Abstract
Rhizosphere microbes benefit plant growth and health. How plant-microbe interactions regulate fruit quality remains poorly understood. Here, we elucidate the multi-level modulation of vitamin accumulation in tomato by flavonoid-mediated crosstalk between host plants and rhizosphere microbes. SlMYB12-overexpressing plants with up-regulated flavonoid biosynthesis accumulate higher levels of vitamins C and B6 in fruits compared to wild-type plants grown in natural soil. Flavonoid-mediated improvement of fruit quality depends on the presence of soil microbiomes and relates to rhizosphere enrichment of key taxa (e.g. Lysobacter). Multi-omics analyses reveal that flavonoids attract Lysobacter soli by stimulating its twitching motility and spermidine biosynthesis, which in turn boosts vitamin accumulation in fruits across tomato cultivars and soil types. RpoN acts as a dual regulator in L. soli that is responsive to flavonoids, controlling bacterial motility and spermidine production. Our study provides insight into flavonoid-mediated rhizosphere signalling and underscores plant-microbiome orchestration for improved tomato fruit quality.
Similar content being viewed by others
Introduction
Tomato (Solanum lycopersicum Mill.) is one of the most popular horticultural crops worldwide1, offering a unique flavour and substantial nutritional value2. The tomato fruit is a favourable source of bioactive compounds, especially vitamins involved in various metabolic processes3. Owing to their antioxidant capacity, vitamins can scavenge free radicals in the human body, lowering the risk of cardiovascular diseases and cancers4,5,6. Therefore, substantial effort has been made to develop cost-effective and environmentally friendly methods (e.g., microbial agents) for enhancing vitamin accumulation in tomato fruits.
Plant growth-promoting rhizobacteria (PGPR) are a diverse group of beneficial microbes that can modify plant traits. Numerous PGPR are able to stimulate plant growth or strengthen stress resistance in crops at the seedling stage by releasing auxins, siderophores and 1-aminocyclopropane-1-carboxylate (ACC) deaminases7,8,9. There are insufficient studies on the potential role of PGPR in fruit quality regulation of crops at the later growth stages8,9. Among common PGPR, Lysobacter species exhibit strong antagonistic effects against a broad range of pathogenic bacteria, fungi, oomycetes and nematodes10,11. Previous studies have demonstrated the role of Lysobacter in plant disease control10,11. An interesting but unsolved question is whether Lysobacter species contribute to fruit quality in crops, such as tomato.
Root exudates, as mediators of plant-microbe communication, regulate plant growth, development and health12. Many root exudates play a role in improving the rhizosphere micro-environment by enhancing nutrient availability and recruiting plant-beneficial microbes13,14,15. Flavonoids, one of the major groups of root exudates14, are active molecules mediating plant-PGPR interactions16. Flavonoids can alleviate nutrient deficiencies17,18 and drought stress19,20 to maintain normal plant growth. These effects are achieved by mediating rhizosphere microbiome restructuring and beneficial taxa enrichment. However, the feedback mechanism of flavonoid-responsive microbes to regulate fruit quality in tomato has not been deciphered. It remains unclear whether flavonoids modulate secondary metabolism in key rhizosphere taxa, leading to fruit metabolic reprogramming and consequently promoting bioactive constituent accumulation.
In this work, we demonstrate that tomato plants recruit specific rhizosphere microbes (e.g., Lysobacter) and modulate microbial metabolic patterns through root exudates (e.g., flavonoids), thereby enhancing fruit vitamin accumulation. We show that flavonoids reshape the rhizosphere microbiome and stimulate bacterial spermidine biosynthesis via the regulator RpoN. This microbial response subsequently increases vitamin C and B6 levels in tomato fruits across different cultivars and soil types. Our study identifies a flavonoid-mediated signalling pathway that underpins plant-microbiome coordination for improved fruit quality.
Results
Flavonoids mediate fruit vitamin accumulation driven by the soil microbiome
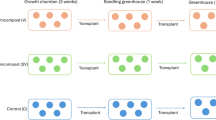
To determine whether flavonoid-mediated variations in the rhizosphere microbiome contribute to vitamin accumulation in tomato fruits, we conducted independent pot experiments under different soil conditions (Fig. 1A–C). First, wild-type plants of Micro-Tom tomato and SlMYB12-overexpressing plants with up-regulated flavonoid biosynthesis21 were grown in natural soil from Yangling, China. Flavonoid-targeted metabolomic analysis revealed that 19 root metabolites and three rhizosphere soil metabolites were differentially accumulated (DAMs; fold-change ≥ 10, P < 0.01) in SlMYB12-overexpressing plants compared to wild-type plants. Among them, rutin, naringenin chalcone (NC) and kaempferol-3-O-rutinoside (K3OR) showed the most prominent differences in their relative abundance between groups (Supplementary Fig. 1). Based on widely targeted metabolomic analysis, we compared the metabolite profiles of tomato fruits between wild-type and SlMYB12-overexpressing plants. A total of 3,092 metabolites were identified in all fruit samples, representing 13 chemical classes. The most predominant classes were “flavonoids” (14.46 %), “amino acids and its derivatives” (13.69 %) and “alkaloids” (13.53 %; Supplementary Fig. 2A). There were 679 DAMs (fold-change ≥ 2, P < 0.05), with 409 up-regulated and 270 down-regulated (Supplementary Fig. 2B). To identify the physiological processes responsible for the DAMs, we performed KEGG pathway analysis. “Vitamin B6 metabolism” and “ascorbate and aldarate metabolism” were predominantly enriched for the DAMs (P < 0.01; Supplementary Fig. 2C). The relative abundances of vitamin C (ascorbic acid) and vitamin B6 (pyridoxine, pyridoxal and pyridoxamine) in fruit samples were significantly higher in SlMYB12-overexpressing plants compared to wild-type plants (≥ 7-fold, P < 0.001; Supplementary Fig. 2D). This pattern was confirmed by hydrophilic interaction chromatography (HILIC) and high-performance liquid chromatography (HPLC) of vitamins C and B6 in fruit samples (P < 0.05; Supplementary Fig. 3A–D), but not in sterilised soil (Supplementary Fig. 3E–H). These results allowed us to posit that flavonoids promote vitamin accumulation in tomato fruits depending on the presence of soil microbiomes.
A Assay 1: Determination of differential flavonoid components in root exudates of tomato. Assay 2: Exploration of how exogenous flavonoids affect rhizosphere microbiome structure and fruit vitamin accumulation of tomato in natural soil (rutin: naringenin chalcone: kaempferol-3-O-rutinoside = 1:1:1, m/m/m, A: 10 µmol g−1; B: 3 µmol g−1; C: 1 µmol g−1; D: 0.1 µmol g−1; R: rutin, 1 µmol g−1; N: naringenin chalcone, 1 µmol g−1; K: kaempferol-3-O-rutinoside, 1 µmol g−1; Con & SIMYB12: wild-type and SIMYB12-overexpressing plants as controls). Assay 3: Evaluation of the effects of exogenous flavonoids on microbiomes in unplanted natural soil. Assay 4: Analysis of fruit vitamin accumulation in wild-type plants grown in sterilised soil without (Con) and with (C) exogenous flavonoids and in SlMYB12-overexpressing plants. B Isolation key bacteria enriched by flavonoids and evaluation of their role in fruit vitamin accumulation. C Integrated transcriptomic and widely targeted metabolomic analyses, as well as growth, motility and colonisation assays of key bacterial strain under flavonoid treatment. Created in BioRender. fu, w. (https://BioRender.com/y3ukref).
To verify the involvement of soil microbiomes in fruit vitamin accumulation across different tomato cultivars, we devised a flavonoid addition experiment with wild-type Micro-Tom and Sheng Nv-Guo plants under natural and sterilised soil conditions. The experiment included a negative control (wild-type plants with no exogenous flavonoids, group Con) and a positive control (SlMYB12-overexpressing plants with up-regulated flavonoid biosynthesis, group O). In natural soil, exogenous addition of flavonoids, either in combination or individually, increased the vitamin contents in fruit samples of Micro-Tom plants compared to those of group Con. Equal mass combinations of rutin, NC and K3OR at different concentrations (A: 10 µmol g−1, B: 3 µmol g−1, C: 1 µmol g−1, D: 0.1 µmol g−1) showed greater effects on fruit vitamin accumulation than individual flavonoids (R: rutin, N: NC, K: K3OR; 1 µmol g−1 each). The highest vitamin contents in fruit samples were observed for Micro-Tom plants of groups C and O (P < 0.05; Supplementary Fig. 4). Comparable results were obtained from Sheng Nv-Guo plants in natural soil, where the combined and individual flavonoid treatments increased the vitamin contents in fruit samples compared to group Con, and the highest values were observed in groups C (P < 0.05; Supplementary Fig. 4). In sterilised soil, exogenous addition of flavonoids did not affect fruit vitamin contents in Micro-Tom and Sheng Nv-Guo plants (Supplementary Fig. 5). Moreover, we analysed soil chemical properties and found no significant differences in key soil variables (pH and nutrient contents) between the control and flavonoid-treated groups (Supplementary Table 1). These results supported that flavonoids facilitate fruit vitamin accumulation across tomato genotypes in a soil microbiome-dependent manner.
Flavonoids broadly contribute to rhizosphere bacterial diversity
To explore flavonoid-induced variations in rhizosphere microbial diversity, we performed 16S rRNA gene amplicon sequencing. Exogenous addition of flavonoids prominently enhanced bacterial alpha-diversity in the rhizosphere of wild-type Micro-Tom and Sheng Nv-Guo plants under natural soil conditions. The bacterial richness (Chao1) and diversity (Shannon) in rhizosphere soils of Micro-Tom plants were greater in groups B, C, D and N than group Con (P < 0.05), with peak values in group C (Fig. 2A, B). Soil samples from SlMYB12-overexpressing plants (group O) also showed greater bacterial richness and diversity compared to group Con (P < 0.05), and no significant differences were observed between groups O and C (Supplementary Fig. 6A, B). The fungal alpha-diversity in rhizosphere soils of Micro-Tom plants did not respond to exogenous flavonoids (Fig. 2D, E). Similar patterns were observed in alpha-diversity of bacteria and fungi when flavonoids were added to natural soil with Sheng Nv-Guo plants (Supplementary Figs. 6D, E and 7A–D). Furthermore, we added exogenous flavonoids to unplanted natural soil and observed microbial alpha-diversity patterns comparable to those seen in planted natural soil (Supplementary Fig. 8 A–D).
