Abstract
Radio-resistance of cancer stem-like cells (CSCs) is associated with the failure of radiation therapy. ZsGreen1-positive (ZsG⁺) cells, which exhibit low proteasome activity, have been used to enable the detection and isolation of CSCs. However, the mechanisms of radio-resistance in canine tumor cells with low proteasome activity remain unclear. This study aimed to elucidate the radio-resistance mechanisms of ZsG+ cells by identifying a potential target of canine CSCs. ZsG+ cells, isolated using flow cytometric cell sorting, were compared with ZsG− cells. Sulfasalazine was used to suppress glutathione (GSH) synthesis by inhibiting xCT. In vitro experiments demonstrated a significantly higher radio-resistance in ZsG+ cells than in ZsG− cells. After X-irradiation, ZsG+ cells had fewer p53‑binding protein 1 (53BP1) foci, low reactive oxygen species (ROS) accumulation, and high GSH content. Sulfasalazine caused radiosensitization of ZsG+ cells with an increased number of 53BP1 foci by decreasing GSH contents and increasing ROS accumulation. The low proteasome activity played a role in xCT upregulation. In conclusion, canine tumor cells with low proteasome activity are radio-resistant due to high GSH content and low ROS accumulation. Sulfasalazine causes radiosensitization of the tumor cells by altering redox balance by inhibiting GSH synthesis for effective targeting of canine CSCs.
Similar content being viewed by others
Introduction
Cancer stem-like cells (CSCs) or tumor-initiating cells are a distinct subpopulation with specific surface markers that possess the metastatic capability of undergoing unlimited cell growth, self-renewal, asymmetric cell division, and differentiation1 The difference in radiosensitivity may be attributed to tumor heterogeneity, which is responsible for the inconsistent effect of radiation therapy on tumor cells2 Furthermore, the radio-resistance of tumor cells, particularly CSCs, makes it difficult to eradicate the tumor, suggesting that it is essential to understand the mechanism of radio-resistance3,4.
In a previous study, a CSC visualization system was developed for the isolation and identification of CSCs5 In this system, canine tumor cells were engineered to express a ZsGreen1 (ZsG) fluorescent protein connected to a proteasome degron, and canine CSCs were visualized because of their low proteasome activity; these cells were defined as ZsG+ cells. Finally, ZsG+ cells that exhibited CSC-like properties were successfully isolated using flow cytometric cell sorting. However, the radio-resistance of canine tumor cells with low proteasome activity and its mechanism remain unclear.
Various factors influence the radio-resistance of CSCs, including the degree of DNA damage, detection of reactive oxygen species (ROS), and glutathione (GSH) contents4,6,7 DNA damage is one of the most effective mechanisms by which ionizing radiation leads to tumor cell death,8 and DNA double-strand breaks (DSBs) are believed to cause major DNA damage in radiation therapy9 p53‑binding protein 1 (53BP1) is an important component of DSB signaling and facilitates the nonhomologous end-joining (NHEJ)-mediated DSB repair. 53BP1 has been employed as a marker for DNA damage and repair because of the rapid recruitment of 53BP1 to the DNA break sites10,11,12.
The generation of ROS plays the role of indirect damage to tumor cells, and this is observed in a large portion of DNA damage caused by low linear energy transfer radiation, such as X- and gamma-rays13,14 Thus, ROS is an important factor in controlling tumor cell radio-resistance15 To counteract ROS production and survive radiation therapy, radio-resistant tumor cells maintain increased levels of reductants, such as GSH, which protect cells from oxidative damage through reaction with electrophiles or ROS6,16,17.
The radio-resistance of CSCs is associated with their low proteasome activity, which reduces the proteasome-dependent degradation of antioxidant-related molecules such as xCT, Nrf2, and ATF4, thereby leading to their intracellular accumulation18,19,20 The cystine/glutamate antiporter xCT, also known as SLC7A11, is involved in GSH biosynthesis and maintains tumor redox status21 In addition, the accumulation of transcription factors including Nrf2 and ATF4 promotes xCT expression, enhancing the cellular antioxidant capacity19.
Inhibition of xCT using sulfasalazine, which is used for inflammatory bowel disease in dogs, can disrupt the redox balance of tumor cells by suppressing GSH synthesis22,23 GSH deprivation using sulfasalazine induces free radical accumulation and tumor cell death via ferroptosis and apoptosis24 Moreover, sulfasalazine targets tumor cells with high expression levels of CSC markers, such as CD133 or CD44v,25,26 and prolongs the survival of rat brain tumor models by inducing CSC death27 A clinical study of gastric tumors showed that sulfasalazine can reduce the population of CSCs in gastric tumor tissues28 Furthermore, in a study of canine liver tumors, the xCT of canine CSCs was upregulated and sulfasalazine decreased the sphere-forming capacity of the cells29 Our previous report also demonstrated that visualized canine tumor cells with low proteasome activity have upregulated CD133 and possess CSC-like properties5 These findings suggest that sulfasalazine may target canine tumor cells with low proteasome activity, making it a promising radiosensitizer candidate for eliminating canine CSCs.
In this study, the radio-resistance of canine tumor cells with low proteasome activity and its mechanism were explored by investigating the degree of DNA damage, accumulation of ROS, and GSH contents after X-irradiation. Further, the radiosensitizing effects of xCT inhibition were evaluated using sulfasalazine in canine CSCs.
