Abstract
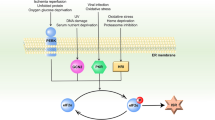
An increased level of phosphorylation of eukaryotic translation initiation factor 2 subunit-α (eIF2α, encoded by EIF2S1; eIF2α-p) coupled with decreased guanine nucleotide exchange activity of eIF2B is a hallmark of the ‘canonical’ integrated stress response (c-ISR)1. It is unclear whether impaired eIF2B activity in human diseases including leukodystrophies2, which occurs in the absence of eIF2α-p induction, is synonymous with the c-ISR. Here we describe a mechanism triggered by decreased eIF2B activity, distinct from the c-ISR, which we term the split ISR (s-ISR). The s-ISR is characterized by translational and transcriptional programs that are different from those observed in the c-ISR. Opposite to the c-ISR, the s-ISR requires eIF4E-dependent translation of the upstream open reading frame 1 and subsequent stabilization of ATF4 mRNA. This is followed by altered expression of a subset of metabolic genes (for example, PCK2), resulting in metabolic rewiring required to maintain cellular bioenergetics when eIF2B activity is attenuated. Overall, these data demonstrate a plasticity of the mammalian ISR, whereby the loss of eIF2B activity in the absence of eIF2α-p induction activates the eIF4E–ATF4–PCK2 axis to maintain energy homeostasis.
Similar content being viewed by others
Main
Protein synthesis is a highly controlled process essential for cellular homeostasis, and its suppression under stress contributes to maintaining cellular energy balance3. The integrated stress response (ISR) is an integral arm of the unfolded protein response that reprograms translation under a broad range of stresses1. It is thought that the ISR encompasses a linear chain of events triggered by stress-induced eIF2α-p, followed by suppression of eIF2B activity. eIF2α-p-dependent reduction in eIF2B activity limits initiatior methionine transfer RNA (Met-tRNAi) delivery and decreases global protein synthesis1,4. Ergo, the current mode of regulation of protein synthesis under the ISR is: eIF2α-p, leading to reduction in eIF2B activity, leading to suppression of protein synthesis and translational reprogramming1. This model supports similar translational control mechanisms for the ISR in yeast and mammalian cells, except that mammals have four eIF2α kinases, whereas yeast has only one5.
Temporal translational regulation in the c-ISR is dynamically controlled through the eIF2α-p–eIF2B axis6,7 (details in Supplementary Notes). However, several stress response mechanisms that involve alternative translation initiation and seem not to be directly linked to eIF2α-p have been documented8. Moreover, eIF2α-p and eIF2B activity are not linearly correlated with translational perturbations during the c-ISR6. eIF2B activity is similarly decreased during both acute and chronic endoplasmic reticulum (ER) stress6. By contrast, eIF2α-p is induced during acute ER stress but decreased during the chronic phase of ER stress, which is accompanied by reduction and partial recovery of global protein synthesis, respectively6. This raised a question of whether stress-induced eIF2α-p is synonymous with decreased eIF2B activity in the context of translational reprogramming during the ISR. Addressing this question is important as eIF2B is inactivated through mutations in the absence of stress and induction of eIF2α-p in human leukodystrophies (for example, vanishing white matter disease2 (VWMD)). VWMD is characterized by a low level of eIF2B activity that is paralleled by compensatory changes including a decrease in eIF2α-p that renders affected oligodendrocytes and astrocytes vulnerable to stress2.
In addition to the reduced level of global protein synthesis, the ISR is characterized by translational activation of a subset of mRNAs6 including activating transcription factor 4 (ATF4)1, which regulates transcription during stress. The ATF4 5′ untranslated region harbours two upstream open reading frames (uORF1 and uORF2); uORF2 overlaps out-of-frame with the main ATF4 ORF such that translation beginning at uORF2 represses ATF4 protein synthesis5. ATF4 mRNA translation and protein levels are elevated in acute and chronic phases of ER stress6. The mechanism of the ATF4 ORF translation during ER stress involves translation of uORF1 followed by delayed reinitiation that promotes skipping of uORF2 and translation of the main ATF4 ORF5. Similarly to the case for yeast GCN4 mRNA5, delayed reinitiation is thought to be a consequence of limited ternary eIF2–GTP–Met-tRNAi complex levels caused by eIF2α-p-dependent attenuation of eIF2B guanine nucleotide exchange factor (GEF) activity9,10. Although this has been a prevailing paradigm of translational control of ATF4 (ref. 5), more recently an additional mechanism emerged11. Notably, these studies used reporter vectors9,10, and were thus restricted in establishing the effects of uORF1 on endogenous ATF4 mRNA translation and its physiological consequences.
eIF2B suppression induces s-ISR
To investigate whether decreased eIF2B activity is synonymous with stress-induced eIF2α-p (Fig. 1a), we first compared the effects of acute thapsigargin (Tg)-induced ER stress (coinciding with maximal induction of eIF2α-p6) versus suppression of eIF2B activity through short hairpin RNA (shRNA)-mediated depletion of the eIF2Bε (encoded by Eif2b5) catalytic subunit in mouse embryonic fibroblasts (MEFs) (Fig. 1b). Although both Tg treatment (1 h) and depletion of eIF2Bε resulted in a comparable reduction in eIF2B activity (Fig. 1c), protein synthesis was more strongly reduced by Tg than by eIF2Bε depletion (Fig. 1d). We therefore set out to dissect the mechanisms that distinguish eIF2α-p-dependent suppression of eIF2B activity versus attenuation of eIF2B function in the absence of stress-induced eIF2α-p. We first examined stress granule assembly, which is in part mediated by an increased level of eIF2α-p12. Both arsenite (positive control) and Tg (1 h) induced stress granules, as illustrated by the punctate cytosolic pattern of the stress granule markers DEAD-box helicase 3 X-linked (DDX3X) and rasGAP SH3-binding protein 1 (G3BP1; Fig. 1e). By contrast, eIF2Bε depletion (Extended Data Fig. 1a) did not induce stress granule formation (Fig. 1e). This indicates that, unlike ER stress, low eIF2B activity under basal, non-stressed conditions seemed to alter cell shape but did not stimulate stress granule formation (Fig. 1e). Moreover, hallmarks of the ER stress response that were induced by Tg, including PERK activation and increased eIF2α-p, BiP, GADD34 and CHOP levels, were largely absent in eIF2Bε-depleted cells (Fig. 1f and Extended Data Fig. 1a). In turn, eIF2Bε depletion resulted in comparable ATF4 upregulation to Tg (1 h), despite the lack of eIF2α-p induction (Fig. 1f and Extended Data Fig. 1a). Expression of the active part of GADD34 (GADD34(ΔNT))7 led to dephosphorylation of eIF2α-p but failed to alter ATF4 protein levels in eIF2Bε-depleted cells (Extended Data Fig. 1b). We and others have previously shown that during the c-ISR, ATF4 induction is independent of the cap-binding protein eIF4E6,13. By contrast, eIF4E was required for ATF4 induction in eIF2Bε-depleted cells (Fig. 1f). Overall, these findings demonstrate that a decrease in eIF2B activity, in the absence of an increased level of eIF2α-p, engages distinct mechanisms as compared to the eIF2α-p-dependent c-ISR.
a, The prevailing view is that ISR induction downstream of the eIF2α kinases represents a linear response comprising increased eIF2α-p and subsequent decrease in eIF2B activity (c-ISR, orange). It is not clear whether decreased eIF2B activity without an increase in eIF2α-p such as in leukodystrophies (s-ISR, pink) induces comparable ISR to the eIF2α-p induction (orange). b,f, Western blot analysis of the denoted proteins, in MEFs treated with the indicated shRNAs for specified times or Tg (400 nM). In f, Tg (0 h) refers to control MEFs, not treated with control shRNAs. Representative images (n = 3 independent experiments) are shown. c,d, eIF2B GEF activity (c) and protein synthesis measured by [35S]methionine and cysteine incorporation (d) in MEFs expressing control shRNA or Eif2b5 shRNA, or treated with Tg (400 nM, 1 h), as indicated. Statistical significance was determined by two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 3 independent experiments). e, Fluorescence micrographs of MEFs treated with a vehicle (control), sodium arsenite (1 mM, 1 h) or Tg (400 nM, 1 h) or expressing control or Eif2b5 shRNAs (day 4), and stained with the indicated antibodies. Scale bar, 20 μm for all images. Representative images are shown (n = 3 independent experiments). g, Left, polysome profile tracings obtained by monitoring absorbance (254 nm) across 10–50% sucrose gradients. Right, distribution of the indicated mRNAs on polysomes isolated from MEFs expressing control, Eif2b5 or Eif2b5 + Eif4e shRNAs, as indicated. Representative data (n = 3 independent experiments) are shown.
We next examined the effect of eIF2Bε depletion or eIF2Bε and eIF4E co-depletion on translation of uORF-containing mRNAs (Extended Data Fig. 1c) that are translationally activated under the c-ISR (that is, ATF4, ATF5 (ref. 14), PPP1R15A (also known as GADD34)15 and DDIT3 (also known as CHOP16)). In agreement with the corresponding protein levels (Fig. 1f), Atf4 mRNA was translationally activated in eIF2Bε-depleted MEFs, which was reversed when eIF4E was co-depleted (Fig. 1g). Translation of Atf5 mRNA followed a similar pattern (Extended Data Fig. 1d). Conversely, translation of Gadd34 and Chop mRNAs was insensitive to depletion of eIF2Bε or co-depletion of eIF2Bε and eIF4E (Fig. 1g), which was consistent with the lack of alteration in the corresponding protein levels (Fig. 1f). Translation of α-tubulin (encoded by Tuba1a) mRNA, which is devoid of uORFs, was not strongly dependent on the eIF2Bε status in the cell (Extended Data Fig. 1d). Therefore, abrogation of eIF2B function in unstressed cells caused translational upregulation of only a subset of mRNAs that are translationally activated under the c-ISR6. These findings further show that distinct mechanisms are engaged during the eIF2α-p-dependent c-ISR and when eIF2B function is abrogated under unstressed conditions in the absence of eIF2α-p induction, which we refer to as the s-ISR.
s-ISR and c-ISR programs are distinct
As the eIF4E-dependent ATF4 induction in the s-ISR contrasts with the eIF4E-independent expression of ATF4 during the c-ISR, we next catalogued the relative changes in transcriptome-wide alterations in mRNA levels and polysome association in the s-ISR. To this end, we performed total- and polysome-RNA-sequencing analysis6,17,18 in MEFs expressing lentiviruses encoding control shRNA (shCon), Eif2b5 shRNA (shEif2b5) or both Eif2b5 and Eif4e shRNAs (shEif2b5 + shEif4e; Fig. 2a). Of note, we used the same quantity of RNA from the heavy polysome fraction from each cell line, which allows reliable comparison between relative changes in translation of specific mRNAs despite pronounced differences in global mRNA translation between conditions18. The resulting data were of sufficient quality as judged by sequencing depth and number of detected genes (Extended Data Fig. 1e). Furthermore, principal component analysis indicated high reproducibility, as samples clustered according to conditions (Extended Data Fig. 1e). Changes in translational efficiency and mRNA abundance (referred to as ‘transcriptome’) were then identified using anota2seq19. Expression patterns of selected genes were validated by quantitative PCR with reverse transcription (RT–qPCR) (Extended Data Fig. 1f). As compared to the control, depletion of eIF2Bε or co-depletion of eIF2Bε and eIF4E induced marked perturbations in mRNA levels that were accompanied by relatively modest changes in translational efficiencies (Fig. 2b,c and Supplementary Table 1). Notably, these perturbations in gene expression programs were markedly different from those observed in the c-ISR6 (Fig. 2d).
a, Absorbance profiles (254 nm) of 10–50% sucrose gradients from indicated cell lines. b–d, Scatter plots comparing fold change (FC) in total and polysome-associated mRNA quantified using RNA sequencing in shEif2b5 versus shCon (b), shEif2b5 + shEif4e versus shEif2b5 (c) and Tg 16 h versus control6 (d) MEFs. Differentially regulated genes are colour-coded (legend under d). All genes are shown in Supplementary Table 1. e–p, Comparisons of how genes controlled at the level of mRNA abundance (e–j) or translation (k–p) are regulated in shEif2b5 versus shCon (e–g,k–m) and shEif2b5 + shEif4e versus shEif2b5 (h–j,n–p) MEFs. For each gene set and comparison, a scatter plot (e,h,k,n) and empirical cumulative distribution plots for log2[FC] in total (f,i,l,o) and polysome-associated (g,j,m,p) mRNA are shown. Gene sets are colour-coded as indicated in d, and background (non-affected) genes are shown in grey. Shifts in the distribution of fold changes for gene sets relative to the background were assessed for total (f,i,l,o) and polysome-associated (g,j,m,p) mRNA using the Mann–Whitney U-test and indicated P < 0.01 for all comparisons. q, Gene Ontology (GO) enrichment analysis for regulated genes (through mRNA abundance or translation) from b. The number of genes (No.) identified in selected GO pathways is indicated, and all GO pathways are shown in Supplementary Table 2. r, Proliferation of MEFs expressing the indicated shRNAs was determined by counting live cells (trypan blue exclusion). P value was determined by the two-tailed Student’s t-test. Data are presented as mean ± s.e.m. Inset shows representative western blot analysis of levels of the indicated proteins. (n = 3 independent experiments).
