Abstract
Human bone marrow mesenchymal stem cells (hBMSCs) are adult stem cells residing in the bone marrow, characterized by their capacity for multi-directional differentiation, self-renewal, migration, and engraftment. Serving as seed cells, BMSCs play a pivotal role in the regeneration of bone defects. Hence, investigating the transcription factors and signaling pathways involved in the regulation of osteogenic differentiation in BMSCs holds significant importance. Recent research has unveiled that certain circular RNAs (circRNAs) can function as molecular sponges, influencing the osteogenic differentiation process of mesenchymal stem cells. However, many circRNAs remain undiscovered, and their precise mechanisms remain elusive. Therefore, the objective of this study is to construct an osteogenic differentiation-related circRNA-miRNA-mRNA network in hBMSCs. Subsequently, through bioinformatics analysis, we constructed a ceRNA network related to the osteogenic differentiation ability of hBMSCs, comprising 22 circRNAs, 17 miRNAs, and 15 mRNAs. The potential circRNA-miRNA-mRNA axes, including the role of hsa_circ_0001600 in promoting the osteogenic differentiation of hBMSCs through the targeted regulation of hsa-miR-542-3p, were validated through in vitro experiments.
Similar content being viewed by others
Introduction
Mesenchymal stem cells (MSCs) hold a prominent position in regenerative medicine due to their multipotency, self-renewal capabilities, and low immunogenicity. MSCs possess the remarkable ability to differentiate into various cell types, including osteoblasts, adipocytes, and chondrocytes, both in vivo and in vitro1. Among MSCs, human bone marrow mesenchymal stem cells (hBMSCs) are extensively utilized in clinical trials for bone tissue regeneration and show promising therapeutic outcomes for skeletal disorders such as bone defects and osteoporosis2.
Glucocorticoids and dexamethasone (DEX) are commonly employed clinically to induce osteogenic differentiation in BMSCs3. In recent years, there has been significant attention directed towards understanding the osteogenic differentiation of BMSCs, which involves a myriad of transcription factors and signaling pathways4.
Competing endogenous RNA (ceRNA), proposed in 2011, introduced a novel mechanism of RNA interaction, expanding the biological functions of mRNA and non-coding RNA (ncRNA)5,6. Besides, circular RNAs (circRNAs) or long non-coding RNAs (lncRNAs) primarily act as molecular sponges for miRNAs, adsorbing miRNAs and relieving their suppressive effects on target mRNA7. CircRNA, characterized by a covalently closed, circular structure, is derived from the reverse splicing of mRNA precursors and has emerged as a significant component of the transcriptome8,9. In recent years, the functional and clinical significance of ceRNA networks in various diseases, including cancer, cardiovascular diseases, and neurological disorders, has been increasingly recognized, offering potential avenues for disease treatment research10,11. In 2017, Yu et al. first reported the correlation between circRNAs and stem cell differentiation, confirming that downstream circRNAs regulate stem cell differentiation through the ceRNA mechanism12. Recent studies have identified numerous circRNAs that exhibit differential expression during MSC osteogenesis, some of which have been shown to regulate the osteogenic differentiation process13,14,15,16. However, the specific roles of many circRNAs and their associated ceRNA networks in BMSC osteogenic differentiation remain unexplored. Therefore, to gain a comprehensive understanding of the impact of the ceRNA mechanism on BMSC osteogenic differentiation, it is imperative to elucidate the circRNA-miRNA-mRNA regulatory network.
In this study, we employed bioinformatic analysis to analyze RNA expression data and establish a competitive regulatory network of circRNA-miRNA-mRNA during early osteogenic differentiation of hBMSCs. Furthermore, we verified the key circRNAs’ important binding sites through in vitro studies. The objective of this research is to uncover detailed molecular mechanisms and potential biomarkers involved in BMSCs osteogenic differentiation.
Results
Identification of differentially expressed circRNAs, miRNAs and mRNAs
To investigate the early molecular events underlying osteogenic differentiation, we obtained three microarray datasets which were listed in Table S3. These datasets profiled the expression patterns at 7 days post chemical induction of osteogenic differentiation in hBMSCs. We included mRNA, miRNA, and circRNA expression profiles of BMSCs undergoing osteogenesis and normal cells in our study.
A total of 80 differentially expressed circRNAs (DE-circRNAs) were identified in the osteogenic induction group compared to the uninduced group in BMSCs at 7 days, with a significance threshold of P < 0.05 and |log2 fold change|> 0.5 (Fig. 1a). Among these, 41 circRNAs were upregulated, while 39 were downregulated. Additionally, 98 differentially expressed miRNAs (DE-miRNAs) were detected during osteogenic differentiation (P < 0.05, |log2 fold change|> 0.5) (Fig. 1b), with 41 upregulated and 57 downregulated DE-miRNAs. Furthermore, a total of 814 differentially expressed mRNAs (DE-mRNAs) were identified using a significance threshold of P < 0.05 and |log2 fold change|> 1.2, comprising 478 upregulated and 336 downregulated DE-mRNAs (Fig. 1c).
The volcano plot shows the differential expression of circRNAs, miRNAs, and mRNAs between the normal culture group and the osteogenic induction group hBMSCs. (a) DE-circRNAs (P < 0.05, |log2 fold change|> 0.5); (b) DE-miRNAs (P < 0.05, |log2 fold change|> 0.5); (c) DE-mRNAs(P < 0.05, |log2 fold change|> 1.2); Red dots represent upregulated gene expression, blue dots represent downregulated gene expression.
