Abstract
Emerging evidence indicates that cancer cells are susceptible to ferroptosis, a form of cell death that is triggered by uncontrolled lipid peroxidation1,2,3. Despite broad enthusiasm about harnessing ferroptosis as a novel anti-cancer strategy, whether ferroptosis is a barrier to tumorigenesis and can be leveraged therapeutically remains unknown4,5. Here, using genetically engineered mouse models of lung adenocarcinoma, we performed tumour-specific loss-of-function studies of two key ferroptosis suppressors, GPX46,7 and ferroptosis suppressor protein 1 (FSP1)8,9, and observed increased lipid peroxidation and robust suppression of tumorigenesis, suggesting that lung tumours are highly sensitive to ferroptosis. Furthermore, across multiple pre-clinical models, we found that FSP1 was required for ferroptosis protection in vivo, but not in vitro, underscoring a heightened need to buffer lipid peroxidation under physiological conditions. Lipidomic analyses revealed that Fsp1-knockout tumours had an accumulation of lipid peroxides, and inhibition of ferroptosis with genetic, dietary or pharmacological approaches effectively restored the growth of Fsp1-knockout tumours in vivo. Unlike GPX4, expression of FSP1 (also known as AIFM2) was prognostic for disease progression and poorer survival in patients with lung adenocarcinoma, highlighting its potential as a viable therapeutic target. To this end, we demonstrated that pharmacologic inhibition of FSP1 had significant therapeutic benefit in pre-clinical lung cancer models. Our studies highlight the importance of ferroptosis suppression in vivo and pave the way for FSP1 inhibition as a therapeutic strategy for patients with lung cancer.
Similar content being viewed by others
Main
Ferroptosis is a non-apoptotic, oxidative stress-dependent mechanism of cell death that is uniquely distinguished by lipid peroxidation of polyunsaturated fatty acids of membrane phospholipids (PUFA-PLs)1,10. Lipid peroxidation involves the formation of lipid and lipid peroxyl radicals, which, in auto-oxidative propagation reactions, generate lipid hydroperoxides. Aberrant, unrestricted lipid peroxidation results in altered membrane integrity, cell swelling, and ultimately membrane rupture11,12. In recent years, growing interest in ferroptosis has revealed that cells depend on several enzymes to protect against ferroptosis6,7,8,9,13,14,15, of which GPX4 and FSP1 have key roles. GPX4 catalyses the reduction of PUFA-PL hydroperoxides to non-toxic alcohols. FSP1 catalyses the reduction of extramitochondrial coenzyme Q10 (CoQ), a highly potent lipid radical-trapping antioxidant (RTA). Conversely, cells can be sensitized to ferroptosis through PUFA-PL composition16 and biosynthesis by ACSL417,18,19. In addition to these cellular pathways, exogenous lipid RTAs, such as LIP1, FER1, vitamin E and vitamin K, have also been shown to specifically inhibit ferroptosis1,6,20,21,22.
Emerging evidence indicates that cancer cells, including drug-resistant cells that have adopted a mesenchymal state2,3, are highly sensitive to lipid peroxidation in vitro, suggesting a potential for harnessing ferroptosis as a novel anti-cancer strategy. Although there have been extensive studies characterizing GPX4 and more recently FSP1 in various diseases4,23, whether ferroptosis constitutes a barrier to tumorigenesis and the functional roles of GPX4 and FSP1 in in vivo cancer models remain poorly characterized. To date, FSP1 has been shown to be a critical ferroptosis suppressor only in the absence of GPX48,9,24. However, given the high toxicity, poor selectivity and low-to-limited bioavailability of GPX4 inhibitors in vivo25,26, as well as the only recent development of FSP1 inhibitors, the majority of which do not have in vivo efficacy9,24,27,28,29,30, much work remains to demonstrate that ferroptosis induction could indeed be a therapeutic strategy for cancer5.
Here we show that genetic knockout of either Gpx4 or Fsp1 (also known as Aifm2) in genetically engineered mouse models (GEMMs) of autochthonous KRAS-driven lung adenocarcinoma (LUAD)31,32,33,34,35 results in marked restriction of lung tumorigenesis. We demonstrate that disruption of FSP1, often considered a backup axis of ferroptosis suppression, is sufficient to trigger exhibit enhanced lipid peroxidation in lung tumours. We confirm that Fsp1 deletion across several human cancer cell lines with various oncogenic drivers, co-mutations and tissue lineages consistently lead to in vivo tumour restriction. As ferroptosis can be functionally defined as a modality of cell death resulting from increased lipid peroxides and rescued only with inhibitors of lipid peroxidation, we demonstrate that Fsp1-knockout (Fsp1KO) tumours have increased oxidized PUFA-PLs and that administration of lipid RTAs, as well as genetic loss of Acsl4, effectively restores lung tumorigenesis. Finally, we report that FSP1 is upregulated as LUAD tumours progress and show that FSP1 inhibition suppresses tumour growth and prolongs survival in multiple pre-clinical cancer models. Thus, our work utilizes novel in vivo models to characterize and test ferroptosis induction, particularly via FSP1 perturbation, as a therapeutic approach for lung cancer.
GPX4 prevents ferroptosis in lung tumours
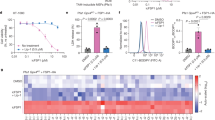
GPX4 is considered the primary mediator of ferroptosis suppression, and has been shown to be essential across multiple mouse and human cancer cell types7,26,36,37. Therefore, we sought to interrogate whether GPX4, and more broadly ferroptosis suppression, is a requirement for lung tumorigenesis in vivo. Using a well-established GEMM of LUAD (KrasLSL-G12D/+; Tp53fl/fl; Rosa26LSL-Cas9/LSL-Cas9), we initiated KrasG12D/+; Tp53−/− (KP) tumours by intratracheal delivery of lentiviruses expressing Cre recombinase and dual single guide RNAs (sgRNAs)38 targeting Gpx4 (sgGpx4) or non-targeting control sgRNAs (sgNeo), enabling CRISPR–Cas9-mediated Gpx4-knockout (Gpx4KO) or wild-type control tumours, respectively. Concurrently, we treated a cohort of mice with each tumour genotype with LIP1, a potent lipid RTA and ferroptosis inhibitor, throughout the entire course of tumorigenic progression (Fig. 1a). Tumour-specific Gpx4 deletion (sgGpx4) led to a significant decrease in lung tumour burden that was not observed in sgGpx4 tumour-bearing mice treated with LIP1 (Fig. 1b–d). Efficient Gpx4 deletion was verified by immunohistochemistry, with less than 15% of tumours in the sgGpx4 group staining positive for GPX4 (Extended Data Fig. 1a). Immunohistochemistry for 4-hydroxy-2-noneal (4-HNE), a marker for lipid peroxidation in tissues20 (Extended Data Fig. 1b), revealed significantly higher levels of lipid peroxidation in sgGpx4 tumours compared with control sgNeo lung tumours, which was blunted with LIP1 treatment (Fig. 1e,f).
a, Schematic of KP LUAD GEMMs intratracheally infected with pUSEC lentiviruses containing dual sgRNAs targeting Neo (control; n = 13) or Gpx4 (n = 13). Mice were dosed with LIP1 (sgNeo: n = 6, sgGpx4: n = 7) or vehicle (sgNeo: n = 7, sgGpx4: n = 6) every other day, starting from tumour initiation to experiment endpoint. b, Tumour burden of KP LUAD tumours with control (sgNeo) or Gpx4 (sgGpx4) knockout, treated with vehicle (Veh) or LIP1. c, Representative MRI of KP LUAD tumours with control (sgNeo) or Gpx4 (sgGpx4) knockout, treated with vehicle or LIP1. d, Representative haematoxylin and eosin (H&E) staining of KP LUAD tumours with control (sgNeo) or Gpx4 (sgGpx4) knockout, treated with vehicle or LIP1. Scale bars, 2,000 µm. e, Immunohistochemical detection of 4-HNE in KP LUAD tumours with control (sgNeo) or Gpx4 (sgGpx4) knockout, treated with vehicle or LIP1. f, Representative images of 4-HNE immunohistochemical staining from e. Scale bars, 20 µm. Data are mean ± s.e.m. One-way ANOVA with multiple comparisons (b,e). Drawing in a created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Similar to our in vivo observations, CRISPR–Cas9-mediated Gpx4KO in KP LUAD cells resulted in a near complete loss of viability and clonogenicity that was fully rescued by the addition of LIP1 (Extended Data Fig. 1c). Targeted lipidomics to profile oxidized phosphatidylcholine and phosphatidylethanolamine species in KP LUAD cells treated with RSL3, a covalent GPX4 inhibitor and inducer of ferroptosis, revealed a higher abundance of oxidized phospholipids in RSL3-treated cells that was decreased in cells treated with concomitant LIP1 (Extended Data Fig. 1d). GPX4 overexpression in KP LUAD cells had no effect on cell viability or clonogenicity basally (Extended Data Fig. 1e), but did confer greater resistance to ferroptosis in vitro (Extended Data Fig. 1f). Similarly, supplementation of KP LUAD cells with sodium selenite (Na2SeO3) to increase translation of GPX439, a selenoprotein, resulted in a corresponding increase in GPX4 expression and enhanced ferroptosis resistance in vitro (Extended Data Fig. 1g). Together, these results demonstrate that GPX4 is essential for lung tumours to evade ferroptosis and provide evidence that ferroptosis induction may be an effective anti-cancer strategy.
FSP1 is upregulated in LUAD
Given our observation that GPX4 was required for lung tumorigenesis in a GEMM that closely recapitulates human LUAD development and progression, we next explored whether there was clinical evidence suggesting that ferroptosis regulators in general are altered in lung cancer40. Although GPX4 expression was modestly increased in KRAS-mutant LUAD tumours compared with normal lung tissue, there was no correlation with tumour stage or overall survival (Extended Data Fig. 1h,i). Concordantly, GPX4 protein levels also did not change during autochthonous LUAD progression in KP GEMM tumours (Extended Data Fig. 1j).
