Abstract
Despite the considerable success of clinically approved immune-based therapies for treating advanced melanoma, a significant fraction of patients are not responsive owing to mechanisms engaged by the tumour to evade the immune system. Here we report the surprising finding that a clinically validated and tunable self-therapeutic ultrasmall silica nanoparticle prolongs survival in a highly resistant melanoma model in combination with interleukin-6 and PD-L1 inhibition through activation of the stimulator of interferon genes/interleukin-6/PD-L1 axis and reprogramming of the tumour microenvironment towards a pro-inflammatory phenotype. In a murine model, induction of significant cytotoxic and antitumour inflammatory responses leads to differential activation of immune cell populations in a CD8-dependent manner via type I/II interferon pathways after systemic particle injection. Importantly, these immunostimulatory responses accompany significant reductions in cell populations and receptors driving suppressive activities. Mechanistic insights highlight the potential clinical utility of this platform to maximize antitumour immunity and efficacy by subverting suppressive components in the tumour microenvironment.
Similar content being viewed by others
Main
Cancer immunotherapies (ITs) using immune checkpoint blockade (ICB) have transformed treatment for advanced malignancies1,2. Although ICB can elicit durable—sometimes curative—responses in subsets of patients with solid tumours3,4, ~87% do not achieve long-term benefit5. This shortfall reflects tumour mechanisms that evade immune recognition and reduce inherent immunogenicity4. Tumour-associated myeloid cells and their suppressive secretomes are key drivers of limited antitumour immunity, leading to T cell dysfunction and poor outcomes in melanoma6,7,8.
Cancer IT strategies aim to deplete, block or reprogramme suppressive immune effectors9. Resistance to anti-CTLA-4 and anti-PD-1/PD-L1 can be mitigated by agonists targeting pattern recognition receptors (PRRs)10, notably Toll-like receptors (TLRs) and the cyclic GMP-AMP synthase (cGAS)–stimulator of interferon genes (STING) pathway11. PRRs may also be activated by endogenous damage-associated molecular patterns (DAMPs)—such as heat-shock proteins (HSPs), adenosine triphosphate (ATP) and calreticulin—generated during cellular stress or death12. PRR engagement promotes co-stimulation, pro-inflammatory gene expression, antigen presentation and phagocytosis9.
The STING pathway plays a central role in melanoma and other advanced cancers, mediating innate immunity through cytosolic DNA sensing, senescence induction and type I/II interferon (IFN) production. Pleiotropic cytokines, including interleukin-6 (IL-6), exert pro-tumorigenic effects by upregulating PD-L1, promoting STING degradation and suppressing immunity13. Elevated IL-6 correlates with poor anti-PD-L1 responses14, making the IL-6/PD-L1 axis an attractive therapeutic target. Despite ongoing trials targeting IL-6/PD-1/PD-L1 and/or PRR pathway inhibitors15, no agents have yet been approved.
An alternative approach is to tailor physicochemical properties of engineered materials—critical determinants of immune responsiveness16,17. Here we highlight an FDA Investigational New Drug (IND)-cleared, ultrasmall poly(ethylene) glycol (PEG)-coated fluorescent silica nanoparticle, Cornell prime dots (C′ dots), with unexpectedly broad adjuvant-therapeutic activity, now in therapeutic18 and image-guided surgical trials19. Without conjugated agonists or cytotoxic drugs, C′ dots elicit pro-inflammatory and cytotoxic responses in ICB-resistant melanomas localized to the tumor microenvironment. Synthesized in aqueous solution, they exhibit ultrasmall size, controlled surface chemistry and neutral charge20,21,22,23—lacking a protein corona—improving biocompatibility, stability and circulation time24,25. Together with favourable pharmacokinetics (PK)26,27,28 and renal clearance24,29, their efficient tumour penetration and diffusion30,31 leads to high efficacy30,31,32,33,34,35,36 and potent antitumour immunity.
Here we exploit these properties to target the novel STING/IL-6/PD-L1 axis in the highly suppressive B16-GMCSF (B16-GM) melanoma model, achieving survival gains beyond traditional IT. C′ dots diminish immune suppression, partly by activating cGAS–STING, triggering immune-related cell death, senescence and cell-cycle arrest, while upregulating PD-L1 and IL-6 on tumour and myeloid cells. This drives tumour immunogenicity, activation of tumour-infiltrating lymphocytes (TILs), macrophage polarization and reduced suppressive immune populations. By contrast, in the less suppressive B16-F10 model, C′ dots primarily induce ferroptosis, an iron-dependent cell death programme. To our knowledge, this is the first study to fully elucidate the adjuvant-therapeutic activity of a clinically validated ultrasmall nanoparticle in combination with IL-6 and PD-L1 blockade to improve treatment outcomes in ICB-resistant models, underscoring the broader potential of ultrasmall particle-based adjuvant therapies to reprogramme the tumour microenvironment (TME) and transform cancer management.
Ferroptotic immunomodulation inhibits B16-F10 tumour growth
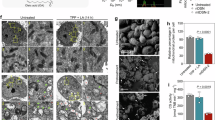
In previous xenograft studies, systemically administered C′ dots functionalized with alpha melanocyte-stimulating hormone (αMSH) peptides (αMSH-PEG-Cy5-C′ dots/αMSH-C′ dots) induced ferroptosis, tumour regression, iron-related gene upregulation and macrophage recruitment37. To assess ferroptosis- and/or inflammation-driven contributions to TME modulation in immunocompetent settings, we used syngeneic B16-F10 and genetically engineered B16-GM melanoma models, the latter exhibiting greater immune suppression and checkpoint resistance38,39—barriers to effective treatment40. Subcutaneously implanted B16-F10 tumours (150–200 mm3) in C57BL/6 mice received three intravenous (i.v.) doses (12 nmoles per dose every 3 days, Q3D×3) of saline, αMSH-C′ dots (60 μM; 36 nmol; Supplementary Fig. 1a–d) or αMSH-C′ dots + the ferroptosis inhibitor, liproxstatin-1 (liprox)41. Particle treatment reduced tumour volumes by ~65% versus vehicle (Fig. 1a), an effect significantly attenuated by liproxstatin-1, confirming ferroptosis as the principal cytotoxic driver with an added immune-mediated component. Haematoxylin-and-eosin staining showed marked necrosis within αMSH-C′ dot-treated tumours that was abolished by liproxstatin-1, while αMSH-C′ dots + liproxstatin-1 treatment resembled controls (Fig. 1b). Expression of T cell and myeloid cell markers, quantified by immunohistochemistry (IHC) (Supplementary Fig. 2), revealed elevated CD3+, CD8+ T cells and Iba1+ macrophage numbers per unit area over controls post-particle treatment, while CD3+/CD8+ T cells decreased and Iba1+ cells rose with liproxstatin-1 (Fig. 1b,c), suggesting ferroptosis-driven lymphocyte recruitment and macrophage-mediated debris clearance. Combining αMSH-C′ dots with anti-programmed cell death protein 1 (anti-PD-1) antibody yielded near-complete responses, with tumour volumes decreasing ~90% versus 63% for particles alone (Fig. 1d), accompanied by the highest CD3+/CD8+ T cell infiltration per unit area across groups (Fig. 1e). In non-obese diabetic/severe combined immunodeficiency (NOD-SCID; T-/B-cell-deficient) and NOD-SCID gamma (NSG; T-/B-/myeloid-deficient) animals, growth inhibition persisted only in NOD-SCID animals (Fig. 1f,g), indicating that the non-ferroptotic component of efficacy is immune driven and primarily mediated by host myeloid cells.
a–c, Ferroptosis study: B16-F10 tumour growth inhibition in mice following i.v. injection of αMSH-C′ dots (12 nmol per dose, Q3D×3 (60 μM), red), as against vehicle (blue) and αMSH-C′ dots + liproxstatin (liprox, green) on days 7, 10 and 14 post-implantation (a). Haematoxylin-and-eosin staining of tumour tissue specimens (scale bars, left panel, 1 mm; right panel, 100 μm) (b). Plots of pan T cell (CD3+), helper T cell (CD4+), cytotoxic (CD8+) T cell and macrophage (Iba1+) populations in the TME by IHC (c). d, Tumour growth inhibition in B16-F10 mice injected, as in a ± anti-PD-1 antibody (250 μg, n = 3 doses), using αMSH-C′ dots + anti-PD-1 (green), administered concomitantly, αMSH-C′ dots alone (red), anti-PD-1 alone (grey) or saline vehicle (blue). e, Plots of pan (CD3+), helper (CD4+) and cytotoxic (CD8+) T cell populations. f,g, Tumour growth in particle-treated B16-F10 tumour-bearing NOD-SCID (f) and NSG (g) mice using the regimen in a. Data reflect n = 4 mice per group for all animal studies, mean ± s.e.m. Non-parametric two-way ANOVA with Sidak’s post hoc test was performed for growth inhibition, and one-way ANOVA with Tukey’s test using multiple comparisons was performed for IHC quantification. All statistical tests were two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001.
To determine whether TME changes arose from the base particle, PEG-Cy5-C′ dots (Supplementary Fig. 3a) were tested. Bulk RNA sequencing (RNA-seq) of particle-exposed B16-F10 cells (15 µM, 24 h) revealed upregulation of inflammatory genes (Cd68, Oas1, Gsdma and Cd40) and downregulation of DNA-replication/mismatch-repair genes (Msh2 and Pola1) (Supplementary Fig. 3b). The Kyoto Encyclopedia of Genes and Genomes (KEGG)-based gene set enrichment analysis (GSEA) indicated enrichment of pro-inflammatory (TH17 cell differentiation, IL-17 signalling) and metabolic pathways, including drug metabolism–cytochrome P450 (CYP) and cyclic adenosine 3,5-monophosphate (cAMP) (Supplementary Fig. 3c). Cytokine assays confirmed increased TH1-type cytokines and chemokines over 72 h (Supplementary Fig. 3d) and elevated PD-L1 surface expression (Supplementary Fig. 3e), consistent with ferroptosis-linked inflammatory changes.
C′ dots induce immune-related cell death in B16-GM cells
To define how uniformly sized, sub-8-nanometre C′ dots (Fig. 2a,b) regulate pro-inflammatory and immune-related signatures, we used the myeloid-rich, dual-ICB-resistant B16-GM model. Bulk RNA-seq of C′ dot-treated cells (15 µM, 72 h; Supplementary Fig. 1e–g) revealed induction of inflammation/PRR/senescence-associated genes (for example, Tmem173, Tlr2/3/4, Ddx58 and Ctsl) and downregulation of cell-cycle and DNA-repair regulators (for example, Cdc25, Top2a and Rad51b) relative to controls (Fig. 2c). KEGG/Reactome GSEA (Fig. 2d,e) indicated activation of cAMP, NOD-like receptor (NLR) and major histocompatibility complex (MHC)-II antigen-presentation pathways with suppression of DNA replication and cell-cycle progression.
a, Schematic of non-targeted C′ dots. b, A representative TEM image of C′ dots (scale bar, 50 nm) captured more than 3 independent times yielding similar results. c, Volcano plot depicting differentially expressed genes for C′ dot and vehicle-treated tumour cells. d, KEGG pathway analysis showing representative upregulated and downregulated pathways. e, Reactome pathway gene set enrichment analysis shows overrepresented (enriched) pathways. f, Cytoplasmic dsDNA following exposure to 15 μM C′ dots for 72 h using IF staining (bright puncta indicate cytoplasmic dsDNA) (scale bars, 8 μm). g, B16-GM cells untreated or exposed to C′ dots over a range of concentrations for 72 h and analysed for Cy5 median fluorescence intensity (MFI) by flow cytometry. h, Quantification of the percentage (%) of cytosolic dsDNA using images in f. i, Ratio of intracellular to extracellular cGAMP. j, Quantification of nuclear and cytoplasmic genomic DNA via Quant-iT assay after subcellular fractionation. Data represent mean ± s.e.m. of three biological replicates. Non-parametric one-way ANOVA with Tukey’s test for g and i and unpaired Student’s t-test for h and j for multiple comparisons were performed. All statistical tests were two-sided. Analyses for d and e were also two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001. conc, concentration.
