Abstract
Papillary thyroid cancer (PTC) is often characterized by indolent behavior, small tumors with slow cell proliferation and a tendency to metastasize to cervical lymph node simultaneously, and the molecular mechanisms underlying that remain poorly understood. In this study, FN1 was the hottest gene of PTC and distinctive expression in PTC cells. FN1 deficiency severely inhibited the p53 signaling pathway, especially cyclin proteins, resulting in increased cell growth but hampered invasion. The alternatively splicing EDB region of FN1 was exclusively expressed in tumors, which impacted integrin β1 (ITGB1) bonding FN1 and its secretion process, resulting in completely distinct roles of two isoforms that FN1 including and skipping EDB domain. The isoform EDB(–)FN1 intracellularly inhibited tumor proliferation by upregulating p21 expression, whereas extracellular EDB(+)FN1 promoted lymph node metastasis via the VEGF signaling pathway in vitro and in vivo. Moreover, the alternative splicing EDB region of FN1 was modulated by p53-targeted protein ZMAT3 which activated cell migration and lymphoangiogenesis. Collectively, combined with p53-induced proteins, FN1 played both anti- and pro-cancer roles owing to EDB domain alternative splicing. FN1 is a potential determinant behind the characteristic behavior of PTC, which may contribute to a deeper understanding of the peculiarity of PTC and provide a promising target for regional lymph node metastasis.
Similar content being viewed by others
Introduction
The incidence of thyroid cancer, particularly papillary thyroid carcinoma (PTC), has doubled in the past three decades and continues to increase at an annual rate of approximately 5%, which poses significant threats to human health1,2,3. PTC typically exhibits slow overall progression and a favorable prognosis, with a 5-year relative survival rate exceeding 95%, far beyond other solid carcinomas4,5. However, PTC has a tendency to early-stage metastasis to the cervical lymph node and prophylactic central compartment neck dissection has revealed occult metastases ranging from 24 to 82%, which increased recurrence rates and surgical complexity6,7. The underlying mechanisms of paradoxical and distinct behavior of PTC remain unknown.
Fibronectin 1 (FN1) is a large secretory protein with a molecular weight from 250 to 270 kDa. It serves as an essential component of the extracellular matrix (ECM), and is primarily synthesized by fibroblasts, endothelial and epithelial cells8. FN1 high expression in colorectal cancer (CRC) promoted lymphatic metastasis and indicated poorer prognosis9. Conversely, in melanoma, high expression of FN1 has been found to inhibit progression and metastasis, showing a tumor suppressor role10. FN1 exists in multiple isoforms as a result of alternative splicing of the precursor mRNA (pre-mRNA), occurring at three sites: EDA, EDB, and IIICS domains11. The inclusion of EDB and EDA domains in FN1 is recognized as a crucial marker of angiogenesis, and essential for some physiological processes such as wound healing, hepatic fibrosis development, and carcinoma progression12.
There are frequent gene alterations in PTC, such as point mutations in BRAF and RAS genes, as well as fusions involving the RET and NTRK1 kinases13,14. However, TP53 gene as a well-known critical tumor suppressor, could transactivate downstream genes and respond to stress signaling pathways, and the mutation is rarely reported in PTC15,16,17. P21 is one of the primary downstream targets of p53, also known as CDKN1A, WAF1, or CIP1, which suppresses cell proliferation and malignancy progression by arresting the G1 phase of cell cycle18,19,20. ZMAT3, also called Wig-1 or PAG608, as a direct transcriptional target gene of p53, exerts post-transcriptional control by binding to 3’ untranslated regions (3’UTRs) of diverse mRNA21,22,23. Recently, ZMAT3 was found as a key regulator of alternative splicing events in the p53 transcriptional program by directly binding the 3’splice site of specific intron or indirectly impacting the splicing of broad transcripts network24.
Here, we focused on elucidating the molecular mechanisms involved in PTC tumors that exhibit slow growth but a high tendency for lymph node metastasis. We found that the reciprocal regulation of ECM protein FN1 and p53 signaling pathways was distinctive and characteristic in PTC cells. FN1 two isoforms that include or skip EDB region were modulated by ZMAT3, which played distinct roles in PTC.
Results
ECM protein FN1 is highly enriched in PTC and positively correlated with the p53 signaling pathway
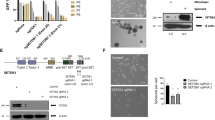
Through seeking the critical signaling pathway and genes of PTC from TCGA dataset, the result uncovered that ECM-receptor interaction and p53 signaling pathway were significantly enriched in PTC tissues (Fig. S1A). Among obtaining 99 mutual differentially expressed genes from GSE60542, GSE33630, and the TCGA three datasets, ECM protein FN1 was ranked as the hottest one via protein-protein interaction analysis (Fig. S1B). FN1 displayed significantly higher expression in PTC than in normal tissues (Fig. S2A-B). Moreover, from the GEPIA2 database, FN1 expression was highest in PTC compared to other tumor tissues (Fig. S2B). Interestingly, lung, liver, and breast solid carcinoma expressed FN1 extracellularly, however, PTC tissues exhibited both intracellular and extracellular expression of FN1 (Fig. S2C). Semiquantitative PCR analysis revealed FN1 didn’t express in A549 lung cancer cell, HeLa cervical cancer cell, KYSE150 and TE-1 esophageal cancer cell, MDA-MB-231 breast cancer cell, SKOV3 ovarian cancer cell, LOVO colon cancer cell, MGC-803 and BGC-823 gastric cancer cell, FRO and BHT101 undifferentiated thyroid cancer cell (Fig. 1A-B). FN1 was only expressed in PTC cells K1, KTC-1, and BCPAP, even normal thyroid cell Nthy-ori-3-1 but very low expression, and the intensity in K1 and KTC-1 cells was comparable to cancer-associated fibroblast (CAF) cells (Fig. 1A-B). Our finding was consistent with the study reporting distinctive expression of FN1 in PTC cells25. Furthermore, we found that ECM protein FN1 expression was positively and preferentially correlated with p53 signaling pathway via Gene Set Enrichment Analysis (GSEA) analysis (Fig. 1C). The mutation rate of TP53 presents very low, approximately 0.4% in PTC based on TCGA data (Fig. 1D). After knocking down FN1 in PTC cells (Fig. 1E), p53 signaling pathway was significantly compromised by GSEA analysis from RNA-seq data (Fig. 1F). These results suggest that FN1 related to p53 signaling pathway played important role in PTC.
