Abstract
The mature retrovirus capsid consists of a variably curved lattice of capsid protein (CA) hexamers and pentamers. High-resolution structures of the curved assembly, or in complex with host factors, have not been available. By devising cryo-EM methodologies for exceedingly flexible and pleomorphic assemblies, we have determined cryo-EM structures of apo-CA hexamers and in complex with cyclophilin A (CypA) at near-atomic resolutions. The CA hexamers are intrinsically curved, flexible and asymmetric, revealing the capsomere and not the previously touted dimer or trimer interfaces as the key contributor to capsid curvature. CypA recognizes specific geometries of the curved lattice, simultaneously interacting with three CA protomers from adjacent hexamers via two noncanonical interfaces, thus stabilizing the capsid. By determining multiple structures from various helical symmetries, we further revealed the essential plasticity of the CA molecule, which allows formation of continuously curved conical capsids and the mechanism of capsid pattern sensing by CypA.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Cryo-EM density maps for CA A92E hexamers are deposited at the EMDB under accession codes EMD-10240 for (−13, 11), EMD-10228 for (−13, 12) and EMD-10239 for (−12, 11). Cryo-EM density maps for CypA-stabilized CA WT hexamers are deposited at the EMDB under accession codes EMD-10226 for (−13, 8), EMD-10246 for (−13, 12) and EMD-10229 for (−13,8) tube map. Cryo-EM density maps for the CypA/CA complexes are deposited at the EMDB under accession codes EMD-10740 (−13, 7), EMD-10739 (−13, 8), EMD-10742 (−13, 9), EMD-11176 (−13, 10), EMD-10738 (−8, 13) and EMD-10741 (−7, 13). Real-space refined atomic models of the CA hexamers are deposited at the Protein Data Bank under accession codes PDB 6SLU (−13, 11), PDB 6SKM (−13, 12), PDB 6SLQ (−12, 11), PDB 6SKK (−13, 8), PDB 6SMU (−13, 12) and PDB 6SKN (−13,8) tube. Real-space refined atomic models of the CypA/CA complexes are deposited at the Protein Data Bank under accession codes PDB 6Y9X (−13, 7), PDB 6Y9W (−13, 8), PDB 6Y9Z (−13, 9), PDB 6ZDJ (−13, 10), PDB 6Y9V (−8, 13) and PDB 6Y9Y (−7, 13). Source data are provided with this paper.
Code availability
The software is freely available from GitHub: https://github.com/kyledent/ArbitrEM.git.
References
James, L. C. & Jacques, D. A. The human immunodeficiency virus capsid is more than just a genome package. Annu. Rev. Virol. 5, 209–225 (2018).
Li, Y.-L. et al. Primate TRIM5 proteins form hexagonal nets on HIV-1 capsids. Elife 5, 582 (2016).
Smaga, S. S. et al. MxB restricts HIV-1 by targeting the tri-hexamer interface of the viral capsid. Structure 27, 1234–1245 (2019).
Gres, A. T. et al. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349, 99–103 (2015).
Pornillos, O. et al. X-ray structures of the hexameric building block of the HIV capsid. Cell 137, 1282–1292 (2009).
Pornillos, O., Ganser-Pornillos, B. K. & Yeager, M. Atomic-level modelling of the HIV capsid. Nature 469, 424–427 (2011).
Price, A. J. et al. Host cofactors and pharmacologic ligands share an essential interface in HIV-1 capsid that is lost upon disassembly. PLoS Pathog. 10, e1004459 (2014).
Yant, S. R. et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 25, 1377–1384 (2019).
Mallery, D. L. et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. Elife 7, e1002762 (2018).
Dick, R. A. et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 560, 509–512 (2018).
Zhao, G. et al. Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013).
Mattei, S., Glass, B., Hagen, W. J. H., Kräusslich, H.-G. & Briggs, J. A. G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437 (2016).
Pertel, T. et al. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 (2011).
Sayah, D. M., Sokolskaja, E., Berthoux, L. & Luban, J. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430, 569–573 (2004).
Li, S., Hill, C. P., Sundquist, W. I. & Finch, J. T. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407, 409–413 (2000).
Perilla, J. R. & Schulten, K. Physical properties of the HIV-1 capsid from all-atom molecular dynamics simulations. Nat. Commun. 8, 15959 (2017).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zhang, P., Meng, X. & Zhao, G. in Electron Crystallography of Soluble and Membrane Proteins (eds Schmidt-Krey, I. & Cheng, Y.) 381–399 (Humana Press, 2012).