Only three shared mRNAs encoding ATF4, ATF5 and SOX5 exhibited an increase in translational efficiency in the s-ISR (triggered by eIF2Bε depletion) and the c-ISR (induced by Tg; Fig. 2b–d). Out of 20 mRNAs whose translational efficiency was reduced in both the s-ISR and the c-ISR, 16 encode ribosomal proteins. Comparison of cells co-depleted of eIF2Bε and eIF4E versus those in which c-ISR was induced by Tg (Fig. 2c,d) revealed 34 (out of 137) and 41 (out of 170) overlapping mRNAs whose translational efficiency was increased or decreased, respectively (Extended Data Fig. 1g). Consistent with polysome profiling and RT–qPCR data (Fig. 1g), co-depletion of eIF2Bε and eIF4E repressed the increase in translational efficiency of Atf4 mRNA observed in cells in which the s-ISR was induced by eIF2Bε depletion (Fig. 2b,c). These data therefore support the existence of distinct translational control mechanisms in the c-ISR and s-ISR.
In contrast to changes in translational efficiency, alterations in total mRNA levels (that is, ‘transcriptome’) were more pronounced in the s-ISR than in the c-ISR (Fig. 2b–d). Moreover, the s-ISR consists of an increase in the levels of known c-ISR target mRNAs including those encoding CHOP and GADD34 (Extended Data Fig. 1f). The increase in the levels of these mRNAs was congruent with their augmented polysome association, thus resulting in no net changes in their translational efficiencies (Fig. 2b). This is consistent with polysome profiling and RT–qPCR experiments in which no major alterations in Chop and Gadd34 mRNA translational efficiencies were observed following eIF2Bε depletion (Fig. 1g). Notably, co-depletion of eIF2Bε and eIF4E resulted in decreased Chop and Atf3 mRNA levels, but Gadd34 and Ibtkα mRNA abundance remained higher than in controls (Fig. 2b,c and Extended Data Fig. 1f). As we did not observe accumulation of CHOP and GADD34 proteins in eIF2Bε-depleted cells or cells co-depleted for eIF2Bε and eIF4E, this suggested that the main ORF of these mRNAs is not translated during the s-ISR, and that the congruent increase in mRNA levels and polysome association is probably due to sustained uORF translation. In addition, in the c-ISR we identified 221 upregulated mRNAs (Extended Data Fig. 2a) that overlapped with about 10% of those increased in the s-ISR. This indicates that the s-ISR and c-ISR entail distinct gene expression reprogramming.
To obtain a more global view of the eIF4E-dependent reprogramming of the transcriptome and translatome in the s-ISR, we compared alterations in total mRNA abundance (Fig. 2e–j) and translational efficiencies (Fig. 2k–p) between eIF2Bε-depleted MEFs and MEFs co-depleted for eIF2Bε and eIF4E. These comparisons revealed that depletion of eIF4E largely reverted the changes in the transcriptome and translatome caused by eIF2Bε depletion (compare Fig. 2e–g versus Fig. 2h–j, and Fig. 2k–m versus Fig. 2n–p). Therefore, in the s-ISR, decreased eIF2B activity reprograms the transcriptome and the translatome in an eIF4E-dependent manner. This conclusion was further strengthened by the identification of 64 transcription factors showing eIF4E-dependent changes in mRNA abundance or translation following eIF2Bε depletion (Fig. 2b,c and Supplementary Table 1). Total mRNA levels of 80 positively regulated ATF4 target genes20 were also induced in eIF2Bε-depleted MEFs (Extended Data Fig. 2a–c). Similarly to above, these effects were eIF4E dependent (Extended Data Fig. 2d,e). These findings are consistent with eIF4E-dependent regulation of ATF4 following s-ISR induction through eIF2Bε depletion. GO enrichment analysis revealed that mRNAs whose abundance increased following eIF2Bε depletion encode proteins involved in cellular response to environmental cues (Fig. 2q and Supplementary Table 2). Among proteins encoded by downregulated mRNAs, there was a strong enrichment for factors regulating cell cycle progression (Fig. 2q and Supplementary Table 2). Except for a few functions related to the immune system, genes whose expression level was increased following eIF2Bε depletion functionally overlapped with those whose expression level was decreased when eIF4E was co-depleted (Extended Data Fig. 2f and Supplementary Table 3). Similarly, cellular processes enriched among genes downregulated in eIF2Bε-depleted MEFs overlapped with those whose expression level was increased following eIF4E co-depletion (Extended Data Fig. 2g). Therefore, s-ISR gene expression programs triggered by eIF2Bε depletion and reversed by abrogation of eIF4E consist of a subset of overlapping and functionally related genes, including cell cycle regulators that were enriched in the downregulated group in eIF2Bε-depleted cells. This is consistent with a decreased level of proliferation of eIF2Bε-depleted relative to control MEFs (Fig. 2r).
VWMD eIF2B alteration triggers s-ISR
VWMD is a leukodystrophy caused by alterations in the eIF2B subunits leading to decreased eIF2B activity21. Among the identified alterations, the mouse substitution R191H in eIF2Bε recapitulated aspects of the human disease, including progressive ataxia, motor skill deficits and shortened lifespan22. Development of these pathologies was associated with persistent ISR induction in the central nervous system of eIF2Bε(R191H) mutant mice22,23,24. Furthermore, treatment of these mice with a small-molecule activator of eIF2B markedly reduced VWMD-like phenotypes and attenuated the ISR2,24. Notably, VWMD is characterized by selective damage of oligodendrocytes and astrocytes25, which suggests that eIF2B alterations may result in metabolic reprogramming that is particularly toxic to these but not other cell types. This may contribute to the increased sensitivity of these cells to ER stress24. We thus introduced the eIF2Bε(R191H) alteration into mouse embryonic stem (ES) cells by mutating the CGC codon to CAC in the genomic region of exon 4 of the Eif2b5 gene (Fig. 3a). We used mouse ES cells as non-specialized cells that facilitated testing of the effect of the R191H alteration on metabolic reprogramming. The eIF2Bε(R191H) alteration was previously shown to cause a 40% decrease in the level of eIF2B GEF activity24. Homozygous Eif2b5R191H/R191H mouse ES cells showed a slight decrease in protein synthesis rates (Fig. 3b) and induction of ATF4, but not GADD34 or CHOP (Fig. 3c), which are characteristics of the s-ISR. Notably, the eIF2Bε(R191H) alteration did not cause induction in eIF2α-p, but rather decreased eIF2α-p levels, a phenomenon also observed in VWMD2,25. To identify differentially expressed genes in Eif2b5R191H/R191H cells, we used RNA sequencing (Fig. 3d and Extended Data Fig. 3a–c). Differential expression (DESeq2) analysis26 revealed that Eif2b5R191H/R191H mouse ES cells increased 886 and decreased 343 mRNA levels as compared to wild-type (WT) mouse ES cells (false discovery rate (FDR) < 0.05 and fold change > 1.2; Fig. 3d and Supplementary Table 1). Pathway analysis of the genes upregulated in Eif2b5R191H/R191H versus WT cells revealed enrichment in similar pathways to those observed in MEFs depleted of eIF2Bε, including response to bacterium, interferon response, adaptive immune response and regulation of defence response (Fig. 2q, Extended Data Fig. 3d and Supplementary Table 4). Of relevance to VWMD pathology, the predominant pathway enriched among downregulated genes is the cellular response to leukaemia inhibitory factor, a known positive regulator of oligodendrocyte survival, proliferation and myelination27. In agreement with this observation, the downregulated pathways included myelination, oligodendrocyte differentiation and stem cell population maintenance and differentiation (Extended Data Fig. 3d). Indeed, mouse induced pluripotent stem cell-derived oligodendrocyte progenitor cells that carry the pathogenic eIF2Bε(R132H) mutation produced markedly fewer mature oligodendrocytes as compared to WT cultures (Extended Data Fig. 3e).
a, Diagram of Eif2b5 exon 4. Eif2b5R191H/R191H mutation is indicated. b, Protein synthesis was monitored by [35S] methionine and cysteine labelling in WT and Eif2b5R191H/R191H mouse ES cells. P values, two-tailed Student’s t-test (n = 5 independent experiments, mean ± s.e.m.). c,e, Western blot of the indicated proteins from WT and Eif2b5R191H/R191H mouse ES cells and MEFs treated with a vehicle or Tg (400 nM) for the specified durations (n = 3 independent experiments) (c), and quantification (e). PCK2 levels were normalized to α-tubulin and quantified. P values, two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 3 independent experiments). d, DESeq2 analysis comparing WT and Eif2b5R191H/R191H mouse ES cell mRNA levels quantified by RNA sequencing. An ATF4-regulated gene signature20 is shown in red. Green arrows indicate selected differentially expressed genes (all regulated genes are shown in Supplementary Table 1). f, Schematic of [13C]glucose tracing in the CAC (black-filled circles indicate [13C]-labelled carbons) showing that serine m + 2 can be produced via oxaloacetate (OAA) m + 2, and serine m + 3 can be produced from 3-phosphoglycerate (3PG) m + 3. g, Steady-state levels of indicated metabolites in Eif2b5R191H/R191H cells compared to WT (n = 3 independent experiments). Data depict fold change of Eif2b5R191H/R191H versus WT. h, Fractional enrichment of serine m + 2 and serine m + 3 in WT mouse ES cells or Eif2b5R191H/R191H mouse ES cells treated with CPA (200 μM, 16 h) or after 48-h washout. P values, two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 6 independent experiments). DMSO, dimethylsulfoxide. i,j, Schematic of CPA treatment and washout (i), and quantification (j). Fold change of bioenergetic capacity of WT and Eif2b5R191H/R191H cells with or without exposure to CPA (200 μM) for 16 h, followed by 48 h washout. P values, two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 4 independent experiments). k, Venn diagrams of comparisons of regulated genes under indicated experimental conditions and published datasets24. A total of 26 identified genes are involved in amino acid metabolism (KEGG (Kyoto Encyclopedia of Genes and Genomes) analysis).
Notably, we observed that phosphoenolpyruvate carboxykinase 2 (Pck2) mRNA and protein levels were induced both during the s-ISR triggered by eIF2Bε depletion in MEFs (Fig. 2b and Extended Data Fig. 4a) and in Eif2b5R191H/R191H mouse ES cells (Fig. 3c–e). These effects were eIF4E dependent (Fig. 2c and Extended Data Fig. 4a). PCK2 is a known ATF4 target28, and was one of four ATF4 target genes with mitochondrial functions29 identified in both MEFs and mouse ES cells (Extended Data Fig. 4b). PCK2 converts oxaloacetate to phosphoenolpyruvate (PEP), a precursor to both pyruvate and 3-phosphoglycerate30. In turn, 3-phosphoglycerate is an intermediate of glycolysis and a precursor to serine and glycine biosynthesis (Fig. 3f). Previous reports showed that increased levels of PCK2 in cancer cells promote metabolic adaptation31,32. Compared to WT controls, Eif2b5R191H/R191H mouse ES cells presented with increased steady-state levels of PEP, serine and glycine (Fig. 3g). On this basis, we tested whether PCK2 contributes to metabolic flux of mitochondrial PEP towards serine biosynthesis. Fractional enrichment of [13C]carbon from uniformly labelled [13C]glucose to [13C]serine was determined through either glycolysis (m + 3 [13C]serine) or mitochondrial PEP (m + 2 [13C]serine; Fig. 3f,h). In agreement with their elevated PCK2 levels, Eif2b5R191H/R191H mouse ES cells exhibited a higher flux to m + 2 [13C]serine than WT mouse ES cells, whereas the level of m + 3 [13C]serine was similar between the two cell lines (Fig. 3h). These data suggest that the s-ISR triggered by R191H alteration in eIF2Bε induces metabolic adaptations that are at least in part mediated by PCK2.
