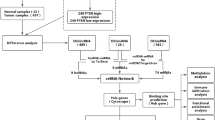
Abstract
Purpose
Aberrant activation/overexpression of RNF126 is implicated as a driving event in tumor progression. However, although some functions of RNF126 in prostate cancer (PCa) cell lines has been reported, more biological functions and in-depth mechanisms should be further clarified in PCa.
Methods
Here, we provide evidence that RNF126 expression is elevated in human PCa tissues and cell lines, which is associated with tumor grades and prognosis. Cell proliferation was measured by the CCK8 and colony-formation assays. Cell migration was performed by Transwell and wound-healing assays. RNF126 target proteins were investigated via proteomic, co-immunoprecipitation and western blot methods. Additionally, we knock-downed MBNL1 expression to perform rescue experiments. In vivo, xenograft mice assay was used to verify the effect of RNF126 on the growth of PCa cell.
Results
Here, we showed that RNF126 was highly expressed in PCa and its higher expression was associated with worse patients’ prognosis. Expression modulation of RNF126 affects PCa cells proliferation, migration, EMT and docetaxel (DTX) resistance in vitro or in vivo. Additionally, RNF126 involves in the regulation of PI3K/AKT, MEK/ERK and EMT pathways. Mechanistically, immunoprecipitation (IP) and coimmunoprecipitation (co-IP) assays indicated that RNF126 could bind to MBNL1 directly. Our data also suggested that MBNL1 was a critical downstream event in RNF126-mediated tumorigenesis and chemo-resistance and played a crucial role in driving the PI3K/AKT, MEK/ERK and EMT pathways.
Conclusion
Taken together, our findings reveal a novel biological and molecular functions of RNF126 and may provide a new treatment option for PCa patients.
Similar content being viewed by others
Introduction
Prostate cancer is one of the most common male-related malignant tumor worldwide and remains the second leading cause of cancer-related deaths among men in Western countries1. Surgical resection, androgen deprivation therapy and chemotherapy remain as the mainstays for PCa patients2. Although PCa progresses slowly, most PCa patients will inevitably progress to advanced stages that requires endocrine therapy, which is likely to lead to the emergency of a castration-resistant state3. Docetaxel is a first-line chemotherapeutic agent for advanced PCa4. However, most PCa patients required inherent or acquired drug resistance to docetaxel5. Therefore, an understanding of mechanisms by which contributes to chemoresistance will be important when development new treatment strategies for PCa.
Ring-finger protein 126 (RNF126) belongs to RING E3 ubiquitin ligase subfamily and plays broad functions in maintaining the protein stability by ubiquitinating misfolded proteins6. RNF126 has been reported to be involved in diverse biological processes, including drug resistance7, DNA damage response8, homologous recombination9, immunoregulatory effect10 and cell cycle11. RNF126 seems to be associated with a series of cellular processes in which its E3 ligase activity may or may not be involved12. RNF126 and BCA1 E3 ligases have similar protein structures, containing a RING finger at the C-terminal and a Zn finger at the N-terminal13. RNF126 has been reported to correlated with many malignant cancer, such as breast cancer8,12, bladder cancer14, and nasopharyngeal cancer10. Although Xu et al. has reported that RNF126 could promote the viability and cell cycle of PCa cell lines13, more functional phenotypes and a deeper mechanism should be further illuminated in PCa.
The alternative splicing of pre-mRNA is a key mechanism regulating eukaryotic gene expression by expanding genome coding diversity15. The Muscleblind-like (MBNL) family of sequence-specific pre-mRNA splicing factors bind RNA through pairs of highly conserved zinc finger, recognizing YGCY and similar motifs16. MBNL1 was originally found to modulate transcriptomes by stabilizing, splicing, polyadenylating, and locating its target mRNAs17,18. Dysfunctional MBNL1 causes myotonic dystrophy as well as defective erythrocyte and myofibroblast development19. Abnormal RNA processing, especially splicing, is associated with cancer, including PCa20. In addition to the molecular mechanisms involved in regulating MBNL1-mediated impacts on tissue development, its influence on carcinogenesis was recently investigated. Recent study has also reported that MBNL1 regulates alternative splicing of several genes including those essential for MLL-rearranged leukemogenesis, such as DOT1L and SETD1A21. MBNL1 inhibits glioblastoma tumor initiation and progression by reducing hypoxia-induced stemness22. MBNL1 also plays a suppressive role in breast cancer progression by promoting transcripts stability of several onco-suppressors23. Moreover, MBNL1 could destabilize snail transcripts and, in turn, suppress the EMT process of colorectal cancer cells through the snail/E-cadherin axis24.