Since GPX4 expression did not appear to correlate with any clinical prognostic factors in patients with KRAS-mutant LUAD, we next investigated FSP1, a second axis of ferroptosis surveillance (Fig. 2a). We found that FSP1 expression was robustly increased in tumours from patients with KRAS-mutant LUAD compared with normal lung, and exhibited a positive correlative trend with higher tumour stages (Extended Data Fig. 2a). Additionally, high FSP1 expression correlated with poor survival in patients with KRAS-mutant LUAD (Fig. 2b). Unlike GPX4, FSP1 protein levels increased temporally as lung tumours progressed from adenomas to adenocarcinomas in KP GEMMs (Fig. 2c,d). Together, these findings suggest that FSP1 may constitute a major suppressor of ferroptosis in LUAD and is potentially a more viable therapeutic target than GPX4 for patients with lung cancer.
a, Schematic depicting mechanisms of FSP1- and GPX4-mediated buffering against cellular lipid peroxidation and ferroptosis. GSH, reduced glutathione; GSSG, glutathoine disulfide; PUFA, polyunsaturated fatty acid; α-TOH, tocopherol (vitamin E). b, Overall survival of patients with KRAS-mutant LUAD (n = 280) from The Cancer Genome Atlas (TCGA), stratified by high versus low primary tumour FSP1 expression. Median survival times for FSP1-high and FSP1-low tumours were 1,215 days and 1,498 days, respectively (hazard ratio 1.51 [1.01–2.25]). c, Representative immunohistochemistry for FSP1 and GPX4 in KP LUAD GEMM adenomas versus adenocarcinomas (AdenoCA). Scale bars, 100 µm. d, Immunohistochemical detection of FSP1 expression in KP LUAD GEMM tumours at 10 weeks and 14 weeks post-tumour initiation. e, Schematic of KP LUAD GEMMs intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting control (Neo; n = 4), Fsp1 (n = 7) or Gpx4 (n = 6). f, Tumour burden of KP LUAD tumours with knockout of either control (Neo), Fsp1 or Gpx4. g, Representative H&E staining of KP LUAD tumours with knockout of either control (Neo), Fsp1 or Gpx4. Scale bars, 2,000 µm. h, Relative proportion of KP adenomas and adenocarcinomas with knockout of either control (Neo), Fsp1 or Gpx4. i, Representative TUNEL staining of KP, Fsp1KO and Fsp1WT orthotopic lung tumours. Scale bars, 50 µm j, Schematic depicting generation of isogenic Fsp1WT versus Fsp1KO cells for paralleled in vitro assays and in vivo syngeneic orthotopic transplantation studies. k, Representative images of crystal violet clonogenic growth assay in isogenic KP, Fsp1KO and Fsp1WT cells treated with RSL3 (0.5 µM) with or without LIP1 (100 nM). l, Tumour burden of KP, Fsp1KO (n = 8) and Fsp1WT (n = 7) orthotopic lung tumours. m, Representative H&E staining of KP, Fsp1KO and Fsp1WT orthotopic lung tumours. Scale bars, 2,000 µm. Data are mean ± s.e.m. Two-sided Student’s t-test (d,l), one-way ANOVA with multiple comparisons (f) or Kaplan–Meier simple survival analysis (b). Drawings in a,e,j created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Since prior studies have suggested that FSP1 may be regulated by KRAS signalling41, to further determine whether FSP1 upregulation was specific to KRAS-mutant LUAD, we stratified patients with LUAD by major oncogenic mutations. Again, FSP1 expression was robustly increased in tumours relative to normal lung tissue, and this upregulation was observed irrespective of driver mutation or KRAS variant (Extended Data Fig. 2b,c). Although FSP1 expression was more robustly upregulated in KRAS-mutant tumours compared with EGFR-mutant tumours, LUAD tumours with neither mutation also exhibited upregulation of FSP1 to the same extent as KRAS-mutant tumours. Next, we investigated whether the MAPK–ERK pathway regulates FSP1 expression. Both in vitro pathway inhibition with RMC-042, a pan-RAS inhibitor, in a panel of KRAS-mutant LUAD cell lines, as well as immunofluorescence of FSP1 and phosphorylated ERK in KP GEMM tumours revealed a lack of correlation between FSP1 expression and ERK activation (Extended Data Fig. 2d,e). Thus, these data indicate that FSP1 expression, at least in the context of LUAD, may not be exclusively dependent on KRAS-mediated oncogenic signalling.
Given that patients with KRAS-mutant LUAD tumours often have concomitant mutations in STK11 and KEAP1, which indicate more advanced disease and poorer prognosis34,42, we also tested whether FSP1 expression was impacted by these mutations. We observed that FSP1 was significantly increased in LUAD tumours with co-mutation of either STK11 or KEAP1 (Extended Data Fig. 2f). Additionally, treatment of a panel of human LUAD cell lines with the NRF2 activator KI696 led to significantly increased FSP1 gene expression (Extended Data Fig. 2g). Of note, these findings are in accordance with recent studies that describe a role for NRF2, which is activated by KEAP1 mutations, in FSP1 upregulation43.
FSP1 is required for lung tumorigenesis
To systematically investigate the functional role of FSP1 in lung cancer, we initiated autochthonous KP LUAD tumours, this time with tumour-specific CRISPR–Cas9-mediated deletion of either Fsp1 or Gpx4 (Fig. 2e). Of note, we found that genetic deletion of Fsp1 (sgFsp1) in KP LUAD tumours strongly suppressed lung tumorigenesis, resulting in significantly decreased tumour burden to the same extent as Gpx4KO (sgGpx4) tumours (Fig. 2f,g). Efficient Fsp1 deletion was verified by immunohistochemistry, with less than 5% of tumours in the in sgFsp1 group still staining positive for FSP1 (Extended Data Fig. 3a). We did not observe significant changes to the total number or size of tumours formed across the three biological groups (Extended Data Fig. 3b,c), which suggested that loss of Fsp1 or Gpx4 primarily affects tumour progression rather than tumour initiation. We also observed that Fsp1 or Gpx4 deletion resulted in higher proportions of adenomas to adenocarcinomas compared with control tumours (Fig. 2h), indicating that FSP1 and GPX4 are functionally important for malignant disease progression. Additionally, there were no notable changes to tumour cell proliferation or apoptotic cell death (Extended Data Fig. 3d,e) in sgFsp1 or sgGpx4 tumours. By contrast, TUNEL staining of lung tumours demonstrated increased cell death with loss of FSP1 (Fig. 2i), suggesting that the decreased lung tumour burden with Fsp1 or Gpx4 genetic deletion is likely to be due to increased tumour cell ferroptosis. Perhaps most notable is the extent to which Fsp1 deletion phenocopied Gpx4 loss in vivo, as this phenotype, to our knowledge, has not previously been reported in any other cell lines, tissues or disease models.
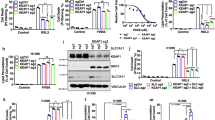
a–g, Longitudinal tumour growth and endpoint tumour mass from indicated cell lines with CRISPR–Cas9-mediated knockout of FSP1 (sgFSP1) or control (sgTOM, sgNeo), transplanted as subcutaneous xenograft tumours into NSG mice. a, H2009 cells. sgFSP1: n = 8, control: n = 8. b, H1299 cells. sgFSP1: n = 9, control: n = 9. c, PC9 cells. sgFSP1: n = 9, control: n = 9. d, H1975 cells. sgFSP1: n = 8, control: n = 9. e, A549 cells. sgFSP1: n = 9, control: n = 10. f, 16645 cells. sgFsp1: n = 7, control: n = 9. g, KPC7 (mouse PDAC cells). sgFsp1: n = 9, control: n = 10. Data are mean ± s.e.m. Two-sided Student’s t-test (a–g).
FSP1 is required uniquely in vivo
The identification of FSP1 as a key regulator of ferroptosis arose from genetic screens to identify ferroptosis suppressors in the absence of functional GPX48,9. Thus, FSP1 is generally thought to control a secondary anti-ferroptotic axis whose function, at least in vitro, is masked when GPX4 is intact. Accordingly, we observed that CRISPR–Cas9-mediated Fsp1 deletion in KP LUAD cells had no effect on cell viability and clonogenicity in vitro (Extended Data Fig. 4a). As expected, Fsp1KO cells were more sensitive to RSL3 treatment than isogenic Fsp1-wild-type (Fsp1WT) cells and were protected from killing following RSL3 administration by LIP1 (Fig. 2k). Despite the remarkable restriction of KP LUAD tumorigenesis by Fsp1 loss in vivo, our in vitro studies indicated that the anti-ferroptotic function of FSP1 was not required by KP LUAD cells in the presence of GPX4.
We sought to dissect this dichotomy and better understand the distinct in vivo dependency of lung tumours on FSP1. We first performed a series of syngeneic transplantation experiments using isogenic KP, Fsp1KO and Fsp1WT cell lines with no proliferation differences in vitro to systematically characterize the impact of FSP1 loss on tumour growth and further interrogate whether FSP1-mediated tumour differences were specific to in vivo growth conditions (Fig. 2j and Extended Data Fig. 4b). Consistently, we found that Fsp1KO cells formed significantly smaller tumours in both immunocompetent subcutaneous xenografts (Extended Data Fig. 4c,d) as well as orthotopic lung tumours (Fig. 2l,m and Extended Data Fig. 4e,f). Robust suppression of Fsp1KO orthotopic lung tumour growth was similarly observed in immunodeficient athymic (NU/J) mice (Extended Data Fig. 4g), as well as in an immunogenic mouse model42 (Extended Data Fig. 4h). We also observed the same phenotype in age- and sex-matched Fsp1WT versus Fsp1KO mice (Extended Data Fig. 4i). These studies collectively provide strong evidence that the pro-tumorigenic effect of FSP1 is primarily cell intrinsic. Fsp1 overexpression (Fsp1OE) in KP LUAD cells exhibited no in vitro growth advantage (Extended Data Fig. 4j) but had enhanced resistance to killing by RSL3 (Extended Data Fig. 4k) and accelerated growth of xenograft tumours in vivo (Extended Data Fig. 4l,m). Altogether, these results suggest that FSP1 is essential for survival of LUAD cells in vivo and that increased FSP1 expression is sufficient to promote Kras-mutant tumorigenesis in vivo.
FSP1 dependency is not mutation-specific
To more thoroughly interrogate the functional importance of FSP1 for tumorigenesis and the translational potential for targeting FSP1 in cancer, we assessed the requirement of FSP1 for growth of xenograft tumours from human LUAD cell lines with a variety of driver mutations, including KRAS, NRAS, EGFR, TP53, STK11 and KEAP1. We found that CRISPR–Cas9-mediated FSP1 genetic deletion consistently and markedly decreased tumour growth in all models tested. Specifically, knockout of FSP1 (sgFSP1) in H2009 (KRAS, TP53 mutant), H1299 (NRAS, TP53 mutant), PC9 and H1975 (EGFR, TP53 mutant) LUAD cells led to robust suppression of subcutaneous tumour growth (Fig. 3a–d), but, analogous to aforementioned studies, did not affect proliferation or viability of cells in vitro unless they were treated with RSL3 (Extended Data Fig. 5a–d). To assess whether KRAS-mutant tumours with STK11 (LKB1) and/or KEAP1 mutations also depend on FSP1, we performed CRISPR–Cas9-mediated FSP1 deletion in A549 (KRAS, KEAP1, STK11 mutant) and 16645 (Kras mutant, Stk11-null) LUAD cells. Again, we observed a dependency for FSP1 in vivo (Fig. 3e,f) but not in vitro (Extended Data Fig. 5e,f). Together these data provide further evidence that FSP1 is indeed required for in vivo LUAD tumour growth irrespective of driver and co-mutations.
