Abstract
Ineffective antibody-mediated responses are a key characteristic of chronic viral infection. However, our understanding of the intrinsic mechanisms that drive this dysregulation are unclear. Here, we identify that targeting the epigenetic modifier BMI-1 in mice improves humoral responses to chronic lymphocytic choriomeningitis virus. BMI-1 was upregulated by germinal center B cells in chronic viral infection, correlating with changes to the accessible chromatin landscape, compared to acute infection. B cell-intrinsic deletion of Bmi1 accelerated viral clearance, reduced splenomegaly and restored splenic architecture. Deletion of Bmi1 restored c-Myc expression in B cells, concomitant with improved quality of antibody and coupled with reduced antibody-secreting cell numbers. Specifically, BMI-1-deficiency induced antibody with increased neutralizing capacity and enhanced antibody-dependent effector function. Using a small molecule inhibitor to murine BMI-1, we could deplete antibody-secreting cells and prohibit detrimental immune complex formation in vivo. This study defines BMI-1 as a crucial immune modifier that controls antibody-mediated responses in chronic infection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
RNA-seq, ATAC-seq and LCMV BCR sequencing data has been deposited to the Gene Expression Omnibus under accession code GSE163365. Source data are provided with this paper.
Code availability
Code for RNA-seq and ATAC-seq analysis followed typical pipelines from public R packages. RNAsik pipeline102 was used to analyze RNA-seq data, Bowtie2/MACS2 was used to analyze ATAC-seq data. All codes are available upon request.
References
Zohar, T. et al. Compromised humoral functional evolution tracks with SARS-CoV-2 mortality. Cell 183, 1508–1519 (2020).
Woodruff, M. C. et al. Extrafollicular B cell responses correlate with neutralizing antibodies and morbidity in COVID-19. Nat. Immunol. 21, 1506–1516 (2020).
Dominguez-Sola, D. et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 13, 1083–1091 (2012).
Calado, D. P. et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat. Immunol. 13, 1092–1100 (2012).
Finkin, S., Hartweger, H., Oliveira, T. Y., Kara, E. E. & Nussenzweig, M. C. Protein amounts of the MYC transcription factor determine germinal center B cell division capacity. Immunity 51, 324–336 (2019).
Hunziker, L. et al. Hypergammaglobulinemia and autoantibody induction mechanisms in viral infections. Nat. Immunol. 4, 343–349 (2003).
Recher, M. et al. Deliberate removal of T cell help improves virus-neutralizing antibody production. Nat. Immunol. 5, 934–942 (2004).
Lane, H. C. et al. Abnormalities of B cell activation and immunoregulation in patients with the acquired immunodeficiency syndrome. N. Engl. J. Med. 309, 453–458 (1983).
Cooper, L. & Good-Jacobson, K. L. Dysregulation of humoral immunity in chronic infection. Immunol. Cell Biol. 98, 456–466 (2020).
Wieland, A. et al. Antibody effector functions mediated by Fcγ-receptors are compromised during persistent viral infection. Immunity 42, 367–378 (2015).
Yamada, D. H. et al. Suppression of Fcγ-receptor-mediated antibody effector function during persistent viral infection. Immunity 42, 379–390 (2015).
Appel, G. B. Immune-complex glomerulonephritis–deposits plus interest. N. Engl. J. Med. 328, 505–506 (1993).
Oldstone, M. B. & Dixon, F. J. Pathogenesis of chronic disease associated with persistent lymphocytic choriomeningitis viral infection. II. Relationship of the anti-lymphocytic choriomeningitis immune response to tissue injury in chronic lymphocytic choriomeningitis disease. J. Exp. Med. 131, 1–19 (1970).
Lu, L. L., Suscovich, T. J., Fortune, S. M. & Alter, G. Beyond binding: antibody effector functions in infectious diseases. Nat. Rev. Immunol. 18, 46–61 (2018).
Straub, T. et al. Nucleoprotein-specific nonneutralizing antibodies speed up LCMV elimination independently of complement and FcγR. Eur. J. Immunol. 43, 2338–2348 (2013).
Chung, A. W. et al. Dissecting polyclonal vaccine-induced humoral immunity against HIV using systems serology. Cell 163, 988–998 (2015).
Wang, T. T. et al. Anti-HA glycoforms drive B cell affinity selection and determine influenza vaccine efficacy. Cell 162, 160–169 (2015).