To validate and refine pathway candidates, the top differentially expressed genes were analysed with Enrichr42. Gene Ontology biological/molecular pathways (Supplementary Fig. 4a,b) showed enrichment of pro-inflammatory, senescence-associated secretory phenotype (SASP) and cytokine-signalling programmes—specifically IL-6 receptor binding, chemokine production and leukocyte chemotaxis—while cell-cycle progression, DNA damage repair and metabolic pathways were inhibited (for example, mitotic division; ion symporter/transporter). Early (24 h) RNA-seq revealed upregulation of tumour necrosis factor (TNF) superfamily, nuclear factor (NF)-κB and TH1/TH2 pathways, with suppression of mismatch repair (Supplementary Fig. 5a). Gene programmes underlying these phenotypes were probed by conducting reverse transcription-quantitative polymerase chain reaction (RT-qPCR) of 20 representative genes across 5 classes (iron metabolism, ferroptosis, MHC-I presentation, DAMPs and IFNs) confirmed dose- and time-dependent increases (Supplementary Fig. 5b,c), particularly in immune-related genes (Cxcl10, Tnfa and Ifi44), MHC-I components (B2m, Tap2 and H2k1) and DAMPs (Hspa1a). Among ferroptosis-linked genes, Gpx4 (ferroptosis/glutathione regulator43) and Slc3a244 (partner of SLC7A11 in xc⁻ cystine–glutamate antiporter) were upregulated. Bulk RNA-seq comparing B16-GM and B16-F10 cells corroborated suppression of oxidative phosphorylation and glutathione metabolism in B16-GM, indicating that ferroptosis was not dominant, while TLR/NLR and cytosolic DNA-sensing (PRR) pathways were activated (Supplementary Fig. 5d). Collectively, these data show that C′ dots engage multiple mechanisms driving pro-inflammatory and immune-mediated cytotoxicity, cell-cycle/DNA-repair inhibition and senescence induction—highlighting their adjuvant-therapeutic potential beyond canonical ferroptotic programmes.
C′ dots upregulate the cGAS–STING pathway in B16-GM cells
Given the transcriptional activation of innate immune pathways, we investigated whether C′ dot-exposed B16-GM cells released double-stranded DNA into the cytoplasm—a trigger of cGAS–STING-mediated inflammation45. Using optimized exposure conditions (15 μM, 72 h) based on uptake kinetics and fluorescence microscopy, treated cells showed marked cytoplasmic dsDNA accumulation versus vehicle (Fig. 2f,g and Supplementary Fig. 5b,c). These findings, graphically depicted (Fig. 2h), are confirmed by PicoGreen staining and confocal microscopy (Supplementary Fig. 6a), with quantitative increases in PicoGreen-positive cells (Supplementary Fig. 6b).
A key component of cGAS–STING-mediated signalling, cancer cell-derived 2′3′-cyclic-GMP-AMP (cGAMP)46, acts as a DAMP signal to antigen-presenting cells. Measured extracellular-to-intracellular cGAMP ratios in B16-GM cells rose dose dependently (0.5–15 μM), indicating active secretion without inhibitory protein (ENPP1)-mediated degradation47 (Fig. 2i). To further confirm DNA leakage and exclude replication-linked expansion, we performed subcellular fractionation48 of particle-exposed B16-GM cells, which showed significantly increased cytosolic dsDNA without nuclear increases relative to controls (Fig. 2j). Collectively, data demonstrated that C′ dots induce oxidative stress, releasing cytosolic dsDNA and activating the cGAS–cGAMP–STING axis and interferon stimulated gene (ISG) signalling—a previously unrecognized nanoparticle-driven pathway.
Whole-transcriptomic profiling revealed coordinated activation of immune-regulatory, metabolic and inflammatory networks downstream of PRRs in particle-treated B16-GM cells (Fig. 3a). Total reactive oxygen species (ROS) rose in a concentration-dependent manner over 72 h, paralleling particle uptake (Fig. 3b), while mitochondrial ROS declined at 15 µM (Fig. 3c), indicating a shift towards endolysosomal ROS generation favoured by particle trafficking. Upregulated DAMPs engaging PRRs likely further amplified ROS-driven antigen uptake and cross-presentation. As oxidative stress can activate cGAS–STING via cytosolic DNA release in stressed/dying cells49, we quantified ISG induction, including Tmem173, Mb21d1, Irf1/3/7 and Nfkb1, in particle-exposed (0.1–15 μM) cells (Fig. 3d–i). ISG expression rose in a concentration-/time-dependent manner, peaking at 15 µM and 72 h; Nfkb1 showed the greatest upregulation (>1,000-fold; Fig. 3i), while Irf1 and related genes increased >10-fold at 24 h before tapering by 72 h, except at 15 μM (Fig. 3f). Tmem173 mirrored Irf3/7 and NfkB kinetics, while other PRR-linked factors (for example, MAVS/MDA5) were unchanged. Together, these data define a particle-driven cascade in which oxidative stress and cytosolic DNA release activate cGAS–STING and downstream ISG programmes, positioning C′ dots as intrinsic inducers of tumour-cell-autonomous immune signalling.
a, Schematic of key PRR pathways activated by non-targeted C′ dots. b,c, Concentration-dependent changes in total (b) and mitochondrial (c) ROS in vehicle- and C′ dot-exposed B16-GM cells by flow cytometry. d–i, Time-dependent particle-driven fold changes for murine STING (d), cGAS (e), IRF1 (f), IRF3 (g), IRF7 (h) and NF-κB (i) normalized to control values. Red dotted lines indicate no fold change and blue bars indicate cut-off levels used to establish significance (that is, 2-fold changes) over vehicle. Particle concentrations indicated by grey bars (0.1 μM), purple bars (0.5 μM) and orange bars (15 μM). RT-qPCR via TaqMan assays performed in triplicate for each gene and normalized to internal housekeeping gene GAPDH, according to markers in Supplementary Table 1. j–l, Time-dependent changes in extracellular ATP (RLU) (j), extracellular HMGB1 (OD) (k) and % calreticulin-expression in vehicle- and C′ dot-treated (15 μM) B16-GM cells (l). m,n, Percentage of phosphorylated STING (p-STING) (m) and IRF3 (p-IRF3) (n) in vehicle- and C′ dot-treated B16-GM cells. o,p, Concentration-dependent OD values for type I and II IFN using supernatants from C′ dot-treated B16-GM cells (72 h) and B16-Blue type I IFN (o) and type II IFNγ (p) reporter cell lines. q, Surface PD-L1 expression in C′ dot-treated B16-GM cells (72 h) by flow cytometry. r, Media from o and p were analysed using mouse cytokine/chemokine proteome profiler arrays. Data represent the mean ± s.e.m. of three biological replicates for treated and control groups. Unpaired Student’s t-test and non-parametric one-way ANOVA with Tukey’s test for multiple comparisons were performed. All statistical tests were two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001. RLU, relative luminescence; OD, optical density; a.u., arbitrary units; TIMP-1, tissue inhibitor of metalloproteinase 1; M-CSF, macrophage-stimulating colony. Schematic in a created with BioRender.com.
Given cGAS–STING activation and accompanying pro-inflammatory signatures, we next examined hallmarks of immune-related cytotoxicity50. Particle-exposed (15 µM, 72 h) B16-GM cells significantly increased secretion of DAMPs over controls—extracellular ATP and high mobility group box 1 (HMGB1) (Fig. 3j,k)—with a higher fraction of cells showing the ‘eat-me’ surface marker, calreticulin (Fig. 3l), suggesting immune-related necrosis. Functional activation of STING and IRF3 via phosphorylation51 was also assessed as differentially expressed genes indicated enhanced cGAS–STING activity that can be highly regulated via post-transcriptional/translational alterations52. After exposing B16-GM cells to C′ dots (15 μM, 72 h), we observed near uniform p-STING positivity and elevated p-IRF3 that markedly exceeded the small-molecule STING agonist ADU-S100 (~20% positivity; Fig. 3m,n) over the same interval. Supernatants showed significantly increased type I (IFN-α/β; Fig. 3o) and type II (IFNγ; Fig. 3p) IFNs. We further assessed particle modulation of PD-L1 expression, which correlates with PRR activity10. Unexpectedly, concentration-dependent declines in the percentage (%) of PD-L1+ tumour cells were observed (Fig. 3q), potentially reflecting post-translational modification53,54 that further amplifies oxidative stress55. Cytokine/chemokine profiling revealed a mixed, but predominantly pro-inflammatory, signature, with elevated myeloid and T cell chemoattractants56 (for example, M-CSF, CCL2 and CCL5; Fig. 3r).
These particle-driven findings, integrated with previous datasets, align with canonical PRR activation of the dsDNA–STING axis (Fig. 3a). As type I/II IFNs were upregulated, we examined feedback regulation through the IFN alpha receptor subunit 1 (IFNAR1). IFNAR1 increased markedly at 15 µM C′ dots (Supplementary Fig. 7a), indicating autocrine/paracrine amplification, alongside increases in other immunomodulatory receptors (CD73, CD38, MHC-I and MHC-II; Supplementary Fig. 7b–e and Supplementary Table 2). Although CD38 and CD73 possess suppressive potential, concurrent upregulation of IFNAR1 and MHC molecules denotes a dominant type I/II IFN-driven antitumour response.
Cell-cycle arrest and senescence induction in B16-GM cells
Beyond driving oxidative stress, pro-inflammatory signalling and canonical/alternative cytotoxic pathways, C′ dots also induce cell-cycle arrest and senescence (schematic, Fig. 4a). Consistent with DNA damage, phospho-γH2AX (p-γH2AX) increased in a concentration-dependent manner (Fig. 4b). Cell-cycle profiling revealed G1 arrest at 24 h and 72 h, with G2–M arrest and S-phase suppression at 72 h (Fig. 4c). A significant fraction of treated cells (15 µM, 72 h), versus vehicle, expressed beta-galactosidase (β-gal+) by flow cytometry and colorimetry (Fig. 4d,e), suggesting cGAS–STING-driven senescence45. We next examined cell-cycle inhibitor genes—hallmarks of senescence and growth arrest57. As B16-GM (and B16-F10) lack the tumour suppressor gene, p16INK4a/ARF58, we analysed Cdkn1a (p21)59, which was strongly upregulated in treated but not control cells (Fig. 4f), confirming senescence-associated cell-cycle inhibition.
a, Schematic depicting particle-induced cell-cycle arrest and senescence in B16-GM cells. b, Percentage (%) of p-H2AX + STING-competent B16-GM cells detected by flow cytometry after particle exposure (15 μM, 72 h) relative to vehicle. c, Cell-cycle analysis using propidium iodide and phosphorylated histone 3 (pHH3) staining after C′ dot or vehicle treatment. d, Proportion of beta-galactosidase (β-Gal+)-expressing B16-GM cells with or without C′ dots (15 μM, 72 h) measured by flow cytometry, and e, in a concentration-dependent manner by colorimetric analysis with microscopy. f,g, RT-qPCR of p21 expression (f) and cytosolic DNA quantification in vehicle- and 15 μM C′ dot-treated cells. Genomic-to-mitochondrial DNA ratios (g) were determined using nuclear DNA (Tert) and mitochondrial DNA (Mt-DNA; D-loop) markers. h, Mean fluorescence intensity (MFI) of autophagosome formation during autophagy in particle- and vehicle-treated cells labelled with DALGreen dye by flow cytometry. i, Western blot analysis of B16-GM (WT), B16-GM STING-KO and B16-GM STING scrambled (scr) cells (right) confirming STING-KO efficiency, with β-actin (left) as a loading control. j, Cell-cycle profiling of STING-KO cells, as in c. k, β-Gal+ staining in B16-GM STING-KO cells exposed to vehicle or C′ dots (15 μM, 72 h). Data reflect analyses of three biological replicates for particle- and vehicle-treated (control) groups computed as the mean ± s.e.m. Unpaired Student’s t-test and non-parametric one-way ANOVA with Tukey’s test for multiple comparisons were performed. All statistical tests were two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001. Schematic in a created with BioRender.com.