Finally, we tested whether KRAS-driven tumours of a different lineage also exhibited a requirement for FSP1 in vivo. Transplantation of pancreatic adenocarcinoma (PDAC) Kras, Tp53 mutant (KPC7) cells with CRISPR–Cas9-mediated Fsp1 deletion also led to a robust tumour suppression in vivo (Fig. 3g and Extended Data Fig. 5g), mirroring the phenotype observed consistently in LUAD models and suggesting that FSP1 dependency in vivo may extend to other tissue lineages.
Ferroptosis inhibition rescues Fsp1 loss
We hypothesized that defective regulation of lipid peroxidation and subsequent induction of ferroptosis was the mechanism underlying Fsp1KO tumorigenic suppression. To assess this, we first performed epilipidomic analysis of Fsp1WT and Fsp1KO orthotopic lung tumours and observed that Fsp1KO tumours exhibited increased abundance of oxidized and truncated phosphatidylcholine and phosphatidylethanolamine species, indicative of increased cellular lipid peroxidation (Fig. 4a). We then explored whether ectopic overexpression of FSP1 or GPX4 (Extended Data Fig. 6a,b) was sufficient to restore the growth of Gpx4KO or Fsp1KO tumours, which would shed light on the capacity for each ferroptosis defence arm to compensate for the loss of the other. Indeed, we found that overexpression of FSP1 and GPX4 effectively restored the growth of Gpx4KO and Fsp1KO tumours, respectively (Fig. 4b,c and Extended Data Fig. 6c,d). These data implicated that tumours require extensive buffering capacity against lipid peroxidation, and that although GPX4 and FSP1 act through distinct pathways, each can compensate for the loss of the other.
a, Heat map of indicated oxidized phosphatidylethanolamine (oxPE) and oxidized phosphatidylcholine (oxPC) lipid species detected via LC–MS from KP, Fsp1KO and Fsp1WT orthotopic lung tumours. b, Longitudinal growth of KP, Gpx4KO subcutaneous xenograft tumours with Fsp1OE (n = 10) versus control (empty vector (emptyOE); n = 9) in C57BL/6 J mice. c, Longitudinal growth of KP, Fsp1KO subcutaneous xenograft tumours with either Fsp1 restoration (Fsp1WT, n = 9), GPX4 overexpression (Gpx4OE; n = 8) or controls (emptyOE: n = 9; Fsp1KO: n = 8) in C57BL/6 J mice. d, Ratio of CoQ9H2/CoQ9 detected via LC–MS in KP, Fsp1KO and Fsp1WT orthotopic lung tumours and cells treated with DMSO or RSL3 (0.5 µM) for 8 h. e, Schematic depicting the pro-ferroptotic function of ACSL4. VitE, vitamin E. f, Tumour burden in C57BL/6 J mice with KP, Fsp1KO and Fsp1WT orthotopic lung tumours with CRISPR–Cas9-mediated Acsl4 or control (Neo) deletion (wild type: n = 5, Acsl4KO: n = 4, Fsp1KO: n = 7, Fsp1KOAcsl4KO: n = 7). g, Schematic depicting dietary vitamin E manipulation studies in h,i. h, Longitudinal lung tumour growth (measured via bioluminescence normalized to first timepoint) in C57BL/6 J mice orthotopically transplanted with KP, Fsp1KO cells and receiving high (n = 8) or low (n = 7) vitamin E diets ad libitum 5 days before tumour initiation. i, Longitudinal lung tumour growth (measured via bioluminescence) in C57BL/6 J mice orthotopically transplanted with KP, Fsp1KO cells and placed on high (n = 6) or low (n = 6) vitamin E diets ad libitum on day 18 after tumour establishment. j, Schematic of KP LUAD GEMMs intratracheally infected with pUSEC lentiviruses containing double sgRNAs targeting Neo (control; n = 12) or Fsp1 (n = 12). Mice were dosed with LIP1 (n = 6 per genotype) or vehicle (n = 6 per genotype) daily starting 5 weeks after tumour initiation. k, Tumour burden of mice described in j. l, Representative H&E staining of tumours from experiment in j. Scale bars, 100 µm. Data are mean ± s.e.m. Two-sided Student’s t-test (b,d,f,h,i,k) or two-way ANOVA with Tukey’s multiple comparison test (c). Drawings in e,g,j created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Mechanistically, FSP1 has been shown to suppress ferroptosis by reducing the endogenous RTA CoQ, and this function is dependent on its localization to the plasma membrane. We utilized a liquid chromatography–mass spectrometry (LC–MS)-based, quantitative method to determine the abundance of CoQ in tumours, which revealed that Fsp1KO tumours had a significantly decreased ratio of reduced CoQ9H2 to oxidized CoQ9 (Fig. 4d). This decreased ratio was not present in genotypically matched cells at baseline in vitro, but upon treatment with RSL3, Fsp1KO cells mimicked the decreased CoQ9H2/CoQ9 observed in tumours. Previous studies have described that mutation of the myristoylation sequence that targets FSP1 to the plasma membrane renders FSP1 unable to protect against lipid peroxidation in vitro8,9, which we also observed in KP LUAD cells (Extended Data Fig. 6e,f). We anticipated that expression of this mutant FSP1 (Fsp1mut) would be similarly unable to restore Fsp1KO tumour growth. Indeed, whereas Fsp1WT-restored tumours grew quickly, Fsp1mut and Fsp1KO tumours exhibited decreased growth (Extended Data Fig. 6g). These data suggest that tumours require Fsp1 enzymatic activity and subcellular localization in vivo to specifically buffer against ferroptosis.
Next, we explored whether ferroptosis suppression, using several orthogonal approaches including tumour-specific Acsl4 knockout, dietary vitamin E supplementation and LIP1 treatment would restore Fsp1KO tumour growth. Loss or inhibition of ACSL4, which limits the supply of PUFA-PL and restricts lipid peroxidation (Fig. 4e), has previously been shown to be protective against ferroptosis in Gpx4KO cells in vitro17,18,19. We therefore sought to determine whether genetic deletion of Acsl4 would suppress ferroptosis and rescue the growth of Fsp1KO tumours in vivo. We performed CRISPR–Cas9-mediated genetic deletion of Acsl4 (sgAcsl4) in isogenic KP, Fsp1KO and Fsp1WT cells. We observed that loss of Acsl4 did not affect cell viability but robustly rescued RSL3-induced ferroptosis in vitro, in both Fsp1WT and Fsp1KO cells (Extended Data Fig. 6h). Of note, Acsl4 deletion significantly restored Fsp1KO lung tumour growth in vivo (Extended Data Fig. 6i) and mice with Acsl4- and Fsp1-double-knockout tumours were found to have increased endpoint disease burden (Fig. 4f and Extended Data Fig. 6j) and decreased overall survival (Extended Data Fig. 6k). Furthermore, tumour-specific knockout of Acsl4 led to a basal acceleration of tumour growth (Extended Data Fig. 6i) and decreased overall survival (Extended Data Fig. 6k), suggesting that decreased accumulation of PUFA-PL in lung tumours may also protect against ferroptosis in vivo.
Vitamin E is nature’s primary lipophilic antioxidant and is specifically able to scavenge lipid radicals and protect against ferroptosis (Fig. 4e). Therefore, we tested whether dietary vitamin E supplementation could rescue the growth of Fsp1KO lung tumours. Mice were fed diets that were high or low in vitamin E ad libitum either before or after tumour initiation (Fig. 4g). We found that increased dietary vitamin E accelerated Fsp1KO tumour growth (Fig. 4h,i), with no effect on growth of Fsp1WT tumours (Extended Data Fig. 6l).
Finally, since LIP1 suppressed tumour lipid peroxidation and entirely mitigated the impact of Gpx4 loss on KP LUAD tumorigenesis (Fig. 1b–d), we conducted parallel studies to investigate whether LIP1 also blunts ferroptosis in and restores Fsp1KO tumorigenesis. In the orthotopic lung tumour model, we observed a tendency for Fsp1KO tumours to grow more quickly with daily LIP1 administration (Extended Data Fig. 6m). In the KP LUAD GEMM (Extended Data Fig. 6n), which was conducted in accordance with experimental parameters for Gpx4KO GEMM studies, LIP1 treatment significantly increased autochthonous Fsp1KO (sgFsp1) tumour burden (Extended Data Fig. 6o). Moreover, sgFsp1 tumours stained positively for 4-HNE, which was effectively suppressed by LIP1 treatment (Extended Data Fig. 6p). As observed with vitamin E supplementation, timing of LIP1 treatment did not appear to have a differential impact, as LIP1 treatment after tumours were already established was equally effective at restoring sgFsp1 lung tumorigenesis (Fig. 4j–l). Together, these studies provide clear evidence that LUAD tumours require FSP1 to protect against ferroptosis in vivo and suggest that increased lipid peroxidation is a barrier for tumour growth and progression.
FSP1 inhibition extends overall survival
As a result of the growing interest in harnessing ferroptosis to kill tumour cells, several FSP1 inhibitors have been developed9,28, although the majority are effective only in vitro against human FSP1 in the context of GPX4 loss or inhibition (Extended Data Fig. 7a–d). icFSP1 was recently developed as the first inhibitor of human FSP1 with in vivo stability and efficacy, albeit solely in tumours with concomitant GPX4 loss24. Accordingly, we observed that icFSP1 treatment of KP LUAD cells in vitro did not affect cell viability (Extended Data Fig. 7e) unless RSL3 was added (Extended Data Fig. 7f). Given that mouse and human FSP1 are highly conserved in sequence and structure, we generated a hybrid tumour model in which human FSP1 was expressed in mouse KP, Fsp1KO tumours. We observed that expression of either mouse or human FSP1 resulted in similar acceleration of tumour growth compared with Fsp1KO tumours (Extended Data Fig. 7g,h), which indicated that human FSP1 can functionally compensate for mouse FSP1 in vivo.
