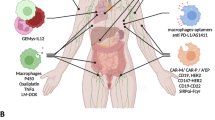
Abstract
Monocyte-derived macrophages (mo-macs) often drive immunosuppression in the tumour microenvironment (TME)1 and tumour-enhanced myelopoiesis in the bone marrow fuels these populations2. Here we performed paired transcriptome and chromatin accessibility analysis over the continuum of myeloid progenitors, circulating monocytes and tumour-infiltrating mo-macs in mice and in patients with lung cancer to identify myeloid progenitor programs that fuel pro-tumorigenic mo-macs. We show that lung tumours prime accessibility for Nfe2l2 (NRF2) in bone marrow myeloid progenitors as a cytoprotective response to oxidative stress, enhancing myelopoiesis while dampening interferon response and promoting immunosuppression. NRF2 activity is amplified during monocyte differentiation into mo-macs in the TME to regulate stress and drive immunosuppressive phenotype. NRF2 genetic deletion and pharmacological inhibition significantly reduced the survival and immunosuppression of mo-macs in the TME, restoring natural killer and T cell anti-tumour immunity and enhancing checkpoint blockade efficacy. Our findings identify a targetable epigenetic node of myeloid progenitor dysregulation that sustains immunoregulatory mo-macs in the lung TME and highlight the potential of early interventions to reprogram macrophage fate for improved immunotherapy outcomes.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
Accession numbers for re-analysed published datasets are listed in Supplementary Table 7. Processed matrix files and metadata for the mouse tissue scRNA-seq and scATAC-seq generated in this study are made publicly available on GEO (GSE270148). Processed matrix files and metadata for human tissue scATAC-seq and 10x Multiome data generated in this study are also made publicly available at the time of publication on GEO (GSE270148). Source data are provided with this paper.
Code availability
Notable software package versions are listed in the Methods and in Supplementary Table 7. No new pipelines were used in the study beyond those described in the relevant Methods sections. Exemplar data objects and code for notable analyses in this study are provided at github.com/Merad-Lab/Hegde_Myelopoiesis_Epigenetics. Any additional information required to interpret data reported in this paper is available from the corresponding author upon request.
References
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382 (2019).
Swann, J. W., Olson, O. C. & Passegue, E. Made to order: emergency myelopoiesis and demand-adapted innate immune cell production. Nat. Rev. Immunol. 24, 596–613 (2024).
Goswami, S., Anandhan, S., Raychaudhuri, D. & Sharma, P. Myeloid cell-targeted therapies for solid tumours. Nat. Rev. Immunol. 23, 106–120 (2023).
Sica, A., Guarneri, V. & Gennari, A. Myelopoiesis, metabolism and therapy: a crucial crossroads in cancer progression. Cell Stress 3, 284–294 (2019).
Giles, A. J. et al. Activation of hematopoietic stem/progenitor cells promotes immunosuppression within the pre-metastatic niche. Cancer Res. 76, 1335–1347 (2016).
Casbon, A. J. et al. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc. Natl Acad. Sci. USA 112, E566–E575 (2015).
Wu, W. C. et al. Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients. Proc. Natl Acad. Sci. USA 111, 4221–4226 (2014).
Porembka, M. R. et al. Pancreatic adenocarcinoma induces bone marrow mobilization of myeloid-derived suppressor cells which promote primary tumor growth. Cancer Immunol. Immunother. 61, 1373–1385 (2012).
Trzebanski, S. et al. Classical monocyte ontogeny dictates their functions and fates as tissue macrophages. Immunity 57, 1225–1243 (2024).
Ikeda, N. et al. The early neutrophil-committed progenitors aberrantly differentiate into immunoregulatory monocytes during emergency myelopoiesis. Cell Rep. 42, 112165 (2023).
LaMarche, N. M. et al. An IL-4 signalling axis in bone marrow drives pro-tumorigenic myelopoiesis. Nature 625, 166–174 (2024).
Hao, X. et al. Osteoprogenitor–GMP crosstalk underpins solid tumor-induced systemic immunosuppression and persists after tumor removal. Cell Stem Cell 30, 648–664 (2023).
Gerber-Ferder, Y. et al. Breast cancer remotely imposes a myeloid bias on haematopoietic stem cells by reprogramming the bone marrow niche. Nat. Cell Biol. 25, 1736–1745 (2023).
Dey, S., Curtis, D. J., Jane, S. M. & Brandt, S. J. The TAL1/SCL transcription factor regulates cell cycle progression and proliferation in differentiating murine bone marrow monocyte precursors. Mol. Cell. Biol. 30, 2181–2192 (2010).
Pham, T. H. et al. Dynamic epigenetic enhancer signatures reveal key transcription factors associated with monocytic differentiation states. Blood 119, e161–e171 (2012).
Mandula, J. K. & Rodriguez, P. C. Tumor-related stress regulates functional plasticity of MDSCs. Cell Immunol. 363, 104312 (2021).
Paul, F. et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell 163, 1663–1677 (2015).
Kwart, D. et al. Cancer cell-derived type I interferons instruct tumor monocyte polarization. Cell Rep. 41, 111769 (2022).
Alicea-Torres, K. et al. Immune suppressive activity of myeloid-derived suppressor cells in cancer requires inactivation of the type I interferon pathway. Nat. Commun. 12, 1717 (2021).
