Abstract
Glycolysis and mitochondrial fatty acid oxidation (FAO) regulate CD8+ T cell differentiation, but how this metabolic balance regulates T cell exhaustion is unclear. PD-1 signaling inhibits glycolysis and enhances FAO. Here, we show that CD8+ T cells in tumors adhere to glycolysis with attenuated FAO despite high PD-1 expression. Active aldehydes, final products of lipid peroxidation, accumulate in CD8+ T cells in proportion to their level of exhaustion, defined by mitochondrial mass and potential. Aldehydes promote glycolysis and inhibit FAO in T cells. Mice deficient in an FAO enzyme in T cells generate more acrolein, a representative aldehyde, enhancing T cell exhaustion and attenuating antitumor immunity. Acrolein is generated partly from mitochondria and damages mitochondrial architecture. Inhibitors of lipid peroxidation or aldehydes enhanced PD-1-blockade by rectifying metabolic imbalance. Therefore, active aldehydes resulting from FAO impairment can cause a vicious cycle of metabolic imbalance that leads to T cell exhaustion.
Similar content being viewed by others
Main
A major challenge of PD-1-blockade cancer immunotherapy is that many patients do not respond to such treatment. The therapeutic effects of cancer immunotherapy are largely influenced by exhaustion of tumor-infiltrating CD8+ T cells1,2,3,4. Although the exhaustion stage of CD8+ T cells is a spectrum, exhausted CD8+ T (TEX) cells are roughly divided into progenitor TEX cells and terminal TEX cells5,6,7. Progenitor TEX cells are characterized by longevity and proliferative capacity and can give rise to effector T cells. On the other hand, terminal TEX cells have lost their proliferation capacity and the majority of them undergo cell death4,5,6,7. Recent studies reveal that the antitumor effect of PD-1-blockade therapy is mediated not by the rejuvenation of terminal TEX cells but by the expansion of functional effector T cells from progenitor TEX cells4,8,9. Therefore, mechanisms and methodologies by which progenitor TEX cells are generated or maintained in the tumor microenvironment (TME) have been studied7. The manipulation of T cell metabolism is a practical strategy for inducing the progenitor TEX cells in tumor-bearing host10,11,12,13,14,15.
The balance of glycolysis and mitochondrial metabolism in T cells is finely controlled to fulfill their energy demands according to their differentiation stages such as naive, effector and memory T cells16,17. Although researchers in general agree that mitochondria in TEX cells are dysfunctional13,18,19,20, there are different conclusions of the glycolysis usage in TEX cells. Some reports indicate that TEX cells decrease the expression of glucose transporter-1 (Glut1) and/or decrease the glucose uptake21,22,23,24, whereas others indicate that TEX cells favor glycolysis with upregulated HIF-1α, glucose transporters and/or glucose uptake25,26. This unresolved discrepancy might be attributed to differences in the definition of T cell exhaustion among researchers.
Although glycolysis is critical for the differentiation of naive T cells to effector T cells, extreme reliance on glycolysis paradoxically impairs the effector function and boosts exhaustion of T cells26,27,28,29. Indeed, glycolysis inhibitors such as 2-deoxy-D-glucose (2-DG) and Akt inhibitors delay T cell exhaustion, increase the number of long-surviving T cells, and consequently, enhance the antitumor immunity10,30. Upon glycolysis inhibition, T cells prefer to utilize mitochondrial metabolism and upregulate fatty acid oxidation (FAO) activity, which is important for the longevity of T cells30,31. Consistently, FAO activators, such as bezafibrate, fenofibrate, metformin and spermidine, reduce the generation of terminal TEX cells and increase the number of long-survival T cells12,15,32,33. This growing evidence suggests that the balance between glycolysis and FAO is crucial for determining antitumor immune responses.
Recent studies have indicated that terminal TEX cells accumulate lipid peroxides and dampen their mitochondrial activity, which further accelerates exhaustion34,35,36. As lipid peroxidation leads to ferroptosis, terminal TEX cells could be subjected to oxidative stress-induced cell death35,37. The lipid accumulation is associated with a reduced expression of very-long-chain acyl-CoA dehydrogenase, a FAO-related enzyme, in CD8+ TEX cells, suggesting an important role of FAO in the robust function of CD8+ T cells in the TME34.
PD-1 is a checkpoint for T cells to avoid overactivation-induced cell death by inhibiting T cell receptor and CD28 signaling38,39. PD-1 signaling is known to inhibit glycolysis and enhance FAO, which are advantageous for T cell longevity21,40. Nevertheless, PD-1+ TEX cells in the TME are usually short-lived and carry impaired mitochondrial metabolism, suggesting the dysregulated balance of glycolysis and FAO. How the glycolysis and FAO balance are associated with the T cell exhaustion process is not fully understood.
In this paper, we show that the mitochondrial status of CD8+ T cells, characterized by the mitochondrial mass and potential, correlates with exhaustion depth of these cells. This mitochondrial compartmentation distinguishes terminal TEX cells into living and early apoptotic populations. Despite high expression of PD-1, living terminal TEX cells have high glycolytic activity and low FAO, which we define as ‘metabolic exhaustion’. TEX increased accumulation of active aldehydes, final products of lipid peroxidation in an exhaustion depth-dependent manner. The accumulation of active aldehydes is more prominent in FAO enzyme (HADHA)-deficient T cells. These results indicate that FAO activity is critical to prevent active aldehyde accumulation during T cell exhaustion. Acrolein, a representative active aldehyde generated from mitochondria, impairs FAO activity and accelerates the glycolysis signal transduction. Therefore, active aldehyde shapes the metabolic exhaustion of T cells in the TME.
Results
Mitochondrial status is associated with metabolic exhaustion of T cells in TME
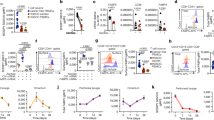
Naive CD8+ T cells could be divided into four populations (D1–D4) according to their mitochondrial potential and mitochondrial mass (Fig. 1a; day 0). Using the in vitro CD8+ T cell exhaustion model of repetitive stimulation with anti-CD3 antibody and interleukin (IL)-2 (ref. 41), we found that these stimulations promoted the sequential differentiation of CD8+ T cells from D1–D4 in vitro (Fig. 1a). To know the mitochondrial status of exhausted CD8+ TEX cells in tumors, we examined the mitochondrial potential and mass of PD-1+ CD8+ T cells among the tumor-infiltrating lymphocytes (TILs) after gating on a CD45+Zombie− population (Extended Data Fig. 1a). We confirmed that the T cells, which have depolarized mitochondria and are recognized as ‘terminal’ TEX cells in the literature, were located in D4 in our mitochondrial compartmentation definition (Extended Data Fig. 1a)13,18,22. D4 cells were mostly small (FSC-Alow) and Zombie−annexin V+, suggesting that they were early-stage apoptotic T cells (Fig. 1b and Extended Data Fig. 1a)42. To trace the T cell differentiation trajectory in vivo, we intravenously transferred CD45.2+ naive OT-1 T cells into CD45.1 recipient mice with MC38-OVA tumor. Over time, tumor-infiltrating donor PD-1+CD8+ T cells progressively differentiated from D1 to D3 (Extended Data Fig. 1b). Although D4 cells did not change drastically, they were presumably eliminated via rapid dead-cell clearance mechanisms such as efferocytosis in response to phosphatidylserine exposure on the cell surface (Fig. 1b)43.
a, Schema of repetitive stimulation. Naive (CD44low) CD8+ T cells isolated from mouse spleens and lymph nodes (LNs) were stimulated by anti-CD3/CD28 coated plates. Cells were collected and re-stimulated every 2 days. Representative flow cytometry profiles and frequencies of D1–D4 cells, based on mitochondrial mass and mitochondrial potential on day 0, 2 and 8. Technical replicates were day 0 and 2, n = 3; day 8, n = 5. b–d, Frequencies of annexin V+ (b), Tim-3+, LAG-3+, TIGIT+ (c) and Slamf6+ (d) cells in D1–D4 populations among PD-1+CD8+ T cells in MC38 tumors on day 14 (n = 8 mice). e–i, scRNA-seq analysis of D1, D2 and D3 populations sorted from PD-1+ CD8+ T cells in MC38 tumors on day 14. e, Uniform Manifold Approximation and Projection (UMAP) plot according to scRNA-seq profiles of each cell. Colors of dots indicate D1, D2 or D3 category. f, Violin plots of exhausted or stem-like signature scores in D1–D3. g, GO enrichment analysis of genes that were significantly upregulated in D3 compared to D1 (adjusted P < 0.05 and log2 fold change (D3/D1) > 1). h, Pseudotime and trajectory analysis mapped onto the UMAP. Violin plots of pseudotime score in D1–D3. i, Clonal distribution of TCR clonotypes in D1–D3. T cell clones were categorized as rare (range, 0 to ≤0.003), small (range, 0.003 to ≤0.01), medium (range, 0.01 to ≤0.03) and large (range, 0.03 to ≤0.1), hyperexpanded (range, 0.1 to ≤1) based on the proportion of each clonotype in D1–D3. j, D1, D2 and D3 of PD-1+CD8+ T cells were sorted from MC38 tumors on day 25, labeled with CellTrace Violet and stimulated with anti-CD3 and anti-CD28 coated beads for 49 h. An unstimulated sample from D1 group was included to assess baseline dye intensity. The figure shows a representative CellTrace histogram with CellTracelow frequencies shown next to the sample annotations, along with their quantification (D1 and D2, n = 12; D3, n = 8 technical replicates). Data are mean ± s.e.m. (j). P values were determined using paired two-tailed Student’s t-tests (b–d), one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons tests (f,h,j) or a one-sided hypergeometric test and adjusted for multiple testing using the Benjamini–Hochberg (BH) method (g). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results, except e–i.
To avoid analytic limitations of the color combination in flow cytometry for further analysis, we sought to find surrogate markers of D1–D4. If the D4 population was excluded, the fractions determined by the mitochondrial potential intensity alone (D1’ D2’ and D3’) precisely corresponded to D1, D2 and D3, respectively (Extended Data Fig. 1c). We therefore used D1’–D4’ as a surrogate marker of D1–D4 hereafter. The capacity of cytokine production and cytotoxic function of T cells in MC38 sequentially increased from D1’ to D3’ (Extended Data Fig. 1d). In accordance with the increase in annexin V positivity (Fig. 1b), exhaustion markers gradually increased from D1 to D3 (Fig. 1c and Extended Data Fig. 1e). Most of these exhaustion markers were reduced in D4, possibly due to the death process of these cells. Accordingly, the expression of Slamf6, a progenitor TEX cell marker known to be correlated with Tcf1 (ref. 4), decreased from D1 to D3 (Fig. 1d). Similar cytokine production capacity and surface exhaustion marker results were obtained from the in vitro exhaustion model on day 8 (Extended Data Fig. 1f,g).
To further analyze the role of mitochondrial status in the definition of TEX cells, we conducted single-cell RNA sequencing (scRNA-seq) on the isolated D1, D2 and D3 populations from PD-1+CD8+ T cells in the MC38 tumor. Cluster analysis demonstrated six distinctive populations, where cluster 0, 1 and 2 exhibited exhaustion signatures and cluster 3 showed high stem-like gene signatures (Extended Data Fig. 1h,i). The D1–D3 populations were broadly distributed with a degree of polarization: D2 and D3 populations were enriched for exhausted signatures, whereas the D1 population showed higher stem-like signatures (Fig. 1e,f). Gene Ontology (GO) enrichment analysis revealed that pathways associated with cell division were significantly upregulated in D3 compared to D1, indicating the differentiated or proliferating state of D3 in the TME (Fig. 1g and Supplementary Table 1). The cell differentiation trajectory analysis revealed a clear gradient in the pseudotime score, reflecting the progressive differentiation from D1 to D3 (Fig. 1h). Results of analyzing the scRNA-seq dataset for the CDR3 region of T-cell receptor (TCR) demonstrated directional clonal expansion from D1 to D3 (Fig. 1i). Although the cell cycle-related genes were transiently upregulated in D3 in the TME (Fig. 1g), their proliferation capacity was gradually declined from D1 to D3 upon ex vivo re-stimulation, indicating that D3 cells have already entered a state in which they can no longer effectively divide by further stimulation (Fig. 1j). Although D2 and D3 showed little or only marginal difference in their exhaustion markers reported (Fig. 1c,f), they are clearly different in their differentiation trajectories, clonal expansions and proliferative capacities (Extended Data Fig. 1b and Fig. 1h–j). Therefore, mitochondria-based classification provides a higher resolution definition of exhaustion, which remains ambiguous due to their heterogeneity44,45. The mitochondrial status of TEX cells in the TME also demonstrates that terminal TEX cells existed in either a live state with high membrane potential or an apoptotic state with depolarized mitochondria, a previously undefined feature.