In the present study, we found RNF126 was highly expressed and could be a prognostic biomarker in PCa. In addition, targeting RNF126 inhibits PCa progression and docetaxel resistance both in vitro and in vivo. By further exploration, we determined that RNF126 could regulate MBNL1 protein expression via direct interaction. Therefore, our study describes a novel RNF126/MBNL1 axis that might be a promising therapeutic target for PCa.
Methods and materials
Cell culture and reagents
BPH1, LNCaP, 22Rv1, DU145, PC3 and 293T cells were purchased from American Type Culture Collection (ATCC, USA). DTX-resistant DU145 cell (DU145-DR) and DTX-resistant PC3 cell (PC3-DR) were kindly provided by Minghao Yu. Except for 293T cells cultured in DMEM medium, all the other cells were cultured in 1640 medium supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, USA). PI3K inhibitor LY294002 (HY-10108), ERK inhibitor SCH772984 (HY-50846) and Docetaxel (HY-B0011) were purchased from MedChemExpress (Monmouth Junction, NJ, USA). The study was conducted in accordance with the principles set forth in the Declaration of Helsinki. All patients gave written informed consent before the study.
Transient and stable transfection
Transient transfection of siRNA or plasmids were performed using Lipofectamine 2000 (Invitrogen, USA) when the cells reached 50–70% confluence. The shRNA sequences targeting RNF126 are shown below: shRNF126-1: 5’-TGCATGGTTTGTGGCGGAAGA-3’; shRNF126-2: 5’-GCAACGAGAACGCCACATGGT-3’. The siRNA sequences targeting MBNL1 are shown below: 5’-GCCAACCAGATACCCATAATA-3’. For lentivirus packaging, 293T cells were transfected with shNC and shRNF126 plasmids and viral packaging plasmids (psPAX-2 and pMD2.G) using Lipofectamine 2000. After 48 h, the supernatant was collected and filtered. To generate DU145 and PC3 cell lines with stable RNF126 knockdown, lentivirus particles were infected into DU145 or PC3 cells. After 24 h’ infection, both cells were treated with puromycin (Beyotime, China) for 14 days to select stable cell lines.
Western blot
After whole cell lysate was separated by SDS-PAGE gel, proteins were transferred to PVDF membrane and blocked in 5% non-fat milk, then incubate with corresponding primary antibodies. The membranes were than incubated with anti-rabbit or anti-mouse secondary antibodies (1:2000; CST). Protein content was detected using Amersham Imager 600 (GE Healthcare, USA). Primary antibodies used in this study were shown in Supplementary Table S1.
qRT-PCR
RNA was isolated using a total RNA isolation kit (Life technologies; USA). cDNA synthesis was performed using reverse transcriptase kit and qRT-PCR was carried out using SYBR Green master mix (TAKARA; Japan). Data were analyzed using the ΔΔCt method. The sequences of the primers used for this assay are in Supplementary Table S2.
Immunoprecipitation
The cells were lysed in precipitation lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4) supplemented with protease inhibitors. Cell lysates were immunoprecipitated and analyzed via western blot assay.
Immunofluorescence
The cells were fixed with 4% PFA (10 min) and then incubated in 1% BSA in 0.1% PBS-Tween for 1.0 h to permeabilize the cells and block non-specific protein-protein interactions. The cells were then incubated with the antibody for MBNL1 (Abcam; ab45899) and RNF126 (Abcam; ab234812) overnight at 4 °C. The secondary antibody was Alexa Fluor® 488 Goat anti-Rabbit IgG (H + L) (Abcam; ab150077) used at a 1/1000 dilution for 1.0 h. DAPI (Beyotime; P0131) was used to stain the cell nuclei. The fluorescence was visualized by a confocal microscope.
Cell migration assays
For wound-healing assay, PCa cells were seeded in six-well plates. Cells were wounded with a 200-µL pipette tip and cultured in medium with 1% serum for 24 h. Then we measured the narrowing of the wound via ImageJ software. For Transwell migration assay, a total of 5 × 104 cells were seeded in the upper chambers of 24-well plates. Serum-free medium was added in the upper chambers when complete medium was added to the lower chamber. After 24 h, the cells were fixed with 4% paraformaldehyde, stained by 0.2% crystal violet, and counted.
Cell proliferation assays
For CCK8 assay, cells were seeded in 96-well plates at a density of 3 × 103 cells per well and cultured for 24, 48 and 72 h. Cell viability was determined using Cell Counting Kit-8 kit (Dojindo, Japan) according to the manufacturer’s instructions. For colony-formation assay, PCa cells were seeded in 6-well plates at 1500 cells/well, then incubated in complete medium for 14 days. Colonies were fixed with 4% paraformaldehyde and then were stained with 0.2% crystal violet for 15 min, respectively, followed by washing with PBS, photographing, and counting.