Ilca, S. L. et al. Localized reconstruction of subunits from electron cryomicroscopy images of macromolecular complexes. Nat. Commun. 6, 8843 (2015).
Liu, C. et al. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat. Commun. 7, 10714 (2016).
Shah, V. B. et al. The host proteins transportin SR2/TNPO3 and cyclophilin A exert opposing effects on HIV-1 uncoating. J. Virol. 87, 422–432 (2013).
Gamble, T. R. et al. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87, 1285–1294 (1996).
Schaller, T. et al. HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7, e1002439 (2011).
Morger, D. et al. The three-fold axis of the HIV-1 capsid lattice is the species-specific binding interface for TRIM5α. J. Virol. 92, e01541-17 (2018).
Fricke, T. et al. MxB binds to the HIV-1 core and prevents the uncoating process of HIV-1. Retrovirology 11, 68 (2014).
Jiang, J. et al. The interdomain linker region of HIV-1 capsid protein is a critical determinant of proper core assembly and stability. Virology 421, 253–265 (2011).
Noviello, C. M. et al. Second-site compensatory mutations of HIV-1 capsid mutations. J. Virol. 85, 4730–4738 (2011).
Byeon, I.-J. L. et al. Structural convergence between cryo-EM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell 139, 780–790 (2009).
Saito, A. et al. Roles of capsid-interacting host factors in multimodal inhibition of HIV-1 by PF74. J. Virol. 90, 5808–5823 (2016).
Rankovic, S., Ramalho, R., Aiken, C., Rousso, I. & Kirchhoff, F. PF74 reinforces the HIV-1 capsid to impair reverse transcription-induced uncoating. J. Virol. 92, e00845-18 (2018).
Márquez, C. L. et al. Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. Elife 7, e1002762 (2018).
Momany, C. et al. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Mol. Biol. 3, 763–770 (1996).
Bayro, M. J. & Tycko, R. Structure of the dimerization interface in the mature HIV-1 capsid protein lattice from solid state NMR of tubular assemblies. J. Am. Chem. Soc. 138, 8538–8546 (2016).
Braaten, D., Franke, E. K. & Luban, J. Cyclophilin A is required for an early step in the life cycle of human immunodeficiency virus type 1 before the initiation of reverse transcription. J. Virol. 70, 3551–3560 (1996).
Kim, K. et al. Cyclophilin A protects HIV-1 from restriction by human TRIM5α. Nat. Microbiol. 4, 2044–2051 (2019).
Skorupka, K. A. et al. Hierarchical assembly governs TRIM5α recognition of HIV-1 and retroviral capsids. Sci. Adv. 5, eaaw3631 (2019).
Selyutina, A. et al. Cyclophilin A prevents HIV-1 restriction in lymphocytes by blocking human TRIM5α binding to the viral core. Cell Rep. 30, 3766–3777.e6 (2020).
Obal, G. et al. Conformational plasticity of a native retroviral capsid revealed by X-ray crystallography. Science 349, 95–98 (2015).
Schorb, M., Haberbosch, I., Hagen, W. J. H., Schwab, Y. & Mastronarde, D. N. Software tools for automated transmission electron microscopy. Nat. Methods 16, 471–477 (2019).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Tang, G. et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157, 38–46 (2007).
Egelman, E. H. Reconstruction of helical filaments and tubes. Meth. Enzymol. 482, 167–183 (2010).
He, S. & Scheres, S. H. W. Helical reconstruction in RELION. J. Struct. Biol. 198, 163–176 (2017).
de la Rosa-Trevín, J. M. et al. Scipion: a software framework toward integration, reproducibility and validation in 3D electron microscopy. J. Struct. Biol. 195, 93–99 (2016).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 (2007).
Goddard, T. D. et al. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018).
Wang, W. et al. Inhibition of HIV-1 maturation via small-molecule targeting of the amino-terminal domain in the viral capsid protein. J. Virol. 91, e02155-16 (2017).
Aiken, C. Pseudotyping human immunodeficiency virus type 1 (HIV-1) by the glycoprotein of vesicular stomatitis virus targets HIV-1 entry to an endocytic pathway and suppresses both the requirement for Nef and the sensitivity to cyclosporin A. J. Virol. 71, 5871–5877 (1997).
Jurado, K. A. et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. Proc. Natl Acad. Sci. USA 110, 8690–8695 (2013).
Hultquist, J. F. et al. CRISPR–Cas9 genome engineering of primary CD4+ T cells for the interrogation of HIV-host factor interactions. Nat. Protoc. 14, 1–27 (2019).