We next investigated metabolic flows of D1–D4 PD-1+CD8+ T cells in tumor tissues, focusing on glycolysis and FAO. Glucose transporter-1 (Glut1) expression and 2-NBDG, a fluorescent glucose analog, uptake increased from D1’ to D3’ but decreased in D4’ (Fig. 2a). To measure the precise glycolysis activity, we incubated isolated D1–D3 with 13C-labeled glucose for 3 h and measured the 13C-labeled lactic acid. D4 was excluded from this 13C-tracing assay because this population was mostly trypan blue-positive (dead) after incubation. As shown in Fig. 2b, the glycolytic activity increased from D1 to D3 progressively. These data suggest that live ‘terminal’ TEX cells (D3) had high glycolysis activity, whereas early apoptotic ‘terminal’ TEX cells (D4) did not, which might explain the discrepancy in glycolysis usage of TEX cells between reports21,22,23,24,25,26. Regarding mitochondrial metabolism, the central FAO enzyme HADHA gradually decreased in response to exhaustion depth (Fig. 2c). A similar reduction in HADHA expression was also observed in human cancers, relating with exhaustion depth (Fig. 2d). The carnitine-acylcarnitine translocase SLC25A20 on mitochondrial inner membrane, which is critical for FAO in shuttling acylcarnitine into the mitochondrial matrix and exporting free carnitine into the intermembrane space46,47, also gradually decreased from D1’ to D4’ (Fig. 2e). Furthermore, because elaborated and densely folded cristae is associated with greater oxidative phosphorylation and FAO activity31,48, we analyzed the complexity of cristae architecture in mitochondria using transmission electron microscopy (TEM). TEM analysis revealed a loosed cristae structure with progressively increased spacing between folds from D1 to D3 (Fig. 2f). These data suggest impaired mitochondrial FAO activity according to exhaustion depth.
a, Mean fluorescence intensity (MFI) of Glut1 and 2-NBDG (a glucose analog) in D1’–D4’ populations among PD-1+CD8+ T cells in tumors on day 14 (Glut1, n = 6 mice; 2-NBDG, n = 8 mice). b, Sorted D1, D2 and D3 of PD-1+CD8+ T cells from MC38 tumors on day 17 were incubated in medium containing 10 mM 13C glucose for 3 h. Data shows the levels (peak area) of 13C-labeled lactic acid produced by each subset (D1, n = 4; D2, n = 5; D3, n = 6 technical replicates). c, Representative flow cytometry profiles (including corresponding isotype control) and MFI of HADHA in PD-1−Tim-3− and PD-1+Tim-3+ cells among CD8+ T cells in MC38 tumors on day 9 (n = 10 mice), and MFI of HADHA in D1’–D4’ cells among PD-1+CD8+ T cells in MC38 tumors on day 14 (n = 8 mice). d, MFI of HADHA in D1’–D4’ populations among PD-1+CD8+ T cells in human tumors (four RCC and three lung cancer). e, Representative flow cytometry profiles (including corresponding isotype control) and MFI of SLC25A20 in D1’–D4’ cells among PD-1+CD8+ T cells in MC38 tumors on day 15 (n = 5 mice). f, Quantification of mitochondrial inter-cristae distances (nm) based on TEM images of D1–D3 of PD-1+CD8+ T cells from MC38 tumors on day 20. For D1, D2 and D3, n = 12, 18 and 11 cristae distances were measured from n = 5, 9 and 4 mitochondria derived from n = 2, 4 and 2 cells, respectively. g, MFI of mitochondrial superoxide (MitoSOX) in D1–D4 populations and lipid radical in D1’–D4’ populations among PD-1+CD8+ T cells in tumors on day 14 (MitoSOX, n = 6 mice; lipid radical, n = 8 mice). h, Schema of the lipid peroxidation pathway. ROS create lipid radicals from cellular lipids and then lipid radicals produce active aldehydes, for example, acrolein, in a chain reaction. Active aldehydes cause damage to DNA, proteins and other biomolecules. Created with BioRender.com, elements adapted and assembled using PowerPoint. i, MFI of acrolein in D1–D4 populations and protein-conjugated acrolein (PC-Acro) in D1’–D4’ populations among PD-1+CD8+ T cells in tumors on day 14 (n = 6 mice). j, MFI of lipid radical in D1’–D4’, acrolein in D1–D4 and PC-Acro in D1’–D4’ populations among PD-1+CD8+ T cells in human tumors (four RCC and three lung cancer for lipid radical, six RCC and four lung cancer for acrolein and six RCC and four lung cancer for PC-Acro). Data are mean ± s.e.m. (b,f). P values were determined using paired two-tailed Student’s t-tests (a,c,e,g,i) one-way ANOVA with Tukey’s multiple comparisons tests (b,f), or two-tailed Wilcoxon matched-pairs signed-rank tests (d,j). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results, except d and j.
The PD-1 signal is reported to inhibit glycolysis and enhance FAO21,40; however, these data suggested that live ‘terminal’ TEX cells (D3) adhered to glycolysis with an attenuated mitochondrial function, such as FAO, despite PD-1 expression, which is partly supported by previous reports13,18,19,20,25,26. This unbalanced metabolism in TEX cells in the TME is referred to as ‘metabolic exhaustion’ in this work hereafter. We focused on live TEX cells (D1–D3) for the following mechanistic analysis.
Active aldehydes accumulate in proportion to exhaustion depth
Lipid peroxidation induced by reactive oxygen species (ROS) is one of the exhaustion processes in T cells34,35,36. Mitochondrial ROS (mROS) and lipid peroxides (lipid radical) accumulated in PD-1+CD8+ T cells in the TME in proportion to the exhaustion depth from D1’ to D3’ (Fig. 2g and Extended Data Fig. 2a). Although active aldehydes are the final products of lipid peroxidation and are more than ten times more toxic than ROS49,50, the role of active aldehydes in T cell exhaustion has not been extensively addressed (Fig. 2h). We measured the level of acrolein, a representative active aldehyde, by AcroleinRED, which reacts specifically to the free acrolein in live cells. We also used an antibody to protein-conjugated acrolein (PC-Acro) to detect the reacted acrolein in the cells. Acrolein levels measured by both methods were elevated in correspondence with exhaustion depth (Fig. 2i and Extended Data Fig. 2b). Like acrolein, levels of other active aldehydes, including malonaldehyde (MDA) and 4-hydroxynonenal (4-HNE), were also elevated in highly TEX cells (Extended Data Fig. 2c). Elevated levels of lipid radical, acrolein and PC-Acro were similarly observed among D1’ to D4’ in the in vitro exhaustion model by repetitive stimulation (Extended Data Fig. 2d). The acrolein levels in CD8+ T cells were also increased by repetitive stimulation (Extended Data Fig. 2e) Moreover, in human carcinomas, a greater accumulation of acrolein was observed in accordance with the exhaustion depth in tumor-infiltrating CD8+ T cells (Fig. 2j). Therefore, the gradual accumulation of active aldehydes in accordance with exhaustion depth is a general phenomenon.
Acrolein enhances glycolysis and attenuates FAO
Although acrolein was known to be a detrimental metabolite, how acrolein contributed to metabolic exhaustion was unknown. The D3’ population showed high glycolytic activity (Fig. 2a,b), which is reported as a hallmark metabolic feature of TEX cells25,26. To investigate the effect of acrolein on glycolysis, we stimulated mouse total CD8+ T cells with CD3/CD28 beads with or without acrolein and examined the activity of metabolic pathways. Although free acrolein concentration was approximately 10 μM in MC38 tumor tissues (Extended Data Fig. 3a), we optimized and used acrolein (ACR) at a 2–5 μM range in vitro, which was appropriate for the proliferative capacity assay for primary CD8+ T cells. We found that the Akt–mTOR pathway, which promotes glycolysis during T cell differentiation, was quickly upregulated in CD8+ T cells within 3 h of acrolein stimulation (Fig. 3a,b)40,51. Seahorse analysis revealed that acrolein increased the level of glycolysis, indicated by the extracellular acidification rate (ECAR) (Fig. 3c). Moreover, acrolein stimulation significantly enhanced the glucose uptake, PD-1 expression, and proliferation of CD8+ T cells, indicating accelerated T cell differentiation (Fig. 3d–f). Indeed, acrolein boosted T cell differentiation from D1 to D4 within 7 and 21 h (Fig. 3g and Extended Data Fig. 3b). Increases in Akt–mTOR pathway and PD-1+Tim-3+ TEX cells by ACR stimulation were more obvious in the in vitro exhaustion model on day 8 (Extended Data Fig. 3c,d). In terms of mitochondrial metabolism, we investigated the two main pathways FAO and glutaminolysis using CpT1a inhibitor (Etomoxir) and glutaminase inhibitor (BPTES), respectively. Acrolein stimulation reduced mitochondrial respirations within 3 h in an etomoxir-dependent but BPTES-independent manner, demonstrating that acrolein selectively attenuated FAO activity over glutaminolysis (Fig. 3h).
a, The Akt–mTOR and glycolysis boost the differentiation of CD8+ T cells from naive T cells to exhausted T cells. b, MFI of pAkt, pmTOR and pS6 in primary CD8+ T cells stimulated for 3 h with anti-CD3 and anti-CD28 coated beads in the presence or absence of 2 μM acrolein (ACR) evaluated by flow cytometry (n = 5 technical replicates). c, Seahorse measurements of real-time ECAR of CD8+ T cells 3 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 5 μM ACR (Ctrl, n = 6; ACR, n = 7 technical replicates). d, MFI of 2-NBDG in CD8+ T cells 3 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 2 μM ACR measured by flow cytometry (n = 5 technical replicates). e, Frequencies of PD-1+ cells among CD8+ T cells 3 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 2 μM ACR measured by flow cytometry (n = 5 technical replicates). f, Thymidine incorporation of CD8+ T cells 7 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 5 μM ACR (n = 4 technical replicates). g, Frequencies of D1–D4 populations among CD8+ T cells 7 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 2.5 μM ACR (n = 12 technical replicates). h, Seahorse measurements of oxygen consumption rate (OCR) of CD8+ T cells 3 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 5 μM ACR. 5 μM etomoxir or 3 μM BPTES was added approximately 10 min before running the assay (all Ctrl groups, n = 3; all ACR groups, n = 4 technical replicates). To measure the oxidation activity of endogenous fatty acids or glutamine, low-nutrient medium (RPMI + 2.5 mM glucose) was used during the assay. i,j, Phosphoproteomic analysis of total CD8+ T cells stimulated with anti-CD3/CD28 beads for 1 h in the presence or absence of 5 μM ACR (n = 5 technical replicates). Volcano plot of canonical pathways analyzed by IPA (i). For visualization purposes, pathways with P values equal to 1 or with undefined z-scores were excluded from the figure. A total of 545 pathways are shown in the figure (upregulated, z-score > 0.8 and P value < 0.05; downregulated, z-score < −0.8 and P value < 0.05; the others, −0.8 ≤ z-score ≤ 0.8 or P value ≥ 0.05). Bar graph showing the top five upregulated and downregulated gene names based on phosphorylation changes among approximately 20,000 phosphopeptides (j). k, Immunoblot images of SLC25A20 in CD8+ T cells stimulated with anti-CD3/CD28 beads for 3 h in the presence or absence of 5 μM ACR. l, Quantification of mitochondrial inter-cristae distances (nm) based on TEM images of naive CD8+ T cells stimulated with anti-CD3/CD28 beads for 3 h in the presence or absence of 5 μM ACR. For Ctrl and ACR, n = 125 and 136 cristae distances were measured from n = 33 and 41 mitochondria derived from n = 14 and 18 cells, respectively. Data are mean ± s.e.m. P values were determined using unpaired two-tailed Student’s t-tests (b–g,l) or one-way ANOVA with Tukey’s multiple comparisons test (h) or right-tailed Fisher’s exact test (i). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results, except i and j.
We next investigated the molecular mechanisms by which acrolein stimulation enhanced glycolysis and inhibited FAO. Phosphoproteomic analysis of CD8+ T cells stimulated by acrolein for 1 h revealed the activation of TCR-driven Akt–mTOR-related signaling pathways (including Rho GTPase cycle and phospholipase C pathways) and suppression of mitochondrial biogenesis, as predicted by Ingenuity Pathway Analysis (IPA) software (Fig. 3i and Supplementary Tables 2 and 3). TCR signal is known to activate pyruvate dehydrogenase kinase 1 (Pdk1), leading to phosphorylation and inactivation of pyruvate dehydrogenase E1 α1 (Pdha1), thereby promoting anaerobic glycolysis52. Among ~20,000 phosphopeptides analyzed, Pdha1 was one of the most significantly increased by ACR, ranking third in upregulation (Fig. 3j and Supplementary Table 2). We further investigated the expression levels of glycolysis and FAO-related enzymes by immunoblotting. Among the enzymes examined, only the acylcarnitine transporter SLC25A20 was reduced in a 3-h stimulation (Fig. 3k and Extended Data Fig. 3e). TEM analysis demonstrated that 3 h of ACR stimulation loosened the density of mitochondrial cristae or degraded their architecture (Fig. 3l). These data underpin our findings that acrolein upregulates glycolysis and inhibits FAO.
Treating CD8+ T cells with another active aldehyde, 4-HNE, upregulated glycolysis and proliferation (Extended Data Fig. 3f,g). In contrast to active aldehydes, treating T cells in vitro with hydrogen peroxide (H2O2), a ROS, did not enhance glycolysis or differentiation (Extended Data Fig. 3h–j). Therefore, active aldehydes promoted the differentiation of CD8+ T cells toward an exhausted state with glycolysis upregulation and FAO downregulation despite the high expression of PD-1 in these cells21,40.
