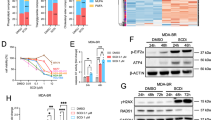
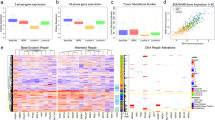
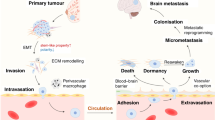
Abstract
Brain metastasis (BM) carries a poor prognosis, yet the molecular basis of brain tropism remains unclear. Analysis of breast cancer BM (BCBM) revealed pervasive p53 inactivation through mutations and/or aneuploidy, with pathway disruption already present in primary tumors. Functionally, p53 inactivation markedly increased BCBM formation and growth in vivo, causally linking p53 perturbation to BM. Mechanistically, p53 inactivation upregulated SCD1 and fatty acid synthesis (FAS), essential for brain-metastasizing cells; SCD1 knockout abolished the p53-dependent growth advantage. Molecularly, p53 suppressed SCD1 directly through promoter binding and indirectly by downregulating its co-activator DEPDC1. Astrocytes further enhanced FAS by secreting factors that were metabolized in a p53-dependent manner, promoting tumor survival, proliferation and migration. Finally, p53-deficient tumors were sensitive to FAS inhibition ex vivo and in vivo. Thus, we identify p53 inactivation as a driver of BCBM, reveal p53-dependent and astrocyte-dependent FAS modulation and highlight FAS as a therapeutically targetable BCBM vulnerability.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
TCGA4,15, METABRIC5, MBC17,29,30 and MSK16,17,19 data were obtained from cBioPortal73. GSE14017 (ref. 28), GSE14018 (ref. 28), GSE125989 (ref. 76), GSE12276 (ref. 74) and GSE76714 (ref. 75) were obtained from the GEO.
CAL51 TP53-KO RNA sequencing data were from ref. 36. RNA sequencing data for the hTERT-HMEC engineered cell lines were deposited to SRA under accession PRJNA1246204. RNA sequencing data of metastases were deposited to GEO under accession GSE245414. RNA sequencing data from patient autopsy samples are available upon request from BROCADE, subject to Access Committee and ethical approvals. These restrictions are in place to ensure that the data are used in compliance with ethical guidelines for patient confidentiality and informed consent. Application details are provided at https://www.petermac.org/research/cohort-studies/brocade/information-on-brocade-for-researchers. Applications are typically reviewed within 4–6 weeks.
RNA sequencing data of MDA-MB231 cells after siRNA-mediated knockdown of DEPDC1 were deposited to GEO under accession GSE281411. P53 CUT&Tag data were deposited to SRA, under accession PRJNA1247024. Source data are provided with this paper.
References
Harbeck, N. et al. Breast cancer. Nat. Rev. Dis. Primers 5, 66 (2019).
Chen, W., Hoffmann, A. D., Liu, H. & Liu, X. Organotropism: new insights into molecular mechanisms of breast cancer metastasis. NPJ Precis. Oncol. 2, 4 (2018).
Achrol, A. S. et al. Brain metastases. Nat. Rev. Dis. Primers 5, 5 (2019).
Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70 (2012).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352 (2012).
Liu, Y. et al. Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531, 471–475 (2016).
Ferraro, G. B. et al. Fatty acid synthesis is required for breast cancer brain metastasis. Nat. Cancer 2, 414–428 (2021).
Lorger, M. Tumor microenvironment in the brain. Cancers (Basel) 4, 218–243 (2012).
Adler, O. et al. Reciprocal interactions between innate immune cells and astrocytes facilitate neuroinflammation and brain metastasis via lipocalin-2. Nat. Cancer 4, 401–418 (2023).
Schwartz, H. et al. Incipient melanoma brain metastases instigate astrogliosis and neuroinflammation. Cancer Res. 76, 4359–4371 (2016).
Priego, N. et al. STAT3 labels a subpopulation of reactive astrocytes required for brain metastasis article. Nat. Med. 24, 1024–1035 (2018).
Doron, H. et al. Inflammatory activation of astrocytes facilitates melanoma brain tropism via the CXCL10-CXCR3 signaling axis. Cell Rep. 28, 1785–1798 (2019).
Zou, Y. et al. Polyunsaturated fatty acids from astrocytes activate PPARγ signaling in cancer cells to promote brain metastasis. Cancer Discov. 9, 1720–1735 (2019).
Jin, X. et al. A metastasis map of human cancer cell lines. Nature 588, 331–336 (2020).
Aaltonen, L. A. et al. Pan-cancer analysis of whole genomes. Nature 578, 82–93 (2020).
Razavi, P. et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell 34, 427–438 (2018).
Zehir, A. et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23, 703–713 (2017).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Nguyen, B. et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell 185, 563–575 (2022).
De Mattos-Arruda, L. et al. The genomic and immune landscapes of lethal metastatic breast cancer. Cell Rep. 27, 2690–2708 (2019).
Schrijver, W. A. M. E. et al. Mutation profiling of key cancer genes in primary breast cancers and their distant metastases. Cancer Res. 78, 3112–3121 (2018).
Siegel, M. B. et al. Integrated RNA and DNA sequencing reveals early drivers of metastatic breast cancer. J. Clin. Invest. 128, 1371–1383 (2018).
Saunus, J. M. et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J. Pathol. 237, 363–378 (2015).
Ding, L. et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature 464, 999–1005 (2010).
Piccirilli, C. et al. Allelic deletions on chromosome-17 and mutations in the p53 gene in tumors metastatic to brain. Int. J. Oncol. 4, 37–42 (1994).
Wang, Z. et al. Loss-of-function but not gain-of-function properties of mutant TP53 are critical for the proliferation, survival and metastasis of a broad range of cancer cells. Cancer Discov. 14, 362–379 (2024).
Xu, J. et al. 14-3-3ζ turns TGF-β’s function from tumor suppressor to metastasis promoter in breast cancer by contextual changes of Smad partners from p53 to Gli2. Cancer Cell 27, 177–192 (2015).
Zhang, X. H.-F. et al. Latent bone metastasis in breast cancer tied to Src-dependent survival signals. Cancer Cell 16, 67–78 (2009).
Jain, E. The Metastatic Breast Cancer Project: leveraging patient-partnered research to expand the clinical and genomic landscape of metastatic breast cancer and accelerate discoveries. Preprint at medRxiv https://doi.org/10.1101/2023.06.07.23291117 (2023).
Parry, M. Introducing the Metastatic Breast Cancer Project: a novel patient-partnered initiative to accelerate understanding of MBC. ESMO Open 3, e000452 (2018).
Riihimäki, M., Thomsen, H., Sundquist, K., Sundquist, J. & Hemminki, K. Clinical landscape of cancer metastases. Cancer Med. 7, 5534–5542 (2018).
Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014).
Xiong, Z., Gharagozlou, S., Vengco, I., Chen, W. & Ohlfest, J. R. Effective CpG immunotherapy of breast carcinoma prevents but fails to eradicate established brain metastasis. Clin. Cancer Res. 14, 5484–5493 (2008).
Rockwell, S. C., Kallman, R. F. & Fajardo, L. F. Characteristics of a serially transplanted mouse mammary tumor and its tissue-culture-adapted derivative. J. Natl Cancer Inst. 49, 735–749 (1972).
Gioanni, J. et al. Establishment and characterisation of a new tumorigenic cell line with a normal karyotype derived from a human breast adenocarcinoma. Br. J. Cancer 62, 8–13 (1990).
Redman-Rivera, L. N. et al. Acquisition of aneuploidy drives mutant p53-associated gain-of-function phenotypes. Nat. Commun. 12, 5184 (2021).
Quail, D. F. & Joyce, J. A. The microenvironmental landscape of brain tumors. Cancer Cell 31, 326–341 (2017).
Savino, A. M. et al. Metabolic adaptation of acute lymphoblastic leukemia to the central nervous system microenvironment depends on stearoyl-CoA desaturase. Nat. Cancer 1, 998–1009 (2020).
Schild, T., Low, V., Blenis, J. & Gomes, A. P. Unique metabolic adaptations dictate distal organ-specific metastatic colonization. Cancer Cell 33, 347–354 (2018).
Perelroizen, R. et al. Astrocyte immunometabolic regulation of the tumour microenvironment drives glioblastoma pathogenicity. Brain 145, 3288–3307 (2022).
Elahi, L. S. et al. Valproic acid targets IDH1 mutants through alteration of lipid metabolism. NPJ Metab. Health Dis. 2, 20 (2024).
Ferris, H. A. et al. Loss of astrocyte cholesterol synthesis disrupts neuronal function and alters whole-body metabolism. Proc. Natl Acad. Sci. USA 114, 1189–1194 (2017).
Moore, S. A. Polyunsaturated fatty acid synthesis and release. J. Mol. Neurosci. 16, 195–200 (2001).
Medina, J. M. & Tabernero, A. Astrocyte-synthesized oleic acid behaves as a neurotrophic factor for neurons. J. Physiol. Paris 96, 265–271 (2002).
Bernoud, N. et al. Astrocytes are mainly responsible for the polyunsaturated fatty acid enrichment in blood–brain barrier endothelial cells in vitro. J. Lipid Res. 39, 1816–1824 (1998).
Altea-Manzano, P. et al. A palmitate-rich metastatic niche enables metastasis growth via p65 acetylation resulting in pro-metastatic NF-κB signaling. Nat. Cancer 4, 344–364 (2023).
Koundouros, N. & Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 122, 4–22 (2020).