Kwak, H. J. et al. Myeloid cell-derived reactive oxygen species externally regulate the proliferation of myeloid progenitors in emergency granulopoiesis. Immunity 42, 159–171 (2015).
Pizzato, H. A. et al. Mitochondrial pyruvate metabolism and glutaminolysis toggle steady-state and emergency myelopoiesis. J. Exp. Med. 220, e20221373 (2023).
Pietras, E. M. et al. Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J. Exp. Med. 211, 245–262 (2014).
Molgora, M. et al. TREM2 modulation remodels the tumor myeloid landscape enhancing anti-PD-1 immunotherapy. Cell 182, 886–900 (2020).
Katzenelenbogen, Y. et al. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell 182, 872–885 (2020).
Matusiak, M. et al. Spatially segregated macrophage populations predict distinct outcomes in colon cancer. Cancer Discov. 14, 1418–1439 (2024).
Mulder, K. et al. Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54, 1883–1900 (2021).
McGinnis, C. S. et al. The temporal progression of lung immune remodeling during breast cancer metastasis. Cancer Cell 42, 1018–1031 (2024).
Beury, D. W. et al. Myeloid-derived suppressor cell survival and function are regulated by the transcription factor Nrf2. J. Immunol. 196, 3470–3478 (2016).
Namgaladze, D., Fuhrmann, D. C. & Brune, B. Interplay of Nrf2 and BACH1 in inducing ferroportin expression and enhancing resistance of human macrophages towards ferroptosis. Cell Death Discov. 8, 327 (2022).
Kobayashi, E. H. et al. Nrf2 suppresses macrophage inflammatory response by blocking proinflammatory cytokine transcription. Nat. Commun. 7, 11624 (2016).
Ryan, D. G. et al. Nrf2 activation reprograms macrophage intermediary metabolism and suppresses the type I interferon response. iScience 25, 103827 (2022).
Olagnier, D. et al. Nrf2 negatively regulates STING indicating a link between antiviral sensing and metabolic reprogramming. Nat. Commun. 9, 3506 (2018).
Ting, K. K. Y. et al. Oxidized low-density lipoprotein accumulation suppresses glycolysis and attenuates the macrophage inflammatory response by diverting transcription from the HIF-1alpha to the Nrf2 pathway. J. Immunol. 211, 1561–1577 (2023).
Park, M. D. et al. TREM2 macrophages drive NK cell paucity and dysfunction in lung cancer. Nat. Immunol. 24, 792–801 (2023).
Alaluf, E. et al. Heme oxygenase-1 orchestrates the immunosuppressive program of tumor-associated macrophages. JCI Insight 5, e133929 (2020).
Leader, A. M. et al. Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 39, 1594–1609 (2021).
Hu, J. et al. Tumor microenvironment remodeling after neoadjuvant immunotherapy in non-small cell lung cancer revealed by single-cell RNA sequencing. Genome Med. 15, 14 (2023).
Zelenay, S. et al. Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270 (2015).
Taniguchi, S. et al. In vivo induction of activin A-producing alveolar macrophages supports the progression of lung cell carcinoma. Nat. Commun. 14, 143 (2023).
Gomez-Chou, S. B. et al. Lipocalin-2 promotes pancreatic ductal adenocarcinoma by regulating inflammation in the tumor microenvironment. Cancer Res. 77, 2647–2660 (2017).
Li, Z. et al. Proinflammatory S100A8 induces PD-L1 expression in macrophages, mediating tumor immune escape. J. Immunol. 204, 2589–2599 (2020).
Uccellini, M. B. & Garcia-Sastre, A. ISRE-reporter mouse reveals high basal and induced type I IFN responses in inflammatory monocytes. Cell Rep. 25, 2784–2796 (2018).
Singh, A. et al. Small molecule inhibitor of NRF2 selectively intervenes therapeutic resistance in KEAP1-deficient NSCLC tumors. ACS Chem. Biol. 11, 3214–3225 (2016).
Schaer, D. J. et al. Hemorrhage-activated NRF2 in tumor-associated macrophages drives cancer growth, invasion, and immunotherapy resistance. J. Clin. Invest. 134, e174528 (2023).
Liu, Z. et al. Fate mapping via Ms4a3-expression history traces monocyte-derived cells. Cell 178, 1509–1525 (2019).
Ren, D. et al. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. Proc. Natl Acad. Sci. USA 108, 1433–1438 (2011).
Perrone, M. et al. ATF3 reprograms the bone marrow niche in response to early breast cancer transformation. Cancer Res. 83, 117–129 (2023).
Zhang, M. et al. Selective activation of STAT3 and STAT5 dictates the fate of myeloid progenitor cells. Cell Death Discov. 9, 274 (2023).
Laurenti, E. et al. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell 3, 611–624 (2008).
Villar, J. et al. ETV3 and ETV6 enable monocyte differentiation into dendritic cells by repressing macrophage fate commitment. Nat. Immunol. 24, 84–95 (2023).
Ratajczak, M. Z. & Kucia, M. Hematopoiesis and innate immunity: an inseparable couple for good and bad times, bound together by an hormetic relationship. Leukemia 36, 23–32 (2022).
Ng, M. S. F. et al. Deterministic reprogramming of neutrophils within tumors. Science 383, eadf6493 (2024).
Zhao, Y. et al. Neutrophils resist ferroptosis and promote breast cancer metastasis through aconitate decarboxylase 1. Cell Metab. 35, 1688–1703 (2023).