Tumorigenicity assay in nude mice
A total of 10 male, 4-week-old BALB/C nude mice were bought from the Center of Comparative Medicine of Yangzhou University in China (Quality certificate number: SCXK(Su)2012-0004). They were used to examine tumorigenicity. The mice were randomly divided into two groups (n = 5/group). shNC or shRNF126 PC3 cells were injected subcutaneously into the flanks of nude mice. At the ending days, the xenograft tumors were excised, then the tumor weight and volume were measured. The tumor tissues were subjected to western blot. In order to minimize any suffering of the animals, the mice were immediately sacrificed by intraperitoneal injection of an excess of 2% pentobarbital sodium. All experimental procedures followed the strict the guidelines of the Baoying People’s Hospital Animal Experimentation Committee on Animals in Research and ARRIVE guidelines and acquired ethical approval from the Ethics Committee of Baoying People’s Hospital (NO. BYRY20221202) for Animal Research.
Statistical analyses
The data are presented as the means ± standard deviations (SDs). Tukey’s test or Student’s t-test for the unpaired results was used to evaluate the differences among more than three groups or between two groups, respectively. Differences were considered significant for values of p < 0.05.
Results
RNF126 expression is upregulated in PCa tissues and is correlated with poor prognosis
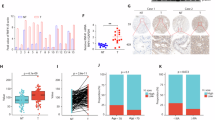
To explore the role of RNF126 in PCa, the expression pattern of RNF126 in PCa samples was assessed. RNF126 mRNA level was higher in PCa tissues than in normal prostate tissues based on TCGA-PRAD and GDS4824 datasets (Fig. 1A, B). Furthermore, higher expression of RNF126 was also observed in PCa tissues compared with normal prostate tissues by western blot and qRT-PCR assays (Fig. 1C, D). In addition, we performed IHC assay to evaluate the difference of RNF126 protein expression and found that the expression of RNF126 in human PCa tissues was higher than that in non-tumor tissues (Fig. 1E). To evaluate the clinical relevance of RNF126 expression, we analyzed its relationship with pathological features. As shown in Supplementary Table S3, RNF126 expression in PCa was correlated with N classification and T classification. Kaplan-Meier analysis showed that patients with low expression of RNF126 had a longer survival time than those with high expression Fig. 1F. These results revealed that RNF126 expression was upregulated in PCa and correlated with poor prognosis.
RNF126 expression is upregulated in PCa tissues and correlated with poor prognosis. A RNF126 levels in PCa tissues and normal prostate tissues from TCGA-PRAD dataset. B GEO profiling of RNF126 levels in PCa tissues and normal prostate tissues extracted from GEO dataset GDS4824. C A western blot showed the relative expression of RNF126 protein in 6 PCa tissues compared with adjacent non-cancerous normal tissues. D A qRT-PCR result showed the relative expression of RNF126 mRNA in 6 PCa tissues compared with adjacent non-cancerous normal tissues. E IHC staining of RNF126 was performed in 8 in PCa tissues and 8 normal prostate tissues (bar, 50 μm, left panel). Comparative analysis of RNF126 expression among PCa tissues and normal prostate tissues was shown (right panel). F The relationship between RNF126 levels and the prognosis of PCa patients based on TCGA-PRAD database. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
RNF126 knockdown inhibits PCa cells proliferation, migration and EMT in vitro
To explore the function of RNF126 in PCa cells, western blot was used to examine the protein expression of RNF126 in different PCa cell lines. As shown in Fig. 2A, RNF126 exhibits an obviously high expression in DU145 and PC3 cells, two more malignant cell lines, while the prostate hyperplastic cell BPH1 had the lowest RNF126 protein expression. To detect the putative malignant functions of RNF126, DU145 and PC3 cells were transfected with shRNA-expression vector targeting RNF126 and the knockdown efficiencies were assessed via qRT-PCR and western blot assays (Fig. 2B, C). Next, we performed CCK8 assays to assess the effect of RNF126 on PCa cells proliferation and the results showed that RNF126 knockdown reduced the cell viabilities of PCa cells (Fig. 2D), which was consistent with the colony-formation assays (Fig. 2E). In subsequent experiments, Transwell migration and wound-healing assays displayed that RNF126 knockdown inhibited the migration capabilities of PCa cells (Fig. 2F, G).