Guthmann, F., Haupt, R., Looman, A. C., Spener, F. & Rüstow, B. Fatty acid translocase/CD36 mediates the uptake of palmitate by type II pneumocytes. Am. J. Physiol. 277, L191–L196 (1999).
Pascual, G. et al. Targeting metastasis-initiating cells through the fatty acid receptor CD36. Nature 541, 41–45 (2017).
Vallvé, J.-C. et al. Unsaturated fatty acids and their oxidation products stimulate CD36 gene expression in human macrophages. Atherosclerosis 164, 45–56 (2002).
Yang, P. et al. Dietary oleic acid-induced CD36 promotes cervical cancer cell growth and metastasis via up-regulation Src/ERK pathway. Cancer Lett. 438, 76–85 (2018).
Feng, W. W., Zuppe, H. T. & Kurokawa, M. The role of CD36 in cancer progression and its value as a therapeutic target. Cells 12, 1605 (2023).
Röhrig, F. & Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 16, 732–749 (2016).
Sen, U., Coleman, C. & Sen, T. Stearoyl coenzyme A desaturase-1: multitasker in cancer, metabolism, and ferroptosis. Trends Cancer 9, 480–489 (2023).
Li, H. et al. The landscape of cancer cell line metabolism. Nat. Med. 25, 850–860 (2019).
Kaya-Okur, H. S., Janssens, D. H., Henikoff, J. G., Ahmad, K. & Henikoff, S. Efficient low-cost chromatin profiling with CUT&Tag. Nat. Protoc. 15, 3264–3283 (2020).
Kirschner, K. et al. Phenotype specific analyses reveal distinct regulatory mechanism for chronically activated p53. PLoS Genet. 11, e1005053 (2015).
Mirza, A. et al. Global transcriptional program of p53 target genes during the process of apoptosis and cell cycle progression. Oncogene 22, 3645–3654 (2003).
Bené, H., Lasky, D. & Ntambi, J. M. Cloning and characterization of the human stearoyl-CoA desaturase gene promoter: transcriptional activation by sterol regulatory element binding protein and repression by polyunsaturated fatty acids and cholesterol. Biochem. Biophys. Res. Commun. 284, 1194–1198 (2001).
Girardini, J. E. et al. A Pin1/mutant p53 axis promotes aggressiveness in breast cancer. Cancer Cell 20, 79–91 (2011).
Zhao, H. et al. High expression of DEPDC1 promotes malignant phenotypes of breast cancer cells and predicts poor prognosis in patients with breast cancer. Front. Oncol. 9, 262 (2019).
Brigandi, R. A., Zhu, J., Murnane, A. A., Reedy, B. A. & Shakib, S. A phase 1 randomized, placebo-controlled trial with a topical inhibitor of stearoyl-coenzyme A desaturase 1 under occluded and nonoccluded conditions. Clin. Pharmacol. Drug Dev. 8, 270–280 (2019).
Menendez, J. A. & Lupu, R. Fatty acid synthase (FASN) as a therapeutic target in breast cancer. Expert Opin. Ther. Targets 21, 1001–1016 (2017).
Tracz-Gaszewska, Z. & Dobrzyn, P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers (Basel) 11, 948 (2019).
Cheng, Y.-J., Fan, F., Zhang, Z. & Zhang, H. Lipid metabolism in malignant tumor brain metastasis: reprogramming and therapeutic potential. Expert Opin. Ther Targets 27, 861–878 (2023).
Tsherniak, A. et al. Defining a cancer dependency map. Cell 170, 564–576 (2017).
Sivanand, S. et al. Cancer tissue of origin constrains the growth and metabolism of metastases. Nat. Metab. 9, 1668–1681 (2024).
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nat. Genet. 33, 49–54 (2003).
Sanghvi, N. et al. Charting the transcriptomic landscape of primary and metastatic cancers in relation to their origin and target normal tissues. Sci. Adv. 10, eadn0220 (2024).
Sammarco, A. et al. Targeting SCD triggers lipotoxicity of cancer cells and enhances anti-tumor immunity in breast cancer brain metastasis mouse models. Commun. Biol. 8, 562 (2025).
Zou, Z., Ohta, T. & Oki, S. ChIP-Atlas 3.0: a data-mining suite to explore chromosome architecture together with large-scale regulome data. Nucleic Acids Res. 52, W45–W53 (2024).
Zou, Z., Ohta, T., Miura, F. & Oki, S. ChIP-Atlas 2021 update: a data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 50, W175–W182 (2022).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012).
Zhang, X. H.-F. et al. Selection of bone metastasis seeds by mesenchymal signals in the primary tumor stroma. Cell 154, 1060–1073 (2013).
Duchnowska, R. et al. Brain metastasis prediction by transcriptomic profiling in triple-negative breast cancer. Clin. Breast Cancer 17, e65–e75 (2017).
Iwamoto, T. et al. Distinct gene expression profiles between primary breast cancers and brain metastases from pair-matched samples. Sci. Rep. 9, 13343 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005).
Reich, M. et al. GenePattern 2.0. Nat. Genet. 38, 500–501 (2006).
Smid, M. et al. Subtypes of breast cancer show preferential site of relapse. Cancer Res. 68, 3108–3114 (2008).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Van Neerven, S. et al. Inflammatory cytokine release of astrocytes in vitro is reduced by all-trans retinoic acid. J. Neuroimmunol. 229, 169–179 (2010).
Agnese, S. T., Spierto, F. W. & Hannon, W. H. Evaluation of four reagents for delipidation of serum. Clin. Biochem. 2, 98–100 (1983).
Segal, E. et al. Targeting angiogenesis-dependent calcified neoplasms using combined polymer therapeutics. PLoS ONE 4, e5233 (2009).
Sanjana, N. E., Shalem, O. & Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Wang, B. et al. ATXN1L, CIC, and ETS transcription factors modulate sensitivity to MAPK pathway inhibition. Cell Rep. 18, 1543–1557 (2017).
Doench, J. G. et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR–Cas9. Nat. Biotechnol. 34, 184–191 (2016).
Tanaka, A., To, J., O’Brien, B., Donnelly, S. & Lund, M. Selection of reliable reference genes for the normalisation of gene expression levels following time course LPS stimulation of murine bone marrow derived macrophages. BMC Immunol. 18, 43 (2017).
Pozzi, S. et al. MCP-1/CCR2 axis inhibition sensitizes the brain microenvironment against melanoma brain metastasis progression. JCI Insight 7, e154804 (2022).
Folch, J., Lees, M. & Sloane Stanley, G. H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 (1957).
Leikin-Frenkel, A. et al. Dietary α linolenic acid in pregnant mice and during weaning increases brain docosahexaenoic acid and improves recognition memory in the offspring. J. Nutr. Biochem. 91, 108597 (2021).
Leikin-Frenkel, A. et al. The effect of α-linolenic acid enrichment in perinatal diets in preventing high fat diet-induced SCD1 increased activity and lipid disarray in adult offspring of low density lipoprotein receptor knockout (LDLRKO) mice. Prostaglandins Leukot. Essent. Fatty Acids 184, 102475 (2022).
Malitsky, S. et al. Viral infection of the marine alga Emiliania huxleyi triggers lipidome remodeling and induces the production of highly saturated triacylglycerol. New Phytol. 210, 88–96 (2016).
Zheng, L. et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 6, 6001 (2015).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based Analysis of ChIP–Seq (MACS). Genome Biol. 9, R137 (2008).
Ramírez, F. et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165 (2016).
Stuart, T., Srivastava, A., Madad, S., Lareau, C. A. & Satija, R. Single-cell chromatin state analysis with Signac. Nat. Methods 18, 1333–1341 (2021).
Zerbib, J. et al. Human aneuploid cells depend on the RAF/MEK/ERK pathway for overcoming increased DNA damage. Nat. Commun. 15, 7772 (2024).
Theodoropoulos, P. C. et al. Discovery of tumor-specific irreversible inhibitors of stearoyl CoA desaturase. Nat. Chem. Biol. 12, 218–225 (2016).
Zhu, L. et al. A clinically compatible drug-screening platform based on organotypic cultures identifies vulnerabilities to prevent and treat brain metastasis. EMBO Mol. Med. 14, e14552 (2022).
Iorio, F. et al. A landscape of pharmacogenomic interactions in cancer. Cell 166, 740–754 (2016).
Acknowledgements
The authors thank J. Pietenpol (Vanderbilt University Medical Center) for generously sharing isogenic CAL51 cells with various TP53 perturbations; N. Erez (Tel Aviv University), A. Sonneblick (Schneider Children's Medical Center), S. Izraeli (Tel Aviv University) and O. Shoshani (Weizmann Institute) for helpful discussions; M. Itkin and A. Felder from the Life Science Core facilities at WIS for their assistance with metabolomics; P. Ofek, E. Yeini and Y. Zilberstein (Tel Aviv University) for assistance with intracranial and intracardiac injections, monitoring and imaging of mice; I. Shur and D. Makarovsky (Tel Aviv University) for assistance with FACS sorting; S. Israeli (Schneider Children's Medical Center), A. Savino (Schneider Children's Medical Center), A. Ruban (Tel Aviv University) and Y. Goldshmit (Tel Aviv University) for the sharing of reagents; P. Cassoni (University of Turin) for funding support of the Bertero lab; and R. Serwa from the IMol Proteomic Core Facility for assistance with proteomics.
