Abstract
Cell types in the hippocampus with unique morphology, physiology and connectivity serve specialized functions associated with cognition and mood. These cell types are spatially organized, necessitating molecular profiling strategies that retain cytoarchitectural organization. Here we generated spatially-resolved transcriptomics (SRT) and single-nucleus RNA-sequencing (snRNA-seq) data from anterior human hippocampus in ten adult neurotypical donors. Using non-negative matrix factorization (NMF) and label transfer, we integrated these data by defining gene expression patterns within the snRNA-seq data and then inferring expression in the SRT data. These patterns captured transcriptional variation across neuronal cell types and indicated spatial organization of excitatory and inhibitory postsynaptic specializations. Leveraging the NMF and label transfer approach with rodent datasets, we identified putative patterns of activity-dependent transcription and circuit connectivity in the human SRT dataset. Finally, we characterized the spatial organization of NMF patterns corresponding to pyramidal neurons and identified regionally-specific snRNA-seq clusters of the retrohippocampus, subiculum and presubiculum. To make this molecular atlas widely accessible, raw and processed data are freely available, including through interactive web applications.
Similar content being viewed by others
Main
Spatio-molecular organization in the brain reflects cellular composition and circuit connectivity patterns that underlie fundamental structure–function relationships. Mapping patterns of gene expression onto brain topography using spatially-resolved transcriptomics (SRT) has facilitated new biological insights into the molecular mechanisms that link structure to function, and enabled better understanding of these relationships over development and disease progression1,2,3. In the human brain, these techniques have enabled molecular delineation of cortical laminae beyond classic histological definitions4,5, identified new cell types and their topographical organization in the noradrenergic locus coeruleus6 and described differentiation trajectories at multiple gestational stages of fetal brain7. These technologies have been deployed to better understand disease states in the brain, including mapping the local microenvironment of multiple sclerosis lesions8,9 and infiltration patterns of malignant glioblastoma10.
Comprehensive spatio-molecular mapping of the human hippocampus (HPC) is critical to understand how its unique organizational structure supports many fundamental biological processes11,12. The HPC includes the dentate gyrus (DG) and the cornu ammonis (CA) regions, subdivided into CA1–CA4, each of which contains specialized cell types and distinct laminar organization. The organization of these specialized neuronal cell types into neuropil-enriched layers, including the DG molecular layer (ML), stratum lucidum (SL) and stratum radiatum (SR), supports well-defined functions within the canonical HPC circuitry. The trisynaptic loop, which supports various features of learning, memory and the stress response, initiates with inputs from the entorhinal cortex (ENT), which traverse from DG to CA3 to CA1, culminating with a relay to the subiculum (SUB), the major output nucleus of the HPC13,14. Output circuits from the SUB to numerous cortical and subcortical regions control important cognitive and motivated behaviors11,15 and are implicated in neuropsychiatric and neurodevelopmental disorders16,17.
Defining the molecular composition of cell types that have specialized roles in HPC circuit function is a prerequisite to targeting their function for therapeutic interventions. However, available transcriptomic profiles generated using single-nucleus RNA-sequencing (snRNA-seq) from postmortem human HPC tissue18,19,20,21 lack important spatial information and do not retain cytosolic or synaptic transcripts22. Additionally, many existing transcriptomic datasets have focused specifically on the DG given its importance in development and aging23,24,25,26,27,28, or have inconsistently sampled across HPC subregions, resulting in cellular composition differences between donors. To investigate gene expression at cellular resolution across the human HPC, we curated postmortem human tissue specimens with well-defined HPC neuroanatomy that systematically encompassed all subfields and sampled across the structure’s diverse longitudinal axis. We deployed a discovery-based experimental design using a well-validated platform to measure gene expression transcriptome-wide in a spatial context and generated paired snRNA-seq data from adjacent tissue sections to investigate gene expression at cellular resolution. To maximize the utility and value of this data resource for the community, we sourced HPC tissue from the same adult, neurotypical brain donors for which we recently provided comprehensive, paired SRT and snRNA-seq data in the dorsolateral prefrontal cortex (dlPFC)5.
We used spot-level deconvolution and non-negative matrix factorization (NMF) to integrate the SRT and snRNA-seq datasets, providing biological insights about the molecular organization of HPC cell types, cell states and spatial domains in the human brain. We also deployed computational strategies to overcome the inherent limitations of postmortem human tissue by incorporating functional molecular data from model organisms. Specifically, we used the human gene expression data to identify latent factors, then incorporated existing rodent datasets that feature information on circuit connectivity and neural activity induction to make predictions about axonal projection targets and likelihood of ensemble recruitment in spatially-defined cellular populations of the human HPC. The ability to infer functional roles for human cell types within the context of intact circuitry has profound potential for understanding how function in the human brain is disrupted in disease.
Results
SRT in the human HPC
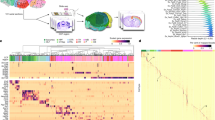
We performed Visium Spatial Gene Expression (Visium-H&E, 10x Genomics) and 3′ Single Cell Gene Expression (10x Genomics) experiments to generate paired SRT maps and snRNA-seq data of the postmortem human anterior HPC (Fig. 1a–c and Supplementary Table 1). Using the same ten neurotypical adult brain donors included in our previous dlPFC study5, we generated Visium-H&E data from two to five capture areas per donor to encompass all major HPC subfields (DG, CA1–CA4 and SUB; Supplementary Fig. 1 and for abbreviations see Supplementary Table 2). Following quality control (QC; Supplementary Figs. 2 and 3), we retained 150,917 spots from n = 36 capture areas, hereafter referred to as the SRT dataset.
a, Postmortem human tissue blocks containing the anterior HPC were dissected from ten adult neurotypical brain donors. b, Tissue blocks were cryosectioned for snRNA-seq assays (gold), and placed on Visium slides (Visium-H&E, blue). Tissue sections (n = 2–4, 100 μm cryosections per donor) from all ten donors were collected from the same tissue blocks for measurement with the 10x Genomics Chromium 3′ gene expression platform. For each donor, two samples were generated, one sorted based on PI+ (purple) and the second sorted based on PI+ and NeuN+ (green). Replicate samples were collected from three donors for a total of n = 26 total snRNA-seq libraries. Panels a and b are created with BioRender.com. c, Tissue blocks were scored with a razor blade to demarcate regions of interest and 10 μm tissue sections were collected for measurement with the 10x Genomics Visium-H&E platform. For all ten donors, two to five tissue sections were collected to include the extent of the HPC (n = 36 total capture areas). Dashed outlines indicate the approximate location of score marks, for example, donor, with color indicating capture areas obtained from consecutive sections. Orientation was verified based on the expression of known marker genes, such as SNAP25 (dashed outline color corresponding to the capture area location on the tissue section). d, Canonical marker genes were identified as SVGs using nnSVG29. Spots are colored by log2-normalized counts. e, SRT data were clustered using PRECAST30 with k = 18, and clusters were annotated (columns) based on expression of known marker genes (rows). Cluster groupings indicated at the top of the heatmap define which clusters contributed to the broad domains of neuron, neuropil, WM and vasc/CSF. Hippocampal region abbreviations are also presented in Supplementary Table 2.
To identify HPC spatial domains in our SRT dataset, we leveraged spatially aware feature selection (nnSVG29 and clustering (PRECAST30) methods. nnSVG and PRECAST were chosen for their ability to improve on nonspatially aware feature selection and clustering methods and their computational efficiency31,32. We evaluated a range of clustering resolutions (k) and used Akaike Information Criterion (AIC), marker gene expression, and comparison with histological annotations to inform our selection of k = 18 PRECAST clusters (Fig. 1d,e, Supplementary Fig. 4 and Supplementary Fig. 5). Due to similar gene expression, multiple clusters corresponding to CA2–CA4 and to CA1 were collapsed into their respective domains (Supplementary Fig. 6a,b). The final spatial domains (Fig. 2a and Extended Data Fig. 1) identified the DG granule cell layer (GCL), CA2–CA4, CA1, SUB, cortical neurons of other retrohippocampal (RHP) regions, and a domain that appeared to be transitional between the SUB and RHP (SUB.RHP). To finalize spatial domains for neuropil clusters, we compared gene expression and anatomical localization to the histologically annotated SL, SR, DG ML, stratum lacunosum-moleculare (SLM) and subgranular zone of the DG (SGZ; Supplementary Fig. 6c,d), resulting in final neuropil spatial domains including ML, SL.SR, SR.SLM and SLM.SGZ. Although the SLM and SGZ are comprised of distinct cell types and exhibit distinct functions, the combinatorial nature of the SLM.SGZ spatial domain was consistent with other neuropil regions SL.SR and SR.SLM, where the k = 18 PRECAST clusters were most accurately annotated to a combination of adjacent regions. We also annotated for clusters of cell types that are not traditionally associated with fixed HPC anatomical regions—GABA, vasculature (Vasc) and CP (Extended Data Fig. 1). We compared these results with other clustering algorithms and found high concordance.
a, Integrated and merged spot plot of four Visium capture areas from the same donor (Br3942) with spots colored by the 16 spatial domains annotated from k = 18 PRECAST clusters. CA1.1/CA1.2 were collapsed to CA1 and CA2-4.1/CA2-4.2 were collapsed to CA2–CA4. See Supplementary Table 2 for abbreviations. b, Schematic illustrating pseudobulking approach, which collapses spot-level data to the spatial domain level within each capture area by summing the total UMIs for each group. c, PCA of the pseudobulked samples captures variation across broad spatial domains. Neuron cell body-enriched (greens and light blue (GABA)), neuropil-enriched (grays), WM-enriched (purples) and Vasc/CSF (dark blue). d, Heatmap showing DEG expression (rows) across the spatial domains (columns). Grouping across the top shows broad domain annotations. Spot plots are filled by log2-normalized counts. Spot borders are colored by a broad domain. e–h, Spot plots demonstrate spatial expression of DEGs—PPFIA2, a known marker for the GCL (e), PRKCG, which is known to be enriched in CA1–CA4 domains (f), APOC1, a known astrocyte cell marker (g) and SFRP2, which was specifically increased in the SLM-subgranular zone (SGZ) domain (h). i, Volcano plots illustrate results from DE analysis for each broad-level domain, with log2(FC) on the x axis and FDR-adjusted, −log10-transformed P values on the y axis. Genes colored red pass both FDR and log2(FC) thresholds (FDR-adjusted P < 0.01 and log2(FC) > 1). Top DEGs for each broad domain grouping are labeled. j, Violin plots of pseudobulk expression of DEGs for neuron-enriched regions (CLSTN3), neuropil-enriched regions (SLC1A3), white matter regions (SHTN1) and vasc/CSF regions (TPM2). Spatial domains are on the x axis and normalized gene expression in log2(CPM) is on the y axis. Boxes are colored by broad cluster grouping.
To identify differentially expressed genes (DEGs) across the spatial domains, we employed a ‘layer-enriched’ linear mixed-effects modeling strategy performed on pseudobulked SRT data, as previously described4,5 (Fig. 2b–d, Extended Data Fig. 2, Supplementary Fig. 7 and Supplementary Table 4). We confirmed canonical marker genes for spatial domains, including PPFIA2 for GCL (Fig. 2e), PRKCG for pyramidal neuron layers CA1–CA4 (Fig. 2f) and MOBP for WM. We also identified several new marker genes, particularly with respect to the SUB and HPC neuropil-enriched domains (Fig. 2g,h (APOC1 and SFRP2, respectively)). Using the same modeling strategy, we identified genes specific to broad domains (neuron, neuropil, WM and Vasc/CSF; Fig. 2i,j and Supplementary Table 5). Altogether, these findings reveal widespread differences in gene expression across the HPC corresponding with both discrete spatial subregions and broad spatial domains.
Paired snRNA-seq in the human HPC
While SRT captures spatial organization that unlocks circuit-level information based on extensive prior histological and anatomical research, snRNA-seq provides improved cellular resolution molecular profiling. Using cryosections adjacent to those collected for the SRT dataset, we collected two populations of nuclei for snRNA-seq—a NeuN+ population to enrich for neurons and a PI+ population (Fig. 1b). After QC and preprocessing (Supplementary Figs. 8 and 9), we retained 75,411 high-quality nuclei across the ten donors. We implemented graph-based clustering on batch-corrected reduced dimension representations of gene expression patterns and annotated the resulting 60 high-resolution clusters based on known marker gene expression (Supplementary Figs. 10–12).
We identified DEGs across snRNA-seq clusters using the pseudobulk enrichment modeling approach above and used these results to collapse our clusters into 24 cell types (Fig. 3a,b and Supplementary Table 6). Through investigation of robust DEGs, we were further able to resolve minute differences between cell classes (Fig. 3c,d and Extended Data Fig. 3). Intriguingly, we observed subsets of GCs that express distinct forms of activin receptors (ACVR1, ACVR2A and ACVR1C), suggesting a stable heterogeneity within the GCL during adulthood (Extended Data Fig. 3g).
a, UMAP representation of snRNA-seq data. Individual nuclei are represented as points, colored and labeled according to their cell type. Cell type abbreviations are listed in Supplementary Table 2. b, Left, stacked bar plots of cell types indicated in a showing proportions of nuclei for each donor (columns). Right, stacked bar plots of cell types indicated in a, with columns indicating nuclei grouped by sort strategy (PI+ or PI+NeuN+), and across the overall dataset (all nuclei). c, Violin plots showing log2-normalized expression (y axis) of select DEGs identified with both spatial domain and n = 60 snRNA-seq cluster DE analyses. Nuclei are grouped based on cell types (x axis) for improved visibility, and the fill color also corresponds to cell type as indicated in a. d, Heatmap showing a selection of DEGs (rows) from snRNA-seq DE analysis across all n = 60 clusters (columns). Heatmap is colored by mean log2-normalized counts. Cell cluster abbreviations are also presented in Supplementary Table 2. UMAP, uniform manifold approximation and projection; GC, DG granule cell; CA1/ProS; CA1 and prosubiculum; Sub, subiculum; HATA, HPC–amygdala transition area; Amy, amygdala; Thal, thalamus; Cajal, Cajal–Retzius cells; GABA.PENK, PENK positive GABAergic neurons; GABA.MGE, medial ganglionic eminence-derived GABAergic neurons; GABA.LAMP5, LAMP5-positive GABAergic neurons; GABA.CGE, central ganglionic eminence-derived GABAergic neurons; Micro/Macro/T, microglia and macrophages and T cells; Astro, astrocytes; Oligo, oligodendrocytes; OPC, oligodendrocyte progenitor cells; Ependy, ependymal cells; AHi, amygdala-hippocampal region; CXCL14, CXCL14 positive GABAergic neurons; HTR3A, HTR3A positive GABAergic neurons; VIP, VIP positive GABAergic neurons; CRABP1, CRABP1 positive GABAergic neurons; C1QL1, C1QL1 positive GABAergic neurons; PV.FS, PVALB positive fast-spiking GABAergic neurons; SST, SST positive GABAergic neurons; CORT, CORT positive GABAergic neurons; COP, committed oligodendrocyte precursor; CP, choroid plexus tissue; Endo, endothelial cells; PC/SMC, pericytes/smooth muscle cells; VLMC, vascular leptomeningeal cells.
Correlating the gene-level enrichment model t statistics of snRNA-seq and SRT revealed strong agreement between the two profiling approaches (Extended Data Fig. 3h). We compared specific DEGs that were significant in pyramidal neuron groups of both data modalities (Figs. 2d and 3c). KIT, a CA3 marker in SRT33, is expressed in pyramidal cell types from CA3 and CA1, as well as in a specific GABAergic population. We observed that POU3F1 is expressed at similar levels in CA1 and Sub.1 clusters in snRNA-seq data, but is restricted to the CA1 in SRT. We identified new SUB markers COL24A1 and PART1 in both modalities, while KCNH5 and TESPA1 were specific to SUB in SRT but were expressed more broadly in snRNA-seq clusters (Figs. 2d and 3c).
The HPC is implicated in many pathological conditions, but the specific cell types and spatial domains driving these associations remain unclear16,34,35,36. To identify cell types and spatial domains associated with genetic risk for diseases and disorders, we used stratified linkage disequilibrium score regression (S-LDSC)37,38,39. S-LDSC regression coefficients represent the contribution of a given annotation (for example, spatial domain or cell type) to the heritability of a given trait, including neuropsychiatric conditions40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55. We observed many expected associations, including enrichment of Alzheimer’s disease risk in microglia41 (Supplementary Fig. 13a). Spatial domain analyses revealed enrichment of genetic risk for schizophrenia (SCZ) in the CA1 domain, and for multiple disorders, including SCZ, in the RHP (Supplementary Fig. 13b). We thus sought additional integration strategies for our snRNA-seq and SRT data to enable better understanding of the diversity of HPC cellular populations and their spatial organization.
