Abstract
Many animals have a conserved adaptive genome defence system known as the Piwi-interacting RNA (piRNA) pathway, which is essential for germ cell development and function. Disruption of individual mouse Piwi genes results in male but not female sterility, leading to the assumption that PIWI genes play little or no role in mammalian oocytes. Here, we report the generation of PIWI-defective golden hamsters, which have defects in the production of functional oocytes. The mechanisms involved vary among the hamster PIWI genes, whereby the lack of PIWIL1 has a major impact on gene expression, including hamster-specific young transposon de-silencing, whereas PIWIL3 deficiency has little impact on gene expression in oocytes, although DNA methylation was reduced to some extent in PIWIL3-deficient oocytes. Our findings serve as the foundation for developing useful models to study the piRNA pathway in mammalian oocytes, including humans.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The raw sequence data generated during this study are available from the Gene Expression Omnibus repository under accession number GSE164356. Requests for materials should be addressed to H. Siomi. Source data are provided with this paper. All data presented in this study are available in the main text or the supplementary materials.
References
Iwasaki, Y. W., Siomi, M. C. & Siomi, H. PIWI-Interacting RNA: its biogenesis and functions. Annu. Rev. Biochem. 84, 405–433 (2015).
Ozata, D. M., Gainetdinov, I., Zoch, A., O’Carroll, D. & Zamore, P. D. PIWI-interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20, 89–108 (2019).
Pillai, R. S. & Chuma, S. piRNAs and their involvement in male germline development in mice. Dev. Growth Differ. 54, 78–92 (2012).
Deng, W. & Lin, H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002).
Aravin, A. A. et al. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol. Cell 31, 785–799 (2008).
Carmell, M. A. et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev. Cell 12, 503–514 (2007).
Kuramochi-Miyagawa, S. et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev. 24, 887–892 (2010).
Reuter, M. et al. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011).
De Fazio, S. et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011).
Cheng, E. C., Kang, D., Wang, Z. & Lin, H. PIWI proteins are dispensable for mouse somatic development and reprogramming of fibroblasts into pluripotent stem cells. PLoS ONE 9, e97821 (2014).
Lim, A. K. et al. The nuage mediates retrotransposon silencing in mouse primordial ovarian follicles. Development 140, 3819–3825 (2013).
Kabayama, Y. et al. Roles of MIWI, MILI and PLD6 in small RNA regulation in mouse growing oocytes. Nucleic Acids Res. 45, 5387–5398 (2017).
Hirano, T. et al. Small RNA profiling and characterization of piRNA clusters in the adult testes of the common marmoset, a model primate. RNA 20, 1223–1237 (2014).
Sasaki, T., Shiohama, A., Minoshima, S. & Shimizu, N. Identification of eight members of the Argonaute family in the human genome. Genomics 82, 323–330 (2003).
Roovers, E. F. et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 10, 2069–2082 (2015).
Ishino, K. et al. Hamster PIWI proteins bind to piRNAs with stage-specific size variations during oocyte maturation. Nucleic Acids Res. 49, 2700–2720 (2021).
Yang, Q. et al. Single-cell CAS-seq reveals a class of short PIWI-interacting RNAs in human oocytes. Nat. Commun. 10, 3389 (2019).
Sia, S. F. et al. Pathogenesis and transmission of SARS-CoV-2 in golden hamsters. Nature 583, 834–838 (2020).
Hirose, M. et al. Acrosin is essential for sperm penetration through the zona pellucida in hamsters. Proc. Natl Acad. Sci. USA 117, 2513–2518 (2020).
Fan, Z. et al. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS ONE 9, e109755 (2014).
Vourekas, A. et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat. Struct. Mol. Biol. 19, 773–781 (2012).
Korsholm, L. M. et al. Recent advances in the nucleolar responses to DNA double-strand breaks. Nucleic Acids Res. 48, 9449–9461 (2020).
Tanaka, H. et al. The SETD8/PR-Set7 methyltransferase functions as a barrier to prevent senescence-associated metabolic remodeling. Cell Rep. 18, 2148–2161 (2017).
Molaro, A. et al. Two waves of de novo methylation during mouse germ cell development. Genes Dev. 28, 1544–1549 (2014).
Zoch, A. et al. SPOCD1 is an essential executor of piRNA-directed de novo DNA methylation. Nature 584, 635–639 (2020).
Watanabe, T., Cui, X., Yuan, Z., Qi, H. & Lin, H. MIWI2 targets RNAs transcribed from piRNA-dependent regions to drive DNA methylation in mouse prospermatogonia. EMBO J. 37, e95329 (2018).
