Abstract
Fatty acids are involved in disease risk and aging processes. In the US National Health and Nutrition Examination Survey (1999–2002), we tested for associations of total, saturated (SFA), monounsaturated (MUFA), polyunsaturated (PUFA), and subtypes of dietary fatty acids with DNA methylation-based aging biomarkers, adjusting for age, BMI, total energy intake, and sociodemographic and behavioral factors (N = 2260). Higher SFA and MUFA were associated with greater GrimAge2, an aging biomarker of mortality; PUFA was associated with lower Horvath1, Hannum, and PhenoAge (p < 0.05). Omega-3 and the PUFA:SFA ratio were negatively associated with Horvath1, Hannum, Vidal-Bralo, and PhenoAge. Notably, a one-unit increase in PUFA:SFA was associated with 1.05 years lower PhenoAge (95% CI = −1.87, −0.22). We found consistent positive associations for SFA subtypes and negative associations for PUFA subtypes with epigenetic aging; associations of MUFA subtypes varied. Future studies, including randomized controlled trials, are needed to investigate causality and downstream clinical outcomes.
Similar content being viewed by others
Introduction
As the global population undergoes a demographic shift to a greater proportion of older adults, it is crucial to understand factors that contribute to healthy aging. Beyond genetic determinants, health behaviors throughout life, including physical activity, socializing, and dietary patterns, can help to delay the onset of age-related decline or morbidities and maintain functional longevity1. Diets characterized by high intake of vegetables, fruits, whole grains, plant-based protein, fish, and unsaturated fats have been associated with downregulation of aging biomarkers and greater health in later life2,3. Nutrition may contribute to healthy aging or, conversely, increased risk of disease, by impacting pathways related to inflammation, oxidative stress, DNA repair, regenerative potential, and insulin sensitivity4. The purpose of this study is to examine associations of fatty acid intake with DNA methylation-based aging biomarkers.
As one of the major macronutrient groups, lipids, or fatty acids, are necessary to support health through critical biological roles as energy sources, constituents of cell membranes, regulators of signaling pathways and gene expression, and building blocks of hormones5. Fatty acids are classified based on their hydrocarbon chain length and structure and are grouped into saturated (SFAs), monounsaturated (MUFAs), and polyunsaturated fatty acids (PUFAs). SFAs and MUFAs can be synthesized by humans; however, many PUFAs are essential fatty acids which cannot be synthesized or are limited by being derived from other essential fatty acids. SFAs, found in animal products and tropical oils, upregulate the synthesis of cholesterol, triglycerides, and phospholipids, and increase the expression of genes involved in inflammatory pathways and insulin sensitivity5. MUFAs, found in vegetable oils and some nuts and seeds, may be anti-inflammatory and reduce low-density lipoprotein (LDL) cholesterol levels6. PUFAs, which have high levels in some vegetable oils, nuts, seeds, and fatty fish, include the essential fatty acids omega-6 (ω-6) and omega-3 (ω-3). Omega-6 fatty acids are major constituents of cell membranes and can lower low-density lipoprotein (LDL) cholesterol levels through the downregulation of biosynthesis7. Omega-3 fatty acid subtypes are abundant in the cell membranes of the brain and eye and influence membrane protein signal transduction5. Omega-3 fatty acids in cell membranes are also involved in controlling inflammation, blood clotting, and constriction of blood vessels.
Recent advancements in the field of epigenetics have enabled the study of age-related health risks and the healthspan through proxies, or biomarkers, of biological aging. Epigenetic clocks, calculated from weighted combinations of DNA methylation levels at age-associated loci, have been developed to estimate chronological age (i.e., first generation clocks), phenotypic age (i.e., second generation clocks), and pace of aging8. Epigenetic clocks are valuable in their ability to capture broad changes in physiological pathways, functional decline, chronic diseases, and damage on the cellular and molecular levels. The difference between chronological age and epigenetic age, known as epigenetic age deviation or epigenetic age acceleration, is associated with morbidities and all-cause mortality9,10. Second-generation clocks, in particular, are strong predictors of cardiometabolic diseases, cancer, and time to death11,12,13. Leveraging epigenetic clocks may allow for the identification of factors that increase the risk of age-related decline and disease, as well as factors that promote healthy aging and extend functional longevity. Moreover, epigenetic clocks hold promise as predictive and prognostic biomarkers in clinical settings and to inform individual-level diet and behavioral interventions.
There is increasing evidence linking diet quality and fatty acids to disease risk and aging processes5,14,15,16. Notably, a recent randomized controlled trial found that omega-3 supplementation decreased epigenetic age acceleration and the pace of aging17. In United States (US) National Health and Nutrition Examination Survey (NHANES), the cohort in which the current study is based, omega-3 intake has been associated with decreased acceleration of phenotypic age, a composite of blood chemistry measures18. However, further research is needed to understand how dietary intake of total fatty acids and fatty acid subtypes is related to epigenetic aging. We investigated cross-sectional associations of fatty acid dietary intake and epigenetic aging biomarkers in NHANES, a representative sample of the US adult population. In particular, we tested for associations of total fatty acids, SFAs, MUFAs, PUFAs, omega-6, omega-3, the ratio of PUFA:SFA (P:S), and 19 fatty acid subtypes. Based on research linking fatty acids to molecular pathways involved in chronic disease development and aging, we hypothesized that higher intake of SFAs would be associated with greater epigenetic aging and that higher intake of PUFAs and the P:S ratio would be associated with lower epigenetic aging.
Results
Characteristics of participants included in our primary analyses of complete cases (N = 1771) are shown in Table 1. Included participants were ≥ 50 years old and had a mean (SD) age of 64.8 (9.3) years. Half of participants were male (n = 952; 53.8%). Dietary data, including fatty acid intake, was estimated by NHANES from 24 h dietary recall questionnaires. Participants reported consuming an average of 1860 (764) kcal/day, and the mean (SD) percentage of total energy from total fatty acids, SFAs, MUFAs, and PUFAs was 32.9% (9.3), 10.3% (3.9), 12.1% (4.1), and 7.0% (3.2), respectively. Approximately half of participants (n = 895; 51%) met the US Department of Agriculture’s (USDA) guidelines for saturated fat supplying less than 10% of daily calories19. Although our estimates of fatty acid intake do not include supplements, only 38.1% of male participants and 45.8% of female participants reported meeting the adequate intake levels for omega-3s of 1.6 and 1.1 g, respectively20. Male participants had lower BMI, were more likely to report ever smoking, and had greater alcohol intake compared to female participants (Table 1). Male participants also reported greater energy intake (mean (SD) = 2090 (822) kcal/day) than female participants (mean (SD) = 1600 (589) kcal/day) and greater fatty acid intake, although the percentage of total energy from fatty acids was similar between sexes.