Subsequently, volcano plots were generated using the pheatmap package, and 80 DE-circRNAs, 98 DE-miRNAs, and 814 DE-mRNAs were selected for further analysis.
Function enrichment analysis
To explore the role of circRNAs as miRNA sponges in modulating the osteogenic differentiation of BMSCs, we utilized an online database to predict circRNAs corresponding to the identified miRNAs. Our analysis revealed that 61 DE-circRNAs interacted with 3184 miRNAs from circbank. Furthermore, by intersecting the circRNAs-targeted miRNAs with the DE-miRNAs from GEO, we identified 56 overlapping miRNAs (Fig. 2a). Subsequently, we predicted that 7,648 mRNAs contained binding sites for the overlapped miRNAs. Through further analysis, we detected 215 overlapping mRNAs by intersecting the miRNAs-targeted mRNAs with the DE-mRNAs (Fig. 2b).
Construction of the circRNA–miRNA–mRNA Interaction Network. (a) Venn diagram of the intersection of molecules predicted to be combined with circRNAs and DE-miRNAs by circBank database; (b) Venn diagram of the intersection of molecules predicted to be combined with miRNAs and DE-mRNAs by mirTarBase database. (c) GO function annotation bar graph(GO enrichment analysis includes Cellular Component (CC), Molecular Function (MF), and Biological Process (BP); P < 0.05.). (d) ceRNA interaction network diagram of circRNA-miRNA-mRNA (Green ellipses represent circRNA, blue rectangles represent miRNA, and orange rhombuses represent mRNA).
To elucidate the functional roles of these 215 mRNAs, we conducted GO enrichment analysis using the DAVID online database. Our analysis revealed that these mRNAs were significantly enriched in biological processes related to transcriptional regulation, proliferation, and differentiation (P < 0.05) (Fig. 2c). Notably, 35 of these mRNAs were specifically enriched in biological processes associated with osteogenesis and cell proliferation, including GO: 0,008,284 ~ positive regulation of cell proliferation, GO: 0,001,503 ~ ossification, GO: 0,008,285 ~ negative regulation of proliferation, and GO: 0,002,053 ~ positive regulation of mesenchymal cell proliferation. These processes are closely associated with osteogenesis, calcification, and cell differentiation. Consequently, we selected the 35 mRNAs enriched in these processes for further investigation.
Construction of the circRNA–miRNA–mRNA interaction network related to osteogenic differentiation
Based on the analysis of differential expression in microarray data and the ceRNA mechanism, we identified a set of interconnected molecules to construct a circRNA-miRNA-mRNA network relevant to osteogenic differentiation. Utilizing published literature, we selected 15 mRNAs closely associated with stem cell differentiation (TableS1). Among the 56 overlapping miRNAs, 17 were found to interact with the previously mentioned 15 mRNAs. Furthermore, out of the 61 differentially expressed circRNAs, 22 were found to interact with the 17 miRNAs. Consequently, we posit that these 22 circRNAs, 17 miRNAs, and 15 mRNAs mutually interact and are closely linked to hBMSCs osteogenic differentiation. Employing Cytoscape software, we constructed a ceRNA network diagram containing:
circRNAs: hsa_circ_0001063, hsa_circ_0001600, hsa_circ_0002415, hsa_circ_0002474, hsa_circ_0003552, hsa_circ_0003563, hsa_circ_0003611, hsa_circ_0004418, hsa_circ_0005991, hsa_circ_0006006, hsa_circ_0007933, hsa_circ_0008621, hsa_circ_0016956, hsa_circ_0022502, hsa_circ_0034293, hsa_circ_0057104, hsa_circ_0057105, hsa_circ_0063756, hsa_circ_0068465, hsa_circ_0072387, hsa_circ_0072678, hsa_circ_0088062
miRNAs: hsa-miR-199b-5p, hsa-miR-15a-5p, hsa-miR-424-5p, hsa-miR-504-5p, hsa-miR-383-5p, hsa-miR-335-5p, hsa-miR-20b-5p, hsa-miR-200b-3p, hsa-miR-30c-2-3p, hsa-miR-30a-5p, hsa-miR-203a-3p, hsa-let-7i-5p, hsa-miR-942-5p, hsa-miR-9-3p, hsa-miR-335-3p, hsa-miR-3200-3p, hsa-miR-542-3p
mRNAs: LIF, ZBTB16, VEGFA, SOST, SOD2, SMAD1, SFRP4, LRRC17, IL6, IGF2, FOXP1, FGF9, CLEC3B, BMP7, AREG (Fig. 2d).
Validation of key circRNAs and miRNAs in the ceRNA network
In the control group, hBMSCs were cultured in complete medium without osteogenic induction. In contrast, the experimental group hBMSCs were treated with osteogenic induction medium for 7 days, corresponding to the matrix maturation stage. This time point is critical for capturing early differentiation events17. RT-qPCR was performed to assess the expression levels of 11 molecules corresponding to differentially expressed circRNAs identified through bioinformatics analysis. Compared to the control group, the expression levels of hsa_circ_0001600, hsa_circ_0002415, hsa_circ_0063756, hsa_circ_0072678, and hsa_circ_0005991 were significantly upregulated in the 7-day osteogenic induction group, while the expression of hsa_circ_0008621 was downregulated (P < 0.05) (Fig. 3a). Similarly, compared to the control group, the expression levels of hsa-miR-20b-5p, hsa-miR-335-3p, hsa-miR-942-5p, hsa-miR-424-5p, and hsa-miR-542-3p were significantly decreased in the osteogenic induction group, whereas the expression of hsa-miR-203a-3p, hsa-miR-30a-5p, hsa-miR-30c-2-3p, and hsa-miR-199b-5p was increased (P < 0.05) (Fig. 3b).