Results
Ratio of ZsG+ cells after X-Irradiation
This study evaluated the ratio of ZsG+ cells using confocal microscopy and flow cytometry after X-irradiation with different doses. The ratio of ZsG+ cells increased in a dose-dependent manner at 3 days after X-irradiation in HMPOS–ZsG and MegTCC–ZsG, which are canine tumor cell lines engineered to visualize ZsG+ cells by green fluorescence under low proteasome activity (Fig. 1A and B). In addition, the ratio of ZsG+ cells increased in a time-dependent manner after treatment with 7.5 Gy of X-irradiation in HMPOS–ZsG (Supplementary Fig. 1A and 1B).
Radio-Resistance and Accumulation of 53BP1 Foci after X-Irradiation in ZsG+ Cells.
When 2 and 5 Gy of X-irradiation was applied to the cells at 6 h after cell sorting, ZsG+ cells exhibited a higher survival fraction than ZsG− cells in HMPOS–ZsG (2 Gy, 0.62 ± 0.02 and 0.79 ± 0.00, p = 0.015; 5 Gy, 0.25 ± 0.06 and 0.44 ± 0.01, p = 0.008 for ZsG− and ZsG+ cells, respectively) and MegTCC–ZsG (2 Gy, 0.45 ± 0.03 and 0.73 ± 0.04, p = 0.000; 5 Gy, 0.05 ± 0.00 and 0.15 ± 0.00, p = 0.023 for ZsG− and ZsG+ cells, respectively). When 5 Gy of X-irradiation was applied to the cells at 12 h after cell sorting, ZsG+ cells exhibited a higher survival fraction than ZsG− cells in HMPOS–ZsG (0.27 ± 0.01 and 0.43 ± 0.01, p = 0.045 for ZsG− and ZsG+ cells, respectively). In contrast, when 2 and 5 Gy of X-irradiation was applied to the cells at 12 h after cell sorting, ZsG+ cells exhibited a higher survival fraction than ZsG− cells in MegTCC–ZsG (2 Gy, 0.54 ± 0.02 and 0.79 ± 0.06, p = 0.001; 5 Gy, 0.09 ± 0.02 and 0.23 ± 0.00, p = 0.022 for ZsG− and ZsG+ cells, respectively) (Fig. 2A).
Different levels of DSB were confirmed by evaluating the accumulation of 53BP1 foci after X-irradiation in ZsG− and ZsG+ cells. The background level of 53BP1 foci per nucleus was < 5 in all groups. After 0.5 h of X-irradiation, ZsG+ cells exhibited a smaller number of 53BP1 foci than ZsG− cells in HMPOS–ZsG (12.80 ± 5.61 and 19.12 ± 3.99 for ZsG− and ZsG+ cells, respectively, p = 0.000) and MegTCC–ZsG (14.80 ± 3.01 and 20.12 ± 2.96 for ZsG− and ZsG+ cells, respectively, p = 0.000). After 1 h of X-irradiation, ZsG+ cells showed a smaller number of 53BP1 foci than ZsG− cells in MegTCC–ZsG (9.88 ± 2.57 and 13.36 ± 2.12 for ZsG− and ZsG+ cells, respectively, p = 0.000) (Fig. 2B and C).
Level of ROS after X-Irradiation and GSH content in ZsG+ cells
Cellular ROS levels were assessed using CellROX® Deep Red staining after X-irradiation. The CellROX fluorescence intensity of ZsG+ cells was lower than that of ZsG− cells in HMPOS–ZsG and MegTCC–ZsG after X-irradiation (Fig. 3A). The relative fluorescence intensity values are presented in Fig. 3B. The relative CellROX fluorescence intensity values in ZsG+ cells were significantly lower than those in ZsG− cells after X-irradiation in HMPOS–ZsG (p = 0.040) and MegTCC–ZsG (p = 0.025) (Fig. 3B).
Following the evaluation of cellular ROS level, total GSH content was measured in ZsG− and ZsG+ cells. The total GSH content was significantly lower in ZsG− cells than in ZsG+ cells in HMPOS–ZsG (6 h, p = 0.000; 12 h, p = 0.003) and MegTCC–ZsG (6 h, p = 0.002; 12 h, p = 0.001) (Fig. 3C).
Changes in radiosensitivity and DSB after the application of sulfasalazine
In ZsG− cells, the suppression of GSH synthesis via 1,000 µM sulfasalazine exhibited a tendency of radiosensitization; however, no significant difference was observed between HMPOS–ZsG and MegTCC–ZsG. In contrast, the suppression of GSH synthesis by 1,000 µM sulfasalazine significantly radiosensitized ZsG+ cells at 2 (p = 0.014) and 5 Gy (p = 0.016) of X-irradiation after 6 h of incubation with sulfasalazine and at 5 Gy (p = 0.040) of X-irradiation after 12 h of incubation with sulfasalazine in HMPOS–ZsG. The ZsG+ cells of MegTCC–ZsG were radiosensitized at 2 Gy of X-irradiation after 12 h of incubation with 1,000 µM sulfasalazine (p = 0.004) (Fig. 4A).