Caskey, M., Klein, F. & Nussenzweig, M. C. Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic. Nat. Med. 25, 547–553 (2019).
Zhang, Y. & Good-Jacobson, K. L. Epigenetic regulation of B cell fate and function during an immune response. Immunol. Rev. 288, 75–84 (2019).
Kealy, L. et al. The histone methyltransferase DOT1L is essential for humoral immune responses. Cell Rep. 33, 108504 (2020).
Sen, D. R. et al. The epigenetic landscape of T cell exhaustion. Science 354, 1165–1169 (2016).
Pauken, K. E. et al. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354, 1160–1165 (2016).
Beguelin, W. et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013).
Caganova, M. et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J. Clin. Invest. 123, 5009–5022 (2013).
Tanaka, S. et al. Tet2 and Tet3 in B cells are required to repress CD86 and prevent autoimmunity. Nat. Immunol. 21, 950–961 (2020).
Beguelin, W. et al. EZH2 and BCL6 cooperate to assemble CBX8–BCOR complex to repress bivalent promoters, mediate germinal center formation and lymphomagenesis. Cancer Cell 30, 197–213 (2016).
Raaphorst, F. M. et al. Cutting edge: Polycomb gene expression patterns reflect distinct B cell differentiation stages in human germinal centers. J. Immunol. 164, 1–4 (2000).
Tarte, K., Zhan, F., De Vos, J., Klein, B. & Shaughnessy, J. Jr. Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B cell differentiation. Blood 102, 592–600 (2003).
Cao, R., Tsukada, Y. & Zhang, Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell 20, 845–854 (2005).
Wang, H. et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431, 873–878 (2004).
Jacobs, J. J., Kieboom, K., Marino, S., DePinho, R. A. & van Lohuizen, M. The oncogene and Polycomb-group gene BMI-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397, 164–168 (1999).
Cantor, D. J. et al. Impaired expression of rearranged immunoglobulin genes and premature p53 activation block B cell development in BMI1 null mice. Cell Rep. 26, 108–118 (2019).
Yamashita, M. et al. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J. Exp. Med. 205, 1109–1120 (2008).
Jagani, Z. et al. The Polycomb group protein BMI-1 is essential for the growth of multiple myeloma cells. Cancer Res. 70, 5528–5538 (2010).
Lessard, J. & Sauvageau, G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255–260 (2003).
Oguro, H. et al. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the Polycomb protein Bmi1. Cell Stem Cell 6, 279–286 (2010).
Haupt, Y., Alexander, W. S., Barri, G., Klinken, S. P. & Adams, J. M. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell 65, 753–763 (1991).
van Lohuizen, M. et al. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell 65, 737–752 (1991).
Dierks, C. et al. Essential role of stromally induced hedgehog signaling in B cell malignancies. Nat. Med. 13, 944–951 (2007).
De Vos, J. et al. Comparison of gene expression profiling between malignant and normal plasma cells with oligonucleotide arrays. Oncogene 21, 6848–6857 (2002).
Teijaro, J. R. et al. Persistent LCMV infection is controlled by blockade of type I interferon signaling. Science 340, 207–211 (2013).
Louten, J., van Rooijen, N. & Biron, C. A. Type 1 IFN deficiency in the absence of normal splenic architecture during lymphocytic choriomeningitis virus infection. J. Immunol. 177, 3266–3272 (2006).
Odermatt, B., Eppler, M., Leist, T. P., Hengartner, H. & Zinkernagel, R. M. Virus-triggered acquired immunodeficiency by cytotoxic T cell-dependent destruction of antigen-presenting cells and lymph follicle structure. Proc. Natl Acad. Sci. USA 88, 8252–8256 (1991).
Bergthaler, A. et al. Impaired antibody response causes persistence of prototypic T cell-contained virus. PLoS Biol. 7, e1000080 (2009).
Mahan, A. E. et al. Antigen-specific antibody glycosylation is regulated via vaccination. PLoS Pathog. 12, e1005456 (2016).
Francica, J. R. et al. Innate transcriptional effects by adjuvants on the magnitude, quality, and durability of HIV envelope responses in NHPs. Blood Adv. 1, 2329–2342 (2017).