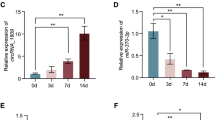
Certain circRNA molecules involved in the regulation of BMSC osteogenic differentiation. (a) qPCR detection of the expression level changes of circRNAs screened by bioinformatics analysis after osteogenic differentiation induction. (b) qPCR detects the expression level changes of miRNAs screened by bioinformatics analysis after osteogenic differentiation induction. (c) qPCR detects the expression level changes of hsa_circ_0001600 and hsa-miR-542-3p during osteogenic differentiation at 0 days, 3 days, and 7 days. (d) qPCR detects the expression level changes of hsa_circ_0005991 and hsa-miR-424-5p during osteogenic differentiation at 0 days, 3 days, and 7 days. (e) Transfection of hsa_circ_0001600 expression in hBMSCs at a siRNA concentration of 50 nM for 48 h. (f) qPCR detects the changes in hsa-miR-542-3p levels in the si-hsa_circ_0001600 2 days after transfection. Compared with the si-NC, the expression of hsa-miR-542-3p increased in the si-hsa_circ_0001600. (si-hsa_circ_0001600 is the hsa_circ_0001600 siRNA transfected knockdown group, si-NC is the negative control group; * represents P < 0.05; ** represents p < 0.01; *** represents p < 0.001, **** represents p < 0.0001).
The interaction between circRNAs and miRNAs within the ceRNA network
During the osteogenic differentiation of hBMSCs, the expression of hsa_circ_0005991 and hsa_circ_0001600 shows an increasing trend, indicating that the two molecules may be involved in the regulation of cell osteogenic differentiation. hsa_circ_0001600 and hsa_circ_0005991 were selected as candidate circRNAs. According to the circBank database prediction, hsa_circ_0001600 (circBank ID: hsa_circFKBP5_002) has one binding site (position: 739) with hsa-miR-542-3p in the miRanda algorithm, and four binding sites (positions: 3624, 752, 3629, 758) in the Targetscan algorithm. hsa_circ_0005991 (circBank ID: hsa_circAPBB2_013) has one binding site (position: 584) with hsa-miR-424-5p in the miRanda algorithm prediction, and two binding sites (positions: 598, 604) according to the Targetscan algorithm. hBMSCs were osteogenically induced for 0, 3, and 7 days, and qPCR was employed to assess the expression levels of hsa_circ_0001600, hsa-miR-542-3p, hsa_circ_0005991, and hsa-miR-424-5p. The findings revealed a progressive increase in the expression of hsa_circ_0001600 throughout osteogenic induction (Fig. 3c), while the expression of hsa-miR-542-3p decreased gradually with prolonged osteogenic induction. Additionally, the expression of hsa_circ_0005991 exhibited a gradual increase over the osteogenic induction period (Fig. 3d), whereas the expression of hsa-miR-424-5p decreased progressively with the extension of osteogenic induction time.
The experimental design included the use of siRNA to suppress the expression of hsa_circ_0001600. Initial investigation was conducted to determine the optimal siRNA, transfection concentration, and duration. The determined concentration of the target gene siRNA was 50 nM, and the transfection duration was 48 h. qPCR analysis indicated a significant reduction in hsa_circ_0001600 expression with si-hsa_circ_0001600 2 in hsa_circ_0001600 siRNA, achieving a knockdown efficiency of 70% (Fig. 3e). The hsa_circ_0001600 siRNA transfection knockdown group exhibited an increase in hsa-miR-542-3p expression compared to the negative control group (Fig. 3f).
Downregulation of hsa_circ_0001600 expression reduces the osteogenic differentiation capacity of hBMSCs
The circular RNA hsa_circ_0001600 is generated through the reverse splicing of the FKBP5 gene, representing a newly discovered circular molecule with unknown biological function and molecular mechanism (Fig. 4a). Compared to linear RNAs, circRNAs are more stable and longer-lasting because they lack a free end for RNA enzyme-mediated degradation. Circular RNAs are generated by the back-splicing process, and they are covalently closed loops, keeping them highly stable to RNase R digestion. Using qPCR, the expression changes of hsa_circ_0001600 and its parental gene FKBP5 in RNase R-digested total RNA (RNase R +) were analyzed. The non-digested group (RNase R-) served as the control group, with the internal reference in the RNase R- group used as the calculation standard. The qPCR results demonstrated no significant alteration in the expression level of hsa_circ_0001600 in the experimental group compared to the control group. However, there was a notable reduction in the expression of FKBP5, indicating that hsa_circ_0001600 may be resistant to RNase R, whereas its parental gene FKBP5 is predominantly digested by RNase R (Fig. 4b).
