Abstract
Genome wide association studies of schizophrenia reveal a complex polygenic risk architecture comprised of hundreds of risk variants; most are common in the population, non-coding, and act by genetically regulating the expression of one or more gene targets (“eGenes”). It remains unclear how the myriad genetic variants that are predicted to confer individually small effects combine to yield substantial clinical impacts in aggregate. Here, we demonstrate that convergence (i.e., the shared downstream transcriptomic changes with a common direction of effect), resulting from one-at-a-time perturbation of schizophrenia eGenes, influences the outcome when eGenes are manipulated in combination. In total, we apply pooled and arrayed CRISPR approaches to target 21 schizophrenia eGenes in human induced pluripotent stem cell-derived glutamatergic neurons, finding that functionally similar eGenes yield stronger and more specific convergent effects. Points of convergence constrain additive relationships between polygenic risk loci: consistent with a liability threshold model, combinatorial perturbations of these same schizophrenia eGenes reveal that pathway-level convergence predicts when observed effects will fail to sum to levels predicted by an additive model. Targeting points of convergence as novel therapeutic targets may prove more efficacious than individually reversing the effects of multiple risk loci.
Similar content being viewed by others
Introduction
The genetic architecture of schizophrenia is complex and polygenic. Highly penetrant rare mutations underlie only a fraction of cases1. Rather, genome-wide association studies (GWAS) indicate that schizophrenia is predominantly associated with genetic variation that is common in the population2. These risk loci have small effect sizes, are typically found in non-coding regions, and regulate the expression of one or more genes3,4,5. Mapping GWAS loci to their target genes (termed “eGenes”, as defined by significant genetic regulation of expression) remains challenging, but is informed by expression quantitative trait loci (eQTL)6,7,8,9, chromatin accessibility10,11,12, enhancers13,14,15,16, and 3D chromatin architecture17,18,19,20,21,22. The regulatory activity of risk loci can be empirically evaluated using massively parallel reporter assays23,24,25 and pooled CRISPR screens26, and causal gene targets and functions definitively resolved by genetic engineering in human induced pluripotent stem cells (hiPSCs)10,11,17,27,28.
Schizophrenia eGenes are particularly expressed during fetal cortical development29,30,31 and in glutamatergic neurons (as well as medium spiny neurons, and certain interneurons)31,32,33. They are highly co-expressed in human brain tissue34 and cultured neurons17, show high connectivity in protein-protein interaction networks17,35,36,37, and are enriched for roles in synaptic function and gene regulation2,7,17,38,39,40,41. Likewise, transcriptomic studies of post-mortem brains from schizophrenia cases also identify aberrant expression of genes associated with synaptic function and chromatin dynamics in neurons42,43,44. The mechanism by which hundreds of distinct eGenes lead to shared molecular pathology is unknown.
We predicted that eGenes linked to schizophrenia would share substantial downstream transcriptomic changes with a common direction of effect (termed “convergence”). Although convergence has been described in the context of loss-of-function autism spectrum disorder risk genes45,46,47,48,49,50,51,52,53,54,55, these rare mutations almost never co-occur in the same individual. The convergent impact of common variants—which are frequently inherited together, and the impacts of which are apparent only in aggregate—remains unknown. We targeted twenty-one schizophrenia eGenes in hiPSC-derived induced glutamatergic neurons (iGLUTs) using pooled and arrayed CRISPR-based approaches, significantly perturbing seventeen (CALN1, CLCN3, DOC2A, FES, FURIN, GATAD2A, NAGA, PCCB, PLCL1, THOC7, TMEM219, SF3B1, SNAP91, SNCA, UBE2Q2L, ZNF823, ZNF804A), and resolving convergent impacts robust to experimental and donor effects. To test if convergence influenced the outcome when eGenes were inherited in combination (i.e. if eGene effects sum linearly according to the additive model26), we compared manipulation of eGenes one at a time and in groups defined by annotated functions at the synapse (“synaptic”: SNAP91, CLCN3, PLCL1, DOC2A, SNCA), or regulating transcription (“regulatory”: ZNF823, INO80E, SF3B1, THOC7, GATAD2A), or with un-related non-synaptic, non-regulatory biology (“multi-function”: CALN1, CUL9, TMEM219, PCCB, FURIN), and random combinations thereof. Altogether, with broad relevance across complex polygenic disease56,57, our work begins to experimentally determine answers to the long-standing question of how risk variants interact in human neurons.
Results
Convergence of downstream transcriptomic impacts across schizophrenia eGene perturbations
We27,58,59,60 and others11,61,62,63,64,65,66,67 demonstrated that iGLUTs are >95% glutamatergic neurons, robustly express glutamatergic genes, release neurotransmitters, produce spontaneous synaptic activity, and recapitulate the impact of psychiatric trait-associated genes. iGLUTs express most schizophrenia eGenes, including all eGenes prioritized herein27.
eGenes whose brain expression was predicted to be up-regulated by GWAS loci2 were prioritized for a pooled CRISPR activation (CRISPRa) experiment, which is currently restricted to one direction of effect. eGenes that were non-coding, located in the MHC locus, or poorly expressed in iGLUTs were excluded. First, transcriptome and epigenome imputation (EpiXcan68) of schizophrenia GWAS2 risk loci from post-mortem brain42,69 prioritized seven schizophrenia eGenes (SCZ1: CALN1, FES, NAGA, NEK4, PLCL1, UBE2Q2L, and ZNF804A) (Table 1; Fig. 1A). Second, transcriptomic imputation (prediXcan70,71,72, p < 6 × 10−6) of SCZ GWAS2 identified ~250 eGenes (SI Table 1), subsequently narrowed by considering colocalization (COLOC73,74, PP4 > 0.8) between schizophrenia GWAS2 and post-mortem brain expression quantitative loci (eQTL) peaks6, which identified 25 eGenes (SI Table 1). 22 eGenes overlapped between approaches, ten of which were coding genes associated with increased expression in schizophrenia (SCZ2: CALN1, CLCN3, CUL9, DOC2A, PLCL1, INO80E, SF3B1, SNAP91, TMEM219, ZNF823) (Table 2; Fig. 1A). Of note, our eGene selection, derived in bulk post-mortem brain, is largely preserved using an excitatory neuron-specific PrediXcan analysis (ExN-PrediXcan, Tables 1A, B).
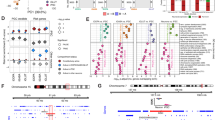
A Schematic of schizophrenia eGene identification and prioritization. Schizophrenia eGenes were prioritized by fine-mapping (COLOC), transcriptomic imputation (PrediXcan), and/or epigenomic imputation (EpiXcan) schizophrenia GWAS using post-mortem brain expression data. B Effect sizes of significant eGenes from either dorsolateral prefrontal cortex (DLPFC) EpiXcan (blue), DLPFC S-PrediXcan (green) or excitatory neuron (ExN) S-PrediXcan (purple) transcriptomic imputation studies. The size of circles corresponds with the −log10(adjusted p value). C Log2(fold change) of all eGenes in the arrayed experiment following single (teal) and joint perturbations across all 15 eGenes (yellow) or functional (orange) or random (maroon) sets of five eGenes in D21 hiPSC-NPC derived iGLUTs, using individual vectors. The size of circles corresponds with the −log(adjusted p value) from a one-tailed t test. D Log2(fold change) of all eGenes in the pooled experiments SCZ1 and SCZ2, comparing all perturbed cells of one target eGene identity to all other cells of different eGene identities (blue) or compared to only Scramble gRNA (teal). The size of circles corresponds with the −log(adjusted p value) from a one-way pairwise Wilcox Rank Sum. Created with BioRender.com.
Pooled CRISPR screening combined single-cell RNA sequencing readouts and direct detection of sgRNAs75. Two independently designed, constructed, and validated pooled CRISPRa libraries (SCZ1 and SCZ2) were transduced into iGLUTs from two donors in independent experiments at unique developmental time-points (DIV7 or DIV21, SI Fig. 1F). Non-perturbed cells from both SCZ1 and SCZ2 demonstrated gene expression patterns that correlated with expression in the adult postmortem DLPFC in neurotypical controls (SI Fig. 12A–C) and cortical neurons. Specifically, these cells were most strongly correlated with fetal cells transitioning to neuronal fate, fetal excitatory neurons, and cortical adult neurons (SI Fig. 13). The large number of presumably wildtype neurons in the population expressing either a scramble gRNA or no detectable gRNA at all (>60% of all pooled cells, see SI Fig. 4A), mitigates the possibility that results were confounded by non-cell autonomous effects. Likewise, there was no significant difference in the degree of variance in maturity of the cell population between experiments and imputed cell fractions were not correlated with perturbation status (SI Figs. 2 and 3). An unsupervised framework, Weighted Nearest Neighbor Analysis76, assigned successful perturbations; in total, we resolved perturbations of six of seven SCZ1 eGenes (SCZ1: CALN1, FES, NAGA, PLCL1, UBE2Q2L, and ZNF804A; ten gRNAs each) and four of ten SCZ2 eGenes (SCZ2: CLCN3, SF3B1, TMEM219, ZNF823; three gRNAs each). For 5401 and 6352 cells, respectively, we identified the sgRNA in each cell, the cis target gene with differential expression, and the downstream trans alterations to pathways resulting from initial cis up-regulation. Following QC, normalization, and removal of doublets (cells containing more than one sgRNA), an average of 316 cells per sgRNA were successfully perturbed (ranging from 93 to 552) for a total of 3640 perturbed cells and 210 scramble controls (SI Fig. 4−7). Upregulation of eGenes by CRISPRa ranged from 0.2 to 3 log2 fold-change (Log2FC), comparable to the predicted effect sizes [SCZ1 (0.08–0.35); SCZ2 (0.2–0.77)] and eGene expression changes (Log2FC range 0.3–5.2) in the post-mortem dorsolateral prefrontal cortex (Fig. 1B, C; SI Tables 2,3; Fig. 8). Effects of different gRNAs targeting the same eGenes were highly concordant, even when the degree of perturbation varied (SI Fig. 15). Differentially expressed genes (DEGs, pFDR < 0.05, Supplementary Data 2) were enriched for neuroactive ligand-receptor interaction, protein processing in the endoplasmic reticulum, proteasome, and spliceosome Gene Ontology and KEGG Pathways terms (Supplementary Data 3), suggesting that diverse eGenes might impact similar neural processes and pathways.
We define “convergence” as the independent development of transcriptomic changes in the same direction resulting from all eGene perturbations. DEGs were meta-analyzed (using METAL), and “convergent” genes were defined as those with shared direction of effect across all eGene perturbations and with non-significant heterogeneity between eGenes (Cochran’s heterogeneity Q-test pHet > 0.05). Across all schizophrenia eGenes, 363 convergent genes (Bonferroni meta p-value<=0.05) were identified by meta-analysis, with 77 passing multiple testing correction (SI Data 2).
To identify groups of genes with similar expression patterns across eGene perturbations, we define “convergent networks” as relationships between genes that are co-regulated by shared biological mechanisms. Unsupervised Bayesian bi-clustering77 and gene co-expression network reconstruction from the pooled CRISPRa single cell RNAseq (n = 3850 cells, 16,851 genes, donor/batch corrected and normalized to adjust for covariates such as cell heterogeneity) identified high-confidence co-expressed gene networks. Across the pooled single-cell experiments, 1048 protein-coding source node genes (>5 edges) were identified, with a total network membership of 1869 genes that clustered together in at least 20% of the runs (Fig. 2A, Supplementary Data 3), and significant enrichments for gene targets of schizophrenia GWAS loci as well as transcription factors (AP4 (TFAP4)62, NFAT63, ERR164, and TCF478,79) and miRNAs (miR-30) that regulate schizophrenia GWAS loci (Fig. 2A, i–iii). The cross-target convergent network was enriched in biological pathways implicated in schizophrenia etiology (SI Fig. 9); overrepresentation analysis revealed schizophrenia, bipolar disorder, intellectual disability, and autism spectrum disorder common and rare risk genes to be significantly over-represented in node genes shared across all eGene perturbations (Fig. 2A, i; Supplementary Data 3).
A Convergent network resolved across the downstream transcriptomic impacts of all 10 target perturbations in the pooled experiments SCZ1 and SCZ2 identified 1869 convergent genes with enrichments for (i) brain-related GWAS genes, (ii) transcription factor binding sites of known schizophrenia-associated TFs (TFAP4, NFAT and ERR1), and (iii) common and rare variant target genes. B Convergent networks resolved across the downstream transcriptomic impacts of all fifteen target perturbations in the arrayed assay identified 255 convergent genes with enrichments for (i) miRNA targets and (ii) transcription factor binding sites of known schizophrenia-associated TFs (TFAP4, NFAT and ERR1), and (iii) common and rare variant target genes. C While largely distinct, the resolved convergent networks from the arrayed and pooled experiments shared 16 significant enrichments for miRNA targets and 4 significant enrichments for TF targets – many of which are thought to play a role in the regulation of schizophrenia-associated genes. Ai, 2Bi Overrepresentation analysis using one-tailed Fisher’s exact test for gene enrichment in curated disorder gene lists with Benjamini-Hochberg FDR multiple testing correction. Aii-iii, Bii-iii, C Geneset enrichment based on one-tailed hypergeometric test P values with Benjamini-Hochberg FDR multiple testing correction using FUMA GENE2FUNC. D Overlapping nodes between the two networks were often involved in neuronal proliferation and differentiation. Created with BioRender.com.
To study the strength and composition of convergent networks, we define “network convergence” as the sum of the network connectivity score (i.e., networks with fewer nodes and more interconnectedness have increased convergence). We endeavored to identify the biological factors (e.g., number of eGenes, functional similarity of eGenes, and eGene co-expression) that influenced network convergence. eGene number tested the number of eGenes used to generate a convergent network. Functional similarity (i.e., the degree of shared biological functions amongst eGenes) was calculated in two ways: Gene Ontology semantic similarity scoring (within biological pathway, cellular component, and molecular function)80, and synaptic/signaling score (proportion of eGenes with annotated function as either “signaling” for pooled or “synaptic” for arrayed). The brain expression correlation was calculated as the strength of the correlation of eGene expression in the post-mortem dorsolateral prefrontal cortex6 (see Methods, SI Fig. 10). Bayesian reconstruction81 was performed across all random combinations of eGene perturbations from the pooled experiment (1003 unique eGene-Convergent Network sets) and arrayed experiment described in the following section (32,752 sets) and resolved distinct networks (Fig. 3B, E). Principal components analysis tested the effect of biological factors on the network convergence scores (Fig. 3C, D, F, G; SI Fig. 10–11). Only brain expression correlation and the proportion of synaptic/signaling genes were significantly positively correlated with network convergence across all sets in both the pooled [brain expression correlation: Pearson’s r = 0.24, adj. p value < 0.001, signaling proportion: Pearson’s r = 0.14, adj. p value < 0.01, n = 826] and arrayed experiments [brain expression correlation: Pearson’s r = 0.083, Bonferroni adjusted p value < 0.001, signaling proportion: Pearson’s r = 0.25, adj. p value < 0.001, n = 16319] (Fig. 3D, G). The average expression of perturbed eGenes was positively correlated with network convergence but was only significantly associated in the arrayed experiment (Fig. 3D, G). Finally, although SCZ1 and SCZ2 pooled CRISPR screens were generated from distinct differentiation timepoints, the proportion of eGene perturbations by experiment did not correlate with the degree of network convergence, indicating that we have adequately controlled for variation in neuronal maturation (Fig. 3D; Pearson’s r = 0.062, Bonferroni p value = 1).