Hessell, A. J. et al. Fc receptor but not complement binding is important in antibody protection against HIV. Nature 449, 101–104 (2007).
Nimmerjahn, F. & Ravetch, J. V. Divergent immunoglobulin g subclass activity through selective Fc receptor binding. Science 310, 1510–1512 (2005).
Shinkawa, T. et al. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 (2003).
Wieland, A. et al. Enhancing FcγR-mediated antibody effector function during persistent viral infection. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aao3125 (2018).
Kao, D. et al. IgG subclass and vaccination stimulus determine changes in antigen specific antibody glycosylation in mice. Eur. J. Immunol. 47, 2070–2079 (2017).
Zotos, D. et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J. Exp. Med. 207, 365–378 (2010).
Good, K. L., Bryant, V. L. & Tangye, S. G. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J. Immunol. 177, 5236–5247 (2006).
Sarosiek, K. A. et al. Novel IL-21 signaling pathway up-regulates c-Myc and induces apoptosis of diffuse large B-cell lymphomas. Blood 115, 570–580 (2010).
Elsaesser, H., Sauer, K. & Brooks, D. G. IL-21 is required to control chronic viral infection. Science 324, 1569–1572 (2009).
Bhattacharya, D. et al. Transcriptional profiling of antigen-dependent murine B cell differentiation and memory formation. J. Immunol. 179, 6808–6819 (2007).
Kong, Y. et al. Targeting of BMI-1 with PTC-209 inhibits glioblastoma development. Cell Cycle 17, 1199–1211 (2018).
Dey, A. et al. Evaluating the mechanism and therapeutic potential of PTC-028, a novel inhibitor of BMI-1 function in ovarian cancer. Mol. Cancer Ther. 17, 39–49 (2018).
Hangartner, L., Zinkernagel, R. M. & Hengartner, H. Antiviral antibody responses: the two extremes of a wide spectrum. Nat. Rev. Immunol. 6, 231–243 (2006).
Lofano, G. et al. Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aat7796 (2018).
Wang, H. B. et al. Sp1 and c-Myc regulate transcription of BMI1 in nasopharyngeal carcinoma. FEBS J. 280, 2929–2944 (2013).
Huang, R. et al. MYCN and MYC regulate tumor proliferation and tumorigenesis directly through BMI1 in human neuroblastomas. FASEB J. 25, 4138–4149 (2011).
Cho, J. H., Dimri, M. & Dimri, G. P. A positive feedback loop regulates the expression of polycomb group protein BMI1 via WNT signaling pathway. J. Biol. Chem. 288, 3406–3418 (2013).
Harker, J. A., Lewis, G. M., Mack, L. & Zuniga, E. I. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science 334, 825–829 (2011).
Wilson, E. B. et al. Blockade of chronic type I interferon signaling to control persistent LCMV infection. Science 340, 202–207 (2013).
Fallet, B. et al. Chronic viral infection promotes efficient germinal center B cell responses. Cell Rep. 30, 1013–1026 (2020).
Arrowsmith, C. H., Bountra, C., Fish, P. V., Lee, K. & Schapira, M. Epigenetic protein families: a new frontier for drug discovery. Nat. Rev. Drug Discov. 11, 384–400 (2012).
Morera, L., Lubbert, M. & Jung, M. Targeting histone methyltransferases and demethylases in clinical trials for cancer therapy. Clin. Epigenetics 8, 57 (2016).
Hiepe, F. & Radbruch, A. Plasma cells as an innovative target in autoimmune disease with renal manifestations. Nat. Rev. Nephrol. 12, 232–240 (2016).
Low, M., Infantino, S., Grigoriadis, G. & Tarlinton, D. Targeting plasma cells: are we any closer to a panacea for diseases of antibody-secreting cells? Immunol. Rev. 270, 78–94 (2016).
Kwon, K. et al. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity 28, 751–762 (2008).
Mich, J. K. et al. Prospective identification of functionally distinct stem cells and neurosphere-initiating cells in adult mouse forebrain. eLife 3, e02669 (2014).
Sangiorgi, E. & Capecchi, M. R. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 40, 915–920 (2008).
Srinivas, S. et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev. Biol. 1, 4 (2001).
Nowak, K. et al. BMI1 is a target gene of E2F-1 and is strongly expressed in primary neuroblastomas. Nucleic Acids Res. 34, 1745–1754 (2006).
