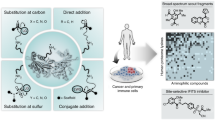
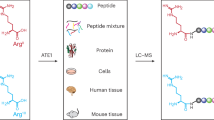
Abstract
Despite the crucial biological functions of arginine, its reactivity and ligandability within the human proteome remain largely unexplored. Here we apply activity-based protein profiling (ABPP) with phenylglyoxal-based chemical probes to map arginine reactivity globally. Screening phenylglyoxal derivatives identified a probe with enhanced coverage and selectivity, enabling quantification of 4,606 arginine sites across human cell lines. Among these, critical residues regulate liquid–liquid phase separation. Arginine reactivity was further assessed by on-beads reductive dimethylation proteomics, revealing a subset of hyper-reactive sites. Competitive fragment screening using data-independent acquisition ABPP (DIA-ABPP) generated a ligandability map of arginine residues across 60 dicarbonyl compounds. This dataset revealed ligandable arginines that modulate protein activity, in particular protein–protein interactions, highlighting potential covalent drug targets. Together, this work provides a proteome-wide profile of arginine reactivity and ligandability, offering insights into the functional landscape of arginines and expanding the scope of covalent drug discovery to include arginine-targeting molecules.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The MS data generated in this study have been deposited to the ProteomeXchange Consortium via the iProX74 partner repository with the dataset identifier PXD056202 (Arginine_Profiling_MS dataset). Source data are provided with this paper.
Code availability
The Python code and the corresponding dataset have been deposited to Zenodo at https://doi.org/10.5281/zenodo.15493178 (ref. 75). Alternatively, it is available from the corresponding author upon request.
References
Jin, Y., Jana, S., Abbasov, M. E. & Lin, H. Antibiotic target discovery by integrated phenotypic and activity-based profiling of electrophilic fragments. Cell Chem. Biol. 32, 434–448.e9 (2025).
Zhang, X. & Cravatt, B. F. Chemical proteomics–guided discovery of covalent ligands for cancer proteins. Annu. Rev. Cancer Biol. 8, 155–175 (2024).
Niphakis, M. J. & Cravatt, B. F. Ligand discovery by activity-based protein profiling. Cell Chem. Biol. 31, 1636–1651 (2024).
Offensperger, F. et al. Large-scale chemoproteomics expedites ligand discovery and predicts ligand behavior in cells. Science 384, eadk5864 (2024).
Liu, Y., Patricelli, M. P. & Cravatt, B. F. Activity-based protein profiling: the serine hydrolases. Proc. Natl Acad. Sci. USA 96, 14694–14699 (1999).
Weerapana, E. et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 (2010).
Shi, Y., Fu, L., Yang, J. & Carroll, K. S. Wittig reagents for chemoselective sulfenic acid ligation enables global site stoichiometry analysis and redox-controlled mitochondrial targeting. Nat. Chem. 13, 1140–1150 (2021).
Fu, L. et al. Nucleophilic covalent ligand discovery for the cysteine redoxome. Nat. Chem. Biol. 19, 1309–1319 (2023).
Qin, W. et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat. Chem. Biol. 15, 983–991 (2019).
Hacker, S. M. et al. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 9, 1181–1190 (2017).
Abbasov, M. E. et al. A proteome-wide atlas of lysine-reactive chemistry. Nat. Chem. 13, 1081–1092 (2021).
Hahm, H. S. et al. Global targeting of functional tyrosines using sulfur-triazole exchange chemistry. Nat. Chem. Biol. 16, 150–159 (2020).
Sun, F., Suttapitugsakul, S. & Wu, R. An azo coupling-based chemoproteomic approach to systematically profile the tyrosine reactivity in the human proteome. Anal. Chem. 93, 10334–10342 (2021).
Chen, Y. et al. Direct mapping of ligandable tyrosines and lysines in cells with chiral sulfonyl fluoride probes. Nat. Chem. 15, 1616–1625 (2023).
Lin, S. X. et al. Redox-based reagents for chemoselective methionine bioconjugation. Science 355, 597–602 (2017).
Bach, K., Beerkens, B. L. H., Zanon, P. R. A. & Hacker, S. M. Light-activatable, 2,5-disubstituted tetrazoles for the proteome-wide profiling of aspartates and glutamates in living bacteria. ACS Cent. Sci. 6, 546–554 (2020).
