Abstract
Most of what we know about adaptive immunity has come from inbred mouse studies, using methods that are often difficult or impossible to confirm in humans. In addition, vaccine responses in mice are often poorly predictive of responses to those same vaccines in humans. Here we use human tonsils, readily available lymphoid organs, to develop a functional organotypic system that recapitulates key germinal center features in vitro, including the production of antigen-specific antibodies, somatic hypermutation and affinity maturation, plasmablast differentiation and class-switch recombination. We use this system to define the essential cellular components necessary to produce an influenza vaccine response. We also show that it can be used to evaluate humoral immune responses to two priming antigens, rabies vaccine and an adenovirus-based severe acute respiratory syndrome coronavirus 2 vaccine, and to assess the effects of different adjuvants. This system should prove useful for studying critical mechanisms underlying adaptive immunity in much greater depth than previously possible and to rapidly test vaccine candidates and adjuvants in an entirely human system.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Requests for raw data, analyzed data and materials will be reviewed by Stanford University to determine if the request is subject to intellectual property or confidentiality obligations. Data and materials that can be shared will be released using a Material Transfer Agreement. Targeted single-cell RNA-seq data are available in the Gene Expression Omnibus under GSE158459.
References
Behring, E. Untersuchungen uber das Zustandekommen der Diphtherie-Immunitat bei Thieren. Dtsch. Med. Wschr 16, 1145–1147 (1890).
Behring, E. & Kitasato, S. Ueber das Zustandekommen der Diphtherie-Immunitat und der Tetanus-Immunitat bei Thieren. Dtsch. Med. Wschr 16, 1113–1114 (1890).
Kaufmann, S. H. E. Emil von Behring: translational medicine at the dawn of immunology. Nat. Rev. Immunol. 17, 341–343 (2017).
Kaufmann, S. H. Immunology’s foundation: the 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nat. Immunol. 9, 705–712 (2008).
Miller, J. F. The discovery of thymus function and of thymus-derived lymphocytes. Immunol. Rev. 185, 7–14 (2002).
Miller, J. F. & Mitchell, G. F. The thymus and the precursors of antigen reactive cells. Nature 216, 659–663 (1967).
Zinkernagel, R. M. & Doherty, P. C. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature 248, 701–702 (1974).
Allen, C. D., Okada, T. & Cyster, J. G. Germinal-center organization and cellular dynamics. Immunity 27, 190–202 (2007).
De Silva, N. S. & Klein, U. Dynamics of B cells in germinal centres. Nat. Rev. Immunol. 15, 137–148 (2015).
MacLennan, I. C. et al. Extrafollicular antibody responses. Immunol. Rev. 194, 8–18 (2003).
Victora, G. D. & Nussenzweig, M. C. Germinal centers. Annu. Rev. Immunol. 30, 429–457 (2012).
Itano, A. A. et al. Distinct dendritic cell populations sequentially present antigen to CD4 T cells and stimulate different aspects of cell-mediated immunity. Immunity 19, 47–57 (2003).
Radtke, A. J. et al. Lymph-node resident CD8α+ dendritic cells capture antigens from migratory malaria sporozoites and induce CD8+ T cell responses. PLoS Pathog. 11, e1004637 (2015).
McCloskey, M. L., Curotto de Lafaille, M. A., Carroll, M. C. & Erlebacher, A. Acquisition and presentation of follicular dendritic cell-bound antigen by lymph node-resident dendritic cells. J. Exp. Med. 208, 135–148 (2011).
Gerner, M. Y., Casey, K. A., Kastenmuller, W. & Germain, R. N. Dendritic cell and antigen dispersal landscapes regulate T cell immunity. J. Exp. Med. 214, 3105–3122 (2017).
Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542 (2014).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Cyster, J. G. & Allen, C. D. C. B cell responses: cell interaction dynamics and decisions. Cell 177, 524–540 (2019).
Qi, H., Kastenmuller, W. & Germain, R. N. Spatiotemporal basis of innate and adaptive immunity in secondary lymphoid tissue. Annu. Rev. Cell Dev. Biol. 30, 141–167 (2014).
Vinuesa, C. G., Linterman, M. A., Goodnow, C. C. & Randall, K. L. T cells and follicular dendritic cells in germinal center B cell formation and selection. Immunol. Rev. 237, 72–89 (2010).
Jameson, S. C. & Masopust, D. What is the predictive value of animal models for vaccine efficacy in humans? Reevaluating the potential of mouse models for the human immune system. Cold Spring Harb. Perspect. Biol. 10, a029132 (2018).