Russ, B. E. et al. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity 41, 853–865 (2014).
Lux, A., Yu, X., Scanlan, C. N. & Nimmerjahn, F. Impact of immune complex size and glycosylation on IgG binding to human FcγRs. J. Immunol. 190, 4315–4323 (2013).
Quinn, K. M. et al. Metabolic characteristics of CD8(+) T cell subsets in young and aged individuals are not predictive of functionality. Nat. Commun. 11, 2857 (2020).
Battegay, M. et al. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J. Virol. Methods 33, 191–198 (1991).
Lopez, E. et al. Low pH exposure during immunoglobulin G purification methods results in aggregates that avidly bind fcγ receptors: implications for measuring Fc dependent antibody functions. Front. Immunol. 10, 2415 (2019).
Mahan, A. E. et al. A method for high-throughput, sensitive analysis of IgG Fc and Fab glycosylation by capillary electrophoresis. J. Immunol. Methods 417, 34–44 (2015).
Scharer, C. D. et al. ATAC-seq on biobanked specimens defines a unique chromatin accessibility structure in naive SLE B cells. Sci. Rep. 6, 27030 (2016).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Zhang, Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008).
Lawrence, M. et al. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Powell, D. Degust: interactive RNA-seq analysis. Zenodo https://doi.org/10.5281/zenodo.3258932 (2015).
Kim, J. et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell 143, 313–324 (2010).
Schuhmacher, M. et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 29, 397–406 (2001).
Gargiulo, G. et al. In vivo RNAi screen for BMI1 targets identifies TGF-β/BMP-ER stress pathways as key regulators of neural- and malignant glioma-stem cell homeostasis. Cancer Cell 23, 660–676 (2013).
Douglas, D. et al. BMI-1 promotes Ewing sarcoma tumorigenicity independent of CDKN2A repression. Cancer Res. 68, 6507–6515 (2008).
Wiederschain, D. et al. Contribution of Polycomb homologues Bmi-1 and Mel-18 to medulloblastoma pathogenesis. Mol. Cell. Biol. 27, 4968–4979 (2007).
Nuytten, M. et al. The transcriptional repressor NIPP1 is an essential player in EZH2-mediated gene silencing. Oncogene 27, 1449–1460 (2008).
Piovesan, D. et al. c-Myb regulates the T-Bet-dependent differentiation program in B cells to coordinate antibody responses. Cell Rep. 19, 461–470 (2017).
Staupe, R. P. et al. Chronic viral infection promotes early germinal center exit of B cells and impaired antibody development. Preprint at bioRxiv https://doi.org/10.1101/849844 (2019).
Rosenfeld, A. M., Meng, W., Luning Prak, E. T. & Hershberg, U. ImmuneDB: a system for the analysis and exploration of high-throughput adaptive immune receptor sequencing data. Bioinformatics 33, 292–293 (2017).
Zhang, B., Meng, W., Luning Prak, E. T. & Hershberg, U. Discrimination of germline V genes at different sequencing lengths and mutational burdens: a new tool for identifying and evaluating the reliability of V gene assignment. J. Immunol. Methods 427, 105–116 (2015).
Giudicelli, V., Brochet, X. & Lefranc, M. P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011, 695–715 (2011).
Yaari, G., Uduman, M. & Kleinstein, S. H. Quantifying selection in high-throughput Immunoglobulin sequencing data sets. Nucleic Acids Res. 40, e134 (2012).
Gupta, N. T. et al. Change-O: a toolkit for analyzing large-scale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358 (2015).
Good-Jacobson, K. L., O’Donnell, K., Belz, G. T., Nutt, S. L. & Tarlinton, D. M. c-Myb is required for plasma cell migration to bone marrow after immunization or infection. J. Exp. Med. 212, 1001–1009 (2015).
Tsyganov, K., Perry, A. J., Archer, S. K. & Powell, D. RNAsik: a pipeline for complete and reproducible RNA-seq analysis that runs anywhere with speed and ease. J. Open Source Softw. 3, 583 (2018).