Ma, N. et al. 2H-Azirine-based reagents for chemoselective bioconjugation at carboxyl residues inside live cells. J. Am. Chem. Soc. 142, 6051–6059 (2020).
Xie, X. et al. Oxidative cyclization reagents reveal tryptophan cation–π interactions. Nature 627, 680–687 (2024).
Zhai, Y. et al. Global profiling of functional histidines in live cells using small-molecule photosensitizer and chemical probe relay labelling. Nat. Chem. 16, 1546–1557 (2024).
Sharma, H. A. et al. Proteomic ligandability maps of phosphorus(V) stereoprobes identify covalent TLCD1 inhibitors. J. Am. Chem. Soc. 147, 15554–15566 (2025).
Backus, K. M. et al. Proteome-wide covalent ligand discovery in native biological systems. Nature 534, 570–574 (2016).
Wang, Q. et al. Quantitative chemoproteomics reveals dopamine’s protective modification of Tau. Nat. Chem. Biol. 21, 1341–1350 (2025).
Wang, C., Weerapana, E., Blewett, M. M. & Cravatt, B. F. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat. Methods 11, 79–85 (2014).
Hodges, A. J. et al. Histone sprocket arginine residues are important for gene expression, DNA repair and cell viability in Saccharomyces cerevisiae. Genetics 200, 795–806 (2015).
Bogan, A. A. & Thorn, K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280, 1–9 (1998).
Bartlett, G. J., Porter, C. T., Borkakoti, N. & Thornton, J. M. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 324, 105–121 (2002).
Gupta, M. N. & Uversky, V. N. Biological importance of arginine: a comprehensive review of the roles in structure, disorder, and functionality of peptides and proteins. Int. J. Biol. Macromol. 257, 128646 (2024).
Fuhrmann, J., Clancy, K. W. & Thompson, P. R. Chemical biology of protein arginine modifications in epigenetic regulation. Chem. Rev. 115, 5413–5461 (2015).
Qamar, S. et al. FUS phase separation is modulated by a molecular chaperone and methylation of arginine cation-π interactions. Cell 173, 720–734.e15 (2018).
Hofweber, M. et al. Phase separation of FUS is suppressed by its nuclear import receptor and arginine methylation. Cell 173, 706–719.e13 (2018).
Hellwig, M. & Henle, T. Baking, ageing, diabetes: a short history of the Maillard reaction. Angew. Chem. Int. Ed. 53, 10316–10329 (2014).
Takahashi, K. The reaction of phenylglyoxal with arginine residues in proteins. J. Biol. Chem. 243, 6171–6179 (1968).
Dovgan, I. et al. Arginine-selective bioconjugation with 4-azidophenyl glyoxal: application to the single and dual functionalisation of native antibodies. Org. Biomol. Chem. 16, 1305–1311 (2018).
Jones, A. X. et al. Improving mass spectrometry analysis of protein structures with arginine-selective chemical cross-linkers. Nat. Commun. 10, 3911 (2019).
Lewallen, D. M. et al. Chemical proteomic platform to identify citrullinated proteins. ACS Chem. Biol. 10, 2520–2528 (2015).
Bicker, K. L., Subramanian, V., Chumanevich, A. A., Hofseth, L. J. & Thompson, P. R. Seeing citrulline: development of a phenylglyoxal-based probe to visualize protein citrullination. J. Am. Chem. Soc. 134, 17015–17018 (2012).
Zanon, P. R. A. et al. Profiling the proteome-wide selectivity of diverse electrophiles. Nat. Chem. 17, 1712–1721 (2025).
Wang, Q. et al. Global profiling of arginine dimethylation in regulating protein phase separation by a steric effect-based chemical-enrichment method. Proc. Natl Acad. Sci. USA 119, e2205255119 (2022).
Liu, Z., Wang, K. & Ye, M. Photoreactive probe-based strategy enables the specific identification of the transient substrates of methyltransferase at the proteome scale. Anal. Chem. 95, 12580–12585 (2023).
Shi, Y. et al. Enabling global analysis of protein citrullination via biotin thiol tag-assisted mass spectrometry. Anal. Chem. 94, 17895–17903 (2022).
