Abstract
Osteosarcoma (OS) is the most common bone malignancy in children and adolescents. The 5-year survival rate is only approximately 20% in patients with metastatic and recurrent OS. There is an urgent need to explore novel therapeutic targets to improve the survivorship of OS patients. It has been reported that dysfunction of programmed cell death (PCD) plays an essential role in tumor malignancy. In this study, differentially expressed PCD genes (PCD-DEGs) in OS were identified. Functional enrichment and somatic mutation analysis were performed. Then we established a novel indicator, the cell death index (CDI), to predict therapeutic interventions, immunity responses, and prognosis concerning OS. The expression of PCD genes in osteosarcoma and normal tissues was determined by immunohistochemistry assays. A total of 265 PCD-DEGs were identified in OS, with 141 genes upregulated and 124 genes downregulated. After functional enrichment and somatic mutation analysis, a final PCD-related gene signature, involving the expression levels of CLTCL1, FADS2, and PLEKHF1, demonstrated significant predictive value. A nomogram including clinical characteristics and CDI was developed, which was verified to perform well. Further investigation showed CDI was strongly correlated with immunomodulators, tumor microenvironment, and drug sensitivity in OS. Immunohistochemistry assays illustrated that FADS2, CLTCL1, and PLEKHF1 were suppressed in OS tissues. Further experiments demonstrated that FADS2 inhibits migration and invasion of osteosarcoma cells, while FADS2 induces ferroptosis collaborating with Erastin in osteosarcoma cells. In conclusion, the PCD-related gene signature proposed in this study is a practical prognostic predictor for OS patients, which can make a notable difference in the assessment of clinical outcomes.
Similar content being viewed by others
Introduction
Osteosarcoma (OS) represents the most common bone malignancy for children and adolescents worldwide1. The 5-year survival of OS patients remains unchanged for decades under the primary treatments of tumor resection and chemotherapy using a combination of cisplatin, doxorubicin, and methotrexate2. Those who respond poorly to chemotherapy have an unfavorable outcome. The 5-year survival rate is only approximately 20% in patients with metastatic and recurrent OS3. Given the poor prognosis of advanced OS, there is an urgent need to explore novel therapeutic targets to improve the survivorship of OS patients, and practical models predicting the survival of OS patients can help to make targeted therapy more viable.
It has been reported that dysfunction of programmed cell death (PCD) plays an essential role in tumor malignancy4. PCD involves various mechanisms and elaborate regulations of cell death, whereas accident cell death is a biological process that is out of control. PCD includes apoptosis, necroptosis, pyroptosis, ferroptosis, cuproptosis, entotic cell death, netotic cell death, parthanatos, autophagy-dependent cell death, lysosome-dependent cell death, oxeiptosis, and alkaliptosis5. Apoptosis is the process of apoptotic vesicles removing unnecessary cells, and an inflammatory response is not triggered6. Necroptosis is a multi-step process of controlled necrosis, which is featured by the formation of necrosomes7,8. Pyroptosis is regulated cell death characterized by cell tumefaction, lysis, and the release of abundant pro-inflammatory factors9. When the iron-dependent accumulation of lipid hydroperoxides reaches fatal levels, ferroptosis occurs10,11. Cuproptosis is a type of copper-triggered cell death, which was discovered to be associated with malignant tumor12. Entotic cell death is a regulated, non-apoptotic process of cell death occurring only within living cells and adjacent areas13. The release of neutrophil extracellular traps causes netotic cell death and usually occurs under injury or infection14. Parthanatos is a highly programmed cell death that occurs through the excessive activation of the nuclease PARP-115. Lysosome-dependent cell death is accommodated by hydrolases and delivered to the cytosol following membrane permeation16. Autophagy-dependent cell death is associated with multistep lysosomal degradation, metabolic adaptation, and nutrient cycling17. Alkaliptosis operates a cell death program by intracellular alkalinization18. Oxeiptosis is a novel form of PCD utilizing the reactive oxygen sensing capabilities of KEAP1, which is likely to work with other cell death pathways together19. Advances in PCD research have provided increasing therapeutic targets for several tumors. The BCL-2 inhibitor was approved for treating lymphoma by regulating the cell apoptosis process20. GSDME-mediated pyroptosis has been reported as a new type of anti-tumor immunotherapy21. Additionally, studies have shown that TYRO3 inhibits tumoral ferroptosis and induces cellular anti-PD-1/PD-L1 resistance22.
PCD has been demonstrated to be deeply involved in the progression of various types of malignant tumors. However, there is a lack of literature regarding a comprehensive understanding of the relationship between PCD and OS. Meanwhile, recognizing the survival-associated genes for prognostic prediction in OS patients is needed from the perspective of PCD. In this study, we established a novel indicator, the cell death index (CDI), to predict therapeutic interventions, immunity responses, and prognosis concerning OS. Our study may help identify potential therapeutic targets for OS patients.
Methods
Data processing and identification of differentially expressed genes (DEGs)
Gene sets related to programmed cell death (PCD) were obtained from previous literature (Supplementary Table S1). The TARGET database’s expression profiles and clinical information data of OS patients were downloaded from the Genomic Data Commons (GDC) website (https://portal.gdc.cancer.gov/). Muscle expression profiles from the Genotype-Tissue Expression (GTEx) database were downloaded from the GTEx portal (https://gtexportal.org/home/). The two datasets were standardized, and batch effects were removed using the limma package. Subsequently, the data were merged. The limma package was employed for differential gene expression analysis between OS and normal samples, yielding Log2FC and corresponding differential P values for all genes. The specific Log2FC and P values for PCD genes in OS are presented in Supplementary Table S2. PCD genes with an absolute Log2FC (TARGET: osteosarcoma vs normal tissue) greater than two and an adjusted P value less than 0.05 were defined as Differentially Expressed PCD Genes (PCD-DEGs).
Functional enrichment analysis and somatic mutation analysis
Enrichment analysis of PCD-DEGs was conducted using the DAVID online tool (https://david.ncifcrf.gov/home), including Gene Ontology (GO) pathways and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Masked somatic mutation data of TARGET-OS patients were downloaded from GDC. The maftools package was utilized to analyze the mutation characteristics.
Construction of a prognostic PCD gene signature based on TARGET-OS cohort
Analysis focusing on prognosis was performed using data from the TARGET project and the GSE21257 database. We extracted the expression profiles of PCD genes from the TARGET-OS patients. The PCD-DEGs from the TARGET-OS patients were subjected to univariable Cox survival analysis. The Hazard Ratio (HR), 95% Confidence Interval (CI), and p-values for each gene were calculated using the Cox proportional hazards regression model. The results are presented in Supplementary Table S3. PCD-DEGs with p-values less than 0.1 were considered potentially significant. Fifty-two genes meeting this criterion were identified and documented in Supplementary Table S4. We performed a similar univariable Cox analysis to validate the findings using the GSE21257 database (Osteosarcoma tissue, samples did not undergo preoperative treatment, Sequencing platform: Affymetrix Human Genome U133 Plus 2.0 Array). The HR, 95% CI, and p-values for all PCD-DEGs are presented in Supplementary Table S5. Among them, 34 genes with p-values less than 0.1 were selected (Supplementary Table S6). We identified seven PCD-related DEGs (CLTCL1, FADS2, FAIM, NPC2, CHMP4C, S100A8, and PLEKHF1) by intersecting genes with p-values < 0.1 in both datasets. These candidate genes were then subjected to multivariable Cox regression analysis using the rms package, employing a stepwise variable selection approach (direction = “both”) to optimize the model. The final model was further assessed.
