Abstract
Cholangiocarcinoma (CCA), a neoplasm arising from biliary epithelial cells, is particularly widespread in Southeast Asia, with northeastern Thailand exhibiting the greatest prevalence attributed to Opisthorchis viverrini infection. This malignancy exhibits considerable molecular heterogeneity, leading to therapeutic resistance and recurrence. Comprehending its molecular mechanisms is essential for enhancing diagnostic and treatment approaches. Our research utilized multi-region LC-MS/MS proteomic analysis to investigate intratumor heterogeneity (ITH) in CCA. We examined 52 tumor areas and 13 neighboring tissues from 13 patients, concentrating on protein profiling, pathway analysis, differential protein expression, and the identification of shared and unique protein signatures. The findings indicated considerable inter-patient proteome variability, characterized by markedly distinct protein expressions among individuals, aligning with prior cancer research. Intra-tumor heterogeneity was apparent, with merely 18 proteins common to all tumor areas and patients, underscoring the intricacy of CCA. Significantly, the common proteins were associated with metabolic reprogramming and oxidative stress pathways, indicating possible indicators and therapeutic targets. This work highlights the significant proteome variability in CCA at both intra-tumor and inter-patient levels, underscoring the necessity for customized therapeutic approaches to tackle the disease’s complexity and improve treatment outcomes.
Similar content being viewed by others
Introduction
Cholangiocarcinoma (CCA) is a disease characterized by the presence of several types of tumours that exhibit similarities to the development of cholangiocytes1. CCA can be classified based on its anatomical location, which includes intrahepatic (iCCA), perihilar (pCCA), or distal (dCCA)2. Over the last forty years, there has been a consistent rise in the global prevalence of CCA, positioning it as the second most prevalent form of liver cancer, behind hepatocellular carcinoma (HCC)3,4,5. CCA, although the second most frequent kind of primary liver cancer, is considered a rare disorder, with a worldwide prevalence of less than 6 cases per 100,000 individuals. CCA is quite prevalent in Southeast Asian nations, especially in the northeastern region of Thailand6. Although several risk factors have been proposed for the initiation of CCA, the precise cause of this disease remains unidentified. Predominant sclerosing cholangitis is the predominant risk factor for CCA in the Western nations. Nevertheless, the occurrence of this disease is closely linked to the infestation of small human liver parasites, such as Opisthorchis viverrini and Clonorchis sinensis, in Southeast Asian nations7. Patients with CCA are typically detected at advanced stages, resulting in a limited selection of treatment choices and a bleak prognosis. In addition, the efficacy of surgical excision, which is the sole dependable remedy for CCA, might be hindered by delayed detection. Therefore, achieving a complete solution for CCA is quite difficult8,9.
Intratumor heterogeneity (ITH) is the presence of many cell populations inside a single tumor tissue. This heterogeneity significantly contributes to the lack of efficiency of anticancer pharmaceutical therapy. ITH leads to variations in the rate of cancer growth, capacity to infiltrate neighbouring tissues, sensitivity to treatments, and overall prognosis. The variations stem from alterations in the genetic and phenotypic traits of the several cell populations inside the tumor10. Tumor heterogeneity in CCA has previously been investigated using genomic approaches, including our earlier study employing whole exome sequencing of multi-regional tumor samples, which revealed significant intratumoral diversity in Opisthorchis viverrini-associated CCA11. Although genomic studies have significantly advanced our understanding of the mutational landscape in CCA, the proteomic aspect of its intratumoral heterogeneity remains insufficiently investigated. Proteomics, by enabling the direct quantification of protein abundance and activity, offers a more functionally relevant perspective on tumor biology and may reveal actionable therapeutic targets12. To address this knowledge gap, the present study employs LC-MS/MS-based proteomic profiling to characterize spatial variability in the tumor proteome across distinct intratumoral regions.
Proteomics is the scientific field that focuses on the analysis of protein composition and modifications in cells, organs, and organisms. Proteomics is extensively utilized in fundamental and clinical medical research13 to detect biomarkers14, investigate posttranslational protein modifications15, and study the regulation of signaling pathways16. Proteomics facilitates an extensive examination of the molecular environment in tumors, offering insights into protein expression patterns that influence cancer growth and therapeutic response. This methodology is especially beneficial in diseases such as CCA, where tumor heterogeneity hinders therapeutic options17,18. Through the analysis of proteome variations both intra- and inter-tumoral heterogeneity, researchers can discern critical proteins that influence tumor dynamics, metastasis, and therapeutic resistance19.
To address these issues, we investigated and assessed the multisite proteomics heterogeneity of CCA by proteome profiling, employing four locations and their adjacent tissues from tumors excised from 13 patients. We focused on ITH (inside a tumor resected from a single patient) and inter-patient heterogeneity (among tumors resected from different patients) of CCA.