The authors acknowledge the patients who donated samples, as well as their families, who supported their wish to donate to the BROCADE Rapid Autopsy Program. The authors are grateful for the support of BROCADE, initially supported by the Australian National Breast Cancer Foundation (NBCF) under an infrastructure grant (IF-14-001). The authors would like to thank the study co-PIs, R. Anderson (ONJCRI/PMCC) and A. Swarbrick (Garvan Institute of Medical Research) and L. Devereux, who manages BROCADE and helped with the identification, collection and processing of samples. The authors also acknowledge the Victorian Institute of Forensic Medicine that conducted the rapid autopsies; P. MacCallum, Kinghorn Centre, Garvan Institute and Tobin Brothers Funeral Directors for providing transport of donors. The BROCADE study operates under Ethics and Governance oversight at the PMCC.
This work was supported by the European Research Council Starting Grant (945674 to U.B.-D.), the Israel Cancer Research Fund Project Award (to U.B.-D.), the Azrieli Foundation Faculty Fellowship (to U.B.-D.), the Israel Science Foundation (grants 1805/21 and 3755/21 to U.B.-D.), the BSF (project grant 2019228 to U.B.-D.), the Schreiber Foundation of Tel Aviv University Faculty of Medicine (to U.B.-D.) and the Tel Aviv University/LaTrobe University/ONJCRI Joint Collaboration Research Program in Cancer Research (to U.B.-D. and D.M.). D.M. is supported by the Love Your Sister Foundation and the Victorian Cancer Agency Mid-Career Research Fellowship (MCRF21011) and the Australian NBCF (IIRS0049). E.V.W. is supported by the Breast Cancer Alliance Young Investigator Grant. H.M., M.C. and T.B. acknowledge support from LOEWE Center Frankfurt Cancer Institute, supported by the Hessen State Ministry for Higher Education, Research, and the Arts (III L 5-519/03/03.001-(0015)). H.M. additionally acknowledges support from the Federal Ministry of Education and Research and is cofunded by the European Commission (01KT2304A). G.D.S. acknowledges support from the Fondazione AIRC IG (grants 22174 and 30570), the Fondazione AIRC Special Program Molecular Clinical Oncology ‘5 per 1,000’ (grant 22759), Worldwide Cancer Research (grant 24-0361), the NRRP NextGenerationEU Project CN00000041-National Center for Gene Therapy and Drugs based on RNA Technology to GDS. R.B. was supported by an AIRC fellowship for Italy. R.A.-P. is supported by Israel Science Foundation (700/24), DFG (706375), the Yoran Institute for Human Genome Research, the Zimin Institute, Claire and Amédée Maratier Institute for the Study of Blindness and Tel Aviv University. R.S.-F. is supported by the European Research Council (ERC) Advanced Grant (835227), 3DBrainStrom and the ERC PoC grant (862580), 3DCanPredict, the Israel Science Foundation (ISF 1969/18, ISF 3706/24), the Israel Cancer Research Fund Professorship Award (PROF-18-602) and the Morris Kahn Foundation.
Author information
Authors and Affiliations
Contributions
K.L. and U.B.-D. conceived the study. K.L. performed most of the computational analyses and in vitro experiments, assisted with in vivo experiments and led the data analysis of all experiments. S. Pozzi performed most of the in vivo animal experiments, spheroid experiments and astrocyte derivation. G.L., R.K. and S.I.D. assisted with animal experiments. J.Z., T.W., Y.C.-S., Y.E. and R. Slutsky performed in vitro experiments. T.W., Y.E. and R. Saad assisted with genomic data analyses. R.Y.M., F.N.K. and E.V.W. contributed the hTERT-HMEC model and its gene expression analysis. K.S. shared proteomics data of MCF7 cells. R.K. and A.I.L.-F. performed the GC experiments. L.A. and A.E. performed the isotope tracing experiments with the help of S.M. and A.B. A.A.R. and L.B. contributed clinical samples of BCBM and performed IHC experiments. I.B. contributed clinical samples of BCBM. A. Sacconi and G.B. provided RNA-seq of clinical samples of BCBM. W.S., Y.L., J.B. and D.M. provided RNA-seq of clinical samples of metastases from various organs. Y.E., S.O. and R.A.-P. performed CUT&Tag experiments. M. Czabanka, T.B., A. Schäffer, A.A.R. and H.M. contributed clinical BM samples and performed drug treatments of patient-derived organotypic cultures. S. Piazza performed bioinformatic analyses of the DEPDC1 RNA-seq data. R.B., A.R. and M. Caputo performed cell culture and biochemical analyses. G.D.S. oversaw the RNA-seq and biochemical analyses of DEPDC1 in BC cells. R.S.-F. directed and supervised the ex vivo spheroid experiments and in vivo work. U.B.-D. directed and supervised the overall study. K.L. and U.B.-D. wrote the manuscript with input from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
R.S.-F. is a board director at Teva Pharmaceutical Industries Ltd. and receives unrelated research funding from Merck KGaA. U.B.-D. received consulting fees from Accent Therapeutics and received unrelated research funding from Novocure. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks S. Bakhoum and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Clinical data analysis of TP53 perturbation in BCBM.
a, Distribution of TP53 perturbation modes in primary BC across three clinical datasets (TCGA, METABRIC, MSK-IMPACT). del17p prevalence: TCGA 86%, METABRIC 81%, MSK-IMPACT 71%. See Supplementary Table 1. b, Distribution of TP53 biallelic perturbation modes in primary BC from the TCGA cohort: 92% of biallelic inactivation cases combine del17p and a point mutation. c, The effect of TP53 status on its gene expression. TP53 mRNA levels are lower in BCs with TP53 copy-number losses (regardless of TP53 mutation status) in TCGA (left) and METABRIC (right) datasets. Deletions of TP53 occurred almost exclusively through del17p, whereas focal deletions were rare (2%). TCGA, two-sided t-test, TP53-WT vs. TP53 monoallelic inactivation, ****p = 1.2 × 10−22, TP53-WT vs. TP53-WT/del ****p = 3 × 10−12, TP53-WT vs. TP53-WT/mut ****p = 1.4 × 10−7, TP53-WT vs. TP53-mut/del ****p = 4 × 10−8. Number of tumors TP53-WT n = 266, TP53 monoallelic inactivation n = 337, TP53-WT/del n = 268 (focal n = 15, del17p n = 271), TP53 TP53-WT/mut n = 41, TP53 TP53-mut/del n = 279. METABRIC, two-sided t-test, TP53-WT vs. TP53 monoallelic inactivation ****p = 3.8 × 10−19, TP53-WT vs. TP53-WT/del ****p = 1.2 × 10−25, TP53-WT vs. TP53-mut/del ****p= 5.1 × 10−10. Number of tumors TP53-WT n = 695, TP53 monoallelic inactivation n = 462, TP53-WT/del n = 311 (all through del17p), TP53-WT/mut n = 127, TP53-mut/del n = 364. Boxplots: bar = median; box = 25–75th percentile; whiskers = range. d, Gene set enrichment analysis (GSEA) confirms that the p53 pathway activity is lower in TP53-perturbed vs. TP53-WT primary BCs, irrespective of TP53 perturbation mode (METABRIC, ‘P53_DN.V2.DN’; p-values and FDR-corrected q-values indicated). e, Proteomics-based ssGSEA scores of p53 targets in MCF7 cell strains show lower p53 activity in del17p vs. 17p-WT MCF7 cells. TMT multiplexing mass spectrometry-based quantification, ‘Oncogenic signatures, P53_DN.V2_DN’, two-sided t-test *p = 0.03. 17p-WT: n = 2, del17p: n = 3. f, Generation of human mammary epithelial (HMEC) cells with TP53 alterations: diploid HMECs spontaneously gained a heterozygous p53 mutation (Y163S) in culture, impairing p53 function. The TP53-WT/mut clone subsequently lost the remaining TP53-WT allele through del17p, resulting in TP53 biallelic inactivation (TP53-mut/del17p). In parallel, CRISPR/Cas9 was used to generate a knockout line on the TP53-WT/mut background, also resulting in TP53 biallelic inactivation (TP53-mut/KO). g, RNA-seq-based comparison of mRNA levels of several p53 targets (CDKN1A, MDM2, TP53INP1) shows their decrease in TP53-mut/del17p cells and TP53-mut/KO vs. TP53-WT; intermediate levels were observed in TP53-WT/mut cells. One-sided t-test, TP53-WT/mut vs. TP53-mut/del17p: CDKN1A ***p = 2×10−4, MDM2 **p = 0.003, TP53INP1 **p = 0.01; TP53-WT/mut vs. TP53-WT: CDKN1A **p = 0.005, MDM2 ****p = 2.9 × 10−5, TP53INP1 **p = 0.007; TP53-mut/del17p vs. TP53-WT: CDKN1A ****p = 3.8 × 10−6, MDM2 ****p = 1 × 10−5, TP53INP1 ****p = 4.3 × 10−5; TP53-mut/KO vs. TP53-WT: CDKN1A ****p = 1.3 × 10−6, MDM2 ****p = 1.4 × 10−6, TP53INP1 ****p = 4 × 10−7. n = 3. Error bars, mean ± s.d. h, GSEA of hTERT-HMECs with various TP53 alterations (TP53-WT/mut, TP53-mut/del17p, or TP53-mut/KO) reveals reduced p53 pathway expression (‘p53-targets Nutlin_UP, U2OS’ gene set) in comparison to TP53-WT cells, which is lower in cells with biallelic vs. monoallelic inactivation. P-values and FDR-corrected q-values indicated. i, Comparison of the prevalence of TP53 alterations in primary breast tumors and across metastatic sites. TP53 alterations are more frequent in BMs vs. primary tumors or other metastatic sites (MSK 2018). Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BMs vs. primary tumors, ****p = 1 × 10−4; BMs vs. other metastatic sites, ****p = 4 × 10−5; primary tumors vs. others, p = 0.19. Primary n = 477, brain n = 26, lung n = 66, liver n = 218, bone n = 116, others n = 322. Panels f and i were created with BioRender.com. j, Comparison of the prevalence of TP53 biallelic perturbations in primary breast tumors and across metastatic sites. TP53 biallelic perturbations are more frequent in BMs vs. primary breast tumors or other metastatic sites (MSK 2018). Two-sided Fisher’s exact test, TP53 biallelic inactivation vs. TP53-WT: BMs vs. primary tumors, ***p = 1.8 × 10−4; BMs vs. others, ***p = 4 × 10−4; primary tumors vs. others, p = 0.33. Primary n = 477, brain n = 26, lung n = 66, liver n = 218, bone n = 116, others n = 322. k, ssGSEA scores of the p53 pathway in brain metastases (brain) and metastases from other organs (others), derived from the same single patient. p53 pathway activity was lower in brain vs. other metastatic sites from the same patient (brain: n = 1, others: n = 6; gray: ‘P53_DN.V1_DN’, purple: ‘P53_DN.V1_UP’). Error bars, mean ± s.d.