RNF126 knockdown inhibits PCa cells proliferation, migration and EMT in vitro. A The protein level of RNF126 was detected by western blot in PCa cell lines and BPH1 cell. B Western blot confirmed that the protein levels of RNF126 in DU145 and PC3 cells were downregulated. C qRT-PCR confirmed that the mRNA levels of RNF126 in DU145 and PC3 cells were downregulated. D, E CCK8 and colony-formation assays were performed to assess the effects of RNF126 on cell proliferation abilities. F, G Wound-healing and Transwell assays were performed to assess the effects of RNF126 on cell migration abilities. H, I The expression of EMT-related markers was detected by western blot and qRT-PCR. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
It has been well known that migration and metastasis of PCa cells are largely dependent on EMT25, which prompts us to investigate whether EMT is involved in RNF126-mediated migration of PCa cells. As expected, the mesenchymal biomarkers Vimentin and Slug were downregulated significantly, while the epithelial marker E-cadherin was upregulated upon RNF126 knockdown via qRT-PCR and western blot assays (Fig. 2H, I), suggesting RNF126’s inductive function in EMT.
RNF126 activates PI3K/AKT and MEK/ERK pathways to induce cell proliferation, migration and EMT in PCa cells
To explore the signaling pathways by which RNF126 affects PCa cells proliferation and migration, we performed GSEA analysis based on TCGA-PRAD databset and found that RNF126 was correlated with PI3K/AKT pathway and MAPK pathway, which indicated that RNF126 was most likely to regulate tumor progression by both pathways (Fig. 3A, B). Indeed, the above two pathways are classical pathways associated with cell proliferation and migration in a various of tumors26,27. Next, the protein levels of p-PI3K, t-PI3K, p-AKT and t-AKT were detected. As a result, RNF126 knockdown remarkably reduced phosphorylated PI3K and AKT amounts, while leaving total protein amounts intact (Fig. 3C). Similarly, levels of p-MEK and p-ERK were decreased while that of t-MEK and t-ERK still unchanged after RNF126 knockdown (Fig. 3D).
RNF126 activates PI3K/AKT and MEK/ERK pathways to induce cell proliferation, migration and EMT in PCa cells. A, B GSEA analysis showing differential enrichment of gene related to PI3K/AKT and MAPK pathways. C, D Total and phosphorylated levels of PI3K, AKT, MEK and ERK were detected in PCa cells after RNF126 knockdown. E, F CCK-8 and colony-formation assays were performed to detect cell proliferation of PCa cells after RNF126 overexpression with LY294002 or SCH772984 treatment. G, H Wound-healing and Transwell migration assays were performed to detect cell migration of PCa cells after RNF126 overexpression with LY294002 or SCH772984 treatment. I EMT-related markers in PCa cells treated with both RNF126 overexpression and LY294002 or SCH772984. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Subsequently, treatment with PI3K inhibitor LY294002 or ERK inhibitor SCH772984 markedly abrogated the enhanced proliferation, migration abilities and phosphorylation levels of AKT or ERK, as well as upregulated Vimentin, Slug and downregulated E-cadherin expression induced by RNF126 overexpression Fig. 3E–I . These results suggested that RNF126 promotes PCa cells growth, migration and EMT, at least partially, through activation of PI3K/AKT and MEK/ERK signaling pathways.
MBNL1 is the downstream target of RNF126
To further explore the molecular association of RNF126 action in PCa, we analyzed differentially expressed genes (DEGs) based on TCGA-PRAD dataset (Supplementary Table S4). Among all the alterations, the MBNL1 constituted the most negatively correlated gene in PCa tissues (Fig. 4A). This observation was further confirmed by other databases, GEPIA and UALCAN (Fig. 4B, C). Moreover, our results showed that the protein and mRNA levels of MBNL1 increased after RNF126 knockdown (Fig. 4D, E), indicating MBNL1 as a downstream target of RNF126. Although MBNL1 has been reported to act as a tumor-suppressor in several cancers22,23,24,28, its role in PCa has not been established. We first mined the TCGA and GEO databases and found that compared with normal prostate tissues, the expression of MBNL1 in PCa tissues was significantly reduced (Fig. 4F, G), which was consistent with our western blot and IHC results (Fig. 4H, I).
MBNL1 is a downstream target of RNF126. A–C Correlation analysis between RNF126 and MBNL1 was conducted based on TCGA, GEPIA and UALCAN databases, respectively. D, E The protein and mRNA levels of MBNL1 after RNF126 knockdown were detected via western blot or qRT-PCR. F, G MBNL1 expression levels in PCa tissues and normal prostate tissues were assessed based on TCGA and GEO databases. H MBNL1 protein levels in PCa tissues and normal prostate tissues from six patients. I IHC analysis of MBNL1 levels in human PCa tissues and normal prostate tissues. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
RNF126 interacts with MBNL1 directly
To explore the mechanism of these changes in MBNL1 above, the co-IP analysis was utilized to determine the association between RNF126 and MBNL1. First, we confirmed that endogenous RNF126 could directly interact with MBNL1 (Fig. 5A). Moreover, we co-transfected exogenous Flag-tagged MBNL1 plasmid and Myc-tagged RNF126 plasmid into 293T cells. The results showed that RNF126 and MBNL1 could also interact with each other exogenously (Fig. 5B, C). More importantly, immunofluorescence assays also verified that RNF126 and MBNL1 were colocalized in DU145 and PC3 cells (Fig. 5D). Collectively, the above data demonstrated that RNF126 serves as a binding partner of MBNL1 and regulates the expression of MBNL1.