To further characterize metabolic perturbations triggered by the s-ISR, we tested how transient ER stress and subsequent recovery affect the bioenergetics of WT and Eif2b5R191H/R191H mouse ES cells. Herein, cells were exposed to a reversible ER-stress inducer, cyclopiazonic acid6 (CPA), for 16 h, followed by compound washout (Fig. 3i). At the baseline, Eif2b5R191H/R191H mouse ES cells exhibited higher oxygen consumption rates (OCRs) and extracellular acidification rates (ECARs) (Extended Data Fig. 4c,d) than WT cells. However, Eif2b5R191H/R191H mouse ES cells demonstrated a larger decrease in OCR and ECAR following treatment and removal of CPA relative to WT mouse ES cells (Extended Data Fig. 4c,d). This pattern was primarily associated with a sharp decline in ATP production from oxidative phosphorylation in Eif2b5R191H/R191H mouse ES cells (Extended Data Fig. 4e). Mutant cells also exhibited a strong decrease in maximal bioenergetic capacity (Fig. 3j and Extended Data Fig. 4f) and reduced flux of [13C]glucose to m + 2 [13C]serine (Fig. 3h) following exposure and removal of CPA as compared to WT mouse ES cells. In summary, these findings suggest that the R191H alteration in eIF2Bε induces metabolic adaptations that are abrogated by episodes of ER stress.
To further support the physiological importance of s-ISR-mediated metabolic perturbations, we determined whether key metabolic genes induced in the brain of VWMD mice24 (2–7 months old) are also stimulated by induction of the s-ISR through depletion of eIF2Bε in MEFs (Fig. 3k). We identified 26 common genes, including Pck2 and Atf4, which we then grouped according to biological functions. The predominant biological functions for these genes were glycine, serine, cysteine and methionine metabolism (Fig. 3k). These results further support the hypothesis that alterations causing VWMD engender PCK2-mediated metabolic adaptation through the s-ISR that are probably disrupted by episodes of ER stress that trigger the c-ISR. This, at least in part, may explain the development of pathologies during persistent c-ISR in the central nervous system and the beneficial effects of abrogating the c-ISR by applying an eIF2B activator24.
Physiological functions of Atf4 uORF1
The prevailing ISR paradigm is that translational induction of the Atf4 mRNA depends on uORF1 translation10. Considering the discrepancy in eIF4E dependency of Atf4 mRNA translational control in the s-ISR versus c-ISR6,13 (Fig. 1f,g), we tested the function of uORF1 in the translational control of the Atf4 mRNA, in mouse ES cells. We first confirmed that the regulation of ATF4 protein levels is similar between mouse ES cells and MEFs under conditions in which the c-ISR was triggered by Tg (Extended Data Fig. 5a) as well as when the s-ISR was induced through eIF2Bε depletion (Figs. 1f and 4a). As in MEFs, eIF2Bɛ depletion in mouse ES cells did not affect eIF2α-p, GADD34 protein levels or PERK activity (Fig. 4a). Indeed, eIF2α-p was even reduced in eIF2Bε-depleted versus control mouse ES cells (Fig. 4a). Moreover, as in MEFs, co-silencing of eIF4E and eIF2Bε strongly reduced ATF4 levels compared to eIF2Bε depletion alone (Fig. 4a). We next used CRISPR–Cas9 gene editing to generate mouse ES cells wherein the AUG initiation codon of Atf4 uORF1 is mutated into AUA (ΔuORF1 mouse ES cells; Fig. 4b and Extended Data Fig. 5b) in its chromosomal location. Unexpectedly, during the chronic phase of the c-ISR (Tg (9 h)), ATF4 protein was induced in both WT and ΔuORF1 mouse ES cells (Fig. 4c, left panel). This was accompanied by elevated PERK activity and an increase in GADD34 and CHOP protein levels (Fig. 4c, left panel). Of note, the c-ISR program induced by chronic ER stress remains stable during a 6–18 h period6. During acute c-ISR (Tg; 1 h), ATF4 was also elevated in both WT and ΔuORF1 mouse ES cells in the absence of GADD34 and CHOP induction (Fig. 4c, middle panel). A decrease in ATF4 in ΔuORF1 relative to WT cells during acute c-ISR is explained by reduced baseline Atf4 mRNA stability in ΔuORF1 cells (see below). In response to the Tg-induced c-ISR, ΔuORF1 mouse ES cells exhibited reduction in protein synthesis and polysome assembly that was comparable to that in WT mouse ES cells (Extended Data Fig. 5c,d). Opposite to the c-ISR, induction of the s-ISR through depletion of eIF2Bε upregulated ATF4 protein levels in WT but not ΔuORF1 mouse ES cells (Fig. 4c, right panel). Similarly to the case in MEFs, eIF2Bε depletion did not induce GADD34 or CHOP protein abundance in either cell line (Fig. 4c, right panel). These data suggest an unanticipated mechanism whereby uORF1 drives induction of ATF4 protein in the s-ISR but seems not to be essential under the c-ISR.
a,l, Representative western blot of the indicated proteins in mouse ES cells expressing control (shCon), eIF2Bε or eIF2Bε + eIF4E shRNAs (a) or in MEFs treated with glucose (0–25 mM, 16 h) (l). LiCl (10 mM) or Torin 1 (250 nM) was used for the specified final hours of the 16 h treatment with glucose-free medium (l) (n = 3 independent experiments). b, Schematic of the Atf4 uORF1 start codon mutation in mouse ES cells. c, Representative western blot of the indicated proteins in WT or ΔuORF1 mouse ES cells treated with vehicle or Tg (400 nM) for the specified times (left and middle) or expressing control or Eif2b5 shRNAs (right). (n = 3 independent experiments). d,e, OCR (d) and ECAR (e) of WT and ΔuORF1 mouse ES cells. Oligomycin (Oligo), FCCP, rotenone (Rot) and antimycin A (AA), and monensin (Mon) were injected as indicated. P values, two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 5 independent experiments). f,g, Quantification of ATP production from oxidative phosphorylation (J ATP ox) or glycolysis (J ATP glyc) (f) and fold change in bioenergetic capacity (g) in WT versus ΔuORF1 mouse ES cells. P values, two-tailed Student’s t-test. Data are presented as mean ± s.e.m. (n = 5 independent experiments). h, J ATP glyc and J ATP ox values in WT and ΔuORF1 mouse ES cells for basal (square), FCCP (triangle) or monensin (circle) conditions. Maximum theoretical boundaries for J ATP ox (horizontal line) and J ATP glyc (vertical line) are indicated. P values, two-tailed Student’s t-test. Data represent mean ± s.e.m. (n = 5 independent experiments). i, Schematic of [13C]glutamine tracing in the CAC. PEP m + 3 and asparagine m + 4 are labelled by carbons derived from [13C]glutamine. CS, citrate synthase. j,k, Fractional enrichment (j) and relative ion abundance (k) of PEP m + 3 in ΔuORF1 cells versus WT. P values, two-tailed Student’s t-test. Data represent mean ± s.e.m. (n = 3 independent experiments).
Consistent with the accumulation of the ATF4 protein in WT mouse ES cells, both the s-ISR caused by eIF2Bε depletion and the c-ISR triggered by Tg induced translation of Atf4 mRNA, as evidenced by an increased level of association of Atf4 mRNA with heavy polysomes relative to that in control cells (Extended Data Fig. 5e). In ΔuORF1 mouse ES cells, Tg induced polysome association of Atf4 mRNA to a similar extent as in WT mouse ES cells (Extended Data Fig. 5e). By contrast, although the s-ISR induced by depletion of eIF2Bε partially shifted Atf4 mRNA towards heavier polysomes in ΔuORF1 mouse ES cells (Extended Data Fig. 5e), this was not accompanied by the increase in ATF4 protein levels (Fig. 4c, right panel). This discordance between Atf4 mRNA translation efficiency and protein levels may stem from increased translation of uORF2 in the absence of uORF1, which leads to out-of-frame translation and suppression of ATF4 protein synthesis. As expected, the polysomal distribution of Gapdh mRNA was not affected in the s-ISR triggered through eIF2Bε depletion, whereas the Tg-induced c-ISR shifted Gapdh mRNA towards lighter polysomes in both WT and ΔuORF1 mouse ES cells (Extended Data Fig. 5e). Consistent with the observed differences between the c-ISR and s-ISR programs, Tg but not eIF2Bε depletion induced translation of Gadd34 mRNA (Extended Data Fig. 5e). Collectively, these findings suggest that Atf4 mRNA translation in the s-ISR occurs through reinitiation following eIF4E-dependent translation of uORF1 and bypassing of uORF2, as described for the GCN4 mRNA in yeast5.
To establish the functional consequences of ATF4 regulation in the absence of stress-induced eIF2α-p, we examined whether uORF1 integrity affects metabolism and bioenergetics. To measure activity of the citric acid cycle (CAC), we performed [3-13C]pyruvate tracing (Extended Data Fig. 6a–c). Relative to WT mouse ES cells, ΔuORF1 mouse ES cells exhibited a decreased level of tracing of pyruvate into glutamine, succinate, malate and aspartate throughout one turn of the CAC (Extended Data Fig. 6b), with larger decreases observed through two CAC turns (Extended Data Fig. 6c). These data suggest that ΔuORF1 mouse ES cells exhibit a decreased level of pyruvate oxidation throughout the CAC as compared to WT mouse ES cells. Furthermore, ΔuORF1 mouse ES cells showed a marked decrease in OCR (Fig. 4d) but only a modest reduction in ECAR (Fig. 4e) relative to WT mouse ES cells. Basal ATP generation from oxidative phosphorylation (J ATP ox, white bars) was diminished in ΔuORF1 mouse ES cells as compared to WT mouse ES cells, whereas there were no significant differences in ATP generation from glycolysis (J ATP glyc, grey bars) between these cells (Fig. 4f). Maximal ATP production from oxidative phosphorylation (treatment with carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP)) was reduced in ΔuORF1 mouse ES cells as compared to WT mouse ES cells (Fig. 4d), suggesting a decreased mitochondrial bioenergetic capacity (Fig. 4g). By contrast, ΔuORF1 and WT mouse ES cells exhibited similar rates of maximal ATP generation from glycolysis (monensin; Fig. 4h). In summary, ΔuORF1 mouse ES cells exhibit decreased CAC activity, resulting in reduced bioenergetic capacity as compared to control WT cells. This demonstrates that uORF1-dependent regulation of ATF4, which is activated during the s-ISR, supports mitochondrial metabolism and bioenergetics.
To identify Atf4-regulated genes that mediate these metabolic effects, we performed RNA sequencing on WT and ΔuORF1 mouse ES cells. Notably, ΔuORF1 cells had lower ATF4 levels (Fig. 4c) and no change in global protein synthesis (Extended Data Fig. 6d). The resulting RNA-sequencing dataset was of optimal quality (Extended Data Fig. 3f–h). We used DESeq2 to identify differentially expressed genes26, which revealed 857 mRNAs with increased and 1,263 mRNAs with decreased levels in ΔuORF1 as compared to WT mouse ES cells (FDR < 0.01 and fold change > 1.5; Extended Data Fig. 6e and Supplementary Table 1). Among the downregulated mRNAs in ΔuORF1 cells was Pck2 mRNA, in agreement with the decreased ATF4 levels in these cells. To monitor PCK2 activity, we used [13C]glutamine labelling in cells deprived of glucose for 6 h (Fig. 4i), as described previously32. ΔuORF1 mouse ES cells showed decreased tracing of glutamine into PEP (Fig. 4j) and decreased absolute abundance of PEP m + 3 (all three carbons are labelled) as compared to WT mouse ES cells (Fig. 4k). Finally, serine and glycine basal levels were also lower in mutant ΔuORF1 versus WT cells, in agreement with decreased PEP abundance (Extended Data Fig. 6f). Therefore, uORF1-driven ATF4 expression and a consequent increase in PCK2 levels underpins s-ISR-dependent metabolic adaptations.