Integrating snRNA-seq and SRT data
One approach to integrating SRT and snRNA-seq data involves estimating the cell type composition within individual Visium spots via spot-level deconvolution algorithms56. To provide a robust spot deconvolution reference dataset for postmortem human anterior HPC, we generated a Visium Spatial Proteogenomics (Visium-SPG) dataset, which replaces H&E histology with immunofluorescence (IF) staining to label proteins of interest. Visium-SPG was conducted with tissue from two donors (one male/one female) with clear anatomical orientation (Supplementary Fig. 14). By labeling cell-type-specific proteins (NEUN for neurons, OLIG2 for oligodendrocytes, GFAP for astrocytes and TMEM119 for microglia), the fluorescence intensity in each IF channel provides an orthogonal validation for predictions of cell type composition (Extended Data Fig. 4a,b). We benchmarked the ability of three spot-level deconvolution algorithms57,58,59 to leverage cell type identity, learned from marker genes in our snRNA-seq data and validated by IF staining, to predict the cell type composition of individual Visium-SPG spots (Extended Data Fig. 4c,d). We found the most consistent performance with RCTD57 and applied this method to the Visium-H&E SRT dataset (Extended Data Fig. 4d,e and Supplementary Note 1).
While spot-level deconvolution methods are valuable, most rely on a priori classification of reference data into discrete clusters and do not allow for the integrated representation of transcriptional variation beyond cell type. Another integration approach involves the consolidation of reference snRNA-seq gene expression information into reduced dimensions with NMF, and the subsequent transfer of these NMF patterns onto a query dataset (for example, SRT)60,61 (Extended Data Fig. 5). Unlike spot-level deconvolution, NMF patterns represent any source of gene expression variation and may be specific to cell types, cell states, other biological processes, or technical artifacts62,63. Additionally, NMF provides interpretable gene-level weights for each pattern, theoretically allowing for the direct biological interpretation of NMF patterns. Here we used NMF to integrate snRNA-seq and SRT datasets by first defining gene expression patterns within the snRNA-seq data and then mathematically projecting the patterns to transfer the weights onto SRT data to probe the spatial organization of these patterns (Extended Data Fig. 5).
We used NMF to distill normalized snRNA-seq counts into 100 NMF patterns. Unsurprisingly, many of these patterns were specific to HPC cell types (Extended Data Fig. 6a). Following pattern transfer to the SRT dataset and removal of poorly represented or donor-specific patterns, we observed 47 patterns that corresponded strongly to specific spatial domains (Extended Data Fig. 6c,d). We compared the ability of RCTD spot-level deconvolution and NMF to classify Visium-H&E spots, and found strong correlations with spot-level predictions for cell type proportion via RCTD (nmf77-Oligo ρ = 0.94, nmf81-Astro ρ = 0.86; Fig. 4a,b,d,e and Supplementary Fig. 15c–f). We performed gene set enrichment analysis (GSEA) using the ordinal rank of gene-level NMF weights and found that genes contributing to the transcription patterns captured by oligodendrocyte- and WM-dominant pattern nmf77 (ANK3 (ref. 64), RHOA65 and DPYSL2 (ref. 66)) and astrocyte-dominant pattern nmf81 (NRXN1 (ref. 67), NCAN68 and PLEC69) were consistent with the known biological role of these cell types (Fig. 4c,f). These findings highlight the agreement of the two integration approaches and the benefit of gene-level weights provided with NMF.
For all spot plots of example capture areas from donor Br3942, spot borders are colored by spatial domain (see Supplementary Table 2 for abbreviations). a, RCTD prediction of proportion of oligodendrocytes (Oligo, fill color) per Visium spot. b, Spot plots displaying spot-level weights (fill color) for NMF pattern nmf77. c, GSEA results showed that genes with higher nmf77 weights contributed to the significant enrichment of biological pathways associated with oligodendrocytes. For GSEA results, the x axis shows the ranked gene-level NMF weight. Each vertical black bar indicates the rank of genes for the indicated Reactome gene set. GSEA statistics are presented to the right of the plot. Top, NES; middle, numerator indicates the number of genes present in the experimental gene set, denominator indicates the total number of genes in the Reactome gene set; bottom, FDR-adjusted one-tailed P value (Padj). d, RCTD prediction of proportion of astrocytes (Astro, fill color) per spot. e, Spot plots displaying spot-level weights (fill color) for NMF pattern nmf81 (specifically elevated in astrocyte snRNA-seq clusters). f, Genes with higher nmf81 weights contributed to the significant enrichment of biological pathways associated with astrocytes. g, Spot plots displaying spot-level weights (fill color) for NMF pattern nmf13 (elevated in neuronal snRNA-seq clusters). h, Spot plots displaying spot-level weights (fill color) for NMF pattern nmf7 (elevated in neuronal snRNA-seq clusters). i, Genes with higher nmf13 weights contributed to the significant enrichment of biological pathways related to neuronal signaling, with increased representation of transcriptional variation highly relevant to excitatory postsynaptic response. j, Genes with higher nmf7 weights contributed to the significant enrichment of biological pathways related to neuronal signaling, with increased representation of transcriptional variation highly relevant to the structure and function of inhibitory synaptic connections. NES, normalized enrichment score.
Another advantage of NMF is the capacity to capture transcriptional variation beyond cell type definition. We observed 19 patterns that were present across all cell types and spatial domains (Extended Data Fig. 6a,d). We examined two of these general patterns (nmf13 and nmf7), both of which exhibited increased spot-level weights in multiple neuronal cell types and HPC spatial domains but were anti-correlated with one another (Fig. 4g,h and Supplementary Fig. 16). Examination of GSEA results suggested that nmf13 and nmf7 encapsulate distinct synaptic transcriptional profiles. Top nmf13-weighted genes include CAMKK1 (ref. 70), LRFN2 (ref. 71) and DLGAP3 (ref.72), and the enrichment of genes related to the activation of NMDA receptors and postsynaptic events (Padj. = 0.01) suggested that nmf13 represents gene expression highly relevant to excitatory postsynaptic response (Fig. 4i). In contrast, top nmf7-weighted genes, like GABRA1, KIF5A73,74,75 and DYNLL2 (ref.76), and the enrichment of genes related to GABA synthesis, release, reuptake and degradation (Padj. = 1.2 × 10−4) indicate that nmf7 more strongly represents the structure and function of inhibitory postsynaptic specializations (Fig. 4j). These data demonstrate that in practice, NMF identifies subfield-specific differences in transcription related to neuronal structure and function.
NMF captures activity-dependent transcription programs
We identified two NMF patterns (nmf91 and nmf20) that captured stimulus-dependent transcriptional programs, exemplified by the expression genes with known roles in modulating synaptic response to activity77,78,79,80 like FOS, JUN and NR4A1 (ref. 81) in nmf91 and SORCS3 (ref. 82), PDE10A83 and HOMER1 (ref. 84) in nmf20 (Extended Data Fig. 7). To further investigate the ability of NMF patterns to capture neuronal activity-related gene expression, we transferred the NMF patterns from our human snRNA-seq dataset onto a mouse snRNA-seq dataset containing HPC neurons activated by electroconvulsive seizures (ECS) or HPC neurons under control conditions (Sham)85. As we observed when transferring human snRNA-seq nuclei-level weights into human SRT data, NMF patterns mathematically projected onto mouse snRNA-seq nuclei were both general and cell-type specific (Extended Data Fig. 8).
We found that nmf91 and nmf20 were more highly weighted onto nuclei from GCs in the ECS condition than the sham-activated group, supporting our hypothesis that these patterns capture activity-dependent transcriptional programs (Fig. 5a). We further found that many of the top-weighted genes to nmf91 and nmf20 were significantly increased in ECS GCs compared to sham. For nmf91, these genes included many canonical activity-dependent transcripts (Fig. 5b). Although highly-weighted genes in nmf20 are less clear, the high weight and increased expression of BDNF86 and intracellular BDNF signaling mediators SORCS3 and SORCS1 (refs. 82,87) suggests that this pattern represents genes involved in secondary activity response (Fig. 5c). We found 98.0% of spots with non-zero nmf20 weights were located in neuron (91.0%) or neuropil (7.0%) broad domains, while 68.9% of spots with non-zero nmf91 weights occurred in these regions (42.8% in neuron and 26.1% in neuropil), suggesting enrichment of these patterns in synapse-rich domains.
a, Select NMF patterns projected onto a mouse snRNA-seq dataset of hippocampal neurons activated by ECS or under control conditions (Sham; y axis, by cell type). The dot color indicates scaled average nuclei NMF weights. Dot size indicates the proportion of y axis group with non-zero pattern weight for the given x axis value. DE analysis was performed on mouse GC nuclei, testing for differences in the expression by activity condition. For all volcano plots (b,c,f,g), the y axis presents the −log10 FDR-adjusted P value and the x axis presents log2(FC), where negative values indicate greater expression in sham-activated GCs and positive values indicate greater expression in ECS GCs. b,c,f,g, Points are colored for the gene-level NMF weight for nmf91 (b), nmf20 (c), nmf10 (f) and nmf14 (g). d,e, Spot plots isolating the DG GCL spatial domain (green outlined spots) demonstrate the differing spatial organization of nmf10 (d) and nmf14 (e) weights in an example capture area from donor Br3942. Spot fill indicates spot-level NMF pattern weight. h, Left, UMAP plot of all nuclei present in our human snRNA-seq, highlighting the GC clusters. Right, zoomed UMAP plot of only GC nuclei from our human snRNA-seq dataset with color indicating cluster identity. i, UMAP plot of human GC nuclei showing (left) nmf10 nuclei-level weights and (right) log2-normalized counts of highly-weighted nmf10 gene CHST9. j, UMAP plot of human GC nuclei showing (left) nmf14 nuclei-level weights and (right) log2-normalized counts of highly-weighted nmf14 gene SORCS3. PS/Sub, prosubiculum and subiculum neurons; L5/Po, layer 5 and polymorphic layer.
Compared to other HPC cell types, mouse GC nuclei exhibited specific increases in nmf14 and nmf10 weights, which were also associated with GC clusters and the GCL in our human datasets (Fig. 5a and Extended Data Fig. 6a,d). We observed that the transcriptional patterns encapsulated by nmf10 and nmf14 represented distinct responses to neuronal activity in the mouse (Fig. 5a) and unique spatial organization in the human (Fig. 5d,e). nmf10 mapped principally to unstimulated GC nuclei, and many top nmf10-weighted genes were significantly increased in baseline conditions (Fig. 5f). Notably, these genes (CNTN1, ADAM22, FAT4, CDH10, ST6GALNAC5 and CHST9) encode for critical synaptic adhesion proteins and proteins that stabilize established synapses.
In contrast, nmf14 was elevated in stimulated GCs. Key genes with higher nmf14 weights that were significantly increased in ECS GCs included GC.4 cluster markers BDNF and ACVR1C (Extended Data Fig. 3g), as well as SORCS3 and SGK1 (Fig. 5g). Given the functional importance of BDNF88, ACVR1C89, SORCS3 (ref. 77) and SGK1 (refs. 90,91) expression in activity-dependent synaptic plasticity, we hypothesize that nmf14 represents gene expression patterns that promote synaptic scaling. The elevated weight of nmf14 in the superficial human GCL in unstimulated conditions (Fig. 5e) suggests that this subset of GCs may be uniquely poised to promote activity-dependent synaptic scaling. Examining the nuclei-level weights of nmf10 and nmf14 and the expression of top-weighted nmf10 and nmf14 genes indicated that the GC.4 cluster fits the criteria for poised GCs (Fig. 5h–j).
These data indicate that NMF can identify activity-regulated gene expression in the context of cell-type-specific recruitment. Activity-regulated transcription is intimately related to physiological function, recruitment of cellular ensembles and synaptic connectivity, all of which fundamentally contribute to cellular behavior. However, these properties are not often considered when annotating cell types based on transcriptomics. NMF enables the transfer of information from animal models to human brain datasets, allowing predictions about the functional properties of human cells across species. This approach can be leveraged to understand cell-type function in the context of HPC circuit connectivity.
Spatial mapping elucidates pyramidal neuron diversity
Although the HPC and RHP contain multiple classes of pyramidal neurons with distinct molecular profiles18,19,92, physiological properties93 and axonal projection targets94, it has historically been difficult to distinguish subfield-specific RHP populations (for example, SUB versus ENT) with snRNA-seq alone. To address this gap, we integrated SRT and snRNA-seq using NMF to better define these cellular populations.
Mapping Sub.1-specific nmf40 and Sub.2-specific nmf54 to SRT revealed distinct laminar organization in situ (CA1-proximal versus CA1-middle, respectively; Fig. 6a). To probe if these differences corresponded to distinct efferent target regions13,14, we used data from a recent study coupling retrograde viral circuit tracing with single-cell methylation sequencing (snmC-seq) in mouse95. Our human snRNA-seq derived NMF patterns were transferred onto the mouse snmC-seq data from excitatory HPC and RHP neurons (n = 2,004 nuclei) and after removing sparsely mapping patterns (<45 nuclei) unfortunately nmf54/Sub.2 was excluded (Extended Data Fig. 9a). However, patterns specific to CA1, SUB and RHP cell types mapped to mouse HPC neurons with distinct efferent targets (Fig. 6b). These results recapitulated known projections from the SUB to the thalamus96 and hypothalamus97, and from the ENT to the HPC and prefrontal cortex98.
All spot plots are filled by the spot-level weights for the indicated NMF pattern, with scales corresponding to the maximum spot-level weight of any spot in the SRT dataset. Spot border color indicates the spatial domain. See Supplementary Table 2 for abbreviations. a, Spot plots of example areas from donor Br3942 highlight the laminar organization of nmf40 (snRNA-seq cluster Sub.1) and nmf54 (snRNA-seq cluster Sub.2), as well as the clear distinction from the CA1 (nmf15). b, Dot plot of NMF weights (dot color) after transfer to mouse single-cell methylation sequencing (snmC-seq) with retroviral tracing (n = 2004 HPC and RHP nuclei)95. Rows indicate nuclei axonal projection target region. Dot size indicates the number of nuclei with non-zero pattern weights. c, Spot plots of example capture area from donor Br2743 show ENT-specific NMF patterns. d, TSNE plots of pyramidal nuclei from the snRNA-seq dataset colored by the NMF patterns in c. e, Spot plots of example capture area from donor Br2743 show RHP NMF patterns that exhibit spot-level weights distributed across the ENT and subicular complex. f, TSNE plots of pyramidal nuclei from the snRNA-seq dataset colored by the NMF patterns in e. g, Spot plots (for example, donor Br3942) exemplify the anatomical location of nmf17 to the PreS, indicated by the asterisk. h, Violin plots show snRNA-seq log2-normalized counts (y axis) across HPC and RHP clusters (x axis, reflecting spatially-informed annotations; Supplementary Table 2) for traditional cortical layer markers (SATB2, TLE4 and CUX2), canonical subiculum marker FN1, and new subiculum markers (COL24A1, TOX). i, new DEGs distinguish the superficial, middle and deep subiculum (Sub.1, Sub.2, Sub.3, respectively), and the PreS. Dot size indicates the proportion of nuclei in each cluster (column) with non-zero expression for each gene (row). The dot color indicates average log2-normalized gene counts. TSNE, t-distributed stochastic neighbor embedding; HPF, hippocampal field; MOB, main olfactory bulb; RSP, retrosplenial cortex; PTLp, posterior parietal cortex; ACA, anterior cingulate cortex; PIR, piriform cortex; STR, striatum; TH, thalamus; AMY, amygdala; HY, hypothalamus; MOp, primary motor cortex; PFC, prefrontal cortex.
NMF patterns enriched in distinct clusters of non-CA pyramidal nuclei corresponded to nonoverlapping SRT spots with stereotypical laminar organization throughout the RHP (Fig. 6c–f). NMF patterns nmf84, nmf45, nmf27 and nmf51 were spatially restricted to the distal and superficial RHP, suggesting that these patterns and associated snRNA-seq clusters are ENT-specific (Fig. 6c,d). NMF patterns corresponding to deeper layer neurons (nmf68, nmf22 and nmf53) were not restricted along the CA1 transverse axis (distal to proximal) but were instead present throughout the SUB-ENT transition (Fig. 6e). One pattern (nmf65), enriched in deep pyramidal clusters (L6.1 and half of L6b), ran along the border of WM and SUB, adjacent to the middle layer of SUB spots highlighted by nmf54 (Fig. 6e,f). Based on the expression of SUB marker genes and top NMF pattern genes, including TOX, TSHZ2 and ZNF385D, we believe that the L6.1 cluster and nmf65 capture the transcriptional profile of the deep SUB (Extended Data Fig. 9e,f). Altogether, spatial registration of pyramidal NMF patterns agreed with broader classification of the snRNA-seq clusters as superficial (L2/3), L5, and deep (L6/6b) and facilitated improved annotation of snRNA-seq clusters.