Using this hybrid tumour model, we next tested whether icFSP1 had therapeutic benefit against lung cancer. We found that FSP1 inhibition as a monotherapy improved overall survival of mice bearing lung tumours, almost to the same extent as genetic Fsp1 deletion (Fig. 5a). To determine whether the therapeutic effect of icFSP1 was indeed due to the induction of ferroptosis, we tested whether LIP1, which we expected to restrict lipid peroxidation induced by FSP1 inhibition, would rescue tumour suppression in icFSP1-treated mice. In correlation with the survival data, we observed that icFSP1 treatment significantly decreased tumour growth compared with vehicle-treated mice, and concomitant LIP1 treatment abrogated the tumour-suppressive effect of icFSP1 treatment (Fig. 5b and Extended Data Fig. 7i). Further, to test whether icFSP1 was exerting an on-target, tumour-specific effect, we repeated the treatment study with an internal control—lung tumours expressing FSP1(Q319K), a mutant that is resistant to icFSP1 but maintains anti-ferroptotic function (Extended Data Fig. 7a,b). We found that icFSP1 treatment specifically extended overall survival of mice bearing tumours expressing wild-type human FSP1 but not those bearing tumours expressing FSP1(Q319K) (Fig. 5c), indicating that indeed the tumour suppression seen with icFSP1 treatment was due to inhibitor activity directly on tumour cells.
a, Overall survival of C57BL/6 J mice with orthotopic lung tumours with Fsp1KO (n = 7) or re-expression of human FSP1, treated with either icFSP1 (n = 8) or vehicle (n = 7). b, Longitudinal lung tumour growth (measured via bioluminescence) in C57BL/6 J mice orthotopically transplanted with FSP1WT cells and treated with either vehicle (n = 8), LIP1 (n = 7), icFSP1 (n = 8) or icFSP1 plus LIP1 (n = 8). c, Overall survival of C57BL/6 J mice with Fsp1KO (n = 5) tumours expressing wild-type human FSP1 (FSP1WT) or human FSP1(Q319K) (FSP1Q319K), treated with icFSP1 (FSP1WT: n = 5, FSP1Q319K: n =7) or vehicle (FSP1WT: n = 7; FSP1Q319K: n = 7). d, Longitudinal growth of PDX LX465 tumours treated with icFSP1 (n = 10) or vehicle (n = 10) in NSG mice. Data are mean ± s.e.m. Two-way ANOVA with Tukey’s multiple comparisons test (b), two-sided Student’s t-test (d) or Kaplan–Meier simple survival analysis (a,c). Drawing in d created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
We considered whether icFSP1 treatment also affected the tumour microenvironment, but we found no differences in the proportion of total T cells, neutrophils, alveolar or interstitial macrophages between the treatment groups (Extended Data Fig. 7j). These data further suggest that icFSP1 primarily exerts on-target, tumour-specific effects, although more granular immune profiling is needed to determine the potential effects, and thus potential side effects, of FSP1 inhibition on the tumour microenvironment. Finally, we utilized a patient-derived xenograft (PDX) model (LX465; KRASG12D, TP53-mutant) to further assess the therapeutic potential of FSP1 inhibition. We found that icFSP1 treatment significantly decreased PDX tumour growth (Fig. 5d), further bolstering the translational potential of targeting FSP1 in patients with LUAD. These pre-clinical studies with icFSP1 are the first to demonstrate that FSP1 is a promising therapeutic target, and our work highlights FSP1 inhibition as a novel therapeutic strategy for improving disease outcome in patients with lung cancer.
Discussion
Cancer is a disease of hyperproliferation, and cancer cells increase their metabolic output to support sustained growth. This occurs at the cost of increased production of reactive oxygen species that can damage macromolecules such as lipids and have deleterious effects44,45. One such effect is excessive lipid peroxidation of membrane-associated PUFA-PLs, which can lead to ferroptosis, a unique non-apoptotic mechanism of cell death. Through our systematic interrogation of the key mechanisms that regulate aberrant lipid peroxidation in vivo, we demonstrated that ferroptosis constitutes a barrier to lung cancer and that lung tumours rely on GPX4 and uniquely, FSP1, to overcome ferroptosis and sustain tumorigenesis (Extended Data Fig. 7k,l).
Our work specifically highlights the dependence of lung tumours on FSP1 in vivo, even in the presence of functional GPX4 (Extended Data Fig. 7k). The companion study by Palma et al.46 reveals that this dependency extends beyond lung tumours, as they show that metastasizing melanoma cells in the lymph also require FSP1 expression to buffer ferroptosis. These studies are the first reports of a context in which FSP1 and GPX4 are independently required for ferroptosis suppression and therefore either can affect disease outcome. Moreover, analysis of GEMM and human lung tumours demonstrated that FSP1 levels increase during tumour progression and correlate with poor survival. By contrast, GPX4 is ubiquitous in tumours and most tissues. Given that germline Gpx4KO mice are not viable47, whereas Fsp1KO mice are viable with no notable physiological defects20,48,49, the therapeutic window for targeting FSP1 with fewer toxic side effects is expected to be much greater than for GPX4. Furthermore, loss of GPX4 is toxic to T cells, which would suppress any anti-tumour immune responses50,51. Leveraging this unique requirement for FSP1 in lung cancer, our studies demonstrate the therapeutic benefit of FSP1 inhibition in promoting ferroptosis, suppressing tumour growth and extending survival as a monotherapy in aggressive pre-clinical lung cancer models. Similarly, the therapeutic potential of FSP1 inhibition in suppressing melanoma lymph node metastases is demonstrated in the accompanying Article46. Finally, our studies also underscore the value of in vivo pre-clinical models in capturing physiologically relevant metabolic dependencies that may be missed in vitro and pave the way for development of FSP1 inhibitors for clinical testing in patients with lung cancer.
Methods
Cell lines
KP LUAD cell lines were obtained from the laboratory of T. Jacks. KP, Fsp1KO cell lines were generated by transient transfection of PX458 (Addgene #48138) expressing an sgRNA targeting Fsp1. Single GFP-positive clones were selected and FSP1 loss was validated by western blot. The 16645 cell line was developed from KrasG12D Stk11−/− GEMM as previously described52. KPC7 cells were obtained from the laboratory of D. Simeone. All cell lines were maintained in DMEM or RPMI 160 (Corning) supplemented with 10% FBS (Sigma Aldrich) and gentamicin (Invitrogen) and were tested for mycoplasma regularly (PlasmoTest, InvivoGen). All mouse cell lines were authenticated by PCR genotyping. All human cell lines used were purchased from ATCC and were authenticated by short-tandem-repeat profiling. Genetic manipulation of cell lines was performed via lentiviral transduction of plasmids, detailed in next section, followed by either puromycin (7 μg ml−1) or hygromycin (800 μg ml−1) selection for 1 week.
Cloning/lentivirus generation
CRISPR–Cas9-mediated knockout of target genes was achieved by cloning sgRNAs into pLenti-USEC or lentiCRISPRv2-puro vectors, as previously described31. In short, backbones were digested with Esp3I (New England Biosciences) and purified with a gel extraction kit (QIAGEN). sgRNAs were designed using CRISPick (Broad Institute), obtained from Integrated DNA Technologies (Coralville), annealed, phosphorylated, Esp3I-digested, and ligated into the purified digested backbones using Quick Ligase (New England Biosciences). Double sgRNA ultramers were designed and generated as previously described38. In brief, ultramers were Gibson-assembled to digested pDonor_sU6 (Addgene #69351), Esp3I-digested, and ligated to purified digested backbones. Guide RNA sequences used to make gene knockouts can be found in Supplementary Data 4. GPX4, mouse FSP1 and human FSP1 expression plasmids were generated using Gibson assembly of the respective cDNA into pLenti-v2-filler.
Lentivirus was generated by co-transfection of HEK293 cells with a viral vector and packaging plasmids psPAX2 (Addgene #12260) and pMD2.G (Addgene #12259) using PEI transfection reagent. Cell supernatant containing lentivirus was collected 72 h after transfection and filtered through 0.45-µm PVDF filters. For in vivo experiments, lentivirus was concentrated by ultracentrifugation at 25,000 rpm for 2 h at 4 °C. The viral pellet was resuspended in PBS and stored at −80 °C until use. Viral titre was quantified with the use of a Cre-dependent GreenGo reporter cell line. For in vitro experiments, medium containing virus was collected, filtered, and added directly to recipient cells with polybrene at 8 µg ml−1 for 48 h before selection.
Mouse models
All mouse experiments described in this study were approved by the NYU Institutional Animal Care and Use Committee (IACUC). Mice were housed according to IACUC guidelines in ventilated caging in a specific pathogen-free animal facility. For all mouse studies, ≥4 mice were used for each experimental condition. KrasLSL-G12D/+; Tp53fl/fl; Rosa26LSL-Cas9/LSL-Cas9 (KPC GEMMs) mice were bred as previously described31,32,33,34,35. C57BL/6 J (JAX strain 000664) mice with the appropriate genotype, aged 8 to 12 weeks were randomly selected to begin tumour initiation studies with pUSEC lentivirus. Care was taken to ensure each experimental arm had an equal number of male and female mice. Mice were intratracheally infected with lentiviruses as described and monitored until experimental endpoint. Tumour burden was quantified by H&E staining and analysed using QuPath software as a measurement of total tumour area/total lung lobe area. All quantifications were done with investigator blinded to the respective sample genotypes. All transplantation experiments were performed using nude (JAX strain 002019), NOD SCID Gamma (NSG; JAX strain 005557 F), C57BL/6 J Fsp1-knockout (Conrad group16) or C57BL/6 J wild-type (JAX strain mice aged 8 to 12 weeks old). For mouse cell xenograft experiments, 100,000 cells in 100 µl of phosphate-buffered saline (PBS) was injected subcutaneously into each flank of the mouse. For the xenograft studies in Fig. 3 the number of cells injected per flank and whether they were injected with 50:50 PBS and Matrigel (Corning) are indicated as follows: H2009 (2 million cells + Matrigel), H1299 (1 million cells), PC9 (1 million cells), H1975 (2 million cells + Matrigel), A549 (1 million cells), 16645 (500,000 cells), KPC7 (250,000 cells). All human cell line xenograft experiments were carried out in male NSG mice unless specified. For the PDX experiment, tumours were implanted subcutaneously in male NSG mice as previously described34. Tumours were measured with callipers, and volume was calculated based on 0.5 × length × width2. The maximum tumour diameter permitted by our IACUC protocol was 2 cm, and this was not exceeded in any experiment. For orthotopic lung tumour experiments, 100,000 luciferase-expressing cells in 200 μl of PBS were injected intravenously into tail vein of male mice unless specified in the legend. Tumour growth was measured by bioluminescence (PerkinElmer IVIS Spectrum In Vivo Imaging System; D-luciferin, PerkinElmer 122799). Data were analysed using Living Image software.
Antioxidant and drug treatments
For LIP1 treatment, mice were dosed with 10 mg kg−1 LIP1 (BOC Sciences) or vehicle (2% DMSO + 40% PEG300 + 2% Tween 80 in sterile H2O) by intraperitoneal injection for frequency and duration indicated in figure schematics. For icFSP1 treatment, mice were dosed with 50 mg kg−1 icFSP1 (WuXi LabNetwork) or vehicle (45% PEG300 in sterile PBS) by intraperitoneal injection twice daily. High (TD.2108412) and low (TD.210841) irradiated vitamin E diets were obtained from Inotivco and provided ad libitum for length of time indicated in figure legends. In all experiments, mice were randomly assigned to treatment group.