The immunofluorescence staining of 53BP1 revealed that xCT inhibition using 1000 µM sulfasalazine altered the DSB levels via X-irradiation in ZsG−and ZsG+ cells. After incubating tumor cells with sulfasalazine and treating with X-irradiation, the DSB levels of ZsG− and ZsG+ cells significantly increased in HMPOS–ZsG and MegTCC–ZsG. In addition, at 0.5 h after X-irradiation, the initial effects of sulfasalazine on DSB levels in ZsG+ cells were more significant than those in ZsG− cells (Fig. 4B). Consistent with these findings, a direct comparison of DSB levels between sulfasalazine-treated ZsG− and ZsG+ cells revealed that the levels were nearly indistinguishable at 0.5 and 1 h post-irradiation, except after 12 h of incubation in HMPOS–ZsG and 6 h of incubation in MegTCC–ZsG (Fig. 4C).
Change in redox status after the application of sulfasalazine
The cellular ROS levels were measured after incubation with 1,000 µM sulfasalazine for 6–12 h with or without X-irradiation. There was no difference in the CellROX fluorescence intensity between ZsG− and ZsG+ cells in the control and sulfasalazine groups for HMPOS–ZsG and MegTCC–ZsG. After X-irradiation, ZsG+ cells exhibited lower CellROX fluorescence intensity than ZsG− cells in the 6 h incubation group in HMPOS–ZsG (p = 0.000) and MegTCC–ZsG (p = 0.035). Moreover, ZsG+ cells showed a similar pattern of fluorescence intensity in the 12 h incubation group in HMPOS–ZsG (p = 0.018). Using the combination of sulfasalazine and X-irradiation, CellROX fluorescence intensity significantly increased in HMPOS–ZsG and MegTCC–ZsG (Fig. 5A).
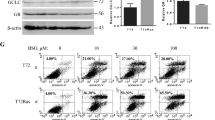
In the GSH content analysis, a sulfasalazine dose-dependent decrease in GSH content was observed in HMPOS–ZsG and MegTCC–ZsG. After adding ≥ 500 µM sulfasalazine to the cells for 6 h, the differences in GSH contents between ZsG− and ZsG+ cells disappeared in HMPOS–ZsG and MegTCC–ZsG. After adding 1,000 or 1,500 µM sulfasalazine to the cells for 12 h, the differences in GSH contents between ZsG− and ZsG+ cells disappeared in HMPOS–ZsG and MegTCC–ZsG (Fig. 5B). The relationship between GSH content and proteasome activity was demonstrated by evaluating xCT protein levels in HMPOS–ZsG (Supplementary Fig. 2). Densitometric analysis of the xCT protein levels revealed that xCT expression in ZsG− cells was markedly lower than in ZsG+ cells, which were shown to exhibit low proteasome activity in a previous study5 In addition, xCT protein levels in adherent cells increased following treatment with 1.0 µM MG-132, compared with both adherent cells treated with 0.5 µM MG-132 and ZsG− cells (Supplementary Fig. 3).
Discussion
Our previous study revealed that canine tumor cells with low proteasome activity possess CSC-like properties. According to another research, tumor cells with low proteasome activity are radio-resistant in human tumor cells; however, its mechanism remains unclear in both human and canine tumor cells31,32,33 The findings of this study unveiled the mechanism of radio-resistance in canine tumor cells with low proteasome activity.
After X-irradiation, the ratio of the survived ZsG+ cells increased in adherent cells in a dose-dependent manner, indicating that ZsG+ cells are radio-resistant. In addition, the actual radio-resistance was evaluated using clonogenic survival assay in ZsG− and ZsG+ cells. In this assay, ZsG+ cells showed higher radio-resistance than ZsG− cells. Since the clonogenic survival assay is the gold standard for measuring tumor cell radiosensitivity, these results further support the radio-resistance of ZsG+ cells.34 The increased ratio of the survived ZsG+ cells and their radio-resistance were consistent with the results of previous reports of human cancers, such as osteosarcoma, squamous cell carcinoma, prostate cancer, and colorectal cancer, in which tumor cells with CSC phenotype exhibited radio-resistance31,32,33,35,36.
In the present study, ZsG+ cells exhibited a small number of 53BP1 foci per nucleus than ZsG− cells after 0.5–1 h of X-irradiation. The accumulation of 53BP1 in the nucleus is associated with DSB because of the activation of RNF8–RNF168-mediated ubiquitylation cascade, which acts downstream of γH2AX, an indicator of DSB12,37 Thus, the results of this study show a low level of initial DNA damage in ZsG+ cells, which is consistent with the results of previous studies showing low foci formation of 53BP1 in CSCs after radiation therapy12,30 A low level of initial DNA damage was observed in radio-resistant human squamous cell carcinoma cells,38 and the radiosensitivity was correlated with initial DNA damage rather than residual DNA damage in 11 cell lines39 This could be due to the high GSH contents and low ROS accumulation38,40 Additionally, early responsive DNA repair such as nonhomologous end-joining (NHEJ) might be associated with the small number of 53BP1 foci in ZsG+ cells because most initial DSBs caused by radiation are repaired via the NHEJ process with a short half-life of 5–30 min41,42 Future studies on the DNA repair system of ZsG+ cells are needed to support this notion.
In the present study, ZsG+ cells exhibited lower accumulation of cellular ROS than ZsG− cells after exposure to X-irradiation in HMPOS–ZsG and MegTCC–ZsG. Mildly upregulated ROS levels in tumor cells contribute to tumor proliferation, survival, oncogene expression, and antitumor gene suppression by modulating several signaling pathways43 However, excessive ROS accumulation results in DSB and cell death with oxidative cellular damage caused by radiation40,44 To prevent this after X-irradiation, CSCs maintain low ROS levels by regulating the production and scavenging of ROS through altered GSH expression, increased antioxidant protein levels, and activation of ROS-dependent signaling pathways45,46 Therefore, maintaining low ROS levels after X-irradiation is associated with the survival and radio-resistance of ZsG+ cells.