A, B Bacterial richness (Chao1) and diversity (Shannon) under combined and individual flavonoid treatments. D, E Fungal richness (Chao1) and diversity (Shannon) under different flavonoid treatments. Tops and bottoms of boxes represent 25th and 75th percentiles, respectively. Horizontal bars within boxes denote medians, and the upper and lower whiskers represent the range of non-outliers. Different letters above the error bars indicate significant differences between treatments. All plots are mean ± SEM (n = 3 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). C, F Bacterial and fungal beta-diversity revealed by principal coordinate analysis (PCoA) based on unweighted UniFrac distances. G List of rhizosphere biomarkers in planted soil identified by linear discriminant analysis (LDA) effect size (n = 3 biological replicates). H List of rhizosphere biomarkers in unplanted soil (n = 3 biological replicates). Flavonoids were applied in combination (rutin: naringenin chalcone: kaempferol-3-O-rutinoside = 1:1:1, m/m/m; A: 10 µmol g−1, B: 3 µmol g−1, C: 1 µmol g−1 and D: 0.1 µmol g−1) and individually (N: naringenin chalcone, 1 µmol g−1; R: rutin, 1 µmol g−1; and K: kaempferol-3-O-rutinoside; 1 µmol g−1). Wild-type (Con) and SlMYB12-overexpressing (O) plants served as controls. P-values are shown in source data. Source data are provided as a Source Data file.
Beta-diversity analysis by principal coordinate analysis (PCoA) revealed distinct variations in rhizosphere bacterial communities among the flavonoid treatment, Con and O groups (P < 0.05; Fig. 2C and Supplementary Fig. 6C, F). However, there was no significant difference in bacterial community composition between groups O and C (Fig. 2C and Supplementary Fig. 6C). The bacterial community composition in unplanted natural soil also shifted following exogenous addition of flavonoids (Supplementary Fig. 8E). Irrespective of the presence or absence of plants, fungal beta-diversity did not respond to any exogenous flavonoid treatments (Fig. 2F, Supplementary Fig. 7F and Supplementary Fig. 8F). The results provided robust evidence for flavonoid-mediated enhancement of soil bacterial diversity, which is not interfered by other metabolites from root exudates.
Flavonoids induce rhizosphere bacterial community restructuring
To disentangle the effect of flavonoids on rhizosphere microbial community structure, we focused on the composition and relative abundance of bacterial taxa at the phylum and genus levels. Exogenous addition of flavonoids altered the structure of rhizosphere bacterial communities associated with tomato plants, causing changes in the composition and relative abundance of major taxa (Supplementary Fig. 9). At the phylum level, Proteobacteria, Acidobacteria, Bacteroidetes, Chloroflexi, Planctomycetes, Gemmantimonadetes and Actinobacteria showed noticeable responses to flavonoid treatment. At the genus level, linear discriminant analysis (LDA) effect size (LEfSe) analysis identified 26 flavonoid-responsive biomarkers in rhizosphere soils of wild-type Micro-Tom plants (LDA > 3). Among them, six genera were enriched in group O, with eight genera enriched in group C. In both groups O and C, Lysobacter and Bacillus were two common genera that increased in relative abundance under flavonoid treatment (Fig. 2G). Furthermore, we identified 32 flavonoid-responsive genera in rhizosphere soils of Sheng Nv-Guo plants (LDA > 3), including Lysobacter and Bacillus that were also enriched in group C (Supplementary Fig. 7G).
Exogenous addition of flavonoids led to shifts in the microbial community structure in unplanted natural soil (Supplementary Fig. 10). LEfSe analysis uncovered the enrichment of both Lysobacter and Bacillus in soil samples of group C after flavonoid treatment (LDA > 3; Fig. 2H). Moreover, we performed quantitative PCR (qPCR) assays targeting the 16S rRNA genes to quantify taxa-specific abundances (Supplementary Method 1, Supplementary Table 2 and 3). Both Lysobacter and Bacillus showed significantly higher absolute abundances in soil samples of groups C and O compared to the control samples (P < 0.05; Supplementary Fig. 11). The results indicated that flavonoids preferentially attract Lysobacter and Bacillus in the rhizosphere microbiome.
Mantel’s test unravelled the relationship between differentially abundant taxa in rhizosphere soils and bioactive compounds in tomato fruits. Fruit vitamin contents were significantly positively correlated with the relative abundances of both Bacillus (P < 0.05) and Lysobacter (P < 0.01; Supplementary Fig. 12). These results provided important clues that flavonoids likely mediate rhizosphere enrichment of key taxa to boost fruit vitamin accumulation.
Flavonoids increase fruit vitamin content through Lysobacter enrichment
To ascertain the potential role of Bacillus and Lysobacter in regulating tomato fruit quality, we obtained a total of 104 bacterial isolates from rhizosphere soils treated with flavonoids. Using 16S rRNA gene sequencing, 54 species representing 27 genera were identified (Fig. 3A). Among them, three dominant Lysobacter strains (designated P18, P25 and P58) and three dominant Bacillus strains (designated P54, P64 and P66) were purified (Fig.3A). Based on high-throughput sequencing data of operational taxonomic units (OTUs), Lysobacter OTU69 and Bacillus OTU341 showed the highest relative abundance in rhizosphere soils, with significant enrichment in groups O and C (Supplementary Fig. 13). We aligned these two representative OTU sequences to the full-length 16S rRNA gene sequences of the dominant bacterial strains. Lysobacter OTU69 shared high similarity to L. soli P18 (95.8%), L. fragariae P25 (94.1%) and L.niastensis P58 (92.8%). Bacillus OTU341 matched to B. subtilis P66, B. amyloliquefaciens P64 and B. cereus P54 (91.7–94.4 % similarity). Therefore, these six strains were selected as key bacteria for plant growth assay, and all of them showed growth-promoting effect on wild-type Micro-Tom tomato seedlings (P < 0.05; Supplementary Fig. 14, Supplementary Table 4).
A Phylogenetic tree of culturable bacterial isolates from flavonoid-treated rhizosphere soils based on the neighbour-joining method. Six key bacteria are marked with an asterisk (*). B Growth of tomato plants treated with cell suspensions of key bacteria. C–F Vitamin contents in fruit samples treated with bacterial cell suspensions. Tops and bottoms of boxes represent 25th and 75th percentiles, respectively. Horizontal bars within boxes denote medians, and the upper and lower whiskers represent the range of non-outlier data values. Different letters above the error bars indicate significant differences between treatments. All plots are mean ± SEM (n = 3 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). G Vitamin contents in fruit samples of tomato plants grown in geographically distant soils that had been sterilised and treated with L. soli P18 cell suspensions. (n = 5 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). The map of China is sourced from the National Platform for Common GeoSpatial Information Services (https://www.tianditu.gov.cn/). SO represents SlMYB12 overexpression, SCon represents sterile 1× phosphate-buffered saline (control) and SP18–SP66 represent cell suspensions of key bacterial strains diluted with 1× phosphate-buffered saline. Source data are provided as a Source Data file.
To validate the function of key bacteria in fruit vitamin accumulation, we conducted a pot experiment in sterile soil with wild-type Micro-Tom and Sheng Nv-Guo plants (Fig. 3B). Compared to the control group, the vitamin C, pyridoxine, pyridoxal and pyridoxamine contents in fruit samples of Micro-Tom plants increased across the six experimental groups treated with different bacterial cell suspensions (P < 0.05; Fig. 3C–F). The highest vitamin contents in fruit samples were observed for the treatment group of L. soli P18 (2.13–5.00-fold). Consistently, the treatment with L. soli P18 increased vitamin contents in fruit samples of Sheng Nv-Guo plants (P < 0.05; Supplementary Fig. 15), confirming that the positive effects of L. soli P18 on fruit vitamin levels are robust across plant genotypes. Moreover, L. soli P18 treatment significantly elevated vitamin contents in fruit samples of Micro-Tom plants in sterilised soils from Kashgar, Ordos, Chengdu and Nanning. There was no significant difference in fruit vitamin contents between L. soli P18-treated plants and SlMYB12-overexpressing plants (P < 0.05; Fig. 3G). These results indicated that the observed effects of L. soli P18 on fruit vitamin levels are consistent across geographically distinct soils. We therefore selected L. soli P18 as a flavonoid-enriched key bacterial strain for subsequent assays.
Flavonoids enhance twitching motility of Lysobacter soli P18
To decipher the mechanism of flavonoid-mediated Lysobacter enrichment in the rhizosphere, we performed metagenomic sequencing. A total of 22,759 and 22,355 unigenes of Lysobacter were identified in rhizosphere soils of groups C/Con and O/Con, respectively. There were 1,374 and 1,297 differentially expressed genes (DEGs, P < 0.05) in groups C/Con and O/Con, respectively (Supplementary Fig. 16A, B). Based on Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathway analysis, 190 DEGs of group C/Con and 185 DEGs of group O/Con were both enriched in the two-component system (Supplementary Fig. 16C, D). Among them, 34 and 32 up-regulated DEGs were identified as related to bacterial twitching motility (P < 0.05; Supplementary Fig. 17). These DEGs comprised genes that encode bacterial type IV pilus assembly protein, twitching motility protein, twitching motility response regulator and chemosensory pili system protein (Supplementary Fig. 17).
We subsequently delved into the transcriptomic changes of L. soli P18 induced by flavonoids (treatment C: 1 µmol g−1). Principal component analysis (PCA) showed a clear separation of flavonoid-treated and control samples along the first principal component (PC1, 75.05 %; Supplementary Fig. 18A). A total of 509 DEGs (P < 0.05) were identified between the two groups (Supplementary Fig. 18B). Categorisation of the up- and down-regulated DEGs indicated that flavonoids broadly influenced a plethora of cellular processes in L. soli P18 (Supplementary Fig. 18 C). Importantly, 27 up-regulated DEGs were identified as related to bacterial twitching motility (P < 0.05; Fig. 4A). These DEGs included genes that encode pilus assembly protein, pilus biogenesis protein, twitching motility response regulator, type IV pilus twitching motility protein, type IV pilus modification protein and chemosensory pili system protein.