uORF1 attenuates Atf4 nonsense-mediated decay
Atf4 mRNA is a nonsense-mediated decay (NMD) target33. To establish the effect of disruption of uORF1 on Atf4 mRNA NMD, we compared the effects of induction of the s-ISR (eIF2Bε depletion) versus the c-ISR (Tg) on Atf4 mRNA levels in WT and ΔuORF1 mouse ES cells. Atf4 mRNA levels were 5.4- and 9.8-fold higher in WT as compared to ΔuORF1 mouse ES cells under basal conditions and following eIF2Bε depletion, respectively (Extended Data Fig. 7a). By contrast, Tg (9 h) resulted in a comparable increase in Atf4 mRNA levels in both WT and ΔuORF1 mouse ES cells (Extended Data Fig. 7a), which is consistent with previously reported transcriptional induction of the Atf4 gene under these conditions34. Notably, under basal conditions, the half-life (t1/2) of the Atf4 mRNA was reduced in ΔuORF1 (t1/2 = 1.2 h) as compared to WT mouse ES cells (t1/2 = 2 h; Extended Data Fig. 7b). The RNA helicase UPF1 is a central factor in NMD35. As expected, UPF1 depletion in mouse ES cells (Extended Data Fig. 7c) stabilized known NMD substrates, Atf3 and Gadd45b mRNAs (Extended Data Fig. 7d,e,h,i). UPF1 depletion also increased Atf4 mRNA half-life and levels (Extended Data Fig. 7f,g) in WT and ΔuORF1 mouse ES cells (compare Extended Data Fig. 7b and Extended Data Fig. 7f). Altogether, these findings indicate a role for NMD in reducing Atf4 mRNA levels when uORF1 is abrogated, and suggest that at least in part, translation of uORF2 may drive NMD of Atf4 mRNA. This was further supported by a comparable increase in Atf4 mRNA stability in Tg-treated WT (from 2 h to 3.6 h) and ΔuORF1 (from 1.2 h to 3.3 h) mouse ES cells (Extended Data Fig. 7j), which is consistent with the Tg-induced c-ISR in which translation of the main Atf4 ORF is expected to clear the exon junction complex and suppress NMD. We also generated uORF2-mutated NIH 3T3 cells in which both the translational repression of the main Atf4 ORF under unstressed conditions and its induction during the c-ISR were disrupted (Extended Data Fig. 7k). This shows that uORF2 has a major role in translational suppression of Atf4 mRNA under basal conditions and induction of ATF4 protein synthesis during the c-ISR5. By contrast, during the s-ISR, induction of Atf4 mRNA translation is mostly dependent on the integrity of uORF1 (Fig. 4c). Collectively, these findings support a model whereby eIF4E-mediated translation of uORF1 suppresses NMD and thus stabilizes the Atf4 mRNA (Extended Data Fig. 7l, top panel). Our results also suggest that opposite to the prevailing model10, uORF1 may not have a prominent role in the induction of ATF4 protein synthesis under the c-ISR induced by Tg. This suggests that ATF4 induction during the s-ISR and c-ISR occurs by distinct mechanisms (Extended Data Fig. 7l, middle and bottom panels, respectively).
S-ISR mechanisms and activators
To further dissect the mechanisms distinguishing the s-ISR and c-ISR, we induced eIF2α-p by salubrinal, which inhibits the PP1:GADD34 phosphatase36. Salubrinal induced the c-ISR and increased ATF4 levels independently of uORF1 integrity (Extended Data Fig. 8a). This corroborated that the mechanisms governing Atf4 mRNA translation in the s-ISR (highly uORF1 dependent) versus the c-ISR (largely uORF1 independent) are distinct. Similarly to inactivating eIF2B, depleting eIF2β triggered the s-ISR, while cells maintained their ability to phosphorylate eIF2α and induce the c-ISR in response to Tg (Extended Data Fig. 8b). This further confirmed that reduction in ternary complex availability in the absence of eIF2α-p drives the s-ISR. Moreover, the RNA helicase DDX3X, which cooperates with eIF4E in promoting translation of a subset of mRNAs37, is required for ATF4 induction during the s-ISR but not the c-ISR (Extended Data Fig. 8c,d). Furthermore, the mTOR inhibitor Torin 1 abolished ATF4 induction during the s-ISR, but not the c-ISR, which is consistent with eIF4E-dependent translational reprogramming in the s-ISR, but not the c-ISR (Extended Data Fig. 8e,f). Disruption of the s-ISR-dependent induction of ATF4 was also observed with the eIF4E inhibitor 4EGI-1 (Extended Data Fig. 8g). Collectively, these data show that the eIF4E–DDX3X axis has a specific role in uORF1-mediated induction of ATF4 in the s-ISR, whereas this mechanism seems not to be substantially engaged during the c-ISR. By contrast, CK2 inhibition caused induction of c-ISR (Extended Data Fig. 8h), which is consistent with previous findings17 and CK2-dependent phosphorylation of eIF3d38.
These mechanistic distinctions between the s-ISR and the c-ISR motivated us to identify physiological and pathological conditions that may activate the s-ISR, but not the c-ISR. As expected, complete starvation of glucose induced the c-ISR38 (Fig. 4l). By contrast, moderate hypoglycaemia (2.5 mM glucose) increased ATF4 but not GADD34 protein levels, which is representative of the s-ISR (Fig. 4l). Moderate hypoglycaemia (2.5 mM glucose) stimulated GSK3, as evidenced by reduced inhibitory site phosphorylation on GSK3β (Ser9)39. GSK3 inhibition by LiCl attenuated ATF4 induction during mild hypoglycaemia (s-ISR) but not complete glucose starvation (c-ISR; Fig. 4l and Extended Data Fig. 8i). As GSK3 suppresses eIF2B activity40, these findings suggest that sensing moderate hypoglycaemia through GSK3 is one of the mechanisms that specifically trigger the s-ISR. Moreover, experiments in the isogenic BT474 breast cancer cell model of Herceptin resistance revealed that following ER stress, the s-ISR is induced in Herceptin-resistant (BT474-R) but not parental drug-sensitive (BT474-P) cells (Extended Data Fig. 9a–d). Induction of ATF4 in BT474-R but not BT474-P cells was dependent on mTOR activity and the eIF4E–DDX3X axis41 (Extended Data Fig. 9e,f). Collectively, these findings illustrate clear mechanistic distinctions between the s-ISR and c-ISR, while providing initial insights into the potential physiological importance of the s-ISR.
Discussion
Our results challenge the prevailing tenet that the ISR represents a series of singular mechanisms that is activated in a similar manner irrespective of the type or duration of stress and/or cellular context. In general, the c-ISR is thought to be characterized by an increased level of eIF2α-p and a subsequent decrease in eIF2B activity, which in turn leads to the global inhibition of protein synthesis and selective translational and transcriptional reprogramming1. More recently, it became apparent that the ISR involves dynamic temporal regulation whereby acute repression of global protein synthesis is followed by partial recovery of mRNA translation6,7. Herein, we show that the programs engaged in response to decreased eIF2B activity differ depending on the eIF2α phosphorylation status (Extended Data Fig. 9g). A decrease in eIF2B activity in the absence of eIF2α-p induction leads to relatively limited, eIF4E-dependent changes in the translatome paralleled by alterations in the transcriptome that are distinct from those observed in the c-ISR6,42,43,44 (Extended Data Fig. 10). Indeed, attenuated eIF2B activity in the absence of induction of eIF2α-p stimulated ATF4 but not GADD34 protein synthesis. To emphasize that under these conditions the regulation of ATF4 and GADD34 are split, we introduced the term s-ISR (Extended Data Fig. 9g). This suggests that the ISR in mammals may be more plastic than previously appreciated, whereby tuning of eIF2B activity and/or eIF2α-p is likely to result in a variety of different cellular outcomes. Indeed, the s-ISR is triggered under mild stress caused by moderate hypoglycaemia and is substituted by the c-ISR under severe stress (that is, complete glucose deprivation). This implies that plasticity of ISR provides mammalian cells with the flexibility to adjust their response on the basis of the intensity of stress (details in Supplementary Notes).
Although the physiological role of the s-ISR remains to be fully characterized, we show that cells carrying a leukodystrophy-causing inactivating mutation in Eif2b5R191H/R191H (for example, VWMD) exhibit s-ISR characteristics (details in Supplementary Notes).
In summary, we describe the s-ISR mechanism that is triggered by a disease-relevant eIF2B alteration. The s-ISR is mechanistically distinct from the c-ISR in respect to translational and transcriptional reprogramming and metabolic outcomes. These findings show plasticity of the ISR that underpins neurodegenerative diseases and probably other pathologies in which the ISR is implicated. Accordingly, future studies are warranted to decipher the full spectrum of ISR plasticity in mammals and its implications in homeostatic stress adaptation and disease.
Methods
Cell lines, cell culture and shRNA treatments
MEFs and HEK293T cells were grown in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco number 11960044) supplemented with 10% defined fetal bovine serum (FBS; Gibco number 26140079), 2 mM l-glutamine, 100 units ml−1 penicillin and 100 μg ml−1 streptomycin (Gibco number 10378016). Mouse ES cells were grown in Iscove’s modified Dulbecco’s medium (Gibco number 12440053) supplemented with 0.1 mM 2-mercaptoethanol (Gibco number 21985023), 1× MEM non-essential amino acids solution (Gibco number 11140050), 50 units ml−1 penicillin and 50 µg ml−1 of streptomycin (Gibco number 15070063), 1,000 units ml−1 ESGRO leukaemia inhibitory factor supplement for mouse ES cell culture (Sigma ESG1106) and 20% Oneshot ES cell heat-inactivated FBS (Gibco number 16141079). Herceptin-sensitive parental (BT474-P) and Herceptin-resistant (BT474-R) human breast cancer cells45 were grown in high-glucose DMEM (Gibco number 11960044) supplemented with 10% heat-inactivated FBS (Gibco number 10082147), 2 mM l-glutamine (Gibco number 10378016), 100 units ml−1 penicillin, 100 μg ml−1 streptomycin and 1 mM sodium pyruvate (Gibco number 11360070). Mouse induced pluripotent stem cells (iPSCs) carrying the eIF2Bε(R132H) alteration (targeting G2723A in the eIF2B5 gene) in the eIF2Bε subunit of eIF2B, alongside WT iPSCs, were differentiated into oligodendrocyte precursor cells (OPCs) as previously described46,47,48. iPSC-derived OPCs were grown in DMEM/F12 (Thermo Fisher Scientific number 11320082), 1× N2 supplement (R&D Systems AR009), 1× B-27 without vitamin A supplement (Thermo Fisher Scientific number 12587010) and 1× Glutamax, supplemented with 20 ng ml−1 fibroblast growth factor 2 (R&D Systems 233-FB) and 20 ng ml−1 platelet-derived growth factor-AA (R&D Systems 221-AA). Mouse ES cells containing the Eif2b5R191H/R191H mutation or NIH 3T3 cells containing a point mutation in ΔuORF2 (ATG to ATA) were generated through CRISPR–Cas9 genome editing technology in the Case Western Transgenic and Targeting Facility. NIH 3T3 cells containing a point mutation in ΔuORF2 (ATG to ATA) were generated through CRISPR–Cas9 genome editing technology in SYNTHEGO.
For glucose-limitation experiments, cells were grown in no-glucose DMEM (Gibco number 11966025) supplemented with 10% FBS (Gibco number 26140079), 100 units ml−1 penicillin, 100 μg ml−1 streptomycin (Gibco number 15070063) and indicated glucose concentrations (Sigma G8644). All cells were maintained at 37 °C with 5% CO2 for all experiments before specific treatments. For shRNA knockdown experiments, lentiviral particles expressing shRNA against target mRNAs were prepared and propagated in HEK293T cells as described previously49,50 using the second-generation pLKO.1, psPAX2 and pMD2.G vectors. After two rounds of lentiviral infection, cells were selected under puromycin (30 μg ml−1 in MEFs, 2 μg ml−1 in mouse ES cells and 1.5 μg ml−1 in BT474) for 3 days. The last-selection-day cells were passaged for experimentation. Day 4 denotes 1 day after passage of the cells in the presence of puromycin. Puromycin was removed 3 h before collection or treatment of the cells. Proliferation was monitored by cell counting with trypan blue exclusion of dead cells as described previously51. In addition, proliferation was estimated using CellTiter-Glo Luminescent Cell Viability Assay kit (Promega G7572) according to the manufacturer’s instructions.
shRNAs and RT–qPCR primers
Plasmid expressing shRNA against Eif2b5 (TRCN0000109990), Eif4e (TRCN0000077474), Upf1 (number 1: TRCN0000009663; number 2: TRCN0000274486), Eif2s2 (TRCN0000096876), Ddx3x (TRCN0000287239) and MISSION pLKO.1-puro Non-Target shRNA Control Plasmid DNA (Sigma-Aldrich, SHC016) were purchased from Sigma-Aldrich.