Total GSH content was measured in the cells, and ZsG+ cells showed greater GSH content than ZsG− cells. According to a previous report, GSH depletion leads to the accumulation of reactive species such as ROS and activates apoptotic signaling pathways, resulting in apoptotic cell death47 In human breast cancer cells, the reduction in GSH levels induced radiosensitization with increased intracellular ROS levels23 In addition, the upregulated GSH level is an important factor for maintaining cancer stemness in CSCs, and inhibition of GSH synthesis decreased cancer stem-like properties and viability of CSCs in human pancreatic cancer cells48 Therefore, these data support the fact that GSH can be a promising target of canine CSCs in radiation therapy.
Subsequently, the effects of sulfasalazine on the radiosensitivity, DSB after X-irradiation, and redox status of ZsG+ cells were evaluated. In this study, sulfasalazine enhanced the radiosensitivity of ZsG+ cells and increased DSB formation after X-irradiation, particularly amplifying the initial DSB of ZsG+ cells compared to ZsG− cells, which resulted in similar initial DSB levels between the two cell types. In addition, sulfasalazine increased the accumulation of intracellular ROS after X-irradiation and decreased GSH content in ZsG+ cells. The protein level of xCT was high in HMPOS–ZsG cells exhibiting low proteasome activity, such as ZsG+ cells and adherent cells treated with MG-132, indicating a relationship between the xCT protein level and proteasome activity. According to previous studies, the inhibition of xCT using sulfasalazine suppressed the synthesis of the reductant GSH,23 and this was associated with the sensitivity of canine tumor cells to oxidative damage caused by ROS such as H2O249 These effects of sulfasalazine on ZsG+ cells may be caused by the upregulated protein levels of xCT, which appear to be associated with the degradation of xCT via the proteasome activity following ubiquitination. This degradation is stabilized by forming a tri-protein complex with the CD44 variant and OTUB1, a member of the deubiquitinase family, thereby contributing to the protein stability of xCT50 Sulfasalazine also exerts antitumor effects on tumor cells expressing CSC markers such as CD133 by increasing their sensitivity to oxidative stress25,51 The use of sulfasalazine enhances CSC sensitivity to antitumor drugs, reduces CSC population, and improves host survival25,26,27 In light of these studies, the high level of xCT expression in ZsG+ cells due to low proteasome activity may have contributed to their radio-resistance by increasing GSH levels, preventing oxidative damage, and reducing DSB levels induced by X-irradiation. Sulfasalazine may have suppressed xCT function in ZsG+ cells, leading to a decrease in GSH levels. This reduction was more pronounced in ZsG+ cells, bringing GSH levels down to those observed in ZsG⁻ cells. These changes may have led to increased post-irradiation ROS accumulation and DSB levels in ZsG+, comparable to those in ZsG− cells, thereby reducing the radio-resistance of ZsG+ cells. Lastly, the elevated xCT protein level following MG-132 treatment supports our hypothesis that high xCT expression in ZsG+ cells is associated with low proteasome activity. This suggests a connection between low proteasome activity and the radio-resistance of canine CSCs. Altogether, these findings support the potential of xCT as a therapeutic target in CSCs, particularly in the context of radiosensitization.
A potential limitation of the present study is that the ratio of ZsG+ cells decreases with incubation time after cell sorting; therefore, incubation time of tumor cells with sulfasalazine was set to 6 and 12 h5. Thus, we could not evaluate a pure population of ZsG+ cells, and experiments following cell sorting should have been performed within a limited period. Specific strategies to prevent this phenomenon are needed in future studies. Additionally, although our findings suggest a role for GSH in the radio-resistance of CSCs, further investigation into GSH metabolic pathways and associated enzymes will be necessary to fully elucidate the contribution of GSH metabolism to radio-resistance.
In conclusion, the present study elucidated that the upregulated GSH contents contribute to the radio-resistance of canine CSCs with low proteasome activity, allowing the cells to tolerate oxidative damage and DSB caused by X-irradiation. Furthermore, sulfasalazine causes radiosensitization of canine tumor cells with low proteasome activity by altering the redox balance, suggesting that xCT is a promising target for the radio-resistance of canine CSCs.
Materials and methods
Cell culture conditions
A previous study engineered canine osteosarcoma cell line (HMPOS) and transitional cell carcinoma cell line (MegTCC) to express ZsGreen1-cODC for generating HMPOS–ZsG and MegTCC–ZsG. The accumulation of ZsGreen1-cODC due to low proteasome activity allowed the visualization of canine CSCs, referred to as ZsG+ cells5. All tumor cells were cultured in RPMI-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Sigma-Aldrich, St. Louis, MO, USA), 100 units/mL penicillin G, 100 µg/mL streptomycin (Wako Pure Chemical Industries, Osaka, Japan), and 10 µg/mL blasticidin (Wako) in a humidified incubator at 37 °C and 5% CO2.