RNF126 interacts with MBNL1 directly. A The endogenous binding between RNF126 and MBNL1 was detected via co-IP assay. B, C The interaction between RNF126 and MBNL1 was observed by co-IP assays after indicated plasmids were transfected into 293T cell. D Immunofluorescent staining of RNF126 and MBNL1 in DU145 and PC3 cells. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
RNF126 promotes PCa cells proliferation, migration and EMT in a MBNL1-dependent manner
To further examine whether RNF126 facilitates tumor progression through MBNL1 in PCa cells, we co-transfected shRNF126 and siMBNL1 into DU145 and PC3 cells. The CCK8 assays showed that MBNL1 silencing could rescue the decrease in cell viability caused by RNF126 knockdown Fig. 6A. Moreover, the colony-formation assays indicated that MBNL1 silencing could reverse the RNF126 knockdown-induced inhibition of proliferation (Fig. 6B). Furthermore, we examined the influence of MBNL1 silencing on RNF126 knockdown-induced migration inhibition. Silencing MBNL1 impaired the RNF126 knockdown-mediated decrease in PCa cells migration (Fig. 6C, D). Western blot confirmed that RNF126 and MBNL1 were decreased in shRNF126 and siMBNL1 groups and demonstrated protein expression of several markers in PI3K/ANK, MEK/ERK and EMT pathways (Fig. 6E). All in all, RNF126 knockdown suppresses tumor cell phenotypes, in part by regulating the expression of MBNL1, therefore silencing MBNL1 on top of RNF126 knockdown rescues the inhibition in tumor phenotypes by shRNF126.
RNF126 promotes PCa cells proliferation, migration and EMT in a MBNL1-dependent manner. A, B CCK-8 and colony-formation assays were performed to detect cell proliferation of PCa cells after RNF126 knockdown with or without siMBNL1 treatment. C, D Wound-healing and Transwell migration assays were performed to detected cell migration of PCa cells after RNF126 knockdown with or without siMBNL1 treatment. E The protein levels of EMT-, PI3K/AKT pathway- and MEK/ERK pathway-related markers were detected after RNF126 knockdown with or without siMBNL1 treatment in DU145 cell. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
RNF126 enhances docetaxel resistance in PCa cells via MBNL1
The GESA analysis showed that RNF126 was associated with multidrug resistance (Fig. 7A). Moreover, as mentioned above, the PI3K/AKT pathway, a docetaxel-related pathways29, was also correlated with RNF126 and MBNL1. On this basis, we estimated that the RNF126/MBNL1 axis may involve in docetaxel resistance. Indeed, the protein expression of RNF126 in docetaxel-resistant cells (DU145-R, PC3-R) was higher than that in their parental cells (DU145-P, PC3-P) (Fig. 7B). Results from the cell viability assays revealed that RNF126 knockdown led to resistance reduction to docetaxel in DU145 and PC3 cells (Fig. 7C, D). Furthermore, when RNF126 was knocked down, cell viability decreased in the presence of docetaxel (Fig. 7E). To further explore whether the RNF126 knockdown-induced PCa cells sensitivity to docetaxel was dependent on MBNL1, we performed rescue experiments. RNF126-knockdown DU145 cells were transfected with siMBNL1 or not. IC50 value of docetaxel was decreased in the RNF126-knockdown group when compared with the control group. However, when combined with siMBNL1, this resistance to docetaxel reactivated markedly (Fig. 7F). Similar trend was also observed in colony-formation assay (Fig. 7G).
RNF126 enhances docetaxel resistance in PCa cells via MBNL1. A GSEA analysis showing differential enrichment of genes related to multiple-drug resistance. B The protein expression of RNF126 in docetaxel-resistant cells (DU145-R, PC3-R) and in their parental cells (DU145-P, PC3-P). C, D CCK-8 assays were used to measure docetaxel sensitivity in DU145 and PC3 cells after transfected with shRNF126. E CCK-8 assay was used to examine cell viability after RNF126 knockdown with or without docetaxel treatment. F CCK-8 assay was used to examine cell viability after RNF126 knockdown with or without siMBNL1 treatment at different docetaxel doses. G DU145 cell after co-transfected with shRNF126 and siMBNL1 with or without docetaxel treatment was determined by colony-formation assay. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
RNF126 knockdown repressed tumor growth in vivo
To confirm whether RNF126 was involved in the malignant growth of PCa cells in vivo, we first established xenograft tumor models by subcutaneous injection of shNC- or shRNF126-treated PC3 cell into nude mice respectively. We found that the tumor growth of RNF126-knockdown group was markedly inhibited, when compared to the control group (Fig. 8A–C). WB analysis showed that the protein levels of RNF126, Vimentin and Slug in shRNF126 group were lower than that in control group, while the protein levels of MBNL1 and E-cadherin showed a contrary trend (Fig. 8D).