Cell clonogenic and viability assays
Cell clonogenic assays were conducted by seeding 2,000 cells per well into 12-well dishes (BD/Falcon) in RPMI-1640 medium. After 12–16 h, medium containing RSL3 and/or LIP1 was added to wells. After 5 days of growth, plates were washed twice with PBS and stained with 0.5% crystal violet (Fisher Scientific) solution in 20% methanol. Plates were dried, scanned, and crystal violet was quantified by solubilization with 10% acetic acid and measurement of absorbance at 592 nm by spectrometer (Molecular Devices). Cell viability assays were conducted by seeding 2,000 cells per well into white-walled, clear-bottom, 96-well plates (Corning) in RPMI-1640 medium. After 12–16 h, medium containing RSL3, LIP1 and/or FSEN1, iFSP1 or icFSP1 was added. After three days, cell viability was assessed by CellTiter-Glo (Promega) and luminescence was measured by spectrometer (Molecular Devices).
Immunoblotting
Cells were plated to 75% confluency in a 6-well dish, and the following day cells were lysed on ice with Pierce RIPA buffer (ThermoScientific) containing 1× protease/phosphatase inhibitor cocktail (Thermo Fisher Scientific). Samples were sonicated in cooled 4 °C water (12 rounds, 30 s on and 30 s off) and then centrifuged at 14,000 rpm at 4 °C for 15 min. The supernatant was collected, and protein was quantified using the DC Rad Protein Assay kit. Protein was diluted to 1 μg μl−1 with water and 4× NuPage LDS sample buffer, then boiled at 95 °C for 10 min. Twenty micrograms of protein per well was loaded into Invitrogen 4–12% Bis-Tris gels and then transferred onto PVDF membranes using a standard protocol. PVDF membranes were then blocked using 5% BSA in TBST for 60 min at room temperature and incubated with primary antibodies in 5% BSA overnight at 4 °C. Primary antibodies were obtained as follows: GPX4 (Abcam), FSP1 (Proteintech), ACSL4 (Santa Cruz Biotechnologies), phospho-ERK (Cell Signaling), ERK (Cell Signaling) and Hsp90 (BD Bioscences). The following day, membranes were washed in TBST and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Enhanced chemiluminescent horseradish peroxidase substrate (ThermoScientific SuperSignal West PICO Plus) was added to the membrane for 1 min, and the resulting membrane was imaged using the General Electric Amersham Imager 680. For gel source data, see Supplementary Data 1.
Oxidized lipidomics
For the Gpx4-knockout experiment, cells were plated to 75% confluency in 6-well dishes. The following day, cells were treated with DMSO, RSL3 (0.5 µM), or RSL3(0.5 µM) plus LIP1(100 nM). After 8 h, cells were collected and washed in an antioxidant solution (PBS containing dibutylhydroxytoluene (100 µM) and diethylenetriamine pentaacetate (100 µM)) and centrifuged. Supernatent was discarded and cell pellets were immediately frozen in liquid nitrogen and stored at –80 °C. Frozen samples were sent on dry ice to Wayne State Lipidomics Core for metabolite extraction and LC–MS analysis.
Fsp1 wild-type and knockout tumour and cell lipidomic analysis were performed in the laboratory of Y.P.K. Fsp1 wild-type and knockout cells were plated at 75% confluency in 10 cm plates and treated as stated above. After euthanasia, mice were perfused with the antioxidant solution (+3.8% trisodium citrate), orthotopic tumours were microdissected, frozen in liquid nitrogen and stored at –80 °C. Mouse lung cancer tissues were cryopulverized and extracted with chloroform:methanol (2:1, v/v) at a tissue concentration of 25 mg mL−1. For the cells, 1.0 × 107 cells were extracted by 1 ml of chloroform:methanol (2:1, v/v). EquiSPLASH LIPIDOMIX internal standard (Avanti Polar Lipids) was added to the extraction solvent at 0.1 µg ml−1 per lipid class. Samples were sonicated on ice for 1 min using a VCX 130 probe sonicator (5 s on/off cycles), incubated for 30 min, and centrifuged at 17,000 × g for 20 min at 4 °C. The supernatant was was dried under vacuum (EZ-2 Elite, Genevac), reconstituted in 10% of the original volume with isopropanol, and transferred to glass autosampler vials for LC–MS analysis. Chromatographic separation was performed using a Waters ACQUITY UPLC CSH C18 column (100 × 2.1 mm, 1.7 µm) with a VanGuard precolumn (5 × 2.1 mm, 1.7 µm). Mobile phase A consisted of acetonitrile/water (60:40, v/v), and mobile phase B of isopropanol/acetonitrile (90:10, v/v), both containing 5 mM ammonium formate and 0.1% formic acid. The column was maintained at 65 °C with a flow rate of 0.6 ml min−1. The gradient was as follows: 0–2 min, 15–30% B; 2–2.5 min, 30–48% B; 2.5–11 min, 48–82% B; 11–11.5 min, 82–99% B; 11.5–12 min, 99% B; 12–12.1 min, 99–15% B; and 12.1–16 min, hold at 15% B for re-equilibration. Mass spectrometry was performed on a Q Exactive Plus Quadrupole-Orbitrap (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) source and operated in negative ion mode. Source settings were: sheath gas, 60 a.u.; auxiliary gas, 25 a.u.; sweep gas, 2 a.u.; spray voltage, 3.0 kV; capillary temperature, 320 °C; S-lens RF level, 50%; and auxiliary gas heater temperature, 370 °C. Parallel reaction monitoring (PRM) was carried out with the following parameters: resolution, 17,500 at m/z 200; AGC target, 2 × 105; maximum injection time, 50 ms; isolation window, m/z 1.2; and stepped normalized collision energies of 20, 30, and 40. The oxidized lipid analysis was adapted by previous PRM based analysis53. To generate the PRM inclusion list, pooled sample extracts were first analysed in DDA mode to identify the most abundant polyunsaturated phosphatidylcholine and phosphatidylethanolamine species using Lipostar254. These precursors were subjected to in silico oxidation using LPPtiger2 to predict candidate oxidized lipids. A semi-targeted DDA experiment was then conducted to confirm precursor detectability and finalize the inclusion list53. Data were analysed using Skyline (v24.1)55. Quantification was based on fragment anions derived from oxidized fatty acyl chains, with peak areas normalized to phosphatidylcholine(15:0/18:1(d7)) or phosphatidylethanolamine(15:0/18:1(d7)) internal standards from the EquiSPLASH LIPIDOMIX mixture.
CoQ9 and CoQ9H2 analysis
CoQ9 and CoQ9H2 analysis was conducted by modification of previous study56. The D9-CoQ10 standard was purchased from IsoSciences (Ambler, PA, USA). The D6-CoQ10H2 standard was synthesized from D6-CoQ10, which was obtained from Good Laboratory Practice Bioscience (Montclair, CA, USA). The CoQ9 standard was purchased from Tokyo Chemical Industry (Tokyo, Japan), and CoQ9H2 was synthesized from CoQ9. D6-CoQ10H2 and D9-CoQ10 were used as internal standards for the CoQ9H2 and CoQ9, respectively. Chloroform: methanol (2:1, v/v), containing internal standards of 0.5 µM of D6-CoQ10H2 and D6-CoQ10 was used to extract mouse lung cancer tissue at a tissue concentration of 25 mg ml−1. For the cells, 1.0 × 107 cells were extracted by 1 ml of same extraction solvent. Samples were then sonicated on ice for 1 min using a VCX 130 probe sonicator (5 s on/off cycles), incubated for 30 min, and centrifuged at 17,000g for 20 min at 4 °C. A 100 µl aliquot of the supernatant was transferred to glass autosampler vials for LC–MS/MS analysis. The liquid chromatography conditions were identical to those used for oxidized lipid analysis. Q Exactive Plus MS was operated in positive ion mode. Source settings were as follows: sheath gas, 60 a.u. (arbitrary units); auxiliary gas, 25 a.u.; sweep gas, 2 a.u.; spray voltage, 3.0 kV; capillary temperature, 320 °C; S-lens RF level, 50%; and auxiliary gas heater temperature, 370 °C. The mass range was m/z 120–1,200; resolution, 70,000 at m/z 200; AGC target, 1 × 106; and maximum injection time, 100 ms. By using EL–MAVEN (v0.12.0), the LC–MS peaks of CoQ9, CoQ9H2, D6-CoQ10H2, D6-CoQ10, and D9-CoQ10 were identified by matching with standard library and their peak areas were extracted with 10 ppm error range. Standard curves were generated using known concentrations of CoQ9H2 and CoQ9, and used to calculate the concentrations of CoQ9H2 and CoQ9 in the samples following an algorithm described in a previous study56.
Immunohistochemistry
Tumour-bearing mice were euthanized by carbon dioxide asphyxiation, after which the lungs were dissected and fixed in 4% PFA solution overnight. Fixed lungs were washed with PBS 2 times, transferred, and stored in 70% ethanol, until cassette loading and paraffin embedding. Sections were cut and stained with H&E. For immunohistochemistry with the exception of TUNEL staining, sections were immunostained on a Leica BondRX automated stainer according to the manufacturer’s instructions. In brief, tissues underwent deparaffinization online, followed by epitope retrieval for 20 min at 100° with Leica Biosystems ER2 solution (pH9, AR9640), endogenous peroxidase activity blocking with H2O2, and non-specific binding site blocking with Rodent Block M (Biocare, RBM961L) and Bond Primary Antibody Diluent (Leica Biosystems, AR9352). Sections were then incubated with primary antibodies against GPX4 (Abcam), FSP1 (obtained from M. Conrad), 4-HNE (JaICA), Ki67 and cleaved caspase-3 for 60 min at room temperature. Primary antibodies were detected with anti-rat HRP-conjugated polymer (Biocare, BRR4016H), 3,3′-diaminobenzidine (DAB) substrate (provided in the Leica BOND Polymer Refine Detection System, DS9800), and for 4-HNE staining Bond DAB Enhancer (Leica Biosystems, AR9432). Following counter-staining with haematoxylin, slides were scanned at 40× on a Hamamatzu Nanozoomer (2.0HT). TUNEL staining was performed by Histowiz according to their protocol.