Garner, H. et al. Understanding and reversing mammary tumor-driven reprogramming of myelopoiesis to reduce metastatic spread. Cancer Cell 43, 1279–1295 (2025).
Daman, A. W. et al. Microbial cancer immunotherapy reprograms hematopoiesis to enhance myeloid-driven anti-tumor immunity. Cancer Cell https://doi.org/10.1016/j.ccell.2025.05.002 (2025).
Singh, A. et al. NRF2 activation promotes aggressive lung cancer and associates with poor clinical outcomes. Clin. Cancer Res. 27, 877–888 (2021).
Chirnomas, D., Hornberger, K. R. & Crews, C. M. Protein degraders enter the clinic—a new approach to cancer therapy. Nat. Rev. Clin. Oncol. 20, 265–278 (2023).
Mohamed, E. et al. The unfolded protein response mediator PERK governs myeloid cell-driven immunosuppression in tumors through inhibition of STING signaling. Immunity 52, 668–682 (2020).
Raines, L. N. et al. PERK is a critical metabolic hub for immunosuppressive function in macrophages. Nat. Immunol. 23, 431–445 (2022).
Kress, J. K. C. et al. The integrated stress response effector ATF4 is an obligatory metabolic activator of NRF2. Cell Rep. 42, 112724 (2023).
Boumelha, J. et al. An immunogenic model of KRAS-mutant lung cancer enables evaluation of targeted therapy and immunotherapy combinations. Cancer Res. 82, 3435–3448 (2022).
Jackson, E. L. et al. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65, 10280–10288 (2005).
Bankhead, P. et al. QuPath: open source software for digital pathology image analysis. Sci. Rep. 7, 16878 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Martin, J. C. et al. Single-cell analysis of Crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell 178, 1493–1508 (2019).
Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Calcagno, D. M. et al. The myeloid type I interferon response to myocardial infarction begins in bone marrow and is regulated by Nrf2-activated macrophages. Sci. Immunol. 5, eaaz1974 (2020).
Agrawal, A. et al. WikiPathways 2024: next generation pathway database. Nucleic Acids Res. 52, D679–D689 (2024).
Crowell, H. L. et al. muscat detects subpopulation-specific state transitions from multi-sample multi-condition single-cell transcriptomics data. Nat. Commun. 11, 6077 (2020).
Granja, J. M. et al. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 53, 403–411 (2021).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Schep, A. N., Wu, B., Buenrostro, J. D. & Greenleaf, W. J. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017).
Yoshida, H. et al. The cis-regulatory atlas of the mouse immune system. Cell 176, 897–912 (2019).
McLean, C. Y. et al. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 (2010).
Ma, S. et al. Chromatin potential identified by shared single-cell profiling of RNA and chromatin. Cell 183, 1103–1116 (2020).
Kartha, V. K. et al. Functional inference of gene regulation using single-cell multi-omics. Cell Genom. 2, 100166 (2022).
Lee, J. J. Early transcriptional effects of inflammatory cytokines reveal highly redundant cytokine networks. J. Exp. Med. 222, e20241207 (2025).
Chen, Y. et al. Spatiotemporal single-cell analysis decodes cellular dynamics underlying different responses to immunotherapy in colorectal cancer. Cancer Cell 42, 1268–1285 (2024).
Bassez, A. et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 27, 820–832 (2021).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Gayoso, A. et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40, 163–166 (2022).
Bravo Gonzalez-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
McDavid, A. et al. Data exploration, quality control and testing in single-cell qPCR-based gene expression experiments. Bioinformatics 29, 461–467 (2013).
Acknowledgements
M.M. was supported by National Institutes of Health (NIH) grants CA257195, CA254104 and CA154947. S.H. was supported by the National Cancer Institute (NCI) fellowship K00CA223043. B.Y.S. was supported by NIH Medical Scientist Training grant T32GM146636. R.M. was supported by the AACR-AstraZeneca Immuno-oncology fellowship 21-40-12-MATT. This work was supported by grant no. R24-072073 to the Immunological Genome Project (ImmGen) consortium. We thank the patients and their families for participating in the clinical study; members of the Merad laboratory and Brown laboratory at the Marc and Jennifer Lipschultz Precision Immunology Institute at Mount Sinai for insightful discussions and feedback; and E. Bernstein, D. Hasson and D. Filipescu for their technical advice and feedback. We acknowledge the Human Immune Monitoring Center, the Mount Sinai Biorepository and Pathology Core and the Mount Sinai Cytometry Core for extensive support and resources. This work was supported in part through the computational resources and staff expertise provided by the Bioinformatics for Next Generation Sequencing (BiNGS) Shared Resource Facility funded by NCI P30 Cancer Center and NIH/NIAMS P30 Skin Biology Disease Resource-based Center support grants. This work was also supported in part through the computational and data resources and staff expertise provided by Scientific Computing and Data at the Icahn School of Medicine at Mount Sinai and supported by the Clinical and Translational Science Awards (CTSA) grant UL1TR004419, and the Office of Research Infrastructure award S10OD026880 and S10OD030463. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.H. and M.M. Methodology: S.H., B.G., S.M., J.D.B., A.M.T., B.D.B. and M.M. Acquisition and analysis of data: S.H., J.L.B., R.M., M.D.P., A. Marks, M.B, P.H., T.C., K.N., L.T., K.A. and G.C. Computational investigation of data: S.H., B.G., B.Y.S., L.H., M.M.S., B.K., M.D.P., A. Magen, D.D., K.F., J.J.L., D.A. and M.J.B. Writing—original draft: S.H. Writing—review and editing: S.H. and M.M. Resources: S.K.-S., R.M.F., A.J.K., F.G., J.D.B., S.Z.J., T.U.M., B.D.B. and M.M. Funding and supervision: M.M.