Characteristics of participants included in imputation analyses (N = 2220) are summarized in Supplemental Table S1 and were similar to those of participants in the primary analyses. Means and SDs of fatty acid subtypes are shown in Supplemental Table S2.
Survey-weighted correlations of the intake of total fatty acids and fatty acid subtypes are shown in Supplementary Figs. S2 and S3. As expected, there were moderate-to-strong correlations between the intake of total fatty acids, SFAs, MUFAs, and PUFAs (r = 0.54–0.97; p < 0.001). The P:S ratio was negatively correlated with total fatty acids, SFAs, and MUFAs (r = −0.06, −0.35, and −0.09, respectively; p < 0.05), and positively correlated with PUFAs (r = 0.43; p < 0.01).
In Supplemental Table S3, we summarize DNA methylation biomarkers of aging analyzed in this study. Scatter plots of chronological age and each of the epigenetic aging biomarkers analyzed are shown in Supplementary Fig. S4. Epigenetic age estimates were strongly correlated with chronological age (r-range = 0.62–0.91). DNAmTL was negatively correlated (r = −0.59), epiTOC was positively correlated (r = 0.22), and DunedinPoAm had a weak correlation with chronological age (r = 0.06).
Associations of total fatty acid intakes with epigenetic aging biomarkers
We evaluated the associations of fatty acid intake with epigenetic aging biomarkers using adjusted survey-weighted linear models, adjusting for age, age2, sex, race and ethnicity, BMI, education level, occupation, poverty to income ratio, smoking status, alcohol intake, physical activity, and total energy intake. Fatty acid intake variables were log2 transformed prior to analysis.
Total fatty acid intake was not significantly associated with any epigenetic aging biomarker. However, a log2 increase, or doubling, of SFA and MUFA intake was associated with 0.42 years (95% CI = 0.02, 0.83) and 0.54 years (95% CI = 0.04, 1.04) greater GrimAge2, respectively (Fig. 1; effect sizes, 95% confidence intervals (CIs), and p-values are shown in Supplementary Table S4). Conversely, a doubling in PUFA intake was associated with 0.68 years lower Horvath1 (95% CI = −1.26, −0.09), 0.60 years lower Hannum (95% CI = −1.15, −0.06), and 0.59 years lower PhenoAge (95% CI = −1.14, −0.03). We did not observe any significant associations of fatty acid intake with DNAmTL, epiTOC, or DunedinPoAm.
Effect estimates (95% confidence intervals (CIs)) are shown for a log2 increase, or doubling, of fatty acid intake. Units are in years for the Horvath1, Horvath2, Hannum, Lin, Zhang, Vidal-Bralo, PhenoAge, and GrimAge2 clocks; kilobases for DNAmTL, and standard deviations for epiTOC and DunedinPoAm. Results are from weighted generalized linear regression models adjusted for age, age2, sex, race and ethnicity, BMI, education level, occupation, poverty to income ratio, smoking status, alcohol intake, physical activity, and total energy intake. Significant associations (p < 0.05) are plotted in red.
Associations of omega-6 and omega-3 with epigenetic aging biomarkers
Across most first- and second-generation epigenetic clocks, there was a trend toward negative associations of omega-6 and omega-3 intake with epigenetic aging (Fig. 2 and Supplementary Table S4). Intake of omega-6 was significantly associated with lower epigenetic aging as measured by the Horvath1 and Hannum clocks, while omega-3 intake was significantly associated with lower epigenetic aging as measured by Horvath1, Hannum, Vidal-Bralo, and PhenoAge clocks (p < 0.05). The greatest effect size was observed for omega-3 intake and PhenoAge, with a doubling of omega-3 intake associated with 0.77 years lower PhenoAge (95% CI = −1.33, −0.21).
Effect estimates (95% confidence intervals (CIs)) are shown for a log2 increase, or doubling, of omega-6 and omega-3 intake and for a one-unit increase in the P:S ratio, which approximates the interquartile range. Units are in years for the Horvath1, Horvath2, Hannum, Lin, Zhang, Vidal-Bralo, PhenoAge, and GrimAge2 clocks; kilobases for DNAmTL, and standard deviations for epiTOC and DunedinPoAm. Results are from weighted generalized linear regression models adjusted for age, age2, sex, race and ethnicity, BMI, education level, occupation, poverty to income ratio, smoking status, alcohol intake, physical activity, and total energy intake. Significant associations (p < 0.05) are plotted in red.
Considering that omega-6 and omega-3 were highly correlated (r = 0.79, Supplementary Fig. S2), we conducted analyses simultaneously modeling both variables. The variance inflation factor (VIF) indicated low-to-moderate multicollinearity, with values ranging from 2.9–3.9 and 2.2–3.4 for omega-6 and omega-3, respectively, across the epigenetic aging biomarker models. In jointly adjusted models, omega-6 intake was not significantly associated with any epigenetic aging biomarker (Supplementary Table S5). However, the associations of omega-3 intake with Vidal-Bralo and PhenoAge remained significant (B (95% CI) = −0.57 years (−1.13, −0.01) and −0.84 years (−1.64, −0.05), respectively).
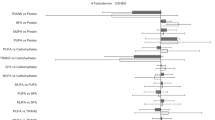
Associations of the ratio of polyunsaturated:saturated fatty acids with epigenetic aging biomarkers
The P:S ratio was significantly associated with lower epigenetic aging as measured by three first-generation clocks and PhenoAge (Fig. 2 and Supplemental Table S4). A one-unit increase in the P:S ratio was associated with 0.87 years lower Horvath1 (95% CI = −1.61, −0.31), 0.89 years lower Hannum (95% CI = −1.62, −0.16), and 0.80 years lower Vidal-Bralo age (95% CI = −1.56, −0.04). However, like the associations with omega-3 intake, the greatest association was with PhenoAge (B (95% CI) = −1.02 years per unit increase (−1.80, −0.25)). A one-unit difference in the P:S ratio approximates the interquartile range (IQR) of 0.92. The associations of the P:S ratio with the Horvath2, Lin, Zhang, and GrimAge2 clocks and with DunedinPoAm were negative but not statistically significant.
