prism 9.0 software (GraphPad Software Inc., San Diego CA). Two-tailed Student’s t-test was used to compare differences between two groups, while analysis of variance (ANOVA) followed by the Dunnett’s test was used to assess differences between more than two groups. P-value of less than 0.05 was considered statistically significant, indicated as *p < 0.05, **p < 0.01, ***p < 0.001.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data generated in this study have been deposited in the ProteomeXchange Consortium under accession code PXD067720 (http://proteomecentral.proteomexchange.org). All the other data supporting the findings of this study are included in the article and its Supplementary Information files. All key raw data deposited are also included in the source data file provided with this paper. Source data are provided with this paper.
References
Cheng, L. & Hill, A. F. Therapeutically harnessing extracellular vesicles. Nat. Rev. Drug Discov. 21, 379–399 (2022).
van Niel, G., D’Angelo, G. & Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 19, 213–228 (2018).
Pegtel, D. M. & Gould, S. J. Exosomes. Annu. Rev. Biochem. 88, 487–514 (2019).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Ponpuak, M. et al. Secretory autophagy. Curr. Opin. Cell Biol. 35, 106–116 (2015).
Ganesan, D. & Cai, Q. Understanding amphisomes. Biochem. J. 478, 1959–1976 (2021).
Fader, C. M. & Colombo, M. I. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 16, 70–78 (2009).
Solvik, T. A. et al. Secretory autophagy maintains proteostasis upon lysosome inhibition. J. Cell Biol. 221, e202110151 (2022).
Minakaki, G. et al. Autophagy inhibition promotes SNCA/alpha-synuclein release and transfer via extracellular vesicles with a hybrid autophagosome-exosome-like phenotype. Autophagy 14, 98–119 (2018).
Leidal, A. M. et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199 (2020).
Wang, T. et al. Extracellular vesicles originating from autophagy mediate an antibody-resistant spread of classical swine fever virus in cell culture. Autophagy 18, 1433–1449 (2022).
Wan, Y. et al. PPRV-induced autophagy facilitates infectious virus transmission by the exosomal pathway. J. Virol. 96, e0024422 (2022).
van der Grein, S. G. et al. The encephalomyocarditis virus Leader promotes the release of virions inside extracellular vesicles via the induction of secretory autophagy. Nat. Commun. 13, 3625 (2022).
Cloherty, A. P. M. et al. Dengue virus exploits autophagy vesicles and secretory pathways to promote transmission by human dendritic cells. Front. Immunol. 15, 1260439 (2024).
Xing, H., Tan, J., Miao, Y., Lv, Y. & Zhang, Q. Crosstalk between exosomes and autophagy: a review of molecular mechanisms and therapies. J. Cell. Mol. Med. 25, 2297–2308 (2021).
Choi, Y., Bowman, J. W. & Jung, J. U. Autophagy during viral infection—a double-edged sword. Nat. Rev. Microbiol. 16, 341–354 (2018).
Aman, Y. et al. Autophagy in healthy aging and disease. Nat. Aging 1, 634–650 (2021).
Welsh, J. A. et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J. Extracell. Vesicles 13, e12404 (2024).
Fader, C. M., Sanchez, D., Furlan, M. & Colombo, M. I. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic 9, 230–250 (2008).
Marie, P. P. et al. Accessory ESCRT-III proteins are conserved and selective regulators of Rab11a-exosome formation. J. Extracell. Vesicles 12, e12311 (2023).
Wollert, T. & Hurley, J. H. Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 (2010).
McCullough, J., Frost, A. & Sundquist, W. I. Structures, functions, and dynamics of ESCRT-III/Vps4 membrane remodeling and fission complexes. Annu. Rev. Cell Dev. Biol. 34, 85–109 (2018).
Ostrowski, M. et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 12, 19–30 (2010).
Pleet, M. L. et al. Autophagy, EVs, and infections: a perfect question for a perfect time. Front. Cell. Infect. Microbiol. 8, 362 (2018).
Chaudhari, P., Ghate, V., Nampoothiri, M. & Lewis, S. Multifunctional role of exosomes in viral diseases: from transmission to diagnosis and therapy. Cell. Signal. 94, 110325 (2022).
Zhang, R. et al. Enterovirus 71-induced autophagosome fusion with multivesicular bodies facilitates viral RNA packaging into exosomes. Microb. Pathog. 173, 105875 (2022).
Shelke, G. V., Williamson, C. D., Jarnik, M. & Bonifacino, J. S. Inhibition of endolysosome fusion increases exosome secretion. J. Cell Biol. 222, e202209084 (2023).
Visnovitz, T. et al. A ‘torn bag mechanism’ of small extracellular vesicle release via limiting membrane rupture of en bloc released amphisomes (amphiectosomes). eLife 13, RP95828 (2025).
Zhang, Q., Jeppesen, D. K., Higginbotham, J. N., Franklin, J. L. & Coffey, R. J. Comprehensive isolation of extracellular vesicles and nanoparticles. Nat. Protoc. 18, 1462–1487 (2023).
Gao, J. et al. Recent developments in isolating methods for exosomes. Front. Bioeng. Biotechnol. 10, 1100892 (2022).
Purushothaman, A. Exosomes from cell culture-conditioned medium: isolation by ultracentrifugation and characterization. Methods Mol. Biol. 1952, 233–244 (2019).
Théry, C. et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 7, 1535750 (2018).
Lucotti, S., Kenific, C. M., Zhang, H. & Lyden, D. Extracellular vesicles and particles impact the systemic landscape of cancer. EMBO J. 41, e109288 (2022).