Methods
Patients and samples collection
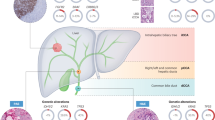
In order to identify ITH in CCA, we obtained fresh frozen tissue samples from 65 distinct tumour locations in 13 patients who had undergone curative excision at Srinagarind Hospital, Faculty of Medicine, Khon Kaen University. The tumour locations varied from area 4 of the tumour to adjacent areas in every instance. In order to investigate the different types of CCA, we gathered data on individuals who were diagnosed with CCA from the Department of Pathology, Faculty of Medicine, Khon Kaen University. The study included patients who satisfied the following criteria: individuals who had undergone surgical procedures and had tumours larger than 6 cm in size. All procedures were executed in accordance with applicable norms and regulations, including the Declaration of Helsinki and appropriate national and institutional guidelines. Authorization for the research was obtained from the Human Research Ethics Committee of Khon Kaen University (Approval No. HE671356). Informed consent was obtained from all participants and/or their legal guardians after a thorough explanation of the study. We collected a total of 52 samples of tumour tissue and non-cancerous surrounding tissue from 13 individuals diagnosed with CCA, as indicated in Table 1. Simultaneously, we gathered comprehensive pathology and clinical data, which encompassed the patients’ prognosis as well. Protein extracted from tissues was used for proteomics profiling by the application of LC-MS/MS. The workflow for this study is illustrated in Fig. 1.
Schematic representation of the overall workflow for identifying common and region-specific protein signatures using LC-MS/MS analysis. The figure was created using BioRender (https://biorender.com) and Adobe Photoshop 2021 (https://www.adobe.com/products/photoshop.html).
Protein extraction and Label-free protein quantification by mass spectrometry
sample preparation for shotgun proteomics.
The tissues were cryogenically frozen using liquid nitrogen and subsequently pulverised into a fine powder. A quantity of 100 milligrammes of this powder was mixed with an equivalent amount of a solution containing 0.5% sodium dodecyl sulphate (SDS). The mixture was rapidly agitated for 1 h, and thereafter subjected to centrifugation at 10,000 g for 15 min. The liquid supernatant was transferred cautiously to a fresh tube and thoroughly blended with an amount of cold acetone equal to double its volume. Subsequently, the combination was subjected to overnight incubation at a temperature of −20 °C. Following the incubation period, the mixture was subjected to centrifugation with a force of 10,000 times the acceleration due to gravity (10,000 g) for a duration of 15 min. The liquid portion above the sediment, known as the supernatant, was then removed and discarded. The residual pellet was dehydrated and preserved at −80 °C until required for subsequent use.
Liquid Chromatography-Tandem mass spectrometry (LC/MS-MS)
The tryptic peptide samples were prepared for injection into an Ultimate3000 Nano/Capillary LC System (manufactured by Thermo Scientific, UK) connected to a Hybrid quadrupole Q-Tof Impact II mass spectrometer (manufactured by Bruker Daltonics) that was equipped with a nano-electrospray ionization source. In summary, a volume of 1 µl from each peptide digest was concentrated using a C18 trap column (300 μm x 5 mm, 5 μm particle size, 100Å pore size from Thermo Scientific). The peptides were subsequently isolated using a 75 μm × 15 cm C18 nano-analytical column filled with 2 μm, 100Å particles (specifically, the Acclaim PepMap RSLC column manufactured by Thermo Scientific). The temperature of the column was maintained at 60 °C. The mobile phases A and B were composed of 0.1% formic acid in water and 0.1% formic acid in 80% acetonitrile, respectively. The elution of peptides was performed using a linear gradient of solvent B, ranging from 5 to 55%, over a period of 30 min. The flow rate during this process was set at 0.30 µL/min. The process of electrospray ionisation was conducted at a voltage of 1.6 kilovolts utilizing the CaptiveSpray source. The drying gas used was nitrogen, with a flow rate of around 50 L per hour. Product ion spectra were obtained using collision-induced dissociation (CID), with nitrogen as the collision gas. Mass spectrometry (MS) and tandem mass spectrometry (MS/MS) data were obtained using positive ion mode at a frequency of 2 Hz, covering the mass-to-charge ratio (m/z) range of 150–2200. The collision energy was increased gradually to 10 electron volts (eV) in relation to the mass-to-charge (m/z) value. The nanoLC-MS/MS approach was used to analyze each sample three times.
Bioinformatics and data analysis
The MaxQuant 2.2.0.0 programs was employed to quantify the proteins in each individual sample. The Andromeda search engine, incorporated within MaxQuant, was utilized to align the obtained tandem mass spectra (MS/MS) with the Uniprot protein database for the Homo sapiens species20. This facilitated the determination and measurement of proteins in the samples based on the identified peptide sequences. MaxQuant standard parameters were used for label-free quantification, allowing a maximum of two missed trypsin cleavages and a mass tolerance of 0.6 Da for the primary search. Trypsin was designated as the enzyme responsible for digestion. The fixed modification chosen was carbamidomethylation of cysteine, whereas the variable modifications chosen were oxidation of methionine and N-terminal protein acetylation. The Uniprot Homo sapiens reference proteome database was utilized for the purpose of conducting the database search.