Associations of fatty acid subtypes with epigenetic aging biomarkers
In analyses of fatty acid subtypes, we found consistent trends in associations across the epigenetic aging biomarkers (Fig. 3 and Supplemental Table S6). Notably, all PUFA subtypes had negative directions of association with first-generation clocks, PhenoAge, and epiTOC, and positive directions of association with DNAmTL. PUFA 22:6 (DHA) was significantly associated with the greatest number of epigenetic aging biomarkers: Horvath1, Horvath2, Hannum, Zhang, PhenoAge, and DNAmTL (p < 0.05). The association of PUFA 18:3 (ALA) with PhenoAge had the greatest effect size (B (95% CI) = −0.69 years (−1.31, 0.06)).
Results are from weighted generalized linear regression models adjusted for age, age2, sex, race and ethnicity, BMI, education level, occupation, poverty to income ratio, smoking status, alcohol intake, physical activity, and total energy intake. Effect estimates per standard deviation increase in epigenetic aging biomarkers for a log2 increase, or doubling, of each fatty acid subtype are represented by tile shading. *p < 0.05; **p < 0.01.
Two SFA subtypes (SFA 16:0 and SFA 18:0) had significant positive associations with GrimAge2 (B (95% CI) = 0.55 years (0.07, 1.04) and 0.43 years (0.06, 0.80), respectively). The directions of association of MUFA subtypes with epigenetic aging biomarkers varied; however, MUFA 16:1 was significantly associated with greater Horvath2 and DunedinPoAm (B (95% CI) = 0.26 years (0.02, 0.49) and 0.08 SDs (0.01, 0.15), respectively).
Effect modification by sex
Sex hormones may affect lipid metabolism21, and epigenetic aging has been shown to differ by biological sex22,23. Therefore, we also conducted analyses including a sex × fatty acid interaction term. We did not find evidence of effect modification for the associations of intake of fatty acids or the P:S ratio with epigenetic aging biomarkers (all pinteraction > 0.05). Interaction p-values and sex-stratified analyses are presented in Supplemental Tables S7 and S8.
Sensitivity analyses
We conducted sensitivity analyses using imputed covariate data (N = 2220). The directions of association and effect sizes were largely consistent with those from analyses of complete cases (Supplemental Table S9). Associations of SFA and MUFA intake with GrimAge2 remained positive but were not statistically significant (B (95% CI) = 0.39 years (−0.03, 0.82) and 0.40 years (−0.05, 0.86), respectively). Similarly, associations of omega-6 intake with Horvath1 and Hannum remained negative but were not statistically significant (B (95% CI) = −0.53 years (−1.08, 0.02) and −0.46 years (−0.98, 0.06), respectively), as did the association of omega-3 intake with Vidal-Bralo (B (95% CI) = −0.47 years (−0.98, 0.04)) and associations of the P:S ratio with Hannum and Vidal-Bralo (B (95% CI) = −0.72 years (−1.46, 0.01) and −0.70 years (−1.41, 0.02), respectively). However, higher omega-3 intake was significantly associated with lower Horvath2 (B (95% CI) = −0.37 (−0.73, −0.02)) and greater DNAmTL (B (95% CI) = 0.02 (0.00, 0.03)). Overall, analyses of associations with fatty acid subtypes using imputed covariate data were consistent with our primary analyses (Supplemental Table S10).
Discussion
Using a nationally representative sample of US adults older than 50, we investigated the associations of fatty acid intake with epigenetic aging biomarkers, which are measures of biological aging. This study builds on previous research linking diet quality, macronutrients, and micronutrients with epigenetic aging24,25,26,27. Across multiple epigenetic clocks, we found that higher intake of total PUFAs, omega-6, and omega-3, and the P:S ratio was associated with lower epigenetic aging after adjusting for BMI, total energy intake, demographics, and behavioral and lifestyle factors. Conversely, higher intake of SFAs and MUFAs was associated with increased GrimAge2, a strong biomarker of mortality risk. Furthermore, analyses of fatty acid subtypes showed distinct patterns of association: PUFA subtypes were negatively associated with epigenetic aging, SFA subtypes were positively associated with epigenetic aging, and associations of MUFA subtypes varied by aging biomarker. Overall, associations with DNAmTL, epiTOC, and DunedinPoAm were null; however, we did observe several significant associations of fatty acid subtypes with these biomarkers.
The most consistent and robust associations were observed for PUFAs. Although omega-3 fatty acids constitute a small proportion of total PUFA intake (mean omega-3 = 1.4 g/day; mean PUFA = 13.0 g/day in our sample), these fatty acids appeared to drive many of the observed associations. Higher omega-3 intake was associated with significantly lower epigenetic aging as measured by the Horvath1, Hannum, Vidal-Bralo, and PhenoAge clocks. For example, a participant with omega-3 intake of 1.4 g/day, or approximately the recommended adequate intake level20, had on average 0.77 years lower PhenoAge than a participant with intake of 0.7 g/day, the 25th percentile in our study. We also found negative but not statistically significant associations with the Horvath2, Lin, Zhang, and GrimAge2 clocks, and a positive association with DNAmTL. Although it is difficult to distinguish between the effects of total PUFA, omega-6, and omega-3 intake due to the high correlation of these variables, the hypothesis that omega-3 fatty acids have the greatest contribution to associations with lower epigenetic aging is supported by results from models jointly including both omega-6 and omega-3 intake, as well as analyses of fatty acid subtypes. In jointly adjusted models, omega-3 intake was significantly associated with lower Vidal-Bralo and PhenoAge, whereas associations of omega-6 intake were null. In addition, three omega-3 subtypes (PUFA 18:3 (ALA), PUFA 22:5 (DPA), and PUFA 22:6 (DHA)) were significantly and negatively associated with PhenoAge; associations of omega-6 subtypes with PhenoAge were not statistically significant.