A Flavonoid-induced changes in twitching motility-related genes revealed by prokaryotic transcriptomic sequencing (n = 3 biological replicates, two-sided Student’s t-test, P < 0.05). B Increased rhizosphere colonisation of L. soli P18 in SlMYB12-overexpressing plants and flavonoid-treated wild-type plants (C) of Micro-Tom tomato compared to untreated wild-type plants (Con; n = 10 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). CFU, colony-forming units. C–E Effects of flavonoids on twitching (left), swarming (middle) and swimming (right) motility of L. soli P18 (n = 10 biological replicates, two-sided Student’s t-test, P < 0.05). F Growth curves of L. soli P18 with flavonoids (C) or not (Con; n = 6 biological replicates, two-sided Student’s t-test, P < 0.05). OD600, optical density at 600 nm. G A hub gene identified in the co-expression network with edges carrying weights > 0.9. The hub gene is shown in red colour, with twitching motility-related genes in purple colour and spermidine biosynthetic-related genes in green colour. Positive correlations between RpoN and other genes are indicated by grey lines. H, I Effects of flavonoids on twitching motility and colonisation of L. soli strains (n = 10 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). P18 represents the wild-type strain, ΔRpoN is a RpoN mutant strain, and ΔRpoN-C is a complemented strain. J Bacterial twitching motility-related pathways in the two-component system (n = 3 biological replicates, two-sided Student’s t-test, P < 0.05). Bacterial growth, colonisation and motility data are presented as means ± SEM, and different letters above the error bars indicate significant differences between treatments. Source data are provided as a Source Data file.
We next verified the mechanism of rhizosphere colonisation by L. soli P18 in response to flavonoids (treatment C: 1 µmol g−1). Heightened bacterial colonisation was observed in rhizosphere soils of SlMYB12-overexpressing plants and flavonoid-treated wild-type Micro-Tom plants compared to the control group (Fig. 4B). Furthermore, bacterial motility assays unveiled that flavonoids enhanced the twitching motility, rather than the swarming or swimming motility, in L. soli P18 on agar plates (Fig. 4C–E). Cell growth assay demonstrated minimal effects of flavonoids on the growth rate of L. soli P18 (Fig. 4F). Among the up-regulated genes related to bacterial twitching motility, RpoN emerged as the hub node in the gene co-expression network. There was a significant positive correlation between the expression levels of RpoN and other genes related to twitching motility (P < 0.01, weight > 0.9; Fig. 4G).
To verify the role of RpoN as a hub gene in response to flavonoid signalling and a regulator of bacterial twitching motility, we constructed a RpoN knockout mutant (ΔRpoN) and a complemented strain (ΔRpoN-C). ΔRpoN showed significantly lower twitching motility and colonisation ability under flavonoid treatment compared to its wild-type strain (P < 0.01), and this phenotype was restored in ΔRpoN-C (Fig. 4H, I). In summary, these results indicated that flavonoids stimulate the twitching motility of L. soli P18 in a RpoN-dependent manner, which enables bacterial enrichment in the rhizosphere (P < 0.05; Fig. 4J).
Flavonoids stimulate spermidine biosynthesis in Lysobacter soli P18
To unravel whether flavonoids regulate bacterial secondary metabolism in the rhizosphere, we carried out widely targeted metabolomic profiling of L. soli P18. Based on PCA, the culture samples treated with flavonoids (treatment C: 1 µmol g−1) were clearly distinguished from the control samples by PC1 (43.05 %; Supplementary Fig. 19A). A total of 1,708 metabolites were identified in all culture samples. The most predominant classes were “amino acids”, “alkaloids”, “aldehyde” and “lipids”. A total of 584 metabolites were differentially accumulated between groups (P < 0.05). Specifically, 354 metabolites were up-regulated and 230 metabolites were down-regulated by flavonoids (Supplementary Fig. 19B).
We then looked at the relationship between flavonoid-induced metabolites in L. soli P18 and vitamins in tomato fruits. Mantel’s test revealed that the relative abundance of bacterial alkaloids was significantly correlated with fruit vitamin contents (P < 0.01; Supplementary Fig. 20). Among alkaloids, spermidine emerged as the greatest contributor to fruit vitamin accumulation based on random forest analysis. Based on the cross-validation results, the random forest model achieved optimal stability and accuracy when selecting the top 20 metabolites (Supplementary Fig. 21). Widely targeted metabolomic profiling unveiled the up-regulation of spermidine (1.89-fold) and its precursors (cysteine: 1.67-fold; methionine: 1.81-fold) in the metabolome of flavonoid-treated L. soli P18 (P < 0.05; Fig. 5A). Therefore, we focused on spermidine as a DAM of L. soli P18 in response to flavonoids.
A Relative abundance of spermidine, cysteine and methionine in fruits of flavonoid treatment (C: 1 µmol g−1) vs. control groups based on widely targeted metabolomic profiling. B Spermidine biosynthetic pathway. C, D Expression levels (fold change, FC) of spermidine biosynthetic enzyme genes in fruits of flavonoid-treated plants (C: rutin: naringenin chalcone: kaempferol-3-O-rutinoside = 1:1:1, m/m/m, 1 µmol g−1; R: rutin, 1 µmol g−1; N: naringenin chalcone, 1 µmol g−1; K: kaempferol-3-O-rutinoside, 1 µmol g−1; C: 1 µmol g−1) and SlMYB12-overexpressing plants vs. wild-type Micro-Tom plants based on metagenomic sequencing. E Expression levels of spermidine biosynthesis-related genes in fruits of flavonoid treatment (C: 1 µmol g−1) vs. control groups based on prokaryotic transcriptomic sequencing. F Spermidine content in broth culture of L. soli P18 treated without and with flavonoids (C: 1 µmol g−1, n = 10 biological replicates, mean ± SEM, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). G Vitamin contents in fruits treated with cell suspensions of L. soli strains (n = 5 biological replicates, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). P18 represents the wild-type strain, ΔRpoN is a RpoN mutant strain, and ΔRpoN-C is a complemented strain. H Spermidine content in broth cultures of L. soli strains treated without and with flavonoids (C: 1 µmol g−1, n = 10 biological replicates, mean ± SEM, one-way ANOVA with correction by Tukey’s HSD test, P < 0.05). I–L Vitamin contents in fruits of tomato plants treated with spermidine or not. Tops and bottoms of boxes represent 25th and 75th percentiles, respectively. Horizontal bars within boxes denote medians, and the upper and lower whiskers represent the range of non-outlier data values. Different letters above the error bars indicate significant differences between treatments. All plots are mean ± SEM (n = 5 biological replicates, one-way ANOVA with correction by Tukey’s HSD test P < 0.05). M Integrated transcriptomic and widely targeted metabolomic analysis of vitamin B6/C metabolic pathways. Source data are provided as a Source Data file.
Spermidine biosynthesis is controlled by a set of genes that encode S-adenosylmethionine decarboxylase (SpeD) and spermidine synthase (SpeE)22 (Fig. 5B). Metagenomic sequencing uncovered that the relative abundances of SpeD and SpeE genes were 1.60–1.95-fold up-regulated in soil samples of groups C and O (Fig. 5C and D). Consistently, prokaryotic transcriptomic profiling revealed 1.50–1.65-fold up-regulation of SpeD and SpeE in L. soli P18 under flavonoid treatment (Fig. 5E). To validate the effect of flavonoids on spermidine biosynthesis in L. soli P18, we performed chemical extraction and quantitative analysis. The spermidine content in culture samples added with combined flavonoids (treatment C: 1 µmol g−1) was 2.8-fold higher than that of the control group (P < 0.05). Combined flavonoids exhibited a greater effect on spermidine production in L. soli P18 compared to individual flavonoids (treatments R, K, N; P < 0.05; Fig. 5F). The results indicated that flavonoids induce up-regulation of genes related to spermidine biosynthetic enzymes in L. soli P18, stimulating spermidine production. Interestingly, the gene co-expression network revealed a positive correlation between the expression levels of RpoN and spermidine biosynthesis related genes, SpeD and SpeE (P < 0.01, weight > 0.9; Fig. 4G).
Further, the function of RpoN was validated by gene knockout in L. soli P18. We found that fruit vitamin contents were significantly higher in Micro-Tom plants treated with cell suspensions of the wild-type strain and the complemented strain ΔRpoN-C compared to the control group (P < 0.01). Cell suspension of the mutant strain ΔRpoN had no significant effect on fruit vitamin contents (Fig. 5G). Under flavonoid treatment, ΔRpoN showed a significantly lower ability to produce spermidine than its wild-type strain (P < 0.01), and this phenotype was restored in ΔRpoN-C; Fig. 5H). In summary, these results indicated that flavonoids enhance spermidine biosynthesis of L. soli P18 under the control of RpoN.
Bacterially produced spermidine boosts fruit vitamin accumulation
To elucidate the role of spermidine in fruit vitamin accumulation, we added exogenous spermidine (1 μmol g−1) to sterilised soil with wild-type Micro-Tom plants. The spermidine treatment significantly increased vitamin C, pyridoxine, pyridoxal and pyridoxamine contents in tomato fruits (P < 0.05; Fig. 5I–L). We then profiled the metabolome of tomato fruits after spermidine addition (Supplementary Fig. 22). A total of 2,476 metabolites were identified in all fruit samples, representing 13 chemical classes. The most predominant classes were “amino acids and its derivatives” (15.11 %), “alkaloids” (12.0 %) and “lipids” (12.4 %; Supplementary Fig. 23A). PCA uncovered that the treatment and control groups were well separated along PC1 (44.49%; Supplementary Fig. 23B), suggestive of a remarkable difference in the fruit metabolome between groups. The heatmap shows that some metabolites were differentially accumulated in response to spermidine treatment. A total of 500 DAMs were identified (fold-change ≥ 1.6, P < 0.05), with 267 up-regulated and 233 down-regulated (Supplementary Fig. 23C). To identify the physiological processes responsible for the DAMs, we performed KEGG pathway analysis. “Vitamin B6 metabolism” and “ascorbate and aldarate metabolism” were predominantly enriched for the DAMs (P < 0.05; Supplementary Fig. 23D).