Endogenous acrolein controls metabolic exhaustion
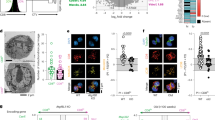
Acrolein is a primary active aldehyde showing the highest toxicity, it reacts immediately with neighboring cysteine and lysine and denatures those proteins13,18,19,20,53. Therefore, we addressed the role of active aldehydes in forming metabolic exhaustion, focusing on acrolein. We stimulated CD8+ T cells with CD3/CD28 beads in the presence of an acrolein scavenger, N-benzylhydroxylamine (NBHA)50. NBHA reduced glycolysis (ECAR), increased the D1 population, and reduced D2 and D3 populations, suggesting that endogenous acrolein enhanced the differentiation of T cells (Fig. 4a,b). This NBHA effect of inhibiting differentiation was more obvious in the in vitro exhaustion model after a long culture; NBHA treatment significantly reduced the frequency of PD-1+Tim-3+ TEX cells on day 8 (Extended Data Fig. 4a). We therefore examined the relationship between cellular acrolein levels and metabolic features in tumor-infiltrating PD-1+CD8+ T cells. As expected, compared to the CD8+ T cell population with a low amount of acrolein, that with a high amount of acrolein exhibited a stronger Akt–mTOR signal transduction, Glut1 expression and 2-NBDG incorporation (Fig. 4c,d). Similarly, Akt–mTOR signal transduction, Glut1 expression and 2-NBDG incorporation were increased in TEX cells infiltrating human renal cell carcinoma (RCC) and lung cancer with high amounts of acrolein (Fig. 4e).
a, Seahorse measurement of real-time ECAR of CD8+ T cells 3 h after stimulation with anti-CD3/CD28 beads in the presence or absence of 10 μM NBHA (n = 4 technical replicates). The control data are shared with those in Extended Data Fig. 3h to compare results under the same conditions in the same experiment, as supportive data. b, Frequencies of D1–D4 cells among CD8+ T cells in the same condition as a measured by flow cytometry (n = 6 technical replicates). c, Representative flow cytometry profiles and MFI of pAkt, pmTOR and pS6 in PC-Acro low and PC-Acro high populations among PD-1+CD8+ T cells in MC38 on day 14 (n = 5 mice). A fluorescent antibody for PC-Acro was used instead of staining dye for acrolein due to the limitation of staining condition. d, Representative flow cytometry profiles and MFI of Glut1 and 2-NBDG in PC-Acro and acrolein, respectively, low and high populations among PD-1+CD8+ T cells in MC38 on day 14 (Glut1, n = 6 mice; 2-NBDG, n = 5 mice). e, MFI of pAkt, pmTOR, pS6, Glut1 and 2-NBDG between PC-Acro or acrolein low and high populations of PD-1+CD8+ T cells in human tumors (six RCC and four lung cancer). f, Representative fluorescence microscopy image and Pearson’s correlation between mitochondria and PC-Acro in sorted PD-1+CD8+ T cells from MC38 on day 19 (n = 12 cells). An experimental control group was not included, as the experiment aimed to examine the colocalization of mitochondria and PC-Acro. g, MFI of MitoPeDPP (mitochondrial membrane peroxidation) in D1’–D4’ cells among PD-1+CD8+ T cells in MC38 on day 14 (n = 6 mice). Data are mean ± s.e.m. (a,b). P values were determined using unpaired two-tailed Student’s t-tests (a,b), paired two-tailed Student’s t-tests (c,d,g) or two-tailed Wilcoxon matched-pairs signed-rank test (e). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results, except e.
Given that acrolein impacted T cell metabolic regulation, we investigated the intracellular location of acrolein production. Super-resolution fluorescence microscopy analysis of PD-1+CD8+ TILs demonstrated that signals of mitochondria (PhenoVue 551) and PC-Acro exhibited approximately 40% spatial overlap (Fig. 4f). In addition, mitochondrial membrane peroxidation progressively increased in accordance with the hierarchal accumulation of acrolein from D1 to D3 (Fig. 2i,j and Fig. 4g), supporting the acrolein production being partly from mitochondria54. CD8+ T cells might be inefficient in detoxifying acrolein due to low expression of aldehyde dehydrogenase 2 (Aldh2); we could not detect the messenger RNA of Aldh2 in scRNA-seq analysis (Extended Data Fig. 4b). These data suggested that the intrinsic acrolein contributed to the dysregulation of mitochondrial function and metabolic exhaustion.
FAO dysfunction accelerates metabolic exhaustion
To determine the role of FAO in metabolic exhaustion in T cells, we analyzed Hadhaflox/flox Cd4-Cre mice that were deficient of HADHA in T cells (HADHA-T KO)15. HADHA-T KO mice develop without any apparent defects and show a similar frequency of CD44high population in CD8+ T cells compared to wild-type mice15. Also, naive peripheral CD8+ T cells in HADHA-T KO mice did not increase the expression of FAO-related molecules or functions, including lipid radicals, acrolein, Akt–mTOR signal transduction, 2-NBDG incorporation, fatty acid transporter CD36 and fatty acid uptake, compared to those in wild-type mice (Extended Data Fig. 5a–c); however, tumor-infiltrating PD-1+CD8+ T cells in HADHA-T KO mice upregulated the expression of the fatty acid transporter CD36 and increased the fatty acid uptake compared to those cells from wild-type mice (Fig. 5a). These HADHA-deficient T cells accumulated more lipid radicals and acrolein as well as presented stronger Akt–mTOR signal transduction and 2-NBDG incorporation than wild-type cells (Fig. 5b,c). To investigate the metabolic balance of glycolysis and mitochondrial pathways in T cells from HADHA-T KO mice, we measured metabolic flow of naive CD8+ T cells during CD3/CD28 bead stimulation in vitro. The Real-Time ATP Rate Assay in Seahorse revealed that ATP demands during stimulation were fulfilled mostly by glycolysis in HADHA-deficient T cells, in contrast to mainly by mitochondrial pathways in control T cells (Fig. 5d). This dependence on glycolysis rather than mitochondria metabolism in CD8+ T cells from HADHA-T KO mice resembled metabolic exhaustion. Indeed, more tumor-infiltrating CD8+ T cells from HADHA-T KO mice expressed exhaustion markers Tim-3 and TOX and the cell death marker annexin V than those from wild-type mice (Fig. 5e,f). Functionally, tumor-infiltrating PD-1+CD8+ T cells from HADHA-T KO mice had less capacity of proliferation than those from wild-type Ctrl mice (Fig. 5g). Accordingly, tumor growth was faster and the frequency of tumor-infiltrating CD8+ T cells was lower in HADHA-T KO mice than in wild-type mice (Fig. 5h,i).
a, CD36 expression and BODIPY C5, C12 and C16 (fatty acid uptake) in PD-1+CD8+ T cells from MC38 tumors grown in Hadhaflox/flox (Ctrl) and Hadhaflox/flox Cd4-Cre (KO) mice on day 14 (CD36, Ctrl n = 8 mice, KO n = 7 mice; BODIPY C5, n = 5 mice; BODIPY C12, n = 5 mice; BODIPY C16, Ctrl n = 8 mice, KO n = 7 mice). b, MFI of lipid radical, acrolein and PC-Acro in PD-1+CD8+ T cells in MC38 tumors from Ctrl and KO mice on day 14 (lipid radical, Ctrl n = 8 mice, KO n = 6 mice; acrolein, n = 5 mice; PC-Acro, Ctrl n = 8 mice, KO n = 6 mice). c, MFI of pAkt, pS6 and 2-NBDG in PD-1+CD8+ T cells in MC38 tumors from Ctrl and KO mice on day 14 (pAkt and pS6, n = 5 mice; 2-NBDG, Ctrl n = 8 mice, KO n = 6 mice). d, ATP productions derived from glycolysis and mitochondrial OXPHOS in naive CD8+ T cells from Ctrl and KO mice before and after 3-h stimulation with anti-CD3/CD28 beads measured by Seahorse (n = 6 technical replicates). e, Frequencies of PD-1−Tim-3−, PD-1+Tim-3+ and TOX+ cells among CD8+ T cells in MC38 tumors from Ctrl and KO mice on day 14 (n = 5 mice). f, Frequencies of annexin V+ cells among PD-1+CD8+ T cells in MC38 tumors from Ctrl and KO mice on day 14 (Ctrl, n = 8 mice; KO, n = 7 mice). g, PD-1+CD8+ T cells sorted from MC38 tumors of Ctrl and KO mice on day 14 were labeled with CellTrace Violet and stimulated with anti-CD3/CD28 beads for 49 h. An unstimulated sample from Ctrl group was included to assess baseline dye intensity. The figure shows a representative CellTrace histogram with CellTracelow frequencies shown next to the sample annotations, along with their quantification (n = 10 technical replicates). h, MC38 tumor growth in Ctrl and KO mice (Ctrl, n = 5 mice; KO, n = 7 mice). Statistical analysis on day 20. i, Representative flow cytometry profiles on day 14 and frequencies of CD8+ T cells among tumor-infiltrating CD45+ cells from Ctrl and KO mice on day 10, 14 and 20 (day 10, Ctrl n = 6 mice, KO n = 8 mice; day 14, Ctrl n = 8 mice, KO n = 7 mice; day 20, Ctrl n = 5 mice, KO n = 6 mice). Statistical analysis on day 14 and day 20. Data are mean ± s.e.m. P values were determined using unpaired two-tailed Student’s t-tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Female mice were used in panel (d), and all other data were obtained from male mice. All data are representative of two or more independent experiments with similar results.
Fatty acids were highly incorporated in CD8+ TIL from HADHA-T KO mice compared to wild-type mice (Fig. 5a). As terminal TEX cells exhibited FAO dysregulation in wild-type mice (Fig. 2c–f), we investigated the fatty acid uptake in the terminal TEX cells. PD-1+Tim-3+ TEX cells from wild-type mice increased the fatty acid incorporation compared to PD-1+Tim-3− T cells (Extended Data Fig. 5d), which might be linked to further accumulation of lipid radical and acrolein in the terminal TEX cells (Fig. 2g,i). These data demonstrate that FAO dysfunction exacerbates the exhaustion state and increases oxidative stress by promoting acrolein accumulation in T cells through a feedback loop mechanism (Extended Data Fig. 5e).
A lipid radical or acrolein scavenger enhances antitumor immunity
We finally examined whether lipid radical or acrolein scavengers could reduce exhaustion and enhance antitumor immunity. The lipid radical scavenger α-tocopherol (vitamin E; VtE) and the aldehyde-specific scavenger NBHA reversed the low antitumor activity of HADHA-T KO mice (Fig. 6a,b). While glycolytic activity was increased in effector CD8+ T cells from HADHA-T KO mice (Fig. 5c,d), NBHA administration suppressed the Akt–mTOR pathway, increased the D1 population, and decreased the D2 population in PD-1+CD8+ TIL in HADHA-T KO mice (Fig. 6c,d). These data validated that acrolein accumulation in T cells of HADHA-T KO mice was one of the primary reasons for the low antitumor immunity, as acrolein promotes their differentiation.
a, MC38 tumor growth in littermate Ctrl (Hadhaflox/flox) and KO (Hadhaflox/flox Cd4-Cre) male mice intraperitoneally (i.p.) treated with α-tocopherol (VtE) (2 mg kg−1) or dimethylsulfoxide (DMSO) (Ctrl) every 3 days from day 7 (n = 5 mice). Statistical analysis on day 28. b, MC38 tumor growth in littermate Ctrl and KO female mice i.p. treated with NBHA (100 mg kg−1) or PBS (Ctrl) every 3 days from day 7 (n = 5 mice). Tumor growth in female mice is usually slower than that in male mice. Statistical analysis on day 16. c,d, MFI of pAkt and pS6 (c) and frequencies of D1–D4 cells (d) on day 10 among PD-1+CD8+ T cells in the TME from KO male mice treated with NBHA or PBS (Ctrl) (n = 5 mice). Data are mean ± s.e.m. P values were determined using unpaired two-tailed Student’s t-tests (a,b). *P < 0.05; **P < 0.01. All data are representative of two or more independent experiments with similar results.