Data analysis in proteomics
The ProteinGroups.txt file from MaxQuant was imported into Perseus version 1.6.6.021. Any putative contaminants that did not match any UPS1 protein were excluded from the dataset. The maximum intensities underwent a log2 transformation. Perseus was used to impute missing data, using a constant value of zero. The dataset underwent normalisation, statistical analysis, and visualisation using Metaboanalyst 6.022,23. The protein interaction networks were predicted using STITCH 5.024.
Results
Patient characteristics
This group comprised patients who underwent resectable surgeries at Srinagarind Hospital, Khon Kaen, Thailand as indicated in Table 1. Samples from 13 CCA patients were obtained, comprising 4 tissue sections per patient, resulting in a total of 52 samples, in addition to neighboring tissues. The patients’ ages varied from 53 to 77 years, with a mean age of 64 years. The cohort comprised 7 males and 6 females. Their post-operative survival duration ranged from 54 to 1694 days, averaging 755 days. Supplementary Table 1 displays the H&E staining results for each patient area.
Volcano plot and ANOVA reveal differential protein expression and inter-patient heterogeneity in cholangiocarcinoma
The findings are displayed in Fig. 2, illustrating two comparative assessments of protein expression in CCA patients’ tumor and adjacent non-tumor tissues. The volcano graphic in Fig. 2 A illustrates the contrast in protein expression levels between tumor and surrounding non-tumor tissues across all patients. Proteins that show substantial differences in expression are depicted as dots, with the color red representing an increase in expression and blue suggesting a decrease in expression. The x-axis depicts the logarithm base 2 of the fold change (FC) in expression levels between tumor and neighboring tissues. The y-axis represents the negative logarithm base 10 of the p-value, which indicates the statistical significance of the observed changes. Several crucial proteins, namely P21695, P05062, and P34896, were identified as being greatly increased in tumor tissues, while P29084 was observed as a noteworthy decreased protein in nearby tissues. In Fig. 2B, the ANOVA analysis examined the protein expression levels in various tumor areas among different patients. The x-axis corresponds to the retention time (rt) and mass-to-charge ratio (m/z) of each identified protein, while the y-axis shows the negative logarithm (base 10) of the p-value obtained from the ANOVA analysis. The color gradient, spanning from yellow to red, denotes escalating statistical significance, while larger dot sizes correspond to higher p-values. The clear delineation of protein expression among the various tumor areas indicates substantial variation in protein expression patterns among different patients. These results highlight the substantial differences in protein expression between tumor and surrounding tissues, as well as the large variations in proteomic profiles among distinct tumor areas in patients.
Volcano plot and ANOVA reveal differential protein expression and inter-patient heterogeneity in cholangiocarcinoma. (A) Volcano plot showing differential protein expression between CCA tumor and adjacent tissues. Upregulated proteins are highlighted in red, downregulated proteins in blue. (B) ANOVA analysis demonstrating inter-patient heterogeneity in protein expression across tumor regions. The color gradient (yellow to red) indicates increasing statistical significance, with dot size reflecting p-value magnitude.
Volcano plot analysis identifies tumor-specific proteins in cholangiocarcinoma patients
The volcano plots as shown in Fig. 3 illustrate the disparity in protein expression between tumor and adjacent non-tumor tissues for each CCA patient. Each graphic corresponds to an individual patient, with proteins that are only present in tumor tissues being emphasized. The x-axis depicts the log2 fold change (FC) in protein expression, while the y-axis represents the -log10(p-value), indicating the statistical significance of the observed changes. Proteins that show a significant increase in expression in tumor tissues are indicated in red, whereas those that show a decrease in expression in nearby tissues are indicated in blue for each patient. The study identifies distinct proteins that are found only in tumor locations among various patients, indicating both shared characteristics and individualized protein expression patterns. This curated catalog of tumor-specific proteins enables the identification of prospective biomarkers and therapeutic targets that are unique to the tumor microenvironment, excluding proteins that are also present in neighboring non-tumor areas. The findings demonstrate distinct expression patterns across tumor and non-tumor tissues, as well as identify specific proteins that are constantly present exclusively in the tumor. The proteins identified as significant in the tumor regions of each patient were further analyzed to assess ITH.
Volcano plots for each of the 13 CCA patients show differential protein expression between tumor tissues and adjacent tissues. The x-axis represents the log2 fold change (FC) of protein expression, and the y-axis displays the -log10(p-value), indicating the statistical significance of the changes. Upregulated proteins in tumor tissues are highlighted in red, while downregulated proteins in adjacent tissues are shown in blue.