Transcriptomic analysis yielded 34,688 annotated unigenes. Among them, 4,976 genes were differentially expressed (fold-change ≥ 1.6, P < 0.05), with 3,065 up-regulated and 1,911 down-regulated in spermidine treatment (Supplementary Fig. 24A). PCA showed a distinct separation between the treatment and control groups along PC1 (60.84%; Supplementary Fig. 24B), indicating a notable difference in the fruit transcriptome between groups. To identify the putative functions of potential transcripts, we performed KEGG enrichment analysis of DEGs. The DEGs were mainly enriched in “ascorbate and aldarate metabolism” and “vitamin B6 metabolism” (P < 0.05; Supplementary Fig. 24C).
By integrating transcriptomics and metabolomics, we selected DAMs and DEGs enriched in the pathways of “ascorbate and aldarate metabolism” and “vitamin B6 metabolism”. The results indicated that gene-regulated changes in the metabolites and associated metabolic pathways could alter vitamin accumulation in spermidine-treated fruits (Supplementary Figs. 23D, 24C). Specifically, the relative abundances of pyridoxamine, pyridoxal, pyridoxine, 4-pyridoxic acid, pyridoxal-5P and L-ascorbate were 1.80–8.97-fold up-regulated by spermidine (P < 0.05; Fig. 5M and Supplementary Fig. 25). Consistently, transcriptomic data showed 1.61–3.27-fold up-regulation of genes modulating EC1.1.3.8 (L-gulonolactone oxidase), EC1.2.3.1 (aldehyde oxidase), EC 3.1.3.74 (pyridoxal phosphatase) and EC4.3.3.6 (pyridoxal 5′-phosphate synthase) in spermidine treatment (P < 0.05; Fig. 5M). The collective results indicated that spermidine produced by L. soli P18 promotes vitamin accumulation in tomato fruits, conferring benefits to human health.
Discussion
We have demonstrated the contribution of flavonoids to the accumulation of bioactive compounds (vitamins) in tomato fruits and linked this improvement of fruit quality to microbiome restructuring in the rhizosphere. We found that root-derived flavonoids mediated higher accumulation of fruit vitamins depending on the presence of soil microbiomes. Flavonoids additionally heightened bacterial alpha-diversity in rhizosphere soil, with an advantage in Lysobacter enrichment. We identified L. soli P18 as a key bacterial strain that contributed prominently to vitamin accumulation in tomato fruits. Through multi-omics and sub-segment analyses, a dual regulator in L. soli P18, RpoN, was identified as being responsive to flavonoids. This regulator participated in rhizosphere recruitment of L. soli P18 and subsequent enhancement of microbial spermidine metabolism, consequently facilitating vitamin accumulation in fruits. These findings underscore the role of flavonoid-mediated plant-microbiome crosstalk in regulating fruit quality through a multi-level mechanism.
The interplay between rhizosphere microbes and host plants is a reciprocal feedback process. Soil microbes perceive signals from the roots and colonise the rhizosphere, establishing an intimate relationship with the plant host23,24. This process is mediated by a large variety of root exudates25,26, which can influence microbial motility to recruit or exclude certain taxa, altering rhizosphere microbiome structure27,28,29. In particular, many plants release large amounts of flavonoids into the rhizosphere soil16. Flavonoids can regulate rhizosphere microbiome restructuring and attract beneficial taxa (e.g., Oxalobacteraceae, Aeromonas, Pseudomonas)17,18,19,20, alleviating abiotic stresses in plants. We found that the soil microbiome played an indispensable role in flavonoid-mediated enhancement of vitamin accumulation in tomato fruits. The diversity and richness of rhizosphere microbial communities have long been recognised as key factors in the integrity, functionality and sustainability of agro-ecosystems30. Higher microbial alpha-diversity is favourable for the ecological health of rhizosphere micro-environment31. Exogenous addition of flavonoids increased bacterial alpha-diversity in the rhizosphere of tomato plants, with the greatest effect observed in the combined treatment at 1 µmol g−1. This result is likely due to the superimposed effect of mixed flavonoids, which aligns with previous studies showing that flavonoid mixture induced legume nodulation more effectively than individual compounds32,33. Together these findings suggest that flavonoids selectively facilitate the development of rhizosphere bacterial communities in a healthy and stable manner. We additionally found that a plethora of PGPR dominated by the genus Lysobacter were highly responsive to flavonoids. Microbiomic analysis and qPCR-based absolute quantification underscored the uniqueness of Lysobacter, which was not only enriched in the rhizosphere of SlMYB12-overexpressing tomato plants. The enrichment of Lysobacter was also detected in the rhizosphere of wild-type tomato plants and unplanted soil treated with flavonoids. The specific recruitment of Lysobacter by flavonoids is supported by both differential relative abundance and absolute abundance. These results confirm that the increased relative abundance of Lysobacter in flavonoid-treated samples reflects a true growth in their absolute cell numbers, rather than compositional artifacts from sequencing or quantitative changes in other taxa. The function of Lysobacter in biocontrol of soil-borne diseases has been well documented34,35,36. Our study presents solid evidence that Lysobacter can regulate fruit quality by promoting vitamin accumulation in tomato in response to flavonoids. This provides an additional perspective on the use of Lysobacter to improve agricultural productivity.
Bacterial motility plays a pivotal role in rhizosphere colonisation17,37,38. We found that flavonoids recruited plant-beneficial microbes to the rhizosphere by a mechanism similar to the enhancement of flagellar motility in Aeromonas19. Here, we propose a regulatory mechanism specific to L. soli P18: flavonoids selectively activate pilus-dependent twitching motility through type IV pili—a surface appendage critical for solid-surface colonisation39,40,41. Metagenomic and prokaryotic transcriptomic profiling of flavonoid-treated L. soli P18 revealed up-regulation of key pilus-assembly genes (PilA, PilE, PilH) within the two-component system. Crucially, we identified RpoN (σ42), a global transcriptional regulator that links environmental cues to bacterial survival strategies in L. soli P18. RpoN is known to perform functions in bacterial virulence, motility, metabolic regulation, and stress response41,43,44,45. As a central hub node in the gene co-expression network, RpoN was highly associated with up-regulated genes related to bacterial twitching motility. Functional validation by gene knockout confirmed the essential role of RpoN in twitching motility and its direct effect on rhizosphere colonisation of Lysobacter under flavonoid treatment. As flavonoids did not affect the growth of Lysobacter, we surmise that flavonoids act as signalling molecules to modulate the bacterial two-component system, where RpoN acts as a key regulatory hub to up-regulate the expression of genes related to twitching motility in Lysobacter. This study reports RpoN as a flavonoid-responsive regulator of bacterial twitching motility, explaining competitive rhizosphere colonisation of Lysobacter.
Rhizosphere microbes play a key role in improving fruit quality46,47,48, as microbial metabolites regulate plant growth and developmental processes46,49. We obtained three flavonoid-responsive Lysobacter strains from the rhizosphere soil, all of which promoted vitamin accumulation in tomato fruits. This provides direct evidence to support the role of Lysobacter in regulating tomato fruit quality and offers PGPR resources for fruit production. L. soli P18 exhibited a universal effect on fruit vitamin accumulation across different soil types and tomato cultivars. PGPR-derived secondary metabolites (e.g., auxins, siderophores) have been shown to accelerate plant growth7,8,9. This raises the question of whether flavonoids mediate the secondary metabolism of rhizosphere microbes to regulate fruit quality. We found that the levels of spermidine and its biosynthetic precursors (cysteine and methionine) were up-regulated in the metabolome and transcriptome of L. soli P18 under flavonoid treatment. Importantly, spermidine was the bacterial metabolite contributing the most to vitamin accumulation in tomato fruits. A key finding of the present study was that flavonoids modulated the metabolic patterns of key microbial taxa represented by Lysobacter in the rhizosphere soil, stimulating bacterial spermidine biosynthesis. After exogenous addition, combined flavonoids promoted spermidine production in L. soli P18 more effectively than individual flavonoids, most likely due to the synergistic effect of mixed flavonoids32,33. Meanwhile, we observed elevated expression of spermidine biosynthesis related genes (speD, speE) in flavonoid-treated L. soli P18, and RpoN was identified as a flavonoid-responsive regulator of bacterial spermidine biosynthesis. These results lay the groundwork for the artificial use of gene editing technology to synthesise spermidine in the future.
Spermidine is a microbial secondary metabolite with low-molecule-weight nitrogenous bases widely present in prokaryotic and eukaryotic cells. Spermidine participates in plant growth, development and stress resistance by alleviating metabolic disorders, reducing oxidative damage and maintaining photosynthetic efficiency50,51,52. When sterilised soil was added with exogenous spermidine, the gene expression of enzymes involved in vitamin biosynthesis was up-regulated in tomato fruits, contributing to vitamin accumulation. Furthermore, we observed higher vitamin contents in fruits of SlMYB12-overexpressing plants compared to those of wild-type Micro-Tom plants in natural soil, but not in sterilised soil. This means that the advantage of SlMYB12-overexpressing plants in fruit vitamin accumulation necessitates the presence of soil microbiomes. To sum up, flavonoids stimulate L. soli P18 to secrete spermidine by inducing the up-regulation of key enzyme genes and precursor biosynthesis pathways, which ultimately enhances vitamin accumulation in tomato fruits. Four alternate routes for vitamin C biosynthesis have been identified in plants: L-galactose, L-glucose, D-galacturonate and myoinositol pathways42,53,54,55. We found that spermidine regulated vitamin C biosynthesis in tomato fruits mainly through the L-galactose pathway. However, some genes (e.g., Pdx1, GULA) for key enzymes (e.g., pyridoxal 5’-phosphate synthase, L-gulonolactone oxidase) involved in vitamin biosynthesis pathways are considered as core hub genes56,57. Knockout of such core hub genes can lead to plant death58,59. Therefore, it is currently difficult to pinpoint the target genes of bacterially produced spermidine for regulating vitamin biosynthesis in tomato fruits by gene knockout.