Extended Data Fig. 2 Gene set enrichment analysis of gene expression data from primary human breast tumors and BC cell lines.
a–c, GSEA comparing primary tumors of patients who developed BMs and those who developed metastases in other organs. a, GSEA plot showing downregulation of a p53 expression signature in primary tumors that metastasized to the brain vs. bone (MBC cohort). Oncogenic signature ‘p53_DN.V1_DN’, brain vs. bone p = 0.005. Brain n = 7, bone = 24. b, GSEA plot showing upregulation of a signature indicative of decreased p53 signaling in primary BCs that metastasized to the brain in comparison to other sites in a second dataset (GSE12276). Brain vs. others, oncogenic signature ‘P53_DN.V2_UP’, p = 0.003. Brain n = 7, others n = 121. c, GSEA plots showing downregulation of a signature indicative of increased p53 signaling (left) and upregulation of a signature indicative of decreased p53 signaling (right) in triple-negative primary breast tumors that metastasized to the brain vs. those that did not (data source GSE76714)75. Brain vs. others, oncogenic signature: ‘P53_DN.V1_DN’ p = 0.018, oncogenic signatures: P53_DN.V2_UP p = 0.035, brain n = 21, others n = 38. See Supplementary Table 3. d, GSEA of primary BCs (TCGA cohort), comparing tumors with TP53 inactivation to TP53-WT tumors. ‘Smid_breast_cancer_relapse_in_brain_dn’, p = 0.001. TP53 inactivation: n = 623, TP53-WT: n = 259. e, ssGSEA of brain-relapse signatures in primary BCs with TP53 inactivation compared to TP53-WT tumors (MBC cohort). Tumors with TP53 inactivation showed increased expression of signatures positively correlated with BM. TP53 inactivation vs. TP53-WT, ‘Smid_breast_cancer_relapse_in_brain’ **p = 0.01, ‘Smid_breast_cancer_relapse_in_brain_up’ **p = 0.003, _DN p = 0.07. See Supplementary Table 4. f, Correlation between BM potential and p53 pathway activity levels in human BC. High potential for BM is strongly correlated with low p53 pathway activity (METABRIC cohort). Readout for p53 pathway activity: ssGSEA scores for ‘Kannan_TP53_targets’ gene set. Readout for brain-metastatic potential: ssGSEA scores for ‘Smid_breast_cancer_relapse_in_brain’ gene set. Two-sided Pearson’s correlation r = −0.41, p = 2.9 × 10−54. n = 1334 tumor samples. g, ssGSEA of brain-relapse signatures in primary BCs that metastasized to the brain compared to those that metastasized elsewhere. GSE12276. Brain vs. others, ‘Smid_breast_cancer_relapse_in_brain’ **p = 0.007, rank #1, ‘Smid_breast_cancer_relapse_in_brain_up’ **p = 0.008, rank #8, ‘Smid_breast_cancer_relapse_in_brain_dn’ ***p = 0.01, rank #8. Brain: n = 7, others: n = 121. See Supplementary Table 5. h,i, GSEA of BM gene expression gene sets in tumors with TP53 mutations, hotspot mutations or copy-number loss, compared to TP53-WT tumors, in the METABRIC cohort. GSEA showed an increase in gene expression that is upregulated in BM and a decrease in gene expression that is downregulated in BM, in (h) tumors with monoallelic TP53 inactivation vs. TP53-WT tumors; and (i) tumors with biallelic TP53 inactivation vs. TP53-WT tumors, independent of the mode of p53 inactivation. P-values and q-values indicated in the respective panels. TP53-WT: n = 693, TP53-WT/mut: n = 168, TP53-WT/Hotspot: n = 30, TP53-WT/del: n = 505, TP53-mut/del: n = 405, TP53-Hotspot/del: n = 60, TP53-del/del: n = 5.
Extended Data Fig. 3 TP53 inactivation is associated with BM across primary carcinomas.
a, Correlation of TP53 inactivation with BM prevalence across carcinomas, for female (left) and male (right) tumors. Each dot represents a primary carcinoma (for lung and kidney, the highest-ranking subtypes were considered). Female: one-sided Spearman’s ρ = 0.58, p = 0.04. Male: one-sided Spearman’s ρ = 0.80, p = 0.003. b, Pan-cancer analysis of TP53 perturbation prevalence: TP53 alterations, and biallelic TP53 inactivation in particular, are enriched in BM in comparison to primary tumors and to other metastatic sites (MSK-IMPACT cohort17). Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BM vs. primary tumor, ****p = 1 × 10−5, BM vs. other metastasis, ****p = 1 × 10−5. TP53 biallelic inactivation vs. TP53-WT: BM vs. primary tumor, ****p = 1 × 10−5, BM vs. other metastasis ****p = 1 × 10−5. Primary n = 2947, brain n = 126, chest wall n = 79, liver n = 658, lymph node n = 571, pleura n = 111, soft tissue n = 80, lung n = 265, bone n = 252, peritoneum n = 48, ovary, skin n = 47, pleural fluid n = 33, adrenal gland n = 34. c–e, Association between TP53 perturbation BM potential in carcinoma cell lines. c, In total, 367 carcinoma cell lines were grouped by their BM potential (top vs. low 25%). Panel c was created with BioRender.com. d, High-BM carcinoma cell lines show enrichment of TP53 biallelic inactivation (two-sided Fisher’s exact test *p = 0.03). e, High-BM carcinoma cell lines show decreased p53 signaling, as evaluated by GSEA. Oncogenic signatures: ‘P53_DN.V1_DN’ p = 0.015, rank #3. ‘P53_DN.V1_UP p = 0.027’, rank #4. See Supplementary Table 3. f–i, TP53 alterations are enriched in BM vs. other metastases from primary lung tumors. f, Non-small cell lung cancer metastasis. Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BM vs. primary tumors ****p = 1 × 10−5, BM vs. other metastasis ***p = 8 × 10−4. TP53 biallelic inactivation vs. TP53-WT: BM vs. primary tumors ****p = 1 × 10−5, BM vs. other metastasis ****p = 1 × 10−5. Primary n = 915, brain n = 59, chest wall n = 11, liver n = 68, lung n = 10, soft tissue n = 12, lymph node n = 211, bone n = 48, adrenal gland n = 19, pleura n = 73, pleural fluid n = 25. g, Lung adenocarcinoma metastasis. Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BM vs. primary tumors ****p = 1 × 10−5, BM vs. other metastasis **p = 0.0017. TP53 biallelic inactivation vs. TP53-WT: BM vs. primary tumors ****p = 1 × 10−5, BM vs. other metastasis ***p = 1 × 10−4. Primary n = 781, brain n = 53, chest wall n = 10, liver n = 65, lung n = 10, lymph node n = 152, bone n = 40, soft tissue n = 10, pleural fluid n = 24, pleura n = 68, adrenal gland n = 16, other n = 425. h, Lung squamous cell carcinoma metastasis. Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BM vs. primary tumors p = 0.13, BM vs. other metastasis p = 0.55. TP53 biallelic inactivation vs. TP53-WT: BM vs. primary tumor p = 0.57, BM vs. other metastasis **p = 0.01. Primary n = 122, brain n = 3, pleura n = 4, bone n = 7, lymph node n = 14. i, Poorly differentiated, non-small cell lung cancer. Two-sided Fisher’s exact test, TP53 inactivation vs. TP53-WT: BM vs. primary tumors p = 1, BM vs. other metastasis p = 1. TP53 biallelic inactivation vs. TP53-WT: BM vs. primary tumors p = 0.4, BM vs. other metastasis *p = 0.045. Primary n = 12, brain n = 3, bone n = 1, lymph node n = 5, pleura n = 1, adrenal gland n = 1, liver n = 1. j,k, GSEA of primary lung adenocarcinomas. j, Comparison of tumors based on their TP53 status. Panel j was created with BioRender.com. k, TP53-deficient primary lung adenocarcinomas show increased expression of BM signatures (TCGA PanCancer Atlas). TP53 inactivation vs. TP53-WT, ‘Smid_breast_cancer_relapse_in_brain_up’ p = 0.002, ‘Smid_breast_cancer_relapse_in_brain_dn’ p = 0. TP53 inactivation: n = 366, TP53-WT: n = 185 tumor samples. l, ssGSEA comparison shows that TP53 inactivation is associated with increased BM expression signatures. Two-sided t-test, ‘Smid_breast_cancer_relapse_in_brain’ ****p = 2.1 × 10−23, ‘Smid_breast_cancer_relapse_in_brain_up’ ****p = 1.5 × 10−11, ‘Smid_breast_cancer_relapse_in_brain_dn’ ****p = 2.7 × 10−18. m, ssGSEA comparison shows that TP53 inactivation is associated with decreased bone-metastasis expression signatures. Two-sided t-test, ‘Smid_breast_cancer_relapse_in_bone’ ****p = 1.2 × 10−9, ‘Smid_breast_cancer_relapse_in_bone_up’ ****p = 4.8 × 10−8, ‘Smid_breast_cancer_relapse_in_bone_dn’ ****p = 5.9 × 10−5.. n, TP53-deficient primary lung adenocarcinomas with biallelic inactivation show increased expression of BM signatures (TCGA PanCancer Atlas). ‘Smid_breast_cancer_relapse_in_brain_up’ p = 0.01, ‘Smid_breast_cancer_relapse_in_brain_dn’ p = 1 × 10−4. TP53-null n = 169, TP53-WT n = 185. o, ssGSEA comparison shows that TP53 biallelic inactivation is associated with increased BM expression signatures. Two-sided t-test, ‘Smid_breast_cancer_relapse_in_brain’ ****p = 2.3 × 10−27, ‘Smid_breast_cancer_relapse_in_brain_up’ ****p = 8.2 × 10−15, ‘Smid_breast_cancer_relapse_in_brain_dn’ ****p = 1.1 × 10−19. p, ssGSEA comparison shows that TP53 biallelic inactivation is associated with decreased bone-metastasis expression signatures. Two-sided t-test, ‘Smid_breast_cancer_relapse_in_bone’ ****p = 1.69 × 10−10, ‘Smid_breast_cancer_relapse_in_bone_up’ ****p = 1.75 × 10−8, ‘Smid_breast_cancer_relapse_in_bone_dn’ ****p = 1.7 × 10−5. See Source Data.