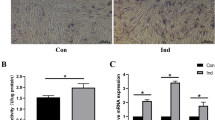
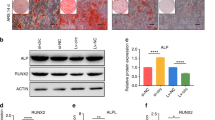
(a) hsa_circ_0001600 generation model diagram; (b) qPCR detects the expression of hsa_circ_0001600 and FKBP5 before and after RNase R treatment. RNase R + represents the group treated with RNase R enzyme, RNase R- represents the group without RNase R enzyme treatment; (c) The left side shows alizarin red staining after 14 days of osteogenic induction, and the right side shows ALP staining after 3 days of osteogenic induction. (i) and (iii) are the control group, and (ii) and (iv) are the hsa_circ_0001600 siRNA transduction knockdown group; the results showed that compared with the control group, the degree of alizarin red and ALP staining was significantly weakened in the hsa_circ_0001600 siRNA transduction knockout group. (d) qPCR detection of changes in osteogenesis-related gene expression levels after 3 days of osteogenic induction. In comparison to the si-NC, the expression of COL1A1 and RUNX2 was downregulated in the si-hsa_circ_0001600. (e) Western Blot detection of protein changes after 3 days of osteogenic induction; the band diagram shows that the bands of COL1A1, OCN, and RUNX2 in the si-hsa_circ_0001600 are noticeably lighter compared to the si-NC. (f) The protein quantification diagram shows reduced protein expression levels of COL1A1, OCN, and RUNX2 in the si-hsa_circ_0001600 compared to the si-NC. (si-hsa_circ_0001600 is the hsa_circ_0001600 siRNA transfected knockdown group, si-NC is the negative control group; * represents P < 0.05; ** represents p < 0.01; *** represents p < 0.001, **** represents p < 0.0001).
hBMSCs were seeded into 6-well plates and induced with osteogenic induction medium for 14 days. Throughout this period, siRNA (si-hsa_circ_0001600 and si-NC) was refreshed every 3 days. Alizarin Red staining was performed for both the control and experimental groups. Microscopic observation revealed a significant reduction in red staining in the hsa_circ_0001600 siRNA knockdown group compared to the control group. Furthermore, after transfecting hBMSCs with hsa_circ_0001600 siRNA for 2 days, the medium was switched to osteogenic induction medium for 3 days. The control group comprised hBMSCs transfected with negative control siRNA for 2 days, followed by a 3-day osteogenic induction. ALP staining was conducted for both the control and experimental groups. Microscopic observation revealed a significant reduction in red staining in the hsa_circ_0001600 siRNA knockdown group compared to the control group, indicating a clear decrease in ALP expression (Fig. 4c). qPCR analysis was carried out for both the control and experimental groups, demonstrating reduced expression of osteogenic-related genes COL1A1 and RUNX2 in the knockdown group with hsa_circ_0001600 siRNA transfection, as compared to the negative control group (Fig. 4d). Furthermore, Western Blot analysis showed decreased expression of osteogenic-related proteins COL1A1, OCN, and RUNX2 in the knockdown group with hsa_circ_0001600 siRNA transfection, as compared to the negative control (Fig. 4e, 4f). The raw bands are shown in Figure S1.
Discussion
Osteogenic differentiation is a vital step in bone formation, during which BMSCs undergo a complex progression from osteoprogenitor cells to pre-osteoblasts, osteoblasts, and ultimately osteocytes. This intricate process involves a variety of intercellular and intracellular signaling pathways18. Increasing evidence suggests that circRNAs and miRNAs play a crucial regulatory role in bone remodeling by directly or indirectly modulating the expression of osteogenesis-related genes19. In the bioinformatics analysis section of this study, we gathered gene expression matrix data of circRNAs, miRNAs, and mRNAs during the early osteogenic differentiation of hBMSCs induced by osteogenic induction medium for 7 days. Through predictive modeling, we identified 22 circRNAs, 17 miRNAs, and 15 mRNAs predicted to interact with each other, successfully constructing a ceRNA network relevant to hBMSCs osteogenic differentiation.
Members of the Bone Morphogenetic Proteins (BMPs) family, such as BMP7, known for their upregulation during BMSCs osteogenic differentiation, were included20. BMPs are multifunctional transcription factors and cytokines that belong to the Transforming Growth Factor-β (TGF-β) family. In humans, BMPs primarily regulate osteogenesis, bone formation, and chondrogenesis. Of the over 20 BMPs identified, BMP2, 4–7, and 9 are strongly linked to the osteogenic process21. Research on in vitro models of MSCs has shown that circRNAs regulate several genes involved in the BMP pathway. Specifically, hsa_circ_0000020 enhances osteogenic differentiation and mitigates osteoporosis by acting as a sponge for microRNA miR-142-5p, which leads to the upregulation of Bone Morphogenetic Protein BMP-222. hsa_circ_0007059 is crucial for osteoclastogenesis through the miR-378/BMP-2 signaling pathway. Targeting the circ_0007059/miR-378/BMP-2 axis may represent a novel approach for treating osteoporosis23. Among the predicted 17 miRNAs, studies have shown that downregulation of miR-542-3p can target BMP7 to promote mouse vascular smooth muscle cell and osteoblast osteogenic differentiation24,25. In vitro cell experiments proved that hsa_circ_0001600 may play an important role in the osteogenic differentiation of hBMSCs, and its possible mechanism is the targeted regulation of hsa-miR-542-3p. hsa_circ_0001600 is a circular RNA formed by the reverse cleavage of the FKBP5 gene, with a length of 4534 base pairs. CircRNA can also directly regulate the expression of the host gene. The host gene FKBP5 of hsa_circ_0001600 is a molecular associated with heat shock proteins and has a strong stress response26. Mutations in FKBP5 are significantly associated with the risk of mental disorders such as anxiety, depression, and post-traumatic stress disorder (PTSD)27. Studies have shown that FKBP5 also plays a positive role in the early osteogenic differentiation of bone marrow mesenchymal stem cells and adipose stem cells, as well as in cartilage and adipogenic differentiation28,29. Furthermore, FKBP5 mutations may lead to Paget’s disease by affecting osteoclast formation30. Overall, hsa_circ_0001600 may regulate osteogenic differentiation by directly regulating the host gene. Meanwhile, qPCR proves that hsa-miR-542-3p is negatively correlated with BMSCs osteogenic differentiation and knocking down hsa_circ_0001600 will increase the expression of hsa-miR-542-3p. Previous studies have shown that hsa-miR-542-3p inhibits the proliferation, migration, and invasion of tumor cells such as osteosarcoma and glioma cells31,32,33. On the other hand, miR-542-3p positively regulates differentiation and regulates osteogenesis under conditions of bone loss24. At the same time, hsa-miR-542-3p has strong osteogenic potential by bioinformatics prediction targeting the highly bone-inducing BMP7. Therefore, we speculate that hsa-miR-542-3p is regulated by hsa_circ_0001600, inhibiting the osteogenic differentiation of hBMSCs by targeting BMP7.