RNF126 knockdown repressed tumor growth in vivo. A PC3 cells transfected with shRNF126 were subcutaneously injected into nude mice. B, C The volumes and weights of xenograft tumors in two groups. D Western blot assay was performed to measure RNF126, MBNL1 and EMT-related genes in different groups. E Schematic diagram illustrated that RNF126/MBNL1 axis regulates tumor progression and docetaxel resistance in PCa. The data are shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
PCa is one of the most common cancer diagnoses in men can progress rapidly after diagnosis30,31. Disease heterogeneity and the emergence of therapeutic resistance remain significant barriers, and there is still a long way to go to identify and apply new molecular biomarkers to guide treatment selection32. The ubiquitin-proteasome system (UPS) plays a central role in regulating a variety of biological functions33. UPS consists of ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and ubiquitin ligase (E3)34. There is ample evidence that abnormal expression of E3 ligases can exert significant influence in cancer occurrence and progression, suggesting that E3 ligases may help to explain the underlying mechanism of cancer potential35,36. RNF126 is a RING domain-containing E3 ligase, with the capability to interact with both RNA and protein and cause their RING-dependent degradation37. For the first time, we highlight the importance and functional relevance of RNF126 in PCa progression. In this study, we found that the expression level of RNF126 was upregulated in PCa tissues when compared with non-tumor tissues, and that relatively high expression of RNF126 was associated with poorer prognosis of PCa patients. And in vitro and in vivo studies demonstrated that RNF126 facilitated PCa cells growth, migration, EMT and chemo-resistance via direct regulation of MBNL1.
It is well-known that the PI3K/AKT and MEK/ERK pathways are regulated by receptor tyrosine kinases. It has been reported that phosphorylation level of AKT decreased after RNF126 silencing in tongue cancer38. In addition, RNF126 could facilitate the ubiquitin-mediated degradation of PTEN, an inhibitor of the PI3K/AKT pathway14. Consistently, in this study, we found that RNF126 activated PI3K/AKT pathway via elevating the phosphorylation levels of PI3K and AKT. Additionally, RNF126 also enhanced MEK/ERK pathway confirmed by western blot assay and GSEA analysis.
Despite the well-defined role of MBNL1 as a tumor suppressor that functions in several cancer progression23,39,40, little is known about its function and regulation mechanism in PCa. Here, we revealed that MBNL1 acted as a tumor suppressor in PCa cells and knockdown of MBNL1 exerted a promoting-cancer role in PCa cells. In addition, Correlation analysis based on several databases revealed that MBNL1 was a potential downstream target of RNA126 and could interact with each other. In this study, our experimental data suggested that RNF126 weaken the expression level of MBNL1 via direct binding to augment the viability and migratory ability of PCa cells.
However, it is worth mentioning that there are also several limitations in our study. Further investigation is required to determine the precise binding domains between RNF126 and MBNL1. In addition, it is unknown whether RNF126, as an E3 ligase, mediates the ubiquitination degradation of MBNL1 by directly binding to the latter. Further research is needed to address these issues. In conclusion, our results demonstrated that RNF126 functions as a tumor oncogene, affecting PCa development and progression by altering malignant phenotypes and docetaxel resistance through the directly modulation of MBNL1 (Fig. 8E).
Data availability
The datasets generated and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- RNF126:
-
Ring finger protein 126
- MBNL1:
-
Muscleblind like splicing regulator 1
- PCa:
-
Prostate cancer
- DTX:
-
Docetaxel
- IP:
-
Immunoprecipitation
- co-IP:
-
Coimmunoprecipitation
- Ub:
-
Ubiquitin
- DOT1L:
-
DOT1 like histone lysine methyltransferase
- SETD1A:
-
SET domain containing 1 A
- EMT:
-
Epithelial-mesenchymal transition
- TCGA:
-
The Cancer Genome Atlas
- GEO:
-
Gene Expression Omnibus
- IHC:
-
Immunohistochemistry
- CCK8:
-
Cell Counting Kit-8
- GSEA:
-
Gene Set Enrichment Analysis
- MAPK:
-
Mitogen activated kinase-like protein
- DEGs:
-
Differentially expressed genes
- GEPIA:
-
Gene expression profiling interactive analysis
- UPS:
-
Ubiquitin proteasome system
- PTEN:
-
Phosphatase and tensin homolog
- PFA:
-
Paraformaldehyde
References
Liu, Z. et al. SLC4A4 promotes prostate cancer progression in vivo and in vitro via AKT-mediated signalling pathway. Cancer Cell. Int. 22 (1), 127 (2022).