For OPAL imaging, coronal 5-µm sections were immunostained on a Leica BondRx auto-stainer according to the manufacturer’s instructions. In brief, sections were deparaffiinized online and then treated with 3% H2O2 to inhibit endogenous peroxidases, followed by antigen retrieval with either ER1 (Leica, AR9961; pH6) or ER2 (Leica, AR9640; pH9) retrieval buffer at 100 °C for 20 min. After blocking with either Rodent Block M (Biocare, RBM961L) or Primary Antibody Diluent (Leica, AR93520), slides were incubated with the first primary antibody (FSP1 (obtained from M. Conrad); phospho-ERK1/2, CST; GPX4, Abcam) and secondary HRP polymer pair, followed by HRP-mediated tyramide signal amplification with a specific Opal fluorophore. Once the Opal fluorophore was covalently linked to the antigen, primary and secondary antibodies were removed with a heat retrieval step. This sequence was repeated three more times with subsequent primary and secondary antibody pairs, using a different Opal fluorophore with each primary antibody (see table below for primary antibody sequence and reagent details). After antibody staining, sections were counterstained with spectral DAPI (Akoya Biosciences, FP1490) and mounted with ProLong Gold Antifade (ThermoFisher Scientific, P36935). Semi-automated image acquisition was performed on an Akoya Vectra Polaris (PhenoImagerHT) multispectral imaging system. Slides were scanned at 20× magnification using PhenoImagerHT 2.0 software in conjunction with Phenochart 2.0 and InForm 3.0 to generate unmixed whole slide qptiff scans. All image files were uploaded to the NYUGSoM’s OMERO Plus image data management system (Glencoe Software).
Flow cytometry
Lung tissue was processed into a single-cell suspension for flow cytometry as previously described42,57. In brief, prior to euthanasia, mice were injected with 2 μg of Anti-mouse CD45-APC conjugated antibody (Biolegend, Clone 30-F11, 103111) retro-orbitally. Lungs were harvested and digested with Collagenase (Sigma-Aldrich, C5138) and deoxyribonuclease I (Sigma-Aldrich, DN25) followed by red blood cell lysis. Single cells were then resuspended in fluorescence-activated cell sorting (FACS) buffer and stained using the following antibodies: CD45 (Biolegend, 103132), CD11b (Biolegend, 101216), CD11c (Biolegend, 117324), Ly6G (Biolegend, 127622), MHCII (BD, 748708), CD103 (Biolegend, 121433), CD64 (Biolegend, 139309), SiglecF (BD, 740956), MertK (R&D, BAF591), CD45 (BD, 748371), CD3e (BD, 740854), CD4 (Invitrogen, MCD0428), CD8a (EBioscience, 563152) and Secondary (Streptavidin) (BD, 564176). Samples were run on the BD LSRFortessa and analysed on FlowJo version 10. Gating strategy is presented in Supplementary Data 2.
TCGA analyses
Gene expression profiles of primary tumours and relevant clinical data of 515 patients with LUAD were obtained from The Cancer Genome Atlas6 (TCGute.org). GPX4 and FSP1 (AIFM2) mutational status of TCGA tumour samples was retrieved from cBioPortal151 using the TCGA PanCancer Atlas collection (https://gdc.cancer.gov/about-data/publications/pancanatlas). For survival data, patients were stratified based on GPX4 or FSP1 expression and overall survival rates were plotted to compare patients with high-GPX4 or FSP1 expression (top 50% above median expression) with the rest of the cohort (n = 464 patients). All survival analyses were conducted using the survival curve analyses in GraphPad Prism v9.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism v9. All data are expressed as mean plus standard error of the mean, unless otherwise specified. Data were analysed by statistical tests as indicated in figure legends. All tests were two-tailed and replicates are biological unless otherwise stated. All western blots were replicated at least three times with results reproducible of data shown in figures. All in vitro assays were replicated at least three times with a minimum of n = 3 biological replicates per group for statistical power. For all in vivo experiments, the minimum sample size was four independent mice or tumours and respective sample size per genotype or condition is further specified in the figure legend. Sample size was not calculated but was chosen in each experiment based on previous experience with various models and to ensure that there were enough samples for statistical power. All in vivo experiments were replicated at least twice with results reproducible of data shown in figures. When representative images are shown, a minimum of three samples from the larger cohort were stained from each group. In the case of representative MRI images, all mice from the cohort were imaged. During sample processing and analysis for the lipidomic the samples were given numeric IDs which after analysis, were unblinded and graphed. For histological and immunohistochemistry analysis, researcher was blinded to the sample condition. The investigators were not blinded during most other data collection or analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data and raw gel images are included with the paper. Raw lipidomic data have been deposited in the MassIVE data base (https://massive.ucsd.edu/) under accession number MSV000098883. Analysed lipidomic data are available in Supplementary Data 3. Gene expression profiles of primary tumours and relevant clinical data of 515 patients with LUAD were obtained from The Cancer Genome Atlas (https://www.cancer.gov/ccg/research/genome-sequencing/tcga). GPX4, FSP1 (AIFM2), KRAS, EGFR, KEAP1 and STK11 mutational status of TCGA tumour samples was retrieved from cBioPortal151 using the TCGA PanCancer Atlas collection (https://gdc.cancer.gov/about-data/publications/pancanatlas). All other materials are available upon request from T.P. Source data are provided with this paper.
References
Dixon, S. J. et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell 149, 1060–1072 (2012).
Viswanathan, V. S. et al. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature 547, 453–457 (2017).
Hangauer, M. J. et al. Drug-tolerant persister cancer cells are vulnerable to GPX4 inhibition. Nature 551, 247–250 (2017).
Berndt, C. et al. Ferroptosis in health and disease. Redox Biol 75, 103211 (2024).
Nakamura, T. & Conrad, M. Exploiting ferroptosis vulnerabilities in cancer. Nat. Cell Biol. 26, 1407–1419 (2024).
Friedmann Angeli, J. P. et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 16, 1180–1191 (2014).
Yang, W. S. et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 156, 317–331 (2014).
Bersuker, K. et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 575, 688–692 (2019).
Doll, S. et al. FSP1 is a glutathione-independent ferroptosis suppressor. Nature 575, 693–698 (2019).
Wiernicki, B. et al. Excessive phospholipid peroxidation distinguishes ferroptosis from other cell death modes including pyroptosis. Cell Death Dis. 11, 922 (2020).
Conrad, M. & Pratt, D. A. The chemical basis of ferroptosis. Nat. Chem. Biol. 15, 1137–1147 (2019).
Dixon, S. J. & Olzmann, J. A. The cell biology of ferroptosis. Nat. Rev. Mol. Cell Biol. 25, 424–442 (2024).
Kraft, V. A. N. et al. GTP cyclohydrolase 1/tetrahydrobiopterin counteract ferroptosis through lipid remodeling. ACS Cent. Sci. 6, 41–53 (2020).
Soula, M. et al. Metabolic determinants of cancer cell sensitivity to canonical ferroptosis inducers. Nat. Chem. Biol. 16, 1351–1360 (2020).
Garcia-Bermudez, J. et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature 567, 118–122 (2019).
Qiu, B. et al. Phospholipids with two polyunsaturated fatty acyl tails promote ferroptosis. Cell 187, 1177–1190.e1118 (2024).
Kagan, V. E. et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat. Chem. Biol. 13, 81–90 (2017).
Doll, S. et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat. Chem. Biol. 13, 91–98 (2017).
Dixon, S. J. et al. Human haploid cell genetics reveals roles for lipid metabolism genes in nonapoptotic cell death. ACS Chem. Biol. 10, 1604–1609 (2015).
Mishima, E. et al. A non-canonical vitamin K cycle is a potent ferroptosis suppressor. Nature 608, 778–783 (2022).
Seiler, A. et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent- and AIF-mediated cell death. Cell Metab. 8, 237–248 (2008).
Zilka, O. et al. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent. Sci. 3, 232–243 (2017).
Jiang, X., Stockwell, B. R. & Conrad, M. Ferroptosis: mechanisms, biology and role in disease. Nat. Rev. Mol. Cell Biol. 22, 266–282 (2021).
Nakamura, T. et al. Phase separation of FSP1 promotes ferroptosis. Nature 619, 371–377 (2023).
Eaton, J. K. et al. Selective covalent targeting of GPX4 using masked nitrile-oxide electrophiles. Nat. Chem. Biol. 16, 497–506 (2020).
Yang, W. S. & Stockwell, B. R. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem. Biol. 15, 234–245 (2008).
Xavier da Silva, T. N., Schulte, C., Alves, A. N., Maric, H. M. & Friedmann Angeli, J. P. Molecular characterization of AIFM2/FSP1 inhibition by iFSP1-like molecules. Cell Death Dis. 14, 281 (2023).
Hendricks, J. M. et al. Identification of structurally diverse FSP1 inhibitors that sensitize cancer cells to ferroptosis. Cell Chem. Biol. 30, 1090–1103.e1097 (2023).
Nakamura, T. et al. Integrated chemical and genetic screens unveil FSP1 mechanisms of ferroptosis regulation. Nat. Struct. Mol. Biol. 30, 1806–1815 (2023).
Yoshioka, H. et al. Identification of a small molecule that enhances ferroptosis via inhibition of ferroptosis suppressor protein 1 (FSP1). ACS Chem. Biol. 17, 483–491 (2022).
Sanchez-Rivera, F. J. et al. Rapid modelling of cooperating genetic events in cancer through somatic genome editing. Nature 516, 428–431 (2014).
Ding, H. et al. Activation of the NRF2 antioxidant program sensitizes tumors to G6PD inhibition. Sci. Adv. 7, eabk1023 (2021).
Lignitto, L. et al. Nrf2 activation promotes lung cancer metastasis by inhibiting the degradation of Bach1. Cell 178, 316–329.e318 (2019).
Romero, R. et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 23, 1362–1368 (2017).
Best, S. A. et al. Distinct initiating events underpin the immune and metabolic heterogeneity of KRAS-mutant lung adenocarcinoma. Nat. Commun. 10, 4190 (2019).
Takahashi, N. et al. 3D culture models with CRISPR screens reveal hyperactive NRF2 as a prerequisite for spheroid formation via regulation of proliferation and ferroptosis. Mol. Cell 80, 828–844.e826 (2020).
Zou, Y. et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 10, 1617 (2019).
Vidigal, J. A. & Ventura, A. Rapid and efficient one-step generation of paired gRNA CRISPR–Cas9 libraries. Nat. Commun. 6, 8083 (2015).
Ingold, I. et al. Selenium utilization by GPX4 is required to prevent hydroperoxide-induced ferroptosis. Cell 172, 409–422.e421 (2018).
Cancer Genome Atlas Research, N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Muller, F. et al. Elevated FSP1 protects KRAS-mutated cells from ferroptosis during tumor initiation. Cell Death Differ. 30, 442–456 (2023).
Zavitsanou, A. M. et al. KEAP1 mutation in lung adenocarcinoma promotes immune evasion and immunotherapy resistance. Cell Rep. 42, 113295 (2023).
Koppula, P. et al. A targetable CoQ–FSP1 axis drives ferroptosis- and radiation-resistance in KEAP1 inactive lung cancers. Nat. Commun. 13, 2206 (2022).
Wu, K., El Zowalaty, A. E., Sayin, V. I. & Papagiannakopoulos, T. The pleiotropic functions of reactive oxygen species in cancer. Nat. Cancer 5, 384–399 (2024).
Cheung, E. C. & Vousden, K. H. The role of ROS in tumour development and progression. Nat. Rev. Cancer 22, 280–297 (2022).