Buchbinder, S. P. et al. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372, 1881–1893 (2008).
Watkins, D. I., Burton, D. R., Kallas, E. G., Moore, J. P. & Koff, W. C. Nonhuman primate models and the failure of the Merck HIV-1 vaccine in humans. Nat. Med. 14, 617–621 (2008).
Tameris, M. D. et al. Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial. Lancet 381, 1021–1028 (2013).
James, K., Skibinski, G. & Hoffman, P. A comparison of the performance in vitro of precision cut tissue slices and suspensions of human spleen with special reference to immunoglobulin and cytokine production. Hum. Antibodies Hybrid. 7, 138–150 (1996).
Edwards, K. M., Snyder, P. N., Stephens, D. S. & Wright, P. F. Human adenoid organ culture: a model to study the interaction of influenza A with human nasopharyngeal mucosa. J. Infect. Dis. 153, 41–47 (1986).
Ferro, L. M., Weedon, H. M., Flego, L. R., Beroukas, D. & Zola, H. An organ fragment culture model to study lymphocyte activation in human lymphoid tissue. Immunobiology 188, 51–61 (1993).
Hoffmann, P., Skibinski, G. & James, K. Organ culture of human lymphoid tissue. I. Characteristics of the system. J. Immunol. Methods 179, 37–49 (1995).
Skibinski, G., Hoffmann, P., Radbruch, A. & James, K. Organ culture of human lymphoid tissue. II. Marked differences in cytokine production and proliferation between slice and suspension cultures of human spleen. J. Immunol. Methods 205, 115–125 (1997).
Soto-Rivera, J. et al. Study of HIV-1 transmission across cervical mucosa to tonsil tissue cells using an organ culture. Am. J. Reprod. Immunol. 69, 52–63 (2013).
Klinman, N. R. The mechanism of antigenic stimulation of primary and secondary clonal precursor cells. J. Exp. Med. 136, 241–260 (1972).
Skibinski, G., Skibinska, A., Deckers, M. & James, K. Tonsil stromal-cell lines expressing FDC-like properties: isolation, characterization and interaction with B lymphocytes. Dev. Immunol. 6, 273–284 (1998).
van Laar, J. M. et al. Sustained secretion of immunoglobulin by long-lived human tonsil plasma cells. Am. J. Pathol. 171, 917–927 (2007).
Grivel, J. C. & Margolis, L. Use of human tissue explants to study human infectious agents. Nat. Protoc. 4, 256–269 (2009).
Jenkinson, E. J. & Owen, J. J. T cell differentiation in thymus organ cultures. Semin. Immunol. 2, 51–58 (1990).
Robinson, J. H. & Owen, J. J. Generation of T cell function in organ culture of foetal mouse thymus. II. Mixed lymphocyte culture reactivity. Clin. Exp. Immunol. 27, 322–327 (1977).
Whittle, J. R. et al. Flow cytometry reveals that H5N1 vaccination elicits cross-reactive stem-directed antibodies from multiple Ig heavy-chain lineages. J. Virol. 88, 4047–4057 (2014).
Tan, Y. C. et al. High-throughput sequencing of natively paired antibody chains provides evidence for original antigenic sin shaping the antibody response to influenza vaccination. Clin. Immunol. 151, 55–65 (2014).
Brinkman, D. M. et al. Vaccination with rabies to study the humoral and cellular immune response to a T cell dependent neoantigen in man. J. Clin. Immunol. 23, 528–538 (2003).
Jackson, K. J. et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe 16, 105–114 (2014).
Jego, G. et al. Plasmacytoid dendritic cells induce plasma cell differentiation through type I interferon and interleukin 6. Immunity 19, 225–234 (2003).
Wolf, A. I. et al. Plasmacytoid dendritic cells are dispensable during primary influenza virus infection. J. Immunol. 182, 871–879 (2009).
Radbruch, A. et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 6, 741–750 (2006).
Mesin, L. et al. Restricted clonality and limited germinal center reentry characterize memory B cell reactivation by boosting. Cell 180, 92–106 (2020).
Giese, C. et al. A human lymph node in vitro—challenges and progress. Artif. Organs 30, 803–808 (2006).
Kuzin, I. et al. Long-term immunologically competent human peripheral lymphoid tissue cultures in a 3D bioreactor. Biotechnol. Bioeng. 108, 1430–1440 (2011).
Byers, A. M., Tapia, T. M., Sassano, E. R. & Wittman, V. In vitro antibody response to tetanus in the MIMIC system is a representative measure of vaccine immunogenicity. Biologicals 37, 148–151 (2009).