Acknowledgements
We thank J. Groom (Walter and Eliza Hall Institute), I. Parish (Peter MacCallum Cancer Centre), C. Zaph and J. Rossjohn for critical reading of this manuscript and/or discussions; M. Pellegrini (Walter and Eliza Hall Institute), M. Degli-Esposti (Monash University) and A. Papa (Monash University) for generously providing, respectively, LCMV stocks, IL-2 and c-Myc antibody; J. Sun and members of the Good-Jacobson laboratory for technical assistance; O. Chernyavskiy and the staff of Monash Micro Imaging for the provision of instrumentation training and technical support; Monash FlowCore, Bioinformatics and Animal Research Platforms; and MHTP Medical Genomics Facility. This work was supported by a Bellberry-Viertel Senior Medical Research Fellowship (K.L.G.-J.); National Health and Medical Research Council (NHMRC) Career Development Fellowships 1108066 (K.L.G.-J.) and 1140509 (A.W.C.); American Association of Immunologists Careers in Immunology Fellowship and Travel for Techniques program (A.D.P. and K.L.G.-J.); Australian Research Council Future Fellowship FT170100174 and Discovery Project DP200102776, NHMRC Ideas grant 1182086 (N.L.L.G.); NHMRC Program grant 1054925 and Investigator award 1175411 (D.M.T. and K.O.D.); Australian Research Council Discovery Project DP170102020 and NHMRC Ideas grant 1183478 (S.J.T.); Monash University Biomedicine Discovery Institute Scholarship (L.C.); Monash University Scholarship for Excellence (J.P.); Monash University Graduate Scholarship and Monash International Postgraduate Research Scholarship (T.H.); Postgraduate Scholarship from the Bonn University – Melbourne University joining PhD program (V.U.); National Institutes of Health P01 AI106697 and the European Union’s Horizon 2020 Research and Innovation program 825821 (U.H. and A.S.). The contents of this document can under no circumstances be regarded as reflecting the position of the European Union.
Author information
Authors and Affiliations
Contributions
K.L.G.-J. conceived the study. K.L.G.-J., A.D.P., N.L.L.G. and A.W.C. designed research. A.D.P., J.P., L.C., L.H., T.D., V.U., K.O.D. and T.H. performed the research. S.J.T., N.L.L.G., A.W.C., C.D.S. and D.M.T. provided additional intellectual input. T.M., A.S., U.H., A.D.P. and C.D.S. analyzed deep-sequencing data. S.P. provided reagents and technical expertise. D.M.T. provided mice. K.L.G.-J. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
K.L.G.-J. has received funding from GSK for a separate project. The remaining authors declare no competing interests.
Additional information
Peer review information Nature Immunology thanks Deepta Bhattacharya, Tri Phan and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. L. A. Dempsey was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Quality control for deep-sequencing dataset and overlap between differentially chromatin accessible regions and differentially expressed genes.
a, Data quality for the ATAC-seq was evaluated by calculating the Fraction of read in peaks (FRIP Score). b, Chromatin accessibility regions pie chart. c, Volcano plot for all significantly differentially accessible regions (DARs). d, Metrics for RNA-seq showing the sequencing library size of assigned reads. e, PCA plot in two dimensions of differential expressed genes in acute (salmon) and chronic (green) GC B cells. f, Histogram of the nominal p-values calculated by DESeq for synthetic data RNA-seq. g, Volcano plot for differentially expressed genes (DEGs) identified by RNA-seq. h, Within Sum of Square (WSS) plot for the optimal number of clusters determined by the K-mean analysis. i, Dot plot showing differential chromatin accessibility regions (DARs) or expressed genes (DEGs) (yellow dots) and the overlap between the two datasets (black dots). j, Heat maps for overlapped DARs (ATAC-Seq, left panel) and DEGs (RNA-Seq, right panel). k, PRC2 targets assessed within DARs (upper panels) or DEGs (lower panels); ****P < 0.0001,Wilcoxon matched-pairs signed rank test, two-tailed p-value. l, Plots of normalised counts for non-canonical PRC1 (top), PRC2 and PRC2 co-factors (bottom); n = 2 mice per group, data represent mean ± SEM.
