Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) represents a serious public health threat. The effectiveness of existing antibiotics to treat MRSA infections is diminishing. This study aims to develop novel antimicrobial alternatives to fight MRSA infections. We conducted an anti-virulence screening targeting the L-lectin module (SLL) of the S. aureus cell wall protein serine-rich adhesin for platelets (SraP) against a database consisting of >14,000 natural products. The top candidates identified, including the polyphenol punicalagin (PA), were validated using surface plasmon resonance and a range of microbiological assays, including cell adhesion and invasion, biofilm formation, checkboard assays, microbiome studies, and RNAseq, for their properties to combat MRSA. The efficacy of punicalagin in treating MRSA infections, alone or in combination with existing antibiotics, was investigated using mouse models. PA was bound to the virulence factor SraP with high affinity and was highly effective in inhibiting MRSA colonization. PA significantly reduced the expression of multiple genes associated with ß-lactam resistance and disrupted biofilm formation. The synergistic effects between PA and cefoperazone (CF), a well-known bactericidal, were remarkable. The combination of PA and CF reduced the bacterial load in key murine organs by >98.9% and completely protected mice infected by MRSA. PA restored native gut microbiota disrupted by antibiotics, enriched butyrate-producing species, and repressed tissue inflammation. PA acted as a potent anti-virulence agent in addition to its intrinsic anti-inflammatory properties. Taking advantage of the strong synergism between PA and CF represents a promising strategy for combating MRSA infection and antibiotic resistance.
Similar content being viewed by others
Introduction
Staphylococcus aureus is a common cause of infections in community and hospital settings. The worldwide prevalence of methicillin-resistant Staphylococcus aureus (MRSA) ranges from 13% to 74%1. MRSA is associated with skin and joint infections as well as pneumonia and can lead to sepsis2. Moreover, the effectiveness of currently available antibiotics to treat MRSA infections, such as clindamycin, daptomycin, linezolid, sulfamethoxazole, teicoplanin, tigecycline, and vancomycin, is rapidly diminishing3. The excessive usage of MRSA-fighting antibiotics is accompanied by an emergence and rapid spread of antibiotic resistance4,5. The challenge posed by antimicrobial resistance (AMR) has evolved into a severe public health concern6. As a result, there is a pressing unmet need for innovative anti-MRSA agents able to decrease antibiotic usage and mitigate the development of AMR.
Adhesion to epithelial surfaces is of crucial significance for bacterial colonization7,8. The interaction of bacterial surface proteins with host receptors plays an important role in bacterial adhesion, invasion, and pathogenicity9. A serine-rich repeat glycoprotein, serine-rich adhesion for platelets (SraP), present on the surface of Staphylococcus aureus, is responsible for mediating S. aureus adhesion to and invasion of host epithelial cells. This occurs via specifically binding to the monosaccharide N-acetylneuraminic acid (Neu5Ac) on host cells through its N-terminal L-Lectin module (SraP L-lectin or SLL)10,11. The adherence and invasion of S. aureus are significantly reduced by targeting SLL using antibodies12. Therefore, SLL likely represents an excellent target for novel anti-virulence drug development.
Natural bioactive compounds have been extensively utilized in the treatment of various infections throughout human civilizations, due to their remarkable structural diversity, ready availability, and excellent biocompatibility13. Over the years, a growing number of natural products that possess anti-MRSA activities have been identified, including alkaloids, polyphenols, quinones, and terpenoids14. These bioactive compounds combat MRSA through multi-target actions, such as reducing cell wall synthesis, disrupting bacterial membrane integrity, affecting virulence-related factors, and inhibiting protein synthesis15,16. As a result, natural bioactive compounds are being recognized as a promising reservoir of novel antibacterial agents15. To date, ~14,000 natural bioactive compounds, including ellagitannins and flavonoids from pomegranate, a widely used traditional medicine, have been collected in the Chinese medicine systems pharmacology database (TCMSP)17. For example, as one of the most abundant polyphenols in pomegranate, punicalagin (PA) is able to inhibit multiple bacterial pathogens, such as E. coli and Streptococcus pyogenes18. S. aureus clinical isolates are also susceptible to PA19. PA also inhibits the growth of Salmonella typhimurium and destroys the integrity of its cell membrane20. However, molecular targets to which PA exerts its antibiotic effects remain elusive, and PA and many other natural products have yet to be thoroughly characterized for their anti-MRSA potential.
In this study, we conducted a systematic in-silico screening of all natural bioactive compounds in TCMSP for their ability to block MRSA adhesion by targeting the virulence factor SraP using Autodock and surface plasmon resonance (SPR) assays. The anti-MRSA effects and molecular mechanisms of selected anti-virulence agents were further investigated using in vitro and in vivo models. We were particularly interested in small molecules that display dual properties for simultaneous reduction in bacterial growth and tissue inflammation caused by MRSA infection. We attempted to harness the strong synergism between bioactive compounds and existing antibiotics in fighting MRSA infections, aiming at reducing the abundance of antibiotic-resistant genes (ARG) in one of their key reservoirs, the gut microbiota, concurrently. ARGs are widespread in both natural and engineered habitats. The presence of ARGs in MRSA strains negates the effect of many widely used antibiotics. Alarmingly, ~24% of over 2500 known ARGs pose a public health risk21.
Results
In silico screening identified bioactive compounds against MRSA
Gene sequences of SLL modules of 48 newly isolated MRSA strains, including 24 clinically associated and 24 community-associated, were compared with S. aureus strain NCTC8325, a generally regarded prototypical strain for genetic manipulation, and MW2, a well-known MRSA strain. At the amino acid (AA) level, the 48 strains shared a high degree of homology, between 96% and 100%, with the strain NCTC8325 (Fig. S1A), whose L-lectin protein structure and function have been well studied22. For example, the similarity between NCTC8323 and MW2 is 98.8%, while the similarity between NCTC8325 and the newly isolated 2016258 strain is 100%. The six key AAs that interact with Neu5Ac in the binding pocket, Ser293, Asn347, Tyr367, Asn369, Gly477, and Ala478, were present (Fig. S1B); and the gene encoding SLL was stably expressed in all MRSA strains tested, with its expression values varying within a narrow range (data not shown), indicating that SLL are structurally conserved among MRSA strains. For example, the SLL structure of the MRSA strain 2016211, recently isolated from clinical samples in our study, shared a 97% similarity with NCTC8325, as predicted by the SWISS-MODEL algorithm (Fig. S1C).
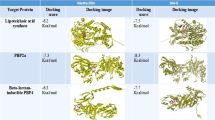
Approximately 14,000 natural compounds in the TCMSP database were in silico screened for their abilities to bind SLL using Autodock Vina. Among them, 10 compounds were predicted to act as SLL antagonists based on their potential binding sites and relative binding free energy (BFE), including corilagin, PA, bilobetin, timosaponin A-III, 8-gingerol, isorhamnetin-3-O-β-rutinoside, (‒)‒Epigallocatechin-3-gallate (EGCG), neoeriocitrin, neohesperidin, and isoviolanthin (Fig. 1A, B). These compounds, all with BFE ≤ −8.0 kcal/mol, were docked at a position known to bind to Neu5Ac, a natural ligand of SLL. They stacked against at least three of the six key AAs via polar interactions. The findings from molecular docking simulations were further verified using surface plasmon resonance (SPR) assays. The bioactivity of an enriched preparation of SLL (Fig. S2A) was first confirmed by the binding of Neu5Ac to SLL (Fig. S2B), consistent with a previous report22. The analysis of SPR steady-state equilibrium curves of the 10 compounds at varying concentrations suggests that seven of the 10 compounds had strong interactions with SLL (Fig. S2C–S2L). Seven of the 10 compounds, PA, timosaponin A-III, 8-gingerol, isorhamnetin-3-O-β-rutinoside, EGCG, neoeriocitrin, and isoviolanthin, were verified to possess strong binding signals in a dose-dependent manner, whereas bilobetin did not appear to bind to SLL, and corilagin and neohesperidin seemingly had non-specific binding to SLL (not dose-dependent). Among the seven compounds with strong binding affinity to SLL, PA showed a good minimum inhibitory concentration (MIC) for a complete prevention of colony growth on agar plates (100% inhibition) of the MRSA strain MW2 (MIC100 = 271 µg/mL, Fig. 1C), while its MIC value for 90% inhibition (MIC90) was ~10.8 µg/mL.
A The putative binding sites of the 10 compounds to SraP predicted by Autodock Vina. B Binding free energy and putative binding sites. C The minimum inhibitory concentration (MIC) of the 10 natural compounds, defined as the lowest concentration that completely (100%) prevents visible growth of methicillin-resistant Staphylococcus aureus (MRSA) test strain MW2 under in vitro conditions (MIC100).
Anti-MRSA activities of PA and its hydrolytic products in vitro
PA displayed an excellent growth inhibition against the MW2 strain across a broad concentration range in vitro (Fig. 2A). PA was also potent against other clinically associated MRSA strains with growth inhibitions ranging from 81.2% to 95.9% at 6 h incubation at 10 µg/mL dose level (Fig. 2B). For example, the inhibition rates of PA at 10 µg/mL on the MW2 strain as represented by the reduction in colony counts after 4, 6, and 8 h incubations were 91.05%, 95.06%, and 97.64%, respectively. Moreover, PA showed a potent dose-dependent biofilm inhibition; and >90% biofilm inhibition was reached at 10 µg/mL PA for MW2 (p < 0.001, Fig. 2C). Of note, the 11 MRSA strains tested displayed a varying degree of biofilm formation capabilities in the presence of PA at 10 µg/mL, with inhibition rates ranging from 48.9% to 78.4%, as judged by bacterial biofilm formation index (BFI) assayed using crystal violet staining (Fig. 2D). Further, our data show that PA significantly blocked the adhesion and invasion of MRSA to human lung epithelial A549 cells. The number of MRSA MW2 strain adhered to A549 cells decreased by 41% in response to PA at 10 µg/mL (p < 0.001, Fig. 2E), compared to that of untreated control cells. PA also significantly prevented bacterial invasion in A549 cells (Fig. 2F).