FN1 is distinctive expression in PTC cells and positively correlated with p53 signaling pathway. (A, B) FN1 mRNA and protein levels in pan-cancer cell lines analyzed by semiquantitative PCR (A) and western blotting (B) respectively. (C) GSEA pathway enrichment plot showing the positive correlation between FN1 expression and p53 signaling pathway in PTC patients from TCGA dataset. (D) The TP53 mutation rate of PTC tissues of TCGA dataset from cBioPortal database. (E) Knockdown of FN1 in K1 and KTC-1 cells verified by western blotting with GAPDH loading control. (F) GSEA pathway enrichment plot showing significant downregulation of p53 signaling pathway following FN1 knockdown in PTC cells.
The isoform EDB(-)FN1 and EDB(+)FN1 are correlated with the p53 signaling pathway and ECM-receptor interaction signaling pathway respectively
As a result of alternative splicing domains EDB, EDA, and IIICS, FN1 could present posttranscriptional different isoforms (Fig. 2A). So, we further examined the expression of the three domains in tissues and cells. The alternative splicing EDB domain was exclusively expressed in tumors and not detected in normal thyroid tissues and cells, while EDA and IIICS regions were expressed in both (Fig. 2B-C and Fig. S3A-B). Next, we established FN1 two isoforms, containing and skipping the EDB domain, EDB(+)FN1 and EDB(-)FN1 overexpression cells after knocking down FN1 (Fig. 2D). To further study the different roles of two isoforms in PTC, RNA-seq analysis was performed. Interestingly, results shed light on that the ECM-receptor interaction pathway was enrichment into EDB(+)FN1 overexpression cells, not into EDB(-)FN1 isoform (Fig. 2E and Fig. S3C), while the p53 signaling pathway was upregulated in EDB(-)FN1 overexpression cells, not EDB(+)FN1 isoform (Fig. 2F and Fig. S3D). These results suggest that two isoforms EDB(-)FN1 and EDB(+)FN1 may play different functions via different behind mechanisms in PTC.
EDB region is exclusively expressed in tumor, EDB(+)FN1 and EDB(−)FN1 two isoforms enriched in different signaling pathways. (A) The simple schematic diagram showing FN1 containing three alternative splicing domains, EDB, EDA and IIICS. (B, C) Semiquantitative PCR analysis of EDB domain expression in normal and papillary thyroid tissues (B) and cells (C) respectively, EDB(+) representing FN1 including EDB domain and EDB(−) representing FN1 skipping EDB domain. (D) Overexpression of two isoforms of FN1, EDB(+)FN1 and EDB(−)FN1, verified by western blotting and Semiquantitative PCR with GAPDH loading control. (E) KEGG enrichment showing ECM-receptor interaction upregulated after co-culture with conditional medium (CM) from OE-EDB(+)FN1 cells. (F) GSEA pathway enrichment plot showing significant upregulation of p53 signaling pathway following EDB(−)FN1 overexpression in shFN1 cells.
EDB(-)FN1 upregulates the expression of p53-induced protein p21, inhibiting cell proliferation
We next explored the exact regulatory gene of EDB(-)FN1 isoform on the p53 tumor suppressor pathway. In RNA-seq result, 18 genes downregulated after FN1 knockdown, including CDKN1A, CCND3, CCND1, CDK4 and CCNG1 cyclin genes, and EDB(-)FN1 overexpression re-upregulated CDKN1A (also known p21), ZMAT3, BAX and MDM2 genes (Fig. 3A and Table S4), but only p21 expression was consistent by q-PCR (Fig. 3B), as was the protein level by western blot(Fig. 3D-E). Besides, we established EDB domain knockdown (shEDB) cells, which did not influence FN1 expression (Fig. 3C), and p21 expression was not decreased, even slightly increased (Fig. 3D-E). P21 serves as an important anti-oncogene and inhibits cell proliferation by controlling the G1/S phase transition of cell cycle19. DNA-damaging drug cis-platinum was used to treat cells, which concentration-dependently provoked p53 expression (Fig. 3F-G). Notably, cis-platinum significantly increased p21 levels of shEDB cells, however, no significant up-regulation in shFN1 cells (Fig. 3F-G), which suggests that the activation of p21 is partly dependent on EDB(-)FN1 in addition to p53 in PTC cells.
EDB(−)FN1 inhibits cell proliferation by upregulating p53-targeted protein p21 expression. (A) The Venn plot showing the genes in p53 signaling pathway downregulated after knockdown of FN1 and re-upregulated after overexpression of EDB(−)FN1. (B) qRT-PCR analysis verifying P21, BAX, ZMAT3 and MDM2 expression in indicated cells. (C) Knockdown of alternative splicing EDB domain of FN1 verified by western blotting and semiquantitative PCR with GAPDH loading control in K1 and KTC-1 cells. (D, E) Western blot analysis of P53 and p21 expression level in different transfection cells. (F, G) PTC cells treated with DNA-induced damage drug cis-platinum for 24 h, then Western blotting analyzed P53 and p21 expression in shFN1 and shEDB cells. (H) Cell proliferation ability of shFN1 cell overexpressed EDB(−)FN1 or EDB(+)FN1 isoform measured by CCK8 assays. (I, J) After shEDB and shFN1 cells transduced with sgRNAs targeting p53 and p21, the cell growth capability tested by CCK8 assays.
We further examined the role of FN1 isoforms in cell growth. EDB(-)FN1 isoform exhibited slower growth ability than FN1 knockdown and EDB(+)FN1 overexpression cells (Fig. 3H), and shEDB cells demonstrated significantly slower growth compared to shFN1 cells (Fig. S4A-B). EDB(-)FN1 similarly inhibited the proliferation of FRO, BGC-823, and TPC-1 cells with wild TP53, but had no effect on MDA-MB-231 cells with mutated TP53 (Fig. S4C-G). Furthermore, p53 and p21 were knockout in shEDB and shFN1 cells respectively (Fig. S4H). After knocking out p53, the growth ability of shFN1 and shEDB cells was significantly accelerated (Fig. 3I-J), however, the two groups had no significant difference (Fig. S4I). Notably, p21 knockout had no marked impact on shFN1 cells, while significantly promoting shEDB cell proliferation (Fig. 3I-J). These findings reveal that p53 and FN1 are parallel to control cell growth, and the isoform EDB(-)FN1 exerts an antitumor effect via repressing p21 expression in PTC cells.