We also investigated whether acrolein undermines the response to the PD-L1 monoclonal antibody (mAb) therapy. Administrating either VtE or NBHA enhanced the antitumor activity of the PD-L1 mAb (Fig. 7a,b). Although the frequency of PD-1+CD8+ TILs among CD45+ cells was increased by PD-L1 mAb treatment with or without NBHA (Fig. 7c), their metabolic phenotypes were altered. Consistent with the previous report21, PD-L1 mAb treatment increased the tendency of glycolysis-associated signaling and significantly reduced FAO activity measured by tracing the metabolites (acylcarnitine) derived from 13C-labeled palmitate (Extended Data Fig. 6 and Fig. 7d). Of note, adding NBHA to PD-L1 mAb treatment abolished effects of PD-L1 mAb treatment on glycolysis-related signals and FAO activity in PD-1+CD8+ TILs (Extended Data Fig. 6 and Fig. 7d). Because D1–D4 defined by mitochondrial states correlated with exhaustion depth (Fig.1), we examined D1–D4 population changes upon PD-L1 mAb treatment alone or in combination with NBHA. While PD-L1 mAb treatment alone reduced the D1 population and increased the D3 population among the PD-1+CD8+ TILs, adding NBHA to PD-L1 mAb treatment abolished these effects (Fig. 7e). Moreover, NBHA recovered the reduction of proliferative capacity induced by PD-L1 mAb alone (Fig. 7f). These data indicate that acrolein scavenger enhances antitumor immunity by inhibiting metabolic alteration and differentiation of CD8+ T cells in TME.
a, MC38 tumor growth in wild-type (WT) female mice i.p. treated with anti-PD-L1 mAb every 6 days and α-tocopherol (VtE) (2 mg kg−1) every 3 days from day 7 (Ctrl, n = 8 mice; anti-PD-L1, n = 7 mice; anti-PD-L1 + VtE, n = 7 mice). Statistical analysis on day 22. b, MC38 tumor growth in WT female mice i.p. treated with anti-PD-L1 mAb every 6 days and NBHA (100 mg kg−1) every 3 days from day 7 (Ctrl, n = 7 mice; anti-PD-L1, n = 6 mice; anti-PD-L1 + NBHA, n = 7 mice). Statistical analysis on day 22. c–f, Analysis of tumor-infiltrating PD-1+CD8+ T cells from MC38 tumors on day 14 in WT mice treated with anti-PD-L1 mAb and NBHA as indicated in b. Frequencies of PD-1+CD8+ T cells among tumor-infiltrating CD45+ cells (anti-PD-L1, n = 8 mice; the other groups, n = 9 mice) (c). Sorted PD-1+CD8+ T cells were incubated in medium containing 200 μM BSA-conjugated palmitate-13C16 for 3 h (d). Data show the levels (fmol) of 13C-labeled acylcarnitines (C16, palmitoylcarnitine; C14, myristoylcarnitine; C12, lauroylcarnitine; C10, decanoylcarnitine; C8, octanoylcarnitine; C6, hexanoylcarnitine) (n = 6 technical replicates). Frequencies of D1–D4 among PD-1+CD8+ T cells in tumors (anti-PD-L1, n = 8 mice; all other groups, n = 9 mice) (e). Sorted PD-1+CD8+ T cells were labeled with CellTrace Violet and stimulated with anti-CD3/CD28 beads for 45 h (f). An unstimulated sample from Ctrl group was included to assess baseline dye intensity. The figure shows a representative CellTrace histogram with CellTracellow frequencies shown next to the sample annotations, along with their quantification (Ctrl, n = 4; anti-PD-L1, n = 12, anti-PD-L1 + NBHA, n = 10; NBHA, n = 12 technical replicates). Data are mean ± s.e.m. P values were determined using unpaired two-tailed Student’s t-tests (a,b), one-way ANOVA with selected column comparisons between the untreated and anti-PD-L1 groups or anti-PD-L1 and anti-PD-L1 + NBHA groups (c–f). *P < 0.05; **P < 0.01; ****P < 0.0001. All data are representative of two or more independent experiments with similar results.
Discussion
In this paper, we demonstrate two key points. First, the mitochondrial status of CD8+ T cells characterizes the exhaustion stages in TME, which the conventional exhaustion markers can not capture. Terminally differentiated TEX cells are divided into living and early apoptotic populations: living terminal TEX cells increase mitochondrial potential and glycolysis activity, and apoptotic terminal TEX cells reduce metabolic fitness characterized by depolarized mitochondria and low glycolysis activity. These results may explain the different conclusions on the utility of glycolysis in terminal TEX cells among the literature21,22,23,24,25,26. Second, T cells with attenuated FAO accelerate their lipid peroxidation and active aldehyde production, which are prominently observed in the terminal TEX cells. Active aldehydes, partly produced by mitochondria, enhance glycolysis and reduce FAO activity in TEX cells, leading to metabolic exhaustion, despite high PD-1 expression (Extended Data Fig. 7a–c).
The metabolic balance of glycolysis and FAO regulates the differentiation fate of T cells toward short- or long-lived cells with low or high proliferation capacity, respectively10,30,31. Effector T cells use both glycolysis and FAO to meet their anabolic and catabolic demands16,17,32; however, chronic stimulation by tumor antigens in TME gradually attenuates the mitochondrial FAO activity, potentially via the mechanism that acrolein, generated from mitochondria as a final product of lipid–ROS reactions, reduces mitochondrial biogenesis and damages mitochondrial membrane structure directly. The HADHA level decreases progressively from D1 to D4, but its level does not decrease immediately after acrolein treatment, unlike SLC25A20. These results suggest that the reduction of HADHA may result from the acrolein-associated signal pathway rather than the direct damage. Although mitophagy is critical for the clearance of damaged mitochondria55, active aldehydes might inhibit autophagy and mitophagy through the activation of the Akt–mTOR pathway56. Active aldehydes would, therefore, reduce the ability to clear damaged mitochondria, resulting in the accumulation of more aldehydes. A robust FAO activity shunts this loop and renders T cell longevity.
Our data indicate that D1 and D2 T cells use both glycolysis and FAO in the TME, whereas highly exhausted D3 T cells rely mostly on glycolysis. As the Slamf6 expression in D1 and D2 populations is higher than in the D3 population, the majority of Tcf1+ progenitor TEX cells should be located around D1–D2. These data, therefore, support that progenitor TEX cells utilize balanced glycolysis and FAO metabolism to retain their self-renewal ability57. Considering the report that Tcf1-knockdown T cells carry dysfunctional and depolarized mitochondria58, the transition from D1–D2 to D3–D4 partly reflects the differentiation from progenitor to terminal TEX cells4. Because the frequency of progenitor TEX cells correlates to the higher response to the PD-1-blockade therapy4,8,9, the frequency of D1–D2 in TME might be a good biomarker for better responsiveness. Further studies are warranted.
In the TME with low glucose and high fatty acid levels, T cells are known to rely on FAO to survive32. Tumor-infiltrating CD8+ T cells in HADHA-T KO mice incorporate fatty acids greater than those of wild-type mice. A possible reason is that FAO dysfunction reduces the ATP production of T cells, and the consequent alteration of AMP:ATP ratio upregulates the AMPK–CD36 pathway59. Therefore, FAO plays an important role in not only consuming fatty acids but also preventing the uptake of unnecessary fatty acids, contributing to the retention of T cell function and their longevity.
Some groups have indicated that ROS accelerates exhaustion of T cells. ROS scavengers or inhibitors such as N-acetylcysteine and mitochondrial antioxidants inhibit T cell exhaustion19,20,60. As active aldehydes are the final products of lipids peroxidation, these ROS scavengers could also inhibit the production of active aldehydes. Given that the reactive toxicity of active aldehydes is higher than that of ROS49,50, active aldehydes are likely to be a major factor for exhaustion in addition to ROS. As the reaction mode of active aldehydes is different from that of ROS, the subsequent signal cascades between ROS and reactive carbonyl species (RCS), such as active aldehydes, could be different61. As we did not find a quick upregulation of ECAR by ROS, ROS and RCS may differently contribute to the exhaustion steps. Further studies on the signal cascade of active aldehydes, especially on glycolysis and FAO, are needed.
Our in vitro analysis was conducted using extracellular acrolein (ACR) treatment, which may not fully replicate the intracellular signaling environment driven by endogenously generated acrolein from mitochondria; however, acrolein, a small molecule, can act as a diffusive agent through the cell membrane62. Mitochondrial acrolein, therefore, should diffuse into the cytoplasm, which is highly likely to activate Akt–mTOR signaling. Acrolein is highly reactive toward cysteine residues through the addition reaction63. Acrolein may not only affect Akt–mTOR signaling-associated molecules in the way of post-translational regulation, but also covalently modify and inactivate PTEN, thereby activating the PI3K–Akt pathway and promoting phosphorylation of Akt substrates64. For SLC25A20 reduction by acrolein, several cysteine residues have been reported to be critical for its functional regulation47. Thus, the rapid reduction of SLC25A20 following acrolein exposure may result from direct cysteine modification, potentially leading to protein denaturation and functional impairment.
Although the active aldehyde-specific scavenger NBHA enhanced the antitumor immune activity in HADHA-T KO mice and during PD-L1 mAb therapy, NBHA alone rarely affected the antitumor activity. A few potential reasons may explain these observations. (1) The FAO dysfunction-exhaustion feedback loop (vicious cycle) is more prominent in these two models, and NBHA may attenuate such a severe vicious cycle. (2) PD-1 blockade therapy is known to promote clonal replacement, where terminally exhausted T cells are replaced by newly recruited, less-differentiated clones8. NBHA may exert a more pronounced effect for the effector T cell differentiation, but not the already exhausted T cells.
In conclusion, we have clarified the mechanism by which active aldehydes, the final products of lipid peroxidation, accelerate CD8+ T cell exhaustion through formatting metabolic exhaustion in TME. Mitochondrial FAO dysfunction leads to the production of active aldehydes from mitochondria, further impairs FAO activity and enhances glycolysis. Our findings shed light on the development of a strategy to improve cancer immunotherapy by preventing metabolic exhaustion.
Methods
Mice
C57BL/6N (5–11 weeks old) mice were obtained from Charles River Laboratories Japan or Japan SLC. OT-1 TCR-transgenic mice (CD45.2) were obtained from The Jackson Laboratory (originally developed by M. B. Bevan at the University of Washington). C57BL/6N-background Hadhaflox mice were obtained from Cyagen Biosciences. Hadhaflox/flox Cd4-Cre mice15 were generated by crossing Hadhaflox mice with Cd4-Cre mice65. C57BL/6-background CD45.1 congenic mice have been maintained at the Institute of Laboratory Animals, Graduate School of Medicine, Kyoto University. Littermate or age/sex-matched mice were used for analysis. Female mice were used except in the experiments in which the usage of male mice was described explicitly. All mice were used under protocols approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University (protocol approval number 25086). They were maintained under specific-pathogen-free conditions. They were fed with F-2 from Funabashi Farm Co. They were kept in the dark from 20:00 to 8:00, in the light from 8:00 to 20:00, at 25 °C and at 45% humidity.
Cell lines
Details of the MC38 murine colon adenocarcinoma cell line have been described previously66. The identity of the cells was confirmed based on morphology, functional characteristics and gene expression profiles. MC38 cells were cultured in RPMI medium (Gibco) with 10% (v/v) heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich) and 1% (v/v) penicillin–streptomycin mixed solution (Nacalai Tesque). MC38-OVA cells were generated by transfection of ovalbumin (OVA) complementary DNA into parental MC38 cells and maintained under Geneticin selection (0.5 mg ml−1 G418; InvivoGen) in the same culture medium as the parental cells.
Chemical reagents
The following chemical reagents were used for the mouse therapies and the in vitro assays. Acrolein (100 µg ml−1 in water) (AccuStandard), 4-hydroxynonenal (MCE) prepared in DMSO (Nacalai Tesque), Etomoxir (Sigma-Aldrich) prepared in MilliQ, BPTES (AdipoGen) prepared in DMSO, N-Benzylhydroxylamine hydrochloride (TCI), α-tocopherol (Wako) prepared in DMSO, IL-2 (PeproTech) prepared in MilliQ and 3H thymidine (PerkinElmer). Other chemical reagents used were equipped with the kits. The concentration of the reagents in vitro was determined following experimental optimization.
Mouse therapy model
MC38 cells (5 × 105) were injected intradermally into the right flank of C57BL/6N mice on day 0. For anti-PD-L1 combination treatment, the mice were injected intraperitoneally (i.p.) with 25–30 µg of an anti-PD-L1 mAb (clone 1-111A, made in-house) every 6 days67, i.p. with 2 mg of NBHA every 3 days, or i.p. with 40 µg of α-tocopherol (VtE) every 3 days from day 7. Tumor measurement was performed every 2 or 3 days, and tumor volume was calculated using the formula for a typical ellipsoid: 3.14 × (length × breadth × height)/6. Accidentally dead mice were excluded. All mice experiments were performed under protocols approved by the Animal Research Committee, Graduate School of Medicine, Kyoto University (protocol approval number 25086).
Cell preparation and culture for analysis of primary CD8+ T cells
For in vitro analysis, axillary, brachial and inguinal LNs on both sides of mice without tumors were collected, homogenized and pooled. Spleens were collected, homogenized and then treated with ACK lysing buffer for 2 min. After pooling the cells of LNs and spleens, total or naive (CD44low) CD8+ T cells were purified using MojoSort system (BioLegend) according to the manufacturer’s instructions. Isolated mouse CD8+ T cells were directly analyzed, or cultured in RPMI with 10% (v/v) heat-inactivated FCS, 1% (v/v) penicillin–streptomycin, 25–50 µM 2-mercaptoethanol (Nacalai Tesque) and other reagents or materials as necessary.
Tumor collection
Tumor-bearing mice were killed on the indicated days. Tumor samples were collected and minced into 1–2-mm pieces with scissors and digested with collagenase type IV (Thermo Fisher Scientific) using a gentleMACS Dissociator (Miltenyi Biotec).
PBMC collection
Blood was sampled from a retro-orbital sinus of mice. It was treated with ammonium–chloride–potassium (ACK) lysing buffer for 2 min twice to lyse red blood cells.
Human samples
Patients admitted to Kyoto University Hospital for cancer surgeries consented to the collection of a part of resected tumors. Obtained samples were minced into 1–2-mm pieces with scissors and digested with Dri Tumor & Tissue Dissociation Reagent (BD Biosciences) using a gentleMACS Dissociator. For CD45+ cell enrichment, tumor digestions were processed by Manual MACS Magnetic Separators (Miltenyi Biotec) according to the manufacturer’s instructions. All human samples were obtained from participants who provided informed consent in accordance with the Declaration of Helsinki and with approval from the Ethics Committee of Kyoto University (G1012).