A Dose curve of PA at various concentrations on the growth of the MRSA strain MW2. B The sensitivity of 12 MRSA strains (the X-axis) to PA at 10 µg/mL, as expressed as the reduction in log10 CFU (the Y-axis). CFU colony-forming unit. C The impact of PA at varying concentrations on biofilm formation index (BFI, the Y-axis). Inlet: temporal responses of BFI (PA at 10 µg/mL) as a percentage of untreated controls. D The effect of PA at 10 µg/mL on BFI among different MRSA strains. PA significantly affected adhesion (E) and invasion (F) of the MRSA strain MW2 on human epithelial cells A549 in vitro. G PA and its hydrolytic products, ellagic acid (EA) and punicalin (PN). H Punicalin retained a majority of the inhibitory property of its parent molecule PA, while the growth inhibition of EA and a bio-transformed metabolite of PA, urolithin A (UA), on the strain MW2 was somewhat reduced, regardless of dose levels. I PN is predicted to bind to the active binding sites in the L-lectin module of SraP with a high affinity, analogous to that of its parent compound, PA. ***P < 0.001; **P < 0.01; *P < 0.05. Four replicates per group.
PA is readily hydrolyzed into ellagic acid (EA) and punicalin (PN, Fig. 2G) and can be biotransformed into urolithin A (UA) by gut microbiota23. The results from the bacterial inhibitory assay showed that PA at 10.84 µg/mL inhibited growth by 94% (Fig. 2H). PN displayed a similar level of growth inhibition at the same concentration, while the anti-MRSA activities of EA and UA were significantly subdued, with only 73.6% and 68.7% inhibition, respectively. PN shared binding characteristics to SLL in a manner similar to PA, with a BFE value of −7.7 kcal/mol (Fig. 2I). Like what was observed with PA, there were four key AAs, Asn347, Tyr367, Gly477, and Ala478, that interact with PN in the binding pocket (Fig. 2I).
Validation of high-affinity binding of punicalagin to SLL using SPR assays
We next validated the binding affinity of PA to SLL using SPR titrations. The SLL recombinant protein was expressed by transforming recombinant plasmid pET28a-SraP L-lectin into E. coli BL21 cells. Relative SPR responses were induced by PA in a dose-dependent manner from 0.08 to 5.42 µg/mL (Fig. 3A, B). The equilibrium dissociation constant Kd of PA was 5.66 × 10−7 M, much lower than that of Neu5Ac (1.31 × 10−5 M, Fig. 3B), suggesting that PA had a stronger binding affinity to SLL than Neu5Ac.
Time (A) and dose (B) dependent binding kinetics and affinity of punicalagin (PA) to the purified L-lectin module from wild-type (WT) MRSA strain MW2 in vitro, as detected using surface plasmon resonance (SPR). The color of dots in B represents the PA concentrations as described in (A). RU: response or resonance unit. C Differences in cell morphology and cell wall structure of WT and ΔL-lectin MW2 strains in response to PA at 10 µg/mL as observed by transmission electron microscopy (TEM). The morphology and cell wall structure between WT and ΔL-lectin mutant were indistinguishable without PA treatment (WT vs. ΔL-lectin mutant). The L-Lectin module deletion (ΔL-lectin) did not appear to affect growth characteristics of the mutant, compared to its WT counterpart (D). However, the mutant compromised MRSA sensitivity to PA as judged by its effect on overall growth inhibition (E), adhesion (F) to host cells, and biofilm formation (G) in vitro. **P < 0.01; *P < 0.05. The number represents the mean ± SD of four replicates.
To verify the structural association between anti-MRSA abilities of PA and SLL, we constructed a SLL mutant strain (ΔL-lectin). Transmission electron microscopy shows that the cell wall of the wild-type (WT) MRSA strain MW2 had a spherical contour and a smooth surface (Fig. 3C). Moreover, the mutant displayed a cell morphology indistinguishable from its WT MW2 strain (Fig. S3). SLL deletion did not appear to alter the growth characteristics of the MRSA strain. Compared to its WT counterpart, ΔL-lectin had a statistically similar growth curve (Fig. 3D; p > 0.05). PA treatment in the WT strain resulted in a rough surface and a waved contour, while the cell wall of the PA-treated mutant strain reverted to that observed in the untreated WT MW2 strain (Fig. 3C). Furthermore, compared to WT, the PA-induced growth inhibition on the mutant ΔL-lectin strain was significantly decreased, from 93.9% in WT to 90.8%, at the 10 µg/mL dose level (p < 0.05, Fig. 3E). Expectedly, SLL deletion had a significant effect on adhesion. Compared to WT, the adhesion rate in the mutant strain was 39.3%, a significant reduction from 65.9% in WT in the absence of PA treatments (Fig. 3F; P < 0.001). PA treatments further decreased the bacterial adhesion rate to 38.8% in WT and 30.2% in the SLL mutant (Fig. 3F). Similarly, biofilm formation was also impacted by SLL deletion in the absence of PA, at 8 h, but not at 4 h incubations (Fig. S3C). PA treatment in the mutant ΔL-lectin strain was also significantly compromised after 4 h (P < 0.05) and 8 h (P < 0.01) incubations, compared to its WT counterparts (Fig. 3G). Together, these data suggest that PA exerts its antibiotic effect at least partially via its interaction with the SLL of SraP.
Punicalagin inhibited MRSA proliferation via pyrimidine metabolism pathway
To understand the potential mechanism underlying the inhibition of PA on bacterial growth, we examined the change in the MW2 transcriptome in response to PA treatment at 10 µg/mL. Among the >2700 genes detected in the transcriptome, 286 genes were significantly down-regulated, whereas 264 genes were significantly up-regulated by PA (FDR < 0.05 and fold change ≥2.0, Fig. 4A). Genes belonging to pyrimidine metabolism, sulfur-relay system, and cell cycle were significantly enriched among the genes regulated by PA (Fig. 4B). For example, the expression of multiple genes in pyrimidine metabolism pathway, including carbamoyl phosphate synthase A and B (carA/B), DNA polymerase III subunit delta (holA), ribonucleotide-diphosphate reductase subunit beta (nrdF), DNA-directed RNA polymerase subunit alpha (rpoA), DNA-directed RNA polymerase subunit beta (rpoC), and thymidylate kinase (tmk), was significantly repressed by PA (Fig. 4C). The expression of select genes was further validated using qRT-PCR. For example, the expression level of aspartate carbamoyltransferase catalytic subunit (pyrB) decreased by 4.84 fold in response to PA, compared to untreated control cells (p < 0.01, Fig. 4D). Similarly, PA also significantly inhibited the expression of carA, pyrC, pyrE, and pyrF (p < 0.01), which was consistent with RNAseq data and showed good correlations between the two platforms (Fig. 4E). Further, the genes involved in the key steps of biofilm formation, including bacterial attachment, multiplication, maturation, and dispersal were examined using RNAseq and qRT-PCR (Fig. 5A). Notably, several genes that play a critical role in bacterial attachment, including clumping factor B (clfB), fibronectin-binding protein B (fnbB), Ser-Asp-rich fibrinogen-binding D (sdrD), and immunoglobulin G binding protein A (spa), was significantly downregulated while the expression of accessory gene regulator B (agrB), a key gene in virulence and bacterial biofilm dispersal, was upregulated by PA treatment (p < 0.01, Fig. 5B), suggesting the primary role of PA on biofilm inhibition may be the disruption of its initial stage, i.e., bacterial attachment, via downregulating several cell wall-anchored genes.
A Volcano plot of up- and down-regulated genes due to the treatment of MRSA strain MW2 with punicalagin (PA) at 10 μg/mL. B Pathways impacted by PA in the transcriptome of the MRSA strain MW2 in the presence of PA at 10 μg/mL. The color represents the significance (−log10 P value) of the changes. The Gene Ratio denotes the enrichment factor, i.e., the ratio of the number of differentially expressed genes to the total number of genes in a pathway. The size of the circle represents the number of differentially expressed genes mapped to the indicated pathway. C The diagram shows the pyrimidine metabolism pathway that was significantly affected by PA. D qRT-PCR verification of the expression of select genes, including carbamoyl phosphate synthase A (carA) and pyrB, pyrC, pyrE, and pyrF, that were significantly repressed by PA as detected by RNAseq. E correlations between qRT-PCR and RNAseq. The number represents the mean relative expression (or fold changes ± SD) of four replicates.
A Key genes involved in various stages of biofilm formation in the MRSA MW2 strain, particularly those cell-wall-associated genes. B The expression of five genes critical to bacterial attachment, clfB, fnbB, sdrD, sraP, and spa, was significantly downregulated, while the expression of agrB was significantly upregulated by PA. C The mRNA expression of pbp2, pbp3, pbp4, mecA, and murF was also significantly downregulated by PA. ***P < 0.001; **P < 0.01; *P < 0.05. The number represents the mean relative expression (or fold changes ± SD) of four replicates.
Punicalagin increased MRSA sensitivity to the antibiotic cefoperazone in vitro
PA significantly inhibited the expression of genes associated with ß-lactam resistance, such as penicillin-binding protein (pbp)2, pbp3, and pbp4 (p < 0.001; Fig. 5C), which play a significant role in remodeling bacterial cell wall peptidoglycan and mediating β-lactam resistance. Additionally, PA induced a significant reduction in the gene expression levels of mecA (1.93-fold, p < 0.01) and murF (3.02-fold, p < 0.01) (Fig. 5C) and other genes related to virulence, such as α-hemolysin (hly) and a gene encoding for Panton-Valentine leukocidin toxin subunit S (lukS-PV) (Fig. S4). However, the expression of extracellular adherence protein Eap/Map (eap) was significantly increased by the PA treatment (Fig. S4).