Taken together, our study underscores that root-derived flavonoids boost vitamin accumulation in tomato fruits through multi-level crosstalk with rhizosphere microbes. (1) Flavonoids mediate the reshaping of rhizosphere microbial communities: Lysobacter is recruited to colonise the rhizosphere through the up-regulation of twitching motility-related genes, forming a dominant PGPR population. (2) Flavonoids mediate the metabolic reprogramming of rhizosphere key taxa: spermidine biosynthesis in Lysobacter is stimulated through the up-regulation of spermidine-related genes, which promotes fruit vitamin accumulation (Fig. 6). These findings elucidate fruit quality improvement with microbial secondary metabolites mediated by root exudates and unlock possibilities for the use of microbial-derived biostimulants in fruit production. Our experiments support a microbiome-dependent mechanism as the dominant pathway driving fruit vitamin accumulation. However, we acknowledge that flavonoids may also exert indirect effects that were not directly assessed in this study. For instance, flavonoids could alter vitamin biosynthesis pathways in tomato fruits potentially by modulating root architecture, redox balance, or secretion of other plant metabolites. Thus, flavonoid–microbiome interactions represent a dominant but not necessarily exclusive pathway influencing fruit vitamin accumulation. This recognition highlights an important direction for future research—assessing the relative contributions of direct microbiome effects versus indirect flavonoid-driven plant/soil responses to vitamin biosynthesis in tomato fruits.
1 Plant roots release flavonoid signals into the rhizosphere, where they interact with soil microbes. 2 Flavonoids recruit Lysobacter to the rhizosphere by enhancing bacterial twitching motility, which reshapes the rhizosphere microbiome. 3 Lysobacter responds to flavonoids in a RpoN-dependent manner and produces more spermidine. 4 Spermidine up-regulates fruit vitamin metabolism through the L-galactose pathway and thereby boosts the accumulation of vitamins B6 and C in fruits. Created in BioRender. fu, w. (https://BioRender.com/7k06qpx).
Methods
Soil and plants
Different types of natural soil were collected from cropland fields in Yangling, Nanning, Chengdu, Ordos, and Kashgar, China. After removing plant debris, worms and stones, the soil was homogenised manually using a 2.5-mm sieve. The chemical properties of the soils are provided in Supplementary Table 5.
Two wild-type tomato cultivars (Micro-Tom, Sheng Nv-Guo) and a SlMYB12-overexpressing transgenic line that over-accumulates flavonoids (Micro-Tom)21 were used as plant materials. Surface sterilisation of seeds was conducted by soaking in 75% (v/v) ethanol for 30 s followed by sodium hypochlorite solution (10% active chlorine) for 1 min. After three washes with sterile water, clean seeds were transferred onto Petri dishes and incubated at 4 °C in the dark for 2 d of germination. Then, the germinated seeds were transferred into a humidity chamber and incubated at 22 °C and 60–70% relative humidity for 3 d until the length of roots reached ~1 cm. Seedlings were transferred to a 45-hole tray for 30 d and watered once a week. After that, 35-d-old seedlings were transplanted to plastic pots (90 mm × 130 mm × 100 mm) containing 5 kg of natural soil from Yangling with an initial moisture content of 100% (v/w). The pots (one plant per pot) were kept in a humidity chamber with a day/night temperature of 26 °C/18 °C, a relative humidity of 60–70%, a light intensity of 12,000 Lux and a photoperiod of 18 h light/6 h dark. In the seedling, flowering and fruiting stages, 10 mL of Hoagland’s nutrient solution was supplied to each pot without chemical fertiliser application.
Flavonoid-targeted metabolomic analysis
Samples were collected from wild-type and SlMYB12-overexpressing plants at 90 d after transplanting (DAT). A 20-g sample of rhizosphere soil was obtained from each pot. The soil samples from two pots were pooled as one biological replicate, with three biological replicates per treatment. Each soil sample was extracted with 50 mL of sterile water shaken at 120 rpm for 60 min. The soil extract was centrifuged at 4 °C and 12,000 × g for 10 min, and the supernatant was collected. Additionally, tomato roots were washed with running water and then rinsed with sterile water five times. The root samples from two pots were pooled as one biological replicate, and three biological replicates were included for each treatment. All samples (soil extracts and tomato roots) were stored at -80 °C until use for targeted metabolomic analysis.
After thawing and vortexing, each root sample was vacuum-lyophilised and ground to powder using a grinding mill at 30 Hz for 1.5 min. Then, 2 mL of soil extract or 20 mg of root powder was transferred to a 10-mL centrifuge tube containing liquid nitrogen. The sample was completely freeze-dried in a lyophiliser and then added with 200 µL of 70% methanol internal standard extract. The mixture was oscillated for 15 min followed by centrifugation at 4 °C and 12,000 × g for 3 min. The supernatant was filtered through a 0.22-μm microporous filter membrane before use. The sample extracts were analysed by targeted metabolomics (Supplementary Method 2) based on ultra-performance liquid chromatography (UPLC, Nexera X2; Shimadzu, Kyoto, Japan) coupled to tandem mass spectrometry (MS/MS, Applied Biosystems 6500 Q-TRAP; Applied Biosystems, Framingham, MA, USA). Declustering potential and collision energy were optimised for individual MRM transitions. A specific set of MRM transitions were monitored for each period based on the eluted metabolites. Identification of metabolites was performed by comparison of the m / z values, retention time, and searching an internal custom database of MetWare Biotechnology Co., Ltd (Wuhan, China). DAMs were identified by based on the variable importance in projection (VIP ≥ 1), P < 0.05 (Two-sided Student’s t-test) and fold-change ≥ 1.6.
Flavonoid addition experiment
An exogenous flavonoid addition assay was carried out with potted tomato plants grown in natural soil from Yangling. Flavonoids (Solarbio, Beijing, China) were dissolved in 85% (v/v) methanol with a dilution factor of 50 and passed through a 0.22-μm sterile microporous filter membrane. Seven flavonoid treatments were established for wild-type plants based on the design of previous studies60,61 and our targeted metabolomics data of flavonoids (Supplementary Fig. 1). Four treatment groups received a mixture of rutin, NC and K3OR at different concentrations (1:1:1, m/m/m; A: 10 µmol g−1, B: 3 µmol g−1, C: 1 µmol g−1 and D: 0.1 µmol g−1). The other three treatment groups received individual flavonoids at the same concentration (R: rutin, N: NC and K: K3OR; 1 µmol g−1 each). At 15 DAT, a sterile bamboo stick was used to puncture holes into the soil around the plant stem at a distance of 1 cm. The prepared flavonoid solutions were repeatedly injected into the holes with a sterile disposable syringe (10 mL per pot, 90 mm × 130 mm × 100 mm) at 22 and 29 DAT. Two controls (wild-type Miro-Tom and SlMYB12-overexpressing plants) received an equal volume of sterile 50-fold diluted 85% methanol. In the seedling, flowering and fruiting stages, 10 mL of Hoagland’s nutrient solution was supplied to each pot. All other management practices were consistent for the treatment and control groups. At 90 DAT, rhizosphere soil samples (50 g each) were collected by gently shaking the roots and kept in a -80 °C freezer until use for DNA extraction. Another set of rhizosphere soil samples (50 g each) were obtained and stored in a 4 °C refrigerator before bacterial isolation. Fruit samples were harvested to measure vitamin contents. Samples from two pots (one plant per pot) were collected as one biological replicate, with three biological replicates per treatment.
The same procedures were followed to conduct a second flavonoid addition assay with tomato plants grown in soil from Yangling that had been sterilised by γ-irradiation (20 kGy, 10 MeV γ-ray). Only one combined flavonoid treatment (1 µmol g−1) was applied to wild-type Micro-Tom and Sheng Nv-Guo plants at 15, 22 and 29 DAT, with two controls (wild-type Micro-Tom and SlMYB12-overexpressing plants) receiving an equal volume of sterile 50-fold diluted 85% methanol. Pots were managed as described for the natural soil experiment. At 90 DAT, rhizosphere soil and fruit samples from three pots (one plant per pot) were collected as one biological replicate, and three biological replicates per treatment were prepared for vitamin C and B6 analysis.
A third flavonoid addition assay was performed in plastic pots (50 mm × 60 mm × 60 mm) containing 1 kg of natural soil from Yangling without plants. Exogenous flavonoids were applied at 15, 22 and 29 days for seven treatments and one control as performed in the natural soil experiment, and pots were managed as described for the tomato growth test. At 90 days, soil samples (5 g each) from three pots were collected as one biological replicate, and three biological replicates were included for each treatment. The samples were frozen at -80 °C until DNA extraction for 16S rRNA gene sequencing and metagenomic profiling.
Soil 16S rRNA gene sequencing
The Power Lyzer Power Soil DNA Isolation Kit (Qiagen GmbH, Hilden, Germany) was used to isolate total genomic DNA from natural soil samples (0.5 g each) as per the manufacturer’s protocol. DNA quality and quantity were respectively assessed using 1% agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). For taxonomic profiling, the V3-V4 region of the bacterial 16S rRNA genes was amplified by PCR with the universal primers 515 F (5’-GTGYCAGCMGCCGCGGTAA-3’) and 806 R (5’-GGACTACNVGGGTWTCTAAT-3’)62, yielding 292-bp amplicons. PCR reactions, product purification, and library construction are described in Supplementary Method 3. Paired-end libraries (2 × 250 bp) were sequenced on an Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA).
After raw data processing and taxonomic assignment (Supplementary Method 4), bioinformatic analysis was carried out using the Magigene Cloud platform (http://cloud.magigene.com/). Based on OTU data, rarefaction curves were created and alpha-diversity metrics (Chao1 and Shannon) were estimated using Mothur v1.30.163. Community similarities among samples were determined by PCoA based on unweighted UniFrac distances using Vegan v2.5-3 (https://CRAN.R-project.org/package =vegan/). Permutational multivariate analysis of variance was performed to assess the percentage of variation explained by the treatment and its statistical significance using Vegan v2.5-3 (https://CRAN.R-project.org/package=vegan/). LEfSe64 was used to identify differentially abundant bacterial taxa (phyla to genera) among the different treatment groups (LDA > 3). LDA scores were calculated using R v4.2.065 to maximise between-class variance while minimising within-class variance. Data were standardised prior to analysis (mean-centred and scaled to unit variance). The discriminant axes were ranked by their associated eigenvalues, and scores were projected onto the first two axes for visualisation.
