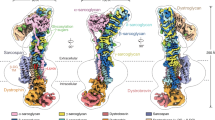
Abstract
Duchenne muscular dystrophy (DMD) is a severe X-linked recessive disorder marked by progressive muscle wasting leading to premature mortality1,2. Discovery of the DMD gene encoding dystrophin both revealed the cause of DMD and helped identify a family of at least ten dystrophin-associated proteins at the muscle cell membrane, collectively forming the dystrophin–glycoprotein complex (DGC)3,4,5,6,7,8,9. The DGC links the extracellular matrix to the cytoskeleton, but, despite its importance, its molecular architecture has remained elusive. Here we determined the native cryo-electron microscopy structure of rabbit DGC and conducted biochemical analyses to reveal its intricate molecular configuration. An unexpected β-helix comprising β-, γ- and δ-sarcoglycan forms an extracellular platform that interacts with α-dystroglycan, β-dystroglycan and α-sarcoglycan, allowing α-dystroglycan to contact the extracellular matrix. In the membrane, sarcospan anchors β-dystroglycan to the β-, γ- and δ-sarcoglycan trimer, while in the cytoplasm, β-dystroglycan’s juxtamembrane fragment binds dystrophin’s ZZ domain. Through these interactions, the DGC links laminin 2 to intracellular actin. Additionally, dystrophin’s WW domain, along with its EF-hand 1 domain, interacts with α-dystrobrevin. A disease-causing mutation mapping to the WW domain weakens this interaction, as confirmed by deletion of the WW domain in biochemical assays. Our findings rationalize more than 110 mutations affecting single residues associated with various muscular dystrophy subtypes and contribute to ongoing therapeutic developments, including protein restoration, upregulation of compensatory genes and gene replacement.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The cryo-EM maps and atomic coordinates have been deposited to the EMDB (EMD-45165) and PDB (9C3C) databases, respectively. Other structures used in this study were retrieved from the PDB with accession codes 6DLH for endo-fucoidan hydrolase MfFcnA4 and 1EG4 for dystrophin’s WW and EF-hand domains with the PPXY peptide of β-DG. The gene IDs of the DGC components were obtained from NCBI: 100009208 for β-SG, 100009214 for γ-SG, 100351233 for δ-SG, 100009178 for α-SG, 100009278 for DG, 100355731 for dystrophin and 100351412 for α-dystrobrevin. All other data are available within the main text or the Extended Data. Raw data and source images are available in the Supplementary Information. Source data are provided with this paper.
References
Hoffman, E. P., Brown, R. H. Jr. & Kunkel, L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51, 919–928 (1987).
Mercuri, E., Bonnemann, C. G. & Muntoni, F. Muscular dystrophies. Lancet 394, 2025–2038 (2019).
Koenig, M. et al. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell 50, 509–517 (1987).
Campbell, K. P. & Kahl, S. D. Association of dystrophin and an integral membrane glycoprotein. Nature 338, 259–262 (1989).
Yoshida, M. & Ozawa, E. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 108, 748–752 (1990).
Ervasti, J. M. & Campbell, K. P. Membrane organization of the dystrophin–glycoprotein complex. Cell 66, 1121–1131 (1991).
Ibraghimov-Beskrovnaya, O. et al. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature 355, 696–702 (1992).
Gao, Q. Q. & McNally, E. M. The dystrophin complex: structure, function, and implications for therapy. Compr. Physiol. 5, 1223–1239 (2015).
Guiraud, S. et al. The pathogenesis and therapy of muscular dystrophies. Annu. Rev. Genomics Hum. Genet. 16, 281–308 (2015).
Le, S. et al. Dystrophin as a molecular shock absorber. ACS Nano 12, 12140–12148 (2018).
Pilgram, G. S., Potikanond, S., Baines, R. A., Fradkin, L. G. & Noordermeer, J. N. The roles of the dystrophin-associated glycoprotein complex at the synapse. Mol. Neurobiol. 41, 1–21 (2010).
Mirouse, V. Evolution and developmental functions of the dystrophin-associated protein complex: beyond the idea of a muscle-specific cell adhesion complex. Front. Cell. Dev. Biol. 11, 1182524 (2023).
Jayasinha, V. et al. Inhibition of dystroglycan cleavage causes muscular dystrophy in transgenic mice. Neuromuscul. Disord. 13, 365–375 (2003).