Downey, G. P. et al. Retention of leukocytes in capillaries: role of cell size and deformability. J. Appl. Physiol. 69, 1767–1778 (1990).
Milo, R. What is the total number of protein molecules per cell volume? A call to rethink some published values. Bioessays 35, 1050–1055 (2013).
Schmid-Schönbein, G. W., Shih, Y. Y. & Chien, S. Morphometry of human leukocytes. Blood 56, 866–875 (1980).
Bhattacharya, A. et al. Structural basis of HIV-1 capsid recognition by PF74 and CPSF6. Proc. Natl Acad. Sci. USA 111, 18625–18630 (2014).
Acknowledgements
We thank C. Liu and J. Liu for help with initial manual data collection, Y. Cheng for access to the UCSF Polara and K2 detector, J. Dong and R. Esnouf for computer system support, J. Perilla for scientific discussion and T. Brosenitsch for critical reading of the manuscript. We thank D. Mastronarde for discussions regarding ArbitrEM development. This work was supported by the National Institutes of Health (P50AI150481, P.Z., C.A., A.N.E.), the UK Wellcome Trust Investigator Award 206422/Z/17/Z (P.Z.) and the UK Biotechnology and Biological Sciences Research Council grant BB/S003339/1 (P.Z.). S.G. is funded by a Wellcome Trust PhD Studentship. We acknowledge Diamond for access and support of the Cryo-EM facilities at the UK Electron Bio-Imaging Centre (eBIC, proposal EM14856 and NT21004, P.Z.), funded by the Wellcome Trust, MRC and BBSRC.
Author information
Authors and Affiliations
Contributions
P.Z. conceived the research and, with A.N.E. and C.A., designed the experiments. K.D. developed ArbitrEM for automated targeted data collection. T.N. and J.N. prepared samples for cryo-EM. T.N., K.D. and G.Z. collected cryo-EM data. T.N., S.G. and G.Z. analyzed cryo-EM data. T.N. refined the atomic models. J.N. performed biochemical analysis and negatively stained EM. J.Z., J.S., J.A.-D. and W.L. performed virus infectivity assays and sectioning and imaging particle production. S.J. prepared primary CD4+ T cells and determined the intracellular concentration of CypA. T.N. and P.Z. analyzed atomic models and wrote the paper with support from all the authors.
Corresponding author
Ethics declarations
Competing interests
A.N.E. over the past 12 months has received fees from ViiV Healthcare, Co. for work unrelated to this topic. No other authors declare the potential for competing financial interests.
Additional information
Peer review information Peer reviewer reports are available. Inês Chen was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Automated targeted data collection flowchart.
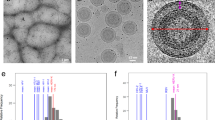
a, The workflow of ArbitrEM, including two steps: target selection and automated data collection. (i) Low-magnification images are firstly acquired to identify holes where tubes are present, and these holes are selected by marking the holes in the images (red crosses); (ii) The images of selected holes are acquired at hole-magnification, centered, and converted into anchor maps; (iii) The acquisition points are marked on anchor maps (boxes), and the beam-image shifts to target the individual acquisition points are calculated. (iv) After all the targets are marked on the hole magnification images, the microscope uses the anchor maps and applies the total beam-image shifts (the stage shift combined with the target-specific beam-image shift) to acquire the high magnification movies. b, A typical low-magnification cryoEM image of CypA-stabilized WT CA assemblies. Scale bar, 2 μm. c, High-magnification image of CypA-stabilized WT CA tubular assembly, illustrating tubes are variable in diameter and easily deformed. d, Gallery of targeted images collected using ArbitrEM. Scale bars, 50 nm.
Extended Data Fig. 2 Data processing and 3D reconstruction flowchart for mature CA hexamer and CypA/CA complex.
a, Individual tubes are cropped from motion-corrected micrographs in EMAN2. b, Tubes are sorted based on their diameters. c, The FFT of each tube is calculated and used to determine helical symmetry. In these data, 12 different helical symmetries exist within a 3 nm diameter variation (red box in b). d, Segments from the same helical symmetry tubes are classified in RELION. e, Iterative Real Space Helical Reconstruction and RELION refinement for each helical symmetry are performed. f, Symmetry expansion and localized reconstruction of individual hexamers (dashed circle) from each tube segment are performed, and C2 symmetry is applied. g, After symmetry expansion, the CA density was subtracted, leaving only the local region of CA and CypA for further classification, as highlighted in the dashed magenta box in e). h, Localized reconstruction was performed to calculate the position of each sub-particle and the extracted sub-particles were subject to 3D-classification into 10 classes without alignment, with a regularization T factor of 10 and resolution limit of 8 Å. i, Classes showing clear features of CypA and CA density were selected, further classified into Mode 1 (one CypA, blue or orange) and Mode 2 (2 CypAs, blue-cyan or orange-pink). j, The Mode 2 classes were aligned and averaged (right), whereas CypA above dimer interface from both Mode 1 and Mode 2 were aligned and averaged (left) to yield the final density maps from each helical symmetry.