Picca, A. et al. Circulating extracellular vesicles: friends and foes in neurodegeneration. Neural Regen. Res. 17, 534–542 (2022).
Yin, Y., Chen, H., Wang, Y., Zhang, L. & Wang, X. Roles of extracellular vesicles in the aging microenvironment and age-related diseases. J. Extracell. Vesicles 10, e12154 (2021).
Vinik, Y. et al. Proteomic analysis of circulating extracellular vesicles identifies potential markers of breast cancer progression, recurrence, and response. Sci. Adv. 6, eaba5714 (2020).
Henne, W. M., Buchkovich, N. J. & Emr, S. D. The ESCRT pathway. Dev. Cell 21, 77–91 (2011).
Lu, Y., Zhang, Z., Sun, D., Sweeney, S. T. & Gao, F. B. Syntaxin 13, a genetic modifier of mutant CHMP2B in frontotemporal dementia, is required for autophagosome maturation. Mol. Cell 52, 264–271 (2013).
Lee, J. A., Liu, L. & Gao, F. B. Autophagy defects contribute to neurodegeneration induced by dysfunctional ESCRT-III. Autophagy 5, 1070–1072 (2009).
Hanson, P. I., Roth, R., Lin, Y. & Heuser, J. E. Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J. Cell Biol. 180, 389–402 (2008).
Bertin, A. et al. Human ESCRT-III polymers assemble on positively curved membranes and induce helical membrane tube formation. Nat. Commun. 11, 2663 (2020).
Lee, I.-H., Kai, H., Carlson, L.-A., Groves, J. T. & Hurley, J. H. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc. Natl. Acad. Sci. USA 112, 15892–15897 (2015).
Wei, D. et al. RAB31 marks and controls an ESCRT-independent exosome pathway. Cell Res. 31, 157–177 (2021).
Hirvonen, M. J. et al. Novel interaction of Rab13 and Rab8 with endospanins. FEBS Open Bio 3, 83–88 (2013).
Ioannou, M. S., Girard, M. & McPherson, P. S. Rab13 traffics on vesicles independent of prenylation. J. Biol. Chem. 291, 10726–10735 (2016).
Nokes, R. L., Fields, I. C., Collins, R. N. & Fölsch, H. Rab13 regulates membrane trafficking between TGN and recycling endosomes in polarized epithelial cells. J. Cell Biol. 182, 845–853 (2008).
Sahgal, P. et al. GGA2 and RAB13 promote activity-dependent β1-integrin recycling. J. Cell Sci. 132, jcs233387 (2019).
Su, W. et al. Identification of autophagic target RAB13 with small-molecule inhibitor in low-grade glioma via integrated multi-omics approaches coupled with virtual screening of traditional Chinese medicine databases. Cell Prolif. 54, e13135 (2021).
Hinger, S. A. et al. Rab13 regulates sEV secretion in mutant KRAS colorectal cancer cells. Sci. Rep. 10, 15804 (2020).
Bryant, K. L. et al. Combination of ERK and autophagy inhibition as a treatment approach for pancreatic cancer. Nat. Med. 25, 628–640 (2019).
Kinsey, C. G. et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med. 25, 620–627 (2019).
Baggen, J., Thibaut, H. J., Strating, J. & van Kuppeveld, F. J. M. The life cycle of non-polio enteroviruses and how to target it. Nat. Rev. Microbiol. 16, 368–381 (2018).
Feng, Z. et al. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496, 367–371 (2013).
Gu, J. et al. Exosomes cloak the virion to transmit Enterovirus 71 non-lytically. Virulence 11, 32–38 (2020).
Takahashi, M. et al. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J. Clin. Microbiol. 48, 1112–1125 (2010).
Santiana, M. et al. Vesicle-cloaked virus clusters are optimal units for inter-organismal viral transmission. Cell Host Microbe 24, 208–220.e208 (2018).
Robinson, S. M. et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 10, e1004045 (2014).
Chen, Y. H. et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell 160, 619–630 (2015).
van der Grein, S. G. et al. Picornavirus infection induces temporal release of multiple extracellular vesicle subsets that differ in molecular composition and infectious potential. PLoS Pathog. 15, e1007594 (2019).
Sin, J., McIntyre, L., Stotland, A., Feuer, R. & Gottlieb, R. A. Coxsackievirus B escapes the infected cell in ejected mitophagosomes. J. Virol. 91, e01347-17 (2017).
Acknowledgements
This work was supported by the National Natural Science Foundation of China under Grant No. 32270162 to Y.F., Priority Academic Program Development of Jiangsu Higher Education Institutions, China; Jiangsu Natural Science Foundation, China (No. BK20231313) to Y.F., Suzhou Science and Technology Development Plan, China (Nos. SKJY2021045, ZXL2023202) to Y.F.
Author information
Authors and Affiliations
Contributions
Y.X.F., K.D.M., and F.F.H. carried out and analyzed the data for most of the in vitro experiments. F.R.W. and K.D.M. collected clinical samples and performed the experiments. K.D.M. and H.Q.W. performed the animal experiments. K.D.M. and Y.X.F. wrote and revised the manuscript. Y.X.F. and S.D.X. supervised the study. All authors reviewed and discussed the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mao, K., Huo, F., Wei, H. et al. Autophagic extracellular vesicles (AEVs) are distinct from exosomes and play crucial roles in viral infections. Nat Commun 17, 1095 (2026). https://doi.org/10.1038/s41467-025-67860-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67860-9