To investigate the effect of PA on the emergence of β-lactam resistance, we tested the growth of the first to tenth generations of the MRSA MW2 under a series of PA concentrations in vitro. As Fig. 6A shows, PA at concentrations up to 256 µg/mL did not appear to induce any resistance as demonstrated by the lack of changes in PA-induced growth inhibition, regardless of the MRSA generation (Fig. 6B) and PA-antibiotic combinations (Fig. 6C). PA did not appear to affect the expression of the virulence factor srtA at the mRNA level, regardless of the presence or absence of SLL (Fig. S5). Expectedly, CF at 20 µg/mL did induce significant MRSA resistance starting at the 7th generation (Fig. 6D). We also compared resistance development at a total compound concentration of 20 µg/mL, whether as monotherapy or combination treatment.
The MRSA strain MW2 cultured for 10 consecutive generations under PA dose levels between 8 and 256 µg/mL did not appear to induce antibiotic resistance, as no statistically significant changes (ns) in optical density value at OD600 (A) and percentage inhibition values (B) were observed (p > 0.05). PA treatment alone at dose levels up to 256 µg/mL in vitro did not appear to induce the development of antibiotic resistance. C The β-lactam antibiotic cefoperazone (CF), alone or in combination with either PA or sulbactam (SB), significantly inhibited the growth of the MW2 strain. D CF at 20 µg/mL indeed induced the antibiotic resistance starting at the 7th generation, while no antibiotic resistance development was observed for the combination therapy, either PA-CF or CF-SB at 10 µg/mL each. The vertical dash lines indicate that statistical comparisons were made. Not all statistically significant differences were marked with three asterisks due to space limitations. ***P < 0.001. The number represents mean ± SD for six replicates.
While the growth of all 10 generations of MW2 can be effectively inhibited by either CF-PA or CF-sulbactam at a combined dose of 20 µg/mL (10 µg/mL each), no β-lactam resistance was observed in vitro at this combined dose level (Fig. 6D). Sulbactam (SB) possesses limited antibacterial activity and is not considered an antibiotic. However, it can prevent bacteria from destroying antibiotics, thus enhancing the efficacy of many β-lactam antibiotics in the combination therapy. Our data also provided evidence to support its role in the combination therapy. Moreover, SB did not appear to affect the growth of the MW2 strain, regardless of the presence or absence of SLL (Fig. S6).
We next examined 17 existing antibiotics to determine if they had any synergistic effect with PA using a checkerboard assay. The sensitivity of 12 MRSA strains to 10 of the 17 antibiotics, as judged by their minimum inhibitory concentrations (µg/mL) for the complete inhibition of visible growth, was first determined (Table S1). Our findings also show that PA significantly enhanced the antibacterial effect of at least eight ß-lactam antibiotics in the combination therapy, including ampicillin, aztreonam, cefathiamidine, cefazolin, cefixime, cefoperazone, ceftriaxone sodium, and cephradine, with a fractional inhibitory concentration index (FICI) value < 0.5. The FICI value for the PA and CF combination was the lowest (FICI = 0.25), suggesting that there may exist a strong synergy between these two (Table S2). We further investigated the growth-inhibitory effects of PA in combination with CF in fighting MRSA strains. The effect of CF on growth inhibition was dose-sensitive. While used alone, CF at 20 µg/mL was significantly more inhibitory than CT at 10 µg/mL at 12 h incubation (p < 0.001; Fig. 6C). Moreover, the CF dose in the CF–PA combination therapy was also sensitive. For example, the growth inhibition of 2.5 µg/mL CF combined with 10 µg/mL PA was significantly different than CF at 5, 7.5, or 10 µg/mL in combination with PA at 10 µg/mL, respectively (p < 0.001; Fig. 6C). Even at the lowest CF concentration tested, at 2.5 µg/mL, the combination therapy with PA was significantly better than that of 20 µg/mL CF alone (p < 0.001; Fig. 6C). The synergy was also dose-dependent. The difference in the growth inhibition between CF 7.5 µg/mL + PA at 10 µg/mL and CF + PA at 10 µg/mL each was significant starting at 4 h incubation (p < 0.001, Fig. 6C). Furthermore, a commonly used antibiotic combination, CF–SB, at a 1:1 concentration ratio (10 µg/mL each), reduced MRSA growth from 6.00 ± 0.00 to 3.49 ± 0.01 log10 CFU/mL during 12 h incubation (p < 0.001), which was indistinguishable than the CF + PA combination at the same dose level (Fig. 6C).
In a THP-1 in vitro cell assay, MRSA infection reduced cell viability by 71.4% (Fig. S7A). PA treatment at 10 µg/mL significantly improved the cell survival rate from 28.6% to 49.9% (p < 0.001; Fig. S7A). The combination treatments also significantly improved cell survival in an in vitro infection model (Fig. S7A). For example, the co-treatment of 2.5 µg/mL CF and 10 µg/mL PA resulted in a survival rate similar to 20 µg/mL CF alone (p > 0.05). Moreover, the CF and PA combination at 10 µg/mL each further improved cell viability to 94.5% (p < 0.05), reaching the same protective effect as the CF–SB combination at the same ratio and dose level but exceeding the protective effect of CF alone at 20 µg/mL (Fig. S7A). As expected, MRSA infection upregulated multiple M1 macrophage marker genes, such as IL1β (6.22-fold change, p < 0.001), IL6 (4.77-fold, p < 0.001), TNFα (4.57-fold change, p < 0.001), CD86 (3.91-fold, p < 0.001), and CD80 (3.64-fold, p < 0.001) (Fig. S7B). PA treatment (10 µg/mL) resulted in a significant reduction in the mRNA expression of these proinflammatory cytokines, to an extent similar to the treatment by CF at 20 µg/mL, whereas the inhibitory effect of PA at 10 µg/mL on CD80 was more profound, exceeding that of CF at 20 µg/mL (p = 0.04). Combining 10 µg/mL CF with either PA (10 µg/mL) or SB (10 µg/mL) displayed a stronger anti-inflammatory effect than either compound alone. While both CF–PA and CF–SB combinations at 10 µg/mL each had a strong yet similar effect in reducing the mRNA expression of pro-inflammatory markers, PA-related treatments promoted the expression of M2 macrophage markers, including IL-10, CD163, and CD206 (Fig. S7B). For example, the CF–PA combination (10 µg/mL each) enhanced the expression of CD163 and CD206 significantly more than the CF–SB combination at the same dose level (p < 0.05; Fig. S7B).
Punicalagin in combination with cefoperazone completely protected mice infected with MRSA
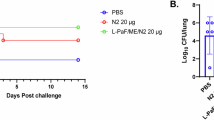
To further confirm the synergistic effect between PA and CF, we investigated the efficacy of CF alone or in combination with PA on MRSA infection in vivo at a dose level of 8 × 108 CFU per mouse. Without any treatments, 90% mice infected with the MRSA strain MW2 at this dose level died within 7 days post-infection. Intraperitoneal administrations of 10 and 20 mg/kg bodyweight (BW) CF for 3 days improved the survival rate to 30% and 70%, respectively (Fig. 7A). The survival rate of MRSA-infected mice treated with either CF (2.5 mg/kg) and PA (10 mg/kg) was also 70%. Remarkably, all MRSA-infected mice treated with either a combination of CF (10 mg/kg BW) and PA (10 mg/kg BW) or CF and SB (10 mg/kg BW each) survived. MRSA infection induced a significant BW loss in untreated mice, but all treated groups showed a recovery in BW at 24, 48, and 72 h post-infection (p < 0.001, Fig. S8A). The effect of dual treatments, either CF–PA or CF–SB combinations at 10 mg/kg each, on BW was indistinguishable. The data on the disease activity index (DAI), calculated based on composite changes in BW, fecal viscosity, and fecal occult blood, also confirmed the efficacy of the dual treatments (Fig. S8B). The effect of the treatment on colon length (Fig. S8C), as described in a published report24, and organ coefficients, the ratio of organ weight to BW (Fig. S8D), followed similar trends. Three days post MRSA infection, the number of bacterial colonies in the major mouse organs was determined (Fig. 7B). Compared to untreated groups, PA treatment at 10 mg/kg BW for three consecutive days reduced the mean number of bacterial colonies recovered from five organs, the liver, spleen, colon, kidney, and lung, by 69.4% (n = 10; p < 0.001), while CF alone at 20 mg/kg BW was able to eliminate up to 94.3% bacterial colonies from these organs. Remarkably, PA (10 mg/kg BW) in combination with CF (2.5 mg/kg BW) was more efficacious in eliminating MRSA than CF alone at 20 mg/kg BW (p = 0.0064; Fig. 7B). A combination of either CF-PA or CF-SB, at a dose level of 10 mg/kg BW each, eliminated >98.9% of mean viable bacterial colonies from the major organs (p < 0.001). Moreover, the organ coefficients exhibited a trend similar to the bacterial burden (Fig. S8D). Concomitant with the reduction in organ MRSA bacterial load were the changes in mRNA expression of pro-inflammatory cytokines and chemokines. For example, MRSA infection increased the gene expression of IL1β, IL6, and TNFα by 4.00-, 3.80-, and 4.60-fold, respectively, as compared to in the uninfected control mice (p < 0.05, Fig. 7C). The treatment with CF alone reduced the inflammatory response in a dose-dependent manner. Moreover, the CF–PA combination at 10 mg/kg BW each reversed the expression of IL1β, IL6, and TNFα to the baseline, better than the CF–SB combination at the same dose level (p < 0.05; Fig. 7C). Furthermore, PA-related treatment elevated the expression of anti-inflammatory IL10 (p < 0.05). The reduction in the number of inflammatory infiltrates in the liver tissue due to the PA-related treatments was also evident (Fig. 7D). The liver pathology damaged by MRSA infections was reverted to the baseline by dual compound treatments of either CF–PA or CF–SB combinations, at a dose level of 10 mg/kg BW each (Fig. 7D).