One NMF pattern (nmf17) enriched in another superficial snRNA-seq cluster (L2/3.1) exhibited a dramatically different spatial location than the other ENT-specific L2/3 patterns (Fig. 6g and Supplementary Fig. 17). The distribution of nmf17 follows previous anatomical descriptions of the presubiculum (PreS)98,99; in more anterior samples (donors Br6423 and Br2743) nmf17 is more adjacent to the SUB. In more posterior samples (donors Br3942 and Br8325), it is present at the curve of the SUB. In some donors, nmf17 is present in an island-like pattern, again consistent with the description of the PreS anatomical studies. Unfortunately, to our knowledge, no molecular markers have been identified for the PreS to provide crucial supporting evidence that cluster L2/3.1 and nmf17 label this specific brain region.
Recognizing that the expression of common cortical layer markers may not be sufficient to identify these spatially-annotated populations in future experiments (Fig. 6h), we revisited our DEG results to identify high-confidence gene markers specifically associated with subpopulations of the SUB (Fig. 6h). Although the canonical SUB marker gene FN1 is helpful to isolate superficial SUB neurons (Sub.1), our data indicated that COL24A1—identified as a SUB DEG in both SRT and snRNA-seq datasets (Fig. 3c)—defines both superficial and middle SUB neurons. TOX, another DEG for the SUB in both SRT and snRNA-seq data, is a more general subicular complex indicator, labeling the PreS as well (Fig. 6h).
We performed additional differential expression (DE) analysis to distinguish our spatially-annotated SUB clusters from transcriptionally similar local neuron populations. We set a robust threshold and identify multiple genes that, in combination with traditional regional and laminar markers, will be useful for future research either in annotating these populations or in biological discovery (Fig. 6i and Extended Data Fig. 10). For example, markers like MDFIC (PreS) and NDST4 (superficial SUB) could elucidate region-specific circuit functions as the products of these genes contribute to glucocorticoid response100 and synaptic specificity101, respectively. Other markers like SCN7A (deep SUB)102 and TRPC3 (middle SUB)103 may correspond to region-specific neuronal firing properties.
Discussion
By integrating SRT and snRNA-seq data, we generated a comprehensive transcriptomic atlas of the adult human HPC. We characterized the molecular organization of the human HPC with spatial and cellular resolution, identifying discrete spatial domains and a rich repertoire of HPC cell types. Spot-level deconvolution and NMF revealed gene expression patterns that represent cell-type-specific molecular signatures. NMF enabled the capture of gene expression profiles shared across biological processes, such as synaptic signaling (Fig. 4). Integration of these patterns with cross-species functional genomics data (Fig. 5) provides spatial context to behaviorally-relevant cell types, cell states and molecular pathways. To molecularly define the organization of the subicular complex, we leverage cross-species neuronal circuit data and anatomical insights from our SRT dataset (Fig. 6).
Measuring transcriptomic changes in response to experimental manipulations is not possible in human postmortem brain tissue, while functional studies in rodents that can test causality of cellular and molecular associations may lack direct relevance to human brain function and behavior. Therefore, integrating human transcriptomic data with functional genomics data from animal models has the potential to facilitate the interpretation of cellular responses to various stimuli. To illustrate the biological insights that can be gleaned using this approach, we mapped NMF patterns from the human HPC onto snRNA-seq data from a mouse model of induced ECS85 to identify gene expression patterns that are putatively associated with neural activity in the human brain (Fig. 5). Identifying and localizing expression patterns associated with activity-regulated genes in the human HPC is important because expression of these genes is critical for recruitment of HPC neuronal ensembles controlling learning, memory and other cognitive processes104. NMF patterns that reflected processes known to be regulated by activity, including immediate-early gene expression and synaptic signaling, were enriched in activated mouse neurons (Fig. 5). In the rodent, the majority of GCs are relatively silent under both baseline conditions105 and during exploration of new environments106,107. However, a small fraction of GCs rapidly increases their firing activity during behavior, supporting a sparse coding scheme that facilitates precision in the detection of newness and the animal’s location during learning108. NMF patterns that differentially map to control versus activated neurons may represent the population of GCs that are more quiescent versus those that are either actively firing or intrinsically primed for activity85. Indeed, active and quiescent mouse GCs have substantially different transcriptomic profiles85. Differences in GC activity levels may reflect differences in their ability to be recruited into neuronal ensembles in response to behavioral experiences109,110,111,112,113,114. These analyses provide insight into the putative spatial and molecular organization of cellular activity states in the human DG, which is critical to better understand circuit function in the HPC. Circuit activity in the HPC flows from the DG, extending through the pyramidal, subicular and RHP regions, which have not been well-characterized in the human at the molecular scale. Since annotations for the RHP transition region rely on a priori knowledge of spatial organization and no molecular profiles are available, we were unable to manually annotate subdivisions in this region. While analysis of the snRNA-seq data enabled identification of many individual cell types along this transition region, unbiased clustering strategies were also unable to fully identify and differentiate across subdivisions. However, application of NMF afforded a more biologically meaningful interpretation of these cell types by mapping their spatial organization in the SRT dataset. With this approach we were able to identify gene signatures attributable to specific subdivisions of the ENT and subicular complex (Fig. 6). These findings highlight the future potential of these approaches for incorporating human transcriptomic data with rapidly emerging viral circuit labeling tools, which have recently enabled the profiling of neuron populations in rodent models based on their innervation patterns115,116.
We demonstrate the effectiveness of NMF for integrating transcriptomics data with other datasets, particularly across species and data modalities. This approach has some limitations. Namely, NMF can be sensitive to initialization values, patterns corresponding with noise may be incorrectly attributed to distinct biological processes, and NMF weights can be scaled differently across different factorizations. Despite these limitations, the illustrated strategy can be iterated upon to map existing and forthcoming datasets to our atlas. NMF is able to transfer continuous patterns of expression, such as cell types (Figs. 4 and 6) and transcriptional activity (Fig. 5), while other tools, including principal component analysis (PCA) and spot-level deconvolution, are not able to find these gradients of expression. NMF-based integration of our snRNA-seq and SRT data leveraged the spatial information provided by the SRT dataset and the cellular resolution of snRNA-seq to enable the discovery of molecular signatures in human tissue. Ultimately, these signatures can be extended with targeted gene panels at single-cell resolution for validation, or reverse translated to test causality in rodent or cellular models. Notably, snRNA-seq and SRT data from dlPFC are available from the same donors as those used in this study5, and emerging computational methods may enable modeling molecular connectivity patterns across this clinically-relevant circuit in the future. This is an important endeavor with substantial potential to improve clinical outcomes given strong scientific rationale and precedence for normalizing circuit-level dysfunction to improve symptoms of neuropsychiatric disorders117,118,119. Understanding functional dynamics in HPC circuits is important because their dysregulation is implicated in various neuropsychiatric and neurodevelopmental disorders120,121. Defining the human HPC at cellular resolution with spatial fidelity is necessary for designing molecular approaches that can facilitate more precise circuit manipulation. Ultimately, identifying unique molecular identities for spatially organized HPC cell types based on their innervation targets in the human brain is necessary for the development of circuit-specific therapeutics.
This highly integrated, well-annotated single-cell and spatial transcriptomics dataset of the human HPC provides unique biological insight into hippocampal molecular neuroanatomy, including activity-dependent transcriptomic profiles in the GCL and molecular definition of the RHP transition zone between the ENT and SUB. Our innovative approach can be effectively used to map external data to this and other transcriptomic atlases, as illustrated. To facilitate wide access to our data, we provided the human HPC snRNA-seq and SRT atlas as a resource to the scientific community through multiple avenues, including interactive web applications for visualization and exploration.
Methods
Postmortem human tissue samples, processing and QC
Postmortem human brain tissues from ten neurotypical adult donors of European ancestry were obtained after autopsy following informed consent from legal next-of-kin, through the Maryland Department of Health IRB protocol 12–24, and from the Western Michigan University Homer Stryker MD School of Medicine, Department of Pathology, the Department of Pathology, University of North Dakota School of Medicine and Health Sciences and the County of Santa Clara Medical Examiner-Coroner Office in San Jose, CA, all under the WCG protocol 20111080, as previously described5,122 (Supplementary Table 1). No statistical methods were used to predetermine sample sizes, but our donor cohort includes the same donors described in a previous publication5.
Anterior hippocampal dissections were performed by the same neuroanatomist (T.M.H.) using anatomical landmarks to include the anterior portion of the HPC proper plus the subicular complex36. A total of 10 μm cryosections were mounted on standard microscopy slides for staining with hematoxylin and eosin (H&E) for orientation and QC. Following QC, 2–4 100 μm cryosections, totaling approximately 50 mg of tissue, were transferred into a low-adhesion tube (Eppendorf) kept on dry ice and reserved from the anterior portion of the HPC for snRNA-seq (Fig. 1a). Then, two 10 μm sections were mounted on standard microscopy slides and reserved for H&E staining and RNAscope to guide reassembly of Visium capture areas and histological annotation of canonical subfields, using previously defined markers of the DG (PROX1), CA3 (NECAB1), CA1 (MPPED1) and SUB (SLC17A6).
The tissue block was then scored using a razor blade, so that tissue sections approximately the size of a Visium capture area could be positioned and placed onto chilled Visium Spatial Gene Expression slides (2000233, 10x Genomics). In some cases, multiple Visium experiments were performed to ensure the inclusion of relevant subfields. In these cases, the distance along the anterior-posterior axis between experiments did not exceed 500 μm. Following the successful completion of Visium experiments, an additional 100 μm cryosections were collected for snRNAseq.
SRT data generation and analysis
Visium Spatial Gene Expression slides were processed as previously described5,28. Experimenters were blind to the age and sex of donors. For each Visium slide, H&E staining was performed (protocol CG000160, revision B, 10x Genomics) and imaged on a Leica CS2 slide scanner equipped with a ×20/0.75NA objective and a ×2 doubler. Following tissue permeabilization (18 min, empirically determined), on-slide cDNA synthesis was performed, and sequencing libraries were generated for all Visium samples following the manufacturer’s protocol (CG000239, revision C, 10x Genomics). Libraries were sequenced on a NovaSeq 6000 System (Illumina) at the Johns Hopkins Single Cell Transcriptomics Core according to the manufacturer’s instructions at a minimum depth of 60,000 reads per spot. Images were processed using VistoSeg for preprocessing and nuclei segmentation123. FASTQ and image data were integrated using the 10x SpaceRanger pipeline124 and reads were aligned to the reference genome GRCh38 2020-A. Spatial coordinates were rotated to reflect the orientation of the intact tissue section (Supplementary Fig. 1). For all SRT analysis, we used R (version 4.3.2, unless otherwise noted) and Bioconductor (version 3.17) for analysis of genomics data125.
Visium QC and count normalization
After removing undetected genes and spots with zero counts, spot-level QC was performed with scuttle to calculate mitochondrial expression rate, library size, and the number of detected genes126,127. We applied a 3× median absolute deviation (MAD) threshold to discard spots with low library sizes and/or low numbers of detected genes (Supplementary Fig. 2). Mitochondrial expression rate was not used to define low-quality spots as this metric appeared to correlate with true biological variation attributable to hippocampal neuropil-enriched layers (Supplementary Fig. 3). Neuropil is characterized by large numbers of synapses and a paucity of neuronal cell bodies, and mitochondria are known to be enriched at both presynaptic and postsynaptic sites128,129. After applying 3MAD thresholds, the SRT dataset consisted of 31,483 genes and 150,917 spots. Counts were normalized by library size and log2-transformed using computeSumFactors() from scran126 and logNormCounts() from scuttle127.
Feature selection: spatially variable genes (SVGs)
Features were selected as input for unsupervised clustering by selecting the 2,000 SVGs identified with nnSVG29. Traditional methods of selecting highly variable genes do not necessarily capture spatial expression patterns. The nnSVG package uses a nearest-neighbor Gaussian process model and measures up against other SVG detection methods in improving spatial domain detection in RNA-seq data, compared to nonspatial feature selection approaches31. nnSVG aims to identify genes with varying expression patterns across different regions within the tissue by estimating gene-specific spatial ranges and retaining detailed spatial correlation information.
Before feature selection, we filtered out genes with fewer than 100 counts across all spots across all capture areas, according to nnSVG documentation. For each capture area, we also filtered out genes with less than three counts in at least 0.5% of spots in that capture area. We did not exclude the mitochondrial genome. nnSVG was run separately on each capture area using the default parameters, generating one result per capture area. To consolidate SVGs across capture areas, we ranked all genes within each capture area by spatial variance, then averaged gene ranks across all capture areas. We retained the top 2,000 genes (based on mean rank) for clustering analysis, further constraining to genes ranked in the top 1,000 genes in at least two different capture areas (Supplementary Table 3 and Fig. 1d).
Clustering and spatial domain annotation
For spatially-aware unsupervised clustering of SRT data, we used PRECAST v1.5 (ref. 30), with the 2,000 SVGs as input features (Supplementary Fig. 4a). PRECAST was chosen based on its ability to leverage spatial information when performing clustering, its use of joint embeddings that allow for integration across samples and batches, and its relatively fast runtime which allows for optimization of the number of desired clusters32. We ran PRECAST using k = 5 through k = 20 and focused k = 15 through k = 20 as these had the lowest AIC values (Supplementary Fig. 4b). Visual inspection indicated similar clusters for k = 15 through k = 20. Focusing on k = 16, k = 17 and k = 18, we examined marker gene expression and the cohesion with histologically defined annotations performed by experienced neurobiologists (Supplementary Fig. 5). We selected k = 18 clustering results as the basis for our HPC spatial domains due to minor differences including a cluster that appeared to map to SLM and SGZ, which results from k = 16 and k = 17 lacked (Supplementary Fig. 4c–e).
Histological annotations
Before unsupervised clustering, experienced neurobiologists (E.D.N. and S.C.P.) generated histologically defined annotations of tissue regions present in the SRT capture areas to assist in later annotation of unsupervised clusters. To facilitate histological annotations, we built a temporary shiny application using the spatialLIBD package4,130. The following anatomical regions were defined (Supplementary Fig. 5)—GCL, subgranular zone (SGZ), ML, CA4 pyramidal cell layer (PCL-CA4), PCL-CA3, PCL-CA1, SUB, cortex, stratum oriens (SO), SR, SL, SLM and white matter (WM). Expert neuroanatomists also recognized choroid plexus (CP) and thalamus tissue in a small number of capture areas.
To define these regions, we visualized log2 counts per million (CPM) expression of marker genes from snRNA-seq studies in both human HPC18,19 and mouse HPC13,92 and used H&E images for histological reference. To identify GCL, we used PROX1 and SEMA5A and were also guided by high levels of hematoxylin staining due to densely packed granule cell bodies. To identify SGZ, we used SEMA5A, but not PROX1, expression. We also used GAD1/GAD2 due to the presence of GABAergic neurons in SGZ131. To identify ML, we noted low unique molecular identifiers (UMIs) and detected genes in areas between GCL and SLM. To identify PCL-CA4, we used NECTIN3, AMPH, SEMA5A, SLC17A7, and observed the presence of higher UMIs and more detected genes compared to adjacent SGZ. For PCL-CA3, we used NECTIN3, NECAB1, AMPH and MPPED1. For PCL-CA1, we used FIBCD1, MPPED1, CLMP and SLC17A7. For SUB, we used FN1, NTS and SLC17A6. For SO, we used PVALB, SST, GAD1/GAD2. For SR, we searched for regions between SLM and PCL-CA1/PCL-CA3 that exhibited high levels of GFAP, but lower levels of MBP/MOBP compared to SLM and WM. For SL, we identified regions between SR and PCL-CA3 with low UMI counts and a high number of detected genes, as well as higher levels of GFAP and MBP/MOBP compared to adjacent PCL-CA3. For SLM, we observed a strip of darker staining compared to adjacent ML and SR, along with the expression of MBP/MOBP and GFAP. To identify WM regions, we looked for very high levels of oligodendrocyte markers such as MBP, MOBP and PLP1. To identify CP, we used TTR, PRLR and MSX1. CP tissue also had a distinctive sponge-like appearance, making visual identification easy. For the thalamus, we used TCF7L2. Finally, for cortex, we used CUX2, RORB and BCL11B.
Annotation of SRT clusters
The expression of well-established marker genes for HPC regions and cell types (Supplementary Fig. 5c) was used to annotate PRECAST k = 18 clusters to the DG GCL, the CA pyramidal cell layers (CA2–CA4 and CA1), the SUB, the RHP, and as GABAergic neuron-rich spots (GABA). Multiple clusters that were annotated to the same HPC domain were collapsed together, differing only in the number of nuclei per spot (Supplementary Fig. 6a,b). One cluster was labeled as SUB.RHP based on the expression of both SUB and RHP genes and its positioning between the two regions. Clusters with distinct non-neuronal gene expression were annotated as WM, CP and Vasc tissue.
To assist in annotation of neuropil-rich domains, we referenced the manual annotations (Supplementary Fig. 6c). We also examined the expression of genes enriched in the CA1 (MPPED1, FIBCD1), CA3 (TSPAN18, NECTIN3, AMPH), DG (PROX1, SEMA5A) and astrocytic genes since we had a cluster with SGZ-proximal spatial organization (Supplementary Fig. 6d). With these aids we annotated domains mapping to DG ML, SL/SR, SR/SLM and SLM/SGZ. Domains mapping to WM (WM.1, WM.2 and WM.3) were grouped as WM domains at the broad domain level. Domains mapping to vascular and CP tissue were grouped as vascular/cerebrospinal fluid (vasc/CSF) domains at the broad domain level. The final HPC domain annotations used throughout the manuscript are presented in Extended Data Fig. 1.