Soil metagenomic sequencing
The E.Z.N.A.® Soil DNA Kit (Omega Bio-tek) was used for genomic DNA isolation from natural rhizosphere soil samples (5 g each) following the manufacturer’s instructions. DNA concentration was measured using TBS-380 (Turner BioSystems, Sunnyvale, CA, USA). DNA purity was evaluated with NanoDrop 2000 (NanoDrop Technologies Inc., Wilmington, DE, USA), and DNA quality was checked by 1% agarose gel electrophoresis. Then, DNA extracts were fragmented to an average size of ~400 bp using Covaris M220 (Gene Company Limited, China), and paired-end libraries were generated using NEXTFLEX® Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). Adaptors containing the full complement of sequencing primer hybridisation sites were ligated to the blunt-end of DNA fragments. Paired-end sequencing was accomplished on Illumina NovaSeq 6000 (Illumina Inc.) at Meige Gene Technology Co., Ltd. (Guangzhou, Guangdong Province, China). Sequencing data were analysed on the Magigene Cloud platform (http://cloud.magigene.com/). Detailed data analyses are provided in Supplementary Method 5. All unigenes of Lysobacter species were selected from the sequenced genes for DEG identification and KEGG enrichment analysis. Genes with fold-change ≥1.3 and P < 0.05 (two-sided Student’s t-test) were identified as DEGs. KEGG annotation was conducted against the KEGG database with an e-value cutoff of 10-5 using Diamond v0.8.3566.
Bacterial isolation and identification
Culturable bacteria were isolated from fresh natural rhizosphere soil samples by serial dilution and spread plate techniques67. Briefly, a 10-g soil sample was added to a flask containing 90 mL of sterile water with glass beads and shaken at 120 rpm for 30 min. The soil suspension was serially diluted to 10-5–10-7, and 10-μL aliquots of each dilution were evenly spread on beef extract peptone (BEP) agar plates. The plates were incubated at 28 °C for 3 d, with five replicates for each dilution. Representative colonies of the dominant bacteria were purified and preserved on BEP agar slants at low temperature. Identification of purified bacteria was based on PCR amplification and 16S rRNA gene sequencing68. Details of DNA extraction, PCR amplification and sequencing have been provided in Supplementary Method 6. The obtained sequences were blasted in the GenBank database (http://blast.ncbi.nlm.nih.gov/). Phylogenetic trees were constructed using the neighbour-joining method with MEGA v1169.
Tomato growth test
A 72-h broth culture of key bacteria in BEP medium was centrifuged at 12,000 ×g (4 °C, 15 min) and passed through a 0.22-μm sterile microporous filter membrane. The cell-free filtrate was diluted 50-, 100-, 200- and 400-fold with sterile water before use. Spermidine aqueous solutions were formulated at gradient concentrations of 0.1, 1.0 and 10.0 μmol g−1. Surface-sterilised seeds of wild-type Micro-Tom were evenly placed in Petri dishes with double layers of sterilised filter paper, followed by the addition of 10 mL of cell-free filtrate or spermidine solution. During the incubation period, each dish received 5 mL of cell-free filtrate or spermidine solution every day. An equal volume of sterile BEP medium at corresponding dilutions and sterile water served as the control for bacterial and spermidine treatments, respectively, with five biological replicates per treatment. All dishes were incubated in a light incubator for 15 d under the following conditions: day/night temperature, 26 °C/18 °C; photoperiod, 14 h/10 h light/dark; relative humidity, 65%; and light intensity, 12,000 Lux.
Single colonies of key bacteria were transferred from 24-h cultures to fresh BEP medium and incubated at 36 °C shaken at 160 rpm for 24 h. Bacterial cultures were harvested by centrifugation at 12,000 × g (4 °C, 10 min). The cell pellet was resuspended in 1 × phosphate-buffered saline (PBS), with an effective viable count of 1×108 colony-forming units (CFU)·mL−1. In the treatment group, pot sterile soil was mixed with 5 mL of cell suspension using a pipette before transplanting. At 15, 30 and 50 DAT, cell suspension (5 mL per pot) was added to the soil around roots. The control group received an equal volume of 1 × PBS. Fruit samples from three pots (one plant per pot) were collected as one biological replicate, and three biological replicates of each treatment were included for determination of vitamin contents.
Construction of L. soli RpoN mutant
A RpoN knockout mutant of L. soli P18 was constructed by homologous recombination using the sucrose lethal factor70. First, upstream and downstream homology arms were obtained using the primer pair RpoN-F1/R1 and RpoN-F2/R2 (Supplementary Table 3 and 6). All oligonucleotide primers were designed based on the genome sequence of L. soli P18 (GenBank accession PRJNA1177803) using Primer Premier 6.0 software. The primers were synthesised by Sangon Biotech (Shanghai, China). These two fragments were purified and inserted into the suicide vector pEX1871, which was subsequently transformed into Escherichia coli DH5α (Carlsbad, CA, USA, D2500-02). The recombinant vector was introduced into the wild-type strain of L. soli P18 by triparental hybridisation using the helper plasmid pRK201372. Positive clones were screened on Luria-Bertani (LB) agar plates containing kanamycin (100 μg/mL) and gentamicin (150 μg/ mL). Positive colonies with single exchange success were inoculated on LB agar plates containing 10% (w/v) sucrose and kanamycin (100 μg/mL) to screen double exchange mutants. PCR validation and sequencing of the RpoN knockout mutant were performed using the primer pair RpoN-F1 and RpoN-R2. The mutant strain with correct results was designated P18ΔRpoN. To obtain the complemented strain, DNA fragments containing the gene coding region and upstream promoter region were amplified by PCR and cloned into the wide host vector pBBR73. Positive clones were introduced into P18ΔRpoN by electroporation, and positive transformants were screened on LB agar plates containing gentamicin (150 μg/ mL).
Bacterial growth and motility assays
The seed solutions of L. soli strains (P18, ΔRpoN, ΔRpoN-C, 1 × 108 CFU mL−1) were prepared using BEP medium as described for the tomato growth test. A 10-μL aliquot of the seed solution was inoculated to 100 mL of BEP medium. The treatment group was added with 5 mL of flavonoid solution to give the final concentration of 1 µmol g−1 (treatment C). An equal volume of sterile 50-fold diluted 85% methanol was added to the control group. All cultures were incubated at 36 °C shaken at 160 rpm. At the start of incubation, samples were taken to measure absorbance at 540 nm. During the incubation period, absorbance measurements were performed every 2 or 6 h, with a total of 12 times. Six biological replicates were tested for each treatment.
The effect of flavonoids (treatment C: 1 µmol g−1) on the motility of L. soli strains (P18, ΔRpoN, ΔRpoN-C) was determined following the method of Rashid and Kornberg74 with minor modifications. Briefly, for swimming and swarming motility assays, 1 μL of seed solution was dispensed at the centre of agar plates containing 10 g L−1 peptone, 5 g L−1 NaCl and 1 µmol g−1 flavonoids with 0.3% and 0.5% agar, respectively. For twitching motility assay, 1 μL of seed solution was spotted into holes punctured by a sterile pipette tip in agar plates containing 10 g L−1 peptone, 5 g L−1 NaCl and 1 µmol g−1 flavonoids with 1.0% agar. After the seed solution was absorbed, the plates for swimming and swarming motility assays were respectively incubated at 30 °C and 37 °C for 12 h. The plates for twitching motility assay were incubated at 30 °C for 24 h followed by air-drying and staining with 0.1% crystal violet for 20 min. Diameters of the swimming and swarming halos and the stained area were measured with a vernier caliper. Ten biological replicates were tested for each assay.
Root colonisation assay
Tomato seedlings aged 35 days were transplanted to plastic pots (90 mm × 130 mm × 100 mm) containing 5 kg of sterilised soil with an initial moisture content of 100% (v/w). A 5-mL aliquot of cell suspension from overnight cultures of L. soli strains (P18, ΔRpoN, ΔRpoN-C; 1.0 × 108 CFU mL−1) was uniformly blended into the soil in each pot before transplanting. At 3 DAT, flavonoids (treatment C: 1 µmol g−1) were applied to wild-type Micro-Tom plants as described for the flavonoid addition experiment. In the control groups, 50-fold diluted 85% methanol was added to wild-type and SlMYB12-overexpressing plants. All pots were incubated in a moisture chamber as described for the flavonoid addition experiment and watered weekly with sterile water. Rhizosphere soil samples were collected at 60 DAT and the effective viable count of L. soli strains (P18, ΔRpoN, ΔRpoN-C) was estimated by spread plate technique. Samples from two pots (one plant per pot) were collected as one biological replicate, with 10 biological replicates per treatment.
Spermidine addition experiment
Wild-type Micro-Tom seedlings aged 35 days were transplanted to plastic pots (90 mm × 130 mm × 100 mm) containing 5 kg sterilised soil with an initial moisture content of 100% (v/w). At 15 DAT, spermidine solution (1 µmol g−1) was added to the soil with an equal volume of sterile w ater as the control, and pots were managed as described for the flavonoid addition experiment. each containing one plant as a biological replicate (three biological replicates per treatment). The samples were used for detecting vitamins, conducting RNA-seq, and performing widely targeted metabolomic profiling.
Fruit vitamin and bacterial spermidine detection
The content of vitamin C (ascorbic and dehydroascorbic acids) in tomato fruits was determined using a validated hydrophilic interaction chromatography (HILIC) method75. The determination of vitamin B6 (pyridoxine, pyridoxal and pyridoxamine) in tomato fruits was performed by validated high performance liquid chromatography (HPLC) assay76. Validated HILIC assay was used to measure the spermidine content in overnight cultures of L. soli strains (P18, ΔRpoN, ΔRpoN-C)77. The analytical details are provided in Supplementary Method 7.