A Daily injection with punicalagin (PA) at 10 mg/kg bodyweight (BW) and the β-lactam antibiotic cefoperazone (CF) at 2.5–20 mg/kg BW, alone or in combination, for 7 days, markedly improved the survival rate of mice challenged with MRSA. The combination of PA and CF at 10 mg/kg BW each completely protected mice starting at 3 days of treatment (mortality = 0%), which was also achieved by the CF and sulbactam (SB) combination at 10 mg/kg BW each. N = 8 per group. B Mean MRSA burden in major organs expressed as the reduction in log10 colony-forming units (CFU, Y-axis), counted on the agar plate after overnight incubation. C The combination therapy between PA and antibiotics for 3 days (72 h) significantly downregulated pro-inflammatory marker genes in the liver tissue. D The combination treatment significantly reduced the number of inflammatory infiltrates in the liver tissue. Arrow shows a pattern of hepatic sinusoid dilatation. Negative control = healthy uninfected and untreated. Control = MRSA infected but untreated. ***P < 0.001; **P < 0.01; *P < 0.05 (one-way ANOVA with Tukey’s HSD as post hoc test). N = 10 per group.
Punicalagin–cefoperazone combinations enriched gut microbial diversity repressed by antibiotics and promoted the expansion of beneficial bacterial species
MRSA challenge infection and ensuing antibiotic treatment by CF significantly depleted gut microbial diversity, particularly the Shannon index (Fig. 8A). Compared to the mice treated with either 10 or 20 mg/kg BW of CF alone, the treatment by the CF–PA combination at 10 mg/kg BW each for 3 days significantly increased the Shannon index (Fig. 8A), whereas the classic combination, CF and SB, failed to improve gut microbial diversity. Compared to untreated mice challenged with the MW2 strain, the combination therapy, either CF–PA or CF–SB, induced a significant difference in β diversity in the gut microbiota (Fig. 8B). Intriguingly, the CF–SB combination significantly reduced the abundance of two beneficial bacterial species in the gut microbiota, including Faecalibaculum rodentium (Fig. 8C), which remodels retinoic acid signaling to govern eosinophil-dependent intestinal epithelial homeostasis25, and Akkermansia muciniphila (Fig. 8D), whereas the CF–PA combination treatment significantly increased the abundance of the former. Conversely, the CF–SB combination tended to increase the abundance of Lactobacillus vaginalis (Fig. 8E), a species isolated from patients suffering from trichomoniasis, and Parabacteroides goldsteinii strain dnLKV18, unlike the CF–PA combination (Fig. 8F).
A Shannon Index was calculated based on full-length 16S rRNA gene sequencing in the gut microbiota of mice in response to various treatments by punicalagin, alone or in combination with cefoperazone, for 3 days. B Beta diversity using PERMANOVA based on the Bray–Curtis similarity matrix calculated based on species/strain level abundance value. Changes in the relative abundance of select species/strains in response to the combination treatment at a single daily dose of 10 mg/kg BW each for 3 days. The effect of the combination therapies on the abundance of select bacterial species/strains in the gut microbiota: C Faecalibaculum rodentium. D Akkermansia muciniphila. E Lactobacillus vaginalis. F Parabacteroides goldsteinii strain dnLKV18. G PA treatment, alone or in combination with CF, repressed the relative abundance of four antibiotic-resistant genes, pbpA, pbp2, mecA, and mecR1, which are critical to the development of β-lactam resistance in mice infected with the MW2 strain. The abundance of these genes was detected using qPCR from total DNA extracted from fecal samples. Negative control = healthy uninfected and untreated. Control = MRSA-infected but untreated. ***P < 0.001; **P < 0.01; *P < 0.05. ns: no significance. N = 10 per group.
In the MRSA infection model, the collective abundance of several β-lactam resistance genes was examined by quantitative PCR using total DNA extracted from fecal samples. CF alone induced a significant increase in the relative abundance of several ARGs in the gut, including pbpA, pbp2, mecA, and mecR1 (Fig. 8G). PA treatment for 3 days, alone or in combination with CF, not only significantly reduced MRSA burden in key organs, but also lowered the abundance of ARGs in the gut (p < 0.001, Fig. 8G). In contrast, the CF–SB combination did not appear to have any significant effect on the abundance of the genes related to β-lactam resistance, suggesting that the CF–PA combination likely had more advantages than CF–SB in mitigating ARGs.
Discussion
MRSA is the leading cause of deaths related to AMR and is responsible for 121,000 deaths globally in 201926. Among residents of elderly care centers, the global prevalence of MRSA colonization reaches an alarming 14.97%, ten times higher than in the general population27. While standard-of-care and last-resort antibiotics, such as vancomycin and daptomycin, are effective in fighting some MRSA strains, their efficacy is diminishing due to the emergence or risk of the spread of AMR. Conventional antibiotics have been designed to target genes or pathways essential to bacterial survival and viability, including bacterial cell wall synthesis and the biosynthesis pathways related to nucleic acids or proteins28. Further, the bacterial envelope has been advocated as a target for anti-MRSA therapy29. In the past decades, increased attention has been focused on targeting virulence as an antimicrobial therapy due to its potential for reduced development of resistance30,31.
MRSA produces a broad array of virulence factors, which can be loosely classified into two major categories, secreted factors and microbial surface components recognizing adhesive matrix molecules (MSCRAMMs). The latter plays a critical role in MRSA colonization and attachment to the host, via recognizing various extracellular matrix (ECM) components, such as fibronectins and laminins. MSCRAMMs represent one of the ideal classes of anti-virulence targets. For example, sortases, a class of enzymes essential for MRSA colonization, have been proposed as a vaccine candidate32. SraP, one of the major virulence factors of MSCRAMM in the SRRP protein family, also has the potential to be an excellent target for drug development. A prior study using a monoclonal antibody (mAb) against SLL provided proof-of-principle results12. The mAb was able to inhibit adherence and invasion of MRSA into A549 cells in vitro and resulted in an ~50% reduction in blood MRSA load in a mouse challenge model12. In this study, we focused on targeting SraP SLL using natural products. Among the 10 natural products identified using in silico screening, PA binding to the active site of SLL was verified using SPR. Our findings show that PA alone was efficacious in fighting MRSA. In a mouse infection model, the treatment using a physiologically relevant dose of PA reduced mean organ bacterial burden by 69%. Moreover, in vivo treatments with PA led to concurrent inhibition of multiple key genes related to quorum sensing and biofilm formation, providing additional benefits.
PA is a versatile molecule with a wide range of biological activities23,33. It is abundant in pomegranate with an excellent safety and bioavailability profile. PA is not cytotoxic. Its cytotoxicity half-maximal inhibitory concentration values (IC50) against human larynx epithelial cancer cell line (Hep2) and small cell lung carcinoma cell line (A549) are 740.0 and 830.0 µM, respectively34, much higher than the physiologically relevant dose level. In our preliminary studies, PA did not have any significant effect on the survival of normal human THP1 cells without MRSA infections at dose levels up to 270 µg/mL (data not shown). Its absorption and metabolism processes in the gut are also well understood. Moreover, therapeutically relevant dose levels can be readily obtained from commercial preparations of pomegranate whole fruit juice, which typically contain ~200 mg/L or 184 μM of PA23,33. The uniqueness of PA as a potent alternative antimicrobial lies in its ability to target bacterial virulence factors. PA is known to bind with high affinity to peptidoglycan (PGN) from Gram+ bacteria. Exposure of S. aureus to a sub-MIC level of PA results in collapsed cell walls and loss of cytoplasmic contents35,36. The direct disruptive effect of PA on MRSA cell walls was also confirmed in our study (Fig. 3C). A recent study has demonstrated direct binding between PA and sortase A37. PA inhibits sortase A activity with an IC50 value of 4.23 µg/mL, resulting in a reduced binding of S. aureus to fibronectin in vitro and subsequent adherence to host cells. Further, in an MRSA-induced lethal pneumonia model, a PA treatment of S. aureus-infected mice doubled the survival rate, from 20% to 40%. These findings suggest that PA likely acts as a sortase A inhibitor. In the present study, our data demonstrate that the deletion of SLL (ΔL-lectin) did not appear to affect the expression of the gene encoding for sortase A (srtA), compared to its WT counterpart (Fig. S5). In addition, a PA treatment on the WT strain had no statistically significant effects on the srtA transcript abundance (Fig. S5), suggesting that the interactions between PA and sortase A or SraP were two independent events. Moreover, PA binds to SraP with high affinity, resulting in a significant reduction in the MRSA burden in the key target organs and increasing the survival of mice challenged with MRSA. Together, these data demonstrated that PA exerts its antimicrobial effect via multiple targets, thus improving its therapeutic value38.