Nagamori, I. et al. Comprehensive DNA methylation analysis of retrotransposons in male germ cells. Cell Rep. 12, 1541–1547 (2015).
Yang, F. et al. TEX15 associates with MILI and silences transposable elements in male germ cells. Genes Dev. 34, 745–750 (2020).
Sasaki, H. & Matsui, Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat. Rev. Genet. 9, 129–140 (2008).
Demond, H. & Kelsey, G. The enigma of DNA methylation in the mammalian oocyte. F1000 Res. https://doi.org/10.12688/f1000research.21513.1 (2020).
Loubalova, Z. et al Formation of spermatogonia and fertile oocytes in golden hamsters requires piRNAs. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00746-2 (2021).
Zheng, K. et al. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Natl Acad. Sci. USA 107, 11841–11846 (2010).
Frost, R. J. A. et al. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl Acad. Sci. USA 107, 11847–11852 (2010).
Zhang, H. et al. The piRNA pathway is essential for generating functional oocytes in golden hamsters. Nat. Cell Biol. https://doi.org/10.1038/s41556-021-00750-6 (2021).
Bourc’his, D., Xu, G. L., Lin, C. S., Bollman, B. & Bestor, T. H. Dnmt3L and the establishment of maternal genomic imprints. Science 294, 2536–2539 (2001).
Suetake, I., Shinozaki, F., Miyagawa, J., Takeshima, H. & Tajima, S. DNMT3L stimulates the DNA methylation activity of Dnmt3a and Dnmt3b through a direct interaction. J. Biol. Chem. 279, 27816–27823 (2004).
Kaneda, M. et al. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429, 900–903 (2004).
Smallwood, S. A. et al. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat. Genet. 43, 811–814 (2011).
Monk, D., Sanchez-Delgado, M. & Fisher, R. NLRPs, the subcortical maternal complex and genomic imprinting. Reproduction 154, R161–R170 (2017).
Flemr, M. et al. A retrotransposon-driven dicer isoform directs endogenous small interfering RNA production in mouse oocytes. Cell 155, 807–816 (2013).
Ludwig, T. E., Squirrell, J. M., Palmenberg, A. C. & Bavister, B. D. Relationship between development, metabolism, and mitochondrial organization in 2-cell hamster embryos in the presence of low levels of phosphate. Biol. Reprod. 65, 1648–1654 (2001).
Naito, Y., Hino, K., Bono, H. & Ui-Tei, K. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123 (2015).
Iwasaki, Y. W. et al. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol. Cell 63, 408–419 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12 (2011).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S. L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Kinsella, R. J. et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database 2011, bar030 (2011).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Iwasaki, Y. W., Ishino, K. & Siomi, H. Deep sequencing and high-throughput analysis of PIWI-associated small RNAs. Methods 126, 66–75 (2017).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Su, W., Sun, J., Shimizu, K. & Kadota, K. TCC-GUI: a Shiny-based application for differential expression analysis of RNA-seq count data. BMC Res. Notes 12, 133 (2019).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017).
Pimentel, H., Bray, N. L., Puente, S., Melsted, P. & Pachter, L. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017).
Miura, F., Enomoto, Y., Dairiki, R. & Ito, T. Amplification-free whole-genome bisulfite sequencing by post-bisulfite adaptor tagging. Nucleic Acids Res. 40, e136 (2012).
Au Yeung, W. K. et al. Histone H3K9 methyltransferase G9a in oocytes Is essential for preimplantation development but dispensable for CG methylation protection. Cell Rep. 27, 282–293 e284 (2019).
Toh, H. et al. Software updates in the Illumina HiSeq platform affect whole-genome bisulfite sequencing. BMC Genomics 18, 31 (2017).
Acknowledgements
We thank all members of the Siomi Laboratory, especially H. Ishizu and K. Murano, for discussions and comments on this work, I. Ishimatsu for preparing paraffin sections, and H. Nishihara (Tokyo Institute of Technology) for his comments on hamster TEs. We also thank J. Oishi of the Sasaki Laboratory and T. Akinaga (Laboratory for Research Support, Medical Institute of Bioregulation, Kyushu University) for their help with whole-genome bisulfite sequencing. We are grateful to P. Svoboda (IMG, Czech Academy of Sciences), J. Li (Model Animal Research Center, Nanjing Medical University) and L. Wu (Shanghai Institute of Biochemistry and cell Biology, CAS) for sharing unpublished data. We are also grateful to M. Okabe and M. Ikawa (Osaka University) for their comments on the manuscript. Funding included support from a Grant-in-Aid for the Japan Society for the Promotion of Science (JSPS) KAKENHI grant no. 20H03175 (to H.H.), the Takeda Science Foundation (to H.H.), a JSPS Fellows (18J22025) (to K.I.), from JSPS KAKENHI grant nos 19H05268 and 18H02421, and from JST PRESTO grant no. JPMJPR20E2 (to Y.W.I.), for Scientific Research (S) (25221003) and the Program of totipotency (19H05753) (to H. Siomi). H. Siomi is also a recipient of funding for the project for elucidating and controlling mechanisms of ageing and longevity (1005442) from the Japan Agency for Medical Research and Development (AMED). This work was also supported by JSPS KAKENHI grant no. JP18H05214 (H. Sasaki).