Extended Data Fig. 4 Organotropism of mouse Trp53-WT and Trp53-null EMT6 isogenic cells following intra-cardiac injection into the same immune-competent mice (competition assay).
a, Experimental design and timeline of an intra-cardiac injection competition assay of EMT6 Trp53-WT and Trp53-null isogenic cells. Fluorescently-labeled Trp53-WT (red) and Trp53-null (green) EMT6 isogenic cells were injected into the same mice. The entire cohort was sacrificed after the first mouse died. In a first experiment only brains were analyzed, whereas in an additional experiment 5 organs (brain, lung, liver, bone and spleen) were collected and analyzed for fluorescence. b, Fluorescence images of dissected organs analyzed for the presence of Trp53-WT (red) and Trp53-null (green) cells. Fluorescence was calibrated to an organ from an uninjected control mouse to account for autofluorescence. Trp53-null tumors were present in 4/5 brains, but only one brain contained a tumor derived from Trp53-WT cells. The opposite trend was observed in bone samples (femur and tibia): Trp53-WT tumors were present in the bone of all mice, whereas no bone contained a tumor derived from Trp53-null cells. Scale bar = 10 mm. c, Fluorescence-based quantification of metastasis incidence to brain, lung, liver, bone and spleen following intra-cardiac injection of Trp53-WT and Trp53-null cells into the same mice. Increased number of brain lesions was detected with Trp53-null eGFP-labeled cells in comparison to Trp53-WT (sgRNA control) mCherry-labeled cells. Two-sided Fisher’s exact test, brain *p = 0.02. n = 10 mice. The opposite trend was observed for bone tumors that only formed from Trp53-WT cells, two-sided Fisher’s exact test, bone **p = 0.007. n = 5 mice. d, Increased tumor burden in the brain in mice from Trp53-null cells following intra-cardiac co-injection of Trp53-WT and Trp53-null BC cells. Tumor burden was defined as fluorescent signal over area. Two-sided t-test, Trp53-null vs. Trp53-WT: brain *p = 0.04, lung *p = 0.02, liver p = 0.86, bone **p = 0.003, spleen p = 0.35. Trp53-WT: n = 5, Trp53-null: n = 5. e, Increased number of fluorescent foci in brain, but not in other organs, from Trp53-null cells in mice co-injected with isogenic Trp53-WT and Trp53-null BC cells. Number of fluorescent foci per organ. Two-sided t-test, Trp53-null vs. Trp53-WT brain *p = 0.04, lung p = 0.17, liver p = 0.55, bone **p = 0.01, spleen p = 0.67. Trp53-WT n = 5, Trp53-null n = 5. f, Mouse brains were processed, and tissue sections were obtained from 5 areas of each brain. Panels a and f were created with BioRender.com. g, Immunofluorescence images of brain sections visualizing isogenic cancer cells. Trp53-null are labeled in green and Trp53-WT cells in red, sections were counterstained with DAPI to visualize cell nuclei. Scale bar = 100 µm. h–j, Fluorescence-based quantification of micrometastases derived from Trp53-WT and Trp53-null cells. h, The number of Trp53-null cells throughout the brain was significantly higher. Two-sided t-test **p = 0.006. i, The number of fluorescent Trp53-null cells per slide was significantly higher. Two-sided t-test **p = 0.003. j, Mouse brains contained bigger cell clusters of Trp53-null cells following competitive injection of labeled Trp53-null BC cells and their isogenic WT controls. Quantification of Trp53-null and Trp53-WT cell cluster size in brain sections from five brain areas. Fold change Trp53-null over Trp53-WT as % cell population. Two-sided t-test cluster size 2–5 cells **p = 0.002, 6-10 cells **p = 0.006, 11-15 cells **p = 0.003, 16-30 cells *p = 0.02. Mice n = 5. Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Extended Data Fig. 5 Mouse astrocytes infiltrate p53-null BMs and promote the survival, proliferation and migration of p53-null mouse BC cells in the brain.
a, Fluorescence-based quantification of microglia in the EMT6-derived tumors. Iba1 staining intensity was similar in tumors growing from Trp53-null and Trp53-WT BC cells. Two-sided t-test p = 0.08. Tumor sections evaluated: Trp53-WT n = 9, Trp53-null n = 9. Mice n = 3. b, Fluorescence-based quantification of endothelial cells in the EMT6-derived tumors. CD31 staining intensity was similar in tumors growing from Trp53-null and Trp53-WT BC cells. Two-sided t-test p = 0.23, Tumor sections evaluated: Trp53-WT n = 11, Trp53-null n = 13. Mice n = 3. c, Live imaging-based proliferation curves of EMT6 BC cells co-cultured with mouse astrocytes. The effect of co-culture on the growth of Trp53-null BC cells is stronger than that of Trp53-WT cells. Two-sided t-test, Trp53-null vs. Trp53-WT: 48 h **p = 0.002, 60 h ***p = 7 × 10−4; 72 h ****p = 2 × 10−5. Trp53-WT n = 8, Trp53-null n = 8. Error bars, mean ± s.e.m. d, Live imaging-based proliferation curves of BC EMT6 cells cultured with ACM. The effect of ACM on the growth of Trp53-null BC cells is stronger than its effect on Trp53-WT cells. Two-sided t-test, Trp53-null vs. Trp53-WT: 24 h ****p = 1.2 × 10−10; 36 h ****p = 1 × 10−10; ****p, 48 h ****p = 4.5 × 10−7; 60 h ****p = 1 × 10−5; 72 h ***p = 2 × 10−4. Trp53-WT n = 7, Trp53-null n = 7. Error bars, mean ± s.e.m. e, Live imaging-based proliferation curves of EMT6 BC cells cultured with ACM. The effect of ACM on the growth of Trp53-null BC cells is stronger than its effect on Trp53-WT cells (shown as cell numbers). Two-sided t-test, Trp53-null vs. Trp53-WT, 60 h *p = 0.04, 72 h *p = 0.02. Error bars, mean ± s.e.m. f, Flow cytometry analysis of BC cells cultured with ACM. The fraction of cells in apoptosis is lower in Trp53-null than in Trp53-WT cells. Fraction of AnnexinV+ cells is shown as fold change relative to SFM controls. Two-sided t-test, Trp53-null vs. Trp53-WT, **p = 0.01, n = 4. See Supplementary Fig. 16a. g, Transwell migration assay to assess the migration of BC cells towards ACM. BC cells were seeded in SFM in the upper compartment and ACM or astrocytes were placed in the lower compartment. h, Quantification of cell migration in the transwell assay after 20 h of co-culture of BC cells and astrocytes, showing all conditions analyzed (see Fig. 3l for relative ACM/SF values). The effect of astrocytes on cell migration was stronger in Trp53-null cells. Two-sided t-test, Trp53-WT SFM vs. Trp53-WT astrocytes ***p = 9 × 10−4; Trp53-null SFM vs. Trp53-null astrocytes ****p = 2.5 × 10−7; Trp53-WT astrocytes vs. Trp53-null astrocytes ****p = 1.7 × 10−6. i, Quantification of cell migration in the transwell assay after 20 h of co-culture of BC cells and ACM. The effect of ACM on cell migration was significantly stronger in Trp53-null cells. Two-sided t-test, Trp53-null SFM vs. Trp53-null ACM p = 0.004, Trp53-WT ACM vs. Trp53-null ACM ****p = 6 × 10−8. The fold increase of migrating cells in ACM over SFM was significant at **p = 0.005. Fields counted TP53-WT n = 17, TP53-null n = 12. j, Live imaging-based quantification of the effect of ACM on cell velocity of Trp53-WT and Trp53-null cells. Cell velocity shown as fold increase in ACM over SFM. Two-sided t-test ****p = 1 × 10−6. Each data point represents a velocity measurement; n = 47. Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Extended Data Fig. 6 p53-dependent fatty acid metabolism in BC cells.