Corresponding author
Ethics declarations
Competing interests
M.M. serves on the scientific advisory board and holds stock from Compugen Inc., Dynavax Inc., Innate Pharma Inc., Morphic Therapeutics, Asher Bio Inc., Dren Bio Inc., Nirogy Inc., Genenta Inc., Oncoresponse, Inc. and Owkin Inc. M.M. also serves on the ad hoc scientific advisory board of DBV Technologies Inc. and Genentech Inc., and on the foundation advisory board of Breakthrough Cancer. M.M. receives funding for contracted research from Genentech, Regeneron and Boehringer Ingelheim. T.U.M. has served on advisory and/or data safety monitoring boards for Rockefeller University, Regeneron Pharmaceuticals, Abbvie, Bristol-Meyers Squibb, Boehringer Ingelheim, Atara, AstraZeneca, Genentech, Celldex, Chimeric, Glenmark, Simcere, Surface, G1 Therapeutics, NGMbio, DBV Technologies, Arcus and Astellas, and receives contracted grants from Regeneron, Bristol-Myers Squibb, Merck and Boehringer Ingelheim. The above interests are not directly relevant to this manuscript. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature thanks Tim Greten, Renato Ostuni and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Lung cancer promotes changes in the chromatin state of BM myeloid progenitors.
a, Averaged heatmap for abundance of LT-HSCs, progenitors, and mature populations in bone marrow (BM) of KP tumor-bearing mice at different timepoints, normalized to tumor-naïve mice (left to right, n = 10,7,12,12). Pooled from two independent experiments. b, Granulocytic-monocytic colony forming units (CFU-GM) relative to erythroid blasts (BFU-E) after 8 days of incubation. n = 4 replicates. Pooled from two independent experiments. c, Longitudinal expansion of sorted GMPs from naïve and KP tumor-bearing mice. n = 3 replicates from one experiment. d, Number of Ly6Chi monocytes and Ly6Ghi neutrophils in blood of naïve and KP tumor-bearing mice at different time points, correlated with tumor burden. Data are individual data points with confidence intervals for linear regression (left to right, n = 5,7,12,12). Pooled from two independent experiments. e, Number of GMPs, cMoPs, and CD157+ Ly6Chi monocytes in BM, and number of Ly6Chi monocytes in blood of KP GEMM at 12 weeks post tumor initiation, compared to tumor-free mice. n = 3 mice per group, representative of two independent experiments. f, scRNA-seq heatmap of per-cell UMI counts across indicated myeloid cell subclusters in BM of tumor-bearing and naïve mice. Pooled over n = 3 mice per group. g, Gene ontology (GO) terms enriched in KP tumor-bearing mouse GMPs (left) and cMoPs (right) compared to naïve counterparts. Curated terms arranged by adjusted p-value (log q-value). h, Gene ontology (GO) terms enriched in PyMT tumor-bearing mouse GMPs compared to naïve counterparts from Gerber-Ferder et al.13 (left) and Hao et al. 2023 (right). Curated terms arranged by adjusted p-value (log q-value). i, exemplar scATAC-seq UMAP and heatmap of column-normalized gene scores across indicated myeloid cell subclusters in BM of tumor-bearing and naïve mice. Pooled over n = 3 mice per group. j, scATAC-seq heatmap of normalized transcription factor (TF) motif accessibility enrichment in marker peak regions of indicated myeloid cell states. Pooled from n = 3 mice per group. k, GREAT pathway terms enriched in indicated H3K4me3 regions from BM GMPs and Ly6Chi monocytes of KP tumor-bearing mice (Fig. 1f). Curated terms arranged by adjusted p-value (q-value). l–m, H3K4me1, H3K27ac, and H3K27me3 CUT&RUN signal clustering (l) and relative DORC scores for exemplar genes (m) in KP tumor-associated GMP relative to naïve GMP. n, Mean difference of ChromVAR-scores for TFs enriched in tumor-associated H3K4me3 signal versus H3K4me1 (left) and H3K27ac (right). o, scRNA-seq per-cell gene expression heatmap scaled across indicated myeloid subclusters in peripheral blood of human patients with NSCLC. Pooled from n = 4 patients. p, GREAT pathway terms enriched in indicated differentially accessible cisTopic region #5 of CD14+ monocytes from blood of patients with NSCLC (Fig. 1j). q, ChIP-X Enrichment Analysis (ChEA) calculated TF regulators for differentially expressed genes in CD14+ monocytes from blood of patients with NSCLC (n = 3) compared to healthy donors (n = 2). Dot color indicate major known biological pathways. Statistics computed by two-tailed Welch’s t-test (b), unpaired t-test with Holm-Sidak multiple comparison (c), unpaired two-tailed Student’s t-test (e), hypergeometric test with multiple test correction (g,h,k,j,p,q). error bars represent mean +/– SEM (c), colorscale indicates significance of motif enrichment based on hypergeometric test (j).