Author information
Authors and Affiliations
Contributions
H. Siomi and H.H. designed the experiments. H.H. generated the PIWIL1-deficient and PIWIL3-deficient hamsters (with the help of H.M.). K.I. and Y.W.I. conducted small RNA-seq and RNA-seq and analysed the obtained data. W.K.A.Y. and H. Sasaki conducted whole-genome bisulfite sequencing and analysed the obtained data. H. Siomi, H.H., Y.W.I. and H. Sasaki wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Cell Biology thanks René Ketting, Jeremy Wang and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
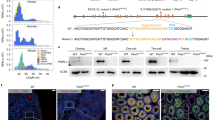
Extended Data Fig. 1 Strategy for the production of PIWIL1-deficient golden hamsters by CRISPR/Cas9.
a, Structure of the golden hamster PIWIL1 gene and target locus for sgRNA. b, The guide RNA sequence for the golden hamster PIWIL1 gene. c, Production of PIWIL1-deficient golden hamster lines. d, The PIWIL1 nucleic acid and amino acid sequences of the targeted site in wild-type hamsters. Upper case underlined denotes the exon sequence, bold with a red box denotes the target sequence, and red denotes the deleted sequence in the 15Adel4 line. e, The PIWIL1 nucleic acid and amino acid sequence of the targeted site in the mutant line (15 A del4). The amino acid sequences in red indicate sequences changed by deletion.
Extended Data Fig. 2 Strategy for the production of PIWIL3-deficient golden hamster by CRISPR/Cas9.
a, Structure of the golden hamster PIWIL3 gene and target loci for sgRNAs. b, The guide RNA sequences for the golden hamster PIWIL3 gene. c, Production of PIWIL3-deficient golden hamster lines. d, The PIWIL3 nucleic acid and amino acid sequence of targeted site in wild-type hamsters. Upper case underlined denotes the exon sequences, bold with a red box denotes the target sequences, and red denotes the deleted sequence in the 26Adel192 line. e, The PIWIL1 nucleic acid and amino acid sequence of targeted site in the mutant line (26 A del192). The amino acid sequences in red indicate sequences changed by deletion.
Extended Data Fig. 3 Defects in spermatogenesis in PIWIL1-deficient male golden hamsters.
a, Testis and cauda epididymis of 1-week-old (1w), 4-week-old (4w) and 10-week-old (10w) from wild-type and PIWIL1-deficient golden hamsters. Scale bars = 10 mm. b, Sperm from the 10w wild-type and PIWIL1-deficient cauda epididymis. Scale bars = 50 µm. c, d, Hematoxylin and eosin-stained 4w (c) and 15w (d) testis sections of wild-type and PIWIL1m/m hamsters, respectively. Scale bars = 100 µm. e, anti-PIWIL1 antibody staining of testis sections of 15-w wild-type and PIWIL1m/m hamsters. Scale bars = 100 µm.
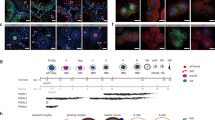
Extended Data Fig. 4 Lack of PIWI proteins in oocytes of PIWIL1- or PIWIL3-deficient golden hamsters.
a, Total lysates of 20 MII oocytes each from heterozygous mutant (w/m) and homozygous mutant (m/m) golden hamsters were subjected to western blotting. TUBB was used as the loading control. Spectra™ Multicolor Broad Range Protein Ladder (Thermo Fisher Scientific) was used as size marker. b, Immunological staining of PIWIL1- and PIWIL3-deficient ovaries with an anti‐PIWIL1 and PIWIL3 antibodies. The arrowheads in the mutants indicate the ovarian follicles. The zona pellucida fixed with paraformaldehyde showed autofluorescence. In particular, PIWIL1, which requires a long exposure time, has strong ring-shaped fluorescence both in the wild type and PIWIL1m/m. Scale bars = 50 µm. c, Ovarian sections from 8‐week‐old wild-type and PIWIL1- and PIWIL3-deficient golden hamsters. Hematoxylin and eosin‐stained ovarian sections of wild-type, PIWIL1- and PIWIL3-deficient golden hamsters. Scale bars = 200 µm. Note: By fixing PIWIs, the antigenicity is weakened. PIWIL1 antigenicity is particularly weak even after antigen retrieval treatment: therefore, the exposure time is long and the background of the zona pellucida becomes visible.