Giese, C. et al. Immunological substance testing on human lymphatic micro-organoids in vitro. J. Biotechnol. 148, 38–45 (2010).
Giese, C. & Marx, U. Human immunity in vitro - solving immunogenicity and more. Adv. Drug Deliv. Rev. 69-70, 103–122 (2014).
Purwada, A. et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 63, 24–34 (2015).
Purwada, A. & Singh, A. Immuno-engineered organoids for regulating the kinetics of B cell development and antibody production. Nat. Protoc. 12, 168–182 (2017).
Beguelin, W. et al. EZH2 enables germinal centre formation through epigenetic silencing of CDKN1A and an Rb-E2F1 feedback loop. Nat. Commun. 8, 877 (2017).
Purwada, A. et al. Ex vivo synthetic immune tissues with T cell signals for differentiating antigen-specific, high-affinity germinal center B cells. Biomaterials 198, 27–36 (2019).
Wagar, L. E., DiFazio, R. M. & Davis, M. M. Advanced model systems and tools for basic and translational human immunology. Genome Med. 10, 73 (2018).
Monto, A. S. et al. Comparative efficacy of inactivated and live attenuated influenza vaccines. N. Engl. J. Med. 361, 1260–1267 (2009).
He, T. C. et al. A simplified system for generating recombinant adenoviruses. Proc. Natl Acad. Sci. USA 95, 2509–2514 (1998).
Kim, L. et al. Safety and immunogenicity of an oral tablet norovirus vaccine, a phase I randomized, placebo-controlled trial. JCI Insight 3, e121077 (2018).
Liebowitz, D. et al. Efficacy, immunogenicity and safety of an oral influenza vaccine: a placebo-controlled and active-controlled phase 2 human challenge study. Lancet Infect. Dis. 20, 435–444 (2020).
Toebes, M. et al. Design and use of conditional MHC class I ligands. Nat. Med. 12, 246–251 (2006).
Nakajima, R. et al. Protein microarray analysis of the specificity and cross-reactivity of influenza virus hemagglutinin-specific antibodies. mSphere 3, e00592–18 (2018).
de Assis, R. R., et al. Analysis of SARS-CoV-2 antibodies in COVID-19 convalescent plasma using a coronavirus antigen microarray. Preprint at bioRxiv https://doi.org/10.1101/2020.04.15.043364 (2020).
Shah, H. B. & Koelsch, K. A. B cell ELISpot: for the identification of antigen-specific antibody-secreting cells. Methods Mol. Biol. 1312, 419–426 (2015).
Ellebedy, A. H. et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat. Immunol. 17, 1226–1234 (2016).
Roskin, K. M. et al. IgH sequences in common variable immune deficiency reveal altered B cell development and selection. Sci. Transl. Med. 7, 302ra135 (2015).
Blum, L. K. et al. Circulating plasmablasts are elevated and produce pathogenic anti-endothelial cell autoantibodies in idiopathic pulmonary arterial hypertension. Eur. J. Immunol. 48, 874–884 (2018).
Kinslow, J. D. et al. Elevated IgA plasmablast levels in subjects at risk of developing rheumatoid arthritis. Arthritis Rheumatol. 68, 2372–2383 (2016).
Nair, N. et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci. Transl. Med. 9, eaam5434 (2017).
Edgar, R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998 (2013).
Alamyar, E., Duroux, P., Lefranc, M. P. & Giudicelli, V. IMGT tools for the nucleotide analysis of immunoglobulin and T cell receptor V–(D)–J repertoires, polymorphisms and IG mutations: IMGT/V-QUEST and IMGT/HighV-QUEST for NGS. Methods Mol. Biol. 882, 569–604 (2012).
Wendel, B. S. et al. Accurate immune repertoire sequencing reveals malaria infection driven antibody lineage diversification in young children. Nat. Commun. 8, 531 (2017).
Joyce, M. G. et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166, 609–623 (2016).
Mallajosyula, V. V. et al. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc. Natl Acad. Sci. USA 111, E2514–E2523 (2014).