To determine the source of increased cytoplasmic dsDNA linked to particle-induced inflammatory activation in B16-GM cells, we distinguished genomic (gDNA) from mitochondrial DNA (mtDNA)48. We observed elevated gDNA/mtDNA ratios (Fig. 4g), consistent with genomic DNA leakage and cGAS–STING activation. Increased autophagy and autophagosome formation (Fig. 4h) likely reflect adaptive responses to lysosomal stress, potentially driven by cGAS–STING or genotoxic-stress-induced autophagy60,61,62. These findings support a DAMP–PRR–ROS axis in STING-competent B16-GM cells linking nanoparticle uptake to genomic DNA release, cGAS–STING signalling and immune-related cell death, revealing a previously unrecognized interconnected antitumour network.
In STING-deficient B16-GM cells (western blot-validated; Fig. 4i), C′ dots induced pronounced G1 arrest with reduced G2 and a transient 24 h S-phase rise (Fig. 4j). Senescence was pronounced (~80% higher SPiDER-βGal expression over vehicle and ~2-fold that of STING-expressing cells; Fig. 4k). Thus, in vitro stress-induced senescence can proceed without STING—potentially via ROS, lysosomal/DAMP signalling or alternate sensors (TLRs, MAVS/MDA5/RIG-I). In STING-competent cells, cGAS–STING chiefly reinforces G2–M arrest and IFN/ISG induction, suggesting a modulatory but non-essential role for tumour-intrinsic STING.
C′ dots promote TILs and inhibit suppressive immune cells
After establishing particle-induced immunostimulatory and cytotoxic programmes in melanoma cells, we determined whether particles enhanced immune responsiveness within the B16-GM TME. As myeloid cells mediate suppression and checkpoint resistance63, bone marrow-derived macrophages (BMDMs) from untreated tumour-bearing mice were expanded and exposed to C′ dots (15 µM, 48 h; Fig. 5a). qRT-PCR revealed upregulation of M1 genes (Nos2 and Il6), inflammatory cytokines (Il12b and Tnfa), the PRR sensor, Mb21d1 (cGAS) and IFN-pathway transcripts (Irf1, Ifnb and Cd274), with concurrent Il10 suppression (Fig. 5b). PD-L1 regulation diverged from that in B16-GM cells, reflecting context-specific IFN signalling. To characterize phenotypic remodelling, we profiled M1/M2 myeloid subsets by flow cytometry and quantified shifts in CD11b+ cell populations and surface markers using treatments in Fig. 5a. Multiplex assays confirmed increased IL-12, TNF-α and IL-6, without elevation of suppressive cytokines (Fig. 5c). Flow cytometry revealed pronounced skewing of CD11b+ BMDMs from M2 to M1, complete ablation of myeloid-derived suppressor cells (MDSCs) and expanded neutrophil/monocyte subsets (Fig. 5d). In tumour–macrophage co-cultures, particle-treated CD11b+ cells showed enhanced phagocytosis over controls (Fig. 5e) and elevated type I IFN and cGAMP (Fig. 5f,g), indicating secretion of potent antitumour mediators.
a, Schematic of experimental procedure to isolate/culture BMDMs and splenocytes (T cells) from B16-GM tumour-bearing mice for treatment. b, Differential fold changes of select genes in BMDMs exposed to either particle (15 μM) or saline vehicle for 48 h and assayed using RT-qPCR; largest differentials are boxed. c, Cytokine secretion analysis of supernatants from b. d, Flow cytometric analysis of CD11b+ cells 48 h post-treatment. e, Percentage (%) of tumour cells phagocytosed by co-cultured BMDMs using flow cytometry 48 h after exposing the former to particles or vehicle. f,g, Analysis of type I IFN (f) and cGAMP (g) production via B16-Blue cells and ELISA, respectively. Data represent mean ± s.e.m. for particle-treated and control groups (n = 3 mice per group) reflecting 2 independent experiments. Unpaired Student’s t-test and non-parametric one-way ANOVA with Tukey’s test for multiple comparisons were performed. All statistical tests were two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001. Schematics in a created with BioRender.com.
To examine adaptive immune activation, we evaluated particle-driven T cell responses using the gp100-specific (TCRVβ13+) pmel-1 model64. C′ dot exposure augmented proliferation of anti-CD3/CD28-stimulated T cells relative to bead stimulation alone (Supplementary Fig. 8a). With pmel-1 T cells, C′ dot incubation drove significantly greater expansion and proliferation than either unstimulated or bead-exposed cells (Supplementary Fig. 8b,c). C′ dot-treated pmel-1 T cells also secreted more IFNγ than cells stimulated by beads alone (Supplementary Fig. 8d), while maximal tumour-cell killing occurred with both anti-CD3/CD28 beads and C′ dots (Supplementary Fig. 8e). In summary, these results suggest that particle-exposed immune cell populations potentiate antitumour immune phenotypes with potent cytotoxic capabilities.
C′ dots have shown efficacy in preclinical models31,32,34,35,36,65, but largely in immunodeficient hosts or as drug conjugates30,34, limiting assessment of intrinsic TME effects. To test this directly, immunocompetent B16-GM tumours received i.v. injected non-targeted or αMSH-C′ dots (12 nmol per dose, Q3D×3). Unlike B16-F10 (Fig. 1a), both regimens significantly reduced tumour volume without liproxstatin-1 rescue, indicating ferroptosis independence (Fig. 6a), and body weights remained stable (Supplementary Fig. 9). In NSG mice lacking adaptive immunity, efficacy was abolished (Fig. 6b), implicating immune effectors—particularly CD8+ T cells and myeloid cells—in tumour control.
a,b, Tumour growth was monitored over a 10-day period following i.v. injection of C′ dots or αMSH-C′ dots (12 nmol per dose, Q3D×3), as against vehicle, in immunocompetent C57BL/6J (n = 3 per cohort) (a) or immunodeficient (NSG, n = 3 per cohort) mice (b). For αMSH-C′ dots, liproxstatin-1 was concurrently administered as a third cohort (a). c,d, Time-dependent changes in tumour volumes for C57BL/6J mice (n = 3 per cohort) i.p. injected with depleting antibodies for CD8+ (blue line), CD4+ (red line), NK+ (green line) (c) or myeloid cells (red line) (d), all treated with C′ dots in a similar manner to a. Arrows indicate timing of C′ dot dose administration. e, Multiplex IF of PD-L1, MELAN A, CD8, CD4, F4/80, Foxp3 and DAPI 10 days after final C′ dot treatment (scale bars, 1st column, 1 mm; middle and right columns, 200 μm). f–j, Quantification of % MELAN A+ PD-L1+ cells (f), PD-L1 intensity score (g), PD-L1+ cells (h), F4/80+ cells (i) and F4/80+ PD-L1+ (j) using HALO AI software (n = 3). k, IF staining for STING (scale bars, 200 μm) with and without C’ dots. l, Percentage of STING-positive cells calculated from images in k (n = 18, 3 biological replicates × 6 different regions from each mouse). m, Schematic illustrating treatment arms (n = 6) used for monitoring survival in B16-GM mice. n, Kaplan–Meier survival analysis treated as in a using non-targeted C′ dots, anti-IL-6 and anti-PD-L1 mAbs (200 mg each, Q3D×3 beginning at the second C′ dot dose). Data in a–d, f–j and l are presented as mean ± s.e.m. Non-parametric two-way ANOVA with Sidak’s post hoc test was performed for growth inhibition and unpaired Student’s t-test for IF quantification was performed. All statistical tests were two-sided. *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001. Schematic in m created with BioRender.com.
Immune-cell depletion at treatment onset after intraperitoneally (i.p.) injected anti-CD8, anti-CD4, anti-NK1.1 or anti-colony-stimulating factor 1 receptor (anti-CSF1R) mAbs revealed tumour regrowth upon depletion of CD8+ T cell (Fig. 6c) or myeloid cells (Fig. 6d). Flow cytometry confirmed selective loss of targeted subsets 10 days post-dosing (Supplementary Fig. 10a–d). Spatial distributions of key immune cell populations/markers (PD-L1, MELAN A, F4/80, CD8, CD4 and FoxP3) showed that particle-treated tumours had higher overall and tumour-surface PD-L1+ cells, along with increased F4/80+ macrophages and intratumoural CD8+ T cells over controls (Fig. 6e–i). PD-L1 was expressed predominantly by tumour rather than myeloid cells (Fig. 6j). STING expression was markedly higher in particle- versus vehicle-treated tumours (Fig. 6k,l), mirroring in vitro findings.
In STING-knockout (KO) hosts or mice bearing STING-deficient B16-GM tumours (Supplementary Fig. 11a,b), antitumour responses were abolished, demonstrating that both host and tumour STING signalling are required for efficacy. BMDMs from B16-GM STING-KO hosts, treated similarly to wild-type (WT) BMDMs, demonstrated upregulation of antitumour genes (Nos2, Irf1/3 and Mavs), but not Mb21d1 (Supplementary Fig. 11c), indicating reliance on alternative pathways. The absence of tumour control likely reflects elevated IL-6, which can regulate PD-L1 expression66, myeloid differentiation/suppression67 and antitumour immunity68. Relative to WT, KO-derived BMDMs showed greater M2 polarization, no MDSC depletion (Supplementary Fig. 11d) and unchanged cytokines/chemokines, except for modest IL-12 increases (Supplementary Fig. 11e)—which may implicate other PRR contributions (that is, MAVS). Together, these data explain the lack of antitumour immunity in STING-KO versus WT hosts.
In survival studies combining C′ dots with ICB (that is, anti-PD-L1; Supplementary Fig. 12), no clear survival benefit emerged, likely owing to rapid resistance. Guided by previous findings that C′ dots increase tumour STING, IL-6 and PD-L1 expression on both tumour and myeloid cell populations (Supplementary Fig. 13), a new regimen combining multi-dose C′ dots (12 nmol per dose, Q3D×3) with IL-6 and PD-L1 blockade markedly improved survival (P < 0.001; Fig. 6n). This triple therapy regimen highlights the essential role that adaptive immune cells play in achieving antitumour efficacy via particle targeting of the STING/IL-6/PD-L1 axis.
We analysed TILs and myeloid populations 4 and 10 days after multi-dose C′ dot treatment (12 nmol per dose, Q3D×3) by flow cytometry using markers in Supplementary Tables 3 and 4. At day 4, CD8+ and CD4+ T cells expressing PD-1, TIM3, LAG3, their co-expressing subsets and regulatory T (Treg) cells were significantly reduced (Extended Data Fig. 1a,b). Concurrently, CD8+CXCR3+ T cells, CD8/CD4 and CD8/Treg ratios increased (Extended Data Fig. 1c), consistent with enhanced antitumour activity. The myeloid compartment showed polarization towards an activating phenotype: although total antigen-presenting cells were modestly reduced over controls (Extended Data Fig. 1d,f), MHC-II+ macrophages, dendritic cells (DCs) and monocytes significantly increased, while MDSCs sharply declined (Extended Data Fig. 1e). Similar trends persisted at day 10 (Supplementary Fig. 14a–e), with additional increases in recruited DCs and monocytes (Supplementary Fig. 14f,g).
Bulk RNA-seq of particle-treated B16-GM tumour specimens revealed upregulation of tumour-associated antigens (Pmel and Tyrp1) and antigen-presentation/DAMP genes (Calr, B2m and H2-K1) (Extended Data Fig. 1h) over controls. KEGG analysis confirmed enrichment of antigen processing/presentation, proteasome, spliceosome, phagosome and endocytosis pathways (Extended Data Fig. 1i), with few downregulated networks.