For RT–qPCR analysis, we used the following primer sets: ATF4 (forward (−) GTTTGACTTCGATGCTCTGTTTC; reverse (+) GGGCTCCTTATTAGTCTCTTGG); GADD34 (forward (−) TACCCCTGTCTCTGGTAACCT; reverse (+) TGGCTTTGCATTGTACTCATCA); IBTKα (forward (−) CCACCGTCTGCAGGATTATT, reverse (+) CTCGACCTTATCCGAATGGA); ATF5 (forward (−) AAGCTTGTAAGGCCCCCTGT, reverse (+) GTGCGCTTGATGTAGGGATT); BiP (forward (−) ACTTGGGGACCACCTATTCCT, reverse (+) ATCGCCAATCAGACGCTCC); α-tubulin (forward (−) CACTTACCACGGAGATAGCGA, reverse (+) ACCTTCTGTGTAGTGCCCCTT); GAPDH (forward (−) CGCCTGGAGAAACCTGCCAAGTATG, reverse (+) GGTGGAAGAGTGGGAGTTGCTGTTG); CHOP (forward (−) CTGGAAGCCTGGTATGAGGAT, reverse (+) CAGGGTCAAGAGTAGTGAAGGT); XBP1s (forward (−) GAGTCCGCAGCAGGTG, reverse (+) CTGGGAGTTCCTCCAGACTA); β-actin (forward (−) CTGGCACCACACCTTCTACAATG, reverse (+) GGTCATCTTTTCACGGTTGGC); GADD45a (forward (−) GAGGAATTCTCGGCTGCAGA, reverse (+) CACGTTATCGGGGTCTACGT).
Chemicals, reagents and antibodies
Chemicals used in this study: Tg (400 nM, Sigma-Aldrich T9033); sodium arsenite (1 mM, Sigma S7400); CPA (100 μM (BT474) and 200 μM (MEFs and mouse ES cells) Tocris number 1235); actinomycin D (10 μg ml−1, Sigma-Aldrich A9415); cycloheximide (100 μg ml−1, Sigma C7698); salubrinal (15 μM, Tocris number 3657); Torin 1 (250 nM, Tocris number 4247); LiCl (10 mM, Sigma); Herceptin (20 μg ml−1, Genentech); 4EGI-1 (200 μM, Med Chem Express HY-19831); SGC-CK2-1 (5 μM, Cayman number 34103).
Antibodies used in this study: anti-PERK (1:1,000, Cell Signaling Technology number 3192); anti-eIF4E (1:1,000, Cell Signaling Technology number 9742); anti-eIF2Bε (1:1,000, Cell Signaling Technology number 3595); anti-eIF2α (1:1,000, Cell Signaling Technology number 9722); anti-eIF2α-phospho(Ser51) (1:3,000, Abcam ab32157); anti-ATF4 (1:1,000, Cell Signaling Technology number 11815); anti-α-tubulin (1:4,000, Sigma T9026); anti-citrate synthase (1:1,000, Sino Biological 14083-T46); anti-GADD34 (1:3,000, Proteintech 10449-1-AP); anti-BiP (1:1,000, Cell Signaling Technology number 3177); anti-DDX3X (1:1,000, Cell Signaling Technology number 2635); anti-CHOP (1:1,000, Cell Signaling Technology number 2895); anti-PCK2 (1:1,000, Cell Signaling Technology number 6924); anti-UPF1 (1:1,000, Cell Signaling Technology number 12040); anti-S6 ribosomal protein (1:1,000, Cell Signaling Technology number 2217); anti-phospho-S6 ribosomal protein (Ser240/244) (1:1,000, Cell Signaling Technology number 5364); anti-GSK3β (1:1,000, Cell Signaling Technology number 9315); anti-phospho-GSK3β (Ser9) (1:1,000, Cell Signaling Technology number 9323); anti-4E-BP1 (1:1,000, Cell Signaling Technology number 9644), anti-phospho-4E-BP1 (Ser65) (1:1,000, Cell Signaling Technology number 9451) and anti-eIF2β (1:1,000, Santa Cruz sc-9978). For immunostaining: anti-G3BP1 (1:200, Santa Cruz sc-81940); anti-DDX3X (1:200, Bethyl A300-474A); anti-O1 (1:100, CCF Hybridoma Core Facility); anti-MBP (1:100, Abcam ab7349); anti-beta actin (1:4,000, Abcam ab6276). Hoechst (Thermo Fisher Scientific number 3570) was used to detect nuclei in immunostaining experiments.
Cell extract preparation for western blotting
Cells were washed twice with ice-cooled 1× PBS before lysis. Ice-cooled (4 °C) lysis buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 2 mM EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate), supplemented with EDTA-free protease inhibitor (Sigma number 34693159001) and PhosSTOP phosphatase inhibitor (Sigma number 4906837001) was added to cells. Cells were scraped off, collected and sonicated on ice. Protein lysates were centrifuged for 5 min at 10,000g and 4 °C. Supernatant was collected and quantified using the DC Protein Assay Kit (Bio-Rad number 5000112). Lysate was diluted to 1 μg μl−1 using lysis buffer. The diluted lysates were mixed with 5× sample loading buffer (300 mM Tris-HCl pH 6.8, 50% glycerol, 10% (v/v) β-mercaptoethanol, 10% (w/v) SDS and 50 mg bromophenol blue) for western blot analysis. Protein lysates were separated by SDS–PAGE before electrotransfer to Immobilon-P PVDF membrane (Sigma-Aldrich). When possible, membranes were stripped and re-probed, but in the cases in which this was not feasible (for example, antibodies from the same species in different dynamic ranges), the same lysates were simultaneously resolved on duplicate gels. Representative corresponding loading controls are shown.
Measuring in vitro GEF activity of eIF2B
eIF2B activity was measured as previously described6. In brief, cells were washed and scraped off in homogenization buffer (45 mM HEPES-KOH pH 7.4, 0.375 mM MaOAc, 75 mM EDTA, 95 mM KOAc, 10% glycerol, 1 mM dithiothreitol (DTT), 2.5 mg ml−1 digitonin), supplemented with EDTA-free protease inhibitor (Sigma number 4693159001) and PhosSTOP phosphatase inhibitor (Sigma number 4906837001). Cell lysates were homogenized and quantified for protein concentration. eIF2B activity was calculated as the rate of exchange from [3H]eIF2α GDP to non-radioactive GDP at each time point.
Measurement of global protein synthesis
Protein synthesis rates were measured as previously described6. In brief, cells were treated with designated chemicals for the indicated durations. At the end of treatments, [35S]Met and Cys (30 μCi ml−1 EXPRE35S Protein Labeling Mix (PerkinElmer NEG072002MC) was added to the cells for an additional 30 min. After labelling, cells were washed and lysed, and the radioactivity incorporated into proteins was determined by liquid scintillation counter. The protein synthesis rate was calculated as the rate of [35S]Met and Cys incorporation to total cellular protein from the same lysate.
Polysome profile analysis and mRNA distribution
Cells were seeded in 150-mm culture dishes and grown up to 70% confluence (about 1.0 × 107 cells). Cells were washed twice with cold PBS containing CHX (100 μg ml−1), scraped off and pelleted at 4,000 r.p.m. for 10 min. The cell pellets were suspended in 500 μl of lysis buffer (10 mM HEPES-KOH at pH 7.4, 2.5 mM MgCl2, 100 mM KCl, 0.25% NP-40, 100 μg ml−1 CHX, 1 mM DTT), 200 units ml−1 of RNase inhibitor (NEB number M0314) and EDTA-free protease inhibitor (Sigma number 693159001), kept on ice for 20 min and then passed 15 times through a 23-gauge needle. Lysates were cleared at 14,000 r.p.m. for 15 min, and supernatants (cell extracts) were collected and measured at absorbance of 260 nm. An equal amount (approximately 500 μg of lysate) was layered over 10–60% of cold sucrose gradients prepared in buffer (10 mM HEPES-KOH at pH 7.4, 2.5 mM MgCl2, 100 mM KCl). Gradients were centrifuged at 35,000 r.p.m. in a Beckman SW41Ti rotor for 3 h at 4 °C. After centrifugation, 12 equal-sized fractions (1 ml per fraction) were collected. RNA from each fraction was isolated using TRIzol LS reagent (Invitrogen number 10296028) and an equal volume of RNA from each fraction was cDNA-synthesized using the SuperScript III First-Strand Synthesis SuperMix (Thermo Fisher Scientific number 18080044). The relative quantity of specific mRNAs was measured by RT–qPCR using the VeriQuest SYBR Green qPCR Master Mix (Thermo Fisher Scientific 756002000RXN) with the StepOnePlus Real-Time PCR System (Applied Biosystem). For conventional measurement of total RNA levels, cells were seeded in 60-mm culture dishes and grown up to 70% confluence (1.0–1.5 × 106 cells) before treatment. Following indicated treatments, total intracellular RNA was isolated using TRIzol reagent (Invitrogen number 15596018). cDNAs were synthesized and relative RNA levels were measured by RT–qPCR as described above.
RNA-sequencing data preprocessing and quality control
RNA-sequencing libraries were prepared according to the TruSeq Stranded Total RNA protocol (Illumina) following the manufacturer’s instructions, and paired-end reads were obtained using a HiSeq2500 system (Illumina). The quality of sequencing reads was confirmed using FastQC (v0.11.4; http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). For the removal of Illumina TruSeq adaptor sequences and low-quality base calls, BBmap (v36.59; https://www.osti.gov/servlets/purl/1241166) was used with the following parameters: k = 13, ktrim = n, useshortkmers = t, mink = 5, qtrim = t, trimq = 10, minlength = 25. Subsequently, resulting reads were mapped to the mm10 genome assembly using HISAT (v2.0.4, in addition to default parameters, –no-mixed and –no-discordant were applied)52. The aligned reads were summarized using htseq-count53. Data quality was assessed using principal component analysis on trimmed mean of M values (TMM)-log2-normalized counts using the PCAtools R package (v2.4.0; https://github.com/kevinblighe/PCAtools; parameters removeVar = 0.75 and scale = T).
Raw fastq files for the dataset from ref. 24 were obtained from the National Center for Biotechnology Information Gene Expression Omnibus repository (GSE128092) and prepared the same way as described above, with a difference that the aligned reads were summarized using the featureCounts function of the RSubread (v2.6.4) R/Bioconductor package54.
Analysis of differential translation in datasets using anota2seq
Genes with 0 mapped RNA-sequencing reads in one or more samples were discarded. The data were TMM-log2-normalized and analysed using anota2seq19,55 (v1.14.0, parameters: minSlopeTranslation = −1, minSlopeBuffering = −1, maxSlopeTranslation = 1.5, maxSlopeBuffering = 1.5, deltaPT = deltaTP = deltaP = deltaT = log2(1.5) and FDR < 0.05). To classify genes into translation, offsetting or mRNA abundance gene expression modes, the anota2seqRegModes function within anota2seq was used.
Analysis of differentially expressed genes in the Eif2b5 R191H/R191H, ΔORF1 mouse ES cells and ref. 24 datasets
The datasets were analysed using DESeq2 (ref. 26; v1.38.2). To identify differentially expressed genes, an FDR threshold of <0.05 was applied to the Eif2b5R191H/R191H data and the datasets (all comparisons) from ref. 24. Additionally, changes with an absolute fold change of <log2(1.2) were filtered out in the Eif2b5R191H/R191H dataset. In the ΔORF1 dataset, an FDR threshold of <0.01 and an absolute fold change of >log2(1.5) were applied.
GO analysis
GO analysis was performed using GOstats (v2.68.0) using a hypergeometric test for categories from biological process ontology terms56.
Bioenergetic analysis
OCR and ECAR were measured using a Seahorse XFe24 analyser (Agilent Technologies), as previously shown57. In brief, mouse ES cells were fed with growth medium for 1 h, and then trypsinized and resuspended in growth medium. Cells were washed twice in Seahorse XF medium (10 mM glucose, 2 mM glutamine, 1 mM sodium pyruvate, pH 7.4) following centrifugation. Cells were subsequently seeded at a density of 100,000 cells in a volume of 200 μl per well on plates coated with 22.4 μg ml−1 Cell-TAK (Corning number 354240). The plate was centrifuged at 200g for 1 min to allow cells to adhere and subsequently placed in a 37 °C non-CO2 incubator for 1 h. Three measurements of OCR and ECAR were recorded under basal conditions, and following the injection of each compound (25 μl per injection): oligomycin (1 μM; Sigma O4876-25MG), FCCP (1.5 μM; Sigma C2920-10MG), rotenone (1 μM; Sigma R8875-1G) and antimycin A (1 μM; Sigma A8674-25MG), and monensin (20 μM; Sigma M5273-1G). Rates of oxidative ATP production (J ATP ox) and glycolytic ATP production (J ATP glyc), as well as bioenergetic capacity, were calculated as previously described58. Bioenergetic capacity is defined by the maximum values of J ATP glyc and J ATP ox in cells58. Buffering power of the Seahorse XF medium was measured as previously described59. All values were normalized to protein content.