We carefully examined the synergistic effect between PA and 17 existing antibiotics. Various MRSA strains display a wide range of sensitivity to PA. The MRSA strain used in this study, MW2, had a higher MIC100, at ~256 µg/mL, than other commonly used strains35. Nevertheless, PA alone displayed a strong anti-MRSA effect with a significant reduction, 69.4 ± 4.7%, in multi-organ mean bacterial burden (MOBB), compared to untreated control mice. In contrast, CF treatment alone was able to reduce MOBB by 91.7 ± 1.1%. The strong synergistic effect between PA and CF at a 1:1 ratio (10 µg/mL each) was supported by a 98.8% reduction in MOBB, which is comparable with a 99.3% reduction in MOBB using the conventional combination (CF + SB at 1:1 ratio) in our study. Concurrent with the reduction in MRSA burden was the improvement in the survival rate of mice challenged with MRSA. A 96-h treatment with PA and CF combination completely protected the mice from MRSA infection, i.e., 100% mice survived MRSA challenge, which was also achieved by the CF and SB combination therapy, compared to a 12.5% survival rate for challenged mice without any treatment. In contrast, the PA and cefotaxime combination only improved the survival rate by 70% in a previous study37. Our data suggest that PA has the potential to replace SB in the combination therapy without compromising the efficacy. Reduced antibiotic usage is one of the simplest approaches for mitigating antibiotic resistance.
Multiple other natural products possess potent anti-MRSA activities. For example, at least 51 medicinal plant species with anti-MRSA activities have been recorded39. Among them stands out curcumin, a bioactive component in turmeric. The MIC of curcumin against several MRSA clinical isolates ranged from 125 to 250 μg/mL40. In a mouse pneumonia model, administration of 100 mg/kg of curcumin reduced the mortality rate to 30–40% at 72 h post-infection41. While curcumin has been shown to be synergistic with multiple classical antibiotics, including cefotaximin and oxacillin, its cytotoxicity, low water solubility, and poor bioavailability restrain its clinical applications42. These challenges remain even with the recent application of nanotechnology in improving its delivery43. Our data shows that PA possesses several advantages over curcumin in fighting MRSA infections, including greater efficacy and higher specificity.
One of the most popular anti-virulence strategies has been targeting the accessory gene regulator (agr) quorum-sensing system (QS) in MRSA. This system is a global regulator of virulence factor production. The potential of chemical inhibition of the agr system, such as agrC and agrA, for developing better therapeutic alternatives to conventional antibiotics has been extensively reviewed in the past few years31,44. The major advantage of this approach has been targeting a wide array of virulence mechanisms rather than a single or limited number of virulence factors45. Multiple QS inhibitors or quorum quenchers have been shown to be efficacious in some animal models. For example, savarin, a small molecule targeting the agr:P3 activation, attenuated weight loss resulting from the MRSA challenge infection in a mouse model46. Bioactive peptides or metabolites from probiotics (e.g., lactic acid bacteria) disrupt QS and have the potential to sensitize MRSA to β-lactam antibiotics47. However, QS dysfunction has been associated with increased biofilm formation48 and likely results in unfavorable clinical outcomes49. Further, the impact of QS inhibition on the native gut microbiota has not been extensively evaluated. Our findings show that PA, alone or in combination therapy with conventional antibiotics, can overcome the limitations associated with QS inhibition. First, PA administration alone downregulated the expression of key genes critical to biofilm formation and disrupted this process in its early stage. PA inhibition of biofilm formation directly contributes to its ability to target MRSA colonization. PA is known to affect gut microbial diversity in a chemical-induced colitis mouse model23, in consistent with the findings from this study. However, the difference was also obvious. A 50-day PA supplementation further repressed Phylogenetic diversity (PD whole-tree) as well as the Shannon index from reduced levels in colitic mice as reported in the previous study, whereas PA tended to improve the Shannon index from the reduced levels resulting from antibiotic treatments in the present study. In both studies, PA demonstrated its gut microbiota-modulating properties. In a colitis mouse model, PA supplementation increases the abundance of the gut commensals, such as Coprococcus and Clostridium23, while in the present study, PA restored the abundance of gut beneficial bacteria, such as F. rodentium and A. muciniphila, which act as the sentinel of the gut, either as direct butyrate producers or species critical to gut functions. PA also enhances beneficial gut bacterial interactions in a colitis model induced by Citrobacter rodentium23, in which PA promotes Candidatus Arthromitus (segmented filamentous bacteria) to serve as a hub species, likely contributing to the effect of PA on host immunity, particularly on its role in differentiation and maturation of Th17 cells.
During MRSA infection, macrophages play a key role in pathogen killing and the resolution of inflammation50. M1 macrophages kill pathogens by secreting pro-inflammatory factors and releasing lysosomal enzymes. Excessive inflammatory response induced by M1 macrophages, however, can lead to tissue destruction51. M2 macrophages, on the other hand, facilitate tissue repair by secreting anti-inflammatory factors52. PA possesses potent anti-inflammatory properties that are independent of its anti-microbial activities33. PA binds to TLR4 with high affinity and downregulates several pattern recognition receptors, such as TLR4 and NOD2. PA strongly dampens multiple pro-inflammatory signaling pathways, such as Toll-like receptor signaling, TNF signaling, Chemokine signaling, NF-κB signaling, Jak-STAT signaling, and IL17 signaling33. In this study, we also provided evidence that PA upregulated the expression of IL10, which activates the STAT3 pathway to inhibit inflammatory response53. Thus, PA possesses the desired properties of being an effective antimicrobial agent as well as an anti-inflammatory compound that is beneficial to epithelial remodeling.
In summary, developing anti-virulence agents represents one of the promising strategies for mitigating antibiotic resistance. Here, we provided experimental evidence that PA acts as a high-affinity ligand to a key cell surface-associated protein, SraP, essential for bacterial attachment, disrupting MRSA colonization of host cells. In a murine model, physiologically relevant doses of PA significantly reduced organ MRSA burden and increased mouse survival. The strong synergism between PA and CF is evident as the combination therapy reduces the bacterial load in major organs by 98.9% and completely protects mice from MRSA challenge infection. PA inhibits biofilm formation of the MRSA strain NW2. PA also partially restores the gut microbiota adversely impacted by classical antibiotics and increases the abundance of beneficial species, such as butyrate-producing bacteria. As a natural product, PA, in combination with conventional antibiotics, is a promising alternative to antimicrobials for its potent prophylactic and curative effects.
Methods
Bacterial strains and culture conditions
The MRSA strain MW2 was obtained from Biosea (Beijing, China). 24 food-associated MRSA strains were isolated from raw meat samples collected in 2015 in Zhengzhou (Henan, China), while 24 clinical-associated MRSA strains were isolated from hospital samples in 2018 (Zhengzhou, Henan, China). All strains were grown in Mueller–Hinton (MH) broth. Briefly, frozen stocks of the primary cultures were streaked out on agar plates. A single colony was used to inoculate media, followed by incubation in 10 mL MH broth at 37 °C with 200 rpm shaking for 3 h. The bacteria were collected by centrifugation and then resuspended in MH media at a concentration of 2 × 106 CFU per mL. MRSA genomic DNA was then extracted for further assays.
A549 human respiratory epithelial cells were grown in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS), 5 mM glutamine, penicillin (5 µg/mL), and streptomycin (100 µg/mL) and were used for cell adhesion and invasion assays. Human THP-1 cells used in this study were obtained from MeisenCTCC (Zhejiang, China) and cultured as previously described54. THP-1 cells were then differentiated as a model for human macrophages using phorbol 12-myristate 13-acetate (PMA) (Solarbio, Beijing, China). The effects of PA and CF, individually or in combination, on cell survival and inflammation were evaluated using the PMA-differentiated THP-1 cells. After 6 h of incubation with individual compounds and their combinations, cells were then harvested for cell survival assays and total RNA isolation.
SraP L-lectin (SLL) domain analysis
SLL modules were amplified from all 49 strains, including MW2, 24 food-associated and 24 clinical strains, using the primer pair, GTGCCGCAGTAGGTATTGG (F) and ACGTTGTCGCAACACCATAA (R). The PCR products were purified and sequenced using an ABI 3700 sequencer. SLL amino acid sequences were then compared using Mega v6.0. The final comparative result was output using Clustal X (v2.1).
Construction of SraP L-lectin deletion mutants (ΔL-lectin)
The amplicons flanking the SLL locus were cloned into the S. aureus pKOR1 vector as previously published55. Briefly, DNA fragments of SLL were amplified by PCR using primers L-lectin-up-F/L-lectin-up-R and L-lectin-down-F/L-lectin-down-R (Table S3). The PCR reactions were as follows: 2 μL of cDNA (100 ng), 2 μL of each primer (forward and reverse, 20 nM each), 25 μL of 2X pfu PCR mix, and 19 μL of nuclease-free water. The amplification profiles were: an initial denaturation at 95 °C for 5 min, and then 94 °C for 30 s, 50 °C for 30 s, 72 °C for 40 s for 2 cycles, followed by 30 cycles at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 40 s. Left and right fragments were ligated using primer L-lectin-up-F/L-lectin-down-R. The ligated fragments were inserted into the pUX-T vector and then transformed into E. coli DH5α and verified by resequencing. L-lectin-UD-pUX-T and pKOR1 plasmids were amplified with primer L-lectin-pKOR1-F/L-lectin-pKOR1-R and pKOR1-L-lectin-F/pKOR1-L-lectin-R, respectively. The plasmid pKOR1ΔL-lectin was constructed as follows: 2 μL 5× infusion mix, 2 μL pKOR1 PCR products, 6 μL L-lectin PCR products, transformed into E. coli DH5α, and then electroporated into the strain MW2. Next, 5–10 μL of pKOR1-ΔSraP L-lectin-positive MW2 culture was transferred to pre-warmed Tryptic Soy Broth (TSB) media with 10 μg/mL chloramphenicol and grown at 42 °C overnight with vigorous shaking. The resultant culture was streaked onto Tryptic Soy agar (TSA) plates with 10 μg/mL chloramphenicol pre-warmed at 42 °C and incubated overnight. A colony was inoculated into 5 mL TSB without any drug and incubated at 30 °C overnight or until growth was evident. The culture was then diluted 10,000 times with sterile water, and 10–100 μL was spread onto TSA plates containing 100 ng/mL anhydrotetracycline (Sigma) and incubated at 37 °C. Ten large colonies were then inoculated in 5 mL TSB and grown at 37 °C with shaking overnight. DNA from the SraP L-lectin deletion mutant (ΔL-lectin) was extracted and purified with a miniprep kit (Qiagen) and verified by quantitative PCR.