Principal component analysis and heatmap reveal intra-tumor proteomic heterogeneity in cholangiocarcinoma samples
To assess proteomic diversity within tumors, proteins identified as significant in CCA were analyzed using principal component analysis (PCA) and visualized alongside heatmaps. Each panel in Fig. 4 corresponds to one patient and includes both a 3D PCA plot and a heatmap representing differential protein expression across four spatially distinct tumor regions. In the PCA plots, different colored points represent different tumor regions (R1–R4) within the same tumor, and the spatial separation between points in 3D space reflects the degree of variation in proteomic profiles. The heatmaps provide complementary visualization, revealing the expression patterns of key proteins across the sampled tumor regions. The results highlight notable ITH, with some patients such as K061 and J122 exhibiting tighter clustering in PCA and more uniform expression in the heatmap, suggesting low ITH. Conversely, patients such as K151 and L096 show greater separation between tumor regions in PCA and more variable protein expression patterns, indicating high ITH. This integrated visualization reinforces the heterogeneity landscape in CCA and the need for spatially resolved proteomic analysis.
PCA plots and heatmaps illustrate intra-tumor proteomic heterogeneity across four tumor regions from 13 cholangiocarcinoma (CCA) patients. Each 3D PCA plot represents the proteomic profiles of four spatially distinct tumor regions (R1–R4) within a single patient. Each point corresponds to the proteome of a specific region, with the distance between points reflecting differences in protein expression. Accompanying heatmaps display the expression patterns of differentially expressed proteins across the same regions, providing complementary visualization of intra-tumor heterogeneity. Together, these plots highlight the varying degrees of proteomic diversity within individual CCA tumors.
Venn diagram analysis reveals region-specific and shared proteins in tumor samples of cholangiocarcinoma patients
Significant proteins identified in CCA tumors were analyzed using Venn diagrams to determine shared and unique proteins across different tumor sites within each patient. The Venn diagrams depicted in Fig. 5 demonstrate the comparison of proteins that are exclusively found in the tumor regions of each individual CCA patient. Each graphic depicts a patient and compares different tumor locations to determine protein expression patterns that overlap or are distinct. The various hues in each diagram correspond to separate tumor regions, and the points where these regions intersect indicate the quantity of proteins that are common to numerous regions, whilst areas that do not overlap signify proteins that are exclusive to specific tumor locations. The quantity of proteins that are present in all tumor locations differs among patients, indicating the presence of both shared and region-specific protein expression patterns. Patients J122 and K151 demonstrate a greater level of overlap between regions, indicating a more uniform protein expression profile across various tumor parts. On the other hand, patients like K002 and K061 have more pronounced variations in protein expression patterns across different locations, suggesting a higher level of diversity within the tumor.
This research highlights the variety of protein expression in various parts of the same tumor, indicating the intricate nature of tumor heterogeneity in CCA. Identifying proteins that are shared among different regions and those that are specific to certain regions can offer valuable understanding of the molecular mechanisms that contribute to tumor progression. This knowledge can also aid in the identification of new treatment targets that are specifically applicable to particular tumor regions. The list of proteins commonly found in all regions of each patient, as well as region-specific proteins unique to individual tumor areas, is provided in Supplementary Table 2. Additionally, the expression levels of the 18 proteins consistently identified across all patients and tumor regions are illustrated in Supplementary Fig. 1, demonstrating their variability across different regions and individuals.
Venn diagrams for 13 CCA patients display the overlap and uniqueness of significant proteins identified across four tumor regions within each patient. Each colored area represents a tumor region, and the numbers indicate the count of significant proteins that are unique to a specific region or shared among multiple regions.
ANOVA analysis reveals tumor-specific protein expression patterns in individual cholangiocarcinoma patients
Significant proteins identified in CCA tumors were subjected to ANOVA analysis to evaluate differences in protein expression across the four tumor regions within each patient. The ANOVA analysis findings (Fig. 6) compared the protein expression levels that are specific to tumor tissues among individual CCA patients. Each plot displays the statistical significance (-log10(p-value)) of proteins that are expressed differently, along with their corresponding retention time (rt) and mass-to-charge ratio (m/z). The color gradient, which spans from yellow to red, serves as an indication of the progressive increase in statistical significance. Additionally, the size of the dots corresponds to the magnitude of the p-value. The red and orange dots in each patient’s figure indicate proteins with the most significant variations in expression, indicating their probable importance to tumor biology. The presence of certain proteins in tumor regions exhibits variable levels of significance. Patients J087 and K002 demonstrate a higher number of proteins with strong relevance (red and orange), whereas patients K061 and L014 exhibit a more moderate degree of variation. This study highlights the variation in tumor composition within each patient, as well as the existence of specific protein expression patterns that are exclusive to different parts of the tumor. Discovering these proteins that are particular to tumors could provide valuable information about biomarkers for CCA and offer possible targets for therapeutic intervention. The ANOVA results demonstrate the heterogeneity of protein expression within tumors, indicating the intricate molecular characteristics of CCA.