Omega-3 intake may act to slow age-related decline through pathways including regulation of lipid metabolism and inflammation via activation of peroxisome proliferator-activated receptors (PPAR)16, regulation of immune responses such as cytokine production28, and maintenance of DHA in brain cell membranes29. There has been particular interest in the effects of omega-3 fatty acids in reducing cardiometabolic risks. Higher omega-3 intake has been associated with decreased triglyceride levels30 and lower risk of heart failure31, coronary heart disease32, myocardial infarction33, ischemic events34, and all-cause mortality35; although other trials and meta-analyses have reported null associations with cardiovascular diseases36,37 or effects limited to omega-3 fatty acids from marine sources35. However, our results and previous studies support the hypothesis of health-promoting effects of omega-3 intake through impacts on biological aging. In NHANES (1999–2018), higher omega-3 intake was associated with lower Phenotypic Age acceleration, a composite blood-based biomarker18. Self-reported omega-3 intake has also been associated with lower GrimAge1 in the Framingham Heart Study12. Moreover, in a recent randomized controlled trial among older adults of 1 g/day omega-3 supplementation, 2000 IU/day vitamin D, and/or exercise (N = 777), participants receiving omega-3 had significantly lower epigenetic aging as measured by PhenoAge, GrimAge2, and DunedinPACE compared to the placebo group17. The greatest effect was observed among participants receiving all three interventions with lower PhenoAge.
Considering the opposing associations of SFAs and PUFAs with the regulation of LDL cholesterol levels and chronic disease risks, we also tested for associations of the P:S ratio with epigenetic aging. A higher P:S ratio was significantly associated with lower epigenetic aging as measured by Horvath1, Hannum, Vidal-Bralo, and PhenoAge. Notably, a one-unit increase in the P:S ratio, which approximates the IQR, was associated with 1.05 years lower PhenoAge. These results suggest that the P:S ratio may be a more informative indicator of disease-related fatty acid status than SFAs or PUFAs alone. Higher P:S ratios have previously been associated with lower LDL cholesterol38. The P:S ratio may also contribute to age-related disease risk through its effect on prandial, or post-meal, levels of blood lipids. A cross-over study of meals with varying P:S ratios found a greater increase in LDL cholesterol following low P:S ratio meals compared to meals with a P:S ratio ≥ 139.
Although we found only two statistically significant associations of MUFA subtypes with epigenetic aging biomarkers (MUFA 16:1 associated with greater Horvath2 and DunedinPoAm), there were notable trends in associations across MUFA subtypes. Most associations of MUFA 16:1 indicated increased epigenetic aging, associations of MUFA 18:1 were varied, and associations of MUFA 20:1 indicated decreased epigenetic aging. This heterogeneity in associations may be due to variation in food sources and prevalence in the US diet. MUFA 16:1 (hexadecenoic acid), obtained from animal and plant food sources, and MUFA 20:1 (eicosenoic acid), found in small amounts in plant oils, constituted a small proportion of total MUFA intake in our sample (mean = 1.19 and 0.18 g/day, respectively). However, the majority of MUFA intake was in the form of MUFA 18:1 (octadecenoic acid; mean = 24.0 g/day). MUFA 18:1 is common in olive and other vegetable oils, with the top food sources among NHANES participants (2005–2006) being desserts, chicken dishes, processed meats, and nuts/seeds40.
Overall, total fatty acid, SFA, and MUFA intake were not significantly associated with epigenetic aging. However, we did find significant positive associations of SFA and MUFA intake with GrimAge2. Among epigenetic aging biomarkers, GrimAge2 may be particularly sensitive to fatty acids with obesogenic or proinflammatory properties. GrimAge2 is calculated as a composite of sex, age, and DNA methylation surrogates for smoking pack years and nine plasma proteins, including markers of appetite regulation (leptin), insulin sensitivity (hemoglobin A1C), and inflammation (C-reactive protein (CRP), beta-2 microglobulin (B2M), and growth differentiation factor 15 (GDF15)). Feeding trials in humans have found that diets high in saturated fat are associated with increased expression of genes involved in inflammatory pathways41 and insulin resistance42. Unexpectedly, with the exception of MUFA 16:1, we did not observe significant associations of fatty acid intake with DunedinPoAm. DunedinPoAm was trained on longitudinal changes in 18 biomarkers from adults aged 26–38, including lipoprotein(a), triglycerides, total cholesterol, and HDL cholesterol43. Although DunedinPoAm has been associated with physical and cognitive decline and overall mortality, it may be less sensitive to variation in individual nutritional factors, cross-sectional associations, and in older populations.
Overall, our study included eleven epigenetic aging biomarkers available for NHANES: six first-generation clocks, four second-generation clocks (including predictors of telomere length and cell divisions), and the pace of aging. We found that PUFA and PUFA subtype intake and the P:S ratio were associated with lower epigenetic aging as measured by the Horvath144 and Hannum45 first-generation clocks, seminal epigenetic clocks that have been associated with a broad range of health outcomes and dietary, behavioral, and environmental factors46,47. However, we also identified significant associations of PUFA subtypes or the P:S ratio with the newer, and less widely used, Zhang48 and Vidal-Bralo49 clocks. The Zhang clock relied on a large training set (>13,000 samples) to increase precision48 and demonstrated the strongest correlation with chronological age in our NHANES sample (r = 0.91). To our knowledge, and as summarized in a recent systematic review, the Zhang clock has not previously been associated with dietary, lifestyle, or behavioral factors46. The Vidal-Bralo clock, which is based on eight CpGs, was developed for use with low-cost DNA methylation assays49, and found to be independent of smoking status49,50. Although this clock has previously been associated with physical activity in NHANES51, its application has been limited. Our results demonstrate that a diversity of epigenetic clocks may be sensitive to modifiable factors such as diet.