Molecular docking
The structures of the MRSA SraP protein were selected and retrieved from the Protein Data Bank (PDB, ID# 4M00). The raw proteins were further prepared by removing water molecules and ligands and adding hydrogen to polar groups and charges using Chimera. All 14,000 bioactive natural compounds were downloaded from the TCMSP database (https://tcmsp-e.com/tcmsp.php)17. All compounds were prepared using Chem3D to minimize energy and then saved as .mol2 format to select optimal ligand conformations. Molecular docking was carried out using AutoDock Vina56. The size of the docking grid was 30 Å × 30 Å × 30 Å along the x, y, and z axes, respectively, and the exhaustiveness was set to 8. The energy range was 3. The number of modes tested was set to 9. The default values were used for all other parameters. The results were sorted by binding affinity and visually analyzed using PyMOL.
Surface plasmon resonance assays
SPR was used to determine the binding affinities of SLL with natural bioactive compounds. SPR experiments were performed at 25 °C using a Biacore T200 instrument (GE Healthcare). The following buffers were purchased from Cytiva (Marlborough, MA, USA): Concentrated 10× stock HBS-P+ (containing: 0.1 M of 4-(2-Hydroxyethyl)piperazine-1-ethanesulfonic acid or HEPES, 1.5 M NaCl, 0.5% (v/v) Surfactant Tween 20, pH 7.4); screening buffer: 1.0× HBS-P+ (pH 7.4) with 5% DMSO, and regeneration solution (Glycine 3.0 containing 10 mM Glycine–HCl pH 3.0). SLL was covalently immobilized on the carboxymethyl dextran surface of a CM5 sensor chip using a Biacore amine coupling kit. The chip was activated with the N-ethyl-N-[3-dimethylaminopropyl] carbodi-imide/N-hydroxysuccinimide) solution, and SLL in 10 mM acetate buffer (pH 5.5) was injected into the flow channel. The flow rate was set at 5 mL/min. The sensor surface was then blocked with 1 M ethanolamine. The blank channel was treated in the same way, but without the SLL protein injected. Each natural bioactive compound in the running buffer was incubated for 1 min in the flow cells. Both injection and dissociation steps lasted for 5 min. The sensor surface was regenerated with 50 mM NaOH. The natural ligand of SLL, Neu5AC, served as a positive control. All analyses were performed with GE BiaEvaluate software (v4.1). Briefly, SLL-natural product interaction kinetics were analyzed by monitoring the interaction as a function of time over a range of analyte concentrations and then fitting the whole data set to a preset kinetic fit model provided by Biacore T200. The BiaEvaluate software calculated an on-rate ka and an off-rate kd values according to the kinetic parameters and resultant sensorgrams. The equilibrium dissociation constant Kd is defined as kd/ka.
Minimal inhibitory concentration (MIC) assay
The MICs of bioactive natural compounds were determined using a microdilution method as well as agar plating, as described57, and are in compliance with the US National Committee for Clinical Laboratory Standards. Briefly, 100 μL bacterial strains in Mueller–Hinton Broth (MHB) at a concentration of 5 × 105 CFU/mL were added to 96-well plates. 100 μL of culture media containing different concentrations of test compounds and reference antibiotics was then added to the respective wells on the plates. The MIC in this study is defined as the lowest concentration that confers a complete inhibition of colony growth on agar plates (i.e., 100% reduction in colony counts in all replicates or MIC100). In addition, MIC90, i.e., 90% inhibition, was also calculated using microdilution assays.
Bacterial growth curve analysis
For in vitro growth experiments, PA (Push Bio-tech, Chengdu, China) at a given dose range, from 0 to 550 μg/mL, was added to the media for 2, 4, 6, and 8 h, respectively. The MRSA colonies were counted by retrospective plating. The bacterial cells were then harvested for total RNA extraction.
A serial passage or adaptive laboratory evolution approach was used to select the MW2 strain for possible development of resistance to PA. The MW2 strain was cultured in the presence of PA at 10 μg/mL at 37 °C for 12 h. Approximately 100 μL of a bacterial culture was then plated onto an agar plate, and a single colony was streaked out after overnight incubation. The stock was saved as the 1st generation. The procedure was repeated 10 times, and a total of 10 consecutive generations of the MRSA MW2 strain in the presence of PA were obtained. The growth characteristics of these 10 consecutive generations of MW2 under various PA concentrations were compared by measuring optical density (OD) values at 600 nm wavelength or by agar plating.
Cell adhesion and invasion assays
The effect of PA on bacterial adhesion and invasion was evaluated using human lung epithelial A549 cells. Approximately 2.5 × 105 cells were seeded onto 24-well culture plates and grown in 5% CO2 at 37 °C. The monolayers were first washed twice with PBS. For adhesion assays, the final concentration of S. aureus was adjusted to 6.25 × 108 CFU/mL and pre-treated with either PBS or PA (10 µg/mL) for 6 h. The bacteria were then diluted to a concentration of 6.25 × 106 CFU/mL and used to infect the confluent cell monolayers at 37 °C for 2 h. After incubation, the infected monolayers were washed five times with PBS to remove non-adhered bacteria, and then treated with 200 µL trypsin (2.5 mg/mL) at 37 °C for 2 min to release the adhered bacteria. Similarly, for invasion assays, the monolayer cells were infected with either PA or PBS-pretreated S. aureus and incubated at 37 °C for 2 h. After infection, the cells were washed three times with cold PBS and incubated with the medium containing a broad-spectrum antibiotic, gentamicin, at 100 μg/mL for 2 h. Gentamicin kills extracellular bacteria that are not engulfed by cells, while engulfed intracellular bacteria are not affected by it. After incubation, the infected monolayers were washed five times with PBS to remove non-adhered bacteria, and treated with 200 µL trypsin (2.5 mg/mL) at 37 °C for 2 min. The numbers of adhered and invasive bacteria were determined by plating serial dilutions of the recovered bacterial suspensions onto LB agar plates.
Cell viability (survival) assays
THP-1-derived macrophage cells were seeded in 96-well plates at a density of 1–2 × 105 cells per well. A range of PA concentrations (0–250 μg/mL) was added and cultured at 37 °C for 24 h. After the culture, 10 μL of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution at 500 μg/mL (Beyotime, Shanghai, China) was added to each well and incubated for an additional 4 h. DMSO was then added as a solvent. Viable cells metabolite MTT to a colored product. The absorption at 510 nm was measured using an M5/i3X microplate reader (Molecular Devices, San Jose, CA, USA).
Biofilm formation assay
2 × 106 CFU/mL S. aureus MW2 WT and ΔL-lectin strains were prepared in TSB media containing 3% NaCl and 0.5% glucose. 100 μL of the bacterial solution was then added to each well of a 96-well plate. Subsequently, 100 μL PA stock solution, at 500, 250, 100, 20, 4, and 0 μg/mL, was added to the corresponding wells and cultured at 37 °C for 24 h. Biofilm formation was evaluated using the crystal violet assay to calculate BFI. Briefly, the OD630 values of bacterial culture were measured with a microplate reader. The culture medium was then discarded, and 200 μL crystal violet (1%, W/V) was added to each well and stained for 20 min. PBS was used to wash away the excess dye. After air drying, 200 μL anhydrous ethanol was added to each well to elute the binding dye. 100 μL of the eluate was then transferred to a new 96-well plate. The absorbance value, OD570, was measured using a microplate reader. The BFI was calculated as follows: BFI = (S−SC)/(G−GC), where S and SC refer to the OD570 value of PA and control groups, respectively, while G and GC refer to the OD630 value of PA and control groups, respectively.
MRSA morphological analysis
WT and ΔL-lectin MW2 strains were cultured to the logarithmic phase. The cultures were then treated with 10 μg/mL PA at 37 °C for 8 h. Subsequently, 2 mL of the culture medium was collected by centrifugation at 10,000×g for 10 min, and the pellets were fixed in modified Karnovsky’s fixative. The specimens were then examined using a Hitachi transmission electron microscope (Hitachi, Minato-ku, Tokyo, Japan).
Gene expression analysis using qRT-PCR and RNAseq
Total RNA was extracted from MRSA cells using a hot TRIzol procedure. Briefly, 1 mL of pre-heated TRIzol at 60 °C was added to bacterial pellets (per 1–5 × 108 cells) and mixed by vortexing. The mixture was placed on a 60 °C heat-block for 5 min to facilitate the disruption of the protein-RNA complex. All other steps were carried out using the standard TRIzol protocol from the manufacturer. Crude total RNA was further purified using an RNA Cleanup & Concentration kit (Sangon, Shanghai, China) with DNase digestion as described. RNA integrity was verified using a BioAnalyzer 2100 (Agilent, Palo Alto, CA). An iScript advanced cDNA synthesis kit was used for cDNA synthesis from total RNA (Bio-Rad, Hercules, CA). Quantitative (q) RT-PCR reactions were carried out using a CFX Connect Real-Time PCR Detection System (Bio-Rad) as previously described58. The reactions were performed in duplicate in a total volume of 22 μL containing the following: 2 μL of cDNA (100 ng), 0.5 μL of each primer (forward and reverse, 20 nM each, Table S3), 11 μL of SsoAdvanced Universal SYBR Green Supermix, and 8 μL of nuclease-free water. The amplification reactions were subjected to an initial denaturation at 95 °C for 3 min, followed by 40 cycles of 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 30 s.