Immunofluorescent staining
Cells were plated on glass microscope coverslips (Thermo Fisher) in 6-cm culture dishes and were allowed to grow for 48 h. After the designated treatments, cells were washed twice with ice-cooled 1× PBS on ice. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature on a shaker at 50 r.p.m. Fixed cells were washed twice with ice-cooled 1× PBS and incubated in PBST (1× PBS + 0.02% Triton X-100) for 15 min, PBST with 10% FBS for 30 min, and PBST with 10% FBS and primary antibodies at 4 °C for 16 h. After being washed with ice-cooled PBST twice, cells were incubated in PBST with 10% FBS and secondary antibodies for 2 h in the dark. This was followed by washing with ice-cooled PBST twice and nuclei staining with Hoechst 33342 for 5 min in the dark. After being washed with ice-cooled PBST twice, cells were mounted in Fluoromount-G (Electronic Microscope Sciences) and sealed with clear nail polish on microscope slides. The images were captured using a Leica SP8 confocal microscope. Imaging areas were randomly selected in a single-blind manner by a microscope specialist. Three areas were imaged in each condition, and a representative image is shown.
Metabolic labelling
Cells were plated onto 12-well plates and cultured in the cell growth medium. Cells were starved for 6 h with DMEM (Thermo Fisher A1443001) without glucose, glutamine and pyruvate. Next, cells were washed with 1× PBS and incubated with labelling medium containing 4 mM glutamine, 2× non-essential amino acids, 20% ES cell FBS, 1× penicillin–streptomycin and 4 mM [3-13C]pyruvate for 6 h. Finally, labelling medium was removed, and cells were washed with cold saline twice. Metabolites were quenched by the addition of 80% methanol/water (v/v), chilled on dry ice, and stored at −80 °C until processing.
For metabolite extraction, samples were thawed on ice and mixed well, followed by centrifugation at 4 °C for 10 min at 14,000 r.p.m. to pellet protein. Supernatant was removed into gas chromatography–mass spectrometry (GC–MS) vials. Pellets were extracted again with cold 80% methanol in water (v/v), and supernatants were combined and derivatized as described elsewhere60. In brief, 10 µl of 1 N NaOH was added to methanol supernatants and vortexed. Next, 15 µl of NaB2H4 (10 mg ml−1 in 50 mM NaOH) was added to reduce keto bonds and convert them into their respective deuterated hydroxyl groups. Next, samples were vortexed for 20 s and allowed to react at room temperature for 1 h. Reaction was stopped by the addition of 1 N HCl, and samples were evaporated to dryness. To remove boric acid, 50 µl of methanol was added, and samples were left to stand. After methanol was evaporated to dryness, samples were further derivatized by the addition of 60 µl of N-(tert-butyldimethylsilyl)-N-methyltrifluoroacetamide (MTBSTFA) to form the tri-tert-butyldimethylsilyl (t-BDMS), and the reactions were performed for 1 h at 60 °C. Derivatized samples were transferred to inserts and 1 µl of each sample was analysed by GC–MS.
GC–MS analyses were carried out on an Agilent 5973 mass spectrometer equipped with a 6890 Gas Chromatograph. A DB17-MS capillary column (30 m × 0.25 mm × 0.25 μm) was used in all assays with a helium flow of 1 ml min−1. Oven temperature was set to 100 °C, held for 1 min, and ramped at 7.5 °C min−1 until 260 °C, then 30 °C min−1 until 300 °C, and held for 10 min. Injector temperature was set at 250 °C and detector temperature at 280 °C. Samples were analysed in selected ion monitoring mode using electron impact ionization. Ion dwell time was set to 10 ms. Metabolite abundances were collected, and natural abundance was normalized using matrix analyses. Fractional enrichment was determined by dividing the abundance of labelled species by the sum of all of the molecular species for each particular metabolite.
Differentiation and imaging of OPCs
To differentiate WT and eIF2Bε(R132H) OPCs, cells were plated and grown for 72 h in differentiation medium that consisted of DMEM/F12, 1× N2 supplement and 1× B-27 without vitamin A supplement, supplemented with 100 ng ml−1 noggin (R&D Systems 3344-NG), 10 ng ml−1 neurotrophin-3 (NT-3; R&D Systems 267-N3), 50 μM cAMP (Sigma D0260), 100 ng ml−1 insulin-like growth factor 1 (R&D Systems 291-G1) NT-3 and 40 ng ml−1 triiodothyronine (thyroid hormone; Sigma T-6397).
After 72 h of differentiation, cells were live-stained with mouse anti-O1 antibody, then fixed with 4% paraformaldehyde (PFA) and immunostained using rat anti-MBP antibody, secondary antibodies conjugated to an Alexa Fluor (4 μg ml−1, Thermo Fisher) and the nuclear stain DAPI (Sigma, 1 μg ml−1). Cells were then imaged using the Operetta High Content imaging system (PerkinElmer) and analysed using automated scripts in Columbus v2.8.1.141347 software (PerkinElmer). Both outlier tests and P value calculation were performed in GraphPad Prism v10.4.1. Outliers were statistically identified and removed using ROUT test with Q = 1%, and P values were calculated using a Welch’s unpaired t-test.
Quantification and statistical analysis
For quantification of band intensities from western blot, ImageJ software was used (https://imagej.net/ij/). For statistical analysis, unless noted otherwise, data shown in this study are presented as the mean of triplicate determinations ± s.e.m. Unless otherwise indicated in the figure legends, statistical significance between groups was evaluated using Student’s t-test. In the figure legends (n = X) indicates the number of independent experiments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The datasets generated and analysed during this study are available in the Gene Expression Omnibus repository under the accession numbers GSE268132 (shEif2b5 and shEif4e), GSE268130 (Eif2b5R191H/R191H) and GSE268127 (Atf4 uORF1 mutant). All data supporting the findings of this study are included in this article and its Supplementary Information. Source data are provided with this paper.
References
Costa-Mattioli, M. & Walter, P. The integrated stress response: from mechanism to disease. Science 368, eaat5314 (2020).
Abbink, T. E. M. et al. Vanishing white matter: deregulated integrated stress response as therapy target. Ann. Clin. Transl. Neurol. 6, 1407–1422 (2019).
Advani, V. M. & Ivanov, P. Translational control under stress: reshaping the translatome. Bioessays 41, e1900009 (2019).
Wang, X. & Proud, C. G. The role of eIF2 phosphorylation in cell and organismal physiology: new roles for well-known actors. Biochem. J. 479, 1059–1082 (2022).
Dever, T. E., Ivanov, I. P. & Hinnebusch, A. G. Translational regulation by uORFs and start codon selection stringency. Genes Dev. 37, 474–489 (2023).
Guan, B. J. et al. A unique ISR program determines cellular responses to chronic stress. Mol. Cell 68, 885–900 (2017).
Novoa, I. et al. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 22, 1180–1187 (2003).
Mahe, M., Rios-Fuller, T., Katsara, O. & Schneider, R. J. Non-canonical mRNA translation initiation in cell stress and cancer. NAR Cancer 6, zcae026 (2024).
Lu, P. D., Harding, H. P. & Ron, D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell Biol. 167, 27–33 (2004).
Vattem, K. M. & Wek, R. C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA 101, 11269–11274 (2004).
Smirnova, A. M. et al. Stem-loop-induced ribosome queuing in the uORF2/ATF4 overlap fine-tunes stress-induced human ATF4 translational control. Cell Rep. 43, 113976 (2024).
Riggs, C. L., Kedersha, N., Ivanov, P. & Anderson, P. Mammalian stress granules and P bodies at a glance. J. Cell Sci. 133, jcs242487 (2020).
Mukhopadhyay, S., Amodeo, M. E. & Lee, A. S. Y. eIF3d controls the persistent integrated stress response. Mol. Cell 83, 3303–3313 (2023).
Watatani, Y. et al. Stress-induced translation of ATF5 mRNA is regulated by the 5′-untranslated region. J. Biol. Chem. 283, 2543–2553 (2008).
Lee, Y. Y., Cevallos, R. C. & Jan, E. An upstream open reading frame regulates translation of GADD34 during cellular stresses that induce eIF2α phosphorylation. J. Biol. Chem. 284, 6661–6673 (2009).
Palam, L. R., Baird, T. D. & Wek, R. C. Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 (2011).
Gandin, V. et al. mTORC1 and CK2 coordinate ternary and eIF4F complex assembly. Nat. Commun. 7, 11127 (2016).
Gandin, V. et al. Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J. Vis. Exp. https://doi.org/10.3791/51455 (2014).
Oertlin, C. et al. Generally applicable transcriptome-wide analysis of translation using anota2seq. Nucleic Acids Res. 47, e70 (2019).
Han, J. et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 15, 481–490 (2013).
Fogli, A. et al. Decreased guanine nucleotide exchange factor activity in eIF2B-mutated patients. Eur. J. Hum. Genet. 12, 561–566 (2004).
Hanson, F. M., Hodgson, R. E., de Oliveira, M. I. R., Allen, K. E. & Campbell, S. G. Regulation and function of elF2B in neurological and metabolic disorders. Biosci. Rep. 42, BSR20211699 (2022).
Bugiani, M., Vuong, C., Breur, M. & van der Knaap, M. S. Vanishing white matter: a leukodystrophy due to astrocytic dysfunction. Brain Pathol. 28, 408–421 (2018).
Wong, Y. L. et al. eIF2B activator prevents neurological defects caused by a chronic integrated stress response. eLife 8, e42940 (2019).
Bugiani, M. et al. Defective glial maturation in vanishing white matter disease. J. Neuropathol. Exp. Neurol. 70, 69–82 (2011).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Deverman, B. E. & Patterson, P. H. Exogenous leukemia inhibitory factor stimulates oligodendrocyte progenitor cell proliferation and enhances hippocampal remyelination. J. Neurosci. 32, 2100–2109 (2012).
Seenappa, V., Joshi, M. B. & Satyamoorthy, K. Intricate regulation of phosphoenolpyruvate carboxykinase (PEPCK) isoforms in normal physiology and disease. Curr. Mol. Med. 19, 247–272 (2019).
Calvo, S. E., Clauser, K. R. & Mootha, V. K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2016).
Yu, S., Meng, S., Xiang, M. & Ma, H. Phosphoenolpyruvate carboxykinase in cell metabolism: roles and mechanisms beyond gluconeogenesis. Mol. Metab. 53, 101257 (2021).
Mendez-Lucas, A., Hyrossova, P., Novellasdemunt, L., Vinals, F. & Perales, J. C. Mitochondrial phosphoenolpyruvate carboxykinase (PEPCK-M) is a pro-survival, endoplasmic reticulum (ER) stress response gene involved in tumor cell adaptation to nutrient availability. J. Biol. Chem. 289, 22090–22102 (2014).
Vincent, E. E. et al. Mitochondrial phosphoenolpyruvate carboxykinase regulates metabolic adaptation and enables glucose-independent tumor growth. Mol. Cell 60, 195–207 (2015).
Park, Y., Reyna-Neyra, A., Philippe, L. & Thoreen, C. C. mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 19, 1083–1090 (2017).
Dey, S. et al. Both transcriptional regulation and translational control of ATF4 are central to the integrated stress response. J. Biol. Chem. 285, 33165–33174 (2010).
Kim, Y. K. & Maquat, L. E. UPFront and center in RNA decay: UPF1 in nonsense-mediated mRNA decay and beyond. RNA 25, 407–422 (2019).
Boyce, M. et al. A selective inhibitor of eIF2α dephosphorylation protects cells from ER stress. Science 307, 935–939 (2005).
Soto-Rifo, R. et al. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. EMBO J. 31, 3745–3756 (2012).
Lamper, A. M., Fleming, R. H., Ladd, K. M. & Lee, A. S. Y. A phosphorylation-regulated eIF3d translation switch mediates cellular adaptation to metabolic stress. Science 370, 853–856 (2020).
Fang, X. et al. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl Acad. Sci. USA 97, 11960–11965 (2000).
Welsh, G. I., Miller, C. M., Loughlin, A. J., Price, N. T. & Proud, C. G. Regulation of eukaryotic initiation factor eIF2B: glycogen synthase kinase-3 phosphorylates a conserved serine which undergoes dephosphorylation in response to insulin. FEBS Lett. 421, 125–130 (1998).
Adjibade, P. et al. DDX3 regulates endoplasmic reticulum stress-induced ATF4 expression. Sci. Rep. 7, 13832 (2017).
Andreev, D. E. et al. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 16, 90 (2015).
Baird, T. D. et al. Selective mRNA translation during eIF2 phosphorylation induces expression of IBTKα. Mol. Biol. Cell 25, 1686–1697 (2014).
Kaspar, S. et al. Adaptation to mitochondrial stress requires CHOP-directed tuning of ISR. Sci. Adv. 7, eabf0971 (2021).
Merry, C. R. et al. Transcriptome-wide identification of mRNAs and lincRNAs associated with trastuzumab-resistance in HER2-positive breast cancer. Oncotarget 7, 53230–53244 (2016).