EDB(+)FN1 promotes cell invasion and tube formation via the VEGF signaling pathway
Overexpressing EDB(+)FN1 isoform had no significant effect to cell proliferation ability (Fig. 3H). RNA-seq results demonstrated that EDB(+)FN1 cell medium up-regulated the ECM-receptor interaction pathway (Fig. 2E), which could accelerate tumorigenic progression26. By cell invasion assays, the results showed that EDB(+)FN1 overexpression significantly enhanced cell migration and invasion compared to EDB(-)FN1 and shFN1 cells (Fig. 4A-B). Deletion of the EDB region suppressed cell invasiveness, similar to FN1 deletion cells (Fig. S5A). Culturing LECS or HECV cells with the conditional medium (CM) from PTC cells, EDB(+)FN1 facilitated tube formation ability, while EDB(-)FN1 attenuated it as well as shEDB and shFN1 cells (Fig. 4C and Fig. S5B). Moreover, the VEGF signaling pathway was upregulated after overexpression of EDB(+)FN1 by GSEA analysis of RNA-seq data (Fig. 4D). Additionally, angiogenesis factors TGFB1, TIE1, GDF15, HAS2, and IGFBP5 were decreased after FN1 knockdown, and re-raised after overexpression of EDB(+)FN1 (Fig. S5C), and epithelial-mesenchymal transition (EMT) markers vimentin and N-cadherin expression showed similar changes (Fig. S5D). The lymphangiogenesis marker VEGFC was downregulated after the knockdown of EDB and FN1, it re-upregulated in EDB(+)FN1, not EDB(-)FN1 overexpression cells (Fig. 4E-F). Our results demonstrate that EDB(+)FN1 isoform plays an oncogenic role and promotes cell invasion and lymphatic and vascular forming via the VEGF signaling pathway in PTC cells.
EDB(+)FN1 promotes migration, invasion and tube formation extracellularly via VEGF signaling pathway. (A, B) After overexpressing EDB(+)FN1 or EDB(−)FN1 in shFN1 cells, the migration and invasion ability examined by wound healing and transwell assays. (C) Representative images and quantifications of tube formation by LECS and HECV cultured with conditional medium (CM) collected from two isoforms EDB(+)FN1 and EDB(−)FN1 cells. Scale bars: 50 μm. (D) GSEA pathway enrichment plot showing significant upregulation of VEGF signaling pathway following EDB(+)FN1 overexpression in shFN1 cells. (E, F) Western blot analysis of VEGFC expression in LECS and HECV cells after co-culture with CM collected from the indicated PTC cells.
EDB domain deficiency influences the binding of ITGB1 and FN1 and inhibits its secretion
FN1 two isoforms playing quite distinct roles, to investigate the underlying reason, IP-MS assays were performed, EDB(+)FN1 with a Flag epitope tag and EDB(-)FN1 with a His epitope tag. The results uncovered that integrin β1 (ITGB1) bound to EDB(+)FN1 but not to EDB(-)FN1, which was further verified by immunoprecipitation and western blotting (Fig. 5A-C and Table. S5). Endogenous ITGB1 was less co-precipitated with FN1 in OE-EDB(-)FN1 cells than OE-EDB(+)FN1 group (Fig. 5D). FN1 is via binding with ITGB1 and then secreted and assembled into the ECM, where it interacts with collagen and adhesins to play extracellular roles27,28. By testing FN1 level in a cultured medium, we found that EDB(+)FN1 overexpression increased extracellular FN1 level, but EDB(-)FN1 had no influence, moreover, EDB region deficiency severely reduced FN1 secreted expression (Fig. 5E-F). As expected, LECS and HECV cells were co-cultured with ITGB1 knockdown cell medium, and the VEGFC expression was significantly decreased (Fig. 5G-H). These results suggest that EDB domain deficiency affects the localization and assembly of FN1 in subcellular compartments, which leads to FN1 two isoforms with distinct roles in PTC. FN1 isoform containing EDB domain plays an oncogenic role extracellularly while skipping EDB region plays a tumor-suppressed effect intracellularly.
EDB region deficiency inhibits the binding of ITGB1 and FN1 and its secretion. (A) Flag tag with EDB(+)FN1 and His tag with EDB(−)FN1, using anti-flag and anti-his antibodies to pull down corresponding bound proteins for PAGE. (B, C) After transfecting Flag-EDB(+)FN1 or His-EDB(−)FN1 plasmids, then using FN1, flag and his antibodies for immunoprecipitation and western blotting assays. (D) After transfecting Flag-EDB(+)FN1 or His-EDB(−)FN1 plasmids, then using ITGB1 antibody for immunoprecipitation and western blotting assays. (E, F) Western blot analysis of secreted FN1 expression in cultured medium collected from the indicated cells with transferrin loading control. (G) Knockdown of ITGB1 in OE-EDB(+)FN1 cells verified by western blotting with GAPDH loading control. (H) Western blot analysis of VEGFC expression level in LECS cells after co-culture with CM from the indicated cells.
EDB domain is regulated by p53-targeted splicing factor ZMAT3 which facilitates lymph node metastasis
Alternative splice is generally regulated by splicing factors. We found that a splicing factor ZMAT3 was positively correlated with the ECM-receptor interaction pathway beside the p53 signaling pathway by GSEA analysis of the TCGA dataset (Fig. 6A-B). The robust expression of ZMAT3 relies on direct induction by p53, and the enrichment of p53 response elements (REs) in ZMAT3 locus, including the first intron and the promoter of ZMAT324,29. After ZMAT3 knockdown, rMATS analysis of RNA-seq data identified 254 ZMAT3-dependent alternative splicing events, with the majority of skipped exon (SE) events (Table. S6). Notably, FN1 exhibited significant changes in EDB domain splicing, and the inclusion level score (Inclevel) was decreased from 0.3 to 0.19 following suppression of ZMAT3 (Fig. 6C). In other words, the expression of EDB(+)FN1 isoform was reduced, while EDB(-)FN1 had no significant change (Fig. 6D-E). Furthermore, p53 knockout also resulted in more frequent skipping of the EDB domain of FN1 (Fig. 6F). The finding indicates that p53 targeted-ZMAT3 could modulate the alternative splice of the EDB region of FN1 in PTC cells.
EDB region of FN1 is modulated by splicing factor ZMAT3 which promotes cell migration and invasion. (A) GSEA pathway enrichment plot showing the positive correlation between ZMAT3 expression level and P53 signaling pathway from TCGA database. (B) GSEA pathway enrichment plot showing the positive correlation between ZMAT3 expression and ECM-receptor interaction from TCGA database. (C) rMATS plot showing the decrease in Including level (InLevel) of the alternative splicing EDB region of FN1 upon ZMAT3 silencing. (D) The expression of EDB(+)FN1 and EDB(−)FN1 isoforms in shZMAT3 and control groups from RNA-seq data and verified by qRT-PCR. (E) Semiquantitative PCR analysis of EDB(+)FN1 and EDB(−)FN1 isoforms expression of K1 and KTC-1 cells following ZMAT3 knockdown. (F) Western blot analysis of ZMAT3 expression and Semiquantitative PCR analysis of EDB(+)FN1 and EDB(−)FN1 expression following p53 knockout. (G) Representative images and quantifications of tube formation by LECS and HECV cultured with CM collected from the indicated cells. Scale bars: 50 μm. (H) qRT-PCR analysis of VEGFR3 expression of LECS and HECV after co-cultured with CM derived from indicated cells. (I, J) Western blot analysis of VEGFC expression of LECS after co-cultured with CM from the indicated PTC cells.