Due to the rich vascularization of bones, angiogenesis is closely linked to osteogenesis, and both processes can be mutually influenced by shared regulatory factors. Vascular Endothelial Growth Factor A (VEGFA) as an inducer of angiogenesis, has been shown to regulate bone repair and regeneration by affecting the generation of bone vasculature34. Hsa_circ_0074834 functions as a ceRNA to regulate the expression of ZEB1 and VEGF by sponging miR-942-5p, thereby promoting both osteogenic differentiation and vasculogenesis in BMSCs35. Fibroblast Growth Factor 9 (FGF9) is a factor regulating skeletal development, playing a significant role in skeletal development and cartilage formation. lncRNA-H19 (H19) positively regulates the odontoblastic differentiation of human dental pulp stem cells (hDPSCs). The research indicated that H19 exerts its regulatory function through the miR-140-5p/BMP-2/FGF9 axis, suggesting its potential role as a stimulatory regulator in odontogenesis36. However, the role of circRNAs in targeting FGF9 via miRNA remains unknown and warrants further investigation.
In addition to the BMP pathway, the Wnt/β-catenin and TGF-β signaling cascades are key pathways that drive the osteogenic differentiation of MSCs. In the ceRNA network, miR-30a-5p can regulate hBMSCs osteogenic differentiation by targeting SMAD1. During the phenotypic switch of vascular smooth muscle cells, the increased expression of circCOL1A1 counteracts the inhibitory influence of miR-30a-5p on its target, SMAD1, thereby reducing the suppression of the TGF-β signaling pathway37. Secreted Frizzled-Related Protein 4 (SFRP4) can directly activate the canonical Wnt signaling pathway, and through regulating osteoblasts and osteoclasts, it plays a key role in bone development and remodeling, and reducing its expression can prevent age-related bone loss38. The ceRNA network highlights an interaction between SFRP4 and miR-942-5p, which is closely tied to the osteogenic differentiation of BMSCs39. Investigating how circRNA influences SFRP4 through miR-942-5p to enhance osteogenesis could provide valuable insights for future studies.
Alternatively, a single circRNA can negatively regulate several miRNAs that are involved in pathways related to osteogenesis. For example, hsa_circ_0001600 functions as a molecular sponge for specific miRNAs: hsa-miR-542-3p and hsa-miR-20b-5p and hsa-miR-200b-3p. Research has indicated a close association of hsa-miR-335-5p, hsa-miR-200b-3p, and hsa-miR-335-3p with osteoarthritis40. Among the 22 circRNAs unearthed in this study, hsa_circ_0008621 and hsa_circ_0072387 can inhibit glioma progression through miR-338-5p/IKBIP41. hsa_circ_0072387 is a novel circRNA that can serve as a valuable biomarker for oral squamous cell carcinoma42,43. However, the regulatory roles of the screened circRNAs and its ceRNA network in osteogenic differentiation are not yet clear, and further research on their regulatory mechanisms is needed.
We are confident that our research will significantly deepen our knowledge of the molecular mechanisms driving human bone remodeling and related bone disorders. These findings on circRNAs are expected to stimulate the development of innovative preclinical and clinical studies, potentially leading to new therapeutic approaches for treating bone diseases. Zhang et al. demonstrated the stable presence of ncRNA in human and animal serum and plasma. This stability makes ncRNAs valuable as biomarkers in liquid biopsies, providing strong theoretical support for their application in clinical diagnostics44. Extracellular vesicles hold great potential in the treatment of orthopedic diseases. These vesicles can transport proteins, ncRNA, and other biomolecules, facilitating intercellular communication and promoting tissue repair45,46. Research indicates that exosomal circ_0001236 is essential for chondrogenic differentiation, promoting cartilage-specific gene and protein expression via the miR-3677-3p/Sox9 axis. This suggests that circRNA_0001236-enriched exosomes could prevent cartilage degradation and enhance repair in osteoarthritis, offering a potential therapeutic strategy47. These findings suggest that the circRNAs we identified could greatly improve diagnostic and therapeutic applications for bone diseases.
Methods
Dataset collection
The microarray data involved in this study were obtained from GEO(Gene Expression Omnibus) database((http://www.ncbi.nlm.nih.gov/geo/). The circRNA, miRNA and mRNA expression profiles downloaded from GSE135883 (GPL21825 Arraystar Human CircRNA microarray V2,6 pairs of hBMSCs), GSE135586 (GPL18573 Illumina NextSeq 500 (Homo sapiens), 6 pairs of hBMSCs) and GSE18043 (GPL570, Affymetrix Human Genome U133 Plus 2.0 Array, 3 pairs of hBMSCs), respectively(TableS2). The inclusion criteria for these three groups are hBMSCs chemically induced chemically osteogenic differentiation for 7 days48.