Palma, M. et al. Lymph node environment drives FSP1 targetability in metastasizing melanoma. Nature https://doi.org/10.1038/s41586-025-09709-1 (2025).
Yant, L. J. et al. The selenoprotein GPX4 is essential for mouse development and protects from radiation and oxidative damage insults. Free Radic. Biol. Med. 34, 496–502 (2003).
Mei, J., Webb, S., Zhang, B. & Shu, H. B. The p53-inducible apoptotic protein AMID is not required for normal development and tumor suppression. Oncogene 25, 849–856 (2006).
Nguyen, H. P. et al. Aifm2, a NADH oxidase, supports robust glycolysis and is required for cold- and diet-induced thermogenesis. Mol. Cell 77, 600–617.e604 (2020).
Drijvers, J. M. et al. Pharmacologic screening identifies metabolic vulnerabilities of CD8+ T cells. Cancer Immunol. Res. 9, 184–199 (2021).
Matsushita, M. et al. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J. Exp. Med. 212, 555–568 (2015).
Winslow, M. M. et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature 473, 101–104 (2011).
Criscuolo, A. et al. Analytical and computational workflow for in-depth analysis of oxidized complex lipids in blood plasma. Nat. Commun. 13, 6547 (2022).
Goracci, L. et al. Lipostar, a comprehensive platform-neutral cheminformatics tool for lipidomics. Anal. Chem. 89, 6257–6264 (2017).
Adams, K. J. et al. Skyline for small molecules: a unifying software package for quantitative metabolomics. J. Proteome Res. 19, 1447–1458 (2020).
Kang, Y. P., Kim, T. H., Ngoc Nguyen, C. T., Kim, S. M. & Kwon, S. W. Robust determination of coenzyme Q10 redox status using two isotope-labeled internal standards. Preprint at https://doi.org/10.2139/ssrn.4982487 (2024).
Pillai, R. et al. Glutamine antagonist DRP-104 suppresses tumor growth and enhances response to checkpoint blockade in KEAP1 mutant lung cancer. Sci. Adv. 10, eadm9859 (2024).
Acknowledgements
We thankG. Ward and C. Loomis from the Experimental Pathology Core (RRID:SCR_017928) at NYU Langone Health, which is partially supported by the Cancer Center Support Grant P30CA016087 at NYU Langone’s Laura and Isaac Perlmutter Cancer Center, for support with immunohistochemistry staining and imaging. The PhenoImagerHT Mutlispectral imaging system was initially purchased through a Shared Instrumentation Grant S10OD021747. We thank the Wayne State University School of Medicine Lipidomics Core Facility, which is supported in part by National Center for Research Resources, National Institutes of Health Grants S10RR027926 and S10OD032292, for support with LC–MS. The analysis of FSP1-oxidized lipids and CoQ9 were conducted by the Proteomics and Metabolomics Core at the College of Pharmacy, Seoul National University, South Korea. We thank R. Possemato and S. Kotschi for critical reading of this manuscript. We acknowledge J. Teixeira for supporting and motivating this work. T.P. is supported by NIH grants (R37CA222504, R01CA227649, R01CA283049 and R01CA262562) and an American Cancer Society Research Scholar Grant (RSG-17-20001–TBE). K.W. is supported by the Ruth L. Kirschstein Individual Predoctoral NRSA fellowship (F30CA275258) and NIH training grants (T32GM136573 and T32GM136542). A.J.V. is supported by the NIH training grant (T32GM136542). M.C. received funding from the Deutsche Forschungsgemeinschaft (DFG) (CO 291/7-1), the Priority Program SPP 2306 (CO 291/9-1, 461385412 and CO 291/10-1, 461507177) and the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (grant agreement GA 884754).
Author information
Authors and Affiliations
Contributions
T.P., K.W. and A.J.V. designed and directed the study. K.W., A.J.V., J.P.B., A.Z., M.N.N., and M.M. performed various in vitro and in vivo experiments reported in the study. R.P. performed FACS for lung tumour immune profiling. Y.H. performed TCGA patient data analyses. S.M.K. performed FSP1 epilipidomics and CoQ measurements, T.H.K. assisted with data analysis, and Y.P.K. supervised. L.W. and D. Simeone provided guidance for the pancreatic cancer experiment. S.C. performed unbiased tumour burden quantification. M.M. and S.R. maintained and genotyped all experimental mice. M.C., T.N., M.H. and D. Shackelford provided conceptual advice. T.P., K.W. and A.J.V. wrote the manuscript with comments and input from all authors.
Corresponding author
Ethics declarations
Competing interests
T.P. received funding from Pfizer Medical Education Group, Dracen Pharmaceuticals, Kymera Therapeutics, Bristol Myers Squibb and Agios Pharmaceuticals not related to the submitted work. M.C. is a co-founder and shareholder of ROSCUE Therapeutics GmbH. M.C. and T.N. have filed a patent application (WO2024115673A1) for some of the FSP1 inhibitor compounds described here. The other authors declare no competing interests.
Peer review
Peer review information
Nature thanks Jiyeon Kim, Hozumi Motohashi and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Gpx4 is required by lung cancer cells.
a, Quantification and representative images of Gpx4 IHC in KP LUAD GEMM tumours with knockout of either control (Neo, n = 11) or Gpx4 (n = 12). Scale bars: 200 µm. b, Representative 4-hydroxy-2-noneal (4-HNE) IHC of liver tissue from conditional Gpx4-knockout mice. Scale bars: 100 µm. c, Top: Western blot of KP LUAD cells with CRISPR/Cas9-mediated genetic deletion of Gpx4 with either two individual or duplexed sgRNAs. Bottom: representative images of crystal violet clonogenic assay of KP LUAD cells with knockout of either control (Neo) or Gpx4. Cells were treated with 100 nM LIP1. d, Heatmap of LC-MS detection of oxidized phospholipids in KP LUAD cells treated with DMSO control, RSL3 (0.5 µM), and RSL3 (0.5 µM) + LIP1 (100 nM) for 8 h. e, Schematic of Gpx4 ectopic overexpression (OE) method in KP LUAD cells. Western blot and representative images of crystal violet clonogenic assay of KP LUAD cells with wildtype (WT) or OE of Gpx4. f, CellTiter-Glo Luminescence viability assay of KP, Gpx4WT or Gpx4OE cells upon increasing concentrations of RSL3 (n = 5 per group). g, Western blot of KP LUAD cells treated with 20 nM Na2SeO3 or DMSO. CellTiter-Glo Luminescence viability assay of KP LUAD cells treated with 20 nM Na2SeO3 or DMSO with increasing RSL3 addition (n = 5 per group). h, GPX4 expression in KRAS-mutant primary LUAD tumours from TCGA, divided into early and late tumour stages (normal lung, n = 54; stage I/II, n = 354; stage III/IV, n = 98). i, Overall survival of KRAS-mutant LUAD patients (n = 464) from TCGA, stratified by high vs low tumour GPX4 expression. j, Tumour Gpx4 IHC quantification of KP LUAD GEMM tumours at 10 weeks (n = 4) vs 14 weeks (n = 4) post-tumour initiation. Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers). Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA with multiple comparisons (panel h), two-sided student’s t-test (panel j) or Kaplan-Meier simple survival analysis (panel i). For gel source data, see Supplementary Data 1. Drawing in d created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Extended Data Fig. 2 FSP1’s anti-ferroptotic function is not dependent on oncogenic signalling.
a, FSP1 (AIFM2) expression of KRAS-mutant primary LUAD tumours from TCGA, divided into early and late tumour stages (normal lung, n = 54; stage I/II, n = 354; III/IV, n = 98). b, FSP1 (AIFM2) expression of KRAS-mutant primary LUAD tumours from TCGA, separated by the KRAS mutation (G12C n = 51; G12V n = 32; G12D n = 17; G12A n = 16; other n = 20). c, FSP1 (AIFM2) expression of primary LUAD tumours from TCGA, separated by oncogenic driver mutation (normal lung, n = 59; EGFR, n = 71; KRAS, n = 135; other n = 307). d, Western blot of KP LUAD human cell lines treated with 50 nM RMC-042 for indicated durations. e, representative multi-IF images of KP tumours for markers indicated. Panels are 10X, scale bars: 50 µm; insets are 20X, scale bars: 20 µm. f, FSP1 (AIFM2) expression of KRAS-mutant primary LUAD tumours from TCGA, separated by tumour co-mutation status (KEAP1/STK11 WT n = 88; STK11 mutant n = 20; KEAP1 mutant n = 16; KEAP1/STK11 mutant n = 11). g, FSP1 (AIFM2) expression in KP LUAD human cell lines treated with an Nrf2 activator, KI696, for 5 days (n = 3 per group). Box plots indicate median (middle line), 25th, 75th percentile (box) and 5th and 95th percentile (whiskers). Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA with multiple comparisons (panels a–c, f, g). For gel source data, see Supplementary Data 1.
Extended Data Fig. 3 Neither Fsp1 nor Gpx4 have an impact on tumour initiation, proliferation, or apoptosis.
a, Quantification and representative images of tumour Fsp1 IHC in KP LUAD GEMMs with knockout of either control (Neo, n = 11) or Fsp1 (n = 14). Scale bars: 100 µm. b, Tumour number quantification in KP LUAD GEMMs with tumour-specific knockout of either control (Neo, n = 4), Fsp1 (n = 7), or Gpx4 (n = 6). c, Individual area of KP LUAD GEMM tumours with knockout of either control (Neo, n = 465), Fsp1 (n = 453), or Gpx4 (n = 384). d, Quantification of Ki67 IHC of KP LUAD GEMM tumours with knockout of either control (Neo, n = 355), Fsp1 (n = 268), or Gpx4 (n = 291). e, Quantification of cleaved caspase-3 IHC of KP LUAD GEMM tumours with knockout of either control (Neo, n = 46), Fsp1 (n = 22), or Gpx4 (n = 52). Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA with multiple comparisons (panels b–e).