Flow cytometry and cell sorting
HMPOS–ZsG and MegTCC–ZsG were washed with phosphate-buffered saline (PBS) and were suspended using 0.025% trypsin–ethylenediaminetetraacetic acid for subsequent fluorescence-activated cell sorting analysis using a FACSVerse flow cytometer (BD Biosciences, San Jose, CA, USA) or FACSAria II flow cytometer (BD Biosciences). Based on the fluorescence intensity of ZsG, ZsG− and ZsG+ cells were defined, as described in a previous study5.
Irradiation
Cultured tumor cells were X-irradiated using a Titan 320s X-ray generator (Shimadzu Corporation, Kyoto, Japan) at room temperature (dose rate of 4.62 Gy/min, 200 kVp, 20 mA with a 1.5 mm aluminum filter).
Clonogenic survival assay
The sorted ZsG− and ZsG+ cells were plated on 60-mm cell culture dishes and incubated for 6–12 h. Next, the cells were X-irradiated, and the cell culture medium was replaced by a fresh medium, forming colonies for 6 or 7 days at 37 °C and 5% CO2. Methanol fixation and staining were performed using Giemsa solution (Wako), and colonies containing > 50 cells were quantified using ImageJ software (National Institutes of Health). Plating efficiency was calculated and used to normalize each survival fraction.
Immunofluorescence microscopy
Immunofluorescence microscopy was performed using a modified method described in a previous study30 Briefly, cancer cells were plated on glass coverslips and incubated overnight in cell culture medium at 37 °C and 5% CO2. Then, the cells were exposed to 1 Gy of X-irradiation, and each group was incubated for different time periods. Next, the cells were fixed using 4% paraformaldehyde (Wako) and permeabilized using PBS containing 0.5% Triton X-100. After washing with PBS and incubation with blocking buffer, the cells were stained with a primary antibody rabbit anti-53BP1 (1:2,000; Abcam Inc., Cambridge, UK) and a secondary antibody goat antirabbit Alexa Fluor 555 (1:2,000; Invitrogen, Carlsbad, CA, USA). At the end of the experiment, the cells were counterstained with NucBlue (Invitrogen), according to the manufacturer’s instructions, and were mounted with ProLong Diamond Antifade Mountant (Invitrogen) for microscopic fluorescence analysis under an Olympus BX50 microscope (Olympus, Tokyo, Japan). The number of 53BP1 foci, which appeared as fluorescent dots, was estimated.
Detection of reactive oxygen species
The accumulation of ROS was measured using CellROX® Deep Red Reagent (Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. Cells were plated on 60-mm cell culture dishes and incubated for 3 days at 37 °C and 5% CO2, followed by treatment with 5 Gy of X-irradiation. At 24 h after X-irradiation, CellROX® Deep Red Reagent was added to the cells (final concentration, 5 µM), and the cells were incubated for 30 min at 37 °C and 5% CO2. Then, the cells were washed three times with PBS and were resuspended for analysis using the FACSVerse flow cytometer (BD Biosciences).
Glutathione assay
The GSH content was measured using a GSH-Glo™ Glutathione Assay Kit (Promega, Tokyo, Japan) following the manufacturer’s protocol. Briefly, the sorted ZsG− and ZsG+ cells were plated onto 96-white well plates (Greiner Bio-One, Frickenhausen, Germany) at a concentration of 5 × 103 cells/well, and the cells were incubated for 6–12 h at 37 °C and 5% CO2. After removing the culture medium, the cells were incubated with 100 µL of GSH-Glo™ Reagent for 30 min at room temperature. At 15 min after the addition of 100 µL of reconstituted Luciferin Detection Reagent, the luminescence intensity was measured using a microplate reader (Infinite 200, Tecan Japan, Kanagawa, Japan). A standard curve was generated to ensure that the luminescence signal was within the linear detection range under the experimental conditions. Background luminescence was subtracted from the GSH signal, which was then normalized to the values obtained from ZsG⁻ cells.
Statistical analysis
Data are presented as mean ± standard deviation from three independent biological replicates, each performed with three technical replicates. Statistical analysis was conducted using IBM SPSS Statistics version 26.0 (IBM). The normality of the results was evaluated using the Shapiro–Wilk test. The statistical significance between groups was determined using one-way analysis of variance, followed by Tukey’s multiple comparison test. A p-value of < 0.05 (*p), 0.01 (**p) or 0.001 (***p) was considered statistically significant.
Ratio of ZsG+ cells after exposure to X-irradiation. (A) Fluorescence images of HMPOS–ZsG and MegTCC–ZsG showing a change in the ratio of ZsG+ cells after exposure to X-irradiation. Images were captured at a magnification of x100. (B) Dose-dependent change in the ratio of ZsG+ cells after X-irradiation in HMPOS–ZsG and MegTCC–ZsG. Error bars correspond to the standard deviation of the mean from three independent experiments. Gray circles indicate the value from an individual biological replicate. Tukey’s multiple comparison test was used for statistical analysis. *p < 0.05 and ***p < 0.001.
Radio-resistance and accumulation of 53BP1 foci after X-irradiation in ZsG+ cells. (A) Radiation survival curves of ZsG− and ZsG+ cells were evaluated using the clonogenic survival assay at 6–12 h of incubation after cell sorting. (B) Fluorescence images of ZsGreen1, DAPI, and 53BP1 in ZsG− and ZsG+ cells after treatment with 1 Gy of X-irradiation for 0.5 h. Images were captured at a magnification of x200. (C) Time-dependent changes of the average number of 53BP1 foci in ZsG− and ZsG+ cells after treatment with 1 Gy of X-irradiation. Error bars correspond to the standard deviation of the mean from three independent experiments. Tukey’s multiple comparison test was used for statistical analysis. *p < 0.05, **p < 0.01, and ***p < 0.001. 53BP1, p53‑binding protein 1; DAPI, 4′,6-diamidino-2-phenylindole.