ANOVA plots for 13 CCA patients display the distribution of proteins with significant differences in expression across four tumor regions within each patient. The x-axis represents the peaks (m/z/rt), while the y-axis shows the -log10(p-value), indicating statistical significance. Proteins with higher significance are shown in red and orange, while the size of the dots corresponds to the p-value magnitude.
STITCH pathway analysis reveals interactions between oxidative stress, metabolism, and chemotherapeutic agents in cholangiocarcinoma
The common proteins identified across all regions and all patients were analyzed for pathway enrichment using STITCH. The proteins including AIFM1, FTCD, ADH1B, SODC, CATA, ALDOB, ARGI1, FABPL, F16P1, C1 TC, AK1 A1, EST1, S10 AB, PGM1, THIM, ACY1, DPYS, UGPA (Supplementary Table 3). The STITCH pathway analysis (Fig. 7) depicts the network of interacting proteins that are consistently present in all tumor locations and patients with CCA. This network focuses on essential proteins associated with oxidative stress, metabolism, and their interactions with chemotherapy drugs. Important components such as SOD1 (superoxide dismutase 1) and CAT (catalase) play a crucial part in the body’s reaction to oxidative stress. These components have direct associations with hydrogen peroxide and oxygen, which suggests that they are involved in controlling reactive oxygen species (ROS). SOD2, also known as superoxide dismutase 2, plays an additional role in this network that responds to stress. Chemotherapeutic drugs such as cisplatin, gemcitabine, and FOLFOX have been observed to interact within the network, suggesting that they may have an impact on oxidative stress pathways and their connection with important proteins like SOD1 and CAT. Metabolic proteins, such as ADH1B (alcohol dehydrogenase 1B), PGM1 (phosphoglucomutase 1), and ALDOB (aldolase B), are associated with the metabolism of ethanol and carbohydrates, highlighting their role in metabolic pathways. The inclusion of metal ions such as Zn(II) (zinc) and copper in the network emphasizes the importance of these cofactors in controlling enzymatic activity associated with oxidative stress and metabolism. The involvement of UBC (ubiquitin C) with numerous proteins in the network highlights its function in protein breakdown and cellular stress responses. Proteins such as HADHA (hydroxyacyl-CoA dehydrogenase) and FABP1 (fatty acid-binding protein 1) indicate that lipid metabolism is involved in the tumor’s metabolic reprogramming. The STITCH pathway analysis reveals the interconnectedness of oxidative stress, metabolism, and the response to chemotherapeutic drugs in CCA. This study offers valuable information on possible therapeutic targets that could impact tumor growth and treatment response.
The STITCH network analysis illustrates the interactions among proteins consistently present across all tumor regions and patients with cholangiocarcinoma (CCA). The network highlights proteins involved in oxidative stress responses (e.g., SOD1, CAT, SOD2), lipid metabolism (e.g., FABP1, HADHA), carbohydrate metabolism (e.g., ALDOB, PGM1, ADH1B), and protein metabolism (e.g., FTCD, ARG1, CAD).
Pathway enrichment analysis of common proteins identified across all tumor regions and patients in cholangiocarcinoma
The common proteins identified across all regions and all patients were subjected to pathway enrichment analysis. Additionally, proteins found in all regions within each individual patient were analyzed for pathway enrichment to compare the differences between common proteins shared across all patients and proteins consistently present in all regions within individual patients. The pathway enrichment analysis utilizing shared proteins found across all areas and patients demonstrated substantial enrichment in many metabolic and biosynthetic pathways as shown in Fig. 8. The predominant enriched pathways encompass the pentose phosphate route, galactose metabolism, fructose and mannose metabolism, starch and sucrose metabolism, and the production of nucleotide sugars. Furthermore, pathways associated with one-carbon metabolism, arginine biosynthesis, glycolysis/gluconeogenesis, and oxidative stress responses, including chemical carcinogenesis-related reactive oxygen species, were discovered. This indicates a pivotal function of metabolic reprogramming in the advancement of CCA and its therapeutic response. The pathway enrichment analysis revealed diversity in pathway enrichment among tumor areas for individual individuals. Some patients, including J087, J122, and J176, demonstrated significant enrichment in pathways associated with fatty acid degradation and amino acid metabolism, whereas others, such as K002, K061, and L133, displayed notable enrichment in pathways related to arginine biosynthesis, protein processing in the endoplasmic reticulum, and xenobiotic metabolism. This diversity highlights the ITH of CCA at the proteome level, where various locations within the same tumor may utilize diverse metabolic pathways. The extensive pathway analysis of shared proteins across all tumor locations underscores the significance of metabolic processes, oxidative stress, and biosynthetic pathways in CCA, offering prospective targets for therapeutic intervention.