ZMAT3 knockdown showed a slight reduction in cell proliferation but significantly weakened cell invasion ability (Fig. S6A-D). LECS and HECV cells were co-cultured with ZMAT3 knockdown CM, which inhibited the tube formation by downregulating VEGFR3 and VEGFC expression, which could be rescued by the addition of EDB(+)FN1 (Fig. 6G-J). Similarly, the EDB domain knockdown decreased VEGFR3 and VEGFC expression, which could be reversed by overexpression of ZMAT3 (Fig. 6H-J). Our results suggest that in PTC cells ZMAT3 facilitates cell invasion and lymph tube formation by promoting the inclusion of the EDB domain of FN1.
The opposing functions of EDB(-)FN1 and EDB(+)FN1 in tumorigenesis in vivo
We further verified the function of FN1 two isoforms in vivo. The tumor burden in shFN1 cohorts was higher than in shEDB groups in subcutaneous tumorigenesis models (Fig. 7A), moreover, overexpressing EDB(-)FN1 hampered the tumor proliferation (Fig. S7A). Ki67, a cell proliferation marker, was significantly reduced in tumors with EDB knockdown and EDB(-)FN1 overexpression groups (Fig. S7B). The expression of p53 and p21 was significantly increased in EDB(-)FN1 overexpression groups compared to OE-EDB(+)FN1 groups (Fig. 7B and Fig. S7C). Footpad lymph node metastasis models displayed that FN1 knockdown significantly inhibited lymph node metastasis (Fig. S7D), and overexpressing EDB(+)FN1 aggravated metastatic burden of popliteal lymph nodes (Fig. 7C-D), and VEGFC was remarkably increased (Fig. 7E). Cytoskeletal 7 (CK7) was usually expressed only in epithelial cell, as a marker of lymph nodes metastasis, significantly higher expression in EDB(+)FN1 than shFN1 group (Fig. 7E).
EDB(−)FN1 impairs tumor growth while EDB(+)FN1 promotes LNM in mice models. (A) Tumor formation by shFN1 or shEDB cells injected into the subcutaneous tissue of nude mice, and tumor growth curves are shown. (B) The expression of p53 and p21 in subcutaneous tumor tissues were evaluated by IHC. Scale bars: 50 μm. (C) Representative images of popliteal lymph node (LN) metastasis model of nude mouse, the indicated K1 cells were injected into the footpads, and images of popliteal LNs and histogram analysis of the LN volume in the indicated cells (n = 3). (D) The image of LYVE-1 expression in popliteal LNs tested by IHC. (E) The expression of VEGFC and CK7 in popliteal LNs of indicated groups. Scale bars: 50 μm.
In PTC tissues from the TSVdb database, EDB(-)FN1 isoform presents a higher percentage in PTC tumors without lymph node metastasis (LNM, N0 stage), conversely, EDB(+)FN1 isoform presents a higher percentage in tumors with LNM (N1 stage) (Fig. 8A). The ratio of EDB(-)FN1 had no significant difference in T1, T2 and T3 stage, but present a decrease in T4 stage tumor, while EDB(+)FN1 showed an increased percentage in T4 stage (Fig. 8B). Immunohistochemistry (IHC) revealed EDB region in tumor was significantly higher expressed compared to paracancerous tissues (Fig. S7E), and significantly higher in tumors with LNM than without LNM (Fig. 8C, Fig. S7F). Furthermore, we constructed ROC curve to evaluate EDB expression levels in predicting cervical LNM of PTC, the area under curve (AUC) with 0.784 value (Fig. 8D). ZMAT3 expression in PTC tissues was evaluated, and results indicated that it was higher in tumors compared to normal thyroid tissues, significantly higher in metastatic lymph nodes compared to normal lymph nodes, and higher in metastatic PTC compared to non-metastatic PTC tissue from TCGA and GSE60542 data (Fig. 8E-F), which were also verified in our collected samples at both mRNA and protein level (Fig. 8G-H). There was a positive correlation between ZMAT3 and FN1 expression (Spearman correlation = 0.48, Fig. 8I), and high FN1 and ZMAT3 expression groups both exhibited poorer recurrence-free survival of PTC in the TCGA dataset (Fig. 8J-K). These results indicate that FN1 isoform including the EDB region modulated by p53 targeted-ZMAT3 promotes LNM, and isoform skipping EDB restricts carcinoma growth, which is consistent with the biological characteristics of PTC that indolent or small size tumors usually metastasize to the cervical lymph node.
The expression of EDB(−)FN1, EDB(+)FN1 and ZMAT3 in PTC patients. (A) The percentage of EDB(−)FN1 and EDB(+)FN1 accounting for FN1 in tumors with-LNM (N1) and without-LNM (N0) respectively in TCGA dataset from the TSVdb database. (B) The ratio of EDB(−)FN1 and EDB(+)FN1 in different tumor size stage (T1–T4) in TCGA dataset from the TSVdb database. (C) Representative images of EDB region expression in PTC tumors with-LNM (N1) and without-LNM (N0), and statistical diagram was shown. Scale bars: 50 μm. (D) Diagnostic efficacy of EDB region expression in PTC with LNM evaluated by ROC curve. (E) The differential expression of ZMAT3 in normal and tumor tissues, tumor with-LNM and without-LNM PTC tissues from TCGA dataset. (F) The differential expression of ZMAT3 in normal and tumor tissues, normal LN and metastatic LN from GSE60542 dataset. (G) qRT-PCR analysis of ZMAT3 mRNA expression in paired tumor and paracancerous tissues (n = 10). (H) ZMAT3 protein expression in normal and PTC tissues from the Protein Atlas database. (I) The correlation analysis of FN1 and ZMAT3 expression levels from cBioPortal database. (J, K) Kaplan-Meier analyses of the correlations between FN1 (I), ZMAT3 (J) expression and recurrence-free survival of PTC patients from Kmplot database.