Extended Data Fig. 2 BMI-1 expression in B cell subsets.
a, Schematic of NP−KLH in alum immunization and tamoxifen administration of Bmi1CreERT2Rosa26EYFP reporter mice. b, Flow cytometric representative plots EYFP expression in CD19+IgDloCD95hiCD38lo (GC) and B220loCD138hi cells from immunized Bmi1Rosa26eYFP reporter mice. c, EYFP- and EYFP+ frequencies within B220loCD138hi ASC of either IgG, IgA or IgM immunoglobulin isotypes were assessed by flow cytometry; n = 6 mice per group; data represent mean ± SEM; mLN: *P = 0.0411, Spleen: *P = 0.04152, **P = 0.0043 (Mann-Whitney U test, two-tailed p-value). d, Frequency of EYFP+ PB (B220intCD138hi) and PC (B220−CD138hi) in mesenteric lymph nodes and spleen in immunized mice; n = 6 mice per group, combined from two independent experiments; data represent mean ± SEM; **P < 0.01 (Mann-Whitney U test, two-tailed p-value). e-i, Bmi1CreERT2Rosa26EYFP reporter mice and wild-type controls were infected with either LCMV-WE or LCMV-Docile as per schematic in Fig. 1. e, GC B cells were segregated into DZ and LZ subsets and assessed for EYFP+ frequency; WE: n = 5 mice, Docile: n = 3 mice; Data represent mean ± SEM. *P = 0.0357 (Mann-Whitney U test, two-tailed p-value). f-i, Assessment of Bmi1 by RT-qPCR in sort-purified populations as follows: f, EYFP−and EYFP+ lymphocytes in LCMV-Docile-infected mice (2-𝜟𝜟Ct method relative to EYFP−); n = 7 mice per group; data represent mean ± SEM; ***P = 0.0006 (Mann-Whitney U test, two-tailed p-value). g, EYFP−and EYFP+ GC B cells responding to either LCMV-WE or LCMV-Docile (relative to acute EYFP− GC B cells); n = 3 mice per group; data represent mean ± SEM; **P < 0.01 (Welch’s U test, two-tailed p-value). h, GC B cells from wild-type mice infected with either LCMV-WE or LCMV-Docile (relative to acute GC B cells), d14 post-infection; n = 10 mice per group; data represent mean ± SEM; ****P < 0.0001 (Mann-Whitney U test, two-tailed p-value). i, EYFP+ GC B cells and ASCs (relative to GC B cells); n = 3 mice per group; data represent mean ± SEM. *P = 0.0211 (Welch’s U test, two-tailed p-value). j-k, Kinetics of (j) Bmi1 and (k) BMI-1 expression in GC B cells isolated from wild-type mice infected with either LCMV-WE or LCMV-Docile. H3 loading control in bottom panel. RT-qPCR values for (j) combined from 1-3 experiments per timepoint (d7: WE n = 5, Docile n = 6; d14: n = 10 per group; d28: WE n = 4, Docile n = 3; d14 data also shown in h). Data represent mean ± SEM. **P < 0.01 (Mann-Whitney U test, two-tailed p-value). Gels and blots for (k) were processed in parallel, using the same samples.
Extended Data Fig. 3 Quantitation of innate and adaptive cell subsets.
Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice were infected with LCMV-Docile. a, Mouse weights assessed at the indicated time points post-infection (n = 18 Cd23Cre/+ and n = 19 Bmi1f/fCd23Cre/+ mice; data represent mean ± SEM; **P = 0.0058, Mann-Whitney U test, two-tailed p-value). b, Innate immune cell subsets; c, CD4+ T cells; d, CD8+ T cells were assessed at d7. b-d, n = 8 mice per group, combined from two independent experiments (Mann-Whitney U test, two-tailed p-value). e, CD4+ T cells and f, CD8+ T cells were assessed at d21. n = 6 mice per group, combined from two independent experiments (Mann-Whitney U test, two-tailed p-value). g, Sort-purified CD44hiCD8+ cells from Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice at d14 post-infection were seeded in wells with splenocytes so that effector (gp33+CD8+) to target ratio was either 2.5:1 or 1:1; n = 4 mice per group (Mann-Whitney U test, two-tailed p-value). h, Mean fluorescence intensity of IFNγ in gp33+CD8+ T cells; n = 5 mice per group (Mann-Whitney U test, two-tailed p-value). i, Histological analysis of uninfected spleens from Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice from an individual experiment; B220 (cyan), CD3 (magenta). Scale bar = 100μm.
Extended Data Fig. 4 Humoral responses to infection of BMI-1-deficient B cells.
Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice were infected with LCMV-Docile. a, Relative abundance of specific types of N-glycan structures (G0, agalactosylated; G1, monogalactosylated; G2, digalactosylated; F, fucosylated; Z, sialylated; M, mannose) of serum antibody at d14 post-infection. Data represent mean ± SEM, n = 5 per group, combined from two independent experiments. b, Frequency and c, number of unswitched B cells at the indicated time points; d, Frequency of GC B cells within the CD19+IgDlo population and e, number of GC B cells in LCMV-Docile-infected mice at indicated time points. f, Frequency of IgG2c+ cells within the GC B cell population in LCMV-Docile-infected mice at the indicated time points. b-f, Data represent mean ± SEM; d7: n = 6-10 mice per group, d14: n = 12-16 Cd23Cre/+ and n = 12-15 Bmi1f/fCd23Cre/+ mice, d21: n = 6 mice per group, d28: n = 6-8 Cd23Cre/+ and n = 6-9 Bmi1f/fCd23Cre/+ mice; combined from at least two experiments per time point; *P < 0.05, **P < 0.01 (Mann-Whitney U test, two-tailed p-value). g, LCMV-specific serum IgG2c at d14 and d28 post-infection with LCMV-Docile; d14: n = 12 Cd23Cre/+ and n = 15 Bmi1f/fCd23Cre/+ mice, d28: n = 10 Cd23Cre/+ and n = 11 Bmi1f/fCd23Cre/+ mice; combined from at least four experiments per time point. h, Bcl2l11 and Pmaip1 expression in GC B cells isolated from Cd23Cre/+ or Bmi1f/fCd23Cre/+ mice, d14 post-infection. Data represent mean ± SEM, n = 4 per group.
Extended Data Fig. 5 Additional images of Myc expression within GCs.
a, Cd23Cre/+ and b, Bmi1f/fCd23Cre/+ mice were infected with LCMV-Docile and GCs assessed at d7 post-infection as detailed in Fig. 4. Panels show individual staining (IgD, CD3, PNA and Myc; combined top row). Representative images of n = 4 mice per genotype from two individual experiments. Scale bar = 100μm. c, Number of unique synonymous and non-synonymous mutations per clone in CDR and FW regions of GC B cell VH gene repertoire. GC B cells were isolated from Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice infected with either LCMV-WE or LCMV-Docile, d14 post-infection. d, Log ratio of non-synonymous to synonymous mutations in total CDR and total FW regions; median values shown. The boxplots (c, d) visualize the median (middle hinge), two hinges (first and third quartiles, 25th and 75th, respectively), two whiskers (values no larger than the inter-quartile range, 1.5*IQ), and all individual outlying points. e-f, Selection pressure in VH CDR and FW regions; (e) shows individual mice, (f) selection pressure of pooled mice per genotype and infection.
Extended Data Fig. 6 GC B cells are unaffected in Bmi1f/fPrdm1CreERT2 mice.
a, Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice were infected with either LCMV-WE or LCMV-Docile, GC B cells sort-purified 14d post-infection, and gene expression assessed by RNA-seq. Shown are DEGs identified across the four groups. b, Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice were immunized with NP−KLH in alum and serum NP-binding IgG1 and IgM assessed at d7 post-immunization; n = 8 Cd23Cre/+ and n = 7 Bmi1f/fCd23Cre/+ mice, combined from two independent experiments. c, Antigen-specific GC B cells were assessed over time; d7: n = 8 Cd23Cre/+ and n = 7 Bmi1f/fCd23Cre/+ mice, d14: n = 5 mice per group, d28: n = 9 Cd23Cre/+ and n = 10 Bmi1f/fCd23Cre/+ mice; combined from at least two experiments per time point. d, Frequency of IgG1+ within the GC B cell population at d14 and d28 post-immunization; d14: n = 5 mice per group, d28: n = 9 Cd23Cre/+ and n = 10 Bmi1f/fCd23Cre/+ mice. e, DZ and LZ GC B cell frequencies were assessed in immunized Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice; d7: n = 8 Cd23Cre/+ and n = 7 Bmi1f/fCd23Cre/+ mice, d14: n = 5 mice per group, d28: n = 6 mice per group. Data represent mean ± SEM. f, Bmi1CreERT2Rosa26EYFP reporter mice and wild-type controls were immunized and treated as per schematic in Extended Data Fig. 2. EYFP- and EYFP+ GC B cells were isolated and assessed for Myc as per Fig. 4; n = 9 mice per group, combined from two independent experiments (**P = 0.0019, Mann-Whitney U test, two-tailed p-value). g, CDR3 length of VH186.2 sequences from GC B cells; n = 42 cells from Cd23Cre/+ and n = 44 cell from Bmi1f/fCd23Cre/+ mice, combined from n = 3 mice per group. h, Naïve Bmi1f/fPrdm1CreER/+ and controls were administered tamoxifen as per schematic in Fig. 6a. Representative flow cytometric plots of GC B cells in mesenteric lymph nodes. i, Assessment of GC B cell number by flow cytometry; n = 5 mice per group, combined from two independent experiments per time point. Data represent mean ± SEM. j, Deletion analysis within B220loCD138hi ASC in Bmi1f/fPrdm1CreER/+ mice; representative of two independent experiments.