Bacterial and fruit transcriptomic analysis
Single colonies of L. soli P18 were transferred to fresh BEP medium with and without flavonoids (treatment C: 1 µmol g−1) and cultured for 16 h. Samples from three fresh BEP mediums were collected as one biological replicate, with three biological replicates per treatment. Total bacterial RNA was extracted from the culture sample using the RNeasy Protect Bacteria Mini Kit (Qiagen GmbH), and aliquots containing 1 μg of RNA were used for library preparation with the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs), as per the manufacturer’s instructions. Libraries were generated and sequenced at Majorbio Bio-Pharm Technology Co., Ltd. (Shanghai, China). The raw data were pre-processed using Trimmomatic v0.3978. Subsequently, the clean reads were mapped to the L. soli P18 genome using HISAT v2.1.079 with default parameters. The number of reads mapped to each gene was calculated with the htseq-count script in HTSeq v0.5.480. DEGs were identified using R ‘EBSeq’ package81 (fold-change ≥1.3, adjusted P-value < 0.05 for two-sided Student’s t-test) and subjected to KEGG pathway enrichment analysis based on the KEGG database. Three biological replicates of each sample were tested.
Total RNA extraction from tomato fruits was performed using TRIzol® Reagent (Thermo Fisher Scientific, Waltham, MA, USA). The extracted RNA was checked for quality using an Agilent 5300 Bioanalyser (Agilent Technologies) and quantified using ND-2000 (NanoDrop Technologies). RNA purification, reverse transcription, library construction and sequencing were performed at Majorbio Bio-pharm Biotechnology Co., Ltd (Supplementary Method 8). The raw reads were trimmed and quality filtered using fastp82 with default parameters. Then, the clean reads were aligned to the reference genome with an orientation mode using HISAT283. The mapped reads of each sample were assembled by StringTie84 using a reference-based approach. Gene expression levels were estimated based on the fragments per kilobase of transcript per million fragments mapped method85. Multivariate analysis of gene expression profiles was conducted using PCA implemented in R v4.3.265. The EBSeq R package81 was used for differential gene expression analysis (fold-change ≥1.6 and adjusted P-value < 0.05 for two-sided Student’s t-test). KEGG pathway enrichment analysis of DEGs was performed using scipy in Python v3.11.
Bacterial and fruit widely targeted metabolomic analysis
Single colonies of L. soli. P18 were cultured in BEP medium with and without flavonoids (treatment C: 1 µmol g−1) for 16 h. Samples from three fresh BEP mediums were collected as one biological replicate, with three biological replicates per treatment. Fruit samples from three pots (one plant per pot) were collected as one biological replicate, with three biological replicates per treatment. Metabolite extraction from bacterial cultures and tomato fruits was performed with a solvent containing methanol, acetonitrile and water (2:2:1, v/v/v)86. Briefly, a 100-mL culture or 100-mg fruit sample was mixed with 800 µL of the extraction solvent by vortexing for 30 s and then homogenised in a grinding mill at 60 Hz for 4 min. The mixture was ultrasonically shaken in an ice-water bath for 10 min and incubated at -20 °C for 2 h, followed by centrifugation at 12,000 × g for 15 min. The supernatant (600 µL) was vacuum concentrated and then re-dissolved in 50 µL of acetonitrile-water mixture (1:1, v/v).
The sample extracts were analysed by widely targeted metabolomics based on ultra-performance liquid chromatography–electrospray ionisation–tandem mass spectrometry (UPLC–ESI–MS/MS; Metware Biotechnology Co., ltd., Wuhan, Hubei Province, China). This technique integrates the high resolution and wide coverage untargeted MS with the high sensitivity and quantitative accuracy of targeted MRM assay. Detailed procedures are provided in Supplementary Method 9.
Identification of metabolites was based on the accurate precursor ions, product ions, retention times and fragmentation patterns of primary and secondary mass spectra. Three levels of metabolite annotation were set based on previous studies87,88,89. MultiQuant v3.0.2 (AB Sciex) was used for MRM-based quantification of metabolites. The signal intensities of characteristic ions were obtained for each metabolite in the samples, and their chromatographic peak areas were integrated to indicate the metabolite’s relative abundance. The peak areas were corrected with retention time and peak type to facilitate quantitative comparisons of metabolites between samples. PCA was performed to analyse the differences in metabolite composition using R v4.3.265. DAMs were identified based on fold-change ≥1.5 and P < 0.05 (two-sided Student’s t-test). Functional pathway enrichment analysis of DAMs was performed using the KEGG database (https://www.genome.jp/kegg/).
Statistical analysis
All statistical analyses were performed using Excel 2021 (Microsoft Corp., Redmond, WA, USA), R v4.3.2 (R Core Team, R Foundation for Statistical Computing, Vienna, Austria), Origin v2024 (OriginLab Corp., Northampton, MA, USA) and SPSS Statistics 21.0 (IBM, Chicago, IL). One-way ANOVA with correction by Tukey’s HSD test and two-sided Student’s t-test were used to compare group means, with P-value < 0.05 considered significant. Mantel’s correlations between the metabolite profile of L. soli P18 and fruit vitamin composition were analysed based on Euclidean distance matrices using the R package ‘vegan’90. Pearson’s correlation coefficients were used to assess the relationship between key rhizosphere bacteria and fruit vitamin accumulation. Random forest analysis was performed using the R ‘RandomForest’ package to identify the most important metabolite predictors for fruit vitamin accumulation91. Co-expression network analysis was conducted using R v4.3.2 and network visualisation was achieved using Gephi v0.10.1 (Web Atlas, Paris, France). The experimental flow chart and the schematic presentation of the mechanism for this study were created using Biorender (https://app.biorender.com/).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Raw 16S rRNA gene amplicon sequencing and metagenomic data generated in this study have been deposited in the NCBI under BioProject PRJNA1303084 and PRJNA1182265, respectively. The genomic data for strains P18 generated in this study have been deposited in the NCBI under BioProject PRJNA1177803. The transcriptome data have been deposited in NCBI Sequence Read Archive database under BioProject PRJNA1182113 (bacteria) and PRJNA1182117 (tomato fruit). The flavonoid-targeted metabolomic and widely targeted metabolomic data have been deposited in the MetaboLights under MetaboLights accession MTBLS13507 and MTBLS13506, respectively. Source data are provided with a paper. Source data are provided with this paper.
References
Sato, S. et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 7400 (2012).
Zhu, G. et al. Rewiring of the fruit metabolome in tomato breeding. Cell 172, 249–261 (2018).
Slimestad, R. & Verheul, M. Review of flavonoids and other phenolics from fruits of different tomato (Lycopersicon esculentum Mill.) cultivars. J. Sci. Food Agric. 89, 1255–1270 (2009).
Mooney, S., Leuendorf, J. E., Hendrickson, C. & Hellmann, H. Vitamin B6: a long known compound of surprising complexity. Molecules 14, 329–351 (2009).
Spinneker, A. et al. Vitamin B6 status, deficiency and its consequences-an overview. Nutrición Hospitalaria 22, 7–24 (2007).
Capanoglu, E., Beekwilder, J., Boyacioglu, D., De Vos, R. C. H. & Hall, R. D. The effect of industrial food processing on potentially health-beneficial tomato antioxidants. Crit. Rev. Food Sci. Nutr. 50, 919–930 (2010).
Ansari, F. A., Ahmad, I. & Pichtel, J. Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10. Appl. Soil. Ecol. 143, 45–54 (2019).
Olanrewaju, O. S., Glick, B. R. & Babalola, O. O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 33, 197 (2017).
Chandran, H., Meena, M. & Swapnil, P. Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustainability 13, 10986 (2021).
Xie, Y., Wright, S., Shen, Y. & Du, L. Bioactive natural products from Lysobacter. Nat. Prod. Rep. 29, 1277–1287 (2012).
Vasilyeva, N. V. et al. Lytic peptidase L5 of Lysobacter sp. XL1 with broad antimicrobial spectrum. J. Mol. Microbiol. Biotechnol. 24, 59–66 (2014).
De Vries, F. T., Griffiths, R. I. & Knight, C. G. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 368, 270–274 (2020).
Sasse, J., Martinoia, E. & Northen, T. Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41 (2018).
Vives-Peris, V., de Ollas, C., Gómez-Cadenas, A. & Pérez-Clemente, R. M. Root exudates: from plant to rhizosphere and beyond. Plant Cell Rep. 39, 3–17 (2020).
Zhao, M. et al. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 44, 613–628 (2021).
Wang, L. et al. Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 10, 233 (2022).
Yu, P. et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 7, 1–16 (2021).
Wu, J. D. et al. Flavones enrich rhizosphere Pseudomonas to enhance nitrogen utilization and secondary root growth in Populus. Nat. commun. 16, 1461 (2025).
He, D. et al. Flavonoid attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. ISME J. 16, 1427–1441 (2022).
Xu, F. Y. et al. Auxin-producing Pseudomonas recruited by root flavonoids increases rice rhizosheath formation through the bacterial histidine kinase under soil drying. Adv. Sci. 12, e00607 (2025).
Wang, S. et al. SlMYB12 regulates flavonol synthesis in three different cherry tomato varieties. Sci. Rep. 8, 1582 (2018).
Igarashi, K. & Kashiwagi, K. Modulation of cellular functions by polyamines. Nat. Rev. Mol. Cell Bio. 1, 440–448 (2000).
Cole, B. J. et al. Genome-wide identification of bacterial plant colonization genes. PLoS Biol. 15, e2002860 (2017).
Wheatley, R. M. & Poole, P. S. Mechanisms of bacterial attachment to roots. FEMS Microbiol. Rev. 42, 448–461 (2018).
Hirsch, P. R., Miller, A. J. & Dennis, P. G. Molecular microbial ecology of the rhizosphere Ch. Do root exudates exert more influence on rhizosphere bacterial community structure than other rhizodeposits? New Jersey: Wiley Press; 229 (2013).
Tian, T., Reverdy, A., She, Q., Sun, B. & Chai, Y. The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep. 12, 160–172 (2020).
Lebeis, S. L. et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015).
Feng, H. et al. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant-growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant. Microbe . 31, 995–1005 (2018).
Tian, T. et al. Sucrose triggers a novel signaling cascade promoting Bacillus subtilis rhizosphere colonization. ISME J. 9, 2723–2737 (2021).
Mahajan, N. C., Mrunalini, K. & Prasad, K. S. K. Soil quality indicators, building soil organic matter and microbial derived inputs to soil organic matter under conservation agriculture ecosystem: a review. J. Curr. Microbiol. Appl. Sci. 8, 1859–1879 (2019).