Extended Data Fig. 2 Representative gating strategies for mouse and human experiments.
a, Representative gating strategy for mouse bone marrow stem cell, progenitors, and differentiated monocyte and neutrophil populations. b, Gating strategy for mouse blood myeloid populations, and CD157+ phenotype of Ly6Chi monocytes. c, Representative plots for GFP in tumor-naïve and GFP-expressing KP-burdened bone marrow (BM) and lung. d, Representative broad sorting strategy for myeloid populations in mouse BM and blood. e, Representative gating strategy for human blood stem cell, progenitors, and myeloid populations. f, Representative sorting strategy for mouse BM GMPs in adoptive transfer experiments. g, Representative broad sorting strategy for myeloid populations in mouse lung. LT-HSC, long-term hematopoietic stem cell. MPP3, multipotent progenitor 3. LMPP, lymphoid-myeloid primed multipotent progenitors. CMP, common myeloid progenitor. MDP, myeloid dendritic cell progenitor. MEP, megakaryocyte-erythroid progenitor. GMP, granulocytic-monocytic progenitor. GP, committed granulocytic progenitor. cMoP, common monocytic progenitor.
Extended Data Fig. 3 Tumor-induced changes in BM progenitors impact mo-mac fate and function in TME.
a, Representative gating strategy for mouse lung myeloid populations. AM, alveolar macrophages. DC, dendritic cells. mo-mac, monocyte-derived macrophages. b, Representative plots for intratumoral TREM2hi mo-macs and CD86+MHCII+ mo-macs, overlaid on phenotypic markers Arg1, CD206, PDL1, CD73, CD11c. c, CD64+MERTK+ mo-macs differentiated from donor CD45.2+ GMPs of naïve or tumor-primed origin (related to Fig. 2a). d, Frequency of BMDMs expressing Arg1, PDL1, MHCII, CD86 upon in vitro differentiation with KP tumor conditioning early (48 hr) or no conditioning (no prime). n = 4 per group. e, Frequency of BMDMs expressing Arg1, MHCII, CD86, and Arg1hi mo-macs expressing PDL1 upon in vitro differentiation with KP tumor conditioning early (48 hr prime) followed by washout. n = 3 per group. f, Frequency of CellROX-, LiperFluo- (n = 5 per group), and MitoSOX-expressing Ly6Chi monocytes (n = 3 per group) in KP tumor-bearing lung at day 21. g, Median Fluorescence intensity (MFI) for CellROX in KP tumor-infiltrating mo-macs. n = 4 per group. h, Representative cytometry plots and frequency of NRF2+ and HO-1+ cells in KP tumor-infiltrating mo-mac subsets. n = 4 per group. i, NRF2 downstream gene activation per-patient score in lung-infiltrating myeloid cells from independent cohorts of NSCLC (Leader et al., n = 35 and Hu et al. n = 15 patients). j, Frequency of BMDMs expressing NRF2 and HO-1 after 48 h prime and washout of KP CM, related to Extended Data Fig. 3e, n = 3 per group. k, Frequency of intratumoral donor mo-macs expressing NRF2 and HO-1 four days after GMP transfer from naïve (n = 3) or tumor-primed (n = 5) origin into congenic tumor-bearing hosts (related to Fig. 2a). One experiment. l, Browser plots for H3K4me3 signal at known NRF2-associated gene loci in KP tumor-associated and naïve GMPs. Highlighted regions indicate differential signal near known cis-regulatory elements and open chromatin (OCR) containing Nfe2l2 motif. Individual data points with bar denoting mean, representative of two independent experiments. Statistics computed by one-way ANOVA with Tukey’s multiple comparison (d), one-way ANOVA with Dunnett’s multiple comparison (i), and unpaired two-tailed Student’s t-test (e)–(h),(j)–(k).
Extended Data Fig. 4 Multiomic analyses of mouse and human lung cancer-infiltrating myeloid cells.
a, scRNA-seq heatmap of per-cell UMI counts (left) and relative abundance (right) of myeloid sub-clusters in lung tissue of naïve and KP tumor-bearing mice. n = 3 pooled. b, Normalized UCell score for gene module# M24 across KP lung tumor-infiltrating myeloid cell clusters (left), with ENCODE/ChEA calculated TF regulators (right). c, exemplar scATAC-seq UMAP and heatmap of column-normalized gene scores across indicated myeloid cell clusters in lung of naïve and tumor-bearing mice. n = 4 pooled. d, scATAC-seq heatmap of normalized transcription factor (TF) motif accessibility enrichment in marker peak regions of indicated myeloid cell states. n = 4 pooled. e, snRNA-seq heatmap of per-cell UMI counts for indicated myeloid clusters in human NSCLC primary lung tumors. n = 5 patients pooled. f, UMAP of myeloid cells in snRNA-seq data from human NSCLC primary lung tumors annotated by marker genes (left, n = 5 pooled). Relative frequency of WCGNA gene programs (colors) in indicated clusters of macrophage-subsetted dataset (right). AM, alveolar macrophages. IM, interstitial macrophages. g–h, scATAC-seq heatmap depicting column-normalized gene scores (g) and normalized TF motif accessibility enriched in marker peaks (h) of indicated myeloid clusters in tumors of patients with NSCLC. n = 14 patients pooled. i, Candidate TF regulators in KP tumor-infiltrating monocyte and mo-mac clusters, prioritized by maximum TF motif deviation (Δ) across clusters. Dot color indicate known curated biological pathways. j, Reactome pathway terms enriched for conserved TF regulators from mouse and human analysis, ranked by adjusted p-value (log q-value). Red color indicates association to stress-associated cytoprotective signaling. Colorscale indicates significance of motif enrichment based on hypergeometric test (d,h).