Extended Data Fig. 3 3D reconstructions of CA hexamer from five helical symmetries.
a, Electron density maps of CA tubes in the absence and presence of CypA and with different helical symmetries, as indicated. b, Fourier-shell-correlation (FSC) plots of CA hexamer density maps reconstructed from five helical symmetries. The highest resolution at FSC=0.143 is 3.6 Å from (-13, 8) symmetry. c, The FSC plot between the refined CA hexamer model and the 3.6 Å cryoEM density map from (-13, 8) symmetry. d, Representative density map from (-13, 8) symmetry overlapped with the refined atomic model, shown are the helices between adjacent CA monomers in different color. The map was contoured at 2𝜎.
Extended Data Fig. 4 Comparison of hexamers from CA A92E mutant and CypA-stabilized WT CA tubes with the same helical symmetry (-13, 12).
a, Overlay of rigid-body refined CA hexamers from CA A92E (cyan) and CypA-stabilized CA WT (gold) tubes. RMSD between two hexamers is 0.354 Å. CANTD and CACTD were rigid-body fitted into the density map independently. b, Overlay of a seven-hexamer lattice from a CA A92E tube with (-13, 12) symmetry (cyan) and that of a CypA-stabilized CA WT tube with the same symmetry (gold).
Extended Data Fig. 5 B-factors of the mature CA hexamer.
a–c, A sausage representation of B-factors mapped onto a CA monomer (a), hexamer (b), and tri-hexamer (c). The CypA-loop, H4-H5 loop, β-hairpin and H10 display high B-factors. d, e, Close-up views of a dimer interface (d) and a trimer interface (e) circled in (c). The width and coloring of the sausage are directly proportional to the B-factor, from blue (-30) to red (-100). CANTD, CA helices H1-3, CypA-loop, H4-H5 loop, β-hairpin and CACTD H10 are labeled.
Extended Data Fig. 6 Asymmetric and flexible configuration of CA in helical assemblies.
a, Distances between opposing CACTD domains in CA hexamer (measured between Cα of N195) from helical symmetry (-13, 8). b, Correlation plot of distances between CACTD domains versus hexamer array angles from five different helical assemblies. c, d, Close-up views of asymmetric β-hairpins (c, orange) and Arg18 residues adopting two conformations at the hexamer center (d). e, Overlay of electron density map with atomic model, showing two Arg18 side chain positions. f, Overlay of three Arg18 side chains in CA tubular structure (gold) with those in crystal structures in the absence of IP6 (blue, PDB 4WYM, PDB 4XFX, PDB 4XFY, PDB 4XFZ, PDB 5HGL). g, Overlay of Arg18 in crystal structures in complex with IP6 (PDB 6bht), same view as in f. 12 CA molecules from two hexamers (colored in pink and cyan, respectively) in one asymmetric unit were aligned on CANTD.
Extended Data Fig. 7 Binding of PF74 to CA hexamer and its effects on CA tubular assembly.
a, Light scattering of CA assemblies in the presence of PF74 at the indicated concentrations. b, Average number of tubes in each EM micrograph taken at the same magnification, with representatives shown in d–f. c, Distribution of the length of tubular assemblies in the presence of 0 µM (black), 10 µM (green) and 50 µM (pink) PF74. d–f, Representative images of negatively stained CA assemblies in the presence of 0 µM (d), 10 µM (e) and 50 µM (f) PF74. Scale bars, 0.5 µm. g, Atomic model of CA hexamer, clipped to show CANTD. Each CA monomer is labeled. The vertical dashed line indicates the tube axis. h–j, Overlay of density map with atomic model, showing the position of Arg173, which is critical for the binding of PF74 to hexamer57. Three CANTD-CACTD interfaces within the CA hexamer are shown between chains A-F (h), chains B-A (i) and chains C-B (j). Intermolecular hydrogen bonds between CACTD and CANTD are marked by red dashed lines with corresponding distances indicated. k, Overlay of WT and PF74-bound CA crystal structures, colored in grey and cyan, respectively. l, Overlay of three different PF74 pockets from the asymmetric hexamer in (-13,8) helical symmetry with crystal structures (PDB 4XFX and PDB 4XFZ). Arg173 side chains are shown in gold, red and blue, corresponding to panels h–j. The chain C-B interface is more similar to the crystal structure than are the chain A-F and chain B-A interfaces.