Prognostic CDI validation
To validate the PCD gene predictive model internally in the TARGET-OS dataset, we initiated the cell death index (CDI) calculation for each patient. Patients were categorized into high and low CDI groups based on the median CDI value, facilitating a comprehensive evaluation of the model’s performance. Survival curves for these groups were meticulously constructed using the survminer package, and the statistical significance between the high and low CDI cohorts was rigorously assessed via the log-rank test. ROC curves, generated with the timeROC and pROC packages, visually depicted the model’s discriminative ability over different time points. Furthermore, CDI was assessed using the calculation of the Area Under the Curve (AUC) at one, three, and five years. Additionally, calibration curves were fitted using the calibrate function, aligning predicted and observed survival rates at one, three, and five years.
The external validation was performed using the GSE21257 dataset, aiming to validate the generalizability of the PCD gene prognostic model. Like the internal validation, CDI calculations were performed, and patients were categorized into high and low CDI groups based on the median CDI value. The survival curves for these groups were constructed, and the log-rank test was employed to scrutinize the differences in survival rates between the two cohorts. In addition, the robustness of the model was further evaluated by AUC values at one, three, and five years and ROC curves. Calibration curves were generated to assess the alignment between predicted and observed survival rates.
Relationship between CDI and clinical characteristics
Relevant clinical data were used for subsequent analysis. Statistical tests were applied to investigate the significance of CDI variations across different subgroups, such as age, gender, metastasis status, and pathological characteristics. Graphical representations were generated to illustrate the observed associations visually.
GSVA analysis
GSVA analysis was conducted using the GSVA package and GSEABase package to examine the biological functional changes in KEGG pathways between high and low CDI groups in both TARGET-OS and GSE21257 datasets.
Unsupervised clustering of PCD-related model genes
The ConsensusClusterPlus package in R was employed for unsupervised clustering analysis of the unidentified subtypes of OS patients. The consensus matrix was generated to evaluate the stability of the clustering at varying cluster numbers (k). The parameter “maxK” was selected as “10”, “clusterAlg” was set as “km”, and “distance” was set as “pearson”.
Single-cell analysis
We used the Seurat package for the processing of single-cell RNA sequencing data. The analytical workflow encompassed quality control, standardization, high variable gene analysis, normalization, principal component analysis (PCA), and cell clustering. The GSE162454 single-cell sequencing database was selected. Cell classification into various categories was performed, and the CDI for each cell was calculated. CDI mean values across the cell types were then analyzed for significant differences. Additionally, the expression and distribution of PCD model genes were examined for each cell type. A second single-cell sequencing database, GSE152048, was utilized to validate the results.
Exploration of immune microenvironment associated with CDI and PCD model genes
We conducted a correlation analysis between CDI and the expression of numerous immune regulatory factors. To predict the composition of 22 immune cell types in the immune microenvironment of TARGET-OS patients, we utilized the CIBERSORT package. Differential analysis of immune cell levels between the high CDI and low CDI groups was performed, and we further explored the correlation between CDI as a continuous variable and the levels of 22 immune cells. Survival prognosis analysis was performed on immune cells, showing strong CDI correlations.
Establishment of the nomogram
Multivariable regression analysis was employed to assess the association of CDI with clinical variables, including age and gender. The results are presented as HR with corresponding p-values. Visualization of the findings was achieved through the construction of line graphs, illustrating the interplay between CDI, age, and gender. Survival predictions were derived from the line graph, and their accuracy was internally validated using the TARGET-OS database. ROC curves were generated to evaluate the predictive performance, with corresponding AUC values calculated for 1-year, 3-year, and 5-year survival predictions. External validation was performed using the GSE21257 database.
Prediction of response to immunotherapy
The Tumor Immune Dysfunction and Exclusion (TIDE) algorithm was employed for the analysis, generating each patient’s TIDE scores, Dysfunction scores, and Exclusion scores. The distribution of predicted responders and non-responders to immunotherapy was examined qualitatively. Comparative analyses were conducted between non-responders and responders, assessing CDI levels and the expression of PCD model genes. Qualitative trends in CDI levels and PCD gene expression were summarized. TIDE scores were further stratified based on CDI levels and the expression of three PCD genes. Subsequent analyses focused on T-cell dysfunction and exclusion. Qualitative outcomes of T-cell dysfunction analysis were described, and similar qualitative observations were made for T-cell exclusion.
Drug sensitivity analysis
A thorough examination of drug sensitivity in osteosarcoma was performed, utilizing the oncoPredict package in R. The expression matrix of osteosarcoma patients from the TARGET-OS dataset was employed to predict IC50 values for 198 drugs, facilitating the evaluation of sensitivity in 88 patients. Stratification of patients into high and low CDI groups was based on median CDI values. Differential drug sensitivity analysis uncovered significant variations in IC50 values between the two groups, with the top 20 drugs exhibiting the most pronounced differences. Statistical analyses were conducted to determine significance, and correlation coefficients were calculated to explore the relationship between IC50 values and continuous CDI values for each patient. Furthermore, a screening of commonly used chemotherapy drugs and targeted therapies from the 2023 CACA osteosarcoma guideline was conducted using oncoPredict, identifying 13 drugs for further analysis.
Immunohistochemistry
We conducted immunohistochemical staining analysis to measure the protein expression of FADS2, CLTCL1, and PLEKHF1 in osteosarcoma tissues and adjacent normal tissues according to the standard immune-peroxidase staining procedure. Paraffin-embedded tissues were cut into 4 μm sections, deparaffinized and blocked with avidin/biotin for 20 min. The slides were incubated with primary antibodies overnight at 4 °C. Next, the slides were treated with secondary antibody (Invitrogen) and HRP conjugate for 1 h and then developed with DAB (Invitrogen). Finally, the slides were counterstained with hematoxylin. The IHC staining results of FADS2, CLTCL1, and PLEKHF1 were independently evaluated by two pathologists.
Cell culture
Human osteosarcoma cell lines, HOS, MG-63, 143B, and SW1353 cells, were purchased from INDIT Bio-Technology Co., Ltd (Hangzhou, China). The HOS and MG-63 cells were cultured in MEM medium (GIBCO BRL, Grand Island, NY, USA, C11095500BT) with NEAA (GIBCO BRL, 11140-050). The 143B or SW1353 cells were cultured in a Dulbecco’s modified eagle medium (DMEM, BDBIO, Hangzhou, China, L100-500) or Leibovitz’s L-15 medium (GIBCO BRL, 11415064), respectively. All the medium was added with 10% FBS (SERANA, Brandenburg, Germany, S-FBS-SA-015) and 1% penicillin–streptomycin (Pen-Strep, GIBCO BRL, 15140122), and all the cells were maintained at 37 °C in humidified air with 5% CO2. When the cells grew to 80–90%, they were passaged at a 1:3 ratio, and the logarithmic growth stage cells were used for subsequent experiments. Additionally, Ferrostatin-1 (HY-100579) and Erastin (HY-15763) were purchased from MedChem Express (Monmouth Junction, NJ, USA).