Yu, C., Fan, Y., Zhang, Y., Liu, L. & Guo, G. LINC00893 inhibits the progression of prostate cancer through miR-3173-5p/SOCS3/JAK2/STAT3 pathway. Cancer Cell. Int. 22 (1), 228 (2022).
Chen, Q., Fu, L., Hu, J., Guo, G. & Xie, A. Silencing of PSMC2 inhibits development and metastasis of prostate cancer through regulating proliferation, apoptosis and migration. Cancer Cell. Int. 21 (1), 235 (2021).
Wang, X. et al. Inhibitory effect of roburic acid in combination with docetaxel on human prostate cancer cells. J. Enzyme Inhib. Med. Chem. 37 (1), 542–553 (2022).
Ganju, A. et al. Nanoways to overcome docetaxel resistance in prostate cancer. Drug Resist. Updat. 17 (1–2), 13–23 (2014).
Zhang, R., Liu, W., Sun, J., Kong, Y. & Chen, C. Roles of RNF126 and BCA2 E3 ubiquitin ligases in DNA damage repair signaling and targeted cancer therapy. Pharmacol. Res. 155, 104748 (2020).
Yoshino, S. et al. The ERK signaling target RNF126 regulates Anoikis resistance in cancer cells by changing the mitochondrial metabolic flux. Cell. Discov. 2, 16019 (2016).
Liu, W. et al. RNF126-Mediated MRE11 ubiquitination activates the DNA damage response and confers resistance of Triple-Negative breast Cancer to radiotherapy. Adv. Sci. (Weinh). 10 (5), e2203884 (2023).
Wang, Y. et al. RNF126 promotes homologous recombination via regulation of E2F1-mediated BRCA1 expression. Oncogene 35 (11), 1363–1372 (2016).
Yu, C., Xue, B., Li, J. & Zhang, Q. Tumor cell-derived exosome RNF126 affects the immune microenvironment and promotes nasopharyngeal carcinoma progression by regulating PTEN ubiquitination. Apoptosis 27 (7–8), 590–605 (2022).
Fa, P., Qiu, Z., Wang, Q. E., Yan, C. & Zhang, J. A novel role for RNF126 in the promotion of G2 arrest via interaction with 14-3-3sigma. Int. J. Radiat. Oncol. Biol. Phys. 112 (2), 542–553 (2022).
Yang, X. et al. RNF126 as a biomarker of a poor prognosis in invasive breast Cancer and CHEK1 inhibitor efficacy in breast Cancer cells. Clin. Cancer Res. 24 (7), 1629–1643 (2018).
Zhi, X. et al. E3 ubiquitin ligase RNF126 promotes cancer cell proliferation by targeting the tumor suppressor p21 for ubiquitin-mediated degradation. Cancer Res. 73 (1), 385–394 (2013).
Xu, H. et al. E3 ubiquitin ligase RNF126 affects bladder cancer progression through regulation of PTEN stability. Cell. Death Dis. 12 (3), 239 (2021).
Brett, D., Pospisil, H., Valcarcel, J., Reich, J. & Bork, P. Alternative splicing and genome complexity. Nat. Genet. 30 (1), 29–30 (2002).
Cheng, A. W. et al. Muscleblind-like 1 (Mbnl1) regulates pre-mRNA alternative splicing during terminal erythropoiesis. Blood 124 (4), 598–610 (2014).
Batra, R. et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol. Cell. 56 (2), 311–322 (2014).
Wang, E. T. et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell 150 (4), 710–724 (2012).
Bugg, D. et al. MBNL1 drives dynamic transitions between fibroblasts and myofibroblasts in cardiac wound healing. Cell. Stem Cell. 29 (3), 419–433e410 (2022).
Paschalis, A. et al. Alternative splicing in prostate cancer. Nat. Rev. Clin. Oncol. 15 (11), 663–675 (2018).
Itskovich, S. S. et al. MBNL1 regulates essential alternative RNA splicing patterns in MLL-rearranged leukemia. Nat. Commun. 11 (1), 2369 (2020).