Endo, T. Glycobiology of α-dystroglycan and muscular dystrophy. J. Biochem. 157, 1–12 (2015).
Esapa, C. T., Bentham, G. R., Schroder, J. E., Kroger, S. & Blake, D. J. The effects of post-translational processing on dystroglycan synthesis and trafficking. FEBS Lett. 555, 209–216 (2003).
Johnson, K. et al. Detection of variants in dystroglycanopathy-associated genes through the application of targeted whole-exome sequencing analysis to a large cohort of patients with unexplained limb-girdle muscle weakness. Skelet. Muscle 8, 23 (2018).
Waite, A., Brown, S. C. & Blake, D. J. The dystrophin–glycoprotein complex in brain development and disease. Trends Neurosci. 35, 487–496 (2012).
Williamson, R. A. et al. Dystroglycan is essential for early embryonic development: disruption of Reichert’s membrane in Dag1-null mice. Hum. Mol. Genet. 6, 831–841 (1997).
Noguchi, S. et al. Mutations in the dystrophin-associated protein γ-sarcoglycan in chromosome 13 muscular dystrophy. Science 270, 819–822 (1995).
Roberds, S. L. et al. Missense mutations in the adhalin gene linked to autosomal recessive muscular dystrophy. Cell 78, 625–633 (1994).
Duclos, F. et al. Progressive muscular dystrophy in α-sarcoglycan-deficient mice. J. Cell Biol. 142, 1461–1471 (1998).
Lim, L. E. et al. β-Sarcoglycan: characterization and role in limb-girdle muscular dystrophy linked to 4q12. Nat. Genet. 11, 257–265 (1995).
Bonnemann, C. G. et al. β-Sarcoglycan (A3b) mutations cause autosomal recessive muscular dystrophy with loss of the sarcoglycan complex. Nat. Genet. 11, 266–273 (1995).
Nigro, V. et al. Autosomal recessive limb-girdle muscular dystrophy, LGMD2F, is caused by a mutation in the δ-sarcoglycan gene. Nat. Genet. 14, 195–198 (1996).
Noguchi, S., Wakabayashi, E., Imamura, M., Yoshida, M. & Ozawa, E. Formation of sarcoglycan complex with differentiation in cultured myocytes. Eur. J. Biochem. 267, 640–648 (2000).
Shi, W. et al. Specific assembly pathway of sarcoglycans is dependent on β- and δ-sarcoglycan. Muscle Nerve 29, 409–419 (2004).
Hemler, M. E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 19, 397–422 (2003).
Crosbie, R. H., Heighway, J., Venzke, D. P., Lee, J. C. & Campbell, K. P. Sarcospan, the 25-kDa transmembrane component of the dystrophin–glycoprotein complex. J. Biol. Chem. 272, 31221–31224 (1997).
Lebakken, C. S. et al. Sarcospan-deficient mice maintain normal muscle function. Mol. Cell. Biol. 20, 1669–1677 (2000).
Marshall, J. L. et al. Dystrophin and utrophin expression require sarcospan: loss of α7 integrin exacerbates a newly discovered muscle phenotype in sarcospan-null mice. Hum. Mol. Genet. 21, 4378–4393 (2012).
Marshall, J. L. et al. Sarcospan-dependent Akt activation is required for utrophin expression and muscle regeneration. J. Cell Biol. 197, 1009–1027 (2012).
Peter, A. K., Miller, G. & Crosbie, R. H. Disrupted mechanical stability of the dystrophin–glycoprotein complex causes severe muscular dystrophy in sarcospan transgenic mice. J. Cell Sci. 120, 996–1008 (2007).
Suzuki, A. et al. Molecular organization at the glycoprotein-complex-binding site of dystrophin. Three dystrophin-associated proteins bind directly to the carboxy-terminal portion of dystrophin. Eur. J. Biochem. 220, 283–292 (1994).
Jung, D., Yang, B., Meyer, J., Chamberlain, J. S. & Campbell, K. P. Identification and characterization of the dystrophin anchoring site on β-dystroglycan. J. Biol. Chem. 270, 27305–27310 (1995).
Ponting, C. P., Blake, D. J., Davies, K. E., Kendrick-Jones, J. & Winder, S. J. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci 21, 11–13 (1996).