Western blot (WB)
The treated cell total proteins were extracted utilizing RIPA lysis buffer (Merck, Darmstadt, Germany, 20-188). The protein concentration was measured by a Pierce BCA Protein Assay kit (Thermo Fisher, Waltham, MA, USA, 23228) and adjusted to a consistent level. Subsequently, protein samples were loaded in SDS–polyacrylamide gel electrophoresis and shifted onto the 0.45 μm polyvinylidene fluoride (PVDF) membranes (Millipore, Massachusetts, USA, IPVH00010). The membranes were blocked with 5% skim milk prepared with TBST buffer solution and then incubated with the primary antibodies at 4 °C followed by incubating with the corresponding secondary antibodies at room temperature. Finally, after washes with TBST, the target protein bands were visualized using an enhanced chemiluminescence (ECL) kit (Biosharp, Hefei, China, BL520B). The antibodies used for WB are shown in Table 1.
Cell transfection
The plasmids of the FADS2 overexpression and the pcDNA 3.l empty vector were purchased from YouBio (Hunan, China). The overexpressing plasmid of FADS2 was inserted into pcDNA 3.l. Once cells reached 70% confluence, pcDNA3.1-FADS2 and the empty pcDNA3.1 plasmid were separately transfected into SW1353 and 143B cells for 24 h using Lipofectamine 2000 (Invitrogen, Waltham, MA, USA, 11668-019) and Opti-MEN (Gibco, Semitic, NY, USA, 31985-070) according to the manufacturer’s description. The transfection efficiency was validated through WB analysis.
Transwell assay
Transwell inserts (COSTAR, Corning, NY, USA, 3422) were used in 24-well plates. After treatment, SW1353 and 143B cells (2 × 104 cells) were seeded on the upper chamber with serum-free medium for migration (uncoated) and invasion (Matrigel-coated) assays. DMEM with 10% FBS was added to the bottom chamber. After 24 h incubation, non-migratory/invasive cells were removed, and migrated/invaded cells were fixed with methanol and stained with crystal violet (Aladdin, Shanghai, China, C110703) for 20 min. Cell counting under the microscope (Olympus Corp, Tokyo, Japan, Olympus IX73) was conducted at × 100 magnification. Each experiment was performed in triplicate.
MTT assay
The cell proliferation of SW1353 and 143B cells was detected by the MTT assay. Cells after indicated treatment were first seeded into the 96-well plates at a density of 3 × 103 cells/well and cultured in an incubator for 0, 24, 48, and 72 h. Then 1 mg/mL MTT (Beyotime, Shanghai, China, C0009S) was added to the plate (50 μL/well) and cells were incubated for 3 h at 37 °C. The resulting formazan crystals in the plate were dissolved in DMSO (150 μL/well), and the relative cell proliferation was measured using a spectrophotometer (Molecular Device, Silicon Valley, CA, USA) at the absorbance of 570 nm. Each experiment was performed in triplicate.
Flow cytometry
The flow cytometry was performed to examine the lipid reactive oxygen species (ROS) of SW1353 and 143B cells. Firstly, BODIPY 581/591 C11 (Abclonal, Wuhan, China, RM02821, for lipid ROS was diluted using the serum-free DMEM medium (diluted to 2 μmol/L). After the indicated treatment, SW1353 and 143B cells were collected and then separately co-incubated with diluted BODPY 581/591 C11 for 30 min at 37 °C. Then, cells were washed with serum-free DMEM medium. All samples were analyzed under a flow cytometer (Invitrogen, Waltham, MA, USA, Attune NxT) to detected the level of ROS.
Statistical analysis
All statistical analyses were performed using R software (v.4.3.0). Students’ t-tests were used for normally distributed data and Wilcoxon tests for non-parametric data. The normality was verified by Shapiro–Wilk tests as indicated when p > 0.05. Comparisons among more than two groups were analyzed using Kruskal–Wallis’s test. Survival curves were plotted using the Kaplan–Meier method and compared using the log-rank test. The experiments in this study were repeated at least three times. Representative data were analyzed by GraphPad Prism 6.0 (GraphPad Software, La Jolla, CA, USA). Statistical analysis for comparison between the two groups was t-test and that for comparison among multiple groups was one-way ANOVA. P < 0.05 was considered statistically significant.
Results
Variant landscape of programmed cell death genes in OS patients
A total of 265 PCD-DEGs were identified in OS, with 141 genes upregulated and 124 genes downregulated. Figure 1A presents a heatmap illustrating the expression patterns of these PCD-DEGs in TARGET-OS samples, while Fig. 1B displays a volcano plot depicting Log2FC and differential P values of all genes in OS compared to normal samples. Enrichment analysis of PCD-DEGs revealed significant associations with KEGG pathways (Fig. 1C) and GO pathways (Fig. 1D). Somatic mutation analysis of TARGET-OS patients’ data showed prevalent missense mutations and single nucleotide polymorphisms (SNPs) (Fig. 1E). The top 10 mutated genes, including TP53, RB1, MET, PTEN, etc., were further analyzed, and their distribution across all 88 OS samples is presented in Fig. 1F; TP53’s mutation types and base position distribution within the gene framework are detailed in Fig. 1G. Across all genes, C > T mutations were the most common, followed by C > A mutations, as depicted in Fig. 1H.
Programmed cell death genes variation in osteosarcoma. (A) Heatmap illustrating the expression patterns of 265 differentially expressed genes (DEGs) related to programmed cell death (PCD) in TARGET-OS samples. (B) Volcano plot displaying Log2FC and differential P values of all genes in osteosarcoma versus normal samples. (C) Enrichment analysis of PCD-DEGs with KEGG pathways (TARGET-OS)[a-c]. (D) Enrichment analysis of PCD-DEGs with gene ontology (GO) pathways (TARGET-OS). (E) Somatic mutation analysis of TARGET-OS patients’ data revealing prevalent missense mutations and single nucleotide polymorphisms (SNPs). (F) The top 10 mutated genes were distributed across all 88 OS samples (TARGET-OS). (G) Detailed mutation types and base position distribution within the TP53 gene (TARGET-OS). (H) Overview of common mutation patterns across all genes (TARGET-OS).
PCD-related gene signature
Seven genes including CLTCL1, FADS2, FAIM, NPC2, CHMP4C, S100A8, and PLEKHF1 were identified by intersecting genes with p-values < 0.1 in both TARGET-OS and GSE21257 datasets. The optimal model by the stepwise approach selected three genes, i.e., CLTCL1, FADS2 and PLEKHF1. The Hazard Ratios (HR) and p-values are presented in Fig. 2A. The formula is CDI = CLTCL1*(− 0.7021) + FADS2*(0.457) + PLEKHF1*(− 0.2645), which provides a quantitative measure of the predicted impact of CLTCL1, FADS2, and PLEKHF1 gene expression levels on the cell death index. The input values for CLTCL1, FADS2, and PLEKHF1 in the CDI formula represent log2-transformed TPM (Transcripts Per Million) values, normalized using the DESeq2 pipeline. Raw RNA-seq counts were preprocessed with variance-stabilizing transformation (VST) to minimize batch effects.