Chaparro, J. M., Sheflin, A. M., Manter, D. K. & Vivanco, J. M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fert. Soils 48, 489–499 (2012).
Bolanos-Vasquez, M. C. & Warner, D. Effects of Rhizobium tropici, R. etli, and R. leguminosarum bv. phaseoli on nod gene-inducing flavonoids in root exudates of Phaseolus vulgaris. Mol. Plant. Microbe. 10, 339–346 (1997).
Begum, A. A., Leibovitch, S., Migner, P. & Zhang, F. Specific flavonoids induced nod gene expression and pre-activated nod genes of Rhizobium leguminosarum increased pea (Pisum sativum L.) and lentil (Lens culinaris L.) nodulation in controlled growth chamber environments. J. Exp. Bot. 52, 1537–1543 (2001).
Gómez Expósito, R., Postma, J., Raaijmakers, J. M. & De Bruijn, I. Diversity and activity of Lysobacter species from disease suppressive soils. Front. Microbiol. 6, 1243 (2015).
Laborda, P., Ling, J., Chen, X. & Liu, F. ACC deaminase from Lysobacter gummosus OH17 can promote root growth in Oryza sativa nipponbare plants. J. Agric. Food Chem. 66, 3675–3682 (2018).
Ling, J. et al. LbDSF, the Lysobacter brunescens quorum-sensing system diffusible signaling factor, regulates anti-Xanthomonas XSAC biosynthesis, colony morphology, and surface motility. Front. Microbiol. 10, 1230–1230 (2019).
Chen, L. & Liu, Y. The function of root exudates in the root colonization by beneficial soil rhizobacteria. Biology 13, 92 (2024).
Liu, Y. et al. Root colonization by beneficial rhizobacteria. FEMS Microbiol. Rev. 48, fuad066 (2024).
Xia, J., Chen, J., Chen, Y., Qian, G. & Liu, F. Type IV pilus biogenesis genes and their roles in biofilm formation in the biological control agent Lysobacter enzymogenes OH11. Appl. Microbiol. Biotechnol. 102, 833–846 (2018).
Lin, L. et al. A non-flagellated biocontrol bacterium employs a PilZ-PilB complex to provoke twitching motility associated with its predation behavior. Phytopathol. Res. 2, 12 (2020).
Fulano, A. M., Shen, D., Kinoshita, M., Chou, S. & Qian, G. The homologous components of flagellar type III protein apparatus have acquired a novel function to control twitching motility in a non-flagellated biocontrol bacterium. Biomolecules 10, 733 (2020).
Wolucka, B. A. & Van Montagu, M. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J. Biol. Chem. 278, 47483–47490 (2003).
Schulz, S. et al. Elucidation of sigma factor-associated networks in Pseudomonas aeruginosa reveals a modular architecture with limited and function-specific crosstalk. PLoS Pathog. 11, e1004744 (2015).
Lloyd, M. G., Vossler, J. L., Nomura, C. T. & Moffat, J. F. Blocking RpoN reduces virulence of Pseudomonas aeruginosa isolated from cystic fibrosis patients and increases antibiotic sensitivity in a laboratory strain. Sci. Rep. 9, 6677 (2019).
Lundgren, B. R. et al. Utilization of L-glutamate as a preferred or sole nutrient in Pseudomonas aeruginosa PAO1 depends on genes encoding for the enhancer-binding protein AauR, the sigma factor RpoN and the transporter complex AatJQMP. BMC Microbiol 21, 83 (2021).
Lugtenberg, B. & Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 63, 541–556 (2009).
Mendes, R. et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 332, 1097–1100 (2011).
Berendsen, R. L., Pieterse, C. M. J. & Bakker, P. A. H. M. The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012).
Kumar, A., Tripti, Maleva, M., Bruno, L. B. & Rajkumar, M. Synergistic effect of ACC deaminase producing Pseudomonas sp. TR15a and siderophore producing Bacillus aerophilus TR15c for enhanced growth and copper accumulation in Helianthus annuus L. Chemosphere 276, 130038 (2021).
Mustafavi, S. H., Badi, H. N., Sekara, A. & Al, E. Polyamines and their possible mechanisms involved in plant physiological processes and elicitation of secondary metabolites. Acta Physiol. Plant 40, 102 (2018).
Chen, D., Shao, Q., Yin, L., Younis, A. & Zheng, B. Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9, 1945 (2019).
Krysenko, S. & Wohlleben, W. Polyamine and ethanolamine metabolism in bacteria as an important component of nitrogen assimilation for survival and pathogenicity. Med. Sci. 10, 40 (2022).
Wheeler, G. L., Jones, M. A. & Smirnoff, N. The biosynthetic pathway of vitamin C in higher plants. Nature 393, 365–369 (1998).
Agius, F. et al. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 21, 177–181 (2003).
Lorence, A., Chevone, B. I., Mendes, P. & Nessler, C. L. Myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 134, 1200–1205 (2004).
Saito, K. & Matsuda, F. Metabolomics for functional analysis of plant secondary metabolites. Plant J. 61, 306–320 (2010).
Smirnoff, N. Ascorbic acid: physiology and metabolism. : Plant Ascorbic Acid 1, 23 (2000).
Dong, H. et al. Loss of the L-galactose pathway for ascorbic acid biosynthesis in plants. Nat. Plants 1, 15032 (2015).
Zhang, Y. et al. Characterization of a mutant deficient in vitamin B6 biosynthesis in Arabidopsis. J. Exp. Bot. 66, 1647–1660 (2015).
Yang, K. et al. RIN enhances plant disease resistance via root exudate-mediated assembly of disease-suppressive rhizosphere microbiota. Mol. Plant 16, 1379–1395 (2023).
Wen, T. et al. Tapping the rhizosphere metabolites for the prebiotic control of soil-borne bacterial wilt disease. Nat. Commun. 14, 4497–4497 (2023).
Liu, C. et al. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 100, 1421–1426 (2016).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537 (2009).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria (2023).
Buchfink, B., Xie, C. & Huson, D. H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60 (2015).
Jett, B. D., Hatter, K. L., Huycke, M. M. & Gilmore, M. S. Simplified agar plate method for quantifying viable bacteria. Bio. Tech. 23, 648–650 (1997).
Zhang, J. et al. NRT1.1B is associated with root microbiota composition and nitrogen use in field-grown rice. Nat. Biotechnol. 37, 676–684 (2019).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Qian, G. et al. Selection of available suicide vectors for gene mutagenesis using chiA (a chitinase encoding gene) as a new reporter and primary functional analysis of chiA in Lysobacter enzymogenes strain OH11. World J. Microbiol. Biotechnol. 28, 549–557 (2012).
Davidson, M. S. & Summers, A. O. Wide-host-range plasmids function in the genus Thiobacillus. Appl. Environ. Microbiol. 46, 565–572 (1983).
Qian, G. et al. Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616 (2013).
Tao, L., Jackson, R. E. & Cheng, Q. Directed evolution of copy number of a broad host range plasmid for metabolic engineering. Metab. Eng. 7, 10–17 (2005).
Rashid, M. H. & Kornberg, A. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. P. Natl. Acad. Sci. Usa. 97, 4885–4890 (2000).
Barros, A. I. R. N. A., Silva, A. P., Goncalves, B. & Nunes, F. M. A fast, simple, and reliable hydrophilic interaction liquid chromatography method for the determination of ascorbic and isoascorbic acids. Anal. Bioanal. Chem. 396, 1863–1875 (2010).
Islam, M. A. et al. Validation of vitamin B5 (pantothenic acid) and B6 (pyridoxine, pyridoxal, and pyridoxamine) analyses in seafood. J. Food Compos. Anal. 109, 104518 (2022).
Ni, Y. et al. Spermidine ameliorates nonalcoholic steatohepatitis through thyroid hormone-responsive protein signaling and the gut microbiota-mediated metabolism of bile acids. J. Agric. Food Chem. 70, 6478–6492 (2022).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner for RNA-Seq. Bioinformatics 31, 393–400 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-A Python framework to work with high-throughput sequence data. Bioinformatics 31, 166–169 (2015).
Leng, N. et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29, 1035–1043 (2013).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, 884–890 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. methods 12, 357–360 (2015).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L. & Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628 (2008).
Dunn, W. B. et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 6, 1060–1083 (2011).
Chen, W. et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: application in the study of rice metabolomics. Mol. Plant 6, 1769–1780 (2013).
Alseekh, S. et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat. Methods 18, 747–756 (2021).
Liu, P. X. et al. Distinct quality changes of asparagus during growth by widely targeted metabolomics analysis. J. Agric. Food Chem. 70, 15999–16009 (2022).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003).
Liaw, A. & Wiener, M. Classification and regression by random forest. R. N. 2, 18–22 (2002).
Acknowledgements
H.L. and Q.G. were supported by grants from the Special Project of Scientific and Technological Innovation of Xinjiang Research Institute of Arid Area Agriculture (XJHQNY-2025-6). Q.G. was supported by grants from the China National Tobacco Corporation Yunnan Branch Major Science and Technology Programme Special Project (2024530000241022) and the “Scientists + Engineers” Team Project of Xianyang (L2024-CXNL-KJRCT-DWJS-0005). H.L. was supported by grants from the Key Research and Development Programme of Shaanxi Province (S2024-YF-ZDXM-NY-0223; S2024-YF-ZDCXL-ZDLNY-0162).
Author information
Authors and Affiliations
Contributions
W.F. and C.S. designed the experiments. P.L. and W.F. collected and analysed the data. W.F. and B.S. performed the experiments. W.F. interpreted the results and draughted the manuscript. D.X. provided experiment materials. H.L., Q.G. and J.Y. revised the manuscript and provided critical suggestions. H.L. and Q.G. obtained funding and conceived the study. All authors edited and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Mengcen Wang, who co-reviewed with Hongfu Li; Mohammadhossein Ravanbakhsh and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fu, W., Sun, C., Sun, B. et al. Flavonoid-mediated bacterial spermidine biosynthesis enhances vitamin accumulation in tomato fruits. Nat Commun 17, 1524 (2026). https://doi.org/10.1038/s41467-025-68244-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-68244-9