Discussion
The prevalence of thyroid tumors, mainly PTC, is increasing2, however, molecular mechanisms about its characteristic behavior still have no report. In the study, the interplay of FN1 alternative splice variant and p53 targeted proteins could perfectly interpret the behavior of PTC that indolent carcinoma with LNM frequently. High expression of FN1 and a very low mutation rate of TP53 may underlie slow progression and improved overall prognosis of PTC. FN1 and p53 are active responders of cell internal and external environmental stresses, which thyroid tissue is susceptible to and lead to the formation of neoplasm16,30. In addition to the p53 signaling pathway, FN1 expression refers to multiple metabolism-related pathways, such as the biosynthesis of nucleotide sugar, amino acid pathways, AMPK signaling pathway, and steroid biosynthesis (not shown).
The function of FN1 was controversial in cancers, for example, FN1 interacting with Mena/α5 integrin increased tumor stiffness through matrix remodeling, which promotes breast cell invasion31. While, in melanoma, the increasing expression of FN1 secreted by Natural killer cells resulted in reduced metastases formation32. In PTC tumors, FN1 played both pro- and anti-cancer roles owing to the alternative splicing of the EDB domain. EDB deficiency inhibited FN1 bound to ITGB1 and its secretion, which resulted in the isoform EDB(+)FN1 being in extracellular and EDB(-)FN1 being in intracellular playing different roles. A previous study reported that EDB domain deficiency affected the assembly of FN1 into fibrils but not secretion in fibroblasts33. EDB(+)FN1 isoform that interacted with collagen and adhesins in ECM promoted lymph node metastasis, whereas EDB(-)FN1 hampered cell growth by activating the p53/p21 signaling pathway in PTC.
Although the role of FN1 remains controversial in different tumors, the EDB region has been verified as a marker of neo-angiogenesis, and targeting EDB has been applied in tumor therapy and medical imageology34,35. T cells expressing an EDB-CAR based on the L19 mAb could kill EDB-positive tumor vasculature36. In the tumor microenvironment, EDB-FN is a promising target to leverage in the design of a small peptide-targeted contrast agent utilized in magnetic resonance molecular imaging (MRMI)37. In our study, tumors with EDB(+)FN1 expression tended to be cervical lymph node metastasis, and the isoform had well predictive ability for LNM with 0.784 AUC value, so it had great potential as a biological marker for the diagnosis and treatment of PTC.
EDB(-)FN1 isoform has no obvious impact on p53 expression, while it significantly affects the trans-activating p21 gene level. Apart from p53, the research about regulating p21 expression is few. It was reported that RNA polymerase II phosphorylation and recruitment of P-TEFb initiated transcription and activation of p21 gene38. We were surprised to discover FN1’s distinctive expression in the PTC model, and the isoform EDB(-)FN1 could immensely upregulate the p21 level and slow tumor proliferation. It is very important and will provide potentially valuable treatment options especially for high-grade malignancies, but the in-depth mechanism requires further study.
TP53 is a classical-tumor suppressor gene, and its downstream target ZMAT3 was reported as a crucial anti-oncogenic factor inhibiting cell proliferation in lung, liver, and colon cancer24,32,39. While another research uncovered depletion of ZMAT3 in colon cancer cell HCT116 increased cell death by binding FAS mRNA 3’-UTR independently of p5340. ZMAT3 regulates the cell cycle and DNA replication as shown in our RNA-seq data, but it tends to be an oncogenic factor promoting cell invasion in PTC, since it may play a predominant role in modulating the alternative splicing of the EDB domain. The EDB region alternative splicing of FN1 was regulated by splicing factor SRSF5 (SRp40) by recognizing the intronic splicing enhancer11,41, verified in our experiments (not shown). However, ZMAT3 could bind to the 3’splice site and CDS of SRSF5 and target spliceosome by eCLIP assays24.
Conclusion
P53-induced protein ZMAT3 modulates the alternative splicing EDB region promoting lymph node metastasis, and FN1 isoform skipping EDB domain upregulates p53-targeted p21 expression suppressing cell proliferation, as shown in Fig. 9. FN1-p53-p21/ZMAT3 axis is closely related to the biological characteristics of PTC with an indolent tumor but proneness of LNM especially in the early stage. High FN1 expression exerts both pro- and anti-tumor effects via the alternative splicing of the EDB domain, in which one isoform that takes predominance may determine the trend of development of PTC, which provides a rationale for a deeper understanding of the biological features of PTC and the alternative strategies for individualized treatment in the future.
A simple signaling pathway diagram. FN1 plays both oncogenic and tumor suppressive roles combined with p53 signaling pathway owing to the alternative splicing of EDB domain and its subcellular location. The isoform EDB(−)FN1 intracellularly inhibited tumor proliferation by upregulating p21 expression, whereas extracellular EDB(+)FN1 that was up-regulated by ZMAT3 promoted lymph node metastasis via the VEGF signaling pathway.
Materials and methods
Clinical samples
PTC specimens were collected from Xiangya Hospital of Central South University, and the final pathological diagnosis was determined by the Department of Pathology, at Xiangya Hospital. Ethical approval (No. 2019030140) for human research was obtained from the Committees for Ethical Review at Xiangya Hospital, Central South University. Informed consent was obtained from each patient before sample collection. We confirmed that all experiments were performed by relevant guidelines and regulations.
Cell lines and cell culture
Nthy-ori-3, K1, TPC-1, BCPAP, KTC-1, FRO, BHT101, MGC-803, BGC-823, LOVO, MDA-MB-231, SKOV3, A549, HeLa, KYSE50, TE-1, and 293T cells were procured from the Cell Bank of Chinese Academy of Sciences in Shanghai, China. LECS and HECV cells were sourced from the American Type Culture Collection (ATCC) in Manassas, VA, USA. Tumor-associated fibroblast (CAF) cells were obtained through primary culture from tumor tissues. RPMI-1640 medium or DMEM medium supplemented with 10% fetal bovine serum, and 1% penicillin and streptomycin was employed for cells. All cell lines were incubated at 37 °C with 5% CO2 in a controlled environment.
Antibodies and reagents
The following antibodies were used for immunoblotting at 1:1000 and immunohistochemistry at 1:100 and immunoprecipitation at 1:100: Rabbit anti-FN1 (Cat. #BM4460, Boster, Cat. #15613-1-AP, Proteintech), Rabbit anti-EDB (Cat. # ab154210, Abcam), Rabbit anti-Flag (Cat. #ab205606, Abcam), Rabbit anti-His (Cat. #ab245114, Abcam), Rabbit anti-ITGB1(Cat. #12594-1-AP, Proteintech), Rabbit anti-P53 (Cat. #10442-1-AP, Proteintech), Rabbit anti-P21 (Cat. #10355-1-AP, Proteintech), Rabbit anti-LYVE-1 (Cat. #51011-1-AP, Proteintech), Rabbit anti-VEGFC (Cat. #22601-1-AP, Proteintech), Rabbit anti-CK7 (Cat. #17513-1-AP, Proteintech), Mouse anti-GAPDH (Cat. #60004-1-Ig, Proteintech), Mouse anti-Transferrin (Cat. #66171-1-Ig, Proteintech), Rabbit anti-ZMAT3 (Cat. #10504-1-AP, Proteintech).