Adoptive transfer using mouse OT-1 naive CD8+ T cells
MC38-OVA cells (2 × 10⁶) were injected intradermally into the right flank of CD45.1 congenic male and female mice on day 0. Male and female mice were equally distributed across groups. On day 9, naive (CD44low) OT-1 CD8⁺ T cells (CD45.2⁺) were isolated from spleens and LNs using MojoSort system. Isolated cells (1.25 × 106 cells per mouse) were intravenously transferred into the tumor-bearing mice. The isolated cells were also analyzed by flow cytometry before transfer. Tumors were collected on days 13, 16 and 20 after tumor inoculation (days 4, 7 and 11 after transfer) for flow cytometric analysis.
In vitro exhaustion by repetitive stimulation
Naive CD8+ T cells were resuspended at a concentration of 1 × 106 cells per ml in RPMI with 10% FCS, 1% penicillin–streptomycin, 50 µM 2-mercaptoethanol and 10 ng ml−1 IL-2. They were seeded on a flat plate coated with 1 µg ml−1 anti-CD3 (eBioscience) and 1 µg ml−1 anti-CD28 (eBioscience). The plate was kept in a 37 °C CO2 incubator. Cells were passaged at 1 × 106 cells per ml and re-stimulated every 2 days. From day 2, plates were coated with only anti-CD3, in the absence of anti-CD28. As necessary, acrolein or NBHA was added at a concentration of 2 µM (until day 4) or 10 µM (from day 4). The cells were analyzed on either day 0, 2, 6, 8 or 10.
Thymidine incorporation
A total of 2 × 105 CD8+ cells were stimulated with anti-CD3- and anti-CD28-coated Dynabeads (Gibco) in 200 µl of RPMI with 10% FCS, 1% penicillin–streptomycin, 25 µM 2-mercaptoethanol and 12.3 kBq of 3H thymidine, supplied with 5 µM acrolein or 1 µM 4-hydroxynonenal, in a 37 °C CO2 incubator for 7 h. These cells were collected by Filtermate 196 (Packard) and irradiation of 3H thymidine was measured by MicroBeta2 (PerkinElmer). Thymidine incorporation assay was performed at Radioisotope Research Center, Agency for Health, Safety and Environment, Kyoto University.
Flow cytometry
The following antibodies recognizing murine surface antigens were used: CD8a (53-6.7), CD45 (30-F11), CD45.2 (104), CD11b (M1/70), PD-1 (29F.1A12), PD-1 (RMP1-30), Tim-3 (RMT3-23), LAG-3 (C9B7W), TIGIT (1G9), CD38 (90), CD73 (TY/11.8), CD44 (IM7) and CD36 (HM36) from BioLegend; CD8a (53-6.7), CD45 (30-F11), CD45.2 (104), Tim-3 (5D12/TIM-3), TCR β chain (H57-597), PD-1 (29F.1A12) and Slamf6 (13G3) from BD Biosciences; and CD45.1 (A20), PD-1 (RMP1-30), CD39 (24DMS1), CD62L (MEL-14), CD44 (IM7) and CD8a (53-6.7) from eBioscience. The following antibodies recognizing murine intracellular targets, corresponding isotype controls and secondary antibodies for the nonfluorescent primary antibodies were used: IFNγ (XMG1.2), TNF (MP6-XT22) and Granzyme B (GB11) from BioLegend; Phospho-Akt (SDRNR), Phospho-mTOR (MRRBY), Phospho-S6 (cupk43k) and Perforin (eBioOMAK-D) from eBioscience; TOX (REA473) from Miltenyi Biotec; acrolein (10A10, used for protein-conjugated acrolein (PC-Acro)) and malondialdehyde (6H6, used for protein-conjugated malondialdehyde (PC-MDA)) from Novus Biologicals; HADHA (Polyclonal), SLC25A20 (Polyclonal) from Proteintech; Glut1 (EPR3915), HADHA (EPR17940), anti-rabbit IgG H&L (Polyclonal) and rabbit IgG isotype control (EPR25A) from Abcam; 4-hydroxynonenal (polyclonal, used for protein-conjugated 4-hydroxynonenal (PC-4-HNE)) from Bioss; Phospho-Akt (Polyclonal) from Cell Signaling Technology; and anti-rabbit IgG H + L (polyclonal) from Invitrogen. The following antibodies recognizing human surface antigens were used: CD45 (HI30), CD11b (ICRF44) and CD8 (SK1) from BioLegend; CD8 (RPA-T8), CD45 (HI30), PD-1 (MIH4) and PD-1 (EH12.1) from BD Biosciences. The following antibodies recognizing human intracellular targets and a secondary antibody for the nonfluorescent primary antibody were used: Phospho-Akt (SDRNR), Phospho-mTOR (MRRBY) and Phospho-S6 (cupk43k) from eBioscience; acrolein (10A10, used for protein-conjugated acrolein (PC-Acro)) from Novus Biologicals; HADHA (Polyclonal) from Proteintech; Glut1 (EPR3915) and anti-rabbit IgG H&L (Polyclonal) from Abcam. Mitochondrial potential, mitochondrial mass and mitochondrial superoxide were determined by 1.2 nM MitoTracker Deep Red, 15.6 nM MitoTracker Green and 5 µM MitoSOX Red, respectively (all Invitrogen). Lipid radical and acrolein were determined by 1 µM LipiRADICAL Green and 0.5 µM AcroleinRED, respectively (both Funakoshi). Glucose uptake was determined by 100 μg ml−1 2-NBDG (Abcam). Mitochondrial membrane peroxidation was determined by 0.5 μM MitoPeDPP (Dojindo). Fatty acid uptake of C5, C12 and C16 was determined by 1 μM BODIPY FL C5, 1 μM BODIPY FL C12 and 1 μM BODIPY FL C16, respectively (all Invitrogen). Dead cells were excluded by Zombie NIR, Zombie Violet or Zombie Aqua (all BioLegend). Then, 2 × 10⁵ to 1 × 106 cells were used for staining. For MitoTracker Deep Red, MitoTracker Green or MitoSOX Red staining, cells were incubated with appropriately diluted dyes in 2% FCS in PBS for 30 min in a 37 °C CO2 incubator. After incubation, cells were washed twice with 2% FCS in PBS. For staining with LipiRADICAL Green, AcroleinRED, 2-NBDG, BODIPY or MitoPeDPP, cells were washed twice with 1% BSA in PBS, then stained with appropriately diluted dyes in 1% BSA in PBS for 30 min in a 37 °C CO2 incubator. After incubation, cells were washed twice with 1% BSA in PBS. Subsequently, cells were stained with appropriately diluted Zombie dye and Fc-blocked by anti-CD16/32 antibody (BioLegend or BD Biosciences) for mice or by normal mouse serum (Invitrogen) for humans, and then stained with appropriately diluted antibodies for 12 min at 4 °C and washed with 2% FCS in PBS. To measure apoptosis, cells were stained with PE-Cy7 conjugated annexin V (BioLegend, cat. no. 640950) diluted in annexin V Binding Buffer (BioLegend) for 15 min at 4 °C, and then washed with annexin V Binding Buffer. To measure cytokine production, cells were stimulated with 50 ng ml−1 phorbol 12-myristate 13-acetate (PMA) (Selleck) and 500 ng ml−1 ionomycin calcium (Cosmo Bio) in the presence of brefeldin A Solution (eBioscience) for 4 h in a 37 °C CO2 incubator before starting staining. For intracellular staining, cells were processed with Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions, and subsequently stained with antibodies appropriately diluted with permeabilization buffer (eBioscience) for 30 min at 4 °C, and then washed with 2% FCS in PBS. For further detection of unconjugated intracellular primary antibodies, cells were stained with a fluorophore-conjugated secondary antibody diluted in permeabilization buffer for 15 min at 4 °C and then washed with 2% FCS in PBS. All flow cytometry experiments were performed on a LSRFortessa X-20 or a FACSymphony A5 (both BD Biosciences) and analyzed using FlowJo software (v.10.7.2 or v.10.10.0). Please refer to the reporting summary for detailed information about the antibodies.
Cell sorting of CD8+ TIL subsets by flow cytometry
Murine tumor samples were digested as described above, and CD8+ TILs were enriched by Manual MACS Magnetic Separators. Enriched cells were stained with Zombie dye, Fc-blocked by anti-CD16/32 antibody and stained with antibodies. The following antibodies were used: CD8a (53-6.7), CD45.2 (104), PD-1 (29 F.1A12) from BioLegend; CD45.2 (104) from BD Biosciences; PD-1 (RMP1-30) from eBioscience. If D1, D2 and D3 CD8+ TIL subsets were needed, cells were stained with MitoTracker Deep Red and MitoTracker Green for 30 min in a 37 °C CO2 incubator before Zombie staining. Among these stained cells, target CD8+ T cell subsets were sorted by FACSMelody (BD Biosciences). Please refer to the reporting summary for detailed information about the antibodies.
Flow cytometric analysis of CD8⁺ TIL proliferation
To assess the proliferative capacity of mouse CD8+ TIL subsets sorted by flow cytometry, they were labeled with 5 μM CellTrace Violet (Invitrogen) in PBS following the manufacturer’s instructions. Labeled cells were stimulated with anti-CD3/CD28 coated Dynabeads for 2 days in RPMI with 10% FCS, 1% penicillin–streptomycin, 50 μM 2-mercaptoethanol, 1 mM sodium pyruvate (Gibco) and 1% Non-Essential Amino Acids (Gibco). Cells were collected, stained with Zombie NIR, Fc-blocked by anti-CD16/32 antibody and stained with surface antibodies, which were identical to those used for the previous sorting. The fluorescence of stained cells was measured by LSRFortessa X-20. Highly proliferative cells were defined as CellTracelow cells among live and singlet CD8⁺ TILs. An unstimulated sample was also measured to assess baseline CellTrace dye intensity.
Immunofluorescence imaging
For immunostaining, isolated cells were incubated with 0.3 µM PhenoVue 551 (PerkinElmer) and 5.5 µg ml−1 Hoechst 33342 (Invitrogen) in 2% FCS in PBS for 30 min in a 37 °C CO2 incubator. After washing twice with 2% FCS in PBS, cells were processed with Foxp3/Transcription Factor Staining Buffer Set, and subsequently incubated with APC conjugated acrolein antibody (Novus Biologicals) appropriately diluted with permeabilization buffer (eBioscience) at 4 °C for 30 min. Following incubation, cells were washed with 2% FCS in PBS. Immunofluorescence images were acquired and analyzed using a Nikon N-SIMS super-resolution fluorescence microscope system. Please refer to the reporting summary for detailed information about antibodies.
Immunoblotting
Cultured CD8+ T cells were lysed in RIPA buffer (150 mM NaCl, 5 mM EDTA, 30 mM Tris-HCl (pH 7.4), 0.05% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate and 10% glycerol in distilled H2O) supplemented with a phosphatase inhibitor cocktail (Nacalai Tesque) and protease inhibitor cocktail (Roche) for 30 min at 4 °C. They were subsequently centrifuged and the supernatants containing the soluble protein fractions were collected. The protein amount was determined by DC protein assay (Bio Rad) and the cell lysates were diluted to appropriate protein concentration. The lysates were denatured with sample buffer with reducing reagent (Nacalai Tesque) and incubated at 95 °C for 5 min. The proteins were resolved by a 10% polyacrylamide gel and subjected to immunoblotting assay. The transblotted membranes were blocked with a Blocking One (Nacalai Tesque) and incubated with a primary antibody followed by a secondary antibody as below: antibody to hexokinase Ⅱ (C64G5), β-actin (13E5), HRP-linked anti-rabbit IgG secondary antibody (polyclonal) and HRP-linked anti-mouse IgG secondary antibody (polyclonal) from Cell Signaling Technology; antibody to HADHA (polyclonal), SLC25A20 (polyclonal) and HRP-linked GAPDH antibody (1E6D9) from Proteintech; antibody to CPT1A (8F6AE9) and ACADVL/VLCAD (polyclonal) from Abcam. The HRP-linked antibodies were diluted with 5% skim milk in Tris buffered saline with Tween 20 (TBS-T), and the others were diluted with 5% BSA in TBS-T supplemented with 0.02% sodium azide. The indicated antibody-incubated membranes were reacted with a Chemi-Lumi One Super (Nacalai Tesque) and the images were acquired with Amersham ImageQuant 800 (Cytiva). Please refer to the reporting summary for detailed information about antibodies.
Transmission electron microscopy
For TEM observation, cells were embedded in iPGell (GenoStaff) following the manufacturer’s protocol. The resulting cell blocks were fixed with 4% formaldehyde and 2% glutaraldehyde in 0.1 M phosphate buffer (PB) (pH 7.4) overnight at 4 °C, then post-fixed with 1% osmium tetroxide in 0.1 M PB for 2 h. After dehydration using a graded ethanol series, the samples were embedded in epoxy resin (Luveak-812; Nacalai Tesque). Ultrathin sections (70-nm thickness) were prepared using an ultramicrotome (EM UC7; Leica). The sections were stained with uranyl acetate and lead citrate, and examined using a transmission electron microscope (H-7650; Hitachi). TEM was performed at the Division of Electron Microscopic Study, Center for Anatomical Studies, Graduate School of Medicine, Kyoto University. Cristae distance was calculated by using open-source software ImageJ (v.1.53 m).