Identification of the thalamus and amygdala spots
During manual annotation, we identified the presence of a small amount of thalamus tissue in one capture area using thalamus-specific marker TCF7L2 (refs. 132,133; Supplementary Fig. 18a–c). In this capture area, the thalamus tissue was incorrectly annotated as SUB and SUB.RHP PRECAST domains. The thalamic SUB/SUB.RHP spots were the only spots from SUB or SUB.RHP spatial domains in this capture area. These spots were therefore excluded from DE analyses. In another donor, we identified a large, homogeneous expanse of tissue that comprised the entire capture area and was annotated as RHP. Based on the anatomy of the region, we believed this region was in fact amygdalar tissue based on the expression of several genes known to be enriched in the amygdala—SLC17A6, CDH22 (ref. 21), OPRM1 (ref. 134) and CACNG4 (ref. 135; Supplementary Fig. 18e–h). We identified three total capture areas from two donors containing tissue from the adjacent amygdala, based on the expression of these genes, all of which were annotated to the RHP spatial domain. These spots were not included in the DE analyses.
Comparison with other unsupervised spatial clustering algorithms
To ensure robustness of spatial domain predictions across computational algorithms, we compared PRECAST results with those generated from two other leading clustering methods specifically designed for spatial transcriptomics. We examined a domain detection algorithm using graph-based autoencoders, specifically GraphST136. Following the GraphST tutorial, features were selected by the highly variable gene method, and GraphST was performed for each individual slide. Samples were integrated with Harmony137, and we generated k = 16 clusters (Supplementary Fig. 19). We also compared BayesSpace138, which is similar to PRECAST in that it implements a hidden Markov random field but does so within a Bayesian model framework. For BayesSpace implementation, we used the same set of SVGs input to PRECAST to generate PCs that were mutual nearest neighbor (MNN) corrected for donor identity and used k = 18 clusters (Supplementary Fig. 20).
We compared the manual annotations with the results from the unsupervised clustering results by examination of the spot plots (Supplementary Figs. 5, 19 and 20), visualization of clustering results with alluvial plots (Supplementary Fig. 21a–c), and by quantitative comparison of the number of spots assigned to each cluster using the Adjusted Rand Index (ARI). PRECAST k = 18 clusters produced with highest agreement with manual annotations, followed closely by BayesSpace clusters (ARI with manual annotation—PRECAST = 0.192, GraphST = 0.112, BayesSpace = 0.173). When we collapsed clusters into the domains for PRECAST and similarly collapsed CA2.4 for BayesSpace, PRECAST still exhibited higher ARI (ARI with manual annotation—PRECAST domains = 0.231, BayesSpace domain = 0.179). We observed strong agreement between PRECAST and BayesSpace results (Extended Data Fig. 1 and Supplementary Figs. 20 and 21d), and the ARI for these two clustering methods was 0.398, which increased to 0.427 when the domains for both methods were used. We further examined whether GraphST or BayesSpace produced clusters that corresponded to the amygdala and thalamus spots identified manually, and found no improvement on the performance of PRECAST (Supplementary Fig. 21e). Altogether, we did not find evidence to suggest that selecting one of these alternative methods to derive our spatial domain annotations would improve the quality of our data and we continued to use PRECAST-derived spatial domains throughout the remainder of the study.
Pseudobulked SRT DE
Following unsupervised clustering, pseudobulk samples were generated by summing the raw gene expression counts across all Visium spots within a given capture area and spatial domain. We discarded pseudobulk samples that were composed of <50 spots and samples of spots identified as the thalamus or amygdala. We removed lowly expressed genes using the filterByExpr() function from the edgeR package139,140. We followed edgeR recommendations, and counts were normalized by the product of the library size and size factor, then converted to CPM and log2-transformed. After normalization, PCA and examination of the percent of gene-level variance confirmed that pseudobulk samples primarily captured biological variation (Supplementary Figs. 22 and 23). DE analysis was implemented using limma and accounted for block-specific (that is, capture area) variation141. Our DE approach followed a one-vs-rest enrichment model at either the domain level or broad domain level, in each case adjusting for donor age, donor sex and Visium slide as covariates. DE models computed fold change (FC), Student’s t-test statistics and two-tailed P values for each gene within each domain/broad domain were corrected for the false discovery rate (FDR)142 (Supplementary Tables 4 and 5). Genes were considered statistically significant if log2(FC) > 1 or log2(FC) < −1 and FDR < 0.01 (Fig. 2i,j, Extended Data Fig. 2 and Supplementary Fig. 7).
snRNA-seq data generation and analysis
Using 100 μm cryosections collected from each donor, we conducted snRNA-seq using 10x Genomics Chromium Single Cell Gene Expression v3.1 technology. Following standard procedures for nuclei isolation4, nuclei from each donor were split into two equal samples for fluorescence-activated nuclear sorting (FANS) based on propidium iodide (PI+) and Alexa Fluor 488-conjugated anti-NeuN (MAB377X, MilliporeSigma) fluorescence. One sample per donor was sorted based on PI+ fluorescence (neuronal and non-neuronal nuclei), and the second sample was sorted based on both PI+ and NeuN+ fluorescence to facilitate enrichment of neurons. This approach initially gave n = 20 for snRNA-seq (1 PI+ and 1 PI+NeuN+ sample for all ten donors) with a total of 18,000 sorted nuclei per donor (9,000 per Chromium sample). Samples were collected over multiple rounds, each containing one to three donors for two to six samples per round. We obtained poor yields of nuclei from one sequencing round (round 3), which included both PI+ and PI+NeuN+ samples from three donors. We collected additional PI+ and NeuN+ samples from these three donors and performed the nuclei sorting steps again, this time achieving better final yields of nuclei. Therefore, we had a final n = 26 for snRNA-seq (1 P1+ and 1 PI+NeuN+ sample for seven donors, 2 PI+ and 2 PI+NeuN+ samples for three donors).
All samples were sorted into reverse transcription reagents from the 10x Genomics Single Cell 3′ Reagents kit (without enzyme). Enzyme and water were added to bring the reaction to full volume. cDNA synthesis and subsequent sequencing library generation were performed according to the manufacturer’s instructions for the Chromium Next GEM Single Cell 3′ v3.1 (dual-index) kit (CG000315, revision E, 10x Genomics). Samples were sequenced on a NovaSeq (Illumina) at the Johns Hopkins University Single Cell and Transcriptomics Sequencing Core at a minimum read depth of 50,000 reads per nucleus.
snRNA-seq QC
Following sequencing, reads were mapped to Genome Reference Consortium Human Build 38 (GRCh38 2020-A) using cellranger (version 7.0.0). A Bioconductor pipeline was used for downstream analysis125. Empty droplets were detected and removed with dropletUtils143.
Because neurons have larger library sizes and a greater number of expressed genes compared to non-neuronal cells in human brain tissue144, and because our PI+NeuN+ samples are enriched for neurons, initial QC thresholds (3MAD) were computed on a per-sample basis (Supplementary Fig. 8a–c). The data-driven mitochondrial fraction thresholds from two sequencing rounds (round 1 and round 3—previously recognized to contain lower nuclei yields) were dramatically higher than the remaining samples (Supplementary Fig. 8a), resulting in exclusion of fewer low-quality nuclei (Supplementary Fig. 8d). We calculated new per-sample 3MAD thresholds for mitochondrial fraction by restricting threshold calculation to the higher quality samples with original thresholds of <5% expression from the mitochondrial genome (Supplementary Fig. 8g). This increased the number of nuclei excluded based on mitochondrial fraction, particularly in samples from sequencing rounds 1 and 3 (Supplementary Fig. 8j). The initial 3MAD thresholds for the other two QC metrics resulted in no nuclei being excluded from almost all PI+ sorted samples and from many PI+NeuN+ sorted samples (Supplementary Fig. 8e,f). For all samples with no nuclei excluded from the initial 3MAD threshold, we manually set the library size and number of detected genes threshold to 1,000 counts and 1,000 detected genes (Supplementary Fig. 8h,i), thereby increasing the number of excluded nuclei (Supplementary Fig. 8k,l). Following nuclei QC, the dimensions of our dataset were 36,601 genes × 86,905 nuclei, with the kept nuclei exhibiting consistent QC metrics across samples.
We observed that one sample of PI+NeuN+ nuclei remaining after QC filters had noticeably lower library sizes and fewer detected genes than all other PI+NeuN+ samples (Supplementary Fig. 9b,c). To see if this discrepancy was likely the result of a neuronal population unique to that sample (17c-scp), we performed rudimentary clustering with the quickCluster() function from scran and estimated the number of neurons present in each cluster based on raw expression of SYT1 > 1 count (Supplementary Fig. 9d). We found that across neuronal clusters, sample 17c-scp displayed an increased number of nuclei with low library size and few detected genes, but that there remained 17c-scp nuclei that matched the distribution of these QC metrics present in the other PI+NeuN+ samples (Supplementary Fig. 9e). We reasoned that many of the nuclei in sample 17c-scp were low-quality neurons. To prevent a detrimental impact on downstream analyses, we took a conservative approach and removed all nuclei with fewer than 5,000 detected genes in this sample only. Following this removal step, the dimensions of our dataset were 36,601 genes × 80,594 nuclei (Supplementary Fig. 9f).
snRNA-seq feature selection, dimensionality reduction and clustering
Using raw counts, scry identified features based on deviance (rather than variance) and generated Pearson residuals that were used as input for dimension reduction and clustering, avoiding biases introduced by log-normalization of UMI count data145. Subsequent PCA and MNN correction with batchelor146 produced low-dimensional embeddings that were not influenced by donor and sequencing round (Supplementary Fig. 10). Gene expression count data were then normalized and log2 transformed with scran126, using cluster-specific size factors generated from rough cluster assignments.
We implemented graph-based clustering to identify cell types present in the snRNA-seq dataset. A nearest neighbor graph was generated with scran126 based on the 50 MNN-corrected PCs using k = 5 nearest neighbors and Jaccard weights, followed by igraph147 implementation of Louvain clustering148. This resulted in 59 clusters. We annotated these clusters as neuronal or non-neuronal based on expression of neuron-specific genes, such as SYT1 (Supplementary Fig. 11a). We identified three low-quality neuron clusters based on fewer detected genes (Supplementary Fig. 11b). We also identified one cluster of likely doublets, which were marked by coexpression of oligodendrocyte, astrocyte, OPC and microglia markers in the same nuclei (Supplementary Fig. 11c). These four clusters were removed, together accounting for 5% of all nuclei.
After removing low-quality neurons and likely doublets, we repeated feature selection, dimensionality reduction, batch correction and graph-based clustering using the same parameters as before. This resulted in 62 clusters, which we classified as neuronal and non-neuronal based on SYT1 expression (Supplementary Fig. 11d). Using the number of detected genes and coexpression of distinct glial markers, we identified one low-quality neuron cluster (Supplementary Fig. 11e) and one low-quality non-neuronal cluster, respectively (Supplementary Fig. 11f). These two clusters contained 1.6% of all nuclei, and so we did not reprocess the dataset after removal. Our final dataset contained 75,411 nuclei classified into 60 clusters.
Preliminary snRNA-seq cluster annotation
Before DE analysis, we used markers from published snRNA-seq data in humans18 to label broad cell types including glia cell types (Supplementary Fig. 11f), inhibitory/GABAergic neurons and excitatory/glutamatergic neurons (Supplementary Fig. 12a). Within excitatory neurons, we were able to identify distinct clusters of CA pyramidal cells, DG granule cells and SUB pyramidal cells based on marker genes like PROX1, CALB1, FNDC1, TSPAN18, CARTPT and FN1. We knew that we should have a small thalamus population and identified this population by looking for a cluster with elevated TCF7L2 expression based on our SRT data (Supplementary Figs. 18a–c and 12b). Most of the remaining excitatory cell clusters likely comprise the RHP regions which exhibit laminar organization akin to cortical layers and are often annotated based on the expression of CUX2 (superficial marker), SATB2 (deep callosal projection marker), TLE4 (deep subcortical projection marker; Supplementary Fig. 12a). We also knew that there should be some amygdalar neurons present, but were unable to identify these clusters based on the expression of genes used to classify amygdalar domains in the SRT dataset (SLC17A6, CDH22, OPRM1 and CACNG4; Supplementary Fig. 18d–h). To identify amygdala neuron clusters in the snRNA-seq data, we isolated the RHP spots from SRT data and used scran::findMarkers() to implement a binomial test for gene markers that distinguished the spots annotated to the amygdala (n = 4,519 spots) from the true RHP domain (n = 3,940 spots). Evaluating markers increased in either population at an FDR < 0.05 and log2(FC) > 0.3, we identified 73 genes that were increased in amygdala spots and 86 genes that were increased in RHP spots. We examined the expression of these genes in all excitatory cells annotated to RHP layers and identified five clusters that we relabeled as amygdalar neurons (n = 5,524 nuclei; Supplementary Fig. 12c). Notably, these nuclei overwhelmingly came from the donors with amygdala domains identified in SRT (Br6432 n = 3,380 nuclei, Br6423 n = 1,338 nuclei), although we did annotate 721 nuclei from Br8667 to these amygdala clusters.
Pseudobulked snRNA-seq DE
Following unsupervised clustering, nuclei were pseudobulked by cluster (n = 60) and sample. We dropped all genes with zero counts across all nuclei and summed the raw gene expression counts for across all nuclei in a given sample in a given cluster using the aggregateAcrossCells() function from scran126. We discarded pseudobulk samples that were composed of less than ten nuclei (864 remaining pseudobulk samples). This resulted in the complete removal of one cluster (cluster 58), which had already been identified as Cajal–Retzius cells by the expression of RELN. As with SRT pseudobulk samples, snRNA-seq pseudobulk samples were processed with edgeR to remove lowly expressed genes and normalize gene expression. We performed DE analysis on the n = 60 clusters using pseudobulk log2-transformed CPM values. As with SRT DE analysis, we used a limma framework to conduct enrichment DE models, accounting for block-specific (that is, FANS sample) variation and adjusting for age, sex and sequencing round covariates. For each gene and cluster, this model computed FC, Student’s t-test statistics and two-tailed P values. P values were corrected for the FDR142 (Supplementary Table 6). We correlated the gene-level enrichment model t statistics for each of the 60 snRNA-seq clusters with the t statistics from the enrichment model of the SRT domains and found concordance where appropriate (Extended Data Fig. 3h).
Detailed snRNA-seq cluster annotation
We used the statistically significant (log2(FC) > 2 and FDR < 0.0001) markers from pseudobulk DE analysis to provide greater detail to the cluster labels and identified DEGs that distinguished nearly all 60 clusters as unique in some way (Extended Data Fig. 3 and Fig. 3d). With this approach we were able to annotate small populations of distinct non-neuronal cell types through the cluster-specific expression of LAMA2 (fibroblast population of vascular leptomeningeal cells (VLMCs))149, DLC1 (pericytes/smooth muscle cells (PC/SMC))150, MECOM (endothelial cells)151 and GRP17 (committed oligodendrocyte precursors (COPs))152 (Extended Data Fig. 3a). DEGs across the six superficial RHP pyramidal neuron clusters indicated the grouping of two clusters (L2/3.1 and L2/3.5) based on the expression of TESPA1 and TSHZ2 (Extended Data Fig. 3b and Fig. 3d). Even across the amygdala neurons we identified genes distinguishing the clusters from one another, although the number of donors contributing to this population was small as the amygdala was not systematically targeted for dissection (Extended Data Fig. 3c). We were able to annotate two SUB clusters based on CNTN6 expression13 and all HPC pyramidal neurons exhibited a gradient of NUP93 expression that may serve to drive HPC identity153 (Extended Data Fig. 3f).
GABAergic neurons aggregated into cell types based on the expression of cell type-specific gene expression (Extended Data Fig. 3d) and gene expression that indicated broader GABAergic families (Extended Data Fig. 3e). Using these DEGs, we annotated GABAergic families corresponding to central ganglionic eminence (CGE) origin (consisting of VIP and HTR3A cell types) based on ADARB2 expression144 and lack of NXPH1, a LAMP5+ family of ADARB2 expressing interneurons154 (consisting of CXCL14, LAMP5.CGE and LAMP5.MGE cell types), a group of GABAergic neurons from the medial ganglionic eminence (consisting of CRABP1, C1QL1, PV.FS, CORT and SST cell types) based on LHX6 expression144,154, and a population of PENK+ interneurons that originates before the major GABAergic/glutamatergic split154. We noted that the CXCL14 cluster exhibited high RELN expression, but confirmed that these were not mislabeled Cajal–Retzius neurons based on the lack of TP73 expression (Extended Data Fig. 3e) and likely represent a population of LAMP5+ interneurons that continue to express RELN into adulthood155.