Extended Data Fig. 4 Fsp1 is required for cell-autonomous tumour growth in vivo.
a, Top: Western blot of KP LUAD cells with CRISPR/Cas9-mediated genetic deletion of Fsp1 with either two individual or duplexed sgRNAs. Bottom: representative images of crystal violet clonogenic assay of KP LUAD cells with knockout of either control (Neo) or Fsp1. Cells were treated with 100 nM LIP1. b, Schematic depicting all transplantation models performed using isogenic KP, Fsp1KO and Fsp1WT. c,d, Longitudinal growth and endpoint tumour weights of KP sgFsp1 (n = 8) versus control (sgNeo n = 10) subcutaneous (subQ) xenograft tumor transplanted into C57BL/6 J male mice. e, Longitudinal lung tumour growth (measured via bioluminescence) in C57BL/6 J male mice orthotopically transplanted with KP LUAD cells with CRISPR/Cas9-mediated knockout of Fsp1 (n = 8) or control (n = 7). f, Longitudinal lung tumour growth (measured via bioluminescence normalized to first timepoint (day 7)) in C57BL/6 J male mice orthotopically transplanted with isogenic KP, Fsp1KO (n = 6) and Fsp1WT (n = 7) cells. g, Longitudinal lung tumour growth (measured via absolute bioluminescence) in NU/J immunocompromised mice orthotopically transplanted with isogenic KP, Fsp1KO (n = 6) and Fsp1WT (n = 7) cells. h, Longitudinal lung tumour growth (measured via absolute bioluminescence) in C57BL/6 J female mice with orthotopic transplantation of isogenic KP, Fsp1KO (n = 8) and Fsp1WT (n = 8) cells. i, Longitudinal tumour growth and endpoint tumour weights of either isogenic KP, Fsp1KO or Fsp1WT subcutaneous (subQ) xenograft tumours transplanted in either C57BL/6 J WT (Fsp1KO, male n = 10 female n = 10; Fsp1WT, male n = 8, female n = 10) or C57BL/6 J Fsp1-knockout mice (Fsp1KO, males n = 6, females n = 6; Fsp1WT, male n = 4, female n = 6). j, Western blot of KP LUAD cells with wildtype (WT) or overexpression (OE) of Fsp1. Representative images of crystal violet clonogenic assay of KP LUAD cells with Fsp1WT or Fsp1OE. k, CellTiter-Glo Luminescence viability assay of KP, Fsp1WT or Fsp1OE cells (5 biological replicates per group) with increasing concentrations of RSL3. l,m, Longitudinal growth and endpoint weights of KP Fsp1WT (n = 10) or Fsp1OE (n = 10) subcutaneous (subQ) xenograft tumour transplanted into C57BL/6 J male mice. Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA with multiple comparisons (panel i), two-way ANOVA with Tukey’s multiple comparisons test (panels c, e–h, l), or two-sided student’s t-test (panels d, m). For gel source data, see Supplementary Data 1. Drawings in b,c,e–h,l created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Extended Data Fig. 5 FSP1 loss in vitro does not induce ferroptosis without RSL3.
a, Top: Western blot; bottom: baseline growth (normalized to control); and right: CellTiter-Glo Luminescence viability assay of H2009 cells with the lentiviral addition of Cas9 and guide RNAs targeting FSP1 or a non-targeting control (TOM) (at least 4 biological replicates per group). Viability assay is upon increasing concentrations of RSL3. b, As in a, but with H1299 cells. c, As in a, but with PC9 cells. d, As in a, but with H1975 cells. e, As in a, but with A549 cells. f, As in a, but with 16645 cells. g, As in a, but with KPC7 cells. Data are represented as mean values, error bars represent SEM, significance determined via two-sided student’s t-test (panels a–g). For gel source data, see Supplementary Data 1.
Extended Data Fig. 6 Fsp1 loss promotes tumour cell ferroptosis.
a, Western blot of KP, Gpx4KO LUAD cells with either empty vector (—) or Fsp1OE. b, Western blot of KP, Fsp1KO LUAD cells with re-expression of either empty vector (—), Fsp1WT, Gpx4OE. c, CellTiter-Glo Luminescence viability assay of KP, Fsp1KO LUAD cells in b upon increasing concentrations of RSL3. d, Endpoint tumour weights of subcutaneous (subQ) xenograft tumours from indicated cell lines transplanted in C57BL/6 J female mice (control n = 10; Fsp1WT, n = 9; Gpx4OE; n = 8). e, Top: Western blot of KP, Fsp1KO LUAD cells with re-expression of either empty vector (—), wildtype (WT) Fsp1, or mutant (mut) Fsp1. Bottom: representative images of crystal violet clonogenic assay. f, CellTiter-Glo Luminescence viability assay of KP, Fsp1KO LUAD cells with re-expression of either empty vector (Fsp1KO), Fsp1WT, or Fsp1mut upon increasing concentrations of RSL3. LIP1 used at 100 nM. g, Longitudinal tumour growth (measured via bioluminescence normalized to initial imaging timepoint (day14)) in C57BL/6 J male mice orthotopically transplanted with isogenic KP, Fsp1KO LUAD cells with re-expression of either empty control vector (Fsp1KO, n = 7), Fsp1WT (n = 7), or Fsp1mut (n = 7). h, Representative images of crystal violet clonogenic assay of KP, Fsp1WT or Fsp1KO LUAD cells with CRISPR/Cas9- mediated knockout of control (Neo) or Acsl4. i, Longitudinal tumour growth (measured by bioluminescence) of C57BL/6 J male mice with orthotopic transplantation of isogenic KP, Fsp1KO and Fsp1WT cells with CRISPR/Cas9-mediated knockout of control (Neo) or Acsl4 (WT, n = 5; Acsl4KO, n = 4; Fsp1KO, n = 7; Fsp1KOAcsl4KO, n = 7). j, Representative H&E of experiment in i. Scale bars: 1000 µm. k, Overall survival of C57BL/6 J male mice with orthotopic transplantation of isogenic KP, Fsp1KO and Fsp1WT cells with CRISPR/Cas9-mediated knockout of control (Neo) or Acsl4 (n = 5 per group). l, Longitudinal tumour growth (measured via bioluminescence normalized to first imaging timepoint (day7)) of C57BL/6 J mice orthotopically transplanted with KP, Fsp1WT cells and receiving high (n = 6) or low (n = 5) VitE diets ad libitum 5 days pre-tumor initiation. m, Tumour bioluminescence signal normalized to first imaging timepoint (day7) of C57BL/6 J mice orthotopically transplanted with KP, Fsp1KO cells and receiving Liproxstatin-1 (LIP1, n = 7) or vehicle (Veh, n = 5) daily after tumour establishment. n, Schematic of KP LUAD GEMMs intratracheally infected with pUSEC lentiviruses containing dual sgRNAs targeting Fsp1. Mice were dosed with LIP1 (n = 6) or Vehicle (Veh, n = 6) every other day starting from tumour initiation. o, Tumour burden of KP LUAD GEMMs with CRISPR/Cas9-mediated knockout of Fsp1 and treated with Veh or LIP1. p, Representative H&E and 4-HNE IHC of KP LUAD GEMM tumours with knockout of Fsp1 and treated with Veh or LIP1. Scale bars: 1000 µm. Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA (panel d), two-way ANOVA with Tukey’s multiple comparisons (panels g, i, l, m), two-sided student’s t-test (panel o), or Kaplain-Meier simple survival analysis (panel k). For gel source data, see Supplementary Data 1. Drawing in n created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Extended Data Fig. 7 icFSP1 exerts on-target tumor-suppressive effect.
a, Top: Western blot of KP, Fsp1KO LUAD cells with re-expression of either empty vector (—), wildtype (WT) hFSP1, or Q319K-mutant hFSP1. Bottom: representative images of crystal violet clonogenic assay. b, Cell-TiterGlo Luminescence viability assay of KP, Fsp1KO LUAD cells with re-expression of either empty vector (—), WT hFSP1, or Q319K-mutant hFSP1. c, CellTiter-Glo Luminescence viability assay of human or mouse Fsp1WT-expressing cells upon RSL3 (0.2 µM) plus increasing concentrations of iFSP1. d, CellTiter-Glo Luminescence viability assay as in c but with FSEN1. e, CellTiter-Glo Luminescence viability assay of human or mouse Fsp1WT-expressing cells upon increasing concentrations of icFSP1. f, CellTiter-Glo Luminescence viability assay of WT or Q319K-mutant hFSP1-expressing cells upon RSL3 (0.2 µM) plus increasing concentrations of icFSP1. g, Longitudinal tumour growth (measured via bioluminescence normalized to first imaging timepoint (day7)) in C57BL/6 J mice orthotopically transplanted with isogenic KP, Fsp1KO cells with re-expression of either human FSP1 (hFSP1, n = 5), mouse Fsp1 (mFsp1, n = 6), or control (n = 6) vector. h, Tumour burden and representative H&E of KP, Fsp1KO (n = 4) and hFSP1WT (n = 4) orthotopic lung tumours. Scale bars: 1000 µm. i, Representative H&E of KP, hFSP1WT orthotopic lung tumours from indicated treatment groups. Scale bar: 2000 µm. j, Percent of T cells, alveolar macrophages, interstitial macrophages, and neutrophils of total CD45+ cells in tumour bearing lungs from mice treated with the indicated compounds for two weeks (Veh, n = 8; LIP1, n = 7; icFSP1, n = 9; icFSP1 + LIP1, n = 6). k, Graphical summary depicting ferroptosis as a barrier to lung cancer, as loss of either Fsp1 or Gpx4 induces tumour cell ferroptosis and restricts disease progression. l, Summary table of cell lines, their respective tumour lineage, and mutations where Fsp1 loss resulted in tumour suppression in vivo. For all Cell-TiterGlo assays, n = 5 biological replicates per group. Data are represented as mean values, error bars represent SEM, significance determined via one-way ANOVA with multiple comparisons (panels g, j) or student’s t test (panel h). For gel source data, see Supplementary Data 1. Drawings in k created in BioRender. Vaughan, A. (2025) https://BioRender.com/99qhixq.
Supplementary information
Supplementary Data 1 and 2
Supplementary Figure 1: Gel source data. All uncropped western blot images with protein ladder markers. Supplementary Figure 2: Representative gating strategy. Figure from FlowJo that indicates the separation of cells using the various antibodies and fluorophores in the FACS experiment shown in Extended Data Fig. 7j.
Supplementary Data 3
Analysed lipidomic results. Sheet 1: Results of the oxidized lipidomic experiment of Fsp1 wild-type versus knockout tumours. Values are normalized peak areas that were converted to z-scores for the heat map shown in Fig. 4a. Sheet 2: CoQ9 concentration measurements from LC–MS experiment of Fsp1 wild-type and knockout tumours and cell lines treated with RSL3 or DMSO. Data includes raw measurements and those after correction algorithm as described in the methods section.
Supplementary Data 4
sgRNA sequences used to knock out indicated genes
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, K., Vaughan, A.J., Bossowski, J.P. et al. Targeting FSP1 triggers ferroptosis in lung cancer. Nature 649, 487–495 (2026). https://doi.org/10.1038/s41586-025-09710-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41586-025-09710-8
This article is cited by
-
Triggering ferroptosis to tackle cancer
Nature Reviews Drug Discovery (2026)
-
In vivo models bring FSP1 inhibitors to life
Nature Cell Biology (2026)
-
Cell death: targeting ferroptosis in cancer
Signal Transduction and Targeted Therapy (2026)
-
Multi-layered integrated shielding: engineering ferroptosis-resistant mesenchymal stem cells for precision therapy of intervertebral disc degeneration
Apoptosis (2026)
-
Lymph node environment drives FSP1 targetability in metastasizing melanoma
Nature (2026)