a–d, ssGSEA comparisons show upregulation of adipogenesis, FA synthesis, uptake, and activation in BMs vs. other metastatic sites in several cohorts: a, GSE14018; b, GSE245414; c, BROCADE patient 6071 autopsy data; and d, in BMs compared to their matched primary breast tumors (GSE125989)76. Significance is indicated as: *p < 0.05, ** p < 0.01, *** p < 0.001, ****<1 × 10−4. For the list of gene sets and exact p-values, see Supplementary Tables 7–9. Panels a–d were created with BioRender.com. e, ssGSEA comparisons show strong upregulation of FA synthesis/uptake in tumors with biallelic TP53 inactivation; tumors with monoallelic TP53 perturbation show weaker but significant effects (METABRIC cohort). Significance is indicated as: *p < 0.05, ** p < 0.01, *** p < 0.001, ****p <1 × 10−4. For the list of gene sets and exact p-values, see Supplementary Table 7. f, GSEA comparing gene expression signatures related to lipid metabolism in HMEC cells with monoallelic (TP53-WT/mut) or biallelic (TP53-mut/del17p and TP53-mut/KO) TP53 inactivation, in comparison to their parental TP53-WT cells. Shown as a heatmap, upregulated signatures in red and downregulated signatures in blue. TP53-WT/mut cells showed intermediate p53 pathway expression levels and elevated lipid metabolism expression levels. g, Metabolomics analysis of OA-containing triglycerides reveals enrichment in ACM vs. SFM (two-sided t-test ****p = 2.3 × 10−5). SFM n = 3, ACM n = 3. OA-containing triglycerides n = 11. See Supplementary Table 13. h, Live-imaging-based proliferation curves show that Trp53-null EMT6 cells grow better than their isogenic Trp53-WT controls when lipid levels are limited, in line with increased FA synthesis in Trp53-null cells. Two-sided t-test, Trp53-null vs. Trp53-WT cells (delipidated conditions): 24 h **p = 0.005, 36 h **p = 0.001, 48 h **p = 0.009, 60 h *p = 0.02. Trp53-WT n = 4, Trp53-null n = 4. Error bars, mean ± s.e.m. i, Viability quantification shows that Trp53-null cells survive better under lipid starvation conditions (two-sided t-test ***p = 3 × 10−4). Trp53-WT n = 4, Trp53-null n = 4. j, High PA (50 µM) was better tolerated by Trp53-null vs. -WT BC cells (doubling time relative to control). Paired one-sided t-test *p = 0.03). n = 3. k,l, CD36 protein quantification. k, Representative gel. l, Western blot quantification shows CD36 protein upregulation in TP53-null CAL51 cells, especially when cultured in ACM (one-sided t-test *p = 0.02, n = 3). CD36 protein levels are shown as fold change of ACM over WT SFM conditions. Bar = median; whiskers = range highest to lowest data points. m, Comparison of CD36 mRNA expression levels in primary BCs of the luminal B subtype, with monoallelic or biallelic TP53 inactivation vs. TP53-WT tumors, independent of inactivation mode (TCGA and METABRIC cohorts combined). Tumors with biallelic TP53 inactivation expressed lower levels of CD36, whereas tumors with monoallelic TP53 inactivation showed intermediate CD36 mRNA levels. One-sided t-test, monoallelic TP53 inactivation vs. TP53-WT *p = 0.02, biallelic TP53 inactivation vs. TP53-WT ***p = 3 × 10−4, TP53-WT/mut vs. TP53-WT *p = 0.03, TP53-WT/del vs. TP53-WT *p = 0.03, TP53-WT/mut vs. TP53-WT/del p = 0.33 ns. TP53-WT n = 159, TP53 monoallelic inactivation n = 179, TP53 biallelic inactivation n = 126, TP53-WT/mut: n = 20, TP53-WT/del: n = 159. n, Comparison of CD36 mRNA expression levels in human primary BCs with TP53 biallelic inactivation vs TP53-WT of the Luminal A molecular subtype (TCGA). TP53-null tumors exhibited higher CD36 mRNA expression levels. Shown as fold-increase CD36 expression relative to the average expression of TP53-WT tumors. One-sided t-test **p = 0.01, TP53-WT n = 212, TP53-null n = 49. Boxplots: bar = median; box = 25–75th percentile; whiskers = range. See Source Data.
Extended Data Fig. 7 SCD1 and FAS are upregulated in p53-null BC cells and are further increased by ACM.
a, SCD1 expression is higher in BMs vs. their matched primary tumors (GSE125989). Paired two-sided t-test *p = 0.04. Matched primary tumors and BMs: n = 16. b,c, SCD1 and FASN mRNA expression levels are elevated in BMs vs. other metastases. b, GSE1418: one-sided t-test **p = 0.003, FASN *p = 0.03. BM: n = 7, metastases in other sites: n = 29. c, GSE245414: one-sided t-test, SCD1 **p = 0.01, FASN **p = 0.01. BM: n = 6, other metastatic sites: n = 26. d,e, SCD1 mRNA levels were higher in human BCs with monoallelic or biallelic TP53 inactivation, irrespective of their mode of p53 inactivation (mutation or copy-number loss). d, TCGA one-sided t-test: TP53-monoallelic inactivation vs. TP53-WT*p = 0.01, TP53-WT/del vs. TP53-WT *p = 0.02, TP53-WT/mut vs. TP53-WT *p = 0.046; biallelic TP53 inactivation vs. TP53-WT ****p = 1.4 × 10−5. TP53-WT: n = 266, TP53-WT/del: n = 287, TP53-WT/mut: n = 50, biallelic TP53 inactivation: n = 279. e, METABRIC, one-sided t-test: TP53-monoallelic inactivation vs. TP53-WT **p = 0.009; TP53-WT/del vs. TP53-WT *p = 0.02; TP53-WT/mut vs. TP53-WT *p = 0.01; biallelic TP53 inactivation vs. TP53-WT ***p = 6 × 10−4. TP53-WT: n = 592, biallelic TP53 inactivation: n = 324, TP53-WT/del: n = 280, TP53-WT/mut: n = 138. f, FASN mRNA levels were higher in primary BCs with biallelic TP53 inactivation vs. TP53-WT tumors (TCGA; LumB subtype). One-sided t-test *p = 0.02. TP53-WT: n = 31, biallelic TP53 inactivation: n = 63. g, FASN mRNA levels were higher in tumors with biallelic TP53 inactivation vs. TP53-WT tumors (METABRIC; LumB subtype). One-sided t-test, **p = 0.002. TP53-WT: n = 128, TP53-mut/del: n = 63. h, FASN mRNA levels were higher in human BCs with monoallelic or biallelic TP53 inactivation, irrespective of their mode of p53 inactivation (TCGA and METABRIC cohorts combined; LumA subtype). One-sided t-test: TP53-WT/del vs. TP53-WT **p = 0.004; TP53-WT/mut vs. TP53-WT *p = 0.03; biallelic TP53 inactivation vs. TP53-WT ****p = 3 × 10−5. TP53-WT: n = 159, TP53-WT/del: n = 157, TP53-WT/mut: n = 21, TP53-mut/del: n = 126. i, Correlation between FASN expression levels and p53 pathway activity. Low p53 pathway activity (ssGSEA score, ‘Kannan_p53_targets’) correlates with high FASN mRNA levels across primary BCs that metastasized to the brain (GSE125989). Two-sided Spearman’s ρ = –0.47, p = 0.035, n = 16. j, Scd1 mRNA levels by qPCR: Scd1 expression was higher in Trp53-null vs. -WT EMT6 cells grown in full medium, delipidated medium, or SFM (One-sided t-test *p = 0.01, **p = 0.003, **p = 0.003). Full: n = 4, delipidated: n = 3, SFM: n = 4. k, Fasn mRNA levels by qPCR. Fasn expression was higher in Trp53-null EMT6 grown in full medium, delipidated medium or ACM. One-sided t-test p = 0.11, ***p = 7 × 10−4, p = 0.05. Full: n = 3, delipidated: n = 3, ACM: n = 6 and n = 3 for Trp53-WT and Trp53-null, respectively. l,m, ACM increased SCD1 protein levels more in TP53-null and TP53-mut (R175H, R273H) vs. TP53-WT cells in an additional CAL51 isogenic cell system. l, Representative gel. m, Quantification. One-sided one-sample t-test, TP53-WT ACM vs. SFM *p = 0.02; TP53-null ACM vs. SFM **p = 0.002; TP53-mut (R175H) ACM vs. SFM **p = 0.01; TP53-mut (273H) ACM vs. SFM *p = 0.02. Paired one-sided t-test ACM/SFM: TP53-null vs. TP53-WT *p = 0.03; TP53-mut (R175H) vs. TP53-WT *p = 0.03, TP53-mut (R273H) vs. TP53-WT p = 0.07. TP53-WT: n = 9, TP53-null: n = 9. TP53-mut (R175H): n = 9, TP53-mut (R273H): n = 6. n,o, ACM increased FASN protein levels more in TP53-null and TP53-mut (R175H, R273H) vs. TP53-WT cells in an additional CAL51 isogenic cell system. n, Representative gel. o, Quantification. One-sided one-sample t-test, TP53-WT ACM vs. SFM p = 0.32; TP53-null ACM vs. SFM *p = 0.01; TP53-mut (R175H) ACM vs. SFM **p = 0.002; TP53-mut (273H) ACM vs. SFM *p = 0.04. Paired one-sided t-test ACM/SFM: TP53-null vs. TP53-WT **p = 0.008; TP53-mut (R175H) vs. TP53-WT *p = 0.01, TP53-mut (R273H) vs. TP53-WT p = 0.07. TP53-WT: n = 9, TP53-null: n = 9, TP53-mut: (R175H) n = 9, TP53-mut (R273H): n = 7. Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Extended Data Fig. 8 Astrocyte-derived metabolites increase SCD1 and contribute to FA synthesis.