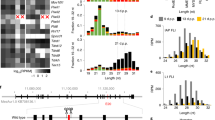
Extended Data Fig. 5 Changes in the expression levels of each unique small RNA sequence in PIWI-deficient mutants.
An MA plot showing the expression level of each unique small RNA sequence. To calculate log2 fold change and expression levels, 1 was added to each value to calculate the levels of small RNA sequences with 0 in either sample.
Extended Data Fig. 6 Gene ontology analysis of MII oocytes in PIWIL1-deficient golden hamsters.
a, b, Enriched gene ontology (biological process) terms using DEGs obtained in PIWIL1-deficient MII oocyte transcriptome analysis. Upregulated genes in the PIWIL1-deficient MII oocytes (a). Downregulated genes in the PIWIL1-deficient MII oocytes (b). Terms with p-values lower than 0.001 are listed.
Extended Data Fig. 7 Nucleolus abnormalities in 2 C embryos derived from PIWIL1-deficient oocytes.
a, Pro-nucleus morphology of the zygotes. There were no pro-nucleus morphology differences in maternally PIWIL1- and PIWIL3-deficient zygotes. b, Nucleus morphology of the 2 C embryos. PIWIL1m/m showed a single large nucleolus, while PIWIL1w/m and PIWIL3m/m formed multiple small nucleoli. Enlarged view of box showed nucleus and arrows indicate the nucleolus. Scale bars = 50 µm.
Extended Data Fig. 8 DNA methylation in PIWIL1- and PIWIL3-deficient GV oocyte.
a, b, 5mC staining of PIWIL1-deficient (a) and PIWIL3-deficient GV oocytes (b). The amounts of 5mC were determined by measuring the fluorescence intensity using image J software (right). n indicates the number of independent GV oocytes collected from three females and used for analysis (Extended Data Fig. 9), and error bars indicate ± s.d. (***P=4.01e-8 by 2-sided t-test) Scale bars = 50 µm.
Extended Data Fig. 9 Representative GV oocytes stained with anti-5mC antibody.
GV oocytes were stained with a 5mC antibody (33D3) and the fluorescence GV nuclei were imaged. The fluorescence was analyzed by ImageJ and used to be described in Extended Data Fig. 8. Scale bars = 25 µm.
Extended Data Fig. 10 Meiosis and MII spindle formation in ovulated oocytes of PIWIL3-deficient golden hamsters.
a, PIWIL3w/m and PIWIL3m/m oocytes were collected, and the stages of meiosis at 7 and 10 am in the morning were analyzed following TUBA and DAPI staining. b, MII oocytes at 10 am in the morning were stained with anti-TUBA antibody (green) and DAPI (blue). Scale bars = 5 µm. c, The size of the MII spindles (A and B in Extended Data Fig. 10b) was measured and compared between PIWL3w/m and PIWL3m/m. n indicates the number of independent MII oocytes collected from three females and used for analysis, error bars indicate ± s.d. (A: ***P=0.000417, B: *P=0.0377 by 2-sided t-test).
Supplementary information
Supplementary Tables
Supplementary Table 1: Sequencing and mapping summary of whole-genome bisulfate sequencing. Supplementary Table 2: Primers and gRNA.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 4
Gel image.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 10
Statistical source data.
Rights and permissions
About this article
Cite this article
Hasuwa, H., Iwasaki, Y.W., Au Yeung, W.K. et al. Production of functional oocytes requires maternally expressed PIWI genes and piRNAs in golden hamsters. Nat Cell Biol 23, 1002–1012 (2021). https://doi.org/10.1038/s41556-021-00745-3
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41556-021-00745-3
This article is cited by
-
A mammalian-like piRNA pathway in Axolotl reveals the origins of piRNA-directed DNA methylation
The EMBO Journal (2025)
-
PIWI-Interacting RNAs in brain health and disease: biogenesis, mechanisms, and therapeutic horizons
Psychopharmacology (2025)
-
Conserved genes regulating human sex differentiation, gametogenesis and fertilization
Journal of Translational Medicine (2024)
-
Divergent composition and transposon-silencing activity of small RNAs in mammalian oocytes
Genome Biology (2024)
-
PIWI-interacting RNAs: who, what, when, where, why, and how
The EMBO Journal (2024)