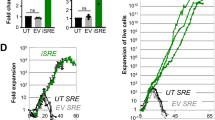
Extended Data Fig. 7 PC are decreased in response to a small molecule inhibitor to BMI-1.
a, b, Representative flow cytometric plots of CTV-labelled wild-type B cells cultured with LPS and IL-4, and in the presence of PTC-209 at the indicated concentrations. Cells were assessed by flow cytometry for (a) ASC markers and (b) cell division, as determined by the dilution of CTV. c, Representative images of histological analysis of spleens 12d post-infection from Cd23Cre/+ and Bmi1f/fCd23Cre/+ mice treated with either the vehicle control or PTC-028; B220 (cyan), CD3 (magenta). Two examples of B cell follicles per individual mouse; two individual mice within each experimental group shown. Scale bar = 100μm.
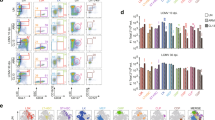
Extended Data Fig. 8 Flow cytometry gating scheme.
a, Gating strategy for sort purification of GCB for RNA-seq and ATAC-seq analysis (Figs. 1, 5; ED Figs. 1, 6). b, Gating strategy for GCB, GCB DZ and LZ distribution, and ASC (“Plasmacells”) used throughout the manuscript, and EYFP gating strategy for Bmi1 reporter mice (Fig. 1; Extended Data Fig. 2). c, Gating strategy for innate cell and T cell subsets (Extended Data Fig. 3). Note either B220 or CD19 were used to gate B cells.
Supplementary information
Supplementary Table
Supplementary Tables 1–4.
Source data
Source Data Fig. 1
Statistical Source Data.
Source Data Fig. 2
Statistical Source Data.
Source Data Fig. 2c
Image Source Data: uncropped spleen image.
Source Data Fig. 3
Statistical Source Data.
Source Data Fig. 4
Statistical Source Data.
Source Data Fig. 5
Statistical Source Data.
Source Data Fig. 6
Statistical Source Data.
Source Data Fig. 7
Statistical Source Data.
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data.
Source Data Extended Data Fig. 2
Image Source Data: unprocessed immunoblots.
Source Data Extended Data Fig. 3
Statistical Source Data.
Source Data Extended Data Fig. 4
Statistical Source Data.
Source Data Extended Data Fig. 5
Statistical Source Data.
Source Data Extended Data Fig. 6
Statistical Source Data.
Source Data Extended Data Fig. 6j
Image Source Data: uncropped gel image.
Rights and permissions
About this article
Cite this article
Di Pietro, A., Polmear, J., Cooper, L. et al. Targeting BMI-1 in B cells restores effective humoral immune responses and controls chronic viral infection. Nat Immunol 23, 86–98 (2022). https://doi.org/10.1038/s41590-021-01077-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-021-01077-y
This article is cited by
-
Spatial transcriptomics unveils estrogen-modulated immune responses and structural alterations in the ectocervical mucosa of depot medroxyprogesterone acetate users
Scientific Reports (2025)
-
CRISPR/Cas9-mediated deletion of a GA-repeat in human GPM6B leads to disruption of neural cell differentiation from NT2 cells
Scientific Reports (2024)
-
Tumors evade immune cytotoxicity by altering the surface topology of NK cells
Nature Immunology (2023)
-
Robust and prototypical immune responses toward COVID-19 vaccine in First Nations peoples are impacted by comorbidities
Nature Immunology (2023)