Acknowledgements
The authors thank the participants for donating their tissues for this study; J. Cyster, J. Idoyaga and B. Pulendran for thoughtful comments on the manuscript; M. Leipold, C. Tato and R. DiFazio for insightful discussions and troubleshooting suggestions; A. Messner, C. Lopez Angel, R. Cadman, A. Nguyen and H. Song for their assistance with collecting tonsillectomy samples; the Gift of Hope Organ and Tissue Donor Network and J. Solway for tissue procurement through the Gift of Hope; Mike Laine for materials development advice; Mary Rieck for sorting assistance; X. Ji, M. Miranda, I. Goncharov and H. Maecker for single-cell RNA-seq help; and the Cell Sciences Imaging Facility and the Stanford Shared FACS Facility for instrument access. This work was supported by a Bill and Melinda Gates Foundation pilot grant (OPP1113682; to M.M.D.), the National Institutes of Health (NIH) grants (5U19AI05722915 to M.M.D.; 1R01AI127877 and 1R01AI130398 to S.D.B.; U54CA224081, U19AI116484, U01CA217851 and U01DK085527 to C.J.K.) and the Howard Hughes Medical Institute (to M.M.D.). Sorting and analyzing were performed in the Stanford Shared FACS facility on instruments supported by the NIH S10 Shared Instrument Grants (S10RR025518-01 and S10RR027431-01) and the Parker Institute for Cancer Immunotherapy.
Author information
Authors and Affiliations
Contributions
L.E.W., A.S., C.J.K. and M.M.D. conceived the study and guided it throughout; L.E.W., A.S., C.M.C., B.S.W., M.M.L., V.M., L.P.J., J.Z.A., L.K.B., N.G., F.Y., K.J.L.J., K.R. and K.M.R. performed experiments and/or analyzed and interpreted data; L.E.W., A.I.S., K.M.B., K.D.M., I.N.A., G.B.H., P.S.K., W.H.R., S.D.B., C.J.K. and M.M.D. contributed to sample acquisition efforts, assay development or conceptual design of the study; M.M.L., M.C., E.G.D. and S.N.T. developed and produced antigens; L.E.W. and M.M.D. wrote the paper, and all authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
S.N.T., M.C. and E.G.D. are employed and have stock options with the biotechnology company Vaxart. P.L.F. and D.H.D. have shares in Nanommune, a company that uses Sino Biological’s proteins on commercially available protein microarrays. L.E.W., A.S., C.M.C., B.S.W., M.M.L., V.M., L.P.J., J.Z.A., L.K.B., N.G., K.J.L.J., F.Y., K.R., K.M.R., K.M.B., K.D.M., I.N.A., A.I.S., A.J., G.B.H., P.S.K., W.H.R., S.D.B., C.J.K. and M.M.D. declare no competing interests.
Additional information
Peer review information Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
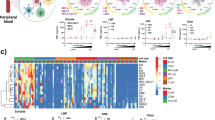
Extended Data Fig. 1 Characteristics of B cells from tonsil organoid cultures.
a, Flow cytometry gating scheme on representative unstimulated and LAIV-stimulated cultures. b, Representative ELISpot wells for detection of Ab-secreting cells with influenza vaccine specificity. The number of spots detected are shown in the corner of each well. c, Comparison of organoid cultures grown in standard flat-bottom wells vs. transwells. Data shown are from day 7 LAIV-stimulated cultures (n = 6 donors). p values shown were determined with paired Wilcoxon signed-rank two-sided tests. Boxplots show median values with hinges representing first and third quartiles and whiskers representing the highest and lowest value that is within 1.5X the interquartile range of the hinges. d, Correlation between specific Ab secretion and plasmablast frequency in influenza-stimulated and unstimulated cultures on day 7. Detection of influenza-specific IgG antibodies by semi-quantitative indirect ELISA. Optical density was corrected for background using unspent culture medium as a control. Each donor (n = 15) was tested under four conditions: no stimulation, IIV-stimulated, LAIV-stimulated, or H1N1 WT virus-stimulated. Plasmablast (CD19+ CD38+++ CD27+) frequencies were determined by flow cytometry.
Extended Data Fig. 2 Confocal imaging showing distribution of different cell types and their interactions in tonsil tissue and organoid cultures.
Merge does not include DAPI staining for clarity. a, One fresh tonsil tissue was stained with panels of markers to define T and B cell areas and GC structure. b, Day 4 unstimulated organoid stained for T and B cell markers. c, Day 4 LAIV organoid stained for germinal center markers. d, Day 4 unstimulated organoid stained for B and T cell distribution. e, Day 4 LAIV organoid stained for markers of T follicular helper cells. For organoid cultures (b-e), representative examples are shown from organoid cultures derived from three different donors.