Even the smallest clusters (like GC.5, n = 96 nuclei; thalamus, n = 35 nuclei; COP, n = 57 nuclei and AHi.4, n = 73 nuclei) exhibited clear expression of marker genes unique from similar clusters. Some clusters were difficult to distinguish even with DEG expression (for example, Micro.1 versus Micro.2, CP.1 versus CP.2 versus CP.3; Extended Data Fig. 3a). However, our ability to discern gene markers unique to each of the 60 clusters suggested that the high resolution with which we determined our cell clusters did not result in over-clustering.
Validation of snRNA-seq clustering approach with orthogonal computation methods
Given the importance of cluster-derived cell type identity for downstream analysis (especially NMF-based integration with SRT data), we performed a re-analysis of the snRNA-seq data from the initial QC through to clustering. Our rationale with this re-analysis is that, if we identify similar cell clusters using an orthogonal framework with different computational tools, then the conclusions generated from our n = 60 clusters are generalizable. Re-analysis was performed by a co-author (J.R.T.) who was not involved in the original snRNA-seq processing and was blinded to whether individual nuclei were included or excluded therein.
In our re-analysis, instead of identifying empty droplets using Bioconductor tools, we began with the filtered feature matrix from which empty droplets had already been removed during Cell Ranger processing. We removed the samples from sequencing round 3 that had increased debris during sorting and exhibited high mitochondrial fraction (starting nuclei = 118,415; Supplementary Fig. 24a). Because the samples from sequencing round 3 had been re-collected, there were still n = 10 donors represented. After evaluating a maximum initial mitochondrial fraction of 20%, we further filtered out all nuclei with >10% reads from the mitochondrial genome (remaining nuclei = 116,494; Supplementary Fig. 24b,c). Minimum thresholds for library size and the number of detected genes were evaluated separately for PI+ and PI+NeuN+ samples due to enrichment of neurons in the PI+/NeuN+ population, the increased number of reads in the larger neuronal nuclei, and the distinct distribution of these metrics across the two sorting methods (Supplementary Fig. 24d,e). For the PI+ samples, we selected a minimum threshold of 750 detected genes and 2,000 reads (Supplementary Fig. 24f,g). For the PI+NeuN+ data, one sample in sequencing round 2 (sample 17c-scp) exhibited a substantially shifted distribution (Supplementary Fig. 24h,i) and was therefore excluded before determining QC thresholds of a minimum of 1,000 detected genes and 3,500 reads (Supplementary Fig. 24j,k). The total number of nuclei after QC filters was 81,838, with 22.5% of the 36,577 discarded nuclei originating from sample 17c-scp with reduced quality round 2 (all remaining samples constituting 2.85–6.80% of excluded nuclei, consistent with Supplementary Fig. 9f).
Feature selection for dimensionality reduction in re-analysis was conducted using the same binomial deviance approach as before, given the ability of this method to handle raw count data and account for batch effects. For dimensionality reduction, we performed scVI156 analysis in Python to generate latent representations that replaced PCA, followed by MNN batch correction. Our scVI model included the covariates of sequencing round, donor ID, mitochondrial fraction, age and postmortem interval. Scanpy157 was then used to construct a nearest neighbors map of k = 15 neighbors based on the scVI latent representations, and subsequently to generate cell clusters using the Leiden algorithm at a resolution of 0.5 (Supplementary Fig. 25a). scVI latent representations were also used to calculate UMAP embeddings for visualization. Clusters were then refined based on the expression of MALAT1, which has been shown to indicate the quality of nuclei in snRNA-seq data, and based on mitochondrial fraction158 (Supplementary Fig. 25b–f). After filtering out low-quality clusters, new neighbors (k = 10) and clusters were found, and new UMAP embeddings computed (Supplementary Fig. 25g,h). Two small clusters (<100 nuclei) were removed due to size, and one additional small cluster was removed because it comprised nuclei from mostly one sample (110 of 138 nuclei from one sample).
Despite the very different approach to QC, re-analysis within a python framework produced a highly similar set of nuclei—64,251 were nuclei present in both the new set (74,216 total) and our original set (75,411 total; Supplementary Fig. 25i). To examine the similarity of the cell clusters generated, we performed the pseudobulk enrichment model on the re-analysis clusters. Specifically, we found that the clusters identified with scVI re-analysis correlated strongly with our original n = 60 clusters when comparing the gene expression patterns captured by the nuclei groupings (Supplementary Fig. 25j). This shows that the same cell types were found in the re-analysis and that the nuclei that were present in only one of the two datasets were spread across all clusters, suggesting we were not missing any cell types in either analysis.
S-LDSC
We performed S-LDSC to evaluate the enrichment of heritability of brain-related traits for gene sets defined for different cell populations. Gene sets were generated by selecting the top 10% of genes by t statistic for each population (unique SRT domain and snRNA-seq cell type). To construct a genome annotation, we added a 100 kb window upstream and downstream of the transcribed region of each gene in the corresponding gene set. GWAS summary statistics were run for each trait40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55. Following recommendations from the LDSC resource website (https://alkesgroup.broadinstitute.org/LDSCORE), S-LDSC was run for each gene set using the baseline LD model v2.2, which included 97 annotations to control for the LD between variants and other functional annotations in the genome. We used HapMap Project Phase 3 SNPs as regression SNPs, and 1,000 Genomes SNPs of European ancestry samples as reference SNPs, obtained from the LDSC resource website. The z score of per-SNP heritability was used to evaluate the unique contribution of gene sets to trait heritability while accounting for contributions from other functional annotations in the baseline model. The P values are derived from the z score, assuming a normal distribution and FDR was computed from the P values based on the Benjamini–Hochberg procedure.
Spot-level deconvolution algorithm benchmarking
A step-by-step protocol of the spot-level deconvolution benchmarking in the human HPC is presented in Supplementary Note 1. To estimate cellular composition across spatial domains (Extended Data Fig. 4a), we generated a spot deconvolution reference dataset in human anterior HPC using Visium-SPG to provide orthogonal validation of cellular identity. Two high-quality samples (one male/one female) were selected for IF staining of NEUN, OLIG2, GFAP and TMEM119 (Supplementary Fig. 14a). After QC and normalization, 37,845 spots across eight capture areas were retained (Supplementary Fig. 14b,c). As these samples were not included in the original PRECAST clustering, spatial domains were projected into Visium-SPG using RcppML60 (Supplementary Fig. 14d). To establish ground truth cell annotations based on IF staining, nuclei were segmented from DAPI using Cellpose 2.0 (ref. 159), and mean intensities across all IF channels were extracted (Supplementary Fig. 26a). A subset of ~1,000 annotated nuclei trained a CART decision tree model160, for automated classification into four broad cell types (Extended Data Fig. 4b and Supplementary Fig. 26b).
To benchmark the performance of three deconvolution tools (RCTD57, cell2location58 and Tangram59), we generated several different classification levels based on the n = 60 superfine cluster annotations (Supplementary Fig. 27a). We verified that across these resolutions we did not see donor-specific enrichment that was unexpected (for example, amygdala enrichment expected; Supplementary Fig. 27b–d). Marker genes selected via DeconvoBuddies161 were used for cell2location and Tangram (Supplementary Figs. 28 and 29 and Extended Data Fig. 4c); however, RCTD performance required the use of the RCTD default marker set (Supplementary Fig. 30). RCTD showed the highest concordance with IF-based counts (Supplementary Figs. 31–33 and Extended Data Fig. 4d). RCTD was thus applied to the full Visium-H&E dataset (Extended Data Fig. 4e), with results available in Samui162 (‘Integrating snRNA-seq and SRT data’).
Integration of snRNA-seq with NMF and label transfer
We used NMF to identify continuous patterns of gene expression within our snRNA-seq data (Extended Data Fig. 5). NMF is a dimensionality reduction technique for pattern recognition163 that is used across various disciplines, including transcriptomics61,63. NMF decomposes a matrix A into two lower-rank matrices W and H corresponding to gene-level and nucleus-level weight matrices, respectively.
where k is the rank of the factorization and both w and h are constrained to have only non-zero elements. The decomposition is performed iteratively, considering the mean squared error between A and the product of w and h. Iterations continue until the change in mean squared error between successive iterations falls below a specified tolerance of fit, which we set to 10−6 (recommended threshold for high-quality factorization results ≥ 10−5).
NMF analysis was implemented using RcppML, which is significantly faster and more memory efficient than other NMF methods60. Using RcppML to implement NMF, we diagonalize the columns in W and the rows in H to ensure they sum to 1, allowing for L1/L2 regularization and reproducible NMF loadings. In our case, A was the normalized snRNA-seq counts matrix with dimensions 36,601 genes × 75,411 nuclei. We selected k = 100 NMF patterns after using the cross_validate_nmf() function from singlet to perform cross-validation of different NMF ranks164 (Supplementary Fig. 34). The matrix of gene-level NMF weights for k = 100 patterns is presented in Supplementary Table 9, and a simplified version containing the top ten genes weighted to each NMF pattern is presented in Supplementary Table 10 and Supplementary Fig. 35.
Dataset integration with NMF
To integrate gene expression patterns identified with NMF in the source dataset (snRNA-seq) with other target datasets (for example, SRT), we performed transfer learning with the project() function from RcppML, which uses a fast implementation of non-negative least squares165. We mathematically projected the source W matrix with k = 100 NMF patterns into a second, target dataset based on the target matrix (A′) with dimensions I’ genes and j′ observations to generate the target coefficient matrix of H′.
Notably, i′ is the number of genes present in both source snRNA-seq (A) and target (A′) gene expression matrices. To assess the validity of NMF pattern transfer, we examined the frequency of non-zero weighted observations for each NMF pattern in the target coefficient matrix (H′) and used the empirical cumulative distribution function to determine a minimum number of observations for consideration of NMF patterns.
To explore different questions in this manuscript, the source snRNA-seq NMF patterns were transferred to three different target datasets—our SRT dataset (Extended Data Fig. 6d), a mouse electroconvulsive stimulation (ECS) snRNA-seq data85 (Extended Data Fig. 8b,c) and a mouse single-nucleus methylation sequencing (snmC-seq) dataset where retroviral tracing before collection meant that sequenced neurons were identified by both the brain region from which they were collected and the brain region to which their axon projected95 (Extended Data Fig. 9b).
NMF transfer to mouse ECS snRNA-seq dataset
For pattern transfer to the mouse ECS snRNA-seq data85, we downloaded the processed data object from the GitHub repository for the original mouse study and used the full dataset (n = 15,990 nuclei; https://github.com/Erik-D-Nelson/ARG_HPC_snRNAseq/blob/main/processed_data/sce_subset.rda.xz). Information on how these data were processed before download can be found in the same GitHub repository (https://github.com/Erik-D-Nelson/ARG_HPC_snRNAseq/tree/main/code). To enhance interpretability of our human datasets, we identified human orthologs for mouse genes in the ECS dataset and removed all genes without identified orthologs. We matched these orthologs with genes in the W matrix and discarded any genes not included in both datasets. Following the removal of genes without orthologs and genes not included in both datasets, we retained 17,557 genes. We used normalized, log2-transformed counts from the mouse ECS snRNA-seq dataset as the A′ matrix. Following label transfer, patterns were normalized to sum to one in each target dataset. We removed patterns with non-zero weights in <1,000 nuclei in the mouse ECS snRNA-seq dataset (Extended Data Fig. 8a,b). To facilitate interpretation within the context of our human findings, we further subset the data to only NMF patterns that also mapped to >1,050 SRT spots (Extended Data Fig. 8c). We found that the appropriate patterns corresponded to the relevant cell types (Extended Data Fig. 8b,c).
NMF transfer to mouse snmC-seq dataset
We leveraged a mouse model linking snmC-seq in neurons with their axon projection targets through retroviral tracing95 to see if any NMF patterns generated from our human snRNA-seq data corresponded to HPC neurons with specific axonal projection targets. We downloaded the data from GEO (accession code GSE230782). As only the raw data was available, we reproduced the original authors’ pipeline by adapting code found in the GitHub repository for the snmC-seq study (https://github.com/zhoujt1994/EpiRetroSeq2023/blob/main/02.integration_mc_rna/03.process_RS2.ipynb). This required extracting CH gene body methylation counts to approximate gene expression, based on the premise that non-CpG cytosine methylation exhibits a strong inverse relationship with gene expression in neurons166. Following the original authors’ specifications, data were QC processed and cell type clusters were generated by integrating with mouse single-cell RNA-seq data. We then subset the snmC-seq dataset to excitatory HPC and RHP neurons only (n = 2,004 nuclei). Then, following the initial study design, CH gene body methylation counts were extracted, log-scaled and negated. This matrix was used for the A′ matrix for pattern transfer. We identified human orthologs for mouse genes in this dataset and removed all genes without orthologs. We matched these orthologs with genes in the W matrix and discarded any genes not included in both datasets. Following the removal of genes without orthologs and genes not included in both datasets, we retained 14,452 genes. Following label transfer, patterns were normalized to sum to one in each target dataset. We isolated the NMF patterns that we previously identified (Extended Data Fig. 6a) as corresponding to the tissue source of the snmC-seq dataset (CA patterns—nmf11, nmf63, nmf61 and nmf15; prosubiculum/subiculum patterns—nmf32, nmf40 and nmf54; RHP patterns—nmf65, nmf22, nmf53, nmf68, nmf51, nmf45, nmf84, nmf27 and nmf17). We then removed those patterns with non-zero weights in <45 snmC-seq nuclei (Extended Data Fig. 9a). We found that the remaining patterns correctly corresponded to nuclei collected from relevant brain regions (Extended Data Fig. 9b).
Postintegration analyses of human HPC datasets
After NMF transfer to SRT, we excluded patterns with non-zero weights in <1,050 spots (Extended Data Fig. 6c). We identified two sex-based patterns based on the elevated weights of XIST (nmf37) and chrY genes (nmf27; Supplementary Fig. 36). These patterns were also excluded from downstream analysis since sex as a biological variable was not a focus in this study.
We observed that that after transfer to SRT cell type-specific patterns corresponded with the appropriate spatial domains (Extended Data Fig. 6d). Spot-level weights of the general NMF patterns remained diffusely distributed across spatial domains, with the exception of nmf94 that was highly weighted in spots annotated to the CP (Supplementary Fig. 37a). The CP marker gene TTR was the highest weighted gene to nmf94 and was weighted 8.79 times higher than the next highest weighted gene (Supplementary Table 10 and Supplementary Fig. 35). We determined that nmf94 was distinct from the TTR-dominant CP-specific transcriptional program captured by nmf48 (Supplementary Fig. 37b–d) and that observation-level weights reflected the donor bias of CP nuclei without being actually elevated in CP nuclei (Supplementary Fig. 37e,f). It appears that when nmf94 was transferred to the SRT dataset, the nuanced donor-specific effect was not conserved due to the strong importance of TTR expression in determining observation-level weights, and nmf94-weighted spots were nearly identical to nmf48-weighted spots (Supplementary Fig. 37g–j). This highlights the importance of a close examination of NMF patterns to determine the biological or technical significance of the gene expression patterns they represent and whether they can be interpreted in the same way in a dataset to which they are projected.
To ascertain if additional NMF patterns captured donor-specific effects that could confound biological interpretation in downstream analysis, we examined the donor-level representation in non-zero weighted observations across all general and specific NMF patterns (Supplementary Fig. 38). We identified two patterns (nmf91 and nmf20) that were equally represented across snRNA-seq samples but exhibited notable enrichment for two donors when projected to the SRT dataset (Extended Data Fig. 7a,c and Supplementary Fig. 38a,b, asterisk). These patterns were characterized by stimulus- and activity-dependent genes (Extended Data Fig. 7b,d). The increased abundance of these genes and the increased weight of nmf91 and nmf20 in nuclei across donors is likely due to the increased sensitivity of snRNA-seq to detect these sparsely expressed transcripts (Extended Data Fig. 7e,f). Thus, the apparent donor bias seemingly introduced when nmf91 and nmf20 are projected onto the SRT dataset is likely biologically meaningful and highlights specific donors in whom stimulus-dependent transcripts were enriched. This is explored further in Fig. 5.
To validate the biological relevance of genes highly weighted to individual NMF patterns, we performed GSEA on patterns explored in Fig. 4. GSEA was implemented with fgsea167 using reactome.db pathways168, limited to pathways annotated to Homo sapiens. Non-zero gene-level weights were used for the gene score, resulting in 4,661 genes for oligodendrocyte-specific nmf44, 9,030 genes for astrocyte-specific nmf81, 7,616 genes for transneuronal nmf13 and 7,675 genes for transneuronal nmf7. Results were evaluated as significant at a FDR-adjusted P value < 0.05. The FDR-adjusted P value and normalized enrichment score for these tests are reported in Fig. 4c,f,i,j.