Differential expression analysis
The expression matrix used FastQC to complete quality checking. By using R Software 4.2.3 (https://www.R-project.org) limma package Subsequently, the DEcircRNAs, DE-miRNAs, and DE-mRNAs were screened out and drawn with the R package pheatmap. The criteria of P < 0.05 and |log2Fold change (FC)|> 1.2 were identified as significantly DE-mRNAs. For circRNAs and miRNAs, we utilized the criteria of P < 0.05 and |log2Fold change (FC)|> 0.5 to identify significantly DE-circRNAs and DE-miRNAs, respectively. Subsequently. The pheatmap package in R software was used to generate volcano plots for the differentially expressed genes.
Construction of ceRNA network
To discover and establish the potential relationship between the DE-circRNAs, DE-miRNAs, and DE-mRNAs, two databases were selected named circBank (http://www.circbank.cn) and miRTarBase (http://miRTarBase.cuhk.edu.cn/)49 to predict the targeting miRNAs and mRNAs. The circRNA–miRNA–mRNA network was constructed with intersection of miRNA and mRNA predicted by online databases and DE-circRNAs, DE-miRNAs, and DE-mRNAs. A ceRNA network was shown using Cytoscape 3.7.2 software.
Functional enrichment analysis
To find genes that are closely related to our research, we used the DAVID database 2021 (DAVID Functional Annotation Tools (ncifcrf.gov)) to perform Gene Ontology (GO) enrichment analysis of DE-mRNAs. The reference gene sets were performed with GO-BP, GO-CC and GO-MF. The results with adjusted p < 0.05 were regarded as the thresholds for selecting significant GO terms.
BMSCs culture and osteoblast differentiation
The bone marrow-derived mesenchymal stem cells (three donors) were purchased from ScienCell Research Laboratories (Carlsbad, CA, USA). The BMSCs were cultured in mesenchymal stem cell medium (MSCM) (ScienCell, CA, United States) containing 5% fetal bovine serum, 1% penicillin–streptomycin, and 1% mesenchymal stem cell growth supplement. The culture medium was replenished every 2–3 days, and the cells were maintained at 37 °C in a 5% CO2 cell culture incubator. Subsequent experiments utilized hBMSCs from the 3rd to 5th passages.
For osteogenic differentiation, hBMSCs were seeded in six-well plates at a density of 1 × 104 cells/cm2. Upon reaching 80–90% confluency, the routine media was replaced with osteogenic media containing 50 μg/ml ascorbic acid, 10 mM β-glycerophosphate (Sigma, USA), 10 nM dexamethasone(Invitrogen, USA), 10% FBS, 1% penicillin–streptomycin and 1% glutamine. The media was replenished every 2–3 days. After 3 days or 7 days, osteogenic differentiation was evaluated by ALP staining and qPCR.
Alizarin red staining
BMSCs osteogenic capacity was detected using alizarin red staining. The hBMSCs were inoculated into 6-well plates and induced with osteogenic induction medium for 14 days, during which siRNA (si-hsa_circ_0001600 and si-NC) was re-placed every 3 days. The plates were rinsed with PBS, fixed with 4% paraformaldehyde for 30 min, washed with ddH2O, and then stained with 1% alizarin red aqueous solution (Solarbio, Beijing, China) for 15 min at room temperature, then washed with ddH2O to observe the staining under the microscope.
Alkaline phosphatase (ALP) staining
ALP activity was analyzed using the NBT/BCIP staining kit (Beyotime, Shanghai, China). hBMSCs were seeded in 6-well plates and cultured in osteogenic medium for 7 days. Cells were washed with PBS and fixed with 4% polyformaldehyde for 30 min and stained for 20 min at 37 ℃.
Real-time quantitative reverse transcriptase PCR (RT-qPCR) and RNase R enzyme digestion
RT-qPCR was used to check the expression of miRNAs, circRNAs, and mRNAs and to confirm the expression of osteogenic differentiation markers. The hBMSCs nuclear and cytoplasmic RNA was isolated with the TRIzol reagent (Invitrogen, USA) at osteogenic 3 days or 7 days. For cDNA synthesis, a stem loop reverse transcription kit (Vazyme, China) was used for reversing mature miRNA. circRNAs and mRNA were reverse transcribed using HiScript Reverse Transcriptase (Vazyme, China). The qPCR for detecting gene expression levels was performed by SYBR Green PCR Mix (Vazyme, China) and a QuantStudioTM 3 System (Applied Biosystems). The GAPDH (for mRNA and circRNA) and U6 (for miRNA) were used as endogenous controls respectively. All primers used in this study were listed in Table S3. After extracting total circRNA, the circRNA was treated with 2U/μg RNase R(GeneSeed, China)digested at 37 °C for 10 min to digest most of the linear RNA.
Cell transfection
The si-hsa_circ_0001600 and si-hsa_circ_0005991 and their control vector si-NC were designed and synthesized by Ribobio (Guangzhou, China). The siRNA target sequences were listed in Table S4. The BMSCs were transfected with siRNA(50 nM) and si-NC in 6-well plates until 70–80% confluence with riboFECTTM CP kit (Ribobio, Guangzhou, China). The transfection was conducted for up to 48 h, unless specified otherwise, followed by subsequent cell treatments.
Western blot analysis
The BMSCs were harvested and lysed in RIPA lysis buffer (Beyotime, Shanghai, China) after washing twice by PBS and the corresponding proteins were determined by the BCA protein analysis kit (Beyotime), according to the manufacturer’s guide. Subsequently, equal quantities of protein samples were added to 12% SDS-PAGE and onto polyvinylidene difluoride (PVDF) membranes after separation. Then, the PVDF membranes were incubated with primary antibody: against RUNX2 (1:1000, Abcam), OCN (1:1000, Abcam), COL1A1 (1:1000, Proteintech), and β-ACTIN (1:1000, Abcam) overnight at 4 °C. The membranes were incubated with corresponding secondary antibodies for 1 h after washing with PBST three times. The band intensity was measured by Image J software (version 8.0). The signal of all target bands was normalized to that of the β-ACTIN band.