A Defining convergence and calculating convergent network strength. Here, we define convergence as the independent development of transcriptomic similarities between separate gene perturbations that move towards union or uniformity of biological function. B–G Principal component analysis (PCA) of the convergence scores, the three Gene Ontology scores (Molecular Function, M.F.; Biological Process, B.P.; Cellular Component, C.C.), brain expression correlation (B.E.C), and sample size across all resolved networks in both the pooled and arrayed assays revealed that some functional scores have similar influence on variance as convergence (SI Fig. 19). B, E Distribution of the degree of convergence (x axis) of networks across all possible combinations of 2–8 (y axis; number of sets tested within each set) target perturbations from the single-cell pooled experiment (B) and arrayed experiment (E) across all possible combinations of 2–14 target perturbations from the arrayed experiment show an influence of sample size on the ability to resolve a network. Data are represented as median values, lower and upper hinges correspond to the 1st and 3rd quartiles, upper and lower whiskers represent the largest values within 1.5*IQR (inter-quartile range) from the first or third quartile. Each point represents convergence based on biclustering between 2–8 unique combinations of CRISPR perturbations. B N = 4 replicates per condition (2 x donors, 2 x independent replicates per donor). E N = 2 biological replicates, 10 gRNA replicates (SCZ1); N = 2 biological replicates, 2 technical replicates (sequencing batches) and 3 gRNA replicates (SCZ2). C, F For both the pooled (C) and the arrayed (F) experiment, PCs 1 (x axis) and 2 (y axis) explain ~62% of the variance between networks. PC loadings demonstrate the influence of each variable on the variance between networks; within the first two PCs, the influence of brain expression correlation (B.E.C) and proportion of signaling genes perturbed (S.P) on PCs 1 and 2 on variance explained is more strongly related to convergence degree compared to other functional scores. Since degree of convergence is influenced by number of eGenes perturbed, we ran PCA analysis within networks of the same set size and found that the pattern of influence of signaling proportion and brain expression correlation is maintained when convergence is ranked within set size shown in SI Fig. 20. D, G This corresponds to an overall significant positive correlation between network convergence degree, signaling/synaptic proportion of perturbed genes in a set, and brain expression correlation between genes in a set (Bonferroni adjusted p value of Pearson’s correlations: *<0.05, **<0.01, ***<0.001. Created with BioRender.com.
Convergence constrains the total impact of combinatorial perturbations of schizophrenia eGenes
We manipulated eGenes in combination to approximate the polygenic nature of schizophrenia and test whether convergence between eGenes influences observed effects. Given that genes implicated in synaptic biology and epigenetic/transcriptional regulation are enriched for the schizophrenia risk2,7,17,38,39,40,41, we sought to generate three groups of eGenes, linked to synaptic biology, gene regulation, or neither (Fig. 1A, arrayed experiment). Unconstrained by the unidirectionality of pooled CRISPR screens, we did not restrict our list to eGenes with a single direction of effect. From the 18 coding genes prioritized by the intersection of transcriptomic imputation and colocalization, eGenes were separated into discrete functional categories based on gene ontology annotations. Our final gene list included five synaptic genes (SNAP91, CLCN3, PLCL1, DOC2A, SNCA), five regulatory genes (ZNF823, INO80E, SF3B1, THOC7, GATAD2A), and five genes with non-synaptic, non-regulatory functions, termed “multi-function” (CALN1, CUL9, TMEM219, PCCB, FURIN) (Table 2; Fig. 1A).
We applied an arrayed design (i.e., distinct conditions in each well) to manipulate schizophrenia eGenes alone and in combination, allowing us to capture cell autonomous and non-cell autonomous effects in a manner not possible in the pooled design (Fig. 4, SI Table 4, SI Fig. 19). Endogenous expression was increased and decreased (via CRISPRa and shRNAs, respectively) in the direction associated with schizophrenia risk. CRISPRa and shRNA were specifically selected for perturbation due to the potential for simultaneous, bi-directional perturbation of target eGenes in joint perturbation conditions (see Methods). Three to five vectors per gene were tested in 7-day-old (D7) iGLUTs, identifying the single vector that best achieved the level of significant perturbation predicted by eQTL analyses as confirmed by qPCR (SI Fig. 1E). Each eGene was perturbed in 21-day-old (D21) iGLUTs for 72 hours (Fig. 1D, SI Fig. 1F, G, 19A, and 21A), individually and jointly, including appropriate vector and scrambled controls, from two neurotypical donors with average polygenic risk scores (one experimental batch per donor). Three groups of five random genes, one group of ten random genes, and one group of all fifteen genes were also included. Significant (p < 0.05) changes in eGene expression in iGLUTs were confirmed by RNAseq in 13/15 eGenes (SNAP91, CLCN3, PLCL1, DOC2A, SNCA, ZNF823, SF3B1, THOC7, GATAD2A, CALN1, TMEM219, PCCB, FURIN) (SI Fig. 1G, SI Fig. 19A); we validated the magnitude and direction of experimental eGene perturbation relative to the dosage effects of the top predicted causal SNPs (e.g., eQTL effect size) and predicted eGene expression changes (Fig. 1B, D; SI Table 2–4). Across donors, donor status did not significantly impact the degree of eGene perturbation (SI Fig. 1Hp = 0.75, paired t test). Single perturbation of eGenes by CRISPRa ranged from 0.07 to 0.44 log2 fold change, and RNAi ranged from −0.22 to −0.87 log2 fold change, comparable to EpiXcan effect sizes of 0.10 to 0.31 and −0.06 to −0.20 and PrediXcan effect sizes of 0.22 to 0.77 and −0.17 and −0.38 for corresponding eGenes.
A Schematic of differential expression analysis. Individual eGene perturbations, the implementation of the expected additive model based on the latter and the measured combinatorial perturbation permit the detection of interactive effects through comparison with the additive model. B Combinatorial perturbation of synaptic and regulatory eGenes resulted in non-additive effects on expression across 16.8% (synaptic) and 20.2% (regulatory) of the transcriptome. No significant non-additive effects were seen following joint perturbation of non-synaptic, non-transcriptional regulatory eGenes. Teal = proportion of genes showing significant non-additivity (two-tailed FDR < 0.1); blue = proportion of genes showing no significant non-additivity. C GSEA of non-additive genes in the Synaptic eGene set demonstrated significant enrichment for genes relating to brain disorders and synaptic function. GSEA of non-additive genes in the Regulatory eGene set demonstrated significant enrichment for genes relating to brain disorders and synaptic function. SCZ schizophrenia, CNV copy number variant, FMRP Fragile X Mental Retardation Protein, FDR false discovery rate. D Non-additive effects following combinatorial perturbation of sets of five, ten, and fifteen eGenes randomly assigned from the synaptic, regulatory, and multi-function eGene groups. The proportion of the transcriptome exhibiting significant non-additive effects increased with increasing numbers of perturbed eGenes (average of 5.1%, 10.0% and 19.2% of the transcriptome with non-additive FDR < 0.1 after joint perturbations of 5, 10, and 15 eGenes, respectively).
Across the majority of the schizophrenia eGenes in our arrayed experiment, competitive gene-set enrichment analysis using 698 manually curated neural59 gene-sets (SI Fig. 19B, C, SI Fig. 20A, Supplementary Data 1) resulted in DEGs (pFDR < 0.05) that were strongly enriched for gene-sets related to rare and common psychiatric disorder risk genes (11/15) (SI Fig. 20B), pre-synaptic biology (10/15) (SI Fig. 20C), and glutamatergic neurotransmission (10/15) (SI Fig. 20D).
Overall, we again observed robust convergence at the gene- (METAL77, p < 1.92 × 10−06) and network-level (Bayesian network reconstruction81) (Supplementary Data 3). A densely interconnected network of 255 genes (n = 63 samples, 4/sgRNA or shRNA, 25,487 genes, and normalized to adjust for covariates such as donor) was significantly enriched for biological pathways implicated in schizophrenia etiology; over representation analysis revealed that target genes of schizophrenia, intellectual disability, and autism spectrum disorder common and rare variants were significantly over-represented in the network (Fig. 2B, i; Supplementary Data 3), as well as genes regulated by miRNAs and transcription factors implicated in schizophrenia etiology, such as hsa-miR-124a82 and NKX2 83 (Fig. 2B, ii–iii). Separation of schizophrenia eGenes based on either signaling (SI Fig. 9A) or regulatory (SI Fig. 9B) function resolved unique convergent networks with no overlap in node genes, suggesting that the functional similarity of schizophrenia eGenes affects downstream convergence. Each of these networks included neuropsychiatric risk genes as well as those annotated for synaptic and immune signaling function (Supplementary Data 3). Networks derived from arrayed and pooled experiments shared significant enrichments for targets of miRNAs and transcription factors associated with schizophrenia (Fig. 2C), although only nine node genes overlapped (Fig. 2D).
Following combinatorial manipulation of schizophrenia eGenes, most genome-wide effects occurred as predicted by summing differential expression for single eGene perturbations (“expected additive” model, Box 1), yet 16.8% of the total transcriptome for synaptic eGenes and 20.2% for the regulatory eGenes did not (Fig. 4; SI Figs. 21–23; SI Table 4). We term these overwhelmingly sub-additive effects (SI Fig. 21D) as “non-additive” (Bayes moderated t-statistics, FDR p < 0.1) and report π1 synergy coefficients84 of 43.86 (synaptic eGenes), 42.74 (regulatory eGenes), and 0.00 (multi-function eGenes). Non-additive genes resulting from combinatorial synaptic and regulatory eGene perturbations were significantly enriched for SZ risk genes as well as synaptic gene sets (Fig. 4C).
Key controls demonstrate that non-additive effects did not result from technical limitations of our approach. Consistent with single-cell level effects, observations were similar whether tested from independent expression vectors (Fig. 4), a single multiplexed vector expressing all gRNAs27 (SI Fig. 22A–F), or a polycistronic gRNA vector (SI Fig. 22G, H). Likewise, modified ECCITE-seq confirmed a high number of unique gRNA integrations at the single cell level (SI Fig. 22I). Non-additivity could not be attributed to differences in the magnitude of eGene perturbation between individual and combinatorial perturbations across both donors (SI Fig. 21B, combined donors p > 0.05 Wilcoxon ranked sum test, individual donors p > 0.05 two-way ANOVA), reduced fold-change of non-additive genes (SI Fig. 23C), or differences in baseline expression between non-additive and additive genes (SI Fig. 23D).
Although increasing the number of eGenes perturbed increased the degree of interactive effects on transcription (compare joint perturbations of random sets of 5, 10 and 15 eGenes, Fig. 4D, SI Fig. 23A), our data suggested that specific eGenes may drive non-additive effects; for example, log2FC of CLCN3 (synaptic) and INO80E (regulatory) are the most correlated with synergy coefficients (SI Fig. 16). When evaluated across all eGene sets, the proportion of synaptic (Pearson’s r = 0.49) and regulatory (r = 0.45) eGenes in a set positively correlated with non-additivity, while proportion of multifunctional eGenes was strongly negatively correlated (r = −0.94).
Given that >95% of non-additive genes (whether up- or down-regulated, FDR p < 0.1) showed less differential expression than predicted by the additive model (i.e., changes that were “less up” or “less down” than expected) (SI Fig. 21C), we queried whether overlapping downstream transcriptomic effects (e.g., convergence) constrain the total effects observed in combinatorial perturbation.
Across all combinational perturbations, convergence was significantly correlated with the degree of non-additive effects seen (Fig. 5A, Pearson’s r2 = 0.6569, p = 0.0147). The robust gene-level convergence observed for the synaptic (1070 genes) and regulatory (1070 genes) eGene groups was dramatically reduced in the multi-function eGene group (71 genes) (METAL77, p < 1.92 × 10−06) (Fig. 5B–E; Supplementary Data 3). Convergent genes highly overlapped with non-additive genes (Fisher’s exact test, p < 2.2 × 10−16 for both synaptic and regulatory eGene groups). 71% (761 of 1070) and 94% (1000 of 1070) of convergent genes downstream of synaptic and regulatory eGenes, were included in respective non-additive gene lists (Fig. 5C, D). Convergent effects of synaptic eGenes were enriched for synaptic function (e.g., mGluR5 interactors, p = 1.64 × 10−03) and brain disorder (e.g., schizophrenia GWAS, p = 8.41 × 10−05) gene-sets (Fig. 5F); regulatory eGene convergence was also enriched for brain disorder gene-sets (e.g., bipolar disorder, p = 9.92 × 10−06) (Fig. 5G). Taken together, these findings highlight convergent effects between schizophrenia eGenes on synaptic function and brain disorder risk.
A–E Meta-analysis of differentially expressed genes (DEGs) elicited by individual eGene perturbations for each five-gene grouping using METAL to identify DEGs that showed altered expression consistently in the same direction across all five eGene perturbation conditions for each set of eGenes. A Convergence across individual eGene perturbations is correlated with the degree of non-additive effect seen in the corresponding joint perturbation condition. Two-tailed Pearson’s r2 = 0.6569, p = 0.0147. Teal number of genes showing significant non-additivity (two-tailed FDR < 0.1); yellow no. of genes showing significant convergent effects (two-tailed FDR < 0.1) for each perturbation set. B For each joint eGene perturbation group, non-additive impacts on transcription were compared with genes showing significant convergence across individual perturbations for the same eGene set. C Evidence of convergence was found in 1070 genes across the synaptic eGene perturbations, 761 of which also exhibited non-additive effects in the additive-combinatorial comparison for the same set. D Evidence of convergence was found in 1070 genes across the regulatory eGene perturbations, 1000 of which also exhibited non-additive effects in the additive-combinatorial comparison for the same set. E No significant non-additive effects and only minimal convergence could be seen in eGene perturbations across functional pathways. F GSEA of convergent genes in the synaptic and regulatory eGene groups demonstrated significant enrichment for genes relating to brain disorders and synaptic function. SCZ schizophrenia, CNV copy number variant, FMRP Fragile X Mental Retardation Protein, ID intellectual disability, PPI protein-protein interaction, KEGG Kyoto Encyclopedia of Genes and Genomes, FDR false discovery rate.
Convergent signatures represent plausible therapeutic targets
Individually targeting all eGenes with perturbed expression in each patient is an insurmountable challenge. If instead it were possible to reverse the impact of many schizophrenia eGenes by targeting a smaller number of shared downstream targets, convergent networks might represent important therapeutic targets.