Chen, P. et al. Cell-active, arginine-targeting irreversible covalent inhibitors for non-kinases and kinases. Angew. Chem. Int. Ed. 64, e202422372 (2025).
Zhang, Z., Morstein, J., Ecker, A. K., Guiley, K. Z. & Shokat, K. M. Chemoselective covalent modification of K-Ras(G12R) with a small molecule electrophile. J. Am. Chem. Soc. 144, 15916–15921 (2022).
Prosser, L., Emenike, B., Sihag, P., Shirke, R. & Raj, M. Chemical carbonylation of arginine in peptides and proteins. J. Am. Chem. Soc. 147, 10139–10150 (2025).
Kong, A. T., Leprevost, F. V., Avtonomov, D. M., Mellacheruvu, D. & Nesvizhskii, A. I. MSFragger: ultrafast and comprehensive peptide identification in mass spectrometry-based proteomics. Nat. Methods 14, 513–520 (2017).
Sletten, E. M. & Bertozzi, C. R. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 48, 6974–6998 (2009).
Weerapana, E., Speers, A. E. & Cravatt, B. F. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)—a general method for mapping sites of probe modification in proteomes. Nat. Protoc. 2, 1414–1425 (2007).
Chang, J. W., Lee, G., Coukos, J. S. & Moellering, R. E. Profiling reactive metabolites via chemical trapping and targeted mass spectrometry. Anal. Chem. 88, 6658–6661 (2016).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444 (2021).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 381, eadg7492 (2023).
Boersema, P. J., Raijmakers, R., Lemeer, S., Mohammed, S. & Heck, A. J. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 (2009).
Yang, F., Gao, J., Che, J., Jia, G. & Wang, C. A dimethyl-labeling-based strategy for site-specifically quantitative chemical proteomics. Anal. Chem. 90, 9576–9582 (2018).
You, K. et al. PhaSepDB: a database of liquid-liquid phase separation related proteins. Nucleic Acids Res. 48, D354–D359 (2019).
Bryant, P., Pozzati, G. & Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat. Commun. 13, 1265 (2022).
Burke, D. F. et al. Towards a structurally resolved human protein interaction network. Nat. Struct. Mol. Biol. 30, 216–225 (2023).
Dao, T. P. et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell 69, 965–978.e6 (2018).
Yamazaki, T. et al. Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell 70, 1038–1053.e7 (2018).
Yang, F., Jia, G., Guo, J., Liu, Y. & Wang, C. Quantitative chemoproteomic profiling with data-independent acquisition-based mass spectrometry. J. Am. Chem. Soc. 144, 901–911 (2022).
Berkholz, D. S., Faber, H. R., Savvides, S. N. & Karplus, P. A. Catalytic cycle of human glutathione reductase near 1 Å resolution. J. Mol. Biol. 382, 371–384 (2008).
Zhang, R. et al. Pleiotropic effects of a mitochondrion-targeted glutathione reductase inhibitor on restraining tumor cells. Eur. J. Med. Chem. 248, 115069 (2023).
Burger, N. et al. The human zinc-binding cysteine proteome. Cell 188, 832–850.e27 (2025).
Board, P. G. et al. S-(4-nitrophenacyl)glutathione is a specific substrate for glutathione transferase omega 1-1. Anal. Biochem. 374, 25–30 (2008).
Ramkumar, K. et al. Mechanistic evaluation and transcriptional signature of a glutathione S-transferase omega 1 inhibitor. Nat. Commun. 7, 13084 (2016).
Liu, Y. et al. Proteome-wide ligand and target discovery by using strain-enabled cyclopropane electrophiles. J. Am. Chem. Soc. 146, 20823–20836 (2024).
Baltgalvis, K. A. et al. Chemoproteomic discovery of a covalent allosteric inhibitor of WRN helicase. Nature 629, 435–442 (2024).
Uechi, H. et al. Small-molecule dissolution of stress granules by redox modulation benefits ALS models. Nat. Chem. Biol. 21, 1577–1588 (2025).
Tanikawa, C. et al. Citrullination of RGG motifs in FET proteins by PAD4 regulates protein aggregation and ALS susceptibility. Cell Rep. 22, 1473–1483 (2018).