Four days post-treatment, tumours exhibited elevated PRR genes (Mb21d1, Mavs and Ddx58) and IFNs (Ifna1 and Ifnb1), which declined by day 10 (Supplementary Fig. 15a,b). These temporal dynamics aligned with IP data (Extended Data Fig. 1 and Supplementary Fig. 14), reflecting sequential immune activation—early PRR induction precedes durable effector cell and cytokine remodelling. Serum cytokine/chemokine profiles collected during peak PET tracer uptake as demonstrated in our previous studies27 showed broad downregulation over vehicle (Supplementary Fig. 15c,d), including suppression of GM-CSF, a myeloid-suppressive driver63. By day 10, IL-27, CXCL10 and CXCL1 rose, reflecting sustained immunostimulation. This favourable profile, coupled with stable weight, indicates selective TME activity that confines ROS/STING signalling locally, avoiding the systemic cytokine surges of soluble STING agonists. At day 4, immunofluorescence (IF)/IHC confirmed increased p-γH2AX and STING with reduced Ki-67 (Supplementary Fig. 15e–h), consistent with DNA damage and STING activation; PD-L1 remained unchanged (Supplementary Fig. 15i).
Flow cytometry 4 days after the final dose (Supplementary Fig. 16) showed elevated ROS and STING activation across tumour (CD45−Tyrp1+), macrophage (CD45+F4/80+) and T cell (CD45+CD3+) populations, accompanied by higher particle uptake. In macrophages, this axis drove type I IFN and cytokine secretion; in T cells, it correlated with IFNγ production (Supplementary Fig. 16k) and enhanced cytotoxic function; future work will examine the diverse effects of these findings (for example, induction of stem-like phenotypes). Parallel STING activation in both lineages suggested a coordinated circuit in which innate sensing augmented adaptive immunity. Hepatic and splenic tissues collected 3 days after therapy showed only minimal spontaneous inflammation (Supplementary Table 5) and no particle-related toxicity, supporting systemic quiescence and absence of off-target STING activation.
Our data reveal a remarkably broad spectrum of adjuvant-therapeutic properties of C′ dots—an ultrasmall (sub-8 nm), non-toxic and clinically validated silica nanoparticle (Extended Data Fig. 2)—that enhance innate and effector cell functions, drive pro-inflammatory and cytotoxic programmes, induce senescence and suppress immunoinhibitory activity within the TME of ICB-refractory melanoma. These findings define a new, clinically relevant paradigm wherein particle-driven modulation of the STING/IL-6/PD-L1 axis improves survival and antitumour immunity in highly suppressive TMEs.
The potent immune activation observed stems largely from the ultrasmall particle size, consistent with previous reports demonstrating improved immune activation, target specificity16 and antigen presentation with small-core (<15 nm) particles relative to larger (30–100 nm) analogues, which often cause off-target toxicity69,70,71 if not locally administered72,73. Most self-therapeutic nanomaterials remain in early-stage development17,74 and face challenges in manufacturing, PK and toxicity; a few, largely non-oncologic, are being clinically tested for their antimicrobial properties17,75.
Mechanistically, C′ dots broadly modulated PRR, TNF-α and type I/II IFN pathways across tumour, myeloid and T cell compartments. Particle exposure triggered oxidative stress, with elevated ROS, cytosolic DNA and STING activation driving tumour cell death, DNA repair inhibition, G2/M arrest and senescence while reprogramming innate and effector cells towards antitumour immunity. These effects translated into higher CD8+/Treg and M1/M2 ratios and reduced suppressive T cell and MDSC subsets (Extended Data Figs. 1 and 2).
At the tumour-cell level, C′ dots localize to lysosomes (LAMP1+)37, indicating endolysosomal confinement without overt degradation. Downregulation of cathepsin D (Ctsd) and Tpcn1, together with upregulation of cathepsin L (Ctsl) and the lysosomal Na+/Ca2+ channel, Tpcn2 (Fig. 2c), indicates a shift from degradative to immune-related lysosomal signalling76,77. Combined with increased autophagosome formation (Fig. 4h), these changes suggest sub-lethal lysosomal perturbation78,79 that releases cathepsins, ROS and DAMPs into the cytosol79—events driving DNA damage, dsDNA release and cGAS–STING activation51,79,80. Tpcn2-mediated ion flux may further contribute to mitochondrial redox imbalance and genomic instability81. Upregulation of Tlr2/3/4 supports DAMP-driven endosomal PRR engagement and NF-κB-dependent cytokine production80. Together, these findings link particle-induced lysosomal stress to cGAS–STING activation and TME remodelling, with endosomal TLRs contributory but STING dominant.
In the B16-GM model, C′ dots engaged both tumour-intrinsic and host STING pathway. In vitro, tumour STING modulates G2/M arrest and IFN/ISG responses, but is dispensable for senescence, which could proceed via STING-independent stress pathways. Ex vivo, BMDMs exhibited cGAS–STING activation, identifying myeloid STING as a principal mediator of innate reprogramming (Fig. 5b). In vivo, efficacy was lost when either host or tumour STING was absent, confirming dual-compartment requirement for antitumour activity.
Selection of anti-PD-L1 and anti-IL-6 antibodies as combination partners was data driven. In STING-KO BMDMs, C′ dots induced an ~100-fold increase in IL-6 expression (Supplementary Fig. 11c), consistent with C′ dots acting as STING agonists and regulators of IL-6. Disruption of downstream STING reduces type I IFNs and their suppression of IL-682,83, linking the IL-6/PD-L1 axis to therapy resistance84,85. Dual blockade of IL-6 and PD-L1 synergized with C′ dots, unlike PD-L1 alone (Supplementary Fig. 12), reducing PD-L1 expression and enhancing T cell infiltration, TH1 and CD8+ T cells84,85. The clinical significance of this treatment paradigm mirrors ongoing trials combining IL-6 and checkpoint inhibitors to overcome dual ICB resistance86,87.
Distinct molecular pathways underlie responses across melanoma models. Ferroptosis dominated in B16-F10 (Fig. 1), whereas B16-GM exhibited immune-related necrosis, senescence and cytoplasmic DNA accumulation (Figs. 3 and 4). Bulk RNA-seq (Supplementary Fig. 5d) confirmed divergent pathway signatures: B16-GM tumours showed suppression of oxidative phosphorylation and glutathione metabolism while upregulating PRR and antigen-presentation networks.
Several areas remain under active investigation. The enzymatic and organellar origins of particle-induced ROS remain unresolved (Fig. 3b); potential contributors include lysosomal and mitochondrial redox enzymes or iron-catalysed reactions within endolysosomal compartments. The route of cytosolic dsDNA accumulation—nuclear envelope instability, micronuclei rupture or mitochondrial/lysosomal leakage—requires further study88 (Fig. 4g). Finally, divergent cytotoxic responses between B16-F10 and B16-GM may reflect differences in iron metabolism, calcium signalling or other metabolic programmes dictating ferroptotic versus immune-related outcomes.
Conclusion
C′ dots act as potent adjuvant therapies, inducing tumour-intrinsic stress—ROS generation, cytosolic DNA release and cGAS–STING activation—that links lysosomal perturbation to innate immune sensing localized to the tumor microenvironment. While the precise ROS sources and DNA-leakage routes remain unresolved, these pathways likely converge on innate DNA sensing to explain how particles couple stress to immunity. Notably, without a conjugated STING agonist, C′ dots modulated the STING/IL-6/PD-L1 axis and synergized with IL-6 and PD-L1 blockade in B16-GM to reverse suppression and improve efficacy. Their tightly controlled physicochemical properties21,22,23 and favourable PK24,27,28,29,33,36 enable preferential tumour uptake and localized pro-inflammatory signalling. Model-specific responses—immune-phenotypic shifts, cytotoxic programme activation and suppression of inhibitory transcripts—provide a blueprint for rational combinations across cold/heterogeneous TMEs and for future surface-chemical and clinical designs.
Methods
Synthesis of PEG-C′ dots and αMSH-PEG-C′ dots
C′ dots were synthesized in aqueous solution as previously described21,22,23. Both plain (PEGylated) C′ dots and αMSH-PEG-C′ dots encapsulate the fluorescent organic dye, Cyanine 5 (Cy5). αMSH peptides with an N-terminal acetylated cysteine thiol enabled reaction with a maleimido-terminated heterobifunctional PEG-silane, the latter obtained by reacting maleimido-NHS ester heterobifunctional PEG (Sigma) with aminopropyl-triethoxysilane (Sigma). The resulting αMSH-PEG-silane was conjugated to the particle surface during the PEGylation step as previously reported27. Once synthesized, particles were purified using gel permeation chromatography (Bio-Rad) and up-concentrated using Vivaspin ultrafiltration spin columns (Cytiva, molecular weight cut-off 30,000). Absorbance measurements of both plain PEG-C′ dots and αMSH-PEG-C′ dots were obtained using a Varian Cary 5000 spectrophotometer, and the number of αMSH peptides per particle was determined via the absorbance of the peptide and its calculated extinction coefficient. Particles were further characterized by fluorescence correlation spectroscopy (FCS) to determine their hydrodynamic radius, concentration and number of encapsulated dyes per particle21.
Cell culture and cell line authentication
B16F10 and B16-GMCSF melanoma cell lines were maintained in RPMI medium supplemented with 10% fetal calf serum (FCS) and 1% penicillin with streptomycin and Plasmocin (InvivoGen catalogue number ant-mpp). B16-Blue IFNα/β (InvivoGen catalogue number bb-ifnt1) and B16-Blue IFNγ cells (InvivoGen catalogue number bb-ifng) cells were maintained in RPMI1640, 10% heat-inactivated FBS, 50 U ml−1 penicillin, 50 μg ml−1 streptomycin, 100 μg ml−1 Normocin and 100 μg ml−1 Zeocin. Cell lines were routinely mycoplasma tested.
Generation of STING-KO B16-GM cells
STING1 KO and scrambled control (scr) B16-GM cells were generated by lentiviral transduction with pLentiCRISPRv2-mSTING-gRNA_1 (GCTGGATGCAGGTTGGAGTA) or pLentiCRISPRv2-mSTING-scrambled_gRNA_1 (CTTTGCGGAGATTGGAGGGA), respectively (Addgene), using standard protocols89. Cells were selected in fresh puromycin (1 μg ml−1; InvivoGen) for 10 days, with media replenished every 2 days. Knockout efficiency was confirmed by western blot. B16-GM, B16-GM STING-KO and B16-GM STING scr cells (5 × 106) were lysed in 1 ml Pierce RIPA buffer (ThermoFisher) on ice for 15 min. Lysates were centrifuged at 15,000 × g for 20 min at 4 °C, and 20 ml supernatant was mixed with 2× Laemmli sample buffer (Bio-Rad), boiled at 95 °C for 10 min and resolved on Mini-PROTEAN 4–20% pre-cast gels (Bio-Rad). Proteins were transferred to nitrocellulose membranes (Bio-Rad), washed once in TBS and blocked with EveryBlot Blocking Buffer (Bio-Rad) for 15 min. Membranes were incubated overnight at 4 °C with rabbit anti-STING (D2P2F, Cell Signaling Technology) or rabbit anti-β-actin (Poly6221, BioLegend) primary antibodies in blocking buffer, followed by HRP-conjugated secondary antibody for 1 h at room temperature (RT). Bands were visualized using Clarity Western ECL substrate (Bio-Rad) and imaged on a ChemiDoc system (Bio-Rad).
Gene expression analysis
Cells were collected at indicated time points and RNA extracted using the Purelink RNA minikit (ThermoFisher catalogue number 12183020) per the manufacturer’s instructions. DNase removal was performed on a column per the manufacturer’s instructions (ThermoFisher catalogue number 12185-010). RNA was extracted from FFPE sections using the Quick RNA-FFPE kit (Zymo Research catalogue number R1008) when tissue samples were limited. Reverse transcription was carried out using the High-Capacity RNA-to-cDNA Kit (ThermoFisher catalogue number 4387406) using between 100 ng and 1 μg of RNA per the manufacturer’s instructions. Quantitative polymerase chain reaction (qPCR) was performed for analysing 2 μl cDNA in a 20 μl reaction using TaqMan assays (FAM-labelled, ABI), TaqMan Fast Advanced Master Mix (ThermoFisher catalogue number 4444556) and a QuantStudio 3 Real-Time PCR Instrument for analysis using the following conditions: 40 cycles, 50 °C (2 min), 95 °C (2 min), PCR cycling 95 °C (1 s) and 60 °C (20 s). mRNA levels were normalized to a reference gene, GAPDH (VIC dye-labelled) using ΔCt = Ct (gene of interest) – Ct (Gapdh), where Ct is the threshold cycle. Results were reported as the relative fold change in mRNA expression over baseline (that is, untreated cells), or ΔΔCt = 2−(ΔCt (sample) − ΔCT (control)) and normalized using a log2 transformation, with untreated cells as the baseline. TaqMan assays are listed in Supplementary Table 1.