Validation of prognostic PCD gene signature. (A) Final PCD-related gene signature encompassing CLTCL1, FADS2, and PLEKHF1 expression levels, demonstrating significant predictive value. HR > 1 indicates risk-associated genes, while HR < 1 indicates protective genes. (B) Internal validation using the TARGET-OS dataset showing survival rate differences between high and low Cell Death Index (CDI) groups. (C) Prognostic performance of CDI with Area Under the Curve (AUC) values at 1, 3, and 5 years (TARGET-OS). (D) Distribution of CDI scores and expression heatmap of PCD prognostic model genes for all TARGET-OS patients. (E) Calibration curves for 1, 3, and 5 years in internal validation (TARGET-OS). (F) External validation using the GSE21257 dataset confirmed survival rate differences in the high CDI group. (G) Consistent predictive performance of CDI with AUC values at 1, 3, and 5 years in external validation (GSE21257). (H) Distribution of CDI scores and expression heatmap of PCD prognostic model genes for all GSE21257-OS patients. (I) Calibration curves for 1, 3, and 5 years in external validation (GSE21257).
Internal and external validation of the gene signature prediction model
To evaluate the clinical utility of CDI, we performed rigorous validationl across independent cohorts, beginning with the internal validation in the TARGET-OS dataset. The survival analysis revealed a significant difference in survival rates between the high and low CDI groups, with a log-rank test p-value of less than 0.001 (Fig. 2B). The predictive performance of CDI was supported by AUC values of 0.79, 0.784, and 0.804 at one, three, and five years, respectively (Fig. 2C). The distribution of CDI scores and the expression heatmap of PCD prognostic model genes for all patients are illustrated in Fig. 2D. Calibration curves for one, three, and five years demonstrated the alignment between predicted and observed survival rates, with a slight underestimation at one year but good concordance at three and five years (Fig. 2E).
In the external validation using the GSE21257 dataset, the survival analysis confirmed a significantly lower survival rate in the high CDI group compared to the low CDI group, with a log-rank test p-value of 0.002 (Fig. 2F). The predictive performance of CDI was consistent, with AUC values of 0.837, 0.738, and 0.785 at one, three, and five years, respectively (Fig. 2G). The distribution of CDI scores and the expression heatmap of PCD prognostic model genes for all GSE21257-OS patients are depicted in Fig. 2H. Calibration curves for one, three, and five years in external validation mirrored the internal validation results, indicating a slight underestimation at one year but good concordance at three and five years (Fig. 2I). Consistent with our hypothesis, CDI demonstrated robust prognostic performance in both internal and external validations.
Relationship between CDI and clinical characteristics
The analysis of clinical characteristics in TARGET-OS patients revealed significant associations with CDI expression. Deceased patients exhibited markedly higher CDI levels than survivors (P < 0.001, Fig. 3A). Age and gender differences showed no significant correlation with CDI levels (Fig. 3B and Fig. 3C). However, metastatic osteosarcoma patients displayed significantly higher CDI levels than their non-metastatic counterparts (P < 0.05, Fig. 3D). Patients with pathological necrosis below 90% also exhibited an elevated level of CDI compared to those with 91–100% necrosis (P < 0.05, Fig. 3E). The comprehensive heatmap (Fig. 3F) visually depicted CDI distribution across 88 patients based on diverse clinical features.
Associations between CDI and clinical characteristics. (A) Associations between CDI expression and survival outcomes, with deceased patients exhibiting higher CDI levels than survivors. (B) Correlation between age and CDI levels. (C) Correlation between gender and CDI levels. (D) Metastatic osteosarcoma patients display higher CDI levels than their non-metastatic counterparts. (E) Patients with pathological necrosis below 90% exhibit elevated CDI compared to those with 91–100% necrosis. F Comprehensive heatmap visually depicting CDI distribution across 88 patients based on diverse clinical features. (G, H) GSVA analysis of KEGG pathways between high and low CDI groups in TARGET-OS patients (TARGET-OS) and the GSE21257 dataset (GSE21257)[a-c].
GSVA analysis of KEGG pathways between high and low CDI groups in TARGET-OS patients (Fig. 3G,H) and the GSE21257 dataset (Fig. 3I,J) revealed substantial differences. "Primary bile acid biosynthesis" consistently ranked among the top 10 differentially expressed pathways in both datasets. Having established CDI as a robust prognostic marker linked to key clinical features, we next investigated whether these CDI-based patient subgroups could be further stratified into biologically distinct clusters through unsupervised analysis of PCD-related gene expression.
Unsupervised clustering of OS patients based on PCD-related model genes
As illustrated in Fig. 4A, the consensus matrix at k = 3 displayed a clear and significant clustering into three distinct groups. Subsequent survival analysis (Fig. 4B) revealed a highly significant difference in survival curves among the three clusters (P = 0.009), with Cluster 2 displaying the worst prognosis. The distribution of survival/death status among the three clusters and their association with the level of CDI (Fig. 4C) was visually represented. This provided a comprehensive overview of the correlation between the identified clusters and clinical outcomes. Upon validation in the GSE21257 dataset (Fig. 4D), the consensus matrix at k = 3 again exhibited pronounced clustering. The survival analysis (Fig. 4E) demonstrated significant differences in survival curves among the three clusters (P = 0.04). Notably, the prognostic distribution across the clusters in GSE21257 closely mirrored that observed in the TARGET-OS dataset (Fig. 4E,F, reinforcing the robustness and generalizability of our findings.
Unsupervised clustering of OS patients based on PCD-related model genes. (A) Consensus matrix at k = 3 displaying an apparent clustering into three distinct groups (TARGET-OS). (B) Survival analysis revealed significant differences in survival curves among the three clusters (TARGET-OS). (C) Distribution of survival/death status among the three clusters and their association with the level of CDI (TARGET-OS). (D) The consensus matrix at k = 3 exhibits pronounced clustering in external validation using the GSE21257 dataset. (E) Survival analysis demonstrates significant differences in survival curves among the three clusters in GSE21257. (F) Prognostic distribution across the clusters in GSE21257 mirroring that observed in the TARGET-OS dataset.
CDI and PCD model genes in single-cell OS tissue
The single-cell RNA sequencing analysis of osteosarcoma identified seven distinct cell types, as illustrated in Fig. 5A. Subsequent calculation of the CDI revealed statistically significant differences in CDI mean values among these seven cell types, as depicted in Fig. 5B. Further exploration of the expression and distribution of PCD model genes across cell types uncovered intriguing patterns. Figure 5C showcased the heightened expression of the PLEKHF1 gene in NK/T cells, with notable expression in myeloid cells and osteosarcoma cells. Figure 5D,E presented the predominant expression of FADS2 and CLTCL1 genes in osteosarcoma cells, myeloid cells, and cancer-associated fibroblasts (CAFs), providing insights into their potential roles within the tumor microenvironment. Validation of these findings was undertaken using the GSE152048 dataset, demonstrating consistent and similar outcomes across different single-cell sequencing databases, as illustrated in Fig. 5F–J. Figure 5F showed the cell types distribution in osteosarcoma tissues. Figure 5G presented the expression and distribution of PCD model genes across these cell types. Figure 5H–J illustrated the expression and distribution of three PCD-related genes in the signature, including PLEKHF1, FADS2, and CLTCL1.