Statistical analysis
All statistical calculations were performed with GraphPad Prism software (version 8.0). The Student’s t-test, Fisher’s exact test, χ2 test, Pearson correlation, or one-way ANOVA were used for statistical analyses in our search. Meanwhile, data are presented as means ± standard deviation (SD). P < 0.05 were considered significant statistically.
Data availability
Sequence data that support the findings of this study have been deposited in the National Center of Biotechnology Information with the primary accession code GSE18043, GSE135883, GSE135586.
References
Tavakoli, S. et al. Mesenchymal stromal cells; a new horizon in regenerative medicine. J. Cell. Physiol. 235(12), 9185–9210 (2020).
Zhou, W. et al. Single-cell profiles and clinically useful properties of human mesenchymal stem cells of adipose and bone marrow origin. Am. J. Sports Med. 47(7), 1722–1733 (2019).
Li, X., Xu, L., Nie, H. & Lei, L. Dexamethasone-loaded β-cyclodextrin for osteogenic induction of mesenchymal stem/progenitor cells and bone regeneration. J. Biomed. Mater. Res. 109(7), 1125–1135 (2021).
Chen, Q. et al. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts?. Cell Death Differ. 23(7), 1128–1139 (2016).
Sen, R., Ghosal, S., Das, S., Balti, S. & Chakrabarti, J. Competing endogenous RNA: the key to posttranscriptional regulation. Sci. World J. 2014, 1–6 (2014).
Salmena, L., Poliseno, L., Tay, Y., Kats, L. & Pandolfi, P. P. A ceRNA hypothesis: the rosetta stone of a hidden RNA language?. Cell. 146(3), 353–358 (2011).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: evidence and controversy. Nat. Rev. Genet. 17(5), 272–283 (2016).
Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell. Biol. 21(8), 475–490 (2020).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20(11), 675–691 (2019).
Cui, X. et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol. Cancer. 17(1), 123 (2018).
Zhong, Y. et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer. 17(1), 79 (2018).
Yu, C.-Y. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 8(1), 1149 (2017).
Wang, Y., Jiang, Z., Yu, M. & Yang, G. Roles of circular RNAs in regulating the self-renewal and differentiation of adult stem cells. Differentiation. 113, 10–18 (2020).
Zhang, B. et al. circAKT3 positively regulates osteogenic differentiation of human dental pulp stromal cells via miR-206/CX43 axis. Stem Cell. Res. Ther. 11(1), 531 (2020).
Zheng, J. et al. CircCDK8 regulates osteogenic differentiation and apoptosis of PDLSCs by inducing ER stress/autophagy during hypoxia. Ann. NY Acad. Sci. 1485(1), 56–70 (2021).
Zhang, D. et al. CircRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin α5 expression. Cell Death Differ. 28(1), 283–302 (2021).
Chen, Y. et al. Dynamic chromatin accessibility landscapes of osteoblast differentiation and mineralization. Biochim. Biophys. Acta Mol. Basis Dis. 1870(2), 166938 (2024).
Mazziotta, C. et al. Regulatory mechanisms of circular RNAs during human mesenchymal stem cell osteogenic differentiation. Theranostics. 14(1), 143–158 (2024).
Ahmadi, A. et al. Recent advances on small molecules in osteogenic differentiation of stem cells and the underlying signaling pathways. Stem Cell Res. Ther. 13(1), 518 (2022).
Hankenson, K. D., Gagne, K. & Shaughnessy, M. Extracellular signaling molecules to promote fracture healing and bone regeneration. Adv. Drug Del. Rev. 94, 3–12 (2015).
Cao, X. & Chen, D. The BMP signaling and in vivo bone formation. Gene. 357(1), 1–8 (2005).
Zhou, R., Miao, S., Xu, J., Sun, L. & Chen, Y. Circular RNA circ_0000020 promotes osteogenic differentiation to reduce osteoporosis via sponging microRNA miR-142-5p to up-regulate Bone Morphogenetic Protein BMP2. Bioengineered. 12(1), 3824–3836 (2021).
Liu, S. et al. Involvement of circRNA_0007059 in the regulation of postmenopausal osteoporosis by promoting the microRNA-378/BMP-2 axis. Cell Biol. Int. 45(2), 447–455 (2021).
Zhang, Y.-L., Liu, L., Su, Y.-W. & Xian, C. J. miR-542-3p attenuates bone loss and marrow adiposity following methotrexate treatment by targeting sFRP-1 and Smurf2. IJMS. 22(20), 10988 (2021).
Liu, H., Wang, H., Yang, S. & Qian, D. Downregulation of miR-542-3p promotes osteogenic transition of vascular smooth muscle cells in the aging rat by targeting BMP7. Hum Genomics. 13(1), 67 (2019).
Matosin, N., Halldorsdottir, T. & Binder, E. B. Understanding the molecular mechanisms underpinning gene by environment interactions in psychiatric disorders: the FKBP5 model. Biol. Psychiatry. 83(10), 821–830 (2018).
Criado-Marrero, M. et al. Hsp90 and FKBP51: complex regulators of psychiatric diseases. Phil. Trans. R Soc. B. 373(1738), 20160532 (2018).