Bulk RNA-seq and analysis
Following RNA isolation, FFPE total RNA integrity was checked using a 2100 Bioanalyzer with a DV200 reading as 47–61% for all 6 samples (Agilent Technologies). RNA concentrations were measured using the NanoDrop system (Thermo Fisher Scientific). We used 200 ng of FFPE total RNA sample as the input for fragmentation by treating with metal ions present in the 2X Priming Buffer at elevated temperature (Agilent SureSelect XT HS2 RNA Library Preparation and Target Enrichment Protocol number G9989-90000, Agilent Technologies). Fragmented RNA was used to perform the first- and the second-strand cDNA synthesis. After AmpPure XP Beads purification, cDNA samples proceeded to end repair, A-tailing and adapter ligation following the manufacturer’s instructions. The adaptor-ligated cDNA library was then PCR amplified and hybridized by SureSelect XT Mouse All Exon probes (Agilent Technologies, catalogue number 5190-4641). Captured and target-enriched cDNA libraries were amplified and purified. Final libraries were pooled and subjected to sequencing on the NovaSeq6000 sequencer (Illumina) at PE 2 × 100 cycles. The raw sequencing reads in BCL format underwent processing through bcl2fastq v.2.20 (Illumina) for FASTQ conversion and demultiplexing. Following adaptor trimming with Cutadapt v.1.18 (ref. 90), RNA reads were aligned and mapped to the GRCm39 mouse reference genome using STAR v.2.5.2 (ref. 91). Read counts per gene were extracted using HTSeq-count v.0.11.2 (ref. 92). Gene expression profiles were constructed for differential expression, clustering and principal component analyses using the DESeq2 package v.1.30.1 (ref. 93). For differential expression analysis, pairwise comparisons between two or more groups utilized parametric tests where read counts followed a negative binomial distribution with a gene-specific dispersion parameter. Corrected P values were calculated based on the Benjamini–Hochberg method to adjust for multiple testing.
The volcano plot was generated using the R EnhancedVolcano package v.1.16.0 (ref. 94). GSEA was conducted using the R clusterProfiler package v.4.6.2 (ref. 95), and the enrichment results were visualized using the R enrichplot package v.1.18.3.
Type I/II IFN assays
B16-Blue IFNα/β (InvivoGen catalogue number bb-ifnt1) and B16-Blue IFNγ cells (InvivoGen catalogue number bb-ifng) were plated in 96-multi-well plates for 24 h (4.2 × 105 cells per ml), followed by addition of media (180 μl) and supernatant/sera (20 μl), and incubated (37 °C and 5% CO2 for 24 h). To determine secreted embryonic alkaline phosphatase production, supernatants from treated and untreated B16-Blue (50 μl) cells were added to new 96-well plates and used immediately or snap frozen at −80 °C. QuantiBlue assay reagent (150 μl; InvivoGen catalogue number rep-qbs) was added to samples to a final volume of 200 μl, and the mixture was incubated at 37 °C and 5% CO2 for 1–6 h and absorbance (l = 620–655 nm) was quantitatively measured using a SpectraMax iD3 Multi-Mode Microplate Reader (Molecular Devices).
Secreted cytokine/chemokine determination
Cytokine/chemokine secretion from treated specimens was detected using a Proteome profiler array kit (R&D Systems, mouse, catalogue number ARY006), according to the manufacturer’s instructions. Arbitrary values of abundance were calculated as integrated densities (ImageJ software, v1.51w) of each dot plot normalized to a reference spot.
Total cellular and mitochondrial ROS quantification
After plating treated and untreated cells in 6-well dishes, adherent cells were stained with CellROX Green Reagent (ThermoFisher catalogue number C10444) or MitoSOX Red Mitochondrial Superoxide Indicator (Thermo Fisher catalogue number M36008) per the manufacturer’s instructions. ROS generation was assessed by an LSRFortessa flow cytometer (BD Biosciences).
In vitro analysis of BMDMs
Using established protocols, BMDMs were derived from tibias and fibulas collected from B16-GM tumour-bearing mice (WT, STING-KO). In brief, bone marrow cell suspensions were cultured for 7 days in complete medium (RPMI 1640 supplemented with L-glutamine, 10% FBS, 1% penicillin and streptomycin, 1 mM sodium pyruvate and 1% non-essential amino acids) in the presence of 10 ng ml−1 recombinant mouse macrophage colony-stimulating factor (M-CSF; PeproTech catalogue number 315-02), with media replaced every 2–3 days. One week later, attached cells were collected and used as part of stimulation assays. For phenotypic analysis, gene expression and IFN/cGAMP analysis, BMDMs were cultured with or without C′ dots for 48 h in complete media. Phenotypic analysis was performed using antibodies listed in Supplementary Table 3 and the Aurora Spectral Cytometer (Aurora Cytek); gene expression analysis used primers listed in Supplementary Table 1. Phagocytosis assays were performed by labelling tumour cells with carboxyfluorescein succinimidyl ester (eBioscience CFSE, ThermoFisher catalogue number 65-0850-84) per the manufacturer’s instructions after being treated with 15 μM C′ dots or vehicle control for 72 h. This was followed by addition of BMDMs at an effector-to-target cell ratio of 1:1. Cells were collected 24 h later, stained for CD11b and analysed on an LSRFortessa flow cytometer; CD11b+ cells were examined for CFSE positivity.
In vitro analysis of T cells
Spleens from pmel-1 TCR transgenic mice were isolated and ground through 100 μm filters to obtain T cells. After red blood cell lysis, T cells were activated by culturing splenocytes with Dynabeads Mouse T-Activator CD3/CD28 (ThermoFisher catalogue number 11456D) in RPMI 1640 medium containing 1% penicillin/streptomycin (MediaTech), 50 mM β-mercaptoethanol (Gibco, Invitrogen), 1% L-glutamine (Gibco) and 100 U ml−1 IL-2 (PeproTech). Cells were labelled with eBioscience CFSE (Thermo Fisher catalogue number 65-0850-84). Following treatment, cytotoxicity was quantified at a T cell-to-tumour cell ratio of 1:1 using the CyQUANT LDH Cytotoxicity Assay (Thermo Fisher catalogue number C20300); supernatants were analysed for type I/II IFNs. Antigen-specific T cells were quantified via H-2Db gp100 Tetramer-EGSRNQDWL-PE (MBL catalogue number TS-M546-1) analysis. Proliferation, tetramer and surface marker analysis were performed 72 h post-stimulation using the LSRFortessa flow cytometer.
DNA damage, immune-related cell death and inflammasome analysis
All antibodies/assays were used per the manufacturer’s instructions. To quantify the degree of cellular DNA damage, PE anti-H2A.X Phospho (Ser139) Antibody (BioLegend catalogue number 613411) was incubated with particle- and vehicle-treated cells and analysed by flow cytometry (BD Biosciences LSR II). For analysis of immune-related cell death, cell surface calreticulin was assayed using PE-conjugated calreticulin (D3E6) XP rabbit mAb (Cell Signaling Technology catalogue number 19780) and analysed with a BD Biosciences LSR II flow cytometer. Extracellular ATP was quantified using the RealTime-Glo Extracellular ATP Assay (Promega catalogue number GA5010), and HMGB1 was quantified using the Lumit HMGB1 Human/Mouse Immunoassay (Promega catalogue number W6110). To assess caspase-1, the Caspase-Glo 1 Inflammasome Assay (Promega catalogue number G9951) was used. Luminescence values were determined by subtracting background luminescence levels (that is, no cells).
Analysis of senescence induction, cell cycle and autophagy
Senescence was quantified using two independent methods: (i) bright-field analysis using the Senescence β-Galactosidase Staining Kit (Cell Signaling Technology, catalogue number 9860) for particle- and vehicle-treated cells (6-well plate, 3 wells per condition) (100 cells at a minimum were counted for each condition), and (ii) flow cytometric analysis (LSRFortessa, BD Biosciences) using the Cellular Senescence Detection Kit SPiDER-βGal (catalogue number SG03-10; Molecular Technologies) for cells treated in the same manner as (i) (10,000 cells per condition were analysed with data shown representing the mean of 2 independent experiments).
For cell-cycle distribution studies, cells were initially synchronized using the cyclin-dependent kinase 1 (CDK1) inhibitor RO-3306 (Selleck, catalogue number S7747), followed by C′ dot or vehicle treatment. Cells were then fixed with 70% ethanol overnight, washed in triplicate with phosphate-buffered saline (PBS), stained with Alexa Fluor 488 anti-Histone H3 Phospho (Ser10) antibody (BioLegend catalogue number 650803) for 1 h, washed again in triplicate with PBS and resuspended in 200 ml propidium iodide/RNase Staining Solution (Cell Signaling Technology catalogue number 4085). Cells were analysed by flow cytometry (LSRFortessa); gates for each phase of the cell cycle were manually determined. At least 10,000 cells per condition were analysed for 2 independent experiments.
Autophagy was assessed by measuring autophagolysomal formation with DALGreen (catalogue number D675-10; Dojindo Molecular Technologies). In brief, DALGreen (1 mmol l−1), after reconstituting (per the manufacturer’s instructions), was added to 5 × 105 B16-GM cells in RPMI 1640 media (30 min, at 37 °C), washed twice with PBS, followed by addition of fresh media, with or without C′ dots. Cells were collected 72 h post-treatment and analysed by flow cytometry (BD Biosciences LSR II) with gates manually determined. At least 10,000 cells per condition were analysed for 2 independent experiments.
Evaluation of STING pathway activation by C′ dots and a reference agonist
WT B16-GM cells were incubated with 15 μM C′ dots for 72 h, washed and stained intracellularly for p-STING (Supplementary Table 2). ROS levels were assessed in parallel. For comparison, cells were incubated over the same time interval with the molecular STING agonist, ADU-S100 (2′3′-c-di-AM(PS)2 (Rp, Rp); InvivoGen) and stained for pSTING. Flow cytometry was used to quantify ROS production and STING activation.
cGAMP analysis via ELISA
B16-GM cells (5 × 105) were resuspended in 250 μl RIPA lysis buffer (Pierce catalogue number 89901) supplemented with cOmplete Protease Inhibitor Cocktail (Sigma catalogue number 11697498001) and lysed with a 28½ gauge needle. Lysates were incubated on ice (30 min), centrifuged (16,000 × g) and cooled at 4 °C (10 min). 2′3′-cGAMP levels in lysates and supernatants from collected tumour cells or BMDMs were quantified using the 2′3′cGAMP ELISA kit (Arbor Assays catalogue number K067-H1W) and then normalized to total numbers of cells per condition.
Subcellular fractionation and quantification of mitochondrial and nuclear DNA
For analysis and quantification of cytosolic DNA, cell fractionation assays were performed using NE-PER nuclear and cytoplasmic extraction reagents (ThermoScientific, catalogue number 78833) per the manufacturer’s instructions48. Primers specific for mitochondrial DNA (mt-Dloop, forward sequence AATCTACCATCCTCCGTGAAACC and reverse sequence TCAGTTTAGCTACCCCCAAGTTTAA) and genomic (TERT, forward sequence CTAGCTCATGTGTCAAGACCCTCTT and reverse sequence GCCAGCACGTTTCTCTCGTT) were used for analysis normalized to whole cell extract values. qPCR was performed using the PowerUp SYBR Green Master Mix (Applied Biosystems catalogue number A25742) with melt curve analysis on a QuantStudio 3 Real-Time PCR Instrument.