After rRNA depletion to enrich mRNA, RNA-seq libraries were prepared using an Illumina HiSeq RNA sample prep kit following the manufacturer’s instructions (Illumina). The libraries for individual samples were pooled at an equal molar ratio and based on their respective sample-specific barcodes. Paired-end sequences were generated at 100 bp/read using an Illumina HiSeq sequencer. The quality of raw sequences was checked using FastQC (v0.11.9). Raw sequences were then trimmed using Trimmomatic (v0.38). The preprocessed reads were analyzed using the Bowtie2-DESeq2 pipeline with default parameters. A false discovery rate (FDR) < 0.05 was used as a cutoff for determining differentially expressed genes. Gene enrichment analysis was conducted using ClusterProfiler59.
Checkerboard assay
The checkerboard method was used to screen antibiotics for the synergistic effect between PA and β-lactam antibiotics in combating clinically isolated MRSA strains. Briefly, 100 μl of PA at different final concentrations were added to each well in a 96-well plate in a certain sequence. Different concentrations of 17 antibiotics, CF, ceftriaxone sodium, cefazolin, ampicillin, aztreonam, cefixime, cefathiamidine, cephradine, cefoxitin, cefaclor, oxacillin sodium, piperacillin sodium, amoxicillin sodium, cefdinir, cefodizime acid, cefditoren pivoxil, and penicillin G Na, as well as sulbactam, a well-known β-lactamase inhibitor, all purchased from McLean Biochemical Technology (Shanghai, China), were added to the MW2 strain in MH media. The absorbance was measured at 600 nm after incubation at 37 °C for 16 h using a microplate reader. The fractional inhibitory concentration index (FICI) was then calculated according to the formula FICI = FICA + FICB = CA/MICA + CB/MICB. The four categories of interactions60 were predefined as follows: synergism, FICI ≤ 0.5; additivity, 0.5 < FICI ≤ 1.0; irrelevance, 1.0 < FICI ≤ 2.0; and antagonism, FICI > 2.
Animal studies
The animal use protocol (AUP) in this study was reviewed and approved by the ethical committee of Zhengzhou University (AUP no. ZZUIRB2023-151). All procedures specified by Chinese regulations on the Management of Laboratory Animal License System and the Guideline on Humane Treatment of Laboratory Animals were strictly followed. A total of 144 ICR male mice, between 4 and 6 months of age, were obtained from SPF Biotechnology (Beijing, China). Upon arrival, all experimental mice were fed a basal AIN-93M diet, formulated by Henan Medical Experimental Animal Center (Zhengzhou, China), on an ad libitum basis and had free access to water throughout the experiment. Mice were group housed, with four mice per cage, according to the standards described in the Guide for the Care and Use of Laboratory Animals. The room in which the cages were placed had a 12 light/12 dark cycle with temperatures of 23–25 °C and 40–60% humidity. Mice were acclimated for seven days before the two animal studies were initiated. In the first study, 80 mice were randomly divided into eight groups (n = 10 per group): uninfected and treated with PBS, MRSA infected and treated with either PBS, PA (10 mg/kg BW), CF (10 mg/kg BW), CF (20 mg/kg BW), PA (10 mg/kg BW) + CF (2.5 mg/kg BW), PA (10 mg/kg BW) + CF (10 mg/kg BW), or CF (10 mg/kg BW) + SB (10 mg/kg BW), all dissolved in PBS, respectively. Mice in MRSA-infected groups received the MW2 strain at a single dose of 2 × 108 CFU in 0.4 mL of MHB by intraperitoneal administration. Uninfected control mice received the 0.4 mL MH broth without any MRSA bacteria. The mice in each group received a single daily dose of 0.2 mL of the respective treatments via intraperitoneal injection starting 1 h after the MRSA inoculation for three consecutive days. Mice were monitored and weighed daily. Mice were euthanized on day 3 post-infection. CO2 asphyxiation was used as the primary method of euthanasia for mice. Briefly, a standard CO2 euthanasia station, consisting of a compressed CO2 tank, a gas regulator, and a flow meter, was used to connect to a lid that fits on a mouse cage. Mice were euthanized in their home cage, within 10 min after their transfer to the euthanasia station located in a separate room. The flow rate into the euthanasia chamber was controlled at a rate of 30–70% of the total volume of the chamber per minute. The mice were exposed to CO₂ for minimal 3 min or until complete cessation of breathing. Death was verified using cervical dislocation. Five organs, including the liver, spleen, colon, kidney, and lung, were removed at necropsy, carefully weighed, and then homogenized in MH broth and plated on LB agar plates to enumerate the total bacterial load. In addition, the liver tissue was fixed in 10% neutral buffered formalin and then sectioned at 5-µm thickness for H&E staining. In the second survival study, a total of 64 mice were randomly divided into the same eight above-mentioned groups (n = 8 per group) and used to determine the cumulative percentage survival. Mice in MRSA-infected groups received the MW2 strain at a single dose of 2 × 109 CFU in 0.4 mL of MHB by intraperitoneal administration. The treatment schemes and dose levels were conducted exactly as in the 1st study. The survival of mice in each group was monitored for 7 days after infection, and the cumulative percentage survival was then determined.
Full-length 16S rRNA gene sequencing-based microbiome study
Colon contents (feces) were collected at necropsy at the conclusion of the 1st animal study. Total DNA was extracted from the fecal samples using a QIAamp Fast DNA Stool Mini kit (Qiagen) with some modifications54. First, a bead-beating procedure using a FastPrep 5 G instrument and Lysing Matrix E was performed. Second, lysis at 70 °C was extended to 8 min. Both nuclease-free water as a non-DNA template control and the microbiome standard (Zymo Research, Irvine, CA, USA) were processed along with experimental samples following the same protocol and parameters to validate the workflow. The full-length 16S rRNA gene was amplified and sequenced based on LoopSeq long-read chemistry (Element BioSciences, San Diego, CA, USA) using an Illumina NovoSeq 6000 sequencer. The raw reads were first trimmed using Trimmomatic (v0.39) and then de novo assembled into full-length 16S sequences using the SPAdes algorithm in the LoopSeq proprietary pipeline. The resultant full-length 16S rRNA gene sequences were analyzed using the DADA2 algorithm (v1.16) to generate amplicon sequence variant (ASV) feature tables61. The ASV data were then analyzed as previously described23. Raw sequence data have been deposited in the NCBI SRA database (SRA accession: PRJNA1101901).
Statistical analysis
Statistical analyses in this study were performed using SPSS 13.0 software (Chicago, IL, USA). One-way ANOVA was used for multi-group comparisons, followed by a post hoc test by Tukey’s honestly significant difference (HSD) for the data collected for cell survival assays, qRT-PCR, and morphological and pathological indicators, including the disease activity Index. In addition, Bartlett’s test and the Brown–Forsythe test were performed before ANOVA to check if the assumption of equal variances across groups is satisfied. A non-parametric Wilcoxon Rank-Sum test (Mann–Whitney U test) was used for two-group statistical comparisons for the data collected from other in vitro (four to six replicates per group) and in vivo animal studies (N = 10 and 8 per group for Study#1 and Study#2, respectively). The number was expressed as the mean ± SD. P value ≤ 0.05 was considered statistically significant. For RNAseq analysis, the cutoff for differentially expressed genes was false discovery rate (FDR) adjusted q values ≤ 0.05 and fold changes ≥ 2.0.
Data availability
The raw sequences have been deposited in the NCBI SRA database (SRA accession: PRJNA1101901). Supporting information is also available as the online supplementary file. All other supporting materials are available from the lead author upon request.
References
Hassoun, A., Linden, P. K. & Friedman, B. Incidence, prevalence, and management of MRSA bacteremia across patient populations—a review of recent developments in MRSA management and treatment. Crit. Care 21, 211 (2017).
Tong, S. Y., Davis, J. S., Eichenberger, E., Holland, T. L. & Fowler, V. G. Jr Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 28, 603–661 (2015).
Bassetti, M. et al. Current pharmacotherapy for methicillin-resistant Staphylococcus aureus (MRSA) pneumonia. Expert Opin. Pharmacother. 23, 361–375 (2022).
Guo, Y., Song, G., Sun, M., Wang, J. & Wang, Y. Prevalence and therapies of antibiotic-resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 10, 107 (2020).
Vestergaard, M., Frees, D. & Ingmer, H. Antibiotic resistance and the MRSA problem. Microbiol. Spectr. 7, https://doi.org/10.1128/microbiolspec.GPP3-0057-2018 (2019).
Poudel, A. N. et al. The economic burden of antibiotic resistance: a systematic review and meta-analysis. PLoS ONE 18, e0285170 (2023).
Josse, J., Laurent, F. & Diot, A. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front. Microbiol. 8, 2433 (2017).
Baker, E. P. et al. Evolution of host-microbe cell adherence by receptor domain shuffling. Elife 11, https://doi.org/10.7554/eLife.73330 (2022).
Foster, T. J., Geoghegan, J. A., Ganesh, V. K. & Hook, M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat. Rev. Microbiol. 12, 49–62 (2014).
Yang, Y. H. et al. Structural insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 10, e1004169 (2014).
Lizcano, A., Sanchez, C. J. & Orihuela, C. J. A role for glycosylated serine-rich repeat proteins in Gram-positive bacterial pathogenesis. Mol. Oral Microbiol. 27, 257–269 (2012).
Zhou, T. T. et al. Monoclonal antibody against l-lectin module of SraP blocks adhesion and protects mice against Staphylococcus aureus challenge. J. Microbiol. Immunol. Infect. 54, 420–428 (2021).
Atanasov, A. G., Zotchev, S. B. & Dirsch, V. M. International Natural Product Sciences, T. & Supuran, C. T. Natural products in drug discovery: advances and opportunities. Nat. Rev. Drug Discov. 20, 200–216 (2021).