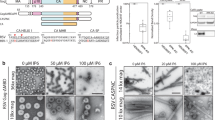
Extended Data Fig. 8 Analysis of CA mutant on virion morphologies and CA assemblies by TEM.
a, Representative images of mature, immature, empty, and eccentric particle morphologies; particles with 1-3 eccentric nucleoids alongside an otherwise electron-lucent core structure were grouped together. Magnification is 30,000X (scale bar, 100 nm). b, Quantitation of core morphology frequencies (average ± SD for n = 2 experiments) for WT and indicated mutant viruses. More than 200 particles were counted for each. Two-tailed Student’s t-test revealed significant differences for indicated categories versus the WT (****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05). For V181T, only 14 particles (predominantly immature) were observed in total. V181T was accordingly omitted from the graph and statistical analyses. c–g, Negative-stained images of WT and mutant CA assemblies. V181C forms two-dimensional crystalline sheets, whereas V181D forms tubular assemblies with much smaller diameters compared to WT tubes (17 nm vs 50 nm). Other mutants failed to form ordered assemblies. h, i, Close-up view of WT (h) and V181D (i) assemblies on the same scale. Scale bars, 200 nm in c–g, 50 nm in h, i.
Extended Data Fig. 9 3D reconstruction of CypA/CA complexes. Local resolution estimation of CypA/CA complex from different helical symmetries.
a, Local resolution estimation of CypA/CA complex from four different helical symmetries, colored by local resolution from blue to red. b, Fourier-shell-correlation (FSC) plots of CypA/CA density maps reconstructed from four helical symmetries. The highest resolution at FSC=0.143 is 4.0Å from (-13, 8) symmetry. c, Gallery of CypA/CA complexes. Eight helical reconstructions are shown in cross-sectional view (left) and face-on view (middle). Density maps are colored by the radius, with regions corresponding to CACTD, CANTD and CypA colored in yellow, green and blue, respectively. The outer diameters of the tubes are indicated. Enlarged views from regions marked by red dashed ovals are shown on the right.
Extended Data Fig. 10 Semi-quantitative analysis of CypA levels in primary CD4+ T cells.
a, CD4+ T cell CypA levels from two different blood donors (labeled D1 and D2) were compared by immunoblotting versus a dilution series of purified recombinant CypA protein. Where indicated, cells were cultured with 5 μg/ml of phytohaemagglutinin (+PHA) for 48 h prior to lysis. Lamin B1 was monitored as a loading control. Approximate ng amount of Cyclophilin A in each of the last four lanes is marked in red color. b, Signal intensity of each band in the dilution series in A was plotted against protein amount to generate a standard curve. Uncropped blot images are available as source data.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Video 1
Overview of CypA/CA complex on top of dimer and trimer interfaces.
Supplementary Video 2
Overview of CypA-decorated tubular assembly of HIV capsid.
Supplementary Video 3
Overview of CypA-decorated conical HIV capsid.
Source data
Source Data Fig. 2
Uncropped SDS–PAGE.
Source Data Extended Data Fig. 10
Uncropped immunoblot images.
Rights and permissions
About this article
Cite this article
Ni, T., Gerard, S., Zhao, G. et al. Intrinsic curvature of the HIV-1 CA hexamer underlies capsid topology and interaction with cyclophilin A. Nat Struct Mol Biol 27, 855–862 (2020). https://doi.org/10.1038/s41594-020-0467-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41594-020-0467-8
This article is cited by
-
Structural basis for HIV-1 capsid adaption to a deficiency in IP6 packaging
Nature Communications (2025)
-
HIV-1 nuclear import is selective and depends on both capsid elasticity and nuclear pore adaptability
Nature Microbiology (2025)
-
Direct visualization of HIV-1 core nuclear import and its interplay with the nuclear pore
EMBO Reports (2025)
-
HIV-1 adapts to lost IP6 coordination through second-site mutations that restore conical capsid assembly
Nature Communications (2024)
-
Open architecture of archaea MCM and dsDNA complexes resolved using monodispersed streptavidin affinity CryoEM
Nature Communications (2024)