Extended Data Fig. 5 NRF2 signaling in tumor-infiltrating monocytic lineage dampens Type I IFN responsiveness.
a, Frequency of NRF2+ Ly6Chi monocytes in BM (left), Ly6Chi monocytes in BM (mid), and frequency of NRF2+ mo-macs in lung (right) of KP GEMM at 12 weeks post tumor initiation, compared to tumor-free mice, n = 3 per group. b, ChromVAR-computed TF motif deviation for Nfe2l2 in tumor condition relative to naïve condition (left) and UCell-computed NRF2 downstream gene activation score (right) across indicated myeloid populations. c, Frequency of NRF2+ cells (top, n = 5 per group) and GFP+ cells (bottom, n = 4 per group) within indicated myeloid cell types from tumor-bearing and naïve (n = 4) Mx1GFP mice. d, Frequency of GFP+ cells within CD64+MERTK+ mo-macs of indicated phenotype in tumor-bearing Mx1GFP mice, n = 4 per group. e, Browser plots at indicated gene loci for H3K27ac signal in KP tumor-associated and naïve GMPs. Highlighted regions indicate differential signal near known cis-regulatory elements and open chromatin (OCR) containing Nfe2l2 motif. Unless noted, data shown as individual data points with bar denoting mean, representative of two independent experiments (a,c,d). Statistics computed by unpaired two-tailed Student’s t-test (a,d), and unpaired t-test with Holm-Sidak multiple comparison (c).
Extended Data Fig. 6 Mitogenic tumor cues can elicit NRF2 activation in BM and TME to support myeloid expansion.
a, Frequency of HO-1+, Arg1+, MHCII+, and CD86+ BMDMs after culture with >3 kDa KP CM protein retentate fraction, and <3 kDa KP CM deproteinated fraction. n = 3 per group. b, Frequency of HO-1+, Arg1+, MHCII+, and CD86+ BMDMs after culture with heat-inactivated KP CM. N = 4 per group. c, Protein abundance of indicated analytes in tumor-associated BM sera, represented as relative fold-change over naïve sera. n = 2 per condition, one experiment. d–e, Analytes upregulated in BM sera (d) and blood sera (e) of tumor-bearing mice compared to naïve mice as assessed by O-link, with relative concentrations of GM-CSF and IL-6 (box). n = 3 per group, one experiment. f, Comparison of analyte concentration from BM sera and blood sera, data from (d) and (e). g, Ex vivo progenitor expansion assay, with frequency of ki67+ and HO-1+ myeloid cells upon culturing BM Kit+ cells with indicated growth factors for 2 days. n = 3 per group. h, Relative expansion (left, n = 4) and frequency of ki67+, HO-1+, and Arg1+ myeloid cells (right, n = 6) upon culturing progenitors with KP CM or indicated growth factors for 2 days. i–j, Relative expansion of myeloid cells with frequency of ki67+, HO-1+, and Arg1+ myeloid cells (i) and median fluorescence intensity (MFI) for CellROX and MitoSOX (j) after culturing progenitors with KP CM and indicated depletion antibodies for 2 days. n = 3 per group. k-l, Relative expansion of myeloid cells with frequency of ki67+, HO-1+, and NRF2+ myeloid cells (k) and MFI for CellROX and MitoSOX (l) after culturing progenitors with KP CM and ML385 for 2 days. n = 3 per group. m–n, Representative histogram and MFI quantification for phospho-p38 MAPK in myeloid progenitors treated with KP CM and indicated neutralizing antibodies (m, n = 3 per group) or KP CM and ML385 (n, n = 4 per group). o–p, Relative expansion of myeloid cells with frequency of ki67+, HO-1+, and Arg1+ myeloid cells (o) and MFI for CellROX and MitoSOX (p) after culturing BM progenitors with indicated growth factors in the presence of SB203580 for 2 days. n = 3 per group. q, Schematic for reactive oxygen species (ROS)-burden dependent poising of NRF2 pathways in growth factor-induced myeloid progenitor expansion in BM which enables NRF2 activation within TME. Individual data points with bar denoting mean, representative of two independent experiments (a,g,h,i–p). Statistics computed by one-way ANOVA with Tukey’s multiple comparison (a,b), Dunnett’s multiple comparison (g)–(p), hypergeometric test for O-link (d,e).
Extended Data Fig. 7 NRF2 signaling in tumor-infiltrating monocytic lineage promotes immunosuppressive program persistence in TME.