Our study was strengthened by the use of a large, nationally representative sample of US adults. We were able to analyze the relationship of fatty acid subtypes with multiple epigenetic aging biomarkers. Consistency of associations across these measures supports the robustness of our findings. However, our study was also limited by its cross-sectional design. Longitudinal analyses are needed to evaluate the association of long-term fatty acid intake, changes in epigenetic aging, and downstream health outcomes. We were also limited by estimates of fatty acid intake based on a 1-day dietary recall, which may not fully capture long-term dietary patterns and is vulnerable to misreporting and recall error. Fatty acid intake estimates also excluded supplement use, which may result in underestimates particularly for omega-3 intake. Furthermore, epigenetic aging biomarker data are only available in NHANES for participants aged ≥ 50 years of age in the 1999–2002 cycles. Therefore, we were not able to evaluate the relationship between fatty acid intake and epigenetic aging in younger populations or more recent NHANES cycles, which may have distinct or shifting dietary patterns. Generalizability may also be impacted by limitations of epigenetic clocks, which may produce biased estimates when applied to diverse populations52. In addition, covariate data were missing for a portion of our sample. While our primary analyses used complete cases, analyses using imputed data were largely consistent. Finally, significance was evaluated at a nominal p-value < 0.05. We did not correct for multiple testing and acknowledge that some of our findings may be due to chance. The use of nominal p-values was due in part to our focus on overall trends in associations, and in part to the use of survey weighting, which decreases statistical power by inflating the variance of parameter estimates53. Interpretation of our results, therefore, should focus on the precision of effect sizes.
To conclude, in this study, we provide evidence that fatty acid intake is associated with epigenetic aging biomarkers. We observed the strongest and most robust associations of higher intake of PUFAs, particularly omega-3, an essential fatty acid found in fatty fish, seeds, and nuts, and the P:S ratio with lower epigenetic aging across multiple clocks. These findings contribute to the growing body of research linking dietary factors to biomarkers of longevity and may inform dietary interventions and the potential utility of epigenetic clocks in clinical settings. Randomized clinical trials are needed to establish causality in the context of various dietary patterns. In addition, future studies should focus on overall dietary patterns, in addition to individual macro- or micronutrients, and include long-term follow-up to identify downstream health outcomes associated with changes in epigenetic aging.
Methods
Study population
Analyses were conducted using data obtained from the NHANES database. NHANES is conducted by the US National Center for Health Statistics (NCHS) to evaluate and monitor health and nutrition among noninstitutionalized individuals in the US, and includes interviews, physical exams, and laboratory measurements. Our study included the 1999–2000 and 2001–2002 survey cycles due to the availability of cross-section dietary and DNA methylation data. Dietary data and blood samples used to measure epigenetic aging biomarkers were collected at the same study visit. Study protocols were approved by the NCHS Research Ethics Review Board, which ensures protection of human subjects consistent with the Declaration of Helsinki, and all participants provided written informed consent.
Epigenetic age estimates were available for 2532 NHANES participants. The ages of participants older than 84 years were top coded as 85 to maintain confidentiality. Participants aged ≥ 85 years old (n = 130) were excluded from our analyses to prevent bias from underestimation of chronological age for older adults. We excluded participants with a mismatch between recorded sex and a DNA methylation-based sex estimate (n = 56). We further excluded 69 participants missing data dietary questionnaire data, 17 participants with unreliable dietary data as classified by NHANES, and 40 participants with extreme total energy intake ( < 500 or > 5000 kcal/day for female participants [n = 27] and < 500 or > 8000 kcal/day for male participants [n = 13]). Our primary analyses included 1771 participants with complete epigenetic age, fatty acid intake, and covariate data, and sensitivity analyses included 2220 with complete epigenetic age and fatty acid intake data (Supplementary Fig. S1).
Fatty acid intake
Total nutrient intakes are included in the NHANES database and were estimated from 24 h dietary recall questionnaires. Methods and data can be accessed on the NHANES website54,55. In 1999–2001, the questionnaire was implemented using the NHANES computer-assisted dietary interview system (CADI). In 2002, the questionnaire was implemented as part of an integration of NHANES and the US Department of Agriculture’s (USDA’s) Continuing Survey of Food Intakes by Individuals (CSFI) using the USDA Automated Multiple Pass Method (AMPM). The 2002 data collection included a 2-day dietary recall; estimated dietary intakes released by NHANES were based on the day-1 recall. The limited release of 2002 data was due to confidentiality concerns; however, restricting analyses to the day-1 recall also ensures consistency across years included in our study. Total energy and nutrient intake per day were calculated by NHANES based on USDA codes assigned to individual foods. This study used data on estimated daily intake in g of total fatty acids, saturated fatty acids (SFA), monounsaturated fatty acids (MUFA), polyunsaturated fatty acids (PUFA), and the following fatty acid subtypes
-
SFA 4:0 (butanoic acid)
-
SFA 6:0 (hexanoic acid)
-
SFA 8:0 (octanoic acid)
-
SFA 10:0 (decanoic acid)
-
SFA 12:0 (dodecanoic acid)
-
SFA 14:0 (tetradecanoic acid)
-
SFA 16:0 (palmitic acid)
-
SFA 18:0 (octadecanoic acid)
-
MUFA 16:1 (hexadecenoic acid)
-
MUFA 18:1 (octadecenoic acid)
-
MUFA 20:1 (eicosenoic acid)
-
MUFA 22:1 (docosenoic acid)
-
PUFA 18:2 (octadecadienoic acid, also known as linoleic acid (LA))
-
PUFA 18:3 (octadecatrienoic acid, also known as alpha-linolenic acid (ALA))
-
PUFA 18:4 (octadecatetraenoic acid, also known as stearidonic acid (SDA))
-
PUFA 20:4 (eicosatetraenoic acid (ETA))
-
PUFA 20:5 (eicosapentaenoic acid (EPA))
-
PUFA 22:5 (docosapentaenoic acid (DPA))
-
PUFA 22:6 (docosahexaenoic acid (DHA)).
Total omega-6 fatty acid intake was calculated as the sum of PUFA 18:2 and PUFA 20:4, and total omega-3 fatty acid intake was calculated as the sum of PUFA 18:3, PUFA 18:4, PUFA 20:5, PUFA 22:5, and PUFA 22:656,57. The PUFA:SFA (P:S) ratio was calculated by dividing PUFA by SFA.