Discussion
The existing treatment modalities for CCA are confined to surgical intervention and adjunctive therapies. Consequently, a comprehensive understanding of the molecular mechanisms governing this heterogeneous disease, especially at the protein level, will facilitate the discovery of novel therapeutic alternatives for CCA. A primary problem in the characterisation of biomarkers and treatment selection for CCA is ITH. This phenomenon denotes the coexistence of diverse subpopulations of cancer cells exhibiting varying genetic and phenotypic characteristics within a specific primary tumor and between that main tumor and its metastases25. The heterogeneity of these cancer cell types can be ascribed to the activation of signaling or metabolic pathways, tumorigenicity, metastasis, migration, and evasion of anti-tumor immunity26,27,28. Our study examined the multi-site proteomics heterogeneity profile of CCA by proteome profiling, utilizing four sites and their adjacent tissue from tumors resected from 13 CCA patients. We concentrated on ITH (inside a tumor excised from a single patient) and inter-patient heterogeneity (between tumors excised from different patients) of CCA. Initially, to investigate inter-patient proteome heterogeneity, an analysis was performed concentrating exclusively on proteins discovered in tumor tissues, and contrasting them with adjacent non-tumorous tissues from all patient. Subsequently, a differential protein analysis was conducted across 13 patients. The results revealed a significant number of differentially expressed proteins among the patients, demonstrating inter-patient heterogeneity. This finding is consistent with previous studies in various cancer types29,30,31. The observed inter-patient proteome heterogeneity, shown by the substantial number of differently expressed proteins among patients, corroborates prior findings in several cancer types, highlighting the complexity and variety in tumor biology among people. To analyze intra-tumor proteome heterogeneity, we identified proteins uniquely present in tumor tissues by contrasting them with adjacent non-tumorous tissues from each patient. The proteins were subsequently analyzed in various tumor areas to evaluate intra-tumor proteome heterogeneity. The findings from all 13 individuals exhibited unequivocal evidence of intra-tumor proteome heterogeneity. Our findings of intra-tumor proteome heterogeneity are consistent with prior studies by Gerlinger et al.32and Lawrence et al.33, which demonstrated significant geographic variability within tumors. Analogous to the branched evolution identified in cancer genomic research, our proteomic analysis demonstrates divergent expression profiles across several tumor locations, underscoring the intricacy of tumor biology and the difficulties it presents for targeted therapeutics. In addition the investigation of differential protein expression, we performed an examination to identify proteins consistently present throughout all tumor areas and in every patient. The findings identified 18 proteins consistently present in all regions and among all patients. The proteins includes: AIFM1, FTCD, ADH1B, SODC, CATA, ALDOB, ARGI1, FABPL, F16P1, C1 TC, AK1 A1, EST1, S10 AB, PGM1, THIM, ACY1, DPYS, UGPA. Previous studies by Cavalloni et al. and Chua-On et al. consistently detected the markedly upregulated proteins AIFM1, FTCD, SODC, CATA, and ACY1 in CCA, which are integral to metabolic reprogramming and oxidative stress modulation. These proteins may function as potential biomarkers or therapeutic targets in CCA because of their involvement in critical pathways that promote tumor development and heterogeneity34,35. The ubiquitous protein demonstrates considerable participation in metabolic pathways associated with nucleotide and amino acid synthesis, encompassing nucleotide sugar production, arginine synthesis, and comprehensive amino acid biosynthesis. The pathway analysis demonstrates notable connections among oxidative stress, metabolic processes, and chemotherapy drugs in CCA. Essential proteins implicated in the oxidative stress response, including as SOD1 and CAT, are associated with the regulation of reactive oxygen species (ROS), whereas chemotherapeutic agents, like cisplatin and gemcitabine, exhibit interactions within this framework. This research identifies possible therapeutic targets that affect tumor growth and treatment response in CCA. The results of this study correspond with prior research indicating the involvement of oxidative stress and metabolic reprogramming in the advancement of CCA. Proteins like as SOD1 and CAT, which modulate reactive oxygen species (ROS), have been demonstrated to play a role in chemoresistance, underscoring prospective therapeutic targets for enhancing treatment efficacy in CCA34,35. Moreover, the studies in hepatocellular carcinoma (HCC) and colorectal cancer have demonstrated that proteins associated with oxidative stress regulation, such as SOD1 and CAT, are pivotal in chemoresistance, underscoring their potential relevance in CCA as well36,37.
This study underscores the notable intra-tumor and inter-patient proteome heterogeneity in CCA. This variability highlights the complexity of CCA at the molecular level, emphasizing the necessity for personalized therapeutic strategies. The identification of common proteins consistently present across all tumor regions and patients indicates potential biomarkers for CCA, especially those associated with oxidative stress and metabolic pathways, which may serve as novel therapeutic targets to enhance treatment outcomes.
In conclusion, our proteomics analysis provides valuable insights into the molecular diversity of CCA, highlighting potential biomarkers and therapeutic targets. The significant inter and ITH observed in this study emphasizes the complexity of CCA and the necessity for personalized treatment strategies. For future research, proteins consistently identified across all tumor locations and patients may function as universal therapeutic targets for CCA, targeting common pathways of disease progression. These proteins have potential for the creation of universal therapeutic targets that could help a wider patient population. Furthermore, the integration of these ubiquitous proteins with individualized protein profiles facilitates customized medicine, customizing treatments to each patient’s distinct molecular composition while considering universal causes. This combined strategy augments therapeutic accuracy as well as assuring enhanced efficacy and results in CCA treatment. Future validation of key proteins identified in this study, along with investigation of targeted therapeutic strategies, will be essential to confirm their functional relevance and to translate these findings into clinical applications for CCA.