We identified drugs predicted to manipulate top node genes85. Across all eGene perturbations, reversers of convergent node signatures were enriched for mechanisms previously associated with psychiatric disorders, including HDAC inhibitors86 (normalized connectivity score (NCS) = −1.63; FDR adjusted p val<0.08), ATPase inhibitors87 (NCS = −1.61; FDR < 0.08), and sodium channel blockers88 (NCS = −1.59; FDR < 0.08). Conversely, mimickers of convergent node signatures were enriched for pathways associated with stress response, including glucocorticoid receptor agonists (NCS = 1.66, FDR < 0.08) and NF-κB pathway inhibitors (NCS = 1.60; FDR < 0.2) (Supplementary Data 4). Finding only nominally significant enrichments in non-neuronal cell lines suggests these may be neuron-specific drug responses.
Three drugs that opposed the transcription signatures of top convergent nodes specifically in neurons or neural progenitor cells (NPCs) were prioritized (see Methods, Box 2): anandamide (reverser of convergent network signature, NCS = −1.59, FDR = 1 as well as CALN1 signature alone, NCS = −1.23, FDR = 0.15), simvastatin (NCS = −1.31, FDR = 1; TMEM219, NCS = −0.8823, FDR = 0.25), and etomoxir (Convergence, NCS = −1.86, FDR < 2.2e-16; CALN1, NCS = −1.42, FDR = 0.0355; TMEM219, NCS = −1.09, FDR = 0.0112) (Supplementary Data 4). These drugs were tested for their ability to reverse, or oppose, the effects of paired schizophrenia eGene perturbations in iGLUTs: CRISPRa for eGenes was followed by treatment with matched reverser drugs (CALN1: anandamide and etomoxir; TMEM219: simvastatin and etomoxir). Downstream transcriptomic (bulk RNA-seq) and phenotypic (high content imaging, multi-electrode array) assays were assessed to resolve eGene-drug effects on neuronal molecular, morphological, and physiological phenotypes (Fig. 6 and SI Figs. 28–30). All drugs reversed or suppressed the transcriptomic impact of the CRISPRa perturbation alone. Notably, simvastatin ameliorated the transcriptomic impact of TMEM219 and blunted an increase in synaptic density caused by TMEM219 perturbation (two-way ANOVA, CRISPRa perturbation p < 0.001; CRISPRa perturbation x drug treatment interaction p < 0.05) (Fig. 6A, B). Etomoxir limited the transcriptomic impact of perturbations of both CALN1 and TMEM219 (SI Fig. 28A, B). Thus, it may be possible to pharmacologically reverse convergent networks rather than targeting schizophrenia eGenes individually.
In vitro validation of drug-eGene phenotypic interactions. A Effects of 48-hour treatment with 10 µM simvastatin on synaptic puncta density in TMEM219 CRISPRa perturbed (teal) or non-perturbed (purple) iGLUT neurons. Syn1-positive puncta values are expressed relative to MAP2-positive neurite length in each well. Perturbation of TMEM219 expression with CRISPRa significantly increased synaptic puncta density; this increase was partially ameliorated by 48 hr treatment with 10 µM simvastatin (two-way ANOVA; CRISPRa variation p < 0.0001; CRISPRa x Drug treatment variation p < 0.05). N minimum of two independent experiments across 2 donor lines with 12 technical replicates per condition. Values for each technical replicate in imaging experiments were averaged from nine separate images per single well. B Treatment of cells perturbed with either TMEM219 CRISPRa with 10 µM Simvastatin reverses or suppresses the transcriptomic impacts of the schizophrenia eGene perturbations alone (SI Figs. 28–30). Treatment of cells with CRISPRa TMEM219-gRNA and 10 µM Simvastatin over 48 hours opposes the transcriptomic impact observed in CRISPRa TMEM219-gRNA + Vehicle-treated cells. Venn diagram of significant DEGs at an (top left) adjusted p val ≤ 0.05 and at an (top right) unadjusted p value of ≤0.05. (bottom) Dot plot demonstrating the logFC of each gene in either the TMEM219 + Vehicle (green) or TMEM219 + 10uM Simvastatin (yellow) condition, ordered by degree of logFC in the TMEM219 + Vehicle-treated cells. Size of the points corresponds to the −log10 (adjusted p value).
Discussion
Shared downstream effects between target genes of schizophrenia GWAS loci were greatest when eGenes had shared biological functions, and enriched for psychiatric risk, brain development and synapse biology genes. Convergent signatures were experimentally robust, detected in three partially overlapping lists of schizophrenia eGenes, whether manipulated in arrayed or pooled experimental designs, and regardless of whether iGLUTs shared a common donor, cell type of origin, or developmental time point. Increased convergence between eGenes with shared biological function correlated to smaller than expected (“sub-additive”) effects following combinatorial perturbations of these same eGenes. Of note, beyond transcription, combinatorial eGene manipulations resulted in phenotypic changes that differed from the summed impacts of individual eGene perturbations (SI Figs. 25–27), reinforcing that polygenic risk cannot be extrapolated from experiments that test one risk gene at a time. Finally, we report that pharmacological manipulation of a convergent hub reversed the effects of multiple eGenes, suggesting that for polygenic disorders, a preferred therapeutic approach may be to target shared downstream effects rather than individual risk loci.
Altogether, the experimental eGene perturbations approximated the magnitude and direction of predicted eGene effect associated with schizophrenia, and generally resulted in downstream gene expression changes related to synaptic biology and psychiatric disorder risk. Nonetheless, further gene set enrichment analysis using 493 inflammation and cell death gene-sets46 revealed enrichments related to cell stress and neurodegenerative diseases across many perturbations (Supplementary Data 1). This enrichment was not seemingly associated with viral burden, being present whether single, combinatorial, or multiplexed vectors were applied. If our in vitro system, defined by repeated lentiviral transduction, antibiotic selection, eGene perturbation, and single cell dissociation, stressed human neurons more than accounted for by the scramble gRNA controls, this would represent a concern of relevance to all CRISPR experiments in human neurons. However, neither high content imaging nor multi-electrode array analyses indicated decreased cell survival or a cessation of neuronal activity (SI Figs. 26 and 27). Moreover, inflammation89 and oxidative stress90, and particularly fetal exposures to inflammation, stress, and hypoxia91,92 are indeed associated with schizophrenia risk.
Mapping GWAS associations to eGene targets is challenging and can yield false positives. How well our three eQTL-based methods prioritized causal eGenes remains a critical question, particularly in that they rely on tissue-specific eQTL data. There are frequent hotspots of multiple TWAS-associated genes in the same locus71, with co-regulation known to underlie pleiotropic TWAS associations93. Here, three eGenes were linked to a single SNP (rs3814883) in schizophrenia-associated copy number variant at 16p11.2, a locus that harbors the greatest excess of psychiatric common polygenic influences94. We posit that a causal GWAS SNP may co-regulate multiple adjacent and distal genes at this locus through chromatin contacts, but it is possible that one or more eGenes at this locus were misidentified. Other schizophrenia GWAS SNPs (e.g., rs2027349) likewise alter expression of multiple genes (VPS45, IncRNA AC244033.2 and a distal gene, C1orf54); indeed, combinatorial perturbation of these eGenes results in non-additive impacts on transcriptomic and cellular phenotypes28.
Given the extent of polygenicity associated with schizophrenia, our conclusions are constrained by the small proportion of eGenes tested here relative to the total number of eGenes impacted by schizophrenia GWAS loci. Technical limitations in testing a larger set of SCZ eGenes include the number of GWAS loci with accurately mapped gene targets; prediction and validation of gRNAs that reliably achieve physiologically relevant gene perturbations across donors and cell types; and the sequencing costs necessary to achieve sufficient gRNA representation to resolve perturbations at scale. Moreover, given that we selected only those schizophrenia eGenes with the very strongest evidence of genetically regulated gene expression, the generalizability of our observations to all schizophrenia eGenes is unclear, particularly if there are non-linear responses to gradual changes in gene dosage95. Thus, future investigation to test across larger gene sets, graded changes in expression95, in vivo brain regions9 and in vitro cell types96, developmental timespans97, drug/environmental contexts98 and donor backgrounds99 will inform the cell-type-specific and context-dependent nature of convergence and non-additivity. Of course, all of this must be considered within the caveat that in vitro perturbations do not exactly recapitulate the physiological impact of possessing multiple genetic variants in human cases and controls. Despite this, it is worth noting that the limited number of perturbations used in our combinatorial conditions is still broadly relevant to studies of common variant interactions. When analyzing the full dataset of 105 S-PrediXcan SCZ eGenes in the post-mortem adult DLPFC6, a median of ten and a maximum of 37 eGenes had outlying expression in the direction of risk association per individual (SI Fig. 17). Across the twenty-one SCZ eGenes targeted in either the pooled or arrayed experiments, a median of two and a maximum of eight eGenes had outlying expression in the direction of risk association per individual (SI Fig. 18). Of course, the present design also falls short of capturing nuances of pleiotropy, incomplete penetrance, and environmental factors.
Whereas population genetics finds very little evidence of non-additive effects in phenotypic variation, molecular biology unequivocally demonstrates the occurrence of gene-gene interactions100. To resolve this seeming contradiction, recall that although the “liability threshold model” assumes that disease risk reflects the total sum of many additive genetic (and/or environmental) effects, the relationship between predisposition and clinical outcome is necessarily binary101. Indeed, the cumulative effect of risk SNPs can exceed observed phenotypic variation. Thus, epistasis at the gene level is consistent with the additivity of complex traits102. Likewise, here we report that convergent perturbations at the pathway level correlated with predominantly sub-additive effects. Our findings indicate that the cumulative effect of gene perturbations is additive only until a downstream pathway is maximally perturbed, after which, additional perturbations yield reduced marginal effects. Our results further suggest that a pathway can be manipulated to the point of saturation effect with only a handful of genes. We posit that there may be many combinations of variants that have the same impact on a pathway. This is supported by emerging evidence of pathway polygenic risk score (PRS) burden103, whereby pathway PRS provided higher prediction power of [disease] than overall genome-wide risk, even in cases with low overall genetic risk104.
We further tested the extent to which our in vitro studies of CRISPR perturbations inform the polygenic architecture of schizophrenia at the population level. First, transcriptomic imputation of brain eGene expression (see Methods,) revealed a dose-dependent effect: schizophrenia case-control status (p < 0.01774, OR > 1.10) was best predicted when three or more eGenes were perturbed (OR3 eGenes = 1.47 vs. OR1 eGene = 1.10) (SI Fig. 1A). Second, transcriptomic risk scores (see Methods) indicated that schizophrenia risk was better predicted from larger (p < 2.2 × 10−16) (SI Fig. 1C) or more biologically diverse (R = 0.19, p < 2.2 × 10−16) (SI Fig. 1D) gene groups. Of note, there was a lack of individuals, either case or control, with strong imputed within-function perturbations, perhaps explaining why population-level schizophrenia risk increased with the number of genes and pathways impacted. Pathway-specific polygenic risk scores (PRSets105) that incorporate biological pathways, co-expression patterns, convergence, and/or non-additivity may improve patient stratification or better predict drug response; consistent with this, non-additive PRSets performed as well as those curated from synaptic genes (SI Fig. 24). Altogether, these studies of transcriptomic imputation and polygenic risk scores suggest that our in vitro studies of CRISPR perturbations indeed inform the polygenic architecture of schizophrenia in vivo.
How does our genetic analysis of convergence advance precision medicine for patients with psychiatric disorders? First, it may inform molecular subtypes of disease. For example, when we cluster individuals based on shared patterns of schizophrenia eGene up-regulation in the post-mortem DLPFC (SI Table 2), diagnosis of included individuals distinguished clusters (Pearson’s Chi-squared; X2 = 140, df = 21, p value = 9.51e-20) (e.g., cluster 8, SCZ, X2 = 3.286; cluster 12, affective disorders (AFF), X2 = 5.57; cluster 16, control, X2 = 3.014) (SI Figs. 31 and 32). A diagnosis of affective disorder (AFF) was significantly associated with up-regulation of FES, NAGA, CALN1, CLCN3, SF3B1 and ZNF804A (cluster 12). Convergence analysis across these six eGenes in iGLUTs identified the central node gene ABCG2, which is a biomarker associated with increased negative symptoms106, down-regulated in a neuroimmune molecular subtype (SCZ Type II)107,108, and associated with SCZ treatment resistance109. Second, points of convergence represent novel therapeutic targets that might be shared across cases; reversing the effects of even a small number of genomic variants could make a substantial difference to an individual’s risk of developing schizophrenia110. We predicted drugs capable of reversing convergent transcriptomic signatures and demonstrated that pharmacological targeting of convergent hubs ameliorated the effects of multiple schizophrenia eGene perturbations. We highlight statins, particularly simvastatin, which crosses the blood-brain barrier and shows promise as an add-on treatment in schizophrenia111. Two double-blind placebo-controlled trials of simvastatin highlighted the possibility that simvastatin may decrease negative symptoms in some patients112,113, potentially predictable based on inflammatory profiles114 and treatment-induced changes in insulin receptor levels115. Targeted shared convergent hubs potentially obviate the need to individually reverse the effects of multiple distinct risk loci in each patient.
That convergent genes were associated with a range of brain disorders indicated that convergent effects may partially explain shared features of psychiatric disorders and pleiotropy of risk. Consistent with this, common and rare risk variants for schizophrenia2,7,17,38,39,40,41,116,117,118, autism spectrum disorder119,120,121 and more broadly across the neuropsychiatric disorder spectrum30,122,123,124 are all highly enriched for genes involved in synaptic biology and gene regulation. Our findings support the hypothesis that common and rare psychiatric risk variants converge on the same biological pathways27. As recently demonstrated for autism125, by combining genetic and clinical data, it may be possible to resolve biologically distinct subtypes of schizophrenia. Our overarching goal is to advance the field towards an era of precision medicine126, whereby patient genetics, in conjunction with clinical evaluation, are used to more accurately predict diagnosis, disorder trajectory, and potential therapeutic interventions.
Methods
Statement of ethics
Yale University Institutional Review Board waived ethical approval for this work. Ethical approval was not required because the hiPSC lines, lacking association with any identifying information and widely accessible from a public repository, are thus not considered to be human subjects research. Post-mortem data are similarly lacking identifiable information and are not considered human subjects research.