Enrichment of phosphopeptides and mass spectrometry
Cultured CD8+ T cells (n = 5 technical replicates) were lysed in 150 µl of a buffer containing 6 M guanidine-HCl, 100 mM HEPES-NaOH (pH 7.5), 10 mM Tris (2-carboxyethyl) phosphine hydrochloride and 40 mM chloroacetamide. The lysates were solubilized by heating and sonication, followed by centrifugation at 20,000g for 15 min at 4 °C. The supernatants were collected and 100 µg of total protein from each sample were purified by methanol–chloroform precipitation and resuspended in 100 µl of 0.1% RapiGest (Waters) in 50 mM triethylammonium bicarbonate solution. Following sonication, the protein solutions were digested with 2 µg of trypsin/Lys-C mix (Promega) at 37 °C overnight. The resulting peptide solutions were acidified with trifluoroacetic acid, centrifuged, and subjected to the High-Select Fe-NTA Phosphopeptide Enrichment kit (Thermo Fisher Scientific). Eluates were acidified, desalted using GL-Tip SDB (GL Sciences), evaporated in a SpeedVac concentrator (Thermo Fisher Scientific) and reconstituted in 0.1% trifluoroacetic acid and 3% acetonitrile. LC–MS/MS analysis of the resultant peptides was conducted using a nanoElute 2 coupled with a timsTOF HT mass spectrometer (Bruker). Peptides were separated on a C18 reversed-phase column (75-μm inner diameter × 150 mm, Nikkyo Technos). The mobile phase consisted of solvent A (0.1% formic acid in water) and solvent B (0.1% formic acid in acetonitrile). Samples were loaded onto the column at a flow rate of 0.2 µl min−1 starting at 3% B, which was linearly ramped to 32% B over 90 min, then raised to 95% B at 91 min and maintained at that level until 101 min. The mass spectrometer was set to operate in data-dependent acquisition mode using parallel accumulation–serial fragmentation (DDA-PASEF). The m/z range for both MS1 and MS2 spectra was 100–1,700 and the ion mobility range was 0.6–1.6 V s−1 cm−3. The ramp time was 100 ms, with a duty cycle of 100%. Each acquisition cycle consisted of ten PASEF MS2 scans. A polygon filter was applied in the m/z and ion mobility space to exclude low m/z, singly charged ions from precursor selection. The raw data were processed using the FragPipe (v.22.0). Database searches were performed using the MSFragger (v.4.1), using the default parameters of the LFQ-phospho workflow against the UniProt mouse database. Carbamidomethylation of cysteine (+57.0215 Da) was set as a fixed modification. The following variable modifications were included: acetylation of protein N terminus (+42.0106 Da); oxidation of methionine (+15.9949 Da); phosphorylation (+79.9663 Da) of serine, threonine or tyrosine. The resulting identifications were filtered using Philosopher with default parameters (MS Booster was disabled) and IonQuant (v.1.10.27) was employed for quantification with default software settings.
Phosphopeptide data analysis
The processed data were analyzed using FragPipe-Analyst68. In this analysis, first contaminants were filtered out. In addition, peptides that were not identified/quantified consistently in same condition have been removed. The quantification values of intensity were converted to log2 scale and biological replicates were grouped by experimental condition. Peptide-wise linear models combined with empirical Bayes statistics were used for the differential expression analyses. The limma package from R Bioconductor was used to generate a list of differentially expressed proteins for each pair-wise comparison. To maintain high data quality and specifically assess phosphorylation changes, phosphopeptides with N-terminal acetylation (+42.0106 Da) were excluded from the analysis. For IPA software (v.01-22-01), identified molecules with log2 fold change (ACR/Ctrl) values >0.5 or <–0.5 were analyzed for canonical pathway analysis.
Metabolic trace analysis of 13C-labeled glucose and palmitate
To assess glycolysis activity, isolated CD8+ T cell subsets (2 ×103 cells per well) were incubated in glucose-free RPMI medium (Gibco) supplemented with 10 mM D-glucose-13C6 (Isotec) for 3 h in a 37 °C CO2 incubator. After incubation, cells were lysed by 80% methanol and mixed by vortex mixer. The lysed solution was then dried by centrifugal evaporation. The dried sample was reconstituted with 50% methanol and subjected to LC–MS analysis. To assess FAO activity, isolated cell subsets (3 × 103 cells per well) were incubated in glucose-free RPMI medium supplemented with 2.5 mM glucose, 0.75 mM carnitine (Sigma-Aldrich) and 200 μM BSA-conjugated palmitate-13C16 (palmitic acid-13C16 (Isotec) bound to BSA (molar ratio of fatty acid to BSA, 5:1)) for 3 h in a 37 °C CO2 incubator. After incubation, cells were lysed with methanol containing stable isotope-labeled acylcarnitine standards (palmitoylcarnitine-d3, myristoylcarnitine-d9, lauroylcarnitine-d3 and octanoylcarnitine-d3; Cambridge Isotope Laboratories). The lysed solution was then dried by centrifugal evaporation. The dried sample was reconstituted with 50% methanol and subjected to LC–MS analysis. The levels of each metabolite were either presented as peak area, or assessed by comparing the peak area ratio, calculated by dividing the peak area of each metabolite by that of the internal standard, and then converted to the absolute amount (fmol).
Acrolein quantification in tumor tissues
For free acrolein detection in tissue, 2,4-dinitrophenylhydrazine (DNPH) (Tokyo Chemical Industry) derivatization was applied. Then, a 450-μl solution of 80% acetonitrile containing 1% formic acid and 1 mg ml−1 of DNPH was added to 50 mg of tissue. The tissue was homogenized three times at 4,000 rpm for 30 s using a bead homogenizer (Micro Smash 100R, TOMY) with 8–10 zirconia beads (1-mm diameter). For the acrolein standard solution, acrolein (AccuStandard) was prepared at concentrations of 0.4, 1, 2, 4, 10 and 20 μg ml−1 and 50 μl was mixed with 450 μl of the same reaction mixture to make acrolein standard solutions. The homogenized tissues or acrolein standard solutions were then incubated for 2 h at 37 °C to complete DNPH derivatization. After centrifugation at 16,000g for 30 min at 4 °C, 50 μl of supernatant was transferred to a glass vial and subjected to LC–MS analysis. Acrolein concentration was quantified by the external standard method. The correlation coefficient of the standard curve was higher than 0.99. Tumor acrolein concentrations were calculated assuming a tumor density to be 1 g ml−1, based on reference data from murine solid tumor models69.
LC–MS analysis
All data acquisition was performed on the LC–MS system consisting of a Nexera UHPLC and LC–MS-8060 (Shimadzu) with the electrospray ionization and selected reaction monitoring mode.
LC separation was conducted on a Shim-pack GIST C18-AQ column (3 μm, 150 mm × 2.1 mm internal diameter; Shimadzu). The mobile phase consisted of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). For palmitate metabolite analysis, the LC gradient was modified to 0 to 3 min, 35% B; 3 to 15 min, linear gradient to 95% B; 15 to 20 min, 95% B; 20.1 min, 95% B; and hold for 4 min. For DNPH-derivatized acrolein, the LC gradient was changed to 0 min, 20% B; 0 to 15 min, linear gradient to 95% B; 15 to 16 min, 95% B; 16.1 min, 20% B; and hold for 4 min.
A Shim-pack Mix-HILIC column (5 μm, 150 mm × 2.1 mm internal diameter; Shimadzu) was adapted, and the mobile phase consisted of 40 mM ammonium bicarbonate in water (pH 9.7) (A) and acetonitrile (B) for glucose metabolite analysis. The gradient program was as follows: 0 to 0.5 min, 95% B; 0.5 to 15.5 min, linear gradient to 40% B; 15.5 to 16.5 min, linear gradient to 0% B; 16.5 to 26.5 min, 0% B; 26.5 to 27.5 min, linear gradient to 95% B; hold for 7.5 min; flow rate, 0.4 ml min−1. The ion transitions (precursor–product ion) for DNPH-derivatized acrolein was m/z 235.05 to 158.30. The targeted metabolites for glycolysis and FAO activity assay are summarized in the Supplementary Table 4.
Single-cell RNA sequencing
Tumor samples from MC38 tumor-bearing mice (n = 20 mice) on day 14 were pooled and digested. CD8+ TILs were enriched by Manual MACS Magnetic Separators. They were subsequently stained with appropriately diluted MitoTracker Deep Red and MitoTracker Green for 30 min in a 37 °C CO2 incubator. After washing with 2% FCS in PBS twice, cells were stained with Zombie Violet, Fc-blocked by anti-CD16/32 antibody and stained for 12 min at 4 °C with appropriately diluted fluorescent-labeled and oligonucleotide-conjugated antibodies (including oligonucleotide-conjugated isotype controls) targeting cell surface antigens as follows: PE CD8a (clone 53-6.7, BioLegend), PE-Cy7 PD-1 (clone 29F.1A12, BioLegend) and BV786 CD45.2 (clone 104, BD Biosciences), along with a panel of oligonucleotide-conjugated antibodies (BioLegend), including CD279 (PD-1, clone RMP1-30), Ly108 (Slamf6, clone 330-AJ), CD226 (clone 10E5), KLRG1 (clone 2F1/KLRG1), CD122 (clone 5H4), CD366 (Tim-3, clone RMT3-23), CX3CR1 (clone SA011F11), TIGIT (clone 1G9), CD39 (clone Duha59), CD49d (clone R1-2), CD183 (clone CXCR3-173), CD185 (clone L138D7), CD103 (clone 2E7), CD107a (clone 1D4B), CD95 (clone SA367H8), CD127 (clone A7R34), CD44 (clone IM7), CD62L (clone MEL-14), CD278 (clone 7E.17G9) and isotype controls (Rat IgG2a κ, clone RTK2758; Rat IgG2b κ, clone RTK4530; Mouse IgG1 κ, clone MOPC-21; Mouse IgG2a κ, clone MOPC-173; Armenian Hamster IgG, clone HTK888). The oligonucleotide-conjugated antibodies were included during sample preparation; however, antibody capture-derived signals from these antibodies were not used for analysis in this study. Corresponding oligonucleotide sequences and feature types are available in the Gene Expression Omnibus submission. Cells were washed with 2% FCS in PBS twice, filtered through a 40-μm cell strainer before sorting. Viable single PD-1−CD45.2⁺CD8a⁺ cells and D1, D2 and D3 subsets of PD-1⁺CD45.2⁺CD8a⁺ cells were sorted using FACSMelody. To identify each subset in scRNA-seq, the sorted cells’ Fc receptor was blocked by anti-CD16/32 antibody (BioLegend) and stained with oligonucleotide-labeled Hashtag 1 antibody for PD-1⁻ population, Hashtag 4 antibody for D1, Hashtag 5 antibody for D2 and Hashtag 6 antibody for D3 (all Hashtag antibodies clone M1/42; 30-F11, BioLegend). After incubation for 30 min at 4 °C, each cell subsets were washed three times with 2% FCS in PBS and pooled. A total of 10,000 pooled cells were loaded into a Chromium controller (10x Genomics). cDNA and library preparation were performed according to the manufacturer’s indications using Chromium Next GEM Single Cell 5’ Reagent Kits v2 (Dual Index) (10x Genomics), and the library was sequenced by a NextSeq 2000 sequencer (Illumina). Please refer to the reporting summary for detailed information about antibodies.
ScRNA-seq data analysis
The sequencing reads were aligned to the GRCm39-2024-A reference genome for 5′ gene expression sequencing data and to vdj_GRCm38_alts_ensembl-7.0.0 for V(D)J data, and assembled using the ‘cellranger multi’ function in Cell Ranger (v.9.0.1, 10x Genomics)70. The resulting raw barcodes, features, and matrix files were loaded into R (v.4.4.0) and processed using Seurat (v.5.2.1)71, along with Hashtag oligonucleotide (HTO) count data and V(D)J data. Low-quality cells were filtered out based on the following criteria: <200 detected genes, >4,000 detected genes and >5% mitochondrial genes. Samples were demultiplexed based on HTO counts using the ‘HTODemux’ function. Cells identified as doublets (containing two Hashtags) or without a Hashtag were removed, and only cells from D1–D3 were retained for further analysis. The D1–D3 singlet data were normalized and log-transformed to mitigate the effects of varying sequencing depths. Dimensional reduction and clustering were performed using principal component analysis and visualized with UMAP via Seurat functions. Gene signature scoring was conducted using the ‘AddModuleScore_UCell’ function in the R package UCell (v.2.8.0)72. Gene signatures for exhausted and stem-like CD8+ T cells were cited from Deak et al.73. Pseudotime was calculated, and trajectory visualization was performed using Monocle 3 (v.1.3.7)74 after transferring the Seurat object via SeuratWrappers (v.0.3.5). T cell clonotypes were analyzed using scRepertoire (v.2.0.7)75 based on V(D)J gene sequences. Differentially expressed genes in D1 versus D3 were identified using the ‘FindMarkers’ function in Seurat. Genes upregulated in D3 were selected based on the criteria: Padjust < 0.05 and log2 fold change (D3/D1) > 1. GO (Biological Process) analysis was performed using clusterProfiler (v.4.12.6)76,77, with annotation provided by org.Mm.eg.db (v.3.19.1). To reduce redundancy in GO terms, the ‘simplify’ function was applied.