DNA methylation
DNA methylation was measured in a subset of adults from the 1999-2000 and 2001–2002 survey cycles. Laboratory methods and data processing are described in detail on the NHANES website58. Briefly, DNA was extracted from whole blood and stored at −80 °C until DNA methylation measurement. Leukocyte DNA methylation was measured in samples from 2532 participants aged ≥ 50 years with available biospecimens. Approximately half of all eligible non-Hispanic White participants were randomly selected, as well as all eligible participants from other racial or ethnic groups. DNA was bisulfite-converted, and DNA methylation was measured using the Illumina Infinium MethylationEPIC BeadChip v1.0 (Illumina, San Diego, CA, US) according to the manufacturer’s instructions. Data preprocessing and quality control were performed in R and included color correction, background subtraction, removal of outlier samples based on median intensity values, and normalization with the beta mixture quantile (BMIQ) method59. For the calculation of the Horvath1, Horvath2, Hannum, GrimAge2, and PhenoAge biomarkers (described below), a modified version of BMIQ was used to normalize data to the gold standard reference44.
Epigenetic age and biomarkers
The NHANES database includes aging biomarkers that were calculated from DNA methylation data. Among available biomarkers, we limited our analyses to those that were trained on microarray data: Horvath1 (pan-tissue)44, Horvath2 (skin & blood)60, Hannum45, Lin61, Zhang48, Vidal-Bralo49, PhenoAge11, GrimAge213, DNA methylation telomere length (DNAmTL)62, epigenetic Timer Of Cancer (epiTOC)63, and Dunedin Pace of Aging Methylation (DunedinPoAm)43. GrimAge2 is an updated version of the GrimAge mortality risk biomarker. GrimAge and GrimAge2 are both available in NHANES and are highly correlated (r = 0.99); therefore, our analyses only included GrimAge2.
Covariates
Demographic and socioeconomic data, anthropometric measures, and health-related behaviors were collected as part of the NHANES questionnaires and physical examination. Chronological age was calculated from self-reported date of birth. Body mass index (BMI) was calculated in kg/m2 from height and weight collected by trained technicians. Participant race and ethnicity, as defined by NHANES, were classified as Non-Hispanic White, Mexican American, other Hispanic, Non-Hispanic Black, or other race, including Multiracial, based on questions about national origin, ancestry, and self-identity. Smoking status was classified as never (having smoked < 100 cigarettes in one’s lifetime), former (having smoked ≥ 100 cigarettes in one’s lifetime but not currently smoking), or current (having smoked ≥ 100 cigarettes in one’s lifetime and currently smoking every day or some days). Alcohol consumption was based on the average number of drinks per day. Leisure-time physical activity was based on the NHANES Physical Activity Questionnaire (PAQ) and was categorized as reporting having performed moderate (causing “only light sweating or a slight to moderate increase in breathing or heart rate”) or vigorous activities (causing “heavy sweating or large increases in breathing or heart rate”) for at least 10 minutes in the past 30 days, compared to not performing or unable to do physical activity. Occupational status was classified as white-collar and professional, white-collar and semi-routine, blue-collar and high-skill, blue-collar and semi-routine, or no work as described in ref. 64. Education was classified as < high school, high school diploma or General Education Development (GED) credential, or > high school. The poverty to family income ratio (PIR) was calculated as the family’s income divided by the poverty guidelines for family size and year based on the Department of Health and Human Services (HHS) guidelines.
Statistical analyses
We calculated unweighted descriptive statistics using means and standard deviations (SDs) for continuous variables and frequencies and proportions for categorical variables. We assessed performance of epigenetic clocks using Person’s correlations and median absolute errors (MAEs) to compare chronological age and estimated epigenetic age. The survey design was specified using the Survey R package65,66 with survey weights provided with the NHANES epigenetic biomarkers dataset. To preserve the study design, the survey design was specified prior to dropping participants ≥ 85 years old, with sex mismatches, or with missing or unreliable dietary data. Correlations between fatty acid variables were calculated using the svycor function using the bootstrap procedure in the jtools R package67.
We evaluated associations of total fatty acids, SFA, MUFA, PUFA, omega-6, and omega-3, fatty acid subtypes, and the P:S ratio with epigenetic aging biomarkers using survey-design weighted generalized linear regression models. PUFA 18:4 was excluded from analyses of individual fatty acid subtypes due to low levels of intake. Prior to analysis, total fatty acids, SFA, MUFA, PUFA, omega-6, omega-3, and fatty acid subtypes in grams were log2 transformed, and for variables containing values of 0, 0.0001 was added prior to transformation. Analyses were adjusted for chronological age, chronological age2, sex, race and ethnicity, BMI, education level, occupation, poverty-to-income ratio, smoking status, alcohol intake, physical activity, and total energy intake. Epigenetic age acceleration, also known as epigenetic age deviation, is commonly calculated as the residuals of regressing epigenetic age on chronological age, which detrends epigenetic age acceleration from chronological age. However, we chose to analyze the raw values of the epigenetic aging biomarkers and adjust models for chronological, which simultaneously detrends age and avoids potential bias if chronological age is correlated with covariates68.
Considering the modifying role of sex hormones on lipid metabolism21 and differences in epigenetic aging by biological sex22,23, we tested for effect modification by including a sex × fatty acid interaction term as well as conducting sex-stratified analyses. We additionally conducted analyses simultaneously modeling omega-6 and omega-3 intake and tested for multicollinearity by calculating the VIF using the function svyvif for survey designs69.
Sensitivity analyses were conducted following imputation of missing covariate data. Data were imputed with multiple imputation by chained equations using the MICE R package70 and 5 iterations. Following imputation, we reanalyzed associations of fatty acid variables with aging biomarkers using fully adjusted weighted generalized linear regression models and pooled estimates from each imputed dataset with the pool function.
We used 95% confidence intervals (95% CIs) to evaluate the precision of associations and p < 0.05 to test for statistical significance. All analyses were conducted in R version 4.4.171.
Data availability
All data sets analyzed in the current study are publicly available from the NHANES website (https://wwwn.cdc.gov/nchs/nhanes).
Code availability
Code for analyses and figures is available at https://github.com/annebozack/NHANES_epigeneticAge_fattyAcids.
References
World Health Organization. Ageing and health. https://www.who.int/news-room/fact-sheets/detail/ageing-and-health (2024).
Tessier, A.-J. et al. Optimal dietary patterns for healthy aging. Nat. Med 31, 1644–1652 (2025).
Mensah, E. O., Danyo, E. K. & Asase, R. V. Exploring the effect of different diet types on ageing and age-related diseases. Nutrition 129, 112596 (2025).
Shannon, O. M. et al. Mediterranean diet and the hallmarks of ageing. Eur. J. Clin. Nutr. 75, 1176–1192 (2021).