Schizophrenia eGene prioritization
eGenes are defined as genes with significant genetic regulation of gene expression levels. In total, across the pooled and arrayed analyses, 20 unique eGenes were prioritized based on statistical and epigenetic evidence supporting genetic (dys)regulation of expression in schizophrenia (see Table 1), rather than GWAS or eQTL effect size; predicted direction and magnitude of eGene effect available in SI Table 1.
i) SCZ1 eGenes: EpiXcan68 was used to impute brain transcriptomes from Psychiatric Genomics Consortium 3 (PGC3)-SCZ GWAS2 at the level of genes and isoforms from the PsychENCODE post-mortem datasets of genotyped individuals (brain homogenate, n = 924)42,69; EpiXcan increases power to identify trait-associated genes under a causality model by integrating epigenetic annotation127 (from REMC128); transcriptomes were imputed at the gene and isoform levels and features with training cross-validation R2 ≥ 0.01 were retained. The epigenetic imputation models were built with the PrediXcan72 method (using a 50kbp window instead of 1Mbp for transcripts), utilizing the recently described ChIPseq datasets21; summary-level imputation was performed with S-PrediXcan70. Peaks were assigned to genes with the ChIPseeker R package129. In addition, PrediXcan72 imputed H3K27ac (brain homogenate, n = 122; neuronal, n = 191) and H3K4me3 (neuronal, n = 163)21 to more confidently identify cis-regulatory elements associated with risk for SCZ. Overall, SCZ eGenes were prioritized from GWAS based on: i) significant genetic up-regulation of expression (z-score >6 for genes), ii) epigenetic support (imputed epigenetic activity (p < 0.01) across at least one of the three assays), iii) exclusion of non-coding genes or those located in the MHC locus, iv) robust expression in our hiPSC neuron RNAseq. Genes were ranked based on the association z-score for imputed gene expression. For pooled experiments (day 7 hiPSC-derived iGLUT), six top coding genes and one top pseudo-gene were selected: NEK4, PLCL1, UBE2Q2L, NAGA, FES, CALN1, and ZNF804 (Table 1).
ii) SCZ2 eGenes: First, transcriptomic imputation (prediXcan70,71,72) identified ~250 significant genes (p < 6 × 10−6) with predicted differential expression between SCZ-cases and controls using SCZ GWAS2 and post-mortem CommonMind Consortium (CMC)6 data (623 samples). Second, colocalization (COLOC73,74) of fine-mapped PGC3-GWAS2 loci (65,205 cases and 87,919 controls) with post-mortem brain6 eQTL (537 EUR samples)6 identified 25 loci with very strong evidence (high posterior probability that a single shared variant is responsible for both signals, PP4 > 0.873). There was significant overlap between the two analyses (binomial test p value 3.03 × 10−112); of the 25 COLOC genes, 22 were also significant by PrediXcan. For each eGene, the magnitude and direction of perturbation associated with SCZ risk were predicted, and expression confirmed in hiPSC neuron RNAseq27. eGenes were further separated into discrete functional categories based on gene ontology annotations (http://geneontology.org/). From these 22, we prioritized the top coding genes across three broad categories: synaptic, regulatory, and multifunctional (defined as not synaptic, regulatory, and seemingly unrelated to each other). To complete the selection of five genes from each category, three additional top-ranked synaptic genes from the prediXcan analysis were included: DOC2A74, CLCN374 and PLCL1124. Overall, 15 SCZ eGenes were prioritized from GWAS based on i) significant genetic regulation by COLOC and/or PrediXcan, ii) exclusion of non-coding genes and those located in the major histocompatibility complex (MHC) locus, iii) robust expression in our hiPSC neuron RNAseq.
For arrayed experiments (day 21 NPC-derived iGLUT), our final gene list for combinatorial perturbations included five synaptic genes (SNAP91, CLCN3, PLCL1, DOC2A, SNCA), five regulatory genes (ZNF823, INO80E, SF3B1, THOC7, GATAD2A), and five genes with non-synaptic, non-regulatory functions, termed “multi-function” (CALN1, CUL9, TMEM219, PCCB, FURIN) (Table 2). For pooled experiments (day 21 NPC-derived iGLUT), the ten coding genes with significant genetic up-regulation were selected: CALN1, CLCN3, CUL9, DOC2A, PLCL1, INO8E0, SF3B1, SNAP91, TMEM219, ZNF823. This list was combined with our eGene set previously evaluated in hiPSC-neurons27; one functionally validated gRNA was included for each of these three genes (SNAP91, TSNARE1, and CLCN3)27.
gRNA design
CRISPRa gRNA design and cloning were conducted as described previously130, using the lentiGuide-Hygro-mTagBFP2 backbone (Addgene, No. 99374). For the fifteen eGenes prioritized by a combination of COLOC and PrediXcan, we designed three gRNAs each. For the seven eGenes prioritized by EpiXcan and PrediXcan, we designed ten gRNAs each. For the three previously tested eGenes27 (intended as a positive control), we used one pre-validated gRNA each. All gRNA sequences and corresponding oligonucleotide sequences used for cloning of gRNA vectors and subsequent experimentation are listed in Supplementary Data 5.
iGLUT induction from hiPSC-derived NPCs27,58,59,60 or hiPSCs65,131
Validated control hiPSCs for eGene perturbation were selected from a previously reported case/control hiPSC cohort of childhood onset schizophrenia132. Informed consent was obtained from all fibroblast donors at the National Institute of Mental Health under the review of the Internal Review Board of the NIMH. All hiPSC work was reviewed by the Internal Review Board of the Icahn School of Medicine at Mount Sinai. This work was also reviewed by the Embryonic Stem Cell Research Oversight Committee at the Icahn School of Medicine at Mount Sinai and Yale University. The following control hiPSC/NPCs were used: NSB553-S1-1 (male), NSB2607-2/NSB2607-1-4 (male), NSB690-2 (male). All fibroblast samples were genotyped by IlluminaOmni 2.5 bead chip genotyping133,134, PsychChip132, and exome sequencing132. Parental hiPSCs were validated by G-banded karyotyping (Wicell Cytogenetics), with ongoing genome stability monitored by Infinium Global Screening Array v3.0 (Illumina). Critically, SNP genotype is inferred from all RNAseq data using the Sequenom SURESelect Clinical Research Exome (CRE) and Sure Select V5 SNP lists to confirm that neuron identity matches the donor.
i) Validated control hiPSC-derived NPCs for CRISPRa/shRNA were selected from a previously reported case/control hiPSC cohort of childhood onset SCZ (COS)132: NSB553-S1-1 (male, average SCZ PRS, European ancestry), NSB2607-1-4 (male, average SCZ PRS, European ancestry). hiPSC-NPCs were generated via dual-SMAD inhibition (0.1 µM LDN193189 and 10 µM SB542431) followed by neural rosette selection and MACS-based purification and validated as previously described)132. hiPSC-NPCs were subsequently transduced with lentiviral vectors for dCas9-VPR-puro (Addgene, No. 99373) and selected with 1 mg/mL puromycin (Sigma, no. P7255) as described previously132. hiPSC-NPCs expressing dCas9-VPR were cultured in hNPC media (DMEM/F12 (Life Technologies #10565), 1x N2 (Life Technologies #17502-048), 1x B27-RA (Life Technologies #12587-010), 1x Antibiotic-Antimycotic, 20 ng/ml FGF2 (Life Technologies)) on Matrigel (Corning, #354230).
At day −2, dCas9-VPR hiPSC-NPCs were seeded as 1.2 × 106 cells/well in a 12-well plate coated with Matrigel. At day −1, cells were transduced with rtTA (Addgene 20342) and NGN2 (Addgene 99378) lentiviruses. Medium was switched to non-viral medium four hours post-infection. At day 0 (D0), 1 µg/ml dox was added to induce NGN2-expression. At D1, transduced hiPSC-NPCs were treated with antibiotics to select for lentiviral integration (300 ng/ml puromycin for dCas9-effectors-Puro, 1 mg/ml G-418 for NGN2-Neo). At D3, NPC medium was switched to neuronal medium (Brainphys (Stemcell Technologies, #05790), 1x N2 (Life Technologies #17502-048), 1x B27-RA (Life Technologies #12587-010), 1 µg/ml Natural Mouse Laminin (Life Technologies), 20 ng/ml BDNF (Peprotech #450-02), 20 ng/ml GDNF (Peprotech #450-10), 500 µg/ml Dibutyryl cyclic-AMP (Sigma #D0627), 200 nM L-ascorbic acid (Sigma #A0278)) including 1 µg/ml Dox. 50% of the medium was replaced with fresh neuronal medium once every second day.
For pooled analysis, on day 5, young hiPSC-NPC NGN2-neurons were replated onto matrigel-coated plates and cells were dissociated with Accutase (Innovative Cell Technologies) for 5–10 min, washed with DMEM/10%FBS, gently resuspended, counted and centrifuged at 1000 × g for 5 min. The pellet was resuspended at a concentration of 1 × 106 cells/mL in neuron media [Brainphys (StemCell Technologies #05790), 1 N2 (ThermoFisher #17502-048), 1 B27-RA (ThermoFisher #12587-010), 1 mg/ml Natural Mouse Laminin (ThermoFisher #23017015), 20 ng/mL BDNF (Peprotech #450-02), 20 ng/mL GDNF (Peptrotech #450-10), 500 mg/mL Dibutyryl cyclic-AMP (Sigma #D0627), 200 nM L-ascorbic acid (Sigma #A0278)] with doxycycline, puromycin, G418 [4 µM Ara-C (Sigma #C6645)] and 1 Thiazovivin (Sigma #420220). Cells were seeded 5 × 105 per 12-well plate. For arrayed analysis, neurons were not replated, owing to the complexity of the conditions involved.
At D13, iGLUTs were treated with 200 nM Ara-C to reduce the proliferation of non-neuronal cells in the culture, followed by half medium changes. At D18, Ara-C was completely withdrawn by full medium change while adding media containing individual shRNA/gRNA vectors or pools of mixed shRNA and gRNA vectors (Addgene 99374), either targeting eGenes or scramble controls. CRISPRa and shRNA vectors were specifically selected for perturbation due to the potential for simultaneous, bi-directional perturbation of target eGenes in joint perturbation conditions. shRNA knockdown was chosen over CRISPRi due to the difficulties in expressing multiple separate CRISPR effectors in the same cell lines (e.g., dCas9-VPR + dCas12a-KRAB). Control conditions were as follows: scramble gRNA vector (for comparing with target gRNA conditions), scramble shRNA vector (for comparing with target shRNA conditions) and scramble gRNA vector + scramble shRNA vector (for comparing with joint perturbation conditions). All control conditions were MOI-matched to their respective target condition. Medium was switched to non-viral medium four hours post-infection. At D19, transduced iGLUTs were treated with corresponding antibiotics to the gRNA lentiviruses (1 mg/ml HygroB for lentiguide-Hygro/lentiguide-Hygro-mTagBFP2), followed by half medium changes until neurons were harvested at D21.
ii) Clonal hiPSCs from two control donors of European ancestry (NSB690-2 (male, average SCZ PRS, European ancestry) and NSB2607-2 (male, average SCZ PRS, European ancestry)132 with lenti-EF1a-dCas9-VPR-Puro (Addgene #99373), pLV-TetO-hNGN2-eGFP-Neo (Addgene #99378), and lentiviral FUW-M2rtTA (Addgene #20342) were maintained in StemFlex™ Medium (ThermoFisher #A3349401) and passaged with EDTA (Life Technologies #15575-020). On day 1, induction media (DMEM/F12 (ThermoFisher #10565), 1 N2 (ThermoFisher #17502-048), 1 B27-RA (ThermoFisher #12587-010), 1 Antibiotic-Antimycotic (ThermoFisher #15240096), and 1 µg/mL doxycycline) was prepared and dispensed 2 mL of suspension at 1.2 × 106 cells/well in induction media onto a 6-well plate coated with Matrigel (Corning #354230). On day 3, media is replaced with induction medium containing 1 μg/mL puromycin and 1 mg/mLG418. On day 5, split neurons were replated onto matrigel-coated plates and cells were dissociated with Accutase (Innovative Cell Technologies) for 5–10 min, washed with DMEM/10%FBS, gently resuspended, counted and centrifuged at 1000 × g for 5 min. The pellet was resuspended at a concentration of 1 × 106 cells/mL in neuron media [Brainphys (StemCell Technologies #05790), 1 N2 (ThermoFisher #17502-048), 1 B27-RA (ThermoFisher #12587-010), 1 mg/ml Natural Mouse Laminin (ThermoFisher #23017015), 20 ng/mL BDNF (Peprotech #450-02), 20 ng/mL GDNF (Peptrotech #450-10), 500 mg/mL Dibutyryl cyclic-AMP (Sigma #D0627), 200 nM L-ascorbic acid (Sigma #A0278)] with doxycycline, puromycin, G418 [4 µM Ara-C (Sigma #C6645)] and 1 Thiazovivin (Sigma #420220). Cells were seeded 5 × 105 per 12-well plate. On day 7, neurons were harvested for scRNA sequencing.
Neuronal pooled CRISPRa screens
Expanded CRISPR-compatible CITE-seq (ECCITE-seq)75, combines Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq) and Cell Hashing for multiplexing and doublet detection135 with direct detection of sgRNAs to enable single cell CRISPR screens with multi-modal single cell readout. By capturing pol III-expressed guide RNAs directly, this approach overcomes limitations of other single-cell CRISPR methods, which detect guide sequences by a proxy transcript, resulting in barcode switching and lower capture rates136,137,138. CRISPRa hiPSC iGLUT neurons (2607 (male) and 690 (male)) were transduced with the pooled gRNA at day −1. After maturation, 7-day-old iGLUT neurons were dissociated into single-cell suspensions with papain, antibody-hashed135, and bar-coded single-cell cDNA generated using 10X Genomics Chromium139. NPC-derived iGLUT neurons (2607 (male) and 553 (male)) were transduced with the mixed-pooled gRNA vectors (Addgene 99374) at day 17. At day 21, media was replaced by 0.5 ml/well Accutase containing 10 μm Rock inhibitor, THX (catalog no. 420220; Millipore) for 1 hour to dissociate neurons. Neurons were spun down (3 mins × 300 g) and resuspended in DMEM/F12 + THX before proceeding to single cell sequencing. Analysis of single-cell CRISPRa screens in DIV 7 and DIV 21 iGLUT Neurons. mRNA sequencing reads were mapped to the GRCh38 reference genome using the Cellranger Software. To generate count matrices for HTO and GDO libraries, the kallisto indexing and tag extraction (kite) workflow was used. Count matrices were used as input into the R/Seurat package140 to perform downstream analyses, including QC, normalization, cell clustering, HTO/GDO demultiplexing, and DEG analysis75,141.