In vivo murine models
All animal studies were performed in accordance with protocols approved by the Weill Cornell Medical College Institutional Animal Care and Use Committee (IACUC) and conformed to NIH guidelines for animal welfare. All mice were obtained from the Jackson Laboratory. Six- to eight-week-old female C57BL/6J, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) and C57BL/6J-Sting1gt/J mice were implanted intradermally with either B16-GM (1.5 × 104) or B16F10 (1 × 105) cells. When tumours reached a size of 150–200 mm3, mice were i.v. injected with a multi-dose regimen (n = 3 doses, every 3 days) of either particles (36 nmol) or saline vehicle. Pmel-1 mice with a gp100-reactive transgenic T cell receptor were used for in vitro studies to assess antigen-specific tumour killing and expansion. For ferroptosis studies, mice were also i.p. injected with liproxstatin-1 (Selleckchem), as previously described37. Tumour volumes were measured daily via calipers, and volumes were calculated using the following formula: volume = (long axis × short axis2)/2. No tumours were allowed to exceed the maximal size of 1,500 mm3, as permitted by our institutional IACUC.
Antibody depletion and survival/treatment studies
Five cohorts of female C57BL/6J mice (6–8 weeks, Jackson Laboratory) were inoculated with 1.5 × 104 B16-GM cells for performing antibody depletion studies 7 days post-implantation. Mice were i.v.-injected with vehicle, C′ dots or C′ dots administered with one of the following i.p. injected mAbs (200 mg each): (i) anti-CD4 (clone GK1.5), (ii) anti-NK1.1 (clone PK136), (iii) anti-CD8 (clone 53-5.8) or (iv) anti-CSF1R (clone AFS98) (BioXCell). mAbs were given on day 1 of treatment, and then biweekly for 2 weeks until study termination. Depletion was also confirmed by flow cytometry using collected tumour tissue and blood samples. For treatment studies, mice received anti-mouse PD-L1 rat IgG2a (clone RMP1-14) or anti-mouse IL-6 (clone MP5-20F3) (BioXCell) antibody by i.p. injection (200 µg per dose, as indicated in treatment schemas). Tumour volumes were measured every 2–3 days until tumours reached 1,500 mm3. Kaplan–Meier survival curves were generated, and the log-rank test was used to investigate the significance of various treatments for survival.
Cellular IF
Cells were grown in 8-well chamber slides (ibidi USA, catalogue number 80841) and then fixed in 4% paraformaldehyde for 20 min at RT. Cells were permeabilized (0.1% Tween 20, 0.01% Triton X-100 in PBS) for 10 min, blocked with 1% BSA in PBS (30 min) and then incubated with 1:500 dilution dsDNA antibody in blocking buffer (clone HYB331-01; Abcam catalogue number ab27156) overnight in a humidified chamber at 4 °C. Cells were triply washed with PBS and then incubated with Alexa Fluor 594 anti-mouse antibody in 0.3 ml blocking buffer (1 h, RT). Cells were again triply washed with PBS, counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and fixed with Invitrogen ProLong Gold Antifade Mountant (ThermoFisher catalogue number P36941) for curing overnight. PicoGreen staining was performed with Quant-iT PicoGreen dsDNA Reagent and Kits (ThermoFisher, catalogue number P7589). For confocal microscopy, cells were incubated with PicoGreen (37 °C, 1 h) (ThermoFisher, catalogue number P11495), which was diluted in cell culture medium (3 ml ml−1). Cells were counterstained with DAPI (Thermo Fisher catalogue number P36941), washed and fixed for microscopy (Invitrogen ProLong Gold Antifade Mountant).
Flow cytometric analysis of collected tumours
Mouse tumour samples were minced with scissors and scalpels before incubation with 1.67 U ml−1 Liberase (Roche) and 0.2 mg ml−1 DNase (Roche) in RPMI for 30 min at 37 °C. Tumour samples were homogenized by repeated pipetting and filtered through a 100 mm nylon filter (BD Biosciences) in RPMI supplemented with 5% FCS to generate single-cell suspensions. Cell suspensions were washed once with PBS and resuspended in FACS buffer (PBS/1% BSA). Cells from mouse spleens were isolated by grinding spleens through 40 mm filters. After RBC lysis (ACK Lysing Buffer, Lonza), all samples were washed and resuspended in FACS buffer (PBS/1% BSA) or RPMI. Cells isolated from mouse tumours and spleens were pre-incubated (15 min, 4 °C) with anti-CD16/32 mAb (Fc block, clone 2.4G, BD Biosciences catalogue number 553142) to block non-specific binding and stained (30 min, 4 °C) with combinations of fluorochrome-conjugated antibodies at a ratio of 1:50 (Supplementary Table 3). Fixation and permeabilization (15 min fixation, RT) were carried out with the Foxp3/Transcription Factor Staining Buffer set (eBioscience catalogue number 00-5523-00), followed by intracellular staining in permeabilization buffer for 30 min at 4 °C. Dead cells were excluded using a Zombie NIR Live/Dead fixable dye (1:2,500 in PBS; BioLegend catalogue number 423105). Flow cytometry was performed (Aurora Spectral Cytometer, Aurora Cytek) and data analysed with FlowJo (v.10) software (FlowJo LLC). A representative gating strategy is shown in Supplementary Fig. 17.
Assessment of ROS and STING activation in collected tumour specimens
Tumours were collected from vehicle- and C′ dot-treated B16-GM-bearing mice (n = 4 per group) 3 days after the final dose, dissociated into single-cell suspensions and subjected to RBC lysis and 70 µm filtration. Cells were stained with anti-CD45 and -TYRP, anti-F4/80 and anti-CD3 mAbs for evaluating tumour and immune cell subsets. ROS was measured with CellROX Green, and STING activation assessed by intracellular staining for p-STING after fixation and permeabilization. Data were acquired by flow cytometry and analysed in FlowJo, with gating on tumour cells (CD45−/TYRP1+), macrophages (CD45+/F4/80+) and T cells (CD45+/CD3+).
IHC and IF for collected tissue specimens
Tissues were collected and incubated for 24 h in 10% buffered formalin and then transferred to 70% ethanol for 24–48 h. Tissues were embedded in paraffin embedding blocks, and 5 μM sections were prepared on slides. The pathology slides were deparaffinized and rehydrated by washing sequentially in xylene 3 times for 3 min each, absolute (100%) ethanol 3 times for 3 min each, 95% ethanol 3 times for 3 min and the final wash with water 2 times for 5 min each. Antigen retrieval was performed by autoclaving the slides in citric acid solution at 121 °C for 20 min in a pH 9.0 buffer. These samples were cooled to RT overnight. After antigen retrieval, slides were washed twice in water for 5 min, PBS for 5 min one time and PBS with 0.1% Tween 20 once for 5 min. Slides were blocked for 1–4 h with 10% normal goat serum in 1% BSA and PBS at RT. IHC for Ki-67 was performed on paraffin tumour sections (3 sections per tumour) using a Leica Bond RX automated stainer. After heat-induced epitope retrieval, the primary antibody, rabbit monoclonal antibody clone D3B5 (Cell Signaling number 12202), was applied at a concentration of 1:500, followed by a polymer detection system, according to the manufacturer’s instructions (DS9800, Novocastra Bond Polymer Refine Detection, Leica Biosystems). The chromogen used was 3,3-diaminobenzidine tetrachloride (DAB), and sections were counterstained with haematoxylin. IF staining was performed for γH2AX, STING and PD-L1 on paraffin tumour sections as follows. After heat-induced epitope retrieval in a pH 9.0 buffer, primary antibodies (anti-γH2AX 0.2 mg ml−1; Abcam, number 11174, anti-STING 0.1 mg ml−1; Abcam, number 178847 or anti-PD-L1 0.5 mg ml−1; Cell Signaling, number 64693) were applied. Secondary biotinylated goat anti-rabbit IgG secondary antibody was subsequently added (5.75 mg ml−1; Vector Labs, number PK6101). Application of streptavidin-HRP D (DAB Map kit, Ventana Medical Systems) was followed by incubation with Tyramide Alexa Fluor 488 (Invitrogen, number T20922), prepared at a 1:150 dilution. Slides were counterstained with DAPI (5 mg ml−1, Sigma-Aldrich, number D9542) and mounted with Mowiol. Quantification was performed using QuPath software (v.0.6.0), and a minimum of 10 fields of view were analysed.
Multiplex IF
Staining
Multiplex IHC was conducted using a tyramide signal amplification approach96,97. Validation and condition optimization for the primary antibody was performed using chromogenic IHC. Slides were heated to 60 °C for 30 min, followed by processing in a Bond Rx Research Stainer (Leica Biosystems), where slides were deparaffinized using the ‘dewax’ option and submitted for antigen retrieval at pH 9 for 25 min at 98 °C (BOND Epitope Retrieval Solution 2, catalogue number AR9640, Leica Biosystems). After several washes with immunostainer buffer 1× (Wash Solution 10× Concentrate, catalogue number AR9590, Leica Biosystems), slides were incubated with the primary antibody anti-AQP1 (30 min, RT) at a specified dilution using an antibody diluent (catalogue number ab64211, Abcam) and then treated with washing buffer. Slides were then incubated with rabbit anti-mouse IgG (Bond Polymer Refine Detection kit, catalogue number DS9800, Leica Biosystems) for 8 min at RT, treated with washing buffer and incubated with polymer anti-rabbit Poly-HRP-IgG. After further washes with buffer and deionized water, slides were incubated with peroxide block for 5 min and 3,3′-diaminobenzidine (10 min, RT). Slides were then counterstained with haematoxylin for 5 min at RT (Bond Polymer Refine Detection). The slide was then dehydrated manually using a series of increasing concentrations of ethanol and Citrisolv (catalogue number 89426-270, Decon Labs). Finally, slides were mounted with a coverslip and Canada balsam.
Multiplex IF staining for six markers was also performed on a Bond Rx Research Stainer, using the Opal 6-plex detection kit (catalogue number NEL821001KT, Akoya Biosciences) for the detection of protein markers, using DAPI as a counterstain for nuclear detection (Opal 6-plex detection kit). All washes were done with BOND wash solution (catalogue number AR9590, Leica Biosystems). After baking, dewaxing and epitope retrieval steps were performed before the first cycle of staining was started. Slides were incubated with the blocking buffer 1× (Opal Antibody Diluent/Block, catalogue number ARD1001EA, Akoya Biosciences) for 6 min at RT, followed by the primary antibody for 30 min at RT, which had been diluted in antibody diluent (catalogue number ab64211, Abcam) to the dilution previously established. After washing, sections were incubated with SuperBoost Goat anti-mouse Poly-HRP (ready-to-use solution, catalogue number B40961, Thermo Scientific) or with anti-rabbit Poly-HRP-IgG (reagent from Bond Polymer Refine Detection, catalogue number DS9800, Leica Biosystems), and then treated again with wash solution. After incubating with Opal 480 reagent (10 min, RT), slides were treated with wash solution and incubated with BOND Epitope Retrieval Solution 1 pH 6.0 (catalogue number AR9961, Leica Biosystems) for 20 min at 98 °C. Following treatment with washing reagent, slides were again incubated with blocking buffer 1×, and another staining cycle started for a total of 6 cycles. Slides were then taken out of the immunostainer and mounted with ProLong Diamond Antifade Mountant (catalogue number P36961, ThermoFisher Scientific).
Image acquisition, processing and analysis
Double IF-stained slides were left to dry and then acquired using the Vectra Polaris instrument (v.1.0, Akoya Biosciences). In brief, slides were mounted on a tray and pre-scanned for the acquisition. After finding the focal plane on the DAPI channel and optimizing the exposure time to avoid saturation for all the channels (DAPI, 6 opal dyes and autofluorescence channel), slides were acquired and then exported into a drive for post-acquisition processing. Spectral unmixing was performed using InForm software (v.2.6, Akoya Biosciences), and tiles produced were stitched and visualized using the Halo software (v.3.6, Indica Labs).
For analysis purposes, 3 tissue areas were considered and defined through annotation: central tumour area, periphery of the tumour (defined as 400 µm of the most external portion of the tumour) and the non-neoplastic peritumoural area. To separate the different tissue compartments, a DenseNet classifier (HALO AI add-on, v.3.6, Indica Labs) was trained to recognize glass background, stroma, melanoma cells, necrotic areas and artefact areas (out-of-focus regions, foldings, foreign bodies and so on). The positivity for each marker was determined manually by an experienced pathologist (F.S.), adjusting the threshold value for each single slide. The analysis module on Halo was used to extract the cytoplasmic positivity for Melan-A, CD8, CD4, PD-L1, F4/80 and nuclear positivity of FOXP3 on all samples, stratifying the results on the basis of areas (that is, central tumour, tumour periphery and non-neoplastic/peritumoural). Double positivity for PD-L1-Melan-A, PD-L1-F4/80 and CD4-FOXP3 was also measured by the module. Results were expressed as the percentage of single- and double-positive cells for each marker in the stroma.