a, Quantification of FA content using gas chromatography, assessing POA over PA enrichment in EMT6 Trp53-WT and Trp53-null cells after ACM exposure for 20 h. Two-sided t-test, ***p = 8 × 10−4, n = 3. Error bars, mean ± s.d. b, 13C-tracing shows higher POA M + 16 fractional enrichment in Trp53-null cells after 13C-PA incubation (20 µM, 25 h). One-sided t-test **p = 0.004, n = 4. c, 13C-PA tracing shows higher POA isotope mean enrichment in Trp53-null vs. WT after 13C-PA incubation. One-sided t-test **p = 0.002, n = 4. d, SCD1 and CD36 mRNA are positively correlated in BCBM (GSE14017). Two-sided Spearman’s ρ = 0.66, p = 0.008, n = 15. e, Schematic illustration of the FA synthesis pathway, with potential carbon sources. f, LC-MS analysis of polar metabolites in ACM shows elevated glutamate but unchanged glucose in ACM vs. SFM. Two-sided t-test, *p = 0.01. SFM: n = 3, ACM: n = 3. See Supplementary Table 13. g, Metabolic profiling55 of human BC cell lines reveals lower glutamate levels in TP53-null vs. TP53-WT BC cells. Two-sided t-test *p = 0.03. TP53-WT: n = 4, TP53-null: n = 20. h, Representative images of GFP+ Trp53-WT and Trp53-null cells after 48 h in SFM, ACM, or glutamate-supplemented SFM (75 mM). Glutamate improved proliferation, more strongly in Trp53-null. Scale bar = 400 µm. i, Live-cell imaging proliferation curves under the same conditions show that glutamate partially phenocopied ACM effect (two-sided t-test *p = 0.02 (36 h); *p = 0.04 (48 h); n = 3). Error bars, mean ± s.e.m. j–l, SCD1 and FASN protein levels in isogenic Trp53-WT and Trp53-null cells cultured in SFM with or without glutamate (75 mM). j, Representative gel. k,l, Western blot quantification shows that ACM exposure elevated SCD1 and FASN protein levels in Trp53-null cells. One-sided t-test SCD1 **p = 0.005 and FASN ***p = 5 × 10−4. Trp53-WT: n = 4, Trp53-null: n = 4. m, 13C-glutamate tracing (24 h) into PA (M + 2) shows higher enrichment in Trp53-null vs. WT. Two-sided t-test *p = 0.03, n = 3. Bar = median; whiskers = range highest to lowest data points. n, 13C-glutamate tracing (24 h) into POA (M + 2) shows higher enrichment in Trp53-null vs. WT. One-sided t-test *p = 0.02, n = 3. Bar = median; whiskers = range highest to lowest data points. o, PA mean enrichment from 13C-glutamate (left) or 13C-glucose (right) after 24-h incubation. Isotope-labeled PA was significantly enriched in Trp53-null cells in comparison to the Trp53-WT controls after 13C-glutamate incubation. One-sided t-test, Trp53-null vs. Trp53-WT, *p = 0.02, n = 3. No difference in PA mean enrichment in Trp53-null vs. Trp53-WT cells was observed after 13C-glucose incubation. Bar = median; whiskers = range highest to lowest data points. p, ¹³C-glutamate tracing (24 h) into POA showed higher enrichment in Trp53-null vs. Trp53-WT. One-sided t-test *p = 0.03, n = 3. Bar = median; whiskers = range highest to lowest data points. q, Schematic illustration of OA isotopologues after 13C-glutamate labeling. Panels a and q were created with BioRender.com. r, Quantification of OA isotopologue distribution after 13C-glutamate incubation (48 h) shows higher M + 2 and M + 4 fractions in Trp53-null vs. Trp53-WT. Two-sided t-test *p = 0.03 (M + 2), *p = 0.02 (M + 4). See Supplementary Table 15. Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Extended Data Fig. 9 p53 regulates FAS by inhibiting SREBP1, SCD1 and DEPDC1.
a, Overlap of CAL51 p53 CUT&Tag peaks with ChIP Atlas p53-bound genes72. A total of 3,524 peaks (54%) overlap; two-sided chi-squared test, p = 1 × 10−5. See Supplementary Table 16. b, CUT&Tag read coverage plot showing p53 binding at CDKN1A promoter in TP53-WT CAL51 cells. c, Overlap of CAL51 p53 CUT&Tag peaks with ChIP Atlas SREBP1-bound genes72. A total of 823 peaks (14%) overlap; two-sided chi-squared test, p = 1 × 10−5. See Supplementary Table 16. d, SREBF1 mRNA by qPCR: Higher SREBF1 mRNA levels in TP53-null vs. TP53-WT CAL51 cells (SFM, two-sided t-test, ***p = 2 × 10−4. n = 5). e, TP53 mRNA by qPCR: siRNA-mediated TP53 knockdown resulted in >90% reduction in TP53 mRNA levels in MCF10A and MCF7 cells (paired one-sided t-test, MCF10A ***p = 9.9 × 10−4, MCF7 ***p = 1.2 × 10−4, n = 3). Error bars, mean ± s.d. f, CDKN1A mRNA by qPCR: elevated CDKN1A mRNA levels following exposure to nutlin-3 in MCF10A and MCF7 cells (paired one-sided t-test, MCF10A *p = 0.02, MCF7 *p = 0.03, n = 3). Error bars, mean ± s.d. g, Cdkn1a mRNA qPCR: elevated Cdkn1a mRNA levels following Trp53 overexpression in 4T1 mouse BC cells (paired one-sided t-test, ***p = 6 × 10−4, n = 5). h, DEPDC1 mRNA (TCGA BC cohort): highest DEPDC1 expression in primary BCs with biallelic TP53 inactivation, intermediate expression in tumors with monoallelic TP53 inactivation, and lowest expression in TP53-WT tumors. Two-sided t-test, monoallelic TP53 inactivation vs. TP53-WT ****p = 8.4 × 10−22; TP53-WT/del vs. TP53-WT ****p = 2.1 × 10−10; TP53-WT/mut vs. TP53-WT ****p = 1.5 × 10−21; biallelic TP53 inactivation vs. TP53-WT ****p = 8.8 × 10−57; monoallelic vs. biallelic TP53 inactivation ****p = 8.3 × 10−22. TP53-WT: n = 266, monoallelic: n = 337, TP53-WT/del: n = 287, TP53-WT/mut: n = 50, biallelic: n = 279. i, ssGSEA of p53 targets: lower p53 pathway activity in DEPDC1-high tumors (top 10%) vs. DEPDC1-low tumors (bottom 10%). Two-sided t-test, ‘KANNAN_p53_targets’, ****p = 9.4 × 10−30, ‘KANNAN_p53_targets_UP’ ****p = 1.9 × 10−23, ‘KANNAN_p53_targets_DN’ ****p = 9.5 × 10−13. DEPDC1-high: n = 90, DEPDC1-low n = 90. j, Transcriptional signatures associated with DEPDC1 by GSEA. DEPDC1-high tumors were enriched for adipogenesis: ‘Burton_Adipogenesis-3’ (red), rank #13/5844; ****p = 5.8 × 10−88). See Supplementary Table 17. k, DEPDC1 mRNA by qPCR: siRNA-mediated DEPDC1 knockdown reduced its mRNA by >90% in MDA-MB-231 cells (paired one-sided t-test, siDEPDC1(I) ****p = 2.5 × 10−7, siDEPDC1(II) ****p = 8.2 × 10−7, n = 4). l,m, DEPDC1 protein by western blot: siRNA-mediated DEPDC1 knockdown resulted in a reduction in SCD1 protein levels in four human BC cell lines. l, Representative gel. m, Quantification. Paired one-sided t-test, siDEPDC1(I), siDEPDC1(II) vs. control: MCF7 **p = 0.0023, **p = 0.001; MDA-MB-231 *p = 0.02, **p = 0.004; MDA-MB-468 **p = 0.009, *p = 0.01; SK-BR-3 **p = 0.008, **p = 0.003. n = 3. Error bars, mean ± s.d. n, DEPDC1 Chromatin IP (ChIP) in MCF10A cells: enrichment for DEPDC1 binding at the proximal promoter of SCD1. ChIP-fold enrichment over IgG, region C (paired one-sided t-test *p = 0.049 n = 3). Error bars, mean ± s.d. o,p, mRNA of SREBP1 targets. o, Higher expression of SREBP1 targets in DEPDC1-high (top 10%) vs. DEPDC1-low (bottom 10%) primary BCs (TCGA). ‘HORTON_SREBP_TARGETS’, one-sided t-test **p = 0.009. DEPDC1-high n = 90, -low n = 90. p, Highest expression of SREBP1 targets in primary BCs with biallelic TP53 inactivation, and intermediate expression in tumors with monoallelic TP53 alterations compared to TP53-WT BCs. ‘Horton_SREBP_targets’ one-sided t-test, TP53 biallelic vs. TP53-WT **p = 0.01; TP53 monoallelic vs. TP53-WT p = 0.09. TP53-WT: n = 266, monoallelic: TP53 n = 337, biallelic: TP53 n = 279. q,r, Metastatic capacity by ssGSEA. q, Reduced bone-metastasis signatures in DEPDC1-high tumors (top 10%, TCGA). Two-sided t-test, ‘Smid_breast_cancer_relapse_in_bone’ ****p = 3.1 × 10−36, ‘Smid_breast_cancer_relapse_in_bone_up’ ****p = 1.4 × 10−37, ‘Smid_breast_cancer_relapse_in_bone_dn’ ****p = 3.9 × 10−29. DEPDC1-high n = 90, DEPDC1-low n = 90. r, Increased brain metastasis signatures in DEPDC1-high tumors (top 25%). Two-sided t-test, ‘Smid_breast_cancer_relapse_in_brain’ ****p = 1.5 × 10−51, ‘Smid_breast_cancer_relapse_in_brain_up’, ****p = 3.6 × 10−50, ‘Smid_breast_cancer_relapse_in_brain_dn’ ****p = 3.4 × 10−48. DEPDC1-high n = 223, DEPDC1-low n = 223. s, Srebf1 mRNA by qPCR: Higher Srebf1 mRNA levels in ACM-exposed Trp53-null vs. Trp53-WT EMT6 cells. Paired one-sided t-test *p = 0.02. TP53-WT: n = 6, TP53-null: n = 6. t, DEPDC1 mRNA by qPCR: Higher DEPDC1 mRNA levels in ACM-exposed TP53-null vs. TP53-WT CAL51 cells. Paired one-sided t-test *p = 0.01. TP53-WT: n = 11, TP53-null: n = 11. u,v, DEPDC1 protein by western blot. u, Representative gel. v, Elevated DEPDC1 protein levels in TP53-null CAL51 cells cultured in SFM, with further increase following ACM exposure. Paired one-sided t-test, TP53-null ACM/SFM vs. TP53-WT ACM/SFM *p = 0.04. n = 4. Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Extended Data Fig. 10 p53 inactivation increases sensitivity to FAS inhibition.