PCD model genes in single-cell of OS tissue. (A) Single-cell RNA sequencing analysis of osteosarcoma identifying seven distinct cell types (GSE162454). (B) Significant differences in CDI mean values among the seven cell types. (C) Expression and distribution of PCD model genes across cell types, showcasing heightened expression of PLEKHF1 in NK/T cells and predominant expression of FADS2 and CLTCL1 in osteosarcoma cells, myeloid cells, and cancer-associated fibroblasts (CAFs). (D, E) Validation of findings using the GSE152048 dataset, demonstrating consistent outcomes across different single-cell sequencing databases.
Exploration of immune microenvironment based on programmed cell death signature
Figure 6A depicts a consistent negative correlation between CDI and the expression levels of various immune regulatory factors, suggesting a potential immune-suppressed state in high CDI patients, particularly those with a poorer prognosis. Utilizing the CIBERSORT package to predict immune cell composition (Fig. 6B), both high and low CDI patient groups exhibited a predominant presence of M0 and M2 macrophages, along with resting memory CD4 T cells. The differential analysis (Fig. 6C) highlighted a significant reduction in resting NK cells in the High CDI group, indicating the impact of CDI on specific immune cell populations. Using CDI as a continuous variable, Fig. 6D illustrates significant correlations with five immune cell types. The survival prognosis analysis (Fig. 6E–H) of these correlated immune cells did not reveal a statistical significance (p values > 0.05). Given the observed link between CDI and immune suppression, we further developed a clinical tool to quantify individual prognostic risk by integrating CDI with key clinical parameters.
Immune microenvironment associated with CDI and PCD model genes. (A) Consistent negative correlation between CDI and the expression levels of various immune regulatory factors. (B) Prediction of immune cell composition in the immune microenvironment of TARGET-OS osteosarcoma patients. (C) Differential analysis highlighting a significant reduction in resting NK cells in the High CDI group. (D) Significant correlations between CDI and five immune cell types. (E–H) Survival prognosis analysis of these correlated immune cells (TARGET-OS), emphasizing potential clinical relevance.
Performance of the prognostic nomogram
As depicted in Fig. 7A, multivariable regression analysis revealed a noteworthy hazard ratio (HR) of 2.881 (P < 0.001) after adjusting for age and gender, establishing CDI as an independent prognostic risk factor. The nomogram representation in Fig. 7B visually delineated the relationship between CDI, age, gender, and prognosis. Internal validation using the TARGET-OS database showcased satisfactory predictive accuracy, with ROC curves (Fig. 7C) indicating AUC values of 0.751, 0.804, and 0.834 for 1-year, 3-year, and 5-year survival predictions, respectively. Calibration curves (Fig. 7D) further affirmed the reliability of the model. Notably, the survival curves (Fig. 7E) demonstrated a significant distinction in survival outcomes between high-risk and low-risk patients identified by the nomogram. External validation using the GSE21257 database (Fig. 7F–H) consistently validated these findings. With the prognostic utility of CDI validated, we then evaluated its potential to predict response to immunotherapy, a critical consideration for personalized treatment strategies.
Prognostic nomogram and performance. (A) Multivariable regression analysis revealing CDI as an independent prognostic risk factor (TARGET-OS). (B) Nomogram visually represents the relationship between CDI, age, gender, and prognosis. (C) Internal validation ROC curves indicating AUC values for 1-year, 3-year, and 5-year survival predictions (TARGET-OS). (D) Calibration curves affirm the reliability of the prognostic nomogram (TARGET-OS). (E) Survival curves demonstrating significant distinctions in survival outcomes between high-risk and low-risk patients identified by the nomogram (TARGET-OS). (F–H) External validation using the GSE21257 database consistently validates these findings.
Prediction of response to immunotherapy based on CDI
Figure 8A provides an overview of the distribution of predicted responders and non-responders to immunotherapy among osteosarcoma patients based on TIDE analysis. The analysis revealed a lower overall count of responders (blue scatter points) compared to non-responders (red scatter points), with subsequent exploration into CDI levels and the expression of three PCD genes in both groups. While non-responders displayed slightly elevated CDI levels, statistical significance was not reached, possibly attributed to the limited number of responder cases (only ten individuals). In Fig. 8B, TIDE scores were stratified based on CDI levels and the expression of three PCD genes. The differences were not substantial, suggesting that CDI and PCD gene expression may not be sufficient predictors for immunotherapy outcomes in osteosarcoma patients. However, further investigation into T-cell dysfunction (Fig. 8D,E) demonstrated that patients with higher CDI had notably lower T-cell dysfunction scores. This finding suggests that despite the poorer survival prognosis associated with high CDI, these patients might still harbor potential benefits from immunotherapy due to a lower degree of T-cell dysfunction. Results for T-cell exclusion (Fig. 8F,G) mirrored those obtained from TIDE analysis, with no significant differences observed. While TIDE scores did not reveal marked distinctions, a deeper analysis illuminated substantial variations in the tumor’s immune microenvironment, particularly regarding T-cell dysfunction.
Prediction of response to immunotherapy. (A) Overview of the distribution of predicted responders and non-responders to immunotherapy among osteosarcoma patients based on TIDE analysis. (B) Exploration into CDI levels and the expression of three PCD genes in both groups. (D, E) T-cell dysfunction analysis demonstrates lower T-cell dysfunction scores in patients with higher CDI. (F, G) T-cell exclusion analysis shows no significant differences between high and low CDI groups. (H–J) Survival curves based on TIDE score median, T-cell dysfunction score median, and T-cell exclusion score median.
Survival curves in Figs. 8H–J were generated based on the TIDE score median, T-cell dysfunction score median, and T-cell exclusion score median as cutoff values. While TIDE and exclusion did not exhibit pronounced differences, the close relationship between T-cell dysfunction score and survival (P-value = 0.053) suggested that lower dysfunction scores were associated with poorer survival. Patients with lower dysfunction scores, identified through CDI, might have potential benefits from immunotherapy, highlighting the clinical significance of identifying this subgroup. Beyond immunotherapy, we explored whether CDI could guide drug selection by analyzing its correlation with sensitivity to both targeted therapies and conventional chemotherapeutics.
Correlation of CDI with drug sensitivity prediction
Differential analysis of IC50 values between high and low CDI groups revealed the top 20 drugs with significant differences (p < 0.01), as illustrated in Fig. 9A. For instance, daporinad exhibited lower IC50 values in the high CDI group, suggesting heightened sensitivity. Correlation analysis further elucidated the relationship between IC50 values of these drugs and continuous CDI, as shown in Fig. 9B. As for analyzing commonly used chemotherapy drugs and targeted therapies, 13 drugs were identified for further investigation. Box plots in Fig. 9C depicted a significant increase in lapatinib sensitivity in the high CDI group, suggesting potential therapeutic benefits in this subgroup. While statistical differences were not observed for other drugs, sorafenib and gemcitabine were close to statistical significance (p ≈ 0.1), as well as cisplatin, paclitaxel, and dactinomycin (p ≈ 0.2–0.25). The heatmap in Fig. 9D also indicated a significant correlation for sorafenib.