Histopathological analysis of treated hepatic and splenic specimens
Hepatic and splenic specimens were collected from particle-treated and control mice, fixed in 10% neutral-buffered formalin and embedded in paraffin. Sections (5 mm) were cut, mounted on glass slides and stained with haematoxylin and eosin following standard protocols. Stained sections were examined by light microscopy, and histopathologic evaluation was performed in a blinded manner by an experienced pathologist (S.M.) to assess tissue architecture and treatment-related changes.
Statistical analysis
Statistical comparisons for double-arm studies in which animals received either the particle treatment or vehicle control were analysed using a two-sided unpaired t-test. Group means and s.e.m. were calculated for time- and/or concentration-dependent changes in cellular uptake, tumour and immune cell markers, functional assays and tumour volumes. Statistical comparisons across multiple experimental groups used a non-parametric one-way analysis of variance (ANOVA) followed by post hoc Tukey’s test. A two-way analysis of variance (ANOVA) followed by Sidak’s multiple comparison test was used for analysing growth inhibition studies. Statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.005 and ****P < 0.001 for all studies, unless otherwise indicated. All graphs were constructed and analysed using GraphPad Prism 7 (GraphPad Software).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The primary data underpinning the findings of this study are available in the Article and its Supplementary Information. Additional data are available from the corresponding author upon request. Source data are provided with this paper.
References
Carlino, M. S., Larkin, J. & Long, G. V. Immune checkpoint inhibitors in melanoma. Lancet 398, 1002–1014 (2021).
Naimi, A. et al. Tumor immunotherapies by immune checkpoint inhibitors (ICIs); the pros and cons. Cell Commun. Signal. 20, 44 (2022).
Khalil, D. N., Smith, E. L., Brentjens, R. J. & Wolchok, J. D. The future of cancer treatment: immunomodulation, CARs and combination immunotherapy. Nat. Rev. Clin. Oncol. 13, 273–290 (2016).
Martin, J. D., Cabral, H., Stylianopoulos, T. & Jain, R. K. Improving cancer immunotherapy using nanomedicines: progress, opportunities and challenges. Nat. Rev. Clin. Oncol. 17, 251–266 (2020).
Haslam, A. & Prasad, V. Estimation of the percentage of US patients with cancer who are eligible for and respond to checkpoint inhibitor immunotherapy drugs. JAMA Netw. Open 2, e192535 (2019).
Coussens, L. M., Zitvogel, L. & Palucka, A. K. Neutralizing tumor-promoting chronic inflammation: a magic bullet? Science 339, 286–291 (2013).
Gajewski, T. F., Schreiber, H. & Fu, Y. X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14, 1014–1022 (2013).
Schmid, M. C. & Varner, J. A. Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation. J. Oncol. 2010, 201026 (2010).
Waldman, A. D., Fritz, J. M. & Lenardo, M. J. A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat. Rev. Immunol. 20, 651–668 (2020).
Shekarian, T. et al. Pattern recognition receptors: immune targets to enhance cancer immunotherapy. Ann. Oncol. 28, 1756–1766 (2017).
Man, S. M. & Jenkins, B. J. Context-dependent functions of pattern recognition receptors in cancer. Nat. Rev. Cancer 22, 397–413 (2022).
Li, D. & Wu, M. Pattern recognition receptors in health and diseases. Signal Transduct. Target. Ther. 6, 291 (2021).
Kang, S., Narazaki, M., Metwally, H. & Kishimoto, T. Historical overview of the interleukin-6 family cytokine. J. Exp. Med. 217, e20190347 (2020).
Huseni, M. A. et al. CD8+ T cell-intrinsic IL-6 signaling promotes resistance to anti-PD-L1 immunotherapy. Cell Rep. Med. 4, 100878 (2023).
Yi, M. et al. Targeting cytokine and chemokine signaling pathways for cancer therapy. Signal Transduct. Target. Ther. 9, 176 (2024).
Xiao, Y., Shi, K., Qu, Y., Chu, B. & Qian, Z. Engineering nanoparticles for targeted delivery of nucleic acid therapeutics in tumor. Mol. Ther. Methods Clin. Dev. 12, 1–18 (2019).
Seaberg, J., Clegg, J. R., Bhattacharya, R. & Mukherjee, P. Self-therapeutic nanomaterials: applications in biology and medicine. Mater. Today 62, 190–224 (2023).
Ma, W. W. et al. Abstract CT255: ELU-FRα-1: a study to evaluate ELU001 in patients with solid tumors that overexpress folate receptor alpha (FRα). Cancer Res. 83, CT255 (2023).
Zanoni, D. K. et al. Use of ultrasmall core-shell fluorescent silica nanoparticles for image-guided sentinel lymph node biopsy in head and neck melanoma: a nonrandomized clinical trial. JAMA Netw. Open 4, e211936 (2021).
Gardinier, T. C. et al. High-performance chromatographic characterization of surface chemical heterogeneities of fluorescent organic–inorganic hybrid core–shell silica nanoparticles. ACS Nano 13, 1795–1804 (2019).
Ma, K. et al. Control of ultrasmall sub-10 nm ligand-functionalized fluorescent core–shell silica nanoparticle growth in water. Chem. Mater. 27, 4119–4133 (2015).
Ma, K. & Wiesner, U. Modular and orthogonal post-PEGylation surface modifications by insertion enabling penta-functional ultrasmall organic-silica hybrid nanoparticles. Chem. Mater. 29, 6840–6855 (2017).
Ma, K., Zhang, D., Cong, Y. & Wiesner, U. Elucidating the mechanism of silica nanoparticle PEGylation processes using fluorescence correlation spectroscopies. Chem. Mater. 28, 1537–1545 (2016).
Chen, F. et al. Ultrasmall renally clearable silica nanoparticles target prostate cancer. ACS Appl. Mater. Interfaces 11, 43879–43887 (2019).
Erstling, J. A. et al. Overcoming barriers associated with oral delivery of differently sized fluorescent core-shell silica nanoparticles. Adv. Mater. 36, e2305937 (2024).
Chen, F. et al. Molecular phenotyping and image-guided surgical treatment of melanoma using spectrally distinct ultrasmall core-shell silica nanoparticles. Sci. Adv. 5, eaax5208 (2019).
Chen, F. et al. Melanocortin-1 receptor-targeting ultrasmall silica nanoparticles for dual-modality human melanoma imaging. ACS Appl. Mater. Interfaces 10, 4379–4393 (2018).
Benezra, M. et al. Multimodal silica nanoparticles are effective cancer-targeted probes in a model of human melanoma. J. Clin. Invest. 121, 2768–2780 (2011).
Chen, F. et al. Target-or-clear zirconium-89 labeled silica nanoparticles for enhanced cancer-directed uptake in melanoma: a comparison of radiolabeling strategies. Chem. Mater. 29, 8269–8281 (2017).
Chen, F. et al. Ultrasmall targeted nanoparticles with engineered antibody fragments for imaging detection of HER2-overexpressing breast cancer. Nat. Commun. 9, 4141 (2018).
Wu, F. et al. Ultrasmall folate receptor alpha targeted enzymatically cleavable silica nanoparticle drug conjugates augment penetration and therapeutic efficacy in models of cancer. ACS Nano 16, 20021–20033 (2022).
Juthani, R. et al. Ultrasmall core-shell silica nanoparticles for precision drug delivery in a high-grade malignant brain tumor model. Clin. Cancer Res. 26, 147–158 (2020).
Madajewski, B. et al. Molecular engineering of ultrasmall silica nanoparticle-drug conjugates as lung cancer therapeutics. Clin. Cancer Res. 26, 5424–5437 (2020).
Urbanska, A. M. et al. A genomic profile of local immunity in the melanoma microenvironment following treatment with α particle-emitting ultrasmall silica nanoparticles. Cancer Biother. Radiopharm. 35, 459–473 (2020).
Zhang, X. et al. Targeted melanoma radiotherapy using ultrasmall 177Lu-labeled α-melanocyte stimulating hormone-functionalized core-shell silica nanoparticles. Biomaterials 241, 119858 (2020).
Zhang, L. et al. Engineered ultrasmall nanoparticle drug-immune conjugates with “hit and run” tumor delivery to eradicate gastric cancer. Adv. Ther. 6, 2200209 (2023).
Kim, S. E. et al. Ultrasmall nanoparticles induce ferroptosis in nutrient-deprived cancer cells and suppress tumour growth. Nat. Nanotechnol. 11, 977–985 (2016).
Soiffer, R. et al. Vaccination with irradiated, autologous melanoma cells engineered to secrete granulocyte-macrophage colony-stimulating factor by adenoviral-mediated gene transfer augments antitumor immunity in patients with metastatic melanoma. J. Clin. Oncol. 21, 3343–3350 (2003).
Hara, I. et al. Rejection of mouse melanoma elicited by local secretion of interleukin-2: implicating macrophages without T cells or natural killer cells in tumor rejection. Int J. Cancer 61, 253–260 (1995).
Lesokhin, A. M. et al. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 72, 876–886 (2012).
Tong, J. et al. Ferroptosis inhibitor liproxstatin-1 alleviates metabolic dysfunction-associated fatty liver disease in mice: potential involvement of PANoptosis. Acta Pharmacol. Sin. 44, 1014–1028 (2023).
Xie, Z. et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 1, e90 (2021).
Ma, T. et al. GPX4-independent ferroptosis—a new strategy in disease’s therapy. Cell Death Discov. 8, 434 (2022).
Cao, J. Y. & Dixon, S. J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. 73, 2195–2209 (2016).
Li, T. & Chen, Z. J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).
Carozza, J. A. et al. Extracellular cGAMP is a cancer-cell-produced immunotransmitter involved in radiation-induced anticancer immunity. Nat. Cancer 1, 184 (2020).
Li, J. et al. Metastasis and immune evasion from extracellular cGAMP hydrolysis. Cancer Discov. 11, 1212–1227 (2021).
Bryant, J. D., Lei, Y., VanPortfliet, J. J., Winters, A. D. & West, A. P. Assessing mitochondrial DNA release into the cytosol and subsequent activation of innate immune-related pathways in mammalian cells. Curr. Protoc. 2, e372 (2022).
Motwani, M., Pesiridis, S. & Fitzgerald, K. A. DNA sensing by the cGAS-STING pathway in health and disease. Nat. Rev. Genet. 20, 657–674 (2019).
Zhang, R., Kang, R. & Tang, D. The STING1 network regulates autophagy and cell death. Signal Transduct. Target. Ther. 6, 208 (2021).
Hopfner, K. P. & Hornung, V. Molecular mechanisms and cellular functions of cGAS-STING signalling. Nat. Rev. Mol. Cell Biol. 21, 501–521 (2020).
Liu, J. et al. The cGAS-STING pathway: post-translational modifications and functional implications in diseases. Cytokine Growth Factor Rev. 68, 69–80 (2022).
Cha, J. H., Chan, L. C., Li, C. W., Hsu, J. L. & Hung, M. C. Mechanisms controlling PD-L1 expression in cancer. Mol. Cell 76, 359–370 (2019).
Feng, C., Zhang, L., Chang, X., Qin, D. & Zhang, T. Regulation of post-translational modification of PD-L1 and advances in tumor immunotherapy. Front. Immunol. 14, 1230135 (2023).
Bailly, C. Regulation of PD-L1 expression on cancer cells with ROS-modulating drugs. Life Sci. 246, 117403 (2020).
Sierra-Filardi, E. et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J. Immunol. 192, 3858–3867 (2014).
Hsu, C. H., Altschuler, S. J. & Wu, L. F. Patterns of early p21 dynamics determine proliferation-senescence cell fate after chemotherapy. Cell 178, 361–373 e312 (2019).