Data availability
The raw proteomics data generated and analyzed during this study are available in the jPOST (Japan Proteome Standard Repository) under the accession number JPST003436.
Abbreviations
- CCA:
-
cholangiocarcinoma
- ITH:
-
intratumoral heterogeneity
- LC-MS:
-
Liquid chromatography mass spectrometry
References
Banales, J. M. et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat. Rev. Gastroenterol. Hepatol. 17 (9), 557–588 (2020).
Rizvi, S., Khan, S. A., Hallemeier, C. L., Kelley, R. K. & Gores, G. J. Cholangiocarcinoma - evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 15 (2), 95–111 (2018).
Saha, S. K., Zhu, A. X., Fuchs, C. S. & Brooks, G. A. Forty-Year trends in cholangiocarcinoma incidence in the U.S.: intrahepatic disease on the rise. Oncologist 21 (5), 594–599 (2016).
Khan, S. A. et al. Changing international trends in mortality rates for liver, biliary and pancreatic tumours. J. Hepatol. 37 (6), 806–813 (2002).
Taylor-Robinson, S. D. et al. Increase in mortality rates from intrahepatic cholangiocarcinoma in England and Wales 1968–1998. Gut 48 (6), 816–820 (2001).
Sripa, B. et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 4 (7), e201 (2007).
Sripa, B. & Pairojkul, C. Cholangiocarcinoma: lessons from Thailand. Curr. Opin. Gastroenterol. 24 (3), 349–356 (2008).
Anderson, C. D., Pinson, C. W., Berlin, J. & Chari, R. S. Diagnosis and treatment of cholangiocarcinoma. Oncologist 9 (1), 43–57 (2004).
Khan, S. A. et al. Guidelines for the diagnosis and treatment of cholangiocarcinoma: an update. Gut 61 (12), 1657–1669 (2012).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168 (4), 613–628 (2017).
Sitthirak, S. et al. Whole exome sequencing of multi-regions reveals tumor heterogeneity in Opisthorchis viverrini-associated cholangiocarcinoma. Sci. Rep. 15 (1), 10886 (2025).
Aebersold, R. & Mann, M. Mass-spectrometric exploration of proteome structure and function. Nature 537 (7620), 347–355 (2016).
Khan, S., Khan, P., Hassan, M. I., Ahmad, F. & Islam, A. Protein stability: determination of structure and stability of the transmembrane protein Mce4A from M. tuberculosis in membrane-like environment. Int. J. Biol. Macromol. 126, 488–495 (2019).
Sung, H. J. et al. Quiescin sulfhydryl oxidase 1 (QSOX1) secreted by lung Cancer cells promotes Cancer metastasis. Int. J. Mol. Sci. 19(10), 3213 (2018).
Chen, R. et al. Quantitative proteomics reveals that long non-coding RNA MALAT1 interacts with DBC1 to regulate p53 acetylation. Nucleic Acids Res. 45 (17), 9947–9959 (2017).
Arshid, S. et al. High performance mass spectrometry based proteomics reveals enzyme and signaling pathway regulation in neutrophils during the early stage of surgical trauma. Proteom. Clin. Appl. 11, 1–2 (2017).
Andrade, J. R., Gallagher, A. D., Maharaj, J. & McClelland, S. E. Disentangling the roles of aneuploidy, chromosomal instability and tumour heterogeneity in developing resistance to cancer therapies. Chromosome Res. 31 (4), 28 (2023).
Dagogo-Jack, I. & Shaw, A. T. Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15 (2), 81–94 (2018).
Le Faouder, J. et al. Proteomic landscape of cholangiocarcinomas reveals three different subgroups according to their localization and the aspect of Non-Tumor liver. Proteom. Clin. Appl. 13 (1), e1800128 (2019).
Tyanova, S., Temu, T. & Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 11 (12), 2301–2319 (2016).
Tyanova, S. et al. The perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 13 (9), 731–740 (2016).
Pang, Z. et al. Using metaboanalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17 (8), 1735–1761 (2022).
Pang, Z. et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 52 (W1), W398–W406 (2024).
Szklarczyk, D. et al. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 44 (D1), D380–D384 (2016).
Marusyk, A., Almendro, V. & Polyak, K. Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer. 12 (5), 323–334 (2012).
Andrechek, E. R. & Nevins, J. R. Mouse models of cancers: opportunities to address heterogeneity of human cancer and evaluate therapeutic strategies. J. Mol. Med. (Berl). 88 (11), 1095–1100 (2010).