a, Gene expression in KP conditioned media (CM)-exposed BMDMs as measured by reverse transcription quantitative PCR (RT–qPCR). Relative values depicted as fold-change compared to control media, after normalization to Hprt expression. n = 3 replicates, representative of two independent experiments. b, Gene expression in KP CM-exposed BMDMs treated with ML385, as measured by RT–qPCR. Relative values depicted as fold-change compared to BMDMs treated with vehicle DMSO, after normalization to Hprt expression. n = 3 replicates, representative of two independent experiments. c, Relative MFI quantification of CellROX, LiperFluo, and MitoSOX in Nfe2l2TKO (KO) or control (WT) BMDMs exposed to KP CM. n = 3 per group. d, Representative cytometry plots for Annexin-V and Propidium iodide (PI) in Nfe2l2TKO (KO) or control (WT) BMDMs exposed to KP CM, with relative frequency of apoptotic cells. n = 3 per group. e, Relative frequency of apoptotic cells in Nfe2l2TKO (KO) or WT BMDMs without any tumor conditioning. n = 3 per group. f–g, Number of LT-HSCs and myeloid progenitors in BM (f) and myeloid cells in blood (g) of tumor-free Nfe2l2ΔMs4a3 mice (n = 7) or Nfe2l2fl/fl controls (n = 5). h, MFI for indicated phenotypic markers in blood-circulating Ly6Chi monocytes from tumor-free Nfe2l2ΔMs4a3 mice (n = 5) or Nfe2l2fl/fl controls (n = 4). i, Fluorescent bead phagocytosis assay using tumor-free Nfe2l2ΔMs4a3 or Nfe2l2fl/fl BM Ly6Chi monocytes at different ratio of cells to beads. n = 4 per group. j, frequency of NRF2+ CD45.2+ donor-derived mo-macs derived from GMPs in Brusatol-treated tumor-bearing mice or N-acetyl cysteine-treated tumor-naïve mice transferred into KP tumor-bearing CD45.1 hosts, four days post transfer. (left to right, n = 3,4,4,3). Individual data points with bar denoting mean, representative of two independent experiments (a,b,f–j) or three independent experiments (c,d,e). Statistics computed by unpaired two-tailed Student’s t-test (c–h), two-way ANOVA with Fisher’s LSD test (a,b) or Sidak’s multiple comparison (i).
Extended Data Fig. 8 Tumor-induced NRF2 signaling in BM impacts myelopoiesis and intratumoral myeloid phenotype.
a, Kaplan-Meier plot for overall survival of KP tumor-bearing Nfe2l2ΔMs4a3 mice (n = 10) or Nfe2l2fl/fl littermates (n = 8). b, Lung tumor burden in LLC1 tumor-bearing (left) and B16-F10 tumor-bearing (right) Nfe2l2ΔMs4a3 mice (n = 9,10) or Nfe2l2fl/fl littermates (n = 8,9). c, Tumor burden in KP tumor-bearing Keap1ΔMs4a3 mice (n = 7) or Keap1fl/fl littermates (n = 8). d, Frequency of Annexin-V+ cells within indicated intratumoral myeloid cells of KP tumor-bearing Nfe2l2ΔMs4a3 mice (n = 7) or Nfe2l2fl/fl littermates (n = 5). e, Number of tumor-infiltrating TREM2hi mo-macs, CD86+MHCII+ mo-macs, and TREM2hi mo-macs expressing PDL1 in Keap1ΔMs4a3 mice or Keap1fl/fl littermates (n = 7 each). f, Representative flow plots (left) and quantification (right) of donor chimerism in blood and lung of Nfe2l2WT 1:1 Nfe2l2ΔMs4a3 chimera mice or Nfe2l2ΔMs4a3-only chimera mice (left). left to right, n = 8,4 and n = 8,5. Individual data points with lines connecting same mouse. g–h, Frequency of BM GMPs and blood CD157+ Ly6Chi monocytes (g), frequency of lung-infiltrating TREM2hi mo-macs, CD86+MHCII+ mo-macs, and lung-infiltrating TREM2hi mo-macs expressing PDL1 (h) derived from indicated BM donors. Individual data points with lines connecting same mouse (n = 8). i, scRNA-seq heatmap of per-cell UMI counts across indicated myeloid cell subclusters in BM of tumor-bearing Nfe2l2ΔMs4a3 and Nfe2l2fl/fl mice. n = 3 mice pooled per group. j–k, Number of differentially expressed genes i.e., nDEGs (j) (|logFC | >1, adjP<0.05) and number of differentially accessible regions i.e., nDARs (k) (|logFC | >0.25 and adjP<0.05) within indicated BM myeloid clusters of Nfe2l2ΔMs4a3 mice. l, Differentially expressed genes in BM Ly6Chi monocytes of tumor-bearing Nfe2l2ΔMs4a3 mice compared to Nfe2l2fl/fl littermates. m, Transcription factor (TF) motifs differentially enriched in BM Ly6Chi monocytes of tumor-bearing Nfe2l2ΔMs4a3 compared to Nfe2l2fl/fl littermates; ranked by FDR. n, scRNA-seq UMAP and heatmap of per-cell UMI counts (left) alongside relative abundance (right) of indicated myeloid sub-clusters in lung tissue of tumor-bearing Nfe2l2ΔMs4a3 and Nfe2l2fl/fl mice. n = 3 mice pooled per group. o, Number of DEGs (|logFC| >1, adjP<0.05) within indicated lung tumoral myeloid clusters of Nfe2l2ΔMs4a3 mice. p, Normalized UCell-computed scores for NRF2 activation and Type I/III Interferon (IFN)-stimulation in lung-infiltrating Ly6Chi monocytes from scRNA-seq of tumor-bearing Nfe2l2ΔMs4a3 and Nfe2l2fl/fl mice. n = 3 per group. Data are per-cell distribution plot. q, Differentially expressed genes in tumor-infiltrating mo-macs (Arg1hi,Trem2hi) of tumor-bearing Nfe2l2ΔMs4a3 mice compared to Nfe2l2fl/fl littermates. Individual data points with bar denoting mean, representative of two independent experiments (a,b,c,d,e). Scale bar, 2 mm (c). Statistics computed by Log-rank (Mantel-Cox) test (a), unpaired two-tailed Student’s t-test (b–e), paired two-tailed t-test (f–h), hypergeometric test with multiple test correction (l,p,q).