Melnikova, V. O., Bolshakov, S. V., Walker, C. & Ananthaswamy, H. N. Genomic alterations in spontaneous and carcinogen-induced murine melanoma cell lines. Oncogene 23, 2347–2356 (2004).
Basit, A. et al. The cGAS/STING/TBK1/IRF3 innate immunity pathway maintains chromosomal stability through regulation of p21 levels. Exp. Mol. Med. 52, 643–657 (2020).
Galati, S., Boni, C., Gerra, M. C., Lazzaretti, M. & Buschini, A. Autophagy: a player in response to oxidative stress and DNA damage. Oxid. Med. Cell Longev. 2019, 5692958 (2019).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262-+ (2019).
Liu, D. et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 26, 1735–1749 (2019).
De Henau, O. et al. Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539, 443–447 (2016).
Overwijk, W. W. et al. gp100/pmel 17 is a murine tumor rejection antigen: induction of “self”-reactive, tumoricidal T cells using high-affinity, altered peptide ligand. J. Exp. Med. 188, 277–286 (1998).
Aragon-Sanabria, V. et al. Ultrasmall nanoparticle delivery of doxorubicin improves therapeutic index for high-grade glioma. Clin. Cancer Res. 28, 2938–2952 (2022).
Zhang, W. et al. IL-6 promotes PD-L1 expression in monocytes and macrophages by decreasing protein tyrosine phosphatase receptor type O expression in human hepatocellular carcinoma. J. Immunother. Cancer 8, e000285 (2020).
Zhang, C. X. et al. STING signaling remodels the tumor microenvironment by antagonizing myeloid-derived suppressor cell expansion. Cell Death Differ. 26, 2314–2328 (2019).
Zhu, Y. et al. STING: a master regulator in the cancer-immunity cycle. Mol. Cancer 18, 152 (2019).
Li, Y. et al. Nanoparticle ferritin-bound erastin and rapamycin: a nanodrug combining autophagy and ferroptosis for anticancer therapy. Biomater. Sci. 7, 3779–3787 (2019).
Zhang, Y. et al. Imidazole ketone erastin induces ferroptosis and slows tumor growth in a mouse lymphoma model. Cell Chem. Biol. 26, 623–633 e629 (2019).
Wang, S. et al. Arginine-rich manganese silicate nanobubbles as a ferroptosis-inducing agent for tumor-targeted theranostics. ACS Nano 12, 12380–12392 (2018).
Liu, Y., Hardie, J., Zhang, X. & Rotello, V. M. Effects of engineered nanoparticles on the innate immune system. Semin. Immunol. 34, 25–32 (2017).
Shao, K. et al. Nanoparticle-based immunotherapy for cancer. ACS Nano 9, 16–30 (2015).
Zhang, Y. N., Poon, W., Tavares, A. J., McGilvray, I. D. & Chan, W. C. W. Nanoparticle-liver interactions: cellular uptake and hepatobiliary elimination. J. Control. Release 240, 332–348 (2016).
Han, R., Xiao, Y., Bai, Q. & Choi, C. H. J. Self-therapeutic metal-based nanoparticles for treating inflammatory diseases. Acta Pharm. Sin. B 13, 1847–1865 (2023).
Mijanovic, O. et al. Cathepsin D—managing the delicate balance. Pharmaceutics 13, 837 (2021).
Wang, X. et al. TPC proteins are phosphoinositide-activated sodium-selective ion channels in endosomes and lysosomes. Cell 151, 372–383 (2012).
Li, P., Gu, M. & Xu, H. Lysosomal ion channels as decoders of cellular signals. Trends Biochem. Sci. 44, 110–124 (2019).
Reinheckel, T. & Tholen, M. Low-level lysosomal membrane permeabilization for limited release and sublethal functions of cathepsin proteases in the cytosol and nucleus. FEBS Open Bio 12, 694–707 (2022).
Ma, M., Jiang, W. & Zhou, R. DAMPs and DAMP-sensing receptors in inflammation and diseases. Immunity 57, 752–771 (2024).
Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Biol. 21, 101–118 (2020).
Tanaka, T., Narazaki, M. & Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 6, a016295 (2014).
Yan, H., Lu, W. & Wang, F. The cGAS-STING pathway: a therapeutic target in chromosomally unstable cancers. Signal Transduct. Target. Ther. 8, 45 (2023).
Tsukamoto, H. et al. Combined blockade of IL6 and PD-1/PD-L1 signaling abrogates mutual regulation of their immunosuppressive effects in the tumor microenvironment. Cancer Res. 78, 5011–5022 (2018).
Hailemichael, Y. et al. Interleukin-6 blockade abrogates immunotherapy toxicity and promotes tumor immunity. Cancer Cell 40, 509–523.e506 (2022).
Soler, M. F., Abaurrea, A., Azcoaga, P., Araujo, A. M. & Caffarel, M. M. New perspectives in cancer immunotherapy: targeting IL-6 cytokine family. J. Immunother. Cancer 11, e007530 (2023).
Delyon, J. & Lebbe, C. IL-6 blockade in cancer patients treated with immune checkpoint blockade: a win-win strategy. Cancer Cell 40, 450–451 (2022).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017).
Jneid, B. et al. Selective STING stimulation in dendritic cells primes antitumor T cell responses. Sci. Immunol. 8, eabn6612 (2023).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 3 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Blighe, K., Rana, S. & Lewis, M. EnhancedVolcano: publication-ready volcano plots with enhanced colouring and labeling. R package. GitHub https://github.com/kevinblighe/EnhancedVolcano (2023).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics 16, 284–287 (2012).
Cohen, R. et al. Optimising multiplex immunofluorescence staining for characterising the tumour immune micro-environment. Methods 219, 48–57 (2023).
McGue, J. J., Edwards, J. J., Griffith, B. D. & Frankel, T. L. Multiplex fluorescent immunohistochemistry for preservation of tumor microenvironment architecture and spatial relationship of cells in tumor tissues. Methods Mol. Biol. 2660, 235–246 (2023).
Acknowledgements
This study was funded by grants from the NIH (R01CA253658 and U54CA199081 to M.S.B. and U.W.) and Cancer Center Support Grant (P30 CA008748CCSG to C. Thompson), Cycle for Survival/Parker Institute for Cancer Immunotherapy grants. We thank T. Quinn, Director of the Missouri Peptide Synthesis Core, and his staff for the peptides used in this work.
Author information
Authors and Affiliations
Contributions
Study conception and design: G.D.L. and M.S.B. Performed experiment or data collection: G.D.L., L.Z., N.A.S., N.N., R.P., R.L., M.O., S.M. and F.S. Computation and statistical analysis: G.D.L., M.O. and T.Z. Data interpretation and biological analysis: G.D.L., L.Z., N.A.S., F.C., T.Z., R.L., S.M., F.S., M.O., T.M., T.P.Q., U.W., J.D.W. and M.S.B. In vitro and in vivo experiments: G.D.L., L.Z., N.A.S., N.N., R.P., F.S., S.M. and M.O. Writing (original drafts): G.D.L., N.A.S., F.C., T.P.Q., M.O., J.D.W., U.W. and M.S.B. Writing (review and editing): G.D.L., N.A.S., F.C., T.P.Q., M.P., M.O., U.W., J.D.W. and M.S.B. Supervision: M.S.B. and U.W. Administration: M.S.B. and U.W.
Corresponding authors
Ethics declarations
Competing interests
M.S.B., M.O. and U.W. are inventors on patent applications related to ferroptosis: Methods of Treatment Using Ultrasmall Nanoparticles to Induce Cell Death of Cancer Cells via Ferroptosis; Methods of Cancer Treatment via Regulated Ferroptosis. M.S.B., U.W., M.O. and G.D.L. are inventors on patent applications related to work on the tumour microenvironment, immunomodulatory nanoparticle conjugates and immunotherapies: Inducing Favorable Effects on Tumor Microenvironment via Administration of Nanoparticle Compositions; Nanoparticle-Mediated Enhancement of Immunotherapy to Promote Ferroptosis-induced Cytotoxicity and Anti-tumor Immune Responses. M.S.B., U.W. and F.C. are listed as inventors on the following patent: Ultrasmall Nanoparticles Labeled with Zirconium-89 and Methods Thereof. J.D.W. is a consultant for Apricity; Ascentage Pharma; AstraZeneca; BeiGene; Bicara Therapeutics; Bristol Myers Squibb; Daiichi Sankyo; Dragonfly; Imvaq; Larkspur; Takeda; Tizona; Trishula Therapeutics; and Immunocore (Data Safety Board). J.D.W. received Grant/Research Support from Bristol Myers Squibb and Enterome. J.D.W. has Equity in Apricity, Arsenal IO/CellCarta; Ascentage; Imvaq; Linneaus, Larkspur; Georgiamune; Maverick; Tizona Therapeutics; and Xenimmune. J.D.W. is an inventor on the following patents: Xenogeneic DNA Vaccines; Newcastle Disease Viruses for Cancer Therapy; Myeloid-derived Suppressor Cell (MDSC) Assay; Prediction of Responsiveness to Treatment with Immunomodulatory Therapeutics and Method of Monitoring Abscopal Effects During Such Treatment; anti-PD1 antibody; anti-CTLA4 antibodies; anti-GITR antibodies and methods of use thereof. T.M. is a consultant for Immunos Therapeutics, Daiichi Sankyo Co., TigaTX, Normunity and Pfizer. T.M. is a co-founder of and equity holder in IMVAQ Therapeutics. T.M. receives research grants funding from Bristol Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals, Peregrine Pharmaceuticals, Adaptive Biotechnologies, Leap Therapeutics and Aprea Therapeutics. T.M. is an inventor on patent applications related to work on oncolytic viral therapy, alpha virus-based vaccine, neoantigen modelling, CD40, GITR, OX40, PD-1 and CTLA-4. The other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks John Wilson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Particle-driven modulation of cell populations and immunogenicity within the TIME of B16-GM tumor mice.
B16-GM tumor-bearing mice were i.v.-injected with a multi-dose regimen of C’ dots (12 nmol/dose, Q3Dx3) or vehicle. Tumors were harvested 4 days post final treatment and subjected to flow cytometric analysis for quantifying (a) inhibitory CD8 + T, (b) inhibitory CD4 + T, and (c) anti-tumor CD8 + T cell populations, as well as (d) macrophages, (e) MDSCs, (f) dendritic cells, and (g) monocytes, according to markers in Tables S3 and S4. (h,i) Volcano plot of bulk RNA-seq data (h) and NES plot reflecting the KEGG pathway enrichment analysis (i) 10 days after treating tumors with the final particle dose. The latter plot displays normalized enrichment scores (NES) for gene sets (represented as bars), their positions corresponding to NES values normalized to those of vehicle-treated cohorts. Data represents the mean ± s.e.m. of particle-treated and control mice for both IP (n = 3 mice/group) and bulk RNA-seq (n = 3 mice/group) reflecting two independent experiments for each study. Unpaired Student’s t-tests were performed. All statistical tests were two-sided. *p < 0.05, **p < 0.01, ***p < 0.005, ****p < 0.001. Schematics in a–g created with BioRender.com.
Extended Data Fig. 2 Multi-therapeutic properties of C’ dot adjuvants lead to diverse responses in the TME of melanoma models.
A diverse array of proinflammatory and cytotoxic responses (upper left) promoted by C’ dots are illustrated, in addition to their induction of cell cycle arrest and senescence (SASP) (upper right). Inhibition of resistance mechanisms in the TME driving upregulation of suppressive cell populations and proteins are shown (lower left) along with factors improving anti-tumor immunity (lower right). Schematic created with BioRender.com.
Supplementary information
Supplementary Information
Supplementary Tables 1–5 and Figs. 1–17.
Source data
Source Data Fig. 4
Uncropped western blots of β-actin and Sting.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
De Leon, G., Zhang, L., Siddiqui, N.A. et al. An ultrasmall core–shell silica nanoparticle improves antitumour immunity and survival by remodelling suppressive melanoma microenvironments. Nat. Nanotechnol. (2025). https://doi.org/10.1038/s41565-025-02083-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41565-025-02083-z