Normalization and downstream analysis of RNA data were performed using the Seurat R package (v.2.3.0), which enables the integrated processing of multimodal single-cell datasets. Each ECCITE-seq experiment was initially processed separately. Cells with RNA UMI feature counts were filtered (200 <nFeature_RNA < 8000), and the percentage of all the counts belonging to the mitochondrial, ribosomal, and hemoglobin genes calculated using Seurat::PercentageFeatureSet. Hashtag and guide-tag raw counts were normalized using centered log ratio transformation, where counts were divided by the geometric mean of the corresponding tag across cells and log-transformed. For demultiplexing based on hashtag, Seurat::HTODemux function was used; and for guide-tag counts Seurat::MULTIseqDemux function within the Seurat package was performed with additional MULTIseq semi-supervised negative-cell reclassification. In both experiments, 8-10% of retained cells contained multiple gRNAs and were assigned as doublets after de-multiplexing. To remove variation related to the cell-cycle phase of individual cells, cell cycle scores were assigned using Seurat::CellCycleScoring, which uses a list of cell cycle markers142 to segregate by markers of G2/M phase and markers of S phase. RNA UMI count data were then normalized, log-transformed, and the percent mitochondrial, hemoglobulin, and ribosomal genes, batch, donor (HTO-maxID; as a biological replicate), and cell cycle scores (Phase) were regressed out using Seurat::SCTransform. The scaled residuals of this model represent a ‘corrected’ expression matrix, that was used for all downstream analyses. To ensure that cells assigned to a guide-tag identity class demonstrated successful perturbation of the target gene, we performed ‘weighted-nearest neighbor’ (WNN) analysis, to assign clusters based on both guide-tag identity class and gene expression76. To identify successfully perturbed cells, we calculated a p value based on the Wilcox rank sum test and Area Under the Curve (AUC) statistic, which reflects the power of each gene (or gRNA) to serve as a marker of a given cluster using Presto. WNN Clusters were then filtered based on two criteria (1) single gRNA-identity with an AUC statistic of ≥ 0.8 (where 1 means the gRNA is a perfect marker of a given cluster) and (2) a logFC ≥ 2 standard deviations of the mean or logFC > 0 and p-val > 0.05, of the target gene (but no other target genes) compared to scramble (non-targeting sgRNAs) controls (SI Fig. 4−8). These clusters were then used for downstream analyses143.
Of note, there was a lower representation of specific gRNAs and fewer gene perturbations resolved in SCZ2 than SCZ1. This likely reflected the use of a single pre-validated gRNA vector per gene from our arrayed experiments for SCZ2, rather than a pooled library comprised of multiple gRNAs targeting each eGene in SCZ1.
Cell fraction imputation and quantification of heterogeneity in the composition of iGLUT neurons
Using CiberSortx, we imputed the cell-faction identity of randomly sampled scramble control cells from each experiment (n = 100/exp) using the PsychEncode scRNAseq dataset as a reference (100 permutations). To determine if the level of heterogeneity of iGLUT neuron maturity and subtype was similar between DIV7 and DIV21 iGLUT neurons in the given experiments, we performed a non-parametric Levene’s Test for Homogeneity of Variance (LT-test) on the imputed cell fraction matrices. Although we observed heterogeneity in relative central and peripheral nervous system marker expression across the cell fractions, this heterogeneity was not due to gRNA identity, and the level of variance in our data due to cellular heterogeneity was not significantly different by time-point. We were underpowered to compare gRNAs between cells with higher expression of different cell markers.
Meta-analysis of gene expression across perturbations144
We performed a meta-analysis and Cochran’s heterogeneity Q-test (METAL77) using the p values and direction of effects (t-statistic), weighted according to sample size across all sets of perturbations in both the arrayed and pooled assays (Target vs. Scramble DEGs). Genes were defined as “convergent” if they (1) had the same direction of effect across all 5, 10, or 15 target combinations, (2) were Bonferroni significant in our meta-analysis (Bonferroni adjusted p value ≤ 0.05), and (3) had a heterogeneity p value = >0.05.
Bayesian Bi-clustering to identify target-convergent networks144
eGene-Convergent gene co-expression Networks (eGCN)81 were built using an unsupervised Bayesian biclustering model, BicMix145, on the log2CPM expression data from all the replicates across each of the 5-target sets and scramble gRNA jointly or all the cells across 10 targets and scramble gRNA jointly for the arrayed and pooled assays, respectively. To account for neuronal maturity differences in the single-cell screen, expression matrices were batch corrected and normalized, and the scramble cells from both experiments (matched scramble gRNA across experiments) were used as a single control population. To perform this as a joint analysis across two experiments, (1) Count matrices from each experiment were combined and RNA transcripts, mitochondrial, ribosomal, and hemoglobin genes were removed ([‘^MT- | ^RP[SL][[:digit:]]|^RPLP[[:digit:]]|^RPSA | ^HB[ABDEGMQZ][[:digit:]]’) as well as genes that had at fewer than 2 read counts in 90% of samples, (2) and limma:voom normalization and transformation was used to compute the log2cpm counts from the effective library sizes of each cell (16851 genes). 40 runs of BicMix were performed on these data, and the output from iteration 400 of the variational Expectation-Maximization algorithm was used. The hyperparameters for BicMix were set based on previous extensive simulation studies146. Convergent networks were identified across all possible combinations of 2–14 as well as all 15 of the targets (n = 32752 combinations) in the arrayed assay, and all possible combinations of 2,3,4,5,6,7 or 8 as well as all 10 of the targets (n = 1003 combinations) in the pooled experiment. Network connections that did not replicate in more than 10% of the runs were excluded. Nodes with fewer than 5 edges or non-coding genes were removed from gene set enrichment analysis (GSEA). (The threshold of >5 edges is based on the likelihood of more than 5 edges being present by chance, with 10% being the percentage of runs where the connection was identified, see refs. 81,145. Duplication thresholds are network-dependent, and a metric of confidence in the connections, including those with especially low duplication rates, was not included in downstream analysis.) Of all random sets tested in the pooled experiment, 64.8% resolved a convergent network passing at least a 10% duplication threshold; of all random sets tested in the arrayed experiment, ~50% resolved a convergent network with a 5–255 threshold of duplication depending on the node-edge connection. Using FUMAGWAS: GENE2FUNC, the protein-coding genes were functionally annotated, and overrepresentation gene-set analysis for each network gene set was performed147. Using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit)148, over-representation analysis (ORA) was performed on all convergent network gene sets against a curated list of common and rare variant target genes across ASD, BIP, SCZ, and ID27. Nodes were annotated using GeneCards149, MalaCards150, and GWAS Catalog151. Specific enrichments were observed with 1’ de novo SCZ-CNV152, SCZ eQTLs153, SCZ brain hub gene106, downregulated in ASD/WS154, SCZ neurons155, and Sox21 neural patterning156.
Influence of Functional Similarity on Convergence Degree
Functionally similarity scores across the eGenes represented in each set was calculated using three metrics: (1) Gene Ontology Scores: the average semantic similarity score based on Gene Ontology pathway membership (within Biological Pathway (BP), Cellular Component (CC), and Molecular Function (MF) between genes in a set80, (2) Brain expression correlation (B.E.C.) score: based on the strength of the correlation in gene expression in the CMC (n = 991 after QC) post-mortem dorsolateral prefrontal cortex (DLPFC) gene expression data6, and (3) Signaling Score: based on the proportion of eGenes whose basic functional annotation was categorized as “signaling” (CALN1, CLCN3, FES, NAGA, PLCL1, TMEM219; with PLCL1 and CLCN3 further separated as specific synaptic genes) or four “epigenetic/regulatory” target genes (SF3B1, UBE2Q2L, ZNF823, ZNF804A; with ZNF823, ZNF804A as specific transcription factors) using FUMAGWAS: GENE2FUNC147 (SI Fig. 10).
Bi-clustering identifies co-expressed genes shared across the downstream transcriptomic impacts of any given set of eGene perturbations, thus, the resolved networks are the transcriptomic similarities between distinct perturbations (convergence). While bi-clustering resolves convergent gene co-expression networks, the strength of convergence within a network can be defined by (i) the degree of network connectivity as defined by two small-world network connectivity coefficients (edge density and average path length) and (ii) the degree of functional similarity or unity between genes represented within the network. Given this definition, (1) represents perturbations with no convergent downstream effects, (2) represents a network with a moderate degree of convergence because it (i) has resolved gene co-expression clusters that can be constructed into a network, (ii) has a moderate degree of network connectivity and (ii) is enriched in biological pathways with some redundancy, while (3) represents a highly convergent network because the degree of network connectivity is stronger and there is greater uniformity in biological pathway gene membership. Overall, we quantify the strength or degree of convergence using the function in (4), where Cp is the edge density (the proportion of edges present given all possible edges) and Lp is the average path length (the mean of the shortest distance between each pair of nodes), MFsc is the average semantic similarity score between each pair of nodes in the network based on Molecular Function Gene Ontology, BPsc is the average semantic similarity score based on Biological Pathway Gene Ontology and CCsc is the average semantic similarity score based on Cellular Component Gene Ontology. Semantic similarity is based on the idea that genes with similar function have similar Gene Ontology annotations. Semantic similarity scores were calculated by aggregating four information content-based methods and one graph structure-based method with the R package GoSemSim.
We assigned each network a “degree of convergence” based on (1) network connectivity and (2) similarity of network genes based on biological pathway membership. We performed a principal components analysis on the functional similarity scores and the degree of network convergence. PCA loadings determined the effect of the included variables on the variability across all resolvable sets (arrayed=16320, pooled=827, variables=6). To quantify this, we calculated two small-world connectivity network coefficients: the cluster connectivity coefficient based on the proportion of edges present out of all possible edges (Cp) and the average path length (Lp)157.
Here, we define convergence as (1) increased connectivity of the resolved networks and (2) functional similarity of genes within the network. Network connectivity was defined by the sum of the clustering coefficient (Cp) and the difference in average length path (Lp) from the maximum average length path resolved across all possible sets [(max)Lp-Lp]. Network functional similarity was scored by taking the sum of the mean semantic similarity scores between all genes in the network. Overall, convergence degree represented the sum of the network connectivity score and the network functional similarity score (1):
Convergent networks with matched patterns of gene expression in the post-mortem brain
We clarify that this approach asks how often eGenes are up-regulated together in individual post-mortem brains. To do this, we ran target-convergent network reconstruction in our scRNA-seq data, not the CMC bulk tissue data, for sets of eGenes defined by the clustering observed in the CMC bulk tissue data. We found zero individuals in the CMC data with significant upregulation of all ten risk eGenes. Instead of only evaluating convergence on the basis of eGene functional similarity as in the first portion of the manuscript, we define eGene pairings more broadly based on the signatures of these eGenes in the post-mortem DLPFC, increasing the relevance to risk at the individual level. Target sets based on gene expression patterns in the CMC (n = 991 after QC) post-mortem dorsolateral prefrontal cortex (DLPFC). We performed K-means clustering to subset the data into clusters based on the Z-scored gene expression of the 10 target genes. Although initial silhouette analysis identified the optimal number of clusters as two, visualization by a scree plot suggested the optimal number to be between 4 and 6 clusters. Given that data clustered by case/control status (2 clusters), and sub-diagnosis of BP, SCZ, AFF, and Controls (4 clusters), to assess clustering based on 10 eGenes, we tested the impact of using 10 clusters and 20 clusters (SI Fig. 32). Perturbation identities were assigned based on average positive Z-scores of ≥0.5 within each cluster. We then assigned our single-cell data to clusters based on the overlap of perturbations and performed network reconstruction to replicate our convergent analysis using groups based on CMC post-mortem data. We retained clusters that resolved networks with at least a 10% duplication rate, calculated convergence scores, and performed GSEA using protein-coding network genes. Of the twenty clusters, networks were recovered for the combination of targets represented in cluster 4 (2 targets; 913 cells; 15% duplication; 13 node genes), cluster 5 (3 targets; 1260 cells; 15% duplication; 13 node genes), cluster 6 (6 targets; 2035 cells; 15% duplication; 34 node genes), cluster 9 (6 targets, 1822 cells, 20% duplication, 108 node genes), cluster 11 (5 targets; 1640 cells; 15% duplication; 25 node genes), cluster 12 (6 targets; 2357 cells; 20% duplication, 152 node genes), cluster 13 (5 targets; 1741 cells; 17.5% duplication, 17 node genes), cluster 18 (6 targets; 1884 cells, 15% duplication, 25 nodes), cluster 19 (6 targets, 2327 cells, 20% duplication, 153 nodes), cluster 20 (6 targets, 2015 cells, 20% duplication, 33 nodes), while low confidence convergence was resolved for cluster 1 (5 targets, 1600 cells; 7.5% duplication; 38 node genes), cluster 8 (3 targets, 1233 cells, 7.5% duplication, 38 node genes), cluster 14 (3 targets, 1020 cells, 5% duplication, 23 nodes) and 16 (4 targets, 1177 cells, 2.5% duplication, 16 nodes). To determine if convergent networks were distinct between diagnostic groups, we first performed a Pearson’s chi-squared test to determine whether there was a significant difference between the expected frequencies and the observed frequencies in the diagnosis of AFF, BIP and SCZ within the clusters and then calculated Jaccard Similarity Indices between clusters based on convergent network gene membership.
Drug prioritization based on perturbation signature reversal in LiNCs Neuronal Cell Lines
To identify drugs that could reverse the resolved convergent perturbation signature across all ten targets, and within each target individually, we used the Query tool from The Broad Institute’s Connectivity Map (Cmap) Server. Briefly, the tool computes weighted enrichment scores (WTCS) between the query set and each signature in the Cmap LINCs gene expression data (dose, time, drug, cell-line), normalizes the WRCS by dividing by the signed mean w/in each perturbation (NCS), and computes FDR as fraction of “null signatures” (DMSO) where the absolute NCS exceeds reference signature158. We prioritized drugs that reversed signatures specifically in neuronal cells (either neurons (NEU) or neural progenitor cells (NPCs) with NCS ≤ −1.00) and filtered for (i) drugs that cross the blood-brain barriers, (ii) drugs that target genes expressed in iGLUT neurons based on bulk RNA-sequencing data from our lab and (ii) drugs that are currently launched or in clinical trial according to the cMAP Drug Repurposing database and without evidence of neurotoxicity (Box 2).
CRISPRa/shRNA validation27
At day −2, dCas9-VPR hiPSC-NPCs were seeded as 0.6 × 106 cells/well in a 24-well plate coated with Matrigel. At day −1, cells were transduced with rtTA (Addgene 20342) and NGN2 (Addgene 99378) lentiviruses. Medium was switched to non-viral medium four hours post infection. At D0, 1 µg/ml dox was added to induce NGN2-expression. At D1, transduced hiPSC-NPCs were treated with the corresponding antibiotics to the lentiviruses (1 mg/ml G-418 for NGN2-Neo) in order to increase the purity of transduced hiPSC-NPCs. At D3, NPC medium was switched to neuronal medium (Brainphys (Stemcell Technologies, #05790), 1× N2 (Life Technologies #17502-048), 1× B27-RA (Life Technologies #12587-010), 1 µg/ml Natural Mouse Laminin (Life Technologies), 20 ng/ml BDNF (Peprotech #450-02), 20 ng/ml GDNF (Peptrotech #450-10), 500 µg/ml Dibutyryl cyclic-AMP (Sigma #D0627), 200 nM L-ascorbic acid (Sigma #A0278)) including 1 µg/ml Dox. 50% of the medium was replaced with fresh neuronal medium once every second day. At D4, individual shRNA/gRNA vectors (Addgene 99374) were used, either targeting eGenes or scramble controls. 3–5 vectors were tested per eGene. Medium was switched to non-viral medium four hours post-infection. At D5, transduced iGLUTs were treated with corresponding antibiotics to the gRNA lentiviruses (1 mg/ml HygroB for lentiguide-Hygro/lentiguide-Hygro-mTagBFP2) before harvesting at D7 in order to assess eGene perturbation efficacy via qPCR.