Rentschler, S. et al. The WW domain of dystrophin requires EF-hands region to interact with β-dystroglycan. Biol. Chem. 380, 431–442 (1999).
Ishikawa-Sakurai, M., Yoshida, M., Imamura, M., Davies, K. E. & Ozawa, E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to β-dystroglycan. Hum. Mol. Genet. 13, 693–702 (2004).
Swiderski, K. et al. Phosphorylation within the cysteine-rich region of dystrophin enhances its association with β-dystroglycan and identifies a potential novel therapeutic target for skeletal muscle wasting. Hum. Mol. Genet. 23, 6697–6711 (2014).
Sadoulet-Puccio, H. M., Rajala, M. & Kunkel, L. M. Dystrobrevin and dystrophin: an interaction through coiled-coil motifs. Proc. Natl Acad. Sci. USA 94, 12413–12418 (1997).
Grady, R. M. et al. Role for α-dystrobrevin in the pathogenesis of dystrophin-dependent muscular dystrophies. Nat. Cell Biol. 1, 215–220 (1999).
Grady, R. M. et al. Maturation and maintenance of the neuromuscular synapse: genetic evidence for roles of the dystrophin–glycoprotein complex. Neuron 25, 279–293 (2000).
Grady, R. M. et al. Tyrosine-phosphorylated and nonphosphorylated isoforms of α-dystrobrevin: roles in skeletal muscle and its neuromuscular and myotendinous junctions. J. Cell Biol. 160, 741–752 (2003).
Ahn, A. H. & Kunkel, L. M. Syntrophin binds to an alternatively spliced exon of dystrophin. J. Cell Biol. 128, 363–371 (1995).
Huang, X. et al. Structure of a WW domain containing fragment of dystrophin in complex with β-dystroglycan. Nat. Struct. Mol. Biol. 7, 634–638 (2000).
Bozic, D., Sciandra, F., Lamba, D. & Brancaccio, A. The structure of the N-terminal region of murine skeletal muscle α-dystroglycan discloses a modular architecture. J. Biol. Chem. 279, 44812–44816 (2004).
Norwood, F. L., Sutherland-Smith, A. J., Keep, N. H. & Kendrick-Jones, J. The structure of the N-terminal actin-binding domain of human dystrophin and how mutations in this domain may cause Duchenne or Becker muscular dystrophy. Structure 8, 481–491 (2000).
Borgert, A., Foley, B. L. & Live, D. Contrasting the conformational effects of α-O-GalNAc and α-O-Man glycan protein modifications and their impact on the mucin-like region of α-dystroglycan. Glycobiology 31, 649–661 (2021).
Nagata, Y. & Burger, M. M. Wheat germ agglutinin. Molecular characteristics and specificity for sugar binding. J. Biol. Chem. 249, 3116–3122 (1974).
Dwyer, T. M. & Froehner, S. C. Direct binding of Torpedo syntrophin to dystrophin and the 87 kDa dystrophin homologue. FEBS Lett. 375, 91–94 (1995).
Steinbacher, S. et al. Crystal structure of P22 tailspike protein: interdigitated subunits in a thermostable trimer. Science 265, 383–386 (1994).
Stummeyer, K., Dickmanns, A., Muhlenhoff, M., Gerardy-Schahn, R. & Ficner, R. Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 (2005).
Chan, Y. M., Bonnemann, C. G., Lidov, H. G. & Kunkel, L. M. Molecular organization of sarcoglycan complex in mouse myotubes in culture. J. Cell Biol. 143, 2033–2044 (1998).
Yis, U. et al. Childhood onset limb-girdle muscular dystrophies in the Aegean part of Turkey. Acta Myol. 37, 210–220 (2018).
Mitraki, A., Papanikolopoulou, K. & Van Raaij, M. J. Natural triple β-stranded fibrous folds. Adv. Protein Chem. 73, 97–124 (2006).
Yoshida, M. et al. Dissociation of the complex of dystrophin and its associated proteins into several unique groups by n-octyl β-d-glucoside. Eur. J. Biochem. 222, 1055–1061 (1994).
Yoshida, M. et al. The fourth component of the sarcoglycan complex. FEBS Lett. 403, 143–148 (1997).