ROS accumulation after X-irradiation and GSH contents in ZsG+ cells. (A) Flow cytometric analysis of ROS accumulation in ZsG− and ZsG+ cells after 0–5 Gy of X-irradiation. (B) Comparative analysis using the results of ROS accumulation normalized to the CellROX fluorescence of ZsG− cells in HMPOS–ZsG without X-irradiation. (C) Relative GSH contents of ZsG− and ZsG+ cells at 6–12 h of incubation after cell sorting were determined in HMPOS–ZsG and MegTCC–ZsG. Error bars correspond to the standard deviation of the mean of three independent experiments. Gray circles indicate the value from an individual biological replicate. Tukey’s multiple comparison test was used for statistical analysis. *p < 0.05, **p < 0.01, and ***p < 0.001. ROS, reactive oxygen species; GSH, glutathione.
Effect of sulfasalazine on the radio-resistance of ZsG+ cells. (A) Radiation survival curves showing radiosensitizing effects of 1,000 µM sulfasalazine. (B) Change in DSB levels due to X-irradiation after incubation with 1,000 µM sulfasalazine in ZsG− and ZsG+ cells. (C) Comparison of X-irradiation–induced DSB levels between ZsG− and ZsG+ cells after incubation with 1,000 µM sulfasalazine. Error bars correspond to the standard deviation of the mean from three independent experiments. Tukey’s multiple comparison test was used for statistical analysis. *p < 0.05, **p < 0.01, and ***p < 0.001. DSB, DNA double-strand break.
Effect of sulfasalazine on ROS accumulation after X-irradiation and GSH contents in ZsG+ cells. (A) Accumulation of intracellular ROS after X-irradiation (RT) with 1,000 µM sulfasalazine (Sulfa) or without sulfasalazine (Con) in ZsG− and ZsG+ cells. (B) GSH contents of ZsG− and ZsG+ cells after treatment with different concentrations of sulfasalazine. Error bars correspond to the standard deviation of the mean from three independent experiments. Gray circles indicate the value from an individual biological replicate. Tukey’s multiple comparison test was used for statistical analysis. *p < 0.05, **p < 0.01, and ***p < 0.001. ROS, reactive oxygen species; GSH, glutathione.
Data availability
Data is provided within the manuscript or supplementary information files.The datasets generated for this study can be found in the https://doi.org/10.6084/m9.figshare.19594354.
References
Walcher, L. et al. Cancer stem Cells—Origins and biomarkers: perspectives for targeted personalized therapies. Front. Immunol. 11, 1280. https://doi.org/10.3389/fimmu.2020.01280 (2020).
Olivares-Urbano, M. A., Griñán-Lisón, C., Marchal, J. A. & Núñez, M. I. CSC radioresistance: A therapeutic challenge to improve radiotherapy effectiveness in Cancer. Cells 9, 1651. https://doi.org/10.3390/cells9071651 (2020).
Li, Y., Wang, Z., Ajani, J. A. & Song, S. Drug resistance and Cancer stem cells. Cell. Commun. Signal. 19, 19. https://doi.org/10.1186/s12964-020-00627-5 (2021).
Arnold, C. R., Mangesius, J., Skvortsova, I. I. & Ganswindt, U. The role of Cancer stem cells in radiation resistance. Front. Oncol. 10, 1–12. https://doi.org/10.3389/fonc.2020.00164 (2020).
Sung, K. et al. Visualizing the cancer stem-like properties of canine tumour cells with low proteasome activity. Vet. Comp. Oncol. 20, 324–335. https://doi.org/10.1111/vco.12779 (2022).
Rycaj, K. & Tang, D. G. Cancer stem cells and radioresistance. Int. J. Radiat. Biol. 90, 615–621. https://doi.org/10.3109/09553002.2014.892227 (2014).
Lyakhovich, A. & Lleonart, M. E. Bypassing Mechanisms of Mitochondria-Mediated Cancer Stem Cells Resistance to Chemo- and Radiotherapy. Oxid. Med. Cell. Longev. 1–10. (2016). https://doi.org/10.1155/2016/1716341 (2016).
Santivasi, W. L., Xia, F. & Ionizing Radiation-Induced, D. N. A. Damage, response, and repair. Antioxid. Redox Signal. 21, 251–259. https://doi.org/10.1089/ars.2013.5668 (2014).
Lomax, M. E., Folkes, L. K. & O’Neill, P. Biological consequences of Radiation-induced DNA damage: relevance to radiotherapy. Clin. Oncol. 25, 578–585. https://doi.org/10.1016/j.clon.2013.06.007 (2013).
Sekhar, K. R., Wang, J., Freeman, M. L. & Kirschner, A. N. Radiosensitization by enzalutamide for human prostate cancer is mediated through the DNA damage repair pathway. PLoS One. 14, e0214670. https://doi.org/10.1371/journal.pone.0214670 (2019).
Shimamura, M., Yamamoto, K., Kurashige, T. & Nagayama, Y. Intracellular redox status controls spherogenicity, an in vitro cancer stem cell marker, in thyroid cancer cell lines. Exp. Cell. Res. 370, 699–707. https://doi.org/10.1016/j.yexcr.2018.07.036 (2018).