Postintegration analyses of the mouse ECS dataset
To investigate whether the genes contributing to the differential weights of select NMF patterns between ECS GCs and sham GCs were associated with neuronal activation85, we performed DE analysis using scran to identify markers using a binomial test. The ECS snRNA-seq data was subset to GCs and limited to the genes with non-zero weights for the NMF pattern being tested. Results were evaluated at a strict threshold of FDR-adjusted two-tailed P value < 1 × 10−30 and an absolute value of the log2(FC) > 1. Genes labeled with text in Fig. 5 volcano plots were further characterized by high NMF pattern weights—nmf91 weight > 0.0015, nmf20 weight > 0.00065, nmf10 weight > 0.00065 and nmf14 weight > 0.0005.
We observed that nmf55 nuclei weights were higher in ECS GCs compared to sham GCs (Extended Data Fig. 8c). However, eight of the top ten genes weighted to nmf55 encode ribosomal subunits (RPS24, RPL26, RPS27A, RPL32, RPS12, RPLP1, RPL34 and RPS8), and the remaining two genes are strongly associated with translation (TPT1 and EEF1A1; Supplementary Table 10). We performed DE analysis on non-zero weighted nmf55 genes and found that although these genes were significantly increased in ECS GCs, the magnitude of log2(FC) and adjusted P values were much attenuated compared to nmf91 and nmf20 which captured activity-dependent transcriptional programs (Extended Data Fig. 8d and Fig. 5). De novo protein production in response to neuronal activity is critical for subsequent changes in synaptic and circuit connectivity169 and production of translational machinery like ribosomes likely facilitates such changes170. Thus, although ribosome transcript-dominant nmf55 is associated with activated mouse GCs, it is unlikely that nuclei or spots highly weighted for nmf55 in our human snRNA-seq or SRT datasets were recently activated. This is supported by the ubiquitous weighting of nmf55 across SRT spatial domains (Extended Data Fig. 8e). This example highlights the need for biological context in interpreting NMF patterns.
Spatial mapping of RHP laminar organization and identification of layer-specific SUB marker genes
Spatial mapping of SUB cluster-specific NMF patterns with SRT revealed laminar distribution in the SUB (Fig. 6a). To capitalize on the enhanced detail present in the integrated NMF representations, we generated binarized thresholds for individual NMF patterns by taking one fifth of the 95% max spot-level weight (Supplementary Fig. 39a). This revised spot-level annotation strategy exhibited strong concordance with the SUB spatial domain identified via unsupervised clustering (Supplementary Fig. 39b) and was used in combination with broad domain labels in Fig. 6c,e to provide visual reference for SUB spots when examining the laminar organization of other RHP-enriched NMF patterns. This binarization strategy was extended to the RHP-enriched NMF patterns to permit the discrete labeling of superficial and L5 ENT spots and was performed for the CA1 and CA3 as a measure of validation (Supplementary Fig. 39d,e). The following NMF patterns were used to generate the binarized domain labels—CA1 (nmf15), CA3 (union of nmf11 and nmf63), ENT_sup (union of nmf84, nmf45, nmf27) and ENT_L5 (nmf51). Only spots that passed a single one of the thresholds were kept for the combined annotation (Supplementary Fig. 39d). The increased detail of these NMF-driven thresholded annotations provided histological context that assisted in the classification of the spots (Supplementary Fig. 17) and nuclei (Supplementary Fig. 39e) labeled by nmf17 as the PreS.
We examined SUB and deep RHP snRNA-seq clusters (Extended Data Fig. 9c) for evidence that nmf65 indicated nuclei corresponding to the deep SUB as suggested by nmf65 laminar organization (Fig. 6e). While the entirety of the L6.1 cluster was highly weighted to nmf65, only a portion of the L6b cluster was highly weighted to this pattern (Fig. 6f). To ascertain if these deep pyramidal nuclei corresponded to the separate populations of RHP and SUB-adjacent spots labeled by nmf65, we split L6b into two portions by thresholding nmf65 weights (L6b_nmf65 with nmf65 weights > 0.00035; Extended Data Fig. 9d). This grouping was used to visualize gene expression of top DEGs that were also highly weighted to the relevant NMF patterns and indicated that L6.1 is more accurately described as deep SUB (Extended Data Fig. 9e,f).
Following NMF-driven assessment of the spatial localization of the clusters corresponding to SUB and RHP pyramidal neurons, the following snRNA-seq clusters were relabeled—L6.1 to Sub.3 (Extended Data Fig. 9e,f); L2/3.1 to PreS (Fig. 6g and Supplementary Figs. 17 and 39e); L2/3.4 to ENT.sup1, L2/3.6 to ENT.sup2a, L2/3.3 to ENT.sup2b, L2/3.2 to ENT.sup3, L5.1 to ENT.L5 (Fig. 6c,d); L5.2 to RHP.CBLN2+ (based on Extended Data Fig. 3b) and L6.2 to RHP.L6 and L6b to RHP.L6b (Fig. 6e,f).
Characterization of SUB layers
Integration of our snRNA-seq and SRT datasets with NMF revealed that four snRNA-seq clusters corresponded to subicular gene expression patterns with laminar organization. Following the identification of the superficial SUB (Sub.1), middle SUB (Sub.2), deep SUB (Sub.3) and presubiculum (PreS), we performed DE analysis (scran-implemented binomial test) in our snRNA-seq dataset focused on identifying genes that could specifically identify these regions (Fig. 6i and Extended Data Fig. 10).
To test for genes that distinguished the Sub.1 and Sub.2 clusters, we subset to regions with similar gene expression and that are spatially adjacent (CA1, ProS, Sub.3, PreS), testing for enrichment across six groups in total and only for genes with counts in >100 nuclei (22,572 genes × 8,783 nuclei). Sub.1 markers were considered statistically significant at −log10(FDR) > 30 and with log2(FC) > 1 versus each of the CA1, ProS, Sub.2, Sub.3 and PreS (146 genes). After filtering to genes with an average log2 CPM expression of > 1 in the Sub.1 nuclei, we identified ten superficial SUB marker genes—AC007368.1, AL138694.1, ATP6V1C2, COL21A1, EBF4, FN1, NDST4, PARD3B, PRKCH, RAPGEF3 (Extended Data Fig. 10a). Sub.2 markers were considered statistically significant at −log10(FDR) > 30 and with log2(FC) > 1 versus each of the CA1, ProS, Sub.1, Sub.3 and PreS (184 genes). After filtering to genes with an average log2 CPM expression of > 1 in the Sub.2 nuclei, we identified seven middle SUB marker genes—GDNF-AS1, LHFPL3, MAMDC2, PCED1B, RORB, SULF1 and TRPC3 (Extended Data Fig. 10b). ProS markers were considered statistically significant at −log10(FDR) > 30 and with log2(FC) > 1 versus each of the CA1, Sub.1, Sub.2, Sub.3 and PreS (17 genes). After filtering for genes with an average log2 CPM expression of >1 in the ProS nuclei, we identified no unique prosubiculum marker genes.
To test for genes that distinguished the Sub.3 cluster from other SUB layers and other deep RHP clusters, we subset to regions with similar gene expression and that are spatially adjacent (Sub.1, Sub.2, RHP.L6b, RHP.L6), testing for enrichment across five groups in total and only for genes with counts in >100 nuclei (21,037 genes × 5,647 nuclei). Sub.3 markers were considered statistically significant at −log10(FDR) > 30 and with log2(FC) > 1 versus each of the Sub.1, Sub.2, RHP.L6b and RHP.L6 (68 genes). After filtering to genes with an average log2 CPM expression of >1 in the Sub.3 nuclei, we identified 21 deep SUB marker genes—AC007100.1, AC010967.1, AC023503.1, AC046195.2, AL356295.1, CD36, COL4A1, COL4A2, DISC1, FSTL5, GUCA1C, LINC01194, LINC01239, LINC01821, NR2F2-AS1, PLEKHG1, RASGEF1B, SCN7A, SCUBE1, SNCAIP and VEGFC (Extended Data Fig. 10c).
To test for genes that distinguished the PreS cluster from other SUB layers and other superficial RHP clusters, we subset to regions with similar gene expression and that are spatially adjacent (Sub.1, Sub.2, ProS, ENT.sup1, ENT.sup2a, ENT.sup2b and ENT.sup3), testing for enrichment across eight groups in total and only for genes with counts in >100 nuclei (22,708 genes × 9,835 nuclei). PreS markers were considered statistically significant at −log10(FDR) > 30 and with log2(FC) > 1 compared to each of the Sub.1, Sub.2, ProS, ENT.sup1, ENT.sup2a, ENT.sup2b and ENT.sup3 (38 genes). After filtering for genes with an average log2 CPM expression of >1 in the PreS nuclei, we identified the following five PreS marker genes: AC008662.1, AL161629.1, FSTL5, MDFIC and WSCD1 (Extended Data Fig. 10d).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
This atlas of integrated single-cell and spatial transcriptomic data is the most comprehensive map of the human HPC to date. To facilitate the exploration of these rich data resources, we have created multiple interactive data portals, available at research.libd.org/spatial_hpc/. The pseudobulked snRNA-seq and SRT data are freely available through iSEE apps, allowing users to visualize gene expression for genes of interest across cell types and spatial domains. To facilitate exploration of SRT data in anatomical context, we merged all Visium capture areas from each donor for visualization of high-resolution images and corresponding gene expression data, NMF patterns and spot-level deconvolution results at the donor level using a web-based interactive tool, Samui Browser162 (ten donors, n = 36 Visium-H&E capture areas; two donors, n = 8 Visium-SPG capture areas. Raw and processed data are available through Gene Expression Omnibus (GEO)171 under accession GSE264624 to facilitate access for method development and in-depth analysis. Finally, to make the data accessible to the broader neuroscience community, we created an ExperimentHub Data package (https://bioconductor.org/packages/humanHippocampus2024) within the Bioconductor framework, allowing for convenient download of processed snRNA-seq and SRT data.
Code availability
The code for this project is publicly available through GitHub at https://github.com/LieberInstitute/spatial_hpc (ref. 172). Analyses were performed using R version 4.3.2 with Bioconductor version 3.17 unless otherwise noted. Image alignment and transformation was performed with ImageJ (version 2.14.0).
References
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Moffitt, J. R., Lundberg, E. & Heyn, H. The emerging landscape of spatial profiling technologies. Nat. Rev. Genet. 23, 741–759 (2022).
Williams, C. G., Lee, H. J., Asatsuma, T., Vento-Tormo, R. & Haque, A. An introduction to spatial transcriptomics for biomedical research. Genome Med. 14, 68 (2022).
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Huuki-Myers, L. A. et al. A data-driven single-cell and spatial transcriptomic map of the human prefrontal cortex. Science 384, eadh1938 (2024).
Weber, L. M. et al. The gene expression landscape of the human locus coeruleus revealed by single-nucleus and spatially-resolved transcriptomics. eLife 12, RP84628 (2024).
Braun, E. et al. Comprehensive cell atlas of the first-trimester developing human brain. Science 382, eadf1226 (2023).
Alsema, A. M. et al. Spatially resolved gene signatures of white matter lesion progression in multiple sclerosis. Nat. Neurosci. 27, 2341–2353 (2024).
Lerma-Martin, C. et al. Cell type mapping reveals tissue niches and interactions in subcortical multiple sclerosis lesions. Nat. Neurosci. 27, 2354–2365 (2024).
Harwood, D. S. L. et al. Glioblastoma cells increase expression of notch signaling and synaptic genes within infiltrated brain tissue. Nat. Commun. 15, 7857 (2024).
Strange, B. A., Witter, M. P., Lein, E. S. & Moser, E. I. Functional organization of the hippocampal longitudinal axis. Nat. Rev. Neurosci. 15, 655–669 (2014).
Lisman, J. et al. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nat. Neurosci. 20, 1434–1447 (2017).
Ding, S.-L. et al. Distinct transcriptomic cell types and neural circuits of the subiculum and prosubiculum along the dorsal-ventral axis. Cell Rep. 31, 107648 (2020).
Cembrowski, M. S. et al. The subiculum is a patchwork of discrete subregions. eLife 7, e37701 (2018).
Bienkowski, M. S. et al. Homologous laminar organization of the mouse and human subiculum. Sci. Rep. 11, 3729 (2021).
Small, S. A., Schobel, S. A., Buxton, R. B., Witter, M. P. & Barnes, C. A. A pathophysiological framework of hippocampal dysfunction in ageing and disease. Nat. Rev. Neurosci. 12, 585–601 (2011).
Genon, S., Bernhardt, B. C., La Joie, R., Amunts, K. & Eickhoff, S. B. The many dimensions of human hippocampal organization and (dys)function. Trends Neurosci. 44, 977–989 (2021).
Ayhan, F. et al. Resolving cellular and molecular diversity along the hippocampal anterior-to-posterior axis in humans. Neuron 109, 2091–2105 (2021).
Franjic, D. et al. Transcriptomic taxonomy and neurogenic trajectories of adult human, macaque, and pig hippocampal and entorhinal cells. Neuron 110, 452–469 (2022).
Siletti, K. et al. Transcriptomic diversity of cell types across the adult human brain. Science 382, eadd7046 (2023).
Tran, M. N. et al. Single-nucleus transcriptome analysis reveals cell-type-specific molecular signatures across reward circuitry in the human brain. Neuron 109, 3088–3103 (2021).
Armand, E. J., Li, J., Xie, F., Luo, C. & Mukamel, E. A. Single-cell sequencing of brain cell transcriptomes and epigenomes. Neuron 109, 11–26 (2021).
Zhou, Y. et al. Molecular landscapes of human hippocampal immature neurons across lifespan. Nature 607, 527–533 (2022).
Tosoni, G. et al. Mapping human adult hippocampal neurogenesis with single-cell transcriptomics: reconciling controversy or fueling the debate? Neuron 111, 1714–1731 (2023).
Yao, J. et al. Deciphering molecular heterogeneity and dynamics of human hippocampal neural stem cells at different ages and injury states. eLife 12, RP89507 (2024).
Lu, Z. et al. Tracking cell-type-specific temporal dynamics in human and mouse brains. Cell 186, 4345–4364 (2023).
Wang, W. et al. Transcriptome dynamics of hippocampal neurogenesis in macaques across the lifespan and aged humans. Cell Res. 32, 729–743 (2022).
Ramnauth, A. D. et al. Spatiotemporal analysis of gene expression in the human dentate gyrus reveals age-associated changes in cellular maturation and neuroinflammation. Cell Rep. 44, 115300 (2025).
Weber, L. M., Saha, A., Datta, A., Hansen, K. D. & Hicks, S. C. nnSVG for the scalable identification of spatially variable genes using nearest-neighbor Gaussian processes. Nat. Commun. 14, 4059 (2023).
Liu, W. et al. Probabilistic embedding, clustering, and alignment for integrating spatial transcriptomics data with PRECAST. Nat. Commun. 14, 296 (2023).
Li, Z. et al. Benchmarking computational methods to identify spatially variable genes and peaks. Preprint at bioRxiv https://doi.org/10.1101/2023.12.02.569717 (2023).
Hu, Y. et al. Benchmarking clustering, alignment, and integration methods for spatial transcriptomics. Genome Biol. 25, 212 (2024).
Kondo, T., Katafuchi, T. & Hori, T. Stem cell factor modulates paired-pulse facilitation and long-term potentiation in the hippocampal mossy fiber-CA3 pathway in mice. Brain Res. 946, 179–190 (2002).
Shi, H.-J., Wang, S., Wang, X.-P., Zhang, R.-X. & Zhu, L.-J. Hippocampus: molecular, cellular, and circuit features in anxiety. Neurosci. Bull. 39, 1009–1026 (2023).
Kohen, R., Dobra, A., Tracy, J. H. & Haugen, E. Transcriptome profiling of human hippocampus dentate gyrus granule cells in mental illness. Transl. Psychiatry 4, e366 (2014).
Jaffe, A. E. et al. Profiling gene expression in the human dentate gyrus granule cell layer reveals insights into schizophrenia and its genetic risk. Nat. Neurosci. 23, 510–519 (2020).
Finucane, H. K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Bulik-Sullivan, B., Loh, P.-R., Finucane, H. K., Ripke, S. & Yang, J. Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. LD score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 47, 291–295 (2015).
Finucane, H. K. et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 50, 621–629 (2018).
Demontis, D. et al. Genome-wide analyses of ADHD identify 27 risk loci, refine the genetic architecture and implicate several cognitive domains. Nat. Genet. 55, 198–208 (2023).
Bellenguez, C. et al. New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022).
Watson, H. J. et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat. Genet. 51, 1207–1214 (2019).
Huang, J. et al. Genomics and phenomics of body mass index reveals a complex disease network. Nat. Commun. 13, 7973 (2022).
Mullins, N. et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat. Genet. 53, 817–829 (2021).
Saunders, G. R. B. et al. Genetic diversity fuels gene discovery for tobacco and alcohol use. Nature 612, 720–724 (2022).
Yengo, L. et al. A saturated map of common genetic variants associated with human height. Nature 610, 704–712 (2022).
Grove, J. et al. Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444 (2019).