Macao, B., Johansson, D. G., Hansson, G. C. & Hard, T. Autoproteolysis coupled to protein folding in the SEA domain of the membrane-bound MUC1 mucin. Nat. Struct. Mol. Biol. 13, 71–76 (2006).
Holm, L., Laiho, A., Toronen, P. & Salgado, M. DALI shines a light on remote homologs: one hundred discoveries. Protein Sci. 32, e4519 (2023).
Vickers, C. et al. Endo-fucoidan hydrolases from glycoside hydrolase family 107 (GH107) display structural and mechanistic similarities to α-l-fucosidases from GH29. J. Biol. Chem. 293, 18296–18308 (2018).
Crosbie, R. H. et al. Molecular and genetic characterization of sarcospan: insights into sarcoglycan–sarcospan interactions. Hum. Mol. Genet. 9, 2019–2027 (2000).
Fraiberg, M., Borovok, I., Bayer, E. A., Weiner, R. M. & Lamed, R. Cadherin domains in the polysaccharide-degrading marine bacterium Saccharophagus degradans 2-40 are carbohydrate-binding modules. J. Bacteriol. 193, 283–285 (2011).
Cao, L. et al. CHDL: a cadherin-like domain in Proteobacteria and Cyanobacteria. FEMS Microbiol. Lett. 251, 203–209 (2005).
Bork, P. & Patthy, L. The SEA module: a new extracellular domain associated with O-glycosylation. Protein Sci. 4, 1421–1425 (1995).
Hemler, M. E. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6, 801–811 (2005).
Charrin, S., Jouannet, S., Boucheix, C. & Rubinstein, E. Tetraspanins at a glance. J. Cell Sci. 127, 3641–3648 (2014).
Susa, K. J., Kruse, A. C. & Blacklow, S. C. Tetraspanins: structure, dynamics, and principles of partner-protein recognition. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2023.09.003 (2023).
Miller, G., Wang, E. L., Nassar, K. L., Peter, A. K. & Crosbie, R. H. Structural and functional analysis of the sarcoglycan–sarcospan subcomplex. Exp. Cell. Res. 313, 639–651 (2007).
Crosbie, R. H. et al. Membrane targeting and stabilization of sarcospan is mediated by the sarcoglycan subcomplex. J. Cell Biol. 145, 153–165 (1999).
Peter, A. K., Marshall, J. L. & Crosbie, R. H. Sarcospan reduces dystrophic pathology: stabilization of the utrophin–glycoprotein complex. J. Cell Biol. 183, 419–427 (2008).
Miller, G., Peter, A. K., Espinoza, E., Heighway, J. & Crosbie, R. H. Over-expression of Microspan, a novel component of the sarcoplasmic reticulum, causes severe muscle pathology with triad abnormalities. J. Muscle Res. Cell Motil. 27, 545–558 (2006).
Peter, A. K. et al. Nanospan, an alternatively spliced isoform of sarcospan, localizes to the sarcoplasmic reticulum in skeletal muscle and is absent in limb girdle muscular dystrophy 2F. Skelet. Muscle 7, 11 (2017).
Yoshida, M. et al. Biochemical evidence for association of dystrobrevin with the sarcoglycan–sarcospan complex as a basis for understanding sarcoglycanopathy. Hum. Mol. Genet. 9, 1033–1040 (2000).
Rafael, J. A. et al. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J. Cell Biol. 134, 93–102 (1996).
Crawford, G. E. et al. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 150, 1399–1410 (2000).
Yoder, M. D., Keen, N. T. & Jurnak, F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science 260, 1503–1507 (1993).
Emsley, P., Charles, I. G., Fairweather, N. F. & Isaacs, N. W. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature 381, 90–92 (1996).
Petersen, T. N., Kauppinen, S. & Larsen, S. The crystal structure of rhamnogalacturonase A from Aspergillus aculeatus: a right-handed parallel β helix. Structure 5, 533–544 (1997).
Gibbs, E. M. et al. High levels of sarcospan are well tolerated and act as a sarcolemmal stabilizer to address skeletal muscle and pulmonary dysfunction in DMD. Hum. Mol. Genet. 25, 5395–5406 (2016).
Parvatiyar, M. S. et al. Sarcospan regulates cardiac isoproterenol response and prevents Duchenne muscular dystrophy-associated cardiomyopathy. J. Am. Heart Assoc. 4, e002481 (2015).