Calder, P. C. Functional roles of fatty acids and their effects on human health. JPEN J. Parenter. Enter. Nutr. 39, 18S–32S (2015).
Pedersen, J. et al. Monounsaturated Fatty Acids in Cardiovascular Disease: Intake, Individual Types, and Content in Adipose Tissue as a Biomarker of Endogenous Exposure. Nutrients 17, 2509 (2025).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest 109, 1125–1131 (2002).
Crimmins, E. M., Klopack, E. T. & Kim, J. K. Generations of epigenetic clocks and their links to socioeconomic status in the Health and Retirement Study. Epigenomics 16, 1031–1042 (2024).
Chen, B. H. et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8, 1844–1865 (2016).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet 19, 371–384 (2018).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018).
Lu, A. T. et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019).
Lu, A. T. et al. DNA methylation GrimAge version 2. Aging 14, 9484–9549 (2022).
Chen, Y. et al. Relationship between fatty acid intake and aging: a Mendelian randomization study. Aging 16, 5711–5739 (2024).
Ma, J. et al. The association between dietary nutrient intake and acceleration of aging: evidence from NHANES. Nutrients 16, 1635 (2024).
Xiong, Y. et al. Omega-3 PUFAs slow organ aging through promoting energy metabolism. Pharmacol. Res. 208, 107384 (2024).
Bischoff-Ferrari, H. A. et al. Individual and additive effects of vitamin D, omega-3 and exercise on DNA methylation clocks of biological aging in older adults from the DO-HEALTH trial. Nat. Aging 5, 376–385 (2025).
Wu, D., Jia, Y., Liu, Y. & Shang, M. Dose-response relationship of dietary Omega-3 fatty acids on slowing phenotypic age acceleration: a cross-sectional study. Front. Nutr. 11, 1424156 (2024).
Dietary Guidelines for Americans, 2020–2025.
NIH Office of Dietary Supplements. Omega-3 Fatty Acids Fact Sheet for Health Professionals. https://ods.od.nih.gov/factsheets/Omega3FattyAcids-HealthProfessional/ (2025).
Decsi, T. & Kennedy, K. Sex-specific differences in essential fatty acid metabolism. Am. J. Clin. Nutr. 94, S1914–S1919 (2011).
Horvath, S. et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 17, 171 (2016).
Yusipov, I., Kalyakulina, A., Trukhanov, A., Franceschi, C. & Ivanchenko, M. Map of epigenetic age acceleration: A worldwide analysis. Ageing Res Rev. 100, 102418 (2024).
Kim, Y. et al. Higher diet quality relates to decelerated epigenetic aging. Am. J. Clin. Nutr. 115, 163–170 (2022).
Reynolds, L. M. et al. Diet quality and epigenetic aging in the Women’s Health Initiative. J. Acad. Nutr. Diet. 124, 1419–1430.e3 (2024).
Chiu, D. T., Hamlat, E. J., Zhang, J., Epel, E. S. & Laraia, B. A. Essential Nutrients, Added Sugar Intake, and Epigenetic Age in Midlife Black and White Women. JAMA Netw. Open 7, e2422749 (2024).
Bozack, A. K. et al. One-carbon metabolism-related compounds are associated with epigenetic aging biomarkers: results from the cross-sectional National Health and Nutrition Examination Survey 1999–2002. Am. J. Clin. Nutr. S0002-9165(25)00317-X (2025) https://doi.org/10.1016/j.ajcnut.2025.05.029.
Gutiérrez, S., Svahn, S. L. & Johansson, M. E. Effects of omega-3 fatty acids on immune cells. Int J. Mol. Sci. 20, 5028 (2019).
Denis, I., Potier, B., Vancassel, S., Heberden, C. & Lavialle, M. Omega-3 fatty acids and brain resistance to ageing and stress: Body of evidence and possible mechanisms. Ageing Res. Rev. 12, 579–594 (2013).
ORIGIN Trial Investigators et al. n-3 fatty acids and cardiovascular outcomes in patients with dysglycemia. N. Engl. J. Med 367, 309–318 (2012).
Djoussé, L., Akinkuolie, A. O., Wu, J. H. Y., Ding, E. L. & Gaziano, J. M. Fish consumption, omega-3 fatty acids and risk of heart failure: a meta-analysis. Clin. Nutr. 31, 846–853 (2012).
Del Gobbo, L. C. et al. ω-3 polyunsaturated fatty acid biomarkers and coronary heart disease: pooling project of 19 cohort studies. JAMA Intern Med 176, 1155–1166 (2016).
Manson, J. E. et al. Marine n−3 fatty acids and prevention of cardiovascular disease and cancer. N. Engl. J. Med 380, 23–32 (2019).
Bhatt, D. L. et al. Cardiovascular Risk Reduction with Icosapent Ethyl for Hypertriglyceridemia. N. Engl. J. Med 380, 11–22 (2019).
Wang, C. et al. n-3 Fatty acids from fish or fish-oil supplements, but not alpha-linolenic acid, benefit cardiovascular disease outcomes in primary- and secondary-prevention studies: a systematic review. Am. J. Clin. Nutr. 84, 5–17 (2006).
Nicholls, S. J. et al. Effect of high-dose omega-3 fatty acids vs corn oil on major adverse cardiovascular events in patients at high cardiovascular risk: the STRENGTH randomized clinical trial. JAMA 324, 2268–2280 (2020).
Aung, T. et al. Associations of omega-3 fatty acid supplement use with cardiovascular disease risks: meta-analysis of 10 trials involving 77 917 individuals. JAMA Cardiol. 3, 225–233 (2018).
Hart, T. et al. Dietary polyunsaturated to saturated fatty acid ratio as a predictor for LDL-C response in adults: a systematic review and meta-analysis of randomized controlled trials. Curr. Dev. Nutr. 8, 102796 (2024).
Karupaiah, T. & Sundram, K. Modulation of human postprandial lipemia by changing ratios of polyunsaturated to saturated (P/S) fatty acid content of blended dietary fats: a cross-over design with repeated measures. Nutr. J. 12, 122 (2013).
National Cander Institute, Epidemiology and Genomics Research Program. Identification of Top Food Sources of Various Dietary Components. https://epi.grants.cancer.gov/diet/foodsources/ (2019).
van Dijk, S. J. et al. A saturated fatty acid–rich diet induces an obesity-linked proinflammatory gene expression profile in adipose tissue of subjects at risk of metabolic syndrome12. Am. J. Clin. Nutr. 90, 1656–1664 (2009).