Panier, S. & Boulton, S. J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell. Biol. 15, 7–18. https://doi.org/10.1038/nrm3719 (2014).
Desouky, O., Ding, N. & Zhou, G. Targeted and non-targeted effects of ionizing radiation. J. Radiat. Res. Appl. Sci. 8, 247–254. https://doi.org/10.1016/j.jrras.2015.03.003 (2015).
Kawamura, K., Qi, F. & Kobayashi, J. Potential relationship between the biological effects of low-dose irradiation and mitochondrial ROS production. J. Radiat. Res. 59, ii91–ii97. https://doi.org/10.1093/jrr/rrx091 (2018).
Wang, H., Jiang, H., Van De Gucht, M. & De Ridder, M. Hypoxic radioresistance: can ROS be the key to overcome it? Cancers (Basel). 11, 112. https://doi.org/10.3390/cancers11010112 (2019).
Diehn, M. et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature 458, 780–783. https://doi.org/10.1038/nature07733 (2009).
Ribas, V., GarcÃa-Ruiz, C. & Fernández-Checa, J. C. Glutathione and mitochondria. Front. Pharmacol. 5, 1–19. https://doi.org/10.3389/fphar.2014.00151 (2014).
Voutsadakis, I. A. Proteasome expression and activity in cancer and cancer stem cells. Tumor Biol. 39, 101042831769224. https://doi.org/10.1177/1010428317692248 (2017).
Ye, P. et al. Nrf2- and ATF4-Dependent upregulation of xCT modulates the sensitivity of T24 bladder carcinoma cells to proteasome Inhibition. Mol. Cell. Biol. 34, 3421–3434. https://doi.org/10.1128/MCB.00221-14 (2014).
Liu, T., Jiang, L., Tavana, O. & Gu, W. The deubiquitylase OTUB1 mediates ferroptosis via stabilization of SLC7A11. Cancer Res. 79, 1913–1924. https://doi.org/10.1158/0008-5472.CAN-18-3037 (2019).
Lim, J. K. M. et al. Cystine/glutamate antiporter xCT (SLC7A11) facilitates oncogenic RAS transformation by preserving intracellular redox balance. Proc. Natl. Acad. Sci. 116, 9433–9442. (2019). https://doi.org/10.1073/pnas.1821323116
Nagane, M. et al. Sulfasalazine, an inhibitor of the cystine-glutamate antiporter, reduces DNA damage repair and enhances radiosensitivity in murine B16F10 melanoma. PLoS One. 13, e0195151. https://doi.org/10.1371/journal.pone.0195151 (2018).
Cobler, L., Zhang, H., Suri, P., Park, C. & Timmerman, L. A. xCT Inhibition sensitizes tumors to γ-radiation via glutathione reduction. Oncotarget 9, 32280–32297. https://doi.org/10.18632/oncotarget.25794 (2018).
Koppula, P., Zhuang, L. & Gan, B. Cystine transporter SLC7A11/xCT in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 12, 599–620. https://doi.org/10.1007/s13238-020-00789-5 (2021).
Song, Y. et al. Sulfasalazine attenuates evading anticancer response of CD133-positive hepatocellular carcinoma cells. J. Exp. Clin. Cancer Res. 36, 38. https://doi.org/10.1186/s13046-017-0511-7 (2017).
Thanee, M. et al. Sulfasalazine modifies metabolic profiles and enhances cisplatin chemosensitivity on cholangiocarcinoma cells in in vitro and in vivo models. Cancer Metab. 9, 11. https://doi.org/10.1186/s40170-021-00249-6 (2021).
Haryu, S. et al. Convection-enhanced delivery of sulfasalazine prolongs survival in a glioma stem cell brain tumor model. J. Neurooncol. 136, 23–31. https://doi.org/10.1007/s11060-017-2621-7 (2018).
Shitara, K. et al. Dose-escalation study for the targeting of CD44v + cancer stem cells by sulfasalazine in patients with advanced gastric cancer (EPOC1205). Gastric Cancer. 20, 341–349. https://doi.org/10.1007/s10120-016-0610-8 (2017).
Itoh, H. et al. Sphere-forming cells display stem cell-like characteristics and increased xCT expression in a canine hepatocellular carcinoma cell line. Res. Vet. Sci. 139, 25–31. https://doi.org/10.1016/j.rvsc.2021.06.013 (2021).
Deguchi, T. et al. Analysis of radiosensitivity of cancer stem-like cells derived from canine cancer cell lines. Vet. Comp. Oncol. 17, 119–129. https://doi.org/10.1111/vco.12452 (2019).
Tamari, K. et al. Identification of chemoradiation-resistant osteosarcoma stem cells using an imaging system for proteasome activity. Int. J. Oncol. 45, 2349–2354. https://doi.org/10.3892/ijo.2014.2671 (2014).
Della Donna, L., Lagadec, C. & Pajonk, F. Radioresistance of prostate cancer cells with low proteasome activity. Prostate 72, 868–874. https://doi.org/10.1002/pros.21489 (2012).
Kano, Y. et al. Novel drug discovery system for cancer stem cells in human squamous cell carcinoma of the esophagus. Oncol. Rep. 31, 1133–1138. https://doi.org/10.3892/or.2013.2952 (2014).