Voss, D. M., Sloan, A., Spina, R., Ames, H. M. & Bar, E. E. The alternative splicing factor, MBNL1, inhibits glioblastoma tumor initiation and progression by reducing Hypoxia-Induced stemness. Cancer Res. 80 (21), 4681–4692 (2020).
Fish, L. et al. Muscleblind-like 1 suppresses breast cancer metastatic colonization and stabilizes metastasis suppressor transcripts. Genes Dev. 30 (4), 386–398 (2016).
Tang, L., Zhao, P. & Kong, D. Muscleblind–like 1 destabilizes snail mRNA and suppresses the metastasis of colorectal cancer cells via the snail/E–cadherin axis. Int. J. Oncol. 54 (3), 955–965 (2019).
Nauseef, J. T. & Henry, M. D. Epithelial-to-mesenchymal transition in prostate cancer: paradigm or puzzle? Nat. Rev. Urol. 8 (8), 428–439 (2011).
Xue, C., Li, G., Lu, J. & Li, L. Crosstalk between circrnas and the PI3K/AKT signaling pathway in cancer progression. Signal. Transduct. Target. Ther. 6 (1), 400 (2021).
Ullah, R., Yin, Q., Snell, A. H. & Wan, L. RAF-MEK-ERK pathway in cancer evolution and treatment. Semin Cancer Biol. 85, 123–154 (2022).
Li, H. et al. STAT3/miR-130b-3p/MBNL1 feedback loop regulated by mTORC1 signaling promotes angiogenesis and tumor growth. J. Exp. Clin. Cancer Res. 41 (1), 297 (2022).
Lu, X. et al. Quercetin reverses docetaxel resistance in prostate cancer via androgen receptor and PI3K/Akt signaling pathways. Int. J. Biol. Sci. 16 (7), 1121–1134 (2020).
Teo, M. Y., Rathkopf, D. E. & Kantoff, P. Treatment of advanced prostate Cancer. Annu. Rev. Med. 70, 479–499 (2019).
Cackowski, F. C. & Heath, E. I. Prostate cancer dormancy and recurrence. Cancer Lett. 524, 103–108 (2022).
Yamada, Y. & Beltran, H. The treatment landscape of metastatic prostate cancer. Cancer Lett. 519, 20–29 (2021).
Narayanan, S. et al. Targeting the ubiquitin-proteasome pathway to overcome anti-cancer drug resistance. Drug Resist. Updat. 48, 100663 (2020).
Ciechanover, A. The ubiquitin-proteasome proteolytic pathway. Cell 79 (1), 13–21 (1994).
Senft, D., Qi, J. & Ronai, Z. A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 18 (2), 69–88 (2018).
Cai, C., Tang, Y. D., Zhai, J. & Zheng, C. The RING finger protein family in health and disease. Signal. Transduct. Target. Ther. 7 (1), 300 (2022).
Kolapalli, S. P. et al. RNA-Binding RING E3-Ligase DZIP3/hRUL138 stabilizes Cyclin D1 to drive Cell-Cycle and Cancer progression. Cancer Res. 81 (2), 315–331 (2021).
Wang, L. et al. E3 ubiquitin ligase RNF126 regulates the progression of tongue cancer. Cancer Med. 5 (8), 2043–2047 (2016).
Ray, D. et al. A tumor-associated splice-isoform of MAP2K7 drives dedifferentiation in MBNL1-low cancers via JNK activation. Proc. Natl. Acad. Sci. U S A. 117 (28), 16391–16400 (2020).
Zhao, Y. et al. The MBNL1/circNTRK2/PAX5 pathway regulates aerobic Glycolysis in glioblastoma cells by encoding a novel protein NTRK2-243aa. Cell. Death Dis. 13 (9), 767 (2022).
Acknowledgements
We thank to the investigators and research laboratories whose original studies were cited in this article.
Funding
This work was supported by the Medical Research Project of Yangzhou Municipal Health Commission (2023-2-23), Yangzhou Basic Research Program (2024-2-23) and the Beijing Medical Award Foundation (YXJL-2024-0299-0065).
Author information
Authors and Affiliations
Contributions
Xin Jiang, Ji Li and Jiali Zhang: Formal analysis, Methodology, Project administration, Supervision, Validation, Writing – review & editing. Yulei Zhao: Methodology. Guoqin He and Xiaohui Yao: Conceptualization, Data curation, Formal analysis, Investigation, Project administration, Writing – original draft. All authors read and approved the final paper.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Baoying People’s Hospital (BYRY20221202).
Competing interests
The authors declare no competing interests.
Patient consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, X., Li, J., Zhang, J. et al. Ring-finger protein RNF126 promotes prostate cancer progression via regulation of MBNL1. Sci Rep 15, 23847 (2025). https://doi.org/10.1038/s41598-025-04629-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04629-6