Elitt, M. S. et al. Chemical screening identifies enhancers of mutant oligodendrocyte survival and unmasks a distinct pathological phase in Pelizaeus-Merzbacher disease. Stem Cell Rep. 11, 711–726 (2018).
Lager, A. M. et al. Rapid functional genetics of the oligodendrocyte lineage using pluripotent stem cells. Nat. Commun. 9, 3708 (2018).
Najm, F. J. et al. Transcription factor-mediated reprogramming of fibroblasts to expandable, myelinogenic oligodendrocyte progenitor cells. Nat. Biotechnol. 31, 426–433 (2013).
Chen, C. W. et al. Adaptation to chronic ER stress enforces pancreatic beta-cell plasticity. Nat. Commun. 13, 4621 (2022).
Krokowski, D. et al. Stress-induced perturbations in intracellular amino acids reprogram mRNA translation in osmoadaptation independently of the ISR. Cell Rep. 40, 111092 (2022).
Dowling, R. J. et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328, 1172–1176 (2010).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Anders, S., Pyl, P. T. & Huber, W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015).
Liao, Y., Smyth, G. K. & Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 47, e47 (2019).
Oertlin, C., Watt, K., Ristau, J. & Larsson, O. Anota2seq analysis for transcriptome-wide studies of mRNA translation. Methods Mol. Biol. 2418, 243–268 (2022).
Beissbarth, T. & Speed, T. P. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics 20, 1464–1465 (2004).
Vlaski-Lafarge, M. et al. Bioenergetic changes underline plasticity of murine embryonic stem cells. Stem Cells 37, 463–475 (2019).
Mookerjee, S. A., Gerencser, A. A., Nicholls, D. G. & Brand, M. D. Quantifying intracellular rates of glycolytic and oxidative ATP production and consumption using extracellular flux measurements. J. Biol. Chem. 292, 7189–7207 (2017).
Mookerjee, S. A., Nicholls, D. G. & Brand, M. D. Determining maximum glycolytic capacity using extracellular flux measurements. PLoS ONE 11, e0152016 (2016).
Mamer, O. et al. The complete targeted profile of the organic acid intermediates of the citric acid cycle using a single stable isotope dilution analysis, sodium borodeuteride reduction and selected ion monitoring GC/MS. Metabolomics 9, 1019–1030 (2013).
Acknowledgements
We thank Z. Gao, Y. Feng and S. McLaughlan for experimental and intellectual contributions; and J. R. Woodgett for advice on GSK3 signalling. Funding sources: National Institutes of Health DK53307 and DK060596 (to M.H.); 5T34GM137792 (to M.A. and M.H.); GM128981 (to A.A.K.); AG000511 (to M.J.P. and M.G.); R3M127089 (to C.V.), CA259386 (to J. A. Steitz—supporting R.J.), the Terry Fox Foundation Oncometabolism Team Grant TFF‐242122 and Canadian Institutes for Health Research PJT-183843 and PJT-451236 (to I.T.); PJT-168864 (to A.E.K.); Swedish Research Council 2020-01665 and Swedish Cancer Society 22 2186 (to O.L.); National Institutes of Health CA230453, GM148662 and Case Comprehensive Cancer Center pilot grant P30CA043703 (to Y.Z.); R35NS116842 and R. Blane & Claudia Walter (to P.J.T.); National Multiple Sclerosis Society TA-2105-37619 (to B.L.L.C.); 2018/30/E/NZ1/00605 NCN, Poland (to D.K.). I.T. is a Canada Research Chair in Regulation of mRNA Translation and Metabolism. O.L. is supported by the Wallenberg Academy Fellow’s programme. F.W. is supported by an American Cancer Society Postdoctoral Fellowship (1144828). D.P. is supported by a Canadian Institutes for Health Research Postdoctoral Fellowship (MFE-171312) and the Cancer Research Society The Next Generation of Scientists Award. L.S.V. was supported by a Grant of Excellence in Basic Research (EXPRO 2019) provided by the Czech Science Foundation (19-25821X) and CZ.02.01.01/00/22_008/0004575 RNA for therapy by ERDF and MEYS. R.J. is a cancer research fellow for HFCR-24-03-03. M.M.A. was supported by National Institute of Health 5T34GM137792 under the supervision of M.H. Metabolic analysis was performed under the supervision of D. Avizonis at The Rosalind and Morris Goodman Cancer Research Institute’s Metabolomics Core Facility, which is supported by the Canada Foundation for Innovation, The Dr. John R. and Clara M. Fraser Memorial Trust, the Terry Fox Foundation (TFF-Oncometabolism Team Grant; TFF‐242122) and McGill University. Plasmids expressing the GADD34 C terminus or N terminus were provided by D. Ron.
Author information
Authors and Affiliations
Contributions
Conceptualization: C.-W.C., D.P., K.J.S., M.H., I.T. and O.L. Methodology: C.-W.C., D.P., K.J.S., M.H., O.L., I.T. and R.J. Software: K.J.S. and O.L. Validation: A.V., W.C.M., A.A.K., A.E.K., M.G., A.S., A.M., E.J., J.D.D., B.T., O.L. and I.T. Formal analysis: C.-W.C., D.P., K.J.S., D.K. and I.B. Investigation: C.-W.C., D.P., B.-J.G., L.S.V., M.A., J.W., R.J., M.M.A., D.K., M.J.P., F.W., B.L.L.C., P.J.T., C.V. and I.B. Data curation: K.J.S. and O.L. Writing: M.H., O.L. and I.T. Visualization: C.-W.C., D.P., K.J.S., D.K. and Y.Z. Supervision: M.H., O.L. and I.T. Project administration: M.H.
Corresponding authors
Ethics declarations
Competing interests
E.J. is an employee of and holds equity in Moderna Inc. All other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Assen Marintchev, Davide Ruggero and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Decreased eIF2B activity in the absence of stress-induced eIF2α-p reprograms the transcriptome in an eIF4E-dependent manner.
(a,b) Western blot analysis of the indicated proteins in cell extracts isolated from MEFs expressing control or Eif2b5 shRNAs (a,b); or active GADD34 (ΔNT, N-terminus truncated protein) or inactive GADD34 (ΔCT, C-terminus truncated protein)7 (b) and treated with Tg (400 nM) or Sodium Arsenite (1 mM) for specified times (a). Representative blots are shown (n=3 independent experiments). (c) Diagrams of 5′ UTR uORFs of mouse Atf410, Atf514, Gadd3415 and Chop16 mRNAs. The stop codon of uORF1 in the Gadd34 5′ UTR overlaps with the initiation codon of uORF2 by one nucleotide15. (d) Polysome profile tracings obtained by monitoring absorbance (254 nm) across the 10–50% sucrose gradients (left) in indicated cells. Distribution of Atf5 and Tuba1a mRNAs on polyribosomes (right) isolated from MEFs expressing control, Eif2b5, or Eif2b5 + Eif4e shRNAs, as indicated. Representative experiment is shown (n = 3 independent experiments). (e) (Top) A bar plot shows an overall number of reads that are assigned to genes after all pre-processing steps and summarization with htseq-count for each sample. (Middle) the number of detected genes from total RNA sequencing libraries (red) and polysome-associated RNA sequencing libraries (brown) across sampled sequencing depths. (Bottom) projection of samples in principal components 1 and 2, with samples shaped according to library type (circle: polysome-associated RNA; triangle: total RNA) and colored according to condition (control samples are grey, shEif2b5+shEif4e ivory and shEif2b5 orange). (f) RT-qPCR analysis of total RNA isolated from MEFs expressing shRNAs for Eif2b5, Eif2b5 + Eif4e or Control (shCon). RT-qPCR data for the indicated genes (Supplementary Table 1) were normalized to the values of the Gapdh mRNA. Statistical significance was determined using the two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 3 independent experiments). (g) Venn diagrams showing overlapping genes between indicated experimental conditions. Data set of Tg 16 h versus Con in MEFs were obtained from raw data analysis of a previous publication6. The experimental conditions and the number of genes in each experimental condition along with the mode of regulation are shown.
Extended Data Fig. 2 Co-depletion of eIF4E reverses expression of genes affected by eIF2Bε depletion that are enriched in overlapping biological processes.
(a) Venn diagrams show the number of overlapping genes from the indicated experimental conditions6,20. (b-e) Scatterplots of fold changes quantified by RNA sequencing data from total and polysome-associated RNA in MEFs expressing shEif2b5 versus shCon (b) and shEif2b5+shEif4e versus shEif2b5 (d). Cumulative distributions for log2 fold changes (log2FC) in mRNA abundance (c, e) of background (grey) and selected Atf4 gene signature20 (blue). Statistical analysis comparing Atf4 gene signature to background using Mann-Whitney U-test indicated p<0.001 for both comparisons (c, e). ATF4-regulated genes are indicated in blue20, (f) Heatmaps comparing the 20 most significant biological processes (gene ontology) enriched among genes upregulated upon eIF2Bε depletion versus those downregulated upon eIF2Bε and eIF4E co-depletion (g) Heatmaps comparing the 20 most significant biological processes (gene ontology) enriched among genes downregulated upon eIF2Bε depletion versus those upregulated upon eIF2Bε and eIF4E co-depletion. A full list of genes can be found in Supplementary Table 3.
Extended Data Fig. 3 Quality control of RNA sequencing data in mouse ES Cells from ΔuORF1 and Eif2b5R191H/R191H cells. Deficiency in oligodendrocyte differentiation in Eif2b5R132H/R132H OPCs.
(a, f) Bar plots showing the total number of reads that are assigned to genes after all pre-processing steps and summarization with htseq-count for each sample in Eif2b5R191H/R191H mouse ES Cells (a) and in ΔuORF1 mouse ES Cells (f). (b, g) Projection of samples in principal components 1 and 2, with samples shaped according to replicate and colored according to condition [control samples in red and Eif2b5R191H/R191H (b) and ΔuORF1(g) in blue]. (c, h) Number of detected genes from total RNA sequencing libraries across sampled sequencing depths in mouse ES Cells from Eif2b5R191H/R191H cells (c), and ΔuORF1 mouse ES Cells (h) colored in brown and controls colored in red. (d) GO enrichment analysis of upregulated (yellow) or downregulated (green) cohorts of genes in Eif2b5R191H/R191H versus WT mouse ES Cells RNA sequencing data shown in Fig. 3d. Number of genes identified in selected GO individual pathways are shown and all GO pathways are shown in Supplementary Table 4. FDR, False Discovery Rate. (e) (Left) iPSCs-derived OPCs from WT or Eif2b5R132H/R132H mice were differentiated for 72 h with thyroid hormone (T3) and stained for the intermediate oligodendrocyte marker protein O1 and late/mature oligodendrocyte marker protein, myelin basic protein (MBP). (Right) Quantification demonstrates dramatically lower mature oligodendrocytes in Eif2b5R132H/R132H cells. Data is presented as the mean +/− standard deviation. Outliers were statistically identified and removed using ROUT test with Q = 1%, and p-values were calculated using a Welch’s unpaired, two tailed t-test. (n = 3 independent experiments, ≥16 wells). Both outlier tests and p-value calculation were performed in GraphPad Prism v10.4.1.
Extended Data Fig. 4 Bioenergetic characterization of the mouse ES Cells harboring VWMD-associated eIF2Bε mutation (Eif2b5R191H/R191H).
(a) Western blot analysis of the indicated proteins in MEFs expressing control, Eif2b5, Eif4e or Eif2b5 + Eif4e shRNAs. Representative experiments (n = 3 independent experiments) are shown (Left two panels). Quantification of PCK2 protein levels [over α-tubulin or mitochondrial protein citrate synthase (CS)] in shRNA-treated MEFs (right two panels) (n = 4 independent experiments). p-values, two-tailed Student’s t-test. Error bars represent S.E.M values (Right panels). (b) Venn diagrams showing numbers of overlapping genes between the indicated experimental conditions. “Split-ISR eIF4E-dependent”: genes whose mRNA abundance or translation mode is increased by sheIF2Bε depletion and reversed by eIF4E co-depletion. “NE-mitos”: nuclear-encoded mitochondrial genes29. “NE-mitos split-ISR”: genes that overlap between the “Split-ISR eIF4E-dependent” and the “NE-mitos” categories. “ATF4 target genes” ATF4 regulated genes20. PCK2 is a nuclear encoded mitochondrial protein31. (c, d) Oxygen consumption rate (OCR, c) and extracellular acidification rate (ECAR, d) in WT and Eif2b5R191H/R191H cells treated with CPA (200 μM) or a vehicle for 16 h, followed by 48 h washing-out of CPA (n = 4 independent experiments). (e) Quantitation of ATP production rate from OXPHOS (J ATP ox) or glycolysis (J ATP glyc) in WT and Eif2b5R191H/R191H cells treated with a vehicle or CPA (200 μM) for 16 h, followed by 48 h washing-out of CPA (n = 4 independent experiments). Data are presented as mean ± SEM (c-e). (f) Bioenergetic plot of WT and Eif2b5R191H/R191H cells treated as described in panels c-e. Data points represent J ATP values (J ATP glyc on the x-axis and J ATP ox on the y-axis) measured under basal conditions (square), and following injections of FCCP (triangle), and Monensin (circle). p-values, two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 4 independent experiments).