Drug sensitivity analysis. (A) Differential analysis of IC50 values between high and low CDI groups revealing the top 20 drugs with significant differences. (B) Correlation analysis elucidates the relationship between IC50 values of these drugs and continuous CDI. (C) Box plots indicate a significant increase in lapatinib sensitivity in the high CDI group. (D) Heatmap indicating a significant correlation for sorafenib.
Immunohistochemistry
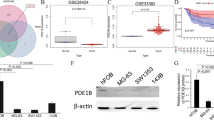
We selected seven pairs of OS tissues and adjacent tissues from different patients with OS to determine whether FADS2, CLTCL1 and PLEKHF1 were down-regulated in OS tissues. Immunohistochemistry assays illustrated that the number of FADS2/CLTCL1/PLEKHF1-positive cells was significantly lower in OS tissues than in adjacent tissues, suggesting that FADS2/CLTCL1/PLEKHF1 was suppressed in OS (p < 0.05, Fig. 10). We prioritized FADS2 for further functional validation because: (1) As an oncogenic driver, its overexpression directly promotes OS progression, making it a more actionable therapeutic target than tumor suppressors (CLTCL1/PLEKHF1); (2) Pharmacological inhibition of FADS2 is clinically feasible, unlike gene-replacement therapies required for tumor suppressor restoration.
FADS2 inhibits migration and invasion of osteosarcoma cells
In this study, we first detected the protein levels of FADS2 in 4 human osteosarcoma cell lines. Subsequently, SW1353 cells and 143B cells whose constitutive expression of FADS2 is lower were selected to define the role of FADS2 in osteosarcoma and explore its mechanism (Fig. 11A). To investigate the role of FADS2, we overexpressed FADS2 in SW1353 cells and 143B cells respectively to perform the subsequent function tests (Fig. 11B). Using the above two FADS2-overexpressed cell lines and their control cell lines, we performed the transwell migration and invasion assay, and found the capacity of migration and invasion were significantly inhibited in FADS2-overexpressed osteosarcoma cells (Fig. 11C), suggesting high expression level of FADS2 could suppress the malignant progression of human osteosarcoma cells.
FADS2 inhibits migration and invasion of osteosarcoma cells. (A) The constitutive expression of FADS2 in multiple human osteosarcoma cells was tested by WB assay; (B) The protein level of FADS2-flag in osteosarcoma cells with FADS2 overexpression was tested by WB assay; (C) Representative images and quantification of the transwell migration and invasion assay in osteosarcoma cells with FADS2 overexpression. Quantitative analysis presented as mean ± SD from n = 3 independent experiments (A–C). Statistical significance was determined by the two-tailed t-test.*P < 0.05; **P < 0.01.
FADS2 induces ferroptosis in osteosarcoma cells
Given FADS2’s established association with ferroptosis pathways in our bioinformatics analysis, we next sought to functionally characterize its role in OS cell migration and drug response, as these processes are known to be modulated by ferroptosis sensitivity in cancer cells. The result of the MTT assay showed that treatment of 2 μM ferrostatin-1 (Fer-1), a common ferroptosis inhibitor, could rescue the proliferation capacity of osteosarcoma cells inhibited by overexpression of FADS2 (Fig. 12A). To further identify whether FADS2 could induce ferroptosis in osteosarcoma cells, we measured lipid ROS using BODIPY 581/591 C11 dye (Fig. 12B), and detected the protein level of a key regulator of inhibiting ferroptosis—GPX4 (Fig. 12C), showing that overexpression of FADS2 promoted lipid ROS and reduced the level of GPX4, both of which could be reversed by Fer-1 in osteosarcoma cells. In summary, these results primarily suggest that high expression of FADS2 induces ferroptosis in osteosarcoma cells by ROS accumulation to reverse the tumor-promoting efficacy of Fer-1.
FADS2 induces ferroptosis in osteosarcoma cells. (A) The viability of FADS2-overexpressed osteosarcoma cells with/without Fer-1 (2 μM) treatment was evaluated by MTT assay; (B) Lipid ROS assessment of FADS2-overexpressed osteosarcoma cells with/without Fer-1 (2 μM) treatment was detected by flow cytometry; (C) The protein levels of GPX4 in FADS2-overexpressed osteosarcoma cells with/without Fer-1 (2 μM) treatment was detected by WB assay. Quantitative analysis presented as mean ± SD from n = 3 independent experiments (A–C). The two-tailed t-test determined statistical significance. *P < 0.05; **P < 0.01.
FADS2 promotes ferroptosis collaborating with Erastin in osteosarcoma cells
To pharmacologically validate these findings, we employed Erastin—a well-characterized ferroptosis inducer that specifically inhibits system Xc- activity, thereby depleting glutathione and sensitizing cells to lipid peroxidation. This approach allowed us to: (1) verify whether FADS2 plays the same role in human osteosarcoma cells, and (2) explore potential therapeutic synergies. Similar to above, an MTT assay was applied to reflect the proliferation capacity of two FADS2-overexpressed cell lines and their control cell lines with or without the treatment of 10 μM Erastin, indicating FADS2 combined with Erastin significantly suppressed the proliferation of osteosarcoma cells (Fig. 13A). Next, we detected the protein level of GPX4 (Fig. 13B), and measured lipid ROS in the above cell lines with different treatments (Fig. 13C), showing that promotion of ferroptosis under FADS2 overexpression and Erastin treatment were similar in osteosarcoma cells. Additionally, the combined action of FADS2 overexpression and Erastin treatment exhibits a remarkable enhancement of ferroptosis in osteosarcoma cells, implying a synergistic mechanism and underscoring the therapeutic potential of this combination for addressing osteosarcoma.
FADS2 promotes ferroptosis collaborating with Erastin in osteosarcoma cells. (A) The viability of FADS2-overexpressed osteosarcoma cells with/without Erastin (10 μM) treatment was evaluated by MTT assay; (B) The protein levels of GPX4 in FADS2-overexpressed osteosarcoma cells with/without Erastin (10 μM) treatment was detected by Immunoblotting; (C) Lipid ROS assessment of FADS2-overexpressed osteosarcoma cells with/without Erastin (10 μM) treatment was detected by flow cytometry. Quantitative analysis presented as mean ± SD from n = 3 independent experiments (A–C). The two-tailed t-test determined statistical significance. *P < 0.05; **P < 0.01. [a] Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. and Ishiguro-Watanabe, M.; KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672-D677 (2025). [pubmed]. [b] Kanehisa, M; Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019). [c] Kanehisa, M. and Goto, S.; KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000).