Franken, N. A. P., Rodermond, H. M., Stap, J., Haveman, J. & van Bree, C. Clonogenic assay of cells in vitro. Nat. Protoc. 1, 2315–2319. https://doi.org/10.1038/nprot.2006.339 (2006).
Lagadec, C. et al. Tumor cells with low proteasome subunit expression predict overall survival in head and neck cancer patients. BMC Cancer. 14, 152. https://doi.org/10.1186/1471-2407-14-152 (2014).
Munakata, K. et al. Cancer stem-like properties in colorectal cancer cells with low proteasome activity. Clin. Cancer Res. 22, 5277–5286. https://doi.org/10.1158/1078-0432.CCR-15-1945 (2016).
Tsvetkova, A. et al. γH2AX, 53BP1 and Rad51 protein foci changes in mesenchymal stem cells during prolonged X-ray irradiation. Oncotarget 8, 64317–64329. https://doi.org/10.18632/oncotarget.19203 (2017).
Hanot, M. et al. Glutathione depletion and carbon ion radiation potentiate clustered DNA lesions, cell death and prevent chromosomal changes in Cancer cells progeny. PLoS One. 7, e44367. https://doi.org/10.1371/journal.pone.0044367 (2012).
Roos, P., Binder, A. & Böhm, L. Determination of the initial DNA damage and residual DNA damage remaining after 12 hours of repair in eleven cell lines at low doses of irradiation. Int. J. Radiat. Biol. 76, 1493–1500. https://doi.org/10.1080/09553000050176252 (2000).
Kim, W. et al. Cellular stress responses in radiotherapy. Cells 8, 1105. https://doi.org/10.3390/cells8091105 (2019).
Ahmed, E. A., Rosemann, M. & Scherthan, H. NHEJ contributes to the fast repair of Radiation-induced DNA Double-strand breaks at late prophase i telomeres. Health Phys. 115, 102–107. https://doi.org/10.1097/HP.0000000000000852 (2018).
Morgan, M. A. & Lawrence, T. S. Molecular pathways: overcoming radiation resistance by targeting DNA damage response pathways. Clin. Cancer Res. 21, 2898–2904. https://doi.org/10.1158/1078-0432.CCR-13-3229 (2015).
Perillo, B. et al. ROS in cancer therapy: the bright side of the Moon. Exp. Mol. Med. 52, 192–203. https://doi.org/10.1038/s12276-020-0384-2 (2020).
Reczek, C. R. & Chandel, N. S. The two faces of reactive oxygen species in cancer. Annu. Rev. Cancer Biol. 1, 79–98. https://doi.org/10.1146/annurev-cancerbio-041916-065808 (2017).
Tuy, K., Rickenbacker, L. & Hjelmeland, A. B. Reactive oxygen species produced by altered tumor metabolism impacts cancer stem cell maintenance. Redox Biol. 44, 101953. https://doi.org/10.1016/j.redox.2021.101953 (2021).
Ding, S. et al. Redox regulation in cancer stem cells. Oxid. Med. Cell. Longev. 1–11. (2015). https://doi.org/10.1155/2015/750798 (2015).
Franco, R. & Cidlowski, J. A. Apoptosis and glutathione: beyond an antioxidant. Cell. Death Differ. 16, 1303–1314. https://doi.org/10.1038/cdd.2009.107 (2009).
Jagust, P., Alcalá, S., Heeschen, C. & Sancho, P. Glutathione metabolism is essential for self-renewal and chemoresistance of pancreatic cancer stem cells. World J. Stem Cells. 12, 1410–1428. https://doi.org/10.4252/wjsc.v12.i11.1410 (2020).
Tanabe, A. et al. Functional analysis of CD44 variants and xCT in canine tumours. Vet. Med. Sci. 7, 577–585. https://doi.org/10.1002/vms3.397 (2021).
Gan, B. Dubbing ferroptosis in cancer cells. Cancer Res. 79, 1749–1750. https://doi.org/10.1158/0008-5472.CAN-19-0487 (2019).
Jang, J. W., Song, Y., Kim, S. H., Kim, J. & Seo, H. R. Potential mechanisms of CD133 in cancer stem cells. Life Sci. 184, 25–29. https://doi.org/10.1016/j.lfs.2017.07.008 (2017).
Acknowledgements
The authors would like to thank Dr. Yuki Hoshino (Department of Small Animal Surgery, Faculty of Agriculture, Iwate University) for providing canine cancer cell line MegTCC.
Funding
This research was supported by Grant-in-Aid for Scientific Research (Grant number: 20K06424) and Grant-in-Aid for Early-Career Scientists (Grant number: 20K15655).
Author information
Authors and Affiliations
Contributions
KS carried out experiments and wrote the original manuscript. TS, SK, and TD contributed to the development of study design and methodologies. KY provided important technical advice and a review of methodologies. KH and HY provided critical insights into the writing manuscript and mentoring. OI and MO supervised the research activities. All authors have reviewed the manuscript and approved it for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sung, K., Hosoya, K., Deguchi, T. et al. Glutathione-associated redox regulation alleviates radio-resistance of canine cancer stem-like cells with low proteasome activity. Sci Rep 15, 18017 (2025). https://doi.org/10.1038/s41598-025-02569-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02569-9
Keywords
This article is cited by
-
Targeting proteostasis for cancer therapy: current advances, challenges, and future perspectives
Molecular Cancer (2025)