Seahorse for ECAR, OCR and ATP
Cultured cells were seeded in an XFe96 plate (2–4 × 105 cells per well) and their ECAR and OCR were measured by the XFe96 Extracellular Flux Analyzer (Seahorse Bioscience) as previously described11. The Seahorse XF Glycolysis Stress Test kit, the Seahorse XF Cell Mito Stress Test kit, and the Seahorse XF Real-Time ATP Rate Assay kit (all Seahorse Bioscience) were used for measurement of ECAR, OCR and ATP production, respectively. For ECAR assays, XF DMEM Base Medium (Agilent Technologies) was used, supplemented with 2 mM glutamine (Agilent Technologies). To measure OCR derived from FAO or glutaminolysis activity, XF RPMI Base Medium (Agilent Technologies) was used, supplemented with 2.5 mM glucose (Agilent Technologies) on the day of the assay. Etomoxir (5 µM) or BPTES (3 µM) was added about 10 min before running the assay. For Real-Time ATP Rate Assay, XF DMEM Base Medium was used, supplemented with 1 mM pyruvate (Agilent Technologies), 2 mM glutamine (Agilent Technologies) and 10 mM glucose (Agilent Technologies). Mitochondrial and glycolytic ATP production rates were automatically calculated by the Seahorse machine operating software Wave Controller in accordance with the manufacturer’s instructions.
Statistical analysis
For phosphoproteomics data, FragPipe-Analyst (v.1.13) and IPA software (v.01-22-01) were used for statistical analysis. Results of IPA were visualized using GraphPad Prism (v.10.3.1 or v.10.4.1). Statistical analyses and visualization of scRNA-seq data were performed using R (v.4.4.0). To perform multiple-group analysis between D1–D3 or six distinct clusters of UMAP, one-way ANOVA was used. Statistical analyses of the other datasets were performed using Prism. For mice, to compare two groups, the two-tailed paired or unpaired Student’s t-test was used, and to perform multiple-group analysis, ANOVA was used. A P value < 0.05 was considered significant. The variations of data were evaluated as the mean or s.e.m. The data distribution was assumed to be normal but this was not formally tested. Three or more samples were thought to be appropriate for the sample size estimate for in vitro experiments, and five or more samples for in vivo experiments. No statistical methods were used to predetermine sample sizes. In some cases, cells were pooled from multiple mice in one experiment. Samples and animals were randomly chosen from the pool and treated. No blinding method was used for the treatment of samples and animals. To compare two groups for humans, a two-tailed Wilcoxon matched-pairs signed-rank test was used, and a P value < 0.05 was considered significant. Seven or more samples were thought to be appropriate for the sample size estimate.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
ScRNA-seq data generated in this study have been deposited in the Gene Expression Omnibus under accession code GSE307051. Source data are provided with this paper.
Code availability
Custom scripts supporting the data analysis were developed using standard computational tools and are available from the corresponding author upon reasonable request.
References
Daud, A. I. et al. Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J. Clin. Invest. 126, 3447–3452 (2016).
Philip, M. et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 (2017).
Koyama, S. et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. Nat. Commun. 7, 10501 (2016).
Miller, B. C. et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20, 326–336 (2019).
Blank, C. U. et al. Defining T cell exhaustion. Nat. Rev. Immunol. 19, 665–674 (2019).
Kallies, A., Zehn, D. & Utzschneider, D. T. Precursor exhausted T cells: key to successful immunotherapy? Nat. Rev. Immunol. 20, 128–136 (2020).
Gebhardt, T., Park, S. L. & Parish, I. A. Stem-like exhausted and memory CD8(+) T cells in cancer. Nat. Rev. Cancer 23, 780–798 (2023).
Yost, K. E. et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25, 1251–1259 (2019).
Liu, B. et al. Temporal single-cell tracing reveals clonal revival and expansion of precursor exhausted T cells during anti-PD-1 therapy in lung cancer. Nat. Cancer 3, 108–121 (2022).
Sukumar, M. et al. Inhibiting glycolytic metabolism enhances CD8+ T cell memory and antitumor function. J. Clin. Invest. 123, 4479–4488 (2013).
Chamoto, K. et al. Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T cell-dependent antitumor activity. Proc. Natl Acad. Sci. USA 114, E761–E770 (2017).
Chowdhury, P. S., Chamoto, K., Kumar, A. & Honjo, T. PPAR-induced fatty acid oxidation in T cells increases the number of tumor-reactive CD8(.) T cells and facilitates anti-PD-1 therapy. Cancer Immunol. Res. 6, 1375–1387 (2018).
Yu, Y. R. et al. Disturbed mitochondrial dynamics in CD8(+) TILs reinforce T cell exhaustion. Nat. Immunol. 21, 1540–1551 (2020).
Tanaka, K. et al. Combination bezafibrate and nivolumab treatment of patients with advanced non-small cell lung cancer. Sci. Transl. Med. 14, eabq0021 (2022).
Al-Habsi, M. et al. Spermidine activates mitochondrial trifunctional protein and improves antitumor immunity in mice. Science 378, eabj3510 (2022).
Geltink, R. I. K., Kyle, R. L. & Pearce, E. L. Unraveling the complex interplay between T cell metabolism and function. Annu. Rev. Immunol. 36, 461–488 (2018).
Rangel Rivera, G. O. et al. Fundamentals of T cell metabolism and strategies to enhance cancer immunotherapy. Front. Immunol. 12, 645242 (2021).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 374–388 (2016).
Vardhana, S. A. et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21, 1022–1033 (2020).
Scharping, N. E. et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 22, 205–215 (2021).
Patsoukis, N. et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 6, 6692 (2015).
Bengsch, B. et al. Bioenergetic insufficiencies due to metabolic alterations regulated by the inhibitory receptor PD-1 are an early driver of CD8(+) T cell exhaustion. Immunity 45, 358–373 (2016).
Scharping, N. E. et al. The tumor microenvironment represses T cell mitochondrial biogenesis to drive intratumoral T cell metabolic insufficiency and dysfunction. Immunity 45, 701–703 (2016).
Siska, P. J. et al. Mitochondrial dysregulation and glycolytic insufficiency functionally impair CD8 T cells infiltrating human renal cell carcinoma. JCI Insight 2, e93411 (2017).
Schurich, A. et al. Distinct metabolic requirements of exhausted and functional virus-specific CD8 T cells in the same host. Cell Rep. 16, 1243–1252 (2016).
Wu, H. et al. Mitochondrial dysfunction promotes the transition of precursor to terminally exhausted T cells through HIF-1α-mediated glycolytic reprogramming. Nat. Commun. 14, 6858 (2023).
Zhang, C. et al. STAT3 activation-induced fatty acid oxidation in CD8(+) T effector cells is critical for obesity-promoted breast tumor growth. Cell Metab. 31, 148–161 e145 (2020).
Pollizzi, K. N. et al. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J. Clin. Invest. 125, 2090–2108 (2015).
Cao, J. et al. Effects of altered glycolysis levels on CD8(+) T cell activation and function. Cell Death Dis. 14, 407 (2023).
Crompton, J. G. et al. Akt inhibition enhances expansion of potent tumor-specific lymphocytes with memory cell characteristics. Cancer Res. 75, 296–305 (2015).
Buck, M. D. et al. Mitochondrial dynamics controls T cell fate through metabolic programming. Cell 166, 63–76 (2016).
Zhang, Y. et al. Enhancing CD8(+) T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 32, 377–391 e379 (2017).
Luo, L. et al. Enhanced immune memory through a constant photothermal-metabolism regulation for cancer prevention and treatment. Biomaterials 270, 120678 (2021).
Manzo, T. et al. Accumulation of long-chain fatty acids in the tumor microenvironment drives dysfunction in intrapancreatic CD8+ T cells. J. Exp. Med. 217, e20191920 (2020).
Ma, X. et al. CD36-mediated ferroptosis dampens intratumoral CD8(+) T cell effector function and impairs their antitumor ability. Cell Metab. 33, 1001–1012 e1005 (2021).
Xu, S. et al. Uptake of oxidized lipids by the scavenger receptor CD36 promotes lipid peroxidation and dysfunction in CD8(+) T cells in tumors. Immunity 54, 1561–1577 e1567 (2021).
Xiao, L. et al. IL-9/STAT3/fatty acid oxidation-mediated lipid peroxidation contributes to Tc9 cell longevity and enhanced antitumor activity. J. Clin. Invest. 132, e153247 (2022).
Okazaki, T., Chikuma, S., Iwai, Y., Fagarasan, S. & Honjo, T. A rheostat for immune responses: the unique properties of PD-1 and their advantages for clinical application. Nat. Immunol. 14, 1212–1218 (2013).
Hui, E. et al. T cell costimulatory receptor CD28 is a primary target for PD-1-mediated inhibition. Science 355, 1428–1433 (2017).
Parry, R. V. et al. CTLA-4 and PD-1 receptors inhibit T-cell activation by distinct mechanisms. Mol. Cell. Biol. 25, 9543–9553 (2005).
Belk, J. A. et al. Genome-wide CRISPR screens of T cell exhaustion identify chromatin remodeling factors that limit T cell persistence. Cancer Cell 40, 768–786 e767 (2022).
Rieger, A. M., Nelson, K. L., Konowalchuk, J. D. & Barreda, D. R. Modified annexin V/propidium iodide apoptosis assay for accurate assessment of cell death. J. Vis. Exp. https://doi.org/10.3791/2597 (2011).
Lang, C. R. et al. Efferocytosis drives myeloid NLRP3 dependent inflammasome signaling secretion of IL-1β to promote tumor growth. Front. Immunol. 13, 993771 (2022).
Song, X., Zhao, G., Wang, G. & Gao, H. Heterogeneity and differentiation trajectories of infiltrating CD8+ T cells in lung adenocarcinoma. Cancers 14, 5183 (2022).
Zheng, L. et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 374, abe6474 (2021).
Qu, Q., Zeng, F., Liu, X., Wang, Q. J. & Deng, F. Fatty acid oxidation and carnitine palmitoyltransferase I: emerging therapeutic targets in cancer. Cell Death Dis. 7, e2226 (2016).
Tonazzi, A., Giangregorio, N., Console, L., Palmieri, F. & Indiveri, C. The mitochondrial carnitine acyl-carnitine carrier (SLC25A20): molecular mechanisms of transport, role in redox sensing and interaction with drugs. Biomolecules 11, 521 (2021).
Klein Geltink, R. I. et al. Mitochondrial priming by CD28. Cell 171, 385–397 e311 (2017).
Yoshida, M. et al. Acrolein toxicity: comparison with reactive oxygen species. Biochem. Biophys. Res. Commun. 378, 313–318 (2009).
Saiki, R. et al. Brain infarction correlates more closely with acrolein than with reactive oxygen species. Biochem. Biophys. Res. Commun. 404, 1044–1049 (2011).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015).
Menk, A. V. et al. Early TCR signaling induces rapid aerobic glycolysis enabling distinct acute T cell effector functions. Cell Rep. 22, 1509–1521 (2018).
Mano, J., Miyatake, F., Hiraoka, E. & Tamoi, M. Evaluation of the toxicity of stress-related aldehydes to photosynthesis in chloroplasts. Planta 230, 639–648 (2009).
Anderson, E. J., Katunga, L. A. & Willis, M. S. Mitochondria as a source and target of lipid peroxidation products in healthy and diseased heart. Clin. Exp. Pharm. Physiol. 39, 179–193 (2012).
Xu, X. et al. Autophagy is essential for effector CD8(+) T cell survival and memory formation. Nat. Immunol. 15, 1152–1161 (2014).
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141 (2011).
Li, F., Liu, H., Zhang, D., Ma, Y. & Zhu, B. Metabolic plasticity and regulation of T cell exhaustion. Immunology 167, 482–494 (2022).
Cai, H. J. et al. Downregulation of TCF1 in HIV infection impairs T-cell proliferative capacity by disrupting mitochondrial function. Front. Microbiol. 13, 880873 (2022).
Wu, W. et al. AMPK facilitates intestinal long-chain fatty acid uptake by manipulating CD36 expression and translocation. FASEB J. 34, 4852–4869 (2020).
Fisicaro, P. et al. Targeting mitochondrial dysfunction can restore antiviral activity of exhausted HBV-specific CD8 T cells in chronic hepatitis B. Nat. Med. 23, 327–336 (2017).
Biswas, M. S. & Mano, J. Lipid peroxide-derived reactive carbonyl species as mediators of oxidative stress and signaling. Front. Plant Sci. 12, 720867 (2021).
Shi, R., Rickett, T. & Sun, W. Acrolein-mediated injury in nervous system trauma and diseases. Mol. Nutr. Food Res. 55, 1320–1331 (2011).
Cai, J., Bhatnagar, A. & Pierce, W. M. Jr Protein modification by acrolein: formation and stability of cysteine adducts. Chem. Res. Toxicol. 22, 708–716 (2009).
Covey, T. M., Edes, K., Coombs, G. S., Virshup, D. M. & Fitzpatrick, F. A. Alkylation of the tumor suppressor PTEN activates Akt and β-catenin signaling: a mechanism linking inflammation and oxidative stress with cancer. PLoS ONE 5, e13545 (2010).
Tanigaki, K. et al. Regulation of αβ/γδ T cell lineage commitment and peripheral T cell responses by Notch/RBP-J signaling. Immunity 20, 611–622 (2004).
Kumar, A., Chamoto, K., Chowdhury, P. S. & Honjo, T. Tumors attenuating the mitochondrial activity in T cells escape from PD-1 blockade therapy. eLife 9, e52330 (2020).
Iwai, Y. et al. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl Acad. Sci. USA 99, 12293–12297 (2002).