Discussion
To the best of our knowledge, this study is the first to analyze twelve diverse PCD patterns in OS comprehensively, construct a cell death signature in the Target-OS cohort, and further validate its excellent performance in two other external cohorts (GSE21257, GSE152048). A nomogram including clinical characteristics and CDI was developed, which was verified to perform well. Finally, we identified whether CDI was correlated with immunomodulators, tumor microenvironment, and drug sensitivity in OS.
Increasing evidence indicates that PCD plays an essential role in various biological processes, which has a close relationship with the development and metastasis of malignant tumors for decades7. We created a signature featuring 3 PCD-related genes (CLTCL1, FADS2, and PLEKHF1) and found that it could predict survival in OS patients. CLTCL1 (clathrin, heavy chain-like 1) is a member of the clathrin family and encodes a protein highly expressed in muscle tissues and consists of 1640 amino acids23,24. It has been reported that the downregulation of CLTCL1 promotes the progression of ductal carcinoma of the breast25, which is consistent with our study. We found that CLTCL1 expression was lower in OS tissues compared with normal tissues, suggesting the expression of CLTCL1 was negatively correlated with survival in OS. FADS2 (fatty acyl desaturase 2) is a key fatty acid desaturase26. The roles of FADS2 in different malignant tumors are controversy in the previous literatures27. Yang et al. showed that FADS2 promoted lipid metabolism and ferroptosis in ovarian cancer cells28. Park et al. demonstrated that FADS2 is weakly expressed in breast cancer, and low FADS2 expression is significantly correlated with poor prognosis29. Tian et al. discovered that FADS2 promotes colorectal cancer30. Our study found that the expression of FADS2 was lower in OS samples compared with normal tissues, suggesting high expression of FADS2 was associated with a better prognosis. Further experiments demonstrated that FADS2 inhibits migration and invasion of osteosarcoma cells, while FADS2 induces ferroptosis collaborating with Erastin in osteosarcoma cells, which are in consistence with the histological findings. Interestingly, our bioinformatic results showed FADS2 seems to be a tumor-progressive role in osteosarcoma. However, bioinformatic results need to be verified by rigorous experiments in vitro. Our experimental results finally demonstrated FADS2 has a tumor-suppressive role in osteosarcoma, again presenting that the roles of FADS2 in different malignant tumors are controversy. Future studies will evaluate kinase inhibitors (e.g., sorafenib/lapatinib) to dissect FADS2’s kinase-independent functions in OS, particularly its crosstalk with ferroptosis and lipid metabolism pathways. PLEKHF1, pleckstrin homology, and Fab1-YotB-Vac1p-EEA1 (FYVE) domain containing 1 is colocalized with and phosphorylated by tyrosine kinase Src, inducing Src-Plekhf1-caveolin complex formation in macrophages31. As far as we know, there has been no literature concerning the role PLEKHF1 plays in malignant tumors. Yan et al. found that the down-regulation of PLEKHF1 attenuated pulmonary fibrosis by modulating macrophage polarization32. Our study firstly indicated that PLEKHF1 expression was lower in OS tissues than in normal tissues, suggesting the expression of PLEKHF1 was associated with a better prognosis in OS. Therefore, it might thus be a prognostic factor for OS.
The tumor microenvironment allows tumor cells to escape immune surveillance and drug interference33. Theoretically, the expression levels of antitumor infiltrating immune cells will decrease in the high-CDI group because of the holistic damage to immune functions34,35. These results were confirmed in our study. Our study revealed a consistent negative correlation between CDI and the expression levels of various immune regulatory cells, suggesting a potential immune-suppressed state in high CDI patients, particularly those with a poorer prognosis. Additionally, the single-cell RNA sequencing analysis of osteosarcoma revealed the expression of the PLEKHF1 increased in NK/T cells, myeloid cells, and osteosarcoma cells, while the expression of FADS2 and CLTCL1 predominated in osteosarcoma cells, myeloid cells, and cancer-associated fibroblasts. Furthermore, despite the poor survival prognosis associated with high CDI, these patients with high CDI might still benefit from immunotherapy due to a lower degree of T-cell dysfunction. We found an elevated lapatinib sensitivity in the high CDI group, suggesting potential therapeutic benefits in this subgroup. Lapatinib is an inhibitor of human epidermal growth factor receptor 2 (HER2) phosphorylation, which has been widely utilized in treating various malignant tumors, such as breast cancer, ovarian cancer, and bladder cancer36,37,38. Long et al. represented that lapatinib can alter the malignant phenotype of osteosarcoma cells via downregulation of the activity of the HER2-PI3K/AKT-FASN axis39. No previous literature suggests the application of lapatinib in patients with OS from the aspects of immunotherapy. These results provide novel insights into the tumor microenvironment of OS and the potential therapeutic targets.
Although our model showed excellent performance in both training and validation cohorts, our study is still with some limitations. First, the lack of statistical significance in some analyses (as shown in Figs. 6E–H and 9) may stem from the relatively small sample size in the subgroups and clinical variability (e.g., treatment heterogeneity inherent to retrospective cohorts). Second, the patients were retrospectively recruited, which may inevitably lead to bias to some extent. The above-mentioned factors highlight the need for larger, prospectively studies to validate these trends. Third, we observed discordance between mRNA expression levels (RNA-seq) and protein abundance (IHC) for CDI components, particularly FADS2, which is often attributable to post-transcriptional regulation, tumor heterogeneity, or technical factors. Future studies employing single-cell multi-omics or targeted proteomics could better resolve these relationships. This highlights the importance of complementing transcriptomic signatures with functional protein-level validation in prognostic model development. Fourth, while our analysis provides computationally validated insights, we acknowledge the lack of experimental validation using techniques like multiplex immunohistochemistry. This limitation primarily stems from the technical challenges in obtaining comprehensive immune profiling data for rare pediatric malignancies like osteosarcoma. Last but not the least, there is still a need for phase 3 randomized controlled trials to verify the decision-making role of this model in selecting patients who could benefit from lapatinib and other potential immunotherapy candidates.
In conclusion, the PCD-related gene signature proposed in this study is a practical prognostic predictor for OS patients, which can make a notable difference in the assessment of clinical outcomes.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Huang, J. et al. Global incidence, mortality and temporal trends of cancer in children: A joinpoint regression analysis. Cancer Med. 12(2), 1903–1911 (2023).
Casali, P. G. et al. Bone sarcomas: ESMO–PaedCan–EURACAN clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 29, 79–95 (2018).
Yuan B, Shi K, Zha J, et al. Nuclear receptor modulators inhibit osteosarcoma cell proliferation and tumour growth by regulating the mTOR signaling pathway. Cell Death & Disease. 2023;14(1).
Liu J, Hong M, Li Y, Chen D, Wu Y, Hu Y. Programmed Cell Death Tunes Tumor Immunity. Frontiers in Immunology. 2022;13.
Tang, D., Kang, R., Berghe, T. V., Vandenabeele, P. & Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 29(5), 347–364 (2019).
Zhao, R. et al. Novel roles of apoptotic caspases in tumor repopulation, epigenetic reprogramming, carcinogenesis, and beyond. Cancer Metastasis Rev. 37(2–3), 227–236 (2018).