Liu, T. M. et al. Identification of common pathways mediating differentiation of bone marrow- and adipose tissue-derived human mesenchymal stem cells into three mesenchymal lineages. Stem Cells. 25(3), 750–760 (2007).
Kuçi, S. et al. Molecular signature of human bone marrow-derived mesenchymal stromal cell subsets. Sci. Rep. 9(1), 1774 (2019).
Lu, B. et al. A FKBP5 mutation is associated with Paget’s disease of bone and enhances osteoclastogenesis. Exp. Mol. Med. 49(5), e336-e (2017).
Cai, W., Xu, Y., Zuo, W. & Su, Z. MicroR-542-3p can mediate ILK and further inhibit cell proliferation, migration and invasion in osteosarcoma cells. Aging. 11(1), 18–32 (2019).
Luo, H. et al. circ_PTN contributes to -cisplatin resistance in glioblastoma via PI3K/AKT signaling through the miR-542-3p/PIK3R3 pathway. Mol. Ther. Nucleic Acids. 26, 1255–1269 (2021).
Jia, Z. et al. Effect of lncRNA XLOC_005950 knockout by CRISPR/Cas9 gene editing on energy metabolism and proliferation in osteosarcoma MG63 cells mediated by hsa-miR-542-3p. Oncol. Lett. 22(3), 669 (2021).
Hu, K. & Olsen, B. R. Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J. Clin. Investig. 126(2), 509–526 (2016).
Ouyang, Z. et al. CircRNA hsa_circ_0074834 promotes the osteogenesis-angiogenesis coupling process in bone mesenchymal stem cells (BMSCs) by acting as a ceRNA for miR-942-5p. Cell Death Dis. 10(12), 932 (2019).
Zhong, J. et al. LncRNA H19 promotes odontoblastic differentiation of human dental pulp stem cells by regulating miR-140-5p and BMP-2/FGF9. Stem Cell Res. Ther. 11(1), 202 (2020).
Ye, M. et al. CircRNA circCOL1A1 acts as a sponge of miR-30a-5p to promote vascular smooth cell phenotype switch through regulation of Smad1 expression. Thromb. Haemost. 123(1), 97–107 (2023).
Haraguchi, R. et al. sFRP4-dependent Wnt signal modulation is critical for bone remodeling during postnatal development and age-related bone loss. Sci. Rep. 6(1), 25198 (2016).
Ji, H., Cui, X., Yang, Y. & Zhou, X. CircRNA hsa_circ_0006215 promotes osteogenic differentiation of BMSCs and enhances osteogenesis-angiogenesis coupling by competitively binding to miR-942-5p and regulating RUNX2 and VEGF. Aging (Albany NY). 13(7), 10275–10288 (2021).
Ali, S. A. et al. Sequencing identifies a distinct signature of circulating microRNAs in early radiographic knee osteoarthritis. Osteoarthr. Cartilage. 28(11), 1471–1481 (2020).
Liang, J. et al. hsa_circ_0072389, hsa_circ_0072386, hsa_circ_0008621, hsa_circ_0072387, and hsa_circ_0072391 aggravate glioma via miR-338-5p/IKBIP. Aging. 13(23), 25213–25240 (2021).
Han, L., Cheng, J. & Li, A. hsa_circ_0072387 suppresses proliferation, metastasis, and glycolysis of oral squamous cell carcinoma cells by downregulating miR-503-5p. Cancer Biother. Radiopharmac. 36(1), 84–94 (2021).
Dou, Z. et al. Decreased expression of hsa_circ_0072387 as a valuable predictor for oral squamous cell carcinoma. Oral Diseases. 25(5), 1302–1308 (2019).
Chen, X. et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18(10), 997–1006 (2008).
Wei, J., Ou, Z., Tong, B., Liao, Z. & Yang, C. Engineered extracellular vesicles as therapeutics of degenerative orthopedic diseases. Front. Bioeng. Biotechnol. 11, 1162263 (2023).
Wu, Y. et al. Exosomes rewire the cartilage microenvironment in osteoarthritis: from intercellular communication to therapeutic strategies. Int. J. Oral Sci. 14(1), 40 (2022).
Mao, G. et al. Exosome-transported circRNA_0001236 enhances chondrogenesis and suppress cartilage degradation via the miR-3677-3p/Sox9 axis. Stem Cell Res. Ther. 12(1), 389 (2021).
Della Bella, E. et al. Differential regulation of circRNA, miRNA, and piRNA during early osteogenic and chondrogenic differentiation of human mesenchymal stromal cells. Cells. 9(2), 398 (2020).
Huang, H.-Y. et al. miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic Acids Res. 2019, 96 (2020).
Acknowledgements
The authors acknowledge other participants in our laboratory for their active help in this study.
Funding
This research was funded by Health and Family Planning Commission of Hunan Province, grant number 20201660; Health Commission of Hunan Province, grant number 202108031817; Beijing Municipal Natural Science Foundation, grant number 7222079.
Author information
Authors and Affiliations
Contributions
Study concept and design, J.Z., Q.Y. and O.L; experiment performed, K.S. and X.C.; data analysis and visualization, K.S. and X.C.; writing—original draft preparation, K.S.; writing—review and editing, J.Z., Q.Y. and O.L.; modification of figures and manuscript, O.L. Besides, K.S. and X.C. contributed equally to this work; Q.Y. and O.L. should be considered joint senior authors. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Su, K., Cui, X., Zhou, J. et al. Construction of an interactome network among circRNA-miRNA-mRNA reveals new biomarkers in hBMSCs osteogenic differentiation. Sci Rep 14, 24507 (2024). https://doi.org/10.1038/s41598-024-76136-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-76136-z