Moreno Cardenas, C. & Cicek, S. S. Structure-dependent activity of plant natural products against methicillin-resistant Staphylococcus aureus. Front. Microbiol. 14, 1234115 (2023).
Liang, M. et al. Phytochemicals with activity against methicillin-resistant Staphylococcus aureus. Phytomedicine 100, 154073 (2022).
Vaou, N., Stavropoulou, E., Voidarou, C., Tsigalou, C. & Bezirtzoglou, E. Towards advances in medicinal plant antimicrobial activity: a review study on challenges and future perspectives. Microorganisms 9, https://doi.org/10.3390/microorganisms9102041 (2021).
Ru, J. et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform 6, 13 (2014).
Kupnik, K., Primozic, M., Vasic, K., Knez, Z. & Leitgeb, M. A Comprehensive study of the antibacterial activity of bioactive juice and extracts from pomegranate (Punica granatum L.) peels and seeds. Plants (Basel) 10, https://doi.org/10.3390/plants10081554 (2021).
Alvarez-Martinez, F. J., Rodriguez, J. C., Borras-Rocher, F., Barrajon-Catalan, E. & Micol, V. The antimicrobial capacity of Cistus salviifolius and Punica granatum plant extracts against clinical pathogens is related to their polyphenolic composition. Sci. Rep. 11, 588 (2021).
Li, G., Xu, Y., Pan, L. & Xia, X. Punicalagin damages the membrane of Salmonella Typhimurium. J. Food Prot. 83, 2102–2106 (2020).
Zhang, Z. et al. Assessment of global health risk of antibiotic resistance genes. Nat. Commun. 13, 1553 (2022).
Yang, Y. H. et al. Structural Insights into SraP-mediated Staphylococcus aureus adhesion to host cells. PLoS Pathog. 10, ARTN e1004169 (2014).
Liu, F. et al. Multi-omics analysis detected multiple pathways by which pomegranate punicalagin exerts its biological effects in modulating host–microbiota interactions in murine colitis models. Food Funct. 14, 3824–3837 (2023).
Zhao, Y. et al. Butyrate protects against MRSA pneumonia via regulating gut–lung microbiota and alveolar macrophage M2 polarization. mBio 14, e0198723 (2023).
Cao, Y. G. et al. Faecalibaculum rodentium remodels retinoic acid signaling to govern eosinophil-dependent intestinal epithelial homeostasis. Cell Host Microbe 30, 1295–1310 e1298 (2022).
Antimicrobial Resistance, C. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655 (2022).
Hasanpour, A. H. et al. The global prevalence of methicillin-resistant Staphylococcus aureus colonization in residents of elderly care centers: a systematic review and meta-analysis. Antimicrob. Resist Infect. Control 12, 4 (2023).
Lade, H. & Kim, J. S. Bacterial targets of antibiotics in methicillin-resistant Staphylococcus aureus. Antibiotics (Basel) 10, https://doi.org/10.3390/antibiotics10040398 (2021).
Van Bambeke, F., Mingeot-Leclercq, M. P., Struelens, M. J. & Tulkens, P. M. The bacterial envelope as a target for novel anti-MRSA antibiotics. Trends Pharm. Sci. 29, 124–134 (2008).
Clatworthy, A. E., Pierson, E. & Hung, D. T. Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 (2007).
Salam, A. M. & Quave, C. L. Targeting virulence in Staphylococcus aureus by chemical inhibition of the accessory gene regulator system in vivo. mSphere 3, https://doi.org/10.1128/mSphere.00500-17 (2018).
Schneewind, O. & Missiakas, D. M. Staphylococcal protein secretion and envelope assembly. Microbiol. Spectr. 7, https://doi.org/10.1128/microbiolspec.GPP3-0070-2019 (2019).
Liu, F. et al. Ellagitannin punicalagin disrupts the pathways related to bacterial growth and affects multiple pattern recognition receptor signaling by acting as a selective histone deacetylase inhibitor. J. Agric. Food Chem. 71, 5016–5026 (2023).
Kulkarni, A. P., Mahal, H. S., Kapoor, S. & Aradhya, S. M. In vitro studies on the binding, antioxidant, and cytotoxic actions of punicalagin. J. Agric. Food Chem. 55, 1491–1500 (2007).
Mun, S. H. et al. Punicalagin suppresses methicillin resistance of Staphylococcus aureus to oxacillin. J. Pharm. Sci. 137, 317–323 (2018).
Mun, S. H. et al. Subinhibitory concentrations of punicalagin reduces expression of virulence-related exoproteins by Staphylococcus aureus. FEMS Microbiol. Lett. 363, https://doi.org/10.1093/femsle/fnw253 (2016).
Song, W. et al. Punicalagin, an inhibitor of Sortase A, is a promising therapeutic drug to combat methicillin-resistant Staphylococcus aureus infections. Antimicrob. Agents Chemother. 66, e0022422 (2022).
Worthington, R. J. & Melander, C. Combination approaches to combat multidrug-resistant bacteria. Trends Biotechnol. 31, 177–184 (2013).
Okwu, M. U., Olley, M., Akpoka, A. O. & Izevbuwa, O. E. Methicillin-resistant Staphylococcus aureus (MRSA) and anti-MRSA activities of extracts of some medicinal plants: a brief review. AIMS Microbiol. 5, 117–137 (2019).
Mun, S. H. et al. Synergistic antibacterial effect of curcumin against methicillin-resistant Staphylococcus aureus. Phytomedicine 20, 714–718 (2013).
Wang, J. et al. Curcumin protects mice from Staphylococcus aureus pneumonia by interfering with the self-assembly process of alpha-hemolysin. Sci. Rep. 6, 28254 (2016).
Teow, S. Y., Liew, K., Ali, S. A., Khoo, A. S. & Peh, S. C. Antibacterial action of curcumin against Staphylococcus aureus: a brief review. J. Trop. Med 2016, 2853045 (2016).
Bugli, F. et al. Curcumin-loaded graphene oxide flakes as an effective antibacterial system against methicillin-resistant Staphylococcus aureus. Interface Focus 8, 20170059 (2018).
Otto, M. Critical assessment of the prospects of quorum-quenching therapy for Staphylococcus aureus infection. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms24044025 (2023).
Sully, E. K. et al. Selective chemical inhibition of agr quorum sensing in Staphylococcus aureus promotes host defense with minimal impact on resistance. PLoS Pathog. 10, e1004174 (2014).
Hall, P. R. et al. Nox2 modification of LDL is essential for optimal apolipoprotein B-mediated control of agr type III Staphylococcus aureus quorum-sensing. PLoS Pathog. 9, e1003166 (2013).
Cella, M. A. et al. Probiotic disruption of quorum sensing reduces virulence and increases cefoxitin sensitivity in methicillin-resistant Staphylococcus aureus. Sci. Rep. 13, 4373 (2023).
He, L. et al. Key role of quorum-sensing mutations in the development of Staphylococcus aureus clinical device-associated infection. Clin. Transl. Med. 12, e801 (2022).
Lee, S. O. et al. Dysfunctional accessory gene regulator (agr) as a prognostic factor in invasive Staphylococcus aureus infection: a systematic review and meta-analysis. Sci. Rep. 10, 20697 (2020).
Chen, Y. et al. The effect of Staphylococcus aureus on innate and adaptive immunity and potential immunotherapy for S. aureus-induced osteomyelitis. Front. Immunol. 14, 1219895 (2023).
Gordon, S. & Martinez, F. O. Alternative activation of macrophages: mechanism and functions. Immunity 32, 593–604 (2010).
Parisi, L. et al. Macrophage polarization in chronic inflammatory diseases: killers or builders?. J. Immunol. Res. 2018, 8917804 (2018).
Lang, R., Patel, D., Morris, J. J., Rutschman, R. L. & Murray, P. J. Shaping gene expression in activated and resting primary macrophages by IL-10. J. Immunol. 169, 2253–2263 (2002).
Liu, F. et al. Mechanistic insights into the attenuation of intestinal inflammation and modulation of the gut microbiome by krill oil using in vitro and in vivo models. Microbiome 8, 83 (2020).
Bae, T. & Schneewind, O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid 55, 58–63 (2006).
Trott, O. & Olson, A. J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31, 455–461 (2010).
Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin. Microbiol. Infect. 9, ix–xv (2003).
Liu, F. et al. Malvidin 3-glucoside modulated gut microbial dysbiosis and global metabolome disrupted in a murine colitis model induced by dextran sulfate sodium. Mol. Nutr. Food Res. 63, e1900455 (2019).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Cambridge) 2, 100141 (2021).
Konate, K. et al. Antibacterial activity against beta-lactamase producing methicillin and ampicillin-resistants Staphylococcus aureus: fractional inhibitory concentration index (FICI) determination. Ann. Clin. Microbiol. Antimicrob. 11, 18 (2012).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Acknowledgements
This work was partially supported by the CSC, China Postdoctoral Science Foundation (2022M722861), and National Natural Science Foundation of China (No 82204036).
Author information
Authors and Affiliations
Contributions
F.L.: Conceptualization, investigation, formal analysis, methodology, visualization, and writing. J.R.: Investigation. P.Z.: Investigation. WZ: Investigation. W.L.: Investigation. A.D.S.: Investigation. R.W.L.: Formal analysis and writing. H.Y.Y.: Conceptualization and supervision. All authors approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, F., Ren, J., Zhang, P. et al. Exploiting strong synergies between punicalagin and cefoperazone to combat methicillin-resistant Staphylococcus aureus infections. npj Biofilms Microbiomes 11, 193 (2025). https://doi.org/10.1038/s41522-025-00822-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41522-025-00822-7