Generation of stable transfectants
The short hairpin RNAs (shRNAs) utilized for genome editing were employed to achieve knockdown of FN1, EDB and ZMAT3 in K1, and KTC-1 cells, as well as ITGB1 knockdown in OE-EDB(+)FN1 cells. Additionally, single guide RNAs (sgRNAs) were used in CRISPR/Cas9 gene editing to induce knockout of P53 and P21 in FN1-knockdown and EDB-knockdown cells, respectively. The sequences of these shRNAs and sgRNAs can be found in Table S1. To initiate viral infection, packaging plasmids were co-transfected into 293T cells along with the intended genes. After 72 h of transfection, viral particles were harvested. Lentiviral plasmids of overexpressing EDB(+)FN1, EDB(-)FN1, and ZMAT3 were obtained from GENE Company. Cells were infected with the viral particles 8–12 h in the presence of polybrene (10 µg/ml) and subsequently selected using puromycin (2 µg/ml) or G-418 (2,000 µg/ml). The efficacy of stable transfection cells was confirmed through q-PCR and immunoblotting.
Immunoblotting
Cellular proteins were extracted using RAPI reagent and a protease inhibitor PMSF mixture in a ratio of 100:1. Medium proteins were obtained by centrifuging serum-free medium to remove impurities, subsequently transferring to an ultrafiltration centrifuge tube (Millipore) and to concentrate the supernatant. The protein concentration was determined using the BCA kit. The proteins, with varying molecular weights, were separated through SDS‒PAGE and subsequently transferred onto PVDF membranes. Following the blocking step, the membranes were incubated overnight at 4 °C with primary antibodies against FN1(1:1000 dilution), P53 (1:5000 dilution), P21 (1:1000 dilution), VEGFC (1:1000 dilution), ZMAT3 (1:1000 dilution) or ITGB1 (1:1000 dilution). The corresponding secondary antibodies were then applied at appropriate dilutions for 2 h. Lastly, an ECL chemiluminescence kit was employed for the detection and visualization of the developed bands.
QRT-PCR and semiquantitative PCR
RNA extraction was performed using the Trizol reagent (Thermo Scientific), followed by reverse transcription using the reverse transcriptase kit (Cat. #AE311, TransGen Biotech). Quantitative PCR was conducted in triplicate using gene-specific primers and SYBR green (Yeasen Biotechnology) in a 7900HT Fast Real-Time PCR machine (Applied Biosystems). GAPDH was as the reference gene, and the relative expression of desired genes was calculated using 2-△△CT method. Semiquantitative PCR was according PCR SuperMix kit (Cat. #AS221, TransGen Biotech), and the PCR products were visualized on a 2% agarose gel and scanned using a Typhoon scanner. The primer sequences can be found in Table S2.
Immunohistochemistry (IHC) staining
IHC staining was performed following previously established protocols42. Briefly, mouse or thyroid tissues were fixed in formalin overnight and subsequently embedded in paraffin. Sections were subjected to baking at approximately 65 °C for 2–3 h and subsequently dewaxed using xylene and gradient ethanol. After antigen repair, sections were incubated in an endogenous peroxidase blocker and normal goat serum. This was followed by overnight incubation at 4 °C with primary antibodies against FN1 (1:100 dilution), EDB (1:50 dilution), P53(1:100 dilution), P21(1:100 dilution), VEGFC (1:80 dilution), CK7 (1:100 dilution) and LYVE-1 (1:100 dilution). Mouse or rabbit IgG secondary antibodies were then applied for 30 min at room temperature. DAB chromogenic solution for reaction, hematoxylin for staining the nuclei, and the samples were dehydrated in gradient alcohol and xylene, sealed in neutral resin, observed under a microscope (Leica Camera), and photographed. Based on the proportion and degree of coloration of positive cells, the scores were categorized into four groups: high (6–7, +++), medium (4–5, ++), low (2–3, +), and negative (0–1, -).
RNA-seq and analysis
K1 cells were transduced with lentiviruses carrying FN1 shRNA and ZMAT3 shRNA and three unique negative control shRNA. Additionally, the cells were transduced with plasmids expressing EDB(+)FN1 and EDB(-)FN1. Following transduction, selected in puromycin and cells were cultured for an additional 10 days. RNA was extracted from the cultured cells using a Trizol reagent. RNA-seq libraries were prepared using the DNBSEQ™ Kit, following the manufacturer’s instructions. Transcripts showing significantly differential expression, with an adjusted p-value below 0.05, were subjected to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analysis43, Gene Ontology (GO) function enrichment analysis, and Gene Set Enrichment Analysis (GSEA) using the phyper functions in Rstudio. The acquired genes’ expression value of each sample of RNA-seq data can be found in Table S3, and the differentially expressed genes identified can be found in Table S4. The RNA-seq data have been submitted to the Gene Expression Omnibus (GEO) database with the accession number GSE285016 for public access.
CoIP and label-free quantitative proteomics
To investigate the protein interactions of FN1 two isoforms, co-immunoprecipitation (CoIP) and Label-free quantitative proteomics were employed. FN1 knockdown cells (approximately 1 × 107 cells) were transfected with Flag-tagged EDB(+)FN1 or His-tagged EDB(-)FN1 plasmids using Lipofectamine 2000 (Invitrogen) as per the manufacturer’s recommended protocols. After 48 h of transfection, the cells were lysated using cell lysis buffer for IP added 1× protease inhibitor cocktail (Roche). The lysates were incubated overnight at 4 °C with specific antibodies (5 µg anti-Flag, 5 µg anti-His) and protein A/G PLUS-agarose (Santa Cruz Biotechnology, Cat. #sc-2003). Subsequently, the protein mixtures were subjected to four rounds of washing with a wash buffer. The bound proteins were eluted using a loading buffer and separated by SDS-PAGE.