Real time-quantitative PCR
Real-time qPCR was performed as previously described130. Specifically, cell cultures were harvested with Trizol, and total RNA extraction was carried out following the manufacturer’s instructions. Quantitative transcript analysis was performed using a QuantStudio 7 Flex Real-Time PCR System with the Power SYBR Green RNA-to-Ct Real-Time qPCR Kit (all Thermo Fisher Scientific). Total RNA template (25 ng per reaction) was added to the PCR mix, including primers listed below. qPCR conditions were as follows; 48 °C for 15 min, 95 °C for 10 min followed by 45 cycles (95 °C for 15 s, 60 °C for 60 s). All qPCR data is collected from at least three independent biological replicates of one experiment. A one-way ANOVA with post hoc Dunnett’s multiple comparisons test was performed on data for the set of targeting vectors for each eGene relative to the scramble control vector. Data analyses were performed using GraphPad PRISM 6 software. For a list of primer sequences used for real-time qPCR, see SI Table 5.
Immunostaining and high-content imaging microscopy, neurite analysis
Immature iGLUTs were seeded as 1.5 × 104 cells/well in a 96-well plate coated with 4x Matrigel at day 3. iGLUTs were plated in media containing individual shRNA/gRNA vectors or pools of mixed shRNA and gRNA vectors (Addgene 99374), either targeting eGenes or scramble controls. Medium was switched to non-viral medium four hours post-infection. At day 4, transduced iGLUTs were treated with corresponding antibiotics to the gRNA lentiviruses (1 mg/ml HygroB for lentiguide-Hygro/lentiguide-Hygro-mTagBFP2) followed by half medium changes until the neurons were fixed at day 7. At day 7, cultures were fixed using 4% formaldehyde/sucrose in PBS with Ca2+ and Mg2+ for 10 minutes at room temperature (RT). Fixed cultures were washed twice in PBS and permeabilized and blocked using 0.1% Triton/2% Normal Donkey Serum (NDS) in PBS for two hours. Cultures were then incubated with primary antibody solution (1:1000 MAP2 anti-chicken (Abcam, ab5392) in PBS with 2% NDS) overnight at 4 °C. Cultures were then washed 3x with PBS and incubated with secondary antibody solution (1:500 donkey anti chicken Alexa 647 (Life Technologies, A10042) in PBS with 2% NDS) for 1 hour at RT. Cultures were washed a further 3x with PBS, with the second wash containing 1 μg/ml DAPI. Fixed cultures were then imaged on a CellInsight CX7 HCS Platform with a 20× objective (0.4 NA), and neurite tracing analysis was performed using the neurite tracing module in the Thermo Scientific HCS Studio 4.0 Cell Analysis Software. 12–24 wells were imaged per condition across a minimum of 2 independent cell lines, with 9 images acquired per well for neurite tracing analysis; each N therefore represents an average of hundreds of neurons per image. A one-way ANOVA with a post hoc Bonferroni multiple comparisons test was performed on data for neurite length per neuron using Graphpad Prism.
Immunostaining and high-content imaging microscopy, synapse analyses
Commercially available primary human astrocytes (pHAs, Sciencell, #1800; isolated from fetal female brain) were seeded on D3 at 0.85 × 104 cells per well on a 4x Matrigel-coated 96 W plate in neuronal media supplemented with 2% fetal bovine serum (FBS). iGLUTs were seeded as 1.5 × 105 cells/well in a 96-well plate coated with 4x Matrigel at day 5. Half changes of neuronal media were performed twice a week until fixation. At day 13, iGLUTs were treated with 200 nM Ara-C to reduce the proliferation of non-neuronal cells in the culture. At day 18, Ara-C was completely withdrawn by full medium change while adding media containing individual shRNA/gRNA vectors or pools of mixed shRNA and gRNA vectors (Addgene 99374), either targeting eGenes or scramble controls. Medium was switched to non-viral medium four hours post-infection. At day 19, transduced iGLUTs were treated with corresponding antibiotics to the gRNA lentiviruses (1 mg/ml HygroB for lentiguide-Hygro/lentiguide-Hygro-mTagBFP2) followed by half medium changes until the neurons were fixed at day 21. At day 21, cultures were fixed and immunostained as described previously, with an additional antibody stain for Synapsin1 (primary antibody: 1:500 Synapsin1 anti-mouse (Synaptic Systems, 106 011); secondary antibody: donkey anti-mouse Alexa 568 (Life Technologies A10037)). Stained cultures were imaged and analyzed as above using the synaptogenesis module in the Thermo Scientific HCS Studio 4.0 Cell Analysis Software to determine SYN1+ puncta number, area, and intensity per neurite length in each image. 20 wells were imaged per condition across a minimum of 2 independent cell lines, with 9 images acquired per well for synaptic puncta analysis. A one-way ANOVA with a post hoc Bonferroni multiple comparisons test was performed on data for puncta number per neurite length using Graphpad Prism. For a list of antibodies used for immunostaining, see SI Table 6.
Multiple Electrode array (MEA)
Commercially available primary human astrocytes (pHAs, Sciencell, #1800; isolated from fetal female brain) were seeded on D3 at 1.7 × 104 cells per well on a 4x Matrigel-coated 48 W MEA plate (catalog no. M768-tMEA-48W; Axion Biosystems) in neuronal media supplemented with 2% fetal bovine serum (FBS). At D5, iGLUTs were detached, spun down, and seeded on the pHA cultures at 1.5 × 105 cells per well. Half changes of neuronal media supplemented with 2% FBS were performed twice a week until day 42. At day 13, co-cultures were treated with 200 nM Ara-C to reduce the proliferation of non-neuronal cells in the culture. At Day 18, Ara-C was completely withdrawn by full medium change. At day 25, a full media change was performed to add media containing individual shRNA/gRNA vectors or pools of mixed shRNA and gRNA vectors (Addgene 99374), either targeting eGenes or scramble controls. Medium was switched to non-viral medium four hours post-infection. If drug treatments were included, D26 neurons were treated for 48 hours with either Anandamide (10 µM), Etomoxir (10 µM), Simvastatin (10 µM), or matched vehicles. Electrical activity of iGLUTs was recorded at 37 °C twice every week from day 28 to day 42 using the Axion Maestro MEA reader (Axion Biosystems). Recording was performed via AxiS 2.4. The batch mode/statistic compiler tool was run following the final recording. Quantitative analysis of the recording was exported as a Microsoft Excel sheet. Data from 6 to 12 biological replicates were analyzed using GraphPad PRISM 6 software or R.
RNAseq
RNA Sequencing libraries were prepared using the Kapa Total RNA library prep kit. Paired-end sequencing reads (100 bp) were generated on a NovaSeq platform. Raw reads were aligned to hg19 using STAR aligner159 (v2.5.2a) and gene-level expression was quantified by featureCounts160 (v1.6.3) based on Ensembl GRCh37.70 annotation model. Genes with over 10 counts per million (CPM) in at least four samples were retained. After filtering, the raw read counts were normalized by the voom161 function in limma and differential expression was computed by the moderated t-test implemented in limma162. Differential gene expression analysis was performed between each CRISPRa/shRNA target group and the scramble control group. Bayes shrinkage (limma::eBayes) estimated modified t and p values and identified differentially expressed genes (DEGs) based on an FDR ≤ 0.05 (limma::TopTable)163. Gene Ontology/pathways were evaluated using Gene-set Enrichment Analysis (GSEA)164, with genes expressed in iGLUTs as our baseline comparison. In these analyses, the t test statistics from the differential expression contrast were used to rank genes in the GSEA using the R package ClusterProfiler165. Permutations (up to 100,000 times) were used to assess the GSEA enrichment P value. Log2 fold changes in expression were calculated across all RNA-seq samples in our arrayed dataset.
Analysis of additive and non-additive effects27
We applied our published approach to resolve distinct additive and non-additive transcriptomic effects after combinatorial manipulation of genetic variants and/or chemical perturbagens, developed27, applied58, and described in detail84. The expected additive effect was modeled through the addition of the individual comparisons; the non-additive effect was modeled by the subtraction of the additive effect from the combinatorial perturbation comparison. Fitting of this model for differential expression identifies genes that show a difference in the expected differential expression computed for the additive model compared to the observed combinatorial perturbation. Briefly, the non-additive effect between eGenes was identified using limma’s linear model analysis. The coefficients, standard deviations and correlation matrix were calculated, using contrasts.fit, in terms of the comparisons of interest. Empirical Bayes moderation was applied using the eBayes function to obtain more precise estimates of gene-wise variability. P values were adjusted for multiple hypothesis testing using false discovery rate (FDR) estimation, and differentially expressed genes were determined as those with FDR ≤ 10%, unless stated otherwise. Two methods were used to compare the extent of synergy between data sets. First, we calculated the fraction of synergistic genes (FDR < 10%) to measure the extent of synergy. Second, we calculated a synergy coefficient, π1, as the fraction of non-null synergistic P values, to inform the existence of a synergistic component, even if the P values themselves are not significant genome-wide.
However, interpretation of the resulting DEGs depends on several factors, such as the direction of fold change (FC) in all three models. To identify genes whose magnitude of change is larger in the combinatorial perturbation vs. the additive model, we categorized all genes by the direction of their change in both models and their log2(FC) in the non-additive model. First, log2(FC) standard errors (SE) were calculated for all samples. Genes were then grouped into ‘positive non-addition’ if their FC was larger than SE and ‘negative non-addition’ if smaller than -SE. If the corresponding additive model log2(FC) showed the same or no direction, the gene was classified as more differentially expressed in the combinatorial perturbation than predicted. GSEA was performed on a curated subset of the MAGMA collection using the limma package camera function, which tests if genes are ranked highly in comparison to other genes in terms of differential expression, while accounting for inter-gene correlation. Due to the small sample size in this study and moderate fold changes in some eGene perturbations, changes in gene expression may be small and distributed across many genes. However, powerful enrichment analyses in the limma package may be used to evaluate enrichment based on genes that are not necessarily genome-wide significant and identify sets of genes for which the distribution of t-statistics differs from expectation. Over-representation analysis (ORA) was performed when subsets of DEGs were of interest; genes of interest were ranked by –log10 (p value), and enrichment was performed against a background of all expressed genes using the WebGestaltR package.
Dataset for population-level analysis of synergy
Individuals from the Sweden-SCZ Population-Based cohort were obtained from the database of Genotypes and Phenotypes, Study Accession: phs000473.v2.p2 (NCases = 5232, NControls = 6468)166.
Pathway polygenic risk scores
Pathway-specific polygenic risk score (PRS) analyses were performed using PRSice-2 (v2.3.5) on individual genotype data for the Sweden-SCZ population-based cohort. A total of 4,834 individuals diagnosed with SCZ and 6,128 controls were included after quality control. To calculate the scores, we used a version of the summary statistics from the PGC SCZ GWAS that excludes the Sweden-SCZ data to prevent inflation of results. SNPs were annotated to genes and pathways based on GTF files obtained from ENSEMBL (GRCh37.75). To include potential gene regulatory elements, gene coordinates were extended 35 kilobases (kb) upstream and 10 kb downstream of each gene. We excluded from analyses the MHC region (chr6:25Mb-34Mb), ambiguous SNPs (A/T and G/C), and SNPs not present in both GWAS summary statistics and genotype data.
To obtain empirical competitive P values, that assess GWAS signal enrichment while accounting for pathway size, we performed the following permutation procedure: first, a background pathway containing all genic SNPs is constructed, and clumping is performed within this pathway. For each pathway with m SNPs, N = 10,000 null pathways are generated by randomly selecting m SNPs from the background pathway. The competitive P value can then be calculated as (2):
where I(.) is an indicator function, taking a value of 1 if the association P value of the observed pathway (P0) is larger than the one obtained from the nth null pathway (Pn), and 0 otherwise (see ref. 105 for additional details).
Pathway-specific polygenic risk scores (PRS) (PRSet105) were calculated from non-additive signatures from synaptic (4306 genes in PRS; R2 = 0.0431), regulatory (5249 genes in PRS; R2 = 0.0419), all fifteen eGenes (4988 genes in PRS, R2 = 0.0425), and genome-wide PRS (19,340 genes plus SNPs in regions outside gene annotations in genome-wide PRS, R2 = 0.0925). For the analyses testing whether non-additive genes from synaptic/regulatory pathways explain larger R2 than the same number of non-additive genes from random combinations (SI Fig. 24), we took 2799 random genes from the non-additive synaptic and regulatory transcriptome, which corresponds to the number of genes with non-additive effects in one of the random joint perturbations. For the GTF NULL permutation analyses, we selected n = 2799 random genes from the GTF file GRCh37.75. Pathway-specific PRS for each sample of 2799 genes was calculated using PRSet105, as described above. This procedure was repeated 1000 times.
Transcriptomic risk score (TRS) analyses
In order to test the impact of non-additive genetic effects in silico, we used transcriptomic imputation methods to calculate genetically-regulated gene expression (GREX) for individuals from the Sweden-SCZ Population-Based cohort (SI Table 3). Brain GREX was calculated using PrediXcan72 with CMC dorsolateral prefrontal cortex (CMC-DLPFC) models6. Predicted GREX levels were calculated for the fifteen eGenes. An initial test of aberrant gene expression was performed by counting the number of genes with dysregulated GREX (defined as predicted GREX in the top or bottom decile of overall expression of that gene, defined in the direction of effect of that gene’s association with SCZ from S-PrediXcan analyses (top decile for positive effect, bottom decile for negative effect) for each of the five-gene groups (synaptic, regulatory, multi-function), and summed the number of aberrant genes present in each individual for each perturbed gene group (Synaptic, Regulatory, and Multi-function). We then looked at the SCZ case/control proportion within each group of individuals with 3 + , 1–2, and any genes with aberrant GREX.
Association of synaptic, regulatory, and multi-function gene-sets with SCZ
We tested for association of each of the fifteen eGene GREX individually with SCZ (SCZ ~ GREX), and then calculated composite scores of group GREX (Synaptic, Regulatory, and Multi-function) using a Transcriptomic Risk Score (TRS), calculated as the sum of each GREX weighted by the direction of gene perturbation (1 for activation, −1 for inhibition) from in vivo experiments, divided by the total number of genes (N) in the gene-set (3):
We then tested for the association of each TRS (Synaptic, Regulatory, and Multi-function) with SCZ status in the Swedish cohort.