Holt, K. H. et al. Functional rescue of the sarcoglycan complex in the BIO 14.6 hamster using δ-sarcoglycan gene transfer. Mol. Cell 1, 841–848 (1998).
Roberts, T. C., Wood, M. J. A. & Davies, K. E. Therapeutic approaches for Duchenne muscular dystrophy. Nat. Rev. Drug Discov. 22, 917–934 (2023).
Yue, Y., Liu, M. & Duan, D. C-terminal-truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double-knockout mice. Mol. Ther. 14, 79–87 (2006).
Mitchell, R. D., Palade, P. & Fleischer, S. Purification of morphologically intact triad structures from skeletal muscle. J. Cell Biol. 96, 1008–1016 (1983).
Ervasti, J. M., Kahl, S. D. & Campbell, K. P. Purification of dystrophin from skeletal muscle. J. Biol. Chem. 266, 9161–9165 (1991).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Kimanius, D., Dong, L., Sharov, G., Nakane, T. & Scheres, S. H. W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 478, 4169–4185 (2021).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Bepler, T. et al. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods 16, 1153–1160 (2019).
Kimanius, D. et al. Data-driven regularization lowers the size barrier of cryo-EM structure determination. Nat Methods 21, 1216-1221, doi:10.1038/s41592-024-02304-8 (2024).
Punjani, A., Zhang, H. & Fleet, D. J. Non-uniform refinement: adaptive regularization improves single-particle cryo-EM reconstruction. Nat. Methods 17, 1214–1221 (2020).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Mirdita, M. et al. ColabFold: making protein folding accessible to all. Nat. Methods 19, 679–682 (2022).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature https://doi.org/10.1038/s41586-024-07487-w (2024).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010).
Buchan, D. W. A. & Jones, D. T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 47, W402–W407 (2019).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 42, W320–W324 (2014).
Edgar, R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Acknowledgements
We thank R. H. Crosbie for comments on the scientific content and T. Nguyen, J. Jih and L. Wang for comments on the writing and presentation of the paper. This project is supported by a grant from the US National Institutes of Health (R01GM071940 to Z.H.Z.). We acknowledge the use of resources at the Electron Imaging Center for NanoSystems, supported by UCLA and grants from the National Institutes of Health (S10RR23057 and S10OD018111) and National Science Foundation (DBI-1338135 and DMR-1548924).
Author information
Authors and Affiliations
Contributions
Z.H.Z. conceived the project and supervised the research. S.L. designed the experimental protocols. S.L. and X.X. prepared samples and collected cryo-EM images. S.L., T.S. and X.X. determined the 3D structures. S.L. and T.S. built atomic models and generated the figures. T.S. and S.L. engineered the recombinant proteins and performed biochemical analyses. S.L. and T.S. interpreted the results, prepared the illustrations and wrote the original draft of the manuscript. S.L., T.S., X.X. and Z.H.Z. contributed to the editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature thanks Jeffrey Chamberlain and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Representative cryo-EM images and 2D analyses.
a,b,Negative staining (a) and drift-corrected cryo-EM (b) micrographs of DGC isolated from rabbit skeletal muscle. Representative side view and top view particles are shown in red and yellow circles, respectively. c, Representative 2D class averages of DGC isolate. Top views and side views of the DGC are labelled with red and yellow boxes, respectively.
Extended Data Fig. 2 Cryo-EM structural determination of DGC.
a, Data processing workflow. Binned 2 (pixel size of 2.72 Å) and binned 1 (pixel size of 1.36 Å, grey shaded) data processing is separated via dashed line. The masks used in focused 3D classification are outlined with coloured dashed lines. Data processing in RELION and cryoSPARC is denoted by a circular inscribed “R” and “C”, respectively. See Methods for more details. b, FSC as a function of spatial frequency demonstrating the resolution of the final reconstruction of DGC. c,d, View direction distribution histogram (c) and posterior precision plot (d) show view diversity of all particles used for the final map of DGC (from cryoSPARC). e, Local resolution estimation (from cryoSPARC) using a local FSC threshold of 0.5. f, FSC coefficients as a functional of spatial frequency between model and cryo-EM density maps. The generally similar appearances between the FSC curves obtained with half maps with (red) and without (blue) model refinement indicate that the refinement of the atomic coordinates did not suffer from severe over-fitting.