Extended Data Fig. 5 uORF1 translation is required for ATF4 induction in s-ISR but is largely dispensable for c-ISR.
(a) Western blot analysis of the indicated proteins in cell extracts isolated from mouse ES Cells or from MEFs, treated with Tg (400 nM) for specified durations. Representative blots are shown (n = 3 independent experiments). (b) Sanger sequencing traces of genomic DNAs from WT and ΔuORF1 mouse ES Cells. Only the uORF1 region is shown. (c) Protein synthesis (incorporation of [35S]-cysteine and methionine into proteins) in ΔuORF1 mouse ES Cells treated with Tg (400 nM) for the indicated times. p-values, two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 5 independent experiments). (d) Polysome profile tracings obtained by monitoring absorbance (254 nm) across the 10-50% sucrose gradients from WT or ΔuORF1 mouse ES Cells expressing control (top panel) or eIF2Bε shRNA (middle panel), or treated with Tg (400 nM, 9 h) (bottom panel). Representative experiments are shown (n = 3 independent experiments). (e) Distribution of indicated mRNAs across polysome fractions (from d) isolated from WT or ΔuORF1 mouse ES Cells expressing control or Eif2b5 shRNAs, or treated with Tg (400 nM, 9 h) was determined by RT-qPCR. Representative experiments are shown (n = 3 independent experiments).
Extended Data Fig. 6 Loss of basal ATF4 levels in ΔuORF1 mouse ES Cells impairs cellular bioenergetics.
(a) Schematic of [3-13C]-pyruvate tracing in the citric acid cycle. Black-filled circles indicated [13C]-labelled carbons. Citrate m+1, Glutamine m+1, Succinate m+1, Malate m+1, and Aspartate m+1 refer to isotopomers of metabolites after incorporation of one [13C]-labelled carbon following one turn of the citric acid cycle. Citrate m+2, Glutamine m+2, Succinate m+2, Malate m+2, and Aspartate m+2, refer to isotopomers of metabolites after incorporation of two [13C]-labelled carbons after two turns of the citric acid cycle. (b, c) Fractional enrichment of the indicated metabolites (m+1, b and m+2, c) in WT and ΔuORF1 mouse ES Cells. p-values, two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 4 independent experiments). (d) Protein synthesis rates in WT and ΔuORF1 mouse ES Cells were measured using the incorporation of [35S]-cysteine and methionine. No statistical significance (n.s.) according to the two-tailed Student’s t-test (n = 3 independent experiments). (e) Differential expression analysis (DESeq2) of comparing mRNA expression quantified by RNA sequencing in WT versus ΔuORF1 mouse ES Cells. Genes belonging to the ATF4-regulated signature20 are shown in red. Selected differentially expressed genes are indicated in green. All regulated genes can be found in Supplementary Table 1. (f) Steady state levels of metabolites (serine, glycine, PEP) in ΔuORF1 mouse ES Cells compared to WT (n = 3 independent experiments). Data depicts fold change of ΔuORF1 mouse ES Cells relative to WT.
Extended Data Fig. 7 uORF1 protects Atf4 mRNA from degradation by nonsense-mediated decay, while uORF2 regulates ATF4 protein synthesis in c-ISR.
(a) RT-qPCR analysis of Atf4 mRNA levels in WT or ΔuORF1 mouse ES Cells expressing control or Eif2b5 shRNAs and treated with vehicle or Tg (400 nM) for specified times. Data were normalized to Gapdh mRNA values. p-values were determined using two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 3 independent experiments). (b, d-f, j) Parental WT or ΔuORF1 mouse ES Cells (b), or WT or ΔuORF1 mouse ES Cells expressing control or UPF1 shRNAs (d-f), or WT or ΔuORF1 mouse ES Cells treated with Tg (400 nM, 9 h) (j) were exposed to actinomycin D (10 μg/ml) for the indicated durations, to determine the half-life (t1/2) of the denoted mRNAs. mRNA levels were quantified by RT-qPCR analysis. Values were normalized to Gapdh mRNA and expressed as percentages of the levels before the addition of actinomycin D. t1/2 of mRNAs was calculated using exponential decay function. P-values; n.s., not significant evaluated by the two-tailed Student’s t-test (n = 3 independent experiments). (c,k) Western blot analysis of the indicated proteins in cell extracts isolated from WT or ΔuORF1 mouse ES Cells expressing control or Upf1 shRNAs (c) or WT and ΔuORF2 NIH3T3 cells treated with Tg (400 nM, 9 h) (k). Representative blots are shown (n = 3 independent experiments). The cartoon in (k) indicates CRSPR-Cas9 mutation in the Atf4 gene introduced in the initiation codon of uORF2. (g-i) RT-qPCR analysis of the indicated mRNAs in mouse ES Cells expressing control (shCon) or Upf1 (shUpf1) shRNAs. P-values, two-tailed Student’s t-test. Data are presented as mean ± SEM (n = 3 independent experiments). (l) Model for regulation of Atf4 mRNA translation and mRNA decay of the Atf4 mRNA via the functions of uORF1: (i) In the absence of stress, eIF4E-mediated translation of uORF1 in part inhibits translation of uORF2 and stabilizes Atf4 mRNA. (ii) When eIF2B activity is decreased in the absence of stress, translation of the Atf4 main ORF increases. Decreased uORF2 translation initiation and translation of the main Atf4 ORF, protects the Atf4 mRNA from NMD. (iii) Under chronic ER stress, Atf4 main ORF translation is independent of eIF4E and dependent on eIF3d6. The ribosomes (40S in red and 60S in yellow) are shown translating the mRNA as an 80S or as recycling/scanning units. The engagement of NMD via the translation of uORF2 is depicted as the mechanism of regulation of Atf4 mRNA abundance (indicated by the black lines). The degradation symbol has been sized as per the expected outcome of Atf4 mRNA decay under each condition.
Extended Data Fig. 8 Distinct mechanisms of regulation of c-ISR and s-ISR in response to inhibitors of eIF4E or CK2 activity.
(a) Representative western blot analysis of the indicated proteins in ΔuORF1 and WT mouse ES Cells treated with the inhibitor of PP1 phosphatase (containing GADD34 as a subunit), Salubrinal (15 μM, 5 h; (n = 3 independent experiments). (b,f) Western blot analysis of the indicated proteins in MEFs expressing Con (shCon) or Eif2s2 (shEif2s2) shRNAs and treated with Tg (400 nM) (b) or Torin1 (250 nM) (f) for the specified times. Representative blots are shown (n = 3 independent experiments). (c-d) Representative western blot analyses of the indicated proteins in MEFs treated with Tg (400 nM) for the specified durations, or MEFs expressing the indicated shRNAs, without (c) or with Tg-treatment (d) (n = 3 independent experiments). Tg-treated MEFs in (c) were used as a positive control for induction of the c-ISR. (e) Representative western blot analysis of the indicated proteins in MEFs expressing control (shCon) or Eif2b5 shRNA (shEif2b5) and treated with (Tg-400 nM) or Torin1 (250 nM) of the specified durations (n = 3 independent experiments). (g-h) Western blot analysis for the indicated proteins in MEFs expressing control (shCon) or Eif2b5 shRNA (shEif2b5) and treated with 4EGI-1 (200 μM) (g) Tg (400 nM) or SGC-CK2-1 (5 μM) (h) for the specified times. Representative western blots are shown (n = 2 independent experiments). Lower eIF2α-p levels are consistent with GADD34 induction (h, 4th lane). (i) Representative western blot analyses of the indicated proteins in MEFs treated with glucose (0 or 2.5 mM, 16 h). LiCl (10 or 25 mM) or Torin1 (250 nM) were used for the specified final hours of the 16 h treatment with medium containing 2.5 mM glucose (n = 3 independent experiments).
Extended Data Fig. 9 S-ISR is induced in Herceptin-resistant but not Herceptin-sensitive human breast cancer cells in response to ER stress. Schematic of the model contrasting s-ISR versus c-ISR.
(a) Relative proliferation rates (estimated by CellTiter-Glo) of Herceptin-sensitive parental (BT474-P, upper) and Herceptin-resistant (BT474-R, lower) human breast cancer cells, treated with Herceptin (20 μg/ml) for the indicated times (n = 3 independent experiments). (b) eIF2B GEF activity was measured in BT474-P and BT474-R cells treated with CPA (100 μM) for the specified durations. Statistical significance in (a,b) was determined by the two-tailed Student’s t-test. n.s., not significant evaluated by the two-tailed Student’s t-test (n = 3 independent experiments). Data are presented as mean ± SEM (n = 3 independent experiments). (c-f) Western blot analysis of the denoted proteins in BT474-P and BT474-R cells treated with CPA (100 μM) and/or control (shCon), EIF4E (shEIF4E) or DDX3X (shDDX3X) shRNAs as indicated. Torin1 was added for the last hour of treatment with CPA for the indicated concentrations and durations (e). Representative western blots are shown (n = 3 independent experiments). (g) Schematics contrasting “split” (s-) and “canonical” (c-) ISR. (Left) S-ISR: Decreased eIF2B activity in the absence of stress-induced eIF2α-p. S-ISR translational reprogramming comprises only a subset of the c-ISR targets including ATF4, but not GADD34. The mechanism of ATF4 mRNA translational control is highly dependent on uORF1 but eIF2α-p independent (purple). (Right) ER stress induces PERK-mediated eIF2α-p which decreases eIF2B activity and strongly suppresses global protein synthesis. This is followed by induction of the ATF4-GADD34 axis, which dephosphorylates eIF2α-p and establishes the chronic c-ISR program. The levels of eIF2α-p control the amplitude of adaptive mRNA translation via mechanisms that include translation of uORF-containing mRNAs (e.g., ATF4 and GADD34-orange). In contrast to s-ISR, the c-ISR is less dependent on the function of eIF4E.
Extended Data Fig. 10 S-ISR gene expression alterations are distinct from those observed during other stress conditions.
(a-c) (Left), Scatter plots from anota2seq analysis of shEif2b5 versus control cells (left panels) wherein genes whose expression was identified as induced (orange) or suppressed (red) following (a) ER stress (polysome-profiling)43, (b) mitochondrial stress (proteomics)44 or (c) oxygen and glucose deprivation (ribosome profiling)42 are indicated together with genes showing unaltered expression (background in grey). (Middle and Right) empirical cumulative distribution function (ECDF) plots of log2fold-changes (log2FC) from the sheIF2Bε versus control comparison for polysome-associated mRNA (middle panels) and total RNA (right panels) for stress-sensitive gene sets and background genes (grey). Significant shifts in the distribution of gene sets relative to the background were assessed using a two sided Wilcoxon rank-sum test and p-values are indicated.
Supplementary information
Supplementary Information
Supplementary Notes (Introduction and Discussion) and References.
Supplementary Fig. 1
Uncropped and unprocessed scans of all western blots: Figs. 1b,f, 2r, 3c,e and 4a,c,l and Extended Data Figs. 1a,b, 4a, 5a, 7c,k, 8a–i and 9c–f.
Supplementary Table 1
Fold changes of gene expression obtained from RNA-sequencing data in WT and eIF2Bε-depleted MEFs and mouse ES cells (WT or mutant).
Supplementary Table 2
Pathway enrichment analysis (GO) based on fold changes of gene expression obtained from RNA-sequencing data in WT and eIF2Bε-depleted MEFs.
Supplementary Table 3
Top enriched biological processes (by FDR) identified among genes whose RNA level was affected by eIF2Bε depletion or co-depletion of eIF2Bε and eIF4E.
Supplementary Table 4
Pathway enrichment analysis (GO) based on fold changes of gene expression obtained from RNA-sequencing data in WT and VWMD mutant mouse ES cells.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, CW., Papadopoli, D., Szkop, K.J. et al. Plasticity of the mammalian integrated stress response. Nature 641, 1319–1328 (2025). https://doi.org/10.1038/s41586-025-08794-6
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-08794-6
This article is cited by
-
Nucleophosmin supports WNT-driven hyperproliferation and tumor initiation
Nature Genetics (2026)
-
Prime editing links the split integrated stress response to pathogenic eIF2B mutations and white matter degeneration
Cell Death & Disease (2025)