Su, Z., Yang, Z., Xu, Y., Chen, Y. & Yu, Q. Apoptosis, autophagy, necroptosis, and cancer metastasis. Mol. Cancer 14(1), 1–14 (2015).
Xie, J. et al. Establishment of a cell necroptosis index to predict prognosis and drug sensitivity for patients with triple-negative breast cancer. Front. Mol. Biosci. 9, 834593 (2022).
Tang, R. et al. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J. Hematol. Oncol. 13(1), 23 (2020).
Stockwell, B. R. et al. Ferroptosis: A regulated cell death nexus linking metabolism, redox biology, and disease. Cell 171(2), 273–285 (2017).
Zou, Y. et al. N6-methyladenosine regulated FGFR4 attenuates ferroptotic cell death in recalcitrant HER2-positive breast cancer. Nat. Commun. 13(1), 2672 (2022).
Tsvetkov, P. et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375(6586), 1254–1261 (2022).
Wen, S., Niu, Y., Lee, S. O. & Chang, C. Androgen receptor (AR) positive vs negative roles in prostate cancer cell deaths including apoptosis, anoikis, entosis, necrosis and autophagic cell death. Cancer Treat. Rev. 40(1), 31–40 (2014).
Zou, Y. et al. Leveraging diverse cell-death patterns to predict the prognosis and drug sensitivity of triple-negative breast cancer patients after surgery. Int. J. Surg. 107, 106936 (2022).
Fatokun, A. A., Dawson, V. L. & Dawson, T. M. Parthanatos: Mitochondrial-linked mechanisms and therapeutic opportunities. Br. J. Pharmacol. 171(8), 2000–2016 (2014).
Sonja Aits, M. J. Lysosomal cell death at a glance. J. Cell Sci. 126(9), 1905–1912 (2013).
Amaravadi, R. K., Kimmelman, A. C. & Debnath, J. Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 9(9), 1167–1181 (2019).
Song, X. et al. JTC801 induces pH-dependent death specifically in cancer cells and slows growth of tumors in mice. Gastroenterology 154(5), 1480–1493 (2018).
Scaturro, P. & Pichlmair, A. Oxeiptosis: A discreet way to respond to radicals. Curr. Opin. Immunol. 56, 37–43 (2019).
Carneiro, B. A. & El-Deiry, W. S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 17(7), 395–417 (2020).
Zhang, Z. et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature 579(7799), 415–420 (2020).
Jiang, Z. et al. TYRO3 induces anti–PD-1/PD-L1 therapy resistance by limiting innate immunity and tumoral ferroptosis. J. Clin. Investig. 131, 8 (2021).
Long, K. R., Trofatter, J. A., Ramesh, V., Mccormick, M. K. & Buckler, A. J. Cloning and characterization of a novel human clathrin heavy chain gene (CLTCL). Genomics 35(3), 466–472 (1996).
Liu, S. H. et al. A novel clathrin homolog that co-distributes with cytoskeletal components functions in the trans-Golgi network. EMBO J. 20(1), 272–284 (2001).
Sens-Abuázar, C. et al. Down-regulation of ANAPC13 and CLTCL1: Early events in the progression of preinvasive ductal carcinoma of the breast. Transl. Oncol. 5(2), 113–118 (2012).
Young, R. S. E. et al. Apocryphal FADS2 activity promotes fatty acid diversification in cancer. Cell Rep. 34, 6 (2021).
Koundouros, N. & Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 122(1), 4–22 (2019).
Xuan, Y. et al. SCD1/FADS2 fatty acid desaturases equipoise lipid metabolic activity and redox-driven ferroptosis in ascites-derived ovarian cancer cells. Theranostics. 12(7), 3534–3552 (2022).
Park, H. G., Kim, J. H., Dancer, A. N., Kothapalli, K. S. & Brenna, J. T. The aromatase inhibitor letrozole restores FADS2 function in ER+ MCF7 human breast cancer cells. Prostaglandins Leukotrienes Essential Fatty Acids 171, 102312 (2021).
Tian, J. et al. Risk SNP-mediated enhancer-promoter interaction drives colorectal cancer through both FADS2 and AP0027542. Cancer Res. 80(9), 1804–1818 (2020).
Li, T., Qin, K., Li, N., Han, C. & Cao, X. An endosomal LAPF is required for macrophage endocytosis and elimination of bacteria. Proc. Natl. Acad. Sci. 116(26), 12958–12963 (2019).
Yan, L. et al. Local administration of liposomal-based Plekhf1 gene therapy attenuates pulmonary fibrosis by modulating macrophage polarization. Sci. China Life Sci. 66(11), 2571–2586 (2023).
Zou, Y. et al. The role of circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: A pan-cancer analysis. Biomolecules 9(9), 429 (2019).
Zou, Y. et al. Discordance of immunotherapy response predictive biomarkers between primary lesions and paired metastases in tumours: A systematic review and meta-analysis. EBioMedicine 63, 262 (2021).
Zou, Y. et al. Efficacy and predictive factors of immune checkpoint inhibitors in metastatic breast cancer: A systematic review and meta-analysis. Therap. Adv. Med. Oncol. 12, 17588 (2020).
Narayan, V. et al. Cisplatin, gemcitabine, and lapatinib as neoadjuvant therapy for muscle-invasive bladder cancer. Cancer Res. Treat. 48(3), 1084–1091 (2016).
Lebedeva, I. V. et al. Lapatinib and poziotinib overcome ABCB1-mediated paclitaxel resistance in ovarian cancer. PLoS ONE 16(8), e0254205 (2021).
Simigdala, N. et al. Loss of Kmt2c in vivo leads to EMT, mitochondrial dysfunction and improved response to lapatinib in breast cancer. Cell. Mol. Life Sci. 80(4), 264 (2023).
Long, X.-H. et al. Lapatinib alters the malignant phenotype of osteosarcoma cells via downregulation of the activity of the HER2-PI3K/AKT-FASN axis in vitro. Oncol. Rep. 31(1), 328–334 (2014).
Acknowledgements
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Funding
This work was supported by the grants from National Natural Science Foundation of China (82372668), the National Science Foundation for Young Scientists of China (82001312), Special Fund for the Incubation of Young Clinical Scientist, Children’s Hospital of Zhejiang University School of Medicine (CHZJU2022YS008) and the Medical Innovation Foundation of Fujian Province (2021CXA032).
Author information
Authors and Affiliations
Contributions
W.C. designed the experiments and directed the study. G.B., S.Z., and L.C. performed the experiments. M.Z. evaluated the clinical specimens by immunohistochemistry. G.B., S.Z., and M.Z. analyzed the data and generated the figures. W.C. provided critical design and analysis support. G.B. and W.C. wrote the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
The study was approved by the Research Ethics Committee of Children’s Hospital of Zhejiang University School of Medicine (2022-IRB-109) and complied with the Helsinki Declaration. Written informed consent was obtained from the patient’s parents/legal guardian for publication and any accompanying images.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bai, G., Zhao, S., Chen, L. et al. Leveraging diverse cell death patterns in osteosarcoma patients and identification of the function of FADS2 in osteosarcoma cells. Sci Rep 15, 20831 (2025). https://doi.org/10.1038/s41598-025-05480-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05480-5