Hsiao, Y. et al. Analysis and visualization of quantitative proteomics data using FragPipe-Analyst. J. Proteome Res. 23, 4303–4315 (2024).
Ng, C. E. & Inch, W. R. Comparison of the densities of clonogenic cells from EMT6 fibrosarcoma monolayer cultures, multicell spheroids, and solid tumors in Ficoll density gradients. J. Natl Cancer Inst. 60, 1017–1022 (1978).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Andreatta, M. & Carmona, S. J. UCell: robust and scalable single-cell gene signature scoring. Comput. Struct. Biotechnol. J. 19, 3796–3798 (2021).
Codarri Deak, L. et al. PD-1-cis IL-2R agonism yields better effectors from stem-like CD8(+) T cells. Nature 610, 161–172 (2022).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502 (2019).
Borcherding, N., Bormann, N. L. & Kraus, G. scRepertoire: an R-based toolkit for single-cell immune receptor analysis. F1000Res 9, 47 (2020).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16, 284–287 (2012).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2, 100141 (2021).
Acknowledgements
We sincerely thank members of the Department of Immunology and Genomic Medicine, Center for Cancer Immunotherapy and Immunobiology, Graduate School of Medicine, Kyoto University for sample preparation and discussion; S. Fagarasan and T. Carvalho for productive discussion; the staff of the Clinical Bio-Resource Center at Kyoto University Hospital for collecting the human samples; and the staff of the Division of Electron Microscopic Study, Center for Anatomical Studies, Graduate School of Medicine, Kyoto University, for preparing TEM slides. We are grateful to the Radioisotope Research Center, Agency for Health, Safety and Environment, Kyoto University, for supporting the experiments involving thymidine incorporation assays. This work was supported by the Japan Agency for Medical Research and Development under grant number JP25ama221330 (K.C.); JSPS KAKENHI under grant number JP23KJ1378 (K.K.); Yanai Fund (T. Honjo), Meiji Holdings Co. (T. Honjo and K.C.), Meiji Seika Pharma Co. (T. Honjo and K.C.) and Shimadzu Corporation (T. Honjo and K.C.). This manuscript was edited by Life Science Editors.
Author information
Authors and Affiliations
Contributions
Y.H., K.K. and K.C. designed research; Y.H., K.K., K. Ichimaru, J.W., K.S., A.M., S.H., Y.W., K.Y., M.K. and H.K. performed research; Y.H., K.K., K. Ichimaru, T. Hirano, J.W., S.H., Y.W., T. Kozuki, T.Y., and K.C. analyzed data; Y.H., K.K., K. Ichimaru, J.W., K. Ito, T.M., H.D., T. Kobayashi, K.O. and T.Y. collected human samples; and Y.H., K.K., T. Honjo and K.C. wrote the paper. All authors reviewed the results and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
T. Honjo and K.C. received research funding from Shimadzu Corporation. All other authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Navdeep Chandel, Ping-Chih Ho and the other anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Nick Bernard, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Surrogate markers and phenotypic characterization of D1–D4 populations.
(A) A representative gating strategy: FSC-A/SSC-A profiles and frequencies of FSC-Alow cells of D1–D4 populations among PD-1+ CD8+ TILs in MC38 tumors on day 14 (n = 6 mice). (B) MC38-OVA cells were injected intradermally into CD45.1 congenic male and female mice on day 0. Male and female mice were equally distributed across groups. On day 9 after tumor inoculation, isolated naïve (CD44low) OT-1 CD8⁺ T cells (CD45.2⁺) were intravenously transferred into the tumor-bearing CD45.1 recipient mice. Prior to transfer, the isolated naïve CD8⁺ T cells (CD45.2⁺) were also analyzed by flow cytometry (representative profiles shown). Tumor-infiltrating donor PD-1⁺ CD8⁺ T cells (CD45.2⁺) were analyzed by flow cytometry on days 13, 16, and 20 after tumor inoculation (that is, days 4, 7, and 11 after transfer). Representative flow cytometry profiles (male recipients) and frequencies of D1–D4 cells, based on mitochondrial mass and mitochondrial potential (all recipients, n = 6 mice). (C) Among PD-1+ CD8+ T cells in MC38 tumors, the FSC-Alow population is regarded as D4’ whereas the FSC-Ahigh population is divided into 3 populations (D1’ to D3’) according to their mitochondrial potential intensity. Distributions of D1’-D4’ populations are compared with D1–D4 gates. (D) Representative histogram and MFI of IFN-γ, TNF-α, granzyme B and perforin in D1’-D4’ populations among PD-1+ CD8+ T cells in MC38 tumors on day 14 (n = 6 mice). (E) Frequencies of CD38+, CD39+ and CD73+ cells in D1–D4 populations among PD-1+ CD8+ T cells in tumors on day 14 (n = 8 mice). (F) MFI of IFN-γ, TNF-α, granzyme B and perforin in D1’-D4’ populations among naïve CD8+ T cells on day 8 of repetitive stimulation (n = 5 technical replicates). (G) MFI of Annexin V, Tim-3 and LAG-3 in D1’-D4’ or D1–D4 populations among naïve CD8+ T cells on day 8 of repetitive stimulation (n = 5 technical replicates). (H) Based on the data shown in Fig. 1e, UMAP projection revealed six distinct clusters. (I) Violin plots showing exhausted or stem-like signature scores for each cluster. Data are means ± SEMs (A and B). P values were determined using paired two-tailed Student’s t tests (D to G) or one-way ANOVA with Tukey’s multiple comparisons tests (B and I). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results, except (H) and (I).
Extended Data Fig. 2 Active aldehyde accumulates in CD8+ T cells in an exhaustion depth-dependent manner in vitro and in vivo.
(A) Flow cytometry profile and MFI of lipid radical in Tim-3− and Tim-3+ populations among PD-1+ CD8+ T cells in tumors on day 14 (n = 8 mice). (B) Flow cytometry profile and MFI of acrolein or PC-Acro in Tim-3− and Tim-3+ populations among PD-1+ CD8+ T cells in tumors on day 14 (acrolein, n = 5 mice; PC-Acro, n = 7 mice). (C) MFI of protein-conjugated malondialdehyde (PC-MDA) and protein-conjugated 4-hydroxynonenal (PC-4-HNE) in Tim-3− and Tim-3+ populations among PD-1+ CD8+ T cells in tumors on day 14 (n = 7 mice) and on day 17 (n = 8 mice), respectively. (D) MFI of lipid radical, acrolein and PC-Acro in D1’-D4’ populations among naïve CD8+ T cells on day 8 of repetitive stimulation (n = 5 technical replicates). (E) MFI of acrolein in naïve-derived CD8+ T cells on day 0, 2, 6 and 10 of repetitive stimulation (n = 5 technical replicates). P values were determined using paired two-tailed Student’s t tests (A to D) or one-way ANOVA with Tukey’s multiple comparisons tests (E). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of three or more independent experiments with similar results.
Extended Data Fig. 3 Active aldehydes, but not hydrogen peroxide (H2O2), enhance glycolysis and proliferation of CD8+ T cells.
(A) Acrolein concentration (μM) in MC38 tumor tissue on day 14 (n = 6 mice). An experimental control group was not included, as the experiment aimed to determine the physiological acrolein levels in tumor tissues. (B) Frequencies of D1–D4 populations among CD8+ T cells 21 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 2 μM acrolein (ACR) (n = 5 technical replicates). (C) MFI of pAkt, pmTOR and pS6 in naïve-derived CD8+ T cells on day 8 of repetitive stimulation in the presence or absence of ACR (n = 5 technical replicates). (D) Frequencies of PD-1− Tim-3− and PD-1+ Tim-3+ cells among naïve-derived CD8+ T cells on day 8 of repetitive stimulation in the presence or absence of ACR (n = 5 technical replicates). (E) Western blotting images of enzymes associated with glycolysis and FAO in CD8+ T cells stimulated with anti-CD3/CD28 beads for 3 hours in the presence or absence of 5 μM ACR. (F) Seahorse measurement of real-time ECAR of naïve CD8+ T cells 3 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 0.5 μM 4-hydroxynonenal (4-HNE) (n = 4 technical replicates). (G) Thymidine incorporation of CD8+ T cells 7 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 0.5 μM 4-HNE (n = 4 technical replicates). The control data are shared with those in Fig. 3f to compare results under the same conditions in the same experiment, as supportive data. (H) Seahorse measurement of real-time ECAR of CD8+ T cells 3 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 120 μM H2O2 (n = 4 technical replicates). (I) Frequencies of D1–D4 populations among CD8+ T cells 7 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 120 μM H2O2 (n = 6 technical replicates). (J) Frequencies of D1–D4 populations among CD8+ T cells 14 hours after stimulation with anti-CD3/CD28 beads in the presence or absence of 60 μM H2O2 (n = 5 technical replicates). Data are means ± SEMs. P values were determined using unpaired two-tailed Student’s t tests. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. All data are representative of two or more independent experiments with similar results.
Extended Data Fig. 4 Suppressed T cells exhaustion by acrolein scavengers and expression level of Aldh2 in D1–D3.
(A) Frequencies of PD-1− Tim-3− and PD-1+ Tim-3+ cells among naïve-derived CD8+ T cells on day 8 of repetitive stimulation in the presence or absence of N-benzylhydroxylamine (NBHA) (n = 5 technical replicates). The control data are shared with those in Extended Data Fig. 3d to compare results under the same conditions in the same experiment, as supportive data. (B) Expression level of Aldh2 in D1–D3 of PD-1+ CD8+ T cells sorted from MC38 tumors on day 14, evaluated by scRNA-seq. Data are means ± SEMs (A). P values were determined using unpaired two-tailed Student’s t tests (A). ****P < 0.0001. Data are representative of two or more independent experiments with similar results, except (B).
Extended Data Fig. 5 Unnecessary fatty acid uptake boosts FAO dysfunction in TME CD8+ T cells.
(A) MFI of lipid radical, acrolein and PC-Acro in naïve (CD62Lhigh CD44low) CD8+ T cells in PBMCs from Hadhaflox/flox (Ctrl) and Hadhaflox/flox Cd4-Cre (KO) male mice (Ctrl, n = 7 mice; KO, n = 5 mice). (B) MFI of pAkt, pS6 and 2-NBDG in naïve CD8+ T cells in PBMCs from Ctrl and KO male mice (Ctrl, n = 7 mice; KO, n = 5 mice). (C) CD36 expression and BODIPY C5, C12, and C16 (fatty acid uptake) in naïve CD8+ T cells in PBMCs from Ctrl and KO male mice. (Ctrl, n = 7 mice; KO, n = 5 mice). (D) BODIPY C5, C12, and C16 in Tim-3− and Tim-3+ populations among PD-1+ CD8+ T cells in MC38 tumors on day 14 (n = 5 mice). (E) The positive loop toward intensive oxidative stress by FAO dysfunction in CD8+ T cells in TME. Acrolein production following lipid peroxidation damages mitochondria and reduces the FAO activity. FAO dysregulation promotes fatty acid uptake and drives further accumulation of lipid peroxidation and acrolein. Created with BioRender.com, elements adapted and assembled using PowerPoint. Data are means ± SEMs (A to C). P values were determined using unpaired two-tailed Student’s t tests (A to C) or paired two-tailed Student’s t tests (D). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. Data are representative of two independent experiments with similar results.
Extended Data Fig. 6 Acrolein scavenger attenuates Akt/mTOR/S6 pathway of CD8+ T cells in TME.
MFI of pAkt, pmTOR, pS6 and Glut1 in PD-1+ CD8+ T cells in MC38 tumors on day 10 from WT female mice treated with anti-PD-L1 mAb and N-benzylhydroxylamine (NBHA) as indicated in Fig. 7b (Ctrl, n = 8 mice; the other groups, n = 6 mice). Data are means ± SEMs. P values were determined using unpaired two-tailed Student’s t tests. *P < 0.05. Data are representative of two independent experiments with similar results.
Extended Data Fig. 7 Vicious loop toward metabolic exhaustion by active aldehyde accumulation in the process of CD8+ T cell exhaustion in TME.
(A) In CD8+ T cells of TME, lipid peroxidation gradually proceeds due to mitochondrial ROS production. Lipid peroxidation generates active aldehydes from mitochondria, attenuating the mitochondrial FAO function. A positive loop of active aldehydes accumulation is then established as shown in Extended Data Fig. 5e. (B) Active aldehydes boost the glycolysis pathway forcedly even if the cell expresses PD-1. (C) Active aldehyde accumulation attenuates FAO and boosts glycolysis, shaping the metabolic exhaustion state. Created with BioRender.com, elements adapted and assembled using PowerPoint.
Supplementary information
Supplementary Table 1
Genes upregulated in D3 compared to D1 (GO analysis).
Supplementary Table 2
Phosphoproteomics data comparing ACR and Ctrl groups.
Supplementary Table 3
Canonical pathway analysis of IPA.
Supplementary Table 4
The targeted metabolite for metabolic trace analysis of 13C-labeled glucose and palmitate.
Source data
Source Data Fig. 3
Uncropped immunoblot data for Fig. 3k.
Source Data Extended Data Fig. 3
Uncropped immunoblot data for Extended Data Fig. 3e.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Haku, Y., Kitaoka, K., Ichimaru, K. et al. Active aldehydes accelerate CD8+ T cell exhaustion by metabolic alteration in the tumor microenvironment. Nat Immunol 27, 281–294 (2026). https://doi.org/10.1038/s41590-025-02370-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-025-02370-w