Extended Data Fig. 9 Tumor-induced NRF2 signaling in BM limits T and NK cell activity In TME.
a–b, Number of blood Ly6Ghi neutrophils in blood (a) and lung (b) of tumor-bearing Nfe2l2ΔMs4a3CreERT2 mice with early (n = 5) vs late deletion (n = 6) of NRF2. c, Representative gating strategy for major mouse lung lymphoid (T, B, NK cell) populations, with exemplar phenotypic markers depicted for CD8+ T cells and NK cells. d–e, Number of lung-infiltrating NK cells (d) and frequency of NK cells expressing CD69, NKG2D, and producing IFNγ (e) in KP tumor-bearing Nfe2l2ΔMs4a3 mice (n = 8) or negative littermates (n = 7) at day 21. f–g, Number of lung-infiltrating CD8+ T cells (f) and frequency of lung-infiltrating CD8+ T cells expressing PD-1, LAG3, CD69, or producing IFNγ, TNFα (g) in KP tumor-bearing Nfe2l2ΔMs4a3 mice (n = 8) or negative littermates (n = 7) at day 21. h, Representative histology and tumor burden for KP tumor-bearing Nfe2l2ΔMs4a3 mice and Nfe2l2fl/fl control that received anti-CSF1R depletion antibodies or isotype. n = 5 mice per group. i, Representative histology and tumor burden for KP tumor-bearing Nfe2l2ΔMs4a3 mice that received anti-NK1.1 antibodies or isotype. n = 7 mice per group. Unless noted, data shown as individual data points with bar denoting mean, representative of two independent experiments (a,b,d–g). Scale bar, 2 mm (h,i). Statistics computed by unpaired two-tailed Student’s t-test (a,b),(d–g),(i), one-way ANOVA with Sidak’s multiple comparison (h).
Extended Data Fig. 10 NRF2 pathway activation in TME myeloid cells is associated with immunotherapy resistance.
a, ChIP-X Enrichment Analysis (ChEA) calculated TF regulators for differentially expressed genes in tumoral mo-macs enriched in non-responders (NR cohort) to neoadjuvant immunotherapy (NAIT) for lung cancer (NSCLC, Hu et al., n = 12 patients), compared to responders (R cohort). arranged by adjusted p-value (log q-value). b, ChEA calculated TF regulators for differentially expressed genes in tumoral mo-macs from patients with no clonal T cell expansion (NE cohort) after immune checkpoint blockade (ICB) in breast cancer (BC, Bassez et al., n = 29 patients) compared to macs from patients with clonal expansion (E cohort). c–d, ChEA calculated TF regulators for differentially expressed genes in tumoral mo-macs (c) and peripheral blood CD14+ monocytes (d) enriched in non-responders exhibiting stable disease (SD) to immune checkpoint blockade (ICB) for colorectal cancer (CRC, Chen et al., n = 22 patients) compared to complete response (CR). e, Kaplan-Meier plot depicting overall survival of KP tumor-bearing mice treated with Brusatol in conjunction with anti-PD-1 immunotherapy (n = 8), Brusatol alone (n = 6), anti-PD-1 alone (n = 7), or vehicle (n = 6). f, Kaplan-Meier plot depicting overall survival of KP tumor-bearing mice treated with ML385 in conjunction with anti-PD-1 immunotherapy (n = 6), ML385 alone (n = 5), or vehicle (n = 5). g, Representative histology and tumor burden for KP tumor-bearing Nfe2l2ΔMs4a3 and Nfe2l2fl/fl mice that received Brusatol in conjunction with anti-PD1 (left to right, n = 6,4,5,4,5,5) h, Frequency of NK cells within CD45+ cells (left), and percentage of NK cells expressing CD69 and producing IFNγ (right) in tumor-bearing WT mice treated as indicated. n = 5 mice per group. i, Frequency of CD8+ T cells within CD45+ cells (left), and percentage of CD8+ T cells expressing PD-1, LAG3 or producing IFNγ, TNFα (right) in tumor-bearing WT mice treated as indicated. n = 5 per group. Individual data points with bar denoting mean (g,h,i), representative of two independent experiments (e,f,g,h,i). Scale bar, 2 mm (g). Statistics computed by hypergeometric test with multiple test correction (a)–(d), Log-rank (Mantel-Cox) test (e,f), and one-way ANOVA with Sidak’s multiple comparison (g) or Dunnett’s multiple comparison (h,i).
Supplementary information
Supplementary Information
Supplementary Tables 1–7.
Source data
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hegde, S., Giotti, B., Soong, B.Y. et al. Myeloid progenitor dysregulation fuels immunosuppressive macrophages in tumours. Nature 646, 1214–1222 (2025). https://doi.org/10.1038/s41586-025-09493-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09493-y
This article is cited by
-
Machine learning-based construction of an oxidative stress-related model reveals molecular subtypes and tumor microenvironment infiltration signatures in colon cancer
Discover Oncology (2025)
-
Nrf2 functions as a biomarker and therapeutic target in lung cancer based on bibliometric analysis and molecular mechanisms
Discover Oncology (2025)