Permutation tests
We performed permutation tests to assess the impact of (1) the number of genes included in our TRS gene group and (2) the number of pathways impacted by those genes on SCZ case status. We used S-PrediXcan to find genes with CMC-DLPFC GREX associated with SCZ in a large SCZ cohort (NCases = 11,260, NControls = 24,542)39. From this resulting list of genes, we assigned genes to two groups: nominally-significant genes (N = 1963, Bonferroni p < 0.05), and tissue-specific significant genes (N = 144, p < 0.05/NGenes in CMC-DLPFC PrediXcan model). We created pathway sets affected by these genes using the overlap with Kyoto encyclopedia of genes and genomes (KEGG)167 and gene ontology (GO)168,169. This gave us a sampling pool of 1465 genes affecting 8324 pathway sets for the nominally-significant group, and 110 genes affecting 2382 pathway sets for the tissue-specific group. We then performed permutation sampling analyses (for nominally-significant and tissue-specific significant gene-pathway set pools) where we randomly sampled sets of five, ten, or fifteen genes from the sampling pool (adjusted for the size of each pathway set), calculated TRS from the sampled gene-set, and looked at the association of TRS with SCZ. We performed sampling 100,000 times for each gene-set size. For this analysis, TRS was calculated by taking the sum of each gene in the gene-sets GREX weighted by the direction of effect of the gene association with SCZ from our S-PrediXcan analysis (1 or −1) (4):
We then looked at the overall association of the number of pathways hit by each TRS (based on the annotated lists) with SCZ variance explained (SI Fig. 1A–C). To determine if the type of pathways hit by our perturbed genes was important to SCZ risk (i.e., is it more important to hit multiple, similar pathways or more diverse pathways to increase SCZ variance explained), we additionally assessed whether the similarity in make-up of pathways affected by the TRS was associated with SCZ. To do this, we used the R GeneOverlap package to calculate the average Jaccard Index of pathways for each TRS, and looked at the association of that index with SCZ.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All source donor hiPSCs have already been deposited at the Rutgers University Cell and DNA Repository (study 160; http://www.nimhstemcells.org/). All vectors are available at https://www.addgene.org/Kristen_Brennand/. Bulk and single-cell RNA sequencing data are available at the Gene Expression Omnibus under accession code GSE200774. Processed data can be accessed through Synapse under Synapse accession code syn27819129 [https://www.synapse.org/Synapse:syn27819129/wiki/623524]. For the pooled and arrayed CRISPR analyses, all raw FASTQ Count files and corresponding processed data are available on the Gene Expression Omnibus under GEO accession code GSE200774). Average expression count matrices and metadata following quality control and normalization of ECCITE-seq data, as well as results of differential gene expression analysis and Target Network Reconstruction of Bayesian Bi-clustering are available on Synapse accession code syn27819129 [https://www.synapse.org/Synapse:syn27819129/wiki/623524. All corresponding code was uploaded to Synapse under accession code syn27819129 [https://www.synapse.org/Synapse:syn27819129/wiki/623524]). DEGs, GSEA tables, synergy sub-categories, and synergy sub-category over-representation analysis for arrayed screen RNA-seq data; individual scRNA-seq perturbation DEGs and pathway enrichments from pooled experiments; reconstructed convergent networks and convergent network enrichment results (FUMA, ClusterProfiler, ORA of common/rare/variants) from arrayed and pooled screens; and CMAP drug prioritization queries and GSEA for 10 targets and each individual perturbation signature used in CMAP query are available in Supplementary Data Files 1–4.
Code availability
Code used for the convergence and additivity analyses presented in this manuscript can be accessed through Synapse under Synapse accession code syn27819129 [https://www.synapse.org/Synapse:syn27819129/wiki/623524].
References
Singh, T. et al. Rare coding variants in ten genes confer substantial risk for schizophrenia. Nature 604, 509–516 (2022).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Roussos, P. et al. A role for noncoding variation in schizophrenia. Cell Rep. 9, 1417–1429 (2014).
Hauberg, M. E. et al. Large-scale identification of common trait and disease variants affecting gene expression. Am. J. Hum. Genet. 101, 157 (2017).
Hauberg, M. E. et al. Differential activity of transcribed enhancers in the prefrontal cortex of 537 cases with schizophrenia and controls. Mol. Psychiatry 24, 1685–1695 (2018).
Fromer, M. et al. Gene expression elucidates functional impact of polygenic risk for schizophrenia. Nat. Neurosci. 19, 1442–1453 (2016).
Jaffe, A. E. et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat. Neurosci. 21, 1117–1125 (2018).
Zeng, B. et al. Multi-ancestry eQTL meta-analysis of human brain identifies candidate causal variants for brain-related traits. Nat. Genet. 54, 161–169 (2022).
Sieberts, S. K. et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci. Data 7, 340 (2020).
Forrest, M. P. et al. Open chromatin profiling in hipsc-derived neurons prioritizes functional noncoding psychiatric risk variants and highlights neurodevelopmental loci. Cell Stem Cell 21, 305–318.e8 (2017).
Zhang, S. et al. Allele-specific open chromatin in human iPSC neurons elucidates functional disease variants. Science 369, 561–565 (2020).
Zhu, K. et al. Multi-omic profiling of the developing human cerebral cortex at the single-cell level. Sci. Adv. 9, eadg3754 (2023).
Huo, Y., Li, S., Liu, J., Li, X. & Luo, X. J. Functional genomics reveal gene regulatory mechanisms underlying schizophrenia risk. Nat. Commun. 10, 670 (2019).
Alver, M., Lykoskoufis, N., Ramisch, A., Dermitzakis, E. T. & Ongen, H. Leveraging interindividual variability of regulatory activity for refining genetic regulation of gene expression in schizophrenia. Mol. Psychiatry 27, 5177–5185 (2022).
Casella, A. M., Colantuoni, C. & Ament, S. A. Identifying enhancer properties associated with genetic risk for complex traits using regulome-wide association studies. PLoS Comput. Biol. 18, e1010430 (2022).
Dong, P., Voloudakis, G., Fullard, J. F., Hoffman, G. E. & Roussos, P. Convergence of the dysregulated regulome in schizophrenia with polygenic risk and evolutionarily constrained enhancers. Mol. Psychiatry 29, 782–792 (2024).
Rajarajan, P. et al. Neuron-specific signatures in the chromosomal connectome associated with schizophrenia risk. Science 362, eaat4311 (2018).
Song, M. et al. Mapping cis-regulatory chromatin contacts in neural cells links neuropsychiatric disorder risk variants to target genes. Nat. Genet. 51, 1252–1262 (2019).
Espeso-Gil, S. et al. A chromosomal connectome for psychiatric and metabolic risk variants in adult dopaminergic neurons. Genome Med. 12, 19 (2020).
Hu, B. et al. Neuronal and glial 3D chromatin architecture informs the cellular etiology of brain disorders. Nat. Commun. 12, 3968 (2021).
Girdhar, K. et al. Chromatin domain alterations linked to 3D genome organization in a large cohort of schizophrenia and bipolar disorder brains. Nat. Neurosci. 25, 474–483 (2022).
Girdhar, K. et al. The neuronal chromatin landscape in adult schizophrenia brains is linked to early fetal development. medRxiv (2023).
Guo, M. G. et al. Integrative analyses highlight functional regulatory variants associated with neuropsychiatric diseases. Nat. Genet. 55, 1876–1891 (2023).
Rummel, C. K. et al. Massively parallel functional dissection of schizophrenia-associated noncoding genetic variants. Cell 186, 5165–5182 e5133 (2023).
McAfee, J. C. et al. Systematic investigation of allelic regulatory activity of schizophrenia-associated common variants. Cell Genom. 3, 100404 (2023).
Yang, X. et al. Functional characterization of gene regulatory elements and neuropsychiatric disease-associated risk loci in iPSCs and iPSC-derived neurons. bioRxiv, 2023.2008.2030.555359 (2023).
Schrode, N. et al. Synergistic effects of common schizophrenia risk variants. Nat. Genet. 51, 1475–1485 (2019).
Zhang, S. et al. Multiple genes in a single GWAS risk locus synergistically mediate aberrant synaptic development and function in human neurons. Cell Genom. 3, 100399 (2023).
Gulsuner, S. et al. Spatial and temporal mapping of de novo mutations in schizophrenia to a fetal prefrontal cortical network. Cell 154, 518–529 (2013).
Schork, A. J. et al. A genome-wide association study of shared risk across psychiatric disorders implicates gene regulation during fetal neurodevelopment. Nat. Neurosci. 22, 353–361 (2019).
Liu, D. et al. Impact of schizophrenia GWAS loci converge onto distinct pathways in cortical interneurons vs glutamatergic neurons during development. Mol. Psychiatry 27, 4218–4233 (2022).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Skene, N. G. et al. Genetic identification of brain cell types underlying schizophrenia. Nat. Genet. 50, 825–833 (2018).
Radulescu, E. et al. Identification and prioritization of gene sets associated with schizophrenia risk by co-expression network analysis in human brain. Mol. Psychiatry 25, 791–804 (2018).
Li, J. et al. Spatiotemporal profile of postsynaptic interactomes integrates components of complex brain disorders. Nat. Neurosci. 20, 1150–1161 (2017).
Jia, P., Chen, X., Fanous, A. H. & Zhao, Z. Convergent roles of de novo mutations and common variants in schizophrenia in tissue-specific and spatiotemporal co-expression network. Transl. psychiatry 8, 105 (2018).
Hsu, Y. H. et al. Using brain cell-type-specific protein interactomes to interpret neurodevelopmental genetic signals in schizophrenia. iScience 26, 106701 (2023).
Bipolar Disorder and Schizophrenia Working Group of the Psychiatric Genomics Consortium Genomic dissection of bipolar disorder and schizophrenia, including 28 subphenotypes. Cell 173, 1705–1715 e1716 (2018).
Pardinas, A. F. et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat. Genet. 50, 381–389 (2018).
Hall, L. S. et al. A transcriptome-wide association study implicates specific pre- and post-synaptic abnormalities in schizophrenia. Hum. Mol. Genet. 29, 159–167 (2020).
Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 18, 199–209 (2015).
Gandal, M. J. et al. Transcriptome-wide isoform-level dysregulation in ASD, schizophrenia, and bipolar disorder. Science 362, eaat8127 (2018).
Gandal, M. J. et al. Shared molecular neuropathology across major psychiatric disorders parallels polygenic overlap. Science 359, 693–697 (2018).
Ruzicka, W. B. et al. Single-cell multi-cohort dissection of the schizophrenia transcriptome. medRxiv, 2022.2008.2031.22279406 (2022).
Cederquist, G. Y. et al. A multiplex human pluripotent stem cell platform defines molecular and functional subclasses of autism-related genes. Cell Stem Cell 27, 35–49 e36 (2020).
Jin, X. et al. In vivo Perturb-Seq reveals neuronal and glial abnormalities associated with autism risk genes. Science 370, eaaz6063 (2020).
Paulsen, B. et al. Autism genes converge on asynchronous development of shared neuron classes. Nature 602, 268–273 (2022).
Lalli, M. A., Avey, D., Dougherty, J. D., Milbrandt, J. & Mitra, R. D. High-throughput single-cell functional elucidation of neurodevelopmental disease-associated genes reveals convergent mechanisms altering neuronal differentiation. Genome Res. 30, 1317–1331 (2020).
Willsey, H. R. et al. Parallel in vivo analysis of large-effect autism genes implicates cortical neurogenesis and estrogen in risk and resilience. Neuron 109, 1409 (2021).
Weinschutz Mendes, H. et al. High-throughput functional analysis of autism genes in zebrafish identifies convergence in dopaminergic and neuroimmune pathways. Cell Rep. 42, 112243 (2023).
Meng, X. et al. Assembloid CRISPR screens reveal impact of disease genes in human neurodevelopment. Nature 622, 359–366 (2023).
Li, C. et al. Single-cell brain organoid screening identifies developmental defects in autism. Nature 621, 373–380 (2023).
Wang, B. et al. A foundational atlas of autism protein interactions reveals molecular convergence. bioRxiv, 2023.2012.2003.569805 (2023).
Pintacuda, G. et al. Protein interaction studies in human induced neurons indicate convergent biology underlying autism spectrum disorders. Cell Genomics 3, 100250 (2023).
Murtaza, N. et al. Neuron-specific protein network mapping of autism risk genes identifies shared biological mechanisms and disease-relevant pathologies. Cell Rep. 41, 111678 (2022).
Wainschtein, P. et al. Assessing the contribution of rare variants to complex trait heritability from whole-genome sequence data. Nat. Genet. 54, 263–273 (2022).
Nag, A., McCarthy, M. I. & Mahajan, A. Large-scale analyses provide no evidence for gene-gene interactions influencing type 2 diabetes risk. Diabetes 69, 2518–2522 (2020). https://doi.org/10.2337/db20-0224
Wang, M. et al. Transformative network modeling of multi-omics data reveals detailed circuits, key regulators, and potential therapeutics for Alzheimer’s disease. Neuron 109, 257–272 e214 (2021).
Flaherty, E. et al. Neuronal impact of patient-specific aberrant NRXN1alpha splicing. Nat. Genet. 51, 1679–1690 (2019).
Ho, S. M. et al. Rapid Ngn2-induction of excitatory neurons from hiPSC-derived neural progenitor cells. Methods 101, 113–124 (2016).
Pak, C. et al. Cross-platform validation of neurotransmitter release impairments in schizophrenia patient-derived NRXN1-mutant neurons. Proc. Natl. Acad. Sci. USA 118, e2025598118 (2021).
Marro, S. G. et al. Neuroligin-4 regulates excitatory synaptic transmission in human neurons. Neuron 103, 617–626 e616 (2019).
Zhang, Z. et al. The fragile X mutation impairs homeostatic plasticity in human neurons by blocking synaptic retinoic acid signaling. Sci. Transl. Med. 10, eaar4338 (2018).
Yi, F. et al. Autism-associated SHANK3 haploinsufficiency causes Ih channelopathy in human neurons. Science 352, aaf2669 (2016).
Zhang, Y. et al. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron 78, 785–798 (2013).
Meijer, M. et al. A single-cell model for synaptic transmission and plasticity in human iPSC-derived neurons. Cell Rep. 27, 2199–2211 e2196 (2019).
Sun, Y. et al. A deleterious Nav1.1 mutation selectively impairs telencephalic inhibitory neurons derived from Dravet Syndrome patients. Elife 5, e13073 (2016).
Zhang, W. et al. Integrative transcriptome imputation reveals tissue-specific and shared biological mechanisms mediating susceptibility to complex traits. Nat. Commun. 10, 3834 (2019).
Wang, D. et al. Comprehensive functional genomic resource and integrative model for the human brain. Science 362, 2314 (2018).=
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Gusev, A. et al. Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 50, 538–548 (2018).
Huckins, L. M. et al. Gene expression imputation across multiple brain regions provides insights into schizophrenia risk. Nat. Genet. 51, 659–674 (2019).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Dobbyn, A. et al. Landscape of conditional eQTL in dorsolateral prefrontal cortex and co-localization with schizophrenia GWAS. Am. J. Hum. Genet. 102, 1169–1184 (2018).
Mimitou, E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529 (2021).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Batiuk, M. Y. et al. Upper cortical layer-driven network impairment in schizophrenia. Sci. Adv. 8, eabn8367 (2022).
Teixeira, J. R., Szeto, R. A., Carvalho, V. M. A., Muotri, A. R. & Papes, F. Transcription factor 4 and its association with psychiatric disorders. Transl. psychiatry 11, 19 (2021).
Yu, G. Gene ontology semantic similarity analysis using GOSemSim. Methods Mol. Biol. 2117, 207–215 (2020).