Extended Data Fig. 3 Quantification of organoid organization and function.
a, Quantification of CXCR4 and CD83 expression levels (mean intensity, left panel) and percent positive (percentage of CD20+B cells, right panel) in day 4 tonsil organoids from one donor. Two areas of an LAIV-stimulated organoid (top GC and bottom GC as also shown in Fig. 3a) and a representative GC-organized area from an unstimulated organoid were used for the calculation. b, Representative intracellular AID flow cytometry staining profiles. Data shown are from a day 4, LAIV-stimulated organoid culture. Total B cells were subsetted based on CD38 and CD27 expression and are shown as individual profiles in red compared to a ‘no AID primary antibody’ FMO control in grey.
Extended Data Fig. 4 B cell receptor sequencing from HA-specific B cells.
a, Phylogenetic trees from two donors are shown (top). Day 0 clones are indicated by black points. Different isotypes (IgM, IgA, IgG) are indicated by color (green, blue, and red respectively). Oligoclonal populations and clonal families from single cell data are represented by larger points (open circles). Clonal families were defined as BCR sequences that use the same V and J genes and have at least 70% amino acid identity in the CDR3 region for heavy and light chains. b, FACS staining and B cell phenotypes of HA-specific B cells compared to the total B cell pool.
Extended Data Fig. 5 Tracking individual heavy-chain BCR lineages before and after LAIV stimulation in tonsil organoids show evidence of isotype switching.
Immunoglobulin heavy-chain gene rearrangements for each isotype were sequenced from total memory B cells, germinal center B cells, and plasmablasts sorted from cultures of four donors on days 0 and 7. Heavy-chain BCR sequences from lineages that contained members as only IgM on day 0 and as only isotype-switched on day 7 were compared for their somatic hypermutation levels. For each lineage, the mean SHM was calculated for day 0 IgM members and for day 7 switched members, and the difference between these means was plotted. Lineages with increased mutation are shown in blue and those with decreased mutation in red.
Extended Data Fig. 6 Depletion of pre-existing HA-specific and non-naive B cells do not prevent production of new high-affinity HA+ B cells after organoid culture.
B cells with high affinity BCRs for A/California 2009 HA+ and all non-naive B cells were depleted by FACS and depletion was confirmed by post-sort analysis. After 10 days in organoid culture, cells were harvested, re-stained for A/California HA+ B cells, and run on a flow cytometer. n = 4 donors were tested.
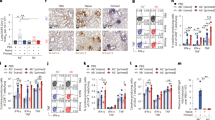
Extended Data Fig. 7 Effects of cell depletion on influenza-specific antibodies and their affinities.
a, Rescue of plasmablast and Ab responses to LAIV stimulation by supplementing pDC-depleted cultures with type I IFNs. b, Frequency of CD4+ T cells (of live cells) in intact and CD4-depleted cultures on day 7. CD4+ cells were depleted by positive selection with magnetic particles. c, Biolayer interferometry data indicating Ab affinity for A/California 2009 H1N1 HA full length, head-specific, and stem domains. Colors are matched to patient samples in (b).
Extended Data Fig. 8 SARS-CoV2-specific Abs detected in organoid cultures stimulated with Ad5 vectored vaccine candidates.
Day 14 post-stimulation culture supernatants were tested for the presence of SARS-CoV2-specific IgM, IgG, and IgA in n = 12 donors. Abs were detected using a protein microarray. Individuals donors are represented by symbols. The signal intensities shown were background subtracted based on unstimulated control cultures from the same donor. Boxplots show median values with hinges representing first and third quartiles and whiskers representing the highest and lowest value that is within 1.5X the interquartile range of the hinges.
Supplementary information
Supplementary Table 1
Lymphoid tissue donor characteristics. Unless otherwise noted in donor name, samples are from palatine tonsillectomy surgeries.
Rights and permissions
About this article
Cite this article
Wagar, L.E., Salahudeen, A., Constantz, C.M. et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med 27, 125–135 (2021). https://doi.org/10.1038/s41591-020-01145-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41591-020-01145-0
This article is cited by
-
Long noncoding RNA regulation of immunity
Nature Immunology (2026)
-
Immune-related actinopathies at the cross-road of immunodeficiency, autoimmunity and autoinflammation
Nature Reviews Immunology (2026)
-
Organoid-Immune Co-Cultures: A Next-Generation approach to disease modeling
Molecular Biology Reports (2026)
-
T cell receptor associated transmembrane adaptor 1 (TRAT1) modulates human Th17 and Treg responses via PI3-kinase and STAT dependent mechanisms
Cell Communication and Signaling (2025)
-
Organoids in respiratory virus research: advances and perspectives
Molecular Biomedicine (2025)