Howard, D. M. et al. Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat. Neurosci. 22, 343–352 (2019).
International League Against Epilepsy Consortium on Complex Epilepsies. GWAS meta-analysis of over 29,000 people with epilepsy identifies 26 risk loci and subtype-specific genetic architecture. Nat. Genet. 55, 1471–1482 (2023).
Trubetskoy, V. et al. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature 604, 502–508 (2022).
Nagel, M. et al. Meta-analysis of genome-wide association studies for neuroticism in 449,484 individuals identifies novel genetic loci and pathways. Nat. Genet. 50, 920–927 (2018).
Nalls, M. A. et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 18, 1091–1102 (2019).
Loh, P.-R., Kichaev, G., Gazal, S., Schoech, A. P. & Price, A. L. Mixed-model association for biobank-scale datasets. Nat. Genet. 50, 906–908 (2018).
Savage, J. E. et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat. Genet. 50, 912–919 (2018).
Morris, A. P. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat. Genet. 44, 981–990 (2012).
Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 19, 662–670 (2022).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021).
DeBruine, Z. J., Melcher, K. & Triche, T. J. Fast and robust non-negative matrix factorization for single-cell experiments. Preprint at bioRxiv https://doi.org/10.1101/2021.09.01.458620 (2021).
Stein-O’Brien, G. L. et al. Enter the matrix: factorization uncovers knowledge from omics. Trends Genet. 34, 790–805 (2018).
Kotliar, D. et al. Identifying gene expression programs of cell-type identity and cellular activity with single-cell RNA-Seq. eLife 8, e43803 (2019).
Stein-O’Brien, G. L. et al. Decomposing cell identity for transfer learning across cellular measurements, platforms, tissues, and species. Cell Syst. 8, 395–411 (2019).
Chang, K.-J. et al. Glial ankyrins facilitate paranodal axoglial junction assembly. Nat. Neurosci. 17, 1673–1681 (2014).
Vale-Silva, R. et al. RhoA regulates oligodendrocyte differentiation and myelination by orchestrating cortical and membrane tension. Glia 73, 381–398 (2025).
Grycel, K. et al. CRMP2 conditional knockout changes axonal function and ultrastructure of axons in mice corpus callosum. Mol. Cell. Neurosci. 126, 103882 (2023).
Uchigashima, M., Cheung, A., Suh, J., Watanabe, M. & Futai, K. Differential expression of neurexin genes in the mouse brain. J. Comp. Neurol. 527, 1940–1965 (2019).
Irala, D. et al. Astrocyte-secreted neurocan controls inhibitory synapse formation and function. Neuron 112, 1657–1675 (2024).
Potokar, M. & Jorgačevski, J. Plectin in the central nervous system and a putative role in brain astrocytes. Cells 10, 2353 (2021).
Kaitsuka, T. et al. Forebrain-specific constitutively active CaMKKα transgenic mice show deficits in hippocampus-dependent long-term memory. Neurobiol. Learn. Mem. 96, 238–247 (2011).
Li, Y. et al. Lrfn2-mutant mice display suppressed synaptic plasticity and inhibitory synapse development and abnormal social communication and startle response. J. Neurosci. 38, 5872–5887 (2018).
Wan, Y., Feng, G. & Calakos, N. Sapap3 deletion causes mGluR5-dependent silencing of AMPAR synapses. J. Neurosci. 31, 16685–16691 (2011).
Nakajima, K. et al. Molecular motor KIF5A is essential for GABA(A) receptor transport, and KIF5A deletion causes epilepsy. Neuron 76, 945–961 (2012).
Maas, C. et al. Synaptic activation modifies microtubules underlying transport of postsynaptic cargo. Proc. Natl Acad. Sci. USA 106, 8731–8736 (2009).
Rathgeber, L. et al. GSK3 and KIF5 regulate activity-dependent sorting of gephyrin between axons and dendrites. Eur. J. Cell Biol. 94, 173–178 (2015).
Fejtova, A. et al. Dynein light chain regulates axonal trafficking and synaptic levels of Bassoon. J. Cell Biol. 185, 341–355 (2009).
Breiderhoff, T. et al. Sortilin-related receptor SORCS3 is a postsynaptic modulator of synaptic depression and fear extinction. PLoS ONE 8, e75006 (2013).
Christiansen, G. B. et al. The sorting receptor SorCS3 is a stronger regulator of glutamate receptor functions compared to GABAergic mechanisms in the hippocampus. Hippocampus 27, 235–248 (2017).
Zhang, Y. et al. The phosphodiesterase 10A ihibitor PF-2545920 enhances hippocampal excitability and seizure activity involving the upregulation of GluA1 and NR2A in post-synaptic densities. Front. Mol. Neurosci. 10, 100 (2017).
Inoue, Y., Udo, H., Inokuchi, K. & Sugiyama, H. Homer1a regulates the activity-induced remodeling of synaptic structures in cultured hippocampal neurons. Neuroscience 150, 841–852 (2007).
Sheng, M. & Greenberg, M. E. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron 4, 477–485 (1990).
Hermey, G. et al. The three sorCS genes are differentially expressed and regulated by synaptic activity. J. Neurochem. 88, 1470–1476 (2004).
Bonate, R. et al. Phosphodiesterase 10A (PDE10A): regulator of dopamine agonist-induced gene expression in the striatum. Cells 11, 2214 (2022).
Bottai, D. et al. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J. Neurosci. 22, 167–175 (2002).
Nelson, E. D. et al. Activity-regulated gene expression across cell types of the mouse hippocampus. Hippocampus 33, 1009–1027 (2023).
Tao, X., West, A. E., Chen, W. G., Corfas, G. & Greenberg, M. E. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron 33, 383–395 (2002).
Subkhangulova, A. et al. SORCS1 and SORCS3 control energy balance and orexigenic peptide production. EMBO Rep. 19, e44810 (2018).
Wang, C. S., Kavalali, E. T. & Monteggia, L. M. BDNF signaling in context: From synaptic regulation to psychiatric disorders. Cell 185, 62–76 (2022).
Keiser, A. A. et al. Specific exercise patterns generate an epigenetic molecular memory window that drives long-term memory formation and identifies ACVR1C as a bidirectional regulator of memory in mice. Nat. Commun. 15, 3836 (2024).
Martin-Batista, E. et al. SGK1.1 limits brain damage after status epilepticus through M current-dependent and independent mechanisms. Neurobiol. Dis. 153, 105317 (2021).
Lian, B. et al. Hippocampal overexpression of SGK1 ameliorates spatial memory, rescues Aβ pathology and actin cytoskeleton polymerization in middle-aged APP/PS1 mice. Behav. Brain Res. 383, 112503 (2020).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241 (2021).
Földy, C. et al. Single-cell RNAseq reveals cell adhesion molecule profiles in electrophysiologically defined neurons. Proc. Natl Acad. Sci. USA 113, 5222–5231 (2016).
Qiu, S. et al. Whole-brain spatial organization of hippocampal single-neuron projectomes. Science 383, eadj9198 (2024).
Zhou, J. et al. Brain-wide correspondence of neuronal epigenomics and distant projections. Nature 624, 355–365 (2023).
Fei, F. et al. Discrete subicular circuits control generalization of hippocampal seizures. Nat. Commun. 13, 5010 (2022).
Viellard, J. M. A. et al. A subiculum-hypothalamic pathway functions in dynamic threat detection and memory updating. Curr. Biol. 34, 2657–2671 (2024).
Insausti, R., Herrero, M. T. & Witter, M. P. Entorhinal cortex of the rat: cytoarchitectonic subdivisions and the origin and distribution of cortical efferents. Hippocampus 7, 146–183 (1997).
Honda, Y., Shimokawa, T., Matsuda, S., Kobayashi, Y. & Moriya-Ito, K. Hippocampal connectivity of the presubiculum in the common marmoset (Callithrix jacchus). Front. Neural Circuits 16, 863478 (2022).
Oakley, R. H., Busillo, J. M. & Cidlowski, J. A. Cross-talk between the glucocorticoid receptor and MyoD family inhibitor domain-containing protein provides a new mechanism for generating tissue-specific responses to glucocorticoids. J. Biol. Chem. 292, 5825–5844 (2017).
Condomitti, G. & de Wit, J. Heparan sulfate proteoglycans as emerging players in synaptic specificity. Front Mol. Neurosci. 11, 14 (2018).
Dolivo, D. et al. The Nax (SCN7A) channel: an atypical regulator of tissue homeostasis and disease. Cell. Mol. Life Sci. 78, 5469–5488 (2021).
Wu, B. et al. TRPC3 is a major contributor to functional heterogeneity of cerebellar Purkinje cells. eLife 8, e45590 (2019).
Ma, H. et al. Excitation-transcription coupling, neuronal gene expression and synaptic plasticity. Nat. Rev. Neurosci. 24, 672–692 (2023).
Pernía-Andrade, A. J. & Jonas, P. Theta-gamma-modulated synaptic currents in hippocampal granule cells in vivo define a mechanism for network oscillations. Neuron 81, 140–152 (2014).
Senzai, Y. & Buzsáki, G. Physiological properties and behavioral correlates of hippocampal granule cells and mossy cells. Neuron 93, 691–704 (2017).
Diamantaki, M., Frey, M., Berens, P., Preston-Ferrer, P. & Burgalossi, A. Sparse activity of identified dentate granule cells during spatial exploration. eLife 5, e20252 (2016).
Borzello, M. et al. Assessments of dentate gyrus function: discoveries and debates. Nat. Rev. Neurosci. 24, 502–517 (2023).
Ohline, S. M. et al. Adult-born dentate granule cell excitability depends on the interaction of neuron age, ontogenetic age and experience. Brain Struct. Funct. 223, 3213–3228 (2018).
Marín-Burgin, A., Mongiat, L. A., Pardi, M. B. & Schinder, A. F. Unique processing during a period of high excitation/inhibition balance in adult-born neurons. Science 335, 1238–1242 (2012).
McHugh, S. B. et al. Adult-born dentate granule cells promote hippocampal population sparsity. Nat. Neurosci. 25, 1481–1491 (2022).
Kirschen, G. W. et al. Active dentate granule cells encode experience to promote the addition of adult-born hippocampal neurons. J. Neurosci. 37, 4661–4678 (2017).
Denny, C. A. et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83, 189–201 (2014).
Parylak, S. L. et al. Neuronal activity-related transcription is blunted in immature compared to mature dentate granule cells. Hippocampus 33, 412–423 (2023).
Wee, R. W. S. & MacAskill, A. F. Biased connectivity of brain-wide inputs to ventral subiculum output neurons. Cell Rep. 30, 3644–3654 (2020).
Gergues, M. M. et al. Circuit and molecular architecture of a ventral hippocampal network. Nat. Neurosci. 23, 1444–1452 (2020).
Johnson, K. A., Okun, M. S., Scangos, K. W., Mayberg, H. S. & de Hemptinne, C. Deep brain stimulation for refractory major depressive disorder: a comprehensive review. Mol. Psychiatry 29, 1075–1087 (2024).
Mayberg, H. S. et al. Deep brain stimulation for treatment-resistant depression. Neuron 45, 651–660 (2005).
Mayberg, H. S. Targeted electrode-based modulation of neural circuits for depression. J. Clin. Invest. 119, 717–725 (2009).
Thompson, S. M. et al. An excitatory synapse hypothesis of depression. Trends Neurosci. 38, 279–294 (2015).
Lieberman, J. A. et al. Hippocampal dysfunction in the pathophysiology of schizophrenia: a selective review and hypothesis for early detection and intervention. Mol. Psychiatry 23, 1764–1772 (2018).
Lipska, B. K. et al. Critical factors in gene expression in postmortem human brain: focus on studies in schizophrenia. Biol. Psychiatry 60, 650–658 (2006).
Tippani, M. et al. VistoSeg: processing utilities for high-resolution images for spatially resolved transcriptomics data. Biol. Imaging 3, e23 (2023).
10x Genomics. Space Ranger count. https://support.10xgenomics.com/spatial-gene-expression/software/pipelines/latest/using/count (2020).
Amezquita, R. A. et al. Orchestrating single-cell analysis with bioconductor. Nat. Methods 17, 137–145 (2020).
Lun, A. T. L., McCarthy, D. J. & Marioni, J. C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with bioconductor. F1000Res. 5, 2122 (2016).
McCarthy, D. J., Campbell, K. R., Lun, A. T. L. & Wills, Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017).
Ly, C. V. & Verstreken, P. Mitochondria at the synapse. Neuroscientist 12, 291–299 (2006).
Devine, M. J. & Kittler, J. T. Mitochondria at the neuronal presynapse in health and disease. Nat. Rev. Neurosci. 19, 63–80 (2018).
Pardo, B. et al. spatialLIBD: an R/Bioconductor package to visualize spatially-resolved transcriptomics data. BMC Genomics. 23, 434 (2022).
Amaral, D. G., Scharfman, H. E. & Lavenex, P. The dentate gyrus: fundamental neuroanatomical organization (dentate gyrus for dummies). Prog. Brain Res. 163, 3–22 (2007).
Lee, M. et al. Tcf7l2 plays crucial roles in forebrain development through regulation of thalamic and habenular neuron identity and connectivity. Dev. Biol. 424, 62–76 (2017).
Nagalski, A. et al. Molecular anatomy of the thalamic complex and the underlying transcription factors. Brain Struct. Funct. 221, 2493–2510 (2016).
Mansour, A., Khachaturian, H., Lewis, M. E., Akil, H. & Watson, S. J. Autoradiographic differentiation of μ, δ, and κ opioid receptors in the rat forebrain and midbrain. J. Neurosci. 7, 2445–2464 (1987).
Letts, V. A., Mahaffey, C. L., Beyer, B. & Frankel, W. N. A targeted mutation in Cacng4 exacerbates spike-wave seizures in stargazer (Cacng2) mice. Proc. Natl Acad. Sci. USA 102, 2123–2128 (2005).
Long, Y. et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat. Commun. 14, 1155 (2023).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296 (2019).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Lun, A. T. L., Chen, Y., Smyth, G. K. & It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol. Biol. 1418, 391–416 (2016).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Methodol. 57, 289–300 (1995).
Lun, A. T. L., Riesenfeld, S., Andrews, T., Dao, T. P. & Gomes, T. participants in the 1st Human Cell Atlas Jamboree, et al. EmptyDrops: distinguishing cells from empty droplets in droplet-based single-cell RNA sequencing data. Genome Biol. 20, 63 (2019).
Hodge, R. D. et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019).
Townes, F. W., Hicks, S. C., Aryee, M. J. & Irizarry, R. A. Feature selection and dimension reduction for single-cell RNA-Seq based on a multinomial model. Genome Biol. 20, 295 (2019).
Haghverdi, L., Lun, A. T. L., Morgan, M. D. & Marioni, J. C. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol. 36, 421–427 (2018).
Csardi, G. & Nepusz, T. The igraph software package for complex network research. InterJ. Complex Syst.1695 (2005).
Blondel, V. D., Guillaume, J.-L., Lambiotte, R. & Lefebvre, E. Fast unfolding of communities in large networks. J. Stat. Mech. 2008, P10008 (2008).
Kearns, N. A. et al. Dissecting the human leptomeninges at single-cell resolution. Nat. Commun. 14, 7036 (2023).
Lu, Y. et al. Role of pericytes in cardiomyopathy-associated myocardial infarction revealed by multiple single-cell sequencing analysis. Biomedicines 11, 2896 (2023).
Lv, J. et al. Epigenetic landscape reveals MECOM as an endothelial lineage regulator. Nat. Commun. 14, 2390 (2023).
Fang, M., Chen, L., Tang, T., Qiu, M. & Xu, X. The committed oligodendrocyte precursor cell, a newly-defined intermediate progenitor cell type in oligodendroglial lineage. Glia 71, 2499–2510 (2023).
Cho, U. H. & Hetzer, M. W. Nuclear periphery takes center stage: the role of nuclear pore complexes in cell identity and aging. Neuron 106, 899–911 (2020).
Tasic, B. et al. Shared and distinct transcriptomic cell types across neocortical areas. Nature 563, 72–78 (2018).
Pesold, C. et al. Reelin is preferentially expressed in neurons synthesizing γ-aminobutyric acid in cortex and hippocampus of adult rats. Proc. Natl Acad. Sci. USA 95, 3221–3226 (1998).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Clarke, Z. A. & Bader, G. MALAT1 expression indicates cell quality in single-cell RNA sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2024.07.14.603469 (2024).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
Huuki-Myers, L. A. et al. Benchmark of cellular deconvolution methods using a multi-assay dataset from postmortem human prefrontal cortex. Genome Biol. 26, 88 (2025).
Sriworarat, C. et al. Performant web-based interactive visualization tool for spatially-resolved transcriptomics experiments. Biol. Imaging 3, e15 (2023).
Lee, D. D. & Seung, H. S. Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791 (1999).
Owen, A. B. & Perry, P. O. Bi-cross-validation of the SVD and the nonnegative matrix factorization. Ann. Appl. Stat. 3, 564–594 (2009).