Luukkonen, P. K. et al. Saturated fat is more metabolically harmful for the human liver than unsaturated fat or simple sugars. Diab. Care 41, 1732–1739 (2018).
Belsky, D. W. et al. Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife 9, e54870 (2020).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell 49, 359–367 (2013).
Chervova, O. et al. Breaking new ground on human health and well-being with epigenetic clocks: A systematic review and meta-analysis of epigenetic age acceleration associations. Ageing Res Rev. 102, 102552 (2024).
Fadadu, R. P., Bozack, A. K. & Cardenas, A. Chemical and climatic environmental exposures and epigenetic aging: A systematic review. Environ. Res 274, 121347 (2025).
Zhang, Q. et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med 11, 54 (2019).
Vidal-Bralo, L., Lopez-Golan, Y. & Gonzalez, A. Simplified assay for epigenetic age estimation in whole blood of adults. Front. Genet. 7, 126 (2016).
Crimmins, E. M., Thyagarajan, B., Levine, M. E., Weir, D. R. & Faul, J. Associations of Age, Sex, Race/Ethnicity, and Education With 13 Epigenetic Clocks in a Nationally Representative U.S. Sample: The Health and Retirement Study. J. Gerontol. A Biol. Sci. Med Sci. 76, 1117–1123 (2021).
You, Y. et al. Relationship between physical activity and DNA methylation-predicted epigenetic clocks. NPJ Aging 11, 27 (2025).
Khodasevich, D. et al. Influence of race, ethnicity, and sex on the performance of epigenetic predictors of phenotypic traits. Clin. Epigenetics 17, 59 (2025).
Bollen, K. A., Biemer, P. P., Karr, A. F., Tueller, S. & Berzofsky, M. E. Are Survey Weights Needed? A Review of Diagnostic Tests in Regression Analysis. Annu Rev. Stat. Appl 3, 375–392 (2016).
National Center for Health Statistics. National Health and Nutrition Examination Survey: Dietary Interview - Total Nutrient Intakes (1999-2000). https://wwwn.cdc.gov/Nchs/Data/Nhanes/Public/1999/DataFiles/DRXTOT.htm (2010).
National Center for Health Statistics. National Health and Nutrition Examination Survey: Dietary Interview - Total Nutrient Intakes (2001-2002). https://wwwn.cdc.gov/Nchs/Data/Nhanes/Public/2001/DataFiles/DRXTOT_B.htm (2010).
Chen, J., Sun, B. & Zhang, D. Association of dietary n3 and n6 fatty acids intake with hypertension: NHANES 2007–2014. Nutrients 11, 1232 (2019).
Dong, X. et al. Association of dietary ω-3 and ω-6 fatty acids intake with cognitive performance in older adults: National Health and nutrition examination Survey (NHANES) 2011–2014. Nutr. J. 19, 25 (2020).
National Center for Health Statistics. NHANES 1999-2002 DNA Methylation Array and Epigenetic Biomarkers. https://wwwn.cdc.gov/Nchs/Nhanes/DNAm/Default.aspx (2024).
Teschendorff, A. E. et al. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450K DNA methylation data. Bioinformatics 29, 189–196 (2013).
Horvath, S. et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 10, 1758–1775 (2018).
Lin, Q. et al. DNA methylation levels at individual age-associated CpG sites can be indicative for life expectancy. Aging 8, 394–401 (2016).
Lu, A. T. et al. DNA methylation-based estimator of telomere length. Aging 11, 5895–5923 (2019).
Yang, Z. et al. Correlation of an epigenetic mitotic clock with cancer risk. Genome Biol. 17, 205 (2016).
Rehkopf, D. H., Berkman, L. F., Coull, B. & Krieger, N. The non-linear risk of mortality by income level in a healthy population: US National Health and Nutrition Examination Survey mortality follow-up cohort, 1988-2001. BMC Public Health 8, 383 (2008).
Lumley, T., Gao, P. & Schneider, B. survey: Analysis of Complex Survey Samples. R package version 4.4. (2024).
Lumley, T. Analysis of complex survey samples. J. Stat. Softw. 9, 1–19 (2004).
Long, J. A. jtools: analysis and presentation of social scientific data. J. Open Source Softw. 9, 6610 (2024).
Krieger, N. et al. Use of correct and incorrect methods of accounting for age in studies of epigenetic accelerated aging: implications and recommendations for best practices. Am. J. Epidemiol. 192, 800–811 (2023).
Valliant, R. svydiags: Regression Model Diagnostics for Survey Data. R package version 0.7. (2024).
Buuren, van, S. & Groothuis-Oudshoorn, K. mice: multivariate imputation by chained equations in R. J. Stat. Softw. 45, 1–67 (2011).
R Core Team. R: A Language and Environment for Statistical Computing. https://www.r-project.org/ (2024).
Acknowledgements
A.K.B. is supported by the National Institutes of Health (NIH) grant K99ES035109. J.N.E. and A.C. are supported by the NIH grant R01ES031259. This research was also supported by the National Institute on Minority Health and Health Disparities (R01MD011721, MPI: BLN and DHR; and R01MD016595). M.G.F. is supported by the Novo Nordisk Foundation (NNF) grant NNF23SA0084103. The funders had no role in the decision to publish, preparation, review, or approval of the manuscript.
Author information
Authors and Affiliations
Contributions
A.K.B. designed and conducted research, analyzed data, performed statistical analysis, and wrote the article; D.K., J.C.N.-E., N.G., H.S., S.D. contributed to the analyses; H.S. calculated the epigenetic ages; D.K., J.C.N.-E., N.G., H.S., S.D., B.L.N., D.H.R., M.G.-F., and A.C. reviewed and edited the manuscript; A.C. supervised the work; A.K.B. has primary responsibility for final content; and all authors: read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bozack, A.K., Khodasevich, D., Nwanaji-Enwerem, J.C. et al. Dietary fatty acids and epigenetic aging in US adults: results from the National Health and Nutrition Examination Survey. npj Aging 12, 3 (2026). https://doi.org/10.1038/s41514-025-00298-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41514-025-00298-x