a, Correlation between p53 pathway activity (ssGSEA score for MSigDB signature ‘Kannan_p53_targets_dn’) and sensitivity of human BC cell lines to CRISPR/Cas9 knockout of SCD1 (negative values represent increased sensitivity). Two-sided Spearman’s rho = 0.29, p = 0.048. BC cell lines: n = 34. b, Comparison of drug sensitivity: Trp53-null vs. Trp53-WT EMT6 cells treated with the SCD1 inhibitor SW203668 (72 h, MTT viability readout). Trp53-null cells showed lower IC50. Paired one-sided t-test, *p = 0.02. Trp53-WT: n = 6, Trp53-null: n = 6. c, Relative viability of Trp53-WT and Trp53-null EMT6 cells treated with SCD1 inhibitor A939572 (75 nM, 72 h). Trp53-null cells were more sensitive to the drug. One-sided t-test, **p = 0.001. Trp53-WT: n = 3, Trp53-null: n = 3. d, Representative images of mCherry-labeled TP53-WT and TP53-null CAL51 cells following treatment with SW203668 (100 nM, 48 h) or vehicle control. TP53-null cells showed increased sensitivity to the drug. Scale bar = 400 µm. e, Proliferation curves of TP53-WT and TP53-null human CAL51 BC cells following treatment with SW203668 (100 nM, 72 h) or vehicle control. TP53-null cells were more sensitive to the drug. One-sided t-test: 48 h, *p = 0.03; 60 h *p = 0.02; 68 h **p = 0.01; 15 fields per experiment. Error bars, mean ± s.e.m. f,g, Relative viability of TP53-WT and TP53-null CAL51 cells treated with the SCD1 inhibitors SW203668 (100 nM, 72 h; f) or A939572 (75 nM, 72 h; g). TP53-null cells were more sensitive to the drug. One-sided t-test, *p = 0.02, **p = 0.005. TP53-WT: n = 3, TP53-null: n = 3. h, Proliferation curves of Trp53-WT and Trp53-null EMT6 cells treated with SW203668 (1 µM) with or without POA (20 µM). POA rescued SCD1i-induced growth reduction (two-sided t-test, Trp53-null SCD1i vs. Trp53-null SCD1i+POA (20 µM): 24 h *p = 0.02; 28 h ***p = 3 × 10−4; 32 h ***p = 3 × 10−4; 36 h ***p = 4 × 10−4; 40 h ***p = 6.5 × 10−4; 44 h ***p = 5 × 10−4, n = 4). Error bars, mean ± s.e.m. i, Sensitivity of TP53-WT and TP53-null BC lines to RNAi-mediated knockdown of FASN. TP53-null cells are more sensitive to FASN knockdown. One-sided t-test *p = 0.02. BC cell lines: TP53-WT: n = 4, TP53-null: n = 14. j, Comparison of drug sensitivity between TP53-WT and TP53-null BC lines treated with the FASN inhibitor C75 (AUC values, GDSC102). TP53-null cells were more sensitive to the drug. One-sided t-test *p = 0.03. BC cell lines, TP53-WT: n = 3, TP53-null: n = 2. k,l, Representative images (k) and proliferation curves (l) of mCherry-labeled Trp53-WT and Trp53-null EMT6 cells treated with C75 (20 µM, 28 h). Trp53-null cells were more sensitive to the drug. One-sided t-test **p = 0.01 at 24 h, **p = 0.005 at 28 h. Error bars, mean ± s.e.m. m, Comparison of drug sensitivity between TP53-WT and TP53-null EMT6 cells treated with C75 (72 h, MTT viability readout). Trp53-null cells were more sensitive to the drug. Paired one-sided t-test, *p = 0.02, n = 6. n, Proliferation curves of Trp53-WT and Trp53-null EMT6 cells cultured as 3D spheroids and treated with SW203668 (0.1 µM or 3 µM) or vehicle control. Trp53-null spheroids were more sensitive to the drug. One-sided t-test 0.1 µM: 36 h *p = 0.02; 48 h *p = 0.03; 60 h and 72 h *p = 0.02; 3 µM: 48 h **p = 0.005; 60 h ***p = 9 × 10−4; 72 h ***p = 9 × 10−4. Trp53-WT: n = 3, Trp53-null: n = 3. Error bars, mean ± s.e.m. o, Quantification of spheroid area of Trp53-WT and Trp53-null EMT6 cells treated with C75 (20 µM, 48 h) or vehicle control. Trp53-null spheroids were smaller following drug treatment. One-sided t-test, ****p = 2 × 10−4. Trp53-WT: n = 3, Trp53-null: n = 3. p, Fraction of apoptotic cells, measured by staining for cleaved-caspase-3, in human BCBM organotypic cultures treated with the SCD1i SW203668 (3 µM) or vehicle control. Treated organotypic cultures showed a higher fraction of apoptotic cells. Paired one-sided t-test, *p = 0.02, n = 3 BCBM samples. q. Fraction of Ki67+ cells in human BM slices treated with the SCD1i SW203668 (3 µM) or vehicle control. Trend towards reduced proliferation was observed (17% decrease, paired one-sided t-test, p = 0.13, n = 5). Boxplots: bar = median; box = 25–75th percentile; whiskers = range.
Supplementary information
Supplementary Information
Supplementary Notes 1–51, Supplementary Figs. 1–16, Supplementary Methods and supporting data for Supplementary Figs. 4, 6, 10 and 14.
Supplementary Tables 1–19
Supplementary Tables 1–19.
Supplementary Data 1
Supporting data for Supplementary Figs. 1–8, 10–15.
Source data
Source Data Figs. 1–7 and Extended Data Figs. 1–10
Source data for Figs. 1a–e,i,m,n, 2a,c,g–i,k,n,p, 3b,d,g,i,l,n,p,r, 4a–f,h,i,l,n–p,r,t–v,x,z, 5a–d,f,g,i,k,l,n–r,t,u,w, 6a,d–h,j,k,m–s,u and 7a,b,d,e,g,j,k,n, and Extended Data Figs. 1a–c,e,g,i–k, 2e–g, 3a,b,d,f–i,l,m,o,p, 4c–e,h–j, 5a–f,h,i,g, 6a–h,j,l–n, 7a–k,m,o, 8a–d,f,g,i,k–p,r, 9d–k,m–t and 10a–c,e–j,l–q.
Source Data Figs. 4–6 and Extended Data Figs. 6–9
Unprocessed western blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Laue, K., Pozzi, S., Zerbib, J. et al. p53 inactivation drives breast cancer metastasis to the brain through SCD1 upregulation and increased fatty acid metabolism. Nat Genet 58, 116–131 (2026). https://doi.org/10.1038/s41588-025-02446-1
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02446-1
This article is cited by
-
Aneuploidy-driven vulnerabilities in breast cancer metastasis
Nature Genetics (2025)