Hofweber, M. & Dormann, D. Friend or foe—post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 294, 7137–7150 (2019).
Communi, D., Lecocq, R. & Erneux, C. Arginine 343 and 350 are two active site residues involved in substrate binding by human type I D-myo-inositol 1,4,5-trisphosphate 5-phosphatase. J. Biol. Chem. 271, 11676–11683 (1996).
Chen, G. & Chen, X. Arginine residues in the active site of human phenol sulfotransferase (SULT1A1). J. Biol. Chem. 278, 36358–36364 (2003).
Yan, J., He, G., Yan, F., Zhang, J. & Zhang, G. The dicarbonylation of indoles via Friedel–Crafts reaction with dicarbonyl nitrile generated in situ and retro-cyanohydrination. RSC Adv. 6, 44029–44033 (2016).
Hirapara, P. et al. CO2-assisted synthesis of non-symmetric α-diketones directly from aldehydes via C–C bond formation. Green Chem. 19, 5356–5360 (2017).
Li, J. et al. ACR-based probe for the quantitative profiling of histidine reactivity in the human proteome. J. Am. Chem. Soc. 145, 5252–5260 (2023).
Ma, J. et al. iProX: an integrated proteome resource. Nucleic Acids Res. 47, D1211–D1217 (2019).
Wang, Y. Global profiling of arginine reactivity and ligandability in the human proteome. Zenodo https://doi.org/10.5281/zenodo.15493178 (2025).
Acknowledgements
We are grateful for financial support of this work from Shenzhen Bay Laboratory Startup (21240041 to G.L.), Guangdong Special Support Plan for Outstanding Young Talents (2023TQ07A238 to G.L.) and the Natural Science Foundation of Guangdong Province (2023A1515111118 to Y.W.). We thank the Multi-omics Mass Spectrometry Core Facility at the Bio-Tech Center of Shenzhen Medical Academy of Research and Translation (SMART) for their technical support. We thank Z. Li (Jinan University) for providing the S-(4-nitrophenacyl)glutathione (4-NPG) substrate. We thank Y. Jiang, L. Fu, W. Xiao, Y. Chen, T. Guo and X. Guo for helpful discussions. The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
G.L. conceived the study and supervised the research. Y.W., T.H., Y.Z., X.H., B.Y. and G.L. designed and analysed the biological experiments. L.Z., C.X. and G.L. designed and synthesized the chemical compounds. X.Y., Y.L. and S.X. developed the Python and R code for data processing, bioinformatics and chemoinformatics analyses. Y.W. and G.L. wrote the paper with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Chemistry thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Specificity analysis of the probe 1 and 2.
a) Distribution of peptide modifications for probe 1 and 2, considering localized peptide spectrum matches (PSMs) exceeding 80. b) Proposed structures of the probe 1-derived adducts corresponding to the mass shifts of +318.1117 Da and its oxidized form (+334.1066 Da). c) Proposed structures of the probe 2-derived adducts corresponding to the mass shifts of +276.0997 Da and its oxidized product (+292.0961 Da). d) Probe selectivity analysis using an open-search method. e) Selectivity of probe 1 and 2 determined through mass-offset search using the mass shifts in (a). f) Number of modification sites identified by a closed-search using the expected differential masses for each probe.
Extended Data Fig. 2 Strategies to enhance arginine labeling sites.
a) Identification of arginine labeling sites in membrane and soluble fractions using probe 1, with two biological replicates. b) Comparison of arginine labeling sites using an alternative workflow where trypsin digestion precedes enrichment, versus the traditional approach where enrichment precedes digestion. Two biological replicates were performed. c) Overlap of the arginine sites labelled in the human proteome across two biological replicates. d) Proposed chemical structure of the PhGO-alkyne derived adducts. e) Comparison of arginine-modified sites identified by probe 1 and PhGO-alkyne in HeLa cell lysates. f) Overlap of arginine sites labelled by probe 1 and PhGO-alkyne across the human proteome.