Extended Data Fig. 3 Representative cryo-EM density maps of DGC.
C-terminal cap of β-, δ-, γ-SG in panel a and dystrophin (EF-hands+WW)–dystrobrevin (EF-hands) heterodimer in panel f shown using the unsharpened 4.3 Å map. CDHL1-SEA1 of α-SG (d) displayed using a gaussian low-pass filtered map. All others are shown using sharpened densities of the 4.1-Å local-refined map or the 4.3-Å map.
Extended Data Fig. 4 Structure prediction of the triplex structure of β-, γ- or δ-SG using artificial intelligence (AI) programs.
Panels from left to right: cryo-EM model of the β-SG–γ-SG–δ-SG trimer (panel 1) and the predicted models of single-component trimers (panels 2-4). All predicted models were generated using AlphaFold. Regions with pLDDT > 90 are expected to be modelled to high accuracy; regions with pLDDT between 70 and 90 are expected to be modelled well; regions with pLDDT between 50 and 70 are modelled with low confidence and should be treated with caution.
Extended Data Fig. 5 Transfected β-, γ- and δ-SG separated by SDS-PAGE and visualized by immunoblot with Myc tag-specific antibody.
For gel source data, see Supplementary Fig. 1.
Extended Data Fig. 6 Identifying the potential calcium (Ca2+) binding site in DGC’s CDHL domains.
a, Superposition of the CDHL domains with focused views of the Ca2+ coordination site in endo-fucoidan hydrolase MfFcnA4 (PDB ID: 6DLH, purple), and potential Ca2+ binding sites in dystroglycans (salmon and grey) and α-sarcoglycan (cyan). Residues possibly involved in Ca2+ coordination are annotated. b, Cross-species multiple sequence alignment of the three CDHL domains in DGC. Red background depicts identical residues in all CDHL domains. Conserved residues are coloured in red and divergent residues are in black. All potential coordination residues shown in a are labelled with red circles. MfFcnA4 residues structurally homologous to CDHL2 positions are numbered; green-filled dash circles (half: not conserved; full: identical) indicate potential Ca2+ binding sites in CDHL2.
Extended Data Fig. 7 Divergent domain orientations between CDHL and SEA in the three CDHL-SEA modules of DGC.
a, Structural superposition of the three CDHL-SEA modules within DGC. SEA-CDHL modules are aligned against the SEA domain. b, Unique orientations are essential for the TM-proximal loop of β-DG (left), α-DG (top right) and α-SG (bottom right) to interact with the β-helix. The circle slash denotes interaction disruption by detachment, while the circle cross indicates interaction disruption via atomic clashes.
Extended Data Fig. 8 Comparison of β-DG binding sarcospan with both partner-binding and apo states of the canonical tetraspanin CD81.
Sarcospan and tetraspanin CD81 are depicted as ribbons, while β-DG and CD81’s binding partner are shown as surfaces. CD81’s unmodelled regions are represented by dashed lines.
Extended Data Fig. 9 Subcellular localization of dystrophin cysteine-rich region and its WW deletion construct.
The fragments used are residues 3042-3426 and 3077-3426 for dystrophin and dystrophin (ΔWW), respectively.
Supplementary information
Supplementary Fig. 1
Uncropped gels used in the preparation of Figs. 1b, 2g, 3h,i and 5e and Extended Data Fig. 5. Page 1 presents the uncropped SDS–PAGE gel for Fig. 1b; pages 2–6 contain uncropped western blot images from the co-immunoprecipitation assays used for Figs. 2g, 3h,i and 5e and Extended Data Fig. 5. Black dashed boxes indicate the cropped areas displayed in the respective figures.
Supplementary Table 1
Pathogenic mutations in DGs, dystrophin and α-dystrobrevin. Mutation residues listed in the third column are numbered according to the sequences of the human proteins. The mutation impacts in bold in the fifth column are inferred from our structural analysis, while the impacts in regular text in the same column are derived from previous reports.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, S., Su, T., Xia, X. et al. Native DGC structure rationalizes muscular dystrophy-causing mutations. Nature 637, 1261–1271 (2025). https://doi.org/10.1038/s41586-024-08324-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-024-08324-w