Marusyk, A. & Polyak, K. Tumor heterogeneity: causes and consequences. Biochim. Biophys. Acta. 1805 (1), 105–117 (2010).
Michor, F. & Polyak, K. The origins and implications of intratumor heterogeneity. Cancer Prev. Res. (Phila). 3 (11), 1361–1364 (2010).
Berl, A. et al. Exploring multisite heterogeneity of human basal cell carcinoma proteome and transcriptome. PLoS One. 18 (11), e0293744 (2023).
Buczak, K. et al. Spatial tissue proteomics quantifies Inter- and intratumor heterogeneity in hepatocellular carcinoma (HCC). Mol. Cell. Proteom. 17 (4), 810–825 (2018).
Hoffmann, O. I. et al. Interpatient heterogeneity in drug response and protein biomarker expression of recurrent ovarian Cancer. Cancers (Basel) 14(9), 2110 (2022).
Gerlinger, M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl. J. Med. 366 (10), 883–892 (2012).
Lawrence, R. T. et al. The proteomic landscape of Triple-Negative breast Cancer. Cell. Rep. 11 (6), 990 (2015).
Cavalloni, G. et al. Proteomic analysis identifies deregulated metabolic and oxidative-associated proteins in Italian intrahepatic cholangiocarcinoma patients. BMC Cancer. 21 (1), 865 (2021).
Chua-On, D. et al. Bioinformatic prediction of novel signaling pathways of Apoptosis-inducing factor, Mitochondrion-associated 3 (AIFM3) and their roles in metastasis of cholangiocarcinoma cells. Cancer Genomics Proteom. 19 (1), 35–49 (2022).
Li, S. et al. The role of oxidative stress and antioxidants in liver diseases. Int. J. Mol. Sci. 16 (11), 26087–26124 (2015).
Liu, Y. et al. The role of oxidative stress in the development and therapeutic intervention of hepatocellular carcinoma. Curr. Cancer Drug Targets. 23 (10), 792–804 (2023).
Funding
The funding for this study was provided by the NSRF under the Basic Research Fund of Khon Kaen University, the National Research Council of Thailand through the Fluke Free Thailand Project and the IMSUT Joint Research Project, the Institute of Medical Science, University of Tokyo, Japan for Watcharin Loilome. Furthermore, Sirinya Sitthirak was granted financial assistance from the Invitation Research Grant (IN67067). Sirinya Sitthirak received a scholarship from the Cholangiocarcinoma Research Institute, specifically under Grant No. CARI01/2565. The funders were not involved in the study’s design, data collection and analysis, decision to publish, or preparation of the publication.
Author information
Authors and Affiliations
Contributions
S.S. conducted the experiments, analyzed the data, created figures and/or tables, contributed to the writing of the paper, and approved the final version. S.R. conducted the experiments, analyzed the data, created visual representations such as figures and tables, contributed to the writing of the paper, and gave final approval to the document.A.W. conducted the experiments, analyzed the data, created visual representations such as figures and tables, contributed to the writing of the paper, and gave final approval to the document. P.K. conducted the experiments, analyzed the data, contributed to the drafting of the article, and gave final approval to the completed version. H.D. conducted the experiments, analyzed the data, contributed to the drafting of the article, and gave final approval to the completed version. N.N. conducted the experiments, analyzed the data, contributed to the writing and reviewing of the paper, and gave final approval to the document. V.T. conducted data analysis, contributed to the writing and revision of article drafts, handled specimen inquiries and visualizations, and gave approval for the final draft. A.T. conducted data analysis, prepared or revised versions of the paper, handled specimen inquiries and visualizations, and gave final approval to the last version. A.J. conducted data analysis, prepared or revised versions of the paper, handled specimen inquiries and visualizations, and gave final approval to the last version. N.K. conducted data analysis, prepared or revised versions of the paper, handled specimen inquiries and visualizations, and gave final approval to the last version. P.S. conducted data analysis, prepared or revised versions of the paper, handled pathological characterization and visualizations, and gave final approval to the last version. L.B. was responsible for the conception and design of the experiments, contributed to the writing and reviewing of versions of the paper, and approved the final text. W.L. was responsible for the conception and design of the experiments, funding acquisition, project administration, execution of the experiments, analysis of the data, creation of figures and/or tables, contribution to the writing and reviewing of versions of the paper, and approval of the final text.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Supplementary Information
The online version contains supplementary material available at submission system.
Ethical approval
The provided information pertains to the ethical approvals, including the authorizing authority and any associated reference numbers. The procedure for specimen collection and study received approval by the Ethic Committee of Human Research at Khon Kaen University (HE671356).
Consent to participate
All authors have given their agreement for the publishing of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sitthirak, S., Roytrakul, S., Wangwiwatsin, A. et al. Proteomic profiling reveals common and region-specific protein signatures underlying tumor heterogeneity in cholangiocarcinoma. Sci Rep 15, 17228 (2025). https://doi.org/10.1038/s41598-025-02713-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-02713-5