For the label-free quantitative proteomic analysis, an equal amount of protein solution from each IP sample, the volume was adjusted to match the consistency of the original lysate (PTM Biolabs, Hangzhou). The resulting peptides were purified using C18 ZipTips (Millipore) according to the manufacturer’s instructions, and the purified peptides were subjected to analysis using LC-MS/MS (ThermoFisher Scientific). The detailed results can be found shown in Table S5. The MS proteomics data have been deposited to the ProteomeXchange Consortium (https://proteomecentral.proteomexchange.org) via the iProX partner repository with the dataset identifier PXD053745.
RNA-seq alternative splicing analysis
Alternative splicing events were analyzed using rMATS software, based on RNA-seq data obtained from shZMAT3 and shControl cells. Differential alternative splicing events were identified based on a criterion of < 5% change in exon inclusion (percent-spliced-in, PSI > 0.05) and a p-value < 0.05 (Table S6).
CCK8 assays
In triplicate, 1–2 × 103 cells per well for both transfected and control groups were seeded in 96-well plates to conduct a growth test. Following a 2-hour incubation with 10% CCK8 (Dojindo, Japan), the absorbance at 450 nm was measured using a microplate reader.
Wound healing and transwell experiments
Cells were seeded in 6-well plates at a density of 90–100%, and a vertical scratch was created using a tip, followed by incubation with serum-free medium for 48–72 h. The wound healing rate = (wound width at time t – wound width at time 0) / wound width at time 0. Cells were incubated in serum-free medium for 24 h and then seeded into the upper transwell chambers (BD Biosciences, MA, USA), while the bottom wells were filled with complete medium or conditioned medium. After 48 h of culture, the cells that invaded the lower surface of the membrane were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Five random fields were microscopically recorded, and the cell count was determined.
Tube formation experiments
Conditioned medium derived from cultured PTC cells was co-cultured with LECS and HECV cells for 24 h. Subsequently, 4 × 104 LECS or 4.5 × 104 HECV cells were seeded into matrix gel (Cat. #356234, Corning, USA) and incubated for 6–8 h. The lymphatic tubes were then photographed and quantified by measuring the length and area of the tubule structures using ImageJ software (Download (imagej.net)).
Animal studies
Female BALB/c nude mice, aged 5 weeks old, were purchased from Hunan SJA Laboratory Animal Co., Ltd and utilized for the animal studies. Cells numbering 2 × 106 were subcutaneously injected into either the left or right flanks of the mice. Tumor growth was monitored every three days using a caliper, and tumor volume was calculated using the formula a × b2 × 0.5 (a, longest diameter; b, shortest diameter). After four weeks of the tumor cell injection, all mice were euthanized via cervical dislocation, and the subcutaneous tumors were excised for subsequent IHC staining. For examining lymph node metastasis, 100 µl PBS suspensions containing PTC cells (4 × 106) were injected footpad. After four weeks, the evaluation of tumor growth and lymphatic metastasis of popliteal were performed. The primary tumors and popliteal LNs were enucleated for subsequent IHC staining. Ethical approval (No. 2019030140) for animal research was obtained from the Committees for Ethical Review at Xiangya Hospital, Central South University. All methods were reported by ARRIVE guidelines, and we confirmed that all experiments were performed by relevant guidelines and regulations.
Statistical analysis
The quantitative data are presented as the means ± standard deviation (sd). Paired t-tests were employed to compare paired specimens, while other significance tests included unpaired two-tailed t-tests, Chi-squared tests, Fisher’s exact tests, or two-sided unpaired Wilcoxon tests. Diagnostic efficacy was assessed by constructing receiver operating characteristic (ROC) curves to calculate the area under the curve (AUC) values. All of the statistical analyses were performed using the R package (version 3.3.3) or Prism version 6.0 (GraphPad). Significance was determined with P-values < 0.05 (ns, nonsignificant; *, P < 0.05; **, P ≤ 0.01; ***, P ≤ 0.001).
Data availability
Data is provided within the manuscript or supplementary information files.
Abbreviations
- PTC:
-
Papillary thyroid cancer
- ECM:
-
Extracellular matrix
- AS:
-
Alternative splicing
- Pre-mRNA:
-
Precursor mRNA
- 3ʹUTRs:
-
3ʹ untranslated regions
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- GSEA:
-
Gene Set Enrichment Analysis
- SFs:
-
Splicing factors
- REs:
-
Response elements
- RBPs:
-
RNA-binding proteins
- Inclevel:
-
Inclusion level score
- GO:
-
Gene Ontology
- ROC:
-
Receiver operating characteristic curve
- AUC:
-
Area under curve
- EMT:
-
Epithelial–mesenchymal transition
- CAF:
-
Cancer associated fibroblasts
- LNM:
-
Lymph node metastasis
- IHC:
-
Immunohistochemistry
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. Ca-a Cancer J. Clin. 72 (1), 7–33 (2022).
Miranda, A. et al. Thyroid cancer incidence trends by histology in 25 countries: a population-based study. Lancet Diabetes Endocrinol. 9 (4), 225–234 (2021).
Schlumberger, M. & Leboulleux, S. Current practice in patients with differentiated thyroid cancer. Nat. Reviews Endocrinol. 17 (3), 176–188 (2021).
Yu, X., Wan, Y., Sippel, R. & Chen, H. J. A. o. s., Should all papillary thyroid microcarcinomas be aggressively treated? An analysis of 18,445 cases. Ann. Surg. 254 (4), 653–660 (2011).
Sciuto, R. et al. Natural history and clinical outcome of differentiated thyroid carcinoma: a retrospective analysis of 1503 patients treated at a single institution. Ann. Oncol. 20 (10), 1728–1735 (2009).
Hughes, D. T. et al. Prophylactic Central Compartment Neck Dissection in Papillary Thyroid Cancer and Effect on Locoregional Recurrence. Ann. Surg. Oncol. 25 (9), 2526–2534 (2018).
Wada, N. et al. Lymph node metastasis from 259 papillary thyroid microcarcinomas - Frequency, pattern of occurrence and recurrence, and optimal strategy for neck dissection. Ann. Surg. 237 (3), 399–407 (2003).
Mosher, D. F., Fogerty, F. J., Chernousov, M. A. & Barry, E. L. Assembly of fibronectin into extracellular matrix. Ann. N. Y. Acad. Sci. 614, 167–180 (1991).
Cai, X. et al. Down-regulation of FN1 inhibits colorectal carcinogenesis by suppressing proliferation, migration, and invasion. J. Cell. Biochem. 119 (6), 4717–4728 (2018).
Liu, W., Cheng, S., Asa, S. L. & Ezzat, S. The melanoma-associated antigen A3 mediates fibronectin-controlled cancer progression and metastasis. Cancer Res. 68 (19), 8104–8112 (2008).