Myre, J. M., Frahm, E., Lilja, D. J. & Saar, M. O. TNT-NN: a fast active set method for solving large non-negative least squares problems. Procedia Comput. Sci. 108, 755–764 (2017).
Mo, A. et al. Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384 (2015).
Korotkevich, G. et al. Fast gene set enrichment analysis. Preprint at bioRxiv https://doi.org/10.1101/060012 (2016).
Ligtenberg, W. reactome.db. Bioconductor https://doi.org/10.18129/B9.bioc.reactome.db (2017).
Richter, J. D. & Klann, E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Genes Dev. 23, 1–11 (2009).
Fusco, C. M. et al. Neuronal ribosomes exhibit dynamic and context-dependent exchange of ribosomal proteins. Nat. Commun. 12, 6127 (2021).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 41, D991–D995 (2013).
Tippani, M. et al. LieberInstitute/spatial_hpc: v1.0.0. Zenodo https://doi.org/10.5281/zenodo.15282862 (2025).
Acknowledgements
We thank the LIBD neuropathology team, particularly J. Tooke and A. Deep-Soboslay, for curation of the brain samples and assistance with tissue dissections. We thank the staff and physicians at the brain donation sites, and the generosity of the brain donors and their families, without whom this work would not be possible. We thank D. R. Weinberger, members of the Hansen-Hicks working group and the Spatial LIBD team for feedback on the manuscript. Finally, we thank the families of C. Lieber and S. Lieber and M. Maltz and T. Maltz for their generous support. This project was supported by U01MH122849 (to K.M.), R21AG083328 (to S.C.P. and S.C.H.), R01MH105592 (to K.M.) and the Lieber Institute for Brain Development (to K.R.M., K.M. and S.C.P.). Portions of Fig. 1a,b and Extended Data Fig. 4a were created with BioRender.com.
Author information
Authors and Affiliations
Contributions
K.M., S.C.P. and S.C.H. conceived and designed the study. E.D.N., A.D.R., H.R.D., E.A.P., S.H.K., S.V.B., U.M.K. and S.C.P performed experiments and collected data. J.R.T., E.D.N., M.T., J.Y., C.H. and S.C.H. applied new computational methodology. J.R.T., E.D.N., M.T., A.D.R., R.A.M., N.J.E., J.Y., C.H., L.C.T., S.H. and S.C.H. helped with software development and implementation. J.R.T., E.D.N. and A.D.R. worked on computational validation. J.R.T., E.D.N., M.T., A.D.R., J.Y. and S.C.H. worked on coding and computational pipeline curation. J.R.T., E.D.N., M.T., A.D.R. and S.C.H. helped with data curation. J.E.K. and T.M.H. performed postmortem brain tissue procurement and maintenance, as well as tissue dissection. J.R.T., E.D.N., M.T., K.R.M., K.M., S.C.P. and S.C.H. helped in writing the original draft of the manuscript. While J.R.T., E.D.N., M.T., A.D.R., H.R.D., R.A.M., S.V.B., S.H., K.R.M., K.M., S.C.P. and S.C.H. also helped in writing, reviewing and editing the manuscript. J.R.T., E.D.N., M.T., A.D.R. and S.C.P. performed data visualization and figure generation. K.R.M., K.M., S.C.P. and S.C.H. supervised the study. K.R.M., K.M., S.C.P. and S.C.H. worked on funding acquisition. K.R.M., K.M., S.C.P. and S.C.H. worked on project administration.
Corresponding authors
Ethics declarations
Competing interests
E.D.N. is now a full-time employee at GSK, which is unrelated to the contents of this manuscript. His contributions to this manuscript were made while previously enrolled as a student at Johns Hopkins University and performing research at the Lieber Institute for Brain Development (LIBD). J.E.K. is a consultant on a Data Monitoring Committee for an antipsychotic drug trial for Merck & Co. The other authors declare no competing interests.
Peer review
Peer review information
Nature Neuroscience thanks Qin Ma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Data-driven HPC domain annotations derived from spatial clustering (PRECAST k = 18) across all n = 36 Visium-H&E samples.
Spatial clustering results from PRECAST k = 18 were annotated to HPC spatial domains by examining marker gene expression and anatomical location. From the 18 PRECAST clusters we generated 16 spatial domains by merging clusters that were annotated to the same HPC subfields. Spot color indicates the final spatial domain annotation (legend in bottom right). See Supplementary Table 2 for abbreviations. Capture areas are grouped by donor number.
Extended Data Fig. 2 Spatial domain-level differential expression analysis of spatially-resolved transcriptomics (SRT) data.
Volcano plotting of pseudobulked differential expression (DE) analysis for each spatial domain, with log2 fold change on the x-axis and FDR adjusted, -log10 transformed p-values on the y-axis. Genes colored red pass both FDR and log2 fold change thresholds (FDR adjusted p-value < 0.01 and absolute value of log2 fold change > 1). Top DE genes are labeled. See Supplementary Table 2 for spatial domain abbreviations.
Extended Data Fig. 3 Differentially expressed snRNA-seq genes and correlation with SRT results.
Dot plots for select differentially expressed genes (y-axis) for n = 60 cell types (x-axis). Dot color indicates the cluster average log2 normalized counts and dot size corresponds to the proportion of cluster nuclei with non-zero expression. Heatmaps are included for (a) glia, (b) retrohippocampus (RHP), (c) amygdala, (d) GABAergic neurons, (e) GABAergic neuron markers with higher expression range (including Cajal–Retzius neurons for RELN expression comparison), (f) hippocampus (HPC), (g) and dentate gyrus granule cells. See Supplementary Table 2 for cluster abbreviations. h, Heatmap of Pearson correlation coefficients (color) showing the relationship between pseudobulk differential expression analysis enrichment model t statistics for the SRT spatial domains (rows) and those of n = 60 snRNAseq clusters (columns).
Extended Data Fig. 4 Spot level deconvolution benchmarking with Visium Spatial Proteogenomics (Visium-SPG).
a, Schematic showing the complexity of cell types in Visium spots. b, Immunofluorescence (IF) images used to train a Classification And Regression Tree (CART) model to identify cell types (indicated by color, legend in image). c, The marker genes detected for selected ‘mid’ cell types (facets) are enriched in Visium-SPG spots from corresponding spatial domains (y-axis). Each data point summarized in boxplot represents an individual spot with the x-axis indicating what proportion of the ‘mid’ cell types marker genes were expressed in each spot. Similar box plots for all ‘mid’ and ‘fine’ cell types are shown in Supplementary Fig. 29. d, Performance of spot-level deconvolution algorithms was benchmarked by comparing to IF-derived assignment of cell type identity in (b). Scatter pie plots (highlighted in inset) illustrate the proportion of cell types predicted in each Visium spot (color legend in b). e, RCTD was applied to the Visium-H&E data, where the orthogonal cell type information is not present. Bar plots illustrate the proportion of spots (x-axis) predicted to each ‘mid’ cell type (fill color) within each spatial domain (y-axis).
Extended Data Fig. 5 Schematic representation of non-negative matrix factorization (NMF).
a, Schematic for how to identify patterns using non-negative matrix factorization (NMF). Specifically, for a given source matrix A with dimensions of i genes and j observations, such as the snRNA-seq data, we use NMF to decompose A into two matrices W, representing the feature-level weights matrix with dimensions of i genes and k NMF patterns, and H, representing the observation-level weights matrix for j observations and k patterns. Gray matrices are known matrices, blue matrices are generated by the analysis step. Panel a is created with BioRender.com. b, Schematic for how we transfer the NMF patterns into other datasets, for example, the SRT data. Specifically, we use the transposed feature-level weights matrix W (learned from the source data, glow) and a target dataset matrix (A’ with dimensions of i’ genes and j’ observations) to obtain a new observation-level weights matrix from the target (H’ with dimensions of k patterns and j’ observations). Therefore, for every observation (spot or nuclei) in the target dataset, we have a weight for every corresponding NMF pattern learned from the source data (snRNA-seq). Gray matrices are known matrices, blue matrices are generated by the analysis step.
Extended Data Fig. 6 Non-negative matrix factorization (NMF) patterns recapitulate transcriptional distinctions by cell type and spatial domain.
a, NMF patterns (x-axis) learned from snRNA-seq log2 normalized gene expression counts correspond well with snRNA-seq clusters (y-axis). For dot color, the nuclei-level weights for each NMF pattern were scaled and then averaged across the y-axis groups. Many of these patterns mapped to a high number of SRT spots after transfer and can be classified as general NMF patterns and specific NMF patterns based on the abundance of non-zero weighted nuclei across snRNA-seq cell type clusters (dot size). See Supplementary Table 2 for y-axis abbreviations. b, All NMF patterns (points) learned from snRNA-seq data produce non-zero weights for thousands of nuclei (x-axis). c, When these NMF patterns are transferred to SRT data, the number of NMF patterns (points) with few non-zero weighted spots (x-axis) increases. NMF patterns that produced non-zero weights in fewer than 1050 spots (34 patterns, red points) were excluded from downstream analysis, as were the sex-based patterns nmf37 and nmf28 (Supplementary Fig. 36). d, After transfer to SRT data, NMF patterns with non-zero weights in >1050 spots (x-axis) correspond well to spatial domains (y-axis). With one exception (nmf94, discussed in Supplementary Fig. 37), the NMF patterns classified as general or specific in snRNA-seq data (a), likewise, are abundant across multiple spatial domains or specific spatial domains. For dot color, the spot-level weights for each NMF pattern were scaled and then averaged across the y-axis groups. Dot size indicates the proportion of spots belonging to each y-axis group that exhibited non-zero weights for the given NMF pattern (x-axis). See Supplementary Table 2 for y-axis abbreviations.
Extended Data Fig. 7 Non-negative matrix factorization (NMF) identifies donor enrichment due to activity-dependent transcription.
a, Pie charts for snRNA-seq nuclei (top) and SRT spots (bottom) show the proportion of observations with non-zero nmf91 weights originating from each donor. Top: snRNA-seq n = 39,991 total non-zero nmf91 nuclei. Bottom: SRT n = 9,380 total non-zero nmf91 spots; Br3942 (pink) n = 4020 non-zero nmf91 spots; Br6423 (orange) n = 2736 non-zero nmf91 spots. b, Nine of the 50 genes with the highest nmf91 weight (y-axis) are canonical immediate early genes (IEGs, red) that are transcribed after strong stimulus in neurons and non-neuronal cells. Average log2 normalized gene expression in all nuclei on x-axis, nmf91 gene-level weight on y-axis. c, Pie charts for snRNA-seq nuclei (top) and SRT spots (bottom) show the proportion of observations with non-zero nmf20 weights originating from each donor. Top: snRNA-seq n = 36,679 total non-zero nmf20 nuclei. Bottom: SRT n = 2109 total non-zero nmf20 spots; Br2743 (green) n = 991 non-zero nmf20 spots; Br6423 (orange) n = 565 non-zero nmf20 spots. d, The three genes (red) with the highest nmf20 weights (y-axis) are known regulators of neuronal function in response to activation. Average log2 normalized gene expression in all nuclei on x-axis, nmf20 gene-level weight on y-axis. e, The log2 CPM expression (dot color) and abundance (proportion of observations, dot size) of three top-ranked nmf91 genes (x-axis) across donor groups (observations grouped into neurons and non-neurons) indicate consistent enrichment of IEGs in 1–2 donors in both the snRNA-seq data (left) and SRT data (right). Expression of JUN further reflects the more equal non-zero weighting of nmf91 in snRNA-seq nuclei compared to the strong donor enrichment of nmf91 observed in SRT (a). f, The log2 CPM expression (dot color) and abundance (proportion of observations, dot size) of three top-ranked nmf20 genes (x-axis) across donor groups (observations grouped into neurons and non-neurons) indicate consistent enrichment of SORCS3, HOMER1, and PDE10A in neurons and equal distribution across all donors in snRNA-seq data (left). The increased expression of HOMER1 and SORCS3 in capture areas from two SRT donors reflects the strong donor enrichment of nmf20 observed in SRT (c).
Extended Data Fig. 8 Non-negative matrix factorization (NMF) patterns across annotated cell types in mouse neuron electroconvulsive stimulation (ECS) snRNA-seq data.
a, Following label transfer of NMF patterns to mouse snRNA-seq electroconvulsive stimulation (ECS) dataset with retroviral tracing dataset85 (n = 15990 nuclei), NMF patterns were removed (red) that mapped to <1000 nuclei (x-axis). b, Dotplot of mouse nuclei-level weights for NMF patterns (x-axis) after filtering, averaged by cluster (y-axis). Dot size indicates the proportion of nuclei in each cluster with non-zero pattern weights and dots are colored by the scaled pattern weight. GC: granule cells, CA2–4: cornu ammonis (CA) regions 2 through 4 (CA2, CA3, CA4), PS/Sub: prosubiculum and subiculum neurons, L5/Po: layer 5 and polymorphic layer. c, Dotplot of nuclei-level weights for NMF patterns (x axis) after further filtering to patterns that were present in >1,050 SRT spots, averaged by cluster and ECS condition (y axis). Dot size indicates the proportion of nuclei in each cluster with non-zero pattern weights and dots are colored by the scaled pattern weight averaged across nuclei in y-axis groups. d, Volcano plot of differential expression results tested on genes with non-zero nmf55 weights. Y-axis of −log10(FDR) values are plotted with a log10 scale. X axis is the log2 fold change (FC), where negative values indicate greater expression in sham-activated GCs and positive values indicate greater expression in ECS GCs. Points are colored by nmf55 weight. Gene names are shown for genes with nmf55 weight > 0.0025, log2(FC) > 0.5, and FDR < 0.05. e, Spot plots for example capture area from donor Br3942 are colored by (left) spatial domain and (right) nmf55 weight, demonstrating the ubiquitous presence of nmf55.
Extended Data Fig. 9 Non-negative matrix factorization (NMF) label transfer to mouse single-nucleus methylation sequencing (snmC-seq) dataset; subiculum annotation via binarization of NMF patterns, and elucidation of new deep subiculum cluster snRNA-seq cluster.
a, Following label transfer of NMF patterns to mouse snmC-seq with retroviral tracing dataset95 (n = 2004 nuclei), hippocampal (HPC)- and retrohippocampal (RHP)-specific patterns were removed that mapped to <45 nuclei (red). X-axis indicates number of nuclei mapping to pattern, y-axis indicates proportion of patterns. b, Verification that NMF patterns (x-axis) identified as HPC- and RHP-specific in our human SRT and human snRNA-seq datasets corresponded with the nuclei collection source (y-axis) for the mouse snmC-seq dataset. Dots are colored by scaled, average pattern weight and dot size indicates the number of mouse nuclei with non-zero pattern weights. ENT: entorhinal cortex, CAa: anterior cornu ammonis, CAp: posterior cornu ammonis, SUB: subiculum. c, t-distributed stochastic neighbor embedding (TSNE) embedding of pyramidal clusters from our human snRNA-seq dataset. Subiculum and deep pyramidal clusters used to characterize nmf65 are labeled and colored. d, nmf65 mapped strongly to a subset of L6b nuclei, so to examine if there were differences in marker expression between these groups, L6b nuclei were thresholded based on nmf65 weights (point color). Each point represents a single nuclei in the L6b cluster, x-axis indicates nmf53 weight (RHP L6; Fig. 6e), y-axis indicates nmf65 weight. e, Dot plot demonstrating the non-overlapping expression of SUB and deep RHP marker genes (y-axis) in snRNA-seq clusters (x-axis). L6.1 exhibits higher expression of SUB markers (orange box) and not deep RHP markers (blue box), while L6b nuclei with high nmf65 weights express both groups and all other L6b nuclei only express deep RHP markers. Bolded y-axis genes are highlighted in (f). Dot size indicates the proportion of nuclei, dot color indicates the average log2 normalized expression. See Supplementary Table 2 for cluster abbreviations. f, TSNE plots of the expression of select marker genes from (e) that support the reassignment of L6.1 (asterisk) as deep SUB nuclei (Sub.3). Pyramidal nuclei of our human snRNA-seq results are represented by individual points that are colored by the log2 normalized expression.
Extended Data Fig. 10 Subiculum-focused differential expression analysis in snRNA-seq.
Violin plots depict the log2 normalized expression (y-axis) across snRNA-seq cell type clusters (x-axis) that were tested. Plots are grouped by the significant cluster: (a) superficial subiculum (Sub.1), (b) middle subiculum (Sub.2), (c) deep subiculum (Sub.3) and (d) presubiculum (PreS).
Supplementary information
Supplementary Information
Supplementary Note and Supplementary Figs. 1–39.
Supplementary Tables 1–10
Supplementary Tables 1–10.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Thompson, J.R., Nelson, E.D., Tippani, M. et al. An integrated single-nucleus and spatial transcriptomics atlas reveals the molecular landscape of the human hippocampus. Nat Neurosci 28, 1990–2004 (2025). https://doi.org/10.1038/s41593-025-02022-0
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41593-025-02022-0