Saha, A. et al. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 27, 1843–1858 (2017).
Namkung, H. et al. The miR-124-AMPAR pathway connects polygenic risks with behavioral changes shared between schizophrenia and bipolar disorder. Neuron 111, 220–235 e229 (2023).
Malt, E. A., Juhasz, K., Malt, U. F. & Naumann, T. A role for the transcription factor Nk2 homeobox 1 in schizophrenia: convergent evidence from animal and human studies. Front Behav. Neurosci. 10, 59 (2016).
Schrode, N., Seah, C., Deans, P. J. M., Hoffman, G. & Brennand, K. J. Analysis framework and experimental design for evaluating synergy-driving gene expression. Nat. Protoc. 16, 812–840 (2021).
Corsello, S. M. et al. The drug repurposing hub: a next-generation drug library and information resource. Nat. Med 23, 405–408 (2017).
Kurita, M. et al. HDAC2 regulates atypical antipsychotic responses through the modulation of mGlu2 promoter activity. Nat. Neurosci. 15, 1245–1254 (2012).
Lichtstein, D. et al. Na(+), K(+)-ATPase signaling and bipolar disorder. Int. J. Mol. Sci. 19, 2314 (2018).
Rees, E. et al. Targeted sequencing of 10,198 samples confirms abnormalities in neuronal activity and implicates voltage-gated sodium channels in schizophrenia pathogenesis. Biol. Psychiatry 85, 554–562 (2019).
Kim, E., Huh, J. R. & Choi, G. B. Prenatal and postnatal neuroimmune interactions in neurodevelopmental disorders. Nat. Immunol. 25, 598–606 (2024).
Hardingham, G. E. & Do, K. Q. Linking early-life NMDAR hypofunction and oxidative stress in schizophrenia pathogenesis. Nat. Rev. Neurosci. 17, 125–134 (2016).
Cannon, M., Jones, P. B. & Murray, R. M. Obstetric complications and schizophrenia: historical and meta-analytic review. Am. J. Psychiatry 159, 1080–1092 (2002).
Estes, M. L. & McAllister, A. K. Maternal immune activation: implications for neuropsychiatric disorders. Science 353, 772–777 (2016).
Wainberg, M. et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet, 51, 592–599 (2019).
Weiner, D. J. et al. Statistical and functional convergence of common and rare genetic influences on autism at chromosome 16p. Nat. Genet. 54, 1630–1639 (2022).
Domingo, J. et al. Non-linear transcriptional responses to gradual modulation of transcription factor dosage. bioRxiv, 2024.2003.2001.582837 (2024).
Bryois, J. et al. Cell-type specific cis-eQTLs in eight brain cell-types identifies novel risk genes for human brain disorders. medRxiv, 2021.2010.2009.21264604 (2021).
Walker, R. L. et al. Genetic control of expression and splicing in developing human brain informs disease mechanisms. Cell 179, 750–771 e722 (2019).
Seah, C. et al. Modeling gene x environment interactions in PTSD using human neurons reveals diagnosis-specific glucocorticoid-induced gene expression. Nat. Neurosci. 25, 1434–1445 (2022).
Dobrindt, K. et al. Publicly available hiPSC lines with extreme polygenic risk scores for modeling schizophrenia. Complex Psychiatry 6, 68–82 (2021).
Carre, C. et al. Next-Gen GWAS: full 2D epistatic interaction maps retrieve part of missing heritability and improve phenotypic prediction. Genome Biol. 25, 76 (2024).
Baselmans, B. M. L., Yengo, L., van Rheenen, W. & Wray, N. R. Risk in relatives, heritability, snp-based heritability, and genetic correlations in psychiatric disorders: a review. Biol. Psychiatry 89, 11–19 (2021).
Barton, N. H. & Keightley, P. D. Understanding quantitative genetic variation. Nat. Rev. Genet. 3, 11–21 (2002).
Choi, S. W. et al. PRSet: Pathway-based polygenic risk score analyses and software. PLoS Genet. 19, e1010624 (2023).
Xu, J. et al. Polygenicity at the pathway level for anorexia nervosa. medRxiv, 2025.2010.2008.25337623 (2025).
Choi, S. W. et al. The power of pathway-based polygenic risk scores. Res. Sq. PPR362752 (2021).
Li, Z. et al. Identification of potential biomarkers and their correlation with immune infiltration cells in schizophrenia using combinative bioinformatics strategy. Psychiatry Res. 314, 114658 (2022).
Childers, E., Bowen, E. F. W., Rhodes, C. H. & Granger, R. Immune-related genomic schizophrenic subtyping identified in DLPFC transcriptome. Genes (Basel) 13, 1200 (2022).
Luo, C. et al. Subtypes of schizophrenia identified by multi-omic measures associated with dysregulated immune function. Mol. Psychiatry 26, 6926–6936 (2021).
Zubiaur, P. et al. Variants in COMT, CYP3A5, CYP2B6, and ABCG2 alter quetiapine pharmacokinetics. Pharmaceutics 13, 1573 (2021).
Visscher, P. M., Gyngell, C., Yengo, L. & Savulescu, J. Heritable polygenic editing: the next frontier in genomic medicine?. Nature 637, 637–645 (2025).
Shen, H. et al. Adjunctive therapy with statins in schizophrenia patients: a meta-analysis and implications. Psychiatry Res 262, 84–93 (2018).
Sommer, I. E. et al. Simvastatin augmentation for patients with early-phase schizophrenia-spectrum disorders: a double-blind, randomized placebo-controlled trial. Schizophr. Bull. 47, 1108–1115 (2021).
Weiser, M. et al. A randomized controlled trial of add-on naproxen, simvastatin and their combination for the treatment of schizophrenia or schizoaffective disorder. Eur. Neuropsychopharmacol. 73, 65–74 (2023).
Aichholzer, M. et al. Inflammatory monocyte gene signature predicts beneficial within group effect of simvastatin in patients with schizophrenia spectrum disorders in a secondary analysis of a randomized controlled trial. Brain Behav. Immun. Health 26, 100551 (2022).
Zaki, J. K. et al. Exploring peripheral biomarkers of response to simvastatin supplementation in schizophrenia. Schizophr. Res. 266, 66–74 (2024).
Purcell, S. M. et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190 (2014).
Fromer, M. et al. De novo mutations in schizophrenia implicate synaptic networks. Nature 506, 179–184 (2014).
Marshall, C. R. et al. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat. Genet. 49, 27–35 (2017).
Satterstrom, F. K. et al. Large-scale exome sequencing study implicates both developmental and functional changes in the neurobiology of autism. Cell 180, 568–584 e523 (2020).
Sanders, S. J. et al. Insights into autism spectrum disorder genomic architecture and biology from 71 risk loci. Neuron 87, 1215–1233 (2015).
De Rubeis, S. et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 515, 209–215 (2014).
Sey, N. Y. A. et al. A computational tool (H-MAGMA) for improved prediction of brain-disorder risk genes by incorporating brain chromatin interaction profiles. Nat. Neurosci. 23, 583–593 (2020).
Watanabe, K. et al. A global overview of pleiotropy and genetic architecture in complex traits. Nat. Genet. 51, 1339–1348 (2019).
Cross-Disorder Group of the Psychiatric Genomics Consortium Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469–1482 e1411 (2019).
Litman, A. et al. Decomposition of phenotypic heterogeneity in autism reveals underlying genetic programs. Nat. Genet. 57, 1611–1619 (2025).
McMahon, F. J. & Insel, T. R. Pharmacogenomics and personalized medicine in neuropsychiatry. Neuron 74, 773–776 (2012).
Cao, C. et al. Power analysis of transcriptome-wide association study: Implications for practical protocol choice. PLoS Genet. 17, e1009405 (2021).
Roadmap Epigenomics, C. et al. Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015).
Yu, G., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Ho, S. M. et al. Evaluating synthetic activation and repression of neuropsychiatric-related genes in hiPSC-derived NPCs, neurons, and astrocytes. Stem Cell Rep. 9, 615–628 (2017).
Powell, S. K. et al. Induction of dopaminergic neurons for neuronal subtype-specific modeling of psychiatric disease risk. Mol. Psychiatry 28, 1970–1982 (2023).
Hoffman, G. E. et al. Transcriptional signatures of schizophrenia in hiPSC-derived NPCs and neurons are concordant with post-mortem adult brains. Nat. Commun. 8, 2225 (2017).
Ahn, K. et al. High rate of disease-related copy number variations in childhood onset schizophrenia. Mol. Psychiatry 19, 568–572 (2014).
Ahn, K., An, S. S., Shugart, Y. Y. & Rapoport, J. L. Common polygenic variation and risk for childhood-onset schizophrenia. Mol. Psychiatry 21, 94–6 (2014).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single-cell genomics. Genome Biol. 19, 224 (2018).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 e1817 (2016).
Adamson, B. et al. A multiplexed single-cell CRISPR screening platform enables systematic dissection of the unfolded protein response. Cell 167, 1867–1882 e1821 (2016).
Datlinger, P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Papalexi, E. et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 53, 322–331 (2021).
Tirosh, I. et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196 (2016).
Tian, R. et al. CRISPR interference-based platform for multimodal genetic screens in human iPSC-derived neurons. Neuron https://doi.org/10.1016/j.neuron.2019.07.014 (2019).
Garcia M. F. et al. Dynamic convergence of neurodevelopmental disorder risk genes across neurodevelopment. Nat. Neuro. https://doi.org/10.1101/2024.08.23.609190 (2025).
Gao, C., McDowell, I. C., Zhao, S., Brown, C. D. & Engelhardt, B. E. Context specific and differential gene co-expression networks via Bayesian biclustering. PLoS Comput. Biol. 12, e1004791 (2016).
Gao C., Brown, C.D., Engelhardt, B.E. A latent factor model with a mixture of sparse and dense factors to model gene expression data with confounding effects. arXiv https://arxiv.org/abs/1310.4792 (2013).
Watanabe, K., Taskesen, E., van Bochoven, A. & Posthuma, D. Functional mapping and annotation of genetic associations with FUMA. Nat. Commun. 8, 1826 (2017).
Wang, J. & Liao, Y. WebGestaltR: gene set analysis toolkit WebGestaltR. R package version 0.4.3., https://CRAN.R-project.org/package=WebGestaltR (2020).
Stelzer, G. et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinformatics 54, 1 30 31–31 30 33 (2016).
Rappaport, N. et al. MalaCards: an amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 45, D877–D887 (2017).
Cerezo, M. et al. The NHGRI-EBI GWAS catalog: standards for reusability, sustainability and diversity. Nucleic Acids Res. 53, D998–D1005 (2025).
Kirov, G. et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol. Psychiatry 17, 142–153 (2012).
Liang, Q. et al. Impact of common variants on brain gene expression from RNA to protein to schizophrenia risk. Nat. Commun. 16, 10773 (2025).
Niego, A. & Benitez-Burraco, A. Autism and Williams syndrome: dissimilar socio-cognitive profiles with similar patterns of abnormal gene expression in the blood. Autism 25, 464–489 (2021).
Hribkova, H. et al. Clozapine reverses dysfunction of glutamatergic neurons derived from clozapine-responsive schizophrenia patients. Front. Cell. Neurosci. 16, 830757 (2022).
Kim, S. K. et al. Individual variation in the emergence of anterior-to-posterior neural fates from human pluripotent stem cells. Stem Cell Rep. 19, 1336–1350 (2024).
Watts, D. J. & Strogatz, S. H. Collective dynamics of ‘small-world’ networks. Nature 393, 440–442 (1998).
Subramanian, A. et al. A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell 171, 1437–1452 e1417 (2017).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Phipson, B., Lee, S., Majewski, I. J., Alexander, W. S. & Smyth, G. K. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann. Appl. Stat. 10, 946–963 (2016).
Subramanian, A., Tamayo, P. & Mootha, V. K. & others. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS 102, 15545–15550 (2005).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Ripke, S. et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat. Genet. 45, 1150–1159 (2013).
Du, J. et al. KEGG-PATH: kyoto encyclopedia of genes and genomes-based pathway analysis using a path analysis model. Mol. Biosyst. 10, 2441–2447 (2014).
Gene Ontology Consortium The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 (2000).
Acknowledgements
Special thanks to Michael Talkowski and Douglas Ruderfer for countless discussions on convergence, and to Naomi Wray for thoughtful insights and contextualizations of our analyses on non-additive effects.
Author information
Authors and Affiliations
Contributions
SCZ eGene lists were prioritized by E.S., L.M.H., W.Z., G.V. and P.R., together with K.J.B. Epigenome data provided by K.G., S.A. and P.R. iGLUT transcriptomic and phenotypic studies were conducted by P.J.M.D., with assistance by E.C. Synergy analysis was conceptualized by N.S. and applied by P.J.M.D.; convergent analyses were conceptualized by K.G.R-T. and applied by C.S. Transcriptomic imputation was conducted by J.J. and L.M.H.; pathway-specific PRS by J.G.G. and P.O. The ECCITE-seq pipeline was adapted to hiPSC-neurons by A.L., supported by P.J.M.D., J.F.F., A.Y., S.C and B.Z. ECCITE-seq iGLUT neuron studies were conducted by A.L. and P.J.M.D. 10x analyses were conducted by J.F.F., and preliminary ECCITE-seq quality control was conducted by M.W. Convergent analyses were conceptualized by K.J.B. and L.H., developed and conducted entirely by K.G.R-T. The paper was written by P.J.M.D., K.G.R-T., L.H., and K.J.B., with input from all authors. The following pairs of authors contributed equally to this manuscript: P.J.M.D. and K.G.R-T; C.S. and A.L.; J.J. and J.G.G. This work was supported by F31MH130122 (K.G.R-T.), R01MH109897 (K.J.B., P.R.), R56MH101454 (K.J.B., E.S., L.M.H.), R01MH123155 (K.J.B.) and R01ES033630 (L.M.H., K.J.B.), R01MH124839 (L.M.H.), R01MH118278 (L.M.H.), R01MH106056 (K.J.B and S.A.), U01DA047880 (K.J.B and S.A), R01DA048279 (K.J.B and S.A), T35DK104689-07 (E.C.), K08MH122911 (G.V.). R01MH125246 (P.R.), U01MH116442 (P.R.), R01MH109677 (P.R.), I01BX002395 (P.R.), and by the State of Connecticut, Department of Mental Health and Addiction Services. This publication does not express the views of the Department of Mental Health and Addiction Services or the State of Connecticut.
Corresponding authors
Ethics declarations
Competing interests
E.S. is today an employee at Regeneron. K.J.B. is on the advisory board of Cell Stem Cell and a scientific advisor to Rumi Scientific Inc. and Neuro Pharmaka Inc. The remaining authors declare that they have no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Michael Deans, P., Retallick-Townsley, K.G., Li, A. et al. Functional implications of polygenic risk for schizophrenia in human neurons. Nat Commun 17, 1355 (2026). https://doi.org/10.1038/s41467-025-67959-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-67959-z
This article is cited by
-
The polygenic, omnigenic and stratagenic models of complex disease risk
Nature Genetics (2026)