White, E. S., Baralle, F. E. & Muro, A. F. New insights into form and function of fibronectin splice variants. J. Pathol. 216 (1), 1–14 (2008).
Kumra, H. & Reinhardt, D. P. Fibronectin-targeted drug delivery in cancer. Adv. Drug Deliv Rev. 97, 101–110 (2016).
Xing, M. Molecular pathogenesis and mechanisms of thyroid cancer. Nat. Rev. Cancer. 13 (3), 184–199 (2013).
Cancer Genome Atlas Research. Integrated genomic characterization of papillary thyroid carcinoma. Cell 159 (3), 676–690 (2014).
Boutelle, A. M. & Attardi, L. D. p53 and Tumor Suppression: It Takes a Network. Trends Cell. Biol. 31 (4), 298–310 (2021).
Levine, A. J. 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer. p53 (8), 471–480 (2020). 20.
Kandoth, C. et al. Mutational landscape and significance across 12 major cancer types. Nature 502 (7471), 333–339 (2013).
el-Deiry, W. S. et al. WAF1, a potential mediator of p53 tumor suppression. Cell 75 (4), 817–825 (1993).
El-Deiry, W. S. p21(WAF1) Mediates Cell-Cycle Inhibition, Relevant to Cancer Suppression and Therapy. Cancer Res. 76 (18), 5189–5191 (2016).
Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 29 (5), 946–960 (2022).
Varmeh-Ziaie, S. et al. Wig-1, a new p53-induced gene encoding a zinc finger protein. Oncogene 15 (22), 2699–2704 (1997).
Vilborg, A., Bersani, C., Wilhelm, M. T. & Wiman, K. G. The p53 target Wig-1: a regulator of mRNA stability and stem cell fate? Cell Death Differ. 18 (9), 1434–1440 (2011).
Bersani, C. et al. Genome-wide identification of Wig-1 mRNA targets by RIP-Seq analysis. Oncotarget 7 (2), 1895–1911 (2016).
Bieging-Rolett, K. T. et al. Zmat3 Is a Key Splicing Regulator in the p53 Tumor Suppression Program. Mol. Cell. 80 (3), 452–469 (2020). e9.
Ryu, S., Jimi, S. & Takebayashi, S. Thyroid carcinoma distinctively expresses intracellular fibronectin in vivo. Cancer Lett. 121 (2), 189–193 (1997).
Sun, H. F. et al. Loss of TMEM126A promotes extracellular matrix remodeling, epithelial-to-mesenchymal transition, and breast cancer metastasis by regulating mitochondrial retrograde signaling. Cancer Lett. 440, 189–201 (2019).
Hynes, R. O. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69 (1), 11–25 (1992).
Obara, M., Kang, M. S. & Yamada, K. M. Site-directed mutagenesis of the cell-binding domain of human fibronectin: separable, synergistic sites mediate adhesive function. Cell 53 (4), 649–657 (1988).
Wilhelm, M. T., Méndez-Vidal, C. & Wiman, K. G. Identification of functional p53-binding motifs in the mouse wig-1 promoter. FEBS Lett. 524 (1–3), 69–72 (2002).
Kuramoto, K., Liang, H. J., Hong, J. H. & He, C. C. Exercise-activated hepatic autophagy via the FN1-05?1 integrin pathway drives metabolic benefits of exercise. Cell Metabol. 35 (4), 620– (2023).
Berger, A. J. et al. Scaffold stiffness influences breast cancer cell invasion via EGFR-linked Mena upregulation and matrix remodeling. Matrix Biol. 85–86, 80–93 (2020).
Glasner, A. et al. NKp46 Receptor-Mediated Interferon-γ Production by Natural Killer Cells Increases Fibronectin 1 to Alter Tumor Architecture and Control Metastasis. Immunity 48 (1), 107– (2018).
Fukuda, T. et al. Mice lacking the EDB segment of fibronectin develop normally but exhibit reduced cell growth and fibronectin matrix assembly < i > in vitro. Cancer Res. 62 (19), 5603–5610 (2002).
Haubner, R., Beer, A. J., Wang, H. & Chen, X. Y. Positron emission tomography tracers for imaging angiogenesis. Eur. J. Nucl. Med. Mol. Imaging. 37, S86–S103 (2010).
Menrad, A. & Menssen, H. D. ED-B fibronectin as a target for antibody-based cancer treatments. Expert Opin. Ther. Targets. 9 (3), 491–500 (2005).
Wagner, J. et al. Antitumor Effects of CAR T Cells Redirected to the EDB Splice Variant of Fibronectin. Cancer Immunol. Res. 9 (3), 279–290 (2021).
Lu, Z. R., Laney, V. & Li, Y. J. Targeted Contrast Agents for Magnetic Resonance Molecular Imaging of Cancer (Accounts of Chemical Research, 2022).
Gomes, N. P. et al. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20 (5), 601–612 (2006).
Janic, A. et al. DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 24 (7), 947– (2018).
Bersani, C., Xu, L. D., Vilborg, A., Lui, W. O. & Wiman, K. G. Wig-1 regulates cell cycle arrest and cell death through the p53 targets FAS and 14-3-3σ. Oncogene 33 (35), 4407–4417 (2014).
Lim, L. & Sharp, P. J. M. biology, c., Alternative splicing of the fibronectin EIIIB exon depends on specific TGCATG repeats. 18 (7), 3900–3906. (1998).
Liu, M. et al. Down-regulating GRP78 reverses pirarubicin resistance of triple negative breast cancer by miR-495-3p mimics and involves the p-AKT/mTOR pathway. Biosci. Rep. 42 (1). (2022).
Minoru, K., Miho, F., Yoko, S., Masayuki, K. & Mari, I. W. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, 0 (2022).
Funding
This work was supported by grants from the National Natural Science Foundation of Changsha, Hunan Province (kq2403002), the National Natural Science Foundation of China (81974423) and the Fundamental Research Funds for the Central Universities of Central South University (CX20210354).
Author information
Authors and Affiliations
Contributions
SC and ML designed the project; LA, NL and YKJ performed animal experiments; ML, YXZ, PC, BW performed in vitro experiments; ML, HLT and JL analyzed the data; SC supervised and founder the project; ML and SJL wrote the draft manuscript and SC made the modification.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Ethical approval was provided by the Committees for Ethical Review of Research involving Human Subjects and Animal experiments at Xiangya Hospital, Central South University (No. 2019030140).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, M., Chen, P., Wei, B. et al. FN1 shapes the behavior of papillary thyroid carcinoma through alternative splicing of EDB region. Sci Rep 15, 327 (2025). https://doi.org/10.1038/s41598-024-83369-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-83369-5