Extended Data Fig. 3 Live cell arginine labeling by probe 1.
a) Optimization of probe 1 concentration and incubation time for live-cell labeling. b) Number of arginine-modified sites identified in live cells via closed-search using the expected mass shift of probe 1. c) Cytotoxicity of probe 1 in HeLa cells measured by CellTiter-Glo assay. d) Intracellular concentration of probe 1 in HeLa cells determined by targeted LC–MS/MS. Data are shown as mean ± SD from three biologically independent replicates.
Extended Data Fig. 4 Quantitative profiling of arginine reactivity in the human proteome.
a) Waterfall plot of log2(R10:1) values from two biological replicates, representing the probe-modified arginine sites used to quantify arginine reactivity across the proteome. b-d) Validation of labeling for proteins containing hyper-reactive arginine residues (GSHR R81, RIR1 R284, and FSCN1 R217). Recombinant wild-type (WT) proteins and corresponding arginine-to-lysine (R-to-K) mutants were expressed in HEK293T proteomes, labelled with probe 1, and analyzed via in-gel fluorescence. e) Western blots were used to confirm protein expressions for LLPS assays. All data are representative of two biologically independent experiments (n = 2).
Extended Data Fig. 5 Arginine-π interaction mediated phase separation.
a, c, e) Fluorescence imaging comparing wild-type and mutant cells, confirming the functional role of R269 in UBQL2 (a), R313 in NONO (c) and H90 in SFPQ (e) for promoting phase separation. Scale bar, 5 μm. n = 3 biological independent replicates. b, d, f) Quantification of puncta number and size in wild-type and mutant cells (UBQL2 R269A in b, NONO R313A in d, and SFPQ H90A in f) after arsenite treatment. Boxes represent the interquartile range (25th-75th percentiles), and center lines denote the median. Statistical significance was assessed using unpaired two-tailed Student’s t-tests; P values are indicated.
Extended Data Fig. 6 Chemical structures of glyoxal compounds used in this study.
Glyoxal compounds are categorized into three structural types: aryl-disubstituted glyoxals (Type 1), alkyl-disubstituted glyoxals (Type 2), and monosubstituted glyoxals (Type 3).
Extended Data Fig. 7 Validation of liganded arginines in cell lysate.
a) Representative MS1 extracted ion chromatograms (XIC) showing decreased probe 1-labelled peptide signal upon competition with CP 1 for KAD1. Average RDMSO/CP values, calculated from biological duplicates, are displayed below the XIC. b) Western blot validation of liganded arginines in HEK293T cell lysates expressing wild-type or arginine-to-alanine mutant KAD1, showing selective blockade of probe 1 labeling by CP 1. c) Structural model of KAD1 (PDB: 2C95) highlighting the liganded arginine residue. d) Bar plots illustrating the effects of CP 1, R81A and R81W mutation on the redox-dependent enzymatic activities of GSHR. Data are shown as mean ± SD from at least three biologically independent replicates.
Supplementary information
Supplementary Information
Supplementary Figs. 1–46 and Table 1.
Supplementary Data
Statistical source data for supplementary figures.
Source data
Source Data Fig. 1
Statistical source data for Fig. 1.
Source Data Fig. 2
Statistical source data for Fig. 2.
Source Data Fig. 3
Unprocessed western blots and gels pdf and statistical source data for Fig. 3.
Source Data Fig. 4
Statistical source data for Fig. 4.
Source Data Fig. 5
Unprocessed western blots and gels pdf and statistical source data for Fig. 5.
Source Data Fig. 6
Unprocessed western blots and gels pdf and statistical source data for Fig. 6.
Source Data Extended Data Fig. 1
Statistical source data for Extended Fig. 1.
Source Data Extended Data Fig. 2
Statistical source data for Extended Fig. 2.
Source Data Extended Data Fig. 3
Unprocessed western blots and gels pdf and statistical source data for Extended Data Fig. 3.
Source Data Extended Data Fig. 4
Unprocessed western blots and gels pdf and statistical source data or Extended Data Fig. 4.
Source Data Extended Data Fig. 5
Statistical source data for Extended Fig. 5.
Source Data Extended Data Fig. 7
Unprocessed western blots and gels pdf and statistical source data for Extended Data Fig. 7.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, Y., Hu, T., Zhu, L. et al. Global profiling of arginine reactivity and ligandability in the human proteome. Nat. Chem. (2026). https://doi.org/10.1038/s41557-025-02012-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41557-025-02012-6